Abstract
Melanoma is a malignant tumor of melanocytes that can spread to other organs of the body, resulting in severe and/or lethal malignancies. Melanocytes are pigment-producing cells found in the deep layer of the epidermis and are originated from melanocytes stem cells through a cellular process called melanogenesis. Several genes and epigenetic and micro-environmental factors are involved in this process via the regulation and maintenance of the balance between melanocytes stem cells proliferation and their differentiation into melanocytes. Dysregulation of this balance through gain or loss of function of key genes implicated in the control and regulation of cell cycle progression and/or differentiation results in melanoma initiation and progression. This review aims to provide a comprehensive overview about the origin of melanocytes, the oncogenic events involved in melanocytes stem cells transformation, and the mechanisms implicated in the perpetuation of melanoma malignant phenotype.
Keywords: Cancer stem cells, Malignant melanoma-initiating cells, HAGE, ABCB5, MicroRNA, Chemoresistance, Melanocyte, RAS signaling
Introduction
Melanocytes begin their journey at the gastrula stage of development where the neural crest generates glial-melanocyte progenitors that commit to the production of melanoblasts [1, 2]. These cells later generate melanocyte stem cells (MSCs), found in the hair follicle where they ensure the continuous generation of melanocytes, the cells responsible for melanin-pigment production throughout the adult life. These cells possess stem cell-like properties associated with self-renewal and differentiation. The process of transformation of MSCs results in the initiation of melanoma and/or its progression. In this regard, several factors involved in the early development and survival of MSCs such as MITF or EDNRB are overexpressed in melanoma and act as “survival” oncogenes [3–5]. Other causes of MSC transformation involve genetic mutations targeting key genes implicated in cell cycle regulation, cell differentiation, and signal transduction. Examples include mutations in tumor suppressor genes such CDKN2A and CDK4 and gain-of-function mutations affecting the RAS-RAF-MAPK pathways responsible for nearly 75 % (BRAF and NRAS mutations) of melanoma cases [6, 7]. The implication of epigenetic factors in melanoma progression is reflected by the hypomethylation or hypermethylation events affecting the expression of oncogenes and tumor suppressors, respectively. Up-regulation or down-regulation of microRNAs expression, which act as tumor suppressors or oncogenes through the regulation of target genes expression, have also been shown to be implicated in melanoma progression [7]. More recently, the identification of malignant melanoma-initiating cells expressing the chemo-resistant ATP-binding cassette (ABC) subfamily B member 5 (ABCB5), responsible for reducing drug accumulation in ABCB5-positive tumor cells allowed better understanding of the chemotherapeutic refractoriness of advanced malignant melanoma [8]. These populations of cancer-initiating cells also express the cancer testis antigen HAGE (DDX43), which plays an important role in their proliferation and survival through the activation of RAS-RAF-MAPK and RAS-Pi3K-AKT pathways [9]. This review attempts to highlight the important molecular and cellular events implicated in melanoma initiation and progression with regard to what is known about melanoma cancer-initiating cells.
From melanocytes to melanoma: the process of transformation
Origin and regulation of melanocytes development
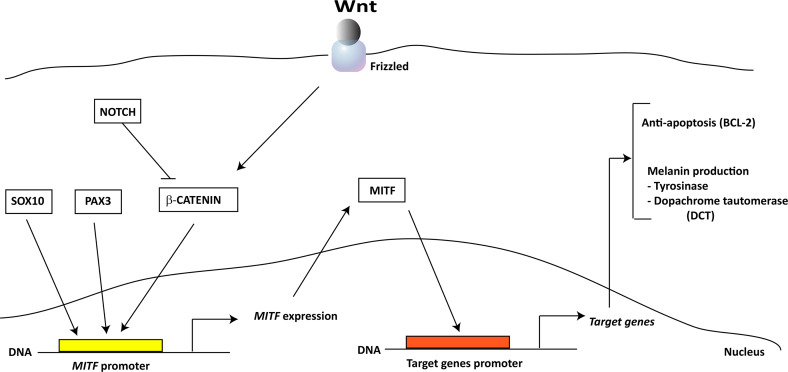
Melanocytes are pigment-producing cells found in the deep layer of the epidermis and are originated from the multipotent and highly migrating neural crest [1]. During embryonic development, neural crest found in the neural tube generates bipotential glial-melanocyte progenitors, which became lineage restricted and produce melanoblasts that originate melanocytes [2]. The commitment of glial-melanocyte progenitors into melanocytes involves Wnt signaling and takes place in the neural tube or after the cells migrating as suggested by Wnt1 and Wnt3a mice knockout studies [10]. In the opposite way, BMP signaling may function through promoting the glial-melanocyte progenitor commitment to neural and glial progenitor cells [11]. Moreover, Wnt signaling reportedly plays a role in the terminal differentiation of melanoblasts, indicating its key involvement in melanocyte development [12, 13]. Apart from Wnt signaling, several genes are important for melanocyte development through their involvement in the different developmental stages of melanogenesis. MITF, a gene in which expression is induced by SOX10 and PAX3 and beta-catenin, promotes melanocyte lineage specification and survival by activating the expression of pigment-producing genes such as DCT (dopachrome tautomerase) and tyrosinase and the anti-apoptotic gene BCL-2 [14] (Fig. 1). The SLUG gene, also induced by SOX10, initiates neural crest migration, a step that is necessary for melanocyte development by repression of E-cadherin and activation of the epithelial–mesenchymal transition (EMT) essential to neural crest migration [15–17]. KIT, a gene that belongs to type III receptor kinase, is another important gene implicated in melanocyte development, migration, and survival. Mutant animal models for this gene presented a fewer melanocytes probably due to a failure in melanocyte migration and enhanced apoptosis [18–21]. Other genes include endothelins, a group of vasoactive proteins that signal by binding to the G-coupled receptor EDNRA or EDNRB and play an essential role for survival and migration of melanoblast. Knock-out models for the endothelin-B recptor (EDNRB) or its ligand endothelin-3 showed a dramatic decrease of melanocyte number [5]. Finally, once melanoblasts arise, they migrate along the dorsolateral pathway between the dermatome and the epidermis toward the ventral midline and further invade the overlying epidermis where they proliferate and migrate extensively to distribute the entire epidermis toward newly developing hair follicles. In the follicles, melanoblasts are segregated into melanocytes and MSCs. The melanocytes contribute to the initial wave of melanogenesis while MSCs migrate in the lower permanent portion of the hair follicle and constitute the MC system in subsequent hair cycles [22, 23]. In conclusion, melanogenesis is a highly regulated spatio-temporal event that involves several developmental factors that lead to the generation of melanocytes found later on in the epidermis skin and other tissues and organ of adult organism.
Fig. 1.
Schematic representation of MITF-related pathway. MITF expression is induced by SOX10, PAX3, and Wnt/β-catenin pathways. The induction of MITF promoter by β-catenin is counteracted by NOTCH signaling. MITF induces the expression of genes coding for anti-apoptotic (BCL-2) and melanin producing (DCT and tyrosinase) molecules
Molecular pathogenesis and progression of melanoma
Common genetic mutations associated with melanoma
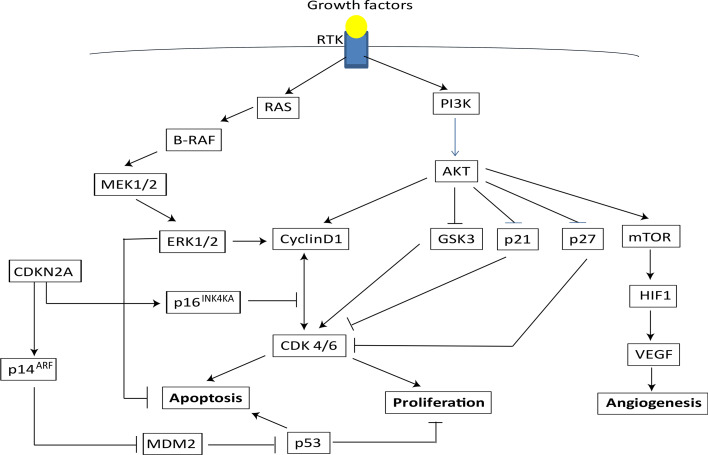
In human skin, melanocytes reside in the basal layer of interfollicular epidermis and are thought to arise from MSCs in the hair follicle [24]. Several genes and pathways involved in the regulation of melanocyte proliferation and differentiation are subverted, leading to an overproliferation of this population of cells resulting in melanoma. For instance, MITF, a molecule that promotes melanocyte lineage specification and survival is found to be overexpressed in many melanomas and has recently been characterized as a ‘lineage survival’ oncogene in melanoma [5]. EDNRB, an essential molecule involved in the developmental survival and migration of MSCs, is also found to be overexpressed in several human melanoma cell lines and EDNRB-specific antagonist inhibition led to the inhibition of the proliferation and the induction of melanoma cell differentiation [3, 4]. However, most of the causes associated with the dysregulation of melanocyte proliferation and differentiation are linked mainly to genetic factors and UV irradiation [25]. Nearly 60 and 15 % of melanomas implicate somatic mutations that overactivate BRAF and NRAS genes, respectively [26, 27]. Other mutations affect tumor suppressor genes such as CDKN2A and CDK4, and genes implicated in DNA repair machinery (Fig. 2) [28].
Fig. 2.
Signaling pathways implicated in the regulation of proliferation, apoptosis, and angiogenesis. Binding of a growth factor to receptor tyrosine kinase (RTK) activate RAS and PI3K. RAS activates a downstream signal involving BRAF-MEK1/2-ERK1/2, which promotes proliferation and prevents apoptosis through the up-regulation of cyclin D1, which controls G1-S transition. AKT also stimulates proliferation by upregulating cyclin D1 and inhibition of p21, p27, which prevents G1/S transition. AKT-mediated inhibition of GSK-3 suppresses apoptosis. AKT-mediated activation of mTOR promotes angiogenesis by activating the transcriptor factor HIF-1 and the vascular endothelial growth factor (VEGF). p16INK4A and p14ARF encoded by CDKN2 gene inhibit the cell cycle progression and induce apoptosis, respectively, through preventing cyclin D1/CDK4/6 interactions and inhibiting p53 degradation through MDM2
BRAF a serine/threonine-protein kinase, is a component of the RAS–RAF–MAPK (mitogen-activated protein kinase) signaling cascade, which is activated by many external stimuli, including growth-factor binding to receptor tyrosine kinases and G-protein-coupled receptors. Activation of this pathway results in the phosphorylation of MAPKs, which phosphorylate and regulate the activities of substrates such as transcription factors, cytoskeletal components, and other kinases involved in the regulation of cell survival, proliferation, differentiation, and motility (Fig. 2) [35]. Several studies have implicated BRAF in the molecular pathogenesis of melanoma most commonly through its mutant form BRAFV600E. This mutation results in 700-fold overactivation of BRAF kinase activity and consequently promotes the oncogenic activation of proliferative and survival signaling pathways in melanoma cells [29, 30]. The NRAS GTPase functions upstream RAF serine-threonine kinases (ARAF, BRAF, CRAF) and upon its activation binds RAF and facilitates RAF induction and initiation of a kinase cascade that signals to the dual specificity kinases, MAPK/ERK kinases 1 and 2 (MEK1/2), and subsequently to ERKs 1 and 2. In a similar fashion, mutations of NRAS gene result in constitutive activation of downstream signaling pathways involving RAF serine-threonine kinases and leading to overproliferation and survival of melanoma cells (Fig. 2) [31, 32]. Finally, other mutations of the MAPK pathways have been recently identified. MAP3K5 and MAP3K9 have been found to be mutated in 24 % of melanoma cell lines while MAP2K1 and MAP2K2 where mutated in 8 % of the melanoma cell lines analyzed [33–35].
Beside the implication of the RAS-RAF-MAPK pathways in melanoma pathogenesis and progression, mutations in tumor suppressor genes have also been reported in the literature. The CDKN2A locus encodes several spliced transcript variants and two of them encode the proteins INK4a and ARF both known to function as CDK4 inhibitors (Fig. 2) [28]. ARF acts as a stabilizer of p53 through its interaction and sequestration of MDM2, a protein involved in the degradation of p53 and therefore enhancing p53-dependent transactivation and apoptosis. INK4a protein directly inhibits the cyclin D-dependent kinases CDK4 and CDK6 responsible for the retinoblastoma protein (pRb) phosphorylation and functional inactivation at G1-S phase of cell cycle progression. Rb phosphorylation by the CDKs prevents its interaction with the pro-proliferation factors E2Fs. Therefore, the loss of the CDKN2A locus in melanoma contributes to overproliferation of melanoma cells due to the absence of counterbalancing mechanisms involving ARF and INK4a [28]. Although, mutations in the p53 gene are a rare event in melanoma, ARF frequent loss in melanoma suggests its important role in melanoma development [36]. Finally, patients with xeroderma pigmentosum (XP), a rare autosomal-recessive disease of mutated DNA repair genes that confer hypersensitivity to ultraviolet light, suggest that DNA repair is involved in the etiology of melanoma and in melanoma progression. The common defect of XP affects the nucleotide excision repair (NER) enzymes (endonucleases and DNA polymerases), which if mutated lead to mutations in tumor suppressor genes such as TP53 or proto-oncogenes resulting in the development of melanoma [37–39].
In conclusion, further understanding of the correlation between the loss of functions of these genes and melanoma progression will greatly contribute to the effort of developing efficient drug-based therapies.
MicroRNA regulation of melanoma progression
The involvement of microRNAs (miRNAs or miRs) in the epigenetic regulation of melanoma progression has been well described in the literature. In this chapter, a brief overview with examples of their role and importance in melanoma pathogenesis is provided. MicroRNAs have been shown to regulate the expression of a variety of tumor suppressor genes and oncogenes associated with melanoma initiation, progression, and invasion [40, 41]. In many cases, the level of expression of miRNAs in melanoma cells is associated with their functions as tumor suppressors or oncogenes. Several miRs, including miR-137, miR-221/222, and miR-182, have been found to be involved in melanoma progression by regulating key genes such as c-KIT, MITF, FOXO3, ITGB3, and CCND1 [40]. For example, Mir-221/222 expression is upregulated in melanoma and correlates with an increased proliferation and activation of melanoma invasion/migration along with anchorage-independent growth. Mir-221/222 functions by down-regulating the expression of Kit and CDKN1B resulting in overproliferation and decreased differentiation of melanoma cells [42, 43]. Mir-214 expression promotes melanoma cells invasion and motility by down-regulating the expression of the integrins member ITGA3 and the transcription factor TFAP2C [42]. In contrast, many microRNAs including miR-211, Let-7a, Let-7b, miR-193b, and miR-205 function as tumor suppressors when over-expressed in melanoma cell lines. MiR-211 prevents melanoma progression by down-regulating the expression of BRN2, TGFBR2, NFAT5, and DPP4 [44, 45]. Let-7a and Let-7b regulate cell cycle progression by down-regulating the expression of ITGB3 and the cyclins proteins CCNA, CCND1, CCND3, and CDK4, respectively [46, 47]. MiR-193b also down-regulated the expression of CCND1 in over-expression experiments while miR-205 down-regulated the expression of proliferation factors E2F1 and E2F5 [48, 49]. Finally, microRNAs are expressed at different stages of melanoma progression and understanding their functions will probably shed new light about the mechanisms that perpetuate the malignant phenotype and will open up the possibility to use miRNAs as diagnostic and prognostic markers as well as potential therapeutic targets for many malignancies.
Malignant melanoma-initiating cells (MMICs)
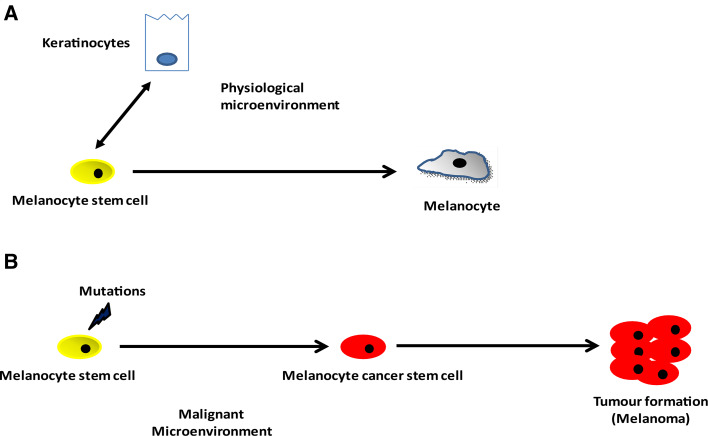
In the skin, normal adult stem cells referred to as MSCs, form the cellular basis for skin homeostasis, maintenance, and repair [24]. These cells reside within a specialized stem cell niche called the bulge, located in the lower permanent portion of the hair follicle known as the outer root sheath [24, 50, 51].These melanocyte stem cells have the ability to replicate themselves through self-renewal, and to give rise to progenitor or transiently amplifying cells which further differentiate into melanin-producing melanocytes, that migrate to the base of the hair follicle (hair bulb) and provide pigment for the growing hair matrix (Fig. 3) [51]. The existence of melanocyte stem cells and their role in self maintenance and generation of melanin-producing cells was demonstrated using melanocyte-tagged transgenic mice [52–54]. These studies have shown that the hair graying observed in these mice, is due to incomplete melanocyte stem cell maintenance and identify Pax3, Bcl2 and Mitf as key molecules that help regulate the balance between MSC maintenance and differentiation (Fig. 1) [55]. In MSCs, the expression of the transcription factor Pax3 and Dct were found up-regulated while the expression of Mitf and another important player Sox10 were found down-regulated providing to melanocyte stem cells a Dct+, Pax3+, Mitf−, Sox10− molecular signature. Thus, this phenotype can maintain an undifferentiated state and promotes quiescence of these lineage-restricted stem cells [23]. More recently, Cheli and colleagues [56] demonstrated further the important role played by Mitf in promoting the differentiation of MSCs. They showed that upon mitf depletion in melanoma cells, the levels of expression of Oct4 and Nanog (two key factors implicated in MSC maintenance) were upregulated, suggesting that oct4 and nanog genes might be direct transcriptional targets of Mitf. Furthermore, the authors reported in the same study an increase of the CDK inhibitor p27 expression which correlated with the exacerbation of the tumorigenic properties of the melanoma cells. Another important player is Notch signaling which has been shown to promote the maintenance of melanocyte stem cells. Conditional ablation of Notch signaling in the melanocyte lineage led to a severe defect in hair pigmentation. Notch signaling through its downstream target HES1 prevented the elimination of MSCs by apoptosis [57]. In the other hand, elevated levels of Wnt signaling molecules, including activated β-catenin, antagonize the regulatory balance between PAX3, SOX10, and MITF towards terminal differentiation (Fig. 1) [58–60]. Finally, the POU domain transcription factor Brn-2 (N-Oct3, POU3F2), expressed in the postnatal hair follicle melanocytes has been shown to be highly expressed in melanomas, suggesting that Brn-2 may function as positive regulator of melanoma survival/proliferation. Brn-2 expression is up-regulated by both MAP kinase and β-catenin signaling and may play a role in the downregulation of Mitf expression and in promoting cell invasion in vitro [61, 62].
Fig. 3.
Melanocyte stem cells and cancer initiation. a Melanocytes reside in the basal layer of interfollicular epidermis and are thought to arise from melanocyte stem cells in the hair follicle. b Somatic mutations in melanocyte stem cells result in their transformation and initiation of melanoma. These cancer stem cells are thought to be responsible for the cases of relapse and chemoresistance
Several studies have shown the importance of micro-environment in the regulation and maintenance of melanocyte stem cells. For instance, keratinocytes in close proximity to melanocyte stem cells, in the bulge in the outer root sheath, have been shown to regulate melanocyte stem cell fate by inducing SCF/c-KIT signaling through continuous secretion of stem cell factor (SCF) [63]. Other studies have shown the involvement of keratinocyte- and/or fibroblast-secreted growth factors in recreating malignant melanoma environment that promotes the switching of cadherin subtypes and the decoupling of melanoma cells from the basement membrane, thereby accelerating melanoma cell proliferation and progression [64–66]. During melanoma development, innate and adaptive immune cells are recruited in the vicinity of the tumor where they play a critical role in inducing angiogenesis, a critical process for the growth, invasion, and metastasis of melanoma. For instance, macrophages induce angiogenesis by releasing granule-storing proteins that directly affect the bioavailability of angiogenic molecules such as the growth factor bFGF (FGF2) and interleukin-8 (IL-8) [67]. Mast cells that arrive first to the site of proliferation in response to tumor-originated SCF and growth factors release vesicle-stored vascular endothelial growth factor (VEGF) [68], several serine proteases including chymase and tryptase [69], and matrix metalloproteinase-9 (MMP-9) [68, 70]. These molecules play an important role in tumor-associated vasculature. The switch to metastatic melanoma is an important process in melanoma progression and has been associated with the loss of expression of the transcription factor AP-2α, which regulates the expression of VEGF, MMP-2 (matrix metalloproteinase-2), c-KIT, MCAM, and the receptors PAR-1 (protease-activated receptor-1) and PAFR (platelet-activating factor receptor) [71].
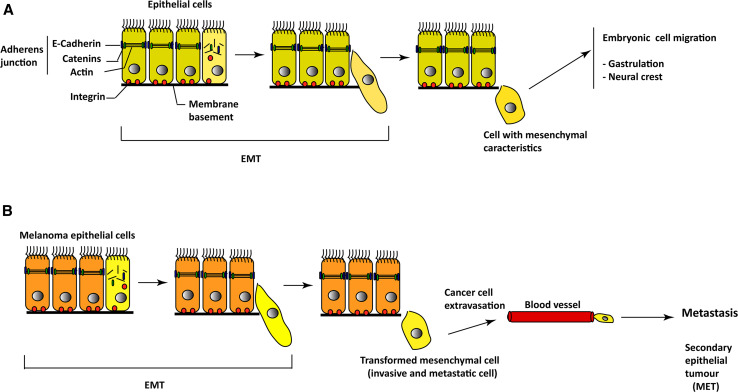
Oncogenic events mediated by genetic mutations or epigenetic factors affecting key molecules such as MITF, SLUG, PAX3, and/or TWIST, involved in melanoma stem cells, quiescence, self-renewal, and differentiation, result in the transformation of normal melanocyte stem cells into populations of cancer cells with inherited properties of self-renewal and differentiation [5, 58, 72, 73]. The involvement of the zinc finger proteins SNAI1 and TWIST in melanoma metastasis through a cellular process called EMT has been reported in the literature (Fig. 4) [74, 75]. EMT is characterized by the expression of mesenchymal molecules such as fibronectin, vimentin, and metalloproteinases and inhibition of E-cadherin. In melanoma, this event has been associated with the constitutive hyperactivation of survival/antiapoptotic pathways such as the MAPK, NF-κB, and PI3K/AKT, which regulate the expression of genes targeting the initiation of the metastatic cascade and involving SNAI1 and TWIST. Moreover, the zinc-finger transcription factor SLUG, which belongs to the SNAIL superfamily, has been shown to function as a melanocyte-specific factor required for the strong metastatic propensity of melanoma [76]. SLUG pro-EMT function has been shown to be regulated by AKT and the secreted matricellular protein SPARC (also known as osteonectin) [77]. In conclusion, EMT involvement in melanoma metastasis appears to involve constitutive hyperactivation of RAS-RAF-MAPK and RAS-Pi3K-AKT pathways.
Fig. 4.
Involvement of epithelial–mesenchymal transition (EMT) in melanoma progression. a During embryonic development (gastrulation and neural crest formation), epithelial cells lose their adherent junctions, their attachments to the membrane basement, and their baso-apical polarity, which result in the production of highly migrating cells with mesenchymal characteristics, necessary for embryogenesis. b In melanoma, carcinoma cells under the influence of EMT signals undergo EMT with the generation of highly invasive and metastatic mesenchymal cells that reach other organs, convert to carcinoma cells in a process called mesenchymal–epithelial transition (MET)
Cancer stem cells (CSCs) are thought to be implicated in melanoma tumor initiation, progression, chemoresistance, and recurrence in human malignant melanoma [78–82]. Corresponding to various data, CSCs represent from 1 to 25 % of total melanoma cell populations [62]. In this regard, Fang and colleagues [73] were able to isolate melanoma populations with stem cell-like properties. Using sphere-forming assays, they were able to show that these subpopulations of human melanoma cells are able of self-renew and to differentiate into multiple cell lineages. Xenotransplantation of these populations into immunodeficient mice, showed a higher tumorigenicity when compared with their adherent counterparts [73].
More recently, Schatton and colleagues [8] isolated from human melanoma patients tissue, populations of malignant melanoma-initiating cells (MMICs) expressing the ATP-binding cassette (ABC) subfamily B member 5 (ABCB5). These populations of cells are highly tumorigenic when xenotransplanted in human to mouse experiments. They have been shown to possess the property of self-renewal and are able to differentiate into ABCB5-positive and ABCB5-negative progeny. The expression of the ABCB5 molecule by MMICs may confer chemo-resistance responsible for recurrence and melanoma progression. High-expression levels of ABC drug efflux transporters reduced drug accumulation in ABCB5-positive tumor cells, which may explain the chemotherapeutic refractoriness of advanced malignant melanoma [83]. Finally, systemic administration of a monoclonal antibody directed to ABCB5, shown to be capable of inducing antibody-dependent cell-mediated cytotoxicity in ABCB5+ MMIC, exerted tumor-inhibitory effects further demonstrating the importance of ABCB5+ MMICs in melanoma initiation, progression, and chemo-resistance [8]. Prior to these studies, several papers reported the presence of cancer stem cells populations in melanoma. The authors used markers such as CD20 (B-lymphocyte antigen CD20), CD133 (prominin-1), and ABCG2 (ATP-binding cassette sub-family G member 2) to isolate those populations [84, 85]. However, the use of CD133 and ABCG2 to isolate stem cell populations raised a big controversy about their status as markers of melanocyte stem cells. For instance, CD133 expression is not restricted to stem cells, and both CD133+ and CD133− metastatic colon cancer cells initiate tumors [86]. ABCG2 is expressed by a smaller subpopulation, but it is not known if this population possesses self-renewal capacity, one of the important properties associated with stem cell populations [87]. In conclusion, better understanding of the molecular mechanisms responsible for melanocytes stem cells transformation will provide key information about melanoma initiation, progression, and chemo-resistance, and will allow specific targeting of these malignant cells.
The helicase HAGE (DDX43), ABCB5+ MMICs, and melanoma progression
Stem and progenitor cell-associated proteins such us cancer testis antigens are encoded by genes that are normally expressed only in the human germ line and the trophoblast, but are also aberrantly expressed in melanoma and a variety of human malignancies [88, 89]. The helicase antigen (HAGE), a non-X-linked cancer testis (CT) antigen of 648 amino acids, was originally identified in a rhabdomyosarcoma cell line [90]. HAGE, also known as DDX43 or CT13, belongs to the DEAD-box family of ATP-dependent RNA helicases characterized by the presence of nine conserved motifs grouped into two domains connected by a polylinker SAT region [91]. The Q motif, motifs I, II (also called the D-E-A-D-box as a single-letter code for Asp-Glu-Ala-Asp), and III, together with motif IV, bind and hydrolyze ATP molecules. The remaining motifs (V, VI, Ia, and Ib) are thought to interact with the RNA substrate. It has been suggested that RNA helicases browse RNA molecules in a bidirectional fashion using the energy gained from ATP hydrolysis until they encounter ribonucleoprotein-RNA complexes [92]. The DEAD-box RNA helicases are ubiquitous, highly conserved enzymes that participate in nearly all aspects of RNA metabolism [92–96]. Their role in tumor growth has been reported previously for some members of this family. For instance, p68 (DDX5), p72 (DDX17), and cancer-associated antigen (CAGE) (DDX53) have been implicated in promoting cell proliferation and survival in cancer cell lines from different tissue origins [97–104]. Members including CAGE and HAGE (DDX43) were found to be highly expressed in different cancer tissues and cancer cell lines, suggesting their role in tumorigenesis [98–105].
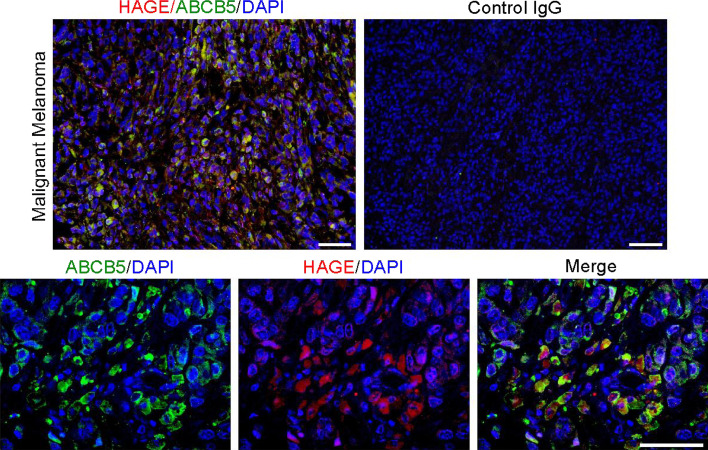
Recently, our group has identified a role of HAGE (DDX43) in promoting the proliferation and survival of ABCB5+ Malignant melanoma-initiating cells (ABCB5+ MMICs) [8]. The implication of HAGE in melanoma ABCB5+ MMICs-mediated initiation and progression was determined by investigating the expression of HAGE and ABCB5 using melanoma tissue from patients, which showed a co-expression of HAGE and ABCB5+ in malignant melanoma-initiating cells. Moreover, HAGE and ABCB5 co-expression was not generalized to the whole tumor and was localized to specific “niches” within melanoma tissues (Fig. 5). In vitro, melanoma cell lines (FM82 and FM55) expressing naturally HAGE (DDX43) co-expressed also ABCB5 and were able to form melanoma-spheres [9]. Upon withdrawal of mitogens, these cells differentiated into ABCB5+ and ABCB5− progeny. HAGE knock-down experiments affected the proliferation of ABCB5+ MMICs, both in vitro using melanoma cell lines and in vivo using the NOD/SCID tumor xenotransplantation assay. Interestingly, the helicase HAGE promoted the proliferation and survival of ABCB5+ MMICs through increasing the expression of NRAS, a molecule involved in melanoma progression through its function as an activator of RAF-MAPK and PI3K-AKT pathways [9, 106–108]. The helicase HAGE appears to promote RAS expression by enhancing NRAS mRNA unwinding, which then increases NRAS mRNA translation in tumor cells [9]. So far, we do not know if the helicase HAGE has other functions in tumor cells or if it plays a direct role in ABCB5+ MMICs-mediated chemo-resistance by promoting the RNA unwinding and expression of ABCB5 in MMICs. Furthermore, hypomethylation of HAGE promoter has previously been reported in the literature, which raises the question as to whether HAGE expression is associated with a specific stage of melanoma progression and whether this expression correlates with the expression of ABCB5 [109, 110]. Finally, HAGE being expressed only by tumor cells suggests that cancer therapies targeting HAGE helicase may have broad applications for treating ABCB5+ malignant melanoma.
Fig. 5.
HAGE (DDX43) and ABCB5 co-expression in patient malignant melanoma sample. ABCB5 have been shown to be expressed by malignant melanoma-initiating cells
Concluding remarks
Substantial advances have been made in our understanding of the molecular mechanisms implicated in melanoma initiation and progression and further dissection of these events will enable the development of efficient drug-based therapies targeting key molecules involved in perpetuating this malignancy. This effort is already taking place with the development of drugs targeting the mutant form of BRAF (BRAFV600E) or in combination with MEK1 inhibitor [111, 112]. It will also allow further development of melanoma stage-specific biomarkers that enable the diagnosis and prediction of clinical outcome of melanoma patients. For instance, microRNAs expression during melanoma expression could be used to differentiate between benign and malignant lesions with poor versus favorable outcome. Finally, as melanoma cancer-initiating cells appear to play an important role in melanoma initiation and chemo-resistance, further investigations of the molecular mechanisms implicated in their regulation will also enable the development of drug-based therapy targeting the molecules involved in the chemotherapeutic refractoriness of advanced malignant melanoma. To this date, this remains an important challenge facing the clinical and basic scientific communities.
References
- 1.Erickson CA, Reedy MV. Neural crest development: the interplay between morphogenesis and cell differentiation. Curr Top Dev Biol. 1998;40:177–209. doi: 10.1016/s0070-2153(08)60367-1. [DOI] [PubMed] [Google Scholar]
- 2.Dupin E, Glavieux C, Vaigot P, Le Douarin NM. Endothelin 3 induces the reversion of melanocytes to glia through a neural crest-derived glial-melanocytic progenitor. Proc Natl Acad Sci. 2000;97(14):7882–7887. doi: 10.1073/pnas.97.14.7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lahav R, Heffner G, Patterson PH. An endothelin receptor B antagonist inhibits growth and induces cell death in human melanoma cells in vitro and in vivo. Proc Natl Acad Sci. 1999;96(20):11496–11500. doi: 10.1073/pnas.96.20.11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24(3):227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 5.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, Lee C, Wagner SN, Li C, Golub TR, Rimm DL, Meyerson ML, Fisher DE, Sellers WR. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436(7047):117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 6.Bonazzi VF, Stark MS, Hayward NK. MicroRNA regulation of melanoma progression. Melanoma Res. 2012;22(2):101. doi: 10.1097/CMR.0b013e32834f6fbb. [DOI] [PubMed] [Google Scholar]
- 7.Haluska FG, Tsao H, Wu H, Haluska FS, Lazar A, Goel V. Genetic alterations in signaling pathways in melanoma. Clin Cancer Res. 2006;12(7):2301s–2307s. doi: 10.1158/1078-0432.CCR-05-2518. [DOI] [PubMed] [Google Scholar]
- 8.Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, Zhan Q, Jordan S, Duncan LM, Weishaupt C, Fuhlbrigge RC, Kupper TS, Sayegh MH, Frank MH. Identification of cells initiating human melanomas. Nature. 2008;451(7176):345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linley AJ, Mathieu MG, Miles AK, Rees RC, McArdle SE, Regad T. The helicase HAGE expressed by malignant melanoma-initiating cells is required for tumor cell proliferation in vivo. J Biol Chem. 2012;287(17):13633–13643. doi: 10.1074/jbc.M111.308973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikeya M, Lee SM, Johnson JE, McMahon AP, Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389(6654):966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- 11.Jin EJ, Erickson CA, Takada S, Burrus LW. Wnt and BMP signaling govern lineage segregation of melanocytes in the avian embryo. Dev Biol. 2001;233(1):22–37. doi: 10.1006/dbio.2001.0222. [DOI] [PubMed] [Google Scholar]
- 12.Dorsky RI, Moon RT, Raible DW. Control of neural crest cell fate by the Wnt signalling pathway. Nature. 1998;396(6709):370–373. doi: 10.1038/24620. [DOI] [PubMed] [Google Scholar]
- 13.Dunn KJ, Williams BO, Li Y, Pavan WJ. Neural crest-directed gene transfer demonstrates Wnt1 role in melanocyte expansion and differentiation during mouse development. Proc Natl Acad Sci. 2000;97(18):10050–10055. doi: 10.1073/pnas.97.18.10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vance KW, Goding CR. The transcription network regulating melanocyte development and melanoma. Pigment Cell Res. 2004;17(4):318–325. doi: 10.1111/j.1600-0749.2004.00164.x. [DOI] [PubMed] [Google Scholar]
- 15.Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2(2):76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 16.Honoré SM, Aybar MJ, Mayor R. Sox10 is required for the early development of the prospective neural crest in Xenopus embryos. Dev Biol. 2003;260(1):79–96. doi: 10.1016/s0012-1606(03)00247-1. [DOI] [PubMed] [Google Scholar]
- 17.Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7(3):291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Geissler EN, Ryan MA, Housman DE. The dominant-white spotting (W) locus of the mouse encodes the c-kit proto-oncogene. Cell. 1988;55(1):185. doi: 10.1016/0092-8674(88)90020-7. [DOI] [PubMed] [Google Scholar]
- 19.Williams DE, Eisenman J, Baird A, Rauch C, Van Ness K, March CJ, Park LS, Martin U, Mochizukl DY, Boswell SH, Burgess GS, Cosman D, Lyman SD. Identification of a ligand for the c-kit proto-oncogene. Cell. 1990;63(1):167–174. doi: 10.1016/0092-8674(90)90297-r. [DOI] [PubMed] [Google Scholar]
- 20.Wehrle-Haller B, Weston JA. Soluble and cell-bound forms of steel factor activity play distinct roles in melanocyte precursor dispersal and survival on the lateral neural crest migration pathway. Development. 1995;121(3):731–742. doi: 10.1242/dev.121.3.731. [DOI] [PubMed] [Google Scholar]
- 21.Parichy DM, Rawls JF, Pratt SJ, Whitfield TT, Johnson SL. Zebrafish sparse corresponds to an orthologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ cell development. Development. 1999;126(15):3425–3436. doi: 10.1242/dev.126.15.3425. [DOI] [PubMed] [Google Scholar]
- 22.Erickson CA. From the crest to the periphery: control of pigment cell migration and lineage segregation. Pigment Cell Res. 1993;6(5):336–347. doi: 10.1111/j.1600-0749.1993.tb00611.x. [DOI] [PubMed] [Google Scholar]
- 23.Nishikawa SI, Osawa M. Generating quiescent stem cells. Pigment Cell Res. 2007;20(4):263–270. doi: 10.1111/j.1600-0749.2007.00388.x. [DOI] [PubMed] [Google Scholar]
- 24.Fuchs E. Scratching the surface of skin development. Nature. 2007;445(7130):834–842. doi: 10.1038/nature05659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uong A, Zon LI. Melanocytes in development and cancer. J Cell Physiol. 2010;222(1):38–41. doi: 10.1002/jcp.21935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhomen N, Reis-Filho JS, da Rocha DS, Hayward R, Savage K, Delmas V, Larue L, Pritchard C, Marais R. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15(4):294. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 27.Daniotti M, Oggionni M, Ranzani T, Vallacchi V, Campi V, Di Stasi D, Torre GD, Perrone F, Luoni C, Suardi S, Frattini M, Pilotti S, Anichini A, Tragni G, Parmiani G, Pierotti MA, Rodolfo M. BRAF alterations are associated with complex mutational profiles in malignant melanoma. Oncogene. 2004;23(35):5968–5977. doi: 10.1038/sj.onc.1207780. [DOI] [PubMed] [Google Scholar]
- 28.Robinson MJ, Cobb MH. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9(2):180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 29.Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004;6(4):313–319. doi: 10.1016/j.ccr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 30.Thomas NE. BRAF somatic mutations in malignant melanoma and melanocytic naevi. Melanoma Res. 2006;16(2):97. doi: 10.1097/01.cmr.0000215035.38436.87. [DOI] [PubMed] [Google Scholar]
- 31.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, Morales T, Aliagas I, Liu B, Sideris S, Hoeflich KP, Jaiswal BS, Seshagiri S, Koeppen H, Belvin M, Friedman LS, Malek S. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464(7287):431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 32.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464(7287):427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stark MS, Woods SL, Gartside MG, Bonazzi VF, Dutton-Regester K, Aoude LG, Chow D, Sereduk C, Niemi NM, Tang N, Ellis JJ, Reid J, Zismann V, Tyagi S, Muzny D, Newsham I, Wu Y, Palmer JM, Pollak T, Youngkin D, Brooks BR, Lanagan C, Schmidt CW, Kobe B, MacKeigan JP, Yin H, Brown KM, Gibbs R, Trent J, Hayward NK. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nat Genet. 2011;44(2):165–169. doi: 10.1038/ng.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C, Harshman K, Guipponi M, Bukach O, Zoete V, Michielin O, Muehlethaler K, Speiser D, Beckmann JS, Xenarios I, Halazonetis TD, Jongeneel CV, Stevenson BJ, Antonarakis SE. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet. 2011;44(2):133–139. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- 35.Kwong LN, Costello JC, Liu H, Jiang S, Helms TL, Langsdorf AE, Jakubosky D, Genovese G, Muller FL, Jeong JH, Bender RP, Chu GC, Flaherty KT, Wargo JA, Collins JJ, Chin L. Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nat Med. 2012;18(10):1503–1510. doi: 10.1038/nm.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Houben R, Hesbacher S, Schmid CP, Kauczok CS, Flohr U, Haferkamp S, Müller CS, Schrama D, Wischhusen J, Becker JC. High-level expression of wild-type p53 in melanoma cells is frequently associated with inactivity in p53 reporter gene assays. PLoS One. 2011;6(7):e22096. doi: 10.1371/journal.pone.0022096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leibeling D, Laspe P, Emmert S. Nucleotide excision repair and cancer. J Mol Histol. 2006;37(5):225–238. doi: 10.1007/s10735-006-9041-x. [DOI] [PubMed] [Google Scholar]
- 38.De Boer J, Hoeijmakers JH. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000;21(3):453–460. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- 39.Daya-Grosjean L, Sarasin A. The role of UV induced lesions in skin carcinogenesis: an overview of oncogene and tumor suppressor gene modifications in xeroderma pigmentosum skintumors. Mut Res Fundam Mol Mech Mutagen. 2005;571(1-2):43–56. doi: 10.1016/j.mrfmmm.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 40.Völler D, Ott C, Bosserhoff A (2013) MicroRNAs in malignant melanoma. Clinic Biochem. doi:10.1016/j.clinbiochem.2013.01.008 [DOI] [PubMed]
- 41.Mueller DW, Bosserhoff AK. Role of miRNAs in the progression of malignant melanoma. Br J Cancer. 2009;101(4):551–556. doi: 10.1038/sj.bjc.6605204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Felicetti F, Errico MC, Segnalini P, Mattia G, Carè A. MicroRNA-221 and-222 pathway controls melanoma progression. Expert Rev Anticancer Ther. 2008;8(11):1759–1765. doi: 10.1586/14737140.8.11.1759. [DOI] [PubMed] [Google Scholar]
- 43.Penna E, Orso F, Cimino D, Tenaglia E, Lembo A, Quaglino E, Poliseno L, Haimovic A, Osella-Abate S, De Pittà C, Pinatel E, Stadler MB, Provero P, Bernengo MG, Osman I, Taverna D. microRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J. 2011;30(10):1990–2007. doi: 10.1038/emboj.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyle GM, Woods SL, Bonazzi VF, Stark MS, Hacker E, Aoude LG, Dutton-Regester K, Cook AL, Sturm RA, Hayward NK. Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Res. 2011;24(3):525–537. doi: 10.1111/j.1755-148X.2011.00849.x. [DOI] [PubMed] [Google Scholar]
- 45.Levy C, Khaled M, Iliopoulos D, Janas MM, Schubert S, Pinner S, Chen PH, Li S, Fletcher AL, Yokoyama S, Scott KL, Garraway LA, Song JS, Granter SR, Turley SJ, Fisher DE, Novina CD. Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol Cell. 2010;40(5):841–849. doi: 10.1016/j.molcel.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Müller DW, Bosserhoff AK. Integrin β3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene. 2008;27(52):6698–6706. doi: 10.1038/onc.2008.282. [DOI] [PubMed] [Google Scholar]
- 47.Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18(5):549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- 48.Chen J, Feilotter HE, Paré GC, Zhang X, Pemberton JG, Garady C, Lai D, Yang X, Tron VA. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am J pathol. 2010;176(5):2520–2529. doi: 10.2353/ajpath.2010.091061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dar AA, Majid S, de Semir D, Nosrati M, Bezrookove V, Kashani-Sabet M. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem. 2011;286(19):16606–16614. doi: 10.1074/jbc.M111.227611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61(7):1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 51.Nishimura EK, Jordan SA, Oshima H, Yoshida H, Osawa M, Moriyama M, Jackson IJ, Barrandon Y, Miyachi Y, Nishikawa SI. Dominant role of the niche in melanocyte stem-cell fate determination. Nature. 2002;416(6883):854–860. doi: 10.1038/416854a. [DOI] [PubMed] [Google Scholar]
- 52.Nishimura EK, Granter SR, Fisher DE. Mechanisms of hair graying: incomplete melanocyte stem cell maintenance in the niche. Science. 2005;307(5710):720–724. doi: 10.1126/science.1099593. [DOI] [PubMed] [Google Scholar]
- 53.Steingrímsson E, Copeland NG, Jenkins NA. Melanocyte stem cell maintenance and hair graying. Cell. 2005;121(1):9–12. doi: 10.1016/j.cell.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 54.Sarin KY, Artandi SE. Aging, graying, and loss of melanocyte stem cells. Stem Cell Rev. 2007;3(3):212–217. doi: 10.1007/s12015-007-0028-0. [DOI] [PubMed] [Google Scholar]
- 55.Moriyama M, Osawa M, Mak SS, Ohtsuka T, Yamamoto N, Han H, Delmas V, Kageyama R, Beermann F, Larue L, Nishikawa SI. Notch signaling via Hes1 transcription factor maintains survival of melanoblasts and melanocyte stem cells. J Cell Biol. 2006;173(3):333–339. doi: 10.1083/jcb.200509084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheli Y, Guiliano S, Botton T, Rocchi S, Hofman V, Hofman P, Hofman V, Hofman P, Bahadoran P, Bertolotto C, Ballotti R. Mitf is the key molecular switch between mouse or human melanoma-initiating cells and their differentiated progeny. Oncogene. 2011;30(20):2307–2318. doi: 10.1038/onc.2010.598. [DOI] [PubMed] [Google Scholar]
- 57.Lang D, Lu MM, Huang L, Engleka KA, Zhang M, Chu EY, Lipner S, Skoultchi A, Millar SE, Epstein JA. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature. 2005;433(7028):884–887. doi: 10.1038/nature03292. [DOI] [PubMed] [Google Scholar]
- 58.Schepsky A, Bruser K, Gunnarsson GJ, Goodall J, Hallsson JH, Goding CR, Steingrimsson E, Hecht A. The microphthalmia-associated transcription factor Mitf interacts with β-catenin to determine target gene expression. Mol Cell Biol. 2006;26(23):8914–8927. doi: 10.1128/MCB.02299-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takeda K, Yasumoto KI, Takada R, Takada S, Watanabe KI, Udono T, Saito H, Takahashi K, Shibahara S. Induction of melanocyte-specific microphthalmia-associated transcription factor by Wnt-3a. J Biol Chem. 2000;275(19):14013–14016. doi: 10.1074/jbc.c000113200. [DOI] [PubMed] [Google Scholar]
- 60.Mak SS, Moriyama M, Nishioka E, Osawa M, Nishikawa SI. Indispensable role of Bcl2 in the development of the melanocyte stem cell. Dev Biol. 2006;291(1):144–153. doi: 10.1016/j.ydbio.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 61.Vance KW, Goding CR. The transcription network regulating melanocyte development and melanoma. Pigment Cell Res. 2004;17(4):318–325. doi: 10.1111/j.1600-0749.2004.00164.x. [DOI] [PubMed] [Google Scholar]
- 62.Hoek KS, Goding CR. Cancer stem cells versus phenotype-switching in melanoma. Pigment cell Melanoma Res. 2010;23(6):746–759. doi: 10.1111/j.1755-148X.2010.00757.x. [DOI] [PubMed] [Google Scholar]
- 63.Haass NK, Smalley KS, Li L, Herlyn M. Adhesion, migration, and communication in melanocytes and melanoma. Pigment Cell Res. 2005;18(3):150–159. doi: 10.1111/j.1600-0749.2005.00235.x. [DOI] [PubMed] [Google Scholar]
- 64.Lee JT, Herlyn M. Microenvironmental influences in melanoma progression. J Cell Biochem. 2006;101(4):862–872. doi: 10.1002/jcb.21204. [DOI] [PubMed] [Google Scholar]
- 65.Hsu MY, Meier F, Herlyn M. Melanoma development and progression: a conspiracy between tumor and host. Differentiation. 2002;70(9–10):522–536. doi: 10.1046/j.1432-0436.2002.700906.x. [DOI] [PubMed] [Google Scholar]
- 66.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, Naber SP, Weinberg RA. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37(10):1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Kempen LC, Ruiter DJ, van Muijen GN, Coussens LM. The tumor microenvironment: a critical determinant of neoplastic evolution. Eur J Cell Biol. 2003;82(11):539–548. doi: 10.1078/0171-9335-00346. [DOI] [PubMed] [Google Scholar]
- 68.Norrby K. Mast cells and angiogenesis. Apmis. 2002;110(5):355–371. doi: 10.1034/j.1600-0463.2002.100501.x. [DOI] [PubMed] [Google Scholar]
- 69.Yong LCJ. The mast cell: origin, morphology, distribution, and function. Exp Toxicol Pathol. 1997;49(6):409–424. doi: 10.1016/S0940-2993(97)80129-7. [DOI] [PubMed] [Google Scholar]
- 70.Fang KC, Raymond WW, Blount JL, Caughey GH. Dog mast cell α-chymase activates progelatinase B by cleaving the Phe88-Gln89 and Phe91-Glu92 bonds of the catalytic domain. J Biol Chem. 1997;272(41):25628–25635. doi: 10.1074/jbc.272.41.25628. [DOI] [PubMed] [Google Scholar]
- 71.Braeuer RR, Zigler M, Villares GJ, Dobroff AS, Bar-Eli M. Transcriptional control of melanoma metastasis: the importance of the tumor microenvironment. Semin Cancer Biol. 2011;21(2):83–88. doi: 10.1016/j.semcancer.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 73.Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, Van Belle PA, Xu X, Elder DE, Herlyn M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65(20):9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- 74.Lin K, Baritaki S, Militello L, Malaponte G, Bevelacqua Y, Bonavida B. The role of B-RAF mutations in melanoma and the induction of EMT via dysregulation of the NF-κB/Snail/RKIP/PTEN circuit. Genes Cancer. 2010;1(5):409–420. doi: 10.1177/1947601910373795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weiss MB, Abel EV, Mayberry MM, Basile KJ, Berger AC, Aplin AE. TWIST1 Is an ERK1/2 effector that promotes invasion and regulates MMP-1 expression in human melanoma cells. Cancer Res. 2012;72(24):6382–6392. doi: 10.1158/0008-5472.CAN-12-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, Naber SP, Weinberg RA. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37(10):1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fenouille N, Tichet M, Dufies M, Pottier A, Mogha A, Soo JK, Rocchi S, Tartare-Deckert S. The epithelial-mesenchymal transition (EMT) regulatory factor SLUG (SNAI2) is a downstream target of SPARC and AKT in promoting melanoma cell invasion. PLoS One. 2012;7(7):e40378. doi: 10.1371/journal.pone.0040378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frank NY, Margaryan A, Huang Y, Schatton T, Waaga-Gasser AM, Gasser M, Sayegh MH, Sadee W, Frank MH. ABCB5-mediated doxorubicin transport and chemoresistance in human malignant melanoma. Cancer Res. 2005;65(10):4320–4333. doi: 10.1158/0008-5472.CAN-04-3327. [DOI] [PubMed] [Google Scholar]
- 79.Monzani E, Facchetti F, Galmozzi E, Corsini E, Benetti A, Cavazzin C, Gritti A, Piccinini A, Porro D, Santinami M, Invernici G, Parati E, Alessandri G, La Porta CA. Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur J Cancer. 2007;43(5):935–946. doi: 10.1016/j.ejca.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 80.Topczewska JM, Postovit LM, Margaryan NV, Sam A, Hess AR, Wheaton WW, Nickoloff BJ, Topczewski J, Hendrix MJ. Embryonic and tumorigenic pathways converge via Nodal signaling: role in melanoma aggressiveness. Nat Med. 2006;12(8):925–932. doi: 10.1038/nm1448. [DOI] [PubMed] [Google Scholar]
- 81.Grichnik JM, Ali WN, Burch JA, Byers JD, Garcia CA, Clark RE, Shea CR. KIT expression reveals a population of precursor melanocytes in human skin. J Invest Dermatol. 1996;106(5):967–971. doi: 10.1111/1523-1747.ep12338471. [DOI] [PubMed] [Google Scholar]
- 82.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2(1):48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 83.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5(8):615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 84.Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, Van Belle PA, Xu X, Elder DE, Herlyn M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65(20):9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- 85.Monzani E, Facchetti F, Galmozzi E, Corsini E, Benetti A, Cavazzin C, Gritti A, Piccinini A, Porro D, Santinami M, Invernici G, Parati E, Alessandri G, La Porta CA. Melanoma contains CD133- and ABCG2-positive cells with enhanced tumourigenic potential. Eur J Cancer. 2007;43(5):935–946. doi: 10.1016/j.ejca.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 86.Shmelkov SV, Butler JM, Hooper AT, Hormigo A, Kushner J, Milde T, St Clair R, Baljevic M, White I, Jin DK, Chadburn A, Murphy AJ, Valenzuela DM, Gale NW, Thurston G, Yancopoulos GD, D’Angelica M, Kemeny N, Lyden D, Rafii S. CD133 expression is not restricted to stem cells, and both CD133+ and CD133− metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118(6):2111. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.La Porta C. Cancer stem cells: lessons from melanoma. Stem Cell Rev. 2009;5(1):61–65. doi: 10.1007/s12015-008-9048-7. [DOI] [PubMed] [Google Scholar]
- 88.Akers SN, Odunsi K, Karpf AR. Regulation of cancer germline antigen gene expression: implications for cancer immunotherapy. Future Oncol. 2010;6(5):717–770. doi: 10.2217/fon.10.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martelange V, De Smet C, De Plaen E, Lurquin C, Boon T. Identification on a human sarcoma of two new genes with tumor-specific expression. Cancer Res. 2000;60:3848–3855. [PubMed] [Google Scholar]
- 90.Caruthers JM, McKay DB. Helicase structure and mechanism. Curr Opin Struct Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 91.Silverman E, Edwalds-Gilbert G, Lin RJ. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene. 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- 92.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 94.Linder P. Dead-box proteins. Family affair—active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Linder P, Jankowsky E. From unwinding to clamping—the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 96.Camats M, Guil S, Kokolo M, Bach-Elias M. p68 RNA helicase (DDX5) alters activity of cis- and trans-acting factors of the alternative splicing of H-Ras. PLoS One. 2008;3:2926. doi: 10.1371/journal.pone.0002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Causevic M, Hislop RG, Kernohan NM, Carey FA, Kay RA, Steele RJ, Fuller-Pace FV. Overexpression and poly-ubiquitylation of the DEAD-box RNA helicase p68 in colorectal tumours. Oncogene. 2001;20:7734–7743. doi: 10.1038/sj.onc.1204976. [DOI] [PubMed] [Google Scholar]
- 98.Shim H, Shim E, Lee H, Hahn J, Kang D, Lee YS, Jeoung D. CAGE, a novel cancer/testis antigen gene, promotes cell motility by activation ERK and p38 MAPK and downregulating ROS. Mol Cells. 2006;21(3):367. [PubMed] [Google Scholar]
- 99.Clark EL, Coulson A, Dalgliesh C, Rajan P, Nicol SM, Fleming S, Heer R, Gaughan L, Leung HY, Elliott DJ, Fuller-Pace FV, Robson CN. The RNA helicase p68 is a novel androgen receptor co-activator involved in splicing and is overexpressed in prostate cancer. Cancer Res. 2008;6:7938–7946. doi: 10.1158/0008-5472.CAN-08-0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim Y, Park H, Park D, Lee YS, Choe J, Hahn JH, Lee H, Kim YM, Jeoung D. Cancer/testis antigen CAGE exerts negative regulation on p53 expression through HDAC2 and confers resistance to anti-cancer drugs. J Biol Chem. 2010;285:25957–25968. doi: 10.1074/jbc.M109.095950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fuller-Pace FV, Moore HC. RNA helicases p68 and p72: multifunctional proteins with important implications for cancer development. Future Oncol. 2011;7:239–251. doi: 10.2217/fon.11.1. [DOI] [PubMed] [Google Scholar]
- 102.Por E, Byun HJ, Lee EJ, Lim JH, Jung SY, Park I, Kim YM, Jeoung DI, Lee H. The cancer/testis antigen CAGE with oncogenic potential stimulates cell proliferation by up-regulating cyclins D1 and E in an AP-1- and E2F-dependent manner. J Biol Chem. 2010;285:14475–14485. doi: 10.1074/jbc.M109.084400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim Y, Jeoung D. Role of CAGE, a novel cancer/testis antigen, in various cellular processes, including tumorigenesis, cytolytic T lymphocyte induction, and cell motility. J Microbiol Biotechnol. 2008;18:600–610. [PubMed] [Google Scholar]
- 104.Liggins AP, Lim SH, Soilleux EJ, Pulford K, Banham AH. A panel of cancer-testis genes exhibiting broad-spectrum expression in haematological malignancies. Cancer Immun. 2010;10:8. [PMC free article] [PubMed] [Google Scholar]
- 105.Mathieu MG, Linley AJ, Reeder SP, Badoual C, Tartour E, Rees RC, McArdle SE. HAGE, a cancer/testis antigen expressed at the protein level in a variety of cancers. Cancer Immun. 2010;10:2. [PMC free article] [PubMed] [Google Scholar]
- 106.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 107.Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. Understanding Ras: “it ain’t over’til it’s over”. Trends Cell Biol. 2000;10:147–154. doi: 10.1016/s0962-8924(00)01740-2. [DOI] [PubMed] [Google Scholar]
- 108.Castellano E, Downward J. Role of RAS in the regulation of PI3-kinase. Curr Top Microbiol Immunol. 2010;346:143–169. doi: 10.1007/82_2010_56. [DOI] [PubMed] [Google Scholar]
- 109.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Castillejo JA, Navarro G, San Jose-Eneriz E, Garate L, Cordeu L, Cervantes F, Prosper F, Heiniger A, Torres A. Epigenetic regulation of human cancer/testis antigen gene, HAGE, in chronic myeloid leukemia. Haematologica. 2007;92(2):153–162. doi: 10.3324/haematol.10782. [DOI] [PubMed] [Google Scholar]
- 110.Chen Q, Lin J, Yao DM, Qian J, Qian W, Yang J, Chai HY, Ma JC, Deng ZQ, Wang CZ, Li Y. Aberrant hypomethylation of DDX43 promoter in myelodysplastic syndrome. Br J Haematol. 2012;158(2):293–296. doi: 10.1111/j.1365-2141.2012.09138.x. [DOI] [PubMed] [Google Scholar]
- 111.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, Jouary T, Schadendorf D, Ribas A, O’Day SJ, Sosman JA, Kirkwood JM, Eggermont AM, Dreno B, Nolop K, Li J, Nelson B, Hou J, Lee RJ, Flaherty KT, McArthur GA. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, Hamid O, Schuchter L, Cebon J, Ibrahim N, Kudchadkar R, Burris HA, Falchook G, Algazi A, Lewis K, Long GV, Puzanov I, Lebowitz P, Singh A, Little S, Sun P, Allred A, Ouellet D, Kim KB, Patel K, Weber J. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367(18):1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]