Abstract
Somatic cell reprogramming consists of the induction of a complex sequence of events that results in the modification of the developmental state of the cell. It is now routinely possible to reprogram fully differentiated cells back to pluripotent cells, and to transdifferentiate cells of a given type in cells of a totally different lineage origin. However, whether there are key initiating factors that are distinct from those that control stem-cell renewal and that can initiate the reprogramming process remains unknown. In contrast, what is clear is that, by modifying the epigenetic status of a cell, its reprogramming can be initiated. Here, we review the current literature that shows how the plasticity of a cell can be modulated by modifying its epigenetic status, and we discuss how epigenetic barriers can be removed, to induce an efficient reprogramming process.
Keywords: Epigenetic modifications, Somatic cell reprogramming, Histone marks, Chromatin remodeling complexes, Cell fusion, iPSCs, Nuclear transfer
Basic transcription versus chromatin structure: the merging of the two fields
The functional cross-talk between basic transcription activation and chromatin structure has remained elusive for many years, and hence research in these two fields remained distinct for a long time. Only about 15 years ago, when chromatin-modifying complexes that can remodel nucleosome structures and modify histone tails were discovered, and shown to play key roles in gene transcription, the two fields converged. As a result, the histone code was recognized as an essential regulator of the basic transcription machinery in transcriptional activation [1].
The key role of the nucleosomes in the regulation of the basic processes of transcription initially opened up novel ways to investigate the differentiation process, and then, secondarily, initiated the study of the process that induces a differentiated cell to return to a stem-cell phenotype, namely, somatic cell reprogramming.
Reprogramming as opposed to the Waddington theory
Another dogma that remained unchanged for many years was cell identity, which has been considered for a long time as a fixed, irreversible state. After fertilization, the zygote in the pre-implantation stage converts into a two-cell embryo, then a four-cell embryo, then a morula, and, finally, a blastocyst. After this, the development of the post-implantation stages follows. Cellular totipotency, i.e. the ability of a cell to differentiate into different cell types, is lost progressively during development, and only a few cell types can maintain a certain degree of potency during adult life, namely, adult stem cells and progenitor cells. Adult stem cells and progenitors are, however, restricted in their potency, and they differentiate towards the specific lineages of the tissue where they are resident.
That the differentiation process is unidirectional has been a doctrine for five or more decades. To describe this unidirectionality, in 1957, Waddington introduced the epigenetic landscape, in which the state of a totipotent cell closes down during its development, until it reaches its final fate [2].
However, already in 1958, John Gurdon demonstrated a change in the paradigm: by transplanting intestinal cell nuclei into enucleated oocytes, he generated adult tadpoles [3]. This was the first demonstration that differentiation is indeed a reversible process, and that differentiated cells are not in a fixed and unchangeable ‘locked’ state. Furthermore, the conversion of differentiated cells into cells of different lineages was also demonstrated very early on in transdifferentiation experiments, where fibroblasts were converted into muscle cells through the ectopic expression of MyoD [4, 5].
Although these important studies were performed some 50 or more years ago, it is only recently, within the last 10 years, that the process of reprogramming of differentiated cells into pluripotent cells has become largely accepted and well perceived. This was when cell-fusion-mediated reprogramming experiments were carried out, and subsequently, when induced pluripotent stem cells (iPSCs) were generated for the first time [6, 7].
Why did somatic cell reprogramming take so long to be recognized in the scientific community as a relevant biological process?
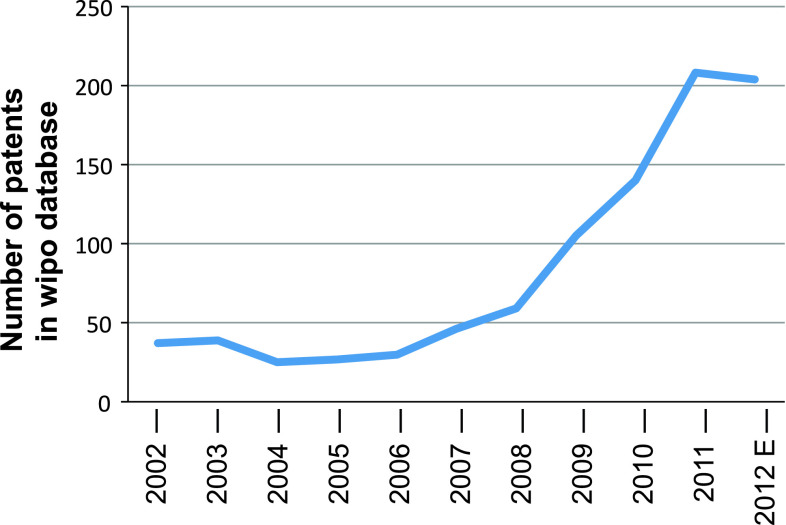
One possible explanation was the need to overcome the ethical issues associated with the use of human embryonic stem cells. Then, with the practicality offered by the use of iPSCs generated from cells isolated from the same patient in need of treatment, this led to the explosion of the field. Importantly, the economic benefits also pushed for fast development of research in this area, for the high number of new scientific journals born in the fields of stem cells and reprogramming, and for the high number of related patents that have been filed in the last 6 years (Fig. 1). Of note, the curve in Fig. 1 starts to increase in the year 2006, when iPSCs were generated for the first time [7], while it reaches a plateau in the year 2012, indicating a saturation of the inventive capabilities at that time. Novel reprogramming approaches will have to be devised in order to meet current expectations.
Fig. 1.
Patents per year from the Wipo database, using the keywords ‘somatic cell reprogramming’. The search was performed in the World intellectual property organization database (http://www.wipo.int/portal/index.html.en), considering only Patent Cooperation Treaty (PCT) patents. For 2012E, this includes the number of expected patents for the year 2012, which was linearly extrapolated based on the data collected on August 20, 2012
However, despite the promising use of iPSCs in the clinic, at present, the pressure is to understand if reprogramming of somatic cells can take place during normal tissue homeostasis in vivo. Now that the proof of principle has clearly been fulfilled, this has become the challenge that scientists have to take on in the coming years.
‘Physiological’ reprogramming
Somatic cell reprogramming can be induced in culture via different methods. However, ‘physiological’ reprogramming occurs in the course of a few days immediately after fertilization in developing embryos. Two important waves of global DNA demethylation have been reported: in the epigenome of pre-implantation mouse embryos, and when the primordial germ cells (PGCs) undergo reprogramming in the first days post-fertilization (Fig. 2) [8–11].
Fig. 2.
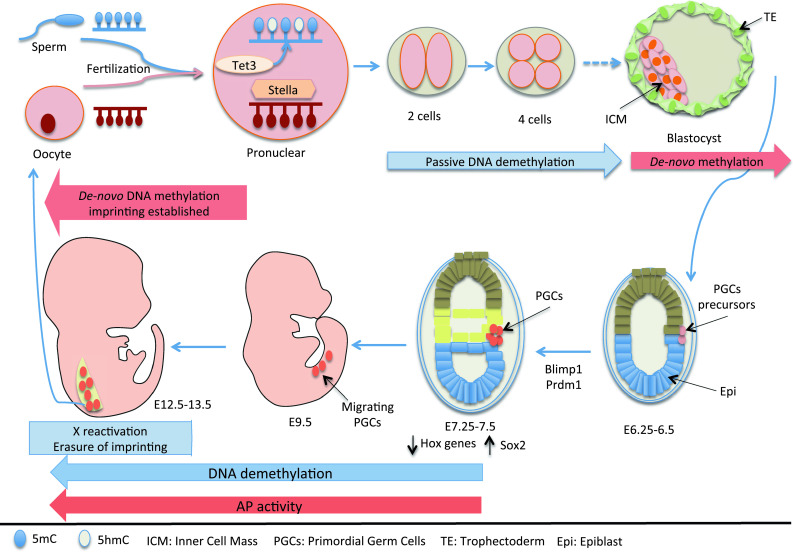
‘Physiological’ somatic cell reprogramming. Epigenetic modifications that include DNA demethylation and expression of specific classes of genes during preimplantation development and during PGC specification
DNA methylation at the 5-position of cytosine (5-methylcytosine; 5mC) is one of the key epigenetic marks that have a crucial role during development and gene transcription [12–14]. It is known that distinct genomic regions are differentially methylated depending on the cell or tissue type, and on the developmental stage [8].
In pre-implantation embryos, upon DNA replication, DNA methylation is passively removed from both the paternal and maternal genomes, from the one-cell stage to the morula/blastocyst stage. Therefore, embryos have lower levels of DNA methylation than zygotes [9, 15]. However, other active processes, which are independent of DNA replication, have been suggested. DNA demethylation might be partially due to a reduction in DNMT1, a DNA methylase, which regulates the maintenance of DNA methylation [16, 17]. On the other hand, 7–8 h after fertilization, at pronuclear stage 3, the paternal and maternal genomes are not demethylated at the same time. The paternal genome undergoes genome-wide DNA demethylation before DNA replication via an active mechanism [18–21], while the maternal genome is protected from demethylation by Stella (also known as DPPA3, developmental pluripotency-associated protein 3), which is a maternal factor that is essential for early development (Fig. 2) [22, 23]. The Tet enzymes that oxidize 5mC to 5-hydroxymethylcytosine (5hmC) have an essential role in this process [9, 14]. Indeed, recent studies have shown that 5hmC levels increase in the paternal genome at pronuclear stage 3, while those of 5mC decrease. This occurs independently of DNA replication, and the 5hmC remains until the two-cell stage [23–25]. Interestingly, this effect is dependent on maternal Tet3 [23, 24], which accounts for the active DNA demethylation of the paternal genome during this developmental stage (Fig. 2). Recently, it was also reported that Tet proteins can convert 5hmC to 5-formylcytosine and 5-carboxylcytosine [26–28], and these bases have been found in embryonic stem cells [29]. Whether the formation of these new bases has any role during physiological reprogramming still remains to be determined. Interestingly, it has been proposed that demethylation in pre-implantation embryos might be important for the creation of the pluripotent genome of the primitive ectoderm [30].
The other physiological reprogramming occurs when PGCs form. PGCs are normally monopotent, and they differentiate during development to finally generate eggs or sperms. However, PGCs can be converted in culture into pluripotent embryonic germ (EG) cells [31], suggesting that they can be reprogrammed in vitro. During development, PGC specification takes place in response to bone morphogenetic protein 4 (BMP4) and Wnt3 signaling, at E6.0 [32]. After expression of the transcription factors BLIMP1 (also known as Prdm1, a known zinc finger transcriptional repressor) and PRDM14 (PR domain containing 14), a cluster of about 40 alkaline-phosphatase-positive PGCs at E7.25 is established (Fig. 2) [33–37]. These established PGCs turn off the somatic genome, including the Hox genes, and activate expression of pluripotent factors, such as Sox2. Finally, the PGCs embark into epigenetic reprogramming, showing large dynamic changes in their epigenetic status [38]. These events take place from E7.5, when they start to migrate, to finally reach the genital ridges. Interestingly, while methylation at imprinted loci is maintained at E7.25, one X chromosome is inactivated and transposable elements are highly methylated; finally, from E7.5 to E13.5, massive epigenome reprogramming occurs, such as genome-wide DNA demethylation that encompasses genic, intergenic, and transposon elements. As a consequence, the inactive X-chromosome is reactivated, imprinted loci are demethylated, and methylation at transposons is removed [39].
The mechanisms accompanying genome-wide DNA demethylation in PGCs remain unclear. It is possible that demethylation is due to a passive process, even if putative active mechanisms have been proposed. It is known that around E7.0, expression of Dnmt1 is transiently down-regulated (even if it is re-expressed at E8.25) [40]. AID, a DNA deaminase, which could be involved in the process of DNA demethylation [41, 42], is also expressed in PGCs [43], implying its possible role in PGC demethylation, although AID-deficient mice are fertile, which is not as would be expected [44].
In addition to DNA demethylation, the migrating PGCs reduce heterochromatin signatures, such as the methylation of lysine 9 of histone H3 (H3K9me2), which coincides with down-regulation of G9a, a H3K9 methyltransferase [45]. In contrast, the levels of trimethylation of lysine 27 of histone 3 (H3K27me3) increase. Regulation of H3K27me3 are mainly due to the polycomb-group proteins (PcGs), which comprise multiprotein complexes that are required for maintaining transcriptional silencing of a subset of repressed genes [46, 47]. Two main repressor complexes, polycomb repressive complexes 1 and 2 (PRC1, 2), have been identified. These have different catalytic properties and core components. PRC2 consists of three core components: embryonic ectoderm development (Eed), suppressor of Zeste 12 (Suz12), and the SET-domain-containing protein enhancer of Zeste homolog 2 (Ezh2). The catalytic subunit, Ezh2 is a SET domain-containing methyltransferase that catalyzes the formation of the H3K27me3 marker, which forms the docking site for recruitment of PRC1 [47]. Interestingly, Ezh2 might be involved in the elevation of H3K27me3 in PGCs, because it is stably expressed in PGCs at least up to E8.25 [48].
Interestingly, the epigenetic markers associated with open chromatin also fluctuate during PGC development. Specifically, H3 lysine 9 acetylation (H3K9ac) and H3 lysine 4 methylation (H3K4me) increase in PGCs by E10.5, and subsequently decrease by E12.5 [40]. In agreement with this, the efficiency to establish pluripotent EG cells from PGCs drops off after E11.5 [49], when markers of open chromatin are not longer present in the forming PGCs.
Reprogramming mechanisms: different approaches, similar outcomes
At present, the process of somatic cell reprogramming is considered the in vitro approach to generate pluripotent cells from somatic cells. However, how much this process resembles physiological reprogramming is not known. On the other hand, since the Yamanaka ‘brand’ of factors (Oct4, Klf4, Sox2, and c-Myc) was defined for the reprogramming of mouse and human fibroblasts [7, 50], a large number of different approaches have been implemented. These have included: alternative factors (reduced numbers of the same four factors, or use of different factors) delivered with a variety of diverse types of viral vectors [51]; delivery of purified recombinant proteins [52]; use of whole-cell extracts isolated from ESCs [53]; and delivery of modified RNA molecules encoding reprogramming factors [54]. In addition, microRNAs [55–59] and large intergenic non-coding RNAs [60] have also been shown to induce reprogramming of somatic cells.
A large number of different somatic cell lines derived from the three germ layers and from different species have now been successfully reprogrammed [61]. Recently, a further approach has also been used, which is known as ‘direct reprogramming’ or ‘transdifferentiation’: to directly convert one somatic cell type into another without passing through the embryonic stage. Studies from the Graf laboratory have demonstrated that the transcription factor C/EBPα can convert nearly 100 % of primary pro-B and pre-B cells into functional macrophages [62, 63]. Also, it was shown more recently that, by overexpression of three specific transcription factors, Ascl1, Brn2, and Myt1l, mouse and human fibroblasts can be directly reprogrammed into induced-neuronal cells, bypassing a pluripotent intermediate state. Overexpression of the same three factors induced direct lineage conversion of hepatocytes into functional neurons [64]. Furthermore, overexpression of Mash1, Nurr1 and Lmx1a induced transdifferentiation of mouse and human fibroblasts into dopaminergic neurons [65–73]. Similarly, exocrine cells have been converted into beta islet cells using Pdx1, Ngn3, and Mafa; moreover, other different sets of factors have been used to convert muscle precursor cells into fat cells, and fibroblasts into cardiomyocytes or into hepatocytes [74–78].
Efficiency of the reprogramming process: the epigenetic barriers
Despite the numerous efforts that have been made, the efficiency of the reprogramming process has remained relatively low, and new ways to improve the procedure will be of great help. This is clearly because most cells undergo reprogramming with different efficiencies and at different speeds. From nuclear transfer experiments, as well as from heterokaryon and iPSC-generation experiments, it is clear that, as more of the cells to be reprogrammed are differentiated, the less efficient the process becomes. For example, already many years ago, Gurdon demonstrated that the transfer of the nucleus of a donor cell at the gastrula stage into a Xenopus egg was much more efficient for the generation of the swimming larva than when tadpole intestinal cell nuclei were transferred [79].
One of the important issues is to overcome the epigenetic barriers to reprogram somatic cells to pluripotency. A long time ago, through the measurement of the expression of a human muscle gene, decreasing reprogramming efficiency was demonstrated when heterokaryons were formed by fusing mouse muscle cells with human lung fibroblasts, with human keratinocytes, and with human hepatocytes, in this order [80].
More recently, generation of iPSCs has also demonstrated that, although the reprogramming efficiency is generally very low, it can be increased when the starting cells to be reprogrammed are precursors. Mature T cells are reprogrammed much less efficiently than thymic progenitor cells, for example [81].
Another layer of complexity is the cell origin of the epigenetic memory. In both nuclear transfer experiments as well as in iPSCs, the reprogrammed nuclei maintain the expression of some genes that were active in the cells of origin [82–85]. Genome-wide methylome analysis of human iPSCs as compared to ESCs showed aberrant reprogramming of DNA methylation in large genomic regions, as well as differences in histone modifications, which clearly indicates that epigenetic memory can be retained in iPSCs [86]. Interestingly, nuclear transfer was shown to be more effective than factor-based reprogramming at establishing the ground state of pluripotency and at erasing the epigenetic memory [82].
Many factors that can open up chromatin and facilitate chromosome decondensation have been shown to increase reprogramming efficiency. Nucleoplasmin, which is a histone H2A and H2B chaperone, and B4, which is an oocyte-specific linker histone, can facilitate decondensation of chromatin and pluripotency gene activation in nuclear transfer experiments [87, 88]. Likewise, the remodeling and opening of chromatin by the BAF remodeling complex components, such as Brg1 and Baf155, has been shown to enhance the efficiency of mouse embryonic fibroblast (MEF) reprogramming (Fig. 3) [89].
Fig. 3.
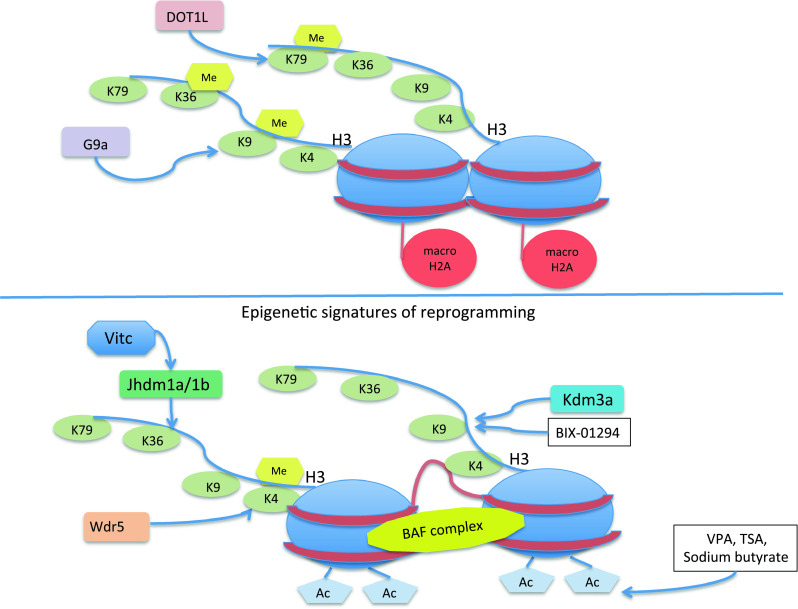
The epigenetic landscape of reprogrammed cells. Somatic cell reprogramming is facilitated by cell cloning, cell fusion, and iPSCs, via the modulation of epigenetic signatures. Nucleosome remodeling and histone tail modifications can be induced genetically or by the use of different drugs
Interestingly, the knock-down of the histone variant macroH2A increased reprogramming efficiency of MEFs (Fig. 3) [90]. MacroH2A is normally associated with heterochromatin and gene silencing [91], and reduces the binding of the remodeling complex SWI/SNF [92].
All these findings indicate that the epigenetic state of a cell is clearly associated with its grade of differentiation. ESCs contain more euchromatin then heterochromatin [93], and FRET-based techniques have shown that chromatin proteins, such as HP1α and histone H1, are hyperdynamic and bind chromatin in ESCs with more rapid kinetics than in somatic cells [94]. Additionally, global transcription in ESCs is very high, and it is considered a hallmark of pluripotency [93]. What does all this mean? It is possible that any event that induces major remodeling of the chromatin structure will be sufficient to induce somatic cell reprogramming. Indeed, precursor cells, which contain chromatin structures that are less enriched in heterochromatin, are more prone to be reprogrammed than fully differentiated cells. In principle, even with a chromatin modification induced by an electric or a mechanical stimulus, if this is effective in modifying the chromatin structure of a cell, it might induce reprogramming and induction of pluripotency. A clear answer to this already strong evidence will be possible when the reprogramming of somatic cells can be studied in single living cells, and when the sequence of events can be dissected out. High-resolution imaging techniques will be essential for these types of studies and will teach us how the process occurs. It is likely that, already in the first few hours of reprogramming, nucleosomes will be largely remodeled and the three-dimensional chromatin structure will be rearranged.
Histone tail modifications and the induction of reprogramming
A different way to modulate reprogramming efficiency is to directly modify the DNA methylation state and the histone ‘tails’.
Many molecules can promote a general acetylation of histones and enhance reprogramming efficiency, including inhibitors of histone deacetylases (Hdacs), such as valproic acid, trichostatin A, vitamin C, and sodium butyrate [95–100]. Interestingly, vitamin C was recently shown to enhance reprogramming through the modulation of the activity of the histone demethylases Jhdm1a/1b (Fig. 3) [101].
MEFs are efficiently reprogrammed by the microRNAs cluster miR302/367 when Hdac2 is also inhibited [55]. Interestingly, the miR302/367 cluster is exclusively expressed at high levels in ESCs, but not in somatic cells [102]. Wdr5, an effector of H3K4 methylation and a core member of the mammalian Trithorax (trxG) complex, has been shown to be essential for ESC self-renewal and the generation of iPSCs [103] (Fig 3). Specifically, it has been shown that Wdr5 interacts with Oct4 to increase H3K4me3 in target genes, thereby maintaining pluripotency in ESCs or facilitating the transcriptional activation of pluripotent genes in somatic cells that undergo reprogramming [103]. In contrast, the methylation of H3K9, a marker of transcriptional silencing, inhibits reprogramming. In agreement with this, the activity of G9a, an H3K9 methyltransferase, restricts reprogramming, while the H3K9 demethylase Kdm3a (JMJD1A/JHDM2A) and the H3K9me3 inhibitor BIX-01294 can facilitate reprogramming, increasing the efficiency of iPSC generation (Fig. 3) [104–106].
Interestingly, the PcG proteins, required for maintaining transcriptional silencing of a subset of genes, are essential for the chromatin-remodeling events necessary for the reprogramming of differentiated cells towards pluripotency in cell-fusion experiments [107]. This demonstrates that maintenance of the H3K27me3 mark is essential for reprogramming of somatic nuclei.
Recently, in a screening where different chromatin-modifying enzymes were silenced, it was shown that inhibition of the H3K79 histone methyltransferase DOT1L accelerated reprogramming, significantly increasing the yield of iPSC colonies [108]. Loss of H3K9me3 heterochromatin foci and the increase in H3Ac were also seen to facilitate the reprogramming of neural precursor cells (NPCs) in cell-fusion experiments and during the generation of iPSCs through the overexpression of Oct4 and Klf4 and the silencing of Tcf3. Furthermore, an increase in genome-wide H3K4me3 was observed, while H3K27me3 remained largely unchanged [109]. Interestingly, these epigenetic modifications occurred very early: already at 5 days post-infection of the reprogramming factors. This indicates that remodeling of the chromatin state is essential to induce the reprogramming process [109].
Similar observations were made after 4-factor-induced reprogramming during the early steps of the process. H3K4me2 changed rapidly at many loci, while the repressive mark H3K27me3 remained largely unchanged. These chromatin events were seen to precede transcriptional changes [110].
At the DNA level, it has been shown that, as well as during PGCs determination, control of DNA methylation is also important for somatic cell reprogramming. Specifically, treatment with 5-aza-cytidine (AZA), a potent inhibitor of Dnmt1, promotes reprogramming efficiency [97]. In addition, AZA has been shown to facilitate the transition to full pluripotency of partially reprogrammed cell lines [111].
It is clear from all these reports that, in addition to nucleosome remodeling, modifications to the histone tails can induce changes in the transcriptional activity of different loci. These ultimately lead to the activation of pluripotent genes and to the silencing of lineage genes, thereby inducing a change in the differentiation status of the cells and their consequent reprogramming. It still remains to be clearly understood how histone tail modifications cross-talk with the nucleosome remodeling to induce the recruitment of the basic transcription machinery during the process of somatic cell reprogramming. Again, high-resolution imaging of single cells might provide us with clear answers. On the other hand, even by studying populations of cells, it is clearly evident that the induction of modifications of histone tails, which ‘open up’ the chromatin, can be sufficient to strongly enhance the reprogramming of somatic cells to pluripotency.
Physiological reprogramming versus in vitro somatic cell reprogramming
PGCs can form pluripotent EG cells in culture; however, this possibility drops off after E11.5 [49]. It is interesting to note that the epigenetic modifications that occur during the process of physiological reprogramming of PGC determination up to E11.5 resemble the ones that occur during the process of factor-induced or cell-fusion-induced reprogramming (Fig. 4).
Fig. 4.
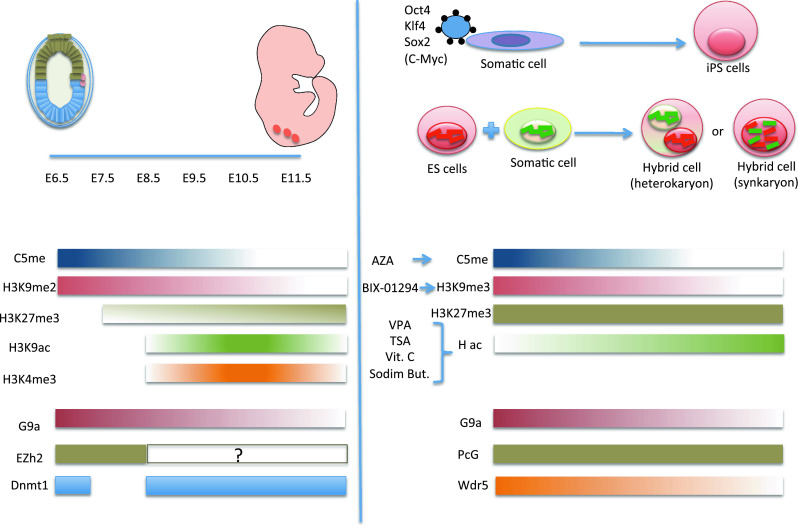
Comparison between the genome-wide epigenetic fluctuations that occur during development of PGCs and the process of factor-induced or cell-fusion-induced reprogramming. Changes in DNA methylation, histone modifications, and epigenetic factor expression during PGC development (left) and somatic cell reprogramming (right) are shown
At the DNA level, during PGC development, there is both intense active and passive DNA demethylation. The active mechanism might be due to either the reduction of Dnmt1 expression or the increase in AID expression [16, 17, 43], although different methylases or demethylases might be involved in the process.
Interestingly, in a bona fide similar fashion, the treatment of somatic cells with AZA, which inhibits Dnmt1 and consequently demethylates DNA, increases reprogramming efficiency in vitro [97, 111]. This is an effect that is also observed when AID is overexpressed in cell-fusion experiments (Fig. 4) [41].
Histone tail modification is also an important regulated process during PGC development [38]. Indeed, H3K9me2 is reduced, while H3K27me3, H3K9ac, and H3K4me3 are increased in embryo development. In a similar way, treatment of somatic cells with BIX-01294 (which reduces H3K9me3) or with drugs that inhibit Hdacs (such as valproic acid or trichostatin A) increases reprogramming (Fig. 4) [95–100, 104, 105, 112].
Overall, modifications that induce genome-wide, open chromatin (such as DNA demethylation or increased acetylation of K9 in histone 3) increase somatic cell reprogramming efficiency (Fig. 4). In contrast, the repressive mark H3K27me3 is associated with the induction of reprogramming. Indeed, reprogramming is not possible without the expression of the PcG proteins [107]. It is important to note that PcG proteins localize to the promoters of a subset of repressed genes that encode transcription factors that are required for specification during late development. This suggests that the silencing of somatic genes through PcG and H3K27me3 is essential in the process of somatic cell reprogramming. Indeed, it has been demonstrated that successful reprogramming of mouse B cells requires silencing of the B cell-specific factor Pax5 [113].
Genome-wide epigenetic modifications are essential in the process of somatic cell reprogramming as discussed above; however, it would be of great interest to study gene specific epigenetic changes. In addition, the development of PGCs, and therefore physiological reprogramming, is a well time-defined process, with several fluctuations in epigenetic modifications occurring at precise times during development. It would be very interesting to investigate whether it is possible to increase reprogramming efficiency of somatic cells by controlling epigenetic modifications in a sequential mode, as in PGC development (Fig. 4).
Conclusions: can cell plasticity be considered a different chromatin state?
More and more excellent studies have indicated that epigenetic barriers largely influence the reprogramming process, which, once released, facilitate the overall process. However, it remains to be established if there is an initial unknown factor that starts the program and induces all the epigenetic modifications, or, alternatively, if the epigenome modifications trigger one or more factor(s) that can silence the lineage genes and activate the stem genes. This is in some way a chicken-and-egg problem, and the solution might need to be found in what we indicate as physiological reprogramming; i.e. in all the events that occur during the preimplantation phase in embryos. Unknown factors, which might well be different from the ones that maintain cell stemness, self-renewal, and pluripotency, might be the important and essential players in reprogramming. If so, in the future, these can hopefully be used to improve the efficiency of the reprogramming process in culture.
Finally, what can be considered as the reprogrammed state of a cell? Excellent reviews have extensively summarized the literature in this area [30, 39, 46, 61, 74, 114–117]. These have classified marker genes and phenotypes that can univocally or satisfactorily define the reprogrammed status of a cell. A provocative definition might also be that the reprogrammed status of a cell can be defined as a different chromatin state of the same cell, which results in a different phenotype and identity (Fig. 3). The plasticity of a cell is indeed all about this: to break the epigenetic barriers, and to change identity.
Acknowledgments
The authors would like to thank F. Aulicino for help with the figures and S. Tortola for suggestions on the patent analysis. We are grateful for support from the European Research Council Grant, StERC 242630-RERE (M.P.C.), from HFSP Grant RGP0011/2010 (to M.P.C.), from Ministerio de Ciencia e Inovación SAF2011-28580 (M.P.C.), from Ministero della Salute GR-2008-1138068 (M.P.C.), and from CP10/00445 Project “Miguel Servet” of the Instituto de Salud Carlos III (F.L.).
Abbreviations
- 5mC
5-Methylcytosine
- 5hmC
5-Hydroxymethylcytosine
- AID
Activation-induced deaminase
- AZA
5-Aza-cytidine
- Blimp1
B lymphocyte-induced maturation protein 1
- BMP
Bone morphogenetic protein
- DNMT1
DNA (cytosine-5)-methyltransferase 1
- Hdacs
Histone deacetylases
- iPSCs
Induced pluripotent stem cells
- MEFs
Mouse embryonic fibroblasts
- NPCs
Neural precursor cells
- PcGs
Polycomb-group proteins
- PGCs
Primordial germ cells
- PRDM14
PR domain containing 14
References
- 1.Cosma MP, Panizza S, Nasmyth K. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol Cell. 2001;7(6):1213–1220. doi: 10.1016/S1097-2765(01)00266-0. [DOI] [PubMed] [Google Scholar]
- 2.Waddington CH. The strategy of the genes. London: Allen & Unwin; 1957. [Google Scholar]
- 3.Gurdon JB, Elsdale TR, Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182(4627):64–65. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- 4.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-X. [DOI] [PubMed] [Google Scholar]
- 5.Lassar AB, Paterson BM, Weintraub H. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell. 1986;47(5):649–656. doi: 10.1016/0092-8674(86)90507-6. [DOI] [PubMed] [Google Scholar]
- 6.Tada M, Tada T, Lefebvre L, Barton SC, Surani MA. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 1997;16(21):6510–6520. doi: 10.1093/emboj/16.21.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 9.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139(11):1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11(9):607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Surani MA. Reprogramming of genome function through epigenetic inheritance. Nature. 2001;414(6859):122–128. doi: 10.1038/35102186. [DOI] [PubMed] [Google Scholar]
- 12.Bestor TH, Coxon A. Cytosine methylation: the pros and cons of DNA methylation. Curr Biol. 1993;3(6):384–386. doi: 10.1016/0960-9822(93)90209-7. [DOI] [PubMed] [Google Scholar]
- 13.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 14.Hackett JA, Zylicz JJ, Surani MA. Parallel mechanisms of epigenetic reprogramming in the germline. Trends Genet. 2012;28(4):164–174. doi: 10.1016/j.tig.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Howlett SK, Reik W. Methylation levels of maternal and paternal genomes during preimplantation development. Development. 1991;113(1):119–127. doi: 10.1242/dev.113.1.119. [DOI] [PubMed] [Google Scholar]
- 16.Branco MR, Oda M, Reik W. Safeguarding parental identity: Dnmt1 maintains imprints during epigenetic reprogramming in early embryogenesis. Genes Dev. 2008;22(12):1567–1571. doi: 10.1101/gad.1690508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22(12):1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403(6769):501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 19.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10(8):475–478. doi: 10.1016/S0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 20.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241(1):172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 21.Wossidlo M, Arand J, Sebastiano V, Lepikhov K, Boiani M, Reinhardt R, Scholer H, Walter J. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J. 2010;29(11):1877–1888. doi: 10.1038/emboj.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, Tanaka S, Shiota K, Nakano T. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9(1):64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- 23.Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 24.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, Iqbal K, Shi YG, Deng Z, Szabo PE, Pfeifer GP, Li J, Xu GL. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477(7366):606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 25.Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci USA. 2011;108(9):3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nabel CS, Kohli RM. Molecular biology. Demystifying DNA demethylation. Science. 2011;333(6047):1229–1230. doi: 10.1126/science.1211917. [DOI] [PubMed] [Google Scholar]
- 29.Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C, Carell T. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew Chem Int Ed Engl. 2011;50(31):7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 30.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128(4):747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 31.Matsui Y, Zsebo K, Hogan BL. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70(5):841–847. doi: 10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- 32.McLaren A, Lawson KA. How is the mouse germ-cell lineage established? Differentiation. 2005;73(9–10):435–437. doi: 10.1111/j.1432-0436.2005.00049.x. [DOI] [PubMed] [Google Scholar]
- 33.Ginsburg M, Snow MH, McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development. 1990;110(2):521–528. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- 34.Ohinata Y, Ohta H, Shigeta M, Yamanaka K, Wakayama T, Saitou M. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137(3):571–584. doi: 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 35.Ohinata Y, Payer B, O’Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, Saitou M, Surani MA. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436(7048):207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 36.Vincent SD, Dunn NR, Sciammas R, Shapiro-Shalef M, Davis MM, Calame K, Bikoff EK, Robertson EJ. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development. 2005;132(6):1315–1325. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- 37.Yamaji M, Seki Y, Kurimoto K, Yabuta Y, Yuasa M, Shigeta M, Yamanaka K, Ohinata Y, Saitou M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet. 2008;40(8):1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- 38.Mochizuki K, Matsui Y. Epigenetic profiles in primordial germ cells: global modulation and fine tuning of the epigenome for acquisition of totipotency. Dev Growth Differ. 2010;52(6):517–525. doi: 10.1111/j.1440-169X.2010.01190.x. [DOI] [PubMed] [Google Scholar]
- 39.Hayashi K, Surani MA. Resetting the epigenome beyond pluripotency in the germline. Cell Stem Cell. 2009;4(6):493–498. doi: 10.1016/j.stem.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol. 2005;278(2):440–458. doi: 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 41.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463(7284):1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends Genet. 2009;25(2):82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279(50):52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 44.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463(7284):1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, Saga Y, Tachibana M, Shinkai Y, Saitou M. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development. 2007;134(14):2627–2638. doi: 10.1242/dev.005611. [DOI] [PubMed] [Google Scholar]
- 46.Boyer LA, Mathur D, Jaenisch R. Molecular control of pluripotency. Curr Opin Genet Dev. 2006;16(5):455–462. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 47.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the polycomb and trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 48.Yabuta Y, Kurimoto K, Ohinata Y, Seki Y, Saitou M. Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biol Reprod. 2006;75(5):705–716. doi: 10.1095/biolreprod.106.053686. [DOI] [PubMed] [Google Scholar]
- 49.Matsui Y, Tokitake Y. Primordial germ cells contain subpopulations that have greater ability to develop into pluripotential stem cells. Dev Growth Differ. 2009;51(7):657–667. doi: 10.1111/j.1440-169X.2009.01125.x. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 51.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322(5903):945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, Siuzdak G, Scholer HR, Duan L, Ding S. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4(5):381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cho HJ, Lee CS, Kwon YW, Paek JS, Lee SH, Hur J, Lee EJ, Roh TY, Chu IS, Leem SH, Kim Y, Kang HJ, Park YB, Kim HS. Induction of pluripotent stem cells from adult somatic cells by protein-based reprogramming without genetic manipulation. Blood. 2010;116(3):386–395. doi: 10.1182/blood-2010-02-269589. [DOI] [PubMed] [Google Scholar]
- 54.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7(5):618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8(4):376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Z, Yang CS, Nakashima K, Rana TM. Small RNA-mediated regulation of iPS cell generation. EMBO J. 2011;30(5):823–834. doi: 10.1038/emboj.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, Saito T, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Yamamoto H, Sekimoto M, Doki Y, Mori M. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8(6):633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 58.Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29(5):443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang CS, Li Z, Rana TM. microRNAs modulate iPS cell generation. RNA. 2011;17(8):1451–1460. doi: 10.1261/rna.2664111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, Manos PD, Datta S, Lander ES, Schlaeger TM, Daley GQ, Rinn JL. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42(12):1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24(20):2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bussmann LH, Schubert A, Vu Manh TP, De Andres L, Desbordes SC, Parra M, Zimmermann T, Rapino F, Rodriguez-Ubreva J, Ballestar E, Graf T. A robust and highly efficient immune cell reprogramming system. Cell Stem Cell. 2009;5(5):554–566. doi: 10.1016/j.stem.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 63.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117(5):663–676. doi: 10.1016/S0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 64.Marro S, Pang ZP, Yang N, Tsai MC, Qu K, Chang HY, Sudhof TC, Wernig M. Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell. 2011;9(4):374–382. doi: 10.1016/j.stem.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ambasudhan R, Talantova M, Coleman R, Yuan X, Zhu S, Lipton SA, Ding S. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9(2):113–118. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Broccoli V, Caiazzo M, Dell’Anno MT. Setting a highway for converting skin into neurons. J Mol Cell Biol. 2011;3(6):322–323. doi: 10.1093/jmcb/mjr029. [DOI] [PubMed] [Google Scholar]
- 67.Caiazzo M, Dell’Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Sotnikova TD, Menegon A, Roncaglia P, Colciago G, Russo G, Carninci P, Pezzoli G, Gainetdinov RR, Gustincich S, Dityatev A, Broccoli V. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476(7359):224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 68.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, Wernig M. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476(7359):220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, Bjorklund A, Lindvall O, Jakobsson J, Parmar M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci USA. 2011;108(25):10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qiang L, Fujita R, Yamashita T, Angulo S, Rhinn H, Rhee D, Doege C, Chau L, Aubry L, Vanti WB, Moreno H, Abeliovich A. Directed conversion of Alzheimer’s disease patient skin fibroblasts into functional neurons. Cell. 2011;146(3):359–371. doi: 10.1016/j.cell.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463(7284):1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang N, Ng YH, Pang ZP, Sudhof TC, Wernig M. Induced neuronal cells: how to make and define a neuron. Cell Stem Cell. 2011;9(6):517–525. doi: 10.1016/j.stem.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476(7359):228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Graf T. Historical origins of transdifferentiation and reprogramming. Cell Stem Cell. 2011;9(6):504–516. doi: 10.1016/j.stem.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 75.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142(3):375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460(7259):1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475(7356):390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 78.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455(7213):627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J Embryol Exp Morphol. 1962;10:622–640. [PubMed] [Google Scholar]
- 80.Blau HM, Pavlath GK, Hardeman EC, Chiu CP, Silberstein L, Webster SG, Miller SC, Webster C. Plasticity of the differentiated state. Science. 1985;230(4727):758–766. doi: 10.1126/science.2414846. [DOI] [PubMed] [Google Scholar]
- 81.Eminli S, Foudi A, Stadtfeld M, Maherali N, Ahfeldt T, Mostoslavsky G, Hock H, Hochedlinger K. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat Genet. 2009;41(9):968–976. doi: 10.1038/ng.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467(7313):285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10(1):102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- 84.Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, Qi Z, Downey SL, Manos PD, Rossi DJ, Yu J, Hebrok M, Hochedlinger K, Costello JF, Song JS, Ramalho-Santos M. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13(5):541–549. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, Natesan S, Wagers AJ, Melnick A, Evans T, Hochedlinger K. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28(8):848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, Downes M, Yu R, Stewart R, Ren B, Thomson JA, Evans RM, Ecker JR. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471(7336):68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cho H, Wolffe AP. Xenopus laevis B4, an intron-containing oocyte-specific linker histone-encoding gene. Gene. 1994;143(2):233–238. doi: 10.1016/0378-1119(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 88.Jullien J, Astrand C, Halley-Stott RP, Garrett N, Gurdon JB. Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation. Proc Natl Acad Sci USA. 2010;107(12):5483–5488. doi: 10.1073/pnas.1000599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Singhal N, Graumann J, Wu G, Aráuzo-Bravo J, Han DW, Greber B, Gentie L, Mann M, Scholer H. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 90.Pasque V, Gillich A, Garrett N, Gurdon JB. Histone variant macroH2A confers resistance to nuclear reprogramming. EMBO J. 2011;30(12):2373–2387. doi: 10.1038/emboj.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Banaszynski LA, Allis CD, Lewis PW. Histone variants in metazoan development. Dev Cell. 2010;19(5):662–674. doi: 10.1016/j.devcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Angelov D, Molla A, Perche PY, Hans F, Cote J, Khochbin S, Bouvet P, Dimitrov S. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol Cell. 2003;11(4):1033–1041. doi: 10.1016/S1097-2765(03)00100-X. [DOI] [PubMed] [Google Scholar]
- 93.Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, Gingeras TR, Misteli T, Meshorer E. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2(5):437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10(1):105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bui HT, Wakayama S, Kishigami S, Park KK, Kim JH, Thuan NV, Wakayama T. Effect of trichostatin A on chromatin remodeling, histone modifications, DNA replication, and transcriptional activity in cloned mouse embryos. Biol Reprod. 2010;83(3):454–463. doi: 10.1095/biolreprod.109.083337. [DOI] [PubMed] [Google Scholar]
- 96.Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J, Li W, Weng Z, Chen J, Ni S, Chen K, Li Y, Liu X, Xu J, Zhang S, Li F, He W, Labuda K, Song Y, Peterbauer A, Wolbank S, Redl H, Zhong M, Cai D, Zeng L, Pei D. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6(1):71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 97.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26(7):795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kishigami S, Mizutani E, Ohta H, Hikichi T, Thuan NV, Wakayama S, Bui HT, Wakayama T. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem Biophys Res Commun. 2006;340(1):183–189. doi: 10.1016/j.bbrc.2005.11.164. [DOI] [PubMed] [Google Scholar]
- 99.Liang G, Taranova O, Xia K, Zhang Y. Butyrate promotes induced pluripotent stem cell generation. J Biol Chem. 2010;285(33):25516–25521. doi: 10.1074/jbc.M110.142059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, Brodsky RA, Ohm JE, Yu W, Baylin SB, Yusa K, Bradley A, Meyers DJ, Mukherjee C, Cole PA, Cheng L. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28(4):713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang T, Chen K, Zeng X, Yang J, Wu Y, Shi X, Qin B, Zeng L, Esteban MA, Pan G, Pei D. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011;9(6):575–587. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 102.Barroso-del Jesus A, Lucena-Aguilar G, Menendez P. The miR-302-367 cluster as a potential stemness regulator in ESCs. Cell Cycle. 2009;8(3):394–398. doi: 10.4161/cc.8.3.7554. [DOI] [PubMed] [Google Scholar]
- 103.Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, Wang J, Rendl M, Bernstein E, Schaniel C, Lemischka IR. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145(2):183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H, Bergman Y. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15(11):1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ma DK, Chiang CH, Ponnusamy K, Ming GL, Song H. G9a and Jhdm2a regulate embryonic stem cell fusion-induced reprogramming of adult neural stem cells. Stem Cells. 2008;26(8):2131–2141. doi: 10.1634/stemcells.2008-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3(5):568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 107.Pereira CF, Piccolo FM, Tsubouchi T, Sauer S, Ryan NK, Bruno L, Landeira D, Santos J, Banito A, Gil J, Koseki H, Merkenschlager M, Fisher AG. ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell. 2010;6(6):547–556. doi: 10.1016/j.stem.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 108.Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, Lander ES, Armstrong SA, Daley GQ. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483(7391):598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lluis F, Ombrato L, Pedone E, Pepe S, Merrill BJ, Cosma MP. T-cell factor 3 (Tcf3) deletion increases somatic cell reprogramming by inducing epigenome modifications. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1017402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Koche RP, Smith ZD, Adli M, Gu H, Ku M, Gnirke A, Bernstein BE, Meissner A. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8(1):96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454(7200):49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2(6):525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 113.Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, Creyghton MP, Steine EJ, Cassady JP, Foreman R, Lengner CJ, Dausman JA, Jaenisch R. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133(2):250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vierbuchen T, Wernig M. Direct lineage conversions: unnatural but useful? Nat Biotechnol. 2011;29(10):892–907. doi: 10.1038/nbt.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465(7299):704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136(4):509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Niwa H. How is pluripotency determined and maintained? Development. 2007;134(4):635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]