Abstract
Reduced levels of survival of motoneuron (SMN) protein lead to spinal muscular atrophy, but it is still unknown how SMN protects motoneurons in the spinal cord against degeneration. In the nucleus, SMN is associated with two types of nuclear bodies denoted as gems and Cajal bodies (CBs). The 23 kDa isoform of fibroblast growth factor-2 (FGF-223) is a nuclear protein that binds to SMN and destabilizes the SMN-Gemin2 complex. In the present study, we show that FGF-223 depletes SMN from CBs without affecting their general structure. FRAP analysis of SMN-EGFP in CBs demonstrated that the majority of SMN in CBs remained mobile and allowed quantification of fast, slow and immobile nuclear SMN populations. The potential for SMN release was confirmed by in vivo photoconversion of SMN-Dendra2, indicating that CBs concentrate immobile SMN that could have a specialized function in CBs. FGF-223 accelerated SMN release from CBs, accompanied by a conversion of immobile SMN into a mobile population. Furthermore, FGF-223 caused snRNP accumulation in CBs. We propose a model in which Cajal bodies store immobile SMN that can be mobilized by its nuclear interaction partner FGF-223, leading to U4 snRNP accumulation in CBs, indicating a role for immobile SMN in tri-snRNP assembly.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-012-1242-8) contains supplementary material, which is available to authorized users.
Keywords: FGF signaling, Fluorescence recovery after photobleaching, FRAP, Cajal body, Nuclear gem, snRNP
Introduction
The survival of motoneuron protein (SMN) is found ubiquitously in the cytoplasm and nucleus of mammalian cells [1, 2] where it binds to or is associated with many proteins mainly involved in pre-mRNA splicing regulation [3, 4]. Deletion or mutation of the SMN1 gene is linked to spinal muscular atrophy (SMA) [5], a disease that results in the loss of motoneurons in the anterior horn of the spinal cord [6]. Only low levels of full-length (FL-SMN) protein are expressed by the human SMN2 gene [7]. However, a higher number of SMN2 gene copies leads to higher FL-SMN levels and milder phenotypes of the disease [8, 9]. The number of SMN-containing nuclear bodies correlates negatively with the severity of SMA [10]. In the nucleus, SMN is concentrated in two kinds of nuclear structures: Cajal bodies (CBs) and gems [1]. These are separate structures in fetal tissues, and their colocalization increases during ontogenesis [11]. CBs were discovered by Santiago Ramon y Cajal and originally termed “nucleolar accessory bodies” [12, 13]. These subnuclear organelles differ in size from less than 0.2 up to 2 μm or larger [14]. CBs are very dynamic structures that can move together and fuse or split into smaller bodies. Their size and number vary during the cell cycle, and they can differ in their specific composition and possible biological roles [14–20]. CBs play a role in the transport and maturation of small nuclear ribonucleoprotein particles (snRNPs) [21, 22]. Spliceosomal snRNPs consist of one small nuclear RNA (snRNA), a ring of seven Sm or Like-Sm (LSm) proteins and several other proteins, and they are named by the snRNA they contain: U1, U2, U3 U4, U5 and U6 [23]. SnRNPs are assembled in the cytoplasm where SMN plays an essential role in the step-wise and ordered ring formation of Sm proteins B/B′, D3, D1, D2, E, F and G [24]. After nuclear import [25], snRNPs are targeted to CBs by SMN [16] because of direct interaction with coilin [26, 27], which is a marker protein for CBs [28]. CBs are assumed to be the place where intermediate U4/U6 di-snRNPs and mature U4/U6•U5 tri-snRNPs are formed, and where U4/U6•U5 tri-snRNP release occurs [18]. The U4/U6•U5 tri-snRNP complex is part of the spliceosome, which is formed by ordered interaction of the tri-snRNP, U1 U2 and U3 snRNPs and numerous other proteins [23]. Gems are nuclear bodies devoid of coilin; however, they contain SMN and its interacting protein Gemin2 [1]. The interaction of SMN with Gemin2 is important for the stability of nuclear gems [4]. We have previously shown that gem formation is affected by a 23-kDa isoform of fibroblast growth factor-2 (FGF-223) because of competition with Gemin2 for binding to SMN [29]. As a consequence of the interaction and colocalization of FGF-223 with SMN [30, 31], the number of gems was decreased in HEK293T cells as well as in motoneurons of transgenic FGF-2 overexpressing mice. In contrast, the total number of CBs was not affected [29].
Distinct FGF-2 isoforms are expressed by alternative initiated translation of FGF-2 mRNA; the 18-kDa low-molecular-weight isoform FGF-218 is translated from a conventional AUG start codon, rodent 21- and 23-kDa (FGF-223) high-molecular-weight isoforms from upstream CUG start codons [32]. FGF-223 is translocated to the nucleus via a nuclear localization signal (NLS) along with its nuclear receptor, fibroblast growth factor receptor 1 (FGFR1). The nuclear uptake of FGFR1 was shown to be mediated by importin-β [33]. Subsequently, nuclear FGFR1/FGF-2 is suggested to promote neuronal differentiation by activation of a pathway designated as integrative nuclear FGFR1 signaling (INFS) [34–36]. Interestingly, the pheochromocytoma (PC12) cells overexpressing high-molecular weight isoforms of FGF-2 showed a stabilization of the endocrine phenotype while FGF-218 overexpressing cells started to differentiate toward the neuronal phenotype [37]. These results indicate differential biological roles of FGF-2 isoforms.
In the present study, we focused on the influence of FGF-223 on the dynamics of SMN at the CB. On a cellular level, we showed that mobility of SMN at CBs was increased by FGF-223. This is mechanistically regulated by a competition of coilin with FGF-223 for binding to SMN. These results demonstrate that FGF-223 promotes the release of SMN from its immobile pool in the CB and further defines the function of this nuclear growth factor. Furthermore, we could show that FGF-223 causes an accumulation of U4 snRNPs in CBs. Besides the function of targeting snRNPs to Cajal bodies [18], immobile SMN appears to have a role in tri-snRNP assembly, since a decrease of immobile SMN leads to an abnormal accumulation of snRNPs.
Results
Localization of SMN to Cajal bodies and gems
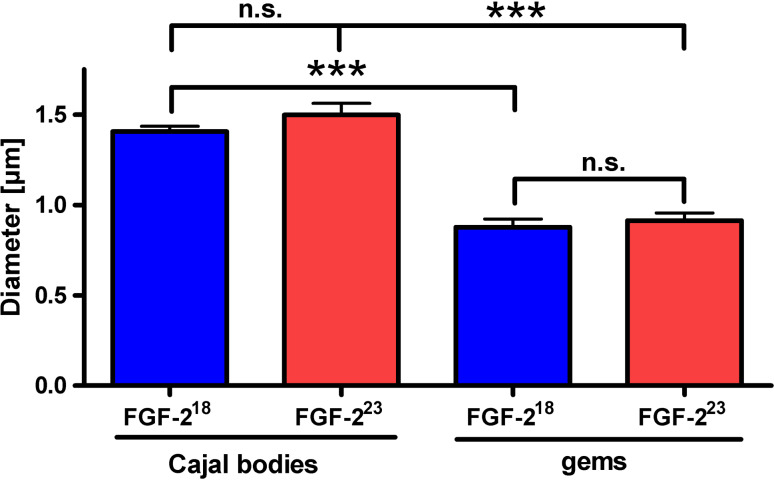
CBs and nuclear gems are present in most tissues [2]. However, unlike CBs, gems lack coilin, fibrillarin and spliceosomal snRNPs [1]. To monitor FGF-2-dependent SMN dynamics in and out of CBs of living cells, we first validated particle size as a reliable method to distinguish between CBs and gems under conditions where we coexpressed SMN-EGFP with FGF-218-DsRed2 or FGF-223-DsRed2. Cells were transfected with the respective plasmids and stained with anti-coilin antibody to determine the differences in the size of the coilin-positive CBs and coilin-negative gems, as well as the influence of FGF-223 on their dimensions (Fig. 1). We avoided CB labeling by overexpressed coilin since it could potentially form abnormal CBs. As expected, the fluorescent-tagged SMN localized to coilin-positive and -negative puncta; however, the diameters of CBs were significantly larger than those of gems. Cells overexpressing FGF-218 or FGF-223 did not display differences in the sizes of CBs or gems. Therefore, CB size could be used as a reliable parameter for their identification without the need of tagged coilin coexpression. This parameter was used in the subsequent live cell imaging experiments.
Fig. 1.
Different size of Cajal bodies and gems in HEK cells. HEK293T cells were cotransfected with pSMN-EGFP and pFGF-223-DsRed2 or, as a control, pFGF-218-DsRed2 24 h before image acquisition. Subsequently, the diameters of coilin-positive (FGF-218: n = 52; FGF-223: n = 24) and coilin-negative (FGF-218: n = 19, FGF-223: n = 16) nuclear bodies were determined. The diameters of SMN- and coilin-positive nuclear bodies (CBs) were significantly larger than nuclear bodies that were only SMN positive (gems). The overexpression of FGF-223 showed no influence on the size of nuclear bodies compared to FGF-218 overexpressing cells (means ± SEM; ***p < 0.001; n.s. non-significant; Mann-Whitney test)
Dynamic change of SMN localization to Cajal bodies
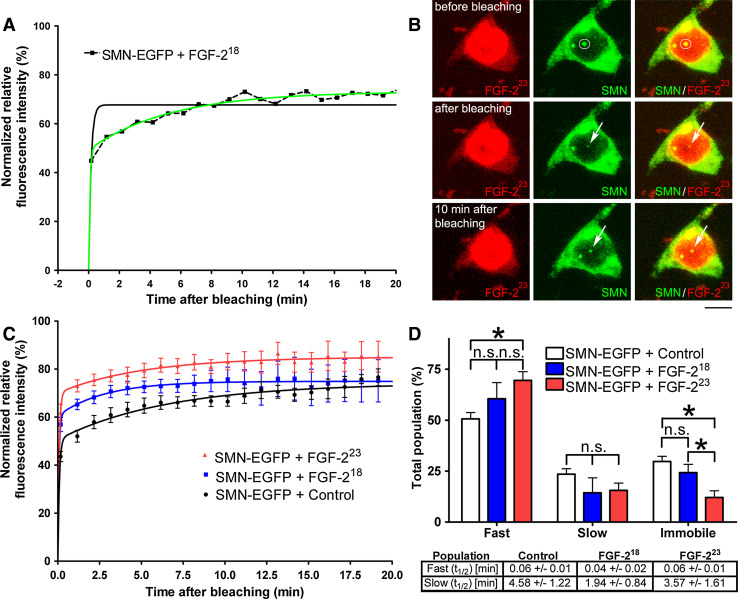
While FGF-223 does not change the quantity of CBs, in contrast to gems [29], we wanted to determine the effect of nuclear FGF-2 on SMN dynamics in and out of CBs by fluorescence recovery after photobleaching (FRAP). Nuclear bodies with a diameter larger than 1.5 μm, indicative of CBs as described above, were chosen for live cell FRAP experiments after cotransfection of SMN-EGFP and FGF-223-DsRed2 (or FGF-218-DsRed2 as a non-SMN-binding control or DsRed2 as a second control) to measure the mobility of SMN localized to CBs. One day after transfection, the largest single CB of a nucleus was bleached, and fluorescence intensity was measured every minute up to 20 min. Since CBs are mobile structures [14–16], several z-sections were scanned unless the bleached nuclear body left the volume under observation (Fig. 2b). Data were fitted to one- and two-exponential recovery kinetics (Fig. 2a). SMN-EGFP showed the best fit for two-exponential recovery kinetics (Fig. 2a), indicating the presence of at least two mobile populations of SMN-EGFP. Fractions of hypermobile (fast) and hypomobile (slow) populations can be derived from the exponential coefficients F slow and F fast of the two-exponential equation describing the best fit. Fast and slow populations are defined by half-life times t1/2 derived from the exponential constants (Fig. 2d). The immobile fraction is represented by the difference between the experimental plateau phase (reached after approximately 10 min) and the relative fluorescence intensity before bleaching (Fig. 2a, b). A significant influence of FGF-223 on the fluorescence recovery of SMN-EGFP was identified after fitting a two-exponential curve to the averaged FRAP measurements (Fig. 2c). The immobile population of SMN-EGFP localized to CBs was significantly decreased in cells transfected with the SMN-binding FGF-223 isoform compared to cells transfected with the control SMN-nonbinding FGF-218 and to a second control where we overexpressed no FGF-2 isoform at all (Fig. 2d). The increase in the fast-moving mobile population of SMN-EGFP in FGF-223-transfected cells was significantly increased compared to the control without FGF-2 overexpression (Fig. 2d). The slow SMN-EGFP population showed no statistically significant change (Fig. 2d). The change of immobile to mobile SMN indicates increased SMN dynamics and shortened average residence time at CBs.
Fig. 2.
FRAP reveals mobilization of SMN from Cajal bodies by FGF-223. For live cell imaging, single CBs of pSMN-EGFP and pFGF-223-DsRed2 or pFGF-218-DsRed2 or pDsRed2 coexpressing HEK293T cells were bleached for 1 s (405 nm). Fluorescence recovery was measured 10 s before and 10 s after photobleaching and then every minute up to 20 min unless the focused nuclear body left the volume as defined by z-sections. a Example of a single FRAP measurement. Fitting of a one-exponential curve is represented by the black line; fitting of a two-exponential curve is shown by the green line. Data display best fit to a two-exponential model. b Example of a SMN-EGFP and FGF-223-DsRed2 coexpressing cell before and after photobleaching. Scale bar 10 μm. c Summary kinetics of averaged FRAP measurements (n = 10). FGF-223-DsRed2 increased the fluorescence recovery of SMN-EGFP to CBs. d The immobile populations of SMN-EGFP was significantly decreased after overexpression of FGF-223-DsRed2. The fast population of SMN-EGFP was significantly increased compared to the DsRed2 control. The slow populations were not significantly altered. The recovery halftimes (t 1/2) of the slow and fast populations of SMN-EGFP were non-significantly changed after overexpression of FGF-223-DsRed2 (means ± SEM; *p < 0.05; n.s. non-significant; unpaired, two-tailed t test)
Photoconversion of Dendra2-tagged SMN confirms an FGF-223-dependent increase of SMN mobility
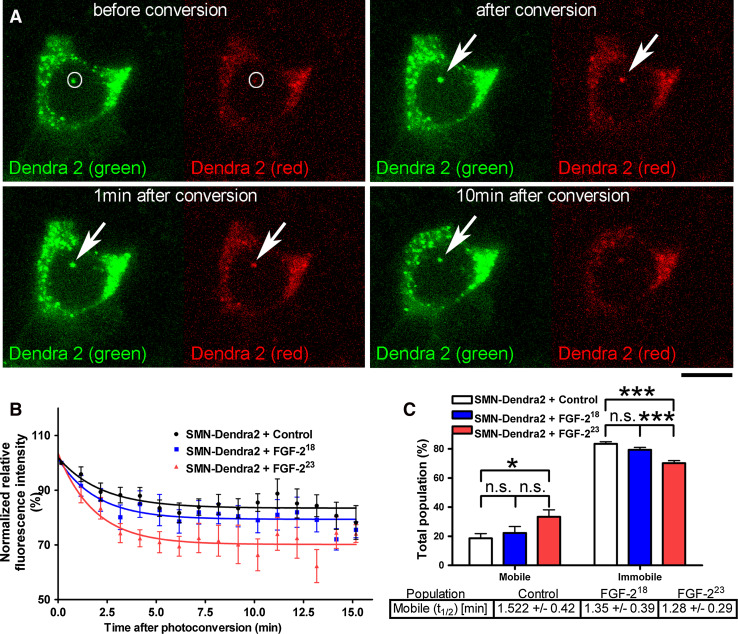
In addition to the FRAP experiments, we analyzed the mobility of SMN at CBs with an independent method employing the photoswitchable protein Dendra2. Dendra2 is an Anthozoa-derived green-to-red photoconvertible fluorescent protein, activated by a 405-nm laser line [38]. We used SMN-Dendra2 to measure the FGF-223-induced loss of SMN from CBs in live cell imaging. HEK293T cells were cotransfected with pSMN-Dendra2 and with pFGF-223-ECFP or controls pFGF-218-ECFP or pECFP. After photoconversion of SMN-Dendra2 in CBs, the loss of red fluorescence was measured for up to 20 min in the CBs (Fig. 3a). For statistical analyses, only the first 16 min was used, as the majority of the nuclear bodies did not remain in the observed volume for 20 min. However, the plateau phase was already reached after approximately 10 min, similar to the FRAP experiments. Fluorescence after photoconversion was normalized to 100 % and the data fitted to one-exponential curves. Similar to the FRAP analyses, the photoconversion experiments demonstrated the influence of FGF-223 on SMN mobility at CBs (Fig. 3b). The rate of fluorescence loss was increased after overexpression of FGF-223 compared to the SMN non-binding FGF-218. Before activation of the 405-nm laser, Dendra2 was already partially photoconverted, when the efficacy of transfection was assessed using laser lines 491 and 552 nm prior to the measurements. For this reason, the plateau of red fluorescence after photoconversion remained above 60 %, and, consequently, the calculated immobile fraction appeared larger to the same degree. In FGF-223-transfected cells, a significant decrease of the immobile population was observed while the mobile population was significantly increased compared to the control without FGF-2 expression (Fig. 3c). Both FRAP and photoconversion data demonstrate FGF-223-induced increase in SMN mobility consistent with the release of SMN from the CB coilin complexes.
Fig. 3.
Photoconversion of Dendra2-tagged SMN confirms mobilization of SMN by FGF-223. Live cell photoconversion experiments were performed using HEK293T cells coexpressing pSMN-Dendra2 and pFGF-223-ECFP or pFGF-218-ECFP or pECFP. SMN-Dendra2 in CBs was photoconverted for 1 s (405 nm). Fluorescence loss was measured 10 s before, 10 s after conversion and then every minute up to 20 min unless the nuclear body left the defined volume. a Example of a SMN-Dendra2 and FGF-223-ECFP (not shown) coexpressing cell before and after photoconversion. Scale bar 10 μm. b Averaged photoconversion measurements were fitted to a one-exponential curve and statistically analyzed (n = 10). Coexpression of FGF-223-ECFP increased the loss of red fluorescence of SMN-Dendra2 from large nuclear bodies after photoconversion. c The immobile population was significantly decreased after overexpression of FGF-223-ECFP; the mobile population of SMN-EGFP was significantly increased compared to the ECFP control. The slow populations were not significantly altered. The recovery halftimes (t 1/2) of the mobile population of SMN-EGFP were non-significantly changed after overexpression of FGF-223-ECFP (means ± SEM; *p < 0.05; ***p < 0.001; n.s. non-significant; unpaired two tailed t test)
To determine whether FGF-223 can influence the CB movement through interaction with SMN, we used HEK293T cells cotransfected with pSMN-EGFP and pFGF-223-DsRed2 or control pFGF-218-DsRed2 and measured velocities of CBs up to 20 min in a defined volume of nuclei. The distribution of velocities showed no differences under the influence of FGF-223 (Supplemental Fig. 1). Also, the size of nuclear bodies showed no correlation to its speed in the nucleus (data not shown).
FGF-223 reduces SMN binding to coilin
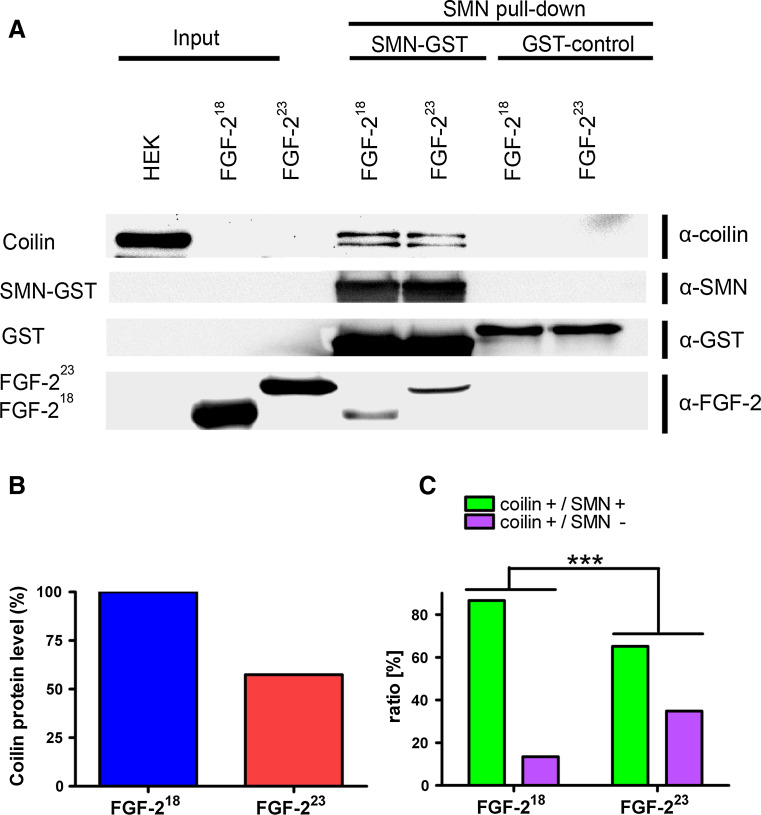
The FRAP and photoconversion studies indicated that the mobility of SMN proteins at CBs is influenced by its binding partner FGF-223. To determine if this may involve a release of SMN from the CB component coilin, we performed pulldown experiments. Recombinantly expressed human FL-SMN tagged with GST at its C-terminus was immobilized on glutathione magnetic beads and incubated with recombinant FGF-223 or FGF-218 as a control, respectively, and lysates of HEK293T cells. After the pulldown, endogenous coilin was detected on Western blots in both lysates but not in the GST controls (Fig. 4a) and analyzed densitometrically (Fig. 4b). Importantly, the coilin protein levels bound to the SMN beads were decreased to ~50 % in FGF-223 incubated pulldowns compared to non SMN-binding FGF-218 control. This indicates a capability of FGF-223 to displace coilin from SMN. To verify this in vivo, we transfected HEK293T cells with either pFGF-223-DsRed2 or control pFGF-218-DsRed2 fusion constructs (Fig. 4d1–e1”) and quantified SMN labeling of coilin-positive nuclear bodies (CBs) (Fig. 4d2–e4”). Thereby, the influence of FGF-2 isoforms on SMN depletion from CBs could be analyzed on a cellular level. Indeed, FGF-223 significantly increased the fraction of SMN-negative CBs ~30 % compared to ~10 % in the FGF-218 control (Fig. 4c–e4”), which is consistent with a displacement model fully supporting our pulldown experiments (Fig. 4b).
Fig. 4.
FGF-223 reduces SMN binding to coilin. a Endogenous coilin was purified from lysates of HEK293T cells using recombinantly expressed FGF-223, FGF-218 and GST fusion of SMN coupled to glutathione beads (SMN-GST) and analyzed by Western blot. GST was used as a control (GST control). FGF-223 decreased binding of coilin to SMN-GST. Because of the high amounts of FGF-218 used, a slight unspecific binding of FGF-218 to SMN-GST was found. After SMN-GST pulldown, also fragments of coilin (anti-coilin) and fragments of SMN-GST (anti-GST) were detected. b The level of endogenous, SMN-bound coilin was decreased in lysates of cells with FGF-223 (densitometrical analysis of the highest band of coilin). c To determine changes of coilin-SMN association in CBs in vivo, HEK293T cells were transfected with pFGF-223-DsRed2 (d–d’) or pFGF-218-DsRed2 (as a control, e–e”). Cells were fixed and immunostained with anti-coilin (d3–e3”) and anti-SMN (d2–e2”). Nuclei were analyzed by counting SMN-positive or -negative CBs (FGF-218: n = 119, FGF-223: n = 132) as demonstrated in the merged pictures (d4–e4”). The distribution of SMN-positive (yellow arrows) and -negative (red arrows) CBs was significantly changed after overexpression of FGF-223-DsRed2 (c, ***p < 0.001, χ 2 test)
We have previously shown that the number of SMN-positive nuclear bodies was decreased in motoneurons of FGF-2 transgenic mice, overexpressing all isoforms of FGF-2 [29]. In this study, we counted SMN-positive and -negative CBs in motoneurons of FGF-2 knockout mice. As expected, no change was detected. We concluded that the FGF-2/SMN interaction has a physiological role only under conditions where endogenous FGF-2 becomes upregulated. Previous studies have shown that this occurs, e.g., after lesions of peripheral nerves [39–41].
FGF-223-induced depletion of immobile SMN in Cajal bodies leads to snRNP accumulation
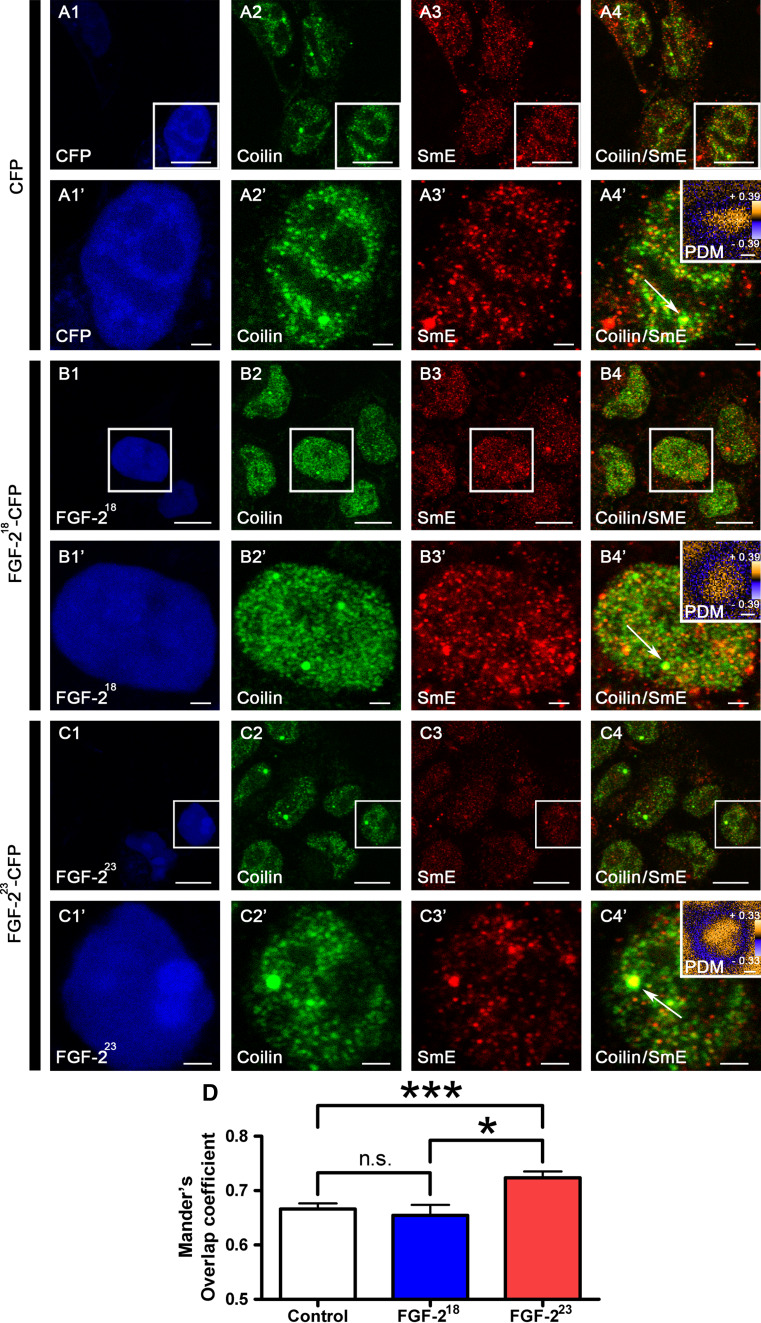
Biophotonic and pulldown experiments demonstrated an influence of FGF-223 on immobilized SMN at Cajal bodies. To further elucidate the function of this population of SMN at CBs, we transfected HEK293T cells with pFGF-223-ECFP or as controls pFGF-218-ECFP or pECFP and performed immunocytochemistry with anti-coilin antibody to detect CBs and anti-SmE antibodies to detect snRNPs (Fig. 5). Intensity correlation analyses at single CBs illustrated a colocalization of snRNPs with CBs (Fig. 5 product of differences from the mean, PDM images). After overexpression of FGF-223, which depleted the immobile SMN, we found a significantly increased accumulation of snRNPs at CBs compared to non-SMN binding FGF-218 and to the non-FGF-2 control (Fig. 5d).
Fig. 5.
SnRNPs accumulate at CBs after reduction of immobile SMN by FGF-223. HEK293T cells were transfected with pFGF-223-ECFP (c1, c1’) or as controls pFGF-218-ECFP (b1, b1’) or pECFP (a1, a1’) 24 h before fixation. Cells were fixed and immunostained with anti-coilin (a2–c2’) and anti-SmE (a3–c3’). Coilin and SmE showed colocalization in the nucleus as demonstrated in the merged images (a4–c4’) primarily in CBs. Arrows point to CBs that were enlarged. Intensity correlation analysis demonstrated in PDM (product of differences from the mean) images (a4’, b4’, c4’; LUT: orange, positive PDM; blue, negative PDM) showed a significant increase in colocalization of coilin and SmE in CBs after overexpression of pFGF-223-ECFP (M means ± SEM; *p < 0.05, ***p < 0.001; Mann-Whitney test; CFP: n = 45, FGF-218: n = 45, FGF-223: n = 45, pooled from 3 independent experiments). Scale bar 10 μm (a, b, c), 2 μm (a’, b’, c’), 0.2 μm (PDM images)
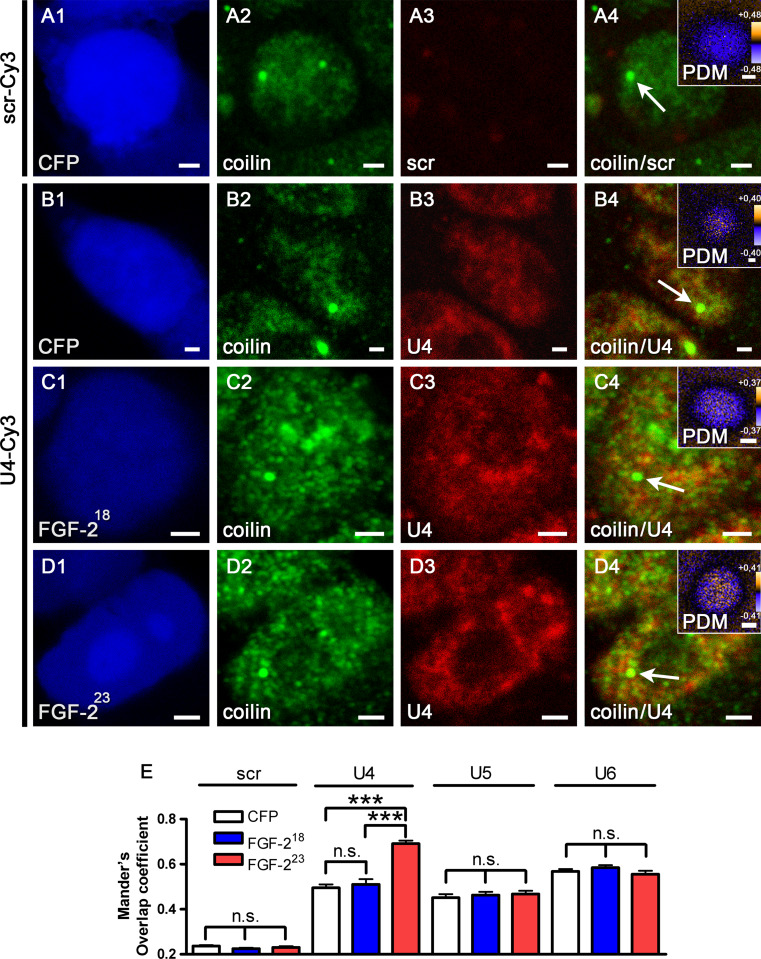
To test whether the accumulated snRNPs at CBs after overexpression of FGF-223 are part of the tri-snRNP, we additionally performed fluorescence in situ hybridization (FISH) experiments in HEK293T cells transfected with pFGF-223-ECFP or as controls pFGF-218-ECFP or pECFP (Fig. 6, Supp. Fig. 2). We used Cy3-labeled antisense oligonucleotides as probes to detect U4, U5 and U6 snRNPs in the nucleus as well as a scrambled probe as control followed by immunocytochemistry with anti-coilin antibodies to identify CBs (Fig. 6, Supp. Fig. 2, arrows). The degree of colocalization was evaluated by intensity correlation analysis for individual CBs (Fig. 6, Supp. Fig. 2, insets). As expected, no signal was obtained with the scrambled control probe. While the distribution of U5 and U6 snRNPs was unaffected by FGF-223 compared to non-SMN binding FGF-218 and ECFP controls, we found a significantly increased accumulation of U4 snRNPs at CBs (Fig. 6e). Amounts of U5 and U6 snRNPs were not significantly altered. Thus, depletion of immobile SMN by FGF-223 likely causes disturbed snRNP assembly leading to increased U4 snRNP accumulation at CBs. Taken together, the results from the FISH and immunocytochemical experiments give rise to a model in which immobile SMN is engaged in tri-snRNP maturation, and, consequently, the depletion of immobile SMN leads to U4 snRNP accumulation at Cajal bodies (Fig. 7).
Fig. 6.
Reduction of immobile SMN by FGF-223 leads to accumulation of U4 snRNPs at CBs. Fluorescence in situ hybridization (FISH, a3–d3) with probes for U4, U5 and U6 snRNAs and immunostainings with anti-coilin (a2–d2) were performed in HEK293T cells transfected with pFGF-223-ECFP (d1) or, as controls, pFGF-218-ECFP (c1) or pECFP (a1, b1) 24 h before fixation. Arrows point to the CBs in the merged images (a4–d4) that were enlarged in the inserted PDM images. Here, intensity correlation analyses are demonstrated (LUT: orange, positive PDM; blue, negative PDM). Colocalized pixels (both channels vary synchronically from the mean pixel intensity) are represented by positive PDM (yellow). A significant increase in colocalization of coilin and U4 snRNP in CBs after overexpression of pFGF-223-ECFP was detected (E means ± SEM; ***p < 0.001; Mann-Whitney test; CFP: n = 45, FGF-218: n = 45, FGF-223: n = 45, pooled from 3 independent experiments). Scale bar, 2 μm (a1–d4), 0.2 μm (insets). Stainings of U5 and U6 snRNPs and additional stainings of U4 snRNPs are shown in Supplemental Fig. 2
Fig. 7.
Molecular model for functional effects of FGF-223/SMN complex formation. Complex formation with FGF-223 results in a release of SMN stored and immobilized in CBs. Different populations of mobile and immobile SMN indicate equilibrium with several other interacting proteins. Additional interaction partners like FGF-223 change the balance of this network, leading to a mobilization of SMN from the CBs. Immobile SMN at CBs appears to have a function in the assembly of tri-snRNPs. Therefore, decreasing the amount of immobile SMN likely results in an accumulation of U4 snRNPs and indicates a dysfunction in tri-snRNP assembly
Discussion
In this study, the FRAP and photoconversion experiments demonstrate that FGF-223 modifies mobility of SMN at CBs. Our analyses revealed three SMN populations with different recovery kinetics. FGF-223 decreased the tightly bound SMN fraction in the CB (immobile population) and augmented the prevalent fast fraction. In earlier FRAP studies, coilin and SMN displayed a different flux through CBs in living HeLa cells. A plateau of signal recovery in a bleached CB was previously observed within 1 min after YFP-coilin bleaching [15]. The recovery of GFP-SMN reached only 50 % 1 h after bleaching, which indicates that the incorporation of SMN into the CB reflects a complex, multi-molecular interaction [15]. In CBs of Xenopus oocytes, three kinetic components of coilin had been identified by FRAP experiments with residence times of several seconds to >30 min [42]. This also indicates complex interactions in CBs for coilin and a dynamic equilibrium of proteins in CBs with proteins in the nucleoplasm. These observations are consistent with the finding that SmB competes with coilin for binding to SMN [27]. Inverse FRAP had been used to detect the residence times of several fluorescently tagged proteins in CBs of living HeLa cells. Three kinetically distinct groups of components within CBs were detected. SMN and coilin belonged to the group of proteins with the longest residence time [43]. Mechanistically, binding of SMN to coilin can be regulated by methylation and/or phosphorylation of coilin [44–46], and these modifications may also be partly responsible for the dynamic equilibrium of SMN between CBs and nucleoplasm. FGF-223 shifts this state toward emigration of SMN from CBs to the nucleoplasm.
FGF-223/SMN complex formation does not generally destabilize CBs since their numbers remain unchanged. In contrast, recruitment of SMN from nuclear gems decreased the number of these nuclear bodies [29]. The present study shows that FGF-223 mobilizes SMN from CBs, resulting in an accumulation of snRNPs at CBs (Fig. 7). Such an impairment of tri-snRNP export from CBs has been shown earlier upon inhibition of tri-snRNP formation. The authors demonstrated that knockdown of hPrp31 or hPrp6 leads to accumulation of U4 and U6 snRNPs at CBs—in contrast to U5 snRNPs that were not found accumulated in CBs [18, 47]. Therefore, accumulation of U4 snRNPs but not U5 snRNPs hints at disturbed tri-snRNP assembly. This indicates that immobile SMN plays a role in tri-snRNP assembly, besides the known function of mobile SMN in targeting snRNPs to CBs [21, 22]. Only the mature tri-snRNP seems to be released from the CB [18]. Especially the U4 snRNPs were shown to accumulate at CBs, indicating that targeting of U4 snRNPs is not affected by decreasing the amounts of immobile SMN, while the di-snRNP assembly could already be inhibited.
Based on the results of the present study, it appears that CBs contain a reservoir of immobile SMN that can be mobilized by its nuclear interaction partner FGF-223. Decreased amounts of immobilized SMN were observed in association with an accumulation of U4 snRNPs, leading to a model in which immobile SMN at CBs is involved in tri-snRNP assembly (Fig. 7). SMN targets snRNPs to CBs and passes snRNPs to coilin [18]. Whether the coilin-bound SMN in CBs is directly involved in tri-snRNP formation is currently not known.
Some specific proteins have been shown to directly influence the localization of SMN to CBs. The zinc-finger protein ZPR1 colocalizes with SMN in CBs and nuclear gems. Decreased ZPR1 expression was shown to prevent SMN localization to nuclear bodies [48]. In cells from SMA patients, the interaction of ZPR1 with SMN was disrupted [49]. Also, an overexpression of ZPR1 promoted accumulation of SMN in nuclear bodies of fibroblasts from SMN patients [50]. In spinal cord neurons from SMA mice, ZPR1 rescued axonal growth defects and induced neurite growth [50], indicating the ability of ZPR1 to be a protective modifier of SMA by positively influencing the localization of SMN to CBs. The WRAP53 protein was found colocalized with SMN in CBs. Depletion of WRAP53 prevented the SMN complex from reaching CBs and led to an accumulation of SMN in the cytoplasm [51]. Moreover, the integrator complex-mediating processing of snRNAs was found to be essential for integrity of the CB. Coilin-positive nucleoplasmic foci were free of SMN after depletion of INTS4, a subunit of the integrator complex [52]. Taken together, the mobilization of nuclear SMN by FGF-223 appears to be antagonistic compared to the accumulation of SMN in CBs caused by other proteins like ZPR1 and WRAP53. Additionally, the FGF-223-induced destabilization of gems, accompanied by a release of SMN to the nucleoplasm [29], indicates that the role of FGF-223 is antagonistic to ZPR1, which accumulates SMN in nuclear bodies.
The storage of readily available SMN in the nucleus appears to be a novel function of CBs in addition to their known roles in transport and maturation of snRNPs [21, 22]. SMN was proposed to have several functions in the nucleus. It was found to be telomerase-associated with a possible role in telomerase biogenesis [53]. Moreover, some studies have indicated a role of SMN in the regulation of gene transcription. For instance, SMN was found to be associated with (1) the transcription corepressor mSin3A and repressed transcription of a downstream luciferase reporter gene [54], (2) papillomavirus E2 enhancing E2-dependent transcriptional activation [55] and (3) interacting with a transcription factor called the Ewing’s sarcoma (EWS) protein, which contains an amino-terminal transcriptional activation domain [56]. However, EWS had no influence on the nuclear distribution of SMN and its association to CBs [56]. A release of stored SMN protein from CBs and consequently increased concentration of SMN in the nucleoplasm could alter transcriptional activity. Targeting of SMN from the nucleoplasm to CBs and nuclear gems could indicate a storage function of these nuclear bodies for SMN. Coilin immobilizes SMN in CBs and restricts its interactions with other proteins. In contrast, an FGF-223-induced increased mobile population of SMN would be able to engage in new, productive interactions. SMN released from the Cajal bodies and gems could render it readily available for its nuclear action outside the CBs.
The physiological role of the FGF-2 isoforms has been analyzed in a number of studies. Transgenic mice overexpressing all isoforms of FGF-2 displayed increased total fiber numbers after lesions compared to wild-type mice, indicating a positive influence of FGF-2 on axonal regrowth [57]. In motoneurons of these transgenic FGF-2 mice, we have previously shown that the number of SMN-positive nuclear bodies was significantly decreased [29]. In a non-transgenic wild-type mouse model, all FGF-2 isoforms have been shown to be expressed in several tissues including dorsal root ganglia and peripheral nerve [58]. After lesion of the sciatic nerve, endogenous FGF-2 was actually found upregulated [59]. After unilateral hypoglossal nerve transection, FGF-2 expression was also increased. Notably, the strongest expression was found for the high-molecular isoforms (FGF-223 and FGF-221) [39]. In this physiological context, FGF-223 becomes specifically regulated. The results pinpoint its specific role in early peripheral nerve regeneration [40, 41]. However, the molecular mechanism for the observations remained elusive. In this and previous studies we have described the interaction between FGF-223 and SMN, thereby providing a molecular mechanism for a specific action of FGF-223 (Fig. 7). Further studies need to reveal whether the FGF-223/SMN interaction has a regulatory role in peripheral nerve regeneration.
Materials and methods
Plasmids
Human full-length SMN (FL-SMN) plasmid pSMN1-294-EGFP and FLAG-tagged FL-SMN pSMN1-294-3XFLAG were described previously [29, 60]. Photoconvertible FL-SMN vector pSMN1-294-Dendra2 was produced by cloning the insert of pSMN1-294-EGFP into the EcoRI/SalI sites of pDendra2 (Evrogen). ECFP-tagged FGF-218 and FGF-223 encoding C-terminal FLAG-tagged rat FGF-2 isoforms were constructed by cloning FGF-223 cDNA into the EcoRI/HindIII sites and FGF-218 cDNA into the EcoRI/NheI sites of pECFP (Clontech).
Cell culture
Cells were cultivated in a humidified atmosphere at 37 °C and 5 % CO2. HEK293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10 % (v/v) fetal calf serum (FCS), 100 U/ml penicillin, 0.1 mg/ml streptomycin, 6 mM l-glutamine and 1 mM sodium pyruvate. Transfections of HEK293T cells were performed using metafectene (Biontex) according to the manufacturer’s recommendations or polyethylenimine (Sigma).
Confocal laser scanning microscopy for FRAP and photoconversion experiments
HEK293T live cell imaging was performed in μ-dishes (Ibidi) using an Olympus Fluoview FV1000 microscope equipped with an incubation chamber (37 °C and 5 % CO2), an oil immersion objective (60×, 1.35 NA), a zoom magnification (sixfold), laser lines 405, 491, 559 nm and a dichroic mirror (DM405/488/559/635). The laser output for FRAP bleaching of SMN-EGFP was set to 93 % (405 nm), for photoconversion of SMN-Dendra2 to 3 % (405 nm). Depending on the size of the CBs, 10–18 optical sections (0.5 μm apart) were taken three times each. The confocal microscope was equipped with a laser for bleaching and a second one for excitation during measurements. A first stack was taken as a pre-bleaching or pre-photoconversion control. Bleaching or photoconversion proceeded while recording the second stack of sections. After bleaching or photoconversion, CBs were observed every minute up to 20 min unless the CB left the volume of the stack. For FRAP measurements, images were used only for subsequent analysis when the relative fluorescence intensity after bleaching was 40–80 % in relation to the fluorescence intensity before bleaching. For photoconversion measurements, images were used when the red fluorescence before photoconversion was 15–40 % related to the green FI before. Red RFI after photoconversion was 140–260 % in relation to the red FI before photoconversion. Data analyses were performed using MBF ImageJ (1.43 m) and Prism 4 (GraphPad Software, San Diego, CA). Normalized relative fluorescence intensities, expressed as percentages (fluorescence intensity after conversion was set to 100 %), were quantified by fitting one- and two-phase exponential associations (for FRAP) or one- and two-phase exponential decay (for photoconversion) to the data.
The two-phase exponential association for FRAP can be described with the equation
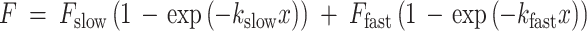 |
1 |
with F slow, the hypomobile, and F fast, the hypermobile fraction. The immobile fraction (F immobile) is defined as
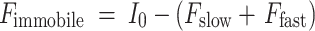 |
2 |
with I0, normalized fluorescence intensity before bleaching. Fluorescence was normalized in FRAP measurements for each time point compared to the whole cell with the equation
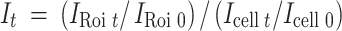 |
3 |
with It, normalized fluorescence intensity at timepoint t, I Roi t, fluorescence intensity in the bleached area at timepoint t, I Roi 0, fluorescence intensity in the bleached area before bleaching, I cell t, fluorescence intensity of the whole cell at timepoint t, and I cell 0, fluorescence intensity of the whole cell before bleaching.
For the one phase exponential decay for photoconversion, fluorescence loss can be described with the equation
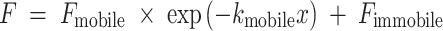 |
4 |
with F mobile, mobile fraction. Fluorescence was normalized for photoconversion measurements as described for Eq. (3) above:
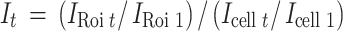 |
5 |
with I Roi 1, fluorescence intensity in the photoconverted area after photoconversion and I cell 1, fluorescence intensity in the whole cell after photoconversion. Recovery half-times and the half-times of fluorescence loss (t 1/2) were obtained from the k values of Eqs. (1) and (4), defined as
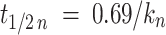 |
6 |
Stastical significance was analyzed by using unpaired, two-tailed t tests. All parameters are reported as arithmetic mean ± standard error of the means (SEMs).
Immonocytochemistry and fluorescence microscopy
Cells were fixed for 5 min with 4 % (w/v) fresh paraformaldehyde in phosphate-buffered saline (PBS), pH 7.4, and subsequently washed with PBS. HEK293T cells were permeabilized with 0.3 % (v/v) Triton X-100 in PBS with 5 % (v/v) normal goat serum or horse serum for anti-SmE staining for 1 h, and incubated with anti-coilin (H-300, rabbit IgG, Santa Cruz) and anti-SMN (mouse IgG, BD Bioscience) in PBS/1 % (v/v) goat serum, 0.3 % (v/v) Triton X-100 or anti-SmE (goat IgG, Santa Cruz) in PBS/1 % (v/v) horse serum, 0.3 % (v/v) Triton X-100 for 1 h. After an additional washing step with PBS, cells were incubated with secondary antibody (Alexa Fluor® 647 goat anti-rabbit IgG, Alexa Fluor® 488 goat anti-mouse IgG or Alexa Fluor® 555 donkey anti-goat IgG, Invitrogen) and mounted in Prolong Gold (Molecular Probes). Confocal images were taken using a Leica TCS SP2, equipped with an oil immersion objective HCX PL APO BL (63×/numeric aperture 1.4) with Leica acquisition software. Spinal cords were stained using the same antibodies as described previously [29].
Fluorescence in situ hybridization (FISH) experiments and fluorescence microscopy
Cells were fixed for 5 min with 4 % (w/v) fresh paraformaldehyde in diethyl pyrocarbonate (DEPC) treated PBS (pH 7.4) and subsequently washed with PBS-DEPC. HEK293T cells were permeabilized with 0.3 % (v/v) Triton X-100 in PBS-DEPC for 1 h and incubated with pre-hybridization mix: 15 % (v/v) formamide in 2xSSC (0.3 M NaCl, 0.03 M citric acid pH 7.0), 10 mM NaH2PO4 for 10 min. Before hybridization, the oligonucleotides were mixed with 200 μl hybridization buffer with a final concentration of the oligonucleotide of 1 pmol/ml in 15 % (v/v) formamide in 2× SSC, 10 mM NaH2PO4, 0.2 % BSA, pH 7.0 and 0.5 mg/ml each E. coli tRNA and herring sperm DNA. Coverslips were incubated at 37 °C with 200 μl hybridization mix for 3 h in a humidified incubator. After hybridization, the coverslips were washed 30 min at 37 °C and 30 min twice at room temperature with 15 % (v/v) formamide in 2×SSC, 10 mM NaH2PO4 and fixed again for 1 min with 4 % (w/v) fresh paraformaldehyde in PBS-DEPC, pH 7.4. After in situ hybridization, the cells were washed twice with PBS, blocked with 0.3 % (v/v) Triton X-100 in PBS with 5 % (v/v) normal goat serum for 10 min and stained with anti-coilin as already described.
The DNA oligonucleotide probes used for FISH were Cy3-labeled at their 3′ ends (MWG Eurofins). Sequences were as follows: U4 snRNA, 5′-TCA CGG CGG GGT ATT GGG AAA AGT TTT CAA TTA GCA ATA ATC GCG CCT-3′; U5 snRNA, 5′-CTC TCC ACG GAA ATC TTT AGT AAA AGG CGA AAG ATT TAT ACG ATT TGA AGA G- 3′; U6 snRNA, 5′-CAC GAA TTT GCG TGT CAT CCT TGC GCA GGG GCC ATG CTA ATC-3′; scrRNA, 5′- AAC TGG ATC CAG ATT GCT GCA ATA TAC TAG TAG TAC GAT GAT AAG TAC GATA- 3′. Confocal images were taken using a Leica DM-R equipped with an oil immersion objective HCX PL APO CS (100×/numeric aperture 1.4) with Leica acquisition software.
Colocalization analysis
Cajal bodies in the nuclei of randomly selected cells (n = 45, pooled from three independent experiments) were analyzed by colocalization analysis using MBF ImageJ (1.43 m) with intensity correlation analysis plug-in [61–63]. PDM (product of differences from the mean) images were generated. Colocalized pixels (both channels vary synchronically from the mean pixel intensity) are represented by positive PDM (yellow). Non-colocalizing pixels (both channels vary asynchronically from the mean pixel intensity) are represented by negative PDM (blue). The ratio of intersecting volume to total object volume with a total pixel intensity >0 is represented by Mander’s overlap coefficient (R), ranging from zero to one illustrating low to high colocalization.
Pulldown assay
Cell lysates of HEK293T cells were prepared by sonication followed by incubation of cells 30 min on ice with modified RIPA buffer [50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 2 mM dithiothreitol (DTT) 1 mM EDTA, 1 % NP-40, 0.25 % (w/v) sodium deoxycholate and 1× Complete protease inhibitor (Roche)]. FGF-2 purification was described before [30]. Recombinant SMN-GST and GST proteins were expressed in E.coli using the pET System (Novagene) as described previously [30]. Proteins were purified and immobilized on glutathione MagBeads (GenScript) after quantification by sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS–PAGE) and Coomassie staining. Recombinant SMN-GST or GST was preincubated for 1 h at 4 °C with 20 μg FGF-223 or FGF-218 in a total volume of 300 μl in modified RIPA buffer. Cell lysates with a total volume of 1 ml were added to immobilized recombinant SMN-GST or GST, incubated for 1 h at 4 °C and washed five times in 750 μl modified RIPA buffer. Analyses were performed by SDS-PAGE and Western blots using the same antibodies as in immunocytochemistry and in addition anti-FGF-2 (mouse IgG, Millipore).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1 The velocities of CBs after overexpression of FGF-2 are not altered For live cell imaging, single CBs of pSMN-EGFP and pFGF-223-DsRed2 or (control) pFGF-218-DsRed2 transfected HEK293T cells were measured every minute up to 20 minutes unless the focused nuclear body left the volume as defined by z-sections. The total distance per minute for each CB was measured. All distances per minute were plotted as a velocity distribution (FGF-223-DsRed2 n = 149 single distances, FGF-218-DsRed2 n = 163 single distances). The overexpression of FGF-223 showed no influence on the distribution of CB velocities compared to FGF-218 overexpressing cells (χ2 -test, n.s., non-significant) (JPEG 529 kb)
Supplemental Fig. 2 Accumulation of U4 snRNPs at CBs after reduction of immobile SMN by FGF-223 HEK293T cells were transfected with pECFP (A, D, G, H, I) or pFGF-218-ECFP (B, E, K, L, M) or pFGF-223-ECFP (C, F, N, O, P) 24 hours before fixation. Cells were examined after fluorescence in situ hybridization (FISH, A3-P3) with probes for U4, U5 and U6 snRNAs and immunostainings with anti-coilin (A2-P2). Arrows point to the CBs in the merged images (A4-P4) that were enlarged in the inserted PDM images. Here, Intensity Correlation Analyses are demonstrated (LUT; orange, positive PDM; blue, negative PDM). Colocalized pixels (both channels vary synchronically from the mean pixel intensity) are represented by positive PDM (yellow). Scale bar, 2 μm (A1-L4), 0.2 μm (insets). For FGF-223 transfected cells and also controls probed against U4 snRNA, three individual cells have been shown each. Supplemental Fig. 2 (JPEG 7747 kb)
Acknowledgments
The authors thank Kerstin Kuhlemann and Hildegard Streich for expert technical help. We are grateful to Dr. R. Bauerfeind and W. Posselt (Central Laser Microscopy Facility, Hannover Medical School) for expert technical advice on laser confocal microscopy. M.K.S. was supported by grants NYSTEM, contracts C026415 and C026714.
Abbreviations
- CB
Cajal body
- FRAP
Fluorescence recovery after photobleaching
- SMN
Survival of motoneuron protein
- SMA
Spinal muscular atrophy
- FGF
Fibroblast growth factor
- SnRNP
Small nuclear ribonucleoprotein particle
References
- 1.Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 2.Matera AG, Frey MR. Coiled bodies and gems: janus or gemini? Am J Hum Genet. 1998;63:317–321. doi: 10.1086/301992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meister G, Eggert C, Fischer U. SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol. 2002;12:472–478. doi: 10.1016/S0962-8924(02)02371-1. [DOI] [PubMed] [Google Scholar]
- 4.Paushkin S, Gubitz AK, Massenet S, Dreyfuss G. The SMN complex, an assemblyosome of ribonucleoproteins. Curr Opin Cell Biol. 2002;14:305–312. doi: 10.1016/S0955-0674(02)00332-0. [DOI] [PubMed] [Google Scholar]
- 5.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 6.Pearn J. Classification of spinal muscular atrophies. Lancet. 1980;1:919–922. doi: 10.1016/S0140-6736(80)90847-8. [DOI] [PubMed] [Google Scholar]
- 7.Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, Coulson SE, Androphy EJ, Prior TW, Burghes AH. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 8.McAndrew PE, Parsons DW, Simard LR, Rochette C, Ray PN, Mendell JR, Prior TW, Burghes AH. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldkotter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70(2):358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 11.Young PJ, Le TT, Dunckley M, Nguyen TM, Burghes AH, Morris GE. Nuclear gems and Cajal (coiled) bodies in fetal tissues: nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp Cell Res. 2001;265:252–261. doi: 10.1006/excr.2001.5186. [DOI] [PubMed] [Google Scholar]
- 12.Cajal Ry (1903) Un sencillo metodo de coloracion selectiva del reticulo protoplasmico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Lab Invest Biol Univ (Madrid) pp 129–143
- 13.Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- 14.Cioce M, Lamond AI. Cajal bodies: a long history of discovery. Annu Rev Cell Dev Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- 15.Sleeman JE, Trinkle-Mulcahy L, Prescott AR, Ogg SC, Lamond AI. Cajal body proteins SMN and Coilin show differential dynamic behaviour in vivo. J Cell Sci. 2003;116:2039–2050. doi: 10.1242/jcs.00400. [DOI] [PubMed] [Google Scholar]
- 16.Sleeman JE, Ajuh P, Lamond AI. snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J Cell Sci. 2001;114:4407–4419. doi: 10.1242/jcs.114.24.4407. [DOI] [PubMed] [Google Scholar]
- 17.Morris GE. The Cajal body. Biochim Biophys Acta. 2008;1783:2108–2115. doi: 10.1016/j.bbamcr.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 18.Stanek D, Neugebauer KM. The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma. 2006;115:343–354. doi: 10.1007/s00412-006-0056-6. [DOI] [PubMed] [Google Scholar]
- 19.Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell. 2009;17:639–647. doi: 10.1016/j.devcel.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hebert MD. Phosphorylation and the Cajal body: modification in search of function. Arch Biochem Biophys. 2010;496:69–76. doi: 10.1016/j.abb.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dundr M, Misteli T. Functional architecture in the cell nucleus. Biochem J. 2001;356:297–310. doi: 10.1042/0264-6021:3560297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sleeman JE, Lamond AI. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr Biol. 1999;9:1065–1074. doi: 10.1016/S0960-9822(99)80475-8. [DOI] [PubMed] [Google Scholar]
- 23.Will CL, Luhrmann R. Protein functions in pre-mRNA splicing. Curr Opin Cell Biol. 1997;9:320–328. doi: 10.1016/S0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- 24.Buhler D, Raker V, Luhrmann R, Fischer U. Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum Mol Genet. 1999;8:2351–2357. doi: 10.1093/hmg/8.13.2351. [DOI] [PubMed] [Google Scholar]
- 25.Mattaj IW, Boelens W, Izaurralde E, Jarmolowski A, Kambach C. Nucleocytoplasmic transport and snRNP assembly. Mol Biol Rep. 1993;18:79–83. doi: 10.1007/BF00986760. [DOI] [PubMed] [Google Scholar]
- 26.Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, Chan EK, Lafarga M, Conlon RA, Matera AG. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol. 2001;154:293–307. doi: 10.1083/jcb.200104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hebert MD, Szymczyk PW, Shpargel KB, Matera AG. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev. 2001;15:2720–2729. doi: 10.1101/gad.908401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrade LE, Chan EK, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruns AF, van Bergeijk J, Lorbeer C, Nolle A, Jungnickel J, Grothe C, Claus P. Fibroblast growth factor-2 regulates the stability of nuclear bodies. Proc Natl Acad Sci USA. 2009;106:12747–12752. doi: 10.1073/pnas.0900122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Claus P, Bruns AF, Grothe C. Fibroblast growth factor-2(23) binds directly to the survival of motoneuron protein and is associated with small nuclear RNAs. Biochem J. 2004;384:559–565. doi: 10.1042/BJ20040801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Claus P, Doring F, Gringel S, Muller-Ostermeyer F, Fuhlrott J, Kraft T, Grothe C. Differential intranuclear localization of fibroblast growth factor-2 isoforms and specific interaction with the survival of motoneuron protein. J Biol Chem. 2003;278:479–485. doi: 10.1074/jbc.M206056200. [DOI] [PubMed] [Google Scholar]
- 32.Sorensen V, Nilsen T, Wiedlocha A. Functional diversity of FGF-2 isoforms by intracellular sorting. BioEssays. 2006;28:504–514. doi: 10.1002/bies.20405. [DOI] [PubMed] [Google Scholar]
- 33.Reilly JF, Maher PA. Importin beta-mediated nuclear import of fibroblast growth factor receptor: role in cell proliferation. J Cell Biol. 2001;152:1307–1312. doi: 10.1083/jcb.152.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng H, Myers J, Fang X, Stachowiak EK, Maher PA, Martins GG, Popescu G, Berezney R, Stachowiak MK. Integrative nuclear FGFR1 signaling (INFS) pathway mediates activation of the tyrosine hydroxylase gene by angiotensin II, depolarization and protein kinase C. J Neurochem. 2002;81:506–524. doi: 10.1046/j.1471-4159.2002.00833.x. [DOI] [PubMed] [Google Scholar]
- 35.Stachowiak MK, Fang X, Myers JM, Dunham SM, Berezney R, Maher PA, Stachowiak EK. Integrative nuclear FGFR1 signaling (INFS) as a part of a universal “feed-forward-and-gate” signaling module that controls cell growth and differentiation. J Cell Biochem. 2003;90:662–691. doi: 10.1002/jcb.10606. [DOI] [PubMed] [Google Scholar]
- 36.Stachowiak MK, Maher PA, Stachowiak EK. Integrative nuclear signaling in cell development—a role for FGF receptor-1. DNA Cell Biol. 2007;26:811–826. doi: 10.1089/dna.2007.0664. [DOI] [PubMed] [Google Scholar]
- 37.Grothe C, Meisinger C, Holzschuh J, Wewetzer K, Cattini P. Over-expression of the 18 kD and 21/23 kD fibroblast growth factor-2 isoforms in PC12 cells and Schwann cells results in altered cell morphology and growth. Brain Res Mol Brain Res. 1998;57:97–105. doi: 10.1016/S0169-328X(98)00076-X. [DOI] [PubMed] [Google Scholar]
- 38.Chudakov DM, Lukyanov S, Lukyanov KA (2007) Using photoactivatable fluorescent protein Dendra2 to track protein movement. Biotechniques 42:553, 555, 557 passim [DOI] [PubMed]
- 39.Huber K, Meisinger C, Grothe C. Expression of fibroblast growth factor-2 in hypoglossal motoneurons is stimulated by peripheral nerve injury. J Comp Neurol. 1997;382:189–198. doi: 10.1002/(SICI)1096-9861(19970602)382:2<189::AID-CNE4>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 40.Jungnickel J, Claus P, Gransalke K, Timmer M, Grothe C. Targeted disruption of the FGF-2 gene affects the response to peripheral nerve injury. Mol Cell Neurosci. 2004;25:444–452. doi: 10.1016/j.mcn.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 41.Jungnickel J, Haastert K, Grzybek M, Thau N, Lipokatic-Takacs E, Ratzka A, Nolle A, Claus P, Grothe C. Mice lacking basic fibroblast growth factor showed faster sensory recovery. Exp Neurol. 2010;223:166–172. doi: 10.1016/j.expneurol.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 42.Handwerger KE, Murphy C, Gall JG. Steady-state dynamics of Cajal body components in the Xenopus germinal vesicle. J Cell Biol. 2003;160:495–504. doi: 10.1083/jcb.200212024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, Meier UT, Neugebauer KM, Matera AG, Misteli T. In vivo kinetics of Cajal body components. J Cell Biol. 2004;164:831–842. doi: 10.1083/jcb.200311121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toyota CG, Davis MD, Cosman AM, Hebert MD. Coilin phosphorylation mediates interaction with SMN and SmB’. Chromosoma. 2010;119:205–215. doi: 10.1007/s00412-009-0249-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hebert MD, Shpargel KB, Ospina JK, Tucker KE, Matera AG. Coilin methylation regulates nuclear body formation. Dev Cell. 2002;3:329–337. doi: 10.1016/S1534-5807(02)00222-8. [DOI] [PubMed] [Google Scholar]
- 46.Tapia O, Bengoechea R, Berciano MT, Lafarga M. Nucleolar targeting of coilin is regulated by its hypomethylation state. Chromosoma. 2010;119:527–540. doi: 10.1007/s00412-010-0276-7. [DOI] [PubMed] [Google Scholar]
- 47.Schaffert N, Hossbach M, Heintzmann R, Achsel T, Luhrmann R. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 2004;23:3000–3009. doi: 10.1038/sj.emboj.7600296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gangwani L, Mikrut M, Theroux S, Sharma M, Davis RJ. Spinal muscular atrophy disrupts the interaction of ZPR1 with the SMN protein. Nat Cell Biol. 2001;3:376–383. doi: 10.1038/35070059. [DOI] [PubMed] [Google Scholar]
- 49.Gangwani L, Flavell RA, Davis RJ. ZPR1 is essential for survival and is required for localization of the survival motor neurons (SMN) protein to Cajal bodies. Mol Cell Biol. 2005;25:2744–2756. doi: 10.1128/MCB.25.7.2744-2756.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahmad S, Wang Y, Shaik GM, Burghes AH, Gangwani L. The zinc finger protein ZPR1 is a potential modifier of spinal muscular atrophy. Hum Mol Genet. 2012;21(12):2745–2758. doi: 10.1093/hmg/dds102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mahmoudi S, Henriksson S, Weibrecht I, Smith S, Soderberg O, Stromblad S, Wiman KG, Farnebo M. WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol. 2010;8:e1000521. doi: 10.1371/journal.pbio.1000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takata H, Nishijima H, Maeshima K, Shibahara K. The integrator complex is required for integrity of Cajal bodies. J Cell Sci. 2012;125:166–175. doi: 10.1242/jcs.090837. [DOI] [PubMed] [Google Scholar]
- 53.Bachand F, Boisvert FM, Cote J, Richard S, Autexier C. The product of the survival of motor neuron (SMN) gene is a human telomerase-associated protein. Mol Biol Cell. 2002;13:3192–3202. doi: 10.1091/mbc.E02-04-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zou J, Barahmand-pour F, Blackburn ML, Matsui Y, Chansky HA, Yang L. Survival motor neuron (SMN) protein interacts with transcription corepressor mSin3A. J Biol Chem. 2004;279:14922–14928. doi: 10.1074/jbc.M309218200. [DOI] [PubMed] [Google Scholar]
- 55.Strasswimmer J, Lorson CL, Breiding DE, Chen JJ, Le T, Burghes AH, Androphy EJ. Identification of survival motor neuron as a transcriptional activator-binding protein. Hum Mol Genet. 1999;8:1219–1226. doi: 10.1093/hmg/8.7.1219. [DOI] [PubMed] [Google Scholar]
- 56.Young PJ, Francis JW, Lince D, Coon K, Androphy EJ, Lorson CL. The Ewing’s sarcoma protein interacts with the tudor domain of the survival motor neuron protein. Brain Res Mol Brain Res. 2003;119:37–49. doi: 10.1016/j.molbrainres.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 57.Jungnickel J, Haase K, Konitzer J, Timmer M, Grothe C. Faster nerve regeneration after sciatic nerve injury in mice over-expressing basic fibroblast growth factor. J Neurobiol. 2006;66:940–948. doi: 10.1002/neu.20265. [DOI] [PubMed] [Google Scholar]
- 58.Grothe C, Nikkhah G. The role of basic fibroblast growth factor in peripheral nerve regeneration. Anat Embryol (Berl) 2001;204:171–177. doi: 10.1007/s004290100205. [DOI] [PubMed] [Google Scholar]
- 59.Grothe C, Meisinger C, Claus P. In vivo expression and localization of the fibroblast growth factor system in the intact and lesioned rat peripheral nerve and spinal ganglia. J Comp Neurol. 2001;434:342–357. doi: 10.1002/cne.1181. [DOI] [PubMed] [Google Scholar]
- 60.van Bergeijk J, Rydel-Konecke K, Grothe C, Claus P. The spinal muscular atrophy gene product regulates neurite outgrowth: importance of the C terminus. FASEB J. 2007;21(7):1492–1502. doi: 10.1096/fj.06-7136com. [DOI] [PubMed] [Google Scholar]
- 61.Baron O, Forthmann B, Lee YW, Terranova C, Ratzka A, Stachowiak EK, Grothe C, Claus P, Stachowiak MK. Cooperation of nuclear fibroblast growth factor receptor 1 and Nurr1 offers new interactive mechanism in postmitotic development of mesencephalic dopaminergic neurons. J Biol Chem. 2012;287:19827–19840. doi: 10.1074/jbc.M112.347831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farias GG, Valles AS, Colombres M, Godoy JA, Toledo EM, Lukas RJ, Barrantes FJ, Inestrosa NC. Wnt-7a induces presynaptic colocalization of alpha 7-nicotinic acetylcholine receptors and adenomatous polyposis coli in hippocampal neurons. J Neurosci. 2007;27:5313–5325. doi: 10.1523/JNEUROSCI.3934-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Q, Lau A, Morris TJ, Guo L, Fordyce CB, Stanley EF. A syntaxin 1, G alpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J Neurosci. 2004;24:4070–4081. doi: 10.1523/JNEUROSCI.0346-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1 The velocities of CBs after overexpression of FGF-2 are not altered For live cell imaging, single CBs of pSMN-EGFP and pFGF-223-DsRed2 or (control) pFGF-218-DsRed2 transfected HEK293T cells were measured every minute up to 20 minutes unless the focused nuclear body left the volume as defined by z-sections. The total distance per minute for each CB was measured. All distances per minute were plotted as a velocity distribution (FGF-223-DsRed2 n = 149 single distances, FGF-218-DsRed2 n = 163 single distances). The overexpression of FGF-223 showed no influence on the distribution of CB velocities compared to FGF-218 overexpressing cells (χ2 -test, n.s., non-significant) (JPEG 529 kb)
Supplemental Fig. 2 Accumulation of U4 snRNPs at CBs after reduction of immobile SMN by FGF-223 HEK293T cells were transfected with pECFP (A, D, G, H, I) or pFGF-218-ECFP (B, E, K, L, M) or pFGF-223-ECFP (C, F, N, O, P) 24 hours before fixation. Cells were examined after fluorescence in situ hybridization (FISH, A3-P3) with probes for U4, U5 and U6 snRNAs and immunostainings with anti-coilin (A2-P2). Arrows point to the CBs in the merged images (A4-P4) that were enlarged in the inserted PDM images. Here, Intensity Correlation Analyses are demonstrated (LUT; orange, positive PDM; blue, negative PDM). Colocalized pixels (both channels vary synchronically from the mean pixel intensity) are represented by positive PDM (yellow). Scale bar, 2 μm (A1-L4), 0.2 μm (insets). For FGF-223 transfected cells and also controls probed against U4 snRNA, three individual cells have been shown each. Supplemental Fig. 2 (JPEG 7747 kb)