Abstract
The cell is a crowded volume, with estimated mean mass percentage of macromolecules and of water ranging from 7.5 to 45 and 55 to 92.5 %, respectively. However, the concentrations of macromolecules and water at the nanoscale within the various cell compartments are unknown. We recently developed a new approach, correlative cryo-analytical scanning transmission electron microscopy, for mapping the quantity of water within compartments previously shown to display GFP-tagged protein fluorescence on the same ultrathin cryosection. Using energy-dispersive X-ray spectrometry (EDXS), we then identified various elements (C, N, O, P, S, K, Cl, Mg) in these compartments and quantified them in mmol/l. Here, we used this new approach to quantify water and elements in the cytosol, mitochondria, condensed chromatin, nucleoplasm, and nucleolar components of control and stressed cancerous cells. The water content of the control cells was between 60 and 83 % (in the mitochondria and nucleolar fibrillar centers, respectively). Potassium was present at concentrations of 128–462 mmol/l in nucleolar fibrillar centers and condensed chromatin, respectively. The induction of nucleolar stress by treatment with a low dose of actinomycin-D to inhibit rRNA synthesis resulted in both an increase in water content and a decrease in the elements content in all cell compartments. We generated a nanoscale map of water and elements within the cell compartments, providing insight into their changes induced by nucleolar stress.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-013-1267-7) contains supplementary material, which is available to authorized users.
Keywords: Water, Ions, Nucleus, Nucleolus, Stress, Correlative imaging
Introduction
Macromolecules are very abundant in all cell compartments, which have total protein and RNA concentrations in the range of 275–450 mg/ml [1]. In the nucleus, macromolecule concentration ranges between 75 in nucleoplasm and 400 mg/ml in condensed chromatin [2, 3]. Due to their specific volume of about 1 ml/g, macromolecules occupy 20–45 % of the total volume of a cell [1, 4]. This high-volume occupancy, which is well illustrated in drawings [5], computer simulations [6], and on cryoelectron tomograms [7], induces molecular crowding [1] also referred to as the excluded volume effect. This effect modifies the activity of many macromolecules [1, 8, 9] and is dependent on small variations in the amount of water present [4]. It is therefore important to take the water content of cell compartments into account when studying molecular crowding. Most of the water in mammalian cells is sequestered by the macromolecular components of the cell. This osmotically unresponsive water accounts for 31–92 % of the total water content in various cell types [10]. Proteins are less hydrated than nucleic acids and DNA is less hydrated than RNA [11, 12]. Hydration modifies the stabilization and folding of nucleic acids [13] and is important for sequence recognition by DNA-binding proteins [11]. The structure of nucleic acids also depends on monovalent and divalent anions [3, 13, 14].
Despite the critical roles of water and ions in the organization and function of proteins and nucleic acids [11, 15–18], only a few studies have considered their levels and distribution in the cytoplasm and nucleus. In some of these studies, macromolecules were identified and quantified within purified cell compartments, whereas in others, the dry mass of cell compartments was quantified at low resolution by interference microscopy [19]. Several approaches have been used to achieve higher-resolution analyses: (1) quantitative scanning transmission electron microscopy (STEM) for the measurement of DNA packing density within condensed chromatin [20], (2) secondary ion mass spectroscopy (SIMS) [14, 21] and energy-dispersive X-ray spectrometry (EDXS) for elementary analysis in cryo-fixed specimens [22].
However, the simultaneous quantification of water and the principal elements present has never been performed in the cytoplasm and nucleus and this lack of information limits our understanding of the cell activity.
We have shown [23–25] that dark-field cryo-STEM can be used to quantify the mass percentages of water, dry matter, and elements in each pixel of classically freeze-dried ultrathin sections of vitrified cells [22]. However, the unequivocal identification of nuclear compartments is difficult in these conditions, due to the absence of a limiting membrane. It is thus difficult to identify clumps of condensed chromatin, uncondensed chromatin, and tangentially sectioned nucleoli. We recently overcame this limitation by developing a new correlative light and electron microscopy imaging approach combining the observation of a given cryo-ultrathin section by fluorescence microscopy (for the localization of nuclear GFP-tagged protein) and by cryo-STEM (for the nanoscale quantification of water and elements) [26].
Here, we applied this new approach to stably transfected HeLa cell lines producing H2B-GFP or NOP52-GFP, for identification of chromatin and the nucleolus, respectively [27, 28]. We first calculated water and element concentrations at the nanoscale in control cells, in: (1) chromatin identified on the basis of fluorescence, either in nuclear compartments (condensed and uncondensed chromatin) or in metaphase chromosomes, (2) nucleoli (and their subcompartments), identified on the basis of their ultrastructure and (3) mitochondria (identified on the basis of their cristae) and the surrounding cytosol. This led to generation of the first nanoscale map of water and elements within the nuclear and cytoplasmic compartments. We then investigated the changes to these water and elements concentrations induced by nucleolar stress resulting from a rapid shutdown of ribosome biogenesis following the inhibition of rRNA synthesis by actinomycin D [29–31].
Results
Nanoscale map of water and ions within control cells
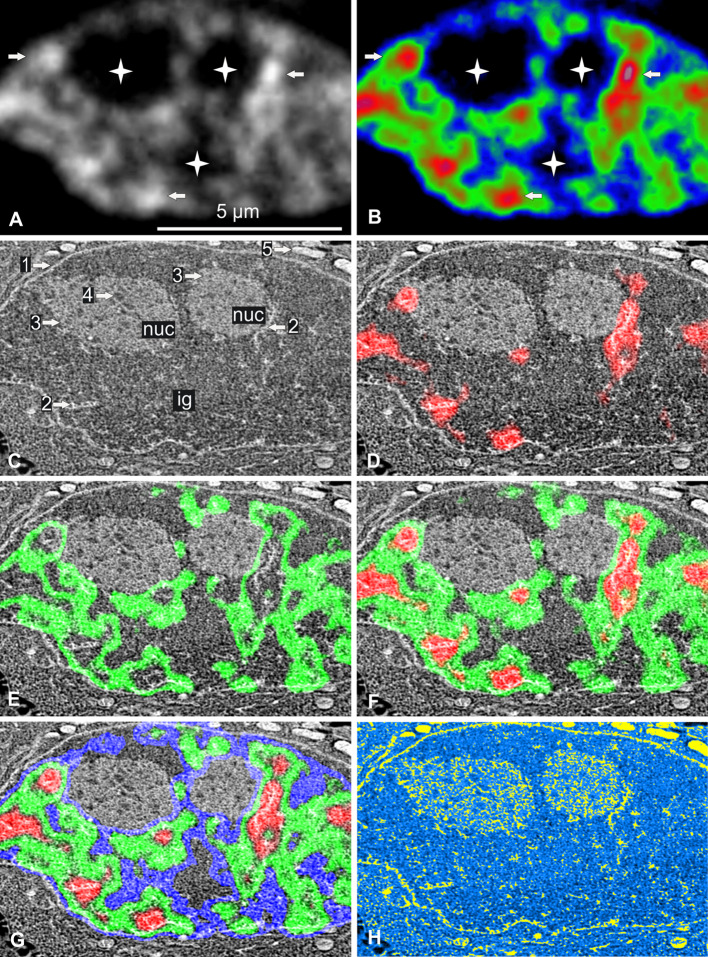
We fixed living H2B-GFP cells without cryoprotectant or chemical fixation by plunging them directly in liquid ethane, as previously described [26]. Ultrathin cryo-sections, 85 nm thick, were cut and collected on a formvar- and carbon-coated indexed grid, which was placed directly and definitively in the EM cryo-holder. By avoiding the need for further careful sample grid manipulation, our approach greatly improved existing cryo-correlative methods [32, 33]. The cryo-holder was inserted into a dedicated dewar, which was then placed on the stage of an epifluorescence microscope that could be kept at −170 °C for at least 1 h. Images from a nucleus and the surrounding cytoplasm are provided as an illustration of the results of this technique (Fig. 1). Nuclear fluorescence, as indicated by gray-scale levels (Fig. 1a) or color-coded in red, green, and blue (Fig. 1b), revealed the presence of both rounded and elongated foci of condensed chromatin (arrows), surrounded by large irregular areas of uncondensed chromatin. Some broad areas were entirely devoid of fluorescence (stars). The EM cryo-holder was transferred directly to the STEM and the sample was slowly freeze-dried for 1 h by allowing the temperature to rise to −80 °C. It was then cooled again and observed at −170 °C. The same nucleus was observed by STEM (Fig. 1c), which revealed the nuclear envelope (arrow 1) and several high-contrasted narrow elongated (arrow 2) and large roundish nuclear domains (arrow 3). We determined the precise location of chromatin within this STEM image by merging the image obtained with the color-coded fluorescence intensity image previously obtained (Fig. 1d–g). The fluorescence and STEM images were perfectly aligned due to the absence of differential shrinkage during freeze-drying. We found that: (1) condensed chromatin was present in the narrow, elongated, high-contrast domains identified by STEM, (2) uncondensed chromatin surrounded these domains, and (3) the areas without chromatin corresponded to structures that we identified as interchromatin granules (ig) and nucleoli (nuc). We were also able to identify nucleolar compartments, such as fibrillar centers (arrow 4) surrounded by a mixture of dense fibrillar and granular components. In the cytoplasm, mitochondria (arrow 5) are clearly identified at higher magnification (results not shown).
Fig. 1.
Correlative cryo-analytical scanning transmission electron microscopy of a control HeLa H2B-GFP interphase cell. a, b Fluorescence mode: a fluorescence intensity imaged as gray-scale levels or b coded in red, green, and blue for high, medium, and low fluorescence intensity, respectively. Chromatin is readily detectable within this 85-nm ultrathin cryo-section. Higher fluorescence intensities identify both rounded and elongated foci of condensed chromatin (arrows) surrounded by large irregular areas of uncondensed chromatin. Several broad areas displayed no fluorescence (stars). c Dark-field STEM image of the same freeze-dried cell. The nuclear envelope (arrow 1), several high-contrast narrow elongated structures (arrow 2), large roundish nuclear domains (arrow 3) containing tiny roundish structures with low levels of contrast (arrow 4) and mitochondria (arrow 5) can be seen. d–g Merging of the STEM image with the fluorescence intensity image: d high (red); e medium (green); f high and medium (red and green); g high, medium, and low intensity (red, green, blue). The fluorescence and STEM images were perfectly aligned, demonstrating an absence of differential shrinkage during freeze-drying. This makes it possible to identify clumps of condensed chromatin (arrow 2). The areas with no fluorescence correspond to two nucleoli (3, nuc) (containing fibrillar centers (arrow 4)) and interchromatin granules (ig). h Parametric water content image computed from the STEM image. Values in % are coded in yellow (0–50 %) and on a linear gradient ranging from light to dark blue (51–100 %). Mitochondria were the least hydrated organelles of the cell. In the nucleus, condensed chromatin (previously identified by fluorescence) was found in both narrow clumps and filaments, which, together with part of the nucleolus, were the least hydrated domains of the nucleus. Nucleolar fibrillar centers and some domains of the nucleoplasm contain the largest amounts of water. Scale bar 5 μm
We then quantified water content (in %) for each pixel (20 nm in size) of the STEM image. We computed parametric images for water content, in which pixels containing 0–50 % water are shown in yellow and those containing 51–100 % water are shown as a linear gradient ranging from light to dark blue (Fig. 1h). The less hydrated cell compartments (yellow and light blue) were identified directly as: (1) mitochondria, (2) narrow clumps of chromatin and filaments (previously identified by fluorescence) and (3) part of the nucleolus. The more highly hydrated cell compartments (dark blue) were identified as nucleolar fibrillar centers (FCs) and particular domains of the nucleoplasm. Interestingly, this image demonstrates that water content may differ considerably between compartments located close together.
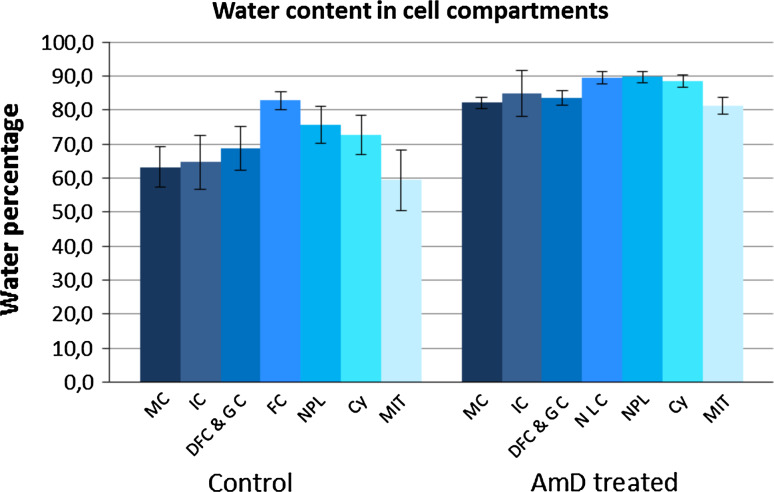
We used these parametric images to calculate water and dry mass content within regions of interest (ROI) of defined cell domains within large numbers of cells analyzed in triplicate: chromatin in mitotic chromosomes and in interphase condensed clumps, nucleolar components (fibrillar centers and surrounding dense fibrillar and granular components), nucleoplasm (nuclear areas that are neither nucleoli nor condensed chromatin), cytosol and mitochondria. Water content was higher (Fig. 2, control) in: (1) nucleoplasm (75.7 %) than in narrow clumps of interphase condensed chromatin (64.8 %) or the chromatin of mitotic chromosomes (63.2 %), (2) FCs (82.9 %) than in dense fibrillar and granular components (68.8 %) and (3) the cytosol (72.7 %) than in mitochondria (59.4 %).
Fig. 2.
Water content was determined in control and actinomycin D-treated HeLa H2B-GFP cells. Quantification was performed for regions of interest in several compartments: MC (mitotic chromatin); IC (interphase chromatin); DFC/GC (nucleolar dense fibrillar component and granular component); FC/NLC (nucleolar fibrillar centers and nucleolar light caps); NPL (nucleoplasm); CY (cytosol); MIT (mitochondria). The data shown are mean ± SD from triplicate analyses (n = 80 and 25 for interphase and mitotic control cells, respectively; n = 33 and 5 for interphase and mitotic AMD-treated cells, respectively)
We were able to determine the quantities of both water and dry matter in a region of interest, making it possible to calculate a hydration index “HI”, corresponding to the ratio of the percentage water to the percentage dry matter in the region considered (Online Resource 1). This HI is similar to the “h” index of proteins [34] and facilitates efficient comparisons of hydration between cell compartments. The HI of the mitochondria, at about 1.5, was the smallest of all the cell compartments considered. The HI values for: (1) condensed chromatin (mitosis and interphase) and both dense fibrillar and granular components of the nucleolus were about 1.2–1.5 times higher, and those of the nucleoplasm and cytosol was about twice as high, whereas the HI of the FCs was around three times higher than that of the mitochondria.
One great advantage of the correlative method we developed [26] is that it can be used for a targeted elementary analysis in exactly the same regions as those in which water and dry mass are quantified. Thus, after the quantification of water and dry mass, regions of interest (ROI) can be identified either by comparison with fluorescence imaging (see above) or on the basis of ultrastructure for elementary analysis by EDXS. ROI were drawn within mitotic chromatin, interphase chromatin, the nucleolus (dense fibrillar plus granular components on the one hand and FCs on the other), the nucleoplasm, cytosol, and mitochondria. The concentrations of nitrogen (N) (within proteins, nucleic acids, and nucleotides), phosphorus (P) (mostly in nucleic acids and nucleotides), sulfur (S), potassium (K), chloride (Cl), and magnesium (Mg) were finally calculated in mmol/l, taking the previously measured water content into account.
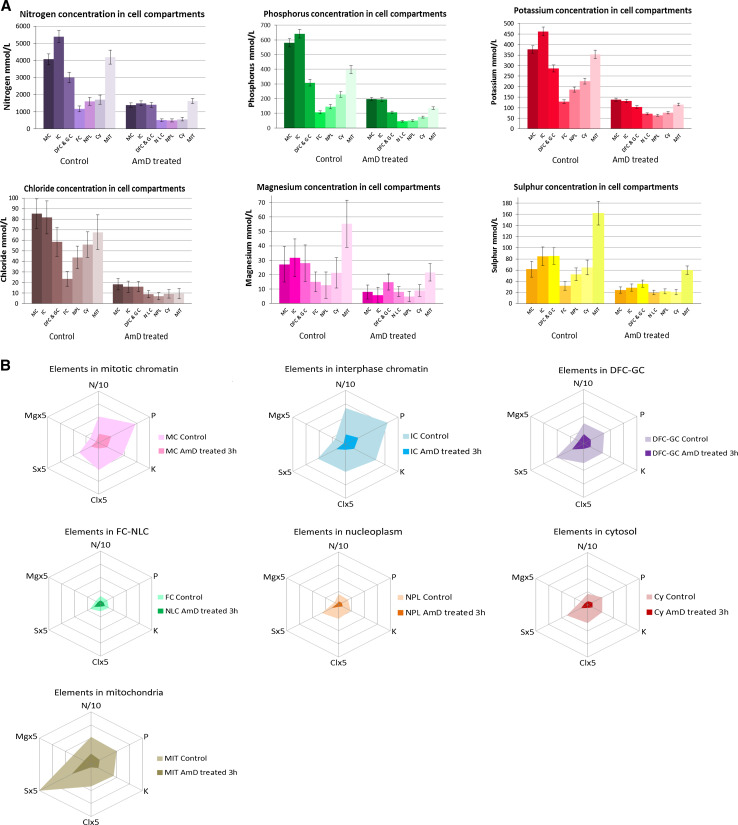
The concentrations of the elements (Fig. 3a; Online Resource 2) ranged from 1,163 to 5,390 mmol/l for N, 106 to 642 mmol/l for P, 128 to 462 mmol/l for K, 23 to 85 mmol/l for Cl, 32 to 162 mmol/l for S, and 13 to 55 mmol/l for Mg. Chromatin (during mitosis or interphase) contained the highest concentration of N, P, K, and Cl, with most other compartments containing intermediate concentrations of these elements. The FCs, nucleoplasm and cytosol contained very low concentrations of each of the elements considered and the mitochondria contained between two and five times more S and Mg than the other compartments. Large differences in K, Mg, and Cl content were also demonstrated between contiguous compartments. Condensed chromatin contained about 2.5, 2.4 and 1.9 times more K, Mg and Cl, respectively, than the nucleoplasm, whereas the fibrillar/granular nucleolar components contained 1.5, 2.1, and 1.3 times more K, Mg, and Cl, respectively, than the FCs. Furthermore, the K/P, Cl/P, Mg/P, and K + Cl + Mg/P ratios were low in nuclear compartments with a low hydration index (condensed chromatin and dense fibrillar/granular nucleolar components) and high in compartments with a high hydration index (FC and nucleoplasm).
Fig. 3.
We identified several elements (C, O, N, P, S, K, Cl, Mg) and we quantified them by EDXS in control and actinomycin D-treated HeLa H2B-GFP cells. We analyzed regions of interest in several compartments: MC (mitotic chromatin); IC (interphase chromatin); DFC/GC (nucleolar dense fibrillar component and granular component); FC/NLC (nucleolar fibrillar centers and nucleolar light caps); NPL (nucleoplasm); CY (cytosol); MIT (mitochondria). The data are mean ± SD from triplicate analyses (n = 80 and 25 for interphase and mitotic control cells, respectively; n = 33 and 5 for interphase and mitotic AMD-treated cells, respectively). Data are presented: (1) as histograms (Fig. 3a), for comparison of the concentration of each element between compartments, (2) as spiderweb diagrams (Fig. 3b), to provide an “element descriptor” for each compartment
We then compiled the data for each cell compartment and plotted them on a single spiderweb diagram (Fig. 3b, light colors) to obtain an “element descriptor” of the compartment concerned. As we used the same scale for all these spiderweb diagrams, they are comparable and can be used to identify similarities and differences between compartments. Interphase chromatin appears to be the compartment with the highest levels of all investigated elements (N, P, K, Cl, S and Mg), and its element descriptor differs from that of mitotic chromatin. The element descriptor of nucleolar DFC and GC is markedly different from that of FCs (the smallest of all compartments), whereas those of the nucleoplasm and cytosol are very similar. Finally, the element descriptor of the mitochondria is unique, with high S and Mg levels.
Nanoscale map of water and elements within stressed cells
Having studied the water and element contents of control cells, we then looked at changes in these contents in stressed cells (i.e., cells not engaged in apoptosis but with major changes to several metabolic activities). We treated cells with a low dose of actinomycin D (AMD), which is known to inhibit rRNA synthesis [30] inducing a rapid shut-down of nucleolar ribosome biogenesis [35, 36] and a clear segregation of nucleolar components [37] in an active process [38]. AMD also simultaneously affects cytoplasmic metabolic activities, by inhibiting mitochondrial RNA polymerases [39], respiration, and glycolysis, resulting in a large decrease in ATP concentration [40].
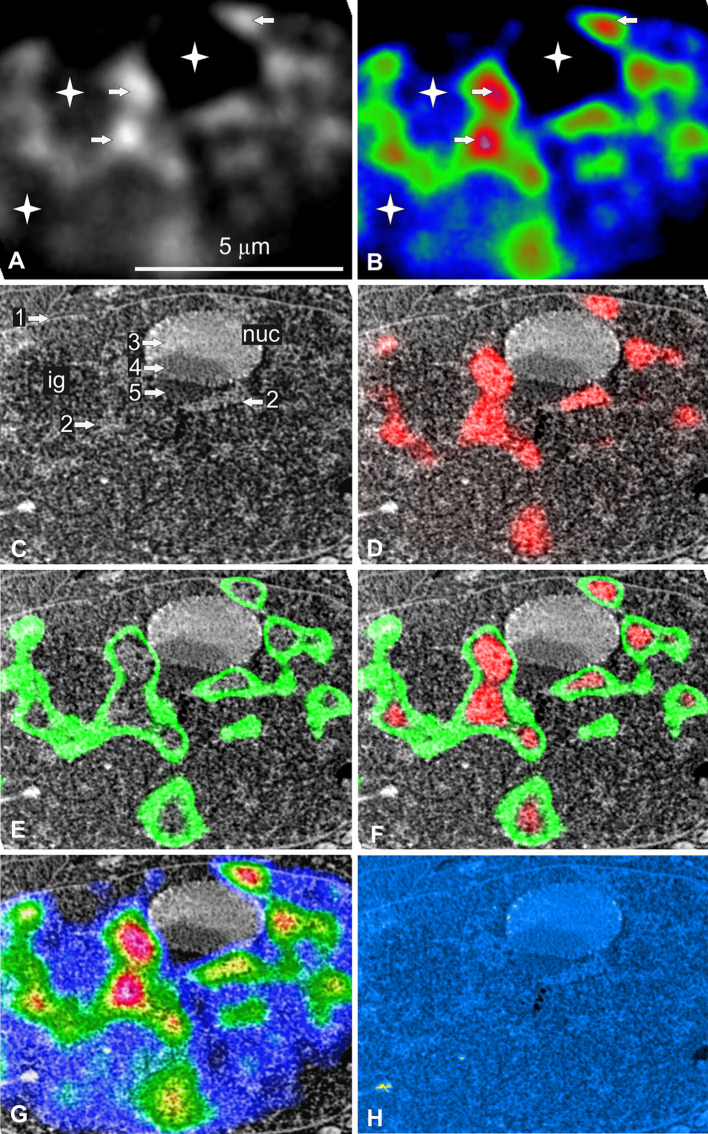
We treated the cells with AMD for 3 h. We imaged living cells by time-lapse confocal microscopy and demonstrated that AMD treatment induced nucleolar components segregation but no change in nuclear volume (3D reconstruction and quantification, results not shown). We then immuno localized the p-53 protein in both control and AMD-treated cells. We found that this protein was present in small amounts in the cytoplasm of control cells and that it was not translocated in the nucleus in response to the nucleolar stress induced by AMD. These results confirm that p53 is present at low levels in HeLa cells and show that, as expected, it is ineffective due to the presence of the HPV-18 virus [41, 42]. We then demonstrated that these cells were not engaged in apoptosis by showing that neither activated caspase 3 nor cleaved PARP was present in the cytoplasm and nucleus (results not shown). Finally, we processed the AMD-treated cells by our cryo-correlative light microscopy and STEM approach. The nuclei of AMD-treated cells differed from those of control cells (Fig. 4) in two ways: (1) they contained regular ovoid nucleoli with typically segregated components and (2) they contained more numerous, larger clumps of condensed chromatin.
Fig. 4.
Correlative cryo-analytical scanning transmission electron microscopy of a HeLa H2B-GFP interphase cell treated with AMD for 3 h. a, b Fluorescence mode: a fluorescence intensity imaged as gray-scale levels or b coded in red, green, and blue for high, medium, and low fluorescence intensity, respectively. High fluorescence intensity identifies both rounded and elongated foci of condensed chromatin (arrows) surrounded by large irregular areas of uncondensed chromatin. Several broad areas were devoid of fluorescence (stars). c Dark-field STEM image of the same freeze-dried nucleus. The nuclear envelope (arrow 1) can be identified, along with several high-contrast irregular and elongated structures (arrow 2), a smooth ovoid nuclear domain (nuc) containing three well-delineated areas with different degrees of contrast (3, 4, and 5). d–g Merging of the STEM and fluorescence intensity images: d high (red); e medium (green); f high and medium (red and green); g high, medium, and low intensity (red, green, blue). Fluorescence and STEM images were perfectly aligned due to the absence of differential shrinkage during freeze-drying. The fluorescence and STEM images can therefore be merged, making it possible to identify clumps of condensed chromatin (arrow 2). The ovoid structure is a nucleolus with the following segregated components: granular (arrow 3), dense fibrillar (arrow 4), and nucleolar light caps (arrow 5). One area without fluorescence contained interchromatin granules (ig). h Parametric water content image computed from the STEM image. Values in % are coded in yellow (0–50) and a linear gradient ranging from light to dark blue (51–100 %). The clumps of condensed chromatin (previously identified by fluorescence) and nucleolar granular components (arrow 3) were the least hydrated domains of the nucleus. Water content was higher in the nucleolar light caps (arrow 5) and some domains of the nucleoplasm contained the largest amounts of water. Scale bar, 5 μm
AMD treatment resulted in a global increase in water content in all cell compartments (Fig. 2, AMD-treated; Online Resource 1). However, water content differed between the cell compartments. It was higher in: (1) the nucleoplasm (90.7 %) than in narrow clumps of interphase condensed chromatin (84.8 %) and the chromatin of mitotic chromosomes (82.2 %), (2) light nucleolar caps (90.7 %) than in dense fibrillar/granular components (84.8 %) and (3) the cytosol (89.1 %) than the mitochondria (81.2 %). We then calculated the hydration index (HI) of the main cell compartments. The HI of treated cell compartments was two to three times higher than that in control cells (range 4–9). Mitochondria and mitotic chromatin had the smallest HI of any of the cell compartments (HI around 4). The HI values of interphase chromatin and of the dense fibrillar/granular components of the nucleolus were about 1.3–1.5 higher than that of mitotic chromatin, whereas those of the nucleoplasm, cytosol, and nucleolar light caps were about twice that of mitotic chromatin.
We then carried out targeted EDXS analysis in exactly the same compartments to calculate the concentrations of the various elements considered (Fig. 3a, b; Online Resource 2) in mmol/l. The concentrations of all elements in all cell compartments were lower after AMD treatment than in the absence of such treatment and ranged from 502 to 1,624 mmol/l for N, 45 to 196 mmol/l for P, 64 to 138 mmol/l for K, 7 to 18 mmol/l for Cl, 20 to 60 mmol/l for S, and 5 to 22 mmol/l for Mg.
However, the relative decrease differed between these elements (Online Resource 2). The concentration of Cl decreased more strongly than for the other elements considered by factors of 2.5 (Nucleolar Light Components, NLC) to 7.5 (mitochondria). Moreover, the decrease in Mg concentration was greater for interphase chromatin than for the nucleolus and nucleoplasm (decrease by factors of 5.3, 1.8, and 2.6, respectively) and the decreases in Cl, Mg and K concentrations were much smaller for the NLC than for the other compartments. We also found that the ratios of each of the three elements (K, Cl, and Mg) to phosphorus content varied differently with respect to the control. Cl/P ratio decrease in all cell compartments with the exception of NLC in response to nucleolar stress, whereas no change in the K/P ratio was observed.
The changes in content of the elements studied in response to AMD stress were then studied by comparing element descriptors (Fig. 3b, dark colors). Major changes with respect to control conditions were observed [for each compartment, compare light (control) and dark colors (AMD)]. Thus, the element descriptors for interphase and mitotic chromatin, nucleolar DFC-GC and mitochondria were similar and were the largest. Conversely, those of NLC, nucleoplasm and the cytosol were the smallest.
Finally, we analyzed another stably transfected HeLa cell line producing NOP52-GFP [28], which is involved in the late stages of rRNA processing and can be used for targeted fluorescence imaging of the dense fibrillar/granular components the nucleolus. It was not possible to identify condensed chromatin (absence of specific fluorescence) and fibrillar centers in the nucleolus (too small) unambiguously in this cell line, but we were able to analyze: dense fibrillar/granular components in the nucleolus, nucleolar light components after segregation induced by AMD treatment, nucleoplasm (non-condensed areas of the nucleus), cytosol, and mitochondria in control and AMD-treated cells. Overall, we confirmed the results obtained in the HeLa cell-line producing H2B-GFP in terms of water and element contents and the changes in these contents induced by nucleolar stress. (Online Resource 3 to 7).
Discussion
It is now accepted that water is a very important but underestimated molecule in cell biology [11, 15, 43, 44], playing an essential role in the organization of functional proteins and macromolecular complexes [8, 45]. However, little is known about the water content of defined cell compartments, and this limits our understanding of cell functions. For example, it was recently suggested that quantification of the water and ions present in the nucleus is an essential perquisite for improvements in our understanding of the function and organization of this organelle [16, 17].
To our knowledge, this study is the first to determine water and elements contents simultaneously in the main cell compartments at the nanoscale level. Improvements to existing correlative cryo methods [32, 33] were required to achieve this [26].
In this study, we first calculated the percentages of water and of dry mass in the various cell compartments, which were perfectly identified by correlative imaging (in stably transfected HeLa cell lines producing H2B-GFP or NOP52-GFP for identification of chromatin and the nucleolus, respectively) or on the basis of ultrastructure (membrane-limited organelles). For each cell compartment, we calculated a hydration index (HI), corresponding to the ratio of water content to dry mass. This ratio is similar to the classical “h” index for proteins [10] and facilitates data comparison. We found that the nuclear domains contained between 60 and 83 % water (HI range 1.4–4.8), values below previous estimates [2, 3]. This discrepancy may be accounted for by improvements in sample preservation due to direct cryofixation and the higher sensitivity of our approach than of chemical assays or interference microscopy. Thus, dry mass content is between 170 and 400 mg/ml, these values being higher than previous estimates for mammalian cells (65–220 mg/ml). This suggests that the so-called “macromolecular crowding” effect is stronger than previously thought [2, 3], with potential implications such as higher levels of thermodynamic activity, more limited molecular diffusion and changes to chromatin packing, and the organization of compartments [4, 46]. However, even condensed chromatin was found to contain only 35 % dry mass, consistent with previous findings suggesting that condensed chromatin does not constitute a physical barrier to the diffusion of transcription factors [47]. The nucleoplasm and nucleolar FCs, which we found to be the most hydrated nuclear compartments, have been shown to be accessible to the smallest dextran molecules [48], suggesting that hydration and the diffusion of molecules are directly linked. Finally, we found that contiguous compartments had very different water contents, consistent with a sponge-like organization within which molecules diffuse [49]. For example, condensed chromatin contains about 65 % water (HI = 1.8), whereas the surrounding nucleoplasm contains 75 % water (HI = 3.1). The same is true for the nucleolar compartments. Thus, the water content of FCs, at about 83 % (HI = 4.8), is much higher than that of dense fibrillar/granular components: at about 69 % (HI = 2.2). As FCs are the most hydrated compartments of the nucleolus and are known to contain actively transcribed uncondensed rDNA genes for rRNA synthesis [50], hydration may serve as a marker of active genes.
We also found that the water content of the cytosol (72 %; HI = 2.6) was close to that of nucleoplasm, and we confirmed that mitochondria were the least hydrated organelles (about 60 %; HI = 1.4) [45].
By combining simultaneous water content and elementary analyses, we were able to determine the physiological concentrations of several elements (expressed in mmol/l) within all cell compartments in situ. This represents a major improvement on classical studies involving quantification of the concentration of either a single element (such as ions) at a low spatial resolution with fluorescent indicators [51] or several elements calculated per kilogram of dry mass by classical EDXS on freeze-dried ultrathin sections [52, 53]. Here, it must be stressed that our approach cannot discern free from bound elements and water. Consequently, what we measured in a region of interest (ROI) is the quantity of a given element (free and bound) and the quantity of free and bound water. We then used these quantities to calculate the concentration of the element in mmol/l within the ROI. Finally, these concentrations do not reflect those in buffers employed to stabilize isolated organelles as stated by Schnell and Hancock for the nucleus [3].
We generated original data on the concentrations of nitrogen (N), phosphorus (P), and sulfur (S) within hydrated cell compartments in situ. Mitochondria were found to contain the highest sulfur concentration, probably due to their high glutathione (GSH and GSS) content [54]. We also showed that N and P were heterogeneously distributed. Their concentrations were: (1) highest in chromatin and mitochondria, due to their high protein, nucleic acid, and nucleotides content, (2) twice as high in the mitochondria as in the cytosol, (3) three times higher in condensed chromatin than in the nucleoplasm which contains both uncondensed chromatin and RNA and (4) about threefold higher in nucleolar dense fibrillar and granular components (DFC/GC) than in fibrillar centers (FC).
We focused on the K, Cl, and Mg ions, because these elements were the most abundant in both control and AMD-treated cells, the concentrations of Na and Ca being at the limit of detection in our experimental conditions. Potassium was the most abundant ion in all compartments, accounting for about 75–80 % of all ions [14, 53]. In hydrated cell compartments, the total concentrations of K, Cl, and Mg in more compact compartments were more than twice those in contiguous less compact compartments: condensed chromatin and nucleoplasm or DFC/GC and FC in the nucleolus, respectively. The ratios of each ion to phosphorus were low for nuclear compartments with a low hydration index (condensed chromatin and DFC/GC nucleolar components) and high for compartments with a high hydration index (FCs and nucleoplasm). However, no such relationship was observed for the cytosol and mitochondria. Our results suggest that low and high ratios of K, Cl, and Mg content to P content may be characteristic of condensed chromatin (clumps of interphasic chromatin and mitotic chromosomes) and uncondensed chromatin (fibrillar centers and nucleoplasm), respectively.
We then investigated the consequences of stress induced by a low dose of actinomycin D (AMD). We found that AMD treatment induced a large increase in the water content of nuclear and cytoplasmic compartments. The relative water content of contiguous compartments remained similar to that in control cells. One consequence of the increase in water content was a strong increase in the hydration index of all cell compartments. It is unclear whether this increase in water content resulted from an increase in bulk water or an increase in the hydration water of proteins and nucleic acids. Changes in salt concentrations and pH are known to induce changes in protein conformation and hydration [55], which have many consequences for molecular crowding and enzymatic reaction rates [1, 4, 9]. These modifications of cell parameters may result from the many consequences of AMD treatment. AMD is a well-known “nucleolar stressor” [29, 56] that rapidly induces: (1) the inhibition of rRNA synthesis [30], (2) the segregation of nucleolar components [37] and (3) the disruption of ribosome biogenesis. It has been estimated that one HeLa cell imports 560,000 ribosomal proteins per minute, generating about 14,000 ribosomal subunits over the same period [57]. Another consequence of the interruption of ribosome biogenesis is the destruction of numerous ribosomal proteins by proteasomes [31, 42, 56]. We can therefore hypothesize that AMD treatment induces major general changes to the molecular environment within the nucleus. Moreover, these modifications are concomitant to changes in the cytoplasm, because AMD also inhibits: (1) both respiration and glycolysis, resulting in a decrease in ATP production [40] and (2) mitochondrial RNA polymerases [39]. It is tempting to link the increase in water content with the cell proliferation arrest induced by AMD within these cells, which contain an inefficient p53 due to the integration of active HPV virus [41]. One hypothesis worthy of further study is that the increase in water content may be an additional mechanism involved in cell proliferation arrest due to impaired ribosome biogenesis in cells lacking active p53 [58]. Finally, we found that nucleolar light caps (NLC), which contain uncondensed rDNA genes [37], and nucleoplasm were the most hydrated cell compartments. However, as low concentration of AMD inhibits RNA pol I in the nucleolus, but not class II gene transcription in the nucleoplasm [59], high levels of hydration in nuclear domains may be a characteristic feature of either active or inactive uncondensed genes.
Elementary analysis showed that the concentrations of ions were much lower after AMD treatment, probably resulting in major changes to the ionic environment of both the cytoplasm and nucleus. Moreover, the ratio of the content of each ion to that of phosphorus varied differently. The Cl/P ratio decreased in all cell compartments except NLC, whereas the K/P ratio was unaffected by AMD treatment. This suggests that the K and Cl ions, which are known to localize within the minor groove of AT and CG duplexes of B-DNA [18], behave differently under AMD stress. Similarly, the Mg/P ratio was largely unaffected, other than displaying an increase in DFC/GC nucleolar components and a decrease in condensed chromatin, which is indicative of a change in chromatin condensation [14].
Finally, in the future, this approach should be useful for determining the actual concentrations of ions and the hydration status necessary for defined cell activities. For example, the concentration of K in the nucleoplasm and cytosol decreased strongly in response to AMD treatment, but remained above 60–70 mmol/l. This concentration is sufficiently high to inhibit procaspase 3 activation, as demonstrated in vitro [60] and confirmed by the absence of cleaved procaspase-3 in the cells studied here. This correlative approach should make it possible to determine the actual concentrations of all ions required to activate the cleavage of procaspase-3 in the various compartments of cells engaged in the apoptotic pathway.
In conclusion, our new correlative approach made it possible to map water, elements, and ions, at the nanoscale level in cell compartments identified by specific GFP-tagged proteins. This study, by providing data on water and elements/ion contents in the nucleus and cytoplasm, sheds new light on the link between cell functions and differences in macromolecular crowding between control and stressed cells.
Materials and methods
Cell culture
Two HeLa cell lines stably expressing H2B-GFP or NOP 52-GFP (courtesy of K. Monier, University of Lyon, France) were cultured in DMEM (Gibco) supplemented with 10 % FBS, in 25-cm2 Nunc flasks, with twice-weekly passaging. For experiments, the cells were used to seed Petri dishes and were cultured to confluence. All cultures tested negative for mycoplasma infection.
Cryofixation
The confluent cells were detached by trypsin and centrifuged to obtain a cell pellet. Cells mounted on home-made metallic small rods were vitrified with a Gatan Cryoplunge CP3, by rapid plunging into liquid ethane cooled by liquid nitrogen (N2) [26]. We avoided the use of cryoprotectants (such as sucrose and dextran), to prevent movements of water and ions. Frozen-hydrated samples were subsequently stored in liquid N2 until vitreous sectioning.
Vitreous sectioning
Small rods are inserted in the ultracryomicrotome chuck. Vitreous sections were cut at a nominal thickness setting of 85 nm (Leica Ultramicrotome UC6-FC6), with 35° diamond knives (Diatome), at a temperature of −160 °C. Vitreous sections were transferred to formvar-carbon TEM finder grids (Agar). The grids were either directly mounted in the cryoholder or stored in liquid nitrogen for use in subsequent experiments.
Cryoholder
For both light and electron microscopy, we used a Gatan 626 cryo-EM holder. This made it possible to measure and control grid temperature. For light microscopy, the holder was placed in a thermally isolated reservoir filled with liquid N2, to maintain a dry N2 atmosphere around the grid.
Light microscopy for correlative microscopy
Light microscopy experiments were performed with a Zeiss Axioscope Vario A1 epifluorescence microscope with a 560 mm stand column. This microscope is equipped for illumination with a LED module (white, blue, red, and green). The EX BP 470-40/EM BP 525-50 filter set was used to detect GFP-tagged-proteins. Reflected and fluorescent light images were recorded with a black-and-white CCD camera (AxioCam MRm, Zeiss). Our setup included a Plan-Neofluar objective (10×; 0.3 NA; 5.2 mm working distance) for rapid screening of the entire EM grid and an Epiplan Neofluar objective (50×; 0.55 NA; 9.1 mm working distance) for higher resolution of the region of interest. The liquid N2 reservoir into which the cryoholder was inserted was placed on the stage of the epi fluorescence microscope. We used a simple procedure to improve the recording of light and electron microscopy images. Two fiducial markers fixed on the stage were first used to position the reservoir and to limit its rotation so that there was no more than a few degrees of difference between experiments. The grid was then imaged (reflected light mode) at low magnification to identify the two triangles marking its center and an image was recorded. The position of the cryosections was determined with reflected light and these sections were imaged in fluorescence mode. Cryosection images were recorded at a higher magnification, to obtain more detailed information. After imaging, the shutter of the cryoholder was closed to protect the cryosections during transfer to the electron microscope. The temperature of the grid was continually monitored to ensure that cryo preservation remained optimal throughout the procedure.
Electron microscopy
We used a CM30 transmission electron microscope (FEI, The Netherlands) operating at an accelerating voltage of 100 kV. The microscope was fitted with a post-viewing screen Gatan STEM Dark-Field/Bright-Field detector. We ensured that the grid was placed in exactly the same position as in the light microscope by obtaining a low-magnification image in STEM mode (500×) and identifying the two triangles marking the center of the grid. This image was compared with that taken in the reflected light mode, on the same screen, and was precisely rotated (to within a few degrees) with the image rotation function of the microscope. Once the two images had been aligned, it was easier to identify the fields of interest previously identified by light microscopy in STEM mode because we were able to ensure that they were placed in roughly similar orientations.
Energy-dispersive X-ray spectrometry (EDXS) was performed with an EDAX 30 mm2 Si(Li) R-SUTW detector. Spectrum quantification was carried out with custom-made dedicated software, according to Hall’s continuum method, taking into account the support film signal and the spurious background signal from uncollimated electrons exciting the copper grid. Mass concentrations were converted from mmol/kg (C D) of dry matter into mmol/l (C H) with the following equation C H = ((100 − L)/L) × C D, where L is the percentage of water determined by quantitative dark-field STEM imaging.
Water-content measurements by quantitative dark-field STEM
This quantitative dark-field STEM method [61] is an indirect method for determining the proportion of water mass in freeze-dried cryosections based on the linear relationship between the sample mass thickness density ρt and the dark-field STEM intensity signal. We have already extensively discussed the sensitivity and limitations of this method elsewhere [23]. The main practical limitation of the method is the potential for differential shrinkage of the different cell compartments during freeze-drying of the cryosections. An interesting result of our correlative approach is to highlight the precision of the process of merging fluorescence images of the hydrated cryosections and the STEM images of the cryosections after freeze-drying: this indicates the isotropy of the shrinkage effect related to freeze-drying, and consequently, also, the relevance of the method [26]. Therefore, the sensitivity of the method is now essentially limited by discrepancies in the thickness of the cryosections. We estimated, in our experimental conditions, an uncertainty of approximately 10 %. However, this uncertainty due to thickness discrepancies can be reduced statistically by analyzing several cryosections for each set of biological conditions. This method is therefore particularly useful for detecting water content variations in different biological conditions.
Finally, quantification on H2B-GFP cells was performed on four different cell cultures for control (a total of 80 interphase and 25 mitotic cells were analyzed on 12 cryosections) and on two different cell cultures for AMD treatment (a total of 33 interphase and five mitotic cells were analyzed on 12 cryosections). Quantification on NOP-52-GFP cells was performed on three different cell cultures for control (a total of 40 interphase cells were analyzed on 30 cryosections) and on three different cell cultures for AMD treatment (a total of 40 interphase cells were analyzed on 30 cryosections).
Image processing
All images were processed with ImageJ software (NIH).
In fluorescence images, the five main levels of intensity [from low (dark) to high (white)] were color-coded in dark, blue, green, red and pink, respectively.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 3 Percentage water in control and actinomycin D-treated HeLa NOP 52-GFP cells. Quantification was carried out for regions of interest in several compartments: DFC/GC (nucleolar dense fibrillar component and granular component); FC/NLC (nucleolar fibrillar centers and nucleolar light caps); NPL (nucleoplasm); CY (cytosol); MIT (mitochondria). Data are means ± SD from triplicate experiments (n = 40 for each condition). (TIFF 2389 kb)
Online Resource 4 Elements (N, P, S, K, Cl, Mg) were identified and quantified by EDXS in control and actinomycin D-treated HeLa NOP 52-GFP cells. Analyses were performed on regions of interest in several compartments: DFC/GC (nucleolar dense fibrillar component and granular component); FC/NLC (nucleolar fibrillar centers and nucleolar light caps); NPL (nucleoplasm); CYT (cytosol); MIT (mitochondria). Data are means ± SD from triplicate experiments (n = 40 for each condition). (TIFF 6637 kb)
Online Resource 5 Elements (N, P, S, K, Cl, Mg) were identified and quantified by EDXS in control and actinomycin D-treated HeLa NOP 52-GFP cells. Analyses were performed on regions of interest in several compartments: DFC/GC (nucleolar dense fibrillar component and granular component); NPL (nucleoplasm); CYT (cytosol); MIT (mitochondria). Data are means ± SD from triplicate experiments (n = 40 for each condition) and are presented as spiderweb diagrams for each cell compartment. (DOC 21 kb)
Acknowledgments
We received funding from: the Agence Nationale pour la Recherche (ANR-07 Nano-CESIWIN), Europe Community (FEDER) and Région Champagne Ardennes.
Footnotes
J. Michel and D. Ploton are co-senior authors.
References
- 1.Ellis RJ. Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr Opin Struct Biol. 2001;11:114–119. doi: 10.1016/S0959-440X(00)00172-X. [DOI] [PubMed] [Google Scholar]
- 2.Hancock R. A role for macromolecular crowding effects in the assembly and function of compartments in the nucleus. J Struct Biol. 2004;146:281–290. doi: 10.1016/j.jsb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Schnell S, Hancock R (2008) The intranuclear environment. In: Hancock R (ed) The nucleus, vol 1. Humana Press, pp 3–19
- 4.Minton AP. How can biochemical reactions within cells differ from those in test tubes? J Cell Sci. 2006;119:2863–2869. doi: 10.1242/jcs.03063. [DOI] [PubMed] [Google Scholar]
- 5.Goodsell DS. Inside a living cell. Trends Biochem Sci. 1991;16:203–206. doi: 10.1016/0968-0004(91)90083-8. [DOI] [PubMed] [Google Scholar]
- 6.Ando T, Skolnick J. Crowding and hydrodynamic interactions likely dominate in vivo macromolecular motion. Proc Natl Acad Sci USA. 2010;107:18457–18462. doi: 10.1073/pnas.1011354107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medalia O, Weber I, Frangakis AS, Nicastro D, Gerish G, Baumeister W. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 2002;298:1209–1213. doi: 10.1126/science.1076184. [DOI] [PubMed] [Google Scholar]
- 8.Zhou HX, Rivas G, Minton AP. Macromolecular crowding and confinement: biochemical, biophysical and potential physiological consequences. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmerman S, Harrison B. Macromolecular crowding increases binding of DNA polymerase to DNA: an adaptive effect. Proc Natl Acad Sci USA. 1987;84:1871–1875. doi: 10.1073/pnas.84.7.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fullerton GD, Kanal KM, Cameron IL. On the osmotically unresponsive water compartment in cells. Cell Biol Int. 2006;30:74–77. doi: 10.1016/j.cellbi.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 11.Ball P. Water as an active constituent in cell biology. Chem Rev. 2008;108:74–108. doi: 10.1021/cr068037a. [DOI] [PubMed] [Google Scholar]
- 12.Feig M, Pettitt M. Modeling high-resolution hydration patterns in correlation with DNA sequence and conformation. J Mol Biol. 1998;286:1075–1095. doi: 10.1006/jmbi.1998.2486. [DOI] [PubMed] [Google Scholar]
- 13.Auffinger P, Hashem Y. Nucleic acid salvation: from outside to insight. Curr Opin Struct Biol. 2007;17:325–333. doi: 10.1016/j.sbi.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Strick R, Strissel PL, Gavrilov K, Levi-Setti R. Cation-chromatin binding as shown by ion microscopy is essential for the structural integrity of chromosomes. J Cell Biol. 2001;155:899–910. doi: 10.1083/jcb.200105026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaplin M. Do we underestimate the importance of water in cell biology? Nat Rev Mol Cell Biol. 2006;7:861–866. doi: 10.1038/nrm2021. [DOI] [PubMed] [Google Scholar]
- 16.Pederson T. The nucleus introduced. Cold Spring Harb Perspect Biol. 2010;2:a000521. doi: 10.1101/cshperspect.a000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodcock CL, Ghosh RP. Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol. 2010;2:a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howard JJ, Lynch GC, Pettitt BM. Ion and solvent density distributions around canonical B-DNA from integral equations. J Phys Chem. 2011;27:547–556. doi: 10.1021/jp107383s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hancock R. Packing of the polynucleosome chain in interphase chromosomes: evidence for a contribution of crowding and entropic forces. Semin Cell Dev Biol. 2007;18:668–675. doi: 10.1016/j.semcdb.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 20.Bohrmann B, Haider M, Kellenberger E. Concentration evaluation of chromatin in unstained resin-embedded sections by means of low-dose ratio-contrast imaging in STEM. Ultramicroscopy. 1993;49:235–251. doi: 10.1016/0304-3991(93)90230-U. [DOI] [PubMed] [Google Scholar]
- 21.Guerquin-Kern JL, Wu TD, Quintana C, Croisy A. Progress in analytical imaging of the cell by dynamic secondary ion mass spectroscopy (SIMS microscopy) Biochem Biophys Acta. 2005;1724:228–238. doi: 10.1016/j.bbagen.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez-Segura E, Warley A. Electron probe X-ray microanalysis for the study of cell physiology. Methods Cell Biol. 2008;88:19–43. doi: 10.1016/S0091-679X(08)00402-0. [DOI] [PubMed] [Google Scholar]
- 23.Terryn C, Michel J, Kilian L, Bonhomme P, Balossier G. Comparison of intracellular water content measurements by dark-field imaging and EELS in medium voltage TEM. The Eur Phys J Appl Phys. 2000;11:215–226. doi: 10.1051/epjap:2000156. [DOI] [Google Scholar]
- 24.Zierold K, Michel J, Terryn C, Balossier G. The distribution of light elements in biological cells measured by electron probe X-ray microanalysis of cryosections. Microsc Microanal. 2005;11:138–145. doi: 10.1017/S1431927605050130. [DOI] [PubMed] [Google Scholar]
- 25.Delavoie F, Molinari M, Milliot M, Zahm JM, Coraux C, Michel J, Balossier G. Salmeterol restores secretory functions in cystic fibrosis airway submucosal gland serous cells. Am J Resp Cell Mol Biol. 2009;40:388–397. doi: 10.1165/rcmb.2008-0037OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nolin F, Ploton D, Wortham L, Tchelidze P, Balossier G, Banchet V, Bobichon H, Lalun N, Terryn C, Michel J. Targeted nano analysis of water and ions using cryocorrelative light and scanning transmission electron microscopy. J Struct Biol. 2012;180:352–361. doi: 10.1016/j.jsb.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 27.Kanda T, Sullivan KF, Wahl G. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/S0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 28.Savino TM, Bastos R, Jansen E, Hernandez-Verdun D. The nucleolar antigen Nop52, the human homologue of the yeast ribosomal RNA processing RRP1, is recruited at late stages of nucleologenesis. J Cell Sci. 1999;112:1889–1900. doi: 10.1242/jcs.112.12.1889. [DOI] [PubMed] [Google Scholar]
- 29.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burger K, Mühl B, Harasim T, Rohrmoser M, Malamoussi A, Orban M, Kellner M, Gruber-Eber A, Kremmer E, Hölzel M, et al. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J Biol Chem. 2010;285:12416–12425. doi: 10.1074/jbc.M109.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersen JS, Lam YW, Leung AKL, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 32.Sartori A, Gatz R, Beck F, Rigort A, Baumeister W, Plitzko JM. Correlative microscopy: bridging the gap between fluorescence light microscopy and cryo-electron tomography. J Struct Biol. 2007;160:135–145. doi: 10.1016/j.jsb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 33.Briegel A, Chen S, Koster AJ, Plitzko JM, Schwartz CL, Jensen GJ. Correlated light and electron cryo-microscopy. Meth Enzymol. 2010;481:317–341. doi: 10.1016/S0076-6879(10)81013-4. [DOI] [PubMed] [Google Scholar]
- 34.Fullerton GD, Cameron IL. Water compartments in cells. Meth Enzymol. 2007;428:1–28. doi: 10.1016/S0076-6879(07)28001-2. [DOI] [PubMed] [Google Scholar]
- 35.Cavanaugh A, Hirschler-Laszkiewiez I, Rothblum LI (2004) Ribosomal DNA transcription in mammals. In: Olson M (ed) The nucleolus. Kluwer Academic/Plenum Publishers, Dordrecht, pp 88–127
- 36.Henras AK, Soudet J, Gérus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puvion-Dutilleul F, Mazan S, Nicoloso M, Pichard E, Bachellerie JP, Puvion E. Alterations of nucleolar ultrastructure and ribosome biogenesis by actinomycin D. Implications for U3 snRNP function. Eur J Cell Biol. 1992;58:149–162. [PubMed] [Google Scholar]
- 38.Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukamachi S, Bartoov B, Freeman KB. Synthesis of ribonucleic acid by isolated rat liver mitochondria. Biochem J. 1972;128:299–309. doi: 10.1042/bj1280299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laszlo J, Miller DS, McCarty KS, Hochstein P. Actinomycin D: inhibition of respiration and glycolysis. Science. 1966;151:1007–1010. doi: 10.1126/science.151.3713.1007. [DOI] [PubMed] [Google Scholar]
- 41.Scheffner M, Münger K, Byrne JC, Howley PM. The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc Natl Acad Sci USA. 1991;88:5523–5527. doi: 10.1073/pnas.88.13.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol. 2007;17:749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bellissent-Funel MC (2011) Protein dynamics and hydration water. In: Le Bihan D (ed) Water: the forgotten biological molecule. Pan Stanford Publishing, Singapore, pp 23–47
- 44.Chaplin M (2011) The water molecule, liquid water, hydrogen bonds, and water networks. In: Le Bihan D (ed) Water: the forgotten biological molecule. Pan Stanford Publishing, Singapore, pp 4–19
- 45.Mentré P. Water in the orchestration of the cell machinery. Some misunderstandings: a short review. J Biol Phys. 2012;38:13–26. doi: 10.1007/s10867-011-9225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bancaud A, Huet S, Daigle N, Mozziconacci J, Beaudoin J, Ellenberg J. Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J. 2009;28:3785–3798. doi: 10.1038/emboj.2009.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verschure PJ, Van der Kraan I, Manders EMM, Hoogstraten D, Houtsmuller AB, Van Driel R. Condensed chromatin domains in the mammalian nucleus are accessible to large macromolecules. EMBO Rep. 2003;4:861–866. doi: 10.1038/sj.embor.embor922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Görisch SM, Richter K, Scheuermann MO, Herrmann H, Lichter P. Diffusion-limited compartmentalization of mammalian cell nuclei assessed by microinjected macromolecules. Exp Cell Res. 2003;289:282–294. doi: 10.1016/S0014-4827(03)00265-9. [DOI] [PubMed] [Google Scholar]
- 49.Handwerger KE, Cordero JA, Gall JG. Cajal bodies, nucleoli and speckles in the Xenopus oocyte nucleus have a low-density, sponge-like structure. Mol Biol Cell. 2005;16:202–211. doi: 10.1091/mbc.E04-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Derenzini M, Pasquinelli G, O’Donohue MF, Ploton D, Thiry M. Structural and functional organization of ribosomal genes within the mammalian cell nucleolus. J Histochem Cytochem. 2006;54:131–146. doi: 10.1369/jhc.5R6780.2005. [DOI] [PubMed] [Google Scholar]
- 51.Bortner CD, Sifre MI, Cidlowski JA. Cationic gradient reversal and cytoskeleton-independent volume regulatory pathway define an early stage of apoptosis. J Biol Chem. 2008;283:7219–7229. doi: 10.1074/jbc.M707809200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warley A, Stephen J, Hockaday A, Appleton TC. X-ray microanalysis of HeLa S3 cells. J Cell Sci. 1983;60:217–229. doi: 10.1242/jcs.60.1.217. [DOI] [PubMed] [Google Scholar]
- 53.Arrebola F, Fernandez-Segura E, Campos A, Crespo PV, Skepper JN, Warley A. Changes in intracellular electrolyte concentrations during apoptosis induced by UV irradiation of human myeloblastic cells. Am J Physiol Cell Physiol. 2006;290:638–649. doi: 10.1152/ajpcell.00364.2005. [DOI] [PubMed] [Google Scholar]
- 54.Lu SC. Regulation of glutathione synthesis. Mol Asp Med. 2009;30:42–59. doi: 10.1016/j.mam.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cameron IL, Kanal KM, Fullerton GD. Role of protein conformation and aggregation in pumping water in and out of a cell. Cell Biol Int. 2006;30:78–85. doi: 10.1016/j.cellbi.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 56.Deisenroth C, Zhang Y. The ribosomal protein-mdm2-p53 pathway and energy metabolism: bridging the gap between feast and famine. Genes Cancer. 2011;2:392–403. doi: 10.1177/1947601911409737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Görlich D, Mattaj W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 58.Donati G, Montanaro L, Derenzini M. Ribosome biogenesis and control of cell proliferation: p53 is not alone. Cancer Res. 2012;72:1602–1607. doi: 10.1158/0008-5472.CAN-11-3992. [DOI] [PubMed] [Google Scholar]
- 59.Bensaude O. Inhibiting eukaryotic transcription. Which compound to choose? How to evaluate its activity? Transcription. 2011;2:103–108. doi: 10.4161/trns.2.3.16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hughes FM, Bortner CD, Purdy GD, Cidlowski JA. Intracellular K+ suppresses the activation of apoptosis in lymphocytes. J Biol Chem. 1997;272:30567–30576. doi: 10.1074/jbc.272.48.30567. [DOI] [PubMed] [Google Scholar]
- 61.Zierold K. The determination of wet weight concentrations of elements in freeze-dried cryosection from biological cells. Scanning Microsc. 1986;2:713–724. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Online Resource 3 Percentage water in control and actinomycin D-treated HeLa NOP 52-GFP cells. Quantification was carried out for regions of interest in several compartments: DFC/GC (nucleolar dense fibrillar component and granular component); FC/NLC (nucleolar fibrillar centers and nucleolar light caps); NPL (nucleoplasm); CY (cytosol); MIT (mitochondria). Data are means ± SD from triplicate experiments (n = 40 for each condition). (TIFF 2389 kb)
Online Resource 4 Elements (N, P, S, K, Cl, Mg) were identified and quantified by EDXS in control and actinomycin D-treated HeLa NOP 52-GFP cells. Analyses were performed on regions of interest in several compartments: DFC/GC (nucleolar dense fibrillar component and granular component); FC/NLC (nucleolar fibrillar centers and nucleolar light caps); NPL (nucleoplasm); CYT (cytosol); MIT (mitochondria). Data are means ± SD from triplicate experiments (n = 40 for each condition). (TIFF 6637 kb)
Online Resource 5 Elements (N, P, S, K, Cl, Mg) were identified and quantified by EDXS in control and actinomycin D-treated HeLa NOP 52-GFP cells. Analyses were performed on regions of interest in several compartments: DFC/GC (nucleolar dense fibrillar component and granular component); NPL (nucleoplasm); CYT (cytosol); MIT (mitochondria). Data are means ± SD from triplicate experiments (n = 40 for each condition) and are presented as spiderweb diagrams for each cell compartment. (DOC 21 kb)