Abstract
The field of epigenetics is expanding rapidly, yet there is persistent uncertainty in the definition of the term. The word was coined in the mid-twentieth century as a descriptor of how intrinsic, yet largely unknown, forces act with genes to channel progenitor cells along pathways of differentiation. Near the end of the twentieth century, epigenetics was defined more specifically as the study of changes in gene activity states. In some definitions, only those activity states that are inherited across cell division were considered. Other definitions were broader, also including activity states that are transient, or occurring in non-dividing cells. The greatest point of disagreement in these current definitions, is if the term should concern only inherited activity states. To alleviate this disparity, an alternative term, ‘memigenetics’, could be used in place of epigenetics to describe inherited chromatin activity states. The advantage of this term is that it is self-defining, and would serve to emphasize the important concept of cell memory. It would also free the term epigenetics to be used in a broader sense in accord with the meaning of the prefix ‘epi’, that is, as a descriptor of what is ‘over’ DNA at any point in time.
Keywords: Epigenetics, Memigenetics, Cell memory, Definition of epigenetics
Introduction
Epigenetics as a discipline has expanded rapidly in recent years, and the term has become very familiar. However, at meetings and in articles, we still feel compelled to define the term. This suggests a persistent uncertainty and lack of concurrence in its definition. Here, some authoritative definitions are provided, and used to point out the areas of disagreement and confusion in the meaning of the term. Also, a new term, ‘memigenetics’, is introduced, that could be used to alleviate some of the confusion.
The definitions of epigenetics
The term epigenetics was originally coined as a broad descriptor of the forces that, in cooperation with the genes, produce a phenotype. These forces were modelled in an ‘epigenetic landscape’ of descending valleys or canals that act with genes to channel the cell’s lineage to a stable and terminally differentiated form [1, 2]. At the time, it was not clear if differentiation proceeded through the discarding of genes, or through differential gene activity. We now know that the large majority of cells have the same complement of genes. Cells differentiating along defined pathways, or ‘canals’, must activate only some genes, for example, neural-specific genes, to reach stable phenotypic states. More specific definitions of the term epigenetics have since been provided. These later adoptions began in the 1980s, inspired by the increasing realization that DNA methylation provides a mechanism for influencing and maintaining gene activity states. For this discussion, five current definitions are provided (Table 1).
Table 1.
Some current definitions of epigenetics
| ‘The study of changes in gene expression, which occur in organisms with differentiated cells, and the mitotic inheritance of given patterns of gene expression’ [11] |
| ‘The study of mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in DNA sequence.’ [3] |
| ‘…the structural adaptation of chromosomal regions so as to register, signal or perpetuate altered activity states.’ [12] |
| ‘An epigenetic trait is a stably heritable phenotype resulting from changes in a chromosome without alterations in the DNA sequence.’ [4] |
| ‘…the inheritance of variation (-genetics) above and beyond (epi-) changes in the DNA sequence.’ [5] |
Heritability in epigenetics: required or optional?
In the essentially equivalent definitions [3–5] (Table 1), an epigenetic state or phenotype is one that is transferred from mother to daughter cells, or one that is ‘remembered’, this being achieved without any change to the genome. An obvious example is the maintenance of a differentiated cellular phenotype on cell division. Other clear examples include mammalian X chromosome inactivation (XCI), and the variant peloric phenotype in the flowering plant, toadflax (Fig. 1a–c). Disease states that presumably involve the heritability of aberrant gene activity include the fetal programming of disease [6, 7], and the inducement of disease states in young through parental or grand-parental exposure to toxins, or to an inadequate diet [8–10]. However, a significant problem with the stipulation of heritability is that the prefix ‘epi’ (from Greek epi (prep.); upon, above, in addition; Oxford dictionary) does not convey this meaning.
Fig. 1.
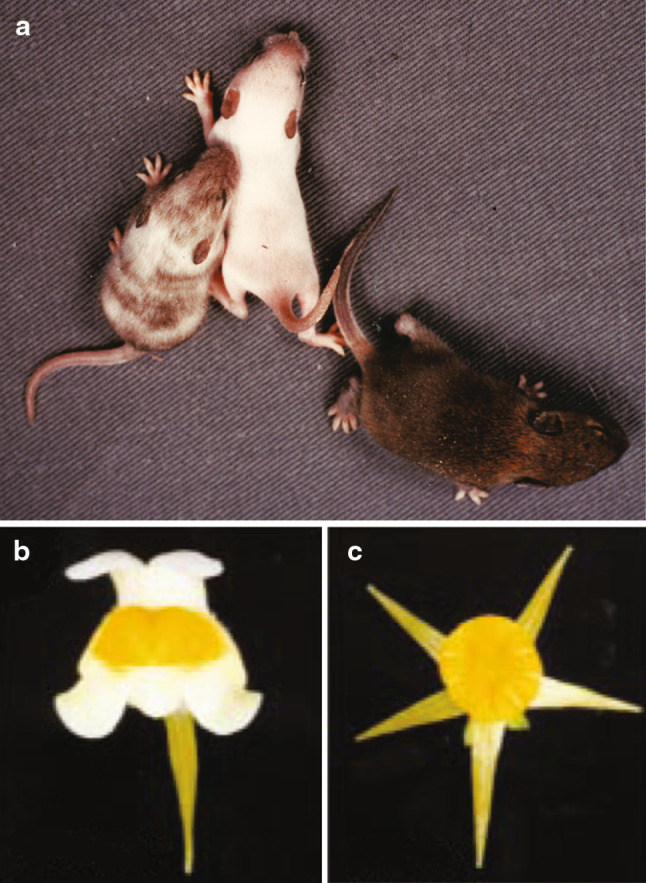
Examples of heritable epigenetic phenomena. a Neonatal mice carrying a mutation of the X-linked Atp7a (ATPase, Cu++ transporting, alpha polypeptide) gene have a low activity of tyrosinase, a copper-dependent enzyme that is required for melanin production. XY males are depigmented (centre) relative to pigmented, wild-type XY males or XX females (right). Random X chromosome inactivation (XCI) and clonal expansion in XX embryos carrying one wild-type and one mutant allele lead to depigmented and pigmented patches, as seen in the mottled female (left) [52]. Reprinted from [53]. b Toadflax wild-type flower, c toadflax peloric flower. The variant peloric flower phenotype, heritable through generations, cosegregates with DNA methylation and inactivation of the cycloidea gene. Here, methylation is probably conferring gene silencing, and maintaining this state through cell division. This type of gene inactivation, involving no change in DNA sequence, is termed an ‘epimutation’ [54]. Figures of toadflax flowers [44] reproduced with the permission of Macmillan Publishers, Ltd
By contrast, the remaining definitions [11, 12] are broader in scope. In these, a gene activity state need not necessarily be heritable across cell division. Indeed, it can be argued that the requirement for heritability or ‘cell memory’ places a severe constraint on the term. Alteration in gene activity states that could occur in terminally differentiated and non-dividing cells (for example, neurons) do not qualify under the definition [13]. Also, changes in activity state that are limited to a particular stage of the cell cycle do not qualify [12]. Examples of the latter are the upregulation of canonical core histone gene expression at S-phase [14] and the cell-cycle-dependent redistribution of the histone variant H2A.Z to centromeres and sub-telomeres [15]. While these broader definitions are more in keeping with the meaning of the prefix epi-, a problem is that they de-emphasize the concept of heritability. That is, heritable and transient changes in gene activity, which are qualitatively different phenomena, are defined by the same term. For example, the clonal inheritance of mammalian XCI, and a transient cell-cycle-specific histone modification that may aid in transcriptional activation, are both defined as epigenetic. The concept of cell memory, central to some definitions [3–5], is a very important one, not the least because the mechanisms that enable the faithful transfer of gene activity states across cell division are little understood [16].
What could help in easing this important disparity in the definitions is to use an alternative term when the heritability of activity states is being considered. The term ‘epigenetic inheritance’ was used to make this distinction [17, 18]. However, the main difficulty with this term is that it now adds to the confusion, given that in some current definitions [3–5], the ‘inheritance’ component of the term is redundant: that is, ‘epigenetics’ and ‘epigenetic inheritance’ have the same meaning. Instead, an alternative term, ‘memigenetics’, could be used to stipulate when an epigenetic state is inherited. The prefix memi- is a contraction of ‘memo-epi-’, and therefore is intended to mean a ‘remembering’ of what is ‘over’ (memo, from Latin memoria; mindful, remembering; Oxford dictionary). This term serves to highlight and delineate when epigenetics does involve ‘The study of mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in DNA sequence.’ [3]. Heritable epigenetic phenomena, such as XCI, genomic imprinting, and fetal programming, can be classified as memigenetic phenomena—when this is helpful. The term should not be easily misused, as the prefix is self-defining. Also, it emphasizes the important concept of cell memory. The use of this word should simultaneously have the effect of freeing the term epigenetics from its oft-perceived constraint of heritability, and allow it to be used in a broader sense as befits its literal meaning.
The nuts and bolts of epigenetics
If we can be accept the broader definition of epigenetics as outlined [11–13], then some further clarification on the use of this term is useful. Two definitions (Table 1) stipulate that an epigenetic state is one pertaining to gene activity or transcription [3, 11]. By contrast, in the other three definitions [4, 5, 12], ‘activity’ is more general, and can be thought of as chromatin activity states, whether this be transcription or some other kind of activity, such as DNA repair. Further examples could include the complex structures of centromeres and telomeres, for which the primary function is kinetochore and chromosome-end protection, respectively. There seems no a priori reason to exclude such non-transcriptional activities of chromatin from the definition, and their inclusion fits well with the broader definition of epigenetics, as provided [12].
This leads to another point—that epigenetics is often perceived somewhat arbitrarily as the study of DNA methylation and histone post-translational modifications (PTMs). Here, the perception of epigenetics has shifted somewhat from a concept to something more limited in scope. A difficulty with this shift is that other molecules that are in chromatin, and that can induce changes in chromatin activity states, are excluded from the perception—for example, core histone variants and linker histones [19, 20]. Other examples include CTCF (CCCTC binding factor), which can form a chromatin insulator and block the interaction between an enhancer and a promoter [21], and the cohesin complex, which can promote the interaction of enhancers with promoters [22]. Further, the inducing properties of transcription factors (TFs) are often forgotten [23, 24]. A classical paradigm is the DNA binding of the lac repressor in bacteria, that efficiently blocks transcription of the lac operon in maintaining a basal activity state [25–27].
Word usage
A chromatin activity or epigenetic state—for example, a repressed promoter, can be viewed as the end-point of an epigenetic mechanism. Within the epigenetic mechanism are epigenetic effectors, these being entities that directly bring about the epigenetic state. Also, there are epigenetic modifiers—entities that bring the effectors into play, or modify the activity of the effectors. For example, DNA methylation is an effector, as it can alter DNA conformation and directly suppress transcription by inhibiting the binding of TFs. DNA methyltransferase (DNMT) enzymes, which catalyse DNA methylation, are modifiers. Polycomb repressor complex 1 (PRC1) is an effector of chromatin compaction, while histone 3 lysine 27 trimethylation (H3K27me3), which signals the binding of PRC1, is a modifier. Further upstream, PRC2 is also a modifier, as it catalyses the formation of H3K27me3. siRNAs processed from RNAs derived from pericentric repeats in fission yeast are modifiers, as they lead to the formation of H3K9me3, a waypoint in the formation of pericentric constitutive heterochromatin [28]. It is debatable if miRNAs are modifiers, as they repress gene activity at the post-transcriptional level. The Xist (inactive X specific transcripts) RNA is thought to attract repressive complexes to inactivate the X, and therefore would be a modifier [29]. Epigenetic marks or modifications can be epigenetic states per se, effectors, and sometimes modifiers. While these terms are used loosely, they are often used as descriptors of methylated sites and histone PTMs, or entities that are relatively fixed to chromatin.
Memigenetic states and mechanisms
If memigenetics is useful as a term to describe the study of heritable chromatin activity states, then epigenetic states will often also be memigenetic states. This is because many, if not the large majority, of chromatin activity states are reassembled in daughter cells. For example, all repressed promoters in mother cells will also be repressed in daughter cells, unless differentiation has resulted in the activation of some promoters. Memigenetic states are maintained by memigenetic mechanisms. These mechanisms, by definition, are those that carry the memory of a chromatin activity state across DNA replication and cell division so that it can be faithfully reassembled in daughter cells.
What do we know of memigenetic mechanisms? As previously emphasized [16], the DNA sequence itself enforces cell memory. For example, in the budding yeast (Saccharomyces cerevisiae), the binding of centromeric proteins is DNA sequence-specific [30]. TFs are memigenetic: as mentioned above, the classical paradigm is the DNA binding of the lac repressor in bacteria. Its repressive activity is maintained through cell division, until lactose is encountered [25–27]. The activity of the lac repressor fulfils all the criteria of the various definitions of epigenetics (Table 1). That TFs are drivers of cellular phenotype in metazoans is strikingly evident in experiments in which a few selected TFs can induce the formation of pluripotent cells from differentiated somatic cells [31]. Also, in cell reconstitution experiments, when the nucleus of one cell type is transplanted to the foreign enucleated cytoplasm of another cell type, it can adopt new gene activity states corresponding to the cell type from which the foreign cytoplasm was obtained, and maintain these states through cell division [32]. The best-known example of this experimental paradigm is Dolly the sheep, cloned by the transplantation of a differentiated somatic cell nucleus to an enucleated oocyte [33]. These reconstitution experiments show that the cell cytoplasm contains factors that can convey cell memory. The memory effectors are undefined, but are likely to include mRNAs [32]. In light of the findings of Takahashi and Yamanaka [31], these mRNAs could include those encoding TFs.
If a promoter is repressed by DNA methylation, then the maintenance DNMT machinery acting at replication fork can be viewed as the mechanism that maintains the memory of this repression. Without maintenance DNMT activity, the large majority of DNA methylation in the mammalian genome is rapidly lost through cell division [34]. ‘Bookmarking’ proteins also appear to be memigenetic. These proteins remain bound to promoters in mitotic chromosomes, prevent compaction of the promoter in an otherwise condensed chromatin milieu, and are required for proper activation of the promoter at the completion of cell division [35–37]. We note that an epigenetic modification—such as a histone variant or histone PTM, even if required for the assembly of a memigenetic state in mother cells and then in daughter cells, would not comprise part of the memigenetic mechanism unless it was needed for transferring the memory of the activity state across cell division. There is no strong evidence to date that histone variants and histone PTMs play a role in this regard. Memigenetics does not apply to cell-cycle transient phenomena such as DNA repair, which can be viewed as only epigenetic. A summary of known and possible memigenetic mechanisms, as discussed above, is provided (Table 2).
Table 2.
Molecular memigenetic mechanisms
| Known |
|---|
| DNA sequence: sequence elements are required for the binding of centromeric proteins in budding yeast. Also, the binding of TFs is dependent on DNA sequence [16, 26, 30] |
| Maintenance DNA methyltransferase activity: during S-phase, a fully methylated CpG dyad replicates to two hemi-methylated CpG dyads. Maintenance DNMT activity then converts hemi-methylated dyads to fully methylated dyads [34] |
| Diffusible TFs: if TFs are evicted from DNA at replication, or from condensed mitotic chomosomes, they can freely diffuse into daughter cells and reassemble into chromatin after chromosome decondensation [23, 24, 26, 31] |
| Bookmark or pioneer TFs: some TFs can remain bound to chromatin through mitosis, and thereby carry the memory of transcription into daughter cells [35–37] |
| Histone variant CENPA/CenH3: once incorporated and functional in specifying a centromere, it can self-propagate its incorporation at a genomic location, and hence centromere specification, through DNA replication [46, 47] |
| Possible |
| Histone variants and histone PTMs: the distribution of histone variants and their PTMs can be correlated with transcriptional activity. These correlations could be secondary to other mechanisms that carry transcriptional and structural memory of chromatin across cell division [15, 16, 48–51] |
Finally, mention should be made of the germ line. In mammals, primordial germ cells (PGCs) undergo a genome-wide loss of DNA methylation, then re-establish global DNA methylation at later stages of germ cell development [38]. In contrast to ‘remembering’, the erasure of DNA methylation in PGCs can be viewed as a ‘forgetting’ of epigenetic information derived from cellular ancestors. This loss of memory is required so that new epigenetic information can be established in gametes, in preparation for the fertilization and development of the new individual [39]. The clearest example is seen in the resetting of genomic imprints. Parental imprints are removed in migrating PGCs (maternal and paternal), so that new germ cell sex-specific imprints can be established in oocytes and spermatogonia [40, 41]. Other sequences are also involved, the most prominent being globally dispersed transposable elements (TEs). This germ line epigenetic re-establishment is best categorized as a de novo renewal of epigenetic states. In mammals, this renewal occurs without cell division, and for TEs, is driven by short non-coding RNAs that guide de novo DNMTs to TE targets. This mechanism for de novo DNA methylation operates only in the germ line [42]. Despite the efficient erasure of epigenetic information in PGCs, and then its de novo renewal, some epigenetic states are transmitted intergenerationally, as seen in some variant phenotypes [43–45] and disease states [8–10]. The memigenetic mechanisms involved in these intergenerational phenomena are still obscure, and are the subject of considerable research.
Conclusion
The term epigenetics is well-suited as a broad descriptor for what is ‘over’ DNA at any point in time, in accord with the literal meaning of the prefix. By contrast, there is a semantic constraint when the term is applied exclusively to inherited chromatin activity states, and this constraint often leads to confusion. For describing inherited activity states, an alternative term, memigenetics, is self-defining, and serves to emphasize the important concept of cell memory. This term does not introduce any new concept, but merely rebrands existing definitions of epigenetics that require cellular inheritance. Memigenetics can be thought of as a subset, or a branch of epigenetics, concerned with the remembering of chromatin activity states across cell division. Often, there may be no need to use the term in place of epigenetics. At the least, the term should serve as a useful device when we try to explain the concept of epigenetics.
Acknowledgments
Damien Hudson, Danielle Irvine, Paul Kalitsis, and Deidre Mattiske are thanked for helpful comments on the text. This work was supported by the National Health and Medical Research Council.
References
- 1.Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- 2.Waddington CH. The strategy of the genes. A discussion of some aspects of theoretical biology. London: George Allen and Unwin Ltd; 1957. [Google Scholar]
- 3.Riggs AD, Martienssen RA, Russo VEA. Introduction. In: Riggs AD, Martienssen RA, Russo VEA, editors. Epigenetic mechanisms of gene regulation. New York: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 4.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandman CA, Davis EP, Buss C, Glynn LM. Prenatal programming of human neurological function. Int J Pept. 2011;2011:837596. doi: 10.1155/2011/837596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjöström M, Golding J, ALSPAC Study Team Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet. 2006;14:159–166. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- 10.Ng SF, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature. 2010;467:963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 11.Holliday R. Epigenetics: an overview. Dev Genet. 1994;15:453–457. doi: 10.1002/dvg.1020150602. [DOI] [PubMed] [Google Scholar]
- 12.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 13.Holliday R. Epigenetics comes of age in the twentyfirst century. J Genet. 2002;81:1–4. doi: 10.1007/BF02715863. [DOI] [PubMed] [Google Scholar]
- 14.Rattray AM, Müller B. The control of histone gene expression. Biochem Soc Trans. 2012;40:880–885. doi: 10.1042/BST20120065. [DOI] [PubMed] [Google Scholar]
- 15.Nekrasov M, Amrichova J, Parker BJ, Soboleva TA, Jack C, Williams R, Huttley GA, Tremethick DJ. Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics. Nat Struct Mol Biol. 2012;19:1076–1083. doi: 10.1038/nsmb.2424. [DOI] [PubMed] [Google Scholar]
- 16.Fisher AG, Brockdorff N. Epigenetic memory and parliamentary privilege combine to evoke discussions on inheritance. Development. 2012;139:3891–3896. doi: 10.1242/dev.084434. [DOI] [PubMed] [Google Scholar]
- 17.Holliday R. Mechanisms for the control of gene activity during development. Biol Rev Camb Philos Soc. 1990;65:431–471. doi: 10.1111/j.1469-185X.1990.tb01233.x. [DOI] [PubMed] [Google Scholar]
- 18.Jablonka E, Lamb MJ. The changing concept of epigenetics. Ann N Y Acad Sci. 2002;981:82–96. doi: 10.1111/j.1749-6632.2002.tb04913.x. [DOI] [PubMed] [Google Scholar]
- 19.Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao B, Freedman BS, Miller KE, Heald R, Marko JF. Histone H1 compacts DNA under force and during chromatin assembly. Mol Biol Cell. 2012;23:4864–4871. doi: 10.1091/mbc.E12-07-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacPherson MJ, Sadowski PD. The CTCF insulator protein forms an unusual DNA structure. BMC Mol Biol. 2010;11:101. doi: 10.1186/1471-2199-11-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riggs AD, Porter TN. Overview of epigenetic mechanisms. In: Riggs AD, Martienssen RA, Russo VEA, editors. Epigenetic mechanisms of gene regulation. New York: Cold Spring Harbor Laboratory Press; 1996. pp. 1–4. [Google Scholar]
- 24.Ptashne M. On the use of the word ‘epigenetic’. Curr Biol. 2007;17:R233–R236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 25.Novick A, Weiner M. Enzyme induction as an all-or-none phenomenon. Proc Natl Acad Sci USA. 1957;43:553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis M. A tale of two repressors. J Mol Biol. 2011;409:14–27. doi: 10.1016/j.jmb.2011.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon AJ, Satory D, Halliday JA, Herman C. Heritable change caused by transient transcription errors. PLoS Genet. 2013;9:e1003595. doi: 10.1371/journal.pgen.1003595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev. 2010;20:134–141. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sado T, Brockdorff N. Advances in understanding chromosome silencing by the long non-coding RNA Xist. Philos Trans R Soc Lond B Biol Sci. 2013;368:20110325. doi: 10.1098/rstb.2011.0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/S0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 32.Shay JW. Cytoplasmic modification of nuclear gene expression. Mol Cell Biochem. 1983;57:17–26. doi: 10.1007/BF00223521. [DOI] [PubMed] [Google Scholar]
- 33.Campbell KH, McWhir J, Ritchie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64–66. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- 34.Chen ZX, Riggs AD. Maintenance and regulation of DNA methylation patterns in mammals. Biochem Cell Biol. 2005;83:438–448. doi: 10.1139/o05-138. [DOI] [PubMed] [Google Scholar]
- 35.Xing H, Wilkerson DC, Mayhew CN, Lubert EJ, Skaggs HS, Goodson ML, Hong Y, Park-Sarge OK, Sarge KD. Mechanism of hsp70i gene bookmarking. Science. 2005;307:421–423. doi: 10.1126/science.1106478. [DOI] [PubMed] [Google Scholar]
- 36.Xing H, Vanderford NL, Sarge KD. The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat Cell Biol. 2008;10:1318–1323. doi: 10.1038/ncb1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 39.Lefèvre C, Mann JR. RNA expression microarray analysis in mouse prospermatogonia: identification of candidate epigenetic modifiers. Dev Dyn. 2008;237:1082–1089. doi: 10.1002/dvdy.21482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szabó PE, Mann JR. Biallelic expression of imprinted genes in the mouse germ line: implications for erasure, establishment, and mechanisms of genomic imprinting. Genes Dev. 1995;9:1857–1868. doi: 10.1101/gad.9.15.1857. [DOI] [PubMed] [Google Scholar]
- 41.Szabó PE, Hübner K, Schöler H, Mann JR. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech Dev. 2002;115:157–160. doi: 10.1016/S0925-4773(02)00087-4. [DOI] [PubMed] [Google Scholar]
- 42.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 43.Wolff GL. Influence of maternal phenotype on metabolic differentiation of agouti locus mutants in the mouse. Genetics. 1978;88:529–539. doi: 10.1093/genetics/88.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401:157–161. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- 45.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 46.Dunleavy EM, Almouzni G, Karpen GH. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G1 phase. Nucleus. 2011;2:146–157. doi: 10.4161/nucl.2.2.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mendiburo MJ, Padeken J, Fülöp S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- 48.Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- 49.Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 50.Jullien J, Astrand C, Szenker E, Garrett N, Almouzni G, Gurdon JB. HIRA dependent H3.3 deposition is required for transcriptional reprogramming following nuclear transfer to Xenopus oocytes. Epigenetics Chromatin. 2012;5:17. doi: 10.1186/1756-8935-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Banaszynski LA, Wen D, Dewell S, Whitcomb SJ, Lin M, Diaz N, Elsässer SJ, Chapgier A, Goldberg AD, Canaani E, Rafii S, Zheng D, Allis CD. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013;155:107–120. doi: 10.1016/j.cell.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 53.Mann JR (1981) Copper metabolism in mouse mutants. MSc Dissertation, The University of Melbourne
- 54.Jeggo PA, Holliday R. Azacytidine-induced reactivation of a DNA repair gene in Chinese hamster ovary cells. Mol Cell Biol. 1986;6:2944–2949. doi: 10.1128/mcb.6.8.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]


