Abstract
The Saccharomyces cerevisiae VPS55 (YJR044c) gene encodes a small protein of 140 amino acids with four potential transmembrane domains. VPS55 belongs to a family of genes of unknown function, including the human gene encoding the obesity receptor gene-related protein (OB-RGRP). Yeast cells with a disrupted VPS55 present normal vacuolar morphology, but exhibit an abnormal secretion of the Golgi form of the soluble vacuolar carboxypeptidase Y. However, trafficking of the membrane-bound vacuolar alkaline phosphatase remains normal. The endocytosis of uracil permease, used as an endocytic marker, is normal in vps55Δ cells, but its degradation is delayed and this marker transiently accumulates in late endosomal compartments. We also found that Vps55p is mainly localized in the late endosomes. Collectively, these results indicate that Vps55p is involved in late endosome to vacuole trafficking. Finally, we show that human OB-RGRP displays the same distribution as Vps55p and corrects the phenotypic defects of the vps55Δ strain. Therefore, the function of Vps55p has been conserved throughout evolution. This study highlights the importance of the multispanning Vps55p and OB-RGRP in membrane trafficking to the vacuole/lysosome of eukaryotic cells.
INTRODUCTION
The vacuole of Saccharomyces cerevisiae, the equivalent of the mammalian lysosome, serves as an important storage reservoir for amino acids, small ions, and polyphosphates, and is essential for osmoregulation and ion homeostasis (for a review, see Jones et al., 1997). The vacuole receives endocytic traffic from the cell surface, biosynthetic traffic from the late Golgi compartments, and elements from the cytoplasm (Bryant and Stevens, 1998; Conibear and Stevens, 1998; Klionsky and Emr, 2000). Newly synthesized vacuolar hydrolases transit through the early stages of the secretory pathway and are actively sorted away from secreted proteins in the late Golgi compartment, the equivalent of the mammalian trans-Golgi network (TGN), before being delivered to the vacuole (Raymond et al., 1992; Vida et al., 1993; Bryant and Stevens, 1998; Conibear and Stevens, 1998). There are at least two routes from the Golgi apparatus to the vacuole. Various proteins, including the vacuolar carboxypeptidase Y (CPY) itself, follow the so-called “CPY pathway” and transit through a prevacuolar/endosomal compartment (PVC; Conibear and Stevens, 1998). The alkaline phosphatase (ALP) follows an alternative route to the vacuole, the adaptor protein-3-mediated pathway (AP-3), which bypasses the PVC, leading directly from the late Golgi to the vacuole (Conibear and Stevens, 1998). Proteins from the plasma membrane that have undergone endocytosis converge with the CPY pathway at the PVC (Piper et al., 1995; Rieder et al., 1996).
The overproduction of CPY or mutations in its gene result in the missorting and secretion of the protein into the medium (Stevens et al., 1986; Johnson et al., 1987). During the past years, classical genetic screens in yeast, notably based on the selection of mutants secreting CPY in the medium, have led to the identification of >50 gene products involved in biosynthetic protein trafficking to the vacuole (Bankaitis et al., 1986; Rothman and Stevens, 1986; Stack et al., 1995; For review, see Conibear and Stevens, 1998; Lemmon and Traub, 2000). These different genes, referred to as VPS (vacuolar protein sorting), have been classified in several complementation groups according to the phenotype and the morphology of endocytic compartments of the cells carrying null mutations (class A, B, etc.; Raymond et al., 1992).
The sequencing of the S. cerevisiae genome permits the use of new strategies to characterize functionally important genes, in particular, those involved in the VPS pathway. A more global approach is the systematic screening of strains disrupted for one given gene. The European Functional Analysis Network (EUROFAN) project has established a collection of 800 strains disrupted for nonessential genes (Oliver, 1996), and a complete library of S. cerevisiae mutant strains is now available (Delneri et al., 2001). To identify potential defects in the vacuolar pathway, i.e., VPS genes, we have performed a systematic analysis of the EUROFAN collection by monitoring the transport of the vacuolar CPY in the secretory pathway and its targeting to the vacuole (Avaro et al., 2002). In this study, we report the characterization of YJR044c, a new VPS gene that we have named VPS55.
VPS55 encodes a small protein of 140 amino acids with four potential transmembrane domains with homologs of unknown function in a large number of species, including human OB-RGRP (obesity receptor gene-related protein). The OB-RGRP transcript is derived by alternative splicing from the same mRNA as OB-R, the leptin receptor that is involved in the regulation of body weight, but it does not display any sequence homology with the leptin receptor itself (Bailleul et al., 1997). OB-R and OB-RGRP have similar patterns of expression, suggesting that their functions may be connected (Bailleul et al., 1997).
This study provides the first clues to the function of a member of the OB-RGRP family, Vps55p. We show that disruption of the VPS55 gene leads to secretion of the CPY precursor into the medium and a delay in the late steps of endocytosis. We report the localization of Vps55p and show that human OB-RGRP produced in yeast complements the disruption of the VPS55 gene.
MATERIALS AND METHODS
Media and Growth Conditions
The S. cerevisiae strains and plasmids used in this study are listed in Table 1. Cells were grown in YPD medium (1% yeast extract, 2% peptone, 2% glucose) or in defined YNB minimal medium containing 0.67% yeast nitrogen base without amino acids (Difco, Detroit, MI) supplemented with appropriate nutrients (Sherman et al., 1986). The carbon source was 2% glucose, lactate, or galactose.
Table 1.
Strain and plasmid list
| Genotype | Source | |
|---|---|---|
| Strains | ||
| FHER001-03B | Matα ura3-52 leu2Δ1 his3Δ200 | EUROFAN |
| FHERBN1 | Matα ura3-52 his3Δ200 leu2-1 kanMX6 VPS55-GFP | This study |
| FHERBN2 | Matα ura3-52 his3Δ200 leu2-1 kanMX6 VPS55-HA | This study |
| FFNS008-07C | Matα his3Δ200 leu2Δ1 ura3 YJR044c∷kanMX4 | EUROFAN |
| RH2605 | Matα leu2 ura3 his bar1 end13-1/vps4 | Munn and Riezman, 1994 |
| RH2180 | Matα ade6 his4 leu2 ura3 pep4 vps1-delta2 | Munn and Riezman, 1994 |
| RPY2 | Matα leu2-3,112 ura3-52 his4-519 ade6 gal2 vps27-1 | Piper et al., 1995 |
| SEY5076 | Matα ura3-52 leu2, 3, -112 sec7-1 | Emr et al., 1984 |
| FY SEC7GFP | Matα his3 leu2 ura3 SEC7-GFP | Séron et al., 1998 |
| FY SEC7GFPBN1 | Matα his3 leu2 ura3 kanMX6 SEC7-GFP VPS55-Myc | This study |
| IW-6A | Matα ura3-1 his3-11,-15 leu2-3 112 trp1 pep4-3 | Volland et al., 1994 |
| RPY2BN1 | Matα leu2-3,112 ura3-52 his4-519 ade6 gal2 vps27-1 kanMX6 VPS55-GFP | This study |
| RPY2BN5 | Matα leu2-3,112 ura3-52 his4-519 ade6 gal2 vps27-1 kanMX6 VPS55-HA | This study |
| MHY1269 | Matα his3-Δ200 leu2-3,112 ura3-52 lys2-801 trp1-1 vps27-Δ1∷LEU2 | Swaminathan et al., 1999 |
| Y01271 | Mat α his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 vps35Δ∷kanMX4 | EUROSCARF |
| Y01271BN1 | Mat α leu2Δ0 met15Δ0 ura3Δ0 vps35Δ∷kanMX4 VPS55-HA | This study |
| FDRN008-05A(A) | Matα ura3-52 trp1Δ 63 apm3Δ∷kanMX4 | EUROFAN |
| Plasmids | ||
| PFL38gF | ARS/CEN, URA3, GAL10-FUR4 | Séron et al., 1999 |
| pFL38gF-GFP | ARS/CEN, URA3, GAL10-FUR4-GFP (S65G, S72A) | Dupré and HaguenauerTsapis, 2001 |
| pgF (PAP3) | 2μ, LEU2, GAL10-FUR4 | Jund et al., 1988 |
| PYEF2 | 2μ, URA3 | Cullin and MinvielleSebastia, 1994 |
| PYEF2-GAL10-VPS55-HA | 2μ, URA3, GAL10-VPS55-HA | This study |
| PYEF2-GAL10-OB-RGRP-HA | 2μ, URA3, GAL10-OB-RGRP-HA | This study |
| pYCG-YJR044C (pYCG-VPS55) | ARS, URA3, VPS55 | EUROFAN |
| Oligonucleotides | Sequence | |
| VPS55-F | 5′-CGC GGA TCC ATC GAT ATG ATG GAA TTC AAA GTG TCA CC-3′ | This study |
| VPS55-R | 5′-CTT TTC CTT TTG CGG CCG CCA CCA AAA AGG GAA TCA TCT TC-3′ | This study |
| OB-RGRP-F | 5′-CGG GAT CCA TCG ATA TGG CGG GCG TTA AAG CTC TC-3′ | This study |
| OB-RGRP-R | 5′-GGA ATT CGC GGC CGC CCC ACT GCT CCC AGC TAA AAT C-3′ | This study |
| F2-VPS55 | 5′-TGG TTT TTC AAA AAA GAT TTC AAC GAA GAT GAT TCC CTT TTT GGT CGG ATC CCC GGG TTA ATT AA-3′ | This study |
| R1-VPS55 | 5′-ACC CAG TTG GCT TAG TCT GTT GAG GGG ATG AGT AAT TGT TAT ATA GAA TTC GAG CTC GTT TAA AC-3′ | This study |
Plasmids and Strains Construction
DNA manipulations, including restriction analysis, ligation, and polymerase chain reaction (PCR) amplification were performed essentially as described by (Maniatis et al., 1982)
To construct pYEF2-GAL10-VPS55-HA and pYEF2-GAL10-OB-RGRP-HA, BamHI/NotI fragments encoding either Vps55 or OB-RGRP were obtained by PCR (oligonucleotides VPS55-F and VPS55-R to amplify Vps55p, and OB-RGRP-F and oligonucleotides OB-RGRP-R to amplify OB-RGRP). The fragments were inserted into the BamHI and NotI sites of pYEF2 (Cullin and Minvielle-Sebastia, 1994) such that they were under the control of the GAL10 promoter and were in frame with the fragment encoding the hemagglutinin (HA) epitope.
For the HA, Myc, and green fluorescent protein (GFP) fusion constructs, the 3′ flanking sequence of VPS55, including the stop codon, was replaced by a PCR-amplified fragment containing the kanMX6 module as a selection marker and the genes encoding the GFP (S65T) variant, the 13Myc, or the 3HA epitopes (Wach et al., 1997; Longtine et al., 1998). The PCR fragments were generated by amplification with oligonucleotides F2-VPS55 and R1-VPS55 using pFA6a-GFPS65T-kanMX6, pFA6a-13Myc-kanMX6, or pFA6a-3HA-kanMX6 as the template (Wach et al., 1997). The fragments were introduced into appropriate strains and clones were selected on appropriate plates.
Uracil Permease Activity
Uracil permease uptake, used to quantify the amount of cell-surface permease, was measured in wild-type and vps55Δ cells transformed with pFL38gF containing the FUR4 gene, encoding uracil permease, under the control of the GAL10 promoter as described in Volland et al. (1994). For complementation studies with yeast and human VPS55, wild-type and vps55Δ cells were transformed with pgF and pYCG-VPS55, pYEF2, pYEF2-GAL10-VPS55-HA, or pYEF2-OB-RGRP-HA vectors. Cells were grown overnight in a medium containing lactate, and uracil permease uptake was measured after 3 h of induction in a medium containing galactose.
Yeast Cell Extracts and Immunoblotting
For Western immunoblotting, cells were grown in YPD (for the detection of CPY, ALP, or Vps10p) or YNB galactose based medium (for detection of Fur4p), and were collected by centrifugation during the exponential phase of growth. Protein extracts were prepared from 1 to 2 ml of culture, and were lysed by incubation with 0.185 N NaOH for 10 min on ice. Proteins were precipitated by incubation with 10% trichloroacetic acid (TCA) for 10 min on ice, and were collected by centrifugation for 10 min at 12,000 × g. Protein pellets were resuspended in 50 μl of SDS sample buffer (4% SDS, 0.3 M Tris-HCl, 4 mM EDTA, 20% glycerol, 2% β-mercaptoethanol, 0.02% bromphenol blue) and were heated for 4 min at 95°C, except for uracil permease (10 min at 37°C). Proteins were separated by SDS-PAGE in 10% Tricine gels and were analyzed by immunoblotting with monoclonal or polyclonal antibodies as described below. The primary antibodies were detected with horseradish peroxidase–conjugated anti-mouse IgG or anti-rabbit IgG secondary antibodies, and the secondary antibodies were detected by enhanced chemiluminescence (ECL; Boehringer Mannheim. Indianapolis, IN).
Membrane Extraction and Subcellular Fractionation
Cells expressing Vps55-HA were spheroplasted as previously described (McNew and Goodman, 1994) and extracts were prepared as described by (Ossig et al., 1991). The homogenate was aliquoted in polyallomer microultracentrifuge tubes. Extractions were performed for 30 min on ice with either lysis buffer alone, 1% Triton-X100 in lysis buffer, or 0.1 M sodium carbonate in water (pH 11.5). The extracts were separated into membrane and soluble fractions by centrifugation at 100,000 × g for 1 h at 4°C in an ultracentrifuge (TL100; Beckman Instruments, Palo Alto, CA). Proteins from the supernatant were precipitated in 10% TCA for 30 min, pelleted, and resuspended in SDS sample buffer, as were the membrane pellets. The organelles from wild-type and vps35Δ cells were fractionated by differential centrifugation as described by Kranz et al. (2001). Proteins from the various fractions containing equal amounts of cells (2 × 108 cells) were separated by SDS-PAGE and analyzed by Western immunoblot as described above.
Detection of Secreted CPY
For the detection of secreted CPY, 10 μl of a culture of exponentially growing cells (107 cells/ml) were collected and spotted onto YPD or YNB + 2% galactose plates, and were grown for 48 h in contact with a 0.45-μm nitrocellulose filter (Schleicher & Schuell, Keene, NH). The filter was then removed from the plate, the cells were washed with distilled water, and we tested for CPY immunologically as previously described (Roberts et al., 1991).
Pulse-Chase Labeling and Immunoprecipitation
Yeast cells were cultured in YNB medium to a density of 2 × 107 cells/ml with glucose as a carbon source. Cells were collected by centrifugation, concentrated to 2 × 108 cells/ml in fresh medium with 50 mM KPO4, pH 5.7, and 2 mg/ml bovine serum albumin (BSA), and incubated for 15 min at 30°C. They were then labeled by incubation for 10 min with 150 μCi [35S]-Translabel (NEN, Boston, MA) per milliliter of culture, followed by a chase with 10 mM unlabeled methionine plus cysteine. Aliquots (0.9 ml) were removed at various times during the chase, and proteins were precipitated either from total cell culture or after separating the cells and medium. Precipitated proteins were processed for CPY immunoprecipitation as previously described (Volland et al., 1992), except that samples were heated for 4 min at 95°C. Pulse-chase labeling and immunoprecipitation of Vps10p and Vps55-HA were done as described for CPY except that the cells were concentrated in fresh medium without KPO4 and BSA, and the proteins were precipitated from total cell culture. Proteins were separated by SDS-PAGE in 8% Tricine gels, and radioactivity was detected by fluorography as previously described (Volland et al., 1992).
Antibodies
The following monoclonal antibodies were used throughout this study: CPY, ALP, Vps10p, and Pep12p (Molecular Probes, Eugene, OR), HA (F-7 from Santa Cruz Biotechnology, Santa Cruz, CA), and Myc (9E10 from Roche Diagnostics, Somerville, NJ). The following polyclonal antibodies were used: CPY, Vps10p, Sss1p, Pma1p (gifts from H. Riezman, A.A. Cooper, A. Kepes, and R. Serrano, respectively), and Fur4p (Volland et al., 1994). Monoclonal and polyclonal CPY antibodies were used for CPY colony immunoblotting and immunoprecipitation, respectively. The secondary antibodies used for immunoblotting were horseradish peroxidase–conjugated anti-mouse IgG or anti-rabbit IgG. Bound secondary antibodies were detected by ECL (Boehringer Mannheim).
Lucifer Yellow (LY) Accumulation Assay
The accumulation of LY carbohydrazide (Sigma, St. Louis, MO) in vacuoles was assessed essentially as described by Dulic et al. (1991). Exponentially growing cells were concentrated to an A600 nm of 20 in fresh medium and were incubated with 8 mg/ml LY in the dark for 30 min at 30°C. Cells were then washed three times in 1 ml of ice-cold buffer (50 mM sodium phosphate, 10 mM sodium azide, 10 mM sodium fluoride, pH 7). Samples were examined under a microscope (Leica, Deerfield, IL) with fluorescein isothiocyanate (FITC) fluorescence optics.
FM4–64 Endocytosis Assay
Exponentially growing cells were collected by centrifugation, resuspended to an A600 nm of 10 in fresh medium with 10 μM FM4–64 (Molecular Probes), and incubated for 30 min at 30°C. Cells were then collected by centrifugation and were resuspended in 1 ml of fresh medium for 1 h. Cells were collected, rinsed, suspended in water, and examined by epifluorescence microscopy with a microscope (Leica) using a rhodamine filter set.
Fur4-GFP Endocytosis Assay
Wild-type and vps55Δ cells transformed with pFL38gF-GFP were grown at 30°C in YNB medium with galactose as a carbon source. Cycloheximide (CHX; 100 μg/ml) was added to exponentially growing cells. Aliquots were withdrawn at the times indicated after the addition of CHX and were washed with 10 mM ice-cold sodium azide. To visualize the vacuolar morphology, cells were collected and incubated for 15 min in 1.2 M sorbitol, 20 mM KPi, pH 7.4. Cells were examined by Nomarski optics and for GFP fluorescence with an FITC filter set.
Immunofluorescence
Yeast cells were grown to a density of 107 cells/ml and were fixed by incubation for 45 min in 3.7% formaldehyde in buffer A (1.2 M sorbitol, 20 mM potassium phosphate, pH 7.4). Cells were collected by centrifugation and were resuspended to a density of 4 × 107 cells/ml in buffer B (10 mM dithiothreitol [DTT], 0.1 M Tris/HCl, pH 9.4). The cells were incubated for 8 min, washed twice in buffer A, resuspended in 2 ml of buffer A supplemented with 0.2 mg/ml Zymolyase 20 T (Seikagaku, Tokyo, Japan), and incubated for 15–30 min at room temperature. The resulting spheroplasts were washed twice in buffer A and were spotted onto polylysine-coated slides for 5 min. They were then permeabilized by incubation in phosphate-buffered saline (PBS; 50 mM potassium phosphate pH 7.5, 150 mM NaCl) supplemented with 1% Triton X-100 for 5 min on ice. The slides were rinsed with PBS and were incubated for 1 h with the primary antibody diluted 1:50 in PBS supplemented with 1% BSA. The slides were then washed three times in PBS and were incubated with rhodamine-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) diluted 1:250 for 30 min. The slides were mounted in Citifluor (Citifluor, London, United Kingdom). Samples were viewed under a microscope (Leica) with FITC and rhodamine filter sets and Nomarski optics.
RESULTS
The Vps55Δ Strain Is Impaired in Trafficking from the Golgi Apparatus to the Vacuole
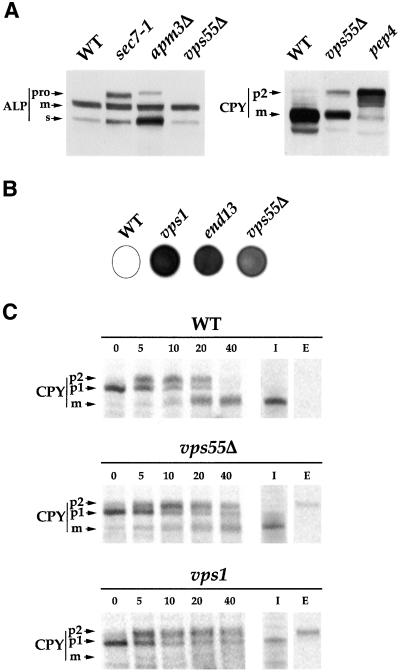
The EUROFAN project consisted of the systematic disruption of 800 genes followed by their functional analysis. Analysis of the vps55Δ mutant showed that the disrupted cells do not present any thermosensitivity (data from the Munich Information Center for Protein Sequences: http://mips.gsf.de/). The cells grow normally at 15°C, 24°C, 30°C, and 37°C (our unpublished results). To determine whether vps55Δ cells present any defect in the secretory pathway, we used Western immunoblots to systematically analyze various marker proteins that undergo posttranslational modifications during their intracellular trafficking: the secreted pheromone α-factor, the periplasmic glycoprotein invertase, and the plasma membrane glycosylphosphatidylinositol (GPI)-anchored protein Gas1p. These three proteins displayed normal electrophoretic patterns, indicating that vps55Δ cells have normal endoplasmic reticulum (ER) translocation, ER-associated modifications, and ER to Golgi trafficking (Avaro et al., 2002). The membrane-bound vacuolar ALP also displayed a normal electrophoretic pattern (Figure 1A). Conversely, the precursor form of ALP (proALP) was detected in the sec7–1 mutant, which is defective in ER to Golgi and intra-Golgi transport (Franzusoff and Schekman, 1989; Wolf et al., 1998), and in the apm3Δ mutant, which lacks one of the four subunits of the heterotetrameric adaptor protein complex AP-3 and is thus defective in the transport of ALP from the TGN to the vacuole (Cowles et al., 1997). These data indicate that vps55Δ cells display normal trafficking of ALP. In contrast, vps55Δ cells accumulate a precursor form of the soluble vacuolar CPY (Figure 1A).
Figure 1.
CPY maturation is delayed and p2-CPY is secreted in the medium in vps55Δ cells. (A) Total protein extracts were prepared from exponentially growing wild-type, vps55Δ, sec7–1, apm3Δ, and pep4 cells. ALP and CPY were analyzed by SDS-PAGE and immunoblotting. The positions of the precursors (p2-CPY and proALP) and the mature (m) forms are indicated. In addition to proALP, all cells also display a lumenal aberrantly processed soluble form of ALP, as described by Stepp et al. (1997). (B) vps55Δ, vps1, and end13–1/vps4 cells were grown on YPD plates (105 cells/spot) for 48 h in contact with a nitrocellulose membrane. Secreted CPY was detected on the membrane by probing with a mAb directed against CPY. (C) Wild-type, vps55Δ, and vps1Δ cells were labeled with [35S]-Translabel for 10 min and chased for the indicated times in the presence of 10 mM unlabeled methionine plus cysteine. CPY was precipitated either from total cell culture (0, 5, 10, 20, and 40 min) or after the separation of cells (I, intracellular) and medium (E, extracellular; 40 min of chase). Aliquots corresponding respectively to 1 vol (total cell culture), 3 vol (I), or 6 vol (E) of original proteins were analyzed by SDS-PAGE, and radioactivity was detected by fluorography. As no p2-CPY was recovered in the intracellular fractions after 40 min of chase, the percentage of CPY secretion was roughly estimated from the overall CPY pattern in total cell culture at 40 min. The recovery of immunoprecipitated proteins was lower for I and E than in T. The ER (p1), Golgi (p2), and mature (m) forms of CPY are indicated.
The prepro-CPY protein is translocated into the lumen of the ER, where it is converted into the p1-CPY glycosylated precursor form of the protein. During its transit through the Golgi apparatus, mannose residues are added to p1-CPY to generate the p2-CPY form. CPY is finally transported via late endosomes to the vacuole, where it is processed to the mature form (Graham and Emr, 1991; Vida et al., 1993). The vps55Δ cells had lower steady-state amounts of the mature form of CPY and accumulated the Golgi precursor form (p2-CPY) of the protein (Figure 1A). A huge accumulation of the p2-CPY form is observed in a control strain, the pep4 mutant, which is deficient in proteinase A, the product of the PEP4 gene, which is necessary for the vacuolar maturation of p2-CPY (Ammerer et al., 1986). Changes in CPY targeting from the Golgi apparatus to the vacuole lead to the secretion of the inactive zymogen into the medium. Colony immunoblotting analysis of the extracellular CPY showed that the protein was secreted by the vps55Δ mutant, but to a lesser extent than in the control strains, vps1 and vps4/end13, which are impaired in Golgi to vacuole targeting (Figure 1B).
We characterized the CPY defect displayed by the vps55Δ strain further by analyzing CPY processing in wild-type, vps1, and vps55Δ cells using pulse-chase experiments followed by the immunoprecipitation of CPY (Figure 1C). After a 5-min pulse, the predominant signal was the p1-CPY and 5 min of chase were necessary to detect p2-CPY in the three strains (Figure 1C). The mature form of CPY could be detected in wild-type cells after a 10-min chase, whereas processing was delayed in vps55Δ cells and 20 min of chase were required before the mature form could be detected. Even after 40 min of chase, processing was incomplete in vps55Δ and no mature form could be detected in vps1 cells, whereas CPY was completely processed in wild-type cells. We estimated that the rate of formation of mCPY was at least a 1.5-fold lower in vps55Δ cells.
To characterize the secreted CPY, we separated the cells, the medium, and immunoprecipitated CPY. After a 40-min chase, p2-CPY was secreted into the culture medium of vps1 cells (∼50% secretion) and vps55Δ cells (∼20% secretion), whereas no CPY was detected in the culture medium of wild-type cells (Figure 1C). These observations indicated that the secreted CPY detected by colony immunoblot probably corresponds to the Golgi form of the protein. Thus, vps55Δ cells are impaired in the CPY pathway.
Vps55p Is Required for Targeting from Late Endosomes to the Vacuole
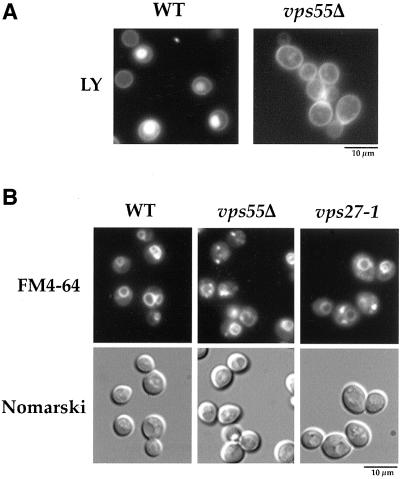
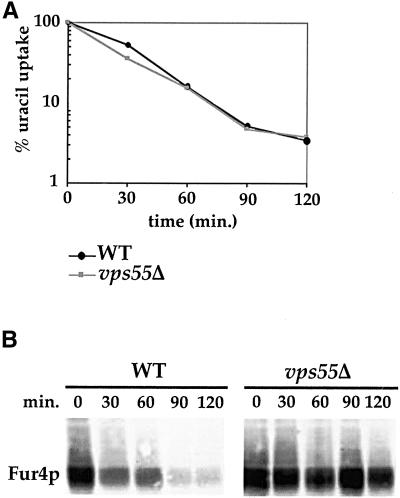
The Golgi to vacuole targeting defect observed in vps55Δ cells may result either from a defect in trafficking from the Golgi apparatus to late endosomes, or a defect in trafficking from late endosomes to the vacuole. We therefore checked whether this mutant displayed a defect in the endocytic pathway that intersects with the Golgi to vacuole trafficking at the level of late endosomes. We first assessed endocytosis using the small LY dye (Figure 2). LY was internalized by fluid phase endocytosis and accumulated in the vacuolar lumen of wild-type cells (Figure 2A; Dulic et al., 1991). After incubating the vps55Δ mutant in the presence of LY for 30 min, most of the cells exhibited a punctuate fluorescent pattern reminiscent of endosomes and unstained vacuoles (Figure 2A). The vps55Δ cells hence display a strong defect in fluid phase endocytosis. Defective vacuolar accumulation of LY can arise from either defective internalization or impairment in later endocytic steps. We thus also monitored the endocytosis of the lipophilic steryl dye FM4–64. In wild-type cells, FM4–64 was incorporated into the plasma membrane from where it was endocytosed and transported to the vacuole, where it accumulated at the limiting vacuolar membrane (Figure 2B; Wendland et al., 1996). When vps55Δ cells were incubated with FM4–64 only, a subset of the cells displayed vacuolar membrane staining and in addition, the dye accumulated in two to three spots close to the vacuolar membrane (Figure 2B). This indicates that FM4–64 was trapped in an endosomal compartment, delaying its targeting to the vacuole. The FM4–64–labeled spots that accumulated in vps55Δ cells appeared to be smaller than those that accumulated at a restrictive temperature in thermosensitive vps27–1 cells, which are known to accumulate FM4–64 in the enlarged prevacuolar “class E” compartment. Thus, endocytosis was considerably altered in vps55Δ cells. However, this test cannot clearly discriminate between a defect in early to late endosomes or in late endosomes to vacuole trafficking. We therefore assessed the endocytosis of a third marker, the plasma membrane uracil permease. Uracil permease undergoes basal endocytosis and vacuolar degradation in exponentially growing cells and accelerated endocytosis in certain conditions such as the inhibition of protein synthesis (Volland et al., 1994; Galan et al., 1996). We followed the fate of plasma membrane uracil permease in a wild-type strain and in the vps55Δ mutant after the addition of CHX (Figure 3, A and B). Uracil uptake, used to assess the amount of permease at the cell surface, was determined at various times (Figure 3A), and uracil permease was analyzed by Western immunoblotting (Figure 3B). The wild-type and vps55Δ strains displayed a similar decrease in uracil permease uptake, showing that internalization occurs at the same rate in the two strains (Figure 3A). In contrast, the rate of permease degradation in vps55Δ cells was strongly reduced compared with that of wild-type cells (Figure 3B). Almost no degradation of uracil permease was observed 120 min after the addition of CHX in the vps55Δ strain, whereas in wild-type cells, almost all the uracil permease had disappeared within this period (Figure 3B).
Figure 2.
Vps55Δ cells are defective in endocytosis of LY and FM4–64. (A) Wild-type and vps55Δ cells were incubated in the presence of 8 mg/ml LY for 30 min at 30°C as described in “Materials and Methods” and were then viewed by fluorescence microscopy using an FITC filter set. (B) Wild-type and vps55Δ cells were incubated in the presence of 10 μM FM4–64 for 30 min at 30°C as described in “Materials and Methods.” The vps27–1 cells were shifted to 37°C for 1 h and were then incubated with FM4–64. The localization of FM4–64 was detected by fluorescence microscopy using the rhodamine filter set.
Figure 3.
Vps55Δ cells are impaired in the uracil permease endocytic pathway. (A) Wild-type and vps55Δ cells transformed with pFL38gF (producing Fur4p under the control of a galactose promoter) were grown to the exponential growth phase in galactose medium. CHX was added to a final concentration of 100 μg/ml, and uracil uptake was measured at the indicated times. Results are presented as a percentage of initial activity. (B) Aliquots were collected at the indicated times after CHX addition and protein extracts were prepared. Proteins from 1.5 × 107 cells were analyzed for uracil permease (Fur4p) by immunoblot.
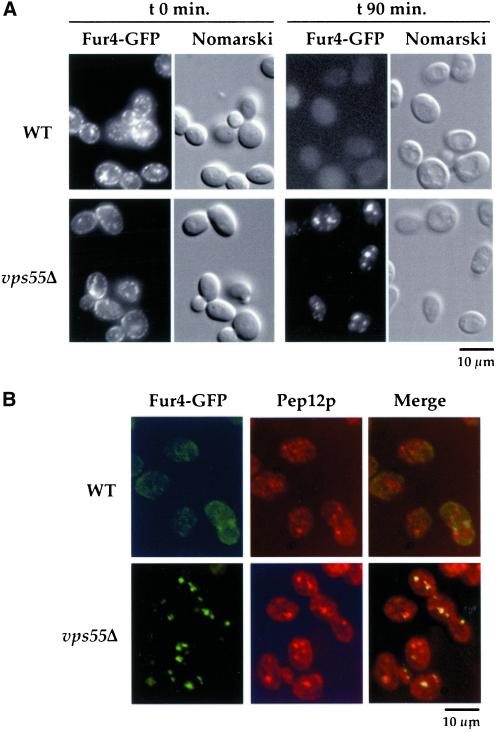
To determine whether impairment of permease degradation results from a deficiency in vacuolar protease activities or from an impairment in permease trafficking, we followed the endocytosis of uracil permease tagged at its C terminus with a brilliant version of the GFP (Dupré and Haguenauer-Tsapis, 2001). Punctuate Fur4-GFP staining was detected at the cell periphery in exponentially growing wild-type and vps55Δ cells (Figure 4A). Some small dots were also found throughout the cytoplasm. After 90 min of treatment with CHX, the fluorescence almost entirely disappeared from wild-type cells, consistent with the permease degradation observed on Western immunoblots. In vps55Δ cells, plasma membrane staining entirely disappeared after 90 min of CHX treatment, in agreement with the loss of permease activity, and Fur4-GFP accumulated in small dots (∼3–4 per cell) often located near the vacuoles, as detected by Nomarski optics (Figure 4A). To characterize these compartments further, cells that had been treated with CHX for 90 min were analyzed simultaneously for Fur4-GFP and for Pep12p, a t-soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein receptor (SNARE) that is a marker of the late endosomal/PVC (Becherer et al., 1996). As above, Fur4-GFP was almost undetectable in wild-type cells (Figure 4B). The Fur4-GFP signal detected in vps55Δ cells partially overlapped with Pep12p, indicating that these cells transiently retained uracil permease in late endosomal-type structures (Figure 4B). These data suggest that Vps55p is required for the correct trafficking of endocytic material from the late endosomes to the vacuole.
Figure 4.
Uracil permease accumulates in late endosomal/prevacuolar compartments in vps55Δ cells. (A) Wild-type and vps55Δ cells transformed with pFL38gF-GFP (expressing Fur4-GFP under the control of a galactose promoter) were grown at 30°C in galactose medium. CHX was added to exponentially growing cells. Aliquots were withdrawn at the times indicated after addition of CHX and were examined by Nomarski optics and for GFP fluorescence using the FITC filter set. (B) Wild-type and vps55Δ cells expressing Fur4-GFP and treated with CHX for 90 min were processed for immunofluorescence using the anti-Pep12p antibody followed by rhodamine-labeled goat anti-mouse IgG as described in “Materials and Methods,” except that cells were permeabilized with Triton-X100 for 10 min. The Pep12p signal was detected using the rhodamine filter set and GFP using the FITC filter set.
Vps55Δ Cells Are Class A Mutant Cells
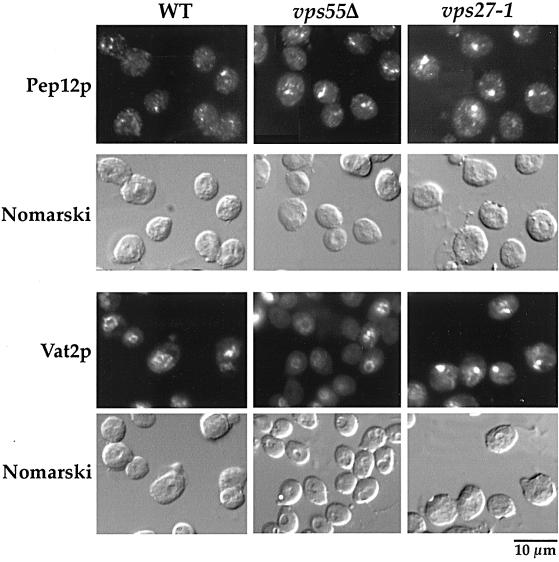
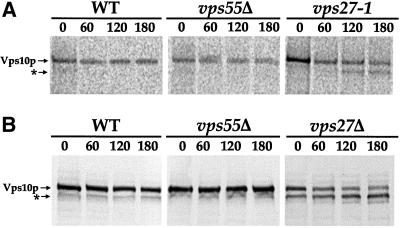
VPS genes that may act at the same step of the trafficking pathway have been grouped into six classes (A through F) based on vacuolar morphology (Raymond et al., 1992; for review, see Conibear and Stevens, 1998; Lemmon and Traub, 2000). Among these mutants, only class A and class E mutants display wild-type vacuolar morphology. The vps55Δ cells present a wild-type vacuolar morphology, as shown by Nomarski optics on unfixed cells (Figure 2B, compare wild-type and Vps55Δ cells), and thus can be considered to be either class A or class E mutants. In contrast to class A mutants, class E mutants accumulate a structure adjacent to the vacuole that corresponds to an exaggerated version of the PVC found in the wild-type cells and is referred to as the “class E compartment.” Class E mutants accumulate late endosomal and vacuolar markers in this class E compartment. We examined vps55Δ cells by immunofluorescence using Pep12p as a marker of the PVC. In wild-type cells, the anti-Pep12p antibody stained the cytoplasm as series of small dots (Figure 4 and 5). In cells lacking Vps55p, the anti-Pep12p gave a slightly different pattern, comma-shaped rather than punctuate (Figure 5), and the Pep12p signal was slightly enhanced. This was most visible after Triton-X100 permeabilization for slightly longer periods (10 min in Figure 4 compared with 5 min in Figure 5). This pattern is similar to, but less pronounced than, that observed in the class E vps27–1 mutant. In this thermosensitive mutant, Pep12p accumulated, at 37°C, in the exaggerated PVC, which appears mostly as a unique spot close to the vacuole (Figure 5; Piper et al., 1995). We also investigated the distribution of Vma2p/Vat2p, the peripheral subunit of the vacuolar ATPase (Figure 5). In wild-type and vps55Δ mutant cells, the anti-Vma2p antibody stained the vacuolar membrane, whereas in vps27–1 cells shifted to 37°C, Vma2p was trapped in the class E compartment (Figure 5). Similar results were obtained with the anti-Vph1p antibody, which binds to the transmembrane subunit of the vacuolar ATPase (our unpublished results), suggesting that vps55Δ cells are a class A mutant. However, some vps class E mutants, like the nhx1Δ mutant, were shown to display impaired localization of Pep12p but normal localization of Vma2p/Vat2p (Bowers et al., 2000). To determine whether vps55Δ cells do accumulate a class E compartment or not, we monitored the intracellular fate of Vps10p. Vps10p binds CPY in the TGN, and CPY is released in the PVC compartment (Vida et al., 1993; Marcusson et al., 1994). Vps10p then recycles from the PVC to the Golgi apparatus (Cereghino et al., 1995; Cooper and Stevens, 1996). Class E mutants that are impaired in PVC to vacuole transport and in the retrograde transport from the PVC to the TGN accumulate Vps10p in the class E compartment where it is proteolytically cleaved, resulting in a lower molecular weight form (Piper et al., 1995). The intracellular fate of Vps10p was followed in wild-type, vps55Δ, and control vps27 cells by classical pulse-chase analysis (Figure 6A). We also followed Vps10p processing by Western immunoblot after the addition of CHX to the culture medium (Figure 6B). Vps10p appeared to be very stable in the wild-type and in the vps55Δ strains and was not cleaved even after a 3 h chase. Conversely, the clipped form of Vps10p appeared after a 2-h chase in the thermosensitive vps27–1 strain (Figure 6A). Both the normal and clipped forms of Vps10p were detected in equal amounts in growing vps27Δ cells, and the proportion of the clipped form gradually increased after CHX treatment (Figure 6B). This shows that in contrast to vps27 cells, proteolytically active class E compartments do not accumulate in vps55Δ cells. Thus, vps55Δ cells can be classified as class A mutants.
Figure 5.
Vps55Δ cells do not accumulate a class E compartment. The distribution of Pep12p and Vat2p in wild-type, vps55Δ, and vps27–1 cells was determined by immunofluorescence. Exponentially growing cells were fixed and permeabilized with 1% Triton-X100 for 5 min to preserve vacuolar morphology. Immunofluorescence was performed with monoclonal anti-Pep12p or anti-Vat2p antibodies. Fluorescence was viewed using rhodamine filter set and cell morphology was observed with Nomarski optics.
Figure 6.
Vps55Δ is not affected in the retrieval of Vps10p from the prevacuolar compartment. (A) Wild-type, vps55Δ, and vps27–1 cells were shifted to 37°C for 1 h before labeling with [35S]-Translabel for 15 min. The chase was performed by adding 10 mM unlabeled methionine plus cysteine. Vps10p was immunoprecipitated from the lysate at the indicated times (min) after the chase and was analyzed by fluorography after SDS-PAGE. Vps10p was clipped (denoted by an asterisk) in the vps27–1 strain. (B) The wild-type, vps55Δ, and vps27Δ strains were grown at 30°C in YPD medium. CHX was added at a final concentration of 100 μg/ml. Protein extracts were prepared at the indicated times after CHX addition and were analyzed by SDS-PAGE and immunoblotting.
Vps55p Is Mainly Localized in Late Endosomes
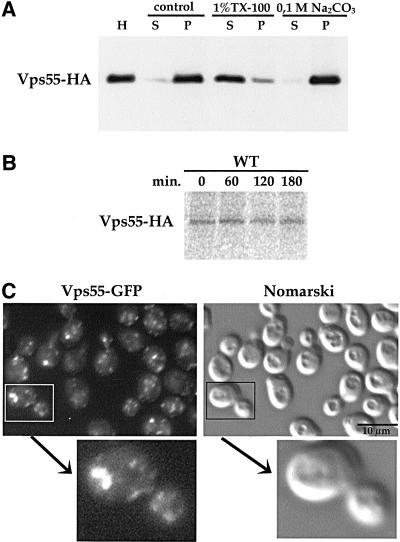
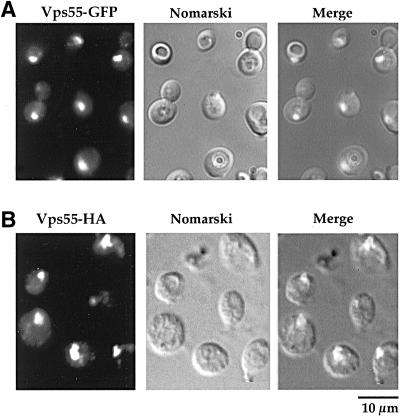
To visualize Vps55p and to determine its subcellular localization, we tagged the 3′ end of the chromosomal VPS55 gene with a DNA fragment encoding GFP (variant S65T), 13Myc, or 3HA tag (Wach et al., 1997; Longtine et al., 1998). Colony immunoblotting was used to detect secreted CPY and it revealed that cells producing the Myc- and HA-tagged versions of Vps55p exhibit a wild-type phenotype. This indicates that Vps55-Myc and Vps55-HA are functional. However, cells producing the GFP-tagged version of Vps55p displayed a very low-level secretion of CPY (our unpublished results). We first investigated the biochemical status of Vps55p by fractionation studies and its stability by pulse-chase experiments (Figure 7, A and B). Vps55-HA from lysed spheroplasts fractionated with the 100,000 × g pellet (Figure 7A). To determine the nature of membrane association, lysates were treated with Na2CO3 and with the nonionic detergent Triton-X100. Vps55-HA was resistant to Na2CO3 extraction, indicating that it is an integral membrane protein, as predicted from sequence data. On the other hand, Vps55-HA was almost completely solubilized by Triton-X100 (Figure 7A). Pulse-chase experiments showed that Vps55-HA is very stable, as the protein was not degraded even after a 3-h chase.
Figure 7.
Vps55p is a stable integral membrane protein localized in small punctuate structures. (A) Cells expressing Vps55-HA were grown to exponential phase, converted to spheroplasts, and lysed with glass beads. Cell lysates were treated with lysis buffer alone (control), 1% Triton-X100, or 0.1 M sodium carbonate (pH 11). The initial homogenate (H) was separated into supernatant (S) and pellet (P) by centrifugation at 100,000 × g. Samples were analyzed by SDS-PAGE and immunoblotting using a monoclonal anti-HA antibody. (B) Wild-type strain expressing Vps55-HA was labeled with [35S]-Translabel for 15 min and was chased for the indicated times in the presence of 10 mM unlabeled methionine plus cysteine. Vps55-HA was immunoprecipitated from the cell lysate at the indicated times using a monoclonal antibody directed against the HA tag. Vps55-HA was analyzed by fluorography after SDS-PAGE. (C) Cells producing the Vps55-GFP fusion protein were viewed with a microscope (Leica) using the FITC filter set and Nomarski optics. An image of a budding cell at higher magnification is presented.
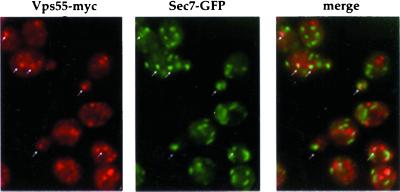
The subcellular localization of Vps55p was investigated using the Myc-, the HA-, and the GFP-tagged proteins. The GFP fusion protein is more readily evidenced in living cells. The chimeric proteins displayed the same pattern of staining consisting of a number of small dots, with occasional larger dots, in the cytoplasm and nascent buds (Figure 7C). This pattern is reminiscent of that of Pep12p (Figures 4 and 5; Nass and Rao, 1998) and of Golgi membrane proteins (Franzusoff et al., 1991; Redding et al., 1991). To determine whether Vps55p was present in the late endosomes or in the late Golgi compartment, we used vps27–1 cells. The distribution of chromosome-encoded Vps55-GFP was altered when this mutant was grown at 37°C and this protein accumulated in the class E compartment (Figure 8). A similar distribution was observed for plasmid-encoded HA-tagged Vps55p produced under the control of a galactose promoter after 3 h of induction in the presence of galactose (Figure 8). The vps27–1 cells are defective in late endosome to vacuole trafficking, but also in retrograde transport from late endosomes to the late Golgi. Thus, proteins that cycle between late endosomes and the late Golgi are trapped in the enlarged PVC of this mutant. The results obtained for the vps27–1 mutant suggest that Vps55p either resides in late endosomes or cycles between late endosomes and the late Golgi. To address this question, we investigated whether Vps55-Myc was colocalized with a marker of the late Golgi, Sec7p (Franzusoff et al., 1991; Rossanese et al., 1999). We tagged VPS55 at the chromosomal locus with the 13Myc epitope in a wild-type strain producing the Sec7-GFP fusion protein (Séron et al., 1998). An apparent partial colocalization of Vps55-Myc and Sec7-GFP was observed (Figure 9). This colocalization was strengthened when Vps55p was overproduced (our unpublished results). These results indicate that Vps55p is not entirely restricted to late endosomes, but could cycle between late endosomes and the late Golgi or could be present at two locations. A major fraction of the protein could be localized in the late endosomes and a minor one in the TGN.
Figure 8.
Vps55p is present in the enlarged PVC of vps27–1 mutant cells. Vps27–1 cells carrying a chromosomal copy of Vps55-GFP (A) or transformed with pYEF2-GAL10-VPS55-HA (B) were grown overnight at 30°C in YPD (A) or in YNB medium supplemented with 2% lactate (B). During logarithmic growth, cells were shifted to 37°C for 3 h in YPD medium to induce the accumulation of an enlarged PVC adjacent to the vacuole. In the case of cells transformed with pYEF2-GAL10-VPS55-HA, galactose (2%) was added to induce Vps55p synthesis when cells were shifted at a restrictive temperature. Vps27–1 cells producing the Vps55-GFP fusion protein were viewed directly using an FITC filter set. Vps27–1 cells producing the Vps55-HA fusion protein were fixed and processed for immunofluorescence using the monoclonal anti-HA antibody followed by rhodamine-labeled goat anti-mouse IgG. Fluorescence was viewed using rhodamine filter set, and cell morphology was observed with Nomarski optics. Note the altered cellular morphology in cells expressing the Vps55-HA fusion protein compared with living cells producing the Vps55-GFP fusion protein.
Figure 9.
Vps55p partially colocalizes with Sec7-GFP. Wild-type cells harboring chromosomal copies of SEC7-GFP and VPS55-MYC were grown overnight in YPD medium. Cells were fixed and processed for immunofluorescence using the monoclonal anti-Myc antibody followed by rhodamine-labeled goat anti-mouse IgG. GFP fluorescence was visualized with an FITC filter set, and HA distribution was observed with a rhodamine filter set. Arrows indicate Myc staining colocalizing with Sec7p-GFP.
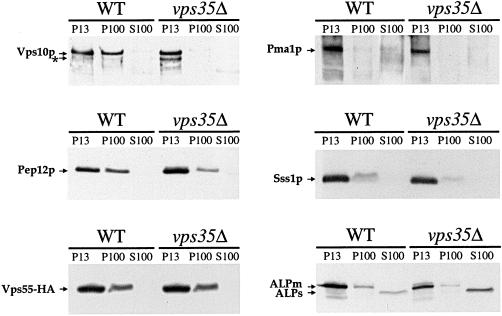
The retrieval from late endosomes back to the Golgi has been characterized for several TGN proteins such as Kex2p and Vps10p (for review, see Nothwehr and Stevens, 1994; Cooper and Stevens, 1996). In vps35Δ cells lacking a retromer protein, both Kex2p and Vps10p are delocalized to the vacuole (Seaman et al., 1997). To check whether this was also the case for Vps55p, we used subcellular fractionation to investigate the localization of Vps55p in wild-type and vps35Δ strains. The lysates were separated into pellet and supernatant fractions by sequential centrifugation at 13,000 and 100,000 × g. Pma1p, the plasma membrane H+ATPase, Sss1p, an ER localized protein, and the vacuolar ALP were mainly localized in the P13 fraction in wild-type cells as previously described (Figure 10; Marcusson et al., 1994; Gerrard et al., 2000; Kranz et al., 2001). Vps10p was found in both the P13 and P100 fractions in the wild-type strain, and only in the P13 fraction, where it displayed proteolytic cleavage, in the vps35Δ mutant (Figure 10; Cereghino et al., 1995; Seaman et al., 1997; Gerrard et al., 2000). In the wild-type strain, Vps55-HA was found in both the P13 and P100 fractions in the same proportions as Pep12p. Vps55-HA was not delocalized to the P13 fraction in the vps35Δ mutant. These data are consistent with Vps55p being mainly localized in late endosomes, as observed by fluorescence in the vps27–1 strain. The small proportion of Vps55p that colocalized with Sec7p, as observed by immunofluorescence, does not correspond to a fraction of the protein that is recycled back to the TGN via the retromer complex.
Figure 10.
Subcellular fractionation of cells expressing tagged Vps55p. Wild-type and vps35Δ cells expressing chromosomal integrated Vps55-HA were lysed with glass beads and extracts were fractionated by differential centrifugation. The proteins were separated by SDS-PAGE and were analyzed by immunoblotting for the presence of Vps55-HA and various marker proteins (an asterisk indicates clipped Vps10p; ALPm, ALP mature; ALPs, ALP soluble).
Human OB-RGRP Is a Functional Homolog of Vps55p
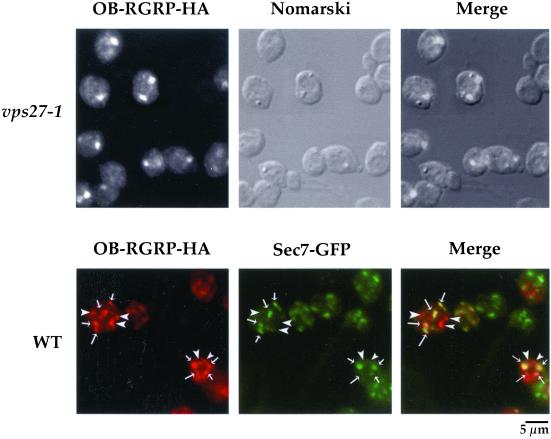
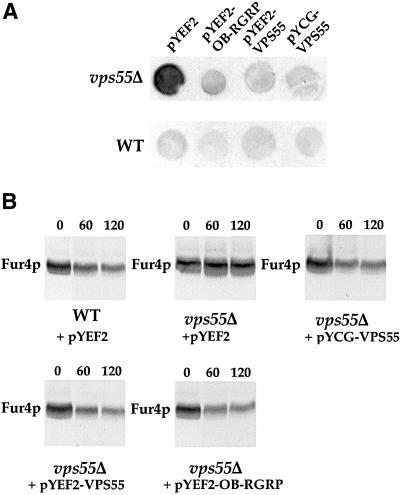
Searches in data banks indicate that VPS55 has homologs of still unknown function in many species. These include human OB-RGRP (GenBank accession no. Y12670), human LEPROTL1 (GenBank accession no. NP-056159.1), mouse OB-RGRP (GenBank accession no. AJ011565), rat OB-RGRP (GenBank accession no. NP-064484), Caenorhabditis elegans C30B5.2 (GenBank accession no. AAC46738.1), Schizosaccharomyces pombe (GenBank accession no. CAB52733.1), Drosophila melanogaster (GenBank accession no. AAD46832), and Arabidopsis thaliana (GenBank accession no. AAF00667.1). Hence, it could be of interest to determine if any of these proteins are functional homologues of Vps55p. Vps55p and human OB-RGRP present 29% amino acid sequence identity. We first examined the distribution of the OB-RGRP-HA fusion protein in the vps27–1 strain and in the wild-type strain expressing the Sec7-GFP marker (Figure 11). As observed for Vps55p, human OB-RGRP was present in the class E compartment of vps27–1 cells and was partially colocalized with Sec7-GFP in wild-type cells (Figure 11). We thus investigated whether OB-RGRP is able to complement the phenotypes observed in the vps55Δ mutant. Therefore, we produced plasmid-encoded human OB-RGRP and yeast Vps55p under the control of a galactose promoter in a wild-type and in the vps55Δ mutant strains. We used colony immunoblotting to analyze CPY secretion (Figure 12). Whereas the wild-type cells plated on galactose medium secreted a very small amount of CPY, vps55Δ cells transformed with the empty control vector (pYEF2) secreted large amounts of CPY (Figure 12). The production of Gal-induced Vps55p or human OB-RGRP clearly abolished the CPY secretion phenotype, as did production of Vps55p expressed under the control of its own promoter from the ARS-CEN pYCG-VPS55 plasmid. These data also indicate that limited overproduction of Vps55p or its mammalian homolog does not result in CPY secretion. We also determined the effect of OB-RGRP production on a second phenotype of vps55Δ cells, the impaired uracil permease trafficking. Vps55Δ cells transformed with pgF, a plasmid producing uracil permease under the control of a galactose promoter, were or were not cotransformed with plasmids encoding Gal-inducible VPS55 or OB-RGRP. Cells were grown in the presence of lactate and galactose was added, resulting in the simultaneous induction of uracil permease and either Vps55p or OB-RGRP. CHX was added after 3 h. The production of either Vps55p or OB-RGRP restored a normal degradation of uracil permease, as did the production of Vps55p from pYCG-VPS55 (Figure 12). These data demonstrate that human OB-RGRP, which presents the same localization pattern as Vps55p, is able to complement the loss of VPS55 function in yeast. Thus, the function of Vps55p has been conserved during evolution from yeast to human.
Figure 11.
Human OB-RGRP displays the same distribution as Vps55p. Vps27–1 and wild-type cells producing the chromosomal encoded Sec7-GFP were transformed with pYEF2-GAL10-OB-RGRP-HA. Cells were cultured overnight in YNB medium supplemented with 2% lactate. Galactose (2%) was added and the cells were shifted to 37°C for 3 h. Cells were fixed and processed for immunofluorescence using the monoclonal anti-HA antibody followed by rhodamine-labeled goat anti-mouse IgG. The FITC filter set was used to visualize GFP fluorescence, and the rhodamine filter set was used to visualize HA fluorescence. Arrows indicate colocalized Vps55-HA and Sec7-GFP, and arrowheads indicate HA staining not colocalized with Sec7p-GFP.
Figure 12.
Human OB-RGRP complements the disruption of VPS55. (A) Wild-type and vps55Δ strains transformed with pYEF2, pYEF2-GAL10-VPS55-HA, or pYEF2-GAL10-OB-RGRP-HA were cultured overnight in YNB medium supplemented with 2% glucose. Exponentially growing cells (105 cells/spot) were plated on YNB supplemented with 2% galactose to induce the production of Vps55p and OB-RGRP, and were cultured for 48 h in contact with a nitrocellulose membrane. Wild-type and vps55Δ cells transformed with the centromeric pYCG-VPS55 were treated in the same way. Secreted CPY bound on the nitrocellulose membrane was probed using a monoclonal antibody raised against CPY. (B) The strains listed above were transformed with pgF (expressing uracil permease under the control of a galactose promoter) and were grown overnight in YNB medium supplemented with 2% lactate. Galactose (2%) was added to the cells for 3 h during exponential growth to induce uracil permease, Vps55p, and OB-RGRP synthesis. Protein extracts were prepared at the indicated times (min) after CHX addition and were analyzed by SDS-PAGE and immunoblotting. Uracil permease (Fur4p) was detected using a polyclonal antibody.
DISCUSSION
We describe here the characterization of a new VPS gene, VPS55, involved in targeting of newly synthesized and endocytosed proteins to the vacuole. Our results suggest that Vps55p may function in protein transport from endosomes to the vacuole and that it is mainly localized in late endosomes. Interestingly, the human homolog of Vps55p, OB-RGRP, is able to complement the disruption of VPS55.
Clearly, Vps55p has a role in protein targeting to the vacuole. Analysis of CPY targeting in the vps55Δ strain shows a delay in the maturation of this protease and a significant missorting of the immature CPY (the Golgi form of the protein) into the culture medium, which is characteristic of mutants impaired in trafficking to the vacuole. Vps55p is most likely involved in protein transport from prevacuolar compartments to the vacuole. The internalization of uracil permease from the plasma membrane is not affected in vps55Δ cells, whereas its degradation is drastically impaired because of its transient accumulation in late endosome/PVC. Furthermore, the small dyes, LY and FM4–64, were detected in structures reminiscent of endosomes after internalization. The vps55Δ cells present a wild-type vacuolar morphology and could thus either be class A or class E vps mutant. There was a small but detectable accumulation of the t-SNARE Pep12p, a protein resident in late endosomes, in elongated structures in the vps55Δ mutant. However, other proteins such as the vacuolar Vma2p/Vat2p, which accumulates in the PVC in class E mutants, displayed a wild-type distribution. Furthermore, Vps10p, which is cleaved when accumulated in the class E compartment, was not degraded in vps55Δ cells, which indicates that vps55Δ cells do not accumulate a class E compartment. Therefore, the vps55Δ mutant is a class A vps mutant. The absence of clipping of Vps10p also indicates that vps55Δ cells are not defective in the retrieval of Vps10p from the late endosomes to the TGN as are the retromer mutants that are specifically impaired in this pathway.
To gain further insight into Vps55p function, we used immunofluorescence and fractionation studies to localize tagged Vps55p. We showed that Vps55p was restricted to the PVC of vps27–1 mutants and was partially colocalized with Sec7-GFP, a late Golgi marker, in wild-type strains. Fractionation studies showed that in contrast to the CPY receptor, Vps10p, Vps55p localization was not modified in vps35Δ cells. This indicates that the minor Golgi pool of Vps55p does not result from retromer-dependent retrieval from late endosomes back to the Golgi. If the only late endosome to TGN trafficking identified to date in yeast is mediated by the retromer, other retrieval routes involving other coats might exist, as in mammalian cells (reviewed in: Pfeffer, 2001). Our fractionation studies showed that Vps55p is not recycled back to the Golgi via the retromer complex, which indicates that there may be two pools of Vps55p, a major one localized in late endosomes and a minor one in the TGN. We cannot exclude the possibility that this minor pool results from a slight delay in Golgi to endosomal trafficking as a result of epitope tagging.
How Vps55p can function in protein trafficking to the vacuole remains an open question that must be addressed in the future. The overall structure of Vps55p might provide some clues. Vps55p, as well as its human homolog OB-RGRP, contains four predicted transmembrane domains (TMDs) and VPS55 is the first VPS gene characterized that encodes a protein with four predicted TMDs. Approximately 80% of the 600 or so small membrane proteins in yeast (molecular mass < 25 kDa, data from YPD: http://www.proteome.com/databases) have not yet been studied. The genes encoding these proteins have probably escaped from the various genetic screens using random mutagenesis because of their small size. For instance, 22 proteins with a molecular mass lower than 25 kDa and containing four predicted TMDs are referenced in the YPD database. Few of these proteins have been characterized. Examples of such small proteins include Vps55p and Sys1p, which suppresses the deficiency of ypt6 a small GTPase involved in the retrieval of proteins from late endosomes to the Golgi (Anraku et al., 1989; Li and Warner, 1996; Tsukada and Gallwitz, 1996; Hirata et al., 1997; Votsmeier and Gallwitz, 2001). Other examples include Shr3p, a chaperone required for the exit of amino acid permeases from the ER, Sft2p and Got1p, required for ER to Golgi transport, and Rer1p, involved in the recycling of membrane proteins from the early Golgi compartment back to the ER (Conchon et al., 1999; Gilstring et al., 1999; Sato et al., 2001). It is noteworthy that a number of these proteins are involved in trafficking processes. In humans, one example of small protein with four TMDs involved in vesicular transport is VIP17/MAL, which is required for Golgi to apical surface trafficking (Zacchetti et al., 1995; Cheong et al., 1999).
Vps55p and human OB-RGRP are homologous proteins that exhibit up to 40% amino acid identity in the second and the third TMDs. This suggests that the function of Vps55p relies on these TMDs. What could be the functional importance of these transmembrane domains? They could be involved in interactions with other transmembrane segments of membrane proteins. Interestingly, Rer1p, a component of COPII vesicles, is a small protein with four TMDs that cycles between the ER and the Golgi apparatus (Sato et al., 2001). Rer1p acts as a sorting receptor in the cis-Golgi and recognizes via its TMD the TMD of Sec12p, a guanine-nucleotide exchange factor (GEF) for Sar1p involved in the formation of ER vesicles, and allows its retrieval from the Golgi apparatus to the ER (Sato et al., 2001). Vps55p could function in a similar manner, interacting with the TMD of other membrane proteins also involved in various aspects of vacuolar targeting. Identification of the interacting partners of Vps55p could help us to elucidate its precise function.
When expressed in yeast, the human OB-RGRP not only presents the same localization as Vps55p, but also complements the loss of function of the VPS55 gene. This suggests that these two proteins function in a similar manner both in yeast and mammalian cells. Functional complementation between human and yeast proteins has been reported for other proteins involved in the secretory pathway. These include Sec23p and Sec13p, two components of the COPII complex (Shaywitz et al., 1995; Paccaud et al., 1996); the v-SNARE Vti1p (Fischer von Mollard and Stevens, 1998); syntaxin 7 (Nakamura et al., 2000); Got1p, a small protein with four TMDs required in ER to Golgi traffic (Conchon et al., 1999); Rer1p (Fullekrug et al., 1997); and the GTPase Ypt6p/Rab6 (Tsukada and Gallwitz, 1996). To our knowledge, heterologous complementation with a human protein has only been observed for one vps mutant, the apg6/vps30 mutant, which is impaired in the CPY vacuolar pathway and in the cytoplasm to vacuole transport (Liang et al., 1999). However, although Beclin 1, the human homolog of Vps30p, restored the autophagic function of autophagy-defective vps30Δ cells, it did not complement their vps phenotype.
OB-RGRP and OB-R, the leptin receptor, share the same promoter but are completely unrelated proteins. OB-R is a single membrane-spanning receptor that belongs to the cytokine superfamily (Tartaglia, 1997). It is still unknown whether OB-RGRP is involved in lysosomal targeting in mammalian cells. If this were true, the conservation of Vps55p function throughout evolution would be indicative of a critical function of these proteins in protein transport to endocytic compartments. In humans, the leptin receptor, OB-R, and OB-RGRP display the same overall expression pattern, suggesting that their functions might be connected (Bailleul et al., 1997). However, OB-RGRP may have a more general role in membrane trafficking than just in OB-R trafficking. This notion is supported by the following observations. First, OB-R homologues are not found in yeast and plants. Second, OB-RGRP mRNA presents a widespread distribution, consistent with the general role of Vps55p in protein trafficking (Mercer et al., 2000). Finally, OB-RGRP expression did not change in conditions that altered OB-R expression (upregulation in the hypothalamus of ob/ob mice, reversed by the administration of exogenous leptin; Mercer et al., 1997). If it remains conceivable that OB-RGRP may be involved in the leptin receptor trafficking, OB-RGRP may be involved in the down-regulation of other membrane proteins and their targeting to lysosomes, as we have observed in this study for Vps55p and the vacuolar targeting of the uracil permease.
ACKNOWLEDGMENTS
We thank E. Schiebel and coworkers (EUROFAN network) for constructing the whole set of strains and plasmids that allowed us to begin this work. We are indebted to K. Bowers and T. Stevens for sending us the Pep12p antibody used for immunofluorescence experiments and the vps27–1 strain and to C. Cullin for sending us pYEF2 plasmid. We also thank M. Hochstrasser for vps27Δ cells, H. Riezman for supplying the polyclonal antibody against CPY and the end13 and vps1 mutant strains, A. A. Cooper for the anti-Vps10p antibody, R. Serrano for the anti-Pma1p antibody, and F. Kepes for the anti-Sss1p antibody. We also thank B. Bailleul for his interest in this study and his encouragement in the study of Vps55p after his discovery of OB-RGRP and its homology with YJR044c/VPS55. We thank Alex Edelman and Associates for editorial assistance. We also thank C. Volland, D. Urban-Grimal, J.M. Galan, and S. Dupré for critical reading of the manuscript and helpful discussion and advice. This work was supported by the Center National de la Recherche Scientifique (UMR 7592) and by the EUROFAN program B104CT97–2294. N. B.-T. received a fellowship from la Société de Secours des Amis des Sciences and S.A. received a fellowship from the above European program.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–12–0597. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–12–0597.
REFERENCES
- Ammerer G, Hunter CP, Rothman JH, Saari GC, Valls LA, Stevens TH. PEP4 gene of Saccharomyces cerevisiaeencodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol Cell Biol. 1986;6:2490–2499. doi: 10.1128/mcb.6.7.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anraku Y, Umemoto N, Hirata R, Wada Y. Structure and function of the yeast vacuolar membrane proton ATPase. J Bioenerg Biomembr. 1989;21:589–603. doi: 10.1007/BF00808115. [DOI] [PubMed] [Google Scholar]
- Avaro S, Belgareh-Touzé N, Sibella-Argüelles C, Volland C, Haguenauer-Tsapis R. Mutants defective in secretory/vacuolar pathways in the EUROFAN collection of yeast disruptants. Yeast. 2002;19:351–371. doi: 10.1002/yea.838. [DOI] [PubMed] [Google Scholar]
- Bailleul B, Akerblom I, Strosberg AD. The leptin receptor promoter controls expression of a second distinct protein. Nucleic Acids Res. 1997;25:2752–2758. doi: 10.1093/nar/25.14.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis VA, Johnson LM, Emr SD. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc Natl Acad Sci USA. 1986;83:9075–9079. doi: 10.1073/pnas.83.23.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherer KA, Rieder SE, Emr SD, Jones EW. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K, Levi BP, Patel FI, Stevens TH. The sodium/proton exchanger nhx1p is required for endosomal protein trafficking in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:4277–4294. doi: 10.1091/mbc.11.12.4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, Stevens TH. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol Mol Biol Rev. 1998;62:230–247. doi: 10.1128/mmbr.62.1.230-247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghino JL, Marcusson EG, Emr SD. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPS gene products regulate receptor stability, function, and localization. Mol Biol Cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong KH, Zacchetti D, Schneeberger EE, Simons K. VIP17/MAL, a lipid raft-associated protein, is involved in apical transport in MDCK cells. Proc Natl Acad Sci USA. 1999;96:6241–6248. doi: 10.1073/pnas.96.11.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conchon S, Cao X, Barlowe C, Pelham HR. Got1p and Sft2p: membrane proteins involved in traffic to the Golgi complex. EMBO J. 1999;18:3934–3946. doi: 10.1093/emboj/18.14.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim Biophys Acta. 1998;1404:211–230. doi: 10.1016/s0167-4889(98)00058-5. [DOI] [PubMed] [Google Scholar]
- Cooper AA, Stevens TH. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Odorizzi G, Payne GS, Emr SD. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell, 1997;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- Cullin C, Minvielle-Sebastia L. Multipurpose vectors designed for the fast generation of N- or C-terminal epitope-tagged proteins. Yeast. 1994;10:105–112. doi: 10.1002/yea.320100110. [DOI] [PubMed] [Google Scholar]
- Delneri D, Brancia FL, Oliver SG. Towards a truly integrative biology through the functional genomics of yeast. Curr Opin Biotechnol. 2001;12:87–91. doi: 10.1016/s0958-1669(00)00179-8. [DOI] [PubMed] [Google Scholar]
- Dulic V, Egerton M, Elguindi I, Raths S, Singer B, Riezman H. Yeast endocytosis assays. Methods Enzymol. 1991;194:697–710. doi: 10.1016/0076-6879(91)94051-d. [DOI] [PubMed] [Google Scholar]
- Dupré S, Haguenauer-Tsapis R. Deubiquitination step in the endocytic pathway of yeast plasma membrane proteins: crucial role of doa4p ubiquitin isopeptidase. Mol Cell Biol. 2001;21:4482–4494. doi: 10.1128/MCB.21.14.4482-4494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emr SD, Schauer I, Hansen W, Esmon P, Schekman R. Invertase β-galactosidase hybrid proteins fail to be transported from the endoplasmic reticulum in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2347–2355. doi: 10.1128/mcb.4.11.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G, Stevens TH. A human homolog can functionally replace the yeast vesicle-associated SNARE Vti1p in two vesicle transport pathways. J Biol Chem. 1998;273:2624–2630. doi: 10.1074/jbc.273.5.2624. [DOI] [PubMed] [Google Scholar]
- Franzusoff A, Redding K, Crosby J, Fuller RS, Schekman R. Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J Cell Biol. 1991;112:27–37. doi: 10.1083/jcb.112.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzusoff A, Schekman R. Functional compartments of the yeast Golgi apparatus are defined by the sec7 mutation. EMBO J. 1989;8:2695–2702. doi: 10.1002/j.1460-2075.1989.tb08410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullekrug J, Boehm J, Rottger S, Nilsson T, Mieskes G, Schmitt HD. Human Rer1 is localized to the Golgi apparatus and complements the deletion of the homologous Rer1 protein of Saccharomyces cerevisiae. Eur J Cell Biol. 1997;74:31–40. [PubMed] [Google Scholar]
- Galan JM, Moreau V, André B, Volland C, Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- Gerrard SR, Levi BP, Stevens TH. Pep12p is a multifunctional yeast syntaxin that controls entry of biosynthetic, endocytic, and retrograde traffic into the prevacuolar compartment. Traffic. 2000;1:259–269. doi: 10.1034/j.1600-0854.2000.010308.x. [DOI] [PubMed] [Google Scholar]
- Gilstring CF, Melin-Larsson M, Ljungdahl PO. Shr3p mediates specific COPII coatomer-cargo interactions required for the packaging of amino acid permeases into ER-derived transport vesicles. Mol Biol Cell. 1999;10:3549–3565. doi: 10.1091/mbc.10.11.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TR, Emr SD. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J Cell Biol. 1991;114:207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata R, Graham LA, Takatsuki A, Stevens TH, Anraku Y. VMA11 and VMA16 encode second and third proteolipid subunits of the Saccharomyces cerevisiae vacuolar membrane H+-ATPase. J Biol Chem. 1997;272:4795–4803. doi: 10.1074/jbc.272.8.4795. [DOI] [PubMed] [Google Scholar]
- Johnson LM, Bankaitis VA, Emr SD. Distinct sequence determinants direct intracellular sorting and modification of a yeast vacuolar protease. Cell. 1987;48:875–885. doi: 10.1016/0092-8674(87)90084-5. [DOI] [PubMed] [Google Scholar]
- Jones EW, Webb GC, Hiller MA. In: In: Biogenesis and Function of the Yeast Vacuole: Vol. 3, Cell Cycle and Cell Biology. Pringle JR, Broach JR, Jones EW, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 363–470. [Google Scholar]
- Jund R, Weber E, Chevallier MR. Primary structure of the uracil transport protein of Saccharomyces cerevisiae. Eur J Biochem. 1988;171:417–424. doi: 10.1111/j.1432-1033.1988.tb13806.x. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz A, Kinner A, Kolling R. A family of small coiled-coil-forming proteins functioning at the late endosome in yeast. Mol Biol Cell. 2001;12:711–723. doi: 10.1091/mbc.12.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon SK, Traub LM. Sorting in the endosomal system in yeast, and animal cells. Curr Opin Cell Biol. 2000;12:457–466. doi: 10.1016/s0955-0674(00)00117-4. [DOI] [PubMed] [Google Scholar]
- Li B, Warner JR. Mutation of the Rab6 homologue of Saccharomyces cerevisiae, YPT6, inhibits both early Golgi function and ribosome biosynthesis. J Biol Chem. 1996;271:16813–16819. doi: 10.1074/jbc.271.28.16813. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch ET, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- McNew JA, Goodman JM. An oligomeric protein is imported into peroxisomes in vivo. J Cell Biol. 1994;127:1245–1257. doi: 10.1083/jcb.127.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer JG, Moar KM, Hoggard N, Strosberg AD, Froguel P, Bailleul B. B219/OB-R 5′-UTR, and leptin receptor gene-related protein gene expression in mouse brain, and placenta. tissue-specific leptin receptor promoter activity. J Neuroendocrinol. 2000;12:649–655. doi: 10.1046/j.1365-2826.2000.00501.x. [DOI] [PubMed] [Google Scholar]
- Mercer JG, Moar KM, Rayner DV, Trayhurn P, Hoggard N. Regulation of leptin receptor and NPY gene expression in hypothalamus of leptin-treated obese (ob/ob) and cold-exposed lean mice. FEBS Lett. 1997;402:185–188. doi: 10.1016/s0014-5793(96)01525-6. [DOI] [PubMed] [Google Scholar]
- Munn AL, Riezman H. Endocytosis is required for the growth of vacuolar H(+)-ATPase-defective yeast: identification of six new END genes. J Cell Biol. 1994;127:373–386. doi: 10.1083/jcb.127.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Yamamoto A, Wada Y, Futai M. Syntaxin 7 mediates endocytic trafficking to late endosomes. J Biol Chem. 2000;275:6523–6529. doi: 10.1074/jbc.275.9.6523. [DOI] [PubMed] [Google Scholar]
- Nass R, Rao R. Novel localization of a Na+/H+exchanger in a late endosomal compartment of yeast. Implications for vacuole biogenesis. J Biol Chem. 1998;273:21054–21060. doi: 10.1074/jbc.273.33.21054. [DOI] [PubMed] [Google Scholar]
- Nothwehr SF, Stevens TH. Sorting of membrane proteins in the yeast secretory pathway. J Biol Chem. 1994;269:10185–10188. [PubMed] [Google Scholar]
- Oliver S. A network approach to the systematic analysis of yeast gene function. Trends Genet. 1996;12:241–242. doi: 10.1016/0168-9525(96)30053-x. [DOI] [PubMed] [Google Scholar]
- Ossig R, Dascher C, Trepte HH, Schmitt HD, Gallwitz D. The yeast SLY gene products, suppressors of defects in the essential GTP-binding Ypt1 protein, may act in endoplasmic reticulum-to-Golgi transport. Mol Cell Biol. 1991;11:2980–2993. doi: 10.1128/mcb.11.6.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paccaud JP, Reith W, Carpentier JL, Ravazzola M, Amherdt M, Schekman R, Orci L. Cloning and functional characterization of mammalian homologues of the COPII component Sec23. Mol Biol Cell. 1996;7:1535–1546. doi: 10.1091/mbc.7.10.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR. Membrane transport. Retromer to the rescue. Curr Biol. 2001;11:R109–R111. doi: 10.1016/s0960-9822(01)00042-2. [DOI] [PubMed] [Google Scholar]
- Piper RC, Cooper AA, Yang H, Stevens TH. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K, Holcomb C, Fuller RS. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. J Cell Biol. 1991;113:527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder SE, Banta LM, Kohrer K, McCaffery JM, Emr SD. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol Biol Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Raymond CK, Yamashiro CT, Stevens TH. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- Rossanese OW, Soderholm J, Bevis BJ, Sears IB, O'Connor J, Williamson EK, Glick BS. Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J Cell Biol. 1999;145:69–81. doi: 10.1083/jcb.145.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JH, Stevens TH. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- Sato K, Sato M, Nakano A. Rer1p, a retrieval receptor for endoplasmic reticulum membrane proteins, is dynamically localized to the Golgi apparatus by coatomer. J Cell Biol. 2001;152:935–944. doi: 10.1083/jcb.152.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN, Marcusson EG, Cereghino JL, Emr SD. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J Cell Biol. 1997;137:79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séron K, Blondel M-O, Haguenauer-Tsapis R, Volland C. Uracil-induced down regulation of the yeast uracil permease. J Bacteriol. 1999;181:1793–1800. doi: 10.1128/jb.181.6.1793-1800.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séron K, Tieaho V, Prescianotto-Bashong C, Aust T, Blondel M-O, Guillaud P, Devilliers G, Rossanese OW, Glick B, Riezman H, Keränen S, Haguenauer-Tsapis R. A yeast t-SNARE involved in endocytosis. Mol Biol Cell. 1998;9:2873–2889. doi: 10.1091/mbc.9.10.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaywitz DA, Orci L, Ravazzola M, Swaroop A, Kaiser CA. Human SEC13Rp functions in yeast and is located on transport vesicles budding from the endoplasmic reticulum. J Cell Biol. 1995;128:769–777. doi: 10.1083/jcb.128.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink G, Hicks JB. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Stack JH, Horazdovsky B, Emr SD. Receptor-mediated protein sorting to the vacuole in yeast: roles for a protein kinase, a lipid kinase and GTP-binding proteins. Annu Rev Cell Dev Biol. 1995;11:1–33. doi: 10.1146/annurev.cb.11.110195.000245. [DOI] [PubMed] [Google Scholar]
- Stepp JD, Huang K, Lemmon SK. The yeast adaptor protein complex, AP-3, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J Cell Biol. 1997;139:1761–1774. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens TH, Rothman JH, Payne GS, Schekman R. Gene dosage-dependent secretion of yeast vacuolar carboxypeptidase Y. J Cell Biol. 1986;102:1551–1557. doi: 10.1083/jcb.102.5.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Amerik AY, Hochstrasser M. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol Biol Cell. 1999;10:2583–2594. doi: 10.1091/mbc.10.8.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia LA. The leptin receptor. J Biol Chem. 1997;272:6093–6096. doi: 10.1074/jbc.272.10.6093. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Gallwitz D. Isolation and characterization of SYS genes from yeast, multicopy suppressors of the functional loss of the transport GTPase Ypt6p. J Cell Sci. 1996;109:2471–2481. doi: 10.1242/jcs.109.10.2471. [DOI] [PubMed] [Google Scholar]
- Vida TA, Huyer G, Emr SD. Yeast vacuolar proenzymes are sorted in the late Golgi complex and transported to the vacuole via a prevacuolar endosome-like compartment. J Cell Biol. 1993;121:1245–1256. doi: 10.1083/jcb.121.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volland C, Garnier C, Haguenauer-Tsapis R. In vivophosphorylation of the yeast uracil permease. J Biol Chem. 1992;267:23767–23771. [PubMed] [Google Scholar]
- Volland C, Urban-Grimal D, Géraud G, Haguenauer-Tsapis R. Endocytosis and degradation of the yeast uracil permease under adverse conditions. J Biol Chem. 1994;269:9833–9841. [PubMed] [Google Scholar]
- Votsmeier C, Gallwitz D. An acidic sequence of a putative yeast Golgi membrane protein binds COPII, and facilitates ER export. EMBO J. 2001;20:6742–6750. doi: 10.1093/emboj/20.23.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Wendland B, McCaffery JM, Xiao Q, Emr SD. A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J Cell Biol. 1996;135:1485–1500. doi: 10.1083/jcb.135.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J, Nicks M, Deitz S, van Tuinen E, Franzusoff A. An N-end rule destabilization mutant reveals pre-Golgi requirements for Sec7p in yeast membrane traffic. Biochem Biophys Res Commun. 1998;243:191–198. doi: 10.1006/bbrc.1998.8084. [DOI] [PubMed] [Google Scholar]
- Zacchetti D, Peranen J, Murata M, Fiedler K, Simons K. VIP17/MAL, a proteolipid in apical transport vesicles. FEBS Lett. 1995;377:465–469. doi: 10.1016/0014-5793(95)01396-2. [DOI] [PubMed] [Google Scholar]