Abstract
The vesicular transport pathways, which shuttle materials to and from the cell surface and within the cell, and the metabolic (growth factor and nutrient) signalling pathways, which integrate a variety of extracellular and intracellular signals to mediate growth, proliferation or survival, are both important for cellular physiology. There is evidence to suggest that the transport and metabolic signalling pathways intersect—vesicular transport can affect the regulation of metabolic signals and vice versa. The Rab family GTPases regulate the specificity of vesicular transport steps in the cell. Together with their interacting proteins, Rabs would likely constitute the points of intersection between vesicular transport and metabolic signalling pathways. Examples of these points would include growth factor signalling, glucose and lipid metabolism, as well as autophagy. Many of these processes involve mechanistic/mammalian target of rapamycin (mTOR) complex 1 (mTORC1) in downstream cascades, or are regulated by TORC signalling. A general functionality of the vesicular transport processes controlled by the Rabs is also important for spatial and temporal regulation of the transmission of metabolic signals between the cell surface and the nucleus. In other cases, specific Rabs and their interacting proteins are known to function in recruiting metabolism-related proteins to target membranes, or may compete with other factors in the TORC signalling pathway as a means of metabolic regulation. We review and discuss herein examples of how Rabs and their interacting proteins can mediate metabolic signalling and regulation in cells.
Keywords: Rab GTPases, Growth factor signalling, Metabolism, Autophagy, TORC, Vesicular trafficking
Introduction
In eukaryotic cells, vesicular trafficking processes convey newly synthesised proteins to various membrane compartments within the cell and recycles proteins such as receptors and signalling molecules from the cell surface. An important set of players in regulating the process of vesicular trafficking is the Rab/Ypt proteins, the biggest subfamily in the Ras superfamily of small GTPases. More than 60 mammalian Rabs and 11 yeast Ypt genes have been identified thus far [1, 2]. These serve multiple cellular processes ranging from structure biogenesis and maintenance, to various aspects of membrane dynamics.
Several sets of interacting proteins work together with Rabs to ensure pathway and membrane-specific function. The localisation of Rab proteins are regulated in part by Rab escort proteins (REPs), which bring newly synthesised Rabs to geranylgeranyl transferases (GGTase) for lipid modification of their two C-terminal cysteines, thus enabling their anchoring onto specific target membranes. Rab GDP dissociation inhibitors (GDI), on the other hand, regulate the recycling of Rabs between the membrane and the cytosol [3, 4]. Rabs are essentially molecular switches that alternate between their active and inactive forms depending on their guanine nucleotide binding status—they are either activated or inactivated via the engagement of GTP or GDP, respectively, with the help of two sets of regulatory proteins. Guanine nucleotide exchange factors (GEFs) catalyse the exchange of GDP for GTP [1]. The thus activated Rabs can then interact with Rab effectors, which are various factors involved in vesicular trafficking, including but not limited to tethering factors, motor proteins, and SNAREs [5–8]. Conversely, GTPase-activating proteins (GAPs) engage GTP-bound Rab and enhance their intrinsic GTPase activity, thus accelerating hydrolysis of GTP to GDP to effectively inactivate it [9].
Rab-mediated vesicular transport processes are important for many aspects of cellular physiology, from key processes of structural turnover to cell cycle progression. However, a perhaps under-appreciated aspect of vesicular transport in eukaryotic cells is its role in metabolic regulation. Growth signals from the cell’s external environment, such as growth factors, hormones and nutrients, are integrated through various cellular signalling pathways triggered by receptors on the cell surface and linked via a cascade of second messenger systems, ultimately resulting in transcriptional and translational processes that culminate in cell proliferation or growth. Findings in the past several years point to the fact that vesicular transport and growth factor signalling/nutrient sensing systems not only work in parallel but also intersect [10, 11]. In the paragraphs below, we shall provide an overview on the intersection between vesicular trafficking and signalling processes in metabolic regulation, with a particular focus on the roles of Rabs (and their interacting partners), which are briefly summarised in Fig. 1.
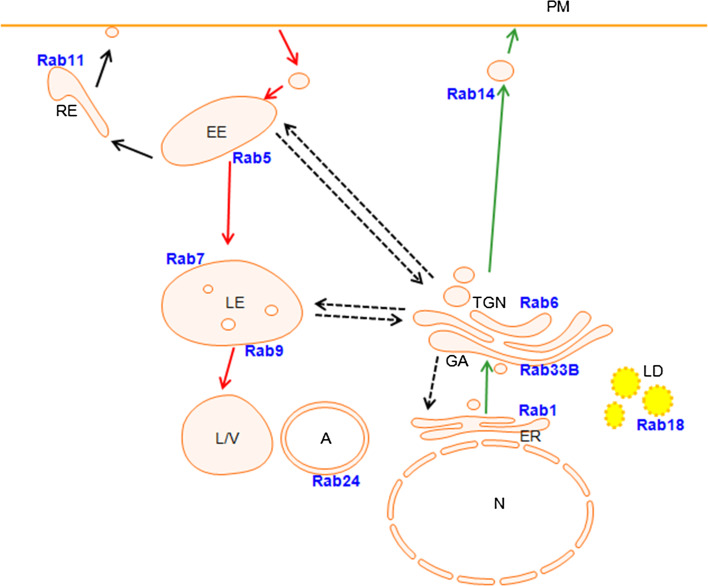
Fig. 1.
Rabs and their unique subcellular localisation. The endocytic pathway involves retrograde transport pathways (red arrows) and the exocytic/biosynthetic pathway involves anterograde transport pathways (green arrows). Cargo at the early endosome can also be recycled to the plasma membrane or returned to the TGN (black arrows). Proteins localised to various membranous compartments are themselves trafficked and recycled to and from the Golgi (dotted arrows). Rabs are found on various membrane compartments in the cell and mediate various trafficking steps. Some representative Rabs acting in these regards are shown. N nucleus, ER endoplasmic reticulum, GA Golgi apparatus, TGN trans-Golgi network, EE early endosome, RE recycling endosome, LE late endosome, L/V lysosome/vacuole, LD lipid droplet, A autophagosome, PM plasma membrane
Rabs in growth factor signal transduction
Rabs affect growth factor signalling via regulation of vesicular trafficking
Growth factors are typically soluble, secreted signals which stimulate cell growth, proliferation and differentiation. Endocytic vesicular transport is one important means by which these external signals are relayed and integrated within the cell. The participation of post-Golgi and endosomal Rabs in growth factor signalling has been well explored and documented. For example, Rab5 mediates early endosomal trafficking steps and plays an essential role in the internalisation and endosomal transport of ligand-bound epidermal growth factor receptors (EGFRs) from the plasma membrane to eventual lysosomal degradation [12, 13]. It is through this endocytic itinerary that signalling from EGF is eventually terminated, but before lysosomal degradation the internalised EGF-EGFR complex continues to signal from the endosomes [14]. It has recently been shown that other members of the Rab5 subfamily, including Rab22A and Rab31, also regulate the internalisation and trafficking of ligand-bound EGFR [15–17]. Rab7 was also shown to play a role in the lysosomal degradation of EGFR by mediating trafficking between the late endosome and the lysosome [18]. Internalised receptors can also be recycled to the cell surface. Rab11 mediates the trafficking and function of a pool of recycling endosomes that can return internalised receptors to the cells’ surface. The Rab11 family interacting protein-2 (FIP2), was shown to aid in the internalisation with EGFR from the cell surface and is also postulated to target EGFR to recycling endosomes via its interaction with Rab11 [19]. Rab4, on the other hand, mediates a fast recycling step involving the transition of cargo from Rab5-positive to Rab4-positive compartments. This is postulated to be mediated by the dual-specificity effector Rabenosyn5, which can bind both Rab5 and Rab4 simultaneously [20]. Rab4 is therefore also believed to affect EGFR recycling dynamics. Together with their effectors, these Rabs modulate the rate and strength of intracellular signalling from ligand-bound EGF receptors on the cell surface and signalling endosomes.
EGF downstream signalling involves the mitogen-activated protein kinase (MAPK) and the Akt kinase/protein kinase B pathways, which culminate in DNA synthesis, migration, and cell proliferation. Consequentially, regulation of growth factor transport and signalling by Rabs impacts on fundamental aspects of pathology such as cellular transformation and cancer. A link with cancer cell proliferation has been documented for several Rabs [21]. Overexpression of Rab31, for example, has been shown to decrease the rate of proliferation in the A431 epidermoid carcinoma line, presumably by increasing the internalisation and hence degradation of the EGF-induced signal [16]. On the other hand, the Rab5 GEF Rin1, which activates Rab5, appears to be important for EGFR signalling and cell proliferation in A549 non-small cell lung adenocarcinomas [22]. Studies of clinical samples of breast tumour have indicated that high levels of Rab31 are associated with a poorer prognosis [23]. These seemingly contrasting observations could perhaps be explained by the fact that altered levels of activity of one Rab may impact the levels and function of other Rabs and their shared regulators/effectors, resulting in compensatory mechanisms that may mask the direct effect of the altered Rab. Moreover, EGF/EGFR signalling is multifaceted, and signalling from internalised EGF/EGFR differs qualitatively and quantitatively from that at the cell surface. Signalling from internalised EGF may play a more dominant role in certain cell types than others [11, 24].
Endosomal signalling was also shown to be crucial for the ability of the nerve growth factor (NGF) receptor TrkA to enhance neurite outgrowth. Rab22 was shown to promote NGF-dependent neurite outgrowth in PC12 cells by aiding internalisation of the ligand-bound receptor into signalling endosomes [25]. This example again suggests that Rabs impact growth factor signalling not only through regulating cell surface signalling, but also subsequent signalling from internalised receptors.
Rabs also regulate growth factor receptors by regulating its cell surface expression. Rab14, for example, was shown to interact with kinesin superfamily proteins (KIFs). Fibroblast growth factor (FGF) signalling is important during mammalian embryogenesis and regulates the development of the epiblast. KIF16B was shown to be recruited to FGFR2-containing vesicles by Rab14, which is responsible for Golgi-to-endosome trafficking, ultimately resulting in the cell surface expression of the receptor that facilitates FGF signal transduction [26].
Rabs affect growth factor signalling via regulation of molecular interactions
Another role for Rabs in mediating growth factor signal transduction was uncovered through the interaction of Rab5 with the Adaptor protein containing PH domain, PTB domain, and Leucine zipper motif 1 and 2 (APPL1 and 2), which binds to GTP-bound Rab5 on a subpopulation of early endosomes. APPL1 and 2 are multifunctional scaffold molecules that interact with a myriad of receptors and signalling intermediates, including adiponectin receptors, follicle stimulating hormone receptor, Akt, and TrkA amongst others [27]. APPL1 is a prominent mediator of Akt substrate specificity. Loss of APPL1 affects the phosphorylation of the Akt substrate GSK3β but not Tsc2, resulting in enhanced cell survival in HeLa and zebrafish embryos. This specific phosphorylation of GSK3β is mediated by APPL1 serving as an anchor for Akt on a subset of Rab5-positive endosomes upon growth factor stimulation [28]. The endosomal partitioning of Akt and its specific substrate GSK3β was proposed as a mechanism by which cells control growth factor signal transduction, and this is in line with the notion that growth factor signalling from endosomes differs qualitatively from that on the cell surface. The specificity of APPL1’s effect on GSK3β but not Tsc2 is a means by which cells could channel growth factor signalling towards cell survival in a manner that is decoupled from cell proliferation. In this way, a Rab protein contributes not merely to vesicular trafficking, but also to the spatial regulation of growth factor signalling. The mechanism by which Rab5/APPL1 enhances the substrate specificity of APPL1 for GSK3β remains to be fully addressed. It is also worth noting that other groups have reported that a loss of APPL1 reduces Akt phosphorylation in mouse embryo, indicating multiplicity of APPL1 action in different cell types and model organisms [29].
The role of Rab5 and APPL in signal transduction does not end with their roles and association at the endosome. It has been shown that after EGF internalisation and upon GTP hydrolysis of Rab5, APPL could be released from membranes and translocated to the nucleus, where it interacts with components of the nucleosome remodelling and histone deacetylase complex (NuRD/MeCP1) to regulate cell proliferation [30, 31]. Silencing of APPL proteins resulted in an inhibition of DNA synthesis, suggesting that binding of APPL to the NuRD complex was important for DNA synthesis and subsequent cell proliferation. Later work has shown that binding of APPL1 to histone deacetylase 1 (HDAC1), a component of the NuRD complex, restricted the repressor potential of HDAC1 by reducing its binding to other NuRD components [30]. In this way, Rab5, together with APPL1, could connect between a growth factor signal at the plasma membrane and the eventual transcriptional profile changes in the nucleus.
APPL1 and 2 have also been implicated in adipokine signalling, which exerts fine control over insulin-mediated glucose uptake, which is described further in the following section.
Rabs in nutrient and energy metabolism
Rabs affect trafficking of glucose transporters
Nutrient status monitoring and energy sensing are key processes in cellular metabolism, and a change in the capacity for glucose uptake is a major response to changes in cellular energy status. Glucose uptake in eukaryotes is mediated by the family of glucose transporters (GLUTs) of the SLC2 (solute carrier) gene family. All 13 members of GLUTs in the mammalian system are integral membrane proteins with 12 membrane spanning domains [32, 33]. Binding of glucose induces a conformational change of the transporter, which then facilitates glucose transport along its concentration gradient across the plasma membrane. The best characterised GLUTs are GLUT1–4. GLUT1 is responsible for basal uptake of glucose that sustains basal respiratory processes in all cells, and its levels at the cell membrane increases in response to low levels of glucose [34]. GLUT2 is a high-capacity and low-affinity isoform found in cell types like the renal tubule epithelium, hepatocytes and pancreatic β cells [35], while GLUT3 is the major neuronal isoform expressed by most neurons [36]. GLUT4 is the insulin-responsive form in adipose tissue and striated muscle, and is important for glucose uptake and storage in response to insulin [37].
GLUT4 at steady state is partitioned between internal membranous compartments [sometimes referred to as GLUT4-storage vesicles (GSVs)] and the cell surface, which are connected by fast endocytosis and slow exocytosis to and from the plasma membrane [38]. Upon insulin stimulation, phosphatidylinositol 3-kinase (PI3K) is activated and phosphorylates Akt, which in turn phosphorylates several targets, ultimately resulting in GLUT4 translocation to the cell surface. This translocation is believed to be mediated, in part, by GLUT4-storage vesicles (GSV)-bound Rabs [39]. Analysis with the expression of dominant-negative Rab mutants in adipocytes or muscle cells (cells which are particularly responsive to insulin signalling) and immunoprecipitation of GLUT4 interacting proteins have identified a number of Rabs that regulate GLUT4 trafficking [38, 40–44]. For example, a role for Rab4 has been attributed to its interaction with its effector, the kinesin family motor protein KIF3 [45], making it important for the movement of GLUT4 vesicles. Rab4 also interacts with another effector syntaxin4, a t-SNARE on the plasma membrane that mediates the fusion of exocytic/recycling vesicles [46]. Rab11, another Rab implicated in the recycling of endosomes, was also shown to regulate the trafficking of GLUT4 from recycling endosomes to GLUT4-specific compartments [38].
Other Rabs with a role in GLUT4 trafficking have been identified as targets of Akt substrate of 160 kDa (AS160). AS160 when phosphorylated stimulates GLUT4 trafficking to the plasma membrane [47, 48]. AS160 (also known as TBC1D4) has a putative Rab-GAP domain, and phosphorylation by insulin-dependent PI3K signalling likely inhibits its GAP activity [38, 48]. It is postulated that high Rab-GAP activity keeps target Rabs in their GDP-bound form, until insulin stimulation inhibits the GAP activity, enabling Rabs to engage in their function, which includes the trafficking of GLUT4 to the plasma membrane. In vitro, AS160 has been shown to act as a Rab-GAP for Rab2A, 8A, 10 and 14 [41, 49]. In muscle cells, overexpression of Rab8A rescued the block in insulin-stimulated GLUT4 translocation caused by an AS160 mutation that renders it non-responsive to phosphorylation (thus rendering the Rab-GAP domain constitutively active) [50]. It is speculated that Rab8A plays a role in endosomal transit of GLUT4 vesicles [51], and recently it was shown that Rab8A engages Myosin Vb [52] and Myosin Va as an effector in this process [53]. More recently, Rab13 was also found to be regulated by AS160 GAP activity, and found to colocalise with GLUT4 near the cell surface in muscle cells [51]. Silencing of Rab8A and 14, but not Rab10, inhibited insulin-dependent GLUT4 translocation in myoblasts [52]. In adipocytes, however, overexpression of Rab10 was shown to increase GLUT4 surface levels, while silencing of Rab10 attenuated the GLUT4 translocation [40, 54–56]. This suggests that Rabs act with varying specificities in muscle cells and adipocytes. In adipocytes, Rab14 was shown to impact the trafficking of GLUT4 from early endosomes to the trans-Golgi network (TGN) and the sorting to GSVs, more than the translocation to the cell surface, again suggesting cell-type specificity of Rab action [56]. As with Rab8A, Rab10 was also shown to interact with Myosin Va, and is postulated to mediate trafficking and fusion of GSVs with the plasma membrane [57].
Subsequently another RabGAP, TBC1D1, was found in greater abundance in muscle tissue and is regulated in a similar fashion to AS160 [52, 58, 59]. Besides insulin-dependent Akt phosphorylation, it was also shown that during skeletal muscle contraction, AMP-activated protein kinase (AMPK) phosphorylates TBC1D1 and results in GLUT4 translocation to the cell surface [60]. Depletion of TBC1D1 affected both the basal levels of GLUT4 at the cell surface, as well as the insulin-induced translocation of GLUT4. Knockdown of both Rab8A and Rab14 reversed this effect, suggesting that Rab8A and Rab14 may also be substrates of this Rab-GAP [52].
Details of how the various Rabs are regulated by AS160 and TBC1D1, and their mechanism of action remains to be elucidated. It is, however, clear that both these Rab-GAPs are important for glucose homeostasis, as various studies have shown that depletion of these Rab-GAPs affect glucose uptake in adipocytes and skeletal muscle [61, 62]. Most recently, a double knockdown of AS160 and TBC1D1 in mice was shown to hinder proper glucose metabolism by impairing insulin responsiveness in both adipose and skeletal muscle tissue [63].
The Rab5 subfamily of Rabs, including Rab5, Rab22A and Rab31 with well characterised roles in membrane trafficking in the early endocytic pathway, may affect GLUT4 trafficking in an opposite manner to the exocytic regulation described above. These Rabs regulate the slow endocytosis step of the GSVs at steady state. The role of Rab31 in this regard was identified through the involvement of its GEF, GAPex5. GAPex5 interacts with the Rho family member TC10 at the cell surface, but also interacts with and activates Rab31 at the TGN or endosomes. Overexpression of GAPex5 decreased insulin-stimulated movement of GLUT4 to the cell surface. Overexpression of wild-type or dominant active Rab31 resulted in a similar phenotype [44]. Lodhi and colleagues thus postulated that Rab31 keeps GLUT4 in a “futile” internalisation cycle, until insulin stimulation generates active TC10, which then recruits GAPex5 to the cell surface, away from Rab31. When Rab31 is inactivated, this internalisation cycle is disrupted, resulting in increased GLUT4 transport to the cell surface in response to insulin [44]. In a similar manner, active Rab5 enabled the internalisation of GSVs via dynein, one of its effector proteins, and insulin stimulation inhibits this process [43]. Rabs have thus been extensively implicated in both the fast endocytosis and slow exocytosis steps responsible for regulating GLUT4 levels at the cell surface.
Rabs affects trafficking of other nutrient sensing-related proteins
Besides a direct role in regulating the movement of glucose transporters, Rabs and their interacting partners have also been implicated in other related regulatory mechanisms in energy metabolism. Adiponectin (or Arcp30) is a key hormone secreted mainly by adipose tissue that regulates several aspects of fatty acid and glucose metabolism [64]. Adiponectin binds to its receptors AdipoR1 (found in most tissues and abundantly in skeletal muscle) and AdipoR2 (mainly in the liver), and stimulates AMPK activity [65]. Together with insulin, it stimulates the translocation of GLUT4 to the cell surface and glucose uptake. APPL1, the Rab5 effector, was first found to interact with AdipoR1 by a yeast 2-hybrid screen. This interaction was enhanced with adiponectin, although it was not clear if APPL1 or Rab5 itself translocates to the plasma membrane upon adiponectin stimulation. Overexpression of APPL1 in C2C12 myocytes increased GLUT4 translocation upon adiponectin stimulation [27]. Rab5 was also shown to colocalise with AdipoR1, and adiponectin increased the interaction between APPL1 and Rab5. In cells expressing mutant forms of APPL1 that do not bind Rab5, the adiponectin-stimulated translocation of GLUT4 was diminished. These results appear to suggest that APPL1, together with Rab5, positively mediates adiponectin signalling. Studies in C2C12 myocytes also showed that dominant-negative Rab5 inhibited the constitutive, ligand-independent endocytosis of the adiponectin receptor, resulting in enhanced AMPK phosphorylation when adiponectin was present [66]. This suggests that a Rab can also be responsible for controlling the steady-state amount of surface receptors as a means of regulating the intensity of signalling. Whether or not APPL1 is also involved in this event remains to be determined. Taken together, the studies highlighted above suggest that Rab5 plays a role in adiponectin signalling, albeit in somewhat contrasting manners. There is crosstalk between the insulin and adiponectin pathways, as APPL1 overexpression was shown to enhance insulin-stimulated Akt phosphorylation [67]. Rab5 and APPL1 could therefore serve as a link between adiponectin and insulin-stimulated pathways in regulating glucose homeostasis. At the same time, APPL2 was shown to be a negative regulator of adiponectin signalling, as depletion of APPL2 in C2C12 myocytes enhanced glucose uptake and fatty acid metabolism stimulated by adiponectin. It is believed to compete with APPL1 for adiponectin binding, and sequestering APPL1 by forming a heterodimer [68]. Recently, it was shown that APPL2 interacts with TBC1D1 in regulating glucose metabolism in skeletal muscle [69]. Phosphorylation of TBC1D1 at Ser-235 enhanced its interaction with APPL2, which in turn inhibited insulin-dependent phosphorylation of Thr-596 on TBC1D1. This results in an inhibition of GLUT4 translocation due to the persistence of GAP activity of TBC1D1. Interestingly, Rab31 was also found to interact with APPL2, although the exact physiological implication of this interaction is not yet known [70].
Rabs in lipid droplet (LD) formation and function
While glucose metabolism is an important part of cellular function, another key aspect of cellular metabolism involves lipids. A particularly prominent aspect of metabolic disorder relates to lipid storage and mobilisation. Lipid droplets (LDs) are organellar reservoirs of neutral lipids bound by a phospholipid monolayer found in most cell types. Their distribution and size change dynamically with cellular energy status [71]. LDs have critical roles in lipid homeostasis, and membrane trafficking processes associated with LDs have received much recent attention because of their metabolic disease implications [72], as well as their role in replication and assembly of viruses such as the hepatitis C virus (HCV) [73].
Several proteomics analyses of LDs have unsurprisingly revealed the association of multiple Rabs with LDs [74–76]. The exact roles of these Rabs in LD biogenesis and function have not been particularly clear [77]. The biogenesis of LDs has been intensively investigated [78], and although many details are still unclear, LD de novo formation is believed to occur at the ER [79, 80]. Rab1, a key regulator of ER-Golgi transport, is recruited with a cognate GAP, TBC1D20, to LDs [81], and dominant-negative Rab1 could inhibit LD formation. This implies that Rab1 may have at least an indirect role in LD biogenesis. Both GDP- and GTP-bound Rab5 could be targeted to LD, but only the latter could recruit the tether EEA1 [82], which implied a possible role for Rab5 in LD fusion. The process of LD macroautophagy, or lipophagy, would likely involve the prelysosomal Rab7 [83]. Therefore, while there is some evidence for association and some functional connections between these Rabs and LD, their exact mechanisms of action are not known.
Rab18 is perhaps the most prominent Rab GTPase at LD membrane [77]. Rab18’s association with the LD monolayer has been shown with both the GFP-tagged protein and immune-electron microscopy analyses. Rab18’s association with LD is increased by enhanced lipolysis and reduced by β-adrenergic antagonists [84]. Rab18 levels are elevated when 3T3-L1 adipocytes are induced to differentiate, and its recruitment to LD could be stimulated by insulin [85]. Conversely, Rab18 expression increases lipogenesis, while its silencing impairs insulin-stimulated lipogenesis [85]. The exact role of Rab18 in LD biogenesis and function is still unclear. It may, as suggested by the evidence above, act in LD biogenesis and triacylglycerol (TG) accumulation. On the other hand, it may also have a role in LD fission and fusion, the latter perhaps through its interaction with LD-associated SNAREs [86]. As mentioned above, LDs are important for HCV replication and assembly, and Rab1 is known to interact with HCV’s nonstructural protein NS5A [81]. Activated Rab18 also interacts with the NS5A protein and promotes interaction between viral assembly sites and LD [87], and appear to facilitate the trafficking of HCV core protein to LDs [88]. Interestingly, an LD binding protein Tip47, also binds NS5A [89]. Tip47 is a key effector of endosomal Rab9 [90], and its interaction with Rab9 is required for viral particle release [91]. The subversion of hepatocytic LD and their associated Rabs for viral replication is more than just an intriguing phenomenon, but could underlie liver steatosis and subsequent cirrhosis and malignancy associated with HCV infection. In a related viral connection, Rab18 is upregulated by the viral tumour promoting factor hepatitis B virus X (HBx) with a consequence of lipogenesis dysregulation and hepatoma cell proliferation [92].
Another very recent finding implicated the involvement of Rab8A in the regulation of LD fusion [93]. The authors were investigating fat-specific protein 27 (Fsp27/Cidec), a member of the Cell death-inducing DFFA-like effect (CIDE) family, which has roles in thermogenesis, lipolysis as well as apoptosis [94]. Fsp27 is known to function in LD fusion and is enriched in contact sites between LDs [95]. Rab8A interacts with Fsp27, and acts as an activator of Fsp27-mediated LD fusion and growth, and liver-specific knockdown of Rab8A in ob/ob mice (a model for Type II diabetes) resulted in smaller LDs and lower liver lipid levels. An interesting aspect of Rab8A action is that LD fusion is promoted by GDP-bound Rab8A, and not the GTP-bound form [93]. Accordingly, the GAP AS160 forms a ternary complex with both Rab8A and Fsp27 in promoting LD fusion, while the Rab8A GEF Mss4 antagonised fusion. In its GDP-bound form, Rab8A is unlikely able to recruit cognate effectors such as tethers or SNAREs, and the mechanism underlying this non-canonical mode of Rab8A action would therefore be interesting.
Rabs in autophagic responses to growth factor and nutrient deprivation
The flip side of nutrient sensing and growth factor signalling is the response to nutrient and/or growth factor deprivation. Autophagy is a key cellular homeostatic process that is induced by growth factor/nutrient deprivation, and enables the recycling of molecules from nonessential components to be used for the synthesis of essential components during times of need [96]. Formation of the autophagosome begins with the formation of a cup-shaped isolation membrane which eventually elongates to form the double-membrane autophagosome. Cytoplasmic content is engulfed by the autophagosome and is eventually degraded in the autolysosome. Microtubule-associated protein light chain 3 (LC3), is a ubiquitin-like protein. During autophagosome formation, LC3 undergoes a lipidation process and is incorporated into the autophagosome membrane, and has a role in remodelling the membranes. It is often used as a marker for induction of autophagy [97].
Several Rabs have been linked to the autophagic process, although not all of them have fully elucidated roles in this regard. Rab1, which is believed to regulate ER-Golgi and Golgi transport, was found to colocalise with LC3-positive autophagosomes, and overexpression of Rab1 increased the number of LC3-positive structures, suggesting that the Rab is important for autophagosome formation. The yeast Rab1 homologue Ypt1 was shown to engage Atg11, a scaffolding protein that is part of the pre-autophagosomal structure. It is likely that Ypt1/Rab1 is involved in engaging autophagy-related proteins onto membranes [98]. Rab11, which is associated with post-Golgi membranes and is involved in recycling membrane traffic, is increased in its colocalisation with LC3 upon nutrient starvation [99]. A dominant-negative Rab11SN loses this colocalisation, suggesting that functional Rab11 is necessary for the above phenomenon. Rab11 may be essential for regulating MVB formation and subsequent movement to the plasma membrane for exosome release. This process is particularly important during terminal differentiation of erythroid cells, which relies on autophagy for the elimination of organelles [99, 100]. Rab24, whose role in conventional vesicular trafficking remains to be defined, also colocalises with LC3 on autophagosomes upon starvation [101]. However, the exact role that Rab24 plays in autophagy remains unclear.
In yeast, the Atg5/12/16L complex found on isolation membranes is a key autophagy initiating complex [102]. Atg16L was shown to be an effector for Rab33B, and GTP-locked Rab33BQL induces lipidation of LC3 even in the absence of starvation signals. As Rab33B was originally identified as a Golgi-resident Rab, it was initially suggested that Rab33B might be important to recruit the Atg5/12/16L complex to membranes and induce subsequent lipidation of LC3 [103]. In further work, it was found that OATL1/TBC1D25 is present on isolation membranes and autophagosomes, and is a GAP for Rab33B that binds LC3 [104]. The authors envisaged a possible feedback loop in which autophagy induction and lipidation of LC3 is paired to a cycle of active and inactive Rab33B. Several other Rab-GAPs have also been found to bind LC3 family proteins, many of which are GAPs for Rabs in the endocytic pathway. These Rab-GAPs serve to link the vesicular transport pathway with autophagic pathway, possibly with a role in remodelling membrane compartments [105].
While it is clear that Rabs can be found on autophagic membranes and their loss of function results in dysregulation of autophagy, their exact mechanisms of action in the autophagic processes remains to be clearly defined. Rabs are clearly involved in the membrane dynamics of the autophagy process, and may act in recruitment and regulation of the sequence of Atg protein action on the various membranes.
Recently, it has also come to light that besides regulating the formation of autophagic membranes, Rabs can also affect autophagy through upstream events such as mTOR signalling. This will be discussed in the following section.
Rabs and TORC signalling
Beyond the sensing of nutrients and receptor trafficking, another emerging aspect in metabolic regulation is the role of Rab-modulated membrane traffic in more downstream events. Metabolic regulation in eukaryotes is very much dependent on a conserved serine/threonine protein kinase—the target of rapamycin (TOR), or mechanistic TOR (mTOR, formerly mammalian TOR) in mammals. mTOR was initially identified through screens of rapamycin-resistant Saccharomyces cerevisiae, which were not susceptible to rapamycin, an antifungal compound isolated from Streptomyces hygroscopicus that inhibited G1 to S phase transition [106]. The TOR kinase stimulates cell growth together in response to nutrients and other anabolic signals [107]. At the same time, activated mTOR is also a negative regulator of autophagic induction [108]. Dietary restriction or rapamycin have both been shown to result in inhibition of mTORC signalling, culminating in the inhibition of translation via decreased ribosome biogenesis and reduced phosphorylation of various proteins essential for translation [109, 110]. Various human pathologies including diabetes, cardiac hypertrophy, malignancies, neurodegenerative syndromes and aging have been linked to defective or aberrant mTORC signalling [111–113].
There are two different mTOR-containing complexes, TORC1 and TORC2 [114, 115]. The TORC1 complex is activated by amino acids, oxygen, energy, and growth signals, and it in turn phosphorylates other substrates to increase mRNA translation and ribosome biogenesis, with a concomitant inhibition of autophagy [113]. Substrates of mTORC1 include S6 kinase (S6K/p70) and eIF4E-binding protein 1 (4EBP1); both important modulators of protein translation. Typically, growth promoting factors such as insulin activate the PI3K/Akt pathway. Akt in turn phosphorylates and inactivates Tsc1/2, which is a GAP for the small GTPase Ras homologue enriched in brain (Rheb) [116, 117]. Rheb in turn activates mTORC1 by a mechanism that is still incompletely understood, but is believed to involve its binding to the mTORC1 kinase domain, and enhancing its interaction with Regulatory associated protein of mTOR (Raptor), another member of the mTORC1 complex. Nutrients such as amino acids can also activate mTORC1. Accumulation of amino acids in the lysosomal lumen results in the recruitment of another small GTPase Rag. Rag is responsible for the recruitment of mTORC1 to Rheb on the lysosomes. Lysosomes appear to have a unique role in amino acid regulated activation of mTORC1 [118, 119].
TORC2 is activated by growth signals and interaction with the ribosome, and phosphorylates substrates such as Akt, SGK, and related AGC kinases [120], but its exact mechanisms of activation are less well understood. Unlike TORC1, TORC2 is traditionally believed to be rapamycin-insensitive, with functions in cell polarisation, lipid synthesis, chromatin metabolism and recovery from DNA damage [121]. As it is responsible for Akt activation [122], it can thus also be indirectly responsible for the activation of TORC1.
TORC signalling intersects with vesicular trafficking pathways. The Rab GTPases are well-poised to also regulate TORC signalling, as signalling pathways may involve lipid modifications to membranes, such as in the PI3K pathway, or their recruitment to specific cellular membranes. Given the central importance of the TORC signalling pathways, the following discussions focus on several examples of how Rabs are involved in TORC pathway regulation.
Rabs as general regulators of localisation and function of TORC1
Emerging evidence suggests that Rabs regulate membrane localisation and function of TORC1. Li and colleagues attempted to identify GTPases that regulate TORC1 in the fly genome, by an RNAi screen in Drosophila S2 cells, using S6K phosphorylation as a readout for TORC1 activity [123]. Interestingly, constitutive activation of several Rabs diminished TORC1 activity. Rab1, 5, 7, 10 and 31 were thus identified as potential TORC1 activity modulators. These results affirmed that Rab-mediated vesicular trafficking steps are important for TORC1 function. Interestingly, overexpression of either the wild-type, dominant-negative or dominant active Rab5 mutants all inhibited TORC1 activity, but not TORC2 activity. This suggests that Rab5 may act downstream of TORC2. Overexpression of the GEF for Rab5, GAPex5, had the same effect as Rab5 overexpression. Rab5 and TORC1 did not co-immunoprecipitate, but they were found to colocalise in the same membrane compartment. A Rab5 mutant, Rab5QL-2CS, which is constitutively active but lacks the domain for geranyl-geranylation and hence cannot be membrane localised, did not inhibit TORC1 activity, suggesting that Rab5 must be localised to the membrane to exert its effect.
Importantly, this Rab5-mediated effect only occurred with amino acid (and Rag) stimulation of the TORC pathway, but not glucose (and Rheb) stimulation, suggesting that this Rab-mediated effect is pathway specific. It is proposed that in the presence of sufficient amino acids, Rag presents TORC1 at the correct vesicular compartment (lysosomes) for activation by Rheb [119, 124]. As such, the amino acid-stimulated TORC1 pathway is particularly sensitive to disruptions in vesicular trafficking. The results point to an overall importance of functional vesicular trafficking to the TORC1 pathway, especially with regard to the amino acid-stimulated pathway involving lysosomes. The Rabs identified have been implicated in autophagosomal and lysosomal trafficking, and as such their role in vesicular trafficking places them in a position to influence TORC signalling, as the latter has been shown to occur at lysosomal membranes.
Bridges and colleagues have further linked the effect of Rab5 on mTORC1 function to its role in the PI3K signalling pathway [125]. Overexpression of Rab5 in 293A cells resulted in enlarged, swollen vacuolar structures, consistent with previous observations [126]. mTORC1 was mislocalised to these structures instead of late endosome/lysosomes, in a PI3P-dependent manner, as the effect could be eliminated by wortmannin. In yeast, loss of Vps9 function (a GEF for the yeast Rab5 homologue Vps21p), but not Tor1 mutants, could be overcome with active Vps21p. This suggests that Rab5/Vps21 acts upstream of Tor1. Besides that, Rab5, via its effector APPL, is also important for PI3K/Akt signalling. As Akt signalling is important for mTORC1 activation, it is conceivable that a disruption of Rab5 function would affect mTORC1 activation.
Knockdown of Rab12 was also shown to increase the phosphorylation of S6K, suggesting that loss of Rab12 increased mTORC1 activity. Unlike the case with Rab5, this was shown to be independent of the role of PI3K/Akt signalling, as Rab12 did not affect phospho-Akt levels. Instead, Rab12 affected mTORC activity through regulation of amino acid levels, via the cell surface amino acid transporter PAT4. It is postulated that Rab12 enhances the vesicular trafficking of PAT4 from the cell surface to the lysosome, in this way regulating intracellular amino acid levels and the level of mTORC1 activation [127].
Another Rab which has been shown to modulate mTOR signalling is Rab8A, which acts in the context of the innate immune response. The activation of Toll-like receptor 4 (TLR4) by exposure of macrophages to lipopolysaccharide (LPS) resulted in recruitment of Rab8A to membrane ruffles, together with PI3Kγ, and components of TLR4 complex. PI3Kγ is subsequently responsible for the activation of mTOR [128]. Although this was shown in immune response, Rab8A is found in other cell types and may well regulate various other physiological aspects of mTOR signalling.
S. pombe Rab GTPase Ryh1 regulates mTORC2 by enhancing its interaction with its substrate Gad8
Other studies have yielded a functional link between a Rab protein and TORC2. TORC2 is responsible for the activation of Akt in mammals, or its orthologue Gad8 in yeast Schizosaccharomyces pombe. S. pombe is particularly suitable for the study of the nonessential aspects of TORC2 function as it has been shown that loss of TORC2 function is non-lethal in S. pombe, unlike in S. cerevisiae. It was shown that a Rab-like GTPase, Ryh1 in S. pombe (orthologous to Rab6 in mammals), regulates TORC2 action by affecting interaction with its substrates [129].
Upon nitrogen starvation, wild-type S. pombe cells arrest in the G1 phase. A screen identified mutants which failed to arrest, and are presumably defective in the TORC-mediated starvation response. Amongst these mutants are Ryh1 and its GEFs Sat1 and Sat4. The authors found that the phosphorylation of Gad8 was reduced in Ryh1 and Sat1 mutants. TORC1 activity was unaffected, as shown by a lack of effect on the phosphorylation of S6K, which suggest that the coupling of TORC2 and TORC1 may not be particularly strong in S. pombe.
The role of Ryh1 on TORC2 signalling does not appear to be directly linked to its role in vesicular trafficking. GDP/GTP exchange, a necessary step for Rab’s function in vesicular trafficking, was not necessary for Gad8 phosphorylation, as a GTP-locked, constitutively active mutant of Ryh, RyhQ70L did not significantly affect TORC2 signalling. Although Ryh1 was shown to interact with TORC2, its apparent predominant Golgi and endosomal localisation appears distinct from that of either TORC2 (which is at the cell cortex), or Gad8 (which is cytoplasmic). These points suggest that Ryh1’s role in TORC2 signalling may be distinct from its role in membrane traffic. Tatebe and colleagues also showed that Ryh1 appears to act by enhancing the interaction of TORC2 with Gad8. How do Ryh1 and TORC2 interact in the first place, and how does this interaction in turn enhance TORC2-Gad8 interaction? The effector domain of Ryh1 is apparently important for the interaction between TORC2 and Ryh1 but the interaction is not completely abrogated by a point mutation to the Ryh1 effector domain. Hence, the actual mechanism by which Ryh1 and TORC2 interact with each other remains to be elucidated [130]. It is notable here that the authors also showed that an indirect perturbation of Ryh1 localisation, for example through the mutation of another Rab, could affect TORC2 signalling. Thus, although the cellular localisation of TORC2 is not affected by Ryh1 localisation, the functionality of TORC2 is perhaps dependent on the integrity of vesicular traffic in general.
Most interestingly, the function of Ryh1 in S. pombe could be replaced by its mammalian orthologue Rab6. This highlights a conservation of function between the two orthologues. In mammalian cells, Rab6 has been shown to modulate both retrograde and anterograde traffic, including intra-Golgi, Golgi-to-PM, endosome-to-Golgi, and Golgi-to-ER transport processes [131]. Similarly, in S. pombe, Ryh1 is responsible for retrograde transport between the endosome and Golgi, but has also been implicated in anterograde transport of the secretory pathway [132]. It would be interesting to see if Ypt6, the homologue of Rab6 in S. cerevisiae, could also rescue the Ryh1 defect. It would be of further interest to check if Rab6 regulates mTORC2 in a similar manner.
Mrs6 and Sfp1: regulation of TORC signalling via a Rab escort protein (REP)-mediated localisation of TORC downstream effector in S. cerevisiae
Besides the role of Rab GTPases in regulating TORC activity, other Rab-interacting proteins have been shown to intersect with TORC signalling pathways. Sfp1 is a transcription factor in yeast Saccharomyces cerevisiae that responds to nutrients by cytoplasm-nuclear translocation, resulting in the transcription of ribosomal proteins (RP) and ribosomal biogenesis (Ribi) genes [133, 134]. Conversely, Sfp1 relocates to the cytoplasm upon nutrient limitation. Likewise, rapamycin treatment results in cytoplasmic localisation of Sfp1, suggesting that Sfp1 localisation is dependent on TORC1. Sfp1 directly interacts with, and is phosphorylated by, TORC1, which facilitates its nuclear localisation [135]. It remains unclear whether this interaction persists in the nucleus.
Singh and Tyers attempted to identify components of the yeast vesicular transport pathways that might be involved in the movement of Sfp1 under different conditions [136]. The authors isolated several yeast mutants that did not show cytoplasmic localisation of Sfp1 upon rapamycin treatment. Three classes of factors were thus identified: those involved in (1) ER-to-Golgi transport, (2) Golgi vesicle trafficking, and (3) structural or mechanical factors of secretion. The result of the screen itself points to multiple points of interconnection between vesicular trafficking and TORC signalling. Co-immunoprecipitation using antibodies against Flag-tagged Sfp1 identified Mrs6, a Rab escort protein (REP), as having a direct physical interaction with not only Sfp1, but also the TORC components. REPs chaperone newly synthesised Rabs to the geranyl-geranyl transferase for the addition of a C-terminal prenyl tail that enables Rab insertion into target membranes. The interaction between Sfp1 and Mrs6 is increased upon nutrient starvation and stress conditions. Overexpressed Ypt1, a substrate for Mrs6, competed with Sfp1 for Mrs6 binding, which suggests that it shares the same binding site on Mrs6. Overexpression of Mrs6 increased, while its depletion decreased, the cytoplasmic localisation of Sfp1 in starvation conditions, indicating that Mrs6 is responsible for Sfp1 cytoplasmic localisation. However, the role of Mrs6 in vesicular trafficking is apparently distinct from its role in TORC regulation. The authors were able to isolate specific mutations of Mrs6 that conferred rapamycin resistance without compromising cell viability, which would have occurred if these mutants have an overall defect in vesicular trafficking. It remains to be understood how Mrs6 is activated directly (or indirectly) by TORC, although evidence suggests that it acts downstream or parallel to the phosphorylation-dependent nuclear localisation of Sfp1 by protein kinase A (PKA). What benefit would the cell derive in allowing for a direct competition between Rab GTPases and TORC for Sfp1? It is conceivable that a coupling between secretory mechanisms and sensing of stress could allow the cell to decrease the activity of the secretory pathway under conditions of stress. This could free Mrs6 to interact with Sfp1 and thus limit TORC signalling and downregulate ribosome biogenesis.
Lempiainen and colleagues showed grossly similar, albeit slightly contrasting results in their study of Sfp1 and Mrs6. Mrs6 was found to promote Sfp1 phosphorylation, and the mutant Mrs6-2 reduced Sfp1 nuclear (rather than cytoplasmic) localisation and its interaction with TORC1 [135]. A reason for the discrepancy between the two reports may be the different allelic mutants used (Mrs6-R in Singh and Tyers [136] versus Mrs6-2 in Lempiainen et al. [135]). Mrs6 mutants also appeared to directly affect TORC1 function, as evidenced by a decrease in phosphorylation of Sch9 (yeast homologue of the mammalian S6K), a well-known substrate of TORC1 that also controls translation of ribosomal proteins and ribosomal biogenesis genes, when Mrs6 is mutated. Although the discrepancies between the two studies and the actual mechanism of action of Mrs6 on Sfp1 and/or TORC1 remains to be resolved, it is clear that an REP could also play an important role in regulating TORC signalling, either as a positive or negative regulator. Although no clear orthologues of Sfp1 has been identified in metazoans, the authors suggest that c-myc, a known mammalian regulator of RP and Ribi gene transcription, may be a functional homologue based on a similarity in its physiological roles (e.g. positive regulation of cell size, link to mTOR and PKA signalling). Future work will shed further light on whether there is a similar mechanism regulating the localisation of c-myc in mammalian cells.
RabGAP TBC proteins in regulating the mTOR-S6K pathway
Another Rab-interacting protein that regulates TORC signalling is TBC1D1. As described earlier, TBC proteins contain a Rab-GAP domain and are believed to be putative GAPs for the Rab GTPases. Zhou and colleagues have identified TBC1D1 as a substrate of Akt in adipocytes. Silencing of TBC1D1 activated the mTOR-S6K pathway and resulted in increased GLUT1 expression, ultimately leading to an increase in basal glucose transport [137]. The authors showed that insulin stimulates phosphorylation of multiple GAPs by Akt, as this phosphorylation is lost with wortmannin treatment. Activated mTOR phosphorylates S6K, leading to its activation. Silencing of TBC1D1, but not TBC1D4, increased the basal levels of phosphorylation, suggesting that TBC1D1 negatively regulates mTOR activity. It is possible that insulin signalling may result in phosphorylation and inactivation of TBC1D1. The consequential activation of S6K results in increased GLUT1 expression. This increase is abolished by rapamycin, reinforcing the notion that TBC1D1 acts via an mTOR-dependent pathway. Likewise, a TBC1D1 mutant lacking the Akt phosphorylation site also lacks an effect on mTOR activity. At present, it is yet unclear if the role of TBC1D1 in the mTOR signalling pathway is Rab-dependent. It is not inconceivable, however, that TBC1D1, by inactivating a particular Rab, would prevent mTOR activation as described in the other cases above.
Concluding remarks
As discussed above, new insights are emerging on the crosstalk between vesicular trafficking and the metabolic response system, many of which involve Rabs and their interacting partners (Table 1; Fig. 2). With regards to growth factor signalling, Rabs have been implicated in the regulation of the strength, duration and type of signalling, by playing a role in the movement of the ligand-bound receptors through the endocytic pathway. In this way, Rabs are responsible in part for the spatial regulation of the signalling from these receptors, and serve to relay signals between the plasma membrane and the nucleus. Glucose transport is an integral part of the metabolic pathways in the cell, and Rabs and their inactivating GAP proteins have important roles in insulin-stimulated trafficking of glucose transporters to the cell surface. This is achieved in two ways, firstly by mediating fast exocytosis and slow endocytosis of the transporters in response to external signals, and secondly by controlling the steady-state amount of surface receptors, to regulate the subsequent intensity of signalling upon addition of the ligand. Rabs have also been found to be associated with the formation and regulation of lipid droplets, which play a role in lipid metabolism. Autophagy is another aspect of the cell’s response to a lack of nutrients. Rabs have been shown to be important for engaging autophagy-related proteins to the relevant membranes. Rabs are also important for TORC signalling, which acts downstream of both nutrient sensing and growth factor signalling. One way in which Rabs are important to TORC signalling is simply in the maintenance of functional vesicular trafficking, as some TORC signalling may occur from membranes. Some Rabs may compete with mTORC for common binding factors, thus enabling a coupling between trafficking mechanisms and stress sensing mechanisms. Other roles of Rabs and/or their interacting proteins may not be directly linked to their function in vesicular trafficking, but may involve the recruitment of relevant effector proteins with overlapping functions in signalling. As more functions of Rabs, and the identity of Rab-interacting proteins are discovered, it is likely that more interconnections between the vesicular transport pathways and nutrient/growth factor signalling systems will be uncovered. This, in turn, may lead to novel mechanisms of targeting various diseases stemming from dysregulated metabolic and/or growth factor signalling.
Table 1.
List of Rabs, their known roles in vesicular trafficking and their links to metabolic signalling and regulation
| Type of metabolic signalling and regulation | Rab | Known role(s) in trafficking step/pathway | Known effect or association with metabolism/metabolic signalling | References |
|---|---|---|---|---|
| Growth factor signal transduction | Rab5 subfamily (Rab5A, B and C, Rab22 and Rab31) | Endosomal trafficking | EGFR trafficking | [12–17] |
| Rab7 | Late endosome and lysosomal trafficking | Lysosomal degradation of EGFR | [18] | |
| Rab11 | Trafficking of recycling endosomes | Internalisation and recycling of EGFR | [19] | |
| Rab4 | Trafficking of recycling endosomes | Internalisation and recycling of EGFR | [20] | |
| Rab22 | Endosomal trafficking | NGF-dependent signalling | [25] | |
| Rab14 | Golgi-endosome trafficking | FGFR2 surface expression | [26] | |
| Rab5 | Endosomal trafficking | APPL1-mediated Akt substrate specificity and HDAC1-regulated transcription | [28, 30] | |
| Nutrient and energy metabolism | Rab4 | Trafficking of recycling endosomes | KIF3-mediated movement of GLUT4 vesicles | [45] |
| Rab11 | Trafficking of recycling endosomes | Recycling of GLUT4 | [38] | |
| Rab8A | Endosomal trafficking | Transit of GLUT4 vesicles | [50, 52] | |
| Rab10 | Varied, including endosomal and ER trafficking | GLUT4 translocation | [40] | |
| Rab14 | Golgi-endosome trafficking | Sorting to GSVs | [56] | |
| Rab31 | Endosomal and Golgi trafficking | Keeps GLUT4 in internalisation cycle | [44] | |
| Rab5 | Endosomal trafficking | GSV internalisation | [43] | |
| Rab5 | Endosomal trafficking | Adiponectin signalling | [27, 66] | |
| Rab1 | ER and Golgi trafficking | LD biogenesis | [81] | |
| Rab5 | Endosomal trafficking | LD fusion | [82] | |
| Rab18 | Varied, including ER, Golgi and endosomal trafficking | Impacts lipogenesis | [85] | |
| Ra8A | Endosomal trafficking | LD fusion | [93] | |
| Autophagic responses | Rab1 | ER and Golgi trafficking | Autophagosome formation | [98] |
| Rab11 | Trafficking of recycling endosomes | MVB formation | [99, 100] | |
| Rab33B | Intra-Golgi trafficking | Lipidation of LC3 | [103, 104] | |
| TORC signalling | Rab5 | Endosomal trafficking | Vesicular trafficking and Akt activation in mTORC1 signalling | [124–126] |
| Rab12 | Unclear, possibly in Golgi trafficking | Regulate intracellular amino acid levels and level of mTORC1 activation | [127] | |
| Ryh1/Rab6 | Golgi traffic | Regulate TORC2 interaction with substrates | [129] |
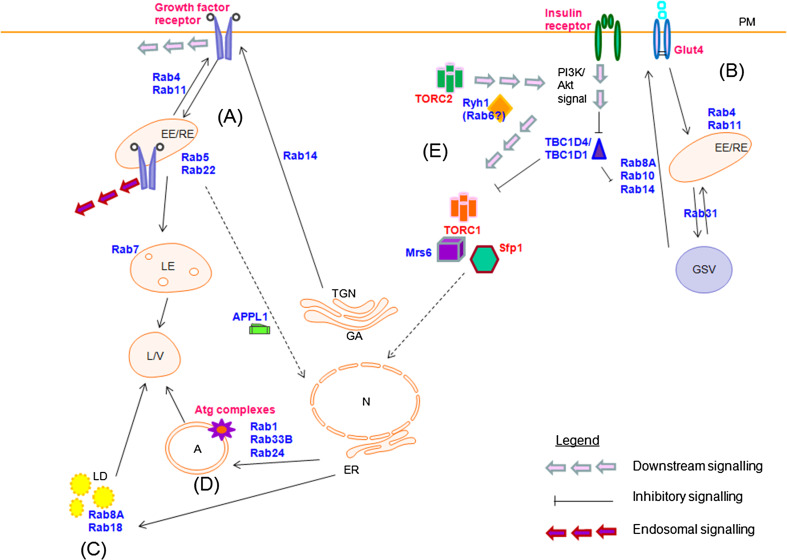
Fig. 2.
Various ways in which Rabs impact metabolic signalling and regulation. a Rabs mediate the signalling of various growth factors by regulating trafficking of the receptors to and from various intracellular compartments to the plasma membrane or lysosome. They also impact downstream signalling from the plasma membrane and endosomes. b Rabs play a role in the movement of glucose storage vesicles to and from the cell surface in response to insulin signalling. c Rabs also play a role in lipid metabolism and d autophagic processes. e Rabs impact TORC signalling by regulating various interacting proteins that play a role in TORC1 and TORC2 function. Rabs and related regulatory or interacting proteins are shown in blue. For simplicity, various intermediate details have been omitted. Refer to text for more description. N nucleus, ER endoplasmic reticulum, GA Golgi apparatus, TGN trans-Golgi network, EE early endosome, RE recycling endosome, LE late endosome, L/V lysosome/vacuole, LD lipid droplet, A autophagosome, GSV glucose storage vesicle, PM plasma membrane
Acknowledgements
Christelle Chua and Bor Luen Tang are supported by the NUS Graduate School for Integrative Sciences and Engineering, National University of Singapore.
References
- 1.Segev N. Ypt/rab gtpases: regulators of protein trafficking. Sci STKE. 2001;2001:RE11. doi: 10.1126/stke.2001.100.re11. [DOI] [PubMed] [Google Scholar]
- 2.Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ullrich O, et al. Rab GDP dissociation inhibitor as a general regulator for the membrane association of rab proteins. J Biol Chem. 1993;268:18143–18150. [PubMed] [Google Scholar]
- 4.Dirac-Svejstrup AB, Sumizawa T, Pfeffer SR. Identification of a GDI displacement factor that releases endosomal Rab GTPases from Rab-GDI. EMBO J. 1997;16:465–472. doi: 10.1093/emboj/16.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohya T, et al. Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature. 2009;459:1091–1097. doi: 10.1038/nature08107. [DOI] [PubMed] [Google Scholar]
- 6.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci USA. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Markgraf DF, Peplowska K, Ungermann C. Rab cascades and tethering factors in the endomembrane system. FEBS Lett. 2007;581:2125–2130. doi: 10.1016/j.febslet.2007.01.090. [DOI] [PubMed] [Google Scholar]
- 8.Seabra MC, Coudrier E. Rab GTPases and myosin motors in organelle motility. Traffic. 2004;5:393–399. doi: 10.1111/j.1398-9219.2004.00190.x. [DOI] [PubMed] [Google Scholar]
- 9.Nottingham RM, Pfeffer SR. Defining the boundaries: Rab GEFs and GAPs. Proc Natl Acad Sci USA. 2009;106:14185–14186. doi: 10.1073/pnas.0907725106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorkin A, von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol. 2009;10:609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howe CL. Modeling the signaling endosome hypothesis: why a drive to the nucleus is better than a (random) walk. Theor Biol Med Model. 2005;2:43. doi: 10.1186/1742-4682-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinneen JL, Ceresa BP. Expression of dominant negative rab5 in HeLa cells regulates endocytic trafficking distal from the plasma membrane. Exp Cell Res. 2004;294:509–522. doi: 10.1016/j.yexcr.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Chen PI, Kong C, Su X, Stahl PD. Rab5 isoforms differentially regulate the trafficking and degradation of epidermal growth factor receptors. J Biol Chem. 2009;284:30328–30338. doi: 10.1074/jbc.M109.034546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ceresa BP. Regulation of EGFR endocytic trafficking by rab proteins. Histol Histopathol. 2006;21:987–993. doi: 10.14670/HH-21.987. [DOI] [PubMed] [Google Scholar]
- 15.Kauppi M, Simonsen A, Bremnes B. The small GTPase Rab22 interacts with EEA1 and controls endosomal membrane trafficking. J Cell Sci. 2002;115:899–911. doi: 10.1242/jcs.115.5.899. [DOI] [PubMed] [Google Scholar]
- 16.Ng EL, Ng JJ, Liang F, Tang BL. Rab22B is expressed in the CNS astroglia lineage and plays a role in epidermal growth factor receptor trafficking in A431 cells. J Cell Physiol. 2009;221:716–728. doi: 10.1002/jcp.21911. [DOI] [PubMed] [Google Scholar]
- 17.Chua CEL, Tang BL. Engagement of the small GTPase Rab31 protein and its effector, early endosome antigen 1, is important for trafficking of the ligand-bound epidermal growth factor receptor from the early to the late endosome. J Biol Chem. 2014;289:12375–12389. doi: 10.1074/jbc.M114.548321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ceresa BP, Bahr SJ. Rab7 activity affects epidermal growth factor: epidermal growth factor receptor degradation by regulating endocytic trafficking from the late endosome. J Biol Chem. 2006;281:1099–1106. doi: 10.1074/jbc.M504175200. [DOI] [PubMed] [Google Scholar]
- 19.Cullis DN, Philip B, Baleja JD, Feig LA. Rab11-FIP2, an adaptor protein connecting cellular components involved in internalization and recycling of epidermal growth factor receptors. J Biol Chem. 2002;277:49158–49166. doi: 10.1074/jbc.M206316200. [DOI] [PubMed] [Google Scholar]
- 20.De Renzis S, Sönnichsen B, Zerial M. Divalent Rab effectors regulate the sub-compartmental organization and sorting of early endosomes. (3) Nat Cell Biol. 2002;4:124–133. doi: 10.1038/ncb744. [DOI] [PubMed] [Google Scholar]
- 21.Chia WJ, Tang BL. Emerging roles for Rab family GTPases in human cancer. Biochim Biophys Acta. 2009;1795:110–116. doi: 10.1016/j.bbcan.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Tomshine JC, et al. Cell proliferation and epidermal growth factor signaling in non-small cell lung adenocarcinoma cell lines are dependent on Rin1. (1) J Biol Chem. 2009;284:26331–26339. doi: 10.1074/jbc.M109.033514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotzsch M, et al. Urokinase receptor splice variant uPAR-del4/5-associated gene expression in breast cancer: identification of rab31 as an independent prognostic factor. Breast Cancer Res Treat. 2008;111:229–240. doi: 10.1007/s10549-007-9782-6. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Pennock S, Chen X, Wang Z. Endosomal signaling of epidermal growth factor receptor stimulates signal transduction pathways leading to cell survival. Mol Cell Biol. 2002;22:7279–7290. doi: 10.1128/MCB.22.20.7279-7290.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Liang Z, Li G. Rab22 controls NGF signaling and neurite outgrowth in PC12 cells. Mol Biol Cell. 2011;22:3853–3860. doi: 10.1091/mbc.E11-03-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ueno H, Huang X, Tanaka Y, Hirokawa N. KIF16B/Rab14 molecular motor complex is critical for early embryonic development by transporting FGF receptor. Dev Cell. 2011;20:60–71. doi: 10.1016/j.devcel.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Mao X, et al. APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nat Cell Biol. 2006;8:516–523. doi: 10.1038/ncb1404. [DOI] [PubMed] [Google Scholar]
- 28.Schenck A, et al. The endosomal protein Appl1 mediates Akt substrate specificity and cell survival in vertebrate development. Cell. 2008;133:486–497. doi: 10.1016/j.cell.2008.02.044. [DOI] [PubMed] [Google Scholar]
- 29.Tan Y, You H, Wu C, Altomare DA, Testa JR. Appl1 is dispensable for mouse development, and loss of Appl1 has growth factor-selective effects on Akt signaling in murine embryonic fibroblasts. J Biol Chem. 2010;285:6377–6389. doi: 10.1074/jbc.M109.068452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banach-Orlowska M, Pilecka I, Torun A, Pyrzynska B, Miaczynska M. Functional characterization of the interactions between endosomal adaptor protein APPL1 and the NuRD co-repressor complex. Biochem J. 2009;423:389–400. doi: 10.1042/BJ20090086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miaczynska M, et al. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. (1) Cell. 2004;116:445–456. doi: 10.1016/s0092-8674(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 32.Scheepers A, Joost H, Schurmann A. The glucose transporter families SGLT and GLUT: molecular basis of normal and aberrant function. J Parenter Enter Nutr. 2004;28:364–371. doi: 10.1177/0148607104028005364. [DOI] [PubMed] [Google Scholar]
- 33.Wood IS, Trayhurn P. Glucose transporters (GLUT and SGLT): expanded families of sugar transport proteins. Br J Nutr. 2003;89:3–9. doi: 10.1079/BJN2002763. [DOI] [PubMed] [Google Scholar]
- 34.Olson AL, Pessin JE. Structure, function, and regulation of the mammalian facilitative glucose transporter gene family. Annu Rev Nutr. 1996;16:235–256. doi: 10.1146/annurev.nu.16.070196.001315. [DOI] [PubMed] [Google Scholar]
- 35.Thorens B, Sarkar HK, Kaback HR, Lodish HF. Cloning and functional expression in bacteria of a novel glucose transporter present in liver, intestine, kidney, and beta-pancreatic islet cells. Cell. 1988;55:281–290. doi: 10.1016/0092-8674(88)90051-7. [DOI] [PubMed] [Google Scholar]
- 36.Nagamatsu S, et al. Glucose transporter expression in brain. cDNA sequence of mouse GLUT3, the brain facilitative glucose transporter isoform, and identification of sites of expression by in situ hybridization. J Biol Chem. 1992;267:467–472. [PubMed] [Google Scholar]
- 37.Birnbaum MJ. Identification of a novel gene encoding an insulin-responsive glucose transporter protein. Cell. 1989;57:305–315. doi: 10.1016/0092-8674(89)90968-9. [DOI] [PubMed] [Google Scholar]
- 38.Zeigerer A, et al. GLUT4 retention in adipocytes requires two intracellular insulin-regulated transport steps. Mol Biol Cell. 2002;13:2421–2435. doi: 10.1091/mbc.E02-02-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rea S, James DE. Moving GLUT4: the biogenesis and trafficking of GLUT4 storage vesicles. Diabetes. 1997;46:1667–1677. doi: 10.2337/diab.46.11.1667. [DOI] [PubMed] [Google Scholar]
- 40.Sano H, et al. Rab10, a target of the AS160 Rab GAP, is required for insulin-stimulated translocation of GLUT4 to the adipocyte plasma membrane. Cell Metab. 2007;5:293–303. doi: 10.1016/j.cmet.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 41.Mîinea CP, et al. AS160, the Akt substrate regulating GLUT4 translocation, has a functional Rab GTPase-activating protein domain. Biochem J. 2005;391:87–93. doi: 10.1042/BJ20050887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larance M. Characterization of the role of the Rab GTPase-activating protein AS160 in insulin-regulated GLUT4 trafficking. J Biol Chem. 2005;280:37803–37813. doi: 10.1074/jbc.M503897200. [DOI] [PubMed] [Google Scholar]
- 43.Huang J, Imamura T, Olefsky JM. Insulin can regulate GLUT4 internalization by signaling to Rab5 and the motor protein dynein. Proc Natl Acad Sci. 2001;98:13084–13089. doi: 10.1073/pnas.241368698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lodhi IJ, et al. Gapex-5, a Rab31 guanine nucleotide exchange factor that regulates Glut4 trafficking in adipocytes. Cell Metab. 2007;5:59–72. doi: 10.1016/j.cmet.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imamura T, et al. Insulin-induced GLUT4 translocation involves protein kinase C-lambda-mediated functional coupling between Rab4 and the motor protein kinesin. Mol Cell Biol. 2003;23:4892–4900. doi: 10.1128/MCB.23.14.4892-4900.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L. Direct interaction of Rab4 with syntaxin 4. J Biol Chem. 2001;276:5265–5273. doi: 10.1074/jbc.M003883200. [DOI] [PubMed] [Google Scholar]
- 47.Kane S, et al. A method to identify serine kinase substrates. Akt phosphorylates a novel adipocyte protein with a Rab GTPase-activating protein (GAP) domain. J Biol Chem. 2002;277:22115–22118. doi: 10.1074/jbc.C200198200. [DOI] [PubMed] [Google Scholar]
- 48.Sano H. Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J Biol Chem. 2003;278:14599–14602. doi: 10.1074/jbc.C300063200. [DOI] [PubMed] [Google Scholar]
- 49.Leney SE, Tavaré JM. The molecular basis of insulin-stimulated glucose uptake: signalling, trafficking and potential drug targets. J Endocrinol. 2009;203:1–18. doi: 10.1677/JOE-09-0037. [DOI] [PubMed] [Google Scholar]
- 50.Ishikura S, Bilan PJ, Klip A. Rabs 8A and 14 are targets of the insulin-regulated Rab-GAP AS160 regulating GLUT4 traffic in muscle cells. Biochem Biophys Res Commun. 2007;353:1074–1079. doi: 10.1016/j.bbrc.2006.12.140. [DOI] [PubMed] [Google Scholar]
- 51.Sun Y, Bilan PJ, Liu Z, Klip A. Rab8A and Rab13 are activated by insulin and regulate GLUT4 translocation in muscle cells. Proc Natl Acad Sci. 2010;107:19909–19914. doi: 10.1073/pnas.1009523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishikura S, Klip A. Muscle cells engage Rab8A and myosin Vb in insulin-dependent GLUT4 translocation. Am J Physiol Cell Physiol. 2008;295:C1016–C1025. doi: 10.1152/ajpcell.00277.2008. [DOI] [PubMed] [Google Scholar]
- 53.Sun Y, Chiu TT, Foley KP, Bilan PJ, Klip A. Myosin Va mediates Rab8A-regulated GLUT4 vesicle exocytosis in insulin-stimulated muscle cells. Mol Biol Cell. 2014;25:1159–1170. doi: 10.1091/mbc.E13-08-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ishikura S, Koshkina A, Klip A. Small G proteins in insulin action: Rab and Rho families at the crossroads of signal transduction and GLUT4 vesicle traffic. Acta Physiol (Oxf) 2008;192:61–74. doi: 10.1111/j.1748-1716.2007.01778.x. [DOI] [PubMed] [Google Scholar]
- 55.Sadacca LA, Bruno J, Wen J, Xiong W, McGraw TE. Specialized sorting of GLUT4 and its recruitment to the cell surface are independently regulated by distinct Rabs. Mol Biol Cell. 2013;24:2544–2557. doi: 10.1091/mbc.E13-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reed SE, et al. A role for Rab14 in the endocytic trafficking of GLUT4 in 3T3-L1 adipocytes. J Cell Sci. 2013;126:1931–1941. doi: 10.1242/jcs.104307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y, et al. Rab10 and myosin-Va mediate insulin-stimulated GLUT4 storage vesicle translocation in adipocytes. J Cell Biol. 2012;198:545–560. doi: 10.1083/jcb.201111091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chavez JA, Roach WG, Keller SR, Lane WS, Lienhard GE. Inhibition of GLUT4 translocation by Tbc1d1, a Rab GTPase-activating protein abundant in skeletal muscle, is partially relieved by AMP-activated protein kinase activation. J Biol Chem. 2008;283:9187–9195. doi: 10.1074/jbc.M708934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szekeres F, et al. The Rab-GTPase-activating protein TBC1D1 regulates skeletal muscle glucose metabolism. Am J Physiol Endocrinol Metab. 2012;303:E524–E533. doi: 10.1152/ajpendo.00605.2011. [DOI] [PubMed] [Google Scholar]
- 60.Peck GR, et al. Insulin-stimulated phosphorylation of the Rab GTPase-activating protein TBC1D1 regulates GLUT4 translocation. J Biol Chem. 2009;284:30016–30023. doi: 10.1074/jbc.M109.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dokas J, et al. Conventional knockout of Tbc1d1 in mice impairs insulin- and AICAR-stimulated glucose uptake in skeletal muscle. Endocrinology. 2013;154:3502–3514. doi: 10.1210/en.2012-2147. [DOI] [PubMed] [Google Scholar]
- 62.Lansey MN, Walker NN, Hargett SR, Stevens JR, Keller SR. Deletion of Rab GAP AS160 modifies glucose uptake and GLUT4 translocation in primary skeletal muscles and adipocytes and impairs glucose homeostasis. Am J Physiol Endocrinol Metab. 2012;303:E1273–E1286. doi: 10.1152/ajpendo.00316.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chadt A, et al. Deletion of both Rab-GTPase-activating proteins TBC1D1 and TBC1D4 in mice eliminates insulin- and AICAR-stimulated glucose transport. Diabetes. 2014 doi: 10.2337/db14-0368. [DOI] [PubMed] [Google Scholar]
- 64.Yamauchi T, et al. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat Med. 2002;8:1288–1295. doi: 10.1038/nm788. [DOI] [PubMed] [Google Scholar]
- 65.Yamauchi T, et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature. 2003;423:762–769. doi: 10.1038/nature01705. [DOI] [PubMed] [Google Scholar]
- 66.Ding Q, Wang Z, Chen Y. Endocytosis of adiponectin receptor 1 through a clathrin- and Rab5-dependent pathway. Cell Res. 2009;19:317–327. doi: 10.1038/cr.2008.299. [DOI] [PubMed] [Google Scholar]
- 67.Deepa SS, Dong LQ. APPL1: role in adiponectin signaling and beyond. AJP Endocrinol Metab. 2008;296:E22–E36. doi: 10.1152/ajpendo.90731.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang C, et al. Yin-Yang regulation of adiponectin signaling by APPL isoforms in muscle cells. (1) J Biol Chem. 2009;284:31608–31615. doi: 10.1074/jbc.M109.010355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng KKY, et al. The adaptor protein APPL2 inhibits insulin-stimulated glucose uptake by interacting with TBC1D1 in skeletal muscle. Diabetes. 2014;63:3748–3758. doi: 10.2337/db14-0337. [DOI] [PubMed] [Google Scholar]
- 70.King GJ, et al. Membrane curvature protein exhibits interdomain flexibility and binds a small GTPase. J Biol Chem. 2012;287:40996–41006. doi: 10.1074/jbc.M112.349803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thiam AR, Farese RV, Walther TC. The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol. 2013;14:775–786. doi: 10.1038/nrm3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krahmer N, Farese RV, Walther TC. Balancing the fat: lipid droplets and human disease. EMBO Mol Med. 2013;5:905–915. doi: 10.1002/emmm.201100671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stehr M, Elamin AA, Singh M. Cytosolic lipid inclusions formed during infection by viral and bacterial pathogens. Microbes Infect. 2012;14:1227–1237. doi: 10.1016/j.micinf.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 74.Liu P, et al. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem. 2004;279:3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- 75.Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- 76.Turró S, et al. Identification and characterization of associated with lipid droplet protein 1: a novel membrane-associated protein that resides on hepatic lipid droplets. Traffic. 2006;7:1254–1269. doi: 10.1111/j.1600-0854.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 77.Kiss RS, Nilsson T. Rab proteins implicated in lipid storage and mobilization. J Biomed Res. 2014;28:169–177. doi: 10.7555/JBR.28.20140029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilfling F, Haas JT, Walther TC, Farese RV. Lipid droplet biogenesis. Curr Opin Cell Biol. 2014;29:39–45. doi: 10.1016/j.ceb.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Robenek MJ, et al. Lipids partition caveolin-1 from ER membranes into lipid droplets: updating the model of lipid droplet biogenesis. FASEB J Off Publ Fed Am Soc Exp Biology. 2004;18:866–868. doi: 10.1096/fj.03-0782fje. [DOI] [PubMed] [Google Scholar]
- 80.Robenek H, et al. Adipophilin-enriched domains in the ER membrane are sites of lipid droplet biogenesis. J Cell Sci. 2006;119:4215–4224. doi: 10.1242/jcs.03191. [DOI] [PubMed] [Google Scholar]
- 81.Nevo-Yassaf I, et al. Role for TBC1D20 and Rab1 in hepatitis C virus replication via interaction with lipid droplet-bound nonstructural protein 5A. J Virol. 2012;86:6491–6502. doi: 10.1128/JVI.00496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu P, et al. Rab-regulated interaction of early endosomes with lipid droplets. Biochim Biophys Acta. 2007;1773:784–793. doi: 10.1016/j.bbamcr.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weidberg H, Shvets E, Elazar Z. Lipophagy: selective catabolism designed for lipids. Dev Cell. 2009;16:628–630. doi: 10.1016/j.devcel.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 84.Martin S, Driessen K, Nixon SJ, Zerial M, Parton RG. Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J Biol Chem. 2005;280:42325–42335. doi: 10.1074/jbc.M506651200. [DOI] [PubMed] [Google Scholar]
- 85.Pulido MR, et al. Rab18 dynamics in adipocytes in relation to lipogenesis, lipolysis and obesity. PLoS One. 2011;6:e22931. doi: 10.1371/journal.pone.0022931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boström P, et al. SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity. Nat Cell Biol. 2007;9:1286–1293. doi: 10.1038/ncb1648. [DOI] [PubMed] [Google Scholar]
- 87.Salloum S, Wang H, Ferguson C, Parton RG, Tai AW. Rab18 binds to hepatitis C virus NS5A and promotes interaction between sites of viral replication and lipid droplets. PLoS Pathog. 2013;9:e1003513. doi: 10.1371/journal.ppat.1003513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dansako H, Hiramoto H, Ikeda M, Wakita T, Kato N. Rab18 is required for viral assembly of hepatitis C virus through trafficking of the core protein to lipid droplets. Virology. 2014;462–463:166–174. doi: 10.1016/j.virol.2014.05.017. [DOI] [PubMed] [Google Scholar]
- 89.Vogt DA, et al. Lipid droplet-binding protein TIP47 regulates hepatitis C Virus RNA replication through interaction with the viral NS5A protein. PLoS Pathog. 2013;9:e1003302. doi: 10.1371/journal.ppat.1003302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aivazian D, Serrano RL, Pfeffer S. TIP47 is a key effector for Rab9 localization. J Cell Biol. 2006;173:917–926. doi: 10.1083/jcb.200510010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ploen D, et al. TIP47 is associated with the hepatitis C virus and its interaction with Rab9 is required for release of viral particles. Eur J Cell Biol. 2013;92:374–382. doi: 10.1016/j.ejcb.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 92.You X, et al. Hepatitis B virus X protein upregulates oncogene Rab18 to result in the dysregulation of lipogenesis and proliferation of hepatoma cells. Carcinogenesis. 2013;34:1644–1652. doi: 10.1093/carcin/bgt089. [DOI] [PubMed] [Google Scholar]
- 93.Wu L, et al. Rab8a-AS160-MSS4 regulatory circuit controls lipid droplet fusion and growth. Dev Cell. 2014;30:378–393. doi: 10.1016/j.devcel.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 94.Xu L, Zhou L, Li P. CIDE proteins and lipid metabolism. Arterioscler Thromb Vasc Biol. 2012;32:1094–1098. doi: 10.1161/ATVBAHA.111.241489. [DOI] [PubMed] [Google Scholar]
- 95.Gong J, et al. Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites. J Cell Biol. 2011;195:953–963. doi: 10.1083/jcb.201104142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 97.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 98.Lipatova Z, et al. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc Natl Acad Sci. 2012;109:6981–6986. doi: 10.1073/pnas.1121299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fader CM, Sánchez D, Furlán M, Colombo MI. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- 100.Longatti A, et al. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol. 2012;197:659–675. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Munafo DB, Colombo MI. Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic. 2002;3:472–482. doi: 10.1034/j.1600-0854.2002.30704.x. [DOI] [PubMed] [Google Scholar]
- 102.Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Itoh T, et al. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol Biol Cell. 2008;19:2916–2925. doi: 10.1091/mbc.E07-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Itoh T, Kanno E, Uemura T, Waguri S, Fukuda M. OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J Cell Biol. 2011;192:839–853. doi: 10.1083/jcb.201008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Popovic D, et al. Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol Cell Biol. 2012;32:1733–1744. doi: 10.1128/MCB.06717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 107.Zoncu R, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Avruch J, et al. Activation of mTORC1 in two steps: rheb-GTP activation of catalytic function and increased binding of substrates to raptor1. Biochem Soc Trans. 2009;37:223–226. doi: 10.1042/BST0370223. [DOI] [PubMed] [Google Scholar]
- 110.Sato T, Nakashima A, Guo L, Tamanoi F. Specific activation of mTORC1 by Rheb G-protein in vitro involves enhanced recruitment of its substrate protein. J Biol Chem. 2009;284:12783–12791. doi: 10.1074/jbc.M809207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dazert E, Hall MN. mTOR signaling in disease. Curr Opin Cell Biol. 2011;23:744–755. doi: 10.1016/j.ceb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 112.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Robida-Stubbs S, et al. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 2012;15:713–724. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–176. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 115.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 116.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cai SL. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J Cell Biol. 2006;173:279–289. doi: 10.1083/jcb.200507119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zoncu R, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. (1) Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kamada Y, et al. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol Cell Biol. 2005;25:7239–7248. doi: 10.1128/MCB.25.16.7239-7248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cybulski N, Hall MN. TOR complex 2: a signaling pathway of its own. Trends Biochem Sci. 2009;34:620–627. doi: 10.1016/j.tibs.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 122.Sarbassov DD, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 123.Li L, et al. Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem. 2010;285:19705–19709. doi: 10.1074/jbc.C110.102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Durán RV, Hall MN. Regulation of TOR by small GTPases. EMBO Rep. 2012;13:121–128. doi: 10.1038/embor.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bridges D, et al. Rab5 proteins regulate activation and localization of target of rapamycin complex 1. (1) J Biol Chem. 2012;287:20913–20921. doi: 10.1074/jbc.M111.334060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Roberts RL, Barbieri MA, Ullrich J, Stahl PD. Dynamics of rab5 activation in endocytosis and phagocytosis. J Leukoc Biol. 2000;68:627–632. [PubMed] [Google Scholar]
- 127.Matsui T, Fukuda M. Rab12 regulates mTORC1 activity and autophagy through controlling the degradation of amino-acid transporter PAT4. EMBO Rep. 2013;14:450–457. doi: 10.1038/embor.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Luo L, et al. Rab8a interacts directly with PI3Kγ to modulate TLR4-driven PI3K and mTOR signalling. Nat Commun. 2014;5:4407. doi: 10.1038/ncomms5407. [DOI] [PubMed] [Google Scholar]
- 129.Tatebe H, Morigasaki S, Murayama S, Zeng CT, Shiozaki K. Rab-family GTPase regulates TOR complex 2 signaling in fission yeast. Curr Biol. 2010;20:1975–1982. doi: 10.1016/j.cub.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tatebe H, Shiozaki K. Rab small GTPase emerges as a regulator of TOR complex 2. Small GTPases. 2010;1:180–182. doi: 10.4161/sgtp.1.3.14936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Martinez O, Schmidt A. The small GTP-binding protein rab6 functions in intra-Golgi transport. J Cell Biol. 1994;127:1575–1588. doi: 10.1083/jcb.127.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.He Y, et al. Genetic and functional interaction between Ryh1 and Ypt3: two Rab GTPases that function in S. pombe secretory pathway. Genes Cells. 2006;11:207–221. doi: 10.1111/j.1365-2443.2006.00935.x. [DOI] [PubMed] [Google Scholar]
- 133.Marion RM. Inaugural article: Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci. 2004;101:14315–14322. doi: 10.1073/pnas.0405353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fingerman I, Nagaraj V, Norris D, Vershon AK. Sfp1 plays a key role in yeast ribosome biogenesis. Eukaryot Cell. 2003;2:1061–1068. doi: 10.1128/EC.2.5.1061-1068.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lempiäinen H, et al. Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol Cell. 2009;33:704–716. doi: 10.1016/j.molcel.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 136.Singh J, Tyers M. A Rab escort protein integrates the secretion system with TOR signaling and ribosome biogenesis. Genes Dev. 2009;23:1944–1958. doi: 10.1101/gad.1804409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhou QL, et al. Akt substrate TBC1D1 regulates GLUT1 expression through the mTOR pathway in 3T3-L1 adipocytes. Biochem J. 2008;411:647–655. doi: 10.1042/BJ20071084. [DOI] [PMC free article] [PubMed] [Google Scholar]