Abstract
Reduction of nutrient intake without malnutrition positively influences lifespan and healthspan from yeast to mice and exerts some beneficial effects also in humans. The AMPK-FoxO axis is one of the evolutionarily conserved nutrient-sensing pathways, and the FOXO3A locus is associated with human longevity. Interestingly, FoxO3A has been reported to be also a mitochondrial protein in mammalian cells and tissues. Here we report that glucose restriction triggers FoxO3A accumulation into mitochondria of fibroblasts and skeletal myotubes in an AMPK-dependent manner. A low-glucose regimen induces the formation of a protein complex containing FoxO3A, SIRT3, and mitochondrial RNA polymerase (mtRNAPol) at mitochondrial DNA-regulatory regions causing activation of the mitochondrial genome and a subsequent increase in mitochondrial respiration. Consistently, mitochondrial transcription increases in skeletal muscle of fasted mice, with a mitochondrial DNA-bound FoxO3A/SIRT3/mtRNAPol complex detectable also in vivo. Our results unveil a mitochondrial arm of the AMPK-FoxO3A axis acting as a recovery mechanism to sustain energy metabolism upon nutrient restriction.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-012-1244-6) contains supplementary material, which is available to authorized users.
Keywords: Glucose restriction, FoxO3A, AMPK, SIRT3, OXPHOS
Introduction
Reduction of nutrient intake without malnutrition has been proven to increase lifespan and/or healthspan across species, from yeast to mammals [1–3]. Glucose restriction (GR) extends lifespan in C. elegans and delays senescence of human primary cells [4–6]. In nematodes, GR exerts this effect through the AMP-activated kinase (AMPK) AAK-2 [4]. The nutrient-sensing AMPK pathway is evolutionarily conserved from invertebrates to mammals. In C. elegans, AAK-2, together with the Forkhead-box O (FoxO) transcription factor DAF-16, is also required for lifespan and healthspan extension induced by other different dietary regimens [7, 8]. In mammals, the AMPK-FoxO3A axis has a central role in the balance between energy-consuming and energy-producing pathways [9]. Recently, the FOXO3A locus was shown to be associated with longevity in genetically diverse groups of European and Asian descent, indicating that FOXO3A is a modifier of general relevance that may play a role in many human populations [10–14].
FoxO3A plays a role in oxidative stress response and regulation of energy metabolism in a number of tissues by inducing specific activation of a coordinated transcriptional program encoding for genes involved in cell cycle arrest, cell death, cell metabolism, autophagy and stress resistance [9, 15, 16]. Interestingly, in C. elegans the AMPK-dependent dietary restriction pathway does not stimulate nuclear accumulation of the FoxO homolog DAF-16 [7], indicating a functional cytoplasmic role for this transcription factor. In mammalian cells, FoxO3A has been reported to precipitate within the mitochondrial fraction together with SIRT3 [17]. However, the functional significance of such localization is still unclear. As SIRT3 is a mitochondrial sirtuin, reported to be the primary mitochondrial protein deacetylase [18], the evidence reported above suggests the existence of a previously unrecognized FoxO3A-SIRT3 interaction involved in mitochondrial signaling.
Here we report that FoxO3A mitochondrial import is regulated in response to glucose availability in an AMPK-dependent manner. Into the mitochondria, SIRT3 mediates FoxO3A binding to mitochondrial DNA (mtDNA) and transcription of mitochondrial-encoded core or catalytic subunits of the OXPHOS machinery to increase respiration, thus sustaining energy metabolism. Our results reveal a novel mitochondrial arm of the AMPK-FoxO3A axis acting as a nutrient-sensing mechanism to sustain cellular metabolism.
Results
GR-dependent FoxO3A mitochondrial import leads to increased mitochondrial respiration
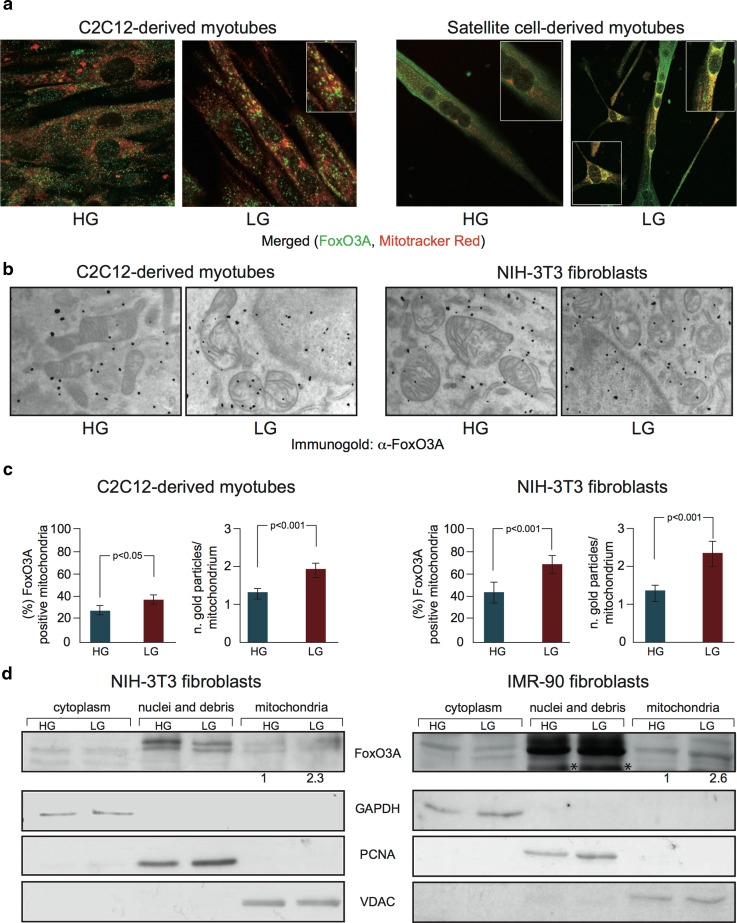
GR induces FoxO3A accumulation into the mitochondria of several established and primary cells, including murine C2C12-derived terminally differentiated skeletal muscle cells (myotubes), primary myotubes derived from adult skeletal muscle stem cells, human IMR90 primary fibroblasts, and NIH-3T3 mouse embryonic fibroblasts (Fig. 1 and Supplementary Fig. 1), a phenomenon that was not observed after glutamine deprivation or oxidative stress (Supplementary Fig. 2).
Fig. 1.
FoxO3A accumulates into the mitochondria upon GR. a, b, c Upon GR (LG), FoxO3A accumulates into the mitochondria as shown by immunofluorescence analysis (a) in murine C2C12-derived (LG 24 h) and primary satellite cell-derived myotubes (LG 36 h), and by immunogold labeling (b) in murine C2C12-derived myotubes and NIH-3T3-fibroblasts (LG 24 h) (black dots represent gold particles recognizing FoxO3A immunocomplexes). These data (n = 3) were quantified by scoring the percentage of FoxO3A-positive cells and counting the number of gold particles per single mitochondria. Data are presented as mean ± SEM and significance was calculated with Student’s t test (c). d Cellular fractionation of murine NIH-3T3 and human primary IMR90 fibroblasts shows mitochondrial enrichment of FoxO3A under low glucose conditions (LG 16 and 24 h, respectively). The values indicated are the results of the densitometric analysis of FoxO3A normalized against the mitochondrial fractionation control VDAC (arbitrary units, HG = 1). Cellular fractionation controls: nuclei: PCNA; cytoplasm: GAPDH; mitochondria: VDAC. Asterisks non-specific band
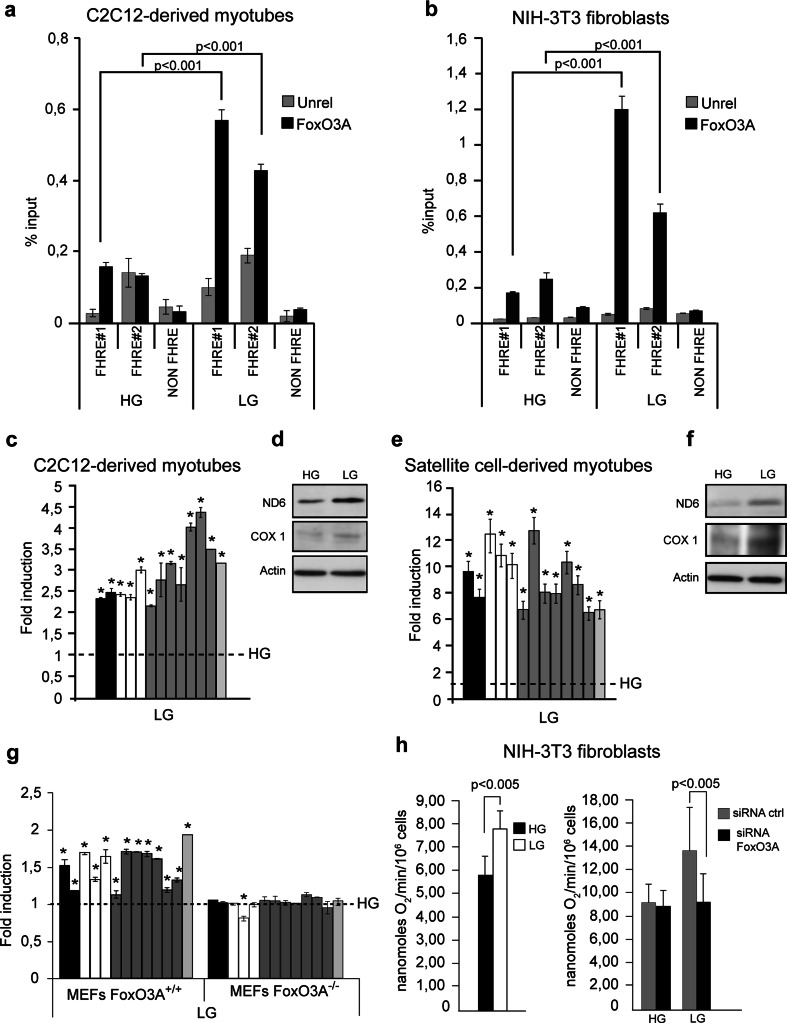
These findings prompted us to evaluate the possibility that FoxO3A may bind mtDNA and regulate transcription of the mitochondrial genome. Indeed, two putative FHRE binding sites (FoxO binding sites) were identified from the in silico analysis of both mitochondrial DNA strands. Chromatin immunoprecipitation (ChIP) experiments revealed that upon reduction of glucose levels, FoxO3A was specifically enriched at these consensus sites located at the D-Loop regulatory region of both the light (FHRE#1) and the heavy strand (FHRE#2) of mitochondrial DNA in both C2C12-derived myotubes and NIH-3T3 fibroblasts (Fig. 2a, b).
Fig. 2.
GR-dependent FoxO3A mitochondrial import leads to increased mitochondrial respiration. a, b Under GR conditions (LG 12 h), FoxO3A binds to two different target sites (FHRE#1: 14,963–15,110 bp; FHRE#2: 15,400–15,469 bp) on mitochondrial DNA in C2C12 myotubes (n = 5) (a) and NIH-3T3 fibroblasts (n = 5) (b). Signal specificity is confirmed by the analysis of a distal region not containing FHRE sites (NON FHRE, 8,653–8,768 bp). Unrel unrelated antibody (anti-IgG). Data are presented as mean ± SEM and significance was calculated with Student’s t test. c–g GR (LG) induces the upregulation of mitochondrial genes both at the RNA (c, e, n = 5) and the protein level (d, f, n = 3) in C2C12-derived (c LG 12 h, d LG 24 h) and primary satellite cell-derived myotubes (e LG 36 h, f LG 48 h). Upregulation of mitochondrial genes upon GR (LG 24 h) is blunted by the absence of FoxO3A in primary MEFs (n = 5) (g). h Upon genetic ablation of FoxO3A using a specific siRNA, GR (LG 36 h) fails to induce an increase in DNP-uncoupled mitochondrial endogenous respiration in murine NIH-3T3 fibroblasts (left panel n = 9, right panel n = 6). Data are presented as mean ± SEM and significance was calculated with Student’s t test. (c, e, g) black bars ATPase 6 and 8 genes, white bars COI, COII and COIII genes, grey bars ND1, ND2, ND3, ND4, ND4L, ND5, and ND6 genes, light grey bar cytochrome b gene. The dotted line corresponds to the expression levels detected in cells cultured in standard glucose conditions (HG). Data are presented as mean ± SEM, *p < 0.05
The mitochondrial genome encodes 13 polypeptides, which are essential components of the OXPHOS system as they constitute core or catalytic subunits of Complexes I (ND1, ND2, ND3, ND4, ND4L, ND5, and ND6), III (cytochrome b) and IV (COI, COII, and COIII) or are needed for the assembly of Complex V (ATPases 6 and 8), along with 22 tRNAs and 2 rRNA subunits necessary for the translation of these polypeptides. Transcription of the mtDNA initiates at two sites, both located at the D-Loop region of the heavy and light strands, yielding polycistronic messages. Mature mRNAs, rRNAs, and tRNAs are then released by cleavage [19]. FoxO3A signal-dependent mitochondrial localization and DNA binding coincided with the upregulation of all 13 mitochondrial-encoded genes in glucose-restricted rodent C2C12-derived myotubes, primary myotubes, NIH-3T3 fibroblasts, and human IMR90 primary fibroblasts (Fig. 2c, e, and Supplementary Fig. 3a, c, d). Increase of gene expression was time-dependent and was not associated with mitochondrial biogenesis, as revealed by quantification of the total amount of mitochondrial DNA at all time-points tested (Supplementary Fig. 4a). Glucose-restricted cell lines and primary cells showed increased protein levels of the ND6 and COX I mitochondrial-encoded catalytic subunits of OXPHOS complexes (Fig. 2d, f, and Supplementary Fig. 3b), indicating that these transcripts were effectively translated into proteins. Of note, genetic deficiency of the FoxO3A gene prevented GR-dependent increased transcription of mitochondrial-encoded genes, as shown by gene expression analysis of glucose restricted primary mouse embryonic fibroblasts (MEFs) derived from FoxO3A+/+ and FoxO3A−/− mice (Fig. 2g). Consistent with these findings, RNAi-mediated ablation of FoxO3A significantly impaired cell adaptation to GR by preventing induction of maximal mitochondrial respiratory capacity (Fig. 2h and Supplementary Fig. 4b).
AMPK mediates GR-dependent FoxO3A mitochondrial enrichment
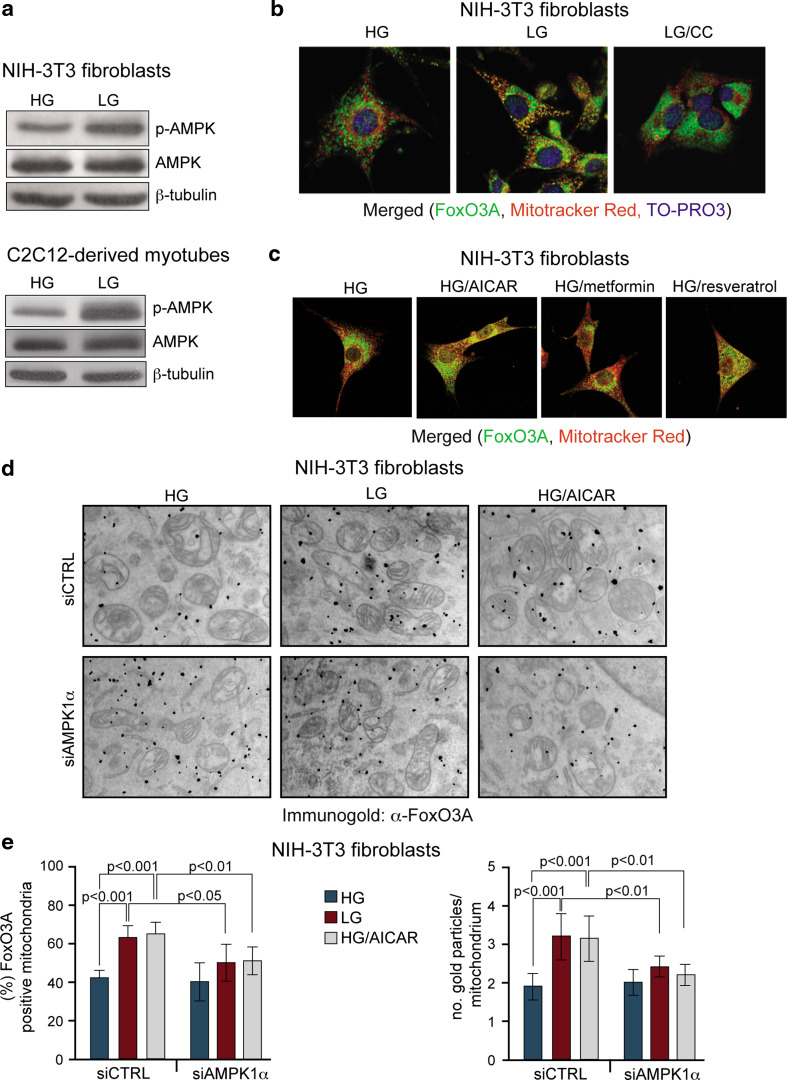
Since the low-energy sensor AMPK is activated in response to decreased glucose levels (Fig. 3a), we tested whether its kinase activity was required for the mitochondrial import of FoxO3A. Of note, pharmacological blockade of AMPK by the chemical inhibitor compound C, or its genetic ablation by a specific siRNA targeted against AMPK1α (Supplementary Fig. 5a), prevented GR-dependent FoxO3A mitochondrial accumulation (Fig. 3b, d, e).
Fig. 3.
AMPK mediates GR-dependent FoxO3A mitochondrial enrichment. a GR (LG) induces phospho-activation of AMPK in NIH-3T3 fibroblasts (LG 24 h) and C2C12-derived myotubes (LG 12 h). b GR-dependent mitochondrial accumulation of FoxO3A significantly decreases upon pharmacological inhibition of AMPK using compound C(CC) in NIH-3T3 fibroblasts (LG 16 h). c Exogenous activation of AMPK using AICAR, metformin or resveratrol induces FoxO3A accumulation into the mitochondria in NIH-3T3 fibroblasts. d FoxO3A accumulates into the mitochondria in response to GR and AMPK exogenous activation (AICAR), while AMPK ablation by a specific siRNA prevents FoxO3A accumulation even in low glucose (LG 24 h) or in the presence of an AMPK-activator (AICAR) in high glucose (HG 24 h), as shown by immunogold labeling of NIH-3T3 fibroblasts (black dots represent gold particles recognizing FoxO3A immunocomplexes). e These data (n = 3) were quantified by scoring the percentage of FoxO3A-positive cells and counting the number of gold particles per single mitochondria. Data are presented as mean ± SEM and significance was calculated with Student’s t test
The role of AMPK was further evaluated by the use of AMPK agonists. Even in the presence of high glucose, AICAR, metformin or resveratrol triggered FoxO3A mitochondrial enrichment (Fig. 3c). AICAR ability to force FoxO3A into the mitochondria in high-glucose conditions required proper expression of AMPK, as shown by quantification of the immunogold analysis performed on NIH-3T3 fibroblasts (Fig. 3d, e).
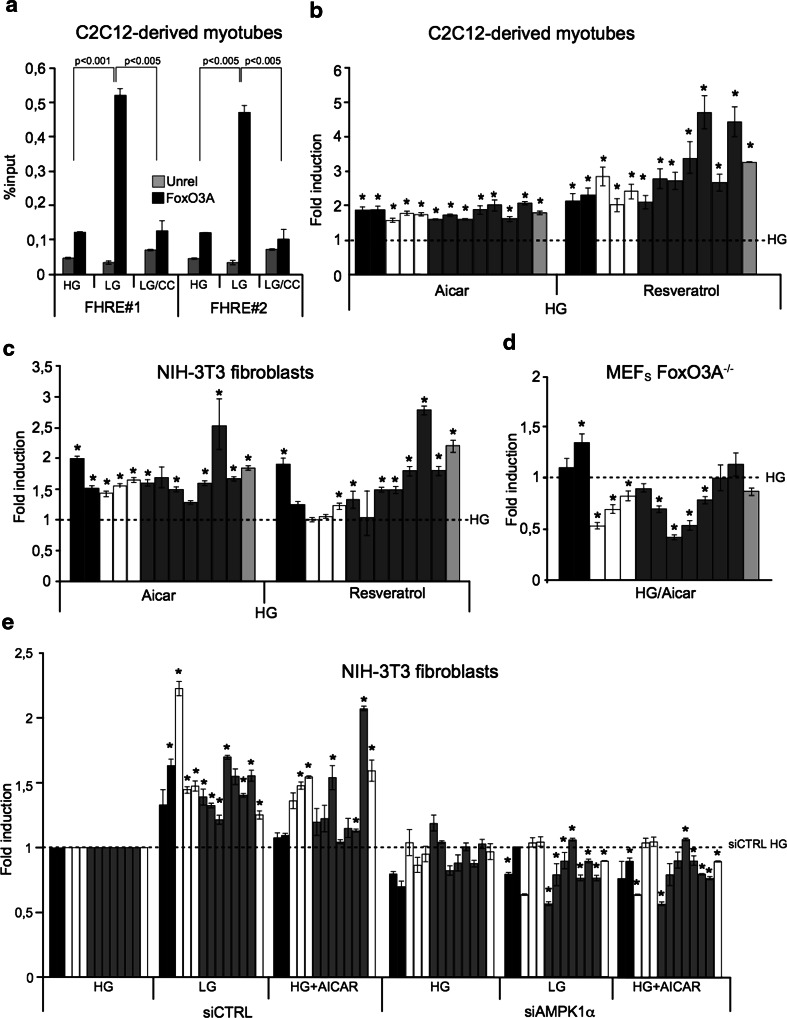
Concordant with the requirement of functional AMPK for proper GR-dependent FoxO3A accumulation into the mitochondria, the chemical inhibitor compound C prevented FoxO3A enrichment at both mtDNA consensus sequences FHRE#1 and #2 in myotubes (Fig. 4a). Moreover, genetic depletion of AMPK1α abrogated GR-dependent upregulation of mitochondrial genes (Fig. 4e and Supplementary Fig. 5a).
Fig. 4.
AMPK mediates the GR-dependent FoxO3A mitochondrial transcriptional program. a Inhibition of AMPK using compound C(CC) abrogates GR-dependent FoxO3A binding to mitochondrial DNA at both its target sites (FHRE#1: 14,963–15,110 bp; FHRE#2: 15,400–15,469 bp) in myotubes (n = 3). Unrel unrelated antibody (anti-IgG). Data are presented as mean ± SEM and significance was calculated with Student’s t test. b, c Exogenous activation of AMPK using AICAR or resveratrol promotes the upregulation of mitochondrial genes in C2C12-derived myotubes (b n = 3) and NIH-3T3 fibroblasts (c n = 3) in standard culture conditions (HG 24 h). d The AMPK activator AICAR (HG 24 h) fails to induce mitochondrial gene expression in primary MEFs derived from FoxO3A−/− mice (n = 3). e siRNA-mediated AMPK knock-down interferes with FoxO3A-dependent upregulation of mitochondrial genes induced by low glucose (LG 24 h) and addition of AICAR (HG 24 h) in NIH-3T3 fibroblasts (n = 3). The dotted line corresponds to the expression levels detected in cells transfected with a control siRNA and cultured in high-glucose conditions. b–e black bars ATPase 6 and 8 genes, white bars COI, COII and COIII genes, grey bars ND1, ND2, ND3, ND4, ND4L, ND5, and ND6 genes, light grey bar cytochrome b gene. The dotted line corresponds to the expression levels detected in cells cultured in standard glucose conditions (HG). Data are presented as mean ± SEM, *p < 0.05
Chemical activation of AMPK by AICAR and resveratrol in fibroblasts and myotubes cultured in high glucose induced mitochondrial transcription (Fig. 4b, c) in the absence of significant changes in terms of mitochondrial biogenesis (Supplementary Fig. 5b, c). Importantly, AMPK activation failed to induce mitochondrial gene expression in FoxO3A−/− cells (Fig. 4d). Similarly, siRNA-mediated AMPK knock-down interfered with AICAR-dependent upregulation of mitochondrial genes (Fig. 4e and Supplementary Fig. 7a, b).
A FoxO3A/SIRT3/mtRNAPol protein complex is assembled on mtDNA to regulate gene expression upon GR
The activity of FoxO proteins is modulated by the interaction with cofactors, including sirtuins. Previous data showed that FoxO3A co-precipitates with the primary mitochondrial protein deacetylase SIRT3 in mitochondrial fractions of mammalian cells [17]. SIRT3 has been shown to efficiently deacetylate FoxO3A. This activity does not appear to be required for FoxO3A mitochondrial localization, but is needed for the regulation of FoxO3A DNA binding ability [17, 20], thus resembling the role of SIRT1 in the positive regulation of FoxO-dependent transcription in the nucleus [21].
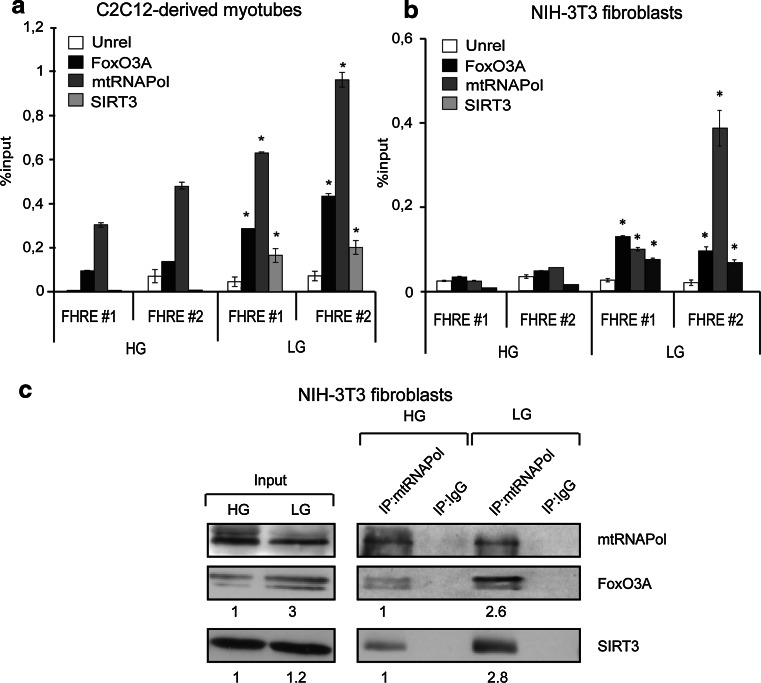
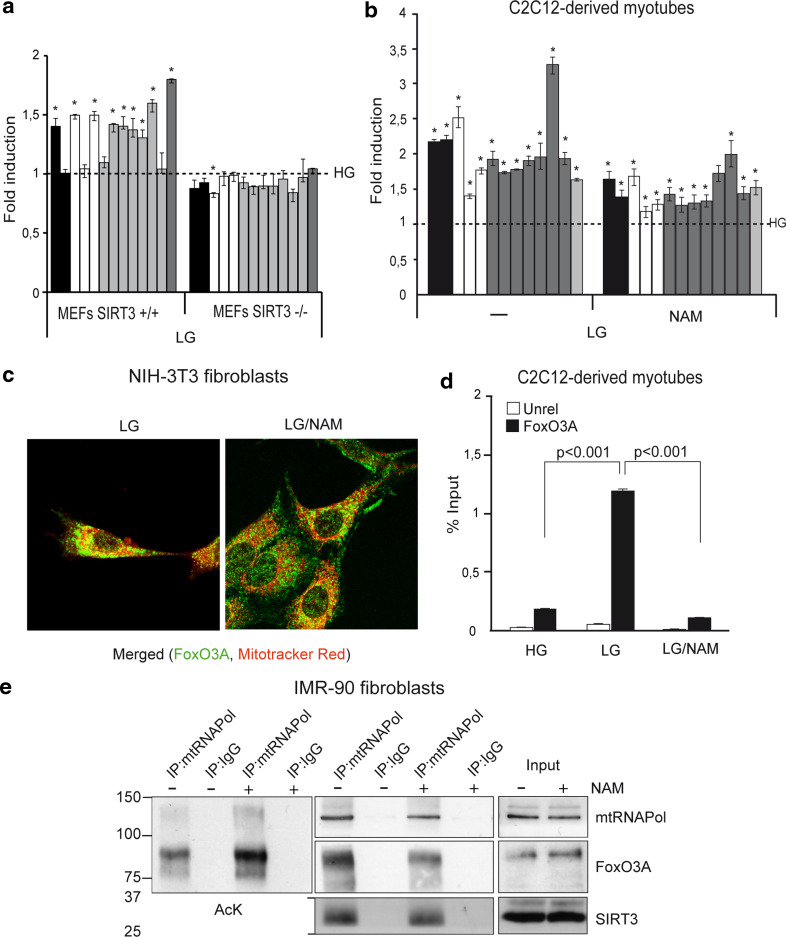
ChIP experiments performed in myotubes and fibroblasts showed that FoxO3A and SIRT3 were co-recruited together with mtRNAPol to mtDNA in a GR-dependent manner (Fig. 5a, b). Accordingly, co-immunoprecipitation studies revealed that mtRNAPol immunocomplexes are enriched in both FoxO3A and SIRT3 in glucose-restricted fibroblasts (Fig. 5c). The relevance of SIRT3 recruitment on mtDNA in response to GR was supported by genetic studies. Indeed, glucose restricted SIRT3-deficient MEFs failed to upregulate mitochondrial transcription (Fig. 6a). Of note, sirtuin activity was required for GR-dependent expression of the mitochondrial genome (Fig. 6b and Supplementary Fig. 5d), but not for FoxO3A mitochondrial localization (Fig. 6c). Indeed, nicotinamide (NAM) significantly prevented FoxO3A binding to mitochondrial DNA (Fig. 6d and Supplementary Fig. 6), and increased its acetylation levels (Fig. 6e, left panel) without affecting the amount of FoxO3A and SIRT3 bound to mtRNAPol (Fig. 6e, right panel).
Fig. 5.
A FoxO3A/SIRT3/mtRNAPol protein complex is assembled on mtDNA upon GR. a, b FoxO3A, mtRNAPol, and SIRT3 are co-recruited on mitochondrial DNA upon GR as shown by chromatin immunoprecipitation (ChIP) experiments in C2C12 myotubes (a n = 3, LG 12 h) and NIH-3T3 fibroblasts (b n = 3, LG 16 h). Unrel unrelated antibody (anti IgG). Data are presented as mean ± SEM * p < 0.05. c mtRNAPol immunocomplexes are enriched in both FoxO3A and SIRT3 in glucose-restricted NIH-3T3 fibroblasts upon GR (LG 16 h) as shown by co-immunoprecipitation analysis
Fig. 6.
SIRT3 activity was required for GR-dependent expression of the mitochondrial genome. a Upregulation of mitochondrial genes upon GR (LG, 24 h) is blunted by the absence of SIRT3 in primary MEFs (n = 5). b–d Sirtuin inhibition using nicotinamide (NAM) reduces the upregulation of mitochondrial genes (b n = 5) and FoxO3A binding to mtDNA (i.e., FHRE#1) (d n = 3) in myotubes (LG 12 h), but not its accumulation into the mitochondria in NIH-3T3 fibroblasts (c) upon low-glucose conditions (LG 24 h). Data in d are presented as mean ± SEM and significance was calculated with Student’s t test. e Co-IP performed in glucose-restricted (LG 24 h) primary IMR90 human fibroblasts. mtRNAPol immunocomplexes were blotted with an anti-acetyl lysine antibody (AcK), then stripped and re-blotted with anti-mtRNAPol, anti-FoxO3A, and anti-SIRT3 antibodies. a, b black bars ATPase 6 and 8 genes, white bars COI, COII, and COIII genes, grey bars ND1, ND2, ND3, ND4, ND4L, ND5, and ND6 genes, light grey bar cytochrome b gene. The dotted line corresponds to the expression levels detected in cells cultured in standard glucose conditions (HG). Data are presented as mean ± SEM, *p < 0.05
FoxO3A binds to mtDNA in muscle of fasted mice
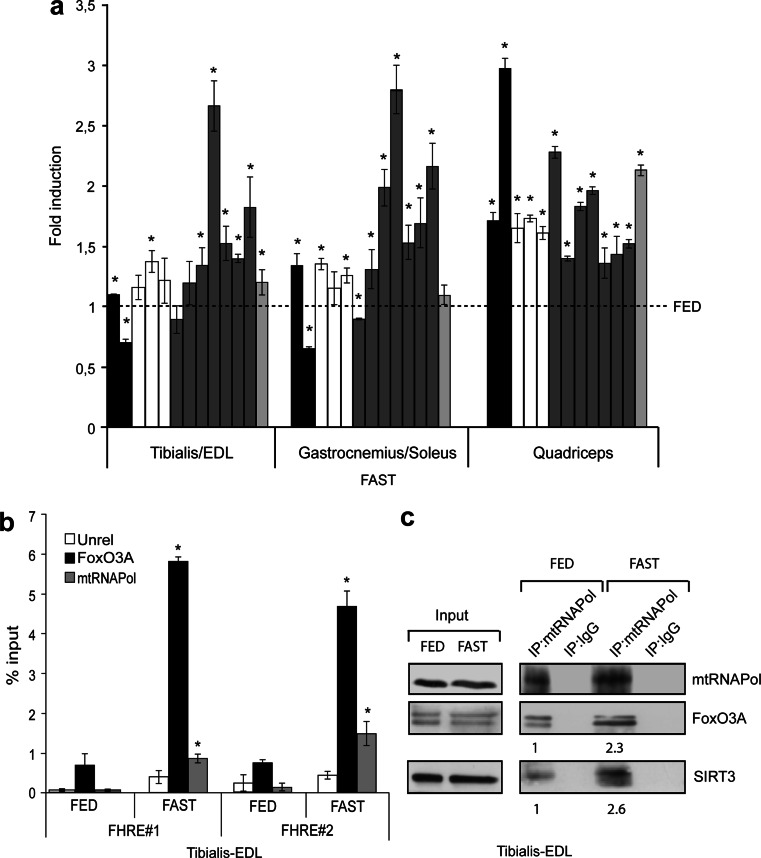
To get further insight into the GR-dependent mechanism described in muscle cells and fibroblasts in vitro, we subjected mice to overnight fasting (18 h), a procedure known to significantly lower plasma glucose levels [22]. Indeed, skeletal muscles represent the “storage” for post-prandial peripheral glucose disposal, being responsible for the uptake of the majority of the ingested carbohydrates [23, 24]. Consistent with our findings in both established cell lines and primary cultures, overnight fasting drives activation of AMPK in Tibialis-EDL muscles (Supplementary Fig. 8), and we detected upregulation of mitochondrial-encoded genes in glycolytic, oxidative and mixed type skeletal muscles of fasted mice (Fig. 7a). Importantly, FoxO3A and mtRNAPol were both significantly enriched at FHRE#1 and FHRE#2 of mtDNA in skeletal muscle of fasted mice (Fig. 7b), and co-immunoprecipitation studies revealed that mtRNAPol immunocomplexes are enriched in both FoxO3A and SIRT3 during fasting (Fig. 7c).
Fig. 7.
FoxO3A binds to mtDNA in muscle of fasted mice. a The transcription of mitochondrial genes increases in fasted mice (FAST 18 h) in both glycolytic (tibialis/EDL, n = 5), oxidative (gastrocnemius/soleus, n = 5) and mixed type (quadriceps, n = 10) skeletal muscles. black bars ATPase 6 and 8 genes, white bars COI, COII, and COIII genes, grey bars ND1, ND2, ND3, ND4, ND4L, ND5, and ND6 genes, light grey bar cytochrome b gene. The dotted line corresponds to the expression levels detected in fed mice. b FoxO3A and mtRNAPol are both enriched at regulatory sites of mtDNA in tibialis and EDL skeletal muscles of fasted mice, as shown by ChIP experiments (n = 3). Data are presented as mean ± SEM, *p < 0.05. c In vivo co-immunoprecipitation analyses revealed that mtRNAPol immunocomplexes are enriched in both FoxO3A and SIRT3 in tibialis and EDL skeletal muscles during fasting (FAST 18 h)
These data suggest a physiological role for the mtFoxO3A-SIRT3 complex in skeletal muscles and are in agreement with the observation that this tissue increases its oxidative metabolism in fasting conditions.
Discussion
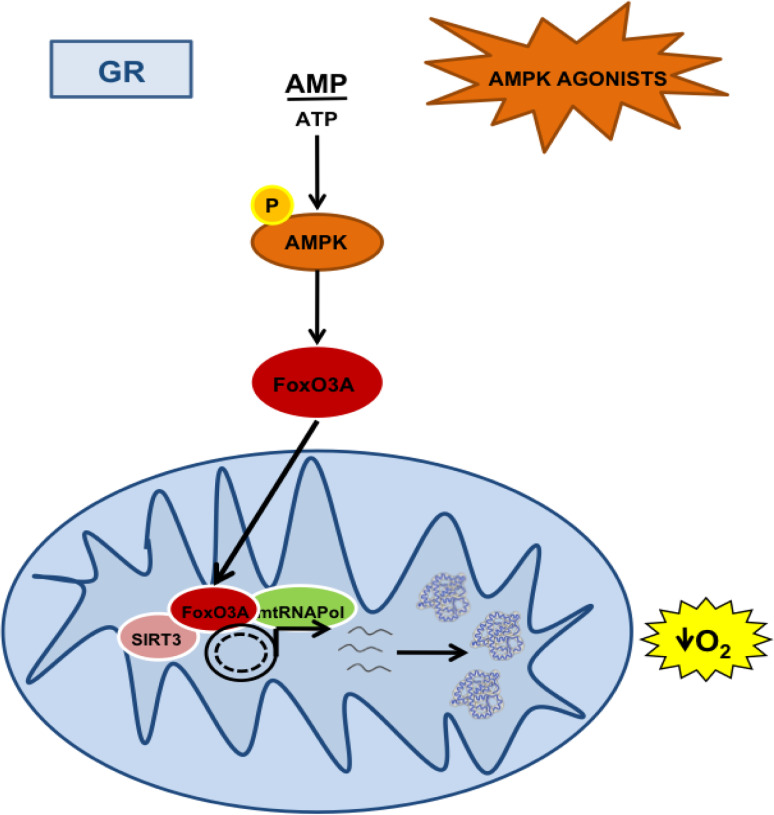
The results presented in this paper support a model where GR triggers AMPK activation, which is required for FoxO3A mitochondrial enrichment and SIRT3-dependent FoxO3A transactivation of mitochondrial-encoded core or catalytic subunits of the OXPHOS machinery, finally leading to increased respiration to sustain energy metabolism (Fig. 8). Interestingly, GR has been proven to increase lifespan in worms by inducing mitochondrial respiration in an AMPK-dependent manner [4] and primary human cells [5, 6]. Moreover, recent studies have shown that restriction of nutrients increases mitochondrial activity in several organisms, and that blocking respiration prevents nutrient restriction-driven lifespan extension [25].
Fig. 8.
The AMPK-FoxO3A-SIRT3 pathway induces mitochondrial transcription and respiration upon GR. A representative scheme depicted to summarize the findings of this work. Activation of AMPK by glucose restriction (GR) or AMPK agonists lead to FoxO3A mitochondrial enrichment. Into the mitochondria, SIRT3 mediates FoxO3A binding to mtDNA and transcription of mitochondrial-encoded core or catalytic subunits of the OXPHOS machinery, resulting in increased mitochondrial respiration
Our findings uncover an unprecedented mechanism that supports mitochondrial energy production during nutrient restriction and links the mitochondrial AMPK-FoxO3A-SIRT3 pathway to the beneficial effects of limiting nutrient intake in mammals. The intracellular trafficking of FoxO proteins is regulated by phosphorylation, a mechanism that is evolutionarily conserved from invertebrates to humans [15]. Indeed, Akt, SGK, CK1, DYRK1A, and IKKα, directly phosphorylate FoxOs allowing their binding to the 14-3-3 nuclear export protein. On the other hand, JNK1 phosphorylates the 14-3-3 protein thus inhibiting its binding to FoxOs and promoting their nuclear localization [15].
As a further mechanism of regulation, AMPK phosphorylates FoxO in response to the activation of the energy sensor pathway both in nematodes and mammalian cells [26]. In nematodes, AMPK targets six residues of FoxO/DAF-16 (T166, S202, S314, S321, T463, S466) and the AMPK-FoxO axis is sufficient and required for dietary restriction-dependent healthspan and lifespan extension [7, 27]. Indeed, these effects are suppressed in AMPK-defective strains and strongly reduced in FoxO mutant nematodes. Furthermore, this pathway is also required for oxidative stress resistance, which is tightly linked to lifespan extension. Of note, in C. elegans the AMPK-mediated FoxO-dependent beneficial effects of dietary restriction are not reliant on a transcriptional role of FoxO, which shows a cytoplasmic localization and fails to localize into the nuclei. Conversely, the oxidative stress response requires FoxO-dependent transcription of sod-3 [27]. These evidences indicate that in nematodes FoxO exerts its role in lifespan extension in two different ways, one requiring cytoplasmic localization and the other implying nuclear translocation, which are both regulated by the activity of AMPK. Mammalian FoxO3A also has six residues targeted by AMPK in vitro (T179, S399, S413, S555, S588, S626), five of which are located to the transactivation domain [7, 27]. Importantly, it has been shown that a FoxO3A construct carrying substitution of all six residues with unphosphorylatable amino acids partially retains the ability (16 %) to be phosphorylated by AMPK, suggesting the existence of additional phosphor-sites in its sequence. This FoxO3A mutant was found to be severely impaired in nuclear transcriptional activity, but maintained the ability to move between the nucleus and the cytoplasm depending on external cues. Our previous studies unveiled that AMPK activity is also required for FoxO3A nuclear accumulation in response to increased energy demand, at least in colorectal and ovarian cancer cells [9, 28–30].
These findings suggest that AMPK might directly phosphorylate additional FoxO3A sites, which may be important for mitochondrial import in response to restriction of nutrient availability across species.
Once into the mitochondria, FoxO3A forms a complex with SIRT3 and mtRNAPol to activate transcription. SIRT3 is the primary mitochondrial protein deacetylase [18] and has been shown to efficiently deacetylate FoxO3A [20]. Our data revealed that SIRT3 activity is not required for FoxO3A mitochondrial localization nor is it necessary for its interaction with mtRNAPol and SIRT3; however it is needed for mtDNA binding. Indeed, NAM significantly increased FoxO3A acetylation and prevented its recruitment on FHRE sites in the D-loop regulatory region. These data are in accordance with the proposed role of acetylation in the impairment of FoxO-chromatin interaction [21].
This newly identified FoxO3A function could be part of a coordinated nuclear-mitochondrial gene expression program involved in nutrient sensing. Indeed, a number of nuclear receptors and a few other nuclear transcription factors have been detected into the mitochondria. As a general principle, it seems that the activity of mitochondrially localized transcription factors affects mitochondrial transcription, energy yield and mtDNA integrity in a way that complements their known nuclear role rather than disclosing new or unrelated functions. The emerging picture suggests the existence of coordinated nuclear/mitochondrial activities that regulate transcription of genes serving common functions [31]. In agreement with this view, nuclear FoxO3A appears as a master regulator of the expression of fundamental genes required for proper mitochondrial activities and functions, such as PGC1α, SOD and catalase [9].
Our results might have important implications from a pharmacological point of view. Indeed, they reveal that AMPK activators, now proposed as calorie restriction (CR) mimetics [32] or exercise mimetics [33], might also act through this new arm of the AMPK biological functions. CR, a dietary regimen restricting calorie intake without malnutrition, is able to enhance animal lifespan. CR not only increases average lifespan, but also prolongs maximum lifespan and extends healthspan—defined as the number of years an organism can live without any major chronic disease. The positive effects of CR are conserved throughout evolution from nematodes to mice [1–3]. Of note, circumstantial evidences indicate that some of the beneficial effects of CR also apply to humans [34].
CR mimetics are drugs or chemical compounds which reproduce the effects of calorie restriction by activating physiological mediators, such as AMPK [32, 35]. One of the most important CR mimetics is the anti-diabetic drug metformin, which is considered a receptor sensitizer in muscle and fat cells and causes the drop of circulating glucose levels and the balancing of fat metabolism, with a final effect of body weight loss. Furthermore, metformin also reduces gluconeogenesis and inhibits excessive absorption of glucose by the gut, thus contributing to the overall glucose-lowering effect. Another AMPK activator proven to be a potent CR mimetic is resveratrol [36], a polyphenol plant chemical. In yeast, it stimulates Sir2 and extends lifespan by 70 %. In humans, it reduces the risk of age-related chronic pathologies, such as cardiovascular diseases and cancer.
Based on our findings, we speculate that pharmacological targeting of the AMPK-mtFoxO3A-SIRT3 pathway to induce expression of the mitochondrial genome could thus be exploited to help preventing and/or treating age-related diseases, since aging is also characterized by the accumulation of heteroplasmic mtDNA mutations [37].
Materials and methods
Cell culture and reagents
NIH-3T3, IMR90, and primary mouse embryonic fibroblasts (MEFs) were maintained in DMEM high glucose (HG), without pyruvate; C2C12 myoblasts were cultured as previously described [38]. All cell lines were glucose restricted by using DMEM 0.75 mM (LG). The final glucose concentration in the HG and LG medium should be considered as 40 and 15.75 mM, respectively, according to Li and Tollefson [6]. All culture media were supplemented with 10 % FBS and penicillin–streptomycin (Sigma). Primary satellite cells were obtained as previously described [39]. Compound C (5 μM), Nicotinamide (10 mM), AICAR (5 mM), Metformin (4 mM), Resveratrol (150 μM), and H2O2 (100 μM) were all from Sigma.
Immunofluorescence and immunogold labeling
For immunofluorescence analyses, cells were seeded on glass coverslips and treated as indicated. At the end of the treatment Mitotracker-red (Invitrogen) was added directly to the culture medium following manufacturer’s instructions. Cells were fixed in 4 % paraformaldehyde, permeabilized using 0.1 % Triton X-100 and incubated with the primary antibody (anti-FKHRL1, Santa Cruz Biotech or Millipore) at room temperature. The secondary antibody was Alexa Fluor 488 (Invitrogen); nuclei were counterstained using TO-PRO3 (Invitrogen). Images were acquired using a Zeiss LSM-5 Pascal confocal microscope. For pre-embedding gold labeling, cells were fixed with 4 % formaldehyde and 0.005 % glutaraldehyde, washed, incubated with the primary antibody (anti-FKHRL1, Santa Cruz Biotech) overnight, and then with Nanogold conjugated Fab fragments of the secondary antibodies (Nanoprobes) for 2 h. Nanogold particles were developed using the Gold-enhance kit (Nanoprobes). EM images were acquired from thin sections under a Philips Tecnai 10/12 electron microscope (Philips) using an ULTRA VIEW CCD digital camera (Soft Imaging Systems GmbH). Quantification of gold particles was carried out using the AnalySIS software (Soft Imaging Systems GmbH) and systematic random sampling methods [40]. Briefly, for each experimental condition 20 cells were analyzed. In each cell, 15 areas (mean 3 × 106 nm2) were randomly selected. Data presented in the Results section were obtained by scoring the percentage of FoxO3A-positive cells and counting the number of gold particles per single mitochondria.
Cellular fractionation, immunoblot analysis and co-immunoprecipitation studies
Cellular fractionation was performed by using the Qproteome Mitochondria Isolation KIT (Qiagen) following manufacturer’s instructions. Immunoblots were carried out as previously described [28]. For immunoprecipitation (IP) studies, mitochondria were lysed in IP lysis buffer (150 mM NaCl, 50 mM Tris pH 7.5, 0.3 % CHAPS, 1 mM EDTA, 1 % NP40, 10 % glycerol, 1 % Triton X-100 with the protease inhibitors 1 mM phenylmethylsulphonylfluoride, 5 mM NaF, 1 mM Na3VO4, 1.5 μM pepstatin, 2 μM leupeptin, 10 μg/ml aprotinin, 1 mM NaB, 40 μM bestatin). The lysates were cleared by centrifugation and then incubated overnight at 4 °C with 1 μg of anti-mtRNAPol (Santa Cruz Biotechnology) covalently bound to Protein G-Sepharose (GE Healthcare). Immunocomplexes were washed extensively and boiled in Laemmli sample buffer before SDS-PAGE and western blot analysis. Anti-actin (Sigma), anti-phosphoAMPK (Thr172), anti-AMPK (all from Cell Signaling), anti-β-tubulin, anti-mtRNAPol (all from Santa Cruz Biotechnology), anti-SIRT3 (Abcam or Cell Signaling), anti-ND6, anti-COX I (all from Invitrogen) and anti-FKHRL1 (Santa Cruz Biotechnology or Upstate or Cell Signaling) were used as primary antibodies. HRPO-conjugated antibodies (GE Healthcare) were used as secondary antibodies and revealed using the ECL-plus chemiluminescence reagent (GE Healthcare). The densitometric evaluation was performed by ImageJ software.
Quantitative real-time PCR
Total RNA was extracted with TRIzol reagent (Invitrogen) following manufacturer’s instructions. Samples were then treated with DNAase-1 (Ambion). Total RNA (1–4 μg) was retro-transcribed using High Capacity DNA Archive Kit (Applied Biosystems) following manufacturer’s instructions. PCRs were carried out in triplicate using an ABI 7500HT machine (Applied Biosystems). The following PCR conditions were used for all experiments: 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 60 s. Quantitative normalization of the cDNA in each sample was carried out by amplification of β-actin, GAPDH and 18S as internal controls. Relative quantification was done using the ΔΔCT method. Primer sequences are described in the Online Supplementary Material.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation analysis was conducted as previously described [38, 39] with minor modifications. Briefly, formaldehyde (Sigma) was added directly to tissue culture medium to a final concentration of 1 %. Cross-linking was allowed to proceed for 15 min at room temperature and then stopped by the addition of glycine to a final concentration of 0.125 M (Sigma). Cross-linked cells were scraped, washed with PBS, and then lysed in CLB (10 mM Tris pH 8.0, 10 mM NaCl, 0.2 % NP40) plus protease inhibitors (1 mM phenylmethylsulphonylfluoride, 5 mM NaF, 1 mM Na3VO4, 1,5 μM pepstatin, 2 μM leupeptin, 10 μg/ml aprotinin, 1 mM NaB, 40 μM bestatin). The mitochondrial fraction was enriched by differential microcentrifugation and lysed by incubation in mitochondrial lysis buffer (50 mM Tris–Cl pH 8.1, 10 mM EDTA, 1 % SDS) or purified using the Qproteome™ Mitochondria Isolation KIT (QIAGEN). The chromatin solution was sonicated, cleared by centrifugation and the supernatant was divided into aliquots. 1 % of the supernatant was taken as input. Four micrograms of antibody per point were used to immunoprecipitate chromatin-bound complexes. Immunoprecipitation was performed on a rotating platform overnight at 4 °C. Anti-FoxO3A and anti-mtRNAPol were purchased from Santa Cruz, the SIRT3 antibody was raised in rabbits against the (C)DLMQRERGKLDGQDR epitope conjugated to KLH at the added cysteine (Covance Research Products); monoclonal anti-HA (12CA5) or IgG were used as unrelated antibodies. Immunocomplexes were pulled down using protein G (GE-Healthcare). Following extensive washing, bound DNA was reverse cross-linked, purified using phenol:chloroform (Sigma) and analyzed by quantitative real-time PCR as previously described. Primer sequences are described in the Online Supplementary Material.
RNA interference
NIH3T3 or IMR90 cells were seeded in a 30-mm dish and transfected with 50 nM siRNA using RNAiMAX (Invitrogen) following manufacturer’s instructions. Specific siRNAs (Validated Stealth-oligos, Invitrogen) against FoxO3A and AMPK [28] were used:
- siFoxO3A
- 5′CCAAGACCUGCUUGCUUCAGACUCA3′;
- 5′UGAGUCUGAAGCAAGCAGGUCUUGG3′
- siAMPK1alpha
- 5′GAGGAGAGCUAUUUGAUUA 3′
- 5′UAAUCAAAUAGCUCUCCUC 3′
On-TARGET-plus control siRNAs (Thermo Scientific) were used as control.
Measurement of respiration rates in intact cells
Respiration of intact cells was measured polarographically with a Clark-type oxygen electrode in a water-jacketed chamber (Hansatech Instruments) and magnetically stirred at 37 °C as previously described [41]. Briefly, exponentially growing cells were collected by trypsinization and transferred into the polarographic chamber at 2 × 106 cells/ml in TD Buffer (0.137 M NaCl, 5 mM KCl, 0.7 mM Na2HPO4, 25 mM Tris/HCl, pH 7.4), previously air-equilibrated at 37 °C. Resuspended cell samples were read directly (basal endogenous respiration) and after addition of 45 μM 2,4-dinitrophenol (DNP) (uncoupled endogenous respiration). These rates were inhibited by over 95 % with antimycin A (50 nM). Optimal DNP concentration was determined after titrating the effect of the uncoupler on cellular respiration as in Villani et al. [42].
Animal care and use
Male pure strain C57/Bl6J mice were bred on a 12-h light/dark cycle and fed standard diet. Mice between 8 and 10 weeks of age were assigned to two experimental groups: one with free access to food, the other fasted for 18 h. Each different experimental group (fasted/fed) included at least five mice. To minimize the effect of circadian variation on gene expression profile, mice were sacrificed at the beginning of the light cycle during three consecutive days; on each separate day mice from the different experimental groups were sacrificed simultaneously. Muscles were isolated and snap frozen in liquid nitrogen and stored at −80 °C until RNA extraction. EDL, anterior tibialis, soleus, gastrocnemius and quadriceps femoris skeletal muscles were chopped into small pieces with a razor blade and then disaggregated using a Medimachine (Becton–Dickinson). Cells were harvested and processed for IP and ChIP experiments as described above. Procedures involving animals and their care were conducted in conformity with the institutional guidelines that are in compliance with national and international laws and policies.
Statistical analysis
Results are expressed as mean ± SEM and n ≥ 3. Student’s t test was used to define p values. A p value <0.05 was considered statistically significant. Multiple comparisons are accounted for via the Hochberg’s method using PROC MULTTEST in SAS [43].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1 FoxO3A accumulates into the mitochondria upon GR (LG 24h), as shown by immunogold labeling of murine C2C12-derived and murine NIH-3T3 fibroblasts (black dots represent gold particles recognizing FoxO3A immunocomplexes). (TIFF 15990 kb)
Supplementary Fig. 2. Upon GR (LG 24h), but not glutamine deprivation or oxidative stress, FoxO3A localizes to mitochondria in primary IMR90 human fibroblasts, as shown by immunofluorescence analysis using the indicated antibodies and probes. (TIFF 15991 kb)
Supplementary Fig. 3 a-d GR induces the time-dependent upregulation of mitochondrial genes both at the RNA (a, c, d) and the protein level (b) in murine NIH-3T3 (a, b), C2C12-derived terminally differentiated myotubes (c) and primary IMR90 human fibroblasts (d). (a, c, d) black bars: ATPase 6 and 8 genes; white bars: COI, COII, and COIII genes; diagonally hatched bars: ND1, ND2, ND3, ND4, ND4L, ND5, and ND6 genes; checkered bars: cytochrome b gene. The dotted line corresponds to the expression levels detected in cells cultured in standard glucose conditions (HG). Data are presented as mean ± SEM. (TIFF 19782 kb)
Supplementary Fig. 4 a GR-induced time-dependent upregulation of mitochondrial genes occurs in the absence of mitochondrial biogenesis in C2C12-derived myotubes (LG 24h), primary IMR90 (LG 48h), and NIH-3T3 cells (LG 48h). Data are presented as mean ± SEM. b The efficacy of FoxO3A genetic ablation by a specific siRNA was confirmed by densitometric analysis of Western-blot signals (FoxO3A protein levels were reduced 62% to 79% in both HG and LG conditions compared to control siRNAs). (TIFF 14984 kb)
Supplementary Fig. 5 a The efficacy of AMPK genetic ablation by a specific siRNA was confirmed by Western-blot analysis. b, c AMPK activation in HG does not affect mitochondrial biogenesis in C2C12-derived myotubes (b) and NIH-3T3 cells (c), as revealed by quantification of the total amount of mitochondrial DNA 24h after the addition of AICAR, metformin or resveratrol. d Nicotinamide does not influence mitochondrial content in myotubes upon low-glucose conditions (LG 12h). Data are presented as mean ± SEM. (TIFF 15990 kb)
Supplementary Fig. 6. ChIP experiments showing that sirtuin inhibition using nicotinamide (NAM) reduces FoxO3A binding to mtDNA in NIH-fibroblasts (FHRE#1, LG 16h). Unrel: unrelated antibody (anti-IgG). Data are presented as mean ± SEM. (TIFF 15986 kb)
Supplementary Fig. 7. a, b Immunogold labeling of NIH-3T3 fibroblasts showing that (a) FoxO3A accumulates into the mitochondria in response to GR and AMPK exogenous activation (AICAR) and (b) AMPK ablation by a specific siRNA prevents FoxO3A accumulation even in the presence of low glucose conditions or addition of AICAR (black dots represent gold particles recognizing FoxO3A immunocomplexes). (TIFF 15954 kb)
Supplementary Fig. 7. a, b Immunogold labeling of NIH-3T3 fibroblasts showing that (a) FoxO3A accumulates into the mitochondria in response to GR and AMPK exogenous activation (AICAR) and (b) AMPK ablation by a specific siRNA prevents FoxO3A accumulation even in the presence of low-glucose conditions or addition of AICAR (black dots represent gold particles recognizing FoxO3A immunocomplexes). (TIFF 15954 kb)
Supplementary Fig. 8. Overnight fasting activates AMPK in tibialis and EDL skeletal muscles (18h FAST). (TIFF 19780 kb)
Acknowledgments
We thank Dr. Francesco Paolo Jori for his helpful discussion during the preparation of the manuscript and editorial assistance, Dr. Roberta Ledonne for preparing the illustrations and editing this manuscript, Drs. Michele Petruzzeli and Daniele Di Giandomenico for technical assistance, Dr. Antonio Moschetta for discussion, Dr. Karen Arden for generously providing Foxo3A−/− MEFs and Drs. Lucisano and Pellegrini (Unit of Biostatistics, Consorzio Mario Negri Sud) for statistical analysis. Image acquisition and image data analysis were performed at the Advanced Light and Electron Microscopy Facility of the Consorzio Mario Negri Sud. This work was partially supported by a ‘My First Grant 2007’ and an ‘Investigator Grant 2010’ (IG10177) (to C.S.) from the Italian Association for Cancer Research (AIRC).
Abbreviations
- GR
Glucose restriction
- FoxO
Forkhead-box O
- AMPK
AMP-activated kinase
- FoxO binding sites
FHRE
- mtDNA
Mitochondrial DNA
- MEFs
Mouse embryonic fibroblasts
- mtRNAPol
Mitochondrial RNA polymerase
- CR
Calorie restriction
- HG
High glucose medium
- LG
Low glucose medium
- CC
Compound C
- NAM
Nicotinamide
- DNP
2,4-Dinitrophenol
- FED
Feeding
- FAST
Fasting
- AcK
Anti-acetyl lysine
References
- 1.McCay CM, Crowell MF, Maynard LA. The effect of retarded growth upon the length of life span and upon the ultimate body size. J Nutr. 1935;10:63–79. [PubMed] [Google Scholar]
- 2.Kirkwood TB, Shanley DP. Food restriction, evolution and ageing. Mech Ageing Dev. 2005;126:1011–1016. doi: 10.1016/j.mad.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 3.Fontana L, Partridge L, Longo VD. Extending healthy life span—from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Liu L, Tollefsbol TO. Glucose restriction can extend normal cell lifespan and impair precancerous cell growth through epigenetic control of hTERT and p16 expression. FASEB J. 2010;5:1442–1453. doi: 10.1096/fj.09-149328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Tollefsbol TO. p16INK4a suppression by glucose restriction contributes to human cellular lifespan extension through SIRT1-mediated epigenetic and genetic mechanism. PLOS One. 2011;6:e17421, 1–13. doi: 10.1371/journal.pone.0017421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, Gygi SP, Brunet A. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. 2007;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams DS, Cash A, Hamadani L, Diemer T. Oxaloacetate supplementation increases lifespan in Caenorhabditis elegans through an AMPK/FOXO-dependent pathway. Aging Cell. 2009;8:765–768. doi: 10.1111/j.1474-9726.2009.00527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiacchiera F, Simone C. The AMPK-FoxO3A axis as a target for cancer treatment. Cell Cycle. 2010;6:1091–1096. doi: 10.4161/cc.9.6.11035. [DOI] [PubMed] [Google Scholar]
- 10.Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci USA. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flachsbart F, Caliebe A, Kleindorp R, Blanché H, von Eller-Eberstein H, Nikolaus S, Schreiber S, Nebel A. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci USA. 2009;106:2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anselmi CV, Malovini A, Roncarati R, Novelli V, Villa F, Condorelli G, Bellazzi R, Puca AA. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12:95–104. doi: 10.1089/rej.2008.0827. [DOI] [PubMed] [Google Scholar]
- 13.Pawlikowska L, Hu D, Huntsman S, Sung A, Chu C, Chen C, Joyner AH, Schork NJ, Hsueh WC, Reiner AP, Psaty BM, Atzmon G, Barzilai N, Cummings SR, Browner WS, Kwok PY, Ziv E. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009;8:460–472. doi: 10.1111/j.1474-9726.2009.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Wang WJ, Cao H, Lu J, Wu C, Hu FY, Guo J, Zhao L, Yang F, Zhang YX, Li W, Zheng GY, Cui H, Chen X, Zhu Z, He H, Dong B, Mo X, Zeng Y, Tian XL. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet. 2009;18:4897–4904. doi: 10.1093/hmg/ddp459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27(16):2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 16.Van der Horst A, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol. 2007;8:440–450. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs KM, Pennington JD, Bisht KS, Aykin-Burns N, Kim HS, Mishra M, Sun L, Nguyen P, Ahn BH, Leclerc J, Deng CX, Spitz DR, Gius D. SIRT3 interacts with the daf-16 homolog FOXO3a in the mitochondria, as well as increases FOXO3a-dependent gene expression. Int J Biol Sci. 2008;4:291–299. doi: 10.7150/ijbs.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Jr, Fareser RV, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clayton DA. Transcription of the mammalian mitochondrial genome. Annu Rev Biochem. 1984;53:573–594. doi: 10.1146/annurev.bi.53.070184.003041. [DOI] [PubMed] [Google Scholar]
- 20.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 22.Andrikopoulos S, Blair AR, Deluca N, Fam BC, Proietto J. Evaluating the glucose tolerance test in mice. Am J Physiol Endocrinol Metab. 2008;295:E1323–E1332. doi: 10.1152/ajpendo.90617.2008. [DOI] [PubMed] [Google Scholar]
- 23.Meyer C, Dostou JM, Welle SL, Gerich JE. Role of human liver, kidney, and skeletal muscle in postprandial glucose homeostasis. Am J Physiol Endocrinol Metab. 2002;282:E419–E427. doi: 10.1152/ajpendo.00032.2001. [DOI] [PubMed] [Google Scholar]
- 24.DeFronzo RA. Pathogenesis of type 2 diabetes: metabolic and molecular implications for identifying genes. Diabetes Rev. 1997;5:177–269. [Google Scholar]
- 25.Guarente L. Mitochondria-a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132:171–176. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 27.Greer EL, Banko MR, Brunet A. AMP-activated protein kinase and FoxO transcription factors in dietary restriction-induced longevity. Ann NY Acad Sci. 2009;1170:688–692. doi: 10.1111/j.1749-6632.2009.04019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiacchiera F, Matrone A, Ferrari E, Ingravallo G, Lo Sasso G, Murzilli S, Petruzzelli M, Salvatore L, Moschetta A, Simone C. p38alpha blockade inhibits colorectal cancer growth in vivo by inducing a switch from HIF1alpha- to FoxO-dependent transcription. Cell Death Differ. 2009;16:1203–1214. doi: 10.1038/cdd.2009.36. [DOI] [PubMed] [Google Scholar]
- 29.Chiacchiera F, Simone C. Inhibition of p38alpha unveils an AMPK-FoxO3A axis linking autophagy to cancer-specific metabolism. Autophagy. 2009;5:1030–1033. doi: 10.4161/auto.5.7.9252. [DOI] [PubMed] [Google Scholar]
- 30.Matrone A, Grossi V, Chiacchiera F, Fina E, Cappellari M, Caringella AM, Di Naro E, Loverro G, Simone C. p38alpha is required for ovarian cancer cell metabolism and survival. Int J Gynecol Cancer. 2010;20:203–211. doi: 10.1111/IGC.0b013e3181c8ca12. [DOI] [PubMed] [Google Scholar]
- 31.Psarra AM, Sekeris CE. Nuclear receptors and other nuclear transcription factors in mitochondria: regulatory molecules in a new environment. Biochim Biophys Acta. 2008;1783:1–11. doi: 10.1016/j.bbamcr.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 32.Ingram DK, Zhu M, Mamczarz J, Zou S, Lane MA, Roth GS, deCabo R. Calorie restriction mimetics: an emerging research field. Aging Cell. 2006;5:97–108. doi: 10.1111/j.1474-9726.2006.00202.x. [DOI] [PubMed] [Google Scholar]
- 33.Narkar VA, Downes M, Yu RT, Embler E, Wang YX, Banayo E, Mihaylova MM, Nelson MC, Zou Y, Juguilon H, Kang H, Shaw RJ, Evans RM. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134:405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, Deutsch WA, Smith SR, Ravussin E. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith DL, Jr, Nagy TR, Allison DB. Calorie restriction: what recent results suggest for the future of ageing research. Eur J Clin Invest. 2010;40:440–450. doi: 10.1111/j.1365-2362.2010.02276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park CB, Larsson NG. Mitochondrial DNA mutations in disease and aging. J Cell Biol. 2011;193:809–818. doi: 10.1083/jcb.201010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet. 2004;36:738–743. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- 39.Serra C, Palacios D, Mozzetta C, Forcales SV, Morantte I, Ripani M, Jones DR, Du K, Jhala US, Simone C, Puri PL. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol Cell. 2007;28:200–213. doi: 10.1016/j.molcel.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lucocq JM, Habermann A, Watt S, Backer JM, Mayhew TM, Griffiths G. A rapid method for assessing the distribution of gold labeling on thin sections. J Histochem Cytochem. 2004;52:991–1000. doi: 10.1369/jhc.3A6178.2004. [DOI] [PubMed] [Google Scholar]
- 41.Villani G, Attardi G. Polarographic assays of respiratory chain complex activity. Methods Cell Biol. 2007;80:121–133. doi: 10.1016/S0091-679X(06)80005-1. [DOI] [PubMed] [Google Scholar]
- 42.Villani G, Greco M, Papa S, Attardi G. Low reserve of cytochrome c oxidase capacity in vivo in the respiratory chain of a variety of human cell types. J Biol Chem. 1998;273:31829–31836. doi: 10.1074/jbc.273.48.31829. [DOI] [PubMed] [Google Scholar]
- 43.Hochberg Y. A sharper Bonferroni procedure for multiple tests of significance. Biometrika. 1988;75:800–803. doi: 10.1093/biomet/75.4.800. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 FoxO3A accumulates into the mitochondria upon GR (LG 24h), as shown by immunogold labeling of murine C2C12-derived and murine NIH-3T3 fibroblasts (black dots represent gold particles recognizing FoxO3A immunocomplexes). (TIFF 15990 kb)
Supplementary Fig. 2. Upon GR (LG 24h), but not glutamine deprivation or oxidative stress, FoxO3A localizes to mitochondria in primary IMR90 human fibroblasts, as shown by immunofluorescence analysis using the indicated antibodies and probes. (TIFF 15991 kb)
Supplementary Fig. 3 a-d GR induces the time-dependent upregulation of mitochondrial genes both at the RNA (a, c, d) and the protein level (b) in murine NIH-3T3 (a, b), C2C12-derived terminally differentiated myotubes (c) and primary IMR90 human fibroblasts (d). (a, c, d) black bars: ATPase 6 and 8 genes; white bars: COI, COII, and COIII genes; diagonally hatched bars: ND1, ND2, ND3, ND4, ND4L, ND5, and ND6 genes; checkered bars: cytochrome b gene. The dotted line corresponds to the expression levels detected in cells cultured in standard glucose conditions (HG). Data are presented as mean ± SEM. (TIFF 19782 kb)
Supplementary Fig. 4 a GR-induced time-dependent upregulation of mitochondrial genes occurs in the absence of mitochondrial biogenesis in C2C12-derived myotubes (LG 24h), primary IMR90 (LG 48h), and NIH-3T3 cells (LG 48h). Data are presented as mean ± SEM. b The efficacy of FoxO3A genetic ablation by a specific siRNA was confirmed by densitometric analysis of Western-blot signals (FoxO3A protein levels were reduced 62% to 79% in both HG and LG conditions compared to control siRNAs). (TIFF 14984 kb)
Supplementary Fig. 5 a The efficacy of AMPK genetic ablation by a specific siRNA was confirmed by Western-blot analysis. b, c AMPK activation in HG does not affect mitochondrial biogenesis in C2C12-derived myotubes (b) and NIH-3T3 cells (c), as revealed by quantification of the total amount of mitochondrial DNA 24h after the addition of AICAR, metformin or resveratrol. d Nicotinamide does not influence mitochondrial content in myotubes upon low-glucose conditions (LG 12h). Data are presented as mean ± SEM. (TIFF 15990 kb)
Supplementary Fig. 6. ChIP experiments showing that sirtuin inhibition using nicotinamide (NAM) reduces FoxO3A binding to mtDNA in NIH-fibroblasts (FHRE#1, LG 16h). Unrel: unrelated antibody (anti-IgG). Data are presented as mean ± SEM. (TIFF 15986 kb)
Supplementary Fig. 7. a, b Immunogold labeling of NIH-3T3 fibroblasts showing that (a) FoxO3A accumulates into the mitochondria in response to GR and AMPK exogenous activation (AICAR) and (b) AMPK ablation by a specific siRNA prevents FoxO3A accumulation even in the presence of low glucose conditions or addition of AICAR (black dots represent gold particles recognizing FoxO3A immunocomplexes). (TIFF 15954 kb)
Supplementary Fig. 7. a, b Immunogold labeling of NIH-3T3 fibroblasts showing that (a) FoxO3A accumulates into the mitochondria in response to GR and AMPK exogenous activation (AICAR) and (b) AMPK ablation by a specific siRNA prevents FoxO3A accumulation even in the presence of low-glucose conditions or addition of AICAR (black dots represent gold particles recognizing FoxO3A immunocomplexes). (TIFF 15954 kb)
Supplementary Fig. 8. Overnight fasting activates AMPK in tibialis and EDL skeletal muscles (18h FAST). (TIFF 19780 kb)