Abstract
Protein 4.1B/DAL-1 is a membrane skeletal protein that belongs to the protein 4.1 family. Protein 4.1B/DAL-1 is localized to sites of cell–cell contact and functions as an adapter protein, linking the plasma membrane to the cytoskeleton or associated cytoplasmic signaling effectors and facilitating their activities in various pathways. Protein 4.1B/DAL-1 is involved in various cytoskeleton-associated processes, such as cell motility and adhesion. Moreover, protein 4.1B/DAL-1 also plays a regulatory role in cell growth, differentiation, and the establishment of epithelial-like cell structures. Protein 4.1B/DAL-1 is normally expressed in multiple human tissues, but loss of its expression or prominent down-regulation of its expression is frequently observed in corresponding tumor tissues and tumor cell lines, suggesting that protein 4.1B/DAL-1 is involved in the molecular pathogenesis of these tumors and acts as a potential tumor suppressor. This review will focus on the structure of protein 4.1B/DAL-1, 4.1B/DAL-1-interacting molecules, 4.1B/DAL-1 inactivation and tumor progression, and anti-tumor activity of the 4.1B/DAL-1.
Keywords: Protein 4.1B/DAL-1, Tumor suppressor, Interacting molecules, Solid tumor
Introduction
Protein 4.1B/DAL-1 (differentially expressed in adenocarcinoma of the lung) is an important membrane skeletal protein that belongs to the protein 4.1 family, and the protein 4.1 family belongs to the protein 4.1 superfamily. Protein 4.1B/DAL-1 links the plasma membrane to the cytoskeleton or associated cytoplasmic signaling effectors and facilitating their activities in various pathways. It has been validated that protein 4.1B/DAL-1 served as a broad-spectrum tumor suppressor in a variety of cancers. Promoter methylation and loss of heterozygosity (LOH) at the 4.1B/DAL-1 locus on 18p11.3 are responsible for the inactivation of 4.1B/DAL-1 gene expression. Abnormal expression of protein 4.1B/DAL-1 may lead to tumorigenesis and/or promote tumor progression. Protein 4.1B/DAL-1 can be involved in different mechanisms that modulate cell growth, motility, adhesion, and cytoskeleton organization. Therefore, protein 4.1B/DAL-1 is likely to become an important prognostic indicator of human cancers and a potential target molecule for antitumor therapies.
Overview of subfamilies of protein 4.1 superfamily in tumorigenesis and tumor progression
The protein 4.1 superfamily comprises a group of proteins that share a conserved FERM (4.1/ezrin/radixin/moesin) domain at the N-terminal. On the basis of protein sequence similarity, this superfamily can be divided into five subfamilies: protein 4.1 family, merlin/ERM (ezrin/radixin/moesin) protein family, talin-related molecules, PTPH (protein tyrosine phosphatase) proteins and NBL4 (novel band 4.1-like 4) proteins. This protein superfamily serves as membrane cytoskeleton linkers between transmembrane proteins and participates in a wide variety of cellular events, such as proliferation, survival, motility, and cell–cell/cell–substrate adhesion and cell shape [1, 2]. Moreover, the protein 4.1 family and merlin/ERM family have been shown to be involved in tumorigenesis and tumor progression. The protein 4.1 family and merlin are negative growth regulators (tumor suppressors), however, high expression of ERM proteins has been observed in a variety of epithelial cancers to promote tumor progression [3].
Protein 4.1 family: structure, modification, and functions
The protein 4.1 family contains four homologous proteins in mammalian non-erythroid cells: 4.1R (erythrocyte type), 4.1N (neuron type), 4.1G (general type) and 4.1B (brain type). The protein 4.1 family members are encoded by distinct paralogous genes, and a variety of tissue- and development-specific protein isoforms are generated by pre-mRNA alternative splicing and alternative first exons [4–6]. Despite the enormous variety of splice variants, each member of the protein 4.1 family is characterized by three highly conserved structural and functional domains (Fig. 1a). The unique domains act as modulators of protein 4.1 interactions mediated by the conserved domains and that the tissue- and cell-specific splicing patterns of these domains confer the unique characteristics upon each 4.1 proteins [7, 8]. Through three highly conserved domains, 4.1 proteins can bind to membrane molecules, thereby playing important roles in membrane organization and signal transduction. The binding capacity of 4.1 proteins is regulated by calmodulin and/or phosphorylation. Both calmodulin/Ca2+ and phosphorylation can result in the down-regulation of the membrane- and/or spectrin-actin binding activities of 4.1 proteins [9–11]. Interestingly, the U2 domain contains a Ser312 and an Arg309 residue that are the primary substrates for PKC (protein kinase C). These two residues are conserved in four protein 4.1 members. It is possible that PKC-dependent phosphorylation plays a key regulatory role in the function of 4.1 proteins [12]. The protein 4.1 family is an important component of the membrane skeleton. It can stabilize the membrane skeleton and assemble protein complexes. This family of proteins is necessary to maintain cellular morphology, cell polarity, and the mechanical properties of the cell membrane [13]. The protein 4.1 family also plays important roles in membrane organization and signal transduction in tumorigenesis and tumor progression. 4.1R and 4.1N are negative regulators in the tumorigenesis and progression [13]. Moreover, 4.1B/DAL-1 serves as a broad-spectrum tumor suppressor in a variety of cancers [14–16].
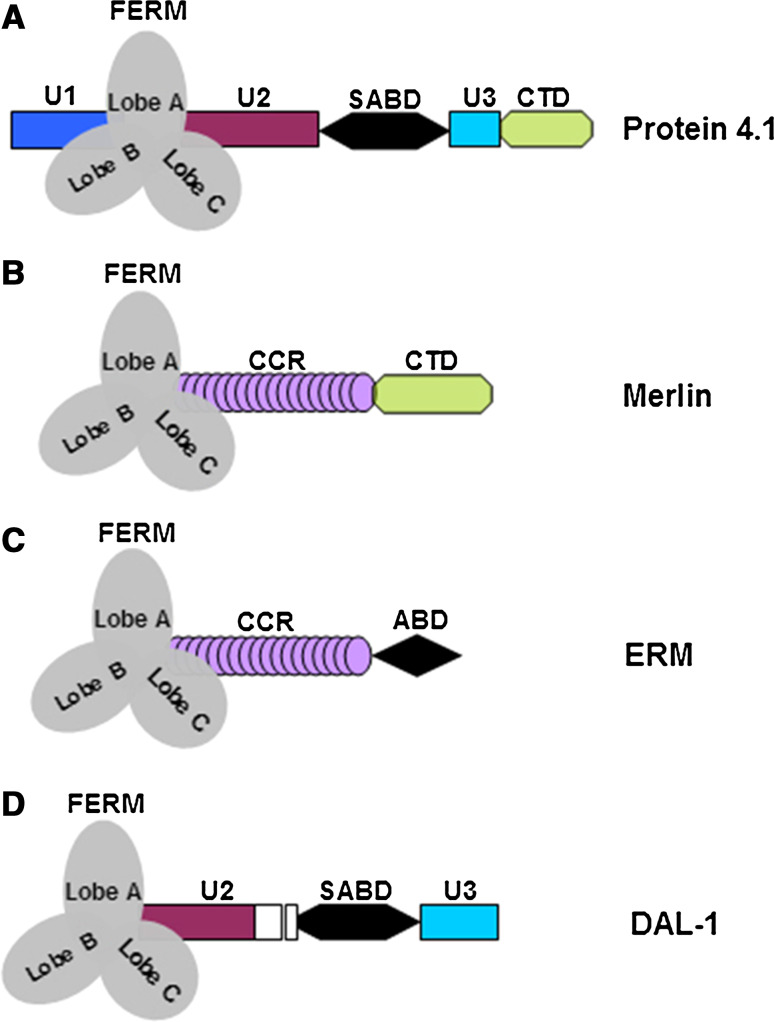
Fig. 1.
Structural domains of protein 4.1, merlin/ERM, and DAL-1. The protein 4.1 family (4.1R, 4.1N, 4.1G, and 4.1B) contains three conserved structural domains (FERM, SABD, and CTD) and three unique domains (U1, U2, and U3). The U1 domain is an N-terminal headpiece (HP), the U2 domain is situated between the FERM and the SABD, and the U3 domain lies between the SABD and the CTD. As compared with protein 4.1R, the sequence identity of protein 4.1B in the FERM and SAB domains is 74 and 50 %, respectively. The CTD is highly homologous to the NuMA binding domain of 4.1R (93 % identity). Merlin and ERM members display a similar structural organization: merlin is composed of an N-terminal FERM domain, a coiled-coil region (CCR), and a carboxy-terminal domain. Unlike the merlin proteins, ERM members lack the CTD, while containing an actin-binding domain (ABD) in its C-terminal domain. DAL-1 is a protein fragment of 4.1B. The white box denotes internal sequences that are absent in DAL-1
Merlin/ERM family: differences with the protein 4.1 family
Similar to the protein 4.1 family, merlin/ERM proteins have been shown to be involved in tumorigenesis and tumor progression [17, 18]. Merlin (moesin, ezrin, radixin-like protein) has a relatively broad tumor suppressor function, however, ERM proteins have been indicated to confer oncogenic activity. Merlin and ERM display a similar structural organization: merlin is composed of an N-terminal FERM domain, a coiled-coil region (CCR), and a carboxy-terminal domain (Fig. 1b). Unlike the merlin proteins, ERM members lack the CTD while containing an actin-binding domain (ABD) in its C-terminal domain (Fig. 1c). It is possible that conserved functions of the protein 4.1 and merlin/ERM family are correlated with the FERM domain, whereas their specific functions reside within less conserved domains. The reverse effects on tumor cells between ERM family and protein 4.1 family may be attributable to the different effects of post-transcriptional modifications. In this regard, phosphorylation of protein 4.1 negatively regulates its interaction with membrane proteins and the cytoskeleton. While ERM proteins upon are phosphorylated, they are activated and interact with transmembrane proteins and F-actin [17, 19]. Furthermore, the distinct subcellular localization in polarized epithelial cells (ERMs/the apical domain versus protein 4.1/the basolateral domain) may also be responsible for their reverse effects [12, 20, 21].
Protein 4.1B/DAL-1: gene, structure, and localization
Protein 4.1B, or type II brain 4.1 (genes KIAA0987, EPB41L3), is a neuronal-enriched protein 4.1 homolog [22]. EPB41L3 is located on human chromosome 18p11.3, and the large EPB41L3 gene is approximately 240 kb in length. EPB41L3 contains exons encoding full-length FERM (exons 4–11), full-length CTD (exons 18–21), U1 (exon 2 and part of exon 4), U2 (exons 13–14) and U3 (exon 17C) [23]. With regard to the SAB domain, while exon 17 is present, exon 16 is missing [24]. Among the conserved domains of protein 4.1B, only exon 16 in the SABD and exon 21 in the CTD exhibit tissue-specific alternative splicing [8, 22]. DAL-1 is a splice variant of protein 4.1B. Significantly reduced expression (>50 %) of DAL-1 was detected in primary non-small cell lung cancer (NSCLC) tissues as compared with patient-matched normal lung tissues [25]. DAL-1 contains the amino acid residues spanning from Met110 to Ser853 in protein 4.1B, and it lacks a U1 structural domain at the N-terminal, a CTD domain at the C-terminal, a partial region of the U2 and SABD domains [25, 26] (Fig. 1d). Although DAL-1 lacks a partial actin-binding domain, the necessary residues remain to function as a tumor suppressor, and may also associate with the actin skeleton indirectly by binding to all known 4.1B-binding proteins [27]. In addition, a study identified a Golgi-specific 200-kDa protein 4.1B variant and found that it is required for both the structural integrity of the Golgi complex and for the assembly of a subset of plasma membrane proteins. Depletion of this variant in HBE cells led to disruption of the Golgi structure and impaired membrane trafficking of Na+/K+-ATPase, ZO-1, and ZO-2 [28].
4.1B/DAL-1 is located beneath the plasma membrane and is distributed to cell–cell junctions [29] (Fig. 2), as well as cell-basement membrane contacts [25]. In the PNS and CNS, 4.1B is concentrated at internodes, paranodes, and juxtaparanodes in myelinated axons. This protein contributes to the stabilization of membrane proteins at paranodes, the clustering of juxtaparanodal proteins, and the regulation of the internodal axon caliber [30–33]. In addition to the nervous system, protein 4.1B is also expressed in the lungs [25], kidneys [34], intestines [35], pancreas [36], testes [37], prostate [38], ovaries [39], and breasts [40]. In these tissues, 4.1B/DAL-1 maintains cell–cell and cell–matrix interactions and stabilizes the organization of the cytoskeleton by linking transmembrane proteins to the cytoskeleton [41].
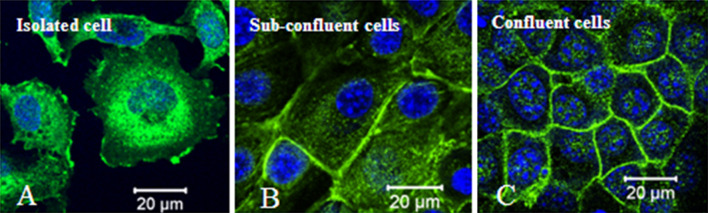
Fig. 2.
Sub-cellular localization of protein 4.1B in primary mouse embryonic fibroblasts (MEF). a In isolated MEF cells, protein 4.1B is predominantly located at peri-nuclear region and also distributed in cytoplasm and membrane ruffling edges. b Protein 4.1B is recruited to areas of cell–cell contact in sub-confluent conditions. c Protein 4.1B is very strongly enriched at cell–cell contacts in completely confluent cells [29]
Protein 4.1B/DAL-1-interacting molecules
To elucidate the tumor suppressor functions of protein 4.1B/DAL-1, the binding ligands that directly interact with protein 4.1B/DAL-1 in various tumors must be identified (Fig. 3). To date, a series of potential protein 4.1B/DAL-1-interacting molecules have been described specifically (Table 1). Furthermore, protein 4.1B/DAL-1 is also known to interact with ERM proteins, as well as the amino terminus of merlin in vitro and two known merlin interactor proteins, CD44 and βII-spectrin [27].
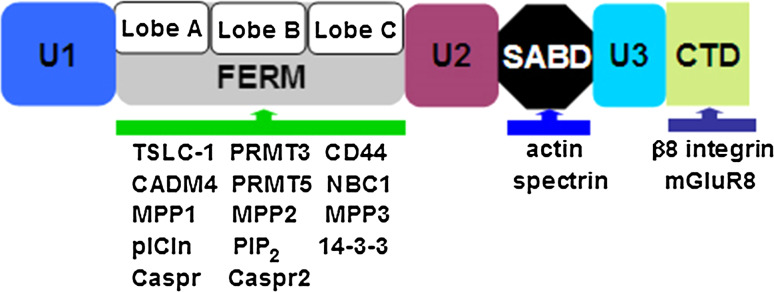
Fig. 3.
Schematic representation of protein 4.1B and interacting molecules. Protein 4.1B binds to interacting molecules through FERM, SABD, and CTD. Binding ligands: TSLC-1 tumor suppressor in lung cancer 1, CADM cell adhesion molecule, PRMT protein arginine methyltransferase, NBC1 Na+ bicarbonate cotransporter 1, MPP membrane palmitoylated protein, pICln chloride channel, PIP 2 phosphatidylinositol 4,5-bisphosphate, 14-3-3 tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, Caspr contactin-associated protein, and mGluR8, metabotropic glutamate receptor isoform 8
Table 1.
Protein 4.1B/DAL-1-interacting molecules
| Molecules | Characteristics | Binding domain | Functions of binding protein-4.1B/DAL-1 complex | Note | References |
|---|---|---|---|---|---|
| Spectrin, actin | The cytoskeleton | SABD | Provides cytoskeletal linkage and modulates important cytoskeletal functions in a wide variety of cells and tissues | 1, 2 | [42] |
| 14-3-3 proteins | Adapter proteins | FERM | Not determined | 3, 4, 5, 6 | [43] |
| Caspr/paranodin, Caspr2 | Neuronal transmembrane glycoprotein | FERM | At paranodes and juxtaparanodes, 4.1B associates with Caspr/paranodin and Caspr2, respectively, and anchors them to the axonal cytoskeleton | 5, 6, 7 | [30] |
| pICln | Swelling-activated anion channel | FERM | Participating in the regulation of cell volume and cellular RNA splicing processes | 3, 5 | [44] |
| NBC1 | Na+–HCO3 − cotransporter | FERM | Maintaining normal intracellular homeostasis | 5, 6 | [45] |
| PRMT3, PRMT5 | Protein arginine methyltransferase | FERM | 4.1B/DAL-1 is not itself a substrate for PRMT3/5, but rather modulates PRMT3/5-mediated methylation activity | 3, 4, 5, 6 | [44, 46] |
| β8-integrin | Cell adhesion receptor | CTD | 4.1B/DAL-1 involves in β8-integrin adhesion and signaling pathways | 6, 7 | [47, 48] |
| CD44 | Transmembrane receptor and stem cell marker | FERM | Binding of 4.1B/DAL-1 to CD44 is likely implicated in 4.1B/DAL-1 growth suppression | 8 | [14, 26] |
| PIP2 | Membrane phospholipids | FERM | PIP2 has been suggested to play a regulatory role in the association of 4.1B/DAL-1 with membrane proteins | 9 | [49] |
| TSLC-1, CADM4 | Immunoglobulin superfamily cell adhesion molecules | FERM | Organization of the actin cytoskeleton; construction of epithelial-like cell structure | 5, 6, 7 | [50, 51] |
| MPP1/2/3 | Membrane-associated guanylate kinase family members | FERM | Construction of epithelial-like cell structure | 5, 6, 7 | [52] |
| MGluR8 | Metabotropic glutamate receptors | CTD | Proper targeting and/or stabilization of mGluR8 at the plasma membrane and inhibition of mGluR8-mediated reductions in intracellular cAMP concentrations | 3, 6 | [53] |
Supported by 1 co-sedimentation assay, 2 resonant mirror detection, 3 yeast two-hybrid interaction, 4 in vitro binding assays, 5 co-immunoprecipitation, 6 GST fusion protein pull-down, 7 subcellular co-localization, 8 in vivo interaction studies, 9 PIP2-binding motifs were obtained from the UniProt database
Spectrin and actin
The SAB domain of protein 4.1B mediates the formation of spectrin-4.1B-actin ternary complexes. It is less efficient than the 4.1R SABD due to a decrease in the actin-binding activity, while its binding affinity for spectrin is the same as that of the 4.1R SAB domain [42]. The fully functional SABD is encoded by the alternatively spliced exon 16 plus the constitutively expressed exon 17. Exon 16 encodes one spectrin-binding motif, while exon 17 encodes the actin-binding motif and the second spectrin-binding motif [24]. The majority of the nonerythroid protein 4.1 expressed in mammals lacks a high affinity SAB domain (no exon 16 paralog in 4.1N; splicing out of exon 16 from 4.1R, 4.1G, and 4.1B in many tissues) [23]. Although 4.1B does not retain exon 16, exon 17 alone could confer physiologically relevant cytoskeleton-binding activity. Protein 4.1B serves as a key membrane-cytoskeleton linker that likely tethers transmembrane proteins in the plasma membrane and functions in organizing the cytoskeleton. It has been shown that a 4.1B/spectrin II cytoskeletal complex stabilized the expression of multiple FERM-binding adhesion molecules in axonal membrane. While the loss of 4.1B resulted in a significant reduction of spectrin II, Nectin-like (Necl)-1 and -2 expression along the internodal membrane of the axon [33]. In addition, a recent report showed that 4.1B-deficient mouse embryo fibroblasts (MEF) cells failed to form actin stress fibers [29].
14-3-3 family members
The 14-3-3 proteins are a family of acidic regulatory proteins that consist of seven isoforms: β, γ, ε, ζ, η, τ and θ. The 14-3-3 proteins serve as adapter proteins that regulate protein–protein interactions and the subcellular localization of proteins. This protein family is also involved in regulating many biologically important processes, including the cell growth cycle, apoptosis, signal transduction, polarity, and skeletal structure [54]. Due to its unique binding interaction with 4.1B/DAL-1, but not merlin, ezrin, or radixin, 14-3-3 is believed to be involved in the control of 4.1B/DAL-1-mediated cell growth. Disruption of the interaction of 14-3-3 with the 4.1B FERM domain did not impair the growth-inhibitory effects of 4.1B/DAL-1 in meningioma cells, suggesting that 14-3-3 may not be an indispensable molecule for 4.1B/DAL-1-mediated growth inhibition [26, 43].
Caspr/paranodin and Caspr2
Several 4.1 proteins are expressed in the nervous system. However, only 4.1B is enriched at paranodes and juxtaparanodes in myelinated axons. 4.1B associates with the neural cell adhesion molecules, Caspr/paranodin and Caspr2 (a protein closely related to Caspr/paranodin), at paranodes and juxtaparanodes, respectively. Both Caspr and Caspr2 bind to the N-terminal FERM domain of protein 4.1B through their GNP motifs [55]. Caspr is required for the generation of a membrane barrier at the paranodal junction (PNJ), which prevents juxtaparanodal components from entering the paranodal space. The interaction of Caspr with protein 4.1B is necessary for its stabilization at the PNJ and the generation of an efficient membrane barrier at this site. Loss of 4.1B in the PNS leads to mislocalization of Caspr at the paranodes and destabilization of paranodal axo-glial septate junctions. Caspr2 is necessary for clustering Kv1 channels at the juxtaparanodal region (JXP) [32, 56]. As for Caspr2, 4.1B is necessary for the accumulation of Caspr2 and Kv1 channels at juxtaparanodal axonal membrane. In 4.1B mutant mice, Caspr2 and Kv1 channels are not clustered at the JXP [30, 31]. Thus, 4.1B is a critical component of the paranodal and juxtaparanodal domains and play an important role in the organization of myelinated axons.
pICln and NBC1
pICln, a cell-swelling activated chloride channel that is primarily present in the cytosol of resting cells, is targeted to and inserted into the plasma membrane in response to cell swelling. Once inserted into the membrane, pICln forms a channel-like structure, resulting in Cl− efflux and regulatory volume decrease [57–59]. pICln has been found to bind to the FERM domain of 4.1B and 4.1R, both of which may anchor pICln to the cell membrane and link pICln to the spectrin/actin skeleton to regulate cell volume [12, 60]. A high metabolic rate is a prominent characteristic of the majority of malignant tumors. Tumor cells have very efficient acid extrusion mechanisms that help dispose of excess acid to the point of actually decreasing cytosolic [H+] levels below those of normal cells. Membrane acid–base transport represents a key pathway for the disposal of cellular acid and can be mediated by a number of ion transport proteins, such as NHE1 (Na+/H+ exchanger 1) and NBC1. NHE1, a 4.1R-binding molecule, is significantly activated in 4.1R-deficient erythrocytes [61]. Interestingly, over-activation of NHE1 can cause cytoplasmic alkalization in breast cancer cells and generate an acidic hypoxic tumor microenvironment, thereby promoting tumor cell proliferation and metastasis [21]. 4.1B can interact with NBC1 in the basolateral membrane of the renal proximal tubules [45]. NBC1 has been found to be expressed in renal cell carcinomas and may be involved in the establishment of an acidic microenvironment [62]. Evidence suggests that an increase in cell volume and intracellular alkalinization occurs during cell proliferative processes [63, 64]. Therefore, 4.1 proteins may maintain normal cell volume and intracellular homeostasis by stabilizing the cytoskeleton and regulating their binding proteins, such as pICln, NHE1, and NBC1. Loss of 4.1 proteins may lead to dysregulation of critical ion transport proteins and thus promote tumor development.
PRMT family
PRMT family members can catalyze the sequential transfer of methyl groups from S-adenosyl-l-methionine to the guanidino nitrogens of arginine residues in proteins. Protein methylation by PRMTs has been implicated in the regulation of various processes, including signal transduction, transcriptional regulation, and RNA splicing [65]. 4.1B/DAL-1 is not only a substrate for PRMT3/5, but it can also suppress PRMT3-mediated protein methylation and enhance or inhibit PRMT5-mediated protein methylation in a substrate-specific manner [44, 46]. 4.1B/DAL-1 may regulate the intracellular mRNA splicing process by inhibiting PRMT5-mediated spliceosomal Sm proteins, including B/B′, D1 and D3, methylation. Both PRMT5 and pICln are components of the 20S methyl transfectase enzyme complex (methylosome). In the presence of pICln, pICln interacts with PRMT5 and stimulates PRMT5 methylation of Sm proteins [66]. Methylation of Sm proteins by the methylosome is necessary for directing Sm proteins from methylosome to the SMN (survival of motor neuron) complex for assembly into snRNP (small nuclear ribonucleoprotein) core particles in the cytoplasm [67, 68]. Subsequently, snRNPs transport into the nucleus and participate in pre-mRNA splicing events. It is possible that a 4.1B/DAL-1-PRMT5-pICln complex may either interfere with PRMT5 protein and pICln protein assembly on methylosome or prevent PRMT5 from methylating the spliceosomal Sm B/B′, D1, and D3 proteins. Abnormal functions of 4.1B/DAL-1 may lead to a disruption of cells’ ability to form functional SMN complexes, thereby causing abnormal pre-mRNA splicing [44].
Integrins
Integrins are heterodimeric transmembrane receptors that consist of α and β glycoprotein subunits. Integrins are involved in mediating cellular interactions with extracellular matrix (ECM) and cell–cell interactions. Studies have identified that 4.1B associated with αvβ8 integrin through direct interaction between cytoplasmic tail of β8 and CTD of 4.1B. 4.1B-αvβ8 integrin-mediated adhesion and signaling pathways are important for the normal formation and function of the heart and the proper development of the CNS [47, 48]. It has been reported that αvβ8 integrin expression was generally absent in human lung cancers, suggesting that dysregulation of 4.1B-αvβ8 integrin signaling pathways may be involved in the lung tumor progression [69]. Moreover, although 4.1B has been found to not directly interact with β1 integrin, β1 integrin and its heterodimer partner α5 integrin also showed decreased surface expression in 4.1B-deficient MEF cells. Fluorescence pulse-chase studies found that after 4 h of chase, β1 integrin in 4.1B WT-MEF cells could undergo continuous internalization and recycling. However, in 4.1B-deficient MEF cells, the internalized β1 integrin vesicles were mainly accumulated at the perinuclear region and could not recycle to the membrane, suggesting that 4.1B can control cell surface expression of β1 integrin by mediating its trafficking [29].
CD44
CD44 is a transmembrane glycoprotein that binds to hyaluronic acid (HA) in the extracellular matrix. It is involved in cell adhesion, migration, and signaling. Merlin, ERM proteins, and 4.1R and 4.1B have all been shown to bind to CD44. Merlin and ERM proteins maybe have different or even opposite effects on CD44 signaling events. CD44 has been proposed to be involved in the formation and elongation of microvilli in various cell types by concentrating activated ERM proteins at the plasma membrane [70]. However, Merlin may compete with ERM proteins for binding to CD44, which depends on the cell proliferative status. At high cell density, hypophosphorylated/activated merlin displaces ERM proteins from membrane components and mediates contact inhibition signals of growth from the extracellular matrix through its interaction with CD44. Whereas at low cell density phosphorylated/activated ERM proteins associate with CD44 and phosphorylated/inactivated merlin, this complex might serve to prevent merlin activation and facilitate cell proliferation [17, 71]. Protein 4.1B/DAL-1 and 4.1R also interact with CD44 via their FERM domain. It has been demonstrated that incremental expression of both protein 4.1B and 4.1R was similar to merlin in the cell membrane fractions under growth arrest conditions [27, 72]. It is tempting to speculate that under growth arrest conditions, protein 4.1B and 4.1R are redistributed to the cell membrane where they may interact with critical effector molecules like CD44 to negatively regulate cell growth, as has been described for merlin.
PIP2
Phosphatidylinositol 4,5-biphosphate (PIP2) is a low-abundance phospholipid in the plasma membrane inner leaflet. PIP2 interacts with the FERM domain and regulates the activities of protein 4.1 superfamily proteins, especially their membrane and actin binding activities. For example, PIP2 plays a regulatory role in the association of the ERM proteins with membrane proteins and with the actin cytoskeleton. PIP2 binding triggers conformational changes that unmask the membrane protein-binding sites in the FERM domain. Such conformational changes are critical for the association of ERM proteins with cytoplasmic tail of CD44 and the recruitment of ERM proteins to the plasma membrane in vivo and in vitro [73, 74]. In addition, the sequence of events consisting of binding to PIP2 followed by the phosphorylation is critical for the proper activation and correct apical localization of ezrin in epithelial cells [75]. Binding of PIP2 to 4.1R selectively modulated the ability of 4.1R to interact with its different binding partners. It has been reported that 4.1R bound to PIP2-containing liposomes through its FERM domain. PIP2 binding induced a conformational change in this domain accompanied by an increase in glycophorin C (GPC) binding and a decrease in band3 binding. The potential PIP2-binding motifs are highly conserved in the protein 4.1 family. It is likely that the functions of 4.1B are similarly regulated by PIP2 in many different cell types [49].
TSLC-1
TSLC-1 is an immunoglobulin superfamily cell-adhesion molecule and also termed CADM1/Necl-2/SgIGSF/RA175/IGSF4/SynCAM1. TSLC-1 can maintain the phenotype of epithelial cells and inhibit EMT (epithelial–mesenchymal transition) through the formation of mature cell–cell adhesion [52, 76]. TSLC-1 is frequently absent in many types of epithelial tumors, particularly in some invasive or metastatic tumors [77, 78]. In culture, NSCLC cells lacking TSLC-1 display a typical transformed morphology with immature cell adhesions [79]. The cytoplasmic transmembrane domain of TSLC-1 contains a 4.1-binding motif, which is homologous to neurexin IV, paranodin, syndecan-2, and GPC. TSLC-1 can bind to the lobe C in the FERM domain of 4.1B/DAL-1 via the 4.1-binding motif (Fig. 4) [50]. The fact has been confirmed that TSLC-1 interacted with the actin cytoskeleton and participated in organizing the actin cytoskeleton through 4.1B/DAL-1 [50, 80, 81]. The PDZ (PSD-95/Dlg/ZO-1) binding motif at the C-terminus of TSCL-1 is linked to MAGUK family members, including MPP1/2/3, CASK, Pals2, and syntenin [52, 79]. Both the 4.1- and PDZ-binding motifs may play important roles in the lateral localization of TSLC-1 in human epithelia [76]. Moreover, the 4.1- and PDZ-binding motifs are likely involved in the anti-tumor activity of TSLC-1. Since deletion of the 4.1- or PDZ-binding motifs significantly enhanced the tumorigenicity of the A549 NSCLC cell line in a nude mouse model [82]. In addition, the presence of the 4.1- and PDZ-binding motifs is essential for the ability of TSLC-1 to inhibit cell proliferation and induce apoptosis in A549 cells [83]. NSCLC and breast cancers frequently display a loss of TSLC-1 and/or 4.1B/DAL-1 [81, 84, 85]. Interestingly, no significant difference in clinicopathological features was observed between the tumors with low expression of either 4.1B/DAL-1 or TSLC-1 versus both 4.1B/DAL-1 and TSLC-1 in primary breast cancer [78]. This evidence strongly suggests that dysfunction of the TSLC-1-4.1B/DAL-1 cascade is involved in the pathogenesis of breast cancers.
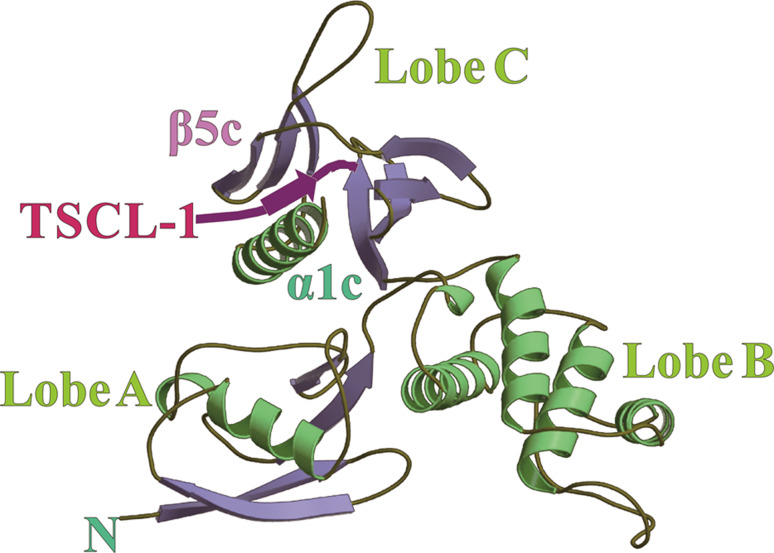
Fig. 4.
Molecular graphic illustrating the interaction of the DAL-1 FERM domain with TSLC-1.The DAL-1 FERM domain consists of three lobes (Lobe A, B, and C), interacting with the TSLC-1 peptide (pink) in a cleft between helix α1C and β-strand β5C [50]
Membrane-associated guanylate kinases (MAGUKs)
MAGUKs are a large family of scaffold proteins that are located at sites of epithelial cell–cell contact and at the synaptic junctions of neuronal cells [86]. Three MAGUK proteins, including MPP1/p55 [87], CASK (calcium/calmodulin-dependent serine protein kinase) [88], and hDlg (human discs large)/SAP97 [89], share a conserved protein 4.1-binding site in their HOOK domain. 4.1 proteins can form a 4.1-transmembrane protein-MAGUK ternary complex. This ternary complex provides a functional unit serving as a clustering apparatus at the plasma membrane. MPP3, Drosophila discs large tumor suppressor homologue, has been demonstrated to form a ternary complex with TSLC-1 and 4.1B/DAL-1 at epithelial cell–cell attachment sites. However, MPP3 expression has been detected in all NSCLC and SCLC (small-cell lung cancer) cell lines. Whether MPP3 is implicated in the tumorigenesis of lung cancer still needs further studies [81, 90].
4.1B/DAL-1 inactivation and tumor progression
Mechanisms of 4.1B/DAL-1 inactivation
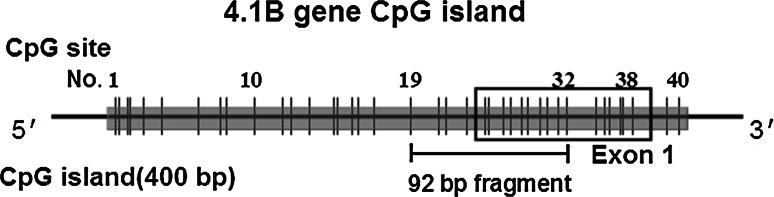
Many studies have confirmed that protein 4.1B/DAL-1 is lost or down-regulated in a variety of malignant and benign tumors (Table 2), such as NSCLC [25, 85, 91], breast cancer [78, 84, 92], meningiomas [93, 94], ependymomas [95, 96], renal cell carcinomas [34], prostate cancer [38, 97], ovarian cancer [39], liver cancer [98], nasal NK/T cell lymphomas [99], and colon cancer [35]. Biallelic promoter methylation and loss of heterozygosity (LOH) at the 4.1B/DAL-1 locus on 18p11.3 are the main mechanisms responsible for the inactivation of 4.1B/DAL-1 gene expression (Tables 3, 4). An approximately 400-bp fragment around the promoter and exon 1 of the 4.1B/DAL-1 gene contains 38 CpG sites with a GC content of 75 %, matching the criteria for a CpG island. Among them, hypermethylation of the 14 CpG sites (CpG sites 19-32) within the 92-bp fragment around the 4.1B/DAL-1 promoter (Fig. 5) played a critical role in 4.1B/DAL-1 gene silencing [34, 85]. Monosomy of chromosome 18 or large deletions on chromosome 18p are the main mechanisms for LOH of 4.1B/DAL-1 gene [92, 94]. In addition, point mutations have not been demonstrated to be the main mechanism responsible for 4.1B/DAL-1 inactivation [100]. There are additional mechanisms leading to the silencing of 4.1B/DAL-1 that do exist, such as histone deacetylation and loss of transcription factor, which require further investigation.
Table 2.
4.1B/DAL-1 deficiency in tumors
| Tumor type | Number of cases/cell lines | Number of cases/cell lines with 4.1B/DAL-1 deficiency (%) | References |
|---|---|---|---|
| NSCLC cell lines | 16 | 14 (87) | [81, 91] |
| Adenocarcinoma | 11 | 9 (82) | |
| Squamous cell carcinoma | 1 | 1 (100) | |
| Large cell carcinoma | 3 | 3 (100) | |
| Nonspecified NSCLCs | 1 | 1 (100) | |
| Pleural fluid NSCLC cells | 10 | 9 (90) | |
| SCLC cell lines | 11 | 4 (36) | |
| Primary NSCLC tumors | 39 | 21 (54) | [25] |
| Adenocarcinoma | 15 | 6 (40) | |
| Squamous cell carcinoma | 16 | 11 (69) | |
| Large cell carcinoma | 1 | 1 (100) | |
| Nonspecified NSCLCs | 7 | 3 (43) | |
| Breast cancer | |||
| Breast cancer cell lines | 8 | 6 (75) | [84] |
| Primary breast cancers | 67 | 49 (73) | [78] |
| Lymph node metastasis | |||
| 0 | 46 | 29 (63) | |
| 1–3 | 17 | 16 (94) | |
| ≥4 | 4 | 4 (100) | |
| Lymphovascular invasion | |||
| − | 50 | 33 (66) | |
| + | 17 | 16 (94) | |
| pT stage | |||
| 1 | 31 | 18 (58) | |
| 2 | 33 | 28 (85) | |
| 3 | 3 | 3 (100) | |
| Pathological stage | |||
| I | 23 | 11 (48) | |
| II | 40 | 34 (85) | |
| III | 4 | 4 (100) | |
| Renal clear cell carcinoma | |||
| RCC cell lines | 19 | 10 (53) | [34] |
| Primary RCCC | 19 | 12 (63) | |
| Meningioma | |||
| Benign, non-recurring (grade I) | 20 | 12 (60) | [77] |
| Benign, recurring (grade I) | 16 | 10 (63) | |
| Atypical (grade II) | |||
| High proliferative | 23 | 15 (65) | |
| Brain invasive, low proliferative | 22 | 17 (77) | |
| Anaplastic (grade III) | 7 | 6 (86) | |
| Ovarian cancer | |||
| Ovarian cancer cell lines | 19 | 15 (78) | [39] |
| Benign ovarian tumors | 33 | 8 (24) | |
| Invasive ovarian tumors | 794 | 524 (66) | |
| Clear cell | 61 | 42 (69) | |
| Serous | 454 | 300 (66) | |
| Mucinous | 70 | 43 (61) | |
| Endometrioid | 150 | 86 (57) | |
| Undifferentiated | 38 | 10 (26) | |
| Papillary | 21 | 8 (38) | |
Table 3.
4.1B/DAL-1 methylation in tumors
| Tumor type | Number of cases/cell lines | Number of cases/cell lines with 4.1B/DAL-1 methylation (%) | References |
|---|---|---|---|
| Lung cancer | |||
| NSCLC cell lines | 39 | 17 (44) | [81, 91] |
| Adenocarcinoma | 28 | 12 (43) | |
| Squamous cell carcinoma | 5 | 3 (60) | |
| Large cell carcinoma | 6 | 2 (33) | |
| Pleural fluid NSCLC cells | 9 | 6 (67) | |
| SCLC cell lines | 11 | 3 (27) | |
| Primary lung tumors | [81, 85] | ||
| NSCLC | |||
| Adenocarcinoma | 133 | 75 (56) | |
| Squamous cell carcinoma | 109 | 60 (55) | |
| Large cell carcinoma | 8 | 4 (50) | |
| Adenosquamous cells carcinoma | 18 | 8 (44) | |
| SCLC | 4 | 0 (0) | |
| Corresponding nonmalignant lung tissue samples | 30 | 3 (10) | |
| Pleural fluid NSCLC | 103 | 59 (57) | |
| Breast cancer | |||
| Breast cancer cell lines | 8 | 6 (75) | [84] |
| Primary breast cancers | 95 | 26 (27) | |
| Ductal carcinoma | 81 | 21 (26) | |
| Lobular carcinoma | 13 | 4 (31) | |
| Other | 1 | 1 (100) | |
| Renal clear cell carcinoma | |||
| RCC cell lines | 19 | 9 (47) | [34] |
| Primary RCCC | 55 | 25 (45) | |
| Prostate cancer | |||
| Prostate carcinoma cell lines | 4 | 3 (75) | [97] |
| Primary tumor tissues | 113 | 89 (79) | |
Table 4.
4.1B/DAL-1 LOH in tumors
| Tumor type | Number of cases/cell lines | Number of cases/cell lines with 4.1B/DAL-1 LOH (%) | References |
|---|---|---|---|
| Lung cancer | |||
| Primary NSCLC tumors | 100 | 39 (39) | [85] |
| Adenocarcinoma | 66 | 26 (39) | |
| Squamous cell carcinoma | 25 | 8 (32) | |
| Large cell carcinoma | 7 | 3 (43) | |
| Adenosquamous cells Carcinoma | 2 | 2 (100) | |
| Pleural fluid NSCLC | 100 | 39 (39) | |
| Breast cancer | |||
| Breast cancer tissues | 43 | 27 (63) | [92] |
| Ductal carcinoma | 16 | 9 (56) | |
| Invasive ductal carcinoma | 21 | 14 (67) | |
| Metastatic disease | 6 | 4 (67) | |
| Renal clear cell carcinoma | |||
| RCC cell lines | 19 | 9 (47) | [34] |
| Primary RCCC | 54 | 4 (7.4) | |
| Sporadic meningioma | |||
| Sporadic meningioma | 62 | 12 (19) | [94] |
| Benign | 31 | 1 (3) | |
| Malignant | 5 | 3 (60) | |
Fig. 5.
Results of the methylation analysis of the 5′ upstream region of the 4.1B promoter and schematic. Vertical bars indicate CpG sites numbered 1–40. The box indicates exon1 of the 4.1B/DAL-1 gene. The sequence traces correspond to the 92-bp fragment containing the 14 CpG sites numbered 19 through 32 within the upstream region of the transcription initiation site and the beginning of exon 1
The relationship between 4.1B/DAL-1 and tumor progression
In squamous cell lung carcinomas, DAL-1 methylation seemed to be a relatively early event; the incidence was 90 % in stage I tumors, but it was not altered with tumor progression to advanced stages. In contrast, DAL-1 methylation may be a late event in lung adenocarcinomas, since the incidence increased significantly as the tumor stage advanced from stage I to stage IV. Preferential DAL-1 methylation was also observed in invasive, metastatic, and pleural lung adenocarcinomas. Furthermore, a significant correlation between DAL-1 methylation in tumors and shorter disease-free and overall survival was observed for patients with lung adenocarcinomas. Therefore, DAL-1 methylation may be used as a potential prognostic indicator for NSCLC, particularly for lung adenocarcinomas [85]. In breast cancer, the incidence of TSLC-1 and/or DAL-1 methylation in patients with grade 3 tumors was more frequent than in patients with grade 1 and 2 tumors [84]. Yuka et al. confirmed that as compared to non-invasive lesions, invasive lesions generally display low and abnormal 4.1B expression in breast cancer tissue samples. Additionally, low or abnormal 4.1B expression in tumors was significantly correlated with lymph node metastasis, advanced pT stages of pT2 and pT3, and advanced pathological stages of II and III [78]. In kidney, 4.1B is located in the basolateral membrane of the renal proximal tubules, while RCCC primarily originate from the proximal tubule cells [45, 51]. Abnormal 4.1B methylation likely occurs during the early stages of RCCC. Since 4.1B hypermethylation occurred in 42 % of tumors with pathological stage I and 47 % of tumors with stage pT1a, while the incidence of 4.1B hypermethylation did not increase significantly in tumors with more advanced stages. In addition, 4.1B hypermethylation was significantly associated with higher nuclear grade (an indicator of abnormal nucleus of tumor cells) and shorter recurrence-free survival [34]. In meningiomas, LOH of DAL-1 at chromosome 18p11.3 was originally reported in 71 % (12/17) of sporadic meningiomas, regardless of histological grade, suggesting that loss of DAL-1 was an early event in the tumorigenesis of meningiomas [93]. However, subsequently, Nunes et al. reported that LOH of 4.1B/DAL-1 occurred only in 19 % (12/62) of meningiomas. LOH of 4.1B/DAL-1 was more common in malignant meningiomas (three of five tumors, 60 %) than that in benign meningiomas (1 of 31, 3.2 %), indicating that loss of 4.1B/DAL-1 may be involved in the development of meningiomas rather than an initiating event [94]. In ovarian cancers, the expression of 4.1B was found in normal ovarian epithelia tissues. However, there is absent expression of 4.1B in 24 % of benign ovarian tumors and in 66 % of invasive ovarian tumors. This suggests that 4.1B loss is a late rather than an initiating event during the ovarian cancer development process [39]. In prostate cancer, Wong et al. [38] established an orthotopic xenotransplant model of prostate cancer by using PC3 cells, a human prostate adenocarcinoma cell line, and separated the highly, moderately, and poorly metastatic cell sublines. This study found that 4.1B expression was significantly down-regulated in highly metastatic cell sublines as compared to parental PC3 cells and the poorly metastatic cell sublines, and showed an intermediate level in moderately metastatic cell sublines. Down-regulation of 4.1B level in poorly metastatic cell sublines enhanced their metastatic propensity in an orthotopic model of prostate cancer, suggesting that down-regulation of 4.1B promotes prostate cancer progression to metastatic phenotype. Furthermore, 4.1B-deficient mice displayed increased predisposition to developing aggressive, undifferentiated carcinomas relative to 4.1B-heterozygous mice. In both models, tumorigenic cells lacking 4.1B displayed reduced rates of apoptosis.
Antitumor function of protein 4.1B/DAL-1
Metastasis is a complex multistep process. Tumor cells require a series of molecular changes to assist them to progress through every step of the metastatic cascade, including cytoskeletal rearrangements and alteration in adhesiveness, increased motility and invasion, and enhanced survival and proliferation. Recent studies have shown that 4.1B/DAL-1 can maintain cell adhesion and F-actin cytoskeleton organization, participate in regulating cells differentiation, growth, motility, and apoptosis, suggesting that 4.1B/DAL-1 might act as an important tumor suppressor in tumor progression.
Protein 4.1B/DAL-1 and cell differentiation
4.1B/DAL-1 plays a crucial regulatory role in cell growth and differentiation. In normal epithelia, 4.1B/DAL-1 is related to cell proliferation and differentiation and its high expression could be a marker for the maturity of epithelial cells [101]. In the intestinal epithelium, 4.1B/DAL-1 strongly expresses in the basolateral membrane of the simple columnar epithelium in villi [35, 101]. Intestinal epithelial cells originate from the stem cells in the crypts and subsequently undergo functional and morphological differentiation as they migrate along the crypt–villus axis. Interestingly, 4.1B/DAL-1 expression gradually increased during the epithelial cells’ migration from the crypt to the top of villi, with the crypt showing no expression of 4.1B [35, 101]. However, the membranous expression of protein 4.1B/DAL-1 was reduced during the malignant transformation of the intestinal epithelium [35]. In addition, two studies reported a concomitant loss of 4.1B and E-cadherin at cell–cell contact regions upon the phenotypic transition from adenoma (benign tumor) to carcinoma (invasive malignant tumor) in the pancreatic and colonic epithelia [36, 101]. This evidence suggests that 4.1B/DAL-1 may function in maintaining an epithelial and differentiated organization, and normal cellular proliferation and adhesion, thereby preventing the malignant transformation of epithelial cells.
Protein 4.1B/DAL-1 maintains cell adhesion
The desquamation of cells from the primary tumor caused by disruption of intercellular adhesion allows metastatic cells to escape from their original site and to acquire a more motile and invasion phenotype. To date, concomitant loss of both TSLC-1 and 4.1B/DAL-1 has been frequently observed in NSCLC, breast cancer, meningiomas, and nasal NK/T-cell lymphomas [81, 84, 85, 99]. Sakurai-Yageta et al. [52] reported that 4.1B-TSLC-1-MPPs complexes were distributed at cell–cell adhesion sites in HEK293 cells. Inhibition of TSLC-1 expression in HEK293 cells by siRNA caused abrogation of epithelial-like structure and displayed a flat morphology with immature cell adhesions. In addition, HEK293 cells lacking TSLC-1 showed the mislocalization of 4.1B, MPP2, E-cadherin, and ZO-1 from the membrane. 4.1B/DAL-1 is not only an adhesion molecule but the TSLC-1-4.1B/DAL-1 cascade appears to be involved in building stable adhesions between adjacent epithelial cells and organizing the actin cytoskeleton [80]. Functional loss of TSLC-1 and/or 4.1B/DAL-1 may disrupt intercellular adhesion and the association between the membrane and the cytoskeleton, which may lead tumor cells to metastasis or invasion. Furthermore, a recent study found that 4.1B/DAL-1 was associated with CADM4, a protein homology to TSLC-1, in human proximal tubules. Loss of CADM4 is involved in the tumorigenicity of human renal cell carcinoma. Interestingly, disruption of the CADM4-4.1B cascade frequently occurs in human RCC cells and RCCC tumors. The average size of tumors lacking CADM4 or 4.1B or both was significantly larger than that of the tumors expressing both CADM4 and 4.1B. In addition, no pathological changes were observed between the tumors lacking both CADM4 and 4.1B and those lacking only CADM4 or 4.1B, suggesting that CADM4-4.1B cascade participated in normal cell adhesion of the proximal tubules [51].
Protein 4.1B/DAL-1 suppresses cell motility
Metastatic tumor cells are characterized by more rapid cell migration and the presence of more pseudopodia-like protrusions than primary tumor cells [102]. The morphological and dynamic changes in these highly motile structures originate from the continuous reorganization of the actin cytoskeleton in response to extracellular signals [103]. However, 4.1B/DAL-1 can anchor F-actin to the cell membrane, inhibiting cell motility by supporting orderly arrangements of actin stress fibers. 4.1B/DAL-1 loss may promote F-actin reorganization and enhance cell motility. For example, Cavanna et al. [104] demonstrated that the depletion of 4.1B caused the loss of stress fibers in non-metastatic sarcoma cells and was accompanied by enhanced cell motility, while metastatic sarcoma cells exogenously expressing 4.1B migrated at half the speed of control metastatic cells. Furthermore, the oncogenic transcription factor E-26 related gene (ERG) is frequently overexpressed in prostate cancer, which results in the increased invasiveness and metastasis of prostate cancer cells. Significantly lower levels of 4.1B expression were exhibited by prostate tumors overexpressing ERG compared with tumors that did not overexpress ERG. It is speculated that 4.1B down-regulation may promote ERG-mediated prostate cancer progression [105].
Protein 4.1B/DAL-1 inhibits cell proliferation
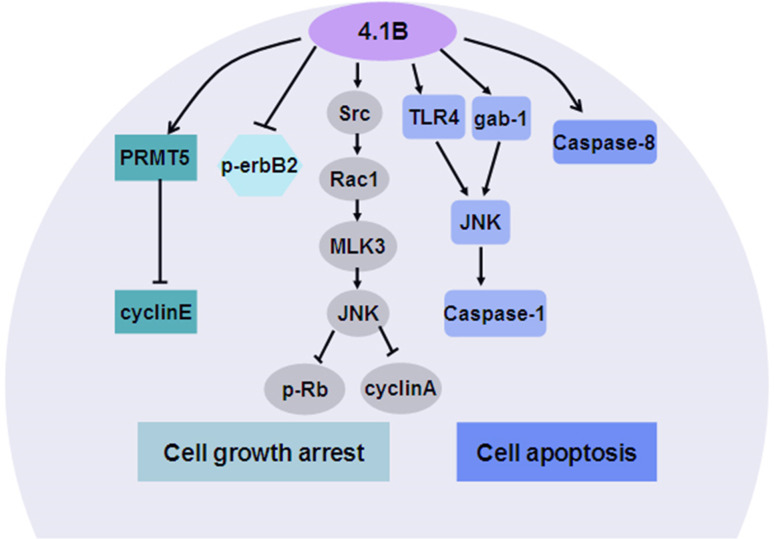
The proper membrane localization of the U2 domain seems to be necessary for protein 4.1B/DAL-1-mediated growth suppression. Gerber et al. found that when the U2 domain was properly localized to the plasma membrane, 4.1B/DAL-1 suppressed meningioma cell growth by sequentially activating Src, Rac1, MLK3, and JNK, resulting in reduced expression levels of cyclinA and decreased retinoblastoma (Rb) protein hyperphosphorylation (see Fig. 6). However, deletion of the U2 domain of DAL-1 abolished the ability for suppressing meningioma cell growth [14, 15]. Kuns et al. [40] reported that 4.1B loss significantly enhanced mammary epithelial cell proliferation of pregnant 4.1B-null mice, while 4.1B overexpression resulted in reduced cyclinA expression and Rb phosphorylation, which was accompanied by decreased tyrosine kinase receptor erbB2 phosphorylation, causing the mammary epithelial cells to arrest in G1 phase. However, the G1 cell cycle arrest-related signaling molecules JNK, MAPK, and Akt were not activated, which suggested that specific signaling pathways involved in 4.1B/DAL-1-mediated growth inhibition may exist in different types of cells. Furthermore, 4.1B/DAL-1 may mediate its growth-inhibitory effects via PRMT5, its binding partner. PRMT5 has an inhibitory effect on cyclinE1 promoter activity through methylating histone H4 on the nucleosome that surrounds the cyclinE1 regulatory element [106]. The abnormal expression of 4.1B/DAL-1 likely alters the association of cyclinE1 promoter with histone H4 by affecting PRMT5 methylation, and then results in cell cycle change.
Fig. 6.
Mechanism underlying the effects of protein 4.1B/DAL-1 on tumor cell growth arrest and apoptosis. In the process of cell growth arrest, 4.1B/DAL-1 can suppress cell growth by sequentially activating Src, Rac1, MLK3, and JNK, resulting in reduced cyclinA levels and decreased retinoblastoma (Rb) protein hyperphosphorylation. 4.1B can also inhibit erbB2 phosphorylation, causing cell cycle arrest in the G1 phase. Furthermore, 4.1B/DAL-1 can bind to PRMT5, thereby inhibiting cyclinE1 transcription. In the process of cell apoptosis, 4.1B/DAL-1 can enhance Caspase-8 activation levels to induce cell apoptosis. Alternatively, 4.1B/DAL-1 can activate successively activate TLR4/gab1, JNK, and Caspase-1 signaling to induce apoptosis
Protein 4.1B/DAL-1 promotes apoptosis
4.1B/DAL-1 inhibits tumor cell growth by not only suppressing cell proliferation but also inducing apoptosis (see Fig. 6). 4.1B/DAL-1 induced apoptosis in MCF-7 breast cancer cells through significantly enhancing Caspase-8 activation, but without activation of downstream effecter Caspases (Caspase-3, -6, and -7), suggesting that 4.1B/DAL-1-induced apoptosis in MCF-7 may involve a Caspase-8-dependent pathway that functions independently of the major effector Caspase pathways [107, 108]. Modulation of post-translational protein methylation may also be a potential mechanism underlying 4.1B/DAL-1-induced apoptosis in MCF-7 cells. It has been observed that the apoptosis levels in the 4.1B/DAL-1-null MCF-7 cells were not influenced by the hypomethylating treatment of cells with protein methylation inhibitor adenosine dialdehyde (AdOX). However, AdOX treatment specifically increases the mortality of the 4.1B/DAL-1-induced MCF-7 cells [107]. Thus, the association of 4.1B/DAL-1 with protein methylation pathway components, such as PRMTs protein, is important for controlling tumorigenesis. In addition, Robb et al. [14] reported that in IOMM-Lee meningioma cells, JNK phosphorylation levels and apoptosis-related genes, including Caspase-1, Toll-like receptor (TLR) 4 and grb2-associated binder 1 (gab1), were up-regulated upon DAL-1 expression. JNK is the downstream mediator of TLR4 and gab1 signaling. Several studies have found that TLR4 and gab1 appeared to induce Caspase-mediated apoptosis via stimulating JNK activation [109, 110]. Therefore, it is possible that 4.1B activates JNK activation through TLR4 and/or gab1 signaling, thereby initiating Caspase-mediated apoptosis.
Summary and perspective
Down-regulation or loss of protein 4.1B/DAL-1 expression has been observed in many types of human tumors, which can promote tumor development during the relatively early or late stages. 4.1B/DAL-1 expression is gradually decreased during the development process in lung adenocarcinomas, breast cancers, meningiomas, prostate cancers, and ovarian cancers. In other words, as compared with normal tissues, 4.1B/DAL-1 expression was down-regulated during the early stages. Its expression was then further reduced or absent, promoting the progression of tumors to invasive or metastatic phenotypes. 4.1B/DAL-1 participates in the organization of transmembrane proteins and membrane–cytoskeletal interaction. Loss of it likely alters the assembly of transmembrane proteins, their activation states, and cytoskeletal organization, which may upset membrane-mediated signaling pathways or cell cycle regulation and culminate in malignant metastasis and invasion phenotype. Given that 4.1B/DAL-1 may serve as a negative modulator of tumor progression across a broad-spectrum of tumor types, demethylation of the 4.1B/DAL-1 promoter through DNA methyltransferase inhibitors or delivery of 4.1B/DAL-1 into tumor cells lacking 4.1B/DAL-1 by adenovirus/lentivirus or plasmids may be used as cancer treatment strategies.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 81270576, 81301774, 81301997), New Century Excellent Talents in University (NCET-11-0518), Doctoral Fund of Ministry of Education of China (No. 20120162110054), the fundamental research funds for the central universities (No. 2011JQ015).
Conflict of interest
The authors declare no conflicts of interest.
Footnotes
Z. Wang, J. Zhang, and M. Ye contributed equally to this work.
Contributor Information
Jing Liu, Phone: +86-731-84805026, Email: jingliucsu@hotmail.com.
Xiuli An, Email: xan@nybloodcenter.org.
References
- 1.Sun CX, Robb VA, Gutmann DH. Protein 4.1 tumor suppressors: getting a FERM grip on growth regulation. J Cell Sci. 2002;115:3991–4000. doi: 10.1242/jcs.00094. [DOI] [PubMed] [Google Scholar]
- 2.Diakowski W, Grzybek M, Sikorski AF. Protein 4.1, a component of the erythrocyte membrane skeleton and its related homologue proteins forming the protein 4.1/FERM superfamily. Folia Histochem Cytobiol. 2006;44:231–248. [PubMed] [Google Scholar]
- 3.Clucas J, Valderrama F. ERM proteins in cancer progression. J Cell Sci. 2014;127:267–275. doi: 10.1242/jcs.133108. [DOI] [PubMed] [Google Scholar]
- 4.Conboy JG, Chan J, Mohandas N, Kan YW. Multiple protein 4.1 isoforms produced by alternative splicing in human erythroid cells. Proc Natl Acad Sci USA. 1988;85:9062–9065. doi: 10.1073/pnas.85.23.9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conboy JG, Chan JY, Chasis JA, Kan YW, Mohandas N. Tissue- and development-specific alternative RNA splicing regulates expression of multiple isoforms of erythroid membrane protein 4.1. J Biol Chem. 1991;266:8273–8280. [PubMed] [Google Scholar]
- 6.Tan JS, Mohandas N, Conboy JG. Evolutionarily conserved coupling of transcription and alternative splicing in the EPB41 (protein 4.1R) and EPB41L3 (protein 4.1B) genes. Genomics. 2005;86:701–707. doi: 10.1016/j.ygeno.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Schischmanoff PO, Yaswen P, Parra MK, Lee G, Chasis JA, Mohandas N, Conboy JG. Cell shape-dependent regulation of protein 4.1 alternative pre-mRNA splicing in mammary epithelial cells. J Biol Chem. 1997;272:10254–10259. doi: 10.1074/jbc.272.15.10254. [DOI] [PubMed] [Google Scholar]
- 8.Ramez M, Blot-Chabaud M, Cluzeaud F, Chanan S, Patterson M, Walensky LD, Marfatia S, Baines AJ, Chasis JA, Conboy JG, Mohandas N, Gascard P. Distinct distribution of specific members of protein 4.1 gene family in the mouse nephron. Kidney Int. 2003;63:1321–1337. doi: 10.1046/j.1523-1755.2003.00870.x. [DOI] [PubMed] [Google Scholar]
- 9.Nunomura W, Takakuwa Y. Regulation of protein 4.1R interactions with membrane proteins by Ca2+ and calmodulin. Front Biosci. 2006;11:1522–1539. doi: 10.2741/1901. [DOI] [PubMed] [Google Scholar]
- 10.Nunomura W, Jinbo Y, Isozumi N, Ohki S, Izumi Y, Matsushima N, Takakuwa Y. Novel mechanism of regulation of protein 4.1G binding properties through Ca2+/calmodulin-mediated structural changes. Cell Biochem Biophys. 2013;66:545–558. doi: 10.1007/s12013-012-9502-7. [DOI] [PubMed] [Google Scholar]
- 11.Pinder JC, Gardner B, Gratzer WB. Interaction of protein 4.1 with the red cell membrane: effects of phosphorylation by protein kinase C. Biochem Biophys Res Commun. 1995;210:478–482. doi: 10.1006/bbrc.1995.1685. [DOI] [PubMed] [Google Scholar]
- 12.Calinisan V, Gravem D, Chen RP, Brittin S, Mohandas N, Lecomte MC, Gascard P. New insights into potential functions for the protein 4.1 superfamily of proteins in kidney epithelium. Front Biosci. 2006;11:1646–1666. doi: 10.2741/1911. [DOI] [PubMed] [Google Scholar]
- 13.Baines AJ, Lu HC, Bennett PM. The Protein 4.1 family: Hub proteins in animals for organizing membrane proteins. Biochim Biophys Acta. 1838;2014:605–619. doi: 10.1016/j.bbamem.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 14.Robb VA, Gerber MA, Hart-Mahon EK, Gutmann DH. Membrane localization of the U2 domain of Protein 4.1B is necessary and sufficient for meningioma growth suppression. Oncogene. 2005;24:1946–1957. doi: 10.1038/sj.onc.1208335. [DOI] [PubMed] [Google Scholar]
- 15.Gerber MA, Bahr SM, Gutmann DH. Protein 4.1B/differentially expressed in adenocarcinoma of the lung-1 functions as a growth suppressor in meningioma cells by activating Rac1-dependent c-Jun-NH(2)-kinase signaling. Cancer Res. 2006;66:5295–5303. doi: 10.1158/0008-5472.CAN-05-1628. [DOI] [PubMed] [Google Scholar]
- 16.Perry A, Cai DX, Scheithauer BW, Swanson PE, Lohse CM, Newsham IF, Weaver A, Gutmann DH. Merlin, DAL-1, and progesterone receptor expression in clinicopathologic subsets of meningioma: a correlative immunohistochemical study of 175 cases. J Neuropathol Exp Neurol. 2000;59:872–879. doi: 10.1093/jnen/59.10.872. [DOI] [PubMed] [Google Scholar]
- 17.Morrison H, Sherman LS, Legg J, Banine F, Isacke C, Haipek CA, Gutmann DH, Ponta H, Herrlich P. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 2001;15:968–980. doi: 10.1101/gad.189601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClatchey AI. Merlin and ERM proteins: unappreciated roles in cancer development? Nat Rev Cancer. 2003;3:877–883. doi: 10.1038/nrc1213. [DOI] [PubMed] [Google Scholar]
- 19.Bretscher A, Chambers D, Nguyen R, Reczek D. ERM-Merlin and EBP50 protein families in plasma membrane organization and function. Annu Rev Cell Dev Biol. 2000;16:113–143. doi: 10.1146/annurev.cellbio.16.1.113. [DOI] [PubMed] [Google Scholar]
- 20.Hanada T, Takeuchi A, Sondarva G, Chishti AH. Protein 4.1-mediated membrane targeting of human discs large in epithelial cells. J Biol Chem. 2003;278:34445–34450. doi: 10.1074/jbc.M305209200. [DOI] [PubMed] [Google Scholar]
- 21.Fiévet B, Louvard D, Arpin M. ERM proteins in epithelial cell organization and functions. Biochim Biophys Acta. 2007;1773:653–660. doi: 10.1016/j.bbamcr.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 22.Parra M, Gascard P, Walensky LD, Gimm JA, Blackshaw S, Chan N, Takakuwa Y, Berger T, Lee G, Chasis JA, Snyder SH, Mohandas N, Conboy JG. Molecular and functional characterization of protein 4.1B, a novel member of the protein 4.1 family with high level, focal expression in brain. J Biol Chem. 2000;275:3247–3255. doi: 10.1074/jbc.275.5.3247. [DOI] [PubMed] [Google Scholar]
- 23.Parra M, Gee S, Chan N, Ryaboy D, Dubchak I, Mohandas N, Gascard PD, Conboy JG. Differential domain evolution and complex RNA processing in a family of paralogous EPB41 (protein 4.1) genes facilitate expression of diverse tissue-specific isoforms. Genomics. 2004;84:637–646. doi: 10.1016/j.ygeno.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Liu C, Debnath G, Baines AJ, Conboy JG, Mohandas N, An X. Comprehensive characterization of expression patterns of protein 4.1 family members in mouse adrenal gland: implications for functions. Histochem Cell Biol. 2010;134:411–420. doi: 10.1007/s00418-010-0749-z. [DOI] [PubMed] [Google Scholar]
- 25.Tran YK, Bogler O, Gorse KM, Wieland I, Green MR, Newsham IF. A novel member of the NF2/ERM/4.1 superfamily with growth suppressing properties in lung cancer. Cancer Res. 1999;59:35–43. [PubMed] [Google Scholar]
- 26.Robb VA, Li W, Gutmann DH. Disruption of 14-3-3 binding does not impair Protein 4.1B growth suppression. Oncogene. 2004;23:3589–3596. doi: 10.1038/sj.onc.1207445. [DOI] [PubMed] [Google Scholar]
- 27.Gutmann DH, Hirbe AC, Huang ZY, Haipek CA. The protein 4.1 tumor suppressor, DAL-1, impairs cell motility, but regulates proliferation in a cell-type-specific fashion. Neurobiol Dis. 2001;8:266–278. doi: 10.1006/nbdi.2000.0376. [DOI] [PubMed] [Google Scholar]
- 28.Kang Q, Wang T, Zhang H, Mohandas N, An X. A Golgi-associated protein 4.1B variant is required for assimilation of proteins in the membrane. J Cell Sci. 2009;122:1091–1099. doi: 10.1242/jcs.039644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Song J, An C, Dong W, Zhang J, Yin C, Hale J, Baines AJ, Narla M, An X (2013) A 130 kDa protein 4.1B regulates cell adhesion, spreading and migration of mouse embryo fibroblasts by influencing actin cytoskeleton organization. J Biol Chem [DOI] [PMC free article] [PubMed]
- 30.Horresh I, Bar V, Kissil JL, Peles E. Organization of myelinated axons by Caspr and Caspr2 requires the cytoskeletal adapter protein 4.1B. J Neurosci. 2010;30:2480–2489. doi: 10.1523/JNEUROSCI.5225-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cifuentes-Diaz C, Chareyre F, Garcia M, Devaux J, Carnaud M, Levasseur G, Niwa-Kawakita M, Harroch S, Girault JA, Giovannini M, Goutebroze L. Protein 4.1B contributes to the organization of peripheral myelinated axons. PLoS One. 2011;6:e25043. doi: 10.1371/journal.pone.0025043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buttermore ED, Dupree JL, Cheng J, An X, Tessarollo L, Bhat MA. The cytoskeletal adaptor protein band 4.1B is required for the maintenance of paranodal axoglial septate junctions in myelinated axons. J Neurosci. 2011;31:8013–8024. doi: 10.1523/JNEUROSCI.1015-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Einheber S, Meng X, Rubin M, Lam I, Mohandas N, An X, Shrager P, Kissil J, Maurel P, Salzer JL. The 4.1B cytoskeletal protein regulates the domain organization and sheath thickness of myelinated axons. Glia. 2013;61:240–253. doi: 10.1002/glia.22430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamada D, Kikuchi S, Williams YN, Sakurai-Yageta M, Masuda M, Maruyama T, Tomita K, Gutmann DH, Kakizoe T, Kitamura T, Kanai Y, Murakami Y. Promoter hypermethylation of the potential tumor suppressor DAL-1/4.1B gene in renal clear cell carcinoma. Int J Cancer. 2006;118:916–923. doi: 10.1002/ijc.21450. [DOI] [PubMed] [Google Scholar]
- 35.Ohno N, Terada N, Murata S, Yamakawa H, Newsham IF, Katoh R, Ohara O, Ohno S. Immunolocalization of protein 4.1B/DAL-1 during neoplastic transformation of mouse and human intestinal epithelium. Histochem Cell Biol. 2004;122:579–586. doi: 10.1007/s00418-004-0716-7. [DOI] [PubMed] [Google Scholar]
- 36.Terada N, Ohno N, Yamakawa H, Baba T, Fujii Y, Christofori G, Ohara O, Ohno S. Protein 4.1B in mouse islets of Langerhans and beta-cell tumorigenesis. Histochem Cell Biol. 2003;120:277–283. doi: 10.1007/s00418-003-0573-9. [DOI] [PubMed] [Google Scholar]
- 37.Terada N, Ohno N, Yamakawa H, Baba T, Fujii Y, Zea Z, Ohara O, Ohno S. Immunohistochemical study of protein 4.1B in the normal and W/W(v) mouse seminiferous epithelium. J Histochem Cytochem. 2004;52:769–777. doi: 10.1369/jhc.3A6192.2004. [DOI] [PubMed] [Google Scholar]
- 38.Wong SY, Haack H, Kissil JL, Barry M, Bronson RT, Shen SS, Whittaker CA, Crowley D, Hynes RO. Protein 4.1B suppresses prostate cancer progression and metastasis. Proc Natl Acad Sci USA. 2007;104:12784–12789. doi: 10.1073/pnas.0705499104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dafou D, Grun B, Sinclair J, Lawrenson K, Benjamin EC, Hogdall E, Kruger-Kjaer S, Christensen L, Sowter HM, Al-Attar A, Edmondson R, Darby S, Berchuck A, Laird PW, Pearce CL, Ramus SJ, Jacobs IJ, Gayther SA. Microcell-mediated chromosome transfer identifies EPB41L3 as a functional suppressor of epithelial ovarian cancers. Neoplasia. 2010;12:579–589. doi: 10.1593/neo.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuns R, Kissil JL, Newsham IF, Jacks T, Gutmann DH, Sherman LS. Protein 4.1B expression is induced in mammary epithelial cells during pregnancy and regulates their proliferation. Oncogene. 2005;24:6502–6515. doi: 10.1038/sj.onc.1208813. [DOI] [PubMed] [Google Scholar]
- 41.Terada N, Ohno N, Yamakawa H, Ohara O, Ohno S. Topographical significance of membrane skeletal component protein 4.1 B in mammalian organs. Anat Sci Int. 2005;80:61–70. doi: 10.1111/j.1447-073x.2005.00094.x. [DOI] [PubMed] [Google Scholar]
- 42.Gimm JA, An X, Nunomura W, Mohandas N. Functional characterization of spectrin-actin-binding domains in 4.1 family of proteins. Biochemistry. 2002;41:7275–7282. doi: 10.1021/bi0256330. [DOI] [PubMed] [Google Scholar]
- 43.Yu T, Robb VA, Singh V, Gutmann DH, Newsham IF. The 4.1/ezrin/radixin/moesin domain of the DAL-1/Protein 4.1B tumour suppressor interacts with 14-3-3 proteins. Biochem J . 2002;365:783–789. doi: 10.1042/BJ20020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang W, Roemer ME, Newsham IF. The tumor suppressor DAL-1/4.1B modulates protein arginine N-methyltransferase 5 activity in a substrate-specific manner. Biochem Biophys Res Commun. 2005;329:522–530. doi: 10.1016/j.bbrc.2005.01.153. [DOI] [PubMed] [Google Scholar]
- 45.Terada N, Ohno N, Saitoh S, Seki G, Komada M, Suzuki T, Yamakawa H, Soleimani M, Ohno S. Interaction of membrane skeletal protein, protein 4.1B and p55, and sodium bicarbonate cotransporter1 in mouse renal S1-S2 proximal tubules. J Histochem Cytochem. 2007;55:1199–1206. doi: 10.1369/jhc.7A7266.2007. [DOI] [PubMed] [Google Scholar]
- 46.Singh V, Miranda TB, Jiang W, Frankel A, Roemer ME, Robb VA, Gutmann DH, Herschman HR, Clarke S, Newsham IF. DAL-1/4.1B tumor suppressor interacts with protein arginine N-methyltransferase 3 (PRMT3) and inhibits its ability to methylate substrates in vitro and in vivo. Oncogene. 2004;23:7761–7771. doi: 10.1038/sj.onc.1208057. [DOI] [PubMed] [Google Scholar]
- 47.Jung Y, Kissil JL, McCarty JH. beta8 integrin and band 4.1B cooperatively regulate morphogenesis of the embryonic heart. Dev Dyn. 2011;240:271–277. doi: 10.1002/dvdy.22513. [DOI] [PubMed] [Google Scholar]
- 48.McCarty JH, Cook AA, Hynes RO. An interaction between {alpha}v{beta}8 integrin and Band 4.1B via a highly conserved region of the Band 4.1 C-terminal domain. Proc Natl Acad Sci USA. 2005;102:13479–13483. doi: 10.1073/pnas.0506068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.An X, Zhang X, Debnath G, Baines AJ, Mohandas N. Phosphatidylinositol-4,5-biphosphate (PIP2) differentially regulates the interaction of human erythrocyte protein 4.1 (4.1R) with membrane proteins. Biochemistry. 2006;45:5725–5732. doi: 10.1021/bi060015v. [DOI] [PubMed] [Google Scholar]
- 50.Busam RD, Thorsell AG, Flores A, Hammarstrom M, Persson C, Obrink B, Hallberg BM. Structural basis of tumor suppressor in lung cancer 1 (TSLC1) binding to differentially expressed in adenocarcinoma of the lung (DAL-1/4.1B) J Biol Chem. 2011;286:4511–4516. doi: 10.1074/jbc.M110.174011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagata M, Sakurai-Yageta M, Yamada D, Goto A, Ito A, Fukuhara H, Kume H, Morikawa T, Fukayama M, Homma Y, Murakami Y. Aberrations of a cell adhesion molecule CADM4 in renal clear cell carcinoma. Int J Cancer. 2012;130:1329–1337. doi: 10.1002/ijc.26160. [DOI] [PubMed] [Google Scholar]
- 52.Sakurai-Yageta M, Masuda M, Tsuboi Y, Ito A, Murakami Y. Tumor suppressor CADM1 is involved in epithelial cell structure. Biochem Biophys Res Commun. 2009;390:977–982. doi: 10.1016/j.bbrc.2009.10.088. [DOI] [PubMed] [Google Scholar]
- 53.Rose M, Dutting E, Enz R. Band 4.1 proteins are expressed in the retina and interact with both isoforms of the metabotropic glutamate receptor type 8. J Neurochem. 2008;105:2375–2387. doi: 10.1111/j.1471-4159.2008.05331.x. [DOI] [PubMed] [Google Scholar]
- 54.Obsilova V, Silhan J, Boura E, Teisinger J, Obsil T. 14-3-3 proteins: a family of versatile molecular regulators. Physiol Res. 2008;57(Suppl 3):S11–S21. doi: 10.33549/physiolres.931598. [DOI] [PubMed] [Google Scholar]
- 55.Denisenko-Nehrbass N, Oguievetskaia K, Goutebroze L, Galvez T, Yamakawa H, Ohara O, Carnaud M, Girault JA. Protein 4.1B associates with both Caspr/paranodin and Caspr2 at paranodes and juxtaparanodes of myelinated fibres. Eur J Neurosci. 2003;17:411–416. doi: 10.1046/j.1460-9568.2003.02441.x. [DOI] [PubMed] [Google Scholar]
- 56.Duflocq A, Chareyre F, Giovannini M, Couraud F, Davenne M. Characterization of the axon initial segment (AIS) of motor neurons and identification of a para-AIS and a juxtapara-AIS, organized by protein 4.1B. BMC Biol. 2011;9:66. doi: 10.1186/1741-7007-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ritter M, Ravasio A, Jakab M, Chwatal S, Furst J, Laich A, Gschwentner M, Signorelli S, Burtscher C, Eichmuller S, Paulmichl M. Cell swelling stimulates cytosol to membrane transposition of ICln. J Biol Chem. 2003;278:50163–50174. doi: 10.1074/jbc.M300374200. [DOI] [PubMed] [Google Scholar]
- 58.Musch MW, Luer CA, Davis-Amaral EM, Goldstein L. Hypotonic stress induces translocation of the osmolyte channel protein pICln in embryonic skate (Raja eglanteria) heart. J Exp Zool. 1997;277:460–463. doi: 10.1002/(sici)1097-010x(19970415)277:6<460::aid-jez6>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 59.Buyse G, de Greef C, Raeymaekers L, Droogmans G, Nilius B, Eggermont J. The ubiquitously expressed pICln protein forms homomeric complexes in vitro. Biochem Biophys Res Commun. 1996;218:822–827. doi: 10.1006/bbrc.1996.0146. [DOI] [PubMed] [Google Scholar]
- 60.Tang CJ, Tang TK. The 30-kD domain of protein 4.1 mediates its binding to the carboxyl terminus of pICln, a protein involved in cellular volume regulation. Blood. 1998;92:1442–1447. [PubMed] [Google Scholar]
- 61.Rivera A, De Franceschi L, Peters LL, Gascard P, Mohandas N, Brugnara C. Effect of complete protein 4.1R deficiency on ion transport properties of murine erythrocytes. Am J Physiol Cell Physiol. 2006;291:C880–C886. doi: 10.1152/ajpcell.00436.2005. [DOI] [PubMed] [Google Scholar]
- 62.Yamada H, Yamazaki S, Moriyama N, Hara C, Horita S, Enomoto Y, Kudo A, Kawakami H, Tanaka Y, Fujita T, Seki G. Localization of NBC-1 variants in human kidney and renal cell carcinoma. Biochem Biophys Res Commun. 2003;310:1213–1218. doi: 10.1016/j.bbrc.2003.09.147. [DOI] [PubMed] [Google Scholar]
- 63.Lang F, Shumilina E, Ritter M, Gulbins E, Vereninov A, Huber SM. Ion channels and cell volume in regulation of cell proliferation and apoptotic cell death. Contrib Nephrol. 2006;152:142–160. doi: 10.1159/000096321. [DOI] [PubMed] [Google Scholar]
- 64.Lagadic-Gossmann D, Huc L, Lecureur V. Alterations of intracellular pH homeostasis in apoptosis: origins and roles. Cell Death Differ. 2004;11:953–961. doi: 10.1038/sj.cdd.4401466. [DOI] [PubMed] [Google Scholar]
- 65.McBride AE, Silver PA. State of the arg: protein methylation at arginine comes of age. Cell. 2001;106:5–8. doi: 10.1016/s0092-8674(01)00423-8. [DOI] [PubMed] [Google Scholar]
- 66.Pesiridis GS, Diamond E, Van Duyne GD. Role of pICLn in methylation of Sm proteins by PRMT5. J Biol Chem. 2009;284:21347–21359. doi: 10.1074/jbc.M109.015578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M, Dreyfuss G. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol. 2001;21:8289–8300. doi: 10.1128/MCB.21.24.8289-8300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, Pestka S, Clarke S. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem. 2001;276:32971–32976. doi: 10.1074/jbc.M105412200. [DOI] [PubMed] [Google Scholar]
- 69.Cambier S, Mu DZ, O’Connell D, Boylen K, Travis W, Liu WH, Broaddus VC, Nishimura SL. A role for the integrin alphavbeta8 in the negative regulation of epithelial cell growth. Cancer Res. 2000;60:7084–7093. [PubMed] [Google Scholar]
- 70.Yonemura S, Tsukita S. Direct involvement of ezrin/radixin/moesin (ERM)-binding membrane proteins in the organization of microvilli in collaboration with activated ERM proteins. J Cell Biol. 1999;145:1497–1509. doi: 10.1083/jcb.145.7.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bai Y, Liu YJ, Wang H, Xu Y, Stamenkovic I, Yu Q. Inhibition of the hyaluronan-CD44 interaction by merlin contributes to the tumor-suppressor activity of merlin. Oncogene. 2007;26:836–850. doi: 10.1038/sj.onc.1209849. [DOI] [PubMed] [Google Scholar]
- 72.Robb VA, Li W, Gascard P, Perry A, Mohandas N, Gutmann DH. Identification of a third Protein 4.1 tumor suppressor, Protein 4.1R, in meningioma pathogenesis. Neurobiol Dis. 2003;13:191–202. doi: 10.1016/s0969-9961(03)00071-8. [DOI] [PubMed] [Google Scholar]
- 73.Yonemura S, Matsui T, Tsukita S. Rho-dependent and -independent activation mechanisms of ezrin/radixin/moesin proteins: an essential role for polyphosphoinositides in vivo. J Cell Sci. 2002;115:2569–2580. doi: 10.1242/jcs.115.12.2569. [DOI] [PubMed] [Google Scholar]
- 74.Hirao M, Sato N, Kondo T, Yonemura S, Monden M, Sasaki T, Takai Y, Tsukita S. Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association: possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J Cell Biol. 1996;135:37–51. doi: 10.1083/jcb.135.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fievet BT, Gautreau A, Roy C, Del Maestro L, Mangeat P, Louvard D, Arpin M. Phosphoinositide binding and phosphorylation act sequentially in the activation mechanism of ezrin. J Cell Biol. 2004;164:653–659. doi: 10.1083/jcb.200307032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Masuda M, Kikuchi S, Maruyama T, Sakurai-Yageta M, Williams YN, Ghosh HP, Murakami Y. Tumor suppressor in lung cancer (TSLC)1 suppresses epithelial cell scattering and tubulogenesis. J Biol Chem. 2005;280:42164–42171. doi: 10.1074/jbc.M507136200. [DOI] [PubMed] [Google Scholar]
- 77.Surace EI, Lusis E, Murakami Y, Scheithauer BW, Perry A, Gutmann DH. Loss of tumor suppressor in lung cancer-1 (TSLC1) expression in meningioma correlates with increased malignancy grade and reduced patient survival. J Neuropathol Exp Neurol. 2004;63:1015–1027. doi: 10.1093/jnen/63.10.1015. [DOI] [PubMed] [Google Scholar]
- 78.Takahashi Y, Iwai M, Kawai T, Arakawa A, Ito T, Sakurai-Yageta M, Ito A, Goto A, Saito M, Kasumi F, Murakami Y. Aberrant expression of tumor suppressors CADM1 and 4.1B in invasive lesions of primary breast cancer. Breast Cancer. 2012;19:242–252. doi: 10.1007/s12282-011-0272-7. [DOI] [PubMed] [Google Scholar]
- 79.Murakami Y. Involvement of a cell adhesion molecule, TSLC1/IGSF4, in human oncogenesis. Cancer Sci. 2005;96:543–552. doi: 10.1111/j.1349-7006.2005.00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yageta M, Kuramochi M, Masuda M, Fukami T, Fukuhara H, Maruyama T, Shibuya M, Murakami Y. Direct association of TSLC1 and DAL-1, two distinct tumor suppressor proteins in lung cancer. Cancer Res. 2002;62:5129–5133. [PubMed] [Google Scholar]
- 81.Heller G, Fong KM, Girard L, Seidl S, End-Pfutzenreuter A, Lang G, Gazdar AF, Minna JD, Zielinski CC, Zochbauer-Muller S. Expression and methylation pattern of TSLC1 cascade genes in lung carcinomas. Oncogene. 2006;25:959–968. doi: 10.1038/sj.onc.1209115. [DOI] [PubMed] [Google Scholar]
- 82.Mao X, Seidlitz E, Ghosh K, Murakami Y, Ghosh HP. The cytoplasmic domain is critical to the tumor suppressor activity of TSLC1 in non-small cell lung cancer. Cancer Res. 2003;63:7979–7985. [PubMed] [Google Scholar]
- 83.Mao X, Seidlitz E, Truant R, Hitt M, Ghosh HP. Re-expression of TSLC1 in a non-small-cell lung cancer cell line induces apoptosis and inhibits tumor growth. Oncogene. 2004;23:5632–5642. doi: 10.1038/sj.onc.1207756. [DOI] [PubMed] [Google Scholar]
- 84.Heller G, Geradts J, Ziegler B, Newsham I, Filipits M, Markis-Ritzinger EM, Kandioler D, Berger W, Stiglbauer W, Depisch D, Pirker R, Zielinski CC, Zochbauer-Muller S. Downregulation of TSLC1 and DAL-1 expression occurs frequently in breast cancer. Breast Cancer Res Treat. 2007;103:283–291. doi: 10.1007/s10549-006-9377-7. [DOI] [PubMed] [Google Scholar]
- 85.Kikuchi S, Yamada D, Fukami T, Masuda M, Sakurai-Yageta M, Williams YN, Maruyama T, Asamura H, Matsuno Y, Onizuka M, Murakami Y. Promoter methylation of DAL-1/4.1B predicts poor prognosis in non-small cell lung cancer. Clin Cancer Res. 2005;11:2954–2961. doi: 10.1158/1078-0432.CCR-04-2206. [DOI] [PubMed] [Google Scholar]
- 86.Oliva C, Escobedo P, Astorga C, Molina C, Sierralta J. Role of the MAGUK protein family in synapse formation and function. Dev Neurobiol. 2012;72:57–72. doi: 10.1002/dneu.20949. [DOI] [PubMed] [Google Scholar]
- 87.Marfatia SM, Leu RA, Branton D, Chishti AH. Identification of the protein 4.1 binding interface on glycophorin C and p55, a homologue of the Drosophila discs-large tumor suppressor protein. J Biol Chem. 1995;270:715–719. doi: 10.1074/jbc.270.2.715. [DOI] [PubMed] [Google Scholar]
- 88.Cohen AR, Woods DF, Marfatia SM, Walther Z, Chishti AH, Anderson JM. Human CASK/LIN-2 binds syndecan-2 and protein 4.1 and localizes to the basolateral membrane of epithelial cells. J Cell Biol. 1998;142:129–138. doi: 10.1083/jcb.142.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lue RA, Marfatia SM, Branton D, Chishti AH. Cloning and characterization of hdlg: the human homologue of the Drosophila discs large tumor suppressor binds to protein 4.1. Proc Natl Acad Sci USA. 1994;91:9818–9822. doi: 10.1073/pnas.91.21.9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fukuhara H, Masuda M, Yageta M, Fukami T, Kuramochi M, Maruyama T, Kitamura T, Murakami Y. Association of a lung tumor suppressor TSLC1 with MPP3, a human homologue of Drosophila tumor suppressor Dlg. Oncogene. 2003;22:6160–6165. doi: 10.1038/sj.onc.1206744. [DOI] [PubMed] [Google Scholar]
- 91.Zhang Y, Xu R, Li G, Xie X, Long J, Wang H. Loss of expression of the differentially expressed in adenocarcinoma of the lung (DAL-1) protein is associated with metastasis of non-small cell lung carcinoma cells. Tumour Biol. 2012;33:1915–1925. doi: 10.1007/s13277-012-0452-x. [DOI] [PubMed] [Google Scholar]
- 92.Kittiniyom K, Gorse KM, Dalbegue F, Lichy JH, Taubenberger JK, Newsham IF. Allelic loss on chromosome band 18p11.3 occurs early and reveals heterogeneity in breast cancer progression. Breast Cancer Res. 2001;3:192–198. doi: 10.1186/bcr294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gutmann DH, Donahoe J, Perry A, Lemke N, Gorse K, Kittiniyom K, Rempel SA, Gutierrez JA, Newsham IF. Loss of DAL-1, a protein 4.1-related tumor suppressor, is an important early event in the pathogenesis of meningiomas. Hum Mol Genet. 2000;9:1495–1500. doi: 10.1093/hmg/9.10.1495. [DOI] [PubMed] [Google Scholar]
- 94.Nunes F, Shen Y, Niida Y, Beauchamp R, Stemmer-Rachamimov AO, Ramesh V, Gusella J, MacCollin M. Inactivation patterns of NF2 and DAL-1/4.1B (EPB41L3) in sporadic meningioma. Cancer Genet Cytogenet. 2005;162:135–139. doi: 10.1016/j.cancergencyto.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 95.Singh PK, Gutmann DH, Fuller CE, Newsham IF, Perry A. Differential involvement of protein 4.1 family members DAL-1 and NF2 in intracranial and intraspinal ependymomas. Mod Pathol. 2002;15:526–531. doi: 10.1038/modpathol.3880558. [DOI] [PubMed] [Google Scholar]
- 96.Rajaram V, Gutmann DH, Prasad SK, Mansur DB, Perry A. Alterations of protein 4.1 family members in ependymomas: a study of 84 cases. Mod Pathol. 2005;18:991–997. doi: 10.1038/modpathol.3800390. [DOI] [PubMed] [Google Scholar]
- 97.Schulz WA, Alexa A, Jung V, Hader C, Hoffmann MJ, Yamanaka M, Fritzsche S, Wlazlinski A, Muller M, Lengauer T, Engers R, Florl AR, Wullich B, Rahnenfuhrer J. Factor interaction analysis for chromosome 8 and DNA methylation alterations highlights innate immune response suppression and cytoskeletal changes in prostate cancer. Mol Cancer. 2007;6:14. doi: 10.1186/1476-4598-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, Van De Rijn M, Botstein D, Brown PO. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13:1929–1939. doi: 10.1091/mbc.02-02-0023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fu L, Gao Z, Zhang X, Tsang YH, Goh HK, Geng H, Shimizu N, Tsuchiyama J, Srivastava G, Tao Q. Frequent concomitant epigenetic silencing of the stress-responsive tumor suppressor gene CADM1, and its interacting partner DAL-1 in nasal NK/T-cell lymphoma. Int J Cancer. 2009;124:1572–1578. doi: 10.1002/ijc.24123. [DOI] [PubMed] [Google Scholar]
- 100.Kittiniyom K, Mastronardi M, Roemer M, Wells WA, Greenberg ER, Titus-Ernstoff L, Newsham IF. Allele-specific loss of heterozygosity at the DAL-1/4.1B (EPB41L3) tumor-suppressor gene locus in the absence of mutation. Genes Chromosomes Cancer. 2004;40:190–203. doi: 10.1002/gcc.20034. [DOI] [PubMed] [Google Scholar]
- 101.Terada N, Ohno N, Yamakawa H, Baba T, Fujii Y, Ohara O, Ohno S. Immunolocalization of protein 4.1B in the rat digestive system. J Mol Histol. 2004;35:347–353. doi: 10.1023/b:hijo.0000039848.86488.74. [DOI] [PubMed] [Google Scholar]
- 102.Arjonen A, Kaukonen R, Ivaska J. Filopodia and adhesion in cancer cell motility. Cell Adh Migr. 2011;5:421–430. doi: 10.4161/cam.5.5.17723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lambrechts A, Van Troys M, Ampe C. The actin cytoskeleton in normal and pathological cell motility. Int J Biochem Cell Biol. 2004;36:1890–1909. doi: 10.1016/j.biocel.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 104.Cavanna T, Pokorna E, Vesely P, Gray C, Zicha D. Evidence for protein 4.1B acting as a metastasis suppressor. J Cell Sci. 2007;120:606–616. doi: 10.1242/jcs.000273. [DOI] [PubMed] [Google Scholar]
- 105.Schulz WA, Ingenwerth M, Djuidje CE, Hader C, Rahnenfuhrer J, Engers R. Changes in cortical cytoskeletal and extracellular matrix gene expression in prostate cancer are related to oncogenic ERG deregulation. BMC Cancer. 2010;10:505. doi: 10.1186/1471-2407-10-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fabbrizio E, El Messaoudi S, Polanowska J, Paul C, Cook JR, Lee JH, Negre V, Rousset M, Pestka S, Le Cam A, Sardet C. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 2002;3:641–645. doi: 10.1093/embo-reports/kvf136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jiang W, Newsham IF. The tumor suppressor DAL-1/4.1B and protein methylation cooperate in inducing apoptosis in MCF-7 breast cancer cells. Mol Cancer. 2006;5:4. doi: 10.1186/1476-4598-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Charboneau AL, Singh V, Yu T, Newsham IF. Suppression of growth and increased cellular attachment after expression of DAL-1 in MCF-7 breast cancer cells. Int J Cancer. 2002;100:181–188. doi: 10.1002/ijc.10470. [DOI] [PubMed] [Google Scholar]
- 109.Sun Y, Yuan J, Liu H, Shi Z, Baker K, Vuori K, Wu J, Feng GS. Role of Gab1 in UV-induced c-Jun NH2-terminal kinase activation and cell apoptosis. Mol Cell Biol. 2004;24:1531–1539. doi: 10.1128/MCB.24.4.1531-1539.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM (2004) Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430:213–218 [DOI] [PubMed]