Abstract
Discoidin domain receptor 1 (DDR-1)-deficient mice exhibited a high incidence of osteoarthritis (OA) in the temporomandibular joint (TMJ) as early as 9 weeks of age. They showed typical histological signs of OA, including surface fissures, loss of proteoglycans, chondrocyte cluster formation, collagen type I upregulation, and atypical collagen fibril arrangements. Chondrocytes isolated from the TMJs of DDR-1-deficient mice maintained their osteoarthritic characteristics when placed in culture. They expressed high levels of runx-2 and collagen type I, as well as low levels of sox-9 and aggrecan. The expression of DDR-2, a key factor in OA, was increased. DDR-1-deficient chondrocytes from the TMJ were positively influenced towards chondrogenesis by a three-dimensional matrix combined with a runx-2 knockdown or stimulation with extracellular matrix components, such as nidogen-2. Therefore, the DDR-1 knock-out mouse can serve as a novel model for temporomandibular disorders, such as OA of the TMJ, and will help to develop new treatment options, particularly those involving tissue regeneration.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-013-1436-8) contains supplementary material, which is available to authorized users.
Keywords: Temporomandibular joint, Osteoarthritis, Extracellular matrix, Collagen receptor, Chondrocyte signaling
Introduction
Temporomandibular disorders (TMDs) are structural, functional, biochemical, and physiological dysregulations of the muscle or the temporomandibular joint (TMJ). It is estimated that 10–40 % of the population between 18 and 45 years of age present symptoms or signs of TMD, and nearly 10 % are classified as suffering from osteoarthritis (OA) in this joint [1, 2]. Because OA of the TMJ is usually diagnosed only in the later stages of the disease, it is likely that the true incidence of OA of the TMJ may be higher [1]. TMD patients experience severe pain in the mastication muscles, joint clicking, displacement or perforation of the articular disc, and inflammatory or degenerative changes in the joint itself. Untreated TMDs ultimately result in OA of the TMJ [2]. The current therapeutic interventions for OA primarily provide short-term symptomatic relief, and almost all patients ultimately require joint replacement [3]. While the pathogenesis of OA of the TMJ has some special features, the general aspects are similar to those of OA in other joints in which the imbalance between cartilage degradation and matrix synthesis ultimately results in the complete loss of joint function [4]. Within the joint, the articular cartilage is responsible for the smooth transmission of force from one bone to another, thereby allowing painless skeletal movements [5]. The maintenance of articular cartilage extracellular matrix (ECM) is required for biomechanical functions, such as rigidity and resistance to compression and shear forces [6, 7]. Chondrocytes, the cells responsible for cartilage tissue homeostasis, are embedded in a framework of collagens that, together with proteoglycans and glycoproteins [8], act as linking proteins to stabilize the collagen network. The chondrocytes in the articular cartilage do not make direct cell-to-cell contact; instead, they rely on cell–matrix interactions [9] via integrins [10] or DDRs [11] for communication. Cartilage appears to have a low capacity for regeneration, and ECM degradation overrides the well-known tissue regeneration attempts. Recently, we identified chondrogenic progenitor cells (CPCs), which drive these regeneration processes. During the late stages of OA, CPCs are located in the repair tissue of human articular knee cartilage. They exhibit stem cell characteristics such as clonogenicity, multipotency, and migratory activity and exhibit a high chondrogenic potential [12, 13]. Whether CPCs might also play a role in the regeneration attempts of the TMJ is unknown, although this information is of importance in designing new therapies to treat TMD. Because human TMJ specimens are sparse, we aimed to establish a novel mouse model for OA of the TMJ. Histological analyses of the existing mouse models currently used to study OA, including ICR mice [14], Del 1 mice [15], Cho mice [16], mechanically-induced OA via partial discectomy [17], and Col IX knock-out (KO) mice [18], showed a low incidence of TMJ OA. In contrast, we demonstrate that Discoidin domain receptor 1 (DDR-1)-deficient mice exhibit a high incidence of TMJ OA beginning at an early age, and are suitable as a model for TMD. Receptor tyrosine kinases such as DDRs are widely expressed in human and mouse tissues. The binding of collagen to DDR-1 results in tyrosine kinase activation [19]. This activation causes downstream signaling via Shc [20] or FRS2 [21], altering the gene expression levels of ECM molecules [19] that are important for maintaining healthy articular cartilage. Here, we demonstrate that DDR-1-deficient mice display the histological characteristics typical of OA of the TMJ. Furthermore, isolated osteoarthritic DDR-1 KO TMJ cells can be converted to chondrocytes with a more normal phenotype, rendering cell biological interventions possible.
Materials and methods
Tissue sources and preparation
Animals were obtained according to the regulations of the Animal Welfare Act of the County of Lower Saxony, Germany. The generation and genotyping of DDR-1 null mice has been described previously [22]. Cartilage destruction was graded according to the modified Mankin score [23]; however, toluidine blue staining was used instead of safranin-O staining. Accordingly, healthy cartilage received a minimum score of zero points, and late stage OA received a maximum score of 23 points. Three histologists evaluated the tissues independently. The scores of KO and wild-type (WT) mice are shown in Tables 1 and 2.
Table 1.
Grading of TMJ cartilage destruction in KO mice
| Age at examination (weeks) | Number of sacrificed KO mice | Average modified Mankin score (0–23) | Average of OA prevalence (in %) |
|---|---|---|---|
| 6 | 8 | 1 | 50 |
| 9 | 21 | 15 | 83 |
| 12 | 28 | 17 | 89 |
| 24 | 17 | 18 | 91 |
| 28 | 12 | 18 | 91 |
Table 2.
Grading of TMJ cartilage destruction in WT mice
| Age at examination (weeks) | Number of sacrificed WT mice | Average modified Mankin score (0–23) | Average of OA prevalence (in %) |
|---|---|---|---|
| 6 | 6 | 0 | 0 |
| 9 | 15 | 0 | 0 |
| 12 | 20 | 0 | 0 |
| 24 | 20 | 1 | 15 |
| 28 | 15 | 2 | 20 |
Micro-computed tomography
Three DDR-1 knockout and three wild-type (WT) mice were sacrificed at ages ranging from 9 to 12 weeks, and their jaws were prepared. The mineral content was measured in the subchondral bone of the articular surface of the mandible from each group using μ-CT scanning. The bones were scanned with a GE eXplore Locus SP Pre-Clinical Specimen MicroCT instrument (GE Medical Systems, Muenchen, Germany) operated at a 13-m isotropic voxel resolution. The specimens were immersed in water, and hydroxyapatite (1.13 g/cm3) was included in each scan to provide reference values.
Antibodies for immunohistochemistry, immunocytochemistry, immunoblotting and FACS analysis
A monoclonal rat-anti-nidogen-1 antibody (JF4) and a polyclonal rabbit-anti-nidogen-2 antibody (1080+E2) were generously donated by Dr. T. Sasaki (University of Erlangen, Germany). The nidogen-1 and nidogen-2 antibodies have previously been demonstrated not to cross-react [8]. The anti-COMP antibody is an affinity purified polyclonal rabbit-anti-bovine antibody [24]. Goat-anti-rabbit or goat-anti-rat (Dako, Hamburg, Germany) secondary antibodies were used for immunostaining for light microscopy. DDR-1 (h-126, sc-8988), DDR-2 (h-108, sc-8989), aggrecan (4F4: sc-33695), runx-2 (M-70: sc-10758) and sox-9 (H-90: sc-2095) antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Monoclonal anti-type I collagen (M-38) and an anti-type II collagen (CIIC1) antibodies were obtained from the Developmental Studies Hybridoma Bank, University of Iowa, USA. Intracellular FACS analysis was conducted using the Fix & Perm kit® (Invitrogen, Darmstadt, Germany). The MMP-13 antibody (ab39012) was obtained from Abcam (Cambridge, MA, USA). Goat-anti-mouse, goat-anti-rabbit-FITC (Dianova, Hamburg, Germany) and anti-mouse-PE/FITC monoclonal immunoglobulin isotype controls (BD Pharmingen, Mountain View, CA, USA) were used as secondary antibodies. For immunoblotting, we used a goat-anti-mouse antibody coupled with alkaline phosphatase and a pan-β-actin (Dako, Hamburg, Germany) or α-tubulin (mouse monoclonal, DM1A; Sigma-Aldrich, St. Gallen, Switzerland) antibody as gel loading control. The detection of primary cilia was performed with anti-acetylated α-tubulin antibody [6-11B-1] (ab24610), which was purchased from Abcam.
Antibody immunoreactions were also performed in the absence of primary antibody, as a negative control, and images show representatives of three individual experiments.
Light microscopic immunohistochemistry
Immunoperoxidase staining was performed on paraffin-embedded tissue sections as follows. The tissues were deparaffinised, rehydrated, and rinsed for 10 min in PBS. Endogenous peroxidase was blocked by a 45-min treatment with a solution of methanol and 3 % H2O2 in the dark. Each of the reactions was followed by rinsing for 10 min in PBS. The sections were pre-treated for 5 min with 10 μg/ml protease XXIV (P8038; Sigma, Deisenhofen, Germany) and chondroitinase (C3667-5UN; Sigma). The antibodies were applied at a dilution of 1:100 in PBS for 12 h at room temperature. A standard peroxidase–anti-peroxidase procedure followed with the application of a peroxidase-coupled goat-anti-rabbit antibody (Dako;) at a dilution of 1:150 in PBS for 1 h at room temperature. The color reaction was carried out with a DAB (diaminobenzidine) substrate [25].
Electron microscopy
For ultrastructural investigations, 1-mm3 cartilage samples from the condyle were resected. All tissue samples were then fixed and embedded in Epon® (Serva, Heidelberg, Germany). Subsequently, semi-thin (1 μm) and ultra-thin sections (80 nm) were cut. The ultra-thin sections were collected on Formvar®-coated grids and stained as described elsewhere [26].
Cell isolation and culture
Standard explant cultures were established from 1-mm3 tissue specimens taken from the TMJ cartilage tissue of 9-week-old DDR-1 KO mice and their WT littermates. Care was taken to ensure that no bone tissue was included. After 10 days, outgrown chondrocytes were harvested, and 103 cells/cm2 were transferred to cell culture in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10 % fetal bovine serum (Invitrogen; Lot. nr. 41F2061K), gentamycin (50 μg/ml) and l-glutamine (10 mM). Furthermore, we cultured TMJ chondrocytes for 3 weeks in alginate beads, which provides the three-dimensional environment that is important for the phenotypic stability of the chondrocytes [27]. To test the influence of the ECM components, 40,000 cells at P2 were mixed with a 1.2 % alginate solution supplemented with either 125 ng/ml of laminin-1 (Dianova) or nidogen-2 (a kind gift from the late R. Timpl) or with 5 ng/μl of BMP-2 and BMP-4 and grown for 3 weeks in 3D in 6-well plates.
Immunofluorescence microscopy
The primary cells were transferred at P1 in 96-well plates for 16 h, they were fixed with 70 % ethanol and then incubated with 100 μl of primary antibody diluted 1:50 in PBS for 1 h at RT in the dark. When necessary, this step was followed by incubation with a secondary fluorescence-conjugated antibody (diluted 1:500) for 20 min at RT. Two washes with PBS were performed, followed by DAPI staining. The cells were examined under a fluorescence microscope, and the images were captured with a Nikon D90 camera (Düsseldorf, Germany).
Confocal microscopy
Cells were imaged with a FluoView1000 (Olympus) confocal microscope using a ×60 NA1.35 UPLS-APO objective. The excitation/emission wavelengths for DAPI and TRITC were 405/425–520 nm and 561 nm/570–670 nm, respectively. Images were acquired at a resolution of 1,024 × 1,024 with the confocal pinhole set at airy disk 1. Two sequential frames were acquired and averaged.
FACS analysis
The cultured cells were suspended in PBS with the fluorescence-coupled antibodies listed above (1 μl added to 100 μl containing 106 cells) at RT for 1 h in the dark. Two subsequent washing steps were performed (with centrifugation for 10 min at 800 rpm). The cells were analyzed on a FACScan instrument (Becton–Dickinson, Mountain View, CA, USA) as described in detail elsewhere [28]. At least 10,000 living cells were analyzed. The data were evaluated with the aid of WinMDI v.2.9. For cell selection, we applied FACS Vantage SE (Becton–Dickinson). We performed analyses using the Cell Quest Pro 2000 software package.
Immunoblotting
Proteins were extracted using 5 M guanidine hydrochloride and protease inhibitors, precipitated in ethanol, washed in PBS, precipitated again, and finally dissolved in PBS containing 0.4 % SDS. SDS–PAGE was performed with a 6 % acrylamide stacking gel and 12 % separation gel. Gel loading was evaluated using α-actin staining. The proteins were blotted onto nitrocellulose membranes, which were subsequently washed, and blocked, and immunoreactions were performed by exposure to antibodies diluted 1:500 in PBS. The secondary goat-anti-mouse antibody was diluted 1:1,000 and incubated with the membrane for 1 h at RT. Visualization was achieved by applying the ECL Prime Detection Regent (GE Healthcare, Muenchen, Germany), and certain results were quantified using the Image J® program.
siRNA transfections
For silencing experiments, we used the iLenti-GFP siRNA expression vector (Biocat, Heidelberg, Germany) in which the target sequence of the runx-2 siRNA (CAGCACGCTATTAAATCCAAATT) is under the control of the H1 and U6 promoters. For transfection confirmation and efficiency, the reporter gene GFP was placed under the CMV promoter. All control experiments were performed with the vector without the runx-2 silencing insert. Nucleofection of TMJ chondrocytes was performed according to the manufacturer’s instructions (Lonza, Basel, Switzerland). Briefly, the TMJ chondrocytes were trypsinized and counted. The cells were centrifuged at 1,200 rpm for 10 min, and 5 × 105 cells were resuspended in 100 μl of the nucleofection reagent containing 2 μg of plasmid DNA. To increase the transfection efficiency, the U-23 program was selected. Immediately after nucleofection, the TMJ chondrocytes were plated in warm culture medium in a T25 culture flask. The culture medium was replaced the next day to remove dead cells. The cells were harvested after 24 h after medium replacement.
Overexpression
Runx-2 was cloned into the pPM-C-His vector (ABM, Richmond, Canada) using standard procedures. The vector with the insert and the vector without the insert were transfected (PolyFect; Qiagen, Hilden, Germany) into the cells as described above.
RNA extraction and complementary DNA (cDNA) synthesis
Cells in P1 were directly lysed in RLT buffer, and the RNA was isolated according to the manufacturer’s instructions (RNeasy Mini Kit; Qiagen, Chatsworth, CA, USA). RNA was reverse-transcribed into cDNA with the help of the Qiagen QuantiTect Reverse Transcription Kit, as per the manufacturer’s instructions.
Microarray analysis
Quality control and the quantification of total RNA samples was performed prior to the microarray experiments (Agilent 2100 Bioanalyzer; Agilent Technologies, Palo Alto, CA, USA). We used equal amounts of total RNA from the condyles of each of three pairs of DDR-1 KO mice and two samples of their WT littermates. The microarray analysis was conducted at the university transcriptome facility using an Affymetrix whole-mouse genome chip (Affymetrix, Santa Clara, CA, USA). The microarray experiments were performed according to the manufacturer’s protocols. A complete list of the genes present on the chip can be found at http://www.affymetrix.com/analysis/index.affx. The data were analyzed using Affymetrix Microarray Suite 5.0. Gene expression was evaluated using the Affymetrix Data Mining Tool 3.0. The entire dataset is published in a MIAME-compliant format in the GEO database with the accession number GSE35297 (http://www.ncbi.nlm.nih.gov/geo/).
Quantitative rt RT-PCR
PCR was performed in a final volume of 10 μl containing 5 μl Platinum SYBR Green qPCR SuperMixTM (Invitrogen), 20 pmol of each primer and 1 ng of cDNA were added to a final volume of 10. The primers were designed using Primer3® software (http://frodo.wi.mit.edu/cgibin/primer3/primer3). The primer sequences used are shown below:
| Gene ID | Primer forward primer | Primer reverse primer | Accession no. |
|---|---|---|---|
| runx-2 | cagaccagcagcactccata | cagcgtcaacaccatcattc | NM_001271631.1 |
| acan | aggactgaaatcagcggaga | agggacatggttgtttctgc | NM_007424.2 |
| sox-9 | tcagatgcagtgaggagcac | ccagccacagcagtgagtaa | NM_011448.4 |
| col1a1 | tgactggaagagcggagagt | gttcgggctgatgtaccagt | NM_007742.3 |
| IBSP | gcagtagtgactcatccgagaa | gcctcagagtcttcatcttcattc | NM_004967.3 |
| SPP1 | agacctgacatccagtaccctg | gtgggtttcagcactctggt | NM_001040058.1 |
After an initial activation step lasting 3 min at 95 °C, the reaction were performed in 45 cycles of denaturation for 20 s at 95 °C, annealing for 20 s at the primer-specific annealing temperatures and elongation for 20 s at 72 °C. Data acquisition was carried out after each elongation step, and amplification was followed melting curve followed by a melting curve in 0.1 °C steps from 50 to 95 °C. The reaction were performed in a Mastercycler Realplex2 S® instrument (Eppendorf, Hamburg, Germany). Normalization was performed against the mRNA of the appropriate control for each experiment. HPRT-1, MAPK-1, and β2-microglobulin were chosen as housekeeping genes, after we demonstrated that they were expressed at a constant level in individual experimental settings. The PCR products were sequenced (Seqlab, Goettingen, Germany) to confirm their identity. The relative ratios were calculated according to an algorithm published by Pfaffl [29]. Therefore, it was necessary to repeat every PCR-run three times in triplicate, including the housekeeping genes, and a control cDNA for the KO and WT chondrocytes. The intra-test and inter-test variation were both <1 %. The efficiency of each primer was tested with a range of the cDNA dilutions from 1:1 to 1:10,000 and this value was included in the calculation.
PCR array
The transduction pathway array [SAbio Mouse Signal Transduction Pathway Finder PCR Array (PAMM-014; SAbiosciences, Hilden, Germany)] was used according to the manufacturer’s instructions, and the PCR results were calculated and structured using the manufacturer’s online software.
Statistical analyses
We reported representative data from at least three independent experiments, and we statistically tested our results using separate specimens. The analyses were performed using SPSS software 13.0 (SPSS, Chicago, IL, USA). The results are reported as the mean values and standard deviations (SD). After testing for normal distribution and variance homogeneity, we performed a one-way analysis of variance (ANOVA) and a post hoc pairwise comparison of mean values. The Pearson correlation coefficients were calculated to examine the relationships between the parameters. A p value <0.05 was considered significant.
Results
Structural and genomic aspects of OA of the TMJ in DDR-1 KO mice in vivo
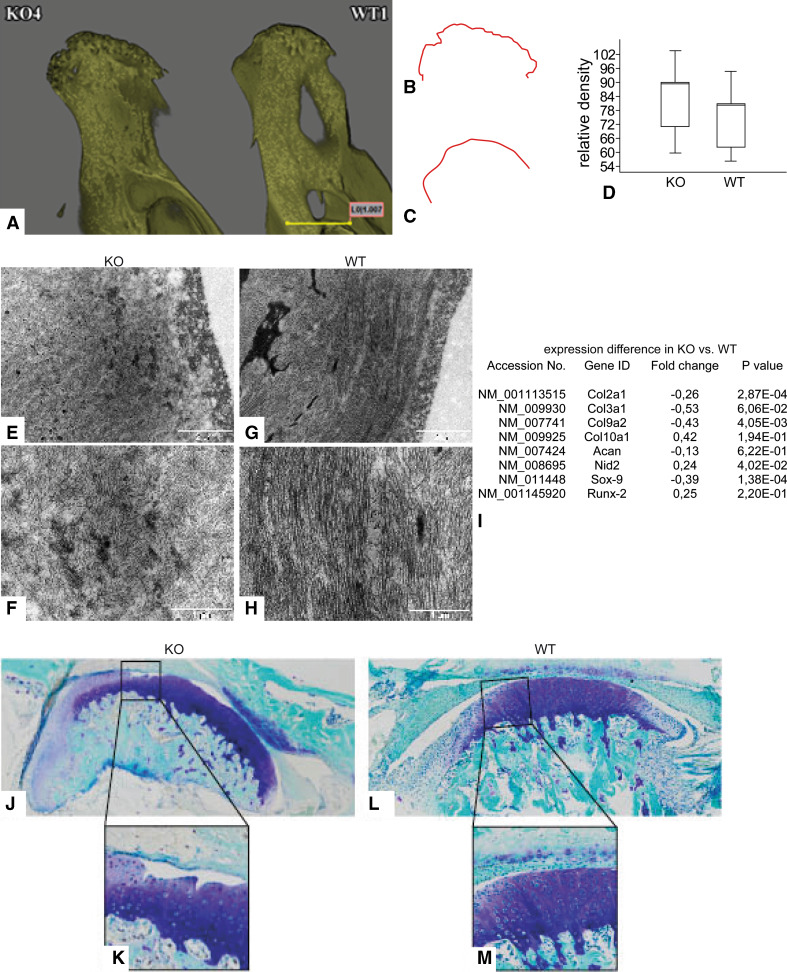
Disordered cell matrix interactions play a central role in the development of OA [9]. Therefore, it is reasonable to hypothesize that collagen receptors, such as integrins [10] and DDRs, are involved in OA pathogenesis. Because severe OA is associated with bone structure alterations [30], we applied microcomputed tomography (μCT) as a straightforward initial approach to examine DDR-1 KO TMJ. Three-dimensional reconstruction of the mandibular condyles of these mice showed a rugged subchondral bone surface and a flattening of the mandibular condyle (Fig. 1a, left, b), structural changes that are typical of TMD [2]. In contrast, the subchondral bone surface of WT mice was smooth and rounded (Fig. 1a, right, c), as expected for a normal joint. Furthermore, the DDR-1 KO mice exhibited a greater relative bone mineral density of the subchondral bone (Fig. 1d). Ultrastructurally, the joint surface of the mandibular condyle of 9-week-old DDR-1 KO mice demonstrated an altered collagen fiber network with loosely packed and randomly arranged collagen fibers in the cartilage (Fig. 1e, f). In comparison, the collagen fiber assembly in WT mice was parallel to the joint surface (Fig. 1g), and the arrangement appeared more compact (Fig. 1h). To investigate the overall changes in gene expression, we performed a microarray analysis of cartilage tissue samples from the mandibles of DDR-1-deficient and WT mice (Fig. 1i). Major changes in the DDR-1 KO mice were associated with ECM components. For example, the DDR-1 KO mice exhibited a lower expression of collagen type II, collagen type III, collagen type IX, aggrecan, and sox-9, while collagen type X, nidogen-2, and runx-2 expression levels were increased; this expression pattern is typical of OA. At the histopathological level, toluidine blue staining (Fig. 1j vs. l) indicated that, by 9 weeks of age, DDR-1 KO mice showed a decreased proteoglycan content at the joint surface of the mandibular condyle compared with their WT littermates. Moreover, loss of the superficial cartilage layer and deep surface fissures were observed in DDR-1 KO mice (Fig. 1k vs. m), but not in WT mice. These OA features were also present in 24-week-old mice, as shown here for toluidine blue staining (Fig. 3i–l).
Fig. 1.
Structural and genomic aspects of OA of the TMJ in DDR-1 KO mice in vivo. a The micro-CT 3D reconstructions of the condylus mandibulae of the DDR-1 KO mice (KO, always on the left side) and the WT (WT, always on the right side). b, c Outline of the subchondral bone surface. The KO mice (b) exhibited a rough surface and an abnormal bone structure compared to the WT (c). d The measurements of the condyle bone mineral density revealed that the KO mice had a higher bone density. e–h Ultrastructural analysis: e, f the collagen fiber arrangement was altered in the superficial layer of DDR-1 KO mice compared to the parallel fiber alignment observed in the WT mice (g, h). i A short list of the fold changes of OA-relevant genes found in the microarray analysis. The complete lists can be found at GEO, GSE35297. j–m Toluidine blue histology of the condyles: j, k the DDR-1 KO condyle exhibits well-known signs of OA such as reduced staining of the superficial zone, cluster formation and surface fissures; l, m normal WT condyles. Three pairs of DDR-1 KO mice condyles and samples from two of their WT littermates were used for microarray analysis. (n = 6, including 3 KO mice and 3 WT mice, for electron microscopy; n = 5, including 3 KO mice and 2 WT mice for the microarray)
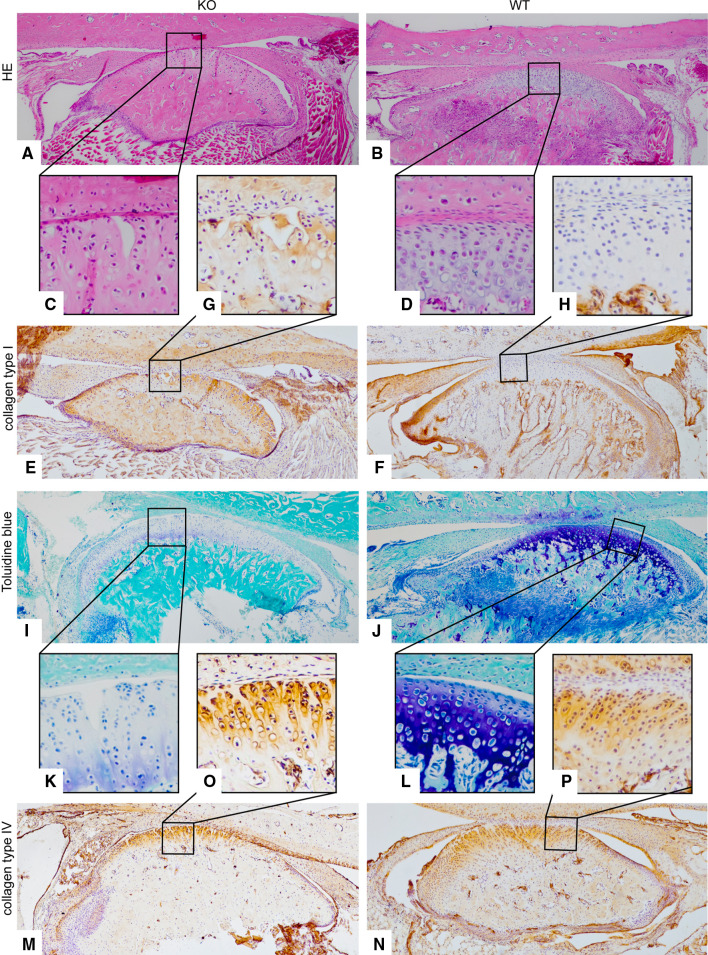
Fig. 3.
The histopathology and immunohistology of 24- and 28-week-old mice. a–d HE staining of 24-week-old mice. c Note the surface fissures in the KO compared with the intact joint surface of the WT mice. d Immunohistochemistry results for collagen type I of staining in 28-week-old mice. There was an increase of collagen type I in the KO (e) versus the WT mice (f). The joint surface of the KO is destroyed (g), while the WT joint was smooth and rounded shaped (h). i–l Toluidine blue histology of 24-weeks old mice: the DDR-1 KO condyle exhibited well-known signs of OA, e.g., reduced staining of the superficial zone (i), cluster formation and surface fissure (k); normal WT condyles were observed in (j, l). m–p Immunohistochemistry results of collagen type IV staining in 28-week-old KO mice: intense staining for collagen type IV was observed at the joint surface (m), especially in the pericellular matrix (o); nNormal WT staining was observed in (n, p). The numbers of the animals evaluated at each time point are shown in Tables 1 and 2
Molecular changes in the ECM of the TMJ during OA in vivo
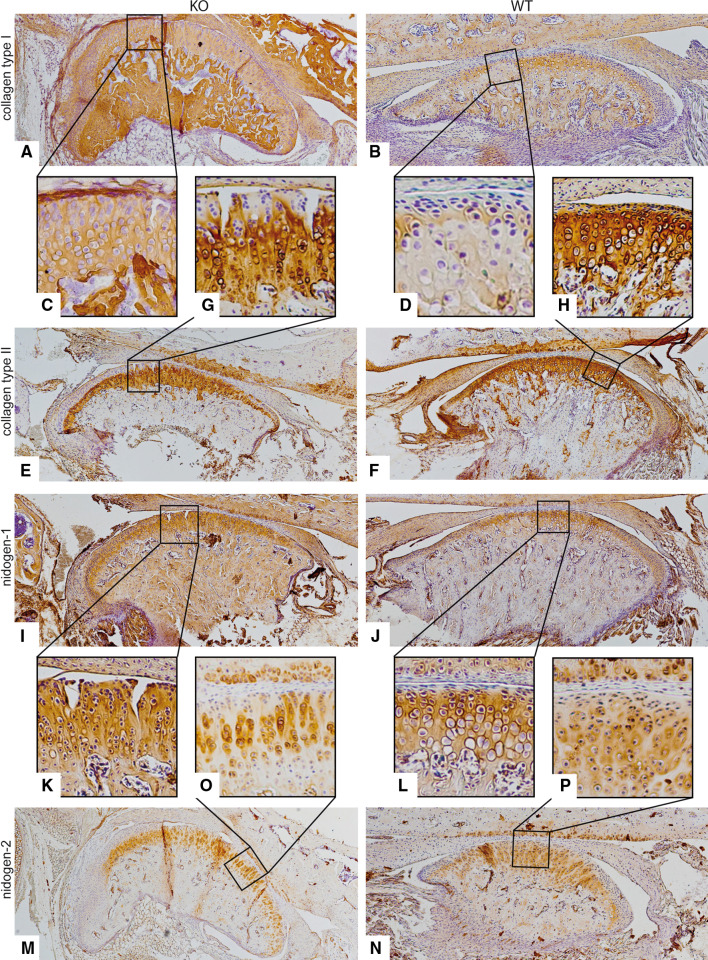
Immunohistochemical analyses of the TMJs of 9-week-old DDR-1 KO mice revealed surface fissures, increased collagen type I, increased collagen fiber fibrillation, and an increased number of cells at the articular surface of the mandibular condyle (Fig. 2a vs. b). This area also exhibited collagen type I staining (Fig. 2c vs. d). Surface fissures also became visible in HE staining of 24-week-old KO mice (Fig. 3a, c) compared with the corresponding WT mice (Fig. 3b, d). These features have been well described and are specific for OA progression [30]. Collagen type I staining was also detected in 28-week-old KO mice (Fig. 3e vs. f). Furthermore, these KO mice also exhibit the typically destroyed joint surface (Fig. 3g vs. h). The expression of collagen type II, the collagen typical of the hyaline cartilage, was decreased in the DDR-1-deficient mice compared with WT controls (Fig. 2e vs. f). This was especially apparent in the superficial layer of the articular cartilage of the TMJ of DDR-1 KO mice, from which collagen type II was absent (Fig. 2g vs. h). The basement membrane proteins [31], most notably the nidogens, are involved as players in the pericellular matrix during the pathogenesis of human OA [6], and an increased amount of nidogen-2 is found primarily around elongated chondrocytes from the late stages of OA [8]. There were no differences in the localization of nidogen-1 in DDR-1 KO mice (Fig. 2i, k) compared with the WT mice (Fig. 2j, l). A pericellular increase in nidogen-2 staining in the middle zone was observed in the TMJ of DDR-1-deficient mice (Fig. 2m vs. n); however, less nidogen-2 staining was observed in the superficial and deeper layers (Fig. 2o vs. p). The same pattern was seen for collagen type IV in 24-week-old mice (Fig. 3m–p). On the basis of these histopathological findings of OA in vivo, we isolated chondrocytes from the TMJs of DDR-1 KO mice for further in vitro studies.
Fig. 2.
The molecular changes in the ECM of the TMJ during OA in vivo of 9-week-old mice: Immunohistochemistry was performed for collagen type I (a–d), collagen type II (e–h), nidogen-1 (i–l) and nidogen-2 (m–p). a, c DDR-1 KO TMJ stained for collagen type I is shown. Note the fibrocartilaginous tissue as a sign of tissue regeneration in (c). b, d The WT TMJ exhibited the well-known collagen type I staining. e, g The KO TMJ showed less collagen type II staining than the WT (f, h). i Nidogen-1 is present in both the KO and the WT mice (j). However, there were no differences between the DDR-1 KO mice (k) and the corresponding WT mice (l). m Nidogen-2 staining was stronger in the TMJs of DDR-1-deficient mice than in the TMJs of WT mice (n). o Note the intense pericellular staining in the deeper zones of the DDR-1 KO TMJ cartilage. Less nidogen-2 was present in the pericellular matrix of the WT cartilage (p). The numbers of the animals evaluated at each time point are shown in Tables 1 and 2
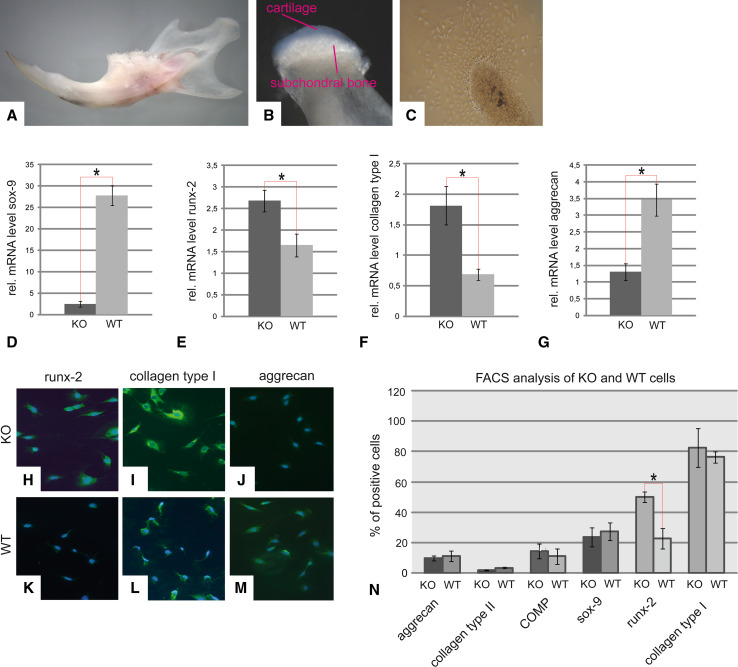
Cell isolation and characterization
Mouse cartilage was separated from the subchondral bone under a stereomicroscope (Fig. 4a, b). After 10 days in culture, the cells from the articular cartilage isolated from DDR-1 KO mice migrated out of the tissue specimens (Fig. 4c). We found that DDR-1 KO TMJ chondrocytes exhibited low levels of sox-9 (a chondrogenic transcription factor; Fig. 4d) and aggrecan (Fig. 4g) mRNA but high levels of runx-2, an osteogenic transcription factor (Fig. 4e), and collagen type I (Fig. 4f) mRNA compared with cells from the WT mice. DDR-1 KO and WT TMJ chondrocytes were positive for runx-2 (Fig. 4h, k), collagen type I (Fig. 4i, l), and aggrecan (Fig. 4j, m) proteins. Chondrocytes from DDR-1 KO mice seemed to express higher levels of runx-2 (Fig. 4h) than WT chondrocytes (Fig. 4k), consistent with the results of our real-time RT-PCR experiments. After passage 6, the phenotype of the TMJ chondrocytes was altered. This dedifferentiation of chondrocytes in culture is a well-known phenomenon and is usually associated with higher passage numbers for cells in culture [32]. We noticed that DDR-1-deficient chondrocytes changed more obviously than WT cells and had smaller cell bodies and numerous cell protrusions compared with WT controls (data not shown). Therefore, we re-evaluated the protein expression levels of the DDR-1-deficient cells at passage 6. We found that both DDR-1 null cells and their WT counterparts were positive for the typical components of cartilage ECM, and both exhibited a chondrocytic nature (Fig. 4n). Even at passage 6, the DDR-1 KO cells and their WT counterparts maintained their differences in runx-2 expression (Fig. 4n, second to last pair of bars). Therefore, up to passage 6, the isolated chondrocytes of the TMJ of the DDR-1 KO and WT cells are appropriate for in vitro investigations of TMD.
Fig. 4.
Cell isolation and characterization. a The dissected mandible of a 9-week-old WT mouse is shown. Care was taken to sample only the translucent cartilage tissue, as shown in (b). c The cells growing out of a tissue sample after 10 days are shown. d–n These cells were identified as chondrocytes. However, DDR-1 KO cells exhibit reduced mRNA expression levels of sox-9 (d) and aggrecan (g) compared with WT cells. In contrast, the KO cells showed an increased expression of runx-2 (e) and collagen type I (f). A similar pattern was observed for the immunocytology; more pronounced staining for runx-2 was present in the DDR-1 KO (h) than in WT cells (k). Collagen type I (i, l), and aggrecan (j, m) were detected in both KO and WT cells. n Intracellular FACS analysis of cells in passage 6 identified aggrecan, collagen type II, COMP, and sox-9, as well as the osteoarthritic markers, runx-2 and collagen type I, in both cell types. A higher percentage of DDR-1 KO chondrocytes expressed runx-2 (second-to-last bars). *Significant differences (p ≤ 0.05); data are mean values with SD of three individual experiments (n = 10, including 4 KO mice, 4 WT mice, and 2 controls, for mRNA measurements; n = 6, including 3 KO mice and 3 WT mice, for immunocytochemistry and FACS-analysis)
Differences of protein patterns of KO and WT cells
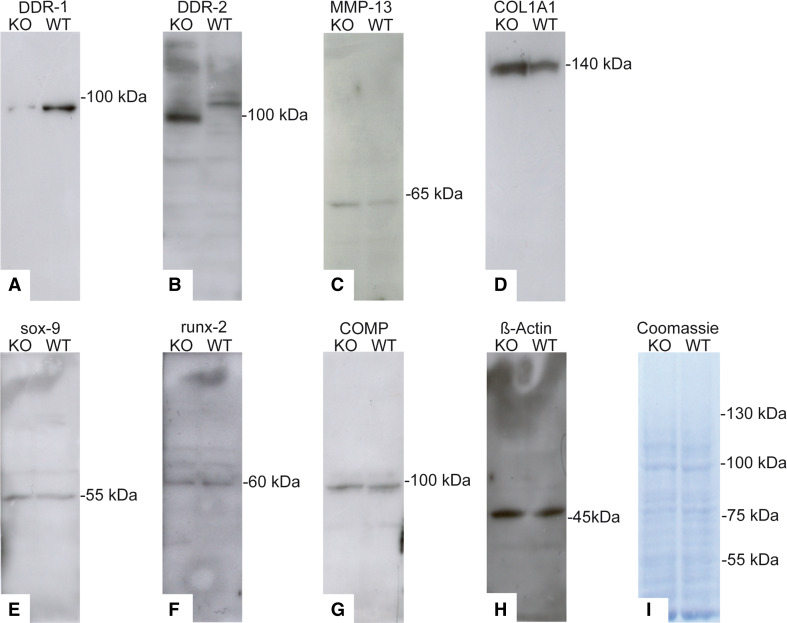
To elucidate ECM and cellular protein expression, we performed western blots at passage 2. In the absence of DDR-1 (Fig. 5a), the expression levels of two key players in OA, DDR-2 (Fig. 5b) and MMP-13 (Fig. 5c), were increased. DDR-1 KO cells produced 1.8× more DDR-2 and 1.6× more MMP-13 than WT cells. Notably, the two described isoforms of DDR-2 [33] were both present in WT cells, whereas the KO chondrocytes exhibited just one stronger band with a lower molecular weight. However, DDR-1 KO cells produced 5.1× more collagen type I than WT cells (Fig. 5d). Sox-9-, runx-2-, and COMP (Fig. 5e–g) were present in nearly equal amounts in DDR-1 KO and WT chondrocytes. The β-actin staining indicated the equal loading of the gels (Fig. 5h). Coomassie blue staining was performed to evaluate the overall protein bands (Fig. 5i).
Fig. 5.
Differences in protein patterns in KO and WT cells. a The western blot for DDR-1 in the DDR-1 KO confirmed its absence; however, DDR-2 (b), MMP-13 (c) and collagen type I (d) were upregulated in the KO chondrocytes. Sox-9 (e), runx-2 (f) and COMP (g) were present in both the KO and WT cells. h β-actin staining confirmed the equal loading of the gels. i Coomassie blue staining was performed to evaluate the overall protein patterns. Protein isolation was performed using cells at passage 2 of the cells. Data are representatives of three individual experiments, or quantified as stated in “Results” (n = 6, including 3 KO mice and 3 WT mice, for western blots)
The influence of the three-dimensional alginate matrix, bone morphogenetic proteins (BMPs), laminin-1, and nidogen-2 and the knockdown of runx-2 on TMJ chondrocytes
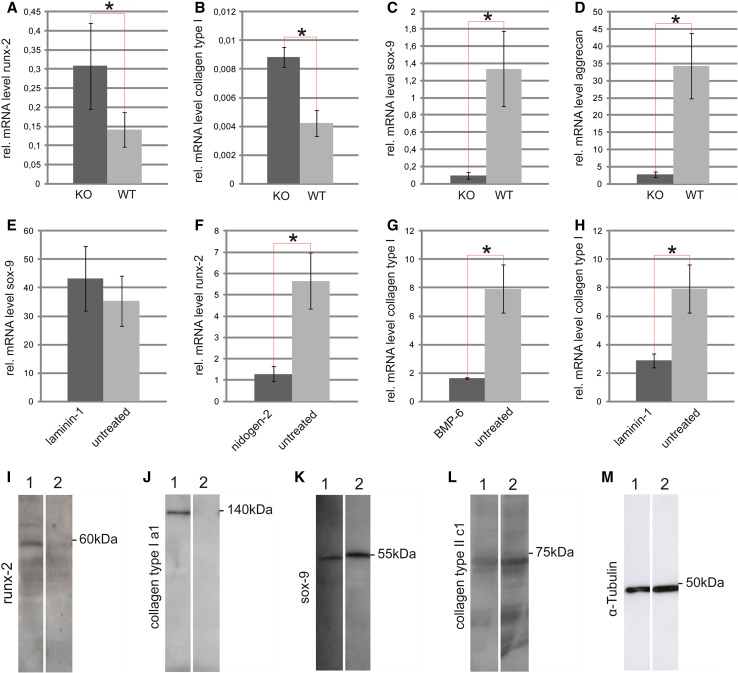
Under 3D conditions [27], DDR-1 KO cells still exhibited high mRNA levels of runx-2 and collagen type I (Fig. 6a, b), and lower amounts of sox-9 and aggrecan (Fig. 6c, d). This observation underscores their osteoarthritic nature. However, we additionally stimulated the cells with the chondrogenic factor BMP-6 [34] and the basement membrane components laminin-1 and nidogen-2 [8], which are present in the normal pericellular chondrocyte matrix [31]. Laminin-1 and nidogen-2 enhanced the chondrogenesis of DDR-1-deficient TMJ chondrocytes, insignificantly increasing mRNA levels of sox-9 (Fig. 6e) and decreasing runx-2 levels (Fig. 6f). Treatment with BMP-6 (Fig. 6g) or laminin-1 (Fig. 6h) reduced collagen type I expression; these expression patterns are all signs of enhanced chondrogenesis. It is known that runx-2 and sox-9 acts as antagonists in the CPCs isolated from late-stage human knee OA [12]. Therefore, we tested whether the knockdown of runx-2 would influence the chondrogenic potential of DDR-1 KO chondrocytes. The transient knockdown of runx-2 mRNA resulted in the complete loss of the runx-2 protein at 24 h (Fig. 6i). The same expression pattern was observed for collagen type I (Fig. 6j). The chondrogenic potential of DDR-1-deficient chondrocytes was improved with the knockdown of runx-2, consistent with the 1.8× higher amounts of sox-9 (Fig. 6k) and 3.4× greater collagen type II expression (Fig. 6l). Tubulin staining was assessed to confirm the equal loading of the gels (Fig. 6m). The key role of runx-2 as a transcription factor of the osteoblastic lineage, was demonstrated by its localization in the nucleus (Supplemental Fig. 1a–c) and the overexpression of runx-2 in the cells (Supplemental Fig. 1d, e). The overexpression resulted in the enhanced gene expression of the downstream mediators Col1A1, SPP1 and IBSP (Supplemental Fig. 1f–i).
Fig. 6.
The influence of three-dimensional alginate matrix, BMPs, laminin-1, nidogen-2 and the knockdown of runx-2 on TMJ chondrocytes: The expression patterns of runx-2 (a), collagen type I (b), sox-9 (c) and aggrecan (d) were similar to the results obtained in 2D-cultured cells (Fig. 3d–g). e Laminin-1 stimulation resulted in an upregulation of sox-9 in DDR-1 KO chondrocytes, while nidogen-2 down-regulates runx-2 (f). Therefore, the two basement membrane components promote the chondrogenesis of these cells. g BMP-6 reduced the relative mRNA levels of collagen type I in DDR-1 KO chondrocytes, as did laminin-1 (h), which is also a marker of chondrogenic differentiation. i Runx-2 protein expression was not detectable 24 h (lane 2) after the knockdown performed by the transient transfection with the siRNA vector. Lane 1 always represents the control. j Collagen type I was not detectable 24 h (lane 2) after runx-2 knockdown, as expected; k the amount of sox-9 was elevated compared with the control cells (lane 1). l Collagen type II was detectable (lane 2), after runx-2 knockdown, but not in the control cells, in which collagen type II was not detectable (lane 1). m Tubulin staining confirmed the equal loading of the gels. k–m are composite figures. *Significant differences (p ≤ 0.05); data are mean values with SD from three individual experiments. (n = 10, including 4 KO mice, 4 WT mice and 2 controls, for mRNA measurements)
Pathways involved in OA of the TMJ and the downstream signaling of DDRs
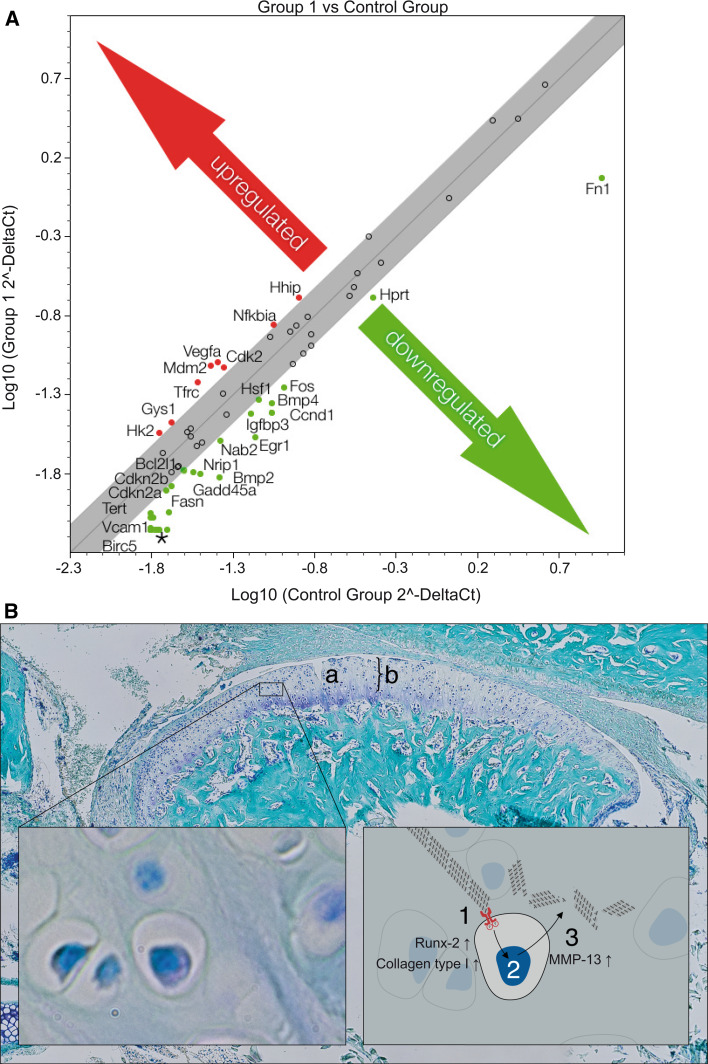
We detected an upregulation of hedgehog interacting protein (HHIP), which is known to be involved in OA pathogenesis [35]. We also found that vascular endothelial growth factor A (VEGFA), which is associated with the Wnt pathway in OA [36], was increased in DDR-1 KO chondrocytes (Fig. 7a). The loss of DDR-1, with its concomitant increase in DDR-2, initiates an upregulation of MMP-13 (Fig. 4c), presumably resulting in the degradation of collagens, mainly type II [17], in TMJ OA. Pit is possible that downstream signaling involves players of the IHH and Wnt pathways (Fig. 7a, b).
Fig. 7.
Possible players downstream of the DDRs in OA of the TMJ. a A PCR array identified the regulated signaling pathway molecules, with the IHH pathway prominently involved in SSH and BMP-2 interactions. VEGFA upregulation was observed. b Surface fissures (a) and proteoglycan degradation (b) of the TMJ of the DDR-1 KO mice revealed typical OA characteristics. The pathomechanism of OA in the TMJ of DDR-1 KO mice results in an upregulation of runx-2, collagen type I and DDR-2 (1). This leads to an increased activation of MMP-13 (2) to enhance matrix degradation, especially of collagens (3). *Also downregulated: Naip1, Brca1, Ccl2, Ccl20, Cd5, Cdh1, Csf2, Cxcl1, Cxcl9, Cyp19a1, En1, Fasl, Fgf4, Greb1, Hoxa1, Icam1, Il1a, Il2, Il2ra, Lef1, Lep, Lta, Mmp10, Mmp7, Nos2, Pparg, Rbp1, Tnf, Wnt1, Wnt2, MGDC, and Selp. Data are representative of three individual experiments. (n = 6, including 3 KO mice and 3 WT mice, for PCR array)
Investigation of the primary cilia in TMJ chondrocytes
It has been established that ECM proteins, mainly collagens, are responsible for the transduction of forces within the cartilage tissue; this is essential for skeletal growth [37]. The main cellular mediator of mechanosensing in chondrocytes is the primary cilium [38]. The microarray data showed that many of the regulated genes with altered expression in the DDR-1 KO were associated with the primary cilia (Supplemental Fig. 2a). Surprisingly, we observed fewer cells with primary cilia among the osteoarthritic DDR-1 KO chondrocytes (Supplemental Fig. 2b, upper panel) than among the WT chondrocytes (Supplemental Fig. 2b, lower panel); however, the differences in numbers did not reach statistical significance (Supplemental Fig. 2c).
Discussion
The present study introduces the DDR-1 null mouse as a new model for OA of the TMJ. These mice develop OA more frequently and at a younger age than other mouse models currently used to study TMD, including the ICR mouse [14], the Del 1 mouse [15], and the Cho mouse [16]. The loss of DDR-1 expression has several significant consequences that influence OA pathogenesis, including increased DDR-2, MMP-13, collagen type I, and runx-2 expression.
The pathomechanism of the DDR-1 KO mice
We applied several approaches to characterize the articular chondrocytes from DDR-1 KO TMJs. Using a combination of real-time RT-PCR, immunocytochemistry, and FACS analysis, we found that DDR1-deficient chondrocytes exhibited characteristics of osteoarthritis. The observed differences between the mRNA and protein levels of certain factors (Fig. 4) are not completely understood. There are a wide variety of post-transcriptional regulatory processes; for example, it is possible that micro RNAs [39] or catabolic enzymes such as RNase [40] play an important role in the cartilage biology of the TMJ cartilage. The KO chondrocytes produced high amounts of collagen type I and runx-2 as well as low levels of collagen type II and sox-9. These changes in gene expression promoted the development of an osteoarthritic phenotype [41]. Most significantly, DDR-1 KO chondrocytes displayed a compensatory increase in the expression of a DDR-2 isoform, and this receptor is linked to elevated MMP-13 (Fig. 5c) expression [42, 43] and articular cartilage degeneration [44]. Using signaling arrays, we determined that DDR-1-null chondrocytes exhibited increased expression of IHH signaling pathway components, as measured by increases in HHIP [35], a protein reported to be overexpressed in human OA and in cartilage from other OA mouse models. Hydrostatic compression of the chondrocytic primary cilia, which is essential for cartilage mechanotransduction [45], upregulates IHH gene expression [46]. It is also known that mechanical stimulation upregulates IHH expression in chondrocytes and is associated with OA [47]. We found a slight tendency towards a reduction of the primary cilia in the osteoarthritic TMJ chondrocytes. We, therefore, only speculate that primary cilia in the DDR-1-deficient chondrocytes might be involved in the pathogenesis.
Why does the lack of DDR-1 result in OA of the TMJ?
Although DDR-1-deficient mice develop OA of the TMJ by 9 weeks of age, we did not detect any signs of OA in the knee joints of mice at this age. In fact, few DDR-1 KO mice developed OA of the large joints. One possible explanation for this finding is the structural difference between the two joints. Unlike the articular cartilage of the knee, the cartilage of the mandibular condyle is considered a secondary cartilage [48, 49] and has a different embryonic origin. In addition, the molecular composition of the TMJ differs from that of the larger synovial joints in that it contains large amounts of collagen type I [50], especially in the marginal areas of the joint where the joint capsule and associated ligaments are found. Furthermore, in contrast to the articular cartilage of other joints, the superficial layer of the mandibular condylar cartilage does not normally express collagen type II, although the functional significance of this difference is unknown [1]. Despite these structural differences, the absence of DDR-1 further reduced the amount of collagen type II and increased the amount of MMP-13 within the TMJ, thereby promoting the early development of OA (Fig. 7b).
DDR-1-deficient chondrocytes and cartilage regeneration
Interestingly, we observed areas of regeneration on the articular surface of the degenerating jaws of DDR-1 KO mice. These areas displayed intense staining for collagen type I, similar to that observed in OA in the human knee joint [51]. In the pericellular space, especially that of cell clusters, an upregulation of nidogen-2 and collagen type IV was observed. This is also an indication of regenerative efforts within the diseased cartilage tissue. TMJ chondrocytes, similar to their counterparts in the knee [12], are regulated by runx-2 and sox-9; here, we were able to enhance the chondrogenic potential of the DDR-1-deficient chondrocytes via runx-2 knockdown. The runx-2 knockdown resulted in an increased expression of sox-9, which then stimulates the enhanced expression of collagen type II and reduced levels of collagen type I. Similar effects were also shown for aggrecan [52] and COMP [53]. Therefore, the chondrogenic potential is enhanced as DDR-1-deficient chondrocytes lose their osteoarthritic character via the runx-2 knockdown. Another target for the regeneration of TMJ cartilage is the DDR-2 receptor. It is well known that DDR-2 is activated via a direct interaction with collagen type II, which does not appear in the pericellular matrix of the chondrocyte in healthy cartilage, but it does in OA. Therefore, DDR-2 is activated during OA. For example, a mutation in DDR-2, that disrupts its binding to collagen type II, reduces the collagen-induced expression of MMP-13 [54].
Taken together, our findings indicate that the DDR-1-deficient mouse is a novel animal model for the in vivo study of human TMD. Furthermore, the isolated TMJ chondrocytes can be used for the in vitro analysis of pathomechanisms of the TMJ. We show that TMJ cartilage regeneration is controlled by the transcription factors sox-9 and runx-2 and is influenced by signals from the pericellular matrix, including BMPs that enhance chondrogenesis. These measures result in a gene expression signature similar to that of normal articular cartilage, suggesting that the DDR-1 KO mouse can serve as a novel model for TMD, such as OA of the TMJ. This model will help to develop and test new treatment options, particularly those involving tissue regeneration.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1 The role of runx-2 as a transcription factor in the osteoblastic lineage. a DAPI staining of a chondrocytes nucleus, b immunocytochemistry of runx-2, localization in the cytoplasm and nucleus of the same chondrocyte was observed. c Merged image of a and b for better visualization of runx-2 in the nucleus. Overexpression of runx-2. Lane 1 control with the vector without insert; lane 2 page ruler; and lane 3 overexpressed runx-2. Coomassie blue staining was performed to determine the overall protein pattern and to confirm equal loading, as observed in d. Western blot of runx-2, as observed in e. Measured mRNA expression levels of osteogenic markers of the cells after overexpression of runx-2. We observed enhanced gene expression of f runx-2, g Col1A1, h SPP1 and i IBSP. *Significant differences (p ≤ 0.05); data are mean values with SD from three individual experiments (JPEG 2551 kb)
Supplemental Fig. 2 Preliminary investigation of primary cilia in TMJ chondrocytes. a Microarray results of genes associated with primary cilia. b DAPI staining (left) of the nuclei of the chondrocytes and acetylated α-tubulin staining (red for better visualization) of the primary cilia (right) are shown. c The quantification of the results indicated a tendency towards a reduction of the percentage of cells with primary cilia in DDR-1 KO chondrocytes. Data are mean values with SD of three individual experiments. (n = 6, including three KO mice and three WT mice, for cilia staining) (JPEG 5676 kb)
Acknowledgments
The authors would like to thank the staff of the animal facilities at the MPI Experimental Medicine, Goettingen and the Medical Faculty of the University of Goettingen for animal care, Dr. Bunt, Molecular & Live Cell Imaging (MOLCI), central imaging facility of the UMG, for the confocal microscopy work, Dr. Salinas-Riester for performing the microarray and Mr. Opitz for statistical evaluation, and Dr. Dullin for help with the micro-CT. We would also like to thank Mr. Hoehne, as parts of the results were taken from his doctoral thesis. We also wish to thank Mr. Menrath for professional assistance with the figure layout.
Footnotes
B. Schminke and H. Muhammad contributed equally to this work.
Contributor Information
Vicki Rosen, Phone: +1-61-74325910, FAX: +1-61-74323246, Email: vicki_rosen@hsdm.harvard.edu.
Nicolai Miosge, Phone: +49-55-1393927, FAX: +49-55-1398621, Email: nmiosge@gwdg.de.
References
- 1.Wadhwa S, Embree M, Ameye L, Young MF. Mice deficient in biglycan and fibromodulin as a model for temporomandibular joint osteoarthritis. Cells Tissues Organs. 2005;181(3–4):136–143. doi: 10.1159/000091375. [DOI] [PubMed] [Google Scholar]
- 2.Carlson GE, Magnusson T. Management of temporomandibular disorders in the general dental practice. Berlin: Quintessenz; 1999. [Google Scholar]
- 3.Lohmander LS, Roos EM. Clinical update: treating osteoarthritis. Lancet. 2007;370(9605):2082–2084. doi: 10.1016/S0140-6736(07)61879-0. [DOI] [PubMed] [Google Scholar]
- 4.Buckwalter JA, Mankin HJ. Articular cartilage: degeneration and osteoarthritis, repair, regeneration, and transplantation. Instr Course Lect. 1998;47:487–504. [PubMed] [Google Scholar]
- 5.Horton WE, Jr, Bennion P, Yang L. Cellular, molecular, and matrix changes in cartilage during aging and osteoarthritis. J Musculoskel Neuronal Interact. 2006;6(4):379–381. [PubMed] [Google Scholar]
- 6.Poole AR. An introduction to the pathophysiology of osteoarthritis. Front Biosci. 1999;4:D662–D670. doi: 10.2741/Poole. [DOI] [PubMed] [Google Scholar]
- 7.Sandell LJ. Modern molecular analysis of a traditional disease: progression in osteoarthritis. Arthr Rheum. 2007;56(8):2474–2477. doi: 10.1002/art.22760. [DOI] [PubMed] [Google Scholar]
- 8.Kruegel J, Sadowski B, Miosge N. Nidogen-1 and nidogen-2 in healthy human cartilage and in late-stage osteoarthritis cartilage. Arthr Rheum. 2008;58(5):1422–1432. doi: 10.1002/art.23480. [DOI] [PubMed] [Google Scholar]
- 9.Kuettner KE. Biochemistry of articular cartilage in health and disease. Clin Biochem. 1992;25(3):155–163. doi: 10.1016/0009-9120(92)90224-G. [DOI] [PubMed] [Google Scholar]
- 10.Loeser RF. Chondrocyte integrin expression and function. Biorheology. 2000;37(1–2):109–116. [PubMed] [Google Scholar]
- 11.Goldring MB, Otero M. Inflammation in osteoarthritis. Curr Opin Rheumatol. 2011;23(5):471–478. doi: 10.1097/BOR.0b013e328349c2b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koelling S, Kruegel J, Irmer M, Path JR, Sadowski B, Miro X, Miosge N. Migratory chondrogenic progenitor cells from repair tissue during the later stages of human osteoarthritis. Cell Stem Cell. 2009;4(4):324–335. doi: 10.1016/j.stem.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Seol D, McCabe DJ, Choe H, Zheng H, Yu Y, Jang K, Walter MW, Lehman AD, Ding L, Buckwalter JA, Martin JA. Chondrogenic progenitor cells respond to cartilage injury. Arthr Rheum. 2012;64(11):3626–3637. doi: 10.1002/art.34613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silbermann M, Livne E. Age-related degenerative changes in the mouse mandibular joint. J Anat. 1979;129(Pt 3):507–520. [PMC free article] [PubMed] [Google Scholar]
- 15.Rintala M, Metsaranta M, Saamanen AM, Vuorio E, Ronning O. Abnormal craniofacial growth and early mandibular osteoarthritis in mice harbouring a mutant type II collagen transgene. J Anat. 1997;190(Pt 2):201–208. doi: 10.1046/j.1469-7580.1997.19020201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu L, Flahiff CM, Waldman BA, Wu D, Olsen BR, Setton LA, Li Y. Osteoarthritis-like changes and decreased mechanical function of articular cartilage in the joints of mice with the chondrodysplasia gene (cho) Arthr Rheum. 2003;48(9):2509–2518. doi: 10.1002/art.11233. [DOI] [PubMed] [Google Scholar]
- 17.Xu L, Polur I, Servais JM, Hsieh S, Lee PL, Goldring MB, Li Y. Intact pericellular matrix of articular cartilage is required for unactivated discoidin domain receptor 2 in the mouse model. Am J Pathol. 2011;179(3):1338–1346. doi: 10.1016/j.ajpath.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lam NP, Li Y, Waldman AB, Brussiau J, Lee PL, Olsen BR, Xu L. Age-dependent increase of discoidin domain receptor 2 and matrix metalloproteinase 13 expression in temporomandibular joint cartilage of type IX and type XI collagen-deficient mice. Arch Oral Biol. 2007;52(6):579–584. doi: 10.1016/j.archoralbio.2006.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogel W. Discoidin domain receptors: structural relations and functional implications. FASEB J. 1999;13(Suppl):S77–S82. doi: 10.1096/fasebj.13.9001.s77. [DOI] [PubMed] [Google Scholar]
- 20.Vogel W, Gish GD, Alves F, Pawson T. The discoidin domain receptor tyrosine kinases are activated by collagen. Mol Cell. 1997;1(1):13–23. doi: 10.1016/S1097-2765(00)80003-9. [DOI] [PubMed] [Google Scholar]
- 21.Foehr ED, Tatavos A, Tanabe E, Raffioni S, Goetz S, Dimarco E, De Luca M, Bradshaw RA. Discoidin domain receptor 1 (DDR1) signaling in PC12 cells: activation of juxtamembrane domains in PDGFR/DDR/TrkA chimeric receptors. FASEB J. 2000;14(7):973–981. doi: 10.1096/fasebj.14.7.973. [DOI] [PubMed] [Google Scholar]
- 22.Vogel WF, Aszodi A, Alves F, Pawson T. Discoidin domain receptor 1 tyrosine kinase has an essential role in mammary gland development. Mol Cell Biol. 2001;21(8):2906–2917. doi: 10.1128/MCB.21.8.2906-2917.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Little C, Smith S, Ghosh P, Bellenger C. Histomorphological and immunohistochemical evaluation of joint changes in a model of osteoarthritis induced by lateral meniscectomy in sheep. J Rheumatol. 1997;24(11):2199–2209. [PubMed] [Google Scholar]
- 24.Hedbom E, Antonsson P, Hjerpe A, Aeschlimann D, Paulsson M, Rosa-Pimentel E, Sommarin Y, Wendel M, Oldberg A, Heinegard D. Cartilage matrix proteins. An acidic oligomeric protein (COMP) detected only in cartilage. J Biol Chem. 1992;267(9):6132–6136. [PubMed] [Google Scholar]
- 25.Koelling S, Clauditz TS, Kaste M, Miosge N. Cartilage oligomeric matrix protein is involved in human limb development and in the pathogenesis of osteoarthritis. Arthr Res Ther. 2006;8(3):R56. doi: 10.1186/ar1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gersdorff N, Kohfeldt E, Sasaki T, Timpl R, Miosge N. Laminin gamma 3 chain binds to nidogen and is located in murine basement membranes. J Biol Chem. 2005;280(23):22146–22153. doi: 10.1074/jbc.M501875200. [DOI] [PubMed] [Google Scholar]
- 27.Hauselmann HJ, Fernandes RJ, Mok SS, Schmid TM, Block JA, Aydelotte MB, Kuettner KE, Thonar EJ. Phenotypic stability of bovine articular chondrocytes after long-term culture in alginate beads. J Cell Sci. 1994;107(Pt 1):17–27. doi: 10.1242/jcs.107.1.17. [DOI] [PubMed] [Google Scholar]
- 28.Diaz-Romero J, Gaillard JP, Grogan SP, Nesic D, Trub T, Mainil-Varlet P. Immunophenotypic analysis of human articular chondrocytes: changes in surface markers associated with cell expansion in monolayer culture. J Cell Physiol. 2005;202(3):731–742. doi: 10.1002/jcp.20164. [DOI] [PubMed] [Google Scholar]
- 29.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldring MB, Goldring SR. Articular cartilage and subchondral bone in the pathogenesis of osteoarthritis. Ann NY Acad Sci. 2010;1192:230–237. doi: 10.1111/j.1749-6632.2009.05240.x. [DOI] [PubMed] [Google Scholar]
- 31.Kvist AJ, Nystrom A, Hultenby K, Sasaki T, Talts JF, Aspberg A. The major basement membrane components localize to the chondrocyte pericellular matrix—a cartilage basement membrane equivalent? Matrix Biol. 2008;27(1):22–33. doi: 10.1016/j.matbio.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 32.Holtzer H, Abbott J, Lash J, Holtzer S. The loss of phenotypic traits by differentiated cells in vitro, I. Dedifferentiation of cartilage cells. Proc Natl Acad Sci USA. 1960;46(12):1533–1542. doi: 10.1073/pnas.46.12.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruiz PA, Jarai G. Collagen I induces discoidin domain receptor (DDR) 1 expression through DDR2 and a JAK2-ERK1/2-mediated mechanism in primary human lung fibroblasts. J Biol Chem. 2011;286(15):12912–12923. doi: 10.1074/jbc.M110.143693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bandyopadhyay A, Tsuji K, Cox K, Harfe BD, Rosen V, Tabin CJ. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006;2(12):e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin AC, Seeto BL, Bartoszko JM, Khoury MA, Whetstone H, Ho L, Hsu C, Ali SA, Alman BA. Modulating hedgehog signaling can attenuate the severity of osteoarthritis. Nat Med. 2009;15(12):1421–1425. doi: 10.1038/nm.2055. [DOI] [PubMed] [Google Scholar]
- 36.Enomoto H, Inoki I, Komiya K, Shiomi T, Ikeda E, Obata K, Matsumoto H, Toyama Y, Okada Y. Vascular endothelial growth factor isoforms and their receptors are expressed in human osteoarthritic cartilage. Am J Pathol. 2003;162(1):171–181. doi: 10.1016/S0002-9440(10)63808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reich A, Maziel SS, Ashkenazi Z, Ornan EM. Involvement of matrix metalloproteinases in the growth plate response to physiological mechanical load. J Appl Physiol. 2010;108(1):172–180. doi: 10.1152/japplphysiol.00821.2009. [DOI] [PubMed] [Google Scholar]
- 38.Whitfield JF. The solitary (primary) cilium—a mechanosensory toggle switch in bone and cartilage cells. Cell Signal. 2008;20(6):1019–1024. doi: 10.1016/j.cellsig.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 39.Martinez-Sanchez A, Dudek KA, Murphy CL. Regulation of human chondrocyte function through direct inhibition of cartilage master regulator SOX9 by microRNA-145 (miRNA-145) J Biol Chem. 2012;287(2):916–924. doi: 10.1074/jbc.M111.302430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mattijssen S, Welting TJ, Pruijn GJ. RNase MRP and disease. Wiley Interdiscip Rev RNA. 2010;1(1):102–116. doi: 10.1002/wrna.9. [DOI] [PubMed] [Google Scholar]
- 41.Kamekura S, Kawasaki Y, Hoshi K, Shimoaka T, Chikuda H, Maruyama Z, Komori T, Sato S, Takeda S, Karsenty G, Nakamura K, Chung UI, Kawaguchi H. Contribution of runt-related transcription factor 2 to the pathogenesis of osteoarthritis in mice after induction of knee joint instability. Arthr Rheum. 2006;54(8):2462–2470. doi: 10.1002/art.22041. [DOI] [PubMed] [Google Scholar]
- 42.Xu L, Peng H, Wu D, Hu K, Goldring MB, Olsen BR, Li Y. Activation of the discoidin domain receptor 2 induces expression of matrix metalloproteinase 13 associated with osteoarthritis in mice. J Biol Chem. 2005;280(1):548–555. doi: 10.1074/jbc.M411036200. [DOI] [PubMed] [Google Scholar]
- 43.Otero M, Plumb DA, Tsuchimochi K, Dragomir CL, Hashimoto K, Peng H, Olivotto E, Bevilacqua M, Tan L, Yang Z, Zhan Y, Oettgen P, Li Y, Marcu KB, Goldring MB. E74-like factor 3 (ELF3) impacts on matrix metalloproteinase 13 (MMP13) transcriptional control in articular chondrocytes under proinflammatory stress. J Biol Chem. 2012;287(5):3559–3572. doi: 10.1074/jbc.M111.265744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sunk IG, Bobacz K, Hofstaetter JG, Amoyo L, Soleiman A, Smolen J, Xu L, Li Y. Increased expression of discoidin domain receptor 2 is linked to the degree of cartilage damage in human knee joints: a potential role in osteoarthritis pathogenesis. Arthr Rheum. 2007;56(11):3685–3692. doi: 10.1002/art.22970. [DOI] [PubMed] [Google Scholar]
- 45.Wann AK, Zuo N, Haycraft CJ, Jensen CG, Poole CA, McGlashan SR, Knight MM. Primary cilia mediate mechanotransduction through control of ATP-induced Ca2+ signaling in compressed chondrocytes. FASEB J. 2012;26(4):1663–1671. doi: 10.1096/fj.11-193649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shao YY, Wang L, Welter JF, Ballock RT. Primary cilia modulate Ihh signal transduction in response to hydrostatic loading of growth plate chondrocytes. Bone. 2012;50(1):79–84. doi: 10.1016/j.bone.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang CF, Ramaswamy G, Serra R. Depletion of primary cilia in articular chondrocytes results in reduced Gli3 repressor to activator ratio, increased Hedgehog signaling, and symptoms of early osteoarthritis. Osteoarthr Cartil. 2012;20(2):152–161. doi: 10.1016/j.joca.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Symons NB. A histochemical study of the secondary cartilage of the mandibular condyle in the rat. Arch Oral Biol. 1965;10(4):579–584. doi: 10.1016/0003-9969(65)90003-8. [DOI] [PubMed] [Google Scholar]
- 49.Shen G, Darendeliler MA. The adaptive remodeling of condylar cartilage—a transition from chondrogenesis to osteogenesis. J Dent Res. 2005;84(8):691–699. doi: 10.1177/154405910508400802. [DOI] [PubMed] [Google Scholar]
- 50.Benjamin M, Ralphs JR. Biology of fibrocartilage cells. Int Rev Cytol. 2004;233:1–45. doi: 10.1016/S0074-7696(04)33001-9. [DOI] [PubMed] [Google Scholar]
- 51.Koelling S, Miosge N. Sex differences of chondrogenic progenitor cells in late stages of osteoarthritis. Arthr Rheum. 2010;62(4):1077–1087. doi: 10.1002/art.27311. [DOI] [PubMed] [Google Scholar]
- 52.Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22(1):85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 53.Tallheden T, Karlsson C, Brunner A, Van Der Lee J, Hagg R, Tommasini R, Lindahl A. Gene expression during redifferentiation of human articular chondrocytes. Osteoarthr Cartil. 2004;12(7):525–535. doi: 10.1016/j.joca.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 54.Xu L, Peng H, Glasson S, Lee PL, Hu K, Ijiri K, Olsen BR, Goldring MB, Li Y. Increased expression of the collagen receptor discoidin domain receptor 2 in articular cartilage as a key event in the pathogenesis of osteoarthritis. Arthr Rheum. 2007;56(8):2663–2673. doi: 10.1002/art.22761. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1 The role of runx-2 as a transcription factor in the osteoblastic lineage. a DAPI staining of a chondrocytes nucleus, b immunocytochemistry of runx-2, localization in the cytoplasm and nucleus of the same chondrocyte was observed. c Merged image of a and b for better visualization of runx-2 in the nucleus. Overexpression of runx-2. Lane 1 control with the vector without insert; lane 2 page ruler; and lane 3 overexpressed runx-2. Coomassie blue staining was performed to determine the overall protein pattern and to confirm equal loading, as observed in d. Western blot of runx-2, as observed in e. Measured mRNA expression levels of osteogenic markers of the cells after overexpression of runx-2. We observed enhanced gene expression of f runx-2, g Col1A1, h SPP1 and i IBSP. *Significant differences (p ≤ 0.05); data are mean values with SD from three individual experiments (JPEG 2551 kb)
Supplemental Fig. 2 Preliminary investigation of primary cilia in TMJ chondrocytes. a Microarray results of genes associated with primary cilia. b DAPI staining (left) of the nuclei of the chondrocytes and acetylated α-tubulin staining (red for better visualization) of the primary cilia (right) are shown. c The quantification of the results indicated a tendency towards a reduction of the percentage of cells with primary cilia in DDR-1 KO chondrocytes. Data are mean values with SD of three individual experiments. (n = 6, including three KO mice and three WT mice, for cilia staining) (JPEG 5676 kb)