Abstract
The gamma (γ)-secretase holoenzyme is composed of four core proteins and cleaves APP to generate amyloid beta (Aβ), a key molecule that causes major neurotoxicity during the early stage of Alzheimer’s disease (AD). However, despite its important role in Aβ production, little is known about the regulation of γ-secretase. OCIAD2, a novel modulator of γ-secretase that stimulates Aβ production, and which was isolated from a genome-wide functional screen using cell-based assays and a cDNA library comprising 6,178 genes. Ectopic expression of OCIAD2 enhanced Aβ production, while reduction of OCIAD2 expression suppressed it. OCIAD2 expression facilitated the formation of an active γ-secretase complex and enhanced subcellular localization of the enzyme components to lipid rafts. OCIAD2 interacted with nicastrin to stimulate γ-secretase activity. OCIAD2 also increased the interaction of nicastrin with C99 and stimulated APP processing via γ-secretase activation, but did not affect Notch processing. In addition, a cell-permeable Tat-OCIAD2 peptide that interfered with the interaction of OCIAD2 with nicastrin interrupted the γ-secretase-mediated AICD production. Finally, OCIAD2 expression was significantly elevated in the brain of AD patients and PDAPP mice. This study identifies OCIAD2 as a selective activator of γ-secretase to increase Aβ generation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-013-1515-x) contains supplementary material, which is available to authorized users.
Keywords: Alzheimer’s disease, OCIAD2, γ-secretase, Nicastrin
Introduction
Alzheimer’s disease (AD) is a major type of dementia that affects at least 35 million people worldwide [1]. The main symptoms of the disease are memory loss, cognitive impairment, and behavioral abnormalities caused by synaptic dysfunction and neuronal loss [2]. Two major features characterize the AD patient brain. One is the presence of amyloid plaques, made up of amyloid beta (Aβ) fibrils surrounded by degenerating neurons [3, 4]. The second are neurofibrillary tangles, which are composed of hyper-phosphorylated Tau protein aggregates inside neurons [5]. Aβ peptides are 37–46 amino acids in length (with the predominant form being Aβ1–40); pathogenicity is mainly associated with the longer form, Aβ1–42 [6]. Aβ causes major toxicity to neurons in the early stage of disease progression (i.e., the mild cognitive impairment stage), and affects Tau-mediated memory loss [7, 8]. Thus, elucidation of the molecular regulation of Aβ generation is important for understanding the pathogenesis of AD and to slow disease progression.
Aβ is produced from sequential cleavage of the Aβ precursor protein (APP) by β-site amyloid precursor protein cleaving enzyme (BACE1) and γ-secretase. γ-Secretase is composed of four core components: presenilin (PS), nicastrin (NCT), anterior pharynx-defective phenotype 1 (APH-1), and presenilin enhancer 2 (PEN2) [9–11]. PS is a catalytic subunit of the enzyme complex, while NCT functions as the gatekeeper of over 80 substrates, including APP, Notch, ErbB4, and CD44. APH-1 functions as a scaffold in complex assembly, forming a sub-complex with NCT, and PEN2 is linked to PS endoproteolysis which generates cleaved PS-NTF and PS-CTF. In most cases, the proteolytic activity of γ-secretase is targeted to type 1 membrane proteins that are shed by other proteases, for example, α-secretase or β-secretase in APP processing. Because γ-secretase processing is the rate-limiting step of Aβ generation, there have been many efforts to modulate the activity of this pathogenic enzyme complex in AD.
At the molecular level, fewer than 10 regulators of γ-secretase have been identified, such as CD147, TMP21, GPR3, and β-arrestin [12–19]. Of these, only GPR3 and β-arrestin1 exhibit Notch-sparing activity without sharing a common mechanism of action. In addition, a small molecule has been identified that can also regulate APP processing by binding to APP, leading to the generation of a non-toxic form of Aβ without causing the side effects associated with Notch deficiency [20]. These results raise the possibility that γ-secretase components, and/or substrate-targeting γ-secretase modulators, are involved. Because a Notch-defective phenotype is toxic during animal development [21–26], the identification of a Notch-sparing, novel modulator of γ-secretase activity towards APP is required, which could also serve as a potential therapeutic target for drug development.
A genome-wide functional screen using a cell-based assay and full-length cDNA library was undertaken to identify γ-secretase activators. A novel protein, OCIAD2, was identified, which stimulates γ-secretase activity to enhance Aβ production.
Materials and methods
Antibodies
The following antibodies were used: anti-Tubulin (Sigma-Aldrich, T6074), anti-Actin (Sigma-Aldrich, A1978), anti-GAPDH (Santa Cruz Biotechnology, sc-365062), anti-GFP (Santa Cruz Biotechnology, sc-8334), anti-HA (HA hybridoma), anti-V5 (Sigma-Aldrich, S2540), anti-LC3 (Novus Biologicals, NB600-1384), anti-PRNP (Abcam, ab52604), anti-FLOT1 (BD Biosciences, 610821), anti-FACL4 (kindly gifted from Dr. S.M. Prescott, University of Utah, USA), anti-FLAG (Sigma-Aldrich, S7425), anti-PS1-NTF (Santa Cruz Biotechnology, sc-7860), anti-BACE1 (Cell Signaling, 5606), anti-APP-CTF (Sigma-Aldrich, A8717), anti-NOTCH1 (Abcam, ab27526), anti-NICD (Novus, NB200-251), anti-NCT (Sigma-Aldrich, N1660), anti-4G8 (Convance, SIG-39220), anti-6E10 (Convance, SIG-39320), anti-OCIAD2 (Sigma-Aldrich, SAB3500119, 3500118), anti-Tom20 (Santa Cruz Biotechnology, sc-17764), anti-DR5 (Abcam, 47179), anti-ADAM10 (Millipore, AB19026), anti-TACE (Santa Cruz Biotechnology, H-300), and anti-APH-1a and anti-PEN2 antibodies (kindly gifted from Dr. T. Tomita, University of Tokyo, Japan).
Cell culture and DNA transfection
SY5Y-APPswe cells, BACE KO MEF (knockout mouse embryonic fibroblast) cells, and HEK-APP695 cells are described in Refs. [27, 28]. HEK293T, HeLa, CHO-7PA2 cells, and stable cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (HyClone, SH30243.01) supplemented with 10 % fetal bovine serum (HyClone, SH30919.03), and 50 μg/ml Gentamicin (GIBCO, 15750-060) at 37 °C under 5 % CO2 (v/v) condition. HeLa/shCtrl and HeLa/shOCIAD2 stable cells were selected with 2 mg/ml G418 (Gold Bio, G418-5) and maintained with 1 mg/ml G418 for experiments. According to the manufacturer’s instructions, transfection was performed using Polyfect (Qiagen, 1015586) or PEI (Sigma, 764647).
Genome-wide functional screening using cDNA
By modifying pC99-GVP and UAS-Luciferase assay [27], we generated C99-TetOn and TRE-GFP fusion constructs to regulate its activity with doxycyclin. HEK293T cells were co-transfected with pC99-TetOn, pTRE-GFP, pDsRed monomer, and either pCtrl or each cDNA for 24 h and then incubated with conditioned media containing 100 ng/ml doxycycline (Sigma-Aldrich) for another 24 h. Once C99-rtTA is cleaved by γ-secretase, the cleavage product AICD-rtTA is transported into the nucleus and induces GFP expression through TRE in the presence of doxycyclin. Compared to control, the putative positive cDNA clones which significantly increased green fluorescence under fluorescence microscope (Olympus) were isolated. After the primary screening, the secondary screening was conducted using pC99-GVP and pUAS-luciferase reporter assay [27]. After confirming the stimulatory effect on the reporter activity, the putative positive cDNA clones were tested for its stimulatory effect on Aβ generation using sandwich ELISA kit. The cDNA library was prepared as previously described [29, 30], and purified by a mini-prep kit (Cosmo Genetech).
Plasmid construction and shRNA sequences
Primer sequences for the construction of OCIAD2 deletion mutants are: pOCIAD2 FL (1–154) (forward: 5′-CCC AAG CTT GGG ACA AAG GGC CGG AAG GAC TCT CCG CTG C-3′, reverse: 5′-CCC AAG CTT GGG GGA AGC TGA AGG CTG AG-3′); pOCIAD2 N120 (1–120) (reverse: 5′-CGG GGT ACC CCG AGC CCC ACG GAG CTG ATC-3′); pOCIAD2 N48 (1–48) (reverse: 5′-CGG GGT ACC CCG TTC CTG ACA TTC TCG CAT-3′). Primer sequences for OCIAD2 Chimera I are: Chimera I (OCIAD2 N120 + OCIAD1 C133) (forward: 5′-CGG GGT ACC CCG GCT TTA CGA TCA GGA CAA-3′, reverse: 5′-CGG GGT ACC CCG CTC ATC CCA AGT ATC TCC-3′). Primer sequences for pAPPswe-FLAG are: (forward: 5′-CCC AAG CTT GGG ATG CTG CCC GGT TTG G-3′, reverse: 5′-TGC TTA GAG CAG TTC TGC ATC TGC TCA A-3′). pNCT-V5, pHA-PEN2, pPS1-Myc and pAPH1-FLAG are previously described in ref. [31]. pFLAG-SGK1/CA was kindly provided by Dr. Park H.S. (Chonnam National University, Korea). PCR was conducted using Pfu polymerase (Neurotics) and the PCR products were subcloned into pcDNA3 (HA) or p3XFLAG CMV14. The sequences for the construction of pSuper-neo-shOCIAD2 are: (forward: 5′-GAT CCC CGA AGA AAG TTT CTG GAA GAT TCA AGA GAT CTT CCA GAA ACT TTC TTC TTT TTA-3′, reverse: 5′-AGC TTA AAA AGA AGA AAG TTT CTG GAA GAT CTC TTG AAT CTT CCA GAA ACT TTC TTC GGG-3′). Primer sequences for the construction of OCIAD2 ER-/Mitochondria-targeting construct are: pER-OCIAD2 (APP signal peptide forward: 5′-AGC TTA TGC TGC CCG GTT TGG CAC TGC TCC TGC TGG CCG CCT GGA CGG CTC GGGCGCCGC-3′, reverse: 5′-GGC GCC CGA GCC GTC CAG GCG GCC AGC AGG AGC AGT GCC AAA CCG GGC AGC ATA-3′); pMito-OCIAD2 (Ds-Mito2 MTS sequence forward: 5′-CCC AAG CTT GGG ATG TCC GTC CTG ACG CC-3′, reverse: 5′-TCC CCG CGG GGA CAA CGA ATG GAT CTT GG-3′); OCIAD2 insertion (OCIAD insertion forward: 5′-CCC AAG CTT GGG TCC CCG CGG GGA ATG GCT TCA GCG TCT GCT CG-3′, reverse: 5′-CGG GGT ACC CCG GGA AGC TGA AGG CTG AG-3′). Primer sequences for the construction of GFP-NCTC19 are: (forward: 5′-AAT TCT AAT GCC AAA GCT GAT GTC CTT TTC ATT GCT CCC CGG GAG CCA GGA GCT GTG TCA TAC TGA GGT AC-3′, reverse: 5′-CTC AGT ATG ACA CAG CTC CTG GCT CCC GGG GAG CAA TGA AAA GGA CAT CAG CTT TGG CAT TAG-3′).
ELISA
Aβ1–40 and Aβ1–42 levels in the culture media were measured by sandwich ELISA kits (Invitrogen and IBL). Each sample was harvested from six-well plates and subjected to assay following the manufacturer’s instruction.
SDS-PAGE and western blot analysis
The harvested cells were resuspended with HEPES buffer (pH 7.4) containing protease inhibitor cocktail (1 mM PMSF, 1 mM Aprotinin, 0.2 mM 1,10-phenanthroline monohydrate and 1 mM Leupeptin) and lysed with sampling buffer (60 mM Tris pH 6.8, 2 % SDS, 20 % Glycerol, 10 % 2-Mercaptoethanol, and 0.04 % Bromophenol blue). For the preparation of membrane protein, cells were solubilized in RIPA or 1 % CHAPS buffer containing protease inhibitor cocktail and centrifuged at 10,000g for 10 min. The soluble supernatants were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and the separated proteins were transferred to polyvinylidene fluoride membrane (PVDF) using a Bio-Rad semi-dry transfer unit (Bio-Rad). Blots were blocked with 3 % (w/v) BSA in TBS-T solution (25 mM Tris pH 7.5, 150 mM NaCl, and 0.05 % Tween-20) and then blots were incubated with primary antibodies, followed by horseradish peroxidase-conjugated secondary antibodies for ECL analysis.
In vitro AICD generation assay
In vitro AICD generation assay was conducted as previously described with minor modification [32]. The harvested cells were lysed by sonication in buffer A (50 mM HEPES pH 7.4, 150 mM NaCl, 5 mM 1,10-phenanthroline monohydrate, 2 mM EDTA, and protease inhibitor cocktail). The homogenate was centrifuged at 1,000g for 10 min and the remaining supernatant was further centrifuged at 10,000g for 15 min. The membrane fraction in pellets was washed once with buffer A and centrifuged again. The final membrane pellet was resuspended with buffer A and total protein was quantified by Bradford assay (Bio-Rad). The same amount of protein was incubated at 37 °C for 2 h with or without 100 μM Compound E (Comp. E).
BN-PAGE
BN-PAGE was performed as previously described [33, 34]. The same amount of microsomal membranes protein was solubilized in BN-PAGE buffer (0.5 % dodecylmaltoside, 20 % glycerol, and 25 mM Bis–Tris pH 7.0) for 60 min on ice. After ultracentrifugation at 100,000g for 30 min, the same volume of soluble protein was separated by BN-PAGE at 4 °C and transferred into PVDF membrane. The transferred blot was destained for 1 h in destaining solution (distilled water:methanol:acetic acid, 6:3:1) and analyzed with western blotting.
Glycerol velocity gradient fractionation
Glycerol velocity gradient fractionation was performed as previously described [14]. HEK293T cells were washed with ice-cold PBS and resuspended in buffer A (5 mM HEPES pH 7.4, 1 mM EDTA, 250 mM sucrose and protease inhibitor cocktail). Cells were homogenized with homogenization buffer (5 mM HEPES pH 7.4, 1 mM EDTA, 250 mM sucrose and protease inhibitor cocktail) and postnuclear supernatant was prepared by centrifugation. After centrifugation at 100,000g for 1 h at 4 °C, the pellet was resuspended in buffer B (50 mM Tris pH 7.5, 2 mM EDTA, 150 mM NaCl, and protease inhibitor cocktail), followed by solubilization process with buffer C (2 % CHAPSO, 50 mM Tris pH 7.5, 2 mM EDTA and 150 mM NaCl). The soluble lysates were centrifuged again at 100,000g for 30 min and the supernatants were subjected to glycerol gradient centrifugation. Total 1 mg of protein extracts was applied to the top of 9.6-ml 10–40 % (w/v) linear glycerol gradient and centrifuged for 15 h at 100,000g and 4 °C using a Beckman SW32.1 Ti rotor. Each fraction was collected from the top to the bottom of the gradient and same volume of each fraction was analyzed by western blotting.
Sucrose gradient fractionation
Sucrose gradient fractionation was performed as previously described with minor modification [35]. HEK293T cells were washed with ice-cold PBS and solubilized in buffer S (25 mM HEPES pH 7.4, 150 mM NaCl, 2 mM EDTA, 1 % CHAPSO and protease inhibitor cocktail). Cells were homogenized by 10 passages through a 25-gauge needle and soluble lysates were adjusted to final concentration of sucrose (45 %) and transferred to a 10-ml ultracentrifuge tube. Then, a discontinuous sucrose gradient was formed by sequentially layering 35 % sucrose (3.2 ml) and 5 % sucrose (3.2 ml), and the tubes were subjected to ultracentrifugation at 100,000g for 19 h in Beckman SW32.1 Ti rotor at 4 °C with no brakes. Twelve 0.8-ml fractions were collected from the top to the bottom of the gradient and the same volume of each fraction was analyzed by western blotting.
Immunoprecipitation (IP) assay
General IP was performed using RIPA buffer (50 mM Tris–Cl, 150 mM NaCl, 2 mM EDTA, 0.1 % SDS, 1 % NP40, 0.5 % sodium deoxycholate, and protease inhibitor cocktail). IP of γ-secretase protein complex was performed using 1 % CHAPS buffer (25 mM HEPES pH 7.4, 150 mM NaCl, 2 mM EDTA, 1 % CHAPS, and protease inhibitor cocktail). Cell lysates were prepared by sonication in RIPA or CHAPS buffer. After brief centrifugation, the supernatants were incubated with anti-HA, anti-APP-CTF, anti-NCT, anti-FLAG, anti-PS1, or anti-OCIAD2 antibody at 4 °C for overnight and then pulled-down by Protein G Sepharose beads (GE Healthcare).
Luciferase reporter γ-secretase activity assay
Luciferase reporter γ-secretase activity assay was sas described in Ref. [27]. HEK293T cells were co-transfected with pC99-GVP, pUAS-Luciferase, pβ-galactosidase, and experimental control genes with or without 1 μM N-[N-(3,5-Difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT). After 24 h, cell extracts were analyzed by luciferase assay following the manufacturer’s instruction (Promega). The luciferase activity was normalized by β-galactosidase activity for transfection efficiency.
α-Secretase assay
α-Secretase assay was performed as previously described with minor modification [46]. When HEK-APP695 cells were confluent (>90 %), medium was changed to serum-free DMEM supplemented with Gentamicin for 6 h. The conditioned medium was collected and concentrated about 50-fold using Amicon Ultra-15 (30,000 molecular weight cutoff) (Millipore). Total 20 μg of concentrated protein was subjected to SDS-PAGE and analyzed by western blotting using anti-sAPPα antibody (anti-6E10). sAPPα signal on the blot was quantified by densitometry analysis and compared to APP level.
β-Secretase assay
β-Secretase activity was measured with β-Secretase assay kit (R&D System). Each sample was subjected to enzyme assay following the manufacturer’s instruction.
General fractionation
General fractionation was performed as described with minor modification [47]. Cells were homogenized in IB-1 buffer (30 mM Tris–Cl pH 7.4, 225 mM Mannitol, 75 mM sucrose, and 0.1 mM EGTA) by sonication. The homogenate was centrifuged at 1,000g for 10 min to remove unbroken cells and nuclei. The supernatant was collected and centrifuged at 10,000g for 10 min to collect the heavy membrane fraction. After centrifugation, the supernatant was collected and centrifuged again at 100,000g for 60 min to collect the light membrane fraction.
AD patient information
AD patient and control brain samples were supplied by the Harvard Brain Tissue Resource Center (McLean Hospital, Boston). Detailed information is described in the supplementary data.
Statistical analysis
The expression data from AD brain is represented as mean ± SEM by the indicated densitometry analysis. Student’s t test was used to calculate the P value of the each independent experiment between OCIAD2 and the control group. Data analysis was conducted with the Excel 2010 software except for the quantification of Aβ (Graph Pad Prism).
Results
OCIAD2 regulates APP processing to increase Aβ generation
To identify a new γ-secretase modulator, a novel cell-based assay using a C99-TatOn fusion construct and TRE–GFP reporter was established and used to screen a human full-length cDNA library consisting of 6,178 genes inserted into a mammalian expression vector. After co-transfection of HEK293T cells cultivated on multi-well culture plates with C99-TatOn, TRE-GFP, DsRed Monomer, and each cDNA, potential positive cDNA clones that had significantly higher green fluorescence than control cells were isolated. Among the primary cDNA clones, an OCIAD2 cDNA clone was identified through quantitative analysis of γ-secretase activity in the secondary screen, and it was highly effective in activating γ-secretase. Therefore, the regulation of γ-secretase by OCIAD2 was characterized in greater detail.
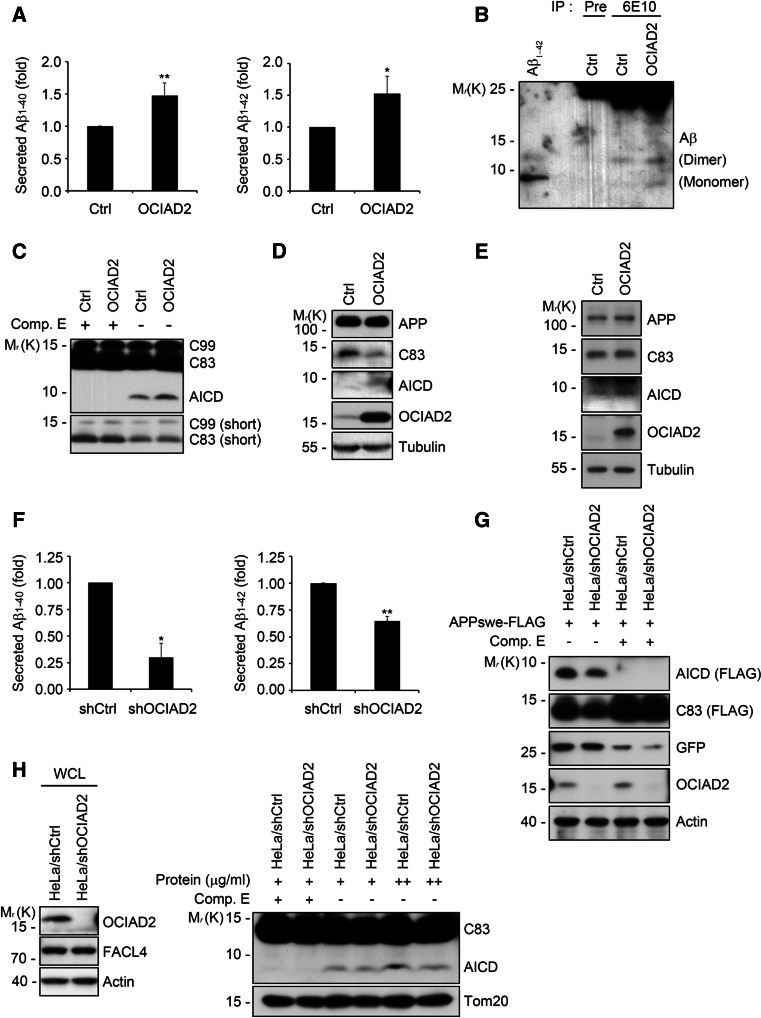
The levels of secreted Aβ1–40 and Aβ1–42 were measured after OCIAD2 overexpression in SH-SY5Y-APPswe neuroblastoma cells. Results from the ELISA showed that the levels of Aβ1–40 and Aβ1–42 in the culture medium increased by 50 % following OCIAD2 overexpression (Fig. 1a). When the amount of Aβ was examined by immunoprecipitation using an anti-Aβ antibody (6E10), a similar result was obtained, demonstrating that OCIAD2 overexpression increased Aβ levels in the culture medium relative to the control (Fig. 1b). Furthermore, in vitro processing of APP was assessed by measuring the production of APP intracellular domain (AICD), a cleavage product of γ-secretase. Compared to the control, a crude membrane fraction prepared from cells overexpressing OCIAD2 showed a significant increase in AICD production (Fig. 1c) and this was confirmed by western blot analysis using whole cell lysates (WCL) (Fig. 1d). AICD production was inhibited by incubating the cells with the γ-secretase inhibitor Comp. E (Fig. 1c). In contrast, this in vitro incubation had no effect on the levels of C99 (β-APP-CTF) and C83 (α-APP-CTF), which are cleavage products of β- and α-secretase, respectively (supplementary Fig. 1a–d). Moreover, OCIAD2 overexpression increased AICD production in BACE knockout MEF cells (Fig. 1e), but did not alter the levels of BACE1 protein in HEK293T cells (supplementary Fig. 1b).
Fig. 1.
OCIAD2 increases Aβ generation. a OCIAD2 overexpression stimulates Aβ1–40 and Aβ1–42 generation in cells. SH-SY5Y-APPswe cells were transfected with pcDNA3 (pCtrl) or OCIAD2 (pOCIAD2), and maintained in the conditioned media for 48 h. The media was measured for Aβ1–40 (left) and Aβ1–42 (right) using ELISA kit. Bars mean ± SD (n = 5). *P < 0.05, **P < 0.01. b OCIAD2 overexpression increases the production of Aβ species. CHO-7PA2 cells were transfected with pCtrl or pOCIAD2 for 36 h and then incubated in serum-free DMEM for another 12 h. The media was subjected to immunoprecipitation (IP) assay using pre-immune serum (Pre) or anti-Aβ antibody (6E10), and the immunoprecipitates were analyzed with western blotting. Monomer and dimer of Aβ are indicated. c, d OCIAD2 overexpression increases the generation of AICD. HEK-APP695 cells were transfected with pCtrl or pOCIAD2 for 48 h and crude membrane fraction was prepared by centrifugation as described in “Materials and methods”. The membrane fraction (10 μg) was then incubated at 37 °C for 2 h with or without 100 μM Compound E (Comp. E) and separated by SDS-PAGE for western blotting (c). The whole cell lysates from the homogenized sample from (c) were subjected to SDS-PAGE and analyzed by western blotting (d). e Ectopic expression of OCIAD2 enhances AICD generation in BACE KO MEF cells. BACE KO MEF cells were transfected with either pCtrl or pOCIAD2 for 48 h and then subjected to western blot analysis. f OCIAD2 knockdown reduces Aβ1–40 and Aβ1–42 production. HeLa/pSuper-neo (shCtrl) and HeLa/pSuper-neo-shOCIAD2 (shOCIAD2) stable cells were transfected with pAPPswe-FLAG and pEGFP for 72 h in the conditioned media, and Aβ1–40 (left) and Aβ1–42 (right) were then measured using ELISA kit. Bars mean ± SD (n = 3). *P < 0.05, **P < 0.01. g, h OCIAD2 knockdown reduces AICD generation. After co-transfection of HeLa/shCtrl and HeLa/shOCIAD2 stable cells with pAPPswe-FLAG and pEGFP for 24 h, cell extracts were analyzed by western blotting (g). Membrane fraction (30 and 50 μg) prepared from HeLa/shCtrl and HeLa/shOCIAD2 stable cells was incubated at 37 °C for 2 h with or without 100 μM Comp. E and then analyzed by western blotting (h, right). Expression of OCIAD2 was determined by western blotting (h, left)
Next, the effects of OCIAD2 knockdown on APP processing and Aβ production were examined. A stable HeLa cell line (HeLa/shOCIAD2) was generated in which OCIAD2 expression was attenuated (Fig. 1h, left). When exogenous APPswe-FLAG was introduced into these cells, the levels of secreted Aβ1–40 and Aβ1–42 in the culture medium were significantly reduced in HeLa/shOCIAD2 cells compared to control HeLa/shCtrl cells (Fig. 1f). When cellular levels of the generated AICD were evaluated by western blot, it was found that AICD production was decreased by OCIAD2 knockdown (Fig. 1g). In contrast, the expression level of EGFP, which was co-expressed as a negative control, was unaffected. An in vitro assay measuring AICD generation in these cells found that a reduction in OCIAD2 expression led to significant suppression of AICD production, which was also inhibited by Comp. E (Fig. 1h, right). Taken together, these results suggest that OCIAD2 regulates APP processing to increase Aβ generation by modulating γ-secretase activity.
OCIAD2 increases γ-secretase activity by regulating the stability of NCT
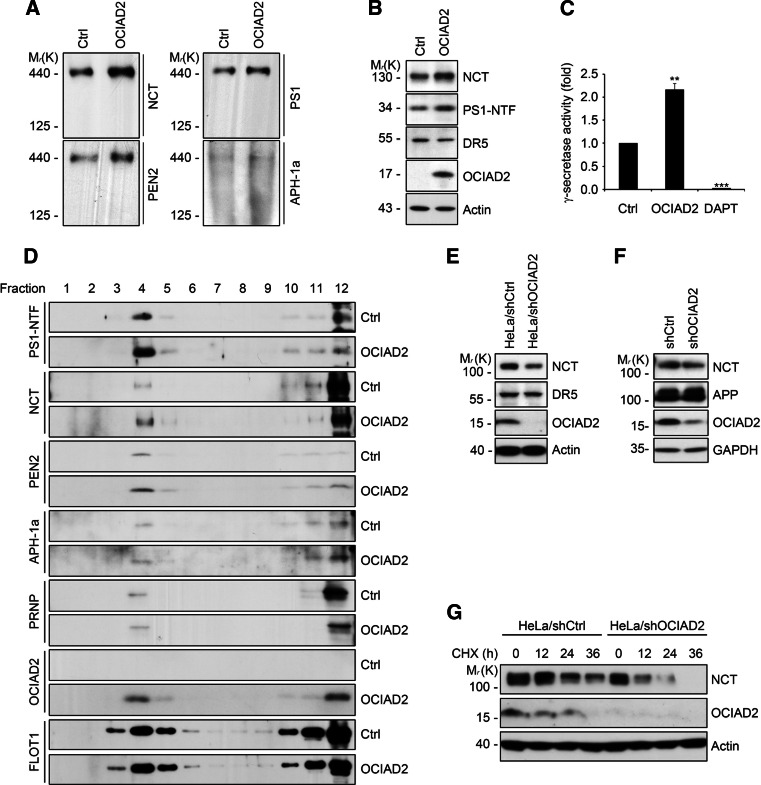
In order to determine whether OCIAD2 affects the formation of the γ-secretase complex and thereby increases the enzymatic activity, a blue-native (BN)-PAGE assay was used [33, 34]. A 1 % digitonin-soluble microsome fraction was purified by centrifugation and separated by BN-PAGE. The western blot revealed a significant increase in the amount of a high molecular weight protein complex of around 440 kDa, which contained NCT, presenilin1 (PS1), PEN2, and APH-1 resulting from OCIAD2 overexpression (Fig. 2a). The expression of each protein was determined by western blot, and it was found that the levels of each of the active components of the complex, including mature NCT, PS1-NTF, and PEN2, were increased (Fig. 2b; supplementary Fig. 2a). In contrast, expression of death receptor 5, which served as a negative control, was unchanged. In addition, a C99-GVP/UAS-luciferase reporter assay of γ-secretase activity revealed that OCIAD2 overexpression induced a two-fold increase in γ-secretase activity (Fig. 2c).
Fig. 2.
OCIAD2 enhances γ-secretase activity and regulates NCT. a Ectopic expression of OCIAD2 increases the amount of γ-secretase complex on blue-native (BN) gel. After transfection of HEK293T cells with either pCtrl or pOCIAD2 for 48 h, membrane fraction (50 μg) solubilized in 1 % digitonin was separated by BN-PAGE and analyzed by western blotting. b Ectopic expression of OCIAD2 increases the amount of mature forms of NCT and PS1-NTF. After transfection of HEK293T cells with either pCtrl or pOCIAD2 for 48 h, cell lysates solubilized in 1 % CHAPS lysis buffer were analyzed by western blotting. c Ectopic expression of OCIAD2 increases reporter activity of γ-secretase. HEK293T cells were co-transfected with pC99-GVP, pUAS-Luciferase, pβ-galactosidase, and either pCtrl or pOCIAD2 with or without 1 μM DAPT. After 24 h, cell extracts were analyzed by luciferase assay. Transfection efficiency was normalized by β-galactosidase activity and the relative activity compared to control is described as a fold unit. Bars mean ± SD (n = 3). **P < 0.01, ***P < 0.001. d Localization of γ-secretase components in lipid raft is enhanced by OCIAD2 overexpression. HEK293T cells transfected with pCtrl or pOCIAD2 were solubilized in 1 % CHAPS lysis buffer. The soluble protein (12 mg) was subjected to discontinuous (5, 35, 45 %) sucrose gradient fractionation assays (top, fraction 1; bottom, fraction 12) and same volumes of each fraction were analyzed by western blotting. e–g OCIAD2 knockdown reduces the stability of NCT protein. HeLa/shCtrl and HeLa/shOCIAD2 cell lysates were analyzed by western blotting (e). After transfection of HEK-APP695 cells with pshCtrl or pshOCIAD2 for 60 h, cell lysates were analyzed by western blotting (f). HeLa/shCtrl or HeLa/shOCIAD2 stable cells were treated with 30 μM cycloheximide for the indicated times. Cell lysates solubilized in RIPA buffer were analyzed by western blotting (g)
Previous reports have shown that lipid rafts are the major location of γ-secretase function [35, 38]. To ascertain whether OCIAD2 functions in lipid rafts with the γ-secretase complex, the subcellular localization of overexpressed OCIAD2 was determined by sucrose gradient fractionation. Around 30 % of the OCIAD2 co-localized with flotillin1 (FLOT1), a lipid raft marker. Interestingly, there was a two-fold increase in γ-secretase components in the same lipid raft fraction upon OCIAD2 overexpression, along with a considerable amount of OCIAD2 (Fig. 2d). These observations suggest that OCIAD2 enhances the presence of γ-secretase complex components in lipid rafts.
The effects of OCIAD2 knockdown on NCT levels were examined in HEK-APP695 and HeLa cells, which expressed higher levels of endogenous OCIAD2 than HEK293T and Hep3B cells (supplementary Fig. 2b). In stable HeLa/shOCIAD2 cells, NCT expression was significantly reduced (Fig. 2e). Similar results were observed in HEK-APP695 and HeLa cells transiently transfected with OCIAD2 shRNA (Fig. 2f; supplementary Fig. 2c). Regulation of NCT expression by OCIAD2 was at the protein and not the mRNA level (supplementary Fig. 2d). In addition, HEK293T and Hep3B cells, which have low endogenous levels of OCIAD2, also showed limited NCT expression (supplementary Fig. 2b). Protein translation was blocked in HeLa/shOCIAD2 cells using cycloheximide to confirm OCIAD2-mediated regulation of NCT protein stability. In the absence of OCIAD2, the half-life of NCT was reduced from about 12 to <6 h (Fig. 2g), indicating that OCIAD2 is required to stabilize NCT. The same result was observed when this experiment was performed in HEK-APP695 cells (supplementary Fig. 2e). In light of recent reports suggesting that the stability of NCT is regulated by SGK1 and ERK1/2 [39, 40], the effect of these kinases on OCIAD2-mediated NCT stabilization was examined. However, OCIAD2 knockdown had no affect the protein level of SGK1 and ERK1/2 (supplementary Fig. 2f, g). Collectively, these results suggest that OCIAD2 regulates the stability of the NCT protein.
OCIAD2 interacts with NCT via its C-terminus to activate γ-secretase
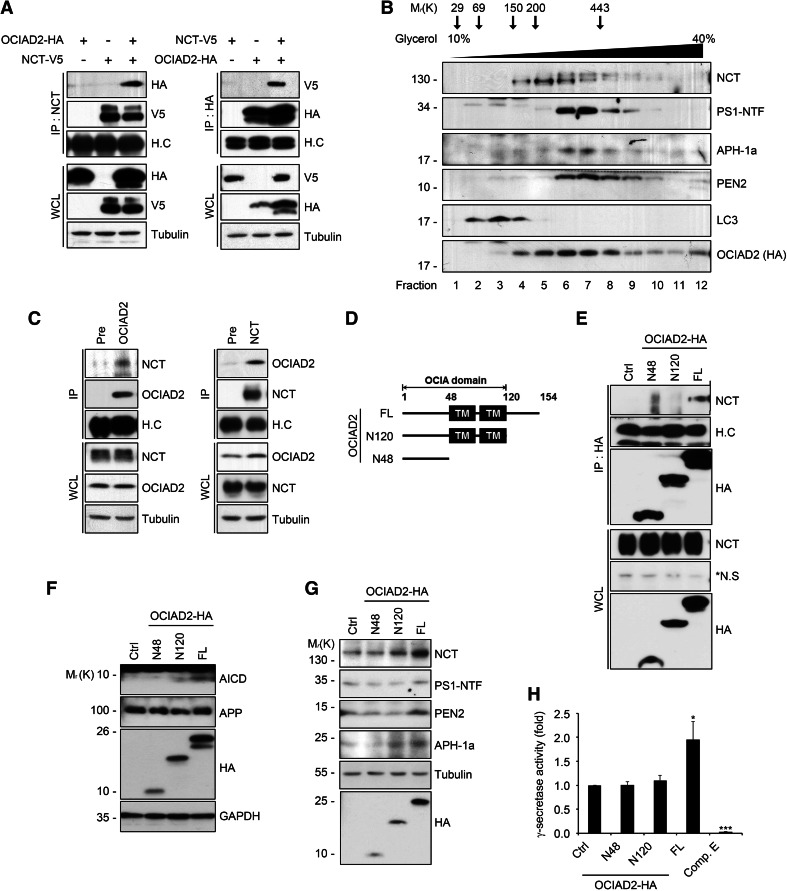
To understand the molecular mechanisms by which OCIAD2 regulates the formation of the γ-secretase complex and its enzymatic activity, a potential interaction between OCIAD2 and γ-secretase components was examined. Immunoprecipitation revealed that OCIAD2 interacts robustly with NCT in HEK293T cells transfected with OCIAD2 (Fig. 3a), but only weakly with PS1, and not with other components of the complex such as APH-1a and PEN2 (supplementary Fig. 3a). In addition, western blot analysis of fractions obtained by glycerol velocity gradient sedimentation revealed that OCIAD2 was mainly present in the same fractions as NCT (Fig. 3b). In contrast, the autophagosome marker LC3 (used as a negative control) was excluded from these fractions. Furthermore, NCT was detected in immunoprecipitates from HEK-APP695 cells containing endogenous OCIAD2 and vice versa (Fig. 3c; supplementary Fig. 3b), indicating that endogenous OCIAD2 directly binds to NCT.
Fig. 3.
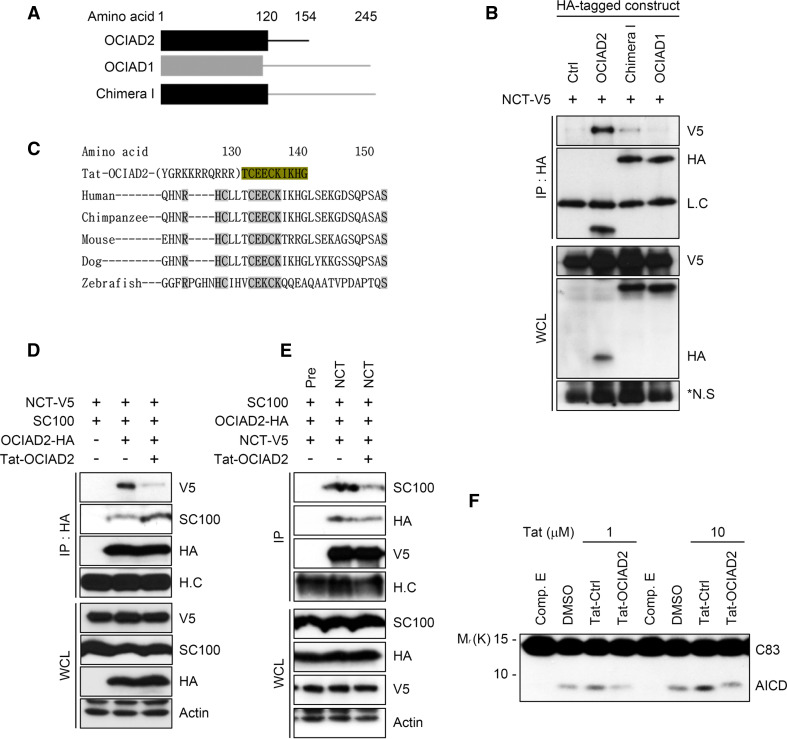
OCIAD2 interacts with NCT through its C-terminal region. a Exogenous OCIAD2 interacts with NCT. After transfection of HEK293T cells with pNCT-V5 and pOCIAD2-HA for 36 h, cell extracts were analyzed by IP assays using anti-NCT (left) or anti-HA (right) antibody followed by western blotting. H.C heavy chain of immunoglobulin. b Co-localization of OCIAD2 with γ-secretase complex in microsomal membrane fraction. After transfection of HEK293T cells with pOCIAD2-HA for 24 h, microsomal membrane fraction prepared by centrifugation and solubilized in 1 % CHAPSO was subjected to glycerol velocity gradient fractionation assay. Each fraction (top, fraction 1; bottom, fraction 12) was analyzed by western blotting. c Endogenous OCIAD2 interacts with NCT. Cell lysates of HEK-APP695 cells were analyzed by IP assays using either anti-OCIAD2 (left) or anti-NCT (right) antibody. d Schematic diagram of OCIAD2 and its deletion mutants. e The C-terminal region of OCIAD2 is required for its interaction with NCT. HEK293T cells were transfected with pOCIAD2 deletion mutant for 48 h and cell extracts were then analyzed by IP assay using anti-HA antibody. The immunoprecipitates and whole cell lysates were proved by western blotting. f Effects of OCIAD2 mutants on AICD generation. HEK-APP695 cells were transfected with pOCIAD2 or pOCIAD2 deletion mutant for 48 h and cell lysates were analyzed by western blotting. g Effects of OCIAD2 mutants on the amount of γ-secretase components. HEK293T cells were transfected with pOCIAD2 or pOCIAD2 deletion mutant for 48 h and cell lysates were analyzed by western blotting. h Effects of OCIAD2 mutants on γ-secretase reporter activity. HEK293T cells were transfected with pC99-GVP, pUAS-Luciferase, pβ-galactosidase, and either pOCIAD2 or pOCIAD2 deletion mutant with or without 3 μM Comp. E for 24 h. Cell extracts were subjected to luciferase assay. The luciferase activity was normalized by β-galactosidase activity. Bars mean ± SD (n = 3). *P < 0.05, ***P < 0.001
OCIAD2 consists of the OCIA domain (residues 1–120) containing the N-terminus (residues 1–48), transmembrane (TM) region (harboring 2 TM region in residues 49–120), and the hydrophilic C-terminal cytosolic domain (residues 121–154) (Fig. 3d). To identify the domain responsible for the interaction of OCIAD2 with NCT, deletion constructs of OCIAD2 were generated based on predictions of hydrophobicity (using TMHMM and Uniprot). Immunoprecipitation experiments in which the various OCIAD2 deletion constructs were expressed revealed that full-length OCIAD2 bound NCT as expected, but neither the OCIAD2 N120 construct lacking the C-terminal 34 residues nor the OCIAD2 N48 construct lacking both the TM domain and the C-terminal 34 residues interacted with NCT (Fig. 3e), indicating that the C-terminal 34 residues of OCIAD2 are essential for the interaction between these two proteins. Moreover, when the ability of each construct to modulate AICD production was examined, full-length OCIAD2 was the most effective at increasing AICD production in HEK-APP695 cells (Fig. 3f). In addition, full-length OCIAD2 increased the levels of mature NCT and PS1-NTF (Fig. 3g), as well as the luciferase reporter activity of γ-secretase (Fig. 3h). These results suggest that OCIAD2 interacts with NCT through its hydrophilic C-terminal domain, which is also critical for the regulation of γ-secretase activity.
OCIAD2 enhances docking of C99 with NCT, but has no effect on Notch∆E processing
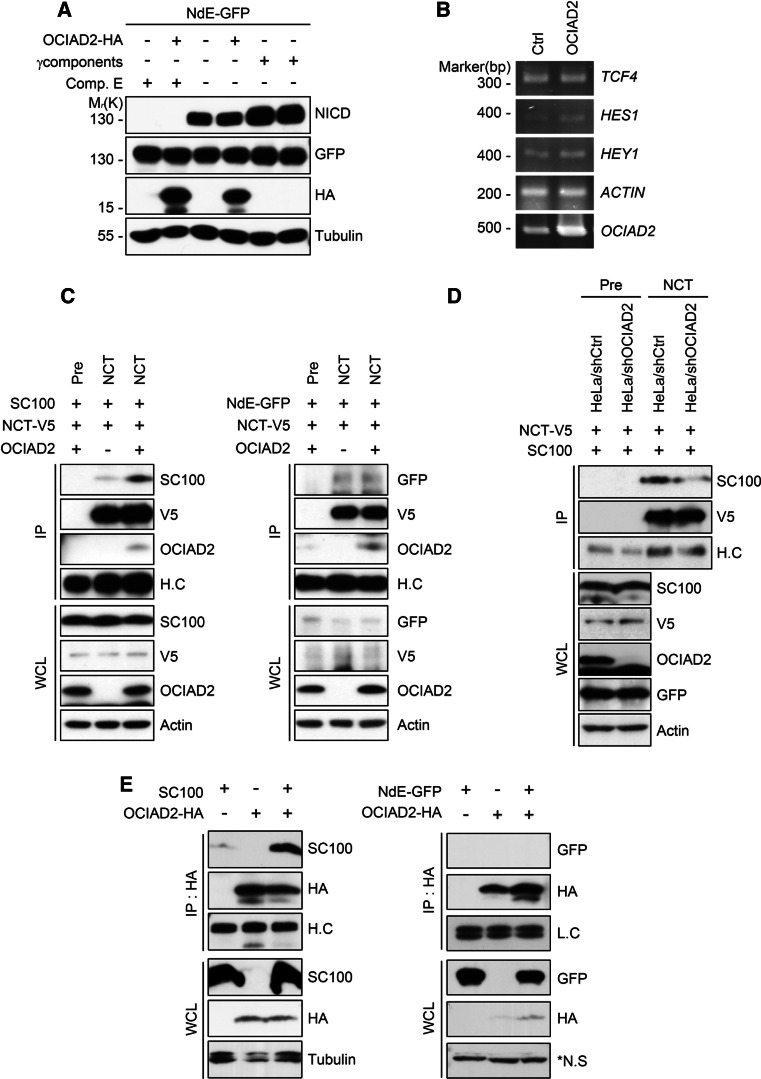
The selectivity of OCIAD2 on the processing of γ-secretase substrates, specifically APP and Notch, was assessed. To address this point, a truncated form of the Notch receptor tagged with EGFP (Notch∆E-GFP) was used. The level of Notch intracellular domain (NICD), generated by γ-secretase cleavage of Notch, was determined by western blot. The overexpression of OCIAD2 did not stimulate the production of NICD in HEK293T cells (Fig. 4a; supplementary Fig. 4). In this assay, NICD generation was significantly increased by co-expression with 4 core γ-secretase components and was inhibited by Comp. E. Furthermore, results obtained from expression analysis of NICD downstream genes using RT-PCR showed that OCIAD2 overexpression had no effect on TCF4, HES1, and HEY1 mRNA levels (Fig. 4b) [41]. These results imply that OCIAD2 does not affect the processing of Notch by γ-secretase.
Fig. 4.
OCIAD2 enhances docking of NCT with C99 but does not affect Notch∆E processing. a, b OCIAD2 overexpression does not affect the cleavage of Notch and the expression of NICD-regulated genes. HEK293T cells were transfected with pNotch∆∆E-GFP (NdE-GFP) and either pCtrl, pOCIAD2-HA, or 4 core γ-secretase components for 24 h with or without 3 μM Comp. E. Cell extracts were analyzed by western blotting (a). After transfection of HEK293T cells with pCtrl or pOCIAD2-HA for 24 h, total RNA was purified and analyzed by RT-PCR using synthetic primers for TCF4, HES1, and HEY1 (b). c OCIAD2 enhances the interaction of NCT with C99 (SC100) but not with NotchΔE. HEK293T cells were co-transfected with pNCT-V5, pOCIAD2, and either pSC100 (left) or pNdE-GFP (right) for 36 h, and cell extracts were subjected to IP assays using anti-NCT antibody. The immunoprecipitates were proved by western blotting. d Down-regulation of OCIAD2 expression reduces the interaction between NCT and C99. HeLa/shCtrl and HeLa/shOCIAD2 cells were transfected with pNCT-V5 and pSC100 for 48 h. The RIPA-soluble cell lysates were subjected to IP assay using anti-NCT antibody, followed by western blot analysis. pEGFP was included in every transfection for the normalization of transfection efficiency. e OCIAD2 interacts with C99 but not with NotchΔE. HEK293T cells were transfected with pOCIAD2-HA and either pSC100 (left) or pNdE-GFP (right) for 36 h, and cell extracts were subjected to IP assays using anti-HA antibody. The immunoprecipitates and whole cell lysates were analyzed by western blotting
NCT functions as a gate keeper of γ-secretase substrate [49]. To test whether OCIAD2 provides substrate selectivity by controlling the docking of NCT with its substrate, immunoprecipitation was used to assess the interaction between NCT and its two major substrates, C99 (SC100) and Notch∆E. The results revealed that overexpression of OCIAD2 increased the interaction of NCT with C99 (Fig. 4c, left), but did not influence the interaction of NCT with Notch∆E (Fig. 4c, right). Conversely, the interaction of NCT with C99 was greatly reduced by OCIAD2 knockdown in HeLa cells (Fig. 4d). In addition, the experiments showed that OCIAD2 could interact with C99, but not with Notch∆E (Fig. 4e). Together, these results suggest that OCIAD2 enhances the docking of NCT with C99 but not with Notch∆E.
Blocking the interaction between OCIAD2 and NCT inhibits AICD generation
A homology search was performed to further assess the functional importance of the C-terminus of OCIAD2. The search identified OCIAD1, which also contains an OCIA domain as well as a more extended C-terminus, and which showed 36 % amino acid sequence homology with OCIAD2 (Fig. 5a). Unlike OCAID2, however, OCIAD1 did not interact with NCT and C99 (data not shown). To further corroborate the importance of the C-terminus of OCIAD2 for its interaction with NCT, a Chimera I construct was generated, containing the N-terminal OCIA domain (residues 1–120) of OCAID2 and the C-terminus (residues 113–245) of OCIAD1 (Fig. 5a). The interaction between Chimera I and NCT was examined; the results showed that replacing the C-terminus of OCIAD2 with that of OCIAD1 abrogated its interaction with NCT (Fig. 5b). Additional C-terminal deletion constructs of OCIAD2 were then generated (encompassing residues 120–154), and it was found that OCIAD2 N131 (lacking residues 132–154) failed to interact with NCT, while OCIAD2 N142 (lacking residues 143–154) was able to bind NCT (supplementary Fig. 5a, b). A search for sequence homology in the C-terminal residues of OCIAD2 in different species, including humans, chimpanzees, mice, dogs, and zebrafish, revealed a highly conserved sequence, 134CEXCK138 (Fig. 5c).
Fig. 5.
Interference of the interaction between OCIAD2 and NCT reduces AICD generation. a Schematic diagram showing OCIAD2, OCIAD1 and Chimera I containing the N-terminus of OCIAD2 (OCIA domain) fused to the C-terminus of OCIAD1 (lacking OCIA domain). Colored boxes represent the OCIA domain. b The C-terminus of OCIAD2, not OCIAD1, is critical for the interaction with NCT. HEK293T cells were co-transfected with pNCT-V5 and pOCIAD2-HA, pOCIAD1-HA, or pChimera I-HA for 36 h. Cell extracts were analyzed by IP assay using anti-HA antibody. c Amino acid sequence alignment in the C-terminus of OCIAD2 among different species. Tat-OCIAD2 synthetic peptide comprises the conserved CEXCK sequence of OCIAD2 and Tat sequence (YGRKKRRQRRR). d, e Treatment with Tat-OCIAD2 peptide prevents the interaction between OCIAD2 and NCT. HEK293T cells were pretreated with 20 μM Tat-control (Tat-Ctrl) or Tat-OCIAD2 peptide for 1 h and then co-transfected with pNCT-V5, pSC100, and pOCIAD2-HA for another 36 h in the presence of Tat-peptides. RIPA-soluble cell extracts were then analyzed by IP assays using anti-HA (d) or anti-NCT (e) antibody, followed by western blotting. f Treatment with Tat-OCIAD2 peptide interferes AICD generation. The crude membrane fraction of HEK-APP695 cells prepared by fractionation method was pretreated with Tat-Ctrl or Tat-OCIAD2 peptides at 4 °C for 10 min and then incubated at 37 °C with or without 100 μM Comp. E for 2 h in the presence of Tat-peptides. The samples were separated by SDS-PAGE and analyzed by western blotting
To directly address the contribution of the interaction between OCIAD2 and NCT to APP processing, a cell-permeable Tat sequence-containing OCIAD2 peptide (Tat-OCIAD2) was synthesized, containing the C-terminal charged amino acid residues (133TCE E CKIKHG142) of OCIAD2 which included the highly conserved CEXCK sequence (Fig. 5c). When HEK293T cells expressing NCT, C99, and OCIAD2 were treated with the peptide, immunoprecipitation revealed that the Tat-OCIAD2 peptide, but not the control Tat-peptide (Tat-Ctrl), abolished the interaction between OCIAD2 and NCT (Fig. 5d). In contrast, the interaction between OCIAD2 and C99 was increased in the presence of the Tat-OCIAD2 peptide. The same experiment conducted using an anti-NCT antibody showed that the Tat-OCIAD2 peptide prevented the association of NCT with C99 (Fig. 5e), consistent with the finding that a reduction in OCIAD2 expression abolished the interaction between NCT and C99 (Fig. 4d). Furthermore, in vitro measurement of a crude membrane fraction of HEK-APP695 cells found that 10 min of pre-incubation with Tat-OCIAD2 peptide was sufficient to attenuate AICD production (Fig. 5f).
The C-terminal domain of NCT is short, containing only 19 amino acids. To address whether this segment is important for the interaction of NCT with OCIAD2, a fusion protein was generated in which GFP was tagged with the C-terminal 19 amino acids of NCT (GFP-NCTC19). Competitive immunoprecipitation assays revealed that overexpression of GFP-NCTC19 weakened the interaction between OCIAD2 and NCT in HEK293T cells (supplementary Fig. 5d), and decreased AICD production in HEK-APP695 cells (supplementary Fig. 5e, f). Taken together, these results suggest that the evolutionarily conserved C-terminus (residues 133–142) of OCIAD2 is important for its interaction with NCT and for the regulation of APP processing by γ-secretase.
OCIAD2 localizes to mitochondria and MAM
To investigate the localization of OCIAD2 in cells, GFP- and RFP-tagged OCIAD2 fusion constructs were generated. When either OCIAD2-GFP or OCIAD2-RFP was expressed in combination with an intracellular marker protein tagged with a fluorescent reporter (ER-RFP, Mito-REP, Rab5-GFP, or Lamp1-GFP) in HeLa cells, OCIAD2 was mainly observed in mitochondria and mitochondria-associated ER membrane (MAM) (supplementary Fig. 6a). Based on our data, it was speculated that MAM-associated OCIAD2, not mitochondrial OCIAD2, modulates γ-secretase activity. We made an attempt to dissect the stimulatory function of OCIAD2 in either mitochondria or MAM on γ-secretase activity. To test this possibility, ER- and mitochondria (Mito)-targeted constructs of OCIAD2 were generated (supplementary Fig. 6b). Because there was no available way to target OCIAD2 to MAM, we took the second best try to send OCIAD2 to ER membrane using the signal peptide of APP. When these constructs were expressed in HEK293T cells, the ER-targeted OCIAD2, but not the mitochondria-targeted OCIAD2, retained its stimulatory effect on γ-secretase activity (supplementary Fig. 6c). This result was confirmed by examining endogenous AICD production in HEK-APP695 cells. ER-OCIAD2 stimulated AICD production to the same degree as WT-OCIAD2, but with just 10 % of the transfected amount, while Mito-OCIAD2 had little effect on AICD level (supplementary Fig. 6d). Together, it appears that OCIAD2 in MAM has the stimulatory effect on γ-secretase activity.
OCIAD2 expression is elevated in the brains of AD patients and AD model mice
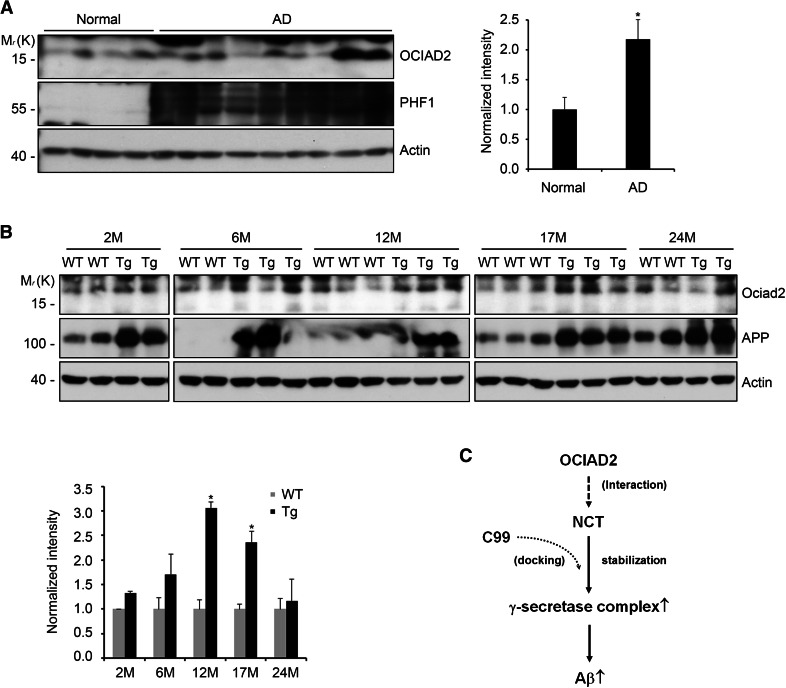
To investigate the pathological relevance of OCIAD2 in Alzheimer’s disease (AD), the expression level of OCIAD2 in the brains of AD patients was examined. Compared to age-matched non-AD patients, OCIAD2 expression was significantly elevated by 2.2-fold in the hippocampus of AD patients (Fig. 6a). When Ociad2 expression was examined in the brains of PDAPP mice (J20 line), which express familial AD-mutant amyloid precursor protein (hAPP) and show learning and memory deficits [42], Ociad2 expression in the cortex was slightly but significantly elevated in 6-month-old mice, and was high in 12- and 17-month-old animals (Fig. 6b). These results indicate that OCIAD2 expression is upregulated in the brains of AD patients and AD model mice, highlighting the potential importance of OCIAD2 in AD.
Fig. 6.
OCIAD2 expression is elevated in the brains of Alzheimer’s disease (AD) patients and PDAPP transgenic mice. a The expression of OCIAD2 is significantly elevated in the hippocampus of AD patients. Hippocampal extracts of control (Normal) and AD patient (AD) brains were analyzed by western blotting. Quantification of the signals on the blots in (a) was performed by densitometry analysis. Bars represent mean value ± SD (n = 4 for normal control; n = 9 for AD). *P < 0.05. b The expression of Ociad2 is elevated in the cortical region of PDAPP mice. Cortical extracts of WT and PDAPP mice brains were analyzed by western blotting. Quantification of the signals on the blots in (b) was performed as in (a). Statistical significance was determined from 12-month- and 17-month-old mouse samples. *P < 0.05. c A proposed model on γ-secretase regulation by OCIAD2. OCIAD2 interacts with NCT and stabilizes it, leading to stimulation of γ-secretase activity in lipid raft and increase in the production of Aβ
Discussion
This study identified OCIAD2 as a novel activator of γ-secretase through a gain-of-function screen (GOFS) of a cDNA expression library. This approach was selected over a loss-of-function screen (LOFS) of a siRNA library because the objective was to identify a γ-secretase activator, and a gain of activity may not have been detected in the cell-based assays of a LOFS. The screening procedure involved three rounds of selection from overexpression experiments; potential positive clones were initially isolated based on C99 cleavage-induced changes in GFP intensity in cells, which was followed by a secondary screening process that measured γ-secretase activity using a C99-GVP/UAS-luciferase reporter. Finally, the clones were screened by directly measuring Aβ levels using ELISA. OCIAD2 was identified as being the most effective in all three assays. OCIAD2 is a membrane protein enriched in mitochondria and MAM, and is mainly expressed in the brain, kidney, and liver [MOPED, PaxDb]. As predicted from the GOFS, the level of OCIAD2 was increased in SH-SY5Y neuroblastoma cells after exposure to oxidative stress (supplementary Fig. 7).
Previous studies have reported that γ-secretase regulators such as CD147, TMP21, and GPR3 interact with core components of the γ-secretase complex to regulate enzymatic activity [12, 14, 16]. In this study, OCIAD2 was observed to interact selectively with NCT, and this interaction was essential for the regulation of γ-secretase activity. However, it is unlikely that OCIAD2 is an auxiliary component of the γ-secretase holoenzyme, because OCIAD2 was not detected in the γ-secretase core complex that was pulled down by the anti-PS1 antibody in immunoprecipitation experiments (data not shown). However, in the fractionation assay, OCIAD2 and NCT were distributed throughout the microsome fraction in which γ-secretase components were also found. Interestingly, OCIAD2 seemed to co-localize primarily with the immature form of NCT in low molecular protein complexes (150–200 kDa). These data indicate that OCIAD2 may be involved in the pre-assembly of the enzyme complex, which includes NCT.
To date, there have been no reports on the function of OCIAD2, which has a relatively low molecular weight of 16 kDa. Deletion mapping revealed that the C-terminus (residues 131–142) is required for its interaction with NCT, as well as for stimulation of γ-secretase activity. Specifically, the highly conserved 134CEXCK138 sequence is found in a variety of species. Structural analyses using in silico modeling indicated that the C-terminus may form a valley that has a high chance of engaging in protein–protein interactions, while the other regions do not possess any distinguishing features (data not shown). The C- and N-termini of OCIAD2 were sensitive to limited proteolysis (supplementary Fig. 5c), suggesting that the C-terminus of OCIAD2 protrudes from the membrane to interact with NCT. NCT may also interact with OCIAD2 through its C-terminus. As in other cases in which proteins are stabilized through interaction with a binding partner [43, 44], the evidence presented here suggests that OCIAD2 interacts with and stabilizes NCT.
Over the past decade, a major obstacle in AD therapeutics research has been the development of a selective inhibitor of γ-secretase against APP. Because a phenotype of Notch deficiency is severe cytotoxic damage and cell death [21–26], the main issue in drug development has been to understand the selectivity of γ-secretase for its substrates, particularly APP and Notch. GPR3 and β-arrestin1 have been shown to modulate Aβ production by regulating γ-secretase function, with Notch-sparing activity [16, 18]. However, it is unclear how interaction with these proteins can confer substrate selectivity to γ-secretase. It is known that NCT is a common substrate receptor for both APP and Notch [49]. Then, interaction of NCT with other proteins, such as OCIAD2, might increase the substrate selectivity of NCT toward APP. In addition, substrate selectivity of γ-secretase could also be regulated by substrate selective γ-secretase modulator which interacts with APP [20]. In this context, OCIAD2 enhanced the interaction of NCT with C99 but not with Notch, and was able to interact with C99 but not with Notch. From these results, it is not clear whether OCIAD2 can directly bind to NCT and C99; however, the interaction between OCIAD2 and NCT apparently does not require C99 because the endogenous level of C99 is very low in most cells, including HEK293T and HeLa cells. The finding that Tat-OCIAD2 peptide prevented the interaction of NCT with C99, and suppressed AICD generation, provides further evidence that the interaction of OCIAD2 with NCT may be critical for determining substrate selectivity.
Another important discovery is that OCIAD2 enriches γ-secretase in lipid rafts. These are the membrane microdomains that stabilize γ-secretase and maintain its activity at a high level while having spatial proximity to the C99 substrate [35–37]. APP is also known to be recruited to lipid rafts, while this is still uncertain in the case of Notch [38]. Thus, enhanced incorporation of γ-secretase into lipid rafts by OCIAD2 may also account for the substrate selectivity of γ-secretase toward APP. OCIAD2 co-localized with γ-secretase components in the FLOT1-positive lipid raft-enriched fraction in sucrose gradient fractionation experiments. Moreover, OCIAD2 seemed to activate γ-secretase in MAM. MAM is a well-known intracellular site representing major γ-secretase activity [48] and consists of intracellular detergent-resistant lipid raft-like domain showing aberrant lipid metabolism in AD model [50]. Therefore, it is possible that OCIAD2 acts in conjunction with γ-secretase in the lipid rafts of MAM. Still, many more questions need to be addressed. First, whether the effects of OCIAD2 on the subcellular localization of NCT, C99, and Notch are direct or indirect needs to be established. Second, the NCT-stabilizing function of OCIAD2 must be more rigorously assessed, particularly as it relates to the subcellular localization of these proteins.
It is worth noting the increases of Ociad2 level at an early age in PDAPP mice. Amyloid plaques are normally detected in PDAPP mice at about 9 months of age [42]. It stands to reason that an elevation in Ociad2 levels might affect APP processing to increase Aβ production in PDAPP mice. In SH-SY5Y neuroblastoma cells, OCIAD2 expression increased upon treatment with H2O2. Not only are reactive oxygen species (ROS) strongly associated with AD but they also stimulate γ-secretase activity [1, 27, 45]. Thus, an increase in OCIAD2 levels may underlie an ROS-induced increase of Aβ production through γ-secretase activation. Interfering with the γ-secretase-activating function of OCIAD2 may provide a means of promoting a non-amyloidogenic pathway for APP processing. Inhibiting APP processing led to a decrease in the production of Aβ and AICD in the absence of OCIAD2. More importantly, overexpressing OCIAD2 enhanced the interaction of NCT with APP (C99), but not with Notch. These findings, combined with the results obtained from Tat-OCIAD2 peptide treatment, suggest that interfering with the interaction between OCIAD2 and NCT could provide a means of modulating γ-secretase activity in a substrate-specific manner. Collectively, this study suggests that Notch-sparing γ-secretase activator, OCIAD2, may function in the pathogenesis of AD, providing an insight into γ-secretase modulation in AD.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors thank Dr. Stephen M. Prescott (University of Utah, Utah) for the anti-FACL4 antibody; Dr. T. Tomita (University of Tokyo, Japan) for the anti-PEN2 antibody; Dr. Dennis J. Selkoe (Harvard Medical School, MA) for CHO and CHO-7PA2 cells; and Dr. W. Araki (National Institute of Neuroscience, Japan) for SH-SY5Y-APPswe cells. AD tissue samples were provided by the Harvard Brain Tissue Resource Center of McLean Hospital (Massachusetts, USA). Mr. J. Han and Ms. S. Jung were supported by the Brain Korea 21 program. This work was supported by a CRI grant (NRF-2013R1A2A1A01016896) and Global Research Laboratory (NRF-2010-00341) funded by the Ministry of Education, Science and Technology; an AD grant (A092058) funded by the Ministry of Human Health and Welfare in Korea; and a grant (to Y.K. Jung) funded by the Alzheimer’s Association (USA).
Abbreviations
- AICD
APP intracellular domain
- AD
Alzheimer’s disease
- Aβ
Amyloid beta
- APP
Aβ precursor protein
- APH-1
Anterior pharynx-defective phenotype 1
- BACE1
β-site amyloid precursor protein cleaving enzyme
- GOFS
Gain-of-function screen
- BN-PAGE
Blue-native-PAGE
- LOFS
Loss-of-function screen
- MAM
Mitochondria-associated ER membrane
- NCT
Nicastrin
- NICD
Notch intracellular domain
- PS
Presenilin
- PEN2
Presenilin enhancer 2
References
- 1.Querfurth HW, LaFerla FM. Alzheimer’s disease. N Engl J Med. 2010;362:329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 2.Greene JD, Baddeley AD, Hodges JR. Analysis of the episodic memory deficit in early Alzheimer’s disease: evidence from the doors and people test. Neuropsychologia. 1996;34:537–551. doi: 10.1016/0028-3932(95)00151-4. [DOI] [PubMed] [Google Scholar]
- 3.Glenner GG, Wong CW. Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120:885–890. doi: 10.1016/S0006-291X(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 4.Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci USA. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardy J, Duff K, Hardy KG, Perez-Tur J, Hutton M. Genetic dissection of Alzheimer’s disease and related dementias: amyloid and its relationship to tau. Nat Neurosci. 1998;1:355–358. doi: 10.1038/1565. [DOI] [PubMed] [Google Scholar]
- 6.Takami M, Nagashima Y, Sano Y, Ishihara S, Morishima-Kawashima M, Funamoto S, Ihara Y. Gamma-Secretase: successive tripeptide and tetrapeptide release from the transmembrane domain of beta-carboxyl terminal fragment. J Neurosci. 2009;29:13042–13052. doi: 10.1523/JNEUROSCI.2362-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 8.Ballatore C, Lee VM, Trojanowski JQ. Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat Rev Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- 9.Roberson ED, Mucke L. 100 years and counting: prospects for defeating Alzheimer’s disease. Science. 2006;314:781–784. doi: 10.1126/science.1132813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selkoe DJ, Wolfe MS. Presenilin: running with scissors in the membrane. Cell. 2007;131:215–221. doi: 10.1016/j.cell.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Periz G, Fortini ME. Functional reconstitution of gamma-secretase through coordinated expression of presenilin, nicastrin, Aph-1, and Pen-2. J Neurosci Res. 2004;77:309–322. doi: 10.1002/jnr.20203. [DOI] [PubMed] [Google Scholar]
- 12.Zhou S, Zhou H, Walian PJ, Jap BK. CD147 is a regulatory subunit of the gamma-secretase complex in Alzheimer’s disease amyloid beta-peptide production. Proc Natl Acad Sci USA. 2005;102:7499–7504. doi: 10.1073/pnas.0502768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ni Y, Zhao X, Bao G, Zou L, Teng L, Wang Z, Song M, Xiong J, Bai Y, Pei G. Activation of beta2-adrenergic receptor stimulates gamma-secretase activity and accelerates amyloid plaque formation. Nat Med. 2006;12:1390–1396. doi: 10.1038/nm1485. [DOI] [PubMed] [Google Scholar]
- 14.Chen F, Hasegawa H, Schmitt-Ulms G, Kawarai T, Bohm C, Katayama T, Gu Y, Sanjo N, Glista M, Rogaeva E, Wakutani Y, Pardossi-Piquard R, Ruan X, Tandon A, Checler F, Marambaud P, Hansen K, Westaway D, St George-Hyslop P, Fraser P. TMP21 is a presenilin complex component that modulates gamma-secretase but not epsilon-secretase activity. Nature. 2006;440:1208–1212. doi: 10.1038/nature04667. [DOI] [PubMed] [Google Scholar]
- 15.Spasic D, Raemaekers T, Dillen K, Declerck I, Baert V, Serneels L, Füllekrug J, Annaert W. Rer1p competes with APH-1 for binding to nicastrin and regulates gamma-secretase complex assembly in the early secretory pathway. J Cell Biol. 2007;176:629–640. doi: 10.1083/jcb.200609180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thathiah A, Spittaels K, Hoffmann M, Staes M, Cohen A, Horré K, Vanbrabant M, Coun F, Baekelandt V, Delacourte A, Fischer DF, Pollet D, De Strooper B, Merchiers P. The orphan G protein-coupled receptor 3 modulates amyloid-beta peptide generation in neurons. Science. 2009;323:946–951. doi: 10.1126/science.1160649. [DOI] [PubMed] [Google Scholar]
- 17.He G, Luo W, Li P, Remmers C, Netzer WJ, Hendrick J, Bettayeb K, Flajolet M, Gorelick F, Wennogle LP, Greengard P. Gamma-secretase activating protein is a therapeutic target for Alzheimer’s disease. Nature. 2010;467:95–98. doi: 10.1038/nature09325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Zhao X, Zeng X, Bossers K, Swaab DF, Zhao J, Pei G. β-arrestin1 regulates γ-secretase complex assembly and modulates amyloid-β pathology. Cell Res. 2012;23:351–365. doi: 10.1038/cr.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thathiah A, Horré K, Snellinx A, Vandewyer E, Huang Y, Ciesielska M, De Kloe G, Munck S, De Strooper B. β-arrestin 2 regulates Aβ generation and γ-secretase activity in Alzheimer’s disease. Nat Med. 2012;19:43–49. doi: 10.1038/nm.3023. [DOI] [PubMed] [Google Scholar]
- 20.Kukar TL, Ladd TB, Bann MA, Fraering PC, Narlawar R, Maharvi GM, Healy B, Chapman R, Welzel AT, Price RW, Moore B, Rangachari V, Cusack B, Eriksen J, Jansen-West K, Verbeeck C, Yager D, Eckman C, Ye W, Sagi S, Cottrell BA, Torpey J, Rosenberry TL, Fauq A, Wolfe MS, Schmidt B, Walsh DM, Koo EH, Golde TE. Substrate-targeting gamma-secretase modulators. Nature. 2008;453:925–929. doi: 10.1038/nature07055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selkoe D, Kopan R. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu Rev Neurosci. 2003;26:565–597. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- 22.Goutte C, Hepler W, Mickey KM, Priess JR. Aph-2 encodes a novel extracellular protein required for GLP-1-mediated signaling. Development. 2000;127:2481–2492. doi: 10.1242/dev.127.11.2481. [DOI] [PubMed] [Google Scholar]
- 23.Goutte C, Tsunozaki M, Hale VA, Priess JR. APH-1 is a multipass membrane protein essential for the Notch signaling pathway in Caenorhabditis elegans embryos. Proc Natl Acad Sci USA. 2002;99:775–779. doi: 10.1073/pnas.022523499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Francis R, McGrath G, Zhang J, Ruddy DA, Sym M, Apfeld J, Nicoll M, Maxwell M, Hai B, Ellis MC, Parks AL, Xu W, Li J, Gurney M, Myers RL, Himes CS, Hiebsch R, Ruble C, Nye JS, Curtis D. Aph-1 and pen-2 are required for Notch pathway signaling, gamma-secretase cleavage of betaAPP, and presenilin protein accumulation. Dev Cell. 2002;3:85–97. doi: 10.1016/S1534-5807(02)00189-2. [DOI] [PubMed] [Google Scholar]
- 25.Kopan R, Goate A. A common enzyme connects notch signaling and Alzheimer’s disease. Genes Dev. 2000;14:2799–2806. doi: 10.1101/gad.836900. [DOI] [PubMed] [Google Scholar]
- 26.Sisodia SS, St George-Hyslop PH. Gamma-Secretase, Notch, Abeta and Alzheimer’s disease: where do the presenilins fit in? Nat Rev Neurosci. 2002;3:281–290. doi: 10.1038/nrn785. [DOI] [PubMed] [Google Scholar]
- 27.Gwon AR, Park JS, Arumugam TV, Kwon YK, Chan SL, Kim SH, Baik SH, Yang S, Yun YK, Choi Y, Kim S, Tang SC, Hyun DH, Cheng A, Dann CE, 3rd, Bernier M, Lee J, Markesbery WR, Mattson MP, Jo DG. Oxidative lipid modification of nicastrin enhances amyloidogenic γ-secretase activity in Alzheimer’s disease. Aging Cell. 2012;11:559–568. doi: 10.1111/j.1474-9726.2012.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yachiyo M, Hiroshi H, Akinori M, Wataru A, Toshiharu S, Shinji T, Masayasu O, Masatoshi T, Ronald R, Masaki N. Human CRB2 inhibits γ-secretase cleavage of amyloid precursor protein by binding to the presenilin complex. J Biol Chem. 2010;285:14920–14931. doi: 10.1074/jbc.M109.038760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee H, Noh JY, Oh Y, Kim Y, Chang JW, Chung CW, Lee ST, Kim M, Ryu H, Jung YK. IRE1 plays an essential role in ER stress-mediated aggregation of mutant huntingtin via the inhibition of autophagy flux. Hum Mol Genet. 2012;21:101–114. doi: 10.1093/hmg/ddr445. [DOI] [PubMed] [Google Scholar]
- 30.Shim SM, Lee WJ, Kim Y, Chang JW, Song S, Jung YK. Role of S5b/PSMD5 in proteasome inhibition caused by TNF-α/NFκB in higher eukaryotes. Cell Rep. 2012;2:603–615. doi: 10.1016/j.celrep.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki Y, Ohta K, Itoh M, Sakoh-Sumitomo Y, Mitsuda T, Ueda M, Hayakawa-Yano Y, Li S, Hida Y, Inuzuka T, Jung YK, Nakagawa T. An alternative spliced mouse presenilin-2 mRNA encodes a novel gamma-secretase inhibitor. FEBS Lett. 2009;583:1403–1408. doi: 10.1016/j.febslet.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 32.Tesco G, Ginestroni A, Hiltunen M, Kim M, Dolios G, Hyman BT, Wang R, Berezovska O, Tanzi RE. APP substitutions V715F and L720P alter PS1 conformation and differentially affect Abeta and AICD generation. J Neurochem. 2005;95:446–456. doi: 10.1111/j.1471-4159.2005.03381.x. [DOI] [PubMed] [Google Scholar]
- 33.Nyabi O, Bentahir M, Horré K, Herreman A, Gottardi-Littell N, Van Broeckhoven C, Merchiers P, Spittaels K, Annaert W, De Strooper B. Presenilins mutated at Asp-257 or Asp-385 restore Pen-2 expression and Nicastrin glycosylation but remain catalytically inactive in the absence of wild type Presenilin. J Biol Chem. 2003;278:43430–43436. doi: 10.1074/jbc.M306957200. [DOI] [PubMed] [Google Scholar]
- 34.Schägger H, Cramer WA, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- 35.Vetrivel KS, Cheng H, Lin W, Sakurai T, Li T, Nukina N, Wong PC, Xu H, Thinakaran G. Association of gamma-secretase with lipid rafts in post-Golgi and endosome membranes. J Biol Chem. 2004;279:44945–44954. doi: 10.1074/jbc.M407986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vetrivel KS, Thinakaran G. Membrane rafts in Alzheimer’s disease beta-amyloid production. Biochim Biophys Acta. 2010;1801:860–867. doi: 10.1016/j.bbalip.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urano Y, Hayashi I, Isoo N, Reid PC, Shibasaki Y, Noguchi N, Tomita T, Iwatsubo T, Hamakubo T, Kodama T. Association of active gamma-secretase complex with lipid rafts. J Lipid Res. 2005;46:904–912. doi: 10.1194/jlr.M400333-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Ehehalt R, Keller P, Haass C, Thiele C, Simons K. Amyloidogenic processing of the Alzheimer beta-amyloid precursor protein depends on lipid rafts. J Cell Biol. 2003;160:113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mo JS, Yoon JH, Hong JA, Kim MY, Ann EJ, Ahn JS, Kim SM, Baek HJ, Lang F, Choi EJ, Park HS. Phosphorylation of nicastrin by SGK1 leads to its degradation through lysosomal and proteasomal pathways. PLoS ONE. 2012;7:e37111. doi: 10.1371/journal.pone.0037111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim SK, Park HJ, Hong HS, Baik EJ, Jung MW, Mook-Jung I. ERK1/2 is an endogenous negative regulator of the gamma-secretase activity. FASEB J. 2006;20:157–159. doi: 10.1096/fj.05-4621fje. [DOI] [PubMed] [Google Scholar]
- 41.Ohta K, Mizuno A, Li S, Itoh M, Ueda M, Ohta E, Hida Y, Wang MX, Furoi M, Tsuzuki Y, Sobajima M, Bohmoto Y, Fukushima T, Kobori M, Inuzuka T, Nakagawa T. Endoplasmic reticulum stress enhances γ-secretase activity. Biochem Biophys Res Commun. 2011;416:362–366. doi: 10.1016/j.bbrc.2011.11.042. [DOI] [PubMed] [Google Scholar]
- 42.Mucke L, Masliah E, Yu G, Mallory M, Rockenstein EM, Tatsuno G, Hu K, Kholodenko D, Johnson-Wood K, McConlogue L. High-level neuronal expression of abeta 1-42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J Neurosci. 2000;20:4050–4058. doi: 10.1523/JNEUROSCI.20-11-04050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prokop S, Shirotani K, Edbauer D, Haass C, Steiner H. Requirement of PEN-2 for stabilization of the presenilin N-/C-terminal fragment heterodimer within the gamma-secretase complex. J Biol Chem. 2004;279:23255–23261. doi: 10.1074/jbc.M401789200. [DOI] [PubMed] [Google Scholar]
- 44.Gu Y, Chen F, Sanjo N, Kawarai T, Hasegawa H, Duthie M, Li W, Ruan X, Luthra A, Mount HT, Tandon A, Fraser PE, St George-Hyslop P. APH-1 interacts with mature and immature forms of presenilins and nicastrin and may play a role in maturation of presenilin nicastrin complexes. J Biol Chem. 2003;278:7374–7380. doi: 10.1074/jbc.M209499200. [DOI] [PubMed] [Google Scholar]
- 45.Shen C, Chen Y, Liu H, Zhang K, Zhang T, Lin A, Jing N. Hydrogen peroxide promotes Abeta production through JNK-dependent activation of gamma-secretase. J Biol Chem. 2008;283:17721–17730. doi: 10.1074/jbc.M800013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park SK, Han SB, Lee K, Lee HJ, Kho YH, Chun H, Choi Y, Yang JY, Yoon YD, Lee CW, Kim HM, Choi HM, Tae HS, Lee HY, Nam KY, Han G. Gelastatins and their hydroxamates as dual functional inhibitors for TNF-alpha converting enzyme and matrix metalloproteinases: synthesis, biological evaluation, and mechanism studies. Biochem Biophys Res Commun. 2006;341(2):627–634. doi: 10.1016/j.bbrc.2005.12.219. [DOI] [PubMed] [Google Scholar]
- 47.Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat Protoc. 2009;4:1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- 48.Area-Gomez E, de Groof AJ, Boldogh I, Bird TD, Gibson GE, Koehler CM, Yu WH, Duff KE, Yaffe MP, Pon LA, Schon EA. Presenilins are enriched in endoplasmic reticulum membranes associated with mitochondria. Am J Pathol. 2009;175(5):1810–1816. doi: 10.2353/ajpath.2009.090219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shah S, Lee SF, Tabuchi K, Hao YH, Yu C, LaPlant Q, Ball H, Dann CE, Südhof T, Yu G. Nicastrin functions as a gamma-secretase-substrate receptor. Cell. 2005;122:435–447. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 50.Area-Gomez E, Del Carmen Lara Castillo M, Tambini MD, Guardia-Laguarta C, de Groof AJ, Madra M, Ikenouchi J, Umeda M, Bird TD, Sturley SL, Schon EA. Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J. 2012;31:4106–4123. doi: 10.1038/emboj.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.