Abstract
Tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) has dual functions mediating both apoptosis and survival of cells. This review focusses on the current regulatory factors that control TRAIL transcription. Here, we also highlight the role of distinct transcription factors that co-operate and regulate TRAIL in different pathological states. A better understanding of the molecular signalling pathways of TRAIL-induced cell death and survival in disease may lead to more sophisticated technologies for novel therapeutic targets.
Keywords: TRAIL, Transcription factor, Regulation, Gene expression, Apoptosis, Cell survival
Introduction
Tumour necrosis factor-related apoptosis-inducing ligand (TRAIL)
Tumour necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL, also known as Apo2L and TNFSF10) is a member of TNF-cytokine family of ligands, originally discovered based on sequence homology of its extracellular domain to Fas ligand (FasL) and TNF, and has the ability to induce apoptosis [1, 2]. TRAIL is a type II transmembrane protein that can be proteolytically processed by metalloproteinases (MMPs) to produce a soluble form [3, 4]. MMP-2, for example, was recently shown to cleave TRAIL in vitro [4, 5]. In humans, TRAIL signalling is the most complex of all TNF ligand members since membrane-bound or soluble TRAIL has the ability to bind five different receptors leading to either apoptosis or survival of cells (Fig. 1). Receptors of TRAIL include two death-domain-containing receptors, TRAIL-R1 (DR4) [6] and TRAIL-R2 (DR5) [7, 8]; two decoy receptors, TRAIL-R3 (DcR1) [7–9] and TRAIL-R4 (DcR2) [10, 11]; and osteoprotegerin (OPG), the only known soluble receptor for TRAIL [12].
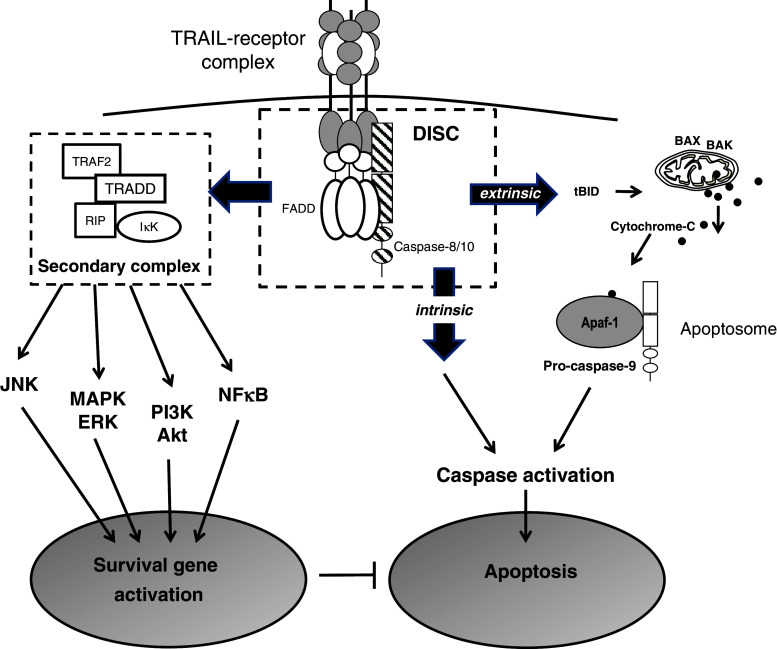
Fig. 1.
TRAIL-receptor signalling. TRAIL initiates cell death by binding to its death receptors. This results in activation of caspases via either the intrisic (mitochondria) or extrinsic pathway, resulting in cell death. By binding its receptors, TRAIL can also activate the JNK, MAPK, PI3K and NFκB pathways to induce expression of survival genes resulting in increased proliferation and migration of cells and inhibition of apoptosis
TRAIL signalling
Apoptosis Depending on the cell type, trimeric TRAIL mediates apoptosis via the extrinsic and intrinsic (mitochondrial) apoptotic pathways upon binding to its death-domain containing receptors, TRAIL-R1 and -R2 (Fig. 1). Formation of the death-inducing signalling complex (DISC), which contains Fas-associated death domain (FADD) and caspase-8 or -10, and subsequent activation of caspase-8 (or -10) and downstream effector caspases-3, -6 and -7, results in apoptosis characteristic of the extrinsic apoptotic pathway. On the other hand, TRAIL-induced apoptosis via the mitochondrial or the intrinsic pathway involves activation of caspase-8, activation of bid (truncated bid), Bax and Bak, and formation of the apoptosome complex, containing apoptotic peptidase activator factor-1 (Apaf-1), cytochrome c and caspase-9. This complex is then able to activate caspase-9 as well as effector caspases resulting in apoptosis [13–15].
Non-apoptotic functions Intriguingly, TRAIL can also mediate non-apoptotic signalling following binding to its receptors, including death-domain containing receptors (Fig. 1). A secondary signalling complex formation has been proposed following DISC assembly. This secondary complex contains TNF-receptor-associated death domain (TRADD), TNF-receptor-associated factor-2 (TRAF2), receptor interacting protein (RIP) and the inhibitor of кB kinase (IKKγ). When this complex is assembled, NFкB, PI3K/Akt and members of mitogen-activated protein kinases (MAPKs), including extracellular signal-regulated kinase (ERK), as well as c-Jun NH2-terminal kinase (JNK) and p38, are activated, leading to survival signals [13, 16, 17].
The human TRAIL gene
The human gene that encodes for TRAIL spans approximately 20 kb [18] and is located on chromosome 3q26 [1]. The genomic organisation of TRAIL includes five exons and four introns [18] (Fig. 2). The first exon encodes a 21-amino acid hydrophobic transmembrane domain, whilst exons 2 and 3 encode products that are 46 and 14 amino acids, respectively. Exons 4 and 5 encode the domain responsible for TRAIL’s interaction with its receptors. Exon 5, being the longest exon, also encodes the 142-amino acid C-terminal domain, a termination codon (TAA), a 3′-untranslated region and a poly-A tail [18].
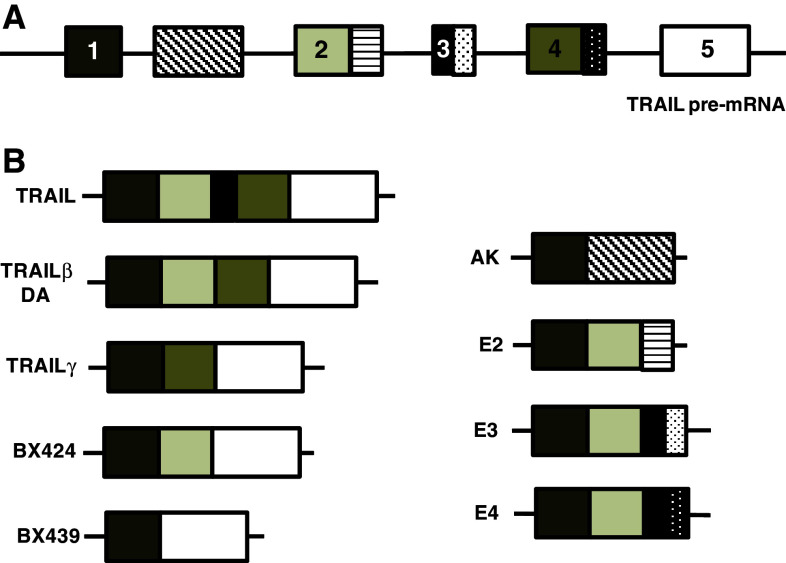
Fig. 2.
Genomic structure of human TRAIL isoforms. Patterned boxes depict novel exons and sequences recently identified
Interestingly, spliced variants of human TRAIL have been identified. The first of these include an RT-PCR study by Kreig et al. [19], identifying two TRAIL spliced variants, termed TRAILβ and TRAILγ. Whilst TRAILβ mRNA lacks exon 3, TRAILγ was found to lack both exons 2 and 3 (Fig. 2). Five years later, a third isoform was identified by the same group and termed TRAILδ [20]. TRAILδ was found to lack both exons 2 and 3 [20] (Fig. 2). These truncated variants appear not to display apoptotic properties, but may neutralise the biological activity of wild-type TRAIL [19]. More recently, additional TRAIL variants were cloned and functional assessment by NFκB activation was determined in vitro [21]. This study identified seven alternatively spliced variants, termed DA, AK, E2, E3, E4, BX424 and BX439 (Fig. 2). While DA encodes a protein product identical to TRAILβ [19], both DA and AK were shown to contain a novel exon [21]. E2, E3 and E4 contain extended sequences at the last exon. BX424, which was previously identified and known as TRAILδ [20], lacks exons 2 and 3, whilst BX439 lacks exons 2, 3 and 4. All the identified variants share common N-terminal sequences encoding the transmembrane domain, but display altered C-terminal regions. Interestingly, Wang et al. [21] showed that all variants were widely expressed in tissues and cells, and could also activate NFκB, except for BX439. Surprisingly, BX424, DA and E4 displayed significantly greater activation of the NFκB promoter, as well as promoter activation of inflammatory cytokines IL-8, CCL4 and CCL20, greater than wild-type TRAIL. Furthermore, these variants failed to induce apoptosis of multiple cancer cell lines [21]. Expression and function of these variants in disease are currently unknown.
Transcriptional regulation
While novel pathways of TRAIL-mediated apoptosis and non-apoptotic functions are emerging, we also need to recognise the control of this gene at the molecular level. In this review, we will discuss transcription factor control of TRAIL.
The human TRAIL promoter lacks a TATA box; however, its transcriptional start site has been identified [18, 22]. TRAIL gene expression is regulated at the transcriptional level by distinct interactions between transcription factors and DNA. To date, a number of transcription factors have been identified to play a role in regulating TRAIL transcription. These include nuclear factor kappa B (NFκB), specificity protein-1 (Sp1), interferon regulatory factors (IRFs), signal transducers and activators of transcription (STATs), FOXO3a, p53, early growth response (Egr) and nuclear factor of activated T cells (NFATc).
Table 1 lists the identified transcription factors and sequences of the elements that control TRAIL transcriptional activity at the promoter level. Here we also discuss TRAIL gene expression in the context of different pathological settings including vascular diseases, cancer, immune regulation and infection, particularly in response to various stimuli such as cytokines, growth factors, pharmacological/chemotherapeutic agents and viral infection. A list of transcription factors controlling TRAIL gene expression in response to various stimuli and in multiple cell types can be found in Table 2, and is discussed in more detail below. Understanding control at this utmost basic level may provide additional clues into the mechanisms of TRAIL’s pleiotropic functions observed in vivo.
Table 1.
Transcription factors with their coordinates and sequences of corresponding cis elements (relative to transcriptional start site) on the human TRAIL promoter
| Transcription factor | Element | Sequence | Site | Functionality | References |
|---|---|---|---|---|---|
| NFκB, Sp1 | Sp1-1 | 5′-GGGAGGG-3′ | −74/−68 | √ | [25] |
| NFκB, Sp1 | Sp1-2 | 5′-GGTGGG-3′ | −43/−37 | √ | [25] |
| NFκB | κB1 | 5′-AGAAAATCCC-3′ | −265/−256 | √ | [80] |
| NFκB | κB2 | 5′-TGGAAGTTTC-3′ | −385/−376 | √ | [80] |
| NFκB | κB3 | 5′-TGACTCAGTGG-3′ | −1,327/−1,317 | – | [80] |
| Sp1 | Sp1-5/6 | 5′-CCTCCCCTCC-3′ | −257/−248 | √ | [31] |
| Sp1 | Sp1-7 | 5′-CCCTCC-3′ | −129/−124 | √ | [31] |
| FOXO3a | FBST | 5′-ATAAATAAAT-3′ | −995/−986 | √ | [52] |
| IRF-1 | IRF-E | 5′ACAACTCATTCGCTTTCATTTCCTCACTGA-3 | −13/+17 | √ | [59] |
| IRF-1 | ISRE | – | −165/−35 | √ | [59] |
| IRF-9a | ISRE | 5′-AGTTTCACTTTTGG-3′ | −57/−44 | – | [87] |
| IRF-3 | ISRE | 5-GCTTCTTTCAGTTTCCCTCCTTT-3 | −143/−121 | √ | [85] |
| NFATc1 | NFAT binding site (N4) | 5′-ATGTTTTTTTCCTTTGCCTT-3′ | −666/−647 | – | [94] |
| NFATc1 | NFAT binding site (N5) | 5′-ATTTCTATTTTCCTTTATCC-3′ | −458/−439 | – | [94] |
| p53 | p53 binding site | 5′-AAACAGGCCT-3′ | −346/−337 | – | [64] |
| p53 | p53 binding site | 5′-AGCCAGGCCA-3′ | −324/−315 | – | [64] |
| HSF-1 | HSE | 5′-GCTTCTTTCAGTTTCCCTCCTTTCCAACG-3′ | −143/−115 | – | [82] |
aMouse TRAIL promoter sequence
Table 2.
Transcription factors regulating TRAIL in response to various stimuli including cytokines, growth factors, pharmacological agents and infection
| Stimuli | Transcription factor | TRAIL expression | Cell type | References |
|---|---|---|---|---|
| Injury, FGF2 | NFкB, Sp1 | ↑ mRNA and protein | VSMC | [25] |
| PMA, PMA + Con A | NFкB | ↑ mRNA and protein | Jurkat (WT and IκBα mutant), human primary T cells | [80] |
| TNF, PMA + Io | NFкB | ↑ mRNA | Jurkat (WT and IKKγ mutant) | [81] |
| Influenza virus | NFкB | ↑ protein | A549 | [88] |
| Tax oncoprotein (human T cells leukemia virus) | NFкB | ↑ mRNA | Jurkat (WT and IKKγ-deficent cells) | [53] |
| 15d-PGJ2 | NFкB, HSF-1 | ↓ mRNA and protein | Jurkat (induced by PMA + Io) | [82] |
| W peptide (FPRL1 agonist) | NFкB | ↑ protein | Human THP-1 monocytes and neutrophils, mouse leukocytes | [83] |
| IFN-β | Stat1 | ↑ mRNA and protein | Colorectal cancer cells, fibrosarcoma | [56] |
| IFN-α | Stat1, IRF-1 | ↑ protein | UM-UC-12 (bladder cancer cell) | [58] |
| IFN-γ | Stat1 | ↑ mRNA and protein | KMS-20 (myeloma) | [57] |
| Retinoic acid + IFN-γ | IRF-1 | ↑ mRNA | Breast cancer cells | [59] |
| Sendai virus infection | IRF-3 | ↑ mRNA | Colon adenocarcinoma, HEC-1B (lacks IFN receptor) | [85] |
| HIV-1 infection | IRF1, IRF-7, STAT1 | ↑ mRNA and protein | Human MDM | [86] |
| PMA + Io | NFAT | ↑ protein | Human intestinal cells | [94] |
| BCR-ABL oncoprotein | FOXO3a | ↓ mRNA | Haematopoietic cells | [52] |
| PolyIC | IRF-9 | ↑ mRNA | Mouse NK cells | [87] |
| Adriamycin | p53 | ↑ protein | Human colon cancer | [64] |
| PDGF-BB | Sp1 | ↑ mRNA and protein | VSMC | [31] |
| TNF-α | Sp1 | ↑ protein | Human breast cancer cells | [65] |
| MS275 + Adriamycin | Sp1 | ↑ protein | Human breast cancer cells | [66] |
| TNF-α | Egr-1 | ↓ mRNA | HUVEC | [39] |
| SEB | Egr-2, Egr-3 | ↑ mRNA | Murine intestinal epithelial cells | [84] |
TRAIL regulation in vascular disease
TRAIL has the ability to kill vascular cells in vitro [23, 24] and may play a role in death of cells in the vasculature. Emerging data indicate a protective role for TRAIL in the development of vascular disorders including restenosis, atherosclerosis and pulmonary hypertension [25–30]. While the TRAIL-dependent mechanism(s) in these pathologies are not fully elucidated, recent studies implicate TRAIL in its ability to promote vascular cell proliferation and migration as opposed to cell death [25, 27, 28, 31–33]. Intriguingly, circulating TRAIL levels are significantly reduced in patients with cardiovascular disease [30, 34], and low TRAIL levels are linked to increased cardiovascular events and mortality [35, 36]. TRAIL transcriptional control and gene expression in the vascular cells are not fully established.
A study by our laboratory demonstrated the involvement of Sp1 and NFκB in regulating TRAIL transcription and expression in vascular smooth muscle cells (VSMCs) in response to vascular injury both in vitro and in vivo following perivascular cuff placement to the femoral artery of wild-type and TRAIL−/− mice [25]. Sp1 and NFκB are transcription factors that are ubiquitously expressed and control a number of essential cellular functions such as proliferation and apoptosis. Mechanical injury increased TRAIL mRNA, protein and promoter activity in an Sp1 and NFκB-dependent manner, a finding also observed with fibroblast growth factor-2 (FGF-2) [25], a potent growth factor for VSMCs released within minutes of injury. Electro mobility shift assays (EMSA) confirmed binding of Sp1 and NFκB to Sp1-1 (−74/−68) and Sp1-2 (−44/−37) elements on the human TRAIL promoter [25], and interestingly, a role for Sp1 phosphorylation at Thr453 following injury and regulation of TRAIL was identified. ChIP analysis confirmed enrichment of phospho-Sp1-Thr453 and NFκB on the endogenous human TRAIL promoter [25].
More recently, we have shown that platelet-derived growth factor-BB (PDGF-BB), another potent growth factor for VSMCs, can positively regulate TRAIL expression to control proliferation and migration inducible by PDGF-BB [31]. PDGF-BB-inducible TRAIL transcription was blocked via introduction of a dominant negative-Sp1 mutant [31], again demonstrating the importance of Sp1 in mediating TRAIL transcription in VSMCs. Along with the originally identified Sp1-1 and Sp1-2 sites [25], two novel Sp1 binding sites were discovered to bind Sp1: Sp1-5/6 (2 overlapping sites) and Sp-7. PDGF-BB-inducible TRAIL promoter activity was blocked in the presence of transverse mutations of each site, suggesting that all sites are essential for PDGF-BB’s actions in VSMCs [31]. Furthermore, PDGF-BB-inducible TRAIL transcription required the cooperation of Sp1, the transcriptional coactivator p300 and acetylated histone-3, demonstrated by co-immunoprecipitation and ChIP assays [31].
In contrast to Sp1 and NFκB in positively regulating TRAIL transcription and expression in VSMCs, in endothelial cells, Egr-1 inhibited TRAIL expression. Egr-1 is a member of the Egr family of zinc finger transcription factors. Other members include Egr-2, Egr-3, Egr-4 and NGF1-B. Egr-1 mRNA is expressed constitutively in many tissues, but not Egr-2 and Egr-3 [37]. Egr-1 is the major isoform expressed in the vasculature and contributes to the pathogenesis of vascular disorders [38]. A study in human umbilical vein endothelial cells (HUVECs) confirmed inhibition of TRAIL mRNA expression via adenovirus overexpression of Egr-1 [39]. TNF-α-inducible Egr-1 expression also repressed TRAIL mRNA. Interestingly, this repression was blocked by NAB2, a co-repressor of Egr-1 [39]. While mechanisms of TRAIL repression by Egr-1 at the transcriptional level are not established, both Egr-1 and TRAIL are involved in VSMC proliferation and neointima formation [25, 40], and it is currently unknown whether Egr-1 overexpression can lead to suppression (or induction) of TRAIL in VSMCs. Taken together, the findings from the abovestudies implicate TRAIL as a potential therapeutic target for vascular proliferative disorders.
TRAIL regulation in cancer
TRAIL is emerging as an attractive anti-tumour agent since multiple studies report its selective cytotoxicity in vitro [2, 41, 42] and in vivo [43–45], which is further strengthened by the ability of normal cells to resist soluble TRAIL’s cytotoxic effects [46]. In support of these, TRAIL and monoclonal antibodies against TRAIL-R1 and R2 are currently in clinical trials for cancer therapy (reviewed in [47]). Resistance to soluble TRAIL-induced cytotoxicity has been observed in some cancer cells [48, 49], and in apoptosis-resistant tumour cell populations, TRAIL is able to stimulate proliferation and inhibit cell death via activation of NFκB [50].
To date, the transcriptional regulation of TRAIL in cancer cells in response to oncoproteins, cytokines, pharmacological and chemotherapeutic agents has been the most extensively researched. However, given the conflict in TRAIL-induced effects in cancer cells, it is of great importance to decipher the mechanisms of TRAIL transcriptional activity and expression.
FOXO3a belongs to a member of the forkhead family of winged helix transcription factors. This family regulates a variety of cellular processes including proliferation, apoptosis, cell cycle, differentiation and DNA repair [51]. FOXO3a can regulate TRAIL transcription in cytokine-dependent haematopoietic cell lines [52]. There are two overlapping FOXO3a consensus-binding sites (FBST), from −995 to −986 on the human TRAIL promoter. Using EMSA, Ghaffari et al. [52] confirmed FOXO3a binding to these sites, which was abolished with the introduction of a mutation. Tranfections using both the full length and a short sequence reporter (−1,023 to −973) demonstrated enhanced TRAIL promoter activity by FOXO3a. Interestingly mutations to the FBST site (−1,023 to −973) did not completely reduce this activation, suggesting the presence of additional non-consensus FBST sites within this region [52]. Inhibition of FOXO3a activation is associated with inhibition of TRAIL mRNA expression, mediated by BCR-ABL expressing haematopoietic cells [52]. BCR-ABL or breakpoint cluster region-abelson is an oncoprotein that causes chronic myeloid leukemia. Inhibition of TRAIL expression by BCR-ABL oncoprotein led to suppression of apoptosis. These results provide novel mechanism(s) by which TRAIL may be potentially used as a therapy against BCR-ABL-induced chronic myeloid leukemia [52].
In contrast to TRAIL’s protective effect against BCR-ABL oncoprotein-induced chronic myeloid leukemia, TRAIL has been shown to play a role in the development of adult T cell leukemia [53]. Tax is encoded by the human T cell leukemia virus (HTLV) and is involved in the development of adult T cell leukemia by promoting apoptosis of HTLV-infected T cells [54, 55]. Rivera-Walsh et al. [53] demonstrated that Tax increased TRAIL mRNA, and this induction was blocked in IKKγ-deficient Jurkat cells, where IKKγ is essential for NFκB activation. Furthermore, Tax-induced Jurkat T cell apoptosis was blocked by an anti-TRAIL antibody [53]. These findings demonstrate that NFκB signalling is critical for inducible TRAIL expression, mediating Tax-induced Jurkat T cell apoptosis [53].
In addition to oncoproteins and associated transcription factors regulating TRAIL as discussed above, TRAIL can also be induced by interferon family members promoting cancer cell apoptosis [56–59]. Interferon-induced TRAIL expression is mediated via STATs, a family of transcription factors that can be activated by a variety of cytokines. Seven structurally and functionally related members of STATs have been identified: STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b and STAT6. STATs are activated via phosphorylation by members of the janus-activated kinase (JAK) family (JAK1, 2, 3 and Tyk2) following binding of cytokines to their cognate receptors. Once activated, STATs dimerise and translocate to the nucleus to induce the expression of target genes [60]. STAT1, together with STAT2 and IRF-9/ISGF3γ, form a heterotrimeric transcription factor complex known as IFN-stimulated gene factor-3 (ISGF3). Upon stimulation with type I IFNs (IFN-α and -β), ISGF3 translocates into the nucleus to bind the interferon-stimulated response element (ISRE), thus activating IFN-inducible genes [61, 62].
In parental human colorectal adenocarcinoma cells, IFN-β treatment resulted in increased TRAIL promoter activity, mRNA, protein expression and apoptosis, in conjunction with increased levels of STAT1, STAT2 and ISGF3γ/IRF-9 protein expression [56]. Interestingly, these inductions were no longer observed in IFN-β-resistant cells [56]. IFN-β-inducible TRAIL mRNA was reduced in STAT1-deficient cells, and these cells did not respond to IFN-β-induced apoptosis, confirming the role of STAT1 in this process [56]. IFN-γ-induced apoptosis in myeloma cells was also associated with increased TRAIL mRNA, protein and STAT1 phosphorylation [57]. Treatment with IFN-γ did not increase STAT1 phosphorylation in myeloma cells resistant to IFN-γ-induced apoptosis [57]. Furthermore, inhibition of the JAK/STAT pathway using AG490, an inhibitor of protein tyrosine kinase, suppressed IFN-γ-induced TRAIL secretion and recovered cell viability in IFN-γ-induced cell death [57]. In conclusion, STAT1 activation following interferon treatment can lead to transcriptional activation of TRAIL, leading to increased cancer cell death [56, 57].
IRF-1 has also been shown to regulate TRAIL expression in response to interferon stimulation. IRF-1 is a member of the IRF family of transcription factors that regulates interferon-stimulated genes. Other family members include IRF-2 to IRF-9, which are characterised by an amino-terminal DNA-binding domain [63]. Following stimulation, they bind to consensus sequences including ISREs, IRF-E and interferon consensus sequence (ICS), thus inducing interferon-stimulated genes [62]. A study by Clarke et al. [59] showed that IRF-1 mediated synergistic induction of TRAIL mRNA, protein and promoter activity following combination treatment of IFN-γ and retinoic acid (RA) in breast cancer cells [59]. Using promoter mapping and mutational studies, two IFN-response elements, ISRE (located between −165 to −35) and IRF-E (located between −35 to +1), were responsible for IRF-1′s actions [59]. Mutation to the IRF-E, or to both IRF-E and ISRE, completely reduced this synergistic effect. Interestingly, this was not observed with mutation to the ISRE site alone [59]. EMSA using the TRAIL IRF-E revealed the existence of an IFN-γ- or RA-inducible complex containing IRF-1. Consistent with this, ChIP assays demonstrated synergistic occupancy of IRF-1 to the TRAIL promoter following IFN-γ/RA treatment, resulting in increased recruitment of the creb-binding protein (CBP) and acetylated histone-3 [59]. Whilst Jurkat cells are insensitive to IFN-γ/RA treatment, breast cancer SK-BR-3 cells die upon treatment. In order to examine apoptosis occurring in a paracrine manner, SK-BR-3 and Jurkat cells were co-cultured, followed by treatment with IFN-γ/RA. IFN-γ/RA treatment dramatically increased cell death of both cell lines, which was prevented by a neutralizing TRAIL antibody [59]. These findings suggest that paracrine apoptosis induced by TRAIL plays a role in this system and may therefore explain the observed synergistic induction of TRAIL by IFN-γ/RA [59].
Although IRF-1 and STAT1 have been shown to individually regulate TRAIL in different cancer cells, both IRF-1 and STAT1 are involved in IFN-α-induced TRAIL expression in human bladder cancer [58]. IFN-α treatment not only resulted in increased IRF-1 protein expression, but also rapid phosphorylation of STAT1. While ChIP assays confirmed IRF-1 and STAT1 binding to the TRAIL promoter between −696/−437 following IFN-α treatment, the exact elements were not verified [58]. In addition, IFN-α-induced apoptosis was inhibited by an anti-TRAIL antibody and TRAIL siRNA, suggesting direct involvement of TRAIL in IFN-α-induced cell death [58]. These results demonstrate that IRF-1- and STAT1-mediated IFN-α-inducible TRAIL expression leads to IFN-α-mediated apoptosis in these cells [58]. Further investigation of the TRAIL promoter is necessary to identify the molecular mechanism(s) regulating TRAIL transcription by STAT1 and IRF-1, and whether a common mechanism in response to interferon exists between the different cancer cells used. Whether STAT1 and IRF-1 cooperate to induce TRAIL transcription in this setting is still unknown.
TRAIL expression can also be induced by DNA-damaging agents, including chemotherapy, adriamycin and 5-fluorouracil in HCT116 human colon cancer cells [64]. Interestingly, induction of TRAIL protein was observed in wild-type HCT116 cells but not in p53−/− HCT116 cells, indicating the involvement of p53. TRAIL mRNA and protein expression was induced in HCT116 p53−/− cells transduced with adenovirus overexpressing p53, confirming the role of p53 as a regulator of TRAIL [64]. Using human TRAIL promoter reporter constructs containing two p53 sites (located between −346 and −625 bp), adenovirus overexpressing p53 enhanced TRAIL promoter activity compared to the control [64]. TRAIL promoter activity was also induced by adriamycin in a dose-dependent manner, and this induction was associated with reduced cell viability in p53-expressing HCT116 cells. Furthermore, TRAIL induction by adriamycin promoted apoptosis, suggesting that TRAIL is required for p53-dependent cell apoptosis [64].
TRAIL is also involved in TNF-α- and MS275-mediated cancer cell sensitisation to chemotherapy adriamycin, and this is mediated via Sp1 [65, 66]. Xu et al. [65] showed that a region between −80 and +1 on the human TRAIL promoter, which contains two Sp1 binding elements, is responsible for TNF-α-induced TRAIL transcriptional activity. These Sp1 elements may correspond to the Sp1-1 and Sp1-2 sites that our group has previously identified [25]. Deletion of the second Sp1 element (from −80 to −65) completely abolished TNF-α-induced TRAIL promoter activity, suggesting that this region is essential for TRAIL induction by TNF-α [65]. Besides TNF-α, TRAIL transcriptional activation by the HDAC inhibitor, MS275 or in combination with adriamycin chemotherapy was abolished when the second Sp1 site was mutated [66]. Consistent with this, Sp1 siRNA inhibited TRAIL expression by MS275 or the combination of MS275 and adriamycin [66]. Caspase activation was significantly increased following combination treatment of MS275 or TNF-α with adriamycin, which was blocked by TRAIL siRNA [65, 66]. These studies suggest that TNF-α and MS275 may sensitise human breast cancer cells to chemotherapy by increasing TRAIL transcription through Sp1 and subsequent cancer cell apoptosis [65, 66].
In addition to the findings described above, Clarke et al. [59] demonstrated that Sp1 may contribute to IRF-1 and retinoic acid-induced TRAIL transcription in breast cancer cells. Basal and retinoic acid-induced TRAIL promoter activity was completely blocked when the GC box in the proximal promoter was mutated. Mutation in the distal GC box completely inhibited basal and reduced RA-induced TRAIL promoter activity compared to wild type [59]. These GC boxes are located between −165/−35 in the human TRAIL promoter, again corresponding to the Sp1-1 and Sp1-2 elements we identified to be functional in VSMCs [25]. Taken together, IRF-1-mediated RA-induced TRAIL transcription may also involve Sp1 [59]. This is not surprising since crosstalk between IRF-1 and Sp1 has been demonstrated with the human CDK2 promoter [67].
TRAIL in immune regulation and infection
TRAIL is expressed on a number of cells in the innate and adaptive immune systems including monocytes, macrophages, dendritic cells, natural killer (NK) cells, T cells and B cells, in response to cytokines such as IFN-β and IFN-γ [68–72]. TRAIL has also been implicated in the regulation of T helper responses, haematopoiesis and homeostasis of T cells [73–75]. Furthermore, once challenged, TRAIL−/− mice display phenotypes of autoimmune diseases including arthritis, diabetes and multiple sclerosis (reviewed in [76]), implicating a role for TRAIL in modulating an autoimmune response. Consistent with this, soluble TRAIL levels are increased in patients with systemic lupus erythaematous, multiple sclerosis and systemic sclerosis [77–79]. However, how TRAIL gene expression and transcription are controlled during an immune response or during infection in cells of the immune system has not been widely studied.
A role for NFκB in T cell activation-induced TRAIL expression has been described [80, 81]. Baetu et al. [80] showed that induction of TRAIL mRNA and protein expression was inhibited following stimulation with T cell receptor (TCR) mimetics, including phorbol 12-myristate 13-acetate (PMA) and PMA/Con A (concanavalin A) in Jurkat T cells expressing the IκBα mutant [80]. These results suggest that inducible TRAIL expression is NFκB dependent [80]. Siegmund et al. [81] used another mutant cell type, IKKγ/NEMO-deficient Jurkat T cells, which also inhibited NFκB activation despite the presence of a variety of NFκB stimuli. The induction of TRAIL mRNA by PMA/ionomycin (PMA/I) and TNF was completely prevented in these mutant cells [81]. In addition, pharmacological inhibitors of NFκB, NaSa1 and Bay 11-7082 (both inhibit IκB phosphorylation), and MG132 (proteosomal inhibitor) dramatically reduced PMA/ConA-induced TRAIL mRNA in primary human T lymphocytes [80]. Collectively, these findings demonstrate a key role of NFκB in the regulation of TRAIL expression in response to T cell activation in both Jurkat T cells and primary T lymphocytes, and this regulation may contribute to TRAIL-mediated apoptosis in T lymphocytes [80, 81]. Baetu et al. [80] further reported three potential NFκB binding elements—κB1 and κB2 (both in the proximal −538 bp) and κB3 (−1,317 to −1,327 bp)—on the human TRAIL promoter. EMSA revealed that only κB1 (−256 to −265) is essential for increased NFκB binding by PMA in Jurkat T cells. This is in agreement with mutation to κB1, which inhibited NIK (an activator of the NFκB pathway)-induced TRAIL promoter activity [80]. Interestingly, mutation to the κB2 element also inhibited NIK-induced TRAIL promoter activity, and further suppression was observed when both elements were mutated. This suggests cooperative binding to these elements for activation of TRAIL transcription [80].
In addition to the studies outlined above, inhibition of TRAIL transcription and expression by the anti-inflammatory 15-deoxy-∆-12,14-prostaglandin J2 (15d-PGJ2) following T cell-activation was associated with inhibition of NFκB binding to the κB1 element in Jurkat T cells [82]. Besides the κB1 site, the region between −165 and −35 contained additional regulatory element(s) essential for 15d-PGJ2-mediated-suppression of TRAIL transcription [82]. Interestingly, this region was essential for heat shock factor protein-1 (HSF-1)-mediated TRAIL transcriptional repression. Consistent with this, putative HSF-1 binding elements are located between −143 and −115 bp [82]. EMSA analysis demonstrated that 15d-PGJ2 increased binding of HSF-1 to the −143/−115 TRAIL oligonucleotide. While functional studies were not carried out, these findings suggest that HSF-1 negatively regulates TRAIL transcription in response to 15d-PGJ2 [82]. In conclusion, this study demonstrated that 15d-PGJ2 exerted its anti-inflammatory properties by inhibiting TRAIL expression via inhibition of NFκB and also via HSF-1. The anti-inflammatory action of 15d-PGJ2 may provide a new therapeutic approach for the treatment of autoimmune diseases mediated by TRAIL [82].
The role of NFκB is not limited in regulating TRAIL expression following T cell activation. NFκB can also regulate TRAIL expression in response to the FPRL1 agonists, e.g., W peptide (peptide of the invading pathogen) [83]. FPRL1 or formyl peptide receptor-like-1 plays a key role in the regulation of host defence against pathogenic infection. In this study, W peptide increased TRAIL protein levels in human THP-1 monocytes, in freshly isolated normal human neutrophils and in mouse leukocytes [83]. Pretreatment with LLnL (N-acetyl l-leucinyl l-leucinyl l-norleucind), an NFκB inhibitor, inhibited W peptide-inducible IκBα phosphorylation and TRAIL expression in THP-1 monocytes and neutrophils [83]. Furthermore, enhanced TRAIL expression by W peptide led to tumour growth suppression by apoptosis in mice bearing tumours. These results suggest that FPRL1 activation-mediated endogenous TRAIL expression plays a role in tumour immunosurveilance and innate immunity against pathogenic infection [83].
In separate studies involving non-lymphoid cells, TRAIL, Egr-2 and Egr-3 mRNA were upregulated in primary intestinal epithelial cells isolated from mice injected with superantigen staphylococcus enterotoxin B (SEB) [84]. Egr-2 and Egr-3 are immediate early genes that can be induced rapidly by various stimuli. Egr-2 and Egr-3 overexpression induced TRAIL mRNA expression in these cells, and this induction was blocked by Nab1 overexpression, a co-repressor of Egr [84]. Although this study did not determine the functionality of the Egr sites in the promoter, sequence analysis identified a putative Egr site, suggesting that Egr family members may directly regulate TRAIL transcription in non-lymphoid cells [84]. These findings implicate a role for TRAIL during an immune response, whereby inducible TRAIL expression in response to an antigen in non-lymphoid tissue may contribute to peripheral lymphocyte deletion by this tissue [84].
TRAIL expression has been shown to be induced following several virus infections. A study by Kirshner et al. [85] reported that Sendai virus (SeV) infection induced TRAIL mRNA expression in a human colon adenocarcinoma cell line. This induction was mediated by the transcription factor IRF-3, since IRF-3 overexpression increased TRAIL mRNA in SeV-infected cells lacking the IFN receptor [85]. IRF-3 overexpression also increased TRAIL promoter activity. Consistent with this, overexpression of the constitutively active IRF-3 construct transactivated the promoter, whilst overexpression of dominant negative IRF-3 inhibited transactivation [85]. IRF-1, -2 and -7 failed to induce promoter activity suggesting that the IRF-3 effect on the TRAIL promoter was specific [85]. Increased IRF-3 bound to the TRAIL ISRE site (−121 to −140), whilst no IRF-7 binding was observed in a no-shift assay (similar to EMSA) in SeV-infected cells [85]. Mutations to the TRAIL ISRE, mutant 1 and 2, reduced IRF-3 binding. Furthermore, both ISRE mutations significantly impaired basal and SeV-induced TRAIL promoter activity [85]. This study implicates a mechanism of TRAIL induction by SeV infection and may suggest TRAIL as a mediator of SeV-induced apoptosis in infected cells [85].
In addition to SeV infection, HIV-1 infection increased TRAIL mRNA and membrane-bound TRAIL in human monocyte-derived macrophages (MDM) [86]. However, in contrast to SeV infection [85], IRF-3 was not required for HIV-1-induced TRAIL expression. Instead, this induction was mediated by IRF-1 and IRF-7, since knockdown of IRF-1 and IRF-7, but not IRF-3, reduced TRAIL mRNA in HIV-infected cells. Silencing IRF-1 and IRF-7 also reduced STAT1 phosphorylation, and STAT1 inhibition blocked HIV-1-induced TRAIL expression. Interestingly, knockdown of IRF-1 but not IRF-7 inhibited HIV replication. The increase in TRAIL expression was also associated with type I IFN activity since TRAIL mRNA was reduced following treatment with IFN-α or IFN-β neutralizing antibody in HIV-1-infected cells with STAT1 phosphorylation also reduced [86]. This study showed that TRAIL induction following HIV-1 infection involves a feedback loop through IRF-1, IRF-7, type I IFNs and STAT1 signalling, thus contributing to the macrophage-induced HIV pathogenesis [86].
Interestingly, TRAIL-mediated cytotoxicity is induced by encephalomyocarditis virus (EMCV) infection, and TRAIL mainly contributed to reducing the encephalomyocarditis replication in vivo through the NK-mediated antiviral response [87]. In this system, IRF-9 was implicated in regulating inducible TRAIL expression. As demonstrated by Sato et al. [87], poly-IC (a potent inducer of IFN-α/β that can mimic viral infection) treatment in IRF-9−/− mice reduced TRAIL mRNA expression on isolated natural killer (NK) cells compared to NK cells isolated from wild-type mice. Furthermore, transient transfections using a murine TRAIL promoter luciferase reporter construct (−220 to +47), containing an ISRE element (−57/−44), demonstrated induction of TRAIL promoter activity by IFN-β in wild-type mouse embryonic fibroblasts (MEFs), which was completely abrogated in IRF-9−/− MEFs [87]. The involvement of IRF-9 was further confirmed using EMSA with TRAIL ISRE oligonucleotides, which showed an inducible ISRE–ISGF3 complex formation following IFN-β treatment. ISGF3 or IFN-stimulated gene factor-3 is a heterotrimeric transcription factor complex consisting of IRF-9, Stat1 and Stat2.
Besides IRFs, NFκB has been demonstrated to regulate TRAIL expression in response to virus infection, and this is associated with TRAIL’s proapoptotic activity [88]. Apoptosis via caspase activation is essential for efficient influenza virus propagation, and TRAIL enhanced-influenza virus propagation required NFκB signalling [88]. In this study, inhibition of NFκB signalling by utilising host cells expressing a dominant negative mutant of IKK2/IKKβ or a non-degradable phosphorylation site mutant of IκBα (IкBαmut) blocked influenza virus-induced TRAIL expression [88]. Virus propagation was also reduced in these mutant cells. Interestingly, recombinant human TRAIL was able to rescue this effect by inducing virus propagation [89].
TRAIL regulation in cellular differentiation
NFATc proteins are a family of transcription factors with 4 members: NFATc1, 2, 3 and 4 [90]. Their activity is controlled by calcineurin, a calcium-dependent phosphatase, and they are involved in a variety of biological processes including regulation of cell differentiation [91–93]. TRAIL gene expression can be indirectly regulated by NFATc1, implicating a role for TRAIL in cell differentiation [94]. Treatment with PMA + calcium ionophore A23187, which is known to activate NFAT, resulted in increased TRAIL protein expression in human intestinal cells. This induction was blocked by pretreatment with cyclosporine A, an antagonist of NFAT signalling [94]. NFATc1 overexpression, but not NFATc2, NFATc3 and NFATc4, strongly activated the TRAIL promoter. NFATc1 overexpression also increased TRAIL mRNA and protein expression [94]. Five putative NFAT-binding elements were identified—N1, N2, N3, N4 and N5—located between −923 and −383 of the human TRAIL promoter. Although EMSA analysis showed that PMA + calcium ionophore increased N4 and N5 binding activity and identified the presence of NFATc1, NFATc1-induced TRAIL promoter activity was not inhibited following deletion of these sites (from −1,371 to −165) [94]. Instead, the region between −165 and −35 was found to be required for TRAIL induction by NFATc1. Although this region lacks an NFAT consensus binding site, further analysis identified two sites containing Sp1-binding elements (−86/−57 and −58/−33), important for NFATc1 induction of TRAIL expression [94]. Knocking down NFATc1 by siRNA or shRNA led to increased Sp1 binding to −86/−57 and −58/−33 sites, also confirmed by ChIP studies [94]. These studies suggest that NFATc1-induced TRAIL transcriptional activity involves inhibition of Sp1 binding to the TRAIL promoter [94].
Challenges
TRAIL’s activity and function(s) are complicated, displaying multiple levels of control dependent on cell type and the cellular environment. In this review we have discussed the transcriptional control of TRAIL in multiple cell types and in multiple pathologies. We know that TRAIL’s activity is also controlled at other levels. For example, the newly identified TRAIL isoforms may mediate the differential functions of TRAIL we observe in many systems. We know that in vascular proliferative disorders, at least in our hands, TRAIL’s role to promote survival is more important than its role to induce cell death [25]. In contrast, TRAIL is more likely to promote apoptosis of cancer cells. It is therefore tantalising to speculate that the TRAIL isoforms lacking cytotoxic activity may be more highly expressed in the vasculature than in tumours and cancer cell lines. We are currently in the process of examining the expression of these isoforms in the vessel wall. However, their function(s) in disease settings are currently unknown.
Conflicting reports demonstrating differential cytotoxicity dependent on membrane-bound versus the soluble form of TRAIL are also implicated in disease and further complicate TRAIL’s activity in vivo. For example, adenoviral-mediated TRAIL expression significantly increased apoptosis in human breast cancer cell lines, whilst these cells appeared to be resistant to apoptosis with the soluble form, even at high concentrations [95]. Membrane-bound TRAIL-expressing CD34+ cells also had the ability to kill myeloma, lymphoma and epithelial cancer cells resistant to soluble TRAIL-mediated killing [96]. Interestingly, TRAIL-R1 could be activated by both membrane-bound and soluble TRAIL, whilst TRAIL-R2 was only activated by membrane-bound TRAIL or via soluble TRAIL secondarily cross-linked by antibodies [97]. While these studies support the notion that membrane-bound TRAIL may be more potent in inducing apoptosis, caution needs to be taken in interpreting these results since different preparations of soluble TRAIL may have variable effects on TRAIL-receptor signals [98].
In addition to these further challenges, defects in TRAIL-receptor signalling and components of DISC may also modulate the differential functions of TRAIL. A characteristic feature in human cancers is their ability to become resistant to cell death. Defects in caspase-8, for example, can promote tumour progression and resistance to treatment [99]. In support of this, TRAIL-induced proliferation of tumour cells may be mediated by defects in TRAIL-receptor signalling at the level of the DISC [100] or loss of cell surface TRAIL-R1 and -R2 [49].
Conclusion
Although the mechanisms regulating TRAIL at the level of transcription have still not been widely explored, this review has highlighted recent understandings of the mechanisms that control TRAIL transcription and expression. Identification of transcription factor(s) regulating TRAIL expression will help better understand TRAIL functions in the context of both physiological and pathological conditions. In fact, targeting TRAIL or specific transcription factors may provide novel therapeutic approaches for treatment of diseases associated with TRAIL.
References
- 1.Wiley SR, Schooley K, Smolak PJ, Din WS, Huang C-P, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA, Goodwin RG. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 2.Pitti RM, Marsters SA, Ruppert S, Donahue CJ, Moore A, Ashkenazi A. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem. 1996;271:12687–12690. doi: 10.1074/jbc.271.22.12687. [DOI] [PubMed] [Google Scholar]
- 3.Mariani SM, Krammer PH. Differential regulation of TRAIL and CD95 ligand in transformed cells of the T and B lymphocyte lineage. Eur J Immunol. 1998;28:973–982. doi: 10.1002/(SICI)1521-4141(199803)28:03<973::AID-IMMU973>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 4.Secchiero P, Gonelli A, Corallini F, Ceconi C, Ferrari R, Zauli G. Metalloproteinase 2 cleaves in vitro recombinant TRAIL: potential implications for the decreased serum levels of TRAIL after acute myocardial infarction. Atherosclerosis. 2010;211:333–336. doi: 10.1016/j.atherosclerosis.2010.02.024. [DOI] [PubMed] [Google Scholar]
- 5.Mariani S, Krammer P. Surface expression of TRAIL/Apo2 ligand in activated mouse T and B cells. Eur J Immunol. 1998;28:1492–1498. doi: 10.1002/(SICI)1521-4141(199805)28:05<1492::AID-IMMU1492>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 6.Pan G, O’Rourke K, Chinnaiyan AM, Gentz R, Ebner R, Ni J, Dixit VM. The receptor for the cytotoxic ligand TRAIL. Science. 1997;276:111–113. doi: 10.1126/science.276.5309.111. [DOI] [PubMed] [Google Scholar]
- 7.Pan G, Ni J, Wei YF, Yu G, Gentz R, Dixit VM. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science. 1997;277:815–818. doi: 10.1126/science.277.5327.815. [DOI] [PubMed] [Google Scholar]
- 8.Sheridan JP, Marsters SA, Pitti RM, Gurney A, Skubatch M, Baldwin D, Ramakrishnan L, Gray CL, Baker K, Wood WI, Goddard AD, Godowski P, Ashkenazi A. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science. 1997;277:818–821. doi: 10.1126/science.277.5327.818. [DOI] [PubMed] [Google Scholar]
- 9.Degli-Esposti M. Cloning and characterization of TRAIL-R3, a novel member of the emerging TRAIL receptor family. J Exp Med. 1997;186:1165–1170. doi: 10.1084/jem.186.7.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marsters SA, Sheridan JP, Pitti RM, Huang A, Skubatch M, Baldwin D, Yuan J, Gurney A, Goddard AD, Godowski P, Ashkenazi A. A novel receptor for Apo2L/TRAIL contains a truncated death domain. Curr Biol. 1997;7:1003–1006. doi: 10.1016/S0960-9822(06)00422-2. [DOI] [PubMed] [Google Scholar]
- 11.Degli-Esposti MA, Dougall WC, Smolak PJ, Waugh JY, Smith CA, Goodwin RG. The novel receptor TRAIL-R4 induces NF-kappaB and protects against TRAIL-mediated apoptosis, yet retains an incomplete death domain. Immunity. 1997;7:813–820. doi: 10.1016/S1074-7613(00)80399-4. [DOI] [PubMed] [Google Scholar]
- 12.Emery JG. Osteoprotegerin is a receptor for the cytotoxic ligand TRAIL. J Biol Chem. 1998;273:14363–14367. doi: 10.1074/jbc.273.23.14363. [DOI] [PubMed] [Google Scholar]
- 13.Kavurma MM, Bennett MR. Expression, regulation and function of trail in atherosclerosis. Biochem Pharmacol. 2008;75:1441–1450. doi: 10.1016/j.bcp.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 14.Holoch PA, Griffith TS. TNF-related apoptosis-inducing ligand (TRAIL): a new path to anti-cancer therapies. Eur J Pharmacol. 2009;625:63–72. doi: 10.1016/j.ejphar.2009.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falschlehner C, Emmerich CH, Gerlach B, Walczak H. TRAIL signalling: decisions between life and death. Int J Biochem Cell Biol. 2007;39:1462–1475. doi: 10.1016/j.biocel.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Varfolomeev E, Maecker H, Sharp D, Lawrence D, Renz M, Vucic D, Ashkenazi A. Molecular determinants of kinase pathway activation by Apo2 ligand/tumor necrosis factor-related apoptosis-inducing ligand. J Biol Chem. 2005;280:40599–40608. doi: 10.1074/jbc.M509560200. [DOI] [PubMed] [Google Scholar]
- 17.Newsom-Davis T, Prieske S, Walczak H. Is TRAIL the holy grail of cancer therapy? Apoptosis. 2009;14:607–623. doi: 10.1007/s10495-009-0321-2. [DOI] [PubMed] [Google Scholar]
- 18.Gong B, Almasan A. Genomic organization and transcriptional regulation of human Apo2/TRAIL gene. Biochem Biophys Res Commun. 2000;278:747–752. doi: 10.1006/bbrc.2000.3872. [DOI] [PubMed] [Google Scholar]
- 19.Krieg A, Krieg T, Wenzel M, Schmitt M, Ramp U, Fang B, Gabbert HE, Gerharz CD, Mahotka C. TRAIL-beta and TRAIL-gamma: two novel splice variants of the human TNF-related apoptosis-inducing ligand (TRAIL) without apoptotic potential. Br J Cancer. 2003;88:918–927. doi: 10.1038/sj.bjc.6600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woods DC, Alvarez C, Johnson AL. Cisplatin-mediated sensitivity to TRAIL-induced cell death in human granulosa tumor cells. Gynecol Oncol. 2008;108:632–640. doi: 10.1016/j.ygyno.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 21.Wang P, Lu Y, Li C, Li N, Yu P, Ma D. Novel transcript variants of TRAIL show different activities in activation of NF-kappaB and apoptosis. Life Sci. 2011;89:839–846. doi: 10.1016/j.lfs.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Wang QD, Ji YS, Wang XF, Evers BM. Isolation and molecular characterization of the 5′-upstream region of the human TRAIL gene. Biochem Biophys Res Commun. 2000;276:466–471. doi: 10.1006/bbrc.2000.3512. [DOI] [PubMed] [Google Scholar]
- 23.Li JH, Kirkiles-Smith NC, McNiff JM, Pober JS. TRAIL induces apoptosis and inflammatory gene expression in human endothelial cells. J Immunol. 2003;171:1526–1533. doi: 10.4049/jimmunol.171.3.1526. [DOI] [PubMed] [Google Scholar]
- 24.Sato K, Niessner A, Kopecky SL, Frye RL, Goronzy JJ, Weyand CM. TRAIL-expressing T cells induce apoptosis of vascular smooth muscle cells in the atherosclerotic plaque. J Exp Med. 2006;203:239–250. doi: 10.1084/jem.20051062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan J, Prado-Lourenco L, Khachigian LM, Bennett MR, Di Bartolo BA, Kavurma MM. TRAIL promotes VSMC proliferation and neointima formation in a FGF-2-, Sp1 phosphorylation-, and NFkappaB-dependent manner. Circ Res. 2010;106:1061–1071. doi: 10.1161/CIRCRESAHA.109.206029. [DOI] [PubMed] [Google Scholar]
- 26.Di Bartolo BA, Chan J, Bennett MR, Cartland S, Bao S, Tuch BE, Kavurma MM. TNF-related apoptosis-inducing ligand (TRAIL) protects against diabetes and atherosclerosis in Apoe (−)/(−) mice. Diabetologia. 2011;54:3157–3167. doi: 10.1007/s00125-011-2308-0. [DOI] [PubMed] [Google Scholar]
- 27.Hameed AG, Arnold ND, Chamberlain J, Pickworth JA, Paiva C, Dawson S, Cross S, Long L, Zhao L, Morrell NW, Crossman DC, Newman CM, Kiely DG, Francis SE, Lawrie A. Inhibition of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) reverses experimental pulmonary hypertension. J Exp Med. 2012;209:1919–1935. doi: 10.1084/jem.20112716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kavurma MM, Schoppet M, Bobryshev YV, Khachigian LM, Bennett MR. TRAIL stimulates proliferation of vascular smooth muscle cells via activation of NF-kappa B and induction of insulin-like growth factor-1 receptor. J Biol Chem. 2008;283:7754–7762. doi: 10.1074/jbc.M706927200. [DOI] [PubMed] [Google Scholar]
- 29.Lawrie A, Waterman E, Southwood M, Evans D, Suntharalingam J, Francis S, Crossman D, Croucher P, Morrell N, Newman C. Evidence of a role for osteoprotegerin in the pathogenesis of pulmonary arterial hypertension. Am J Pathol. 2008;172:256–264. doi: 10.2353/ajpath.2008.070395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoppet M, Sattler AM, Schaefer JR, Hofbauer LC. Osteoprotegerin (OPG) and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) levels in atherosclerosis. Atherosclerosis. 2006;184:446–447. doi: 10.1016/j.atherosclerosis.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 31.Azahri NS, Di Bartolo BA, Khachigian LM, Kavurma MM. Sp1, acetylated histone-3 and p300 regulate TRAIL transcription: mechanisms of PDGF-BB-mediated VSMC proliferation and migration. J Cell Biochem. 2012;113:2597–2606. doi: 10.1002/jcb.24135. [DOI] [PubMed] [Google Scholar]
- 32.Secchiero P, Gonelli A, Carnevale E, Milani D, Pandolfi A, Zella D, Zauli G. TRAIL promotes the survival and proliferation of primary human vascular endothelial cells by activating the Akt and ERK pathways. Circulation. 2003;107:2250–2256. doi: 10.1161/01.CIR.0000062702.60708.C4. [DOI] [PubMed] [Google Scholar]
- 33.Secchiero P, Zerbinati C, Rimondi E, Corallini F, Milani D, Grill V, Forti G, Capitani S, Zauli G. TRAIL promotes the survival, migration and proliferation of vascular smooth muscle cells. Cell Mol Life Sci. 2004;61:1965–1974. doi: 10.1007/s00018-004-4197-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corallini F, Celeghini C, Rizzardi C, Pandolfi A, Di Silvestre S, Vaccarezza M, Zauli G. Insulin down-regulates TRAIL expression in vascular smooth muscle cells both in vivo and in vitro. J Cell Physiol. 2007;212:89–95. doi: 10.1002/jcp.21006. [DOI] [PubMed] [Google Scholar]
- 35.Secchiero P, Candido R, Corallini F, Zacchigna S, Toffoli B, Rimondi E, Fabris B, Giacca M, Zauli G. Systemic tumor necrosis factor-related apoptosis-inducing ligand delivery shows antiatherosclerotic activity in apolipoprotein E-null diabetic mice. Circulation. 2006;114:1522–1530. doi: 10.1161/CIRCULATIONAHA.106.643841. [DOI] [PubMed] [Google Scholar]
- 36.Volpato S, Ferrucci L, Secchiero P, Corallini F, Zuliani G, Fellin R, Guralnik JM, Bandinelli S, Zauli G. Association of tumor necrosis factor-related apoptosis-inducing ligand with total and cardiovascular mortality in older adults. Atherosclerosis. 2011;215:452–458. doi: 10.1016/j.atherosclerosis.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patwardhan S, Gashler A, Siegel MG, Chang LC, Joseph LJ, Shows TB, Le Beau MM, Sukhatme VP. EGR3, a novel member of the Egr family of genes encoding immediate-early transcription factors. Oncogene. 1991;6:917–928. [PubMed] [Google Scholar]
- 38.Khachigian LM, Lindner V, Williams AJ, Collins T. Egr-1-induced endothelial gene expression: a common theme in vascular injury. Science. 1996;271:1427–1431. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- 39.Fu M, Zhu X, Zhang J, Liang J, Lin Y, Zhao L, Ehrengruber MU, Chen YE. Egr-1 target genes in human endothelial cells identified by microarray analysis. Gene. 2003;315:33–41. doi: 10.1016/S0378-1119(03)00730-3. [DOI] [PubMed] [Google Scholar]
- 40.Santiago FS, Lowe HC, Day FL, Chesterman CN, Khachigian LM. Early growth response factor-1 induction by injury is triggered by release and paracrine activation by fibroblast growth factor-2. Am J Pathol. 1999;154:937–944. doi: 10.1016/S0002-9440(10)65341-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Almasan A, Ashkenazi A. Apo2L/TRAIL: apoptosis signaling, biology, and potential for cancer therapy. Cytokine Growth Factor Rev. 2003;14:337–348. doi: 10.1016/S1359-6101(03)00029-7. [DOI] [PubMed] [Google Scholar]
- 42.Wiley SR. A novel TNF receptor family member binds TWEAK and is implicated in angiogenesis. Immunity. 2001;15:837–846. doi: 10.1016/S1074-7613(01)00232-1. [DOI] [PubMed] [Google Scholar]
- 43.Fulda S, Wick W, Weller M, Debatin KM. Smac agonists sensitize for Apo2L/TRAIL- or anticancer drug-induced apoptosis and induce regression of malignant glioma in vivo. Nat Med. 2002;8:808–815. doi: 10.1038/nm735. [DOI] [PubMed] [Google Scholar]
- 44.Mitsiades CS, Treon SP, Mitsiades N, Shima Y, Richardson P, Schlossman R, Hideshima T, Anderson KC. TRAIL/Apo2L ligand selectively induces apoptosis and overcomes drug resistance in multiple myeloma: therapeutic applications. Blood. 2001;98:795–804. doi: 10.1182/blood.V98.3.795. [DOI] [PubMed] [Google Scholar]
- 45.Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T, Smith C, Smolak P, Goodwin RG, Rauch CT, Schuh JC, Lynch DH. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- 46.Ashkenazi A, Pai RC, Fong S, Leung S, Lawrence DA, Marsters SA, Blackie C, Chang L, McMurtrey AE, Hebert A, DeForge L, Koumenis IL, Lewis D, Harris L, Bussiere J, Koeppen H, Shahrokh Z, Schwall RH. Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest. 1999;104:155–162. doi: 10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bellail AC, Qi L, Mulligan P, Chhabra V, Hao C. TRAIL agonists on clinical trials for cancer therapy: the promises and the challenges. Rev Recent Clin Trials. 2009;4:34–41. doi: 10.2174/157488709787047530. [DOI] [PubMed] [Google Scholar]
- 48.Tomek S, Horak P, Pribill I, Haller G, Rossler M, Zielinski CC, Pils D, Krainer M. Resistance to TRAIL-induced apoptosis in ovarian cancer cell lines is overcome by co-treatment with cytotoxic drugs. Gynecol Oncol. 2004;94:107–114. doi: 10.1016/j.ygyno.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Zhang B. TRAIL resistance of breast cancer cells is associated with constitutive endocytosis of death receptors 4 and 5. Mol Cancer Res. 2008;6:1861–1871. doi: 10.1158/1541-7786.MCR-08-0313. [DOI] [PubMed] [Google Scholar]
- 50.Ehrhardt H, Fulda S, Schmid I, Hiscott J, Debatin KM, Jeremias I. TRAIL induced survival and proliferation in cancer cells resistant towards TRAIL-induced apoptosis mediated by NF-kappaB. Oncogene. 2003;22:3842–3852. doi: 10.1038/sj.onc.1206520. [DOI] [PubMed] [Google Scholar]
- 51.Kops GJ, Burgering BM. Forkhead transcription factors are targets of signalling by the proto-oncogene PKB (C-AKT) J Anat. 2000;197(Pt 4):571–574. doi: 10.1046/j.1469-7580.2000.19740571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghaffari S, Jagani Z, Kitidis C, Lodish HF, Khosravi-Far R. Cytokines and BCR-ABL mediate suppression of TRAIL-induced apoptosis through inhibition of forkhead FOXO3a transcription factor. Proc Natl Acad Sci USA. 2003;100:6523–6528. doi: 10.1073/pnas.0731871100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rivera-Walsh I, Waterfield M, Xiao G, Fong A, Sun SC. NF-kappaB signaling pathway governs TRAIL gene expression and human T-cell leukemia virus-I Tax-induced T-cell death. J Biol Chem. 2001;276:40385–40388. doi: 10.1074/jbc.C100501200. [DOI] [PubMed] [Google Scholar]
- 54.Chlichlia K, Moldenhauer G, Daniel PT, Busslinger M, Gazzolo L, Schirrmacher V, Khazaie K. Immediate effects of reversible HTLV-1 tax function: T-cell activation and apoptosis. Oncogene. 1995;10:269–277. [PubMed] [Google Scholar]
- 55.Los M, Khazaie K, Schulze-Osthoff K, Baeuerle PA, Schirrmacher V, Chlichlia K. Human T cell leukemia virus-I (HTLV-I) Tax-mediated apoptosis in activated T cells requires an enhanced intracellular prooxidant state. J Immunol. 1998;161:3050–3055. [PubMed] [Google Scholar]
- 56.Choi EA, Lei H, Maron DJ, Wilson JM, Barsoum J, Fraker DL, El-Deiry WS, Spitz FR. Stat1-dependent Induction of Tumor Necrosis Factor-related Apoptosis-inducing Ligand and the Cell-Surface Death Signaling Pathway by Interferon B in Human Cancer Cells. Cancer Res. 2003;63:5299–5307. [PubMed] [Google Scholar]
- 57.Miura Y, Tsujioka T, Nishimura Y, Sakaguchi H, Maeda M, Hayashi H, Dong M, Hyodoh F, Yata K, Wada H, Sugihara T, Otsuki T. TRAIL expression up-regulated by interferon-gamma via phosphorylation of STAT1 induces myeloma cell death. Anticancer Res. 2006;26:4115–4124. [PubMed] [Google Scholar]
- 58.Papageorgiou A, Dinney CP, McConkey DJ. Interferon-alpha induces TRAIL expression and cell death via an IRF-1-dependent mechanism in human bladder cancer cells. Cancer Biol Ther. 2007;6:872–879. doi: 10.4161/cbt.6.6.4088. [DOI] [PubMed] [Google Scholar]
- 59.Clarke N, Jimenez-Lara AM, Voltz E, Gronemeyer H. Tumor suppressor IRF-1 mediates retinoid and interferon anticancer signaling to death ligand TRAIL. EMBO J. 2004;23:3051–3060. doi: 10.1038/sj.emboj.7600302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene. 2002;285:1–24. doi: 10.1016/S0378-1119(02)00398-0. [DOI] [PubMed] [Google Scholar]
- 61.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 62.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 63.Veals SA, Schindler C, Leonard D, Fu XY, Aebersold R, Darnell JE, Jr, Levy DE. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol Cell Biol. 1992;12:3315–3324. doi: 10.1128/mcb.12.8.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuribayashi K, Krigsfeld G, Wang W, Xu J, Mayes PA, Dicker DT, Wu GS, El-Deiry WS. TNFSF10 (TRAIL), a p53 target gene that mediates p53-dependent cell death. Cancer Biol Ther. 2008;7:2034–2038. doi: 10.4161/cbt.7.12.7460. [DOI] [PubMed] [Google Scholar]
- 65.Xu J, Zhou JY, Wu GS. Tumor necrosis factor-related apoptosis-inducing ligand is required for tumor necrosis factor alpha-mediated sensitization of human breast cancer cells to chemotherapy. Cancer Res. 2006;66:10092–10099. doi: 10.1158/0008-5472.CAN-06-1633. [DOI] [PubMed] [Google Scholar]
- 66.Xu J, Zhou J-Y, Wei W-Z, Philipsen S, Wu GS. Sp1-mediated TRAIL induction in chemosensitization. Cancer Res. 2008;68:6718–6726. doi: 10.1158/0008-5472.CAN-08-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xie RL, Gupta S, Miele A, Shiffman D, Stein JL, Stein G, Svan Wijnen AJ. The tumor suppressor interferon regulatory factor 1 interferes with SP1 activation to repress the human CDK2 promoter. J Biol Chem. 2003;278:26589–26596. doi: 10.1074/jbc.M301491200. [DOI] [PubMed] [Google Scholar]
- 68.Ehrlich S, Infante-Duarte C, Seeger B, Zipp F. Regulation of soluble and surface-bound TRAIL in human T cells, B cells, and monocytes. Cytokine. 2003;24:244–253. doi: 10.1016/S1043-4666(03)00094-2. [DOI] [PubMed] [Google Scholar]
- 69.Fanger NA, Maliszewski CR, Schooley K, Griffith TS. Human dendritic cells mediate cellular apoptosis via tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) J Exp Med. 1999;190:1155–1164. doi: 10.1084/jem.190.8.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Janssen EM, Droin NM, Lemmens EE, Pinkoski MJ, Bensinger SJ, Ehst BD, Griffith TS, Green DR, Schoenberger SP. CD4+ T-cell help controls CD8+ T-cell memory via TRAIL-mediated activation-induced cell death. Nature. 2005;434:88–93. doi: 10.1038/nature03337. [DOI] [PubMed] [Google Scholar]
- 71.Kayagaki N, Yamaguchi N, Nakayama M, Eto H, Okumura K, Yagita H. Type I interferons (IFNs) regulate tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) expression on human T cells: a novel mechanism for the antitumor effects of type I IFNs. J Exp Med. 1999;189:1451–1460. doi: 10.1084/jem.189.9.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zauli G, Secchiero P. The role of the TRAIL/TRAIL receptors system in hematopoiesis and endothelial cell biology. Cytokine Growth Factor Rev. 2006;17:245–257. doi: 10.1016/j.cytogfr.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 73.Oh S, Perera LP, Terabe M, Ni L, Waldmann TA, Berzofsky JA. IL-15 as a mediator of CD4+ help for CD8+ T cell longevity and avoidance of TRAIL-mediated apoptosis. Proc Natl Acad Sci USA. 2008;105:5201–5206. doi: 10.1073/pnas.0801003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Secchiero P, Melloni E, Heikinheimo M, Mannisto S, Di Pietro R, Iacone A, Zauli G. TRAIL regulates normal erythroid maturation through an ERK-dependent pathway. Blood. 2004;103:517–522. doi: 10.1182/blood-2003-06-2137. [DOI] [PubMed] [Google Scholar]
- 75.Zhang XR, Zhang LY, Devadas S, Li L, Keegan AD, Shi YF. Reciprocal expression of TRAIL and CD95L in Th1 and Th2 cells: role of apoptosis in T helper subset differentiation. Cell Death Differ. 2003;10:203–210. doi: 10.1038/sj.cdd.4401138. [DOI] [PubMed] [Google Scholar]
- 76.Falschlehner C, Schaefer U, Walczak H. Following TRAIL’s path in the immune system. Immunology. 2009;127:145–154. doi: 10.1111/j.1365-2567.2009.03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Azab NA, Rady HM, Marzouk SA. Elevated serum TRAIL levels in scleroderma patients and its possible association with pulmonary involvement. Clin Rheumatol. 2012;31:1359–1364. doi: 10.1007/s10067-012-2023-3. [DOI] [PubMed] [Google Scholar]
- 78.Castellino G, Corallini F, Trotta F, Secchiero P. Elevated levels of TRAIL in systemic lupus erythematosus are associated to the presence of anti-SSA/SSB antibodies. Lupus. 2007;16:479–482. doi: 10.1177/0961203307079455. [DOI] [PubMed] [Google Scholar]
- 79.Zai-Xing Y, Yan L, Hao W, Ye Z, Chang L, Ren-Qian Z. Preliminary clinical measurement of the expression of TNF-related apoptosis inducing ligand in patients with ankylosing spondylitis. J Clin Lab Anal. 2008;22:138–145. doi: 10.1002/jcla.20231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baetu TM, Kwon H, Sharma S, Grandvaux N, Hiscott J. Disruption of NF-kappaB signaling reveals a novel role for NF-kappaB in the regulation of TNF-related apoptosis-inducing ligand expression. J Immunol. 2001;167:3164–3173. doi: 10.4049/jimmunol.167.6.3164. [DOI] [PubMed] [Google Scholar]
- 81.Siegmund D, Hausser A, Peters N, Scheurich P, Wajant H. Tumor necrosis factor (TNF) and phorbol ester induce TNF-related apoptosis-inducing ligand (TRAIL) under critical involvement of NF-kappa B essential modulator (NEMO)/IKKgamma. J Biol Chem. 2001;276:43708–43712. doi: 10.1074/jbc.M106421200. [DOI] [PubMed] [Google Scholar]
- 82.Fionda C, Nappi F, Piccoli M, Frati L, Santoni A, Cippitelli M. Inhibition of trail gene expression by cyclopentenonic prostaglandin 15-deoxy-delta12,14-prostaglandin J2 in T lymphocytes. Mol Pharmacol. 2007;72:1246–1257. doi: 10.1124/mol.107.038042. [DOI] [PubMed] [Google Scholar]
- 83.Lin C, Wei W, Zhang J, Liu S, Liu Y, Zheng D. Formyl peptide receptor-like 1 mediated endogenous TRAIL gene expression with tumoricidal activity. Mol Cancer Ther. 2007;6:2618–2625. doi: 10.1158/1535-7163.MCT-07-0286. [DOI] [PubMed] [Google Scholar]
- 84.Droin NM, Pinkoski MJ, Dejardin E, Green DR. Egr family members regulate nonlymphoid expression of Fas ligand, TRAIL, and tumor necrosis factor during immune responses. Mol Cell Biol. 2003;23:7638–7647. doi: 10.1128/MCB.23.21.7638-7647.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kirshner JR, Karpova AY, Kops M, Howley PM. Identification of TRAIL as an interferon regulatory factor 3 transcriptional target. J Virol. 2005;79:9320–9324. doi: 10.1128/JVI.79.14.9320-9324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang Y, Walstrom A, Zhang L, Zhao Y, Cui M, Ye L, Zheng JC. Type I interferons and interferon regulatory factors regulate TNF-related apoptosis-inducing ligand (TRAIL) in HIV-1-infected macrophages. PLoS ONE. 2009;4:e5397. doi: 10.1371/journal.pone.0005397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sato K, Hida S, Takayanagi H, Yokochi T, Kayagaki N, Takeda K, Yagita H, Okumura K, Tanaka N, Taniguchi T, Ogasawara K. Antiviral response by natural killer cells through TRAIL gene induction by IFN-α/β. Eur J Immunol. 2001;31:3138–3146. doi: 10.1002/1521-4141(200111)31:11<3138::AID-IMMU3138>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 88.Wurzer WJ, Ehrhardt C, Pleschka S, Berberich-Siebelt F, Wolff T, Walczak H, Planz O, Ludwig S. NF-kappaB-dependent induction of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and Fas/FasL is crucial for efficient influenza virus propagation. J Biol Chem. 2004;279:30931–30937. doi: 10.1074/jbc.M403258200. [DOI] [PubMed] [Google Scholar]
- 89.Wurzer WJ, Planz O, Ehrhardt C, Giner M, Silberzahn T, Pleschka S, Ludwig S. Caspase 3 activation is essential for efficient influenza virus propagation. EMBO J. 2003;22:2717–2728. doi: 10.1093/emboj/cdg279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang TT, Xiong Q, Enslen H, Davis RJ, Chow CW. Phosphorylation of NFATc4 by p38 mitogen-activated protein kinases. Mol Cell Biol. 2002;22:3892–3904. doi: 10.1128/MCB.22.11.3892-3904.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Delling U, Tureckova J, Lim HW, De Windt LJ, Rotwein P, Molkentin JD. A calcineurin-NFATc3-dependent pathway regulates skeletal muscle differentiation and slow myosin heavy-chain expression. Mol Cell Biol. 2000;20:6600–6611. doi: 10.1128/MCB.20.17.6600-6611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ho IC, Kim JH, Rooney JW, Spiegelman BM, Glimcher LH. A potential role for the nuclear factor of activated T cells family of transcriptional regulatory proteins in adipogenesis. Proc Natl Acad Sci USA. 1998;95:15537–15541. doi: 10.1073/pnas.95.26.15537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Santini MP, Talora C, Seki T, Bolgan L, Dotto GP. Cross talk among calcineurin, Sp1/Sp3, and NFAT in control of p21(WAF1/CIP1) expression in keratinocyte differentiation. Proc Natl Acad Sci USA. 2001;98:9575–9580. doi: 10.1073/pnas.161299698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang Q, Zhou Y, Weiss HL, Chow C-W, Evers BM. NFATc1 regulation of TRAIL expression in human intestinal cells. PLoS ONE. 2011;6:e19882. doi: 10.1371/journal.pone.0019882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin T, Huang X, Gu J, Zhang L, Roth JA, Xiong M, Curley SA, Yu Y, Hunt KK, Fang B. Long-term tumor-free survival from treatment with the GFP-TRAIL fusion gene expressed from the hTERT promoter in breast cancer cells. Oncogene. 2002;21:8020–8028. doi: 10.1038/sj.onc.1205926. [DOI] [PubMed] [Google Scholar]
- 96.Carlo-Stella C, Lavazza C, Di Nicola M, Cleris L, Longoni P, Milanesi M, Magni M, Morelli D, Gloghini A, Carbone A, Gianni AM. Antitumor activity of human CD34+ cells expressing membrane-bound tumor necrosis factor-related apoptosis-inducing ligand. Hum Gene Ther. 2006;17:1225–1240. doi: 10.1089/hum.2006.17.1225. [DOI] [PubMed] [Google Scholar]
- 97.Wajant H, Moosmayer D, Wuest T, Bartke T, Gerlach E, Schonherr U, Peters N, Scheurich P, Pfizenmaier K. Differential activation of TRAIL-R1 and -2 by soluble and membrane TRAIL allows selective surface antigen-directed activation of TRAIL-R2 by a soluble TRAIL derivative. Oncogene. 2001;20:4101–4106. doi: 10.1038/sj.onc.1204558. [DOI] [PubMed] [Google Scholar]
- 98.Lawrence D. Differential hepatocyte toxicity of recombinant Apo2L/TRAIL versions. Nat Med. 2001;7:383–385. doi: 10.1038/86397. [DOI] [PubMed] [Google Scholar]
- 99.Fulda S. Caspase-8 in cancer biology and therapy. Cancer Lett. 2009;281:128–133. doi: 10.1016/j.canlet.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 100.Baader E, Toloczko A, Fuchs U, Schmid I, Beltinger C, Ehrhardt H, Debatin KM, Jeremias I. Tumor necrosis factor-related apoptosis-inducing ligand-mediated proliferation of tumor cells with receptor-proximal apoptosis defects. Cancer Res. 2005;65:7888–7895. doi: 10.1158/0008-5472.CAN-04-4278. [DOI] [PubMed] [Google Scholar]