Abstract
Recent years witnessed increased interest in intrinsically disordered proteins and regions. These proteins and regions are abundant and possess unique structural features and a broad functional repertoire that complements ordered proteins. However, modern studies on the abundance and functions of intrinsically disordered proteins and regions are relatively limited in size and scope of their analysis. To fill this gap, we performed a broad and detailed computational analysis of over 6 million proteins from 59 archaea, 471 bacterial, 110 eukaryotic and 325 viral proteomes. We used arguably more accurate consensus-based disorder predictions, and for the first time comprehensively characterized intrinsic disorder at proteomic and protein levels from all significant perspectives, including abundance, cellular localization, functional roles, evolution, and impact on structural coverage. We show that intrinsic disorder is more abundant and has a unique profile in eukaryotes. We map disorder into archaea, bacterial and eukaryotic cells, and demonstrate that it is preferentially located in some cellular compartments. Functional analysis that considers over 1,200 annotations shows that certain functions are exclusively implemented by intrinsically disordered proteins and regions, and that some of them are specific to certain domains of life. We reveal that disordered regions are often targets for various post-translational modifications, but primarily in the eukaryotes and viruses. Using a phylogenetic tree for 14 eukaryotic and 112 bacterial species, we analyzed relations between disorder, sequence conservation and evolutionary speed. We provide a complete analysis that clearly shows that intrinsic disorder is exceptionally and uniquely abundant in each domain of life.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-014-1661-9) contains supplementary material, which is available to authorized users.
Keywords: Intrinsic disorder, Intrinsically disordered proteins, Intrinsically disordered regions, Cellular localization, Post-translational modifications, Evolutionary speed
Introduction
It is now recognized that in addition to globular, transmembrane and fibrillar proteins that are known to be characterized by unique three dimensional (3D)-structure, there is another tribe of proteins, which, being biologically functional, do not have unique 3D-structures in their native states under the physiologic conditions in vitro and in vivo [1–5]. The members of this novel tribe are known as intrinsically disordered proteins (IDPs). Their structures are defined as highly dynamic ensembles of flexible conformations, where sampling of a large portion of a polypeptide’s available conformational space is allowed. Although IDPs and intrinsically disordered regions (IDRs) in proteins are devoid of stable 3D-structures, they possess crucial biological functions and play multiple important roles in living organisms. In fact, the conformational plasticity associated with intrinsic disorder provides IDPs/IDRs with a wide spectrum of exceptional functional advantages over the functional modes of ordered proteins and ordered protein domains [1, 2, 5–17]. For example, the high accessibility of sites within the disordered proteins simplifies their post-translational modifications, such as phosphorylation, acetylation, lipidation, ubiquitination, sumoylation, etc., allowing for modulation of their biological functions [5]. Many IDRs contain specific identification regions, which they use to participate in various regulation, recognition, signaling and control pathways [11, 12]. As exemplified by the gene ontology analysis, IDPs are involved in crucial biological processes, such as signaling, recognition, and regulation [14, 15, 18–21].
The existence of functional proteins without unique 3D-structures is in apparent conflict with the traditional sequence-structure–function paradigm that relies on the “one protein-one structure-one function” concept [1, 2, 5, 6, 11, 12, 15, 22]. For a long time, cases of protein function without structure or protein function originating from the conformational ensemble were taken as unique and rare exceptions, and the one protein-one structure-one function concept was considered as a general and undisputable rule. However, this has changed recently, leading to the recognition of the importance of IDPs. The concept of protein intrinsic disorder became an important part of modern structural biology and proteomic studies [1, 2, 22].
This (revolutionary) change in the understanding of the molecular bases of protein functions was fueled by growing appreciation of the idea that IDPs and IDRs are not rare and obscure exceptions, but are exceptionally common and fascinating entities. In fact, several efforts were devoted to estimating the abundance of intrinsically disordered proteins in nature [23–31]. In these studies, predictive algorithms were used to estimate the content of intrinsic disorder in various proteomes or specific protein. Although the estimated fractions of disordered residues for any given organism are different in these studies (being dependent on the algorithms used to evaluate the disorder content), the general trend of intrinsic disorder distribution over the tree of life is quite consistent: eukaryotes are systematically predicted to have much higher intrinsic disorder contents than prokaryotes. The number of species analyzed in the studies discussed above ranged from a few to a few hundreds. For example, the abundance of IDPs and IDRs in 53 archaean species was recently evaluated [28]. In another recent study, Burra et al. analyzed 332 prokaryotic proteomes [29], and in still another recent work (which, to the best of our knowledge, is the largest scale intrinsic disorder analysis undertaken so far), the proteomes of 3,484 species were analyzed [30].
In addition to studies on the abundance of protein disorder in various proteomes, the functions of IDPs and IDRs at the proteome/large-protein-database level were also scrutinized. For example, Ward et al. analyzed distribution of IDPs in six archaean, 13 bacterial and five eukaryotic genomes, and studied the function of proteins with long predicted regions of disorder using the gene ontology annotations supplied with the Saccharomyces genome database. They have shown that proteins containing disorder are often located in the cell nucleus and are involved in the regulation of transcription and cell signaling, and are commonly associated with the molecular functions of kinase activity and nucleic acid binding [24]. Based on the bioinformatics analysis of the functional keywords associated with 20 or more proteins in Swiss-Prot, it was concluded that many functions are indeed related to the increased propensity for intrinsic disorder. Specifically, out of 710 Swiss-Prot keywords, 310 functional keywords are associated with ordered proteins, 238 functional keywords are attributed to disordered proteins, and the remainder 162 keywords yield ambiguity in the likely function-structure associations [19–21]. Study of the occurrence of protein disorder in the human proteome and analysis of the ontology categories that are enriched in disordered human proteins revealed that the IDP-specific functions are both length and position dependent, and these observations were used to develop classifiers for human protein function prediction [32]. The inclusion of the disorder features improved the prediction accuracies for 26 Gene Ontology (GO) categories related to signaling and molecular recognition [32]. Recently, analysis of human proteome revealed that disordered regions frequently act as independent functional units [33], and this functional modularity supports the earlier notion that there is an association between disorder and alternative splicing [34].
In spite of this obvious progress in the field, modern studies on the natural abundance and functions of IDPs/IDRs are relatively limited in terms of the number of species analyzed and scope of the analysis, which often targets only one of a handful of aspects. To fill this gap, we performed a large-scale, comprehensive and detailed analysis of 6,438,736 proteins from 965 complete proteomes, using arguably more accurate consensus-based disorder predictions. Since in addition to the analysis of 59 archaean, 471 bacterial and 110 eukaryotic proteomes we studied~20,000 proteins from 325 viral proteomes, our work represents one of the first large-scale analyses of abundance and function of intrinsic disorder in viruses. We seamlessly combined proteome-level analysis that characterizes abundance and differences in profiles of disorder between the domains of life with analysis at the protein level that concerns a detailed, large-scale, and comprehensive characterization of functional roles and cellular localization of intrinsic disorder. We are the first to perform large-scale analysis of enrichment of disorder in functional annotations and post-translational modification sites, to reveal relations between structural coverage and disorder across various domains and kingdoms of life, to annotate the abundance of disorder in cells, and to study interplay between intrinsic disorder, evolutionary pace, and sequence conservation. More specifically, we investigated enrichment of disorder in a broad range of over 1,200 functional annotations, compared to previous “small-scale” studies that investigated a narrower range of functional aspects based on at most a couple dozen of proteomes excluding viruses. We included a similarly comprehensive characterization of enrichment of disorder in cellular components/compartment in archaea, bacteria, eukaryota and viruses, and, for the first time, we mapped intrinsic disorder into archaean, bacterial, eukaryotic cells. We quantified and contrasted enrichment of intrinsic disorder in various types of post-translational modification sites across the four domains of life. We also estimated current structural coverage of the considered proteomes, and found that the abundance of disorder negatively correlates with this coverage for certain kingdoms and phyla.
Materials and methods
We analyzed all 965 complete proteomes, which total to 6,438,736 proteins, from UniProt release 2011_08 [35]. The proteomes were assigned to their taxonomic lineage based on the National Center for Biotechnology Information (NCBI) [36], where the lowest taxonomic level, which we refer to as “species”, could be the genus, family or species. The resulting UniProt Complete Proteome Dataset (UCPD) includes 231,466 proteins (3.6 % of all considered proteins) from 59 species in archaea, 4,285,619 proteins (66.6 %) from 471 species in bacteria, 1,901,810 proteins (29.5 %) from 110 species in eukaryota, and 19,841 proteins (0.3 %) from 325 viral proteomes; see Supplementary Table 1. All 965 proteomes were used to characterize disorder at the taxonomic domain level, while 225 small proteomes (with less than 30 proteins) were excluded when performing analysis at the species level.
Table 1.
Summary of the biological processes, molecular functions, and cellular components, which were annotated based on Gene Ontology (GO), across the four domains of life
| annotation | types of annotations | in Archaea | in Bacteria | in Eukaryota | in Viruses |
|---|---|---|---|---|---|
| biological processes | total # of processes | 12 | 318 | 104 | 2 |
| # of processes with significant depletion in disorder | 0 | 76 | 31 | 0 | |
| # of processes with significant enrichment in disorder | 1 | 14 | 10 | 1 | |
| molecular functions | total # of functions | 34 | 581 | 161 | 4 |
| # of functions with significant depletion in disorder | 1 | 184 | 63 | 0 | |
| # of functions with significant enrichment in disorder | 2 | 20 | 6 | 1 | |
| cellular components | total # of components | 6 | 61 | 50 | 5 |
| # of components with significant depletion in disorder | 0 | 12 | 6 | 0 | |
| # of components with significant enrichment in disorder | 1 | 13 | 3 | 2 |
The numbers in bold indicate the total number of significant sub-functions in a given domain of life that are used to investigate potential depletion or enrichment of the disorder
We applied two fast and accurate disordered predictors, IUPred [37, 38] and Espritz [39], to obtain putative disordered residues and segments. We used two versions of IUPred that were designed for predictions of long and short disordered segments, respectively, and three versions of Espritz that consider disorder annotations based on nuclear magnetic resonance (NMR) structures, X-ray crystal structures, and experimental annotations from DisProt database [40]. Espritz and IUPred are competitive in terms of their predictive quality [38, 41], and they cover the main characteristics of the disorder including the three annotation types and two types of disordered segments. The resulting five predictions were combined together using the majority vote consensus. This is motivated by the fact that consensus-based approaches provide improved predictive quality [42]. Our approach is a marked improvement over the previous studies, where only one [24, 30, 32] or two [28, 29] predictors were used to characterize disorder. The putative disorder was used to calculate the disorder content (fraction of disordered residues in a given chain), the number and size of disordered segments and long disordered segments that consist of at least 30 consecutive disordered amino acids, and to characterize fully disordered proteins. The analysis of long segments is motivated by the fact that they are implicated in protein–protein recognition [43] and serve as functional units [33]. Consistent with previous works [44], we count the disordered segments with at least four consecutive disordered residues. We include such short segments, since they can be predicted with relatively high predictive performance [44] and they were included in some of the similar studies [28, 30]. However, we note that our results may include some artifacts, since these short regions were speculated as being less likely to be functionally relevant compared to long disordered regions [24]. We normalized the count of disordered segments to accommodate for the bias due to differences in chains length between taxonomic domains; see Supplementary Fig. 1. We calculated the number of disordered segments per unit segment of 100 amino acids, by dividing the actual count in a given chain by its length and multiplying the result by 100. Similar to the recent study concerning abundance of disorder in viral proteomes [30], which was limited in the context of functional analysis, viral polyproteins were analyzed as a single polypeptide chain. This potentially affects disorder predictions for only a few residues close to the cleavage sites, and has a negligible effect on the overall proteome-wide results.
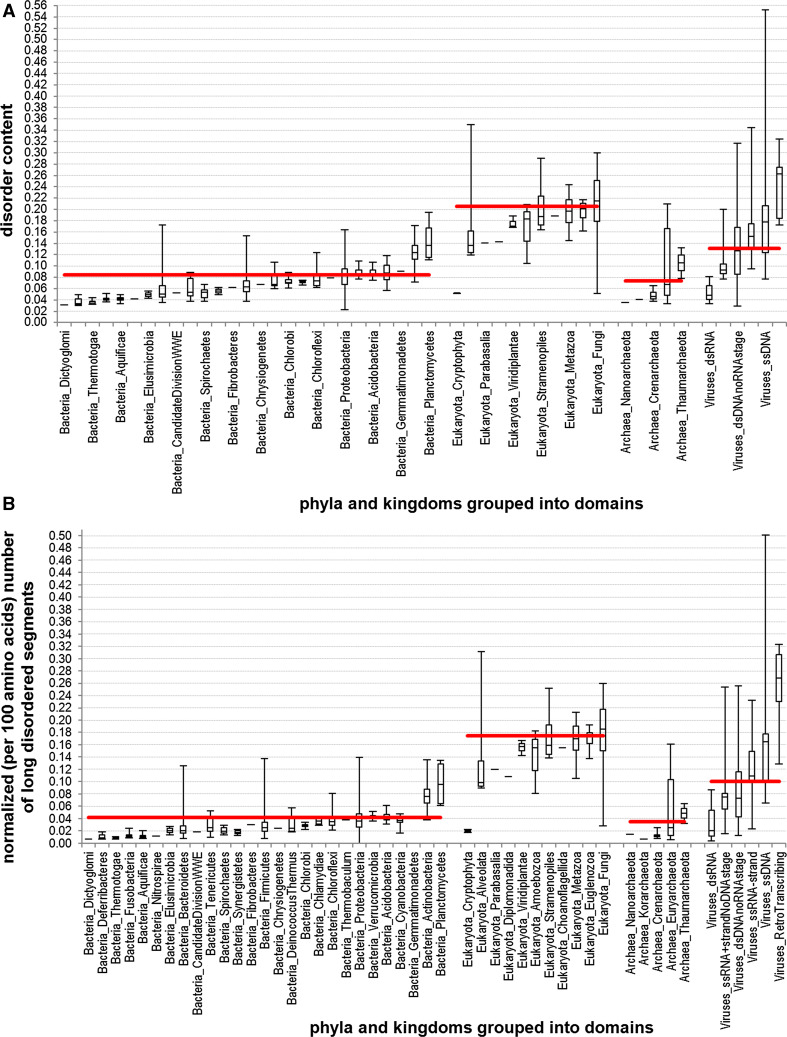
Fig. 1.
Disorder content (panel A) and normalized number of long (30 or more consecutive amino acids) disordered segments across different phyla and kingdoms (second level of the taxonomic lineage). The phyla and kingdoms (x-axis) are grouped into domains of life including bacteria, eukaryota, archaea, and viruses. Solid horizontal red lines denote average disorder content per domain of life. Box plots show the minimum, first quartile, second quartile (median), third quartile, and maximum disorder content (panel A) or normalized number of long disordered segments (panel B) across different species in a given phyla/domain of life; one line is shown for phyla with only one species (e.g., Dictyoglomi)
We investigated disorder in certain cellular components and relations between disorder and protein functions based on the GO terms [45] that are linked in the UniProt, and between disorder and post-translational modifications (PTMs) that are annotated in the UniProt. We consider all annotations for each protein, which means that the same protein may be counted in multiple biological processes, molecular functions, and cellular components. We excluded annotations with qualifiers “potential”, “probable” and “by similarity” that are associated with computer-prediction or indirect experimental evidence. We also removed annotations with insufficient number of samples in a given taxonomic domain; i.e., PTMs with less than 100 annotated residues and function/components with less than 100 chains. In each domain of life, we empirically analyzed whether disorder is significantly enriched/depleted in proteins with a given function, in a given cellular component or in residues with a given type of PTMs. Similar to earlier analysis [24], we evaluated statistical significance of these differences by contrasting disorder content in a given functional or localization-based set of chains or a set of residues with a given PTM with the baseline disorder content in a given domain of life; this accommodates for differences in the abundance of disorder between the domains of life. We randomly selected half of the GO-annotated chains or PTM-annotated residues and compared them with the same number of chains/residues drawn at random from the entire taxonomic domain. This was repeated ten times, and we evaluated significance of the differences in the disorder content between these two vectors. If the measurements were normal, as evaluated with the Anderson–Darling test at 0.05 significance, then we utilized the t test; otherwise, we used the non-parametric Wilcoxon rank sum test. We considered only the differences with sufficiently large magnitude; i.e., the average difference/enrichment must be larger than 50 % of the average disorder content in a given domain of life.
The structural coverage was computed based on method described in Ref. [46]. Briefly, we compared a given protein chain against all sequences from the Protein Data Bank using three rounds of PSI-BLAST. The sequence was considered structured if PSI-BLAST found a hit with an E-value below 0.001 that had at least 50 amino acids in length. The structural coverage of a given proteome was defined as the fraction of (non-redundant) structured sequences in this proteome.
Using the evolutionary tree reconstructed in Ref. [47], we studied relations between the intrinsic disorder and the evolutionary speed that is quantified with the branch length, i.e., longer branches indicate faster pace of the sequence evolution. We mapped 112 bacterial, 14 eukaryotic and two archaea species into our data set from among 191 species that were used in Ref. [47], and compared their disorder content against the branch length. Consequently, we had to exclude viruses that were not considered in Ref. [47] and archaea that had small sample size.
Similarly as in [48, 49], we quantified the sequence conservation using relative entropy [50], which was computed from the Weighted Observed Percentages (WOP) profiles produced by PSI-BLAST [51]. PSI-BLAST was run with default parameters (−j 3, −h 0.001) against the nr database. Due to the high computational cost, we estimated conservation based on results for 100 randomly selected proteins from a given proteome.
Results
Disorder at the proteomic level
First, we analyzed the overall abundance of intrinsic disorder in the 965 complete proteomes. Results of this analysis for selected proteomes are shown in Fig. 1. We analyze the averaged disorder content (Fig. 1A) and the normalized number of long (30 or more consecutive amino acids) disordered segments (Fig. 1B) across different phyla and kingdoms (second level of the taxonomic lineage) in all the domains of life. This analysis reveals that intrinsic disorder is common in all the proteomes studied, and that the eukaryotic proteomes are noticeably more disordered than proteomes from the other domains of life using different disorder measures. In fact, disorder content equals 20.5 % for eukaryotes, 13.2 % for viruses, 8.5 % for bacteria, and 7.4 % for archaea. Furthermore, the normalized number of long disordered segments per 100 amino acids is at 17.4 % for eukaryotes, 10 % for viruses, 4.2 % for bacteria, and 3.6 % for archaea. Note the relatively smaller proportions for the bacteria and archaea, which means that they have relatively fewer long disordered segments. The results of our analysis are consistent (a bit higher, but in the same order) with the results of earlier analysis that was performed for a smaller set of proteomes (six archaean, 13 bacterial, and five eukaryotic proteomes) and reported in Ref. [24]. In this study, the disorder content was estimated to be 18.9 % in eukaryotes, 5.7 % in bacteria, and 3.8 % in archaea; viruses were not considered. Figure 1 also shows that the disorder content in viral species varies to a wide extent, ranging between 3 and 55 %; in eukaryotic species between 5 and 35 %; and in bacterial and archaean species, the disorder contents are below 20 and 21 %, respectively (whiskers/error bars show the range). Also, the fraction of the long disordered segments is proportional to the overall disorder content, with the exception of some viruses that contain relatively more of longer disordered segments, i.e., whiskers are taller when compared to the content whiskers.
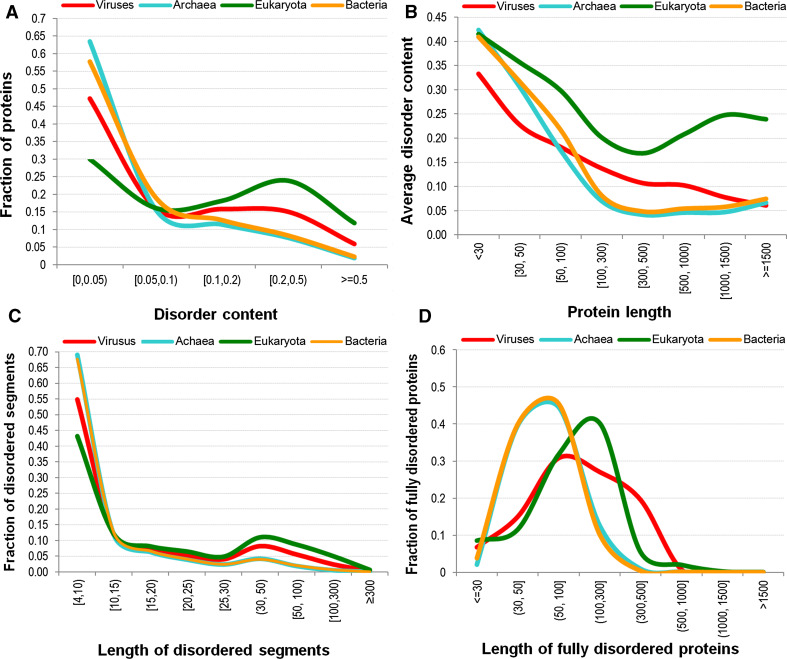
Next, we looked at the peculiarities of disorder distribution in four domains of life, prokaryotes, archaea, eukaryotes and viruses. In our study, viruses were considered as a fourth domain of life, although currently there is no common opinion on whether viruses are a form of life, or organic structures that interact with living organisms. Figure 2A shows that the majority of proteins in viral, bacterial, and archaean species have relatively small amounts of disorder. In fact, 79, 77, and 63 % of chains in archaean, bacterial, and viral proteomes, respectively, have up to 10 % disorder, compared to only 46 % of such proteins in eukaryotes. On the other hand, eukaryotic proteomes are characterized by a large fraction of chains with substantial amounts of disorder. Here, 36 % of eukaryotic chains have > 20 % disorder and 12 % of eukaryotic proteins possess > 50 % disorder.
Fig. 2.
Distribution of disorder content (panel A), disorder content against chain size (panel B), size of the disordered segments (panel C), and size of the fully disordered proteins (panel D) for the four domains of life
Figure 2B illustrates another interesting fact, namely that in the bacterial and archaean species the larger amounts of disorder are present only in short chains (shorter that 100 residues long). Specifically, 12 and 11 % of proteins in archaea and bacteria, respectively, which are shorter than 100 residues, have on average 19 and 24 % of disorder. This is almost threefold higher than their overall average. To compare, chains longer than 100 residues, which account for 88 % of archaean and 89 % of bacterial proteins, have on average below 6 % of disorder. On the other hand, in viruses and eukaryotes, the disorder is more evenly distributed across protein sizes. Specifically, chains longer than 100 residues, which account for 82 and 93 % of proteins in viruses and eukaryotes, respectively, have an average amount of disorder at 12 and 20 %, respectively. This is comparable with their overall disorder content. Chains longer than 500 amino acids in eukaryotes, which total to 32 % of eukaryotic proteins, have on average 22 % of disorder, compared to 9 % in viruses, 6 % in bacteria, and 5 % in archaea.
As is evident from Fig. 2C, short (below ten amino acids) disordered segments account for two-thirds of the disordered segments in archaea and bacteria. This noticeably exceeds the corresponding values of 55 and 43 % evaluated for viruses and eukaryotes, respectively. Only eukaryotes and viruses have relatively large fractions of longer disordered segments, which result in the bimodal distribution in Fig. 2C. More specifically, 25 and 16 % of disordered segments in eukaryotes and viruses, respectively, are longer than 30 residues, compared to just 7 % in bacteria and archaea.
Our analysis reveals that between 0.9 % of proteins in eukaryotes (close to 18 thousand) and 0.2 % of proteins in archaea (around 500 chains) are fully disordered. Figure 2D, which analyzes these fully disordered proteins, shows that in archaea and bacteria, they are relatively short compared to their sizes in the eukaryotic and viral proteomes. In fact, in archaea and bacteria, 86 and 89 % of fully disordered chains are shorter than 100 residues, compared to 53 and 52 % in viruses and eukaryota, respectively. Interestingly, 20 % of fully disordered viral proteins are longer than 300 amino acids, compared to 8, 1, and 1 % for eukaryotes, archaea, and bacteria, respectively.
Functional analysis and cellular localization of disorder at the protein level
We analyzed the functional importance of intrinsic disorder by considering correlations between the intrinsic disorder propensity and biological processes, molecular functions, and cellular components annotated based on the GO terms that are available in UniProt database [35] for many proteins in the completed genomes across the four domains of life. Results of these analyses are summarized in Table 1 and Fig. 3. Globally, Table 1 suggests that number of functional annotations does not reflect the complexity of a given domain; rather, it is correlated with the completeness of its annotations in GO. In each domain of life, there are some processes, functions and cellular components that are enriched in the intrinsic disorder and some other with a significant depletion in the disorder. For example, between 4 and 10 % of processes, functions, and components in eukaryotes are significantly enriched in disorder, whereas in bacteria, about 20 % of GO annotated cellular components are enriched in disorder.
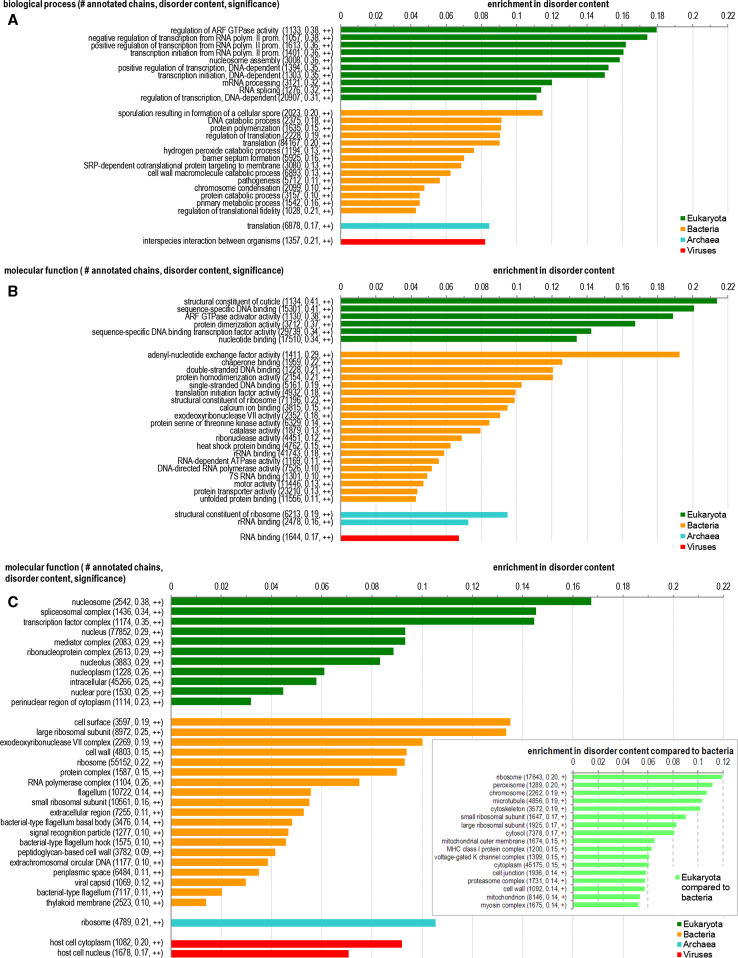
Fig. 3.
Biological processes (panel A), molecular function (panel B), and cellular components (panel C) that are significantly enriched in the disorder across eukaryotic, bacterial, archaea, and viral species. The y-axis gives all significant functions/components, including the number of corresponding proteins, their average disorder content, and significance of the enrichment. The x-axis shows the enrichment in the average disorder content between proteins with a given function/in a given compartment and the baseline disorder content in a given domain of life. Details of the calculation are provided in the Materials and Methods section. The significance of the difference is denoted with “ + ” and “ ++”, which indicate that the P-value is smaller than 0.01 and 0.001, respectively. The functions/cellular components are sorted, within each domain of life, by the values of the enrichment
Figure 3A contains a more detailed representation of a correlation between intrinsic disorder and biological processes in the four domains of life. Among disorder-enriched biological processes in eukaryotes are transcription, regulation of GTPase, nucleosome assembly [52], and RNA splicing. Overall, disorder in eukaryotes seems to be important for protein–RNA, protein–DNA, and protein–nucleotide interactions. In addition to sharing similarities to eukaryotes with respect to disorder-based protein–DNA interactions, bacteria utilize a wider array of biological processes with enriched disorder, with most illustrative examples being sporulation, protein polymerization, translation, catabolic and metabolic processes, pathogenesis, and chromosome condensation. Figure 3B shows that intrinsic disorder is important for several molecular functions, such as DNA and nucleotide binding, protein dimerization, and transcription in eukaryotes and DNA and RNA binding, protein dimerization, translation, etc., in bacteria. Overall, our analysis shows that biological processes that are enriched in disorder are consistent with the corresponding molecular functions, and that these enriched functions/processes carry over across the considered domains of life.
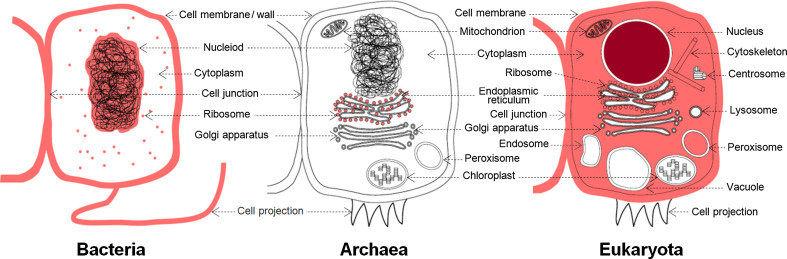
Figure 3C illustrates that among eukaryotic cellular components, the abundance of IDPs/IDRs generally follows the disorder preferences observed in biological processes, with nucleosome, spliceosome, and transcription factor complexes being especially enriched in the disorder. Bacteria also contain a large number of components associated with disorder, such as ribosome, cell wall, and flagellum, to name a few. We also show a substantial number of components in eukaryotic cells that are enriched in disorder when compared with a bacterial cells; see inset in Fig. 3C. In contrast, proteins in Archaea use disorder primarily only for translation, which is why archaean IDPs are commonly involved in RNA binding and are located in the ribosome. In addition to using disorder for the RNA binding, viruses commonly utilize IDPs to implement interactions with other organisms, and their IDPs are often located in the cytoplasm and nucleus. We mapped the components enriched in disorder from Fig. 3C into their cellular compartments, see Fig. 4. The compartments colored in red in the archaea and bacteria cell and in dark red in the eukaryotic cell include at least one component that is significantly enriched in disorder in a given domain of life. The light red in the eukaryotic cell denotes the compartments that are enriched in disorder compared to the bacteria. We observe that disorder is preferentially localized across the three domains of life in the ribosome. Furthermore, disorder is relatively abundant in most of the bacterial cell and several eukaryotic organelles/compartments, including nucleus, mitochondrion, cytoskeleton, peroxisome, and cell membrane and junction. However, some other compartments, such as the majority of intra-cellular membranes, Golgi apparatus, endoplasmic reticulum, endosome, lysosome, centrosome, chloroplast, and vacuole, include mostly structured proteins.
Fig. 4.
Mapping of intrinsic disorder into eukaryotic, bacterial, and archaea cells. The cellular components significantly enriched in disorder from Fig. 3C were mapped into the corresponding organelles/compartments. The light red color in bacteria or archaea cells identifies compartments that include at least one annotation that is enriched by at least 5 % in the disorder content in this domain of life. In the eukaryotic cell, the dark red color shows compartments that include at least one annotation enriched by at least 5 % in eukaryota, while the light red color denotes compartments with annotations enriched by at least 5 % compared to the disorder in bacteria (based on inset in Fig. 3C)
Disorder in post-translational modification sites
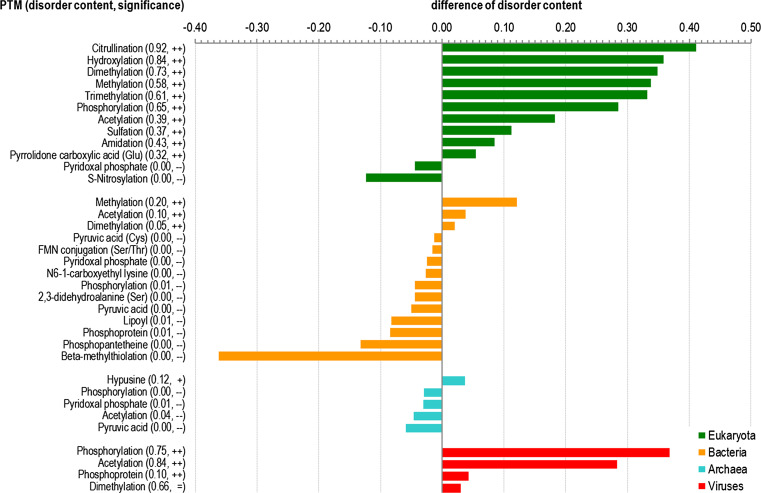
We also considered correlation between disorder and post-translational modifications (PTMs) that are annotated in the UniProt [35]. Figure 5 shows that most PTM sites are significantly enriched in disorder in eukaryota and viruses. This is in contrast to bacteria and archaea, which generally contain fewer PTMs that are associated with disorder. For example, phosphorylation sites are substantially enriched in disorder in eukaryotes (65 % of these sites are in the disordered regions) and viruses (75 % in the disordered regions), but they are depleted in disorder in archaea (virtually no phosphorylation sites are in the disordered regions) and bacteria (only 1 % in the disordered regions). Similarly, acetylation sites are enriched in disorder in eukaryotes (39 %) and viruses (84 %), while their enrichment in bacteria is lower (10 %) and they are depleted in disorder in archaea (4 %). We note only a few exceptions from that generic observation, e.g., a universally disorder-depleted piridoxal phosphate PTM.
Fig. 5.
Post-translational modifications (PTMs) that are significantly enriched/depleted in the disorder across eukaryotic, bacterial, archaea, and viral species. The y-axis gives PTMs, including the average disorder content among the corresponding amino acids and significance of the enrichment/depletion. The x-axis shows the difference in the average disorder content between amino acids with a given PTM and the baseline disorder content in a given domain of life. The significance of the difference is denoted with “–” and “-”, which indicate that the disorder is depleted with a P value smaller than 0.01 and 0.001, respectively; “+” and “++”, which indicate that the disorder is enriched with a P value smaller than 0.01 and 0.001, respectively; and “=”, which shows that disorder is not significantly different. The PTMs are sorted, within each domain of life, by the values of the difference
Disorder and structural coverage
Structural coverage is defined as a fraction of proteins expressed in a given proteome that are similar to a fold with known structure, and was calculated based on an approach proposed in Ref. [46]. Supplementary Fig. 2 demonstrates that structural coverage is modestly negatively correlated with the disorder content for archaean, bacterial and fungal species. This can be explained by the fact that structures of proteins that have disordered segments are usually harder to obtain using the dominant structure determination approach via X-ray crystallography [53]. The differences in the structural coverage are relatively substantial; for instance, the coverage drops by about 15 % for bacteria when comparing organisms with low and high disorder content. Among eukaryotes, animals have the highest coverage values, which likely stems from the focus on these species by the Protein Structure Initiative [54]. Moreover, lack of correlation with disorder for animal species suggests that given sufficient resources, high levels of coverage can be attained even for proteomes with relatively high disorder content. Our analysis also reveals that viral proteomes are characterized on average by the lowest structural coverage that lacks correlation with the disorder content.
Disorder and evolution
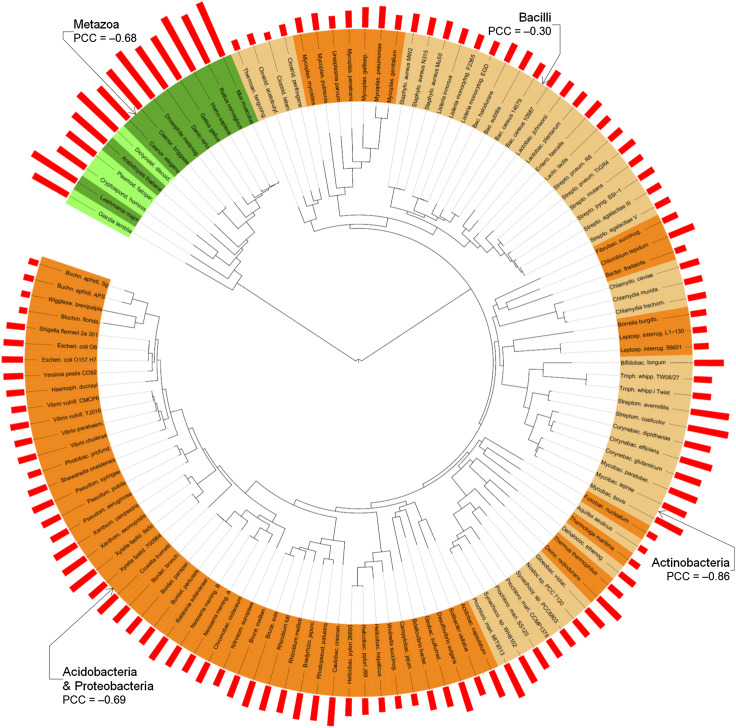
In order to put our observations into evolutionary perspective, we built a phylogenetic tree to include 126 species whose proteomes have been fully sequenced. Results of this analysis are shown in Fig. 6. This Figure represents the evolutionary data for 14 eukaryotic (on green background) and 112 bacterial proteomes (on orange background) in the form of a phylogenetic tree. Our analysis is based on the evolutionary tree presented in Ref. [47], which was reconstructed using a supermatrix of 31 concatenated, universally occurring genes with indisputable orthology in 191 species with completely annotated genomes in the three domains of life. In the original tree, the evolutionary speed of a given genome is proportional to the cumulative branch length from the tip to the root, with faster evolving genomes being characterized by longer branch length [47]. Figure 6 represents the superposition of the intrinsic disorder data on that evolutionary tree. Here, labels indicate individual species and various color shadings indicate subdivisions into phyla. Disorder contents in corresponding proteomes are shown as red bars outside of the tree. For each given genome, the length of the solid black line on the inside is the cumulative branch length from the tip to the root, which was estimated in Ref. [47], indicates the speed of evolution. Phyla containing at least eight species are named outside of the tree, together with the corresponding value of the Pearson correlation coefficient (PCC) between the branch length and the disorder content. We further analyzed these four phyla, one eukaryotic and three bacterial, as the remaining phyla have too few species to obtain conclusive results.
Fig. 6.
The phylogenetic tree based on Ref. [47], with 126 species whose proteomes have been fully sequenced, including 14 in eukaryota (on green background) and 112 in bacteria (on orange background). Labels indicate individual species and color shadings indicate subdivisions into phyla, where alternating light and dark green are for phyla in eukaryotes and light and dark orange are for phyla in bacteria. The red bars on the outside indicate the disorder content. The length of the solid black lines on the inside indicates speed of evolution, as estimated in Ref. [47]. Phyla with at least eight species are named on the outside, together with the corresponding value of the Pearson correlation coefficient (PCC) between the speed of evolution and disorder content
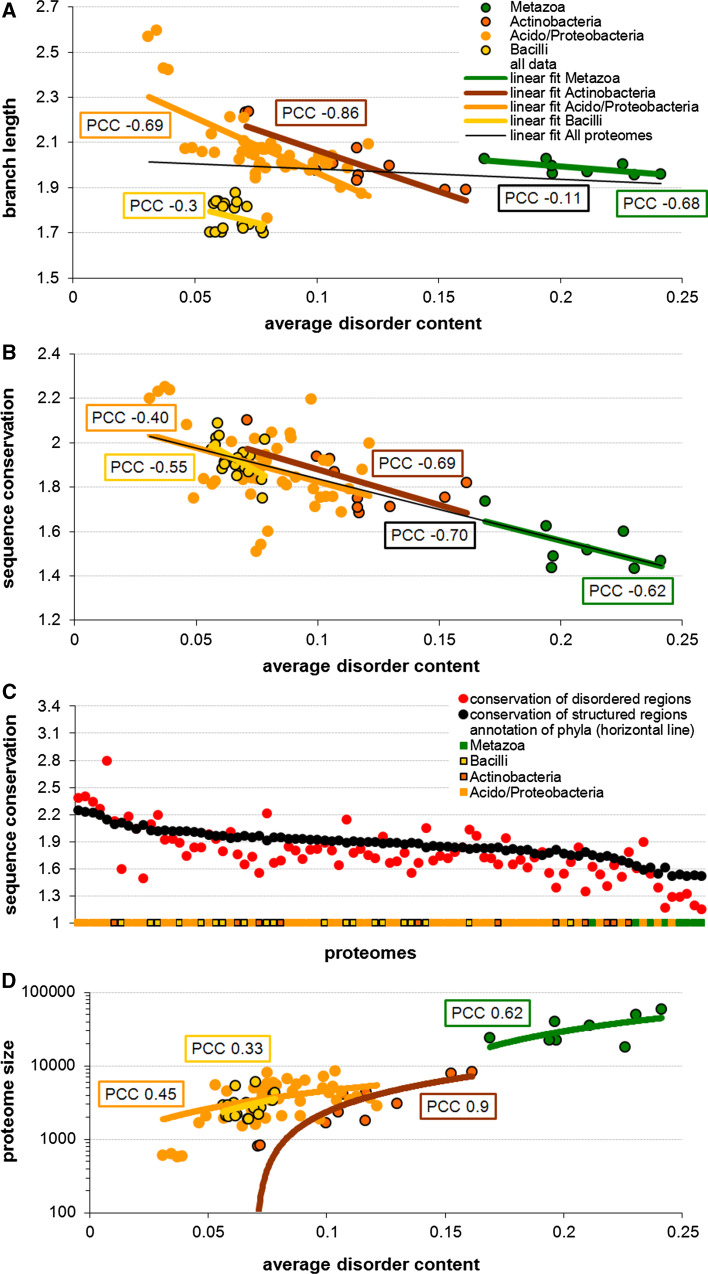
Figure 7 provides analysis of these evolutionary data combined with the analysis of sequence conservation. Figure 7A shows negative correlations between the disorder content and the evolutionary speed (measured as the branch length) within the selected four phyla. The PCC values are consistently negative and range between −0.3 and −0.86, suggesting that proteomes with more disorder evolve slower than proteomes with less disorder. Importantly, this trend holds true only within a given phylum. The correlation across proteomes from the four phyla is low and equals −0.11. Figure 7B shows that proteomes with higher disorder content are less conserved and that this trend is true even across phyla from bacteria and eukaryota, with the PCC value over all considered proteomes of −0.70. We note that our approach to quantify conservation for IDPs based on the sequence alignment could be somehow flawed, since conservation of function could occur in ways that are not discretely alignable (e.g., via compositional conservation). However, our observation agrees with prior observations that disordered regions are more likely to undergo non-conservative changes that lead to the lower sequence conservation compared to the structured regions [55]. Our analysis where we summarize the conservation at the proteome-level corroborates this finding. Furthermore, Fig. 7C reveals that disordered regions have lower sequence conservation than ordered regions for majority of the considered proteomes, irrespective of the overall conservation in a given proteome. For instance, the lower overall conservation of the considered eukaryotes when compared with bacteria (Fig. 7B) is combined with proportionally lower conservation of the corresponding disordered regions (Fig. 7C). The relatively low conservation of the disordered regions does not explain the negative correlation between disorder content and evolutionary speed in a specific phylum. A possible explanation for the latter trend is that disordered regions tend to be enriched in proteins with high connectivity (i.e., hubs) of protein–protein interactions networks [56], and the connectivity of these networks was shown to be negatively correlated with their rate of evolution [57]. Thus, enrichment in disorder could lead to higher connectivity (relative to a group of taxonomically related species in a given phylum), which, in turn, would lead to the reduced evolutionary speed. Another plausible explanation is related to the observation that smaller genomes evolved faster, which was explained by their limited ability to remove mutations by means of recombination or DNA repair [47]. Figure 7D shows a positive correlation between genome size (approximated by the number of proteins expressed by a given genome) and the disorder content within each of the four phyla. This figure, taken together with Fig. 3B reported by Ciccarelli et al., which represents negative correlation between the evolutionary speed and genome size [47], suggests that lower evolutionary speed could be a consequence of the enlarged proteome size that is associated with the enrichment in disorder. Perhaps another reason that the evolutionary speed is lower for proteomes with more disorder is that the proteins enriched in disorder are functionally important, such as by being involved in the protein–protein or protein–DNA interactions. To sum up, based on our empirical results, we hypothesize that there is a correlation between the speed of evolution and the degree of disorderedness, where larger proteomes in the same phyla contain more disorder and evolve slower.
Fig. 7.
Relation between disorder content and evolutionary characteristics, including evolutionary speed, sequence conservation and proteome size, for the bacterial and eukaryotic species. Panels A and B show relationship of the disordered content with the pace of evolution quantified using branch length in an evolutionary tree, and with the sequence conservation, respectively. Panel C compares sequence conservation of disordered (red markers) and structured (black markers) regions across the species grouped by phyla, which are denoted using the horizontal line at the bottom; species are sorted by the conservation of their structured regions. Panel D shows the relation between disorder content and proteome size. Solid lines in panels A, B, and D show linear fits together with the corresponding value of the PCC; y-axis in panel D is in logarithmic scale. The conservation was estimated based on relative entropy of WOP profiles produced by PSI-BLAST that was run against the nr database
Discussion
In agreement with a number of earlier studies, we show that IDPs/IDRs should not be considered as rare and obscure exceptions. Instead, these proteins and regions are very common in all the domains of life, including viruses, and clearly possess specific set of molecular functions. Our analysis reveals that the eukaryotic species have a unique disorder profile compared to the corresponding profiles of viruses and bacterial and archaean species. Here, eukaryotic proteomes are overall substantially more (about 20 %) disordered, contain more disorder in longer/larger proteins, and are characterized by a larger fraction of proteins with larger amounts of disorder. Eukaryotes and viruses have larger number of longer fully disordered proteins and longer disordered segments, compared to bacteria and archaea; particularly, viruses have relatively large number of long (over 300 amino acids), fully disordered chains.
Abundance of intrinsic disorder in eukaryotes and some of the viruses can be connected to the requirement of more profound signaling and regulation of these species. Analysis of the length-dependence of the average disorder content produced rather unexpected outcomes. In fact, based on the simple probability evaluations, one can expect that short proteins would contain less disorder than long proteins, and therefore the disorder content would increase with the protein length. However, dependence of the average disorder content on the protein length obtained in our study possesses an intriguing shape; see Fig. 2B. For example, in eukaryotes, short proteins are predicted to have significant amount of disorder. The amount of the predicted disorder decreases as protein length increases, and reaches minimum at ~15 % for proteins with the length of 300–500 residues. Then, the amount of intrinsic disorder starts to increase, reaches a plateau at the level of 25 % for proteins with length of~1,000–2,000 residues, and then again starts to decrease for longer proteins. Since the number of very long proteins is relatively small, that part of the plot corresponding to proteins longer than 5,000 residues is relatively noisy. Importantly, some long proteins contain very significant amount of predicted disorder, up to 90–95 %. Similarly, short proteins from other domains of life are typically more disordered than longer proteins. The fact that short proteins contain the highest amount of predicted disorder and the fact that long disordered proteins in eukaryotes seem to have some optimal length (1,500–2,000 residues) with relatively high disorder content (25 %) may potentially have some functional explanations.
Functional correlation study shows that disorder is enriched in many key processes, including transcription, translation, nucleosome assembly/chromosome condensation, RNA splicing, protein polymerization and dimerization, catabolic and metabolic processes, and pathogenesis in bacteria. Furthermore, disordered proteins are preferentially located in certain cellular compartments, including nucleosome, spliceosome, transcription factor complexes, ribosome, and cell wall and flagellum in bacteria. Archaean proteins use disorder for translation, whereas viruses use disorder for RNA binding and to implement interactions with other organisms. We also provide a convenient mapping of disorder into archea, bacterial and eukaryotic cells. Interestingly, we show a strong pattern of disorder enrichment in the PTM sites where these sites are significantly enriched in disorder in eukaryotes and viruses, while substantially fewer PTMs are associated with disorder in bacteria and archaea.
The content of disorder in certain domains and phyla, including bacteria, archaea and fungi, is negatively correlated with the structural coverage of these species. This suggests a bias towards solving structures of proteins that are depleted in disorder. This observation is in line with the fact that presence of disordered regions makes crystallization of proteins more difficult [53], while crystallization-based structure determination pipelines account for a significant majority of effort in this area. However, such a trend does not appear for the relatively highly structurally covered animals. This demonstrates that relatively high structural coverage can be attained even for species with a high amount of disorder.
Finally, we expand on the prior observations that linked proteome/genome size with the evolutionary speed by inclusion of the degree of disorder. We observe that among closely related species from the same eukaryotic or bacterial phyla, species with smaller proteomes that evolved faster have less disorder.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Contributor Information
Vladimir N. Uversky, Email: vuversky@health.usf.edu
Lukasz Kurgan, Email: lkurgan@ece.ualberta.ca.
References
- 1.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293(2):321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 2.Dunker AK, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19(1):26–59. doi: 10.1016/S1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 3.Uversky VN. Protein folding revisited. A polypeptide chain at the folding-misfolding-nonfolding cross-roads: which way to go? Cell Mol Life Sci. 2003;60(9):1852–1871. doi: 10.1007/s00018-003-3096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turoverov KK, Kuznetsova IM, Uversky VN. The protein kingdom extended: ordered and intrinsically disordered proteins, their folding, supramolecular complex formation, and aggregation. Prog Biophys Mol Biol. 2010;102(2–3):73–84. doi: 10.1016/j.pbiomolbio.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804(6):1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6(3):197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 7.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12(1):54–60. doi: 10.1016/S0959-440X(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 8.Dunker AK, Uversky VN. Signal transduction via unstructured protein conduits. Nat Chem Biol. 2008;4(4):229–230. doi: 10.1038/nchembio0408-229. [DOI] [PubMed] [Google Scholar]
- 9.Cortese MS, Uversky VN, Dunker AK. Intrinsic disorder in scaffold proteins: getting more from less. Prog Biophys Mol Biol. 2008;98(1):85–106. doi: 10.1016/j.pbiomolbio.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oldfield CJ, et al. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genom. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets: The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272(20):5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 12.Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18(5):343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 13.Dunker AK, Brown CJ, Obradovic Z. Identification and functions of usefully disordered proteins. Adv Protein Chem. 2002;62:25–49. doi: 10.1016/S0065-3233(02)62004-2. [DOI] [PubMed] [Google Scholar]
- 14.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41(21):6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 15.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18(6):756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Dunker AK, Obradovic Z, Romero P, Kissinger C, Villafranca E. On the importance of being disordered. PDB Newsletter. 1997;81:3–5. [Google Scholar]
- 17.Brown CJ, et al. Evolutionary rate heterogeneity in proteins with long disordered regions. J Mol Evol. 2002;55(1):104–110. doi: 10.1007/s00239-001-2309-6. [DOI] [PubMed] [Google Scholar]
- 18.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323(3):573–584. doi: 10.1016/S0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 19.Xie H, et al. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J Proteome Res. 2007;6(5):1882–1898. doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vucetic S, et al. Functional anthology of intrinsic disorder. 2. Cellular components, domains, technical terms, developmental processes, and coding sequence diversities correlated with long disordered regions. J Proteome Res. 2007;6(5):1899–1916. doi: 10.1021/pr060393m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie H, et al. Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J Proteome Res. 2007;6(5):1917–1932. doi: 10.1021/pr060394e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tompa P (2011) Intrinsically unstructured proteins. Trends Biochem Sci 27 (10):527-533 (2002) [DOI] [PubMed]
- 23.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- 24.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337(3):635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Feng ZP, et al. Abundance of intrinsically unstructured proteins in P. falciparum and other apicomplexan parasite proteomes. Mol Biochem Parasitol. 2006;150(2):256–267. doi: 10.1016/j.molbiopara.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Tompa P, Dosztanyi Z, Simon I. Prevalent structural disorder in E. coli and S. cerevisiae proteomes. J Proteome Res. 2006;5(8):1996–2000. doi: 10.1021/pr0600881. [DOI] [PubMed] [Google Scholar]
- 27.Galea CA, et al. Large-scale analysis of thermostable, mammalian proteins provides insights into the intrinsically disordered proteome. J Proteome Res. 2009;8(1):211–226. doi: 10.1021/pr800308v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue B, Williams RW, Oldfield CJ, Dunker AK, Uversky VN. Archaic chaos: intrinsically disordered proteins in Archaea. BMC Syst Biol. 2010;4(Suppl 1):S1. doi: 10.1186/1752-0509-4-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burra PV, Kalmar L, Tompa P. Reduction in structural disorder and functional complexity in the thermal adaptation of prokaryotes. PLoS ONE. 2010;5(8):e12069. doi: 10.1371/journal.pone.0012069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue B, Dunker AK, Uversky VN. Orderly order in protein intrinsic disorder distribution: disorder in 3,500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn. 2012;30(2):137–149. doi: 10.1080/07391102.2012.675145. [DOI] [PubMed] [Google Scholar]
- 31.Yan J, Mizianty MJ, Filipow PL, Uversky VN. Kurgan L (2013) RAPID: fast and accurate sequence-based prediction of intrinsic disorder content on proteomic scale. Biochim Biophys Acta. 1834;8:1671–1680. doi: 10.1016/j.bbapap.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 32.Lobley A, Swindells MB, Orengo CA, Jones DT. Inferring function using patterns of native disorder in proteins. PLoS Comput Biol. 2007;3(8):e162. doi: 10.1371/journal.pcbi.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pentony MM, Jones DT. Modularity of intrinsic disorder in the human proteome. Proteins. 2010;78(1):212–221. doi: 10.1002/prot.22504. [DOI] [PubMed] [Google Scholar]
- 34.Romero PR, et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc Natl Acad Sci U S A. 2006;103(22):8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.UniProt_Consortium (2012) Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res 40 (Database issue):D71-75 [DOI] [PMC free article] [PubMed]
- 36.Geer LY, et al. The NCBI BioSystems database. Nucleic Acids Res. 2010;38:D492–D496. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21(16):3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 38.Dosztanyi Z, Csizmok V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol. 2005;347(4):827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 39.Walsh I, Martin AJ, Di Domenico T, Tosatto SC. ESpritz: accurate and fast prediction of protein disorder. Bioinformatics. 2012;28(4):503–509. doi: 10.1093/bioinformatics/btr682. [DOI] [PubMed] [Google Scholar]
- 40.Sickmeier M, et al. (2007) DisProt: the Database of Disordered Proteins. Nucleic Acids Res 35 (Database issue):D786-793 [DOI] [PMC free article] [PubMed]
- 41.Peng ZL, Kurgan L. Comprehensive comparative assessment of in silico predictors of disordered regions. Curr Protein Pept Sci. 2012;13(1):6–18. doi: 10.2174/138920312799277938. [DOI] [PubMed] [Google Scholar]
- 42.Peng ZL, Kurgan L (2012) On the complementarity of the consensus-based disorder prediction. Pac Symp Biocomput, 176-187 [PubMed]
- 43.Tompa P, et al. Close encounters of the third kind: disordered domains and the interactions of proteins. BioEssays. 2009;31(3):328–335. doi: 10.1002/bies.200800151. [DOI] [PubMed] [Google Scholar]
- 44.Monastyrskyy B, Fidelis K, Moult J, Tramontano A, Kryshtafovych A. Evaluation of disorder predictions in CASP9. Proteins. 2011;79(Suppl 10):107–118. doi: 10.1002/prot.23161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vitkup D, Melamud E, Moult J, et al. Completeness in structural genomics. Nature Struct Mol Biol. 2001;8(6):559–566. doi: 10.1038/88640. [DOI] [PubMed] [Google Scholar]
- 47.Ciccarelli FD, et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311(5765):1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 48.Peng Z, Xue B, Kurgan L, Uversky VN. Resilience of death: intrinsic disorder in proteins involved in the programmed cell death. Cell Death Differ. 2013;20(9):1257–1267. doi: 10.1038/cdd.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng Z, Oldfield CJ, Xue B, Mizianty MJ, Dunker AK, Kurgan L, Uversky VN. A creature with a hundred waggly tails: intrinsically disordered proteins in the ribosome. Cell Mol Life Sci. 2013;71(8):1477–1504. doi: 10.1007/s00018-013-1446-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang K, Samudrala R. Incorporating background frequency improves entropy-based residue conservation measures. BMC Bioinformatics. 2006;7:385. doi: 10.1186/1471-2105-7-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng ZL, Mizianty MJ, Xue B, Kurgan L, Uversky VN. More than just tails: intrinsic disorder in histone proteins. Mol BioSyst. 2012;8(7):1886–1901. doi: 10.1039/c2mb25102g. [DOI] [PubMed] [Google Scholar]
- 53.Oldfield CJ, Xue B, Van YY, Ulrich EL, Markley JL, Dunker AK. Uversky VN (2013) Utilization of protein intrinsic disorder knowledge in structural proteomics. Biochim Biophys Acta. 1834;2:487–498. doi: 10.1016/j.bbapap.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Markley JL, Aceti DJ, Bingman CA, Fox BG, et al. The Center for Eukaryotic Structural Genomics. J Struct Funct Genomics. 2009;10(2):165–179. doi: 10.1007/s10969-008-9057-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown CJ, Johnson AK, Daughdrill GW. Comparing models of evolution for ordered and disordered proteins. Mol Biol Evol. 2010;27(3):609–621. doi: 10.1093/molbev/msp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bertolazzi P, Bock ME, Guerra C. On the functional and structural characterization of hubs in protein–protein interaction networks. Biotechnol Adv. 2013;31(2):274–286. doi: 10.1016/j.biotechadv.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 57.Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. Evolutionary rate in the protein interaction network. Science. 2002;296(5568):750–752. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.