Abstract
Lysophosphatidic acid (LPA) is a bioactive lipid mediator that activates G protein-coupled LPA receptors to exert fundamental cellular functions. Six LPA receptor genes have been identified in vertebrates and are classified into two subfamilies, the endothelial differentiation genes (edg) and the non-edg family. Studies using genetically engineered mice, frogs, and zebrafish have demonstrated that LPA receptor-mediated signaling has biological, developmental, and pathophysiological functions. Computational analyses have also identified several amino acids (aa) critical for LPA recognition by human LPA receptors. This review focuses on the evolutionary aspects of LPA receptor-mediated signaling by comparing the aa sequences of vertebrate LPA receptors and LPA-producing enzymes; it also summarizes the LPA receptor-dependent effects commonly observed in mouse, frog, and fish.
Keywords: Phylogenetic tree, Evolution, Lpar, ATX, LIPH, LIPI, Lipase
Introduction
Lysophosphatidic acid (LPA) is an intercellular lipid mediator that exerts a wide variety of cellular effects through G protein-coupled LPA receptors [1–5]. Thus far, six LPA receptor subtypes (Lpar1–Lpar6) have been identified in various vertebrates including human, mouse, rat, chick, frog, zebrafish, and medaka (Figs. 1, 2; Table 1). In other vertebrates, these LPA receptor genes have been predicted by in silico analysis. In addition, genes for LPA signal-related enzymes, such as the LPA-producing enzyme autotaxin (ATX) and phosphatidic acid-preferring phospholipase A1 (PA-PLA1), and LPA-degrading PA phosphatases (PAPs), have also been isolated or predicted in these vertebrates. PA-PLA1 has two isoforms, α and β, which are encoded by genes for lipase member H (Liph) and I (Lipi), respectively [6]. LPA-producing enzymes that show low amino acid (aa) identities (approximately 28 %) to vertebrate LIPH/LIPI have been identified in Drosophila melanogaster. However, no report has demonstrated the existence of LPA receptors in invertebrates. These observations suggest that extracellular LPA signaling was important in vertebrate evolution and plays common biological roles in a wide variety of vertebrate taxa.
Table 1.
The GenBank accession numbers or EMBL protein IDs for sequences used in the multiple sequence alignment and phylogenetic analysis
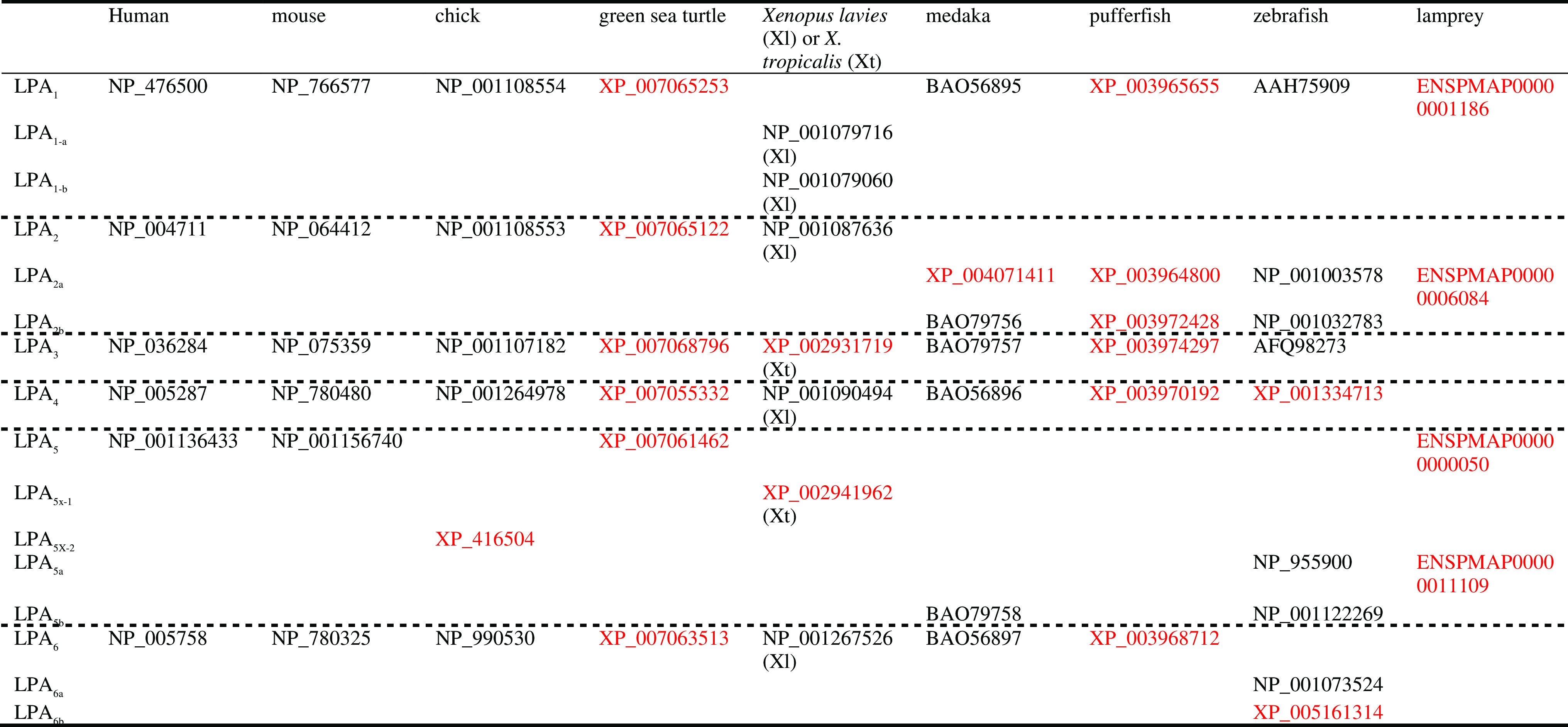
Numbers in black and red indicate validated and predicted sequences in the database, respectively
Table 2.
Tissue distribution of LPA receptors in mouse and medaka
| Receptor subtype | Species | Tissue distribution | References |
|---|---|---|---|
| LPA1 |
Mouse Medaka |
Brain, lung, heart, testes, small intestine, spleen, skeletal muscle Brain, heart, gut, testes, spleen, skeletal muscle |
[27] [25] |
|
LPA2 LPA2a LPA2b |
Mouse Medaka Medaka |
Brain, lung, kidney, testis Gut, testis, ovary, spleen Brain, heart, gut, testis, ovary, spleen, skeletal muscle |
[25] [25] |
| LPA3 |
Mouse Medaka |
Lung, kidney, testes, small intestine Gut, testis, ovary |
[27] [25] |
| LPA4 |
Mouse Medaka |
Brain, heart, lung, kidney, muscle, ovary, thymus Brain, heart, gut, testis, spleen, skeletal muscle |
[22] [25] |
|
LPA5 LPA5b |
Mouse Medaka |
Small intestine, spleen, thymus Heart, kidney, gut, spleen |
[75] [25] |
| LPA6 |
Mouse Medaka |
Ubiquitous including brain Brain, heart, liver, kidney, gut, testis, ovary, spleen, skeletal muscle |
[25] |
Since the first LPA receptor gene Lpar1 was identified in 1996, many studies have explored the expressions of LPA receptor genes and the biological functions of LPA receptor-mediated signaling in vitro and in vivo [1, 2, 7–9] (Table 2). Over the last decade, the biological and pathophysiological roles of each LPA receptor have been clarified using genetically engineered mice or zebrafish as well as in human genetic studies (Table 3). For example, LPA1-mediated signaling is involved in neurogenesis, brain formation, neuropathic pain initiation, and pulmonary fibrosis formation [10–12]. In a zebrafish model, autotaxin-LPA1 signaling is involved in the blood vessel formation [13]. LPA2 affects synaptic transmission and tumorigenesis, and LPA3 is important for the implantation of oocytes [14, 15]. LPA4 mediates blood and lymphatic vessel formation [16]. Like LPA1, LPA5 is involved in neuropathic pain [17]. Lpar6 is a gene associated with autosomal recessive hypotrichosis [18]. Furthermore, transcriptome analyses for LPA receptors, ATX and LIPI in human normal and malignant tissues have demonstrated that different cancer entities display distinct expression profiles of LPA1~6, ATX, and LIPI [19]. For instances, colon carcinomas show higher expression of LPA3 and LPA5, whereas other LPA receptor subtypes and LPA-generating enzymes are expressed at low levels. Ewing’s sarcomas exhibit higher expression of LPA4 and LIPI. Another report demonstrates up-regulation of LIPH in lung cancer tissues [20]. Therefore, distinct LPA signaling might play a different role in cancer development, depending on the cancer types.
Table 3.
LPA receptor-mediated functions revealed in genetically modified mouse, zebrafish, and frog
| Receptor subtype | Species | Function | References |
|---|---|---|---|
| LPA1 |
Mouse Zebrafish |
Brain development, psychiatric function, pain transmission, fibrosis, vascular formation Vascular formation |
|
|
LPA2 LPA2a |
Mouse Zebrafish |
Synaptic transmission, vascular formation in concert with LPA1 Unknown |
[14, 42] |
| LPA3 |
Mouse Zebrafish |
Embryo implantation, spermatogenesis in concert with LPA1 and LPA2 Left–right asymmetry |
[46] |
| LPA4 |
Mouse Zebrafish |
Vasculogenesis, lymphogenesis, osteogenesis Vasculogenesis in concert with LPA1 |
[16] [13] |
| LPA5 |
Mouse Frog |
Pain transmission Embryogenesis (?) |
[17] [24] |
| LPA6 |
Mouse Frog |
Hair development Forebrain development |
[54] [56] |
Although there are many excellent review articles describing the biological and pathophysiological roles of LPA signaling and LPA receptor-mediated signal transduction in mammals [1–3, 5, 11, 12, 19, 21–23], the effects of LPA signaling in non-mammals are not well known, and a comparative analysis of LPA receptor genes is necessary. Recent studies including ours have identified LPA receptor genes expressed in non-mammals, including frog, zebrafish, and medaka, and showed their expression profiles in medaka [13, 24, 25] (Table 2). Therefore, we have performed a comparative analysis of LPA receptors and discussed the evolution of LPA receptor-mediated signaling and its potential biological roles in non-mammals. To this end, we selected validated or predicted LPA receptor genes of human (Homo sapiens), mouse (Mus musculus), chicken (Gallus gallus), green sea turtle (Chelonia mydas), frog (Xenopus laevis and X. tropicalis), teleosts including medaka (Oryzias latipes), pufferfish (Fugu rubripes), and zebrafish (Danio rerio), and lamprey (Petromyzon marinus) (Figs. 1, 2, 3). Lamprey LPA2a showed 65.4 and 63.9 % aa identities to human and zebrafish LPA1, respectively, but 55.8 and 52.6 % identities to human and zebrafish LPA2, respectively. Thus, we considered lamprey LPA2a as a member of the LPA1 family in this review. On the other hand, lamprey LPA5 and LPA5b had about 40 % aa identity to zebrafish LPA4 or LPA6, while they showed about 30 % aa identity to zebrafish LPA5. However, as shown in the phylogenetic tree (Fig. 3b), lamprey LPA5 and LPA5b appeared to belong to the LPA5 cluster, probably due to limited sequence information. Thus, we omitted lamprey LPA5 and LPA5b from the discussion.
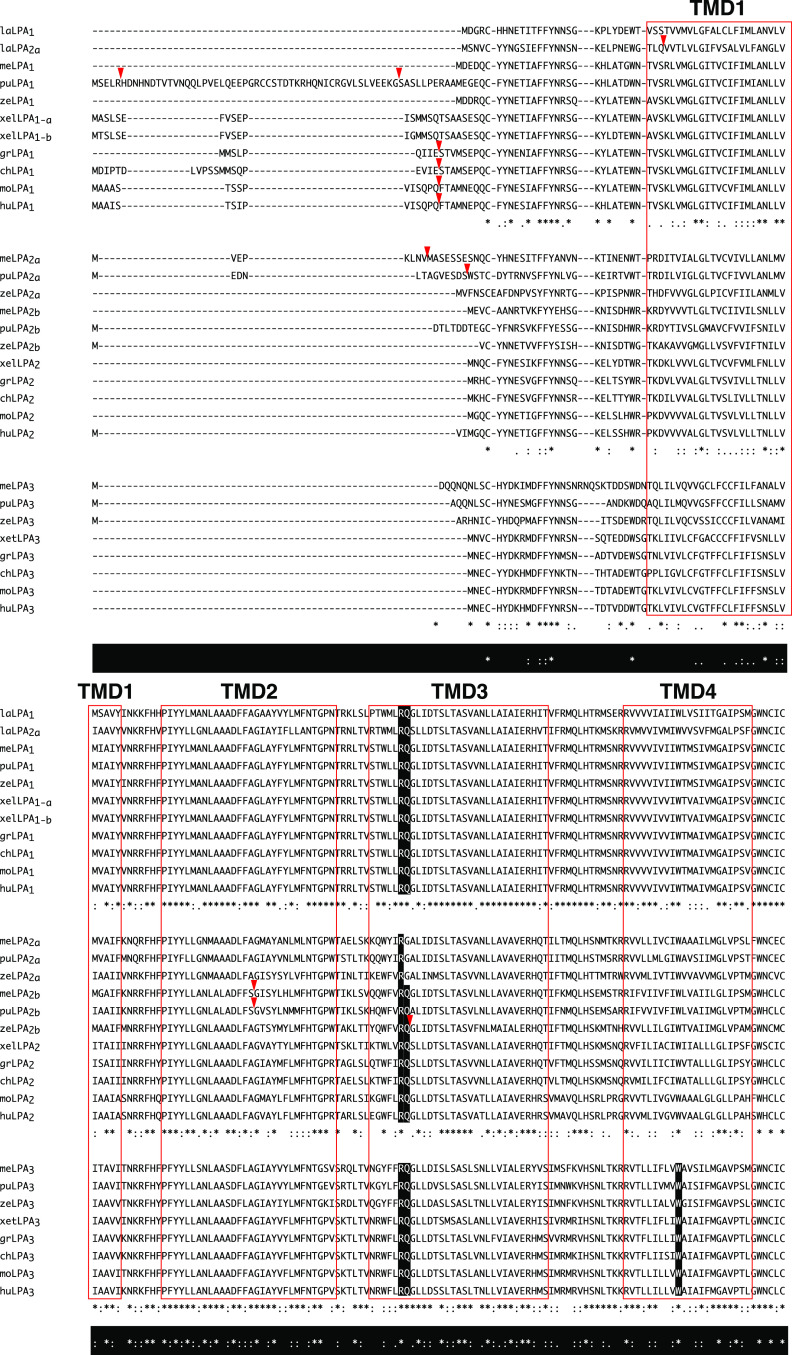
Fig. 1.
Multiple sequence alignment for edg family LPA1–LPA3. The multiple sequence alignment was performed using the MAFFT web service (http://mafft.cbrc.jp/alignment/server/). The GenBank accession numbers for the LPA receptors used in this analysis are shown in Table 1. Red boxes indicate the seven putative transmembrane domains (TMD1–TMD7). Red arrowheads indicate the exon–intron boundary in the coding regions of the genes. Asterisks (*), colons (:), and periods (.) below each human LPA receptor sequence indicate no substitutions (identical), conserved substitutions, and semi-conserved substitutions across all animal species for each LPA receptor subtype, respectively. Asterisks (*), colons (:), and periods (.) in white on a black background below the LPA receptor sequences indicate no substitutions, conserved substitutions, and semi-conserved substitutions, respectively, across all animal species for LPA1–LPA3. Residues involved in LPA binding are shown in white on a black background. me medaka, pu pufferfish, ze zebrafish, xel Xenopus lavies, xet X. tropicalis, gr green sea turtle, ch chick, mo mouse, hu human
Fig. 2.


Multiple sequence alignment for non-edg family LPA4–LPA6. A multiple sequence alignment was performed using the MAFFT web service (http://mafft.cbrc.jp/alignment/server/). The GenBank accession numbers for the LPA receptors used in this analysis are shown in Table 1. Red boxes indicate the seven putative transmembrane domains (TMD1–TMD7). Red arrowheads indicate the exon–intron boundary in the coding region of the genes. Asterisks (*), colons (:), and periods (.) below each human LPA receptor sequence indicate no substitutions, conserved substitutions, and semi-conserved substitutions, respectively, across all animal species for each LPA receptor subtypes. Asterisks (*), colons (:), and periods (.) in white on a black background below the LPA receptor sequences indicate no substitutions, conserved substitutions, and semi-conserved substitutions, respectively, across all animal species for LPA4–LPA6. Residues involved in LPA binding are shown in white on a black background. me medaka, pu pufferfish, ze zebrafish, xel Xenopus lavies, xet Xenopus tropicalis, gr green sea turtle, ch chick, mo mouse, hu human
Fig. 3.

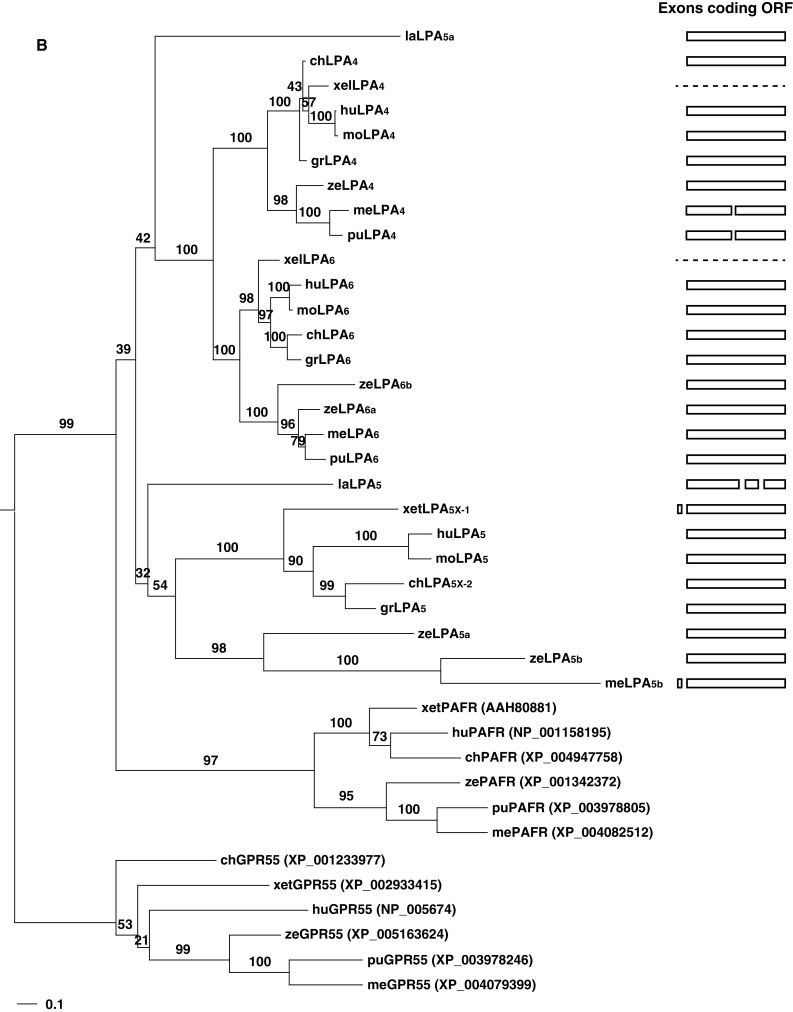
Phylogenetic trees and gene structures of the edg (a) and non-edg LPA receptors (b). For each family, closest outgroups [S1P receptors, GPR55, and platelet-activating factor receptors (PAFR)] were selected based on a preceding analysis by Yanagida and Ishii [69] using human LPA receptor genes. The amino acid sequences of LPA receptors of various vertebrates were aligned by MAFFT [70] with the auto option. The resulting alignments were subjected to PhyML [71] to infer phylogenetic trees based on the maximum likelihood (ML) method assuming the WAG [72] plus discrete Gamma model with four rate categories [73]. The GenBank accession numbers and EMBL protein ID numbers for the LPA receptors used in this analysis are shown in Table 1. The GenBank accession numbers for S1P receptors, GPR55, and PAFRs are shown in the parenthesis on the right of the receptor names. Bootstrap values are indicated at each node of the tree. On the right, each exon corresponding to the open reading frame is illustrated as a box. Dashed lines indicate no information for gene structures existing in the database. la lamprey, me medaka, pu pufferfish, ze zebrafish, xel Xenopus lavies, xet X. tropicalis, gr green sea turtle, mo mouse, hu human
LPA receptor genes
The first LPA receptor gene Lpar1 encoding LPA1 was identified in the developing cerebral cortex of mice in 1996 [8]. The aa sequence showed 40.5 % identity to a product of the endothelial differentiation gene (edg)-1, which was later identified as a receptor for sphingosine 1-phosphate (S1P) [26]. Subsequent studies have further isolated Lpar2 and Lpar3, which encode LPA2 and LPA3, respectively, from mouse tissues and demonstrate that pairwise comparisons between LPA1, LPA2, and LPA3 show over 50 % aa identity [27].
Although they mediate some biological effects of LPA, several cellular activities induced by LPA could not be explained by the activation of these LPA receptors. These findings implied the existence of additional LPA receptor(s), and in 2003, human Lpar4 was identified. It has been classified as a member of the purinergic receptor family and is distantly related to Lpar1–Lpar3 [28]. Subsequently, two additional LPA receptor genes, Lpar5 and Lpar6, have been discovered in mammals, and these three receptors (Lpar4, Lpar5, and Lpar6) have been termed the non-edg family LPA receptors [22].
The LPA receptor family is divided into the edg and non-edg family LPA receptors (Fig. 3), which together can account for most of the biological functions of LPA. Some other G protein-coupled receptor (GPCR) genes in mammals are thought to belong to the LPA receptor family, including GPR87 and GPR35 [29, 30]. However, further research is needed to confirm that these GPCRs function as LPA receptors. Thus, we will not focus on these yet unconfirmed genes in this review.
Edg family LPA receptor genes
Lpar1
A multiple protein sequence alignment of the LPA receptors of various vertebrates revealed that Lpar1 is highly conserved (Figs. 1, 3a). Human LPA1 shared more than 88 % aa identity with medaka LPA1 and 64.5 % aa identity even with lamprey LPA1, indicating that ligand specificity and intracellular signal transduction pathways as well as LPA1-dependent biological functions are common in vertebrates.
A computational analysis of edg family LPA receptors indicated several aa residues critical for LPA recognition and binding within the 3rd and 7th transmembrane domains (TMDs) [31]. Three residues identified by mutagenesis of human LPA1 are important for the activation of LPA1, specifically R3.28, Q3.29, and K7.36. For example, mutations of R3.28 or K7.36 to alanine result in changes in the efficacy and potency of LPA1. A Q3.29 mutation to alanine also decreases ligand interaction. These three residues are conserved across all vertebrates examined, suggesting that they are commonly critical for the activation of LPA1 in animal species (Fig. 1). Indeed, human, mouse, frog, zebrafish, and medaka LPA1 can couple to G proteins and activate intracellular signaling in response to LPA exposure with a comparable degree of efficacy and potency in a heterologous expression system [13, 25, 32–34].
Another unique feature of LPA1 sequences is the class I PDZ (PSD95/Dlg1/ZO-1)-binding motif consisting of SVV at the C-terminal end [35–37]. This motif sequence generally consists of X-(S/T)-X-(V/I/L)-COOH; it interacts with PDZ proteins and is involved in the regulation of receptor-mediated intracellular signaling. Mammalian LPA1 interacts with RhoGEF or GIPC (GAIP-interacting protein, C-terminus), leading to the activation of the Rho pathway or intracellular trafficking of LPA1. The finding that LPA1 of all species except for lamprey LPA1 and LPA2a contains the SVV sequence suggests the existence of common signaling pathways and biological roles (Fig. 1). Lamprey Lpar2a contains the initiation and stop codons, whereas no stop codon is found in the predicted lamprey Lpar1. Thus, the precise function of the lamprey LPA1 sequence remains unclear.
A recent report identified three amino acid residues, Y85, L87, and I325, which are essential for the proper folding and signal transduction of human LPA1 in the endoplasmic reticulum [38]. These resides are also conserved in all vertebrates except for lamprey LPA2a. Therefore, critical amino acid residues for ligand binding, receptor activation and folding, and the PDZ-binding motif are mostly conserved across animal species, strongly suggesting that the functions of LPA1 are largely common in these animals.
Although Lpar1 is involved in brain development, psychiatric and sensory functions, and fibrosis in mice [2, 10, 39, 40] (Table 3), it is not known whether similar LPA1-mediated effects are common in vertebrates. One biological function of Lpar1 commonly observed in mice and zebrafish is blood vessel formation. In Lpar1-deficient mice, a small fraction of infants showed frontal hematoma caused by abnormal blood vessel formation [41]. The incidence of hematoma appearance is increased in Lpar1/Lpar2-double knockout mice [42]. On the other hand, zebrafish embryos co-injected with morpholino antisense oligonucleotides for LPA1 and/or LPA4 have defects in vascular formation [13, 43]. These findings suggest that LPA1-mediated signaling cooperates with other LPA receptor subtypes, leading to normal blood vessel formation in vertebrates. This idea is also supported by the findings that ATX plays a role in vascular formation in both mice and zebrafish [13, 44].
Lpar2
In the phylogenetic analysis, the Lpar2 cluster was clearly divided into two subclusters, one consisting of loci in teleosts and the other consisting of loci in other vertebrates (Fig. 3a). Teleost Lpar2 was further duplicated, generating Lpar2a and Lpar2b. Computational analyses and mutagenesis studies have identified several specific residues, R3.28, Q3.29, R5.38, and K7.36, which are critical for human LPA2 activation [31]. The R3.28, R5.38, and K7.36 residues are conserved in the LPA2 sequences of all species. Q3.29 is conserved in non-teleost LPA2 and teleost LPA2b, but not teleost LPA2a, in which the corresponding aa is replaced by glycine (Fig. 1). Because mutations of Q3.29 to alanine or glutamic acid decrease human LPA2 activation [31], teleost LPA2a may show some unique interactions with LPA compared with teleost LPA2b or non-teleost LPA2. However, zebrafish LPA2a responds to LPA exposure by secreting transforming growth factor-α (TGF-α) in a TGF-α shedding assay, with a similar degree of efficacy and potency as zebrafish LPA2b [13]. Thus, the replacement of glutamine with glycine is unlikely to affect the recognition of LPA by LPA2a.
Similar to LPA1, human and mouse LPA2 contain a PDZ-binding motif consisting of STL at the C-terminal region. This motif of human LPA2 can interact with PDZ proteins, such as Na+–H+ exchanger regulatory factor 2 or RhoGEF, resulting in a change in the activation of intracellular signaling pathways [37, 45]. Although chicken and turtle LPA2 also contain a similar PDZ-binding motif consisting of SSI or SSL, frog and teleost LPA2a and LPA2b have no such motif (Fig. 1). Thus, C-terminal region-mediated signaling of LPA2 varies depending on animal species.
As mentioned above, mice lacking both Lpar1 and Lpar2, but not those lacking just Lpar2, show more severe frontal hematoma and loss of LPA responses that affect cortical development, suggesting a non-essential role for LPA2 in mouse development [42] (Table 3). LPA2 also plays a role in excitatory synaptic transmission [14] (Table 3). However, there is no direct evidence that Lpar2 has the same biological functions in other vertebrates.
Lpar3
Mutagenesis studies have identified several residues involved in human LPA3 activation, including R3.28, Q3.29, W4.64, R5.38, and K7.35 [31]. LPA3 of all vertebrates examined contained R3.28, Q3.29, and W4.64. R5.38, in which arginine was replaced by threonine, was also present in LPA3 of most species, with the exception of medaka (Fig. 1). In the 7th TMD, both R7.36 and K7.35, which are important for LPA recognition by LPA3, were observed in all animal species considered (Fig. 1). Therefore, LPA3-mediated signaling is likely conserved across animal species. Indeed, stimulation of mouse and medaka LPA3 results in the same cytoskeletal responses in a heterologous expression system [25].
Mouse LPA3 is important for embryo implantation in the uterus [15] (Table 3). However, considering that most non-mammals are oviparous and never develop eggs within the mother, LPA3 has distinct biological roles in non-mammals. In zebrafish, it regulates left–right asymmetry. This is based on the finding that knockdown of Lpar3 impaired calcium influxes in dorsal forerunner cells, leading to the inhibition of Kupffer’s vesicle formation and the disruption of organ left–right asymmetries [46] (Table 3). However, abnormal organ formation, which might be caused by disrupted left–right asymmetries, has not been demonstrated in mice lacking Lpar3 or Lpar1–Lpar3 [47]. Another potential biological role for Lpar3 is the regulation of male reproduction. Lpar3 is highly expressed in the testis of human, mouse, and medaka [25, 48, 49]. Indeed, a triple deletion of Lpar1–Lpar3 results in defects in germ cell survival and an increased prevalence of azoospermia in aging mice, although single deletions of Lpar3 have no effect [47] (Table 3). Thus, LPA3 might co-operate with LPA1 and LPA2 to regulate the maintenance of reproductive organs. More detailed analyses of LPA3-mediated effects in the reproduction systems of fish and other animal species are needed.
Non-edg family LPA receptor genes
Lpar4
Lpar4 was originally identified as the orphan GPCR GPR23/p2y9, which belongs to the purinergic receptor family, whose ligands are adenosine, ADP, or ATP. Ligand screening studies identified LPA as an agonist for human GPR23 [28]. In our analysis, the aa identity of human LPA4 to the edg family LPA receptors was about 25 %. There were no critical residues for receptor activation corresponding to R3.28, Q3.29, or K7.36, which each exist in LPA1 and LPA2, and in LPA4 of most animal species (Fig. 2). Computational analyses show that the polar head group of LPA interacts with LPA4 at distinct regions compared with LPA1–LPA3 [50]. S3.28, a threonine in the second extracellular loop (EL2), and Y6.51 are critical residues for human LPA4 activation. S3.28 and the threonine in EL2 were conserved across LPA4 of all animal species, while Y6.51 was present in animal species except for frog, in which tyrosine was replaced by phenylalanine (Fig. 2). This observation indicates that LPA4 has maintained LPA binding activity during evolution. Indeed, mammalian and teleost LPA4 respond to LPA with cellular activation, such as cytoskeletal changes and TGF-α release in a heterologous expression system [13, 25, 51].
In mice, Lpar4 is involved in vasculogenesis, lymphogenesis, and osteogenesis [16] (Table 3). In zebrafish, Lpar4 is involved in vasculogenesis in concert with Lpar1 [13] (Table 3). Further detailed analyses are needed to clarify the biological roles of LPA4-mediated signaling.
Lpar5
Computational analyses have identified four ligand recognition residues, R2.60, H4.64, R6.62, and R7.32, in human LPA5, all of which are predicted to bind to the polar head group of LPA [52]. R2.60 also existed in LPA5 of other vertebrate species examined (Fig. 2). However, H4.64 was replaced by phenylalanine and isoleucine in medaka and zebrafish LPA5b, respectively (Fig. 2). R6.62 was replaced by lysine in chicken and turtle LPA5 and zebrafish LPA5a, and absent from medaka LPA5b and zebrafish LPA5b; these LPA5b sequences were very short (Fig. 2). R7.32 was replaced by lysine in frog LPA5X-1, asparagine in medaka LPA5b, and phenylalanine in zebrafish LPA5 (Fig. 2). Thus, such variation at critical aa residues across various animal species suggests the differential ability of LPA5 to recognize and bind LPA, resulting in variation in LPA5 activation. Indeed, zebrafish LPA5a and LPA5b, and medaka LPA5b fail to show cellular responses after treatment with LPA, such as TGF-α release or actin rearrangement in a heterologous expression system, whereas mammalian LPA5 is activated by LPA treatment [13, 25]. Alternatively, the action of LPA5b may not be coupled to G12/13 because the C-terminal regions are shorter than those of non-teleost LPA5 sequences (Fig. 2).
LPA5-dependent signaling plays an important role in pain transmission as evidenced by experiments using Lpar5-deficient mice [17] (Table 3). However, there are no reports of LPA5-mediated biological functions in other vertebrates, although maternal Lpar5 transcripts are present in the animal pole of frog embryos [24]. The exploration of the biological roles of LPA5 has just commenced.
Lpar6
A recent in silico study on human LPA6 has identified several missense mutations leading to a shift in orientation of LPA at the binding site, which may affect LPA6 activation [53]. These include S3T, D63V, G146R, I188F, E189K, D248Y, and L277P, which have been severely associated with abnormal hair development. LPA6 of other vertebrates contained glutamine and glutamic acid corresponding to D63 and E189 of human LPA6, respectively (Fig. 2). Isoleucine and leucine corresponding to I188 and L277 of human LPA6, respectively, were also conserved in all LPA6 except for zebrafish LPA6b (Fig. 2). However, other aa residues varied among animal species. These analyses employ the docking assay using human LPA6 and 1-acyl-LPA. Because LPA6 preferentially binds 2-acyl LPA [5], other residues that interact with this type of LPA might exist. Nonetheless, medaka LPA6 activation results in cytoskeletal changes similar to those observed for mammalian LPA6, suggesting that LPA6-mediated signaling is highly conserved during evolution [25].
Although Lpar6 plays an important role in mammalian hair development in concert with Liph [18, 54] (Table 3), most tissues seem to ubiquitously express Lpar6 in mammals and fish [25, 55]. Thus, LPA6-mediated signaling might have additional fundamental and ubiquitous biological functions; this is partially supported by a recent finding that Lpar6 is involved in forebrain formation of X. lavies [56] (Table 3).
Phylogenetic analysis of LPA receptor genes
Phylogenetic analysis of non-edg LPA receptor genes
A unique feature of edg family LPA receptor genes is the insertion of an intron in the middle of the coding region for the 6th TMD [27]. The intron insertion within the coding region for the 6th TMD was found not only in mammals but also in all vertebrates examined (Figs. 1, 3a). This observation suggests that the intron insertion occurred in the ancestral edg sequence and has likely been evolutionarily preserved. However, another edg family consisting of the S1P receptor genes (S1pr1–S1pr5) does not contain an intron insertion within the open reading frame [57]. Thus, it is likely that the ancestral edg diverged into two classes, the edg family LPA receptor genes and the edg family S1P receptor genes, and the intron insertion appeared only in the edg family LPA receptor lineage during evolution.
In addition to the conserved intron insertion, human, mouse, chicken, and turtle Lpar1 genes contained an additional intron insertion within the coding region at the N-terminus (Figs. 1, 3a). In pufferfish Lpar1, two intron insertions are likely to be present within the coding region at the N-terminus. By contrast, lamprey Lpar1 and Lpar2a genes contained an intron insertion within the coding region at the C-terminus, although common features are not observed for the insertions in these two genes. However, because the sequences of turtle, pufferfish, and lamprey LPA receptor genes are only predicted by in silico analysis, the precise gene structures need to be determined.
Two Lpar2 genes, Lpar2a and Lpar2b, have been identified or predicted in medaka, zebrafish, and pufferfish, but not in mammals [13, 25, 40], indicating that they may be specific to teleosts. The structures of these teleost Lpar2a and Lpar2b differ from each other (Fig. 3a). In addition to the conserved intron insertion within the coding region for the 6th TMD, medaka and pufferfish Lpar2b had an additional intron insertion within the coding region for the 2nd TMD, while zebrafish Lpar2b contained an intron within the coding region for the 3rd TMD (Figs. 1, 3a). Pufferfish Lpar2b also contained an intron in the coding region for the C-terminus, while zebrafish Lpar2b contained two introns in the C-terminal coding region (Figs. 1, 3a). Likewise, the medaka and pufferfish Lpar2a genes included an intron within the coding region at the C-terminus (Figs. 1, 3a). However, shared features were not observed for the intron insertions in the two teleost Lpar2 genes, probably due to their high aa sequence diversities. Taken together, the two Lpar2 genes Lpar2a and Lpar2b might have undergone a duplication event in the ancestral teleost lineage, and exon usage might have evolved independently.
Phylogenetic analysis of non-edg LPA receptor genes
The medaka and pufferfish Lpar4 genes contained a unique insertion of a short intron consisting of about 80 and 270 bp within the coding region for the 5th TMD, respectively [25] (Gene ID: LOC101078478). This insertion did not exist in non-teleost or zebrafish Lpar4, or in other LPA receptor genes, indicating that it arose in the medaka and pufferfish lineage.
The Lpar5 clusters in the phylogenetic tree were more characteristic than other LPA receptor genes (Fig. 3b). During evolution, an ancestral Lpar5 gene diverged from the Lpar4 and Lpar6 clusters. Furthermore, the Lpar5 cluster was divided into two subclusters, one consisting of teleost loci and the other consisting of other vertebrate loci. Teleost Lpar5 was further duplicated, generating Lpar5a and Lpar5b. Identification of endogenous ligand or signaling pathways for teleost LPA5a and LPA5b will provide a better understanding of the evolutionary significance of Lpar5.
Evolutionary rate
In phylogenetic trees of LPA receptor genes (Fig. 3), the evolutionary rate greatly differs between different subtypes. This tree reveals a possible relationship between evolutionary rate of a gene and its tissue distribution. In LPA1, the evolutionary distance between mammals and teleosts is 0.255 on this tree. On the other hand, the corresponding distance is 1.06 in LPA3 (Fig. 3a). The former is more than four times slower than the latter in evolutionary rate. LPA1 expression in brain is commonly observed in various vertebrates including human, mouse, rat, frog, zebrafish, and medaka (Table 2) [34, 43, 58]. In mouse, LPA1 is known to be involved in brain development and functions, while LPA3 is involved in reproductive functions (Table 3). The observation that evolutionary rate of brain-expressed genes is relatively slow is consistent with the finding by Kuma et al. [59] on protein kinase and immunoglobulin genes. They argued that many proteins are involved in complex biochemical networks in the brain or in central nervous system [60], and thus these proteins interact with a larger number of molecules than in other tissues. Accordingly, brain-expressed genes are under stronger functional constraints to maintain such interactions [61]. As a result, their evolutionary rates at the amino acid level remain low.
A similar tendency is observed in the non-edg LPA receptor family. LPA4 and LPA6, expressed in brain, are more slowly evolving than LPA5 (Fig. 3b). As the functions and tissue distributions of LPA receptors in different species are determined more clearly in the future, it may become possible to interpret the changes in evolutionary rate or specific amino acid substitutions according to functional changes.
Phylogenetic analysis of genes for LPA-generating enzymes
Signaling LPA is extracellularly generated via the catalytic action of two structurally distinct lipases, ATX and PA-PLA1 [6, 62–65]. ATX was firstly identified as ectonucleotide pyrophosphatase/phosphodiesterase encoded by the Enpp2 gene, and later shown to exhibit higher lysophospholipase D activity, which mainly involves cleaving lysophospholipids to produce LPA. Interestingly, among Enpp family members, consisting of Enpp1–7, only ATX/Enpp2 shows lipase activity to produce LPA [62, 65]. Human ATX shows high aa identities to other vertebrate ATXs, and higher aa identities when catalytic domains located in the central region of ATX protein are compared (Fig. 4a). The central catalytic domain is responsible for lipase activity, although the N-terminal somatomedin B-like domain and C-terminal nuclease domain are cooperative for the activity [65]. Thus, it is suggested that lysophospholipase D activity of ATX is highly conserved across animal species. We found two possible ATX-like proteins in Caenorhabditis elegans in the database (accession numbers NP_001041085 and NP_001041087). However, these show about 15 % aa identity to a catalytic domain of human ATX, and are likely to be closer to ENPP1 and ENPP3, respectively.
Fig. 4.
Phylogenetic trees of ATX (a), and LIPH/LIPI (b). For each family, closest outgroups [ENPP, phosphatidylserine-specific PLA1 (PLA1A), hepatic triacylglycerol lipase (LIPC), endothelial lipase (LIPG), and lipoprotein lipase (LPL)] were selected based on preceding analyses [6, 62, 65]. The amino acid sequences of ATX, or LIPH and LIPI of various vertebrates were used to infer phylogenetic trees, as described in Fig. 3 legend. The GenBank accession numbers and EMBL protein ID numbers for ATX or LIPH and LIPI used in this analysis are shown in the parenthesis. Designation of LIPH and LIPI is based on the gene deposited in the database. Bootstrap values are indicated above each branch of the tree. The percentage aa identities shown on the right were determined by using the homology search tool of the software GENETYX-MAC®. For ATX, full length (FL) and a catalytic domain (CD) corresponding to the region of aa 161–540 of human ATX were used for the analysis. fl fly, la lamprey, me medaka, pu pufferfish, ze zebrafish, xel Xenopus lavies, gr green sea turtle, mo mouse, hu human
Liph and Lipi belong to the lipase gene family consisting of phosphatidylserine-specific PLA1, hepatic lipase, lipoprotein lipase, endothelial lipase, pancreatic lipase, and pancreatic lipase-related protein [6]. However, there is no established theory on evolutionary relationship between Liph and Lipi. Holmes and Cox [66] discussed that the mammalian Lipi arose from a recent gene duplication event of an ancestral Liph-like gene during mammalian evolution, but the phylogenetic tree (Fig. 5 in Ref. [66]) does not support this theory in a straightforward way. On the other hand, Hesse et al. [67] performed a careful analysis using syntenic information and showed that the Lipi locus already existed in the common ancestor of mammals and teleosts; however, the syntenic conservation of teleost “Liph” is unclear. Phylogenetic reconstruction of Liph and Lipi is difficult due to a great difference in evolutionary rate between both isoforms. Generally, lineages with high evolutionary rate are often misestimated to have an ancient origin, and it is known that the maximum likelihood (ML) method is relatively robust in such a situation [68].
A phylogenetic tree based on the ML method (Fig. 4b) is easier to interpret than trees in the two above-mentioned papers. Assuming this tree topology and the syntenic information reported by Hesse et al. [67], a possible evolutionary scenario is: (1) Lipi is the ancestral locus in early bony fishes, (2) Liph was duplicated from Lipi in the lineage of ancestral amniote, (3) only in the mammalian lineage, the evolutionary rate of Lipi was greatly increased, (4) in the teleost lineage, an independent gene duplication occurred. The increase in evolutionary rate of mammalian Lipi may be due to the change in the expression pattern; mammalian Lipi is expressed only in testis, but avian Lipi is more broadly expressed [67], although there is a possibility of many other factors. Since the statistical support is not high in our tree, the possibility of ancient duplication between Liph and Lipi is not rejected.
All vertebrate lipases in the LIPH/LIPI cluster contain a catalytic triad consisting of three amino acids, serine, aspartic acid, and histidine, essential for catalytic activity common to the lipase gene family [6]. Unlike LPA receptors or ATX, two LIPH/LIPI-like proteins in D. melanogaster were found in the database (NP_573259 and NP_648652). NP_573259 possess a catalytic triad, whereas NP_648652 lacks serine. However, whether NP_573259 catalyzes PA to produce LPA remains unknown. Even if it is the case, it is unclear whether LPA produced in fly acts as an extracellular signaling mediator. Further comparative studies on LIPH/LIPI structures and functions would help us to understand the biological role of LPA or emergence of signaling LPA during evolution.
Conclusion
In the present study, we performed comparative analyses of LPA receptors across fish, frog, turtle, chicken, and mammals, and discussed the potentially conserved biological roles of LPA receptor-mediated signaling, although limited in vivo studies are available to assess its biological importance in non-mammals. We believe that further analyses that consider non-mammals will reveal more conserved biological roles of LPA signaling, particularly with respect to the development of innate behaviors common to many vertebrates. Recent genome editing technologies, widely applied to many animals including mouse and fish will facilitate the generation of LPA receptor gene deletion animals to explore the functions of LPA signaling.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas (to NF) and Platform for Drug Discovery, Informatics, and Structural Life Science (to KK), from the Ministry of Education, Culture, Sports, Science and Technology, Japan. We thank Dr. D. Standley for reading of the manuscript.
Conflict of interest
The authors declare that there is no conflict of interest.
Contributor Information
Nobuyuki Fukushima, Phone: +81-6-4307-3436, Email: nfukushima@life.kindai.ac.jp.
Kazutaka Katoh, Phone: +81-6-6879-4925, Email: katoh@ifrec.osaka-u.ac.jp.
References
- 1.Choi JW, Chun J. Lysophospholipids and their receptors in the central nervous system. Biochim Biophys Acta. 2013;1831:20–32. doi: 10.1016/j.bbalip.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi JW, Herr DR, Noguchi K, Yung YC, Lee CW, Mutoh T, Lin ME, Teo ST, Park KE, Mosley AN, Chun J. LPA receptors: subtypes and biological actions. Annu Rev Pharmacol Toxicol. 2010;50:157–186. doi: 10.1146/annurev.pharmtox.010909.105753. [DOI] [PubMed] [Google Scholar]
- 3.Lin ME, Herr DR, Chun J. Lysophosphatidic acid (LPA) receptors: signaling properties and disease relevance. Prostaglandins Other Lipid Mediat. 2010;91:130–138. doi: 10.1016/j.prostaglandins.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsujiuchi T, Araki M, Hirane M, Dong Y, Fukushima N. Lysophosphatidic acid receptors in cancer pathobiology. Histol Histopathol. 2014;29:313–321. doi: 10.14670/HH-29.313. [DOI] [PubMed] [Google Scholar]
- 5.Yanagida K, Kurikawa Y, Shimizu T, Ishii S. Current progress in non-Edg family LPA receptor research. Biochim Biophys Acta. 2013;1831:33–41. doi: 10.1016/j.bbalip.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Aoki J, Inoue A, Makide K, Saiki N, Arai H. Structure and function of extracellular phospholipase A1 belonging to the pancreatic lipase gene family. Biochimie. 2007;89:197–204. doi: 10.1016/j.biochi.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 7.Fukushima N, Ishii I, Contos JA, Weiner JA, Chun J. Lysophospholipid receptors. Annu Rev Pharmacol Toxicol. 2001;41:507–534. doi: 10.1146/annurev.pharmtox.41.1.507. [DOI] [PubMed] [Google Scholar]
- 8.Hecht JH, Weiner JA, Post SR, Chun J. Ventricular zone gene-1 (vzg-1) encodes a lysophosphatidic acid receptor expressed in neurogenic regions of the developing cerebral cortex. J Cell Biol. 1996;135:1071–1083. doi: 10.1083/jcb.135.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishii I, Fukushima N, Ye X, Chun J. Lysophospholipid receptors: signaling and biology. Annu Rev Biochem. 2004;73:321–354. doi: 10.1146/annurev.biochem.73.011303.073731. [DOI] [PubMed] [Google Scholar]
- 10.Herr KJ, Herr DR, Lee C-W, Noguchi K, Chun J. Stereotyped fetal brain disorganization is induced by hypoxia and requires lysophosphatidic acid receptor 1 (LPA1) signaling. Proc Natl Acad Sci USA. 2011;108:15444–15449. doi: 10.1073/pnas.1106129108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ueda H, Matsunaga H, Olaposi OI, Nagai J. Lysophosphatidic acid: chemical signature of neuropathic pain. Biochim Biophys Acta. 2013;1831:61–73. doi: 10.1016/j.bbalip.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 12.Yung YC, Mutoh T, Lin M-E, Noguchi K, Rivera RR, Choi JW, Kingsbury MA, Chun J. Lysophosphatidic acid signaling may initiate fetal hydrocephalus. Sci Transl Med. 2011;3:99ra87. doi: 10.1126/scitranslmed.3002095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yukiura H, Hama K, Nakanaga K, Tanaka M, Asaoka Y, Okudaira S, Arima N, Inoue A, Hashimoto T, Arai H, Kawahara A, Nishina H, Aoki J. Autotaxin regulates vascular development via multiple lysophosphatidic acid (LPA) receptors in zebrafish. J Biol Chem. 2011;286:43972–43983. doi: 10.1074/jbc.M111.301093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trimbuch T, Beed P, Vogt J, Schuchmann S, Maier N, Kintscher M, Breustedt J, Schuelke M, Streu N, Kieselmann O, Brunk I, Laube G, Strauss U, Battefeld A, Wende H, Birchmeier C, Wiese S, Sendtner M, Kawabe H, Kishimoto-Suga M, Brose N, Baumgart J, Geist B, Aoki J, Savaskan NE, Brauer AU, Chun J, Ninnemann O, Schmitz D, Nitsch R. Synaptic PRG-1 modulates excitatory transmission via lipid phosphate-mediated signaling. Cell. 2009;138:1222–1235. doi: 10.1016/j.cell.2009.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye X, Hama K, Contos JJ, Anliker B, Inoue A, Skinner MK, Suzuki H, Amano T, Kennedy G, Arai H, Aoki J, Chun J. LPA3-mediated lysophosphatidic acid signalling in embryo implantation and spacing. Nature. 2005;435:104–108. doi: 10.1038/nature03505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sumida H, Noguchi K, Kihara Y, Abe M, Yanagida K, Hamano F, Sato S, Tamaki K, Morishita Y, Kano MR, Iwata C, Miyazono K, Sakimura K, Shimizu T, Ishii S. LPA4 regulates blood and lymphatic vessel formation during mouse embryogenesis. Blood. 2010;116:5060–5070. doi: 10.1182/blood-2010-03-272443. [DOI] [PubMed] [Google Scholar]
- 17.Lin ME, Rivera RR, Chun J. Targeted deletion of LPA5 identifies novel roles for lysophosphatidic acid signaling in development of neuropathic pain. J Biol Chem. 2012;287:17608–17617. doi: 10.1074/jbc.M111.330183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasternack SM, von Kugelgen I, Al Aboud K, Lee YA, Ruschendorf F, Voss K, Hillmer AM, Molderings GJ, Franz T, Ramirez A, Nurnberg P, Nothen MM, Betz RC. G protein-coupled receptor P2Y5 and its ligand LPA are involved in maintenance of human hair growth. Nat Genet. 2008;40:329–334. doi: 10.1038/ng.84. [DOI] [PubMed] [Google Scholar]
- 19.Willier S, Butt E, Grunewald TG. Lysophosphatidic acid (LPA) signalling in cell migration and cancer invasion: a focussed review and analysis of LPA receptor gene expression on the basis of more than 1700 cancer microarrays. Biol Cell Auspices Eur Cell Biol Org. 2013;105:317–333. doi: 10.1111/boc.201300011. [DOI] [PubMed] [Google Scholar]
- 20.Seki Y, Yoshida Y, Ishimine H, Shinozaki-Ushiku A, Ito Y, Sumitomo K, Nakajima J, Fukayama M, Michiue T, Asashima M, Kurisaki A. Lipase member H is a novel secreted protein selectively upregulated in human lung adenocarcinomas and bronchioloalveolar carcinomas. Biochem Biophys Res Commun. 2014;443:1141–1147. doi: 10.1016/j.bbrc.2013.12.106. [DOI] [PubMed] [Google Scholar]
- 21.Yung YC, Stoddard NC, Chun J. LPA receptor signaling: pharmacology, physiology, and pathophysiology. J Lipid Res. 2014;55:1192–1214. doi: 10.1194/jlr.R046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishii S, Noguchi K, Yanagida K. Non-Edg family lysophosphatidic acid (LPA) receptors. Prostaglandins Other Lipid Mediat. 2009;89:57–65. doi: 10.1016/j.prostaglandins.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Leblanc R, Peyruchaud O (2014) New insights into the autotaxin/LPA axis in cancer development and metastasis. Exp Cell Res. doi:10.1016/j.yexcr.2014.11.010 [DOI] [PubMed]
- 24.Masse K, Kyuno J, Bhamra S, Jones EA. The lysophosphatidic acid (LPA) and sphingosine-1-phosphate (S1P) receptor gene families: cloning and comparative expression analysis in Xenopus laevis. Int J Dev Biol. 2010;54:1361–1374. doi: 10.1387/ijdb.103068km. [DOI] [PubMed] [Google Scholar]
- 25.Morimoto Y, Ishii S, Ishibashi J, Katoh K, Tsujiuchi T, Kagawa N, Fukushima N. Functional lysophosphatidic acid receptors expressed in Oryzias latipes . Gene. 2014;551:189–200. doi: 10.1016/j.gene.2014.08.056. [DOI] [PubMed] [Google Scholar]
- 26.Lee MJ, Van Brocklyn JR, Thangada S, Liu CH, Hand AR, Menzeleev R, Spiegel S, Hla T. Sphingosine-1-phosphate as a ligand for the G protein-coupled receptor EDG-1. Science. 1998;279:1552–1555. doi: 10.1126/science.279.5356.1552. [DOI] [PubMed] [Google Scholar]
- 27.Contos JJA, Ishii I, Chun J. Lysophosphatidic acid receptors. Mol Pharmacol. 2000;58:1188–1196. doi: 10.1124/mol.58.6.1188. [DOI] [PubMed] [Google Scholar]
- 28.Noguchi K, Ishii S, Shimizu T. Identification of p2y9/GPR23 as a novel G protein-coupled receptor for Lysophosphatidic acid, structurally distant from the Edg family. J Biol Chem. 2003;278:25600–25606. doi: 10.1074/jbc.M302648200. [DOI] [PubMed] [Google Scholar]
- 29.Zhao P, Abood ME. GPR55 and GPR35 and their relationship to cannabinoid and lysophospholipid receptors. Life Sci. 2013;92:453–457. doi: 10.1016/j.lfs.2012.06.039. [DOI] [PubMed] [Google Scholar]
- 30.Ochiai S, Furuta D, Sugita K, Taniura H, Fujita N. GPR87 mediates lysophosphatidic acid-induced colony dispersal in A431 cells. Eur J Pharmacol. 2013;715:15–20. doi: 10.1016/j.ejphar.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 31.Valentine WJ, Fells JI, Perygin DH, Mujahid S, Yokoyama K, Fujiwara Y, Tsukahara R, Van Brocklyn JR, Parrill AL, Tigyi G. Subtype-specific residues involved in ligand activation of the endothelial differentiation gene family lysophosphatidic acid receptors. J Biol Chem. 2008;283:12175–12187. doi: 10.1074/jbc.M708847200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukushima N, Kimura Y, Chun J. A single receptor encoded by vzg-1/lp A1 /edg-2 couples to G proteins and mediates multiple cellular responses to lysophosphatidic acid. Proc Natl Acad Sci USA. 1998;95:6151–6156. doi: 10.1073/pnas.95.11.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bandoh K, Aoki J, Taira A, Tsujimoto M, Arai H, Inoue K. Lysophosphatidic acid (LPA) receptors of the EDG family are differentially activated by LPA species. Structure-activity relationship of cloned LPA receptors. FEBS Lett. 2000;478:159–165. doi: 10.1016/S0014-5793(00)01827-5. [DOI] [PubMed] [Google Scholar]
- 34.Kimura Y, Schmitt A, Fukushima N, Ishii I, Kimura H, Nebreda AR, Chun J. Two novel Xenopus homologs of mammalian LPA1/EDG-2 function as lysophosphatidic acid receptors in Xenopus oocytes and mammalian cells. J Biol Chem. 2001;276:15208–15215. doi: 10.1074/jbc.M011588200. [DOI] [PubMed] [Google Scholar]
- 35.Shano S, Hatanaka K, Ninose S, Moriyama R, Tsujiuchi T, Fukushima N. A lysophosphatidic acid receptor lacking the PDZ-binding domain is constitutively active and stimulates cell proliferation. Biochim Biophys Acta. 2008;1783:748–759. doi: 10.1016/j.bbamcr.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 36.Varsano T, Taupin V, Guo L, Baterina OY, Jr, Farquhar MG. The PDZ protein GIPC regulates trafficking of the LPA1 receptor from APPL signaling endosomes and attenuates the cell’s response to LPA. PLoS One. 2012;7:e49227. doi: 10.1371/journal.pone.0049227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamada T, Ohoka Y, Kogo M, Inagaki S. Physical and functional interactions of the lysophosphatidic acid receptors with PDZ domain-containing Rho guanine nucleotide exchange factors (RhoGEFs) J Biol Chem. 2005;280:19358–19363. doi: 10.1074/jbc.M414561200. [DOI] [PubMed] [Google Scholar]
- 38.Zhao J, Wei J, Bowser RK, Dong S, Xiao S, Zhao Y. Molecular regulation of lysophosphatidic acid receptor 1 trafficking to the cell surface. Cell Signal. 2014;26:2406–2411. doi: 10.1016/j.cellsig.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukushima N. Neural effects of lysophosphatidic acid (LPA) signaling. In: Chun J, Hla T, Moolenaat W, Spigel S, editors. Lysophospholipid: receptors signaling and biochemistry. London: Wiley; 2013. pp. 399–418. [Google Scholar]
- 40.Contos JJA, Chun J. Genomic characterization of the lysophosphatidic acid receptor gene, lp A2/Edg4, and identification of a frameshift mutation in a previously characterized cDNA. Genomics. 2000;64:155–169. doi: 10.1006/geno.2000.6122. [DOI] [PubMed] [Google Scholar]
- 41.Contos JJA, Fukushima N, Weiner JA, Kaushal D, Chun J. Requirement for the lp A1 lysophosphatidic acid receptor gene in normal suckling behavior. Proc Natl Acad Sci USA. 2000;97:13384–13389. doi: 10.1073/pnas.97.24.13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Contos JJA, Ishii I, Fukushima N, Kingsbury MA, Ye X, Kawamura S, Brown JH, Chun J. Characterization of lpa 2 (Edg4) and lpa 1/lpa 2 (Edg2/Edg4) lysophosphatidic acid receptor knockout mice: signaling deficits without obvious phenotypic abnormality attributable to LPA2 . Mol Cell Biol. 2002;22:6921–6929. doi: 10.1128/MCB.22.19.6921-6929.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SJ, Chan TH, Chen TC, Liao BK, Hwang PP, Lee H. LPA1 is essential for lymphatic vessel development in zebrafish. Faseb J. 2008;22:3706–3715. doi: 10.1096/fj.08-106088. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka M, Okudaira S, Kishi Y, Ohkawa R, Iseki S, Ota M, Noji S, Yatomi Y, Aoki J, Arai H. Autotaxin stabilizes blood vessels and is required for embryonic vasculature by producing lysophosphatidic acid. J Biol Chem. 2006;281:25822–25830. doi: 10.1074/jbc.M605142200. [DOI] [PubMed] [Google Scholar]
- 45.Oh YS, Jo NW, Choi JW, Kim HS, Seo SW, Kang KO, Hwang JI, Heo K, Kim SH, Kim YH, Kim IH, Kim JH, Banno Y, Ryu SH, Suh PG. NHERF2 specifically interacts with LPA2 receptor and defines the specificity and efficiency of receptor-mediated phospholipase C-β3 activation. Mol Cell Biol. 2004;24:5069–5079. doi: 10.1128/MCB.24.11.5069-5079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai SL, Yao WL, Tsao KC, Houben AJ, Albers HM, Ovaa H, Moolenaar WH, Lee SJ. Autotaxin/Lpar3 signaling regulates Kupffer’s vesicle formation and left-right asymmetry in zebrafish. Development. 2012;139:4439–4448. doi: 10.1242/dev.081745. [DOI] [PubMed] [Google Scholar]
- 47.Ye X, Skinner MK, Kennedy G, Chun J. Age-dependent loss of sperm production in mice via impaired lysophosphatidic acid signaling. Biol Reprod. 2008;79:328–336. doi: 10.1095/biolreprod.108.068783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Contos JJA, Chun J. The mouse lp A3/Edg7 lysophosphatidic acid receptor gene: genomic structure, chromosomal localization, and expression pattern. Gene. 2001;267:243–253. doi: 10.1016/S0378-1119(01)00410-3. [DOI] [PubMed] [Google Scholar]
- 49.Bandoh K, Aoki J, Hosono H, Kobayashi S, Kobayashi T, Murakami-Murofushi K, Tsujimoto M, Arai H, Inoue K. Molecular cloning and characterization of a novel human G-protein-coupled receptor, EDG7, for lysophosphatidic acid. J Biol Chem. 1999;274:27776–27785. doi: 10.1074/jbc.274.39.27776. [DOI] [PubMed] [Google Scholar]
- 50.Li G, Mosier PD, Fang X, Zhang Y. Toward the three-dimensional structure and lysophosphatidic acid binding characteristics of the LPA(4)/p2y(9)/GPR23 receptor: a homology modeling study. J Mol Graph Model. 2009;28:70–79. doi: 10.1016/j.jmgm.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee CW, Rivera R, Dubin AE, Chun J. LPA4/GPR23 is a lysophosphatidic acid (LPA) receptor utilizing Gs-, Gq/Gi-mediated calcium signaling and G12/13-mediated Rho activation. J Biol Chem. 2007;282:4310–4317. doi: 10.1074/jbc.M610826200. [DOI] [PubMed] [Google Scholar]
- 52.Williams JR, Khandoga AL, Goyal P, Fells JI, Perygin DH, Siess W, Parrill AL, Tigyi G, Fujiwara Y. Unique ligand selectivity of the GPR92/LPA5 lysophosphatidate receptor indicates role in human platelet activation. J Biol Chem. 2009;284:17304–17319. doi: 10.1074/jbc.M109.003194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raza SI, Muhammad D, Jan A, Ali RH, Hassan M, Ahmad W, Rashid S. In silico analysis of missense mutations in LPAR6 reveals abnormal phospholipid signaling pathway leading to hypotrichosis. PLoS One. 2014;9:e104756. doi: 10.1371/journal.pone.0104756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inoue A, Arima N, Ishiguro J, Prestwich GD, Arai H, Aoki J. LPA-producing enzyme PA-PLA1α regulates hair follicle development by modulating EGFR signalling. EMBO J. 2011;30:4248–4260. doi: 10.1038/emboj.2011.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Regard JB, Sato IT, Coughlin SR. Anatomical profiling of G protein-coupled receptor expression. Cell. 2008;135:561–571. doi: 10.1016/j.cell.2008.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geach TJ, Faas L, Devader C, Gonzalez-Cordero A, Tabler JM, Brunsdon H, Isaacs HV, Dale L. An essential role for LPA signalling in telencephalon development. Development. 2014;141:940–949. doi: 10.1242/dev.104901. [DOI] [PubMed] [Google Scholar]
- 57.Zhang G, Contos JJ, Weiner JA, Fukushima N, Chun J. Comparative analysis of three murine G-protein coupled receptors activated by sphingosine-1-phosphate. Gene. 1999;227:89–99. doi: 10.1016/S0378-1119(98)00589-7. [DOI] [PubMed] [Google Scholar]
- 58.An S, Bleu T, Hallmark OG, Goetzl EJ. Characterization of a novel subtype of human G protein-coupled receptor for lysophosphatidic acid. J Biol Chem. 1998;273:7906–7910. doi: 10.1074/jbc.273.14.7906. [DOI] [PubMed] [Google Scholar]
- 59.Kuma K, Iwabe N, Miyata T. Functional constraints against variations on molecules from the tissue-level—slowly evolving brain-specific genes demonstrated by protein-kinase and immunoglobulin supergene families. Mol Biol Evol. 1995;12:123–130. doi: 10.1093/oxfordjournals.molbev.a040181. [DOI] [PubMed] [Google Scholar]
- 60.Sutcliffe JG. Messenger-RNA in the mammalian central nervous-system. Annu Rev Neurosci. 1988;11:157–198. doi: 10.1146/annurev.ne.11.030188.001105. [DOI] [PubMed] [Google Scholar]
- 61.Kimura M. The neutral theory of molecular evolution. Cambridge: Cambridge University Press; 1985. [Google Scholar]
- 62.Hausmann J, Perrakis A, Moolenaar WH. Structure-function relationships of autotaxin, a secreted lysophospholipase D. Adv Biol Regul. 2013;53:112–117. doi: 10.1016/j.jbior.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 63.Perrakis A, Moolenaar WH. Autotaxin: structure-function and signaling. J Lipid Res. 2014;55:1010–1018. doi: 10.1194/jlr.R046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aikawa S, Hashimoto T, Kano K, Aoki J. Lysophosphatidic acid as a lipid mediator with multiple biological actions. J Biochem. 2014;157(2):81–89. doi: 10.1093/jb/mvu077. [DOI] [PubMed] [Google Scholar]
- 65.Nakanaga K, Hama K, Aoki J. Autotaxin—an LPA producing enzyme with diverse functions. J Biochem. 2010;148:13–24. doi: 10.1093/jb/mvq052. [DOI] [PubMed] [Google Scholar]
- 66.Holmes RS, Cox LA. Comparative structures and evolution of vertebrate lipase H (LIPH) genes and proteins: a relative of the phospholipase A1 gene families, 3 . Biotech. 2012;2:263–275. [Google Scholar]
- 67.Hesse M, Willscher E, Schmiedel BJ, Posch S, Golbik RP, Staege MS. Sequence and expression of the chicken membrane-associated phospholipases A1 alpha (LIPH) and beta (LIPI) Mol Biol Rep. 2012;39:761–769. doi: 10.1007/s11033-011-0796-0. [DOI] [PubMed] [Google Scholar]
- 68.Felsenstein J. Inferring phylogenies. Massachusetts: Sinauer Associates Inc.; 2004. [Google Scholar]
- 69.Yanagida K, Ishii S. Non-Edg family LPA receptors: the cutting edge of LPA research. J Biochem. 2011;150:223–232. doi: 10.1093/jb/mvr087. [DOI] [PubMed] [Google Scholar]
- 70.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 72.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 73.Yang ZH. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 74.Furuta D, Yamane M, Tsujiuchi T, Moriyama R, Fukushima N. Lysophosphatidic acid induces neurite branch formation through LPA3. Mol Cell Neurosci. 2012;50:21–34. doi: 10.1016/j.mcn.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 75.Lee CW, Rivera R, Gardell S, Dubin AE, Chun J. GPR92 as a new G12/13 and Gq coupled lysophosphatidic acid receptor that increases cAMP: LPA5 . J Biol Chem. 2006;281:23589–23597. doi: 10.1074/jbc.M603670200. [DOI] [PubMed] [Google Scholar]