Abstract
In vertebrates, most of the skeleton is formed through endochondral ossification. Endochondral bone formation is a complex process involving the mesenchymal condensation of undifferentiated cells, the proliferation of chondrocytes and their differentiation into hypertrophic chondrocytes, and mineralization. This process is tightly regulated by various factors including transcription factors, soluble mediators, extracellular matrices, and cell–cell and cell–matrix interactions. Defects of these factors often lead to skeletal dysplasias and short stature. Moreover, there is growing evidence that epigenetic and microRNA-mediated mechanisms also play critical roles in chondrogenesis. This review provides an overview of our current understanding of the regulators for the development of growth plate cartilage and their molecular mechanisms of action. A knowledge of the regulatory mechanisms underlying the proliferation and differentiation of chondrocytes will provide insights into future therapeutic options for skeletal disorders.
Keywords: Chondrocyte, Transcription factors, Growth factors, Extracellular matrix, Differentiation
Introduction
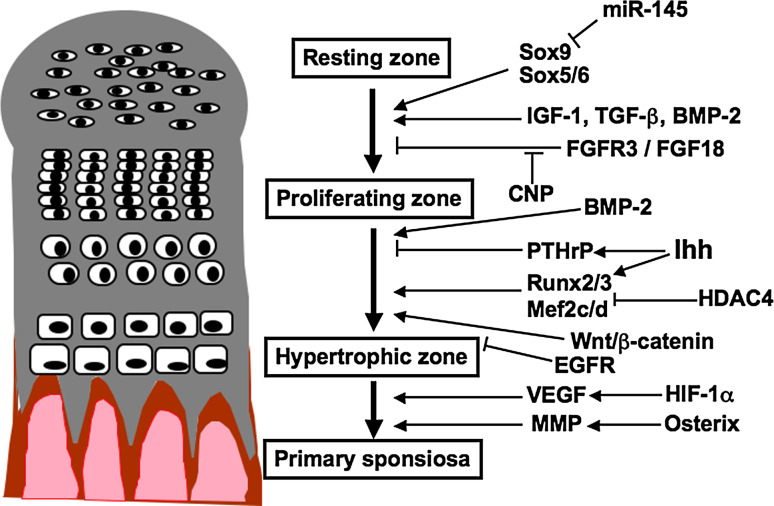
Most of the skeleton, including the long bones of the limbs and the vertebral columns, is formed through endochondral ossification, involving a cartilaginous intermediate [1–3]. Endochondral bone formation starts with the condensation of mesenchymal cells, which differentiate into chondrocytes characterized by the production of specific extracellular matrix (ECM) proteins such as type II collagen (Col II) and aggrecan. The chondrocytes proliferate unidirectionally to form orderly parallel columns which accumulate a cartilaginous matrix [2]. These cells then exit the cell cycle, differentiate further to become hypertrophic, and produce type X collagen (Col X) [4]. In growth plates, maturing chondrocytes are organized into zones, including a resting zone, a proliferating zone, a prehypertrophic zone, and a hypertrophic zone [5]. Once the hypertrophic chondrocytes have terminally differentiated, the cartilaginous matrix is mineralized and the cells undergo apoptosis. These mature chondrocytes express vascular endothelial growth factor (VEGF) to induce blood vessel invasion and matrix metalloproteinases (MMPs) to aid in the degradation of the cartilaginous matrix by chondroclasts, and the primary ossification center is developed [6, 7] (Fig. 1).
Fig. 1.
Schematic representation of molecules involved in the development of growth plate cartilage
The process of chondrocyte proliferation and differentiation is regulated by various transcription factors, growth factors, ECMs, and cell–matrix interactions [2, 8–10]. In addition, recent studies have revealed the importance of epigenetic and microRNA-mediated control in cartilage development. Defects in the factors involved in the development of growth plate cartilage are often associated with skeletal dysplasias and short stature [11]. In this review article, I will mainly address the mechanisms regulating the development of growth plate cartilage.
Transcriptional control of chondrogenesis
Sox9 is a member of the Sox family of transcription factors characterized by a high-mobility-group-box DNA binding motif related to that of the sex determining factor SRY, and plays a central role in chondrogenesis. In chondrocyte lineage cells, the expression of Sox9 starts at the mesenchymal osteochondroprogenitor stage and remains high during differentiation. Mutations in the human SOX9 gene result in camptomelic dysplasia characterized by severe skeletal malformation, indicating the critical role of SOX9 in skeletogenesis [12, 13]. Accumulating evidence in mice also has revealed that Sox9 is indispensable for chondrocyte differentiation [3, 14]. Sox9 transcriptionally controls the expression of cartilage-specific genes such as Col2a1 encoding Col II [15, 16]. Two other Sox family members, Sox5 (L-Sox5) and Sox6, cooperate with Sox9 to activate the chondrocyte-specific enhancers in the genes for ECM components [15, 17, 18]. Other transcription factors, such as members of the activating transcription factor (ATF)/cyclic AMP response element binding protein (CREB) family and the AP1 family member c-Fos, are required to maintain the proliferative capacity of early chondrocytes [19–21]. RhoA is a Rho GTPase, which functions as a regulator of cytoskeletal dynamics. RhoA signaling through its main effector ROCK inhibits chondrogenesis by suppressing the expression of Sox9 [22, 23].
A decrease in the expression and/or activity of the Sox proteins is required for the hypertrophic maturation of chondrocytes. In addition to the negative regulation by Sox proteins, other transcription factors, such as the Runt domain family members Runx2 and Runx3, function to promote chondrocyte hypertrophy [24]. Mice lacking both Runx2 and Runx3 lack hypertrophic chondrocytes [24]. Runx2 directly binds and activates the genes Ihh (Indian hedgehog), Col10a1 encoding Col X, and MMP13 [25–27]. A recent study using a doxycycline-inducible conditional knockout of Sox9 has revealed that Sox9 suppresses the expression of Runx2 and β-catenin signaling and thereby inhibits the progression from proliferation to prehypertrophy of chondrocytes [28]. Twist-1 is a basic helix-loop-helix-type transcription factor, which represses the expression of Runx2 in the perichondrium. Runx2 enhances the expression of fibroblast growth factor 18 (Fgf18) and exerts an indirect negative effect on chondrocyte maturation [29]. Osterix regulates the calcification and degradation of cartilaginous matrix through MMP13 expression in association with Runx2 [30].
MADS-box transcription factors Mef2c and Mef2d (myocyte enhancer factor 2c and 2d) are also involved in chondrocyte hypertrophy. Genetic deletion of Mef2c in endochondral cartilage impairs hypertrophic maturation, while the forced expression of a superactivating form of Mef2c resulted in precocious chondrocyte hypertrophy [31]. The activity of Runx2/3 and Mef2c/d is inhibited by the histone deacetylase HDAC4 [31–33]. Other transcription factors, such as Msx2, the AP1 family member Fra2, and FoxA family transcription factors, also positively control chondrocyte hypertrophy [34–37].
The developmental growth plate is hypoxic, especially in its interior. The transcription factor hypoxia-inducible factor I (HIF-1) is one of the major regulators of the hypoxic response in mammals. Genetic evidence obtained from mice lacking HIF-1α suggests its role in chondrocyte survival and the regulation of Vegf expression [38]. Conditional overexpression of VEGF164 in chondrocytes lacking HIF-1α rescued the phenotype of HIF-1α-deficient growth plate only partially, indicating VEGF-independent functions of HIF-1α in developing growth plate cartilage [39]. It is also reported that HIF-1α regulates collagen hydroxylation and secretion in developing cartilage [40].
Soluble mediators involved in chondrogenesis
Ihh, a member of the hedgehog family of signaling molecules, is expressed in prehypertrophic chondrocytes, and regulates the onset of hypertrophic differentiation through a negative feedback loop with parathyroid hormone-related protein (PTHrP). Ihh increases the expression of PTHrP in perichondrial cells and chondrocytes at the ends of long bones, which inhibits chondrocyte hypertrophy through its cognate receptor expressed in proliferating chondrocytes and keeps the cells in the proliferating stage [41]. Moreover, it is also reported that Ihh stimulates the proliferation and maturation of chondrocytes independently of PTHrP [42, 43]. Activation of Wnt and bone morphogenetic protein (BMP) signaling is suggested to be involved in the PTHrP-independent role of Ihh to regulate chondrocyte hypertrophy [44].
Fibroblast growth factors (FGFs) also play important roles in skeletogenesis by activating signaling through FGF receptors (FGFRs) [45]. Gain-of-function mutations in human FGFR3 result in chondrodysplasias and dwarfism [46–49]. As to FGF ligands, Fgf2 was first identified to be expressed in chondrocytes [50], and is also expressed in periosteal cells and osteoblasts [51, 52]. However, Fgf2-knockout mice demonstrate no defects in chondrogenesis [52, 53]. Fgf9 is also expressed in immature chondrocytes in mesenchymal condensation. In the perichondrium, the expression of Fgf7, Fgf8, Fgf9, Fgf17, and Fgf18 has been reported [54–58]. Evidence obtained from mouse models indicates profound role for Fgf18 in chondrogenesis [56, 57]. Fgf9 was also proven to regulate early hypertrophic chondrocyte differentiation and skeletal vascularization by the defects in chondrogenesis in Fgf9-knockout mice [59]. Among the FGFRs, Fgfr3 is expressed in chondrocytes undergoing mesenchymal condensation and proliferating chondrocytes, whereas Fgfr1 is expressed in prehypertrophic and hypertrophic chondrocytes [60–62]. Genetic and functional studies demonstrated that the signaling through FGFR3 negatively regulates the chondrocyte proliferation and differentiation [63–66]. The effects of FGFR3 in chondrogenesis are partly exerted by direct signaling in chondrocytes, and in part indirectly through the regulation of Ihh/PTHrP/BMP signaling [67]. In achondroplasia, constitutive activation of FGFR3 results in the activation of downstream pathways including STAT1 and ERK signaling [45]. The growth plates of mice lacking Fgf18 have a similar histology to those of Fgfr3-knockout mice, suggesting that FGF18 is a physiological ligand for FGFR3 in chondrocytes [56, 57].
C-type natriuretic peptide (CNP) controls cell behavior through the activation of two transmembrane receptors, NPR1 and NPR2 [68–70]. Since these receptors synthesize cyclic GMP in response to ligand binding, NPR1 and NPR2 are also called guanylyl cyclase A and B (GC-A and GC-B), respectively. CNP exerts its signal mainly through NPR2/GC-B. The importance of CNP signaling in chondrogenesis was shown by the severe dwarfism of CNP-knockout mice and the finding that CNP stimulated the longitudinal growth of cartilage in organ cultures [71, 72]. NPR2-null mice display a similar phenotype to CNP-knockout mice [73]. CNP promotes endochondral bone growth through several mechanisms, including the stimulation of chondrocyte proliferation, an acceleration of chondrocyte hypertrophy, and an increase of ECM production. In humans, loss-of-function mutations in the NPR2 gene cause acromesolic dysplasia, type Maroteaux, characterized by severe dwarfism [74]. We have recently identified a novel gain-of-function type mutation of the NPR2 gene in a family with overgrowth [75]. Such evidence indicates the critical role of CNP/NPR2 signaling in chondrogenesis both in humans and in mice.
The skeletal phenotype in CNP-deficient mice resembles that in cases of achondroplasia. Overexpression of CNP in cartilage rescued the skeletal phenotypes in a mouse model of achondroplasia, suggesting an intimate link between the FGF and CNP signaling [73]. CNP signaling inhibited the activation of the ERK pathway induced by FGF signaling, while FGF signaling blocked CNP-induced cGMP production in a MAPK-dependent manner [73]. In addition to the ERK pathway, recent studies demonstrated the possible involvement of the p38MAPK and PI3K/Akt pathways in the regulation of chondrocyte development by CNP [76]. CNP analogues are promising as new drugs for the dwarfism associated with skeletal dysplasias [77].
Studies have established the involvement of signaling mediated by epidermal growth factor receptor (EGFR) in chondrogenesis. Delayed primary endochondral ossification associated with defective osteoclast recruitment was reported in mice lacking EGFR [78]. Ubiquitous overexpression of betacellulin, a ligand for EGFR, resulted in defects in growth plates characterized by a smaller zone of hypertrophic chondrocytes in mice [79]. In addition, cartilage-specific inactivation of EGFR in mice as well as the administration of an EGFR-specific small-molecule inhibitor, gefitinib, into rats caused hypertrophic cartilage enlargement [80].
Other growth factors such as Wnts, BMPs, transforming growth factor-beta (TGF-β), insulin-like growth factors (IGFs), thyroid hormone, and connective tissue growth factor (CTGF) also play roles in chondrogenesis. There are several excellent review articles on their actions [81–83].
Regulation of chondrogenesis by the ECM
In the early stages of chondrogenesis, cell–cell interaction via adhesion molecules such as N-cadherin and N-CAM plays a role in cellular condensation and the subsequent chondrogenesis [84, 85]. As chondrocytes mature, they produce abundant matrix proteins, and the cell–matrix interactions come to have important roles. Integrins bind various extracellular components such as ECMs and other cell surface proteins [86]. The binding of ligands to integrins leads to the formation of focal adhesion complexes, and transduces the signaling from the ECM to intracellular effectors such as cytoskeleton [87, 88]. Integrins exist as dimers of an α subunit and a β subunit, and chondrocytes express several integrin subunits including fibronectin receptors (α5β1, αnβ3, αnβ5), a laminin receptor (α6β1) and collagen receptors (α1β1, α2β1, α10β1) [89–92]. The importance of β1 integrin-mediated signaling in chondrogenesis was demonstrated by the chondrodysplasia-like phenotype of chondrocyte-specific β1 integrin-knockout mice [93]. Growth plates of these mice exhibited unorganized proliferative columns and an abnormal cell shape due to the loss of adhesion to Col II. The chondrocytes isolated from these mice displayed reduced proliferation caused by a defect in G1/S transition and cytokinesis.
Inactivation of the α10 integrin gene also resulted in growth plate dysfunction, which was associated with an abnormal cell shape and increased apoptosis of chondrocytes [94]. On the other hand, knockout of the gene for α1 integrin resulted in osteoarthritis but no abnormalities in the growth plates, despite this gene’s predominant expression in hypertrophic chondrocytes [95].
Activation of integrin-mediated signaling triggers the formation of a complex consisting of multiple proteins, which regulate various cellular processes. Integrin-linked kinase is one of the components, and the knockout of its gene caused a chondrodysplasia-like phenotype resembling that of chondrocyte-specific β1 integrin-knockout mice [96]. In these mice, reduced proliferation of chondrocytes was the main cause for the skeletal phenotype, and the expression of chondrocyte-specific genes such as Col2a1 was comparable with that in wild-type mice.
CD44 is a cell surface glycoprotein that functions as a receptor for collagens and hyaluronan. It was reported that the blocking of CD44-hyaluronan binding on chondrocytes resulted in degradation of the cartilage matrix, suggesting a role for CD44 in cartilage homeostasis [97]. Annexin V acts as a receptor for collagen, specifically for a fragment of Col II in articular chondrocytes [98]. Antibodies against annexin V inhibited the binding of chondrocytes to Col II [99]. It is also suggested that annexin V is involved in regulating the apoptosis of growth plate chondrocytes [100].
Cartilage contains abundant proteoglycans. The sulfate transporter SLC26A2 is responsible for sulfate uptake by chondrocytes, and mutations in its gene lead to undersulfation of cartilaginous proteoglycans, resulting in a chondrodysplasia called diastrophic dysplasia. In dtd mice with a knock-in Slc26a2 mutation, the resulting undersulfation of glycosaminoglycans such as chondroitin destroys the articular surface and correlates with the rate of chondroitin synthesis across epiphyseal cartilage [101]. Chondroitin sulfate N-acetylgalactosaminyltransferase 1 (CSGalNAcT-1) is an enzyme that participates in the initiation of the biosynthesis of chondroitin sulfate. Mice lacking the gene encoding CSGalNAcT-1 exhibit shorter, disorganized chondrocyte columns in the growth plates with a rapid catabolism of aggrecan [102].
In addition to providing signals to cells by binding to integrins and other ECM receptors, ECM proteins regulate chondrogenesis through the binding, storage, and release of soluble factors. TGF-β is produced by chondrocytes as a high molecular weight macromolecule in association with latent TGF-β binding protein (LTBP), which functions in the storage of TGF-β in the ECM [103, 104]. Proteoglycans such as decorin, biglycan, and fibromodulin also regulate TGF-β activity by sequestering TGF-β in the ECM [105]. Most FGFs bind to heparan sulfate proteoglycans. They bind to cognate receptors in the context of heparan sulfate proteoglycans and evoke signaling into the cells. Genetic evidence obtained with mice lacking sulfate-modifying factor 1 (Sumf1) has suggested that the desulfation of proteoglycans regulates chondrocyte proliferation and differentiation by limiting FGF signaling [106].
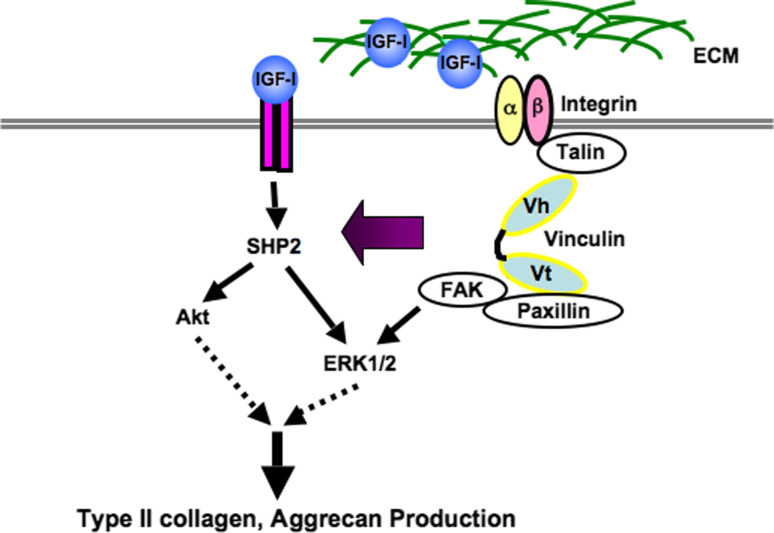
Signals from the ECM itself and those triggered by soluble mediators appear to interplay to regulate chondrogenesis. We have demonstrated that vinculin plays a role in chondrogenesis [107]. Vinculin is a component of multimolecular complexes which function in adhesion and/or signaling between the extracellular microenvironment and the cell, via integrins and cadherins. Impaired functioning of vinculin by knockdown in primary chondrocytes and organ cultures of metatarsal explants resulted in the reduced expression of Col2a1, aggrecan, Col10a1, and Runx2. In addition, knockdown of vinculin in the metatarsals abrogated IGF-I-induced growth, and inhibited the up-regulation of Col2a1 and aggrecan expression by IGF-I. These results suggest that vinculin regulates the expression of chondrocyte-specific genes via the integration of signaling from the ECM and soluble factors such as IGF- I (Fig. 2). It is also reported that cell adhesion via integrin regulates the activation of growth factor receptors. The orchestration of the signaling of soluble factors and the ECM should be considered a factor in the regeneration of cartilage [107].
Fig. 2.
Vinculin regulates the production of type II collagen and aggrecan by orchestrating the signal of extracellular matrix and that of IGF-1
Epigenetic and microRNA-mediated regulation of chondrogenesis
There is growing evidence that epigenetic and microRNA-mediated mechanisms play roles in chondrogenesis as well as in pathogenesis of osteoarthritis [108]. Histone modifications involving acetylation and deacetylation have an impact on the phenotype of chondrocytes. Among the histone deacetylases (HDACs), HDAC4 has been suggested to prevent premature chondrocyte hypertrophy by blocking the activity of Runx2, as described above [32]. HDAC1 and HDAC2 were shown to repress the expression of some cartilage-specific genes including Col2a1, and the Snail transcription factor was identified as a mediator of the repression [109]. The up-regulation of HDAC7 expression was suggested to contribute to the cartilage degradation by promoting the expression of MMP13 [110].
SIRT1 is a NAD+-dependent histone deacetylase, and enhances the expression of cartilage-specific ECM genes, such as Col2a1, by recruiting co-activators to the enhancer and promoter and facilitating Sox9-mediated transcription [111]. In addition, SIRT1 has been suggested to regulate chondrocyte apoptosis [112].
DNA methylation at CpG dinucleotides is commonly associated with gene repression. An in vitro study demonstrated that the induction of COL10A1 during the chondrogenesis of mesenchymal stem cells correlated with the demethylation of 2 CpG sites in the COL10A1 promoter [113]. It was reported that DNA methylation inversely correlated with the expression of cartilage-specific genes including COL9A1, but not catabolic genes such as MMP13, during fetal femur development in human [114]. Moreover, recent reports have suggested that the methylation of a specific CpG site inhibits the transactivation of MMP13 by transcription factors HIF-2α and CREB [115, 116].
MicroRNAs (miRNAs) are a class of ~22 nucleotide noncoding RNAs that regulate the expression of other genes at the posttranscriptional level. Knockout of Dicer, an enzyme required for miRNA synthesis, led to severe skeletal growth defects caused by decreased chondrocyte proliferation and accelerated differentiation in mice, indicating the critical roles of miRNAs in chondrogenesis [117]. Specific miRNAs have been identified to have roles in chondrocyte differentiation. miR-199a was shown to be responsive to BMP and to regulate chondrogenesis by directly targeting Smad1 [118]. Mice lacking miR-140 showed a mild skeletal phenotype with a short stature and age-related OA-like changes associated with the elevated expression of ADAMTS-5, suggesting that miR-140 regulates cartilage development and homeostasis [119]. miR-145 was reported to directly target Sox9 and regulate chondrogenic differentiation of mesenchymal stem cells [120]. miR-675, whose production is positively regulated by Sox9, increases the expression of COL2A1 in human articular chondrocytes [121]. These findings have established the importance of miRNA-mediated regulation in cartilage development.
Conclusion
The development of growth plate cartilage is a complex process, regulated by transcription factors, soluble factors, cell–cell and cell–matrix interactions, and epigenetic factors. These factors interplay to control the proliferation and differentiation of chondrocytes. Failure in the development of growth plate cartilage is often associated with skeletal dysplasias, for which currently there is no effective treatment. Understanding the mechanisms regulating chondrogenesis may lead to new therapeutic drugs for these diseases, such as CNP analogues.
References
- 1.Wagner EF, Karsenty G. Genetic control of skeletal development. Curr Opin Genet Dev. 2001;11:527–532. doi: 10.1016/s0959-437x(00)00228-8. [DOI] [PubMed] [Google Scholar]
- 2.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 3.Lefebvre V, Bhattaram P. Vertebrate skeletogenesis. Curr Top Dev Biol. 2010;90:291–317. doi: 10.1016/S0070-2153(10)90008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beier F. Cell-cycle control and the cartilage growth plate. J Cell Physiol. 2005;202:1–8. doi: 10.1002/jcp.20111. [DOI] [PubMed] [Google Scholar]
- 5.Burdan F, Szumilo J, Korobowicz A, Farooquee R, Patel S, Patel A, Dave A, Szumilo M, Solecki M, Klepacz R, Dudka J. Morphology and physiology of the epiphyseal growth plate. Folia Histochem Cytobiol. 2009;47:5–16. doi: 10.2478/v10042-009-0007-1. [DOI] [PubMed] [Google Scholar]
- 6.Zelzer E, Mamluk R, Ferrara N, Johnson RS, Schipani E, Olsen BR. VEGFA is necessary for chondrocyte survival during bone development. Development. 2004;131:2161–2171. doi: 10.1242/dev.01053. [DOI] [PubMed] [Google Scholar]
- 7.Stickens D, Behonick DJ, Ortega N, Heyer B, Hartenstein B, Yu Y, Fosang AJ, Schorpp-Kistner M, Angel P, Werb Z. Altered endochondral bone development in matrix metalloproteinase 13-deficient mice. Development. 2004;131:5883–5895. doi: 10.1242/dev.01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartmann C. Transcriptional networks controlling skeletal development. Curr Opin Genet Dev. 2009;19:437–443. doi: 10.1016/j.gde.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Nishimura R, Hata K, Matsubara T, Wakabayashi M, Yoneda T. Regulation of bone and cartilage development by network between BMP signalling and transcription factors. J Biochem. 2012;151:247–254. doi: 10.1093/jb/mvs004. [DOI] [PubMed] [Google Scholar]
- 10.Woods A, Wang G, Beier F. Regulation of chondrocyte differentiation by the actin cytoskeleton and adhesive interactions. J Cell Physiol. 2007;213:1–8. doi: 10.1002/jcp.21110. [DOI] [PubMed] [Google Scholar]
- 11.Warman ML, Cormier-Daire V, Hall C, Krakow D, Lachman R, LeMerrer M, Mortier G, Mundlos S, Nishimura G, Rimoin DL, Robertson S, Savarirayan R, Sillence D, Spranger J, Unger S, Zabel B, Superti-Furga A. Nosology and classification of genetic skeletal disorders: 2010 revision. Am J Med Genet A. 2011;155A:943–968. doi: 10.1002/ajmg.a.33909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster JW, Dominguez-Steglich MA, Guioli S, Kwok C, Weller PA, Stevanovic M, Weissenbach J, Mansour S, Young ID, Goodfellow PN, et al. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature. 1994;372:525–530. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- 13.Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, Hustert E, Wolf U, Tommerup N, Schempp W, Scherer G. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 14.Akiyama H, Lefebvre V. Unraveling the transcriptional regulatory machinery in chondrogenesis. J Bone Miner Metab. 2011;29:390–395. doi: 10.1007/s00774-011-0273-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefebvre V, Li P, de Crombrugghe B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 1998;17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol. 1997;17:2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han Y, Lefebvre V. L-Sox5 and Sox6 drive expression of the aggrecan gene in cartilage by securing binding of Sox9 to a far-upstream enhancer. Mol Cell Biol. 2008;28:4999–5013. doi: 10.1128/MCB.00695-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long F, Zhang XM, Karp S, Yang Y, McMahon AP. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development. 2001;128:5099–5108. doi: 10.1242/dev.128.24.5099. [DOI] [PubMed] [Google Scholar]
- 20.Vale-Cruz DS, Ma Q, Syme J, LuValle PA. Activating transcription factor-2 affects skeletal growth by modulating pRb gene expression. Mech Dev. 2008;125:843–856. doi: 10.1016/j.mod.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 21.Wang ZQ, Ovitt C, Grigoriadis AE, Mohle-Steinlein U, Ruther U, Wagner EF. Bone and haematopoietic defects in mice lacking c-fos. Nature. 1992;360:741–745. doi: 10.1038/360741a0. [DOI] [PubMed] [Google Scholar]
- 22.Woods A, Wang G, Beier F. RhoA/ROCK signaling regulates Sox9 expression and actin organization during chondrogenesis. J Biol Chem. 2005;280:11626–11634. doi: 10.1074/jbc.M409158200. [DOI] [PubMed] [Google Scholar]
- 23.Woods A, Beier F. RhoA/ROCK signaling regulates chondrogenesis in a context-dependent manner. J Biol Chem. 2006;281:13134–13140. doi: 10.1074/jbc.M509433200. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida CA, Komori T. Role of Runx proteins in chondrogenesis. Crit Rev Eukaryot Gene Expr. 2005;15:243–254. doi: 10.1615/critreveukargeneexpr.v15.i3.60. [DOI] [PubMed] [Google Scholar]
- 25.Zheng Q, Zhou G, Morello R, Chen Y, Garcia-Rojas X, Lee B. Type X collagen gene regulation by Runx2 contributes directly to its hypertrophic chondrocyte-specific expression in vivo. J Cell Biol. 2003;162:833–842. doi: 10.1083/jcb.200211089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida CA, Yamamoto H, Fujita T, Furuichi T, Ito K, Inoue K, Yamana K, Zanma A, Takada K, Ito Y, Komori T. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18:952–963. doi: 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selvamurugan N, Kwok S, Partridge NC. Smad3 interacts with JunB and Cbfa1/Runx2 for transforming growth factor-beta1-stimulated collagenase-3 expression in human breast cancer cells. J Biol Chem. 2004;279:27764–27773. doi: 10.1074/jbc.M312870200. [DOI] [PubMed] [Google Scholar]
- 28.Dy P, Wang W, Bhattaram P, Wang Q, Wang L, Ballock RT, Lefebvre V. Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev Cell. 2012;22:597–609. doi: 10.1016/j.devcel.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinoi E, Bialek P, Chen YT, Rached MT, Groner Y, Behringer RR, Ornitz DM, Karsenty G. Runx2 inhibits chondrocyte proliferation and hypertrophy through its expression in the perichondrium. Genes Dev. 2006;20:2937–2942. doi: 10.1101/gad.1482906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimura R, Wakabayashi M, Hata K, Matsubara T, Honma S, Wakisaka S, Kiyonari H, Shioi G, Yamaguchi A, Tsumaki N, Akiyama H, Yoneda T. Osterix regulates calcification and degradation of chondrogenic matrices through matrix metalloproteinase 13 (MMP13) expression in association with transcription factor Runx2 during endochondral ossification. J Biol Chem. 2012;287:33179–33190. doi: 10.1074/jbc.M111.337063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12:377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, Karsenty G, Olson EN. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 33.Kozhemyakina E, Cohen T, Yao TP, Lassar AB. Parathyroid hormone-related peptide represses chondrocyte hypertrophy through a protein phosphatase 2A/histone deacetylase 4/MEF2 pathway. Mol Cell Biol. 2009;29:5751–5762. doi: 10.1128/MCB.00415-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satokata I, Ma L, Ohshima H, Bei M, Woo I, Nishizawa K, Maeda T, Takano Y, Uchiyama M, Heaney S, Peters H, Tang Z, Maxson R, Maas R. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat Genet. 2000;24:391–395. doi: 10.1038/74231. [DOI] [PubMed] [Google Scholar]
- 35.Amano K, Ichida F, Sugita A, Hata K, Wada M, Takigawa Y, Nakanishi M, Kogo M, Nishimura R, Yoneda T. MSX2 stimulates chondrocyte maturation by controlling Ihh expression. J Biol Chem. 2008;283:29513–29521. doi: 10.1074/jbc.M803681200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karreth F, Hoebertz A, Scheuch H, Eferl R, Wagner EF. The AP1 transcription factor Fra2 is required for efficient cartilage development. Development. 2004;131:5717–5725. doi: 10.1242/dev.01414. [DOI] [PubMed] [Google Scholar]
- 37.Ionescu A, Kozhemyakina E, Nicolae C, Kaestner KH, Olsen BR, Lassar AB. FoxA family members are crucial regulators of the hypertrophic chondrocyte differentiation program. Dev Cell. 2012;22:927–939. doi: 10.1016/j.devcel.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schipani E, Ryan HE, Didrickson S, Kobayashi T, Knight M, Johnson RS. Hypoxia in cartilage: HIF-1alpha is essential for chondrocyte growth arrest and survival. Genes Dev. 2001;15:2865–2876. doi: 10.1101/gad.934301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maes C, Araldi E, Haigh K, Khatri R, Van Looveren R, Giaccia AJ, Haigh JJ, Carmeliet G, Schipani E. VEGF-independent cell-autonomous functions of HIF-1alpha regulating oxygen consumption in fetal cartilage are critical for chondrocyte survival. J Bone Miner Res. 2012;27:596–609. doi: 10.1002/jbmr.1487. [DOI] [PubMed] [Google Scholar]
- 40.Bentovim L, Amarilio R, Zelzer E. HIF1alpha is a central regulator of collagen hydroxylation and secretion under hypoxia during bone development. Development. 2012;139:4473–4483. doi: 10.1242/dev.083881. [DOI] [PubMed] [Google Scholar]
- 41.Kronenberg HM. PTHrP and skeletal development. Ann N Y Acad Sci. 2006;1068:1–13. doi: 10.1196/annals.1346.002. [DOI] [PubMed] [Google Scholar]
- 42.St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobayashi T, Soegiarto DW, Yang Y, Lanske B, Schipani E, McMahon AP, Kronenberg HM. Indian hedgehog stimulates periarticular chondrocyte differentiation to regulate growth plate length independently of PTHrP. J Clin Invest. 2005;115:1734–1742. doi: 10.1172/JCI24397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mak KK, Kronenberg HM, Chuang PT, Mackem S, Yang Y. Indian hedgehog signals independently of PTHrP to promote chondrocyte hypertrophy. Development. 2008;135:1947–1956. doi: 10.1242/dev.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ornitz DM. FGF signaling in the developing endochondral skeleton. Cytokine Growth Factor Rev. 2005;16:205–213. doi: 10.1016/j.cytogfr.2005.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rousseau F, Bonaventure J, Legeai-Mallet L, Pelet A, Rozet JM, Maroteaux P, Le Merrer M, Munnich A. Mutations in the gene encoding fibroblast growth factor receptor-3 in achondroplasia. Nature. 1994;371:252–254. doi: 10.1038/371252a0. [DOI] [PubMed] [Google Scholar]
- 47.Shiang R, Thompson LM, Zhu YZ, Church DM, Fielder TJ, Bocian M, Winokur ST, Wasmuth JJ. Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell. 1994;78:335–342. doi: 10.1016/0092-8674(94)90302-6. [DOI] [PubMed] [Google Scholar]
- 48.Bellus GA, McIntosh I, Smith EA, Aylsworth AS, Kaitila I, Horton WA, Greenhaw GA, Hecht JT, Francomano CA. A recurrent mutation in the tyrosine kinase domain of fibroblast growth factor receptor 3 causes hypochondroplasia. Nat Genet. 1995;10:357–359. doi: 10.1038/ng0795-357. [DOI] [PubMed] [Google Scholar]
- 49.Tavormina PL, Shiang R, Thompson LM, Zhu YZ, Wilkin DJ, Lachman RS, Wilcox WR, Rimoin DL, Cohn DH, Wasmuth JJ. Thanatophoric dysplasia (types I and II) caused by distinct mutations in fibroblast growth factor receptor 3. Nat Genet. 1995;9:321–328. doi: 10.1038/ng0395-321. [DOI] [PubMed] [Google Scholar]
- 50.Sullivan R, Klagsbrun M. Purification of cartilage-derived growth factor by heparin affinity chromatography. J Biol Chem. 1985;260:2399–2403. [PubMed] [Google Scholar]
- 51.Hurley MM, Abreu C, Gronowicz G, Kawaguchi H, Lorenzo J. Expression and regulation of basic fibroblast growth factor mRNA levels in mouse osteoblastic MC3T3-E1 cells. J Biol Chem. 1994;269:9392–9396. [PubMed] [Google Scholar]
- 52.Montero A, Okada Y, Tomita M, Ito M, Tsurukami H, Nakamura T, Doetschman T, Coffin JD, Hurley MM. Disruption of the fibroblast growth factor-2 gene results in decreased bone mass and bone formation. J Clin Invest. 2000;105:1085–1093. doi: 10.1172/JCI8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colvin JS, Feldman B, Nadeau JH, Goldfarb M, Ornitz DM. Genomic organization and embryonic expression of the mouse fibroblast growth factor 9 gene. Dev Dyn. 1999;216:72–88. doi: 10.1002/(SICI)1097-0177(199909)216:1<72::AID-DVDY9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 54.Finch PW, Cunha GR, Rubin JS, Wong J, Ron D. Pattern of keratinocyte growth factor and keratinocyte growth factor receptor expression during mouse fetal development suggests a role in mediating morphogenetic mesenchymal-epithelial interactions. Dev Dyn. 1995;203:223–240. doi: 10.1002/aja.1002030210. [DOI] [PubMed] [Google Scholar]
- 55.Mason IJ, Fuller-Pace F, Smith R, Dickson C. FGF-7 (keratinocyte growth factor) expression during mouse development suggests roles in myogenesis, forebrain regionalisation and epithelial-mesenchymal interactions. Mech Dev. 1994;45:15–30. doi: 10.1016/0925-4773(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 56.Ohbayashi N, Shibayama M, Kurotaki Y, Imanishi M, Fujimori T, Itoh N, Takada S. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 2002;16:870–879. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Z, Xu J, Colvin JS, Ornitz DM. Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes Dev. 2002;16:859–869. doi: 10.1101/gad.965602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu J, Lawshe A, MacArthur CA, Ornitz DM. Genomic structure, mapping, activity and expression of fibroblast growth factor 17. Mech Dev. 1999;83(1–2):165–178. doi: 10.1016/s0925-4773(99)00034-9. [DOI] [PubMed] [Google Scholar]
- 59.Hung IH, Yu K, Lavine KJ, Ornitz DM. FGF9 regulates early hypertrophic chondrocyte differentiation and skeletal vascularization in the developing stylopod. Dev Biol. 2007;307:300–313. doi: 10.1016/j.ydbio.2007.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 61.Peters KG, Werner S, Chen G, Williams LT. Two FGF receptor genes are differentially expressed in epithelial and mesenchymal tissues during limb formation and organogenesis in the mouse. Development. 1992;114:233–243. doi: 10.1242/dev.114.1.233. [DOI] [PubMed] [Google Scholar]
- 62.Peters K, Ornitz D, Werner S, Williams L. Unique expression pattern of the FGF receptor 3 gene during mouse organogenesis. Dev Biol. 1993;155:423–430. doi: 10.1006/dbio.1993.1040. [DOI] [PubMed] [Google Scholar]
- 63.Chen L, Adar R, Yang X, Monsonego EO, Li C, Hauschka PV, Yayon A, Deng CX. Gly369Cys mutation in mouse FGFR3 causes achondroplasia by affecting both chondrogenesis and osteogenesis. J Clin Invest. 1999;104:1517–1525. doi: 10.1172/JCI6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen L, Li C, Qiao W, Xu X, Deng C. A Ser(365)→Cys mutation of fibroblast growth factor receptor 3 in mouse downregulates Ihh/PTHrP signals and causes severe achondroplasia. Hum Mol Genet. 2001;10:457–465. doi: 10.1093/hmg/10.5.457. [DOI] [PubMed] [Google Scholar]
- 65.Dailey L, Laplantine E, Priore R, Basilico C. A network of transcriptional and signaling events is activated by FGF to induce chondrocyte growth arrest and differentiation. J Cell Biol. 2003;161:1053–1066. doi: 10.1083/jcb.200302075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li C, Chen L, Iwata T, Kitagawa M, Fu XY, Deng CX. A Lys644Glu substitution in fibroblast growth factor receptor 3 (FGFR3) causes dwarfism in mice by activation of STATs and ink4 cell cycle inhibitors. Hum Mol Genet. 1999;8:35–44. doi: 10.1093/hmg/8.1.35. [DOI] [PubMed] [Google Scholar]
- 67.Minina E, Kreschel C, Naski MC, Ornitz DM, Vortkamp A. Interaction of FGF, Ihh/Pthlh, and BMP signaling integrates chondrocyte proliferation and hypertrophic differentiation. Dev Cell. 2002;3:439–449. doi: 10.1016/s1534-5807(02)00261-7. [DOI] [PubMed] [Google Scholar]
- 68.Anand-Srivastava MB. Natriuretic peptide receptor-C signaling and regulation. Peptides. 2005;26:1044–1059. doi: 10.1016/j.peptides.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 69.Baxter GF. The natriuretic peptides. Basic Res Cardiol. 2004;99:71–75. doi: 10.1007/s00395-004-0457-8. [DOI] [PubMed] [Google Scholar]
- 70.Teixeira CC, Agoston H, Beier F. Nitric oxide, C-type natriuretic peptide and cGMP as regulators of endochondral ossification. Dev Biol. 2008;319:171–178. doi: 10.1016/j.ydbio.2008.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chusho H, Tamura N, Ogawa Y, Yasoda A, Suda M, Miyazawa T, Nakamura K, Nakao K, Kurihara T, Komatsu Y, Itoh H, Tanaka K, Saito Y, Katsuki M. Dwarfism and early death in mice lacking C-type natriuretic peptide. Proc Natl Acad Sci USA. 2001;98:4016–4021. doi: 10.1073/pnas.071389098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yasoda A, Ogawa Y, Suda M, Tamura N, Mori K, Sakuma Y, Chusho H, Shiota K, Tanaka K, Nakao K. Natriuretic peptide regulation of endochondral ossification. Evidence for possible roles of the C-type natriuretic peptide/guanylyl cyclase-B pathway. J Biol Chem. 1998;273:11695–11700. doi: 10.1074/jbc.273.19.11695. [DOI] [PubMed] [Google Scholar]
- 73.Yasoda A, Komatsu Y, Chusho H, Miyazawa T, Ozasa A, Miura M, Kurihara T, Rogi T, Tanaka S, Suda M, Tamura N, Ogawa Y, Nakao K. Overexpression of CNP in chondrocytes rescues achondroplasia through a MAPK-dependent pathway. Nat Med. 2004;10:80–86. doi: 10.1038/nm971. [DOI] [PubMed] [Google Scholar]
- 74.Bartels CF, Bukulmez H, Padayatti P, Rhee DK, van Ravenswaaij-Arts C, Pauli RM, Mundlos S, Chitayat D, Shih LY, Al-Gazali LI, Kant S, Cole T, Morton J, Cormier-Daire V, Faivre L, Lees M, Kirk J, Mortier GR, Leroy J, Zabel B, Kim CA, Crow Y, Braverman NE, van den Akker F, Warman ML. Mutations in the transmembrane natriuretic peptide receptor NPR-B impair skeletal growth and cause acromesomelic dysplasia, type Maroteaux. Am J Hum Genet. 2004;75:27–34. doi: 10.1086/422013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miura K, Namba N, Fujiwara M, Ohata Y, Ishida H, Kitaoka T, Kubota T, Hirai H, Higuchi C, Tsumaki N, Yoshikawa H, Sakai N, Michigami T, Ozono K. An overgrowth disorder associated with excessive production of cGMP due to a gain-of-function mutation of the natriuretic peptide receptor 2 gene. PLoS ONE. 2012;7:e42180. doi: 10.1371/journal.pone.0042180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ulici V, Hoenselaar KD, Gillespie JR, Beier F. The PI3K pathway regulates endochondral bone growth through control of hypertrophic chondrocyte differentiation. BMC Dev Biol. 2008;8:40. doi: 10.1186/1471-213X-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lorget F, Kaci N, Peng J, Benoist-Lasselin C, Mugniery E, Oppeneer T, Wendt DJ, Bell SM, Bullens S, Bunting S, Tsuruda LS, O’Neill CA, Di Rocco F, Munnich A, Legeai-Mallet L. Evaluation of the therapeutic potential of a CNP analog in a Fgfr3 mouse model recapitulating achondroplasia. Am J Hum Genet. 2012;91:1108–1114. doi: 10.1016/j.ajhg.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang K, Yamamoto H, Chin JR, Werb Z, Vu TH. Epidermal growth factor receptor-deficient mice have delayed primary endochondral ossification because of defective osteoclast recruitment. J Biol Chem. 2004;279:53848–53856. doi: 10.1074/jbc.M403114200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schneider MR, Mayer-Roenne B, Dahlhoff M, Proell V, Weber K, Wolf E, Erben RG. High cortical bone mass phenotype in betacellulin transgenic mice is EGFR dependent. J Bone Miner Res. 2009;24:455–467. doi: 10.1359/jbmr.081202. [DOI] [PubMed] [Google Scholar]
- 80.Zhang X, Siclari VA, Lan S, Zhu J, Koyama E, Dupuis HL, Enomoto-Iwamoto M, Beier F, Qin L. The critical role of the epidermal growth factor receptor in endochondral ossification. J Bone Miner Res. 2011;26:2622–2633. doi: 10.1002/jbmr.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pogue R, Lyons K. BMP signaling in the cartilage growth plate. Curr Top Dev Biol. 2006;76:1–48. doi: 10.1016/S0070-2153(06)76001-X. [DOI] [PubMed] [Google Scholar]
- 82.Mackie EJ, Ahmed YA, Tatarczuch L, Chen KS, Mirams M. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int J Biochem Cell Biol. 2008;40:46–62. doi: 10.1016/j.biocel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 83.Olney RC, Mougey EB. Expression of the components of the insulin-like growth factor axis across the growth-plate. Mol Cell Endocrinol. 1999;156:63–71. doi: 10.1016/s0303-7207(99)00144-6. [DOI] [PubMed] [Google Scholar]
- 84.Oberlender SA, Tuan RS. Expression and functional involvement of N-cadherin in embryonic limb chondrogenesis. Development. 1994;120:177–187. doi: 10.1242/dev.120.1.177. [DOI] [PubMed] [Google Scholar]
- 85.Tavella S, Raffo P, Tacchetti C, Cancedda R, Castagnola P. N-CAM and N-cadherin expression during in vitro chondrogenesis. Exp Cell Res. 1994;215:354–362. doi: 10.1006/excr.1994.1352. [DOI] [PubMed] [Google Scholar]
- 86.ffrench-Constant C, Colognato H. Integrins: versatile integrators of extracellular signals. Trends Cell Biol. 2004;14:678–686. doi: 10.1016/j.tcb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 87.Clancy RM, Rediske J, Tang X, Nijher N, Frenkel S, Philips M, Abramson SB. Outside-in signaling in the chondrocyte. Nitric oxide disrupts fibronectin-induced assembly of a subplasmalemmal actin/rho A/focal adhesion kinase signaling complex. J Clin Invest. 1997;100:1789–1796. doi: 10.1172/JCI119706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Legate KR, Wickstrom SA, Fassler R. Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev. 2009;23:397–418. doi: 10.1101/gad.1758709. [DOI] [PubMed] [Google Scholar]
- 89.Shakibaei M. Integrin expression on epiphyseal mouse chondrocytes in monolayer culture. Histol Histopathol. 1995;10:339–349. [PubMed] [Google Scholar]
- 90.Loeser RF. Chondrocyte integrin expression and function. Biorheology. 2000;37:109–116. [PubMed] [Google Scholar]
- 91.Loeser RF. Integrins and cell signaling in chondrocytes. Biorheology. 2002;39:119–124. [PubMed] [Google Scholar]
- 92.Egerbacher M, Haeusler G. Integrins in growth plate cartilage. Pediatr Endocrinol Rev. 2003;1:2–8. [PubMed] [Google Scholar]
- 93.Aszodi A, Hunziker EB, Brakebusch C, Fassler R. Beta1 integrins regulate chondrocyte rotation, G1 progression, and cytokinesis. Genes Dev. 2003;17:2465–2479. doi: 10.1101/gad.277003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bengtsson T, Aszodi A, Nicolae C, Hunziker EB, Lundgren-Akerlund E, Fassler R. Loss of alpha10beta1 integrin expression leads to moderate dysfunction of growth plate chondrocytes. J Cell Sci. 2005;118:929–936. doi: 10.1242/jcs.01678. [DOI] [PubMed] [Google Scholar]
- 95.Zemmyo M, Meharra EJ, Kuhn K, Creighton-Achermann L, Lotz M. Accelerated, aging-dependent development of osteoarthritis in alpha1 integrin-deficient mice. Arthritis Rheum. 2003;48:2873–2880. doi: 10.1002/art.11246. [DOI] [PubMed] [Google Scholar]
- 96.Terpstra L, Prud’homme J, Arabian A, Takeda S, Karsenty G, Dedhar S, St-Arnaud R. Reduced chondrocyte proliferation and chondrodysplasia in mice lacking the integrin-linked kinase in chondrocytes. J Cell Biol. 2003;162:139–148. doi: 10.1083/jcb.200302066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nicoll SB, Barak O, Csoka AB, Bhatnagar RS, Stern R. Hyaluronidases and CD44 undergo differential modulation during chondrogenesis. Biochem Biophys Res Commun. 2002;292:819–825. doi: 10.1006/bbrc.2002.6697. [DOI] [PubMed] [Google Scholar]
- 98.Lucic D, Mollenhauer J, Kilpatrick KE, Cole AA. N-telopeptide of type II collagen interacts with annexin V on human chondrocytes. Connect Tissue Res. 2003;44:225–239. [PubMed] [Google Scholar]
- 99.Reid DL, Aydelotte MB, Mollenhauer J. Cell attachment, collagen binding, and receptor analysis on bovine articular chondrocytes. J Orthop Res. 2000;18:364–373. doi: 10.1002/jor.1100180307. [DOI] [PubMed] [Google Scholar]
- 100.Kirsch T. Annexins—their role in cartilage mineralization. Front Biosci. 2005;10:576–581. doi: 10.2741/1553. [DOI] [PubMed] [Google Scholar]
- 101.Mertz EL, Facchini M, Pham AT, Gualeni B, De Leonardis F, Rossi A, Forlino A. Matrix disruptions, growth, and degradation of cartilage with impaired sulfation. J Biol Chem. 2012;287:22030–22042. doi: 10.1074/jbc.M110.116467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sato T, Kudo T, Ikehara Y, Ogawa H, Hirano T, Kiyohara K, Hagiwara K, Togayachi A, Ema M, Takahashi S, Kimata K, Watanabe H, Narimatsu H. Chondroitin sulfate N-acetylgalactosaminyltransferase 1 is necessary for normal endochondral ossification and aggrecan metabolism. J Biol Chem. 2011;286:5803–5812. doi: 10.1074/jbc.M110.159244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pedrozo HA, Schwartz Z, Gomez R, Ornoy A, Xin-Sheng W, Dallas SL, Bonewald LF, Dean DD, Boyan BD. Growth plate chondrocytes store latent transforming growth factor (TGF)-beta 1 in their matrix through latent TGF-beta 1 binding protein-1. J Cell Physiol. 1998;177:343–354. doi: 10.1002/(SICI)1097-4652(199811)177:2<343::AID-JCP16>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 104.Pedrozo HA, Schwartz Z, Mokeyev T, Ornoy A, Xin-Sheng W, Bonewald LF, Dean DD, Boyan BD. Vitamin D3 metabolites regulate LTBP1 and latent TGF-beta1 expression and latent TGF-beta1 incorporation in the extracellular matrix of chondrocytes. J Cell Biochem. 1999;72:151–165. [PubMed] [Google Scholar]
- 105.Hildebrand A, Romaris M, Rasmussen LM, Heinegard D, Twardzik DR, Border WA, Ruoslahti E. Interaction of the small interstitial proteoglycans biglycan, decorin and fibromodulin with transforming growth factor beta. Biochem J. 1994;302:527–534. doi: 10.1042/bj3020527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Settembre C, Arteaga-Solis E, McKee MD, de Pablo R, Al Awqati Q, Ballabio A, Karsenty G. Proteoglycan desulfation determines the efficiency of chondrocyte autophagy and the extent of FGF signaling during endochondral ossification. Genes Dev. 2008;22:2645–2650. doi: 10.1101/gad.1711308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Koshimizu T, Kawai M, Kondou H, Tachikawa K, Sakai N, Ozono K, Michigami T. Vinculin functions as regulator of chondrogenesis. J Biol Chem. 2010;287:15760–15775. doi: 10.1074/jbc.M111.308072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Goldring MB, Marcu KB. Epigenomic and microRNA-mediated regulation in cartilage development, homeostasis, and osteoarthritis. Trends Mol Med. 2012;18:109–118. doi: 10.1016/j.molmed.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hong S, Derfoul A, Pereira-Mouries L, Hall DJ. A novel domain in histone deacetylase 1 and 2 mediates repression of cartilage-specific genes in human chondrocytes. FASEB J. 2009;23:3539–3552. doi: 10.1096/fj.09-133215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Higashiyama R, Miyaki S, Yamashita S, Yoshitaka T, Lindman G, Ito Y, Sasho T, Takahashi K, Lotz M, Asahara H. Correlation between MMP-13 and HDAC7 expression in human knee osteoarthritis. Mod Rheumatol. 2010;20:11–17. doi: 10.1007/s10165-009-0224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dvir-Ginzberg M, Gagarina V, Lee EJ, Hall DJ. Regulation of cartilage-specific gene expression in human chondrocytes by SirT1 and nicotinamide phosphoribosyltransferase. J Biol Chem. 2008;283:36300–36310. doi: 10.1074/jbc.M803196200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gabay O, Oppenhiemer H, Meir H, Zaal K, Sanchez C, Dvir-Ginzberg M. Increased apoptotic chondrocytes in articular cartilage from adult heterozygous SirT1 mice. Ann Rheum Dis. 2012;71:613–616. doi: 10.1136/ard.2011.200504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zimmermann P, Boeuf S, Dickhut A, Boehmer S, Olek S, Richter W. Correlation of COL10A1 induction during chondrogenesis of mesenchymal stem cells with demethylation of two CpG sites in the COL10A1 promoter. Arthritis Rheum. 2008;58:2743–2753. doi: 10.1002/art.23736. [DOI] [PubMed] [Google Scholar]
- 114.de Andres MC, Kingham E, Imagawa K, Gonzalez A, Roach HI, Wilson DI, Oreffo RO. Epigenetic regulation during fetal femur development: DNA methylation matters. PLoS ONE. 2013;8:e54957. doi: 10.1371/journal.pone.0054957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bui C, Barter MJ, Scott JL, Xu Y, Galler M, Reynard LN, Rowan AD, Young DA. cAMP response element-binding (CREB) recruitment following a specific CpG demethylation leads to the elevated expression of the matrix metalloproteinase 13 in human articular chondrocytes and osteoarthritis. FASEB J. 2012;26:3000–3011. doi: 10.1096/fj.12-206367. [DOI] [PubMed] [Google Scholar]
- 116.Hashimoto K, Otero M, Imagawa K, de Andres MC, Coico JM, Roach HI, Oreffo RO, Marcu KB, Goldring MB. Regulated transcription of human matrix metalloproteinase 13 (MMP13) and interleukin-1beta (IL1B) genes in chondrocytes depends on methylation of specific proximal promoter CpG sites. J Biol Chem. 2013;288:10061–10072. doi: 10.1074/jbc.M112.421156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kobayashi T, Lu J, Cobb BS, Rodda SJ, McMahon AP, Schipani E, Merkenschlager M, Kronenberg HM. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci USA. 2008;105:1949–1954. doi: 10.1073/pnas.0707900105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lin EA, Kong L, Bai XH, Luan Y, Liu CJ. miR-199a, a bone morphogenic protein 2-responsive MicroRNA, regulates chondrogenesis via direct targeting to Smad1. J Biol Chem. 2009;284:11326–11335. doi: 10.1074/jbc.M807709200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Miyaki S, Sato T, Inoue A, Otsuki S, Ito Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S, Takada S, Lotz MK, Ueno-Kudo H, Asahara H. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24:1173–1185. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang B, Guo H, Zhang Y, Chen L, Ying D, Dong S. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS ONE. 2011;6:e21679. doi: 10.1371/journal.pone.0021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dudek KA, Lafont JE, Martinez-Sanchez A, Murphy CL. Type II collagen expression is regulated by tissue-specific miR-675 in human articular chondrocytes. J Biol Chem. 2010;285:24381–24387. doi: 10.1074/jbc.M110.111328. [DOI] [PMC free article] [PubMed] [Google Scholar]