Abstract
ALCAM/CD166 is a member of the immunoglobulin superfamily of cell adhesion molecules (Ig-CAMs) which mediates intercellular adhesion through either homophilic (ALCAM–ALCAM) or heterophilic (ALCAM–CD6) interactions. ALCAM-mediated adhesion is crucial in different physiological and pathological phenomena, with particular relevance in leukocyte extravasation, stabilization of the immunological synapse, T cell activation and proliferation and tumor growth and metastasis. Although the functional implications of ALCAM in these processes is well established, the mechanisms regulating its adhesive capacity remain obscure. Using confocal microscopy colocalization, and biochemical and functional analyses, we found that ALCAM directly associates with the tetraspanin CD9 on the leukocyte surface in protein complexes that also include the metalloproteinase ADAM17/TACE. The functional relevance of these interactions is evidenced by the CD9-induced upregulation of both homophilic and heterophilic ALCAM interactions, as reflected by increased ALCAM-mediated cell adhesion and T cell migration, activation and proliferation. The enhancement of ALCAM function induced by CD9 is mediated by a dual mechanism involving (1) augmented clustering of ALCAM molecules, and (2) upregulation of ALCAM surface expression due to inhibition of ADAM17 sheddase activity.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-012-1132-0) contains supplementary material, which is available to authorized users.
Keywords: ALCAM, CD166, Tetraspanins, CD9, ADAM17, TACE
Introduction
ALCAM, also known as CD166 antigen or melanoma metastasis clone D (MEMD), is a 110-kDa transmembrane glycoprotein that belongs to the Ig-CAM superfamily. ALCAM contains five extracellular Ig-like domains, a transmembrane region and a short cytoplasmic tail, and mediates cell–cell adhesion through either homophilic or heterophilic interactions with CD6. Interestingly, ALCAM homophilic interactions have 100-fold lower affinity and faster dissociation rate than heterophilic ALCAM–CD6 interactions [1]. Two distinct structural and functional modules have been defined in the extracellular region of the ALCAM molecule: the first two N-terminal Ig-like extracellular domains control ligand binding affinity, and the three membrane proximal domains constitute an oligomerization module involved in avidity regulation [2, 3].
Although ALCAM is distributed widely in tissues, its expression seems to be restricted to specific subsets of cells involved in dynamic growth and migration processes (see references [2, 4–6] for reviews). Functional studies have implicated ALCAM-mediated adhesion in a variety of physiological and pathological phenomena. The heterophilic interaction between ALCAM on APCs and CD6 on T cells has been shown to be essential for immunological synapse stabilization and optimal T cell activation and proliferation [1, 7, 8]. In addition, ALCAM mediates leukocyte transendothelial migration and trafficking into central nervous system [9, 10], axon fasciculation [11], and invasive growth and metastasis of different tumors, including melanoma and breast, prostate and colorectal carcinomas [2, 4, 5].
ALCAM-mediated homotypic cell adhesion is dynamically regulated by the actin cytoskeleton. Inhibition of actin polymerization by cytochalasin D induces an important increase in the lateral mobility and clustering of ALCAM molecules on the cell membrane and dramatically enhances homotypic ALCAM-mediated cell adhesion, without significantly altering the affinity of these interactions [12, 13].
The 22-kDa sALCAM isoform, containing only the single aminoterminal Ig-like domain, is encoded by an alternative transcript in a variety of cell types after premature poly-adenylation in intron 3 [14, 15]. Another form of sALCAM of 96 kDa, encompassing most of its extracellular region, is generated by ADAM-17-mediated proteolytic shedding [16, 17]. Levels of sALCAM are elevated in patients with different types of tumors, including ovarian carcinomas, which is in agreement with the involvement of this ALCAM soluble ectodomain in the motility of human ovarian carcinoma cells [17]. Furthermore, ALCAM has been recently proposed as a novel biomarker for breast cancer, based on the elevated levels of sALCAM that are detected in the serum of these patients [18].
ADAM17 (also termed TACE, TNF-α converting enzyme) was initially identified as the enzyme cleaving TNF-α from its transmembrane precursor form, and later found to be the “sheddase” that also cleaves the ectodomains of many other cell surface proteins that are critically involved in development, cell growth, adhesion, differentiation and migration of leukocytes and tumor cells (for review see references [19, 20]). ADAM17 seems to be particularly important in the regulation of cell adhesion, since this metalloproteinase can cleave the ectodomains of many critically important adhesion molecules, which belong either to the immunoglobulin superfamily—such as ICAM-1, VCAM-1, NCAM, L1CAM and ALCAM—or to other families of adhesion receptors—such as CD44 and l-selectin. The physiological and pathological relevance of ADAM17 substrates suggests that its proteolytic activity must be finely regulated. However, the mechanisms controlling ADAM17 activity remain largely unknown, although several possibilities have been proposed, including regulation through phosphorylation of its cytoplasmic tail, interaction with regulatory proteins involved in cell signaling or control of substrate accessibility [21, 22]. We have recently reported that the tetraspanin CD9 interacts directly with ADAM17 on the surface of different types of leukocytes and through these interactions CD9 exerts an inhibitory effect on the proteolytic activity of ADAM17 [23].
CD9 was initially identified as a lymphohematopoietic marker and later implicated in diverse functions, including cell signaling, growth, adhesion, motility, tumorigenesis, metastasis and sperm–egg fusion. Like other tetraspanins, CD9 participates in the organization of a type of cell surface microdomain—the tetraspanin web—through lateral association with additional tetraspanins and other transmembrane proteins [24]. It is well established that most lateral interactions of tetraspanins with other membrane proteins are mediated by the variable region within their LEL domain, whose conformation is maintained by distinct disulfide bonds [25]. CD9 associates directly through its LEL domain with ICAM-1 in specialized endothelial adhesive platforms, influencing leukocyte adhesion and extravasation [26, 27].
We report here that ALCAM forms a complex with the tetraspanin CD9 and with the metalloproteinase ADAM17 on the leukocyte surface. Through these associations, CD9 enhances both the homophilic (ALCAM–ALCAM) and the heterophilic (ALCAM–CD6) interactions, resulting in increased ALCAM-mediated leukocyte adhesion, migration and T cell activation. This increased ALCAM adhesion induced by CD9 is regulated through a dual mechanism involving augmented clustering of ALCAM molecules as well as elevated levels of ALCAM surface expression due to inhibition of ADAM17 sheddase activity.
Materials and methods
Cells and antibodies
CHO cells stably expressing ALCAM-Fc [12] were grown in DMEM/10 % fetal calf serum. The Jurkat leukemic T cell line, Raji B cells, BLM melanoma cells and erythroblastic K562 cell line were grown in RPMI-1640/10 % fetal calf serum. Human PBMCs were isolated from buffy coats from normal donors over a Lymphoprep (Nycomed Pharma) gradient according to standard procedures. Monocytes were depleted from PBMCs by magnetic cell sorting using CD14 microbeads (Miltenyi Biotec). Culture media were supplemented with 2 mM glutamine, 50 μg/ml streptomycin and 50 U/ml penicillin.
Anti-β1 integrin (TS2/16), anti-β2 integrin (Lia3/2), anti-CD9 (VJ1/20, PAINS-10 and PAINS-13), anti-CD147 (VJ1/9), anti-CD25 (TP1/6), anti-CD69 (TP1/8) and anti-CD151 (Lia1/1) mAbs have been previously described [28–33]. The mAb PAINS-15 has been produced and characterized in our laboratory as an ALCAM-specific antibody based on (1) identification of the 110-kDa band immunoprecipitated by this antibody as ALCAM by protein fingerprinting/MALDI-TOF MS, (2) its specific reactivity with cell surface ALCAM following transfection of human full-length cDNA in ALCAM-negative K562 cells, and (3) its reduced reactivity following interference with specific commercial ALCAM shRNAs (OriGene Technologies). The anti-CD81 mAb 5A6 was kindly provided by Dr. S. Levy (Stanford University School of Medicine, CA). The anti-ADAM17 polyclonal antibody H-300 was from Santa Cruz Biotechnologies and the anti-GST antibody from Amersham Biosciences. VJ1/20 and PAINS-15 were biotinylated as previously described [28].
RNA silencing and transfections
Jurkat cells were infected with retrovirus containing the following shRNA-coding plasmids (OriGene Technologies) according to manufacturer′s instructions: TR20003 (the control plasmid without shRNA cassette insert), TI356235 (the plasmid with the shRNA insert to silence CD9); TI359388 (the plasmid containing shRNA inserts to knock-down ALCAM), and TI364190 (the plasmid with the shRNA to silence ADAM17). After retroviral transduction, cells were selected in medium containing 1 μg/ml puromycin. Raji B cells were electroporated with CD9 cDNA or with the empty vector pcDNA3 to generate stable transfectants after selection with G418 (0.8 mg/ml), as described previously [23]. Subsequently, cells were electroporated with the shRNA-coding plasmids for ALCAM, ADAM17 or with the control TR20003 plasmid, and selected in medium containing 1 μg/ml puromycin to obtain double-transfected cells. K562 cells were electroporated with empty vector pRc/CMV (K562–vector) or with full-length ALCAM cDNA (K562–ALCAM), and selected with G418 (0.6 mg/ml). In some experiments, CD9 was stably knocked-down in ALCAM-expressing K562 transfectants by electroporation with the TI356235 shRNA and selection with 1 μg/ml puromycin. All experiments employing virally transduced or transfected cells were performed with polyclonal cell populations, and not with individual selected clones.
Flow cytometry
Protein surface expression was analyzed as previously described [28]. For sALCAM-Fc binding assays, cells were incubated with 20 μg/ml ALCAM-Fc for 30 min at 37 °C, washed with warm RPMI-1640 medium and further incubated with fluorescein isothiocyanate-conjugated secondary anti-human Fc antibody (Sigma) for 15 min at 37 °C. In experiments for ALCAM expression on Jurkat and Raji cells transfected with the shRNA plasmid specific for ADAM17, an additional antibody layer between the primary and detection antibodies was included to improve the detection signal. Cells were then washed with warm medium and their fluorescence determined on a FACScan cytometer (Becton-Dickinson).
Immunofluorescence and confocal microscopy
Immunofluorescence experiments were performed as previously described [23]. Quantification of ALCAM clustering was performed as follows. Cells were plated on 12-mm diameter glass coverslips coated with poly-l-lysine (50 μg/ml) and treated or not with 2.5 μg/ml of cytochalasin D for 1 h at room temperature. Cells were then fixed with 4 % formaldehyde in PBS (10 min at room temperature) or with 70 % ethanol (1 h at −20 °C) as an alternative fixation protocol where indicated. After fixation, cells were incubated with the anti-ALCAM mAb PAINS-15 (5 μg/ml) for 1 h at room temperature. Cells were washed with PBS and incubated for 30 min with Alexa Fluor 594-conjugated anti-mouse antibody. Samples were mounted with Mowiol (Calbiochem) and images were obtained with a Leica LSM510 inverted microscope. Images were improved by deconvolution with Huygens 3.2 software. A threshold was fixed for cluster quantification by intensity and size with ImageJ software (NIH). Total cluster area is related to the total area within cell contours.
Co-immunoprecipitation
Jurkat or Raji-CD9 cells were lysed for 15 min at 4 °C in TBS/1 % Brij-97 containing 1 mM CaCl2, 1 mM MgCl2 and protease inhibitors. Cell lysates were centrifuged for 15 min at 13,000 rpm, and aliquots incubated with specific antibodies for 3 h at 4 °C, and finally incubated with protein A-Sepharose overnight at 4 °C. Beads were then washed with 1:10 diluted lysis buffer and immune complexes boiled in nonreducing Laemmli buffer, resolved by 10 % SDS-PAGE, and transferred to nitrocellulose membranes. Blots were blocked with TBS/5 % bovine serum albumin and developed with the corresponding primary antibody followed by washes and incubation with secondary horseradish peroxidase-coupled antibodies (Sigma). Chemiluminescence was detected using an ECL detection kit (Amersham Biosciences). In another set of experiments, cells were incubated for 1 h at 4 °C with the specific antibodies prior to lysis, followed by washing of excess antibody to ensure that only cell surface proteins were engaged for immunoprecipitation. The total protein concentrations in whole cellular lysates were calculated using the DC protein assay (Bio-Rad Laboratories).
Protein–protein interaction assays
The human wt-CD9-LEL-GST, wt-CD151-LEL-GST, wt-CD81-LEL-GST and GST-PAK-CRIB fusion proteins as well as CD9-LEL-GST mutants with Cys152, Cys153, Cys167 or Cys181 replaced by alanine, were produced as described previously [23, 26, 34]. ALCAM-Fc and ICAM-1-Fc fusion proteins were purified from supernatants of stably transfected CHO cells as described previously [12, 33]. Fusion proteins were incubated with ALCAM-Fc overnight at 4 °C in binding buffer (TBS with 1 mM CaCl2, 1 mM MgCl2, 10 % glycerol, 1 % Brij-97 and protease inhibitors). Protein complexes were pulled-down with protein A-Sepharose (2 h at 4 °C), washed in 1:10 diluted binding buffer, fractionated in 12 % SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted using specific antibodies.
ALCAM-Fc adhesion assays
Jurkat and Raji cells transfected with the indicated plasmids or MD-PBMC activated with phytohemagglutinin for 2 days and cultured in medium supplemented with interleukin-2, were loaded with the fluorescent probe BCECF-AM (Sigma). Adhesion experiments were carried out with 1 × 105 cells per well in RPMI-1640. Cells were left to adhere for 40 min at 37 °C in 96-well microplates coated with 10 μg/ml of ALCAM-Fc. For washing, the plate was submerged in warm PBS and turned over to let the non-adherent cells detach by gravity for 20 min at 37 °C. Adherent cells were measured by fluorescence of BCECF-AM with a fluorescence bottom reader (TecanGENios). The percentage adhesion was calculated considering the number of cells prior to washing as 100 % adhesion.
Migration assays
For migration quantification we used 5-μm pore size modified Boyden chambers coated with 70 μg/ml of ALCAM-Fc. Jurkat cells (6 × 105) retrovirally transduced with the indicated plasmids were allowed to migrate towards 6.25 mM CXCL12 for 1 h and quantified as previously described [35]. Assays were run in duplicate.
Soluble ALCAM quantification
The amount of sALCAM released from Jurkat and Raji cells was determined with an ALCAM DuoSet ELISA development kit (R&D Systems). Jurkat or Raji cells (2 × 106), expressing or lacking CD9 and/or ADAM17, were seeded in 24-well plates overnight in 600 μl of medium, and the culture supernatants were collected. ALCAM concentrations in the supernatants were calculated from a standard curve constructed with known amounts of this adhesion molecule. Assays were done in triplicate, and background values were subtracted.
Proliferation assays
For MD-PBMC proliferation assays, 4 × 103 Raji cells, either expressing or lacking CD9 or/and ALCAM, were pulsed with 2.5 μg/ml SEE superantigen for 30 min at 37 °C and then co-cultured with 1 × 106 primary MD-PBMC for 72 h in 96-well plates. Proliferation was quantified by adding 5 μg/ml MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) to the wells and incubating for additional 4 h at 37 °C. MTT was solubilized with 100 μl of 50 % dimethylformamide and 20 % SDS overnight at 37 °C in the dark. Assays were done in triplicate and basal proliferation of Raji and MD-PBMC was subtracted.
T cell activation assays
For T cell activation following antigen presentation, conjugates of Jurkat T cells and Raji B cells (either expressing or lacking CD9) were allowed to form, as described previously [8]. In brief, 2 × 105 Raji cells were pulsed with 2.5 μg/ml of SEE superantigen for 30 min at 37 °C and subsequently cocultured with 4 × 105 Jurkat cells for 72 h. Expression of the activation markers CD69 and CD25 on Jurkat T cells was assessed by flow cytometry.
Statistical analysis
The distribution of data was assessed for normality by the Bonferroni test. One-factor ANOVA was performed to determine the significance of differences using SPSS statistics software (IBM).
Results
ALCAM/CD166 and CD9 associate on the cell surface of leukocytic and tumor cells
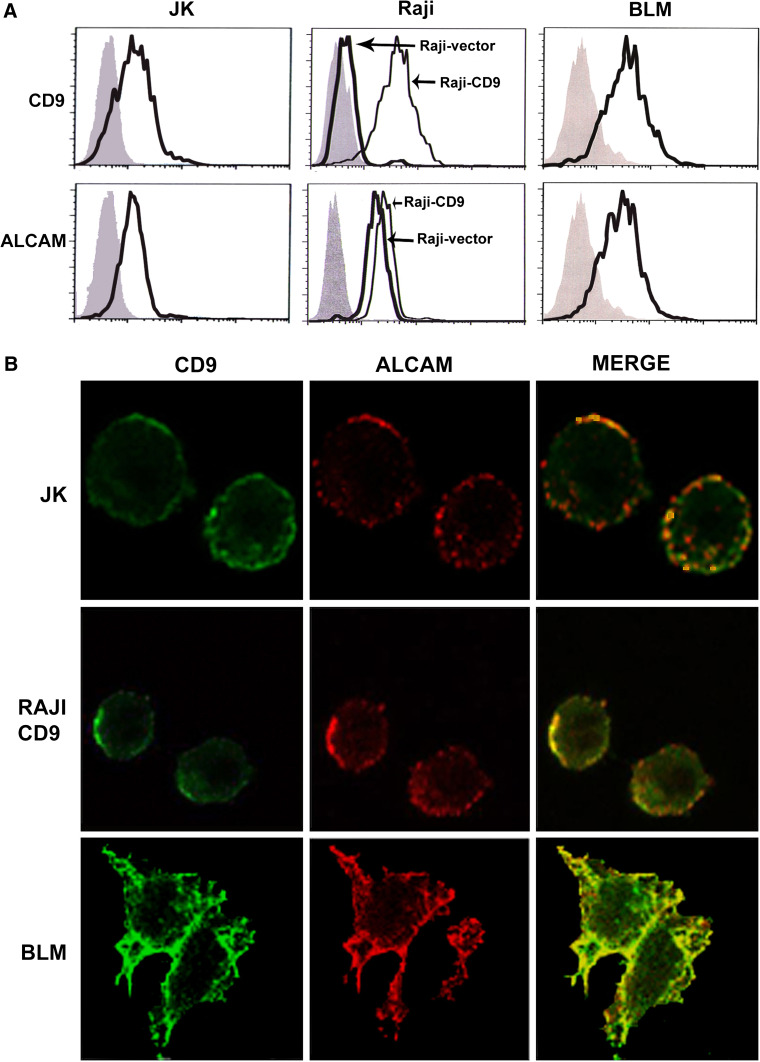
Since the tetraspanin CD9 associates on the cell surface with several members of the immunoglobulin superfamily [24], we hypothesized that CD9 could also functionally interact with ALCAM. The initial evidence indicating an interaction between ALCAM and CD9 on the cell surface was provided by double immunofluorescence staining of these molecules performed on fixed cells, followed by confocal microscopy analysis. These experiments were performed with the lymphoblastic Jurkat T cell line and with the invasive metastatic melanoma BLM cell line, both of which express endogenous CD9, and with the B lymphoblastoid Raji-CD9 stable transfectant cell line. We first checked that these three cell lines display a significant expression of both ALCAM and CD9 on their surface (Fig. 1a). As shown in Fig. 1b, partial colocalization between CD9 and ALCAM could be observed on the plasma membrane of Jurkat, Raji-CD9 and BLM cells. Interestingly, the areas of colocalization between CD9 and ALCAM on the lymphoid cells were distributed in a patched pattern, which agrees with the previously described distribution of ALCAM on other leukocytic cells, especially after being released from cytoskeletal restraints [12]. Colocalization of CD9 and ALCAM on the adherent BLM melanoma cells seems to be particularly enriched in the regions of cell–cell contact.
Fig. 1.
Confocal microscopy analysis of colocalization of ALCAM and CD9. a Flow cytometric detection of cell surface CD9 and ALCAM on Jurkat T cells, Raji B cells and BLM melanoma cells. Gray filled histograms correspond to negative controls; thick black lines correspond to the expression of the indicated molecules. For Raji cells, the thin black lines show the expression of CD9 and ALCAM on stably CD9-transfected cells as indicated (Raji-CD9). b Immunofluorescence analysis of ALCAM and CD9 co-localization. Jurkat T cells, CD9-transfected Raji B cells and BLM melanoma cells were fixed and stained with the anti-ALCAM mAb PAINS-15 followed by the biotinylated anti-CD9 mAb VJ1/20. Representative confocal slides for each channel and merged images are shown
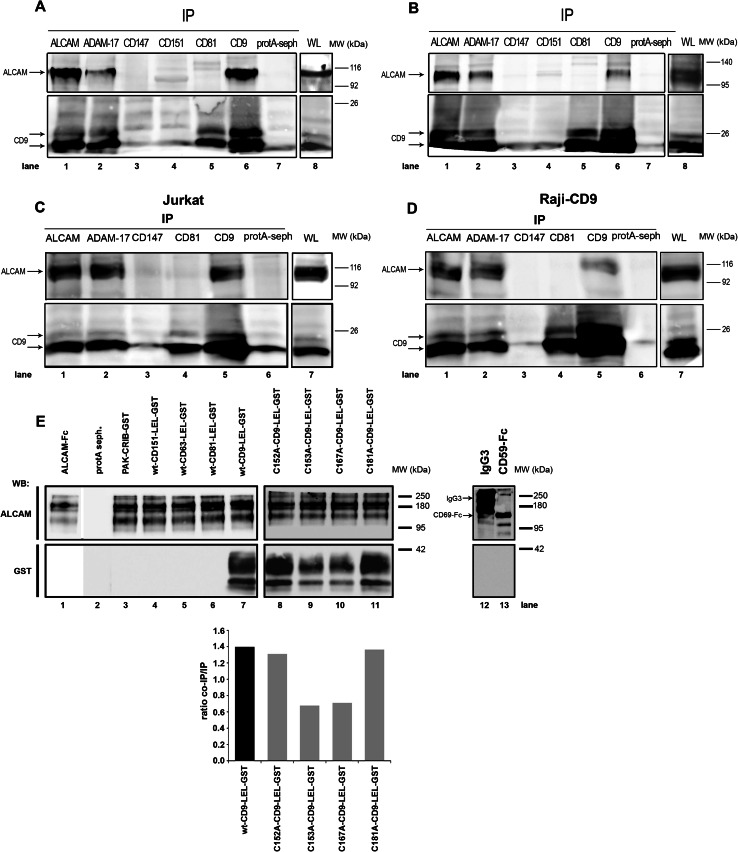
We further explored this possible association between CD9 and ALCAM by coimmunoprecipitation analysis using lysates of Jurkat (Fig. 2a) and Raji-CD9 (Fig. 2b) cells obtained using the relatively mild detergent Brij-97, with the aim of preserving primary as well as higher-order tetraspanin interactions with other proteins [24]. A double 21/24-kDa band corresponding to CD9 was clearly coimmunoprecipitated with ALCAM (Fig. 2a, b; lane 1) and, likewise, a 110-kDa band corresponding to ALCAM was observed when CD9 was immunoprecipitated with a specific antibody (Fig. 2a, b; lane 6). As expected, ALCAM, which has been reported to be processed by ADAM17 [16, 17], was efficiently coimmunoprecipitated with this metalloproteinase (Fig. 2a, b; lane 2). CD9 was also coimmunoprecipitated with ADAM17 (Fig. 2a, b; lane 2), as we have recently reported [23]. In contrast, no detectable ALCAM was coimmunoprecipitated with CD147 (emmprin/basigin; lane 3) or with other tetraspanins such as CD151 (lane 4) or CD81 (lane 5). As expected, CD9 also coimmunoprecipitated with CD81, since these two tetraspanins are known to associate with each other to form heterodimers [36]. Immunoprecipitations were also performed using intact cells incubated with the immunoprecipitating mAbs prior to their lysis. The results of these experiments (Fig. 2c, d) were similar to those starting with whole cell lysates, demonstrating that interactions between CD9 and ALCAM take place on the cell surface.
Fig. 2.
Biochemical analysis of the association between ALCAM and CD9. Jurkat cells (a, c) or CD9-transfected Raji cells (b, d) were lysed using 1 % Brij 97 either before (a, b) or after (c, d) addition of the immunoprecipitating antibodies. Proteins were immunoprecipitated with the following antibodies: anti-ALCAM (PAINS-15), anti-ADAM-17 (2A10), anti-CD9 (VJ1/20), anti-CD81 (5A6), anti-CD151 (Lia1/1) and anti-CD147 (VJ1/9). Precipitated protein complexes were resolved by 10 % SDS-PAGE under nonreducing conditions, and immunoblotted with biotinylated anti-ALCAM (PAINS-15) (upper panels) or anti-CD9 (VJ1/20) (lower panels) mAbs. Blots are representative of six different experiments. e Pull-down assay using the wt-CD9-LEL-GST, its mutant forms (C152A, C153A, C167A and C181A), wt-CD151-LEL-GST, wt-CD63-LEL-GST, wt-CD81-LEL-GST or PAK-CRIB-GST fusion constructs. All these GST fusion proteins were incubated with recombinant purified ALCAM-Fc or with other Ig domain-containing proteins such as CD59-Fc or whole human IgG3, as described in Materials and methods. Upper blots show ALCAM immunodetected with the PAINS-15 mAb or Ig proteins immunodetected with anti-human Fc antibody. Lower blots reveal the fusion proteins immunodetected with a commercial anti-GST antibody. Bars in the bottom panel correspond to a densitometric quantitation with Quantity-One software of pulled-down GST-LELs/ALCAM-Fc ratios calculated from the above bands in lanes 7–11. The experiment shown is a representative one out of four
We further investigated whether the observed CD9–ALCAM interactions were direct or mediated through other proteins. Most CD9 lateral interactions with other surface proteins involve its LEL [25]. Accordingly, fusion proteins encompassing the CD9 LEL fused to GST (wt-CD9-LEL-GST) and the whole extracellular region of ALCAM fused to the Fc of human IgG (ALCAM-Fc) were employed to assess the direct association between CD9 and ALCAM. The wt-CD9-LEL-GST was efficiently pulled-down with ALCAM-Fc (Fig. 2e; lane 7). The interaction between the CD9 LEL and ALCAM-Fc was specific, as neither the recombinant wt LELs of other tetraspanins (wt-CD151-LEL-GST, wt-CD63-LEL-GST, wt-CD81-LEL-GST) (Fig. 2e; lanes 4, 5, 6) nor the unrelated GST-containing protein PAK-CRIB-GST (Fig. 2e; lane 3) were pulled-down by ALCAM-Fc. Interestingly, four CD9 LEL-GST constructs in which cysteines C152, C153, C167 and C181 (that are involved in the formation of two intra-LEL domain disulfide bonds) were mutated to alanines retained—to varying degrees—the ability to pull-down ALCAM-Fc (Fig. 2e; lanes 8, 9, 10, 11), with C153A and C167A mutants clearly showing reduced interaction. To rule out the possibility that the observed interaction between ALCAM-Fc and wt-CD9-LEL-GST recombinant proteins could be mediated through the Fc region of ALCAM-Fc, control pull-down experiments were also carried out with two additional Ig domain-containing proteins, CD59-Fc fusion protein and whole human IgG3, which were unable to interact with wt-CD9-LEL-GST protein (Fig. 2e; lanes 12, 13).
Taken together, all these colocalization and biochemical results reveal a direct interaction between ALCAM/CD166 and CD9 on the cell surface.
Expression of CD9 supports homophilic ALCAM cell adhesion and migration
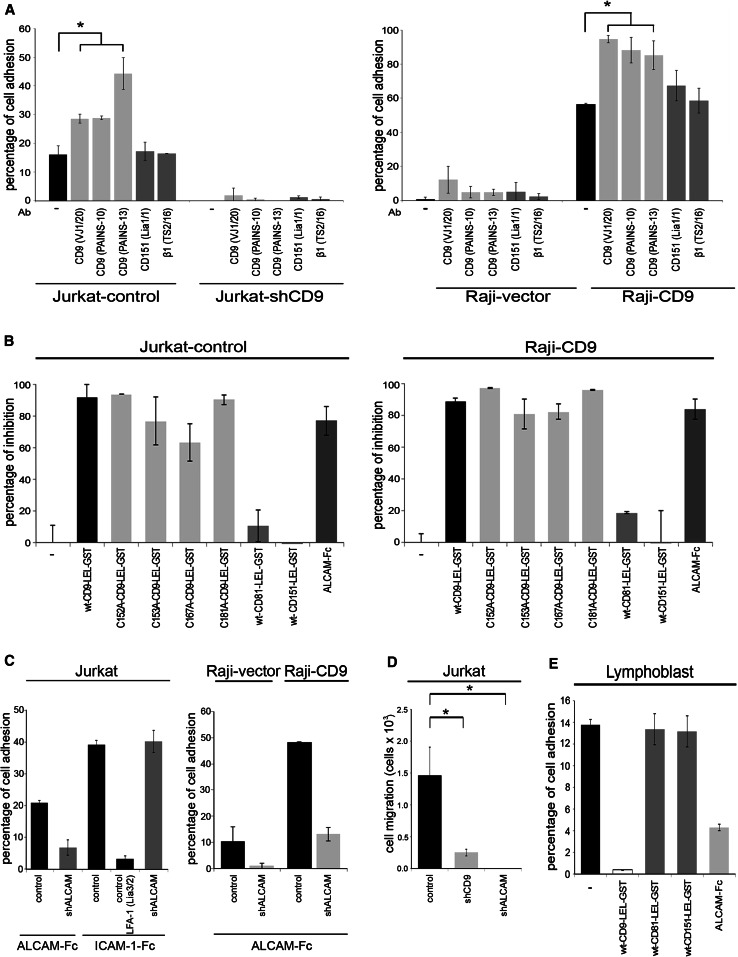
The tetraspanin CD9 regulates integrin-mediated cell adhesion in different cell types [26, 31, 37–39]. Given the observed association between CD9 and ALCAM on the cell surface, we investigated whether the expression of CD9 exerts regulatory effects on ALCAM-mediated homophilic adhesion. For this purpose, we carried out adhesion assays on plastic-immobilized ALCAM-Fc employing two leukocytic cell types: (1) Jurkat cells (which constitutively express endogenous CD9) either stably transduced with a control shRNA (Jurkat control) or with a CD9-specific shRNA (Jurkat shCD9) (Supplementary Fig. 1), and (2) Raji cells (which do not express endogenous CD9) either transfected with empty pcDNA3 vector (Raji-vector) or with CD9 cDNA (Raji-CD9) (Supplementary Fig. 1). CD9 silencing in Jurkat cells abrogated cell adhesion to immobilized ALCAM-Fc (Fig. 3a; left panel), whereas ectopic expression of CD9 in Raji cells promoted their adhesion to ALCAM-Fc (Fig. 3a; right panel). It is noteworthy that CD9 interference in Jurkat cells did not inhibit their LFA-1 integrin-mediated adhesion to immobilized ICAM-1-Fc (data not shown), ruling out the possibility that a general change in the steps after ligand binding, such as cytoskeletal reorganization, is responsible for the observed inhibition in ALCAM-mediated cell adhesion.
Fig. 3.
CD9 regulates cell adhesion and migration on immobilized ALCAM-Fc. a Jurkat cells (left panel) were retrovirally transduced with either control shRNA (Jurkat-control) or shRNA specific for CD9 (Jurkat-shCD9) and Raji cells (right panel) were stably transfected with either empty vector (Raji-vector) or a CD9 cDNA plasmid (Raji-CD9). Prior to adhesion, cells were loaded with the fluorescent probe BCECF-AM (Sigma) and then allowed to adhere to plastic-immobilized ALCAM-Fc (10 μg/ml) for 40 min at 37 °C in the presence or absence of the indicated mAbs (20 μg/ml). Data are presented as the percentages (means ± SD) of adherent cells. The experiment shown is a representative of five. b Adhesion of Jurkat-control cells (left panel) or Raji-CD9 cells (right panel) to immobilized ALCAM-Fc in the presence or absence of sALCAM-Fc, soluble wt-CD9-LEL-GST, its mutant forms (C152A, C153A, C167A and C181A), wt-CD81-LEL-GST or wt-CD151-LEL-GST (all at a final concentration of 20 μg/ml). The percentage inhibition was calculated taking as 0 % the cell adhesion detected in the absence of soluble proteins. Data are presented as means ± SD. The experiment shown is a representative of three. c Adhesion of ALCAM-knocked-down Jurkat cells (Jurkat) to immobilized ALCAM-Fc or ICAM-1-Fc and of Raji cells (Raji-vector and Raji-CD9) to immobilized ALCAM-Fc. The blocking mAb Lia3/2 specific for the β2 subunit (CD18) of integrin LFA-1 was used as a positive control for inhibition of Jurkat adhesion to ICAM-1-Fc. Data are presented as means ± SD. The experiment is representative of three. d Jurkat cells transduced with the shRNA specific for CD9 and ALCAM were allowed to migrate towards 6.2 mM SDF-1α in ALCAM-Fc-coated modified Boyden chambers as specified in Materials and methods. The cells that migrated to the lower chamber were quantified by flow cytometry. Data are presented as means ± SD of transmigrated cells from four independent experiments. e Cell adhesion of PBMC-derived lymphoblasts stimulated for 2 days with phytohemagglutinin and cultured in medium containing interleukin-2. Prior to adhesion, cells were loaded with the fluorescent probe BCECF-AM (Sigma) and then allowed to adhere to plastic-immobilized ALCAM-Fc (10 μg/ml) for 40 min at 37 °C in the presence or absence of 20 μg/ml sALCAM-Fc, soluble wt-CD9-LEL-GST, wt-CD81-LEL-GST or wt-CD151-LEL-GST. Data are presented as the percentages (means ± SD) of adherent cells. The experiment shown is a representative one out of five. *p < 0.01
The adhesion-promoting effect derived from CD9 expression was further enhanced by three different anti-CD9 mAbs (PAINS-10, PAINS-13 and VJ1/20), which have been previously shown to stimulate cellular events mediated by this tetraspanin [23, 31]. In contrast, mAbs directed to a different tetraspanin (CD151) or the stimulatory anti-β1 integrin mAb TS2/16 did not enhance cell adhesion to ALCAM-Fc. These results clearly reveal the specific involvement of CD9 in the induction of ALCAM-mediated cell adhesion. This cell adhesion was greatly inhibited in both cell types by excess sALCAM-Fc, as has been previously described [40], as well as by interfering CD9 function with soluble recombinant wt-CD9-LEL-GST fusion protein. In contrast, an excess of soluble recombinant LELs of two other tetraspanins (wt-CD81-LEL-GST and wt-CD151-LEL-GST) only exerted negligible inhibitory effects on ALCAM-mediated cell adhesion, clearly indicating the specific regulatory role of CD9 in this process. Interestingly, an excess of soluble C152 and C181 CD9 LEL mutants retained the maximal ability to inhibit adhesion to immobilized ALCAM-Fc, similar to that of the wt-CD9-LEL construct, whereas only partial inhibition was attained with the C153 and C167 CD9 LEL mutants (Fig. 3b).
To demonstrate that the observed effects on adhesion are not mediated by the heterophilic interaction of cellular CD6 with immobilized ALCAM-Fc, ALCAM was silenced by shRNA interference in Jurkat and Raji-CD9 cells. This resulted in approximately 50 % reduction in ALCAM cell surface expression in both cell types, without any change in CD6 expression (Supplementary Fig. 1). The adhesion-enhancing effect derived from CD9 expression was greatly reduced in these ALCAM-silenced Jurkat and Raji cells (Fig. 3c), demonstrating that the homophilic interaction between cellular ALCAM and immobilized ALCAM-Fc governs this type of adhesion. In contrast, LFA-1-mediated adhesion of Jurkat cells to ICAM-1-Fc was not affected by ALCAM silencing, confirming the specificity of these adhesion assays.
Homophilic ALCAM interactions mediate monocyte and lymphocyte transendothelial migration towards chemotactic factors [9, 10], and we have confirmed that these homophilic interactions also mediate the migration of Jurkat cells across ALCAM-Fc-coated polycarbonate inserts towards the chemokine SDF-1α (CXCL12), based on its complete abrogation after silencing ALCAM (Jurkat-shALCAM) (Fig. 3d). The functional role of CD9 in regulating this ALCAM-mediated process was evidenced by the significant reduction in the transmigration of CD9-silenced Jurkat cells (Jurkat-shCD9) compared to control cells (Jurkat-control). These functional data are consistent with the abrogation of ALCAM-dependent cell adhesion upon CD9 silencing in Jurkat cells (Fig. 3a).
Similar to the results with Jurkat and Raji cell lines, the adhesion of human MD-PBMC-derived lymphoblasts to immobilized ALCAM-Fc was inhibited by excess soluble wt-CD9-LEL-GST and ALCAM-Fc recombinant proteins, but not by other soluble tetraspanin constructs (wt-CD81-LEL-GST and wt-CD151-LEL-GST), again demonstrating the specific involvement of CD9 in regulating ALCAM-mediated adhesion of primary leukocytic cells (Fig. 3e).
CD9 controls clustering, but not the affinity of cell surface ALCAM
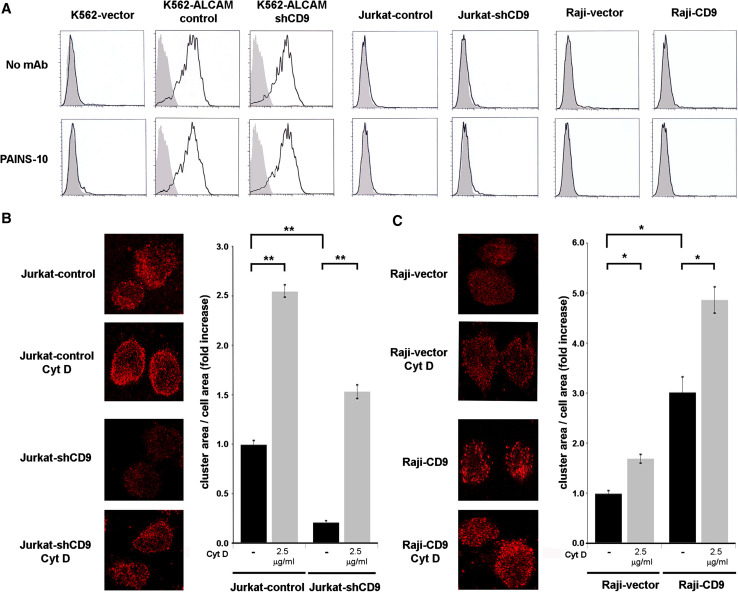
Cell adhesion can be regulated through changes in the affinity of individual adhesion molecules or through their aggregation/clustering which controls avidity [24, 41–43]. To discriminate which of these mechanisms predominates in the CD9-induced increase in ALCAM-mediated adhesion, we first analyzed the affinity of ALCAM homophilic interactions. For this purpose, we generated stable ALCAM transfectants in the human erythroleukemic K562 cell line which lacks endogenous expression of ALCAM [12]. Binding of sALCAM-Fc to cell surface ALCAM was measured by flow cytometry in both empty vector (K562-vector) and ALCAM-transfected cells (K562-ALCAM), as has been previously described [13]. As expected, only K562-ALCAM cells were able to bind sALCAM-Fc (Fig. 4a, upper row), reflecting the high affinity of this homophilic interaction. Interestingly, silencing CD9 in these cells (K562-ALCAM shCD9, Supplementary Fig. 1) did not affect the binding of sALCAM-Fc, implying that expression of CD9 does not modify ALCAM affinity. On the other hand, neither Jurkat nor Raji cells, although they abundantly express ALCAM on their surface, were able to bind sALCAM-Fc. Furthermore, this inability to bind ALCAM-Fc was not affected by the expression or lack of expression of CD9 on these two cell lines. It is noteworthy that incubation of all these cells with a stimulatory anti-CD9 mAb (PAINS-10) (Fig. 4a, lower row), or with other stimulatory anti-CD9 mAbs (PAINS-13, VJ1/20; not shown), had no effect on the binding of sALCAM-Fc to these cells.
Fig. 4.
CD9 regulates clustering but not affinity of ALCAM. a Binding of sALCAM-Fc was assessed as described in Materials and methods. Gray filled histograms correspond to negative controls stained only with secondary antibody; black line histograms represent the binding of ALCAM-Fc in the presence (lower row) or absence (upper row) of the anti-CD9 mAb PAINS-10 (20 μg/ml). b Immunofluorescence ALCAM staining of Jurkat cells, either transduced with specific CD9 shRNA (Jurkat-shCD9) or with control plasmid (Jurkat-control) both in the presence and absence of cytochalasin D (2.5 μg/ml), performed as described in Materials and methods. Maximal projections of confocal stacks are shown on the left and clustering quantifications on the right. Fold-increases in cluster area/cell area ratios are in relation to Jurkat-control cells without treatment, which was considered as 1. Data are presented as means ± SD of five independent experiments. c Clustering of ALCAM in Raji cells. Cells were transfected with the cDNA of CD9 (Raji-CD9) or with empty vector (Raji-vector). Maximal projections of confocal stacks are shown on the left and clustering quantifications on the right. Fold-increases in cluster area/cell area ratios are in relation to Raji-vector cells without treatment, which was considered as 1. Data are presented as means ± SD of cluster area relative to total area of the cell from five independent experiments. *p < 0.01, **p< 0.001
These results clearly indicate that expression of CD9 does not increase the affinity of cell surface ALCAM. We therefore investigated whether CD9 could be affecting the aggregation state (clustering) of cell surface ALCAM, as has been described for integrins [31, 44, 25] and other Ig-CAMs [24, 26, 27]. As shown in Fig. 4b, ALCAM is constitutively aggregated in conspicuous clusters on the surface of Jurkat cells. Interestingly, the size of ALCAM clusters was reduced upon CD9 silencing in these cells (Jurkat-shCD9). Similarly, ectopic expression of CD9 in Raji cells (Raji-CD9) was accompanied by an increase in the size of ALCAM clusters when compared with control Raji cells (Raji-vector) (Fig 4c). Cytochalasin D was used as a positive control for induction of ALCAM clustering [12]. Similar results on ALCAM clustering induction by the presence of CD9 or cytochalasin D on the surface of Jurkat and Raji cells were obtained using 70 % ethanol as an alternative fixation protocol, ruling out an artifactual effect of formaldehyde fixation on ALCAM aggregation (Supplementary Fig. 2). Taken together, these results indicate that clustering of ALCAM, rather than affinity regulation, is a mechanism contributing to CD9-mediated enhancement of ALCAM homophilic cell adhesion.
ALCAM expression on the cell surface is regulated by CD9 through ADAM17-mediated shedding
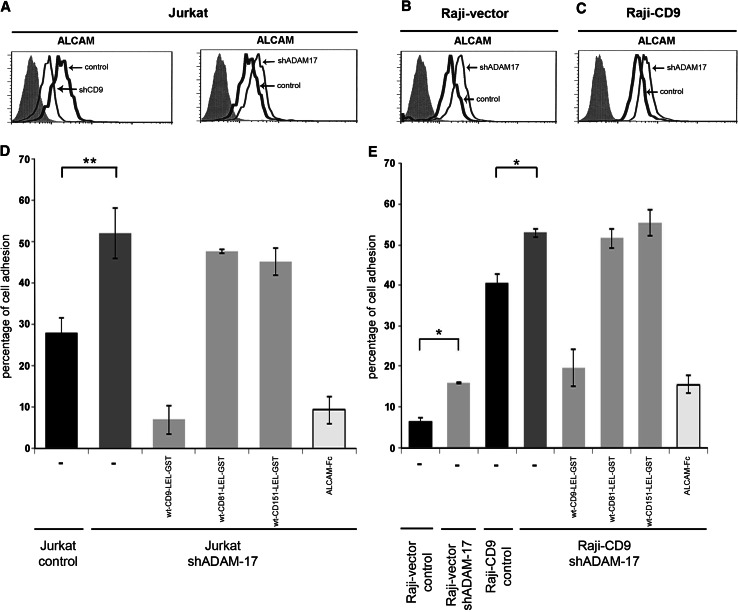
We have recently reported that CD9 interacts with, and negatively regulates the activity of, ADAM17/TACE, which explains why the surface expression levels of different protein substrates of this metalloproteinase (ICAM-1 and TNF-α) are altered depending on the presence or absence of CD9 [23]. ALCAM has been described as a substrate of ADAM17 [16, 17]. Interestingly, we observed that the surface expression of ALCAM in Jurkat cells was markedly reduced upon CD9 silencing (Jurkat-shCD9) (Fig. 5a), which is consistent again with a CD9-mediated inhibition of ADAM17 activity. To demonstrate the role of ADAM17 in controlling the surface expression of ALCAM we silenced this metalloproteinase in Jurkat cells (Jurkat-shADAM17) (Supplementary Fig. 1) and observed that the ALCAM surface expression was greatly increased in these silenced cells (Fig. 5a). This increment in ALCAM expression in ADAM17 knocked-down Jurkat cells correlated with augmented homophilic adhesion to immobilized ALCAM-Fc compared to control Jurkat cells (Fig. 5d). Furthermore, we showed that this enhanced adhesion to immobilized ALCAM-Fc of Jurkat cells after silencing ADAM17 (Jurkat shADAM17) was selectively blocked by competitive inhibitors of either CD9 or ALCAM (an excess of soluble wt-CD9-LEL-GST and ALCAM-Fc, respectively), but not by competitive inhibitors of other tetraspanins (an excess of soluble wt-CD81-LEL-GST or wt-CD151-LEL-GST) (Fig. 5d), which clearly demonstrates the specific involvement of cellular CD9 and ALCAM in the augmented cell adhesion to ALCAM-Fc caused by knocking-down ADAM-17.
Fig. 5.
ADAM17 regulates ALCAM surface expression and adhesion. Surface expression of ALCAM on Jurkat (a), Raji-vector (b) and Raji-CD9 (c) cells, assessed by flow cytometry. Gray filled histograms correspond to negative controls stained only with secondary antibody; thick black line histograms represent the expression of ALCAM on cells transfected with empty vector, and thin black line histograms represent the expression of ALCAM on cells transfected with the indicated shRNAs. Adhesion of Jurkat (d) and Raji (e) cells knocked-down for ADAM17 (shADAM17) to immobilized ALCAM in the presence or absence of 20 μg/ml sALCAM-Fc, wt-CD9-LEL-GST, wt-CD81-LEL-GST, or wt-CD151-LEL-GST. Data are presented as the percentages (means ± SD) of adherent cells. The experiment shown is a representative one out of four. *p < 0.01, **p < 0.001
In agreement with these data, when CD9 was transfected in Raji cells (Raji-CD9) the surface expression of ALCAM was increased over that observed on control cells (Raji-vector) (Fig. 1a). ADAM17 silencing in both control Raji cells (Raji-vector-shADAM17) and Raji-CD9 cells (Raji-CD9-shADAM17) (Supplementary Fig. 1) was similarly accompanied by an increase in ALCAM surface expression (Fig. 5b, c), which also correlated with augmented adhesion to ALCAM-Fc (Fig. 5e). In agreement with these results, inhibition of ADAM-17 activity on Raji-CD9 cells with the pharmacological agent TAPI-2 (20 μM) also enhanced adhesion of these cells to immobilized ALCAM-Fc (data not shown). As for Jurkat cells, the adhesion of ADAM-17-silenced Raji-CD9 cells to immobilized ALCAM-Fc was substantially reduced by competitive inhibitors of CD9 (an excess of soluble wt-CD9-LEL-GST) and ALCAM (an excess of sALCAM-Fc), but not by inhibitors of other tetraspanins (an excess of soluble wt-CD81-LEL-GST or wt-CD151-LEL-GST), again demonstrating the specific involvement of CD9 and ALCAM in this process (Fig. 5e).
The above experiments suggest that the changes in ALCAM surface expression observed upon CD9 silencing (reduced ALCAM expression; Fig. 5a) or transfection (increased ALCAM expression; Fig. 1a) are due to the altered ability of ADAM17 to cleave off this adhesion molecule. To demonstrate this hypothesis, stably CD9-silenced Jurkat cells were transiently cotransfected either with control shRNA plasmid (Fig. 6a, upper panels) or with specific ADAM17 shRNA plasmid (Fig. 6a, lower panels). This led to the complete restoration of the decreased ALCAM surface expression on CD9 silenced cells upon transient ADAM17 knock-down. To definitively confirm that CD9 indeed regulates the ADAM17-mediated ALCAM shedding from Jurkat and Raji cells, secreted sALCAM was measured using a specific ELISA. As shown in Fig. 6b, silencing of CD9 in Jurkat cells increased the secretion of ALCAM, while CD9 transfection into Raji cells decreased ALCAM shedding. Importantly, knocking-down ADAM17 caused in both cell lines a substantial reduction in ALCAM secretion, demonstrating that this metalloproteinase is mainly responsible for ALCAM shedding under these conditions.
Fig. 6.
CD9 regulates ALCAM shedding. a Flow cytometric detection of cell surface ALCAM on Jurkat-shCD9 cells, either transiently transfected with control plasmid (top panels, Jurkat-shCD9-control) or with ADAM17-specific shRNA plasmid (lower panels, Jurkat-shCD9-shADAM17). Gray filled histograms correspond to negative controls; thick black lines correspond to the expression of ALCAM. Mean fluorescence intensities and percentages of positive cells (% pos. cells) are indicated in the boxes. b ELISA quantification of sALCAM released from Jurkat, Raji-vector and Raji-CD9 cells transfected with control plasmid or shRNA plasmid specific for ADAM17 (shADAM17). Released ALCAM concentrations were calculated from a standard curve constructed with known amounts of this molecule. Data are presented as means ± SD of released ALCAM. The experiment shown is a representative one out of four
Collectively, all these results demonstrate that, in addition to cell surface clustering of ALCAM, regulation of ADAM17 activity represents another mechanism contributing to the CD9-mediated modulation of ALCAM surface expression and homophilic adhesion.
CD9 regulates T cell activation through heterophilic ALCAM–CD6 interactions
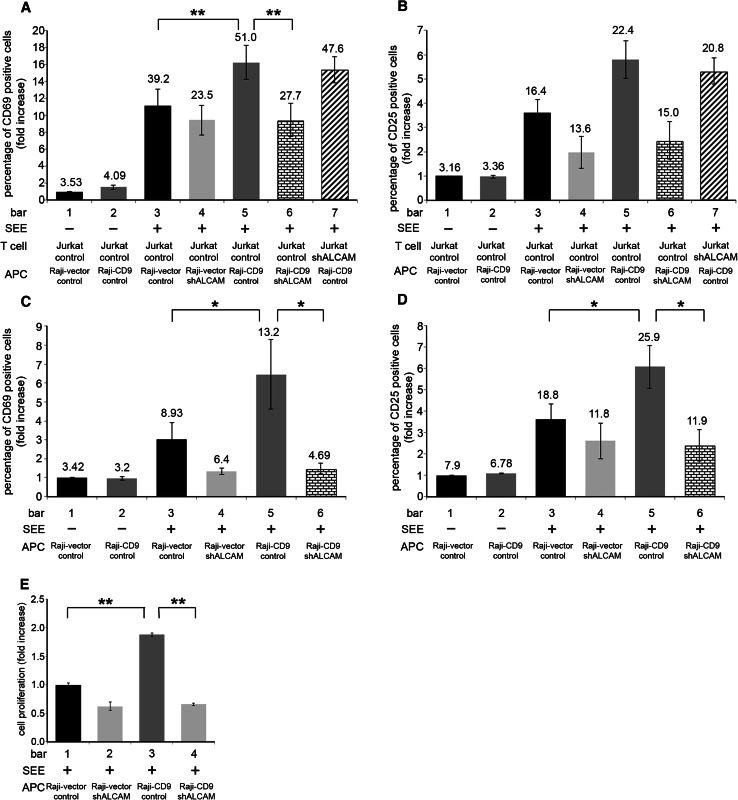
ALCAM is a costimulatory molecule on APCs which interacts with CD6 on T cells; both molecules are redistributed to the center of the immunological synapse and their heterophilic interaction contributes to T cell activation and proliferation [7, 8]. As we established a role for CD9 in regulating homophilic ALCAM–ALCAM interactions (Fig. 3), we decided to investigate whether CD9 also has a functional role in regulating the heterophilic ALCAM–CD6 interactions contributing to T cell activation. For this purpose, we analyzed the expression of CD69 and CD25, two well-established T cell activation markers, on Jurkat T cells following their antigen-dependent conjugation with CD9-deficient Raji cells (Raji-vector) or CD9-expressing Raji cells (Raji-CD9) loaded with the SEE superantigen. SEE-loaded control Raji cells were significantly less efficient (Fig. 7a, b; bar 3) than CD9-expressing Raji cells (Fig. 7a, b; bar 5) at inducing the expression of CD69 and CD25 activation molecules on the surface of Jurkat T cells, as determined by two different flow cytometric parameters: increase (1) in the percentages of cells positive for CD69 and CD25, and (2) in their levels of expression (relative mean fluorescence intensities). It is noteworthy that the induced levels of CD25 and CD69 on Jurkat T cells were similarly decreased after their conjugation with ALCAM-silenced Raji-vector (Raji-vector-shALCAM) (Fig. 7a, b; bar 4) and Raji-CD9 (Raji-CD9-shALCAM( (Fig. 7a, b; bar 6) cells, indicating the functional involvement of ALCAM on the APC side in this heterotypic cell conjugate assay. In contrast, no reduction in the induction of CD25 or CD69 expression was found when ALCAM was silenced in Jurkat T cells (Jurkat-shALCAM) (Fig. 7a, b; bar 7), indicating that the heterophilic interaction of ALCAM on APC with CD6 on T cells was responsible for the observed effects. Taken together, these results indicate that CD9 plays a functional regulatory role on the heterophilic interaction of ALCAM with CD6.
Fig. 7.
CD9 regulates ALCAM-mediated T cell activation. Raji cells expressing CD9 (Raji-CD9) or not (Raji-vector) were loaded or not with 2.5 μg/ml of SEE superantigen and then cocultured with responder cells for 72 h. Expression levels of the T cell activation markers CD69 and CD25 on Jurkat cells (a, b, respectively) and on MD-PBMC (c, d, respectively) were measured by flow cytometry. In all cases, bars represent the percentages of positive cells (mean ± SEM) for surface expression of CD69 or CD25 activation markers and the numbers on top of each bar represent the mean fluorescence intensity of each activation marker as an indication of the level of expression of these surface molecules. Data were calculated from six different experiments for Jurkat cells (a and b) and from nine experiments using primary cells from six different healthy donors. e MD-PBMC proliferation. Raji cells (either expressing or lacking CD9 and/or ALCAM) were pulsed with 2.5 μg/ml of SEE superantigen and then cocultured with primary MD-PBMC for 72 h. Proliferation was quantified using MTT as described in Materials and methods. Assays were done in triplicate and basal proliferation of Raji cells and MD-PBMC was subtracted. Fold-increases in proliferation are in relation to Raji-vector control cells which were considered as 1. Data are presented as proliferation (mean ± SEM). The experiment shown is a representative one of two for each of the six donors. *p < 0.05, **p < 0.001
Very similar CD69 and CD25 surface expression results were obtained when preparations of primary MD-PBMCs were used as responder cells after their conjugation with CD9-deficient Raji cells (Raji-vector) or CD9-expressing Raji cells (Raji-CD9) loaded with the SEE superantigen as APCs (Fig. 7c, d).
In addition to quantifying T cell activation through the induction of CD69 and CD25, we directly measured the proliferation of primary MD-PBMCs induced after their conjugation with either CD9-deficient SEE-loaded Raji cells (Raji-vector) or CD9-expressing SEE-loaded Raji cells (Raji-CD9). Again, the presence of CD9 on Raji APCs enhanced the proliferation of responder MD-PBMCs, which was reduced upon ALCAM silencing in these APCs. These results demonstrate the importance of CD9 in the regulation of the costimulatory heterophilic ALCAM interactions during T cell activation and proliferation.
Discussion
ALCAM/CD166 is a widespread member of the Ig-CAM family which mediates intercellular adhesion through either homophilic (ALCAM–ALCAM) or heterophilic (ALCAM–CD6) interactions. ALCAM-mediated adhesion is crucial in different physiological and pathological processes, with particular relevance in leukocyte extravasation, stabilization of the immunological synapse and T cell activation and tumor growth and metastasis (reviewed in references [2, 4, 5]). Although the functional implication of ALCAM in these phenomena is well established, the mechanisms regulating its adhesive capacity are poorly characterized.
We demonstrate here that ALCAM associates with the tetraspanin CD9 on the leukocyte surface, based on colocalization and coimmunoprecipitation experiments. Furthermore, pull-down experiments with purified recombinant proteins have shown that these CD9–ALCAM interactions are direct and mediated by the CD9 LEL domain. Most tetraspanin interactions with other membrane proteins are known to be mediated by the variable region within their LEL domain, whose conformation is maintained by distinct disulfide bonds [25]. Partial loss of the CD9 LEL conformation in mutants C153A and C167A, which form one disulfide bridge that extends from the beginning (N end) to the central region of the variable portion of the LEL, dramatically reduced the interaction with ALCAM, whereas mutants in cysteines C152A and C181A, that form a second disulfide bond between the N and C ends of the variable region, had only a minor impact. Interestingly, the same C153A and C167A mutations in the CD9 LEL also weakened its interaction with ADAM17 [23]. Therefore, an intact conformation of the variable region within the LEL domain of CD9 seems to be required for optimal lateral interaction with ALCAM on the cell surface.
Our results clearly indicate that the adhesive function of ALCAM is upregulated upon its interaction with CD9. Adhesion experiments have revealed that ectopic expression of CD9 in Raji B cells promotes ALCAM-mediated homophilic cell adhesion to immobilized ALCAM-Fc. In contrast, CD9-silencing in Jurkat T cells abrogates homophilic ALCAM adhesion. Interestingly, soluble wt-CD9-LEL-GST protein almost completely blocked ALCAM-mediated adhesion of these Jurkat and Raji-CD9 leukocytic cells as well as of primary lymphoblasts. In contrast, the CD9 LEL mutants C153A and C167A, which showed reduced ability to interact with ALCAM, also displayed lower capacity to inhibit ALCAM adhesion. It is noteworthy that, consistent with the biochemical data, the soluble LEL domains of other related tetraspanins, such as CD151 and CD81, had minimal inhibitory effect on ALCAM-mediated cell adhesion. These data clearly highlight the specific involvement of the CD9 LEL domain in regulating ALCAM function.
As for other adhesion molecules, particularly integrins [41, 43], increased ALCAM-mediated adhesion can result from changes either in the affinity or in the avidity/clustering of ALCAM molecules. In fact, clustering of ALCAM on the cell surface, which is dynamically controlled by the actin cytoskeleton, has been characterized as an important regulatory mechanism of ALCAM-mediated adhesion [12, 13]. In contrast, changes in the affinity of individual ALCAM molecules have not been implicated in the augmented ALCAM adhesion that is induced by actin-disrupting agents [13]. In agreement with these reports, our results show for the first time that CD9 enhances ALCAM–ALCAM homophilic interactions through a mechanism involving increased clustering of ALCAM molecules on the cell surface, without detectable changes in their affinity. One of the hallmarks of tetraspanin function is their ability to recruit other cell surface proteins into tetraspanin-enriched microdomains, thereby facilitating their clustering [24, 45]. In particular, CD9 has been shown to promote the adhesive function of β1 integrins [31] and other Ig-CAMs, such as ICAM-1 and VCAM-1, through their inclusion into tetraspanin-enriched microdomains and induction of clustering [26, 27]. Although it is established that the actin cytoskeleton regulates ALCAM clustering/avidity, the nature of the molecules linking ALCAM with the actin cytoskeleton has so far remained elusive, although the ezrin/radixin/moesin and α-catenin proteins have been suggested as possible candidates [6, 46]. In this regard, association of ALCAM with CD9 could provide an indirect link with the actin cytoskeleton, as several specific protein partners of CD9, such as EWI-2, EWI-F or CD81, are known to interact directly with ezrin/radixin/moesin proteins [47].
Our group has recently reported that CD9 associates with and inhibits the activity of ADAM17 [23]. This inhibition results in augmented cell surface expression of TNF-α and ICAM-1, two well-characterized substrates of ADAM17. Consistent with these previous findings, we report here that surface expression of ALCAM is also increased in leukocytic cells expressing CD9 (Raji-CD9, Jurkat-control) compared to their counterparts lacking expression of this tetraspanin (Raji-control, Jurkat-shCD9). Furthermore, we demonstrated that these effects of CD9 on ALCAM-surface expression are mediated through regulation of ADAM17 metalloproteinase activity, as evidenced by the restoration, following knocking-down of this enzyme, of the decreased ALCAM expression in CD9-interfered cells. We have additionally confirmed the involvement of ADAM17 in the CD9-mediated regulation of ALCAM shedding by ELISA quantification of sALCAM. Therefore, the increased ALCAM surface expression and reduced ALCAM shedding caused by the presence of CD9 through ADAM17 inhibition also help explain the observed increase in ALCAM-mediated adhesion.
The relative contribution of both mechanisms—facilitation of ALCAM clustering and increased ALCAM surface expression due to inhibition of ADAM17—to the CD9 enhancement of ALCAM-mediated adhesion remains to be characterized in detail. However, our data showing that only a modest increase in ALCAM surface expression and adhesion is observed after silencing ADAM17 in Raji cells compared to the much more evident adhesion increment caused by the presence of CD9 on these cells, would suggest that the contribution of CD9-mediated clustering of ALCAM molecules is greater than its inhibitory effect on ADAM17 proteolytic activity.
The involvement of CD9 in the regulation of migration in different cellular settings is recognized as a key functional feature of this tetraspanin, which indeed was originally termed MRP-1 (motility-related protein-1) [48]. We have shown that the consequences of the observed effects of CD9 on ALCAM-mediated cell adhesion are also clearly reflected in a functional assay of Jurkat cell migration which is dependent on homophilic ALCAM interactions. In this assay, CD9 silencing dramatically reduced the number of cells that transmigrated across ALCAM-Fc-coated filters. In this regard, ALCAM has been implicated in the transendothelial migration of monocytes [10] and as an adhesion molecule on central nervous system endothelial cells involved in leukocyte migration across the blood–brain barrier [9]. Different tetraspanins, including CD9, play an essential role in the assembling of transmigratory cups, which are specialized platforms containing complexes of Ig-CAMs, such as ICAM-1 and VCAM-1, involved in leukocyte adhesion and transmigration [26, 49, 50]. Further experiments should be carried out to elucidate whether CD9 also recruits ALCAM to these platforms. Our current data strongly support the view that the adhesive function of ALCAM is dynamically regulated through its association with CD9, representing an additional example of the functional importance of this tetraspanin in the regulation of cell migration.
ALCAM has been identified as an important costimulatory molecule on APCs which, through its heterophilic interaction with CD6 on T cells, contributes to the antigen-specific induction of T cell activation and proliferation [1, 7, 8]. Using conjugates between SEE-loaded Raji B cells (either lacking or expressing CD9) as APCs and primary MD-PBMCs or Jurkat cells as a model for antigen-induced T cell activation and proliferation, we have also demonstrated the functional role of CD9 in regulating ALCAM-mediated heterotypic APC–T cell interactions.
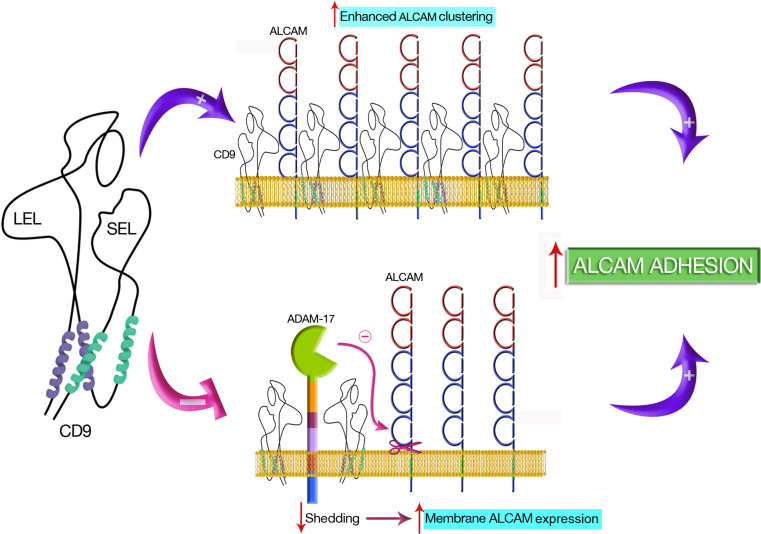
In conclusion, we report here that ALCAM directly associates with the tetraspanin CD9 in a protein complex on the leukocyte surface that also includes the metalloproteinase ADAM17. Through these interactions, CD9 upregulates both homophilic and heterophilic ALCAM interactions, which is functionally reflected in increased ALCAM-mediated leukocyte adhesion and T cell migration, activation and proliferation. The results presented here are depicted in the functional model shown in Fig. 8, and show that a cooperative dual mechanism involving on the one hand increased clustering of ALCAM, possibly resulting in enhanced avidity of this molecule, and on the other hand augmented ALCAM surface expression due to inhibition of ADAM17 metalloproteinase activity, contributes to the enhancement of ALCAM-mediated adhesion and function induced by the tetraspanin CD9.
Fig. 8.
Functional model. A cooperative dual mechanism involving on the one hand increased clustering of ALCAM, possibly resulting in enhanced avidity of this molecule, and on the other hand augmented ALCAM surface expression due to inhibition of ADAM17 metalloproteinase activity, contributes to the enhancement of ALCAM-mediated adhesion and function induced by the tetraspanin CD9
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1. Changes in ALCAM, CD9, ADAM17, and CD6 surface expression following specific silencing with shRNA or ALCAM cDNA transfection. Surface expression of the indicated molecules on Jurkat, Raji-vector, Raji-CD9 and K562 cells was measured by flow cytometry. Grey-filled histograms represent the negative controls, stained only with secondary antibody; thick-black line histograms represent the expression of the indicated molecules on cells transfected with control empty plasmids, and thin-black lines correspond to their expression on cells transfected with the indicated specific shRNAs (Jurkat, Raji-vector, Raji-CD9 and K562) or cDNA-containing (K562) plasmids. (JPEG 262 kb)
Supplementary Fig. 2. ALCAM clustering after ethanol fixation. A) Immunofluorescence ALCAM staining of Jurkat cells, either transduced with specific CD9 shRNA (Jurkat-shCD9) or with control plasmid (Jurkat-control) both in the presence or absence of cytochalasin D (2.5 μg/ml), was performed as described in “Materials and Methods”, using ethanol as fixative. Maximal projections of confocal stacks are shown on the left and clustering quantifications on the right. Fold-increases in clusters area/cell area ratios were related to Jurkat-control cells without treatment, which was considered as 1. Data represent the mean ± SD of three independent experiments. B) Clustering of ALCAM in Raji cells. Cells were transfected with the cDNA of CD9 (Raji-CD9) or with empty vector (Raji-vector). Cells were fixed with 70 % ethanol instead of formaldehyde. Maximal projections of confocal stacks are shown on the left and clustering quantifications on the right. Fold-increases in clusters area/cell area ratios were related to Raji-vector cells without treatment, which was considered as 1. Data represent the mean ± SD of area cluster relative to total area of the cell from three independent experiments. * (p < 0.01) denote statistically significant differences in the clustering size and number. (JPEG 251 kb)
Acknowledgments
We are grateful to Dr. P.N. Monk (University of Sheffield Medical School, UK) for providing all the tetraspanin LEL-GST fusion proteins, to Dr. M. Yáñez-Mó and Dr. Francisco Sánchez-Madrid (Instituto de Investigación Sanitaria Princesa, Hospital de la Princesa, Madrid, Spain) for providing the CD9 cDNA and for their continuous support and helpful discussions, to Dr. J.M. Serrador (CBMSO, Madrid, Spain) for his assistance with APC T cell assays and helpful discussions, to Irene Tejada for technical assistance, and to Belén Capdevila for assistance with artwork model illustration. This work was supported by grants BFU2010-19144/BMC and SAF2012-34561 from MICINN, a grant from Fundación de Investigación Médica Mutua Madrileña and by the RETICS Program RD08/0075-RIER from Instituto de Salud Carlos III (to C.C.), a grant from Fundación MAPFRE (to M.D.G.L.), and grant SAF2007-60578 from MICINN (to E.M.L.). A.G. has been supported by a predoctoral fellowship from Instituto de Salud Carlos III and by grants BFU2007-66443/BMC from MICINN and Fundación de Investigación Médica Mutua Madrileña. L.S.M. has been supported by the RETICS Program RD08/0075-RIER and grant BFU2007-66443/BMC.
Abbreviations
- ALCAM
Activated leukocyte cell adhesion molecule
- APC
Antigen-presenting cell
- Ig-CAM
Immunoglobulin superfamily cell adhesion molecule
- LEL
Large extracellular loop
- mAb
Monoclonal antibody
- MD-PBMCs
Monocyte-depleted peripheral blood mononuclear cells
- PBS
Phosphate-buffered saline
- sALCAM
Soluble ALCAM
- SEE
Staphylococcal enterotoxin E superantigen
- shRNA
Small hairpin RNA
- TBS
Tris-buffered saline
- TNF-α
Tumor necrosis factor-α
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
References
- 1.Hassan NJ, Barclay AN, Brown MH. Frontline: optimal T cell activation requires the engagement of CD6 and CD166. Eur J Immunol. 2004;34(4):930–940. doi: 10.1002/eji.200424856. [DOI] [PubMed] [Google Scholar]
- 2.Swart GW. Activated leukocyte cell adhesion molecule (CD166/ALCAM): developmental and mechanistic aspects of cell clustering and cell migration. Eur J Cell Biol. 2002;81(6):313–321. doi: 10.1078/0171-9335-00256. [DOI] [PubMed] [Google Scholar]
- 3.van Kempen LC, Nelissen JM, Degen WG, Torensma R, Weidle UH, Bloemers HP, Figdor CG, Swart GW. Molecular basis for the homophilic activated leukocyte cell adhesion molecule (ALCAM)–ALCAM interaction. J Biol Chem. 2001;276(28):25783–25790. doi: 10.1074/jbc.M011272200. [DOI] [PubMed] [Google Scholar]
- 4.Swart GW, Lunter PC, Kilsdonk JW, Kempen LC. Activated leukocyte cell adhesion molecule (ALCAM/CD166): signaling at the divide of melanoma cell clustering and cell migration? Cancer Metastasis Rev. 2005;24(2):223–236. doi: 10.1007/s10555-005-1573-0. [DOI] [PubMed] [Google Scholar]
- 5.Ofori-Acquah SF, King JA. Activated leukocyte cell adhesion molecule: a new paradox in cancer. Transl Res. 2008;151(3):122–128. doi: 10.1016/j.trsl.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Weidle UH, Eggle D, Klostermann S, Swart GW. ALCAM/CD166: cancer-related issues. Cancer Genomics Prot. 2010;7(5):231–243. [PubMed] [Google Scholar]
- 7.Zimmerman AW, Joosten B, Torensma R, Parnes JR, van Leeuwen FN, Figdor CG. Long-term engagement of CD6 and ALCAM is essential for T-cell proliferation induced by dendritic cells. Blood. 2006;107(8):3212–3220. doi: 10.1182/blood-2005-09-3881. [DOI] [PubMed] [Google Scholar]
- 8.Gimferrer I, Calvo M, Mittelbrunn M, Farnos M, Sarrias MR, Enrich C, Vives J, Sanchez-Madrid F, Lozano F. Relevance of CD6-mediated interactions in T cell activation and proliferation. J Immunol. 2004;173(4):2262–2270. doi: 10.4049/jimmunol.173.4.2262. [DOI] [PubMed] [Google Scholar]
- 9.Cayrol R, Wosik K, Berard JL, Dodelet-Devillers A, Ifergan I, Kebir H, Haani AS, Kreymborg K, Krug S, Moumdjian R, Bouthillier A, Becher B, Arbour N, David S, Stanimirovic D, Prat A. Activated leukocyte cell adhesion molecule promotes leukocyte trafficking into the central nervous system. Nat Immunol. 2008;9(2):137–145. doi: 10.1038/ni1551. [DOI] [PubMed] [Google Scholar]
- 10.Masedunskas A, King JA, Tan F, Cochran R, Stevens T, Sviridov D, Ofori-Acquah SF. Activated leukocyte cell adhesion molecule is a component of the endothelial junction involved in transendothelial monocyte migration. FEBS Lett. 2006;580(11):2637–2645. doi: 10.1016/j.febslet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 11.Weiner JA, Koo SJ, Nicolas S, Fraboulet S, Pfaff SL, Pourquie O, Sanes JR. Axon fasciculation defects and retinal dysplasias in mice lacking the immunoglobulin superfamily adhesion molecule BEN/ALCAM/SC1. Mol Cell Neurosci. 2004;27(1):59–69. doi: 10.1016/j.mcn.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Nelissen JM, Peters IM, de Grooth BG, van Kooyk Y, Figdor CG. Dynamic regulation of activated leukocyte cell adhesion molecule-mediated homotypic cell adhesion through the actin cytoskeleton. Mol Biol Cell. 2000;11(6):2057–2068. doi: 10.1091/mbc.11.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zimmerman AW, Nelissen JM, van Emst-de Vries SE, Willems PH, de Lange F, Collard JG, van Leeuwen FN, Figdor CG. Cytoskeletal restraints regulate homotypic ALCAM-mediated adhesion through PKCalpha independently of Rho-like GTPases. J Cell Sci. 2004;117(Pt 13):2841–2852. doi: 10.1242/jcs.01139. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda K, Quertermous T. Molecular isolation and characterization of a soluble isoform of activated leukocyte cell adhesion molecule that modulates endothelial cell function. J Biol Chem. 2004;279(53):55315–55323. doi: 10.1074/jbc.M407776200. [DOI] [PubMed] [Google Scholar]
- 15.van Kilsdonk JW, Wilting RH, Bergers M, van Muijen GN, Schalkwijk J, van Kempen LC, Swart GW. Attenuation of melanoma invasion by a secreted variant of activated leukocyte cell adhesion molecule. Cancer Res. 2008;68(10):3671–3679. doi: 10.1158/0008-5472.CAN-07-5767. [DOI] [PubMed] [Google Scholar]
- 16.Bech-Serra JJ, Santiago-Josefat B, Esselens C, Saftig P, Baselga J, Arribas J, Canals F. Proteomic identification of desmoglein 2 and activated leukocyte cell adhesion molecule as substrates of ADAM17 and ADAM10 by difference gel electrophoresis. Mol Cell Biol. 2006;26(13):5086–5095. doi: 10.1128/MCB.02380-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosso O, Piazza T, Bongarzone I, Rossello A, Mezzanzanica D, Canevari S, Orengo AM, Puppo A, Ferrini S, Fabbi M. The ALCAM shedding by the metalloprotease ADAM17/TACE is involved in motility of ovarian carcinoma cells. Mol Cancer Res. 2007;5(12):1246–1253. doi: 10.1158/1541-7786.MCR-07-0060. [DOI] [PubMed] [Google Scholar]
- 18.Kulasingam V, Zheng Y, Soosaipillai A, Leon AE, Gion M, Diamandis EP. Activated leukocyte cell adhesion molecule: a novel biomarker for breast cancer. Int J Cancer. 2009;125(1):9–14. doi: 10.1002/ijc.24292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29(5):258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reiss K, Saftig P. The “a disintegrin and metalloprotease” (ADAM) family of sheddases: physiological and cellular functions. Semin Cell Dev Biol. 2009;20(2):126–137. doi: 10.1016/j.semcdb.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Murphy G. Regulation of the proteolytic disintegrin metalloproteinases, the ‘Sheddases’. Semin Cell Dev Biol. 2009;20(2):138–145. doi: 10.1016/j.semcdb.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Mezyk R, Bzowska M, Bereta J. Structure and functions of tumor necrosis factor-alpha converting enzyme. Acta Biochim Pol. 2003;50(3):625–645. [PubMed] [Google Scholar]
- 23.Gutierrez-Lopez MD, Gilsanz A, Yanez-Mo M, Ovalle S, Lafuente EM, Dominguez C, Monk PN, Gonzalez-Alvaro I, Sanchez-Madrid F, Cabanas C. The sheddase activity of ADAM17/TACE is regulated by the tetraspanin CD9. Cell Mol Life Sci. 2011;68(19):3275–3292. doi: 10.1007/s00018-011-0639-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yanez-Mo M, Barreiro O, Gordon-Alonso M, Sala-Valdes M, Sanchez-Madrid F. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 2009;19(9):434–446. doi: 10.1016/j.tcb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 25.Hemler ME. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu Rev Cell Dev Biol. 2003;19:397–422. doi: 10.1146/annurev.cellbio.19.111301.153609. [DOI] [PubMed] [Google Scholar]
- 26.Barreiro O, Yanez-Mo M, Sala-Valdes M, Gutierrez-Lopez MD, Ovalle S, Higginbottom A, Monk PN, Cabanas C, Sanchez-Madrid F. Endothelial tetraspanin microdomains regulate leukocyte firm adhesion during extravasation. Blood. 2005;105(7):2852–2861. doi: 10.1182/blood-2004-09-3606. [DOI] [PubMed] [Google Scholar]
- 27.Barreiro O, Zamai M, Yanez-Mo M, Tejera E, Lopez-Romero P, Monk PN, Gratton E, Caiolfa VR, Sanchez-Madrid F. Endothelial adhesion receptors are recruited to adherent leukocytes by inclusion in preformed tetraspanin nanoplatforms. J Cell Biol. 2008;183(3):527–542. doi: 10.1083/jcb.200805076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gutierrez-Lopez MD, Ovalle S, Yanez-Mo M, Sanchez-Sanchez N, Rubinstein E, Olmo N, Lizarbe MA, Sanchez-Madrid F, Cabanas C. A functionally relevant conformational epitope on the CD9 tetraspanin depends on the association with activated beta1 integrin. J Biol Chem. 2003;278(1):208–218. doi: 10.1074/jbc.M207805200. [DOI] [PubMed] [Google Scholar]
- 29.Arroyo AG, Sanchez-Mateos P, Campanero MR, Martin-Padura I, Dejana E, Sanchez-Madrid F. Regulation of the VLA integrin-ligand interactions through the beta 1 subunit. J Cell Biol. 1992;117(3):659–670. doi: 10.1083/jcb.117.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yanez-Mo M, Alfranca A, Cabanas C, Marazuela M, Tejedor R, Ursa MA, Ashman LK, de Landazuri MO, Sanchez-Madrid F. Regulation of endothelial cell motility by complexes of tetraspan molecules CD81/TAPA-1 and CD151/PETA-3 with alpha3 beta1 integrin localized at endothelial lateral junctions. J Cell Biol. 1998;141(3):791–804. doi: 10.1083/jcb.141.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ovalle S, Gutierrez-Lopez MD, Olmo N, Turnay J, Lizarbe MA, Majano P, Molina-Jimenez F, Lopez-Cabrera M, Yanez-Mo M, Sanchez-Madrid F, Cabanas C. The tetraspanin CD9 inhibits the proliferation and tumorigenicity of human colon carcinoma cells. Int J Cancer. 2007;121(10):2140–2152. doi: 10.1002/ijc.22902. [DOI] [PubMed] [Google Scholar]
- 32.Cebrian M, Yague E, Rincon M, Lopez-Botet M, de Landazuri MO, Sanchez-Madrid F. Triggering of T cell proliferation through AIM, an activation inducer molecule expressed on activated human lymphocytes. J Exp Med. 1988;168(5):1621–1637. doi: 10.1084/jem.168.5.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez-Martin L, Sanchez-Sanchez N, Gutierrez-Lopez MD, Rojo AI, Vicente-Manzanares M, Perez-Alvarez MJ, Sanchez-Mateos P, Bustelo XR, Cuadrado A, Sanchez-Madrid F, Rodriguez-Fernandez JL, Cabanas C. Signaling through the leukocyte integrin LFA-1 in T cells induces a transient activation of Rac-1 that is regulated by Vav and PI3K/Akt-1. J Biol Chem. 2004;279(16):16194–16205. doi: 10.1074/jbc.M400905200. [DOI] [PubMed] [Google Scholar]
- 34.Higginbottom A, Takahashi Y, Bolling L, Coonrod SA, White JM, Partridge LJ, Monk PN. Structural requirements for the inhibitory action of the CD9 large extracellular domain in sperm/oocyte binding and fusion. Biochem Biophys Res Commun. 2003;311(1):208–214. doi: 10.1016/j.bbrc.2003.09.196. [DOI] [PubMed] [Google Scholar]
- 35.de la Rosa G, Longo N, Rodriguez-Fernandez JL, Puig-Kroger A, Pineda A, Corbi AL, Sanchez-Mateos P. Migration of human blood dendritic cells across endothelial cell monolayers: adhesion molecules and chemokines involved in subset-specific transmigration. J Leukoc Biol. 2003;73(5):639–649. doi: 10.1189/jlb.1002516. [DOI] [PubMed] [Google Scholar]
- 36.Kovalenko OV, Yang X, Kolesnikova TV, Hemler ME. Evidence for specific tetraspanin homodimers: inhibition of palmitoylation makes cysteine residues available for cross-linking. Biochem J. 2004;377(Pt 2):407–417. doi: 10.1042/BJ20031037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masellis-Smith A, Shaw AR. CD9-regulated adhesion. Anti-CD9 monoclonal antibody induce pre-B cell adhesion to bone marrow fibroblasts through de novo recognition of fibronectin. J Immunol. 1994;152(6):2768–2777. [PubMed] [Google Scholar]
- 38.Cook GA, Wilkinson DA, Crossno JT, Jr, Raghow R, Jennings LK. The tetraspanin CD9 influences the adhesion, spreading, and pericellular fibronectin matrix assembly of Chinese hamster ovary cells on human plasma fibronectin. Exp Cell Res. 1999;251(2):356–371. doi: 10.1006/excr.1999.4596. [DOI] [PubMed] [Google Scholar]
- 39.Berditchevski F. Complexes of tetraspanins with integrins: more than meets the eye. J Cell Sci. 2001;114(Pt 23):4143–4151. doi: 10.1242/jcs.114.23.4143. [DOI] [PubMed] [Google Scholar]
- 40.Arai F, Ohneda O, Miyamoto T, Zhang XQ, Suda T. Mesenchymal stem cells in perichondrium express activated leukocyte cell adhesion molecule and participate in bone marrow formation. J Exp Med. 2002;195(12):1549–1563. doi: 10.1084/jem.20011700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanchez-Mateos P, Cabanas C, Sanchez-Madrid F. Regulation of integrin function. Semin Cancer Biol. 1996;7(3):99–109. doi: 10.1006/scbi.1996.0015. [DOI] [PubMed] [Google Scholar]
- 42.Kinashi T. Intracellular signalling controlling integrin activation in lymphocytes. Nat Rev. 2005;5(7):546–559. doi: 10.1038/nri1646. [DOI] [PubMed] [Google Scholar]
- 43.Stewart MP, Cabanas C, Hogg N. T cell adhesion to intercellular adhesion molecule-1 (ICAM-1) is controlled by cell spreading and the activation of integrin LFA-1. J Immunol. 1996;156(5):1810–1817. [PubMed] [Google Scholar]
- 44.Ziyyat A, Rubinstein E, Monier-Gavelle F, Barraud V, Kulski O, Prenant M, Boucheix C, Bomsel M, Wolf JP. CD9 controls the formation of clusters that contain tetraspanins and the integrin alpha 6 beta 1, which are involved in human and mouse gamete fusion. J Cell Sci. 2006;119(Pt 3):416–424. doi: 10.1242/jcs.02730. [DOI] [PubMed] [Google Scholar]
- 45.Charrin S, le Naour F, Silvie O, Milhiet PE, Boucheix C, Rubinstein E. Lateral organization of membrane proteins: tetraspanins spin their web. Biochem J. 2009;420(2):133–154. doi: 10.1042/BJ20082422. [DOI] [PubMed] [Google Scholar]
- 46.Uhlenbrock K, Eberth A, Herbrand U, Daryab N, Stege P, Meier F, Friedl P, Collard JG, Ahmadian MR. The RacGEF Tiam1 inhibits migration and invasion of metastatic melanoma via a novel adhesive mechanism. J Cell Sci. 2004;117(Pt 20):4863–4871. doi: 10.1242/jcs.01367. [DOI] [PubMed] [Google Scholar]
- 47.Sala-Valdes M, Ursa A, Charrin S, Rubinstein E, Hemler ME, Sanchez-Madrid F, Yanez-Mo M. EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins. J Biol Chem. 2006;281(28):19665–19675. doi: 10.1074/jbc.M602116200. [DOI] [PubMed] [Google Scholar]
- 48.Miyake M, Koyama M, Seno M, Ikeyama S. Identification of the motility-related protein (MRP-1), recognized by monoclonal antibody M31-15, which inhibits cell motility. J Exp Med. 1991;174(6):1347–1354. doi: 10.1084/jem.174.6.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barreiro O, Vicente-Manzanares M, Urzainqui A, Yanez-Mo M, Sanchez-Madrid F. Interactive protrusive structures during leukocyte adhesion and transendothelial migration. Front Biosci. 2004;9:1849–1863. doi: 10.2741/1285. [DOI] [PubMed] [Google Scholar]
- 50.Barreiro O, Yanez-Mo M, Serrador JM, Montoya MC, Vicente-Manzanares M, Tejedor R, Furthmayr H, Sanchez-Madrid F. Dynamic interaction of VCAM-1 and ICAM-1 with moesin and ezrin in a novel endothelial docking structure for adherent leukocytes. J Cell Biol. 2002;157(7):1233–1245. doi: 10.1083/jcb.200112126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Changes in ALCAM, CD9, ADAM17, and CD6 surface expression following specific silencing with shRNA or ALCAM cDNA transfection. Surface expression of the indicated molecules on Jurkat, Raji-vector, Raji-CD9 and K562 cells was measured by flow cytometry. Grey-filled histograms represent the negative controls, stained only with secondary antibody; thick-black line histograms represent the expression of the indicated molecules on cells transfected with control empty plasmids, and thin-black lines correspond to their expression on cells transfected with the indicated specific shRNAs (Jurkat, Raji-vector, Raji-CD9 and K562) or cDNA-containing (K562) plasmids. (JPEG 262 kb)
Supplementary Fig. 2. ALCAM clustering after ethanol fixation. A) Immunofluorescence ALCAM staining of Jurkat cells, either transduced with specific CD9 shRNA (Jurkat-shCD9) or with control plasmid (Jurkat-control) both in the presence or absence of cytochalasin D (2.5 μg/ml), was performed as described in “Materials and Methods”, using ethanol as fixative. Maximal projections of confocal stacks are shown on the left and clustering quantifications on the right. Fold-increases in clusters area/cell area ratios were related to Jurkat-control cells without treatment, which was considered as 1. Data represent the mean ± SD of three independent experiments. B) Clustering of ALCAM in Raji cells. Cells were transfected with the cDNA of CD9 (Raji-CD9) or with empty vector (Raji-vector). Cells were fixed with 70 % ethanol instead of formaldehyde. Maximal projections of confocal stacks are shown on the left and clustering quantifications on the right. Fold-increases in clusters area/cell area ratios were related to Raji-vector cells without treatment, which was considered as 1. Data represent the mean ± SD of area cluster relative to total area of the cell from three independent experiments. * (p < 0.01) denote statistically significant differences in the clustering size and number. (JPEG 251 kb)