Abstract
The retinal circadian clock is crucial for optimal regulation of retinal physiology and function, yet its cellular location in mammals is still controversial. We used laser microdissection to investigate the circadian profiles and phase relations of clock gene expression and Period gene induction by light in the isolated outer (rods/cones) and inner (inner nuclear and ganglion cell layers) regions in wild-type and melanopsin-knockout (Opn −/−4) mouse retinas. In the wild-type mouse, all clock genes are rhythmically expressed in the photoreceptor layer but not in the inner retina. For clock genes that are rhythmic in both retinal compartments, the circadian profiles are out of phase. These results are consistent with the view that photoreceptors are a potential site of circadian rhythm generation. In mice lacking melanopsin, we found an unexpected loss of clock gene rhythms and of the photic induction of Per1-Per2 mRNAs only in the outer retina. Since melanopsin ganglion cells are known to provide a feed-back signalling pathway for photic information to dopaminergic cells, we further examined dopamine (DA) synthesis in Opn −/−4 mice. The lack of melanopsin prevented the light-dependent increase of tyrosine hydroxylase (TH) mRNA and of DA and, in constant darkness, led to comparatively high levels of both components. These results suggest that melanopsin is required for molecular clock function and DA regulation in the retina, and that Period gene induction by light is mediated by a melanopsin-dependent, DA-driven signal acting on retinal photoreceptors.
Keywords: Clock genes, Photoreceptor, Dopamine, ipRGC
Introduction
The mammalian retina contains a circadian clock that controls a broad range of temporal retinal physiology, including photoreceptor disc shedding and phagocytosis [1, 2], expression of immediate early genes and visual photopigments [3, 4], N-acetyltransferase expression, dopamine/melatonin synthesis [5–8], circadian gene and clock gene expression [9–11] and visual processing [10, 12, 13]. The retinal clock generates molecular circadian rhythms independent of the master clock of the suprachiasmatic nucleus (SCN; [9, 10, 14, 15]) through cell-autonomous autoregulatory transcriptional/translational feedback loops composed of several clock genes, including Per1, Per2, Clock, Bmal1, Cry1, Cry2, Reverbα and Rorβ and their protein products [16, 17].
The presence of an endogenous clock in the retina was first described in Xenopus laevis [18] and subsequently in fish [19], reptiles [20], birds [3] and mammals [5, 21]. In contrast to the amphibian retina, in which photoreceptors are cell-autonomous circadian oscillators [22, 23] that mediate rhythmic melatonin release [24], in mammals, the cellular location of the circadian clock is still controversial, and widespread clock gene expression has been found in different retinal neurons. In rodents, clock genes are widely expressed in the inner nuclear (INL) and ganglion cell (GCL) layers [9, 25–29]. Ruan et al. [9], using real time RT-PCR, have shown that several neurons of the inner retina coordinately express six core clock genes whereas rod photoreceptors do not, and in cultured rd mouse retinas, the circadian rhythm of PER2::LUC is sustained. Although this suggests the absence a circadian clock in the outer retina, other studies have confirmed that photoreceptors contain clock genes [11, 26, 27] that are rhythmically expressed in the isolated photoreceptor layer [11, 30, 31]. Furthermore, Tosini et al. [11] found that the isolated photoreceptors contain a functional clock involved in the regulation of melatonin synthesis. A recent study by Liu et al. [32] demonstrated circadian rhythms of clock proteins in several classes of inner and outer retinal neurons including cones, but nevertheless observed disparities in their temporal profiles. Taken together, these studies suggest distinct cellular clocks in the mammalian retina.
Light acting through retinal photoreceptors resets the retinal circadian clock and exerts acute effects on clock-controlled functions [15, 33, 34], but it is unknown whether photopigment deletion impacts on the molecular function of the clock. Intrinsically, photosensitive melanopsin retinal ganglion cells (ipRGCs) participate in several aspects of retinal function, including the circadian regulation of the light-adapted ERG, a function that has been proposed to be mediated by the upregulation of dopamine (DA) by light and/or by the circadian clock [13, 35]. Indeed, retina-specific Bmal1 clock gene-deficient mice display a similar phenotype of altered light-adapted circadian ERG [10]. ipRGCs are functionally and synaptically connected to dopaminergic (DAergic) amacrine cells [36–38], and play a crucial role as an intraretinal retrograde pathway, providing excitatory sustained light responses to DAergic neurons [12, 39]. In turn, DA signalling is implicated in many aspects of retinal neuromodulation, mediation of light responsiveness, and clock gene regulation [33].
We thus used laser microdissection to investigate the circadian profiles and phase relations of clock gene expression and Period gene induction by light in the isolated outer (rods and cones) and inner (INL plus GCL) regions in wild-type and melanopsin-knockout (Opn −/−4) mouse retinas. In addition, we examined the role of melanopsin in the regulation of tyrosine-hydroxylase (TH) mRNA expression, the key enzyme in DA synthesis, and DA content. We found that melanopsin is essential for circadian retinal function since invalidation of this photopigment leads to an increase in retinal DA coupled with the loss of circadian clock gene expression and light induction of Per1 and Per2 in the outer retina. Furthermore, the light-induced increase in TH activity and DA synthesis are absent in the Opn −/−4 mouse.
Materials and methods
Animals
Male C57Bl6 wild-type and Opn −/−4 mice between 6 and 8 weeks of age were used. Animals were housed in plexiglass cages under a 12-h light/12-h dark cycle (12L/12D, light intensity around 200 lux), with food and water ad libitum. All treatment of animals was in strict accordance with current national and international regulations on animal care, housing, breeding and experimentation.
Laser capture microdissection of the inner and outer retina
Circadian expression of clock and clock-controlled genes and light induction of Per1 and Per2 in the inner and outer retina
Adult wild-type and Opn −/−4 male mice (n = 18 of each genotype) were initially maintained for 2 weeks under a 12L/12D cycle with broad-band white light. Subsequently, animals were kept under constant dark (DD) conditions. To quantify circadian expression of clock genes and clock-controlled genes, eyes were collected during the first circadian cycle every 4 h (CT0, CT4, CT8, CT12, CT16 and CT20). To analyse Per1 and Per2 induction by light, a single pulse of monochromatic light (480 nm) of constant duration and irradiance (15 min, 2.8 × 1014 photons/cm2/s) was administered at CT16 (first day in DD). Animals were sacrificed 30 min after the beginning of the light pulse (n = 5 for each genotype). In both cases, eyes were embedded in OCT (Tissue-Tek), frozen on dry ice, and stored at −80 C. All dissections during the dark phase were carried out under dim red light.
Laser capture microdissection
Frozen eyes were cut at 12-μm thickness and mounted on uncharged glass slides. The frozen sections were thawed for 30 s and were dehydrated in graded ethanol solutions (75, 95, 100 %) of 30 s each. Then, the sections were cleared in three successive baths of xylene (respectively, 3, 3 and 5 min). The slides were kept in a vacuum desiccator for a minimum of 10 min. In order to isolate the outer compartment (outer nuclear layer) of the retina from the inner compartment (INL plus GCL) in a contact- and contamination-free manner, the PixCell II laser capture microdissection system (LCM; Arcturus Engineering) was used. Laser capture was performed by separately lifting these two compartments onto HS-CapSure noncontact laser capture microdissection film. After visual control of the completeness of laser microdissection (caps control), the captured tissue is lysed with buffer RLT (Qiagen Rneasy Micro Kit). We evaluated the isolation of the inner from outer microdissected samples by using specific markers to control for the absence of tyrosine-hydroxylase (TH, 235 bp) in the outer retina and of middle-wavelength (MW, 143 bp, results not shown) opsin and rhodopsin(179 bp) in the inner retina (Fig. 1). Results in Fig. 1B show that rhopdopsin transcripts are amplified from photoreceptor samples, whereas TH is not, and the inverse was found for TH transcripts in the inner retina (Fig. 1A) for wild-type and Opn −/−4 mouse at all circadian times (CT) 0, 4, 8, 12, 16 and 20.
Fig. 1.
The isolation of inner from outer microdissected samples was evaluated by using specific markers to control for the absence of A tyrosine-hydroxylase (TH, 235 bp) transcript in the outer retina, and B of rhodopsin (179 bp) transcript in the inner retina by RT-PCR. Agarose gel electrophoresis of the transcript amplification products show that rhopdopsin transcripts are amplified from photoreceptor samples, whereas TH is not, and the inverse was found for TH transcripts in the inner retina for wild-type and Opn −/−4 mouse at all circadian times (CT) 0, 4, 8, 12, 16 and 20
RNA extraction and amplification of the inner and outer retina samples
RNA was isolated on silica-based columns, DNase I-digested and eluted with water, using RNeasy Micro Kit following the manufacturer’s instructions (Qiagen, France). RNA qualities were assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies, USA); RNA was prepared from microdissected assays have a RIN >8 (RNA integrity number). Samples (10 ng of RNA) were amplified using ExpressArt mRNA linear amplification Nano kit (Amp Tec-Exilone, France), using the manufacturer’s protocol. Reverse transcription of amplified RNA was performed with a iScript cDNA Synthesis Kit (Bio-Rad, France). cDNA synthesis was carried out from 1 μg amplified cRNA.
Whole retina sampling
Circadian expression and light induction of tyrosine-hydroxylase mRNA in the retina of wild-type and Opn−/−4 mice
Mice were exposed to a 12L/12D cycle for 15 days (n = 5 for each genotype). Subsequently, animals were kept under DD conditions. For circadian expression of TH, pools of two retinas from the same animal were directly collected at CT0 and CT12 during the first circadian cycle in DD. To analyse TH mRNA induction by light, a single pulse of monochromatic light (480 nm, 15 min, 2.8 × 1014 photons/cm2/s) was administered at CT16. Animals were sacrificed 30 min after the beginning of the light pulse (n = 5 for each genotype). Pools of two retinas from the same animal were directly collected. In the two experiments, retinas were frozen on dry ice and stored at −80 °C until RNA extraction and quantification.
Circadian expression and SW-, MW- and rhodopsin mRNA in the retina of wild-type and Opn−/−4 mice
To quantify circadian expression of middle-wavelength (MW) and short-wavelength (SW) opsins and rhodopsin, we maintained adult wild-type and Opn −/−4 male mice (n = 24 of each genotype) for 2 weeks under a 12L/12D cycle with broad-band white light. Animals were then kept for one cycle under DD conditions and euthanized during the first circadian cycle every 4 h (CT0, CT4, CT8, CT12, CT16 and CT20). Pooled retinas from the same animal were directly collected under dim red light, frozen on dry ice and stored at −80 °C.
RNA extraction
Total RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer’s instructions and subsequently subjected to DNase digestion. Total RNAs was reverse transcribed using random primers and MMLV Reverse Transcriptase (Invitrogen).
Real-time RT-PCR
Real-time PCR was then performed on a LightCycler™ system (Roche Diagnostics) using the light Cycler-DNA Master SYBR Green I mix. The efficiency and the specificity of the amplification were controlled by generating standard curves and carrying out melting curves and agarose gels of the amplicons, respectively. Relative transcript levels of each gene were calculated using the second derivative maximum values from the linear regression of cycle number versus log concentration of the amplified gene. Amplification of the non cycling control gene HPRT was used for normalization. Primer sequences were the following: Per1 sens, GCGTTGCAAACGGGATGGT and reverse, GAACCTGCAGAGGTGCCAG; Per2 sens, CCACACTTGCCTCCGAAATA and reverse ACTGCCTCTGGACTGGAAGA; Per3 sens, CAGTGGCAGAGACGTGCGT and reverse GACACTGTCGATACAGTTCAT; Clock sens, GTT TGA TCA CAG CCC AAC TC and reverse CTC CGC TGT GTC ATC TTT TC; Bmal1 sens, CTCAGCTGCCTCGTTGCAAT and reverse GCTGTCGCCCTCTGATCTAC; Cry1 sens GCCAGCTGATGTATTTCCCAG and reverse CGCCAGCCTCAGTAGCCAG; Cry2 sens, GAGAGACCTCGGATGAATGC and reverse CTCGCCACAGGAGTTGTCCA; Reverbα sens, GCTCCATCGTTCGCATCAAT and reverse CTAGAGGGCACAGGCTGCT; Rorβ sens, GCG AGC ACA AAT TGA AGT GA and reverse AAC GGT TCC TGT TGG TTC TG; E4Bp4 sens, CGG AAG TTG CAT CTC AGT CA and reverse GCA AAG CTC TCC AAC TCC AC; Dbp sens, CGTGGAGGTGCTAATGACCT and reverse CGGCTCCAGTACTTCTCATC; HPRT sens, ATCAGTCAACGGGGGACATA and reverse AGAGGTCCTTTTCACCAGCA. For the target genes, primer sequences are the following; TH sens, GAAGGGCCTCTATGCTACCC and reverse GGCATGACGGATGTACTGTG SW sens, GCCCAGACGTGTTCAGCG and reverse, GACCATCACCACCACCAT; MW sens, GCTGCATCTTCCCACTCAG and reverse GACCATCACCACCACCAT; Rhodopsin sens, GCCACCACTCAGAAGGCAG and reverse GATGGAAGAGCTCTTAGCAAAG.
Biochemistry
Adult wild-type and Opn −/−4 male mice (n = 9–11 for each genotype) were initially maintained for 2 weeks under a 12L/12D cycle with broad-band white light. Subsequently, animals were kept under DD conditions and euthanized during the first circadian cycle in DD at CT0 and CT12. To analyse DA induction by light, a single pulse of monochromatic light (480 nm, 15 min, 2.8 × 1014 photons/cm2/s) was administered at CT16. Animals were sacrificed 30 min after the beginning of the light pulse (n = 7–10 for each genotype and for dark- and light-stimulated conditions). For quantification of circadian and light induction of DA, pools of two retinas from the same animal were collected under dim red light, frozen on dry ice and stored at −80 °C until analyses. Retinas were homogenised by sonication in 60 μl buffered perchloric acid 0.1 M, and centrifuged at 4 °C, 10.000 rpm for 15 min. The supernatants were injected into the HPLC separation system (column SpheriSorb RP18-5UM, mobile phase 50 mM KH2PO4, 15 mg/ml EDTA-Na, 0.26 mM sodium octyle sulphate, methanol 8 %, adjusted to pH 4.5, flow rate 0.3 ml/min). DA and DOPAC peaks were identified based on retention time, and concentrations were estimated by rationing areas of each substance and their respective external standard (analytical software AZUR; Datalys, St Martin d’Heres, France). DA and DOPAC concentrations were expressed as pg/mg tissue.
Statistical analysis
A one-way ANOVA was used to determine the presence of significantly different gene expression between time points in DD. If significant differences were found, a Student–Newman–Keuls test for multiple comparisons was performed.
To evaluate rhythmicity in gene expression, the data were modelled using R software [40] by estimating the mesor (M midline estimating statistic of rhythm), amplitude A (difference between minimum and maximum of fitted function), and acrophase φ (in hours: time of maximum in fitted function) of the best fitting sine-wave of the 24-h period, by least squares using the equation.
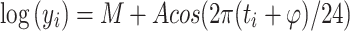 |
1 |
where y i is the ith expression value measured at time t i in hours. A log transformation of the expression values was chosen as the Box–Cox transformation that maximised the profile likelihood function [41, 42]. To facilitate the fitting procedure and estimation of the standard errors of the parameters, Eq. 1 can be re-written as a linear model in which we estimate linear coefficients,
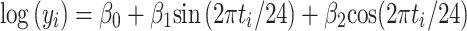 |
2 |
where M = β 0, A = √(β 1 + β 22) and φ = (24/2π tan−1(β 1/β 2). The standard errors for the parameters of Eq. 1 can be estimated from the standard errors of the parameters of Eq. 2 using the Delta method [43]. The standard errors were used to estimate confidence intervals for each parameter assuming normality. This model was directly applied to compare different groups (wild-type and Opn −/−4 knockout mice). To gain sensitivity in the estimates for experiments in which samples were obtained from two sites (inner and outer retinas) in the same retina (wild-type or Opn −/−4 knockout mice), we treated them as matched pairs and analyzed the differences in the logarithms of expression. A p value for the rejection of the zero amplitude assumption (i.e. no rhythm) was calculated, with rhythm detection considered statistically significant at p ≤ 0.05.
Results
Amplitude and phase relationships of clock and clock-controlled gene expression in the inner and outer layers of the wild-type mouse retina
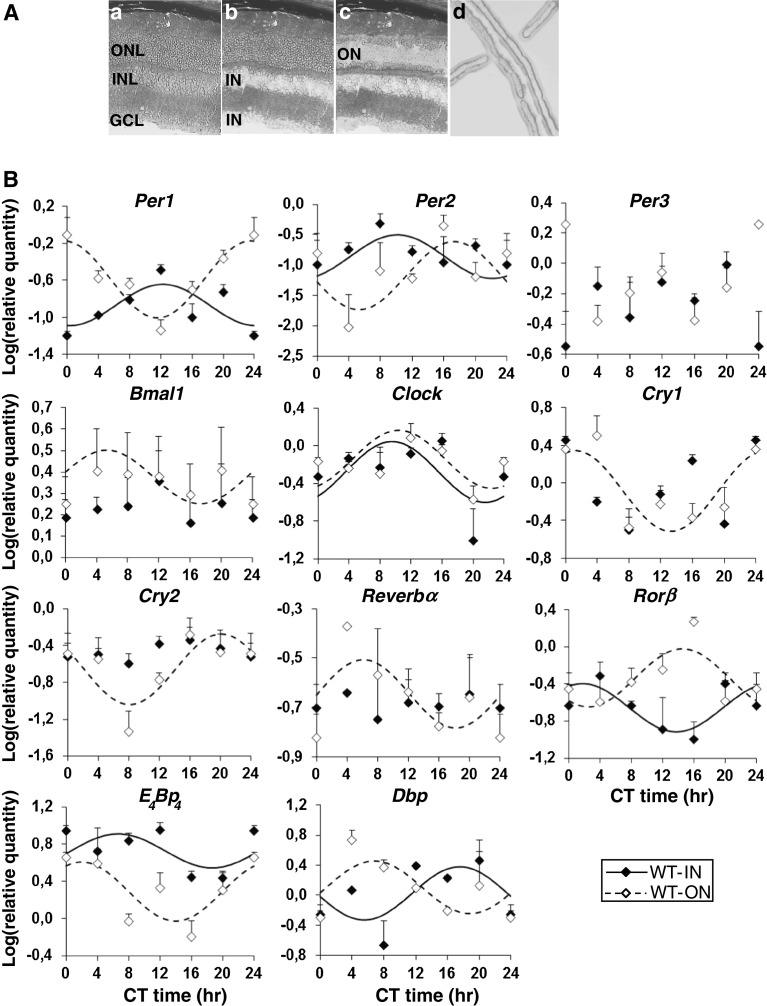
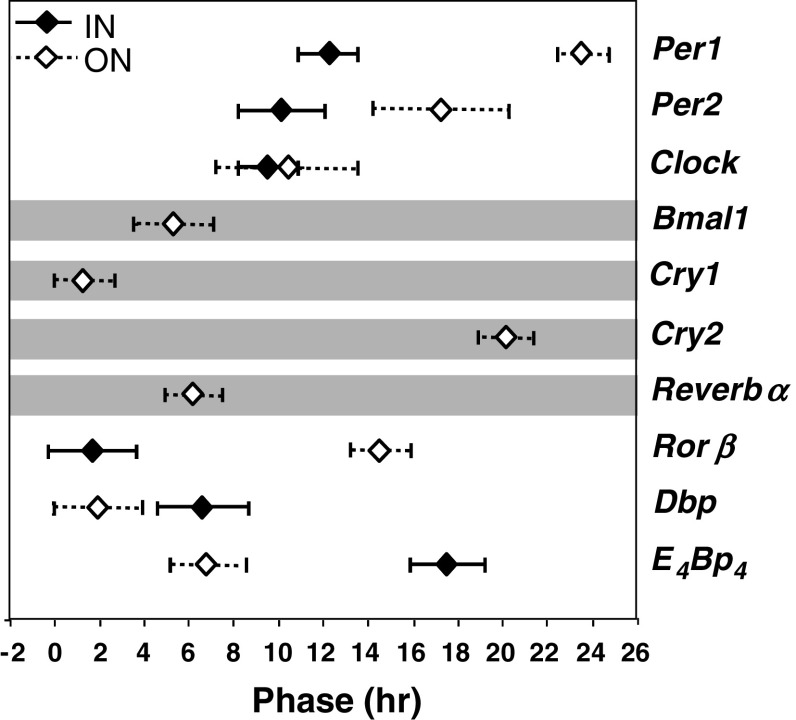
As a first step to characterize the circadian clock in the retina of the wild-type mouse, circadian profiles of clock genes (Per1-3, Clock, Bmal1, Cry1-2, Reverbα and Rorβ) and clock-controlled genes (Dbp, E 4 Bp 4) were analysed in the microdissected inner (INL + GCL) and outer (ONL) layers (Fig. 2A). We found that all clock genes and clock-controlled genes examined were expressed in both inner and outer layers (Fig. 2B). In the outer photoreceptor retina, all clock genes and clock-controlled genes (except Per3) showed circadian variations of transcript levels under a DD cycle (10 of 11 genes tested), whereas in the inner retina, Per3, Bmal1, Cry1-2 and Reverbα did not show a significant 24-h rhythmic component (test of amplitude 0; p > 0.05; 6 of 11 genes tested). No significant differences were observed in the amplitudes (difference between minimum and maximum of fitted cosine function) of the expression of rhythmic genes in the inner or outer retinas. In contrast, for all six clock and clock-controlled genes that showed a circadian profile, the phase of mRNA expression differed in the inner and outer retina of the wild-type mouse (Table 1; Fig. 3) with the exception of Clock.
Fig. 2.
A Photomicrographs of a retinal section of wild-type mouse a before and after laser microdissection of the b inner region (IN) of the retina [including the inner nuclear (INL) + ganglion cell (GCL) layers] and the (c) outer region (ON) which includes the outer nuclear layer (ONL). d Photograph of the captured cell layers onto CapSure™ HS non-contact LCM film. B Circadian expression of clock genes (Per1-3, Clock, Bmal1, Cry1-2, Reverbα and Rorβ) and clock-controlled genes (Dbp, E 4 Bp 4) in the inner (IN) and the outer (ON) retinas of wild-type mouse (n = 18). Mice were exposed to a 12L/12D cycle for 15 days before being released into constant darkness (DD) for one cycle. Animals were euthanized during the next circadian cycle, every 4 h (CT0, CT4, CT8, CT12, CT16 and CT20). The inner and the outer regions of the retina were isolated by laser microdissection and mRNA levels were measured using real-time reverse transcription-polymerase chain reaction. Results are expressed as mean ± SEM (n = 3 for each CT). Data from CT0 are double-plotted at CT24. Continuous (IN retina) and dashed (ON retina) lines represent the periodic sinusoidal functions determined by cosinor analysis. Only periodic sinusoidal functions with amplitude significantly different from zero are represented (p ≤ 0.05; zero amplitude test)
Table 1.
Acrophase (±SD) of mRNA levels of clock and clock-controlled genes in the inner and outer retina, dissected by laser microdissection in the wild-type and Opn −/−4 mouse
| Acrophase (h) | ||||
|---|---|---|---|---|
| WT | Opn −/−4 | |||
| Gene | Inner retina | Outer retina | Inner retina | Outer retina |
| Per1 | 12.23 ± 1.32 | 23.55 ± 0.70* | – | 19.92 ± 1.50 |
| Per2 | 10.13 ± 1.92 | 17.27 ± 1.23* | 22.49 ± 1.57 | – |
| Per3 | – | – | – | – |
| Clock | 9.55 ± 1.36 | 10.42 ± 1.53 NS | 20.56 ± 0.95 | 13.33 ± 2.00# |
| Bmal1 | – | 5.3 ± 2.15 | 17.63 ± 1.66 | – |
| Cry1 | – | 1.26 ± 1.13 | – | – |
| Cry2 | – | 20.12 ± 1.10 | 6.33 ± 2.08 | – |
| Reverbα | – | 6.13 ± 1.75 | – | – |
| Rorβ | 1.71 ± 1.97 | 14.50 ± 1.50* | – | – |
| Dbp | 6.62 ± 2.01 | 1.96 ± 1.23* | 11.91 ± 1.45 | – |
| E 4 Bp 4 | 17.53 ± 1.66 | 6.79 ± 1.58* | 2.95 ± 1.42 | – |
Acrophase value is determined using cosinor analysis and test of amplitude 0 to assess goodness of fit. Only rhythms with a significant value are shown. In the wild-type mouse, for all the genes that present significant circadian expression in both the inner and the outer retina, the acrophases are significantly different between the two regions, with the exception of Clock; * p ≤ 0.05
In the Opn −/−4 mouse, only Clock presents a significant circadian expression in both the inner and the outer retina with an acrophase significantly different between both regions; # p ≤ 0.05
Fig. 3.
Comparison of the phase of mRNA expression for clock genes (Per1-2, Clock, Bmal1, Cry1-2, Reverbα and Rorβ) and clock-controlled genes (Dbp, E 4 Bp 4) in the inner (IN, black diamond) and outer (ON, white diamond) retina in the wild-type mouse. Acrophase (peak of best-fitting 24 h cosine) +95 % confidence limits is shown. Only genes with rhythm detection considered statistically significant (p ≤ 0.05; zero amplitude test) were represented. Horizontal grey shade indicates genes that do not present significant circadian expression in the inner retina (Bmal1, Cry1-2 and Reverbα)
Loss of circadian expression of clock and clock-controlled genes in the photoreceptor layer in the Opn−/−4 mouse
To assess whether melanopsin is required for the generation of endogenous circadian rhythms of clock and clock-controlled genes, we quantified the temporal patterns of expression in the inner and outer retinas of the Opn −/−4 mouse (Fig. 4A). We found that in the inner retina the expression of the majority of genes were unaffected (see Table 1). The circadian expression of Per1 and the clock-controlled gene Rorβ that were rhythmic in the wild-type mouse became arrhythmic in the Opn −/−4 mouse (test of amplitude 0: p ≥ 0.05), whereas Bmal1 and Cry2 that were arrhythmic in the wild-type mouse were rhythmic in the Opn −/−4 mouse (test of amplitude; 0 p ≤ 0.05; see also Fig. 4B). In contrast, all genes (excepting Per1 and Clock) in the outer retina of the wild-type mouse that present a significant circadian rhythm lacked circadian rhythmicity in the Opn −/−4 mouse (8 of 11 genes). Figure 4B presents a schematic summary of clock and clock-controlled genes that show a significant circadian expression in the inner and outer retina in the wild-type mouse and the impact of melanopsin deletion on their rhythmic expression.
Fig. 4.
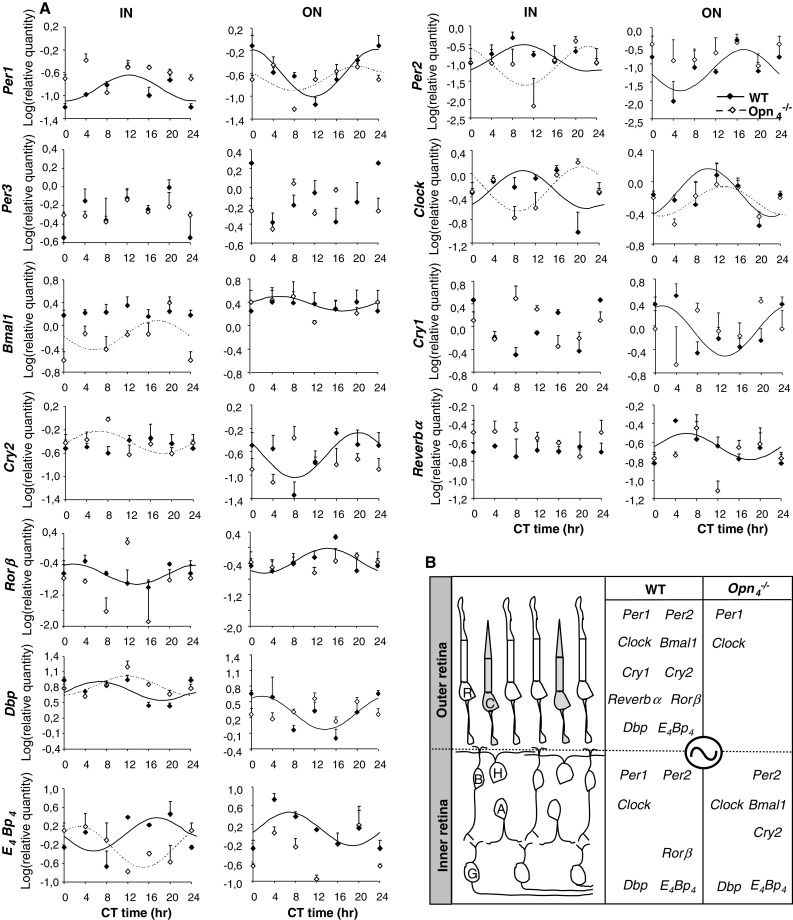
A Circadian expression of clock genes (Per1-3, Clock, Bmal1, Cry1-2, Reverbα and Rorβ) and clock-controlled genes (Dbp, E 4 Bp 4) in the inner (IN) and the outer (ON) retinas of wild-type and Opn −/−4 mouse. Data from the wild-type mouse presented in this figure are the same as shown in Fig. 2. Eyes were collected every 4 h (CT0, CT4, CT8, CT12, CT16 and CT20). The inner and the outer regions of the retina were isolated by laser microdissection and mRNA levels were quantified using real-time reverse transcription-polymerase chain reaction. Results are expressed as mean ± SEM (n = 3 for each CT and each genotype). Data from CT0 are double-plotted at CT24. Continuous (wild-type) and dashed (Opn −/−4 ) lines represent the periodic sinusoidal functions determined by cosinor analysis. Only periodic sinusoidal functions with test of amplitude different from zero were represented. As for the wild-type mouse, no significant differences were observed in the amplitude of clock and clock-controlled gene expression in either the inner or outer regions between the two genotypes. B Schematic summary of clock (Per1-3, Clock, Bmal1, Cry1-2, Reverbα and Rorβ) and clock-controlled (Dbp, E 4 Bp 4) genes that show a significant circadian expression both in the inner and the outer retina in the wild-type mouse and the impact of melanopsin deletion on their rhythmic expression. Genes that do not present a circadian expression (p ≥ 0.05; zero amplitude test) are not represented. We can observe that, in the Opn −/−4 mouse, the majority of genes lose their rhythmicity in the outer retina (except Per1 and Clock), whereas the inner retina is less affected. C cone, R rod, B bipolar cell, H horizontal cell, A amacrine cell, G ganglion cell
Altered light induction of Per1 and Per2 in the outer retina in the Opn−/−4 mouse
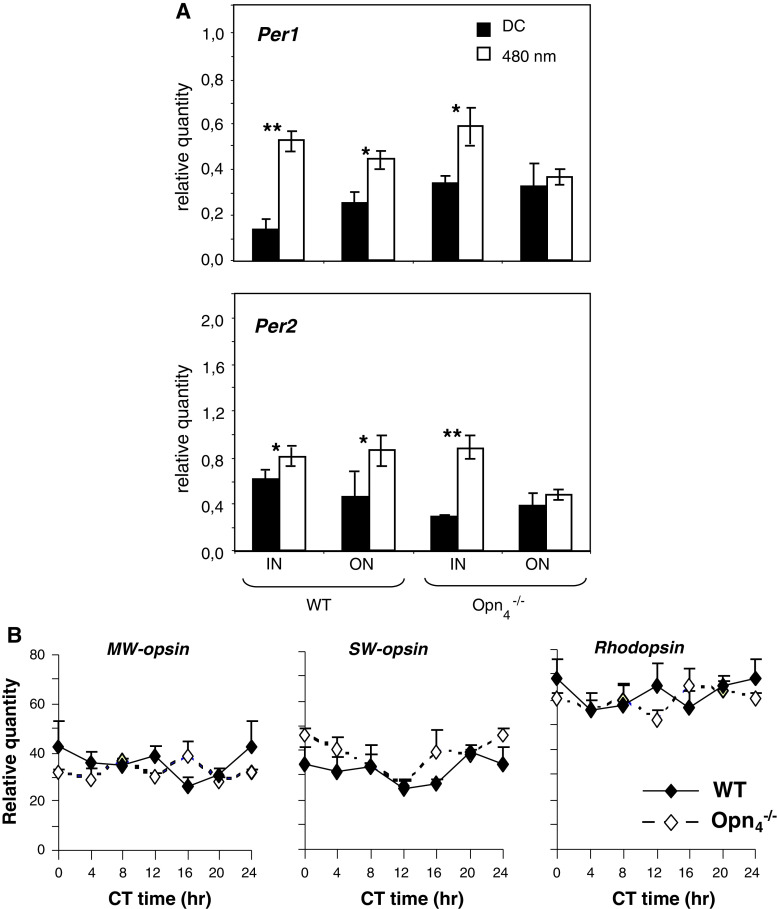
Our results clearly show that, paradoxically, melanopsin is necessary for the maintenance of circadian clock gene expression in the outer retina. In order to further assess whether melanopsin is required for light-induced clock gene expression in the retina, a 480-nm monochromatic light pulse was applied at CT16 (15 min) to wild-type and Opn −/−4 mice. In the wild-type mouse, Per1 and Per2 were significantly induced in both the inner and outer retinal compartments in comparison to dark controls (Fig. 4A). However, in the Opn −/−4 mouse, both clock genes are induced in the inner but not in the outer retina. Since light transduction is mediated by rods, cones and melanopsin, the alteration in the response to light in the outer retina of the Opn −/−4 mouse could result from an indirect effect on outer retinal opsin gene expression. Indeed, retinal photopigment deletion (mutation, transgene) can affect transcriptional regulation of conserved photopigments [44, 45]. This hypothesis was tested by analysing circadian expression of middle- and short-wavelength (MW and SW) opsins and rhodopsin in both genotypes (Fig. 5B). No differences in any opsin gene transcripts (MW, SW and rhodopsin) were found between genotypes for all circadian times suggesting that the deficit in the light-response of the outer retina is not due to down-regulation of cone or rod opsin mRNA that remains unchanged.
Fig. 5.
A Light induction of Per1 and Per2 in the retina of wild-type and Opn −/−4 mice using a pulse of monochromatic light (480 nm, 15 min, 2.8 × 1014 photons/cm2/s) administered at CT16. Animals were sacrificed 30 min after the beginning of the light pulse. The inner (IN) and the outer (ON) regions of the retina were isolated by laser microdissection and mRNA levels were measured using real-time reverse transcription-polymerase chain reaction. Results are expressed as mean ± SEM (n = 5 for each genotype). Asterisks indicate a statistically significant difference (ANOVA: p < 0.05; post hoc Newman-Keuls tests comparing genotypes between the IN and ON retinas (*p < 0.05, **p < 0.01). B Expression of middle-wavelength (MW), short-wavelength (SW) opsins and rhodopsin mRNA in the retina of wild-type (black diamonds) and Opn −/−4 (white diamonds) mouse kept under DD cycle. Transcripts were measured by real-time reverse transcription-polymerase chain reaction, expression was normalized to HPRT control gene. For both genotype, transcripts are not rhythmic over the 24-h cycle in the retina and for each CT, no significant differences between genotype are observed (ANOVA: p > 0.05)
Alteration of TH and DA expression in the Opn−/−4 mouse
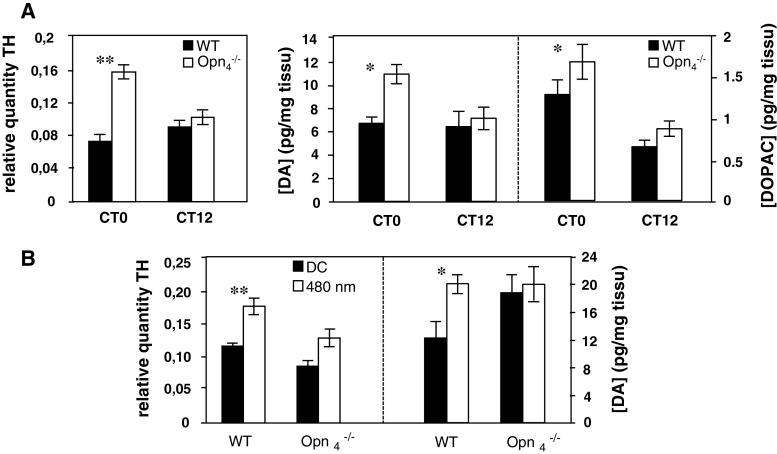
We then explored the hypothesis that in the Opn −/−4 mouse the loss of circadian rhythmicity of clock and clock-controlled genes exclusively in the outer retina results from disrupted DA signalling to the outer retina. This hypothesis is suggested by the excitatory influence of ipRGCs on DAergic amacrine cells [12, 39] and the role of DA on the regulation of clock gene expression in both the retina [33, 46] and the striatum [47]. We thus examined the effect of the absence of melanopsin on TH mRNA expression and found that, in DD conditions, the relative quantity of TH mRNA is significantly increased at CT0 (p = 0.00018) in the Opn −/−4 mouse compared to the wild-type mouse, whereas at CT12 both mouse strains show equivalent expression (Fig. 6A; p = 0.35).
Fig. 6.
A Left panel Circadian expression of tyrosine-hydroxylase (TH) mRNA in wild-type and Opn −/−4 mouse (n = 5 for each genotype). Animals were sacrificed at two circadian times CT0 and CT12 and TH mRNA was quantified by real time RT-PCR. In the wild-type mouse, the level of TH mRNA was constant between CT0 and CT12 whereas in the Opn −/−4, a significantly increase (p ≤ 0.01) was observed at CT0 compared to the control mouse. Right panel Dopamine (DA) and DOPAC concentrations in wild-type and Opn −/−4 mouse (n = 9–11 for each genotype). Post hoc Newman–Keuls test shows significant differences in DA and DOPAC level between genotypes at CT0 (p < 0.05), whereas no significant differences were observed at CT12. B Light induction of TH mRNA (n = 5 for each genotype) and DA content (n = 7–10 for each genotype and for dark and light conditions) in wild-type and Opn −/−4 mouse. A single light pulse (480 nm, 15 min, 2.8 × 1014 photons/cm2/s) was administered at CT16. Animals were sacrificed 30 min after the beginning of the light pulse.TH mRNA induction by light was quantified by real time RT-PCR and DA content by HPLC. In the wild-type mouse, light significantly induces TH mRNA (p < 0.01) and DA concentration (p < 0.05) whereas in the Opn −/−4, no increase was observed compared to the dark control
We then tested whether, in the absence of melanopsin, the increase in TH transcript at CT0 also induced an increase in the retinal level of DA. In the Opn −/−4mouse DA and its main metabolite, 3,4-dihydroxyphenylacetic acid (DOPAC) contents were significantly increased (Fig. 6A; p = 0.037 for DA and p = 0.022 for DOPAC) at CT0, whereas DA and DOPAC concentrations were similar at CT12 in both genotypes. The DOPAC:DA ratio, which is a reliable indicator of DA metabolism, are similar in the two genotypes (data not shown). Previous studies have also shown that TH up-regulation and DA release are increased by light in the wild-type mouse [48]. We confirmed that, following a 480-nm monochromatic light exposure, TH mRNA (p < 0.01) and DA (p < 0.05) are significantly upregulated in the wild-type mouse; however, this was not observed in the Opn −/−4 mouse (Fig. 6B).
Discussion
Mouse photoreceptors are a potential site of endogenous circadian rhythm generation
Based on molecular and physiological data from amphibian and avian retinas, the prevailing view of the circadian organization of the vertebrate retina is that photoreceptors contain self-sustained circadian clocks [23, 24]. In the mammalian retina, the existence of more than one clock has been proposed [49, 50] based on the regional expression of clock genes-clock proteins in the inner nuclear/ganglion cell layers or in the photoreceptors [9, 25–28, 31, 32].
In this study, we examined the endogenous rhythmicity of clock genes in the wild-type mouse retina dissected separately into inner and outer compartments by laser microdissection. This ex vivo approach is essential to understand retinal clock organisation, since the cellular diversity in the mouse retina renders the analysis of the circadian clock at the whole tissue level problematic [51]. Our results show that all core clock genes are expressed in both inner and outer retina, indicating that neurons with the requisite molecular components for circadian pacemakers are present among the major classes of retinal neurons. However, a circadian rhythm of all clock genes was only observed in the photoreceptor layer. Furthermore, for clock genes that show a circadian profile of expression in both inner and outer compartments, the rhythms are out of phase. These results, in agreement with previous laser-dissection and in vitro studies [11, 31], support the view that in the wild-type retina, photoreceptors express endogenous circadian rhythms. A recent study provides evidence that the circadian expression of clock proteins is observed in cones rather than in rods [32], lending support to previous studies that failed to detect core clock genes in rods [9, 29]. One limitation of these in vivo or ex vivo approaches is the difficulty in determining whether the observed rhythm is an autonomous property of the photoreceptor layer itself, or is driven by factors originating elsewhere in the retina.
In the inner retina, the identification of autonomous cellular clocks suffers from similar limitations. The presence of several clock genes or proteins has been documented in the inner retina in several studies [9, 25–29, 32], as well as the coordinate expression of six core clock genes, in particular in DAergic amacrine cells [9, 29]. These clock genes transcripts express circadian rhythms in cultured photoreceptor-deficient isolated retinas and in vertical retinal slice cultures, with PER2::LUC rhythms predominantly localized in the INL [9, 15]. Since the inner retina contains several different cell types (bipolar, horizontal, amacrine and ganglion cells), this does not exclude the possibility of distinct neuronal populations that oscillate with different periods and phases, resulting in low amplitude rhythmicity or masking of coherent clock gene expression [32].
Alteration of photoreceptor clock function in the Opn−/−4 mouse
An unexpected finding is that, in the absence of melanopsin, the circadian expression of the majority of clock and clock-controlled genes is lost in the outer retina. The absence of melanopsin also led to an increase in TH mRNA, the rate-limiting enzyme in DA synthesis, and an increased level of retinal DA and DOPAC compared to wild-type mice at CT0, whereas these levels were similar in both genotypes at CT12 (Fig. 6A). The DOPAC:DA ratio, which is a reliable indicator of DA metabolism, is similar in the two genotypes (data not shown) suggesting that the increased DA in the Opn −/−4 mouse is the result of increased synthesis, turnover and accumulation of this neuromodulator in the retina. DA is known to control the strength of rod-cone photoreceptor coupling and melanopsin mRNA expression [52] through activation of D2–like receptors [53]. Due to a genetic deficit, C57Bl6 mice express a constant, residual level of melatonin, retinal DA content and metabolism are not circadian and DA release is strictly light-driven. In the Opn −/−4 mouse, the abnormally high levels of DA in constant darkness could potentially desynchronize photoreceptors that remain uncoupled, since light is no longer able to reset the clock gene expression of individual oscillators (absence of light-induction Per1 and Per2 mRNAs) and leads to a loss of phase synchrony as has previously been suggested for the loss of circadian regulation of ERG in the Opn −/−4 mouse [13] and for the DA-specific depleted retinal mouse [35]. Loss of clock function results in a similar loss of the light-adapted ERG in the retina-specific Bmal1 invalidated mouse and the Cry1-Cry2 knock-out mice [10, 54], providing further evidence that synchronous clock function is critical. This situation is analogous to the desynchronization of SCN oscillators observed in vivo in the absence of the presumed coupling effect of vasoactive intestinal peptide [55]. Under an LD cycle, SCN neuronal activity is synchronized, whereas in constant conditions, individual SCN neurons continue to oscillate but with broader distribution of individual phases at the population level, resulting in an absence of circadian rhythmicity [55, 56]. Phase coherence between individual oscillators for generation of coherent circadian rhythms at the systems level may be a general principle for both the SCN and for the retina. Taken together, these results suggest that the absence of ipRGC input to DAergic amacrine cells leads to the disruption of DA regulation and consequently may induce a loss of circadian synchronisation among photoreceptor cells. Melanopsin has also been found in some rod/cone photoreceptors in the mouse [57], in human cones [58] and in human and mouse retinal pigment epithelium [59–61]. We cannot rule out the hypothesis that melanopsin present in rods and or cones may play a role in the coupling of photoreceptors through horizontal cell interactions and impact on the circadian oscillations and light-induced expression of outer retinal clock genes [58].
Melanopsin is required for light induced Per gene expression and DA synthesis in the retina
We show for the first time that, in the absence of melanopsin, light fails to induce TH mRNA, DA synthesis and Per1-Per2 mRNA expression in the outer retina. There is substantial evidence that ipRGCs and DAergic cells are functionally and synaptically interconnected in the mammalian retina [36, 62, 63]. ipRGCs receive synaptic inputs from interplexiform DAergic amacrine cells [38]. In turn, ipRGCs provide photic input back to DAergic cells [63] that show transient, sustained or null responses to light [39]. Despite these functional relations between ipRGCs and DAergic amacrine cells, a previous study reported that melanopsin is neither necessary nor sufficient for DA release and c-FOS induction by light in DAergic cells, suggesting that these responses were mediated by rods and/or cones [64]. We find, on the contrary, that the light-induced increase in both TH mRNA and DA requires melanopsin. The differences in the responses observed between the two studies may be related to the light stimulation protocols. Cameron et al. used longer duration and higher intensity white broadband light in comparison to our shorter and monochromatic light stimulation. In our study, the shorter exposure was perhaps insufficient to elicit TH mRNA and dopamine induction in the Opn −/−4 mice. In support of this, Zhang et al. [39] found that, in wild-type mice, ipRGCs provide excitatory sustained light responses to DAergic neurons with a peak sensitivity near 480 nm and that the invalidation of melanopsin eliminates this sustained melanopsin-driven response [12]. However, light induction of c-FOS in DAergic cells is reduced but not eliminated in rd/rd mice, and following pharmacological blockade of rod/cone signaling pathways in wild-type mice, suggesting that rods and/or cones also contribute to the response.
DA plays a critical role in the regulation of many retinal processes including clock gene expression in the inner retina, resetting the phase of the circadian oscillator, the light-induced resetting of PER2::Luc rhythm [15], acute light induction of Per1 [33] and protein phosphorylation in rods and cones [65]. In Xenopus photoreceptors, DA increases Per2 expression and resets the photoreceptor circadian clock [34, 46]. These results emphasize the importance of DA as a retinal signalling and synchronizing factor and suggest that period gene induction by light in the mouse is mediated by a melanopsin-dependent, DA driven signal acting on retinal photoreceptors. In conclusion, melanopsin appears to exert a critical role in the circadian regulation of the mouse retina through its intraretinal retrograde signalling pathway driving DA function.
Acknowledgments
We thank C. Gronfier and G. Gingras for critical reading of the manuscript, S. Hattar for the gift of the Opn −/−4 mice. This research was supported by Rhône-Alpes CMIRA, ANR-09-MNPS-040, Retina France, Volubilis, GDRI Neurosciences, Cluster Handicap Vieillissement Neurosciences. The authors declare no competing financial interests.
References
- 1.Besharse JC, Hollyfield JG, Rayborn ME. Turnover of rod photoreceptor outer segments. II. Membrane addition and loss in relationship to light. J Cell Biol. 1977;75:507–527. doi: 10.1083/jcb.75.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bobu C, Hicks D. Regulation of retinal photoreceptor phagocytosis in a diurnal mammal by circadian clocks and ambient lighting. Invest Ophthalmol Vis Sci. 2009;50:3495–3502. doi: 10.1167/iovs.08-3145. [DOI] [PubMed] [Google Scholar]
- 3.Pierce ME, et al. Circadian regulation of iodopsin gene expression in embryonic photoreceptors in retinal cell culture. Neuron. 1993;10:579–584. doi: 10.1016/0896-6273(93)90161-J. [DOI] [PubMed] [Google Scholar]
- 4.Von Schantz M, Lucas RJ, Foster RG. Circadian oscillation of photopigment transcript levels in the mouse retina. Brain Res Mol Brain Res. 1999;72:108–114. doi: 10.1016/S0169-328X(99)00209-0. [DOI] [PubMed] [Google Scholar]
- 5.Tosini G, Menaker M. Circadian rhythms in cultured mammalian retina. Science. 1996;272:419–421. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- 6.Nir I, Haque R, Iuvone PM. Diurnal metabolism of dopamine in the mouse retina. Brain Res. 2000;870:118–125. doi: 10.1016/S0006-8993(00)02409-4. [DOI] [PubMed] [Google Scholar]
- 7.Doyle SE, Grace MS, McIvor W, Menaker M. Circadian rhythms of dopamine in mouse retina: the role of melatonin. Vis Neurosci. 2002;19:593–601. doi: 10.1017/S0952523802195058. [DOI] [PubMed] [Google Scholar]
- 8.Doyle SE, McIvor WE, Menaker M. Circadian rhythmicity in dopamine content of mammalian retina: role of the photoreceptors. J Neurochem. 2002;83:211–219. doi: 10.1046/j.1471-4159.2002.01149.x. [DOI] [PubMed] [Google Scholar]
- 9.Ruan GX, Zhang DQ, Zhou T, Yamazaki S, McMahon DG. Circadian organization of the mammalian retina. Proc Natl Acad Sci USA. 2006;103:9703–9708. doi: 10.1073/pnas.0601940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Storch KF, et al. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;130:730–741. doi: 10.1016/j.cell.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tosini G, Davidson AJ, Fukuhara C, Kasamatsu M, Castanon-Cervantes O. Localization of a circadian clock in mammalian photoreceptors. FASEB J. 2007;21:3866–3871. doi: 10.1096/fj.07-8371com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang D-Q, Belenky MA, Sollars PJ, Pickard GE, McMahon DG. Melanopsin mediates retrograde visual signaling in the retina. PLoS one. 2012;7:e42647. doi: 10.1371/journal.pone.0042647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnard AR, Hattar S, Hankins MW, Lucas RJ. Melanopsin regulates visual processing in the mouse retina. Curr Biol. 2006;16:389–395. doi: 10.1016/j.cub.2005.12.045. [DOI] [PubMed] [Google Scholar]
- 14.Sakamoto K, et al. Two circadian oscillatory mechanisms in the mammalian retina. Neuroreport. 2000;11:3995–3997. doi: 10.1097/00001756-200012180-00018. [DOI] [PubMed] [Google Scholar]
- 15.Ruan GX, Allen GC, Yamazaki S, McMahon DG. An autonomous circadian clock in the inner mouse retina regulated by dopamine and GABA. PLoS Biol. 2008;6:e249. doi: 10.1371/journal.pbio.0060249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 17.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(Spec No 2):R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 18.Besharse JC, Iuvone PM. Circadian clock in Xenopus eye controlling retinal serotonin N-acetyltransferase. Nature. 1983;305:133–135. doi: 10.1038/305133a0. [DOI] [PubMed] [Google Scholar]
- 19.Whitmore D, Foulkes NS, Strahle U, Sassone-Corsi P. Zebrafish Clock rhythmic expression reveals independent peripheral circadian oscillators. Nat Neurosci. 1998;1:701–707. doi: 10.1038/3703. [DOI] [PubMed] [Google Scholar]
- 20.Tosini G, Menaker M. Multioscillatory circadian organization in a vertebrate, iguana iguana. J Neurosci. 1998;18:1105–1114. doi: 10.1523/JNEUROSCI.18-03-01105.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tosini G, Menaker M. The clock in the mouse retina: melatonin synthesis and photoreceptor degeneration. Brain Res. 1998;789:221–228. doi: 10.1016/S0006-8993(97)01446-7. [DOI] [PubMed] [Google Scholar]
- 22.Zhu H, Green CB. Three cryptochromes are rhythmically expressed in Xenopus laevis retinal photoreceptors. Mol Vis. 2001;7:210–215. [PubMed] [Google Scholar]
- 23.Zhu H, et al. The Xenopus clock gene is constitutively expressed in retinal photoreceptors. Brain Res Mol Brain Res. 2000;75:303–308. doi: 10.1016/S0169-328X(99)00309-5. [DOI] [PubMed] [Google Scholar]
- 24.Cahill GM, Besharse JC. Circadian clock functions localized in xenopus retinal photoreceptors. Neuron. 1993;10:573–577. doi: 10.1016/0896-6273(93)90160-S. [DOI] [PubMed] [Google Scholar]
- 25.Namihira M, et al. Circadian pattern, light responsiveness and localization of rPer1 and rPer2 gene expression in the rat retina. Neuroreport. 2001;12:471–475. doi: 10.1097/00001756-200103050-00010. [DOI] [PubMed] [Google Scholar]
- 26.Witkovsky P, et al. Cellular location and circadian rhythm of expression of the biological clock gene Period 1 in the mouse retina. J Neurosci. 2003;23:7670–7676. doi: 10.1523/JNEUROSCI.23-20-07670.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dinet V, Ansari N, Torres-Farfan C, Korf HW. Clock gene expression in the retina of melatonin-proficient (C3H) and melatonin-deficient (C57BL) mice. J Pineal Res. 2007;42:83–91. doi: 10.1111/j.1600-079X.2006.00387.x. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Fernandez JM, Alvarez-Lopez C, Cernuda–Cernuda R. Cytoplasmic localization of mPER1 clock protein isoforms in the mouse retina. Neurosci Lett. 2007;419:55–58. doi: 10.1016/j.neulet.2007.03.054. [DOI] [PubMed] [Google Scholar]
- 29.Dorenbos R, Contini M, Hirasawa H, Gustincich S, Raviola E. Expression of circadian clock genes in retinal dopaminergic cells. Vis Neurosci. 2007;24:573–580. doi: 10.1017/S0952523807070538. [DOI] [PubMed] [Google Scholar]
- 30.Schneider K, et al. Unique clockwork in photoreceptor of rat. J Neurochem. 2010;115:585–594. doi: 10.1111/j.1471-4159.2010.06953.x. [DOI] [PubMed] [Google Scholar]
- 31.Sandu C, Hicks D, Felder-Schmittbuhl MP. Rat photoreceptor circadian oscillator strongly relies on lighting conditions. Eur J Neurosci. 2011;34:507–516. doi: 10.1111/j.1460-9568.2011.07772.x. [DOI] [PubMed] [Google Scholar]
- 32.Liu X, Zhang Z, Ribelayga CP. Heterogeneous Expression of the Core Circadian Clock Proteins among Neuronal Cell Types in Mouse Retina. PLoS One. 2012;7:e50602. doi: 10.1371/journal.pone.0050602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yujnovsky I, Hirayama J, Doi M, Borrelli E, Sassone-Corsi P. Signaling mediated by the dopamine D2 receptor potentiates circadian regulation by CLOCK:BMAL1. Proc Natl Acad Sci USA. 2006;103:6386–6391. doi: 10.1073/pnas.0510691103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steenhard BM, Besharse JC. Phase shifting the retinal circadian clock: xPer2 mRNA induction by light and dopamine. J Neurosci. 2000;20:8572–8577. doi: 10.1523/JNEUROSCI.20-23-08572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson CR, et al. Retinal dopamine mediates multiple dimensions of light-adapted vision. J neurosci Off J Soci Neurosci. 2012;32:9359–9368. doi: 10.1523/JNEUROSCI.0711-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Contini M, Raviola E. GABAergic synapses made by a retinal dopaminergic neuron. Proc Natl Acad Sci USA. 2003;100:1358–1363. doi: 10.1073/pnas.0337681100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vugler AA, Redgrave P, Hewson-Stoate NJ, Greenwood J, Coffey PJ. Constant illumination causes spatially discrete dopamine depletion in the normal and degenerate retina. J Chem Neuroanat. 2007;33:9–22. doi: 10.1016/j.jchemneu.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 38.Viney TJ, et al. Local retinal circuits of melanopsin-containing ganglion cells identified by transsynaptic viral tracing. Curr Biol. 2007;17:981–988. doi: 10.1016/j.cub.2007.04.058. [DOI] [PubMed] [Google Scholar]
- 39.Zhang DQ, et al. Intraretinal signaling by ganglion cell photoreceptors to dopaminergic amacrine neurons. Proc Natl Acad Sci USA. 2008;105:14181–14186. doi: 10.1073/pnas.0803893105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knoblauch K, Maloney L (2012) Modeling psychophysical data in R. Springer, New York
- 41.Box G, Cox D. An analysis of transformmations (with discussion) J R Stat Soc B. 1964;26:211–252. [Google Scholar]
- 42.Venables WN, Ripley BD (2002) Modern applied statistics with S, 4th edn. Springer, Berlin
- 43.Wasserman LA (2004) All of statistics. A concise course in statistical inference. Springer, New York
- 44.Sakamoto K, Liu C, Tosini G. Classical photoreceptors regulate melanopsin mRNA levels in the rat retina. J Neurosci. 2004;24:9693–9697. doi: 10.1523/JNEUROSCI.2556-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dkhissi-Benyahya O, Gronfier C, De Vanssay W, Flamant F, Cooper HM. Modeling the role of mid-wavelength cones in circadian responses to light. Neuron. 2007;53:677–687. doi: 10.1016/j.neuron.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cahill GM, Besharse JC. Resetting the circadian clock in cultured Xenopus eyecups: regulation of retinal melatonin rhythms by light and D2 dopamine receptors. J Neurosci. 1991;11:2959–2971. doi: 10.1523/JNEUROSCI.11-10-02959.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imbesi M, et al. Dopamine receptor-mediated regulation of neuronal “clock” gene expression. Neuroscience. 2009;158:537–544. doi: 10.1016/j.neuroscience.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Witkovsky P, Gabriel R, Haycock JW, Meller E. Influence of light and neural circuitry on tyrosine hydroxylase phosphorylation in the rat retina. J Chem Neuroanat. 2000;19:105–116. doi: 10.1016/S0891-0618(00)00055-7. [DOI] [PubMed] [Google Scholar]
- 49.Tosini G, Pozdeyev N, Sakamoto K, Iuvone PM. The circadian clock system in the mammalian retina. BioEssays. 2008;30:624–633. doi: 10.1002/bies.20777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Green CB, Besharse JC. Retinal circadian clocks and control of retinal physiology. J Biol Rhythm. 2004;19:91–102. doi: 10.1177/0748730404263002. [DOI] [PubMed] [Google Scholar]
- 51.Peirson SN, et al. Comparison of clock gene expression in SCN, retina, heart, and liver of mice. Biochem Biophys Res Commun. 2006;351:800–807. doi: 10.1016/j.bbrc.2006.10.118. [DOI] [PubMed] [Google Scholar]
- 52.Sakamoto K, et al. Dopamine regulates melanopsin mRNA expression in intrinsically photosensitive retinal ganglion cells. Eur J Neurosci. 2005;22:3129–3136. doi: 10.1111/j.1460-9568.2005.04512.x. [DOI] [PubMed] [Google Scholar]
- 53.Ribelayga C, Cao Y, Mangel SC. The circadian clock in the retina controls rod-cone coupling. Neuron. 2008;59:790–801. doi: 10.1016/j.neuron.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cameron MA, et al. Electroretinography of wild-type and Cry mutant mice reveals circadian tuning of photopic and mesopic retinal responses. J Biol Rhythm. 2008;23:489–501. doi: 10.1177/0748730408325874. [DOI] [PubMed] [Google Scholar]
- 55.Lucassen EA, et al. Role of vasoactive intestinal peptide in seasonal encoding by the suprachiasmatic nucleus clock. Eur J Neurosci. 2012;35:1466–1474. doi: 10.1111/j.1460-9568.2012.08054.x. [DOI] [PubMed] [Google Scholar]
- 56.vanderLeest HT, Rohling JHT, Michel S, Meijer JH. Phase shifting capacity of the circadian pacemaker determined by the SCN neuronal network organization. PLoS One. 2009;4:e4976. doi: 10.1371/journal.pone.0004976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ecker JL et al. ([date unknown]) Melanopsin-expressing retinal ganglion-cell photoreceptors: cellular diversity and role in pattern vision. Neuron 67: 49–60 [DOI] [PMC free article] [PubMed]
- 58.Dkhissi-Benyahya O, Rieux C, Hut RA, Cooper HM. Immunohistochemical evidence of a melanopsin cone in human retina. Invest Ophthalmol Vis Sci. 2006;47:1636–1641. doi: 10.1167/iovs.05-1459. [DOI] [PubMed] [Google Scholar]
- 59.Provencio I, Jiang G, De Grip WJ, Par Hayes W, Rollag MD. Melanopsin: an opsin in melanophores, brain and eye. Proc Natl Acad Sci USA. 1998;95:340–345. doi: 10.1073/pnas.95.1.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Provencio I, et al. A novel human opsin in the inner retina. J Neurosci. 2000;20:600–605. doi: 10.1523/JNEUROSCI.20-02-00600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peirson SN, et al. Expression of the candidate circadian photopigment melanopsin (Opn4) in the mouse retinal pigment epithelium. Brain Res Mol Brain Res. 2004;123:132–135. doi: 10.1016/j.molbrainres.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 62.Wong KY, Dunn FA, Graham DM, Berson DM. Synaptic influences on rat ganglion-cell photoreceptors. J Physiol. 2007;582:279–296. doi: 10.1113/jphysiol.2007.133751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vugler AA, et al. Dopamine neurones form a discrete plexus with melanopsin cells in normal and degenerating retina. Exp Neurol. 2007;205:26–35. doi: 10.1016/j.expneurol.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 64.Cameron MA, et al. Light regulation of retinal dopamine that is independent of melanopsin phototransduction. Eur J Neurosci. 2009;29:761–767. doi: 10.1111/j.1460-9568.2009.06631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pozdeyev N, et al. Dopamine modulates diurnal and circadian rhythms of protein phosphorylation in photoreceptor cells of mouse retina. Eur J Neurosci. 2008;27:2691–2700. doi: 10.1111/j.1460-9568.2008.06224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]