Abstract
In eukaryotic cells, proteasomes are highly conserved protease complexes and eliminate unwanted proteins which are marked by poly-ubiquitin chains for degradation. The 26S proteasome consists of the proteolytic core particle, the 20S proteasome, and the 19S regulatory particle, which are composed of 14 and 19 different subunits, respectively. Proteasomes are the second-most abundant protein complexes and are continuously assembled from inactive precursor complexes in proliferating cells. The modular concept of proteasome assembly was recognized in prokaryotic ancestors and applies to eukaryotic successors. The efficiency and fidelity of eukaryotic proteasome assembly is achieved by several proteasome-dedicated chaperones that initiate subunit incorporation and control the quality of proteasome assemblies by transiently interacting with proteasome precursors. It is important to understand the mechanism of proteasome assembly as the proteasome has key functions in the turnover of short-lived proteins regulating diverse biological processes.
Keywords: Multisubunit protease, Precursor complex, Chaperones, Blm10, Ecm29
Proteasome assembly
Proteasomes are the key protease complexes responsible for the ubiquitin-dependent degradation of proteins and play crucial roles in cellular processes such as the regulation of cell cycle progression, gene expression, transcription and the quality control of newly synthesized proteins [1–4]. Proteasomes are the second-most abundant protein complexes and constitute up to 5 % of the protein content [5]. The biogenesis of proteasomes requires the synthesis of around three dozen different subunits and one dozen transiently auxiliary proteins depending on which protein is considered as an integral component of the proteasome. The auxiliary proteins function like chaperones and orchestrate the incorporation of the multiple subunits into the protease complex. The proteasome holoenzyme consists of the proteolytic core particle (CP, alternatively named 20S proteasome) and the regulatory particle (RP, 19S proteasome, Proteasome Activator PA700). The proteasome with RP–CP configuration is referred to as 26S proteasome, while the RP–CP–RP configuration is mentioned as 30S proteasome [6]. In principle, proteasome holoenzyme complexes are assembled from inactive precursors of the CP and modules of the RP, short-lived intermediate protein complexes. Despite their short half-life, proteasome assembly intermediates represent a considerable protein fraction, particularly during cell division when the number of proteasomes needs to be doubled [7]. With proteasome maturation, the proteolytic active sites within the CP are formed by autocatalytic propeptide cleavage according to the concept of a self-compartmentalizing threonine protease [8].
Throughout all kingdoms of life, the CP consists of four stacked heptameric rings arranged in a α1–7β1–7β1–7α1–7 architecture as shared by the archaea Thermoplasma acidophilus and Archaeoglobus fulgidus, the eubacteria Rhodococcus erythropolis and Mycobacterium tuberculosis, and eukaryotic cells in yeast Saccharomyces cerevisiae, Bos Taurus and Homo sapiens [9–14]. Archaeal and bacterial CP are usually assembled from single α and β subunits, of which the β subunit is synthesized as a precursor protein with an N-terminal propeptide that is autocatalytically cleaved off with the maturation of the proteolytic chamber [15, 16]. In eukaryotes, the CP is composed of seven different α and β subunits, respectively. Five of the seven β subunits, β1, β2, β5, β6 and β7 are synthesized with propeptides. Upon CP maturation, the cleavage of the β-propeptides liberates the active site threonines, which are contributed by the β1, β2 and β5 subunits. The β1, β2 and β5 active sites exhibit different peptide cleavage preferences and confer trypsin-, caspase-, and chymotrypsin-like activities, respectively [17].
The RP exists in eukaryotic cells and is composed of a base and lid subcomplex. The RP base consists of a ring of six AAA ATPases (ATPases associated with different cellular activities) named Rpt1 to Rpt6 (Regulatory particle triple A protein), and four non-ATPase subunits (Rpn1, Rpn2, Rpn10 and Rpn13, Regulatory particle non-ATPase). Rpn1 and Rpn2 receive substrate delivery proteins (Rad23, Dsk2, Ddi1, the proteasome-intrinsic poly-ubiquitin receptors Rpn10 and Rpn13), which recognize the protein substrates by poly-ubiquitin chains [18]. The RP ATPase ring attaches to the CP α ring, regulates α ring gating, and promotes the translocation of unfolded protein substrates into the proteolytic chamber of the CP [19]. In prokaryotic cells, there are AAA ATPases, RP-related base complexes, that deliver unfolded protein substrates for degradation to prokaryotic proteasomes without the aid of poly-ubiquitin chains [20–22].
In eukaryotic cells, prior to or during the translocation of the protein substrate into the proteolytic cavity of the CP, the poly-ubiquitin chain is released. The RP lid contains Rpn11, a subunit with metalloprotease activity, that is involved in recycling ubiquitin moieties from poly-ubiquitin chains [23, 24]. Also Ubp6/UCH14, a proteasome-associated deubiquitinating enzyme, assists in removing (poly-)ubiquitin from protein substrates [25].
This review will focus on molecular mechanisms that govern the assembly of archaeal, bacterial and eukaryotic CP, the RP and RP–CP association.
Archaeal CP
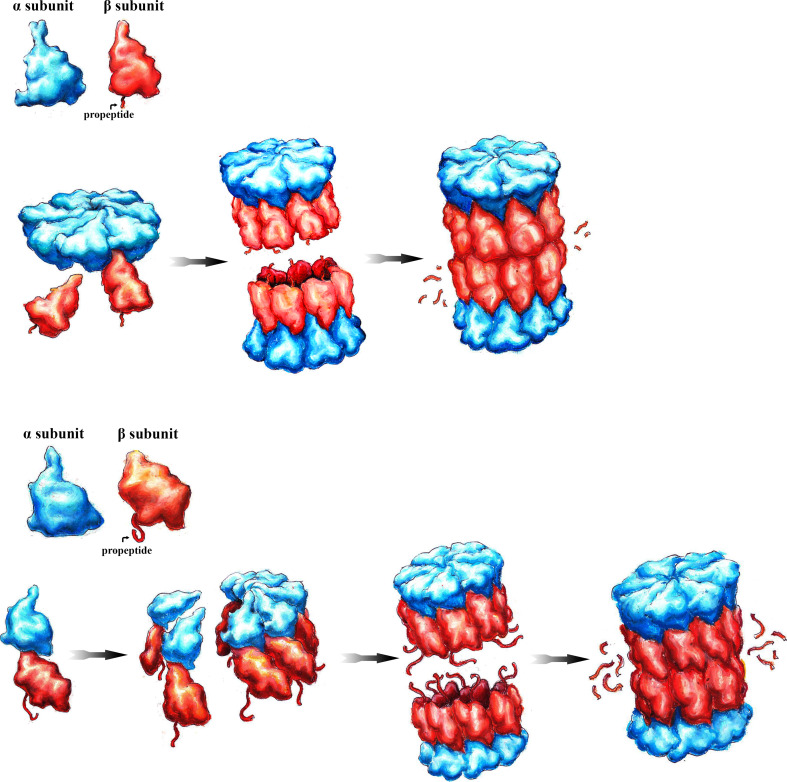
Archaeal CP is simple in composition, with a single α and β subunit stacked in four heptameric rings with symmetric α1–7β1–7β1–7α1–7 arrangement (Fig. 1, upper panel). The β subunits are expressed as precursor proteins with N-terminal propeptides, that are cleaved off by autocatalytic processing upon which the active site threonines are exposed [9].
Fig. 1.
Assembly of archaeal and eubacterial proteasome core particles (CP). The CP consists of α- and β-type subunits. In T. acidophilum, the first step is the assembly of the α ring which serves as template for pro-β subunit incorporation. The dimerization of two half CP yields the pre-holo-CP which is converted into the mature CP by autocatalytic cleavage of the β-propeptides (upper panel). In R. erythropolis, the first step is the formation of an α/pro-β heterodimer. The oligomerization of seven α/pro-β dimers results in the formation of the half-CP. Two half-CP dimerize into the pre-holo-CP in which β-propeptide processing yields CP maturation (lower panel)
In principle, as shown for archaea T. acidophilum, recombinant α and pro-β subunits can self-assemble into CP that is indistinguishable from the in vivo assembled CP [26].
While recombinant archaeal α subunits readily oligomerize into heptameric rings, the pro-β subunits stay monomeric and immature when expressed alone. Expressed together, the α ring serves as platform for pro-β subunit incorporation. The propeptides of the β subunit precursors are not required for incorporation, since the CP is readily assembled with mature recombinant β subunits lacking the propeptides [27].
The divergence in assembly behavior between archaeal α and β subunits is explained through a key difference in the tertiary structure. Although archaeal α and β subunits show striking homology with regard to their tertiary structures, the α-subunit exhibits a specific N-terminal H0 helix. Interactions between neighboring α subunits are mediated through contacts between H0 helices and the loop preceding the H0 helix from the adjacent α subunit. Hence, α subunits with truncated H0 helices no longer oligomerize into rings. Archaeal CP self-assembly from recombinant subunits suggest that no external chaperone is required for the assembly, at least in vitro [27].
Bacterial CP
Eubacterial proteasomes have been identified among the actinomycete lineage such as the M. tuberculosis and R. erythropolis (Fig. 1, lower panel).
Actinomycetes usually have a single α and a single β subunit except for the R. erythropolis genome, which encodes two α and two β subunits.
An important difference between the CP from the eubacteria R. erythropolis and archaea T. acidophilum is that the bacterial α subunits do not self-assemble into rings, since they lack the N-terminal H0 helix. When R. erythropolis α and β subunits are co-expressed in E. coli, self-assembly into functional CP occurred [28]. Instead of the formation of an α ring, bacterial CP assembly starts with the formation of a α/pro-β heterodimer followed by oligomerization into half-CP with α1-7β1-7 configuration [29].
Bacterial β-propeptides are different both in size and function compared to archaeal β-propeptides. As opposed to the propeptides in archaea, bacterial β-propeptides are longer, around 60 amino acid residues. Unlike in T. acidophilum in which the β subunits can be incorporated without propeptides, the β-propeptides in R. erythropolis are required for efficient CP assembly. They assist in the folding of the β precursor protein, control the dimerization of two half-CP precursor complexes with α1–7β1–7 configuration, because they extrude to the interfaces of two opposing β rings, and finally act in trans in β-propeptide processing. Thus, bacterial propeptides are considered as intramolecular chaperones of CP assembly [28].
Bacterial CP readily self-assemble from recombinant subunits without additional chaperones, but dedicated factors might exist which improve the fidelity and efficiency of CP assembly in vivo. Indeed, orthologs of yeast PAC1 and PAC2, two CP-dedicated chaperones, were annotated in archaeal and bacterial genomes [30]. PbaA and PbaB, the archaeal homologs of PAC1 and PAC2 do not play a crucial role in orchestrating CP assembly. PbaB rather forms a homotetramer that functions as an ATP-independent proteasome activator providing insights into the evolutionary relationships between CP assembly factors and CP activators [31].
Eukaryotic CP
The PAC chaperones control α ring formation in early CP assembly
The assembly of eukaryotic CP starts similar to the assembly of archaeal CP with the formation of the α subunit ring serving as template for β subunits incorporation (Fig. 2).
Fig. 2.
The assembly of eukaryotic CP involves CP-dedicated chaperones. The chaperones PAC1–PAC2 assist the assembly of the α ring from seven different α subunits and prevent faulty α–α ring dimerization. The chaperones PAC3–PAC4 support the incorporation of pro-β2. PAC3–PAC4 is released, when β3, β4 and Ump1 join the α ring. With the incorporation of pro-β1, pro-β5 and pro-β6 the half-CP is almost completed. The dimerization of two half-CP into the pre-holo-CP is stabilized by the incorporation of pro-β7, the last β subunit. The half-CP is also found to be associated with Blm10, especially, if the ability to close the α ring gates with CP maturation is impaired. Within the pre-holo-CP, the β-propeptides are removed by autocatalytic cleavage and Ump is degraded by the nascent CP. PAC1–PAC2, Blm10 and the RP are bound to the nascent and mature CP, if the α ring gates are disordered or opened. Usually, the α ring gates are closed within free mature CP. See main text for details
However, unlike the self-assembly of archaeal or bacterial CP which can be reconstituted from purified α and β subunits, eukaryotic CP subunits fail to do so, already at the stage of α ring formation. While human α7 subunits expressed alone in E. coli formed dimerized homoheptameric rings, the physiologically adjacent subunits α1 and α6 do not incorporate unless they were co-expressed with α7. However, these reconstituted α rings were heterogeneous in subunit stoichiometry and did not allow the incorporation of β subunits [32, 33].
In yeast, the deletion of α3, the only non-essential CP subunit, can be compensated by the incorporation of the α4 subunit. However, CP assembly is delayed in yeast mutants lacking α3, highlighting the importance of correct α-interface contacts for efficient assembly [30].
In contrast to archaeal and bacterial CP assembly, correct and efficient eukaryotic CP assembly depends on five dedicated assembly chaperones. Chaperones are defined as proteins that facilitate assembly or folding, but are absent from the final matured protease. These chaperones have been identified bona fide by high throughput genetic arrays in yeast in search for null mutants with phenotypes generally displayed by proteasomal mutants such as cell cycle arrest and hypersensitivity against DNA damage and proteotoxic stress [7, 34]. None of the deletions of a single chaperone is lethal, which may explain why these chaperones remained unidentified for a long time. Thus, CP assembly can occur in vivo by mass interactions of proteasomal subunits without the aid of chaperones [34]. Under stress conditions, however, the combination of deletions in CP-dedicated chaperones exacerbates the phenotype of proteasomal mutants and results in the accumulation of assembly intermediates and dead end products of CP assembly [30, 34].
In 1998, Ump1 was discovered as the first CP-dedicated chaperone in yeast [7] and later identified in human as hUmp1/POMP/protoassemblin [35–37].
Ten years later, four additional chaperones emerged named PAC1, PAC2, PAC3 and PAC4 (also known as Poc1–Poc4 of Pba1–Pba4 in yeast, the yeast orthologs of human PAC1–PAC4; for completeness yeast Dmp1 and Dmp2 are alternative names for Pba3 and Pba4, respectively) [30, 34, 38].
Altogether these CP-dedicated chaperones regulate the timing of subunit incorporation, prevent faulty arrangements of subunits and control the processing of β subunit propeptides.
Meanwhile, the process of CP assembly is well understood. Independently from each other, several laboratories analyzed the composition of CP precursor complexes in eukaryotic cells. The heptameric α ring always serves as template for the incorporation of the seven β subunits. In both yeast and mammals, the so-called 13S CP precursor complex seems to be the most stable intermediate composed of an α ring with pro-β2, β3, and β4 subunits and Ump1 to be associated [39–43]. In yeast, the 15S CP precursor complex adds up pro-β1, pro-β5 and β6 subunits but still lacks pro-β7 [41, 44]. The human 16S CP precursor complex may correspond to the yeast 15S CP precursor complex, but includes pro-β7 [35, 45]. Although CP precursor complexes are often symbolized by half-assembled CP with α1–7β1–7 configuration, half-CP precursor complexes may be rather unstable in vivo, as they tend to dimerize into the pre-holo-CP, alternatively named the nascent CP. In principle, the pre-holo-CP is hardly detected after cell disintegration, unless β-propeptide processing is delayed due to genetic manipulations using tagged versions of proteasomal subunits and Ump1. Thus, it is possible that half-CP deprived of late incorporated β subunits as detected in cell lysates represent decay products of pre-holo-CP [41, 46].
Although the amino acid sequences of PAC1 and PAC2 are not related, their tertiary structures show similarities. PAC1 and PAC2 act as heterodimer and each subunit is only stable in the presence of the other [47]. Specifically, PAC1–PAC2 dimers bind α5 and α7 subunits in vitro [38]. Intriguingly, PAC1–PAC2 is associated stoichiometrically with the earliest CP precursor complexes, the α ring, the 15S CP precursor complex and also found in association with the fully assembled CP, both in yeast and mammalian cells indicating that quality control on α ring functionality extends from CP precursor complexes to the mature CP.
Whereas yeast pac1Δ pac2Δ null mutants display mild phenotypic defects [30, 34, 41], siRNA-mediated knockdown of PAC1 and PAC2 in mammals resulted in the formation of aberrant α rings as well as the faulty dimerization of two α rings suggesting that PAC1–PAC2 prevent α–α ring dimerization [43]. Consistent with this finding, the PAC1–PAC2 dimer binds to the outer surface of the α rings of the CP as resolved by crystal structure analysis [47].
The binding of PAC1 and PAC2 to the α subunit ring involves the C-terminal HbYX/HbYX-like motif, a sequence of a hydrophobic amino acid (Hb) and a penultimate tyrosine (Y), which is a common CP-binding motifs in the C-termini of CP activators. The HbYX motifs interact with a conserved lysine residue located in side pockets between certain α subunits, and induce conformational changes and regulates CP gating (for review see [48, 49]).
Neither PAC1 nor PAC2 activate the CP by binding through the HbYX motif. Instead of activation, their apical conjugation hinders the α rings from inappropriate dimerization, prevents off-pathways of premature interactions with the nascent CP, thus gate opening and the assembly of activators to incompletely matured CP.
Initially, studies in cultured human cells suggested that PAC1–PAC2 are degraded with the nascent CP [38]. However, recombinant PAC1–PAC2 are not degraded by yeast CP raising the possibility that PAC1–PAC2 is released with CP maturation [50]. PAC1–PAC2 is only tightly bound to the CP, if the active sites are inhibited by MG132 [47]. Either conformational changes occur with active site formation that results in the release of PAC1–PAC2 or PAC1–PAC2 remains bound to a fraction of abnormal CP which cannot be fully matured. Possibly, an allosteric mechanism in which the maturation of the active sites determines the binding of PAC1-PAC2 shields assembly intermediates or misassembled complexes from non-productive downstream associations [50].
Just like PAC1 and PAC2, PAC3 and PAC4 form heterodimers and are stable in the presence of its counterpart; PAC3 and PAC4 share negligible sequence homologies, but their tertiary structures with an α-helices β-sheet β-sheet α-helices sandwich are related and remind the structures of α and β subunits [51]. Thus, the PAC3–PAC4 heterodimer occupying the region around the α5 subunit could be place holders for specific α and β subunits [30]. The lack of PAC3–PAC4 resulted in a significant fraction of α rings with two copies of α4 instead of an α3–α4 pair [30], while others found that the lack of PAC3–PAC4 results in α rings without the incorporation of α4 [51].
Cells absent of PAC3 or PAC4, as yielded by siRNA knockdown in mammalian cells and deletion mutants in yeast, exhibited growth defect due to the decrease of properly matured CP, and the decline of proteasomal activity with the result of an accumulation of poly-ubiquitylated protein substrates [30, 34, 51, 52]; the phenotypic defects were stronger than in the absence of PAC1-PAC2, especially in yeast.
Interestingly, unlike PAC1-PAC2 knockdowns or deletions, cells without PAC3-PAC4 do not exhibit inappropriate α ring dimerizations of two outer α ring surfaces. According to the model of the α5-PAC3-PAC4 structure, these chaperones must be located at the inner α ring surface [51].
Following α ring assembly, β subunits and their precursors in the case of β1, β2, β5, β6 and β7 are incorporated by the assistance of N-terminal propeptides, in the case of β2 and β7 by the aid of C-terminal extensions. Ump1, the CP maturation factor, a protein of 17 kDa, guides the incorporation of the β subunits as addressed in detail below. The order of β subunit incorporation as pro-β2, β3, β4, pro-β5, pro-β6, pro-β1 and pro-β7, was initially deduced from the series of identified CP precursor complexes and later confirmed by elegant knockdown experiments of each β subunit in mammalian cells [52]. Pro-β2 is the first β subunit to be recruited to the α ring, which is facilitated by the CP-dedicated maturation factor Ump1. The PAC3–PAC4 heterodimer is associated with the α ring hosting the first β subunit, pro-β2, but is displaced with the incorporation of the subunit β4, as structural studies revealed steric hindrance between the α ring-bound PAC3–PAC4 heterodimer and β4 [51]. With the incorporation of the last β subunit, pro-β7, the CP precursor complexes dimerize into the pre-holo-CP which triggers CP maturation [41, 53]. The order of β-propeptide processing is studied in detail in yeast and mammals with autocatalytic cleavage steps between β subunits in cis- and trans-interactions [42, 54–59].
Intramolecular chaperones of β-subunits guide CP maturation
As mentioned above, the β subunits are incorporated in a specific order, after the α ring is assembled. Five of the seven β subunits are synthesized with propeptides, which are cleaved off during CP maturation. In principle, the propeptides participate in protein folding and assist in β subunit incorporation [60].
In addition to facilitated protein folding and maturation, the propeptides on the catalytically active β subunits protect their N-terminal catalytic threonine from being inhibited by N-acetylation [61].
The β-propeptides from yeast and human vary in length and do not show striking sequence homologies. The deletion of the β2-propeptide deletion is lethal in human cells, because β3 cannot be recruited without pro-β2 [43]. In yeast, the deletion of the β5- propeptide is lethal as it facilitates the β5 incorporation [62], while in human cells the β5-propeptide facilitates the incorporation of pro-β6 into the nascent CP [43]. Intriguingly, in yeast the chromosomal deletion of the β5-propeptide is rescued by plasmid-born overexpression of the β5-propeptide, indicating that the β5-propeptide acts “in trans” as an intramolecular chaperone in CP assembly. The lethality of the β5-propeptide deletion is also suppressed by the deletion of UMP1. The deletion of the β1 and β7 propeptides do not affect the cell growth in yeast suggesting unequal roles of propeptides in CP maturation and a hierarchy in active site formation as β5 >> β2 ≥ β1 [54]. Interestingly, β6 also contains an essential propeptide of which the lethal deletion is suppressed by UMP1 deletion. During CP maturation, the β6-propeptide is partly cleaved off leaving an N-terminal extension. While the β6-propeptide prevents premature dimerization of CP half-mers, the remaining N-terminal extension on β6 promotes pro-β7 incorporation and dimerization of the CP half-mers by overcoming the Ump1-dependent checkpoint in CP maturation [41]. This finding illustrates the sophisticated interplay between intrinsic elements of proteasomal β subunits to guarantee correct and efficient CP maturation.
Not only N-terminal propeptides, but also C-terminal extensions of β subunits play an important role in CP maturation. In yeast and mammals, the β2 and β7 subunits stabilize with their C-terminal extensions the pre-holo-CP. The β7 C-terminal extension inserts between the β1 and β2 subunits on the trans-β subunit ring. As β7 is the last β subunit to be incorporated into the CP half-mers simultaneously with their dimerization, its C-terminal extension stabilizes the nascent CP through trans-interactions between the annealing β rings. This extension also enhances the caspase-like activity, which is conferred by the β2 subunit [41, 43, 53].
The β2 C-terminal extension also serves as an essential intrinsic assembly factor in yeast and human cells, as it controls the successful incorporation of the neighboring β3 subunit [43, 52, 53].
Ump1 has a pivotal function in the hierarchy of CP-dedicated chaperones
The first CP-dedicated chaperone to be discovered was Ump1 by a search for null mutants defective in ubiquitin-mediated proteolysis [7]. In yeast, the ump1Δ null mutant displays the phenotype of proteasomal mutants, the accumulations of poly-ubiquitylated proteins, hypersensitivity to proteolytic stress and cell cycle arrest at elevated temperatures. The siRNA [7] knockdown of Ump1/Pomp in human cells also leads to severe growth defects and decreases cell viability [7, 34, 43, 63]. Human Ump1 is already required for the incorporation of the first β subunit, pro-β2, and Ump1 interacts with certain α subunits and all β subunits at least by yeast-two-hybrid assays [64].
In yeast cells lacking Ump1, the accumulation of CP precursor complexes composed of the α rings with pro-β2, β3 and β4 suggests that Ump1 joins the CP assembly line following the incorporation of β4 [41]. Since the presence of Ump1 facilitates the incorporation of pro-β5 but delays the incorporation of pro-β7, Ump1 was proposed to serve as a checkpoint until all seven β subunits are correctly positioned on the α ring before the dimerization of the two CP half-mers is allowed to take place [43]. Thus, Ump1 guides the dimerization of two CP half-mers into the pre-holo-CP. With the β-propeptide processing, the active site threonines are exposed and Ump1 is degraded by the nascent CP.
Intriguingly, Ump1 has recently shown to be an intrinsically disordered protein [65]. Driven by a newly defined mechanism based on disordered protein domains, Ump1 may benefit from being an intrinsically disordered protein during degradation as the first substrate of the matured CP [66].
The role of Blm10 in CP maturation and CP activation
Blm10, the yeast ortholog of mammalian PA200, is also found in association with CP precursor complexes and mature CP (Fig. 2, Blm10 is colored in green). Blm10/PA200 was initially identified as CP activator, as Blm10/PA200 stimulates the cleavage of chromogenic peptides within single capped Blm10-CP and contains, like three of the Rpt ATPases, the C-terminal HbYX motif for CP binding [67–70]. The closed dome-like structure of the double-capped Blm10-CP-Blm10 revealed that the insertion of the Blm10 C-terminus into the binding pocket between the α5–α6 subunits of the CP α ring induces disordered gates, but the observed sidewise opening of 7 Å does not allow access of unfolded polypeptides. Nevertheless, Blm10 might bind specific substrates before being docked to the CP. N-acetylated histones were proposed to be recognized by Blm10 and degraded by Blm10-bound CP in an ubiquitin-independent manner [71].
Besides the association of Blm10 to the CP, Blm10 is found in nascent pre-holo-CP and CP precursors consisting of the α ring, early incorporated β subunits β2, β3 and β4, PAC1-PAC2 and Ump1 [41, 44, 46]. As Blm10 preferentially binds to CP species with disordered α rings, Blm10 was proposed to function as checkpoint protein on CP assembly and distinguishes between α ring gate conformations. CP precursor complexes and pre-holo-CP exhibit disordered α rings. Once the α ring cannot be properly closed upon CP maturation, Blm10 remains bound to pre-activated and constitutively active CP [72]. Thus, Blm10-bound CP configurations predominate in open gate mutants and CP assembly mutants such as pac3Δpac4Δ, ump1Δ and with C-terminal β7 truncation. Combinations of these mutations with BLM10 deletion result in severe growth defects, while blm10Δ mutants only show mild growth defects [44, 72].
In the case, the BLM10 gene is deleted in yeast mutants with defective CP maturation, free CP with constitutively open gate is capped by the RP suggesting that the RP substitutes the control function of Blm10 in CP maturation [72]. If the RP is defective in rpn2blm10Δ mutants, CP precursor accumulates, because neither a functional RP nor Blm10 monitors the final steps of CP maturation [44]. The protein content of Blm10 increases with proteotoxic stress supporting the conclusion that Blm10 has a checkpoint function in CP α ring gating and correspondingly in CP activation [72].
Blm10 has an additional function in nuclear transport of assembled CP upon exit of quiescence, when CP precursor complexes are not available as import cargoes due to stalled protein synthesis [73].
Assembly of CP isoforms
In vertebrate cells three CP isoforms, the standard CP (CP), the immuno-CP (i-CP) and the thymo-CP (t-CP) exist. So far, we depicted the assembly of the standard CP. The i-CP and t-CP differ from the CP by distinct β subunits. In the i-CP, three β subunits, β1, β2 and β5 can be replaced by interferon-γ inducible isoforms, namely, the β1i, β2i and β5i subunits, respectively. Mixed assortments of standard and immuno-subunits yield CP subtypes with either β1–β2–β5i or β1i–β2–β5i configurations [74]. In the t-CP, the β5i subunit is replaced by a thymus-specific β5t isoform. The i-CP promotes the production of antigenic peptides which are presented on MHC class-I molecules [75, 76].
The assembly of the i-CP differs from the assembly of the standard CP that β1i is incorporated earlier than standard β1 and is able to consequently facilitate the incorporation of β2i and β5i [77, 78]. The i-CP assembles four times faster than the standard CP possibly due to interferon-γ inducible Pomp, the mammalian ortholog of Ump1. Pomp also has a higher affinity toward the β5i-propeptide than the β5-propeptide [79]. The β5t subunit is cooperatively incorporated with β1i and β2i into the t-CP [80]. Isoform-specific chaperones were not identified supporting the assumption that Pomp has a pivotal function in the assembly of CP isoforms. PI31, a 31-kDa protein-inhibiting CP activity, attenuates i-CP assembly, as PI31 overexpression interferes with the maturation of CP precursors containing βi subunits [81, 82]. The lack of PA28αβ, an interferon-γ inducible and ring-like shaped 11S proteasome activator complex, results in impaired i-CP assembly and immune response suggesting that i-CP assembly is influenced by several proteins in vertebrates [83].
Alternative CP also exists in Drosophila melanogaster in which the presence of testis-specific α and β subunits is essential for spermatogenesis [84]. The genome of Arabidopsis thaliana encodes isoforms of α and β subunits and shows a gene duplication of Ump1 [85]. According to the HUGO nomenclature committee, several proteasomal pseudogenes are scattered through the genome of mammalian cells (http://www.genenames.org/genefamilies/PSM). Nothing is known about their expression and incorporation into the CP. An interferon-γ inducible variant of β5i (LMP7E1 instead of LMP7E2) is expressed in human cancer cells, which is unable to interact with Pomp and inefficiently incorporated into the i-CP [63]. The downregulation of i-CP seems to be a strategy of tumor cells to escape immune surveillance.
In response to prolonged exposure to proteasome inhibitors such as bortezomib, the expression of β5i (LMP7E2) but not other immuno-subunits is upregulated and facilitates the assembly of CP subtypes. Downregulation of β5i expression and impaired assembly of i-CP were observed in bortezomib-resistant cell lines, highlighting a unique role for β5i in controlling the assembly of i-CP and providing a therapeutic rationale for targeting β5i in cancer treatment [86, 87]. Mutations in β5i are a worldwide phenomenon associated with autoimmune diseases and disorders in cell differentiation and the maintenance of protein homeostasis (for review see [88]).
Molecular composition of the RP base
The RP is composed of two subcomplexes, the RP base and lid. The RP base contains six different ATPase subunits of the AAA family. These ATPase subunits are numbered as Rpt1 to 6. The base also contains four non-ATPases Rpn1, Rpn2, Rpn10 and Rpn13. Rpn1 and Rpn2 belong to the family of HEAT repeat-like proteins and have eleven proteasome/cyclosome (PC) repeats adopting a toroid structure [89, 90]. Rpn10 and Rpn13 serve as receptors for poly-ubiquitylated protein substrates [18, 91].
The Rpt ATPases assemble in a six-membered ring with Rpt1–Rpt5–Rpt4–Rpt3–Rpt6–Rpt2 configuration, which builds an asymmetric interface between the RP and the seven-membered α ring of the CP [92–95]. Three C-terminal tails with HbYX motifs of distinct Rpt ATPases dock into pockets between two defined α subunits of the CP and regulate the gate opening of the central channels in the CP α ring [96, 97]. The subunits of the RP base can be expressed in E. coli as soluble proteins. The reconstituted RP base confers an equivalent activity to the 26S proteasomes than the endogenous complex [98, 99].
RP base-dedicated chaperones
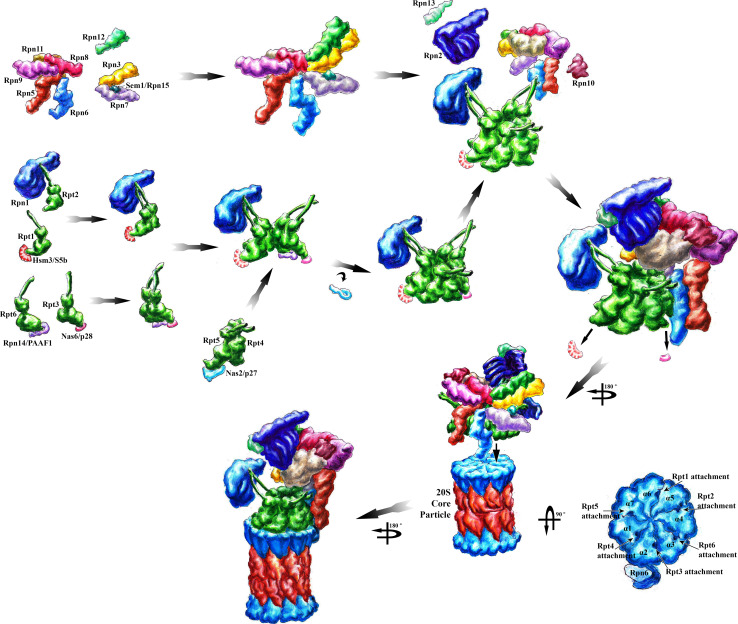
Like the assembly of the CP, the assembly of about 18 different RP subunits into the RP is orchestrated by several chaperones (Fig. 3). In the case of the RP base, the chaperones assist in the incorporation of the six different Rpt ATPase subunits and two non-ATPase subunits, such that each subunit finds its position in the complex with high fidelity and efficiency [100]. No chaperones are known to be required for the assembly of eukaryotic and prokaryotic ATPase ring complexes consisting of a single ATPase subunit, like archaeal PAN and VAT, eubacterial ARC and Mpa, yeast Cdc48 and mammalian p97, suggesting that the hexagonal symmetry of the complex and the demand of space for each subunit does not allow alternative configurations [22, 101].
Fig. 3.
The model of CP-independent RP assembly is depicted. In the CP-independent assembly pathway, four RP-dedicated chaperones join RP base modules: Hsm3/S5b binds to the ATPase subunits Rpt1 and Rpt2, Nas2/p27 to Rpt5, Nas6/p28 to Rpt3 and Rpn14/PAAF1 to Rpt6, respectively, by forming three modules or precursors, namely, the Nas6/p28-Rpt3–Rpt6-Rpn14/PPAF1 module, the Nas2/p27-Rpt5–Rpt4 module and the Hsm3/S5b-Rpt1–Rpt2-Rpn2 module. The chaperones are dislocated with the attachment to the CP. The RP lid is assembled from two modules, the Rpn5, 6, 8, 9 and 11 module and the Rpn3, 7 and Sem1 module. The incorporation of Rpn12 completes RP lid assembly. RP base and lid associate with Rpn10 and bind to the CP. Rpn6 also stabilizes the RP–CP interaction. In the CP-dependent assembly pathway, the RP base modules successively dock onto the CP template. The lid modules are assembled on top of the CP–RP base. In the CP-dependent model, the chaperones are most likely released, when the RP base is docking on the CP template to avoid chaperone collisions with the CP–RP base interface. Aberrant RP–CP assemblies are preferentially recognized by Ecm29, which may contact the RP and CP. The structure of Ecm29-bound RP–CP assemblies is unknown. Structures of the proteasomal subunits and proteasomal chaperones were deduced from single particle electron cryo-microscopy and crystal structures deposited in the PDB database. See main text for details
In 2009, four groups independently identified four RP base-dedicated chaperones, Hsm3/S5b (nomenclature in yeast/mammals), Nas2/p27, Nas6/p28 gankyrin and Rpn14/PAAF1 in yeast [102–106] and mammalian cells [107]. None of the identified RP-dedicated chaperones is essential in yeast. The absence of Hsm3 led to the most severe growth phenotype in yeast and to hypersensitivity toward DNA damage [102, 106]. Defective RP base, but not RP lid assembly was observed for hsm3Δ cells indicating that Hsm3 is an assembly chaperone for the RP base and not the RP lid complex [102].
The combination of deletions of RP-dedicated chaperones in yeast or the knockdown of these chaperones by siRNA in mammalian cells resulted in compromised RP base assembly [102, 103, 105, 106].
The RP-dedicated chaperones are structurally not related to each other, but all bind to the C-terminal region of individual RP ATPases. Hsm3/S5b binds to the ATPase subunits Rpt1 and Rpt2, Nas2/p27 to Rpt5, Nas6/p28 to Rpt3 and Rpn14/PAAF1 to Rpt6, respectively, by forming three modules or precursors, namely, the Nas6/p28-Rpt3–Rpt6-Rpn14/PPAF1 module, the Nas2/p27-Rpt4–Rpt5 module and the Hsm3/S5b-Rpt1-Rpt2-Rpn2 module [104, 105, 108].
These three modules come together to form the heterohexameric ATPase ring with Rpn1, which then assembles with the Rpn2–Rpn13 dimer into the full RP base complex. Based on studies in mammalian cells [107, 109], in which proteasome assembly appears to be slower compared with yeast cells presumably reflecting the longer generation time in mammals compared to yeast, the chaperones seem to define the order of the assembly of the three modules. In general, the chaperones have been suggested to solubilize the ATPase subunits, provide stability, serve as scaffold to bring submodules together and temporally sequence the assembly processes by restricting the access of the Rpt ATP ases’ C-termini to the CP-binding pockets within the α rings. Consistent with this assumption, the major binding site of these chaperones is directed towards the C-termini of the Rpt ATPase subunits. The release of RP-dedicated chaperones was inhibited by mutations in the C-terminal region of the Rpt ATPases that interfere with the binding of the RP ATPase ring to the CP. Thus, docking of the Rpt ATPases into the pocket sites of the CP is required to expel the chaperone [50, 104, 105, 108].
Since biochemical and structural data suggest that RP-dedicated chaperones interfere with the binding of the RP to the CP, specific deletion combinations affecting these four chaperones in yeast cause severe perturbations in RP–CP assembly. The presence of Nas2 and Hsm3 prevents close RP–CP apposition by physically occluding premature RP–CP activation [110].
The inhibition of premature interactions of RP and CP seems to be the general function of these chaperones, certainly for Hsm3 which ranks on top of the hierarchical order of RP-dedicated chaperones [110]. Hsm3 fulfills a key function in RP–CP assembly as it binds to the nascent RP at the interface with the CP. Accordingly, the overexpression of Hsm3 interferes with RP–CP assembly [99, 110].
Two models exist about the RP assembly. One pathway of RP base assembly is independent of the CP. In the other pathway, the CP is used as scaffold for RP base assembly [111].
Mark Hochstrasser’s [103] and Keiji Tanaka’s’s laboratories [107, 112] favored a pathway in which the three above-mentioned chaperone-bound modules coalesce to form the hexameric Rpt ATPase ring including Rpn1. The chaperones Rpn14/PAAF1, Hsm3/S5b, Nas2/p27 and Nas6/p28 sterically protect the RP base modules from premature association with the CP. The chaperones are step-wise replaced by incoming subunit Rpn2, Rpn10/S5a and the independently assembled RP lid. The association of the RP precursor complex with the CP subsequently displaces the remaining chaperones from the complementary surfaces and finally yields the RP–CP holoenzyme. Hsm3 and Nas6 are later displaced than Nas2 [95], consistent with recent structural data showing that Nas2 operates as a proteasome activator blocker during RP assembly prior to its docking onto the CP [113].
Dan Finley’s laboratory [104, 105, 108] favored an alternative pathway in which the CP serves as a template for the assembly of the Rpn2-Rpt6–Rpt3–Rpt4 module. This subcomplex assembles with the Rpt1–Rpt2–Rpt5-Rpn1 module by displacing the S5b chaperone. Then the RP lid and S5a follow to finally form the RP–CP holoenzyme. The absence of Hsm3 in RP–CP assemblies argues in favor for RP formation prior to CP docking. Hsm3 binds directly to the C-terminal part of Rpt1, and the C-terminal HbYX motif of this ATPase subunit is critical for the stabilization of RP–CP assemblies [102, 114]. The Hsm3 release from the Rpt1 subunit coincides with the association of the base and lid. Thus, Hsm3 serves as a checkpoint by preventing premature docking of the Rpt1 HbYX motif into the binding pocket located in the CP α ring. Also, a RP precursor consisting of the CP with Rpn2, Rpn10 and Rpn11 has been proposed to serve as a platform for subsequent association of the ATPase subunits and Rpn1 [115, 116].
Possibly both models of RP assembly describe confluent pathways. According to the first model, independently formed modules are coming together to build up the RP. The finding that the chaperones are protecting the nascent RP from premature assembly with the CP argues for a consecutive incorporation of RP modules/precursors. RP base and lid assembly into the RP would occur prior to its association with the CP.
In the second pathway, the CP serves as a scaffold for the assembly of RP modules which allows the transition of chaperone displacements from the RP modules/precursors. This complex most likely represents the nascent RP–CP assembly intermediate, which might have been stabilized by genetic manipulations required to tag and trap otherwise unstable RP modules and precursors. From this point of view, free existing RP modules/precursors could be considered as decay products of RP–CP assembly intermediates. The observation of mutants that impaired in CP assembly accumulate abnormal RP subcomplexes argues in favor of this interpretation. Also the Rpt ATPase-CP interface is reconfigured, when the RP lid complex joins the nascent proteasome suggesting a communication between the RP and CP during assembly of the holoenzyme complex [108].
Molecular composition of the RP lid and RP lid-specific chaperones
The lid is asymmetrically positioned on the RP base and contacts the CP by Rpn6 providing a molecular clamp between RP base ATPase Rpt6 and CP α subunit Pre8 (α2) [117]. The most-investigated RP lid subunit is Rpn11, which embedded in the RP lid confers the metalloprotease activity to hydrolyze the ubiquitin molecules from the poly-ubiquitin chain prior degradation of the protein substrate [23, 118]. The eight different RP lid subunits can be expressed as soluble recombinant proteins in E. coli and allow the reconstitution of RP lid complexes [93, 119]. There is compelling evidence from the literature that the assembly of the RP lid occurs independently of the RP base mainly from two modules, the Rpn5, 6, 8, 9 and 11 module and the Rpn3, 7 and Sem1 module [102, 114, 120, 121]. The incorporation of Rpn12 couples the completion of RP lid assembly with the binding of the RP lid to the RP base [122]. C-terminal helices of the lid subunits with PCI (Proteasome COP9 Initiation factor 3) domains form a helical bundle that directs the ordered self-assembly of the RP lid [123]. Sem1, an intrinsically disordered protein, further tethers lid subunits suggesting that RP lid assembly is facilitated by a newly defined mechanism involving intrinsically disordered protein domains [124]. Intriguingly, Ump1, the CP-dedicated chaperone, is an intrinsically disordered protein as well suggesting that a disorder-driven mechanism also applies for CP assembly [65, 66].
Regulation of RP–CP assembly
Upon CP maturation, the gates of the α rings are closed yielding a proteolytic chamber with latent enzyme activity [125, 126]. Access to the proteolytic chamber is provided by the association with RP. Thus, the regulation of RP–CP assembly is crucial for the proteasome function in the degradation of poly-ubiquitylated protein substrates.
In the absence of sufficient amounts of CP, fully assembled RP appeared to be unstable in vivo suggesting that stabilizing affects occur during docking of the RP to the CP. The subunit Rpn10, which serves as a receptor for poly-ubiquitylated protein substrates, was assigned with stabilizing functions [127, 128]. Consistent with this finding, RP–CP assemblies are stable entities during the degradation of protein substrates, because premature RP–CP dissociation might result in the release of toxic peptide intermediates produced by interrupted protein degradation. Thus, ongoing protein degradation might suppress RP–CP disassembly. However, the RP could not always be detected as a whole complex upon RP–CP dissociation leading to the so-called “chew and spew” model, by which the RP dissociates into modules, when the ATP-dependent hydrolysis of peptides within the proteolytic chamber is completed. Thus, RP–CP dissociation/reassembly cycles might be inherent to proteasome activation/deactivation [109]. Whether the RP dissociation products reassociate with RP-dedicated chaperones, thus represent RP precursors or modules, is unknown. At least their fate of not being degraded by the nascent RP–CP would allow for a quick reassembly on demand for the degradation of poly-ubiquitylated proteins.
For more than two decades, it is known that RP–CP assembly requires ATP [129, 130]. The decline of ATP upon the transition from cell proliferation to quiescence comes along with the dissociation of RP–CP assemblies into RP and CP [73, 131], which reflects the requirement of ATP for RP–CP stabilization. Possibly, the energy expenditure of quiescent cells dictates that the ATP consumption to generate more poly-ubiquitin chains on protein substrates cannot be afforded or may not be needed, since short-lived proteins regulating cell cycle progression and aberrant newly synthesized proteins will not accumulate in quiescence. This might explain why significantly less RP–CP assemblies exist in quiescence compared with ongoing cell proliferation [73]. RP–CP assemblies are also dissociated in neuronal cells with quiescent synapses [132]. However in yeast, conflicting data exist suggesting that RP–CP assemblies still exist in quiescence [133–135].
Heat-shock protein Hsp90 is also required for the maintenance of stable RP–CP assemblies, as expected for a multisubunit complex exposed to increasing temperatures [136]. It remains to be shown whether heat-shock proteins play a general role in stabilizing RP and CP precursor complexes or have additional functions in delivering misfolded proteins for proteasomal degradation.
Post-translational modifications, such as phosphorylation, influence the assembly/disassembly of RP–CP [137–139]. Interconversion between different phosphorylation states would allow for rapid adjustments to metabolic changes as required upon the transition from proliferation to quiescence and vice versa.
Quality control on RP–CP assemblies by ECM29
Ecm29, a conserved 200-kDa protein belonging to the HEAT repeat-like family [140], is associated with a fraction of RP–CP assemblies and stabilizes their interaction [141–144]. The fraction of Ecm29-bound RP–CP assemblies is enhanced, when proteasome assembly is defective. This is the case, when the CP-dedicated chaperone Ump1 is deleted. Distinct β subunits such as β3 are not fully incorporated leading to pre-holo CP with incomplete β propeptide processing. Remnants of β propeptides might be comparable with active sites that are either inhibited by synthetic compounds or that are engaged in protein degradation. The gates within the α ring of this kind of CP might be constitutively open and covered by the RP. These RP–CP assemblies with incompletely mature CP are sequestered by Ecm29. RP–CP assemblies with faulty RP are also primarily bound by Ecm29 suggesting a control function of Ecm29 in RP–CP assembly [145]. The E3 ligase Not4, which associates with RP–CP assemblies also contributes to the control of RP–CP assemblies stabilized by Ecm29 [146]. Ecm29 is degraded, if the defective RP–CP complex can be converted into the regular enzyme in vitro by replacing the defective or missing subunits [147]. The ECM29 gene expression is augmented by oxidative stress and chronic ethanol feeding, which gives rise to reactive oxygen species and generally affects protein folding and DNA repair. It is also known that oxidative stress affects the correct folding and incorporation of multiple proteasomal subunits into the proteasome holoenzyme [148, 149]. The protein content of Ecm29 increases when aberrant products of RP–CP assembly are recognized [147].
Localization of proteasome assembly
Proteasomes represent up to 5 % of the total protein content of eukaryotic cells [5]. Thus, new proteasome holoenzyme complexes need to be continuously assembled during cell proliferation. Proteasomal subunits are synthesized in the cytoplasm and incorporated into the precursor complexes of the RP and CP. In mammalian cells, CP maturation was proposed to exclusively take place at the ER, which may provide a scaffold for the incorporation of proteasomal subunits [64]. Genetic analysis in yeast identified components of the guided entry of tail-anchored proteins (GET) pathway, to be involved in CP assembly [150]. The GET pathway is responsible for the insertion of tail-anchored proteins into the ER. Its mammalian counterpart is called the transmembrane recognition complex (TRC) pathway (for review see [151]).
Deletion of either GET1, GET2 or GET3 in combination with deletions in UMP1, NAS6 and HSM3 results in synthetic growth defect in yeast. In mammalian cells, the knockdown of TRC4, the Get3 homolog, and BAG6, the most upstream component of the TRC pathway, caused the accumulation of CP precursors, in which β subunits are inefficiently assembled on the α ring template. The tail-anchored protein recruiting the CP α ring to the ER is not yet identified.
However, GFP-labeling techniques and live cell imaging do not reveal a major localization of RP and CP chaperones at the ER in yeast [152]. In contrast, Ump1 and Hsm3, chaperones with pivotal functions in proteasome assembly are localized in the cytoplasm and also in considerable amount in the nucleus suggesting that proteasome assembly can also occur in the nucleus from imported CP precursors and RP modules [106, 120, 153]. CP precursor complexes of i-CP are present in the nucleus of mouse cortical astrocytes, in which i-CP maturation is attenuated [154]. Pomp is also primarily nuclear as shown by indirect immunofluorescence microscopy of human embryonic kidney cells (HEK293) [155]. Thus, future studies are required to gain insight into the localization of proteasome assembly. Possibly, early assembly intermediates are stabilized by a yet unknown tail-anchored protein at the ER. Late assembly intermediates such as the pre-holo-CP are imported into the nucleus. The presence of nuclear Ump1/Pomp argues for nuclear CP maturation in yeast and mammalian cells [153, 154]. However, Blm10-associated CP and RP–CP assemblies are also transported as matured enzymes between the cyto- and nucleoplasm suggesting that multiple nuclear transport pathways exist for proteasomes (for review see [144]).
Acknowledgments
We thank Paula Ramos for critical reading of the manuscript. This work was supported by NSERC.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Hilt W, Wolf DH. The ubiquitin-proteasome system: past, present and future. Cell Mol Life Sci. 2004;61(13):1545. doi: 10.1007/s00018-004-4128-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka K. The proteasome: from basic mechanisms to emerging roles. Keio J Med. 2013;62(1):1–12. doi: 10.2302/kjm.2012-0006-re. [DOI] [PubMed] [Google Scholar]
- 4.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, Bahler J. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151(3):671–683. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enenkel C. Using native gel electrophoresis and phosphofluoroimaging to analyze GFP-tagged proteasomes. In: Dohmen RJ, Scheffner M, editors. Ubiquitin family modifiers and the proteasome: reviews and protocols. New York: Humana Press; 2011. [Google Scholar]
- 7.Ramos PC, Hockendorff J, Johnson ES, Varshavsky A, Dohmen RJ. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell. 1998;92(4):489–499. doi: 10.1016/s0092-8674(00)80942-3. [DOI] [PubMed] [Google Scholar]
- 8.Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92(3):367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 9.Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268(5210):533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 10.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386(6624):463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 11.Kwon YD, Nagy I, Adams PD, Baumeister W, Jap BK. Crystal structures of the Rhodococcus proteasome with and without its pro-peptides: implications for the role of the pro-peptide in proteasome assembly. J Mol Biol. 2004;335(1):233–245. doi: 10.1016/j.jmb.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 12.Hu G, Lin G, Wang M, Dick L, Xu RM, Nathan C, Li H. Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate. Mol Microbiol. 2006;59(5):1417–1428. doi: 10.1111/j.1365-2958.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- 13.Huber EM, Basler M, Schwab R, Heinemeyer W, Kirk CJ, Groettrup M, Groll M. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell. 2012;148(4):727–738. doi: 10.1016/j.cell.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 14.Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure. 2002;10(5):609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- 15.Seemuller E, Lupas A, Stock D, Lowe J, Huber R, Baumeister W. Proteasome from Thermoplasma acidophilum: a threonine protease. Science. 1995;268(5210):579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- 16.Seemuller E, Lupas A, Baumeister W. Autocatalytic processing of the 20S proteasome. Nature. 1996;382(6590):468–471. doi: 10.1038/382468a0. [DOI] [PubMed] [Google Scholar]
- 17.Heinemeyer W, Ramos PC, Dohmen RJ. The ultimate nanoscale mincer: assembly, structure and active sites of the 20S proteasome core. Cell Mol Life Sci. 2004;61(13):1562–1578. doi: 10.1007/s00018-004-4130-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenzweig R, Bronner V, Zhang D, Fushman D, Glickman MH. Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J Biol Chem. 2012;287(18):14659–14671. doi: 10.1074/jbc.M111.316323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bar-Nun S, Glickman MH. Proteasomal AAA-ATPases: structure and function. Biochim Biophys Acta. 2012;1823(1):67–82. doi: 10.1016/j.bbamcr.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 20.Snider J, Thibault G, Houry WA. The AAA + superfamily of functionally diverse proteins. Genome Biol. 2008;9(4):216. doi: 10.1186/gb-2008-9-4-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sauer RT, Baker TA. AAA + proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 22.Striebel F, Kress W, Weber-Ban E. Controlled destruction: AAA + ATPases in protein degradation from bacteria to eukaryotes. Curr Opin Struct Biol. 2009;19(2):209–217. doi: 10.1016/j.sbi.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298(5593):611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 24.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419(6905):403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 25.Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127(1):99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 26.Zwickl P, Grziwa A, Puhler G, Dahlmann B, Lottspeich F, Baumeister W. Primary structure of the Thermoplasma proteasome and its implications for the structure, function, and evolution of the multicatalytic proteinase. Biochemistry. 1992;31(4):964–972. doi: 10.1021/bi00119a004. [DOI] [PubMed] [Google Scholar]
- 27.Zwickl P, Kleinz J, Baumeister W. Critical elements in proteasome assembly. Nat Struct Mol Biol. 1994;1(11):765–770. doi: 10.1038/nsb1194-765. [DOI] [PubMed] [Google Scholar]
- 28.Zuhl F, Seemuller E, Golbik R, Baumeister W. Dissecting the assembly pathway of the 20S proteasome. FEBS Lett. 1997;418(1–2):189–194. doi: 10.1016/s0014-5793(97)01370-7. [DOI] [PubMed] [Google Scholar]
- 29.Sharon M, Witt S, Glasmacher E, Baumeister W, Robinson CV. Mass spectrometry reveals the missing links in the assembly pathway of the bacterial 20 S proteasome. J Biol Chem. 2007;282(25):18448–18457. doi: 10.1074/jbc.M701534200. [DOI] [PubMed] [Google Scholar]
- 30.Kusmierczyk AR, Kunjappu MJ, Funakoshi M, Hochstrasser M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat Struct Mol Biol. 2008;15(3):237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- 31.Kumoi K, Satoh T, Murata K, Hiromoto T, Mizushima T, Kamiya Y, Noda M, Uchiyama S, Yagi H, Kato K. An archaeal homolog of proteasome assembly factor functions as a proteasome activator. PLoS ONE. 2013;8(3):e60294. doi: 10.1371/journal.pone.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerards WL, Enzlin J, Haner M, Hendriks IL, Aebi U, Bloemendal H, Boelens W. The human alpha-type proteasomal subunit HsC8 forms a double ringlike structure, but does not assemble into proteasome-like particles with the beta-type subunits HsDelta or HsBPROS26. J Biol Chem. 1997;272(15):10080–10086. doi: 10.1074/jbc.272.15.10080. [DOI] [PubMed] [Google Scholar]
- 33.Sugiyama M, Kurimoto E, Yagi H, Mori K, Fukunaga T, Hirai M, Zaccai G, Kato K. Kinetic asymmetry of subunit exchange of homooligomeric protein as revealed by deuteration-assisted small-angle neutron scattering. Biophys J. 2011;101(8):2037–2042. doi: 10.1016/j.bpj.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Tallec B, Barrault MB, Courbeyrette R, Guerois R, Marsolier-Kergoat MC, Peyroche A. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol Cell. 2007;27(4):660–674. doi: 10.1016/j.molcel.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 35.Witt E, Zantopf D, Schmidt M, Kraft R, Kloetzel PM, Kruger E. Characterisation of the newly identified human Ump1 homologue POMP and analysis of LMP7(beta 5i) incorporation into 20 S proteasomes. J Mol Biol. 2000;301(1):1–9. doi: 10.1006/jmbi.2000.3959. [DOI] [PubMed] [Google Scholar]
- 36.Burri L, Hockendorff J, Boehm U, Klamp T, Dohmen RJ, Levy F. Identification and characterization of a mammalian protein interacting with 20S proteasome precursors. Proc Natl Acad Sci USA. 2000;97(19):10348–10353. doi: 10.1073/pnas.190268597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffin TA, Slack JP, McCluskey TS, Monaco JJ, Colbert RA. Identification of proteassemblin, a mammalian homologue of the yeast protein, Ump1p, that is required for normal proteasome assembly. Mol Cell Biol Res Commun. 2000;3(4):212–217. doi: 10.1006/mcbr.2000.0213. [DOI] [PubMed] [Google Scholar]
- 38.Hirano Y, Hendil KB, Yashiroda H, Iemura S, Nagane R, Hioki Y, Natsume T, Tanaka K, Murata S. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature. 2005;437(7063):1381–1385. doi: 10.1038/nature04106. [DOI] [PubMed] [Google Scholar]
- 39.Nandi D, Woodward E, Ginsburg DB, Monaco JJ. Intermediates in the formation of mouse 20S proteasomes: implications for the assembly of precursor beta subunits. EMBO J. 1997;16(17):5363–5375. doi: 10.1093/emboj/16.17.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frentzel S, Pesold-Hurt B, Seelig A, Kloetzel PM. 20 S proteasomes are assembled via distinct precursor complexes. Processing of LMP2 and LMP7 proproteins takes place in 13–16 S preproteasome complexes. J Mol Biol. 1994;236(4):975–981. doi: 10.1016/0022-2836(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 41.Li X, Kusmierczyk AR, Wong P, Emili A, Hochstrasser M. Beta-subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. EMBO J. 2007;26(9):2339–2349. doi: 10.1038/sj.emboj.7601681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidtke G, Kraft R, Kostka S, Henklein P, Frommel C, Lowe J, Huber R, Kloetzel PM, Schmidt M. Analysis of mammalian 20S proteasome biogenesis: the maturation of beta-subunits is an ordered two-step mechanism involving autocatalysis. EMBO J. 1996;15(24):6887–6898. [PMC free article] [PubMed] [Google Scholar]
- 43.Hirano Y, Kaneko T, Okamoto K, Bai M, Yashiroda H, Furuyama K, Kato K, Tanaka K, Murata S. Dissecting beta-ring assembly pathway of the mammalian 20S proteasome. EMBO J. 2008;27(16):2204–2213. doi: 10.1038/emboj.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marques AJ, Glanemann C, Ramos PC, Dohmen RJ. The C-terminal extension of the beta7 subunit and activator complexes stabilize nascent 20 S proteasomes and promote their maturation. J Biol Chem. 2007;282(48):34869–34876. doi: 10.1074/jbc.M705836200. [DOI] [PubMed] [Google Scholar]
- 45.Schmidtke G, Schmidt M, Kloetzel PM. Maturation of mammalian 20S proteasome: purification and characterization of 13 S and 16 S proteasome precursor complexes. J Mol Biol. 1997;268(1):95–106. doi: 10.1006/jmbi.1997.0947. [DOI] [PubMed] [Google Scholar]
- 46.Fehlker M, Wendler P, Lehmann A, Enenkel C. Blm3 is part of nascent proteasomes and is involved in a late stage of nuclear proteasome assembly. EMBO Rep. 2003;4(10):959–963. doi: 10.1038/sj.embor.embor938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stadtmueller BM, Kish-Trier E, Ferrell K, Petersen CN, Robinson H, Myszka DG, Eckert DM, Formosa T, Hill CP. Structure of a proteasome Pba1-Pba2 complex: implications for proteasome assembly, activation, and biological function. J Biol Chem. 2012;287(44):37371–37382. doi: 10.1074/jbc.M112.367003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stadtmueller BM, Hill CP. Proteasome activators. Mol Cell. 2011;41(1):8–19. doi: 10.1016/j.molcel.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saeki Y, Tanaka K. Unlocking the proteasome door. Mol Cell. 2007;27(6):865–867. doi: 10.1016/j.molcel.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 50.Kusmierczyk AR, Kunjappu MJ, Kim RY, Hochstrasser M. A conserved 20S proteasome assembly factor requires a C-terminal HbYX motif for proteasomal precursor binding. Nat Struct Mol Biol. 2011;18(5):622–629. doi: 10.1038/nsmb.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yashiroda H, Mizushima T, Okamoto K, Kameyama T, Hayashi H, Kishimoto T, Niwa S, Kasahara M, Kurimoto E, Sakata E, Takagi K, Suzuki A, Hirano Y, Murata S, Kato K, Yamane T, Tanaka K. Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes. Nat Struct Mol Biol. 2008;15(3):228–236. doi: 10.1038/nsmb.1386. [DOI] [PubMed] [Google Scholar]
- 52.Hirano Y, Hayashi H, Iemura S, Hendil KB, Niwa S, Kishimoto T, Kasahara M, Natsume T, Tanaka K, Murata S. Cooperation of multiple chaperones required for the assembly of mammalian 20S proteasomes. Mol Cell. 2006;24(6):977–984. doi: 10.1016/j.molcel.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 53.Ramos PC, Marques AJ, London MK, Dohmen RJ. Role of C-terminal extensions of subunits beta2 and beta7 in assembly and activity of eukaryotic proteasomes. J Biol Chem. 2004;279(14):14323–14330. doi: 10.1074/jbc.M308757200. [DOI] [PubMed] [Google Scholar]
- 54.Jager S, Groll M, Huber R, Wolf DH, Heinemeyer W. Proteasome beta-type subunits: unequal roles of propeptides in core particle maturation and a hierarchy of active site function. J Mol Biol. 1999;291(4):997–1013. doi: 10.1006/jmbi.1999.2995. [DOI] [PubMed] [Google Scholar]
- 55.Heinemeyer W, Fischer M, Krimmer T, Stachon U, Wolf DH. The active sites of the eukaryotic 20S proteasome and their involvement in subunit precursor processing. J Biol Chem. 1997;272(40):25200–25209. doi: 10.1074/jbc.272.40.25200. [DOI] [PubMed] [Google Scholar]
- 56.Groll M, Heinemeyer W, Jager S, Ullrich T, Bochtler M, Wolf DH, Huber R. The catalytic sites of 20S proteasomes and their role in subunit maturation: a mutational and crystallographic study. Proc Natl Acad Sci USA. 1999;96(20):10976–10983. doi: 10.1073/pnas.96.20.10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kingsbury DJ, Griffin TA, Colbert RA. Novel propeptide function in 20 S proteasome assembly influences beta subunit composition. J Biol Chem. 2000;275(31):24156–24162. doi: 10.1074/jbc.M001742200. [DOI] [PubMed] [Google Scholar]
- 58.De M, Jayarapu K, Elenich L, Monaco JJ, Colbert RA, Griffin TA. Beta 2 subunit propeptides influence cooperative proteasome assembly. J Biol Chem. 2003;278(8):6153–6159. doi: 10.1074/jbc.M209292200. [DOI] [PubMed] [Google Scholar]
- 59.Schmidt M, Zantopf D, Kraft R, Kostka S, Preissner R, Kloetzel PM. Sequence information within proteasomal prosequences mediates efficient integration of beta-subunits into the 20S proteasome complex. J Mol Biol. 1999;288(1):117–128. doi: 10.1006/jmbi.1999.2660. [DOI] [PubMed] [Google Scholar]
- 60.Shinde U, Inouye M. Intramolecular chaperones: polypeptide extensions that modulate protein folding. Semin Cell Develop Biol. 2000;11(1):35–44. doi: 10.1006/scdb.1999.0349. [DOI] [PubMed] [Google Scholar]
- 61.Arendt CS, Hochstrasser M. Eukaryotic 20S proteasome catalytic subunit propeptides prevent active site inactivation by N-terminal acetylation and promote particle assembly. EMBO J. 1999;18(13):3575–3585. doi: 10.1093/emboj/18.13.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen P, Hochstrasser M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell. 1996;86(6):961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- 63.Heink S, Fricke B, Ludwig D, Kloetzel PM, Kruger E. Tumor cell lines expressing the proteasome subunit isoform LMP7E1 exhibit immunoproteasome deficiency. Cancer Res. 2006;66(2):649–652. doi: 10.1158/0008-5472.CAN-05-2872. [DOI] [PubMed] [Google Scholar]
- 64.Fricke B, Heink S, Steffen J, Kloetzel PM, Kruger E. The proteasome maturation protein POMP facilitates major steps of 20S proteasome formation at the endoplasmic reticulum. EMBO Rep. 2007;8(12):1170–1175. doi: 10.1038/sj.embor.7401091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sa-Moura B, Simoes AM, Fraga J, Fernandes H, Abreu IA, Botelho HM, Gomes CM, Marques AJ, Dohmen RJ, Ramos PC, Macedo-Ribeiro S. Biochemical and biophysical characterization of recombinant yeast proteasome maturation factor ump1. Comput Struct Biotech J. 2013;7:e201304006. doi: 10.5936/csbj.201304006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uekusa Y, Okawa K, Yagi-Utsumi M, Serve O, Nakagawa Y, Mizushima T, Yagi H, Saeki Y, Tanaka K, Kato K. Backbone 1H, 13C, and 15N assignments of yeast Ump1, an intrinsically disordered protein that functions as a proteasome assembly chaperone. Biomol NMR Assign. 2013 doi: 10.1007/s12104-013-9523-1. [DOI] [PubMed] [Google Scholar]
- 67.Schmidt M, Haas W, Crosas B, Santamaria PG, Gygi SP, Walz T, Finley D. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat Struct Mol Biol. 2005;12(4):294–303. doi: 10.1038/nsmb914. [DOI] [PubMed] [Google Scholar]
- 68.Ustrell V, Hoffman L, Pratt G, Rechsteiner M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002;21(13):3516–3525. doi: 10.1093/emboj/cdf333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dange T, Smith D, Noy T, Rommel PC, Jurzitza L, Cordero RJ, Legendre A, Finley D, Goldberg AL, Schmidt M. Blm10 protein promotes proteasomal substrate turnover by an active gating mechanism. J Biol Chem. 2011;286(50):42830–42839. doi: 10.1074/jbc.M111.300178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lopez AD, Tar K, Krugel U, Dange T, Ros IG, Schmidt M. Proteasomal degradation of Sfp1 contributes to the repression of ribosome biogenesis during starvation and is mediated by the proteasome activator Blm10. Mol Biol Cell. 2011;22(5):528–540. doi: 10.1091/mbc.E10-04-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qian MX, Pang Y, Liu CH, Haratake K, Du BY, Ji DY, Wang GF, Zhu QQ, Song W, Yu Y, Zhang XX, Huang HT, Miao S, Chen LB, Zhang ZH, Liang YN, Liu S, Cha H, Yang D, Zhai Y, Komatsu T, Tsuruta F, Li H, Cao C, Li W, Li GH, Cheng Y, Chiba T, Wang L, Goldberg AL, Shen Y, Qiu XB. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell. 2013;153(5):1012–1024. doi: 10.1016/j.cell.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lehmann A, Jechow K, Enenkel C. Blm10 binds to pre-activated proteasome core particles with open gate conformation. EMBO Rep. 2008;9(12):1237–1243. doi: 10.1038/embor.2008.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weberruss MH, Savulescu AF, Jando J, Bissinger T, Harel A, Glickman MH, Enenkel C. Blm10 facilitates nuclear import of proteasome core particles. EMBO J. 2013 doi: 10.1038/emboj.2013.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guillaume B, Chapiro J, Stroobant V, Colau D, Van Holle B, Parvizi G, Bousquet-Dubouch MP, Theate I, Parmentier N, Van den Eynde BJ. Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc Natl Acad Sci USA. 2010;107(43):18599–18604. doi: 10.1073/pnas.1009778107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kloetzel PM, Ossendorp F. Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr Opin Immunol. 2004;16(1):76–81. doi: 10.1016/j.coi.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 76.Rock KL, York IA, Goldberg AL. Post-proteasomal antigen processing for major histocompatibility complex class I presentation. Nat Immunol. 2004;5(7):670–677. doi: 10.1038/ni1089. [DOI] [PubMed] [Google Scholar]
- 77.Griffin TA, Nandi D, Cruz M, Fehling HJ, Kaer LV, Monaco JJ, Colbert RA. Immunoproteasome assembly: cooperative incorporation of interferon gamma (IFN-gamma)-inducible subunits. J Exp Med. 1998;187(1):97–104. doi: 10.1084/jem.187.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Groettrup M, Standera S, Stohwasser R, Kloetzel PM. The subunits MECL-1 and LMP2 are mutually required for incorporation into the 20S proteasome. Proc Natl Acad Sci USA. 1997;94(17):8970–8975. doi: 10.1073/pnas.94.17.8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heink S, Ludwig D, Kloetzel PM, Kruger E. IFN-gamma-induced immune adaptation of the proteasome system is an accelerated and transient response. Proc Natl Acad Sci USA. 2005;102(26):9241–9246. doi: 10.1073/pnas.0501711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Murata S, Takahama Y, Tanaka K. Thymoproteasome: probable role in generating positively selecting peptides. Curr Opin Immunol. 2008;20(2):192–196. doi: 10.1016/j.coi.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 81.Zaiss DM, Standera S, Holzhutter H, Kloetzel P, Sijts AJ. The proteasome inhibitor PI31 competes with PA28 for binding to 20S proteasomes. FEBS Lett. 1999;457(3):333–338. doi: 10.1016/s0014-5793(99)01072-8. [DOI] [PubMed] [Google Scholar]
- 82.Zaiss DM, Standera S, Kloetzel PM, Sijts AJ. PI31 is a modulator of proteasome formation and antigen processing. Proc Natl Acad Sci USA. 2002;99(22):14344–14349. doi: 10.1073/pnas.212257299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Preckel T, Fung-Leung WP, Cai Z, Vitiello A, Salter-Cid L, Winqvist O, Wolfe TG, Von Herrath M, Angulo A, Ghazal P, Lee JD, Fourie AM, Wu Y, Pang J, Ngo K, Peterson PA, Fruh K, Yang Y. Impaired immunoproteasome assembly and immune responses in PA28−/− mice. Science. 1999;286(5447):2162–2165. doi: 10.1126/science.286.5447.2162. [DOI] [PubMed] [Google Scholar]
- 84.Belote JM, Zhong L. Duplicated proteasome subunit genes in Drosophila and their roles in spermatogenesis. Heredity. 2009;103(1):23–31. doi: 10.1038/hdy.2009.23. [DOI] [PubMed] [Google Scholar]
- 85.Fu H, Doelling JH, Arendt CS, Hochstrasser M, Vierstra RD. Molecular organization of the 20S proteasome gene family from Arabidopsis thaliana. Genetics. 1998;149(2):677–692. doi: 10.1093/genetics/149.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Niewerth D, Kaspers GJ, Assaraf YG, van Meerloo J, Kirk CJ, Anderl J, Blank JL, van de Ven PM, Zweegman S, Jansen G, Cloos J. Interferon-gamma-induced upregulation of immunoproteasome subunit assembly overcomes bortezomib resistance in human hematological cell lines. J Hematol Oncol. 2014;7(1):7. doi: 10.1186/1756-8722-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sijts EJ, Kloetzel PM. The role of the proteasome in the generation of MHC class I ligands and immune responses. Cell Mol Life Sci. 2011;68(9):1491–1502. doi: 10.1007/s00018-011-0657-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tanaka K, Mizushima T, Saeki Y. The proteasome: molecular machinery and pathophysiological roles. Biol Chem. 2012;393(4):217–234. doi: 10.1515/hsz-2011-0285. [DOI] [PubMed] [Google Scholar]
- 89.Effantin G, Rosenzweig R, Glickman MH, Steven AC. Electron microscopic evidence in support of alpha-solenoid models of proteasomal subunits Rpn1 and Rpn2. J Mol Biol. 2009;386(5):1204–1211. doi: 10.1016/j.jmb.2009.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.He J, Kulkarni K, da Fonseca PC, Krutauz D, Glickman MH, Barford D, Morris EP. The structure of the 26S proteasome subunit Rpn2 reveals its PC repeat domain as a closed toroid of two concentric alpha-helical rings. Structure. 2012;20(3):513–521. doi: 10.1016/j.str.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 91.Sakata E, Bohn S, Mihalache O, Kiss P, Beck F, Nagy I, Nickell S, Tanaka K, Saeki Y, Forster F, Baumeister W. Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc Natl Acad Sci USA. 2012;109(5):1479–1484. doi: 10.1073/pnas.1119394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tian G, Park S, Lee MJ, Huck B, McAllister F, Hill CP, Gygi SP, Finley D. An asymmetric interface between the regulatory and core particles of the proteasome. Nat Struct Mol Biol. 2011;18(11):1259–1267. doi: 10.1038/nsmb.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482(7384):186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.da Fonseca PC, He J, Morris EP. Molecular model of the human 26S proteasome. Mol Cell. 2012;46(1):54–66. doi: 10.1016/j.molcel.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 95.Tomko RJ, Jr, Funakoshi M, Schneider K, Wang J, Hochstrasser M. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: implications for proteasome structure and assembly. Mol Cell. 2010;38(3):393–403. doi: 10.1016/j.molcel.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN. Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J Biol Chem. 2008;283(46):31813–31822. doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell. 2007;27(5):731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Beckwith R, Estrin E, Worden EJ, Martin A. Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA + unfoldase. Nat Struct Mol Biol. 2013;20(10):1164–1172. doi: 10.1038/nsmb.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takagi K, Kim S, Yukii H, Ueno M, Morishita R, Endo Y, Kato K, Tanaka K, Saeki Y, Mizushima T. Structural basis for specific recognition of Rpt1p, an ATPase subunit of 26S proteasome, by proteasome-dedicated chaperone Hsm3p. J Biol Chem. 2012;287(15):12172–12182. doi: 10.1074/jbc.M112.345876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Peth A, Uchiki T, Goldberg AL. ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol Cell. 2010;40(4):671–681. doi: 10.1016/j.molcel.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barthelme D, Chen JZ, Grabenstatter J, Baker TA, Sauer RT. Architecture and assembly of the archaeal Cdc48*20S proteasome. Proc Natl Acad Sci USA. 2014;111(17):E1687–E1694. doi: 10.1073/pnas.1404823111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Le Tallec B, Barrault MB, Guerois R, Carre T, Peyroche A. Hsm3/S5b participates in the assembly pathway of the 19S regulatory particle of the proteasome. Mol Cell. 2009;33(3):389–399. doi: 10.1016/j.molcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 103.Funakoshi M, Tomko RJ, Jr, Kobayashi H, Hochstrasser M. Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell. 2009;137(5):887–899. doi: 10.1016/j.cell.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Park S, Roelofs J, Kim W, Robert J, Schmidt M, Gygi SP, Finley D. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature. 2009;459(7248):866–870. doi: 10.1038/nature08065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Roelofs J, Park S, Haas W, Tian G, McAllister FE, Huo Y, Lee BH, Zhang F, Shi Y, Gygi SP, Finley D. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature. 2009;459(7248):861–865. doi: 10.1038/nature08063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Saeki Y, Toh EA, Kudo T, Kawamura H, Tanaka K. Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell. 2009;137(5):900–913. doi: 10.1016/j.cell.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 107.Kaneko T, Hamazaki J, Iemura S, Sasaki K, Furuyama K, Natsume T, Tanaka K, Murata S. Assembly pathway of the Mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell. 2009;137(5):914–925. doi: 10.1016/j.cell.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 108.Park S, Li X, Kim HM, Singh CR, Tian G, Hoyt MA, Lovell S, Battaile KP, Zolkiewski M, Coffino P, Roelofs J, Cheng Y, Finley D. Reconfiguration of the proteasome during chaperone-mediated assembly. Nature. 2013;497(7450):512–516. doi: 10.1038/nature12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Thompson D, Hakala K, DeMartino GN. Subcomplexes of PA700, the 19 S regulator of the 26S proteasome, reveal relative roles of AAA subunits in 26S proteasome assembly and activation and ATPase activity. J Biol Chem. 2009;284(37):24891–24903. doi: 10.1074/jbc.M109.023218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Barrault MB, Richet N, Godard C, Murciano B, Le Tallec B, Rousseau E, Legrand P, Charbonnier JB, Le Du MH, Guerois R, Ochsenbein F, Peyroche A. Dual functions of the Hsm3 protein in chaperoning and scaffolding regulatory particle subunits during the proteasome assembly. Proc Natl Acad Sci USA. 2012;109(17):E1001–E1010. doi: 10.1073/pnas.1116538109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Besche HC, Peth A, Goldberg AL. Getting to first base in proteasome assembly. Cell. 2009;138(1):25–28. doi: 10.1016/j.cell.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 2009;10(2):104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- 113.Satoh T, Saeki Y, Hiromoto T, Wang YH, Uekusa Y, Yagi H, Yoshihara H, Yagi-Utsumi M, Mizushima T, Tanaka K, Kato K. Structural basis for proteasome formation controlled by an assembly chaperone nas2. Structure. 2014;22(5):731–743. doi: 10.1016/j.str.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 114.Godderz D, Dohmen RJ. Hsm3/S5b joins the ranks of 26S proteasome assembly chaperones. Mol Cell. 2009;33(4):415–416. doi: 10.1016/j.molcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 115.Hendil KB, Kriegenburg F, Tanaka K, Murata S, Lauridsen AM, Johnsen AH, Hartmann-Petersen R. The 20S proteasome as an assembly platform for the 19S regulatory complex. J Mol Biol. 2009;394(2):320–328. doi: 10.1016/j.jmb.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 116.Savulescu AF, Shorer H, Kleifeld O, Cohen I, Gruber R, Glickman MH, Harel A. Nuclear import of an intact preassembled proteasome particle. Mol Biol Cell. 2011;22(6):880–891. doi: 10.1091/mbc.E10-07-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pathare GR, Nagy I, Bohn S, Unverdorben P, Hubert A, Korner R, Nickell S, Lasker K, Sali A, Tamura T, Nishioka T, Forster F, Baumeister W, Bracher A. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc Natl Acad Sci USA. 2012;109(1):149–154. doi: 10.1073/pnas.1117648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385(6618):737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 119.Pathare GR, Nagy I, Sledz P, Anderson DJ, Zhou HJ, Pardon E, Steyaert J, Forster F, Bracher A, Baumeister W. Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proc Natl Acad Sci USA. 2014;111(8):2984–2989. doi: 10.1073/pnas.1400546111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Isono E, Nishihara K, Saeki Y, Yashiroda H, Kamata N, Ge L, Ueda T, Kikuchi Y, Tanaka K, Nakano A, Toh-e A. The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol Biol Cell. 2007;18(2):569–580. doi: 10.1091/mbc.E06-07-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fukunaga K, Kudo T, Toh-e A, Tanaka K, Saeki Y. Dissection of the assembly pathway of the proteasome lid in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2010;396(4):1048–1053. doi: 10.1016/j.bbrc.2010.05.061. [DOI] [PubMed] [Google Scholar]
- 122.Tomko RJ, Jr, Hochstrasser M. Incorporation of the Rpn12 subunit couples completion of proteasome regulatory particle lid assembly to lid-base joining. Mol Cell. 2011;44(6):907–917. doi: 10.1016/j.molcel.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Estrin E, Lopez-Blanco JR, Chacon P, Martin A. Formation of an intricate helical bundle dictates the assembly of the 26S proteasome lid. Structure. 2013;21(9):1624–1635. doi: 10.1016/j.str.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 124.Tomko RJ, Jr, Hochstrasser M. The intrinsically disordered Sem1 protein functions as a molecular tether during proteasome lid biogenesis. Mol Cell. 2014;53(3):433–443. doi: 10.1016/j.molcel.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Orlowski M, Wilk S. Catalytic activities of the 20S proteasome, a multicatalytic proteinase complex. Arch Biochem Biophys. 2000;383(1):1–16. doi: 10.1006/abbi.2000.2036. [DOI] [PubMed] [Google Scholar]
- 126.Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. A gated channel into the proteasome core particle. Nat Struct Mol Biol. 2000;7(11):1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 127.Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94(5):615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 128.Kriegenburg F, Seeger M, Saeki Y, Tanaka K, Lauridsen AM, Hartmann-Petersen R, Hendil KB. Mammalian 26S proteasomes remain intact during protein degradation. Cell. 2008;135(2):355–365. doi: 10.1016/j.cell.2008.08.032. [DOI] [PubMed] [Google Scholar]
- 129.Driscoll J, Goldberg AL. The proteasome (multicatalytic protease) is a component of the 1500-kDa proteolytic complex which degrades ubiquitin-conjugated proteins. J Biol Chem. 1990;265(9):4789–4792. [PubMed] [Google Scholar]
- 130.Armon T, Ganoth D, Hershko A. Assembly of the 26 S complex that degrades proteins ligated to ubiquitin is accompanied by the formation of ATPase activity. J Biol Chem. 1990;265(34):20723–20726. [PubMed] [Google Scholar]
- 131.Bajorek M, Finley D, Glickman MH. Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr Biol. 2003;13(13):1140–1144. doi: 10.1016/s0960-9822(03)00417-2. [DOI] [PubMed] [Google Scholar]
- 132.Tai HC, Besche H, Goldberg AL, Schuman EM. Characterization of the brain 26S proteasome and its interacting proteins. Front Mol Neurosci. 2010;3:12. doi: 10.3389/fnmol.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fujimuro M, Takada H, Saeki Y, Toh-e A, Tanaka K, Yokosawa H. Growth-dependent change of the 26S proteasome in budding yeast. Biochemical and biophysical research communications. 1998;251(3):818–823. doi: 10.1006/bbrc.1998.9560. [DOI] [PubMed] [Google Scholar]
- 134.Hanna J, Waterman D, Boselli M, Finley D. Spg5 protein regulates the proteasome in quiescence. J Biol Chem. 2012;287(41):34400–34409. doi: 10.1074/jbc.M112.390294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Saunier R, Esposito M, Dassa EP, Delahodde A. Integrity of the Saccharomyces cerevisiae Rpn11 protein is critical for formation of proteasome storage granules (PSG) and survival in stationary phase. PLoS ONE. 2013;8(8):e70357. doi: 10.1371/journal.pone.0070357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Imai J, Maruya M, Yashiroda H, Yahara I, Tanaka K. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 2003;22(14):3557–3567. doi: 10.1093/emboj/cdg349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Satoh K, Sasajima H, Nyoumura KI, Yokosawa H, Sawada H. Assembly of the 26S proteasome is regulated by phosphorylation of the p45/Rpt6 ATPase subunit. Biochemistry. 2001;40(2):314–319. doi: 10.1021/bi001815n. [DOI] [PubMed] [Google Scholar]
- 138.Bose S, Stratford FL, Broadfoot KI, Mason GG, Rivett AJ. Phosphorylation of 20S proteasome alpha subunit C8 (alpha7) stabilizes the 26S proteasome and plays a role in the regulation of proteasome complexes by gamma-interferon. Biochem J. 2004;378(Pt 1):177–184. doi: 10.1042/BJ20031122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Guo X, Engel JL, Xiao J, Tagliabracci VS, Wang X, Huang L, Dixon JE. UBLCP1 is a 26S proteasome phosphatase that regulates nuclear proteasome activity. Proc Natl Acad Sci USA. 2011;108(46):18649–18654. doi: 10.1073/pnas.1113170108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kajava AV, Gorbea C, Ortega J, Rechsteiner M, Steven AC. New HEAT-like repeat motifs in proteins regulating proteasome structure and function. J Struct Biol. 2004;146(3):425–430. doi: 10.1016/j.jsb.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 141.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10(3):495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 142.Kleijnen MF, Roelofs J, Park S, Hathaway NA, Glickman M, King RW, Finley D. Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat Struct Mol Biol. 2007;14(12):1180–1188. doi: 10.1038/nsmb1335. [DOI] [PubMed] [Google Scholar]
- 143.Gorbea C, Goellner GM, Teter K, Holmes RK, Rechsteiner M. Characterization of mammalian Ecm29, a 26 S proteasome-associated protein that localizes to the nucleus and membrane vesicles. J Biol Chem. 2004;279(52):54849–54861. doi: 10.1074/jbc.M410444200. [DOI] [PubMed] [Google Scholar]
- 144.Enenkel C. Proteasome dynamics. Biochim Biophys Acta. 2014;1843(1):39–46. doi: 10.1016/j.bbamcr.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 145.Park S, Kim W, Tian G, Gygi SP, Finley D. Structural defects in the regulatory particle-core particle interface of the proteasome induce a novel proteasome stress response. J Biol Chem. 2011;286(42):36652–36666. doi: 10.1074/jbc.M111.285924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Panasenko OO, Collart MA. Not4 E3 ligase contributes to proteasome assembly and functional integrity in part through Ecm29. Mol Cell Biol. 2011;31(8):1610–1623. doi: 10.1128/MCB.01210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lehmann A, Niewienda A, Jechow K, Janek K, Enenkel C. Ecm29 fulfils quality control functions in proteasome assembly. Mol Cell. 2010;38(6):879–888. doi: 10.1016/j.molcel.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 148.Bousquet-Dubouch MP, Nguen S, Bouyssie D, Burlet-Schiltz O, French SW, Monsarrat B, Bardag-Gorce F. Chronic ethanol feeding affects proteasome-interacting proteins. Proteomics. 2009;9(13):3609–3622. doi: 10.1002/pmic.200800959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang X, Yen J, Kaiser P, Huang L. Regulation of the 26S proteasome complex during oxidative stress. Sci Signal. 2010;3(151):ra88. doi: 10.1126/scisignal.2001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Akahane T, Sahara K, Yashiroda H, Tanaka K, Murata S. Involvement of Bag6 and the TRC pathway in proteasome assembly. Nat Commun. 2013;4:2234. doi: 10.1038/ncomms3234. [DOI] [PubMed] [Google Scholar]
- 151.Sahara K, Kogleck L, Yashiroda H, Murata S. The mechanism for molecular assembly of the proteasome. Adv Biol Regul. 2014;54:51–58. doi: 10.1016/j.jbior.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 152.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 153.Lehmann A, Janek K, Braun B, Kloetzel PM, Enenkel C. 20 S proteasomes are imported as precursor complexes into the nucleus of yeast. J Mol Biol. 2002;317(3):401–413. doi: 10.1006/jmbi.2002.5443. [DOI] [PubMed] [Google Scholar]
- 154.Kremer M, Henn A, Kolb C, Basler M, Moebius J, Guillaume B, Leist M, Van den Eynde BJ, Groettrup M. Reduced immunoproteasome formation and accumulation of immunoproteasomal precursors in the brains of lymphocytic choriomeningitis virus-infected mice. J Immunol. 2010;185(9):5549–5560. doi: 10.4049/jimmunol.1001517. [DOI] [PubMed] [Google Scholar]
- 155.Hoefer MM, Boneberg EM, Grotegut S, Kusch J, Illges H. Possible tetramerisation of the proteasome maturation factor POMP/proteassemblin/hUmp1 and its subcellular localisation. Int J Biol Macromol. 2006;38(3–5):259–267. doi: 10.1016/j.ijbiomac.2006.03.015. [DOI] [PubMed] [Google Scholar]