Abstract
A study was performed to evaluate the presence of porcine reproductive and respiratory syndrome virus (PRRSV) in pig meat collected at slaughterhouses and its potential transmission to pigs via pig meat. A total of 1039 blood samples were collected from pigs upon their arrival at the abattoir. The following day, meat samples (n = 1027) were collected from the carcasses of these same pigs. Samples originated from 2 Canadian slaughterhouses, 1 situated in the province of Quebec and the other situated in the province of Manitoba. Serum samples were tested for antibodies to PRRSV and both serum and meat samples were also tested for PRRSV nucleic acid by polymerase chain reaction (PCR). Seropositivity to PRRSV for all serum samples was 74.3%. Furthermore 45 (4.3%) of the total serum samples and 19 (1.9%) of the 1027 meat samples were positive for PRRSV by PCR. Sequence analysis of open reading frame (ORF) 5 performed on 15 of the 19 PRRSV strains identified in pig meat indicated that 9 were field strains and 6 were vaccine-like (98% to 99.7% nucleotide homology with the Ingelvac RespPRRS/Repro vaccine). One of these 6 strains presented an intermediate 2-6-2 restriction fragment length polymorphism (RFLP) cut pattern and the others showed the characteristic 2-5-2 RFLP pattern of the vaccine strain. All strains sequenced were determined to be North American strains. In only 1 of the 19 PRRSV-positive meat samples could PRRSV be isolated. To test the potential infectivity of meat samples containing residual PRRSV, 11 of the PCR-positive meat samples (weighing 1.05 to 1.8 kg) were each used in feeding experiments of 2 PRRSV antibody-negative specific pathogen-free pigs of 9 wk of age. Samples were cut into several pieces and fed to each pair of pigs on 2 consecutive days. Each pig pair was housed in a separate cubicle and serum samples were collected at –7, 0, 7, 14, and 20 to 21 days post exposure. Seven pig pairs were found to be infected by PRRSV following ingestion of meat samples, including meat samples containing vaccine-like virus, as judged by the demonstration of PRRSV antibodies and/or PRRSV nucleic acid in the serum. In summary, the present study indicated that low residual quantities of PRRSV may be found in a small percentage of pig meat collected at slaugtherhouses. Furthermore, when this meat was fed raw to pigs in the experimental setting designed, pigs could be infected by PRRSV.
Résumé
Une étude a été réalisée dans le but d’évaluer la présence du virus du syndrome reproducteur et respiratoire du porc (SRRP) dans la viande de porc récoltée à l’abattoir ainsi que la transmission potentielle de ce virus aux porcs via cette viande. Au total, 1039 échantillons de sang ont été récoltés de porcs à leur arrivée à l’abattoir et le jour suivant, des échantillons de viande (n = 1027) ont été prélevés des carcasses de ces porcs. Les échantillons provenaient de 2 abattoirs canadiens, 1 situé au Québec et l’autre au Manitoba. Les échantillons de sérum ont été éprouvés pour la présence d’anticorps envers le virus SRRP et les sérums ainsi que les échantillons de viande ont de plus été éprouvés pour la présence d’acide nucléique du virus SRRP par PCR. Le taux de séropositivité global pour les sérums était de 74,3 %. De plus, 45 (4,3 %) des échantillons sériques et 19 (1,9 %) des échantillons de viande étaient positifs au virus SRRP par PCR. L’analyse des sequences de l’ORF 5, réalisée sur 15 des 19 souches de virus SRRP identifiées dans la viande de porc, a démontré que 9 souches étaient des souches de terrain et 6 souches étaient des souches de type vaccinal (98–99,7 % d’homologie nucléotidique avec la souche vaccinale Ingelvac RespPRRS/Repro). Une de ces 6 souches présentait un patron RFLP intermédiaire 2-6-2 et les autres présentaient le patron RFLP 2-5-2 caractéristique de la souche vaccinale. Toutes les souches séquencées étaient nord-américaines. Le virus SRRP n’a été isolé que dans 1 seul des 19 échantillons de viande positifs au virus SRRP par PCR. Afin d’évaluer le potentiel infectieux des échantillons de viande contenant du virus SRRP résiduel, 11 des échantillons de viande positifs au PCR (pesant 1,05 à 1,8 kg) ont été utilisés dans des experiences visant à nourrir, avec chaque échantillon de viande, 2 porcs de 9 semaines d’âge. Les échantillons furent coupés en plusieurs morceaux et furent donnés à manger à chaque groupe de 2 porcs, deux jours consécutifs. Chaque groupe de 2 porcs, exempts d’organismes pathogens spécifiques, était maintenu dans des cubicules séparés et des échantillons de sérum furent récoltés de chacun des porcs à –7, 0, 7, 14, 20–21 jours post-exposition. Dans 7 des groupes de porcs, l’infection par le virus SRRP suite à l’ingestion des échantillons de viande a pu être démontrée, incluant des échantillons de viande contenant des souches de type vaccinal. L’infection fut déterminée par la demonstration dans le sérum d’anticorps envers le virus SRRP et/ou d’acide nucléique de virus SRRP. En résumé, la présente étude a indiqué que de faibles quantités résiduelles de virus SRRP peuvent se retrouver dans un petit pourcentage de viande de porc récoltée à l’abattoir. De plus, dans le contexte expérimental élaboré, lorsque cette viande fut donnée à manger crue à des porcs, ceux-ci purent être infectés par le virus SRRP.
(Traduit par les auteurs)
Introduction
There are relatively few papers in the literature on porcine reproductive and respiratory syndrome virus (PRRSV) dealing with the potential presence of this virus in pig meat. Experimental transmission studies have demonstrated transient detection only of PRRSV in muscle tissue. Bloemraad et al (1) found low levels of the PRRSV Lelystad isolate in muscle of 6-month-old pigs killed 5 and 10 d post inoculation (dpi). Magar et al (2) found PRRSV in muscle 7 dpi but not 14 dpi of 6-month-old pigs experimentally inoculated with 2 Canadian PRRSV isolates. Following experimental inoculations of 6-week-old pigs with 3 isolates of PRRSV from the United States (US), Mengeling et al (3) found PRRSV in only 1 (ham muscle at 7 dpi) out of all muscle tissue samples collected from 21 pigs between 3 and 70 dpi. When pig meat of slaughtered pigs was tested, no PRRSV was isolated from muscle tissues of 44 abattoir pigs aged between 4 and 8 mo (2). These pigs originated from herds where seropositivity was high suggesting that PRRSV did not appear to persist in muscle tissues of pigs up to the time they arrive at the abattoir. Frey et al (4) found only 6 of 1049 sample pools of fresh pork (from 178 lots) positive for PRRSV often after multiple passages in cell culture and with low virus numbers. In addition, Larochelle and Magar (5) did not detect PRRSV by reverse transcription (RT) polymerase chain reaction (PCR) or virus isolation in muscle tissue samples representing 73 lots of packaged pig meat. Similar findings were later reported in another study on carcasses of market pigs, in which all 472 carcasses specimens tested by PCR were negative (6). These studies suggest that most market-aged pigs are either clear of virus or that the titer of virus is very low.
Previous experimental work by Magar et al (7) has demonstrated that pigs can become infected by PRRSV following oral as well as intranasal inoculations of cell-cultured PRRSV, suggesting that pigs could be infected through the oro-pharynx via infection of tonsils. These latter studies were, however, performed using high virus doses for inoculum. Recently, it was shown that PRRSV can be infectious for pigs through the oral route via infected meat collected from experimentally infected pigs (8). The present work investigated if pig meat obtained from slaughterhouses contained PRRSV and, if so, whether the virus present in a given meat sample was infectious for pigs via oral exposure under the experimental conditions of this study.
Materials and methods
Sample collection and preparation
Blood and meat samples were collected from pigs at 2 Canadian slaughterhouses, 1 situated in the province of Quebec and the other in the province of Manitoba. A total of 1039 blood samples were collected from pigs upon their arrival at the abattoir and the next day meat samples (n = 1027) were collected from the carcasses of these same pigs. Samples were collected during the periods from March to June and from September to December 2002 in Quebec and Manitoba, respectively. The slaughterhouse situated in Quebec also received pigs originating from the province of Ontario and the slaughterhouse in Manitoba occasionally received pigs from Alberta and Saskatchewan. The samples collected from the Quebec abattoir were determined to originate from 156 different pork producers and those from the Manitoba abattoir were determined to originate from no less than 62 different producers.
During 1 d, 14 to 15 blood samples were collected after slaughter in vacutainer tubes and these were kept at 4°C. Blood was randomly collected from about 1 in 30 pigs. Carcasses of pigs from which blood samples were taken were identified and stored separately at 4°C. The next morning, a meat sample of approximately 1 kg from the shoulder region was collected from the respective carcasses and deposited in clean plastic bags. At this time, the pH value of the carcass was recorded from the opposite shoulder using a puncture-type pH electrode and a portable pH meter. Meat samples and corresponding blood samples were deposited in a cooler unit containing ice packs and transported to the laboratory by vehicle (Quebec samples) or by overnight air transport (Manitoba samples). We collected pH data of meat samples since in a previous study (1) it was suggested that the half-life of PRRSV in muscle tissue depended on the pH reached during the cooling and hardening period.
Upon receipt of the samples in the morning, meat and blood samples were further processed. Serum samples were separated from blood by centrifugation and aliquots were prepared and stored at –20°C. Homogenates of meat (20% in minimum essential medium) were prepared, as described previously (5), by placing a small piece of muscle tissue in a sterile plastic bag containing an appropriate quantity of medium and homogenized (Stomacher Lab Blender; Seward Laboratory, London, United Kingdom). Homogenates were then clarified and aliquots were prepared and stored at –70°C. Original meat samples were then frozen at –70°C.
Serological analysis
Serum samples were tested for the presence of antibodies to PRRSV using a commercially available enzyme-linked immunosorbent assay (ELISA) (HerdChek; IDEXX Laboratories, Westbrook, Maine, USA). A sample-to-positive (S/P) ratio of equal or greater than 0.4 was considered as positive for the presence of PRRSV antibodies.
Polymerase chain reaction analysis
Polymerase chain reaction for the detection of PRRSV nucleic acid was performed on serum samples and meat sample homogenates. Briefly, RNA was extracted from 250 μL of meat homogenates or serum using TRIZOL LS reagent (Canadian Life Technologies, Burlington, Ontario) and RT was performed using random primers (Roche Diagnostics, Laval, Quebec) and Superscript II reverse transcriptase (Canadian Life Technologies). The PCR was carried out as previously described using open reading frame (ORF) 5 and ORF 7 primers that could detect North American or both North American and European PRRSV isolates (9,10).
Sequencing and phylogenetic analysis
The PRRSV strains identified in meat samples were sequenced over the ORF 5 region using specific primers. The PCR products were purified using a commercial kit (QIAquick PCR purification kit; Qiagen, Mississauga, Ontario) and were sequenced in both directions using standard automated sequencing methods. Briefly, nucleotide sequences and phylogenetic analyses were performed as previously described (10). The nucleotide sequences were aligned using multiple sequence alignment program (CLUSTAL W, version 1.8) (11). A phylogenetic analysis was performed on the aligned data set and a phylogenetic tree was generated using the distance based neighbor-joining method. All programs were part of the CLUSTAL X package. The predicted restriction fragment length polymorphism (RFLP) cut patterns for enzymes MluI, HincII, and SacII (12) were determined from sequences using a computer program (WEBCUTTER 2.0).
Virus isolation
Meat samples found to be PCR positive for PRRSV were further tested by virus isolation using MARC145 cells and porcine alveolar macrophages (PAM), as previously described (2,7,13). Sample homogenates diluted 1/5, 1/15, 1/45, and 1/135 in medium were inoculated in duplicate on MARC145 and PAM seeded in 96 well plates. The presence of PRRSV following 2 passages on these cells was detected by indirect immunofluorescence using PRRSV monoclonal antibody SR30 (Rural Technologies, Brookings, South Dakota, USA) reacting with both North American and European types of PRRSV.
Experimental transmissions
Eleven PCR-positive meat samples (weighing 1.05 to 1.8 kg) were used in feeding experiments. Each meat sample was fed to 2 PRRSV antibody-negative specific pathogen-free pigs of 9 wk of age. Each pig pair was housed in a separate cubicle. Each meat sample was thawed at 4°C, cut into approximately 4 equal parts, further subdivided into smaller pieces and fed to each pair of pigs on 2 consecutive days. Samples fed the 2nd d were kept at 4°C in the mean-time. To ensure thorough consumption of raw meat samples, pigs were deprived of food but not water for about 36 h prior to feeding. Normal diet was restored the following day. Experiments were conducted in 3 trials with exposure of 3, 3, and 5 PCR-positive meat samples to receiver pigs per trial, including 2 control pigs per trial. The 6 control pigs (2 per cubicle) were maintained on a normal diet throughout the experiments. Negative pressure high efficiency particulate air (HEPA)-filtered cubicles housing the control and exposed pigs were composed of a main animal room and adjacent shower and changing rooms. Biosecurity measures for personnel moving between cubicles included changing of rubber boots and rubber overalls; showering; using boot baths; and wearing disposable gloves, hats, and masks. Blood samples were taken from all pigs at arrival and on days 0, 7, 14, and 20 or 21 post exposure (dpe) to meat samples. Serum samples were tested for the presence of PRRSV antibodies and nucleic acid. Virus isolation, as previously described, was performed on PCR-positive serum samples.
Results
Slaughterhouse samples
Serologic and virologic results from blood and meat samples collected from the 2 slaughterhouses are shown in Table I. A total of 584 blood and 573 meat samples were collected from the Quebec abattoir, and 455 blood samples and 454 meat samples from the Manitoba abattoir. Seropositivity to PRRSV for all serum samples was 74.3%. Samples originating from the Quebec abattoir were 89.9% seropositive and those collected from the Manitoba slaughterhouse were 54.3% seropositive. Furthermore 45 (4.3%) of total serum samples were positive for PRRSV nucleic acid when tested by PCR. Of these, 26 (4.5%) and 19 (4.2%) serum samples originated from the Quebec and Manitoba abattoirs, respectively.
Table I.
Serologic and virologic results from blood and meat samples collected from 2 slaughterhouses, 1 in Quebec and 1 in Manitoba
| Quebec | Manitoba | Total | |
|---|---|---|---|
| Serum samples | 584 | 455 | 1039 |
| PRRSV antibody positive | 525 (89.9%) | 247 (54.3%) | 772 (74.3%) |
| PRRSV positive by PCR | 26 (4.5%) | 19 (4.2%) | 45 (4.3%) |
| Meat samples | 573 | 454 | 1027 |
| PRRSV positive by PCR | 7 (1.2%) | 12 (2.6%) | 19 (1.9%) |
PRRSV — Porcine reproductive and respiratory syndrome virus; PCR — Polymerase chain reaction
A total of 7 (1.2%) of the 573 meat samples collected in Quebec were found positive for PRRSV when tested by PCR. No virus was isolated from these 7 PCR-positive meat samples when tested on MARC145 cells and PAM. Sequence analysis of the PRRSV strains identified in pig meat indicated that 6 were field strains and 1 was vaccine-like (98% nucleotide homology with the Boehringer Ingelheim Ingelvac RespPRRS/Repro vaccine now referred to as Ingelvac PRRS MLV) (Table II). This strain presented a 2-6-2 RFLP pattern, intermediate to the characteristic 2-5-2 RFLP pattern of the vaccine strain. Of the 7 meat samples that tested positive for PRRSV by PCR, 6 originated from animals that also tested PCR positive in serum. All 7 strains were determined to be North American strains.
Table II.
Meat samples positive for porcine reproductive and respiratory syndrome virus (PRRSV) by polymerase chain reaction (PCR), corresponding restriction fragment length polymorphism (RFLP) pattern, and serologic status of pigs of origin. Animals having the same identification numbers are grouped
| Meat sample | PCR | RFLP | ELISAa |
|---|---|---|---|
| Quebec | |||
| 02VD-35 | + | 1-8-2 | + |
| 02VD-293 | + | 2-6-2 | – |
| 02VD-331 | + | 1-4-4 | + |
| 02VD-428 | + | 1-8-3 | + |
| 02VD-437 | + | 1-10-3 | + |
| 02VD-471 | + | 1-9-4 | + |
| 02VD-511 | + | 1-4-4 | + |
| Manitoba | |||
| 02VD-714 | + | 2-5-2 | + |
| 02VD-716 | + | 2-5-2 | + |
| 02VD-760 | + | 1-8-1 | + |
| 02VD-921 | + | 1-8-2 | + |
| 02VD-960 | + | 2-5-2 | – |
| 02VD-962 | + | nd | + |
| 02VD-963 | + | 2-5-2 | + |
| 02VD-964 | + | nd | + |
| 02VD-965 | + | nd | + |
| 02VD-1009 | + | 2-5-2 | – |
| 02VD-1012 | + | nd | – |
| 02VD-1014 | + | 1-8-1 | + |
nd — Sequence not determined; ELISA — Enzyme-linked immunosorbent assay
S/P ratio ≥ 0.4 is positive
Of the 454 meat samples collected from Manitoba, 12 (2.6%) were positive for PRRSV by PCR. In only 1 of these 12 meat samples could virus be isolated. In this case, virus was detected in MARC145 cells in only 1 of the 8 wells inoculated for virus isolation and, in addition, attempts to titrate virus from the original meat homogenate failed, suggesting the quantity of infectious virus in the meat sample was low. The 12 PCR-positive meat samples originated from pigs representing a total of 5 different identification numbers (tattoo numbers) suggesting that several animals may have originated from the same herd. For example, 7 of these 12 PCR-positive meat samples originated from pigs having the same identification number. Sequencing was performed on 8 of the 12 strains originating from pigs representing the 5 different identification numbers (Table II). There were 3 strains having a 99.7% homology with the vaccine (2-5-2 RFLP pattern), 2 strains having a 98.3% homology with the vaccine (2-5-2 RFLP pattern), and the 3 others were different field strains. Four strains were not sequenced because they originated from meat samples of pigs having the same identification number as the 3 strains that had an homology of 99.7% with the vaccine and 100% homology between the strains themselves. Sequencing of the only isolate obtained was 100% homologous to the strain identified in the original meat homogenate. Of the 12 meat samples tested PCR positive for PRRSV, 10 originated from animals that also tested PCR positive in serum. All 12 strains were North American.
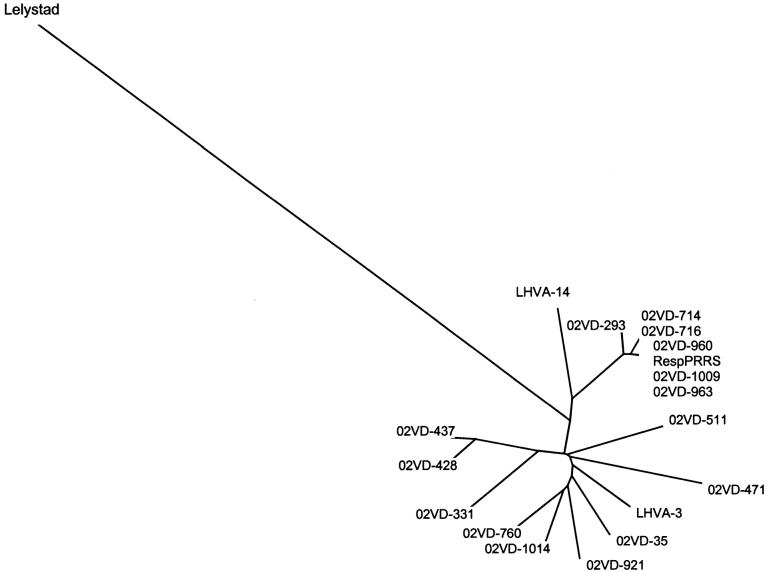
A phylogenetic tree was generated with nucleotide sequences (ORF 5 gene region) of 15 PRRSV strains identified from meat samples, of the Ingelvac RespPRRS vaccine strain, of 2 Canadian laboratory reference isolates from Quebec (LHVA-3) and British Columbia (LHVA-14) as well as of the European Lelystad isolate (Figure 1). It can be seen that all strains identified in meat samples are clustered apart from the European Lelystad isolate and that the 6 vaccine-like strains are grouped together with the RespPRRS vaccine.
Figure 1.
Phylogenetic analysis of strains identified in pig meat samples during the present study (02VD strains) along with Canadian porcine reproductive and respiratory syndrome virus (PRRSV) reference laboratory isolates from Quebec (LHVA-3) and British Columbia (LHVA-14), RespPRRS vaccine strain, and the European Lelystad PRRSV isolate. Note the close clustering of strains 02VD-714, 02VD-716, 02VD-960, 02VD-1009, 02VD-963, and 02VD-293 with the RespPRRS vaccine strain. All PRRSV strains identified in the meat samples were of the North American type.
The mean pH value taken from 502 carcasses in Quebec was 6.07 ± 0.31 and the pH values taken from the carcasses yielding the 7 PRRSV-positive meat samples ranged from 5.40 to 6.21. The mean pH value taken from 454 carcasses in Manitoba was 5.96 ± 0.36 and the pH values registered from the carcasses yielding the 12 PRRSV-positive meat samples ranged from 5.39 to 6.19. The pH values corresponding to the meat samples that resulted or did not result in infection of experimental pigs were similar (5.66 to 6.21 and 5.75 to 6.12, respectively).
Experimental transmissions
Meat samples were generally readily consumed after abundant chewing. In only a few cases were some small pieces left uneaten and later removed. Generally within 30 to 45 min all meat pieces had been eaten by the pigs. Results of PRRSV and antibody detection from pigs fed with PRRSV positive meat samples collected from Quebec are summarized in Table III. The 4 control pigs remained PRRSV negative throughout the experiments. In addition, 2 pig pairs remained negative although they had eaten pig meat samples (437 and 511) found to be positive to PRRSV by PCR. However, 4 pig pairs were found to be infected by PRRSV following ingestion of the other PCR-positive pig meat samples (35, 293, 428, and 471), as judged by the demonstration of PRRSV antibodies starting at 14 dpe, an increased S/P ratio at 21 dpe, and/or the detection of PRRSV nucleic acid in blood starting at 7 dpe. Of these 8 infected pigs, pig number 4 failed to develop detectable antibodies in the time course of the experiment but was PCR-positive at 14 and 21 dpe. None of the PRRSV-infected pigs demonstrated clinical signs of disease.
Table III.
Serologic and virologic results of pigs experimentally fed with porcine reproductive and respiratory syndrome virus (PRRSV)-positive pig meat samples collected in Quebec
| Days post exposure | ||||||
|---|---|---|---|---|---|---|
| –7 | 0 | 7 | 14 | 21 | ||
| Pig number | Sample number | P/E | P/E | P/E | P/E | P/E |
| 1 | C | –/– | –/– | –/– | –/– | –/– |
| 2 | –/– | –/– | –/– | –/– | –/– | |
| 3 | 293 | –/– | –/– | +/– | +/1.309b | +/1.596 |
| 4 | (1.55 kg)a> | –/– | –/– | –/– | +/– | +/– |
| 5 | 437 | –/– | –/– | –/– | –/– | –/– |
| 6 | (1.2 kg) | –/– | –/– | –/– | –/– | –/– |
| 7 | 35 | –/– | –/– | +/– | +/– | +/0.891 |
| 8 | (1.8 kg) | –/– | –/– | +/– | +/0.812 | +/1.229 |
| 9 | C | –/– | –/– | –/– | –/– | –/– |
| 10 | –/– | –/– | –/– | –/– | –/– | |
| 11 | 428 | –/– | –/– | +/– | +/1.171 | +/1.536 |
| 12 | (1.25 kg) | –/– | –/– | +/– | +/0.843 | +/1.232 |
| 13 | 471 | –/– | –/– | +/– | +/1.525 | +/1.862 |
| 14 | (1.25 kg) | –/– | –/– | +/– | +/1.919 | +/2.159 |
| 15 | 511 | –/– | –/– | –/– | –/– | –/– |
| 16 | (1.55 kg) | –/– | –/– | –/– | –/– | –/– |
P/E — Polymerase chain reaction (PCR)/enzyme-linked immunosorbent assay (ELISA) (positive + or negative –);
C — Control pigs
Meat sample weight
Positive ELISA sample-to-positive (S/P) ratio
Results of PRRSV and antibody detection from pigs fed with PRRSV positive meat samples collected from Manitoba are summarized in Table IV. Evidence of infection was demonstrated in 6 of the 10 exposed pigs. These pigs also demonstrated a rise in antibody titers and/or PRRSV nucleic acid in serum. One of the 2 pigs (pig number 7) exposed to meat sample 1014 remained seronegative and only at 20 dpe did this pig demonstrate PRRSV nucleic acid in serum. An example of PCR results on original meat sample 960 containing a vaccine-like strain and on the sera at days 0, 7, 14, and 20 dpe of the 2 pigs fed with this meat are shown in Figure 2. The 2 control pigs remained PRRSV negative throughout the experimental transmission. Virus could be isolated from the serum of 8 of the 14 infected pigs.
Table IV.
Serologic and virologic results of pigs experimentally fed with porcine reproductive and respiratory syndrome virus (PRRSV)-positive pig meat samples collected in Manitoba
| Days post exposure | ||||||
|---|---|---|---|---|---|---|
| –7 | 0 | 7 | 14 | 20 | ||
| Pig number | Sample number | P/E | P/E | P/E | P/E | P/E |
| 1 | C | –/– | –/– | –/– | –/– | –/– |
| 2 | –/– | –/– | –/– | –/– | –/– | |
| 3 | 921 | –/– | –/– | –/– | –/– | –/– |
| 4 | (1.1 kg)a | –/– | –/– | –/– | –/– | –/– |
| 5 | 760 | –/– | –/– | –/– | –/– | –/– |
| 6 | (1.05 kg) | –/– | –/– | –/– | –/– | –/– |
| 7 | 1014 | –/– | –/– | –/– | –/– | +/– |
| 8 | (1.1 kg) | –/– | –/– | +/– | +/– | +/0.667b |
| 9 | 960 | –/– | –/– | +/– | +/0.865 | +/1.224 |
| 10 | (1.3 kg) | –/– | –/– | +/– | +/1.157 | +/1.335 |
| 11 | 714 | –/– | –/– | +/– | +/0.849 | +/1.329 |
| 12 | (1.05 kg) | –/– | –/– | +/– | +/1.763 | +/1.916 |
P/E — Polymerase chain reaction (PCR)/enzyme-linked immunosorbent assay (ELISA) (positive + or negative –);
C — Control pigs
Meat sample weight
Positive ELISA sample-to-positive (S/P) ratio
Figure 2.
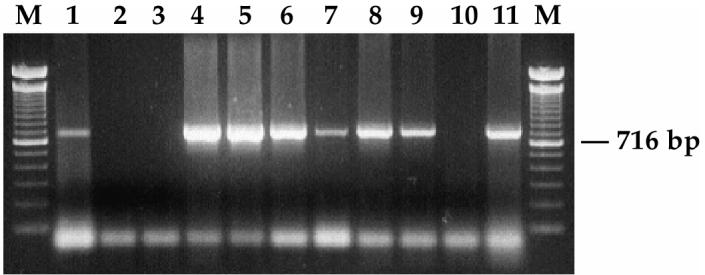
Detection by reverse transcription-polymerase chain reaction (RT-PCR) of porcine reproductive and respiratory syndrome virus (PRRSV) (open reading frame [ORF] 5) in sera collected from pigs orally exposed to PRRSV-positive meat samples. Lanes M: 100 base pairs (bp) DNA ladder; lane 1: original meat sample number 960; lanes 2 and 3: day 0; lanes 4 and 5: day 7 post exposure (pe); lanes 6 and 7: day 14 pe; lanes 8 and 9: day 20 pe; lane 10; negative control; and lane 11: positive PRRSV control.
Finally, in order to confirm infection of pigs by the PRRSV strains identified in the pig meat ingested, strains identified by PCR in serum of experimentally exposed pigs were also sequenced. Sequences of strains identified from the serum of infected pigs were homologous (99.5% to 100%) to the strains identified in the respective meat samples.
Discussion
In the present study, high PRRSV antibody prevalence in market pigs of Eastern and Western Canada was demonstrated. Although PRRSV seropositivity rates were higher in Quebec than in Manitoba samples, the global figure of 74.3% is similar to the results of a study conducted in Quebec between September and December 1993, in which 85.6% of slaughter pigs tested were PRRSV-antibody positive (2) and also to those of a more recent study from Taiwan where seropositivity rate of market pigs was 85.4% (6). The proportion of viremic market pigs from the Quebec and Manitoba abattoirs was found to be similar, averaging 4.3%. Wang (6) found that 11 of 140 (7.9%) serum samples of market pigs contained PCR detectable virus, while all of the 472 tested carcass specimens (primarily muscle, liver, and spleen) were negative. In our study, a low proportion of meat samples (1.9%) were found to be PRRSV-positive. The lower number of PRRSV-positive meat samples in comparison with the number of PRRSV-positive serum samples was expected, since it has been demonstrated by experimental infections, that the virus can be isolated from meat between 5 and 11 dpi (1–3,8), while virus in blood can be detected until at least 35 dpi using virus isolation (14–16).
Of interest in our study, 4 PCR-positive meat samples originated from PRRSV antibody negative pigs (Table II). This may suggest that these animals had been recently infected and had not yet developed detectable antibody levels. Factors, such as sensitivity of the ELISA used or the development of a poor humoral immune response in these pigs, should also be considered. Two (293 and 960) out of the 4 meat samples were fed to pigs and induced an infection.
The results of the present study indicated that low residual quantities of PRRSV may be found in a small percentage (1.9%) of pig meat samples collected at slaughterhouses. These results are in agreement with a previous study performed using virus isolation (4). By comparison, the greater number of PRRSV-positive meat samples found in this study may have resulted from the more sensitive assay used, the PCR. However, it should be noted that the overall positivity rate may have been influenced by a herd effect, since several of the Manitoba PRRSV-positive meat samples may have originated from pigs of the same herd (Table II).
In addition, the present study has demonstrated that if this PRRSV-positive pig meat is fed raw to pigs, these may become infected by PRRSV. In all but 1 PRRSV-positive meat sample tested, no PRRSV could be demonstrated by virus isolation, but by PCR only. Yet the PCR-positive meat samples could, in most cases, induce infection. These findings suggest that PRRSV infectivity is high. Indeed previous studies on PRRSV have demonstrated that as little as 20 infectious units were sufficient to achieve infection by either intramuscular or intranasal routes (17). Recently, a study demonstrated that pig meat collected from PRRSV experimentally-infected pigs could, when fed to receiver pigs, induce infection in spite of the fact that some of these meat samples had undetectable levels of PRRSV (8). In that study, freezing (at –23°C for 10 d) and thawing decreased virus titers in the majority of PRRSV infected muscle samples. In the present study, meat samples prior to feeding were kept at –70°C, which probably contributed to maintaining residual virus infectivity. This low freezing temperature was selected to optimize conditions for preservation of potential virus infectivity in muscle samples used for exposure to receiver pigs in an experimental setting. It is probably not representative of the conditions of meat storage under normal commercial arrangements.
In the present study, PRRSV-positive pig meat samples containing vaccine-like strains were also identified. These strains had RFLP patterns (or intermediate RFLP patterns) typical of the Ingelvac RespPRRS/Repro vaccine strain, demonstrated 98% to 99.7% nucleotide homology (ORF 5 region) with this vaccine and clustered around the RespPRRS vaccine strain following phylogenetic analysis (Figure 1). Furthermore, ingestion by pigs of meat samples containing these vaccine-like strains was capable of inducing infection.
The situation of meat samples 293 and 1014 is interesting since it appears to suggest transmission of PRRSV from meat-infected pigs to penmates. Indeed 1 pig fed with meat sample 293 was PRRSV serum positive starting at 7 dpe and was antibody positive at 14 and 21 dpe. The penmate remained PRRSV antibody negative throughout and was viremic only at 14 and 21 dpe. The situation resulting from exposure to sample number 1014 was similar. While 1 pig was positive for PRRSV at 7, 14, and 20 dpe and seropositive starting at 20 dpe, the penmate remained seronegative throughout and became PCR positive in serum only at 20 dpe. These results suggest that in these 2 cases the penmates may have become infected not through initial absorption of PRRSV-infected pig meat, but later through contact with the infected pig.
In summary, the results of the present study indicated that low residual quantities of PRRSV may be found in a small percentage of pig meat collected at slaughterhouses. Furthermore, pigs could be infected by PRRSV via oral exposure to this raw meat in an experimental setting designed to maximize the probability that such infection would occur.
Acknowledgments
The authors are grateful to Peter Müller, Diane Longtin, Marie-Josée Bernard, Benjamin Delisle for excellent technical assistance; to Avila Croisetière and René Mineau for animal care; to Olymel in St-Valérien and to Maple Leaf in Winnipeg for their participation in supplying the meat samples; and to the Olymel and Maple Leaf staff for their collaboration throughout the project. This project was made possible by the financial support of Canada Pork International through the Matching Investment Initiative Program of the Canadian Food Inspection Agency.
Footnotes
Dr. Magar’s current address is Veterinary Biologics Section, Canadian Food Inspection Agency, 59 Camelot Drive, Ottawa, Ontario K1A 0Y9.
References
- 1.Bloemraad M, de Kluijver EP, Petersen A, et al. Porcine reproductive and respiratory syndrome: temperature and pH stability of Lelystad virus and its survival in tissue specimens from viraemic pigs. Vet Microbiol. 1994;42:361–371. doi: 10.1016/0378-1135(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 2.Magar R, Robinson Y, Dubuc C, Larochelle R. Evaluation of the persistence of porcine reproductive and respiratory syndrome virus in pig carcases. Vet Rec. 1995;137:559–561. doi: 10.1136/vr.137.22.559. [DOI] [PubMed] [Google Scholar]
- 3.Mengeling WL, Lager KM, Vorwald AC. Diagnosis of porcine reproductive and respiratory syndrome. J Vet Diagn Invest. 1995;7:3–16. doi: 10.1177/104063879500700102. [DOI] [PubMed] [Google Scholar]
- 4.Frey ML, Landgraf JG, Schmitt BJ, et al. Recovery of porcine reproductive and respiratory syndrome virus from tissues of slaughter weight pigs. Second Int Symp On Porcine Reproductive And Respiratory Syndrome (PRRS). 1995:28.
- 5.Larochelle R, Magar R. Evaluation of the presence of porcine reproductive and respiratory syndrome virus in packaged pig meat using virus isolation and polymerase chain reaction (PCR) method. Vet Microbiol. 1997;58:1–8. doi: 10.1016/s0378-1135(97)00150-8. [DOI] [PubMed] [Google Scholar]
- 6.Wang F-I. Minimal residues of porcine reproductive and respiratory syndrome virus in pig carcases and boar semen. Proc Natl Sci Counc ROC (B) 1999;23:167–174. [PubMed] [Google Scholar]
- 7.Magar R, Robinson Y, Dubuc C, Larochelle R. Isolation and experimental oral transmission in pigs of a porcine reproductive and respiratory syndrome virus isolate. In: Talbot PJ, Levy GA, eds. Corona and Related Viruses. New York, Plenum Press, 1995:139–144. [DOI] [PubMed]
- 8.Martin RG, Steverink PJGM. Oral transmission of PRRS virus via the feeding of infected muscle to pigs. Proc Am Assoc Swine Vet 2002:45.
- 9.Larochelle R, Magar R, D’Allaire S. Comparative serologic and virologic study of commercial swine herds with and without postweaning multisystemic wasting syndrome. Can J Vet Res. 2003;67:114–120. [PMC free article] [PubMed] [Google Scholar]
- 10.Larochelle R, D’Allaire S, Magar R. Molecular epidemiology of porcine reproductive and respiratory syndrome virus (PRRSV) in Québec. Virus Res. 2003;96:3–14. doi: 10.1016/s0168-1702(03)00168-0. [DOI] [PubMed] [Google Scholar]
- 11.Thompson JD, Gibson TJ, Plewniak F, et al. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wesley RD, Mengeling WL, Lager KM, et al. Differentiation of a porcine reproductive and respiratory syndrome virus vaccine strain from North American field strains by restriction fragment length polymorphism analysis of ORF 5. J Vet Diagn Invest. 1998;10:140–144. doi: 10.1177/104063879801000204. [DOI] [PubMed] [Google Scholar]
- 13.Magar R, Larochelle R, Robinson Y, Dubuc C. Immunohistochemical detection of porcine reproductive and respiratory syndrome by immunohistochemistry using colloidal gold. Can J Vet Res. 1993;57:300–304. [PMC free article] [PubMed] [Google Scholar]
- 14.Wills RW, Zimmerman JJ, Yoon KJ, et al. Porcine reproductive and respiratory syndrome virus: a persistent infection. Vet Microbiol. 1997;55:231–240. doi: 10.1016/s0378-1135(96)01337-5. [DOI] [PubMed] [Google Scholar]
- 15.Chang CC, Yoon KJ, Zimmerman JJ, et al. Evolution of porcine reproductive and respiratory syndrome virus during sequential passages in pigs. J Virol. 2002;76:4750–4763. doi: 10.1128/JVI.76.10.4750-4763.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horter DC, Progranichniy RM, Chang CC, et al. Characterization of the carrier state in porcine reproductive and respiratory syndrome virus infection. Vet Microbiol. 2002;86:213–228. doi: 10.1016/s0378-1135(02)00013-5. [DOI] [PubMed] [Google Scholar]
- 17.Yoon KJ, Zimmerman JJ, Chang CC, et al. Effect of challenge dose and route on porcine reproductive and respiratory syndrome virus (PRRSV) infection in young swine. Vet Res. 1999;30:629–638. [PubMed] [Google Scholar]