Abstract
The human gut represents a highly complex ecosystem, which is densely colonized by a myriad of microorganisms that influence the physiology, immune function and health status of the host. Among the many members of the human gut microbiota, there are microorganisms that have co-evolved with their host and that are believed to exert health-promoting or probiotic effects. Probiotic bacteria isolated from the gut and other environments are commercially exploited, and although there is a growing list of health benefits provided by the consumption of such probiotics, their precise mechanisms of action have essentially remained elusive. Genomics approaches have provided exciting new opportunities for the identification of probiotic effector molecules that elicit specific responses to influence the physiology and immune function of their human host. In this review, we describe the current understanding of the intriguing relationships that exist between the human gut and key members of the gut microbiota such as bifidobacteria and lactobacilli, discussed here as prototypical groups of probiotic microorganisms.
Keywords: Probiotics, Bifidobacteria, Lactobacilli, Gut microbiota, Host–microbe cross-talk, Genomics
Introduction
The human gut is colonized by a myriad of bacteria, which collectively are estimated to outnumber somatic and germinal cells by a factor of ten [1]. Advances in sequencing technologies have allowed researchers unprecedented insights in niche-specific microbiota composition, revealing that the human body harbors dozens of different bacterial species in the stomach, hundreds on the skin, and thousands within the oral cavity and the large intestine [1–3]. Gut bacteria exert a profound impact on human physiology, immunology, and nutrition. The symbiotic relationship of the gut microbiota with the human host is the consequence of a long history of various co-evolutionary processes, where neither partner is disadvantaged, and where unique metabolic activities or other benefits are provided to both partners [4]. Compositional alterations that disturb the normal balance of the gut microbiota, a phenomenon referred to as gut dysbiosis, are considered, at least in part, responsible for metabolic disorders such as obesity, as well as debilitating and life-threatening diseases like inflammatory bowel disease and colon cancer [5]. Hence, ensuring the appropriate compositional balance of the intestinal microbiota is considered to be an effective means to maintain (gut) health.
According to the Food and Agriculture Organization of the United Nations and World Health Organization (FAO)/WHO criteria, probiotic bacteria are microorganisms that are consumed as live dietary supplements and that confer one or more health benefits to the host [6, 7]. Currently, the majority of probiotic bacteria that are commercially exploited belong to two genera, Bifidobacterium and Lactobacillus, both of which are common inhabitants of the human intestine. It is suggested that probiotic bacteria beneficially affect human health through different mechanisms that are typically divided into a number of general categories, involving strengthening of the intestinal barrier, modulation of the immune response, and antagonism of pathogens either by the production of antimicrobial compounds or through competition for mucosal binding sites [4]. Although there is suggestive evidence for each of these functional claims, the precise molecular mechanisms underlying most of these beneficial activities have essentially remained obscure.
The decoding of the genome sequence of a probiotic bacterium is arguably a prerequisite step in the discovery of the genetic basis of probiotic action of such a health-promoting bacterium. Such (comparative) probiotic genome analysis, also referred to as probiogenomics [8, 9], and its subsequent functional characterization has provided important insights into the diversity and evolution of probiotic bacteria, and has in several cases guided the unraveling of the molecular basis for a particular beneficial activity. These “omics” approaches allow the simultaneous analysis of very large numbers of genes, proteins, and/or metabolites, while the integration of genomic information with data on host gene expression in the human gut is expected to further expand our understanding of the roles of (probiotic) microbiota, microbe–microbe, and host–microbe interactions [10].
Here, we will assess how probiotic bacteria interact with the immune system of the host through the action of molecules that may also play a pivotal role in gut colonization and persistence. We will focus on the model gut probiotic bacteria that belong to Bifidobacterium spp. and Lactobacillus spp., which are phylogenetically distant relatives that each possess distinct beneficial attributes.
The human gut microbiota
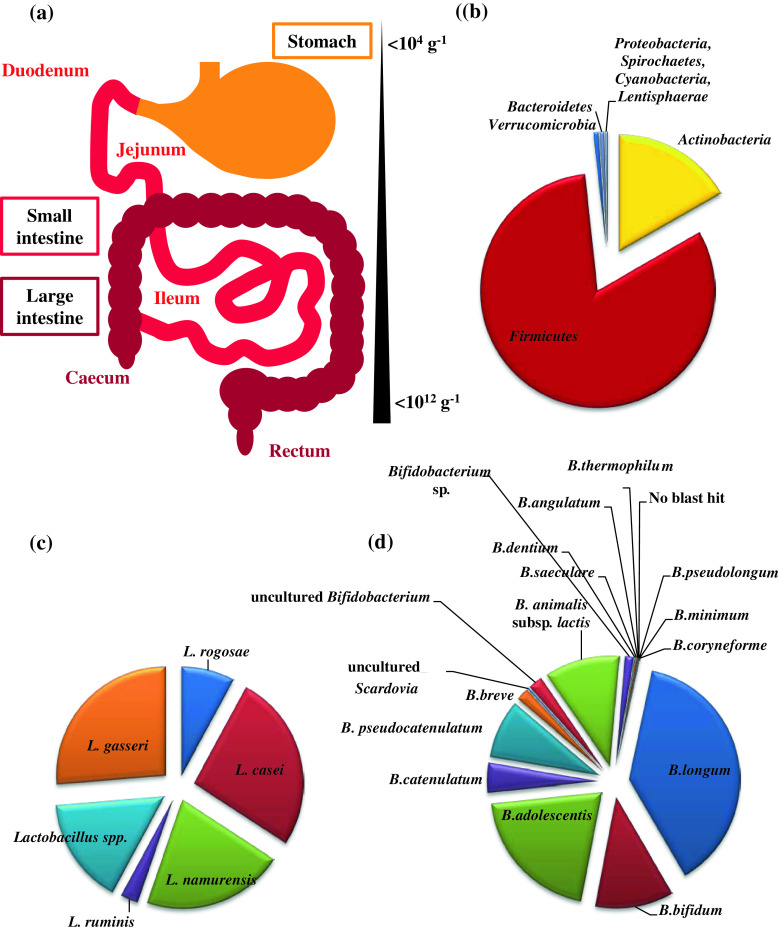
The trillions of commensal bacteria that make up the intestinal microbiota primarily belong to five microbial phyla: Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria and Fusobacteria, which are distributed throughout the gut in different numbers, likely as a result of varying microbial ecosystems (Fig. 1) [11]. The dominant phylum in the human adult microbiota is represented by Firmicutes, which in some individuals represents over 90 % of the total gut microbiota [12]. In contrast, these values are considerably different in the (human) infant intestine, where Actinobacteria and in particular representatives of the genus Bifidobacterium are numerically in the majority [12]. Furthermore, the infant gut microbiota has been shown to be much less complex than its adult equivalent in terms of total number of bacteria and encountered diversity of microbial taxa [1, 12]. Also, a relatively simplified composition (compared to the adult situation) was observed for the gut microbiota of the elderly population [13]. The diversification of microbial gut communities is influenced by several variables such as host factors (e.g., pH, bile acids, transit time, and mucus), environmental factors (e.g., nutrients and medication) and microbial factors (e.g., adhesion capability, bacterial enzymes, metabolic strategies, and bacteriocin production) [14]. Another key force that drives diversity and composition of the gut bacterial community is represented by bacteriophage [15, 16].
Fig. 1.
Schematic representation of the human gastrointestinal tract showing its different compartments and the relative abundance of bacteria (a). b The relative abundance of the main microbial phyla detected in the adult fecal samples [12]. c The aggregate microbiota composition of the genus Lactobacillus as determined from adult fecal sample analysis [12]. d The aggregate microbiota composition of the genus Bifidobacterium as determined from adult fecal sample analysis [12]
At birth, the human intestine is considered to be sterile; however, the gut is rapidly colonized after delivery. Natural sources of gut bacteria are represented by mother’s vaginal and fecal microbiota, as well as other environmental microbes [17]. Multiple factors may thus be responsible for the differences observed between the gut microbiota of infants, such as the delivery mode, local environment, type of infant feeding (breast-fed or formula-fed), gestational age, or antibiotic treatment [18, 19]. The delivery mode plays an important role in shaping the neonatal microbiota: for instance, Lactobacillus spp. dominate the gut microbiota of infants delivered vaginally as compared infants delivered by caesarean section [20, 21]. This is presumed to be a consequence of Lactobacillus spp. being among the dominant inhabitants of the urogenital ecosystem of healthy women [22, 23]. Colonization of the infant gut by L. rhamnosus and L. gasseri was shown to have a sinusoidal wave trend reaching 45 % proportional abundance in neonates up to 6 months of life. L. rhamnosus, L. paracasei, L. fermentum, L. gasseri and, though at lower frequencies, L. plantarum, L. delbrueckii and L. reuteri, were among the most commonly isolated bacteria in fecal samples obtained from Swedish infants during the first year of life [24]. Besides the abundance of lactobacilli, it is usually the bifidobacteria that dominate the intestinal microbiota of breast-fed full-term infants, while formula-fed, full-term infants also harbor bacteroides, clostridia, enterobacteria, and streptococci [25].
Another intriguing source of bacteria in this context is human milk, which has been proposed to act as an inoculum for breast-feeding infants, thus contributing to the establishment of a bifidobacteria-abundant infant gut microbiota [26]. Recently, comparative and functional genomic investigations have highlighted that certain human milk oligosaccharides (HMOs) play a crucial role in the development of the human infant gut microbiota [27]. Genomic analyses of bifidobacterial species that are typically associated with the infant gut microbiota, like Bifidobacterium longum ssp. infantis and Bifidobacterium bifidum, have revealed the existence of a gene set that encodes enzymes dedicated to the metabolism of HMOs [28, 29], which highlights the genetic specialization and presumed co-evolution of these bacteria to the infant gut and associated diet (see below).
Elements of the gut microbiota not only play an important role in the fermentation of indigestible complex plant polysaccharides and host-produced glycans (such as mucin) but are also believed to provide protection against pathogenic microorganisms. In addition, elements of the gut microbiota are thought to be required for the proper development of the host’s immune system [30]. There is evidence that the gut microbiota exerts a key role in inducing IgA production [31, 32], as well as maintaining the homeostasis of several T cell populations in different human body compartments including the gut, including regulatory T cells (Treg), and T helper 1 (TH1) and 17 (TH17) cells [33]. It has, for example, been reported that in vitro lactobacilli can modulate the host immune response suppressing inflammation by inducing CD4+CD25+Foxp3+RORγt− Treg cells [34, 35].
It has recently been demonstrated that commensal bacteria of the human gut produce molecules that mediate healthy immune responses and protect the host from inflammatory disease [36]. In this context, Mazmanian et al. [36] suggested that the human genome does not encode all functions required for immunological development, but, rather, that mammals depend on critical gene/metabolic products of their gut microbiota [36]. This has, for example, led to the discovery of key microbial gut molecules such as capsular polysaccharides that support and maintain immune functions of the human host (see below).
Additional metabolic functions of the gut microbiota include synthesis of vitamins (see review [37]), and bile acid biotransformation [38]. The major short-chain fatty acids produced by elements of the gut microbiota are acetate, butyrate, and propionate, with many other, yet minor, microbial metabolic end products such as lactate, ethanol, succinate, valerate, caproate, isobutyrate, 2-methyl-butyrate, and isovalerate [39]. One of the most important effects of these metabolites is their trophic effect on the intestinal epithelium [40]. In particular, butyrate is the preferred energy source for epithelial cells [41]. Furthermore, such metabolites have been described to have effects well beyond their place of production, as they have been reported to affect brain function [42, 43].
The human gut microbiota and diseases
Several studies have investigated possible relationships between gut microbiota composition and various diseases, such as necrotizing enterocolitis [44], type I and type II diabetes [45, 46], irritable bowel syndrome (IBD) [47, 48], atopic diseases and allergy [49], and colon cancer [50]. Most of these diseases have been associated with dysbiosis, which is a status in which the microbiota behaves abnormally as a consequence of an alteration of its composition, a change in its metabolic activity, and/or a shift in the local distribution of bacterial communities. However, the difficulty in accurately delineating a true core microbiota makes the concept of “dysbiosis” very hard to define.
Several factors may be responsible for microbial alterations of the gastrointestinal ecosystem, including antibiotic treatment, physical injuries or psychological stresses, radiation, altered peristalsis, and dietary shifts [51]. Among the diseases whose aetiology is linked to gut dysbiosis we will discuss: (1) autoimmune diseases, (2) IBD, and (3) allergy.
Autoimmune diseases
Autoimmune diseases occur when the body’s immune system attacks and destroys its own (healthy) cells and tissues as in the case of type I diabetes, celiac disease, inflammatory bowel diseases, and allergic asthma. Although most of these diseases have unknown causes, it has been suggested that dysbiosis may be an underlying cause [14]. Celiac disease is an inflammation of the small intestine, which is triggered by the storage proteins of wheat, barley, and rye. Analysis of the fecal microbiota of celiac patients showed a markedly numerical reduction in Bifidobacterium species, Clostridium histolyticum, C. lituseburense, and Faecalibacterium prausnitzii, coupled to increased proportions of Bacteroidetes/Prevotella [52, 53].
Type 1 diabetes, which is characterized by insulin deficiency caused by immune-mediated destruction of pancreatic beta cells, is thought to be triggered by environmental factors in genetically susceptible individuals. However, recent data suggest that alteration of the gut microbiota in rats is associated with progression of type 1 diabetes. Furthermore, a microbiota survey of diabetes-prone rats versus diabetes-resistant rats revealed a higher abundance of Lactobacillus and Bifidobacterium in diabetes-resistant rats [54]. These observations are supported in studies where L. casei Shirota has been shown to exert a suppressive effect on experimental models of immune disorders, such as arthritis, type I diabetes [55], murine lupus [56], and chronic IBD [57]. However, the molecular mechanisms responsible for such therapeutic effects are still unknown.
Gut microbiota in inflammatory bowel disease (IBD)
The initiating and perpetuating stimuli for immune dysregulation in IBD are not fully explained by genetic predisposition, since the number of known IBD-promoting gene mutations, such as CARD15, largely exceeds the clinical prevalence of the disease [58]. In addition, animal models have demonstrated that clinically identical mucosal inflammation can be initiated by different genetic immune defects [59]. These findings indicate that other modifying factors must be present, and suggest that changes in the luminal environment are crucial to the pathogenesis of IBD. Furthermore, the finding that experimental colitis does not develop in germ-free animals, while introduction of a single bacterial species can induce mucosal inflammation in animal models, coupled to the observation that diversion of the fecal stream is an effective treatment of active Crohn’s disease (CD), further implicates gut bacteria in the pathogenesis of IBD [60, 61]. As mentioned above, dysbiosis is considered the main cause of development of inflammatory diseases. Marked shifts in the luminal bowel microbiota, including loss of population diversity leading to a predominance of (pathogenic) bacterial species, such as Clostridium difficile, Bacteroides vulgatus and Escherichia coli, have been observed before relapse in IBD [62, 63]. Reduced bacterial diversity has been demonstrated in mucosal biopsies of patients with active IBD, with loss of commensal species such as Clostridium leptum, Eubacterium and bifidobacteria [64, 65]. Recent analysis of the fecal microbiota of patients suffering from IBD revealed an under-representation of the Firmicutes phylum, in particular the species Faecalibacterium prausnitzii [66].
Although inappropriate reaction by the innate immune system to intestinal bacteria has been implicated in the pathogenesis of IBD, it has also been demonstrated that the products of bacterial activity, such as butyrate, have a regulatory effect on inflammation in IBD [40, 67, 68].
Toki et al. [69] reported that the probiotic L. casei strain Shirota elicits anti-inflammatory (and anti-tumor) properties, which are linked to a cell envelope-associated polysaccharide–peptidoglycan complex, capable of inhibiting IL-6 production in LPS-stimulated lamina propria mononuclear cells isolated from IBD mice.
The probiotic mixture named VSL#3 is used as a supplement in the treatment of microbiota dysbiosis and a growing body of evidence reveals its ability to reduce colitis scores in IL10-deficient mice [70], to suppress ileitis [71] and colitis [72] in murine models, to prevent colorectal cancer in a DSS-mice model [73], and to ameliorate liver dysfunction [74].
Allergic disorders and human gut microbiota
The prevalence of allergic disorders has been steadily increasing in Western societies, and such conditions now comprise the most common chronic disease of childhood. There is an increase in the prevalence of atopic diseases [75], which represents a related group of conditions including allergic rhinoconjunctivitis, asthma, and atopic eczema. These are frequently associated with the generation of T helper (TH) cell 2-type cytokines, including IL-4, IL-5, and IL-3, which promote IgE production. The TH2 skewed immune type may be balanced by cytokines secreted by TH1, TH3, and T regulatory cells, partially as a result of stimulation by the gut microbiota [76]. The establishment of the gut microbiota provides an initial and impressive source of stimuli to the host. The route for the first allergic responses frequently arises from the gastrointestinal tract, and food allergy is a common problem in infants with atopic eczema. Aberrant barrier functions in the gut mucosa lead to increased antigen transfer across the mucosal barrier with transfer routes being altered, thereby evoking aberrant immune responses and release of proinflammatory cytokines with further impairment of the barrier functions. Such an enhanced inflammation state in turn leads to increases in intestinal permeability and results in a vicious circle of self-promoting allergenic responses and a more permanent dysregulation of the immune responses to ubiquitous antigens in (genetically) susceptible individuals.
The “hygiene hypothesis” proposes that the increased prevalence of allergic disorders in developed countries is due to a reduced early exposure to infectious agents that may alter the immune response and the immunoregulatory compartment, which may then affect the composition of the gut microbiota [77]. An alternative or derivative hypothesis is based on the changes in the intestinal microbiota that are due to increased antibiotic treatments and an altered infant diet [78]. In both cases, the regulatory mechanisms controlling the TH2 responses that are commonly associated with allergic disorders do not seem to function properly. However, it is still unclear to what extent the dysbiosis in the early stages of life can be attributed to antibiotic treatment and/or by human genetics.
Several studies have described the microbiota composition of infants who develop allergic disorders [79–81]. Bifidobacteria are predominant in the intestinal microbiota of a normal, healthy infant, with high numbers of Bifidobacterium breve, B. bifidum and B. longum, whereas bifidobacterial species such as B. adolescentis and B. pseudocatenulatum are more characteristic of an adult-type intestinal microbiota [82].
Recovery of the gut microbiota after perturbation
As described above, alteration of gut homeostasis might be the aetiological cause of a disease. The modification from homeostasis to a disease state can occur in different ways, including physical insult to the gut mucosa, disruption of the autochthonous microbiota through the use of antimicrobials (e.g., antibiotics), and virulence factors (for review, see [83]).
Restoration of the indigenous microbiota is expected to be driven by the action of several species, and is possibly subject to substantial microbial re-organization following the initial disruption of the microbiota. The mechanisms involved in the re-organization of the normal microbiota include co-aggregation, biosurfactant production, bacteriocin, and hydrogen peroxide biosynthesis, and competitive exclusion and modification of the diet composition (for review, see [83]). Co-aggregation of non-pathogens and pathogens interferes with the capability of pathogenic bacteria to infect the host; whereas biosurfactant biosynthesis helps to prevent the adhesion of pathogens to mucosal surfaces and, in the case of vaginal lactobacilli, the production of biosurfactants consisting of a mixture of lipids, proteins, and carbohydrates assist in the displacement of the uropathogenic E. coli, Enterococcus faecalis and Gardnerella vaginalis [84, 85]. Bacteriocin production is considered a key process in restoring homeostasis. Bacteriocins are small molecules synthetized by bacterial ribosomes that interfere with cell wall biosynthesis and that are capable of inhibiting growth of certain (Gram-positive) bacteria [86]. Bacteriocins are typically active against a narrow range of bacteria that are phylogenetically closely related to the producing organism, and they kill these cells by pore formation and leakage of cell contents [87]. There is good evidence that bacteriocin production is important for probiotic effects in the oral cavity, and indirect evidence for the gut environment. A recombinant Streptococcus mutans strain making abnormally high levels of mutacin, but lacking lactate dehydrogenase, out-competed other S. mutans strains to cause long term mono-colonization, yet did not cause caries [88]. In the gut environment, evidence for modulation of the microbiota is mostly indirect, from animal model experiments. For example, bacteriocin Abp118, synthetized by L. salivarius UCC118 [89], was shown to be important for host protection against Listeria monocytogenes infection in mice [90]. In a recent study of bacteriocin effect upon the total murine intestinal microbiota, we found that an Abp118-producing strain caused a relative increase in Bacteroidetes and Proteobacteria, and a decrease in Actinobacteria, as compared with an isogenic, non-bacteriocin-producing control, in a diet-induced obesity model [91]. As noted below, we also detected activity of the Abp118-producing L. salivarius UCC118 against Gram-negative bacteria in the porcine gut, while this antagonistic effect was lacking in pigs that harbored the genetically modified knock-out strain. Although many probiotic lactobacilli produce bacteriocins (e.g., L. casei, L. acidophilus), others apparently do not (e.g., L. rhamnosus LGG does not appear to, though it has the required genes). Furthermore, the ethical barriers to administering genetically modified micro-organisms to humans mean that bacteriocin-producing/non-producing wild-type/knock-out comparisons have not yet been performed in humans, to our knowledge. This is a significant impediment, because such a comparison would provide definitive proof for the importance of bacteriocin production to exert a probiotic effect.
Another critical mechanism of gut microbiota recovery is represented by competitive exclusion. Autochthonous members of the human gut microbiota have been postulated to be capable of effective competition with transient pathogens through the production of specific molecules such as capsular polysaccharides. An example is represented by the multiple capsular polysaccharides produced by Bacteroides fragilis, which are not only essential for persistence and gut colonization but also have an imuno-modulatory function, and help to exclude pathogens and restore homeostasis (see below) [92]. Nevertheless, B. fragilis is currently not considered to represent a probiotic bacterium, while the study carried out by Mazmanian’s group was conducted using a murine model rather than a human clinical trial. A perturbed intestinal microbiome has been associated with an increasing number of gastrointestinal and non-gastrointestinal diseases including C. difficile infection (CDI). It has become recognized that fecal microbiota transplantation can correct the dysbiosis that characterizes chronic CDI, and effect a seemingly safe, relatively inexpensive, and rapidly effective cure in the majority of treated patients. Recent studies involving on fecal microbiota transplantation have resulted in a gut microbiota restoration rate to normal homeostasis of more than 90 % [93–96].
The microbiota-modulating activity of lactic acid bacteria may be clinically beneficial. For example, a mixture of three bacteria (L. casei, L. bulgaricus, and Str. thermophilus) has been shown to reduce the incidence of antibiotic-associated diarrhoea (AAD) and C. difficile-associated diarrhoea (CDAD) [97].
In a recent randomized clinical trial, the supplementation of B. bifidum MIMBb75 revealed a significant reduction of the symptoms linked to irritable bowel syndrome (IBS) [98, 99].
Bifidobacteria as prototypical human gut commensals
Bifidobacteria were originally isolated by Tissier at the beginning of the last century from stool samples of breast-fed infants, and since then more than 38 taxa have been included in this bacterial genus (for a recent review, see [100]). From the very beginning, bifidobacteria attracted the interest of both scientists and food industry because of their perceived health-promoting activities, but they only recently have been subjected to more detailed molecular analyses through genome sequencing and functional genomics efforts [8]. These analyses have significantly expanded our understanding regarding the roles of gut-derived bifidobacteria in both microbe–microbe and host–microbe interactions, and have revealed a number of key molecules, produced by particular Bifidobacterium species, that promote their establishment in the human intestine. The strict co-evolution of many bifidobacterial species with the eukaryotic digestive tract has lead these microorganisms to acquire important colonization factors and metabolic abilities, which render them one of the major microbial players in gut colonization during the first stages of life. Furthermore, one may argue that such a highly developed colonization ability renders bifidobacteria very effective in colonization of the gut when this is covered by a thick microbial biofilm of other enteric commensals or pathogens.
Various sequencing projects have allowed the decoding of complete genome sequences of mainly enteric bifidobacterial species, such as B. longum ssp. longum [101, 102], B. longum ssp. infantis [28], B. bifidum [103], and B. breve [104]. Notably, the subsequent analyses of such genomic data reinforces the notion of strict genetic adaptation of these bifidobacterial taxa to the human gut and has revealed the existence of key bifidobacterial molecules that are responsible for gut colonization and survival of bifidobacteria in the human intestine. A clear example of such genomic adaptation is represented by the identification of a varied arsenal of genes encoding enzymes that are involved in the breakdown of complex carbohydrates derived from the diet (e.g., plant polysaccharides) or from the host (e.g., mucin). These carbohydrates cannot be digested by host-derived enzymes and will thus reach the large intestine in an intact form (for a recent review, see [105]). These specific metabolic properties of bifidobacteria are a clear indication as to how these microorganisms function in the human gut environment and how they positively affect human health status.
Glycans and glycoproteins produced by the host constitute a key carbon and energy source for gut bacteria, in particular in the distal colon where the availability and accessibility of simple carbohydrates is limited. Mucins represent a large polymeric network of carbohydrates that cover the intestinal mucosa, forming the main glycoprotein component of the mucus layer, and thus offering the microbiota a rich source of host-produced carbon and energy [106]. The main monosaccharide constituents of mucin-derived glycoproteins are N-acetylglucosamine, N-acetylgalactosamine, fructose, and galactose, and such glycoproteins are frequently decorated with sialic acid and sulphate groups [107]. Analysis of the chromosomal sequence of B. bifidum PRL2010, a strain isolated from infant stool, revealed a competitive nutrient-utilization strategy targeting mucin-derived O-glycans [103]. In silico analyses of the B. bifidum PRL2010 chromosome, together with functional genome approaches, revealed the existence of a gene set responsible for mucin metabolism. A significant proportion of the predicted proteome of B. bifidum PRL2010 is involved in mucin metabolism and comprises extracellular enzymes that include putative exo-α-sialidases, as well as a predicted 1,2-α-l-fucosidase, 1,3/4-α-l-fucosidase, and a putative cell wall-anchored endo-α-N-acetylgalactosaminidase. This enzymatic mucin breakdown machinery is completed by four N-acetyl-β-hexosaminidases, and four β-galactosidases, which allows the complete hydrolysis of the glycan component of mucin into monosaccharides [103]. A similar enzymatic repertoire has been identified and characterized in other members of the B. bifidum species [108–112], suggesting that this is a common and perhaps a species-defining property of this taxon. Recently, another human gut microbiota member, Akkermansia mucinophila, was identified as an important mucin degrader [113–116]. Mucin-degrading activity as operated by enteric bacteria such as A. mucinophila and B. bifidum may stimulate the secretion of colonic mucin, which constitutes an important feature especially in patients suffering from irritable bowel syndrome [117].
Mucin degradation as observed in B. bifidum seems to be a specialized form of a more general genetic adaptation of bifidobacteria to the human gut, which appears to be geared towards carbohydrate metabolism, where the ability to rapidly retrieve a specific carbon source from a particular environment/diet represents an important feature that would endow a bacterium with an undisputed ecological fitness [118].
Recently, the genetic requirements for carbohydrate uptake in B. bifidum PRL2010 have been investigated [119] and compared to those of other enteric bifidobacterial species, such as B. longum ssp. infantis, B. breve and B. longum ssp. longum [120]. Notably, PRL2010 contains a relatively limited number of such genes, which is thereby restricted to the uptake of a comparatively small number of carbohydrates [119]. These results suggest an interesting genetic strategy for efficient colonization and survival of PRL2010 in its ecological niche.
Another example of adaptation of bifidobacteria to the human gut is represented by the ability of several bifidobacterial species (e.g., B. longum ssp. infantis, B. bifidum, and B. breve) to utilize HMOs present in human milk [121, 122]. The genome sequence of an infant-derived bifidobacterial strain, B. longum ssp. infantis ATCC15697, has revealed the presence of an extensive gene set, located on a 43-Kb genomic island, whose encoded enzymes are involved in HMO breakdown (fucosidases, sialidases, a β-hexosaminidase, and β-galactosidases) and internalization (e.g., extracellular solute binding proteins and permeases of ABC transporter systems) [28]. Furthermore, genome analysis of this strain revealed the presence of an operon predicted to be involved in urea metabolism, which appears to be uniquely present in the genome of strains belonging to B. longum ssp. infantis [28]. Urea represents an important source of nitrogen in human milk and thus the presence of a urease-encoding operon in a bifidobacterial strain residing in the human gut of infants might represent another key sign of adaptation of B. longum ssp. infantis to this environment [122].
Further data concerning bifidobacterial adaptation to the human gut is represented by the ability of these bacteria to survive the stressful conditions that are encountered in the intestine such as exposure to an acid environment during passage through the stomach, to bile salts in the intestine, and osmotic stress as a result of dietary changes. Enteric bifidobacteria are endowed with a repertoire of molecular chaperones, bile efflux transporters, bile salt hydrolases, and ATPases to cope with these stressful conditions (for a review, see [123]). Furthermore, bifidobacteria possess various sensing systems and defence mechanisms against stress thereby allowing them to survive harsh conditions and sudden environmental changes. Microbial stress responses rely on coordinated gene expression in order to alter various cellular processes and structures (e.g., DNA metabolism, housekeeping genes, membrane composition), which act in concert to improve bacterial stress tolerance [123]. The existence of regulatory networks in the bifidobacterial genomes enables tight regulation of the expression of stress-induced molecular chaperones, and consequently allows the cell to rapidly react to various and sometimes complex environmental changes [124].
Lactobacilli as a model probiotic group
The genus Lactobacillus encompasses about 145 species, belongs to the phylum Firmicutes [125, 126], and is recognised for its extensive phylogenetic, phenotypic, and ecological diversity [127]. Enteric lactobacilli detected in adult human fecal samples include strains of the following species: L. acidophilus, L. crispatus, L. gasseri, L. plantarum, L. ruminis, L. casei, L. paracasei, and L. reuteri [128–130], while infant fecal samples have indicated an abundance of L. acidophilus, L. casei, L. paracasei, and L. salivarius [128, 131]. Interestingly, lactobacilli have also been detected at high relative levels in the stomach and in the small intestine of humans. This predominance of lactobacilli is exemplified in a recent analysis of ileal contents from ileostomy patients employing metagenomic and HITChip approaches [132, 133]. Whole genome sequencing efforts of probiotic lactobacilli have resulted in the complete genome sequencing of eight probiotic species including L. acidophilus, L. casei, L. fermentum, L. gasseri, L. johnsonii, L. plantarum, L. reuteri, and L. salivarius (reviewed by [8, 134]). Comparative genomics of the sequenced Lactobacillus strains has demonstrated that, in contrast to bifidobacteria, lactobacilli genomes show a considerable degree of auxotrophy for amino acids, which is compensated by an expanded set of peptide and amino acid transport functions [134]. An extension of this comparative analysis to include lactobacilli from intestinal, plant, or milk environments revealed functions that underscore their particular niche adaptation. In this context, typical milk-adapted lactobacilli such as L. delbrueckii ssp. bulgaricus and L. helveticus contain various genes that sustain their adaptation to growth on lactose, whereas genomes of other lactobacilli revealed the existence of sugar-uptake systems and mucus-binding proteins that are suggestive of a specific intestinal adaptation [135–137]. In addition, the finding that certain lactobacilli (are predicted to) encode typical intestinal enzymes such as bile salt hydrolase (BSH) is also suggestive of an ecological adaptation to the intestine [138]. For example, in silico analysis of the L. salivarius UCC118 genome revealed the presence of a large extra-chromosomal replicon, also known as a megaplasmid, that represents more than 10 % of the overall coding genome capacity of this strain, encoding biologically important features such bacteriocin production, a bile salt hydrolase, and components of the phosphoketolase pathway, reclassifying this organism as a facultative heterofermentative lactic acid bacterium [139].
Genome dissection of the probiotic strain L. rhamnosus GG demonstrated the presence of pili-encoding loci that have been shown to be pivotal in bacterial colonization and persistence within the human gut (see below). Similarly, the L. johnsonii NCC533 genome encodes fimbrial structures that were suggested to play a role in epithelial cell adhesion [140]. Other encoded lactobacilli molecules suggestive to be important for human gut adaptation have been discovered by the analysis of putative secretomes through in silico approaches. The prediction of the secretome of L. plantarum WCFS1 revealed the existence of at least 12 proteins, mainly hydrolases and transglycosylases that are predicted to be involved in the adherence to host components like collagen and mucin [141]. Similarly, putative adhesin proteins have been identified in the genome of L. acidophilus NCFM [142], and further studies confirmed their role in bacterial adherence to a human cell monolayer (Caco2 cells) [143].
The intestinal mucosa and probiotic action
As mentioned above one of the main routes through which probiotic bacteria are believed to act is by means of microbially produced molecules, which can be metabolites, proteins, or polysaccharides, that modulate the host immune system. It is now recognized that the innate and adaptive immune systems in the colon are linked to other important enteric functions such as adsorption of nutrients [144]. Anatomically, the intestinal mucosa is formed by a single-cell thick layer known as the epithelium and by an underlying layer, i.e. the lamina propria, which is virtually sterile and contains various immune cells. The epithelium is largely formed by columnar cells that perform key roles in the adsorption of nutrients and protecting from the passage of “non-self” luminal components like bacteria. Furthermore, Paneth cells and goblet cells in the epithelium contribute to innate immune defenses [145] by producing a vast array of antimicrobials (e.g., defensins and lysozyme) and mucin, the latter constituting a protective layer on the epithelium by preventing a direct epithelial contact with luminal bacteria [146–148]. Adaptive immunity in the intestinal mucosa is exploited by the lymphoid tissues associated with the lamina propria of the gut [gut-associated lymphoid tissue (GALT)] formed by B cells, macrophages, dendritic cells (DCs), and T cells [145]. It is suggested that, for probiotic action, the most important classes of T cells are the T helper (TH) and regulatory T cells (TReg) [149], where the latter regulate the production of the anti-inflammatory interleukin-10 (IL10). Macrophage cells are mainly involved in the elimination of pathogens, whereas DCs regulate both adaptive and innate immunity [150]. DCs are intimately associated with the lamina propria in an immature form that can activate the nuclear factor-kB (NF-kB) pathway, which allows the maturation and subsequent activation of DCs through exposure to so-called microbe-associated molecular patterns (MAMPs) [151].
Complex interaction between intestinal mucosa, macrophages, and DCs are responsible for the development of the intestinal immune homeostasis in response to elements of the gut microbiota [152, 153]. This process is regulated by the recognition of specific pattern recognition receptors (PRRs) recognizing MAMPs derived from bacteria, including probiotics. PRRs encompass a long list of receptors such as the Toll-like receptors (TLRs), NOD-like receptors (NRLs), and C-type lectin receptors (CLRs) [154].
Bacterial cell wall components and probiotic effectors
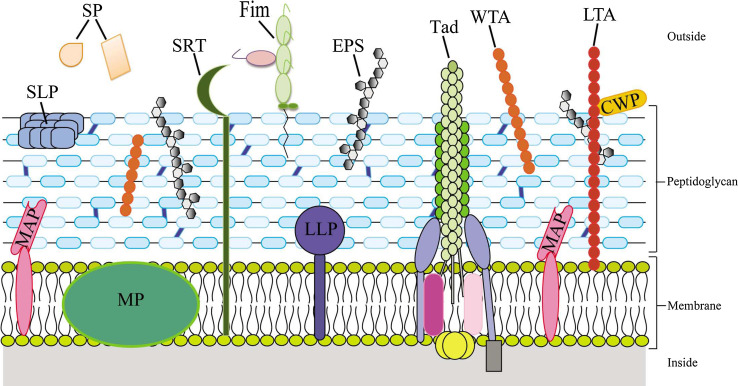
Efficient probiotic molecules are believed to cover or protrude from the microbial cell, thus being exposed to the external environment of the bacterial cell [144]. Many such molecules represent MAMPs that are recognized by specific PRRs, such as TLR, NLR and RIG-I-like receptors (RLR), produced by the host mucosa [148]. Several of the constituents of these extracellular macromolecular structures have been proposed to be directly involved in interactions with mammalian cells and thus in promoting a positive health effect (Fig. 2).
Fig. 2.
Schematic overview of the Gram-positive cell wall together with main macromolecular structures that have been implicated in host–microbe interaction. Specific components of the cell envelope are shown such as peptidoglycan layer, wall and lipoteichoic acids (WTA and LTA), exopolysaccharide (EPS), as well as secreted proteins (SP), membrane proteins (MP), cell wall-associated proteins (CWP), lipoproteins (LPP), membrane-associated proteins (MAP), surface layer proteins (SLP), fimbrial proteins (Fim), and tad proteins (TAD). Furthermore, the sortase-dependent assembly apparatus (SRT) is indicated
Among these microbial extracellular molecules are: (1) peptidoglycan; (2) (lipo)teichoic acids; (3) capsular polysaccharides; (4) fimbrial/pili structures; (5) surface layer proteins; (6) bacteriocins; and (7) sortase-dependent proteins (other than sortase-dependent pili). Notably, most of these cellular structures also occur in pathogenic bacteria, where they are considered to play key roles for survival and colonization within the human body. Apparently, the genetic strategies followed by microorganisms to establish themselves within their eukaryotic host are similar despite the opposing effect on the health status of their host. It is widely accepted that interactions between various bacteria and the human host can be categorized as a continuum, ranging from symbiosis to commensalisms, through to pathogenesis, thus reinforcing the notion that the mechanisms/effector molecules used for bacterial colonization are very similar for both pathogens and probiotic bacteria.
Peptidoglycan
Bacterial cell wall envelope components represent an important source of immune-modulatory action. For example, the cell wall constituent peptidoglycan is known to be the target for TLR2, which preferentially recognizes the diaminopimelic acid or the lysine moieties within the interpeptide bridge of peptidoglycan [155, 156]. Nevertheless, it has been claimed that the observed immune-modulatory action of peptidoglycan is due to lipoprotein contamination [157]. Alternative peptidoglycan-mediated signaling may occur by NOD-like receptors that interact with cell wall components such as the glutamyl-mesodiaminopimelic acid present in the peptidoglycan of Gram negative bacteria or through muramyl peptides of Gram-positive bacteria [158, 159]. Such immune-modulatory activities of peptidoglycan constituents have been demonstrated in the human gut commensal L. salivarius, which was shown by in vivo murine studies to induce local mucosal IL-10 production through the release of specific muropeptides [160]. Nevertheless, these muropeptides are not released by other gut lactobacilli, such as L. acidophilus NCFM, suggesting that the importance of peptidoglycan components as immune-modulators is strain-specific [160].
Peptidoglycan from L. casei has been reported to play a role in the inhibition of IL12p40 mRNA, leading to a reduction in secretion of IL12 and IL23, two cytokines that have been implicated in autoimmune diseases and inflammatory bowel diseases [161]. Furthermore, the probiotic strain L. casei Shirota has been shown to exert anti-inflammatory properties by means of a cell wall-derived polysaccharide–peptidoglycan complex capable of down-regulating IL-6 production in LPS-stimulated lamina propria mononuclear cells [162].
Teichoic acids
Gram-positive bacteria are capable of synthesizing teichoic acids that are organized on the cell surface as lipoteichoic acid (LTA), when anchored to the cytoplasmic membrane with a glycolipid anchor, and/or wall teichoic acid (WTA), when it is covalently linked to the peptidoglycan [163]. A large body of information is available regarding the LTA produced by lactobacilli and their immune-modulatory properties in terms of induction of tumor necrosis factor (TNF) by means of a TLR2-dependent mechanism [164–166]. It has been shown that differences in LTA composition are responsible for differential immunostimulatory effects. For instance, Ryu et al. [167] demonstrated that LTA from L. plantarum KCTC 10887BP causes TNF-α secretion and activates NF-κB transcription to a lower degree than LTA from S. aureus ATCC 6538 or B. subtilis ATCC 6633. The amino acid composition and stereoisomers present in the polyglycerophosphate backbone of LTA are critical for its biological activity; in fact, substitution of d-Alanine with l-Alanine causes significant reduction in cytokine secretion by human PBMCs [168]. Analysis of mutations in the dlt operon, which is responsible for D-alanylation of LTA, have elucidated the important role of dltABCD-encoded proteins on growth rate, morphology, and biofilm formation of L. rhamnosus GG [169] and on gastrointestinal tract colonization by L. reuteri 100–23 [170]. Furthermore, a L. plantarum WCFS1 mutant strain that is unable to D-alanylate LTA induces less TLR2-dependent secretion of pro-inflammatory cytokines but more production of anti-inflammatory cytokines than the wild-type strain, which improves the capability of the mutant to be protective against colitis relative to that of the wild-type [164]. Similar results have been achieved for other lactobacilli, such as L. rhamnosus GG and L. acidophilus NCFM, thereby suggesting that modifications of LTA increase anti-inflammatory immune-modulation [171]. In addition, progression of cancer polyposis in mice is arrested by oral administration of an LTA-deficient L. acidophilus strain NCK2025, due to the ability of this bacterial strain to enhance an anti-inflammatory subset of Treg [35]. In addition, LTA containing a low d-alanine content is responsible for the beneficial effect of L. plantarum EP007 on visceral pain perception in response to colorectal distension (CRD) in rats [172]. Moreover, pre-treatment with LTA from L. plantarum K8 has been shown to reduce LPS-induced TNF-α secretion in vitro and to repress endotoxic shock induced by LPS in BALB/c mice in vivo, suggesting that LTA induces tolerance in the host [173]. Furthermore, WTA in lactobacilli have been shown to induce IL-10 through a TLR2-dependent pathway [174]. Taken together, these studies validate the observation that the immunomodulatory effects of Lactobacillus spp. are indeed species- and strain-dependent, and that the profound range of modulatory effects on the innate immune system warrant further scientific scrutiny.
Capsular polysaccharides
Members of the genus Bacteroides have, furthermore, been shown to be capable of producing multiple capsular polysaccharides [175], several of which, such as polysaccharide A (PSA), possess immuno-modulatory properties [176]. These findings highlight the importance of capsular or exo-polysaccharides produced by gut commensals as important mediators of gut microbiota colonization, host–microbe cross-talk, and/or immune modulation. In this context, much of the current knowledge on capsular polysaccharides has focused on pathogens, and on the role of pathogen-encoded surface polysaccharides in pathogenesis, such as biofilm formation, tissue adherence, and anti-phagocytic activity during immune evasion [177]. In the context of capsular polysaccharides as mediators of host–commensal interactions, many studies have reported on the expression of multiple capsular polysaccharides by B. fragilis, and on attempts to eliminate capsular-mediated protection against the host-immune system, which was shown to result in growth defects of such acapsular mutants with subsequent spontaneous phenotypic reversion [92, 175, 178, 179]. The establishment of the expression of other capsular polysaccharides restores the reduced fitness of acapsular mutants in the gut. This represents an elegant way of identifying essential functions needed for bacterial colonization during host–symbiont mutualism. Shen et al. [180] reported a peculiar mechanism of communication between the commensal B. fragilis and the immune system of its host involving the delivery of PSA through outer membrane vesicles. Such cross-talk enhances IL10-producing Treg cells in vitro and may protect mice from experimental colitis in vivo [36, 180, 181].
The infant gut isolate B. breve UCC2003 was shown to possess a genome containing a gene cluster predicted to specify the production of two different cell surface-associated exopolysaccharides (sEPSs) [104]. Notably, the alternate biosynthesis of each of these sEPSs is directed by either half of this bidirectional gene cluster, which seems to be subject to phase variation by means of an apparent promoter inversion mechanism [182]. It has been shown that sEPS produced by UCC2003 facilitates increased stress tolerance to both bile and low pH, while it was also demonstrated that sEPS production influences gut persistence, but not, at least under the specific conditions tested, initial colonization of this strain. sEPS-producing B. breve UCC2003 cells elicit only a weak adaptive immune response as compared to isogenic mutants lacking this cell envelope-associated structure. Furthermore, it was established, in a murine model, that colonization of B. breve UCC2003 expressing sEPS, as compared to an isogenic EPS-negative derivative, reduced infection levels of the murine pathogen Citrobacter rodentium. These data implicate bifidobacterial sEPS in modulating microbe–host cross-talk, resulting in host-mediated immune tolerance of the commensal, while it also, in an as yet unknown manner, provides protection against a pathogen [182].
The role of capsular polysaccharides in other probiotic bacteria such as lactobacilli has also been investigated [183, 184]. Notably, in the L. casei strain Shirota, the capsular polysaccharide (CPS) has been shown to modulate the suppression of pro-inflammatory responses in macrophages [183]. In addition, down-regulation of CPS production in L. rhamnosus GG appears to be involved in adherence to intestinal epithelial cells. In contrast, up-regulation of CPS has been demonstrated to protect L. rhamnosus GG against intestinal innate immune factors like the antimicrobial peptide LL-37 [185]. However, difficulties related to the analysis of their complicated glycan structure have hindered the identification of their specific roles in the interaction with PRRs [186]. The levan exopolysaccharide from L. reuteri strain 100–23 has been shown to be required for colonization of the mouse gut and to exert an immune-stimulatory role inducing secretion of Treg (Fox3+) in the spleen [187]. As for LTA, the increasing evidence for surface polymers of lactobacilli modulating innate immune cells is an exciting development and merits more investigation.
Pili and fimbrial structures
Non-flagellar appendages were identified in bacteria in the early 1950s, and a relatively abundance of molecular data has been collected on the functions of pili in pathogenic microorganisms (for a review, see [188]). These structures are considered crucial in the initial adhesion of pathogens to host tissues during colonization and thus represent key effector molecules in pathogenesis. However, their presence was only very recently established in lactobacilli and bifidobacteria. Human gut colonization of the probiotic L. rhamnosus GG has been demonstrated to be linked to two so-called sortase-dependent pili, SpaCBA and SpaEF [189, 190]. Pili of L. rhamnosus GG were shown to be essential for efficient adherence to a human intestinal epithelial cell line as well as biofilm formation [191]. This study also showed that an L. rhamnosus GG pilus-defective derivative promoted elevated mRNA levels of the chemokine IL-8 in vitro compared to the wild-type, possibly involving an interaction between LTA and TLR2.
Sortase-dependent pili have also been discovered in bifidobacteria [192]. The publicly available genomes of enteric bifidobacteria, such as B. bifidum, B. longum ssp. longum, B. adolescentis and B. breve, encompass one to three predicted sortase-dependent pilus gene clusters, each of which are predicted to encode one major pilin subunit plus one or two minor pilin subunits, as well as a so-called sortase specifically dedicated to covalently assemble these pilin subunits [192]. In addition, transcriptomic analyses revealed that the genes encompassing the major and minor pilin subunits of each of these sortase-dependent pilus gene clusters are transcribed as a polycistronic mRNA, and that these genes are differentially expressed depending on the available carbohydrate in the growth medium [192]. Apart from sortase-dependent pili, a member of the so-called type IVb or tight-adherence (Tad) pilus family was recently shown to be specifically expressed by B. breve UCC2003 under in vivo conditions in a murine [104]. Mutational analysis of the corresponding tad locus of UCC2003 demonstrated that this cluster is essential for efficient in vivo murine gut colonization. Notably, the tad locus is highly conserved among all sequenced bifidobacterial strains, which supports the notion of a ubiquitous pilus-mediated host colonization and persistence mechanism for intestinal bifidobacteria [104, 144].
Surface layer proteins
The bacterial cell wall is decorated with cell wall-associated proteins that may be covalently anchored, e.g., N- or C-terminally anchored proteins, lipoproteins, and LPxTG-anchored proteins, or non-covalently anchored, e.g., proteins containing LysM, WxL, or SH3 domains, choline-binding domains, peptidoglycan-binding domains, and surface layer proteins, also known as S-layer proteins [134]. S-layer proteins are widely distributed in both Gram-positive and Gram-negative bacteria and are characterized as paracrystalline arrays that completely cover the cell surface [193]. Human gut lactobacilli such as L. acidophilus and L. crispatus have been shown to produce a clear S-layer, displaying typical adhesive properties [194, 195]. Notably, S-layer proteins of lactobacilli display additional roles involving competitive exclusion of intestinal pathogens as well as immune-modulatory activity through the induction of anti-inflammatory cytokines [196, 197]. Recently, the S-layer protein of L. helveticus MIMLh5 has been shown to mediate a stimulatory effect on innate immunity by triggering the expression of pro-inflammatory factors TNF-α and COX-2 in the human macrophage cell line U937 via recognition through TLR-2 [198].
Bacteriocins
Bacteriocins are antibacterial peptides produced by bacteria, which in the case of probiotic microorganisms has been shown to modulate pathogen growth in in vivo models [90]. Bacteriocin gene transcription in L. salivarius UCC118 has recently been demonstrated to be strongly up-regulated following adhesion to epithelial cells in vitro [199], suggesting that quorum sensing in the gut could have ecological aspects and biological consequences. Bacteriocin production may also affect microbiota composition in murine and porcine models [200, 201], although such effects in the latter study appear to be minor, and affecting genera and species not susceptible to the bacteriocin in vitro.
Bacteriocins produced by commensal bacteria may also possess a role in immuno-modulation. For example, the bacteriocin produced by L. plantarum WCFS1, plantaricin, has been shown to be pivotal in the synthesis of the anti-inflammatory cytokine IL-10 [202]. This evidence correlates nicely with publications suggesting that antimicrobial peptides impact on the immune response through changes in cytokine secretion as was shown in the porcine gut [203]. Notably, plantaricin has been shown to be expressed during WCFS1 colonization of mice, thus supporting the importance of this bacteriocin under in vivo conditions [204]. In contrast to the generally anti-inflammatory properties of plantaricin, levels of production of a L. salivarius bacteriocin have been shown to inversely correlate with the ability to restore transepithelial resistance in epithelial cell monolayers that had been treated with H2O2 [205].
Sortase-dependent proteins (other than sortase-dependent pili)
Surface proteins anchored by the classical SrtA sortase proteins may also play a role in the active dialogue between bacteria and host epithelium. For instance, the disruption of the sortase gene srtA negatively influences adhesion to epithelial cells by L. salivarius UCC118 [199, 206] and L. acidophilus NCFM [143]. Similar results were obtained when the sortase-encoding gene of L. casei BL23 was deleted, causing a reduction of over 60 % in the mutant’s ability to in vitro adhere to CaCO2 and HT29 cells [207]. Sortase-anchored proteins have easily recognizable cell wall-interacting motifs and domains, but a large variety of exposed functional domains [208], including proteins containing mucin-binding domains [209], one of which was shown by structural analysis to be an immunoglobulin binding domain [210].
Examples of host–microbe interaction models
Post-genomic approaches involving global genome transcription profiling as well as metabolomics of probiotic bacteria have been used in order to highlight the impact of a specific probiotic strain on the host. Most of these studies have been carried out using animal models, though a few have been conducted directly in humans [211, 212]. Recently, a study aimed at investigating the duodenal transcriptome profiles of health subjects who consumed L. plantarum WCFS1 represents one of the most significant advances in this field [212]. Notably, a clear stimulation of a regulatory network was observed involving JUN and NF-kB signaling cascades. In addition, IL-6, IL-10, TLR, and T cell receptor signaling pathways were considerably modified [212]. Taken together, these results indicate that L. plantarum WCFS1 enhances the alertness of the mucosal immune system while preserving immune homeostasis.
In another in vivo transcriptomics study involving human subjects [212], duodenal biopsies collected in a placebo-controlled cross-over design trial including L. rhamnosus GG consumption by healthy subjects demonstrated a correlation between transcriptional modulation and the observed probiotic effects. Among the human genes that were shown to exhibit enhanced transcription upon colonization with L. rhamnosus GG cells were those that are associated with TH 1 cell development and pathways that attenuate apoptosis, cell proliferation, and epithelial integrity [212].
Recently, a metabolic profiling study was published involving germ-free mice that were inoculated with human gut microbiota as a model for dietary interventions with probiotic L. rhamnosus or L. paracasei strains and/or galacto-oligosaccharides [213]. Apart from changes in the microbiota composition, the interventions were correlated with changes in different host-metabolic pathways including those involved in lipid profiles, gluconeogenesis, and amino acid and methylamine metabolism. Supplementation with a probiotic resulted in considerably different bile acid microbiota correlation networks, which may be linked to BSH activity expressed by lactobacilli.
Other intriguing post-genomics studies aimed at evaluation of the impact of probiotics on the gut microbiota of their host have been carried out using axenic mice as a host model [214, 215]. In this context, global transcription profiling investigations on axenic mice, that were mono-associated with B. thetaiotaomicron and subsequently colonized by B. longum ssp. longum NCC2705, showed that the presence of NCC2705 enhanced the diversity of polysaccharides targeted for breakdown by B. thetaiotaomicron including mannose and xylose-containing glycans [214]. The modifications in the transcriptional profiles of genes associated with polysaccharide utilization by B. longum ssp. longum and B. thetaiotaomicron may imply the existence of symbiosis between these microbes, where each species possesses a complementary set of glycosyl hydrolase activities, which when combined allow both to participate in a synergistic harvest of xylose and mannose-containing sugars. This study was also directed to explore the molecular impact, expressed in terms of transcriptome changes of members of the human microbiota, on a murine host was analyzed by studying the host epithelium mRNA response to co-colonization by B. longum ssp. longum NCC2705 and B. thetaiotaomicron [214]. Interestingly, the presence of B. thetaiotaomicron was shown to enhance the expression of tumor necrosis factor α-centered signaling in the corresponding murine host, whereas B. longum ssp. longum was shown to enhance the expression of the cytokine interferon-γ-mediated response genes and to reduce production of host antibacterial proteins such as regenerating islet-derived-3γ and pancreatitis-associated protein.
Bioengineering of probiotic bacteria
In recent years, bioengineering has been applied to probiotics, with the development of “designer probiotics”, for instance by expressing receptor-mimic structures to circumvent pathogens by blocking crucial ligand–receptor interactions [216]. In this context, a new type of bioengineered probiotic is constituted by an Escherichia coli strain expressing a lipopolysaccharide coupled to a Shiga toxin receptor, which is able to bind and neutralize toxins in the lumen of the intestine, limiting adhesion of pathogens to the gut mucosa [217]. Other bioengineered probiotic bacteria include genetically modified L. jensenii strains, which have been used to reduce the infections of HIV [218].
Nevertheless, despite the improved functionality of such bioengineered probiotics as compared to their natural counterparts, the use of recombinant bacteria in the food chain will meet with significant consumer reluctance, while they will also have to overcome regulatory requirements imposed by governmental authorities.
Conclusions
Most probiotic products that are currently available on the market contain the first generation of probiotic bacteria that were originally selected based on technological stability or on a variety of easily measurable phenotypes, such as ability to tolerate bile salts or survive GIT passage, but where their health-promoting activities were much less well defined and understood. This lack of knowledge has promoted a large body of research that intends to discover the precise mechanisms by which probiotic bacteria influence human health, and which would also lead to the identification of valid biomarkers that could be used for screening and validation processes of the next generation of probiotic microorganisms. All these investigations have benefited from the involvement of so-called “omics” approaches including (meta) genomics and functional analyses. In addition, molecular interaction models are currently being developed, although more are required, that monitor the activation of in vivo cellular and systemic responses in murine models and in diet intervention trials through the measurement of previously validated biomarkers. Together, the verified molecular models with functional and comparative genomics-based approaches should enable selection of the most appropriate probiotic strain, or strain combinations, for a specific health-promoting activity or enhancement of strain-processing and administration regimes that optimize the established health benefit. Ultimately, this may allow the selection of specific probiotics for a particular human genotype, analogous to personalized genomic medicine efforts.
As has been described here, the process of identification of “probiotic genes” or biomarkers for beneficial properties has just started. This has already provided the first results with the identification of a number of key molecules decorating the microbial cell surface (e.g., cell wall components, pili, and capsular surface polysaccharides) that mediate host–microbe interaction in specific human gut commensal bacteria and ultimately might be important in modulating the health status of the host. The rapidly progressing field of human intestinal metagenomics, together with the high-throughput functional screening of the metagenomic libraries (e.g., metatranscriptomics and/or metaproteomics), will no doubt aid in the identification of further probiotic biomarkers.
Regular consumption of probiotic bacteria has, in an increasing number of cases, been shown to be a crucial factor in the maintenance of gut homeostasis as well as in delivering health benefits. Thus, it is essential to identify and characterize those microbial activities and products that are required for the survival in, colonization of, and interaction with the human gut.
The probiotic concept is also changing in accordance with new insights on gut microbiota composition. As a consequence, the so far simplistic view of good and bad microorganisms will evolve in a way such that activity of a consortium or alternative species will be modulated through nutritional interventions (e.g., by supplementation of prebiotics) or otherwise, in order to elicit specific health-promoting effects. The integration of various scientific disciplines to delineate the gut microbiota composition coupled to the precise characterization of human physiological profiles will be crucial in order to improve our understanding of the complex host–microbe and microbe–microbe interactions occurring at the gut mucosal sites. This process will be essential in order to provide molecular criteria to predict the susceptibility of individuals to a particular probiotic therapy, and thus guarantee a positive outcome of probiotic application.
Acknowledgments
This work was financially supported by the Cariparma Bank Foundation to M.V., and by a FEMS Advanced Fellowship 2011 and an IRCSET Embark postdoctoral fellowship to F.T. This work was also financially supported by a PhD fellowship (Spinner 2013, Regione Emilia Romagna) to S.D. D.v.S., M.O.C.M., P.W.O.T., and L.F.B. are members of The Alimentary Pharmabiotic Centre; D.v.S. is also a member of the Alimentary Glycoscience Research Cluster, both funded by Science Foundation Ireland (SFI), through the Irish Government’s National Development Plan (Grant numbers 07/CE/B1368 and 08/SRC/B1393, respectively). M.O.C.M. is the recipient of a HRB postdoctoral fellowship (Grant No. PDTM/20011/9).
References
- 1.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci USA. 2007;104(8):2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marco ML, Pavan S, Kleerebezem M. Towards understanding molecular modes of probiotic action. Curr Opin Biotechnol. 2006;17(2):204–210. doi: 10.1016/j.copbio.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Tamboli CP, Neut C, Desreumaux P, Colombel JF. Dysbiosis as a prerequisite for IBD. Gut. 2004;53(7):1057. [PMC free article] [PubMed] [Google Scholar]
- 6.Organization FaAOotUNaWH . Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Cordoba: FAO/WHO; 2001. [Google Scholar]
- 7.FAO/WHO . Guidelines for the evaluation of probiotics in food. Canada: Ontario; 2002. [Google Scholar]
- 8.Ventura M, O’Flaherty S, Claesson MJ, Turroni F, Klaenhammer TR, van Sinderen D, O’Toole PW. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat Rev Microbiol. 2009;7(1):61–71. doi: 10.1038/nrmicro2047. [DOI] [PubMed] [Google Scholar]
- 9.Ventura M, Turroni F, van Sinderen D. Probiogenomics as a tool to obtain genetic insights into adaptation of probiotic bacteria to the human gut. Bioeng Bugs. 2012;3(2):73–79. doi: 10.4161/bbug.18540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joyce AR, Palsson BO. The model organism as a system: integrating ‘omics’ data sets. Nat Rev Mol Cell Biol. 2006;7(3):198–210. doi: 10.1038/nrm1857. [DOI] [PubMed] [Google Scholar]
- 11.Rajilic-Stojanovic M, Smidt H, de Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol. 2007;9(9):2125–2136. doi: 10.1111/j.1462-2920.2007.01369.x. [DOI] [PubMed] [Google Scholar]
- 12.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O’Toole PW, van Sinderen D, Marchesi JR, Ventura M. Diversity of bifidobacteria within the infant gut microbiota. PLoS One. 2012;7(5):e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, Stanton C, van Sinderen D, O’Connor M, Harnedy N, O’Connor K, Henry C, O’Mahony D, Fitzgerald AP, Shanahan F, Twomey C, Hill C, Ross RP, O’Toole PW. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prakash S, Rodes L, Coussa-Charley M, Tomaro-Duchesneau C. Gut microbiota: next frontier in understanding human health and development of biotherapeutics. Biologics. 2011;5:71–86. doi: 10.2147/BTT.S19099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ventura M, Sozzi T, Turroni F, Matteuzzi D, van Sinderen D. The impact of bacteriophages on probiotic bacteria and gut microbiota diversity. Genes Nutr. 2011;6(3):205–207. doi: 10.1007/s12263-010-0188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466(7304):334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaishampayan PA, Kuehl JV, Froula JL, Morgan JL, Ochman H, Francino MP. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. GenomeBiol Evol. 2010;2:53–66. doi: 10.1093/gbe/evp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penders J, Stobberingh EE, Thijs C, Adams H, Vink C, van Ree R, van den Brandt PA. Molecular fingerprinting of the intestinal microbiota of infants in whom atopic eczema was or was not developing. Clin Exp Allergy. 2006;36(12):1602–1608. doi: 10.1111/j.1365-2222.2006.02599.x. [DOI] [PubMed] [Google Scholar]
- 19.Westerbeek EA, van den Berg A, Lafeber HN, Knol J, Fetter WP, van Elburg RM. The intestinal bacterial colonisation in preterm infants: a review of the literature. Clin Nutr. 2006;25(3):361–368. doi: 10.1016/j.clnu.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arboleya S, Ruas-Madiedo P, Margolles A, Solis G, Salminen S, de Los Reyes-Gavilan CG, Gueimonde M. Characterization and in vitro properties of potentially probiotic Bifidobacterium strains isolated from breast-milk. Int J Food Microbiol. 2011;149(1):28–36. doi: 10.1016/j.ijfoodmicro.2010.10.036. [DOI] [PubMed] [Google Scholar]
- 22.Hyman RW, Fukushima M, Diamond L, Kumm J, Giudice LC, Davis RW. Microbes on the human vaginal epithelium. Proc Natl Acad Sci USA. 2005;102(22):7952–7957. doi: 10.1073/pnas.0503236102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahrne S, Lonnermark E, Wold AE, Aberg N, Hesselmar B, Saalman R, Strannegard IL, Molin G, Adlerberth I. Lactobacilli in the intestinal microbiota of Swedish infants. Microbes Infect. 2005;7(11–12):1256–1262. doi: 10.1016/j.micinf.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Yoshioka H, Iseki K, Fujita K. Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics. 1983;72(3):317–321. [PubMed] [Google Scholar]
- 26.Gueimonde M, Laitinen K, Salminen S, Isolauri E. Breast milk: a source of bifidobacteria for infant gut development and maturation? Neonatology. 2007;92(1):64–66. doi: 10.1159/000100088. [DOI] [PubMed] [Google Scholar]
- 27.Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4653–4658. doi: 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci USA. 2008;105(48):18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshida E, Sakurama H, Kiyohara M, Nakajima M, Kitaoka M, Ashida H, Hirose J, Katayama T, Yamamoto K, Kumagai H. Bifidobacterium longum subsp. infant is uses two different beta-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology. 2012;22(3):361–368. doi: 10.1093/glycob/cwr116. [DOI] [PubMed] [Google Scholar]
- 30.Kaplan JL, Shi HN, Walker WA. The role of microbes in developmental immunologic programming. Pediatr Res. 2011;69(6):465–472. doi: 10.1203/PDR.0b013e318217638a. [DOI] [PubMed] [Google Scholar]
- 31.Klaasen HLBM, Vanderheijden PJ, Stok W, Poelma FGJ, Koopman JP, Vandenbrink ME, Bakker MH, Eling WMC, Beynen AC. Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune-system of mice. Infect Immun. 1993;61(1):303–306. doi: 10.1128/iai.61.1.303-306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67(4):1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31(4):677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 34.Smith JM. Atopy and asthma: an epidemic of unknown cause. J Allergy Clin Immunol. 2005;116(1):231–232. doi: 10.1016/j.jaci.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Khazaie K, Zadeh M, Khan MW, Bere P, Gounari F, Dennis K, Blatner NR, Owen JL, Klaenhammer TR, Mohamadzadeh M. Abating colon cancer polyposis by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc Natl Acad Sci USA. 2012;109(26):10462–10467. doi: 10.1073/pnas.1207230109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 37.Leblanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M (2012) Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol (in press) [DOI] [PubMed]
- 38.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O’Sullivan O, Fitzgerald GF, Deane J, O’Connor M, Harnedy N, O’Connor K, O’Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP, O’Toole PW. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488(7410):178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 40.Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40(3):235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 41.Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70(2):567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- 42.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13(10):701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 43.Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci USA. 2011;108(38):16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mai V, Young CM, Ukhanova M, Wang X, Sun Y, Casella G, Theriaque D, Li N, Sharma R, Hudak M, Neu J. Fecal microbiota in premature infants prior to necrotizing enterocolitis. PLoS One. 2011;6(6):e20647. doi: 10.1371/journal.pone.0020647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, Drew JC, Ilonen J, Knip M, Hyoty H, Veijola R, Simell T, Simell O, Neu J, Wasserfall CH, Schatz D, Atkinson MA, Triplett EW. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5(1):82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sorensen SJ, Hansen LH, Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5(2):e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carroll IM, Ringel-Kulka T, Keku TO, Chang YH, Packey CD, Sartor RB, Ringel Y. Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2011;301(5):G799–G807. doi: 10.1152/ajpgi.00154.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S, Weidler EM, Qin X, Coarfa C, Milosavljevic A, Petrosino JF, Highlander S, Gibbs R, Lynch SV, Shulman RJ, Versalovic J. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141(5):1782–1791. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe J, Fujiwara R, Sasajima N, Ito S, Sonoyama K. Administration of antibiotics during infancy promoted the development of atopic dermatitis-like skin lesions in NC/Nga mice. Biosci Biotechnol Biochem. 2010;74(2):358–363. doi: 10.1271/bbb.90709. [DOI] [PubMed] [Google Scholar]
- 50.Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, Corthier G, Tran Van Nhieu J, Furet JP. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6(1):e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hawrelak JA, Myers SP. The causes of intestinal dysbiosis: a review. Altern Med Rev. 2004;9(2):180–197. [PubMed] [Google Scholar]
- 52.De Palma G, Nadal I, Medina M, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol. 2010;10:63. doi: 10.1186/1471-2180-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nadal I, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J Med Microbiol. 2007;56(Pt 12):1669–1674. doi: 10.1099/jmm.0.47410-0. [DOI] [PubMed] [Google Scholar]
- 54.Roesch LF, Lorca GL, Casella G, Giongo A, Naranjo A, Pionzio AM, Li N, Mai V, Wasserfall CH, Schatz D, Atkinson MA, Neu J, Triplett EW. Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. ISME J. 2009;3(5):536–548. doi: 10.1038/ismej.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsuzaki T, Nagata Y, Kado S, Uchida K, Kato I, Hashimoto S, Yokokura T. Prevention of onset in an insulin-dependent diabetes mellitus model, NOD mice, by oral feeding of Lactobacillus casei. Acta Pathol Microbiol Immunol Scand. 1997;105(8):643–649. doi: 10.1111/j.1699-0463.1997.tb05066.x. [DOI] [PubMed] [Google Scholar]
- 56.Mike A, Nagaoka N, Tagami Y, Miyashita M, Shimada S, Uchida K, Nanno M, Ohwaki M. Prevention of B220+ T cell expansion and prolongation of lifespan induced by Lactobacillus casei in MRL/lpr mice. Clin Exp Immunol. 1999;117(2):368–375. doi: 10.1046/j.1365-2249.1999.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsumoto S, Hara T, Hori T, Mitsuyama K, Nagaoka M, Tomiyasu N, Suzuki A, Sata M. Probiotic Lactobacillus-induced improvement in murine chronic inflammatory bowel disease is associated with the down-regulation of pro-inflammatory cytokines in lamina propria mononuclear cells. Clin Exp Immunol. 2005;140(3):417–426. doi: 10.1111/j.1365-2249.2005.02790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaya DR, Russell RK, Nimmo ER, Satsangi J. New genes in inflammatory bowel disease: lessons for complex diseases? Lancet. 2006;367(9518):1271–1284. doi: 10.1016/S0140-6736(06)68345-1. [DOI] [PubMed] [Google Scholar]
- 59.Bouma G, Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol. 2003;3(7):521–533. doi: 10.1038/nri1132. [DOI] [PubMed] [Google Scholar]
- 60.Uronis JM, Muhlbauer M, Herfarth HH, Rubinas TC, Jones GS, Jobin C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One. 2009;4(6):e6026. doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Willing B, Halfvarson J, Dicksved J, Rosenquist M, Jarnerot G, Engstrand L, Tysk C, Jansson JK. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm Bowel Dis. 2009;15(5):653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 62.Meyer AM, Ramzan NN, Loftus EV, Jr, Heigh RI, Leighton JA. The diagnostic yield of stool pathogen studies during relapses of inflammatory bowel disease. J Clin Gastroenterol. 2004;38(9):772–775. doi: 10.1097/01.mcg.0000139057.05297.d6. [DOI] [PubMed] [Google Scholar]
- 63.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Dore J. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55(2):205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Favier C, Neut C, Mizon C, Cortot A, Colombel JF, Mizon J. Fecal beta-D-galactosidase production and Bifidobacteria are decreased in Crohn’s disease. Dig Dis Sci. 1997;42(4):817–822. doi: 10.1023/a:1018876400528. [DOI] [PubMed] [Google Scholar]
- 65.Mangin I, Bonnet R, Seksik P, Rigottier-Gois L, Sutren M, Bouhnik Y, Neut C, Collins MD, Colombel JF, Marteau P, Dore J. Molecular inventory of faecal microflora in patients with Crohn’s disease. FEMS Microbiol Ecol. 2004;50(1):25–36. doi: 10.1016/j.femsec.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 66.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottiere HM, Dore J, Marteau P, Seksik P, Langella P. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105(43):16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Segain JP, Raingeard de la Bletiere D, Bourreille A, Leray V, Gervois N, Rosales C, Ferrier L, Bonnet C, Blottiere HM, Galmiche JP. Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn’s disease. Gut. 2000;47(3):397–403. doi: 10.1136/gut.47.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sanderson IR. Short chain fatty acid regulation of signaling genes expressed by the intestinal epithelium. J Nutr. 2004;134(9):2450S–2454S. doi: 10.1093/jn/134.9.2450S. [DOI] [PubMed] [Google Scholar]
- 69.Toki S, Kagaya S, Shinohara M, Wakiguchi H, Matsumoto T, Takahata Y, Morimatsu F, Saito H, Matsumoto K. Lactobacillus rhamnosus GG and Lactobacillus casei suppress Escherichia coli-induced chemokine expression in intestinal epithelial cells. Int Arch Allergy Immunol. 2009;148(1):45–58. doi: 10.1159/000151505. [DOI] [PubMed] [Google Scholar]
- 70.Madsen K, Cornish A, Soper P, McKaigney C, Jijon H, Yachimec C, Doyle J, Jewell L, De Simone C. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology. 2001;121(3):580–591. doi: 10.1053/gast.2001.27224. [DOI] [PubMed] [Google Scholar]
- 71.Pagnini C, Saeed R, Bamias G, Arseneau KO, Pizarro TT, Cominelli F. Probiotics promote gut health through stimulation of epithelial innate immunity. Proc Natl Acad Sci USA. 2010;107(1):454–459. doi: 10.1073/pnas.0910307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Uronis JM, Arthur JC, Keku T, Fodor A, Carroll IM, Cruz ML, Appleyard CB, Jobin C. Gut microbial diversity is reduced by the probiotic VSL#3 and correlates with decreased TNBS-induced colitis. Inflamm Bowel Dis. 2011;17(1):289–297. doi: 10.1002/ibd.21366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bassaganya-Riera J, Viladomiu M, Pedragosa M, De Simone C, Hontecillas R. Immunoregulatory mechanisms underlying prevention of colitis-associated colorectal cancer by probiotic bacteria. PLoS One. 2012;7(4):e34676. doi: 10.1371/journal.pone.0034676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Loguercio C, Federico A, Tuccillo C, Terracciano F, D’Auria MV, De Simone C, Del Vecchio Blanco C. Beneficial effects of a probiotic VSL#3 on parameters of liver dysfunction in chronic liver diseases. J Clin Gastroenterol. 2005;39(6):540–543. doi: 10.1097/01.mcg.0000165671.25272.0f. [DOI] [PubMed] [Google Scholar]
- 75.Isolauri E. Dietary modification of atopic disease: use of probiotics in the prevention of atopic dermatitis. Curr Allergy Asthma Rep. 2004;4(4):270–275. doi: 10.1007/s11882-004-0070-9. [DOI] [PubMed] [Google Scholar]
- 76.Rautava S, Ruuskanen O, Ouwehand A, Salminen S, Isolauri E. The hygiene hypothesis of atopic disease–an extended version. J Pediatr Gastroenterol Nutr. 2004;38(4):378–388. doi: 10.1097/00005176-200404000-00004. [DOI] [PubMed] [Google Scholar]
- 77.Okada H, Kuhn C, Feillet H, Bach JF. The ‘hygiene hypothesis’ for autoimmune and allergic diseases: an update. Clin Exp Immunol. 2010;160(1):1–9. doi: 10.1111/j.1365-2249.2010.04139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Noverr MC, Huffnagle GB. The ‘microflora hypothesis’ of allergic diseases. Clin Exp Allergy. 2005;35(12):1511–1520. doi: 10.1111/j.1365-2222.2005.02379.x. [DOI] [PubMed] [Google Scholar]
- 79.Kalliomaki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet. 2001;357(9262):1076–1079. doi: 10.1016/S0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 80.Bjorksten B, Sepp E, Julge K, Voor T, Mikelsaar M. Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol. 2001;108(4):516–520. doi: 10.1067/mai.2001.118130. [DOI] [PubMed] [Google Scholar]
- 81.Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, Adams H, van Ree R, Stobberingh EE. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA birth cohort study. Gut. 2007;56(5):661–667. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ventura M, Canchaya C, Fitzgerald GF, Gupta RS, van Sinderen D. Genomics as a means to understand bacterial phylogeny and ecological adaptation: the case of bifidobacteria. Antonie Van Leeuwenhoek. 2007;91(4):351–372. doi: 10.1007/s10482-006-9122-6. [DOI] [PubMed] [Google Scholar]
- 83.Reid G, Younes JA, Van der Mei HC, Gloor GB, Knight R, Busscher HJ. Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat Rev Microbiol. 2011;9(1):27–38. doi: 10.1038/nrmicro2473. [DOI] [PubMed] [Google Scholar]
- 84.Velraeds MM, van de Belt-Gritter B, van der Mei HC, Reid G, Busscher HJ. Interference in initial adhesion of uropathogenic bacteria and yeasts to silicone rubber by a Lactobacillus acidophilus biosurfactant. J Med Microbiol. 1998;47(12):1081–1085. doi: 10.1099/00222615-47-12-1081. [DOI] [PubMed] [Google Scholar]
- 85.Reid G, Kang YS, Lacerte M, Tieszer C, Hayes KC. Bacterial biofilm formation on the bladder epithelium of spinal cord injured patients. II. Toxic outcome on cell viability. Paraplegia. 1993;31(8):494–499. doi: 10.1038/sc.1993.80. [DOI] [PubMed] [Google Scholar]
- 86.Asaduzzaman SM, Sonomoto K. Lantibiotics: diverse activities and unique modes of action. J Biosci Bioeng. 2009;107(5):475–487. doi: 10.1016/j.jbiosc.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 87.Cotter PD, Hill C, Ross RP. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005;3(10):777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 88.Hillman JD. Genetically modified Streptococcus mutans for the prevention of dental caries. Antonie Van Leeuwenhoek. 2002;82(1–4):361–366. [PubMed] [Google Scholar]
- 89.Flynn S, van Sinderen D, Thornton GM, Holo H, Nes IF, Collins JK. Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology. 2002;148((Pt 4)):973–984. doi: 10.1099/00221287-148-4-973. [DOI] [PubMed] [Google Scholar]
- 90.Corr SC, Li Y, Riedel CU, O’Toole PW, Hill C, Gahan CG. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci USA. 2007;104(18):7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Murphy EF, Clarke SF, Marques TM, Hill C, Stanton C, Ross RP, O’Doherty RM, Shanahan F, Cotter PD. Antimicrobials: strategies for targeting obesity and metabolic health? Gut microbes. 2013;4(1):48–53. doi: 10.4161/gmic.22328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu CH, Lee SM, Vanlare JM, Kasper DL, Mazmanian SK. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc Natl Acad Sci USA. 2008;105(10):3951–3956. doi: 10.1073/pnas.0709266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Aas J, Gessert CE, Bakken JS. Recurrent Clostridium difficile colitis: case series involving 18 patients treated with donor stool administered via a nasogastric tube. Clin Infect Dis. 2003;36(5):580–585. doi: 10.1086/367657. [DOI] [PubMed] [Google Scholar]
- 94.Bakken JS, Borody T, Brandt LJ, Brill JV, Demarco DC, Franzos MA, Kelly C, Khoruts A, Louie T, Martinelli LP, Moore TA, Russell G, Surawicz C, Fecal Microbiota Transplantation Workgroup Treating Clostridium difficile infection with fecal microbiota transplantation. Clin Gastroenterol Hepatol. 2011;9(12):1044–1049. doi: 10.1016/j.cgh.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis. 2011;53(10):994–1002. doi: 10.1093/cid/cir632. [DOI] [PubMed] [Google Scholar]
- 96.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, Speelman P, Dijkgraaf MG, Keller JJ. Duodenal infusion of donor feces for recurrent Clostridium difficile . N Engl J Med. 2013;368(5):407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 97.Hickson M, D’Souza AL, Muthu N, Rogers TR, Want S, Rajkumar C, Bulpitt CJ. Use of probiotic Lactobacillus preparation to prevent diarrhoea associated with antibiotics: randomised double blind placebo controlled trial. BMJ. 2007;335(7610):80. doi: 10.1136/bmj.39231.599815.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guglielmetti S, Mora D, Gschwender M, Popp K. Randomised clinical trial: Bifidobacterium bifidum MIMBb75 significantly alleviates irritable bowel syndrome and improves quality of life–a double-blind, placebo-controlled study. Aliment Pharmacol Ther. 2011;33(10):1123–1132. x. doi: 10.1111/j.1365-2036.2011.04633.x. [DOI] [PubMed] [Google Scholar]
- 99.O’Mahony L, McCarthy J, Kelly P, Hurley G, Luo F, Chen K, O’Sullivan GC, Kiely B, Collins JK, Shanahan F, Quigley EM. Lactobacillus and bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology. 2005;128(3):541–551. doi: 10.1053/j.gastro.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 100.Turroni F, van Sinderen D, Ventura M. Genomics and ecological overview of the genus Bifidobacterium . Int J Food Microbiol. 2011;149(1):37–44. doi: 10.1016/j.ijfoodmicro.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 101.Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, Pridmore RD, Arigoni F. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci USA. 2002;99(22):14422–14427. doi: 10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee JH, Karamychev VN, Kozyavkin SA, Mills D, Pavlov AR, Pavlova NV, Polouchine NN, Richardson PM, Shakhova VV, Slesarev AI, Weimer B, O’Sullivan DJ. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics. 2008;9:247. doi: 10.1186/1471-2164-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, Sanchez B, Bidossi A, Ferrarini A, Giubellini V, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Fitzgerald GF, Mills D, Margolles A, Kelly D, van Sinderen D, Ventura M. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci USA. 2010;107(45):19514–19519. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.O’Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O’Brien F, Flynn K, Casey PG, Munoz JA, Kearney B, Houston AM, O’Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O’Toole PW, van Sinderen D. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci USA. 2011;108(27):11217–11222. doi: 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pokusaeva K, Fitzgerald GF, van Sinderen D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 2011;6(3):285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hansson GC. Role of mucus layers in gut infection and inflammation. Curr Opin Microbiol. 2012;15(1):57–62. doi: 10.1016/j.mib.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Forstner JO, Oliver MG, Sylvester FA. Production, structure and biologic relevance of gastrointestinal mucins. New York: Raven; 1995. [Google Scholar]
- 108.Nagae M, Tsuchiya A, Katayama T, Yamamoto K, Wakatsuki S, Kato R. Structural basis of the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase from Bifidobacterium bifidum . J Biol Chem. 2007;282(25):18497–18509. doi: 10.1074/jbc.M702246200. [DOI] [PubMed] [Google Scholar]
- 109.Ashida H, Maki R, Ozawa H, Tani Y, Kiyohara M, Fujita M, Imamura A, Ishida H, Kiso M, Yamamoto K. Characterization of two different endo-alpha-N-acetylgalactosaminidases from probiotic and pathogenic enterobacteria, Bifidobacterium longum and Clostridium perfringens . Glycobiology. 2008;18(9):727–734. doi: 10.1093/glycob/cwn053. [DOI] [PubMed] [Google Scholar]
- 110.Katayama T, Fujita K, Yamamoto K. Novel bifidobacterial glycosidases acting on sugar chains of mucin glycoproteins. J Biosci Bioeng. 2005;99(5):457–465. doi: 10.1263/jbb.99.457. [DOI] [PubMed] [Google Scholar]
- 111.Miwa M, Horimoto T, Kiyohara M, Katayama T, Kitaoka M, Ashida H, Yamamoto K. Cooperation of beta-galactosidase and beta-N-acetylhexosaminidase from bifidobacteria in assimilation of human milk oligosaccharides with type 2 structure. Glycobiology. 2010;20(11):1402–1409. doi: 10.1093/glycob/cwq101. [DOI] [PubMed] [Google Scholar]
- 112.Kiyohara M, Nakatomi T, Kurihara S, Fushinobu S, Suzuki H, Tanaka T, Shoda S, Kitaoka M, Katayama T, Yamamoto K, Ashida H. alpha-N-acetylgalactosaminidase from infant-associated bifidobacteria belonging to novel glycoside hydrolase family 129 is implicated in alternative mucin degradation pathway. J Biol Chem. 2012;287(1):693–700. doi: 10.1074/jbc.M111.277384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Derrien M, Collado MC, Ben-Amor K, Salminen S, de Vos WM. The Mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl Environ Microbiol. 2008;74(5):1646–1648. doi: 10.1128/AEM.01226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol. 2007;73(23):7767–7770. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Derrien M, van Passel MW, van de Bovenkamp JH, Schipper RG, de Vos WM, Dekker J. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut microbes. 2010;1(4):254–268. doi: 10.4161/gmic.1.4.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.van Passel MW, Kant R, Zoetendal EG, Plugge CM, Derrien M, Malfatti SA, Chain PS, Woyke T, Palva A, de Vos WM, Smidt H. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS One. 2011;6(3):e16876. doi: 10.1371/journal.pone.0016876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Turroni F, Milani C, van Sinderen D, Ventura M. Genetic strategies for mucin metabolism in Bifidobacterium bifidum PRL2010: an example of possible human-microbe co-evolution. Gut Microbes. 2011;2(3):183–189. doi: 10.4161/gmic.2.3.16105. [DOI] [PubMed] [Google Scholar]
- 118.van den Broek LA, Hinz SW, Beldman G, Vincken JP, Voragen AG. Bifidobacterium carbohydrases-their role in breakdown and synthesis of (potential) prebiotics. Mol Nutr Food Res. 2008;52(1):146–163. doi: 10.1002/mnfr.200700121. [DOI] [PubMed] [Google Scholar]
- 119.Turroni F, Strati F, Foroni E, Serafini F, Duranti S, van Sinderen D, Ventura M. Analysis of predicted carbohydrate transport systems encoded by Bifidobacterium bifidum PRL2010. Appl Environ Microbiol. 2012;78(14):5002–5012. doi: 10.1128/AEM.00629-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Parche S, Amon J, Jankovic I, Rezzonico E, Beleut M, Barutcu H, Schendel I, Eddy MP, Burkovski A, Arigoni F, Titgemeyer F. Sugar transport systems of Bifidobacterium longum NCC2705. J Mol Microbiol Biotechnol. 2007;12(1–2):9–19. doi: 10.1159/000096455. [DOI] [PubMed] [Google Scholar]
- 121.Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, Kumagai H, Ashida H, Hirose J, Kitaoka M. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem. 2011;286(40):34583–34592. doi: 10.1074/jbc.M111.248138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sela DA. Bifidobacterial utilization of human milk oligosaccharides. Int J Food Microbiol. 2011;149(1):58–64. doi: 10.1016/j.ijfoodmicro.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 123.Ventura M, Canchaya C, Zhang Z, Bernini V, Fitzgerald GF, van Sinderen D. How high G+ C Gram-positive bacteria and in particular bifidobacteria cope with heat stress: protein players and regulators. FEMS Microbiol Rev. 2006;30(5):734–759. doi: 10.1111/j.1574-6976.2006.00031.x. [DOI] [PubMed] [Google Scholar]
- 124.Zomer A, Fernandez M, Kearney B, Fitzgerald GF, Ventura M, van Sinderen D. An interactive regulatory network controls stress response in Bifidobacterium breve UCC2003. J Bacteriol. 2009;191(22):7039–7049. doi: 10.1128/JB.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Claesson MJ, van Sinderen D, O’Toole PW. Lactobacillus phylogenomics–towards a reclassification of the genus. Int J Syst Evol Microbiol. 2008;58(Pt 12):2945–2954. doi: 10.1099/ijs.0.65848-0. [DOI] [PubMed] [Google Scholar]
- 126.Stiles ME, Holzapfel WH. Lactic acid bacteria of foods and their current taxonomy. Int J Food Microbiol. 1997;36(1):1–29. doi: 10.1016/s0168-1605(96)01233-0. [DOI] [PubMed] [Google Scholar]
- 127.Makarova KS, Koonin EV. Evolutionary genomics of lactic acid bacteria. J Bacteriol. 2007;189(4):1199–1208. doi: 10.1128/JB.01351-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Heilig HG, Zoetendal EG, Vaughan EE, Marteau P, Akkermans AD, de Vos WM. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl Environ Microbiol. 2002;68(1):114–123. doi: 10.1128/AEM.68.1.114-123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Reuter G. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr Issues Intest Microbiol. 2001;2(2):43–53. [PubMed] [Google Scholar]
- 130.Walter J, Hertel C, Tannock GW, Lis CM, Munro K, Hammes WP. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2001;67(6):2578–2585. doi: 10.1128/AEM.67.6.2578-2585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Satokari RM, Vaughan EE, Akkermans AD, Saarela M, de Vos WM. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2001;67(2):504–513. doi: 10.1128/AEM.67.2.504-513.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zoetendal EG, Raes J, van den Bogert B, Arumugam M, Booijink CC, Troost FJ, Bork P, Wels M, de Vos WM, Kleerebezem M. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6(7):1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Booijink CC, El-Aidy S, Rajilic-Stojanovic M, Heilig HG, Troost FJ, Smidt H, Kleerebezem M, De Vos WM, Zoetendal EG. High temporal and inter-individual variation detected in the human ileal microbiota. Environ Microbiol. 2010;12(12):3213–3227. doi: 10.1111/j.1462-2920.2010.02294.x. [DOI] [PubMed] [Google Scholar]
- 134.Kleerebezem M, Vaughan EE. Probiotic and gut lactobacilli and bifidobacteria: molecular approaches to study diversity and activity. Annu Rev Microbiol. 2009;63:269–290. doi: 10.1146/annurev.micro.091208.073341. [DOI] [PubMed] [Google Scholar]
- 135.Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee JH, Diaz-Muniz I, Dosti B, Smeianov V, Wechter W, Barabote R, Lorca G, Altermann E, Barrangou R, Ganesan B, Xie Y, Rawsthorne H, Tamir D, Parker C, Breidt F, Broadbent J, Hutkins R, O’Sullivan D, Steele J, Unlu G, Saier M, Klaenhammer T, Richardson P, Kozyavkin S, Weimer B, Mills D. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci USA. 2006;103(42):15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MW, Stiekema W, Lankhorst RM, Bron PA, Hoffer SM, Groot MN, Kerkhoven R, de Vries M, Ursing B, de Vos WM, Siezen RJ. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci USA. 2003;100(4):1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Siezen R, Boekhorst J, Muscariello L, Molenaar D, Renckens B, Kleerebezem M. Lactobacillus plantarum gene clusters encoding putative cell-surface protein complexes for carbohydrate utilization are conserved in specific gram-positive bacteria. BMC Genomics. 2006;7:126. doi: 10.1186/1471-2164-7-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lambert JM, Siezen RJ, de Vos WM, Kleerebezem M. Improved annotation of conjugated bile acid hydrolase superfamily members in Gram-positive bacteria. Microbiology. 2008;154(Pt 8):2492–2500. doi: 10.1099/mic.0.2008/016808-0. [DOI] [PubMed] [Google Scholar]
- 139.Claesson MJ, Li Y, Leahy S, Canchaya C, van Pijkeren JP, Cerdeno-Tarraga AM, Parkhill J, Flynn S, O’Sullivan GC, Collins JK, Higgins D, Shanahan F, Fitzgerald GF, van Sinderen D, O’Toole PW. Multireplicon genome architecture of Lactobacillus salivarius . Proc Natl Acad Sci USA. 2006;103(17):6718–6723. doi: 10.1073/pnas.0511060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pridmore RD, Berger B, Desiere F, Vilanova D, Barretto C, Pittet AC, Zwahlen MC, Rouvet M, Altermann E, Barrangou R, Mollet B, Mercenier A, Klaenhammer T, Arigoni F, Schell MA. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc Natl Acad Sci USA. 2004;101(8):2512–2517. doi: 10.1073/pnas.0307327101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Boekhorst J, Wels M, Kleerebezem M, Siezen RJ. The predicted secretome of Lactobacillus plantarum WCFS1 sheds light on interactions with its environment. Microbiology. 2006;152(Pt 11):3175–3183. doi: 10.1099/mic.0.29217-0. [DOI] [PubMed] [Google Scholar]
- 142.Barrangou R, Altermann E, Hutkins R, Cano R, Klaenhammer TR. Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus . Proc Natl Acad Sci USA. 2003;100(15):8957–8962. doi: 10.1073/pnas.1332765100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Buck BL, Altermann E, Svingerud T, Klaenhammer TR. Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2005;71(12):8344–8351. doi: 10.1128/AEM.71.12.8344-8351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ventura M, Turroni F, Motherway MO, MacSharry J, van Sinderen D. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol. 2012;20(10):467–476. doi: 10.1016/j.tim.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 145.McCracken VJ, Lorenz RG. The gastrointestinal ecosystem: a precarious alliance among epithelium, immunity and microbiota. Cell Microbiol. 2001;3(1):1–11. doi: 10.1046/j.1462-5822.2001.00090.x. [DOI] [PubMed] [Google Scholar]
- 146.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9(5):356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 147.Salzman NH. Microbiota-immune system interaction: an uneasy alliance. Curr Opin Microbiol. 2011;14(1):99–105. doi: 10.1016/j.mib.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wells JM, Rossi O, Meijerink M, van Baarlen P. Epithelial crosstalk at the microbiota-mucosal interface. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4607–4614. doi: 10.1073/pnas.1000092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liew FY. T(H)1 and T(H)2 cells: a historical perspective. Nat Rev Immunol. 2002;2(1):55–60. doi: 10.1038/nri705. [DOI] [PubMed] [Google Scholar]
- 150.Kelsall BL. A focus on dendritic cells and macrophages as key regulators of mucosal immunity. Mucosal Immunol. 2008;1(6):423–424. doi: 10.1038/mi.2008.66. [DOI] [PubMed] [Google Scholar]
- 151.Rescigno M. Intestinal dendritic cells. Adv Immunol. 2010;107:109–138. doi: 10.1016/B978-0-12-381300-8.00004-6. [DOI] [PubMed] [Google Scholar]
- 152.Sansonetti PJ, Medzhitov R. Learning tolerance while fighting ignorance. Cell. 2009;138(3):416–420. doi: 10.1016/j.cell.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 153.Chung H, Kasper DL. Microbiota-stimulated immune mechanisms to maintain gut homeostasis. Curr Opin Immunol. 2010;22(4):455–460. doi: 10.1016/j.coi.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 154.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8(6):411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 155.Asong J, Wolfert MA, Maiti KK, Miller D, Boons GJ. Binding and cellular activation studies reveal that Toll-like receptor 2 can differentially recognize peptidoglycan from Gram-positive and Gram-negative bacteria. J Biol Chem. 2009;284(13):8643–8653. doi: 10.1074/jbc.M806633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 157.Volz T, Nega M, Buschmann J, Kaesler S, Guenova E, Peschel A, Rocken M, Gotz F, Biedermann T. Natural Staphylococcus aureus-derived peptidoglycan fragments activate NOD2 and act as potent costimulators of the innate immune system exclusively in the presence of TLR signals. FASEB J. 2010;24(10):4089–4102. doi: 10.1096/fj.09-151001. [DOI] [PubMed] [Google Scholar]
- 158.Zeuthen LH, Fink LN, Frokiaer H. Epithelial cells prime the immune response to an array of gut-derived commensals towards a tolerogenic phenotype through distinct actions of thymic stromal lymphopoietin and transforming growth factor-beta. Immunology. 2008;123(2):197–208. doi: 10.1111/j.1365-2567.2007.02687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S, Valvano MA, Foster SJ, Mak TW, Nunez G, Inohara N. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4(7):702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 160.Macho Fernandez E, Valenti V, Rockel C, Hermann C, Pot B, Boneca IG, Grangette C. Anti-inflammatory capacity of selected lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide. Gut. 2011;60(8):1050–1059. doi: 10.1136/gut.2010.232918. [DOI] [PubMed] [Google Scholar]
- 161.Shida K, Kiyoshima-Shibata J, Kaji R, Nagaoka M, Nanno M. Peptidoglycan from lactobacilli inhibits interleukin-12 production by macrophages induced by Lactobacillus casei through Toll-like receptor 2-dependent and independent mechanisms. Immunology. 2009;128(1 Suppl):e858–e869. doi: 10.1111/j.1365-2567.2009.03095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Matsumoto S, Hara T, Nagaoka M, Mike A, Mitsuyama K, Sako T, Yamamoto M, Kado S, Takada T. A component of polysaccharide peptidoglycan complex on Lactobacillus induced an improvement of murine model of inflammatory bowel disease and colitis-associated cancer. Immunology. 2009;128(1 Suppl):e170–e180. doi: 10.1111/j.1365-2567.2008.02942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xia G, Kohler T, Peschel A. The wall teichoic acid and lipoteichoic acid polymers of Staphylococcus aureus. Intern J Med Microbiol. 2010;300(2–3):148–154. doi: 10.1016/j.ijmm.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 164.Grangette C, Nutten S, Palumbo E, Morath S, Hermann C, Dewulf J, Pot B, Hartung T, Hols P, Mercenier A. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc Natl Acad Sci USA. 2005;102(29):10321–10326. doi: 10.1073/pnas.0504084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mohamadzadeh M, Pfeiler EA, Brown JB, Zadeh M, Gramarossa M, Managlia E, Bere P, Sarraj B, Khan MW, Pakanati KC, Ansari MJ, O’Flaherty S, Barrett T, Klaenhammer TR. Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4623–4630. doi: 10.1073/pnas.1005066107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Saber R, Zadeh M, Pakanati KC, Bere P, Klaenhammer T, Mohamadzadeh M. Lipoteichoic acid-deficient Lactobacillus acidophilus regulates downstream signals. Immunotherapy. 2011;3(3):337–347. doi: 10.2217/imt.10.119. [DOI] [PubMed] [Google Scholar]
- 167.Ryu YH, Baik JE, Yang JS, Kang SS, Im J, Yun CH, Kim DW, Lee K, Chung DK, Ju HR, Han SH. Differential immunostimulatory effects of Gram-positive bacteria due to their lipoteichoic acids. Int Immunopharmacol. 2009;9(1):127–133. doi: 10.1016/j.intimp.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 168.Deininger S, Stadelmaier A, von Aulock S, Morath S, Schmidt RR, Hartung T. Definition of structural prerequisites for lipoteichoic acid-inducible cytokine induction by synthetic derivatives. J Immunol. 2003;170(8):4134–4138. doi: 10.4049/jimmunol.170.8.4134. [DOI] [PubMed] [Google Scholar]
- 169.Lebeer S, Verhoeven TL, Perea Velez M, Vanderleyden J, De Keersmaecker SC. Impact of environmental and genetic factors on biofilm formation by the probiotic strain Lactobacillus rhamnosus GG. Appl Environ Microbiol. 2007;73(21):6768–6775. doi: 10.1128/AEM.01393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Walter J, Loach DM, Alqumber M, Rockel C, Hermann C, Pfitzenmaier M, Tannock GW. D-alanyl ester depletion of teichoic acids in Lactobacillus reuteri 100–23 results in impaired colonization of the mouse gastrointestinal tract. Environ Microbiol. 2007;9(7):1750–1760. doi: 10.1111/j.1462-2920.2007.01292.x. [DOI] [PubMed] [Google Scholar]
- 171.Claes IJ, Lebeer S, Shen C, Verhoeven TL, Dilissen E, De Hertogh G, Bullens DM, Ceuppens JL, Van Assche G, Vermeire S, Rutgeerts P, Vanderleyden J, De Keersmaecker SC. Impact of lipoteichoic acid modification on the performance of the probiotic Lactobacillus rhamnosus GG in experimental colitis. Clin Exp Immunol. 2010;162(2):306–314. doi: 10.1111/j.1365-2249.2010.04228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Duncker SC, Wang L, Hols P, Bienenstock J. The d-alanine content of lipoteichoic acid is crucial for Lactobacillus plantarum-mediated protection from visceral pain perception in a rat colorectal distension model. Neurogastroenterol Motil. 2008;20(7):843–850. doi: 10.1111/j.1365-2982.2008.01085.x. [DOI] [PubMed] [Google Scholar]
- 173.Kim HG, Kim NR, Gim MG, Lee JM, Lee SY, Ko MY, Kim JY, Han SH, Chung DK. Lipoteichoic acid isolated from Lactobacillus plantarum inhibits lipopolysaccharide-induced TNF-alpha production in THP-1 cells and endotoxin shock in mice. J Immunol. 2008;180(4):2553–2561. doi: 10.4049/jimmunol.180.4.2553. [DOI] [PubMed] [Google Scholar]
- 174.Kaji R, Kiyoshima-Shibata J, Nagaoka M, Nanno M, Shida K. Bacterial teichoic acids reverse predominant IL-12 production induced by certain lactobacillus strains into predominant IL-10 production via TLR2-dependent ERK activation in macrophages. J Immunol. 2010;184(7):3505–3513. doi: 10.4049/jimmunol.0901569. [DOI] [PubMed] [Google Scholar]
- 175.Krinos CM, Coyne MJ, Weinacht KG, Tzianabos AO, Kasper DL, Comstock LE. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature. 2001;414(6863):555–558. doi: 10.1038/35107092. [DOI] [PubMed] [Google Scholar]
- 176.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 177.Mazmanian SK, Kasper DL. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat Rev Immunol. 2006;6(11):849–858. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]
- 178.Kuwahara T, Yamashita A, Hirakawa H, Nakayama H, Toh H, Okada N, Kuhara S, Hattori M, Hayashi T, Ohnishi Y. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc Natl Acad Sci USA. 2004;101(41):14919–14924. doi: 10.1073/pnas.0404172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Cerdeno-Tarraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, Barron A, Clark L, Corton C, Doggett J, Holden MT, Larke N, Line A, Lord A, Norbertczak H, Ormond D, Price C, Rabbinowitsch E, Woodward J, Barrell B, Parkhill J. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307((5714)):1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- 180.Shen Y, Torchia ML, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. 2012;12(4):509–520. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107(27):12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fanning S, Hall LJ, Cronin M, Zomer A, MacSharry J, Goulding D, Motherway MO, Shanahan F, Nally K, Dougan G, van Sinderen D. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc Natl Acad Sci USA. 2012;109(6):2108–2113. doi: 10.1073/pnas.1115621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lebeer S, Verhoeven TL, Francius G, Schoofs G, Lambrichts I, Dufrene Y, Vanderleyden J, De Keersmaecker SC. Identification of a gene cluster for the biosynthesis of a long, Galactose-Rich Exopolysaccharide in Lactobacillus rhamnosus GG and functional analysis of the priming glycosyltransferase. Appl Environ Microbiol. 2009;75(11):3554–3563. doi: 10.1128/AEM.02919-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lebeer S, Claes IJ, Verhoeven TL, Vanderleyden J, De Keersmaecker SC. Exopolysaccharides of Lactobacillus rhamnosus GG form a protective shield against innate immune factors in the intestine. Microb Biotechnol. 2011;4(3):368–374. doi: 10.1111/j.1751-7915.2010.00199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hafez M, Hayes K, Goldrick M, Warhurst G, Grencis R, Roberts IS. The K5 capsule of Escherichia coli strain Nissle 1917 is important in mediating interactions with intestinal epithelial cells and chemokine induction. Infect Immun. 2009;77(7):2995–3003. doi: 10.1128/IAI.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lebeer S, Vanderleyden J, De Keersmaecker SC. Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol Rev. 2008;72(4):728–764. doi: 10.1128/MMBR.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sims IM, Frese SA, Walter J, Loach D, Wilson M, Appleyard K, Eason J, Livingston M, Baird M, Cook G, Tannock GW. Structure and functions of exopolysaccharide produced by gut commensal Lactobacillus reuteri 100–23. ISME J. 2011;5(7):1115–1124. doi: 10.1038/ismej.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in gram-positive pathogens. Nat Rev Microbiol. 2006;4(7):509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- 189.Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx AP, Lebeer S, De Keersmaecker SC, Vanderleyden J, Hamalainen T, Laukkanen S, Salovuori N, Ritari J, Alatalo E, Korpela R, Mattila-Sandholm T, Lassig A, Hatakka K, Kinnunen KT, Karjalainen H, Saxelin M, Laakso K, Surakka A, Palva A, Salusjarvi T, Auvinen P, de Vos WM. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc Natl Acad Sci USA. 2009;106(40):17193–17198. doi: 10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.von Ossowski I, Reunanen J, Satokari R, Vesterlund S, Kankainen M, Huhtinen H, Tynkkynen S, Salminen S, de Vos WM, Palva A. Mucosal adhesion properties of the probiotic Lactobacillus rhamnosus GG SpaCBA and SpaFED pilin subunits. Appl Environ Microbiol. 2010;76(7):2049–2057. doi: 10.1128/AEM.01958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lebeer S, Claes I, Tytgat HL, Verhoeven TL, Marien E, von Ossowski I, Reunanen J, Palva A, Vos WM, Keersmaecker SC, Vanderleyden J. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl Environ Microbiol. 2012;78(1):185–193. doi: 10.1128/AEM.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Foroni E, Serafini F, Amidani D, Turroni F, He F, Bottacini F, O’Connell Motherway M, Viappiani A, Zhang Z, Rivetti C, van Sinderen D, Ventura M. Genetic analysis and morphological identification of pilus-like structures in members of the genus Bifidobacterium . Microb Cell Fact. 2011;10(Suppl 1):S16. doi: 10.1186/1475-2859-10-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Avall-Jaaskelainen S, Palva A. Lactobacillus surface layers and their applications. FEMS Microbiol Rev. 2005;29(3):511–529. doi: 10.1016/j.femsre.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 194.Schneitz C, Nuotio L, Lounatma K. Adhesion of Lactobacillus acidophilus to avian intestinal epithelial cells mediated by the crystalline bacterial cell surface layer (S-layer) J Appl Bact. 1993;74(3):290–294. doi: 10.1111/j.1365-2672.1993.tb03028.x. [DOI] [PubMed] [Google Scholar]
- 195.Toba T, Virkola R, Westerlund B, Bjorkman Y, Sillanpaa J, Vartio T, Kalkkinen N, Korhonen TK. A Collagen-Binding S-Layer Protein in Lactobacillus crispatus . Appl Environ Microbiol. 1995;61(7):2467–2471. doi: 10.1128/aem.61.7.2467-2471.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Konstantinov SR, Smidt H, de Vos WM, Bruijns SC, Singh SK, Valence F, Molle D, Lortal S, Altermann E, Klaenhammer TR, van Kooyk Y. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci USA. 2008;105(49):19474–19479. doi: 10.1073/pnas.0810305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Johnson-Henry KC, Hagen KE, Gordonpour M, Tompkins TA, Sherman PM. Surface-layer protein extracts from Lactobacillus helveticus inhibit enterohaemorrhagic Escherichia coli O157:h7 adhesion to epithelial cells. Cell Microbiol. 2007;9(2):356–367. doi: 10.1111/j.1462-5822.2006.00791.x. [DOI] [PubMed] [Google Scholar]
- 198.Taverniti VSM, Minuzzo M, Arioli S, De Noni I, Scabiosi C, Cordova ZM, Junttila I, Hämäläinen S, Turpeinen H, Mora D, Karp M, Pesu M, Guglielmetti S. S-Layer Protein Mediates the Stimulatory Effect of Lactobacillus helveticus MIMLh5 on Innate Immunity. Appl Environ Microbiol. 2013;79(4):1221–1231. doi: 10.1128/AEM.03056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.O’Callaghan J, Butto LF, MacSharry J, Nally K, O’Toole PW. Influence of adhesion and bacteriocin production by Lactobacillus salivarius on the intestinal epithelial cell transcriptional response. Appl Environ Microbiol. 2012;78(15):5196–5203. doi: 10.1128/AEM.00507-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Murphy EF, Cotter PD, Hogan A, O’Sullivan O, Joyce A, Fouhy F, Clarke SF, Marques TM, O’Toole PW, Stanton C, Quigley EM, Daly C, Ross PR, O’Doherty RM, Shanahan F. Divergent metabolic outcomes arising from targeted manipulation of the gut microbiota in diet-induced obesity. Gut. 2012;62:220–226. doi: 10.1136/gutjnl-2011-300705. [DOI] [PubMed] [Google Scholar]
- 201.Riboulet-Bisson E, Sturme MH, Jeffery IB, O’Donnell MM, Neville BA, Forde BM, Claesson MJ, Harris H, Gardiner GE, Casey PG, Lawlor PG, O’Toole PW, Ross RP. Effect of Lactobacillus salivarius bacteriocin Abp118 on the mouse and pig intestinal microbiota. PLoS One. 2012;7(2):e31113. doi: 10.1371/journal.pone.0031113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Meijerink M, van Hemert S, Taverne N, Wels M, de Vos P, Bron PA, Savelkoul HF, van Bilsen J, Kleerebezem M, Wells JM. Identification of genetic loci in Lactobacillus plantarum that modulate the immune response of dendritic cells using comparative genome hybridization. PLoS One. 2010;5(5):e10632. doi: 10.1371/journal.pone.0010632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Walsh MC, Gardiner GE, Hart OM, Lawlor PG, Daly M, Lynch B, Richert BT, Radcliffe S, Giblin L, Hill C, Fitzgerald GF, Stanton C, Ross P. Predominance of a bacteriocin-producing Lactobacillus salivarius component of a five-strain probiotic in the porcine ileum and effects on host immune phenotype. FEMS Microbiol Ecol. 2008;64(2):317–327. doi: 10.1111/j.1574-6941.2008.00454.x. [DOI] [PubMed] [Google Scholar]
- 204.Marco ML, Bongers RS, de Vos WM, Kleerebezem M. Spatial and temporal expression of Lactobacillus plantarum genes in the gastrointestinal tracts of mice. Appl Environ Microbiol. 2007;73(1):124–132. doi: 10.1128/AEM.01475-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Miyauchi E, O’Callaghan J, Butto LF, Hurley G, Melgar S, Tanabe S, Shanahan F, Nally K, O’Toole PW. Mechanism of protection of transepithelial barrier function by Lactobacillus salivarius: strain dependence and attenuation by bacteriocin production. Am J Physiol Gastrointest Liver Physiol. 2012;303(9):G1029–G1041. doi: 10.1152/ajpgi.00003.2012. [DOI] [PubMed] [Google Scholar]
- 206.van Pijkeren JP, Canchaya C, Ryan KA, Li Y, Claesson MJ, Sheil B, Steidler L, O’Mahony L, Fitzgerald GF, van Sinderen D, O’Toole PW. Comparative and functional analysis of sortase-dependent proteins in the predicted secretome of Lactobacillus salivarius UCC118. Appl Environ Microbiol. 2006;72(6):4143–4153. doi: 10.1128/AEM.03023-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Munoz-Provencio D, Rodriguez-Diaz J, Collado MC, Langella P, Bermudez-Humaran LG, Monedero V. Functional analysis of the Lactobacillus casei BL23 sortases. Appl Environ Microbiol. 2012;78(24):8684–8693. doi: 10.1128/AEM.02287-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Boekhorst J, de Been MW, Kleerebezem M, Siezen RJ. Genome-wide detection and analysis of cell wall-bound proteins with LPxTG-like sorting motifs. J Bacteriol. 2005;187(14):4928–4934. doi: 10.1128/JB.187.14.4928-4934.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Boekhorst J, Helmer Q, Kleerebezem M, Siezen RJ. Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology. 2006;152(Pt 1):273–280. doi: 10.1099/mic.0.28415-0. [DOI] [PubMed] [Google Scholar]
- 210.Juge N, Muroyama A, Hiasa M, Omote H, Moriyama Y. Vesicular inhibitory amino acid transporter is a Cl-/gamma-aminobutyrate Co-transporter. J Biol Chem. 2009;284(50):35073–35078. doi: 10.1074/jbc.M109.062414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Troost FJ, van Baarlen P, Lindsey P, Kodde A, de Vos WM, Kleerebezem M, Brummer RJ. Identification of the transcriptional response of human intestinal mucosa to Lactobacillus plantarum WCFS1 in vivo. BMC Genomics. 2008;9:374. doi: 10.1186/1471-2164-9-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.van Baarlen P, Troost FJ, van Hemert S, van der Meer C, de Vos WM, de Groot PJ, Hooiveld GJ, Brummer RJ, Kleerebezem M. Differential NF-kappaB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc Natl Acad Sci USA. 2009;106(7):2371–2376. doi: 10.1073/pnas.0809919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wang M, Ahrne S, Jeppsson B, Molin G. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol. 2005;54(2):219–231. doi: 10.1016/j.femsec.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 214.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307(5717):1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 215.Tannock GW, Wilson CM, Loach D, Cook GM, Eason J, O’Toole PW, Holtrop G, Lawley B. Resource partitioning in relation to cohabitation of Lactobacillus species in the mouse forestomach. ISME J. 2012;6(5):927–938. doi: 10.1038/ismej.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Paton AW, Morona R, Paton JC. Designer probiotics for prevention of enteric infections. Nat Rev Microbiol. 2006;4(3):193–200. doi: 10.1038/nrmicro1349. [DOI] [PubMed] [Google Scholar]
- 217.Sleator RD. Probiotics—a viable therapeutic alternative for enteric infections especially in the developing world. Discov Med. 2010;10(51):119–124. [PubMed] [Google Scholar]
- 218.Chang TL, Chang CH, Simpson DA, Xu Q, Martin PK, Lagenaur LA, Schoolnik GK, Ho DD, Hillier SL, Holodniy M, Lewicki JA, Lee PP. Inhibition of HIV infectivity by a natural human isolate of Lactobacillus jensenii engineered to express functional two-domain CD4. Proc Natl Acad Sci USA. 2003;100(20):11672–11677. doi: 10.1073/pnas.1934747100. [DOI] [PMC free article] [PubMed] [Google Scholar]