Abstract
Endoplasmic reticulum-associated degradation (ERAD) is a key cellular process whereby misfolded proteins are removed from the endoplasmic reticulum (ER) for subsequent degradation by the ubiquitin/proteasome system. In the present work, analysis of the released, free oligosaccharides (FOS) derived from all glycoproteins undergoing ERAD, has allowed a global estimation of the mechanisms of this pathway rather than following model proteins through degradative routes. Examining the FOS produced in endomannosidase-compromised cells following α-glucosidase inhibition has revealed a mechanism for clearing Golgi-retrieved glycoproteins that have failed to enter the ER quality control cycle. The Glc3Man7GlcNAc2 FOS species has been shown to be produced in the ER lumen by a mechanism involving a peptide: N-glycanase-like activity, and its production was sensitive to disruption of Golgi-ER trafficking. The detection of this oligosaccharide was unaffected by the overexpression of EDEM1 or cytosolic mannosidase, both of which increased the production of previously characterised cytosolically localised FOS. The lumenal FOS identified are therefore distinct in their production and regulation compared to FOS produced by the conventional route of misfolded glycoproteins directly removed from the ER. The production of such lumenal FOS is indicative of a novel degradative route for cellular glycoproteins that may exist under certain conditions.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-013-1304-6) contains supplementary material, which is available to authorized users.
Keywords: ER-associated degradation, Free oligosaccharides, N-glycosylation, Quality control, MAN2C1, EDEM
Introduction
During N-glycoprotein biosynthesis, preassembled oligosaccharide moieties are transferred en bloc from a dolichol-linked precursor onto asparagine side-chains of newly-synthesised polypeptide chains at the lumenal face of the endoplasmic reticulum (ER) membrane [1]. These N-linked oligosaccharides are transferred in the form of Glc3Man9GlcNAc2 and become subjected to recognition and processing by a variety of ER- and Golgi-resident factors that assist protein folding and assembly, mediate anterograde and retrograde flow of secretory cargo and trigger ER-associated degradation (ERAD) [2].
The ERAD is a cellular pathway that targets misfolded proteins, as well as improperly-assembled multimeric complexes of the endoplasmic reticulum for translocation into the cytosol, ubiquitination and subsequent hydrolysis by the proteasome [3]. The ERAD targeting presumably occurs through a variety of mechanisms, depending on the nature of the substrate as well as the localisation of the misfolded region [4]. For glycoproteins, the consensus route is currently thought to involve demannosylation by ER or Golgi mannosidases, interactions with EDEM1–3 (which could also function as mannosidases themselves), followed by OS-9/XTP3-B-mediated delivery of the substrate to the Hrd1 ubiquitination complex through the interaction with a membrane-spanning adaptor protein, SEL1L [5–7]. The Hrd1 is an E3 ubiquitin ligase that has been additionally shown to mediate retrotranslocation of substrate polypeptides [4, 8]. Glycoproteins degraded in this manner have their glycan portion released prior to the proteasomal destruction in the cytosol by a peptide: N-glycanase (PNGase) [9]. As a result, free oligosaccharides (FOS) are produced. The latter have been employed as markers of ERAD [10–13]. The FOS analysis provides both qualitative and quantitative insight into glycoprotein degradation in the ER on a global cellular level, as opposed to studies limited to a single ERAD substrate, whether endogenous or recombinantly expressed.
The FOS produced by PNGase initially bear two GlcNAc residues at the reducing terminus [9, 14]. The cytosolic pathway for FOS catabolism in normal conditions involves rapid processing by endo-β-N-acetylglucosaminidase (ENGase), which removes the terminal GlcNAc residue, resulting in a mono-GlcNAc form of FOS [14, 15]. This is trimmed by a cytosolic neutral α-mannosidase (NAM, MAN2C1) and subsequently transported into the lysosome for further hydrolysis to monosaccharides [15]. It is likely that the cytosolic FOS degradation pathway provides a feedback mechanism to modulate ERAD, since overexpression of NAM has been shown to increase the rate of glycoprotein degradation [16].
Processing of nascent glycoproteins by ER glucosidases I and II is required for interaction with calnexin (CNX) or calreticulin (CRT), ER-resident lectin chaperones that bind monoglucosylated forms of glycoproteins to facilitate folding [17]. Consequently, inhibition of ER glucosidases results in the inability of glycoproteins to engage with CNX/CRT, which in turn leads to increased rates of protein misfolding and ERAD [17]. As a result, glucosylated FOS are produced in cells treated with glucosidase inhibitors [10, 12]. The latter are processed in the cytosol to the end form of Glc3Man5GlcNAc1 (and slowly to Glc3Man4GlcNAc1) [10]. However, glucosylated FOS are unable to access the lysosome, resulting in their cytosolic accumulation [18, 19]. The use of the ER glucosidase imino sugar inhibitor N-butyl-deoxynojirimycin (NB-DNJ) has demonstrated that the generation of glucosylated FOS is both dose- and treatment time-dependent [10].
In the majority of mammalian cell lines, inhibition of ER glucosidases does not result in a complete loss of deglucosylating capability. Apart from its role in the folding process described above, removal of glucose residues is required for further biosynthetic processing of N-glycans in the Golgi apparatus [20, 21]. Golgi-resident endomannosidase provides a backup mechanism for deglucosylation of glycoproteins escaping the ER prior to complete removal of glucose residues from their asparagine-linked oligosaccharides [22]. Importantly though, endomannosidase-mediated deglucosylation in the Golgi can also precede ERAD [11]. Consequently, FOS analysis in the presence of glucosidase inhibitors underestimates ERAD in endomannosidase-positive cell lines, since endomannosidase-catalysed deglucosylation results in rapid processing of FOS and their eventual destruction in the lysosome [15, 23]. We, therefore, sought to establish a deglucosylation-incompetent cellular model to further explore the mechanisms of ERAD using FOS analysis. To this end, we hereby employ NB-DNJ-treated Madin-Darby bovine kidney (MDBK) and Chinese hamster ovary (CHO)-K1 cells, both of which have been previously shown to lack endomannosidase activity [24]. Using these model systems, we show that in contrast to endomannosidase-positive cells, MDBK and CHO-K1 cells treated with NB-DNJ accumulate glucosylated FOS in the ER lumen in addition to cytosolic species. These lumenal FOS are derived from glycoproteins and as the glucose-bearing forms are unable to be transported to the cytosol and merge with the catabolic pathway for cytosolic FOS, in agreement with previous data [9, 15]. We further demonstrate that glucosylated lumenal FOS constitute a novel marker for ERAD occurring via an alternative route that appears to be partially independent from the pathway in which cytosolic FOS are produced.
Materials and methods
Materials
Tissue culture media were from Gibco/Invitrogen, (Paisley, UK) or Sigma (Gillingham, UK). The AnalaR and HPLC grade solvents were from VWR International. The pcDNA3-NAM was a gift from Prof. Pirkko Heikinheimo and Elina Kuokkanen (University of Helsinki, Finland). The pcDNA3-α1AT and pcDNA3.1(-)-EDEM1 were a gift from Dr. John Christianson (University of Oxford, UK). The CHPV was a gift from Dr. Anthony Marriott (University of Warwick, UK). All other reagents were from Sigma, unless otherwise stated. Water was Milli-Q™ grade.
Inhibitors
The N-butyldeoxynojirimycin (NB-DNJ) was provided by Celltech (Slough, UK). Thapsigargin (TG), Z-VAD-fmk, Q-VD-OPh, MG132 and ALLN were all supplied by Sigma (Gillingham, UK). The N-butylated azepane 1 (NBA1) was a gift from Dr. Yves Blériot (UPMC, Paris, France).
Cell culture
The HL60 and MDBK cells were cultured in RPMI medium containing 10 % foetal calf serum, 2 mM l-glutamine and 1 U/ml penicillin and 1 μg/ml streptomycin (Invitrogen). The CHO-K1 and BHK-21 cells were cultured in DMEM containing 10 % foetal calf serum, 1 U/ml penicillin and 1 μg/ml streptomycin.
Free oligosaccharides analysis
Cells were cultured to high density (1 × 107 cells/ml) prior to growth in fresh medium containing NB-DNJ at varying concentrations as described previously [10]. The FOS were extracted, labelled with 2-anthranilic acid (2-AA) and purified using lectin-affinity chromatography before separation by NP-HPLC as described [25].
Removal of 0.5 M methyl α-d-mannopyranoside prior to enzyme digests
When enzyme digests of the ConA-Sepharose-purified 2-AA-labelled oligosaccharides were required, 0.5 M methyl α-d-mannopyranoside was removed using a 1 ml (25 mg) PGC column (Thermo Electron, Runcorn, UK). The column was pre-equilibrated with 1 ml methanol followed by 1 ml water and 1 ml acetonitrile containing 0.1 % trifluoroacetic acid (TFA) and finally 2 × 0.5 ml water. After sample loading the column was washed with 2 × 0.5 ml water before elution with 2 ml 50 % acetonitrile containing 0.1 % TFA. No loss of FOS was observed.
Concentration of fluorescently-labelled FOS
Free, unconjugated 2-AA was removed following phase splitting using ethyl acetate. Essentially, 2-AA-labelled sample (1 ml in water) was added to 1.5 ml of ethyl acetate and vortexed before separating into two phases by centrifugation at 3,000g for 5 min. The upper phase was removed and a further 1.5 ml ethyl acetate was added and separation was repeated. Following a further addition of ethyl acetate, the lower phase was removed and dried. The sample was then resuspended in 30 μl water before a preparative run by HPLC to allow isolation of individual peaks for further analysis.
Digitonin extraction of cytosol
Madin-Darby bovine kidney cells were grown, treated and harvested as described above. Cell pellets were washed in PBS and resuspended in cold (4 °C) PBS containing 10 μg/ml digitonin (optimal concentration determined empirically). Samples were then incubated at 4 °C for 10 min under gentle agitation, followed by centrifugation at 1,000g, 4 °C, for 5 min. Supernatants were then removed. Pellets were resuspended in water and lysed by sonication (Branson sonifier, set. 3, 50 % duty cycle, 20 bursts, on ice). Equivalent volumes of post-digitonin treatment supernatants and lysed pellets were then subjected to FOS analysis or immunoblotting.
Enzyme digests
The following enzyme digests were performed on the complete FOS populations and individually isolated peaks. The enzymes used were α-glucosidases I and II (purified from rat liver) [26, 27], α-1,2 linkage-specific mannosidase (purified from Aspergillus saitoi) and broad specificity jack bean α-mannosidase (purified from jack bean meal). Following enzyme treatment, all digests were centrifuged through a 10,000 molecular weight cut off filter (pre-washed with 150 μl of water) at 7,000g for 45 min to remove protein before HPLC analysis.
Matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry
Positive-ion MALDI-TOF mass spectra were recorded with a Micromass TofSpec 2E reflectron-TOF mass spectrometer (Waters-Micromass (UK) Ltd., Manchester, UK) fitted with delayed extraction and a nitrogen laser (337 nm). The acceleration voltage was 20 kV; the pulse voltage was 3,000 V; and the delay for the delayed extraction ion source was 500 ns. The FOS, individually isolated by HPLC, were dried and resuspended in 100 μl of water. A 1 μl aliquot of each was mixed with 1 μl of a 50 mg/ml solution of 2,5-dihydroxybenzoic acid (2,5-DHB) in acetonitrile and placed on the MALDI target and allowed to air dry. The samples were re-crystallised from ethanol prior to analysis. The m/z scale was calibrated using a 2-AA-labelled dextran hydrolysate ladder standard.
Protein synthesis rate determination
The [3H]-leucine was added to the cell culture medium at 1 μCi/ml at the beginning of the incubation period. Cell monolayers were washed with 2 × 1 ml PBS followed by 2 × 1 ml ice-cold 10 % (v/v) perchloric acid (PCA)/2 % (w/v) phosphotungstic acid (PTA) followed by 2 × 1 ml ice cold ethanol. Plates were dried for 30 s at 80 °C. Sodium hydroxide (0.5 ml of 0.5 M solution) was added to dried cells and left at room temperature overnight. Aliquots (2.5 μl) were taken for a bicinchoninic acid (BCA) protein assay, and 250 μl samples were taken for radioactivity determination. Protein assay was carried out using the BCA Assay Kit (Sigma, UK), according to the manufacturer’s instructions.
Antibodies
The following antibodies were used: goat polyclonal anti-α1-antitrypsin, biotin conjugate (Abcam), goat polyclonal anti-NAM (Santa Cruz Biotechnology), mouse polyclonal anti-β-actin (Abcam), mouse monoclonal to BiP/GRP78 (BD Biosciences). Goat anti-mouse IgG (Abcam) and mouse anti-goat IgG (Santa Cruz Biotechnology) HRP conjugates were used as secondary antibodies for immunoblotting.
Transfection of plasmids
Cells were seeded into 6-well plates at the densities of 0.3 × 106 cells/well. The following day, plasmids were transfected using polyethylenimine (PEI, Polysciences Inc.). Media were replaced with 4 ml serum-free DMEM. The PEI was mixed with DNA (3 μg DNA per well) at the P:N ratio of 1:14 in a NaCl solution (150 mM) and incubated at room temperature for 10 min. The resulting mix (400 μl) was added dropwise to the wells. Cells were then incubated for 4 h at 37 °C, after which foetal calf serum was added (10 % final concentration). Compound treatment was started in fresh medium following an overnight incubation. For α1-antitrypsin expression, cells were grown in 225-cm2 flasks, and the protocol was adjusted proportionally. In this case, media were not replaced post-transfection, and compound treatment was started immediately in serum-free medium.
Virus purification
The BHK-21, CHO-K1 or MDBK cells were infected with Chandipura virus (CHPV) in serum-free medium. At 4 h post-infection, media were replaced with complete media supplemented with FCS and the appropriate compounds. Viral media were collected following incubation for 24 h and clarified by low-speed centrifugation (2 × 600g, 10 min). Viral particles were precipitated by adding 1/3 volume of a buffer containing PEG-6000 (final concentration 6 %, w/v) and NaCl (final concentration 0.5 M) and incubating the media overnight at 4 °C without agitation, followed by centrifugation (2,000g, 15 min). The pellets were resuspended in 5 ml PBS and sonicated to disrupt clumps (Branson sonifier, set. 1, 50 % duty cycle, 5 bursts, on ice), then centrifuged again (2,000g, 15 min). Supernatants were loaded on a 30/60 % (w/v, PBS) sucrose cushion and centrifuged at 100,000g for 1 h. Viral bands were collected and diluted >10-fold with PBS, filtered through a 0.22 μm filter to remove residual clumps and subjected to ultracentrifugation (100,000g, 2 h) to obtain a purified viral pellet.
Immunopurification
Biotinylated anti-α1AT IgG (1 mg) was immobilised on streptavidin-agarose resin (Thermo Sci., UK) following manufacturer’s instructions. The CHO-K1 cells were transiently transfected with pcDNA3-α1AT as described above. Four days post-transfection, media were harvested, clarified by centrifugation (600g, 10 min) and passed through the IgG column by gravity flow. Following three washes with 10 ml PBS, α1AT was eluted using 0.1 M glycine–HCl, pH 2.8, neutralised by adding 2.5 M Tris–HCl (pH 8.0) and concentrated using Amicon protein concentrators (10 kDa molecular weight cut-off).
Glycan analysis of purified glycoproteins
Purified CHPV or immunopurified α1AT were resolved by SDS-PAGE, under reducing conditions followed by transfer to PVDF membranes which were then stained with Coomassie Blue (0.1 % w/v in 30 % methanol (v/v), 7 % glacial acetic acid (v/v) in water) for 15 min, and destained in 50 % methanol (v/v), 10 % glacial acetic acid (v/v) in water, followed by a wash in water. Membranes were then air-dried, and the bands corresponding to the proteins of interest cut out and subjected to a digest by peptide: N-glycanase F (NEB), at 5,000 U/ml following manufacturer’s instructions, in a total volume of 100 μl for 24 h. Supernatants containing released oligosaccharides were dried, resuspended in 30 μl water and subjected to fluorescent labelling and HPLC analysis.
Western blotting
Following SDS-PAGE of crude cell lysates, proteins were transferred to PVDF membranes, which were then blocked in 5 % (w/v) non-fat dry milk in PBS supplemented with Tween 20 0.2 % (v/v) at 4 °C overnight. Primary and secondary antibodies were diluted in blocking buffer and incubated for 1–2 h, followed by washing in PBS supplemented with 0.05 % (v/v) Tween 20. The ECL substrate (GE Healthcare) was used for detection.
Pulse-chase analysis
The FCS was extensively dialysed against PBS and used throughout. The CHO-K1 cells were pretreated with NB-DNJ (1 mM) for 16 h, followed by a 30 min incubation in DMEM (minus glucose) containing 1 % (v/v) FCS, 1 mM NB-DNJ. Cells were then pulsed using the same medium containing 200 μCi/ml [3H]-mannose with or without MG132 (10 μM) for 15 min, followed by a chase in complete medium containing 10 % FCS, 2 mM cold mannose and 1 mM NB-DNJ, also with or without MG132. At the indicated time points, cells were washed with ice-cold PBS, harvested by scraping on ice and centrifuged at 4 °C to obtain a cell pellet that was then frozen on dry ice. The FOS were extracted as described above, N-linked oligosaccharides bound to protein were released using peptide: N-glycanase F (NEB), at 5,000 U/ml following manufacturer’s instructions and lipid-linked oligosaccharides were obtained following acid release as published [28]. Depending on the experiment, whole pools of glycans or individual HPLC peaks were collected, and the radioactivity associated with pools or peaks was measured. For short-term FOS and protein-bound N-linked oligosaccharide analysis, glycan levels in each experiment were normalised to the levels of total cellular FOS at the 4 h time point.
Results
ER α-glucosidase inhibition reveals production of glucosylated ER-located FOS species
Following α-glucosidase inhibition in HL60 cells, the major FOS species that can be detected by normal-phase high performance liquid chromatography (NP-HPLC) is Glc3Man5GlcNAc1, as a result of the export of glucosylated glycoproteins into the cytosol by the ERAD pathway. The FOS are released from the glycoprotein by cytosolically located PNGase and the free glycans are rapidly processed by ENGase (endo-β-N-acetylglucosaminidase), followed by NAM (neutral α-mannosidase) to Glc3Man5GlcNAc1 (and slowly to Glc3Man4GlcNAc1) [10]. However, in some cell lines, most notably in those deficient in Golgi endomannosidase activity (MDBK and CHO-K1 cells) [24], an additional major peak was observed at 10.12 GU (glucose units) (Fig. 1a).
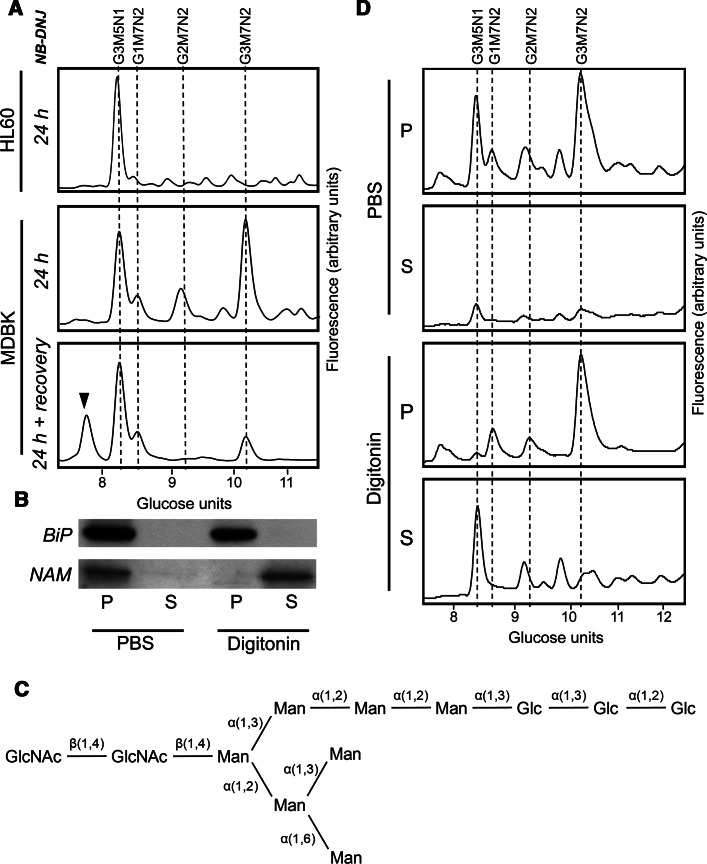
Fig. 1.
Major lumenal free oligosaccharide accumulates in cells following deglucosylation blockage. a NP-HPLC analysis of 2-AA-labelled FOS isolated from HL60 or MDBK cells following treatment with NB-DNJ for 24 h. Cells were harvested either immediately after treatment (24 h) or following incubation for additional 24 h in the absence of NB-DNJ (24 h + recovery). Dashed lines indicate positions of Glc3Man5GlcNAc1 (G3M5N1) or Glc1–3Man7GlcNAc2 (G1–3M7N2). Arrowhead indicates the position of Man7GlcNAc2. The NP-HPLC profiles are representative of experiments carried out and are not drawn to the same scale of fluorescent intensity. b Immunoblotting of BiP and NAM in equivalent fractions of either pellet (P) or supernatant (S) following treatment of MDBK cells with either PBS or digitonin solution. c Structure of the Glc3Man7GlcNAc2 FOS. Labelled lines indicate linkage types. d NP-HPLC analysis of 2-AA labelled FOS extracted from cells fractionated as in b
The additional major free glycan detected following a 24 h NB-DNJ treatment in MDBK and CHO-K1 cells was isolated following NP-HPLC separation and sequenced using a series of enzyme digests, including α-glucosidases I and II, Aspergillus saitoi α-mannosidase (α-1,2-specific) and jack bean α-mannosidase (α1,2/3/6-specific), in addition to MALDI-TOF (matrix-assisted laser desorption/ionisation time-of-flight) MS (mass spectrometry) analysis. This species was determined to be Glc3Man7GlcNAc2 with the structure shown in Fig. 1c. In addition to the described Glc3Man7GlcNAc2 peak, an increase in Glc1–2Man7GlcNAc2 species was detected (Fig. 1a), which was likely due to incomplete ER α-glucosidase inhibition.
The presence of an intact GlcNAc2 core suggests that the accumulating glycans are not cytosolically localised since Glc1-3Man7GlcNAc2 FOS are substrates for rapid processing by ENGase. We speculated that the GlcNAc2-bearing FOS could be localised to the ER. Indeed, free glucosylated glycans in the ER are unable to exit to the cytosol since these species are not substrates for the FOS transporter [23].
To address the hypothesis on the ER localisation of Glc3Man7GlcNAc2, we carried out NB-DNJ treatment in MDBK cells for 24 h as previously, followed by removal of the inhibitor from the growth medium and recovery for a further 24 h. The FOS were then extracted and analysed as before (Fig. 1a). Glc3Man7GlcNAc2 was shown to be reduced by 75 % after a 24 h recovery period and completely absent after 72 h (data not shown). The reduction in detectable species suggests that either the Glc3Man7GlcNAc2 is cleared by bulk flow through the secretory pathway following inhibitor removal or, more likely, by deglucosylation following exposure to ER-localised glucosidases I and II. Consistent with the latter proposal, a Man7GlcNAc2 FOS species (GU = 7.87) was observed after inhibitor removal (Fig. 1a). The glycan was purified and structurally characterised by MS as the deglucosylated form of Glc3Man7GlcNAc2. The Glc2Man7GlcNAc2 glycan was also reduced following glucosidase inhibition recovery. It is noteworthy that following deglucosylation, FOS become substrates for the ER–cytosol FOS transporter, which makes them accessible for further hydrolysis by the cytosolic enzymes ENGase and NAM, and eventually lysosomal hydrolases [9]. Therefore, direct comparison of the amounts of glucosylated and deglucosylated lumenal FOS is difficult.
To further confirm the lumenal location of Glc3Man7GlcNAc2, digitonin extraction of cytosolic FOS was carried out. Extraction of the cytosolic fraction was confirmed by immunoblotting for NAM, using BiP as the protein marker for the lumenal fraction (Fig. 1b). The FOS analysis showed that over 90 % of the Glc3Man7GlcNAc2 species was found in the pellet following extraction with the non-ionic detergent digitonin (Fig. 1d). It is important to note that the digitonin extraction alone does not exclude the possibility of Glc3Man7GlcNAc2 accumulating in a cellular compartment other than ER lumen. However, combined with the fact that following inhibitor recovery Glc3Man7GlcNAc2 is deglucosylated to Man7GlcNAc2, the data strongly suggest ER lumenal accumulation of the FOS species.
Glc3Man7GlcNAc2 species is produced from glycoproteins by ER-localised PNGase activity
The α-glucosidase inhibition in endomannosidase-deficient cell lines results in the build-up of an ER-localised triglucosylated FOS species. Although the glycan appears following α-glucosidase inhibition it is possible that it has not been generated from a glycoprotein. The PNGase responsible for deglycosylation is believed to be active in the cytosol only after translocation of the protein targeted for proteasomal degradation. To elucidate the origin of the Glc3Man7GlcNAc2 FOS, a PNGase-specific inhibitor, Z-VAD-fmk, was utilised [29]. The MDBK cells were treated with NB-DNJ (1 mM) in the presence or absence of the PNGase inhibitor, Z-VAD-fmk (30 μM), for up to 8 h (Fig. 2a). The FOS analysis revealed that following treatment with Z-VAD-fmk, no effect on the levels of Glc3Man5GlcNAc1, a product of ERAD [10], was observed. However, the accumulation of Glc3Man7GlcNAc2 was significantly reduced. In addition to inhibiting PNGase, Z-VAD-fmk is a known caspase inhibitor. We, therefore, employed a related caspase inhibitor, Q-VD-OPh, which does not inhibit PNGase activity, to verify the specificity of the observed effect of Z-VAD-fmk. As expected, Q-VD-OPh (30 μM) did not affect the accumulation of either Glc3Man5GlcNAc1 or Glc3Man7GlcNAc2 in NB-DNJ-treated cells (Fig. 2a). We, therefore, conclude that the Glc3Man7GlcNAc2 FOS was derived from glycoprotein in the ER, suggesting lumenal exposure of glucosylated proteins to a PNGase-like activity. Indeed, this agrees with the previously proposed existence of a lumenal PNGase [30].
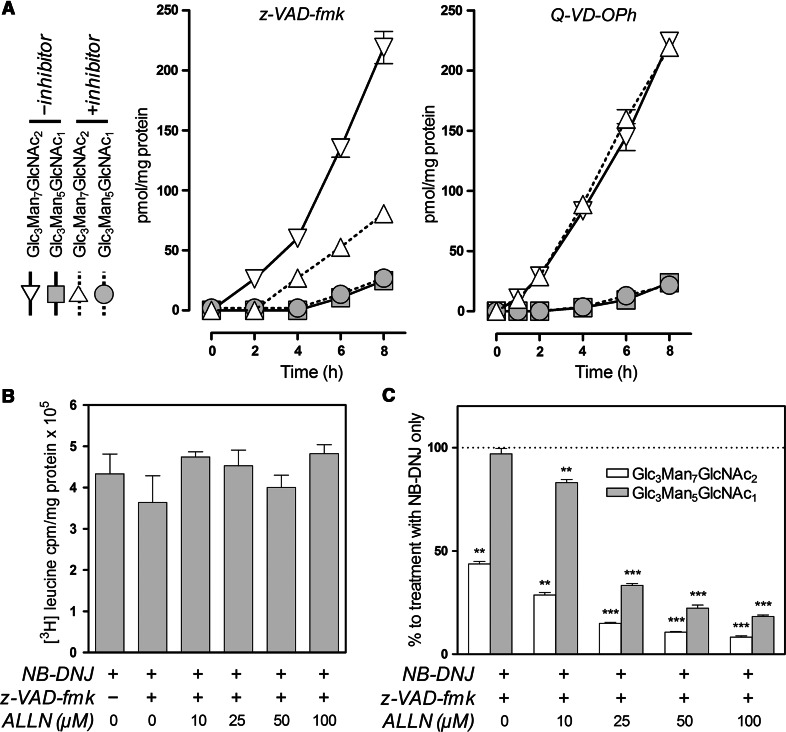
Fig. 2.
Accumulation of Glc3Man7GlcNAc2 is sensitive to PNGase and proteasome inhibitors. a MDBK cells were treated with 1 mM NB-DNJ in the presence (+inhibitor) or absence (−inhibitor) of Z-VAD-fmk or Q-VD-OPh (both: 30 μM). Cells were harvested at the times indicated, FOS were extracted and fluorescently labelled prior to analysis by NP-HPLC as described in the text. Cellular levels of Glc3Man5GlcNAc1 and Glc3Man7GlcNAc2 are plotted as a function of treatment time for each inhibitor. b [3H] leucine incorporation in MDBK cells treated with NB-DNJ (1 mM) in the presence or absence of Z-VAD-fmk (30 μM) and varying amounts of ALLN for 8 h. Differences were not significant (p > 0.05; n = 3). c Levels of Glc3Man5GlcNAc1 and Glc3Man7GlcNAc2 treated as in b, per cent to levels of the respective FOS following treatment with NB-DNJ alone. Results in all plots and histograms are mean ± SEM (n = 3). For panel c: **p < 0.01, ***p < 0.001 (two-tailed unpaired Student’s t tests)
The lack of effect on the levels of Glc3Man5GlcNAc1 following PNGase inhibition is interesting but not completely surprising since it has been previously demonstrated that in the presence of a PNGase inhibitor, the deglycosylation and subsequent proteolysis of glycoproteins such as MHC class I is not prevented unless a proteasomal inhibitor is also present [29]. On the other hand, inhibition of proteasomes itself is known to decrease the levels of glycoprotein-derived cytosolic free oligosaccharides [31]. The empirical observation is that in the presence of a PNGase inhibitor and ALLN, a reduction in ERAD and lumenal FOS is observed. The hydrolysis of a deglycosylated substrate by the proteasome could be compromised by the burden ERAD substrates produced after glucosidase inhibition. The ER-stress related mechanisms or some feedback inhibition of ubiquitinated substrates may be responsible and further insight is being explored by experiment. It would seem premature to present any mechanism at this stage other than that of proteasome involvement.
Data supporting the glycoprotein origin of glucosylated FOS species was obtained using puromycin to inhibit protein synthesis. During a 24 h incubation of inhibitor at 1 μg/ml in CHO cells, the generation of Glc3Man5GlcNAc1 and Glc3Man7GlcNAc2 FOS was reduced by greater than 60 and 90 %, respectively, compared to untreated cells (Supplemental Fig 1).
Consequently, MDBK cells were treated as before for 8 h with Z-VAD-fmk and NB-DNJ in the presence of a proteasome inhibitor, calpain inhibitor I (ALLN), at 10, 25, 50 and 100 μM. We first verified that treatment with the PNGase and proteasome inhibitors did not reduce overall rates of protein synthesis. The amount of [3H]-leucine incorporation was measured over the period of 8 h with a cocktail of inhibitors (Fig. 2b). Radioactive amino acid incorporation was standardised to cellular protein concentration and showed that, even in the presence of the cocktail of inhibitors at the highest concentration (1 mM NB-DNJ, 30 μm Z-VAD-fmk and 100 μM ALLN), the differences were not significant. The FOS were then extracted and analysed (Fig. 2c). As previously, we observed that the combination of Z-VAD-fmk and NB-DNJ only reduced the level of the Glc3Man7GlcNAc2, species. By contrast, further addition of ALLN reduced the level of both glucosylated FOS in a concentration-dependent manner, with greater than 80 % reduction in Glc3Man7GlcNAc2 at the highest ALLN concentration administered.
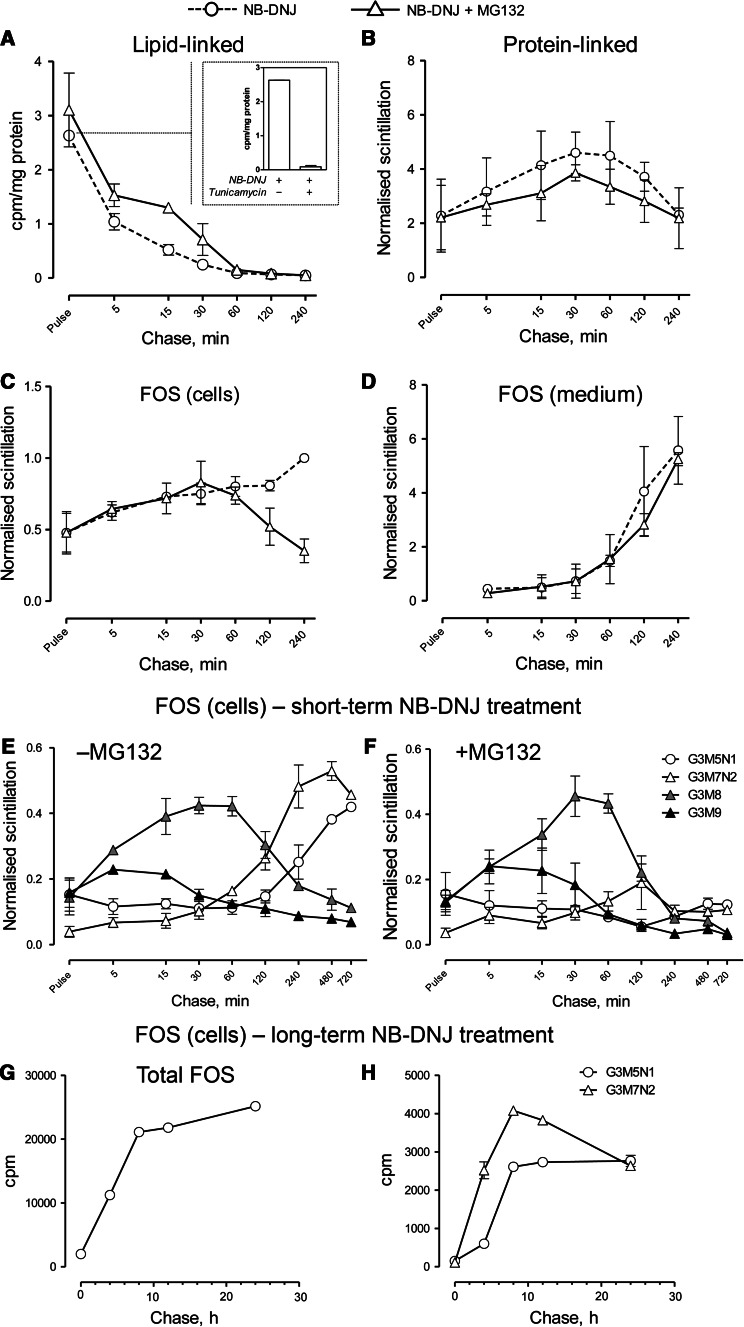
In order to further confirm the connection between ERAD and Glc3Man7GlcNAc2 accumulation in NB-DNJ-treated cells, we studied the kinetics of lipid-linked oligosaccharide release in parallel with the production of protein-linked and free oligosaccharides (Fig. 3). Following a 15-min pulse with [3H]-mannose, the level of dolichol-linked oligosaccharides decreased steadily, with the full discharge occurring between 30 and 60 min (Fig. 3a). The specificity of the analysis was confirmed by including a tunicamycin-treated control at the zero time point (Fig. 3a, inlet). The kinetics of oligosaccharide discharge from the dolichol precursor correlated strongly with those of intracellular protein glycosylation, which reached its peak at approximately 30 min and decreased slowly (Fig. 3b), most likely due to glycoprotein secretion. Neither lipid-linked oligosaccharide release, nor N-linked glycan accumulation were significantly affected by treatment with the potent, cell-permeable proteasome inhibitor, MG132. However, when cellular FOS were analysed under the same conditions, the effect of proteasome inhibition was striking (Fig. 3c). While in cells treated with NB-DNJ alone total FOS began to accumulate immediately after the pulse and continued to build up even after 4 h, in MG132-treated cells the pattern of FOS accumulation mirrored that of N-linked oligosaccharides, i.e., peaking at 30 min with a gradual drop to pulse levels by 4 h (Fig. 3c). At the same time, the secretion of glucosylated FOS into the culture medium remained unaffected following proteasome inhibition (Fig. 3d). These observations suggested the presence of two distinct populations of FOS: “early”, building up between 0 and 30 min, unaffected by proteasome inhibition and secreted into the medium, and “late”, accumulating mainly from 60 min onwards, MG132-sensitive and retained in the cells. Further FOS analysis demonstrated that the former group comprises mainly Glc3Man9GlcNAc2 (almost immediately after the pulse) and Glc3Man8GlcNAc2 (Fig. 3e–f), which are then presumably demannosylated to Glc3Man7GlcNAc2, [explaining a subtle peak at approximately 120 min in MG132-treated cells (Fig. 3f)] and secreted into the medium (Fig. 3d). At the same time, the FOS species dominant after a 24 h NB-DNJ treatment, i.e., Glc3Man5GlcNAc1 and Glc3Man7GlcNAc2, were found in the “late” MG132-sensitive population of FOS (Fig. 3e–f). Their levels continued to increase rapidly for 8 h, followed by a near-plateau (Fig. 3g). Levels of Glc3Man7GlcNAc2 decrease slightly between 8 and 24 h (Fig. 3h), mainly due to residual glucosidase activity mediating its deglucosylation to Glc1–2Man7GlcNAc2 (data not shown). We postulate that the distinction between “early” and “late” FOS and their differential sensitivity to proteasome inhibition reflects the two origins of glucosylated FOS, i.e., dolichol-derived and glycoprotein-derived. However, since the former appear to be transient and secreted into the medium at the same rate as the bulk of the glycoprotein flow (Fig. 3b–c), whereas the latter continue to accumulate for the duration of the 24 h NB-DNJ treatment in our experiments (Fig. 3g), the contribution of lipid-derived FOS to the total pool of free glycans is likely to be negligible. The reasons for cellular retention of ERAD-derived but not lipid-derived lumenal FOS, is unclear. One explanation for such distinction could be in compartmentalisation of ERAD as suggested by Lederkremer’s group [2]. Spatial separation of glycosylation and degradation could potentially account for differential behaviour of the identical free glycan molecules derived from different sources.
Fig. 3.
Biphasic production of FOS in NB-DNJ-treated CHO-K1 cells. a quantitation of lipid-linked oligosaccharide discharge. Cells were pulsed with 3H-mannose for 15 min, followed by chase in media containing NB-DNJ with or without MG132, for the periods indicated. Total LLO levels after the pulse with or without tunicamycin are shown in the inlet. b quantitation of protein-linked glycans as in a. c quantitation of total cellular FOS. d quantitation of total FOS secreted into the culture medium. e–f quantitation of individual cellular FOS. Cells treated with NB-DNJ with (e) or without (f) MG132 were pulsed and chased as in a, FOS purified, fluorescently labelled, followed by the purification of individual peaks by preparative HPLC and radioactivity quantitation. Glycans in b–f are normalised to the control total FOS value at 240 min chase in each experiment. g kinetics of long-term FOS accumulation in NB-DNJ-treated cells. h same for Glc3Man5GlcNAc1 and Glc3Man7GlcNAc2. Cells were treated as in a, followed by a chase for the time periods indicated
We therefore conclude that despite its lumenal localisation, free Glc3Man7GlcNAc2 is generated from glycoproteins by a PNGase-like activity, and the production of the glycan is linked to proteasome function in the same way as that of Glc3Man5GlcNAc1. Taken together, these findings strongly suggest that glucosylated lumenal FOS are markers of an ERAD route characterised by deglucosylation that necessarily precedes retrotranslocation.
Level of ER-localised glycan release is dependent on Golgi to ER transportation
Thapsigargin (TG) is a sesquiterpene lactone that inhibits the ATP-dependent calcium sequestration by the Ca2+/Mg2+ ATPase, causing a decrease in the level of calcium in the ER lumen. The TG is also an inhibitor of the sarco-endoplasmic reticulum Ca2+ ATPase (SERCA) [32, 33]. The SERCA inhibition increases Ca2+ removal from the ER to the cytosol. The TG is also thought to be an inhibitor of retrograde transport (ERGIC-ER) but not anterograde transport (ER-ERGIC and Golgi) [34]. Since the accumulation of Glc3Man7GlcNAc2 appeared to correlate with the deficiency in Golgi endomannosidase activity, we reasoned that the pathway leading to the production of lumenal FOS could be dependent on ER-Golgi-ER recycling of substrate glycoproteins and, hence, be sensitive to the inhibition of retrograde transport. Indeed, certain ERAD substrates have been suggested to require cycling to the Golgi and then retrograde transport to the ER to occur before the substrate is targeted for ERAD [35–37].
To this end, we assessed the effect of TG on the production of Glc3Man7GlcNAc2. We first verified, that following a 24 h treatment, the concentrations of TG used did not significantly alter the rate of protein synthesis in MDBK cells by measuring [3H]-leucine incorporation (Supplemental Table 1). Next, cells were incubated with NB-DNJ for 24 h both in the absence and presence of TG, FOS analysed as described above, and the levels of Glc3Man7GlcNAc2 measured in proportion to those of cytosolic FOS (Glc3Man5GlcNAc1) (Fig. 4a, b). It was revealed that following treatment with TG, levels of Glc3Man7GlcNAc2 were reduced significantly compared to those in cells incubated with NB-DNJ only. The effect of TG was concentration-dependent and significant at a concentration of 0.1 μM, which did not cause any detectable decrease in Glc3Man5GlcNAc1. The results were, therefore, consistent with the assumption that lumenal FOS production is dependent on retrograde Golgi-ER recycling of ERAD substrates. Similar results were obtained using endomannosidase-deficient CHO-K1 cells and an alternative retrograde transport inhibitor, brefeldin A (results not shown). Cells treated with brefeldin at 0.1 μg/ml showed a 50 % decrease in the ratio of Glc3Man7GlcNAc2 to Glc3Man5GlcNAc1 despite a more apparent reduction in protein synthesis, compared to TG treatment, as measured following [3H]-leucine incorporation.
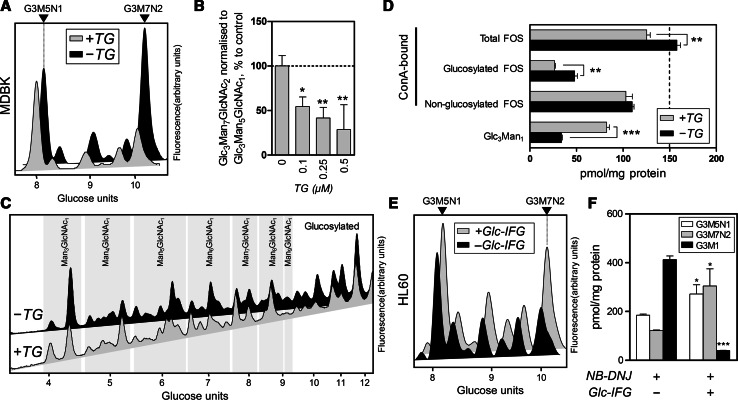
Fig. 4.
Glc3Man7GlcNAc2 accumulation is dependent on Golgi-ER transport. a NP-HPLC analysis of FOS extracted from MDBK cells treated with NB-DNJ in the presence (+TG) or absence (−TG) of thapsigargin (0.5 μM). Arrowheads indicate positions of Glc3Man5GlcNAc1 (G3M5N1) or Glc3Man7GlcNAc2 (G3M7N2). b Levels of Glc3Man7GlcNAc2 normalised to those of Glc3Man5GlcNAc1 in MDBK cells treated with NB-DNJ (1 mM) with or without TG in varying concentrations. Values are presented in per cent to treatment with NB-DNJ only. c NP-HPLC analysis of 2-AA-labelled ConA-binding FOS in BHK-21 cells incubated with (+TG) or without(−TG) thapsigargin, as well as with NB-DNJ and NBA1. Groups of peaks are structurally annotated on the diagram. Arrowheads indicate positions of Glc3Man5GlcNAc1 (G3M5N1) or Glc3Man7GlcNAc2 (G3M7N2). d Quantitation of total, glucosylated and non-glucosylated ConA-binding FOS and Glc3Man1 (non ConA-binding) oligosaccharide in BHK cells incubated with (+TG) or without (−TG) thapsigargin as described above. e NP-HPLC analysis of FOS extracted from HL60 cells treated with NB-DNJ in the presence (+Glc-IFG) or absence (−Glc-IFG) of an endomannosidase inhibitor (1 mM). f Levels of Glc3Man7GlcNAc2 (G3M7N2), Glc3Man5GlcNAc1 (G3M5N1) and Glc3Man1 (G3M1) in HL60 cells treated with NB-DNJ with or without Glc-IFG. Results in all histograms are mean ± SEM (n = 3). For panels b, d and f: *p < 0.05, **p < 0.01, ***p < 0.001 (panel a: single-sample Student’s t tests, panels d and f: two-tailed unpaired Student’s t tests)
A series of experiments was carried out to elucidate the mechanism by which thapsigargin affects FOS. First, to investigate the effect of TG in the presence of endomannosidase, FOS analysis was carried out in BHK-21 cells that have been previously shown to contain a highly active form of endomannosidase [24] (Fig. 4c). The BHK-21 cells were treated with cycloheximide for 8 h to allow complete processing of FOS already present in the cells, followed by a 24 h treatment with or without TG together with NB-DNJ and NBA1, a cytosolic mannosidase inhibitor employed to minimise cytosolic degradation of FOS [38]. Following treatment with TG and NB-DNJ, the levels of the Glc3Man1, a characteristic tetrasaccharide product of Golgi endomannosidase [39], were increased dramatically compared to the cells in the absence of TG (Fig. 4d). At the same time, we observed a decrease in the levels of glucosylated ConA-bound FOS, which contributed to a decrease in total FOS. However, the levels of non-glucosylated FOS were not altered significantly. We, therefore, speculated that since the levels of Glc3Man1 increase in TG/NB-DNJ-treated BHK-21 cells, glycoproteins in the secretory pathway are subjected to more extensive processing by the enzymes in the Golgi apparatus, including endomannosidase. This was in agreement with the known ability of TG to inhibit retrograde transport to the endoplasmic reticulum. The levels of glucosylated FOS as well as the total free oligosaccharides produced decreased, concomitant with the increase in Glc3Man1. Since the levels of deglucosylated FOS did not compensate for the decreased production of glucose-bearing species, we concluded that TG has a “bottleneck” effect on the retrograde trafficking of ERAD substrates from the post-ER compartments, limiting the rate of glycoprotein retrieval, which additionally contributes to their increased residence time in the ERGIC/Golgi and, therefore, processing by the corresponding enzymes.
Although the concentrations of thapsigargin employed in the described experiments did not cause significant alterations in cell viability, the rates of protein synthesis or the glycosylation state of secreted proteins (see below), and, furthermore, the data on BHK-21 FOS supported the inhibition of retrograde transport following thapsigargin/NB-DNJ cotreatment, a more direct confirmation of the dependence of Glc3Man7GlcNAc2 accumulation on Golgi-to-ER transport was sought. Since previous observations suggested correlation between the detectable levels of lumenal FOS and deficiency in endomannosidase activity, HL-60 cells were subjected to a cotreatment with NB-DNJ and α-Glc-1,3-isofagomine (Glc-IFG), a significantly more potent inhibitor of endomannosidase in vitro and in cells [40] than either α-Glc-1,3-deoxymannojirimycin or α-Man-1,3-deoxymannojirimycin, previously synthesised [41]. Following a 24 h incubation with the compounds, the levels of both Glc3Man5GlcNAc1 and Glc3Man7GlcNAc2 were elevated (Fig. 4e, f), supporting the previously demonstrated ability of endomannosidase to deglucosylate ERAD substrates on a global level [11]. However, the levels of lumenal FOS were elevated to a much greater extent than cytosolic species (Fig. 4e). Inhibition of endomannosidase in cells was confirmed by a dramatic drop in the levels of Glc3Man1 (Fig. 4f). While it has been previously shown that overexpressed endomannosidase affects the levels of both Glc3Man5GlcNAc1 and Glc3Man7GlcNAc2, [11], the data presented hereby demonstrate that endogenous endomannosidase preferentially deglucosylates glycoproteins to produce lumenal FOS.
Although Glc-IFG is the most potent in vitro endomannosidase inhibitor available to date [40], its Golgi accessibility is yet to be determined. We, therefore, cannot be certain of the level of inhibition of endomannosidase in Glc-IFG-treated HL60 cells. However, the observed effects of Glc-IFG/NB-DNJ co-treatment on lumenal FOS and Glc3Man1 production were significant, and we, therefore, conclude that the lumenal production of Glc3Man7GlcNAc2 is correlated with increased exposure to enzymes in the ERGIC/Golgi.
Glycosylation state of secreted proteins is unaffected by thapsigargin treatment
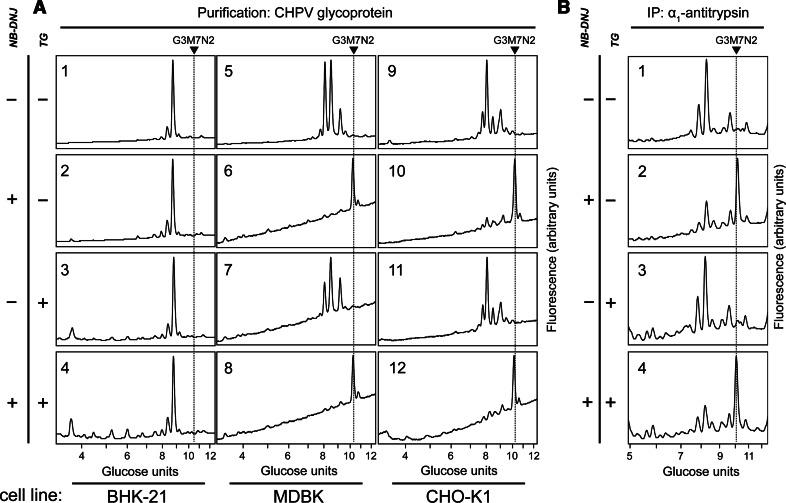
We sought to confirm that the TG-dependent reduction in the levels of Glc3Man7GlcNAc2 in CHO-K1 and MDBK cells is limited to free oligosaccharides rather than reflecting a change in the composition of protein-linked carbohydrates (for instance, by inhibiting demannosylation). For this purpose, we analysed N-linked glycans on the Chandipura virus (CHPV) glycoprotein and α1-antitrypsin following glucosidase inhibition. The CHPV contains one glycoprotein [42] (Supplemental Fig. 2) and bears close homology to the vesicular stomatitis glycoprotein, a classic model for studying the secretory pathway [43, 44]. The α1-antitrypsin is a secretory glycoprotein which has been recently shown to require Golgi-to-ER retrograde transport prior to secretion [45]. In BHK-21 cells, no qualitative changes of the glycoprotein N-linked oligosaccharides were observed for CHPV and α1-antitrypsin, indicating that the endomannosidase-mediated deglucosylation pathway fully compensates for glucosidase inhibition following NB-DNJ treatment (Fig. 5). However, in NB-DNJ treated CHO-K1 and MDBK cells, the glycans of both CHPV glycoprotein and α1-antitrypsin were almost exclusively composed of Glc3Man7GlcNAc2 N-linked oligosaccharide confirming the absence of endomannosidase activity in these cells. Importantly, the glycan composition of both glycoproteins analysed was unaltered following treatment of these cell lines with TG (Fig. 5).
Fig. 5.
Thapsigargin effect on the glycosylation state of secreted proteins. a NP-HPLC analysis of fluorescently labelled PNGase-released CHPV glycoprotein oligosaccharides. Virus was produced in BHK-21 (1–4), MDBK (5–8) or CHO-K1 (9–12) host cells, untreated (1, 5, 9) or treated with NB-DNJ (2, 4, 6, 8, 10, 12) and/or TG (3, 4, 7, 8, 11, 12). Arrows and dotted lines indicate the position of Glc3Man7GlcNAc2 on the chromatograms. b HPLC analysis of α1-antitrypsin glycosylation. CHO-K1 cells were transiently transfected with an α1-antitrypsin-encoding plasmid and incubated for 96 h in control conditions (1) or in the presence of NB-DNJ (2, 4) and/or TG (3, 4). Secreted α1-antitrypsin was immunopurified, its glycans released using PNGase and fluorescently labelled. Arrow and dotted line indicate the position of Glc3Man7GlcNAc2 on the chromatograms
We; therefore, conclude that the TG-dependent decrease in free Glc3Man7GlcNAc2 oligosaccharide is not due to qualitative changes in protein-linked oligosaccharides, but more likely due to a change in the amount of Glc3Man7GlcNAc2 released from degraded glycoproteins.
Cytosolic but not lumenal FOS-producing ERAD is affected by cytosolic mannosidase
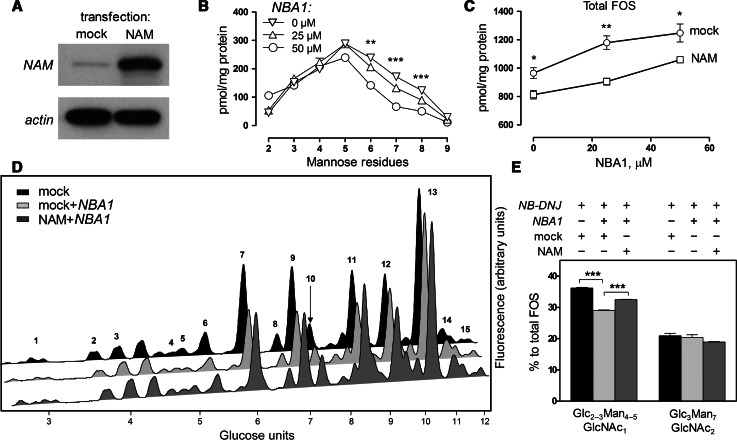
Recent reports suggest that the cytosolic neutral α-mannosidase (NAM) may have a role in ERAD. Overexpression of cytosolic mannosidase has been shown to have a stimulatory effect on glycoprotein degradation as well as the expected increase of the rates of free oligosaccharides processing [16]. Since the alternative pathway we have identified appeared, at least initially, to be confined to the endoplasmic reticulum, we asked if the effect of cytosolic mannosidase on ERAD was limited to the cytosolic FOS-producing route rather than non-selectively affecting protein glycosylation, folding and ERAD targeting. We used NBA1, an inhibitor of cytosolic mannosidase [38], in conjunction with NAM overexpression in CHO-K1 cells (Fig. 6a). We verified the antagonistic effects of NAM inhibition and overexpression by analysing FOS in mock-/NAM-transfected cells over increasing concentrations of NBA1. As expected, NBA1 caused a concentration-dependent increase in larger (Man5 to Man9) oligomannosidic FOS structures (Fig. 6b), which was attenuated following overexpression of NAM (Fig. 6c). Having confirmed this model for studying the role of NAM in manipulating FOS, we performed similar experiments under conditions where glucosidases were inhibited. Following a 48 h treatment with NB-DNJ, the major GlcNAc1-bearing oligosaccharides in the CHO-K1 FOS profile were composed of Glc2–3Man4–5GlcNAc1 (Fig. 6d). A peak at GU = 9.06 contained several glucosylated oligosaccharides, both GlcNAc1- and GlcNAc2-terminating, as well as Man9GlcNAc1–2 and, therefore, was not used for quantitation. The NBA1 caused a significant decrease in the relative levels of Glc2–3Man4–5GlcNAc1. This decrease was not compensated for by an increase in higher-mannose species (Fig. 6d, e). The effect of NBA1 on Glc3Man7GlcNAc2, however, was insignificant. Moreover, the effect of NBA1 on the GlcNAc1-terminating glucosylated FOS was partially rescued by overexpressing NAM in the treated cells, whereas the relative levels of Glc3Man7GlcNAc2 were not altered significantly. We; therefore, conclude that the production of free Glc3Man7GlcNAc2 oligosaccharide is not accessible to the cytosolic mannosidase, further demonstrating the presence of a FOS generation pathway independent from cytosolically-orientated proteasomal degradation of glycoproteins.
Fig. 6.
Cytosolic mannosidase activity affects conventional ERAD but not the alternative degradation pathway. a Immunoblot demonstrating overexpression of NAM in transfected CHO-K1 cells. b Analysis of fluorescently labelled FOS extracted from mock-transfected CHO-K1 cells incubated for 48 h with 0, 25 or 50 μM NBA1. FOS were separated using HPLC, peak areas quantified, and the corresponding amounts plotted in pmol per 1 mg total protein against the number of mannose residues in each species. c Total amounts of FOS in mock- or NAM-transfected CHO-K1 cells treated with increasing concentrations of NBA1, plotted in pmol per mg total protein. d NP-HPLC analysis of fluorescently labelled FOS extracted from mock- or NAM-transfected CHO-K1 cells, treated with NB-DNJ (1 mM) and with or without NBA1 (50 μM). Peaks were identified and structurally annotated as shown in Supplemental Table 2. e Relative quantities of Glc2–3Man4–5GlcNAc1 and Glc3Man7GlcNAc2, per cent to total FOS. Results in all plots and histograms are mean ± SEM (n = 3). For panels b, c and e: *p < 0.05, **p < 0.01, ***p < 0.001 (panel a: one-way ANOVA, panels c and e: two-tailed unpaired Student’s t test)
Lumenal FOS accumulation is unaffected by EDEM1 overexpression
The current view on ERAD attributes a major role in marking glycoproteins for degradation to EDEM1–3 (ER degradation enhancing mannosidase-like). Initially regarded as lectins binding to misfolded glycoproteins [35], EDEM factors as well as their yeast homologue Htm1p are now increasingly viewed as major modulators of ERAD targeting via specific mannose trimming of substrate glycoproteins, which serves as a signal for downstream factors such as OS-9 and XTP3-B (Yos9p in yeast) [5, 46, 47]. The latter appear to deliver the substrate to large membrane-associated complexes mediating dislocation, unfolding, ubiquitination, deglycosylation and transfer to the proteasome. Of these complexes, the most well studied is the complex comprising Hrd1 (Hrd1p) and SEL1L (Hrd3p) alongside a number of additional factors [8].
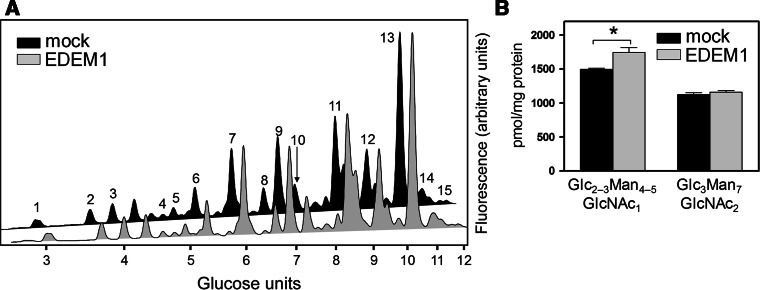
Overexpression of EDEM1 has been previously shown to enhance ERAD [48–50]. We, therefore, asked whether this enhancement would be reflected in the production of either type of FOS generated in our model. The EDEM1 of murine origin was overexpressed in CHO-K1 cells treated with NB-DNJ, and the levels of glucosylated FOS were compared to those in mock-transfected cells (Fig. 7a). The levels of Glc2–3Man4–5GlcNAc1 were significantly increased, indicating an upregulation of the ERAD pathway leading to cytosolic FOS, consistent with the assumption that these species are produced as part of the conventional EDEM/OS-9/Hrd1-dependent glycoprotein degradation route.
Fig. 7.
EDEM1 overexpression upregulates the production of cytosolic but not lumenal FOS. a HPLC analysis of fluorescently labelled FOS extracted from mock- or EDEM1-transfected CHO-K1 cells, treated with NB-DNJ (1 mM). Peaks were identified and structurally annotated as shown in Supplemental Table 2. b Relative quantities of Glc2–3Man4–5GlcNAc1 and Glc3Man7GlcNAc2, per cent to total FOS. For panel b, results in the histogram are mean ± SEM. *p < 0.05
At the same time, the levels of glucosylated GlcNAc2-terminating FOS (Glc3Man7GlcNAc2) were unaffected (Fig. 7b). We, therefore, conclude that the pathway leading to the accumulation of lumenal FOS is not sensitive to EDEM1 overexpression. The observed differences further highlight the deviation of the lumenal FOS-producing pathway from the established properties of conventional ERAD.
Discussion
In the present study, we have employed free oligosaccharides analysis as a means to dissect the pathways for ER-associated glycoprotein degradation in mammalian cells. A deglucosylation-incompetent model (endomannosidase-negative cells treated with glucosidase inhibitors) has been employed to separate and stabilise the cellular pools of FOS generated at different cellular locations. Using this approach, we have identified a novel population of glycoprotein-derived FOS that accumulated in the ER lumen, as opposed to previously characterised [10] cytosolic oligosaccharides that are produced as part of conventional ERAD following glycoprotein dislocation across the ER membrane. The retention of glucose residues on the FOS in our model system is critical for the analysis due to the perturbation in FOS trafficking caused. In particular, glucosylated FOS fail to get transported to the lysosome or traverse the ER/cytosol membrane, therefore, accumulating in the respective subcellular locations of origin [23].
In an attempt to characterise the degradation route leading to the production of lumenal FOS, we have identified several features of the pathway: (1) increased post-ER residence time/processing of the glycoprotein substrates, directly demonstrated by employing an endomannosidase inhibitor in endomannosidase-positive cells, which caused an upregulation of lumenal but not cytosolic FOS production. Increased exposure of lumenal FOS-producing substrates to post-ER compartments was additionally supported by the sensitivity of Glc3Man7GlcNAc2 to thapsigargin treatment; (2) lack of sensitivity to EDEM1 overexpression, which upregulates traffic through the conventional ERAD route; (3) lack of sensitivity to the overexpression of NAM, an ERAD-enhancing cytosolic mannosidase [16], which upregulates production of cytosolic FOS in an enzymatic activity-dependent manner.
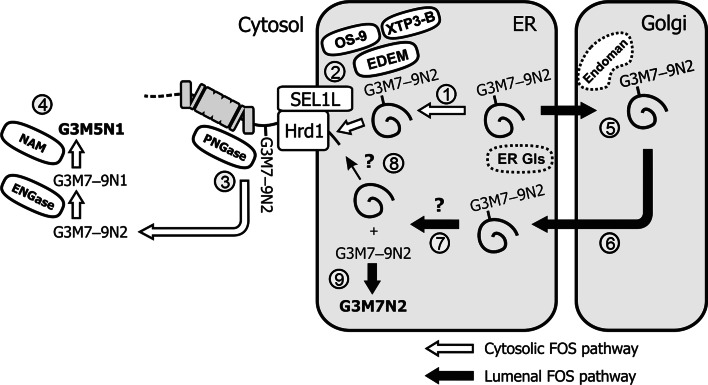
The above findings led us to propose the following model (Fig. 8). Following treatment with NB-DNJ, glucosidases (ER Gls) are unable to remove glucose residues from protein-linked carbohydrates, resulting in the production of Glc3Man7–9GlcNAc2 (G3M7–9N2) forms. Misfolded/unassembled proteins that are predominantly retained in the ER become subject to the EDEM-mediated degradation route (1), which identifies ERAD substrates and targets them to the membrane-associated ubiquitination and retrotranslocation machinery such as the Hrd1/SEL1L complex (2) [2]. Following retrotranslocation, the glycoprotein is deglycosylated by a PNGase-dependent mechanism prior to proteasomal degradation (3), resulting in cytosolic FOS. The latter are then processed first by ENGase and then NAM, resulting in highly demannosylated, GlcNAc1-bearing, glucosylated structures, e.g., Glc3Man5GlcNAc1 (G3M5N1). By contrast, a separate population of misfolded/unassembled glycoproteins is less stringently retained in the ER following glucosidase inhibition. In most cell lines, such substrates are deglucosylated by Golgi-resident endomannosidase (Endoman), but in CHO-K1 and MDBK cells post-ER deglucosylation does not occur (5). If retrogradely targeted to the ER, the described population of ERAD substrates is more likely to undergo lumenal deglycosylation (7). The enzyme responsible is currently unknown, although lumenal PNGase activity has been previously characterised, and treatment with a PNGase inhibitor negatively affects the production of lumenal FOS. The deglycosylated glycoprotein is then likely targeted for retrotranslocation (8). The lumenally-generated free glycans are either already processed by upstream-acting ER/Golgi mannosidases, or get trimmed by ER-resident mannosidase(s) following release from glycoproteins, to the end form of Glc3Man7GlcNAc2 (G3M7N2) (9). Due to the persistence of glucose residues on lumenal FOS, they are unable to escape into the cytosol as part of their catabolic route. This is opposed to FOS in endomannosidase-positive cells, that are produced downstream of Golgi-localised deglucosylation and, therefore, do not accumulate in the ER [11].
Fig. 8.
Proposed model for the lumenal- and cytosolic FOS-producing ERAD pathways. See explanations in the text
By employing FOS analysis coupled with blocked deglucosylation in mammalian cells, we have identified a novel trafficking pathway that precedes ER-associated degradation of glycoproteins. Although certain features of this pathway have been established, several open questions remain. It is currently unclear whether the substrates undergoing both the conventional cytosolic FOS-producing and the novel lumenal FOS-producing pathways represent different glycoproteins, or rather different populations of the same glycoproteins. Furthermore, although current evidence suggests the role for Golgi trafficking in predisposing ERAD substrates for the lumenal FOS route, the mechanisms of such targeting are unclear.
The biological significance of the existence of an alternative ERAD pathway dependent on Golgi recycling of glycoproteins is an additional question that arises from our findings. Although undisputed in yeast [36, 51, 52], post-ER quality control has not been unequivocally demonstrated in mammalian cells. It has been speculated that the absence of Golgi-localised folding checkpoints in higher eukaryotes arose from increasing complexity of quality control in the ER [53]. The presently demonstrated pathway could hypothetically depend on post-ER glycoprotein sorting and therefore represent a route for identification of misfolded ER-escaping glycoproteins in mammalian cells. Whether rudimentary or functional in vivo, this pathway would be induced by employing glucosidase inhibitors, since in these conditions calnexin-mediated ER retention of substrate glycoproteins would be alleviated. The prominence of the lumenal FOS pathway in NB-DNJ-treated CHO-K1/MDBK cells could, therefore, be partially a consequence of glucosidase inhibition itself. Nevertheless, the existence of a recycling route leading to a separate degradation mechanism poses interest from the evolutionary viewpoint, particularly in light of the hypothetical post-ER quality control.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors would like to thank Prof. Pirkko Heikinheimo and Elina Kuokkanen, as well as Dr. John Christianson, for providing plasmids. Dr. Anthony Marriott is thanked for providing CHPV. Amicus Therapeutics are thanked for providing a chemical reagent. The authors would like to thank Oxford Glycobiology Institute and the Australian Research Council for financial support and Clarendon Fund/New College (Robert Lyns) for support of NVK.
Footnotes
D.S. Alonzi and N.V. Kukushkin contributed equally to this work.
References
- 1.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 2.Lederkremer GZ. Glycoprotein folding, quality control and ER-associated degradation. Curr Opin Struct Biol. 2009;19:515–523. doi: 10.1016/j.sbi.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 5.Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006;8:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- 6.Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 7.Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143:579–591. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki T. Cytoplasmic peptide:N-glycanase and catabolic pathway for free N-glycans in the cytosol. Semin Cell Dev Biol. 2007;18:762–769. doi: 10.1016/j.semcdb.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 10.Alonzi DS, Neville DC, Lachmann RH, Dwek RA, Butters TD. Glucosylated free oligosaccharides are biomarkers of endoplasmic- reticulum alpha-glucosidase inhibition. Biochem J. 2008;409:571–580. doi: 10.1042/BJ20070748. [DOI] [PubMed] [Google Scholar]
- 11.Kukushkin NV, Alonzi DS, Dwek RA, Butters TD. Demonstration that endoplasmic reticulum-associated degradation of glycoproteins can occur downstream of processing by endomannosidase. Biochem J. 2011;438:133–142. doi: 10.1042/BJ20110186. [DOI] [PubMed] [Google Scholar]
- 12.Mellor HR, Neville DC, Harvey DJ, Platt FM, Dwek RA, Butters TD. Cellular effects of deoxynojirimycin analogues: inhibition of N-linked oligosaccharide processing and generation of free glucosylated oligosaccharides. Biochem J. 2004;381:867–875. doi: 10.1042/BJ20031824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore SE, Spiro RG. Intracellular compartmentalization and degradation of free polymannose oligosaccharides released during glycoprotein biosynthesis. J Biol Chem. 1994;269:12715–12721. [PubMed] [Google Scholar]
- 14.Spiro RG. Role of N-linked polymannose oligosaccharides in targeting glycoproteins for endoplasmic reticulum-associated degradation. Cell Mol Life Sci. 2004;61:1025–1041. doi: 10.1007/s00018-004-4037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore SE. Oligosaccharide transport: pumping waste from the ER into lysosomes. Trends Cell Biol. 1999;9:441–446. doi: 10.1016/S0962-8924(99)01648-7. [DOI] [PubMed] [Google Scholar]
- 16.Bernon C, Carre Y, Kuokkanen E, Slomianny MC, Mir AM, Krzewinski F, Cacan R, Heikinheimo P, Morelle W, Michalski JC, Foulquier F, Duvet S. Overexpression of Man2C1 leads to protein underglycosylation and upregulation of endoplasmic reticulum-associated degradation pathway. Glycobiology. 2011;21:363–375. doi: 10.1093/glycob/cwq169. [DOI] [PubMed] [Google Scholar]
- 17.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 18.Saint-Pol A, Bauvy C, Codogno P, Moore SE. Transfer of free polymannose-type oligosaccharides from the cytosol to lysosomes in cultured human hepatocellular carcinoma HepG2 cells. J Cell Biol. 1997;136:45–59. doi: 10.1083/jcb.136.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saint-Pol A, Codogno P, Moore SE. Cytosol-to-lysosome transport of free polymannose-type oligosaccharides. Kinetic and specificity studies using rat liver lysosomes. J Biol Chem. 1999;274:13547–13555. doi: 10.1074/jbc.274.19.13547. [DOI] [PubMed] [Google Scholar]
- 20.Gross V, Tran-Thi TA, Schwarz RT, Elbein AD, Decker K, Heinrich PC. Different effects of the glucosidase inhibitors 1-deoxynojirimycin, N-methyl-1-deoxynojirimycin and castanospermine on the glycosylation of rat alpha 1-proteinase inhibitor and alpha 1-acid glycoprotein. Biochem J. 1986;236:853–860. doi: 10.1042/bj2360853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lodish HF, Kong N. Glucose removal from N-linked oligosaccharides is required for efficient maturation of certain secretory glycoproteins from the rough endoplasmic reticulum to the Golgi complex. J Cell Biol. 1984;98:1720–1729. doi: 10.1083/jcb.98.5.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lubas WA, Spiro RG. Golgi endo-alpha-d-mannosidase from rat liver, a novel N-linked carbohydrate unit processing enzyme. J Biol Chem. 1987;262:3775–3781. [PubMed] [Google Scholar]
- 23.Moore SE, Bauvy C, Codogno P. Endoplasmic reticulum-to-cytosol transport of free polymannose oligosaccharides in permeabilized HepG2 cells. EMBO J. 1995;14:6034–6042. doi: 10.1002/j.1460-2075.1995.tb00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karaivanova VK, Luan P, Spiro RG. Processing of viral envelope glycoprotein by the endomannosidase pathway: evaluation of host cell specificity. Glycobiology. 1998;8:725–730. doi: 10.1093/glycob/8.7.725. [DOI] [PubMed] [Google Scholar]
- 25.Neville DC, Coquard V, Priestman DA, Te Vruchte DJ, Sillence DJ, Dwek RA, Platt FM, Butters TD. Analysis of fluorescently labeled glycosphingolipid-derived oligosaccharides following ceramide glycanase digestion and anthranilic acid labeling. Anal Biochem. 2004;331:275–282. doi: 10.1016/j.ab.2004.03.051. [DOI] [PubMed] [Google Scholar]
- 26.Olafson RW, Thomas JR, Ferguson MA, Dwek RA, Chaudhuri M, Chang KP, Rademacher TW. Structures of the N-linked oligosaccharides of Gp63, the major surface glycoprotein, from Leishmania mexicana amazonensis. J Biol Chem. 1990;265:12240–12247. [PubMed] [Google Scholar]
- 27.Karlsson GB, Butters TD, Dwek RA, Platt FM. Effects of the imino sugar N-butyldeoxynojirimycin on the N-glycosylation of recombinant gp120. J Biol Chem. 1993;268:570–576. [PubMed] [Google Scholar]
- 28.Gao N, Lehrman MA. Analyses of dolichol pyrophosphate–linked oligosaccharides in cell cultures and tissues by fluorophore-assisted carbohydrate electrophoresis. Glycobiology. 2002;12:353–360. doi: 10.1093/glycob/12.5.353. [DOI] [PubMed] [Google Scholar]
- 29.Misaghi S, Pacold ME, Blom D, Ploegh HL, Korbel GA. Using a small molecule inhibitor of peptide: N-glycanase to probe its role in glycoprotein turnover. Chem Biol. 2004;11:1677–1687. doi: 10.1016/j.chembiol.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 30.Chantret I, Moore SE. Free oligosaccharide regulation during mammalian protein N-glycosylation. Glycobiology. 2008;18:210–224. doi: 10.1093/glycob/cwn003. [DOI] [PubMed] [Google Scholar]
- 31.Karaivanova VK, Spiro RG. Effect of proteasome inhibitors on the release into the cytosol of free polymannose oligosaccharides from glycoproteins. Glycobiology. 2000;10:727–735. doi: 10.1093/glycob/10.7.727. [DOI] [PubMed] [Google Scholar]
- 32.Kijima Y, Ogunbunmi E, Fleischer S. Drug action of thapsigargin on the Ca2+ pump protein of sarcoplasmic reticulum. J Biol Chem. 1991;266:22912–22918. [PubMed] [Google Scholar]
- 33.Lytton J, Westlin M, Hanley MR. Thapsigargin inhibits the sarcoplasmic or endoplasmic reticulum Ca-ATPase family of calcium pumps. J Biol Chem. 1991;266:17067–17071. [PubMed] [Google Scholar]
- 34.Ying M, Sannerud R, Flatmark T, Saraste J. Colocalization of Ca2+-ATPase and GRP94 with p58 and the effects of thapsigargin on protein recycling suggest the participation of the pre-Golgi intermediate compartment in intracellular Ca2+ storage. Eur J Cell Biol. 2002;81:469–483. doi: 10.1078/0171-9335-00266. [DOI] [PubMed] [Google Scholar]
- 35.Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caldwell SR, Hill KJ, Cooper AA. Degradation of endoplasmic reticulum (ER) quality control substrates requires transport between the ER and Golgi. J Biol Chem. 2001;276:23296–23303. doi: 10.1074/jbc.M102962200. [DOI] [PubMed] [Google Scholar]
- 37.Ahner A, Brodsky JL. Checkpoints in ER-associated degradation: excuse me, which way to the proteasome? Trends Cell Biol. 2004;14:474–478. doi: 10.1016/j.tcb.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 38.Butters TD, Alonzi DS, Kukushkin NV, Ren Y, Bleriot Y. Novel mannosidase inhibitors probe glycoprotein degradation pathways in cells. Glycoconj J. 2009;26:1109–1116. doi: 10.1007/s10719-009-9231-3. [DOI] [PubMed] [Google Scholar]
- 39.Durrant C, Moore SE. Perturbation of free oligosaccharide trafficking in endoplasmic reticulum glucosidase I-deficient and castanospermine-treated cells. Biochem J. 2002;365:239–247. doi: 10.1042/BJ20011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson AJ, Williams RJ, Hakki Z, Alonzi DS, Wennekes T, Gloster TM, Songsrirote K, Thomas-Oates JE, Wrodnigg TM, Spreitz J, Stutz AE, Butters TD, Williams SJ, Davies GJ. Structural and mechanistic insight into N-glycan processing by endo-alpha-mannosidase. Proc Natl Acad Sci USA. 2012;109:781–786. doi: 10.1073/pnas.1111482109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ardron H, Butters TD, Platt FM, Wormald MR, Dwek RA, Fleet GWJ, Jacob GS. Synthesis of 1,5-dideoxy-3-O-(alpha-d-mannoyranosyl)-1,5-imino-d-mannitol and 1,5-dideoxy-3-O-(alpha-d-glucopyranosyl)-1,5-imino-d-mannitol: powerful inhibitors of endomannosidase. Tetrahedron Asymmetry. 1993;4:2011–2024. doi: 10.1016/S0957-4166(00)82250-8. [DOI] [Google Scholar]
- 42.Marriott AC. Complete genome sequences of Chandipura and Isfahan vesiculoviruses. Arch Virol. 2005;150:671–680. doi: 10.1007/s00705-004-0452-2. [DOI] [PubMed] [Google Scholar]
- 43.Lodish HF, Kong N, Snider M, Strous GJ. Hepatoma secretory proteins migrate from rough endoplasmic reticulum to Golgi at characteristic rates. Nature. 1983;304:80–83. doi: 10.1038/304080a0. [DOI] [PubMed] [Google Scholar]
- 44.Zilberstein A, Snider MD, Porter M, Lodish HF. Mutants of vesicular stomatitis virus blocked at different stages in maturation of the viral glycoprotein. Cell. 1980;21:417–427. doi: 10.1016/0092-8674(80)90478-X. [DOI] [PubMed] [Google Scholar]
- 45.Reiterer V, Nyfeler B, Hauri HP. Role of the lectin VIP36 in post-ER quality control of human alpha1-antitrypsin. Traffic. 2010;11:1044–1055. doi: 10.1111/j.1600-0854.2010.01078.x. [DOI] [PubMed] [Google Scholar]
- 46.Clerc S, Hirsch C, Oggier DM, Deprez P, Jakob C, Sommer T, Aebi M. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol. 2009;184:159–172. doi: 10.1083/jcb.200809198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quan EM, Kamiya Y, Kamiya D, Denic V, Weibezahn J, Kato K, Weissman JS. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol Cell. 2008;32:870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hosokawa N, Wada I, Natsuka Y, Nagata K. EDEM accelerates ERAD by preventing aberrant dimer formation of misfolded alpha1-antitrypsin. Genes Cells. 2006;11:465–476. doi: 10.1111/j.1365-2443.2006.00957.x. [DOI] [PubMed] [Google Scholar]
- 49.Oda Y, Okada T, Yoshida H, Kaufman RJ, Nagata K, Mori K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J Cell Biol. 2006;172:383–393. doi: 10.1083/jcb.200507057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ron E, Shenkman M, Groisman B, Izenshtein Y, Leitman J, Lederkremer GZ. Bypass of glycan-dependent glycoprotein delivery to ERAD by upregulated EDEM1. Mol Biol Cell. 2011;22:3945–3954. doi: 10.1091/mbc.E10-12-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vashist S, Kim W, Belden WJ, Spear ED, Barlowe C, Ng DT. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J Cell Biol. 2001;155:355–368. doi: 10.1083/jcb.200106123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vashist S, Ng DT. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arvan P, Zhao X, Ramos-Castaneda J, Chang A. Secretory pathway quality control operating in Golgi, plasmalemmal, and endosomal systems. Traffic. 2002;3:771–780. doi: 10.1034/j.1600-0854.2002.31102.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.