Abstract
Genomes are transcribed well beyond the conventionally annotated protein-encoding genes and produce many thousands of regulatory non-coding RNAs (ncRNAs). In the last few years, ncRNAs, especially microRNAs and long non-coding RNA, have received increasing attention because of their implication in the function of chromatin-modifying complexes and in the regulation of transcriptional and post-transcriptional events. The morphological events and the genetic networks responsible for the development of sensory organs have been well delineated and therefore sensory organs have provided a useful scenario to address the role of ncRNAs. In this review, we summarize the current information on the importance of microRNAs and long non-coding RNAs during the development of the eye, inner ear, and olfactory system in vertebrates. We will also discuss those cases in which alteration of ncRNA expression has been linked to pathological conditions affecting these organs.
Keywords: MicroRNA, Long non-coding RNA, Inner ear, Eye, Olfactory system
Introduction
The continuous improvement of high-throughput technology is providing constantly evolving information on the structure and function of the genome of different species and a high-resolution map of their transcriptional landscape. The resulting picture shows that genomes are at the same time complex and flexible [1], perhaps with the surprise that they are transcribed well beyond the conventionally annotated protein-encoding genes. Indeed, genomes produce many thousands of regulatory non-coding RNAs (ncRNAs). These includes “housekeeping” ncRNAs transcripts (ribosomal RNA, transfer RNA, small nuclear RNA, and small nucleolar RNA), and “regulatory ncRNAs”, including long non-coding RNAs, microRNAs, piRNAs, natural antisense transcripts (NATs), [1–3] and several other poorly characterized ncRNAs that derive from the transcription of expression control sequence elements [4, 5]. Regulatory ncRNAs display dynamic spatial and temporal expression profiles in specific cellular contexts and contribute to tissue patterning and to the control of different cellular programs, such as cell proliferation, differentiation, migration, or apoptosis [6–8]. Table 1 lists a few examples of the main classes of transcripts that have been ascribed to the ever-growing group of regulatory ncRNAs.
Table 1.
Examples of classes of non-coding RNAs
| Class | Description | References |
|---|---|---|
| A) Small ncRNAs | ||
| miRNAs | Small ncRNAs involved with RISC complex in post-transcriptional silencing of protein-coding genes | [182, 183] |
| Piwi-interacting RNA (piRNA) | Small-ncRNA often linked to both post-transcriptional and epigenetic gene silencing of retro-transposons | [184] |
| tiRNAs | Tiny RNAs associated with transcription start sites | [185] |
| Enhancer-like long ncRNAs | lncRNAs displaying enhancer activity, mainly for genes regulating development and differentiation | [186] |
| snoRNAs | Small nucleolar RNAs implicated principally in directing chemical modifications of other RNAs | [187] |
| PROMPTs | Promoter upstream transcripts located upstream of active transcription start sites | [4] |
| TSSa-RNAs | Transcription start site-associated ncRNAs probably linked to the maintenance of gene transcription | [188] |
| B) Long ncRNAs | ||
| T-UCR | Non-coding RNAs transcribed from ultra-conserved regions | [189] |
| NATs | Natural antisense transcripts located in intergenic regions and organized in antisense with protein-coding genes | [190] |
| lincRNAs | Long intergenic non-coding RNAs generally associated post-transcriptional silencing by blocking protein production | [191] |
| ceRNAs | Competing endogenous ncRNAs involved in modulating RNA transcripts by competing for microRNAs | [56] |
ncRNAs have received increasing attention because of their implication in the function of chromatin-modifying complexes and in the regulation of transcriptional and post-transcriptional events [3]. In this review, we will summarize the evidence that ncRNAs contribute to regulate the development and survival of sensory tissues development, paying particular attention to microRNAs and lncRNAs.
Biogenesis and function of non-coding RNAs
MicroRNAs (miRNAs) are a class of small endogenous, single-stranded, non-coding RNAs [9] that were first identified with classical genetic approaches as regulators of Caenorhabditis elegans development [10, 11]. Their existence in eukaryotic organisms was thereafter determined on the basis of sequence conservation across species [12]. miRNAs are transcribed as longer primary transcripts (pri-miRNAs) and processed first into ~70-nucleotide (nt)-long precursors (pre-miRNAs) and then into 20–25 nt mature miRNAs by the subsequent activity of two RNAse III-like endoribonucleases, namely Drosha and Dicer [13]. The best-studied function of mature miRNAs is the control of gene dosage at the post-transcriptional level [14–16]. miRNAs bind specific proteins with catalytic activity, called Argonaute, and form a miRNA-induced silencing complex that impairs either translation or mRNA stability by binding, with imperfect base pairing, to specific sites of the 3′-untranslated regions (3′ UTRs) of their target mRNAs [17]. Occasionally, they have being found also binding to the coding regions or the 5′ UTRs of mRNAs [18–20]. In transcriptional repression events, miRNAs guide heterochromatin formation at promoter regions by both imperfectly and perfectly matched double-stranded RNA–DNA interaction [21, 22]. Notably, miRNAs have been also detected as extracellular nuclease-resistant entities, enclosed in small exosomal vesicles or packaged with RNA-binding proteins [23]. These forms of miRNAs have been proposed to act as secreted signaling molecules that bind and activate receptors to influence the phenotype of the recipient cells [24].
Long non-coding RNAs (lncRNAs; Table 1) are molecules longer than 2 kb, with a coding potential of less than 100 amino acids [2, 25–29]. The biological relevance of these transcripts is supported by their developmental regulation [30–32], cell-specific expression pattern [33, 34], sub-cellular distribution [35–38], and possible association with human diseases [39–41]. lncRNAs can have an intragenic or, more often, an intergenic genomic localization. In many instances, they are localized in the vicinity of protein-coding genes, with respect to which they can be organized in sense or antisense orientation. In the latter case, they are referred to as NATs or opposite-strand (OS) transcripts. The majority of lncRNAs are transcribed from the nucleus or, with less frequency, from the mitochondrial genome [42]. As it happens with coding genes, lncRNAs undergo post-transcriptional processing, including 5′ capping, alternative splicing, RNA editing, and polyadenylation [25, 28]. The activity of only a small fraction of the lncRNAs so far identified has been experimentally defined, revealing a wide variety of functions, which, at present, can be summarized as follows. (1) Modulation of chromatin structure at specific genomic sites by recruitment of histone and chromatin modifying complexes. One of the most representative examples occurs during X chromosome inactivation. The lncRNA named “Xist” physically associates with the Polycomb repressive complex 2 (PRC2), resulting in the localization of the PRC2 and H3K27me3 (histone H3 trimethylated at lysine 27) to the inactive X chromosome [43]. Similarly, other lncRNAs (i.e., Air, Kcnq1ot1, HOTAIR etc.) mediate genomic imprinting in cis and/or in trans, thereby regulating gene expression in a genome-wide scale through the association with chromatin-modifying complexes [44–46]. (2) Recruitment of transcription factors to chromatin, as in the case of Mll1-dependent transcriptional activation of Hoxa6 and Hoxa7 by the lncRNA Mistral in differentiating ES cells during mouse germ layer specification [47]. (3) Modulation of nuclear-cytoplasmic trafficking, as in the case of NRON, the non-coding repressor of nuclear factor of the activated T cells (NFAT), which seems to act as a modulator of NFAT nuclear trafficking [48]. (4) Control of intracellular compartmentalization: the lncRNA nuclear-enriched autosomal transcript 1 contributes to the formation of “paraspeckles”, dynamic structures of the interchromatin space [35, 49]. (5) Modulation of post-transcriptional RNA processing: lncRNAs have been shown to bind with partial base-pairing to complementary sequences present in the 3′ UTR of specific mRNAs, thus creating a recognition site for Staufen, which is a protein that binds double-stranded mRNA and induces its decay. Therefore, lncRNAs represent an unexpected mean to recruit proteins to mRNAs and thus promote its degradation [50]. By contrast, the mRNA of BACE1, a β-secretase responsible β-amyloid production, is stabilized and protected from RNAse cleavage by base-pairing of its antisense (BACE1-AS) [51]. (6) Modulation of local protein synthesis. The antisense AS-Uchl1, transcribed in the opposite strand of the Ubiquitin carboxy-terminal hydrolase L1 (Uchl1) gene, can specifically modulate the translation of Uchl1 through an embedded a short interspersed nuclear element (SINE) B2 repeat and by binding, with perfect base pairing, to the first 73 nucleotides of the sense mRNA [52]. (7) Interference with miRNA action: this is the case of the emerging class of competing endogenous RNAs (ceRNAs) that, by sequestering microRNAs, can regulate mRNA transcripts containing common microRNA recognition elements [53]. Through the above mechanism, ceRNAs (also known as “target mimicry” in plants) [54] have been found to play a role in the genesis of cancer [55] and in cell differentiation [56]. ceRNAs have been also proposed to orchestrate the crosstalk between RNAs [57], which is a so-far-unexplored large-scale regulatory network across the transcriptome.
By means of one or more of the above-described modalities, lncRNAs can play significant roles in the development and differentiation of several organs and tissues, particularly in the CNS. One of the first reported examples is represented by Evf2, which, through an in vivo loss-of-function approach, was found to be essential for the proper development of GABAergic neurons [58]. In non-neural tissues, the muscle-specific ceRNA linc-MD1 acts as an endogenous decoy for miRNAs, thereby controlling the timing of muscle cell differentiation [56], whereas the lncRNA Braveheart (Bvht) is required for the commitment of the cardiovascular lineage [59]. Specific examples of lncRNAs with a role in cell differentiation of sensory organs will be reported below.
ncRNAs associated to the development of the inner ear and to deafness and balance disorders
The vertebrate inner ear is an elaborate organ that mediates different aspects of hearing, motion, and space orientation. The mammalian inner ear is actually composed of six distinct sensory organs: the three cristae of the semicircular canals, the two maculae of the saccule and utricle and the organ of corti in the cochlea. The latter structure mediates hearing, whereas the cristae and the maculae are vestibular structures that respond to angular and linear acceleration, respectively. These three types of organs have a similar and relatively simple epithelial conformation, with two basic cell types: the sensory hair cells and the surrounding non-sensory supporting cells. These epithelia are covered at their apical surface with an extracellular structure known as cupula (cristae), otoconial (macula), or tectorial (organ of corti) membrane. The supporting cells are adhered to a basal lamina and surround with their lateral membranes the hair cells, which are thus isolated from one another and from the basal lamina. The apical portion of the supporting cells are connected one another and with the adjacent hair cells by tight and adherens junctions. A hair bundle, composed of highly specialized microvilli that detect mechanical stimuli and transduce them into electrical signals, occupies the apical surface of the hair cells [60].
All the structures of the inner ear develop from the otic placode, a thickening of the head ectoderm adjacent to the rhombencephalon. The placode invaginates to form the otic pit, which detaches from the ectoderm to form a pear-shaped otocyst. A patch in the ventromedial wall of the otocyst, characterized by the expression of the Notch signaling components Serrate1 and Lunatic fringe [61], acquires pro-sensory characteristics and then differentiate into the six sensory organs described above, under the influence of the transcription factors Pax2, Dlx5, Otx1, and Hmx3, among many other regulators (see [60, 62–64], for up-to-date reviews). The otocyst also gives rise to the neurons that innervate the hair cells of the six organs. These neurons form through a process of delamination that takes place before the formation of the sensory organs. A complex series of morphogenetic events also contributes to the final shape of the inner ear [65, 66].
Most of the events implicated in the development of the inner ear as well as in its adult homeostasis require the activity of a large number of miRNAs (Fig. 1) [67]. Indeed, robust expression of hundreds of miRNAs, distributed with precise cellular and temporal patterns, has been detected in the auditory and vestibular systems at embryonic, postnatal, and adult stages [68–74].
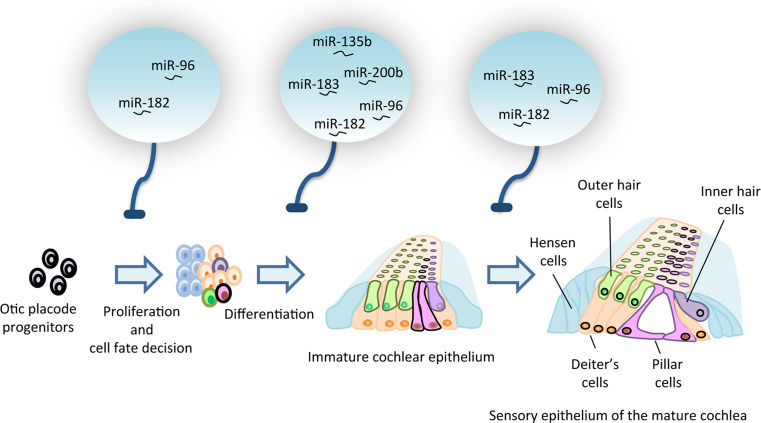
Fig. 1.
Role of miRNAs in inner ear development. The drawing depicts the development of the organ of Corti viewed in the transversal plane, showing the relative position of the inner hair cells (violet) and the three rows of sensory outer hair cells (green). The pale blue circles on the top indicate the main miRNAs implicated in the control of the different steps of inner ear development and its homeostatic role in the mature organ (see text for further details)
Pioneer studies aimed at defining the function of miRNAs in the developing inner ear of the mouse took advantage of the conditional inactivation of Dicer, mediated by Pax2-Cre, which is transiently expressed in the otic placode and thus leads to enzyme depletion in most sensory, supporting, and hair cells [75]. The otic sensory neurons of the resulting embryos formed normally but rapidly underwent apoptosis [76], similarly to what was observed for the hair and sensory cells of the cochlea after Pou4f3- or Atoh1-Cre-mediated inactivation of Dicer [71, 77]. In contrast to this restricted defects, Foxg1-Cre conditional inactivation of Dicer leads to broader defects with an evident disruption of inner ear development marked by extensive cell death [78]. These studies soon led to hypothesize that general miRNA dysfunction could contribute to cochlear dysfunction and disease, such as age-related degeneration and noise- or toxic cochlear damage [79–82].
In a few cases, the role of individual miRNAs in inner ear development/function has also been dissected (Table 2). Among the hundreds of miRNAs expressed in developing and adult ear tissues, the miR-183 cluster is one of the most studied. This cluster is composed of three miRNAs, miRs-183, -182, and -96 that originate from a common primary transcript [68, 74, 77, 83, 84]. Their seed region is identical, suggesting potential common properties in mRNA target recognition. However, the involvement of additional miRNA nucleotides critical for target recognition downstream the seed sequence likely diversifies their cellular functions [85]. The expression of miR-182 and -183 begins at E9.5 in the otic vesicles and then the expression of the entire cluster, including miR-96, becomes localized to the sensory cells of the cochlea, the vestibular end-organs, and the spiral and vestibular ganglia to become finally restricted to the inner sulcus and the spiral limbus of the mature cochlea [77, 83]. After an ENU-induced mutation of the miR-96 seed region, hair cell development begins normally but irregular bundles and persistent clusters of ectopic stereocilia appear at late postnatal stages, followed by progressive cell degeneration and hearing loss [86]. These observations suggest that the functions of the three members of the miR-183 cluster are not completely redundant, although the precise reasons are unclear [86, 87]. It is possible that the mutated miR-96 recognizes novel target and/or that the remaining members of the miR-183 cluster fail to compensate miR-96 activity, despite their similar seed regions. This would imply that a threshold and balanced level of all members of the cluster is needed for proper inner-ear development. This idea is further supported by single and combined knock-down of the expression of members of this cluster in zebrafish embryos, which led to hair cell loss, the magnitude of which is strictly related to the number of affected miRNAs. By contrast, over-expression of miR-182 or -96 causes the growth of extra or ectopic hair cells, whereas injection of miR-183 has no effect [87].
Table 2.
List of the main miRNAs, their proposed target genes, and regulated events during sensory organ development
| MicroRNAs | Target genes | Function |
|---|---|---|
| miRNAs associated to the development of the inner ear | ||
| miR-182; miR-183; miR-96 | T-box1 | Regulation of inner ear progenitor cell differentiation [88] |
| miR-182; miR-183; miR-96 | Sox2 | Hair cell fate specification and differentiation [77] |
| miR-135b | PSIP1 | Regulation of cochlear and vestibular hair cell differentiation and maintenance [70] |
| miRNAs associated to eye development | ||
| miR-24a | Casp9 and Apaf1 | Repression of apoptosis in the developing neural retina [122] |
| miR-124 | Sema3A | Control of the sensitivity of retinal growth cones to the guidance cue Sema3A [125] |
| miR-124a | Lhx2 | Control of the maturation and survival of retinal cone photoreceptors [133] |
| miR-204 | Meis2 | Regulation of lens and retinal development [127] |
| miR-133b | Pitx3 | Control of the maturation and function of dopaminergic amacrine cells [135] |
| miR-218 | Robo1, Robo2, and GLCE | Regulation of normal vascularization of the retina [137] |
| miRNAs associated to olfactory system development | ||
| miR-7a | Pax6 | Regulation of the differentiation of dopaminergic neurons in the olfactory system [178] |
The genes targeted by this cluster in the ear are poorly characterized, but some information has been obtained using cancer cell lines. Aquaporin5, Myosin-, and Rab-interacting protein, a member of the protein kinase A-anchoring family, the outer dense fiber protein, a critical component of the centrosome, and two components of the non-canonical Wnt signaling pathway, Celsr2 and Ryk, have been biologically validated as miR-96 targets [86]. MiR-182 instead modulates T-box1 gene expression and this in turn has a significant impact on the genetic control of otocyst-derived cell differentiation [88]. Targets of the entire miR-183 family are two genes expressed in hair cells: the transcription factor Sox2, which may repress hair cell differentiation [77], and the chloride intracellular channel 5 involved in hair cell function [89].
Besides the miR-183 cluster, other miRNAs have been shown to contribute to inner ear development and function. The auditory and vestibular defects of the Twirler mice, affected by deafness and balance disorders, have been associated to ZEB1/miR-200b activity [90]. Furthermore, miR-135b inhibits the translation of the transcription factor PSIP1, thereby regulating the identity, differentiation, and homeostasis of cochlear and vestibular hair cells [70], whereas miR-181a diminishes hair cell proliferation, which occurs in response to ototoxic injury in cultured avian cochlea [91].
Probably, the most compelling evidence for the relevance of miRNAs in inner ear homeostasis is the identification of a mutation in the seed region of miR-96, which is responsible for autosomal dominant non-syndromic deafness in humans. Notably, this finding also represented the first example of a hereditary disease caused by a mutation in a miRNA [92]. A subsequent mutational screening that took into account the three members of the miR-183 cluster identified an additional mutation but again in miR-96 [93]. MiRNAs likely contribute to other ear pathological conditions. For example, miR-21 has been associated with the development of human vestibular schwannomas [94] and with the growth and proliferation of cholesteatoma [95], epidermal cysts that can lead to loss of auditory functions.
More than a thousand lncRNAs have been identified in the mouse genome and a good proportion of them is specifically expressed in the nervous system [34]. Unfortunately, the information related to their function during inner ear development is limited to the observation that disruption of Rubie, a lncRNA upstream of Bmp4 expression, causes vestibular developmental defects with an undetermined molecular mechanism [96].
Non-coding RNAs in eye development and ocular degenerative diseases
The eye is a bilateral organ that originates from a single field of cells located in the center of the anterior neural plate surrounded rostrally and laterally by telencephalic precursors and caudally and medially by cells that will form the hypothalamus. Cell specification, mostly mediated by Wnt signaling [97] and by the concomitant expression of a few transcription factors, such as Rx, Pax6, Hes1, Otx2, Lhx2, and Six3, coupled to midline convergence of telencephalic precursors results in the formation of the optic pits. The optic pits begin to evaginate and give rise to the optic vesicles, the first morphologically visible eye structures. Folding and progressive specification of the vesicles further originate functionally specialized eye tissues: the optic stalk, the neural retina, and the retinal pigment epithelium (RPE), which, together with the ectodermal-derived distal components (lens, cornea, and iris), compose a functional eye [98]. The neural retina then undergoes proliferation to finally differentiate in six neuronal and one glial cell types: the retinal ganglion cells, the amacrine, horizontal and bipolar cells, the rod and cone photoreceptors, and the Müller glial cells.
The genetic networks that control eye development, composed by the interplay among different morphogenetic signaling pathways and transcription factors, have been intensively investigated. Most of the fundamental regulators have been identified in different species revealing a robust evolutionary conservation of the basic genetic programs and their properties. This extensive information, summarized in several recent reviews [99–102], has provided an excellent ground to identify the role of ncRNAs in eye development.
Indeed, the spatio-temporal distribution of miRNAs suggests their essential contribution to the control of the genetic networks involved in the development of all eye tissues, leading to the proposal that perhaps in the eye each cell type has its own miRNome [84, 103–112]. However, this information is insufficient to predict possible functions because, for example, ubiquitously expressed microRNA may target gene(s) expressed only in a small subset of cells or vice versa.
As in the case of the ear, the broad picture of miRNA activity in the eye derives from the use of conditional inactivation of Dicer in mice (Fig. 2). Mice lacking Dicer expression in the developing lens placode and in the presumptive corneal epithelium are microphthalmic with dystrophic lens and structural defects of the corneal epithelium, likely caused by an increased apoptosis and a reduced cell proliferation [113]. Dicer inactivation in the neural retina has been pursued with different Cre transgenic lines with either a broad and early or a late and more restricted pattern of expression. According to the Cre expression, the resulting mice are characterized by more or less severe phenotypes. With Rx- and Dkk3-Cre lines, in which Dicer inactivation occurred quite early in the development of the eye, embryos presented a pronounced microphthalmia associated with cell autonomous apoptosis that, in the case of Rx, was also extended to the preoptic region [114, 115]. As a consequence, Rx-Cre-mediated Dicer inactivation also led to significant pathfinding defects of the retinal ganglion cell (RGC) axons at the optic chiasm [114]. The use of αPax6- or Chx10-Cre transgenic lines causes instead variable defects in cell differentiation such as a prolonged production of early generated neuronal types followed by a loss of late-born cells [116] or the formation of rosette-like clusters of photoreceptors, which undergo a progressive degeneration, thereby decreasing the retinal response to light stimuli [117]. Similar, although more severe, phenotypes were observed after Dicer inactivation in Xenopus retinal progenitors, which in addition failed to laminate properly [118]. Comparison of Dicer inactivation driven by elements expressed with different spatio-temporal and yet partially overlapping distribution -Tyrp2-Cre, Z/AP, αPax6, and Pou4f3-Cre- demonstrated novel and stage-dependent roles for miRNAs in both the morphogenesis and neurogenesis of the eye [119], whereas forced expression of Cre recombinase into postnatal retinas indicates that miRNAs are required for cell survival [115]. Unexpectedly, generation of a BEST1-Cre conditional Dicer1 knockout, which expresses the recombinase in the RPE, caused the accumulation of dsRNAs of the Alu type. This accumulation, in turn, is responsible for RPE degeneration, demonstrating that Dicer has other RNA processing activities besides the generation of miRNAs [120].
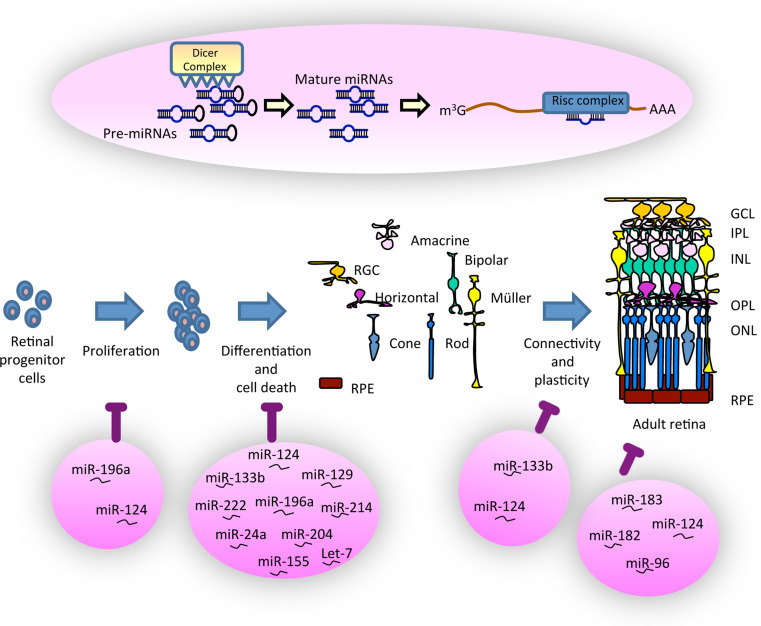
Fig. 2.
MicroRNAs are essential for the development and function of the vertebrate retina. A graphical overview of the canonical pathway of miRNA maturation is represented on the top. MiRNA biogenesis consists of a series of reactions that convert the primary miRNA transcript (pri-miRNA) first into a precursor miRNA (pre-miRNA) and finally into a biologically mature and active miRNA. The main steps of retinal development are depicted in the middle. The six types of neurons of the vertebrate retina originate from a population of neuroepithelial precursor cells in the eye primordium. Pink circle at the bottom indicates the main miRNAs that regulate the different steps of retinal development. Their function extends to homeostasis of the mature retina. Details of the interaction of these miRNAs during eye development are discussed in the text. GCL Ganglion cell layer, IPL inner plexiform layer, INL inner nuclear layer, OPL outer plexiform layer, ONL outer nuclear layer, RPE retinal pigmented epithelium
In addition to this large body of evidence supporting that miRNAs contribute to modulate eye development, studies in fish, frogs, and mice have also provided at least a partial view on the role of individual miRNAs (Fig. 2; Table 2). Studies in Xenopus embryos, for example, have shown that interference with miR-196a function causes microphthalmia [121], whereas miR-24a negatively regulates the pro-apoptotic factors caspase9 and apaf1 in retinal cells [122]. During retinal neurogenesis, miR-124 seems to control cell proliferation and differentiation [123, 124] as well as RGC growth cone sensitivity to Sema3A, an important axon guidance cue [125]. Similarly, differentiation of late-developing retinal cell types seems to require the simultaneous activity of a set of miRNAs composed by miR-129, -155, -214, and -222 [126]. In medaka fish (Oryzias latipes) instead, miR-204 affects both lens and retina development via repression of the Meis2 gene with a consequent alteration of Pax6 expression in both tissues [127].
In mammals, the miR-204/211 family participates to the differentiation of the RPE [128, 129], whereas the miR-183/96/182 cluster is expressed in photoreceptors and retinal interneurons. The latter complex has been functionally inactivated in photoreceptors by generating a “sponge” transgenic mouse carrying ten copies of each of the sequence complementary to the three members of the cluster into the 3′-UTR of the enhanced green fluorescent protein gene under the control of the Opsin promoter. The photoreceptors of the “sponge” mice become extremely sensitive to light-stress, although they showed no alterations under normal light conditions [130]. More recently, the analysis of a mouse deficient in the miR-183 cluster, generated using a gene-trap embryonic stem cell clone, revealed the presence of early onset and progressive synaptic defects of the photoreceptors followed by retinal degeneration [131]. In contrast and despite its abundant expression in the retina, targeted deletion of miR-182 alone in mice has little morphological and molecular consequences [132]. These data together with the work reported above in the inner ear support that the miR-183 cluster has an important protective role for different sensory organs.
Additional studies in mice have shown that miR-124 and the let-7 microRNA signaling pathway are respectively required to control retinal cone survival [133] and Müller glial cell differentiation [134]. In the first case, miR-124 acts by suppressing the expression of Lhx2 [133]. Similarly, miR-133b controls rat retinal dopaminergic amacrine cells differentiation and synaptogenesis by suppressing the expression of its target Pitx3 [135].
The development of the ocular vasculature is also under miRNA control. MiR-31, -150, and -184 are involved in retinal and choroidal neovascularization but their target genes have not been identified [136]. Slit–Robo signaling promotes endothelial cell migration and vessel formation. In the eye, miR-218 controls the levels of Robo receptors and of the heparan sulphate biosynthetic enzymes, thus influencing the organization of the blood vessels [137].
Vision depends on elaborate cellular and molecular events, including, among others, a rapid dark- and light-adaptation. This process is also highly dependent on miRNAs at least in mice [138]. Furthermore, miRNA levels are altered in different mouse models of retinitis pigmentosa, ischemic retina, autoimmune uveoretinitis, and diabetic retinopathy, indicating that a balanced expression of these molecules is required for proper function of the eye [136, 139–143]. This is also supported by the observation that miRNA expression profiles are altered in human pathological conditions such as lens cataract [108, 144]. Notably, mutations in the seed region of miR-184 are responsible for familial keratoconus with cataract [145], a degenerative disease of the cornea that progressively changes its shape. In a mouse model for secondary cataract, this miRNA antagonizes the miR-204-regulated RNA network [146]. This suggests that this network might also be affected in human keratoconus, opening the attractive possibility of treating eye inherited defects with gene transfer mediated by adeno-associated viral vectors facilitated by the relatively easy accessibility of this organ [147–149].
There are other miRNAs that might be useful targets for the treatment of eye-related diseases. Indeed, miR-200b and miR-126 seem to inhibit vascular alteration in diabetic retinopathy and ischemia-induced retinal neovascularization, respectively [150, 151], whereas modulation of miR-328 might be beneficial in Pax6-related high myopia [152].
Similar to miRNAs, the large number of lncRNAs expressed during eye development present cell-specific and spatio-temporal restricted expression profiles [37, 153–157], but their function remains largely unexplored, with limited exceptions. Tug1, one of these exceptions, promotes photoreceptor development and survival likely by altering the chromatin structure and thus activating the expression of the photoreceptor-specific transcription factors Crx and Nrl [158]. The Retinal Non-coding RNA 2 (RNCR2), also known as Gomafu or Miat, is instead expressed in retinal progenitors and later on in Müller, amacrine and retinal ganglion cells [30, 37] as well in other cells of the nervous system [37]. Gomafu/RNCR2 has been shown to control amacrine and Müller cell differentiation [159] and the pluripotency of embryonic stem cells [160], with yet undetermined mechanisms. Notably, however, Gomafu RNA escapes nuclear export and accumulates in the nucleus, in an uncharacterized compartment [37]. Its sequence contains tandem repeats of UACUAAC that bind to the SF1 splicing factor with high affinity, affecting splicing kinetics. These observations suggest that Gomafu could regulates splicing efficiency by changing the local concentration of splicing factors within the nucleus [161], suggesting a possible mechanism for its effect in retinal differentiation.
LncRNAs of the antisense type (NAT or OS) are also relevant to retinal cell differentiation and survival. This is the case of Six3OS and Vax2os1. Manipulation of Six3OS expression interfered with retinal cell specification, possibly because this lncRNA serves as a transcriptional scaffold [155]. Vax2os1 instead is involved in cell cycle progression of photoreceptor progenitors [156].
MiRNAs function in primary olfactory system development
The olfactory system, one of the most ancient sensory systems, serves to discriminate odors and transmit this information from the nose to the brain [162–164]. The primary olfactory system is composed of the olfactory epithelium (OE) and the olfactory bulb (OB). The development of these two structures begins simultaneously but proceeds, at least initially, with independent programs. The OE differentiates from the olfactory placodes, bilateral epithelial thickening localized in the rostro-lateral regions of the head. Soon after the closure of the neural tube, these placodes invaginate to form the nasal pits, the marginal rims of which then fuse originating the nostrils. The nostrils thereafter become more convoluted and transform into the nasal cavity and the vomero-nasal organ that arises from the medial wall of the epithelium [165]. Upon differentiation, the OE becomes pseudo-stratified and composed of basal and supporting cells, among which mature and immature sensory neurons reside. Olfactory sensory neurons are renewed throughout life and derive from a population of self-renewing stem cells, probably a subtype of basal cells, that sequentially give rise to two types of transit-amplifying cells marked by the expression of different neurogenic basic helix-loop-helix transcription factors. Their daughter cells differentiate into the olfactory sensory neurons, which can be identified by a variety of markers, including the olfactory marker protein (OMP) [166].
The OB instead develops from a predetermined region of the rostral telencephalon, likely under the influence of FGF signaling. In mouse embryos, its development begins around E11.5 although the OB becomes morphologically visible as an evagination of the rostral telencephalon only at around E12.5. There is considerable debate on whether further OB development is influenced by the arrival of the olfactory sensor axons derived from the OE. However, the onset of mitral cell differentiation prior to the arrival of the sensory axons and the evidence that in absence of the OE, the OB maintains its differentiation program supports that the OB is independently formed [167].
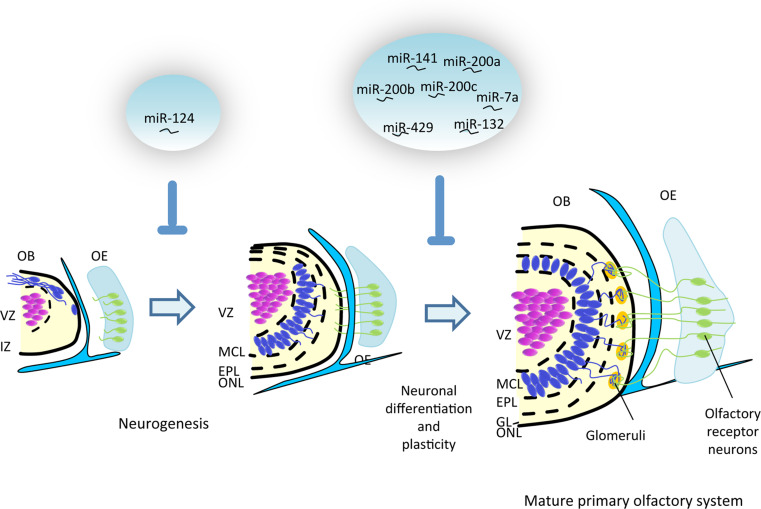
The first information on miRNA expression in the olfactory system derived from studies in zebrafish embryos [73, 168] followed by microarray-based analysis in mice [169]. This analysis was thereafter largely validated by real-time RT-PCR studies [170]. Altogether, these data demonstrate that the OB is particularly enriched in miR-200a, miR-200b, miR-200c, miR-141, and miR-429 expression when compared to the rest of the brain. In both mice and zebrafish, miRNA expression remains relatively stable during postnatal stages [171–174], suggesting that these molecules controls the function of the olfactory system.
The miR-200 family, composed of five members divided into two clusters, has been particularly studied (Fig. 3). In humans, the first cluster, composed of miR-200a, miR-200b, and miR-429, localizes to chromosome 1, whereas the second, formed by miR-200c and miR-141, is found on chromosome 12 [175]. In mice, miR-200 expression starts at E9.5 in the olfactory placodes and then becomes restricted to a subregion of the olfactory pit to finally localize to the immature and mature neuronal cells of the OE [171]. Genetic inactivation of Dicer in mice with a broad (Foxg1-Cre) or more restricted driver (OMP-Cre) showed that miRNAs are required for progression but not initiation of olfactory neurogenesis. However, when the OMP-Cre driver was used to inactivate Dicer at adult stages, both olfactory and vomeronasal neurons appeared normal [171]. Individual or combined inactivation of miR-141/miR-200a or miR-200b/miR-429 using antisense morpholinos had also little effect on the development of the zebrafish olfactory system and only the simultaneous interference with the expression of all the members of the miR-200 family caused abnormal terminal differentiation of olfactory cells and apoptosis. A list of possible target genes, related to neuronal differentiation, was predicted with bioinformatic approaches and two of them, lfng, a regulator of the Notch signaling, and zfhx1, an enhancer of TGFβ pathway, were further validated in zebrafish. Nevertheless, how miR-200-mediated regulation of these genes fits in the molecular pathway underlying olfactory system development is still unclear [171].
Fig. 3.
Emerging roles of miRNAs in olfactory system development. The drawings provide a schematic representation of olfactory system development, staring from the olfactory placode stage to the adult organization. The two blue circles on the top indicate the miRNAs that have been so far described as modulators of these events (see text for further details). VZ ventricular zone, IZ intermediate zone, MCL mitral cell layer, EPL external plexiform, ONL olfactory nerve layer, OE olfactory epithelium, GL glomerular layer
Other miRNAs have important roles in OB development. MiR-124 inactivation impairs neurogenesis and induces the appearance of ectopic cells with astrocyte characteristics [176]. The continuous neurogenesis of OB neurons derived from the subventricular zone involves the function of miR-132 that controls survival of new-born neurons and their dendritic complexity and spine density, likely regulating their plasticity [177]. Furthermore, specific miR-7a-mediated regulation of Pax6 in ventral neural stem cells regulates the molecular network that specify postnatal and adult dopaminergic neurons in the olfactory system [178] (Table 2).
Whether miRNAs are involved in diseases that affect the olfactory system has remained so far unexplored.
Conclusions and perspectives
The recognition of the fundamental and pervasive role of ncRNAs in virtually all biological processes represents one of the most notable outcomes of the Human Genome Project and related efforts [25]. MiRNAs and lncRNAs are now widely recognized as key players in gene regulation processes. As such, their role in the pathogenesis of both monogenic and complex genetic disorders is starting to be unraveled [179]. Nevertheless, there are many unresolved issues concerning both their identity and function. This is particularly evident in the case of lncRNAs, which, despite the remarkable progresses of the past few years [180], still represent a heterogeneous group of transcripts that need to be more finely dissected in the near future.
The extraordinary advances in next-generation sequencing (NGS) technology applied to transcriptome analysis [181] should lead to a more comprehensive definition of the non-coding RNA repertoire of the human and other vertebrates’ transcriptomes. Similarly, the use of whole genome or whole-exome NGS-based approaches will likely shed further light on the pathogenic contribution of ncRNAs to human genetic diseases. Sensory organs are among the structures for which the study of the functional role of ncRNAs has been most rewarding so far. It is remarkable that sensory organs represent the targets of the first two examples of human Mendelian disorders caused by mutations in miRNA sequences [92, 145]. It can be also anticipated that a more precise knowledge of ncRNA function will turn these molecules into therapeutic targets for human disease. Disorders affecting sensory organs, in particular the eye by virtue of its accessibility, may constitute ideal conditions to explore this therapeutic potential in the near future.
Acknowledgements
Work in our laboratories is supported by grants from the Spanish MINECO (BFU2010-16031), Comunidad Autonoma de Madrid (CAM, S2010/BMD-2315), Fundaluce, Fundación ONCE and CIBERER to P.B, from MAE/MOST Israel–Italy Joint Innovation Program to I.C. and from the Italian Telethon Foundation (TGM11SB2) to S.B.
Abbreviations
- lncRNA
Long non-coding RNA
- miRNAs
microRNAs
- NATs
Natural antisense transcripts
- ncRNAs
Non-coding RNAs
- NFAT
Nuclear factor of the activated T cells
- NGS
Next-generation sequencing
- OB
Olfactory bulb
- OE
Olfactory epithelium
- OMP
Olfactory marker proteins
- PRC2
Polycomb repressive complex 2
- RGC
Retinal ganglion cells
- RNCR2
Retinal non-coding RNA 2
- RPE
Retinal pigment epithelium
- Uchl1
Ubiquitin carboxy-terminal hydrolase L1
Contributor Information
Ivan Conte, Phone: +39-081-6132223, FAX: +39-081-5609877, Email: conte@tigem.it.
Sandro Banfi, Phone: +39-081-6132206, FAX: +39-081-5609877, Email: banfi@tigem.it.
Paola Bovolenta, Phone: +34-91-1964718, FAX: +34-91-1964420, Email: pbovolenta@cbm.uam.es.
References
- 1.Jalali S, Jayaraj GG, Scaria V. Integrative transcriptome analysis suggest processing of a subset of long non-coding RNAs to small RNAs. Biol Direct. 2012;7:25. doi: 10.1186/1745-6150-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carninci P, Hayashizaki Y. Noncoding RNA transcription beyond annotated genes. Curr Opin Genet Dev. 2007;17(2):139–144. doi: 10.1016/j.gde.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13(8):528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322(5909):1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 5.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10(12):833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 6.Rosa A, Brivanlou AH. MicroRNAs in early vertebrate development. Cell Cycle. 2009;8(21):3513–3520. doi: 10.4161/cc.8.21.9847. [DOI] [PubMed] [Google Scholar]
- 7.Harfe BD. MicroRNAs in vertebrate development. Curr Opin Genet Dev. 2005;15(4):410–415. doi: 10.1016/j.gde.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 8.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12(2):136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 10.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 11.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans . Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 12.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 13.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 14.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336(6078):233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336(6078):237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 18.Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, Dhanasekaran SM, Chinnaiyan AM, Athey BD. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 2009;19(7):1175–1183. doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 20.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 2007;104(23):9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez S, Pisano DG, Serrano M. Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs. Cell Cycle. 2008;7(16):2601–2608. doi: 10.4161/cc.7.16.6541. [DOI] [PubMed] [Google Scholar]
- 22.Chitnis NS, Pytel D, Bobrovnikova-Marjon E, Pant D, Zheng H, Maas NL, Frederick B, Kushner JA, Chodosh LA, Koumenis C, Fuchs SY, Diehl JA. miR-211 Is a prosurvival microRNA that regulates chop expression in a PERK-dependent manner. Mol Cell. 2012;48(3):353–364. doi: 10.1016/j.molcel.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Horizontal transfer of microRNAs: molecular mechanisms and clinical applications. Protein Cell. 2012;3(1):28–37. doi: 10.1007/s13238-012-2003-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109(31):E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 26.Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol. 2008;4(11):e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frith MC, Bailey TL, Kasukawa T, Mignone F, Kummerfeld SK, Madera M, Sunkara S, Furuno M, Bult CJ, Quackenbush J, Kai C, Kawai J, Carninci P, Hayashizaki Y, Pesole G, Mattick JS. Discrimination of non-protein-coding transcripts from protein-coding mRNA. RNA Biol. 2006;3(1):40–48. doi: 10.4161/rna.3.1.2789. [DOI] [PubMed] [Google Scholar]
- 28.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 29.Saxena A, Carninci P. Long non-coding RNA modifies chromatin: epigenetic silencing by long non-coding RNAs. Bioessays. 2011;33(11):830–839. doi: 10.1002/bies.201100084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, Yung R, Asch E, Ohno-Machado L, Wong WH, Cepko CL. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2(9):E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18(9):1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, Grimmond SM, Hume DA, Hayashizaki Y, Mattick JS. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16(1):11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105(2):716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci. 2007;120(Pt 15):2498–2506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- 38.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19(3):347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. A new frontier for molecular medicine: noncoding RNAs. Biochim Biophys Acta. 2005;1756(1):65–75. doi: 10.1016/j.bbcan.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 40.Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21(1):11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- 41.Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, Zhang Q, Yan G, Cui Q. LncRNAdisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2012;41:D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rackham O, Shearwood AM, Mercer TR, Davies SM, Mattick JS, Filipovska A. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA. 2011;17(12):2085–2093. doi: 10.1261/rna.029405.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol. 2010;7(5):582–585. doi: 10.4161/rna.7.5.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137(15):2493–2499. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- 47.Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43(6):1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309(5740):1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 49.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40(14):6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470(7333):284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat Med. 2008;14(7):723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, Forrest AR, Carninci P, Biffo S, Stupka E, Gustincich S. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491(7424):454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 53.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39(8):1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 55.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, Lieberman J, Rigoutsos I, Pandolfi PP. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147(2):344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12(8):1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, Abo R, Tabebordbar M, Lee RT, Burge CB, Boyer LA. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152(3):570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alsina B, Giraldez F, Pujades C. Patterning and cell fate in ear development. Int J Dev Biol. 2009;53(8–10):1503–1513. doi: 10.1387/ijdb.072422ba. [DOI] [PubMed] [Google Scholar]
- 61.Cole LK, Le Roux I, Nunes F, Laufer E, Lewis J, Wu DK. Sensory organ generation in the chicken inner ear: contributions of bone morphogenetic protein 4, serrate1, and lunatic fringe. J Comp Neurol. 2000;424(3):509–520. doi: 10.1002/1096-9861(20000828)424:3<509::aid-cne8>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 62.Lleras-Forero L, Streit A. Development of the sensory nervous system in the vertebrate head: the importance of being on time. Curr Opin Genet Dev. 2012;22(4):315–322. doi: 10.1016/j.gde.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 63.Groves AK, Fekete DM. Shaping sound in space: the regulation of inner ear patterning. Development. 2012;139(2):245–257. doi: 10.1242/dev.067074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu DK, Kelley MW. Molecular mechanisms of inner ear development. Cold Spring Harb Perspect Biol. 2012;4(8):a008409. doi: 10.1101/cshperspect.a008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brigande JV, Kiernan AE, Gao X, Iten LE, Fekete DM. Molecular genetics of pattern formation in the inner ear: do compartment boundaries play a role? Proc Natl Acad Sci USA. 2000;97(22):11700–11706. doi: 10.1073/pnas.97.22.11700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cantos R, Cole LK, Acampora D, Simeone A, Wu DK. Patterning of the mammalian cochlea. Proc Natl Acad Sci USA. 2000;97(22):11707–11713. doi: 10.1073/pnas.97.22.11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Patel M, Hu BH. MicroRNAs in inner ear biology and pathogenesis. Hear Res. 2012;287(1–2):6–14. doi: 10.1016/j.heares.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006;1111(1):95–104. doi: 10.1016/j.brainres.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 69.Hirai M, Maeda Y, Fukushima K, Sugaya A, Kataoka Y, Nishizaki K. Expression analysis of microRNAs in murine cochlear explants. NeuroReport. 2011;22(13):652–654. doi: 10.1097/WNR.0b013e32834a0273. [DOI] [PubMed] [Google Scholar]
- 70.Elkan-Miller T, Ulitsky I, Hertzano R, Rudnicki A, Dror AA, Lenz DR, Elkon R, Irmler M, Beckers J, Shamir R, Avraham KB. Integration of transcriptomics, proteomics, and microRNA analyses reveals novel microRNA regulation of targets in the mammalian inner ear. PLoS ONE. 2011;6(4):e18195. doi: 10.1371/journal.pone.0018195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Friedman LM, Dror AA, Mor E, Tenne T, Toren G, Satoh T, Biesemeier DJ, Shomron N, Fekete DM, Hornstein E, Avraham KB. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc Natl Acad Sci USA. 2009;106(19):7915–7920. doi: 10.1073/pnas.0812446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hertzano R, Elkon R. High throughput gene expression analysis of the inner ear. Hear Res. 2012;288(1–2):77–88. doi: 10.1016/j.heares.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 73.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309(5732):310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 74.Wang XR, Zhang XM, Zhen J, Zhang PX, Xu G, Jiang H. MicroRNA expression in the embryonic mouse inner ear. NeuroReport. 2010;21(9):611–617. doi: 10.1097/WNR.0b013e328338864b. [DOI] [PubMed] [Google Scholar]
- 75.Ohyama T, Groves AK. Generation of Pax2-Cre mice by modification of a Pax2 bacterial artificial chromosome. Genesis. 2004;38(4):195–199. doi: 10.1002/gene.20017. [DOI] [PubMed] [Google Scholar]
- 76.Soukup GA, Fritzsch B, Pierce ML, Weston MD, Jahan I, McManus MT, Harfe BD. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev Biol. 2009;328(2):328–341. doi: 10.1016/j.ydbio.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weston MD, Pierce ML, Jensen-Smith HC, Fritzsch B, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA-183 family expression in hair cell development and requirement of microRNAs for hair cell maintenance and survival. Dev Dyn. 2011;240(4):808–819. doi: 10.1002/dvdy.22591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kersigo J, D’Angelo A, Gray BD, Soukup GA, Fritzsch B. The role of sensory organs and the forebrain for the development of the craniofacial shape as revealed by Foxg1-cre-mediated microRNA loss. Genesis. 2011;49(4):326–341. doi: 10.1002/dvg.20714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bohne BA, Yohman L, Gruner MM. Cochlear damage following interrupted exposure to high-frequency noise. Hear Res. 1987;29(2–3):251–264. doi: 10.1016/0378-5955(87)90172-9. [DOI] [PubMed] [Google Scholar]
- 80.Fechter LD, Liu Y, Pearce TA. Cochlear protection from carbon monoxide exposure by free radical blockers in the guinea pig. Toxicol Appl Pharmacol. 1997;142(1):47–55. doi: 10.1006/taap.1996.8027. [DOI] [PubMed] [Google Scholar]
- 81.Hu BH, Henderson D, Nicotera TM. Involvement of apoptosis in progression of cochlear lesion following exposure to intense noise. Hear Res. 2002;166(1–2):62–71. doi: 10.1016/s0378-5955(02)00286-1. [DOI] [PubMed] [Google Scholar]
- 82.Sha SH, Taylor R, Forge A, Schacht J. Differential vulnerability of basal and apical hair cells is based on intrinsic susceptibility to free radicals. Hear Res. 2001;155(1–2):1–8. doi: 10.1016/s0378-5955(01)00224-6. [DOI] [PubMed] [Google Scholar]
- 83.Sacheli R, Nguyen L, Borgs L, Vandenbosch R, Bodson M, Lefebvre P, Malgrange B. Expression patterns of miR-96, miR-182, and miR-183 in the development inner ear. Gene Expr Patterns. 2009;9(5):364–370. doi: 10.1016/j.gep.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 84.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282(34):25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 85.Jalvy-Delvaille S, Maurel M, Majo V, Pierre N, Chabas S, Combe C, Rosenbaum J, Sagliocco F, Grosset CF. Molecular basis of differential target regulation by miR-96 and miR-182: the Glypican-3 as a model. Nucleic Acids Res. 2012;40(3):1356–1365. doi: 10.1093/nar/gkr843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lewis MA, Quint E, Glazier AM, Fuchs H, De Angelis MH, Langford C, van Dongen S, Abreu-Goodger C, Piipari M, Redshaw N, Dalmay T, Moreno-Pelayo MA, Enright AJ, Steel KP. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009;41(5):614–618. doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li H, Kloosterman W, Fekete DM. MicroRNA-183 family members regulate sensorineural fates in the inner ear. J Neurosci. 2010;30(9):3254–3263. doi: 10.1523/JNEUROSCI.4948-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang XR, Zhang XM, Du J, Jiang H. MicroRNA-182 regulates otocyst-derived cell differentiation and targets T-box1 gene. Hear Res. 2012;286(1–2):55–63. doi: 10.1016/j.heares.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 89.Gu C, Li X, Tan Q, Wang Z, Chen L, Liu Y. MiR-183 family regulates chloride intracellular channel 5 expression in inner ear hair cells. Toxicol In Vitro. 2013;27(1):486–491. doi: 10.1016/j.tiv.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 90.Hertzano R, Elkon R, Kurima K, Morrisson A, Chan SL, Sallin M, Biedlingmaier A, Darling DS, Griffith AJ, Eisenman DJ, Strome SE. Cell type-specific transcriptome analysis reveals a major role for Zeb1 and miR-200b in mouse inner ear morphogenesis. PLoS Genet. 2011;7(9):e1002309. doi: 10.1371/journal.pgen.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Frucht CS, Santos-Sacchi J, Navaratnam DS. MicroRNA181a plays a key role in hair cell regeneration in the avian auditory epithelium. Neurosci Lett. 2011;493(1–2):44–48. doi: 10.1016/j.neulet.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mencia A, Modamio-Hoybjor S, Redshaw N, Morin M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, Moreno F, Moreno-Pelayo MA. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41(5):609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 93.Solda G, Robusto M, Primignani P, Castorina P, Benzoni E, Cesarani A, Ambrosetti U, Asselta R, Duga S. A novel mutation within the MIR96 gene causes non-syndromic inherited hearing loss in an Italian family by altering pre-miRNA processing. Hum Mol Genet. 2012;21(3):577–585. doi: 10.1093/hmg/ddr493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cioffi JA, Yue WY, Mendolia-Loffredo S, Hansen KR, Wackym PA, Hansen MR. MicroRNA-21 overexpression contributes to vestibular schwannoma cell proliferation and survival. Otol Neurotol. 2010;31(9):1455–1462. doi: 10.1097/MAO.0b013e3181f20655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Friedland DR, Eernisse R, Erbe C, Gupta N, Cioffi JA. Cholesteatoma growth and proliferation: posttranscriptional regulation by microRNA-21. Otol Neurotol. 2009;30(7):998–1005. doi: 10.1097/MAO.0b013e3181b4e91f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Roberts KA, Abraira VE, Tucker AF, Goodrich LV, Andrews NC. Mutation of Rubie, a novel long non-coding RNA located upstream of Bmp4, causes vestibular malformation in mice. PLoS ONE. 2012;7(1):e29495. doi: 10.1371/journal.pone.0029495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Esteve P, Bovolenta P. Secreted inducers in vertebrate eye development: more functions for old morphogens. Curr Opin Neurobiol. 2006;16(1):13–19. doi: 10.1016/j.conb.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 98.Martinez-Morales JR, Rodrigo I, Bovolenta P. Eye development: a view from the retina pigmented epithelium. Bioessays: News Rev Mol Cell Dev Biol. 2004;26(7):766–777. doi: 10.1002/bies.20064. [DOI] [PubMed] [Google Scholar]
- 99.Beccari L, Marco-Ferreres R, Bovolenta P. The logic of gene regulatory networks in early vertebrate forebrain patterning. Mech Dev. 2013;130:95–111. doi: 10.1016/j.mod.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 100.Bassett EA, Wallace VA. Cell fate determination in the vertebrate retina. Trends Neurosci. 2012;35(9):565–573. doi: 10.1016/j.tins.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 101.Shaham O, Menuchin Y, Farhy C, Ashery-Padan R. Pax6: a multi-level regulator of ocular development. Prog Retin Eye Res. 2012;31(5):351–376. doi: 10.1016/j.preteyeres.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 102.Fuhrmann S. Eye morphogenesis and patterning of the optic vesicle. Curr Top Dev Biol. 2010;93:61–84. doi: 10.1016/B978-0-12-385044-7.00003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Frederikse PH, Donnelly R, Partyka LM. MiRNA and Dicer in the mammalian lens: expression of brain-specific miRNAs in the lens. Histochem Cell Biol. 2006;126(1):1–8. doi: 10.1007/s00418-005-0139-0. [DOI] [PubMed] [Google Scholar]
- 104.Karali M, Peluso I, Gennarino VA, Bilio M, Verde R, Lago G, Dolle P, Banfi S. miRNeye: a microRNA expression atlas of the mouse eye. BMC Genomics. 2010;11:715. doi: 10.1186/1471-2164-11-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Karali M, Peluso I, Marigo V, Banfi S. Identification and characterization of microRNAs expressed in the mouse eye. Invest Ophthalmol Vis Sci. 2007;48(2):509–515. doi: 10.1167/iovs.06-0866. [DOI] [PubMed] [Google Scholar]
- 106.Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vis. 2006;12:1175–1184. [PubMed] [Google Scholar]
- 107.Makarev E, Spence JR, Del Rio-Tsonis K, Tsonis PA. Identification of microRNAs and other small RNAs from the adult newt eye. Mol Vis. 2006;12:1386–1391. [PubMed] [Google Scholar]
- 108.Wu C, Lin H, Wang Q, Chen W, Luo H, Zhang H. Discrepant expression of microRNAs in transparent and cataractous human lenses. Invest Ophthalmol Vis Sci. 2012;53(7):3906–3912. doi: 10.1167/iovs.11-9178. [DOI] [PubMed] [Google Scholar]
- 109.Yan N, Ma K, Ma J, Chen W, Wang Y, Cao G, Man-Kit Lam D, Liu X. Profiling microRNAs differentially expressed in rabbit retina. Adv Exp Med Biol. 2010;664:203–209. doi: 10.1007/978-1-4419-1399-9_23. [DOI] [PubMed] [Google Scholar]
- 110.Hackler L, Jr, Wan J, Swaroop A, Qian J, Zack DJ. MicroRNA profile of the developing mouse retina. Invest Ophthalmol Vis Sci. 2010;51(4):1823–1831. doi: 10.1167/iovs.09-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Arora A, Guduric-Fuchs J, Harwood L, Dellett M, Cogliati T, Simpson DA. Prediction of microRNAs affecting mRNA expression during retinal development. BMC Dev Biol. 2010;10:1. doi: 10.1186/1471-213X-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Arora A, McKay GJ, Simpson DA. Prediction and verification of miRNA expression in human and rat retinas. Invest Ophthalmol Vis Sci. 2007;48(9):3962–3967. doi: 10.1167/iovs.06-1221. [DOI] [PubMed] [Google Scholar]
- 113.Li Y, Piatigorsky J. Targeted deletion of Dicer disrupts lens morphogenesis, corneal epithelium stratification, and whole eye development. Dev Dyn. 2009;238(9):2388–2400. doi: 10.1002/dvdy.22056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pinter R, Hindges R. Perturbations of microRNA function in mouse dicer mutants produce retinal defects and lead to aberrant axon pathfinding at the optic chiasm. PLoS ONE. 2010;5(4):e10021. doi: 10.1371/journal.pone.0010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Iida A, Shinoe T, Baba Y, Mano H, Watanabe S. Dicer plays essential roles for retinal development by regulation of survival and differentiation. Invest Ophthalmol Vis Sci. 2011;52(6):3008–3017. doi: 10.1167/iovs.10-6428. [DOI] [PubMed] [Google Scholar]
- 116.Georgi SA, Reh TA. Dicer is required for the transition from early to late progenitor state in the developing mouse retina. J Neurosci. 2010;30(11):4048–4061. doi: 10.1523/JNEUROSCI.4982-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Damiani D, Alexander JJ, O’Rourke JR, McManus M, Jadhav AP, Cepko CL, Hauswirth WW, Harfe BD, Strettoi E. Dicer inactivation leads to progressive functional and structural degeneration of the mouse retina. J Neurosci. 2008;28(19):4878–4887. doi: 10.1523/JNEUROSCI.0828-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Decembrini S, Andreazzoli M, Barsacchi G, Cremisi F. Dicer inactivation causes heterochronic retinogenesis in Xenopus laevis. Int J Dev Biol. 2008;52(8):1099–1103. doi: 10.1387/ijdb.082646sd. [DOI] [PubMed] [Google Scholar]
- 119.Davis N, Mor E, Ashery-Padan R. Roles for Dicer1 in the patterning and differentiation of the optic cup neuroepithelium. Development. 2011;138(1):127–138. doi: 10.1242/dev.053637. [DOI] [PubMed] [Google Scholar]
- 120.Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, Kariko K, Yoo JW, Lee DK, Hadziahmetovic M, Song Y, Misra S, Chaudhuri G, Buaas FW, Braun RE, Hinton DR, Zhang Q, Grossniklaus HE, Provis JM, Madigan MC, Milam AH, Justice NL, Albuquerque RJ, Blandford AD, Bogdanovich S, Hirano Y, Witta J, Fuchs E, Littman DR, Ambati BK, Rudin CM, Chong MM, Provost P, Kugel JF, Goodrich JA, Dunaief JL, Baffi JZ, Ambati J. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471(7338):325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Qiu R, Liu Y, Wu JY, Liu K, Mo W, He R. Misexpression of miR-196a induces eye anomaly in Xenopus laevis . Brain Res Bull. 2009;79(1):26–31. doi: 10.1016/j.brainresbull.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 122.Walker JC, Harland RM. microRNA-24a is required to repress apoptosis in the developing neural retina. Genes Dev. 2009;23(9):1046–1051. doi: 10.1101/gad.1777709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Qiu R, Liu K, Liu Y, Mo W, Flynt AS, Patton JG, Kar A, Wu JY, He R. The role of miR-124a in early development of the Xenopus eye. Mech Dev. 2009;126(10):804–816. doi: 10.1016/j.mod.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu K, Liu Y, Mo W, Qiu R, Wang X, Wu JY, He R. MiR-124 regulates early neurogenesis in the optic vesicle and forebrain, targeting NeuroD1. Nucleic Acids Res. 2011;39(7):2869–2879. doi: 10.1093/nar/gkq904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Baudet ML, Zivraj KH, Abreu-Goodger C, Muldal A, Armisen J, Blenkiron C, Goldstein LD, Miska EA, Holt CE. miR-124 acts through CoREST to control onset of Sema3A sensitivity in navigating retinal growth cones. Nat Neurosci. 2012;15(1):29–38. doi: 10.1038/nn.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Decembrini S, Bressan D, Vignali R, Pitto L, Mariotti S, Rainaldi G, Wang X, Evangelista M, Barsacchi G, Cremisi F. MicroRNAs couple cell fate and developmental timing in retina. Proc Natl Acad Sci USA. 2009;106(50):21179–21184. doi: 10.1073/pnas.0909167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Conte I, Carrella S, Avellino R, Karali M, Marco-Ferreres R, Bovolenta P, Banfi S. miR-204 is required for lens and retinal development via Meis2 targeting. Proc Natl Acad Sci USA. 2010;107(35):15491–15496. doi: 10.1073/pnas.0914785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang FE, Zhang C, Maminishkis A, Dong L, Zhi C, Li R, Zhao J, Majerciak V, Gaur AB, Chen S, Miller SS. MicroRNA-204/211 alters epithelial physiology. FASEB J. 2010;24(5):1552–1571. doi: 10.1096/fj.08-125856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Adijanto J, Castorino JJ, Wang ZX, Maminishkis A, Grunwald GB, Philp NJ. Microphthalmia-associated transcription factor (MITF) promotes differentiation of human retinal pigment epithelium (RPE) by regulating microRNAs-204/211 expression. J Biol Chem. 2012;287(24):20491–20503. doi: 10.1074/jbc.M112.354761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhu Q, Sun W, Okano K, Chen Y, Zhang N, Maeda T, Palczewski K. Sponge transgenic mouse model reveals important roles for the microRNA-183 (miR-183)/96/182 cluster in postmitotic photoreceptors of the retina. J Biol Chem. 2011;286(36):31749–31760. doi: 10.1074/jbc.M111.259028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lumayag S, Haldin CE, Corbett NJ, Wahlin KJ, Cowan C, Turturro S, Larsen PE, Kovacs B, Witmer PD, Valle D, Zack DJ, Nicholson DA, Xu S. Inactivation of the microRNA-183/96/182 cluster results in syndromic retinal degeneration. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1212655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jin ZB, Hirokawa G, Gui L, Takahashi R, Osakada F, Hiura Y, Takahashi M, Yasuhara O, Iwai N. Targeted deletion of miR-182, an abundant retinal microRNA. Mol Vis. 2009;15:523–533. [PMC free article] [PubMed] [Google Scholar]
- 133.Sanuki R, Onishi A, Koike C, Muramatsu R, Watanabe S, Muranishi Y, Irie S, Uneo S, Koyasu T, Matsui R, Cherasse Y, Urade Y, Watanabe D, Kondo M, Yamashita T, Furukawa T. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat Neurosci. 2011;14(9):1125–1134. doi: 10.1038/nn.2897. [DOI] [PubMed] [Google Scholar]
- 134.Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates Muller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat Cell Biol. 2010;12(11):1101–1107. doi: 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li Y, Li C, Chen Z, He J, Tao Z, Yin ZQ. A microRNA, mir133b, suppresses melanopsin expression mediated by failure dopaminergic amacrine cells in RCS rats. Cell Signal. 2012;24(3):685–698. doi: 10.1016/j.cellsig.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 136.Shen J, Yang X, Xie B, Chen Y, Swaim M, Hackett SF, Campochiaro PA. MicroRNAs regulate ocular neovascularization. Mol Ther. 2008;16(7):1208–1216. doi: 10.1038/mt.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Small EM, Sutherland LB, Rajagopalan KN, Wang S, Olson EN. MicroRNA-218 regulates vascular patterning by modulation of Slit-Robo signaling. Circ Res. 2010;107(11):1336–1344. doi: 10.1161/CIRCRESAHA.110.227926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schubeler D, Oertner TG, Schratt G, Bibel M, Roska B, Filipowicz W. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141(4):618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 139.Silva VA, Polesskaya A, Sousa TA, Correa VM, Andre ND, Reis RI, Kettelhut IC, Harel-Bellan A, De Lucca FL. Expression and cellular localization of microRNA-29b and RAX, an activator of the RNA-dependent protein kinase (PKR), in the retina of streptozotocin-induced diabetic rats. Mol Vis. 2011;17:2228–2240. [PMC free article] [PubMed] [Google Scholar]
- 140.Loscher CJ, Hokamp K, Kenna PF, Ivens AC, Humphries P, Palfi A, Farrar GJ. Altered retinal microRNA expression profile in a mouse model of retinitis pigmentosa. Genome Biol. 2007;8(11):R248. doi: 10.1186/gb-2007-8-11-r248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Loscher CJ, Hokamp K, Wilson JH, Li T, Humphries P, Farrar GJ, Palfi A. A common microRNA signature in mouse models of retinal degeneration. Exp Eye Res. 2008;87(6):529–534. doi: 10.1016/j.exer.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ishida W, Fukuda K, Higuchi T, Kajisako M, Sakamoto S, Fukushima A. Dynamic changes of microRNAs in the eye during the development of experimental autoimmune uveoretinitis. Invest Ophthalmol Vis Sci. 2011;52(1):611–617. doi: 10.1167/iovs.10-6115. [DOI] [PubMed] [Google Scholar]
- 143.Kovacs B, Lumayag S, Cowan C, Xu S. MicroRNAs in early diabetic retinopathy in streptozotocin-induced diabetic rats. Invest Ophthalmol Vis Sci. 2011;52(7):4402–4409. doi: 10.1167/iovs.10-6879. [DOI] [PubMed] [Google Scholar]
- 144.Peng CH, Liu JH, Woung LC, Lin TJ, Chiou SH, Tseng PC, Du WY, Cheng CK, Hu CC, Chien KH, Chen SJ. MicroRNAs and cataracts: correlation among let-7 expression, age and the severity of lens opacity. Br J Ophthalmol. 2012;96(5):747–751. doi: 10.1136/bjophthalmol-2011-300585. [DOI] [PubMed] [Google Scholar]
- 145.Hughes AE, Bradley DT, Campbell M, Lechner J, Dash DP, Simpson DA, Willoughby CE. Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am J Hum Genet. 2011;89(5):628–633. doi: 10.1016/j.ajhg.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hoffmann A, Huang Y, Suetsugu-Maki R, Ringelberg CS, Tomlinson CR, Del Rio-Tsonis K, Tsonis PA. Implication of the miR-184 and miR-204 competitive RNA network in control of mouse secondary cataract. Mol Med. 2012;18:528–538. doi: 10.2119/molmed.2011.00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Georgiadis A, Tschernutter M, Bainbridge JW, Robbie SJ, McIntosh J, Nathwani AC, Smith AJ, Ali RR. AAV-mediated knockdown of peripherin-2 in vivo using miRNA-based hairpins. Gene Ther. 2010;17(4):486–493. doi: 10.1038/gt.2009.162. [DOI] [PubMed] [Google Scholar]
- 148.Karali M, Manfredi A, Puppo A, Marrocco E, Gargiulo A, Allocca M, Corte MD, Rossi S, Giunti M, Bacci ML, Simonelli F, Surace EM, Banfi S, Auricchio A. MicroRNA-restricted transgene expression in the retina. PLoS ONE. 2011;6(7):e22166. doi: 10.1371/journal.pone.0022166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pihlmann M, Askou AL, Aagaard L, Bruun GH, Svalgaard JD, Holm-Nielsen MH, Dagnaes-Hansen F, Bek T, Mikkelsen JG, Jensen TG, Corydon TJ. Adeno-associated virus-delivered polycistronic microRNA-clusters for knockdown of vascular endothelial growth factor in vivo. J Gene Med. 2012;14(5):328–338. doi: 10.1002/jgm.2623. [DOI] [PubMed] [Google Scholar]
- 150.Bai Y, Bai X, Wang Z, Zhang X, Ruan C, Miao J. MicroRNA-126 inhibits ischemia-induced retinal neovascularization via regulating angiogenic growth factors. Exp Mol Pathol. 2011;91(1):471–477. doi: 10.1016/j.yexmp.2011.04.016. [DOI] [PubMed] [Google Scholar]
- 151.McArthur K, Feng B, Wu Y, Chen S, Chakrabarti S. MicroRNA-200b regulates vascular endothelial growth factor-mediated alterations in diabetic retinopathy. Diabetes. 2011;60(4):1314–1323. doi: 10.2337/db10-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen KC, Hsi E, Hu CY, Chou WW, Liang CL, Juo SH. MicroRNA-328 may influence myopia development by mediating the PAX6 gene. Invest Ophthalmol Vis Sci. 2012;53(6):2732–2739. doi: 10.1167/iovs.11-9272. [DOI] [PubMed] [Google Scholar]
- 153.Rapicavoli NA, Blackshaw S. New meaning in the message: noncoding RNAs and their role in retinal development. Dev Dyn. 2009;238(9):2103–2114. doi: 10.1002/dvdy.21844. [DOI] [PubMed] [Google Scholar]
- 154.Geng X, Lavado A, Lagutin OV, Liu W, Oliver G. Expression of Six3 Opposite Strand (Six3OS) during mouse embryonic development. Gene Expr Patterns. 2007;7(3):252–257. doi: 10.1016/j.modgep.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rapicavoli NA, Poth EM, Zhu H, Blackshaw S. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev. 2011;6:32. doi: 10.1186/1749-8104-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Meola N, Pizzo M, Alfano G, Surace EM, Banfi S. The long noncoding RNA Vax2os1 controls the cell cycle progression of photoreceptor progenitors in the mouse retina. RNA. 2012;18(1):111–123. doi: 10.1261/rna.029454.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Alfano G, Vitiello C, Caccioppoli C, Caramico T, Carola A, Szego MJ, McInnes RR, Auricchio A, Banfi S. Natural antisense transcripts associated with genes involved in eye development. Hum Mol Genet. 2005;14(7):913–923. doi: 10.1093/hmg/ddi084. [DOI] [PubMed] [Google Scholar]
- 158.Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15(6):501–512. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 159.Rapicavoli NA, Poth EM, Blackshaw S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev Biol. 2010;10:49. doi: 10.1186/1471-213X-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2010;16(2):324–337. doi: 10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tsuiji H, Yoshimoto R, Hasegawa Y, Furuno M, Yoshida M, Nakagawa S. Competition between a noncoding exon and introns: gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells: Devoted Mol Cell Mech. 2011;16(5):479–490. doi: 10.1111/j.1365-2443.2011.01502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mori K, Yoshihara Y. Molecular recognition and olfactory processing in the mammalian olfactory system. Prog Neurobiol. 1995;45(6):585–619. doi: 10.1016/0301-0082(94)00058-p. [DOI] [PubMed] [Google Scholar]
- 163.Yoshihara Y, Mori K. Basic principles and molecular mechanisms of olfactory axon pathfinding. Cell Tissue Res. 1997;290(2):457–463. doi: 10.1007/s004410050953. [DOI] [PubMed] [Google Scholar]
- 164.Mori K, von Campenhause H, Yoshihara Y. Zonal organization of the mammalian main and accessory olfactory systems. Philos Trans R Soc Lond B Biol Sci. 2000;355(1404):1801–1812. doi: 10.1098/rstb.2000.0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Brunjes PC, Greer CA. Progress and directions in olfactory development. Neuron. 2003;38(3):371–374. doi: 10.1016/s0896-6273(03)00263-0. [DOI] [PubMed] [Google Scholar]
- 166.Nicolay DJ, Doucette JR, Nazarali AJ. Transcriptional regulation of neurogenesis in the olfactory epithelium. Cell Mol Neurobiol. 2006;26(4–6):803–821. doi: 10.1007/s10571-006-9058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lopez-Mascaraque L, de Castro F. The olfactory bulb as an independent developmental domain. Cell Death Differ. 2002;9(12):1279–1286. doi: 10.1038/sj.cdd.4401076. [DOI] [PubMed] [Google Scholar]
- 168.Ason B, Darnell DK, Wittbrodt B, Berezikov E, Kloosterman WP, Wittbrodt J, Antin PB, Plasterk RH. Differences in vertebrate microRNA expression. Proc Natl Acad Sci USA. 2006;103(39):14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, Muckenthaler MU. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) RNA. 2006;12(5):913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Choi PS, Zakhary L, Choi WY, Caron S, Alvarez-Saavedra E, Miska EA, McManus M, Harfe B, Giraldez AJ, Horvitz HR, Schier AF, Dulac C. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57(1):41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12(9):735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 173.Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14(3):432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc. 2007;2(6):1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
- 175.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283(22):14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Akerblom M, Sachdeva R, Barde I, Verp S, Gentner B, Trono D, Jakobsson J. MicroRNA-124 is a subventricular zone neuronal fate determinant. J Neurosci. 2012;32(26):8879–8889. doi: 10.1523/JNEUROSCI.0558-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pathania M, Torres-Reveron J, Yan L, Kimura T, Lin TV, Gordon V, Teng ZQ, Zhao X, Fulga TA, Van Vactor D, Bordey A. miR-132 enhances dendritic morphogenesis, spine density, synaptic integration, and survival of newborn olfactory bulb neurons. PLoS One. 2012;7(5):e38174. doi: 10.1371/journal.pone.0038174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.de Chevigny A, Core N, Follert P, Gaudin M, Barbry P, Beclin C, Cremer H. miR-7a regulation of Pax6 controls spatial origin of forebrain dopaminergic neurons. Nat Neurosci. 2012;15(8):1120–1126. doi: 10.1038/nn.3142. [DOI] [PubMed] [Google Scholar]
- 179.Meola N, Gennarino VA, Banfi S. microRNAs and genetic diseases. Pathogenetics. 2009;2(1):7. doi: 10.1186/1755-8417-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Morozova O, Hirst M, Marra MA. Applications of new sequencing technologies for transcriptome analysis. Annu Rev Genomics Hum Genet. 2009;10:135–151. doi: 10.1146/annurev-genom-082908-145957. [DOI] [PubMed] [Google Scholar]
- 182.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 183.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297(5589):2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 184.Kim VN. Small RNAs just got bigger: piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev. 2006;20(15):1993–1997. doi: 10.1101/gad.1456106. [DOI] [PubMed] [Google Scholar]
- 185.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, Irvine K, Arakawa T, Nakamura M, Kubosaki A, Hayashida K, Kawazu C, Murata M, Nishiyori H, Fukuda S, Kawai J, Daub CO, Hume DA, Suzuki H, Orlando V, Carninci P, Hayashizaki Y, Mattick JS. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41(5):572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 186.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109(2):145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 188.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322(5909):1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, Shimizu M, Tili E, Rossi S, Taccioli C, Pichiorri F, Liu X, Zupo S, Herlea V, Gramantieri L, Lanza G, Alder H, Rassenti L, Volinia S, Schmittgen TD, Kipps TJ, Negrini M, Croce CM. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12(3):215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 190.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C. Antisense transcription in the mammalian transcriptome. Science. 2005;309(5740):1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 191.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]