Abstract
Changes in the structure and number of synapses modulate learning, memory and cognitive disorders. Ubiquitin-mediated protein modification is a key mechanism for regulating synaptic activity, though the precise control of this process remains poorly understood. RING finger protein 13 (RNF13) is a recently identified E3 ubiquitin ligase, and its in vivo function remains completely unknown. We show here that genetic deletion of RNF13 in mice leads to a significant deficit in spatial learning as determined by the Morris water maze test and Y-maze learning test. At the ultrastructral level, the synaptic vesicle density was decreased and the area of the active zone was increased at hippocampal synapses of RNF13-null mice compared with those of wild-type littermates. We found no change in the levels of SNARE (soluble N-ethylmaleimide-sensitive factor-attachment protein receptor) complex proteins in the hippocampus of RNF13-null mice, but impaired SNARE complex assembly. RNF13 directly interacted with snapin, a SNAP-25-interacting protein. Interestingly, snapin was ubiquitinated by RNF13 via the lysine-29 conjugated polyubiquitin chain, which in turn promoted the association of snapin with SNAP-25. Consistently, we found an attenuated interaction between snapin and SNAP-25 in the RNF13-null mice. Therefore, these results suggest that RNF13 is involved in the regulation of the SNARE complex, which thereby controls synaptic function.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-012-1103-5) contains supplementary material, which is available to authorized users.
Keywords: RNF13, Ubiquitin ligase, Snapin, Synaptic vesicle, SNARE complex
Introduction
Cognitive impairment can result from synaptic dysfunction and subsequent deficiency in neuronal communication of the mammalian central nervous system [1]. During the process of neuronal communication, synaptic vesicles are loaded with neurotransmitters and translocate to the plasma membrane of axon terminals where they selectively dock close to the active zone [2, 3]. Docked vesicles undergo a priming step, during which vesicles become fusion-competent. Intracellular calcium binds to calcium sensors, triggering vesicle fusion and release of neurotransmitters into the synaptic cleft [4]. Neurotransmitters then bind to receptors located at sites on the postsynaptic membrane [5]. These processes are mediated by dynamic assembly and disassociation of several protein complexes [6]. Modulation of the activity of such protein complexes is also important in the control of synaptic plasticity. However, it remains unclear how the activity of these synaptic proteins is regulated.
Recent studies have highlighted the importance of the ubiquitin pathway in the regulation of synaptic proteins [7–10]. Targeted ubiquitination of synaptic proteins affects multiple facets of the synapse, from synaptogenesis and synapse elimination to activity-dependent synaptic plasticity and remodeling [11]. The F-box protein SCRAPPER can target active zone protein RIM1 for degradation through the ubiquitin–proteasome pathway (UPS) [12]. The RING-type ubiquitin ligase staring ubiquitinates and facilitates the proteasome-dependent degradation of syntaxin 1 [13]. In addition, parkin predominantly promotes monoubiquitination of the PDZ domain-containing protein PICK1 to regulate ion channel activity [14], while a synapse-associated E3 ubiquitin ligase PDZRN3 promotes muscle-specific receptor tyrosine kinase (MuSK) ubiquitination and regulates clustering of nicotinic acetylcholine receptor [15]. These indicate that both degradative and non-degradative manners of ubiquitination may also modulate synaptic vesicle release.
The novel E3 ubiquitin ligase RNF13 belongs to a new protein family, PA-TM-RING, which is characterized by an N-terminal protease-associated (PA) domain and a C-terminal RING finger domain (RING) separated by a middle transmembrane region (TM) [16, 17]. Our previous studies confirmed that the transmembrane RING-type ubiquitin ligase RNF13 is overexpressed in human pancreatic cancer and precancerous lesions [18] and regulates myoblast proliferation [17, 19]. RNF13 protein itself is tightly regulated at the post-translational level in cells, including multiple modifications, rapid protein turnover, proteolytic release of both the N-terminal PA and C-terminal RING finger fragments, as well as the translocation to the inner nuclear membrane through recycling endosomes [20, 21]. Those results suggest that under different cell physiological conditions, RNF13 may exert an important function by ubiquitinating specific substrate(s) at different cellular locations.
Interestingly, the functional role of RNF13 has been suggested to be involved in neuronal development by different studies. Increased expression of chicken RNF13 (c-RZF) was first reported in the embryonic brain cells in response to treatment of extracellular matrix protein Tenascin-C [22]. RNF13 was identified as one of five genes in a screen for proteins whose ectopic expression promotes neurite outgrowth of PC12 cells in vitro [23]. Recently, a higher level of RNF13 expression was observed in the adult mouse brain as compared to the embryonic brain, as well as during the differentiation of stimulated B35 neuroblastoma after treatment with an analog of cAMP, dibutyryl-cAMP [20]. Here in this study, we show that RNF13 functions as a novel regulator of SNARE complex assembly, and its genetic disruption leads to a learning and memory defect in mice.
Materials and methods
Generation of RNF13 knockout mice
Mice were housed in a SPF facility on a 12-h light/12-h dark schedule with access to food and water ad libitum. RNF13-null mice were generated with a target construct with exon 5 flanked by loxP sites (Fig. 1a). RNF13 floxed mice were then crossed with EПA-Cre transgenic mice to produce heterozygous mutant mice. To reduce the variation caused by different genetic backgrounds, RNF13 +/− mice were backcrossed to C57BL/6 mice for five to six generations. Then RNF13-null mice and WT mice were produced by crossing RNF13 +/− heterozygous mice. For genotyping, the genomic DNA of each mouse was extracted from the tip of the tail. DNA amplification for genotyping was performed using rTaq (Takara, Dalian). Three oligonucleotides were used as two sets of PCR primers to identify the mutant and WT amplified fragments (8F: 5′-CCAGAGTGCATGGCATTTCT–3′, 8R: 5′-ATCTGGAGAGCAGCAGTTAG–3′, and 9R: 5′-CCTGAGTTGTGGAAACTAAATTGAC–3′). Fragments of 700 and 432 bp were amplified by using genomic DNA from RNF13-null mice and WT mice, respectively, while both fragments were in RNF13 +/− (Fig. 1a).
Fig. 1.
Generation of RNF13−/− mice. a Targeted disruption of mouse RNF13 exon 5. Mouse RNF13 gene structure, RNF13 conditional knockout construct and primers used for screening and genotyping. 8F/8R and 9F/9R are used to check the first and second loxP in the construct, respectively; 13F/Tpcr-L2 and FRT-2/10R are used to screen the targeting cells. b Genomic DNA-PCR and c Western blot analysis of RNF13 gene and proteins. β-Tubulin is a loading control. d Appearance of RNF13-null (left panel) and WT (right panel) mice
Antibodies
Mouse monoclonal RNF13 antibody (1:200) was raised in our institute. The mouse monoclonal antibodies against SNAP-25 (1:500), syntaxin 1 (1:500), dynamin I (1:500) as well as rabbit polyclonal anti-complexin 1/2 (1:500) were obtained from Santa Cruz Biotechnology. Other antibodies used in this study included mouse monoclonal anti-synaptotagmin 1 (1:500; Assay Designs), rabbit polyclonal anti-VAMP-2 and chicken polyclonal anti-snapin (ab37496) (1:1,000; Abcam), mouse monoclonal anti-munc18-1 (1:1,000; BD Biosciences), and mouse monoclonal anti-β–actin and β–tubulin (1:5,000; Sigma-Aldrich).
Immunohistochemistry and immunofluorescence
Immunostaining was adapted from Schneider Gasser et al. [24]. Briefly, fresh hippocampal slices trimmed from coronal brain slices were fixed in 4 % PFA/PBS on ice for 20 min, washed three times in PBS and incubated “free-floating” in blocking solution (10 % goat serum, 0.1 % Triton X-100, PBS) for 1 h. RNF13 antibody in blocking solution was incubated with slices for 16 h under gentle agitation at 4 °C. Slices were washed three times in PBS and incubated with secondary antibody for 2 h at room temperature. Following incubation, slices were washed three times with PBS, transferred and mounted on to glass slides with Immun-Mount (Thermo Scientific).
Co-localization of RNF13 and snapin in cultured primary hippocampal cells and primary chromaffin cells was identified by using two antibodies for 2 h at room temperature, followed by incubation with FITC- and TRITC-labeled secondary antibodies. Images were acquired using a Zeiss LSM 510 confocal microscope.
Immunoprecipitation and in vitro pull-down assay
Immunoprecipitation of mouse hippocampal samples was performed as described [25]. COS-7 cells were lysed 48 h after transfection with a solution containing 50 mM Tris-HCl (pH 7.4), 100 mM NaCl, 1 % (v/v) Triton X-100 and a cocktail of protease inhibitors (Complete EDTA-free, Roche). Cell lysates were incubated with 2 μg of antibody in 20 μl of protein G-Sepharose beads (Amersham) for 4 h at 4 °C. Immunoprecipitates were washed four times with ice-cold lysis buffer and blotted with appropriate antibodies. In vitro binding using GST fusion proteins and transfected cell extract was performed as described [25].
Ubiquitination assays
The in vitro ubiquitination assay was performed by incubating the GST-fused C-terminal domain of RNF13 with immunoprecipitated FLAG-snapin, 5 nM yeast E1, 100 nM UbcH5a (E2) and ubiquitin (or its mutants) in ubiquitination buffer (50 mM Tris-HCl, pH 7.5, 4 mM MgCl2, 1 mM DTT, 1 mM ATP, 10 mM creatine phosphate and 16 IU/ml creatine phosphokinase) to a final volume of 40 μl. Samples were incubated at 30 °C for 1 h and subsequently analyzed by Western blot. To detect ubiquitination of snapin in cells, transfected COS-7 cells were harvested in lysis buffer containing 2 % SDS. After incubating at 4 °C for 1 h, samples were diluted 1:20 using lysis buffer without SDS and FLAG-tagged snapin was immunoprecipitated and analyzed by Western blot with an anti-HA antibody (Sigma-Aldrich).
Morris water maze test
The swimming pool was a circular metal tank, 122 cm in diameter. The hidden platform, 10 cm in diameter, was clear Plexiglas and submerged 1 cm below the surface of the water. To make the platform invisible, the water was colored white by using powdered milk. Adult male mice, 6 months old, were trained on the water maze task for 5 consecutive days, and each training day included four trials per mouse, separated by 10-min intervals. The starting points for mouse placement in each training episode were varied. Mice were allowed to search for the underwater platform for 120 s. After landing on the hidden platform, the mice were allowed to remain for 30 s before being returned to their cage. Mice that failed to land on the platform by themselves within the time limit were manually guided to it.
Y-maze test
The maze used was a three-arm Y-maze with equal angles between all arms. Each mouse was placed in the center of the “Y” and allowed to explore freely during an 8-min session. The sequence and total number of arms entered were recorded. Arm entry was considered to be complete when the hind paws of the mouse were completely within the arm. Percentage alternation is defined as the number of trials containing entries into all three arms divided by the maximum possible number of alternations (the total number of arms entered minus 2) × 100 %.
Electron microscopy
Electron microscopy was performed as described previously [26]. For hippocampal electron microscopy examination, 8-week-old mice (three WT and three RNF13-null mice) under deep pentobarbital anesthesia were perfused through the heart with 2.5 % glutaraldehyde and 2 % paraformaldehyde in 0.1 M phosphate buffer, pH 7.4. The brains were removed, and the hippocampus was sliced transverse to its longitudinal axis at 1 mm thickness. The blocks were trimmed to contain the hippocampal CA1 pyramidal cell bodies and their apical dendrites in the stratum radiatum. The blocks were immersed in 4 % glutaraldehyde in 0.1 M phosphate buffer, pH 7.4, overnight at 4 °C. Ultrathin sections (~60 nm) were stained with uranyl acetate and lead citrate. Only asymmetrical, i.e., glutamatergic, synapses with clearly identifiable PSDs were analyzed. The quantitative analysis was performed according to the previously published literature [27].
Statistical analysis
Values are given as mean ± SE (standard error). Statistical significance was determined by Student’s t test.
Results
Targeted disruption of RNF13 in mice impairs learning function
To investigate the in vivo function of RNF13 in mice, we genetically targeted RNF13 exon 5 by crossing the RNF13-floxed mice with EПA-Cre transgenic mice (Fig. 1a), which leads to exon 5 deletion at past zygote stage during development. RNF13-null mice were generated with deletion of exon 5 by introducing frame-shift mutated RNF13 mRNA (Fig. 1a). PCR genotyping and Western blot were performed to verify the efficiency of RNF13 deletion (Fig. 1b, c). RNF13-null mice were born with expected Mendelian frequency and showed normal growth, fertility and lifespan, with no overt physiological abnormality (Fig. 1d). As RNF13 has been implicated in neuronal development in vitro [20, 22, 23] and its expression could be detected in hippocampal neurons (Fig. S1 A–D), we first sought to investigate whether animals lacking RNF13 exhibit sub-optimal neuronal phenotypes and behavioral defects.
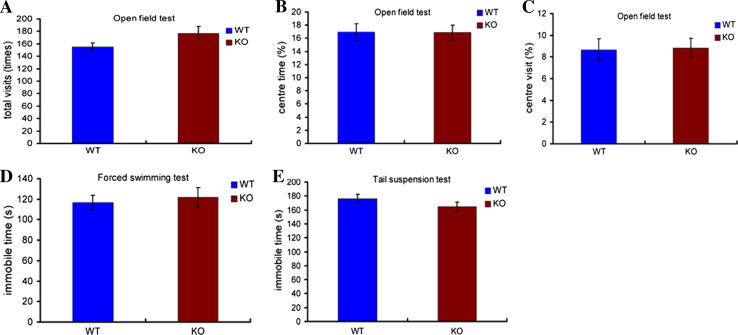
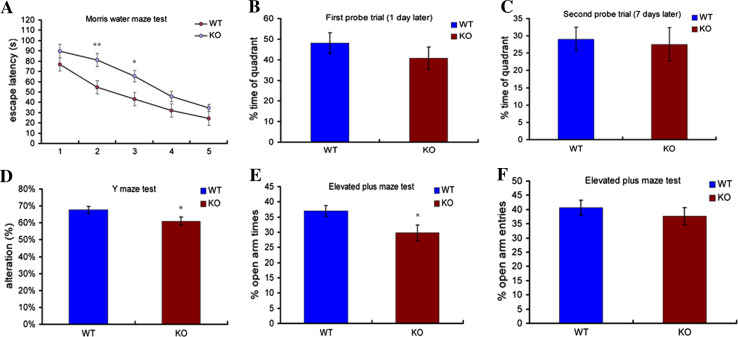
To minimize behavioral variation caused by different genetic backgrounds, RNF13-null mice were backcrossed into the C57BL/6 background. The RNF13-null mutants do not display abnormalities in open field, swimming and tail suspension tests (Fig. 2 a–e), which indicates that RNF13 deletion has no effect on mouse spontaneous activity, as well as anxiety and depression-like behaviors. To further examine hippocampal-dependent learning and memory functions, we performed the Morris water maze (MWM) test and the Y maze test. In the MWM test, both RNF13-null and WT mice showed a reduction of escape latency during the five sessions of the acquisition phase. However, the escape latency of RNF13-null mice differed significantly from that of the wild-type in the second and third sessions (Fig. 3a WT n = 16, 54.67 ± 6.42 s; KO n = 19, 81.14 ± 6.39 s; p = 0.0065 for the second session, and WT 43.27 ± 5.47 s; KO 65.44 ± 6.43 s; p = 0.0148 for the third session). To test memory retention, each trained mouse was tested in probe trials 1 and 7 days after training. Both WT and RNF13-null mice performed well in the probe trial 1 day after training. Although RNF13-null mice showed a slightly decreased time spent in the quadrant containing the platform relative to WT mice, the difference did not reach statistical significance (Fig. 3b). To assess long-term memory, the probe trial was performed 7 days after training. There was no difference between the two groups (Fig. 3c). Moreover, we carried out the Y maze to assess the working memory (without food deprivation or other aversive procedures). RNF13-null mice exhibited a lower rate of alternation compared with WT mice, indicating that mice lacking RNF13 were unable to remember the last arm entered (Fig. 3d WT n = 20, 67.65 ± 2.11 %; KO n = 17, 60.94 ± 2.24 %; p = 0.0433). Additionally, although RNF13-null mice were more anxious than WT in terms of the percentage of time spent in the elevated plus-maze (EPM) open arms, but not the number of entries into those arms (Fig. 3e, f; WT n = 19, 37.02 ± 1.80 %; KO n = 16, 29.79 ± 2.59 %; p = 0.0272), neither RNF13-null mice nor WT mice exhibited any alteration in locomotor activity or central tendency in the open-field test (Fig. 2a–c; p > 0.05 in each case). Taken together, our findings from the behavioral studies indicate that RNF13-null mice have deficiencies in spatial learning in the tasks examined.
Fig. 2.
The effect of RNF13 on mouse spontaneous activity and depression level. a–c Neither RNF13-null mice nor WT exhibited any alteration in spontaneous activity or central tendency in the open-field test (p > 0.05 in each case) (WT, n = 17; RNF13-null mice, n = 19). d Immobile time during forced swimming test (WT, n = 14; RNF13-null mice, n = 13). Immobile time was unaffected by genotype. e Immobile time during tail suspension test (WT, n = 21; RNF13-null mice, n = 18). Immobile time was unaffected by genotype. Data are shown as mean ± SE
Fig. 3.
RNF13-null mice have impaired learning and memory. a The escape latency from Morris water maze analysis throughout the training sessions (WT (n = 16), RNF13-null mice (n = 19); p < 0.01 for the second session, p < 0.05 for the third session). b, c The probe trial tested 1 day and 7 days after training. No significant difference was observed during the probe trial. d Y maze performance of WT (n = 19) and RNF13-null mice (n = 19). RNF13-null mice displayed reduced alternation rates, p < 0.05. e The percentage of time spent in open arms in the elevated-plus maze for anxiety-like behaviors (RNF13-null mice (n = 16) and WT (n = 19), p < 0.05). f Number of open arm entries in the elevated-plus maze. There was no significant difference between groups. Data are shown as mean ± SE. *p < 0.05, **p < 0.01, Student’s t test
RNF13-null mice show structural alteration of synapses in hippocampus
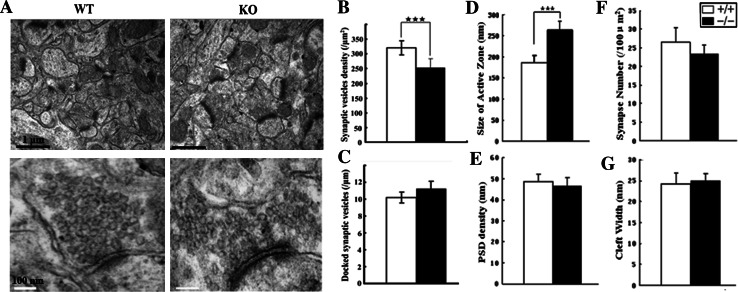
Learning and memory deficiency has been shown to depend primarily upon hippocampal function (HF) [28]. Abnormal behavior, like spatial learning deficits, of RNF13-null mice may be attributed to abnormal hippocampal structure or defective neurotransmitter release in hippocampi. We therefore examined whether the morphological structure of the RNF13-null hippocampal formation was altered. Hematoxylin and eosin staining revealed no obvious histopathological changes in the HF of RNF13-null mice (Fig. S2 A–H). However, as determined by transmission electron microscopy (TEM), abnormalities in the asymmetry of excitatory synapses of the hippocampal CA1 region were observed in mutant mice as compared with WT mice (Fig. 4a). First, in an attempt to investigate changes in the vesicle pool in RNF13-null mice, we observed decreased synaptic vesicle density in the RNF13-null mice (n = 3, WT, 319.6 ± 28.3/μm2; KO, 252.4 ± 25.4/μm2), while total vesicle number and the number of docked vesicles (~50 nm next to the active zone membrane) remained unchanged (Fig. 4b, c). The decrease in synaptic vesicle density would cause a significant reduction in synaptic activity in RNF13-null mice. Interestingly, quantitative analysis also revealed that the absence of RNF13 was correlated with increased size of synaptic active zones (n = 3, WT, 186.23 ± 17.52 nm; KO, 264.12 ± 20.67 nm) (Fig. 4d). In contrast, the postsynaptic density (PSD) (n = 3, WT, 48.6 ± 3.5 nm; KO, 46.3 ± 4.2 nm), the synapse number (n = 3, WT, 26.43 ± 3.86; KO, 23.27 ± 2.45) and cleft width (n = 3, WT, 24.3 ± 2.5 nm; KO, 24.9 ± 1.8 nm) remained unchanged (Fig. 4e–g). Collectively, these results indicate that a lack of RNF13 leads to a lower synaptic vesicle density, which may be structurally correlated with learning impairments in RNF13-null mice.
Fig. 4.
Alteration of hippocampal ultrastructure in RNF13-null mice. a Representative TEM images of hippocampal CA1 ventral region in WT and RNF13-null mice. Upper bar (black): 1 μm; lower bar (white): 100 nm. b–g Comparison of parameters: synaptic vesicle density, number of docked synaptic vesicles in the active zone, size of active zone, PSD density, synaptic number and cleft width. ***p < 0.001, Student’s t test. Error bars indicate the mean ± SE. Active zone here is defined by electron microscopy as the electron dense thickening of the presynaptic where synaptic vesicles accumulate and dock and the area opposing the postsynaptic specialization. Quantization was analyzed by NIH Image J software
RNF13-null mice exhibit decreased SNARE complex assembly
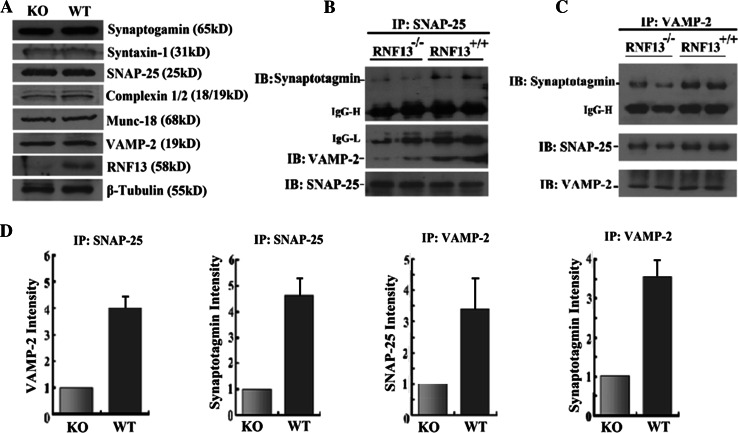
As RNF13 is an E3 ubiquitin ligase and may target specific protein substrates for degradation, we then investigated whether the steady-state levels of key proteins in the synaptic vesicle release machinery were affected in RNF13-null mice. We analyzed the protein levels in the hippocampus homogenates by immunoblot and found that the absence of RNF13 had no effect on the overall abundance of t-SNARE proteins (SNAP-25 and syntaxin 1), vesicle membrane protein VAMP-2, calcium sensor protein synaptotagmin 1 and vesicle-associated proteins (Munc18 and complexin1/2) (Fig. 5a). We also performed co-immunoprecipitation (Co-IP) experiments to corroborate whether RNF13 depletion affected SNARE complex assembly. We examined the interaction of SNAP-25 with VAMP-2, two core SNARE complex proteins and the association of synaptotagmin 1 with this complex, in the hippocampal lysates from RNF13-null and WT mice. Decreased levels of VAMP-2 protein were detected in the immunoprecipitates pulled down by SNAP-25 antibody in RNF13-null mice (Fig. 5b). In addition, synaptotagmin 1 also exhibited an attenuated interaction with SNAP-25 (Fig. 5b). Consistently, use of the VAMP-2 antibody for reciprocal immunoprecipitation in RNF13-null mice showed reduced levels of immunoprecipitated SNAP-25 and synaptotagmin 1 (Fig. 5c). More hippocampal samples from independent mice were examined to detect synaptic protein levels and associations, and the experiments showed consistent results. Furthermore, we quantified the binding affinity of SNARE complex proteins between wild-type and mutant and found several-fold reduction of the affinity in all examined samples when RNF13 is depleted (Fig. 5d). Therefore, our findings revealed that ubiquitin ligase RNF13 modulated the assembly of SNARE complex in vivo.
Fig. 5.
The regulation of RNF13 on SNARE complex formation. a Immunoblotting analysis of hippocampus homogenates (30 μg) from WT and RNF13-knockout mice showed no major changes in the expression levels of several known synaptic vesicle proteins. b Hippocampal lysate immunoprecipitation by SNAP-25 antibody to detect the related SNARE complex proteins, synaptotagmin I and VAMP-2. c Reciprocal immunoprecipitation by VAMP-2 antibody for checking the SNARE complex proteins SNAP-25 and synaptotagmin I. d Histograms showing normalized levels of different immunoprecipitated synaptic proteins from RNF13-null (gray bars) hippocampal homogenate relative to those in WT mice (black bars)
RNF13 directly interacts with the SNARE complex-associated protein snapin
Identification of RNF13 substrates is crucial for deciphering its function in the brain. To further elucidate the molecular relationship between RNF13 and spatial learning, we screened a human fetal brain cDNA library via a yeast two-hybrid system to identify RNF13-interacting proteins. Since RNF13 may target substrates for degradation in yeast and produce false negatives, we utilized the ligase-deficient RING domain mutant (C258A/H260A) RNF13 as the bait protein. A clone encoding snapin, the SNARE complex-associated protein, was identified as a potential RNF13 interaction protein by our yeast two-hybrid screen (Fig. S3). Snapin is a 17-kD protein containing an N-terminal hydrophobic domain (HD) and two coiled-coil domains [29]. Snapin has been verified to interact with SNAP-25 and facilitate the association of SNAP-25 with synaptotagmin 1 to regulate synaptic vesicle fusion [29, 30]. Snapin knockout mice were reported to exhibit impaired exocytosis in chromaffin cells and cortical neurons [31, 32]. Decreased levels of snapin protein in dysbindin-1 spontaneous mutant mice (Sandy mice) are also associated with cognitive deficits [33].
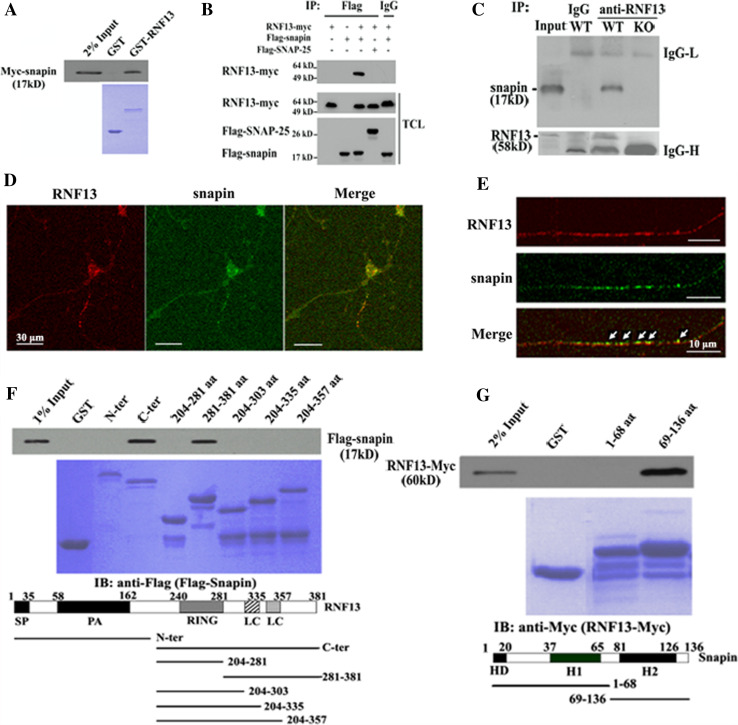
Subsequently, we evaluated the specificity of the interaction between RNF13 and snapin both in vitro and in vivo. As shown in Fig. 6a, the interaction between RNF13 and snapin was verified by a GST pull-down assay. In addition, FLAG-snapin was able to co-immunoprecipitate myc-RNF13 in COS-7 cells whereas SNAP-25 failed to bind RNF13 in the co-IP assay (Fig. 6b). In order to verify the interaction of RNF13 and snapin in vivo, we performed a co-IP assay with hippocampal lysates from both RNF13-null and WT mice. As expected, endogenous RNF13 and snapin only co-immunoprecipitated in WT mice (Fig. 6c), indicating that snapin is a binding partner for RNF13. Consistent with these findings, RNF13 partially colocalized with snapin in primary hippocampal neurons in culture, in both cell bodies and vesicles of neurites (Fig. 6d, e). In primary chromaffin cells of the peripheral nervous system, RNF13 also showed colocalization with snapin in vesicles (Fig. S4). Next, we purified different deletion mutants of GST-RNF13 and GST-snapin. We then performed an in vitro GST pull-down assay and observed that amino acids 281–381 of RNF13 were responsible for interaction with snapin (Fig. 6f). Further, the C-terminus including the second coiled-coil domain (a.a. 69–136) of snapin was involved in its interaction with RNF13 (Fig. 6g). Taken together, these findings indicate that RNF13 associates with snapin in vitro and in vivo.
Fig. 6.
RNF13 associates with SNARE complex-associated protein snapin. a GST pull-down experiment shows a direct interaction between RNF13 and snapin. Purified GST-RNF13 or GST was incubated with 293T cell lysate expressing Myc-snapin. The samples were washed and blotted with Myc antibody. b Immunoblots of immunoprecipitates from COS-7 cells co-transfected with myc-tagged RNF13 and FLAG-tagged snapin. As negative controls, COS-7 cells were transfected with FLAG-snapin or RNF13-myc, respectively. In addition, FLAG-SNAP-25 was also used as a negative control. TCL, total cell lysate. c Interaction of endogenous RNF13 and snapin in mouse hippocampal lysates. Total brain lysates of WT and RNF13-null mice were immunoprecipitated with RNF13 antibody, and immunoprecipitates were assayed by snapin antibody. Immunoprecipitates were generated using the non-specific IgG as negative control. d Immunostaining pattern of endogenous RNF13 (red) and snapin (green) in primary hippocampal neurons (scale bars, 30 μm). e Colocalization of endogenous RNF13 (red) and snapin (green) in the neurites of primary hippocampal neuron. Arrows indicate overlapping signals (scale bars, 10 μm). f, g Mapping of interaction domains responsible for the interaction of RNF13 and snapin. Different deletion mutants of RNF13 or snapin were fused to GST. FLAG-snapin or RNF13-myc was transfected into COS-7 cells and pulled down by different GST fusion proteins. SP signal peptide, PA protease-associated domain, RING RING finger domain, LC low complexity, HC hydrophobic domain H1(2) helical region 1(2)
RNF13 catalyzes the formation of K29-polyubiquitin chains for snapin
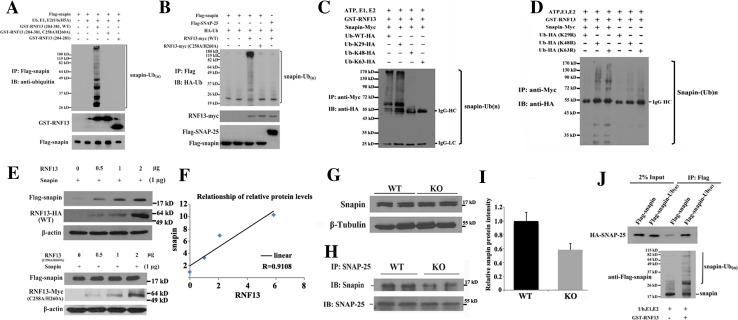
Next we examined whether the interaction of snapin and RNF13 led to ubiquitination of snapin in vitro. Snapin was incubated with recombinant WT or RING domain mutant (C258A/H260A) RNF13 at 30 °C for 1 h in an in vitro ubiquitination system (ubiquitin, E1, UbcH5a). Snapin was modified into a polyubiquitinated, high-molecular-weight species when incubated with WT RNF13, but not with RNF13 RING finger domain mutant and deletion mutant (without snapin binding region) (Fig. 7a). Moreover, FLAG-tagged snapin was co-expressed with WT or RING mutant Myc-tagged RNF13 in COS-7 cells. FLAG-snapin present in the cell lysate was precipitated by anti-FLAG beads, solubilized and analyzed via Western blot using anti-HA antibodies to detect ubiquitinated snapin. As shown in lane 3 in Fig. 7b, snapin was found to undergo polyubiquitination in the presence of WT rather than ubiquitin ligase-mutated (C258A/H260A) RNF13. SNAP-25 was used as a negative control, and no ubiquitinated SNAP-25 was observed in this assay.
Fig. 7.
RNF13 ubiquitinates snapin through ubiquitin lysine-29. a RNF13 ubiquitinates snapin in vitro. Recombinant wild-type, RING domain mutant and C-terminal deletion mutant (without snapin binding region) RNF13 incubated with FLAG-tagged snapin immunoprecipitate together with ubiquitin, E1 and E2 (UbcH5a). Samples were then immunoprecipitated under denatured condition with 2 μg FLAG antibodies and blotted with ubiquitin antibody (P4D1). b RNF13 ubiquitinates snapin in vivo. COS-7 cells were cotransfected with FLAG-snapin, HA-ubiquitin and RNF13-Myc. After immunoprecipitation with FLAG antibody, the immunoprecipitates were denatured with boiling water and re-immunoprecipitated by FLAG antibody. The second immunoprecipitates were blotted with HA antibody. SNAP-25 was used as a negative control. c Ubiquitin lysine-29 was responsible for the ubiquitination of snapin by RNF13. In vitro ubiquitination assay for snapin and RNF13 was carried out with FLAG-snapin immunoprecipitates, GST-RNF13, E1, UbcH5a and HA-tagged ubiquitins with different ubiqutin mutations (K29, K48, K63). The reaction mixture was detected with HA antibody on Western blot. d The same in vitro ubiquitination assay as (c) except that the ubiquitin mutants (K29R, K48R, K63R) carried only one lysine mutant site, respectively. e The expression level of ectopically expressed RNF13 and snapin in COS-7 cells. COS-7 cells were co-transfected with FLAG-snapin and different amounts of wild-type and RING domain mutant RNF13. Snapin protein level was analyzed by FLAG antibody. f Quantitative analysis shows a linear relationship of protein levels between ectopically expressed RNF13 and snapin. R = 0.9108 g Snapin expression level in hippocampi from WT and RNF13-null mice. h Immunoblot of snapin in immunoprecipitates from hippocampi of WT and RNF13-null mice using SNAP-25 antibody. When the amount of snapin associated with SNAP-25 was quantitated, decreased snapin protein levels were shown in RNF13-null mice compared with WT mice (i). j In vitro binding assay of snapin or ubiquitinated snapin with SNAP-25. HA-SNAP-25 was first immunoprecipitated with HA antibody in transfected COS-7 and eluted by HA peptide. The purified HA-SNAP-25 was then incubated with FLAG-snapin or ubiquitinated FLAG-snapin (in vitro ubiquitinated snapin by RNF13) at 4 °C for 4 h. Co-immunoprecipitated HA-SNAP-25 was detected by using HA antibody
To further analyze the type of isopeptide linkage catalyzed by RNF13, we simultaneously mutated ubiquitin at six of seven of the lysine residues, leaving only one lysine residue intact. As a result, only a single type of polyubiquitin chain would form for a given mutant. When immunoprecipitated FLAG-tagged snapin was incubated with three kinds of ubiquitin lysine mutants (K29, K48 and K63) in the in vitro ubiquitination assay, ubiquitination of snapin was predominantly detected when ubiquitin (Ub) K29 (lane 2) was included as compared to Ub K48 or K63 (lanes 3, 4) (Fig. 7c). By using additional ubiquitin mutants in which only one lysine was mutated to an arginine residue (K29R, K48R or K63R), thus blocking only one type of chain to be polymerized, we were able to consistently observe preferential ubiquitination of snapin through K29 chains (Fig. 7d). Therefore, these results demonstrate that snapin is a substrate of RNF13 for ubiquitination in a lysine-29 isopeptide form.
The non-canonical lysine-29 polyubiquitin chains have been verified to both promote protein substrate for degradation [34] and regulate protein activity [35]. To further reveal the fate of K29 polyubiquitination chain linked snapin by RNF13, snapin was co-transfected with different amounts of wild-type or RING domain mutant RNF13. We surprisingly found that more snapin protein was correlated with higher wild-type RNF13 expression, while snapin protein was unaffected by ubiquitin ligase activity-deficient RNF13 (Fig. 7e). The expression levels of the two proteins demonstrated a linear relationship (Fig. 7f). Then we sought to examine the steady-state level of snapin protein in hippocampal lysates of RNF13-null and WT mice. Intriguingly, we found that the snapin protein level did not differ significantly between RNF13-null and WT mice (Fig. 7g). These results suggest that ubiquitinated snapin mediated by RNF13 is not subject to proteasomal degradation. Since snapin is a direct binding partner of SNAP-25 in vivo, we next investigated whether the association of snapin and SNAP-25 was altered in mice lacking RNF13. To this end, hippocampal lysates from RNF13-null and WT mice were subjected to immunoprecipitation with an antibody against SNAP-25, and the precipitates were immunoblotted with anti-snapin antibody. As shown in Fig. 7h, i, snapin protein was decreased in precipitates from the hippocampal lysates of RNF13-null mice relative to WT mice, indicating that RNF13 may regulate the binding affinity of snapin to the SNARE complex. In addition, ubiquitinated snapin mediated by RNF13 enhances its ability to interact with SNAP-25 in vitro (Fig. 7j), which indicated that RNF13 directly and positively affected the association of snapin and SNARE proteins. Thus, we provide evidence both in vivo and in vitro to support the involvement of RNF13 in assisting snapin-SNARE complex interaction.
Discussion
In this study, we show that ablation of RNF13 in mice leads to spatial learning defects, alterations in synaptic structure and impaired SNARE complex assembly in the hippocampus. The hippocampus is the primary region of the brain, which controls learning behavior and memory [36, 37] and has been implicated in both spatial and contextual learning in rodents [38]. The “hidden platform”-based method we performed in the MWM test addresses abnormalities of spatial learning/memory and has proven useful to detect hippocampal-dependent cognitive deficits in mice [39]. In the learning phase of the MWM test, RNF13-null mice showed reduced efficiency in the execution of a standard spatial learning task, revealed by their decreased ability to reach the platform at days 2 and 3 (Fig. 3a). In the less stressful Y maze test, which reflects the working memory of mice, RNF13-null mice demonstrated reduced alteration in exploratory behavior (Fig. 3d). These defects are likely related to impairment of the hippocampus, as revealed by the fact that RNF13-null mice exhibited the decreased density of synaptic vesicles, which may possibly contribute to the reduced release of vesicles in synaptic boutons. Interestingly, we also observed a simultaneously increased size of the active zone in RNF13-null mice. The proposed model suggests that, by limiting the number of docked synaptic vesicles, the increased size of the active zone is potentially advantageous for synaptic transmission [40, 41]. We should theoretically predict that the increased size of the synapse causes an elevated synaptic activity. However, the reduced synaptic vesicle density may have a more profound effect on neurotransmitter release. On the basis of our results, it is conceivable that the two ultrastructural alterations may contribute in concert with each other to the behavior defects. The decrease in synaptic vesicle density in the presynaptic bouton also suggests the possibility that synaptic vesicles might not be transported and/or formed properly to synaptic terminals in RNF13-null mice. Further electrophysiological studies and exocytosis analysis are needed to underpin the molecular role of RNF13 in regulation of synaptic vesicle pools. In addition, as RNF13 is a ubiquitously expressed protein, depletion of RNF13 in areas of the brain other than the hippocampus (e.g., hypothalamus, amygdale and cortex) may also contribute to the behavior abnormality, as we also observed increased anxiety in RNF13-null mice in the elevated plus maze test.
We demonstrate that RNF13 binds to snapin in vitro and in vivo, indicating that the essential role of RNF13 for SNARE complex assembly may, at least in part, be achieved through snapin, as snapin has been shown to interact with SNAP-25 and potentiate the interaction between synaptotagmin I and SNAP-25 [29, 32]. This assumption is supported by our observation of the decreased interaction of SNAP-25 and synaptotagmin I in RNF13-null mice. In the synapses of snapin-null mice, reduced densities of total and active presynaptic vesicles and reduced numbers of docked vesicles at active zones were observed [31], which is also partly consistent with the reduced synaptic vesicle density in RNF13-null mice in this report. The impaired interaction of core SNARE complex proteins SNAP-25 and VAMP-2 is detected in RNF13-null mice, whereas the interaction is not affected in snapin-deficient mice. The possible explanations for the partial discrepancy between these two genetic targeting mice include (1) RNF13 regulates SNAP-25 and VAMP-2 interaction in a snapin-independent manner; (2) RNF13-null mice demonstrate some alterations that snapin knockout mice do not display, such as the alteration of the size of active zone (no change in snapin−/− mice). The active zone is the region where t-SNARE (SNAP-25, syntaxin) and v-SNARE (VAMP-2) mediate synaptic vesicle exocytosis. The increased size of the active zone in RNF13-null mice may lead to the impaired binding of SNAP-25 and VAMP-2 at the molecular level. Therefore, we cannot currently rule out the possibility that RNF13 targets other proteins in its regulation of SNARE complex assembly. In addition, we found an attenuated binding of snapin to SNAP-25 in RNF13-null mice (Fig. 7h). Consistently, in vitro assay suggested the increased binding of ubiquitinated snapin to SNAP-25 (Fig. 7j). It is possible that the interaction of RNF13 and snapin in vivo may lead to the increased binding affinity of snapin to SNAP-25, as we failed to find the difference of snapin protein level (Fig. 7g) and snapin ubiquitination in the hippocampus between RNF13-null mice and WT mice. Post-translational modifications of snapin, both C66-mediated dimerization and S50 phosphorylation by PKA, have been reported to significantly enhance its interaction with SNAP-25 [30, 31, 42]. Here, we show that RNF13 catalyzed snapin K29 polyubiquitination is a novel post-translational modification of the regulation of snapin association with SNARE complex proteins.
Recent studies from Erickson’s group indicated PKC agonist PMA treatment caused RNF13 protein to translocate on the inner nuclear membrane through recycling endosomes [21]. INM-anchored RNF13 would have its RING finger domain facing the nucleoplasm, which suggests RNF13 potentially ubiquitinates substrates inside the nucleus. Therefore, other RNF13 unknown substrates beyond snapin may also contribute to the deficits of RNF13-null mice. On the other hand, RNF13 belongs to PA-TM-RING protein family and has eight paralogs [17]. In RNF13-null mice, there is a possibility that some deficiency could be compensated by its one or more paralogs, which might rescue some deficits in synaptic formation and behavior in mice lacking RNF13.
It is notable that, in addition to regulating SNARE proteins, snapin binds to a plethora of cellular proteins, such as receptors and channel proteins. Further studies are needed to examine the possible role of RNF13 in regulating other snapin-binding partners. In addition, snapin has also been demonstrated as one of eight known subunits of the biogenesis of lysosome-related organelle complex (BLOC)-1 [43]. BLOC-1 is essential for normal biogenesis of specialized organelles, such as melanosomes and platelet-dense granules [44]. It is possible that RNF13 has potential function to regulate BLOC-1 under certain physiological and pathological conditions.
By using different ubiquitin mutants, our study confirmed that lysine-29 of ubiquitin provides the linkage site for snapin by RNF13 in vitro. The isopeptide linkage of a polyubiquitin chain is an important determinant for a certain fate of protein substrate. All seven lysines of ubiquitin have been verified to participate in the ubiquitin modification of proteins [45]. It is generally considered that K48-linked polyubiquitin chains target protein substrates for proteasomal degradation, while proteins modified by non-canical polyubiquitin chains are involved in various cellular functions. K63-linked chains regulate protein trafficking, signal transduction and transcriptional regulation [46]. K11 chains have been reported to modulate mitotic protein degradation by the ubiquitin ligase anaphase-promoting complex (APC/C) and are involved in endoplasmic reticulum-associated degradation (ERAD) [45, 47]. Linear ubiquitin chain-assembly complex (LUBAC) catalyzes the formation of head-to-tail polyubiquitin chains to activate the nuclear factor-κB (NF-κB) pathway [48]. To date, little is known about the consequences of K29 ubiquitin chains on protein function or stability. It has been reported that HECT domain-containing ubiquitin ligase AIP4 directs polyubiquitination and degradation through a linkage involving residue K29, both on itself and on a heterologous substrate, DTX [34]. AMPK-related kinases can form atypical Lys29/Lys33-linked polyubiquitin chains through an unknown ubiquitin ligase that regulates its kinase activities [35]. Here, we show that RNF13 ubiquitinates snapin by forming K29-linked chains in vitro, which seems not to mediate snapin degradation, as we observed the comparable snapin protein levels in RNF13-null mice and WT mice and increased snapin levels when co-transfected with RNF13 in cells (Fig. 7e). In addition, we applied proteasome inhibitors, MG-132 and AdaAhx(3)L(3)VS, to detect snapin protein levels in cultured primary hippocampal neurons and did not observe any difference. Interestingly, another PA-TM-RING family member GRAIL (gene related to anergy in lymphocytes) promotes stabilization of its substrate Rho guanine dissociation inhibitor via K63-linked ubiquitin chains [49]. Whether RNF13-mediated snapin K29 chains occur in vivo and its role for snapin remain to be elucidated.
In summary, we have highlighted the potential importance of E3 ubiquitin ligase RNF13 for its role in learning/memory and SNARE complex assembly. We provide experimental evidence for aiming the elucidation of RNF13 molecular mechanisms in these events. Other key targets of RNF13 involved in the regulation of SNARE complex assembly remain to be identified. Further studies may provide more evidence for RNF13 as a potential drug target for neuronal disease.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Dr. Wei Li, Dr. Jiajia Liu, Yan Li and Dr. Yaqin Feng (Institute of Genetics & Developmental Biology, Chinese Academy of Sciences) for providing technical assistance for this study. We also thank Prof. Jian Zhang (Institute of Genetics & Developmental Biology, Chinese Academy of Sciences) for critical reading of this manuscript and comments on manuscript preparation. This work was supported by the following grants: National Basic Research Program of China (2005CB522405, 2005CB522500, 2007CB946903, 2009CB825403, 2006BAI23B00, and 2006CB943500); National Natural Science Foundation of China (30721063 and 30825024); and Chinese National Programs for High Technology Research and Development (2006AA10A121, 2007AA02Z109).
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Qiang Zhang, Yanfeng Li and Lei Zhang contributed equally to this work.
Contributor Information
Xiang Gao, Phone: +86-25-58641598, FAX: +86-25-58641500, Email: gaoxiang@nju.edu.cn.
Dahai Zhu, Phone: +86-10-65296949, FAX: +86-10-6510-5083, Email: dhzhu@pumc.edu.cn.
References
- 1.Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 2.Rizo J, Rosenmund C. Synaptic vesicle fusion. Nat Struct Mol Biol. 2008;15:665–674. doi: 10.1038/nsmb.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sudhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pang ZP, Sudhof TC. Cell biology of Ca2 + triggered exocytosis. Curr Opin Cell Biol. 2010;22:496–505. doi: 10.1016/j.ceb.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaskolski F, Henley JM. Synaptic receptor trafficking: the lateral point of view. Neuroscience. 2009;158:19–24. doi: 10.1016/j.neuroscience.2008.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cajigas IJ, Will T, Schuman EM. Protein homeostasis and synaptic plasticity. EMBO J. 2010;29:2746–2752. doi: 10.1038/emboj.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haas KF, Broadie K. Roles of ubiquitination at the synapse. Biochim Biophys Acta. 2008;1779:495–506. doi: 10.1016/j.bbagrm.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mabb AM, Ehlers MD. Ubiquitination in postsynaptic function and plasticity. Annu Rev Cell Dev Biol. 2010;26:179–210. doi: 10.1146/annurev-cellbio-100109-104129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segref A, Hoppe T. Think locally: control of ubiquitin-dependent protein degradation in neurons. EMBO Rep. 2009;10:44–50. doi: 10.1038/embor.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi JJ, Ehlers MD. Ubiquitin and protein turnover in synapse function. Neuron. 2005;47:629–632. doi: 10.1016/j.neuron.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 11.Cremona O, Collesi C, Raiteri E. Protein ubiquitylation and synaptic function. Ann N Y Acad Sci. 2003;998:33–40. doi: 10.1196/annals.1254.006. [DOI] [PubMed] [Google Scholar]
- 12.Yao I, Takagi H, Ageta H, Kahyo T, Sato S, Hatanaka K, Fukuda Y, Chiba T, Morone N, Yuasa S, Inokuchi K, Ohtsuka T, Macgregor GR, Tanaka K, Setou M. SCRAPPER-dependent ubiquitination of active zone protein RIM1 regulates synaptic vesicle release. Cell. 2007;130:943–957. doi: 10.1016/j.cell.2007.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin LS, Vavalle JP, Li L. Staring, a novel E3 ubiquitin-protein ligase that targets syntaxin 1 for degradation. J Biol Chem. 2002;277:35071–35079. doi: 10.1074/jbc.M203300200. [DOI] [PubMed] [Google Scholar]
- 14.Joch M, Ase AR, Chen CX, MacDonald PA, Kontogiannea M, Corera AT, Brice A, Séguéla P, Fon EA. Parkin-mediated monoubiquitination of the PDZ protein PICK1 regulates the activity of acid-sensing ion channels. Mol Biol Cell. 2007;18:3105–3118. doi: 10.1091/mbc.E05-11-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Z, Je HS, Young P, Gross J, Lu B, Feng G. Regulation of synaptic growth and maturation by a synapse-associated E3 ubiquitin ligase at the neuromuscular junction. J Cell Biol. 2007;177:1077–1089. doi: 10.1083/jcb.200610060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bocock JP, Carmicle S, Sircar M, Erickson AH. Trafficking and proteolytic processing of RNF13, a model PA-TM-RING family endosomal membrane ubiquitin ligase. FEBS J. 2011;278:69–77. doi: 10.1111/j.1742-4658.2010.07924.x. [DOI] [PubMed] [Google Scholar]
- 17.Jin X, Cheng H, Chen J, Zhu D. RNF13: an emerging RING finger ubiquitin ligase important in cell proliferation. FEBS J. 2011;278:78–84. doi: 10.1111/j.1742-4658.2010.07925.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q, Meng Y, Zhang L, Chen J, Zhu D. RNF13: a novel RING-type ubiquitin ligase over-expressed in pancreatic cancer. Cell Res. 2009;19:348–357. doi: 10.1038/cr.2008.285. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, Wang K, Zhang Y, Meng J, Yu F, Chen Y, Zhu D. The myostatin-induced E3 ubiquitin ligase RNF13 negatively regulates the proliferation of chicken myoblasts. FEBS J. 2010;277:466–476. doi: 10.1111/j.1742-4658.2009.07498.x. [DOI] [PubMed] [Google Scholar]
- 20.Bocock JP, Carmicle S, Chhotani S, Ruffolo MR, Chu H, Erickson AH. The PA-TM-RING protein RING finger protein 13 is an endosomal integral membrane E3 ubiquitin ligase whose RING finger domain is released to the cytoplasm by proteolysis. FEBS J. 2009;276:1860–1877. doi: 10.1111/j.1742-4658.2009.06913.x. [DOI] [PubMed] [Google Scholar]
- 21.Bocock JP, Carmicle S, Madamba E, Erickson AH. Nuclear targeting of an endosomal E3 ubiquitin ligase. Traffic. 2010;11:756–766. doi: 10.1111/j.1600-0854.2010.01060.x. [DOI] [PubMed] [Google Scholar]
- 22.Tranque P, Crossin KL, Cirelli C, Edelman GM, Mauro VP. Identification and characterization of a RING zinc finger gene (C-RZF) expressed in chicken embryo cells. Proc Natl Acad Sci U S A. 1996;93:3105–3109. doi: 10.1073/pnas.93.7.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito S, Honma K, Kita-Matsuo H, Ochiya T, Kato K. Gene expression profiling of cerebellar development with high-throughput functional analysis. Physiol Genomics. 2005;22:8–13. doi: 10.1152/physiolgenomics.00142.2004. [DOI] [PubMed] [Google Scholar]
- 24.Schneider Gasser EM, Straub CJ, Panzanelli P, Weinmann O, Sassoe-Pognetto M, Fritschy JM. Immunofluorescence in brain sections: simultaneous detection of presynaptic and postsynaptic proteins in identified neurons. Nat Protoc. 2006;1:1887–1897. doi: 10.1038/nprot.2006.265. [DOI] [PubMed] [Google Scholar]
- 25.Yao I, Hata Y, Hirao K, Deguchi M, Ide N, Takeuchi M, Takai Y. Synamon, a novel neuronal protein interacting with synapse-associated protein 90/postsynaptic density-95-associated protein. J Biol Chem. 1999;274:27463–27466. doi: 10.1074/jbc.274.39.27463. [DOI] [PubMed] [Google Scholar]
- 26.Huang HP, Wang SR, Yao W, Zhang C, Zhou Y, Chen XW, Zhang B, Xiong W, Wang LY, Zheng LH, Landry M, Hokfelt T, Xu Z-QD, Zhou Z. Long latency of evoked quantal transmitter release from somata of locus coeruleus neurons in rat pontine slices. Proc Natl Acad Sci USA. 2007;104:1401–1406. doi: 10.1073/pnas.0608897104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pozzo-Miller LD, Gottschalk W, Zhang L, McDermott K, Du J, Gopalakrishnan R, Oho C, Sheng ZH, Lu B. Impairments in high-frequency transmission, synaptic vesicle docking, and synaptic protein distribution in the hippocampus of BDNF knockout mice. J Neurosci. 1999;19:4972–4983. doi: 10.1523/JNEUROSCI.19-12-04972.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki WA. Making new memories: the role of the hippocampus in new associative learning. Ann N Y Acad Sci. 2007;1097:1–11. doi: 10.1196/annals.1379.007. [DOI] [PubMed] [Google Scholar]
- 29.Ilardi JM, Mochida S, Sheng ZH. Snapin: a SNARE-associated protein implicated in synaptic transmission. Nat Neurosci. 1999;2:119–124. doi: 10.1038/5673. [DOI] [PubMed] [Google Scholar]
- 30.Chheda MG, Ashery U, Thakur P, Rettig J, Sheng ZH. Phosphorylation of Snapin by PKA modulates its interaction with the SNARE complex. Nat Cell Biol. 2001;3:331–338. doi: 10.1038/35070000. [DOI] [PubMed] [Google Scholar]
- 31.Pan PY, Tian JH, Sheng ZH. Snapin facilitates the synchronization of synaptic vesicle fusion. Neuron. 2009;61:412–424. doi: 10.1016/j.neuron.2008.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian JH, Wu ZX, Unzicker M, Lu L, Cai Q, Li C, Schirra C, Matti U, Stevens D, Deng C, Rettig J, Sheng ZH. The role of Snapin in neurosecretion: snapin knock-out mice exhibit impaired calcium-dependent exocytosis of large dense-core vesicles in chromaffin cells. J Neurosci. 2005;25:10546–10555. doi: 10.1523/JNEUROSCI.3275-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng YQ, Zhou ZY, He X, Wang H, Guo XL, Hao CJ, Guo Y, Zhen XC, Li W. Dysbindin deficiency in sandy mice causes reduction of Snapin and displays behaviors related to schizophrenia. Schizophr Res. 2008;106:218–228. doi: 10.1016/j.schres.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 34.Chastagner P, Israel A, Brou C. Itch/AIP4 mediates Deltex degradation through the formation of K29-linked polyubiquitin chains. EMBO Rep. 2006;7:1147–1153. doi: 10.1038/sj.embor.7400822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al-Hakim AK, Zagorska A, Chapman L, Deak M, Peggie M, Alessi DR. Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochem J. 2008;411:249–260. doi: 10.1042/BJ20080067. [DOI] [PubMed] [Google Scholar]
- 36.Dierssen M, Ramakers GJ. Dendritic pathology in mental retardation: from molecular genetics to neurobiology. Genes Brain Behav. 2006;2:48–60. doi: 10.1111/j.1601-183X.2006.00224.x. [DOI] [PubMed] [Google Scholar]
- 37.Kingsbury MA, Yung YC, Peterson SE, Westra JW, Chun J. Aneuploidy in the normal and diseased brain. Cell Mol Life Sci. 2006;63:2626–2641. doi: 10.1007/s00018-006-6169-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galdzicki Z, Siarey R, Pearce R, Stoll J, Rapoport SI. On the cause of mental retardation in Down syndrome: extrapolation from full and segmental trisomy 16 mouse models. Brain Res Brain Res Rev. 2001;35:115–145. doi: 10.1016/S0926-6410(00)00074-4. [DOI] [PubMed] [Google Scholar]
- 39.Stasko MR, Costa AC. Experimental parameters affecting the Morris water maze performance of a mouse model of Down syndrome. Behav Brain Res. 2004;154:1–17. doi: 10.1016/j.bbr.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Matz J, Gilyan A, Kolar A, McCarvill T, Krueger SR. Rapid structural alterations of the active zone lead to sustained changes in neurotransmitter release. Proc Natl Acad Sci USA. 2010;107:8836–8841. doi: 10.1073/pnas.0906087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weyhersmüller A, Hallermann S, Wagner N, Eilers J. Rapid active zone remodeling during synaptic plasticity. J Neurosci. 2011;31:6041–6052. doi: 10.1523/JNEUROSCI.6698-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Navarro A, Encinar JA, López-Méndez B, Aguado-Llera D, Prieto J, Gómez J, Martínez-Cruz LA, Millet O, González-Ros JM, Fernández-Ballester G, Neira JL, Ferrer-Montiel A. Mutation of Ser-50 and Cys-66 in snapin modulates protein structure and stability. Biochemistry. 2012;51:3470–3484. doi: 10.1021/bi201574t. [DOI] [PubMed] [Google Scholar]
- 43.Starcevic M, Dell’Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J Biol Chem. 2004;279:28393–28401. doi: 10.1074/jbc.M402513200. [DOI] [PubMed] [Google Scholar]
- 44.Dell’Angelica EC. The building BLOC(k)s of lysosomes and related organelles. Curr Opin Cell Biol. 2004;16:458–464. doi: 10.1016/j.ceb.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 45.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, Phu L, Kirkpatrick DS, Hymowitz SG, Rape M, Kelley RF, Dixit VM. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol Cell. 2010;39:477–484. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 48.Iwai K, Tokunaga F. Linear polyubiquitination: a new regulator of NF-kappaB activation. EMBO Rep. 2009;10:706–713. doi: 10.1038/embor.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su L, Lineberry N, Huh Y, Soares L, Fathman CG. A novel E3 ubiquitin ligase substrate screen identifies Rho guanine dissociation inhibitor as a substrate of gene related to anergy in lymphocytes. J Immunol. 2006;177:7559–7566. doi: 10.4049/jimmunol.177.11.7559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.