Abstract
B chromosomes (Bs) are dispensable components of the genome exhibiting non-Mendelian inheritance and have been widely reported on over several thousand eukaryotes, but still remain an evolutionary mystery ever since their first discovery over a century ago [1]. Recent advances in genome analysis have significantly improved our knowledge on the origin and composition of Bs in the last few years. In contrast to the prevalent view that Bs do not harbor genes, recent analysis revealed that Bs of sequenced species are rich in gene-derived sequences. We summarize the latest findings on supernumerary chromosomes with a special focus on the origin, DNA composition, and the non-Mendelian accumulation mechanism of Bs.
Keywords: B chromosome, Genome evolution, Drive, Selfish chromosome, Next generation sequencing
Introduction
The vast majority of a eukaryotic genome do not code for proteins and are composed of so-called ‘non-coding’ DNA. Why evolution would maintain large amounts of ‘useless’ DNA remained a mystery for many years. However, results from the ENCODE project show that most of the presumedly useless DNA is functional and involved in complex regulatory networks. In addition, widespread transcription from non-coding DNA potentially acts as a reservoir for the creation of new functional molecules [2]. It is becoming clear that the genome is much more complex, and gene activity is influenced by multiple stretches of regulatory DNA located both near and far from the gene itself, as well as by RNA molecules that are not translated into proteins: so-called non-coding RNA. Many studies have revealed that much of the so-called junk DNA is active in regulation, thus questioning our gene-centric view of the genome.
B chromosomes (Bs) represent an extraordinary example of extra, apparently inert, DNA. Bs are chromosomes that are additional to the normal set of chromosomes (called A chromosomes or As) (Fig. 1). Bs exhibit non-Mendelian inheritance and have been widely reported in several thousands of animal, plant, and fungi species, but they have remained evolutionary mysteries ever since their first discovery over a century ago. Bs are found in some but not all individuals within a population and can vary in number between (and within) individuals (e.g., Picea glauca, 2n = 24 As + 0–6 Bs; Vulpes vulpes, 2n = 34 As + 0–8 Bs; Rattus rattus, 2n = 42 + 0–5 Bs). In many species, different morphological types of Bs exist within a single species. In some species, they can even exceed the number of As (e.g. Zea mays, 2n = 20 As + 0–34 Bs). Some of the so-called small human supernumerary marker chromosomes, which are found in addition to the normal chromosome complement in some patients with a weak medical phenotype, show similarities to B chromosomes [3]. Initially, Bs were considered non-functional and without any essential genes, as they are dispensable for normal growth. As a result, Bs follow their own species-specific evolutionary pathways. Because most Bs do not confer any advantages on the organisms that harbor them, they may be thought of as parasitic, selfish elements that persist in populations by making use of the cellular machinery required for the inheritance and maintenance of A chromosomes. When present in low numbers, Bs generally have little or no impact on the hosts. However, increased numbers of Bs cause phenotypic differences and may reduce fertility (reviewed in [4–8]).
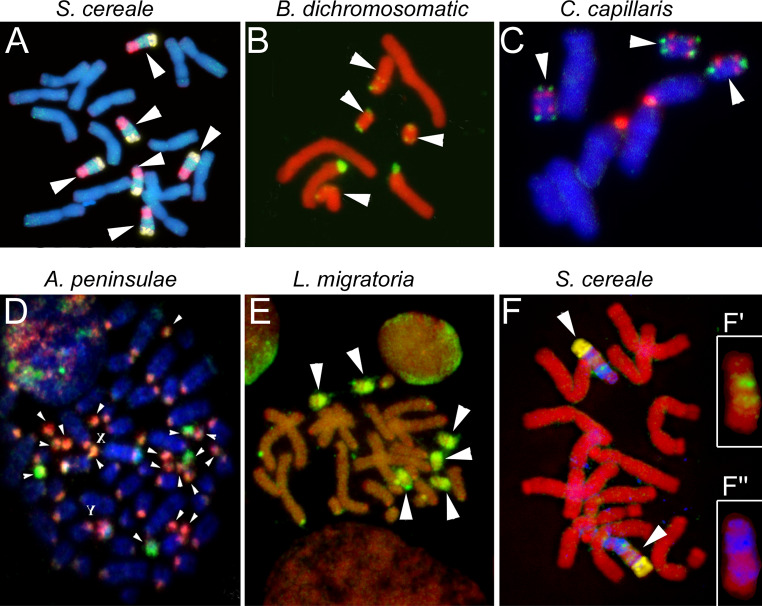
Fig. 1.
Examples of plant and animal species possessing B chromosomes. a The Bs in rye (Secale cereale). Arrowed Bs showing cross-hybridization with the B-specific repeat ScCl11 (red), Sc36c82 (green), and D1100 (yellow). (Picture provided by A. Marques, Gatersleben, Germany.) b The micro and standard Bs (arrowed) in Brachycome dichromosomatica. Metaphase chromosomes showing cross-hybridization with 45S rDNA (green). c The Bs (arrowed) in Crepis capillaris. Metaphase chromosomes showing cross-hybridization with 45S rDNA (red) and Arabidopsis-type telomere probes (green) probe obtained from microdissected Bs. d The Bs in Korean field mouse (Apodemus peninsulae): two-color FISH of microdissected DNA probes derived from centromeric C-positive region of autosome (red) and from C-negative region of the arm of one of the Bs (green) with metaphase chromosomes of specimens containing 18 Bs. Arrows indicate the dot-like Bs; sex chromosomes are marked with X and Y. (Picture provided by N.B. Rubtsov, Novosibirsk, Russia.) e The Bs in migratory locust (Locusta migratoria). Mitotic metaphase from a female embryo showing cross-hybridization with a DNA probe obtained from microdissected Bs. Note the intense painting of the B chromosomes (arrowed) and the pericentromeric regions of several of the 24 A chromosomes. (Picture provided by J.P.M. Camacho, Granada, Spain.) f The Bs in rye (Secale cereale). Arrowed Bs showing cross-hybridization with mitochondrial DNA (blue), plastid DNA (green) and the B-specific D1100 repeat (yellow). Insets show Bs after hybridization with (f′) plastid DNA (green) or (f″) mitochondrial DNA (blue). Note that only 12 of 14 A chromosomes are shown
In most species which carry Bs, the mitotic transmission of Bs during growth and development is normal and hence all cells carry the same number of Bs within the individual. However, there are some exceptions in which the Bs show mitotic instability and therefore they are present in variable numbers, sometimes characterizing specific tissues and/or organs. For example, in the grasses Aegilops speltoides and Ae. mutica, Bs exist in aerial organs but not in roots [9, 10]. In grasshoppers, variation in Bs has been observed to occur specifically among follicles of the testis [11]. The mechanism underlying this latter variation was shown to be nondisjunction of the B chromosome sister chromatids during anaphase of mitosis leading to an absence of Bs in one daughter cell and accumulation in the other.
Frequent B polymorphisms are expected among populations due to the nonessential nature of this type of chromosome. Indeed, besides the characteristic B numerical polymorphisms, several cases of B structural polymorphisms have been reported in plants, e.g., Brachycome dichromosomatica [12], Scilla autumnalis [13], Allium schoenoprasum [14], and Ae. speltoides [15], and in animals including the grasshopper Eyprepocnemis plorans [16]. However, in other species, including geographically distinct populations of cultivated and weedy rye, Bs have maintained a similar molecular and cytological structure at the level of subspecies. The conserved chromosome structure suggests that, after a period of rapid B development, the process of chromosome modification has slowed during more recent evolution. On the other hand, rye is a young species and the separation of the different genotypes analyzed evolved rather recently in evolutionary terms [17].
Despite the wide taxonomic distribution of Bs, our knowledge of the origin, composition, regulation, and accumulation mechanisms of B chromosomes was limited until very recently. Triggered by technological advances in sequencing and genome analysis [18, 19], our knowledge of the biology of B chromosomes has improved significantly in the last few years. The aim of this review is to summarize the latest findings on supernumerary chromosomes with a special focus on the Bs of plants.
How does a B chromosome evolve?
As selfish entities, and because they do not participate in meiotic recombination with the As, Bs take a distinct evolutionary path, and their sequence composition may differ from that of the As. Because Bs are under little or no selection pressure, mobile elements and other DNA species may insert, spread, and amplify, as in Bs of maize [20], Brachycome dichromosomatica [21], and Plantago lagopus [22].
One of the best models for a parasitic chromosome is the B of rye [23]. Bs in this species are found in both cultivated rye (Secale cereale ssp. cereale) and in weedy rye (S. cereale ssp. segetale) from diverse countries [24]. Based on similar morphology and meiotic pairing of Bs derived from weedy and cultivated rye lines of different origins in F1 hybrids, it has been concluded that the rye B has a monophyletic origin [17, 25]. Recent comparative sequence analysis of the A and B chromosomes of rye challenged our view on the biology of supernumerary chromosomes [26]. The most unexpected observation in this study is that rye Bs are rich in gene fragments that are derived from multiple A chromosome fragments. A multi-chromosomal origin of B-chromosome sequences is further supported by the many short sequences that are similar to other regions of the rye A chromosomes [26]. Similarly, analysis of evolutionarily conserved chromosome segments in wild canid species identified several regions of domestic dog sequences that share sequence similarity with canid B chromosomes [27]. But, how did these diverse A chromosome relicts enter the B chromosome? Did gene trafficking occur during double-strand break repair or via hitchhiking of genomic fragments with transposable elements, as demonstrated for non-collinear genes of Triticeae [28]? Or, as postulated by Becker et al. [27], could Bs represent an evolutionary mechanism to sequester additional copies of genes that are generated at the chromosome breakpoints associated with speciation? In addition to this basic A-derived architecture, rye Bs were found to have accumulated large amounts of B-specific repeats and insertions of cytoplasmic organellar DNA. It seems that the B acts like a “genomic sponge” which collects and maintains sequences of diverse origin [26].
Analysis of the composition and distribution of rye B-located high-copy sequences revealed that Bs contain a similar proportion of repeats to A chromosomes, but differ substantially in repeat composition [29]. The most abundant mobile elements (Gypsy, Copia) in the genome of rye are similarly distributed along As and Bs, while the ancient retroelement Sabrina [30], is less abundant on Bs than on As. In contrast, the active element Revolver [31], as well as the predicted Copia retrotransposon Sc36c82, are disproportionately abundant on the B. A B-specific accumulation of Gypsy retrotransposons or other repeated sequences has also been reported in the fish Alburnus alburnus [32] and the fungus Nectria haematococca [33]. The accumulation of active elements might have its cause in relaxed selection pressure on Bs, where the integration of a mobile element does not interrupt essential gene functions. Reduced crossing-over might facilitate retroelement accumulation as proposed for Y chromosomes [34]. A less likely option could be an advantageous transposition to the B, although targeted transposition has been shown for yeast [35] and Arabidopsis lyrata [36]. To explain depletion of an element, a possible scenario might involve transposition of elements predominantly in plants without B. The element could still transpose in 0B individuals, thus allowing accumulation on As exclusively. Such a behavior has been proposed for the Y chromosome-depleted Ogre-element in Silene latifolia [37].
A model that explains depletion and accumulation of B-located retrotransposons on the rye B in particular is based on different transposition activity of these elements [29]. In the Triticeae ancestor, Sabrina transposed and spread over the entire genome. After inactivation of Sabrina [30] before or during speciation of rye, the B was formed from the A chromosomes with Sabrina still present. The newly evolving elements such as Revolver then became active and transposed throughout the rye genome. The dispensable nature of the B and the lack of selection pressure allowed for stronger accumulation of Revolver on the B, diluting even further the remnants of inactive elements which can no longer increase copy numbers.
In maize [20, 38, 39] and rye [29, 40], the majority of B-enriched tandem repeats map to regions important for the accumulation mechanisms of the Bs. Langdon et al. [41] suggested that the rye B-enriched tandem repeat sequences E3900 and D1100 evolved via amplification of ancestral A-located sequences within the dynamic nondisjunction control region on rye B. The B-enriched repeats could have amplified via unequal crossing over [42]. The presence of B-enriched sequences within evolutionarily diverged species indicates that B-specific amplification occurred after separation from the standard chromosome complement.
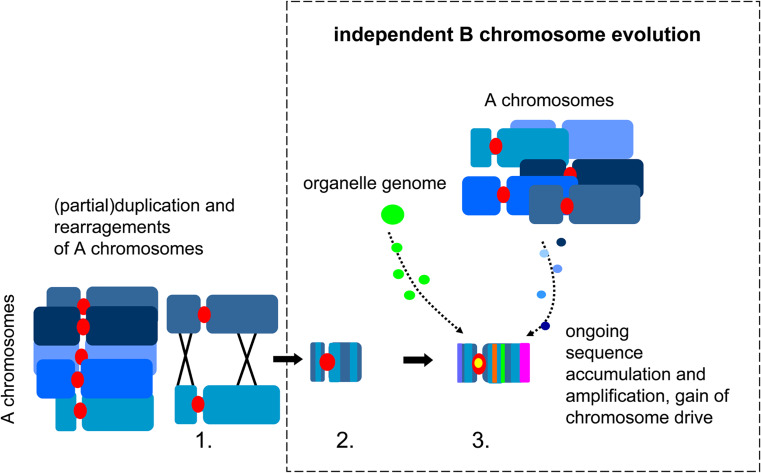
We propose a multi-step model for the origin of B chromosomes (Fig. 2). Initially, a proto-B chromosome was formed by segmental or whole-genome duplication, followed by reductive chromosome translocations, unbalanced segregation of a small translocation chromosome, and subsequent sequence insertions. Recombination with donor A chromosomes became restricted, probably due to multiple rearrangements and illegitimate recombination involving different A chromosomes, which precluded extended pairing with the formerly homologous A regions. This restriction of recombination may be considered as the starting point for the independent evolution of B chromosomes. The presence of fast-evolving repetitive sequences, along with reduced selective pressure on gene integrity, would predispose a nascent B to accumulate the further rapid structural modifications required to establish a drive mechanism. Because any increased gene dosage may affect gene expression, the expression of paralogues on B chromosomes might be reprogrammed (potentially through epigenetic mechanisms) early during the evolution of the Bs. Thus, proto-B genes might first be suppressed and then degenerate due to mutations and the insertion of sequences derived from other A-chromosomal regions and cytoplasmic organellar genomes. Exceptions to this scenario could include those sequences that provide meiotic drive and an advantage for the maintenance of B chromosomes in populations. This hypothesis suggests that B chromosomes, due to their dispensable nature, provide a kind of safe harbor for genes and sequences without immediate selective benefit. In our model, we expect B chromosomes to be found primarily in taxa with elevated levels of chromosomal rearrangement and phylogenetic groups with unstable chromosome numbers.
Fig. 2.
Model of the evolution of a B chromosome. 1 Translocation chromosome derived from duplicated A chromosome fragments results in 2 a decay of meiotic A–B pairing and the formation of a proto-B. 3 The accumulation of organellar and A chromosome-derived DNA fragments, amplification of B-specific repeats, erosion and inactivation of A-derived genes (Muller’s ratchet) and the gain of chromosome drive forms a B chromosome
Considering the similar age of the genus Secale (1.7 Mya) and the age of its B chromosomes (1.1–1.3 Mya), it is tempting to speculate that B chromosomes originated as a by-product of chromosome rearrangement events. Especially considering that the rye genome, in contrast to the otherwise pronounced genome colinearity in the Triticeae, underwent a series of rearrangements since its split from the wheat and barley lineages [43]. An analogous association between chromosome rearrangements and the formation of Bs has been demonstrated in Drosophila. Analysis of B-linked sequences suggest that D. albomicans Bs may have originated as a by-product from centromeric/telomeric fragments created by a fusion of the ancient third autosome and the ancestral sex chromosome [44].
B-located organelle DNA
B chromosomes of rye have accumulated significantly greater amounts of chloroplast- and mitochondrion-derived sequences (NUPTs and NUMTs, respectively) than the A chromosomes [26] (Fig. 1f). All parts of the chloroplast and mitochondrial genomes are found on the Bs, indicating that all sequences are transferable. The higher amount of organelle-derived DNA inserts in B than in A chromosomes and an increased mutation frequency of B-located organellar DNA suggests a reduced selection against the insertion of organellar DNA in supernumerary chromosomes. What mechanism could account for the accumulation of organellar DNA on the B chromosomes? The first possibility is that insertion into B DNA has fewer deleterious genetic consequences than their counterparts on As. Simply expressed, this may reflect the generally inert B chromosome which may tolerate essentially uncontrolled DNA insertions of all sorts. In contrast, insertions into A chromosomal DNA may disrupt gene expression with lethal consequences, particularly when they become homozygous. Plastid DNA fragments are clearly very numerous in some tissues such as the developing pollen gametophyte [45] or after stress [46], so uncontrolled insertion would be expected to result in the accumulation of NUPTs and, by analogy, NUMTs. Alternatively, the mechanisms that prevent nuclear genome expansion may be impaired on the Bs, even though they are normally amongst the smaller chromosomes in karyotypes. Transfer of organellar DNA to the nucleus is very frequent [47], but much of the ‘promiscuous’ DNA is also rapidly lost again within one generation by a partially counterbalancing, but little understood, removal process [48]. This makes about 50 % of de novo NUPTs so unstable that partial or total deletion may be observed by genetic and molecular analyses. If this expulsion mechanism is impaired in B chromosomes, the high turnover rates together with lower degradation that prevent such sequences from accumulating on the A chromosomes would allow for sequence decay. Thus, the dynamic equilibrium between frequent integration and rapid elimination of organellar DNA could be impaired for B chromosomes.
There is no doubt that endosymbiotic evolution, initiated by NUPT (nuclear integrants of plastid DNA) and NUMT (nuclear integrants of mitochondrial DNA) formation, has been a major driver of genetic complexity, with past and present mitochondrial and chloroplastic sequences contributing a large proportion of nuclear genes [47]. Therefore, the presence of a disproportionately large amount of extant organellar DNA on the rye B chromosome makes it tempting to suggest a long-term evolutionary role. Assuming that the sequences of the majority of NORGs (nuclear integrants of cytoplasmic organellar DNA) are not under any selection pressure, they are free to undergo sequence decay without constraint, providing the potential to produce novel beneficial or deleterious genes or, most frequently, genomic garbage. All that is then necessary to complete the birth of a novel functional gene is transfer of B chromosome DNA to the A chromosomes. While such transfers have not been characterized, there is ample evidence of DNA transfer in the other direction, from A to B chromosomes [4, 26], and it would be surprising if B DNA did not rarely or even frequently transfer to the As. This mechanism could apply to all the sequences on the B, but the far-reaching and well-characterized effects of endosymbiotic evolution certainly provide a challenging area for future research. It is difficult to address such transfers experimentally with current techniques, but an unambiguous demonstration would suggest a strong raison d’etre for the very common occurrence of Bs in natural populations. Future analyses of other B-bearing species will be needed to address the question whether organelle-to-nucleus DNA transfer is an important mechanism that drives the evolution of B chromosomes.
Gene content of B chromosomes
Although Bs are not essential, some phenotypic effects have been reported to be associated with their presence. These effects are usually cumulative, depending upon the number and not the presence or absence of Bs (reviewed in [8, 49–51]. For instance, under drought stress conditions, seeds carrying Bs have an advantage concerning germination over 0B seeds in Allium schoenoprasum [52]. In cichlid fishes [53] and in the frog Leiopelma hochstetteri, Bs play a role in sex determination, and, in the fungus Nectria haematococca, Bs account for antibiotic resistance and pathogenicity [33]. In addition, Bs were reported to be associated with their achene color in Haplopappus gracilis [54], meiotic pairing in Ae. mutica [55] and in hybrids between common wheat and Ae. variabilis [56], leaf striping in maize [57], and crown rust resistance in Avena sativa [58].
Considering the intra- or interspecific origin of Bs and the above-listed B-associated effects, it has been of interest for a long time to address whether Bs carry genes. Studies have led to different conclusions regarding the transcriptional activity of Bs. For instance, weak or completely lacking transcriptional activity was concluded for Bs using labeled uridine in the grasshoppers Myrmeleotettix maculatus and Chorthippus parallelus [59], as well as the mouse Apodemus peninsulae [60]. The B-located histone H3 and H4 genes of the migratory locust are likely to be functionally inactive, as higher sequence variations were found compared to their A-located sequences [61]. Inactive 18S rDNA was described in telomeric and centromeric regions of the Bs of the fish Haplochromis obliquidens [62]. Both B chromosome types of B. dichromosomatica contain 45S rDNA [63, 64]. But, using reverse transcriptase PCR of the equivalent region within the 40S precursor rRNA, suggested that the rDNA of the large B is not transcribed. Although Bs with active NORs have also been observed [65], and at least low level transcription seems likely given the global transcription observed after RNA seq studies of plants with and without Bs [66].
There are other examples of gene-possessing Bs. Indirect evidence for transcription was revealed for the frog Leiopelma hochstetteri [67] and the fly Simulium juxtacrenobium [68] based on lateral loops observed in lampbrush chromosomes of Bs. A few B-specific cDNA fragments were identified after comparison of gene expression profiles of the mouse Apodemus flavicollis with and without Bs [69]. In the canids Vulpes vulpes and Nyctereutes procyonoides, the proto-oncogene C-KIT with intron–exon boundaries has been mapped on their Bs [70, 71]. Although the activity of the B-located C-KIT was not analyzed in these studies, the presence of this gene in Bs of different canids argues for its biological significance. In the ascomycete fungus Nectria haematococca, several functional genes conferring resistance to an antimicrobial compound produced by its host (garden pea Pisum sativum), were mapped on a dispensable chromosome [72–78]. These findings suggested the idea that some chromosomes comparable to the bacterial plasmids can define the habits of their carrier. In contrast to single or low copy genes which were rarely found on Bs in early studies, rRNA genes have been frequently identified on Bs of many species (for review, see [8, 79]). This is most likely to be due to the fact that their detection is rather easy by cytogenetic techniques in contrast to unique genes. In the smooth hawksbeard Crepis capillaris and the grasshopper Eyprepocnemis plorans, weak transcription of B-located rRNA genes was demonstrated [65, 80].
A comparative cDNA-AFLP analysis indicated that rye Bs are able to modulate the transcription of corresponding gene copies on A chromosomes [81] and, from these studies, regulatory interactions between A- and B-located coding sequences have been proposed. It is likely that Bs may influence A-localized sequences through epigenetic mechanisms, such as homology-dependent RNA interference pathways [82] as has been proposed for the modulation of gene-activity in newly formed hybrids and allopolyploids [83]. The effects of Bs on the spatial organization of As in interphase nuclei is another possible way that Bs could exert control on As, and it has been suggested that spatial positioning of genes and chromosomes can influence gene expression [84]. A similar effect was shown for the essentially gene-deficient Y chromosome of Drosophila melanogaster, which is able to regulate the activity of hundreds of genes located on other chromosomes [85].
Recent application of next generation sequencing-based approaches revealed that Bs contain an unexpected high number of genic sequences. Analysis of rye Bs resulted in the identification of more than 4000 putative B-located genic sequences [26]. These genic sequences showed elevated sequence polymorphism compared to their A-located counterparts indicating that they had undergone pseudogenization [66]. Sequencing of Nectria haematococca revealed that its supernumarary chromosomes are enriched in unique and duplicated genes [33]. In another analysis of this type, the genome of a male Drosophila albomicans which contained a B and inbred female flies derived from the same strain but without Bs were sequenced and compared. Besides the fact that the B of this species was shown to originate from As, the authors were able to detect one actively transcribed unit on B [44].
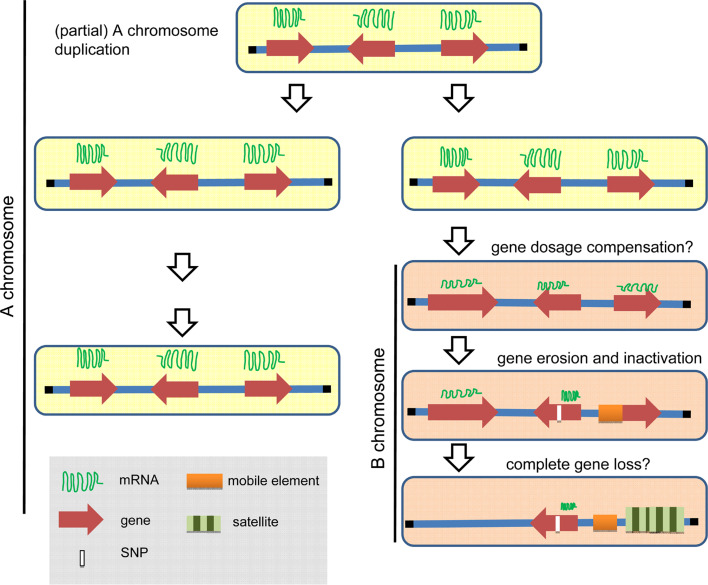
Depending on the process involved in B chromosome de novo origin and the ability of the host genome to compensate for additional copies of A-derived genes, it is likely that Bs of other species are rich in genic reads. What is the evolutionary fate of B-located genic sequences? As Bs are dispensable it is expected that they are prone to accumulate mutations as they undergo pseudogenization (Fig. 3). Depending on the species, the half-life of an active duplicated gene becoming mutated or lost was estimated to be 2–7 million years [86]. Considering this finding, dependent on the age of a B, it is possible that not all B-located genes are inactive. If Bs share many almost identical genic sequences with As, why is the presence of Bs not associated with more severe phenotypes, particularly assuming that some sequence variants may still have a biological effect? Bearing in mind that the relative dosage of a chromosome is critical for normal development (addition of a single human A chromosome to the genome nearly always results in detrimental effects), it is striking that organisms with an additional B are little affected. During early evolution of a proto B, A-derived genes are likely to be downregulated by dosage compensation. The role of micro-RNAs in affecting gene dosage balance is unknown, but it is potentially important in terms of modulating the expression of transcription factors, because micro- RNA is known to operate in a dosage-sensitive manner [87]. While the mechanisms of dosage compensation differ markedly between taxa, each well-studied case involves the recruitment of a chromatin regulatory complex to modulate gene expression (reviewed in [88]).
Fig. 3.
Model of the evolution of B-located pseudogenes. The B chromosome descends from A chromosome(s) after spontaneous whole or partial genome duplication. Proto-B still shows sequence similarity to the parental A chromosome(s). A proposed gene dosage compensation mechanism suppresses activity of B-located genes. Finally, B-located gene sequences became pseudogenized by mutations and accumulation of mobile and satellite DNA
Besides the activity of coding sequences, transcription of B-enriched repetitive sequences has been demonstrated. In maize, portions of a retrotransposon-derived high-copy element were shown to be transcriptionally active [20]. In rye, several B-repeats are active in a tissue-dependent manner [29, 89].
What could be the consequence of B-derived transcripts? Transcripts coming from a B in combination with their related A-located genes provide additional complexity to the transcriptome of their host and this may explain in part the phenotypes and effects associated with the presence of Bs. The transcriptional activity of Bs could form regulatory transcripts such as siRNAs which have the potential to modulate the level of A-derived transcripts or to change the chromatin status of a target region by DNA or chromatin modification [90, 91]. Also, transcripts from Bs similar to pseudogenes may dictate their effects indirectly by competing with A-derived transcripts for regulatory factors such as miRNAs [92]. If truncated proteins or higher representation of functional proteins are produced in the presence of Bs, it could cause overloading of the proteasome machinery for processing these unfolded, misfolded, aggregated, and/or uncomplexed proteins, thus causing an energetic burden for the host [93]. But Bs may also produce functional proteins which may have some role in maintenance of B chromosomes.
How does a nonessential chromosome survive over time?
The maintenance of Bs in natural populations is possible by their transmission at higher than Mendelian frequencies, and this enables the maintenance of Bs in populations [94]. The variety of mechanisms, including segregation failure, by which B chromosomes gain heritable advantage in transmission, are known as accumulation or drive mechanisms. Depending on the species, B chromosome drive can be premeiotic, meiotic, or post meiotic. Drive is the key to understanding B chromosomes and it occurs in many ways, but the molecular mechanisms remain far from clear [95, 96].
In animals, where the gametic nuclei do not replicate, the accumulation mechanism effectively acts either before or during meiosis. Premeiotic drive mechanisms in animals occur in the spermatogonial mitosis in the testes [96]. Post-meiotic drive is frequent in flowering plants during gametophyte maturation. The drive mechanisms of maize and rye Bs are well-studied examples that result in B chromosome accumulation in the next generation. In maize, at least three properties allow the Bs to increase in numbers: nondisjunction at the second pollen grain mitosis, preferential fertilisation of the egg by sperm containing B chromosomes [97–99], and suppression of meiotic loss when the Bs are unpaired [100]. One A-located factor seems to codetermine maize B accumulation by preferential fertilization while another factor(s) determines the meiotic loss of Bs [101]. Sperm nuclei containing deletion derivatives of B-9 (translocation lines involving the B and chromosome 9), which lack the centric heterochromatin and possibly some adjacent euchromatin of the B chromosome, no longer have the capacity for preferential fertilization [102].
The behavior of rye Bs during pollen mitosis was first studied by Hasegawa [103], who described how the two chromatids of the B chromosome do not separate at anaphase of first pollen grain mitosis and in most cases are included in the generative nucleus. In the second pollen grain mitosis, the generative nucleus divides to produce two sperm nuclei, each with an unreduced number of Bs. A similar nondisjunction process may occur in the female gametophytes as well [104].
Deficient Bs lacking the heterochromatic terminal region of the long arm undergo normal disjunction at first pollen anaphase. Therefore, it seems that the accumulation mechanism of the B by nondisjunction requires factors located at the end of its long arm [105–107]. This factor can act in trans because, if a standard B [108] or the terminal region of the long arm of the B [105] is present in the same cell containing a deficient B, nondisjunction occurs for both the standard and the deficient B. The nondisjunction control region is enriched in B-specific repeats which are highly transcriptionally active in anthers. In addition, the distal heterochromatin is marked with the euchromatin-specific histone modification mark H3K4me3 [89].
On the basis of a recent analysis on the cellular mechanism of B chromosome drive in the male gametophyte of rye, a model was proposed to explain how the accumulation mechanism works for the B chromosome [109]. At all mitotic stages of microgametogenesis, the centromeres of As and Bs are active. However, sister chromatid cohesion differs between As and Bs at first pollen mitosis. The B-specific pericentromeric repeats are involved in the formation of pericentric heterochromatin, which is known to play a role in chromosome segregation [110]. In particular, it has been suggested that heterochromatin is required in sister chromatid cohesion. In fission yeast, repeats flanking the kinetochore are essential for cohesion [111]. The failure to resolve the pericentromeric cohesion is under the control of the B-specific nondisjunction control element. The asymmetry of this division plays a critical role in the determination and subsequent fate of the two unequal mitotic products: the vegetative and the generative cells [112]. Due to unequal spindle formation, joined B chromatids become preferentially located towards the generative pole. In the second pollen mitosis, the generative nucleus divides to produce two sperm nuclei, each with an unreduced number of Bs. Hence, a combination of nondisjunction and of unequal spindle formation at first pollen mitosis results in the directed accumulation of Bs to the generative nucleus, which consequently ensures their transmission at a higher than Mendelian rate to the next generation. Nondisjunction works equally well when the rye B is introduced as an addition chromosome into hexaploid wheat [105, 113–115], hypo-pentaploid Triticale [116], or Secale vavilovii [117]. Thus, the B controls the process of nondisjunction autonomously [118, 119]. Whether a comparable mechanism exists in other species remains to be determined. However, it seems that asymmetrical spindles are also a key component of the premeiotic drive in, e.g., the Asteraceae Crepis capillaris [120] and of the meiotic drive of Bs in, e.g., the grasshopper Myrmeleotettix maculatus [121]. Hence, the asymmetry of the mitotic spindle seems to be a major component of diverse B accumulation mechanisms.
The discovery that some of the nondisjunction control region-specific repeats produce noncoding RNA predominantly in anthers of rye [89] suggests an intriguing possibility, namely that the nondisjunction of Bs occurs because the control region provides RNA that somehow maintains cohesion in key regions of B-sister chromatids. One might imagine that the failure in mitotic segregation reflects a failure to properly resolve the pericentromeric heterochromatin during first pollen mitosis. The cell cycle type-specific segregation failure of Bs triggers the question: in which aspect does the first pollen mitosis differ from other mitotic events in other cell types? We argue that either a haploid tissue-type specific expression of nondisjunction controlling transcripts [89] and/or the formation of a contrasting chromatin composition during first pollen mitosis [122] ensure cell type-specific accumulation.
As in rye, the accumulation mechanism in maize Bs requires a factor located on the end of the long arm of the B that can act in trans [123–125], and the B centromeric heterochromatin, irrespective of centromere function, is required for efficient nondisjunction [126, 127]. As the Bs of rye and maize originated independently, comparable drive mechanisms in both species evolved separately. Although much of repetitive DNA evolution is governed by neutral evolutionary processes [128], we propose that some B-located repeats, like those located in the Ab10 maize chromosome involved in neocentromere meiotic drive [129], the satellites involved in segregation distortion of Drosophila melanogaster [130] or in centromere-associated drive in female meiosis [131], are functionally involved in the regulation of chromosome segregation to ensure the maintenance of Bs in natural populations.
Outlook
Recent advances in sequencing and bioinformatics have provided efficient tools for the analysis of extra chromosomes. Applying these methods will shed new light on Bs and thus result in an improvement of our knowledge on genome dynamics. These unique chromosomes will teach us about the evolution of genomes and genes under varying amounts of selection pressure. Likely, because the Bs of different organisms clearly arose in various ways, novel mechanisms of B chromosome formation will be found. A detailed analysis of centromere regulation will be a prerequisite for a better understanding of the drive mechanisms, which are essential for the survival of a nonessential chromosome. The application of RNA seq technology to analyze the effects of Bs on the transcriptome of their host will be another interesting direction for B chromosome research. Hence, uncovering and understanding B chromosome biology will break into uncharted territory and have implications for genome evolution and gene regulation. Further analysis of Bs will provide exciting results on generation of rapid genome changes in higher eukaryotes, with particular relevance to selfish elements.
Acknowledgments
This work was supported by the DFG Germany (HO 1779/14-1) and the IPK. We would like to thank Juan Pedro M. Camacho (Granada, Spain), Nikolay B. Rubtsov (Novosibirsk, Russia) and Andre Marques (Gatersleben, Germany) for providing photomicrographs.
Abbreviations
- As
Standard chromosomes
- Bs
Supernumerary B chromosomes
- FISH
Fluorescence in situ hybridization
- miRNA
microRNA
- NUPT
Nuclear integrants of plastid DNA
- NUMT
Nuclear integrants of mitochondrial DNA
- NORG
Nuclear integrants of cytoplasmic organellar DNA
- SNP
Single nucleotide polymorphism
References
- 1.Wilson EB. The supernumerary chromosomes of Hemiptera . Science. 1907;26:870. [Google Scholar]
- 2.Pennisi E. Genomics encode project writes eulogy for junk DNA. Science. 2012;337(6099):1159–1161. doi: 10.1126/science.337.6099.1159. [DOI] [PubMed] [Google Scholar]
- 3.Liehr T, Mrasek K, Kosyakova N, Ogilvie CM, Vermeesch J, Trifonov V, Rubtsov N. Small supernumerary marker chromosomes (sSMC) in humans; are there B chromosomes hidden among them. Mol Cytogenet. 2008;1(1):12. doi: 10.1186/1755-8166-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones N, Houben A. B chromosomes in plants: escapees from the A chromosome genome? Trends Plant Sci. 2003;8(9):417–423. doi: 10.1016/S1360-1385(03)00187-0. [DOI] [PubMed] [Google Scholar]
- 5.Camacho JPM, Sharbel TF, Beukeboom LW. B-chromosome evolution. Philos Trans R Soc Lond B. 2000;355(1394):163–178. doi: 10.1098/rstb.2000.0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houben A, Banaei Moghaddam AM, Klemme S. Biology and evolution of B chromosomes. In: Leich IK, editor. Plant genome diversity physical structure behaviour and evolution of plant genomes. Wien: Springer; 2013. pp. 149–166. [Google Scholar]
- 7.Camacho JPM. B chromosomes. In: Gregory TR, editor. The evolution of the genome. San Diego, CA: Elsevier; 2005. pp. 223–285. [Google Scholar]
- 8.Bougourd SM, Jones RN. B chromosomes: a physiological enigma. New Phytol. 1997;137(1):43–54. [Google Scholar]
- 9.Mendelson D, Zohary D. Behavior and transmission of supernumerary chromosomes in Aegilops speltoides . Heredity. 1972;29 (Dec):329–339. [Google Scholar]
- 10.Ohta S. Mechanisms of B-chromosome accumulation in Aegilops mutica Boiss. Genes Genet Syst. 1996;71(1):23–29. [Google Scholar]
- 11.Nur U. Mitotic instability leading to an accumulation of B chromosomes in grasshoppers. Chromosoma. 1969;27(1):1–19. doi: 10.1007/BF00326108. [DOI] [PubMed] [Google Scholar]
- 12.Houben A, Thompson N, Ahne R, Leach CR, Verlin D, Timmis JN. A monophyletic origin of the B chromosomes of Brachycome dichromosomatica (Asteraceae) Plant Syst Evol. 1999;219(1–2):127–135. [Google Scholar]
- 13.Parker JS, Jones GH, Edgar LA, Whitehouse C. The population cytogenetics of Crepis capillaris. 4. The Distribution of B-chromosomes in British populations. Heredity. 1991;66:211–218. [Google Scholar]
- 14.Bougourd SM, Parker JS. B-chromosome system of Allium schoenoprasum 2 Stability, inheritance and phenotypic effects. Chromosoma. 1979;75(3):369–383. [Google Scholar]
- 15.Belyayev A, Raskina O. Chromosome evolution in marginal populations of Aegilops speltoides: causes and consequences. Ann Bot. 2013;111(4):531–538. doi: 10.1093/aob/mct023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bakkali M, Cabrero J, Lopez-Leon MD, Perfectti F, Camacho JPM. The B chromosome polymorphism of the grasshopper Eyprepocnemis plorans in north Africa. I. B variants and frequency. Heredity. 1999;83:428–434. doi: 10.1038/sj.hdy.6885950. [DOI] [PubMed] [Google Scholar]
- 17.Marques A, Banaei-Moghaddam AM, Klemme S, Blattner FR, Niwa K, Guerra M, Houben A. B chromosomes of rye are highly conserved and accompanied the development of early agriculture. Ann Bot. 2013;112(3):527–534. doi: 10.1093/aob/mct121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novak P, Neumann P, Macas J. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinforma. 2010;11:378. doi: 10.1186/1471-2105-11-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayer KF, Martis M, Hedley PE, Simkova H, Liu H, Morris JA, Steuernagel B, Taudien S, Roessner S, Gundlach H, Kubalakova M, Suchankova P, Murat F, Felder M, Nussbaumer T, Graner A, Salse J, Endo T, Sakai H, Tanaka T, Itoh T, Sato K, Platzer M, Matsumoto T, Scholz U, Dolezel J, Waugh R, Stein N. Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell. 2011;23(4):1249–1263. doi: 10.1105/tpc.110.082537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb JC, Riddle NC, Cheng YM, Theuri J, Birchler JA. Localization and transcription of a retrotransposon-derived element on the maize B chromosome. Chromosome Res. 2007;15(3):383–398. doi: 10.1007/s10577-007-1135-0. [DOI] [PubMed] [Google Scholar]
- 21.Houben A, Verlin D, Leach CR, Timmis JN. The genomic complexity of micro B chromosomes of Brachycome dichromosomatica . Chromosoma. 2001;110(7):451–459. doi: 10.1007/s00412-001-0173-1. [DOI] [PubMed] [Google Scholar]
- 22.Dhar MK, Friebe B, Koul AK, Gill BS. Origin of an apparent B chromosome by mutation, chromosome fragmentation and specific DNA sequence amplification. Chromosoma. 2002;111(5):332–340. doi: 10.1007/s00412-002-0214-4. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez-Sanchez M, Chiavarino M, Jimenez G, Manzanero S, Rosato M, Puertas MJ. The parasitic effects of rye B chromosomes might be beneficial in the long term. Cytogenet Genome Res. 2004;106:386–393. doi: 10.1159/000079316. [DOI] [PubMed] [Google Scholar]
- 24.Niwa K, Sakamoto S. Detection of B chromosomes in rye collected from Pakistan and China. Hereditas. 1996;124(3):211–215. [Google Scholar]
- 25.Niwa K, Sakamoto S. Origin of B-chromosomes in cultivated rye. Genome. 1995;38(2):307–312. doi: 10.1139/g95-038. [DOI] [PubMed] [Google Scholar]
- 26.Martis MM, Klemme S, Banaei-Moghaddam AM, Blattner FR, Macas J, Schmutzer T, Scholz U, Gundlach H, Wicker T, Simkova H, Novak P, Neumann P, Kubalakova M, Bauer E, Haseneyer G, Fuchs J, Dolezel J, Stein N, Mayer KF, Houben A. Selfish supernumerary chromosome reveals its origin as a mosaic of host genome and organellar sequences. Proc Natl Acad Sci USA. 2012;109(33):13343–13346. doi: 10.1073/pnas.1204237109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker SE, Thomas R, Trifonov VA, Wayne RK, Graphodatsky AS, Breen M. Anchoring the dog to its relatives reveals new evolutionary breakpoints across 11 species of the Canidae and provides new clues for the role of B chromosomes. Chromosome Res. 2011;19(6):685–708. doi: 10.1007/s10577-011-9233-4. [DOI] [PubMed] [Google Scholar]
- 28.Wicker T, Mayer KF, Gundlach H, Martis M, Steuernagel B, Scholz U, Simkova H, Kubalakova M, Choulet F, Taudien S, Platzer M, Feuillet C, Fahima T, Budak H, Dolezel J, Keller B, Stein N. Frequent gene movement and pseudogene evolution is common to the large and complex genomes of wheat, barley, and their relatives. Plant Cell. 2011;23(5):1706–1718. doi: 10.1105/tpc.111.086629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klemme S, Banaei-Moghaddam AM, Macas J, Wicker T, Novak P, Houben A. High-copy sequences reveal distinct evolution of the rye B chromosome. New Phytol. 2013;199:550–558. doi: 10.1111/nph.12289. [DOI] [PubMed] [Google Scholar]
- 30.Shirasu K, Schulman AH, Lahaye T, Schulze-Lefert P. A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res. 2000;10(7):908–915. doi: 10.1101/gr.10.7.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomita M, Shinohara K, Morimoto M. Revolver is a new class of transposon-like gene composing the triticeae genome. DNA Res. 2008;15(1):49–62. doi: 10.1093/dnares/dsm029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ziegler CG, Lamatsch DK, Steinlein C, Engel W, Schartl M, Schmid M. The giant B chromosome of the cyprinid fish Alburnus alburnus harbours a retrotransposon-derived repetitive DNA sequence. Chromosome Res. 2003;11(1):23–35. doi: 10.1023/a:1022053931308. [DOI] [PubMed] [Google Scholar]
- 33.Coleman JJ, Rounsley SD, Rodriguez-Carres M, Kuo A, Wasmann CC, Grimwood J, Schmutz J, Taga M, White GJ, Zhou S, Schwartz DC, Freitag M, Ma LJ, Danchin EG, Henrissat B, Coutinho PM, Nelson DR, Straney D, Napoli CA, Barker BM, Gribskov M, Rep M, Kroken S, Molnar I, Rensing C, Kennell JC, Zamora J, Farman ML, Selker EU, Salamov A, Shapiro H, Pangilinan J, Lindquist E, Lamers C, Grigoriev IV, Geiser DM, Covert SF, Temporini E, Vanetten HD. The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet. 2009;5(8):e1000618. doi: 10.1371/journal.pgen.1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charlesworth D. Plant sex chromosomes. Genome Dyn. 2008;4:83–94. doi: 10.1159/000126008. [DOI] [PubMed] [Google Scholar]
- 35.Zhu Y, Dai J, Fuerst PG, Voytas DF. Controlling integration specificity of a yeast retrotransposon. Proc Natl Acad Sci USA. 2003;100(10):5891–5895. doi: 10.1073/pnas.1036705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukahara S, Kawabe A, Kobayashi A, Ito T, Aizu T, Shin IT, Toyoda A, Fujiyama A, Tarutani Y, Kakutani T. Centromere-targeted de novo integrations of an LTR retrotransposon of Arabidopsis lyrata . Genes Dev. 2012;26:705–713. doi: 10.1101/gad.183871.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kejnovsky E, Vyskot B. Silene latifolia: the classical model to study heteromorphic sex chromosomes. Cytogenet Genome Res. 2010;129(1–3):250–262. doi: 10.1159/000314285. [DOI] [PubMed] [Google Scholar]
- 38.Alfenito MR, Birchler JA. Molecular characterization of a maize B chromosome centric sequence. Genetics. 1993;135(2):589–597. doi: 10.1093/genetics/135.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stark EA, Connerton I, Bennett ST, Barnes SR, Parker JS, Forster JW. Molecular analysis of the structure of the maize B-chromosome. Chromosome Res. 1996;4(1):15–23. doi: 10.1007/BF02254939. [DOI] [PubMed] [Google Scholar]
- 40.Sandery MJ, Forster JW, Blunden R, Jones RN. Identification of a family of repeated sequences on the rye B-chromosome. Genome. 1990;33(6):908–913. doi: 10.1139/g93-095. [DOI] [PubMed] [Google Scholar]
- 41.Langdon T, Seago C, Jones RN, Ougham H, Thomas H, Forster JW, Jenkins G. De novo evolution of satellite DNA on the rye B chromosome. Genetics. 2000;154(2):869–884. doi: 10.1093/genetics/154.2.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith GP. Evolution of repeated DNA sequences by unequal crossover. Science. 1976;191(4227):528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- 43.Devos KM, Atkinson MD, Chinoy CN, Francis HA, Harcourt RL, Koebner RMD, Liu CJ, Masojc P, Xie DX, Gale MD. Chromosomal rearrangements in the rye genome relative to that of wheat. Theor Appl Genet. 1993;85(6–7):673–680. doi: 10.1007/BF00225004. [DOI] [PubMed] [Google Scholar]
- 44.Zhou Q, Zhu HM, Huang QF, Zhao L, Zhang GJ, Roy SW, Vicoso B, Xuan ZL, Ruan J, Zhang Y, Zhao RP, Ye C, Zhang XQ, Wang J, Wang W, Bachtrog D. Deciphering neo-sex and B chromosome evolution by the draft genome of Drosophila albomicans . Bmc Genomics. 2012;13:109. doi: 10.1186/1471-2164-13-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheppard AE, Ayliffe MA, Blatch L, Day A, Delaney SK, Khairul-Fahmy N, Li Y, Madesis P, Pryor AJ, Timmis JN. Transfer of plastid DNA to the nucleus is elevated during male gametogenesis in tobacco. Plant Physiol. 2008;148(1):328–336. doi: 10.1104/pp.108.119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang D, Lloyd AH, Timmis JN. Environmental stress increases the entry of cytoplasmic organellar DNA into the nucleus in plants. Proc Natl Acad Sci USA. 2012;109(7):2444–2448. doi: 10.1073/pnas.1117890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5(2):123. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 48.Sheppard AE, Timmis JN. Instability of plastid DNA in the nuclear genome. PLoS Genet. 2009;5(1):e1000323. doi: 10.1371/journal.pgen.1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones RN, Rees H. B chromosomes. 1. London, New York: Academic Press; 1982. [Google Scholar]
- 50.Jones RN. Tansley Review No 85, B chromosomes in plants. New Phytol. 1995;131:411–434. doi: 10.1111/j.1469-8137.1995.tb03079.x. [DOI] [PubMed] [Google Scholar]
- 51.Carlson W (2009) The B chromosome of maize. In: Bennetzen JL, Hake S (eds) Maize Handbook. Genetics and Genomics, vol II. Springer, Heidelberg, pp 459–480
- 52.Holmes DS, Bougourd SM. B-Cchromosome selection in Allium schoenoprasum. 2. experimental populations. Heredity. 1991;67:117–122. [Google Scholar]
- 53.Yoshida K, Terai Y, Mizoiri S, Aibara M, Nishihara H, Watanabe M, Kuroiwa A, Hirai H, Hirai Y, Matsuda Y, Okada N. B Chromosomes have a functional effect on female sex determination in lake Victoria cichlid fishes. Plos Genetics. 2011;7(8):e1002203. doi: 10.1371/journal.pgen.1002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jackson RC, Newmark P. Effects of supernumerary chromosomes on production of pigment in Haplopappus gracilis . Science. 1960;132(3436):1316–1317. doi: 10.1126/science.132.3436.1316. [DOI] [PubMed] [Google Scholar]
- 55.Dover GA, Riley R. Prevention of pairing of homoeologous meiotic chromosomes of wheat by an activity of supernumerary chromosomes of Aegilops . Nature. 1972;240(5377):159. [Google Scholar]
- 56.Kousaka R, Endo TR. Effect of a rye B chromosome and its segments on homoeologous pairing in hybrids between common wheat and Aegilops variabilis . Genes Genet Syst. 2012;87(1):1–7. doi: 10.1266/ggs.87.1. [DOI] [PubMed] [Google Scholar]
- 57.Staub RW. Leaf striping correlated with the presence of B-chromosomes in maize. J Hered. 1987;78(2):71–74. [Google Scholar]
- 58.Dherawat A, Sadanaga K. Cytogenetics of a crown rust-resistant hexaploid Oat with 42 + 2 fragment chromosomes. Crop Sci. 1973;13(6):591–594. [Google Scholar]
- 59.Fox DP, Hewitt GM, Hall DJ. DNA-replication and RNA transcription of euchromatic and heterochromatic chromosome regions during grasshopper meiosis. Chromosoma. 1974;45(1):43–62. doi: 10.1007/BF00283829. [DOI] [PubMed] [Google Scholar]
- 60.Ishak B, Jaafar H, Maetz JL, Rumpler Y. Absence of transcriptional activity of the B-chromosomes of Apodemus peninsulae during pachytene. Chromosoma. 1991;100(4):278–281. [Google Scholar]
- 61.Teruel M, Cabrero J, Perfectti F, Camacho JPM. B chromosome ancestry revealed by histone genes in the migratory locust. Chromosoma. 2010;119(2):217–225. doi: 10.1007/s00412-009-0251-3. [DOI] [PubMed] [Google Scholar]
- 62.Poletto AB, Ferreira IA, Martins C. The B chromosomes of the African cichlid fish Haplochromis obliquidens harbour 18S rRNA gene copies. BMC Genet. 2010;11:1. doi: 10.1186/1471-2156-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Houben A, Leach CR, Verlin D, Rofe R, Timmis JN. A repetitive DNA sequence common to the different B chromosomes of the genus Brachycome . Chromosoma. 1997;106(8):513–519. doi: 10.1007/pl00007689. [DOI] [PubMed] [Google Scholar]
- 64.Donald TM, Leach CR, Clough A, Timmis JN. Ribosomal RNA genes and the B chromosome of Brachycome dichromosomatica . Heredity. 1995;74(5):556–561. doi: 10.1038/hdy.1995.77. [DOI] [PubMed] [Google Scholar]
- 65.Leach CR, Houben A, Field B, Pistrick K, Demidov D, Timmis JN. Molecular evidence for transcription of genes on a B chromosome in Crepis capillaris . Genetics. 2005;171:269–278. doi: 10.1534/genetics.105.043273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Banaei-Moghaddam AM, Meier K, Karimi-Ashtiyani R, Houben A (2013) Formation and expression of pseudogenes on the B chromosome of rye. Plant Cell (in press) [DOI] [PMC free article] [PubMed]
- 67.Green DM. Cytogenetics of the endemic New Zealand frog, Leiopelma hochstetteri - Extraordinary supernumerary chromosome variation and a unique sex chromosome system. Chromosoma. 1988;97(1):55–70. [Google Scholar]
- 68.Brockhouse C, Bass JAB, Feraday RM, Straus NA. Supernumerary chromosome evolution in the Simulium vernum Group (Diptera, Simuliidae) Genome. 1989;32(4):516–521. [Google Scholar]
- 69.Tanic N, Vujosevic M, Dedovic-Tanic N, Dimitrijevic B. Differential gene expression in yellow-necked mice Apodemus flavicollis (Rodentia, Mammalia) with and without B chromosomes. Chromosoma. 2005;113(8):418–427. doi: 10.1007/s00412-004-0327-z. [DOI] [PubMed] [Google Scholar]
- 70.Graphodatsky AS, Kukekova AV, Yudkin DV, Trifonov VA, Vorobieva NV, Beklemisheva VR, Perelman PL, Graphodatskaya DA, Trut LN, Yang FT, Ferguson-Smith MA, Acland GM, Aguirre GD. The proto-oncogene C-KIT maps to canid B-chromosomes. Chromosome Res. 2005;13(2):113–122. doi: 10.1007/s10577-005-7474-9. [DOI] [PubMed] [Google Scholar]
- 71.Yudkin DV, Trifonov VA, Kukekova AV, Vorobieva NV, Rubtsova NV, Yang F, Acland GM, Ferguson-Smith MA, Graphodatsky AS. Mapping of KIT adjacent sequences on canid autosomes and B chromosomes. Cytogenet Genome Res. 2007;116(1–2):100–103. doi: 10.1159/000097424. [DOI] [PubMed] [Google Scholar]
- 72.Covert SF, Enkerli J, Miao VP, VanEtten HD. A gene for maackiain detoxification from a dispensable chromosome of Nectria haematococca . Mol Gen Genet: MGG. 1996;251(4):397–406. doi: 10.1007/BF02172367. [DOI] [PubMed] [Google Scholar]
- 73.Han Y, Liu X, Benny U, Kistler HC, VanEtten HD. Genes determining pathogenicity to pea are clustered on a supernumerary chromosome in the fungal plant pathogen Nectria haematococca . Plant J. 2001;25(3):305–314. doi: 10.1046/j.1365-313x.2001.00969.x. [DOI] [PubMed] [Google Scholar]
- 74.Covert SF. Supernumerary chromosomes in filamentous fungi. Curr Genet. 1998;33(5):311–319. doi: 10.1007/s002940050342. [DOI] [PubMed] [Google Scholar]
- 75.VanEtten H, Funnell-Baerg D, Wasmann C, McCluskey K. Location of pathogenicity genes on dispensable chromosomes in Nectria haematococca MPVI. Antonie van Leeuwenhoek. 1994;65(3):263–267. doi: 10.1007/BF00871955. [DOI] [PubMed] [Google Scholar]
- 76.Funnell DL, VanEtten HD. Pisatin demethylase genes are on dispensable chromosomes while genes for pathogenicity on carrot and ripe tomato are on other chromosomes in Nectria haematococca . Mol Plant Microbe In. 2002;15(8):840–846. doi: 10.1094/MPMI.2002.15.8.840. [DOI] [PubMed] [Google Scholar]
- 77.Funnell DL, Matthews PS, VanEtten HD. Identification of new pisatin demethylase genes (PDA5 and PDA7) in Nectria haematococca and non-Mendelian segregation of pisatin demethylating ability and virulence on pea due to loss of chromosomal elements. Fungal Genet Biol. 2002;37(2):121–133. doi: 10.1016/s1087-1845(02)00503-0. [DOI] [PubMed] [Google Scholar]
- 78.Rodriguez-Carres A, White G, Tsuchiya D, Taga M, VanEtten HD. The supernumerary chromosome of Nectria haematococca that carries pea-pathogenicity-related genes also carries a trait for pea rhizosphere competitiveness. Appl Environ Microbiol. 2008;74(12):3849–3856. doi: 10.1128/AEM.00351-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Green DM. Muller`s rachet and the evolution of supernumerary chromosomes. Genome. 1990;33:818–824. [Google Scholar]
- 80.Ruiz-Estevez M, Lopez-Leon MD, Cabrero J, Camacho JPM. B-Chromosome Ribosomal DNA Is Functional in the Grasshopper Eyprepocnemis plorans . PLoS ONE. 2012;7(5):e36600. doi: 10.1371/journal.pone.0036600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Carchilan M, Kumke K, Mikolajewski S, Houben A. Rye B chromosomes are weakly transcribed and might alter the transcriptional activity of A chromosome sequences. Chromosoma. 2009;118(5):607–616. doi: 10.1007/s00412-009-0222-8. [DOI] [PubMed] [Google Scholar]
- 82.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8(4):272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 83.Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6(11):836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- 84.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128(4):787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 85.Lemos B, Araripe LO, Hartl DL. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science. 2008;319(5859):91–93. doi: 10.1126/science.1148861. [DOI] [PubMed] [Google Scholar]
- 86.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 87.Guo L, Lu ZH. The fate of miRNA* strand through evolutionary analysis: Implication for degradation as merely carrier strand or potential regulatory molecule? PLoS ONE. 2010;5(6):e11387. doi: 10.1371/journal.pone.0011387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Prestel M, Feller C, Becker PB. Dosage compensation and the global re-balancing of aneuploid genomes. Genome Biol. 2010;11(8):216. doi: 10.1186/gb-2010-11-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Carchilan M, Delgado M, Ribeiro T, Costa-Nunes P, Caperta A, Morais-Cecilio L, Jones RN, Viegas W, Houben A. Transcriptionally active heterochromatin in rye B chromosomes. Plant Cell. 2007;19(6):1738–1749. doi: 10.1105/tpc.106.046946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dalakouras A, Wassenegger M (2013) Revisiting RNA-directed DNA methylation. RNA Biol (in press) [DOI] [PMC free article] [PubMed]
- 91.Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol. 2005;15(3):331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 92.Pink RC, Wicks K, Caley DP, Punch EK, Jacobs L, Carter DR. Pseudogenes: pseudo-functional or key regulators in health and disease? . RNA. 2011;17(5):792–798. doi: 10.1261/rna.2658311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev. 2012;13(3):189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 94.Kimura M, Kayano H. The maintenance of supernumerary chromosomes in wild populations of Lillium callosum by preferential segregation. Genetics. 1961;46:1699–1712. doi: 10.1093/genetics/46.12.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Burt A, Trivers R. Genes in conflict : The biology of selfish genetic elements. Cambridge: Belknap; 2006. [Google Scholar]
- 96.Jones RN. B-chromosome drive. Am Nat. 1991;137:430–442. [Google Scholar]
- 97.Roman H. Selective fertilization in maize. Genetics. 1948;33(1):122. [PubMed] [Google Scholar]
- 98.Carlson WR. Factors affecting preferential fertilization in maize. Genetics. 1969;62(3):543–554. doi: 10.1093/genetics/62.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rusche ML, Mogensen HL, Shi L, Keim P, Rougier M, Chaboud A, Dumas C. B chromosome behavior in maize pollen as determined by a molecular probe. Genetics. 1997;147(4):1915–1921. doi: 10.1093/genetics/147.4.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Carlson WR, Roseman RR. A new property of the maize B-chromosome. Genetics. 1992;131(1):211–223. doi: 10.1093/genetics/131.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gonzalez-Sanchez M, Gonzalez-Gonzalez E, Molina F, Chiavarino AM, Rosato M, Puertas MJ. One gene determines maize B chromosome accumulation by preferential fertilisation; another gene(s) determines their meiotic loss. Heredity. 2003;90(2):122–129. doi: 10.1038/sj.hdy.6800185. [DOI] [PubMed] [Google Scholar]
- 102.Carlson WR. Locating a site on the maize B chromosome that controls preferential fertilization. Genome. 2007;50(6):578–587. doi: 10.1139/g07-035. [DOI] [PubMed] [Google Scholar]
- 103.Hasegawa N. A cytological study on 8-chromosome rye. Cytologia. 1934;6:68–77. [Google Scholar]
- 104.Hakansson A. Behaviour of accessory rye chromosomes in the embryo sac. Hereditas. 1948;34:35–59. [Google Scholar]
- 105.Endo TR, Nasuda S, Jones N, Dou Q, Akahori A, Wakimoto M, Tanaka H, Niwa K, Tsujimoto H. Dissection of rye B chromosomes, and nondisjunction properties of the dissected segments in a common wheat background. Genes Genet Syst. 2008;83(1):23–30. doi: 10.1266/ggs.83.23. [DOI] [PubMed] [Google Scholar]
- 106.Müntzing A. Cytological studies of extra fragment chromosomes in rye. V. A new fragment type arisen by deletion. Hereditas. 1948;34:435–442. [Google Scholar]
- 107.Håkanson A. Behaviour of different small accessry rye chromosomes at pollen mitosis. Hereditas. 1959;45:623–631. [Google Scholar]
- 108.Lima-de-Faria A. Genetic interaction in rye expressed at chromosome phenotype. Genetics. 1962;47:1455–1462. doi: 10.1093/genetics/47.10.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Banaei-Moghaddam AM, Schubert V, Kumke K, Weibeta O, Klemme S, Nagaki K, Macas J, Gonzalez-Sanchez M, Heredia V, Gomez-Revilla D, Gonzalez-Garcia M, Vega JM, Puertas MJ, Houben A. Nondisjunction in favor of a chromosome: the mechanism of rye B chromosome drive during pollen mitosis. Plant Cell. 2012 doi: 10.1105/tpc.112.105270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yamagishi Y, Sakuno T, Shimura M, Watanabe Y. Heterochromatin links to centromeric protection by recruiting Shugoshin. Nature. 2008;455(7210):251–255. doi: 10.1038/nature07217. [DOI] [PubMed] [Google Scholar]
- 111.Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294(5551):2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 112.Twell D. Male gametogenesis and germline specification in flowering plants. Sex Plant Reprod. 2010 doi: 10.1007/s00497-010-0157-5. [DOI] [PubMed] [Google Scholar]
- 113.Niwa K, Horiuchi G, Hirai Y. Production and characterization of common wheat with B chromosomes of rye from Korea. Hereditas. 1997;126(2):139–146. [Google Scholar]
- 114.Müntzing A. Chromosomal variation in the Lindström strain of wheat carrying accessory chromosomes in rye. Hereditas. 1970;66:279–286. [Google Scholar]
- 115.Lindström J. Transfer to wheat of accessory chromosomes from rye. Hereditas. 1965;54:149–155. [Google Scholar]
- 116.Kishikawa H, Suzuki A. Cytological study on hypo-pentaploid Triticale with four B chromosomes of rye. Jpn J Genet. 1982;57:17–24. [Google Scholar]
- 117.Puertas MJ, Romera F, Delapena A. Comparison of B-chromosome effects on Secale cereale and Secale vavilovii . Heredity. 1985;55 (Oct):229–234. [Google Scholar]
- 118.Matthews RB, Jones RN. Dynamics of the B chromosome polymorphism in rye II. Estimates of parameters. Heredity. 1983;50:119–137. [Google Scholar]
- 119.Romera F, Jimenez MM, Puertas MJ. Factors controlling the dynamics of the B-chromosome polymorphism in Korean rye. Heredity. 1991;67:189–195. [Google Scholar]
- 120.Rutishauser A, Rothlisberger E. Boosting mechanism of B-chromosomes in Crepis capilaris . Chromosomes Today. 1966;1:28–30. [Google Scholar]
- 121.Hewitt GM. Meiotic drive for B-chromosomes in the primary oocytes of Myrmeleotettix maculatus (Orthopera: Acrididae) Chromosoma. 1976;56(4):381–391. doi: 10.1007/BF00292957. [DOI] [PubMed] [Google Scholar]
- 122.Houben A, Kumke K, Nagaki K, Hause G. CENH3 distribution and differential chromatin modifications during pollen development in rye (Secale cereale L.) Chromosome Res. 2011;19(4):471–480. doi: 10.1007/s10577-011-9207-6. [DOI] [PubMed] [Google Scholar]
- 123.Roman H. Mitotic nondisjunction in the case of interchanges involving the B-type chromosome in maize. Genetics. 1947;32(4):391–409. doi: 10.1093/genetics/32.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Carlson WR. B-chromosome of corn. Annu Rev Genet. 1978;12:5–23. doi: 10.1146/annurev.ge.12.120178.000253. [DOI] [PubMed] [Google Scholar]
- 125.Lamb JC, Han F, Auger DL, Birchler J. A trans-acting factor required for non-disjunction of the B chromosome is located distal to the TB-4Lb breakpoint on the B chromosome. Maize Genet Cooper Newsl. 2006;80:51–54. [Google Scholar]
- 126.Carlson WR. Unstable inheritance of maize B-type chromosomes that lack centric heterochromatin. Genome. 2006;49(5):420–431. doi: 10.1139/g05-127. [DOI] [PubMed] [Google Scholar]
- 127.Han FP, Lamb JC, Yu WC, Gao Z, Birchler JA. Centromere function and nondisjunction are independent components of the maize B chromosome accumulation mechanism. Plant Cell. 2007;19(2):524–533. doi: 10.1105/tpc.106.049577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371(6494):215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 129.Mroczek RJ, Melo JR, Luce AC, Hiatt EN, Dawe RK. The maize Ab10 meiotic drive system maps to supernumerary sequences in a large complex haplotype. Genetics. 2006;174(1):145–154. doi: 10.1534/genetics.105.048322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Frank SA. Polymorphism of attack and defense. Trends Ecol Evol. 2000;15(4):167–171. doi: 10.1016/s0169-5347(99)01814-5. [DOI] [PubMed] [Google Scholar]
- 131.Fishman L, Saunders A. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science. 2008;322(5907):1559–1562. doi: 10.1126/science.1161406. [DOI] [PubMed] [Google Scholar]