Abstract
Adult skeletal muscle can regenerate in response to muscle damage. This ability is conferred by the presence of myogenic stem cells called satellite cells. In response to stimuli such as injury or exercise, these cells become activated and express myogenic regulatory factors (MRFs), i.e., transcription factors of the myogenic lineage including Myf5, MyoD, myogenin, and Mrf4 to proliferate and differentiate into myofibers. The MRF family of proteins controls the transcription of important muscle-specific proteins such as myosin heavy chain and muscle creatine kinase. Different growth factors are secreted during muscle repair among which insulin-like growth factors (IGFs) are the only ones that promote both muscle cell proliferation and differentiation and that play a key role in muscle regeneration and hypertrophy. Different isoforms of IGFs are expressed during muscle repair: IGF-IEa, IGF-IEb, or IGF-IEc (also known as mechano growth factor, MGF) and IGF-II. MGF is expressed first and is observed in satellite cells and in proliferating myoblasts whereas IGF-Ia and IGF-II expression occurs at the state of muscle fiber formation. Interestingly, several studies report the induction of MRFs in response to IGFs stimulation. Inversely, IGFs expression may also be regulated by MRFs. Various mechanisms are proposed to support these interactions. In this review, we describe the general process of muscle hypertrophy and regeneration and decipher the interactions between the two groups of factors involved in the process.
Keywords: Regeneration, IGF, MyoD, Myogenin, Myogenesis, Hypertrophy
Introduction
About one-half of our body mass is made up skeletal muscle. Besides its obvious role in locomotor activity and postural behavior, skeletal muscle also functions as a metabolic regulator, for example by storing glycogen. Loss of muscle tissue observed in aging or in degenerative muscle diseases therefore alters physical abilities and leads to metabolic problems such as insulin resistance or osteoporosis. This constitutes a major public-health problem.
Adult muscle consists of multinucleated myofibers that can undergo changes in size (atrophy/hypertrophy) and type (slow-contracting, fatigue-resistant type/fast-contracting, fatigable type). Due to its daily mechanical work, muscle undergoes small tears and minor lesions. This leads to a slow turnover of constituent fibers, replacing no more than 1–2 % of myonuclei per week [1]. In normal conditions, adult skeletal muscle is therefore a stable tissue. However, in response to severe injury due to a direct trauma, to an extensive physical activity or to a genetic muscle disease, skeletal muscle has the remarkable ability to regenerate itself, preventing loss of muscle mass and/or delaying the appearance of clinical symptoms of muscle diseases. This is due to the presence of quiescent satellite cells, which are mononucleated cells localized in a niche constituted by the sarcolemma of the adjacent muscle fiber and the basal lamina. The initial phase of muscle repair is characterized by necrosis of damaged fibers and activation of an inflammatory response. Then, quiescent satellite cells reenter the cell cycle and proliferate to renew the quiescent satellite cell pool or to differentiate in new myofibers. The final phase of regeneration consists of a maturation of newly formed myofibers and in a remodeling of regenerated muscle.
Different factors play critical roles in skeletal muscle regeneration. Among them, myogenic regulatory factors (MRFs) and insulin-like growth factors (IGFs) are critical for muscle repair. MRF family proteins constitute the key factors that determine the progression of satellite cell activation during myogenesis and muscle regeneration. However, the mechanisms by which the expression and activity of these factors are regulated during the processes are not completely clarified. Specifically, it is not clear whether the IGF pathway acts upstream/downstream or in parallel to MRFs.
In this review, we present the general mechanisms of muscle repair and hypertrophy with emphasis on MRFs and IGFs and then discuss the molecular mechanisms underlying the cooperation between the two groups of factors.
Animal models of muscle degeneration
Degeneration/regeneration processes can be obtained by different methods. The amplitude and kinetics of the repair depend on the extent of degeneration, the muscle injured, the animal model studied, and the method used to induce injury [2, 3] (review in [4, 5]). It is important to note that, in higher vertebrates, muscle regeneration depends on whether the injured tissue retains an extracellular matrix scaffold that serves as a template for muscle fibers formation [6].
So far, the easiest and the most reproducible technique to induce muscle degeneration seems to be a muscular injection of a myotoxin such as cardiotoxin, bupivacaine (marcain), or notexin [7] (review in [4]). The other methods used to induce myotrauma are muscle crushing or freezing, BaCl2 and muscle denervation, and devascularization obtained by muscle transplantation [7, 8]. Finally, intensive muscle exercise and in particular eccentric exercise (muscle stretch during contraction) can induce muscle lesions and constitutes a physiological mechanism of muscle degeneration [9–11].
On the other hand, muscle repair is also frequently investigated in pathological models with abnormal muscle degeneration such as the mdx (X-linked muscular dystrophy) mouse. This mouse is a model of Duchenne muscular dystrophy (DMD), the most common form of human muscular dystrophy characterized by a progressive and continuous muscle degeneration accompanied by inadequate muscle regeneration [12–15]. DMD is due to a mutation in the p21 locus of chromosome X [16, 17]. The mutation results in the complete lack of expression of dystrophin, a 427-kDa protein localized at the cytoplasmic face of the sarcolemma [16]. Dystrophin-deficient muscle has an increase in muscle membrane fragility making it particularly sensitive to eccentric contractions (significant loss of muscle strength induced by repeated cycles of lengthening contractions) [18–21].
Phases of muscle repair
Injured muscles show a rapid necrosis of myofibers. Disruption of their sarcolemma results in increased serum levels of muscular cytosolic proteins such as creatine kinase [22]. Different kinetics of muscle regeneration have been observed. For example, after cardiotoxin-induced muscle degeneration, muscle repair is achieved in 3–4 weeks. Indeed, 3 days after injection, muscle force production and histochemical analysis show that degeneration is almost complete. Myogenic cell differentiation and new myotube formation are already observed 5–6 days post-injection. By 10 days post-injection, the overall muscle architecture is restored, although most regenerated myofibers are smaller and display central myonuclei; after 3–4 weeks, muscle is morphologically and histochemically indistinguishable from pre-injury muscle [23, 24].
Different phases play critical roles during skeletal muscle regeneration. They include inflammation reaction, activation of satellite cells, and muscle remodeling. These different phases overlap during the process of muscle repair.
Inflammation
Muscle inflammation is generally a beneficial response as it promotes muscle repair and growth (review in [25]). After muscle damage, bone marrow cells rapidly invade zones of necrosis in response to chemotactic factors and then play different roles during muscle repair [26]. Indeed, the inflammatory response occurs within 1 h of muscle damage with invasion by neutrophils, raises a peak in 48 h with invasion by monocytes and macrophages, and persists throughout the process of muscle repair (review in [25, 27]). These inflammatory cells play a key role in phagocytosis of cellular debris and cleaning disrupted myofilaments, cytosolic structures, and damaged sarcolemma of damaged muscle [28].
Independently of the main role they play in cleaning, it has been shown that macrophages actively promote muscle regeneration by releasing growth factors and cytokines [25]. Indeed, two populations of macrophages colonize injured muscle. The first one is present within 24 h after muscle injury and is characterized by expression of CD68 surface marker but absence of CD163 marker. These macrophages are involved in phagocytosis of muscle debris by secreting pro-inflammatory cytokines such as tumor necrosis factor alpha (TNFα) and interleukin-1 beta (IL-1β). Then, they decline rapidly and are replaced about 2–4 days after injury by a second population of macrophages that are CD68−/CD163+ [6]. These CD163+ macrophages secrete IL-10 to resolve inflammation but also release factors enhancing myogenic precursors proliferation, growth, and differentiation [29–31].
The important role of macrophages in the regeneration process is supported by the fact that the suppression of their activity impairs muscle regeneration and leads to muscle fibrosis [32]. However, if growing evidence indicates that macrophages allow muscle repair, the beneficial role of neutrophils during muscle regeneration is much less clear (review in [25]).
Activation, proliferation, and differentiation of satellite cells
In adult skeletal muscle, satellite cells are quiescent and localized in a niche between the sarcolemma of the myofiber and the basal lamina. Satellite cells can be identified by the presence of specific markers, such as CD34, M-cadherin, and Pax-7, a transcription factor involved in embryonic myogenesis [33, 34]. At birth, this cellular population represents a substantial proportion of muscle nuclei, up to 30 %, but this progressively decreases with ageing.
During muscle regeneration, satellite cells become activated along damaged muscle. Indeed, satellite cells migrate from distant undamaged areas to the injured site [35] and proliferate [36] in a process similar to fetal muscle development [37]. The division of satellite cells occurs in an apical–basal asymmetric mode: the basal daughter cell supporting self-renewal by producing a stem cell population that is prevented from differentiation, and the apical daughter cell giving rise to more committed myogenic progeny [38–40]. Committed myogenic progeny proliferate as myoblasts, migrate, and fuse with each other to form novel myofibers or with pre-existing fibers to repair damaged fibers.
The behavior of satellite cells depends on multiple factors that come from their microenvironment. Their niche, the neighboring cells and the extracellular matrix, constitute this microenvironment.
Different signaling pathways have been implicated in the satellite cell activation process. For example, Wnt and Notch signaling, which are mainly involved in embryogenesis, are also shown to play an important role in satellite cell fate, proliferation, and differentiation during muscle regeneration. During myogenesis, Delta-1 (a notch ligand) binding to notch (a transmembrane receptor) activates the notch signaling pathway. This leads to the cleavage of the intracellular portion of notch and its translocation to the nucleus where it activates a family of transcription factors and induces stem cell proliferation [41]. Interestingly, during the asymmetric division of satellite cells, numb, the antagonist of notch is localized asymmetrically, indicating divergent cell fate of daughter cells, the numb + daughter cells becoming committed myoblasts expressing MRFs [42]. The Wnt pathway is activated by the binding of Wnt to its receptor composed of frizzled and low-density lipoprotein-related protein (LRP). This induces the recruitment of Dishevelled (Dvl), which releases the inhibition of glycogen synthase kinase-beta (GSKβ) on β-catenin. β-catenin then translocates to the nucleus and promotes gene transcription. While notch signaling induces satellite cell proliferation and specification, Wnt signaling allows satellite cell differentiation as illustrated by the temporal switch observed from notch to Wnt signaling during myogenesis [43].
The proliferative capacity of satellite cells also depends on muscle innervation. Experiments show that denervation of 6-day-old neonatal rat drastically reduces satellite cell proliferative capacity [44].
Other factors such as growth factors are secreted in the satellite cell microenvironment and play a critical role in their activation. In particular, the role of IGF-induced satellite cell activation during muscle regeneration has been largely reported. However, the involvement of satellite cell activation on IGF-induced muscle hypertrophy is still debated. In both cases, the mechanisms underlying IGF-induced activation of satellite cells remain poorly understood. This will be discussed in detail below.
In turn, activated satellite cells secrete different compounds involved in chemotaxis of bone marrow cells. Indeed, DNA-microarray analysis of genes expressed during regeneration revealed an increase of at least six chemotactic factors: monocyte chemoattractant protein-1, macrophage-derived chemokine (MDC), fractalkine (FKN), vascular endothelial growth factor (VEGF), urokinase type plasminogen-activator receptor (μPAR) and urokinase [26]. Interestingly, their expression was correlated with the activation of satellite cells and decreased at the differentiation phase [45, 46].
Finally, it has been shown that cells derived from lineages other than muscle also possess myogenic potential. This is the case of bone marrow-derived progenitors, mesoangioblasts, pericytes, blood-derived AC133+ progenitors, and interstitial cells that are capable to participate in the formation of multinucleated myotubes [47–49]. In all cases, all potential myogenic cells express Pax7 [50]. Indeed, the use of diphtheria toxin to eliminate Pax7-expressing cells in adult muscle results in the loss of the replenishment of satellite cell pool and muscle repair following injury [51]. The different steps of myogenesis, i.e., myoblast proliferation, migration, alignment, and fusion into myotubes are under the control of diverse regulatory factors such as Pax genes, MRFs, and myocyte enhancer binding factors 2 (Mef2), growth factors, calcium, and calpains [24, 52, 53].
Muscle remodeling
As mentioned above, muscle regeneration crucially depends on the maintenance of the basal lamina [6]. Within the intact basal lamina, satellite cell proliferation, fusion, and differentiation operate a reconstruction of normal muscle fibers in a very short period of time. These steps of muscle regeneration do not require innervation.
However, growth and remodeling of regenerated muscle depends on the development of blood vessels and on the formation of myotendinous connections and neuromuscular junction. In the absence of innervation, myofibers remain atrophic and newly formed myotubes express, by default, embryonic, neonatal, and fast MHC, as in developing muscles, but are not able to express slow MHC. Motor neuron activity is required for both muscle growth and fiber-type specification [54].
Muscle treatment with rapamycin, a specific inhibitor of the mammalian target of rapamycin (mTOR), blocks growth of regenerating muscle, suggesting that nerve activity exerts its effect on muscle growth by selectively activating the Akt–mTOR pathway. Interestingly, it does not interfere with the slow-type expression program [55].
Triggering the expression of slow-type MHC seems to involve two other signaling pathways, namely the ERK and calcineurin–NFAT pathways [56]. This illustrates that Ca2+ transients obtained by the nerve stimulation of slow-type muscle pattern more efficiently activates calcineurin than a fast-type pattern [57].
Extracellular matrix plays an important role in nerve growth. Indeed, interaction between Schwann cells and the ECM are essential for peripheral nerve regeneration and the collagen present in the ECM participates in the process by acting as a driving force in nerve growth and differentiation [58]. The remodeling of the ECM is also involved in the process of muscle repair by activating satellite cells and promoting their migration and fusion into myotubes. This happens through metalloproteinases, which allow the turnover of the ECM by cleaving its components and which participate in the activation of latent growth factors [59, 60].
MRFs pathway
MRFs are muscle-specific basic helix–loop–helix transcription factors. They include Myf5, MyoD, myogenin, and Mrf4 proteins that act as nodal points for myogenic information [61, 62]. MRFs contain a conserved basic DNA binding domain that binds to the E-box consensus sequences (CANNTG) of the promoters of muscle-specific genes [63, 64]. Myf5 and MyoD are mainly involved in muscle specification and trigger conversion of non-muscle cells, such as fibroblasts, into muscle [65, 66], whereas myogenin and Mrf4 act later during myogenesis and allow myotube formation and maturation [67–69].
In response to muscle injury, satellite cells become activated along the damaged myofibers, proliferate and migrate to the injury site plugging the breaches, or by forming new myotubes [35]. Activated satellite cells first express Myf5, which promotes their proliferation. They then start expressing MyoD that induces their withdrawn from the cell cycle [70–72].
MyoD constitutes the key myogenic transcription factor in muscle regeneration [4, 73]. Indeed, primary myogenic cells derived from adult skeletal muscle of MyoD−/− mice fail to exit the cell cycle and to enter the differentiation process. Interestingly, transfection of a MyoD-expression cassette in these cells rescues the differentiation deficit [73]. Expression of MyoD during muscle regeneration controls that of several muscle-specific genes such as developmental myosin heavy chain (MHCd, only present in regenerated fibers) and myogenin [63, 74, 75].
Expression of myogenin occurs at the beginning of myotube formation and is the key factor leading to myoblast fusion [76]. Finally, Mrf4 is involved in the maturation of myotubes allowing reorganization of myofilaments and central nuclei migration to the periphery of the cell [67] (Fig. 1). However, recent evidence points to the role of Mrf4 in skeletal muscle determination. Indeed, it has been shown that muscle formation may occur in Myf5/MyoD double-null mice if Mrf4 is expressed [77]. The effect of MRFs is potentiated by the Mef2 [78, 79]; they act synergistically with them to allow myotube formation [80, 81]. The expression of MRF family proteins is under the control of Pax genes, essentially Pax3 and Pax7. All satellite cells are marked by expression of Pax7 and in many muscles, of its paralogue Pax3. This cell population comes from a somite-derived population of Pax3/Pax7-positive cells responsible for muscle growth during development [82].
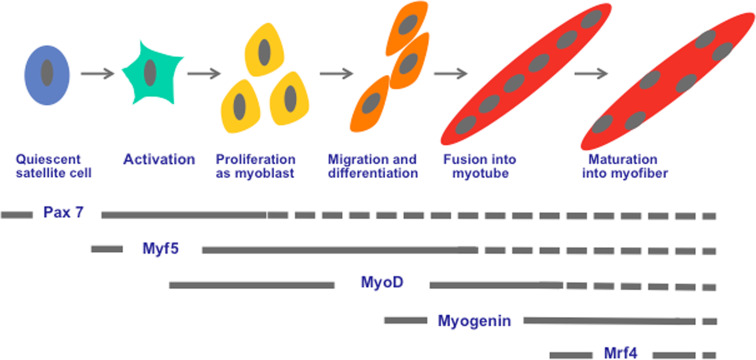
Fig. 1.
The myogenic regulatory factors pathway during skeletal muscle regeneration. Satellite cells are normally quiescent expressing Pax 7 involved in their survival. Upon muscle damage, they become activated and express Myf5 to proliferate as myoblast, then expressing MyoD. MyoD constitutes the key MRF, which regulates myoblast differentiation during muscle regeneration. Indeed, the MyoD-positive cells exit from the cell cycle and express myogenin to initiate the differentiation and fusion into myotube. These myotubes present central nuclei. The expression of Mrf4 allows maturation of myotube in myofiber that present peripheric nuclei
In developing muscles, at late embryonic and fetal stages, most of these cells proliferate and do not express skeletal markers. At this stage, Pax3 and Pax7 control both myogenic cell fate and ensure cell survival. Indeed, in Pax3/Pax7 double-mutant mice, an important increase in satellite cell apoptosis is observed [83, 84].
The situation is different in adult muscle. Many satellite cells are Pax3 negative, and Pax7 (but not Pax3) ensures cell survival. These cells enter the myogenic program by expressing two key myogenic determination genes Myf5 and MyoD. In quiescent satellite cells, Myf5 is expressed at a low level and is not under Pax control. In satellite cells depleted in Myf5, myogenesis occurs but transfection of these cells with a dominant-negative form of Pax3/Pax7 prevents the expression of myogenin and blocks differentiation [51, 77, 85]. This suggests that in adult satellite cells, Pax3 and Pax7 exert their action on myogenesis by inducing the expression of MyoD. In muscle regeneration, Pax7 seems to play an essential role in allowing the formation of myogenic progenitors. Indeed, in mutant mice expressing Pax7 carrying a knock in of a β-galactosidase cassette (Pax7lacz/lacz) but still expressing Pax3, acute regeneration (consecutive to degeneration induced by cardiotoxin injection in TA muscle) showed a severe deficit with only rare centrally nucleated fibers [86]. Inversely, it has been demonstrated that Pax7-expressing satellite cells can allow muscle repair after several bouts of acute injury [51]. Moreover, MyoD-positive cells were observed among Pax7-positive cells in regenerated muscles from wild-type mice but these cells did not express Pax3. In contrast, the mononuclear cells associated with fibers in Pax7lacz/lacz mice were all negative for MyoD, indicating an absence of functional satellite cells in Pax7lacz/lacz mice.
IGFs and other growth factors released during regeneration
Muscle injury causes the release of different growth factors in the extracellular environment. These factors may be released by damaged fibers, newly formed myotubes, inflammatory cells like macrophages, or they can come from the bloodstream. They play an important role at different steps of muscle regeneration (reviewed in [87]).
Expression of IGFs during myogenesis and muscle regeneration
Autocrine secretion of IGFs
In response to muscle injury or exercise, muscle cells express IGF-I and IGF-II [88]. IGF-I is the only growth factor clearly involved in both proliferation and differentiation of myoblasts [89–93]. Different isoforms of IGF-I generated by alternative splicing are involved in muscle regeneration: IGF-IEa and MGF (mechano-growth factor), corresponding to IGF-IEb (in rat) and IGF-IEc (in human).
MGF is only expressed by damaged or loaded skeletal muscle [94]. Following muscle damage, its expression increases quite rapidly and is correlated with the activation of satellite cells; then splicing switches expression to IGF-IEa, the sustained action of which enhances myoblasts fusion [94–96]. Moreover, autocrine secretion of IGF-I during muscle regeneration limits muscle damage and allows recruitment of bone marrow cells to the injury site [94]. Indeed, within 1 h after muscle injury, there is an invasion of neutrophils into the site of the injury, which contributes to muscle damage by releasing free radicals or other oxidants that cause a lysis of muscle cell membranes [25]. Interestingly, the expression of MGF increases the activity of superoxide dismutase, known to decrease the level of free radicals, and so limits muscle damage [97] (for review see [27]). Moreover, the chemoattractive effect of muscle satellite cells on bone marrow stem cells is enhanced by the expression of MGF during muscle regeneration [98]. This increases the number of monocytes/macrophages into the injured muscle and favors muscle repair.
On the other hand, the expression of IGF-II by regenerating muscle cell plays a role in the later step of myoblast differentiation while allowing myotube formation [99–103].
Paracrine secretion of IGFs
Paracrine secretion of IGFs is essentially assumed by two groups of bone marrow-derived cells: monocytes/macrophages, and endothelial cells that induce a rapid increase in IGF-I around damaged myofibers. The paracrine secretion of IGF-I speeds up the inflammatory response and allows muscle architecture restoration [103–106].
Endocrine secretion of IGFs
Circulating IGFs from blood vessels also take part in muscle regeneration. The isoform involved in the process is IGF-IEa. However, several studies indicate a predominant action of locally produced IGFs during myogenesis and muscle repair [72, 94, 98, 99, 101, 104].
Role of IGFs in muscle repair and hypertrophy (Fig. 2)
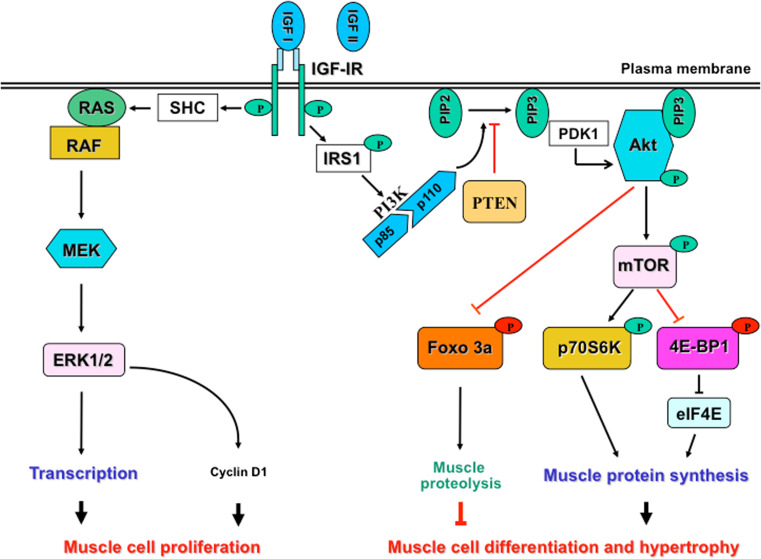
Fig. 2.
Models of IGF-induced muscle cell proliferation and differentiation. The binding of IGF-I to its receptor activates two main pathways: the MAPK pathway that allows myoblast proliferation through the activation of ERK and cyclin, and the PI3K/Akt pathway that promotes muscle cell differentiation and hypertrophy through activation of muscle protein synthesis and inhibition of muscle proteolysis. IGF-II also mediates the effects of IGF-I by its binding to the IGF-IR. These signaling pathways are detailed in the text
IGFs are involved at different steps of myogenesis and muscle regeneration: they activate satellite cell proliferation (hyperplasia), increase protein synthesis (hypertrophy), and promote differentiation [28, 93, 107–109].
IGFs and muscle cell proliferation
The mitogenic action of IGFs on muscle cell is essentially due to IGF-I and mediated by the mitogen-activated protein kinase (MAPK/ERK1/2) pathway [89, 110, 111]. MAPKs are serine/threonine kinases involved in cell cycle progression and cell survival. By altering the levels and activities of transcription factors, MAPK leads to altered transcription of genes involved in the cell cycle. For example, upon IGF-I stimulation, expression of cyclins D1 and D2 increases, accelerating cell cycle progression [111]. This pathway mediates its mitogenic activity essentially through phosphorylation of ERK1/2, allowing cell proliferation [112].
IGFs and muscle cell differentiation
As described above, IGF-I and II are both involved in muscle cell differentiation. IGF-II exerts its effect by binding to the IGFI-R. The effect of these growth factors on myoblasts differentiation in vivo is mainly mediated by the PI3K/Akt/mTOR/p70S6K pathway [72, 101, 111, 113–119].
Phosphatidylinositol 3-kinase (PI3K) contains two subunits: a catalytic subunit called p110 and a regulatory subunit called p85. IGF-I stimulation results in IGF-I receptor (IGF-IR) autophosphorylation. This leads to phosphorylation of insulin receptor substrates (IRS) on tyrosine residues and a subsequent binding of the SH2 domain of the p85 subunit of PI3K to phosphotyrosine residues that releases p110 for PI3K activation. The activation of PI3K generates PIP3 responsible for Akt targeting to the plasma membrane where it is activated through phosphorylation by its activating kinase, phosphoinositide-dependent kinase 1 (PDK1). Negative regulators of Akt are phosphatase and TENsin homolog (PTEN) and SH2-domain-containing inositol 5′ phosphate (SHIP2) that dephosphorylate PIP3 and prevents translocation of Akt to the plasma membrane and its subsequent phosphorylation [120].
Akt, also called protein kinase B (PKB), is a 65-kDa and serine/threonine kinase localized in the cytosol. Three isoforms of Akt are described: Akt 1, 2, 3. In skeletal muscle, two of them are predominant: Akt1 and Akt2. However, Akt mediated-growth effect of IGF-I in skeletal muscle has been attributed essentially to Akt1 [113, 114]. Akt2 seems more involved in muscle glucose metabolism [121]. Insulin and IGF-I stimulation leads to Akt translocation to the plasma membrane where it becomes phosphorylated on threonine 308 and serine 473 by PDK1. After phosphorylation, Akt returns to the cytosol or to the nucleus to target its downstream effectors. Akt mediates the effect of IGF-I in two ways: through anabolic effects and through anti-catabolic effects [115, 117, 122]. Akt promotes protein synthesis and cell growth principally by phosphorylating and activating mTOR and p70S6K. Inhibition of mTOR by rapamycin treatment prevents both myogenic differentiation and myofibers growth during development [123, 124]. The anti-catabolic effect of Akt is due to the inactivation of proteolysis, mediated by inhibition of Foxo3a and GSK 3 by phosphorylation. Phosphorylation of Foxo3a leads to its exclusion from the nucleus where it plays a role in the transcriptional activation of genes that negatively regulate muscle development such as MuRf-1 and Atrogin1 [also called muscle atrophy F (MAF) box] [125, 126].
IGFs and muscle cell hypertrophy
If the role of the IGF-I-induced satellite cell activation and proliferation is well reported in muscle regeneration, its involvement is less clear in muscle hypertrophy.
Using a genetically modified mouse model expressing an inducible Akt1 gene, the group of Reggiani showed that Akt increases skeletal muscle mass and force without satellite cell activation [127]. It is well known that Akt activation allows muscle protein synthesis and muscle cell differentiation [111] and that satellite cell activation occurs before myoblast differentiation. Moreover, the IGF-I pathway-induced muscle cell proliferation is essentially mediated by the MAPK pathway, which does not depend on activation of Akt [110, 111]. Thus, it is expected that muscle hypertrophy induced by Akt stimulation may be more due to a better differentiation of committed myoblasts than to an activation of quiescent satellite cells.
On the other hand, several experiments show that various models of muscle hypertrophy in which IGF-I receptor activation is required [128–130], such as overload-dependent muscle hypertrophy or follistatin-induced muscle hypertrophy, were blocked by X-ray or gamma-ray irradiation (used to inhibit satellite cell activation) [107, 131–133].
These results strongly emphasize the obligatory role of satellite cell activation during these processes, although a possible interference of irradiation with muscle protein synthesis cannot be ruled out.
The importance of satellite cell activation is also supported by the fact that MAPK activation precedes that of the PI3K/Akt pathway during overload-induced muscle hypertrophy and that, at the early phase of the process, mTORC1 activation occurs independently of PI3K/Akt signaling. This suggests a significant contribution of the MAPK pathway to mTORC1 activation [134].
Together, all these results suggest that IGF-induced muscle hypertrophy requires both MAPK and PI3K/Akt and that myoblasts proliferation may precede differentiation during the process.
Other secreted factors involved in muscle repair and hypertrophy
Besides IGFs, other secreted factors modulate the kinetics of muscle repair. The most important ones are briefly mentioned below.
Hepatocyte growth factor (HGF) is released from the extracellular matrix upon muscle injury and is secreted by newly formed myotubes. It leads quiescent satellite cells to reenter the cell cycle and promotes their proliferation by activating p38 MAPK and PI3K [135]. It also inhibits differentiation by upregulating Twist protein, an inhibitor of MyoD expression, and by repressing p27, a cyclin-dependent kinase inhibitor [136].
Platelet-derived growth factor (PDGF) is released by activated platelets and activated macrophages. It stimulates proliferation, inhibits differentiation, and promotes angiogenesis.
Fibroblast growth factor-6 (FGF-6) stimulates proliferation of satellite cells but may also interfere with the expression of MyoD and myogenin, therefore playing a role in differentiation [137].
Interleukin-6 (IL-6) and TNFα produced by activated leukocytes induce proteolysis of damaged myofibers and therefore take part in the degenerative phase of muscle regeneration [138, 139]. However, they also promote satellite cell proliferation and differentiation, favoring replacement of the destroyed tissue [140–142].
Transforming growth factor-beta (TGFβ) is a family of proteins including myostatin (MSTN) and activin/inhibin factors, which bind to the extracellular matrix and are released upon muscle injury. In vitro, they inhibit satellite cell proliferation and differentiation [143–145]. The role of myostatin in muscle growth control has been emphasized by the fact that mutation in MSTN gene causes a very impressive increase of muscle mass [145, 146].
Leukemia inhibitor factor (LIF) is produced by regenerating fibers and plays an important role in remodeling of extracellular matrix in late muscle repair [147].
Follistatin also plays a role during muscle repair and hypertrophy. Myofibers and inflammatory cells secrete it. Follistatin acts by inhibiting the effects of myostatin through its binding with a high affinity to Activin and in a lesser extent to Inhibin [148].
Progranulin (PGRN) is an emergent factor that cooperates with IGF-I in the regulation of skeletal muscle hypertrophy and regeneration. Indeed, PGRN is co-induced with IGF-I and can substitute it during muscle hypertrophy [149]. It is also involved in activation of muscle progenitor cells during muscle regeneration [149, 150].
Interplay between MRFs and IGFs pathways
As described in Fig. 3, the MRFs and IGF pathways constitute important signaling pathways that control skeletal muscle regeneration and hypertrophy [101, 151–153]. However, we only begin to understand how these pathways interact with each other [127, 154].
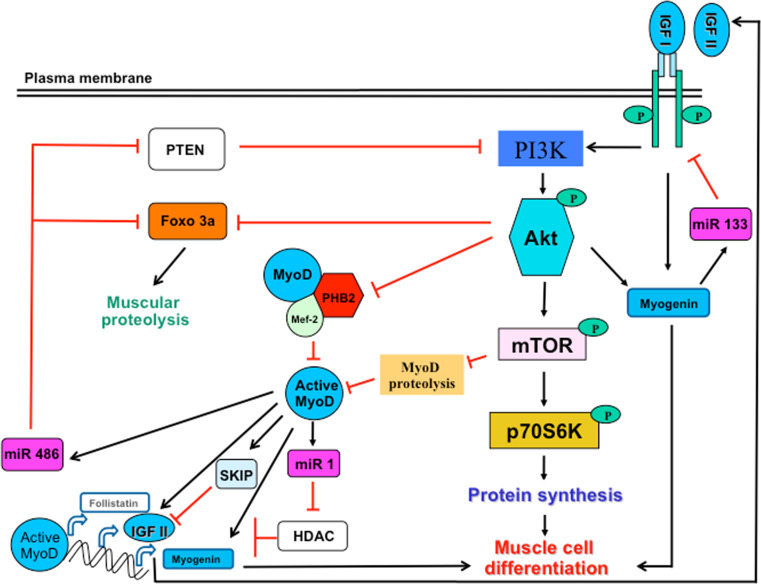
Fig. 3.
Interplay between the MRFs and IGFs pathways during myogenesis and muscle regeneration. The IGF pathway regulates MRFs expression and activity. Indeed, the activity of MyoD, the key regulator of muscle regeneration, is enhanced by the activation of the PI3K/Akt pathway in two ways: (i) interaction of active Akt with PHB2 that releases MyoD from the inhibitory effects of PHB2 (known to complex MyoD and Mef2, inhibiting their activity), (ii) inhibition of MyoD proteolysis by active mTOR, the downstream target of Akt. Active MyoD then induces the transcription of myogenin (to allow myoblast differentiation) or other muscle-specific genes and remove the inhibition of HDAC on the transcription of follistatin, which inhibits the negative regulator of myogenesis, myostatin. MRFs may also modulate the IGF pathway through different ways: (i) induction of the transcription of IGF-II (known to be involved in myoblast differentiation), (ii) inhibition of Foxo 3a that induces muscle proteolysis and PTEN known to inhibit PI3K, both regulations mediated by miR 486, (iii) slowdown in myogenesis through regulation of the expression of IGF-II by SKIP or induction of miR 133 to block IGF-R, each way constituting a negative feedback of MRFs on IGF expression and myogenesis
Induction of MRFs genes by IGFs stimulation
A series of experimental investigations indicate that the IGF pathway might regulate the MRF family proteins, in particular MyoD and myogenin.
As described above, MyoD is expressed by activated satellite cells. There is now a considerable amount of experimental evidences suggesting that IGF might regulate the protein level or the activity of MyoD during myogenesis and muscle regeneration [113, 151, 155–157]. Indeed, in C2C12 cells, IGF-I stimulation increases the transcriptional activity of MyoD [156]. Moreover, mechanical damage or bupivacaine-induced muscle injury is followed by autocrine secretion of MGF in the early phase of muscle repair. This has been shown to control MyoD expression and satellite cell activation [95, 158]. Besides, in vivo treatment of dy dystrophic mice with IGF-I improves restitution of muscle architecture and is accompanied by a higher expression of MyoD [157]. MKR mice that express a dominant negative IGF-I receptor specifically in skeletal muscle also present a marked muscle hypoplasia from birth to 3 weeks of age [159]. Interestingly, this hypoplasia is associated with a decrease in ERK immunoreactivity level and in MyoD and myogenin expression. Muscle hypoplasia has also been observed in IGF-I−/− mice [160]. Moreover, the use of inducible IGF-R knockout showed impairment of satellite cell activation and muscle regeneration [161].
All these experiments establish a probable link between IGF signaling and MRFs, in particular MyoD.
However, the mechanisms by which IGF regulates MyoD expression or activity have not been clearly established yet. Different mechanisms have been proposed.
Prohibitin 2 (PHB2) has been identified as interacting with Akt during myogenesis. PHB2 is a highly conserved and ubiquitously expressed protein, also called repressor of estrogen receptor activity (REA) localized in various intracellular compartments. It plays an important role in maintaining mitochondrial inheritance and morphology and acts as scaffolding protein at the plasma membrane [162]. Sun et al. [151] have shown that PHB2 binds to MyoD and Mef2 and acts as a transcriptional repressor of muscle cell differentiation. Indeed, stable expression of PHB2 in C2C12 inhibits myogenic differentiation. Interestingly, when co-expressed with Akt, the repressive effect of PHB2 is relieved, suggesting a possible crosstalk between Akt and PHB2. The authors identified the central region of PHB2 as interacting with the N-terminus of Akt. Thus, Akt partially reduces the binding of PHB2 to MyoD and consequently relieves its repressive effect on myogenic reporters.
Another mechanism by which IGF may control MyoD activity seems to be a mTORC1-dependent control of its stability by suppressing its proteasome-dependent degradation [163]. Interestingly, MyoD protein protection from degradation allows the expression of muscle-specific micro-RNA 1 (miR 1), which in turn releases the inhibitory effect of histone deacetylase 4 (HDAC4) on follistatin production and myocyte fusion. miRs are 23-nucleotides-long noncoding RNAs that regulate protein expression by targeting the 3′ untranslated region of messenger RNAs [164, 165]. Different miRs are expressed in skeletal muscle. Among them, miR 1 is a conserved muscle-specific miR that is essential for myogenesis. Several studies have revealed that miR-1 regulates numerous biological processes including the development of cardiac and skeletal muscle. Interestingly, repression of miR 1 in skeletal muscle impairs myoblast differentiation [166].
Finally, growing arguments support an increase in MyoD at the mRNA level upon activation of the IGF-I pathway [94, 157, 167].
Taken together, these observations show that IGF-I maintains MyoD activity by stabilizing the protein in a mTORC1-miR 1-dependent way and by increasing its transcription.
One the other hand, several reports indicate that IGF stimulation also induces myogenin expression [155, 156, 167]. For example, a significant correlation is observed between muscle hypertrophy in response to resistance training and mRNA expression of MyoD, myogenin, and IGF-I, suggesting a possible interaction between MRFs and IGF-I in the control of muscle hypertrophy during resistance training [167]. During myogenesis, myogenin expression is essentially under the control of MyoD. It is therefore expected that IGF-I that regulates MyoD expression or activity may in turn control the expression of myogenin and induce muscle differentiation. However, IGF-I may also regulate myogenin expression independently of MyoD, as the effect has been observed in L6 cells reported not to express MyoD [155]. Interestingly, three cis-elements have been identified within the 133-base pair mouse proximal promoter of myogenin, namely E-box, MEF2, and MEF3 that would be regulated by the PI3K/Akt pathway [156].
These results show that IGF-I controls myogenin expression by both MyoD-dependent and MyoD-independent processes.
If the interaction between the IGF signaling pathway and MyoD and myogenin is well established, less is known about Myf-5. Most satellite cells express Myf5 but remain quiescent. Indeed, recent data show that in these quiescent satellite cells, Myf5 mRNA is sequestered in messenger of ribonucleoprotein particles (mRNP) granules together with miR 31, which negatively regulates its translation. Upon activation, mRNP granules are dissociated, the miR 31 level is reduced, and Myf5 protein therefore accumulates [168]. Myf5 activation in these cells promotes both self-renewal and myogenic commitment [169].
So far, only a few studies have reported an induction of Myf5 by IGF [170, 171]. This point needs further investigation.
Induction of IGFs genes by MRFs
Several observations also suggest that the IGF pathway acts downstream of MRFs [101, 119, 152, 172]. The most direct demonstration was provided by Wilson et al. [101]. Indeed, in C3H 10T1/2 fibroblasts acutely converted in myoblasts by MyoD transfection, they observed an increase in the expression of IGF-II at mRNA and protein levels during the differentiation phase. Interestingly, interference with IGF-II production or repression of Akt in those cells inhibited the production of muscle-specific proteins and myoblasts fusion into myotubes. This result indicates a possible induction of IGF-II expression by MyoD, with the subsequent activation of IGF/Akt signaling during muscle cell differentiation. This is corroborated by the fact that in rat muscle cells, PI3K/Akt signaling is enhanced by MyoD, an effect related to the regulation of miR 486 expression that inhibits PTEN (a negative regulator of the PI3K/Akt pathway) and Foxo1a (involved in muscle proteolysis) [101, 152].
Moreover, recent findings indicate a negative regulation of autocrine expression of IGF-II by the skeletal muscle-and kidney-enriched inositol polyphosphate phosphatase (SKIP) and a subsequent decrease of the PI3K/Akt/mTOR pathway during C2C12 myoblasts differentiation [172]. SKIP is known to be a negative regulator of insulin-dependent Akt activation in skeletal muscle through inhibition of the expression of IGF-II. Indeed, repression of SKIP increases IGF-II mRNA and muscle differentiation but co-transfection of SKIP with IGF-II siRNA blocks muscle differentiation [172–174]. SKIP expression is lower at the earlier stages of differentiation and increases at the end of differentiation under the control of MyoD. MyoD-induced SKIP expression could therefore be a way of limiting muscle differentiation through an auto-regulation loop mechanism of the IGF-II/PI3K/Akt/mTOR. It is also known that myogenin, the expression of which is increased by IGF-I stimulation, transactivates miR 133 and negatively regulates IGF-IR expression. This constitutes another negative feedback loop [175].
IGF-I expression might also be under the control of MRFs during myogenesis. Indeed, it has been shown that a mutation in the MRFs binding domain E-box of IGF-1 DNA specifically reduces the expression of IGF-I mRNA during myogenesis [176].
Conclusions
MRFs and IGFs constitute key regulatory factors mediating skeletal muscle growth and regeneration. It has long been known that these factors interact but the molecular mechanisms involved are not completely elucidated. In this review, we report many observations supporting the regulation of MyoD and myogenin by IGFs and inversely. However, the precise mechanisms by which IGFs induce the switch of satellite cells from quiescence to activation are not completely understood. A deeper knowledge of these mechanisms would open new therapeutic horizons for the management of muscle loss related to aging or to dystrophies.
Acknowledgments
Our research is supported by the “Association française contre les myopathies” (AFM), the “Association belge contre les maladies neuro-musculaires” (ABMM), the “Fonds national de la recherche scientifique” (FNRS, Belgium), by grant ARC 10/15-029 from the General Direction of Scientific Research of the French Community of Belgium and by the Interuniversity Attraction Poles program initiated by the Belgian Science Policy Office.
References
- 1.Schmalbruch H, Lewis DM. Dynamics of nuclei of muscle fibers and connective tissue cells in normal and denervated rat muscles. Muscle Nerve. 2000;23:617–626. doi: 10.1002/(sici)1097-4598(200004)23:4<617::aid-mus22>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 2.Irintchev A, Wernig A. Muscle damage and repair in voluntarily running mice: strain and muscle differences. Cell Tissue Res. 1987;249:509–521. doi: 10.1007/BF00217322. [DOI] [PubMed] [Google Scholar]
- 3.Papadimitriou JM, Robertson TA, Mitchell CA, Grounds MD. The process of new plasmalemma formation in focally injured skeletal muscle fibers. J Struct Biol. 1990;103:124–134. doi: 10.1016/1047-8477(90)90016-6. [DOI] [PubMed] [Google Scholar]
- 4.Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 5.Carlson BM, Faulkner JA. The regeneration of skeletal muscle fibers following injury: a review. Med Sci Sports Exerc. 1983;15:187–198. [PubMed] [Google Scholar]
- 6.Ciciliot S, Schiaffino S. Regeneration of mammalian skeletal muscle. Basic mechanisms and clinical implications. Curr Pharm Des. 2010;16:906–914. doi: 10.2174/138161210790883453. [DOI] [PubMed] [Google Scholar]
- 7.Czerwinska AM, Streminska W, Ciemerych MA, Grabowska I. Mouse gastrocnemius muscle regeneration after mechanical or cardiotoxin injury. Folia Histochem Cytobiol. 2012;50:144–153. doi: 10.2478/18710. [DOI] [PubMed] [Google Scholar]
- 8.Sakamoto K, Nosaka K, Shimegi S, Ohmori H, Katsuta S. Creatine kinase release from regenerated muscles after eccentric contractions in rats. Eur J Appl Physiol Occup Physiol. 1996;73:516–520. doi: 10.1007/BF00357673. [DOI] [PubMed] [Google Scholar]
- 9.Jackson RC. Exercise-induced renal failure and muscle damage. Proc R Soc Med. 1970;63:566–570. [PMC free article] [PubMed] [Google Scholar]
- 10.Darr KC, Schultz E. Exercise-induced satellite cell activation in growing and mature skeletal muscle. J Appl Physiol. 1987;63:1816–1821. doi: 10.1152/jappl.1987.63.5.1816. [DOI] [PubMed] [Google Scholar]
- 11.Tsatalas T, Giakas G, Spyropoulos G, Sideris V, Lazaridis S, Kotzamanidis C, Koutedakis Y. The effects of eccentric exercise-induced muscle damage on running kinematics at different speeds. J Sports Sci. 2013;31:288–298. doi: 10.1080/02640414.2012.729135. [DOI] [PubMed] [Google Scholar]
- 12.Vandebrouck C, Martin D, Colson-Van Schoor M, Debaix H, Gailly P. Involvement of TRPC in the abnormal calcium influx observed in dystrophic (mdx) mouse skeletal muscle fibers. J Cell Biol. 2002;158:1089–1096. doi: 10.1083/jcb.200203091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gailly P. TRP channels in normal and dystrophic skeletal muscle. Curr Opin Pharmacol. 2012;12(3):326–334. doi: 10.1016/j.coph.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 14.De Backer F, Vandebrouck C, Gailly P, Gillis JM. Long-term study of Ca2+ homeostasis and of survival in collagenase-isolated muscle fibres from normal and mdx mice. J Physiol Lond. 2002;542:855–865. doi: 10.1113/jphysiol.2002.020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gailly P, De Backer F, Van Schoor M, Gillis JM. In situ measurements of calpain activity in isolated muscle fibres from normal and dystrophin-lacking mdx mice. J Physiol. 2007;582:1261–1275. doi: 10.1113/jphysiol.2007.132191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monaco AP, Neve RL, Collettifeener C, Bertelson CJ, Kurnit DM, Kunkel LM. Isolation of candidate cDNAs for portions of the Duchenne muscular-dystrophy gene. Nature. 1986;323:646–650. doi: 10.1038/323646a0. [DOI] [PubMed] [Google Scholar]
- 17.Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 18.Moens P, Baatsen PHWW, Marechal G. Increased susceptibility of EDL muscles from mdx mice to damage-induced by contractions with stretch. J Muscle Res Cell Motil. 1993;14:446–451. doi: 10.1007/BF00121296. [DOI] [PubMed] [Google Scholar]
- 19.Petrof BJ, Shrager JB, Stedman HH, Kelly AM, Sweeney HL. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc Natl Acad Sci USA. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zanou N, Iwata Y, Schakman O, Lebacq J, Wakabayashi S, Gailly P. Essential role of TRPV2 ion channel in the sensitivity of dystrophic muscle to eccentric contractions. FEBS Lett. 2009;583:3600–3604. doi: 10.1016/j.febslet.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 21.Ducret T, Vandebrouck C, Cao ML, Lebacq J, Gailly P. Functional role of store-operated and stretch-activated channels in murine adult skeletal muscle fibres. J Physiol. 2006;575:913–924. doi: 10.1113/jphysiol.2006.115154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiu D, Wang HH, Blumenthal MR. Creatine phosphokinase release as a measure of tourniquet effect on skeletal muscle. Arch Surg. 1976;111:71–74. doi: 10.1001/archsurg.1976.01360190073013. [DOI] [PubMed] [Google Scholar]
- 23.Park CY, Pierce SA, von Drehle M, Ivey KN, Morgan JA, Blau HM, Srivastava D. skNAC, a smyd1-interacting transcription factor, is involved in cardiac development and skeletal muscle growth and regeneration. Proc Natl Acad Sci USA. 2010;107:20750–20755. doi: 10.1073/pnas.1013493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zanou N, Schakman O, Louis P, Ruegg UT, Dietrich A, Birnbaumer L, Gailly P. Trpc1 channel modulates PI3K/Akt pathway during myoblast differentiation and muscle regeneration. J Biol Chem. 2012;287(18):14524–14534. doi: 10.1074/jbc.M112.341784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tidball JG. Inflammatory processes in muscle injury and repair. Am J Physiol Regul Integr Comp Physiol. 2005;288:R345–R353. doi: 10.1152/ajpregu.00454.2004. [DOI] [PubMed] [Google Scholar]
- 26.Chazaud B, et al. Satellite cells attract monocytes and use macrophages as a support to escape apoptosis and enhance muscle growth. J Cell Biol. 2003;163:1133–1143. doi: 10.1083/jcb.200212046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mourkioti F, Rosenthal N. IGF-1, inflammation and stem cells: interactions during muscle regeneration. Trends Immunol. 2005;26:535–542. doi: 10.1016/j.it.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Grounds MD. Reasons for the degeneration of ageing skeletal muscle: a central role for IGF-1 signalling. Biogerontology. 2002;3:19–24. doi: 10.1023/a:1015234709314. [DOI] [PubMed] [Google Scholar]
- 29.Cantini M, Carraro U. Macrophage-released factor stimulates selectively myogenic cells in primary muscle culture. J Neuropathol Exp Neurol. 1995;54:121–128. doi: 10.1097/00005072-199501000-00014. [DOI] [PubMed] [Google Scholar]
- 30.Cantini M, et al. Macrophage-secreted myogenic factors: a promising tool for greatly enhancing the proliferative capacity of myoblasts in vitro and in vivo. Neurol Sci. 2002;23:189–194. doi: 10.1007/s100720200060. [DOI] [PubMed] [Google Scholar]
- 31.Sonnet C, et al. Human macrophages rescue myoblasts and myotubes from apoptosis through a set of adhesion molecular systems. J Cell Sci. 2006;119:2497–2507. doi: 10.1242/jcs.02988. [DOI] [PubMed] [Google Scholar]
- 32.Segawa M, et al. Suppression of macrophage functions impairs skeletal muscle regeneration with severe fibrosis. Exp Cell Res. 2008;314:3232–3244. doi: 10.1016/j.yexcr.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 33.Beauchamp JR, et al. Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J Cell Biol. 2000;151:1221–1234. doi: 10.1083/jcb.151.6.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 35.Schultz E, Jaryszak DL, Valliere CR. Response of satellite cells to focal skeletal muscle injury. Muscle Nerve. 1985;8:217–222. doi: 10.1002/mus.880080307. [DOI] [PubMed] [Google Scholar]
- 36.Seale P, Polesskaya A, Rudnicki MA. Adult stem cell specification by Wnt signaling in muscle regeneration. Cell Cycle. 2003;2:418–419. [PubMed] [Google Scholar]
- 37.Parker MH, Seale P, Rudnicki MA. Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat Rev Genet. 2003;4:497–507. doi: 10.1038/nrg1109. [DOI] [PubMed] [Google Scholar]
- 38.Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinin V, Gayraud-Morel B, Gomes D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 40.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 41.Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 42.Conboy IM, Rando TA. The regulation of notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- 43.Brack AS, Conboy IM, Conboy MJ, Shen J, Rando TA. A temporal switch from notch to Wnt signaling in muscle stem cells is necessary for normal adult myogenesis. Cell Stem Cell. 2008;2:50–59. doi: 10.1016/j.stem.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 44.Kelly AM. Satellite cells and myofiber growth in the rat soleus and extensor digitorum longus muscles. Dev Biol. 1978;65:1–10. doi: 10.1016/0012-1606(78)90174-4. [DOI] [PubMed] [Google Scholar]
- 45.Chen YW, Nader GA, Baar KR, Fedele MJ, Hoffman EP, Esser KA. Response of rat muscle to acute resistance exercise defined by transcriptional and translational profiling. J Physiol. 2002;545:27–41. doi: 10.1113/jphysiol.2002.021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barash IA, Mathew L, Ryan AF, Chen J, Lieber RL. Rapid muscle-specific gene expression changes after a single bout of eccentric contractions in the mouse. Am J Physiol Cell Physiol. 2004;286:C355–C364. doi: 10.1152/ajpcell.00211.2003. [DOI] [PubMed] [Google Scholar]
- 47.Ferrari G, Cusella-De Angelis G, Coletta M, Paolucci E, Stornaiuolo A, Cossu G, Mavilio F. Muscle regeneration by bone marrow-derived myogenic progenitors. Science. 1998;279:1528–1530. doi: 10.1126/science.279.5356.1528. [DOI] [PubMed] [Google Scholar]
- 48.Gussoni E, Soneoka Y, Strickland CD, Buzney EA, Khan MK, Flint AF, Kunkel LM, Mulligan RC. Dystrophin expression in the mdx mouse restored by stem cell transplantation. Nature. 1999;401:390–394. doi: 10.1038/43919. [DOI] [PubMed] [Google Scholar]
- 49.Torrente Y, et al. Human circulating AC133(+) stem cells restore dystrophin expression and ameliorate function in dystrophic skeletal muscle. J Clin Invest. 2004;114:182–195. doi: 10.1172/JCI20325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuang S, Rudnicki MA. The emerging biology of satellite cells and their therapeutic potential. Trends Mol Med. 2008;14:82–91. doi: 10.1016/j.molmed.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 51.Lepper C, Partridge TA, Fan CM. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138:3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Louis M, Zanou N, Van Schoor M, Gailly P. TRPC1 regulates skeletal myoblast migration and differentiation. J Cell Sci. 2008;121:3951–3959. doi: 10.1242/jcs.037218. [DOI] [PubMed] [Google Scholar]
- 53.Zanou N, et al. Role of TRPC1 channel in skeletal muscle function. Am J Physiol Cell Physiol. 2010;298:C149–C162. doi: 10.1152/ajpcell.00241.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jerkovic R, Argentini C, Serrano-Sanchez A, Cordonnier C, Schiaffino S. Early myosin switching induced by nerve activity in regenerating slow skeletal muscle. Cell Struct Funct. 1997;22:147–153. doi: 10.1247/csf.22.147. [DOI] [PubMed] [Google Scholar]
- 55.Murgia M, Serrano AL, Calabria E, Pallafacchina G, Lomo T, Schiaffino S. Ras is involved in nerve-activity-dependent regulation of muscle genes. Nat Cell Biol. 2000;2:142–147. doi: 10.1038/35004013. [DOI] [PubMed] [Google Scholar]
- 56.Serrano AL, Murgia M, Pallafacchina G, Calabria E, Coniglio P, Lomo T, Schiaffino S. Calcineurin controls nerve activity-dependent specification of slow skeletal muscle fibers but not muscle growth. Proc Natl Acad Sci USA. 2001;98:13108–13113. doi: 10.1073/pnas.231148598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li L, Stefan MI, Le Novere N. Calcium input frequency, duration and amplitude differentially modulate the relative activation of calcineurin and CaMKII. PLoS One. 2012;7:e43810. doi: 10.1371/journal.pone.0043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu HM. The role of extracellular matrix in peripheral nerve regeneration: a wound chamber study. Acta Neuropathol. 1992;83:469–474. doi: 10.1007/BF00310022. [DOI] [PubMed] [Google Scholar]
- 59.Saksela O, Laiho M. Growth factors and the extracellular matrix. Duodecim. 1990;106:297–306. [PubMed] [Google Scholar]
- 60.Lijnen HR, Van Hoef B, Lupu F, Moons L, Carmeliet P, Collen D. Function of the plasminogen/plasmin and matrix metalloproteinase systems after vascular injury in mice with targeted inactivation of fibrinolytic system genes. Arterioscler Thromb Vasc Biol. 1998;18:1035–1045. doi: 10.1161/01.atv.18.7.1035. [DOI] [PubMed] [Google Scholar]
- 61.Weintraub H, et al. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 62.Sassoon D, Lyons G, Wright WE, Lin V, Lassar A, Weintraub H, Buckingham M. Expression of two myogenic regulatory factors myogenin and MyoD1 during mouse embryogenesis. Nature. 1989;341:303–307. doi: 10.1038/341303a0. [DOI] [PubMed] [Google Scholar]
- 63.Davis RL, Cheng PF, Lassar AB, Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990;60:733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- 64.Le Grand F, Rudnicki MA. Skeletal muscle satellite cells and adult myogenesis. Curr Opin Cell Biol. 2007;19:628–633. doi: 10.1016/j.ceb.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choi J, Costa ML, Mermelstein CS, Chagas C, Holtzer S, Holtzer H. MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proc Natl Acad Sci USA. 1990;87:7988–7992. doi: 10.1073/pnas.87.20.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 67.Megeney LA, Rudnicki MA. Determination versus differentiation and the MyoD family of transcription factors. Biochem Cell Biol. 1995;73:723–732. doi: 10.1139/o95-080. [DOI] [PubMed] [Google Scholar]
- 68.Rudnicki MA, Jaenisch R. The MyoD family of transcription factors and skeletal myogenesis. BioEssays. 1995;17:203–209. doi: 10.1002/bies.950170306. [DOI] [PubMed] [Google Scholar]
- 69.Buckingham M, Houzelstein D, Lyons G, Ontell M, Ott MO, Sassoon D. Expression of muscle genes in the mouse embryo. Symp Soc Exp Biol. 1992;46:203–217. [PubMed] [Google Scholar]
- 70.Crescenzi M, Fleming TP, Lassar AB, Weintraub H, Aaronson SA. MyoD induces growth arrest independent of differentiation in normal and transformed cells. Proc Natl Acad Sci USA. 1990;87:8442–8446. doi: 10.1073/pnas.87.21.8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Halevy O, Novitch BG, Spicer DB, Skapek SX, Rhee J, Hannon GJ, Beach D, Lassar AB. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 72.Wilson EM, Rotwein P. Control of MyoD function during initiation of muscle differentiation by an autocrine signaling pathway activated by insulin-like growth factor-II. J Biol Chem. 2006;281:29962–29971. doi: 10.1074/jbc.M605445200. [DOI] [PubMed] [Google Scholar]
- 73.Sabourin LA, Girgis-Gabardo A, Seale P, Asakura A, Rudnicki MA. Reduced differentiation potential of primary MyoD−/− myogenic cells derived from adult skeletal muscle. J Cell Biol. 1999;144:631–643. doi: 10.1083/jcb.144.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beylkin DH, Allen DL, Leinwand LA. MyoD, Myf5, and the calcineurin pathway activate the developmental myosin heavy chain genes. Dev Biol. 2006;294:541–553. doi: 10.1016/j.ydbio.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 75.Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell. 2002;9:587–600. doi: 10.1016/s1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- 76.Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- 77.Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5: MyoD double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- 78.Weintraub H. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- 79.Buckingham M. Skeletal muscle formation in vertebrates. Curr Opin Genet Dev. 2001;11:440–448. doi: 10.1016/s0959-437x(00)00215-x. [DOI] [PubMed] [Google Scholar]
- 80.Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 81.Molkentin JD, Olson EN. Combinatorial control of muscle development by basic helix–loop–helix and MADS-box transcription factors. Proc Natl Acad Sci USA. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Buckingham M. Skeletal muscle progenitor cells and the role of Pax genes. C R Biol. 2007;330:530–533. doi: 10.1016/j.crvi.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 83.Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- 84.Buckingham M, Bajard L, Daubas P, Esner M, Lagha M, Relaix F, Rocancourt D. Myogenic progenitor cells in the mouse embryo are marked by the expression of Pax3/7 genes that regulate their survival and myogenic potential. Anat Embryol (Berl) 2006;211(Suppl 1):51–56. doi: 10.1007/s00429-006-0122-0. [DOI] [PubMed] [Google Scholar]
- 85.Buckingham ME. Muscle: the regulation of myogenesis. Curr Opin Genet Dev. 1994;4:745–751. doi: 10.1016/0959-437x(94)90142-p. [DOI] [PubMed] [Google Scholar]
- 86.Kuang S, Charge SB, Seale P, Huh M, Rudnicki MA. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J Cell Biol. 2006;172:103–113. doi: 10.1083/jcb.200508001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Karalaki M, Fili S, Philippou A, Koutsilieris M. Muscle regeneration: cellular and molecular events. In Vivo. 2009;23:779–796. [PubMed] [Google Scholar]
- 88.Jiao S, Ren H, Li Y, Zhou J, Duan C, Lu L. Differential regulation of IGF-I and IGF-II gene expression in skeletal muscle cells. Mol Cell Biochem. 2013;373:107–113. doi: 10.1007/s11010-012-1479-4. [DOI] [PubMed] [Google Scholar]
- 89.Florini JR, Ewton DZ, Coolican SA. Growth hormone and the insulin-like growth factor system in myogenesis. Endocr Rev. 1996;17:481–517. doi: 10.1210/edrv-17-5-481. [DOI] [PubMed] [Google Scholar]
- 90.Engert JC, Berglund EB, Rosenthal N. Proliferation precedes differentiation in IGF-I-stimulated myogenesis. J Cell Biol. 1996;135:431–440. doi: 10.1083/jcb.135.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Allen RE, Boxhorn LK. Regulation of skeletal muscle satellite cell proliferation and differentiation by transforming growth factor-beta, insulin-like growth factor I, and fibroblast growth factor. J Cell Physiol. 1989;138:311–315. doi: 10.1002/jcp.1041380213. [DOI] [PubMed] [Google Scholar]
- 92.Doumit ME, Cook DR, Merkel RA. Fibroblast growth factor, epidermal growth factor, insulin-like growth factors, and platelet-derived growth factor-BB stimulate proliferation of clonally derived porcine myogenic satellite cells. J Cell Physiol. 1993;157:326–332. doi: 10.1002/jcp.1041570216. [DOI] [PubMed] [Google Scholar]
- 93.Vandenburgh HH, Karlisch P, Shansky J, Feldstein R. Insulin and IGF-I induce pronounced hypertrophy of skeletal myofibers in tissue culture. Am J Physiol. 1991;260:C475–C484. doi: 10.1152/ajpcell.1991.260.3.C475. [DOI] [PubMed] [Google Scholar]
- 94.Owino V, Yang SY, Goldspink G. Age-related loss of skeletal muscle function and the inability to express the autocrine form of insulin-like growth factor-1 (MGF) in response to mechanical overload. FEBS Lett. 2001;505:259–263. doi: 10.1016/s0014-5793(01)02825-3. [DOI] [PubMed] [Google Scholar]
- 95.Hill M, Goldspink G. Expression and splicing of the insulin-like growth factor gene in rodent muscle is associated with muscle satellite (stem) cell activation following local tissue damage. J Physiol. 2003;549:409–418. doi: 10.1113/jphysiol.2002.035832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang SY, Goldspink G. Different roles of the IGF-I Ec peptide (MGF) and mature IGF-I in myoblast proliferation and differentiation. FEBS Lett. 2002;522:156–160. doi: 10.1016/s0014-5793(02)02918-6. [DOI] [PubMed] [Google Scholar]
- 97.Dobrowolny G, et al. Muscle expression of a local Igf-1 isoform protects motor neurons in an ALS mouse model. J Cell Biol. 2005;168:193–199. doi: 10.1083/jcb.200407021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Musaro A, et al. Stem cell-mediated muscle regeneration is enhanced by local isoform of insulin-like growth factor 1. Proc Natl Acad Sci USA. 2004;101:1206–1210. doi: 10.1073/pnas.0303792101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Florini JR, Magri KA, Ewton DZ, James PL, Grindstaff K, Rotwein PS. “Spontaneous” differentiation of skeletal myoblasts is dependent upon autocrine secretion of insulin-like growth factor-II. J Biol Chem. 1991;266:15917–15923. [PubMed] [Google Scholar]
- 100.Ge Y, Sun Y, Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J Cell Biol. 2011;192:69–81. doi: 10.1083/jcb.201007165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wilson EM, Hsieh MM, Rotwein P. Autocrine growth factor signaling by insulin-like growth factor-II mediates MyoD-stimulated myocyte maturation. J Biol Chem. 2003;278:41109–41113. doi: 10.1074/jbc.C300299200. [DOI] [PubMed] [Google Scholar]
- 102.Ge Y, et al. mTOR regulates skeletal muscle regeneration in vivo through kinase-dependent and kinase-independent mechanisms. Am J Physiol Cell Physiol. 2009;297:C1434–C1444. doi: 10.1152/ajpcell.00248.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Levinovitz A, Jennische E, Oldfors A, Edwall D, Norstedt G. Activation of insulin-like growth factor II expression during skeletal muscle regeneration in the rat: correlation with myotube formation. Mol Endocrinol. 1992;6:1227–1234. doi: 10.1210/mend.6.8.1406701. [DOI] [PubMed] [Google Scholar]
- 104.Edwall D, Schalling M, Jennische E, Norstedt G. Induction of insulin-like growth factor I messenger ribonucleic acid during regeneration of rat skeletal muscle. Endocrinology. 1989;124:820–825. doi: 10.1210/endo-124-2-820. [DOI] [PubMed] [Google Scholar]
- 105.Huard J, Li Y, Fu FH. Muscle injuries and repair: current trends in research. J Bone Jt Surg Am. 2002;84-A:822–832. [PubMed] [Google Scholar]
- 106.Barton ER, Morris L, Musaro A, Rosenthal N, Sweeney HL. Muscle-specific expression of insulin-like growth factor I counters muscle decline in mdx mice. J Cell Biol. 2002;157:137–148. doi: 10.1083/jcb.200108071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Barton-Davis ER, Shoturma DI, Sweeney HL. Contribution of satellite cells to IGF-I induced hypertrophy of skeletal muscle. Acta Physiol Scand. 1999;167:301–305. doi: 10.1046/j.1365-201x.1999.00618.x. [DOI] [PubMed] [Google Scholar]
- 108.Semsarian C, Sutrave P, Richmond DR, Graham RM. Insulin-like growth factor (IGF-I) induces myotube hypertrophy associated with an increase in anaerobic glycolysis in a clonal skeletal-muscle cell model. Biochem J. 1999;339(Pt 2):443–451. [PMC free article] [PubMed] [Google Scholar]
- 109.Musaro A, et al. Localized Igf-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nat Genet. 2001;27:195–200. doi: 10.1038/84839. [DOI] [PubMed] [Google Scholar]
- 110.Clemmons DR. Role of IGF-I in skeletal muscle mass maintenance. Trends Endocrinol Metab. 2009;20:349–356. doi: 10.1016/j.tem.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 111.Coolican SA, Samuel DS, Ewton DZ, McWade FJ, Florini JR. The mitogenic and myogenic actions of insulin-like growth factors utilize distinct signaling pathways. J Biol Chem. 1997;272:6653–6662. doi: 10.1074/jbc.272.10.6653. [DOI] [PubMed] [Google Scholar]
- 112.Fuentes EN, Bjornsson BT, Valdes JA, Einarsdottir IE, Lorca B, Alvarez M, Molina A. IGF-I/PI3K/Akt and IGF-I/MAPK/ERK pathways in vivo in skeletal muscle are regulated by nutrition and contribute to somatic growth in the fine flounder. Am J Physiol Regul Integr Comp Physiol. 2011;300:R1532–R1542. doi: 10.1152/ajpregu.00535.2010. [DOI] [PubMed] [Google Scholar]
- 113.Wilson EM, Rotwein P. Selective control of skeletal muscle differentiation by Akt1. J Biol Chem. 2007;282:5106–5110. doi: 10.1074/jbc.C600315200. [DOI] [PubMed] [Google Scholar]
- 114.Rotwein P, Wilson EM. Distinct actions of Akt1 and Akt2 in skeletal muscle differentiation. J Cell Physiol. 2009;219:503–511. doi: 10.1002/jcp.21692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 116.Bodine SC, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 117.Jiang BH, Aoki M, Zheng JZ, Li J, Vogt PK. Myogenic signaling of phosphatidylinositol 3-kinase requires the serine–threonine kinase Akt/protein kinase B. Proc Natl Acad Sci USA. 1999;96:2077–2081. doi: 10.1073/pnas.96.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sasai N, Agata N, Inoue-Miyazu M, Kawakami K, Kobayashi K, Sokabe M, Hayakawa K. Involvement of PI3K/Akt/TOR pathway in stretch-induced hypertrophy of myotubes. Muscle Nerve. 2010;41:100–106. doi: 10.1002/mus.21473. [DOI] [PubMed] [Google Scholar]
- 119.Wilson EM, Tureckova J, Rotwein P. Permissive roles of phosphatidyl inositol 3-kinase and Akt in skeletal myocyte maturation. Mol Biol Cell. 2004;15:497–505. doi: 10.1091/mbc.E03-05-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Leevers SJ, Vanhaesebroeck B, Waterfield MD. Signalling through phosphoinositide 3-kinases: the lipids take centre stage. Curr Opin Cell Biol. 1999;11:219–225. doi: 10.1016/s0955-0674(99)80029-5. [DOI] [PubMed] [Google Scholar]
- 121.Cleasby ME, Reinten TA, Cooney GJ, James DE, Kraegen EW. Functional studies of Akt isoform specificity in skeletal muscle in vivo; maintained insulin sensitivity despite reduced insulin receptor substrate-1 expression. Mol Endocrinol. 2007;21:215–228. doi: 10.1210/me.2006-0154. [DOI] [PubMed] [Google Scholar]
- 122.Glass DJ. PI3 kinase regulation of skeletal muscle hypertrophy and atrophy phosphoinositide 3-kinase in health and disease. Curr Top Microbiol Immunol. 2010;346:267–278. doi: 10.1007/82_2010_78. [DOI] [PubMed] [Google Scholar]
- 123.Park IH, Erbay E, Nuzzi P, Chen J. Skeletal myocyte hypertrophy requires mTOR kinase activity and S6K1. Exp Cell Res. 2005;309:211–219. doi: 10.1016/j.yexcr.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 124.Willett M, Cowan JL, Vlasak M, Coldwell MJ, Morley SJ. Inhibition of mammalian target of rapamycin (mTOR) signalling in C2C12 myoblasts prevents myogenic differentiation without affecting the hyperphosphorylation of 4E-BP1. Cell Signal. 2009;21:1504–1512. doi: 10.1016/j.cellsig.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 125.Hribal ML, Nakae J, Kitamura T, Shutter JR, Accili D. Regulation of insulin-like growth factor-dependent myoblast differentiation by Foxo forkhead transcription factors. J Cell Biol. 2003;162:535–541. doi: 10.1083/jcb.200212107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Glass DJ. Skeletal muscle hypertrophy and atrophy signaling pathways. Int J Biochem Cell Biol. 2005;37:1974–1984. doi: 10.1016/j.biocel.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 127.Blaauw B, et al. Inducible activation of Akt increases skeletal muscle mass and force without satellite cell activation. FASEB J. 2009;23:3896–3905. doi: 10.1096/fj.09-131870. [DOI] [PubMed] [Google Scholar]
- 128.Kalista S, Schakman O, Gilson H, Lause P, Demeulder B, Bertrand L, Pende M, Thissen JP. The type 1 insulin-like growth factor receptor (IGF-IR) pathway is mandatory for the follistatin-induced skeletal muscle hypertrophy. Endocrinology. 2012;153:241–253. doi: 10.1210/en.2011-1687. [DOI] [PubMed] [Google Scholar]
- 129.Adams GR, McCue SA. Localized infusion of IGF-I results in skeletal muscle hypertrophy in rats. J Appl Physiol. 1998;84:1716–1722. doi: 10.1152/jappl.1998.84.5.1716. [DOI] [PubMed] [Google Scholar]
- 130.Awede B, Thissen J, Gailly P, Lebacq J. Regulation of IGF-I, IGFBP-4 and IGFBP-5 gene expression by loading in mouse skeletal muscle. FEBS Lett. 1999;461:263–267. doi: 10.1016/s0014-5793(99)01469-6. [DOI] [PubMed] [Google Scholar]
- 131.Rosenblatt JD, Yong D, Parry DJ. Satellite cell activity is required for hypertrophy of overloaded adult rat muscle. Muscle Nerve. 1994;17:608–613. doi: 10.1002/mus.880170607. [DOI] [PubMed] [Google Scholar]
- 132.Li P, Akimoto T, Zhang M, Williams RS, Yan Z. Resident stem cells are not required for exercise-induced fiber-type switching and angiogenesis but are necessary for activity-dependent muscle growth. Am J Physiol Cell Physiol. 2006;290:C1461–C1468. doi: 10.1152/ajpcell.00532.2005. [DOI] [PubMed] [Google Scholar]
- 133.Gilson H, Schakman O, Kalista S, Lause P, Tsuchida K, Thissen JP. Follistatin induces muscle hypertrophy through satellite cell proliferation and inhibition of both myostatin and activin. Am J Physiol Endocrinol Metab. 2009;297:E157–E164. doi: 10.1152/ajpendo.00193.2009. [DOI] [PubMed] [Google Scholar]
- 134.Miyazaki M, McCarthy JJ, Fedele MJ, Esser KA. Early activation of mTORC1 signalling in response to mechanical overload is independent of phosphoinositide 3-kinase/Akt signalling. J Physiol. 2011;589:1831–1846. doi: 10.1113/jphysiol.2011.205658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shi X, Garry DJ. Muscle stem cells in development, regeneration, and disease. Genes Dev. 2006;20:1692–1708. doi: 10.1101/gad.1419406. [DOI] [PubMed] [Google Scholar]
- 136.Leshem Y, Spicer DB, Gal-Levi R, Halevy O. Hepatocyte growth factor (HGF) inhibits skeletal muscle cell differentiation: a role for the bHLH protein twist and the cdk inhibitor p27. J Cell Physiol. 2000;184:101–109. doi: 10.1002/(SICI)1097-4652(200007)184:1<101::AID-JCP11>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 137.Floss T, Arnold HH, Braun T. A role for FGF-6 in skeletal muscle regeneration. Genes Dev. 1997;11:2040–2051. doi: 10.1101/gad.11.16.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fujita J, Tsujinaka T, Yano M, Ogawa J, Morita T, Taniguchi H, Shiozaki H, Monden M. Participation of interleukin-6 to skeletal muscle proteolysis: the effect of IL-6 administration on mRNA expression by the skeletal muscle cell proteolytic system. Nihon Geka Gakkai Zasshi. 1998;99:332. [PubMed] [Google Scholar]
- 139.Langen RC, Van Der Velden JL, Schols AM, Kelders MC, Wouters EF, Janssen-Heininger YM. Tumor necrosis factor-alpha inhibits myogenic differentiation through MyoD protein destabilization. FASEB J. 2004;18:227–237. doi: 10.1096/fj.03-0251com. [DOI] [PubMed] [Google Scholar]
- 140.Warren GL, Hulderman T, Jensen N, McKinstry M, Mishra M, Luster MI, Simeonova PP. Physiological role of tumor necrosis factor alpha in traumatic muscle injury. FASEB J. 2002;16:1630–1632. doi: 10.1096/fj.02-0187fje. [DOI] [PubMed] [Google Scholar]
- 141.Alvarez B, Quinn LS, Busquets S, Quiles MT, Lopez-Soriano FJ, Argiles JM. Tumor necrosis factor-alpha exerts interleukin-6-dependent and -independent effects on cultured skeletal muscle cells. Biochim Biophys Acta. 2002;1542:66–72. doi: 10.1016/s0167-4889(01)00167-7. [DOI] [PubMed] [Google Scholar]
- 142.Baeza-Raja B, Munoz-Canoves P. p38 MAPK-induced nuclear factor-kappaB activity is required for skeletal muscle differentiation: role of interleukin-6. Mol Biol Cell. 2004;15:2013–2026. doi: 10.1091/mbc.E03-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Florini JR, Ewton DZ, Magri KA. Hormones, growth factors, and myogenic differentiation. Annu Rev Physiol. 1991;53:201–216. doi: 10.1146/annurev.ph.53.030191.001221. [DOI] [PubMed] [Google Scholar]
- 144.Langley B, Thomas M, Bishop A, Sharma M, Gilmour S, Kambadur R. Myostatin inhibits myoblast differentiation by down-regulating MyoD expression. J Biol Chem. 2002;277:49831–49840. doi: 10.1074/jbc.M204291200. [DOI] [PubMed] [Google Scholar]
- 145.McFarlane C, et al. Human myostatin negatively regulates human myoblast growth and differentiation. Am J Physiol Cell Physiol. 2011;301:C195–C203. doi: 10.1152/ajpcell.00012.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Grobet L, et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet. 1997;17:71–74. doi: 10.1038/ng0997-71. [DOI] [PubMed] [Google Scholar]
- 147.White JD, Davies M, Grounds MD. Leukaemia inhibitory factor increases myoblast replication and survival and affects extracellular matrix production: combined in vivo and in vitro studies in post-natal skeletal muscle. Cell Tissue Res. 2001;306:129–141. doi: 10.1007/s004410100432. [DOI] [PubMed] [Google Scholar]
- 148.Phillips DJ, de Kretser DM, Pfeffer A, Chie WN, Moore LG. Follistatin has a biphasic response but follicle-stimulating hormone is unchanged during an inflammatory episode in growing lambs. J Endocrinol. 1998;156:77–82. doi: 10.1677/joe.0.1560077. [DOI] [PubMed] [Google Scholar]
- 149.Hu SY, Tai CC, Li YH, Wu JL. Progranulin compensates for blocked IGF-1 signaling to promote myotube hypertrophy in C2C12 myoblasts via the PI3K/Akt/mTOR pathway. FEBS Lett. 2012;586:3485–3492. doi: 10.1016/j.febslet.2012.07.077. [DOI] [PubMed] [Google Scholar]
- 150.Li YH, et al. Progranulin regulates zebrafish muscle growth and regeneration through maintaining the pool of myogenic progenitor cells. Sci Rep. 2013;3:1176. doi: 10.1038/srep01176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sun L, Liu L, Yang XJ, Wu Z. Akt binds prohibitin 2 and relieves its repression of MyoD and muscle differentiation. J Cell Sci. 2004;117:3021–3029. doi: 10.1242/jcs.01142. [DOI] [PubMed] [Google Scholar]
- 152.Small EM, O’Rourke JR, Moresi V, Sutherland LB, McAnally J, Gerard RD, Richardson JA, Olson EN. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc Natl Acad Sci USA. 2010;107:4218–4223. doi: 10.1073/pnas.1000300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Schultz E, Jaryszak DL. Effects of skeletal muscle regeneration on the proliferation potential of satellite cells. Mech Ageing Dev. 1985;30:63–72. doi: 10.1016/0047-6374(85)90059-4. [DOI] [PubMed] [Google Scholar]
- 154.Miyabara EH, et al. Mammalian target of rapamycin complex 1 is involved in differentiation of regenerating myofibers in vivo. Muscle Nerve. 2010;42:778–787. doi: 10.1002/mus.21754. [DOI] [PubMed] [Google Scholar]
- 155.Florini JR, Ewton DZ, Roof SL. Insulin-like growth factor-I stimulates terminal myogenic differentiation by induction of myogenin gene expression. Mol Endocrinol. 1991;5:718–724. doi: 10.1210/mend-5-5-718. [DOI] [PubMed] [Google Scholar]
- 156.Xu Q, Wu Z. The insulin-like growth factor-phosphatidylinositol 3-kinase-Akt signaling pathway regulates myogenin expression in normal myogenic cells but not in rhabdomyosarcoma-derived RD cells. J Biol Chem. 2000;275:36750–36757. doi: 10.1074/jbc.M005030200. [DOI] [PubMed] [Google Scholar]
- 157.Hsu HH, Zdanowicz MM, Agarwal VR, Speiser PW. Expression of myogenic regulatory factors in normal and dystrophic mice: effects of IGF-1 treatment. Biochem Mol Med. 1997;60:142–148. doi: 10.1006/bmme.1997.2570. [DOI] [PubMed] [Google Scholar]
- 158.Imanaka M, et al. Growth hormone stimulates mechano growth factor expression and activates myoblast transformation in C2C12 cells. Kobe J Med Sci. 2008;54:E46–E54. [PubMed] [Google Scholar]
- 159.Fernandez AM, Dupont J, Farrar RP, Lee S, Stannard B, Le Roith D. Muscle-specific inactivation of the IGF-I receptor induces compensatory hyperplasia in skeletal muscle. J Clin Invest. 2002;109:347–355. doi: 10.1172/JCI13503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Miyake M, et al. Myostatin and MyoD family expression in skeletal muscle of IGF-1 knockout mice. Cell Biol Int. 2007;31:1274–1279. doi: 10.1016/j.cellbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 161.Zhang L, Wang XH, Wang H, Du J, Mitch WE. Satellite cell dysfunction and impaired IGF-1 signaling cause CKD-induced muscle atrophy. J Am Soc Nephrol. 2010;21:419–427. doi: 10.1681/ASN.2009060571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nijtmans LG, et al. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 2000;19:2444–2451. doi: 10.1093/emboj/19.11.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sun Y, Ge Y, Drnevich J, Zhao Y, Band M, Chen J. Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. J Cell Biol. 2010;189:1157–1169. doi: 10.1083/jcb.200912093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 165.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Aguiar AF, Vechetti-Junior IJ, Alves de Souza RW, Castan EP, Milanezi-Aguiar RC, Padovani CR, Carvalho RF, Silva MD (2012) Myogenin, MyoD and IGF-I regulate muscle mass but not fiber-type conversion during resistance training in rats. Int J Sports Med (Epub ahead of print) [DOI] [PubMed]
- 168.Crist CG, Montarras D, Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell. 2012;11:118–126. doi: 10.1016/j.stem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 169.Gayraud-Morel B, et al. Myf5 haploinsufficiency reveals distinct cell fate potentials for adult skeletal muscle stem cells. J Cell Sci. 2012;125:1738–1749. doi: 10.1242/jcs.097006. [DOI] [PubMed] [Google Scholar]
- 170.Mangiacapra FJ, Roof SL, Ewton DZ, Florini JR. Paradoxical decrease in myf-5 messenger RNA levels during induction of myogenic differentiation by insulin-like growth factors. Mol Endocrinol. 1992;6:2038–2044. doi: 10.1210/mend.6.12.1337140. [DOI] [PubMed] [Google Scholar]
- 171.Perez-Ruiz A, Gnocchi VF, Zammit PS. Control of Myf5 activation in adult skeletal myonuclei requires ERK signalling. Cell Signal. 2007;19:1671–1680. doi: 10.1016/j.cellsig.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 172.Ijuin T, Takenawa T. Role of phosphatidylinositol 3,4,5-trisphosphate (PIP3) 5-phosphatase skeletal muscle- and kidney-enriched inositol polyphosphate phosphatase (SKIP) in myoblast differentiation. J Biol Chem. 2012;287:31330–31341. doi: 10.1074/jbc.M112.388785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ijuin T, Takenawa T. SKIP negatively regulates insulin-induced GLUT4 translocation and membrane ruffle formation. Mol Cell Biol. 2003;23:1209–1220. doi: 10.1128/MCB.23.4.1209-1220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ijuin T, Yu YE, Mizutani K, Pao A, Tateya S, Tamori Y, Bradley A, Takenawa T. Increased insulin action in SKIP heterozygous knockout mice. Mol Cell Biol. 2008;28:5184–5195. doi: 10.1128/MCB.01990-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Huang MB, Xu H, Xie SJ, Zhou H, Qu LH. Insulin-like growth factor-1 receptor is regulated by microRNA-133 during skeletal myogenesis. PLoS One. 2011;6:e29173. doi: 10.1371/journal.pone.0029173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.McLellan AS, Kealey T, Langlands K. An E box in the exon 1 promoter regulates insulin-like growth factor-I expression in differentiating muscle cells. Am J Physiol Cell Physiol. 2006;291:C300–C307. doi: 10.1152/ajpcell.00345.2005. [DOI] [PubMed] [Google Scholar]