Abstract
Muscarinic acetylcholine receptors (mAChRs) play a central role in the mammalian nervous system. These receptors are G protein-coupled receptors (GPCRs), which are activated by the agonists acetylcholine and muscarine, and blocked by a variety of antagonists. Mammals have five mAChRs (m1–m5). In this study, we cloned two structurally related GPCRs from the fruit fly Drosophila melanogaster, which, after expression in Chinese hamster ovary cells, proved to be muscarinic acetylcholine receptors. One mAChR (the A-type; encoded by gene CG4356) is activated by acetylcholine (EC50, 5 × 10−8 M) and muscarine (EC50, 6 × 10−8 M) and blocked by the classical mAChR antagonists atropine, scopolamine, and 3-quinuclidinyl-benzilate (QNB), while the other (the B-type; encoded by gene CG7918) is also activated by acetylcholine, but has a 1,000-fold lower sensitivity to muscarine, and is not blocked by the antagonists. A- and B-type mAChRs were also cloned and functionally characterized from the red flour beetle Tribolium castaneum. Recently, Haga et al. (Nature 2012, 482: 547–551) published the crystal structure of the human m2 mAChR, revealing 14 amino acid residues forming the binding pocket for QNB. These residues are identical between the human m2 and the D. melanogaster and T. castaneum A-type mAChRs, while many of them are different between the human m2 and the B-type receptors. Using bioinformatics, one orthologue of the A-type and one of the B-type mAChRs could also be found in all other arthropods with a sequenced genome. Protostomes, such as arthropods, and deuterostomes, such as mammals and other vertebrates, belong to two evolutionarily distinct lineages of animal evolution that split about 700 million years ago. We found that animals that originated before this split, such as cnidarians (Hydra), had two A-type mAChRs. From these data we propose a model for the evolution of mAChRs.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-013-1334-0) contains supplementary material, which is available to authorized users.
Keywords: Acetylcholine, Evolution, G protein-coupled receptor, mAChR
Introduction
Most animals belong to two lineages of animal evolution, the Protostomia (such as insects and most other invertebrates), and the Deuterostomia (such as vertebrates and a minor group of invertebrates), which split about 700 million years (MYR) ago [1]. Many basic biological processes are similar in both Proto- and Deuterostomia, suggesting that these processes have evolved more than 700 MYR ago. The close similarities of the biology of proto- and deuterostomes also implicate that one might use members of the Protostomia (for example the fruit fly Drosophila melanogaster) as model organisms for basic biological questions arising in members of the Deuterostomia (for example humans).
Insects use many neurotransmitters and neurotransmitter receptors that are identical or very similar to those found in mammals. One such transmitter is the biogenic amine acetylcholine that, as in mammals, can activate nicotinic acetylcholine receptors (nAChRs), which are ligand-gated ion channels, and muscarinic acetylcholine receptors (mAChRs), which are G protein-coupled receptors (GPCRs). As in mammals, the insect nAChRs are pentameric structures, belonging to the large family of cys-loop receptors [2]. A large number of nAChR subunits have been cloned from insects, of which many are structurally and functionally closely related to the various mammalian α, β, γ, δ, and ε nAChR subunits [3–5]. The nAChRs from insects are mainly expressed in their central nervous systems, where they mediate fast synaptic transmission [5]. Acetylcholine is generally regarded as the major excitatory transmitter in the brain of insects [5]. In accordance with this, components of the acetylcholine signaling cascade, such as cholinesterase and the nAChRs, have been extensively studied as targets for insecticides [4, 5].
The mAChRs from insects, however, have been less well studied. In mammals, five mAChRs exist (named m1–m5), which play crucial roles both in the peripheral parasympathetic nervous system (e.g., by stimulating intestinal smooth muscle contractions or decreasing heart rate), and in the central nervous system [6, 7]. All five mammalian receptors can be fully activated by the classical agonist muscarine and blocked by the classical mAChR antagonist atropine [7, 8].
Twenty-four years ago, an insect mAChR was cloned and characterized from D. melanogaster, which was also sensitive to atropine [9]. In this early work, however, the activating effects of the natural ligand, acetylcholine, were not measured and, also, atropine apparently only partially inhibited the receptor. In the current paper, therefore, we recloned this receptor (named A-type mAChR) and found that the cDNA sequence published in Shapiro et al. [9] was incomplete. We also found that the D. melanogaster A-type mAChR could be activated by low concentrations of acetylcholine (EC50, 5 × 10−8 M) and fully inhibited by 10−6 M atropine. In addition, we cloned a second, structurally related mAChR (named B-type) from D. melanogaster and, most surprisingly, found that this receptor was insensitive for atropine and other classical mAChR antagonists. We also cloned similar A- and B-type mAChRs from the red flour beetle Tribolium castaneum and, using bioinformatics, identified A- and B-type orthologues in all arthropods and most other invertebrates with a sequenced genome.
Materials and methods
Animals and culture conditions
All the D. melanogaster strains were cultured on either standard yeast, banana, or corn meal fly food (Nutri-Fly Lot# P4041) at 25 °C under daytime (12 h light, 12 h dark) photoperiod. The D. melanogaster strain used for cloning of the two mAChRs was Canton S. For D. melanogaster RNAi mutants, three different transgenic UAS-RNAi lines corresponding to the A-type mAChR (VDRC-33123, VDRC-101407), or B-type mAChR (VDRC-101217) were obtained from the Vienna Drosophila RNAi Center (VDRC). All RNAi lines targeted regions outside the sites where alternative splicing occurred, so all transcripts were targeted. The insertion in VDRC-33123 corresponds to nucleotide position 362–614 in CG4356, VDRC-101407 corresponds to nucleotide position 1099–1100 in CG4356, and VDRC-101217 corresponds to nucleotide position 1024–1438 in CG7918. The UAS/Gal4 system was used to activate these transgenic lines. These lines were crossed with two different Gal4 drivers: Daughterless-Gal4 (w[w*]; P{w[+mW.hs] = GAL4-da.G32}UH1), and Actin-Gal4 (w[*]; P{w[+mC] = Act5C-GAL4}25FO1/CyO, y[+]) for expressing short hairpin iRNAs that interfere with the target gene transcripts for mutant phenotype studies and only with Actin-Gal4 for the experiments described in Fig. S12. Strain w1118/w1118 (VDRC) was used as a control for genetic analysis and observation of development. To observe a possible phenotype when in mutant flies the A-type mAChR mRNA was downregulated to 50 % or the B-type mAChR mRNA was down-regulated to 20 % of their original values (Fig. S12, Actin-Gal4 system), we measured the times that were needed for 60 freshly laid eggs to develop into first-instar larvae (22–24 h), second-instar larvae (46–48 h), third-instar larvae (72–74 h) and the transition from wandering larvae to pupae (142–146 h). We also measured the weights and the sizes of 60 wild-type and mutant third-instar wandering larvae, as well as their survival rate from eggs to larvae. T. castaneum (strain GA-1) was a kind gift from Prof. Martin Klingler (University of Erlangen-Nürnberg, Germany) and kept on full-grain flour at 29 °C.
PCR
For PCR, total RNA from mixed adult (male and female) flies was isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA). cDNA was synthesized and amplified using the SuperScript III First-Strand Synthesis SuperMix (Invitrogen) or, for RACE, the FirstChoice RLM-RACE kit (Ambion).
Primers (see Table 1) were designed based on the sequence data for Drosophila melanogaster genes CG4356 (Flybase gene name: mAcR-60C) and CG7918 published in (http://www.flybase.org), and from the annotations of their orthologues in Tribolium castaneum (Tc7 and Tc8). A Kozak consensus sequence (highlighted in yellow in Table 1) was included in the 5′ end of the sense primer, which began at the first nucleotide of the CDS. For Tc8, the XhoI restriction site was introduced in the sense primer and SacII in the antisense primer (marked in red type) to facilitate subcloning into the expression vector.
Table 1.
Primer sequences used to amplify the full-length coding sequence of the different mAChR cDNAs
A Kozak consensus sequence (highlighted in yellow) was included in the 5′ end of the sense primers, which begins at the first nucleotide of the CDS. For Tc8, an XhoI restriction site was introduced in the sense primer and a SacII restriction site in the antisense primer (marked in red type) to facilitate subcloning into the expression vector. The underlined nucleotide sequences can be retrieved from Figs. S2, S8, S9, and S10
All PCR products were run on a gel to check their sizes, and subsequently cloned into pCR4-TOPO (Invitrogen) using the TOPO TA cloning kit (Invitrogen) and sequenced. The PCR products of the receptor coding sequences were subcloned into the pIRES2-ZsGreen1 expression vector (Clontech Laboratories, Inc., Mountain View, CA, USA) using the Rapid DNA Ligation Kit (Roche Applied Science, Penzberg, Germany) and sequenced. The cloned cDNA sequences were submitted to GenBank with the following accession numbers: CG4356 Short, JQ922420; CG4356 Long, JG922421; CG7918 short, JX028234; CG7918 Long, JX028235; Tc7, JX 174094; Tc8 short, JQ860106; Tc8 Long, JQ860107.
qPCR
Collection of developmental stages of D. melanogaster: after a 4-h egg lay, the adult flies were removed and the eggs were allowed to develop. Samples were then taken at 20–24 h after egg laying, at 44–48 h after egg laying (2 days), 3, 4, and 5 days. These samples were all of mixed sex. When the larvae reached the third instar larval stage (5 days), it became possible to distinguish between male and female. The sexes were separated and pupae were collected from these flies 2 days after pupariation and 3 days after pupariation. On the same day, the 3-day pupae samples were collected, the remaining pupae hatched. For body parts, 30 male or 30 female adults of mixed age were dissected into head, thorax, and abdomen while under CO2 anesthesia, while six of each were left whole.
Total RNA was isolated using Trizol reagent (Invitrogen), diluted to 500 ng/μl, and DNase treatment following the Rigorous protocol with DNA-Free (Ambion, Austin, TX, USA) to remove genomic DNA. cDNA was synthesized and amplified for either 200 ng of the RNA sample in a 20-μl reaction (for the developmental stages) or 1 μg of the RNA in a 20-μl reaction (for the body parts) using the SuperScript III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen).
qPCR was performed in 25-μl reactions in 96-well plates using Brilliant II SYBR Green QPCR Master Mix (Agilent Technologies, Santa Clara, CA, USA) and the Mx3000P instrument. The qPCR program was 1 cycle for 1 min at 95 °C, followed by 40 times the following cycle: 1 step for 30 s at 95 °C, one step at 55 °C for 1 min, one step at 72 °C for 1 min. A control with no template and a control with no reverse transcriptase were included to check for background or genomic contributions to readings. Gel and melting curve analysis showing a single product of the correct size confirmed specificity of amplification product. PCR efficiencies were calculated from a standard curve made from a dilution series of a mix of the cDNAs from all samples, and were always close to 100 %, so 100 % was used as the standard in all calculations in qBASE Plus (Biogazelle NV, Zwijnaarde, Belgium).
The primers for the two genes of interest [D. melanogaster mAChR A (CG4356) and mAChR B (CG7918)] were designed such that one primer is a standard primer while the other spans an exon/exon-boundary to avoid amplification of residual genomic DNA (Table 2). Reference gene primers (Table 2) did not span an intron–exon boundary. However, their specificity for cDNA was validated by gel electrophoresis and melting curve analyses showing a single product of the correct size.
Table 2.
Primer sequences used in qPCR
| Gene | Direction | Sequence 5′–3′ | Tm (°C) | Nucleotide position (see table legend) |
|---|---|---|---|---|
| CG4356 | Sense | CCCAAGGACGAGTGCTACAT | 54 | 757–776 |
| Anti-sense | GTGTTTTCGTCACTGCTGTTTG | 53 | 947–968 | |
| CG7918 | Sense | GATCACCGACTGGAGCCTTA | 54 | 93–112 |
| Anti-sense | CGCCATCAGAGGATTGATTT | 50 | 303–322 | |
| RNApolII | Sense | CAATCAGAGTCCGCGTAACA | 52 | 2308–2327 |
| Anti-sense | GCTGGTAGTTCTCGGAAACG | 54 | 2462–2481 | |
| RpL32 | Sense | CAAGAAGCTAGCCCAACCTG | 54 | 232–251 |
| Anti-sense | ACGTTGTGCACCAGGAACTT | 52 | 481–500 | |
| RpL11 | Sense | CGATCCCTCCATCGGTATCT | 54 | 442–461 |
| Anti-sense | AACCACTTCATGGCATCCTC | 52 | 563–582 |
The nucleotide positions can be retrieved from Figs. S2, S8, and GenBank accession numbers NM_057358, NM_170460, and NM_057706.4
Raw data from MxPro were imported into qBASE Plus (Biogazelle) for analysis. Reference gene stability was calculated with the GeNorm program in qBASE, and was within acceptable limits for heterogeneous samples. For the developmental stages, M = 1.135 and CV = 0.446. For the body parts, the values are M = 0.811 and CV = 0.3. For definitions of M and CV see [10]. Expressions of target genes CG4356 and CG7918 were normalized against the reference genes (we used the following reference genes: RNA polII, RpL32, and RpLII; see Table 2) and presented as relative values normalized to adult female expression = 1.
Cell culture, transfection, and bioassays
Chinese Hamster Ovary (CHO) cells stably expressing the human G-protein G16 (CHO/G16) were grown as described previously [10–12] and transfected using JetPEI transfection reagent (Polyplus, Illkirch, France) and pIRES2-ZsGreen1 (Invitrogen) vector containing a geneticin-resistance gene and one of the receptor cDNAs described above. After antibiotics selection, using 1 mg Geneticin (Life Technologies, Carlsbad, CA, USA) per ml cell culture medium, a single cell clone from several stably transfected cell clones was selected that gave the highest response in our bioluminescence assay (see below). The bioluminescence assay was performed as described earlier [11–13]. In short, the stably transfected clonal cells were transiently transfected with pcDNA-1/mtAEQ [11], which codes for apoaequorin. To reconstitute the coelenterazine-apoaequorin complex that gives bioluminescence after binding to Ca2+, when Ca2+ increases in a second messenger cascade [11], coelenterazin (Molecular Probes) was added to a final concentration of 5 μM 3 h prior to the assay [11–13]. Bioluminescence was measured on a Victor2 (PerkinElmer, Waltham, MA, USA) [12, 13]. We tested acetylcholine, serotonin, octopamine, tyramine, dopamine, tryptamine, Drosophila-neuropeptide F, and perisulfakinin (all at 10−6 M) and found that the transfected cells were only activated by acetylcholine. The mAChR agonists (acetylcholine and muscarine) and antagonists (atropine, scopolamine, and 3-quinuclidinylbenzinate) were tested at various concentrations (see Figs. 1, 2, S4–S6). When an antagonist was tested together with an agonist, the antagonist was added to the wells 5 min prior to the addition of the agonist.
Fig. 1.
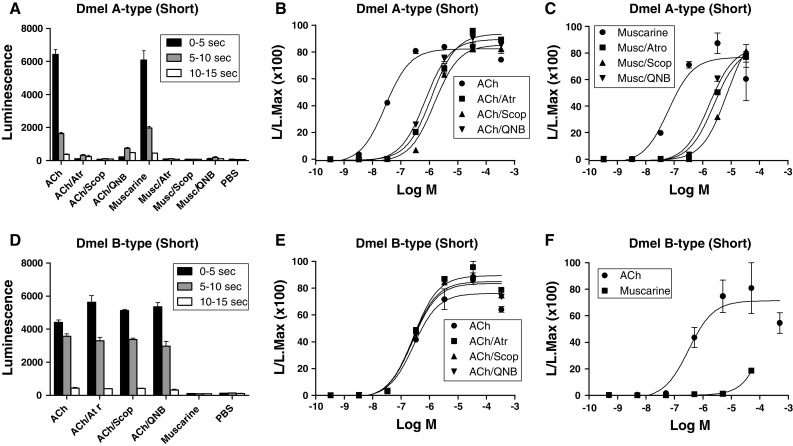
Bioluminescence responses of cloned CHO/G-16 cell lines transfected with DNA coding for the D. melanogaster A- and B-type mAChRs. The vertical bars represent SEM (n = 2 or 3), which sometimes are smaller than the symbols used. In these cases, only the symbols are given. Log M written at the abscissae means log concentration given in M. L/L Max written at the ordinates means luminescence divided by maximal luminescence. A Bioluminescence response of CHO/G-16/mAChR A-type (short version) cells after addition of ACh or muscarine alone (final concentration, 3 × 10−7 M), or in the presence of the antagonists atropine (Atr), scopolamine (Scop), or QNB (final concentration, 3 × 10−7 M), or phosphate-buffered saline (PBS). B Dose–response curves of the effects of ACh with or without antagonists. The transfected cells were activated by ACh with an EC50 of 3 × 10−8 M, while in the presence of 3 × 10−7 M of atropine, scopolamine, or QNB, 100-fold more ACh was needed for activation. C Dose–response curves of the effects of muscarine on these cells, with and without antagonists. The transfected cells were activated by muscarine with an EC50 of 6 × 10−8 M, while the antagonists at 3 × 10−7 M, again, shifted the activation of the receptor by 100-fold. D Bioluminescence response of CHO/G-16/muscarinic acetylcholine receptor B-type (short version) cells after addition of ACh or muscarine alone (final concentration, 3 × 10−7 M), or in the presence of antagonists atropine (Atr), scopolamine (Scop), QNB (final concentration 3 × 10−7 M), or phosphate-buffered saline (PBS). In contrast to the A-type receptor, muscarine does not activate, while the antagonists do not block the B-type receptor. E Dose–response curves of the effects of ACh on these cells, with or without antagonists. The transfected cells were activated by ACh with an EC50 of 3 × 10−7 M, while the antagonists (at 3 × 10−7 M) did not block activation of the receptor. F Dose–response curves of the effects of muscarine or ACh on these cells. The transfected cells were only activated by muscarine at about 1,000-fold higher concentrations than ACh
Fig. 2.
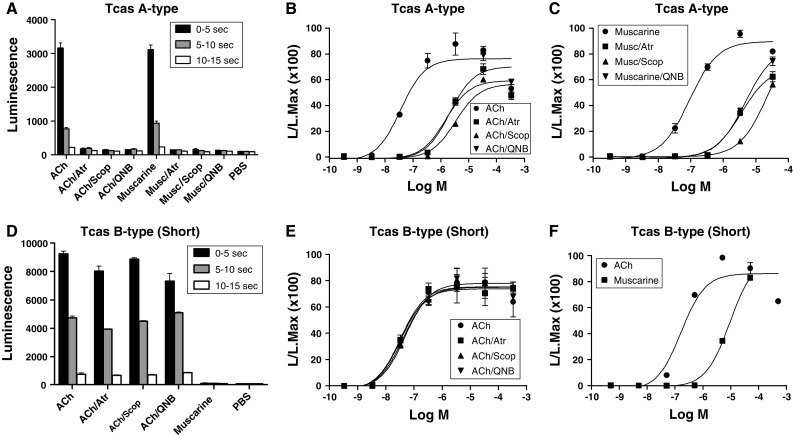
Bioluminescence responses of cloned CHO/G-16 cell lines transfected with DNA coding for the T. castaneum A- and B-type mAChRs. The vertical bars represent SEM (n = 2 or 3), which sometimes are smaller than the symbols used. In these cases, only the symbols are given. A Bioluminescence response of CHO/G-16/mAChR A-type cells after addition of ACh or muscarine alone (final concentration, 3 × 10−7 M), or in the presence of the antagonists atropine (Atr), scopolamine (Scop) or QNB (final concentration, 3 × 10−7 M), or phosphate-buffered saline (PBS). B Dose–response curves of the effects of ACh with or without antagonists. The transfected cells were activated by ACh with an EC50 of 3 × 10−8 M, while the antagonists at 3 × 10−7 M shifted the activation of the receptor by 100-fold. C Dose–response curves of the effects of muscarine on these cells, with and without antagonists. The transfected cells were activated by muscarine with an EC50 of 9 × 10−8 M, while the antagonists at 3 × 10−7 M again shifted the activation of the receptor by 100-fold. D Bioluminescence response of CHO/G-16/mAChR B-type (short version) cells after addition of ACh or muscarine alone (final concentration, 3 × 10−7 M), or in the presence of antagonists atropine (Atr), scopolamine (Scop), QNB (final concentration 3 × 10−7 M), or phosphate-buffered saline (PBS). In contrast to the A-type receptor, muscarine does not activate, and the antagonists do not block the receptor. E Dose–response curves of the effects of ACh on these cells, with or without antagonists. The transfected cells were activated by ACh with an EC50 of 3 × 10−8 M, while the antagonists (at 3 × 10−7 M) did not block activation of the receptor. F Dose–response curves of the effects of muscarine or ACh on these cells. The transfected cells were only activated by muscarine at concentrations 100-fold higher than that of ACh
Software
DNA sequence comparisons were done using CLC Main Workbench Version 6.2 (CLCBio). Protein sequence alignments were carried out using ClustalW (http://www.genome.jp/tools/clustalw/). Phylogenetic tree analyses were performed using Lasergene (DNASTAR). Prediction of transmembrane helices of the receptor protein was done using the TMHMM server (http://www.cbs.dtu.dk/services/TMHMM/). EC50 values were calculated using Prism software.
Results
Recloning and assaying the D. melanogaster A-type mAChR
We recloned the mAChR from D. melanogaster based on the sequence data for gene CG4356 published in http://www.flybase.org. This yielded two splicing variants, differing in 51 nucleotides (Supplementary Material: Figs. S1, S2). Stained gels of the PCR products using primers spanning the alternatively spliced regions showed two bands of about equal intensity (data not shown), indicating that the two splicing variants were about equally expressed. A comparison of the longer sequence with the cDNA sequence published by Shapiro et al. [9] shows that the Shapiro et al. sequence lacks a region of 240 nucleotides, corresponding to the first 80 amino acid residues of the N terminus of the GPCR (Fig. S3), showing that these authors probably assumed a wrong start codon (methionine). This incomplete receptor structure was used by Shapiro et al. [9] and also in later studies [14] for the functional characterization of the receptor. The shorter sequence originates by using an alternative donor splice site in exon 3 (Fig. S1) and corresponds to a sequence published by Onai et al. [15], which was not subsequently functionally characterized. A protein alignment of our currently cloned D. melanogaster receptor sequences, the receptor sequence cloned by Shapiro et al. [9] and that of Onai et al. [15], is given in Fig. S3.
We transfected our cloned cDNAs into CHO cells, selected clones that stably expressed the presumed mAChR cDNAs and tested them for Ca2+ responses, using coelenterazine/aequorin-mediated bioluminescence assays [12, 13]. Figure 1A–C shows that the cDNA for the short variant codes for a receptor (now called the A-type mAChR) that is activated by low concentrations of acetylcholine (EC50 = 3 × 10−8 M) and the agonist muscarine (EC50 = 6 × 10−8 M). The activation by the natural ligand acetylcholine (at its EC50 value 3 × 10−8 M) can be inhibited 100-fold by 3 × 10−7 M atropine, scopolamine, or 3-quinuclidinyl-benzinate (QNB), showing that the pharmacological profile of this receptor resembles that of the mammalian m1–m5 mAChRs (Fig. 1B). Higher concentration (3 × 10−5 M) of the antagonists inhibited the A-type mAChR much more strongly or even fully blocked it (Figs. S5, S6). The longer variant has similar pharmacological properties to the shorter variant (Figs. S4, S5).
Cloning and functional expression of the D. melanogaster B-type mAChR
An earlier bioinformatical analysis of biogenic amine GPCRs in insects showed that the D. melanogaster genome contained another gene, CG7918, that was structurally closely related to the A-type mAChR gene (Fig. S7) [16–18]. We cloned the CG7918 cDNA and, again, found two splicing variants (Fig. S8). Stained gels of the PCR products showed two bands of the same intensity showing, again, that the two splice variants are about equally expressed in intact animals. We expressed both variants in CHO cells, cloned stably expressing cell lines and tested them, using our bioluminescence bioassays, but could only find biological activities in cells transfected with the shorter variant. Figure 1D–F shows that the CG7918 cDNA (shorter variant) codes for a GPCR (now named B-type mAChR) that also is activated by low concentrations of acetylcholine (EC50, 3 × 10−7 M). The classical agonist muscarine could also stimulate the receptor, although 1,000-fold higher concentrations are needed (Fig. 1F). Also in contrast to the A-type mAChR, the B-type mAChR (at 3 × 10−7 M acetylcholine) could not be blocked by 10−6 M atropine or other classical mAChR antagonists, such as scopolamine or QNB (Fig. 1E). Even at 10−4 M, none of the antagonists were able to block the B-type mAChR significantly (Fig. S6C).
Cloning of the A- and B-type mAChRs from T. castaneum
In addition to the D. melanogaster A- and B-type mAChRs, we also cloned their orthologues from the red flour beetle T. castaneum (Figs. S7, S9, S10). For the T. castaneum A-type receptor cDNA, we only found one single transcript (Fig. S9), while for the B-type receptor cDNA we found, again, a shorter and a longer variant (Fig. S10). When stably expressed in CHO cells, the T. castaneum A-type mAChR was activated by low concentrations of acetylcholine (EC50, 3 × 10−8 M) and muscarine (EC50, 9 × 10−8 M) and this activation (at 3 × 10−8 M acetylcholine) was inhibited 100-fold by 3 × 10−7 M atropine, scopolamine, or QNB (Fig. 2A–C). The shorter variant of the B-type mAChR (EC50 for acetylcholine, 3 × 10−8 M) was, again, only stimulated by much higher concentrations of muscarine compared to acetylcholine (about 100-fold higher) and was not blocked (at 3 × 10−6 M acetylcholine) by 10−6 M scopolamine, atropine, or QNB (Fig. 2D–F). Even higher concentrations of the antagonists (10−4 M) did not block the T. castaneum B-type mAChR significantly (Fig. S6F). The longer variant B-type mAChR had similar pharmacological properties to the shorter variant (Fig. S4, S6).
Quantitative PCR (qPCR) of the A- and B-type mAChRs
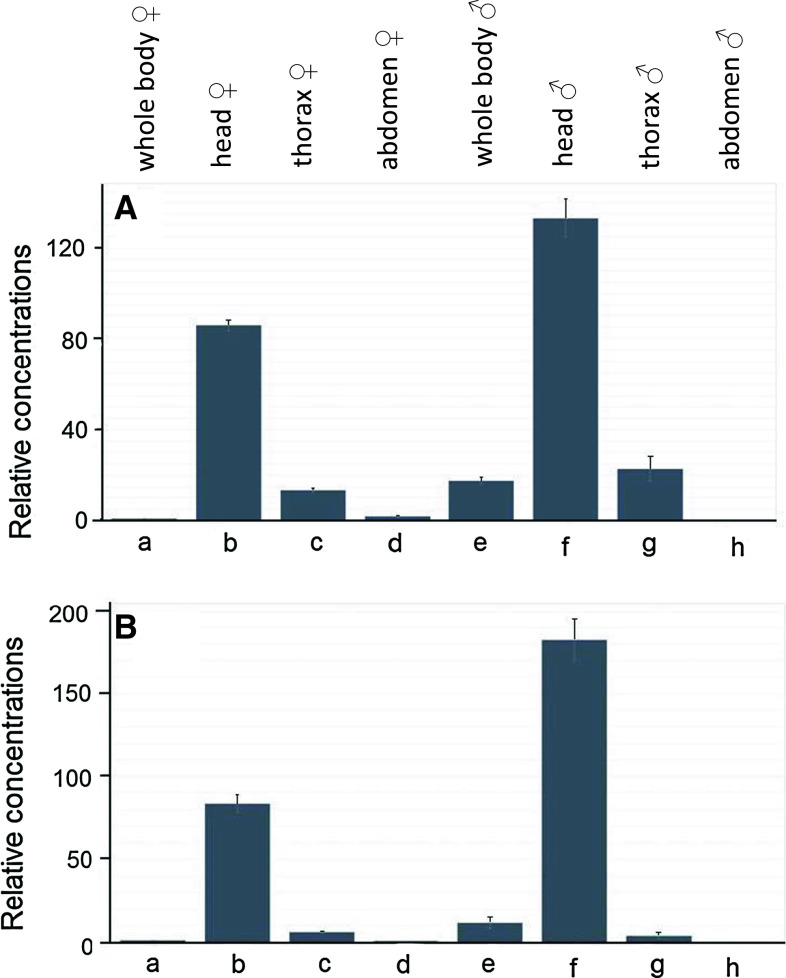
We carried out qPCR of cDNA isolated from different body parts of D. melanogaster. We found a similar distribution pattern for both types of mAChRs, with the head (presumably the brain) in females having 80 times higher concentrations of the receptor mRNAs compared to that of the whole body (Fig. 3). Also in males, expression of both mAChRs was found mainly in the head (Fig. 3).
Fig. 3.
qPCR data for the expression of the D. melanogaster A-type mAChR gene CG4356 (A), and B-type mAChR gene CG7918 (B), in different body parts of adult male and female flies. The combined expression (long and short splicing variants) of the A-type and the combined expression (long and short splicing variants) of the B-type receptor were measured. At least 30 body parts were pooled for each mRNA isolation. These pools are the same for each body part column in A and B. The qPCR experiments were run in triplicate. The vertical bars in each column (which are sometimes smaller than the lines of the column) represent SEM. The mRNA concentrations given are relative to column a (a = 1). a female whole body; b female head; c female thorax; d female abdomen; e male whole body; f male head; g male thorax; h male abdomen. Note that both receptor genes are strongly expressed in the head (presumably the brain) of both males and females
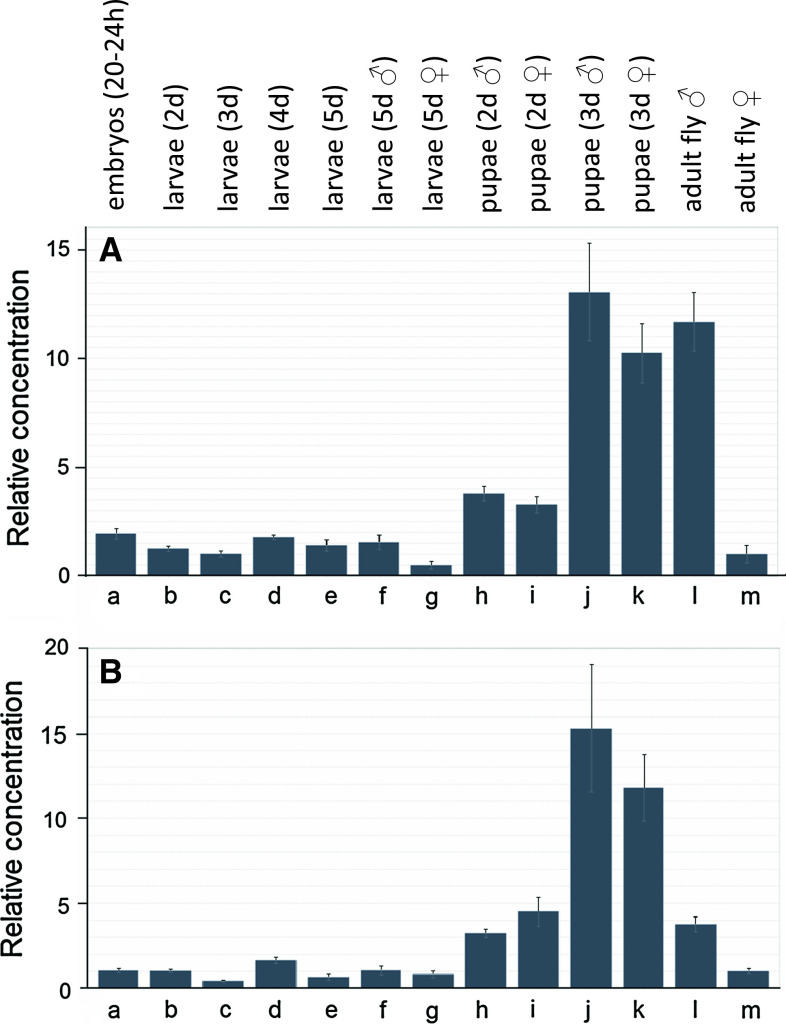
When different developmental stages (eggs, various larval stages, pupae, adults) were compared, we found a high expression of both mAChRs in 3-day-old male and female pupae, and in male adults, while female adults contained about 4–12 times lower concentrations of the mRNAs for the two mAChRs compared to males (Fig. 4).
Fig. 4.
qPCR data for the expression of the D. melanogaster A-type mAChR gene CG4356 (A) and B-type mAChR gene CG7918 (B) in different developmental stages. The combined expression (long and short splicing variants) of the A-type and the combined expression (long and short splicing variants) of the B-type receptor were measured. At least ten animals were pooled for each developmental stage. These pools are the same for each developmental stage in A and B. The sexes are mixed in columns a–e (they could not be discriminated), otherwise they are separated. The mRNA concentrations given are relative to column m (m = 1). Other conditions are as in Fig. 3. a 20–24 h old eggs; b larvae 2 days after egg laying(= first instar); c larvae 3 days after egg laying (= second instar); d larvae 4 days after egg laying (= second instar); e larvae 5 days after egg laying (= third instar); f male third-instar larvae; g female third-instar larvae; h 2-day-old male pupae; i 2-day-old female pupae; j 3-day-old male pupae; k 3-day-old female pupae; l adult male flies; m adult female flies. Note that both receptor genes are strongly expressed in older pupae of both sexes and in adult males
We also looked for the time course of receptor mRNA expression after eclosion (emergence) from pupae and found that the expression for the two receptors stayed high for males, while it strongly dropped by a factor 15–20 within 2 days for females. This dramatic drop in female receptor expression was independent from the absence or presence of mating after eclosion (Fig. S11).
Transgenic fly lines with down-regulated concentrations of mRNAs coding for the A- and B-type mAChRs
We obtained several transgenic D. melanogaster RNAi lines that, after crossing with Gal4 driver fly lines, should down-regulate the mRNAs for either the A- or B-type mAChR in all tissues. Using qPCR, we found that in two such RNAi lines the mRNA concentrations of the A-type mAChR were down-regulated to 50 % of their original values. In another RNAi line, the mRNA concentrations of the B-type mAChR was down-regulated to 20 % of its original value (Fig. S12). A gross inspection of the phenotypes of both RNAi lines, however, did not reveal any differences compared to the wild type. For example, the reproduction rate (=number of eggs), the developmental timing of eggs and larvae, the survival rate from eggs to larvae, and the sizes and weight of the third-instar wandering larvae were the same in both A- and B-type receptor mutants and wild type.
Presence of A- and B-type mAChRs in other arthropods
Using TBLASTN, we found that all the insects with a sequenced genome that we investigated contained both an A- and B-type mAChR (Table S1, Supplementary Material). Also other arthropods, such as the water flea Daphnia pulex (Crustacea), and the tick Ixodes scapularis (Chelicerata) contained both types of mAChRs, suggesting that the presence of an A- and B-type mAChR is a general phenomenon in arthropods.
Discussion
In this paper, we have found that the fruit fly D. melanogaster (order: Diptera, or flies), the red flour beetle T. castaneum (order: Coleoptera, or beetles), and probably all other insects and arthropods contain an A- and a B-type mAChR (Table S1). The insect A-type mAChRs can be activated by low concentrations (10−8 M) of acetylcholine and muscarine, and blocked by the classical mAChR antagonists atropine, scopolamine, and QNB (Fig. 1). They have, thus, the same overall pharmacological properties as the m1–m5 mAChRs from mammals. The insect B-type mAChRs, however, are pharmacologically different from the A-type and m1–m5 receptors, because, although they are activated by low concentrations of acetylcholine, they cannot be effectively activated by muscarine and cannot be blocked by the above-mentioned classical antagonists (Figs. 1, 2). These pharmacological differences between the two types of receptors must originate from structural differences. This idea is confirmed by phylogenetic tree analyses of the overall receptor sequences, where the insect A-type and mammalian m1–m5 receptors cluster closely together, while the B-type receptors form a different cluster (Fig. S13). One could imagine that the binding pocket for the mAChR antagonists is different in the A- and B-type mAChRs, because atropine, scopolamine, and QNB fit in the binding pocket for the A-type receptors, while they apparently do not fit very well in the binding pocket for the B-type receptors. A recent X-ray study of the crystallized human m2 receptor bound to its antagonist QNB showed that the binding pocket for QNB is a water-filled funnel-shaped vestibule formed by the TM3-TM7 transmembrane helices of the receptor [19] and that 14 amino acid residues are involved in the antagonist binding (14 individually numbered residues highlighted in red and grey in Fig. 5). Modeling of acetylcholine into the QNB binding pocket revealed a subpopulation of six amino acid residues that might be the binding sites for this natural agonist [19] (six numbered residues highlighted in red in Fig. 5). Also, the structure of the m3 receptor bound to its antagonist tiotropium has recently been reported [20]. The binding pocket for tiotropium is identical to that of QNB in the m2 receptor [20].
Fig. 5.
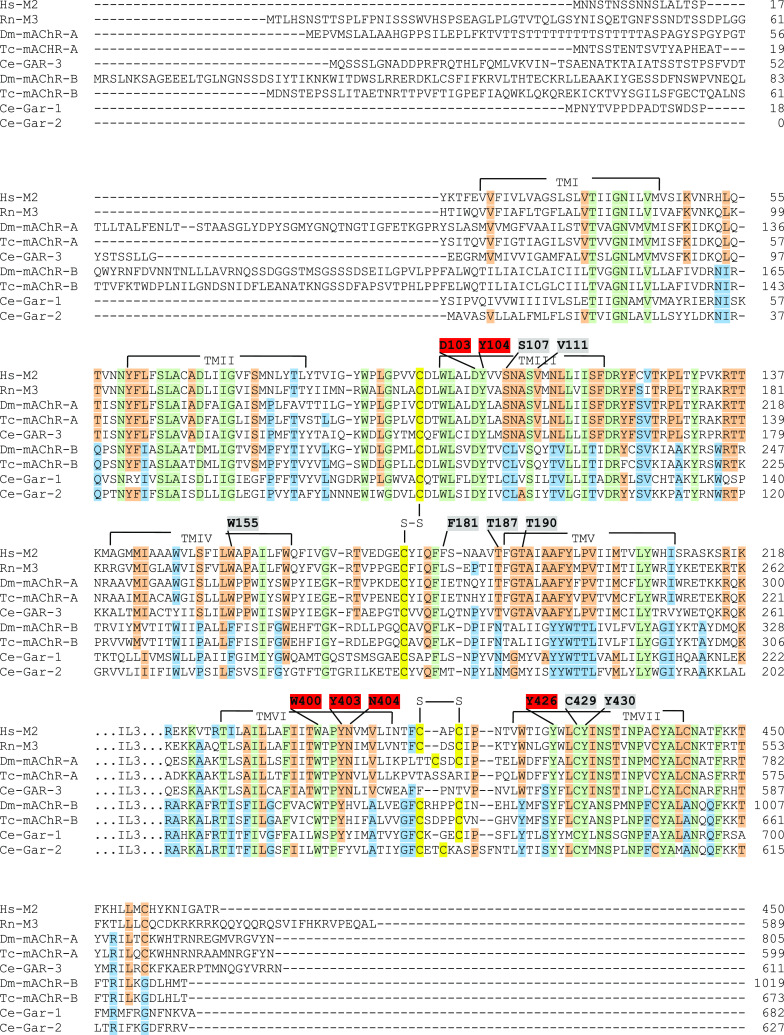
Sequence alignments of the human m2 mAChR (Hs-M2), rat m3 (Rn-M3), D. melanogaster A-type (Dm-mAChR-A), T. castaneum A-type (Tc-mAChR-A), C. elegans A-type (Ce-GAR-3), D. melanogaster B-type (Dm-mAChR-B), T. castaneum B-type (Tc-mAChR-B), and the two C. elegans B-type mAChRs (Ce-GAR-1 and -2). The 14 QNB binding residues of the human m2 receptor [19] are indicated by numbers above the m2 sequence and highlighted in both red and grey. The red numbers are the proposed (“modeled”) binding residues for acetylcholine [19]. The transmembrane helices are indicated by TM1-TMVII. Cystine bridges are highlighted in yellow and indicated by “S–S”. The amino acids residues that are identical in all five A-type mAChRs are highlighted in orange; those that are identical in all B-type mAChRs are highlighted in blue. Green indicates residues that are identical in both A- and B-type mAChRs
What would one expect when the acetylcholine (agonist) and QNB (antagonist) binding pockets are compared between the A- and B-type mAChRs? One would expect that (1) the six amino acid residues forming the acetylcholine binding pockets are the same in both types of mAChRs, because acetylcholine is the natural agonist, having comparable efficacy for both receptors (Figs. 1, 2); (2) about eight (14 minus 6) amino acid residues, binding QNB but not acetylcholine, should be the same in all A-type mAChRs, because these receptors are blocked by QNB; (3) these eight amino acid residues, binding QNB but not acetylcholine, however, should be mostly different between the A- and B-type mAChRs, because QNB blocks the A- but not the B-type receptors.
Impressively, this prediction is exactly what we find (Fig. 5): (1) the acetylcholine-binding residues D103, Y104, W400, Y403, and Y426 [19], highlighted by red numbered residues in Fig. 5, are identical in the A- and B-type mAChRs from D. melanogaster and T. castaneum. N404 (Fig. 5) was also proposed to be involved in acetylcholine binding in the A-type m2 receptor [19], but in the B-type AChR binding pocket of insects this residue is replaced by histidine, making it unlikely that it is involved in acetylcholine binding. Instead, N404 might only be involved in the hydrogen bonding of the ester group of QNB [19]. This conclusion is in agreement with N404A mutations in the m1 and m3 receptors, which greatly affected QNB binding, while they had little or no effect on the binding or activation by acetylcholine [19, 21, 22]; (2) those residues that bind QNB, but are not involved in the binding of acetylcholine (see above) are S107, V111, W155, F181, T187, T190, C429, and Y430 (highlighted by grey numbers in Fig. 5) [19]. These QNB-specific binding residues are identical in the Drosophila and Tribolium A-type, the human m2, and the rat m3 mAChRs with the exception of F181, C429, and Y430. Thus, our prediction fits for five out of eight residues. F181 is known to be varying between the m1–m5 receptors [19, 20]. Therefore, it might be sufficient that this residue is aromatic or aliphatic, enabling van der Waals interactions with one of the two phenyl rings of QNB [19]. C429 and Y430 are present in both the A- and B-type mAChRs (Fig. 5) and might be essential for both acetylcholine and QNB binding, i.e., they might be regarded as acetylcholine binding sites; (3) finally, also in accordance to our predictions, the QNB-specific binding residues S107, V111, W155, F181, T187, and T190 are different in the A- and B-type receptors (Fig. 5). Again, C429 and Y430 are the same, suggesting that they are acetylcholine binding sites.
When searching the literature, we found that mAChRs with a pharmacology different from the m1–m5 receptors also exist in the nematode Caenorabditis elegans, where three mAChRs have been cloned and functionally expressed in CHO cells, named GAR-1, -2, and -3 [23–25]. GAR-3 activation by the agonist carbachol is fully blocked by atropine [25], showing that it has a similar pharmacology to the m1–m5 receptors and, thus, is an A-type mAChR. GAR-1 and GAR-2 activation by agonists, however, could not be blocked by atropine and scopolamine [23, 24], suggesting that they are B-type mAChRs. These conclusions are confirmed when we look at the acetylcholine and antagonist binding sites in the same way as we did for the D. melanogaster mAChRs (Fig. 5): (1) the “modeled” acetylcholine binding sites corresponding to the m2 residues D103, Y104, W404, Y426, C429, and Y430 are the same in all three C. elegans mAChRs. Again, there is an exception for the proposed acetylcholine binding site Y404, which instead might be only involved in antagonist binding; (2) the QNB binding residues are the above-mentioned residue plus S107, V111, W155, F181, T187, T190, C429, and Y430. These are the same between m2 and GAR-3, meaning that GAR-3 is an A-type mAChR. Again, the only exception is F181, but this residue is varying in the m1–m5 receptors [19, 20]; (3) when S107, V111, W155, F181, T187, T190, C429, and Y430 from m2 are compared between m2 and GAR-1 and -2, these residues are different, meaning that GAR-1 and -2 are B-type mAChRs. The residues corresponding to C429 and Y430 do not fit in this conclusion because they are identical in GAR-1, -2, and m2 arguing, again, that they could be acetylcholine binding sites.
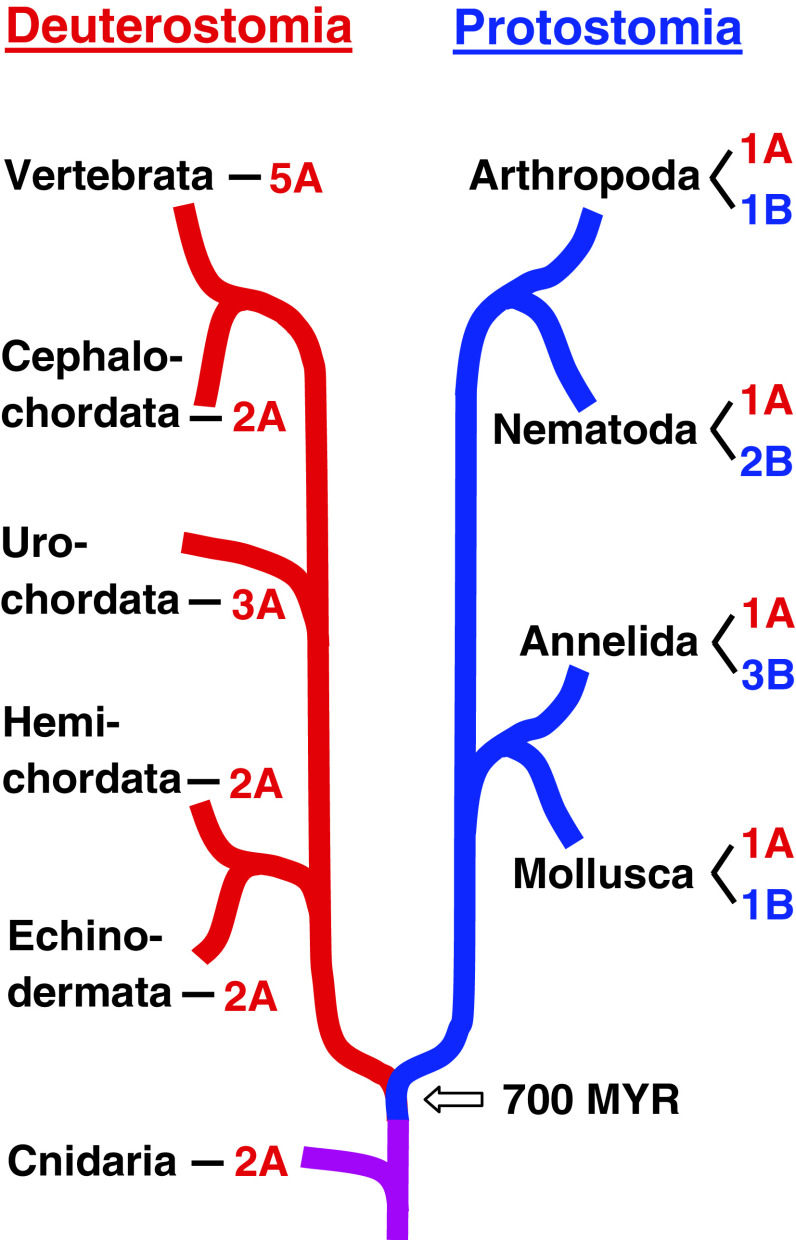
Insects and nematodes belong to the Protostomia, while humans and other vertebrates belong to the Deuterostomia. We, therefore, wondered whether perhaps all protostomes contain A- and B-type mAChRs, while deuterostomes perhaps would only have A-type receptors. Therefore, we investigated selected proto- and deuterostomian species where the genome sequences are known, and also other genome-sequenced animals that originated before the split of Proto- and Deuterostomia. Our criteria for assigning the receptors as A- or B-type were (1) comparisons of their overall protein sequences, using phylogenetic tree analyses (Fig. S13) and (2) the presence (A-type) or absence (B-type) of residues identical to the m2 QNB binding sites (see discussion above). We found that all investigated protostomes contained one A-type and one, or more B-type mAChRs, (Fig. 6, Fig. S13, Table S1). In the deuterostomian lineage, we found that basal deuterostomes contained 2–3 A-type receptors, whereas all vertebrates contain five A-type receptors. No B-type receptors could be found in deuterostomes (Fig. 6, S13, Table S1).
Fig. 6.
An overview of the emergence of A- and B-type mAChRs in Protostomia (highlighted by blue lines) and Deuterostomia (highlighted by red lines). Cnidarians evolved before the split of Proto- and Deuterostomia, which is indicated by a purple line. The B-type mAChRs appear to be confined to the Protostomia, whereas the A-type receptors occur in both evolutionary lineages and in cnidarians. Our criteria for assigning the receptors as A- or B-type were (1) comparisons of their overall protein sequences, using phylogenetic tree analyses (Fig. S13) and (2) the presence (A-type) or absence (B-type) of residues identical to the m2 QNB-specific binding residues S107, V111, W155, T187, and T190 (See Fig. 5 and “Discussion”). Both criteria should be fulfilled
Animals that originated before the split of Proto- and Deuterostomia, such as cnidarians (Hydra) contained two A-type AChRs (Fig. 6, S13). These results show that mAChRs originated during early animal evolution and suggest that A-type mAChRs are the most basal form of mAChRs. This constellation has been conserved in the deuterostomian lineage, where several duplications of the A-type receptor gene led to the m1–m5 receptors that we know from mammals today (Fig. 6). In the protostomian lineage, one of the two A-type receptor genes probably mutated into a B-type receptor gene. During the evolution of annelids and nematodes additional gene duplications occurred, leading to two (nematodes) and three (annelids) B-type receptors further to the single A-type receptor that is always present in protostomes (Fig. 6).
Conclusions
We have discovered that Drosophila and other arthropods contain two different mAChRs: (1) the A-type mAChR that closely resembles the mammalian m1–m5 mAChRs, and (2) the B-type that is different from the A-type in that it is not sensitive to the classical mAChR antagonists atropine, scopolamine, and QNB. Also, the QNB binding pocket in the B-type mAChRs is quite different from that of the A-type mAChRs, while the QNB binding pocket in the A-type mAChRs is identical to those in the mammalian m1–m5 receptors. Furthermore, we have investigated the deep evolutionary roots of the mAChRs and found that they must have evolved before the split of Proto- and Deuterostomia about 700 MYR ago.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Johannes Thomsen for typing the manuscript, and the Danish Research Agency, Novo Nordisk Foundation, and Carlsberg Foundation for financial support.
Abbreviations
- CHO
Chinese hamster ovary
- mAChR
Muscarinic acetylcholine receptor
- MYR
Million years
- QNB
3-Quinuclidinyl-benzylate
- qPCR
Quantitative PCR
Footnotes
References
- 1.Douzéry EJ, Snell EA, Bapteste E, Delsuc F, Philippe H. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc Natl Acad Sci USA. 2004;101:15386–15391. doi: 10.1073/pnas.0403984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yakel JL. Gating of nicotinic Ach receptors: latest insights into ligand binding and function. J Physiol. 2010;588:597–602. doi: 10.1113/jphysiol.2009.182691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Millar NS, Harkness PC. Assembly and trafficking of nicotinic acetylcholine receptors (Review) Mol Membr Biol. 2008;25:279–292. doi: 10.1080/09687680802035675. [DOI] [PubMed] [Google Scholar]
- 4.Jones AK, Sattelle DB. Diversity of insect nicotinic acetylcholine receptor subunits. Adv Exp Med Biol. 2010;683:25–43. doi: 10.1007/978-1-4419-6445-8_3. [DOI] [PubMed] [Google Scholar]
- 5.Dupuis J, Louis T, Gauthier M, Raymond V. Insights from honeybee (Apis mellifera) and fly (Drosophila melanogaster) nicotinic acetylcholine receptors: from genes to behavioral functions. Neurosci Biobehav Rev. 2012;36:1553–1564. doi: 10.1016/j.neubiorev.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Bubser M, Byun N, Wood MR, Jones CK. Muscarinic receptor pharmacology and circuitry for the modulation of cognition. Hanb Exp Pharmacol. 2012;208:121–166. doi: 10.1007/978-3-642-23274-9_7. [DOI] [PubMed] [Google Scholar]
- 7.Tobin G, Giglio D, Lundgren O. Muscarinic receptor subtypes in the alimentary tract. J Physiol Pharmacol. 2009;60:3–21. [PubMed] [Google Scholar]
- 8.Harvey RD. Muscarinic receptor agonists and antagonists: effects on cardiovascular function. Hanb Exp Pharmacol. 2012;208:299–316. doi: 10.1007/978-3-642-23274-9_13. [DOI] [PubMed] [Google Scholar]
- 9.Shapiro RA, Wakimoto BT, Subers EM, Nathanson NM. Characterization and functional expression in mammalian cells of genomic and cDNA clones encoding a Drosophila muscarinic acetylcholine receptor. Proc Natl Acad Sci USA. 1989;86:9039–9043. doi: 10.1073/pnas.86.22.9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-CPR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:R0034.1–R0034.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stables J, Green A, Marshall F, Fraser N, Knight E, Sautel M, Milligan G, Lee M, Rees S. A bioluminescent assay for agonist activity at potentially any G-protein-coupled receptor. Anal Biochem. 1997;252:115–126. doi: 10.1006/abio.1997.2308. [DOI] [PubMed] [Google Scholar]
- 12.Secher T, Lenz C, Cazzamali G, Sørensen G, Williamson M, Hansen GN, Svane P, Grimmelikhuijzen CJP. Molecular cloning of a functional allatostatin gut/brain receptor and an allatostatin preprohormone from the silkworm Bombyx mori . J Biol Chem. 2001;276:47052–47060. doi: 10.1074/jbc.M106675200. [DOI] [PubMed] [Google Scholar]
- 13.Staubli F, Jørgensen TJD, Cazzamali G, Williamson M, Lenz C, Søndergaard L, Roepstorff P, Grimmelikhuijzen CJP. Molecular identification of the insect adipokinetic hormone receptors. Proc Natl Acad Sci USA. 2002;99:3446–3451. doi: 10.1073/pnas.052556499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millar NS, Baylis HA, Reaper C, Bunting R, Mason WT, Sattelle DB. Functional expression of a cloned Drosophila muscarinic acetylcholine receptor in a stable Drosophila cell line. J Exp Biol. 1995;198:1843–1850. doi: 10.1242/jeb.198.9.1843. [DOI] [PubMed] [Google Scholar]
- 15.Onai T, FitzGerald MG, Arakawa S, Gocayne JD, Urquhart DA, Hall LM, Fraser CM, McCombie WR, Venter JC. Cloning, sequence analysis and chromosome localization of a Drosophila muscarinic acetylcholine receptor. FEBS Lett. 1989;255:219–225. doi: 10.1016/0014-5793(89)81095-6. [DOI] [PubMed] [Google Scholar]
- 16.Brody T, Cravchik A. Drosophila melanogaster G protein-coupled receptors. J Cell Biol. 2000;150:F83–F88. doi: 10.1083/jcb.150.2.F83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hauser F, Cazzamali G, Williamson M, Blenau W, Grimmelikhuijzen CJP. A review of neurohormone GPCRs present in the fruitfly Drosophila melanogaster and the honey bee Apis mellifera . Prog Neurobiol. 2006;80:1–19. doi: 10.1016/j.pneurobio.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Hauser F, Cazzamali G, Williamson M, Park Y, Li B, Tanaka Y, Predel R, Neupert S, Schachtner J, Verleyen P, Grimmelikhuijzen CJP. A genome-wide inventory of neurohormone GPCRs in the red flour beetle Tribolium castaneum . Front Neuroendocrinol. 2008;29:142–165. doi: 10.1016/j.yfrne.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Haga K, Kruse AC, Asada H, Yurugi-Kobayashi T, Shiroishi M, Zhang C, Weis WI, Okada T, Kobilka BK, Haga T, Kobayashi T. Structure and function of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, Rosemond E, Green HF, Liu T, Chae PS, Dror RO, Shaw DE, Weis WI, Wess J, Kobilka BK. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blüml K, Mutschler E, Wess J. Functional role in ligand binding and receptor activation of an asparagine residue present in the sixth transmembrane domain of all muscarinic acetylcholine receptors. J Biol Chem. 1994;269:18870–18876. [PubMed] [Google Scholar]
- 22.Ward SD, Curtis CA, Hulme EC. Alanine-scanning mutagenesis of transmembrane domain 6 of the M(1) muscarinic acetylcholine receptor suggests that Tyr381 plays key roles in receptor function. Mol Pharmacol. 1999;56:1031–1041. doi: 10.1124/mol.56.5.1031. [DOI] [PubMed] [Google Scholar]
- 23.Lee Y-S, Park Y-S, Chang DJ, Hwang JM, Min CK, Kaang B-K, Cho NJ. Cloning and expression of a G protein-linked acetylcholine receptor from Caenorhabditis elegans . J Neurochem. 1999;72:58–65. doi: 10.1046/j.1471-4159.1999.0720058.x. [DOI] [PubMed] [Google Scholar]
- 24.Lee Y-S, Park Y-S, Nam S, Suh S-J, Lee J, Kaang B-K, Cho NJ. Characterization of GAR-2, a novel G protein-linked acetylcholine receptor from Caenorhabditis elegans . J Neurochem. 2000;75:1800–1809. doi: 10.1046/j.1471-4159.2000.0751800.x. [DOI] [PubMed] [Google Scholar]
- 25.Park Y-S, Cho T-J, Cho NJ. Stimulation of cyclic AMP production by the Caenorhabditis elegans muscarinic acetylcholine receptor GAR-3 in Chinese hamster ovary cells. Arch Biochem Biophys. 2006;450:203–207. doi: 10.1016/j.abb.2006.03.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.