Abstract
The merging of the maternal and paternal genomes into a single pronucleus after fertilization is accompanied by a remarkable reconfiguration of chromatin in the newly formed zygote. The first stages of embryonic chromatin remodeling take place in the absence of ongoing transcription, during a species-specific developmental time-frame. Once post-fertilization chromatin states are organized, zygotic genome activation (ZGA) is initiated, and embryonic transcripts gradually take control of development. We review here transitions in chromatin modifications associated with the onset of ZGA, and the role of transcription factors and DNA motifs in the regulation of ZGA. We propose a model of sequential chromatin remodeling events preceding ZGA, leading to the onset of embryonic transcription.
Keywords: Chromatin, DNA motif, Gene expression, Transcription factor, Zygote
Introduction
Fertilization represents a clash of two differentiated haploid genomes which, after a remarkable reconfiguration, merge to form a single pronucleus in the newly formed zygote. A unique feature of early embryos is the ability to develop for a pre-determined, species-dependent, number of cell division cycles essentially without transcriptional activity. Maternally stored transcripts and proteins support this initial phase of development. They also ensure proper remodeling of the maternal and paternal genomes, in the context of chromatin, in preparation for the initiation of the developmental program. This remodeling consists of the exchange of sperm-specific DNA-associated proteins for embryonic and somatic proteins, together with the addition, removal, or replacement of biochemical modifications on the DNA itself and on associated proteins such as histones. We refer in this review to modifications on DNA and histones as epigenetic modifications. Arguably the best characterized modification of DNA involved in early embryonic development is DNA methylation, which consists of the addition of a methyl group to the 5 position of a cytosine in a CpG dinucleotide. Post-translational modifications of histone proteins, such as acetylation, methylation, ubiquitination, or phosphorylation, constitute another major category of epigenetic modifications. Emerging evidence suggests that at least some of these modifications are inheritable, in the sense that they may be passed from one cell cycle to the next, or from one generation to the next. In order to enable timely transcriptional activation of the embryonic genome, the female and male epigenomes must be rebuilt.
Following the establishment of post-fertilization chromatin states, zygotic genome activation (ZGA) is initiated, and zygotic transcripts gradually take control of development as maternal transcripts are degraded. The developmental period during which maternal mRNAs are cleared and embryonic transcription is activated is referred to as the maternal-to-zygotic transition, or MZT (reviewed in [1]). These two events are interdependent through interplay between maternal and early embryonic gene products and mRNA clearance: in short, early zygotic transcription generates proteins and micro-RNAs with a feedback effect that enhances maternal mRNA destruction. The mechanisms of regulation of maternal mRNA clearance at the MZT have recently been reviewed elsewhere [2].
In this review, we focus on transitions in the chromatin landscape associated with the onset of ZGA during embryonic development through the MZT. We discuss changes in post-translational histone modifications and DNA methylation, the importance of maternal and early embryonic transcription factors (TFs) in the regulation of ZGA, and place these events in a DNA sequence context.
Genomic enrichment in post-translationally modified histones during the maternal-to-zygotic transition
Histone marks associated with gene regulatory regions
The transition from transcriptional quiescence to activation at the time of ZGA is associated with intense biochemical and physical remodeling of the genome, and evidence highlights chromatin-associated processes involved in the establishment of the embryo’s own transcriptional program. The developmental period around the time of ZGA is associated with dynamic post-translational histone modification processes in regions of the genome of particular importance for early development. Among histone modifications which have been most extensively characterized (reviewed in [3]) are lysine 4 trimethylation on histone H3 (H3K4me3). H3K4me3 marks the transcription start site (TSS) of transcriptionally active, but also inactive, genes and is often regarded as a transcriptionally permissive modification. H3K27me3, on the other hand, is associated with the promoter of transcriptionally inactive genes. Similarly to H3K4me3, H3K27me3 overlaps the TSS of inactive genes; however, its occupancy often extends further 5′ of the TSS than H3K4me3, to cover a broader domain in the promoter and enhancer regions. Notably, in embryonic stem (ES) cells [4, 5], progenitor cells [6] and differentiated cells [7, 8], H3K4me3 and H3K27me3 may be co-enriched over developmentally regulated genes “poised” for later activation, creating a “bivalent” chromatin domain. H3K9me3 is also associated with inactive promoters, but it can also occupy the body of active genes, presumably to inhibit spurious transcription from cryptic intragenic start sites [9]. Whether H3K9me3 and H3K27me3 can together form a co-repressive module on inactive promoters is a matter of current debate, as evidence suggests such co-enrichment may occur both in embryonic and somatic cells [10, 11]. H3K36me3 is localized on gene bodies and parallels RNA polymerase II (RNAPII) occupancy; thus it is commonly considered as a mark of transcriptional elongation [12].
Chromatin marks of enhancer regions have only recently been identified. Monomethylation of H3K4 (H3K4me1) and acetylation of H3K27 (H3K27ac) have proven useful in identifying distal regulatory elements such as enhancers [13]. Enhancer-bound H3K27ac has in ES cells been correlated with transcriptional activity of the nearest gene, whereas enhancer-associated H3K27me3 has been linked to inactivity [13, 14]. Analysis of CD4+ T cells, however, shows no correlation between enhancer-bound histone marks and enhancer activity [7]. Thus, unlike promoter regions for which a relationship between enrichment in specific histone marks and gene activity has been well established, the correlation between histone modifications and enhancer activity is not straightforward.
Fancy wrapping: packaging of the sperm genome in multivalent chromatin domains
Prior to fertilization, the maternal and paternal gametes contain terminally differentiated genomes, which in most (albeit not all) species are condensed into a metaphase plate (female genome) or packaged into the sperm head (male genome). Sperm DNA is tightly wrapped around protamines and histones, whose ratio varies between species. For instance, whereas human [15] and frog [16, 17] sperm contain protamines and histones, zebrafish sperm contains essentially only histones and no detectable protamines [18].
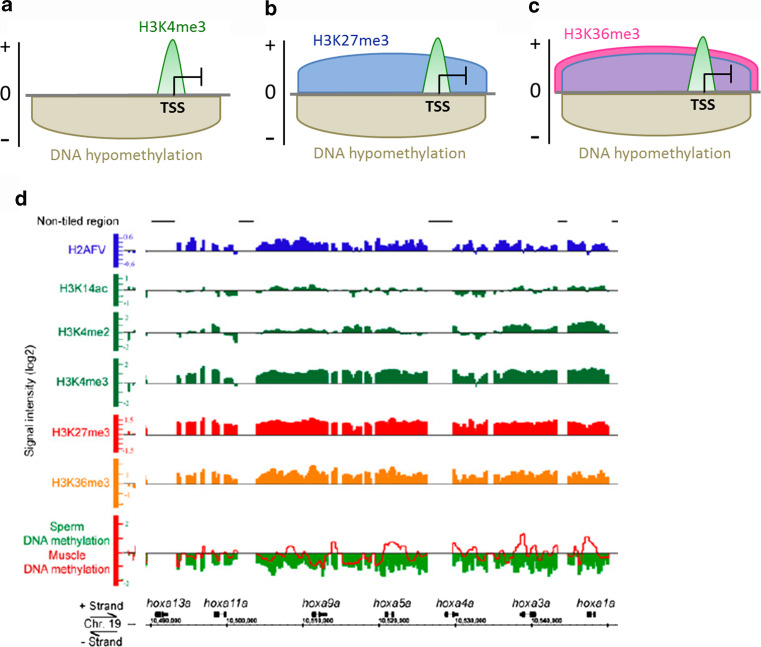
In a context of globally condensed chromatin within the sperm head, a fine tuning of genome packaging appears to be regulated at the level of distinct chromatin domains by modified histones, notably in mouse, human, and zebrafish sperm [15, 18–20] (Fig. 1). These domains contain genes with critical functions in early development, with genes ensuring cellular homeostatic (“housekeeping”) functions marked by H3K4me3 (Fig. 1a), and developmentally regulated genes involved in transcription regulation, embryo patterning, and morphogenesis apparently co-enriched in permissive H3K4me3 (or H3K4me2) and repressive H3K27me3 (Fig. 1b). The promoters of these developmentally regulated genes in human and zebrafish sperm are notably DNA hypomethylated [15, 18, 21, 22] (Fig. 1a,b), presumably conferring “permissiveness” to gene activation in the embryo. An emerging feature of sperm chromatin is the uniqueness of combinations of histone marks identified, some of which have to our knowledge not been described in somatic cells. For instance, H3K36me3, classically reported on the coding regions of active genes, is found in zebrafish sperm on inactive developmental gene promoters, either of stand-alone genes or as part of entire clusters; these areas of H3K36me3 often coincide with blocks of H3K27me3 [18] (Fig. 1c), as illustrated over the hoxa locus in a recent study (Fig. 1d). Thus, epigenetic marking of developmentally critical genes by specific combinations of histone marks in sperm creates “multivalent” chromatin domains presumed to be important for the establishment of gene expression patterns after onset of ZGA in the embryo [15, 18].
Fig. 1.
Marking of sperm chromatin by DNA methylation, post-translational histone modifications and histone variants. a–c DNA methylation and histone modification patterns detected on housekeeping and developmental gene promoters in zebrafish sperm. a H3K4me3 enrichment at the TSS of DNA hypomethylated genes involved in cellular homeostasis. b, c Promoters of developmentally regulated genes are also DNA hypomethylated and marked by H3K4me3 together with b H3K27me3, or c H3K27me3 and H3K36me3. d Multivalent marking of the hoxa locus by modified histones, histone variant and DNA (hypo)methylation in zebrafish sperm. These chromatin immunoprecipitation and array hybridization profiles depict occupancy in histone variant H2AFV (top track), and in the following histone modifications: H3K14ac (absent from the hoxa locus), H3K4me2, H3K4me3 (these three marks are transcriptionally permissive), H3K27me3 (transcriptionally repressive) and H3K36me3 (commonly associated with transcriptionally active genes in somatic cells). Note the overlapping domains marked by H3K4me3, H3K27me3, and H3K36me3. This domain is depleted of DNA methylation (bottom track, beige). Panel d is reproduced and modified from Ref. [18] with permission
Remodeling of chromatin states at the time of zygotic genome activation
Fertilization is accompanied by alterations in the composition of male chromatin as sperm DNA decondenses: DNA is rapidly demethylated, protamines are exchanged for histones, and changes in histone modifications take place. In mammals, the maternal and paternal genomes harbor distinct chromatin modifications, or “marks”, and during the cleavage stages of mouse development (from the one-cell to eight-cell stage), the parental genomes remain epigenetically asymmetrical [23]. Protamine replacement by maternally provided histones and histone variants in the male genome is accompanied by changes in histone methylation states [23–26]. The mouse male genome also becomes rapidly hyperacetylated, consistent with the acquisition of a transcriptionally permissive state [27, 28].
In the mouse, each parental genome contributes differently to the establishment of heterochromatic marks in the zygote. Maternal constitutive heterochromatin is enriched in H3K9me2 and H3K9me3 marks that are inherited from the oocyte, while paternal pericentric heterochromatin is devoid of H3K9me3 [29]. This deficiency is compensated for by the maternal Polycomb repressor complex (PRC)1, which is targeted to paternal heterochromatin [29]. Unlike in somatic cells, PRC1 targeting is independent of the PRC2 complex, yet it is nonetheless accompanied by H3K27 trimethylation of the paternal genome. This process is dependent on the RNF2 component of the PRC1 complex and is functionally critical for the regulation of transcription: in zygotes deficient in maternal Rnf2, PRC1 is non-functional, and paternal (but not maternal) pericentric major satellite transcripts are markedly upregulated [29]. The parent-of-origin-specific states of H3K9 and H3K27 methylation are asymmetrically inherited in the female and male genome during the first three cleavage divisions; however, at the eight-cell stage, both epigenomes are indistinguishable [29]. Transition through the MZT is therefore accompanied by nucleus-wide remodeling of chromatin of the paternal genome from maternally inherited components.
Chromatin immunoprecipitation assays have, over recent years, provided a landscape of histone modifications in relation to transcriptional activity around the time of ZGA, notably in zebrafish, Xenopus, and Drosophila. These studies show genomic enrichment in H3K4me3 and H3K27me3 at the time of ZGA in zebrafish [30–32]. Whereas H3K4me3 marks the promoters of genes with primarily housekeeping functions, H3K27me3 occupies developmentally important genes [30]. Additionally, in line with transcriptional activation occurring upon ZGA, H3K36me3 is detected on the bodies of genes activated at that time. Similar findings have been reported in embryos of Xenopus [33] and Drosophila [34, 35]. In Xenopus, H3K27me3 notably occupies promoters of developmentally important genes that are spatially and temporally regulated during the establishment of body axis [33]. These H3K27me3-marked promoters in Xenopus are devoid of H3K4me3, arguing against “bivalent” co-enrichment by transcriptionally permissive and repressive marks. Whether developmentally regulated promoters halted by H3K27me3 are also marked by H3K4me3 in embryos remains a matter of debate [30, 33, 34], which has been recently addressed [36].
Epigenetic pre-patterning of developmental gene expression by histone marks
Patterning of gene activity by promoter marks
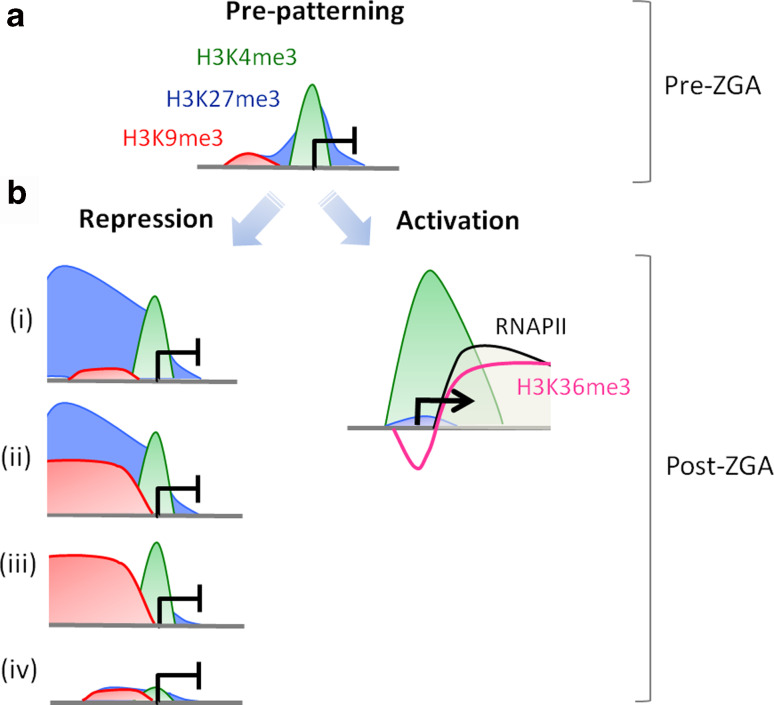
A peculiarity of anamniote vertebrate development is the relatively long window of embryonic development in the absence of transcription before ZGA onset at the MZT. In zebrafish, this window lasts for 3.3 h after fertilization and is accompanied by rapid cell divisions cycles (~20 min) during which the embryo merely replicates its DNA. These rapid cell cycles that precede the MZT make it unlikely that a chromatin signature of embryonic development be established in a locus-specific fashion concomitantly with ZGA onset as initially proposed [30]. Recent evidence indeed demonstrates occupancy of the zebrafish genome by post-translationally modified histones prior to ZGA onset [10], suggesting an epigenetic pre-patterning of developmental gene expression before the onset of transcription (Fig. 2a).
Fig. 2.
Epigenetic pre-patterning of developmental gene expression by histone modifications in zebrafish embryos. a Prior to ZGA, developmentally important genes are marked by trimethylated H3K4, H3K9, and/or H3K27 in the promoter region. These genes are transcriptionally inactive during this developmental period. b At the time of ZGA, activated genes (right) lose any repressive mark they might have acquired prior to ZGA and become enriched in H3K36me3 and RNAPII downstream of the TSS, while H3K4me3 tends to extend towards the coding region. A number of genes marked by H3K4me3 (with or without any other modification) pre-ZGA also remain transcriptionally inactive at the time of ZGA, and are only activated later in development (left). These genes retain or acquire various combinations of repressive modifications, such as H3K27me3 alone (i), H3K27me3 and H3K9me3 (ii), H3K9me3 alone (iii), or lose all marks altogether, including H3K4me3 (iv)
A sensitive chromatin immunoprecipitation assay tailored for early zebrafish embryos has uncovered the marking of specific gene sets in H3K4me3, H3K9me3, and H3K27me3 two cell cycles prior to ZGA [10]. This study identified ~200 and ~500 genes, respectively, enriched in H3K27me3 and H3K9me3 on the promoter at the 256-cell stage, and more than 1,000 promoters marked by H3K4me3 over the TSS at this stage. Gene ontology terms enriched for these genes include cellular homeostatic functions (H3K4me3-marked genes), signaling functions (H3K9me3-marked genes), and transcriptional regulation and developmental functions (H3K27me3-marked genes). Nonetheless, the most significant enrichment of promoters in H3K4me3 is detected at the time of ZGA together with the enrichment of a gradually increasing number of genes in H3K27me3 through the MZT. In contrast, H3K9me3 marking of promoters increases dramatically after ZGA onset, and H3K36me3 enrichment on gene bodies is detected only after onset of ZGA [10]. Thus, developmental transition through the MZT is accompanied by a differential time-course of genomic enrichment in modified histones, reflecting the high dynamics of the reshaping of the chromatin landscape at this stage of development.
Despite the current lack of formal demonstration of the existence, at the protein level, of histone modifying enzymes in pre-ZGA zebrafish embryos, detection of histone marks in these embryos is supported by the loading of the unfertilized egg with polyadenylated transcripts for histone methyltransferases for H3K4, H3K9 and K3K27 [10, 37], and by the immunodetection of other histone modifications (such as acetylated H4) before ZGA [38]. It is therefore becoming increasingly clear that the zebrafish genome is already marked by modified histones on developmentally important loci before the onset of embryonic transcription.
Because embryonic transcription is not initiated before ZGA, genomic occupancy by modified histones suggests a role for these modifications other than regulatory on on-going transcription. To address this issue, histone modification enrichment data on promoters genome-wide [10] have been analyzed in the context of transcriptomic data collected by RNA-sequencing throughout the MZT period and beyond [37]. It emerges from this study that genes pre-marked by H3K4me3 pre-ZGA have a stronger propensity to be activated after ZGA than all RefSeq genes [10]. As expected, these genes retain H3K4me3 at the TSS and acquire both H3K36me3 and RNAPII on the coding region (Fig. 2b). Moreover, the median expression level of pre-ZGA H3K4me3-marked genes is stronger than that of all transcriptionally activated genes at that time. Thus, H3K4me3 marking of promoters pre-ZGA is strongly suggestive of transcriptional activation potential after ZGA. As equally interesting is the observation that pre-ZGA H4 acetylation mediates preferential binding of the Brd4 transcription factor [38, 39], suggesting that acetylated H4 may pre-mark genes for Brd4 targeting and transcription after ZGA [38].
Nevertheless, not all genes marked by H3K4me3 pre-ZGA are destined to be activated at the time of ZGA. Following the nature of post-translational modifications associated with pre-ZGA H3K4me3-marked genes during development through the ZGA period in zebrafish [10] indicates that a number of these genes become enriched in H3K27me3, H3K9me3, or apparently in both repressive modifications (Fig. 2b, i–iii), while retaining H3K4me3 at the TSS (Fig. 2b). A fourth class of genes loses the marks detected pre-ZGA (Fig. 2b, iv). These genes collectively include late developmentally regulated genes, or genes harboring highly specialized functions of terminally differentiated cells, and which therefore are not activated at the time of ZGA [10, 37]. Altogether, these results are consistent with an epigenetic pre-patterning of embryonic gene expression, and with an instructive role of histone modifications established pre-ZGA on the developmental transcription program.
Enhancer marks predictive of spatio-temporal activity in embryos
A tissue-specific analysis of histone modifications and RNAPII occupancy on enhancer elements in Drosophila embryos has recently extended our appreciation of the spatio-temporal orchestration of histone modifications in relation to gene activation during development [35]. Interestingly, active enhancers display a wide array of combinations of histone modifications and RNAPII occupancy. Whereas H3K4me1 marks enhancers irrespective of activity (consistent with earlier observations [13]), H3K27me3 is enriched on enhancers that are in a repressed state (e.g., enhancers of genes driving mesoderm differentiation in non-mesodermal tissues). In contrast, active mesodermal enhancers (in mesodermal cell types) are marked by H3K27ac and H3K79me3, and are occupied by RNAPII; in non-mesodermal cell types, these enhancers are however not enriched in any of these marks or in RNAPII. Moreover, RNAPII recruitment to enhancers is predictive of developmental timing of enhancer activity in Drosophila embryos and is related to TF occupancy [35].
As a result of these observations, a model of stepwise developmental enhancer activation involving histone marks is emerging [35]. In this model, H3K4me1 marks developmentally important enhancers, which are prone to repression by H3K27 trimethylation or activation by H3K27 acetylation and H3K79 trimethylation. The transcriptionally permissive state of these enhancers is in turn compatible with local RNAPII recruitment, TF occupancy, and nucleosome remodeling, which together may determine the active state. It will be important to determine whether enhancers are, in addition to promoters, marked by modified histones prior to ZGA onset. Likewise, it will be informative to assess whether the concept of developmental spatio-temporal enhancer activation also holds true in other organisms and importantly, at earlier stages of development than tissue-specific cellular differentiation [35], for example when body axis is determined.
Dynamics and role of DNA methylation at the time of ZGA
DNA methylation has long been considered as a negative regulator of gene expression. In vertebrates, DNA methylation occurs on cytosines in a cytosine-phosphate-guanine (CpG) context. CpG methylation is catalyzed by DNA methyltransferases (DNMTs), which convert S-adenosylmethionine to S-adenosylhomocysteine by the addition of a methyl group to the 5th position of a cytosine on DNA. DNA methylation can repress local gene expression in several ways: directly by blocking access of transcriptional activators to promoters, or indirectly through the recruitment of methyl-binding proteins; these in turn may recruit transcriptional co-repressors, such as histone deacetylases. Several methyl-binding proteins have been identified, including methyl-CpG-binding domain proteins 1–4, MeCP2 and Kaiso [40]. In addition, the DNMTs themselves may inhibit transcription through physical interactions with several repressive chromatin modifiers. DNA methylation orchestrates several important processes during development, such as long-term gene silencing, X chromosome inactivation, genomic imprinting and silencing of repetitive elements. However, the role and importance of DNA methylation in regulation of ZGA, is still debated.
In the mouse, the phase after fertilization is characterized by a global loss of DNA methylation. During this phase, the maternal genome is passively demethylated as a result of absent maintenance methylation during the mitotic cell divisions [41]. In contrast, the paternal genome, which is hypermethylated relative to the maternal genome [42], becomes actively demethylated, possibly by oxidation of methylated cytosines to oxidized forms such as 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine, which are then eliminated by replication-dependent dilution [43]. The lowest levels of methylation are reached at the blastocyst stage [42]. Methylation marks are then gradually re-established by de novo DNMTs during epiblast development in a cell type-specific manner [44]. Interestingly, lineage commitment (e.g., extraembryonic vs. embryonic lineage determination) already appears to have been decided at this stage [45]. Thus, the primary role of DNA methylation at this stage might not be to define gene expression patterns and cell fate, but rather to ‘lock’ cell fate.
Post-fertilization demethylation is suggested as a mechanism that restores pluripotency in an embryo arising from terminally differentiated gametes. In addition, global demethylation is important for establishment of parental imprints and for elimination of possible epimutations that have occurred in the gametes [46]. However, the repressive effect of promoter DNA methylation on gene expression has led to the hypothesis of a DNA methylation-dependent mechanism of pre-ZGA gene silencing [47]. As a loss of DNA methylation can result in the activation of silent genes, the wave of genome-wide DNA demethylation taking place after fertilization may establish a transcriptionally permissive state favoring gene activation at the time of ZGA [47]. In fact, depletion of the maintenance DNA methyltransferase Dnmt1 in Xenopus embryos leads to gene activation up to two cell cycles prior to ZGA [48]. This premature gene activation only takes place on genes normally expressed at ZGA, while genes scheduled for later expression seem to be unaffected [48]. Localized gene activation in a context of global DNA demethylation would be consistent with a local pre-patterning of gene expression, e.g., by histone modifications, already established on the cohort of genes scheduled for expression at the time of ZGA.
During development, reduced levels of maintenance methylation by DNMT1 are associated with embryonic lethality and cell death in various species, including zebrafish, mouse, and frog [48–52]. Mice mutants expressing a catalytically inactive form of DNMT1 show profound developmental defects including developmental arrest after gastrulation, loss of DNA methylation, and distorted gene expression patterns [51]. However, a similar catalytically inactive Dnmt1 mutant in Xenopus embryos is viable [53]. This may suggest that embryonic DNA methylation is mostly important for phenomena which do not exist in Xenopus, such as X chromosome inactivation and imprinting. However, DNA methylation seems to have no or only a little effect on gene regulation during ZGA.
The latter view is further supported by studies in Xenopus, where methylated DNA is efficiently transcriptionally repressed in oocytes [54, 55], while in gastrula stage embryos, the potential for DNA methylation-dependent transcriptional repression is severely reduced [54]. However, during organogenesis and terminal differentiation, the repressive effect of DNA methylation is restored [54]. Further, DNA methylation profiles identified for single genes by methylated DNA immunoprecipitation and hybridization to promoter arrays reveal remarkably little change in DNA methylation patterns during the developmental progression through ZGA in zebrafish [56]. Methylation profiles of both promoters and gene bodies are overall maintained between pre-ZGA, ZGA, and early post-ZGA stages. Thus, at least in Xenopus and zebrafish, there seems to be an uncoupling of DNA methylation patterns on upstream regulatory regions and gene expression changes taking place upon ZGA.
This does not exclude a regulatory role of DNA methylation on transcriptional events occurring at the time of ZGA. In addition to DNMT1, one of the candidates involved in DNA-methylation-related regulation of the ZGA is Kaiso. Kaiso is a methylated CpG-specific repressor protein that has proven to be crucial for pre-ZGA transcriptional repression in Xenopus and zebrafish [57, 58] where it preferentially binds sequences containing two consecutive symmetrically methylated CpG dinucleotides. Knock-down of Kaiso in Xenopus embryos induces premature transcriptional onset and developmental aberrations [58]. Involvement of DNMT1 (which binds hemimethylated DNA through its binding partner UHRF1) and Kaiso (which binds symmetrically methylated DNA) in the regulation of gene expression at ZGA, suggests an important role of DNA methylation during ZGA. Yet it is important to keep in mind that methylated CpG-binding factors, although dependent on DNA methylation for binding, may not necessarily be active and can be differentially regulated during ZGA without changes in the DNA methylation states on the DNA itself.
DNA methylation functionally interacts with histone modifications [52, 59, 60], so the primary role of DNA methylation in the regulation of ZGA may also include priming of pre-ZGA chromatin for acquiring a transcription competent state through interactions with histone marks. For example, DNA methylation and H3K9me3 can associate on the same genomic regions [52, 61], while H3K4me3 is enriched on regions of unmethylated DNA [62]. In zebrafish embryos, DNA methylation patterns of promoters are essentially established prior to ZGA and do not markedly change during ZGA [56]. Histone modifications, on the other hand, are dynamically regulated during this phase and in particular, a large cohort of DNA hypomethylated genes acquires the H3K4me3 mark prior to ZGA [56]. It is therefore tempting to speculate that DNA methylation state might target histone modifications to specific loci prior to ZGA to pattern developmental gene expression.
Taken together, these observations suggest that DNA methylation per se may not be involved in directly regulating gene activity at the time of ZGA. Rather, it may have an indirect priming function through interactions with other factors including Dnmt1 and Kaiso, post-translationally modified histones, transcription factors or chromatin modifiers.
Origins of chromatin marks detected prior to ZGA: inherited or acquired de novo?
An outstanding question is where histone marks detected in embryos prior to ZGA onset originate from. Based on current observations, two non-exclusive models can be put forward. One model favors an inheritance of histone marks from parental gametes, including sperm, to the embryo. A significant proportion of genes in pre-ZGA zebrafish embryos has been found to carry the same histone modifications in sperm [18, 56], and before ZGA, H3K4me3, and H3K27me3 mark genes with similar gene ontology terms as in mouse, human, or zebrafish sperm [15, 18, 20]. Additionally, the detection of histone marks on male and female chromatin immediately after fertilization in the mouse (as discussed above; [20, 29]) and in Caenorhabditis elegans [63], are consistent with a transmission of these marks through fertilization.
These observations suggest that developmental instructions may be pre-set by histones marks already in the gametes. Remarkably, transcriptional activity in the adult animal correlates well with chromatin states (notably H3K4 and H3K36 methylation) in the embryo also in C. elegans [63], reinforcing the idea of epigenetic pre-patterning of developmental gene expression. In addition, histone modifications in the embryo are not only apparently templated by the nature and genomic location of these marks in gametes; they also reflect transcriptional activity in the parental gemline in the previous generation [63]. Thus, inheritance of modified histone marks through fertilization may constitute a mechanism of “epigenetic memory” of gene expression patterns from one generation to the next. It will be interesting to take these analyses from a low-resolution nucleus-wide level to a high-resolution gene-specific level on a genome-wide scale, to firmly establish the extent of inheritance of chromatin states through fertilization, and refine our understanding of the developmental significance of this process.
An alternative model is that of a removal of histone marks from the gametal genomes and their and re-establishment after fertilization. In this model, histone marks are removed from parental chromatin and modified histones are de novo deposited on target genes in the pre-ZGA embryo. Support for a removal of histone marks comes from observations that genes marked by sets of modified histones in zebrafish sperm are not marked by these modifications in pre-ZGA embryos sperm [10, 18]. This also holds true for DNA methylation marks: a large number of genes DNA methylated in sperm are unmethylated in early stage pre-ZGA embryos [56], likely as a result of the genome-wide demethylation process taking place after fertilization [64]. The view of a re-establishment of histone marks is supported by the ability of maternally inherited histone modifications (e.g., H3K27me3) to be targeted to the male genome post-fertilization [20], implying a de novo epigenetic marking in the early embryo. Furthermore, a large number of genes enriched in histone modifications in pre-ZGA embryos are not marked by these modifications in sperm [10, 18], indicating again a de novo marking of genes. A model of removal/re-deposition of chromatin marks is therefore not exclusive of an inheritance model.
In a context of de novo marking of genes by modified histones after fertilization, whether histones are modified after their assembly into nucleosomal chromatin, or whether they are modified in a non-nucleosomal form prior to assembly into chromatin on replicated DNA, remains unknown. One can only at present speculate that the rapidity of cell divisions in zebrafish, Xenopus, and Drosophila embryos prior to the MZT favors a view of assembly of histones in chromatin from a pre-modified pool stored in the egg. Nevertheless, it is also tempting to speculate that re-deposition of modified histones on target genes, on the basis of DNA sequence rather than of a copy of modified histones on the replicated DNA, might follow an inherent set of instructions from DNA. Novel cues might emerge from recent studies uncovering an instructive role of DNA sequence on the establishment of DNA methylation patterns [65], and on the genomic targeting of developmentally critical TFs, notably of some involved in the regulation of ZGA (see below).
A model of re-establishment of histone marks on genes harboring the same marks in sperm nevertheless arguably suggests some perpetuation of “instructive memory” from sperm [10]. This memory may be encoded in sperm at the level of DNA methylation [18, 54, 66], small RNAs or transposons [67]. Interestingly, the DNA sequence itself may also encode instructions for the establishment of histone modifications, as shown recently by the requirement for hypomethylated CG-rich regions for targeting H3K4 trimethylation [62, 68, 69], and by the identification of genetic elements that autonomously determine cytosine methylation states [65].
Pioneers in the regulation of ZGA onset
Transcription factors are adaptor proteins that detect specific DNA sequences and target the assembly of protein complexes that control gene expression. They are therefore critical components in the orchestration of transcription during development. Embryonic TFs are translated from maternally stored mRNAs following mRNA polyadenylation [37] or encoded by the zygotic genome. Many essential maternally encoded products [70] are involved in chromatin remodeling, modification of histones, maintenance of DNA methylation, and de novo DNA methylation. In contrast, most genes activated at ZGA encode developmentally regulated TFs implicated in the regulation of downstream developmental genes [37, 71].
Recent evidence suggests that “stand-by” occupancy of regulatory regions by TFs prior to ZGA onset may, in addition to chromatin states, determine the activation of developmentally regulated genes at or after ZGA. So-called pioneer TFs have been identified as factors which not only play a role in the activation of transcription of their target genes but can also bind the genome prior to transcription initiation and impart transcriptional competence [72]. For instance, tissue-specific genes activated after induction of ES cell differentiation can already be occupied by pioneer TFs in undifferentiated ES cells where these genes are silent [73]. Some pioneer TFs harbor chromatin remodeling properties and can displace linker histones, thereby relaxing chromatin and enabling binding of additional co-factors [74]. Others can notably be retained on chromosomes at mitosis, providing a “bookmarking” mechanism [75], which may perpetuate site-specific information on transcription induction in subsequent cell cycles.
During development, binding of TFs such as Zelda, STAT92E, or β-catenin to DNA is temporally uncoupled from transcription initiation of their target genes [76–78] and thereby may pre-set transcription potential in a spatio-temporal manner. Zelda is a DNA-binding zinc-finger protein critical for ZGA in Drosophila. Zelda (zld) mutation leads to abnormal early embryonic mitoses and severe cellularization defects at the MZT [79]. Severity and early onset of the effects of the zld mutation have prompted a genome-wide determination of Zelda binding sites in the genome prior to ZGA [77]. Expectedly, at the MZT, Zelda targets promoters and enhancers of genes activated at ZGA. However, as early at the eighth cell cycle (i.e., before ZGA), Zelda also binds the promoter of ~1,000 genes whose transcription is not initiated until ZGA, as well as the enhancer of nearly all genes bound at ZGA onset by TFs essential for their activation [77]. These findings lead to the view of Zelda as a developmental transcription pre-patterning factor.
STAT92E is a transcriptional activator mediating the Jak/Stat pathway, which synergistically with Zelda is required for ZGA and the up-regulation of zygotic genes in Drosophila [78]. Evidence suggests that STAT92E primarily regulates the transcription level of a subset of its target genes, but not their spatial expression patterns in the embryo. However, Zelda is essential for both level and spatial regulation of expression of these genes [80]. Orchestration of Zelda and STAT92E therefore appears to be paramount for proper dosage and distribution of gene products in the embryo.
Another factor with a pre-setting function in developmental gene expression is β-catenin [76]. In Xenopus, the dorsal–ventral axis is specified during the first cell cycle, when cortical rotation results in dorsal enrichment of β-catenin, an activator of Wnt-dependent transcription of dorsal genes [81]. β-catenin itself does not bind DNA; the DNA motif readers enabling β-catenin binding are T cell factor (TCF) proteins, which via their high-mobility group domain bind in a sequence-specific manner on target genes [82–84]. Dorsal-specifying genes in Xenopus are transcribed during the minor wave of ZGA in the 12th cell cycle [85]. Multiple TCF response elements on dorsal gene promoters are occupied by TCFs as early as the four-cell stage [86]. In addition, depletion of TCF leads to a phenocopy of β-catenin deletion [86]. This altogether suggests a central direct mediator function of TCF and β-catenin in the Wnt signaling pathway for the activation of dorsal genes.
A role of β-catenin in specifying dorsal gene expression prior to ZGA onset has been elegantly demonstrated by highlighting an interplay between β-catenin and chromatin remodeling in Xenopus embryos [76]. In pre-ZGA embryos, β-catenin has been shown to recruit the arginine methyltransferase Prmt2 to promoters, establishing symmetrical dimethylation on H3 arginine 8. In addition, β-catenin is implicated in the establishing H3K4 trimethylation on target genes [76], favoring a transcriptionally permissive state. At the molecular level, β-catenin bound to TCF exposes a C-terminal domain, which serves as a platform for the recruitment and sequential exchange of transcriptional co-activators [87]. Some of these factors (e.g., CBP, P300, TIIP60, MLL1/2, HDACs) are in turn able to modify histones, regulate nucleosome positioning (e.g., SWI/SNF), and recruit the RNAPII complex. β-catenin-mediated transcription initiation seems therefore to involve a stepwise recruitment of transcriptional co-activators, rather than the binding of a multifunctional machinery in which co-activators bind β-catenin simultaneously [88]. This stepwise assembly takes place in the context of a permissive chromatin landscape to which β-catenin also contributes [76].
DNA sequence determinants of TF binding and developmental control
The DNA motif-specificity of TF binding argues that target DNA sequence may also be a central determinant in the site-specific action of TFs. In Drosophila, the major wave of zygotic transcription initiation in the 14th cell cycle is preceded by a minor genome activation phase six cell cycles earlier [89]. Many genes transcribed during this period are involved in sex determination, embryonic patterning, and cellularization. Examination of sex-determination genes transcribed at the eighth cell stage has identified a common heptanucleotide motif (CAGGTAG) over-represented in the 500-bp region upstream of the TSS [90]. This motif is not unique to sex-determination genes and was subsequently found to be overrepresented in the upstream regulatory sequence of other genes expressed upon ZGA [90]. The CAGGTAG motif is a member of a family of consensus albeit degenerate motifs termed “TAGteam motifs” [90].
The role of TAGteam motifs in gene regulation has been identified by mutational analysis to show that TAGteam clusters are a key determinant in the timing of transcription initiation in embryos [90]. The number of TAGteam motifs engineered into reporter transgenes is directly correlated with timing of transcription initiation of the transgenes: more TAGteam motifs mean earlier activation of transcription, whereas activation of genes not containing TAGteam motifs is unaltered [90]. A degenerate VBRGGTA motif homologous to Drosophila TAGteam has recently been identified in the yellow fever mosquito, which is also capable of promoting transcription from a heterologous promoter before onset of ZGA [91].
Association of TFs with target DNA motifs may also define the timing of transcriptional onset of the target genes. For instance, during organogenesis, activation of genes involved in pharyngeal development is regulated by PHA-4, the C. elegans homolog of the pioneer TF FOXA, and depends on PHA-4 affinity for its target sites [92]. Genes with motifs containing the strongest similarity to the binding consensus sequence for PHA-4 have been found to be expressed early in development, whereas genes with motifs of weaker similarity are induced later [92]. This suggests that affinity for DNA is a primary determinant of PHA-4 binding. Thus, highly conserved DNA motifs may “encode” timing information for hierarchical gene activation during development.
Keeping chromatin in a transcriptionally primed yet inactive state
Essential for timely initiation of transcription during development and at the time of ZGA in particular is the maintenance of genes in a transcriptionally inactive, yet “primed” state, for activation. This must occur despite the pre-loading of these genes with TFs and, for a small subset, RNAPII [10]. In addition to the “poising” thought to be elicited by combinations of histone marks (e.g., H3K4me3 and H3K27me3) as mentioned earlier, several processes may be involved in the regulation of this transcriptional stand-by.
One possible mechanism is that DNA binding proteins, in the absence of a proper transcriptional co-activator, act as transcriptional repressors. For example, without β-catenin, TCFs act as repressors by forming a complex with the Groucho/TLE repressor, which in turns recruits histone deacetylases and the C-terminal binding co-repressor protein CtBP [93, 94]. A second mechanism is the silencing of transcriptional activators by binding to a repressor. For instance, Reptin and Fhit associate with the TCF/β-catenin complex and repress β-catenin-mediated transcription [95, 96]. A third mechanism may involve binding kinetics and residence time of transcriptional co-activators on chromatin. Transcription output elicited by the Rap1 TF in Saccharomyces cerevisiae depends of long-lasting residence of Rap1 on target promoters and correlates with nucleosomal depletion at Rap1 binding sites [97]. In contrast, fast Rap1 binding turnover is associated with reduced transcriptional output. Thus, the residence time of a TF at its binding site may not necessarily be compatible with transcription initiation. In a developmental context, overall chromatin de-compaction occurring in the pre-MZT period [98, 99] may facilitate long-term occupancy of promoters by TFs.
A model of sequential chromatin-associated events leading to ZGA
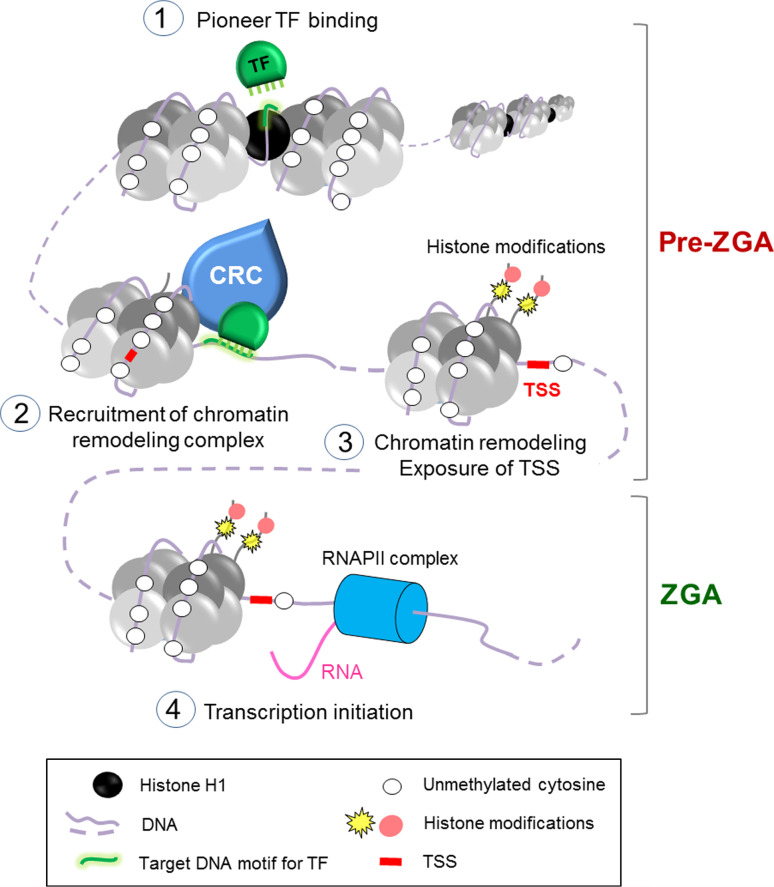
A current view of chromatin-linked processes leading to initiation of transcription at the onset of ZGA is illustrated in Fig. 3. As highlighted in this review, data from a range of vertebrate and invertebrate species indicate that spatial and temporal control of gene expression during early development requires a tight coordination between many factors acting at several regulatory levels. Each level encodes a “layer” of information, being a DNA sequence motif, a DNA methylation state, a set of post-translationally modified histones, or binding of a transcription factor (Fig. 3). These elements need to communicate to pattern the genome for onset of ZGA.
Fig. 3.
Unifying model of sequential chromatin remodeling events preceding ZGA during early development. (1) Pioneer transcription factors (TF) can recognize a DNA sequence motif (green mark) and interfere with linker histone H1 (black). (2) DNA-bound pioneer TFs recruit chromatin remodeling complexes (CRC) that (3) remodel surrounding nucleosomes. This includes a repositioning of nucleosomes, post-translational modifications of histones, and leads to exposure of the transcription start site of the target gene (TSS, red mark). DNA methylation states on the promoters of developmentally regulated genes do not significantly change during the pre-ZGA to ZGA transition; these promoters are largely unmethylated, enabling a transcriptionally permissive state. (4) This remodeling is permissive for recruitment of the RNAPII complex, and transcription initiation at the time of ZGA. Dashed line represents a developmental time interval between two chromatin states
Prior to ZGA onset, DNA motifs within the regulatory sequences of developmentally controlled genes are recognized by pioneer TFs, which are believed to confer transcriptional competence of their target genes (Fig. 3, step 1). Thus, DNA sequence appears as a key determinant of early steps in the initiation of ZGA. TF binding interferes with the linker histone H1 and mediates recruitment of chromatin remodeling complexes (step 2). These in turn remodel surrounding nucleosomes through nucleosome repositioning and changes in post-translational histone modifications, leading to uncovering of the TSS of the target gene (step 3). These events take place on an unmethylated DNA background, whose state does not vary during the pre-ZGA to ZGA transition during the MZT. These remodeling events together create a transcriptionally permissive chromatin state for the recruitment of RNAPII and subsequent ZGA onset (step 4).
Concluding remarks
A critical question in developmental and stem cell biology is how lineage-specificity of TF binding is achieved. Binding site accessibility, modulated by chromatin structure, emerges as a major determinant of binding of key lineage-specific TFs in neurogenic and myogenic cells [100]. Extending a model recently proposed for NEUROD2 and MYOD [100], one may envisage that multiple TFs are likely to be active in multiple cell types in the developing embryo; yet the cell type may determine the target genes that will be activated. In addition to a specific DNA sequence that may determine TF binding regardless of cell type, a key determinant of cell type-specific TF binding may therefore be chromatin accessibility dictated by pre-established chromatin states. The race is on for the identification of a hierarchy in the molecular determinants of transcriptional gene regulation and cell fate.
Acknowledgments
The authors wish to acknowledge their sources of funding: the Carlsberg Foundation (O.Ø.), and The Research Council of Norway, The Norwegian Cancer Society, South East Health Norway and the University of Oslo (P.C.).
Abbreviations
- ac
Acetylated
- DNMT
DNA methyltransferase
- ES
Embryonic stem
- me
Methylated
- MZT
Maternal zygotic transition
- PRC
Polycomb repressor complex
- RNAPII
RNA polymerase II
- TCF
T cell factor
- TF
Transcription factor
- TSS
Transcription start site
- ZGA
Zygotic genome activation
References
- 1.Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- 2.Walser CB, Lipshitz HD. Transcript clearance during the maternal-to-zygotic transition. Curr Opin Genet Dev. 2011;21:431–443. doi: 10.1016/j.gde.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 5.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Lindeman LC, Reiner AH, Mathavan S, Alestrom P, Collas P. Tiling histone H3 lysine 4 and 27 methylation in zebrafish using high-density microarrays. PLoS ONE. 2010;5:e15651. doi: 10.1371/journal.pone.0015651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 10.Lindeman LC, Andersen IS, Reiner AH, Li N, Aanes H, Østrup O, Winata CL, Mathavan S, Müller F, Aleström P, Collas P. Pre-patterning of developmental gene expression by modified histones before zygotic genome activation. Dev Cell. 2011;21:993–1004. doi: 10.1016/j.devcel.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Sørensen AL, Jacobsen BM, Reiner AH, Andersen IS, Collas P. Promoter DNA methylation patterns of differentiated cells are largely programmed at the progenitor stage. Mol Biol Cell. 2010;21:2066–2077. doi: 10.1091/mbc.E10-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicklay JJ, Shechter D, Chitta RK, Garcia BA, Shabanowitz J, Allis CD, Hunt DF. Analysis of histones in Xenopus laevis. II. Mass spectrometry reveals an index of cell type-specific modifications on H3 and H4. J Biol Chem. 2009;284:1075–1085. doi: 10.1074/jbc.M807274200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shechter D, Nicklay JJ, Chitta RK, Shabanowitz J, Hunt DF, Allis CD. Analysis of histones in Xenopus laevis. I. A distinct index of enriched variants and modifications exists in each cell type and is remodeled during developmental transitions. J Biol Chem. 2009;284:1064–1074. doi: 10.1074/jbc.M807273200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu SF, Zhang H, Cairns BR. Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 2011;21:578–589. doi: 10.1101/gr.113167.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, Platts AE, Saida M, Steger K, Tedder P, Miller D. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 2009;19:1338–1349. doi: 10.1101/gr.094953.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters AH. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- 21.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, Thorne NP, Backdahl L, Herberth M, Howe KL, Jackson DK, Miretti MM, Marioni JC, Birney E, Hubbard TJ, Durbin R, Tavare S, Beck S. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farthing CR, Ficz G, Ng RK, Chan CF, Andrews S, Dean W, Hemberger M, Reik W. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arney KL, Bao S, Bannister AJ, Kouzarides T, Surani MA. Histone methylation defines epigenetic asymmetry in the mouse zygote. Int J Dev Biol. 2002;46:317–320. [PubMed] [Google Scholar]
- 24.van der Heijden GW, Dieker JW, Derijck AA, Muller S, Berden JH, Braat DD, van der Vlag J, de Boer P. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech Dev. 2005;122:1008–1022. doi: 10.1016/j.mod.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 25.van der Heijden GW, Derijck AA, Ramos L, Giele M, van der Vlag J, de Boer P. Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Dev Biol. 2006;298:458–469. doi: 10.1016/j.ydbio.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 26.van der Heijden GW, Ramos L, Baart EB, van den Berg IM, Derijck AA, van der Vlag J, Martini E, de Boer P. Sperm-derived histones contribute to zygotic chromatin in humans. BMC Dev Biol. 2008;8:34. doi: 10.1186/1471-213X-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eberharter A, Becker PB. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 2002;3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiekowski M, Miranda M, Depamphilis ML. Requirements for promoter activity in mouse oocytes and embryos distinguish paternal pronuclei from maternal and zygotic nuclei. Dev Biol. 1993;159:366–378. doi: 10.1006/dbio.1993.1248. [DOI] [PubMed] [Google Scholar]
- 29.Puschendorf M, Terranova R, Boutsma E, Mao X, Isono K, Brykczynska U, Kolb C, Otte AP, Koseki H, Orkin SH, van Lohuizen M, Peters AH. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet. 2008;40:411–420. doi: 10.1038/ng.99. [DOI] [PubMed] [Google Scholar]
- 30.Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464:922–926. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindeman LC, Winata CL, Aanes H, Mathavan S, Alestrom P, Collas P. Chromatin states of developmentally-regulated genes revealed by DNA and histone methylation patterns in zebrafish embryos. Int J Dev Biol. 2010;54:803–813. doi: 10.1387/ijdb.103081ll. [DOI] [PubMed] [Google Scholar]
- 32.Aday AW, Zhu LJ, Lakshmanan A, Wang J, Lawson ND. Identification of cis regulatory features in the embryonic zebrafish genome through large-scale profiling of H3K4me1 and H3K4me3 binding sites. Dev Biol. 2011;357:450–462. doi: 10.1016/j.ydbio.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Francoijs KJ, Stunnenberg HG, Veenstra GJ. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell. 2009;17:425–434. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G. Functional anatomy of Polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 2009;7:e13. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EE. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 36.Vastenhouw NL, Schier AF. Bivalent histone modifications in early embryogenesis. Curr Opin Cell Biol. 2012;24:374–386. doi: 10.1016/j.ceb.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aanes H, Winata CL, Lin CH, Chen JP, Srinivasan KG, Lee SG, Lim AY, Hajan HS, Collas P, Bourque G, Gong Z, Korzh V, Aleström P, Mathavan S. Zebrafish mRNA sequencing decifers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res. 2011;21:1328–1338. doi: 10.1101/gr.116012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toyama R, Rebbert ML, Dey A, Ozato K, Dawid IB. Brd4 associates with mitotic chromosomes throughout early zebrafish embryogenesis. Dev Dyn. 2008;237:1636–1644. doi: 10.1002/dvdy.21576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci USA. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 41.Albert M, Peters AH. Genetic and epigenetic control of early mouse development. Curr Opin Genet Dev. 2009;19:113–121. doi: 10.1016/j.gde.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194–195. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakanishi MO, Hayakawa K, Nakabayashi K, Hata K, Shiota K, Tanaka S. Trophoblast-specific DNA methylation occurs after the segregation of the trophectoderm and inner cell mass in the mouse periimplantation embryo. Epigenetics. 2012;7:173–182. doi: 10.4161/epi.7.2.18962. [DOI] [PubMed] [Google Scholar]
- 46.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 47.Stancheva I, El-Maarri O, Walter J, Niveleau A, Meehan RR. DNA methylation at promoter regions regulates the timing of gene activation in Xenopus laevis embryos. Dev Biol. 2002;243:155–165. doi: 10.1006/dbio.2001.0560. [DOI] [PubMed] [Google Scholar]
- 48.Stancheva I, Meehan RR. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes Dev. 2000;14:313–327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- 50.Stancheva I, Hensey C, Meehan RR. Loss of the maintenance methyltransferase, xDnmt1, induces apoptosis in Xenopus embryos. EMBO J. 2001;20:1963–1973. doi: 10.1093/emboj/20.8.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, Jaenisch R. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 52.Rai K, Nadauld LD, Chidester S, Manos EJ, James SR, Karpf AR, Cairns BR, Jones DA. Zebra fish Dnmt1 and Suv39h1 regulate organ-specific terminal differentiation during development. Mol Cell Biol. 2006;26:7077–7085. doi: 10.1128/MCB.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dunican DS, Ruzov A, Hackett JA, Meehan RR. xDnmt1 regulates transcriptional silencing in pre-MBT Xenopus embryos independently of its catalytic function. Development. 2008;135:1295–1302. doi: 10.1242/dev.016402. [DOI] [PubMed] [Google Scholar]
- 54.Bogdanovic O, Long SW, van Heeringen SJ, Brinkman AB, Gomez-Skarmeta JL, Stunnenberg HG, Jones PL, Veenstra GJ. Temporal uncoupling of the DNA methylome and transcriptional repression during embryogenesis. Genome Res. 2011;21:1313–1327. doi: 10.1101/gr.114843.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 56.Andersen IS, Reiner AH, Aanes H, Aleström P, Collas P. Developmental features of DNA methylation during activation of the embryonic zebrafish genome. Genome Biol. 2012;13:R65. doi: 10.1186/gb-2012-13-7-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruzov A, Savitskaya E, Hackett JA, Reddington JP, Prokhortchouk A, Madej MJ, Chekanov N, Li M, Dunican DS, Prokhortchouk E, Pennings S, Meehan RR. The non-methylated DNA-binding function of Kaiso is not required in early Xenopus laevis development. Development. 2009;136:729–738. doi: 10.1242/dev.025569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruzov A, Dunican DS, Prokhortchouk A, Pennings S, Stancheva I, Prokhortchouk E, Meehan RR. Kaiso is a genome-wide repressor of transcription that is essential for amphibian development. Development. 2004;131:6185–6194. doi: 10.1242/dev.01549. [DOI] [PubMed] [Google Scholar]
- 59.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 60.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, Turner DJ, Illingworth R, Bird A. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arico JK, Katz DJ, van der Vlag J, Kelly WG. Epigenetic patterns maintained in early Caenorhabditis elegans embryos can be established by gene activity in the parental germ cells. PLoS Genet. 2011;7:1–15. doi: 10.1371/journal.pgen.1001391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mhanni AA, McGowan RA. Global changes in genomic methylation levels during early development of the zebrafish embryo. Dev Genes Evol. 2004;214:412–417. doi: 10.1007/s00427-004-0418-0. [DOI] [PubMed] [Google Scholar]
- 65.Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schubeler D. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet. 2011;43:1091–1097. doi: 10.1038/ng.946. [DOI] [PubMed] [Google Scholar]
- 66.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 67.Aravin AA, Bourc’his D. Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev. 2008;22:970–975. doi: 10.1101/gad.1669408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Branciamore S, Chen ZX, Riggs AD, Rodin SN. CpG island clusters and pro-epigenetic selection for CpGs in protein-coding exons of HOX and other transcription factors. Proc Natl Acad Sci USA. 2010;107:15485–15490. doi: 10.1073/pnas.1010506107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr AR, James KD, Turner DJ, Smith C, Harrison DJ, Andrews R, Bird AP. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 2010;6:e1001134. doi: 10.1371/journal.pgen.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abrams EW, Mullins MC. Early zebrafish development: it’s in the maternal genes. Curr Opin Genet Dev. 2009;19:396–403. doi: 10.1016/j.gde.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, Brown JB, Cherbas L, Davis CA, Dobin A, Li R, Lin W, Malone JH, Mattiuzzo NR, Miller D, Sturgill D, Tuch BB, Zaleski C, Zhang D, Blanchette M, Dudoit S, Eads B, Green RE, Hammonds A, Jiang L, Kapranov P, Langton L, Perrimon N, Sandler JE, Wan KH, Willingham A, Zhang Y, Zou Y, Andrews J, Bickel PJ, Brenner SE, Brent MR, Cherbas P, Gingeras TR, Hoskins RA, Kaufman TC, Oliver B, Celniker SE. The developmental transcriptome of Drosophila melanogaster . Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liber D, Domaschenz R, Holmqvist PH, Mazzarella L, Georgiou A, Leleu M, Fisher AG, Labosky PA, Dillon N. Epigenetic priming of a pre-B cell-specific enhancer through binding of Sox2 and Foxd3 at the ESC stage. Cell Stem Cell. 2010;7:114–126. doi: 10.1016/j.stem.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 74.Taube JH, Allton K, Duncan SA, Shen L, Barton MC. Foxa1 functions as a pioneer transcription factor at transposable elements to activate Afp during differentiation of embryonic stem cells. J Biol Chem. 2010;285:16135–16144. doi: 10.1074/jbc.M109.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zaidi SK, Young DW, Montecino M, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Bookmarking the genome: maintenance of epigenetic information. J Biol Chem. 2011;286:18355–18361. doi: 10.1074/jbc.R110.197061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blythe SA, Cha SW, Tadjuidje E, Heasman J, Klein PS. beta-Catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2. Dev Cell. 2010;19:220–231. doi: 10.1016/j.devcel.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harrison MM, Li XY, Kaplan T, Botchan MR, Eisen MB. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 2011;7:e1002266. doi: 10.1371/journal.pgen.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsurumi A, Xia F, Li J, Larson K, LaFrance R, Li WX. STAT is an essential activator of the zygotic genome in the early Drosophila embryo. PLoS Genet. 2011;7:e1002086. doi: 10.1371/journal.pgen.1002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila . Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila . Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heasman J. Patterning the Xenopus blastula. Development. 1997;124:4179–4191. doi: 10.1242/dev.124.21.4179. [DOI] [PubMed] [Google Scholar]
- 82.van Beest M, Dooijes D, van De Wetering M, Kjaerulff S, Bonvin A, Nielsen O, Clevers H. Sequence-specific high mobility group box factors recognize 10–12-base pair minor groove motifs. J Biol Chem. 2000;275:27266–27273. doi: 10.1074/jbc.M004102200. [DOI] [PubMed] [Google Scholar]
- 83.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/S0092-8674(00)81925-X. [DOI] [PubMed] [Google Scholar]
- 84.Cadigan KM. TCFs and Wnt/beta-catenin signaling: more than one way to throw the switch. Curr Top Dev Biol. 2012;98:1–34. doi: 10.1016/B978-0-12-386499-4.00001-X. [DOI] [PubMed] [Google Scholar]
- 85.Shiokawa K, Misumi Y, Tashiro K, Nakakura N, Yamana K, Oh-uchida M. Changes in the patterns of RNA synthesis in early embryogenesis of Xenopus laevis . Cell Differ Dev. 1989;28:17–25. doi: 10.1016/0922-3371(89)90019-1. [DOI] [PubMed] [Google Scholar]
- 86.Yang J, Tan C, Darken RS, Wilson PA, Klein PS. Beta-catenin/Tcf-regulated transcription prior to the midblastula transition. Development. 2002;129:5743–5752. doi: 10.1242/dev.00150. [DOI] [PubMed] [Google Scholar]
- 87.Mosimann C, Hausmann G, Basler K. Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol. 2009;10:276–286. doi: 10.1038/nrm2654. [DOI] [PubMed] [Google Scholar]
- 88.Valenta T, Hausmann G, Basler K. The many faces and functions of beta-catenin. EMBO J. 2012;31:2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schejter ED, Wieschaus E. Functional elements of the cytoskeleton in the early Drosophila embryo. Annu Rev Cell Biol. 1993;9:67–99. doi: 10.1146/annurev.cb.09.110193.000435. [DOI] [PubMed] [Google Scholar]
- 90.ten Bosch JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006;133:1967–1977. doi: 10.1242/dev.02373. [DOI] [PubMed] [Google Scholar]
- 91.Biedler JK, Hu W, Tae H, Tu Z. Identification of early zygotic genes in the yellow fever mosquito Aedes aegypti and discovery of a motif involved in early zygotic genome activation. PLoS ONE. 2012;7:e33933. doi: 10.1371/journal.pone.0033933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fakhouri TH, Stevenson J, Chisholm AD, Mango SE. Dynamic chromatin organization during foregut development mediated by the organ selector gene PHA-4/FoxA. PLoS Genet. 2010;6:e1001060. doi: 10.1371/journal.pgen.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell. 2007;28:291–303. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bhambhani C, Chang JL, Akey DL, Cadigan KM. The oligomeric state of CtBP determines its role as a transcriptional co-activator and co-repressor of Wingless targets. EMBO J. 2011;30:2031–2043. doi: 10.1038/emboj.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weiske J, Albring KF, Huber O. The tumor suppressor Fhit acts as a repressor of beta-catenin transcriptional activity. Proc Natl Acad Sci USA. 2007;104:20344–20349. doi: 10.1073/pnas.0703664105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bauer A, Chauvet S, Huber O, Usseglio F, Rothbacher U, Aragnol D, Kemler R, Pradel J. Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. EMBO J. 2000;19:6121–6130. doi: 10.1093/emboj/19.22.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature. 2012;484:251–255. doi: 10.1038/nature10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Deshmukh RS, Ostrup O, Strejcek F, Vejlsted M, Lucas-Hahn A, Petersen B, Li J, Callesen H, Niemann H, Hyttel P. Early aberrations in chromatin dynamics in embryos produced under in vitro conditions. Cell Reprogram. 2012;14:225–234. doi: 10.1089/cell.2011.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ahmed S, Brickner JH. A role for DNA sequence in controlling the spatial organization of the genome. Nucleus. 2010;1:402–406. doi: 10.4161/nucl.1.5.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fong AP, Yao Z, Zhong JW, Cao Y, Ruzzo WL, Gentleman RC, Tapscott SJ. Genetic and epigenetic determinants of neurogenesis and myogenesis. Dev Cell. 2012;22:721–735. doi: 10.1016/j.devcel.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]