Abstract
Formation of metastasis is the most important and lethal step in cancer progression. Circulating and disseminated cancer cells (CTCs/DTCs) in blood and bone marrow are considered as potential metastases-inducing cells. Their detection and characterization has, therefore, become a field of major interest in translational and clinical research in oncology. The main strategy to detect these cells relies thus far on the epithelial characteristics of carcinoma cells and epithelial cell adhesion molecule (EpCAM) represents the most commonly used epithelial marker to capture CTCs/DTCs. Recent data, however, demonstrated a dynamic expression of EpCAM associated with a loss during epithelial-to-mesenchymal transition. The present review summarizes the potential mechanisms and reasons for a dynamic expression of EpCAM.
Keywords: EpCAM, CTCs, DTCs, EMT, MET, Metastases
EpCAM provides access to (early) systemic cancer
Over the last decade, we have learned that dissemination of metastatic precursor cells can occur early during tumor development and that the evolutionary dynamics underlying systemic progression can be very complex, resulting in heterogeneous and rather inefficient metastatic potential [1–4]. Concomitantly, the introduction of rationally designed molecular therapeutics addressing specific aberrant pathways in cancer cells has led to the need for biomarkers for effective patient selection and accurate assessment of pharmacodynamics [5–7]. To address these needs and to enable more successful anti-metastatic therapies, early detection, precise monitoring and molecular characterization of (early) systemic cancer would be required. In this context circulating tumor cells (CTCs) gained much attention since they might provide direct access to systemic cancer.
CTCs are cancer cells that have left the primary tumor site and that can be detected as rare events within the blood circulation, correlated with progressive disease and poor outcome in metastatic and non-metastatic cancer, suggesting an active role for these cells in driving metastatic disease [8–10]. Despite enormous recent technical advancements, the detection and isolation of CTCs remain to be difficult. One problem relates to the low blood volume that is commonly analyzed making CTC detection unreliable, especially in patients with very low CTC numbers in the circulation [11, 12]. This problem could be overcome using high blood volume techniques, such as leukapheresis [11] or a functionalized Seldinger guidewire, which is inserted in the cubital vein of the cancer patient [13]. A more complex problem relates to the detection of the extremely rare CTCs in the absence of any tumor-specific marker. So far, epithelial CTCs are mainly identified by their histogenetic difference to the mesenchymal hematopoietic cells. The group of Gert Riethmüller pioneered this strategy almost 30 years ago, applying immunoassays with antibodies directed against cytokeratins and the transmembrane epithelial cell adhesion molecule EpCAM to trace clinically occult single disseminated cancer cells in bone marrow of non-metastatic cancer patients [14]. In the meantime, EpCAM became one of the most commonly used membrane-associated proteins for capturing CTCs from blood samples (reviewed in [15]), while cytokeratins commonly serve to identify CTCs after isolation. Alternatives such as tumor-specific antigen 9, keratin 19 and pre-progastrin-releasing peptides have been reported [16]. The rationale to use EpCAM as a capturing antigen is based on its strong expression in most carcinomas and the specificity for epithelial cells [17, 18]. Expression of EpCAM in non-epithelial cells is restricted to embryonic stem cells and rare subsets of adult stem and precursors cells [18–22]. Another factor promoting EpCAM as prime marker for CTCs was clearly the success of the CellSearch system, the only U.S. Food and Drug Administration (FDA) cleared CTC detection assay. The CellSearch system relies on the automated immuno-magnetic enrichment of EpCAM+ cancer cells [23], which are then further qualified as CTCs based on expression of cytokeratins (CK), nuclear counterstain with 4′,6-diamidino-2-phenylindole (DAPI), and absence of CD45 expression [24]. Clinical studies demonstrated the validity of the prognostic relevance of CTC baseline counts (≥5 CTC/7.5 mL blood) in metastasized breast [24–26], prostate [27], and (≥3 CTC/7.5 mL blood) colorectal cancer [28]. More recently, large studies revealed the prognostic value of CTCs even in non-metastatic breast cancer, although CTCs were detected markedly less frequently than in metastatic disease [29, 30].
Since, the extracellular domain of EpCAM provides a target for antibodies to capture CTCs without the need to permeabilize cells, vital CTCs can be isolated. Using this opportunity, Bacelli et al. [31] could recently reveal that FACS-sorted EpCAM+ CTCs comprise indeed metastases-inducing cells (MICs) in metastatic breast cancer patients.
Down-regulation of EpCAM on cancer cells impedes effective CTC capturing
At the time of its introduction into CTC assays, EpCAM was perceived as an epithelial cell adhesion molecule [32]. Its steady expression throughout cancer progression remained unquestioned for a long time, until observations of a dynamic EpCAM expression were reported and concerns were raised to miss relevant CTCs if EpCAM is used as exclusive CTC capturing antigen. One of the first comparative studies of EpCAM-based isolation platforms, including CellSearch, OncoCEE microchannel, and On-Q-ity’s CTC chip, demonstrated a comparable performance but the capture efficiency of all assays was clearly related to the EpCAM expression level on the target cells. Consequently, more “aggressive” mesenchymal cancer cell phenotypes could escape EpCAM-dependent capturing [33]. Sieuwerts et al. [34] reported that cells of the normal-like breast cancer subtype, a mesenchymal, vimentin-positive phenotype with aggressive features, were not efficiently isolated using CellSearch in comparison to the basal, HER2-positive, and luminal A/B subtypes. Besides EpCAM-negativity of specific cancer entities/subtypes, data indicated that EpCAM becomes down-regulated on cancer cells during dissemination into the blood stream [35]. Here, the molecule expression per cell was found to be approximately tenfold lower in CTCs compared to primary tumors or metastasis (49,700 vs. 400,000). A recent transcriptome profiling in colorectal cancer validated EpCAM down-regulation on CTCs compared to primary tumors [36]. Similarly, down-regulation was noted in disseminated cancer cells (DTC) detected in bone marrow of esophageal carcinoma patients, where two-thirds of patients harbored EpCAM-negative DTCs despite a strong expression of EpCAM in primary tumors [37]. Interestingly, EpCAM+ DTCs associated with the occurrence of lymph node metastases and poor survival, which possibly reflects the role or even requirement of EpCAM in proliferation of micrometastases. Zhang et al. [38] recently underscored the importance of EpCAM− CTCs by generating cell lines from EpCAM−/ALDH+/CD45− CTCs with pronounced metastatic capacity in mice. Interestingly, EpCAM+/ALDH+/CD45− CTCs could be also kept in vitro but did not survive more than 14 days under the applied culture conditions.
EMT as important trigger for EpCAM down-regulation in CTCs and DTCs
A first experimental evidence for dynamic expression of EpCAM on human cancer cells in the blood circulation came from xenograft experiments. Gorges and colleagues established metastasizing xenograft mouse models with EpCAM-expressing breast cancer cell lines. While tumors at the injection site and metastasis expressed EpCAM, its mRNA expression was absent on CTCs and became down-regulated within the first hour after injection into the blood stream [39]. Interestingly, genes associated with epithelial-to-mesenchymal transition (EMT) were concomitantly up-regulated in EpCAM− circulating cancer cells [39].
EMT is a tightly controlled process occurring physiologically during embryonic development and wound healing, allowing cells to switch phenotypes between epithelial and mesenchymal states. It is believed that EMT is essential for metastasis by promoting motility, invasion, and dissemination in epithelial cancer cells [40–42]. The reverse process, termed mesenchymal-to-epithelial transition (MET), allows then cancer cells to regain their epithelial features including adhesion and proliferation [43, 44], thereby promoting the development of macro-metastases from (single) DTCs. Both, paracrine and autocrine signals including activation of TGFβ receptors [45–47] and WNT-dependent signaling [48] are involved in the transition from the epithelial to the mesenchymal state. These central pathways impact on the expression and function of essential transcription factors such as ZEB1, snail, slug and twist, which finally execute EMT programs [40]. Additional mechanisms, which are instrumental during EMT, became recently better understood and rely mainly on epigenetic changes. Here, polycomb group factors operate at the level of chromatin remodeling, e.g. histone modifications, including the placement of activating and inhibiting methylation marks at histone 3 lysine residues (H3K4 and H3K27 methylation) (reviewed in [49] On the contrary, Rho family GTPases such as rac1 and cdc42 are required for the reverse conversion to an epithelial state [50]. Furthermore, microRNAs of the mir200 and 205 cluster were demonstrated to promote MET [51]. These regulatory mechanisms have also been shown for EpCAM in prostate cancer cells. Induction of chemoresistance was associated with EMT and loss of EpCAM, whereas treatment of chemoresistant cells with miRNA 200 or 205 induced MET and restored EpCAM expression [52]. Similarly, treatment of lung and esophageal carcinoma cells with TGFβ resulted in substantial loss of EpCAM membrane expression, but obviously based on different molecular mechanisms [37]. While reduction of EpCAM expression was seen at the transcriptional and post-translational level in A549 lung cancer cells, post-translational EpCAM down-regulation was seen in esophageal cancer cells.
It is important to stress that in EMT, epithelial and mesenchymal phenotypes must be seen as the two extremes of a broad range of cellular states that can be partly or completely acquired by tumors cells. In other words, carcinoma cells undergo EMT or MET in a gradual manner, with epithelial cells partially acquiring mesenchymal but retaining selected epithelial features, and vice versa [49]. Interestingly, this plasticity might not necessarily reflect targeted, cell-autonomous changes, but could be the result of stochastic changes occurring in cells under given conditions and depending upon inputs from the microenvironment they reside in [53]. In this model, tumor heterogeneity reflects an equilibrium between various states of phenotypes with the proclivity to metastasize. In fact, epithelial/mesenchymal phenotypes can be mixed in CTC populations as demonstrated by Yu et al. [54], who applied a quantifiable, dual-colorimetric RNA-in situ hybridization to assess epithelial and mesenchymal transcripts of individual CTCs. Not only CTC phenotypes were mixed within the patients, but also individual CTCs showed a mixed epithelial/mesenchymal phenotype. Interestingly, an increase of CTCs with mesenchymal phenotype was observed in patients with progressive disease after systemic therapy [54]. In accordance with these findings, previous studies indirectly demonstrated the expression of EMT-associated markers and of the matrix metalloproteinase 1 in CTCs [33, 55, 56], thus underlining the important contribution of phenotypic changes during disease progression and the need to include CTCs with mesenchymal phenotypes into screening and therapeutic approaches.
The molecular mechanisms leading to rapid down-regulation of EpCAM in response to EMT remain incompletely understood. Indications for a potential mechanism came from in vitro observations of human migrating EpCAM+ cancer cells. Here, a progressive loss of EpCAM expression at the membrane along with the appearance of EpCAM-positive speckles in the cytoplasm was observed. These findings were suggestive of endocytosis and subsequent degradation of EpCAM in intracellular compartments [37]. Endocytosis of EpCAM was already indicated by studies on the internalization of fusions of toxins with EpCAM-specific antibodies [57]. Similarly, a loss of EpCAM at the plasma membrane and increased cytoplasmic staining was reported in invading colorectal cancer cells budding from the primary tumors [58] as well as in advanced breast cancer [59], further pinpointing to endocytosis as a relevant mechanism for EpCAM withdrawal from the cell surface. From the mechanistic and molecular point of view, the presence of an NPXY consensus motif within the intracellular domain of EpCAM was mentioned and suggested a clathrin-dependent endocytosis of the molecule [18]. Intriguingly, this stated NPXY motif cannot be found in the currently available amino acid sequence of human EpCAM. Hence, endocytosis of EpCAM and its molecular basis remain as open questions to be addressed in more detail.
Other mechanisms leading to EpCAM down-regulation
The observed loss of EpCAM expression on the cell surface of cancer cells could be further explained based upon the published mode of activation of the protein [60]. EpCAM undergoes a regulated intramembranous proteolysis (RIP), beginning with cleavage of the extracellular ectodomain of EpCAM (EpEX) by sheddases of the ADAM family [61, 62]. Additional insight came from work by Hachmeister et al. [63] who demonstrated the additional cleavage of murine EpCAM by the protease BACE-1, which is enzymatically active in endo- and lysosomes at low pH of 4–5. Accordingly, inhibition of a pH shift in lysosomes using bafilomycin as well as inhibition of clathrin-dependent endocytosis resulted in stabilization of EpCAM in lysosomes or retention at the plasma membrane, respectively (OG, unpublished data). Thus, endocytosis and RIP of EpCAM might represent separate as well as converging routes of EpCAM withdrawal from the plasma membrane.
A more classical understanding of loss of EpCAM expression is based on the transcriptional down-regulation of the EPCAM gene through hypermethylation of its promoter, which was observed in breast cancer cells [64] and human embryonic stem cells [65]. In line with these findings, Tai et al. [66] reported on the hypermethylation of the EPCAM promoter and decreased expression of EpCAM with increasing invasiveness of lung adenocarcinoma cell lines. At the molecular level, silencing of the EPCAM gene in differentiating human embryonic stem cells was reported to depend on the loss of transcription activating methylation marks on the tails of histone three molecules (H3K4me3) and an increase of inhibitory H3K27 trimethylation via the polycomb protein SUZ12 [65]. ZEB1, a transcription factor that regulates EMT induction, has further been reported to bind to the promoter of the EPCAM gene in zebrafish to down-regulated EpCAM expression during gastrulation [67]. Comparable results were obtained in human cancer cell lines, suggesting a role for ZEB1 in the regulation of EpCAM during tumor progression too [67]. In addition, mutations of the EPCAM gene might further contribute to the perturbed expression of the protein. However, so far mutations of the EPCAM gene involved in cancer phenotypes have rarely been described and actually impacted on the expression of the DNA mismatch repair protein MSH2, encoded by a gene located 3-prime of EPCAM. Mutation of EPCAM resulted in genetic silencing of MSH2, increased genomic instability and strongly increased incidence of cancer—a phenotype known as the Lynch syndrome [68, 69].
Initial understanding of EpCAM´s role in systemic tumor progression
Conceptually, a dynamic expression of EpCAM could either be a result of EMT/MET (“passenger”) or represent a driving force of phenotypic changes (“driver”). The major functions assigned to EpCAM speak in favor of a “driver” role in the regulation of the epithelial phenotype, i.e. cell–cell adhesion [32], maintenance of epithelial integrity [70], regulation of contractility and morphogenic movements [71, 72], and regulation of proliferation [60, 62] (see Fig. 1 for a schematic view of EpCAM expression in tumor progression). EpCAM-mediated cell adhesion is associated with the full-length molecule, where the extracellular domains of EpCAM molecules on opposing cells interact, while the intracellular domains connect the protein to the cytoskeleton [73]. It must, however, be noted, that EpCAM, at the same time, was reported to interfere with E-cadherin-dependent functions and thereby appeared to weaken cell adhesion [74]. On the contrary, EpCAM was also demonstrated to cooperate with proteins of the claudin family and to contribute to the formation of tight junctions in the intestine [75]. In full accordance, knock-out of EpCAM in the mouse resulted in embryonic or perinatal lethality, which was associated with severe hemorrhagic diarrhea [76] and intestinal dysfunction [75]. Thus, EpCAM’s role in cell adhesion remains somewhat inconclusive and would profit from experimental revision. Especially, a competition of EpCAM with cadherins appears questionable in the light of present publications. Rather, EpCAM seems to foster actomyosin-dependent contractility and cell adhesion at the level of inhibition of protein kinase C (PKC) signaling [70–72]. A function of EpCAM in the increase of cell adhesion might explain the observed decrease in expression during EMT in migrating cells. However, the reported EpCAM-dependent increase in motility observed under physiological conditions in early gastrulation of Xenopus laevi appears controversial and speaks in favor of a migratory phenotype of EpCAM-positive cells [71, 72] In line with this notion, siRNA-mediated knock-down and p53-dependent down-regulation of EpCAM in breast cancer cell lines resulted in substantially reduces proliferation, migration and invasion [77, 78]. Controversially, reduction of EpCAM expression in esophageal cancer cells induced migration and invasion under conditions of low nutrition [37]. Similar observations were made in head and neck CSCs [37, 79]. In accordance with a potentially dual role in adhesion and proliferation, expression of EpCAM was associated with the formation of larger tumors and an epithelial phenotype of cells [37, 62, 79]. Reinforcing these contrasting results, down-regulation of EpCAM resulted in loss of proliferation and tumor formation capacity in breast epithelial-type cells, whereas induction of EpCAM expression had no impact on mesenchymal-type cells [80], arguing for a differential expression and role of EpCAM in cellular subtypes. Furthermore, modulation of EpCAM expression was oppositely affecting proliferation in Her2-positive versus Her2-negative breast cancer cell lines and correlated with favorable or unfavorable prognosis, respectively [81]. Thus, the role of EpCAM in the regulation of proliferation and migration appears somewhat conflicting throughout different tumor cell systems and certainly requires further experimental approaches. At present, no valid explanation can be given for the observed opposing effects of EpCAM on migration and invasion, except for a potential role of nutrition availability and a mixed phenotype due to additional effects of EpCAM on proliferation and cell metabolism [82–84].
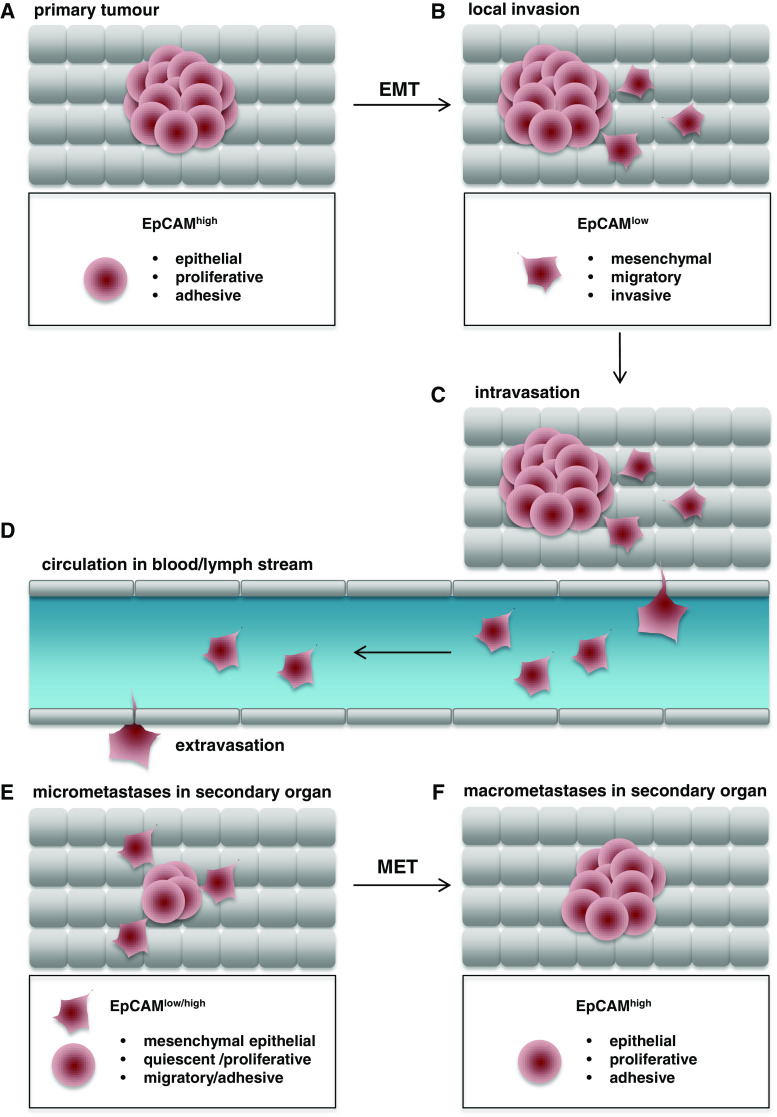
Fig. 1.
Schematic representation of EpCAM expression throughout tumor progression. a EpCAM is strongly expressed in most carcinomas, associates with an epithelial phenotype, and regulates proliferation. b Initial phases of local invasion are characterized by epithelial-to-mesenchymal transition with a loss of EpCAM expression at the plasma membrane (EpCAMhigh to EpCAMlow), a gain of migratory and invasive capacity, and a mesenchymal phenotype. c, d Invasive tumor cells gain access to the blood or lymph stream upon intravasation and begin to circulate in the body. These circulating tumor cells have a mesenchymal, rather quiescent phenotype. Circulating tumor cells leave the blood or lymph stream (extravasation) and invade secondary organs at locoregional or distant sites. e Disseminated tumor cells generate novel malignant seeds and develop into micrometastases. f Micrometastases perform a mesenchymal-to-epithelial transition, re-express EpCAM, regain proliferative capacity, and grow to form macro-metastases
The molecular basis for EpCAM’s direct involvement in the regulation of proliferation was described long time after its role in cell–cell adhesion [77, 83] and was demonstrated to depend on RIP [62]. Proteolytic cleavage results in the generation of extracellular EpEX domains and intracellular domains, EpICD, which translocate into the nucleus in combination with FHL2 and β-catenin to bind Lef-1 consensus sites and induce the transcription of target genes such as cyclin D1 [62, 85] and stemness genes [65]. Implication of EpCAM in the regulation of WNT signaling was also reported to occur in an indirect manner. Here, EpCAM sequestered Kremen-1/Dickkopf2 complexes, inhibitors of the Wnt receptor Lrp6 during hepatocytic differentiation. Thereby, EpCAM fosters Lrp6 retention at the membrane and its signaling through Wnt2bb, representing a licensing factor for the endodermal differentiation towards hepatocytes in zebrafish [86]. This function of EpCAM as a de-repressor of Wnt signaling and induction of endodermal differentiation is of great interest with respect to the loss of EpCAM during EMT, as it might represent a means to repress endodermal differentiation to allow mesodermal differentiation.
Proteolysis of EpCAM was analyzed in a cohort of different cancer types using staining of EpEX and EpICD on consecutive sections. The authors defined a membranous, full-length EpCAM when both subdomains of EpCAM were present at the membrane, and a truncated variant, when EpICD was lacking at the membrane as compared to EpEX. This study revealed strongest cleavage in cancers of the endometrium and the bladder, intermediate cleavage in gastric, colorectal, esophageal, and pancreatic cancers, and low cleavage in lung, ovarian, breast and prostate cancers [87]. Owing to the compulsory initial extracellular shedding of EpCAM to release EpICD [62, 63], the presence of EpCAM molecules composed of extracellular and transmembrane domains but lacking EpICD appears counterintuitive, but has been reported by various groups [62, 87, 88]. Such staining patterns might reflect retention of EpEX at the plasma membrane or in the interstitium in the vicinity of cells or, alternatively, a RIP-independent and selective degradation of EpICD from intact EpCAM molecules. Interestingly, cytoplasmic and nuclear localization of EpICD correlated with an aggressive phenotype of thyroid cancers [88–90]. Additional evidence for a role of cleavage products of EpCAM in the regulation of migration and invasion is, to the best of our knowledge, currently lacking.
In summary, the present data support a role of EpCAM in actively supporting an epithelial phenotype at the expense of mesenchymal differentiation (Fig. 1). The main contribution of EpCAM to tumor progression appears at the moment to relate to proliferation [62, 77, 85, 91] and the response to growth factors [92], but direct effects on differentiation and pluripotency of cells are conceivable in light of the repeatedly observed effects of EpCAM and EpICD in embryonic stem cells [21, 22, 65]. Alternatively, EpCAM could only induce proliferation and be phenotypically associated with epithelial cells, whilst functionally unrelated to mesenchymal cells. Future studies including cancer and stem cells should aim at the elucidation of the actual role of EpCAM in differentiation, which could potentially differ in stem cells, normal and malignant epithelia.
Consequences for CTC-research and concluding remarks
Circulating tumor cells appear currently as promising surrogate markers for the actual targets of systemic cancer therapy, i.e. metastases-inducing cells. Assuming that EMT-CTCs reflect early steps of progression including detachment from the primary tumor and circulation of tumor cells, it seems imperative to use EMT markers in isolation protocols to include EMT-CTCs in monitoring procedures. Interesting marker candidates for EpCAM-independent capturing, are the epidermal growth factor receptor EGF-R and the hepatocyte growth factor receptor c-Met [31, 33]. However, expression profiles of these membrane proteins on CTCs remain to be further defined and might eventually harbor similar problems and drawbacks as EpCAM, including a loss of expression during EMT/MET. Alternatively, marker-independent enrichment of CTCs using selection by size with filtration techniques [11, 93, 94] seems to be a very promising approach. Efforts to characterize the phenotype and capacities of EMT-CTCs might generate new options to inhibit early progression. On the other hand, the expression of EpCAM on CTCs (and DTCs) was generally strongly associated with metastasis and reduced overall survival [24–26, 28, 37, 95–97]. Assuming that EpCAM-positive CTCs/DTCs reflect a rather late stage within the metastatic cascade, potentially associated with MET and the re-expression of epithelial proteins, EpCAM gains impact as a therapeutic target to suppress metastatic outgrowth. Therefore, future work should address the role of EpCAM in non-EMT-CTCs (MET-CTCs) with regard to proliferation and adhesion. Detailed knowledge on EpCAM’s implication in the functionality of proliferation versus migration/invasion is of utmost importance when aiming to counteract such functions in a therapeutic setting. Potential therapeutic interventions cover a full range of methods and agents, which have been reviewed elsewhere [19, 20, 98, 99]. Traditionally, EpCAM could be targeted by humanized monoclonal, bi-specific and tri-functional antibodies or recombinant variants such as BiTEs [19, 98]. However, in view of the EpCAM signaling described here, novel options emerge. These include delivery of protease inhibitors to EpCAM-positive CTCs/DTCs in form of antibody conjugates, but also combinatorial therapies composed of EpCAM-specific antibodies and protease inhibitors. Antagonizing antibodies and small molecule inhibitors interfering with RIP and nuclear translocation of EpICD, respectively, could be further interesting options to inhibit the outgrowth of MET-CTC/DTC.
Acknowledgements
O. Gires and N. H. Stoecklein were funded by the Wilhelm-Sander-Stiftung (2009.083.1); O. Gires was funded by the Deutsche Forschungsgemeinschaft (DFG GI 540/3-1) and the Wilhelm-Sander-Stiftung (2012.051.1); N. H. Stoecklein was funded by the Deutsche Forschungsgemeinschaft (DFG STO464/2-2), and the Deutsche Krebshilfe e.V. (109600).
References
- 1.Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009;9(4):302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 2.Klein CA, Stoecklein NH. Lessons from an aggressive cancer: evolutionary dynamics in esophageal carcinoma. Cancer Res. 2009;69(13):5285–5288. doi: 10.1158/0008-5472.CAN-08-4586. [DOI] [PubMed] [Google Scholar]
- 3.Stoecklein NH, Klein CA. Genetic disparity between primary tumours, disseminated tumour cells, and manifest metastasis. Int J Cancer. 2010;126(3):589–598. doi: 10.1002/ijc.24916. [DOI] [PubMed] [Google Scholar]
- 4.Coumans FA, Siesling S, Terstappen LW. Detection of cancer before distant metastasis. BMC Cancer. 2013;13(1):283. doi: 10.1186/1471-2407-13-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aprile G, Giuliani F, Cordio S, Sartore-Bianchi A, Bencardino K, Ongaro E, Martines C, Giampieri R, Bordonaro R, Siena S, Cascinu S, Scartozzi M. Translational challenges from the 2014 Gastrointestinal Cancers Symposium: toward a true tailored therapy through effective research. Future Oncol. 2014;10(7):1125–1128. doi: 10.2217/fon.14.54. [DOI] [PubMed] [Google Scholar]
- 6.Lehmann BD, Pietenpol JA. Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J Pathol. 2014;232(2):142–150. doi: 10.1002/path.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yap TA, Lorente D, Omlin A, Olmos D, de Bono JS. Circulating tumor cells: a multifunctional biomarker. Clin Cancer Res. 2014;20(10):2553–2568. doi: 10.1158/1078-0432.CCR-13-2664. [DOI] [PubMed] [Google Scholar]
- 8.Pantel K, Alix-Panabieres C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. 2010;16(9):398–406. doi: 10.1016/j.molmed.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Pantel K, Alix-Panabieres C, Riethdorf S. Cancer micrometastases. Nat Rev Clin Oncol. 2009;6(6):339–351. doi: 10.1038/nrclinonc.2009.44. [DOI] [PubMed] [Google Scholar]
- 10.Becker TM, Caixeiro NJ, Lim SH, Tognela A, Kienzle N, Scott KF, Spring KJ, de Souza P. New frontiers in circulating tumor cell analysis: a reference guide for biomolecular profiling toward translational clinical use. Int J Cancer. 2014;134(11):2523–2533. doi: 10.1002/ijc.28516. [DOI] [PubMed] [Google Scholar]
- 11.Fischer JC, Niederacher D, Topp SA, Honisch E, Schumacher S, Schmitz N, Zacarias Fohrding L, Vay C, Hoffmann I, Kasprowicz NS, Hepp PG, Mohrmann S, Nitz U, Stresemann A, Krahn T, Henze T, Griebsch E, Raba K, Rox JM, Wenzel F, Sproll C, Janni W, Fehm T, Klein CA, Knoefel WT, Stoecklein NH. Diagnostic leukapheresis enables reliable detection of circulating tumor cells of nonmetastatic cancer patients. Proc Natl Acad Sci USA. 2013;110(41):16580–16585. doi: 10.1073/pnas.1313594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coumans FA, Ligthart ST, Uhr JW, Terstappen LW. Challenges in the enumeration and phenotyping of CTC. Clin Cancer Res. 2012;18(20):5711–5718. doi: 10.1158/1078-0432.CCR-12-1585. [DOI] [PubMed] [Google Scholar]
- 13.Saucedo-Zeni N, Mewes S, Niestroj R, Gasiorowski L, Murawa D, Nowaczyk P, Tomasi T, Weber E, Dworacki G, Morgenthaler NG, Jansen H, Propping C, Sterzynska K, Dyszkiewicz W, Zabel M, Kiechle M, Reuning U, Schmitt M, Lucke K. A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire. Int J Oncol. 2012;41(4):1241–1250. doi: 10.3892/ijo.2012.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlimok G, Gottlinger H, Funke I, Swierkot S, Hauser H, Riethmuller G. In vivo and in vitro labelling of epithelial tumor cells with anti 17-1A monoclonal antibodies in bone marrow of cancer patients. Hybridoma. 1986;5(Suppl 1):S163–S170. [PubMed] [Google Scholar]
- 15.Barradas AM, Terstappen LW. Towards the biological understanding of CTC: capture technologies, definitions and potential to create metastasis. Cancers. 2013;5(4):1619–1642. doi: 10.3390/cancers5041619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu L, Liao GQ, He P, Zhu H, Liu PH, Qu YM, Song XM, Xu QW, Gao Q, Zhang Y, Chen WF, Yin YH. Detection of circulating cancer cells in lung cancer patients with a panel of marker genes. Biochem Biophys Res Commun. 2008;372(4):756–760. doi: 10.1016/j.bbrc.2008.05.101. [DOI] [PubMed] [Google Scholar]
- 17.Went PT, Lugli A, Meier S, Bundi M, Mirlacher M, Sauter G, Dirnhofer S. Frequent EpCam protein expression in human carcinomas. Hum Pathol. 2004;35(1):122–128. doi: 10.1016/j.humpath.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 18.Balzar M, Winter MJ, de Boer CJ, Litvinov SV. The biology of the 17-1A antigen (Ep-CAM) J Mol Med. 1999;77(10):699–712. doi: 10.1007/s001099900038. [DOI] [PubMed] [Google Scholar]
- 19.Baeuerle PA, Gires O. EpCAM (CD326) finding its role in cancer. Br J Cancer. 2007;96(3):417–423. doi: 10.1038/sj.bjc.6603494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imrich S, Hachmeister M, Gires O. EpCAM and its potential role in tumor-initiating cells. Cell Adh Migr. 2012;6(1):30–38. doi: 10.4161/cam.18953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez B, Denzel S, Mack B, Conrad M, Gires O. EpCAM is involved in maintenance of the murine embryonic stem cell phenotype. Stem Cells. 2009;27(8):1782–1791. doi: 10.1002/stem.97. [DOI] [PubMed] [Google Scholar]
- 22.Ng VY, Ang SN, Chan JX, Choo AB. Characterization of epithelial cell adhesion molecule as a surface marker on undifferentiated human embryonic stem cells. Stem Cells. 2009 doi: 10.1002/stem.221. [DOI] [PubMed] [Google Scholar]
- 23.Terstappen LW, Rao C, Gross S, Kotelnikov V, Racilla E, Uhr J, Weiss A. Flow cytometry–principles and feasibility in transfusion medicine. Enumeration of epithelial derived tumor cells in peripheral blood. Vox Sang. 1998;74(Suppl 2):269–274. doi: 10.1111/j.1423-0410.1998.tb05431.x. [DOI] [PubMed] [Google Scholar]
- 24.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351(8):781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 25.Wallwiener M, Hartkopf AD, Baccelli I, Riethdorf S, Schott S, Pantel K, Marme F, Sohn C, Trumpp A, Rack B, Aktas B, Solomayer EF, Muller V, Janni W, Schneeweiss A, Fehm TN. The prognostic impact of circulating tumor cells in subtypes of metastatic breast cancer. Breast Cancer Res Treat. 2013;137(2):503–510. doi: 10.1007/s10549-012-2382-0. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L, Riethdorf S, Wu G, Wang T, Yang K, Peng G, Liu J, Pantel K. Meta-analysis of the prognostic value of circulating tumor cells in breast cancer. Clin Cancer Res. 2012;18(20):5701–5710. doi: 10.1158/1078-0432.CCR-12-1587. [DOI] [PubMed] [Google Scholar]
- 27.Danila DC, Heller G, Gignac GA, Gonzalez-Espinoza R, Anand A, Tanaka E, Lilja H, Schwartz L, Larson S, Fleisher M, Scher HI. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13(23):7053–7058. doi: 10.1158/1078-0432.CCR-07-1506. [DOI] [PubMed] [Google Scholar]
- 28.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen LW, Meropol NJ. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(19):3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 29.Lucci A, Hall CS, Lodhi AK, Bhattacharyya A, Anderson AE, Xiao L, Bedrosian I, Kuerer HM, Krishnamurthy S. Circulating tumour cells in non-metastatic breast cancer: a prospective study. Lancet Oncol. 2012;13(7):688–695. doi: 10.1016/S1470-2045(12)70209-7. [DOI] [PubMed] [Google Scholar]
- 30.Rack B, Schindlbeck C, Juckstock J, Andergassen U, Hepp P, Zwingers T, Friedl TW, Lorenz R, Tesch H, Fasching PA, Fehm T, Schneeweiss A, Lichtenegger W, Beckmann MW, Friese K, Pantel K, Janni W, Group SS (2014) Circulating tumor cells predict survival in early average-to-high risk breast cancer patients. J Natl Cancer Inst 106(5). doi:10.1093/jnci/dju066 [DOI] [PMC free article] [PubMed]
- 31.Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, Klein C, Saini M, Bauerle T, Wallwiener M, Holland-Letz T, Hofner T, Sprick M, Scharpff M, Marme F, Sinn HP, Pantel K, Weichert W, Trumpp A. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol. 2013;31(6):539–544. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- 32.Litvinov SV, Bakker HA, Gourevitch MM, Velders MP, Warnaar SO. Evidence for a role of the epithelial glycoprotein 40 (Ep-CAM) in epithelial cell–cell adhesion. Cell Adhes Commun. 1994;2(5):417–428. doi: 10.3109/15419069409004452. [DOI] [PubMed] [Google Scholar]
- 33.Punnoose EA, Atwal SK, Spoerke JM, Savage H, Pandita A, Yeh RF, Pirzkall A, Fine BM, Amler LC, Chen DS, Lackner MR. Molecular biomarker analyses using circulating tumor cells. PLoS ONE. 2010;5(9):e12517. doi: 10.1371/journal.pone.0012517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sieuwerts AM, Kraan J, Bolt J, van der Spoel P, Elstrodt F, Schutte M, Martens JW, Gratama JW, Sleijfer S, Foekens JA. Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J Natl Cancer Inst. 2009;101(1):61–66. doi: 10.1093/jnci/djn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rao CG, Chianese D, Doyle GV, Miller MC, Russell T, Sanders RA, Jr, Terstappen LW. Expression of epithelial cell adhesion molecule in carcinoma cells present in blood and primary and metastatic tumors. Int J Oncol. 2005;27(1):49–57. [PubMed] [Google Scholar]
- 36.Steinert G, Scholch S, Niemietz T, Iwata N, Garcia SA, Behrens B, Voigt A, Kloor M, Benner A, Bork U, Rahbari NN, Buchler MW, Stoecklein NH, Weitz J, Koch M. Immune escape and survival mechanisms in circulating tumor cells of colorectal cancer. Cancer Res. 2014;74(6):1694–1704. doi: 10.1158/0008-5472.CAN-13-1885. [DOI] [PubMed] [Google Scholar]
- 37.Driemel C, Kremling H, Schumacher S, Will D, Wolters J, Lindenlauf N, Mack B, Baldus SA, Hoya V, Pietsch JM, Panagiotidou P, Raba K, Vay C, Vallbohmer D, Harreus U, Knoefel WT, Stoecklein NH, Gires O. Context-dependent adaption of EpCAM expression in early systemic esophageal cancer. Oncogene. 2013 doi: 10.1038/onc.2013.441. [DOI] [PubMed] [Google Scholar]
- 38.Zhang L, Ridgway LD, Wetzel MD, Ngo J, Yin W, Kumar D, Goodman JC, Groves MD, Marchetti D. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med. 2013;5((180)):180ra148. doi: 10.1126/scitranslmed.3005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorges TM, Tinhofer I, Drosch M, Rose L, Zollner TM, Krahn T, von Ahsen O. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer. 2012;12:178. doi: 10.1186/1471-2407-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 41.Thiery JP, Lim CT. Tumor dissemination: an EMT affair. Cancer Cell. 2013;23(3):272–273. doi: 10.1016/j.ccr.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5(9):744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 43.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14(6):818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 44.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22(6):725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, Bell G, Guo W, Rubin J, Richardson AL, Weinberg RA. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145(6):926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heldin CH, Landstrom M, Moustakas A. Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr Opin Cell Biol. 2009;21(2):166–176. doi: 10.1016/j.ceb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 47.Moustakas A, Heldin CH. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 2007;98(10):1512–1520. doi: 10.1111/j.1349-7006.2007.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim K, Lu Z, Hay ED. Direct evidence for a role of beta-catenin/LEF-1 signaling pathway in induction of EMT. Cell Biol Int. 2002;26(5):463–476. doi: 10.1006/cbir.2002.0901. [DOI] [PubMed] [Google Scholar]
- 49.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19(11):1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakaya Y, Kuroda S, Katagiri YT, Kaibuchi K, Takahashi Y. Mesenchymal-epithelial transition during somitic segmentation is regulated by differential roles of Cdc42 and Rac1. Dev Cell. 2004;7(3):425–438. doi: 10.1016/j.devcel.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 52.Massoner P, Thomm T, Mack B, Untergasser G, Martowicz A, Bobowski K, Klocker H, Gires O, Puhr M. EpCAM is overexpressed in local and metastatic prostate cancer, suppressed by chemotherapy and modulated by MET-associated miRNA-200c/205. Br J Cancer. 2014 doi: 10.1038/bjc.2014.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 54.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, Brachtel E, Sgroi D, Baselga J, Ramaswamy S, Toner M, Haber DA, Maheswaran S. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339(6119):580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009;11(4):R46. doi: 10.1186/bcr2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cierna Z, Mego M, Janega P, Karaba M, Minarik G, Benca J, Sedlackova T, Cingelova S, Gronesova P, Manasova D, Pindak D, Sufliarsky J, Danihel L, Reuben JM, Mardiak J. Matrix metalloproteinase 1 and circulating tumor cells in early breast cancer. BMC Cancer. 2014;14:472. doi: 10.1186/1471-2407-14-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winkler J, Martin-Killias P, Pluckthun A, Zangemeister-Wittke U. EpCAM-targeted delivery of nanocomplexed siRNA to tumor cells with designed ankyrin repeat proteins. Mol Cancer Ther. 2009;8(9):2674–2683. doi: 10.1158/1535-7163.MCT-09-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gosens MJ, van Kempen LC, van de Velde CJ, van Krieken JH, Nagtegaal ID. Loss of membranous Ep-CAM in budding colorectal carcinoma cells. Mod Pathol. 2007;20(2):221–232. doi: 10.1038/modpathol.3800733. [DOI] [PubMed] [Google Scholar]
- 59.Alberti S, Ambrogi F, Boracchi P, Fornili M, Querzoli P, Pedriali M, La Sorda R, Lattanzio R, Tripaldi R, Piantelli M, Biganzoli E, Coradini D. Cytoplasmic Trop-1/Ep-CAM overexpression is associated with a favorable outcome in node-positive breast cancer. Jpn J Clin Oncol. 2012;42(12):1128–1137. doi: 10.1093/jjco/hys159. [DOI] [PubMed] [Google Scholar]
- 60.Munz M, Baeuerle PA, Gires O. The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res. 2009;69(14):5627–5629. doi: 10.1158/0008-5472.CAN-09-0654. [DOI] [PubMed] [Google Scholar]
- 61.Carpenter G, Red Brewer M. EpCAM: another surface-to-nucleus missile. Cancer Cell. 2009;15(3):165–166. doi: 10.1016/j.ccr.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 62.Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, Kieu C, Papior P, Baeuerle PA, Munz M, Gires O. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol. 2009;11(2):162–171. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- 63.Hachmeister M, Bobowski KD, Hogl S, Dislich B, Fukumori A, Eggert C, Mack B, Kremling H, Sarrach S, Coscia F, Zimmermann W, Steiner H, Lichtenthaler SF, Gires O. Regulated intramembrane proteolysis and degradation of murine epithelial cell adhesion molecule mEpCAM. PLoS ONE. 2013;8(8):e71836. doi: 10.1371/journal.pone.0071836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spizzo G, Gastl G, Obrist P, Fong D, Haun M, Grunewald K, Parson W, Eichmann C, Millinger S, Fiegl H, Margreiter R, Amberger A. Methylation status of the Ep-CAM promoter region in human breast cancer cell lines and breast cancer tissue. Cancer Lett. 2006;246(1):253–261. doi: 10.1016/j.canlet.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 65.Lu TY, Lu RM, Liao MY, Yu J, Chung CH, Kao CF, Wu HC. Epithelial cell adhesion molecule regulation is associated with the maintenance of the undifferentiated phenotype of human embryonic stem cells. J Biol Chem. 2010;285(12):8719–8732. doi: 10.1074/jbc.M109.077081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tai KY, Shiah SG, Shieh YS, Kao YR, Chi CY, Huang E, Lee HS, Chang LC, Yang PC, Wu CW. DNA methylation and histone modification regulate silencing of epithelial cell adhesion molecule for tumor invasion and progression. Oncogene. 2007;26(27):3989–3997. doi: 10.1038/sj.onc.1210176. [DOI] [PubMed] [Google Scholar]
- 67.Vannier C, Mock K, Brabletz T, Driever W. Zeb1 regulates E-cadherin and Epcam expression to control cell behavior in early zebrafish development. J Biol Chem. 2013 doi: 10.1074/jbc.M113.467787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lynch HT, Lynch JF, Snyder CL, Riegert-Johnson D. EPCAM deletions, lynch syndrome, and cancer risk. Lancet Oncol. 2011;12(1):5–6. doi: 10.1016/S1470-2045(10)70291-6. [DOI] [PubMed] [Google Scholar]
- 69.Kastrinos F, Stoffel EM. History, genetics, and strategies for cancer prevention in lynch syndrome. Clin Gastroenterol Hepatol. 2014;12(5):715–727. doi: 10.1016/j.cgh.2013.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Slanchev K, Carney TJ, Stemmler MP, Koschorz B, Amsterdam A, Schwarz H, Hammerschmidt M. The epithelial cell adhesion molecule EpCAM is required for epithelial morphogenesis and integrity during zebrafish epiboly and skin development. PLoS Genet. 2009;5(7):e1000563. doi: 10.1371/journal.pgen.1000563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maghzal N, Vogt E, Reintsch W, Fraser JS, Fagotto F. The tumor-associated EpCAM regulates morphogenetic movements through intracellular signaling. J Cell Biol. 2010;191(3):645–659. doi: 10.1083/jcb.201004074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maghzal N, Kayali HA, Rohani N, Kajava AV, Fagotto F. EpCAM controls actomyosin contractility and cell adhesion by direct inhibition of PKC. Dev Cell. 2013;27(3):263–277. doi: 10.1016/j.devcel.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 73.Balzar M, Briaire-de Bruijn IH, Rees-Bakker HA, Prins FA, Helfrich W, de Leij L, Riethmuller G, Alberti S, Warnaar SO, Fleuren GJ, Litvinov SV. Epidermal growth factor-like repeats mediate lateral and reciprocal interactions of Ep-CAM molecules in homophilic adhesions. Mol Cell Biol. 2001;21(7):2570–2580. doi: 10.1128/MCB.21.7.2570-2580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Litvinov SV, Balzar M, Winter MJ, Bakker HA, Briaire-de Bruijn IH, Prins F, Fleuren GJ, Warnaar SO. Epithelial cell adhesion molecule (Ep-CAM) modulates cell–cell interactions mediated by classic cadherins. J Cell Biol. 1997;139(5):1337–1348. doi: 10.1083/jcb.139.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lei Z, Maeda T, Tamura A, Nakamura T, Yamazaki Y, Shiratori H, Yashiro K, Tsukita S, Hamada H. EpCAM contributes to formation of functional tight junction in the intestinal epithelium by recruiting claudin proteins. Dev Biol. 2012;371(2):136–145. doi: 10.1016/j.ydbio.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 76.Guerra E, Lattanzio R, La Sorda R, Dini F, Tiboni GM, Piantelli M, Alberti S. mTrop1/Epcam knockout mice develop congenital tufting enteropathy THROUGH dysregulation of intestinal E-cadherin/beta-catenin. PLoS ONE. 2012;7(11):e49302. doi: 10.1371/journal.pone.0049302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Osta WA, Chen Y, Mikhitarian K, Mitas M, Salem M, Hannun YA, Cole DJ, Gillanders WE. EpCAM is overexpressed in breast cancer and is a potential target for breast cancer gene therapy. Cancer Res. 2004;64(16):5818–5824. doi: 10.1158/0008-5472.CAN-04-0754. [DOI] [PubMed] [Google Scholar]
- 78.Sankpal NV, Willman MW, Fleming TP, Mayfield JD, Gillanders WE. Transcriptional repression of epithelial cell adhesion molecule contributes to p53 control of breast cancer invasion. Cancer Res. 2009;69(3):753–757. doi: 10.1158/0008-5472.CAN-08-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Biddle A, Liang X, Gammon L, Fazil B, Harper LJ, Emich H, Costea DE, Mackenzie IC. Cancer stem cells in squamous cell carcinoma switch between two distinct phenotypes that are preferentially migratory or proliferative. Cancer Res. 2011;71(15):5317–5326. doi: 10.1158/0008-5472.CAN-11-1059. [DOI] [PubMed] [Google Scholar]
- 80.Martowicz A, Spizzo G, Gastl G, Untergasser G. Phenotype-dependent effects of EpCAM expression on growth and invasion of human breast cancer cell lines. BMC Cancer. 2012;12:501. doi: 10.1186/1471-2407-12-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soysal SD, Muenst S, Barbie T, Fleming T, Gao F, Spizzo G, Oertli D, Viehl CT, Obermann EC, Gillanders WE. EpCAM expression varies significantly and is differentially associated with prognosis in the luminal B HER2(+), basal-like, and HER2 intrinsic subtypes of breast cancer. Br J Cancer. 2013;108(7):1480–1487. doi: 10.1038/bjc.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Munz M, Hofmann T, Scheibe B, Gange M, Junghanns K, Zeidler R, Gires O. The carcinoma-associated antigen EpCAM induces glyoxalase 1 resulting in enhanced methylglyoxal turnover. Cancer Genomics Proteomics. 2004;1(3):241–247. [PubMed] [Google Scholar]
- 83.Munz M, Kieu C, Mack B, Schmitt B, Zeidler R, Gires O. The carcinoma-associated antigen EpCAM upregulates c-myc and induces cell proliferation. Oncogene. 2004;23(34):5748–5758. doi: 10.1038/sj.onc.1207610. [DOI] [PubMed] [Google Scholar]
- 84.Munz M, Zeidler R, Gires O. The tumor-associated antigen EpCAM up-regulates the fatty acid binding protein E-FABP. Cancer Lett. 2005;225(1):151–157. doi: 10.1016/j.canlet.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 85.Chaves-Perez A, Mack B, Maetzel D, Kremling H, Eggert C, Harreus U, Gires O. EpCAM regulates cell cycle progression via control of cyclin D1 expression. Oncogene. 2013;32(5):641–650. doi: 10.1038/onc.2012.75. [DOI] [PubMed] [Google Scholar]
- 86.Lu H, Ma J, Yang Y, Shi W, Luo L. EpCAM is an endoderm-specific Wnt derepressor that licenses hepatic development. Dev Cell. 2013;24(5):543–553. doi: 10.1016/j.devcel.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 87.Fong D, Moser P, Kasal A, Seeber A, Gastl G, Martowicz A, Wurm M, Mian C, Obrist P, Mazzoleni G, Spizzo G. Loss of membranous expression of the intracellular domain of EpCAM is a frequent event and predicts poor survival in patients with pancreatic cancer. Histopathology. 2014;64(5):683–692. doi: 10.1111/his.12307. [DOI] [PubMed] [Google Scholar]
- 88.Ralhan R, Cao J, Lim T, Macmillan C, Freeman JL, Walfish PG. EpCAM nuclear localization identifies aggressive thyroid cancer and is a marker for poor prognosis. BMC Cancer. 2010;10(1):331. doi: 10.1186/1471-2407-10-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He HC, Kashat L, Kak I, Kunavisarut T, Gundelach R, Kim D, So AK, Macmillan C, Freeman JL, Ralhan R, Walfish PG. An Ep-ICD based index is a marker of aggressiveness and poor prognosis in thyroid carcinoma. PLoS ONE. 2012;7(9):e42893. doi: 10.1371/journal.pone.0042893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ralhan R, He HC, So AK, Tripathi SC, Kumar M, Hasan MR, Kaur J, Kashat L, MacMillan C, Chauhan SS, Freeman JL, Walfish PG. Nuclear and cytoplasmic accumulation of Ep-ICD is frequently detected in human epithelial cancers. PLoS ONE. 2010;5(11):e14130. doi: 10.1371/journal.pone.0014130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maaser K, Borlak J. A genome-wide expression analysis identifies a network of EpCAM-induced cell cycle regulators. Br J Cancer. 2008;99(10):1635–1643. doi: 10.1038/sj.bjc.6604725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yoshida GJ, Saya H. EpCAM expression in the prostate cancer makes the difference in the response to growth factors. Biochem Biophys Res Commun. 2014;443(1):239–245. doi: 10.1016/j.bbrc.2013.11.093. [DOI] [PubMed] [Google Scholar]
- 93.Coumans FA, van Dalum G, Beck M, Terstappen LW. Filtration parameters influencing circulating tumor cell enrichment from whole blood. PLoS ONE. 2013;8(4):e61774. doi: 10.1371/journal.pone.0061774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lin HK, Zheng S, Williams AJ, Balic M, Groshen S, Scher HI, Fleisher M, Stadler W, Datar RH, Tai YC, Cote RJ. Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin Cancer Res. 2010;16(20):5011–5018. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schulze K, Gasch C, Staufer K, Nashan B, Lohse AW, Pantel K, Riethdorf S, Wege H. Presence of EpCAM-positive circulating tumor cells as biomarker for systemic disease strongly correlates to survival in patients with hepatocellular carcinoma. Int J Cancer. 2013;133(9):2165–2171. doi: 10.1002/ijc.28230. [DOI] [PubMed] [Google Scholar]
- 96.Peeters DJ, van Dam PJ, Van den Eynden GG, Rutten A, Wuyts H, Pouillon L, Peeters M, Pauwels P, Van Laere SJ, van Dam PA, Vermeulen PB, Dirix LY. Detection and prognostic significance of circulating tumour cells in patients with metastatic breast cancer according to immunohistochemical subtypes. Br J Cancer. 2014;110(2):375–383. doi: 10.1038/bjc.2013.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Riethdorf S, Fritsche H, Muller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Janicke F, Jackson S, Gornet T, Cristofanilli M, Pantel K. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the Cell Search system. Clin Cancer Res. 2007;13(3):920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 98.Simon M, Stefan N, Pluckthun A, Zangemeister-Wittke U. Epithelial cell adhesion molecule-targeted drug delivery for cancer therapy. Expert Opin Drug Deliv. 2013;10(4):451–468. doi: 10.1517/17425247.2013.759938. [DOI] [PubMed] [Google Scholar]
- 99.Armstrong A, Eck SL. EpCAM: a new therapeutic target for an old cancer antigen. Cancer Biol Ther. 2003;2(4):320–326. doi: 10.4161/cbt.2.4.451. [DOI] [PubMed] [Google Scholar]