Abstract
Changes in nucleolar morphology and function are tightly associated with cellular activity, such as growth, proliferation, and cell cycle progression. Historically, these relationships have been extensively examined in cancer cells, which frequently exhibit large nucleoli and increased ribosome biogenesis. Recent findings indicate that alteration of nucleolar activity is a key regulator of development and aging. In this review, we have provided evidences that the nucleolus is not just a housekeeping factor but is actively involved in the regulation of cell proliferation, differentiation, and senescence both in vitro and in vivo. In addition, we have discussed how alteration of nucleolar function and nucleolar proteins induces specific physiological effects rather than widespread effects.
Keywords: Nucleolus, Ribosome biogenesis, Stem cells, Differentiation, Longevity
Introduction
The nucleolus is the most prominent subnuclear structure where ribosomal RNA (rRNA) and ribosomal subunits are synthesized. These events are highly coordinated and the most energy-consuming cellular processes. Within the nucleolus, ribosomal DNA (rDNA) is transcribed by RNA polymerase I (Pol I). The precursor rRNA (pre-rRNA) obtained is then processed and modified to generate 28S, 18S, and 5.8S rRNAs. The mature rRNAs are then assembled with ribosomal proteins and are exported to the cytoplasm for protein synthesis [1, 2]. The size of the nucleolus varies among cells and reflects the rate of ribosome biogenesis. Large nucleoli are found in actively proliferating cells that require continuous ribosome biosynthesis, whereas the size of the nucleolus decreases upon cell cycle arrest [3].
The relationship between nucleolar size and cell activity has been extensively examined in cancer biology for many years. In as early as the late 19th century, a study reported that the large, irregular nucleoli were the hallmarks of progressive cancer cells [2]. Since then, many studies have been performed to determine the molecular mechanisms underlying nucleolar dynamics in cancer progression. Some studies suggest that nucleolar changes are just the consequence of enhanced ribosome biogenesis in cancer cells [4]. This view is supported by the evidence that the rate of ribosome biogenesis is regulated by oncogenes and tumor suppressors [5]. According to this view, changes in oncogenes or tumor suppressors increase ribosome biogenesis to satisfy the increasing demand for protein synthesis in rapidly proliferating cancer cells, thus resulting in hypertrophic nucleoli. In contrast, many studies indicate that nucleolar dynamics play a causative role in tumorigenesis. In fact, some studies have shown that mutations in genes encoding ribosomal proteins, which are observed in patients with inherited genetic disease, increase the incidence of cancer [4]. Moreover, in vitro studies have shown that dysregulation of nucleolar function induces malignant transformation [6, 7]. Thus, nucleolar dynamics is not only the consequence of malignant transformation but also is actively involved in cancer progression.
Recent studies have shown that nucleoli are actively involved in stem cell maintenance [8–11, 75] and lifespan regulation in model organisms [12–14] and play a role in various cellular functions in addition to cancer progression. In this review, we discuss the functions of nucleoli in cancer cell progression, stem cell maintenance, and lifespan regulation, with an emphasis on how nucleoli regulate specific cellular processes rather than the overall cellular homeostasis. In addition, we discuss the findings of recent studies highlighting the specific roles of ribosomal proteins in the regulation of gene expression, which challenges the classical view that nucleoli and ribosome only perform housekeeping functions.
Ribosome biogenesis and cancer
Cancer cells require continuous ribosome biogenesis and protein translation to maintain their high proliferation rate. Therefore, nucleolar hypertrophy with enhanced ribosome biogenesis is one of the characteristic features of cancer cells. In rapidly proliferating cancer cells, nucleolar proteins, which are involved in rRNA synthesis and processing, become more abundant, leading to nucleolar hypertrophy (Fig. 1). In fact, the size of the nucleolus, the amount of nucleolar proteins, and rate of rRNA synthesis are closely associated with the doubling time of various cancer cells [3].
Fig. 1.
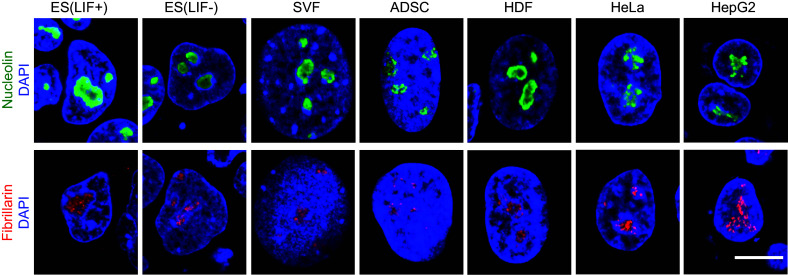
Differently sized nucleoli of pluripotent, differentiated, and cancer cells. Mouse ES cells cultured in the presence or absence of LIF, mouse SVF (stromal vascular fraction), human ADSCs (adipose derived stem cells), HDF (human dermal fibroblasts), HeLa cells, and HepG2 cells were stained with antibody to nucleolin (green), fibrillarin (red), and DAPI (blue). Note that pluripotent ES cells (+LIF) and cancer cells (HeLa and HepG2) have large nucleoli compared to differentiated cells (−LIF ES cells, SVF, ADSC, and HDF). Scale bar 10 μm
Changes in oncogenes and tumor suppressors in cancer cells result in the hyperactivation of ribosome biogenesis. The c-Myc proto-oncogene enhances the transcription of rDNA by directly controlling the expression of upstream binding factor (UBF), an essential component of Pol I transcription machinery, and by interacting with rDNA [15]. Moreover, c-Myc directly regulates the expression of genes involved in pre-rRNA processing and increases 5S rRNA biosynthesis by activating the transcription of RNA polymerase III [16]. These data clearly indicate that overexpression of c-Myc enhances ribosome biogenesis in cancer cells. RAS/RAF/ERK signaling pathway, an oncogenic signaling pathway, activates rRNA synthesis by regulating the phosphorylation and activation of UBF and transcription initiation factor IA (TIF-IA) [17]. Similarly, PI3 K/AKT/mTORC1 signaling pathway also modulates the phosphorylation of UBF and TIF-IA [18]. These data illustrate that activating mutations in these signaling pathways could enhance ribosome biogenesis and trigger cancer cell proliferation. In contrast, tumor suppressor protein p53 inhibits the transcriptional activity of Pol I by binding to SL-1 complex, which is necessary for the formation of the Pol I complex on the rDNA promoter [19]. In addition, retinoblastoma-associated protein represses the transcriptional activity of Pol I by preventing the recruitment of UBF to the rDNA promoter [20]. Thus, inactivation of these tumor suppressors, which is frequently observed in cancer cells, prevents the repression of rRNA transcription, leading to the hyperactivation of ribosome biogenesis.
In addition to transcriptional regulation, epigenetic networks control rRNA expression in cancer cells. The promoter region of the human rRNA gene contains a CpG island encompassing 19 CpGs in the upstream control element and six CpGs in the core promoter region. Bisulfite genomic sequencing has shown that most CpGs in the upstream control element are significantly hypomethylated in human hepatocellular carcinoma cells compared with those in pair-matched liver tissue cells. Luciferase reporter assay using the rRNA promoter showed that hypermethylation of CpG islands inhibits the activity of the rRNA promoter and that methyl-CpG-binding protein MBD2 mediates the silencing of the rRNA promoter, suggesting that CpG methylation density is inversely correlated with the activity of the rRNA promoter [21]. These data imply that hypomethylation of the rRNA promoter enhances the transcription of rRNA genes, thus increasing ribosome biosynthesis in cancer cells.
Although the above results suggest that alteration of ribosome biogenesis can be explained as a consequence of malignant transformation, many evidences show that dysregulation of ribosomal proteins itself increases the susceptibility to tumorigenesis. Several ribosomal proteins are overexpressed in tumor cells and clinical tissue samples obtained from cancer patients [22–24]. For example, RPL36A, a tumor-associated ribosomal protein, is highly expressed in hepatocellular carcinoma. Ectopic overexpression of RPL36A in Chang liver cells enhances colony formation and increases cell proliferation by accelerating the cell cycle [ 7 ], suggesting that increased expression of ribosomal proteins plays a key role in tumor cell proliferation. Similarly, overexpression of another ribosomal protein RPS3a in NIH3T3 cells induces the characteristic features of malignant transformation, such as foci formation, anchorage independent growth, and tumorigenicity in nude mice, suggesting that increased expression of ribosomal proteins may lead to malignant transformation [6]. In addition to enhanced production of ribosomal subunits, reduced expression of ribosomal proteins also leads to tumor formation, which has long been examined in patients with inherited genetic diseases such as ribosomopathies. For example, heterozygous mutations in RPS19 encoding a small ribosomal subunit protein RPS19 are observed in 25 % patients with Diamond–Blackfan anemia (DBA), a bone marrow failure syndrome associated with increased incidence of acute myeloid leukemia and osteogenic sarcoma. These mutations inhibit the processing of pre-rRNA to 18S rRNA and formation of the 40S rRNA subunit, suggesting that defects in ribosome biogenesis are associated with DBA pathogenesis [25]. Other ribosomal proteins, including RPS24, RPS7, RPS15, RPS17, RPL35A, RPL5, and RPL11, are also mutated in patients with DBA [26]. The 5q-syndrome is a type of myelodysplastic syndrome characterized by severe anemia, dysmegakaryopoiesis, and increased susceptibility to hematopoietic tumors [27]. In patients with the 5q-syndrome, an allele of the gene encoding ribosomal protein S14 (RPS14) is deleted, resulting in the haploinsufficient expression of RPS14. Knockdown experiments showed that decreased expression of RPS14 inhibited pre-rRNA processing and decreased the level of mature rRNA, which was similar to the RPS19 deficiency observed in patients with DBA, thus supporting the association between dysregulation of rRNA synthesis and tumor formation [27]. Besides mutations in genes encoding ribosomal proteins, mutations in genes encoding nucleolar proteins responsible for pre-rRNA maturation cause inherited genetic diseases associated with an increased risk of tumor formation. X-linked dyskeratosis congenita (DC) is a bone marrow failure syndrome characterized by ectodermal dysplasia, hematopoietic failure, and increased tumor susceptibility. Pathogenesis of DC is attributed to a mutation in DKC1 that encodes dyskerin, a component of small nucleolar ribonucleoprotein particles (snoRNPs) that are involved in pre-rRNA processing [28, 29]. In addition, mutations in the gene encoding RMRP, an RNase involved in pre-rRNA cleavage, are observed in patients with cartilage–hair hypoplasia that predisposes these patients to cancer [30]. Although molecular mechanisms underlying increased susceptibility to tumor formation remain poorly understood, the above evidences strongly suggest that defects in ribosome biogenesis actively contribute to tumorigenesis.
Non-ribosomal nucleolar proteins in cancer cells
The nucleolus acts as a key stress sensor and controls the cellular level of p53. Under normal condition, p53 is maintained at a low level through continuous degradation by MDM2-mediated ubiquitination. In response to cellular stress, nucleolar proteins, including ribosomal proteins L11 and ARF, are released from the nucleolus into the cytoplasm where they interact with MDM2 and inhibit MDM2-mediated ubiquitination of p53, stabilizing the level of p53. Thus, the nucleolus is a crucial regulator that links stress signaling with cellular p53 level and allows cells to respond to stress by inducing p53-mediated cell cycle arrest [31, 32]. Furthermore, the nucleolus is involved in multiple biological processes, including cell cycle regulation, DNA repair, and telomere replication [33]. Nucleolar proteins that facilitate these cellular processes other than ribosome biogenesis are also involved in cancer progression. Nucleophosmin (NPM) is a non-ribosomal protein, and its dysregulation is tightly associated with tumorigenesis. NPM shuttles between the nucleolus and the cytoplasm during cell cycle and performs diverse cellular functions, including centrosome duplication, protein folding, and genome stability maintenance [34]. Because of its multifunctionality, it is unclear whether NPM has oncogenic or tumor suppressive properties even though its functions have been extensively studied [35]. In fact, NPM is frequently overexpressed in various cancers, whereas mutations in the gene encoding NPM are also observed in patients with acute myeloid leukemia [36]. In NPM-mutant cells, ARF, a tumor suppressor protein, is mislocalized to the cytoplasm where it cannot bind to MDM2, leading to the impairment in ARF-dependent p53 activation [35, 37]. In addition, NPM inactivation induces DNA damage and unrestricted centrosome duplication, resulting in aberrant oncogenesis [38]. These data indicate that NPM is critical for maintaining proper p53 function and genome stability, thus implying its tumor suppressive role. In contrast, some studies reported that NPM overexpression is associated with cell proliferation and tumorigenesis. Clinical samples show that cancer tissues express extremely high levels of NPM compared with normal tissues, and these levels are closely correlated with cancerous growth [39]. In chronic myelogenous leukemia-derived cells, NPM interacts with a tumor suppressor protein IRF-1 and interferes with its DNA-binding and transcriptional activity. Furthermore, overexpression of NPM in NIH3T3 cells induces malignant transformation in vivo and tumorigenicity in nude mice by possibly inhibiting the activity of IRF-1 [40]. These findings imply the oncogenic role of NPM. Nucleostemin, a GTP-binding protein, is another non-ribosomal protein that is highly expressed in various cancer cells and cancer stem cells. Nucleostemin regulates the cell cycle and proliferation by interacting with multiple proteins, including p53, MDM2, and ARF [41]. In cancer cells, overexpression of nucleostemin increases the expression of several markers of tumor-initiating cells and tumorigenicity thorough the formation of a complex containing a telomerase catalytic subunit and Brg1 [42]. Together, these findings illustrate that non-ribosomal nucleolar proteins contribute to tumorigenesis.
Nucleolar dynamics in stem cells
In addition to cancer cells, a large nucleolus is a characteristic feature of stem cells and progenitor cells. For example, embryonic stem (ES) cells possess large, condensed nucleoli, which are the hallmark of pluripotency (Fig. 1). These nucleoli change their morphology from large to small foci during differentiation [43]. In addition, during Drosophila eye disc development, nucleoli are much larger in proliferating progenitor cells of the anterior eye disc than in differentiating and post-mitotic cells of the posterior eye disc [44]. These data show that nucleolar size reflects the stemness and proliferation potential of the cells, which are consistent with the finding that proliferating cells require active nucleolar function with increased ribosome biogenesis.
The proto-oncogene c-Myc plays a critical role in the regulation of nucleolar size during transition from progenitor to post-mitotic differentiated state in stem cells as well as in cancer cells. Actively proliferating stem cells and progenitor cells express high levels of c-Myc. During cell differentiation, downregulation of c-Myc leads to the loss of Pol I transcription initiation factor UBF, which represses rRNA synthesis [45]. Furthermore, lineage-specific differentiation factors such as MyoD, myogenin, Runx2, and C/EBP-β bind to the rDNA loci and decrease c-Myc binding, thus downregulating rRNA transcription [46]. These findings clearly indicate that cell growth regulatory factors and lineage commitment factors regulate nucleolar activity and rRNA synthesis in a mutually exclusive manner.
Novel nucleolar function in stem cell maintenance and differentiation
One fundamental question is whether the change in nucleolar size is just an indicator of stem cell properties or nucleolar activity itself controls the proliferation and differentiation of stem cells. Recent studies by us and by other researchers showed that the nucleolus is a critical regulator of the unique properties of stem cells. In ES cells, non-ribosomal nucleolar proteins such as nucleolin and nucleostemin are highly expressed, which is similar to that observed in cancer cells. Depletion of these nucleolar proteins results in reduced cell proliferation, abnormal cell cycle, and enhanced differentiation, suggesting that proper nucleolar function is required for the self-renewal of ES cells [47, 48]. Mechanistically, knockdown of nucleostemin in ES cells downregulates the expression of cell cycle regulators and decreases the progression through the G1 phase of the cell cycle [48]. This elongated G1 phase allows ES cells to respond to MEK/ERK signaling and promotes the differentiation of ES cells. Depletion of nucleolin in ES cells increases the stability of p53 and activates p53 signaling, resulting in the elongation of the G1 phase and induction of differentiation [47]. These data imply that nucleostemin and nucleolin have similar molecular functions and contribute to the maintenance of ES cell identity by sustaining its unique cell cycle profiles.
Nucleolar proteins involved in ribosome biogenesis are also the critical regulators of stem cell maintenance. Fibrillarin (FBL), a critical methyltransferase involved in rRNA processing, is highly expressed in ES cells and its expression rapidly decreases upon the differentiation of ES cells. Stable expression of FBL prolonged the pluripotent state of mouse ES cells cultured without leukemia inhibitory factor (LIF). Partial knockdown of the gene encoding FBL decreases rRNA synthesis and inhibits the growth of ES cells. This in turn promotes the differentiation of ES cells even in the presence of LIF. Microarray analysis showed that abnormal differentiation caused by a decrease in FBL expression is attributed to the activation of p53 signaling [11]. These data suggest that proper ribosome biogenesis is a critical regulator for the pluripotency of ES cells and that alteration of ribosome production induces the differentiation of ES cells in a p53-dependent manner. In mouse hematopoietic stem cells, inhibition of rRNA production by actinomycin D or siRNA against the gene encoding TIF-IA, an essential factor for Pol I transcription, decreases pre-rRNA levels and induces cell differentiation [10]. Importantly, inhibition of cell cycle without the inhibition of ribosome biogenesis does not induce cell differentiation, suggesting that changes in ribosome biogenesis positively regulate cell differentiation independently of cell cycle progression. Deletion of Notchless, which is known to be involved in pre-60S maturation, leads to defective ribosome biogenesis and p53 activation in hematopoietic stem cells and in turn causes their rapid exhaustion [75]. The role of ribosome biogenesis in stem cells was also investigated in vivo in the development of Drosophila ovary. While screening for mutations that affected early oogenesis, Fichelson el al. observed that wicked (wcd) mutant induced the premature differentiation of germ-line stem cells. Wcd encodes a yeast homolog of U3 snoRNA-associated protein UTP18, a nucleolar protein responsible for pre-rRNA maturation. Consistent with this, knockdown of wcd in S2 cells induced the accumulation of long forms of pre-rRNA, suggesting the role of wcd in rRNA maturation. Intriguingly, Wcd was asymmetrically segregated into the daughter cells during germ stem cell mitosis, and the daughter cells that inherited wcd maintained their stem cell property [8]. These data indicate that modulation of ribosome biosynthesis mediated by the asymmetrical segregation of Wcd is required for stem cell maintenance and germ cell differentiation. Recently, another group showed the requirement of ribosome biogenesis for germ cell maintenance and identified the underlying molecular mechanisms. They searched for mutants on the basis of sterile phenotypes and identified a mutant under-developed (udd) which exhibits germ cell loss. Decreased rRNA production induced by the disruption of udd, a component of Pol I regulatory complex, promoted germ cell differentiation into multicellular cysts while overexpression of TIF-IA, a mediator of rRNA transcription by Pol I, delayed germ cell differentiation. Interestingly, reduction in rRNA production specifically downregulated the level of Mad protein, a component of BMP signaling, but did not affect the level of another BMP signaling component Medea or histone H2B [9]. Thus, this study suggests that alteration of rRNA production regulates cell differentiation by modulating specific proteins but not by reducing the overall protein synthesis. It will be interesting to examine whether downregulation of Mad proteins is dependent on p53 activation, as observed during ES cell differentiation and, if it is not, then how does ribosome biogenesis regulate the expression of specific proteins.
Nucleolar proteins in development
Ribosome biogenesis not only regulates stem cell differentiation, but also controls cell survival and growth during the development of an organism. Deletion or mutation of nucleolar proteins involved in ribosome biogenesis leads to growth defect phenotypes. For example, in Drosophila, RNAi knockdown of NS1, which is the closest homolog of human nucleostemin, inhibits the release of the large ribosomal subunit from the nucleolus, induces loss of adult precursor cells in the midgut epithelium, and retards cell growth in the salivary gland [49]. Loss of Nopp140, which functions as a molecular chaperon for snoRNA [50], disrupts ribosome biogenesis and induces apoptosis in wing discs during larval development [51]. The apoptotic phenotype cannot be rescued by a p53 gene deletion, indicating that depletion of Nopp140 induces apoptosis in a p53-independent manner. Instead of p53, activated JNK and its downstream pro-apoptotic factor Hid accumulate in Nopp140-depleted larvae, suggesting that reduced ribosome biogenesis activates the JNK signaling pathway. In zebrafish, mutation in the nucleolar protein NOM1 results in decreased proliferation of pancreatic cells during the development of pancreas. RNA-seq analysis revealed that NOM1 deficiency affects the expression of ribosomal-related genes, leading to the decreased production of 18S rRNA [52]. These data suggest that the regulation of ribosome biogenesis is essential for the proper development of various organs.
Several studies have revealed that Myc and Tor are involved in the regulation of ribosome biogenesis during Drosophila development. Overexpression of dMyc in larvae increases ribosome biogenesis, whereas the level of rRNA is decreased in dMyc mutant larvae. Using microarray analysis, Grewal et al. revealed that dMyc upregulates the expression of genes encoding Pol I regulators in the developing larvae. Expression of dMyc increases cell size in the wing disc, but the cell size effect is diminished in cells carrying a mutation in Rpl135, a component of the Pol I complex. This suggests that dMyc controls cell growth through the regulation of ribosome biogenesis [53]. Demontis et al. revealed the role of dMyc in skeletal muscles. While inhibition of dMyc activity leads to smaller muscles, dMyc overexpression promotes endoreplication, induces enlarged nucleoli, and upregulates the expression of genes involved in ribosome biogenesis. Under conditions that inhibit Insulin receptor (InR)/Tor signaling, dMyc expression does not promote endoreplication, suggesting that InR/Tor signaling is required for dMyc activity [54]. Grewal et al. also reported the involvement of the Tor pathway in the regulation of ribosome biogenesis in vivo. Drosophila TIF-IA regulates pre-rRNA synthesis through the recruitment of Pol I to the rDNA promoter. While Tif-IA−/− mutants exhibit a growth arrest phenotype, TIF-IA overexpression increases ribosome biogenesis in larvae. Treatment with rapamycin, an inhibitor of Tor, inhibits the association of TIF-IA with rDNA. In addition, TIF-IA overexpression can increase the levels of pre-RNA even in rapamycin-treated larvae, suggesting that TIF-IA functions downstream of Tor [55]. These studies suggest a link between the Tor pathway, Myc, and ribosome biogenesis, which plays an important role in controlling cell growth during development.
Nucleolar proteins in aging
Because the nucleolus functions as a regulator of cell proliferation, cell cycle progression, and telomerase replication, the nucleolus could also be associated with senescence. Sir2, a key regulator of replicative senescence in yeast, is localized in the nucleolus [56] where it regulates rDNA silencing [57, 58] and inhibits homologous recombination at the rDNA locus [59]. Sirt1, the mammalian homolog of Sir2, complexes with nucleomethylin and SUV39H1 to form energy-dependent nucleolar silencing complex that inhibits the transcription of rRNA during nutrient starvation [60, 61]. TORC1 signaling, whose reduction is associated with the extension of lifespan in Drosophila and Caenorhabditis elegans [62], also regulates ribosome biogenesis [63]. These data imply that changes in nucleolar function regulate lifespan. Consistent with these findings, recent in vivo studies have clearly shown that ribosome biogenesis modulates aging and longevity of organisms. For example, yeast strains lacking genes encoding different 60S ribosomal proteins show extended replicative lifespan. Moreover, deletion of 60S-specific ribosomal processing factors or inhibition of 60S subunit biogenesis by treatment with small molecule inhibitors increased the replicative lifespan of yeast [13], suggesting that 60S ribosome biogenesis is a negative regulator of longevity. Using C. elegans as a model organism, Hansen et al. showed that reducing the levels of ribosomal proteins by RNAi inhibited protein synthesis and increased lifespan [12]. Furthermore, C. elegans subjected to RNAi knockdown of nog-1, which encodes a nucleolar GTPase required for 60S ribosome biogenesis, show increased lifespan, whereas those overexpressing nog-1 show decreased adult longevity [14]. These data indicate that ribosome biogenesis regulates lifespan in vivo. A very recent study by Demontis et al. showed a fascinating association among myokine, nucleolar function, and aging in Drosophila. Mnt, a basic helix-loop-helix transcription factor, regulates the expression of genes involved in ribosome biogenesis. Overexpression of Mnt in the skeletal muscle decreases age-related climbing defects and prolongs lifespan. Gene expression analysis showed that Mnt overexpression downregulated the expression of genes encoding nucleolar proteins, which in turn decreased rRNA levels. RNAi knockdown of proteins involved in rRNA synthesis and ribosome biogenesis extended the medial lifespan, suggesting that decreased nucleolar function regulates longevity. Interestingly, changes in nucleolar function by Mnt overexpression were observed not only in the skeletal muscle but also in the adipose tissue. This intertissue control of nucleolar function depends on the activity of myoglianin, a myokine whose expression is induced by Mnt. Similar to Mnt, myoglianin overexpression in the skeletal muscle extends the lifespan and decreases nucleolar size in the adipose tissue [64]. These data suggest that myokine-mediated intertissue connection between the skeletal muscle and adipose tissue regulates lifespan at a whole-body level by modulating ribosome biogenesis.
Although these findings clearly demonstrate the relationship between the rate of ribosome biogenesis and aging, it is unclear how the reduction in ribosome biogenesis extends lifespan. Because increased levels of Arf and p53 induce cancer resistance and extend the lifespan of mice [65], it can be suggested that p53 activation induced by reduced ribosome biogenesis, which is observed in stem cells (see above), may be involved in longevity. Moreover, reduced metabolic rate with a corresponding decrease in free radical production is associated with aging and lifespan [66], suggesting an association between ribosome biogenesis and aging. It will be interesting to examine whether p53 activation or decreased oxidative stress contributes to lifespan extension induced by reduced ribosome biogenesis.
Specific functions of ribosomal proteins
Although ribosome is an essential cellular component that facilitates protein synthesis in all cells, recent evidences indicate that changes in the rate of ribosome biogenesis regulate tumorigenesis, stem cell maintenance, and aging. This raises a question as to why dysregulation of ribosomal proteins induces cell type-specific phenotypes rather than widespread dysfunction. For example, decreased expression of ribosomal proteins in ribosomopathies mainly affects hematopoietic cells in the bone marrow but not other cells in the body (described above). Moreover, knockdown of proteins involved in pre-rRNA processing induces the apoptosis of ES cells but does not significantly affect differentiated cells [11]. Furthermore, in some cases reduced ribosomal biogenesis delays aging but in other cases they induce tumorigenesis. Given the increased demand for protein synthesis in proliferating cells, this specific phenomenon could be attributed to intrinsic differences in cellular activity. Rapidly proliferating ES cells are more sensitive to changes in ribosome biogenesis than differentiated cells. However, the mechanisms underlying cell type-specific dysfunction induced by ribosome dysregulation are poorly understood thus far.
Increasing evidences of unique functions of multiple ribosomal proteins in a wider context have important implications for understanding the cell type-specific roles of ribosome biogenesis. Two studies showed a transient increase in the expression of specific ribosomal proteins in a developmental stage- and stimulus-dependent manner. In the context of sexual differentiation of zebra finch song system, Tang et al. showed that genes encoding ribosomal proteins L17 and L37 were specifically upregulated in the song control nuclei of the forebrain of developing males [67]. To determine the genes involved in freeze tolerance of wood frog, which can survive after freezing in winter, Wu et al. analyzed the gene expression profiles of cold- and warm-acclimated frogs and observed that expression of the gene encoding ribosomal large subunit protein 7 (RPL7) was specifically upregulated in the skin of cold-acclimated frogs [68].
In addition to gene expression analysis, knockdown experiments have clearly shown the requirement of specialized functions of ribosomal proteins for tissue regeneration and development. Translation of ribosomal protein L4 (Rpl4) is increased in PC12 cells during neurite regeneration after neurite injury but is inefficient in differentiated PC12 cells [69]. Knockdown of Rpl4 blocks neurite regeneration, suggesting that translational control of Rpl4 is required for rapid axonal regeneration. In addition to in vitro experiments, in vivo studies have shown that ribosomal proteins perform distinct functions during specific developmental processes. In zebrafish, knockdown of multiple ribosomal proteins gives rise to phenotypes specific to each gene rather than non-specific defects, with abnormalities in the brain, body trunk, eye, and ears [70], suggesting the specific functions of ribosomal proteins. Similarly, loss-of-function analysis showed that ribosomal protein L22 (Rpl22) and its paralog Rpl22l1 have distinct functions in zebrafish. Rpl22 morphants exhibited arrested T cell development in a p53-dependent manner, and knockdown of Rpl22l1 impaired the emergence of HSCs in a p53-independent manner. Mechanistically, both Rpl22 and Rpl22l1 bind to smad1 mRNA, which is an important regulator for HSC emergence, but play distinct roles in smad1 expression, with Rpl22l1 facilitating smad1 expression and Rpl22 repressing it [71]. In yeast, mutant analysis showed that cells lacking ribosomal protein paralogs exhibited different phenotypes with distinct gene expression changes and that translation of specific mRNAs required a specific subset of ribosomal proteins [72]. A very recent study showed an unexpected role of ribosomal proteins in mouse embryonic development. Mice with mutation in Rpl38 exhibited homeotic transformation along the anterior–posterior axis of skeletal patterning. In Rpl38-mutant embryos, global protein synthesis was not changed but translation of 8 out of 39 Hox-encoding genes was inhibited, suggesting that Rpl38 facilitated the expression of a specific subset of Hox proteins [73]. This specialized translation was mediated by structured RNA elements embedded in the 5′-UTR of Hox, through which RPL38 regulated 80S ribosome complex formation and facilitated cap-independent translation. An additional regulatory element in the 5′-UTR of Hox blocked cap-dependent translation and enabled Rpl38-dependent specialized translation [74], suggesting that Rpl38 and 5′-UTR elements control gene expression. These data suggest that ribosomal proteins are expressed in a tissue-specific manner and could regulate developmental processes.
Concluding remarks
Recent evidences suggest that the nucleolus functions not only as a housekeeping factor but also as an active regulator of disease progression, stem cell maintenance, and longevity both in vitro and in vivo (Table 1). Although the exact molecular mechanisms underlying the regulation of these cellular processes by the nucleolus are largely unknown, increasing evidences suggest that nucleoli and ribosome mediate these cellular processes by regulating the expression of specific genes in a cell type-dependent manner. We anticipate that advancements in our understanding of nucleolar functions will not only reveal the novel molecular mechanisms underlying tumorigenesis, stem cell maintenance, and aging but also offer new therapeutic targets for diseases such as cancer, ribosomopathies, and age-associated diseases.
Table 1.
Novel functions of nucleolar proteins
| Protein name | Protein function | Change in protein expression | Organism | Condition | Newly identified phenotype | References |
|---|---|---|---|---|---|---|
| BRF | Involved in the transcription of 5S rRNA | Knockdown | Drosophila | In vivo | Extended lifespan | [64] |
| CG5033 | Involved in pre-rrna processing and ribosome maturation | Knockdown | Drosophila | In vivo | Extended lifespan | [64] |
| Dyskerin | A component of small nucleolar ribonucleoprotein particles involved in pseudouridylation | Mutation | Human | In vivo | Bone marrow failure, abnormal skin pigmentation, cancer (X-linked dyskeratosis congenita) | [28, 29] |
| Fibrillarin | A methyltransferase for ribosomal RNA processing | Overexpression | Mouse | In vitro | Prolongation of pluripotent state of embyonic stem cell | [11] |
| Knockdown | Mouse | In vitro | Embryonic stem cell differentiation, decreased self-renewal ability | |||
| Multiple ribosomal proteins | Components of 40S and 60S subunit | Knockdown | C. elegans | In vivo | Increased lifespan | [12] |
| Multiple ribosomal proteins | Components of 60S subunit | Deletion | Yeast | In vivo | Increased replicative lifespan | [13] |
| Multiple ribosomal proteins | Ribosomal protein | Knockdown | Zebrafish | In vivo | Abnormalities in the brain, body trunk, eyes, and ears | [70] |
| NOG-1 | A nucleolar GTPase required for 60S ribosome biogenesis | Knockdown | C. elegans | In vivo | Increased lifespan | [14] |
| Overexpression | Decreased adult longevity | |||||
| Nom1 | Involved in pre-rRNA processing | Mutation | zebrafish | In vivo | Defects in endoderm development | [52] |
| Nopp140 | A nucleolar phosphoprotein which functions as a snoRNP chaperone | Knockdown | Drosophila | In vivo | Apoptosis induction in larval wing discs | [51] |
| Knockdown | Drosophila | In vivo | Extended lifespan | [64] | ||
| Nucleolin | Involved in various cellular processes (e.g., ribosome biogenesis, cell cycle, proliferation) | Knockdown | Mouse | In vitro | Embryonic stem cell differentiation, decreased self-renewal ability | [47] |
| Nucleophosmin | Involved in various cellular processes (e.g., centrosome duplication, protein folding) | Overexpression of mutant protein | Mouse | In vitro | p53 activation | [37] |
| Heterozygous knockout | Mouse | In vitro, in vivo | Increased tumorigenesis | [38] | ||
| Overexpression | Human | In vivo | Malignant transformation | [40] | ||
| Nucleostemin | A GTP-binding protein involved in the regulation of cell cycle and proliferation | Overexpression | Human | In vitro | Increase in tumor-initiating properties of cells | [42] |
| Knockdown | Mouse | In vitro | Embryonic stem cell differentiation, decreased self-renewal ability | [48] | ||
| Knockdown | Drosophila | In vivo | Loss of imaginal island cells | [49] | ||
| RMRP | A component of the RNase MRP complex involved in pre-rRNA processing | Mutation | Human | In vivo | Cartilage/skeletal defects, hypoplastic anemia, cancer (cartilage-hair hypoplasia) | [30] |
| RPL4 | Ribosomal protein | Knockdown | Rat | In vitro | Impairment of neurite regeneration | [69] |
| RPL7 | Ribosomal protein | Upregulation | Wood frog | In vivo | Involved in natural freezing survival? | [68] |
| RPL17, RPL37 | Ribosomal protein | Upregulation | Zebra finches | In vivo | Involved in induction of song circuit? | [67] |
| RPL22 | Ribosomal protein | Knockdown | Zebrafish | In vivo | Impairment of the development of T lineage progenitors | [71] |
| RPL22L1 | Ribosomal protein | Knockdown | Zebrafish | In vivo | Impairment of the emergence of hematopoietic stem cells | [71] |
| RPL36A | Ribosomal protein | Overexpression | Human | In vitro | Enhanced colony formation | [7] |
| RPL38 | Ribosomal protein | Deletion | Mouse | In vivo | Homeotic transformations of the axial skeleton | [73], [74] |
| RpLP0 | Ribosomal protein | Knockdown | Drosophila | In vivo | Extended lifespan | [64] |
| RPS14 | Ribosomal protein | One allele deletion | Human | In vivo | Macrocytic anemia, acute myeloid leukemia (5q- syndrome) | [27] |
| RPS19 | Ribosomal protein | Heterozygous mutation | Human | In vivo | Bone marrow failure, congenital anomalies, cancer (Diamond-Blackfan anemia) | [25] |
| SSRP | Involved in rDNA transcription | Knockdown | Drosophila | In vivo | Extended lifespan | [64] |
| TIF-IA | RNA polymerase I transcription initiation factor | Knockdown | Mouse | In vitro | Differentiation of hematopoietic stem cells | [10] |
| Knockdown | Drosophila | In vivo | Growth arrest | [55] | ||
| Udd | A component of the Pol I regulatory complex | Heterozygous deletion | Drosophila | In vivo | Germ cell loss | [9] |
| Wicked | A U3 snoRNA-associated protein required for pre-rRNA maturation | Mutation | Drosophila | In vivo | Premature differentiation of germ-line stem cells | [8] |
Abbreviations
- ES cells
Embryonic stem cells
- pre-rRNA
Precursor ribosomal RNA
- Pol I
RNApolymerase I
- rDNA
Ribosomal DNA
- snoRNPs
Small nucleolar ribonucleoprotein particles
- TIF-IA
Transcription initiation factor IA
- UBF
Upstream binding factor
References
- 1.Boulon S, Westman BJ, Hutten S, et al. The nucleolus under stress. Mol Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Sluis M, McStay B. Ribosome biogenesis: achilles heel of cancer? Genes Cancer. 2014;12:710–716. doi: 10.18632/genesandcancer.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derenzini M, Trere D, Pession A, et al. Nucleolar function and size in cancer cells. Am J Pathol. 1998;152:1291–1297. [PMC free article] [PubMed] [Google Scholar]
- 4.Montanaro L, Treré D, Derenzini M. Nucleolus, ribosomes, and cancer. Am J Pathol. 2008;173:301–310. doi: 10.2353/ajpath.2008.070752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hein N, Hannan KM, George AJ, et al. The nucleolus: an emerging target for cancer therapy. Trends Mol Med. 2013;19:643–654. doi: 10.1016/j.molmed.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Naora H, Takai I, Adachi M, et al. Altered cellular responses by varying expression of a ribosomal protein gene: sequential coordination of enhancement and suppression of ribosomal protein S3a gene expression induces apoptosis. J Cell Biol. 1998;141:741–753. doi: 10.1083/jcb.141.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J-H, You K-R, Kim IH, et al. Over-expression of the ribosomal protein L36a gene is associated with cellular proliferation in hepatocellular carcinoma. Hepatology. 2004;39:129–138. doi: 10.1002/hep.20017. [DOI] [PubMed] [Google Scholar]
- 8.Fichelson P, Moch C, Ivanovitch K, et al. Live-imaging of single stem cells within their niche reveals that a U3snoRNP component segregates asymmetrically and is required for self-renewal in Drosophila . Nat Cell Biol. 2009;11:685–693. doi: 10.1038/ncb1874. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Q, Shalaby NA, Buszczak M. Changes in rRNA transcription influence proliferation and cell fate within a stem cell lineage. Science. 2014;343:298–301. doi: 10.1126/science.1246384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi Y, Kuroda T, Kishimoto H, et al. Downregulation of rRNA transcription triggers cell differentiation. PLoS ONE. 2014;9:e98586. doi: 10.1371/journal.pone.0098586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe-Susaki K, Takada H, Enomoto K, et al. Biosynthesis of ribosomal RNA in nucleoli regulates pluripotency and differentiation ability of pluripotent stem cells. Stem Cells. 2014;32:3099–3111. doi: 10.1002/stem.1825. [DOI] [PubMed] [Google Scholar]
- 12.Hansen M, Taubert S, Crawford D, et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 13.Steffen KK, MacKay VL, Kerr EO, et al. Yeast life span extension by depletion of 60S ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y-I, Bandyopadhyay J, Cho I, et al. Nucleolar GTPase NOG-1 regulates development, fat storage, and longevity through insulin/IGF signaling in C. elegans . Mol Cells. 2014;37:51–58. doi: 10.14348/molcells.2014.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai M-S, Lu H. Crosstalk between c-Myc and ribosome in ribosomal biogenesis and cancer. J Cell Biochem. 2008;105:670–677. doi: 10.1002/jcb.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 17.Hannan KM, Sanij E, Hein N, et al. Signaling to the ribosome in cancer-It is more than just mTORC1. IUBMB Life. 2011;63:79–85. doi: 10.1002/iub.428. [DOI] [PubMed] [Google Scholar]
- 18.Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 19.Zhai W, Comai L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol Cell Biol. 2000;20:5930–5938. doi: 10.1128/MCB.20.16.5930-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voit R, Schäfer K, Grummt I. Mechanism of repression of RNA polymerase I transcription by the retinoblastoma protein. Mol Cell Biol. 1997;17:4230–4237. doi: 10.1128/MCB.17.8.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghoshal K. Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J Biol Chem. 2003;279:6783–6793. doi: 10.1074/jbc.M309393200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henry JL, Coggin DL, King CR. High-level expression of the ribosomal protein L19 in human breast tumors that overexpress erbB-2. Cancer Res. 1993;53:1403–1408. [PubMed] [Google Scholar]
- 23.Vaarala MH, Porvari KS, Kyllönen AP, et al. Several genes encoding ribosomal proteins are over-expressed in prostate-cancer cell lines: confirmation of L7a and L37 over-expression in prostate-cancer tissue samples. Int J Cancer. 1998;78:27–32. doi: 10.1002/(SICI)1097-0215(19980925)78:1<27::AID-IJC6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q, Yang C, Zhou J, et al. Cloning and characterization of full-length human ribosomal protein L15 cDNA which was overexpressed in esophageal cancer. Gene. 2001;263:205–209. doi: 10.1016/S0378-1119(00)00570-9. [DOI] [PubMed] [Google Scholar]
- 25.Choesmel V, Bacqueville D, Rouquette J, et al. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 2006;109:1275–1283. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shenoy N, Kessel R, Bhagat TD, et al. Alterations in the ribosomal machinery in cancer and hematologic disorders. J Hematol Oncol. 2012;5:32. doi: 10.1186/1756-8722-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebert BL, Pretz J, Bosco J, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knight SW, Heiss NS, Vulliamy TJ, et al. X-linked dyskeratosis congenita is predominantly caused by missense mutations in the DKC1 gene. Am J Hum Genet. 1999;65:50–58. doi: 10.1086/302446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He J, Navarrete S, Jasinski M, et al. Targeted disruption of Dkc1, the gene mutated in X-linked dyskeratosis congenita, causes embryonic lethality in mice. Oncogene. 2002;21:7740–7744. doi: 10.1038/sj.onc.1205969. [DOI] [PubMed] [Google Scholar]
- 30.Ridanpää M, van Eenennaam H, Pelin K, Chadwick R. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104:195–203. doi: 10.1016/S0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayer C, Grummt I. Cellular stress and nucleolar function. Cell Cycle. 2014;4:1036–1038. doi: 10.4161/cc.4.8.1925. [DOI] [PubMed] [Google Scholar]
- 33.Hein N, Ganley A, Sanij E, et al. (2012) The nucleolus and ribosomal genes in aging and senescence. InTech. doi:10.5772/34581. ISBN: 978-935051-0144-4
- 34.Lim MJ, Wang XW. Nucleophosmin and human cancer. Cancer Detect Prev. 2006;30:481–490. doi: 10.1016/j.cdp.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Fiore PP. Playing both sides: nucleophosmin between tumor suppression and oncogenesis. J Cell Biol. 2008;182:7–9. doi: 10.1083/jcb.200806069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352:254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 37.Colombo E. Delocalization and destabilization of the Arf tumor suppressor by the leukemia-associated NPM mutant. Cancer Res. 2006;66:3044–3050. doi: 10.1158/0008-5472.CAN-05-2378. [DOI] [PubMed] [Google Scholar]
- 38.Grisendi S, Bernardi R, Rossi M, et al. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 39.Bertwistle D, Sugimoto M, Sherr CJ. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol Cell Biol. 2004;24:985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kondo T, Minamino N, Nagamura-Inoue T, et al. Identification and characterization of nucleophosmin/B23/numatrin which binds the anti-oncogenic transcription factor IRF-1 and manifests oncogenic activity. Oncogene. 1997;15:1275–1281. doi: 10.1038/sj.onc.1201286. [DOI] [PubMed] [Google Scholar]
- 41.Pederson T, Tsai RYL. In search of nonribosomal nucleolar protein function and regulation. J Cell Biol. 2009;184:771–776. doi: 10.1083/jcb.200812014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamoto N, Yasukawa M, Nguyen C, et al. Maintenance of tumor initiating cells of defined genetic composition by nucleostemin. Proc Natl Acad Sci USA. 2011;108:20388–20393. doi: 10.1073/pnas.1015171108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 44.Baker NE. Developmental regulation of nucleolus size during Drosophila eye differentiation. PLoS ONE. 2013;8:e58266. doi: 10.1371/journal.pone.0058266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poortinga G, Wall M, Sanij E, et al. c-MYC coordinately regulates ribosomal gene chromatin remodeling and Pol I availability during granulocyte differentiation. Nucleic Acids Res. 2011;39:3267–3281. doi: 10.1093/nar/gkq1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ali SA, Zaidi SK, Dacwag CS, et al. Phenotypic transcription factors epigenetically mediate cell growth control. Proc Natl Acad Sci USA. 2008;105:6632–6637. doi: 10.1073/pnas.0800970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang A, Shi G, Zhou C, et al. Nucleolin maintains embryonic stem cell self-renewal by suppression of p53 protein-dependent pathway. J Biol Chem. 2011;286:43370–43382. doi: 10.1074/jbc.M111.225185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qu J, Bishop JM. Nucleostemin maintains self-renewal of embryonic stem cells and promotes reprogramming of somatic cells to pluripotency. J Cell Biol. 2012;197:731–745. doi: 10.1083/jcb.201103071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosby R, Cui Z, Rogers E, Robinson VL. Knockdown of the Drosophila GTPase nucleostemin 1 impairs large ribosomal subunit biogenesis, cell growth, and midgut precursor cell maintenance. Mol Biol Cell. 2009;20:4424–4434. doi: 10.1091/mbc.E08-06-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCain J, Danzy L, Hamdi A, et al. Tracking nucleolar dynamics with GFP-Nopp140 during Drosophila oogenesis and embryogenesis. Cell Tissue Res. 2005;323:105–115. doi: 10.1007/s00441-005-0044-9. [DOI] [PubMed] [Google Scholar]
- 51.James A, Cindass R, Jr, Mayer D, et al. Nucleolar stress in Drosophila melanogaster. Nucleus. 2014;4:123–133. doi: 10.4161/nucl.23944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qin W, Chen Z, Zhang Y, et al. Nom1 mediates pancreas development by regulating ribosome biogenesis in zebrafish. PLoS One. 2014;9:e100796. doi: 10.1371/journal.pone.0100796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grewal SS, Li L, Orian A, et al. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol. 2005;7:295–302. doi: 10.1038/ncb1223. [DOI] [PubMed] [Google Scholar]
- 54.Demontis F, Perrimon N. Integration of insulin receptor/Foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila . Development. 2009;136:983–993. doi: 10.1242/dev.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grewal SS, Evans JR, Edgar BA. Drosophila TIF-IA is required for ribosome synthesis and cell growth and is regulated by the TOR pathway. J Cell Biol. 2007;179:1105–1113. doi: 10.1083/jcb.200709044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guarente L. Link between aging and the nucleolus. Genes Dev. 1997;11:2449–2455. doi: 10.1101/gad.11.19.2449. [DOI] [PubMed] [Google Scholar]
- 57.Smith JS, Brachmann CB, Pillus L, Boeke JD. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics. 1998;149:1205–1219. doi: 10.1093/genetics/149.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Machín F, Paschos K, Jarmuz A, et al. Condensin regulates rDNA silencing by modulating nucleolar Sir2p. Curr Biol. 2004;14:125–130. doi: 10.1016/S0960-9822(04)00002-8. [DOI] [PubMed] [Google Scholar]
- 59.Guarente L. Diverse and dynamic functions of the Sir silencing complex. Nat Genet. 1999;23:281–285. doi: 10.1038/15458. [DOI] [PubMed] [Google Scholar]
- 60.Murayama A, Ohmori K, Fujimura A, et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 61.Yang L, Song T, Chen L, et al. Regulation of SirT1-nucleomethylin binding by rRNA coordinates ribosome biogenesis with nutrient availability. Mol Cell Biol. 2013;33:3835–3848. doi: 10.1128/MCB.00476-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Evans DS, Kapahi P, Hsueh W-C, Kockel L. TOR signaling never gets old: aging, longevity and TORC1 activity. Ageing Res Rev. 2011;10:225–237. doi: 10.1016/j.arr.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin DE, Powers T, Hall MN. Regulation of ribosome biogenesis: where is TOR? Cell Metab. 2006;4:259–260. doi: 10.1016/j.cmet.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 64.Demontis F, Patel VK, Swindell WR, Perrimon N. Intertissue control of the nucleolus via a myokine-dependent longevity pathway. Cell Rep. 2014;7:1481–1494. doi: 10.1016/j.celrep.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matheu A, Maraver A, Klatt P, et al. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- 66.Hulbert AJ, Pamplona R, Buffenstein R, Buttemer WA. Life and death: metabolic rate, membrane composition, and life span of animals. Physiol Rev. 2007;87:1175–1213. doi: 10.1152/physrev.00047.2006. [DOI] [PubMed] [Google Scholar]
- 67.Ping Tang Y, Wade J. Sexually dimorphic expression of the genes encoding ribosomal proteins L17 and L37 in the song control nuclei of juvenile zebra finches. Brain Res. 2006;1126:102–108. doi: 10.1016/j.brainres.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu S, De Croos JNA, Storey KB. Cold acclimation-induced up-regulation of the ribosomal protein L7 gene in the freeze tolerant wood frog, Rana sylvatica. Gene. 2008;424:48–55. doi: 10.1016/j.gene.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 69.Twiss JL, Smith DS, Chang B, Shooter EM. Translational control of ribosomal protein L4 mRNA is required for rapid neurite regeneration. Neurobiol Dis. 2000;7:416–428. doi: 10.1006/nbdi.2000.0293. [DOI] [PubMed] [Google Scholar]
- 70.Uechi T, Nakajima Y, Nakao A, et al. Ribosomal protein gene knockdown causes developmental defects in zebrafish. PLoS ONE. 2006;1:e37. doi: 10.1371/journal.pone.0000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Duc A-CE, Rao S, et al. Control of hematopoietic stem cell emergence by antagonistic functions of ribosomal protein paralogs. Dev Cell. 2013;24:411–425. doi: 10.1016/j.devcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kondrashov N, Pusic A, Stumpf CR, et al. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xue S, Tian S, Fujii K, et al. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature. 2015;517:33–38. doi: 10.1038/nature14010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Le Bouteiller M, Souilhol C, Beck-Cormier S, et al. Notchless-dependent ribosome synthesis is required for the maintenance of adult hematopoietic stem cells. J Exp Med. 2013;210:2351–2369. doi: 10.1084/jem.20122019. [DOI] [PMC free article] [PubMed] [Google Scholar]


