Abstract
Chromosomal translocations are characteristic features of many cancers, especially lymphoma and leukemia. However, recent reports suggest that many chromosomal translocations can be found in healthy individuals, although the significance of this observation is still not clear. In this review, we summarize recent studies on chromosomal translocations in healthy individuals carried out in different geographical areas of the world and discuss the relevance of the observation with respect to oncogenesis.
Keywords: Leukemia, Lymphoma, Neoplasia, Carcinoma, Sarcoma, Genomic instability, V(D)J recombination
Background
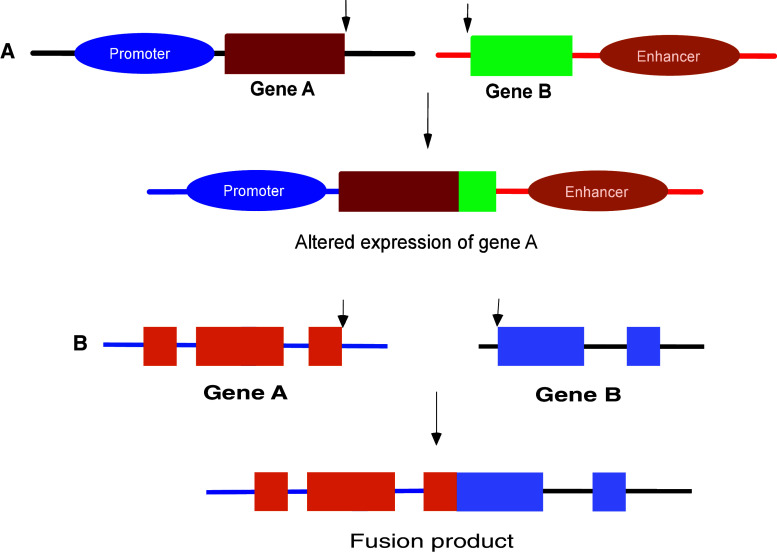
Chromosomal translocations are genomic alterations that result in the joining of heterologous chromosomes. Such a misjoining can lead to either the juxtaposition of certain oncogenes to the enhancer/promoter elements of other loci or novel fusions resulting in the formation of activated oncoproteins (Fig. 1) [1–3]. Many chromosomal translocations have been reported to date; however, an unusual and interesting observation has been their frequent incidence in cancers of the hematopoietic origin, namely leukemia and lymphoma [4–15]. Together, they constitute only around 8–10 % of the total cancer cases, yet more than 500 different translocations have been described thus far. Lack of in-depth studies and appropriate techniques hampered detection of such translocations in solid tumors. However, recently a tremendous advancement has been made in the identification of such chromosomal abnormalities in carcinomas and sarcomas. Studies have shown that common epithelial cancers like breast, prostate, thyroid cancer, and renal carcinoma also possess gene fusions as a result of translocations [16–20]. This has opened a new window in the use of such translocations as biomarkers in diagnosis and prognosis of cancers like carcinoma and sarcoma, which so far was the case only in leukemia and lymphoma.
Fig. 1.
Chromosomal translocations resulting in juxtaposition of promoter/enhancer elements to oncogenes or chimeric fusion proteins. In hematological cancers, like leukemia and lymphoma, translocations can result in the juxtaposition of the coding region of a gene (gene A) to enhancer/promoter elements of another gene (gene B) (a). This results in the enhanced expression of the gene A under the influence of either the enhancer or alternative promoters. Alternatively, translocations can also result in the formation of a fusion/chimeric protein, which may have a novel or enhanced function (b). For example, upon translocation, gene A can join with gene B, resulting in a fusion product. Altogether, these alterations can lead to changes in the cellular physiology and morphology, thereby resulting in malignant transformation
Translocations in cancer
The Philadelphia chromosome was the first translocation to be identified and described in the literature [4]. It results in the fusion of chromosome 22 and 9, which brings together the BCR and ABL genes, leading to the formation of an activated tyrosine kinase [21–23]. The t(14;18) translocation, between the BCL2 gene on chromosome 18 and the immunoglobulin heavy chain (IgH) locus on chromosome 14, is the most common translocation in human cancer. It is characteristically present in follicular lymphoma (FL) and some cases of diffuse large B cell lymphoma (DLBL) [3]. It juxtaposes the BCL2 gene along with its promoter to the IgH enhancer, thereby leading to its overexpression [24–28]. The majority of the breaks in chromosome 18 occur in the BCL2 major breakpoint region or minor breakpoint cluster region and recently the mechanism of fragility of these regions was identified [28–30]. Some other well-known translocations are the c-MYC-IgH translocation in Burkitt’s lymphoma, NPM-ALK translocation in anaplastic large cell lymphoma, and the diverse translocations involving the BCL6 gene in DLBL [31–36]. As mentioned above, not many translocations had been described in carcinomas and sarcomas, until recently. Some of the translocations that have been identified are TMPRSS2-ETS translocations in prostate cancer, EWS translocations in Ewing’s sarcoma, ETV6-NTRK3 translocations in breast carcinoma, and others, as described previously [16–20, 37, 38]. Recently, the incidence and mechanism of these translocations in cancer have been extensively reviewed and summarized [11, 39–41].
Translocations in healthy individuals
In the recent past, an important yet puzzling observation has changed the outlook towards translocations being used as biomarkers in cancer. Multiple studies have shown the presence of some of these translocations in lymphocytes of peripheral blood of healthy individuals [42–46]. Though the number of such cells in circulation is low, it nevertheless leads to ambiguity during diagnosis of these cancers using chromosomal translocations as markers and while studying the progression of the disease. The translocations detected among the healthy individuals include those involving the BCL2-IgH, BCR-ABL, NPM-ALK, BCL1-IgH, and BCL6 loci.
The BCR-ABL translocation or the Philadelphia chromosome is the genetic hallmark of chronic myelogenous leukemia (CML) [1]. During follow-up studies on CML patients, the presence of the BCR-ABL fusion mRNA was detected even in those patients who were cured following treatment [47]. Further studies found blood cells from 23 out of 117 healthy individuals, including both children and adults to have BCR-ABL translocation [47]. Moreover, the occurrence seemed to be age-dependent, being more in adults, which could be explained since CML occurs rarely in children [47]. Alternatively, increased probability of accumulating mutations or exposure to stimulating agents could augment the development of translocations in adults. Similar incidence of this translocation was also reported by another group (Table 1) [48]. However, there are no studies to identify the molecular mechanism responsible for the development of the BCR-ABL translocation in healthy individuals. Since the number of positive cases in healthy individuals for this translocation exceeds that of the disease, it remains to be seen how many of them will actually be at risk of developing CML.
Table 1.
Leukemia/lymphoma-associated gene fusions in healthy individuals
| Translocation | Assay system | Cells | Frequency | Reference |
|---|---|---|---|---|
| t(9;22) | Nested RT-PCR | Peripheral blood leukocytes |
p190 BCR/ABL 11/16 (69 %) p210 BCR/ABL 4/15 (27 %) |
Bose et al. [48] |
| Nested RT-PCR | Peripheral blood leukocytes | 23/117 (19.7 %) | Biernaux et al. [47] | |
| t(2;5) | RT-PCR, Southern hybridization | Peripheral blood leukocytes | 14/29 (48.3 %) | Trumper et al. [49] |
| Real-time PCR | Lymph nodes and spleen | 20/31 (64.5 %) | Maes et al. [50] | |
| t(11;14) | Real-time qPCR | Peripheral blood leukocytes | 1/100 (1 %) | Hirt et al. [51] |
| t(14;18) | Nested PCR | Peripheral blood leukocytes | 19/230 (8.3 %) | Paltiel et al. [102] |
| Nested PCR | Peripheral blood mononuclear cells | 40/254 (15.7 %) | Zignego et al. [71] | |
| Nested PCR | Peripheral blood leukocytes | 31/55 (56.4 %) | Henriksson et al. [103] | |
| Nested qPCR | Peripheral blood mononuclear cells | 10/23 (43.5 %) | Scheerer et al. [104] | |
| Semi nested PCR |
Bone marrow aspirates Peripheral blood lymphocytes |
89/224 (39.7 %) | Rauzy et al. [105] | |
| Nested qPCR | Peripheral blood mononuclear cells, umbilical cord cells | 69/127 (54.3 %) | Liu et al. [106] | |
| Seminested qPCR | Peripheral blood mononuclear cells | 26/57 (45.6 %) | Dolken et al. [56] | |
| Nested qPCR | Peripheral blood leukocytes | 30/34 (88.2 %) | Fuscoe et al. [57] | |
| PCR Southern blot/seminested PCR | Lymph node tissue | 19/48 (39.6 %) | Molina et al. [107] | |
| Nested qPCR | Peripheral blood mononuclear cells | 25/64 (39.1 %) | Cole et al. [108] | |
| Seminested PCR | Peripheral blood mononuclear cells, granulocytes, lymphocytes | 6/9 (66.7 %) | Limpens et al. [58] | |
| Seminested qPCR, P32-labelling | Peripheral blood mononuclear cells | 70/146 (47.9 %) | Ji et al. [68] | |
| Nested qPCR | Peripheral blood leukocytes | 57/122 (46.7 %) | Bell et al. [69] | |
| PCR | Lymph nodes | 18/108 (16.6 %) | Corbally et al. [109] | |
| Nested qPCR | Peripheral blood lymphocytes | 40/84 (47.6 %) | Liu et al. [55] | |
| Seminested PCR and Southern blot | Lymphoid tissue | 10/25 (40 %) | Aster et al. [62] | |
| PCR and Southern blot | Lymph nodes and tonsils | 13/73 (17.8 %) | Limpens et al. [110] | |
| Real-time qPCR | Peripheral blood mononuclear cells | 287/644 (45 %) | Dolken et al. [67] | |
| Nested PCR, real-time PCR | Peripheral blood mononuclear cells | 16/125 (12.8 %) | Ladetto et al. [111] |
The t(2;5) translocation involving the NPM and ALK genes is characteristically found in anaplastic large cell lymphoma (ALCL) [49, 50]. Similar to the Philadelphia chromosome, this gene fusion also results in the formation of a constitutively activated tyrosine kinase, which has oncogenic potential. The t(2;5) translocation alone is insufficient to cause the lymphoma, which was reiterated by the fact that it was found to be circulating in the peripheral blood of healthy individuals [49, 50]. In one study, which used a combination of RT-PCR and Southern blotting techniques, 14 of 29 healthy volunteers (48 %) showed the presence of the t(2;5) translocations in lymphocytes [49]. Another study followed, showing similar results using both cytogenetic and sensitive molecular biology methods [50]. However, it is not yet clear whether the cells containing this translocation persist due to a survival advantage or arise due to multiple, independent translocations. More studies involving analysis of a larger number of healthy individuals as well as follow-up on positive cases is a pre-requisite to better understand the origin of translocation and its prognosis in ALCL.
Another translocation detected in healthy individuals is the t(11;14) translocation, involving the BCL1 gene present in mantle cell lymphoma (MCL) [13, 14]. However, the frequency at which it was found was very low (1 in a group of 100 individuals) from a European population [51]. Studies from an Indian population also showed that the incidence of this translocation is rare [52]. Out of the 210 healthy Indian volunteers studied, none was positive for this translocation [52]. Both these studies have used sensitive detection assays of quantitative real-time PCR and nested PCR followed by Southern hybridization, respectively. It is hypothesized that this translocation may not be the initiating event in the pathogenesis of MCL and additional preceding mutations may be required. This would explain its very low incidence in healthy individuals. A follow-up study on healthy individuals carrying this translocation showed that these t(11;14)-positive cells could persist for a long period of time and probably expand to acquire further aberrations before transformation [53]. Since only limited studies have analyzed this translocation, further studies from different populations with an increased sample size and more sensitive techniques are required.
In another study, inverse PCR was employed to detect the presence of mixed lineage leukemia (MLL) translocations in the peripheral blood of healthy individuals [46]. Of the healthy individuals, 49 % were shown to harbor the MLL translocations. Two main types of rearrangements, t(4;11) and t(9;11), were found upon sequencing, which constituted 66 and 20 % of the total translocations, respectively. It was suggested that the frequent breaks in the 11q23 locus could be due to extended exposure to various exogenous and endogenous chemical agents.
The t(14;18) translocation in healthy individuals
Among the different translocations studied, t(14;18) or the BCL2-IgH translocation is the most commonly reported, even among the healthy population. It is detected in nearly 90 % of the FL patients and around 20 % of the DLBL patients. Using various types of PCR assays, this translocation has been detected in the healthy individuals. The prevalence of t(14;18) translocation in the healthy population from Europe and America was determined to be around 40–60 %, depending on the sensitivity of the assay system, and the sample size of the donors (Table 1) [54–60]. In a recent study, it was shown that the incidence of t(14;18) translocation in Japanese population was lower (16 %) when compared to that in the German population (52 %) [61]. However, a previous study comparing the frequency of t(14;18) translocation between Japanese and American population did not find much difference between the two [62]. This could probably be due to the small number of samples analyzed during this study. More recently, we have determined the incidence of t(14;18) translocation among the healthy population in India [52]. We employed nested PCR followed by Southern hybridization using BCL2 specific probes and the sensitivity of the assay was estimated to detect one translocation bearing cell among 107 cells [52]. Upon analysis of the blood samples from 253 healthy donors, we found that 87 individuals were positive for t(14;18) [52]. The observed incidence of around 34 % was lower than that seen in the American and European countries. It is worth pointing out that the incidence of FL itself varies across geographic regions. In the Western population, FL has a prevalence of around 30–40 % among all the non-Hodgkin’s lymphoma [63], while in India its occurrence has been estimated to be only around 13 % [64, 65]. Hence, it could suggest a direct correlation between occurrence of FL and the presence of t(14;18) in healthy people, as the incidence of FL in Western countries is much higher than that in India as well as other Asian countries.
Previous studies on the transgenic mice expressing the BCL2-IgH translocation showed accumulation of the B cells, but development to the malignant lymphoma occurred only after a long latency period [24, 66]. This indicated that the t(14;18) translocation alone was not sufficient for initiating the development of FL and that further oncogenic mutations may be required for transformation into a cancerous cell.
Factors influencing t(14;18) in healthy individuals
The role of additional elements other than those responsible for causing the t(14;18) translocation like RAGs or DNA DSB repair proteins, in inducing translocations, has been a subject of debate. Some studies, including ours, have shown that with an increase in age, the frequency of t(14;18) translocation increases [52, 55, 67, 68]. However, a number of studies did not find any such correlation [57, 59–61]. This discrepancy could be due to the fact that in the majority of the studies that did not find the correlation, even though the number of individuals analyzed were more than 200, the frequency of incidence of t(14;18) positives was very low [59–61]. This could probably be due to the smaller amount of DNA analyzed for each individual in these studies. Moreover, in case of the study that analyzed 481 healthy subjects (one of the largest samples sizes studied), real-time PCR assay could increase the chances of false negatives, since this assay requires the presence of at least ten copies of the rearranged allele for minimum detection [59]. Therefore, an under-representation of the frequency of t(14;18) translocation in healthy individuals in these studies could be responsible for the inability to find its correlation with age.
With respect to gender, one study suggested a higher incidence of this translocation in males as compared to females [68], which could explain why males are more prone to non-Hodgkin’s lymphoma than females. However, due to the insufficient sample size, this correlation could not be proved to be statistically significant. Besides, in a few other studies, no significant correlation between gender and the occurrence of t(14;18) translocation could be observed [52, 60]. In our study, we had analyzed for a possible correlation between the ethnicity and the prevalence of the translocation in the Indian population. For this, we classified all the 253 healthy volunteers, but were unable to find any significant correlation (Table 2). In a different study, a positive correlation between smoking and the occurrence of t(14;18) translocation was observed [69]. However, more studies are required in this direction to draw any major conclusions. Another interesting observation has been that in patients having chronic hepatitis, liver cirrhosis, or hepatocellular carcinoma due to hepatitis C virus (HCV) infection, the incidence of t(14;18) was much higher (26 %) as compared to HCV-negative patients having similar liver abnormalities (3.6 %) [70, 71]. It is possible that HCV infection may provide the necessary antigenic stimuli for the maintenance of these cells in circulation.
Table 2.
Geographic distribution of t(14;18) translocation in the healthy individuals in the Indian population
| Healthy individuals | North | South | East | West | ||||
|---|---|---|---|---|---|---|---|---|
| No. of individuals | % positive | No. of individuals | % positive | No. of individuals | % positive | No. of individuals | % positive | |
| Male | 17 | 7 (41.2 %) | 115 | 42 (36.5 %) | 22 | 8 (36.4 %) | 9 | 2 (22.2 %) |
| Female | 9 | 1 (11.1 %) | 66 | 23 (34.8 %) | 12 | 3 (25 %) | 6 | 2 (33.3 %) |
| Total | 26 | 8 (30.8 %) | 181 | 65 (35.9 %) | 34 | 11 (32.4 %) | 15 | 4 (26.6 %) |
All the healthy individuals studied have been broadly classified into four categories on the basis of their native places being in the northern, southern, eastern, or western parts of India. The percentage of people positive for the t(14;18) translocation has been calculated. The number of males and females in each region along with the percentage positive for t(14;18) have also been determined
Exposure to environmental pollutants or carcinogens is known to play a role in the generation of translocations in normal individuals. In an interesting study, it has been shown that the frequency of t(14;18) translocation in healthy individuals exposed to benzene was lower than normal age-matched controls [72]. By quantitative PCR analysis, this study found that 37 workers with benzene exposure had a decreased level of t(14;18) in their blood cells. 16.2 % of these workers had more than ten copies of the t(14;18) junctions as compared to 55 % of 20 controls that were not exposed to benzene [72]. This data suggests that the t(14;18)-bearing cells, a subset of B cells, could be more susceptible to the toxicity of benzene, resulting in their reduced numbers. However, studies have also shown that increased incidence of t(14;18) translocation occurs in farmers exposed to pesticides [73]. Previously, there has been evidence to suggest pesticide exposure as one of the major factors responsible for increased incidence of NHL [74–76]. In particular, exposure to a commonly used fumigant, phosphine, enhanced the development of chromosomal rearrangements involving chromosome 14 [77]. In an interesting study, it was found that a correlation could be drawn between chromosomal instability at specific fragile sites in human genome and certain specific chemicals, pesticides, and herbicides, suggesting a site-specific cytogenetic effect within cells [75]. This is in tandem with diverse chemicals having such site-specific consequences in vitro [78]. It was observed that increased breaks at all loci did not always lead to increased rearrangements as was seen for fragile sites 1q21, 3p14, and 9q12 [75]. However, in many cases, such an effect could be seen. In particular, it was noted that individuals exposed to herbicides like eradicane and 2,4-D had an elevated number of breaks at chromosomal loci 18q21, which harbors the BCL2 oncogene and subsequently had its increased rearrangement with 14q32, unlike the control and unexposed individuals [75]. Hence, it is possible that increased exposure of such chemicals could lead to increased double-strand breaks in the genome, especially at fragile sites like the BCL2 major breakpoint region and many others, thereby accounting for the higher frequency of translocations in such exposed individuals. More recently, a long-term study was carried out to understand the relation between pesticide exposure and follicular lymphomagenesis [79]. This study showed that the t(14;18) clones persisted and expanded, particularly in farmers exposed to pesticides rather than the unexposed farmers. In addition, these cells represented bona fide FL precursors at different stages of tumor progression. Therefore, now it remains to be seen whether one can predict how many of these t(14;18) harboring healthy individuals, would actually go on to develop the follicular lymphoma.
Chromosomal translocations and the immune system
The majority of the chromosomal translocations in cancer are largely confined to cells of the immune system and have been shown to be caused by lymphoid-specific processes such as V(D)J recombination or class switch recombination (CSR). Interestingly, from our studies on a healthy Indian population, it was observed that individuals under the age of 20 years had a very high frequency of incidence of the t(14;18) translocation as compared to all other age groups. Even more interesting was the higher incidence of the translocation in females as compared to males. Although this phenomenon needs to be investigated further, one can speculate the role of the immune system in this regard. At a younger age, one can envisage a more actively developing immune system and perhaps more of the immunological processes like V(D)J recombination or CSR to occur. It is known that with an increase in age there is reduced V(D)J recombination activity and an overall decreased immune response [80–82]. A lower Rag2 expression in pro-B cells has been shown in aged mice, which could directly diminish the V(D)J recombinase activity [83]. Another study showed that newborns displayed an increased nucleolytic processing, especially with respect to Artemis protein (associated with V(D)J recombination), indicating that it could be important for modulating the immune system in children [84]. Hence, at a younger age, these processes could increase the susceptibility of the developing B and T cells to generate chromosomal translocations. Interestingly, an increasing number of translocations have been identified in leukemic cells from children, which seem to be caused by the RAG proteins during early development [85]. A remarkable increase in the RAG-mediated HPRT deletions at functional cryptic RSS at the later stages of fetal development also suggests that cryptic RSS sites at various genomic loci could be susceptible to RAG cleavage during specific periods of pediatric development [86].
It is also becoming increasingly clear that there are significant differences in the immune responses between male and females. There are reports suggesting that females mount a more vigorous immune response against identical antigens and also carry larger numbers of resident immune cells, thereby eliciting a stronger response [87]. Moreover, it was observed that females exhibit a higher activity of TdT, a key enzyme involved in V(D)J recombination, suggesting that modulation of the processing machinery could affect the immune repertoire [84]. This can probably explain the bias towards more females carrying the translocation in younger ages. However, further studies are required to get a clear picture for this observation.
Origin of chromosomal translocations
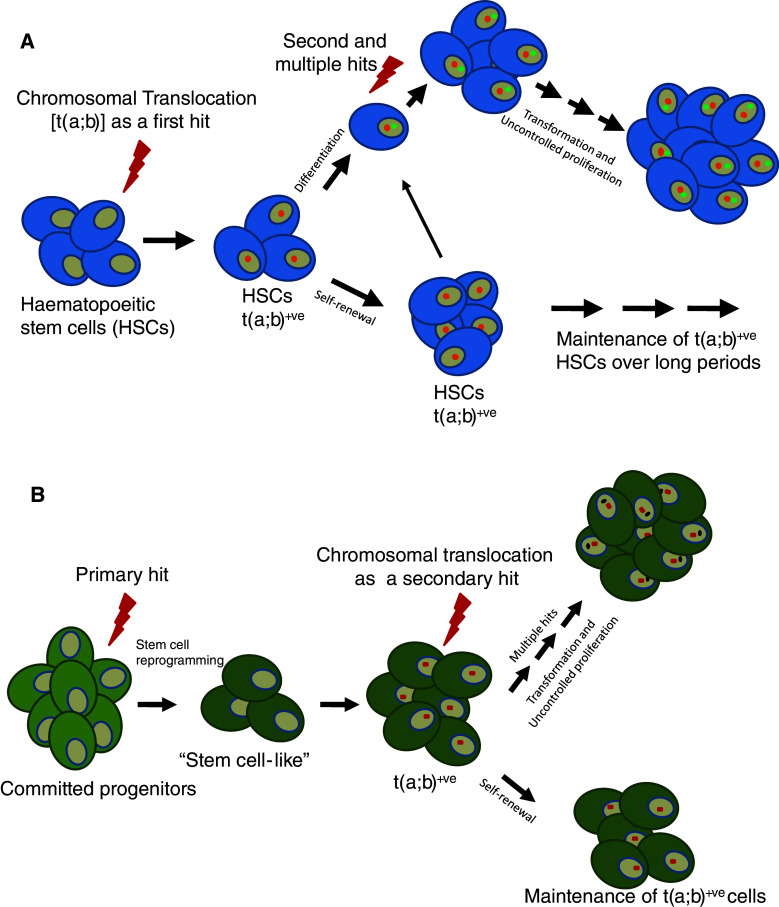
In the recent past, cancer stem cells have caught the attention of researchers in the field of cancer biology [88]. Although initially elusive, recent studies have been successful in isolating this population from leukemic cells and have been shown to be sufficient for the initiation, regeneration, and maintenance of leukemia in mice [89, 90]. More recently their presence has been detected in other forms of cancers like those in the central nervous system, breast, lungs, and colon [91–93]. However, the cellular origins of the cancer stem cells have remained elusive thus far. Since hematopoietic stem cells (HSCs) and leukemic stem cells (LSCs) are thought to be closely related and are similar in many ways, the genesis of LSCs has always been an intriguing question. HSCs have the capacity for self-renewal and are maintained for longer periods of time in an organism. Hence, it can be envisaged that LSCs could be derived from HSCs upon induction of several mutations, especially chromosomal translocations (Fig. 2a). Alternatively, various committed progenitor cells could also be a source of LSCs, after accumulation of mutations that render these cells “stem-cell like” by reactivation of the stem cell program machinery (Fig. 2b) [94]. Different studies have now shown that some of these stem cells or early progenitor cells could indeed harbor pathogenic chromosomal translocations. It was reported that in childhood acute lymphoblastic leukemia, the leukemic-specific rearrangements t(9;22) and t(4;11) could be detected in a high percentage of progenitor/stem cells, which were CD34+CD19− [95]. In another study, it was shown that ETV6-RUNX1 (TEL1-AML1) fusions were confined to B cell progenitor cells CD34+CD38−CD19+ and there was no effect on the size of the normal HSC population by the LSC expansion [96]. In contrast, the major breakpoint BCR-ABL1 fusions, encoding the P210 BCR-ABL fusion protein, were found to have an HSC origin and were present in the CD34+CD33−CD19+, CD34+CD33+CD19− and CD34−CD33+CD19− cells. Interestingly, the minor fusion protein was shown to have a B cell progenitor origin, which suggests that the two types of BCR-ABL fusion represented distinct tumor entities [96]. All these studies suggest that there are different compartments in which cancer-initiating cells responsible for causing different types of leukemia and lymphoma can exist. Therefore, it is important to identify the leukemia/lymphoma initiating cells in various cancers so that these cells can be targeted more effectively and specifically. Moreover, it is believed that HSCs are generally more resistant to radiation and other therapies, therefore targeting the translocation-bearing tumor initiating cells will be more appropriate during development of therapeutic strategies.
Fig. 2.
Models for transformation of a normal cell into cancer cell. a Hematopoietic stem cells can acquire chromosomal translocation as a primary hit and these cells can either differentiate or be maintained in stem cell compartments over long periods of time. The replicating, differentiating cells (leukemic stem cells) can then acquire secondary mutations over a period of time and develop into cancerous cells. b Alternatively, committed progenitors cells might undergo cellular reprogramming in order to get converted into stem cell-like cells, which upon acquiring secondary hits could become like cancer stem cells, responsible for the generation of the tumor over a period of time
Role of translocations in oncogenesis
Chromosomal translocations generally lead to deregulation of genes and hence, differential expression of respective proteins. The t(14;18) translocation results in the overexpression of the anti-apoptotic BCL2 protein thereby providing the cells with a survival advantage. It is possible that this could help in the persistence of such translocation-bearing cells in circulation for a longer time, which upon further oncogenic hits may get transformed into a malignant cell. Recent studies suggest that the t(14;18)-bearing cells in the healthy individuals might act as FL-like B cells (Fig. 3) [97]. The translocation-bearing cells seemed to be enriched in IgM memory cells (IgD+), further showing that such t(14;18) bearing B cells were not naive [97, 98]. These cells, being CD27+, would have transited through the germinal center. They also have additional features that are more FL-like and therefore could act as novel intermediates during the early steps of lymphomagenesis; but how do such cells originate? The sequences of the breakpoint junctions from both healthy individuals and patients are mostly similar, suggesting that the translocations arising in healthy individuals are not mechanistically different from those in patients [7, 54]. The translocation-bearing healthy cells are also thought to overexpress BCL2; however, no studies have been performed to confirm this. Since the t(14;18)-bearing normal cells are memory cells, they would persist for a long time in circulation and upon appropriate stimulation, by antigens (like the HCV antigen described before), could proliferate more and attain further oncogenic hits. Few studies have tried to establish this long-term clonal persistence of such t(14;18)-bearing cells in healthy individuals, by performing follow-up studies on the positive cases over a period of few years [54, 97, 98]. Sequence analysis has also shown that while the majority of the positive cases possess only one breakpoint, some do show the presence of more than one breakpoint region [54]. It can therefore be suggested that though initially there is a clonal expansion and persistence of a single t(14;18)-bearing clone, with time, independent hits may lead to the formation of multiple cells bearing different t(14;18) translocation breakpoint junctions. Further long-term and follow-up studies are needed to understand the step-by-step progression of translocation-bearing cells into tumor cells.
Fig. 3.
Model of initiation and progression of t(14;18)-bearing cell into follicular lymphoma. The pre-B cells, in the bone marrow, undergoing V(D)J recombination could develop the t(14;18) translocation and generate t(14;18)+ immature B cells. These cells upon antigen presentation in the germinal center undergo somatic hypermutation and class switch recombination, further acquiring possibly pathogenic mutations and could get selected upon antigenic stimulation. Such cells may evade apoptosis due to overexpression of the BCL2 protein and may be released in circulation or homed into undefined niches. These cells, which are more follicular lymphoma-like, now act as intermediates and may accumulate secondary aberrations in order to develop into the lymphoma
In mouse models of the MLL-ENL translocation in MLL, it was observed that DNA damage response (DDR) induces a pro-senescence program to prevent MLL-ENL oncogene-induced leukemogenesis [99]. This suggests that the gene fusion alone, although causing myeloproliferation, cannot promote progression into acute leukemia and requires DDR inhibition. It was also noted that preventing the initiation of senescence or DDR leads to augmentation in the LSC population, suggesting that the fusion does not directly induce tumorigenesis and requires an interplay of several other partners. Recently, it was shown that in case of CML, the stem cell population could survive independent of the BCR-ABL fusion and was not oncogene addicted [100]. The BCR-ABL-depleted stem cell population had the ability to maintain their population in vivo and upon BCR-ABL expression could reinitiate transformation into leukemia. This shows that unlike the MLL-ENL fusion, in some cases the development of translocation could be a secondary event, and may be required to trigger the process of leukemia progression. In an interesting study, it was shown that MLL fusions, in particular MLL-ENL, could influence hematopoietic lineage commitment when occurring in the T cell progenitor cell and cause a switch from lymphoid to myeloid lineage during leukemogenesis by reprogramming [101]. Therefore, it suggests that depending on the type of cell, the translocation originates in, the gene fusion can exhibit alternative functions and promote tumorigenesis. However, this also provides a plethora of proteins that could be identified and act as novel therapeutic targets in the future.
Conclusions
It is evident that many chromosomal translocations can also be seen in healthy individuals, besides their occurrence in patients. In almost all of these cases, the frequency of translocation in healthy individuals is many fold higher to the incidence of the respective cancer. This implies that only a small fraction of chromosomal translocation-bearing cells present in healthy individuals can undergo malignant transformation. Therefore, in the coming years, the focus of research may shift to the identification of the environmental and physiological aspects responsible for development of cancer, after a single cell in the body acquires a particular translocation.
Perspective
The long-held view that chromosomal translocations are genetic hallmarks of leukemia, lymphoma, and other types of cancers, and can be used as biomarkers for identification of tumor cells, is fast disappearing. It is increasingly becoming clear that chromosomal translocations are not sufficient to cause transformation of normal to cancer cells, but requirement of either additional preceding or succeeding mutations is essential. Since many such genetic lesions do not lead to any specific changes on the cell surface, expulsion of such cells from the system by our immune response is not possible. Evidence for presence of translocation-bearing HSCs or progenitor cells can possibly explain the persistence of such cells over long periods of time. It will be important to study whether such stem/progenitor cells are more susceptible to development of genomic rearrangements and thus strategies can be developed to specifically target and eliminate them in patients. At the molecular level, it is seen that the translocation breakpoint junctions in healthy individuals are similar to that in patients. However, as in the case of BCL2-IgH translocation, it is not proven whether the differential expression of the affected protein like BCL2, indeed occurs. It is possible that there could be some mechanism by which the aberrant expression of those oncoproteins, despite the translocation, is suppressed in normal cells. However, with the gradual acquiring of certain mutations, the oncoproteins get expressed and activated. The microenvironment around the translocation-bearing normal cells can also play a critical role in transforming the cells more towards cancer-like. Identification of specific niches where such translocation-bearing cells reside could be a challenging yet critical aspect in the future.
Acknowledgments
We thank Mr. Kohal Das, Ms. Nishana M. and Ms. Divyaanka Iyyer for critical reading of the manuscript. Our work is supported by grants from DBT, India BT/PR13722/BRB/10/781/2010 for SCR. MN is supported by IISc Research Associate Fellowship.
References
- 1.Rowley JD. Chromosome translocations: dangerous liaisons revisited. Nat Rev Cancer. 2001;1:245–250. doi: 10.1038/35106108. [DOI] [PubMed] [Google Scholar]
- 2.Rabbitts TH. Chromosomal translocations in human cancer. Nature. 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 3.Korsmeyer SJ. Chromosomal translocations in lymphoid malignancies reveal novel proto-oncogenes. Annu Rev Immunol. 1992;10:785–807. doi: 10.1146/annurev.iy.10.040192.004033. [DOI] [PubMed] [Google Scholar]
- 4.Nowell PC, Hungerford DA. A minute chromosome in human chronic granulocytic leukemia. Science. 1960;132:1497. doi: 10.1126/science.144.3623.1229. [DOI] [PubMed] [Google Scholar]
- 5.Bakhshi A, Jensen JP, Goldman P, Wright JJ, McBride OW, Epstein AL, Korsmeyer SJ. Cloning the chromosomal breakpoint of t(14;18) human lymphomas: clustering around JH on chromosome 14 and near a transcriptional unit on 18. Cell. 1985;41:899–906. doi: 10.1016/S0092-8674(85)80070-2. [DOI] [PubMed] [Google Scholar]
- 6.Armstrong SA, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 7.Jager U, et al. Follicular lymphomas’ BCL-2/IgH junctions contain templated nucleotide insertions: novel insights into the mechanism of t(14;18) translocation. Blood. 2000;95:3520–3529. [PubMed] [Google Scholar]
- 8.Taub R, Kirsch I, Morton C, Lenoir G, Swan D, Tronick S, Aaronson S, Leder P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci USA. 1982;79:7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cleary ML, Sklar J. DNA rearrangements in non-Hodgkin’s lymphomas. Cancer Surv. 1985;4:331–348. [PubMed] [Google Scholar]
- 10.Lieber MR (1993) In: Kirsch I (ed) The Causes and Consequences of Chromosomal Aberrations. CRC Press, USA, pp 239–275
- 11.Nambiar M, Kari V, Raghavan SC. Chromosomal translocations in cancer. Biochim Biophys Acta. 2008;1786:139–152. doi: 10.1016/j.bbcan.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Nambiar M, Choudhary B, Rao CR, Raghavan SC. Amplification of chromosomal translocation junctions from paraffin-embedded tissues of follicular lymphoma patients. Biomed Mater. 2008;3:034103. doi: 10.1088/1748-6041/3/3/034103. [DOI] [PubMed] [Google Scholar]
- 13.Tsujimoto Y, Yunis J, Onorato-Showe L, Erikson J, Nowell PC, Croce CM. Molecular cloning of the chromosomal breakpoint of B-cell lymphomas and leukemias with the t(11;14) chromosome translocation. Science. 1984;224:1403–1406. doi: 10.1126/science.6610211. [DOI] [PubMed] [Google Scholar]
- 14.Tsujimoto Y, Jaffe E, Cossman J, Gorham J, Nowell PC, Croce CM. Clustering of breakpoints on chromosome 11 in human B-cell neoplasms with the t(11;14) chromosome translocation. Nature. 1985;315:340–343. doi: 10.1038/315340a0. [DOI] [PubMed] [Google Scholar]
- 15.Raghavan SC, Lieber MR. DNA structures at chromosomal translocation sites. BioEssays. 2006;28:480–494. doi: 10.1002/bies.20353. [DOI] [PubMed] [Google Scholar]
- 16.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 17.Tomlins SA, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 18.Tomlins SA, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 19.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Evidence of recurrent gene fusions in common epithelial tumors. Trends Mol Med. 2006;12:529–536. doi: 10.1016/j.molmed.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Heisterkamp N, Stam K, Groffen J, de Klein A, Grosveld G. Structural organization of the bcr gene and its role in the Ph’ translocation. Nature. 1985;315:758–761. doi: 10.1038/315758a0. [DOI] [PubMed] [Google Scholar]
- 22.Groffen J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36:93–99. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- 23.Groffen J, Heisterkamp N. The BCR/ABL hybrid gene. Bailliere’s Clin Haematol. 1987;1:983–999. doi: 10.1016/S0950-3536(87)80035-5. [DOI] [PubMed] [Google Scholar]
- 24.McDonnell TJ, Deane N, Platt FM, Nunez G, Jaeger U, McKearn JP, Korsmeyer SJ. Bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell. 1989;57:79–88. doi: 10.1016/0092-8674(89)90174-8. [DOI] [PubMed] [Google Scholar]
- 25.Korsmeyer SJ. BCL-2 gene family and the regulation of programmed cell death. Cancer Res. 1999;59:1693s–1700s. [PubMed] [Google Scholar]
- 26.Graninger W, Seto M, Boutain B, Goldman P, Korsmeyer SJ. Expression of Bcl-2 and Bcl-2-Ig fusion transcripts in normal and neoplastic cells. J Clin Invest. 1987;80:1512–1515. doi: 10.1172/JCI113235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bakhshi A, et al. Mechanism of the t(14;18) chromosomal translocation: structural analysis of both derivative 14 and 18 reciprocal partners. Proc Natl Acad Sci USA. 1987;84:2396–2400. doi: 10.1073/pnas.84.8.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raghavan SC, Swanson PC, Wu X, Hsieh CL, Lieber MR. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- 29.Nambiar M, Raghavan SC. Mechanism of fragility at BCL2 gene minor breakpoint cluster region during t(14;18) chromosomal translocation. J Biol Chem. 2012;287:8688–8701. doi: 10.1074/jbc.M111.307363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nambiar M, Goldsmith G, Moorthy BT, Lieber MR, Joshi VM, Choudhary B, Hosur RV, Raghavan SC. Formation of a G-quadruplex at the BCL2 major breakpoint region of the t(14;18) translocation in follicular lymphoma. Nucleic Acids Res. 2011;39:936–948. doi: 10.1093/nar/gkq824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalla-Favera R, et al. Identification of genetic lesions associated with diffuse large-cell lymphoma. Ann Oncol. 1994;5(Suppl 1):55–60. doi: 10.1093/annonc/5.suppl_1.S55. [DOI] [PubMed] [Google Scholar]
- 32.Cesarman E, Dalla-Favera R, Bentley D, Groudine M. Mutations in the first exon are associated with altered transcription of c-myc in Burkitt lymphoma. Science. 1987;238:1272–1275. doi: 10.1126/science.3685977. [DOI] [PubMed] [Google Scholar]
- 33.Gelmann EP, Psallidopoulos MC, Papas TS, Dalla-Favera R. Identification of reciprocal translocation sites within the c-myc oncogene and immunoglobulin mu locus in a Burkitt lymphoma. Nature. 1983;306:799–803. doi: 10.1038/306799a0. [DOI] [PubMed] [Google Scholar]
- 34.Ye BH, Chaganti S, Chang CC, Niu H, Corradini P, Chaganti RS, Dalla-Favera R. Chromosomal translocations cause deregulated BCL6 expression by promoter substitution in B cell lymphoma. EMBO J. 1995;14:6209–6217. doi: 10.1002/j.1460-2075.1995.tb00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamlyn PH, Rabbitts TH. Translocation joins c-myc and immunoglobulin gamma 1 genes in a Burkitt lymphoma revealing a third exon in the c-myc oncogene. Nature. 1983;304:135–139. doi: 10.1038/304135a0. [DOI] [PubMed] [Google Scholar]
- 36.Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, Palanisamy N, Chinnaiyan AM. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomlins SA, Bjartell A, Chinnaiyan AM, Jenster G, Nam RK, Rubin MA, Schalken JA. ETS gene fusions in prostate cancer: from discovery to daily clinical practice. Eur Urol. 2009;56:275–286. doi: 10.1016/j.eururo.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 39.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brenner JC, Chinnaiyan AM. Translocations in epithelial cancers. Biochim Biophys Acta. 2009;1796:201–215. doi: 10.1016/j.bbcan.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nambiar M, Raghavan SC. How does DNA break during chromosomal translocations? Nucleic Acids Res. 2011;39:5813–5825. doi: 10.1093/nar/gkr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janz S, Potter M, Rabkin CS. Lymphoma- and leukemia-associated chromosomal translocations in healthy individuals. Genes Chromosomes Cancer. 2003;36:211–223. doi: 10.1002/gcc.10178. [DOI] [PubMed] [Google Scholar]
- 43.Rabkin CS, Hirt C, Janz S, Dolken G. t(14;18) Translocations and risk of follicular lymphoma. J Natl Cancer Inst Monogr. 2008;39:48–51. doi: 10.1093/jncimonographs/lgn002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuler F, Hirt C, Dolken G. Chromosomal translocation t(14;18) in healthy individuals. Semin Cancer Biol. 2003;13:203–209. doi: 10.1016/S1044-579X(03)00016-6. [DOI] [PubMed] [Google Scholar]
- 45.Basecke J, Griesinger F, Trumper L, Brittinger G. Leukemia- and lymphoma-associated genetic aberrations in healthy individuals. Ann Hematol. 2002;81:64–75. doi: 10.1007/s00277-002-0427-x. [DOI] [PubMed] [Google Scholar]
- 46.Brassesco MS, Montaldi AP, Gras DE, de Paula Queiroz RG, Martinez-Rossi NM, Tone LG, Sakamoto-Hojo ET. MLL leukemia-associated rearrangements in peripheral blood lymphocytes from healthy individuals. Genet Mol Biol. 2009;32:234–241. doi: 10.1590/S1415-47572009000200005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biernaux C, Loos M, Sels A, Huez G, Stryckmans P. Detection of major bcr-abl gene expression at a very low level in blood cells of some healthy individuals. Blood. 1995;86:3118–3122. [PubMed] [Google Scholar]
- 48.Bose S, Deininger M, Gora-Tybor J, Goldman JM, Melo JV. The presence of typical and atypical BCR-ABL fusion genes in leukocytes of normal individuals: biologic significance and implications for the assessment of minimal residual disease. Blood. 1998;92:3362–3367. [PubMed] [Google Scholar]
- 49.Trumper L, Pfreundschuh M, Bonin FV, Daus H. Detection of the t(2;5)-associated NPM/ALK fusion cDNA in peripheral blood cells of healthy individuals. Br J Haematol. 1998;103:1138–1144. doi: 10.1046/j.1365-2141.1998.01097.x. [DOI] [PubMed] [Google Scholar]
- 50.Maes B, Vanhentenrijk V, Wlodarska I, Cools J, Peeters B, Marynen P, de Wolf-Peeters C. The NPM-ALK and the ATIC-ALK fusion genes can be detected in non-neoplastic cells. Am J Pathol. 2001;158:2185–2193. doi: 10.1016/S0002-9440(10)64690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirt C, Schuler F, Dolken L, Schmidt CA, Dolken G. Low prevalence of circulating t(11;14)(q13;q32)-positive cells in the peripheral blood of healthy individuals as detected by real-time quantitative PCR. Blood. 2004;104:904–905. doi: 10.1182/blood-2004-02-0738. [DOI] [PubMed] [Google Scholar]
- 52.Nambiar M, Raghavan SC. Prevalence and analysis of t(14;18) and t(11;14) chromosomal translocations in healthy Indian population. Ann Hematol. 2010;89:35–43. doi: 10.1007/s00277-009-0755-1. [DOI] [PubMed] [Google Scholar]
- 53.Lecluse Y, Lebailly P, Roulland S, Gac AC, Nadel B, Gauduchon P. t(11;14)-positive clones can persist over a long period of time in the peripheral blood of healthy individuals. Leukemia. 2009;23:1190–1193. doi: 10.1038/leu.2009.31. [DOI] [PubMed] [Google Scholar]
- 54.Roulland S, Lebailly P, Lecluse Y, Heutte N, Nadel B, Gauduchon P. Long-term clonal persistence and evolution of t(14;18)-bearing B cells in healthy individuals. Leukemia. 2006;20:158–162. doi: 10.1038/sj.leu.2404035. [DOI] [PubMed] [Google Scholar]
- 55.Liu Y, Hernandez AM, Shibata D, Cortopassi GA. BCL2 translocation frequency rises with age in humans. Proc Natl Acad Sci USA. 1994;91:8910–8914. doi: 10.1073/pnas.91.19.8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dolken G, Illerhaus G, Hirt C, Mertelsmann R. BCL-2/JH rearrangements in circulating B cells of healthy blood donors and patients with nonmalignant diseases. J Clin Oncol. 1996;14:1333–1344. doi: 10.1200/JCO.1996.14.4.1333. [DOI] [PubMed] [Google Scholar]
- 57.Fuscoe JC, Setzer RW, Collard DD, Moore MM. Quantification of t(14;18) in the lymphocytes of healthy adult humans as a possible biomarker for environmental exposures to carcinogens. Carcinogenesis. 1996;17:1013–1020. doi: 10.1093/carcin/17.5.1013. [DOI] [PubMed] [Google Scholar]
- 58.Limpens J, Stad R, Vos C, de Vlaam C, de Jong D, van Ommen GJ, Schuuring E, Kluin PM. Lymphoma-associated translocation t(14;18) in blood B cells of normal individuals. Blood. 1995;85:2528–2536. [PubMed] [Google Scholar]
- 59.Summers KE, Goff LK, Wilson AG, Gupta RK, Lister TA, Fitzgibbon J. Frequency of the Bcl-2/IgH rearrangement in normal individuals: implications for the monitoring of disease in patients with follicular lymphoma. J Clin Oncol. 2001;19:420–424. doi: 10.1200/JCO.2001.19.2.420. [DOI] [PubMed] [Google Scholar]
- 60.Schmitt C, et al. The bcl-2/IgH rearrangement in a population of 204 healthy individuals: occurrence, age and gender distribution, breakpoints, and detection method validity. Leuk Res. 2006;30:745–750. doi: 10.1016/j.leukres.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 61.Yasukawa M, Bando S, Dolken G, Sada E, Yakushijin Y, Fujita S, Makino H. Low frequency of BCL-2/J(H) translocation in peripheral blood lymphocytes of healthy Japanese individuals. Blood. 2001;98:486–488. doi: 10.1182/blood.V98.2.486. [DOI] [PubMed] [Google Scholar]
- 62.Aster JC, Kobayashi Y, Shiota M, Mori S, Sklar J. Detection of the t(14;18) at similar frequencies in hyperplastic lymphoid tissues from American and Japanese patients. Am J Pathol. 1992;141:291–299. [PMC free article] [PubMed] [Google Scholar]
- 63.Biagi JJ, Seymour JF. Insights into the molecular pathogenesis of follicular lymphoma arising from analysis of geographic variation. Blood. 2002;99:4265–4275. doi: 10.1182/blood.V99.12.4265. [DOI] [PubMed] [Google Scholar]
- 64.Sahni CS, Desai SB. Distribution and clinicopathologic characteristics of non-Hodgkin’s lymphoma in India: a study of 935 cases using WHO classification of lymphoid neoplasms (2000) Leuk Lymphoma. 2007;48:122–133. doi: 10.1080/10428190601043351. [DOI] [PubMed] [Google Scholar]
- 65.Naresh KN, Srinivas V, Soman CS. Distribution of various subtypes of non-Hodgkin’s lymphoma in India: a study of 2773 lymphomas using R.E.A.L. and WHO Classifications. Ann Oncol. 2000;11:63–67. doi: 10.1023/A:1008325827059. [DOI] [PubMed] [Google Scholar]
- 66.McDonnell TJ, Korsmeyer SJ. Progression from lymphoid hyperplasia to high-grade malignant lymphoma in mice transgenic for the t(14;18) Nature. 1991;349:254–256. doi: 10.1038/349254a0. [DOI] [PubMed] [Google Scholar]
- 67.Dolken G, Dolken L, Hirt C, Fusch C, Rabkin CS, Schuler F. Age-dependent prevalence and frequency of circulating t(14;18)-positive cells in the peripheral blood of healthy individuals. J Natl Cancer Inst Monogr. 2008;39:44–47. doi: 10.1093/jncimonographs/lgn005. [DOI] [PubMed] [Google Scholar]
- 68.Ji W, Qu G, Ye P, Zhang X-Y, Halabi S, Ehrlich M. Frequent detection of Bcl-2/JH translocations in human blood and organ samples by a quantitative polymerase chain reaction assay. Cancer Res. 1995;55:2876–2882. [PubMed] [Google Scholar]
- 69.Bell DA, Liu Y, Cortopassi GA. Occurrence of bcl-2 oncogene translocation with increased frequency in the peripheral blood of heavy smokers. J Natl Cancer Inst. 1995;87:223–224. doi: 10.1093/jnci/87.3.223. [DOI] [PubMed] [Google Scholar]
- 70.Zuckerman E, Zuckerman T, Sahar D, Streichman S, Attias D, Sabo E, Yeshurun D, Rowe JM. The effect of antiviral therapy on t(14;18) translocation and immunoglobulin gene rearrangement in patients with chronic hepatitis C virus infection. Blood. 2001;97:1555–1559. doi: 10.1182/blood.V97.6.1555. [DOI] [PubMed] [Google Scholar]
- 71.Zignego AL, et al. T(14;18) translocation in chronic hepatitis C virus infection. Hepatology. 2000;31:474–479. doi: 10.1002/hep.510310230. [DOI] [PubMed] [Google Scholar]
- 72.McHale CM, et al. Chromosome translocations in workers exposed to benzene. J Natl Cancer Inst Monogr. 2008;39:74–77. doi: 10.1093/jncimonographs/lgn010. [DOI] [PubMed] [Google Scholar]
- 73.Roulland S, Lebailly P, Lecluse Y, Briand M, Pottier D, Gauduchon P. Characterization of the t(14;18) BCL2-IGH translocation in farmers occupationally exposed to pesticides. Cancer Res. 2004;64:2264–2269. doi: 10.1158/0008-5472.CAN-03-3604. [DOI] [PubMed] [Google Scholar]
- 74.Zahm SH, Blair A. Pesticides and non-Hodgkin’s lymphoma. Cancer Res. 1992;52:5485s–5488s. [PubMed] [Google Scholar]
- 75.Garry VF, Tarone RE, Long L, Griffith J, Kelly JT, Burroughs B. Pesticide appliers with mixed pesticide exposure: G-banded analysis and possible relationship to non-Hodgkin’s lymphoma. Cancer Epidemiol Biomarkers Prev. 1996;5:11–16. [PubMed] [Google Scholar]
- 76.McDuffie HH, et al. Non-Hodgkin’s lymphoma and specific pesticide exposures in men: cross-Canada study of pesticides and health. Cancer Epidemiol Biomarkers Prev. 2001;10:1155–1163. [PubMed] [Google Scholar]
- 77.Garry VF, Griffith J, Danzl TJ, Nelson RL, Whorton EB, Krueger LA, Cervenka J. Human genotoxicity: pesticide applicators and phosphine. Science. 1989;246:251–255. doi: 10.1126/science.2799386. [DOI] [PubMed] [Google Scholar]
- 78.Yunis JJ, Soreng AL, Bowe AE. Fragile sites are targets of diverse mutagens and carcinogens. Oncogene. 1987;1:59–69. [PubMed] [Google Scholar]
- 79.Agopian J, et al. Agricultural pesticide exposure and the molecular connection to lymphomagenesis. J Exp Med. 2009;206:1473–1483. doi: 10.1084/jem.20082842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller RA. The aging immune system: primer and prospectus. Science. 1996;273:70–74. doi: 10.1126/science.273.5271.70. [DOI] [PubMed] [Google Scholar]
- 81.Szabo P, Zhao K, Kirman I, Le Maoult J, Dyall R, Cruikshank W, Weksler ME. Maturation of B cell precursors is impaired in thymic-deprived nude and old mice. J Immunol. 1998;161:2248–2253. [PubMed] [Google Scholar]
- 82.Ghia P, Melchers F, Rolink AG. Age-dependent changes in B lymphocyte development in man and mouse. Exp Gerontol. 2000;35:159–165. doi: 10.1016/S0531-5565(99)00095-9. [DOI] [PubMed] [Google Scholar]
- 83.Labrie JE, 3rd, Borghesi L, Gerstein RM. Bone marrow microenvironmental changes in aged mice compromise V(D)J recombinase activity and B cell generation. Semin Immunol. 2005;17:347–355. doi: 10.1016/j.smim.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 84.Murray JM, et al. V(D)J recombinase-mediated processing of coding junctions at cryptic recombination signal sequences in peripheral T cells during human development. J Immunol. 2006;177:5393–5404. doi: 10.4049/jimmunol.177.8.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yoshioka M, O’Neill JP, Vacek PM, Finette BA. Gestational age and gender-specific in utero V(D)J recombinase-mediated deletions. Cancer Res. 2001;61:3432–3438. [PubMed] [Google Scholar]
- 86.Finette BA, Kendall H, Vacek PM. Mutational spectral analysis at the HPRT locus in healthy children. Mutat Res. 2002;505:27–41. doi: 10.1016/S0027-5107(02)00119-7. [DOI] [PubMed] [Google Scholar]
- 87.Scotland RS, Stables MJ, Madalli S, Watson P, Gilroy DW. Sex differences in resident immune cell phenotype underlie more efficient acute inflammatory responses in female mice. Blood. 2011;118:5918–5927. doi: 10.1182/blood-2011-03-340281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 89.Lapidot T, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 90.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 91.Al-Hajj M, Clarke MF. Self-renewal and solid tumor stem cells. Oncogene. 2004;23:7274–7282. doi: 10.1038/sj.onc.1207947. [DOI] [PubMed] [Google Scholar]
- 92.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 93.Liu R, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 94.Passegue E, Jamieson CH, Ailles LE, Weissman IL. Normal and leukemic hematopoiesis: are leukemias a stem cell disorder or a reacquisition of stem cell characteristics? Proc Natl Acad Sci USA. 2003;100(Suppl 1):11842–11849. doi: 10.1073/pnas.2034201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hotfilder M, et al. Leukemic stem cells in childhood high-risk ALL/t(9;22) and t(4;11) are present in primitive lymphoid-restricted CD34+CD19− cells. Cancer Res. 2005;65:1442–1449. doi: 10.1158/0008-5472.CAN-04-1356. [DOI] [PubMed] [Google Scholar]
- 96.Castor A, et al. Distinct patterns of hematopoietic stem cell involvement in acute lymphoblastic leukemia. Nat Med. 2005;11:630–637. doi: 10.1038/nm1253. [DOI] [PubMed] [Google Scholar]
- 97.Roulland S, et al. Follicular lymphoma-like B cells in healthy individuals: a novel intermediate step in early lymphomagenesis. J Exp Med. 2006;203:2425–2431. doi: 10.1084/jem.20061292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hirt C, Dolken G, Janz S, Rabkin CS. Distribution of t(14;18)-positive, putative lymphoma precursor cells among B-cell subsets in healthy individuals. Br J Haematol. 2007;138:349–353. doi: 10.1111/j.1365-2141.2007.06671.x. [DOI] [PubMed] [Google Scholar]
- 99.Takacova S, Slany R, Bartkova J, Stranecky V, Dolezel P, Luzna P, Bartek J, Divoky V. DNA damage response and inflammatory signaling limit the MLL-ENL-induced leukemogenesis in vivo. Cancer Cell. 2012;21:517–531. doi: 10.1016/j.ccr.2012.01.021. [DOI] [PubMed] [Google Scholar]
- 100.Hamilton A, et al. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood. 2012;119:1501–1510. doi: 10.1182/blood-2010-12-326843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Drynan LF, Pannell R, Forster A, Chan NM, Cano F, Daser A, Rabbitts TH. Mll fusions generated by Cre-loxP-mediated de novo translocations can induce lineage reassignment in tumorigenesis. EMBO J. 2005;24:3136–3146. doi: 10.1038/sj.emboj.7600760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Paltiel O, Zelenetz A, Sverdlin I, Gordon L, Ben-Yehuda D. Translocation t(14;18) in healthy individuals: preliminary study of its association with family history and agricultural exposure. Ann Oncol. 2000;11(Suppl 1):75–80. doi: 10.1093/annonc/11.suppl_1.S75. [DOI] [PubMed] [Google Scholar]
- 103.Henriksson G, Brant M, Sandor Z, Manthorpe R, Bredberg A. Sjögren's syndrome: lymphoma predisposition coupled with a reduced frequency of t(14;18) translocations in blood lymphocytes. Mol Carcinog. 1999;24:226–231. doi: 10.1002/(SICI)1098-2744(199903)24:3<226::AID-MC9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 104.Scheerer JB, Xi L, Knapp GW, Setzer RW, Bigbee WL, Fuscoe JC. Quantification of illegitimate V(D)J recombinase-mediated mutations in lymphocytes of newborns and adults. Mutat Res. 1999;431:291–303. doi: 10.1016/S0027-5107(99)00173-6. [DOI] [PubMed] [Google Scholar]
- 105.Rauzy O, Galoin S, Chale JJ, Adoue D, Albarede JL, Delsol G, al Saati T. Detection of t(14;18) carrying cells in bone marrow and peripheral blood from patients affected by non-lymphoid diseases. Mol Pathol. 1998;51:333–338. doi: 10.1136/mp.51.6.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu Y, Cortopassi G, Goedert JJ, Rabkin CS. Frequency of Bcl-2 rearrangements in peripheral blood of HIV-infected individuals. Br J Haematol. 1997;99:465–466. [PubMed] [Google Scholar]
- 107.Molina TJ, Devez F, Bigorgne C, Le Tourneau A, Joulin V, Audouin J, Diebold J. Detection of bcl-2 rearrangement in HIV-related follicular lymphoid hyperplasia. Br J Haematol. 1996;94:705–708. doi: 10.1046/j.1365-2141.1996.d01-1837.x. [DOI] [PubMed] [Google Scholar]
- 108.Cole J, Green MH, Bridges BA, Waugh AP, Beare DM, Henshaw D, Last R, Liu Y, Cortopassi G. Lack of evidence for an association between the frequency of mutants or translocations in circulating lymphocytes and exposure to radon gas in the home. Radiat Res. 1996;145:61–69. doi: 10.2307/3579196. [DOI] [PubMed] [Google Scholar]
- 109.Corbally N, Grogan L, Keane MM, Devaney DM, Dervan PA, Carney DN. Bcl-2 rearrangement in Hodgkin's disease and reactive lymph nodes. Am J Clin Pathol. 1994;101:756–760. doi: 10.1093/ajcp/101.6.756. [DOI] [PubMed] [Google Scholar]
- 110.Limpens J, Jong D, Krieken JH, Price CG, Young BD, Ommen GJ, Kluin PM. Bcl-2/JH rearrangements in benign lymphoid tissues with follicular hyperplasia. Oncogene. 1991;6:2271–2276. [PubMed] [Google Scholar]
- 111.Ladetto M, Mantoan B, De Marco F, Drandi D, Aguzzi C, Astolfi M, Vallet S, Ricca I, Dell' Aquila M, Pagliano G, Monitillo L, Pollio B, Santo L, Cristiano C, Rocci A, Francese R, Bodoni CL, Borchiellini A, Schinco P, Boccadoro M, Tarella C. Cells carrying nonlymphoma-associated bcl-2/IgH rearrangements (NLABR) are phenotypically related to follicular lymphoma and can establish as long-term persisting clonal populations. Exp Hematol. 2006;34:1680–1686. doi: 10.1016/j.exphem.2006.08.008. [DOI] [PubMed] [Google Scholar]