Abstract
Transgenic animal technology is one of the fastest growing biotechnology areas. It is used to integrate exogenous genes into the animal genome by genetic engineering technology so that these genes can be inherited and expressed by offspring. The transgenic efficiency and precise control of gene expression are the key limiting factors in the production of transgenic animals. A variety of transgenic technologies are available. Each has its own advantages and disadvantages and needs further study because of unresolved technical and safety issues. Further studies will allow transgenic technology to explore gene function, animal genetic improvement, bioreactors, animal disease models, and organ transplantation. This article reviews the recently developed animal transgenic technologies, including the germ line stem cell-mediated method to improve efficiency, gene targeting to improve accuracy, RNA interference-mediated gene silencing technology, zinc-finger nuclease gene targeting technology and induced pluripotent stem cell technology. These new transgenic techniques can provide a better platform to develop transgenic animals for breeding new animal varieties and promote the development of medical sciences, livestock production, and other fields.
Keywords: Transgenic technologies, Gene targeting, RNA interference, Induced pluripotent stem cells
Introduction
Transgenic technology first emerged in 1974 by microinjection into blastocysts with viral DNA to produce transgenic mice [1]. Gordon et al. [2] first described the introduction of a foreign gene into mice using pronuclear injection into oocytes, an approach which had since been widely employed to study the molecular and cellular functions of many genes. Since then, transgenic methods, such as microinjection [3], sperm vector [4, 5], ICSI-mediated transgenesis [6–8], transfection-mediated transgenesis of male germ stem cell in vitro [9] and in vivo [10, 11], as well as female germ stem cell [12, 13], transgenic by somatic cell nuclear transfer (SCNT) [14–16], lentiviral vectors [17–19] and primordial germ cells (PGCs) [20, 21], have been used successfully to produce transgenic mice and other transgenic animal species such as rats, fish, pigs, sheep, cattle, rabbits, dogs, chickens, and monkey. As new technologies and techniques are developing, transgenic animal technologies will also be improved. The accurate modulation of gene expression will likely come after the improvement of efficiency of transgenic animal production by means of the transgenic technologies mediated by germ line stem cells [9], site-specific integration of gene targeting against embryonic stem cells and somatic cells [22–24], gene silencing by RNA interference [25–27], zinc-finger nuclease gene targeting [28], and pluripotent stem cell induction [29, 30]. It is likely that many more innovative transgenic technologies will significantly broaden the application of transgenic animals, such as the production of bioreactors for increasing the production of domestic animals and improving the quality of meat and milk, production of disease-resistant animals, production of animal organs for human use, and establishment of disease models. These technologies may produce many economic benefits.
Germline stem cell technique
Spermatogonial stem cell technique
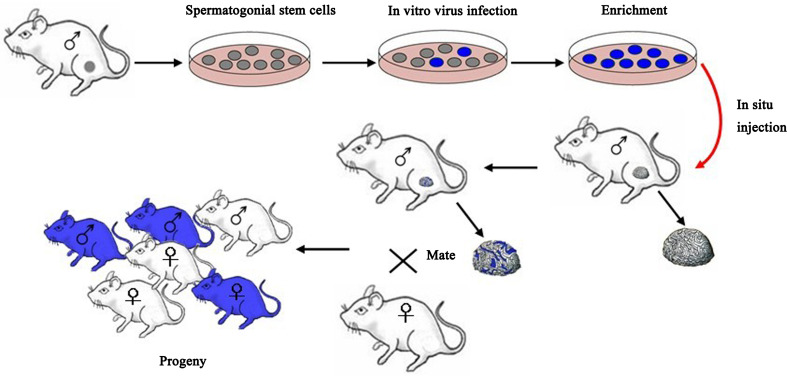
Stem cells in the male germ line (spermatogonial stem cells [SSCs]) are an important target for male fertility restoration and germ line gene modification. Spermatogonial stem cells are a population of cells in mammalian testes that have the potential to self-renew and differentiate, similarly to embryonic stem cells. SSCs transplantation is a recently developed animal reproduction technique that involves the injection of in vitro cultured spermatogonial stem cells from an age-matched male donor into the seminiferous tubule of age-matched host animal to produce germ cells. During the in vitro culture, positive spermatogonial stem cells which will be transferred can be screened and, thus, the transgenic efficacy can be significantly increased [31] (Fig. 1).
Fig. 1.
Generation of transgenic animals using spermatogonial stem cells. The testis from a fertile male is digested to generate a single-cell suspension. The SSCs can be cultured, transfected, and then microinjected into the lumen of seminiferous tubules of an infertile recipient mouse. Only a spermatogonial stem cell can generate a colony of spermatogenesis in the recipient testis. Mating the recipient male to a wild-type female produces progeny that carry foreign genes
Brinster et al. (1994) first established the SSCs transplantation technique and succeeded in spermatogenesis of donor mouse SSCs and monoploid reproductive genetics in the recipient [32]. Using retroviruses, Nagano et al. (2001) transfected mouse SSCs in vitro with a transfection efficiency of 2–20 % and found that if transfected SSCs were transplanted into seminiferous tubule of recipient mice, 4.5 % of offspring were stable transgenic mice and expressed extrinsic genes [33]. This study suggested that exogenous genes could be stably integrated into the stem cell genome. Kanatsu-Shinohara et al. (2006) demonstrate successful gene trapping and homologous recombination in spermatogonial stem cells. Cultured spermatogonial stem cells were transfected with gene trap or gene targeting vectors. Using this strategy, the efficiency of homologous recombination for the occludin gene locus was 1.7 % using a nonisogenic DNA construct. Their results demonstrate the feasibility of altering genes in tissue-specific stem cells in a manner similar to embryonic stem cells and have important implications for gene therapy and animal transgenesis [34]. Kanatsu-Shinohara et al. (2008) attempted to use a lentiviral vector carrying enhanced green fluorescent protein (EGFP) to transduce rat SSCs cultured in vitro and then transplanted these SSCs into seminiferous tubule of immunodeficient mice. This study found that the transduced rat SSCs could produce spermatogenic cells expressing EGFP, and sperm cells were capable of producing transgenic offspring [35]. Adeno-associated virus (AAV)-transduced germ cells from goats were transplanted to recipient males in which endogenous germ cells had been depleted by fractionated testicular irradiation. Transgenic germ cells colonized recipient testes and produced transgenic sperm. When semen was used for in vitro fertilization (IVF), 10 % of embryos were transgenic [36]. This study was the first attempt to produce large transgenic domestic animal embryos via SSCs transplantation and is encouraging for the production of large transgenic animals. We developed a convenient, economic, and efficient method for gene transfer by transfection of male spermatogonial stem cells. The pIRES2-EGFP-Thanatin plasmid was injected into the testes of male mice by a minimally invasive operation. Among those 52 F1 mice produced, 38.46 % were found to be positive for the Thanatin gene by PCR and 30.77 % by Southern blotting. Of an F2 generation, 36.36 % was found to be positive. The study provides a useful method for the future development of disease-resistant animals and production of antibacterial peptides through transgenic animals [37].
Disrupting genes in the rat on a genome-wide scale will allow the investigation of many biological processes linked to human health. Transposon-mediated mutagenesis was used to knock out genes in rat spermatogonial stem cells [38]. This approach paves the way for high-throughput functional genomics studies in the laboratory rat. Recently, mutant alleles in rat SSCs were generated using Sleeping Beauty (SB) transposon-mediated insertional mutagenesis [39]. This simple, economic and user-friendly methodological pipeline enables screens for functional gene annotation in the rat, with applicability in other vertebrate models where germ line-competent stem cells have been established.
With the improvement of culture systems, screening, and transplantation procedures, transgenic animal efficacy will also likely be improved.
Primordial germ cell technique
Primordial germ cells refer to the ancestral cells that can develop into sperm or ovum cells. PGCs reside in the recipient’s gonads, migrate and proliferate in the recipient embryonic gonads. Because PGCs at different stages of development can serve as transgenic recipient cells [40], transgenic studies using PGCs as vector are simple and highly effective, and will likely gain favor for the production of transgenic animals.
The LacZ gene was introduced into PGCs from early chicken embryonic blood using liposome methods and these cells were injected into recipient embryos. Fifty-three chicken embryos were successfully developed until the third day and expressed the LacZ gene in the genital crest [41]. PGCs carrying the GFP gene were injected into the 3-day chicken embryo. Chickens carrying an extrinsic gene and GFP in the sexual glands were obtained [42]. Using a high number of 5.5-day PGCs infected with a lentiviral vector, transgenic chimeras (G(0)) with an acceptable efficiency for germline transmission were obtained. G(0) female chickens produced transgenic progeny (G(1)) with higher efficiency compared to G(0) male chickens. In G(1) transgenic chickens obtained by this method, EGFP was effectively expressed under the control of the actin promoter [43]. In addition, isolated PGCs from transgenic pigs had the capacity of integration and it was possible to transfer genes using PGCs into pigs or other large domestic animals [44]. Up to now, numerous attempts to isolate and culture pluripotent stem cells in domestic animals have been reported [45], which have the potential of such cells for clinical applications.
Using PGCs to produce transgenic animals is a new technique developed because of its advantages in excellent operability, high production, and an induced germline modification [46–48]. In combination with gene targeting technology, it will significantly enhance transgenic efficacy and accuracy, and has broad application in transgenic animal research. However, exactly how to optimize these techniques to enhance transgenic efficacy in transgenic domestic animals still needs to be further explored.
Gene targeting
Gene targeting is a technology that specifically modifies a particular gene in the chromosome through homologous recombination and integrates extrinsic gene into the specific target site. Gene targeting technology overcomes random integration events and is therefore ideal for the modification and reconstruction of biological genetic materials.
Embryonic stem cell gene targeting
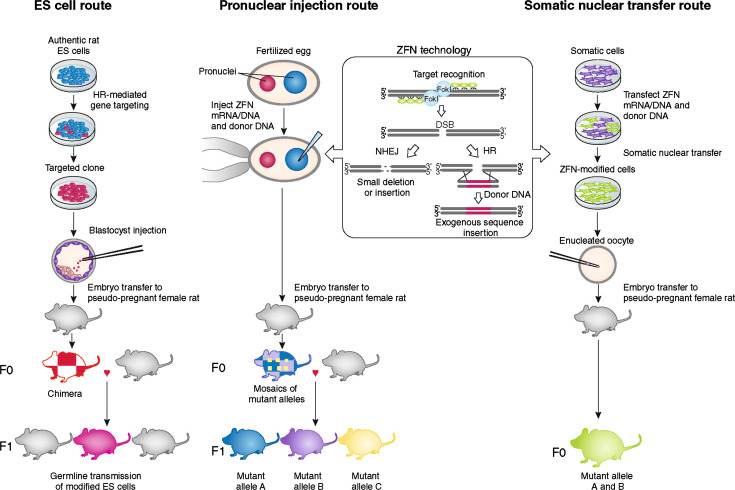
Embryonic stem cell (ESC) gene targeting utilizes the totipotency of embryonic stem cells to introduce modified extrinsic genes into ESCs that can be injected into animal blastula/blastocyst. The ESCs will participate in the formation of host embryo, construct chimera until species chimerism, and transmit the extrinsic gene to the next generation and produce transgenic animals (Fig. 2) [49]. A mouse model with hypoxanthine–guanine phosphoribosyltransferase (HPRT) knockout was established using ESC gene targeting technology [50]. Through gene targeting, current techniques can not only knock out specific genes but also transfer extrinsic genes into the animal genome to obtain gene knock-in animals. The 2007 Nobel Prize in physiology or medicine was awarded to Capecchi, Evans, and Smithies for their series of discoveries in introducing ESC gene targeting in mice. Okada et al. (2009) used an integration-deficient lentiviral vector to reduce the random integration probability and evaluated the practicability of gene targeting in mouse ESCs mediated by chronic viral vector. The success of the development of mouse chimeras suggested the totipotency of homologous recombined ESCs and subsequent migration of the reproductive system [51]. Transgenic, knockin, and knockout gene manipulations are available in mouse ESCs, enabling the production of genetically modified animals. Rats have important advantages over mice as an experimental system for physiological and pharmacological investigations. However, the production of genetically modified rats has been difficult due to problems in rat ESCs after gene introduction [52]. Gene-targeting technology via homologous recombination in rat ES cells demonstrated its use through production of a protease-activated receptor-2 gene (Par-2) KO rat [53]. Targeted disruption of myostatin (mstn) gene in yellow catfish was performed using engineered zinc-finger nucleases (ZFNs), and six mstn nju6/+ and 14 mstn nju7/+ offspring were obtained [54]. p53 (also known as Tp53) gene knockout rats were generated by homologous recombination in embryonic stem (ES) cells. The entire process requires ∼1 year to complete, from derivation of ES cells to generation of knockout rats [55, 56].
Fig. 2.
The three principal routes of ES cell, pronuclear injection, and somatic cell gene targeting technology to achieve targeted genome modifications in the rat and other mammalian species. ES cell embryonic stem cell, HR homologous recombination, ZFN zinc-finger nuclease, DSB double-strand break, NHEJ non-homologous end joining
ES cells provide a system for studying the whole cellular population. By the utilization of ES cells as carrier and the modification of ES cells in vitro, all issues including the numbers, the loci, the expression extent of integrated genes, the stability of inserted genes, and the screening of genes can be finished on cellular level. Currently, homologous recombination in ES cells becomes one common technique to modify any gene site in the mouse chromosome and is applied to studying gene function and disease models. However, ES cells from domestic animals have not been obtained for gene targeting so far, which thereby limits the application of gene targeting for production such as mammary gland bioreactor.
Somatic cell gene targeting
After the successful development of cloning technology for somatic cells, scientists combined gene targeting that utilized the ES cells with somatic cell nuclear transfer to produce transgenic animals, which is an alternative to current techniques.
McCreath et al. (2000) described the efficient and reproducible gene targeting in fetal fibroblast cells to place the therapeutic AATC2 transgene that encodes the recombinant human α1-antitrypsin (rhAAT) at the ovine α1(I) procollagen (COL1A1) locus and the production of live sheep by nuclear transfer. Recombinant human α1-antitrypsin was detected at a concentration of 650 μg ml−1 in milk samples. This is the first production of gene-targeted sheep by nuclear transfer from cultured somatic cells [16].
Kuroiwa et al. [57] used the combination of gene targeting and somatic cell nuclear transfer to produce cattle free of bovine spongiform encephalopathy (BSE), Wall et al. [58] used it to produce cows without mastitis, and Lai et al. [59] used it to produce pigs that were able to synthesize polyunsaturated fatty acids. The presence of recombinant human butyrylcholinesterase in goat milk can reach 1–5 g/l [60]. Since there is a low insertion efficiency of extrinsic DNA into specific gene sites mediated by homologous DNA sequence or gene targeting in mammalian somatic cells, Bertolini et al. (2009) studied the reductive effect of gene targeting on the non-homologous end junction (NHEJ) protein level. This study found that through RNA interference (RNAi) in human colon cancer HCT116 cells where the integrated NHEJ proteins Ku70 and Xrcc4 were transiently knocked-down, the ratio of gene targeting and random insertion was significantly influenced. Timely, the transfection of targeting vector based on HPRT gene after RNAi can reduce the rate of random integration by 70 % and increase the targeting of HPRT gene 33-fold. These results indicated that the transient repression of NHEJ protein by RNAi is an effective pathway to increase gene targeting efficiency [61]. By combining RNAi technology with SCNT method, one live-born transgenic calf with knocked down gene encoding bovine pathogenic (multimer) prion protein (bPrPSc/bPRNP) was produced [62], and using fetal liver-derived cells to perform gene targeting, the transgenic homozygous pig displaying biallelic (double-copy) knockout of α-1,3-galactosyltransferase (α-1,3-GT/GGTA1) gene was created [63]. This technology will also find applications in a multitude of other species, which hitherto have required somatic cell reprogramming to achieve directed modifications of their genomes. Zinc-finger nucleases (ZFNs) have been shown to stimulate gene targeting in a variety of species and can therefore be used in combination with somatic nuclear transfer, removing a bottleneck in achieving directed modification by this route (Fig. 2) [49]. The promise of this technology will stimulate numerous applications.
The somatic cell gene targeting technique avoids the requirement of ESCs for stem cell gene targeting, directly recombines the homologous genes in somatic cells, screens targeted cells in vitro and produces transgenic animals through nuclear transfer. However, unlike ES cells that retain the capacity to enter the reproductive system after targeting modification, screening, and amplification, somatic cells for cloning domestic animals have a limited lifespan in vitro, although some of them retain totipotency after gene targeting. These aged cells reduce the efficacy of gene targeting, resulting in a high frequency of abnormalities in cloned embryos, fetuses, and offspring, which is the main limitation of gene targeting in somatic cell cloning of mammals. Moreover, in many species, especially large farm animals, ES cells have not been successfully isolated or cultured. The elucidation and solving of these problems, and thereby improvement in the technology of somatic cell nuclear transfer (SCNT), could allow increasing the practical application of SCNT to the generation of transgenic animals for different biomedical, veterinary, and agricultural purposes. It is reasonable that the gene targeting efficacy will be enhanced following the development of new techniques, the passage of targeting cell, and the resolution of targeting and nuclear transplantation issues.
Conditional gene targeting
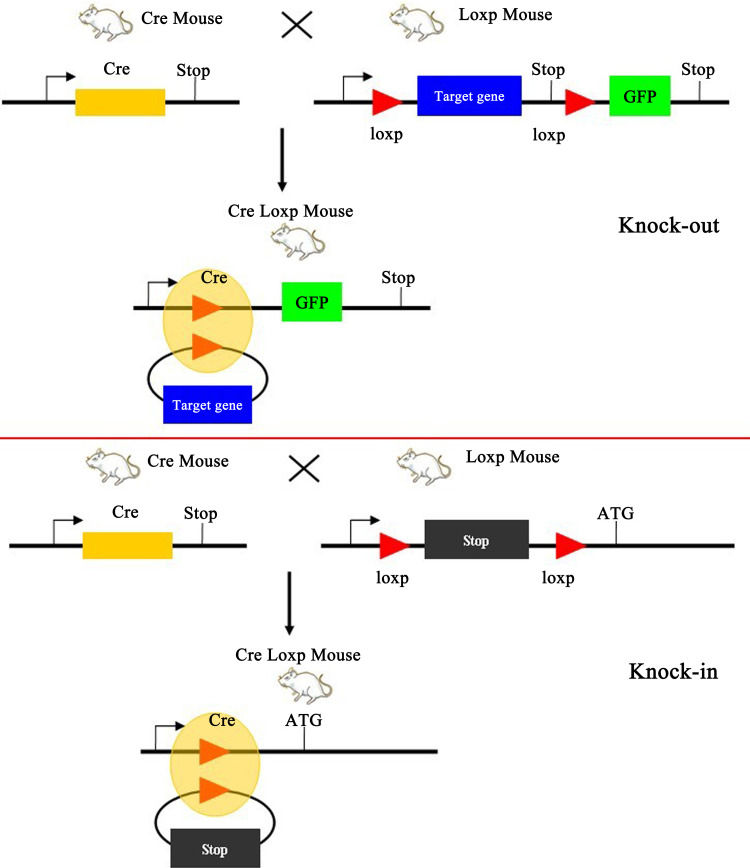
The randomness of extrinsic gene integration into genome makes the expression of inserted gene uncontrollable under some conditions. However, the practical operation usually requires the target gene to be expressed in specific tissues, cellular types, or at special stages during the development. Therefore, the spatiotemporal control of extrinsic genes is an important problem that needs to be resolved. Conditional gene targeting is one strategy and has significant applicable values. This technique is based on the Cre recombinase/locus of X-over P1 (Cre/LoxP) system to achieve the spatiotemporal specificity in gene targeting (Fig. 3).
Fig. 3.
Diagrammatic representation for conditional gene targeting by Cre-loxp recombination. Two different strategies for production of knock-out and knock-in transgenic mice using Cre-loxp recombination system. The Cre-loxp system could be used to delete loxP-flanked DNA and to control site-specific recombination events in genomic DNA
The conditional gene targeting system consists of Cre recombinase and loxP site [64–68]. Cre recombinase can recognize the LoxP site, and cut and replace the DNA segment between the two LoxP sites. The Cre/LoxP system can produce a conditional gene knockout, i.e., two LoxP sequences in the same direction are added to the ends of the target gene, respectively, during the construction of the target vector and then transgenic mouse is produced by gene targeting. At the same time, another transgenic mice strain carrying the Cre recombinase gene is produced. Finally, these two mice are mated and transgenic mice carrying the two sets of genes will be produced. The Cre recombinase gene can be fused with an inducible promoter or ligand binding domain (LBD) of the steroid hormone receptor, matched with suitable tissue-specific promoter, and be activated conditionally after birth; removal of the segment between two LoxP sites can achieve a local knockout of the target gene in specific tissues or organs [69]. In addition, using the Cre/LoxP can also produce conditional gene repair. For example, two LoxP sites can be inserted into a functional gene and the Cre recombinase can be activated by drug induction according to the time requirement and recover the function of the target gene by removal of the segment between the LoxP sites [70].
The Cre/LoxP system with embryonic stem cells was combined to introduce a single-copy gene into a specific gene site and strengthen the expression of the introduced gene controlled by an intrinsic modulating component. Because of the limited cellular absorption of the Cre enzyme, the application of the Cre/LoxP system is limited; therefore, the improvement of cellular absorption efficiency becomes an important component of the application of Cre/LoxP system [71]. The fusion of a nuclear localization signal, Cre recombinase and TAT peptide can enhance the absorption of Cre by fibroblast in mouse embryonic stem cells by 95 % [72].
Taken together, this system guarantees spatial specificity by utilizing tissue-specific promoters to express transgenes and the temporal controllability by utilizing drug induction system. Therefore, this system achieves the accurate spatiotemporal modulation of extrinsic genes and the artificial control of expression. This technique can be used to establish a transgenic modulation system where the spatiotemporal expression can be controlled and is likely to have significant value for the research of gene function.
Gene silencing mediated by RNA interference
RNA interference (RNAi) is the silencing of specific gene expression mediated by the formation of double-stranded RNA that results in the inhibition of gene expression by degrading mRNA [73]. Therefore, RNAi can achieve the spatiotemporality and reversibility of gene expression modulation.
Transgenically supplied siRNA can silence ubiquitously expressed enhanced green fluorescent protein in every part of the mouse and rat body [74]. The results suggest that transgenic RNAi could function as an alternative method of gene silencing by applying homologous recombination to embryonic stem (ES) cells, and should be successful even in species where ES cell lines remain unestablished. Gene targeting via homologous recombination in murine embryonic stem (ES) cells has been the method of choice for deciphering mammalian gene function in vivo. Despite improvements in this technology, it still remains a laborious method. Advances in RNA interference (RNAi) technology have provided a rapid loss-of-function method for assessing gene function in a number of organisms. Because of the dominant nature of the knockdown, embryonic phenotypes could be directly assessed in embryos completely derived from ES cells by the tetraploid aggregation method. Such embryos, in which endogenous p120-Ras GTPase-activating protein (RasGAP), Rasa1 gene (also known as RasGAP), was silenced, had the same phenotype as did the previously reported Rasa1 null mutation [75].
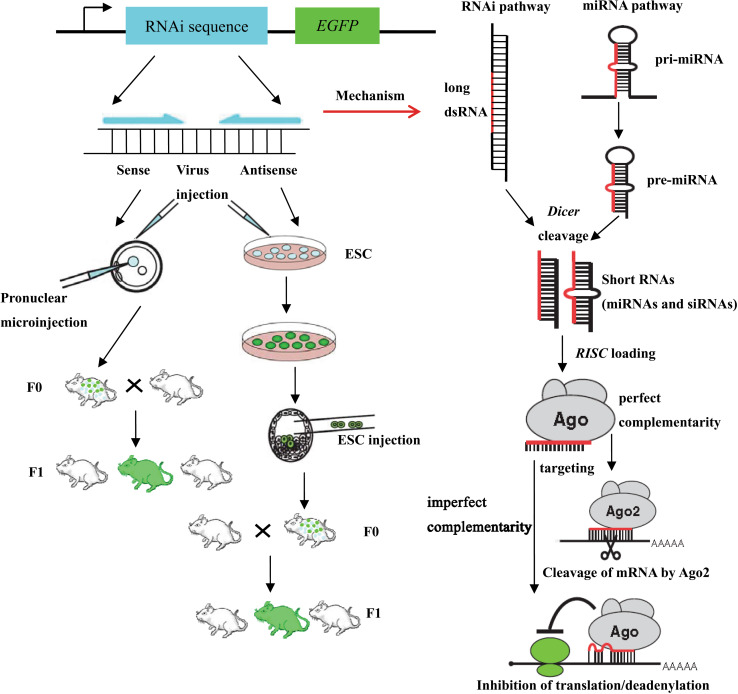
RNA interference (RNAi)-mediated gene knockdown has developed into a routine method to assess gene function in cultured mammalian cells in a fast and easy manner. For the use of RNAi in mice, short hairpin (sh) RNAs expressed stably from the genome are a fast alternative to conventional knockout approaches (Fig. 4). Complete or conditional gene knockdown (the Cre/loxP system) in mice is used to activate RNAi in a time- and tissue-dependent manner. The site-specific insertion of single-copy shRNA vectors allows the expedite and reproducible production of knockdown mice and provides an easy and fast approach to assess gene function in vivo [76].
Fig. 4.
A schematic diagram of RNAi and miRNA pathways and strategies for RNAi-induced gene silencing in living mammals. Mammalian RNAi and miRNA pathways use different RNA substrates processed by the same Dicer protein into short RNAs. These processed RNAs are incorporated into the RNA-induced silencing complex (RISC), which targets messenger RNA to prevent translation. RNAi in animals can be induced from a transgene. Transgenic animals with RNAi knockdown are usually produced by one of the three depicted strategies (pronuclear injection, viral infection, and ESC route). Different strategies for delivering an RNAi agent are available to induce RNAi in adult animals
Spatiotemporal RNAi techniques have been employed to achieve the control of RNAi by controlling transcription of the interfering gene. Transgenic mice were produced by transferring the combined tetracycline-responsive element (TRE)-tight promoter and RNAi genes. Afterwards, they were crossed with another transgenic mouse carrying recombinant tetracycline-controlled transcription factor (rtTA), which is also called a tetracycline-dependent transcriptional activator/transactivator (tTA) protein. The offspring of this pairing produced transgenic mice containing the TRE, RNAi gene, and tTA. The triple transgenic mice were administered tetracycline to activate the tTA system to bind with the promoter and TRE, and ultimately activate RNAi [77]. Highly effective small interfering RNA (siRNA) corresponding to the high conservative region of porcine endogenous retrovirus (PERV) DNA was selected to inhibit the expression of all PERV subtypes in human somatic cells infected with PERV and the original donor cells from pig. All seven pigs that carried the integrated transferred gene expressed shRNA in all the analyzed tissues, and the expression of PERV was significantly inhibited [78]. We used highly effective RNA interference (RNAi) to knockdown the inhibin alpha subunit (INHA) expression in Small Tail Han sheep to produce transgenic sheep with high fecundity using a gonad-injection transgenic method. Four of nine lambs produced were proved to be transgenic. The results suggested that transgenic manipulation of Small Tail Han sheep through an RNAi approach using gonad-injection is feasible [27]. Myostatin (MSTN) is a well-known negative regulator of muscle growth. Microinjection of recombinant lentiviral particles, and expressing transgenes encoding shRNAs targeting an endogenous gene (myostatin) into the perivitelline space of in vitro-produced bovine zygotes was utilized to produce 40 transgenic blastocysts that were transferred into 14 recipient cows, resulting in seven pregnancies. Five transgenic calves were produced, of which three expressed the transgene [79]. These are promising approaches for efficient production of transgenic animals.
Since some siRNA do not have an inhibitory effect, an important issue in the application of RNAi is how to design effective RNAi sequence and produce constitutively stable expression in cells. Although the mechanism of RNAi is not clear and many proteins and enzymes that are involved have an undefined function, this technique is still promising for the application in studies of genetic function analysis and disease therapy.
Zinc-finger nuclease gene targeting technique
Recently, the emergence of the zinc-finger nuclease (ZFN) technique signified a qualitative leap in gene targeting techniques. ZFN is comprised of one DNA binding domain and one non-specific endonuclease domain. ZFNs bind and cut DNA at specific sites, introduce double-stranded DNA break at a specific location, and transfer extrinsic DNA by induction of the endogenous DNA repair procedure, homology-directed repair or non-homology terminal junction, and then modify the cellular endogenous gene (Fig. 2) [49]. This technique breaks through the limitation of gene targeting efficiency, which is enhanced by five orders of magnitude [80]. The ZFN was designed, and subsequently the mRNA coding ZFN of homologous gene for zebrafish-specific receptor of vascular endothelial growth factor-2 was injected into the 1-cell stage zebrafish embryo (zygote). About 10 % of the target genes contained mutation [81]. In the ZFN technique, designing highly specific ZFN is the bottleneck. Maeder et al. [82] produced effective ZFN using Oligomerized Pool Engineering (OPEN) source and established a 66-zinc-finger library, each has one specific zinc-finger structure data file and every zinc-finger structure can bind one specific DNA site. The first site-specific gene knock-out mammals using the ZFN technique were produced. In the study, the ZFN DNA construction or the coded mRNA from ZFN DNA was microinjected into rat embryos and obtained knock-out rats at two endogenous IgM and Rab38 sites with a knock-out efficiency of 25–100 % [83]. X-linked severe combined immunodeficiency (X-SCID) gene disease model by a site-specific knock-out of the interleukin two receptor γ gene (Il-2rg) were produced using the ZFN technique [84]. Using the ZFN technique, target-modified animals were produced in less than 4 months, which significantly reduced the experiment time and raised the efficiency [85]. Gene targeting is indispensable for reverse genetics and the generation of animal models of disease. The mouse has become the most commonly used animal model system owing to the success of embryonic stem cell-based targeting technology, whereas other mammalian species lack convenient tools for genome modification. Microinjection of engineered zinc-finger nucleases (ZFNs) in embryos was used to generate gene knockouts in the rat and the mouse by introducing nonhomologous end joining (NHEJ)-mediated deletions or insertions at the target site. Using ZFN technology in embryos to introduce sequence-specific modifications (knock-ins) by means of homologous recombination in Sprague–Dawley and Long-Evans hooded rats and FVB mice, enables precise genome engineering to generate modifications such as point mutations, accurate insertions and deletions, and conditional knockouts and knock-ins [86]. The same strategy can potentially be applied to many other species for which genetic engineering tools are needed. Zinc-finger nucleases (ZFNs) are powerful tools for producing gene knockouts (KOs) with high efficiency. Whereas ZFN-mediated gene disruption has been demonstrated in laboratory animals such as mice, rats, and fruit flies, ZFNs have not been used to disrupt an endogenous gene in any large domestic species. Recently, cloned pigs carrying a biallelic ZFN-induced knockout of the porcine α1,3-galactosyltransferase (GGTA1) gene were produced [87]. This approach opens a unique avenue toward the creation of gene KO pigs, which could benefit both agriculture and biomedicine.
The ZFN-gene targeting technique significantly enhances the ability of accurate modification of the animal genome and the efficiency of gene integration, and allows the establishment of animal models and the functional gene research. ZFN-based strategies may provide a new method for treating monogenic human diseases by correcting the causative genetic defect. If one could insert the corrected version of the mutation at the precise location of the genetic defect in human genome, most of the difficulties appearing in gene therapy are likely to be overcome. However, there are some defects in the ZFN because of its early stage in development. For example, designing high-affinity and exquisite sequence specificity for the desired target sites is hard work. ZFN can occur off-target during gene modification and induce dosage-dependent toxicity in cells [88]. Therefore, further studies are needed to determine the usefulness of the ZFN gene targeting system.
Induced pluripotent stem cell technique
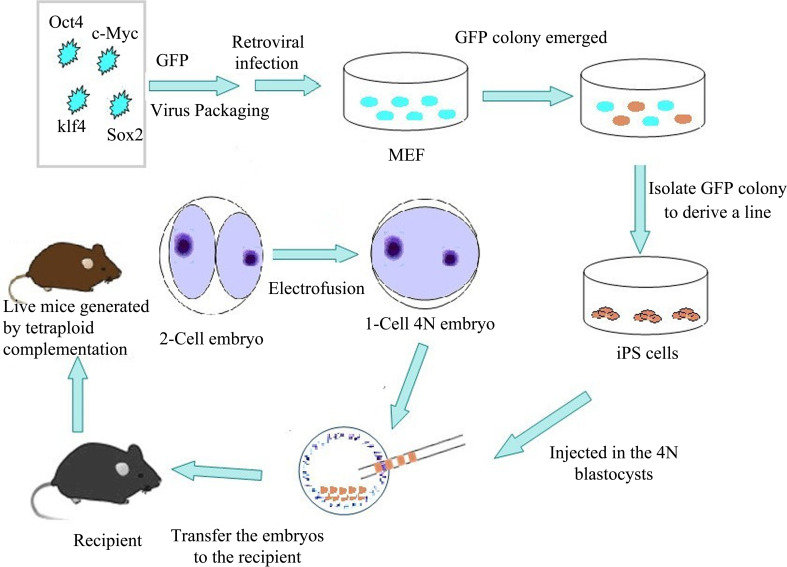
Induced pluripotent stem cell (iPSC) is a type of dedifferentiated somatic cell that is descended from the differentiated counterpart after either its transfection with the genes encoding several transcription factors or its exposure to the combination of a few transcription factor proteins and its subsequent in vitro reprogramming to the state of an embryonic-like stem cell [89]. Similarly, iPSC has the totipotency of self-renewal and differentiation, which resembles the embryonic stem cell. Therefore, it is unnecessary to produce embryos and cells from any organs that can be used to produce cells having the function of stem cells, which can avoid the ethical problems faced by transgenes and simplify the procedure to produce transgenic animals (Fig. 5).
Fig. 5.
Schematic depiction of production of mice using iPS cells and tetraploid complementation. Retrovirus-mediated transfection with four transcription factors (Oct4, Sox2, Klf4, and c-Myc) into mouse embryonic fibroblasts (MEF) has resulted in the generation of induced pluripotent stem (iPS) cells. Then iPS cells are injected into tetraploid blastocysts to generate iPSC mice
These iPS cells were previously constructed by integration into the genomic vector while the integrated vector may produce mutations and limit the application of iPSCs. Therefore, human iPS cells were derived using the nonintegrating episomal vectors [90]. After the removal of the episome, iPS cells completely freed of vectors and transgene sequences were derived, which were similar to human embryonic stem (ES) cells in proliferative and developmental potential. Hiratsuka et al. [91] generated integration-free iPS cells engineered using human artificial chromosome vectors. Warren et al. [92] described a simple, non-integrating strategy for reprogramming cell fate based on administration of synthetic mRNA modified to overcome innate antiviral responses. Expression of the microRNA302/367 (miR302/367) cluster rapidly and efficiently reprograms mouse and human somatic cells to an iPSC state without a requirement for exogenous transcription factors. This microRNA (miRNA)-based reprogramming approach is two orders of magnitude more efficient than standard Octamer-binding transcription factor 4 (Oct-4)/SRY-related high-mobility-group-box protein-2 (Sox2)/Kruppel-like factor-4 (Klf4)/Myelocytomatosis viral oncogene (c-Myc)-mediated methods [93]. The reprogramming of human somatic cells was facilitated by using standard techniques to transfect expression plasmids that encode OCT4, Nanog homeobox (NANOG), SOX2, and Homo sapiens lin-28 homolog (LIN28) without the need for episomal stability or selection. The resulting human iPS cells are free of DNA integration, express pluripotent markers, and form teratomas in immunodeficient animals [94]. Stable iPSCs from human fibroblasts were generated by directly delivering four reprogramming proteins (Oct4, Sox2, Klf4, and c-Myc) fused with a cell-penetrating peptide (CPP). These protein-induced human iPSCs (p-hiPSCs) exhibited similarity to human embryonic stem cells (hESCs) in morphology, proliferation, and expression of characteristic pluripotency markers [95].
These results demonstrated that reprogramming human somatic cells did not require genomic integration or the continued presence of exogenous reprogramming factors, which removed one obstacle to the clinical application of human iPS cells. These findings significantly increase the safety of iPS cells and may have important implications for their applications.
Induced pluripotent stem cells can be used as target cells for transgenes. They can be transferred with extrinsic genes through transgenic techniques and can be genetically modified through gene targeting or gene knock-out. These manipulations will result in the intracellular genetic transformation in iPS cells and the transferred iPS cells can be injected into the blastocoel to obtain chimeric offspring and finally to produce targeted transgenic animals, effectively and directionally. Hockemeyer et al. [96] describe transcription activator-like effector nucleases (TALENs) employing the specific architectures that mediate site-specific genome modification in human pluripotent cells with similar efficiency and precision as do ZFNs. A protocol has been developed and established that allows efficient gene targeting in hPSC lines. This protocol will find broad applications in generating lineage-specific reporter lines and point mutations in genetic repair in disease models using hPSCs [97]. For the first time in a nonrodent species, germline transmission of iPSCs with the live birth of a transgenic piglet that possessed genome integration of the human POU domain, class 5, transcription factor 1 (POU5F1) and NANOG genes was demonstrated. In addition, gross and histological examination of necropsied porcine chimeras at 2, 7, and 9 months showed that these animals lacked tumor formation and demonstrated normal development. Tissue samples positive for human POU5F1 DNA showed no C-MYC gene expression, further implicating C-MYC as a cause of tumorigenicity. The development of germline-competent porcine iPSCs that do not produce tumors in young chimeric animals presents an attractive and powerful translational model to study the efficacy and safety of stem cell therapies and perhaps to efficiently produce complex transgenic animals [98, 99]. Moreover, it will be one good choice to apply iPS cells in somatic cell nuclear transfer. The iPS cells can be used as donor cells where somatic cell nuclei and suitable recipient somatic cells can fuse to produce transgenic animals.
Compared to ES cells, iPS cells have obvious advantages including the avoidance of wasting high-quality embryos during the isolation of ES cells and the availability that it can be obtained from common somatic cells. In addition, the plasticity of iPS cells makes these genetic modifications more effective and the production of transgenic animals simpler and faster. Compared with the special technical expertise required for SCNT, the iPS cell technique is easier to perform in any modern molecular biology laboratory. The therapeutic potential of iPS cells shows amazing prospects, which generate patient-specific and disease-specific iPS cells. It avoids producing human embryos or collecting human oocytes. However, since the iPS cell technique is still in the early stages of development, many problems need to be resolved, such as how to raise the induction efficiency of iPS cells, how to retain pluripotency, and how to induce iPS cells with controllable targeting. As further research is performed, these problems will likely be resolved, and thereby iPSC technology may possibly promote the development of transgenic techniques [100, 101].
Problems and prospects
Transgenic animals have potentially broad applications in the improvement of animal production quality, the enhancement of production capacity, the studies of human disease models, and the production of biomedical materials. However, there are many pressing problems that need to be resolved for transgenic animal studies.
Firstly, the transgenic technique is imperfect, resulting in low success rates and survival rates of transgenic animals. These are the main limiting factors in the development of transgenic animals. Secondly, the integration efficiency of extrinsic genes at the target site is low and unstable, and what affects the intrinsic gene, damages the host’s genome, or activates the closed gene in normal conditions to express and subsequently produces abnormalities in animals is unclear. Thirdly, gene targeting techniques are still in their infancy and require further studies. Therefore, it is necessary for researchers to utilize comprehensive basic transgenic theories and technical procedures to refine these methods. Finally, there are safety concerns for the production of transgenic animals. For example, inserted extrinsic genes may affect the host, contaminate other genes, and produce a variety of threats to ecological balance and species diversity. Transgenic animal production may also lead to food-safety problems such as inadvertent allergies or toxicities. Therefore, serious considerations of the safety of transgenic animals and modification or legislation of related laws and regulations will be helpful for the human fortune with the safety premise.
Since their emergence, transgenic techniques have developed rapidly and have provided more and improved platforms for the preparation of transgenic animals. The transgenic efficiency is increased by germ line stem cells, the site-specific integration of extrinsic gene is achieved by gene targeting, and the spatiotemporal control of target gene expression is possible because of the combination of gene targeting and RNAi. These techniques provide an entirely new pathway for the accurate modulation of genes. In addition, transgenic animals may provide the tools for a series of research hotspots like microRNA function and iPS cells. All of these developments will provide new ideas and bring forth important changes in fields like medicine, health, and livestock improvement. In particular, the economic and social benefits from the production of bioreactors, drug production, and organ culture for human transplantation will be great.
Since the completion of the livestock genome project, the genetic merit of livestock could be improved more conveniently by using transgenic technologies. For example, appropriate extrinsic genes could be transfected into DNA segments exactly to avoid the effects on animal growth and overcome the health problems, which are caused by random integration and abnormal expression. In summary, reliable and efficient transgenic techniques play a critical role in the research and production of transgenic animals. It is conceivable that the development of more simple and novel animal transgenic techniques will lead to more transgenic animals and related products that will likely improve our livelihood and well-being.
Acknowledgments
This work was supported by a grant from The Major Science and Technology Project of New Variety Breeding of Genetically Modified Organisms (Nos. 2009ZX08008-004B and 2008ZX08008-003), the National High Technology Research Development Program of China (863 Program No. 2008AA10Z140), the National Natural Science Foundation of China (No. 30571339) and the Innovation Research Foundation of CAAS (No. 2004-CAAS-1).
References
- 1.Janeisch R, Mintz B. Simian virus 40 DNA sequences in DNA healthy adult mice derived from preimplantation blastocysts injected with viral DNA. Proc Natl Acad Sci USA. 1974;71(4):1250–1254. doi: 10.1073/pnas.71.4.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon JW, Scangos GA, Plotkin DJ, Barbosa JA, Ruddle FH. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc Natl Acad Sci USA. 1980;77:7380–7384. doi: 10.1073/pnas.77.12.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon JW, Ruddle FH. Integration and stable germ line transmission of genes injected into mouse pronuclei. Science. 1981;214:1244–1246. doi: 10.1126/science.6272397. [DOI] [PubMed] [Google Scholar]
- 4.Lavitrano M, Camaioni A, Fazio VM, Dolci S, Farace MG, Spadafora C. Sperm cells as vectors for introducing foreign DNA into eggs: genetic transformation of mice. Cell. 1989;57:717–723. doi: 10.1016/0092-8674(89)90787-3. [DOI] [PubMed] [Google Scholar]
- 5.Hirabayashi M, Hochi S. Generation of transgenic rats by ooplasmic injection of sperm cells exposed to exogenous DNA. Methods Mol Biol. 2010;597:127–136. doi: 10.1007/978-1-60327-389-3_9. [DOI] [PubMed] [Google Scholar]
- 6.Perry AC, Wakayama T, Kishikawa H, Kasai T, Okabe M, Toyoda Y, Yanagimachi R. Mammalian transgenesis by intracytoplasmic sperm injection. Science. 1999;284:1180–1183. doi: 10.1126/science.284.5417.1180. [DOI] [PubMed] [Google Scholar]
- 7.Chan AWS, Chong KY, Martinovich C, Simerly C, Schatten G. Transgenic monkeys produced by retroviral gene transfer into mature oocytes. Science. 2001;291(5502):309–312. doi: 10.1126/science.291.5502.309. [DOI] [PubMed] [Google Scholar]
- 8.García-Vázquez FA, Ruiz S, Matás C, Izquierdo-Rico MJ, Grullón LA, De Ondiz A, Vieira L, Avilés-López K, Gutiérrez-Adán A, Gadea J. Production of transgenic piglets using ICSI-sperm-mediated gene transfer in combination with recombinase RecA. Reproduction. 2010;140(2):259–272. doi: 10.1530/REP-10-0129. [DOI] [PubMed] [Google Scholar]
- 9.Brinster RL. Germline stem cell transplantation and transgenesis. Science. 2002;296:2174–2176. doi: 10.1126/science.1071607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhup S, Majumdar SS. Transgenesis via permanent integration of genes in repopulating spermatogonial cells in vivo. Nat Methods. 2008;5:601–603. doi: 10.1038/nmeth.1225. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Miao XY, Yin XH, Ma YF, Qu ZJ, Zhang QT. Study of transgenic efficiency in different spermatogenic stages in mice. Prog Biochem Biophys. 2009;36(8):1019–1024. doi: 10.3724/SP.J.1206.2008.00839. [DOI] [Google Scholar]
- 12.Zou K, Yuan Z, Yang Z, Luo H, Sun K, Zhou L, Xiang J, Shi L, Yu Q, Zhang Y, Hou R, Wu J. Production of offspring from a germline stem cell line derived from neonatal ovaries. Nat Cell Biol. 2009;11(5):631–636. doi: 10.1038/ncb1869. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Yang Z, Yang Y, Wang S, Shi L, Xie W, Sun K, Zou K, Wang L, Xiong J, Xiang J, Wu J. Production of transgenic mice by random recombination of targeted genes in female germline stem cells. J Mol Cell Biol. 2011;3(2):132–141. doi: 10.1093/jmcb/mjq043. [DOI] [PubMed] [Google Scholar]
- 14.Cibelli JB, Stice SL, Golueke PJ, Kane JJ, Jerry J, Blackwell C, Ponce de Leon FA, Robl JM. Cloned transgenic calves produced from nonquiescent fetal fibroblasts. Science. 1998;280:1256–1258. doi: 10.1126/science.280.5367.1256. [DOI] [PubMed] [Google Scholar]
- 15.Park KW, Cheong HT, Lai L, Im GS, Kuhholzer B, Bonk A, Samuel M, Rieke A, Day BN, Murphy CN, Carter DB, Prather RS. Production of nuclear transfer-derived swine that express the enhanced green fluorescent protein. Anim Biotechnol. 2001;12:173–181. doi: 10.1081/ABIO-100108344. [DOI] [PubMed] [Google Scholar]
- 16.McCreath KJ, Howcroft J, Campbell KH, Colman A, Schnieke AE, Kind AJ. Production of gene-targeted sheep by nuclear transfer from cultured somatic cells. Nature. 2000;405:1066–1069. doi: 10.1038/35016604. [DOI] [PubMed] [Google Scholar]
- 17.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann A, Kessler B, Ewerling S, Weppert M, Vogg B, Ludwig H, Stojkovic M, Boelhauve M, Brem G, Wolf E, Pfeifer A. Efficient transgenesis in farm animals by lentiviral vectors. EMBO Rep. 2003;4(11):1054–1060. doi: 10.1038/sj.embor.7400007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasaki E, Suemizu H, Shimada A, Hanazawa K, Oiwa R, Kamioka M, Tomioka I, Sotomaru Y, Hirakawa R, Eto T, Shiozawa S, Maeda T, Ito M, Ito R, Kito C, Yagihashi C, Kawai K, Miyoshi H, Tanioka Y, Tamaoki N, Habu S, Okano H, Nomura T. Generation of transgenic non-human primates with germline transmission. Nature. 2009;459:523–527. doi: 10.1038/nature08090. [DOI] [PubMed] [Google Scholar]
- 20.Motono M, Yamada Y, Hattori Y, Nakagawa R, Nishijima K, Iijima S. Production of transgenic chickens from purified primordial germ cells infected with a lentiviral vector. J Biosci Bioeng. 2010;109(4):315–321. doi: 10.1016/j.jbiosc.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Kim JN, Park TS, Park SH, Park KJ, Kim TM, Lee SK, Lim JM, Han JY. Migration and proliferation of intact and genetically modified primordial germ cells and the generation of a transgenic chicken. Biol Reprod. 2010;82(2):257–262. doi: 10.1095/biolreprod.109.079723. [DOI] [PubMed] [Google Scholar]
- 22.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 23.Bu L, Gao XL, Jiang X, Chien KR, Wang Z. Targeted conditional gene knockout in human embryonic stem cells. Cell Res. 2010;20:379–382. doi: 10.1038/cr.2010.23. [DOI] [PubMed] [Google Scholar]
- 24.Toledo F, Liu CW, Lee CJ, Wahl GM. RMCE-ASAP: a gene targeting method for ES and somatic cells to accelerate phenotype analyses. Nucleic Acids Res. 2006;34(13):e92. doi: 10.1093/nar/gkl518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasuwa H, Kaseda K, Einarsdottir T, Okabe M. Small interfering RNA and gene silencing in transgenic mice and rats. FEBS Lett. 2002;532:227–230. doi: 10.1016/S0014-5793(02)03680-3. [DOI] [PubMed] [Google Scholar]
- 26.Seibler J, Schwenk F. Transgenic RNAi applications in the mouse. Methods Enzymol. 2010;477:367–386. doi: 10.1016/S0076-6879(10)77019-1. [DOI] [PubMed] [Google Scholar]
- 27.Miao XY, Zhang RJ. Production of transgenic sheep by inhibin α shRNA. Transgenic Res. 2011;20(5):1185. [Google Scholar]
- 28.Porteus MH. Mammalian gene targeting with designed zinc-finger nucleases. Mol Ther. 2006;13(2):438–446. doi: 10.1016/j.ymthe.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Zhao XY, Li W, Lv Z, Liu L, Tong M, Hai T, Hao J, Wang X, Wang L, Zeng F, Zhou Q. Viable fertile mice generated from fully pluripotent iPS cells derived from adult somatic cells. Stem Cell Rev. 2010;6(3):390–397. doi: 10.1007/s12015-010-9160-3. [DOI] [PubMed] [Google Scholar]
- 30.Lee AY, Lloyd KC. Rederivation of transgenic mice from iPS cells derived from frozen tissue. Transgenic Res. 2011;20(1):167–175. doi: 10.1007/s11248-010-9390-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miao XY. Production of transgenic animals using spermatogonial stem cells. Agri Sci China. 2011;10(5):101–105. doi: 10.1016/S1671-2927(11)60060-6. [DOI] [Google Scholar]
- 32.Brinster RL, Avarbock MR. Germline transmission of donor haplotype following spermatogonial transplantation. Proc Natl Acad Sci USA. 1994;91(24):11303–11307. doi: 10.1073/pnas.91.24.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagano M, Brinster CJ, Orwig KE, Ryu BY, Avarbock MR, Brinster RL. Transgenic mice produced by retroviral transduction of male germ-line stem cells. Proc Natl Acad Sci USA. 2001;98(23):13090–13095. doi: 10.1073/pnas.231473498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanatsu-Shinohara M, Ikawa M, Takehashi M, Ogonuki N, Miki H, Inoue K, Kazuki Y, Lee J, Toyokuni S, Oshimura M, Ogura A, Shinohara T. Production of knockout mice by random or targeted mutagenesis in spermatogonial stem cells. Proc Natl Acad Sci USA. 2006;103(21):8018–8023. doi: 10.1073/pnas.0601139103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanatsu-Shinohara M, Kato M, Takehashi M, Morimoto H, Takashima S, Chuma S, Nakatsuji N, Hirabayashi M, Shinohara T. Production of transgenic rats via lentiviral transduction and xenogeneic transplantation of spermatogonial stem cells. Biol Reprod. 2008;79(6):1121–1128. doi: 10.1095/biolreprod.108.071159. [DOI] [PubMed] [Google Scholar]
- 36.Honaramooz A, Megee S, Zeng W, Destrempes MM, Overton SA, Luo J, Galantino-Homer H, Modelski M, Chen F, Blash S, Melican DT, Gavin WG, Avres S, Yang F, Wang PJ, Echelard Y, Dobrinski I. Adeno-associated virus (AAV)-mediated transduction of male germ line stem cells results in transgene transmission after germ cell transplantation. FASEB J. 2008;22(2):374–382. doi: 10.1096/fj.07-8935com. [DOI] [PubMed] [Google Scholar]
- 37.Miao XY, Zhang X. Production of transgenic mice carrying the Thanatin gene by intratesticular injection. Biochem Biophys Res Commun. 2011;415(3):429–433. doi: 10.1016/j.bbrc.2011.10.044. [DOI] [PubMed] [Google Scholar]
- 38.Izsvák Z, Fröhlich J, Grabundzija I, Shirley JR, Powell HM, Chapman KM, Ivics Z, Hamra FK. Generating knockout rats by transposon mutagenesis in spermatogonial stem cells. Nat Methods. 2010;7:443–445. doi: 10.1038/nmeth.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ivics Z, Izsvák Z, Medrano G, Chapman KM, Hamra FK. Sleeping Beauty transposon mutagenesis in rat spermatogonial stem cells. Nat Protoc. 2011;6(10):1521–1535. doi: 10.1038/nprot.2011.378. [DOI] [PubMed] [Google Scholar]
- 40.Honaramooz A, Yang Y. Recent advances in application of male germ cell transplantation in farm animals. Vet Med Int. 2011;2011:657860. doi: 10.4061/2011/657860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naito M, Matsubara Y, Harumi T, Tagami T, Sakurai M, Kuwana T. Foreign gene expression in the gonads of chimaeric chicken embryos by transfer of primordial germ cells transfected in vitro by lipofection for 24 hours. Anim Sci J. 2000;71(3):308–311. [Google Scholar]
- 42.Van de Lavoir MC, Diamond JH, Leighton PA, Mather-Love C, Heyer BS, Bradshaw R, Kerchner A, Hooi LT, Gessaro TM, Swanberg SE, Delany ME, Etches RJ. Germline transmission of genetically modified primordial germ cells. Nature. 2006;441(7094):766–769. doi: 10.1038/nature04831. [DOI] [PubMed] [Google Scholar]
- 43.Motono M, Yamada Y, Hattori Y, Nakagawa R, Nishijima K, Iijima S. Production of transgenic chickens from purified primordial germ cells infected with a lentiviral vector. J Biosci Bioeng. 2010;109(4):315–321. doi: 10.1016/j.jbiosc.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 44.Mueller S, Prelle K, Rieger N, Petznek H, Lassnig C, Luksch U, Aigner B, Baetscher M, Wolf E, Mueller M, Brem G. Chimeric pigs following blastocyst injection of transgenic porcine primordial germ cells. Mol Reprod Dev. 1999;54(3):244–254. doi: 10.1002/(SICI)1098-2795(199911)54:3<244::AID-MRD5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 45.Nowak-Imialek M, Kues W, Carnwath JW, Niemann H. Pluripotent stem cells and reprogrammed cells in farm animals. Microsc Microanal. 2011;17(4):474–497. doi: 10.1017/S1431927611000080. [DOI] [PubMed] [Google Scholar]
- 46.Leighton PA, van de Lavoir MC, Diamond JH, Xia C, Etches RJ. Genetic modification of primordial germ cells by gene trapping, gene targeting, and phiC31 integrase. Mol Reprod Dev. 2008;75(7):1163–1175. doi: 10.1002/mrd.20859. [DOI] [PubMed] [Google Scholar]
- 47.Suraeva NM, Baryshnikov AIu, Fisinin VI, Prokofev MI. Efficacy of various methods of a reporter gene transfer to chicken embryonic cells. Izv Akad Nauk Ser Biol. 2008;1:18–23. [PubMed] [Google Scholar]
- 48.Shin SS, Kim TM, Kim SY, Kim TW, Seo HW, Lee SK, Kwon SC, Lee GS, Kim H, Lim JM, Han JY. Generation of transgenic quail through germ cell-mediated germline transmission. FASEB J. 2008;22(7):2435–2444. doi: 10.1096/fj.07-101485. [DOI] [PubMed] [Google Scholar]
- 49.Li MA, Bradley A. Crafting rat genomes with zinc fingers. Nat Biotechnol. 2011;29:39–41. doi: 10.1038/nbt.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51(3):503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 51.Okada Y, Ueshin Y, Hasuwa H, Takumi K, Okabe M, Ikawa M. Targeted gene modification in mouse ES cells using integrase-defective lentiviral vectors. Genesis. 2009;47(4):217–223. doi: 10.1002/dvg.20469. [DOI] [PubMed] [Google Scholar]
- 52.Kawamata M, Ochiya T. Gene-manipulated embryonic stem cells for rat transgenesis. Cell Mol Life Sci. 2011;68(11):1911–1915. doi: 10.1007/s00018-011-0669-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto S, Nakata M, Sasada R, Ooshima Y, Yano T, Shinozawa T, Tsukimi Y, Takeyama M, Matsumoto Y, Hashimoto T. Derivation of rat embryonic stem cells and generation of protease-activated receptor-2 knockout rats. Transgenic Res. 2011 doi: 10.1007/s11248-011-9564-0. [DOI] [PubMed] [Google Scholar]
- 54.Dong Z, Ge J, Li K, Xu Z, Liang D, Li J, Li J, Jia W, Li Y, Dong X, Cao S, Wang X, Pan J, Zhao Q. Heritable targeted inactivation of myostatin gene in yellow catfish (Pelteobagrus fulvidraco) using engineered zinc-finger nucleases. PLoS One. 2011;6(12):e28897. doi: 10.1371/journal.pone.0028897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tong C, Li P, Wu NL, Yan Y, Ying QL. Production of p53 gene knockout rats by homologous recombination in embryonic stem cells. Nature. 2010;467(7312):211–213. doi: 10.1038/nature09368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tong C, Huang G, Ashton C, Li P, Ying QL. Generating gene knockout rats by homologous recombination in embryonic stem cells. Nat Protoc. 2011;6(6):827–844. doi: 10.1038/nprot.2011.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuroiwa Y, Kasinathan P, Matsushita H, Sathiyaselan J, Sullivan EJ, Kakitani M, Tomizuka K, Ishida I, Robl JM. Sequential targeting of the genes encoding immunoglobulin-μ and prion protein in cattle. Nat Genet. 2004;36(7):775–780. doi: 10.1038/ng1373. [DOI] [PubMed] [Google Scholar]
- 58.Wall RJ, Powell AM, Paape MJ, Kerr DE, Bannerman DD, Pursel VG, Wells KD, Talbot N, Hawk HW. Genetically enhanced cows resist intramammary Staphylococcus aureus infection. Nat Biotechnol. 2005;23(4):445–451. doi: 10.1038/nbt1078. [DOI] [PubMed] [Google Scholar]
- 59.Lai L, Kang JX, Li R, Wang J, Witt WT, Yong HY, Hao Y, Wax DM, Murphy CN, Rieke A, Samuel M, Linville ML, Korte SW, Evans RW, Starzl TE, Prather RS, Dai Y. Generation of cloned transgenic pigs rich in omega-3 fatty acids. Nat Biotechnol. 2006;24(4):435–436. doi: 10.1038/nbt1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baldassarre H, Hockley DK, Dore M, Brochu E, Hakier B, Zhao X, Bordiqnon V. Lactation performance of transgenic goats expressing recombinant human butyryl-cholinesterase in the milk. Transgenic Res. 2008;17(1):73–84. doi: 10.1007/s11248-007-9137-4. [DOI] [PubMed] [Google Scholar]
- 61.Bertolini LR, Bertolini M, Maga EA, Madden KR, Murray JD. Increased gene targeting in Ku70 and Xrcc4 transiently deficient human somatic cells. Mol Biotechnol. 2009;41(2):106–114. doi: 10.1007/s12033-008-9098-8. [DOI] [PubMed] [Google Scholar]
- 62.Wongsrikeao P, Sutou S, Kunishi M, Dong YJ, Bai X, Otoi T. Combination of the somatic cell nuclear transfer method and RNAi technology for the production of a prion gene-knockdown calf using plasmid vectors harboring the U6 or tRNA promoter. Prion. 2011;5(1):39–46. doi: 10.4161/pri.5.1.14075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waghmare SK, Estrada J, Reyes L, Li P, Ivary B, Sidner RA, Burlak C, Tector AJ. Gene targeting and cloning in pigs using fetal liver derived cells. J Surg Res. 2011;171(2):e223–e229. doi: 10.1016/j.jss.2011.07.051. [DOI] [PubMed] [Google Scholar]
- 64.Sakahara M, Ohkawara H, Nakao K, Yokozaki H, Aiba A. The simultaneous induction of tumorigenesis and Cre-loxP recombination in mice. Kobe J Med Sci. 2009;54(6):279–281. [PubMed] [Google Scholar]
- 65.Kyoungmi K, Hwain K, Daekee L. Site-specific modification of genome with cell-permeable Cre fusion protein in preimplantation mouse embryo. Biochem Biophys Res Commun. 2009;388(1):122–123. doi: 10.1016/j.bbrc.2009.07.132. [DOI] [PubMed] [Google Scholar]
- 66.Bouvier J, Cheng JG (2009) Recombineering-based procedure for creating Cre/loxP conditional knockouts in the mouse. Curr Protoc Mol Biol Chapter 23: Unit 23.13 [DOI] [PubMed]
- 67.Friedel RH, Wurst W, Wefers B, Kühn R. Generating conditional knockout mice. Methods Mol Biol. 2011;693:205–231. doi: 10.1007/978-1-60761-974-1_12. [DOI] [PubMed] [Google Scholar]
- 68.Yamamoto M, Takeda K. A method for the generation of conditional gene-targeted mice. Methods Mol Biol. 2012;757:399–410. doi: 10.1007/978-1-61779-166-6_23. [DOI] [PubMed] [Google Scholar]
- 69.Wakita T, Taya C, Katsume A, Kato J, Yonekawa H, Kanegae Y, Saito I, Hayashi Y, Koike M, Kohara M. Efficient conditional transgene expression in hepatitis C virus cDNA transgenic mice mediated by the Cre/loxP system. J Biol Chem. 1998;273(15):9001–9006. doi: 10.1074/jbc.273.15.9001. [DOI] [PubMed] [Google Scholar]
- 70.Metzger D, Chambon P. Site- and time-specific gene targeting in the mouse. Methods. 2001;24(1):71–80. doi: 10.1006/meth.2001.1159. [DOI] [PubMed] [Google Scholar]
- 71.Rendahl KG, Ouiroz D, Ladner M, Covne M, Seltzer J, Manning WC, Escobedo JA. Tightly regulated long-term erythropoietin expression in vivo using tet-inducible recombinant adeno-associated viral vectors. Hum Gene Ther. 2002;13(2):335–342. doi: 10.1089/10430340252769842. [DOI] [PubMed] [Google Scholar]
- 72.Chenuaud P, Larcher T, Rabinowitz JE, Provost N, Joussemet B, Bujard H, Samulski RJ, Favre D, Moullier P. Optimal design of a single recombinant adeno-associated virus derived from serotypes 1 and 2 to achieve more tightly regulated transgene expression from nonhuman primate muscle. Mol Ther. 2004;9(3):410–418. doi: 10.1016/j.ymthe.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 73.Rettig GR, Rice KG. Quantitative in vivo imaging of non-viral-mediated gene expression and RNAi-mediated knockdown. Methods Mol Biol. 2009;574:155–171. doi: 10.1007/978-1-60327-321-3_13. [DOI] [PubMed] [Google Scholar]
- 74.Hasuwa H, Kaseda K, Einarsdottir T, Okabe M. Small interfering RNA and gene silencing in transgenic mice and rats. FEBS Lett. 2002;532(1–2):227–230. doi: 10.1016/S0014-5793(02)03680-3. [DOI] [PubMed] [Google Scholar]
- 75.Kunath T, Gish G, Lickert H, Jones N, Pawson T, Rossant J. Transgenic RNA interference in ES cell-derived embryos recapitulates a genetic null phenotype. Nat Biotechnol. 2003;21(5):559–561. doi: 10.1038/nbt813. [DOI] [PubMed] [Google Scholar]
- 76.Kleinhammer A, Deussing J, Wurst W, Kühn R. Conditional RNAi in mice. Methods. 2011;53(2):142–150. doi: 10.1016/j.ymeth.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 77.Dickins RA, McJunkin K, Hernando E, Premsrirut PK, Krizhanovsky V, Burgess DJ, Kim DY, Cordon-Cardo C, Zender L, Hannon GJ, Lowe SW. Tissue-specific and reversible RNA interference in transgenic mice. Nat Genet. 2007;39(7):914–921. doi: 10.1038/ng2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dieckhoff B, Petersen B, Kues WA, Kurth R, Niemann H, Denner J. Knockdown of porcine endogenous retrovirus (PERV) expression by PERV-specific shRNA in transgenic pigs. Xenotransplantation. 2008;15(1):36–45. doi: 10.1111/j.1399-3089.2008.00442.x. [DOI] [PubMed] [Google Scholar]
- 79.Tessanne K, Golding MC, Long CR, Peoples MD, Hannon G, Westhusin ME. Production of transgenic calves expressing an shRNA targeting myostatin. Mol Reprod Dev. 2012;79(3):176–185. doi: 10.1002/mrd.22007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Luo QM, Miao XY, Zhang RJ. An update on the development of transgenic animal technology. Yi Chuan (Hereditas) 2011;33(5):449–458. doi: 10.3724/SP.J.1005.2011.00449. [DOI] [PubMed] [Google Scholar]
- 81.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26(6):695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, Unger-Wallace E, Sander JD, Müller-Lerch F, Fi Fu, Pearlberg J, Göbel C, Dassie JP, Pruett-Miller SM, Porteus MH, Sgroi DC, Iafrate AJ, Dobbs D, McCray PB, Jr, Cathomen T, Voytas DF, Joung JK. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31(2):294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, Vincent A, Lam S, Michalkiewicz M, Schilling R, Foeckler J, Kalloway S, Weiler H, Ménoret S, Anegon I, Davis GD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jacob HJ, Buelow R. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325(5939):433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mashimo T, Takizawa A, Voigt B, Yoshimi K, Hiai H, Kuramoto T, Serikawa T. Generation of knockout rats with X-linked severe combined immunodeficiency(X-SCID) using zinc-finger nucleases. PLOS One. 2010;5(1):e8870. doi: 10.1371/journal.pone.0008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carbery ID, Ji D, Harrington A, Brown V, Weinstein EJ, Liaw L, Cui X. Targeted genome modification in mice using zinc-finger nucleases. Genetics. 2010;186:451–459. doi: 10.1534/genetics.110.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cui X, Ji D, Fisher DA, Wu Y, Briner DM, Weinstein EJ. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol. 2011;29(1):64–67. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- 87.Hauschild J, Petersen B, Santiago Y, Queisser AL, Carnwath JW, Lucas-Hahn A, Zhang L, Meng X, Gregory PD, Schwinzer R, Cost GJ, Niemann H. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc Natl Acad Sci USA. 2011;108(29):12013–12017. doi: 10.1073/pnas.1106422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gupta A, Meng X, Zhu L, Lawson ND. Zinc-finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc-finger nucleases. Nucleic Acids Res. 2011;39(1):381–392. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 90.Yu JY, Hu KJ, Smuga-Otto K, Tian SL, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324(5928):797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hiratsuka M, Uno N, Kurosaki H, Imaoka N, Kazuki K, Ueno E, Akakura Y, Katoh M, Osaki M, Kazuki Y, Nakagawa M, Yamanaka S, Oshimura M. Integration-free iPS cells engineered using human artificial chromosome vectors. PLoS One. 2011;6(10):e25961. doi: 10.1371/journal.pone.0025961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7(5):618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8(4):376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Si-Tayeb K, Noto FK, Sepac A, Sedlic F, Bosnjak ZJ, Lough JW, Duncan SA. Generation of human induced pluripotent stem cells by simple transient transfection of plasmid DNA encoding reprogramming factors. BMC Dev Biol. 2010;10:81. doi: 10.1186/1471-213X-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4(6):472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29(8):731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu Y, Rao M. Gene targeting in human pluripotent stem cells. Methods Mol Biol. 2011;767:355–367. doi: 10.1007/978-1-61779-201-4_26. [DOI] [PubMed] [Google Scholar]
- 98.West FD, Terlouw SL, Kwon DJ, Mumaw JL, Dhara SK, Hasneen K, Dobrinsky JR, Stice SL. Porcine induced pluripotent stem cells produce chimeric offspring. Stem Cells Dev. 2010;19(8):1211–1220. doi: 10.1089/scd.2009.0458. [DOI] [PubMed] [Google Scholar]
- 99.West FD, Uhl EW, Liu Y, Stowe H, Lu Y, Yu P, Gallegos-Cardenas A, Pratt SL, Stice SL. Brief report: chimeric pigs produced from induced pluripotent stem cells demonstrate germline transmission and no evidence of tumor formation in young pigs. Stem Cells. 2011;29(10):1640–1643. doi: 10.1002/stem.713. [DOI] [PubMed] [Google Scholar]
- 100.Qin T, Miao XY. Current progress and application prospects of induced pluripotent stem cells. Yi Chuan (Hereditas) 2010;32(12):1205–1214. [PubMed] [Google Scholar]
- 101.Miao XY, Chen XY. Study and application of induced pluripotent stem cells. Scientia Agricultura Sinica. 2012;45(2):369–375. [Google Scholar]