Abstract
Understanding genetic regulation is a problem of fundamental importance. Recent studies have made it increasingly evident that, whereas the cellular genetic regulation system embodies multiple disparate elements engaged in numerous interactions, the central issue is the genuine function of the DNA molecule as information carrier. Compelling evidence suggests that the DNA, in addition to the digital information of the linear genetic code (the semantics), encodes equally important continuous, or analog, information that specifies the structural dynamics and configuration (the syntax) of the polymer. These two DNA information types are intrinsically coupled in the primary sequence organisation, and this coupling is directly relevant to regulation of the genetic function. In this review, we emphasise the critical need of holistic integration of the DNA information as a prerequisite for understanding the organisational complexity of the genetic regulation system.
Keywords: Holistic methodology, DNA supercoiling, Chromosome structure, Gene order, Transcriptional regulation, Nucleoid-associated proteins
Introduction
Transcriptional regulation of gene expression is an exceedingly complex phenomenon having many facets, but the prevalent view, in keeping with the Jacob–Monod paradigm, is that it is primarily about the interactions between the regulatory proteins acting as transcription factors (TFs) and their target genes (TGs). This view clearly separates the gene regulatory context (intergenic regions comprising the gene promoters with cognate TF binding sites) and the genetic information (gene coding sequences) in the chromosome. The TF–TG interactions have been studied in great detail and at different levels of complexity, ascending from individual regulatory interactions between a TF and its TG, to combinations of interactions forming topologically distinct network motifs, to the global level of an integrated transcriptional regulatory network (TRN) of a cell described as a hierarchy of directional communications between the global regulators, the more specialised regulators and the structural genes [1]. Consequently, several studies have proposed that the organisation of chromosomal structure on the evolutionary time scale is largely determined by the need of spatial optimisation of TF–TG interactions [2–4]. However, experimental manipulations of the chromosomal DNA dynamics in the bacterial organisms showed that the patterns of gene transcription and metabolism are determined by structural dynamics of the chromosomal DNA, rather than by the TF–TG interactions [5, 6]. Recent data suggesting a highly ordered dynamic structure of the bacterial nucleoid have provided additional support for the notion that modulation of the global chromosomal structure might be determinative for major DNA transactions, including gene transcription [7–9]. It thus appears that we face a “chicken or egg” dilemma—on the one hand the TF–TG interactions are determinative for the chromosomal structure, and on the other hand this very same structure determines the regulatory interactions. In this review, we attempt to resolve this apparent contradiction by proposing that the structural and genetic organisation of a chromosome is essentially inseparable. It is noteworthy that, whereas the development of new technologies encouraged the application of cutting-edge molecular biology, network and polymer theory approaches to model the chromosomes, the fundamental role of the DNA as a coding device has been largely ignored. Under the premise that the DNA polymer evolved primarily for the coding purpose, we argue here that any holistic methodology aiming at comprehensive understanding of gene regulation must be focused on the information carrier function of the DNA as its essential spotlight.
Holistic methodology
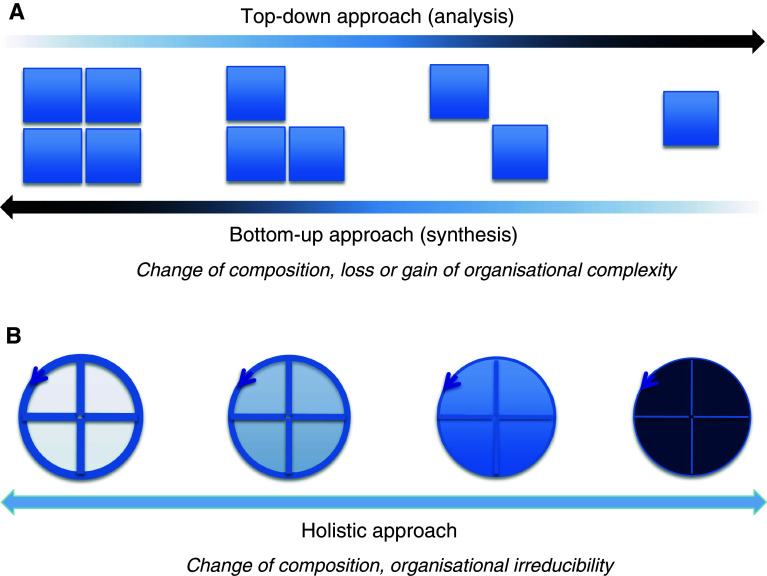
In the first place, a generic difference between the holistic approach and both the conventional top–down and bottom–up approaches is of note. Holistic methodology does not imply decomposition from general to more detailed (analysis) as in the former, or ascendance from the detailed to more general (synthesis) as in the latter. Indeed, by the very fact of the assumption of disparate levels of organisational complexity, both the top–down and the bottom–up modes are essentially reductionist approaches. In contrast, the holistic approach assumes an organisational invariance (Fig. 1). But what does this organisational invariance imply?
Fig. 1.
Cartoon showing the generic difference between the top–down, bottom–up and holistic approaches. a The withdrawal/addition of the squares indicates the gain and loss of the organisational complexity in the top–down and bottom–up approaches, respectively. b The arrow closing on itself indicates an irreducible organisational complexity (circularity) despite compositional changes (indicated by alterations of color intensity) in the holistic approach
Genetic regulation is crucial not only for sustaining the self-reproduction of a cell but also for substituting its worn-out constituents. This implies that a genetic regulation system, as a system consisting of physical elements, must be able not only to perform its primary function but also to perceive any internal changes of state so that it retains the potential, for example, to replenish its own components. In other words, it has to be self-referential. This peculiarity of organisation becomes conspicuous when compared to information coding in natural language, the syntactic and semantic properties of which provide logically different types of information. Syntax determines the structure of the rules of language and, thus, the way in which the words are assembled in sentences, whereas semantics determine the meaning of the words and so the available vocabulary. However, the structural rules of language cannot determine the meanings of the words, and nor is the vocabulary determinative for the structural rules of the language (we do not concern ourselves with any generative mechanisms relevant to the formal language theory here). Therefore, viewed as a coding system composed of two non-convertible types of information, natural language is not self-referential. By the same token, the Jacob–Monod paradigm separating the gene regulatory context from the genetic information is at variance with self-referential organisation. Notably, we do not use this term in the sense of elaborated mathematical concepts of distinction, circulation, feedback, re-entry, recursion, etc. [10]. Self-referential organisation, as we put it here, implies inter-conversion of information between logically distinct coding systems specifying each other reciprocally [11]. Thus, the holistic approach assumes self-referentiality (completeness of the contained information and full consistency of the different codes) as an irreducible organisational complexity of the genetic regulation system of any cell. Put another way, this implies that the structural dynamics of the chromosome must be fully convertible into its genetic expression and vice versa. Since the DNA is an essential carrier of genetic information, the fundamental question is how this self-referential organisation is encoded in the sequence of the DNA polymer.
Two types of information in the DNA
Recent studies have made it increasingly evident that the primary sequence of DNA in addition to the linear genetic code also provides three-dimensional information by means of spatially ordered supercoil structures relevant to all DNA transactions, including transcriptional control [12–15]. In this review, we adopt the previously introduced terms “analog” and “digital” with regard to the two logically distinct types of information provided by the DNA [2, 11, 15, 16]. By digital information, we mean the unique DNA sequence written in distinct succession of individual letters. On this view, any DNA gene is a carrier of digital information by virtue of its unique base sequence. Moreover, a gene conceived as an isolated piece of linear code (no matter whether this isolation occurs at the level of transcription or posttranscriptional processing), is a discontinuous entity that can be expressed or not, thus principally consistent with an “on-or-off” logic and, therefore, belonging to digital information type. Conversely, the physicochemical properties of DNA, as exemplified by supercoiling and mechanical stiffness, are determined not by individual base pairs but by the additive interactions of successive base steps. Supercoiling is by definition a continuous parameter ranging between positive and negative values (you can have more or less of it), and so belongs to analog information type. More specifically, the chirality of the DNA molecule underlies its ability to partition superhelicity between twist (roughly the average inclination angle of the base pairs integrated over the entire polymer length) and writhe (approximated by the average number of crossings of the DNA helical axis with itself). However, the DNA is a heterogeneous polymer, and the partition between twist and writhe (i.e. preferred DNA geometry) is dependent in part on the DNA base composition and sequence.
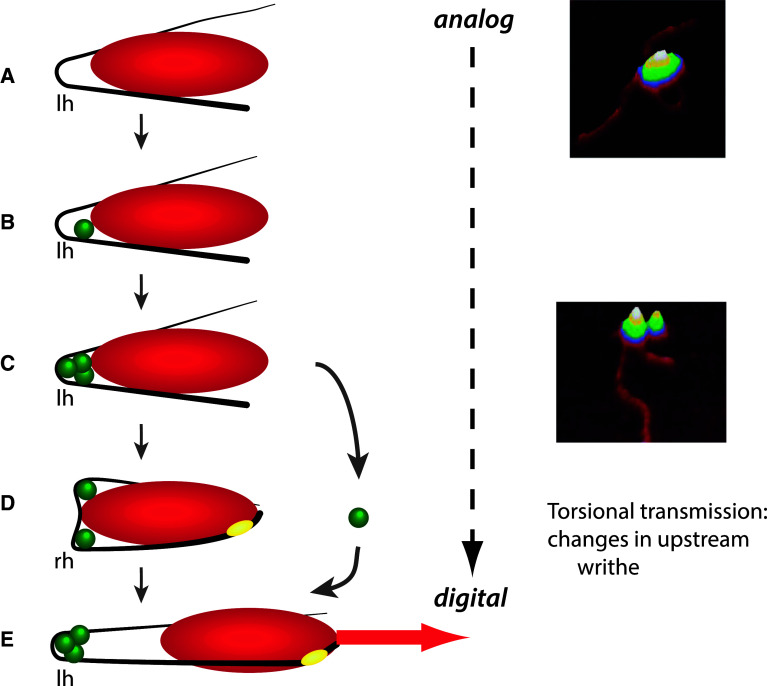
The organisational principle of coupling between the analog DNA structures and the digital patterns of gene expression on a genome-wide scale is just coming into view, but for individual genes it is understood in some detail. This coupling mechanism involves local binding effects of the chromatin architectural factors stabilising distinct DNA configurations in the gene promoter regions [17, 18]. The Escherichia coli tyrT promoter is activated by the nucleoid-associated protein (NAP) FIS, interacting with both the RNA polymerase and three high-affinity binding sites arranged on the same face of the DNA helix in the upstream activating sequence (UAS) of the promoter (Fig. 2). Binding of FIS at these consecutive sites bends the UAS DNA coherently, thus accumulating the DNA torsional energy in a topologically isolated “toroidally” (spiral-like) coiled microloop attached to RNA polymerase [19, 20]. Subsequent conformational alterations of the complex, driven by repartitioning of twist and writhe and resultant change of the exit/entry angles of the DNA microloop, transmit the torsional energy to the transcriptional start site activating tyrT expression [17]. Such dynamic three-dimensional DNA structures, mediating directional channelling of the stored supercoil energy into promoter opening and gene transcription, themselves depend on the superhelical density [21–24]. Accordingly, recent studies of the promoter sequences of supercoiling-responsive gene classes have revealed different base periodicities [25, 26], and have also shown that different periodicities of the toroidally coiled and planar DNA configurations affect the TF binding affinity [27].
Fig. 2.
Conversion of the DNA analog information into the digital code by transmission of the torque stored in the microloop stabilised at the tyrT gene promoter. Left-handed (lh) toroidal coils are metastable to right-handed (rh) plectonemic coils. Activation involves conversion of the left-handed into a right-handed coil thus transmitting the supercoil energy to the promoter region and facilitating DNA untwisting at the transcription startpoint. a RNA polymerase (red ellipse) binding constrains a DNA microloop in the upstream region. b Cooperative binding of transcriptional activator FIS (green spheres) to site I proximal to polymerase. c Cooperative binding of FIS to the upstream sites II and III stabilises the closed polymerase complex. d Untwisting of the transcription start site and promoter opening (yellow ellipse) is coupled with ejection of FIS from binding site II. e Escape of polymerase from the promoter and initiation of transcription generating the digital linear message (red arrow). Right panel shows the AFM images of nucleoprotein complexes assembled at the E. coli tyrT promoter corresponding to depicted stages (a) (RNAP binding alone) and (c) (RNAP and FIS binding) (courtesy S. Maurer)
In general, the regions of chromosomes that are sites for topological manipulation (such as, e.g., transcription and replication initiation sites) correlate strongly with low base stacking energies and high flexibility [28]. Indeed, the sequences at the start sites of transcription and replication are prone to localised untwisting, whereas the termination sites—and especially the regions between two converging translocases (be it a replisome or RNA polymerase)—appear to easily adopt a writhed configuration acting as supercoil repositories. The emerging view is that manipulation of superhelical density and regulation of partitioning between twist and writhe is a fundamental property of both the bacterial and eukaryotic chromosomes. Furthermore, in both the yeast and bacterial chromosomes, the genetic and chromatin organisation was found to be highly dependent on, and likely specified by, the stacking/melting energies of DNA sequences [29–31]. On this view, chromosomes act as machines in which coordinated topological transitions operating at local (e.g. transcription initiation sites), regional (constrained superhelical domains) and global (entire chromosomes) levels specify the genetic activity. How is this coordinated process linked to metabolic demands of a cell?
Circular organisation of the transcriptional regulation system
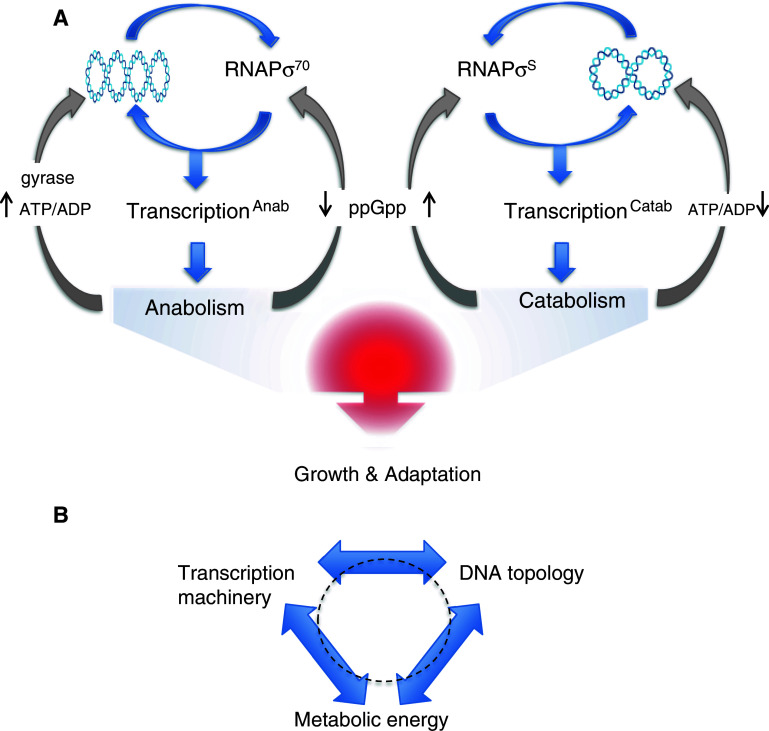
At the bottom line, global transcriptional regulation occurs on the interface of interactions between the transcription machinery and the chromosomal DNA. These interactions mediate the conversion of the DNA analog information into the digital pattern of gene expression [15, 16] specifying the nucleoid shape [32, 33] and eventually the cellular metabolic profile [4]. Investigations of the relationships between the chromosomal DNA topology and the composition of the transcriptional machinery in Escherichia coli indicate that any substantial alteration of the transcription machinery has a potential to induce adaptive changes in DNA topology and vice versa [11, 38, 39]. This implies that the DNA topology and the transcription machinery, as essential components of the genetic system coordinating the gene expression with available metabolic energy, vary interdependently (Fig. 3a). Input of metabolic energy (availability of ATP) in turn changes the energy levels in DNA mediated either via gyrase (the major enzyme introducing negative supercoils into the DNA by an ATP-dependent mechanism) or the processive DNA translocases (replisomes and RNA polymerases which transiently introduce both negative and positive supercoils), and this in turn facilitates the conversion of DNA sequence information into “readable” form, i.e. into specific structures governing the gene transcription [34–37]. A tight coupling between the gene transcription, chromatin structure and metabolism was also observed in eukaryotes, including cancer cells [40–44]. Notably, since these three variables are assumed to determine each other reciprocally [11], this interdependence forms an operationally closed self-referential circuit (Fig. 3b). It is noteworthy that exploration of this peculiar organisation requires an adequate mathematical formalism, and provoked novel self-consistent multiscale analyses for integrating the top–down and bottom–up approaches in order to capture the ensuing circular causality [45]. In mechanistic terms, such interdependence implies a tight coordination of the chromosomal structure, DNA-transactions and the cellular metabolism.
Fig. 3.
Structural coupling between the composition of RNA polymerase holoenzyme, DNA topology and metabolism in E. coli. a The chromosomal DNA is depicted as an interwound “plectonemic” coil. Preference of the vegetative and stationary phase RNA polymerase holoenzymes (RNAPσ70 and RNAPσS, respectively) for correspondingly high or low DNA superhelical density (indicated by changing number of crossings in the DNA plectoneme) specifies either anabolic or catabolic gene expression. The coordination of transcriptional response is mediated by structural coupling between RNAP composition and the DNA topology, whereas both respond to changes of metabolism: increased ATP/ADP ratio supports gyrase activity and high negative superhelicity, whereas increased concentration of the “stringent response” regulator ppGpp supports RNAPσS formation and so the preference for relaxed DNA templates [39]. b Three basic components underlying the irreducible organisational complexity of any living cell. Note that the organisation is essentially circular with all three basic components standing in relationship to reciprocal determination
Structural-organisational complexity of the bacterial chromosome
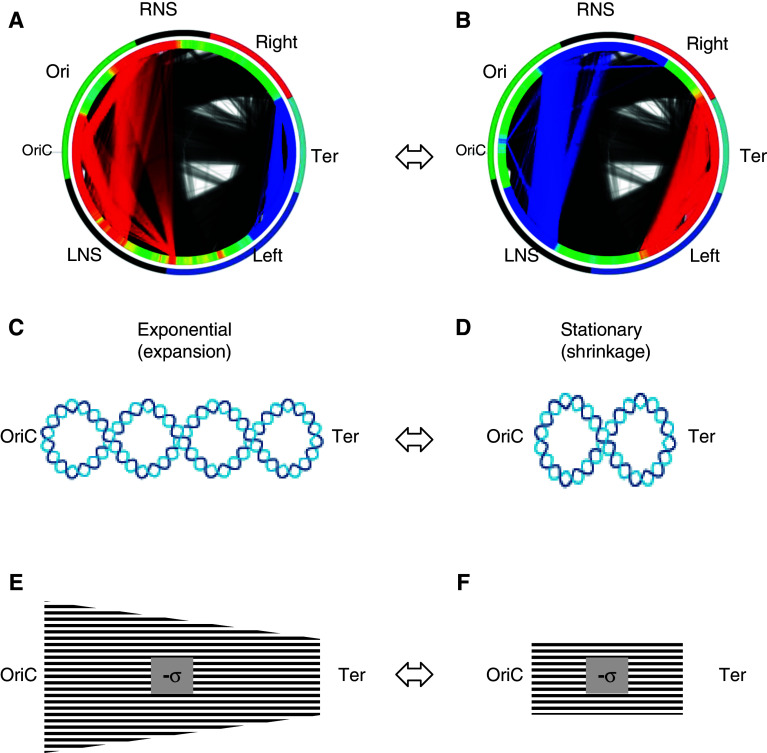
Organisation of the DNA in the bacterial nucleoid has been observed at several levels, from chromosomal macrodomains encompassing up to one megabase of DNA to several hundreds of isolated topological domains of 10-kb average size [4, 46–48]. Recent studies employing 5C analysis of the Caulobacter crescentus chromosome [49], and analyses of functional interconnections within the Escherichia coli chromosome [28], concluded that the highest order of the chromosome folding is best described as a simple plectoneme (see Fig. 5c, d). Organisation of the two chromosomal arms from the origin (OriC) to the terminus (Ter) of replication (the right and left replichores) is less clear, suggesting either toroidally coiled or a branched plectonemic structure [49–52]. However, the evidence for toroidal coiling is based solely on periodicity patterns in certain chromosomal regions and would be equally compatible with a branched plectonemic structure.
Fig. 5.
Changes of the E. coli chromosome morphology, the supercoiling gradient and intra-chromosomal communications during the growth cycle. a, b The outer circle shows chromosomal macrodomains coded in colors: Ori green, Right red, Ter light blue, Left blue. The left (LNS) and right (RNS) non-structured domains are indicated in black. The second circle shows increased (red) and decreased (blue) chromosomal density distributions of the expressed genes during exponential growth (a) and on transition to stationary phase (b). The connecting lines inside the circles accordingly indicate increased (red) and decreased (blue) communications between the chromosomal domains harboring functionally related genes [29, 30]. The origin (OriC) and terminus (Ter) of chromosomal replication are indicated. c, d The changes of the bacterial chromosome morphology. The chromosome is depicted as a simple plectoneme with interwound arms aligned along the OriC–Ter axis. Decrease of the overall superhelicity (indicated by reduction of crossings) and shrinkage along the OriC to Ter axis occur during transition from exponential (c) to stationary phase (d) (Mircea Petrescu and G.M., in preparation). e The proposed spatial gradient of DNA negative superhelical density (-σ) extending along the OriC–Ter axis during the exponential phase, and f the flattening of gradient (indicated by change of shape) on transition to stationary phase. In principle, the level of superhelicity assumed in stationary phase need not correspond to the lowest level in exponential phase
Supercoiling of the DNA is of paramount importance for the compaction of the nucleoid. Compaction is facilitated by the highly abundant NAPs—counterparts of eukaryotic histones constraining the DNA supercoils and regulating global chromosomal transcription [53–59]. Available data indicate that, during the bacterial growth cycle, the crosstalk between the NAPs and the DNA topoisomerases regulates the overall chromosomal supercoil density homeostatically [13, 16, 60, 61]. Indeed, changes in the abundance and composition of the NAPs, as well as of cellular topoisomerase activities, can concertedly modulate the chromosomal compaction, the topological domain boundaries and the spatial transcription patterns [16, 62–69]. Since the superhelical density of chromosomal DNA is coupled to the metabolic state of the cell [34, 37], and can also change instantly under the influence of environmental factors, supercoiling acts as a mediator of the genomic transcriptional response to challenge [3, 34–36, 66, 70]. Optimisation of this adaptive response involves cooperative binding effects of the NAPs competing for distinct supercoil structures [55, 71–75]. Notably, the abundant NAPs predominantly recognise local DNA conformations rather than the DNA bases per se, exhibiting a wide and quasi-continuous range of DNA sequence-dependent affinity [53, 59, 74, 76] and, thus, modulating the chromosomal supercoil dynamics and transcription in a continuous, or analog, mode [2]. Such graded transcriptional responses have also been implicated in the physiological adjustment of both the yeast and mammalian cell systems to environmental challenge [77–79].
Spatiotemporal organisation of genetic function in the chromosome
While the temporal alteration of supercoiling level during the bacterial growth cycle has been long known, recent observations suggest that negative superhelicity is not evenly distributed in the bacterial chromosome. In Escherichia coli, the frequency distribution of the binding sites for DNA gyrase demonstrates a gradient from OriC to Ter along both replichores and, most importantly, this spatial gradient correlates with temporal expression of supercoiling-dependent gene classes during the growth cycle, with early genes transcribed at high superhelicity [28, 31, 65]. Furthermore, the strong ribosomal RNA operons organised on both chromosomal arms are all pointing away from the origin, suggesting that during fast growth their transcription contributes to the accumulation of negative superhelicity in the Ori end of the chromosome [28, 32]. These observations imply gradients of superhelical density on both replichores from the origin to the terminus. Intriguingly, the chromosomal DNA sequences exhibit, on average, a gradient of DNA stacking/melting energy in the same direction. This pattern of DNA thermodynamic stability is strongly conserved in all α- and γ-proteobacteria [29, 30]. It is likely that this gradient in the physicochemical properties of DNA along the OriC–Ter axis integrates the functional response to changes in superhelical density and to regulation by abundant NAPs, constraining the DNA supercoils. Interestingly, similar inferences were drawn from studies of the obligate symbiotic bacterium Buchnera, characterised by drastic genome shrinkage and very low diversity of specific TFs, but conservation of several NAPs and topoisomerases [80]. Also, the organisation of essential bacterial genes along the OriC to Ter axis is biased [81]. Recent study has revealed a distinct organisation of functional gene classes in the Escherichia coli chromosome with regard to their distance to OriC, preferred orientation with respect to OriC, and their DNA physicochemical properties. More specifically, the early expressed anabolic genes show leading strand preference, require high levels of supercoiling, are encoded by DNA of high average negative melting energy and are located in close vicinity to the replication origin; conversely, catabolic genes are expressed later, largely anti-correlate with both the leading strand utilisation and high superhelicity, and are also more distant from the replication origin [30]. These observations are wholly consistent with previous studies [16, 29, 38, 81] and strongly suggest that the spatial organisation of the chromosomal transcriptons, physicochemical properties of the transcribed DNA and the metabolic function of the produced transcripts are tightly coupled.
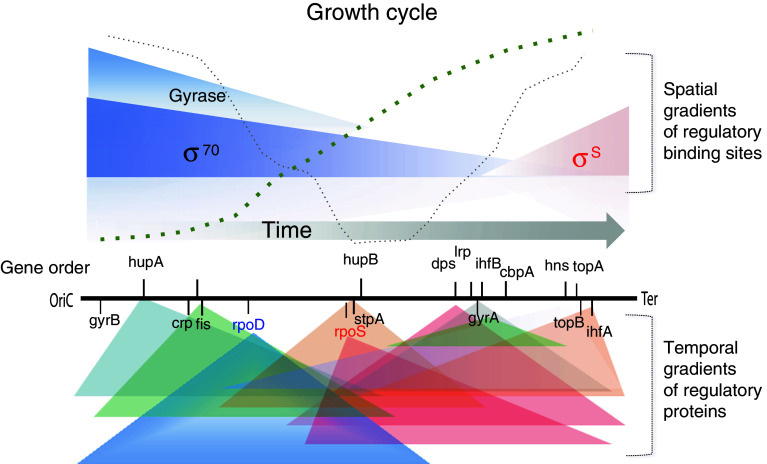
How is the DNA thermodynamic stability related to the dynamic gene expression profile during the growth cycle? Relevant hints are provided by recent observation of correspondence between the chromosomal order of regulatory genes along the OriC–Ter axis and their temporal expression pattern [29], in conjunction with the finding that translation of the gene products occurs in the vicinity of cognate genes [82, 83]. This implies formation of concentration gradients of proteins diffusing from their sites of production. Importantly, the frequency distributions of binding sites, not only for DNA gyrase but also for the major NAPs and RNA polymerase holoenzymes (RNAPσ70 and RNAPσS), form spatial gradients along the OriC–Ter axis in the genome (Fig. 4). Under conditions of changing DNA superhelicity during the growth cycle, it is likely that these spatial binding site gradients distinctly interact with temporal gradients of various NAPs and RNA polymerase holoenzymes, thus coordinating the transcription program with the growth environment [29, 32, 54, 65]. Indeed, analysis of the Escherichia coli growth cycle showed that, after commitment of the cells to growth under conditions of the predominance of RpoD impact and high levels of FIS and HUα [54], the average negative melting energy of the activated gene sequences in the Ori-proximal end of the chromosome increased continuously with oxygen consumption [30] and featured anabolic genes that required high negative superhelicity for transcription. In contrast, with the onset of more anaerobic conditions characterised by increased impact of RpoS, Dps, Lrp and IHF [39, 54] both the average melting energy of the expressed sequences and their average distance to OriC decreased, whereby the transcripts of catabolic genes requiring DNA relaxation showed a major peak at minimal partial oxygen pressure in the medium [30]. These observations support the model in which the bacterial growth phase-dependent transcription program is determined by high oxygen utilisation, generating supercoiling gradients along both replichores from the origin to the terminus of replication [29].
Fig. 4.
Spatiotemporal gradient model of gene regulation. The chromosomal arms aligned along the OriC–Ter axis are indicated by horizontal black line. The order of global regulatory genes (encoding NAPs, topoisomerases and the RNAP sigma factors rpoD and rpoS) is indicated along the OriC–Ter axis. This spatial order correlates with their temporal expression pattern. The colored triangles, labelled Gyrase, σ 70 and σ S, approximate the genomic distribution frequencies of the DNA binding sites for DNA gyrase, the vegetative RNAPσ70, and the stationary phase RNAPσS along the OriC–Ter axis (binding sites of other regulators are omitted for simplicity) [29]. The gradients of regulators diffusing from their production sites [81, 82] are indicated by colored triangles below the ordered genes. The size of the triangles is arbitrary and the color distinguishes the regulators produced from the early (blue/green) or at the late (red/pink) stages of growth. The thick and thin dotted curves, respectively, approximate the cell growth and oxygen consumption during the growth cycle [30]. The time flow is indicated by the grey horizontal arrow. The model proposes that changing interactions between the temporal gradients of regulatory proteins and spatial gradients of genomic binding sites determine the growth phase-dependent chromosomal dynamics and thus the gene expression patterns
Thus, spatiotemporal integration of the analog (syntactic) and digital (semantic) properties of the chromosomal DNA code appears as a basic device coordinating the bacterial growth program. This coordination is facilitated by organising genes in a highly conserved order and orientation on the Ori–Ter axis alongside the gradients of DNA thermodynamic stability and superhelicity in both chromosomal replichores. Accordingly, the communications between the chromosomal regions harboring functionally related genes are also coordinated both in space and time (Fig. 5). The structure of the underpinning network—as opposed to the hierarchical structure of the TRN—is lacking any major organising entity and is correspondingly heterarchical (essentially circular), wholly consistent with the demand of self-referential organisation [11, 29].
Further insights into this organisation are provided by recent observation of dynamically appearing functional domains (FDs) of variable gene expression densities with boundaries separating distinct functional groups of the genes on the chromosome [30]. These spatially delimited domains vary in size, but can extend over hundreds of kilobases and harbour sets of coherently expressed sequences which demonstrate characteristic couplings of the DNA thermodynamic stability, preferred supercoiling regime and encoded gene function. Thus, at the level of the entire chromosome, the separation of the regulatory and coding sequences assumed in the Jacob–Monod paradigm becomes somewhat blurred. Notwithstanding the distinctive role of the gene promoters in initiating transcription, they appear as local supercoil-channelling devices largely subordinated to, and coordinated by, the structural dynamics of extended chromosomal functional domains. However, the promoters of major regulatory genes often carry local signatures, such as, e.g., curvatures or GC-rich regions upstream of the transcription initiation sites (discriminators). These signatures can both act as topology sensors and determinants of the expression strength [13]. Given that the chromosomal order of major regulatory genes corresponds to their temporal expression [29], it is conceivable that the transient domains of coherent gene expression (FDs) reflect the temporally changing inputs of the global regulators in the protein gradients cooperating and competing for binding the spatial gradients of the DNA sites in the chromosome. On this view, the FDs manifest spatiotemporally coordinated chromosomal domains selectively expressing particular sequences with characteristic analog and digital properties in response to changing growth environment (Fig. 6).
Fig. 6.
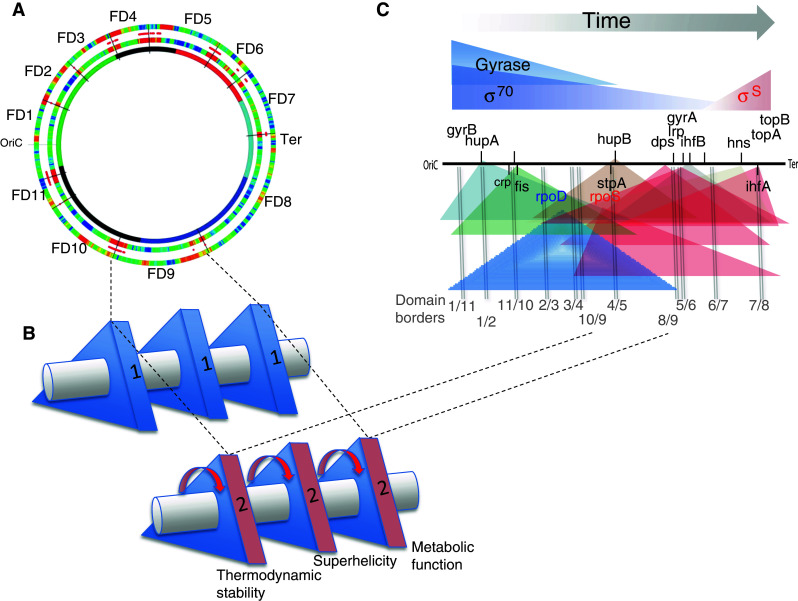
Organisation of the dynamic functional domains (FDs) in the E. coli chromosome. a The chromosomal wheel showing the 11 detected functional domains (FD1–FD11) with approximated boundaries (black lines) as indicated [30]. The outermost ring represents the static borders (red) separating functionally related groups of genes in the chromosome. The middle ring represents the distribution of structural domains with dynamic borders (red) delimited by distributions of DNA analog information in the chromosome. The innermost ring shows the static chromosomal macrodomains indicated as in Fig. 5. b A rod with three triangles representing the three basic coupled elements of the genetic system. In each triangle the three sides (1, 2 and 3) correspond to different values of the same single DNA information type in the expressed sequences—either thermodynamic stability, negative superhelicity or metabolic function (note that the first two belong to analog, while the third to digital information type). Coordination of analog and digital information (shown on the example of in the arbitrarily chosen domain FD9) occurs by virtue of inherent structural coupling, as all these three parameters are “inscribed” in the primary sequence of the DNA. This structural coupling is indicated by an imaginary symmetry axis connecting the triangles (horizontal rod). The alteration of coupled parameter values during the growth is indicated by concerted rotation of triangles around the symmetry axis (red arrows; in principle, the distinct parameter couplings can be conceived as different shapes of the same symmetry group). c Gradient model of domain stabilisation. The depiction of chromosome, temporal gradients of the global regulators, spatial gradients of regulator binding sites and color-coding are as in Fig. 4. The borders between the 11 FDs are indicated by vertical grey lines and labelled by numbers (separated by a slash) corresponding to the neighbouring domains. Note that the positions of many regulatory genes closely coincide with the domain boundaries. The model suggests that, during growth, the changing interactions between the temporal gradients of regulators and the spatial gradients of chromosomal binding sites distinctly modulate the DNA supercoil dynamics leading to stabilisation of FDs. Since FDs harbour functionally related genes, their transient stabilisation during the growth cycle serves as a device instantly sensing and converting the supercoil energy into a corresponding metabolic profile
Dynamic changes of the chromosome structure as a means of controlling the genomic expression is consistent with models in which the gene organisation and overall superhelicity are determined in large part by the translocase activity of the replisomes. During periods of rapid replication when each replichore may contain at least three replication forks, the boundaries between macrodomains are necessarily disrupted, if only transiently, by the passage of a replisome. Similarly, because the affinity of certain “domainin” NAPs (such as FIS and H-NS, which stabilise topological domains in the chromosome) [67] for DNA depends on superhelical density, their activity and target sites will potentially vary with growth phase and act in concert with changes in their intracellular concentration. Such considerations suggest that models invoking a purely static compartmentalisation of the bacterial chromosome are at best an oversimplification.
It is noteworthy that, whereas in Escherichia coli the changing chromosomal dynamics and spatial organisation of transcription correspond to different successive regimes of oxygen consumption, in cyanobacteria the circadian rhythms also appear to be regulated by supercoiling dynamics [84, 85]. Furthermore, a similar organisational logic is observed in regulation of the respiratory oscillation of the transcriptome in the yeast chromosomes where the two gene superclusters, one for anabolic and another for catabolic function, are used successively during high and low oxygen uptake, respectively [43]. Each supercluster has a characteristic sequence organisation and is subject to different effects of ATP-dependent remodelling machineries, whereby the genome-wide oscillations of transcription are assumed to gate synchronous bursts of DNA replication [44]. More specifically, the temporal separation between the oxidative and reductive phases in a continuous yeast culture was shown to propagate throughout the entire transcriptome coordinately with the initiation of DNA replication.
Conclusion
Observations in both the bacterial and eukaryotic systems suggest that chromosomes act as thermodynamic machines converting the supercoil energy into genetic information [11, 15, 28]. In both cases, structural coupling of the two—analog and digital—information types in the very same DNA primary sequence organisation enables a unique response to metabolic demand mediated by stabilisation of appropriate spatially delimited chromosomal domains with corresponding physiologically relevant genetic activity. Thus, it appears that, for both the bacterial and eukaryotic chromosomes, the genetic and chromatin organisation are inseparable and coming to light as one integrated whole [81, 86, 87]. This device inherent in the primary sequence organisation of the chiral DNA polymer is sufficient to specify a self-referential organisation and explain the coordination of the genetic programs in space and time. On this view, the DNA architectural proteins acting according to the cellular physiological state, such as NAPs in bacteria and remodelling complexes in eukaryotes, serve as auxiliary factors optimising the delivery of the integrated DNA information.
Holistic methodology impinges on the experimental approach. It suggests seeking self-reference instead of lineal causality in exploring the interdependences between the chromosome-structuring, chromosome-reading and metabolic–enzymatic components of the genetic regulation system. Can a coordinated change of all these coupled components be induced by alteration of any single component? Is it possible to discover “synthetic lethality” by combining mutations in two non-essential but interdependent elements? And can we identify any specialised structures formed by interactions between the coupled components? These questions are the predictions from the assumed organisation (see Fig. 3) that can be readily tested. In this respect, previous observations associating the mutations in metabolic enzymes with changes of the chromosomal topological domains [67], and mutations altering the kinetic properties of RNA polymerase with changes of both DNA supercoil dynamics and the metabolic profile [38, 88], are revealing. Communications between distinct molecular components of the coupled system could be facilitated by their consolidation in “hyperstructures” [89, 90], as suggested, for example, by the observed cooperation between the nucleoid-associated protein HU and RNA polymerase in forming the transcription foci [32]. We propose that the interactions between the spatiotemporal gradients of the global regulators and spatial gradients of the chromosomal binding sites can specify the transient boundaries of the FDs. Whereas different constellations of the FDs appear to mediate the coordination of the chromosomal supercoil energy with genetic function during the growth cycle, the extent to which the FDs can retain their observed characteristics on transplantation into different chromosomal environments remains an open question. Overall, their organisational properties closely resemble those of the horizontally acquired genomic pathogenicity islands [91], suggesting variations on a common theme.
Acknowledgements
We thank Rainer Machne and the anonymous reviewers for helpful comments. This work was in part supported by the research grant of the Deutsche Forschungsgemeinschaft (InKoMBio) to G.M.
Abbreviations
- Dps
DNA protection during starvation protein
- FD
Functional domain
- FIS
Factor for inversion stimulation
- H-NS
Histone-like nucleoid-structuring protein
- HU
Heat-stable protein from the strain U13
- IHF
Integration host factor
- Lrp
Leucine responsive protein
- NAP
Nucleoid-associated protein
- OriC
Origin of chromosomal replication
- RNAP
RNA polymerase
- RpoD
RNAP vegetative sigma factor (σ70)
- RpoS
RNAP stationary phase sigma factor (σS)
- Ter
Chromosomal replication terminus
- TF
Transcription factor
- TG
Target gene
- Transcripton
Transcription unit
- TRN
Transcriptional regulatory network
- tyrT
Tyrosyl transfer RNA gene
- UAS
Upstream activating sequence
References
- 1.Babu MM. Computational approaches to study transcriptional regulation. Biochem Soc Trans. 2008;36:758–765. doi: 10.1042/BST0360758. [DOI] [PubMed] [Google Scholar]
- 2.Janga SC, Salgado H, Martínez-Antonio A. Transcriptional regulation shapes the organization of genes on bacterial chromosomes. Nucleic Acids Res. 2009;37:3680–3688. doi: 10.1093/nar/gkp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fritsche M, Li S, Heermann DW, Wiggins PA. A model for Escherichia coli chromosome packaging supports transcription factor-induced DNA domain formation. Nucleic Acids Res. 2012;40:972–980. doi: 10.1093/nar/gkr779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babu MM, Janga SC, de Santiago I, Pombo A. Eukaryotic gene regulation in three dimensions and its impact on genome evolution. Curr Opin Genet Dev. 2008;18:571–582. doi: 10.1016/j.gde.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Marr C, Geertz M, Hütt M-T, Muskhelishvili G. Dissecting the logical types of network control in gene expression profiles. BMC Syst Biol. 2008;2:18. doi: 10.1186/1752-0509-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonnenschein N, Geertz M, Muskhelishvili G, Hütt M-T. Analog regulation of metabolic demand. BMC Syst Biol. 2011;5:40. doi: 10.1186/1752-0509-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dame RT, Kalmykowa OJ, Grainger DC (2011) Chromosomal macrodomains and associated proteins: implications for DNA organization and replication in gram negative bacteria. PLoS Genet (6):e1002123 [DOI] [PMC free article] [PubMed]
- 8.Benza VG, Bassetti B, Dorfman KD, Scolari VF, Bromek K, Cicuta P, Lagomarsino MC. Physical descriptions of the bacterial nucleoid at large scales, and their biological implications. Rep Prog Phys. 2012;75(7):076602. doi: 10.1088/0034-4885/75/7/076602. [DOI] [PubMed] [Google Scholar]
- 9.Ptacin JL, Shapiro L. Chromosome architecture is a key element of bacterial cellular organisation. Cell Microbiol. 2013;15:45–52. doi: 10.1111/cmi.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaufman LH. Self-reference and recursive forms. J Soc Biol Struct. 1987;10:53–72. doi: 10.1016/0140-1750(87)90034-0. [DOI] [Google Scholar]
- 11.Muskhelishvili G, Sobetzko P, Geertz M, Berger M. General organisational principles of the transcriptional regulation system: a tree or a circle? Mol BioSyst. 2010;6:662–676. doi: 10.1039/b909192k. [DOI] [PubMed] [Google Scholar]
- 12.Dorman CJ. H-NS: a universal regulator for a dynamic genome. Nat Rev Microbiol. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 13.Travers A, Muskhelishvili G. DNA supercoiling—a global transcriptional regulator for enterobacterial growth? Nat Rev Microbiol. 2005;3:157–169. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- 14.Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 15.Travers AA, Muskhelishvili G, Thompson JMT. DNA Information: from digital code to analogue structure. Philos Trans R Soc Lond A. 2012;370:2960–2986. doi: 10.1098/rsta.2011.0231. [DOI] [PubMed] [Google Scholar]
- 16.Blot N, Mavathur R, Geertz M, Travers A, Muskhelishvili G. Homeostatic regulation of supercoiling sensitivity coordinates transcription of the bacterial genome. EMBO Rep. 2006;7:710–715. doi: 10.1038/sj.embor.7400729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Travers AA, Muskhelishvili G. DNA microloops and microdomains—a general mechanism for transcription activation by torsional transmission. J Mol Biol. 1998;279:1027–1043. doi: 10.1006/jmbi.1998.1834. [DOI] [PubMed] [Google Scholar]
- 18.Levens D, Benham CJ. DNA stress and strain, in silico, in vitro and in vivo. Phys Biol. 2011;8:035011. doi: 10.1088/1478-3975/8/3/035011. [DOI] [PubMed] [Google Scholar]
- 19.Lazarus LR, Travers AA. The Escherichia coli FIS protein is not required for the activation of tyrT transcription on entry into exponential growth. EMBO. 1993;12:2483–2494. doi: 10.1002/j.1460-2075.1993.tb05903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maurer S, Fritz J, Muskhelishvili G, Travers A. RNA polymerase and an activator form discrete subcomplexes in a transcription initiation complex. EMBO J. 2006;25:3784–3790. doi: 10.1038/sj.emboj.7601261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowater RP, Chen D, Lilley DM. Modulation of tyrT promoter activity by template supercoiling in vivo. EMBO J. 1994;13:5647–5655. doi: 10.1002/j.1460-2075.1994.tb06903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Free A, Dorman CJ. Escherichia coli tyrT gene transcription is sensitive to DNA supercoiling in its native chromosomal context: effect of DNA topoisomerase IV overexpression on tyrT promoter function. Mol Microbiol. 1994;14:151–161. doi: 10.1111/j.1365-2958.1994.tb01275.x. [DOI] [PubMed] [Google Scholar]
- 23.Pemberton I, Muskhelishvili G, Travers A, Buckle M. FIS modulates the kinetics of successive interactions of RNA polymerase with the core and upstream regions of the E. coli tyrT promoter. J Mol Biol. 2002;318:651–663. doi: 10.1016/S0022-2836(02)00142-0. [DOI] [PubMed] [Google Scholar]
- 24.Rochman M, Aviv M, Glaser G, Muskhelishvili G. Promoter protection by a transcription factor acting as a local topological homeostat. EMBO Rep. 2002;3:355–360. doi: 10.1093/embo-reports/kvf067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kravatskaya GI, Kravatsky YV, Chechetkin VR, Tumanyan VG. Coexistence of different base periodicities in prokaryotic genomes as related to DNA curvature, supercoiling, and transcription. Genomics. 2011;98:223–231. doi: 10.1016/j.ygeno.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Kravatskaya GI, Chechetkin VR, Kravatsky YV, Tumanyan VG. Coexistence of different base periodicities in prokaryotic genomes as related to DNA curvature, supercoiling, and transcription. Genomics. 2013;101:1–11. doi: 10.1016/j.ygeno.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Holmquist PC, Holmquist GP, Summers ML. Comparing binding site information to binding affinity reveals that Crp/DNA complexes have several distinct binding conformers. Nucleic Acids Res. 2011;39:6813–6824. doi: 10.1093/nar/gkr369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Travers A, Muskhelishvili G. DNA thermodynamics shape chromosome organisation and topology. Biochem Soc Trans. 2013;41:548–553. doi: 10.1042/BST20120334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sobetzko P, Travers A, Muskhelishvili G. Gene order and chromosome dynamics coordinate gene expression during the bacterial growth cycle. Proc Natl Acad Sci USA. 2012;109:E42–E50. doi: 10.1073/pnas.1108229109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobetzko P, Glinkowska M, Travers A, Muskhelishvili G (2013) DNA thermodynamic stability and supercoil dynamics determine the gene expression program during the bacterial growth cycle. Mol Biosyst. doi:10.1039/C3MB25515H [DOI] [PubMed]
- 31.Travers AA, Vaillant C, Arneodo A, Muskhelishvili G. DNA structure, nucleosome placement and chromatin remodeling—a perspective. Biochem Soc Trans. 2012;40:335–340. doi: 10.1042/BST20110757. [DOI] [PubMed] [Google Scholar]
- 32.Berger M, Zhelyaskova P, Brix K, Travers A, Muskhelishvili G. Coordination of genomic structure and function by the main bacterial nucleoid-associated protein HU. EMBO Rep. 2010;11:59–64. doi: 10.1038/embor.2009.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cagliero C, Jin DJ. Dissociation and re-association of RNA polymerase with DNA during osmotic response in Escherichia coli . Nucleic Acids Res. 2013;41:315–326. doi: 10.1093/nar/gks988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McClellan JA, Boublíková P, Palecek E, Lilley DM. Superhelical torsion in cellular DNA responds directly to environmental and genetic factors. Proc Natl Acad Sci USA. 1990;87:8373–8377. doi: 10.1073/pnas.87.21.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsieh LS, Burger RM, Drlica K. Bacterial DNA supercoiling and [ATP]/[ADP]. Changes associated with a transition to anaerobic growth. J Mol Biol. 1991;219:443–450. doi: 10.1016/0022-2836(91)90185-9. [DOI] [PubMed] [Google Scholar]
- 36.Hsieh LS, Rouviere-Yaniv J, Drlica K. Bacterial DNA supercoiling and [ATP]/[ADP] ratio: changes associated with salt shock. J Bacteriol. 1991;173:3914–3917. doi: 10.1128/jb.173.12.3914-3917.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Workum M, van Dooren SJ, Oldenburg N, Molenaar D, Jensen PR, Snoep JL, Westerhoff HV. DNA supercoiling depends on the phosphorylation potential in Escherichia coli . Mol Microbiol. 1996;20:351–360. doi: 10.1111/j.1365-2958.1996.tb02622.x. [DOI] [PubMed] [Google Scholar]
- 38.Geertz M, Travers A, Mehandziska S, Janga SC, Shimamoto N, Muskhelishvili G (2011) Structural coupling between RNA polymerase composition and DNA supercoiling in coordinating transcription: a global role for the omega subunit? mBio 2(4). doi:10.1128/mBio.00034-11 [DOI] [PMC free article] [PubMed]
- 39.Bordes P, Conter A, Morales V, Bouvier J, Kolb A, Gutierrez C. DNA supercoiling contributes to disconnect sigmaS accumulation from sigmaS-dependent transcription in Escherichia coli . Mol Microbiol. 2003;48:561–571. doi: 10.1046/j.1365-2958.2003.03461.x. [DOI] [PubMed] [Google Scholar]
- 40.Ladurner A. Chromatin places metabolism center stage. Cell. 2009;138:18–20. doi: 10.1016/j.cell.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 41.Bellet M, Sassone-Corsi P. Mammalian circadian clock and metabolism—the epigenetic link. J Cell Sci. 2010;123:3837–3848. doi: 10.1242/jcs.051649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knight J, Milner J. SIRT1, metabolism and cancer. Curr Opin Oncol. 2012;24:68–75. doi: 10.1097/CCO.0b013e32834d813b. [DOI] [PubMed] [Google Scholar]
- 43.Machne R, Murray D. The yin and yang of yeast transcription: elements of a global feedback system between metabolism and chromatin. PLoS ONE. 2012;7:e37906. doi: 10.1371/journal.pone.0037906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klevecz RR, Bolen J, Forrest G, Murray DB. A genomewide oscillation in transcription gates DNA replication and cell cycle. Proc Natl Acad Sci USA. 2004;101:1200–1205. doi: 10.1073/pnas.0306490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lesne A (2013) Multiscale analysis of biological systems. Acta Biotheor. doi:10.1007/s10441-013-9170-z. [Epub ahead of print] [DOI] [PubMed]
- 46.Valens M, Penaud S, Rossignol M, Cornet F, Boccard F. Macrodomain organization of the Escherichia coli chromosome. EMBO J. 2004;23:4330–4341. doi: 10.1038/sj.emboj.7600434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng S, Stein RA, Higgins NP. Organization of supercoil domains and their reorganization by transcription. Mol Microbiol. 2005;57:1511–1521. doi: 10.1111/j.1365-2958.2005.04796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Umbarger MA, Toro E, Wright MA, Porreca GJ, Bau D, Hong SH, Fero MJ, Zhu LJ, Marti-Renom MA, McAdams HH, Shapiro L, Dekker J, Church GM. The three-dimensional architecture of the bacterial genome and its alteration by genetic perturbation. Mol Cell. 2011;44:252–264. doi: 10.1016/j.molcel.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiggins PA, Cheveralls KC, Martin JS, Lintner R, Kondev J. Strong intranucleoid organize the Escherichia coli chromosome into a nucleoid filament. Proc Natl Acad Sci USA. 2010;107:4991–4995. doi: 10.1073/pnas.0912062107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Junier I, Herrison J, Képès F. Genomic organization of evolutionarily correlated genes in bacteria: limits and strategies. J Mol Biol. 2012;419:369–386. doi: 10.1016/j.jmb.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 52.Hong SH, Toro E, Mortensen KI, de la Rosa MA, Doniach S, Shapiro L, Spakowitz AJ, McAdams HH. Caulobacter chromosome in vivo configuration matches model predictions for a supercoiled polymer in a cell-like confinement. Proc Natl Acad Sci USA. 2013;110:1674–1679. doi: 10.1073/pnas.1220824110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Azam TA, Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J Biol Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- 54.Ali Azam TA, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dame RT. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol Microbiol. 2005;56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 56.Murphy LD, Zimmerman SB. Isolation and characterization of spermidine nucleoids from Escherichia coli . J Struct Biol. 1997;119:321–335. doi: 10.1006/jsbi.1997.3883. [DOI] [PubMed] [Google Scholar]
- 57.Browning DF, Grainger DC, Busby SJ. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr Opin Microbiol. 2010;13:773–780. doi: 10.1016/j.mib.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 58.Martínez-Antonio A, Medina-Rivera A, Collado-Vides J. Structural and functional map of a bacterial nucleoid. Genome Biol. 2009;10(12):247. doi: 10.1186/gb-2009-10-12-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rimsky S, Travers A. Pervasive regulation of nucleoid structure and function by nucleoid-associated proteins. Curr Opin Microbiol. 2011;14:136–141. doi: 10.1016/j.mib.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 60.Schneider R, Travers A, Kutateladze T, Muskhelishvili G. A DNA architectural protein couples cellular physiology and DNA topology in Escherichia coli . Mol Microbiol. 1999;34:953–964. doi: 10.1046/j.1365-2958.1999.01656.x. [DOI] [PubMed] [Google Scholar]
- 61.Weinstein-Fischer D, Altuvia S. Differential regulation of Escherichia coli topoisomerase I by Fis. Mol Microbiol. 2007;63:1131–1144. doi: 10.1111/j.1365-2958.2006.05569.x. [DOI] [PubMed] [Google Scholar]
- 62.Stuger R, Woldringh CL, van der Weijden CC, Vischer NO, Bakker BM, van Spanning RJ, Snoep JL, Westerhoff HV. DNA supercoiling by gyrase is linked to nucleoid compaction. Mol Biol Rep. 2002;29:79–82. doi: 10.1023/A:1020318705894. [DOI] [PubMed] [Google Scholar]
- 63.Spurio R, Dürrenberger M, Falconi M, La Teana A, Pon CL, Gualerzi CO. Lethal overproduction of the Escherichia coli nucleoid protein H-NS: ultramicroscopic and molecular autopsy. Mol Gen Genet. 1992;231:201–211. doi: 10.1007/BF00279792. [DOI] [PubMed] [Google Scholar]
- 64.Frenkiel-Krispin D, Ben-Avraham I, Englander J, Shimoni E, Wolf SG, Minsky A. Nucleoid restructuring in stationary-state bacteria. Mol Microbiol. 2004;51:395–405. doi: 10.1046/j.1365-2958.2003.03855.x. [DOI] [PubMed] [Google Scholar]
- 65.Jeong KS, Ahn J, Khodursky AB. Spatial patterns of transcriptional activity in the chromosome of Escherichia coli . Genome Biol. 2004;5:R86. doi: 10.1186/gb-2004-5-11-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peter BJ, Arsuaga J, Breier AM, Khodursky AB, Brown PO, Cozzarelli NR. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli . Genome Biol. 2004;5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hardy CD, Cozzarelli NR. A genetic selection for supercoiling mutants of Escherichia coli reveals proteins implicated in chromosome structure. Mol Microbiol. 2005;57:1636–1652. doi: 10.1111/j.1365-2958.2005.04799.x. [DOI] [PubMed] [Google Scholar]
- 68.Ohniwa RL, Morikawa K, Kim J, Ohta T, Ishihama A, Wada C, Takeyasu K. Dynamic state of DNA topology is essential for genome condensation in bacteria. EMBO J. 2006;25:5591–5602. doi: 10.1038/sj.emboj.7601414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferrándiz MJ, Martín-Galiano AJ, Schvartzman JB, de la Campa AG. The genome of Streptoccus pneumoniae is organized in topology-reacting gene clusters. Nucleic Acids Res. 2010;38:3570–3581. doi: 10.1093/nar/gkq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheung KJ, Badarinarayana V, Selinger DW, Janse D, Church GM. A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli . Genome Res. 2003;13:206–215. doi: 10.1101/gr.401003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kar S, Edgar R, Adhya S. Nuceloid remodeling by an altered HU protein: reorganization of a transcription program. Proc Natl Acad Sci USA. 2005;102:16397–16402. doi: 10.1073/pnas.0508032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat Struct Mol Biol. 2007;14:441–448. doi: 10.1038/nsmb1233. [DOI] [PubMed] [Google Scholar]
- 73.Maurer S, Fritz J, Muskhelishvili G. A systematic in vitro study of nucleoprotein complexes formed by bacterial nucleoid associated proteins revealing novel types of DNA organization. J Mol Biol. 2009;387:1261–1276. doi: 10.1016/j.jmb.2009.02.050. [DOI] [PubMed] [Google Scholar]
- 74.Prieto AI, Kahramanoglou C, Ali RM, Fraser GM, Seshasayee AS, Luscombe NM. Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12. Nucleic Acids Res. 2011;40:3524–3537. doi: 10.1093/nar/gkr1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Travers A, Muskhelishvili G. Bacterial chromatin. Curr Opin Genet Dev. 2005;15:507–514. doi: 10.1016/j.gde.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 76.Lang B, Blot N, Bouffartigues E, Buckle M, Geertz M, Gualerzi CO, Mavathur R, Muskhelishvili G, Pon C, Rimsky S, Stella S, Madan Babu M, Travers A. High affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucl Acids Res. 2007;35:6330–6337. doi: 10.1093/nar/gkm712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Biggar SR, Crabtree GR. Cell signaling can direct either binary or graded transcriptional responses. EMBO J. 2001;120:3167–3176. doi: 10.1093/emboj/20.12.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hazzalin CA, Mahadevan LC. MAPK-regulated transcription: a continuously variable gene switch? Nat Rev Mol Cell Biol. 2002;3:30–40. doi: 10.1038/nrm715. [DOI] [PubMed] [Google Scholar]
- 79.Giorgetti L, Siggers T, Tiana G, Caprara G, Notarbartolo S, Corona T, Pasparakis M, Milani P, Bulyk ML, Natoli G. Noncooperative interactions between transcription factors and clustered DNA binding sites enable graded transcriptional responses to environmental inputs. Mol Cell. 2010;37:418–428. doi: 10.1016/j.molcel.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 80.Brinza L, Calevro F, Charles H. Genomic analysis of the regulatory elements and links with intrinsic DNA structural properties in the shrunken genome of Buchnera. BMC Genomics. 2013;14:73. doi: 10.1186/1471-2164-14-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rocha EP, Danchin A. Gene essentiality determines chromosome organisation in bacteria. Nucleic Acids Res. 2003;31:6570–6577. doi: 10.1093/nar/gkg859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Montero Llopis P, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, Jacobs-Wagner C. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–82. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kuhlman TE, Cox EC. Gene location and DNA density determine transcription factor distributions in Escherichia coli . Mol Syst Biol. 2012;8:610. doi: 10.1038/msb.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Woelfle M, Xu Y, Qin X, Johnson C. Circadian rhythms of superhelical status of DNA in cyanobacteria. Proc Natl Acad Sci USA. 2007;104:18819–18824. doi: 10.1073/pnas.0706069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vijayan V, Zuzow R, O’Shea E. Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 2009;106:22564–22568. doi: 10.1073/pnas.0912673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rovinskiy N, Agbleke AA, Chesnokova O, Pang Z, Higgins NP. Rates of gyrase supercoiling and transcription elongation control supercoil density in a bacterial chromosome. PLoS Genet. 2012;8(8):e1002845. doi: 10.1371/journal.pgen.1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Naughton C, Avlonitis N, Corless S, Prendergast JG, Mati IK, Eijk PP, Cockroft SL, Bradley M, Ylstra B, Gilbert N. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol. 2013;20:387–395. doi: 10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Conrad TM, Frazier M, Joyce AR, Cho BK, Knight EM, Lewis NE, Landick R, Palsson BØ. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci USA. 2010;107:20500–20505. doi: 10.1073/pnas.0911253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Norris V, Blaauwen T, Doi RH, Harshey RM, Janniere L, Jiménez-Sánchez A, Jin DJ, Levin PA, Mileykovskaya E, Minsky A, Misevic G, Ripoll C, Saier M, Skarstad K, Thellier M. Toward a hyperstructure taxonomy. Annu Rev Microbiol. 2007;61:309–329. doi: 10.1146/annurev.micro.61.081606.103348. [DOI] [PubMed] [Google Scholar]
- 90.Wang W, Li GW, Chen C, Xie XS, Zhuang X. Chromosome organization by a nucleoid-associated protein in live bacteria. Science. 2011;333:1445–1449. doi: 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]