Abstract
In addition to their critical roles in embryonic development, cell fate decision, and differentiation, members of Sox (Sry-related high-mobility group box) family of transcription factors including Sox4 have been implicated in various cancers. Multiple studies have revealed an increased expression along with specific oncogenic function of Sox4 in tumors, while others observed a reduced expression of Sox4 in different types of malignancies and suppression of tumor initiation or progression by this protein. More interestingly, the prognostic value of Sox4 is debated due to obvious differences between various reports as well as inconsistencies within specific studies. This review summarizes our current understanding of Sox4 expression pattern and its transcription-dependent, as well as transcription-independent, functions in tumor initiation or progression and its correlation with patient survival. We also discuss the existing discrepancies between different reports and their possible explanations.
Keywords: Cancer, Sox4, Oncogene, Tumor suppressor
Introduction
Sox family of transcription factors
The Sox (SRY-related HMG box) gene family is found throughout the animal kingdom. In humans, at least 20 members of this family have so far been identified [1], showing a diverse and dynamic pattern of expression throughout embryogenesis and in a variety of adult tissue types [2]. Sry (for sex-determining region Y), the founding member of this family, was identified through searches for conserved sequences among translocated Y chromosomal DNA from XX male patients [3], and was later confirmed to be involved in male sex differentiation [4].
All Sox proteins are characterized by possession of a high mobility group (HMG) DNA-binding domain. The 79-amino-acid HMG domains bind to the consensus target sequence (A/T)ACAA(T/A) in the minor grooves of DNA [2] and modify the chromatin structure to generate a conformation that facilitates various DNA-dependent activities. This domain is shared with other DNA binding proteins, including those that bind DNA without sequence specificity, such as HMG-1 protein and ubiquitous binding factor (UBF), as well as several sequence-specific DNA-binding proteins, such as the T cell-specific factors TCF/LEF [5]. The DNA-binding domains of Sox proteins are at least 60 % similar or 50 % identical to the HMG box domain of SRY [6]. The nomenclature of this family is based on the order of gene discovery [7]. Nine of the 20 human Sox genes contain a single exon, likely reflecting the mechanism of expansion of this ancient gene family via non-tandem duplication and retroposition [7]. This family is sub-grouped into six distinct classes (A–F), based on homology within the HMG domain and other structural motifs as well as functional properties. These classes include: A, Sry; B, Sox1, -2, -3, -14, -15, and -19; C, Sox4, -11, -12, and -20; D, Sox5, -6, and -13; E, Sox8, -9, and -10; F, Sox7, -17, and -18 [6].
Despite this common feature, each Sox protein selectively interacts with and regulates a unique set of target genes [8]. It appears that Sox proteins are able to bind to a large number of transcription factors [9] and on many occasions cooperate with them to exert their regulatory function. For instance, Sox6 was shown to suppress cyclin-D1 promoter activity by interacting with β-catenin and HDAC1 in pancreatic β-cells [10]. Recruitment of HDAC1 to the promoter regions of the target genes such as DCT and MITF by Sox5 and suppression of their expression was also observed in melanocytic cells [11]. On the other hand, Sox10 physically interacts with MEF2C and cooperatively activates the promoter of the Mef2c gene [12].
Since the discovery of SRY, many other Sox family members have also been implicated in the regulation of critical functions in various developmental processes, such as sex differentiation, neurogenesis, skeletogenesis, hematopoiesis, angiogenesis, cardiogenesis, melanogenesis, and hair development. Interested readers are referred to recent reviews on this topic [13–15].
Sox4
The human Sox4 gene was first identified based on homology with its mouse homologue [5]. The location of Sox4 was determined by metaphase fluorescence in situ hybridization (FISH) at chromosome 6p.23 [5]. Sox4 contains a single exon and its open reading frame (ORF) encodes a protein of 474 amino acids with a molecular weight of 47 kD [6]. The encoded protein is particularly rich in serine residues (18 % overall) with several poly-serine stretches. It also includes multiple stretches of glycine and alanine residues [5].
Sox4 belongs to the class C (SoxC), which contains two other members; Sox11 and Sox12 [1]. In 2- to 3-day-old mice, SoxC genes are expressed at high levels in the brain. Sox4 and Sox12 are also expressed at a high level in the heart and lung. While Sox11 is expressed in developing limbs, face, and kidneys of the mouse embryo, it is only detectable in considerable amounts in neuronal tissues [16] and is not expressed in detectable amounts in tissues of adult mouse, perhaps due to a general decrease in Sox11 expression during late embryogenesis [17].
All SoxC proteins have a high degree of similarity in the HMG domain. The HMG box is 84 % identical and 95 % similar among SoxC proteins across all vertebrates, whereas their TADs share 67 % identity and 94 % similarity [16]. The structure of the sequence-specific HMG domain of Sox4 provided some insight into its mode of function [18]. This domain has an L-shaped structure consisting of three α-helices connected by loop regions, which are stabilized by a highly conserved cluster of mainly aromatic residues. These hydrophobic cores are mostly conserved within the HMG box family. Helices I and II are positioned in an anti-parallel mode and form one arm of the HMG box, while helix III, which is less rigid, forms an average angle of 90° with the other two helices and constitutes the other arm of the molecule [18]. The N-terminus of the HMG box interacts with the first 6 base pairs of the target sequence, and binding of the HMG box to the minor groove of a straight DNA helix in this manner introduces a sharp bend (on the order of 90°) in the DNA helix and alters the local chromatin conformation [18], which is crucial for its transcriptional activity.
Another distinctive feature of the SoxC class is a conserved proline, serine, and acidic residues-rich transactivation domain (TAD) at the C-terminal [16]. Protein secondary structure modeling predicted that 20 residues in the Sox4 C-terminal domain form an α-helix which is interrupted in the middle of its sequence by three randomly coiled residues, and have the last two residues in extended conformation rather than in helical conformation [16]. This domain is crucial and sufficient for the activity of SoxC proteins and its deletion completely abrogates their transactivation capacity [16]. The Sox4 TAD shows stronger transactivating capacity than Sox12 but weaker than that of Sox11 [16, 17] due to the more stable α-helix structure of Sox11 TAD [16]. The transactivating function of Sox4 TAD is independent of its HMG domain which is responsible for DNA binding, as evident from its sustained activity upon grafting onto a GAL4 DNA-binding domain [19]. SoxC proteins seem to compete with each other to bind to their target sequences, since the activity of any full-length SoxC protein is inhibited by expressing equivalent amounts of Sox4, -11 or -12 proteins lacking the TAD [16]. The exact mechanism by which TAD regulates transcriptional activation of target genes is not fully understood. One possible mechanism is that it is achieved through direct interaction with other transcription factors such as p53 [20] and β-catenin/TCF4 complex [21]. In melanoma cells, Sox4 TAD also interacts with syntenin resulting in prevention of proteasomal degradation of Sox4, therefore enhancing Sox4 stability [22].
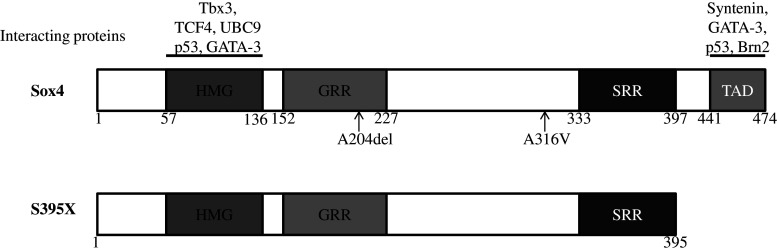
In addition to HMG (aa 57–136) and TAD (aa 441–474) domains, Sox4 also contains a glycine-rich region (aa 152–227) at the center and a serine-rich region (SRR, aa 333–397) adjacent to the TAD (Fig. 1). A study of HEK293 cells revealed that the central domain containing a glycine rich region has pro-apoptotic activity which is independent of the transcriptional activity of Sox4 [23].
Fig. 1.
Structure of the human Sox4 protein, its binding partners, mutations and truncated isoform (S395X) detected in human lung cancer. GRR glycine-rich region, SRR serine-rich region, TAD transactivation domain
Sox4 in development; jack of many trades
Similar to many other members of the family, the SoxC class has also been identified as a necessary factor in embryonic development. In developing mice, Sox4, -11, and -12 are co-expressed at high levels in neuronal and mesenchymal tissues [16]. The absence of overt phenotype in Sox12−/− mice suggests that at least some functions of Sox4, -11, and -12 are redundant and Sox4 and -11 may compensate the loss of Sox12 during mouse development [24]. Consistently, Bhattaram and colleagues showed that concomitant loss of SoxC genes confers a worsened phenotype in mouse embryo than loss of each single gene [25]. Indeed, the more SoxC genes that are deleted, the more severe and widespread is organ hypoplasia [25]. SoxC proteins control neural and mesenchymal cell survival as shown by increased cell death in the neural tube, branchial arches, and somites of the Sox4 −/− 11 −/− embryos. The pro-survival role of SoxC proteins in these tissues is probably through direct activation of Tead2, a transcriptional mediator of the Hippo signalling pathway which can promote cell survival during organogenesis [25]. However, not all functions of the SoxC proteins seem to be redundant. Accordingly, Sox4 knockout mice die halfway through gestation (E14) due to severe heart defect as a result of impaired development of the endocardial ridges into the semilunar valves and the outlet portion of the muscular ventricular septum [26] which indicates that Sox11 or -12 cannot compensate for the lack of Sox4 expression in this tissue. The critical role of Sox4 in development of cardiac tissues may at least partially be due to its role in regulation of expression of the gap junction protein, connexin 43 (Cx43), in cooperation with the Tbx3 transcription factor [27].
Sox11 −/− mice die at birth from ventricular septation defects and outflow tract malformations resulting in heart defects, in addition to malformation in other organs such as skeleton, lung, stomach, and pancreas [28]. The fact that Sox4 knockout embryos die at E14 and the Sox11 −/− mice complete the fetal stage indicates that Sox4 has more important roles in early and Sox11 in late embryogenesis. Interestingly, SoxC-null embryos will arrest in development at E8.5 [25], earlier than the embryos lacking each individual SoxC member, further confirming that these genes have partial overlap in their function which results in exaggerated phenotype upon further loss of each gene.
Sox4 is expressed in the thymus, bone marrow, and gonads of adult mice [29], which highlights its possible function(s) in hematopoiesis. Early studies showed that Sox4 is highly expressed in thymocytes and induces their differentiation while promoting pro-B lymphocyte expansion [29, 30]. Study of hemopoiesis in lethally irradiated mice reconstituted with Sox4 −/− fetal liver cells demonstrated that the absence of Sox4 specifically blocks development of B cells at the pro-B cell stage [26]. Consistently, Sox4-null fetal liver progenitors give rise to all hemopoietic lineages except the B-cell [29]. The pro-B cells in these mice are moderately reduced in number, but their capacity to proliferate in response to IL-7 is strongly abrogated [29]. Sox4 regulates expression of the λ5 and VpreB1 genes which form part of the surrogate light chain and are expressed in pro- and pre-B cells. λ5 and VpreB1 expression is initially regulated by Sox2 in the pluripotent cells of the inner cell mass (ICM) of the developing mouse blastocyst, which is then replaced by Sox4 as the cells differentiate toward a very early progenitor for the hematopoietic and endothelial lineages, the hemangioblasts [31].
The absence of Sox4 expression may also have some subtle effects on thymocyte development. In fact, thymi from Sox4-null embryos contain two- to four-fold fewer cells than thymi from their wild-type or heterozygous counterparts [29]. Furthermore, maturation in fetal thymic organ culture of mutant thymi is impaired, indicating that absence of Sox4 blocks development of T cells from their progenitors [29]. A recent study by Kuwahara and colleagues [32] revealed that Sox4 inhibits the function of GATA-3, the master regulator of T helper type 2 (Th2) cells, therefore suppressing their differentiation and Th2 cell-mediated inflammation. In these cells, Sox4 acts as a downstream factor of TGF-β and upon activation directly binds to GATA-3 and prevents its binding to GATA-3 consensus DNA sequences. In addition, Sox4 binds to the promoter region of the interleukin 5 (IL-5) gene and prevents binding of GATA-3 to this promoter, thereby suppressing its expression [32]. It had been previously observed that, during hematopoiesis, the IL-5 receptor activates Sox4 through syntenin-1 [33], perhaps by suppressing proteasomal degradation of Sox4 protein [22]. It is not clear whether these observations, which were made in different cell types, could indicate the possible existence of a negative feedback loop between Sox4, IL-5, and IL-5R or could simply represent different modes of interaction between these factors in different cell types.
Sox4 is important for the development of the central nervous system. In fact, expression of Sox4 and Sox11 is critical for the establishment of pan-neuronal protein expression [34]. These proteins act as transcriptional activators downstream of proneural basic helix-loop-helix (bHLH) proteins to exert this function. Sox4 and Sox11 exhibit highly overlapping expression in the mouse hippocampal neurogenic lineage, and overexpression of either suffices to induce neuronal marker expression in adult neural stem cells. Consistently, loss of Sox4/Sox11 expression results in loss of expression of neuron-specific protein DCX, a microtubule-associated protein [35]. Sox4 and Sox11 are also critical for specification of corticospinal neurons partially due to the important role of their downstream transcription factor Fezf2 in this process [36]. Interestingly, this function of Sox4 and Sox11 is mediated by direct competition with Sox5 which acts as a repressor of Fezf2 expression [36]. It is also suggested that Sox4 expression in oligodendrocyte precursor cells may induce premature differentiation of these cells. In fact, prolonged expression of Sox4 in vivo reduces myelin gene expression in oligodendrocyte linage which consequently interferes with normal myelination in the central nervous system [37]. Interestingly, expression of Sox4 and Sox11 in neural progenitor cells is restricted by REST/NRSF [34]. REST/NRSF is a transcriptional repressor which restrains the neurogenic program in progenitor cells [38], and repression of Sox4 and Sox11 by this protein may contribute to the mechanism by which REST/NRSF prevents expression of neuronal proteins in these cells.
Other than the nervous system, Sox4 has a number of important roles in other developmental processes. During mouse development, Sox4 expression is more confined to the pancreatic epithelium and later islet cells along with Sox9 expression [39]. Sox4b (an isoform of the zebrafish homologue of Sox4) is also a key player in pancreatic alpha cell differentiation [40]. In adult mice, Sox4 is broadly expressed in the early pancreatic buds and in the nuclei of all islet cells [41]. Homozygous deletion of Sox4 did not show any abnormalities in pancreas development up to embryonic day 12.5; however, a significantly reduced number of endocrine cells were found to be scattered through the culture site, causing failure to form normal islets [41]. This phenomenon significantly affects normal pancreatic function, as it has been proven that Sox4 deletion in adult mouse results in a considerable defect in insulin secretion and leads to impaired glucose tolerance [42]. The other important role of Sox4 in the developmental process was described in the musculoskeletal system. Indeed, 3-month-old Sox4 +/− mice have a 64 % lower bone formation rate compared to wild-type, probably due to significant defects in osteoblast proliferation, differentiation, and mineralization, which eventually lead to lower bone mass and to reduced trabecular and cortical thickness and growth plate in these mice [43].
The SoxC proteins, including Sox4, are also required for the survival of the multipotent neural and mesenchymal stem cells. These cells give rise to various differentiated cell types and are crucial for organogenesis and embryo growth [25]. Importantly, Sox4 is required for maintenance of the stemness of the glioma initiating cells (GICs) through integration of the Oct4/Sox2 axis [44]. The Oct4/Sox2 axis is crucial for the continuous self-renewal and developmental potential of normal stem cells, specially the embryonic stem cells [45]. However, it is not clear whether the Sox4-mediated regulation of stemness is a general phenomenon or is limited to the GICs and multipotent neural and mesenchymal cells.
Regulation of Sox4 expression
Regulation of Sox4 expression, especially in the pathological contexts, is a field that has remained largely unknown and requires further investigation. Initially, Clarke and colleagues [46] reported that Sox4 is a progesterone-regulated gene in breast cancer cells and that its expression is induced by progestins. Consistently, treatment of T-47D breast cancer cells with the synthetic progestin ORG-2058 increased Sox4 transcription within few hours of treatment. Notably, ORG-2058 had no detectable effect on the expression of other Sox genes, suggesting that in this system the observed phenomenon was specific to Sox4 [46]. In osteoblast-like cells, physiological concentrations of human parathyroid hormone stimulate Sox4 mRNA expression in a time-dependent manner, indicating involvement of the PTH/PTHrP receptor [47]. Prostaglandin A2 and delta12-PGJ2 also induce Sox4 mRNA expression in hepatocarcinoma cells [48]. Del Giacco and colleagues found that IFN-beta/all-trans retinoic acid treatment induced the expression of Sox4 in a thioredoxin reductase 1 (TrxR1) enzyme-dependent manner in hepatocarcinoma cells [49]. However, in none of these observations is it clear whether these hormones affect the activity of the Sox4 promoter or whether they exert their function at other levels (e.g., mRNA stability).
Nonetheless, other studies have revealed details on the regulation of the Sox4 promoter. In hematopoietic progenitor cells, Sox4 expression is regulated at transcription level by the HOXB4 transcription factor [50]. The HOX gene family members are important regulators of hematopoiesis. In T cells, expression of Sox4 is upregulated at both mRNA and protein levels by TGF-β. This regulation is mediated by downstream mediators of TGF-β, Smad2 and Smad3, which bind to the regulatory region of the Sox4 gene [32]. Overexpression of the Sox4 transcript upon binding of the Smad2/3 complex to its promoter after TGF-β treatment has also been observed in glioma cell lines [51]. On the other hand, inhibitors of differentiation 2 (ID2) and REST/NRSF repress expression of Sox4 in neural progenitor cells [34]. Since REST/NRSF is a transcriptional repressor, it is pertinent to assume that it could suppress Sox4 promoter activity, although this assumption requires further experimental verification. The retinoic acid receptor-related orphan receptor α (RORα) is also suggested to upregulate Sox4 transcription [52]. RORα plays a critical role in regulation of the circadian clock and enhances Sox4 transcription by direct binding to the promoter sequences after activation by its agonist [52].
Sox4 is transcriptionally regulated by Sox7 in endometrial cancer cells [53]. Accordingly, overexpression of Sox7 activates the Sox4 promoter, leading to the increased expression at both mRNA and protein levels, seemingly due to the presence of Sox7-responsive elements in the promoter region of Sox4 gene. Interestingly, Sox7 inhibits the activity of its own promoter [53], further confirming that the same Sox protein can simultaneously enhance and suppress the activity of different promoters in the same cell, consistent with the context-dependent manner of function for Sox proteins. Sox4 expression may further be influenced by other members of the SoxC class. In fact, whereas ablation of Sox4 in the sympathoadrenal lineage did not have any significant effect on expression of Sox11, knockdown of Sox11 markedly reduced Sox4 protein expression [54], although it is not clear whether or not this effect of Sox11 is exerted through direct binding to the Sox4 promoter. In addition, Sox4 may regulate its own expression as is evident by binding of the Sox4 protein to its promoter sequence in prostate cancer cells [55].
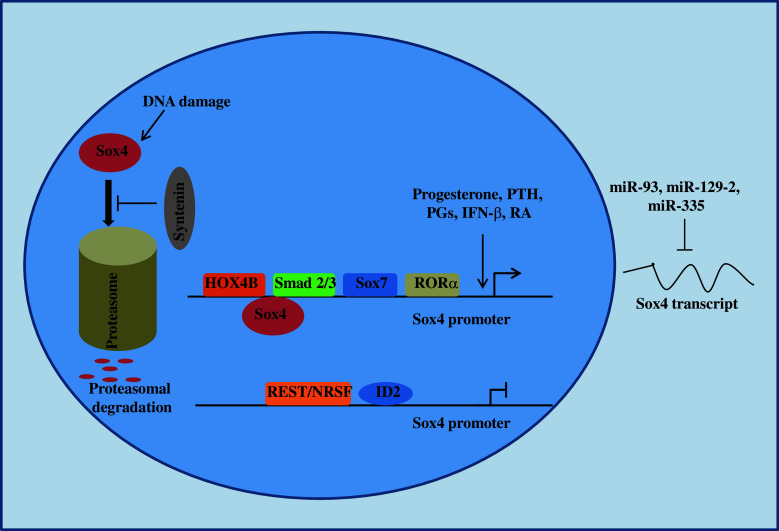
Other mechanisms also contribute to the regulation of Sox4 expression at post-transcriptional and post-translational levels, which are summarized in Fig. 2 and will be discussed in detail throughout this review.
Fig. 2.
Regulation of Sox4 expression. Expression of Sox4 is regulated at three distinct levels; transcriptional, post-transcriptional, and post-translational. PGs prostaglandin A2 and delta12-prostaglandin J2
Role of Sox4 in tumorigenesis
In retrospect, many genes that are involved in developmental processes have also been found to play key roles in the formation of malignancies. For instance, the hedgehog pathway was originally discovered in Drosophila, in which its members encode segment polarity proteins responsible for determining the anterior–posterior orientation of the Drosophila embryo and larva [56], and was later found to be implicated in tumorigenesis [57, 58]. Involvement of the HMG protein family in cancer was already known [59, 60] before the emergence of evidence that the developmentally critical Sox proteins may also play a role in some cancers. Several lines of evidence pointed to the fact that Sox proteins may be implicated in cancer. For instance, amplification of Sox2 at chromosome 3q has been detected in prostate cancer [61]. The chromosomal region 20q13, which contains Sox18 coding gene, is also amplified in some cases of breast [62] and colon cancers [63]. Deregulated expression of other members of the Sox family in malignancies has also been demonstrated [64–66]. Keeping in mind that in many cases the expression status of Sox genes differs in various cancers, it would be implausible to assume a ubiquitous oncogenic or tumor suppressive function for any given Sox gene. This kind of inconsistency in the expression pattern and mode of function (oncogenic vs. tumor suppressive) in different types of cancer exists for several members of the Sox family, especially Sox4, which will be described below in more detail.
Oncogenic functions of Sox4
Several lines of evidence suggest an oncogenic role for Sox4 in a wide range of cancers. Some of the earliest indirect evidences highlighting a role for Sox4 in cancer came from studies on colon carcinoma by Miyamoto and colleagues [67]. In this study, Sox4 was found to enhance transcription of p56lck, a member of the family of Src tyrosine kinases that is aberrantly expressed in colon and small lung carcinoma cell lines. p56lck is also expressed in T cells and is among the first signaling molecules to be activated downstream of the T cell receptor and plays a significant role in T cell differentiation, survival, and activation [68]. In Jurkat and HeLa cells, expression of p56lck is controlled by Myb, in synergy with Ets factors [69]. However, in colon carcinoma, aberrant expression of p56lck arises from transcriptional activation mediated by cooperation between Ets-1 and Sox4 [67]. Amplification of chromosomal region 6p22.3 in at least some tumor types [70, 71] was another indirect evidence for a tumorigenic function of Sox4. Nevertheless, in some cases, there is no significant correlation between expression of Sox4 and amplification of 6p22.3 [73, 74], suggesting that amplification of Sox4 coding sequences might be a bystander effect and not a growth advantage for some cancer cells. This suggestion is in line with existence of important proto-oncogenes such as E2F3 [72], ID4 [73] and, PRL [74] in the 6p22.3 chromosomal region, which might provide the actual benefit to the cancer cells upon amplification. This notion is consistent with an independent observation in bladder cancers. In this cancer, 30.8 % of the patients had chromosomal amplification of this region. The mean E2F3 expression of amplified tumors was a 1.8-fold increase compared with not-amplified tumors, whereas no correlation was found between chromosomal amplification and Sox4 expression [75]. Nonetheless, the inconsistency between 6p amplification and Sox4 expression may not be a ubiquitous phenomenon. In fact, another study by Medina et al. [76] found an increased expression of Sox4 in lung primary tumors and lung cancer cell lines with chromosome 6p amplification. Interestingly, they also reported identification of a somatic mutation at the 395 residue of Sox4 that introduces a premature stop codon at the C-terminal domain, producing a shorter Sox4 protein, S395X, which is devoid of the region immediately after the serine-rich domain and does not possess transactivation potential (Fig. 1). While neither wild-type Sox4 nor its truncated isoform could induce oncogenic transformation of the NIH3T3 mouse fibroblasts, wild-type Sox4 enhances tumorigenicity of the NIH3T3 cells expressing the activated HRAS. On the other hand, the truncated isoform significantly reduced the tumorigenicity of the HRAS-expressing cells [76], which suggests that transcriptional activity of Sox4 supports the HRAS-transforming ability in lung cancer cells. However, it is not clear why cancer cells with a truncated Sox4 that could reduce the tumorigenicity would have a selective advantage.
Increased expression of Sox4 at mRNA or protein levels in several types of epithelial and hematopoietic cancers further confirmed its putative oncogenic function (Table 1). However, this observed correlation between Sox4 overexpression and cancer progression does not seem to always be consistent with patient survival outcome or even functional role of Sox4 in those cells. For instance, Sox4 protein is overexpressed in human hepatocellular carcinomas (HCC) [77], yet there is an inverse correlation between expression of Sox4 and recurrence of cancer as well as a positive correlation between Sox4 expression and overall patient survival which does not fit into a possible oncogenic role for Sox4 in this type of cancer. This study also found that the HMG box domain of Sox4 interacts with p53. This interaction, which happens at the p53-responsive promoters, results in inhibition of p53-mediated activation of Bax promoter, leading to repression of p53-induced Bax expression and subsequent repression of p53-mediated apoptosis induced by gamma irradiation. Therefore, Sox4 suppresses p53-mediated apoptosis induced by gamma irradiation in HCC cells [77]. Of note, this function of Sox4 in HCCs is in sharp contrast with its role in colorectal cancer in which Sox4 interacts with and stabilizes p53 activity and enhances p53-mediated apoptosis, cell cycle arrest, and inhibits tumorigenesis [20].
Table 1.
Expression pattern and biological functions of Sox4 in different types of malignancies and its correlation with patient survival
| Type of tumor | Sox4 expression | Method of detection | Suggested function | Correlation with survival | Reference |
|---|---|---|---|---|---|
| Acute myeloid leukemia | ↑ | Western blot | Promotion of cell growth | ND | [96] |
| Adenoid cystic carcinoma | ↑ | cDNA microarray | ND | ND | [82] |
| Adenoid cystic carcinoma | ↑ | Tissue microarray | Suppression of apoptosis | ND | [81] |
| Bladder carcinoma | ↑ | cDNA microarray/tissue microarray | Induction of apoptosis | Positive | [75] |
| Colon adenocarcinomas | ↓ | cDNA microarray | ND | ND | [131] |
| Colorectal cancer | ↑ | Tissue microarray | ND | ND | [112] |
| Colorectal cancer | ↑ | cDNA microarray | ND | Negative | [112] |
| Cutaneous melanoma | ↓ | Tissue microarray | Suppression of invasion and migration | Positive | [104] |
| Endometrial cancer | ↑ | Tissue microarray | Promotion of cell growth | ND | [85] |
| Hepatocellular carcinoma | ↑ | Tissue microarray | Suppression of apoptosis | Positive | [77] |
| Gallbladder carcinoma | ↓ | IHC | ND | Positive | [110] |
| Gastric cancer | ↑ | Tissue microarray | Suppression of apoptosis | Negative | [86] |
| Glioblastoma | ↑ | Parallel signature sequencing/RT-PCR/IHC | ND | ND | [99] |
| Lung cancer | ↑ | RT-PCR/western blot | Induce transformation of NIH3T3 cells | ND | [76] |
| Medulloblastoma | ↑ | RT-PCR/ISH | ND | ND | [65] |
| Medulloblastoma | ↑ | RT-PCR | ND | ND | [132] |
| Pediatric medulloblastoma | ↑ | cDNA microarray | ND | Positive | [111] |
| Prostate cancer | ↑ | cDNA microarray | ND | ND | [78] |
| Prostate cancer | ↑ | Tissue microarray/cDNA microarray | Suppression of apoptosis | ND | [79] |
| Uveal melanoma | ↓ | Suppressive subtractive hybridization | ND | ND | [105] |
ND not determined, ISH in situ hybridization, IHC immunohistochemistry
A tissue microarray analysis of 2,360 samples revealed an increased expression of Sox4 protein in bladder tumors, suggesting an oncogenic role for Sox4 in this type of tumor [75]. Nevertheless, this notion is in contrast with survival studies in the same set of patients where a significant correlation between strong Sox4 expression and increased patient survival was found, probably due to the role of Sox4 in induction of apoptosis in bladder carcinoma cells. Indeed, in these cells, Sox4 overexpression impairs cell viability and enhances cell death [75]. This study also found that, in bladder cancer cells, Sox4 regulates a plethora of genes including transcription factors, such as MEF2C and zinc finger protein 6 (ZNF6), and signaling molecules such as phosphoinositide-3-kinase regulatory subunit polypeptide 3 (PIK3R3), that promotes cell cycle arrest, and mitogen-activated protein kinase kinase 5 (MAP2K5) [75].
An oncogenic role for Sox4 in prostate cancer and its progression is well established [78, 79]. Moreno and colleagues reported that Sox4 is the only member of the Sox family that is overexpressed in prostate cancer, and its knockdown induces apoptosis in the LNCaP prostate cancer cell line. Consistently, Sox4 overexpression induced transformation of RWPE-1 prostate cancer cells. Furthermore, depletion of Sox4 expression resulted in an increased expression of p53 protein and loss of survivin expression [79], possibly explaining the mechanism by which Sox4 suppresses apoptosis and enhances transformation of prostate cancer cells. However, the mechanism by which Sox4 regulates p53 expression in these cells is not understood. Further analysis by cDNA microarray of LNCaP cells treated with Sox4 siRNAs or overexpressing Sox4 revealed 466 genes which were consistently responsive both to Sox4-knockdown and overexpression. Among the Sox4 target genes identified in this study were genes with roles in Wnt signaling (TLE-1), apoptosis (BCL10 and PUMA), and inflammation (CSF1) [79].
The same group later performed a genome-wide promoter analysis of the Sox4 transcriptional network in prostate cancer cells and identified 3,470 different genes whose promoter sequences contain direct binding sites of Sox4. However, cDNA microarray analysis revealed that, among these genes, expression of only 282 of the 3,470 binding targets is significantly influenced by Sox4 [80]. This observation suggests that a change in expression of Sox4 is not sufficient to alter the expression status of the other 3,188 genes, reflecting the requirement for other regulatory components such as binding partners in this process. Among the genes which were shown to be directly regulated by Sox4 in this study are imminent proto-oncogenes such as Tenascin-C, Frizzled-3, 5 and 8, Patched-1, Delta-like1, and EGFR, which are positively regulated by Sox4 and reduced after Sox4-knockdown in prostate cancer cells [80]. The extent of the Sox4 network encompasses various cancer-related pathways and mechanisms including the TGF-β, Wnt, hedgehog, and Notch pathways, as well as growth factor signaling and tumor metastasis. Notably, this study revealed differential regulation of expression of target genes by Sox4 in different prostate cancer cell lines. For instance, Sox4 binds to the promoter sequences of EGFR and enhances its expression in RWPE-1 cells, yet it does not bind to these sequences in LNCaP cells [80]. Interestingly, Sox4 overexpression increases expression of the transcription factor SON by 1.8-fold, whereas its knockdown increases expression of purine biosynthetic enzyme GART by threefold. These two genes, located on chromosome 21, are regulated by a common bidirectional promoter but transcribed in opposite directions. Differential regulation of expression of these two genes by Sox4 suggests that it regulates the directionality of this promoter [80]. This study also identified co-occurring binding sites of several transcription factors such as E2F1, E2F4, PAX5, LEF1/TCF1, and MYC along with Sox4 in its target promoter [80], which highlights the possibility of cooperation between Sox4 and one or more of these factors in regulation of the expression of target genes.
Sox4-knockdown causes a considerable reduction in adenoid cystic carcinoma (ACC) cell viability due to induction of apoptosis in a caspase-dependent manner [81]. In concert with this observation, both Sox4 transcript and protein were found to be upregulated in ACC [81, 82]. Microarray gene expression profiling of Sox4-knockdown ACC cells confirmed Sox4-modulated expression of critical genes involved in apoptosis and cell cycle control such as cyclin G2 (CCNG2), DUSP4, and BNIP3, indicating the contribution of Sox4 to the malignant phenotype of ACC cells by promoting cell survival. This study, along with other cDNA microarray analyses in human prostate cancer [79, 80], bladder cancer [75], and small cell lung cancer [83] cells, identified a large number of genes, the expression of which is affected by Sox4. Although each study identified a considerable number of target genes, the overlaps between these datasets are almost negligible (Fig. 3). In fact, there is no gene whose expression is at least two-fold changed upon overexpression or knockdown of Sox4 in all five datasets. Even more surprisingly, comparison of two studies in the same prostate cancer cell line (LNCap) only returned three common genes (Fig. 3). Bearing in mind that these comparisons should be considered carefully due to differences in the technical aspects, such as the type of microarray platforms or the time points after overexpression or knockdown of Sox4 at which the RNA samples were collected, these results are also another indication of the highly tissue- and cell-specific network of Sox4.
Fig. 3.
Comparative analysis of cDNA microarray studies of Sox4 target genes in prostate cancer, adenoid cystic carcinoma, bladder, and small cell lung cancer cell lines. Target genes whose expression was >two-fold changed after Sox4 overexpression [75, 79, 80] or knockdown [81, 83] and represented in at least two datasets were included in this analysis
As mentioned earlier, expression of Sox4 is regulated at post-transcriptional level. For instance, miR-129, which is overexpressed in bladder carcinoma and inversely correlated with patient survival, targets Sox4 in these cells [84]. Similarly, expression of miR-129-2 is lost in the majority of primary endometrial tumors, probably due to hypermethylation of the miR-129-2 regulatory regions, with a concomitant gain of Sox4 expression [85]. Restoration of miR-129-2 expression in these cells leads to decreased Sox4 expression and reduced proliferation of cancer cells. Consistently, Sox4 protein is overexpressed in endometrial tumors [85], and its knockdown suppresses endometrial cancer cell growth. Downregulation of Sox4 by miR-129-2 as well as hypermethylation-mediated suppression of this miRNA is also described in gastric cancers, in which both Sox4-knockdown and overexpression of miR-129-2 induce apoptosis [86].
In a search for miRNAs which regulate cancer growth and metastasis, Tavazoie et al. [87] found that expression of miR-335 is lost in the majority of primary breast tumors, and that this loss of expression is associated with poor distal metastasis-free survival. In this type of cancer, miR-335 regulates a set of genes, including Sox4 and the extracellular matrix component Tenascin C, whose expression in a large cohort of human breast tumors is associated with risk of distal metastasis. Consistently, knockdown of Sox4 or Tenascin C significantly abolishes lung colonization by LM2 breast cancer cells. Interestingly, in the genome-wide promoter analysis in prostate carcinoma cells, Sox4 was shown to bind to the promoter sequences of Tenascin C [80]. The study by Tavazoie and colleagues showed that miR-335 directly targets Sox4. However, it is not clear whether downregulation of Tenascin C by miR-335 is a direct event or is achieved through initial downregulation of Sox4, resulting in reduced expression of Tenascin C.
In addition to miR-335, in breast cancer cells miR-93 also targets Sox4. This effect may be important for the observed induction of mesenchymal–epithelial transition (MET) associated with downregulation of TGF-β signaling in breast cancer cells and resulting in cancer stem cell depletion [88]. In line with this observation, Sox4 may contribute to invasion and metastasis of breast cancer cells by induction of epithelial-mesenchymal transition (EMT). In fact, overexpression of Sox4 in immortalized human mammary epithelial cells is sufficient for acquisition of mesenchymal traits, enhanced cell migration, and invasion [89]. Moreover, based on tumorigenesis assay in mice, Sox4 cooperates with activated Ras to promote tumorigenesis in vivo [89]. In addition to breast cancer, Sox4 also augments the migration, invasion, and metastasis of hepatocellular carcinoma cells [90]. This function of Sox4 is probably exerted by upregulation of two downstream factors, neuropilin 1 (NRP1) and semaphorin 3C (SEMA3C), knockdown of which drastically reduces cell migration [90].
By a sophisticated approach, using oncogenic retrovirus-induced insertional mutagenesis, Copeland and colleagues [91] showed that replication-defective retrovirus carrying Sox4 can induce myeloid leukemias after transplanting into mice bone marrow cells in cooperation with Mef2c, a transcription factor involved in regulation of homing and invasiveness of leukemic cells. Sox4 can integrate at the site of Sfpi1, an Ets1-related transcription factor essential for normal myeloid and lymphoid development, and reduce Sfpi1 expression [92]. The same group later revealed that Sox4 represses Sfpi1 transcription by binding to a critical Sfpi1 upstream DNA element [93]. Additionally, by analyzing 285 acute myeloid leukemia (AML) patient samples, they found a significant negative correlation between Sox4 and Sfpi1 mRNA expression providing convincing evidence for the involvement of Sox4 in promotion of at least a subset of myeloid malignancies [93]. Sox4 is the site of the Evi1 (ectopic viral integration site 1) proviral insertions in myeloid leukemias. This insertion results in transcriptional activation and increased Sox4 expression [91]. Interestingly, Sox4 expressing myeloid cells have a higher level of transcripts associated with proliferation including Evi1, despite no measurable effect on cell proliferation or differentiation after overexpression of Sox4 [91].
Sox4 is also reported to be a common retrovirus insertion site in myeloid and monocytic cells [94], suggesting a role of Sox4 in promoting malignant disease in these cells. Consistently, retroviral tagging strategy in combination with high throughput inverse-PCR identified Sox4 as one of the most common retroviral integration sites in a tumor panel composed primarily of B cell lymphomas [95]. In myeloid cells, Sox4 binds to the promoter sequences of CREB and enhances its expression [96]. Transduction of CREB transgenic mouse bone marrow cells with a Sox4 retrovirus increases the in vitro survival and self-renewal. In addition, leukemic blasts from the majority of AML patients have higher CREB, phosphorylated CREB, and Sox4 protein expression, indicating that Sox4 and CREB cooperate and contribute to increased proliferation of hematopoietic progenitor cells and myelogenesis.
Sox4 also binds to and activates the promoter of CD56 in primary myeloma cells [97]. CD56 is a member of the immunoglobulin superfamily that was initially characterized in the cells of the nervous system and is overexpressed in more than 80 % of myeloma patients as well as some other types of malignancies [98]. Thus, it would be of interest to investigate the possible role of Sox4 in regulation of CD56 expression in other types of cancer.
Promotion of tumorigenesis by sustainment of stemness
In another study delineating the molecular details of Sox4’s involvement in tumorigenesis, Ikushima et al. [51] found that Sox4 is highly expressed in GICs and required for the TGF-β-induced expression of Sox2 that is essential for retention of stemness in GICs. Consistently, Sox4-knockdown cells show less sphere-forming ability and self-renewal capacity, two characteristic features of GICs. Sox4 regulates Sox2 expression at transcriptional level by binding to an enhancer element located at its 3′ flanking region. This activity is increased after TGF-β stimulation. In addition, Sox4 mRNA expression is immediately induced after TGF-β stimulation through binding of Smad2/3, the DNA-binding mediators of TGF-β signaling. Consistently, inhibitors of TGF-β signaling drastically deprived tumorigenicity of GICs by promoting their differentiation, while these effects are attenuated in GICs overexpressing Sox2 or Sox4 [51]. The same group later demonstrated that Oct4 physically interacts with Sox4 and cooperatively activates the Sox2 enhancer region to maintain stemness properties of GICs [44]. Supporting this notion, consensus sequences of Sox proteins and Oct4 exist in close proximity of each other in the Sox2 enhancer region, and ChIP re-IP (back-to-back immune-precipitation of chromatin with two different antibodies) assay demonstrated that Sox4 and Oct4 exist in the same transcription complex in the Sox2 enhancer region and enhance the activity of Sox2 promoter [44], further confirming the role of Sox4 in regulation of Sox2 expression and maintenance of GICs’ tumorigenicity. A separate study identified elevated expression of Sox4 and TGF-β in human glioblastoma compared with normal brain tissues at both RNA and protein levels, and confirmed that Sox4 expression is increased by TGF-β stimulation [99].
In breast cancer, Sox4 overexpression induces EMT, resulting in a cancer stem cell-like phenotype in breast cancer cells. These cells exhibited an increase in the CD44 high/CD24 low sub-population and enhanced size and number of mammospheres, indicating that the Sox4-induced EMT generates mesenchymal cells with stem cell-like phenotype [89]. This function of Sox4 may also be mediated by the TGF-β pathway as is evident by increased expression of TGF-β1 and TGF-β2 mRNAs and the phospo-Smad2 protein in Sox4-expressing cells [89].
Sox4 as a tumor suppressor
The first evidence to pinpoint a possible role for Sox4 in suppression of tumorigenesis came from a study by Ahn et al. [48]. Using mRNA differential display and northern blot analysis, they revealed that expression of Sox4 mRNA is enhanced during apoptosis induced by prostaglandin (PG)A2 and Delta12-PGJ2 in human hepatocellular carcinoma cells, Hep3B, suggesting the possible involvement of Sox4 in the process of apoptosis. Nevertheless, Sox4 was also highly expressed in subcutaneous tumors grown in nude mice as a xenograft from Hep3B cells [48], which is not consistent with a tumor suppressor role for Sox4. Unfortunately, the authors did not investigate this contradiction in their results in further detail. The same group later observed that overexpression of Sox4 induces significant apoptosis in Hep3B and HepG2 cell lines, probably through the caspase-mediated pathway [100]. In addition, Sox4-knockdown blocks the apoptosis induced by PGA(2) and delta(12)-PGJ(2) in these cells [100]. The caspase-dependent manner of Sox4 for induction of apoptosis was later confirmed by Kim et al. [101]. Interestingly, although several prominent apoptosis-related factors such as Bax and PUMA are among the direct transcriptional targets of Sox4 (Table 2), the pro-apoptosis function of Sox4 can be dissociated from its transcriptional activity [23]. Indeed, the central domain (aa 166–342) of Sox4 is critical for induction of apoptosis in HEK293 cells. Accordingly, deletion of the DNA-binding domain or trans-activation domain in Sox4 does not significantly affect its pro-apoptotic activity in these cells, whereas overexpression of a construct containing the central domain induces the apoptotic activity comparable to that of the full-length protein [23].
Table 2.
Validated direct transcriptional targets of Sox4
| Tissue/cell type | Gene symbol | Mode of regulation | Function | Assay | References |
|---|---|---|---|---|---|
| Breast cancer | ZEB1 | ↑ | Transcription repressor | Reporter assay | [89] |
| Breast cancer | Snail | ↑ | Transcription repressor | Reporter assay | [89] |
| Colon carcinoma | p56lck | ↑ | Protein tyrosine kinase | Reporter assay | [67] |
| COS1 | Tubb3 | ↑ | β-tubulin isotype III | EMSA | [34] |
| Endometrial carcinomas | TCF4 | ↑ | Transcription factor | Reporter assay | [53] |
| Glioma | Sox2 | ↑ | Transcription factor | ChIP | [51] |
| HCC | SLC2A1 | ↑ | Agmatine transport | ChIP | [90] |
| HCC | Bax | ↓ | Apoptosis | ChIP | [77] |
| HCC | CREB3L1 | ↑ | Axon guidance | ChIP | [90] |
| HCC | MYEF2 | ↑ | Axon guidance | ChIP | [90] |
| HCC | NAV3 | ↑ | Axon guidance | ChIP | [90] |
| HCC | NRP1 | ↑ | Axon guidance | ChIP | [90] |
| HCC | SEMA3C | ↑ | Axon guidance | ChIP | [90] |
| HCC | NEIL3 | ↑ | Base excision DNA repair | ChIP | [90] |
| HCC | CHFR | ↑ | Cell cycle checkpoints | ChIP | [90] |
| HCC | NPNT | ↑ | Extracellular matrix protein | ChIP | [90] |
| HCC | GPRC5B | ↑ | G-protein signaling pathway | ChIP | [90] |
| HCC | HUNK | ↑ | Kinase | ChIP | [90] |
| HCC | PFKFB4 | ↑ | Kinase | ChIP | [90] |
| HCC | AKR1B10 | ↑ | Metabolism | ChIP | [90] |
| HCC | ALDH18A1 | ↓ | Metabolism | ChIP | [90] |
| HCC | DHRS13 | ↑ | Metabolism | ChIP | [90] |
| HCC | GYG | ↑ | Metabolism | ChIP | [90] |
| HCC | PAM | ↑ | Metabolism | ChIP | [90] |
| HCC | RCC2 | ↓ | Metabolism | ChIP | [90] |
| HCC | MAP4 | ↑ | Microtubule-associated protein | ChIP | [90] |
| HCC | SMG5 | ↑ | mRNA decay | ChIP | [90] |
| HCC | HSPBAP1 | ↓ | Protein folding | ChIP | [90] |
| HCC | LCK | ↑ | Protein tyrosine kinase | ChIP | [90] |
| HCC | CSPG2 | ↑ | Proteoglycan | ChIP | [90] |
| HCC | CTSC | ↑ | Proteolysis | ChIP | [90] |
| HCC | RBM10 | ↓ | RNA binding | ChIP | [90] |
| HCC | SERPINE2 | ↑ | Serine protease inhibitor | ChIP | [90] |
| HCC | INST8 | ↑ | Small nuclear RNAs processing | ChIP | [90] |
| HCC | HIC2 | ↓ | Transcription factor | ChIP | [90] |
| HCC | VGLL4 | ↑ | Transcription factor | ChIP | [90] |
| HCC | FOXQ1 | ↑ | Transcription factor | ChIP | [90] |
| HCC | TEAD2 | ↑ | Transcription factor | ChIP | [90] |
| HCC | CCDC97 | ↓ | Unknown | ChIP | [90] |
| HCC | UBAP2L | ↓ | Unknown | ChIP | [90] |
| HCC | DKK1 | ↑ | Wnt signaling pathway | ChIP | [90] |
| Heart | Connexin 43 | ↑ | Gap junction protein | ChIP | [27] |
| Hemangioblasts | λ5 | ↑ | Component of precursor of BCR | ChIP | [31] |
| Hemangioblasts | VpreB1 | ↑ | Component of precursor of BCR | ChIP | [31] |
| Melanoma | NF-κB p50 | ↓ | Inflamation, cell growth | ChIP | [104] |
| Melanoma | Dicer | ↑ | miRNA biogenesis | ChIP | [107] |
| Mouse embryonic mesenchymal cells | Tead2 | ↑ | Transcription factor | ChIP | [25] |
| Myeloid Leukemia | CREB | ↑ | Transcription factor | ChIP | [96] |
| Myeloid leukemias | Sfpi1 | ↓ | Transcription factor | ChIP | [93] |
| Myeloma | CD56 | ↑ | Immunoglobulin superfamily | ChIP | [97] |
| Neural cells | DCX | ↑ | Microtubule-associated protein | ChIP | [35] |
| Neural cells | GnRH | ↑ | Sexual development | ChIP | [133] |
| Neural cells | FEZF2 | ↑ | Transcription factor | ChIP | [36] |
| Prostate | PUMA | ↑ | Apoptosis | ChIP | [79] |
| Prostate | MLL | ↑ | Histone methyl-transferase | ChIP | [80] |
| Prostate | ADAM10 | ↑ | Metalloproteinase | ChIP | [80] |
| Prostate | AGO1 | ↑ | miRNA biogenesis | ChIP | [80] |
| Prostate | DHX9 | ↑ | miRNA biogenesis | ChIP | [55, 80] |
| Prostate | Dicer | ↑ | miRNA biogenesis | ChIP | [55, 80] |
| Prostate | DLL1 | ↑ | Notch signaling | ChIP | [80] |
| Prostate | HES2 | ↑ | Notch signaling | ChIP | [80] |
| Prostate | RBL1 | ↑ | Regulation of cell cycle | ChIP | [80] |
| Prostate | EGFR | ↑ | Signaling | ChIP | [80] |
| Prostate | ELF5 | ↓ | Transcription factor | ChIP | [80] |
| Prostate | Sox4 | ↑ | Transcription factor | ChIP | [55] |
| Prostate | ZNF281 | ↑ | Transcription repressor | ChIP | [80] |
| Prostate | TLE-1 | ↑ | Transcription repressor | ChIP | [79, 80] |
| Prostate | FZD3 | ↑ | Wnt receptor | ChIP | [80] |
| Prostate | FZD5 | ↑ | Wnt receptor | ChIP | [80] |
| Prostate | FZD8 | ↑ | Wnt receptor | ChIP | [80] |
| Prostate | AXIN2 | ↑ | Wnt signaling | ChIP | [55] |
| SCLC | VASH2 | ↑ | Angiogenic factor | ChIP | [83] |
| SCLC | PCDHB | ↑ | Protocadherin | ChIP | [83] |
| SCLC | Tead2 | ↑ | Transcription factor | ChIP | [83] |
| SCLC | MYB | ↑ | Transcription factor | ChIP | [83] |
| SCLC | Tubb3 | ↑ | β-tubulin isotype III | ChIP | [83] |
| T-cells | Interleukin 5 | ↓ | Cytokine | ChIP | [32] |
| T-Cells | CD2 | ↑ | Surface antigen | EMSA | [134] |
EMSA electrophoretic mobility shift assay, ChIP chromatin immunoprecipitation, HCC hepatocellular carcinoma
As an indirect link between Sox4 and regulation of apoptotic cell death, human ubiquitin-conjugating enzyme 9 (Ubc9) interacts with HMG-box of Sox4 and represses Sox4 transcriptional activity in HEK293T cells [102]. The C93S mutant of Ubc9, which abrogates SUMO-1 conjugation activity, does not abolish its ability to inhibit Sox4 activity. Elevated Ubc9 expression has been detected in at least some types of malignancies, such as primary and metastatic melanomas in which Ubc9 plays a crucial role in suppression of apoptosis [102]. On this note, in some malignancies such as breast cancer [103], the ability of Ubc9 to promote cancer initiation and progression is independent of its sumoylation function. The possible involvement of Sox4 inactivation in the sumoylation-independent oncogenic function of Ubc9 remains to be uncovered.
Our own studies on cutaneous malignant melanoma have revealed that Sox4 expression is significantly reduced in metastatic melanomas [104]. Furthermore, Sox4 expression is positively correlated with better patient survival. Consistently, Sox4 suppresses melanoma cell migration and invasion ability through inhibition of NF-κB p50 expression by binding to sequences upstream of the NF-κB p50 promoter [104]. A recent study reported a reduced expression of Sox4 mRNA in uveal melanoma [105]. Overexpression of several members of the NF-κB pathway has also been observed in metastatic uveal melanoma [106]. However, it is not clear whether or not Sox4-mediated inhibition of NF-κB can play a role in suppression of this type of melanoma.
In addition, Sox4 may recruit miRNA machinery to negatively regulate melanoma invasion. In cutaneous melanoma cells, Sox4 regulates transcription of Dicer by binding to its promoter and enhancing its activity which is critical for expression of a considerable number of cancer-related miRNAs [107]. Interestingly, Dicer and Drosha have recently been identified to be necessary to activate the DNA repair mechanism upon exogenous DNA damage and oncogene-induced genotoxic stress [108], although this function appears to be independent from the canonical miRNA-mediated translational repression mechanisms regulated by Dicer. This mechanism may also be responsible for Sox4-mediated suppression of tumorigenesis, considering the regulation of Dicer expression by Sox4 and the fact that Sox4 has also been reported to be a DNA damage sensor and is required for activation of p53 tumor suppressor in response to DNA damage (discussed in further details below).
Regulation of expression of miRNA-related factors such as Dicer, Argonaute 1, and RNA helicase A by Sox4 had been previously reported in prostate cancer cells by Scharer et al. [80]. Although there were some inconsistencies in the pattern of Sox4 target genes between different cell lines, they were able to confirm the regulation of Dicer expression by Sox4 at both transcript and protein levels [80], which was consistent with our observation in melanoma cells [107]. It is noteworthy that, while we observed that Dicer expression is lost in metastatic melanoma and it is required for Sox4-mediated suppression of melanoma cell invasion, expression of Dicer is upregulated in prostate adenocarcinomas [109]. Although the expression status of Dicer is not known in other subtypes of prostate cancer, this observation may be related to overexpression of Sox4 [79] and highlights the importance of Sox4-regulated Dicer expression in these two different types of malignancies.
In primary gallbladder carcinoma, Sox4 expression is reduced compared with the normal epithelium of the gallbladder [110]. Reduced Sox4 expression is associated with high histological grade, high pathologic T stage, and late clinical stage of this malignancy. In addition, expression of Sox4 in tissues with positive nodal metastasis is lower than those without metastasis. In line with these observations, Sox4 expression is positively correlated with a better overall and disease-free patient survival [110]. It is intriguing that, in the majority of studies that investigated the correlation between Sox4 expression and patient survival, Sox4 has been found as a positive marker for survival. For instance, despite an enhanced expression of Sox4 in bladder carcinomas, there is a positive correlation between strong Sox4 expression and increased patient survival [75]. Similarly, there is a trend toward favorable prognosis with increasing Sox4 expression levels in patients with medulloblastoma, despite the enhanced expression of Sox4 in medulloblastoma compared with normal cerebellum [111]. Also, Sox4 is overexpressed in hepatocellular carcinoma, but its expression correlates with diminished risk of recurrence and improved overall survival in HCC patients [77]. Nevertheless, none of these studies addressed the observed discrepancy in the expression level or function of Sox4 and its correlation with survival. In contrast to these reports that suggest Sox4 as a marker for improved prognosis, at least in some cases Sox4 expression was found to be inversely correlated with survival (Table 1). For instance, Sox4 expression inversely correlates with overall patient survival of gastric cancer [86] and recurrence-free survival of microsatellite-stable stage II (no lymph node or distant metastases) colorectal carcinoma [112].
As mentioned earlier, in HCC cells, the HMG box domain of Sox4 interacted with p53, resulting in the inhibition of p53-induced Bax expression and the p53-mediated apoptosis induced by gamma irradiation [77]. However, it is not clear how Sox4 expression would confer a survival advantage to the patient while it can suppress the p53-mediated apoptosis. Also, the observed suppression of p53 activity by Sox4 is in contrast to another report by Pan et al. [20], who demonstrated that Sox4 induces p53 activity in colon cancer cell lines. In these cells, expression of Sox4 protein but not mRNA is increased upon treatment with doxorubicin (DOX) and Sox4 is required for activation of p53 in response to DOX-induced DNA damage. In fact, in these cells, Sox4 interacts with and stabilizes p53 protein by repressing Mdm2-mediated ubiquitination and degradation of p53. Furthermore, Sox4 interacts with p300/CBP and enhances p300/CBP/p53 complex formation and p53 acetylation which increases p53 stability. In this system, Sox4 promotes cell cycle arrest and apoptosis, and inhibits tumorigenesis in a p53-dependent manner [20]. In line with these observations, ionizing radiation induces Sox4 expression, in parallel with induction of p53 protein in medulloblastoma cell lines [113]. In addition, Sox4-knockdown dramatically blocks the radiation-induced increases in p53 (S-15) phosphorylation and XRCC1 protein expression along with an increase in levels of phospho-γ-H2AX compared with cells subjected to radiation alone [113], suggesting that Sox4 functions upstream of p53-mediated DNA repair, an important process in prevention of mutations and tumorigenesis.
Role of Sox4/β-catenin axis in regulation of tumorigenesis
Wnt signaling has long been implicated in tumorigenesis and deregulated expression of its components is a common phenomenon in many types of tumors [114]. One of the main mediators of the wnt signaling pathway is β-catenin, abnormal expression and/or mutation of which is a common feature of almost all types of human malignancies [115]. In HEK293 cells, overexpression of Sox4 enhances β-catenin/TCF activity by increasing the stability of β-catenin, which induces wnt signaling pathway activity and expression of its target genes, cyclin D1 and c-myc. The enhanced β-catenin/TCF activity by Sox4 is caused by stabilization of the β-catenin protein, through induction of CK2, a kinase involved in regulation of β-catenin stability, suggesting that Sox4 may act as an activator of the wnt signaling pathway [116]. In a study that revealed a novel non-transcription related mechanism by which Sox4 may act as an oncogene, Sinner et al. [21] observed that, unlike Sox17 that represses β-catenin/TCF activity and inhibits proliferation of SW480 colon carcinoma cells, Sox4 enhances β-catenin/TCF activity and cell proliferation. In fact, both Sox17 and Sox4 physically interact with TCF/LEF family members via their HMG-box domains and while Sox17 promotes their degradation, Sox4 stabilizes β-catenin and TCF/LEF proteins [21].
γ-catenin, also known as junction plakoglobin (JUP), is a Sox4 binding protein in prostate cancer cells [55]. Interaction between these two proteins is enhanced by wnt3A treatment. However, this interaction could inhibit Sox4 binding to downstream target promoters and may repress its transcriptional activity in addition to modulation of the β-catenin-mediated transcription. These observations led to the suggestion that γ-catenin may compete with β-catenin for binding to Sox4, downregulating wnt-responsive transcription [55]. This model is also consistent with the observations that γ-catenin expression is lost in human prostate cancer through epigenetic and genetic pathways [117], as is its potential to suppress cell migration and tumorigenic potential [118]. In malignant melanoma, Sox4 is reported to activate the wnt/β-catenin signaling pathway [119]. Accordingly, Sox4 knockdown leads to a decreased β-catenin protein expression, resulting in inhibition of wnt/β-catenin signaling pathway activity and reduced expression of survivin as well as attenuated cell proliferation [119]. However, In contrast to this observation, our own studies have shown that knockdown of Sox4 slightly increases melanoma cell growth [104].
As opposed to the aforementioned role of the Sox4/β-catenin axis in the promotion of tumorigenesis, a recent report has revealed that Sox4 could indeed enhance β-catenin/TCF4 transcription, through upregulation of TCF4 at the transcription level, without any direct β-catenin association, resulting in a significant decrease in the proliferation rate, along with increases in expression of p21, as well as of TCF4, in endometrial carcinoma cells [53]. Consistently, Sox4-knockdown increased cell growth in this model. These observations provide evidence that at least in some types of malignancy the Sox4/β-catenin axis may contribute to suppression of tumorigenesis. In concert with this notion, although debated [120], β-catenin is implicated in suppression of melanoma invasion and tumorigenesis [121, 122]. In addition, at least in some cases expression of β-catenin is reduced during melanoma progression [123–125] and decreased β-catenin expression is linked to worsened patient survival [126, 127]. Considering the loss of expression of Sox4 in melanoma and the considerable amount of evidence that Sox4 may regulate β-catenin expression and/or activity, it would be interesting to identify the possible role of Sox4 in regulation of β-catenin in melanoma cells and its significance in tumor progression. It should also be noted that in some cases overexpression of Sox4 may suppress β-catenin expression. In fact, overexpression of Sox4 induces a significant reduction of β-catenin expression in breast cancer cell lines [89], which further complicates the mutual relationship between Sox4 and β-catenin and their functional outcome in different types of tissue.
Concluding remarks
In recent years, numerous publications have described the expression pattern and possible functional significance of Sox4 in various types of malignancies. However, despite this considerable progress, some outstanding questions regarding the mechanistic details of Sox4 functionality and its significance in cancer initiation and progression remain to be answered. For instance, in several types of cancer, while the expression pattern of Sox4 is known to be different than their normal counterparts, the mechanism(s) by which Sox4 contributes to the tumorigenesis (or suppression of tumorigenesis) in those cancers is not known. Also, whereas differential and tissue-dependent regulation of transcription is customary in Sox family, in some cases it is not understood whether or not the mode of Sox4-mediated regulation of activity, but not expression, of other proteins can also be tissue-dependent. For instance, in hepatocellular carcinoma cells, Sox4 interacts with p53, resulting in the inhibition of p53 activity and apoptosis [77], whereas, in colon cancer cell lines, Sox4 interaction results in enhanced p53 activity and apoptosis [20]. Several members of the Sox family have been implicated in tumorigenesis, and in a number of cases some siblings compete with Sox4 in binding to and regulating the activity of the target promoters or binding partners [16, 21, 36]. Nevertheless, except in a few studies, the possible roles of other Sox proteins in Sox4 networks and their putative influence on Sox4’s oncogenic or tumor suppressive functions remain unknown. Despite the recent progress in understanding the mechanisms which regulate expression of Sox4 (Fig. 2), little is known about the mechanisms responsible for the aberrant expression or function of Sox4 in cancers.
In this review, we summarized the progress made over more than two decades after the discovery of Sox4 in defining its role in tumorigenesis, with an emphasis on the tissue-dependent function of Sox4 as well as occasional discrepancies between various reports. Although the actual reason for these discrepancies is not known, in some cases it may be due to the remarkable short half-life of the Sox4 protein and the inability of mRNA expression studies to accurately reflect the level of protein expression. Indeed, it seems that Sox4 mRNA expression may not be able to mirror the expression level of its translation product [22], and is insufficient to permit the drawing of a conclusion regarding the expression levels of the Sox4 protein in different types of cancer. This notion is especially critical in the studies which use RT-PCR or cDNA microarray. It is worth emphasizing that Sox4 contains a single exon and, due to the lack of introns, it is difficult to distinguish between the sequences of the genomic DNA and the complementary DNA (cDNA) in studies that investigate the expression levels of the Sox4 transcript. Therefore, more precautionary measures such as DNase treatment of the extracted mRNAs are required to avoid false positive signals due to genomic DNA contamination in these types of studies.
At least one study has identified three variants, including a nonsense mutation in the coding region of Sox4 that could result in expression of a truncated isoform [76]. Although it is not known whether similar truncated isoforms of Sox4 are also expressed in other types of tissues, the majority of studies regarding the expression status of Sox4 in cancer did not consider its mutation status and possible influences of these mutations in stability and/or functional properties of the Sox4 protein in the tissues of interest. It is intuitive that, due to increased mortality associated with late stages of cancer, factors whose expression positively correlates with tumor progression should have an inverse correlation with patient survival. Nevertheless, in some cases, we observe a contradiction between Sox4 expression pattern and its correlation with survival (Table 1). It is plausible that these contradictions may be due to expression of multiple isoforms and/or mutant forms of Sox4 in these malignant tissues, particularly the mutants that lack the C-terminal domain and are more stable [76], perhaps due to the absence of the degron located in this domain [22]. Future studies are required to empirically validate the expression or lack of expression of such isoforms in other types of cancer and determine what proportion (if any) of the Sox4 staining could be due to these isoforms.
At first glance, the inconsistency in the role of Sox4 in various tumors might seem puzzling. Sox4 expression gives a selective edge to some types of tumors, consistent with an oncogenic role, while it can suppress initiation or progression of other types of cancer, fitting to the definition of a tumor suppressor. As mentioned earlier, this feature is characteristic of Sox family whose function relies on the context of the host cells. In addition, in the same type of cells, Sox4 can impede or augment activity of target promoters. For instance, in melanoma cells, we observed that Sox4 inhibits NF-κB p50 [104] but induces Dicer [107] promoter activity. At the molecular level, this discrepancy might be explained by the requirement for specific partners at the site of the target promoters for Sox4 to either enhance or inhibit their activity. So far, several factors, such as POU proteins Oct4 [44] and Brn2 [16], as well as other factors like Tbx3 [27], p53 [20], GATA-3 [32], TCF4 [21], and JUP [55], have been identified to cooperate with Sox4 in the regulation of target promoters. These binding partners can have a crucial impact on the expression profile of Sox4 downstream targets and its overall function in each type of cell (Fig. 4). The expression pattern of many of these partners is differentially regulated in various cell types or different developmental stages of the cells. For instance, Oct4 is a well-known marker of pluripotency which is expressed in embryonic stem cells, some adult stem cells, and cancer progenitor cells, but not differentiated cells [128]. This phenomenon would restrict the effect of Sox4 on the expression of those targets only to the cell types that express both Sox4 and Oct4. Nevertheless, in most cases, the putative binding partners of Sox4 are not known, which beckons further attention in future studies to this important aspect of the Sox4 network.
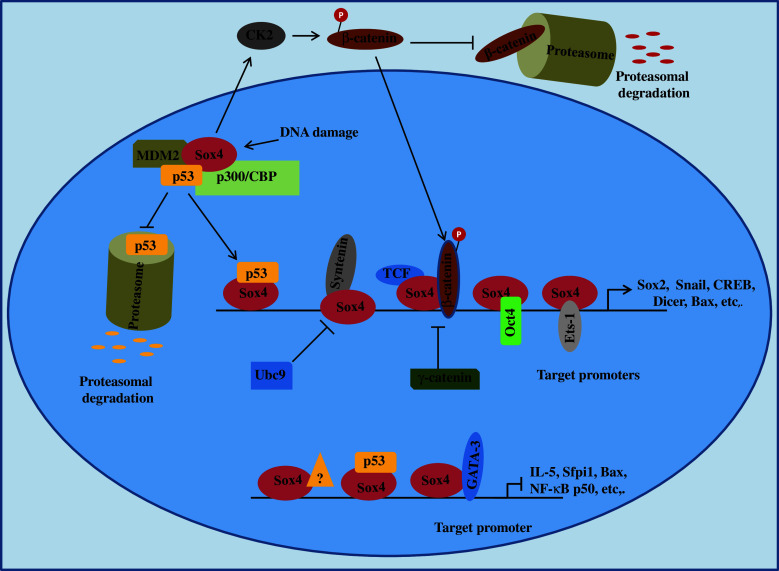
Fig. 4.
Regulation of gene expression by Sox4. So far, two modes of regulation of gene expression by Sox4 have been identified; transcriptional and post-translational. At the transcriptional level, Sox4 can act both as a transcription factor and repressor, depending on the gene or cell type of interest. This differential regulation of gene expression, which is discussed in the text, plays a very important role in oncogenic or tumor suppressive functions of Sox4. It should be noted that not all of the illustrated complexes are present in every type of cell
Sox4 regulates the expression of a large set of genes in different tissues as evidenced by genome-wide cDNA microarray or promoter binding profiling studies. Despite this large number, there seems to be very limited overlap between the Sox4 targets identified in different studies (Fig. 3). This negligible overlap further indicates the existence of distinct Sox4 downstream pathways in each cell type. Also, in the majority of cases, it is not clear whether regulation of target gene expression by Sox4 is achieved by direct binding to their promoters and acting as a transcription factor or via indirect modes such as activation of a mediator. We compiled a list of genes whose expression is demonstrated to be either up- or downregulated in a direct manner by Sox4 (Table 2). This list contains a wide range of proteins involved in different cellular functions, including but not limited to inflammation, axon direction, differentiation, cell growth, and proliferation. But one major group of validated targets consists of prominent transcription factors such as CREB, Sox2, and TCF4. Considering the potential targets of each of these transcription factors, there is an immense network of genes whose regulation is at some level controlled by Sox4. Furthermore, miRNA biogenesis factors are also among the direct transcriptional targets of Sox4. miRNAs are major regulators of gene expression, and it is estimated that at least one-third of all protein-coding genes in the human genome are regulated by miRNAs [129]. These notions highlight the immense number of potential Sox4 target genes, and also indicate that many of the genes whose expression status is changed by loss or overexpression of Sox4, as identified by cDNA microarray studies, may in fact be indirect targets.
Many miRNAs have opposite effects on the biological or pathological state of the cells depending on the type of tissue and the expression profile of their target protein-coding mRNAs. Therefore, regulation of their maturation by Sox4 [80, 107] may explain, at least in part, the pleiotropic role of Sox4. It is notable that regulation of miRNAs expression by Sox4 may not only be limited to their post-transcriptional maturation. Indeed, it is plausible that Sox4, as a transcription factor, can directly regulate the transcription of certain miRNAs. In addition, some downstream targets of Sox4, such as p53, have been shown to control expression of a number of prominent miRNAs involved in tumorigenesis [130]. These notions may contribute to delineating the complexity of Sox4-mediated regulation of normal and pathological processes.
Finally, the pleiotropic and tissue-dependent functions of Sox4 may have some clinical significance. Accordingly, this demands the utmost attention to consider this feature of Sox4 in future studies to design therapeutic strategies involving suppression or activation of Sox4 for treatment of a specific type of malignancy, as the desired effect of the modulation of Sox4 in one tissue may also end up as a major side effect in another type of tissue.
Acknowledgments
This work was supported by Canadian Institutes of Health Research (MOP-84559, MOP-93810 and MOP-110974), Canadian Cancer Society Research Institute (2011-700714) and Canadian Dermatology Foundation to G.L. S.M.J and R.S.A. are recipients of the trainee award from Canadian Institute of Health Research Skin Research Training Centre. S.M.J. is a recipient of Roman M. Babicki Fellowship and the University of British Columbia Graduate Fellowship.
References
- 1.Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 2.Stros M, Launholt D, Grasser KD. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol Life Sci. 2007;64:2590–2606. doi: 10.1007/s00018-007-7162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 4.Berta P, Hawkins JR, Sinclair AH, Taylor A, Griffiths BL, Goodfellow PN, Fellous M. Genetic evidence equating SRY and the testis-determining factor. Nature. 1990;348:448–450. doi: 10.1038/348448A0. [DOI] [PubMed] [Google Scholar]
- 5.Farr CJ, Easty DJ, Ragoussis J, Collignon J, Lovell-Badge R, Goodfellow PN. Characterization and mapping of the human SOX4 gene. Mamm Genome. 1993;4:577–584. doi: 10.1007/BF00361388. [DOI] [PubMed] [Google Scholar]
- 6.Prior HM, Walter MA. SOX genes: architects of development. Mol Med. 1996;2:405–412. [PMC free article] [PubMed] [Google Scholar]
- 7.Schepers GE, Teasdale RD, Koopman P. Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell. 2002;3:167–170. doi: 10.1016/S1534-5807(02)00223-X. [DOI] [PubMed] [Google Scholar]
- 8.van Beest M, Dooijes D, van de Wetering M, Kjaerulff S, Bonvin A, Nielsen O, Clevers H. Sequence-specific high mobility group box factors recognize 10–12-base pair minor groove motifs. J Biol Chem. 2000;275:27266–27273. doi: 10.1074/jbc.M004102200. [DOI] [PubMed] [Google Scholar]
- 9.Wissmuller S, Kosian T, Wolf M, Finzsch M, Wegner M. The high-mobility-group domain of Sox proteins interacts with DNA-binding domains of many transcription factors. Nucl Acids Res. 2006;34:1735–1744. doi: 10.1093/nar/gkl105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iguchi H, Urashima Y, Inagaki Y, Ikeda Y, Okamura M, Tanaka T, Uchida A, Yamamoto TT, Kodama T, Sakai J. SOX6 suppresses cyclin D1 promoter activity by interacting with beta-catenin and histone deacetylase 1, and its down-regulation induces pancreatic beta-cell proliferation. J Biol Chem. 2007;282:19052–19061. doi: 10.1074/jbc.M700460200. [DOI] [PubMed] [Google Scholar]
- 11.Stolt CC, Lommes P, Hillgartner S, Wegner M. The transcription factor Sox5 modulates Sox10 function during melanocyte development. Nucl Acids Res. 2008;36:5427–5440. doi: 10.1093/nar/gkn527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agarwal P, Verzi MP, Nguyen T, Hu J, Ehlers ML, McCulley DJ, Xu SM, Dodou E, Anderson JP, Wei ML, Black BL. The MADS box transcription factor MEF2C regulates melanocyte development and is a direct transcriptional target and partner of SOX10. Development. 2011;138:2555–2565. doi: 10.1242/dev.056804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chew LJ, Gallo V. The Yin and Yang of Sox proteins: activation and repression in development and disease. J Neurosci Res. 2009;87:3277–3287. doi: 10.1002/jnr.22128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris ML, Baxter LL, Loftus SK, Pavan WJ. Sox proteins in melanocyte development and melanoma. Pigment Cell Melanoma Res. 2010;23:496–513. doi: 10.1111/j.1755-148X.2010.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefebvre V, Dumitriu B, Penzo-Mendez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39:2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dy P, Penzo-Mendez A, Wang H, Pedraza CE, Macklin WB, Lefebvre V. The three SoxC proteins—Sox4, Sox11 and Sox12—exhibit overlapping expression patterns and molecular properties. Nucl Acids Res. 2008;36:3101–3117. doi: 10.1093/nar/gkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhlbrodt K, Herbarth B, Sock E, Enderich J, Hermans-Borgmeyer I, Wegner M. Cooperative function of POU proteins and SOX proteins in glial cells. J Biol Chem. 1998;273:16050–16057. doi: 10.1074/jbc.273.26.16050. [DOI] [PubMed] [Google Scholar]
- 18.van Houte LP, Chuprina VP, van der Wetering M, Boelens R, Kaptein R, Clevers H. Solution structure of the sequence-specific HMG box of the lymphocyte transcriptional activator Sox-4. J Biol Chem. 1995;270:30516–30524. doi: 10.1074/jbc.270.51.30516. [DOI] [PubMed] [Google Scholar]
- 19.Vandewetering M, Oosterwegel M, Vannorren K, Clevers H. Sox-4, an Sry-like Hmg box protein, is a transcriptional activator in lymphocytes. EMBO J. 1993;12:3847–3854. doi: 10.1002/j.1460-2075.1993.tb06063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan X, Zhao J, Zhang WN, Li HY, Mu R, Zhou T, Zhang HY, Gong WL, Yu M, Man JH, Zhang PJ, Li AL, Zhang XM. Induction of SOX4 by DNA damage is critical for p53 stabilization and function. Proc Natl Acad Sci USA. 2009;106:3788–3793. doi: 10.1073/pnas.0810147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, Lin SCJ, Jonatan D, Zorn AM, Wells JM. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol. 2007;27:7802–7815. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beekman JM, Vervoort SJ, Dekkers F, van Vessem ME, Vendelbosch S, Brugulat-Panes A, van Loosdregt J, Braat AK, Coffer PJ. Syntenin-mediated regulation of Sox4 proteasomal degradation modulates transcriptional output. Oncogene. 2012;31:2668–2679. doi: 10.1038/onc.2011.445. [DOI] [PubMed] [Google Scholar]
- 23.Hur EH, Hur W, Choi JY, Kim IK, Kim HY, Yoon SK, Rhim H. Functional identification of the pro-apoptotic effector domain in human Sox4. Biochem Bioph Res Co. 2004;325:59–67. doi: 10.1016/j.bbrc.2004.09.215. [DOI] [PubMed] [Google Scholar]
- 24.Hoser M, Potzner MR, Koch JM, Bosl MR, Wegner M, Sock E. Sox12 deletion in the mouse reveals nonreciprocal redundancy with the related Sox4 and Sox11 transcription factors. Mol Cell Biol. 2008;28:4675–4687. doi: 10.1128/MCB.00338-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhattaram P, Penzo-Mendez A, Sock E, Colmenares C, Kaneko KJ, Vassilev A, Depamphilis ML, Wegner M, Lefebvre V. Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat Commun. 2010;1:9. doi: 10.1038/ncomms1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schilham MW, Oosterwegel MA, Moerer P, Ya J, deBoer PAJ, van de Wetering M, Verbeek S, Lamers WH, Kruisbeek AM, Cumano A, Clevers H. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 27.Boogerd CJ, Wong LY, Boogaard M, Bakker ML, Tessadori F, Bakkers J, t Hoen PA, Moorman AF, Christoffels VM, Barnett P. Sox4 mediates Tbx3 transcriptional regulation of the gap junction protein Cx43. Cell Mol Life Sci. 2011;68:3949–3961. doi: 10.1007/s00018-011-0693-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sock E, Rettig SD, Enderich J, Bosl MR, Tamm ER, Wegner M. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol Cell Biol. 2004;24:6635–6644. doi: 10.1128/MCB.24.15.6635-6644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schilham MW, Moerer P, Cumano A, Clevers HC. Sox-4 facilitates thymocyte differentiation. Eur J Immunol. 1997;27:1292–1295. doi: 10.1002/eji.1830270534. [DOI] [PubMed] [Google Scholar]
- 30.Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, van de Wetering M, Verbeek S, Lamers WH, Kruisbeek AM, Cumano A, Clevers H. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 31.Liber D, Domaschenz R, Holmqvist PH, Mazzarella L, Georgiou A, Leleu M, Fisher AG, Labosky PA, Dillon N. Epigenetic priming of a pre-B cell-specific enhancer through binding of Sox2 and Foxd3 at the ESC stage. Cell Stem Cell. 2010;7:114–126. doi: 10.1016/j.stem.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 32.Kuwahara M, Yamashita M, Shinoda K, Tofukuji S, Onodera A, Shinnakasu R, Motohashi S, Hosokawa H, Tumes D, Iwamura C, Lefebvre V, Nakayama T. The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-beta and suppresses T(H)2 differentiation. Nat Immunol. 2012;13:778–786. doi: 10.1038/ni.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geijsen N, Uings IJ, Pals C, Armstrong J, McKinnon M, Raaijmakers JAM, Lammers JWJ, Koenderman L, Coffer PJ. Cytokine-specific transcriptional regulation through an IL-5R alpha interacting protein. Science. 2001;293:1136–1138. doi: 10.1126/science.1059157. [DOI] [PubMed] [Google Scholar]
- 34.Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Gene Dev. 2006;20:3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mu LF, Berti L, Masserdotti G, Covic M, Michaelidis TM, Doberauer K, Merz K, Rehfeld F, Haslinger A, Wegner M, Sock E, Lefebvre V, Couillard-Despres S, Aigner L, Berninger B, Lie DC. SoxC transcription factors are required for neuronal differentiation in adult hippocampal neurogenesis. J Neurosci. 2012;32:3067–3080. doi: 10.1523/JNEUROSCI.4679-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shim SB, Kwan KY, Li MF, Lefebvre V, Sestan N. Cis-regulatory control of corticospinal system development and evolution. Nature. 2012;486:74–79. doi: 10.1038/nature11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potzner MR, Griffel C, Lutjen-Drecoll E, Bosl MR, Wegner M, Sock E. Prolonged Sox4 expression in oligodendrocytes interferes with normal myelination in the central nervous system. Mol Cell Biol. 2007;27:5316–5326. doi: 10.1128/MCB.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao ZL, Ure K, Ding PG, Nashaat M, Yuan LR, Ma J, Hammer RE, Hsieh J. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J Neurosci. 2011;31:9772–9786. doi: 10.1523/JNEUROSCI.1604-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lioubinski O, Muller M, Wegner M, Sander M. Expression of Sox transcription factors in the developing mouse pancreas. Dev Dyn Off Publ Am Assoc Anat. 2003;227:402–408. doi: 10.1002/dvdy.10311. [DOI] [PubMed] [Google Scholar]
- 40.Mavropoulos A, Devos N, Biemar F, Zecchin E, Argenton F, Edlund H, Motte P, Martial JA, Peers B. sox4b is a key player of pancreatic alpha cell differentiation in zebrafish. Dev Biol. 2005;285:211–223. doi: 10.1016/j.ydbio.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 41.Wilson ME, Yang KY, Kalousova A, Lau J, Kosaka Y, Lynn FC, Wang J, Mrejen C, Episkopou V, Clevers HC, German MS. The HMG box transcription factor Sox4 contributes to the development of the endocrine pancreas. Diabetes. 2005;54:3402–3409. doi: 10.2337/diabetes.54.12.3402. [DOI] [PubMed] [Google Scholar]
- 42.Goldsworthy M, Hugill A, Freeman H, Horner E, Shimomura K, Bogani D, Pieles G, Mijat V, Arkell R, Bhattacharya S, Ashcroft FM, Cox RD. Role of the transcription factor sox4 in insulin secretion and impaired glucose tolerance. Diabetes. 2008;57:2234–2244. doi: 10.2337/db07-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nissen-Meyer LS, Jemtland R, Gautvik VT, Pedersen ME, Paro R, Fortunati D, Pierroz DD, Stadelmann VA, Reppe S, Reinholt FP, Del Fattore A, Rucci N, Teti A, Ferrari S, Gautvik KM. Osteopenia, decreased bone formation and impaired osteoblast development in Sox4 heterozygous mice. J Cell Sci. 2007;120:2785–2795. doi: 10.1242/jcs.003855. [DOI] [PubMed] [Google Scholar]
- 44.Ikushima H, Todo T, Ino Y, Takahashi M, Saito N, Miyazawa K, Miyazono K. Glioma-initiating cells retain their tumorigenicity through integration of the Sox axis and Oct4 protein. J Biol Chem. 2011;286:41434–41441. doi: 10.1074/jbc.M111.300863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS, Niwa H. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 46.Graham JD, Hunt SMN, Tran N, Clarke CL. Regulation of the expression and activity by progestins of a member of the SOX gene family of transcriptional modulators. J Mol Endocrinol. 1999;22:295–304. doi: 10.1677/jme.0.0220295. [DOI] [PubMed] [Google Scholar]
- 47.Reppe S, Rian E, Jemtland R, Olstad OK, Gautvik VT, Gautvik KM. Sox-4 messenger RNA is expressed in the embryonic growth plate and regulated via the parathyroid hormone/parathyroid hormone-related protein receptor in osteoblast-like cells. J Bone Miner Res. 2000;15:2402–2412. doi: 10.1359/jbmr.2000.15.12.2402. [DOI] [PubMed] [Google Scholar]
- 48.Ahn SG, Cho GH, Jeong SY, Rhim H, Choi JY, Kim IK. Identification of cDNAs for Sox-4, an HMG-Box protein, and a novel human homolog of yeast splicing factor SSF-1 differentially regulated during apoptosis induced by prostaglandin A(2)/Delta(12)-PGJ(2) in Hep3B cells. Biochem Biophys Res Commun. 1999;260:216–221. doi: 10.1006/bbrc.1999.0856. [DOI] [PubMed] [Google Scholar]
- 49.Gorreta F, Runfola TP, VanMeter AJ, Barzaghi D, Chandhoke V, Del Giacco L. Identification of thioredoxin reductase 1-regulated genes using small interference RNA and cDNA microarray. Cancer Biol Ther. 2005;4:1079–1088. doi: 10.4161/cbt.4.10.1987. [DOI] [PubMed] [Google Scholar]
- 50.Lee HM, Zhang H, Schulz V, Tuck DP, Forget BG. Downstream targets of HOXB4 in a cell line model of primitive hematopoietic progenitor cells. Blood. 2010;116:720–730. doi: 10.1182/blood-2009-11-253872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 52.Wang YJ, Solt LA, Kojetin DJ, Burris TP. Regulation of p53 stability and apoptosis by a ROR agonist. Plos One. 2012;7:e34921. doi: 10.1371/journal.pone.0034921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saegusa M, Hashimura M, Kuwata T. Sox4 functions as a positive regulator of beta-catenin signaling through upregulation of TCF4 during morular differentiation of endometrial carcinomas. Lab Invest. 2012;92:511–521. doi: 10.1038/labinvest.2011.196. [DOI] [PubMed] [Google Scholar]
- 54.Potzner MR, Tsarovina K, Binder E, Penzo-Mendez A, Lefebvre V, Rohrer H, Wegner M, Sock E. Sequential requirement of Sox4 and Sox11 during development of the sympathetic nervous system. Development. 2010;137:775–784. doi: 10.1242/dev.042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lai YH, Cheng J, Cheng DM, Feasel ME, Beste KD, Peng JM, Nusrat A, Moreno CS. SOX4 interacts with plakoglobin in a Wnt3a-dependent manner in prostate cancer cells. BMC Cell Biol. 2011;12:1–11. doi: 10.1186/1471-2121-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila . Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 57.Kinzler KW, Ruppert JM, Bigner SH, Vogelstein B. The Gli gene is a member of the Kruppel family of zinc finger proteins. Nature. 1988;332:371–374. doi: 10.1038/332371a0. [DOI] [PubMed] [Google Scholar]
- 58.Hahn H, Wicking C, Zaphiropoulos PG, Gailani MR, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E, Unden AB, Gillies S, Negus K, Smyth I, Pressman C, Leffell DJ, Gerrard B, Goldstein AM, Dean M, Toftgard R, ChenevixTrench G, Wainwright B, Bale AE. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/S0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 59.Giancotti V, Berlingieri MT, DiFiore PP, Fusco A, Vecchio G, Crane-Robinson C. Changes in nuclear proteins on transformation of rat epithelial thyroid cells by a murine sarcoma retrovirus. Cancer Res. 1985;45:6051–6057. [PubMed] [Google Scholar]
- 60.Bussemakers MJ, van de Ven WJ, Debruyne FM, Schalken JA. Identification of high mobility group protein I(Y) as potential progression marker for prostate cancer by differential hybridization analysis. Cancer Res. 1991;51:606–611. [PubMed] [Google Scholar]
- 61.Sattler HP, Lensch R, Rohde V, Zimmer E, Meese E, Bonkhoff H, Retz M, Zwergel T, Bex A, Stoeckle M, Wullich B. Novel amplification unit at chromosome 3q25-q27 in human prostate cancer. Prostate. 2000;45:207–215. doi: 10.1002/1097-0045(20001101)45:3<207::AID-PROS2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 62.Collins C, Rommens JM, Kowbel D, Godfrey T, Tanner M, Hwang S, Polikoff D, Nonet G, Cochran J, Myambo K, Jay KE, Froula J, Cloutier T, Kuo WL, Yaswen P, Dairkee S, Giovanola J, Hutchinson GB, Isola J, Kallioniemi OP, Palazzolo M, Martin C, Ericsson C, Pinkel D, Albertson D, Li WB, Gray JW. Positional cloning of ZNF217 and NABC1: genes amplified at 20q13.2 and overexpressed in breast carcinoma. Proc Natl Acad Sci USA. 1998;95:8703–8708. doi: 10.1073/pnas.95.15.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Korn WM, Yasutake T, Kuo WL, Warren RS, Collins C, Tomita M, Gray J, Waldman FM. Chromosome arm 20q gains and other genomic alterations in colorectal cancer metastatic to liver, as analyzed by comparative genomic hybridization and fluorescence in situ hybridization. Gene Chromosome Canc. 1999;25:82–90. doi: 10.1002/(SICI)1098-2264(199906)25:2<82::AID-GCC2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 64.Cheng YC, Lee CJ, Badge RM, Orme AT, Scotting PJ. Sox8 gene expression identifies immature glial cells in developing cerebellum and cerebellar tumours. Mol Brain Res. 2001;92:193–200. doi: 10.1016/S0169-328X(01)00147-4. [DOI] [PubMed] [Google Scholar]
- 65.Lee CJ, Appleby VJ, Orme AT, Chan WI, Scotting PJ. Differential expression of SOX4 and SOX11 in medulloblastoma. J Neurooncol. 2002;57:201–214. doi: 10.1023/A:1015773818302. [DOI] [PubMed] [Google Scholar]
- 66.Ueda R, Yoshida K, Kawase T, Kawakami Y, Toda M. Preferential expression and frequent IgG responses of a tumor antigen, SOX5, in glioma patients. Int J Cancer. 2007;120:1704–1711. doi: 10.1002/ijc.22472. [DOI] [PubMed] [Google Scholar]
- 67.McCracken S, Kim CS, Xu Y, Minden M, Miyamoto NG. An alternative pathway for expression of p56(lck) from type I promoter transcripts in colon carcinoma. Oncogene. 1997;15:2929–2937. doi: 10.1038/sj.onc.1201474. [DOI] [PubMed] [Google Scholar]
- 68.Zamoyska R, Basson A, Filby A, Legname G, Lovatt M, Seddon B. The influence of the src-family kinases, Lck and Fyn, on T cell differentiation, survival and activation. Immunol Rev. 2003;191:107–118. doi: 10.1034/j.1600-065X.2003.00015.x. [DOI] [PubMed] [Google Scholar]
- 69.McCracken S, Leung S, Bosselut R, Ghysdael J, Miyamoto NG. Myb and Ets related transcription factors are required for activity of the human lck type I promoter. Oncogene. 1994;9:3609–3615. [PubMed] [Google Scholar]
- 70.Hurst CD, Fiegler H, Carr P, Williams S, Carter NP, Knowles MA. High-resolution analysis of genomic copy number alterations in bladder cancer by microarray-based comparative genomic hybridization. Oncogene. 2004;23:2250–2263. doi: 10.1038/sj.onc.1207260. [DOI] [PubMed] [Google Scholar]
- 71.Grasemann C, Gratias S, Stephan H, Schuler A, Schramm A, Klein-Hitpass L, Rieder H, Schneider S, Kappes F, Eggert A, Lohmann DR. Gains and overexpression identify DEK and E2F3 as targets of chromosome 6p gains in retinoblastoma. Oncogene. 2005;24:6441–6449. doi: 10.1038/sj.onc.1208792. [DOI] [PubMed] [Google Scholar]
- 72.Oeggerli M, Tomovska S, Schraml P, Calvano-Forte D, Schafroth S, Simon R, Gasser T, Mihatsch MJ, Sauter G. E2F3 amplification and overexpression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Oncogene. 2004;23:5616–5623. doi: 10.1038/sj.onc.1207749. [DOI] [PubMed] [Google Scholar]
- 73.Wu QO, Hoffmann MJ, Hartmann FH, Schulz WA. Amplification and overexpression of the ID4 gene at 6p22.3 in bladder cancer. Mol Cancer. 2005;4:16. doi: 10.1186/1476-4598-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stephens BJ, Han HY, Gokhale V, Von Hoff DD. PRL phosphatases as potential molecular targets in cancer. Mol Cancer Ther. 2005;4:1653–1661. doi: 10.1158/1535-7163.MCT-05-0248. [DOI] [PubMed] [Google Scholar]
- 75.Aaboe M, Birkenkamp-Demtroder K, Wiuf C, Sorensen FB, Tbykjaer T, Sauter G, Jensen KME, Dyrskjot L, Orntoft T. SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res. 2006;66:3434–3442. doi: 10.1158/0008-5472.CAN-05-3456. [DOI] [PubMed] [Google Scholar]
- 76.Medina PP, Castillo SD, Blanco S, Sanz-Garcia M, Largo C, Alvarez S, Yokota J, Gonzalez-Neira A, Benitez J, Clevers HC, Cigudosa JC, Lazo PA, Sanchez-Cespedes M. The SRY-HMG box gene, SOX4, is a target of gene amplification at chromosome 6p in lung cancer dagger. Hum Mol Genet. 2009;18:1343–1352. doi: 10.1093/hmg/ddp034. [DOI] [PubMed] [Google Scholar]
- 77.Hur W, Rhim H, Jung CK, Kim JD, Bae SH, Jang JW, Yang JM, Oh ST, Kim DG, Wang HJ, Lee SB, Yoon SK. SOX4 overexpression regulates the p53-mediated apoptosis in hepatocellular carcinoma: clinical implication and functional analysis in vitro. Carcinogenesis. 2010;31:1298–1307. doi: 10.1093/carcin/bgq072. [DOI] [PubMed] [Google Scholar]
- 78.Vanaja DK, Ballman KV, Morlan BW, Cheville JC, Neumann RM, Lieber MM, Tindall DJ, Young CYF. PDLIM4 repression by hypermethylation as a potential biomarker for prostate cancer. Clin Cancer Res. 2006;12:1128–1136. doi: 10.1158/1078-0432.CCR-05-2072. [DOI] [PubMed] [Google Scholar]
- 79.Liu PB, Ramachandran S, Ali Seyed M, Scharer CD, Laycock N, Dalton WB, Williams H, Karanam S, Datta MW, Jaye DL, Moreno CS. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006;66:4011–4019. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 80.Scharer CD, McCabe CD, Ali-Seyed M, Berger MF, Bulyk ML, Moreno CS. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer Res. 2009;69:709–717. doi: 10.1158/0008-5472.CAN-08-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pramoonjago P, Baras AS, Moskaluk CA. Knockdown of Sox4 expression by RNAi induces apoptosis in ACC3 cells. Oncogene. 2006;25:5626–5639. doi: 10.1038/sj.onc.1209566. [DOI] [PubMed] [Google Scholar]
- 82.Frierson HF, Jr, El-Naggar AK, Welsh JB, Sapinoso LM, Su AI, Cheng J, Saku T, Moskaluk CA, Hampton GM. Large scale molecular analysis identifies genes with altered expression in salivary adenoid cystic carcinoma. Am J Pathol. 2002;161:1315–1323. doi: 10.1016/S0002-9440(10)64408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Castillo SD, Matheu A, Mariani N, Carretero J, Lopez-Rios F, Lovell-Badge R, Sanchez-Cespedes M. Novel transcriptional targets of the SRY-HMG box transcription factor SOX4 link its expression to the development of small cell lung cancer. Cancer Res. 2012;72:176–186. doi: 10.1158/0008-5472.CAN-11-3506. [DOI] [PubMed] [Google Scholar]
- 84.Dyrskjot L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, Fristrup N, Jensen JL, Andersen CL, Zieger K, Kauppinen S, Ulhoi BP, Kjems J, Borre M, Orntoft TF. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–4860. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 85.Huang YW, Liu JC, Deatherage DE, Luo JQ, Mutch DG, Goodfellow PJ, Miller DS, Huang THM. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69:9038–9046. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shen R, Pan S, Qi S, Lin X, Cheng S. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 in gastric cancer. Biochem Biophys Res Commun. 2010;394:1047–1052. doi: 10.1016/j.bbrc.2010.03.121. [DOI] [PubMed] [Google Scholar]
- 87.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang QQ, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu S, Patel SH, Ginestier C, Ibarra I, Martin-Trevino R, Bai S, McDermott SP, Shang L, Ke J, Ou SJ, Heath A, Zhang KJ, Korkaya H, Clouthier SG, Charafe-Jauffret E, Birnbaum D, Hannon GJ, Wicha MS. MicroRNA93 regulates proliferation and differentiation of normal and malignant breast stem cells. PLoS Genet. 2012;8:e1002751. doi: 10.1371/journal.pgen.1002751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao X, Feng J, Zhang Y, Gao H, Liu DX, Lu J, Huang B. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012;72:4597–4608. doi: 10.1158/0008-5472.CAN-12-1045. [DOI] [PubMed] [Google Scholar]
- 90.Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL, Horng JT, Hsiao M, Tsou AP. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27:5578–5589. doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]
- 91.Du Y, Spence SE, Jenkins NA, Copeland NG. Cooperating cancer-gene identification through oncogenic-retrovirus-induced insertional mutagenesis. Blood. 2005;106:2498–2505. doi: 10.1182/blood-2004-12-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schwieger M, Schuler A, Forster M, Engelmann A, Arnold MA, Delwel R, Valk PJ, Lohler J, Slany RK, Olson EN, Stocking C. Homing and invasiveness of MLL/ENL leukemic cells is regulated by MEF2C. Blood. 2009;114:2476–2488. doi: 10.1182/blood-2008-05-158196. [DOI] [PubMed] [Google Scholar]
- 93.Aue G, Du Y, Cleveland SM, Smith SB, Dave UP, Liu DL, Weniger MA, Metais JY, Jenkins NA, Copeland NG, Dunbar CE. Sox4 cooperates with PU.1 haploinsufficiency in murine myeloid leukemia. Blood. 2011;118:4674–4681. doi: 10.1182/blood-2011-04-351528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bies J, Sramko M, Fares J, Rosu-Myles M, Zhang S, Koller R, Wolff L. Myeloid-specific inactivation of p15Ink4b results in monocytosis and predisposition to myeloid leukemia. Blood. 2010;116:979–987. doi: 10.1182/blood-2009-08-238360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Suzuki T, Shen HF, Akagi K, Morse HC, Malley JD, Naiman DQ, Jenkins NA, Copeland NG. New genes involved in cancer identified by retroviral tagging. Nat Genet. 2002;32:166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 96.Sandoval S, Kraus C, Cho EC, Cho M, Bies J, Manara E, Accordi B, Landaw EM, Wolff L, Pigazzi M, Sakamoto KM. Sox4 cooperates with CREB in myeloid transformation. Blood. 2012;120:155–165. doi: 10.1182/blood-2011-05-357418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Iqbal MS, Otsuyama K, Shamsasenjan K, Asaoku H, Kawano MM. CD56 expression in human myeloma cells derived from the neurogenic gene expression: possible role of the SRY-HMG box gene, SOX4. Int J Hematol. 2010;91:267–275. doi: 10.1007/s12185-009-0474-3. [DOI] [PubMed] [Google Scholar]
- 98.Gattenlohner S, Stuhmer T, Leich E, Reinhard M, Etschmann B, Volker HU, Rosenwald A, Serfling E, Bargou RC, Ertl G, Einsele H, Muller-Hermelink HK. Specific detection of CD56 (NCAM) Isoforms for the identification of aggressive malignant neoplasms with progressive development. Am J Pathol. 2009;174:1160–1171. doi: 10.2353/ajpath.2009.080647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin BY, Madan A, Yoon JG, Fang XF, Yan XW, Kim TK, Hwang D, Hood L, Foltz G. Massively parallel signature sequencing and bioinformatics analysis identifies up-regulation of TGFBI and SOX4 in human glioblastoma. Plos One. 2010;5:e10210. doi: 10.1371/journal.pone.0010210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ahn SG, Kim HS, Jeong SW, Kim BE, Rhim H, Shim JY, Kim JW, Lee JH, Kim IK. Sox-4 is a positive regulator of Hep3B and HepG2 cells’ apoptosis induced by prostaglandin (PG)A(2) and Delta(12)-PGJ(2) Exp Mol Med. 2002;34:243–249. doi: 10.1038/emm.2002.34. [DOI] [PubMed] [Google Scholar]
- 101.Kim BE, Lee JH, Kim HS, Kwon OJ, Jeong SW, Kim IK. Involvement of Sox-4 in the cytochrome c-dependent AIF-independent apoptotic pathway in HeLa cells induced by Delta(12)-prostaglandin J(2) Exp Mol Med. 2004;36:444–453. doi: 10.1038/emm.2004.56. [DOI] [PubMed] [Google Scholar]
- 102.Moschos SJ, Smith AP, Mandic M, Athanassiou C, Watson-Hurst K, Jukic DM, Edington HD, Kirkwood JM, Becker D. SAGE and antibody array analysis of melanoma-infiltrated lymph nodes: identification of Ubc9 as an important molecule in advanced-stage melanomas. Oncogene. 2007;26:4216–4225. doi: 10.1038/sj.onc.1210216. [DOI] [PubMed] [Google Scholar]
- 103.Zhu S, Sachdeva M, Wu F, Lu Z, Mo YY. Ubc9 promotes breast cell invasion and metastasis in a sumoylation-independent manner. Oncogene. 2010;29:1763–1772. doi: 10.1038/onc.2009.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jafarnejad SM, Wani AA, Martinka M, Li G. Prognostic significance of Sox4 expression in human cutaneous melanoma and its role in cell migration and invasion. Am J Pathol. 2010;177:2741–2752. doi: 10.2353/ajpath.2010.100377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Landreville S, Lupien CB, Vigneault F, Gaudreault M, Mathieu M, Rousseau AP, Guerin SL, Salesse C. Identification of differentially expressed genes in uveal melanoma using suppressive subtractive hybridization. Mol Vis. 2011;17:1224–1233. [PMC free article] [PubMed] [Google Scholar]
- 106.Meir T, Dror R, Yu X, Qian J, Simon I, Pe’er J, Chowers I. Molecular characteristics of liver metastases from uveal melanoma. Invest Ophthalmol Vis Sci. 2007;48:4890–4896. doi: 10.1167/iovs.07-0215. [DOI] [PubMed] [Google Scholar]
- 107.Jafarnejad SM, Ardekani GS, Ghaffari M, Martinka M, Li G (2012) Sox4-mediated Dicer expression is critical for suppression of melanoma cell invasion. Oncogene (in press) [DOI] [PMC free article] [PubMed]
- 108.Francia S, Michelini F, Saxena A, Tang D, De Hoon M, Anelli V, Mione M, Carninci P, Di Fagagna FD. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, Dhir R. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang C, Zhao H, Lu J, Yin J, Zang L, Song N, Dong R, Wu T, Du X. Clinicopathological significance of SOX4 expression in primary gallbladder carcinoma. Diagn Pathol. 2012;7:41. doi: 10.1186/1746-1596-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.de Bont JM, Kros JM, Passier MMCJ, Reddingius RE, Smitt PAES, Luider TM, den Boer ML, Pieters R. Differential expression and prognostic significance of SOX genes in pediatric medulloblastoma and ependymoma identified by microarray analysis. Neuro-Oncology. 2008;10:648–660. doi: 10.1215/15228517-2008-032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Andersen CL, Christensen LL, Thorsen K, Schepeler T, Sorensen FB, Verspaget HW, Simon R, Kruhoffer M, Aaltonen LA, Laurberg S, Orntoft TF. Dysregulation of the transcription factors SOX4, CBFB and SMARCC1 correlates with outcome of colorectal cancer. Br J Cancer. 2009;100:511–523. doi: 10.1038/sj.bjc.6604884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chetty C, Dontula R, Gujrati M, Dinh DH, Lakka SS. Blockade of SOX4 mediated DNA repair by SPARC enhances radioresponse in medulloblastoma. Cancer Lett. 2012;323:188–198. doi: 10.1016/j.canlet.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Polakis P. Wnt signaling and cancer. Gene Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 115.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 116.Lee AK, Ahn SG, Yoon JH, Kim SA. Sox4 stimulates beta-catenin activity through induction of CK2. Oncol Rep. 2011;25:559–565. doi: 10.3892/or.2010.1091. [DOI] [PubMed] [Google Scholar]
- 117.Shiina H, Breault JE, Basset WW, Enokida H, Urakami S, Li LC, Okino ST, Deguchi M, Kaneuchi M, Terashima M, Yoneda T, Shigeno K, Carroll PR, Igawa M, Dahiya R. Functional loss of the gamma-catenin gene through epigenetic and genetic pathways in human prostate cancer. Cancer Res. 2005;65:2130–2138. doi: 10.1158/0008-5472.CAN-04-3398. [DOI] [PubMed] [Google Scholar]
- 118.Rieger-Christ KM, Ng L, Hanley RS, Durrani O, Ma H, Yee AS, Libertino JA, Summerhayes IC. Restoration of plakoglobin expression in bladder carcinoma cell lines suppresses cell migration and tumorigenic potential. Brit J Cancer. 2005;92:2153–2159. doi: 10.1038/sj.bjc.6602651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cai HH, Ni AH, Li W, Li JW. Inhibition of melanoma cell proliferation by targeting Wnt/beta-catenin pathway through Sox4 RNA interference. J Huazhong Univ Sci-Med. 2011;31:565–569. doi: 10.1007/s11596-011-0491-3. [DOI] [PubMed] [Google Scholar]
- 120.Damsky WE, Curley DP, Santhanakrishnan M, Rosenbaum LE, Platt JT, Rothberg BEG, Taketo MM, Dankort D, Rimm DL, McMahon M, Bosenberg M. Beta-catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell. 2011;20:741–754. doi: 10.1016/j.ccr.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Arozarena I, Bischof H, Gilby D, Belloni B, Dummer R, Wellbrock C. In melanoma, beta-catenin is a suppressor of invasion. Oncogene. 2011;30:4531–4543. doi: 10.1038/onc.2011.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chien AJ, Moore EC, Lonsdorf AS, Kulikauskas RM, Rothberg BG, Berger AJ, Major MB, Hwang ST, Rimm DL, Moon RT. Activated Wnt/beta-catenin signaling in melanoma is associated with decreased proliferation in patient tumors and a murine melanoma model. Proc Natl Acad Sci USA. 2009;106:1193–1198. doi: 10.1073/pnas.0811902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Maelandsmo GM, Holm R, Nesland JM, Fodstad O, Florenes VA. Reduced beta-catenin expression in the cytoplasm of advanced-stage superficial spreading malignant melanoma. Clin Cancer Res. 2003;9:3383–3388. [PubMed] [Google Scholar]
- 124.Kageshita T, Hamby CV, Ishihara T, Matsumoto K, Saida T, Ono T. Loss of beta-catenin expression associated with disease progression in malignant melanoma. Brit J Dermatol. 2001;145:210–216. doi: 10.1046/j.1365-2133.2001.04336.x. [DOI] [PubMed] [Google Scholar]
- 125.Pecina-Slaus N, Zigmund M, Kusec V, Martic TN, Cacic M, Slaus M. E-cadherin and beta-catenin expression patterns in malignant melanoma assessed by image analysis. J Cutan Pathol. 2007;34:239–246. doi: 10.1111/j.1600-0560.2006.00601.x. [DOI] [PubMed] [Google Scholar]
- 126.Rothberg BEG, Berger AJ, Molinaro AM, Subtil A, Krauthammer MO, Camp RL, Bradley WR, Ariyan S, Kluger HM, Rimm DL. Melanoma prognostic model using tissue microarrays and genetic algorithms. J Clin Oncol. 2009;27:5772–5780. doi: 10.1200/JCO.2009.22.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bachmann IM, Straume O, Puntervoll HE, Kalvenes MB, Akslen LA. Importance of P-cadherin, beta-catenin, and Wnt5a/frizzled for progression of melanocytic tumors and prognosis in cutaneous melanoma. Clin Cancer Res. 2005;11:8606–8614. doi: 10.1158/1078-0432.CCR-05-0011. [DOI] [PubMed] [Google Scholar]
- 128.Tai MH, Chang CC, Kiupel M, Webster JD, Olson LK, Trosko JE. Oct4 expression in adult human stem cells: evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis. 2005;26:495–502. doi: 10.1093/carcin/bgh321. [DOI] [PubMed] [Google Scholar]
- 129.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 130.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Reichling T, Goss KH, Carson DJ, Holdcraft RW, Ley-Ebert C, Witte D, Aronow BJ, Groden J. Transcriptional profiles of intestinal tumors in Apc(Min) mice are unique from those of embryonic intestine and identify novel gene targets dysregulated in human colorectal tumors. Cancer Res. 2005;65:166–176. [PubMed] [Google Scholar]
- 132.Yokota N, Mainprize TG, Taylor MD, Kohata T, Loreto M, Ueda S, Dura W, Grajkowska W, Kuo JS, Rutka JT. Identification of differentially expressed and developmentally regulated genes in medulloblastoma using suppression subtraction hybridization. Oncogene. 2004;23:3444–3453. doi: 10.1038/sj.onc.1207475. [DOI] [PubMed] [Google Scholar]
- 133.Kim HD, Choe HK, Chung S, Kim M, Seong JY, Son GH, Kim K. Class-C SOX transcription factors control GnRH gene expression via the intronic transcriptional enhancer. Mol Endocrinol. 2011;25:1184–1196. doi: 10.1210/me.2010-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wotton D, Lake RA, Farr CJ, Owen MJ. The high mobility group transcription factor, SOX4, transactivates the human CD2 enhancer. J Biol Chem. 1995;270:7515–7522. doi: 10.1074/jbc.270.13.7515. [DOI] [PubMed] [Google Scholar]