Abstract
Cyclin-dependent kinases (Cdk) are a family of serine/threonine protein kinases that regulate eukaryotic cell cycle progression. Their ability to modulate the cell cycle has made them an attractive target for anti-cancer therapies. Cdk protein function has been studied in a variety of Eukaryotes ranging from yeast to humans. In the social amoebozoan Dictyostelium discoideum, several homologues of mammalian Cdks have been identified and characterized. The life cycle of this model organism is comprised of a feeding stage where single cells grow and divide mitotically as they feed on their bacterial food source and a multicellular developmental stage that is induced by starvation. Thus it is a valuable system for studying a variety of cellular and developmental processes. In this review I summarize the current knowledge of the Cdk protein family in Dictyostelium by highlighting the research efforts focused on the characterization of Cdk1, Cdk5, and Cdk8 in this model Eukaryote. Accumulated evidence indicates that each protein performs distinct functions during the Dictyostelium life cycle with Cdk1 being required for growth and Cdk5 and Cdk8 being required for processes that occur during development. Recent studies have shown that Dictyostelium Cdk5 shares attributes with mammalian Cdk5 and that the mammalian Cdk inhibitor roscovitine can be used to inhibit Cdk5 activity in Dictyostelium. Together, these results show that Dictyostelium can be used as a model system for studying Cdk protein function.
Keywords: Cyclin-dependent kinase, Mitosis, Growth, Development, Cell cycle, Dictyostelium
Cyclin-dependent kinases
The cyclin-dependent kinases (Cdk) belong to a family of serine/threonine protein kinases that were first discovered for their role in regulating eukaryotic cell cycle progression [1, 2]. Cdks are present in all eukaryotic cells and their kinase activity is regulated through their association with cyclins, which also function to recruit substrates [1, 2]. In mammals, the main cell cycle regulator is Cdk1, however additional Cdks (i.e., Cdk2, Cdk4, and Cdk6) are required for normal development and the proliferation of specialized tissues [3]. Due to their ability to regulate the cell cycle and the fact that tumor cells evade anti-growth signals, members of the Cdk protein family are potential targets for anti-cancer therapies [4, 5]. Cdk1 is the most well studied Cdk and its function is best understood in the model organism Saccharomyces cerevisiae where the protein controls the cell cycle by interacting with nine different cyclins (i.e., three G1 cyclins and six B-type cyclins) [3]. In yeast, Cdk1 also regulates DNA replication, chromosome segregation, transcription, and cell morphogenesis and polarization [3]. There are approximately 75 bona fide and over 300 potential Cdk1 targets [6].
Although most Cdks regulate the cell cycle, a subset of Cdks, namely Cdk7, Cdk8, and Cdk9, regulate gene expression through direct interactions with the transcription machinery. In mammals, Cdk8 represses transcription by phosphorylating the carboxy terminal domain of RNA polymerase II [7]. However, Cdk8 has also been reported to activate transcription through the phosphorylation of specific transcription factors [8]. Cdk8 associates with cyclin C to form part of the Mediator complex, which controls both basal and regulated transcription and is composed of at least 36 other proteins that are highly conserved from yeast to human [7, 9]. Like Cdk8, Cdk5 is another member of the Cdk protein family that possesses functions unrelated to the cell cycle. Unlike other members of the mammalian Cdk protein family, Cdk5 is not activated by cyclins. Instead, the protein is activated through its association with the neuron-specific activator molecules p35, p39, and p67 [10]. Cdk5 functions in a diversity of cellular processes including axon guidance, neurite outgrowth, insulin secretion, lens differentiation, membrane transport, myogenesis, and ubiquitin-dependent degradation [11–14]. In addition, unlike other Cdks whose functions have been implicated in tumor growth, Cdk5 dysregulation has been linked to neurodegenerative diseases including amyotrophic lateral sclerosis, Alzheimer’s disease, and Parkinson’s disease [15–17].
The life cycle of Dictyostelium discoideum
Dictyostelium discoideum is a fascinating organism that has been chosen by the National Institutes of Health as part of its model organism initiative for biomedical and human disease research [18, 19]. This model Eukaryote undergoes an asexual life cycle comprised of a vegetative feeding stage where single cells grow and divide mitotically as they chemotactically respond to folic acid, which is secreted by their bacterial food source and a multicellular developmental stage that is induced by starvation [20]. The Dictyostelium cell cycle does not possess a discernible G1 phase during growth and almost all cells are in G2 during both growth and development [21]. Growing cells released from stationary phase enter a short S phase lasting less than 30 min. This is followed by a long G2 phase that lasts 6.5 h on average. Cells then enter M phase, which lasts about 12 min. Upon starvation, cells enter a developmental program that begins with the secretion of 3′-5′-cyclic adenosine monophosphate (cAMP), which acts as a chemoattractant causing cells to aggregate into mounds [20]. Aggregated cells then develop into a motile, multicellular structure known as a pseudoplasmodium or slug. Cells destined to become spores (i.e., pre-spore cells, 80 % of the total cell population) sort to the middle and posterior regions of the slug, while cells destined to become stalk (i.e., pre-stalk cells, 20 % of the total cell population) sort to the anterior region. When conditions are suitable, the slug will develop into a fruiting body comprised of a mass of spores that is supported by a stalk of dead cells. Two periods of cell division occur during Dictyostelium development, one during the early stages of development and the other after multicellular structures have formed [22]. However, only the second period of cell division has been shown to be a result of mitosis [23]. The observed increase in cell number during early development is not due to mitosis, but instead is a result of multinucleated cells undergoing cytokinesis to complete their final growth-stage cell cycle [23].
In addition to its involvement in the regulation of growth, the cell cycle also influences cell type differentiation in Dictyostelium [24–29]. This is an interesting feature of the Dictyostelium life cycle, which has recently been reviewed [30, 31]. Cells within a growing population will be in different phases of the cell cycle (i.e., S, early G2, mid-G2, late G2, M). However, since the G2 phase in Dictyostelium is relatively long compared to the S and M phases, the majority of cells will be in G2 [25]. Upon starvation, cells in M, S, or early G2 tend to form pre-stalk cells, while cells in mid- or late G2 preferentially form pre-spore cells. Unlike growth, there is evidence for a G1 phase during Dictyostelium development [23, 32]. Pre-stalk cells undergo mitosis just before stalk formation (i.e., after 22 h) and arrest in G1 prior to terminal differentiation [23]. Pre-spore cells undergo mitosis between 12 and 20 h of development [23], however whether these cells arrest in G1 or G2 prior to terminal differentiation remains unclear. Chen et al. [23, 32] have reported that pre-spore cells arrest in G1 prior to encapsulation. Their studies, which were based upon cellular DNA content profiles obtained by flow cytometry and quantification of extra-chromosomal and chromosomal DNA, also showed that Dictyostelium cells can carry out the entire developmental sequence in G1 [32]. In contrast, MacWilliams et al. [33] used direct fluorescence measurements of DAPI-stained nuclei to show that the nuclear DNA content of cells and spores do not differ significantly. Since vegetative cells are largely in G2, it follows that the spores under their experimental conditions would also be in G2 [33]. These results were supported by Muramoto and Chubb [34] who introduced a live cell S-phase marker into developing cells (i.e., fluorescently tagged replication factor, proliferating cell nuclear antigen). They reported that germinating spores entered S-phase only after their first round of mitosis, indicating that the spores were in G2 [34]. In addition, while it is generally accepted that DNA replication does occur during development, the source of the DNA synthesis (i.e., nuclear or mitochondrial) is controversial with contradictory results reported in the literature [24, 32–35]. Zimmerman and Weijer [24] labeled developing cells with the thymidine analog 5′-bromodeoxyuridine and reported that 75 % of cells from axenically grown cultures go through S-phase. A later study by Shaulsky and Loomis [35] reported that while DNA synthesis does occur in pre-spore cells, the source of the DNA synthesis is mitochondrial, not nuclear. Unfortunately, more recent studies have yet to clarify this feature of the Dictyostelium life cycle. Chen and colleagues [32] reported that although substantial mitochondrial DNA synthesis does occur in pre-spore cells, chromosomal DNA synthesis is not observed. Using their imaging approach Muramoto and Chubb [34] reported that a large proportion of cells undergo nuclear DNA synthesis during development. As discussed previously, their study also showed that spores arrest in G2 [34]. Since the majority of cells upon starvation are in G2, and it is known that at least one round of mitosis occurs during the later stages of development, their results indicate that S-phase does occur during development [34]. Taken together, it has become increasingly evident that this aspect of the Dictyostelium life cycle requires further study to resolve and clarify the conflicting reports presented in the literature. In addition, although it is well established that the cell cycle influences cell type differentiation in Dictyostelium the function(s) (if any) of Cdks and cyclins during this process is unknown.
The Dictyostelium genome has been sequenced and several homologues of mammalian Cdks have been identified (Table 1). The most well studied of these homologues include Cdk1, Cdk5, and Cdk8, which will be discussed in detail below and will be the focus of this review. The Dictyostelium genome also encodes putative Cdks that have not yet been characterized (e.g., Cdk7, Cdk9, Cdk10, and Cdk11) as well as a number of cyclins and Cdk-binding partners (Tables 1, 2). Some of these binding partners have been verified as bona fide Cdk-binding proteins and will also be discussed in the review. The genome, however, does not contain obvious homologues of mammalian Cdk2, Cdk3, Cdk4, or Cdk6 (Table 1).
Table 1.
List of Cdk homologues in Dictyostelium discoideum
| Gene | Identified in Dictyostelium? | dictyBase gene ID | Function(s) |
|---|---|---|---|
| cdk1 | Yes | DDB_G0272813 | Growth [36, 39] |
| Early development? [36, 39] | |||
| cdk2 | Noa | N/A | N/A |
| cdk3 | No | N/A | N/A |
| cdk4 | No | N/A | N/A |
| cdk5 | Yes | DDB_G0288677 | Growth [42–44] |
| Mid development [42, 44] | |||
| Late development [42, 44] | |||
| cdk6 | No | N/A | N/A |
| cdk7 | Yes | DDB_G0285417 | Uncharacterized |
| cdk8 | Yes | DDB_G0267442 | Growth [84, 86] |
| Early development [84–86] | |||
| Late development [84–86] | |||
| cdk9 | Yes | DDB_G0273207 | Uncharacterized |
| cdk10 | Yes | DDB_G0268480 | Uncharacterized |
| cdk11 | Yes | DDB_G0283279 | Uncharacterized |
a Dictyostelium Cdk1 shares strong sequence similarity with both mammalian Cdk1 and Cdk2, but the genome does not contain separate genes for Cdk1 and Cdk2
Table 2.
List of identified and putative Cdk-binding partners in Dictyostelium discoideum
| Gene | Protein | dictyBase gene ID | Binding partner |
|---|---|---|---|
| cycA | Cyclin A | DDB_G0279085 | Cdk1 (predicted) |
| cycB | Cyclin B | DDB_G0275493 | Cdk1 [36, 40] |
| cycC | Cyclin C | DDB_G0274139 | Cdk8 [87]a |
| cycD | Cyclin D | DDB_G0277439 | Cdk1 (predicted) |
| cycH | Cyclin H | DDB_G0268668 | Cdk7 (predicted) |
| cycK | Cyclin K | DDB_G0286617 | Cdk9 (predicted) |
| cycL | Cyclin L | DDB_G0285553 | Cdk11 (predicted) |
| psaA | Puromycin-sensitive aminopeptidase | DDB_G0270994 | Cdk5 [43] |
| calA | Calmodulin | DDB_G0279407 | Cdk5 [60] |
| cksl | Cyclin-dependent kinases regulatory subunit | DDB_G0271642 | Unknown |
aAlthough a direct interaction between Cdk8 and CycC has not yet been shown, the two proteins are detected in high molecular weight fractions isolated from Dictyostelium nuclei
Cyclin-dependent kinase 1
Cdk1 functions during Dictyostelium growth
In Dictyostelium, Cdk1 functions primarily during vegetative growth (Fig. 1) [36]. cdk1 mRNA and protein is expressed at a constant level during all phases of axenic growth (i.e., exponential and stationary phases) [37]. During development, the level of cdk1 mRNA increases during aggregation and then decreases to low levels by the slug stage, eventually disappearing during the terminal stages of differentiation [36–38] (Fig. 2). The kinase activity of Cdk1 has been verified and the protein level remains constant during development up to 16 h, after which time the amount of Cdk1 protein decreases significantly [36]. Despite the fact that cdk1 mRNA and protein are expressed during development, there is little evidence to suggest that Cdk1 is involved in regulating developmental processes, including the mitotic events that occur during later developmental stages [22, 36, 37, 39]. Although Luo et al. [36] reported an increase in Cdk1 histone H1 kinase activity during early development, a follow-up study by Sharma et al. [39] concluded that this increase in kinase activity is likely not important for Dictyostelium development. This conclusion was based upon observations of cells that expressed a dominant-negative mutant cdk1 gene under the control of the discoidin promoter, which drives gene expression only during growth and early development [38]. Thus the mutant Cdk1 protein was only expressed during these stages of the life cycle. Although the mutant cells showed slow rates of growth, thus supporting a function for Cdk1 during cell proliferation, they were observed to develop and differentiate normally [39]. In other words, expression of the mutated Cdk1 protein during early development did not negatively affect later developmental processes. It is important to note that the cdk1 mutant cells still expressed endogenous Cdk1. Therefore it is likely that the effect of the mutant Cdk1 protein during growth and early development was somewhat mitigated by the presence of the endogenous protein. Furthermore, the absence of an increase in Cdk1 histone H1 kinase activity during later development coupled with the relatively low expression of cdk1 mRNA and protein during this stage of the life cycle, also supports the contention that Cdk1 is not involved in regulating later developmental processes, including the mitotic events that occur during this time [36].
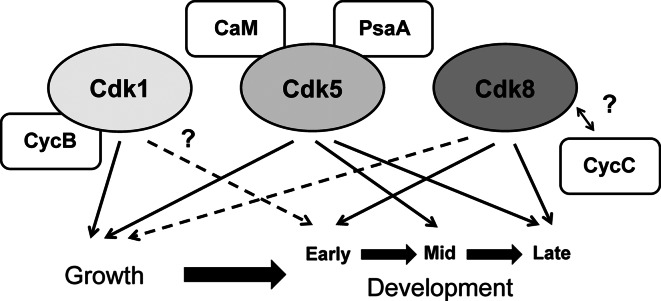
Fig. 1.
The functions of Cdk1, Cdk5, and Cdk8 during Dictyostelium growth and development. Cdk1 interacts with CycB and is required for growth. Its function during early development remains unclear (i.e., indicated by a dashed arrow and question mark). Cdk1 activity is also dependent on CycB expression levels. Cdk5 is a CaMBP that also binds to PsaA. Cdk5 is required for efficient growth and for mid- and late developmental processes. Cdk8 is required for early and late developmental processes. Cdk8 also functions during growth, however its activity is not essential for this cellular process (i.e., indicated by a dashed arrow). Cdk8 and CycC are detected in high molecular weight complexes isolated from Dictyostelium nuclei, however a direct interaction between Cdk8 and CycC has not yet been shown
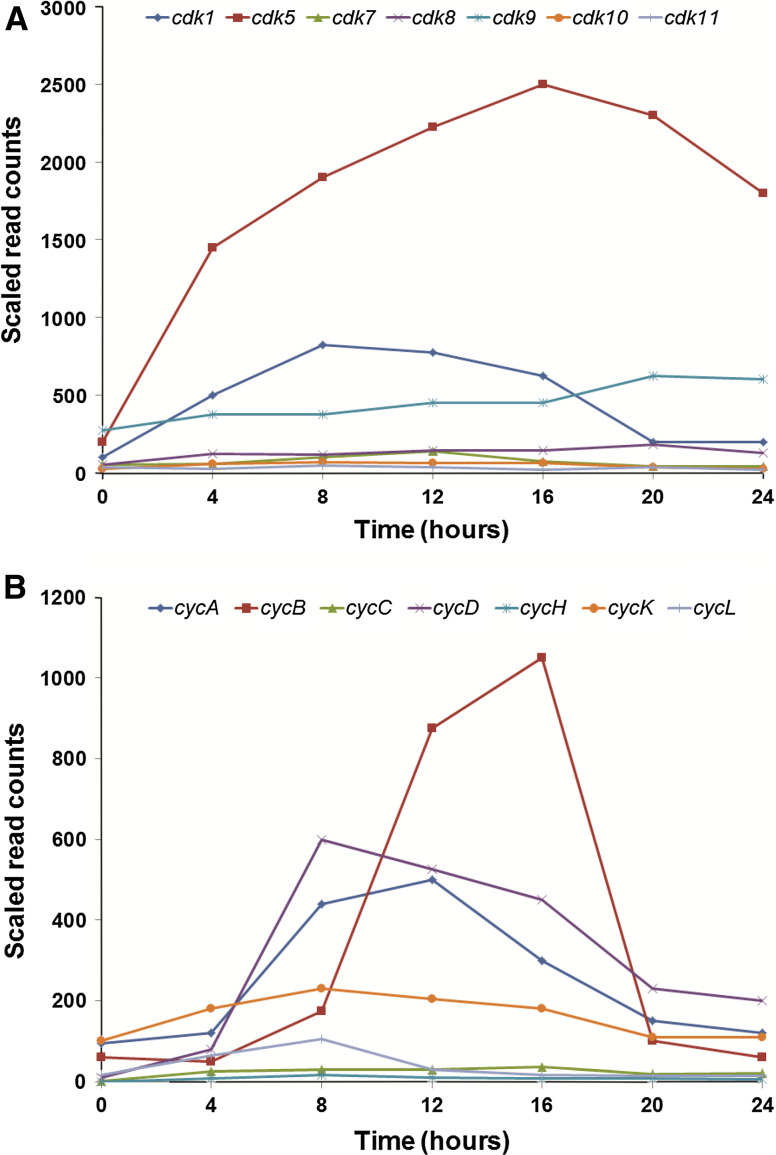
Fig. 2.
Gene expression analysis of Dictyostelium Cdks and cyclins during development. RNA-Seq data was obtained from dictyExpress (www.dictyexpress.biolab.si, [38]) and re-plotted using Microsoft Excel. a Expression profiles of cdk mRNA during development. b Expression profiles of cyclin mRNA during development
Cyclin B interacts with Cdk1 and regulates its activity during Dictyostelium growth
Like its mammalian counterpart, Dictyostelium Cdk1 is activated through its association with cyclin B (CycB) and its activity during growth and development is dependent on CycB expression levels (Fig. 1) [36]. During growth, cycB mRNA and protein expression increases upon entry into mitosis and the Cdk1/CycB complex has been suggested to regulate the transition from G2 to M phase since overexpression of truncated CycB results in mitotic arrest [36, 40]. In addition, CycB degradation is required for vegetative cells to leave mitosis, which has also been reported to occur during the early development of Xenopus [40, 41]. During Dictyostelium development, cycB mRNA and protein expression reaches a maximum between the tipped aggregate and multicellular slug stage (i.e., 12–16 h), which closely adheres to the observed expression profiles of cdk1 mRNA and protein [36, 40] (Fig. 2). Together these findings indicate that Cdk1 and CycB are required for growth, however the function of these proteins during early development (i.e., aggregation) is still unclear.
Cyclin-dependent kinase 5
Cdk5 is required for growth and later development processes in Dictyostelium
Cdk5 is required for growth and for processes that occur during the later stages of Dictyostelium development (Fig. 1) [42–44]. The kinase activity of the protein has been confirmed and Cdk5 protein levels remain constant during all stages of axenic growth (i.e., exponential and stationary phases) [39, 44, 45]. cdk5 mRNA and protein expression increases significantly during development reaching peak expression levels after 16 h and remains relatively high during terminal differentiation [38, 39, 45] (Fig. 2). Overexpression of a dominant negative form of Cdk5 that contains an aspartate to asparagine substitution at amino acid position 144 reduces the rate of growth in suspension culture as well as the rates of fluid-phase endocytosis and phagocytosis [42]. In mammals, this amino acid substitution causes the binding between Cdk5 and its activator p35 to be two to threefold more efficient than the binding of normal Cdk5 to p35 [46]. Thus, when over-expressed, the mutant protein blocks the activation of endogenous Cdk5 by depleting the available pool of p35. During Dictyostelium development, overexpression of this mutant Cdk5 protein delays aggregation and fruiting body formation as well as significantly reduces the number of fruiting bodies and spores that form from these cells indicating that Cdk5 is required for processes that occur during later development [42]. These results also suggest that Cdk5 may regulate cell cycle events during mid- to late development. Although the Dictyostelium genome does not encode an obvious homologue of mammalian p35, Cdk5-binding partners have been identified in Dictyostelium (discussed below).
Cdk5 localizes to the nucleus and cytoplasm in Dictyostelium and translocates from the nucleus to the cytoplasm during mitosis
In Dictyostelium, Cdk5 localizes to both the nucleus and cytoplasm of vegetative cells suggesting a functional role in both cellular locales [43, 44]. Interestingly, this finding mirrors observations in mammalian cells. Unlike other members of the Cdk protein family that function primarily in the nucleus, Cdk5 localizes and functions in both the nucleus and cytoplasm of mammalian cells. Early studies on Cdk5 function in mammals focused on the cytoplasmic functions of the protein, however the nuclear functions of Cdk5 are slowly being revealed [47]. In neurons, Cdk5 has a death-promoting activity when localized to the nucleus and a pro-survival activity when localized to the cytoplasm [48]. In cycling NIH 3T3 cells, the localization of Cdk5 shifts from the nucleus to the cytoplasm suggesting that Cdk5 acts as a cell cycle suppressor when localized to the nucleus [49]. The change in localization occurs before or shortly after the initiation of the cell cycle and blocking the migration of Cdk5 out of the nucleus suppresses the cell cycle. During the early stages of Dictyostelium mitosis, Cdk5 gradually moves from a punctate nucleoplasmic distribution to localize adjacent to the inner nuclear envelope, eventually condensing as an intranuclear ring [43]. During anaphase and telophase, Cdk5 is absent from the nucleoplasm and localizes to the cytoplasm. The protein then returns to the nucleus during cytokinesis. Together these results indicate that the nucleocytoplasmic translocation of Cdk5 is an evolutionarily conserved process. In addition to its localization to the nucleus and cytoplasm, a small amount of Cdk5 has also been detected in the cytoskeleton of Dictyostelium cells [44]. This observation fits with the established role of Cdk5 in regulating fluid-phase endocytosis, phagocytosis, and cytoskeletal dynamics in Dictyostelium and adheres to studies in mammalian cells, which have shown that mammalian Cdk5 and its activators p35 and p39 localize to the actin cytoskeleton possibly playing a role in the regulation of actin-cytoskeletal dynamics [42, 50–52].
Cdk5 binds to a puromycin-sensitive aminopeptidase in Dictyostelium
As discussed above, Cdk5 is one member of the mammalian Cdk protein family that is not activated by cyclins. Instead, Cdk5 is activated through its association with the neuron-specific activator molecules p35, p39, and p67 [10]. The Dictyostelium genome does not encode obvious homologues of mammalian p35, p39, or p67, however immunoprecipitations coupled with immunolocalizations have identified puromycin-sensitive aminopeptidase A (PsaA) as a Cdk5-binding partner in Dictyostelium (Fig. 1) [43]. PSAs are highly conserved metalloproteases that hydrolyze N-terminal amino acids from oligopeptides and are linked to a number of cellular processes [53]. In Dictyostelium, PsaA localizes to the nucleoplasm [54]. Bestatin, which inhibits the final step of intracellular protein degradation (i.e., degradation of small peptides into free amino acids) and represses epidermal growth factor-induced DNA synthesis and cell division in rat hepatocytes, binds to PsaA and inhibits cell division [54–56]. These results thus support a functional role for PsaA in regulating cell proliferation and mitosis in Dictyostelium. The interaction between Cdk5 and PsaA is interesting given that proteolytic activity has been shown to be a critical regulator of the cell cycle [57]. Inhibitors of aminopeptidase activity suppress cell proliferation by arresting the cell cycle and causing an accumulation of cells in the G2/M phase [55, 58, 59]. These findings indicate that PSA activities are required to complete mitosis and are essential for cell division and viability. In Dictyostelium, the association of Cdk5 with PsaA suggests that PsaA proteolytic activity may modulate the cell cycle by regulating the function and/or localization of Cdk5 during mitosis. Several potential phosphorylation sites have been identified in mammalian PSA, however Dictyostelium PsaA does not appear to be phosphorylated in vivo indicating the protein is likely not a target of Cdk5 kinase activity [43, 59].
Cdk5 is a calmodulin-binding protein in Dictyostelium
A recent study showed that Cdk5 is a calmodulin (CaM)-binding protein (CaMBP) in Dictyostelium that binds to apo-CaM (i.e., Ca2+ independent CaM-binding) and associates with PsaA and CaM in Dictyostelium nuclei (Fig. 1) [60]. This was the first evidence in any system for Cdk5 being a CaMBP. CaM is the primary sensor of Ca2+ within the cell and binds to a diversity of CaMBPs that participate in a variety of cellular processes [61–63]. In HEK293T and PC12 cells, p35 activity is regulated by both Cdk5 phosphorylation and CaM-binding, however a direct binding between Cdk5 and CaM has not been shown [64]. Sequence analysis identified two putative CaM-binding domains (CaMBDs) in Dictyostelium Cdk5, however until now, only one of the two putative domains has been verified as a true CaMBD [60]. Deletion of CaMBD2 (132LLINRKGELKLADFGLARAFGIP154) prevented CaM-binding indicating that this region, which contains 1-10, 1-12, 1-14, 1-16, and 1-8-14 hydrophobic motifs, encompasses a functional CaMBD. Interestingly, the two CaMBDs of Dictyostelium Cdk5 are strongly conserved in human Cdk5 indicating that this is an evolutionarily conserved interaction that warrants investigation in human cells. Insight into the function of this interaction was provided when Huber et al. [60] showed that deletion of Cdk5 CaMBD2 significantly increased the nuclear distribution of Cdk5 and that this effect was dramatically enhanced by deletion of both CaMBD1 and CaMBD2. In mammalian cells, CaM functions at multiple points in the cell cycle and is required for cell proliferation [65, 66]. CaM function has also been linked to mitosis and cell proliferation in Dictyostelium [67, 68]. Together, these results suggest that CaM may regulate the nucleocytoplasmic transport of Cdk5, which could ultimately regulate cell proliferation and mitosis in Dictyostelium and other systems. The identification of Cdk5 as a CaMBP opens up new avenues of research to investigate whether Cdk5 binds CaM in other systems and the function of this interaction.
Roscovitine inhibits Cdk5-dependent processes in Dictyostelium
Attempts to generate a cdk5 knockout mutant have been unsuccessful, likely because the protein is essential for cell division and growth [42]. As discussed previously, an early study made use of a mutant strain overexpressing a dominant negative form of Cdk5 [42]. That study showed that Cdk5 is required for optimal growth and differentiation and that the protein is involved in regulating fluid-phase endocytosis, phagocytosis, and cytoskeletal dynamics. The inability to generate a cdk5 knockout mutant has necessitated the use of other approaches to fully elucidate the function of this biomedically important protein. With this in mind, Huber and O’Day [44] showed that the mammalian Cdk inhibitor roscovitine could be used to study the function of Cdk5 during Dictyostelium growth and development.
Roscovitine is a potent, cell-permeable Cdk inhibitor that inhibits kinase activity by binding to the ATP-binding pocket of a number of Cdks (e.g., Cdk1, Cdk2, Cdk5, Cdk7, and Cdk9), however studies have indicated that it preferentially inhibits Cdk5 [69–72]. The ability of roscovitine to inhibit Cdk10 and Cdk11 has not been reported, however it has no effect on the activity of mammalian Cdk4, Cdk6, and Cdk8 [73, 74]. Its effectiveness at treating nasopharyngeal cancer and non-small cell lung cancer is currently being evaluated in phase 2 and 2B clinical trials, respectively, and it is also being investigated for its potential to treat breast cancer, herpes simplex infection, HIV infection, leukemia, and chronic inflammation disorders such as cystic fibrosis and arthritis [75–79]. Roscovitine has been used to show that Cdk5 is required for long-term potentiation induction and NMDA-induced currents in rat hippocampal neurons, morphine tolerance in rats, cell proliferation and apoptosis of MDA-MB-231 cells, and endothelial cell migration [70, 71, 80, 81]. Roscovitine has also been used to inhibit Cdk5 activity in mouse C2C12 myoblasts [82].
In Dictyostelium, roscovitine significantly inhibits kinase activity and axenic growth and this inhibition can be partially rescued by the overexpression of Cdk5-GFP [42, 44]. These results thus support the involvement of Cdk5 in the regulation of axenic growth and indicate that Cdk5 is a primary target of the chemical. Roscovitine does not affect the expression of Cdk5 protein during growth, but it does inhibit its translocation to the nucleus; a result that has not been previously reported for any organism [44]. It also increases the amount of cytoskeletal-associated Cdk5. Development up to the mound stage is unaffected by roscovitine, however treatment does inhibit the later stages of Dictyostelium development, specifically slug and fruiting body formation. The fruiting bodies that do form are small relative to untreated fruiting bodies and contain relatively few spores. In addition, unlike the elliptical shape of untreated spores, roscovitine causes spores to adopt a round shape. These findings mirror the observations of the mutant Cdk5 dominant negative overexpressing strain, thus providing strong evidence for the ability of roscovitine to inhibit Cdk5 activity in Dictyostelium and supporting the involvement of Cdk5 in the regulation of later developmental processes [42]. The relative abundance of cdk5 mRNA during Dictyostelium growth and development compared to other Cdks is also an indication that roscovitine primarily inhibits Cdk5-dependent cellular processes in Dictyostelium (Fig. 2). Since it has been reported that at least one round of mitosis occurs during the later stages of development, these findings also indicate that Cdk5 may be required for the mitotic events that occur during this stage of the developmental cycle [22, 83]. The ability of roscovitine to inhibit Cdk5 activity and its nucleocytoplasmic translocation in Dictyostelium will allow future studies to clarify the function of Cdk5 during cell proliferation and development by identifying the signaling pathways that regulate its activity and localization. Dictyostelium can also be used as a model system to understand the mechanism of action of roscovitine, which could provide important insight for studies aimed at determining its usefulness as a clinical drug. As discussed previously, it is well established that the cell cycle affects cell type differentiation during Dictyostelium development [30, 31]. Therefore, it is also possible that the effect of roscovitine on development is due to the inhibition of the cell cycle during earlier developmental processes rather than roscovitine having a direct effect on later development. In addition, although Huber and O’Day [44] provided strong evidence that roscovitine specifically inhibits Cdk5 activity in Dictyostelium, the ability of the chemical to also inhibit other Cdks during Dictyostelium growth and development remains unclear. This may require future study since Cdk1, Cdk7, and Cdk9 have also been shown to be sensitive to roscovitine in mammals [69].
Cyclin-dependent kinase 8
Cdk8 is required for early and later developmental processes in Dictyostelium
In Dictyostelium, Cdk8 is not absolutely required for growth, but is required for early and later developmental processes (Fig. 1) [84–86]. cdk8 mRNA expression is relatively low during growth (Fig. 2) [84]. Expression increases during early development, reaches peak levels after 12 h, and then quickly decreases between 12 and 15 h of development [84]. Recently published RNA-Seq data supports the increase in cdk8 mRNA expression during early development, however in that analysis expression peaked at 20 h and remained high during the later stages of development (Fig. 2) [38].
cdk8- cells possess a pleiotropic phenotype and there is no obvious effect of Cdk8 overexpression [84, 85]. Mutant cells grow more slowly than parental cells in axenic medium and form small plaques on bacterial lawns indicating that Cdk8 is required for feeding [84, 86]. Upon starvation, cdk8- cells do not aggregate or stream and do not show evidence of pulsatile cell movement [84, 85]. Mutant cells do not express adenyl cyclase A (acaA) transcripts and the expression of 3′-5′-cyclic adenosine monophosphate receptor A (carA) mRNA is strongly reduced indicating that the aggregation defect is likely due to abolished cAMP signaling [84]. Lin et al. [85] confirmed this hypothesis by showing that the aggregation defect could be partially overcome by pulsing cells with cAMP thus indicating an impaired signal relay in cdk8- cells. Lin et al. [85] also reported a dysregulation of gene expression during early development for the catalytic subunit of protein kinase A (PKAc) and discoidin 1 (Disc1). However, overexpression of PKAc could not rescue the aggregation defect indicating that the low expression levels of PKAc are not solely responsible for the aggregation defect [86].
cdk8- cells develop abnormally during the later stages of Dictyostelium development indicating that Cdk8 may be involved in regulating the cell cycle during development [84]. Fruiting bodies form only under certain experimental conditions (i.e., grown axenically in nutrient-rich HL5 medium followed by plating on filters) and most of the spores are not capable of germinating [84]. Follow-up studies confirmed the requirement of Cdk8 for spore cell differentiation [85, 86], however a recent study that generated a cdk8 mutant in a strain lacking detectable gene duplication showed that cdk8- cells are able to form phenotypically normal fruiting bodies [86]. In that study, a slight developmental defect was reported with fruiting body formation occurring 3–4 h later than parental cells. Despite the ability to form normal fruiting bodies, spores that formed were less viable than the parental strain, though they looked morphologically normal. Nonetheless, these findings confirmed earlier observations supporting an involvement of Cdk8 in spore cell differentiation [84, 85].
Cyclin C associates with high molecular weight complexes and regulates the activity of Cdk8 during Dictyostelium development
Dictyostelium Cdk8 localizes primarily to the nucleus and forms part of a high molecular weight complex that has carboxy terminal domain kinase activity [85, 87]. In mammals, the high molecular weight Mediator complex links regulatory proteins to the basal transcription machinery [7, 9]. Dictyostelium cyclin C (CycC) has also been detected in high molecular weight complexes isolated from Dictyostelium nuclei however a direct interaction between Cdk8 and CycC has not yet been shown [87]. CycC protein is expressed in growing cells [87]. During development, the protein level begins to decrease after mound formation [87]. cycC mRNA follows this expression pattern and the decrease in expression after mound formation, which is triggered by extracellular cAMP, reduces the association of Cdk8 with the nuclear high molecular weight complex [87]. Recently published RNA-Seq data shows that cycC mRNA expression increases during early development, but does not decrease after mound formation as reported by Greene et al. [87] (Fig. 2) [38]. Instead, cycC mRNA expression continues to increase after mound formation and reaches its peak level once the multicellular slug has formed (i.e., 16 h) (Fig. 2). Expression then decreases during the later stages of development. While it is not immediately apparent why different expression profiles have been reported research to this point still strongly supports a developmental function for Cdk8 and CycC. Finally, increased levels of either Cdk8 or CycC enhance the rate of early development indicating that the two proteins are rate-limiting for multicellular development [87]. Overexpression of both proteins results in an even greater acceleration of early developmental events. Together, the accumulated evidence indicates that Cdk8 and CycC are not absolutely necessary for growth, but are required during development for aggregation and spore cell differentiation.
Conclusions and future directions
The expression profiles and interacting proteins involved in regulating the activity of Cdk1, Cdk5, and Cdk8 suggest that the proteins likely perform distinct functions during the Dictyostelium life cycle (Fig. 1,2). Accumulated evidence indicates that Cdk1 is important for growth, but does not appear to be essential for development including the regulation of developmental mitosis. In contrast, Cdk5, although also required for growth, is important for the later stages of development, specifically slug and fruiting body formation. Cdk5 activity may also be required for the mitotic events that occur during later development indicating that the regulation of developmental mitosis may be different from the mechanisms that regulate mitosis during vegetative growth. Finally, Cdk8 functions during early and late development, specifically aggregation and spore differentiation, indicating that Cdk8 may also be involved in regulating cell cycle events during development. The research community would benefit from future studies that focus on the identification of signaling pathways that mediate the activity of these Cdks as well as studies that characterize the putative Cdks and cyclins that have also been identified in Dictyostelium to determine their function during growth and development (e.g., Cdk7, Cdk9, Cdk10, and Cdk11) (Tables 1 and 2; Fig. 2). Future research in this area may also be able to shed light on the precise mechanisms that regulate the cell cycle and cell type differentiation during Dictyostelium development. Taken together, these results show that Dictyostelium can be used as a model system to study the function of conserved Cdks in an organism that provides an excellent opportunity to examine their role in a number of cellular and developmental processes. Since the Cdk protein family has been identified as a potential target for anti-cancer therapy, research into the function of Cdks in Dictyostelium could provide valuable new insight for the development of effective treatments and therapies for various forms of cancer. In addition, studies into Cdk5 function in Dictyostelium could provide new insight into the role Cdk5 dysregulation plays in neurodegeneration.
Acknowledgments
The author would like to thank the anonymous reviewers who provided insightful and helpful feedback during the submission process. This review was supported by a Postdoctoral Fellowship from the Canadian Institutes of Health Research (R.J.H.).
Abbreviations
- AcaA
Adenylyl cyclase A
- cAMP
3′-5′-cyclic adenosine monophosphate
- CaM
Calmodulin
- CaMBP
Calmodulin-binding protein
- CaMBD
Calmodulin-binding domain
- carA
3′-5′-cyclic adenosine monophosphate receptor A
- Cdk
Cyclin-dependent kinase
- CycB
Cyclin B
- CycC
Cyclin C
- Disc1
Discoidin 1
- PKAc
Protein kinase A catalytic subunit
- PsaA
Puromycin-sensitive aminopeptidase A
References
- 1.Hochegger H, Takeda S, Hunt T. Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nat Rev Mol Cell Biol. 2008;9:910–916. doi: 10.1038/nrm2510. [DOI] [PubMed] [Google Scholar]
- 2.Doonan JH, Kitsios G. Functional evolution of cyclin-dependent kinases. Mol Biotechnol. 2009;42:14–29. doi: 10.1007/s12033-008-9126-8. [DOI] [PubMed] [Google Scholar]
- 3.Enserink JM, Kolodner RD. An overview of Cdk1-controlled targets and processes. Cell Div. 2010;5:11. doi: 10.1186/1747-1028-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 5.Canavese M, Santo L, Raje N. Cyclin-dependent kinases in cancer: potential for therapeutic intervention. Cancer Biol Ther. 2012;13:451–457. doi: 10.4161/cbt.19589. [DOI] [PubMed] [Google Scholar]
- 6.Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian mediator complex and its role in transcriptional regulation. Trends Biochem Sci. 2005;30:250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Gold MO, Rice AP. Targeting of CDK8 to a promoter proximal RNA element demonstrates catalysis-dependent activation of gene expression. Nucleic Acids Res. 1998;26:3784–3788. doi: 10.1093/nar/26.16.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casamassimi A, Napoli C. Mediator complexes and eukaryotic transcription regulation: an overview. Biochimie. 2007;89:1439–1446. doi: 10.1016/j.biochi.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Amin ND, Albers W, Pant HC. Cyclin-dependent kinase 5 (cdk5) activation requires interaction with three domains of p35. J Neurosci Res. 2002;67:354–362. doi: 10.1002/jnr.10116. [DOI] [PubMed] [Google Scholar]
- 11.Rosales JL, Lee K-Y. Extraneuronal roles of cyclin-dependent kinase 5. Bioessays. 2006;28:1023–1034. doi: 10.1002/bies.20473. [DOI] [PubMed] [Google Scholar]
- 12.Giese KP. Novel insights into the beneficial and detrimental actions of Cdk5. Mol Interv. 2007;7:246–248. doi: 10.1124/mi.7.5.5. [DOI] [PubMed] [Google Scholar]
- 13.Bolin C, Boudra M-T, Fernet M, Vaslin L, Pennaneach V, Zaremba T, Biard D, Cordelieres FP, Favaudon V, Megnin-Chanet F, Hall J. The impact of cyclin-dependent kinase 5 depletion on poly(ADP-ribose) polymerase activity and responses to radiation. Cell Mol Life Sci. 2012;69:951–962. doi: 10.1007/s00018-011-0811-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan Q, Qiao F, Gao C, Norman B, Optican L, Zelenka PS. Cdk5 targets active Src for ubiquitin-dependent degradation by phosphorylating Src(S75) Cell Mol Life Sci. 2011;68:3425–3436. doi: 10.1007/s00018-011-0638-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crews L, Patrick C, Adame A, Rockenstein E, Masliah E. Modulation of aberrant CDK5 signaling rescues impaired neurogenesis in models of Alzheimer’s disease. Cell Death Dis. 2011;2:e120. doi: 10.1038/cddis.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung ZH, Ip NY. Cdk5: a multifaceted kinase in neurodegenerative diseases. Trends Cell Biol. 2012;22:169–175. doi: 10.1016/j.tcb.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Wong AS, Lee RH, Cheung AY, Yeung PK, Chung SK, Cheung ZH, Ip NY. Cdk5-mediated phosphorylation of endophilin B1 is required for induced autophagy in models of Parkinson’s disease. Nat Cell Biol. 2011;13:568–579. doi: 10.1038/ncb2217. [DOI] [PubMed] [Google Scholar]
- 18.Escalante R. Dictyostelium as a model for human disease. Semin Cell Dev Biol. 2011;22:69. doi: 10.1016/j.semcdb.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Mueller-Taubenberger A, Kortholt A, Eichinger L. Simple system - substantial share: the use of Dictyostelium in cell biology and molecular medicine. Eur J Cell Biol. 2013;92:45–53. doi: 10.1016/j.ejcb.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Schaap P. Evolutionary crossroads in developmental biology: Dictyostelium discoideum . Development. 2011;138:387–396. doi: 10.1242/dev.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weijer CJ, Duschl G, David CN. A revision of the Dictyostelium discoideum cell cycle. J Cell Sci. 1984;70:111–131. doi: 10.1242/jcs.70.1.111. [DOI] [PubMed] [Google Scholar]
- 22.Zada-Hames IM, Ashworth JM. The cell cycle and its relationship to development in Dictyostelium discoideum . Dev Biol. 1978;63:307–320. doi: 10.1016/0012-1606(78)90136-7. [DOI] [PubMed] [Google Scholar]
- 23.Chen G, Shaulsky G, Kuspa A. Tissue-specific G1-phase cell-cycle arrest prior to terminal differentiation in Dictyostelium . Development. 2004;131:2619–2630. doi: 10.1242/dev.01151. [DOI] [PubMed] [Google Scholar]
- 24.Zimmerman W, Weijer CJ. Analysis of cell cycle progression during the development of Dictyostelium to differentiation. Dev Biol. 1993;160:178–185. doi: 10.1006/dbio.1993.1296. [DOI] [PubMed] [Google Scholar]
- 25.Weijer CJ, Duschl G, David CN. Dependence of cell-type proportioning and sorting on cell cycle phase in Dictyostelium discoideum . J Cell Sci. 1984;70:133–145. doi: 10.1242/jcs.70.1.133. [DOI] [PubMed] [Google Scholar]
- 26.McDonald SA, Durston AJ. The cell cycle and sorting behaviour in Dictyostelium discoideum . J Cell Sci. 1984;66:195–204. doi: 10.1242/jcs.66.1.195. [DOI] [PubMed] [Google Scholar]
- 27.Ohmori R, Maeda Y. The developmental fate of Dictyostelium discoideum cells depends greatly on the cell-cycle position at the onset of starvation. Cell Differ. 1987;22:11–18. doi: 10.1016/0045-6039(87)90409-X. [DOI] [PubMed] [Google Scholar]
- 28.Gomer RH, Firtel RA. Cell-autonomous determination of cell-type choice in Dictyostelium development by cell-cycle phase. Science. 1987;237:758–762. doi: 10.1126/science.3039657. [DOI] [PubMed] [Google Scholar]
- 29.Weeks G, Weijer CJ. The Dictyostelium cell cycle and its relationship to differentiation. FEMS Microbiol Lett. 1994;124:123–130. doi: 10.1111/j.1574-6968.1994.tb07274.x. [DOI] [PubMed] [Google Scholar]
- 30.Chattwood A, Thompson CR. Non-genetic heterogeneity and cell fate choice in Dictyostelium discoideum . Develop Growth Differ. 2011;53:558–566. doi: 10.1111/j.1440-169X.2011.01270.x. [DOI] [PubMed] [Google Scholar]
- 31.Jang W, Gomer RH. Initial cell type choice in Dictyostelium . Eukaryot Cell. 2011;10:150–155. doi: 10.1128/EC.00219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen G, Kuspa A. Prespore cell fate bias in G1 phase of the cell cycle in Dictyostelium discoideum . Eukaryot Cell. 2005;4:1755–1764. doi: 10.1128/EC.4.10.1755-1764.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacWilliams H, Doquang K, Pedrola R, Dollman G, Grassi D, Peis T, Tsang A, Ceccarelli A. A retinoblastoma ortholog controls stalk/spore preference in Dictyostelium. Development. 2006;133:1287–1297. doi: 10.1242/dev.02287. [DOI] [PubMed] [Google Scholar]
- 34.Muramoto T, Chubb JR. Live imaging of the Dictyostelium cell cycle reveals widespread S phase during development, a G2 bias in spore differentiation and a premitotic checkpoint. Development. 2008;135:1647–1657. doi: 10.1242/dev.020115. [DOI] [PubMed] [Google Scholar]
- 35.Shaulsky G, Loomis WF. Mitochondrial DNA replication but no nuclear DNA replication during development of Dictyostelium . Proc Natl Acad Sci USA. 1995;92:5660–5663. doi: 10.1073/pnas.92.12.5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo Q, Michaelis C, Weeks G. Cyclin B and cdc2 expression and cd2 kinase activity during Dictyostelium differentiation. DNA Cell Biol. 1995;14:901–908. doi: 10.1089/dna.1995.14.901. [DOI] [PubMed] [Google Scholar]
- 37.Michaelis C, Weeks G. Isolation and characterization of a cdc2 cDNA from Dictyostelium discoideum . Biochim Biophys Acta. 1992;1132:35–42. doi: 10.1016/0167-4781(92)90049-6. [DOI] [PubMed] [Google Scholar]
- 38.Rot G, Parikh A, Curk T, Kuspa A, Shaulsky G, Zupan B. dictyExpress: a Dictyostelium discoideum gene expression database with an explorative data analysis web-based interface. BMC Bioinformatics. 2009;10:256. doi: 10.1186/1471-2105-10-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma SK, Michaelis C, Lee K-Y, Wang J-H, Weeks G. Binding and catalytic properties of the cdc2 and crp proteins of Dictyostelium . Eur J Biochem. 1999;260:603–608. doi: 10.1046/j.1432-1327.1999.00236.x. [DOI] [PubMed] [Google Scholar]
- 40.Luo Q, Michaelis C, Weeks G. Overexpression of a truncated cyclin B gene arrests Dictyostelium cell division during mitosis. J Cell Sci. 1994;107:3105–3114. doi: 10.1242/jcs.107.11.3105. [DOI] [PubMed] [Google Scholar]
- 41.Murray AW, Solomon MJ, Kirschner MW. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989;339:280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- 42.Sharma SK, Brock DA, Ammann RR, DeShazo T, Khosla M, Gomer RH, Weeks G. The Cdk5 homologue, Crp, regulates endocytosis and secretion in Dictyostelium and is necessary for optimum growth and differentiation. Dev Biol. 2002;247:1–10. doi: 10.1006/dbio.2002.0684. [DOI] [PubMed] [Google Scholar]
- 43.Huber RJ, O’Day DH. Nucleocytoplasmic transfer of Cdk5 and its binding to puromycin-sensitive aminopeptidase in Dictyostelium discoideum . Histochem Cell Biol. 2011;136:177–189. doi: 10.1007/s00418-011-0839-6. [DOI] [PubMed] [Google Scholar]
- 44.Huber RJ, O’Day DH. The cyclin-dependent kinase inhibitor roscovitine inhibits kinase activity, cell proliferation, multicellular development, and Cdk5 nuclear translocation in Dictyostelium discoideum . J Cell Biochem. 2012;113:868–876. doi: 10.1002/jcb.23417. [DOI] [PubMed] [Google Scholar]
- 45.Michaelis C, Weeks G. The isolation from a unicellular organism, Dictyostelium discoideum, of a highly-related cdc2 gene with characteristics of the PCTAIRE subfamily. Biochim Biophys Acta. 1993;1179:117–124. doi: 10.1016/0167-4889(93)90132-9. [DOI] [PubMed] [Google Scholar]
- 46.Nikolic M, Dudek H, Kwon YT, Ramos YF, Tsai L-H. The cdk5/p35 kinase is essential for neurite outgrowth during neuronal differentiation. Genes Dev. 1996;10:816–825. doi: 10.1101/gad.10.7.816. [DOI] [PubMed] [Google Scholar]
- 47.Yang Q, Mao Z. Regulation and function of Cdk5 in the nucleus. In: Tsai L-H, Ip NY, editors. Cyclin-dependent kinase 5 (Cdk5) New York: Springer; 2008. pp. 107–118. [Google Scholar]
- 48.O’Hare MJ, Kushwaha N, Zhang Y, Aleyasin H, Callaghan SM, Slack RS, Albert PR, Vincent I, Park DS. Differential roles of nuclear and cytoplasmic cyclin-dependent kinase 5 in apoptotic and excitotoxic neuronal death. J Neurosci. 2005;25:8954–8966. doi: 10.1523/JNEUROSCI.2899-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Cicero SA, Wang L, Romito-DiGiacomo RR, Yang Y, Herrup K. Nuclear localization of Cdk5 is a key determinant in the postmitotic state of neurons. Proc Natl Acad Sci USA. 2008;105:8772–8777. doi: 10.1073/pnas.0711355105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Humbert S, Dhavan R, Tsai L. p39 activates cdk5 in neurons, and is associated with the actin cytoskeleton. J Cell Sci. 2000;113:975–983. doi: 10.1242/jcs.113.6.975. [DOI] [PubMed] [Google Scholar]
- 51.Veeranna GJ, Shetty KT, Takahashi M, Grant P, Pant HC. Cdk5 and MAPK are associated with complexes of cytoskeletal proteins in rat brain. Mol Brain Res. 2000;76:229–236. doi: 10.1016/S0169-328X(00)00003-6. [DOI] [PubMed] [Google Scholar]
- 52.He L, Zhang Z, Yu Y, Ahmed S, Cheung NS, Qi RZ. The neuronal p35 activator of Cdk5 is a novel F-actin binding and bundling protein. Cell Mol Life Sci. 2011;68:1633–1643. doi: 10.1007/s00018-010-0562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor A. Aminopeptidases: structure and function. FASEB J. 1993;7:290–298. doi: 10.1096/fasebj.7.2.8440407. [DOI] [PubMed] [Google Scholar]
- 54.Catalano A, Poloz Y, O’Day DH. Dictyostelium puromycin-sensitive aminopeptidase A is a nucleoplasmic nucleomorphin-binding protein that relocates to the cytoplasm during mitosis. Histochem Cell Biol. 2011;136:677–688. doi: 10.1007/s00418-011-0873-4. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi S, Ohishi Y, Kato H, Noguchi T, Naito H, Aoyagi T, Umezawa H. The effects of bestatin, a microbial aminopeptidase inhibitor, on epidermal growth factor induced DNA synthesis and cell division in primary cultured hepatocytes of rats. Exp Cell Res. 1989;183:399–412. doi: 10.1016/0014-4827(89)90400-X. [DOI] [PubMed] [Google Scholar]
- 56.Poloz Y, Catalano A, O’Day DH. Bestatin inhibits cell growth, cell division, and spore cell differentiation in Dictyostelium discoideum . Eukaryot Cell. 2012;11:545–557. doi: 10.1128/EC.05311-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koepp DM, Harper WJ, Elledge SJ. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell. 1999;97:431–434. doi: 10.1016/S0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- 58.Hersh LB. Inhibition of aminopeptidase and acetylcholinesterase by puromycin and puromycin analogs. J Neurochem. 1981;36:1594–1596. doi: 10.1111/j.1471-4159.1981.tb00605.x. [DOI] [PubMed] [Google Scholar]
- 59.Constam DB, Tobler AR, Rensing-Ehl A, Kemler I, Hersh LB, Fontana A. Puromycin-sensitive aminopeptidase: sequence analysis, expression, and functional characterization. J Biol Chem. 1995;270:26931–26939. doi: 10.1074/jbc.270.45.26931. [DOI] [PubMed] [Google Scholar]
- 60.Huber RJ, Catalano A, O’Day DH. Cyclin-dependent kinase 5 is a calmodulin-binding protein that associates with puromycin-sensitive aminopeptidase in the nucleus of Dictyostelium . Biochim Biophys Acta. 2013;1833:11–20. doi: 10.1016/j.bbamcr.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 61.Chin D, Means AR. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. doi: 10.1016/S0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 62.O’Day DH. CaMBOT: profiling and characterizing calmodulin binding proteins. Cell Signal. 2003;15:347–355. doi: 10.1016/S0898-6568(02)00116-X. [DOI] [PubMed] [Google Scholar]
- 63.Shen X, Valencia CA, Szostak JW, Dong B, Liu R. Scanning the human proteome for calmodulin-binding proteins. Proc Natl Acad Sci USA. 2005;102:5969–5974. doi: 10.1073/pnas.0407928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He L, Hou Z, Qi RZ. Calmodulin binding and cdk5 phosphorylation of p35 regulate its effect on microtubules. J Biol Chem. 2008;283:13252–13260. doi: 10.1074/jbc.M706937200. [DOI] [PubMed] [Google Scholar]
- 65.Takuwa N, Zhou W, Takuwa Y. Calcium, calmodulin and cell cycle progression. Cell Signal. 1995;7:93–104. doi: 10.1016/0898-6568(94)00074-L. [DOI] [PubMed] [Google Scholar]
- 66.Kahl CR, Means AR. Regulation of cell cycle progression by calcium/calmodulin-dependent pathways. Endocr Rev. 2003;24:719–736. doi: 10.1210/er.2003-0008. [DOI] [PubMed] [Google Scholar]
- 67.Liu T, Williams JG, Clarke M. Inducible expression of calmodulin antisense RNA in Dictyostelium cells inhibits the completion of cytokinesis. Mol Biol Cell. 1992;3:1403–1413. doi: 10.1091/mbc.3.12.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu Q, Liu T, Clarke M. Calmodulin and the contractile vacuole complex in mitotic cells of Dictyostelium discoideum . J Cell Sci. 1993;104:1119–1127. doi: 10.1242/jcs.104.4.1119. [DOI] [PubMed] [Google Scholar]
- 69.Meijer L, Borgne A, Mulner O, Chong JP, Blow JJ, Inagaki N, Inagaki M, Delcros JG, Moulinoux JP. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases Cdc2, Cdk2 and Cdk5. Eur J Biochem. 1997;243:527–536. doi: 10.1111/j.1432-1033.1997.t01-2-00527.x. [DOI] [PubMed] [Google Scholar]
- 70.Goodyear S, Sharma MC. Roscovitine regulates invasive breast cancer cell (MDA-MB231) proliferation and survival through cell cycle regulatory protein Cdk5. Exp Mol Pathol. 2007;82:25–32. doi: 10.1016/j.yexmp.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 71.Liebl J, Weitensteiner SB, Vereb G, Takács L, Fürst R, Vollmar AM, Zahler S. Cyclin-dependent kinase 5 regulates endothelial cell migration and angiogenesis. J Biol Chem. 2010;285:35932–35943. doi: 10.1074/jbc.M110.126177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jain P, Flaherty PT, Yi S, Chopra I, Bleasdell G, Lipay J, Ferandin Y, Meijer L, Madura JD. Design, synthesis, and testing of an 6-O-linked series of benzimidazole based inhibitors of CDK5/p25. Bioorg Med Chem. 2011;19:359–373. doi: 10.1016/j.bmc.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 73.Bach S, Knockaert M, Reinhardt J, Lozach O, Schmitt S, Baratte B, Koken M, Coburn SP, Tang L, Jiang T, Liang DC, Galons H, Dierick JF, Pinna LA, Meggio F, Totzke F, Schächtele C, Lerman AS, Carnero A, Wan Y, Gray N, Meijer L. Roscovitine targets, protein kinases and pyridoxal kinase. J Biol Chem. 2005;280:31208–31219. doi: 10.1074/jbc.M500806200. [DOI] [PubMed] [Google Scholar]
- 74.Pinhero R, Liaw P, Yankulov K. A uniform procedure for the purification of CDK7/CycH/MAT1, CDK8/CycC and CDK9/CycT1. Biol Proced Online. 2004;6:163–172. doi: 10.1251/bpo86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goh BC, Peh B, Cui C, Soo R, Loh T, Green S, Hsieh W, Lai Y, Salto-Tellez M, Mow B (2005) Seliciclib (R-roscovitine) induces apoptosis in undifferentiated nasopharyngeal cancer (NPC) in vivo and in vitro. In: J Clin Oncol, 2005 ASCO annual meeting proceedings, Vol 23, No. 16S, Part I of II (June 1 Supplement):3145
- 76.Diwan P, Lacasse JJ, Schang LM. Roscovitine inhibits activation of promoters in herpes simplex virus type 1 genomes independently of promoter-specific factors. J Virol. 2004;78:9352–9365. doi: 10.1128/JVI.78.17.9352-9365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pumfery A, de la Fuente C, Berro R, Nekhai S, Kashanchi F, Chao SH. Potential use of pharmacological cyclin-dependent kinase inhibitors as anti-HIV therapeutics. Curr Pharm Des. 2006;12:1949–1961. doi: 10.2174/138161206777442083. [DOI] [PubMed] [Google Scholar]
- 78.Węsierska-Gądek J, Gritsch D, Zulehner N, Komina O, Maurer M. Roscovitine, a selective CDK inhibitor, reduces the basal and estrogen-induced phosphorylation of ER-α in human ER-positive breast cancer cells. J Cell Biochem. 2011;112:761–772. doi: 10.1002/jcb.23004. [DOI] [PubMed] [Google Scholar]
- 79.Rossi AG, Sawatzky DA, Walker A, Ward C, Sheldrake TA, Riley NA, Caldicott A, Martinez-Losa M, Walker TR, Duffin R, Gray M, Crescenzi E, Martin MC, Brady HJ, Savill JS, Dransfield I, Haslett C. Cyclin-dependent kinase inhibitors enhance the resolution of inflammation by promoting inflammatory cell apoptosis. Nat Med. 2006;12:1056–1064. doi: 10.1038/nm1468. [DOI] [PubMed] [Google Scholar]
- 80.Li BS, Sun MK, Zhang L, Takahashi S, Ma W, Vinade L, Kulkarni AB, Brady RO, Pant HC. Regulation of NMDA receptors by cyclin-dependent kinase-5. Proc Natl Acad Sci USA. 2001;98:12742–12747. doi: 10.1073/pnas.211428098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang CH, Lee TH, Tsai YJ, Liu JK, Chen YJ, Yang LC, Lu CY. Intrathecal Cdk5 inhibitor, roscovitine, attenuates morphine antinociceptive tolerance in rats. Acta Pharmacol Sin. 2004;25:1027–1030. [PubMed] [Google Scholar]
- 82.Sahlgren CM, Mikhailov A, Vaittinen S, Pallari H-M, Kalimo H, Pant HC, Eriksson JE. Cdk5 regulates the organization of nestin and its association with p35. Mol Cell Biol. 2003;23:5090–5106. doi: 10.1128/MCB.23.14.5090-5106.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Araki T, Maeda Y. Cell cycle progression during the development of Dictyostelium discoideum and its relation to the subsequent cell-sorting in the multicellular structures. Dev Growth Differ. 1995;37:479–485. doi: 10.1046/j.1440-169X.1995.t01-4-00002.x. [DOI] [PubMed] [Google Scholar]
- 84.Takeda K, Saito T, Ochiai H. A novel Dictyostelium Cdk8 is required for aggregation, but is dispensable for growth. Dev Growth Differ. 2002;44:213–223. doi: 10.1046/j.1440-169X.2002.00636.x. [DOI] [PubMed] [Google Scholar]
- 85.Lin HH, Khosla M, Huang HJ, Hsu DW, Michaelis C, Weeks G, Pears C. A homologue of Cdk8 is required for spore cell differentiation in Dictyostelium . Dev Biol. 2004;271:49–58. doi: 10.1016/j.ydbio.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 86.Greene DM, Bloomfield G, Skelton J, Ivens A, Pears CJ. Targets downstream of Cdk8 in Dictyostelium development. BMC Dev Biol. 2011;11:2. doi: 10.1186/1471-213X-11-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Greene DM, Hsu D-W, Pears CJ. Control of cyclin C levels during development of Dictyostelium . PLoS One. 2010;5:e10543. doi: 10.1371/journal.pone.0010543. [DOI] [PMC free article] [PubMed] [Google Scholar]