Abstract
KIF1Bβ is a kinesin-like, microtubule-based molecular motor protein involved in anterograde axonal vesicular transport in vertebrate and invertebrate neurons. Certain KIF1Bβ isoforms have been implicated in different forms of human neurodegenerative disease, with characterization of their functional integration and regulation in the context of synaptic signaling still ongoing. Here, we characterize human KIF1Bβ (isoform NM015074), whose expression we show to be developmentally regulated and elevated in cortical areas of the CNS (including the motor cortex), in the hippocampus, and in spinal motor neurons. KIF1Bβ localizes to the cell body, axon, and dendrites, overlapping with synaptic-vesicle and postsynaptic-density structures. Correspondingly, in purified cortical synaptoneurosomes, KIF1Bβ is enriched in both pre- and postsynaptic structures, forming detergent-resistant complexes. Interestingly, KIF1Bβ forms RNA–protein complexes, containing the dendritically localized Arc and Calmodulin mRNAs, proteins previously shown to be part of RNA transport granules such as Purα, FMRP and FXR2P, and motor protein KIF3A, as well as Calmodulin. The interaction between KIF1Bβ and Calmodulin is Ca+2-dependent and takes place through a domain mapped at the carboxy-terminal tail of the motor. Live imaging of cortical neurons reveals active movement by KIF1Bβ at dendritic processes, suggesting that it mediates the transport of dendritically localized mRNAs. Finally, we show that synaptic recruitment of KIF1Bβ is activity-dependent and increased by stimulation of metabotropic or ionotropic glutamate receptors. The activity-dependent synaptic recruitment of KIF1Bβ, its interaction with Ca2+ sensor Calmodulin, and its new role as a dendritic motor of ribonucleoprotein complexes provide a novel basis for understanding the concerted co-ordination of motor protein mobilization and synaptic signaling pathways.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-012-1108-0) contains supplementary material, which is available to authorized users.
Keywords: Molecular motors, mRNA transport, Calmodulin, Arc, Synaptic activity, mGluR activation
Introduction
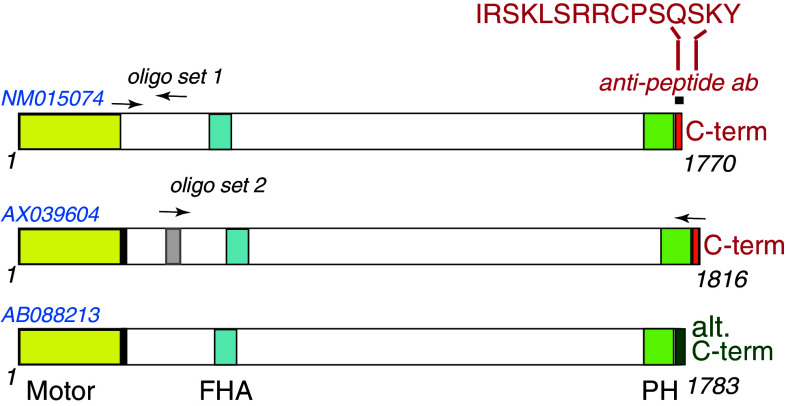
The members of the KIF1B subfamily (Kinesin-3 as per the nomenclature of [1]) of motor proteins have attracted considerable attention because of their important function in anterograde axonal transport in neurons and their putative link to neurodegenerative disease. In human, KIF1B is expressed in two isoform types, KIF1Bα, originally reported as a mitochondrial transporter [2], and also shown to interact with postsynaptic density and synaptic scaffold proteins [3], and at least six, currently known, KIF1Bβ-like forms (NM015074, AB088210–13, and AX039604). The AB088210–13 isoforms share a high level of sequence identity with each other, with small differences over a region of 40 amino acids (aa), and compared to NM015074 have an extra stretch of six aa and, significantly, an “alternative C-terminus” (Fig. 1). AX039604 shares characteristics of both, the AB series and NM015074, being largely identical to AB088210, but with an additional 40-aa stretch and possessing the characteristic C-terminus of NM015074 (Fig. 1).
Fig. 1.
Human KIF1Bβ isoforms. Schematic comparison of isoforms NM015074, AX039604 and AB088213 [the latter being representative of the AB088210–13 subgroup (AB)]. NM015074 (NM) and AX039604 (AX) share a characteristic C-terminus (red) to which an anti-peptide antibody was raised (indicated at the top). The AB series have an alternative C-terminus (denoted by the dark green box) not recognized by the antibody. AX and AB have an additional 6-aa stretch (black box) close to the motor domain, and AX also has a unique additional 40-aa stretch (gray box). The positions of the two sets of oligonucleotides that are each specific to NM or AX only, and can thus discriminate between NM and AX by PCR (Fig. 2b, d–f), are indicated by arrows over the areas to which they hybridize. The sequences of these oligonucleotide pairs are given in Online Resource Table S1. The motor (yellow), FHA (light blue), and PH domains (light green), as well as the two stretches of amino acids (black and gray boxes), unique in certain isoforms, are highlighted. Numbers denote aa positions
The KIF1B family has been linked to several diseases affecting neural cells of the central nervous system. KIF1Bβ (isoform NM015074) is reported as a transporter of synaptic vesicle precursors and, in the case of a single large Japanese family, was implicated in Charcot-Marie-Tooth type 2A (CMT2A) motor neuron disease [4], a disorder commonly associated with mutations in the mitochondrial GTPase mitofusin-2 [5]. KIF1Bβ isoform AB088210, highly enriched in the motor cortex, is significantly downregulated in the motor cortex of patients with sporadic amyotrophic lateral sclerosis, another common type of motor neuron disease [6]. Mechanistic insights into the involvement of KIF1B family members in neuropathies also come from C. elegans, whose KIF1Bβ homologue, UNC-104, is required for the anterograde transport of synaptic vesicle precursors [7], possibly through interactions with the cargo via a lipid-binding pleckstrin homology domain in its stalk [8]. In a genome-wide association study, a single nucleotide polymorphism (SNP) in intron 5 of the KIF1B gene (shared by KIF1Bα and KIF1Bβ) was found to be associated with susceptibility to multiple sclerosis (MS) ([9], but see also [10, 11]). Intriguingly, a tentative explanation for this link is provided by observations in zebrafish, where mutations or RNA interference of KIF1Bα/β prevent correct localization of mRNAs encoding myelin proteins MBP and 36 K to distal processes in myelinating oligodendrocytes, and cause reduction of axonal outgrowth and reduced or ectopic myelination in spinal neurons [12].
Other studies have indicated a wider role for KIF1Bβ in non-neural cell types, such as reports proposing a haploinsufficient tumor suppressor function [13–15]. This link of the KIF1B family to cancer development is further strengthened by a genome-wide association study that assigned high statistical significance to an intronic SNP in the KIF1B gene as a susceptibility locus for hepatocellular carcinoma in carriers of chronic hepatitis B [16].
The existence of multiple isoforms, necessitating isoform-specific characterization of gene expression and function, and use of isoform-specific antibodies, complicate the immediate correlation between the functional analyses offered by different studies. More importantly, despite the established involvement of KIF1Bβ in vesicular transport, less is known about the signaling pathways and mechanisms regulating and modulating the function of KIF1Bβ in neurons. In this work, we identify regulatory signals, including calcium and receptor signaling, that influence the recruitment of KIF1Bβ isoform NM015074 at functional synapses, and identified a KIF1Bβ-containing RNA–protein transport complex. The present study therefore gives fundamental insights into how mobilization and interaction of motor proteins are integrated with synaptic signaling and plasticity.
Materials and methods
Cells
Spinal motor neurons and cortical neurons were obtained from E18–19 mice. Dissected spinal cords or cortex were incubated with 4.5 mL HBSS, containing 0.02 % w/v trypsin and 80 μg/mL DNase I for 15 min at 37 °C, mechanically dissociated and centrifuged at 1,500g for 5 min. Dissociated cells were plated at a density of 12 × 104 cells/cm2 in MEM (Invitrogen), supplemented with 10 % v/v horse serum (for spinal cord), or in Neurobasal A media (Invitrogen), supplemented with B27. Primary neuron cultures were maintained 2–3 weeks in vitro before use.
Mouse NSC-34 neuroblastoma, NIH 3T3 fibroblast cell lines, human SH-SY5Y neuroblastoma and HeLa cells were cultured in DMEM (Gibco/BRL), containing 10 % v/v fetal calf serum (FCS), 2 mM glutamine, 50 U/mL of penicillin/streptomycin, and maintained at 37 °C in 5 % CO2. For induction and differentiation, SH-SY5Y cells were split and maintained for 10–12 days in DMEM with 2 % FCS and 10 μM retinoic acid.
Oligonucleotides and generation of plasmid constructs
All oligonucleotides, used for RT-PCR or plasmid construct generation, were synthesized by MWG Biotech (Germany). Their sequences and further details are given in Online Resource Table S1.
Antibodies
A rabbit polyclonal antibody to synthetic peptide IRSKLSRRCPSQSKY [aa residues 1,756–1,770 in human KIF1Bβ (NM015074)] was custom-made and affinity-purified by Peptide Speciality Laboratories, Germany. It was used at a dilution of 1:500 for immunoblotting and 1:300 for immunofluorescence (IF).
The following primary antibodies were used: mouse anti-dynein (intermediate chain, 74 kDa) monoclonal antibody (mab sc-13524) (Santa Cruz, 1:800 in immunoblot), mouse anti-GST mab (Sigma, 1:4,000 in immunoblot), rat anti-myelin basic protein (MBP) mab (Millipore, 1:2,500 in immunoblot), rabbit anti-glial fibrillary acidic protein (GFAP) polyclonal antibody (Sigma, 1:500 in immunoblot), mouse anti-non-phosphorylated neurofilament H mab (SMI 32, Eurogentech, 1:1,000 in IF), mouse anti-MAP2 mab (Abcam, 1:200 in IF), mouse anti-MAP2 mab (Sigma, 1:300 in IF), mouse anti-GAPDH mab (Chemicon, 1:20,000 in immunoblot), mouse anti-PSD-95 mab (BD Biosciences, 1:100 in IF and 1:1,000 in immunoblot), mouse anti-synaptophysin mab (Abcam, 1:100 in IF and 1:1,000 in immunoblot), mouse anti-SV2 mab (Developmental Studies Hybridoma Bank, 1:100 in IF), mouse anti-syntaxin 1 mab (Synaptic Systems, 1:5,000), mouse anti-SNAP25 mab (Synaptic Systems, 1:1,000), mouse anti-CaM mab (Zymed, 1:30 in IF), rabbit anti-CaM (Abcam, 1:1,000 in immunoblot), rabbit anti-KIF3A (Abcam, 1:1,000 in immunoblot), goat anti-MARCKS (Santa Cruz Biotechnology, 1:400 in immunoblot), rabbit anti-Purα (Millipore, 1:100 for IF, 1:250 in immunoblot), mouse anti-FMRP (IC3, Millipore, 1:100 in IF), rabbit anti-FMRP (rAMII, 1:2,000 in immunoblot) [17], mouse anti-FXR2P (1:1,000 in immunoblot), rabbit anti-FXR2P (Developmental Studies Hybridoma Bank, 1:50 in IF), rabbit anti-KIF5A (Acris, 1:1,000 in immunoblot), and rabbit anti-ELAVs (Santa Cruz, 1:1,000 in immunoblot).
The following secondary antibodies (Molecular Probes) were used for IF: Alexa Fluor (AF) 488 chicken anti-rabbit IgG (1:700), AF568 goat anti-rabbit IgG (1:2,000), AF488 goat anti-mouse IgG (1:700) and AF568 anti-mouse IgG1 (1:2,000). The following secondary antibodies were used for immunobloting: HRP donkey anti-rabbit IgG (Santa Cruz, 1:60,000), HRP sheep anti-mouse IgG (Amersham, 1:8,000) and HRP chicken anti-rat IgG (Santa Cruz, 1:20,000).
Nuclei were stained with Hoechst 33342 (0.5 μg/mL, Invitrogen). Polymerized filamentous actin (F-actin) was visualized at postsynaptic densities in mature dendritic spines of cultured neurons using a phalloidin-AF647 conjugate (1:100, Molecular Probes) with 1 h incubation, followed by three 5-min washes in PBS.
RNA extraction, reverse transcription semi-quantitative PCR (RT-PCR)
For RT-PCR, poly(A) RNA was extracted from human cells (HeLa, SH-SY5Y uninduced or induced with RA) or mouse cells (spinal cord neurons of 20 days in vitro, NSC-34, NIH 3T3) or mouse tissues (cortex, cerebellum, hippocampus from 3-month-old mice) with the RNeasy Purification Kit (Qiagen) and 1 μg used for reverse transcription using the Protoscript Reverse Transcription Kit with a dT23VN primer (New England Biolabs). Human tissue RNA (from the tissues displayed in Fig. 2a, b) was purchased from Ambion Inc. and reverse-transcribed to cDNA in the same manner. RNA from total mouse cortex was extracted with the Trizol reagent (Invitrogen) and digested with RNase-free DNAse (RQ1, Promega). Additionally, mouse RNA from hippocampal neurons in vitro at stages 2 and 5 (staging as per [18]), embryonic stages E13 and E18, juvenile animals (2.5 weeks) and pure astrocytic cultures, derived from newborn mice (Fig. 2d), was obtained as described by [19]. Semi-quantitative PCR reactions from cells, tissues or cell lines (Fig. 2) were set up with equivalent cDNA concentrations (starting with the same amount of RNA) and using cDNA of ribosomal protein L19 as a calibrator for normalization.
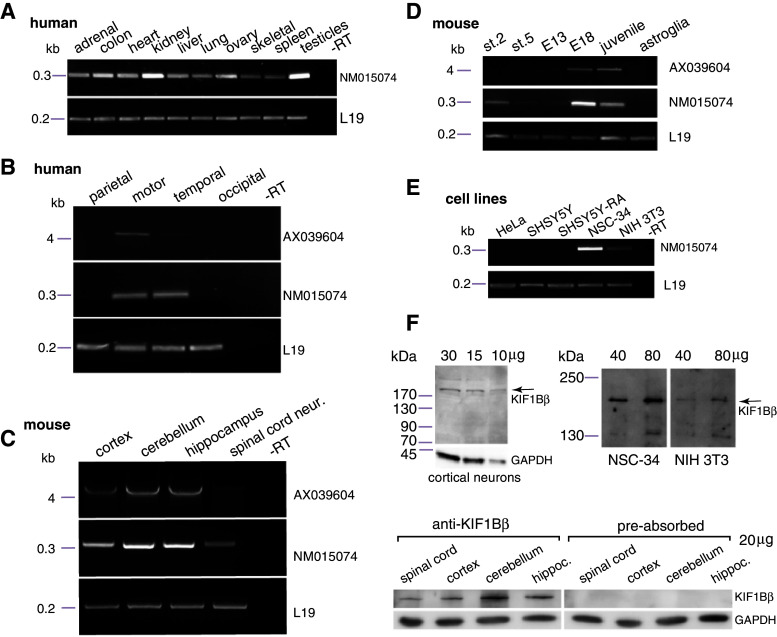
Fig. 2.
a KIF1Bβ (NM015074) mRNA expression pattern in human tissues. Equivalent RT-PCR reactions from various human tissues, including a mock reverse-transcription reaction as negative control (−RT). Equivalent reactions for the mRNA of ribosomal protein L19 were used as internal standards (bottom panel). b–d mRNA expression of KIF1Bβ NM015074 versus AX039604 isoforms in the CNS. Equivalent RT-PCR reactions specific to KIF1Bβ isoforms NM015074 (top panels) and AX039604 (central panels), and housekeeping gene L19 (bottom panels) for b areas of human cortex; c mouse neural tissue, including spinal motor neurons; d mouse stage-2 or stage-5 hippocampal neurons in vitro [18] or the hippocampus of E13, E18 and juvenile mice, and from proliferating astroglial cultures. e KIF1Bβ (NM015074) mRNA expression in human and mouse cell lines. Equivalent RT-PCR reactions reveal a pronounced expression level of KIF1Bβ mRNA in the mouse neuroblastoma NSC-34 line, lower expression levels in mouse NIH 3T3 fibroblasts, and expression below detection levels for other cell lines. Equivalent reactions for L19 (bottom panel). f Protein expression of KIF1Bβ in different brain areas by immunoblot (top panels). Total protein extracts from mouse cultured primary cortical neurons, NSC-34 or NIH 3T3 cells were analyzed by western blotting, using the anti-peptide KIF1Bβ antibody. A signal, consistent with the predicted M r of 199.01 × 103 for KIF1Bβ was detected (e). Also consistent with mRNA level for these cell lines, protein expression of KIF1Bβ in NSC-34 is higher than in NIH 3T3 cells (bottom panels). Expression of KIF1Bβ protein in equivalent samples from different parts of the mouse CNS (left); these signals are abolished if the antibody is pre-absorbed with the antigenic peptide (right). Immunoreactivity for GAPDH serves as internal loading control
siRNA-mediated silencing
Catalogue and custom-made Stealth siRNA duplexes (Invitrogen) were used for siRNA-mediated silencing and scrambled siRNA ‘Low GC’ (a universal control for siRNAs with low GC content; Invitrogen) was used as negative control.
Three pairs of custom-made siRNA duplexes were used for silencing of KIF1Bβ NM015074).
5′gaguguuucacuuucuggugauaaa3′/3′cucgcaaagugaaagaccacuauuu5′ (siRNA1)
5′ccuugugauuguuacagcucccuuu3′/3′ggaacacuaacaaugucgagggaaa5′ (siRNA2)
5′ugagguggaugaagcugcaguugau3′/3′acuccaccuacuucgacgucaacua5′ (siRNA3)
Transfections of NSC-34 cells with siRNAs were performed using Lipofectamine 2000 (Invitrogen) with the three sets of siRNAs used as a cocktail (molar ratios 1:1:1) at a total concentration of 320 pM/transfection. A second transfection was repeated 72 h after initial treatment. At 120 h, coverslips or cells were harvested for microscopic imaging or RNA and protein analysis.
Immunoprecipitation and RT-PCR
Mouse brains (p21) were lysed in 150 mM NaCl, 50 mM Tris–HCl pH 7.4, 1 % v/v Triton X-100, 40 U/mL RNase OUT (Invitrogen), 1 mM DTT, 5 mM β-glycerol phosphate, 0.5 mM Na3VO4, and 10 mg/mL PIC (Sigma). After 5 min incubation on ice, lysates were centrifuged for 8 min at 12,000g (4 °C), and 500 μg of protein from the supernatant were used for the IP. Protein A Sepharose beads (GE Healthcare) were saturated in 10 % w/v BSA and heparin (2 mg/mL) in PBS, incubated with 3 μg of native KIF1Bβ antibody or purified rabbit IgGs (2 h at 4 °C), washed three times (150 mM NaCl, 50 mM Tris–HCl pH 7.40, 1 % v/v Triton X-100), and finally incubated with 500 μg of brain extract plus heparin (1 mg/mL) for 1 h at 4 °C. Stringent washes with 1 M urea and low salt (50 mM NaCl) followed the incubation. Proteins were eluted by boiling of the beads in SDS-PAGE sample buffer, separated by electrophoresis and immunoblotted to a PVDF membrane (Millipore). RNA was eluted from the immunoprecipitates, extracted (phenol/chloroform), and precipitated in 2.5 volumes ethanol. Prior to RNA extraction, and as an internal control for equal RNA extraction from the test and negative control immunoprecipitations, each immunoprecipitate was spiked with 200 ng of human BC200 RNA. First-strand cDNA synthesis ensued, using p(dN)6 and 100 U of M-MuLV RTase (Invitrogen), and was followed by PCR with GoTaq (Promega) to assay for selected cDNAs and the BC200 cDNA internal control (Fig. 8).
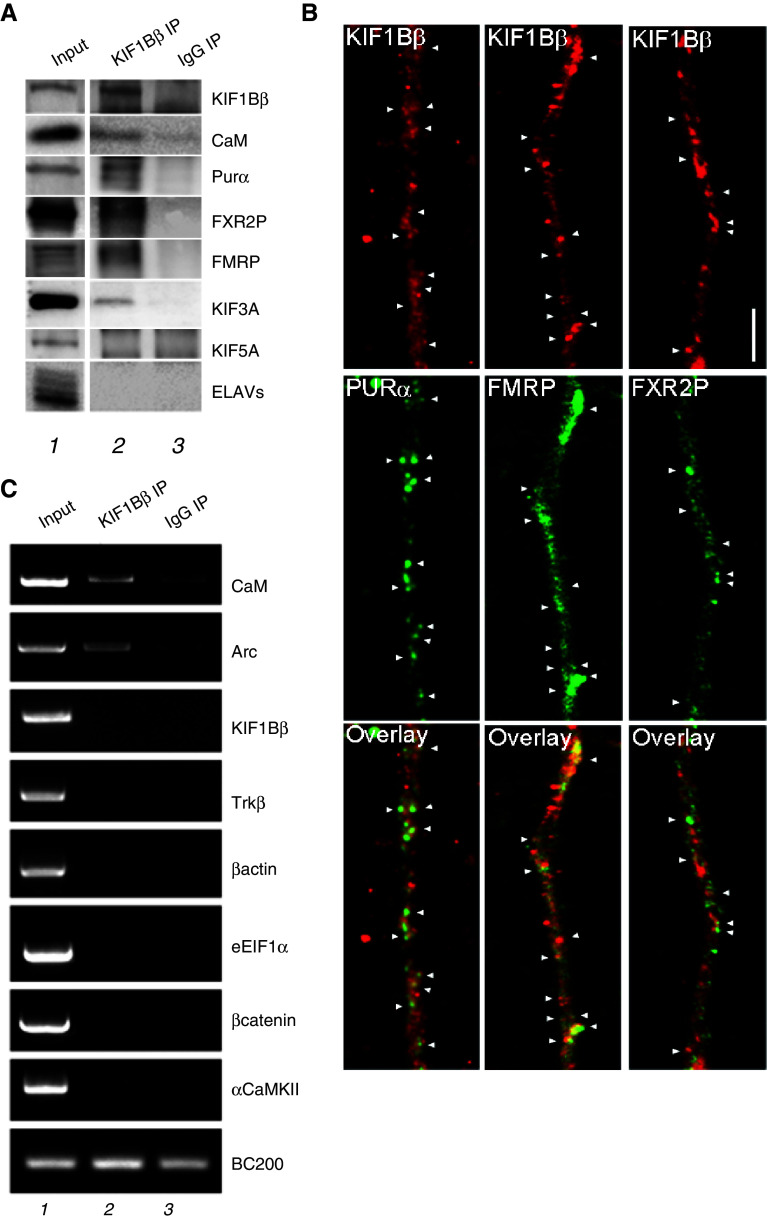
Fig. 8.
a Immunoprecipitation (IP) for KIF1Bβ from mouse brain. Analysis by western blot, using different antibodies, of the anti-KIF1Bβ-immunoprecipitated proteins revealed that KIF1Bβ and its interacting protein Calmodulin (CaM) are part of the same complex. Furthermore, the dendritically-localized RNA-binding proteins Purα, FMRP and FXR2P, as well as the motor protein KIF3A, were also detected in the same KIF1Bβ complex. RNA-binding protein ELAVs and the motor protein KIF5A were absent. Lane 1 represents 1/20 (25 μg) of the original extract used for immunoprecipitation (input), lanes 2 and 3 show the immunoprecipitated protein using, respectively, equal amounts of anti-KIF1Bβ antibody or rabbit IgGs (negative control). b Co-localization of KIF1Bβ and dendritic RNA-binding proteins in mouse cultured cortical neurons. Staining of cortical neurons (12–14 div.) with anti-KIF1Bβ (red panels) and concurrent labeling with antibodies to Purα; FMRP; FXR2P (green panels), reveal extensive pairwise co-localization in puncta (arrows) in a detail of a dendritic process. Scale bar 5 μm. c RT-PCR analysis of the RNA content of the KIF1Bβ IP material. Calmodulin, Arc, KIF1Bβ, TrkB, β-actin, eEIF1α, β-catenin, CamKIIα mRNAs were amplified, following KIF1Bβ immunoprecipitation. Prior to RNA extraction, each sample was spiked with 100 ng of exogenous human BC200 mRNA, which was then tested by PCR to show that RNA extraction, reverse transcription, and amplification efficiency was equal across samples. Lane 1 represents the RT-PCR reaction of 1/20 of the original extract used for immunoprecipitation (input), lanes 2 and 3 represent the RNA immunoprecipitated using equal amounts, respectively, of anti-KIF1Bβ antibody and rabbit IgGs. CaM and Arc mRNAs were detected in the KIF1Bβ complex
Yeast two hybrid screen and yeast mating assays
To screen for proteins that interact with KIF1Bβ, the GAL4-based Matchmaker Two Hybrid System 3 (Clontech) was used in conjunction with a human adult brain cDNA library, constructed in the DNA activation-domain-bearing pACT2 vector (Clontech). A partial cDNA of KIF1Bβ (nucl. 1,980–5,313) was cloned as an in-frame SalI fragment into DNA-binding domain fusion vector pAS2.1 and used as bait. All procedures were performed according to the Matchmaker User’s Manual. After five rounds of selection, putative positive clones were validated by yeast mating, followed by quadruple auxotrophy selection (-Leu, -Trp, -His, -Ade) and β-galactosidase filter assays, and identified by DNA sequencing (MWG Biotech, Germany).
To map the CaM interaction site(s) of KIF1Bβ, deletion constructs of KIF1Bβ (M, C1–C5) were generated in vector pAS2.1 [see Online Resource Table S1; Fig. 7c (see below)] and tested by yeast mating against full-length CaM in vector pACT2. Interaction was defined as the ability of >95 % of resulting clones, containing both constructs, to grow on quadruple auxotrophy selection and also display β-galactosidase activity upon filter assay.
Fig. 7.
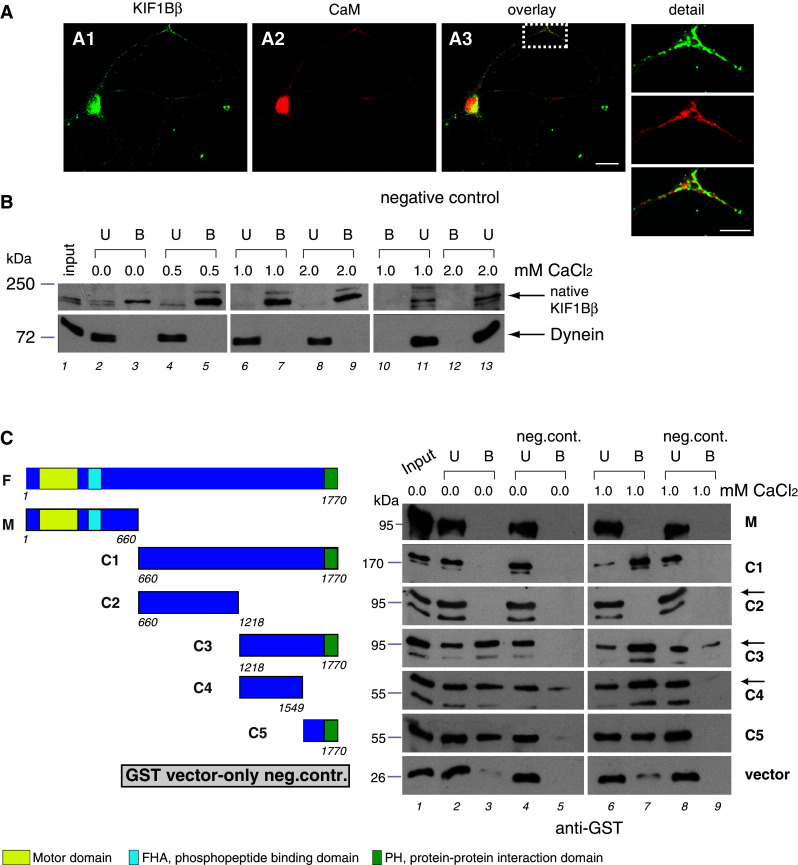
A1–A3 Partial co-localization of KIF1Bβ and CaM in spinal motor neurons. Double immunofluorescence for KIF1Bβ (green) and CaM (red) reveals their co-localization (yellow) in both the cell body and neuronal processes (detail at higher magnif.). Scale bars 20 μm, inset 10 μm. b Pull-down assays with cell extracts confirming the interaction between endogenous KIF1Bβ and CaM and demonstrating its Ca2+-dependence. CaM, immobilized on Sepharose beads, was incubated with equivalent lysates (10 μg total protein) from NSC-34 cells in the presence of increasing Ca2+concentrations. Bound (B; lanes 3, 5, 7, 9) and unbound proteins (U; lanes 2, 4, 6, 8) were probed by immunoblot with anti-KIF1Bβ (top panels). Sepharose beads without CaM were used as negative control (top panels, lanes 10–13; B and U). An anti-dynein antibody (bottom panels) was used as internal control for equal loading. Detection of endogenous KIF1Bβ is obtained only in the presence of CaM (lanes 3, 5, 7, 9) and is enhanced by increased Ca2+ concentration, peaking at about 0.5 mM CaCl2 (lane 5). c Pull-down assays with recombinant proteins confirming a direct interaction between KIF1Bβ and CaM and mapping the interaction site on KIF1Bβ. CaM on Sepharose beads was incubated with equivalent extracts of E. coli expressing GST-tagged deletion constructs of KIF1Bβ (M, C1–C5; shown in schematic form), in the absence (lanes 1–5) or presence of 1 mM Ca2+ (lanes 6–9). Bound (B) and unbound (U) proteins were probed by anti-KIF1Bβ the KIF1Bβ band is indicated by arrows, a lower band appearing in some samples is a degradation product). Sepharose-only beads served as negative control at the same conditions (lanes 4, 5, 8, 9). GST-vector-only was an additional negative control to exclude interaction of the KIF1Bβ constructs and CaM via GST sequences (vector; bottom panels). F full-length KIF1Bβ, M motor-domain-containing construct, C1–C5 non-motor-domain-containing constructs. Constructs are also described in Table 1. Numbers accompanying the construct sketches denote amino-acid residues
Bacterial expression of GST-tagged KIF1Bβ constructs and pull-down assays
For expression and purification of GST-tagged constructs of KIF1Bβ (constructs M, C1–C5 in vector pGEX-4T-1, as shown in Fig. 7c), competent E. coli of the DL21 (DE3) Lys-S strain were transformed with the plasmid construct, and single-colony 1 mL cultures, grown in LB containing 50 μg/mL ampicillin and 30 μg/mL chloramphenicol, were used (at 1:100) to inoculate 10 mL cultures. These cultures were induced at exponential growth with 0.05 mM IPTG at 20 °C and sampled 3–18 h later.
For in vitro pull-down assays, the bacterial pellet was resuspended in 1 mL lysis buffer [PBS containing 1 % v/v Tween 20 and 1 tablet/50 mL of Complete (a protease inhibitor cocktail by Roche)], lysed by ultrasonication and centrifuged at 13,000 rpm for 15 min at 4 °C. Equal parts of the soluble fraction were each mixed with 20 μL pre-equilibrated CaM-Sepharose beads (GE Healthcare) in the absence of Ca2+ (5 mM EDTA to bind all free Ca2+ ions) or at increasing concentrations of Ca2+ (0.5, 1 and 2 mM CaCl2) and incubated for 1 h at 4 °C with mild stirring. Beads were then centrifuged at 2,000 rpm for 2 min at 4 °C, the supernatant kept as the unbound fraction, and the beads (bound fraction) were then washed three times with 10 bead volumes of PBS, 1 % v/v Tween 20 and Complete. Beads and the unbound fraction were each mixed with 5× SDS-PAGE sample buffer and analyzed by electrophoresis, followed by immunoblotting to assess interactions between the different deletion constructs of KIF1Bβ and CaM. Negative control samples, to exclude non-specific binding of KIF1Bβ constructs to the beads, consisted of samples processed in an identical manner but using plain Sepharose beads, rather than CaM-Sepharose beads in the assays.
The same pull-down assay was repeated using native KIF1Bβ. Briefly, a total lysate from three 10 cm-dishes of 80 % confluent NSC-34 cells was prepared by ultrasonication in 0.5 mL of lysis buffer (150 mM NaCl, 10 mM Tris pH = 7.4, 0.5 % v/v Tween-20, 10 % v/v glycerol, 10 mM β-mercaptoethanol, and 1 tablet/50 mL of Complete protease inhibitor cocktail). The lysate was centrifuged at 13,000 rpm for 15 min at 4 °C, and the soluble fraction was mixed with 20 μL pre-equilibrated CaM-Sepharose beads and processed in exactly the same manner as for the assays with recombinant proteins, in the absence or presence of varying concentrations of Ca2+.
Preparation of purified synaptoneurosomes
Synaptoneurosomes were purified from the cortex of 8-weeks-old mice, based on the original protocol of Nagy and Delgado-Escueta [20], as used by Pilo-Boyl et al. [21] and validated by Napoli et al. [22]. Specifically, each cortex was homogenized in 10 mL homogenization buffer (0.32 M sucrose, 1 mM EDTA, 1 mg/mL BSA and 5 mM HEPES pH 7.4), centrifuged at 3,000g for 10 min at 4 °C, the supernatant re-centrifuged at 14,000 rpm for 12 min at 4 °C and the pellet resuspended in 550 μL Krebs–Ringer buffer (140 mM NaCl, 5 mM KCl, 5 mM glucose, 1 mM EDTA and 10 mM HEPES pH 7.4). To this, 450 μL of Percoll (45 % v/v) were added and mixed, and a synaptoneurosomes-enriched top layer was collected after centrifugation at 14,000 rpm for 2 min at 4 °C. The fraction was washed twice in Krebs–Ringer buffer and resuspended in 400 μL Krebs/HEPES solution (140 mM NaCl, 3 mM KCl, 1.25 mM NaH2PO4, 25 mM NaHCO3, 2 mM CaCl2 and 20 mM HEPES pH 7.4). The formulation used provides purified functional synaptoneurosomes (physiologically defined as synapses containing both presynaptic termini, with synaptic vesicles, and post-synaptic termini), as previously demonstrated [21, 22]. Where specified, fractionation of purified synaptoneurosomes into soluble extrasynaptic matrix, presynaptic matrix, and post-synaptic densities was carried out as previously described [23]. Stimulation of purified synaptoneurosomes (100 μg) was carried out in Hepes–Krebs buffer at 37 °C with a final concentration of 100 mM DHPG (S-3,5,-dihydroxyphenylglycine) for 5 min, or with glutamatergic stimulators (100 μM NMDA, or 5 μM AMPA or 100 μM glutamate, or their combinations, all for 30 min), or with PMA (phorbol 12-myristate 13-acetate; 200 μM for 30 min), or with PKC inhibitor Ro320432 (10 μM for 30 min), or with KCl (55 mM for 15 min). Following these treatments, synaptoneurosomes were placed on ice for 2 min and centrifuged at 14,000 rpm for 30 s at 4 °C. Samples of purified synaptoneurosomes or synaptoneurosome fractions (5–15 μg) were analyzed by SDS-PAGE and immunoblotting to probe for the enrichment of KIF1Bβ, using appropriate pre- and post-synaptic markers as positive controls, or non-synaptic, non-neuronal markers as negative controls (Figs. 5, 6).
Fig. 5.
a Detection of KIF1Bβ in purified mouse synaptoneurosomes. KIF1Bβ is detected in total lysates from the cortex (T; 5 μg total protein) and is enriched in a sample of synaptoneurosomes purified from the cortex (S; 5 μg). The level of detection of synaptophysin, a presynaptic marker, and PSD-95, a postsynaptic marker, are evidence of the enrichment of the synaptoneurosomal preparation, which was essentially devoid of non-neuronal proteins, such as myelin basic protein (MBP) and glial fibrillary acidic protein (GFAP). b Detection of KIF1Bβ in further fractionated synaptoneurosomes. The synaptoneurosomal preparation from mouse cortex was further fractionated into its soluble, pre- and postsynaptic components and probed for KIF1Bβ in parallel with compartment-specific protein markers PSD-95 and synaptophysin. KIF1Bβ was detected in both the pre- and postsynaptic, but not in the soluble fraction. An amount of 15 μg of protein was loaded from each fraction
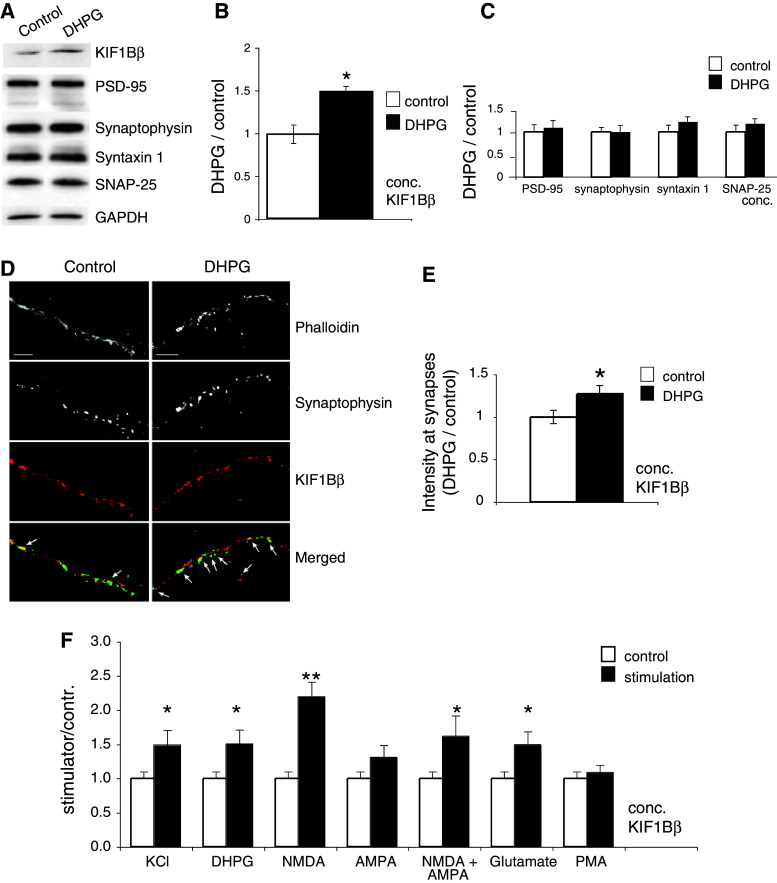
Fig. 6.
a DHPG stimulation increases the amount of KIF1Bβ in synaptoneurosomes. Immunoblot (representative of 8 independent experiments) of protein extracts of cortical synaptoneurosomes either resting (control) or treated with DHPG and probed for KIF1Bβ, PSD-95, synaptophysin, syntaxin1, SNAP-25, and also GAPDH (loading control). b Quantification of the experiments shows a significant increase of KIF1Bβ protein levels in synaptoneurosomes after DHPG stimulation (*p < 0.05; Student’s t test). c Quantification of PSD-95, synaptophysin, syntaxin1 and SNAP-25 levels in the same experiments as in (b) shows no significant change. d, e DHPG stimulation recruits KIF1Bβ to the synapses. d Cortical neurons resting (left panels) or treated with DHPG (right panels), fixed, and stained with phalloidin, anti-synaptophysin or anti-KIF1Bβ antibodies. Merged images show an overlay of the mask that marks vicinity to a synapse (green) and the KIF1Bβ signal in the control or after DHPG, respectively (red). Arrows point to strong KIF1Bβ signals in the vicinity of synapses. Scale bar 10 μm. e Quantification of KIF1Bβ fluorescence that fell into total synaptic areas, as defined by the mask described in “Materials and Methods” (n = 20 neurons; *p < 0.05; Student’s t test). f Quantification of KIF1Bβ signal from 6 independent experiments, showing increases of KIF1Bβ concentration in synaptoneurosomes after stimulation of different types of glutamate receptor (confirmation of PKC stimulation with PMA in Online Resource Fig. S4). The most pronounced increase corresponds to stimulation of the NMDA receptor (n = 20 neurons, 6 independent experiments, **p = 0.0014; Student’s t test)
Protein electrophoresis, immunobloting and quantification of protein expression
SDS-PAGE, immunobloting and ECL visualization (Amersham Pharmacia Biotech) were performed with standard methods. Protein samples for SDS-PAGE were prepared either by direct lysis of cells in 2× SDS-PAGE Laemli sample buffer or by manual homogenization of brain tissue in lysis buffer (100 mM NaCl, 10 mM MgCl2, 10 mM Tris–HCl, pH 7.5, 1 % v/v Triton X-100, 1 mM DTT, 5 mM β-glycerol phosphate, 0.5 mM sodium orthovanadate, a cocktail of protease inhibitors, and 1 U/mL RNAse inhibitor), followed by solubilization with an equal volume of 2× SDS-PAGE Laemli sample buffer. Digital images of immunoblots were obtained with a Fuji LAS-3000 mini camera.
Quantification of immunoblot signals was performed with the AIDA Image Analyzer software v.4.22. For quantification of protein expression (Fig. 6a, b, c), intensity volumes (area × height) of protein signals were first normalized to the GAPDH signal. The ratios of the average of these values from six independent experiments to the average of the control signal, including the standard deviation (STD), were then calculated and plotted. Statistical significance values were assessed by two-sample Student’s t test and assigned as significant with p < 0.05 (*), or very significant with p < 0.01 (**).
Immunolabeling, transient transfections, and quantification of fluorescence images at dendritic processes
Immunolabeling of cultured primary neurons was performed as per [24]. NSC-34 cells were fixed/permeabilized in methanol and processed for immunolabeling as per [25].
NSC-34 cells were transfected with the pEYFP-KIF1Bβ expression vector using Lipofectamine 2000 (Invitrogen) with a transfection efficiency of 30–40 %. Cells were retrieved for immunofluorescence 24–48 h post-transfection.
For the analysis of KIF1Bβ in functional synapses on dendritic processes, we performed triple staining with an anti-synaptophysin antibody (which marks presynaptic sites), with phalloidin (which highlights points of F-actin enrichment in the dendrites and postsynaptic sites), and with the KIF1Bβ antibody (Fig. 6d, e). To assess the association of KIF1Bβ first with the functional synapses, we further developed an approach described earlier [24]. In short, a dedicated algorithm was applied using ImageJ and the JaCoP plugin [26; http://rsb.info.nih.gov/ij/]. With this algorithm, functional synapses were identified by the co-appearance of pre- and postsynaptic markers in close proximity (<0.4 μm). From this co-appearance, an exclusion mask was created for larger aggregations (>650 nm) and cell bodies, so that only medial and distal dendrites (>35 μm distal from the cell body) were considered for co-localization analysis. Co-localization of the KIF1Bβ signal with the sites of functional synapses was then analyzed based on Manders’ coefficients. For the analysis shown on Fig. 6d, e, values were obtained from 4 different neuronal culture experiments, analyzing 20 dendrites in total (from 20 cells in all 4 experiments) with about 120 synapses examined per dendrite. Probability values were obtained with a Student’s t test with a two-tailed distribution and two-sample equal variance.
Fluorescence, confocal and time-lapse microscopy
Immunofluorescence preparations were observed with either a Zeiss Apochromat ×63 1.3 NA oil lens on a Zeiss Axiovert 200 M inverted fluorescence microscope, equipped with a Zeiss AxioCam MRm camera, or with a Nikon Plan Apo ×60 1.4 NA oil lens, fitted on an upright Biorad confocal scan head, mounted on a Nikon Eclipse E800 upright confocal microscope.
Time-lapse microscopy images were acquired from mouse cortical primary neurons (div. 10–12) at 24 h after transfection by calcium-phosphate precipitation [27] with plasmid constructs pEYFP-KIF1Bβ [full-length KIF1Bβ, aa sequence equivalent to construct “F” in Fig. 7c (see below)] or pEGFP-KIF1Bβ-C1 (construct “C1”, lacking the motor domain; Fig. 7c). Neurons were imaged using an Olympus IX81 inverted microscope, equipped with a Hamamatsu ImageEM camera and a 1.3 NA UPLAFLN ×60 objective lens. Temperature was kept constant (37 °C) in a humidity chamber. Images were acquired at a rate of one image per 10 s for at least 10 min. The coordinates of the granules in the time-lapse videos were analyzed in each frame for the distance and speed over 180 s using ImageJ [28], according to [29, 30]. ImageJ was also used to generate kymographs (Fig. 9, see below).
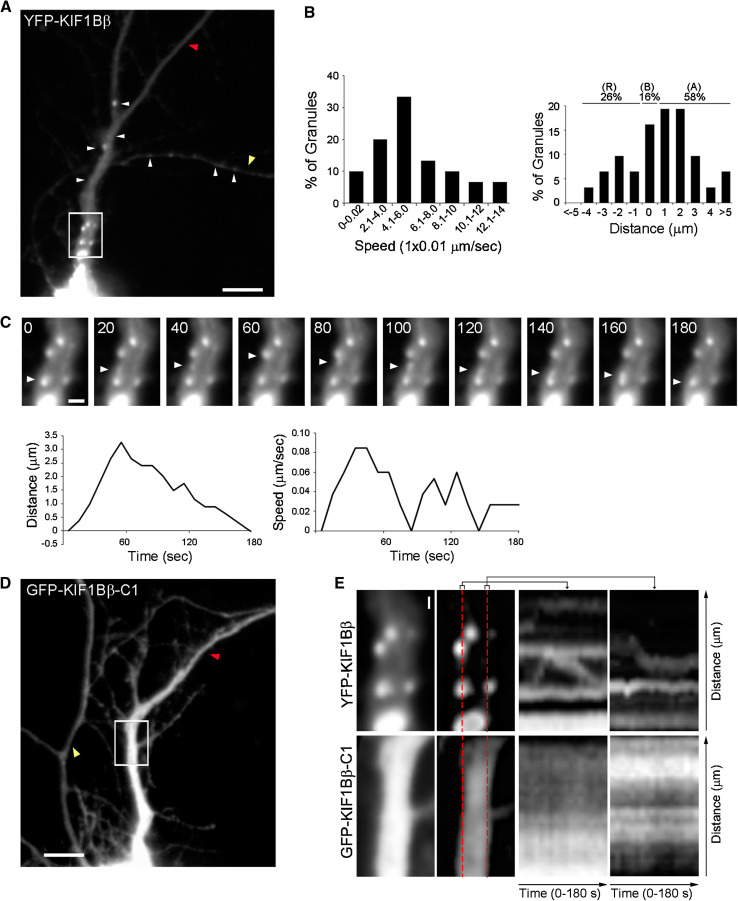
Fig. 9.
Processive movement of YFP-KIF1Bβ granules in the dendrites of living cortical neurons. a Example of a mouse cultured cortical neuron transfected with plasmid pEYFP-KIF1Bβ (full-length construct in Fig. 7c), showing fluorescent granules. The dendrite in this cell is indicated with a red arrowhead and the axon with a yellow arrowhead. Scale bar 10 μm. b Histogram of the maximum speed of 30 granules (average ± SD: 0.060 ± 0.034 μm/s) (left panel); final distances these 30 granules covered in 180 s (average ± SD: 0.9 ± 2.7 μm) (right panel). c Upper panels a series of still images, taken at 20-s intervals, from a time-lapse movie of the rectangular area marked in (a), as an example of the movement of a granule (arrowheads) for a period of 180 s. Scale bar 2 μm. The full video of this sequence is given as an Online Resource video1. (The full video of a sequence derived from a neuron transfected with pEGFP-KIF1Bβ-C1 is given as an Online Resource video2). Bottom panels time course, during 180 s, of the distances covered by this representative granule from the start point, as traced by the arrowheads (upper panels). Corresponding time course of the speeds of the sample granule during the 180 s recording (right panel). d Equivalent example of cortical neuron transfected with pEGFP-KIF1Bβ-C1 (motor-less construct “C1” in Fig. 7c), showing diffuse fluorescent labeling. The dendrite in this cell is indicated with a red arrowhead and the axon with a yellow arrowhead. e Kymographs of neurons transfected with pEYFP-KIF1Bβ or pEGFP-KIF1Bβ-C1. Rectangular areas marked in (a) and (d) indicate, respectively, the region selected to generate the kymographs. Red dashed lines indicate the orthogonal plane represented by the kymographs. Scale bar 1 μm
Animal care
Strict institutional guidelines on animal care were adhered to for experiments that required use of mouse cells or tissues. Institutional guidelines were in compliance with international laws and policies governing the use of animals (European Community Council Directive 86/609, OJa L 358, 1, December 12, 1987 on the protection of animals used for experimental and other scientific purposes).
Results
KIF1Bβ (isoform NM015074) is a motor protein widely expressed in the CNS: isoform specificity and expression within the KIF1B family
To analyze the specific function/s and regulation of KIF1Bβ in neuronal cells, we first studied its gene and protein expression. To address the need for isoform specificity within the KIF1Bβ subfamily in our experiments, we raised and employed an anti-peptide antibody to the distinctive C-terminus of NM015074 (Fig. 1), thus precluding the detection of the AB088210–13 series of isoforms, which will accordingly not be further examined in this work. Additionally, we discriminated between the expression of NM015074 and AX039604 (which share the common C-terminus) by utilizing NM- and AX-specific oligonucleotide pairs (Fig. 1; Online Resource Table 1) that we showed not to cross-hybridize with those two or the additional KIF1Bβ isoforms (as confirmed in the PCR tests of Online Resource Fig. S1).
RT and semi-quantitative PCR analysis of NM015074 mRNA showed a widely spread expression profile with differing abundance across several human tissues (Fig. 2a). Furthermore, of the four regions of human cerebral cortex examined, NM015074 was detected in motor and temporal cortex and AX039604 in the motor cortex (Fig. 2b).
In mouse, both isoforms also appeared in other brain areas such as the cerebellum and hippocampus as well as in the spinal cord (Fig. 2c). The developmental expression profile of both isoforms in the mouse hippocampus was then examined in more detail in hippocampi of E13 and E18 mouse embryos and juvenile animals, and in hippocampal primary cultures (immature stage 2 and fully differentiated stage 5, as per [18]; Fig. 2d). NM015074 mRNA levels appear to be developmentally regulated with an increased expression at late embryogenesis (E18) and during early postnatal development (juvenile). Given that a large proportion of glial cells are present in the in vivo and in vitro samples, and to distinguish possible differences between neurons and glia, the analysis was extended to a pure astrocytic primary culture. Notably, neither of the two isoforms was detectable in astrocytes (Fig. 2d), thus indicating that the detected KIF1Bβ expression came exclusively from neurons.
We further investigated KIF1Bβ expression in different human and mouse cell lines (HeLa, neuroblastoma SH-SY5Y non-induced or induced with retinoic acid, neuroblastoma NSC-34 and fibroblast NIH 3T3). KIF1Bβ NM015074 expression was only detected in the NSC-34 line (high-level expression) and in NIH 3T3 (low-level) (Fig. 2e). Therefore, the NSC-34 line, in addition to primary cultured spinal motor neurons, was also used in some of the subsequent experiments.
The sequence composition of the KIF1B family members prevents the design of oligonucleotide primers for KIF1Bβ that can, at the same time, discriminate KIB1Bβ NM01574 from both, its related beta isoforms (such as the primers used in Fig. 2) and from the known alpha isoform, KIF1Bα (NM008441). Therefore, in order to accurately complete our gene expression analysis, we also designed a set of KIF1Bα-specific primers and selectively tested pertinent tissues (see Online Resource Fig. S2 for validation). The results revealed KIF1Bα expression in the hippocampus and spinal cord motor neurons, but no expression in the cortex and NSC-34 line; however, motor neuronal expression of KIF1Bα does not interfere with our subsequent experiments, because our anti-peptide KIF1Bβ-specific antibody cannot detect the alpha isoform. Indeed, immunoblotting using this anti-peptide antibody, raised against the distinctive 15 aa of the KIF1Bβ C-terminus, revealed a specific protein band of the predicted size for NM015074 (M r of 199.01 × 103) in primary cortical neurons in culture, as well as in the NSC-34 and NIH 3T3 cell lines (Fig. 2f). The intensity of the detected protein bands was at a similar relative ratio between the two lines as their mRNA levels (shown in Fig. 2e). Moreover, the protein was detected in the same parts of the nervous system as the mRNA. Detection of KIF1Bβ was effectively abolished by pre-absorption with the antigenic peptide, as further confirmation of its identity (Fig. 2f). For simplicity, KIF1Bβ isoform NM015074, which is the subject of our study and on which subsequent experiments concentrate, will be referred to as KIF1Bβ hereafter.
KIF1Bβ is enriched at synaptic sites and co-localizes with pre- and postsynaptic structures
Using the anti-peptide antibody against KIF1Bβ, we mapped the intracellular localization of the protein in primary mouse spinal motor neurons (Fig. 3), a selective target for neurodegeneration in CMT2A and a cell type where expression of KIF1Bβ was confirmed by RT-PCR (Fig. 2c). Double staining of mixed co-cultures of spinal neurons (motor neurons, interneurons) and glia with SMI 32, a monoclonal antibody specifically labeling motor neurons, revealed that KIF1Bβ is selectively expressed in motor neurons (Fig. 3a), but not in astroglia (not shown), in accordance with the RT-PCR results of Fig. 2d showing absence of signal in pure astroglial cultures.
Fig. 3.
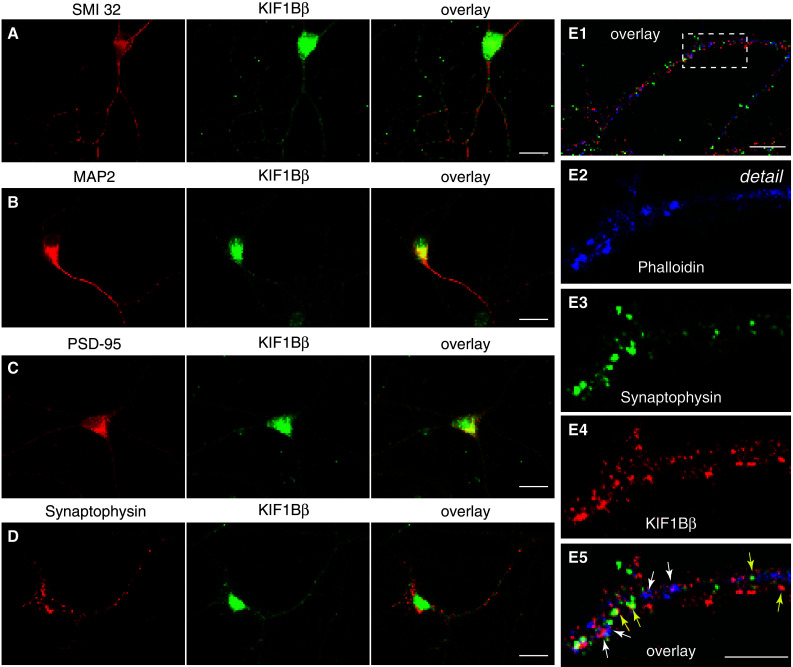
Intracellular localization of KIF1Bβ in mouse primary motor neurons. a–d Red panels spinal motor neurons probed with antibodies against (a) non-phosphorylated neurofilament H, SM132; (b) MAP2; (c) PSD-95; (d) synaptophysin. Corresponding green panels represent labeling for KIF1Bβ. The red and green images are overlaid in the last column. E1–5. Segments of dendritic processes from mouse spinal cord motor neurons stained with AF647-phalloidin (blue, substituted pseudocolour to visualize infrared emission), anti-synaptophysin (green) or anti-KIF1Bβ antibodies (red). E1 A merged image displays an overlay of the three labels. E2–E5 A magnified detail (corresponding to the boxed area in panel E1) for each of the labels and their overlay image is shown in these panels. Examples of localization of KIF1Bβ at postsynaptic sites (co-localization with polymerized F-actin, as revealed by phalloidin staining) are indicated with white arrows (blue and red color merges to purple), while examples of co-localization of KIF1B and synaptophysin (a presynaptic marker) are indicated with yellow arrows (green and red color merges to yellow). Scale bars (E1) 10 μm, (E2–E5) 5 μm
Initially, we examined the overall topology of KIF1Bβ labeling in those motor neurons: KIF1Bβ immunostaining was punctate or granular and detectable both in the cell body, and in the axon and dendrites (Fig. 3a). With the use of an antibody to the dendritic microtubule marker MAP2, we confirmed the expression of KIF1Bβ in dendrites (Fig. 3b) where the motor protein appears co-localized with PSD-95-positive, postsynaptic structures (Fig. 3c). Furthermore, KIF1Bβ also appears partially co-localized with puncta of synaptophysin, a known presynaptic vesicle protein (Fig. 3d). To consolidate these findings, we performed a triple staining (Fig. 3E1, with E2–E5 showing a detail at higher magnification) with fluorescently-labeled phalloidin, which highlights points of filamentous F-actin enrichment and hence mostly dendritic spines (Fig. 2E2, blue), anti-synaptophysin antibody as a presynaptic marker (Fig. 3E3, green), and anti-KIF1Bβ (Fig. 3E4, red). Examination of the labeled patterns consistently revealed colocalization of KIF1Bβ both with synaptophysin puncta, marking presynaptic sites (Fig. 3E5, yellow arrows), and F-actin sites on dendritic spines, indicating postsynaptic density domains (Fig. 3E5, white arrows). Thus, KIF1Bβ-immunoreactive structures were present both at pre- and postsynaptic sites.
Furthermore, as independent confirmation, we also obtained results in clear agreement with this intracellular localization of KIF1Bβ in NSC-34, a spinal cord neuron–neuroblastoma hybrid cell line, which expresses KIF1Bβ (as detected by RT-PCR in Fig. 2e). It is known that NSC-34 cells maintain many of the morphological and physiological characteristics of spinal motor neurons, including the ability to form synapses with myotubes [31], and therefore express synaptic proteins. In NSC-34 cultures, the anti-KIF1Bβ antibody-labeled vesicular structures displaying overlap both with PSD-95 (Fig. 4a) and presynaptic vesicular protein SV2 (Fig. 4b). Consistently, no co-localization of KIF1Bβ puncta was observed when NSC-34 cells were labeled with antibodies specific for other membrane-bound cellular compartments, namely mitochondria (SOD2), lysosomes (LAMP2), or the Golgi (58 K Golgin protein) (Online Resource Fig. S3).
Fig. 4.
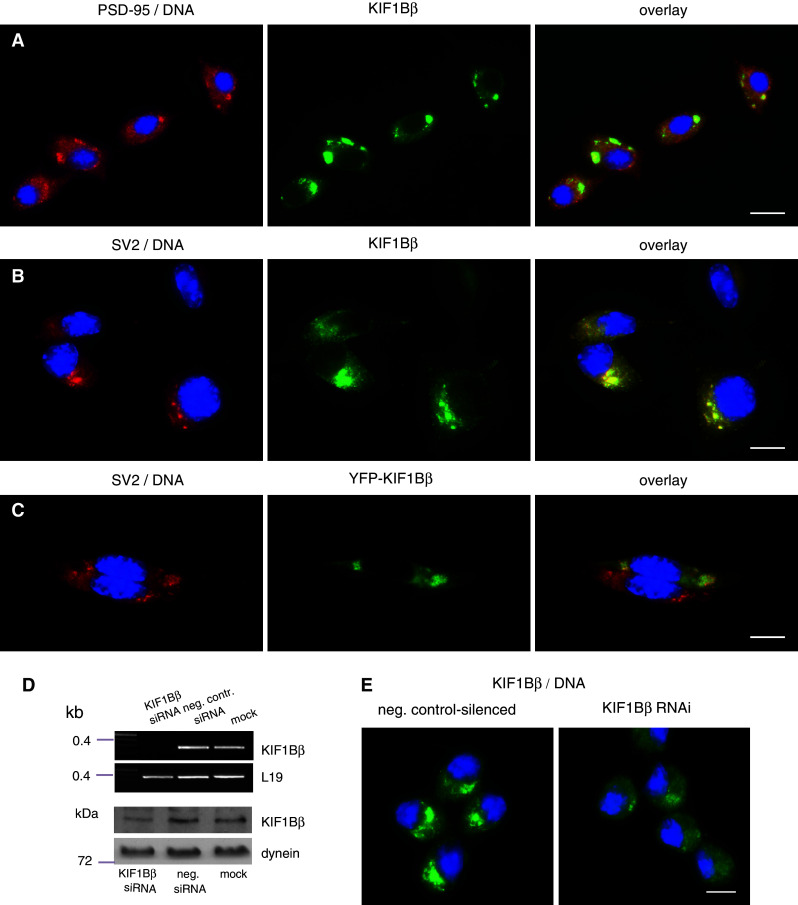
Intracellular localization of KIF1Bβ in mouse neuroblastoma NSC-34 cell line. NSC-34 cells stained with antibodies to a PSD-95 (red), b SV2 (red), with concurrent labeling of nuclei with Hoechst 33342 (blue). Green panels represent labeling with anti-KIF1Bβ. Scale bars (a) 20 μm, (b, c) 10 μm. c NSC-34, transiently transfected with pEYFP-KIF1Bβ, green). Anti-SV2 staining is in red, and DNA in blue. Of the two cells in the field, only the top cell is transfected: YFP-KIF1Bβ localization (green) overlaps extensively with that of SV2 (red), therefore merging to yellow in the overlay. Scale bar 10 μm. d Quantification of silencing efficiency by RT semi-quantitative PCR and immunoblot. Top two panels levels of KIF1Bβ mRNA at 120 h post-transfection in NSC-34 cells silenced with 320 pM of KIF1Bβ-specific siRNA cocktail are compared to control-silenced cells (negative control) and to mock-treated cells (no siRNA). RT-PCR detection of L19 mRNA levels serves as internal control. Bottom two panels KIF1Bβ protein levels are compared in corresponding samples. Detection of the intermediate chain of dynein serves as loading control in the three samples. e KIF1Bβ immunofluorescence in silenced cells versus negative control-silenced cells. NSC-34 cells were immunostained for KIF1Bβ (green) and counterstained for DNA (blue). KIF1Bβ-silenced cells display a marked reduction in the signal of KIF1Bβ-positive vesicular structures when imaged at the same exposure as negative control-silenced cells. Scale bar 10 μm
The co-localization of KIF1Bβ with synaptic structures in NSC-34 was further confirmed upon transient transfection of an YFP-tagged full-length KIF1Bβ construct. In the image shown in Fig. 4c, two neighboring cells are imaged, one untransfected (bottom cell) and one transfected (top cell); a high degree of co-localization of YFP-KIF1Bβ fluorescence and SV2 immunoreactivity can be observed in the transfected cell.
Finally, the native KIF1Bβ-positive structures become less pronounced upon siRNA-mediated silencing of KIF1Bβ (Fig. 4e). It is notable that, although depletion of KIF1Bβ transcript is very efficient upon prolonged silencing (Fig. 4d, top panels), the KIF1Bβ protein appears extremely stable even after 120 h of application of the silencing regime, involving two siRNA transfections (Fig. 4d, bottom panels). This protein stability likely accounts for the residual persistence of the, albeit markedly reduced, vesicular structures of NSC-34 seen in Fig. 4e.
To corroborate biochemically the synaptic localization of KIF1Bβ in neurons, as revealed by immunofluorescence, and further study its synaptic compartmentalization, we subsequently analyzed synaptoneurosomal preparations, isolated from mouse cortex. Synaptoneurosomal preparations consist of purified functional synapses containing both pre- and postsynaptic termini [32] and can also be used to study synaptic function in vitro. The purification protocol we used (see “Materials and Methods”) guaranteed the inclusion of physiologically functional and intact synapses, as previously validated [21, 22]. Consistent with its cellular localization, KIF1Bβ was found enriched in the highly purified synaptoneurosomal fraction, and co-purifying with typical synaptic protein markers PSD-95 and synaptophysin [Fig. 5a, compare total brain (T) vs. synaptoneurosomal (S) fractions]. Further fractionation of the synaptoneurosomal sample to its soluble, pre- and postsynaptic components, as validated by correct segregation of appropriate markers in each case, reproducibly allowed detection of KIF1Bβ only in the pre- and postsynaptic compartments (Fig. 5b). The clear absence of KIF1Bβ from the soluble component after detergent extraction strongly suggests that the motor forms detergent-resistant complexes at the vicinity of synaptic contacts, possibly bound to cytoskeletal or scaffolding structures. The presence of KIF1Bβ in both the pre- and postsynaptic fractions was consistent with the observed co-localization of KIF1Bβ with pre- and postsynaptic markers by immunofluorescence (Fig. 3c, d, E1–E5) and indicated that the motor may be bound to the matrix/structural components on both sides.
Synaptic activation causes increased recruitment of KIF1Bβ to the synaptic area
Given the association of KIF1Bβ with synaptic structures, we next asked the question whether synaptic activity might influence the recruitment of the motor protein to the active synaptic sites. We therefore quantified KIF1Bβ synaptic recruitment with two independent assays, performed in the absence or presence of synaptic stimulation. We conducted: (1) biochemical assays with purified synaptoneurosomes, and (2) morphological assays using mouse cortical neurons in culture.
We employed DHPG (S-3,5,-dihydroxyphenylglycine), a widely used agonist of group I metabotropic glutamate receptors (mGluR1 and mGluR5) implicated in the induction of different forms of synaptic plasticity, to stimulate synaptoneurosomal preparations. We observed, by quantitative immunoblot, a statistically significant (p = 0.023) increase by 50 % in the concentration of KIF1Bβ in the stimulated synaptoneurosomes, compared with unstimulated controls (Fig. 6a, b), while none of the other synaptic markers tested (PSD-95, synaptophysin, syntaxin 1 and SNAP-25) showed significant alteration after stimulation (Fig. 6c).
For independent confirmation, we quantified the effect of DHPG stimulation in living cells. To visualize functional synapses on dendrites, we performed triple staining with phalloidin, which highlights points of polymerized F-actin enrichment and hence dendritic spines, anti-synaptophysin antibody as a presynaptic marker, and anti-KIF1Bβ (Fig. 6d). For KIF1Bβ quantification, we applied a mask that was created with a dedicated algorithm (modified from [24]; see further details in “Materials and Methods”) marking an area in the image lying within 0.4 μm of both synaptophysin puncta and sites of F-actin and thus in close vicinity to a synaptic contact. In this manner, we restricted quantification of KIF1Bβ signal to that associated with functional synapses. In confirmation of our biochemical results, analysis of images from four independent experiments (total of 2400 synapses analyzed) revealed that DHPG stimulation resulted in increased recruitment of KIF1Bβ to the synaptic area (by 25 %), compared with non-stimulated cells (statistical significance p = 0.03; Fig. 6e).
We tested whether a similar activity-dependent increase of the levels of KIF1Ββ at the synapse could occur upon general stimulation with KCl, metabotropic or ionotropic glutamate receptor activation (Fig. 6f). Analysis of the results using KCl or the different agonists, specific for ionotropic and metabotropic glutamate receptors, and from the PMA, a phorbol ester that directly activates PKC (Online Resource Fig. S4), clearly showed that the levels of KIF1Bβ in purified synaptoneurosomes were markedly and statistically very significantly increased by KCl activation and glutamate signaling, but preferentially so upon NMDA receptor activation (p = 0.0014; Fig. 6f).
On the basis of the combined experimental evidence, we therefore conclude that synaptic concentration of motor KIF1Bβ is increased in an activity-dependent manner.
KIF1Bβ interacts directly with Calmodulin (CaM) in a Ca2+-dependent manner and through its carboxy-terminal domain
To obtain insight into the functional significance of the localization and activity-dependent modulation of KIF1Bβ recruitment to the synapse, we employed different approaches to screen for its interacting partners. We first sought to identify interacting protein partners for this motor using a yeast two-hybrid screen. Of the 1.8 × 106 clones of a human adult CNS cDNA library that were tested, using as bait a fragment of KIF1Bβ containing the non-conserved, non-motor domain of the protein (construct “C1”, nt. 1980–5313), 54 positive clones were identified after five rounds of selection (details in “Materials and Methods”). Of these, 25 positive clones contained partial cDNAs derived from the CALM1 (Calmodulin 1) gene, 21 were partial cDNAs derived from the CALM2 gene and 8 from the CALM3 gene. Although these three genes are localized on distinct human chromosomes (chromosomes 14, 2, and 19, respectively; [33, 34]) and exhibit diverse 5’ and 3’ UTRs, because of the degeneracy of the genetic code, their ORFs are synonymous and completely identical at the aa level and thus all encode the same protein, Calmodulin (CaM).
To ensure that the interaction detected between KIF1Bβ and CaM was bona fide and to characterize it further, we carried out several independent sets of experiments. First, we examined the intracellular localization of KIF1Bβ and CaM in mouse primary spinal motor neurons in culture by immunofluorescence. We observed extensive co-localization, as one would likely expect from interacting proteins (Fig. 7A1–A3), both, in the cell body and in neuronal processes (detail at higher magnification in the inset).
Second, we carried out a series of pull-down assays using CaM-Sepharose beads incubated with extracts from NSC-34 neuroblastoma cells in the presence of increasing concentrations of Ca2+ (from 0–2 mM CaCl2) (Fig. 7b). These assays showed that (1) native KIF1Bβ was recovered in the pellet fraction only when incubated in the presence of CaM, consistent with an interaction between the two proteins (Fig. 7b, compare lanes 3, 5, 7, 9 with negative controls in lanes 10, 12), and (2) the interaction was Ca2+-dependent, with recovery of the native KIF1Bβ protein minimal in the absence of Ca2+ and peaking in the presence of 0.5 mM CaCl2 (Fig. 7b, compare lanes 3, 5, 7 and 9 with progressively increasing concentrations of Ca2+).
Third, we demonstrated that the interaction between KIF1Bβ and CaM is direct, confirmed its dependence on Ca2+ using in vitro pull-down assays with bacterially expressed, recombinant proteins (Fig. 7c) and mapped the CaM-binding site on a domain contained within the carboxy-terminal part of KIF1Bβ (Fig. 7c; Table 1). Analytically, Fig. 7c illustrates that the E. coli-expressed GST-tagged C1 fragment of KIF1Bβ (construct “C1”, the fragment originally used in the yeast two-hybrid screen) can bind to CaM immobilized on Sepharose beads, but only in the presence of Ca2+ (Fig. 7c, compare lanes 3 and 7 in panel C1), while the corresponding N-terminus of KIF1Bβ, containing the motor domain (fragment “M”), remains unbound in all circumstances (Fig. 7c, compare lanes 3 and 7 in panel M). By testing progressively smaller GST-tagged subfragments of C1 (constructs C2–C5), we confined the CaM interaction site(s) of KIF1Bβ to within the last 552 carboxy-terminal aa (Fig. 7c, observe lanes 3 and 7 in panels C2, C3). If this carboxy-terminal region is fragmented into two complementary segments of 331 and 221 aa (constructs C4 and C5), both segments retain their CaM binding activity (Fig. 7c, lanes 3 and 7 in panels C4, C5), suggesting that either (1) there is more than one CaM site, or (2) that the CaM binding site extends either side of aa 1549, the position where the subfragments are split. The results of these in vitro pull-down assays are summarized in Table 1 (left part). It is notable that both the native KIF1Bβ (Fig. 7b) and recombinant subfragments C3–C5 (Fig. 3c) retained some CaM binding activity even in the absence of Ca2+, either suggesting a negative regulatory function for the amino-terminal part of KIF1Bβ or merely non-specific interaction with Ca2+-depleted CaM (ApoCaM) as a result of the protein fragmentation.
Table 1.
Mapping of the CaM interaction domain of KIF1Bβ by in vivo and in vitro assays
| In vivo interactions with full-length CaM by mating in S. cerevisiae | In vitro pull-down assays of CaM with native KIF1Bβ or recombinant protein fragments expressed in E. coli | ||||||
|---|---|---|---|---|---|---|---|
| Construct | Result | X gal expos | Construct | 0.0 mM (5 mM EDTA) | 0.5 mM | 1.0 mM | 2.0 mM (Ca2+) |
| pGBK/pGAD7 (pos. control) | + | 2 h | Native KIF1Bβa | + | ++ | ++ | ++ |
| M (aa 1–660) | – | 3 h | M (aa 1–660) | – | NT | – | NT |
| C1 (aa 660–1770) | ++ | 2 h | C1 (aa 660–1770) | – | NT | +++ | NT |
| C2 (aa 660–1218) | – | 3 h | C2 (aa 660-1218) | – | NT | – | NT |
| C3 (aa 1218–1770) | ++ | 3 h | C3 (aa 1218–1770) | + | NT | ++ | NT |
| C4 (aa 1218–1549) | + | 3 h | C4 (aa 1218–1549) | + | NT | + | NT |
| C5 (aa 1549–1770) | + | 3 h | C5 (aa 1549–1770) | + | NT | + | NT |
| pAS2.1 (neg. cont) | – | 3 h | pGEX (neg. cont) | – | NT | – | NT |
The left part displays mating assays with plasmid pACT2.1-CaM, carrying full-length Calmodulin, and the pAS2.1-KIF1Bβ constructs shown (numbers in parentheses denote amino acid (aa) residues in generated KIF1Bβ protein fragments). Interaction was defined as the ability of >95 % of resulting clones, expressing both constructs, to grow on quadruple selective media (-Leu, -Trp, -His, -Ade) and to also display β-galactosidase activity upon filter assay. The right part shows assays conducted with native KIF1Bβ or GST-tagged recombinant proteins from KIF1Bβ deletion constructs, expressed in E. coli and incubated with CaM-Sepharose beads and in the concentration of Ca2+ indicated, with 0.0 mM CaCl2 established by chelation with 5 mM EDTA
NT Not tested
aThe term native KIF1Bβ refers to a reaction using total lysate (500 μg) from NCS-34 cells, incubated with CaM-Sepharose beads
Finally, the same fragments of KIF1Bβ (M and C1–C5) were also subcloned as GAL4 DNA-binding-domain fusions and tested in vivo by yeast mating assays for interaction with full-length CaM, expressed as a GAL4 DNA activation-domain fusion. The results, also summarized in Table 1 (right part), were identical to those obtained with the in vitro assays, thus confirming the position of the CaM binding site(s) within the 552 carboxy-terminal aa of KIF1Bβ.
A KIF1Bβ particle contains neuronal RNA–protein complexes
The identification of CaM as an interacting protein with KIF1Bβ was consistent with the activity-dependent recruitment of KIF1Bβ to the synapse and a role in synaptic signaling. Notably, there are examples of motor proteins shown to mediate the transport of mRNA-containing ribonucleoprotein particles to synapses, for example, mRNA transport to dendrites by motors from the conventional kinesin KIF5 subfamily [29, 35; reviewed by 36]. Dendritic transport granules have been shown to contain several RNA binding proteins [29, 36]. We therefore investigated whether KIF1Bβ is associated with RNA-binding proteins and mRNAs that are synaptically localized [37]. First, we immunoprecipitated the endogenous KIF1Bβ protein complex from mouse brain (Fig. 8a, top panel, compare lanes 2 and 3). The presence of CaM protein in this native complex from brain (Fig. 8a, second panel) confirmed our previous findings using yeast two-hybrid and pull-down assays (Fig. 7) and showed that the complex containing the two interacting proteins is intact. Additionally, we found that KIF1Bβ coimmunoprecipitates with Purα (purine-rich element binding protein alpha) [38, 39], FMRP (fragile X mental retardation protein) [22, 40, 41], and FXR2P (fragile X related protein 2) [42] (Fig. 8a, panels 3–5), all of which are well-characterized components of mRNPs complexes. Interestingly, we found that the immunoprecipitated complex also contained the plus end-directed KIF3A motor protein but not the conventional dendritic kinesin KIF5 (Fig. 8a, panel 6). Of note, other RNA binding proteins, such as ELAVs, are not part of the KIF1Bβ complex (Fig. 8a, panel 7), in agreement with its association to KIF5 [29]. This would indicate that the KIF1Bβ-containing complex is distinct from the well-characterized RNA-containing granule transported by KIF5 [29]. We further investigated the association of KIF1Bβ with the co-imunoprecipitated proteins in the dendrites of cultured cortical neurons by double immunofluorescence. As shown in Fig. 8b, KIF1Bβ-positive puncta extensively co-localize with Purα-, FMRP- and FXR2P-positive granules, consistently with the immunoprecipitation results.
We next investigated the presence of RNA species in the KIF1Bβ-containing complex. The analysis, by RT-PCR, revealed the presence of Arc (activity-regulated cytoskeleton-associated protein) mRNA (Fig. 8c), a well-known, dendritically localized mRNA [43–45]. Interestingly, Calmodulin mRNA was also part of this complex. Both were specifically found in the KIF1Bβ-immunoprecipitated complex, but not in the negative control reaction (Fig. 8c, compare lanes 2 and 3). The specific association of Arc and Calmodulin mRNAs with the KIF1Bβ-containing complex is further supported by the absence of non-synaptically or other synaptically localized mRNAs such as eEIF1α and αCaMKII (Fig. 8c). These observations would suggest the participation of KIF1Bβ in RNP particles in association with at least two dendritically targeted mRNAs, Arc and CaM.
Taken together, our results indicate that motor protein KIF1Bβ forms dendritically localized RNPs, containing some of the previously identified dendritically transported mRNAs and proteins.
Processive dendritic movement mediated by KIF1Bβ in living cortical neurons
Kinesin-driven transport in axons has been extensively studied [reviewed by 46]. Although a few examples of kinesin-driven transport in dendrites have also been described [29, 47, 48], less is known on the functional role of kinesin-like proteins in dendrites. The identification here of an RNA/protein complex associated with KIF1Bβ, and its dendritic localization (Fig. 3b, Online Resource Fig. S5), raised the possibility that KIF1Bβ may mediate the transport of this RNP in dendritic processes. To directly assess this possibility, we employed transient transfection of full-length YFP-KIF1Bβ in mouse cultured cortical neurons, and visualized and recorded the fluorescent protein by time lapse live-imaging over periods of 10 min (Online Resource video 1). Fluorescent mobile YFP-KIF1Bβ appeared in both axons (yellow arrow) and in dendritic processes (red arrow) with a granular expression pattern (Fig. 9a). Axons and dendrites were identified based on their morphology as per [18]. We could observe anterograde movement in axons but also detected slower processive movement in dendritic processes. We focused on the KIF1Bβ dendritic transport and analyzed the directionality and the speed of granule movement through a series of time-lapse images, acquired at 10-s intervals over a period of 180 s (neurons n = 10, granules n = 30) as previously described [29, 30, see “Materials and Methods”] (Fig. 9b). In dendrites, KIF1Bβ showed a speed of up to 0.14 μm/s with average speed of 0.060 ± 0.034 μm/s (Fig. 9b, left panel). This speed is similar to the one observed for the dendritic speed of motor KIF5 [29] and comparable with previously recorded values of mRNA trafficking [47, 49, 50]. When we measured the net movement of the granules over a period of 180 s, we observed that the majority (58 %) moved in the anterograde direction, while the rest moved either retrogradely (26 %) or displayed rapid back-and-forth movement (16 %), covering an average distance of 0.9 ± 2.1 μm (Fig. 9b, right panel). Figure 9c (upper panels) displays a series of time-lapse images to follow the movement of a sample granule (traced by the arrowheads) for 180 s with 20 s intervals. Granule time course distance and speed changes are shown (lower panels). The patterns of movement (explained as a result of the mixed polarity of microtubules at proximal dendrites and competition with retrograde motors, such as dynein) and the measured speeds are again consistent with those detected for RNPs transported by KIF5 [29] or other brain derived, dendritically targeted RNPs [30, 51]. Altogether, these results support our hypothesis that KIF1Bβ-containing granules transport RNA in dendrites.
The complex we identified is not associated to KIF5A (Fig. 8a), thus suggesting an active role of KIF1Bβ in the RNP transport. To rule out the possibility that the movement we detected was not mediated by another motor protein (such as KIF3A that we have identified as part of the complex in Fig. 8a) that carried along KIF1Bβ in a passive, piggy-bag manner, we analyzed the movement of a truncated KIF1Bβ lacking the motor domain (GFP-KIF1Bβ-C1 construct of Fig. 7c). In this case, we did not observe either specific granule formation or movement, but, instead, diffuse fluorescence, as previously shown for KIF5A under the same conditions by Kanai et al. [29] (Fig. 9d; Online Resource video 2). Similar findings were obtained using a GFP-empty plasmid (data not shown). These results indicated that the movement we analyzed by the use of the full-length construct was bona fide, active and specific. To better evaluate moving or immobile particles, we represented in two kymographs the fluorescence changes along the dendritic segment over 180 s in neurons transfected with YFP-KIF1Bβ or GFP-KIF1Bβ-C1, respectively. As expected, kymograph analysis further confirmed the absence of moving granules when the truncated KIF1Bβ was expressed (Fig. 9e). In conclusion, therefore, these combined experiments revealed a novel role for KIF1Bβ as a motor protein mediating transport of RNP particles, which contain RNA-binding proteins and dendritically targeted Calmodulin and Arc mRNAs.
Discussion
The members of the KIF1A/KIF1B/UNC-104 (kinesin-3) family of molecular motors have been shown by a number of studies to transport synaptic vesicle structures [4, 7, 52]. Mechanisms about how signaling pathways may be regulating vesicular transport are gradually emerging. Association of such vesicles with the stalk domain or the PH domains of kinesin-3 family motors (see Fig. 1) via vesicle phosphoinositides is necessary for cargo recognition and binding [8]. Experimental evidence supports a model where KIF1A motors exist in an autoinhibited state in the absence of cargo, and this autoinhibition is relieved following interaction with cargoes [53]. At least one adaptor protein, the DENN/MADD, has been identified as essential in the GTP-dependent axonal transport of Rab3-containing vesicles [54]. In C. elegans, it has been proposed that, upon cargo delivery at the pre-synaptic site, kinesin-3 motor KIF1A/UNC-104 is locally degraded at the synapse, likely via the ubiquitination pathway, and hence is not recycled back retrogradely [55]. Furthermore, in C. elegans, UNC-104 motor clustering along axons as well as their motility is enhanced via multiple interactions with synaptic scaffolding protein SYD-2 [56]. In non-neuronal cells, i.e. in non-polarized epithelial cells, the post-Golgi transport of p75 neurotrophin receptors by kinesin-3 family motors (KIF1Bα and β) appears to also involve interactions with the PH domain [57].
While it is now well understood that a multitude of molecular motors, including KIF1Bβ, act in concert to transport various cellular cargoes to the synapses of neurons, the mechanisms that integrate cargo transport with synaptic activity, or with synaptic assembly in developing neurons or with remodeling of synapses in mature neurons (plasticity), in response to neuronal activity [58] remain largely uncharacterized.
Here, we provide novel findings that functionally characterize KIF1Bβ in the context of synaptic signaling. KIF1Bβ is not neuron-specific but highly expressed in the CNS and particularly prevalent in the motor cortex and spinal cord motor neurons (Fig. 2). It is localized both presynaptically in synaptic boutons and postsynaptically in postsynaptic density structures of dendritic spines (Figs. 3, 4). KIF1Bβ is enriched in isolated synaptoneurosomes, co-fractionates in both pre- and postsynaptic fractions, and is exclusively in a bound and detergent-resistant form, with no detectable soluble KIF1Bβ pool (Fig. 5). Stimulation of metabotrobic or ionotropic glutamate receptors results in increased KIF1Bβ concentration at synaptic sites, revealing an activity-dependent recruitment of the motor to synapses (Fig. 6). In this study, Calmodulin was identified as a direct binding partner of the motor (Fig. 7), via an interaction domain within the 552 amino acids of the carboxy-terminal tail of KIF1Bβ, partly overlapping with the stalk domain where interaction with DENN/MADD protein has previously been localized [54]. The CaM-KIF1Bβ interaction is dependent upon and facilitated by Ca2+, with a possible weak interaction also with the ApoCaM form (Fig. 7). This is an important finding because it directly links KIF1Bβ to Ca2+ via CaM, a universal Ca2+ sensor protein, thus indicating a possible mechanism of KIF1Bβ regulation during depolarization-induced Ca2+ influx in the presynaptic compartment. Our results would predict that, in conditions of facilitation of presynaptic Ca2+ influx, triggered by synaptic depolarization, the interaction between activated CaM and KIF1Bβ is enhanced. Increased strength of the interaction could result in augmented synaptic delivery of vesicles via KIF1Bβ. This mechanism may thus stimulate synaptic vesicle exocytosis by activating bound KIF1Bβ or redistributing bound KIF1Bβ on synaptic vesicles upon presynaptic depolarization, which in turn would tie in with the observed activity-dependent increase of synaptic KIF1Bβ recruitment.
Other examples of interaction of kinesin-like motors with Ca2+-binding proteins have been reported: Ca2+-dependent binding of CaM to the light chains (KLC) has been previously documented in the case of heterotetrameric conventional kinesin [59], and, although the CaM binding site on KLC has not been determined, binding appears to inhibit the ATPase activity of the heavy chains. In rat, the KIF1Bβ2 isoform (most similar to the human AB088210–13 isoform series) interacts with CHP (calcineurin B homologous protein) [60], a Ca2+-binding EF-hand protein with significant similarity to the phosphatase calcineurin, involved in Ca2+-dependent signal transduction pathways [61]. In mammalian neurons, kinesin KIF5-powered mitochondrial mobility is inhibited upon Ca2+ influx in postsynaptic sites, via modulation by Miro1, another Ca2+-binding EF-hand protein [62, 63]. In plants, Ca+2-dependent binding of CaM on a kinesin-like motor (kinesin-like Calmodulin binding protein, KCBP) disrupts this motor’s microtubule binding [64]. It would appear, therefore, that regulation of motor protein properties and activity by Ca2+-sensing proteins, in a Ca2+-dependent manner, is a more generalized mechanism. This strategy would be an effective mechanism for integration of motor protein function and local synaptic activation.
We show here via co-immunoprecipitation experiments that KIF1Bβ forms RNP complexes containing its interacting protein CaM, as well as RNA-binding proteins such as Purα, FMRP, and FXR2P (with which KIF1Bβ co-localizes in dendritic processes; Fig. 8), and Arc and Calmodulin mRNAs (Fig. 8) [43–45]. Both mRNAs have a bona-fide dendritic localization, and motor protein KIF5 has previously been shown to move anterogradely a large protein-mRNA complex that includes Arc mRNAs in dendrites [29]. Although some of the proteins (i.e. Purα and FMRP) identified here are in common to the large RNP granule transported by KIF5A [29], we believe that they are distinct complexes as they have differences both in their mRNA and protein components. The KIF1Bβ complex lacks KIF5A and contains Arc and CaM mRNAs, but not CaMKII mRNA, while the KIF5A complex lacks KIF1Bβ and CaM proteins and includes Arc and CaMKII mRNAs.
Our findings raised the possibility that KIF1Bβ may be one additional motor protein that mediates the dendritic targeting of Arc and CaM mRNA in a distinct RNP complex. Our hypothesis is consistent with all the indirect experimental findings presented here i.e. (1) the localization and enrichment of the motor with synaptic structures, including the postsynaptic density, (2) its co-immunoprecipitation with the two mRNA species, and (3) its activity-dependent recruitment to the synapse, but we also found direct evidence of dendritic transport specifically and actively mediated by KIF1Bβ in mouse cultured cortical neurons (Fig. 9). This is one of the very few examples of a function of KIF1Bβ outside the context of axonal transport, given the requirement for this motor for the localization of myelin mRNA to oligodendrocyte processes in zebrafish [12].
KIF1Bβ-mediated dendritic targeting appears of particular relevance for Arc mRNA, the transcript of an immediate early gene that, upon synaptic stimulation, is rapidly transported to active dendritic spines and is locally translated, thus topically increasing Arc protein concentration at postsynaptic terminals [45, 65, 66]. Arc has a critical role in multiple forms of protein synthesis-dependent synaptic plasticity [reviewed in 67]: it is involved in the endocytosis of AMPA receptors, underlying long-term depression (LTD) [68] and homeostatic plasticity [69, 70], and in the formation of F-actin, underlying the consolidation of long-term potentiation (LTP) [71]. It has emerged from different studies that Arc gene expression can be dynamically regulated by behavioral experience in brain areas related to learning and memory, thus coupling changes in neuronal activity patterns to synaptic plasticity and thereby optimizing information storage in active networks [72, 73; reviewed in 74]. Pertinent to our findings, the translation of Arc is induced within minutes of activation of group-I metabotropic glutamate receptors, a response that is essential to mGluR-dependent LTD [75]. The pathway of Arc translational induction involves the activation of translational elongation factor eEF2 via Ca 2+/CaM [75]. We found DHPG, an agonist of group-I mGluR, to cause mobilization of KIF1Bβ to synaptic sites and identified Arc mRNA, CaM mRNA and CaM protein as parts of KIF1Bβ ribonucleoprotein complexes. It is therefore conceivable that, upon metabotropic glutamatergic stimulation, synaptic signaling, involving CaM, activates the transport of Arc (and CaM) mRNAs to dendritic spines through the motor function of KIF1Bβ. This could generate a reinforcing loop, since local CaM synthesis may facilitate the recruitment of more KIF1Bβ, consequently further increasing Arc mRNA levels. Given that Arc mRNA is probably degraded at the first round of translation, it would need to be continuously supplied over a long period such as during LTD or LTP [76]. The speeds of motor-mediated transport of RNPs in axons range from 0.08 μm/s (slow) to 4 μm/s (fast) [51, 77, 78], while RNPs in dendrites move with lower speeds ranging from 0.01 to 1 μm/s [30, 47–49, 79, 80]. KIF1Bβ granules observed here, under basal (unstimulated) conditions, move within that range with a speed between 0.01 and 0.14 μm/s, consistent with our model of KIF1Bβ-mediated RNA transport.
Of note is the detection of plus end-directed motor protein KIF3A in the ribonucleoprotein complex involving KIF1Bβ (Fig. 8). KIF3A is a motor implicated in many different important processes, such as intraflagellar transport and ciliogenesis [81], signal transduction [82, 83], transport of late endosomes and lysosomes [84, 85], the establishment of neuronal polarity via the targeting of PAR3 to the distal tip of the axon [86, 87], and even mitosis [88]. Of particular relevance to this study, KIF3A in neurons is detected within and probably transports tau-mRNA-containing RNPs, locally translated in axons [89]. The presence of KIF3A in the KIF1Bβ complex therefore raises the possibility of alternative usage or switching to distinct motors (i.e. KIF1Bβ and KIF3A) bound to a common cargo (an mRNP complex) in the transport reaction. This is not an unprecedented concept since motor switching between plus end-directed kinesins and myosin VA during centrifugal vesicular transport or, in the reverse direction, the switching between minus end-directed cytoplasmic dynein and myosin VI have been demonstrated [90, 91].
In conclusion, our combined findings about KIF1Bβ, in particular its established role as an axonal transporter and its current emerging role as a dendritic transporter in mammalian neurons, are direct pointers to a link of motor activity with Ca2+-signaling pathways with dendritic targeting of mRNA, translational regulation, and, eventually, synaptic plasticity, thus providing a firm basis for further characterization of this functional relationship.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Specificity of the oligonucleotide pairs used for the amplification of KIF1Bβ NM015074 vs. AX039604. PCRs with the two plasmid templates (shown at the bottom) using oligonucleotides designed for NM015074 or AX039604. These oligonucleotide sets (used in panels c–g) are specific for the respective isoform in both human and mouse. Negative control was no-template PCR (neg.) (EPS 744 kb)
Expression of KIF1B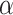 (NM008441) in mouse tissues and the NSC-34 cell line. a Schematic comparison of KIF1B
(NM008441) in mouse tissues and the NSC-34 cell line. a Schematic comparison of KIF1B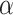 and KIF1Bβ proteins, indicating regions of identity and difference in the primary structure. While the N-terminal parts of the protein are identical, more downstream sequences (starting from the point indicated by an asterisk and stretching over the sequences highlighted in gray in KIF1B1
and KIF1Bβ proteins, indicating regions of identity and difference in the primary structure. While the N-terminal parts of the protein are identical, more downstream sequences (starting from the point indicated by an asterisk and stretching over the sequences highlighted in gray in KIF1B1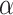 ) are completely different. The oligonucleotide pair (set 3, Online Resource Table S1), hybridizing to the areas indicated on the cartoon, was used to specifically amplify KIF1B
) are completely different. The oligonucleotide pair (set 3, Online Resource Table S1), hybridizing to the areas indicated on the cartoon, was used to specifically amplify KIF1B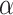 . The motor (yellow) and FHA (light blue) domains are highlighted. Numbers denote amino acid (aa) positions. b Equivalent RT-PCR reactions reveal some expression of this isoform in the hippocampus and spinal motor neurons, but no expression in the mouse neuroblastoma NSC-34 line (top panels). Equivalent reactions for L19 (bottom panel). (EPS 1141 kb)
. The motor (yellow) and FHA (light blue) domains are highlighted. Numbers denote amino acid (aa) positions. b Equivalent RT-PCR reactions reveal some expression of this isoform in the hippocampus and spinal motor neurons, but no expression in the mouse neuroblastoma NSC-34 line (top panels). Equivalent reactions for L19 (bottom panel). (EPS 1141 kb)
KIF1Bβ puncta are not co-localized with markers for mitochondria, lysosomes and the Golgi but with synaptic markers. a–d. Sets of triple fluorescence images of NSC-34 cells and their overlay, showing that the punctate KIF1Bβ labeling does not co-localize with the SOD2 protein (as a marker for mitochondria) (a), or protein LAMP2 (as a marker for lysosomes) (b), or 58K Golgin (Golgi apparatus) (c), but does co-localize with a synaptic marker, post-synaptic protein PSD-95 (d). Images (from left to right): cells labeled with Hoescht (DNA), compartment-specific antibodies (as indicated), anti-KIF1Bβ, and overlay of the set of three images. Scale bars 10 μm. (EPS 3899 kb)
Confirmation of the effect of PMA as a stimulator of PKC. Immunoblot of cortical synaptoneurosomes that were either non-treated, or incubated with phorbol ester PMA, a stimulator of PKC, or incubated with an inhibitor of PKC. The immunoblot was probed with an antibody against the phosphorylated form of MARCKS, one of PKC substrates (top panel). GAPDH was used as internal control to demonstrate equal protein loading in the three samples (bottom panel). (EPS 1005 kb)
KIF1Bβ puncta are co-localized with MAP2 staining. Immunofluorescence images of cortical neurons and their overlay showing that the punctate KIF1Bβ labeling co-localizes with MAP2, a marker for dendrites. Scale bar 10 μm. A detail of the co-localization is shown at higher magnification. Scale bar 5 μm. The histogram illustrates the percentage of KIF1Bβ puncta co-localized with MAP2. (EPS 12359 kb)
This video of a dendrite from a pEYFP-KIF1Bβ-transfected mouse cultured cortical neuron aims at providing a dynamic view to complement the still images from the live imaging shown in Fig. 8D. Fluorescent moving particles can be seen moving in the anterograde direction or in the retrograde direction or in rapid back and forth oscillating movements. Time-lapse frames were captured at a rate of one frame per 10 sec. For the analysis the time period of the first 180 sec was used. Here a movie of 100 frames, assembled with using Prism Video File Converter, is shown at 20 frames per second. (AVI 968 kb)
Equivalent video as video 1 but derived from the pEGFP- KIF1Bβ-C1-transfected mouse cultured cortical neuron shown in Fig. 8B. Fluorescence is spread over all processes but no particle movement can be observed. (AVI 968 kb)
Acknowledgments
We warmly thank Carsten W. Lederer for critically proof reading the manuscript and Teresa Ciotti for her help with primary neuron cultures. D.C.C. was supported by an A.G. Leventis Foundation (France) doctoral studentship and a short-term EMBO Fellowship to visit the laboratory of C.B. for part of this work. Work in this article was included in part in the doctoral thesis of D.C.C. E.P. is supported by an FWO fellowship (aspirant). This work was funded by the University of Cyprus (N.S.), VIB and Telethon (GGP10150)HEALTH-2009-2.1.2-1 EU-FP7 ‘SynSys’ and FWO (C.B.).
Abbreviations
- Arc
Activity-regulated cytoskeleton-associated protein
- CaM
Calmodulin
- CMT2A
Charcot-Marie-Tooth type 2A motor neuron disease
- DHPG
S-3,5,-dihydroxyphenylglycine
- DMEM
Dulbecco’s Modified Eagle’s Medium
- FMRP
Fragile X mental retardation protein
- FXR2P
Fragile X-related protein 2
- HBSS
Hank’s balanced salt solution
- KIF1Bβ
Kinesin family member 1Bβ
- MEM
Minimal essential medium (Eagle’s)
- Purα
Purine-rich element binding protein alpha
- RT-PCR
Reverse-transcription polymerase chain reaction
Footnotes
D. C. Charalambous, E. Pasciuto, C. Bagni, N. Santama contributed equally to the work.
Contributor Information
Claudia Bagni, Phone: +357-22-392757, FAX: +357-22-350557, Email: Claudia.Bagni@cme.vib-kuleuven.be.
Niovi Santama, Phone: +32-16-330944, FAX: +32-16-330939, Email: santama@ucy.ac.cy.
References
- 1.Lawrence CJ, et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nangaku M, et al. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell. 1994;79:1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 3.Mok H, Shin H, Kim S, Lee JR, Yoon J, Kim E. Association of the kinesin superfamily motor protein KIF1Balpha with postsynaptic density-95 (PSD-95), synapse-associated protein-97, and synaptic scaffolding molecule PSD-95/discs large/zona occludens-1 proteins. J Neurosci. 2002;22:5253–5258. doi: 10.1523/JNEUROSCI.22-13-05253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao C, et al. Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bbeta. Cell. 2001;105:587–597. doi: 10.1016/S0092-8674(01)00363-4. [DOI] [PubMed] [Google Scholar]
- 5.Zuchner S, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 6.Pantelidou M, Zographos SE, Lederer CW, Kyriakides T, Pfaffl MW, Santama N. Differential expression of molecular motors in the motor cortex of sporadic ALS. Neurobiol Dis. 2007;26:577–589. doi: 10.1016/j.nbd.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Hall DH, Hedgecock EM. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans . Cell. 1991;65:837–847. doi: 10.1016/0092-8674(91)90391-B. [DOI] [PubMed] [Google Scholar]
- 8.Klopfenstein DR, Vale RD. The lipid binding pleckstrin homology domain in UNC-104 kinesin is necessary for synaptic vesicle transport in Caenorhabditis elegans . Mol Biol Cell. 2004;15:3729–3739. doi: 10.1091/mbc.E04-04-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aulchenko YS, et al. Genetic variation in the KIF1B locus influences susceptibility to multiple sclerosis. Nat Genet. 2008;40:1402–1403. doi: 10.1038/ng.251. [DOI] [PubMed] [Google Scholar]
- 10.Booth DR, et al. Lack of support for association between the KIF1B rs10492972[C] variant and multiple sclerosis. Nat Genet. 2010;42:469–470. doi: 10.1038/ng0610-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hintzen RQ, AulchenkoYS, Ramagopalan S, Ebers, GC, van Duijn CM (2010) Reply to “Lack of support for association between the KIF1B rs10492972[C] variant and multiple sclerosis”. Nat Genet 40:1402–1403 [DOI] [PMC free article] [PubMed]
- 12.Lyons DA, Naylor SG, Scholze A, Talbot WS. Kif1b is essential for mRNA localization in oligodendrocytes and development of myelinated axons. Nat Genet. 2009;41:854–858. doi: 10.1038/ng.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlisio S, et al. The kinesin KIF1Bbeta acts downstream from EglN3 to induce apoptosis and is a potential 1p36 tumor suppressor. Genes Dev. 2008;22:884–893. doi: 10.1101/gad.1648608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeh IT, et al. A germline mutation of the KIF1B beta gene on 1p36 in a family with neural and nonneural tumors. Hum Genet. 2008;124:279–285. doi: 10.1007/s00439-008-0553-1. [DOI] [PubMed] [Google Scholar]
- 15.Munirajan AK, et al. KIF1Bbeta functions as a haploinsufficient tumor suppressor gene mapped to chromosome 1p36.2 by inducing apoptotic cell death. J Biol Chem. 2008;283:24426–24434. doi: 10.1074/jbc.M802316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, et al. Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat Genet. 2010;42:755–758. doi: 10.1038/ng.638. [DOI] [PubMed] [Google Scholar]
- 17.Ferrari F, Mercaldo V, Piccoli G, Sala C, Cannata S, Achsel T, Bagni C. The fragile X mental retardation protein-RNP granules show an mGluR1-dependent localization in the post-synaptic spines. Mol Cell Neurosci. 2007;34:343–354. doi: 10.1016/j.mcn.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 18.Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santama N, Krijnse-Locker J, Griffiths G, Noda Y, Hirokawa N, Dotti CG. KIF2beta, a new kinesin superfamily protein in non-neuronal cells, is associated with lysosomes and may be implicated in their centrifugal translocation. EMBO J. 1998;17:5855–5867. doi: 10.1093/emboj/17.20.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagy A, Delgado-Escueta AV. Rapid preparation of synaptosomes from mammalian brain using nontoxic isoosmotic gradient material (Percoll) J Neurochem. 1984;43:1114–1123. doi: 10.1111/j.1471-4159.1984.tb12851.x. [DOI] [PubMed] [Google Scholar]
- 21.Pilo BP, et al. Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. EMBO J. 2007;26:2991–3002. doi: 10.1038/sj.emboj.7601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Napoli I, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 23.Phillips GR, et al. The presynaptic particle web: ultrastructure, composition, dissolution, and reconstitution. Neuron. 2001;32:63–77. doi: 10.1016/S0896-6273(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 24.Di Penta. A, et al. Dendritic LSm1/CBP80-mRNPs mark the early steps of transport commitment and translational control. J Cell Biol. 2009;184:423–435. doi: 10.1083/jcb.200807033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christodoulou A, Lederer CW, Surrey T, Vernos I, Santama N. Motor protein KIFC5A interacts with Nubp1 and Nubp2, and is implicated in the regulation of centrosome duplication. J Cell Sci. 2006;119:2035–2047. doi: 10.1242/jcs.02922. [DOI] [PubMed] [Google Scholar]
- 26.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 27.Sans N, Wang PY, Du Q, Petralia RS, Wang YX, Nakka S, Blumer JB, Macara IG, Wenthold RJ. mPins modulates PSD-95 and SAP102 trafficking and influences NMDA receptor surphase expression. Nat Cell Biol. 2005;7:1179–1190. doi: 10.1038/ncb1325. [DOI] [PubMed] [Google Scholar]
- 28.Rashbad WS (2007-2009) ImageJ. U.S National Institutes of Health, Bethesda
- 29.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 30.Elvira G, et al. Characterization of an RNA granule from developing brain. Mol Cell Proteom. 2006;5:635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Cashman NR, et al. Neuroblastoma × spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev Dyn. 1992;194:209–221. doi: 10.1002/aja.1001940306. [DOI] [PubMed] [Google Scholar]
- 32.Titulaer MN, Ghijsen WE. Synaptoneurosomes. A preparation for studying subhippocampal GABAA receptor activity. Methods Mol Biol. 1997;72:49–59. doi: 10.1385/0-89603-394-5:49. [DOI] [PubMed] [Google Scholar]
- 33.Berchtold MW, Egli R, Rhyner JA, Hameister H, Strehler EE. Localization of the human bona fide calmodulin genes CALM1, CALM2, and CALM3 to chromosomes 14q24-q31, 2p21.1-p21.3, and 19q13.2-q13.3. Genomics. 1993;16:461–465. doi: 10.1006/geno.1993.1211. [DOI] [PubMed] [Google Scholar]
- 34.Friedberg F, Rhoads AR. Sequence homology of the 3′-untranslated region of calmodulin III in mammals. Mol Biol Rep. 2001;28:27–30. doi: 10.1023/A:1011928702566. [DOI] [PubMed] [Google Scholar]
- 35.Falley K, et al. Shank1 mRNA: dendritic transport by kinesin and translational control by the 5′ untranslated region. Traffic. 2009;10:844–857. doi: 10.1111/j.1600-0854.2009.00912.x. [DOI] [PubMed] [Google Scholar]
- 36.Hirokawa N. mRNA transport in dendrites: RNA granules, motors, and tracks. J Neurosci. 2006;26:7139–7142. doi: 10.1523/JNEUROSCI.1821-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin KC, Zukin RS. RNA trafficking and local protein synthesis in dendrites: an overview. J Neurosci. 2006;26:7131–7134. doi: 10.1523/JNEUROSCI.1801-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallia GL, Darbibian N, Johnson EM, Khalili K. Self-association of Purα is mediated by RNA. J Cell Biochem. 2000;74:334–348. doi: 10.1002/(SICI)1097-4644(19990901)74:3<334::AID-JCB3>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 39.Ohashi S, Koike K, Omori A, Ichinose S, Ohara S, Kobayashi S, Sato TA, Anzai K. Identification of mRNA/protein (mRNP) complexes containing Puralpha, mStaufen, fragile X protein, and myosin VA and their association with rough endoplasmic reticulum equipped with a kinesin motor. J Biol Chem. 2002;277:37804–37810. doi: 10.1074/jbc.M203608200. [DOI] [PubMed] [Google Scholar]
- 40.Brown V, Small K, Lakkis L, Feng Y, Gunter C, Wilkinson KD, Warren ST. Purified recombinant Fmrp exhibits selective RNA binding as an intrinsic property of the fragile X mental retardation protein. J Biol Chem. 1998;273:15521–15527. doi: 10.1074/jbc.273.25.15521. [DOI] [PubMed] [Google Scholar]
- 41.Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassell GJ. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 2005;4:350–359. doi: 10.1111/j.1601-183X.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 42.Ceman S, Brown V, Warren ST. Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the fragile X-related proteins as components of the complex. Mol Cell Biol. 1999;19:7925–7932. doi: 10.1128/mcb.19.12.7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Link W, et al. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci USA. 1995;92:5734–5738. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lyford GL, et al. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- 45.Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/S0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- 46.Vehey KJ, Hammond JW. Traffic control: regulation of kinesin motors. Nat Rev Mol Cell Biol. 2009;10:765–777. doi: 10.1038/nrm2782. [DOI] [PubMed] [Google Scholar]
- 47.Ma B, Savas JN, Yu MS, Culver BP, Chao MV, Tanese N. Huntingtin mediates dendritic transport of β-actin mRNA in rat neurons. Sci Rep. 2011;1:140. doi: 10.1038/srep00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guillaud L, Setou M, Hirokawa N. KIF17 dynamics and regulation of NR2B trafficking in hippocampal neurons. J Neurosci. 2003;23:131–140. doi: 10.1523/JNEUROSCI.23-01-00131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rook MS, Lu M, Kosik KS. CaMKII 3 untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J Neurosci. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Savas JN, Ma B, Deinhardt K, Culver BP, Restituito S, Wu L, Belasco JG, Chao MV, Tanese N. A role for huntington disease protein in dendritic RNA granules. J Biol Chem. 2010;285:13142–13153. doi: 10.1074/jbc.M110.114561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- 52.Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81:769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- 53.Hammond JW, Cai D, Blasius LT, Li Z, Jiang Y, Jih GT, Meyhofer E, Verhey KJ. Mammalian kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. Plos Biol. 2009;7:e72. doi: 10.1371/journal.pbio.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niwa S, Tanaka Y, Hirokawa N. KIF1Bbeta- and KIF1A-mediated axonal transport of presynaptic regulator Rab3 occurs in a GTP-dependent manner through DENN/MADD. Nat Cell Biol. 2008;10:1269–1279. doi: 10.1038/ncb1785. [DOI] [PubMed] [Google Scholar]
- 55.Kumar J, Choudhary BC, Metpally R, Zheng Q, Nonet ML, Ramanathan S, Klopferstein DR, Koushika SP. The Caenorhadditis elegans kinesin-3 motor UNC-104/KIF1A is degraded upon loss of specific binding to cargo. Plos Genetics. 2010;6:1–17. doi: 10.1371/journal.pgen.1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagner OI, Esposito A, Köhler B, Chen C-W, Shen C-P, Wu G-H, Butkevich E, Mandalapu S, Wenzel D, Wouters FS, Klopfenstein DR. Synaptic scaffolding protein SYD-2 clusters and activates kinesin-3 UNC-104 in C. elegans . Proc Natl Acad Sci USA. 2009;106:19605–19610. doi: 10.1073/pnas.0902949106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xue X, Jaulin F, Espenel C, Kreitzer G. PH-domain-dependent selective transport of p75 by kinesin-3 family motors in non-polarized MDCK cells. J Cell Sci. 2010;123:1732–1741. doi: 10.1242/jcs.056366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai Q, Sheng ZH. Molecular motors and synaptic assembly. Neuroscientist. 2009;15:78–89. doi: 10.1177/1073858408329511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matthies HJ, Miller RJ, Palfrey HC. Calmodulin binding to and cAMP-dependent phosphorylation of kinesin light chains modulate kinesin ATPase activity. J Biol Chem. 1993;268:11176–11187. [PubMed] [Google Scholar]
- 60.Nakamura N, Miyake Y, Matsushita M, Tanaka S, Inoue H, Kanazawa H. KIF1Bbeta2, capable of interacting with CHP, is localized to synaptic vesicles. J Biochem. 2002;132:483–491. doi: 10.1093/oxfordjournals.jbchem.a003246. [DOI] [PubMed] [Google Scholar]
- 61.Rusnak F, Mertz P. Calcineurin: form and function. Physiol Rev. 2000;80:1483–1521. doi: 10.1152/physrev.2000.80.4.1483. [DOI] [PubMed] [Google Scholar]
- 62.Macaskill AF, et al. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron. 2009;61:541–555. doi: 10.1016/j.neuron.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X, Schwarz TL. The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell. 2009;136:163–174. doi: 10.1016/j.cell.2008.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reddy VS, Reddy AS. The calmodulin-binding domain from a plant kinesin functions as a modular domain in conferring Ca2+-calmodulin regulation to animal plus- and minus-end kinesins. J Biol Chem. 2002;277:48058–48065. doi: 10.1074/jbc.M205459200. [DOI] [PubMed] [Google Scholar]
- 65.Moga DE, Calhoun ME, Chowdhury A, Worley P, Morrison JH, Shapiro ML. Activity-regulated cytoskeletal-associated protein is localized to recently activated excitatory synapses. Neuroscience. 2004;125:7–11. doi: 10.1016/j.neuroscience.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 66.Rodriguez JJ, et al. Long-term potentiation in the rat dentate gyrus is associated with enhanced Arc/Arg3.1 protein expression in spines, dendrites and glia. Eur J Neurosci. 2005;21:2384–2396. doi: 10.1111/j.1460-9568.2005.04068.x. [DOI] [PubMed] [Google Scholar]
- 67.Bramham CR, Worley PF, Moore MJ, Guzowski JF. The immediate early gene arc/arg3.1: regulation, mechanisms, and function. J Neurosci. 2008;28:11760–11767. doi: 10.1523/JNEUROSCI.3864-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chowdhury S, et al. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rial Verde EM, Lee-Osbourne J, Worley PF, Malinow R, Cline HT. Increased expression of the immediate-early gene arc/arg3.1 reduces AMPA receptor-mediated synaptic transmission. Neuron. 2006;52:461–474. doi: 10.1016/j.neuron.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shepherd JD, et al. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Messaoudi E, et al. Sustained Arc/Arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. J Neurosci. 2007;27:10445–10455. doi: 10.1523/JNEUROSCI.2883-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guzowski JF, et al. Recent behavioral history modifies coupling between cell activity and Arc gene transcription in hippocampal CA1 neurons. Proc Natl Acad Sci USA. 2006;103:1077–1082. doi: 10.1073/pnas.0505519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Plath N, et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–444. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 74.Miyashita T, Kubik S, Lewandowski G, Guzowski JF. A networks of neurons, networks of genes: an integrated view of memory consolidation. Neurobiol Learn Mem. 2008;89:269–284. doi: 10.1016/j.nlm.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Park S, et al. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor elF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 77.Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 78.Hirokawa N. From electron microscopy to molecular cell biology, molecular genetics and structural biology: intracellular transport and kinesin superfamily proteins, KIFs: genes, structure, dynamics and functions. J Electr Microsc (Tokyo) 2011;60(Suppl 1):S63–S92. doi: 10.1093/jmicro/dfr051. [DOI] [PubMed] [Google Scholar]
- 79.Ling SC, Fahner PS, Greenough WT, Gelfand VI. Transport of Drosophila fragile X mental retardation protein-containing ribonucleoprotein granules by kinesin-1 and cytoplasmic dynein. Proc Natl Acad Sci USA. 2004;101:17428–17433. doi: 10.1073/pnas.0408114101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dynes JL, Steward O. Dynamics of bidirectional transport of Arc mRNA in neuronal dendrites. J Comp Neurol. 2007;500:433–447. doi: 10.1002/cne.21189. [DOI] [PubMed] [Google Scholar]
- 81.Nonaka S, et al. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/S0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 82.Jimbo T, et al. Identification of a link between the tumour suppressor APC and the kinesin superfamily. Nat Cell Biol. 2002;4:323–327. doi: 10.1038/ncb779. [DOI] [PubMed] [Google Scholar]
- 83.Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 84.Bananis E, et al. Microtubule-dependent movement of late endocytic vesicles in vitro: requirements for Dynein and Kinesin. Mol Biol Cell. 2004;15:3688–3697. doi: 10.1091/mbc.E04-04-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brown CL, et al. Kinesin-2 is a motor for late endosomes and lysosomes. Traffic. 2005;6:1114–1124. doi: 10.1111/j.1600-0854.2005.00347.x. [DOI] [PubMed] [Google Scholar]
- 86.Nishimura T, Kato K, Yamaguchi T, Fukata Y, Ohno S, Kaibuchi K. Role of the PAR-3-KIF3 complex in the establishment of neuronal polarity. Nat Cell Biol. 2004;6:328–334. doi: 10.1038/ncb1118. [DOI] [PubMed] [Google Scholar]
- 87.Shi SH, Cheng T, Jan LY, Jan YN. APC and GSK-3beta are involved in mPar3 targeting to the nascent axon and establishment of neuronal polarity. Curr Biol. 2004;14:2025–2032. doi: 10.1016/j.cub.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 88.Haraguchi K, Hayashi T, Jimbo T, Yamamoto T, Akiyama T. Role of the kinesin-2 family protein, KIF3, during mitosis. J Biol Chem. 2006;281:4094–4099. doi: 10.1074/jbc.M507028200. [DOI] [PubMed] [Google Scholar]
- 89.Aronov S, Aranda G, Behar L, Ginzburg I. Visualization of translated tau protein in the axons of neuronal P19 cells and characterization of tau RNP granules. J Cell Sci. 2002;115:3817–3827. doi: 10.1242/jcs.00058. [DOI] [PubMed] [Google Scholar]
- 90.Kural C, Serpinskaya AS, Chou YH, Goldman RD, Gelfand VI, Selvin PR. Tracking melanosomes inside a cell to study molecular motors and their interaction. Proc Natl Acad Sci USA. 2007;104:5378–5382. doi: 10.1073/pnas.0700145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Watanabe TM, Higuchi H. Stepwise movements in vesicle transport of HER2 by motor proteins in living cells. Biophys J. 2007;92:4109–4120. doi: 10.1529/biophysj.106.094649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Specificity of the oligonucleotide pairs used for the amplification of KIF1Bβ NM015074 vs. AX039604. PCRs with the two plasmid templates (shown at the bottom) using oligonucleotides designed for NM015074 or AX039604. These oligonucleotide sets (used in panels c–g) are specific for the respective isoform in both human and mouse. Negative control was no-template PCR (neg.) (EPS 744 kb)
Expression of KIF1B (NM008441) in mouse tissues and the NSC-34 cell line. a Schematic comparison of KIF1B
(NM008441) in mouse tissues and the NSC-34 cell line. a Schematic comparison of KIF1B and KIF1Bβ proteins, indicating regions of identity and difference in the primary structure. While the N-terminal parts of the protein are identical, more downstream sequences (starting from the point indicated by an asterisk and stretching over the sequences highlighted in gray in KIF1B1
and KIF1Bβ proteins, indicating regions of identity and difference in the primary structure. While the N-terminal parts of the protein are identical, more downstream sequences (starting from the point indicated by an asterisk and stretching over the sequences highlighted in gray in KIF1B1 ) are completely different. The oligonucleotide pair (set 3, Online Resource Table S1), hybridizing to the areas indicated on the cartoon, was used to specifically amplify KIF1B
) are completely different. The oligonucleotide pair (set 3, Online Resource Table S1), hybridizing to the areas indicated on the cartoon, was used to specifically amplify KIF1B . The motor (yellow) and FHA (light blue) domains are highlighted. Numbers denote amino acid (aa) positions. b Equivalent RT-PCR reactions reveal some expression of this isoform in the hippocampus and spinal motor neurons, but no expression in the mouse neuroblastoma NSC-34 line (top panels). Equivalent reactions for L19 (bottom panel). (EPS 1141 kb)
. The motor (yellow) and FHA (light blue) domains are highlighted. Numbers denote amino acid (aa) positions. b Equivalent RT-PCR reactions reveal some expression of this isoform in the hippocampus and spinal motor neurons, but no expression in the mouse neuroblastoma NSC-34 line (top panels). Equivalent reactions for L19 (bottom panel). (EPS 1141 kb)
KIF1Bβ puncta are not co-localized with markers for mitochondria, lysosomes and the Golgi but with synaptic markers. a–d. Sets of triple fluorescence images of NSC-34 cells and their overlay, showing that the punctate KIF1Bβ labeling does not co-localize with the SOD2 protein (as a marker for mitochondria) (a), or protein LAMP2 (as a marker for lysosomes) (b), or 58K Golgin (Golgi apparatus) (c), but does co-localize with a synaptic marker, post-synaptic protein PSD-95 (d). Images (from left to right): cells labeled with Hoescht (DNA), compartment-specific antibodies (as indicated), anti-KIF1Bβ, and overlay of the set of three images. Scale bars 10 μm. (EPS 3899 kb)
Confirmation of the effect of PMA as a stimulator of PKC. Immunoblot of cortical synaptoneurosomes that were either non-treated, or incubated with phorbol ester PMA, a stimulator of PKC, or incubated with an inhibitor of PKC. The immunoblot was probed with an antibody against the phosphorylated form of MARCKS, one of PKC substrates (top panel). GAPDH was used as internal control to demonstrate equal protein loading in the three samples (bottom panel). (EPS 1005 kb)
KIF1Bβ puncta are co-localized with MAP2 staining. Immunofluorescence images of cortical neurons and their overlay showing that the punctate KIF1Bβ labeling co-localizes with MAP2, a marker for dendrites. Scale bar 10 μm. A detail of the co-localization is shown at higher magnification. Scale bar 5 μm. The histogram illustrates the percentage of KIF1Bβ puncta co-localized with MAP2. (EPS 12359 kb)
This video of a dendrite from a pEYFP-KIF1Bβ-transfected mouse cultured cortical neuron aims at providing a dynamic view to complement the still images from the live imaging shown in Fig. 8D. Fluorescent moving particles can be seen moving in the anterograde direction or in the retrograde direction or in rapid back and forth oscillating movements. Time-lapse frames were captured at a rate of one frame per 10 sec. For the analysis the time period of the first 180 sec was used. Here a movie of 100 frames, assembled with using Prism Video File Converter, is shown at 20 frames per second. (AVI 968 kb)
Equivalent video as video 1 but derived from the pEGFP- KIF1Bβ-C1-transfected mouse cultured cortical neuron shown in Fig. 8B. Fluorescence is spread over all processes but no particle movement can be observed. (AVI 968 kb)