Abstract
Forkhead box class O family member proteins (FoxOs) are highly conserved transcription factors with important roles in cellular homeostasis. The four FoxO members in humans, FoxO1, FoxO3, FoxO4, and FoxO6, are all expressed in skeletal muscle, but the first three members are the most studied in muscle. In this review, we detail the multiple modes of FoxO regulation and discuss the central role of these proteins in the control of skeletal muscle plasticity. FoxO1 and FoxO3 are key factors of muscle energy homeostasis through the control of glycolytic and lipolytic flux, and mitochondrial metabolism. They are also key regulators of protein breakdown, as they modulate the activity of several actors in the ubiquitin–proteasome and autophagy–lysosomal proteolytic pathways, including mitochondrial autophagy, also called mitophagy. FoxO proteins have also been implicated in the regulation of the cell cycle, apoptosis, and muscle regeneration. Depending of their activation level, FoxO proteins can exhibit ambivalent functions. For example, a basal level of FoxO factors is necessary for cellular homeostasis and these proteins are required for adaptation to exercise. However, exacerbated activation may occur in the course of several diseases, resulting in metabolic disorders and atrophy. A better understanding of the precise functions of these transcriptions factors should thus lead to the development of new therapeutic approaches to prevent or limit the muscle wasting that prevails in numerous pathological states, such as immobilization, denervated conditions, neuromuscular disease, aging, AIDS, cancer, and diabetes.
Keywords: Forkhead box class O, Metabolism, Atrophy, Autophagy, Mitophagy, E3 ligase
Introduction
Skeletal muscle mass represents 40–50 % of human body weight, making it the largest tissue mass in the body. Muscle homeostasis is essential to the body’s integrity and maintenance, and the muscle impairment associated with several diseases leads to a poor quality of life. Skeletal muscle exhibits remarkable adaptive capabilities in response to such stimuli as environmental factors (hypoxia), nutritional interventions, loading conditions, and contractile activity. All of these stimuli induce changes in energy metabolism and muscle mass, especially by altering fiber composition or the balance between protein synthesis and protein degradation [1, 2]. In this respect, the forkhead box class O (FoxO) subfamily proteins—which are transcription factors belonging to the forkhead box protein family (Fox proteins)—are increasingly taken into consideration. They are highly conserved through evolution and essential to various cellular processes such as regulating the expression of the genes involved in the cell cycle [3, 4], DNA damage repair [5], oxidative stress resistance [6–8], energy metabolism [9], and apoptosis [10, 11]. Fox proteins contain a sequence of 80–100 amino acids forming a motif that binds to DNA: the forkhead motif. This motif is also known as the winged helix due to the butterfly-like appearance of the loops in the protein structure of the domain [12]. In addition to the forkhead DNA-binding domain motif, FoxO factors possess a nuclear localization sequence (NLS), a nuclear export sequence (NES), and a transactivation domain in their C terminal region in which a helical motif (LXXLL, where L is a leucine and X any amino acid) is quite important for full activation of FoxO transcriptional activity [13] (Fig. 1). Four FoxO family members are expressed in mammals, FoxO1 (also known as FoxO1a), FoxO3 (also known as FoxO3a), FoxO4, and FoxO6, and all of them are expressed in skeletal muscles [14, 15]. Nonetheless, it is notable that, contrary to FoxO1, 3 and 4, which are expressed relatively ubiquitously, FoxO6 is expressed predominantly in the central nervous system [16], although it is also present in oxidative muscles [15]. Also of note, FoxO3- and FoxO4-null mice are viable, but FoxO1-null mice die during embryonic development due to impaired vasculogenesis [17–19]. FoxO3-null mice, although viable, present age-dependent infertility due to abnormal ovarian follicular development [17, 18] and, interestingly, they are characterized by severely impaired muscle regeneration [20]. Indeed, FoxO3-null mice show a downregulation of MyoD transcription, a key regulator of myogenesis that is a direct target of FoxO3 in myoblasts [20]. Moreover, hearts from FoxO3-null mice have a hypertrophic phenotype associated with increased expression of the modulatory calcineurin-interacting protein 1, exon 4 isoform (MCIP1.4) [19]. These data suggest that FoxO3 contributes to cell growth in striated muscle. On the other hand, FoxO4-null mice do not seem to be affected by any consistent abnormalities under physiological conditions [18]. However, under stress conditions like acute colitis, FoxO4 deficiency has been associated with increased nuclear factor-kappa B (NF-κB) activity in colonic epithelial cells [21]. Last, mice lacking FoxO6 are viable but show substantial impairment of contextual and object memory consolidation [16]. However, the precise roles of this last member are largely unknown to date, especially in skeletal muscle.
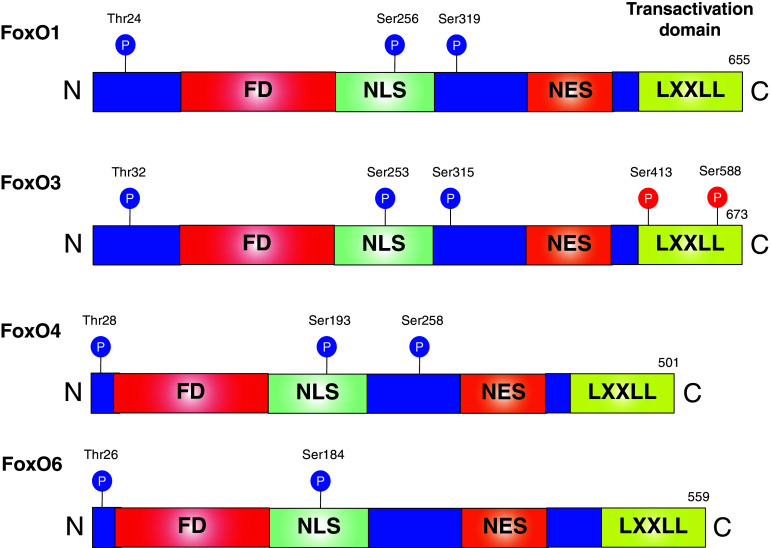
Fig. 1.
Description of mammalian FoxO proteins expressed in skeletal muscle. The following are indicated: locations of the forkhead domain (FD), nuclear localization sequence (NLS), nuclear export sequence (NES) and helical motif (LXXLL), Akt phosphorylation sites (blue circles) and AMPK phosphorylation sites (red circles)
In the present review, we outline the most recent advances on the roles and functions of FoxO proteins in the regulation of skeletal muscle homeostasis. The mode of regulation of these proteins, especially their dynamic control by two major actors in skeletal muscle homeostasis, Akt and AMPK, are detailed, as is the implication of each FoxO member in the regulation of cellular processes in response to physiological and pathophysiological conditions.
Post-translational modifications of FoxO factors in skeletal muscle
Multiple signals converge on FoxO factors via various types of post-translational modifications, including phosphorylation, mono- or polyubiquitination, acetylation, glycosylation and arginine or lysine methylation (for a review, see Ref. [22]). Few of these post-translational modifications of FoxO have been investigated in skeletal muscle, but these changes can differentially regulate FoxO members and modulate their subcellular localization, DNA-binding ability, transcriptional activity, protein–protein interactions, and degradation. Thus, FoxO3 activity in skeletal muscle is negatively regulated by acetylation of lysine 262 and ubiquitination [23]. The histone acetyl-transferase p300 targets and acetylates FoxO3, leading to its cytosolic relocalization and proteasomal degradation through the E3 ligase murine double minute 2 (Mdm2) [23]. Interestingly, overexpression of p300 in muscle alters FoxO3 nuclear localization and FoxO3 and 4 activities but, conversely, increases nuclear localization of FoxO1 [24]. In addition, an increase in p300 histone acetyltransferase activity raises some FoxO1-dependent gene transcription (Gadd45α and cathepsin L) [24]. Thus, acetylation may be an important mechanism to differentially regulate the FoxO homologues.
An essential regulator of FoxO activity is the IGF-1/PI3K/Akt signaling pathway (Fig. 2), which represents the canonical pathway of skeletal muscle hypertrophy. Recently, its implication in muscle protein synthesis and hypertrophy was extended to loading models [25]. Akt, also known as “protein kinase B” (PKB), is a serine/threonine protein kinase that plays a key role in insulin and PI3K/Akt/MTOR signaling pathways, thus contributing to the regulation of energy metabolism and protein synthesis. In the presence of mitogenic growth factors, Akt activates the mechanistic target of rapamycin (MTOR) via phosphorylation and inhibition of tuberous sclerosis complex 2 (TSC2) [26, 27], which is a negative regulator of MTOR. Akt also phosphorylates and inhibits the proline-rich Akt substrate of 40 kilodaltons (PRAS40), leading to increased MTORC1 signaling [28]. The IGF-1/PI3K/Akt pathway critically mediates FoxO inhibition since pharmacological inhibition of phosphatidylinositol 3-kinase (PI3K) reduces Akt phosphorylation and allows FoxO1 protein translocation to the nucleus [29]. Akt has been shown to phosphorylate FoxO3 on residues Thr32, Ser253, and Ser315, leading to its inhibition. Thr32 and Ser253 phosphorylation induces its exclusion from the nucleus and cytoplasmic retention through 14-3-3 chaperone protein binding [10]. Moreover, such phosphorylation promotes the alteration of FoxO binding to target DNA sequences [30, 31]. In non-muscle cells, FoxO1 and FoxO4 are also phosphorylated by Akt [32, 33]. Furthermore, using Akt1 and Akt2 knockout mice, a recent study demonstrated that BCL2/adenovirus E1B 19-kDa protein-interacting protein 3 (BNIP3) and GABA(A) receptor-associated protein-like 1 (GABARAPL1) transcripts, two autophagy-related genes regulated by FoxO3, are significantly elevated in muscle, confirming the importance of Akt in FoxO transcriptional activity regulation in vivo [34].
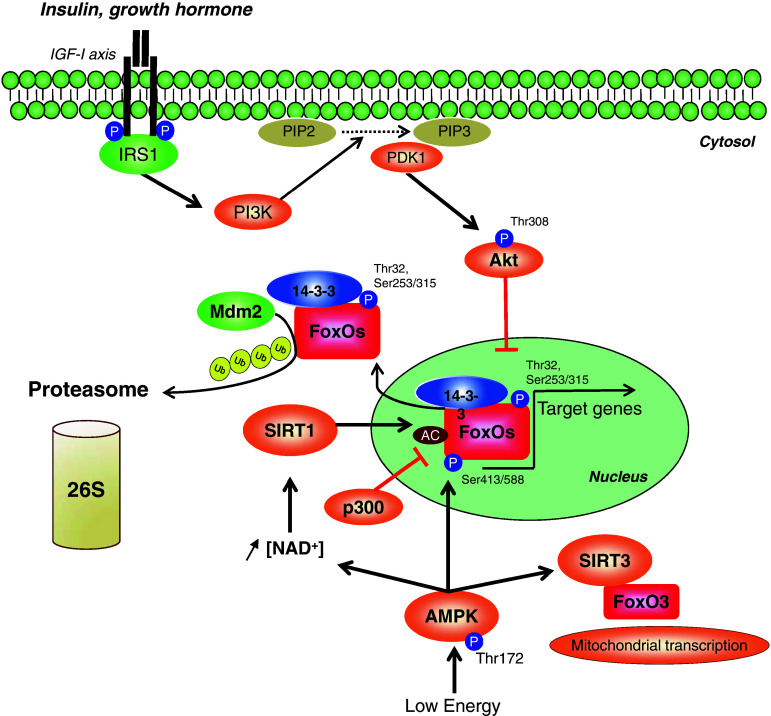
Fig. 2.
Antagonist regulation of FoxO proteins by the IGF-1/PI3K/Akt axis and AMPK. FoxO transcription factors are directly phosphorylated (at Thr32 and Ser253/315 for FoxO3) and inhibited by Akt via 14-3-3 binding in response to insulin/growth factor stimulation, whereas AMPK phosphorylates and activates FoxO3 under conditions of energy stress. AMPK also induces FoxO deacetylation and activation by modulating the activity of the histone deacetylases SIRT1 and SIRT3. Moreover, FoxO3 and FoxO4 can be acetylated by the histone acetyltransferase p300, and cytosolic FoxO3 can be degraded by the murine double minute 2 (Mdm2) via the ubiquitin–proteasome system. IRS1 insulin receptor substrate 1, PIP2 phosphatidylinositol 4,5-bisphosphate, PIP3 phosphatidylinositol 3,4,5-triphosphate, PDK1 phosphoinositide-dependent kinase 1
Unlike Akt, the adenosine monophosphate-activated protein kinase (AMPK), positively regulated by stimuli that decrease cellular energy levels (for a review, see Ref. [35]), phosphorylates FoxO3 at six regulatory sites (Thr-179, Ser-399, Ser-413, Ser-355, Ser-588, and Ser-626) in vitro and promotes its activation [36] (Fig. 2). Importantly, the phosphorylation of Ser-413 and Ser-588 after AMPK activation by AICAR has been found in muscle cells [37, 38] and the AMPK-mediated activation of FoxO3 is associated with myofibrillar proteolysis through the activity of the muscle atrophy F-box (MAFbx/atrogin-1) and the muscle ring-finger protein 1 (MuRF1) [39].
The effects of AMPK on cellular movements of FoxO show discrepancies, depending on the duration of AMPK activation. In C2C12 myotubes, Sanchez et al. [37] showed that the activation of AMPK by AICAR activates FoxO3 and promotes transient nuclear translocation from 30 min to 6 h, but not at 24 h. These data are consistent with the results of Tong et al. [38] who reported that AICAR treatment in the same cell line caused FoxO3 nuclear relocation through a decrease in FoxO3 phosphorylation at Thr-318/321. Nevertheless, Williamson et al. [40] reported that longer AICAR treatment (24 h)—again, in the same cell line—led to a decrease in nuclear FoxO3 content through a peroxisome proliferator activator receptor γ coactivator-1 α (PGC-1α)-dependent mechanism. Taken together, these data suggest that a pool of FoxO3 is rapidly directed toward the nucleus under AMPK activation and thereafter translocates into the cytosol and/or is degraded. Alternatively, Greer et al. suggested that AMPK does not modify FoxO3 localization and affects only the activity of the FoxO3 already present in the nucleus of HEK293T cells. Since FoxO3 cycles constantly in and out of the nucleus, AMPK may activate the fraction of FoxO3 that is always present in the nucleus [36].
Moreover, in a situation of energy stress, AMPK enhances the activity of histone deacetylase sirtuin 1 (SIRT1) by increasing the cellular NAD+ level, which results in the deacetylation and activation of FoxO1 and FoxO3 in muscle [41]. During fasting and after exercise, AMPK promotes SIRT1-dependent deacetylation of FoxO1 and PGC-1-α, leading to the modulation of mitochondrial and lipid utilization genes [42]. Interestingly, the deacetylation of these substrates is reduced in AMPKγ3 KO mice, suggesting that the decreased SIRT1 activity could be consequent to the observed unresponsiveness of intramuscular levels of NAD+ to fasting in these mice [42].
Concerning the transcriptional regulation of FoxO by AMPK, Nystrom and Lang found that 6 h after AICAR injection into mouse skeletal muscle, FoxO1 and FoxO3 mRNA levels increased but not FoxO4, which is also the case in response to sepsis [43]. In the same study, 24 h of AICAR and metformin (another AMPK activator) treatment decreased the mRNA content of FoxO1 and FoxO3 in C2C12 myotubes [43], and 2DG treatment for 8 h led to an increase in FoxO3 but a decrease in FoxO1 and FoxO4 mRNA levels. Furthermore, in the same cell model, AICAR treatment of shorter duration (i.e., 6 h) increased the mRNA content of FoxO1 and FoxO3 [39]. Thus, these data suggest that pharmacological activators of AMPK increase FoxO1 and FoxO3 mRNA levels during short-term treatment in myotubes and in vivo, and that longer treatments lead to a decrease in said content in muscle cells. Concerning protein levels, it has been reported that dominant negative AMPK in muscle cells leads to a critical decrease in FoxO3, whereas AICAR treatment for 6 h elicits an accumulation of FoxO1 and FoxO3 [37, 39], in accordance with previously described AICAR effects on FoxO mRNA content.
In summary, these cumulative data demonstrate that FoxO proteins are regulated on several fronts. In skeletal muscle, the most studied post-translational modifications of FoxOs are acetylation, ubiquitination, and phosphorylation (Fig. 2). AMPK and Akt have opposite effects on FoxO3 localization and activity. While Akt inhibits FoxO1, 3 and 4 by phosphorylation and 14-3-3 binding under mitogenic activation, AMPK phosphorylates FoxO3 at two regulatory sites (Ser-413/588) in skeletal muscle, leading to its activation under stress conditions. Moreover, AMPK activation is associated with increasing levels of FoxO1 and FoxO3 mRNAs and protein content.
Regulation of skeletal muscle homeostasis by FoxO proteins
FoxO1 and FoxO3 regulate energy metabolism
FoxO1 plays a particularly significant role in the regulation of muscle energy homeostasis (Fig. 3). It is an important regulator of glucose metabolism in skeletal muscle, as it is in liver [44], pancreas [45], adipose tissue [45], and bone [46]. First, FoxO1 was shown to affect muscle energy homeostasis through a reduction in carbohydrate catabolism during fasting [47]. FoxO1 can bind directly to the promoter region of the pyruvate dehydrogenase kinase 4 (PDK4) gene, a key factor in maintaining the blood glucose level, to promote its expression during energy deprivation in skeletal muscle [47]. High levels of PDK4 induce a decrease in the activity of pyruvate dehydrogenase (PDH), which catalyzes the reaction from pyruvate to acetyl-CoA and leads to lower use of carbohydrates as an energy substrate [48]. Thus, the increase in PDK4 expression mediated by FoxO1 results in the conservation of glucose and gluconeogenic substrates (lactate, pyruvate, and alanine) and a decrease in glycolytic flux by inactivating the pyruvate dehydrogenase complex (PDC) [49]. Interestingly, it was recently found that FoxO1 activation correlates with PDK4 expression, and consequently with PDC activity, during exercise after several days of high-fat diet in humans [50]. Furthermore, FoxO1 is an important metabolic regulator of muscle fat oxidation, a finding first reported in the 1980s in a study showing that FoxO1 levels are correlated with lipoprotein lipase (LPL) expression, the enzyme that hydrolyses plasma triglycerides into fatty acids and glycerol for uptake by muscle cells under fasting or exercise [51]. Moreover, overexpression of FoxO1 was found to increase LPL expression in C2C12 [52], strengthening these earlier results. FoxO1 also alters the subcellular localization of the membrane fatty acid translocase/cluster of differentiation 36 (FAT/CD36) [49] that permits fatty acid uptake into muscle cells. Last, FoxO1 regulates the expression of adiponectin receptors (AdipR), which transmit a signal for increased fatty acid oxidation in muscle, and adiponectin sensitivity [53]. Taken together, these data demonstrate that FoxO1 acts as a switch for a shift toward the use of lipids instead of glucose as fuel substrate under several stress conditions. Concerning FoxO3, its role in mitochondrial energy metabolism under nutrient restriction has recently been assessed [54]. In this study performed in myotubes, the authors showed that a low-glucose condition raises FoxO3 accumulation into mitochondria in an AMPK-dependent manner. Glucose restriction induces the formation of a multiprotein complex composed of FoxO3, SIRT3 (a mitochondrial sirtuin [55]), and mitochondrial RNA polymerase at mitochondrial DNA-regulatory regions (mtDNA-RR). The association of FoxO3 with mitochondrial DNA was found to correlate with activation of mitochondrial transcription. In this model, SIRT3 mediates FoxO3 binding to mtDNA-RR and the transcription of mitochondrial-encoded core or catalytic subunits of the oxidative phosphorylation machinery, thus increasing mitochondrial respiration. These data extend our understanding of FoxO proteins function into mitochondria, the central governors of energy metabolism, and suggest attractive perspectives for metabolic disorder research.
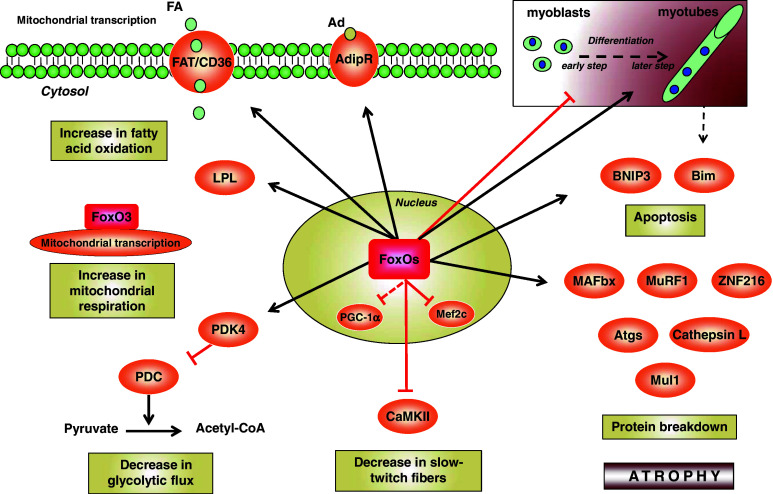
Fig. 3.
Role of FoxO in skeletal muscle homeostasis. FoxO transcription factors negatively regulate glycolysis by increasing the transcription of the pyruvate dehydrogenase kinase 4 (PDK4), which leads to the inhibition of the pyruvate dehydrogenase complex (PDC), and promote fatty acid (FA) degradation by increasing lipoprotein lipase (LPL), fatty acid translocase/cluster of differentiation 36 (FAT/CD36), and adiponectin receptor (AdipR) transcription. FoxO3 regulates transcription of mitochondrial-encoded core or catalytic subunits of the oxidative phosphorylation machinery, thus increasing mitochondrial respiration. FoxO proteins increase the transcription of the muscle atrophy F-box (MAFbx/atrogin-1), muscle ring-finger protein 1 (MuRF1), zinc finger protein 216 (ZNF216), mitochondrial E3 ubiquitin protein ligase 1 (Mul1), autophagic genes (Atgs), Bcl-2 interacting mediator of cell death (Bim) and BCL2/adenovirus E1B 19-kDa protein-interacting protein 3 (BNIP3) in myotubes leading to atrophy. FoxO1 inhibits the differentiation from myoblasts to myotubes in early steps but promotes myotube fusion in later steps of myogenic differentiation. FoxO1 binds and inhibits PGC-1α and the Ca2+/calmodulin-dependent protein kinase II (CaMKII), leading to a decrease in type I fiber expression
In summary, in the FoxO family, FoxO1 appears to be the major regulator of muscle energy homeostasis through the regulation of glycolytic and lipolytic flux. Importantly, the function of FoxO3 in mitochondrial metabolism is emerging. The involvement of FoxO4 and 6 in skeletal muscle energy metabolism still remains to be characterized.
FoxO1 and FoxO3 govern protein breakdown
The role of FoxO transcription factors in the regulation of skeletal muscle protein degradation has been studied over the last 10 years. FoxO has been shown to regulate the two major systems of protein breakdown in skeletal muscle, the ubiquitin–proteasome and the autophagy–lysosomal pathways [29, 56, 57] (Fig. 3).
The ubiquitin–proteasome system is assumed to play a major role in muscle protein degradation since the discovery of two E3 ubiquitin ligases, MAFbx/atrogin-1 and MuRF1, which are overexpressed in various atrophy models like fasting, denervation, immobilization, and hindlimb suspension [58, 59]. The function of E3 ubiquitin ligases is to target specific protein substrates for degradation by the 26S proteasome. FoxO1 and 3 are required for the transcription of MAFbx/atrogin-1 and MuRF1 [56, 60], which leads to the ubiquitination of several proteins involved in skeletal muscle maintenance. MAFbx/atrogin-1 catalyzes the breakdown of the myogenic transcription factors MyoD and myogenin, as well as the eukaryotic initiation factor of translation eIF3f [61–64]. As a result, MAFbx/atrogin-1 inhibits the transcription and translation of muscle genes and prevents the replacement of degraded muscle proteins. MAFbx/atrogin-1 has also recently been shown to ubiquitinate and target sarcomeric proteins (i.e., desmin and vimentin) for degradation, expanding its activities to the degradation of myofibrillar proteins [65]. MuRF1 recognizes myosin heavy chain protein (MyHC) [66] and other myofibrillar proteins such as myosin light chain 1 and 2 (MLC1 and 2, respectively) and myosin-binding protein C (MyBP-C) [67] for breakdown by the proteasome. Interestingly, the thin filament components (actin, tropomyosin, troponins) and Z-band (α-actinin) decrease during atrophy by a mechanism not requiring MuRF1, but rather Trim32 (tripartite motif-containing protein 32), a distinct RING finger ubiquitin ligase [68, 69]. In addition to these E3 ligases, zinc finger protein 216 (ZNF216) also seems to be involved in muscle atrophy. Indeed, ZNF216 expression is upregulated in denervation and fasting-induced models of muscle atrophy, and mice lacking ZNF216 exhibit resistance to atrophy [70]. ZNF216’s ability to directly bind to polyubiquitin chains and its association with the 26S proteasome suggests it may function by shuttling proteins targeted for degradation to the proteasome. Moreover, FoxO4 increases ZNF216 expression in C2C12 myoblasts, suggesting that ZNF216 may also function as a downstream effector of FoxO proteins in muscle atrophy [70]. Further studies are needed to determine the role of this potential FoxO4/ZNF216 axis in skeletal muscle homeostasis and its involvement in both physiological and pathological situations.
The second system is the autophagy–lysosomal pathway, an important mechanism for maintaining cell metabolism and protein turnover. This machinery is implicated in the turnover of organelles and long-lived proteins, as well as in the clearance of damaged cell components and the degradation of cell material in order to allow energy supply and cell survival during starvation and stress [71]. It involves, first, the sequestration of substrates into a vacuolar system called the autophagosome. The autophagosome then fuses with the lysosome to form an autolysosome, in which the content is degraded by lysosomal hydrolases. Autophagy requires the Atg (autophagy-specific gene) proteins implicated in the formation of autophagosomes [72] and two ubiquitin-like conjugation systems. The first one involves the Atg12/Atg5/Atg16 complex [73, 74], which is essential for the formation of the autophagosomal membrane [75, 76]. The second implicates the conjugation system of Atg8, also known as microtubule-associated protein 1 light chain 3 (LC3) in mammals [77]. Pro-LC3 is first cleaved by the cysteine protease Atg4 to its mature form, LC3I. The former is then lipidated to LC3II by Atg7 and Atg3, this event contributing to membrane fusion and the substrate selection for degradation [78]. It is noteworthy that two other complexes (Unc-51-like kinase (Ulk1)/Atg1 and Beclin1/vacuole protein sorting 34 (Vps34)/PI3K) are important for the initiation of the autophagic process [79–82]. These proteins operate in conjunction with other Atgs to mediate the initial assembly of the autophagosomal membrane. Recently, it was shown in vivo during starvation-induced atrophy that FoxO3 regulates the transcription of several Atgs, including LC3B and BNIP3 [57]. FoxO3 controls the skeletal muscle ubiquitin-proteasome and autophagy–lysosomal pathways independently, and BNIP3 appears to mediate the effect of FoxO3 on autophagy. Moreover, FoxO3 regulates the transcription of several other Atgs, Beclin, GABARAPL1, PI3KIII, Atg4B, Atg12 l, and Ulk2, during fasting [83]. This study [83] also reported that expression of a constitutively active form of FoxO3 promotes LC3 lipidation, which could reflect the induction of autophagy. Moreover, FoxO1 directly targets cathepsin L, a lysosomal protease in muscle [84], and induces its expression during fasting [84] or cachectic conditions [85], promoting a role of FoxO factors in lysosomal degradation. Although the implication of FoxO proteins in the transcription of Atg and lysosomal hydrolases has been partially described, their potential role in the genesis of lysosomes has not been investigated. Similarly, the potential role of FoxO1 in skeletal muscle autophagy remains to be characterized.
In order to assess the function of autophagy in skeletal muscle, Masiero and colleagues performed experiments on mice with muscle-specific knockout of Atg7, a gene necessary for the unfolding of the autophagy program. Interestingly, these mice showed obvious signs of muscle weakness and atrophy exacerbated by aging. Indeed, these mice presented an accumulation of degraded proteins and free radicals, deterioration in the internal cellular structures, and activation of the apoptotic program. The authors clearly demonstrated that inhibition of basal autophagy does not protect from the skeletal muscle atrophy induced by denervation or starvation but, on the contrary, contributes greatly to muscle degeneration. A similar atrophic phenotype was also obtained in muscle-specific Atg5 knockout mice [86]. Thus, autophagy in muscle is a complex process that can be beneficial or deleterious, depending on its activation level and cellular environment. Further work is needed to determine whether autophagy during muscle wasting has a protective function, a causative role, or is the result of the disease process itself.
Last, the involvement of FoxO3 in mitochondrial autophagy (also called mitophagy) has recently been characterized. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy, and an increase in mitochondrial fission was reported to be permissive for mitophagy [87]. Enhanced FoxO1 and FoxO3 activity during muscle wasting results in increased transcription of the mitochondrial E3 ubiquitin protein ligase 1 (Mul1), which in turn ubiquitinates and degrades mitofusin-2 (Mfn2) [88], a mitochondrial fusion protein critical for the maintenance and genomic stability of mitochondrial DNA [89]. These events promote fragmentation, depolarization, and clearance of mitochondria through the autophagy–lysosomal pathway [88].
Collectively, these recent studies highlight that FoxO1 and 3 are central regulators of protein breakdown. Although FoxO1 seems to be more involved in the ubiquitin–proteasome pathway, FoxO3 has a role in both the ubiquitin–proteasome and autophagy–lysosomal systems. Importantly, these two proteins were recently identified as having a role in skeletal muscle mitophagy through the regulation of the activity of the mitochondrial E3 ligase Mul1.
Cell proliferation, differentiation, and muscle regeneration are regulated by FoxOs
In addition to the effect of FoxO factors on protein degradation, their role in myogenesis has also been addressed. Muscle progenitor cells, called satellite cells, reside between the sarcolemma and basement membrane of terminally differentiated muscle fibers. These are normally quiescent in adult muscle, but play a major role in muscle cell differentiation and tissue regeneration in response to injury. Overexpressed FoxO3 decreases muscle precursor cell proliferation by increasing expression of p27KIP1, a cyclin-dependent kinase inhibitor [90]. Moreover, a decrease in the activation of the p27KIP1 promoter was found to result from an IGF-I-related inhibition of FoxO1 activity in satellite cells [91]. In addition, the forkhead box protein K1 (Foxk1) interacts with the winged helix of FoxO4 and promotes muscle progenitor cell proliferation by repressing FoxO4 transcriptional activity [92, 93]. Reciprocally, repression of FoxO4 results in decreased FoxO4 target gene (p21CIP, p27KIP1, p57, and Gadd45α) expression and increased cellular proliferation of the muscle progenitor cells [93]. It is noteworthy that the gastrocnemius muscles of FoxO4-null mice, which show increased cellular proliferation, also show altered regeneration and smaller myofibers following cardiotoxin injury [93]. These deleterious effects seem to be linked to the decreased expression of cell cycle inhibitors, especially p21CIP [93], and suggest the prominent role of FoxO4 as a key cell cycle regulator of progenitor cells. Furthermore, the expression of a constitutively active form of FoxO4 controls C2C12 myoblast cycle progression by G(2) arrest [6]. In these cells, oxidative stress activates the stress-inducible gene Gadd45 promoter in a FoxO-dependent manner, resulting in a greater abundance of Gadd45 protein, as well as G(2) arrest [6]. In summary, these data indicate that FoxOs act as negative regulators of cell proliferation.
In addition to their role in the control of the cell cycle, FoxO proteins are also implicated in myogenic differentiation. Illustrating the great complexity of the FoxO system, the studies on myoblast differentiation have shown substantial differences, according to the step of differentiation. For example, Hribal et al. [94] reported that overexpression of a constitutively active form of FoxO1 in C2C12 myoblasts results in the inhibition of muscle differentiation induced by constitutively active Akt, whereas dominant-negative mutant FoxO1 causes a slight but significant increase in the expression of differentiation markers. The results from Kitamura et al. [95] confirmed the inhibitory effect of FoxO1 on myoblast differentiation and demonstrated that FoxO1 and Notch pathways cooperate in the regulation of muscle differentiation. Last, the study by Wu and coworkers also showed that FoxO1 inhibits C2C12 myoblast differentiation, and these authors demonstrated that activation of an inducible mutant of FoxO1 leads to proteasome degradation of MTOR pathway components (i.e., RPTOR, MTOR, p70S6K, and TSC2) [96], which are required for differentiation, by controlling the expression of IGF-II [97]. By contrast, Bois and Grosveld reported that FoxO1 is required for myotube formation of primary mouse myoblasts after the initiation of differentiation [98]. These authors showed that FoxO1 translocates to the nucleus and regulates the fusion of differentiating primary myoblasts during later steps of myogenic differentiation. Taken together, these studies highlight the complex role of FoxO1 during myogenesis. FoxO1 may also have a dual role, depending on the stage of myogenesis.
Notably, inhibition of basal FoxO1 activity decreases myostatin expression and Smad transcriptional activity, whereas it increases MyoD expression, satellite cell proliferation, and fusion, and leads to muscle hypertrophy in normal muscle [95, 99]. Reciprocally, expression of constitutively active FoxO1 in C2C12 myotubes enhances myostatin promoter activity, and mutation of the FoxO-binding sites in the myostatin promoter reduces its activity [100]. Thus, FoxO1 regulates the expression of myostatin, which contributes to the control of muscle cell growth and differentiation [100]. Collectively, these data demonstrate that basal levels of FoxO transcription factors play a role in limiting muscle growth, especially by repressing satellite cell activation.
In summary, while the muscle proteins FoxO1, 3, and 4 inhibit cell proliferation, FoxO1 expression inhibits myoblast differentiation, but only in the early steps. Indeed, it has also been reported that FoxO1 is required for myotube formation of primary myoblasts after the initiation of the differentiation process. Further experiments are needed to clarify the molecular basis of such divergent roles.
Regulation of muscle mass by FoxO1 and 3
Skeletal muscle atrophy occurs under a variety of conditions and can result from alterations in both protein synthesis and protein degradation. The role of FoxO transcription factors in skeletal muscle atrophy has been investigated over the past few years. One study showed that FoxO1 is markedly upregulated in skeletal muscle under energy-deprived states such as fasting and severe diabetes [52]. The study by Kamei et al. [60] demonstrated that FoxO1 transgenic overexpression in skeletal muscle induces a decrease in body size and muscle mass associated with an upregulation of MAFbx/atrogin-1 and MuRF1 expression. It was also established that genetic activation of FoxO3 results in atrophy associated with upregulation of MAFbx/atrogin-1, whereas knockdown of FoxO3 by siRNA prevents muscle atrophy [56]. Similarly, the expression of a dominant-negative form of FoxOs inhibits the MAFbx/atrogin-1, MuRF1, cathepsin L, and BNIP3 mRNA increases associated with cancer cachexia and sepsis, resulting in an inhibition of muscle fiber atrophy [85]. Interestingly, inactivation of autophagic flux by LC3 silencing partially prevents FoxO3-mediated muscle loss, suggesting the major role of the autophagic pathway in FoxO3-mediated atrophy [57]. Concerning the E3 ligases, MuRF1 knockdown attenuates muscle atrophy in response to synthetic glucocorticoid treatment, contrary to MAFbx/atrogin-1 deletion. Moreover, the muscle sparing in MuRF1-null mice is related to the maintenance of protein synthesis rather than the attenuation of muscle protein breakdown [101]. In view of these elements, we can assume that FoxOs/MuRF1 controls both protein synthesis and degradation in skeletal muscle, but the underlying mechanisms remain to be elucidated. Furthermore, recent data have established a link between mitochondrial dysfunction and atrophy, showing that expression of the fission machinery leads to muscle wasting by triggering organelle dysfunction and AMPK activation [102]. Importantly, the essential role of FoxO1- and FoxO3-dependent mitophagy has been characterized during muscle wasting. Thus, the previously described Mul1/mitophagy axis was reported to have a significant deleterious role in denervation and dexamethasone atrophy models [88].
In FoxO-dependent atrophy, FoxO1 activation induces apoptosis in a DNA-binding-dependent manner in mature myotubes [103]. Apoptosis can be directly linked to skeletal muscle atrophy through induction of mitochondria-associated proapoptotic genes and DNA fragmentation during mechanical unloading and denervation [104–107]. Thus, the apoptotic genes, Bcl-2-interacting mediator of cell death (Bim) and BNIP3—which can promote apoptosis through their ability to trigger mitochondrial disruption [108, 109]—are induced by FoxO1 during muscle atrophy [103]. Nonetheless, it is noteworthy that activation of FoxO1 does not lead to apoptosis in primary myoblasts, conversely to what has been found in other cell types [11, 98].
FoxO1 and FoxO3 mRNAs are upregulated during dexamethasone treatment [47, 110] and glucocorticoids seem to induce the activation of FoxO factors by decreasing the activity of the PI3K/Akt pathway. In agreement, FoxO transcription factors in cultured myotubes can be inhibited by IGF-1 following dexamethasone treatment [29]. Interestingly, under the same treatment, IGF-1 is not able to block MAFbx/atrogin-1 or MuRF1 expression in the presence of a constitutively active FoxO1 mutant that cannot be phosphorylated by Akt [10]. This finding demonstrated that IGF-1/PI3K/Akt-related inhibition of atrophy requires the blockade of FoxO1. Recently, it was shown in acute inflammation that the FoxO pathway activation linked to increased glucocorticoid production plays a more crucial role in the induction of proteolytic systems than decreased IGF-I production and an increased tumor necrosis factor α (TNF-α)/NF-κB pathway [111].
The implication of FoxOs in aged-mediated atrophy has also been investigated. FoxO proteins play key roles in the aging process, especially with respect to their ability to reduce the generation of reactive oxygen species and DNA damage [112, 113]. Sandri and coworkers recently established that the modulation of the Akt/FoxO pathway and the proteolytic systems is modest in the aging process and that sarcopenia is not due to FoxO activation [114]. Similarly, it was suggested that apoptosis contribution could be more important than MAFbx/atrogin-1- and MuRF1-linked protein degradation in declining aged-muscle functions [115]. Pardo and colleagues reported a mechanosensitive signaling mechanism in the diaphragm, responsible for the regulation of FoxO transcription factors in aging [116]. Indeed, they found that the alteration in the mechanical properties of the aged diaphragm is associated with a decrease in FoxO1 and FoxO3 nuclear content. Such a decrease is correlated with higher basal activation of Akt during aging. However, in the same model, nuclear FoxO4 content remains unaffected and its stability seems to be dependent on the c-Jun N-terminal kinases (JNKs), which are highly activated in sarcopenia [116]. Thus, these data suggest that FoxO4 is the principal homologue responsible for FoxO-dependent transcriptional activity in the aging diaphragm. In a study investigating the cause of the skeletal muscle mass loss in sarcopenia, Edström et al. [117] found that, contrary to the acute atrophies induced by caloric restriction, chronic atrophies induced by disuse, disease and denervation lead to MAFbx/atrogin-1 and MuRF1 downregulation in the skeletal muscle of 30-month-old rats. The authors suggested that Akt activated by the IGF-1 receptor is responsible for the inactivation of FoxO4 and the subsequent inhibition of MAFbx/atrogin-1 and MuRF1. Moreover, this study confirmed the results of Furuyama et al. [118] and showed that caloric restriction limits sarcopenia as well as the effects of aging by acting on Akt phosphorylation, FoxO4 inhibition, and MAFbx/atrogin-1 and MuRF1 transcript regulation.
Thus, FoxO1 and FoxO3 act through an exacerbation of the ubiquitin-proteasomal and autophagy–lysosomal degradation pathways in muscle atrophy. Moreover, FoxO factors, especially FoxO3, are involved in mitochondrial dysfunction and elimination by regulating the mitophagic pathway under atrophy. Nonetheless, the involvement of apoptosis in the aging process seems to be more important than FoxO-dependent proteolysis.
Regulation of muscle fiber type by FoxOs
Regarding muscle typology, FoxO-mediated atrophy is associated with a decreased expression of type I (slow oxidative) fiber-related genes and proteins (i.e., troponin I (slow) and myoglobin), a diminished number of type I fibers, and smaller type I and type II fibers [60]. A target of FoxO1 is PGC-1α, a metabolic transcriptional coactivator that was originally identified in vitro as a coactivator of the peroxisome proliferator-activated receptor γ (PPAR-γ) [119]. PGC-1α, which is notably induced by endurance exercise [120, 121], promotes fiber-type switching from glycolytic toward more oxidative fibers [122, 123]. Interestingly, FoxO1 expression stimulates the promoter activity of PGC-1α via interaction with insulin response sequences (IRSs) in liver HepG2 cells [124], but to our knowledge, no muscle study has yet supported this finding. However, the study of Kamei et al. [60] showed that PGC-1α mRNA levels are increased in muscle-specific FoxO1 transgenic mice, suggesting that FoxO1 may promote PGC-1α gene expression in muscle. Thus, these studies suggest that the alteration in muscle oxidative capacity mediated by FoxO1 cannot be directly linked to an alteration in PGC-1α mRNA content, but FoxO1 may bind to PGC-1α and inhibit certain functions of this factor. Along the same lines, FoxO6 was recently shown to form a regulatory loop with PGC-1α for setting the oxidative metabolism level in muscle cells [15], but the authors did not investigate whether this mechanism is modulated during muscle wasting. Nevertheless, PGC-1α is downregulated in various types of muscle atrophy, including denervation, streptozotocin-induced diabetes, renal failure, and cancer [125]. Electroporation of PGC-1α in rodent muscles diminished FoxO3 transcriptional activity and reduced denervation- and fasting-induced atrophy by decreasing MAFbx/atrogin-1 and MurF1 expression levels, while increasing several genes involved in glycolysis and oxidative phosphorylation [125]. Moreover, overexpression of PGC-1β or PGC-1α in mouse muscles decreases denervation-induced atrophy, MAFbx/atrogin-1 and MuRF1 induction, and NF-κB activity [126], this last being especially activated during skeletal muscle wasting. In addition to PGC-1α, type I fiber gene expression is also regulated by the transcription factor myocyte enhancer factor 2c (Mef2c) and the Ca2+/calmodulin-dependent kinase II (CaMKII), and levels of both proteins are significantly reduced in transgenic mice overexpressing FoxO1 [60]. It was also reported that FoxO1 is expressed preferentially in fast-twitch fibers and overexpression of FoxO1 induces the formation of fast-twitch fibers [127]. Yuan et al. [127] found that FoxO1 decreases oxidative capacity in C2C12 myotubes, at least in part through inhibition of the calcineurin pathway, a master chief regulatory signaling pathway of slow fiber-selective gene expression [128]. Interestingly, FoxO1 can exert different actions according to the fiber type, and it was shown that FoxO1 mRNA is upregulated in both the soleus and plantaris muscles during hindlimb unloading, but this effect is correlated only with a lower oxidative capacity of soleus muscle [129]. In this study, the percentage of type I fibers is decreased and the percentage of glycolytic fibers is increased in the soleus muscle but not in plantaris, strongly suggesting that slow fibers are affected by FoxO1 more than fast fibers.
Altogether, these studies clearly demonstrate that FoxO transcription factors are negative regulators of type I fiber-related gene expression in muscle atrophy, which results in a shift of muscle phenotype from slow oxidative to fast glycolytic (Fig. 3). The significance of the FoxO6-PGC1-α regulatory loop during muscle wasting should be investigated in order to open new perspectives and assess the importance of this FoxO member in skeletal muscle homeostasis.
FoxO1 and 3 are involved in skeletal muscle adaptation to exercise
Besides the involvement of FoxO proteins in muscle wasting, recent studies have demonstrated that FoxO1 and FoxO3 are involved in skeletal muscle adaptation to exercise. In human skeletal muscle, the FoxO1 gene increased at 3 h and returned to baseline by 48 h during recovery from an exhaustive bout of high-intensity cycling [130]. The FoxO3 gene also increases after a marathon [131]. Consistent with this, it was reported that certain autophagic markers (i.e., LC3bII and Atg12 protein expression), MuRF1 mRNA level, and proteasome β2 subunit activity are increased after ultra-endurance exercise [132]. Furthermore, some of these adaptations also occur during moderate exercise [133–135]. Thus, the rise in catabolic pathways regulated by FoxO1 and 3 could provide amino acids as an alternative energy substrate during prolonged exercise and probably optimizes protein turnover during recovery. The removal or recycling of damaged cell constituents is also needed in skeletal muscle during exercise. In addition, mechanical stimuli induce changes in gene expression that affect metabolism and promote adaptations in muscle function and muscle mass. In support of a role for FoxOs in the adaptations of skeletal muscle to resistance exercise, Goodman et al. [136] showed that total and phosphorylated FoxO1 and 3 were elevated with chronic mechanical overload that induces significant hypertrophy and remodeling. Interestingly, the increase in total FoxO1 was an MTOR kinase-dependent event, suggesting that MTOR is involved in the protein degradation increase observed in response to mechanical overload. Furthermore, in mice, He and coworkers reported that the autophagic process is required for the beneficial metabolic effects of exercise on skeletal muscle [137]. Indeed, mice that have mutations in BCL2 phosphorylation sites, which prevent stimulus-induced disruption of the BCL2-beclin-1 complex and autophagy activation, present decreased endurance and altered glucose metabolism during acute exercise. These emergent studies support the essential role of FoxO transcriptions factors in muscle adaptations to exercise. Future research to further elucidate these events will be key to achieving a better understanding of the molecular adaptations of skeletal muscle to exercise. For example, directions for future research include the possible involvement of FoxO/mitophagy and mitochondrial network remodeling during exercise. Because exercise is associated with improved quality of life and constitutes one of the best approaches to limit atrophy and metabolic disorders, these research directions are crucial in the battle against a wide spectrum of metabolic and muscle diseases.
Summary and conclusions
In summary, significant progress has expanded our understanding of the prominent roles played by FoxO transcription factors in skeletal muscle homeostasis. These proteins control muscle cell growth, differentiation, and apoptosis, as well as energy metabolism, this last one by acting as a switch for a shift toward the use of lipids instead of glucose as fuel substrate and by regulating mitochondrial metabolism. FoxO factors regulate protein breakdown, with FoxO1 seemingly more involved in the regulation of the ubiquitin–proteasome pathway, whereas FoxO3 plays a role in both the ubiquitin–proteasome and autophagy–lysosomal systems. Furthermore, FoxO1 and FoxO3 have recently been implicated in the regulation of mitophagy through activation of the E3 ligase Mul1. A basal level of FoxO1 and 3 is necessary for maintaining cellular homeostasis and responding to the metabolic stress related to exercise, but the exacerbated activation that occurs in several diseases results in atrophy, mitochondrial dysfunction, and a detrimental shift in the muscle phenotype. A basal level of protein breakdown is necessary for maintaining muscle homeostasis, and the complex process of autophagy can be beneficial or deleterious, depending on its activation level. The physiological relevance of the autophagic pathway in skeletal muscle is emerging, as recent studies have demonstrated its deleterious role in wasting and, conversely, its crucial role in cellular homeostasis during exercise. In the light of these results, additional work is necessary to further elucidate the precise role of these proteins, especially in mitochondrial homeostasis, for the development of new therapeutic approaches. This will be important in preventing or limiting the muscle wasting that prevails in numerous physiological and pathological states such as immobilization, aging, denervated conditions, neuromuscular diseases, AIDS, cancer, and diabetes.
Acknowledgments
This work was supported by the Faculty of Sport Sciences of the University of Montpellier 1 and by INRA’s PHASE division. The authors thank Dr. P. Glaviole for helpful discussions.
Conflict of interest
The authors declare no conflicts of interest, financial or otherwise.
References
- 1.Goldberg AL, Etlinger JD, Goldspink DF, Jablecki C. Mechanism of work-induced hypertrophy of skeletal muscle. Med Sci Sports. 1975;7(3):185–198. [PubMed] [Google Scholar]
- 2.Adibi SA, Krzysik BA, Morse EL, Amin PM, Allen ER. Oxidative energy metabolism in the skeletal muscle: biochemical and ultrastructural evidence for adaptive changes. J Lab Clin Med. 1974;83(4):548–562. [PubMed] [Google Scholar]
- 3.Dijkers PF, Medema RH, Pals C, Banerji L, Thomas NS, Lam EW, Burgering BM, Raaijmakers JA, Lammers JW, Koenderman L, Coffer PJ. Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1) Mol Cell Biol. 2000;20(24):9138–9148. doi: 10.1128/mcb.20.24.9138-9148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medema RH, Kops GJ, Bos JL, Burgering BM. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404(6779):782–787. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- 5.Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ, Jr, DiStefano PS, Chiang LW, Greenberg ME. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296(5567):530–534. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- 6.Furukawa-Hibi Y, Yoshida-Araki K, Ohta T, Ikeda K, Motoyama N. FOXO forkhead transcription factors induce G(2)-M checkpoint in response to oxidative stress. J Biol Chem. 2002;277(30):26729–26732. doi: 10.1074/jbc.C200256200. [DOI] [PubMed] [Google Scholar]
- 7.Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH, Burgering BM. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419(6904):316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 8.Nemoto S, Finkel T. Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science. 2002;295(5564):2450–2452. doi: 10.1126/science.1069004. [DOI] [PubMed] [Google Scholar]
- 9.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans . Nature. 1997;389(6654):994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 10.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96(6):857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 11.Dijkers PF, Medema RH, Lammers JW, Koenderman L, Coffer PJ. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr Biol. 2000;10(19):1201–1204. doi: 10.1016/s0960-9822(00)00728-4. [DOI] [PubMed] [Google Scholar]
- 12.Lehmann OJ, Tuft S, Brice G, Smith R, Blixt A, Bell R, Johansson B, Jordan T, Hitchings RA, Khaw PT, John SW, Carlsson P, Bhattacharya SS. Novel anterior segment phenotypes resulting from forkhead gene alterations: evidence for cross-species conservation of function. Invest Ophthalmol Vis Sci. 2003;44(6):2627–2633. doi: 10.1167/iovs.02-0609. [DOI] [PubMed] [Google Scholar]
- 13.Nakae J, Cao Y, Daitoku H, Fukamizu A, Ogawa W, Yano Y, Hayashi Y. The LXXLL motif of murine forkhead transcription factor FoxO1 mediates Sirt1-dependent transcriptional activity. J Clin Investig. 2006;116(9):2473–2483. doi: 10.1172/JCI25518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furuyama T, Nakazawa T, Nakano I, Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J. 2000;349(Pt 2):629–634. doi: 10.1042/0264-6021:3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung SY, Huang WC, Su CW, Lee KW, Chi HC, Lin CT, Chen ST, Huang KM, Tsai MS, Yu HP, Chen SL (2013) FoxO6 and PGC-1alpha form a regulatory loop in myogenic cells. Biosci Rep 33(3). doi: 10.1042/BSR20130031 [DOI] [PMC free article] [PubMed]
- 16.Salih DA, Rashid AJ, Colas D, de la Torre-Ubieta L, Zhu RP, Morgan AA, Santo EE, Ucar D, Devarajan K, Cole CJ, Madison DV, Shamloo M, Butte AJ, Bonni A, Josselyn SA, Brunet A. FoxO6 regulates memory consolidation and synaptic function. Genes Dev. 2012;26(24):2780–2801. doi: 10.1101/gad.208926.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castrillon DH, Miao L, Kollipara R, Horner JW, DePinho RA. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science. 2003;301(5630):215–218. doi: 10.1126/science.1086336. [DOI] [PubMed] [Google Scholar]
- 18.Hosaka T, Biggs WH, III, Tieu D, Boyer AD, Varki NM, Cavenee WK, Arden KC. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc Natl Acad Sci USA. 2004;101(9):2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni YG, Berenji K, Wang N, Oh M, Sachan N, Dey A, Cheng J, Lu G, Morris DJ, Castrillon DH, Gerard RD, Rothermel BA, Hill JA. Foxo transcription factors blunt cardiac hypertrophy by inhibiting calcineurin signaling. Circulation. 2006;114(11):1159–1168. doi: 10.1161/CIRCULATIONAHA.106.637124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu P, Geles KG, Paik JH, DePinho RA, Tjian R. Codependent activators direct myoblast-specific MyoD transcription. Dev Cell. 2008;15(4):534–546. doi: 10.1016/j.devcel.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou W, Cao Q, Peng Y, Zhang QJ, Castrillon DH, DePinho RA, Liu ZP. FoxO4 inhibits NF-kappaB and protects mice against colonic injury and inflammation. Gastroenterology. 2009;137(4):1403–1414. doi: 10.1053/j.gastro.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eijkelenboom A, Burgering BM. FOXOs: signalling integrators for homeostasis maintenance. Nat Rev Mol Cell Biol. 2013;14(2):83–97. doi: 10.1038/nrm3507. [DOI] [PubMed] [Google Scholar]
- 23.Bertaggia E, Coletto L, Sandri M. Posttranslational modifications control FoxO3 activity during denervation. Am J Physiol Cell Physiol. 2012;302(3):C587–C596. doi: 10.1152/ajpcell.00142.2011. [DOI] [PubMed] [Google Scholar]
- 24.Senf SM, Sandesara PB, Reed SA, Judge AR. p300 Acetyltransferase activity differentially regulates the localization and activity of the FOXO homologues in skeletal muscle. Am J Physiol Cell Physiol. 2011;300(6):C1490–C1501. doi: 10.1152/ajpcell.00255.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3(11):1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 26.Dan HC, Sun M, Yang L, Feldman RI, Sui XM, Ou CC, Nellist M, Yeung RS, Halley DJ, Nicosia SV, Pledger WJ, Cheng JQ. Phosphatidylinositol 3-kinase/Akt pathway regulates tuberous sclerosis tumor suppressor complex by phosphorylation of tuberin. J Biol Chem. 2002;277(38):35364–35370. doi: 10.1074/jbc.M205838200. [DOI] [PubMed] [Google Scholar]
- 27.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4(9):648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 28.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9(3):316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 29.Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14(3):395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 30.Cahill CM, Tzivion G, Nasrin N, Ogg S, Dore J, Ruvkun G, Alexander-Bridges M. Phosphatidylinositol 3-kinase signaling inhibits DAF-16 DNA binding and function via 14-3-3-dependent and 14-3-3-independent pathways. J Biol Chem. 2001;276(16):13402–13410. doi: 10.1074/jbc.M010042200. [DOI] [PubMed] [Google Scholar]
- 31.Zhang BH, Tang ED, Zhu T, Greenberg ME, Vojtek AB, Guan KL. Serum- and glucocorticoid-inducible kinase SGK phosphorylates and negatively regulates B-Raf. J Biol Chem. 2001;276(34):31620–31626. doi: 10.1074/jbc.M102808200. [DOI] [PubMed] [Google Scholar]
- 32.Kops GJ, de Ruiter ND, De Vries-Smits AM, Powell DR, Bos JL, Burgering BM. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398(6728):630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 33.Takaishi H, Konishi H, Matsuzaki H, Ono Y, Shirai Y, Saito N, Kitamura T, Ogawa W, Kasuga M, Kikkawa U, Nishizuka Y. Regulation of nuclear translocation of forkhead transcription factor AFX by protein kinase B. Proc Natl Acad Sci USA. 1999;96(21):11836–11841. doi: 10.1073/pnas.96.21.11836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynolds THt, Merrell E, Cinquino N, Gaugler M, Ng L (2012) Disassociation of insulin action and Akt/FOXO signaling in skeletal muscle of older Akt-deficient mice. Am J Physiol Regul Integr Comp Physiol 303(11):R1186–R1194 [DOI] [PMC free article] [PubMed]
- 35.Sanchez AM, Candau RB, Csibi A, Pagano AF, Raibon A, Bernardi H. The role of AMP-activated protein kinase in the coordination of skeletal muscle turnover and energy homeostasis. Am J Physiol Cell Physiol. 2012;303(5):C475–C485. doi: 10.1152/ajpcell.00125.2012. [DOI] [PubMed] [Google Scholar]
- 36.Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282(41):30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 37.Sanchez AM, Csibi A, Raibon A, Cornille K, Gay S, Bernardi H, Candau R. AMPK promotes skeletal muscle autophagy through activation of forkhead FoxO3a and interaction with Ulk1. J Cell Biochem. 2012;113(2):695–710. doi: 10.1002/jcb.23399. [DOI] [PubMed] [Google Scholar]
- 38.Tong JF, Yan X, Zhu MJ, Du M. AMP-activated protein kinase enhances the expression of muscle-specific ubiquitin ligases despite its activation of IGF-1/Akt signaling in C2C12 myotubes. J Cell Biochem. 2009;108(2):458–468. doi: 10.1002/jcb.22272. [DOI] [PubMed] [Google Scholar]
- 39.Nakashima K, Yakabe Y. AMPK activation stimulates myofibrillar protein degradation and expression of atrophy-related ubiquitin ligases by increasing FOXO transcription factors in C2C12 myotubes. Biosci Biotechnol Biochem. 2007;71(7):1650–1656. doi: 10.1271/bbb.70057. [DOI] [PubMed] [Google Scholar]
- 40.Williamson DL, Butler DC, Alway SE. AMPK inhibits myoblast differentiation through a PGC-1alpha-dependent mechanism. Am J Physiol Endocrinol Metab. 2009;297(2):E304–E314. doi: 10.1152/ajpendo.91007.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458(7241):1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Canto C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, Auwerx J. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 2010;11(3):213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nystrom GJ, Lang CH. Sepsis and AMPK Activation by AICAR Differentially Regulate FoxO-1, -3 and -4 mRNA in Striated Muscle. Int J Clin Exp Med. 2008;1(1):50–63. [PMC free article] [PubMed] [Google Scholar]
- 44.Schmoll D, Walker KS, Alessi DR, Grempler R, Burchell A, Guo S, Walther R, Unterman TG. Regulation of glucose-6-phosphatase gene expression by protein kinase Balpha and the forkhead transcription factor FKHR. Evidence for insulin response unit-dependent and -independent effects of insulin on promoter activity. J Biol Chem. 2000;275(46):36324–36333. doi: 10.1074/jbc.M003616200. [DOI] [PubMed] [Google Scholar]
- 45.Nakae J, Biggs WH, III, Kitamura T, Cavenee WK, Wright CV, Arden KC, Accili D. Regulation of insulin action and pancreatic beta-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat Genet. 2002;32(2):245–253. doi: 10.1038/ng890. [DOI] [PubMed] [Google Scholar]
- 46.Rached MT, Kode A, Silva BC, Jung DY, Gray S, Ong H, Paik JH, DePinho RA, Kim JK, Karsenty G, Kousteni S. FoxO1 expression in osteoblasts regulates glucose homeostasis through regulation of osteocalcin in mice. J Clin Investig. 2010;120(1):357–368. doi: 10.1172/JCI39901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furuyama T, Kitayama K, Yamashita H, Mori N. Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochem J. 2003;375(Pt 2):365–371. doi: 10.1042/BJ20030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowker-Kinley MM, Davis WI, Wu P, Harris RA, Popov KM. Evidence for existence of tissue-specific regulation of the mammalian pyruvate dehydrogenase complex. Biochem J. 1998;329(Pt 1):191–196. doi: 10.1042/bj3290191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bastie CC, Nahle Z, McLoughlin T, Esser K, Zhang W, Unterman T, Abumrad NA. FoxO1 stimulates fatty acid uptake and oxidation in muscle cells through CD36-dependent and -independent mechanisms. J Biol Chem. 2005;280(14):14222–14229. doi: 10.1074/jbc.M413625200. [DOI] [PubMed] [Google Scholar]
- 50.Constantin-Teodosiu D, Constantin D, Stephens F, Laithwaite D, Greenhaff PL. The role of FOXO and PPAR transcription factors in diet-mediated inhibition of PDC activation and carbohydrate oxidation during exercise in humans and the role of pharmacological activation of PDC in overriding these changes. Diabetes. 2012;61(5):1017–1024. doi: 10.2337/db11-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rauramaa R, Kuusela P, Hietanen E. Adipose, muscle and lung tissue lipoprotein lipase activities in young streptozotocin treated rats. Horm Metab Res. 1980;12(11):591–595. doi: 10.1055/s-2007-999207. [DOI] [PubMed] [Google Scholar]
- 52.Kamei Y, Mizukami J, Miura S, Suzuki M, Takahashi N, Kawada T, Taniguchi T, Ezaki O. A forkhead transcription factor FKHR up-regulates lipoprotein lipase expression in skeletal muscle. FEBS Lett. 2003;536(1–3):232–236. doi: 10.1016/s0014-5793(03)00062-0. [DOI] [PubMed] [Google Scholar]
- 53.Tsuchida A, Yamauchi T, Ito Y, Hada Y, Maki T, Takekawa S, Kamon J, Kobayashi M, Suzuki R, Hara K, Kubota N, Terauchi Y, Froguel P, Nakae J, Kasuga M, Accili D, Tobe K, Ueki K, Nagai R, Kadowaki T. Insulin/Foxo1 pathway regulates expression levels of adiponectin receptors and adiponectin sensitivity. J Biol Chem. 2004;279(29):30817–30822. doi: 10.1074/jbc.M402367200. [DOI] [PubMed] [Google Scholar]
- 54.Peserico A, Chiacchiera F, Grossi V, Matrone A, Latorre D, Simonatto M, Fusella A, Ryall JG, Finley LW, Haigis MC, Villani G, Puri PL, Sartorelli V, Simone C. A novel AMPK-dependent FoxO3A-SIRT3 intramitochondrial complex sensing glucose levels. Cell Mol Life Sci. 2013;70(11):2015–2029. doi: 10.1007/s00018-012-1244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27(24):8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117(3):399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6(6):458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 58.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294(5547):1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 59.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA. 2001;98(25):14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kamei Y, Miura S, Suzuki M, Kai Y, Mizukami J, Taniguchi T, Mochida K, Hata T, Matsuda J, Aburatani H, Nishino I, Ezaki O. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J Biol Chem. 2004;279(39):41114–41123. doi: 10.1074/jbc.M400674200. [DOI] [PubMed] [Google Scholar]
- 61.Lagirand-Cantaloube J, Cornille K, Csibi A, Batonnet-Pichon S, Leibovitch MP, Leibovitch SA. Inhibition of atrogin-1/MAFbx mediated MyoD proteolysis prevents skeletal muscle atrophy in vivo. PLoS ONE. 2009;4(3):e4973. doi: 10.1371/journal.pone.0004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lagirand-Cantaloube J, Offner N, Csibi A, Leibovitch MP, Batonnet-Pichon S, Tintignac LA, Segura CT, Leibovitch SA. The initiation factor eIF3-f is a major target for atrogin1/MAFbx function in skeletal muscle atrophy. EMBO J. 2008;27(8):1266–1276. doi: 10.1038/emboj.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tintignac LA, Lagirand J, Batonnet S, Sirri V, Leibovitch MP, Leibovitch SA. Degradation of MyoD mediated by the SCF (MAFbx) ubiquitin ligase. J Biol Chem. 2005;280(4):2847–2856. doi: 10.1074/jbc.M411346200. [DOI] [PubMed] [Google Scholar]
- 64.Jogo M, Shiraishi S, Tamura TA. Identification of MAFbx as a myogenin-engaged F-box protein in SCF ubiquitin ligase. FEBS Lett. 2009;583(17):2715–2719. doi: 10.1016/j.febslet.2009.07.033. [DOI] [PubMed] [Google Scholar]
- 65.Lokireddy S, Wijesoma IW, Sze SK, McFarlane C, Kambadur R, Sharma M. Identification of atrogin-1-targeted proteins during the myostatin-induced skeletal muscle wasting. Am J Physiol Cell Physiol. 2012;303(5):C512–C529. doi: 10.1152/ajpcell.00402.2011. [DOI] [PubMed] [Google Scholar]
- 66.Clarke BA, Drujan D, Willis MS, Murphy LO, Corpina RA, Burova E, Rakhilin SV, Stitt TN, Patterson C, Latres E, Glass DJ. The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 2007;6(5):376–385. doi: 10.1016/j.cmet.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 67.Cohen S, Brault JJ, Gygi SP, Glass DJ, Valenzuela DM, Gartner C, Latres E, Goldberg AL. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J Cell Biol. 2009;185(6):1083–1095. doi: 10.1083/jcb.200901052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cohen S, Zhai B, Gygi SP, Goldberg AL. Ubiquitylation by Trim32 causes coupled loss of desmin, Z-bands, and thin filaments in muscle atrophy. J Cell Biol. 2012;198(4):575–589. doi: 10.1083/jcb.201110067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kudryashova E, Kudryashov D, Kramerova I, Spencer MJ. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J Mol Biol. 2005;354(2):413–424. doi: 10.1016/j.jmb.2005.09.068. [DOI] [PubMed] [Google Scholar]
- 70.Hishiya A, Iemura S, Natsume T, Takayama S, Ikeda K, Watanabe K. A novel ubiquitin-binding protein ZNF216 functioning in muscle atrophy. EMBO J. 2006;25(3):554–564. doi: 10.1038/sj.emboj.7600945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klionsky DJ, Codogno P (2013) The mechanism and physiological function of macroautophagy. J Innate Immun 427–433 [DOI] [PMC free article] [PubMed]
- 72.Codogno P. ATG genes and macroautophagy. Med Sci. 2004;20(8–9):734–736. doi: 10.1051/medsci/2004208-9734. [DOI] [PubMed] [Google Scholar]
- 73.Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998;273(51):33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- 74.Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18(14):3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Romanov J, Walczak M, Ibiricu I, Schuchner S, Ogris E, Kraft C, Martens S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31(22):4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walczak M, Martens S. Dissecting the role of the Atg12-Atg5-Atg16 complex during autophagosome formation. Autophagy. 2013;9(3):424–425. doi: 10.4161/auto.22931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130(1):165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 79.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae . Gene. 1997;192(2):245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 80.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150(6):1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2(4):330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20(21):5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6(6):472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 84.Yamazaki Y, Kamei Y, Sugita S, Akaike F, Kanai S, Miura S, Hirata Y, Troen BR, Kitamura T, Nishino I, Suganami T, Ezaki O, Ogawa Y. The cathepsin L gene is a direct target of FOXO1 in skeletal muscle. Biochem J. 2010;427(1):171–178. doi: 10.1042/BJ20091346. [DOI] [PubMed] [Google Scholar]
- 85.Reed SA, Sandesara PB, Senf SM, Judge AR. Inhibition of FoxO transcriptional activity prevents muscle fiber atrophy during cachexia and induces hypertrophy. FASEB J. 2012;26(3):987–1000. doi: 10.1096/fj.11-189977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Raben N, Hill V, Shea L, Takikita S, Baum R, Mizushima N, Ralston E, Plotz P. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet. 2008;17(24):3897–3908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27(2):433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lokireddy S, Wijesoma IW, Teng S, Bonala S, Gluckman PD, McFarlane C, Sharma M, Kambadur R. The ubiquitin ligase Mul1 induces mitophagy in skeletal muscle in response to muscle-wasting stimuli. Cell Metab. 2012;16(5):613–624. doi: 10.1016/j.cmet.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 89.Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, Chan DC. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141(2):280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rathbone CR, Booth FW, Lees SJ. FoxO3a preferentially induces p27Kip1 expression while impairing muscle precursor cell-cycle progression. Muscle Nerve. 2008;37(1):84–89. doi: 10.1002/mus.20897. [DOI] [PubMed] [Google Scholar]
- 91.Machida S, Spangenburg EE, Booth FW. Forkhead transcription factor FoxO1 transduces insulin-like growth factor’s signal to p27Kip1 in primary skeletal muscle satellite cells. J Cell Physiol. 2003;196(3):523–531. doi: 10.1002/jcp.10339. [DOI] [PubMed] [Google Scholar]
- 92.Shi X, Bowlin KM, Garry DJ. Fhl2 interacts with Foxk1 and corepresses Foxo4 activity in myogenic progenitors. Stem Cells. 2010;28(3):462–469. doi: 10.1002/stem.274. [DOI] [PubMed] [Google Scholar]
- 93.Shi X, Wallis AM, Gerard RD, Voelker KA, Grange RW, Depinho RA, Garry MG, Garry DJ. Foxk1 promotes cell proliferation and represses myogenic differentiation by regulating Foxo4 and Mef2. J Cell Sci. 2012;125(Pt 22):5329–5337. doi: 10.1242/jcs.105239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hribal ML, Nakae J, Kitamura T, Shutter JR, Accili D. Regulation of insulin-like growth factor-dependent myoblast differentiation by Foxo forkhead transcription factors. J Cell Biol. 2003;162(4):535–541. doi: 10.1083/jcb.200212107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kitamura T, Kitamura YI, Funahashi Y, Shawber CJ, Castrillon DH, Kollipara R, DePinho RA, Kitajewski J, Accili D. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J Clin Investig. 2007;117(9):2477–2485. doi: 10.1172/JCI32054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu AL, Kim JH, Zhang C, Unterman TG, Chen J. Forkhead box protein O1 negatively regulates skeletal myocyte differentiation through degradation of mammalian target of rapamycin pathway components. Endocrinology. 2008;149(3):1407–1414. doi: 10.1210/en.2007-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Erbay E, Park IH, Nuzzi PD, Schoenherr CJ, Chen J. IGF-II transcription in skeletal myogenesis is controlled by mTOR and nutrients. J Cell Biol. 2003;163(5):931–936. doi: 10.1083/jcb.200307158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bois PR, Grosveld GC. FKHR (FOXO1a) is required for myotube fusion of primary mouse myoblasts. EMBO J. 2003;22(5):1147–1157. doi: 10.1093/emboj/cdg116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu CM, Yang Z, Liu CW, Wang R, Tien P, Dale R, Sun LQ. Effect of RNA oligonucleotide targeting Foxo-1 on muscle growth in normal and cancer cachexia mice. Cancer Gene Ther. 2007;14(12):945–952. doi: 10.1038/sj.cgt.7701091. [DOI] [PubMed] [Google Scholar]
- 100.Allen DL, Unterman TG. Regulation of myostatin expression and myoblast differentiation by FoxO and SMAD transcription factors. Am J Physiol Cell Physiol. 2007;292(1):C188–C199. doi: 10.1152/ajpcell.00542.2005. [DOI] [PubMed] [Google Scholar]
- 101.Baehr LM, Furlow JD, Bodine SC. Muscle sparing in muscle RING finger 1 null mice: response to synthetic glucocorticoids. J Physiol. 2011;589(Pt 19):4759–4776. doi: 10.1113/jphysiol.2011.212845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Romanello V, Guadagnin E, Gomes L, Roder I, Sandri C, Petersen Y, Milan G, Masiero E, Del Piccolo P, Foretz M, Scorrano L, Rudolf R, Sandri M. Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J. 2010;29(10):1774–1785. doi: 10.1038/emboj.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McLoughlin TJ, Smith SM, DeLong AD, Wang H, Unterman TG, Esser KA. FoxO1 induces apoptosis in skeletal myotubes in a DNA-binding-dependent manner. Am J Physiol Cell Physiol. 2009;297(3):C548–C555. doi: 10.1152/ajpcell.00502.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Siu PM, Pistilli EE, Butler DC, Alway SE. Aging influences cellular and molecular responses of apoptosis to skeletal muscle unloading. Am J Physiol Cell Physiol. 2005;288(2):C338–C349. doi: 10.1152/ajpcell.00239.2004. [DOI] [PubMed] [Google Scholar]
- 105.Siu PM, Alway SE. Id2 and p53 participate in apoptosis during unloading-induced muscle atrophy. Am J Physiol Cell Physiol. 2005;288(5):C1058–C1073. doi: 10.1152/ajpcell.00495.2004. [DOI] [PubMed] [Google Scholar]
- 106.Adhihetty PJ, O’Leary MF, Chabi B, Wicks KL, Hood DA. Effect of denervation on mitochondrially mediated apoptosis in skeletal muscle. J Appl Physiol. 2007;102(3):1143–1151. doi: 10.1152/japplphysiol.00768.2006. [DOI] [PubMed] [Google Scholar]
- 107.Allen DL, Linderman JK, Roy RR, Bigbee AJ, Grindeland RE, Mukku V, Edgerton VR. Apoptosis: a mechanism contributing to remodeling of skeletal muscle in response to hindlimb unweighting. Am J Physiol. 1997;273(2 Pt 1):C579–C587. doi: 10.1152/ajpcell.1997.273.2.C579. [DOI] [PubMed] [Google Scholar]
- 108.Vande Velde C, Cizeau J, Dubik D, Alimonti J, Brown T, Israels S, Hakem R, Greenberg AH. BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol Cell Biol. 2000;20(15):5454–5468. doi: 10.1128/mcb.20.15.5454-5468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chipuk JE, Fisher JC, Dillon CP, Kriwacki RW, Kuwana T, Green DR. Mechanism of apoptosis induction by inhibition of the anti-apoptotic BCL-2 proteins. Proc Natl Acad Sci USA. 2008;105(51):20327–20332. doi: 10.1073/pnas.0808036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18(1):39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 111.Schakman O, Dehoux M, Bouchuari S, Delaere S, Lause P, Decroly N, Shoelson SE, Thissen JP. Role of IGF-I and the TNFalpha/NF-kappaB pathway in the induction of muscle atrogenes by acute inflammation. Am J Physiol Endocrinol Metab. 2012;303(6):E729–E739. doi: 10.1152/ajpendo.00060.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303(5666):2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 113.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24(50):7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 114.Sandri M, Barberi L, Bijlsma AY, Blaauw B, Dyar KA, Milan G, Mammucari C, Meskers CG, Pallafacchina G, Paoli A, Pion D, Roceri M, Romanello V, Serrano AL, Toniolo L, Larsson L, Maier AB, Munoz-Canoves P, Musaro A, Pende M, Reggiani C, Rizzuto R, Schiaffino S. Signalling pathways regulating muscle mass in ageing skeletal muscle. The role of the IGF1-Akt-mTOR-FoxO pathway. Biogerontology. 2013;14(3):303–323. doi: 10.1007/s10522-013-9432-9. [DOI] [PubMed] [Google Scholar]
- 115.Whitman SA, Wacker MJ, Richmond SR, Godard MP. Contributions of the ubiquitin-proteasome pathway and apoptosis to human skeletal muscle wasting with age. Pflugers Arch. 2005;450(6):437–446. doi: 10.1007/s00424-005-1473-8. [DOI] [PubMed] [Google Scholar]
- 116.Pardo PS, Lopez MA, Boriek AM. FOXO transcription factors are mechanosensitive and their regulation is altered with aging in the respiratory pump. Am J Physiol Cell Physiol. 2008;294(4):C1056–C1066. doi: 10.1152/ajpcell.00270.2007. [DOI] [PubMed] [Google Scholar]
- 117.Edstrom E, Altun M, Hagglund M, Ulfhake B. Atrogin-1/MAFbx and MuRF1 are downregulated in aging-related loss of skeletal muscle. J Gerontol Ser A Biol Sci Med Sci. 2006;61(7):663–674. doi: 10.1093/gerona/61.7.663. [DOI] [PubMed] [Google Scholar]
- 118.Furuyama T, Yamashita H, Kitayama K, Higami Y, Shimokawa I, Mori N. Effects of aging and caloric restriction on the gene expression of Foxo1, 3, and 4 (FKHR, FKHRL1, and AFX) in the rat skeletal muscles. Microsc Res Tech. 2002;59(4):331–334. doi: 10.1002/jemt.10213. [DOI] [PubMed] [Google Scholar]
- 119.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 120.Goto M, Terada S, Kato M, Katoh M, Yokozeki T, Tabata I, Shimokawa T. cDNA Cloning and mRNA analysis of PGC-1 in epitrochlearis muscle in swimming-exercised rats. Biochem Biophys Res Commun. 2000;274(2):350–354. doi: 10.1006/bbrc.2000.3134. [DOI] [PubMed] [Google Scholar]
- 121.Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J. 2002;16(14):1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- 122.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418(6899):797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 123.Russell AP, Feilchenfeldt J, Schreiber S, Praz M, Crettenand A, Gobelet C, Meier CA, Bell DR, Kralli A, Giacobino JP, Deriaz O. Endurance training in humans leads to fiber type-specific increases in levels of peroxisome proliferator-activated receptor-gamma coactivator-1 and peroxisome proliferator-activated receptor-alpha in skeletal muscle. Diabetes. 2003;52(12):2874–2881. doi: 10.2337/diabetes.52.12.2874. [DOI] [PubMed] [Google Scholar]
- 124.Daitoku H, Yamagata K, Matsuzaki H, Hatta M, Fukamizu A. Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes. 2003;52(3):642–649. doi: 10.2337/diabetes.52.3.642. [DOI] [PubMed] [Google Scholar]
- 125.Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, Goldberg AL, Spiegelman BM. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci USA. 2006;103(44):16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Brault JJ, Jespersen JG, Goldberg AL. Peroxisome proliferator-activated receptor gamma coactivator 1alpha or 1beta overexpression inhibits muscle protein degradation, induction of ubiquitin ligases, and disuse atrophy. J Biol Chem. 2010;285(25):19460–19471. doi: 10.1074/jbc.M110.113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yuan Y, Shi XE, Liu YG, Yang GS. FoxO1 regulates muscle fiber-type specification and inhibits calcineurin signaling during C2C12 myoblast differentiation. Mol Cell Biochem. 2011;348(1–2):77–87. doi: 10.1007/s11010-010-0640-1. [DOI] [PubMed] [Google Scholar]
- 128.Naya FJ, Mercer B, Shelton J, Richardson JA, Williams RS, Olson EN. Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J Biol Chem. 2000;275(7):4545–4548. doi: 10.1074/jbc.275.7.4545. [DOI] [PubMed] [Google Scholar]
- 129.Nagatomo F, Fujino H, Kondo H, Suzuki H, Kouzaki M, Takeda I, Ishihara A. PGC-1alpha and FOXO1 mRNA levels and fiber characteristics of the soleus and plantaris muscles in rats after hindlimb unloading. Histol Histopathol. 2011;26(12):1545–1553. doi: 10.14670/HH-26.1545. [DOI] [PubMed] [Google Scholar]
- 130.Mahoney DJ, Parise G, Melov S, Safdar A, Tarnopolsky MA. Analysis of global mRNA expression in human skeletal muscle during recovery from endurance exercise. FASEB J. 2005;19(11):1498–1500. doi: 10.1096/fj.04-3149fje. [DOI] [PubMed] [Google Scholar]
- 131.Marfe G, Tafani M, Pucci B, Di Stefano C, Indelicato M, Andreoli A, Russo MA, Sinibaldi-Salimei P, Manzi V. The effect of marathon on mRNA expression of anti-apoptotic and pro-apoptotic proteins and sirtuins family in male recreational long-distance runners. BMC Physiol. 2010;10:7. doi: 10.1186/1472-6793-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 132.Jamart C, Francaux M, Millet GY, Deldicque L, Frere D, Feasson L. Modulation of autophagy and ubiquitin-proteasome pathways during ultra-endurance running. J Appl Physiol. 2012;112(9):1529–1537. doi: 10.1152/japplphysiol.00952.2011. [DOI] [PubMed] [Google Scholar]
- 133.Pasiakos SM, McClung HL, McClung JP, Urso ML, Pikosky MA, Cloutier GJ, Fielding RA, Young AJ. Molecular responses to moderate endurance exercise in skeletal muscle. Int J Sport Nutr Exerc Metab. 2010;20(4):282–290. doi: 10.1123/ijsnem.20.4.282. [DOI] [PubMed] [Google Scholar]
- 134.Louis E, Raue U, Yang Y, Jemiolo B, Trappe S. Time course of proteolytic, cytokine, and myostatin gene expression after acute exercise in human skeletal muscle. J Appl Physiol. 2007;103(5):1744–1751. doi: 10.1152/japplphysiol.00679.2007. [DOI] [PubMed] [Google Scholar]
- 135.Jamart C, Naslain D, Gilson H, Francaux M. Higher activation of autophagy in skeletal muscle of mice during endurance exercise in the fasted state. Am J Physiol Endocrinol Metab. 2013;305(8):E964–974. doi: 10.1152/ajpendo.00270.2013. [DOI] [PubMed] [Google Scholar]
- 136.Goodman CA, Frey JW, Mabrey DM, Jacobs BL, Lincoln HC, You JS, Hornberger TA. The role of skeletal muscle mTOR in the regulation of mechanical load-induced growth. J Physiol. 2011;589(Pt 22):5485–5501. doi: 10.1113/jphysiol.2011.218255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, An Z, Loh J, Fisher J, Sun Q, Korsmeyer S, Packer M, May HI, Hill JA, Virgin HW, Gilpin C, Xiao G, Bassel-Duby R, Scherer PE, Levine B. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481(7382):511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]