Abstract
Peroxisome proliferator-activated receptors (PPARs) are members of the nuclear receptor superfamily and function as transcription factors that regulate gene expression in numerous biological processes. Although the PPARβ/δ subtype is highly expressed in the brain, its physiological roles in neuronal function remain to be elucidated. In this study, we examined the presence of PPARβ/δ in the master circadian clock of the Syrian hamster and investigated its putative functional role in this structure. In mammals, the central circadian clock, located in the suprachiasmatic nucleus (SCN), is entrained by the light–dark (LD) cycle via photic6 signals conveyed by a direct pathway whose terminals release glutamate. Using immunocytochemical and qRT-PCR analysis, we demonstrated that the rhythmic expression of PPAR β/δ within the SCN of hamsters raised under an LD cycle was detectable only at the transcriptional level when the hamsters were maintained under constant darkness (DD). The increase in the number of immunoreactive PPARβ/δ cells observed under DD after light stimulation during the early subjective night (CT14), but not during the subjective day (CT06), demonstrated that the expression of PPARβ/δ can be up-regulated according to the photosensitive phase of the circadian clock. All of the PPARβ/δ-positive cells in the SCN also expressed the glutamate receptor NMDAR1. Moreover, we demonstrated that at the photosensitive point (CT14), the administration of L-16504, a specific agonist of PPARβ/δ, amplified the phase delay of the locomotor response induced by a light pulse. Taken together, these data suggest that PPARβ/δ activation modulates glutamate release that mediates entrainment of the circadian clock by light.
Keywords: Circadian clock, PPARβ/δ, Glutamate, Hamster
Introduction
Nuclear receptors represent the largest family of transcription factors. Members of the nuclear receptor superfamily, the peroxisome proliferator-activated receptors (PPARs), have been previously described as lipid sensors, which bind fatty acids and initiate their subsequent metabolism [1]. Long-chain unsaturated fatty acids and their metabolic derivatives are natural PPAR ligands, and their expression levels undergo changes that correspond to the nutritional status. In vertebrates, three different isotypes have been described, PPARα, PPARβ/δ, and PPARγ, with distinct patterns of tissue distribution (liver, adipose tissue, heart, muscle) consistent with their specific functions in the regulation of energy metabolism [2].
In peripheral tissues, PPAR expression exhibits daily rhythms. Moreover, these rhythms display characteristic, yet distinct, tissue-specific patterns [3]. Furthermore, all of the PPARs cycle in the liver [4, 5]. For example, PPARα and δ cycle in brown adipose tissue, and PPARα and γ in white adipose tissue. It has been suggested that these oscillations correlate with a known function of PPARs. For example, in mice, expression levels of PPARα are higher during the light phase when they are in a fasting state that requires ketone bodies and are low at night when feeding provides hexose sugars as a primary fuel source [3]. To our knowledge, such a daily rhythm of PPAR expression has not yet been reported in the brain and notably in the central circadian clock that exhibits daily rhythm in energy metabolism, and in which we previously demonstrated that deficiency in polyunsaturated fatty acids modulates metabolic activity [6].
All of the PPARs are co-expressed in the nervous system during rat embryogenesis. The expression of PPARβ/δ peaks during late development, which coincides with the onset of myelination [7]. Although PPARβ/δ remains highly expressed [8], the expression of PPARα and PPARγ decreases postnatally in brain [7]. In adults, PPARβ/δ is expressed throughout the brain, including the hypothalamus, while PPARα and PPARγ are localized to more restricted brain areas [9]. Because PPARs are able to suppress the inflammatory response in peripheral macrophages [1], most brain-related studies have investigated the putative role of PPARs in inflammatory brain disorders. The sustained expression of PPARβ/δ in the adult central nervous system suggests that it plays a major role in physiological functions. In this study, we looked for the expression of PPARβ/δ in the Syrian hamster master clock.
In mammals, the central circadian clock is located in the suprachiasmatic nuclei (SCN) of the ventral anterior hypothalamus. The SCN, which is entrained directly by the light–dark (LD) cycle, is proposed to synchronize slave oscillators in peripheral tissues by neuronal and hormonal mechanisms [10–12]. This allows animals to adapt their behavioral and physiological activities to daily changes in the environment. The photoperiodic signal is conveyed by the retinohypothalamic tract (RHT), a direct pathway linking a population of retinal ganglion cells to the SCN, whose terminals release glutamate [13, 14]. Under constant conditions (e.g., constant darkness), rhythms persist with a period that is close to 24 h, which demonstrates autonomous functioning of the circadian clock. A hallmark of circadian timing is that the clock responds to resetting signals in a phase-restricted manner. The temporal responses of the clock to light are represented as photic phase shifts.
Based on the intrinsic properties of the central clock, we investigated the putative functional role of PPARβ/δ in this structure. Using qRT-PCR and immunocytochemical analyses, we examined the distribution of PPAR β/δ within the SCN of hamsters raised under the LD cycle or maintained under constant darkness. Furthermore, we investigated whether the PPARβ/δ-positive cells of the SCN were light-responsive. To determine whether PPARβ/δ may be involved in the photic entrainment of the circadian clock, we investigated whether PPARβ/δ and the glutamate receptor NMDAR1 (N-Methyl-d-Aspartate Receptor) were expressed within the same cells of the SCN and studied the effects of a PPARβ/δ agonist on the phase-shifting response to light stimulation.
Materials and methods
Ethical approval
All of the experiments were performed according to the guidelines of the French National Committee for Animal Care and the European Convention of Vertebrate Animals Used for Experimentation, under the European Council Directive 86/609/EEC (dated November, 1986). In addition, all efforts were made to minimize the number, pain and amount of discomfort to the hamsters.
Animals
Syrian hamsters (Mesocricetus auratus) were raised in our colony under a 14/10 h light/dark (LD) cycle, (lights off at 1:00 p.m. and on at 11:00 p.m.) at 23 ± 1 °C. The hamsters had free access to food and water, and only male progeny were used for this study. Fifty-six males maintained under the LD cycle constituted the LD group. The experimental manipulations were timed relative to the light/dark transition (L/D), which was conventionally defined as Zeitgeber time 12 (ZT 12). For the rhythmic expression studies, the hamsters were killed at six different time points during the daily cycle over a 24-h period. Furthermore, 30 other hamsters were housed from weaning and for one month under constant darkness (DD group). At least 2 weeks prior to killing, the hamsters were moved into individual cages that were equipped with running wheels. The wheel rotation was recorded using VitalView software (Mini Mitter Co., Sunriver, OR, USA) to determine the phase of the clock at the time of killing. For each animal kept under DD, the onset of activity was defined as circadian time 12 (CT 12), corresponding to the beginning of the “subjective night” for the nocturnal animals. The rest phase corresponded to the “subjective day.” PPARβ/δ variations under DD were only examined at two time points where the highest and the lowest expressions were observed under the LD cycle. Moreover, to determine whether PPARβ/δ cells may be light-responsive, we performed experimental studies examining PPARβ/δ expression after 1 h of light (100 lux) at ZT14, CT06 and CT 14. At CT14, we also studied the relationship between light-induced c-Fos [13] and PPARβ/δ expression.
Next, we investigated the modulatory effects of the specific PPARβ/δ agonist (L-165,041, Tocris) and antagonist (GSK3787, Tocris) on the photic phase response. The L-165,041 and GSK3787 were dissolved in vehicle (0.1 % hydroxyethyl cellulose with 1 % Tween80). Thirty min before CT14, vehicle, L-165,041 or GSK3787 (10 mg/kg) was orally administered (p.o.). The hamsters were randomly selected and assigned to different groups: vehicle only, vehicle + 15 min light pulse (100 lux), L-165,041 or GSK3787 only, or L-165,041 or GSK3787 + 15 min light pulse. The phase-shift was calculated as the difference between the two lines fitted to the circadian onsets before and after treatment, considering eight cycles before and after treatment, respectively (Clocklab software, Actimetrics, Evanston, IL, USA).
mRNA analysis
The hamsters were killed by decapitation, and their brain was quickly removed. The SCN was dissected on ice and frozen using liquid nitrogen. Total RNA was isolated from 3 to 5 mg of the suprachiasmatic nucleus from the Syrian hamster using the mir Vana miRNA isolation kit (Qiagen, Courtaboeuf, France), in the absence of a DNase digestion step, and was performed according to the manufacturer’s instructions. The concentration and purity of the eluted total RNA were confirmed by the relative UV absorbance as expressed by the A260/A230 and A280/A260 ratios (Biophotometer, Eppendorf SARL, France).
Real-time RT-PCR
The cDNA was prepared from 1 μg total RNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Courtaboeuf, France) and a mixture of oligo-dT and random hexamers as primers, and then diluted with nuclease-free water (1:50, v/v).
The mRNA levels of PPARβ/δ and 18S RNA were measured with real-time RT-PCR using an ABI Prism 7000 Real-Time PCR System (Applied Biosystems, Courtaboeuf, France) and the cDNA was amplified in a 25 μl sample volume containing SYBR Green PCR Master Mix, and 300 nM concentrations of specific primers (Applied Biosystems, Courtaboeuf, France) as recommended by the manufacturer. Each measurement was performed in triplicate and repeated. 18S was used as an internal control to assess the efficiency of the RT-PCR and subsequent normalization. The data were analyzed with the comparative cycle threshold (C
t) method: the expression of PPARβ/δ was normalized to the expression of 18S RNA and normalized to ZT18 as a calibrator to generate the ΔΔC
t values. The relative quantities (RQ) were calculated with the equation: 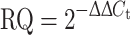 .
. 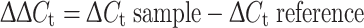 .
.
Primers for PPARβ/δ and the housekeeping gene 18S RNA were designed with Primer Express 2.0 (Applied Biosystems) according to the general guidelines for optimal primer design. All of the selected primers were tested for specificity by BLASTN searches. The primer sequences for PPARβ/δ and 18S RNA were based on sequences obtained from PubMed; NM_013141 and X03205, respectively. The primer sequences were as follows: PPARβ/δ, AGAAGGGCCTGGTGTGGAA (5), AGGGTCACCTGGTCGTTGAG (4) and 18S RNA, CCTGGATACCGCAGCTAGGA (3), CGGCCGTCCCTCTTAATCAT (4). The specificity of the PCR amplification for each primer pair was analyzed on an agarose gel and confirmed to give a single amplicon of the correct size (173 and 91 base pairs, respectively). In addition, the primers were only used when they produced a single amplicon as revealed by a dissociation curve analysis after PCR.
Immunohistochemistry methods
All of the animals used for immunohistochemistry were deeply anesthetized with sodium pentobarbital (120 mg/kg) and intracardially perfused with 0.9 % saline at 37 °C followed by 4 % cold paraformaldehyde in 0.1 M PBS at pH 7.4. The brains were removed, postfixed in the same fixative for 4 h and cryoprotected in 30 % sucrose (in 0.1 M PBS). Serial 30-μm coronal sections were cut on a freezing microtome. Under darkfield illumination, the sections were selected to ensure that they overlapped with the entire SCN.
For ultrastructural examination, the hamsters were perfused with a 4 % paraformaldehyde-0.075 M l-Lysine-0.01 M sodium metaperiodate solution in 0.1 M phosphate buffer. After dissection, the brains were postfixed overnight at 4 °C in the same fixative. Serial 50-μm coronal sections were then obtained using a vibratome (Leica VT1000S) and selected as described above.
PPARβ/δ immunodetection
To inhibit the endogenous peroxidase activity, free-floating tissue sections were placed in a solution of hydrogen peroxide (0.5 % for 30 min). Non-specific binding was blocked by 5 % normal donkey serum in phosphate buffer saline containing 0.3 % Triton X 100 (PBST) for 1 h at room temperature. After washing in PBST, the sections were incubated for 72 h at 4 °C with a polyclonal rabbit PPARβ/δ antibody (1:500 dilution; Santa Cruz sc-7197, CA, USA). The sections were washed extensively with PBST and then incubated overnight at 4 °C with a biotinylated anti-rabbit antibody (1:400 dilution; Jackson). Immunohistochemical localization was determined using the avidin–biotin peroxidase complex method (ABC kit; Vector Laboratories) diluted at 1:100 in PBS for 1 h. The sections were washed for 30 min prior to their reaction with 3,3′-diaminobenzidine tetrahydrochloride as a chromogen (0.02 % in PBS with 0.1 % hydrogen peroxide) for 10 min.
For ultrastructural analysis, the sections were subjected to the same protocol, but the TritonX 100 step was omitted. Once the DAB reaction was completed, the sections were postfixed in 1 % OsO4 in 0.1 M phosphate buffer, contrasted “en bloc” with 1 % uranyl acetate, dehydrated in a graded series of ethanol and flat-embedded in Epoxy resin. Ultrathin sections (70 nm) were collected on copper grids, contrasted with lead citrate and observed with a Zeiss EM902 electron microscope operated at 80 kV (MIMA2–Plateau de Microscopie Electronique, Jouy-en-Josas, France).
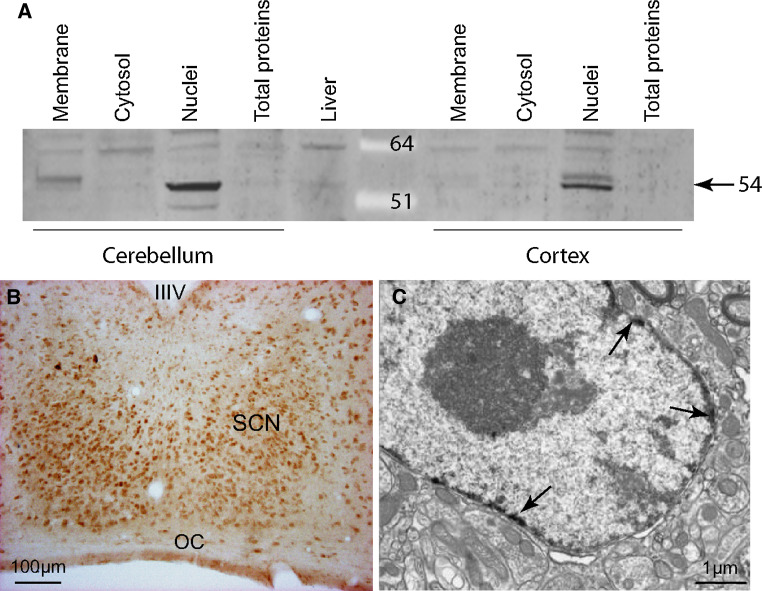
The specificity of the PPARβ/δ antibody was confirmed by omitting the primary antibody and by Western blotting to ensure that the protein was detected and was of the correct size in the subcellular compartments, including the cytoplasm, plasma membrane and cellular nuclei. The antibody recognized a single band at 54 kD, predominantly in the nuclear fraction (Fig. 1a). Moreover, the electron microscopy study localized PPARβ/δ at the neuronal nuclear membrane position (Fig. 1c).
Fig. 1.
Immunodetection of PPARβ/δ in the hamster suprachiasmatic nucleus (SCN). a Specificity for the PPARβ/δ antibody was verified in two hamster cerebral areas (cerebellum and cortex) using Western blotting, which showed the nuclear localization of the protein, and in the liver. b A representative coronal section in the mid-SCN of an adult hamster killed during the light phase. Immunoreactivity was observed throughout the rostro-caudal extent of the SCN with a higher density in the ventro-median part. c An electron microscopy immunolabeling of PPARβ/δ (indicated by the arrows), was only observed in the nuclei of the neurons in the SCN. IIIV third ventricle, OC optic chiasm
Combined immunodetection of PPARβ/δ with NMDAR1 or c-Fos in the SCN
The immunolabeling of PPARβ/δ and NMDAR1 was performed in hamster tissue at ZT06 and ZT14. Furthermore, the immunolabeling of c-Fos and PPARβ/δ was processed in hamsters exposed to a light pulse (100 lux) at CT 14 for 1 h.
Free-floating sections were blocked with a mixture of 10 % horse serum and PBST for 1 h at room temperature. Double-immunolabeling was performed in subsequent steps. The sections were first incubated in the PPARβ/δ antibody (1:500 dilution; Santa Cruz Biotechnology, CA, USA) diluted in PBST for 48 h at 4 °C, followed by a secondary fluorochrome-conjugated antibody (donkey anti-rabbit Alexa-Fluor 488, Invitrogen, France) in PBS for 2–3 h. Next, the sections were processed in the same way with the polyclonal rabbit anti-NMDAR1 (1:400 dilution, Abcam, Cambridge) or polyclonal goat anti-c-Fos (1:100; SantaCruz Biotechnology, CA, USA) and were visualized with secondary fluorochrome-conjugated antibodies (donkey anti-rabbit Alexa-Fluor 589 or donkey anti-goat Alexa-Fluor 556, respectively, Invitrogen, France). Each step was followed by washing with PBS three times for 1 h at room temperature. The sections were then analyzed with a Zeiss ApoTome using Structured Illumination (MIMA2 Platform, INRA Jouy-en-Josas, France).
Quantification of PPARβ/δ-positive cells
In each group, 18 sections per SCN were examined in all of the animals. The number of PPAR-immunopositive cell nuclei was counted using the Mercator software from the Explora Nova image analysis system (La Rochelle, France). Only the uniformly dark-stained nuclei and pycnotic dark-brown nuclei (well above the basal background staining of the surrounding tissue) were mapped and used for quantitative analysis.
The data were treated as follows: (1) the number of PPAR-Ir cells in each SCN was estimated by counting the number of labeled cells in the 18 SCN sections; (2) the number of PPAR-Ir cells, according to their dorso-ventral distribution, was studied throughout each SCN by counting the number of labeled cells in the superior third (dorsal area) and in the inferior two-thirds (ventro-median area) of the SCN; (3) the mean number of PPAR-Ir cells at a given time was calculated by averaging the values obtained from the animals killed at the same circadian time point.
Statistical analysis
The effects of the time of killing or light stimulation were analyzed by a non-parametric one-way ANOVA followed by the post hoc Tukey test to test the linear contrasts. The effects of light and drug on the phase shift were analyzed by a two-way-ANOVA followed by a Tukey test. All of the analyses were performed using SigmaStat software (version 2 for Windows).
Results
PPARβ/δ expression in the hamster SCN
Evidence of a daily rhythm
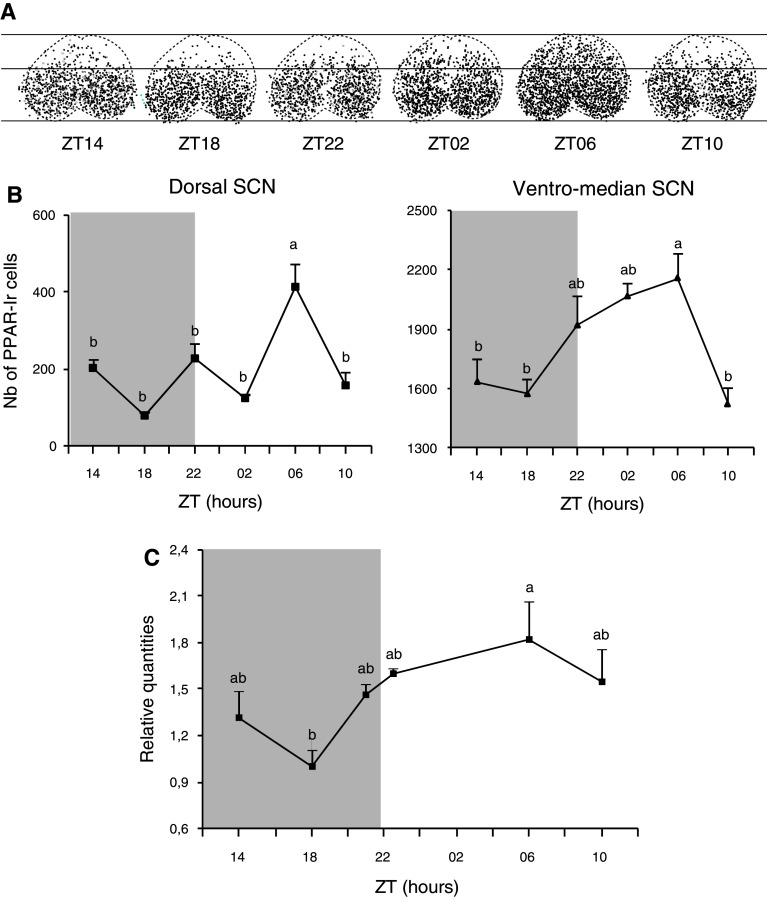
PPARβ/δ-immunoreactivity (Ir) showed a nuclear localization in neurons of the SCN, cerebellum and cerebral cortex (Fig. 1a, b). An electron microscopy study showed that immunolabeling in the SCN was only observed in the nuclei of neurons (Fig. 1c). A dense pool of PPARβ/δ-positive cells was found throughout the rostro-caudal extent of the SCN and particularly in the ventro-median part (Fig. 1b). The animals killed at the six different time points of the 24-h period exhibited various patterns in the number and distribution of the PPAR-Ir cells within the SCN (Fig. 2a). Because the highest contingent of PPARβ/δ cells was in the ventro-median portion of the SCN, we independently counted the cells from the ventral and dorsal areas, as illustrated in Fig. 2a. The number of PPAR cells increased at the light onset only in the ventral part of the SCN, peaked at ZT06 (2,571 ± 342) and decreased at the end of the light phase (Fig. 2b). The number of PPARβ/δ-Ir cells was highest at ZT06 not only in the ventral part (i.e., in the region where VIP cells are found), but also in the dorsal part (i.e., in the region containing vasopressin-positive cells) where the number of labeled cells reached 413 ± 121 and varied between 228 ± 76 and 80 ± 15 at the other time points. The pparβ/δ mRNA expression also exhibited a similar daily profile (Fig. 2c).
Fig. 2.
Spatio-temporal expression of PPARβ/δ in the hamster SCN. a A superimposed map of the immunolabeled neurons plotted in each section throughout the SCN of a representative hamster for each of the six time points over a 24-h period. b The variation in the number of immunoreactive neurons in the dorsal (1/3 higher) and ventro-median part (2/3 lower) of the SCN over the light–dark (LD) cycle as demonstrated by a peak of immunoreactivity observed at ZT06. c The daily mRNA expression exhibited a similar profile to that of proteins. ZT Zeitgeber time, ZT12 = light/dark transition. The grey area delineates the dark phase. The values are presented as the mean ± SEM (n = 4 or 5). a, b Significant difference between the different killing times (p < 0.05, one-way ANOVA)
As previously described [8], the detection of PPARβ/δ was not restricted to the SCN but was also observed in the cerebellar Purkinje cells, hippocampal and cortical pyramidal cells, in the paraventricular and supra-optic nuclei, and was more dispersed in other areas of the hypothalamus. Importantly, the number of labeled cells in the latero-anterior hypothalamic nuclei remained constant over the LD cycle (data not shown).
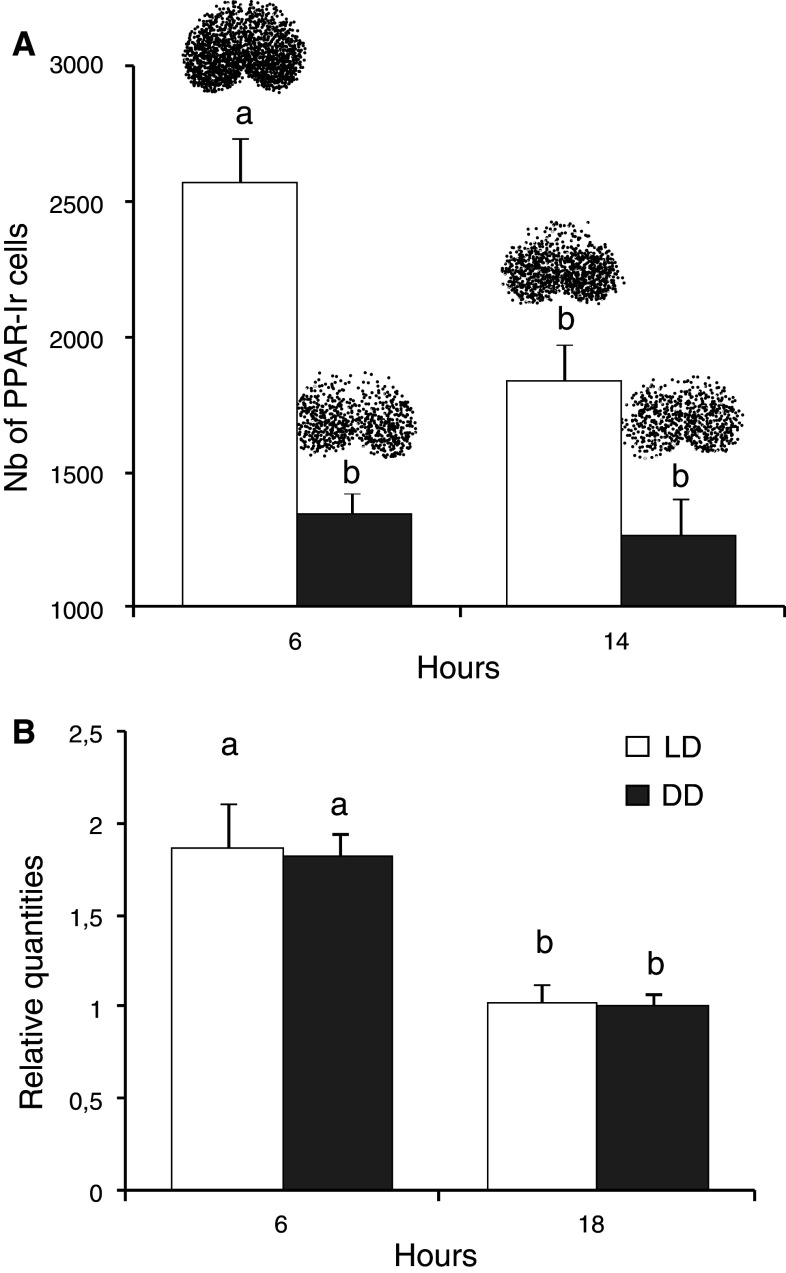
In animals that were maintained from weaning under DD, the number of PPARβ/δ immunoreactive cells was strongly reduced (by 48 and 30 % at CT06 and CT14, respectively), and no difference between CT06 and CT14 was detected (1,343 ± 182 vs. 1,270 ± 227, respectively, Fig. 3a), whereas the mRNA expression exhibited significant variations that were similar to those observed in LD (Fig. 3b). These data suggest that pparβ/δ mRNA expression is a feature related to the endogenous function of the SCN, while PPARβ/δ protein expression is modulated by the light environment.
Fig. 3.
PPARβ/δ expression in the hamster SCN varies with the light condition. The PPARβ/δ expression in the SCN of hamsters maintained under either the light–dark (LD) cycle or constant darkness (DD) was compared in hamsters killed at two time points of the light and dark phase showing significant difference under the LD cycle. a The variation in the number of immunoreactive neurons between ZT06 and ZT14 was abolished under DD; b day-night variations of mRNA expression were still present under DD (i.e., between CT06 and CT18). These results indicate that the PPARβ/δ protein expression is modulated by the presence of light. The values are presented as the mean ± SEM (n = 4 or 5). a, b Significant difference between the different killing times (p < 0.05, one-way ANOVA). Above the bars representative diagram of PPARβ/δ distribution. ZT Zeitgeber time, under the LD cycle, the light/dark transition is conventionally defined as ZT12; CT circadian time, under the DD at the onset of activity is defined as CT12
The effects of light on PPARβ/δ are time-dependent
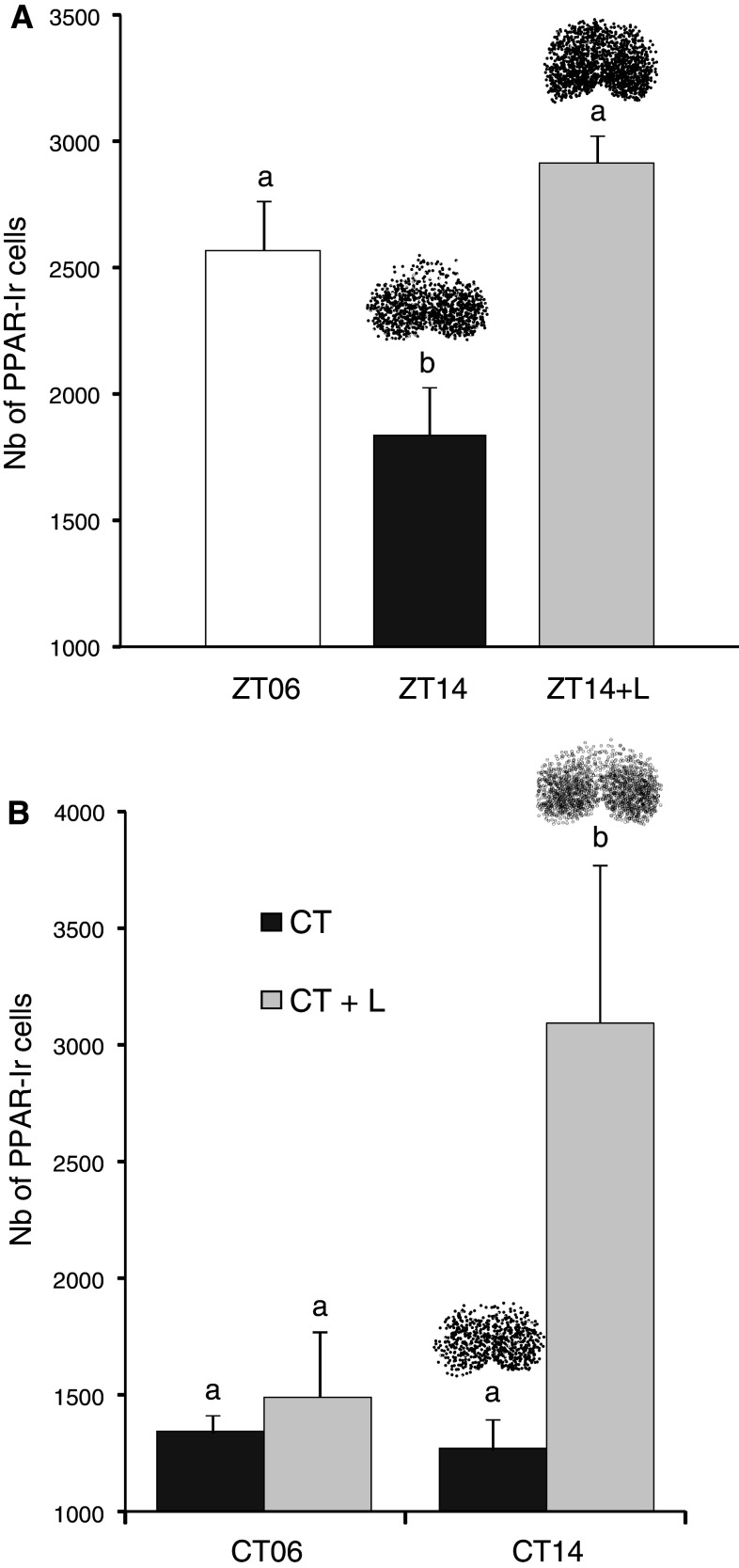
A 1-h light pulse applied at early nighttime induced a dramatic increase [65 % at ZT14 (Fig. 4a) and 116 % at CT14 (Fig. 4b)] in the number of PPARβ/δ cells in the SCN, whereas the same stimulation produced no effect at CT06, where the number of labeled cells remained low (1,495 ± 338) (Fig. 4b). The difference in the light effects observed between CT14 and CT06 (2,755 ± 506 vs. 1,495 ± 338 respectively, Fig. 4b) indicates that PPARβ/δ expression is dependent on the circadian photosensitivity of the SCN.
Fig. 4.
The effect of light (L) on PPARβ/δ immunoreactivity in the SCN is time-dependent. a In the hamsters maintained under the light–dark cycle, a 1-h light pulse given at ZT14 and 2 h after the light/dark transition produced an immunoreactivity that was similar to those observed during the light phase at ZT06 a, b significant difference p < 0.05, one-way ANOVA. b In the hamsters maintained under constant darkness, a light pulse given either during the rest phase (CT06) or 2 h after the onset of the active phase (CT14) exhibited an effect only at CT14 with a dramatic increase of immunolabeling. a, b Significant difference p < 0.05, one-way ANOVA. The values are presented as the mean ± SEM (n = 4 or 5). Above the bars representative diagram of PPARβ/δ distribution
Characterization of PPARβ/δ-positive cells
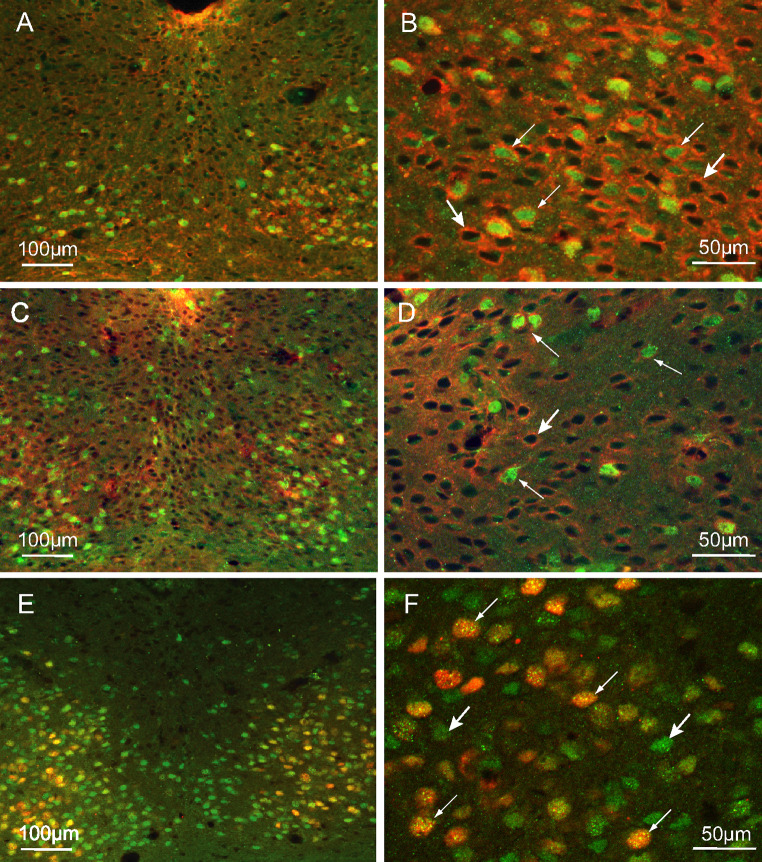
Most PPARβ/δ cells expressed NMDAR1 (Fig. 5a–d), independent of time, ZT06 (Fig. 5c, d) or ZT14 (Fig. 5a, b), which suggests that PPARβ/δ is activated in cells that are sensitive to glutamate stimulation. Moreover, cells that expressed light-induced c-Fos were nearly all PPAR-positive; however, all of the PPAR-cells were not c-Fos-positive (Fig. 5e, f). These observations support a role of PPARβ/δ in the photic entrainment of the circadian clock.
Fig. 5.
Characterization of PPARβ/δ immunoreactive (Ir) neurons. Combined detection of PPARβ/δ (green) and NMDAR1 (red) in the ventral part of the SCN of the hamsters killed at ZT14 (a, b) and in the dorsal part of hamsters killed at ZT06 (c, d); PPARβ/δ is co-expressed with NMDAR1 (thin arrows), there are a number of cells that only expressed NMDAR1 (large arrows). e, f Colocalization of PPARβ/δ (green) and light-induced c-Fos (red) in the SCN of hamsters killed at CT14; the c-Fos-Ir cells are nearly all PPARβ/δ-Ir (thin arrows), and the PPARβ/δ cells are widely distributed within the SCN (large arrows)
A PPARβ/δ agonist (L-165,041) increases the phase shifting effect of light
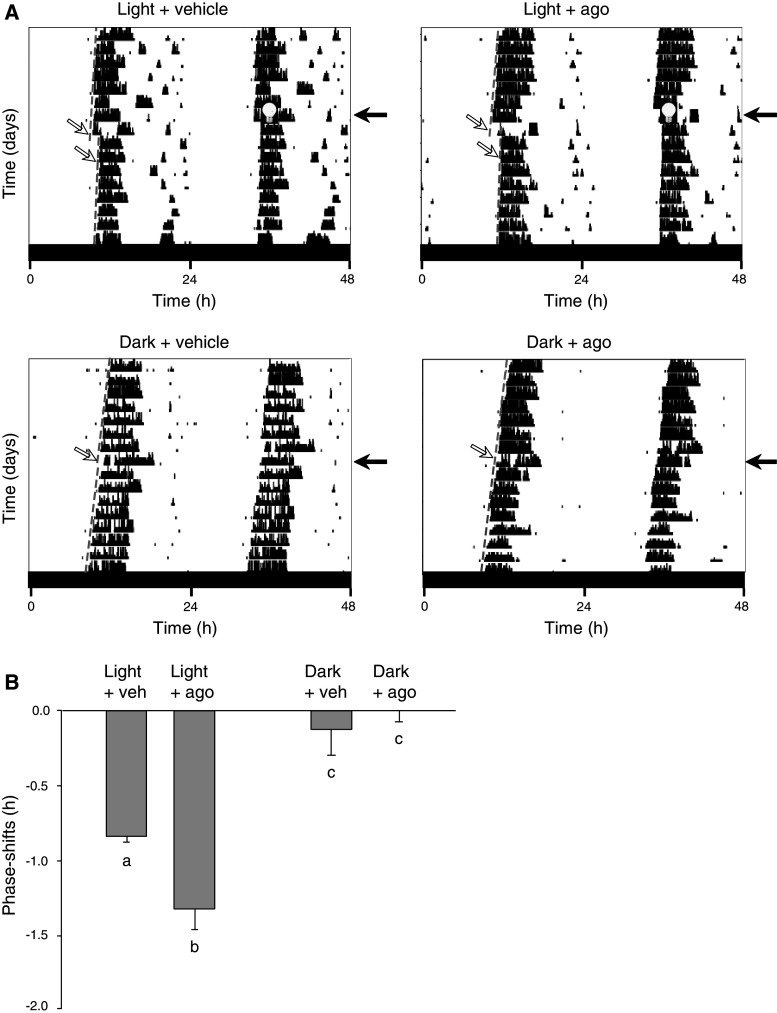
We tested the hypothesis that PPARβ/δ activation modulates the photic signal to the SCN. The magnitude of the phase shifting in response to a 15-min light pulse at CT14 (−0.83 ± 0.1 h) was significantly (+60 %) augmented in the L-165,041-treated group (−1.32 ± 0.34 h). In contrast, the light-induced phase shift was not reduced significantly by the PPARβ/δ antagonist GSK 3787 (−20 % of the control group; data not shown). Administration of vehicle or L-165,041 in the animals maintained in darkness did not produce any significant shifting response (Fig. 6). These results indicate that PPARβ/δ activation amplifies the SCN response to light stimulation, which suggests a role of PPARβ/δ in the regulation of photic stimulation of the circadian clock.
Fig. 6.
Administration of L-165,041, a specific PPARβ/δ agonist, amplifies the light-induced phase shift. a Representative double-plotted actograms of wheel-running activity of four hamsters that were housed in constant darkness DD. The animals were treated with vehicle (“veh”, left panels) or a specific PPARβ/δ agonist (10 mg/kg L-165,041, noted “ago”; right panels), 30 min prior to a 1-h light pulse (100 lux indicated by a bulb) at circadian time 14 (upper panels) or not (lower panels). To calculate the phase shift, the best-fit dashed lines (white arrows) to the circadian onsets have been drawn across 8 days before and after the treatment. Black arrows day of treatment. b The mean-phase-shifts in response to vehicle (n = 4) or PPARβ/δ agonist (n = 4), to vehicle + light (n = 5) or agonist + light (n = 6). The light-induced phase-delays were significantly increased with the PPAR agonist. The values are presented as the mean ± SEM (a, b, c p < 0.05, two-way ANOVA)
Discussion
Our results provide the first evidence for the presence of PPARβ/δ in the hamster SCN and the first demonstration that PPARβ/δ expression can be induced by glutamate stimulation. Previous studies have reported links between the PPAR signaling pathway and clock genes in peripheral clocks [3, 4, 15, 16] and have mainly explored the role of PPARs as key regulators of energy metabolism and potential mediators of the entrainment effect of feeding schedules [17, 18]. In this study, we provide new insights into the functional role of PPARβ/δ in the master clock located in the SCN.
Characteristics of PPARβ/δ expression in the SCN
The rhythmic expression of PPARβ/δ mRNA and protein exhibited similar profiles under LD. The expression was characterized by an increase before the dark/light transition, attained a maximum in the middle of the light phase (i.e., at ZT06) and declined before lights off. PPARβ/δ is predominantly expressed in the ventro-median region of the SCN, where the terminal field of the (glutamatergic) retinohypothalamic afferences project [19]. To investigate whether this rhythmic expression was dependent on the light environment and/or resulted from the endogenous functioning of the circadian clock, the hamsters were maintained under constant darkness. We demonstrated that the ppar β/δ transcript continued to exhibit circadian variations, whereas surprisingly the protein levels did not, and that the number of PPARβ/δ-Ir cells at the two time points studied (i.e., CT06 and CT14) was strongly reduced and became equivalent to the daily trough under LD cycle. By sampling only two CTs, we cannot fully exclude the possibility that the circadian peak of PPARβ/δ-Ir would take place later than CT6 (i.e., with a delay compared to the peak of mRNA occurring at that time-point).
To verify the potential action of light on the expression of PPARβ/δ, we applied a 1-h light pulse in early nighttime (ZT14) and found a 60 % increase in the number of PPARβ/δ-Ir neurons (Fig. 4a). Importantly, this response to light does not result from passive or direct modulation of light on PPARβ/δ expression. First, the same light pulse led to overexpression of SCN PPARβ/δ-Ir in hamsters raised under constant darkness when applied during subjective night, but not during subjective day (i.e., there is a circadian gating in the light-induced up-regulation of PPAR β/δ). Second, the high levels of PPARβ/δ expression in the SCN of hamsters under LD were not strictly in phase with the light period, but instead they anticipated it and finished before its end.
Moreover, the PPARβ/δ-expressing cells in the SCN were also positive for NMDAR1, the principal glutamate receptor that mediates phase shifting of circadian rhythmicity by light [20, 21]. Furthermore, a large number of the cells co-expressed light-induced c-Fos when the hamsters were exposed to light in the early nighttime. Numerous studies demonstrated that c-Fos expression in the SCN was induced under light stimulation only at times associated with the phase shifting. Taken together, these data indicate that PPARβ/δ expression in the SCN is up-regulated by light, and most likely by glutamate release. Thus, we suggest that PPARβ/δ may be involved in the reception/integration of photic signals within the SCN.
PPARβ/δ activation signaling pathway in the SCN
PPARβ/δ is a ligand-activated transcription factor. PPARs are maintained in the nucleus in a repressed state by co-repressors. Ligand binding to PPARs induces a conformational change that is essential to initiate a complex process that results in transcriptional activation [22]. Endogenous ligands include long-chain fatty acids and their derivatives [23–25]. Given the nuclear localization and the increase of PPAR β/δ-Ir under light, this provides further support for the idea that light was causing a conformational change of the protein, and the PPAR β/δ antibody used recognized PPAR β/δ transcriptionally active. Moreover, the signaling cascade induced by photic stimulation in the SCN may explain the potential mechanisms leading to PPARβ/δ activation. Indeed, in the SCN, RHT terminals release glutamate in response to light stimulation [13], which subsequently induces an influx of Ca2+ via the activation of the postsynaptic NMDA receptors [21, 26]. The elevated [Ca2+]i activates and promotes the translocation of the cytosolic phospholipase A2 (cPLA2) to membranes [27] and results in the release of long-chain polyunsaturated fatty acids from membrane phospholipids, which mainly include arachidonic acid (AA) [28–30]. Subsequently, cyclooxygenase (COX)-2 induction [31] then converts AA to prostaglandins. Because COX is highly expressed in the rat SCN [32], and AA and prostaglandins are natural PPAR ligands, the detection of PPARβ/δ in cells that also expressed NMDAR1 is consistent with the activation of PPAR β/δ by light possibly via cPLA2 [33]. Taken together, we propose that photoactivation of PPAR β/δ induces a conformational change in the protein, which then exposes the epitope recognized by the antibody used, and thus the increase in nuclear immunolabeling reflects the activation of PPAR β/δ by light.
However, the overexpression of PPARβ/δ observed after 1 h of light given at CT14 in the hamsters that were maintained under constant darkness was not triggered when the light was applied at CT06 during the rest phase (subjective day). The lack of an effect at CT06 revealed that PPARβ/δ protein expression exhibits a differential sensitivity to light over the 24-h period. Thus, the mere presence of light was not sufficient to explain the variations observed in PPARβ/δ expression over time. In addition, the intrinsic properties of the clock provide some understanding of PPARβ/δ activation specifically in the SCN.
Indeed, NMDARs are considered the principal glutamate receptors that mediate phase shifting of circadian rhythmicity by light [20, 21]. A circadian variation of NMDAR1 expression has been shown in the SCN, with higher protein expression levels during the night, at a time when the light exerts its entraining effects on the circadian clock [34]. A circadian oscillation in NMDA-evoked calcium transients has been demonstrated in the SCN [35].
Under basal conditions, the daily rhythm in resting [Ca2+]i levels in SCN neurons of rats maintained in the LD cycle show values that are significantly higher during the day (150 nM) than during the night (75 nM). When rats are placed in constant darkness, the rhythm continues to be observed [36]. The rhythm in the basal [Ca2+]i accounts for the daily rhythm in Ca2+ influx, which is consistent with the daily rhythm of spontaneous neuronal activity and membrane potential, which peak during the day [37–40]. This peak results in the cells being relatively depolarized, leading to a Ca2+ influx during the day. A light pulse during that period does not cause phase shifts, most likely because the [Ca2+]i is already elevated. Conversely, during the night, the membrane is relatively hyperpolarized, and light-induced depolarization causes a significant Ca2+ influx, resulting in a phase shift of the rhythm [35]. At least in mouse hippocampal cells, ligand-activated PPAR β/δ has been shown to modulate glutamate-induced level of intracellular calcium ions [41].Taken together, these data suggest that (1) under physiological conditions, the rhythm of PPARβ/δ parallels that of basal [Ca2+]i, and (2) the light-induced PPARβ/δ increase observed only during the subjective night when the [Ca2+]i was at the lowest level may originate from the increase in Ca2+ transients induced by glutamate release. We propose that PPARβ/δ activation in the SCN is dependent on the amplitude of Ca2+ transients capable of activating cPLA2 and the subsequent AA release. Close interactions between PPARβ/δ and glutamatergic signaling may similarly occur in other brain areas, albeit probably without circadian gating because our preliminary analysis revealed no daily variations of PPARβ/δ expression in extra-SCN regions. Further investigation will be needed to address this issue in more details.
A putative role for PPARβ/δ activation in photic entrainment
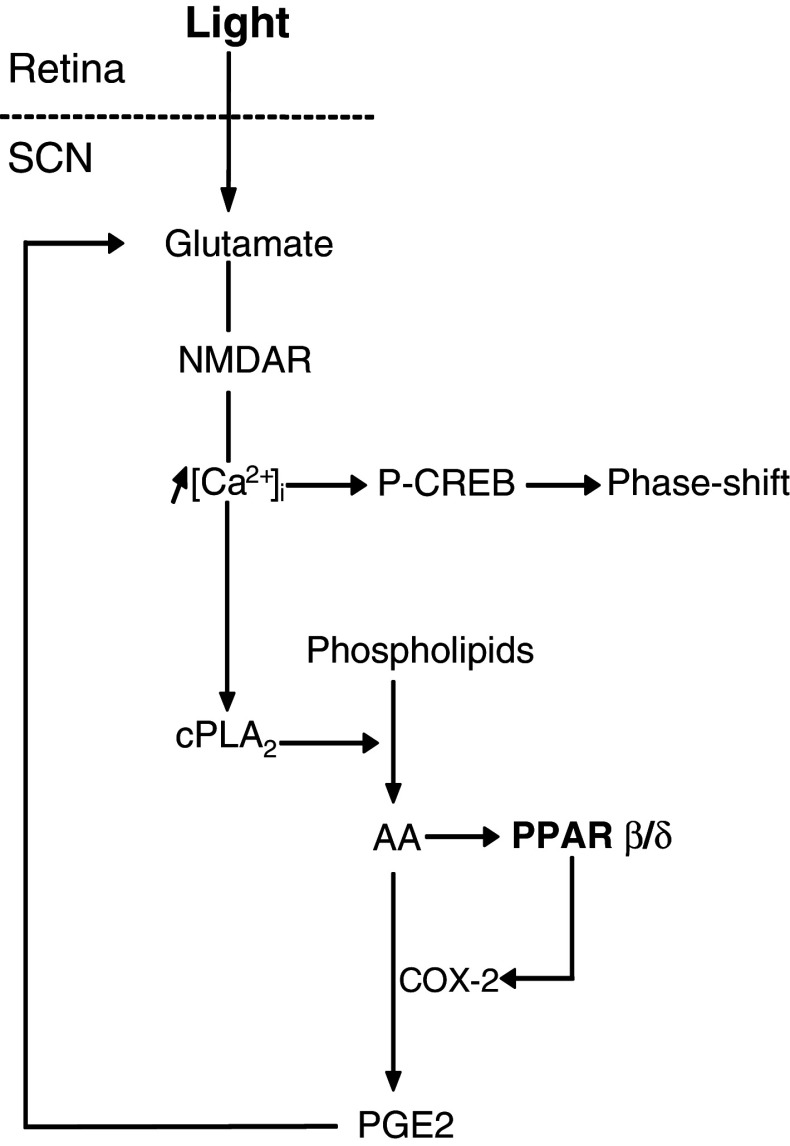
The magnitude of the light-induced phase shifts is linked to the irradiance and intensity of the light stimulus [42, 43]. At CT14, NMDA microinjections into the hamster SCN induce light-like phase shift responses that are dose-dependent [21]. When administered just prior to a light pulse, l-trans-pyrrolidine-2,4-dicarboxylic acid (PDC), a glutamate uptake inhibitor that increases the synaptic glutamate levels, elicits significantly larger phase delays than those after the combination of the light pulse plus vehicle. In contrast, administration of PDC alone does not induce a significant phase shift [44]. In a similar protocol, the PPARβ/δ agonist L-165,041 also magnified the light-induced phase delays, although it has no shifting effect when injected alone. Moreover, the PPARβ/δ antagonist GSK 3787 did not impair the light-induced phase delays. These pharmacological data suggest that PPARβ/δ activation contributes to the increased extracellular glutamate when coupled with a light stimulus, by generating a reinforcing loop on photic resetting under physiological conditions (Fig. 7). Some known PPARβ/δ-regulated genes may be involved in the enhancement of extracellular glutamate, including COX-2 [45, 46], which promotes prostaglandin E2 (PGE2) synthesis from AA. Several studies have shown that PGE2 is a signaling molecule that modulates synaptic transmission [47], in particular by stimulating glutamate release. PGE2 serves as a retrograde messenger that is synthesized from the postsynaptic neurons and acts on the specific membrane receptors of the pre- and postsynaptic neurons and astrocytes [47–49], which may stimulate glutamate release [49, 50]. In astrocytes, PGE2-evoked Ca2+ elevations cause the release of glutamate, which activates neuronal ionotropic receptors [51]. Inhibition of AA synthesis or COX activity prevents glutamate release from the astrocytes, while PGE2 promotes it [52, 53].
Fig. 7.
Hypothetical diagram showing the involvement of PPARβ/δ in regulation of glutamate release and photic resetting within the SCN. The light signal triggers glutamate release within the SCN (i.e., acute effects). Glutamate binds to the NMDA receptor, and as a consequence, an influx of calcium occurs. Calcium-mediated activation of the cytoplasmic phospholipase A2 (cPLA2) results in the release of arachidonic acid (AA) from membrane phospholipids for further PPAR β/δ activation and prostaglandin E2 (PGE2) synthesis via cyclooxygenase 2 (COX-2). PGE2 potentiates light-induced glutamate release. Our pharmacological data indicate that PPARβ/δ activation generates a positive feedback loop on light-induced glutamate release, but blocking PPARβ/δ activation will not impair light-induced phase-shifts (i.e., this is a reinforcing, but not necessary, pathway for photic resetting)
These data are consistent with the observations that although the PPARβ/δ agonist does not induce phase shifts on its own, it can enhance the light-induced phase shift, thus suggesting that an increase in glutamatergic stimulation is possibly due to an increase in COX-2-derived PGE2. The role of PPARβ/δ in photic resetting is further supported by the observation that light induced the overexpression of PPARβ/δ only when applied during the early subjective night.
Taken together, the present data suggest that PPARβ/δ are potential modulators of light-induced glutamatergic activation within the master circadian clock of the SCN (Fig. 7).
While extensive studies have demonstrated disparate roles in a variety of tissues [54], its ubiquitous expression makes it difficult to assign specific functions to PPARβ/δ. In this study our findings highlight an interaction between PPARβ/δ and glutamatergic signaling in the modulation of photic resetting of the master clock in mammals (Fig. 7).
Acknowledgments
The authors would like to thank Paul Pévet for helpful comments on the manuscript, Claire Maudet for animal care, Stéphanie Dumont for preparing drug solutions, Gaëlle Champeil-Potokar for confocal images. This work was supported by the Institut National de la Recherche Agronomique (INRA), Centre National de la Recherche Scientifique (CNRS) and University of Strasbourg. All authors declare no conflict of interest.
References
- 1.Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev. 1999;20(5):649–688. doi: 10.1210/er.20.5.649. [DOI] [PubMed] [Google Scholar]
- 2.Evans RM, Barish GD, Wang Y-X. PPARs and the complex journey to obesity. Nat Med. 2004;10(4):355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- 3.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126(4):801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 4.Lemberger T, Saladin R, Vazquez M, Assimacopoulos F, Staels B, Desvergne B, Wahli W, Auwerx J. Expression of the peroxisome proliferator-activated receptor alpha gene is stimulated by stress and follows a diurnal rhythm. J Biol Chem. 1996;271(3):1764–1769. doi: 10.1074/jbc.271.3.1764. [DOI] [PubMed] [Google Scholar]
- 5.Patel DD, Knight BL, Wiggins D, Humphreys SM, Gibbons GF. Disturbances in the normal regulation of SREBP-sensitive genes in PPARalpha-deficient mice. J Lipid Res. 2001;42(3):328–337. [PubMed] [Google Scholar]
- 6.Ximenes da Silva A, Lavialle F, Gendrot G, Guesnet P, Alessandri JM, Lavialle M. Glucose transport and utilization are altered in the brain of rats deficient in n-3 polyunsaturated fatty acids. J Neurochem. 2002;81(6):1328–1337. doi: 10.1046/j.1471-4159.2002.00932.x. [DOI] [PubMed] [Google Scholar]
- 7.Braissant O, Wahli W. Differential expression of peroxisome proliferator-activated receptor-alpha, -beta, and -gamma during rat embryonic development. Endocrinology. 1998;139(6):2748–2754. doi: 10.1210/en.139.6.2748. [DOI] [PubMed] [Google Scholar]
- 8.Hall MG, Quignodon L, Desvergne B. Peroxisome proliferator-activated receptor β/δ in the brain: facts and hypothesis. PPAR Res. 2008;2008:780452. doi: 10.1155/2008/780452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moreno S, Farioli-Vecchioli S, Cerù MP. Immunolocalization of peroxisome proliferator-activated receptors and retinoid × receptors in the adult rat CNS. Neuroscience. 2004;123(1):131–145. doi: 10.1016/j.neuroscience.2003.08.064. [DOI] [PubMed] [Google Scholar]
- 10.Dickmeis T. Glucocorticoids and the circadian clock. J Endocrinol. 2009;200(1):3–22. doi: 10.1677/JOE-08-0415. [DOI] [PubMed] [Google Scholar]
- 11.Pevet P, Challet E. Melatonin: both master clock output and internal time-giver in the circadian clocks network. J Physiol Paris. 2011;105(4–6):170–182. doi: 10.1016/j.jphysparis.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418(6901):935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 13.Ebling FJ. The role of glutamate in the photic regulation of the suprachiasmatic nucleus. Progr Neurobiol. 1996;50(2–3):109–132. doi: 10.1016/S0301-0082(96)00032-9. [DOI] [PubMed] [Google Scholar]
- 14.Bright JJ, Kanakasabai S, Chearwae W, Chakraborty S. PPAR regulation of inflammatory signaling in CNS diseases. PPAR Res. 2008;2008:658520. doi: 10.1155/2008/658520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue I, Shinoda Y, Ikeda M, Hayashi K, Kanazawa K, Nomura M, Matsunaga T, Xu H, Kawai S, Awata T, Komoda T, Katayama S. CLOCK/BMAL1 is involved in lipid metabolism via transactivation of the peroxisome proliferator-activated receptor (PPAR) response element. J Atheroscler Thromb. 2005;12(3):169–174. doi: 10.5551/jat.12.169. [DOI] [PubMed] [Google Scholar]
- 16.Canaple L, Rambaud J, Dkhissi-Benyahya O, Ba Rayet, Tan NS, Michalik L, Delaunay F, Wahli W, Laudet V. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor alpha defines a novel positive feedback loop in the rodent liver circadian clock. Mol Endocrinol. 2006;20(8):1715–1727. doi: 10.1210/me.2006-0052. [DOI] [PubMed] [Google Scholar]
- 17.Corton JC, Apte U, Anderson SP, Limaye P, Yoon L, Latendresse J, Dunn C, Everitt JI, Voss KA, Swanson C, Kimbrough C, Wong JS, Gill SS, Chandraratna RA, Kwak MK, Kensler TW, Stulnig TM, Steffensen KR, Gustafsson JA, Mehendale HM. Mimetics of caloric restriction include agonists of lipid-activated nuclear receptors. J Biol Chem. 2004;279(44):46204–46212. doi: 10.1074/jbc.M406739200. [DOI] [PubMed] [Google Scholar]
- 18.Teboul M, Guillaumond F, Grechez-Cassiau A, Delaunay F. Minireview: the nuclear hormone receptor family round the clock. Mol Endocrinol. 2008;22(12):2573–2582. doi: 10.1210/me.2007-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavialle M, Begue A, Papillon C, Vilaplana J. Modifications of retinal afferent activity induce changes in astroglial plasticity in the hamster circadian clock. Glia. 2001;34:88–100. doi: 10.1002/glia.1044. [DOI] [PubMed] [Google Scholar]
- 20.Shibata S, Watanabe A, Hamada T, Ono M, Watanabe S. N-methyl-d aspartate induces phase shifts in circadian rhythm of neuronal activity of rat SCN in vitro. Am J Physiol. 1994;267:R360–R364. doi: 10.1152/ajpregu.1994.267.2.R360. [DOI] [PubMed] [Google Scholar]
- 21.Mintz EM, Marvel CL, Gillespie CF, Price KM, Albers HE. Activation of NMDA receptors in the suprachiasmatic nucleus produces light-like phase shifts of the circadian clock in vivo. J Neurosci. 1999;19(12):5124–5130. doi: 10.1523/JNEUROSCI.19-12-05124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan H-P, Ishizuka T, Chui PC, Lehrke M, Lazar MA. Corepressors selectively control the transcriptional activity of PPARgamma in adipocytes. Genes Dev. 2005;19(4):453–461. doi: 10.1101/gad.1263305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc Natl Acad Sci USA. 1997;94(9):4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krey G, Braissant O, L’Horset F, Kalkhoven E, Perroud M, Parker MG, Wahli W. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol Endocrinol. 1997;11(6):779–791. doi: 10.1210/me.11.6.779. [DOI] [PubMed] [Google Scholar]
- 25.Xu HE, Lambert MH, Montana VG, Parks DJ, Blanchard SG, Brown PJ, Sternbach DD, Lehmann M, Jr, Wisely GB, Willson TM, Kliewer SA, Milburn MV. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol Cell. 1999;3(3):397–403. doi: 10.1016/S1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- 26.Irwin RP, Allen CN. Calcium response to retinohypothalamic tract synaptic transmission in suprachiasmatic nucleus neurons. J Neurosci. 2007;27(43):11748–11757. doi: 10.1523/JNEUROSCI.1840-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshihara Y, Watanabe Y. Translocation of phospholipase A2 from cytosol to membranes in rat brain induced by calcium ions. Biochem Biophys Res Commun. 1990;170(2):484–490. doi: 10.1016/0006-291X(90)92117-I. [DOI] [PubMed] [Google Scholar]
- 28.Lazarewicz JW, Salinska E, Wroblewski JT. NMDA receptor-mediated arachidonic acid release in neurons: role in signal transduction and pathological aspects. Adv Exp Med Biol. 1992;318:73–89. doi: 10.1007/978-1-4615-3426-6_7. [DOI] [PubMed] [Google Scholar]
- 29.Strokin M, Sergeeva M, Reiser G. Docosahexaenoic acid and arachidonic acid release in rat brain astrocytes is mediated by two separate isoforms of phospholipase A2 and is differently regulated by cyclic AMP and Ca2+ . Br J Pharmacol. 2003;139(5):1014–1022. doi: 10.1038/sj.bjp.0705326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basselin M, Chang L, Bell JM, Rapoport SI. Chronic lithium chloride administration attenuates brain NMDA receptor-initiated signaling via arachidonic acid in unanesthetized rats. Neuropsychopharmacology. 2006;31(8):1659–1674. doi: 10.1038/sj.npp.1300920. [DOI] [PubMed] [Google Scholar]
- 31.Kolko M, Rodriguez de Turco EB, Diemer NH, Bazan NG. Neuronal damage by secretory phospholipase A2: modulation by cytosolic phospholipase A2, platelet-activating factor, and cyclooxygenase-2 in neuronal cells in culture. Neurosci Lett. 2003;338(2):164–168. doi: 10.1016/S0304-3940(02)01385-X. [DOI] [PubMed] [Google Scholar]
- 32.Li SR, Wu KK, Anggard E, Ferns G. Localization of prostaglandin G/H synthase gene expression in rat brain by in situ hybridization. Biol Signals. 1993;2(2):77–83. doi: 10.1159/000109479. [DOI] [PubMed] [Google Scholar]
- 33.Xu L, Han C, Lim K, Wu T. Cross-talk between peroxisome proliferator-activated receptor delta and cytosolic phospholipase A2alpha/cyclooxygenase-2/prostaglandin E2 signaling pathways in human hepatocellular carcinoma cells. Cancer Res. 2006;66(24):11859–11868. doi: 10.1158/0008-5472.CAN-06-1445. [DOI] [PubMed] [Google Scholar]
- 34.Bendová Z, Sumová A, Mikkelsen JD. Circadian and developmental regulation of N-methyl-d-aspartate-receptor 1 mRNA splice variants and N-methyl-d-aspartate-receptor 3 subunit expression within the rat suprachiasmatic nucleus. Neuroscience. 2009;159(2):599–609. doi: 10.1016/j.neuroscience.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 35.Colwell CS. NMDA-evoked calcium transients and currents in the suprachiasmatic nucleus: gating by the circadian system. Eur J Neurosci. 2001;13:1420–1428. doi: 10.1046/j.0953-816x.2001.01517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colwell CS. Circadian modulation of calcium levels in cells in the suprachiasmatic nucleus. Eur J Neurosci. 2000;12:571–576. doi: 10.1046/j.1460-9568.2000.00939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shibata S, Oomura Y, Kita H, Hattori K. Circadian rhythmic changes of neuronal activity in the suprachiasmatic nucleus of the rat hypothalamic slice. Brain Res. 1982;247(1):154–158. doi: 10.1016/0006-8993(82)91041-1. [DOI] [PubMed] [Google Scholar]
- 38.Jiang ZG, Yang Y, Liu ZP, Allen CN. Membrane properties and synaptic inputs of suprachiasmatic nucleus neurons in rat brain slices. J Physiol. 1997;499(Pt 1):141–159. doi: 10.1113/jphysiol.1997.sp021917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meijer JH, Schaap J, Watanabe K, Albus H. Multiunit activity recordings in the suprachiasmatic nuclei: in vivo versus in vitro models. Brain Res. 1997;753(2):322–327. doi: 10.1016/S0006-8993(97)00150-9. [DOI] [PubMed] [Google Scholar]
- 40.Schaap J, Bos NPA, deJeu MTG, Geurtsen AMS, Meijer JH, Pennartz CMA. Neurons of the rat suprachiasmatic nucleus show a circadian rhythm in membrane properties that is lost during prolonged whole-cell recording. Brain Res. 1999;815(1):154–166. doi: 10.1016/S0006-8993(98)01025-7. [DOI] [PubMed] [Google Scholar]
- 41.Jin H, Ham SA, Kim MY, Woo IS, Kang ES, Hwang JS, Lee KW, Kim HJ, Roh GS, Lim DS, Kang D, Seo HG. Activation of peroxisome proliferator-activated receptor-delta attenuates glutamate-induced neurotoxicity in HT22 mouse hippocampal cells. J Neurosci Res. 2012;90(8):1646–1653. doi: 10.1002/jnr.23053. [DOI] [PubMed] [Google Scholar]
- 42.Nelson DE, Takahashi JS. Sensitivity and integration in a visual pathway for circadian entrainment in the Hamster (Mesocricetus-Auratus) J Physiol. 1991;439:115–145. doi: 10.1113/jphysiol.1991.sp018660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muscat L, Morin LP. Binocular contributions to the responsiveness and integrative capacity of the circadian rhythm system to light. J Biol Rhythms. 2005;20(6):513–525. doi: 10.1177/0748730405280458. [DOI] [PubMed] [Google Scholar]
- 44.Kallingal GJ, Mintz EM. Glutamatergic activity modulates the phase-shifting effects of gastrin-releasing peptide and light. Eur J Neurosci. 2006;24(10):2853–2858. doi: 10.1111/j.1460-9568.2006.05165.x. [DOI] [PubMed] [Google Scholar]
- 45.Glinghammar B, Skogsberg J, Hamsten A, Ehrenborg E. PPARdelta activation induces COX-2 gene expression and cell proliferation in human hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2003;308(2):361–368. doi: 10.1016/S0006-291X(03)01384-6. [DOI] [PubMed] [Google Scholar]
- 46.Xu L, Han C, Wu T. A novel positive feedback loop between peroxisome proliferator-activated receptor-delta and prostaglandin E2 signaling pathways for human cholangiocarcinoma cell growth. J Biol Chem. 2006;281(45):33982–33996. doi: 10.1074/jbc.M600135200. [DOI] [PubMed] [Google Scholar]
- 47.Yang H, Chen C. Cyclooxygenase-2 in synaptic signaling. Curr Pharm Des. 2008;14(14):1443–1451. doi: 10.2174/138161208784480144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C, Bazan NG. Lipid signaling: sleep, synaptic plasticity, and neuroprotection. Prostaglandins Other Lipid Mediat. 2005;77(1–4):65–76. doi: 10.1016/j.prostaglandins.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 49.Sang N, Zhang J, Marcheselli V, Bazan NG, Chen C. Postsynaptically synthesized prostaglandin E2 (PGE2) modulates hippocampal synaptic transmission via a presynaptic PGE2 EP2 receptor. J Neurosci. 2005;25(43):9858–9870. doi: 10.1523/JNEUROSCI.2392-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bazan NG. Synaptic lipid signaling: significance of polyunsaturated fatty acids and platelet-activating factor. J Lipid Res. 2003;44(12):2221–2233. doi: 10.1194/jlr.R300013-JLR200. [DOI] [PubMed] [Google Scholar]
- 51.Sanzgiri RP, Araque A, Haydon PG. Prostaglandin E2 stimulates glutamate receptor-dependent astrocyte neuromodulation in cultured hippocampal cells. J Neurobiol. 1999;41(2):221–229. doi: 10.1002/(SICI)1097-4695(19991105)41:2<221::AID-NEU5>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 52.Bezzi P, Carmignoto G, Pasti L, Vesce S, Rossi D, Rizzini BL, Pozzan T, Volterra A. Prostaglandins stimulate calcium-dependent glutamate release in astrocytes. Nature. 1998;391(6664):281–285. doi: 10.1038/34651. [DOI] [PubMed] [Google Scholar]
- 53.Dave KA, Platel J-C, Huang F, Tian D, Stamboulian-Platel S, Bordey A. Prostaglandin E2 induces glutamate release from subventricular zone astrocytes. Neuron Glia Biol. 2011;7:1–7. doi: 10.1017/S1740925X12000166. [DOI] [PubMed] [Google Scholar]
- 54.Wagner K-D, Wagner N. Peroxisome proliferator-activated receptor beta/delta (PPARbeta/delta) acts as regulator of metabolism linked to multiple cellular functions. Pharmacol Ther. 2010;125(3):423–435. doi: 10.1016/j.pharmthera.2009.12.001. [DOI] [PubMed] [Google Scholar]