Abstract
MMP23 is a member of the matrix metalloprotease family of zinc- and calcium-dependent endopeptidases, which are involved in a wide variety of cellular functions. Its catalytic domain displays a high degree of structural homology with those of other metalloproteases, but its atypical domain architecture suggests that it may possess unique functional properties. The N-terminal MMP23 pro-domain contains a type-II transmembrane domain that anchors the protein to the plasma membrane and lacks the cysteine-switch motif that is required to maintain other MMPs in a latent state during passage to the cell surface. Instead of the C-terminal hemopexin domain common to other MMPs, MMP23 contains a small toxin-like domain (TxD) and an immunoglobulin-like cell adhesion molecule (IgCAM) domain. The MMP23 pro-domain can trap Kv1.3 but not closely-related Kv1.2 channels in the endoplasmic reticulum, preventing their passage to the cell surface, while the TxD can bind to the channel pore and block the passage of potassium ions. The MMP23 C-terminal IgCAM domain displays some similarity to Ig-like C2-type domains found in IgCAMs of the immunoglobulin superfamily, which are known to mediate protein–protein and protein–lipid interactions. MMP23 and Kv1.3 are co-expressed in a variety of tissues and together are implicated in diseases including cancer and inflammatory disorders. Further studies are required to elucidate the mechanism of action of this unique member of the MMP family.
Keywords: Matrix metalloprotease, MMP23 Pro-domain, Potassium channel, Kv1.3, Trans-membrane domain, Toxin domain
Introduction
Matrix metalloprotease 23 (MMP23) belongs to a large family of zinc-dependent endopeptidases that function at neutral pH to degrade extracellular matrix proteins, cleave cell-surface receptors, release apoptotic ligands, and activate chemokines and cytokines [1–3]. The 23 MMPs in humans are encoded by 24 genes, with two identical genes on chromosome 1 encoding MMP23 (MMP23A and MMP23B) [4]. Matrix metalloproteases (MMPs) are involved in a wide range of cellular processes, including tissue remodeling, cell proliferation, cell migration, differentiation, angiogenesis, apoptosis, and the immune response [5–10]. Besides these normal physiological processes, MMPs have been implicated in a large number of pathological processes such as arthritis, Alzheimer’s disease, atherosclerosis, vascular disease, central nervous system disease, liver cirrhosis, and various cancers [11, 12].
MMP activity is regulated at a number of different levels, including biosynthesis (transcription/translation) [13], zymogen activation [11, 13], compartmentalization [11, 14, 15], and inactivation [16–19]. Some MMPs are required during development and normal physiology and may play a role in homeostasis [8, 10, 20–23], while others are produced in response to tissue injury and infection [5, 8]. In chronically inflamed tissues and most cancers, MMPs contribute to a number of pathological processes, including tissue degradation, tumor progression and invasion [24–31].
MMPs are synthesized as zymogens and usually secreted or anchored to the plasma membrane, confining their activity to the extracellular environment or the cell surface. Recent evidence suggests that secreted MMPs bind to specific cell-surface receptors, membrane-anchored proteins, and cell-associated extracellular matrix (ECM) molecules, and function pericellularly at focused locations [15]. MMPs are also found in the cell nucleus (MMP2, 3, 9, 13, and MT1-MMP), cytoplasm (MMP1, 2, 26, and 23), and various organelles, where their localization is mediated by interactions with other proteins, proteoglycan core proteins, and/or their glycosaminoglycan chains and other molecules [11, 14, 32–43].
This review focuses on the structural and functional features that distinguish MMP23 from other MMPs. MMP23 is a membrane-anchored protein consisting of 391 amino acid residues and four separate domains, three of which differ significantly from those of other MMPs. The N-terminal pro-domain of MMP23 lacks the enzymatic inhibitory sequence motif and folded globular structure characteristic of other MMP pro-domains, and has been shown to modulate voltage-gated potassium (Kv1) activity by regulating the intracellular trafficking of these channels. MMP23 also contains a toxin-like domain (TxD) that has been shown to block Kv1 channels. In addition, the C-terminal hemopexin domain common to other MMPs is replaced by an immunoglobulin-like cell adhesion molecule (IgCAM) domain of unknown function in MMP23.
Domain structure
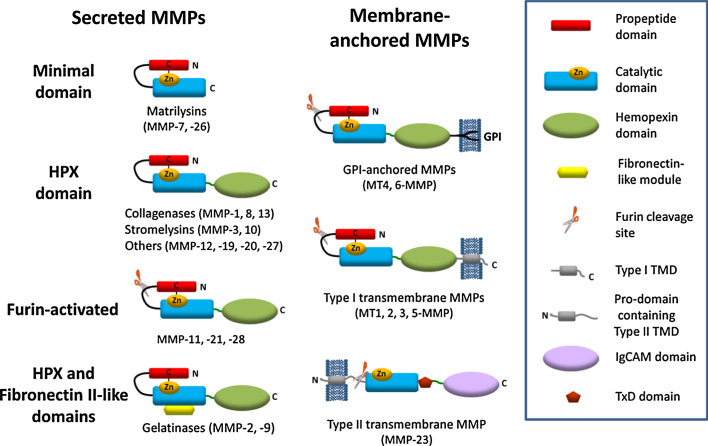
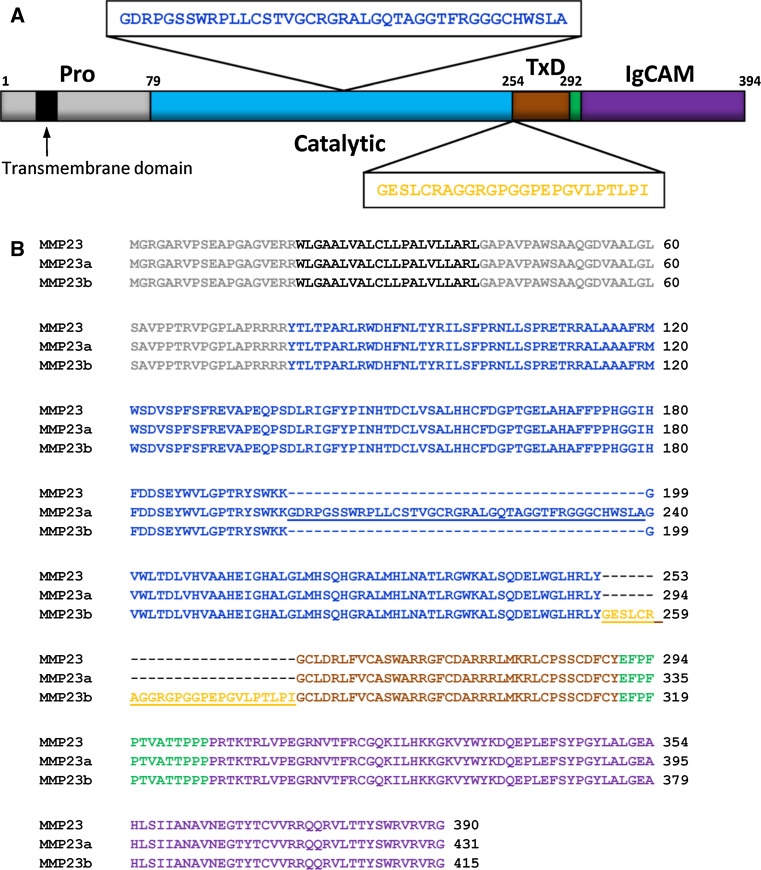
MMPs typically contain a short signal sequence followed by a pro-domain, a central zinc- and calcium-dependent catalytic domain, a linker region (also called the ‘hinge region’), and a C-terminal hemopexin domain (Fig. 1) [11, 44–46]. MMP23, by contrast, contains an N-terminal pro-domain that includes a membrane-anchoring transmembrane domain (TMD), a cysteine-rich TxD located immediately after the catalytic domain, and a C-terminal IgCAM domain.
Fig. 1.
Schematic of the domain architecture of human matrix metalloproteases. Most MMPs contain a pro-domain (red), a catalytic domain (blue), a linker (hinge region, dark green) and a hemopexin domain (green). Furin-activated MMPs, including the membrane-anchored MMPs, possess a basic RX{K/R}R sequence motif at the C-terminal end of the pro-domain (where X represents any amino acid residue). Cleavage at this site results in release of the pro-domain and activation of the enzyme. Two MMPs (MMP2 and MMP9) contain three fibronectin (FnII)-like repeats (yellow) in the catalytic domain prior to the catalytic Zn2+ ion-binding site. Four MMPs (MT1, 2, 3, 5-MMP) are anchored to the cell membrane via a C-terminal type-I transmembrane domain (TMD) and two MMPs (MT4, 6-MMP) are tethered by a glycosylphosphatidylinositol (GPI)-anchor. In contrast, MMP23 is anchored via a N-terminal type-II transmembrane domain (gray). The two minimal domain MMPs (MMP7, 26) and MMP23 lack the hemopexin domain, and in MMP23 this domain is replaced by a C-terminal cysteine-rich toxin-like (TxD, brown) domain and an immunoglobulin-like cell adhesion molecule (IgCAM) domain (purple)
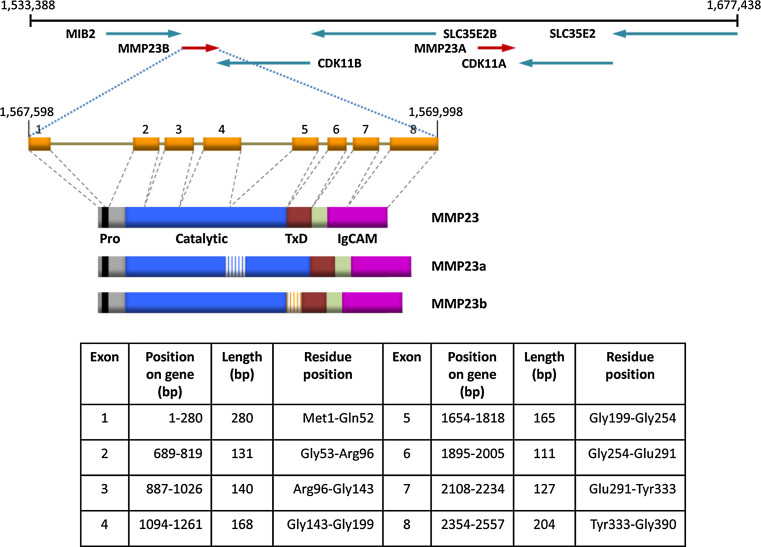
MMP23 is located in a duplicated region of human chromosome 1p36.3 (Fig. 2). The two MMP23 genes, referred to as mmp23a (pseudogene, MMP23A) and mmp23b (MMP23B denoted as MMP23 in this review), each produce three alternatively spliced products, MMP23, MMP23a, and MMP23b (also known as MMP21/22A, MMP21/22B, and MMP21/22C, respectively) (Fig. 2). Rodent genomes contain a single copy of the MMP23 gene, implying that the two copies in humans evolved from a recent duplication event that occurred after the divergence of the rodent and primate linages [47]. Comparative genomics analyses indicate that the TxD and IgCAM domains were introduced during the early evolution of tetrapods, since zebrafish has an orthologue of this gene that lacks these C-terminal domains [48].
Fig. 2.
Structure of the human MMP23 loci and the corresponding gene product [4]. Schematic representation of the ~150-kb region of chromosome 1p36.3 that contains the two metalloprotease 23 genes (mmp23a and mmp23b). The genes are represented by arrows orientated to highlight their transcriptional orientation relative to the neighboring CDK11 (cyclin-dependent kinase 11), MIB2 (mindbomb E3 ubiquitin protein ligase 2), and SLC35E2B (solute carrier family 35, member E2B) genes. The exon/intron structure and protein domain architecture for the MMP23 gene and isoforms (MMP23, MMP23a, and MMP23b) formed by alternative splicing are also illustrated. The location of exons constituting the mmp23 gene and protein (MMP23) are listed in the table
Pro-domain
Most MMPs are expressed as zymogens (inactive enzymes) where the N-terminal pro-domain adopts a folded globular structure that interacts with the catalytic domain and maintains the enzyme in a latent state (Fig. 3). The highly conserved pro-domain possesses a ‘cysteine-switch’ motif (PRCGXPD) containing a cysteine residue that coordinates with the catalytic zinc ion (Fig. 3b, c) [49]. Destabilization or removal of the pro-domain unblocks the active site, allowing it to process substrate [46]. Protease activation can occur either by direct cleavage of the pro-domain by itself (autolysis) or by another protease, or by modification of the cysteine that binds to the catalytic zinc ion [11, 49, 50].
Fig. 3.
MMP pro- and catalytic domain structures. a Superposition of the catalytic (blue) and pro- (red) domains of MMP1 (PDB id: 1SU3), MMP2 (1CK7), MMP3 (1SLM), MMP9 (1L6J). The fibronectin-related domains in MMP2 and MMP9 have been removed for clarity. Zinc and calcium ions are colored magenta and green, respectively. b Pro- and catalytic domains of MMP3 where residues of the conserved ‘cysteine-switch’ (PRCGXPD, where X is any amino acid residue) are colored yellow. c Magnification of (b) where side chains for residues comprising the ‘cysteine-switch’ motif are labeled. The figure was generated using the program Pymol [173]
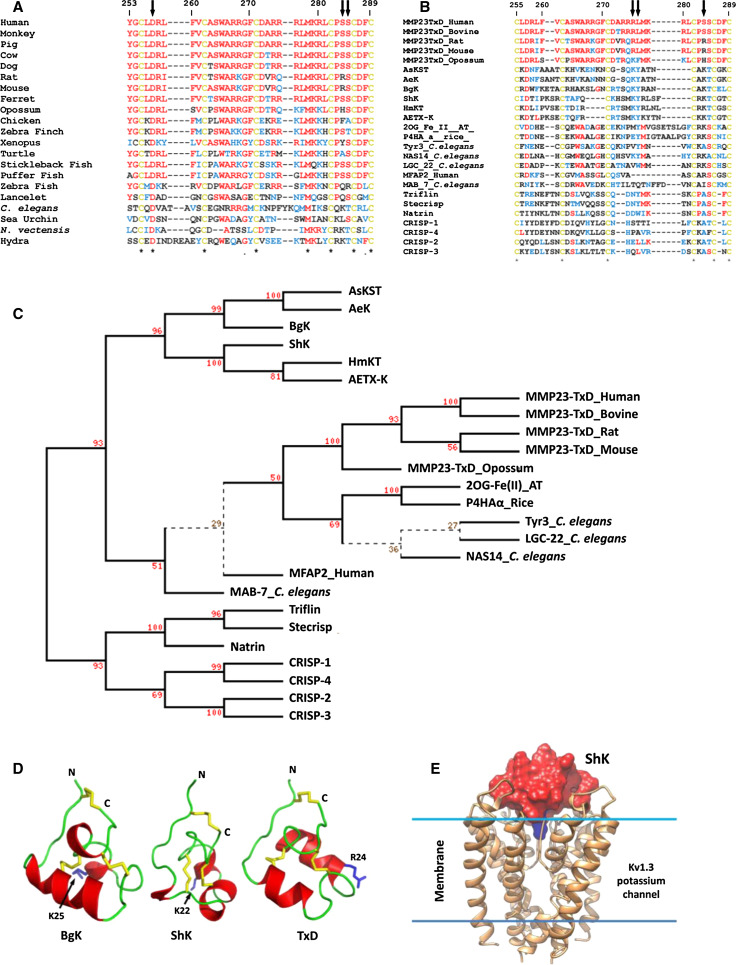
The 80-residue MMP23 pro-domain sequence, by contrast, is shorter and significantly different from those of other MMP pro-domains. It lacks the classical cysteine-switch motif and displays only limited sequence homology among different species [51, 52]. Rather than the folded globular structure formed by other MMP pro-domains, the MMP23 pro-domain contains a N-terminal type-II TMD (20WLGAALVALCLLPALVLL37) that anchors the protein to the membrane [39, 51–54]. In contrast, other membrane-tethered MMPs (MT-MMPs) are anchored to the membrane by a C-terminal TMD (MMP14, 15, 16, and 24) or a glycosylphophatidylinositol (GPI) anchor (MMP17 and 25) [55, 56]. The MMP23 TMD acts as a signal peptide that directs MMP23 to the ER/Golgi prior to secretion [7, 9–11], in a manner similar to the N-terminal cleavable signal peptide of other MMPs, which is absent in MMP23 [54].
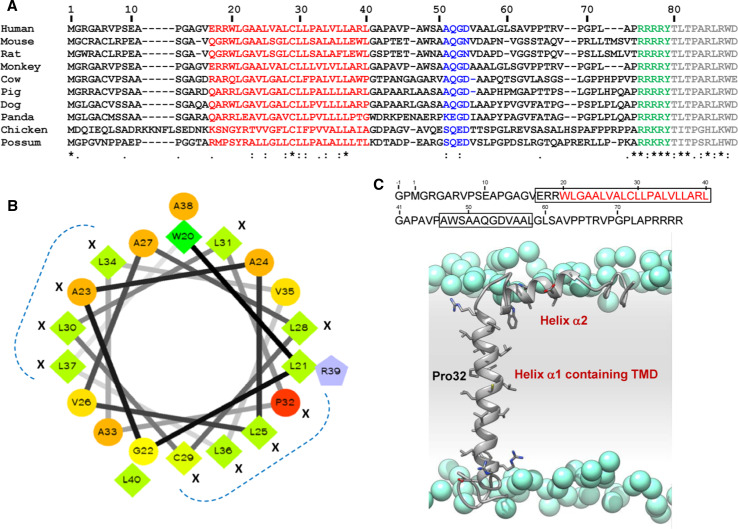
The MMP23 TMD sequence displays only limited homology across various species, although there is strong conservation of several hydrophobic residues (Fig. 4a). Helical wheel analysis of residues in the MMP23 TMD shows that these conserved residues cluster in two regions on the surface of this transmembrane helix (Fig. 4b), suggesting that they may play a functional role [38].
Fig. 4.
Conservation of residues within the TMD of the pro-domain of MMP23. a Sequence alignment of pro-domain regions of MMP23 from various species where residues comprising the TMD are colored red, the furin recognition motif is green and the catalytic domain is gray. The accession numbers for each MMP23 are: human, UniProt 075900; mouse, 088676; rat, 088272; monkey, 10FJ41; cow, Q2TBM7; pig, F1RJC6; dog, E2R4V4; panda, G1M504; chicken, E1BX58; and possum, F7CDE4. Residues corresponding to the conserved astacin FXGD motif are colored blue. The sequence alignment was performed using ClustalW [174]. Residues are numbered according to human MMP23. The level of conservation is illustrated along the lower row: Asterisk identical, colon highly conserved, dot weakly conserved. b Helical wheel representation of the alignment of the TMD in (a) generated using the web-based program WHEEL (http://rzlab.ucr.edu/scripts/wheel/wheel.cgi). Identical and conserved residues are denoted by X and residues that cluster on separate faces of the helical wheel are denoted by a dotted blue line. Residues are represented as follows: hydrophilic—circles, hydrophobic—diamonds, and potentially positively charged—pentagons. Residues are color-coded according to hydrophobicity/hydrophilicity, from red (most hydrophilic) through yellow (moderately hydrophilic) to green (most hydrophobic). Potentially charged residues are colored light blue. c Secondary structure of pro-domain for human MMP23. Model of the TMD (α1 helix) and juxta-membrane helix (α2 helix) of the pro-domain for human MMP23 in a lipid membrane (residues 1–74). Atoms of the side-chain groups of residues in the two helices are highlighted. Phosphate groups of the lipid are represented as blue spheres; lipid choline, glycerol and acyl chains, and water are omitted for clarity. The amino acid sequence of the MMP23 pro-domain is shown at the top where α-helical regions are boxed and residues of the TMD are colored red. Adapted from Nguyen et al. [38]
NMR studies of the MMP23 pro-domain in dodecylphosphocholine (DPC) micelles show that it contains a relatively long α-helix (α1) (residues Glu17–Leu40), incorporating the membrane-anchoring TMD, connected by a 6-residue linker to a short juxta-membrane α-helix (α2) (residues Ala47–Leu58) (Fig. 4c) [38]. Paramagnetic relaxation enhancement and solvent exchange measurements indicate that the N-terminal capping region of helix α1 (residues Glu17–Arg19) is exposed to solvent while the remaining portion of the helix is buried within the DPC micelles and not accessible to solvent [38]. The juxta-membrane helix (α2) appears to adopt a partially flexible structure that interacts with the surface of the micelle and is partially buried [38].
The short N-terminal cytoplasmic tail of the MMP23 pro-domain is unstructured [38], and it is not known whether this region plays a regulatory role as in other MT-MMPs. MT-MMPs can modulate intracellular processes through interactions of their short unstructured cytoplasmic C-terminal tail with various intracellular proteins, leading to the induction of intracellular signaling pathways [57, 58]. Most of these interactions occur at the plasma membrane but they can also take place in other organelles such as the Golgi [58–66].
Some MMPs, including all MT-MMPs, contain a furin-recognition motif between the pro- and catalytic domains (Figs. 1, 4) [13, 50]. Cleavage at this site releases the inhibitory pro-domain and activates the enzyme in the secretory pathway [50]. MMP23 also contains a furin recognition motif 76RRRRY80 between the propeptide and catalytic domain [52, 54], and cleavage between residues Arg79 and Tyr80 has been shown to release the active enzyme [67–70]. However, it is not known whether furin is the principal protease involved in the activation of MMP23 [51].
The pro-domain of MMP23 differs significantly from those of other MMPs, and it is unclear how it is maintained in a latent state during passage through the secretory pathway. The MMP23 pro-domain contains a poly-proline motif immediately prior to the furin recognition motif (i.e. 68RVPGPLAPRRRRY80) that exhibits limited similarity to various SH3 domain ligands [71, 72]. This motif may bind to the catalytic active site and inhibit activity, although this region is poorly conserved across species.
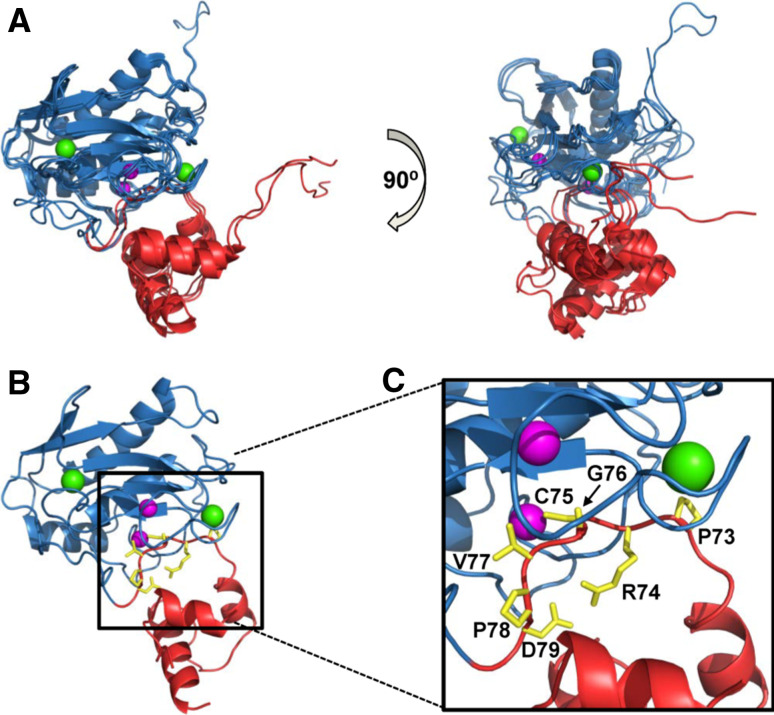
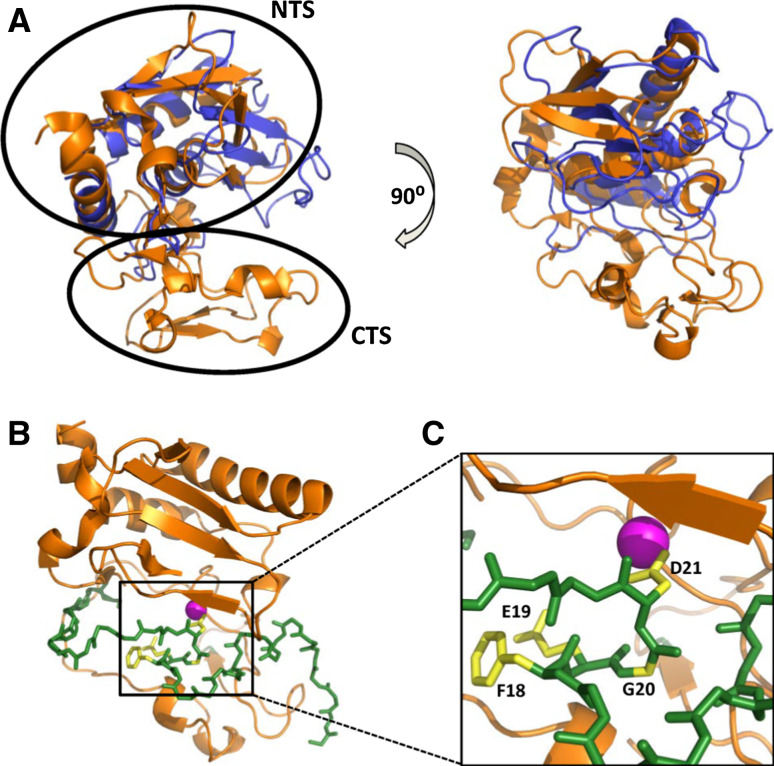
MMPs generally display many similarities with the astacins including (1) structural homology between the MMP catalytic domain and upper N-terminal subdomain (NTS) of the astacins (Fig. 5a, b), (2) similar zinc-binding motifs, (3) a partially unstructured N-terminal pro-segment, and (4), in several astacins, the presence of one or more toxin-like (ShK-like) domains similar to the TxD of MMP23 [73–75]. The MMP23 pro-domain displays several characteristics similar to the pro-segment of astacin zinc-dependent metalloproteases (Fig. 5) [73]. The astacin pro-segment is variable in length (34 residues for crayfish astacin to 393 residues for the Drosophila tolloid-related 1 protease) [76] and contains only a short FXGD consensus sequence [73, 76]. The aspartate residue within this FXGD motif binds the catalytic zinc and inhibits protease activity via an ‘aspartate-switch’ mechanism. The structure of crayfish pro-astacin shows that the pro-segment extends across the front surface of the enzyme in the opposite direction to that of the substrate, with a short helical motif located towards the N-terminus (Fig. 5c). MMP23 contains a short sequence (51AQGD54; Fig. 4a), similar to the FXGD motif of the astacins, which is located within the juxta-membrane helix 47AWSAAQGDVAAL58 of the MMP23 pro-domain. However, this motif varies among MMP23s from different species (Fig. 4a), suggesting that the pro-domain may inhibit the catalytic domain via a novel mechanism.
Fig. 5.
Structure of pro-astacin. a Two orthogonal views of a superposition of the X-ray crystal structures of the activated catalytic domains of crayfish astacin (PDB ID: 1AST, orange) and MMP3 (1SLM, blue). The N- (NTS) and C-terminal (CTS) subdomains of crayfish astacin are denoted by open circles. b Crystal structure for pro-astacin (3LQ0) depicting the backbone of the pro-segment as sticks (green) and the enzyme moiety as a ribbon (orange). c Magnification of (b) where side-chains for residues comprising the conserved FXGD motif are shown (yellow) and labeled while the catalytic zinc ion is illustrated as a magenta sphere
Catalytic domain
MMP catalytic domains, including MMP23, share a high degree of structural (Fig. 3a) and sequence homology (Fig. 6a) [77–79]. The MMP23 catalytic domain structure has not yet been determined, although it is likely to be similar to that for other MMPs since they share such a high degree of sequence and structural homology.
Fig. 6.
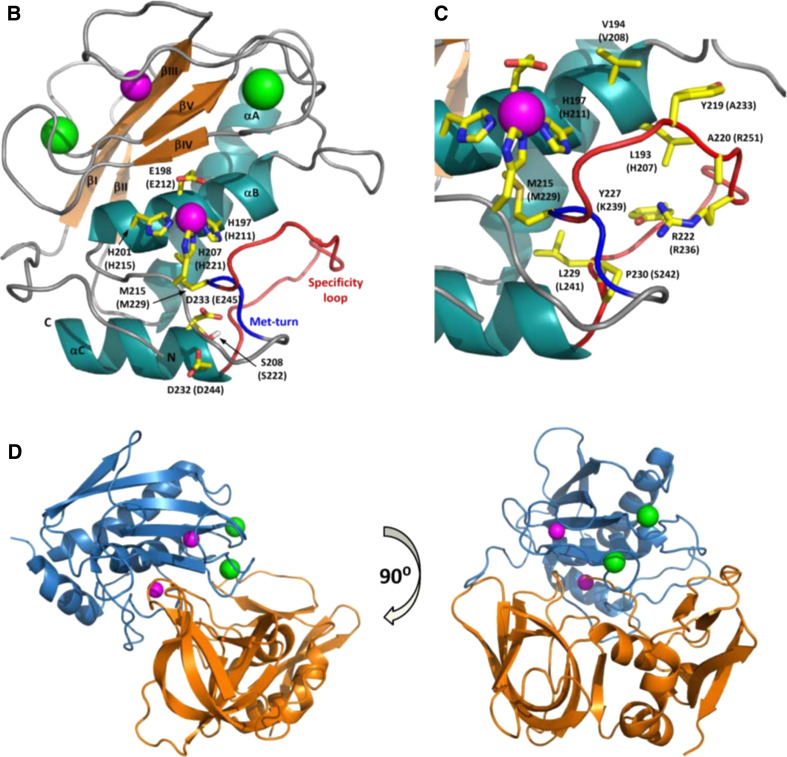
MMP catalytic domain. a Sequence alignment of the catalytic domains of MMP23 with soluble MMPs (MMP1, UniProt P03956; MMP3, P08254; MMP7, P09237; MMP8, P22894; MMP10, P09238; MMP11, P24347; MMP12, P39900; MMP13, P45452; MMP19, Q99542; MMP20, O60882; MMP21, Q8N119; MMP27, Q9H306; MMP28, Q9H239), MT-MMPs (MMP14, P50281; MMP15, P51511; MMP16, P51512; MMP17, Q9ULZ9; MMP23, O75900; MMP24, Q9Y5R2; MMP25, Q9NPA2) and fibronectin-like domain containing MMPs (MMP2, P08253; MMP9, P14780). Residues binding to calcium (C1 and C2) and zinc (Z1 and Z2) ions are colored blue and red, respectively, while those located in the specificity loop (SL) are colored green. The conserved Met residue of the ‘Met-turn’ (MetT) is colored purple, and the conserved Asp or Glu residues involved in stabilizing electrostatic interactions (NA) are brown. Secondary structure elements based on the X-ray crystal structure of human MMP1 (PDB ID: 1SU3) are shown above the sequence alignment where arrows represent β-strands and cylinders are α-helices. b, c MMP catalytic domain structure. b Catalytic domain of MMP8 (1JAN) showing secondary structure elements (orange arrows for β-strands, β1-βV; dark green helices for α-helices, αA-αC) and the four metal ions (zinc ions are colored magenta and calcium ions are light green). Side-chains for residues binding the catalytic zinc ion and forming salt-bridges that stabilize the Met-turn and N-terminus are illustrated with yellow carbon atoms and labeled for MMP8 and MMP23 (in brackets). The Met-turn is colored blue and the specificity loop is red. c Expanded view of active site region depicting side-chains for residues engaged in zinc binding and residues forming the specificity pocket (labeled). The figure was prepared using the program Pymol [173]. d Two orthogonal views of the crystal structure (PDB ID: 1UEA) of MMP3 (blue) bound to TIMP1 (orange)
MMP catalytic domains possess a globular conformation with a diameter of ~40 Å and a shallow active site cleft that binds substrates in an approximately extended conformation [78]. They have a common architecture comprised of three α-helices and a five-stranded β-sheet, and typically contain four metal ions (two calcium and two zinc) in highly conserved calcium- and zinc-binding sites (Fig. 6b, c) [44, 77, 78]. The extended zinc-binding consensus sequence, HEXXHXXGXXH (where X represents any residue) [80, 81], ‘Met-turn’ and, N-terminal anchoring residues typically found in MMPs are conserved in MMP23, while the ‘specificity loop’ that contributes to substrate specificity is significantly different (Fig. 6a, b).
The MMP23 catalytic domain contains two inserts that are not found in other MMPs (Fig. 7a). The first is located in the loop between strands βIII and βIV and is flanked by two Cys residues (153CLVSAVHHC161) that may form a disulfide bond [52]. Residues on either side of this loop interact with the structural zinc ion and one of the calcium ions, and it is possible that a disulfide bond within this loop may help to restrain these residues so that they can ligate these metal ions. The second insert (193TRYSWKKGV201) is positively charged and lies in the loop between strand βV and helix αB, which contains the fibronectin type II repeats that are important for substrate recognition in MMP2 and MMP9 (Fig. 7a) [52, 82]. Interestingly, the MMP23 isoform MMP23a contains a 41-residue insert at this site although the insert displays no sequence homology to the fibronectin type II domain (Fig. 7).
Fig. 7.
MMP23 isoforms. a Schematic of domain architecture of MMP23 showing the sequence and location of inserted sequences for isoforms MMP23a and MMP23b. b Sequence alignment for various human MMP23 isoforms (MMP23, O75900; MMP23a, O75900-2; MMP23b, O75900-3). Residues corresponding to the pro-domain are colored gray (TMD colored black), catalytic domain are blue, TxD are brown, linker domain are green and IgCAM domain are purple. Residues of the insert between the catalytic domain and TxD in the MMP23 isoform MMP23b are colored orange
MMP proteolytic activities are tightly regulated by several endogenous inhibitors since uncontrolled proteolytic activity following activation would result in tissue damage and functional alterations. Plasma-associated MMPs are inhibited by liver-secreted α2-macroglobulin, while tissue or extracellular MMPs are regulated by a family of four tissue inhibitors of metalloproteases (TIMP1-4) [7, 13, 16, 17, 83]. Human α2-macroglobulin is a tetrameric 725-kDa glycoprotein that entraps MMPs via a ‘venus flytrap’-like mechanism [84]; this bound complex is rapidly cleared by receptor (low density lipoprotein receptor-related protein-1)-mediated endocytosis [85, 86]. The in vitro proteolytic activity of MMP23 is inhibited by the tissue inhibitors of metalloproteases TIMP1 [54] and TIMP2 [52]. TIMPs (TIMP1–4) are comprised of 184–194 amino acid residues that form structured N- and C-domains that are stabilized by six disulfide bonds. The TIMP N-terminal domain binds tightly to the MMP catalytic domain, blocking the substrate binding site and inhibiting protease activity (Fig. 6d) [19, 87].
Immunoglobulin-like cell adhesion molecule (IgCAM) domain
The C-terminal immunoglobulin-like cell adhesion molecule (IgCAM) domain of MMP23 is unique among the MMPs. All other MMPs either possess a hemopexin C-domain or contain no C-terminal domain (Fig. 1) [7, 44, 46]. The hemopexin domain acts as a secondary specificity site or “exosite” located outside the active site, which binds and helps to correctly orientate the substrate [88]. This additional protein-binding domain increases the affinity of the protease for the substrate and can modify the specificity of the MMP catalytic domain [89, 90]. Exosites can also play a role in preparing substrates [91] such as localized unwinding of triple helical native collagen prior to cleavage [92]. The C-terminal immunoglobulin-like cell adhesion molecule (IgCAM) domain of MMP23 may also mediate protein–protein interactions similar to the hemopexin domain of other MMPs.
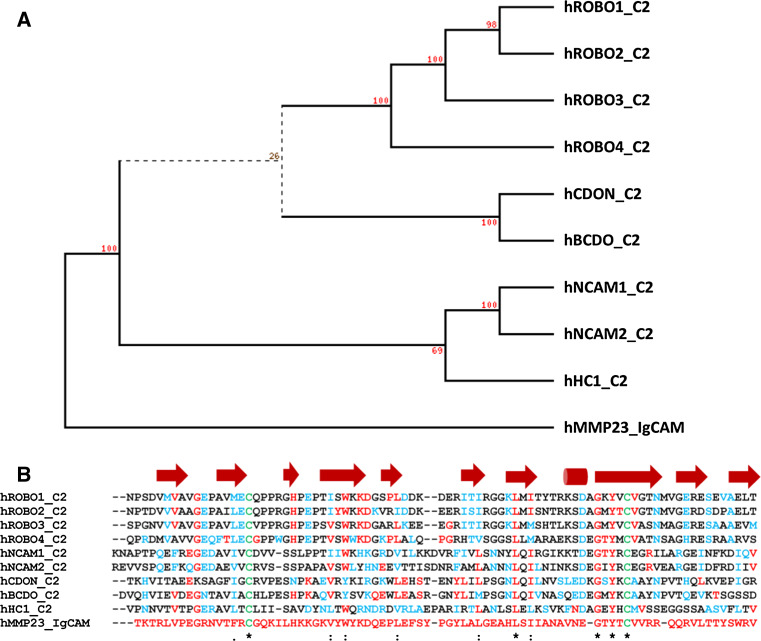
The MMP23 IgCAM domain displays sequence similarity to the Ig-like C2 type domains of the immunoglobulin superfamily (Fig. 8a, b). These domains mediate protein–protein interactions among themselves and with other types of molecules such as components of the ECM, signaling molecules, and several membrane-bound proteins (e.g., growth-factor receptors, integrins and cadherins) [93–97], as well as mediating direct membrane interactions (e.g., perforin) [98–100]. Protein or lipid interactions mediated by the MMP23 IgCAM domain may help to target and position protein substrates for proteolytic cleavage, similar to the hemopexin domain of other MMPs.
Fig. 8.
The Ig-CAM domain of human MMP23 shares sequence homology with the Ig-CAM and Ig-like C2 type domains from various proteins. Phylogenetic (PHYLIP) [175] analysis (a) and sequence alignment (b) of the IgCAM domains of human MMP23 (UniProt 075900) with rat CDON (XP_003751100), human Brother of CDO (Q9BWV1), human ROBO1-4 (ROBO1, Q1RMC7; ROBO2,Q9HCK4; ROBO3, Q96MS0; ROBO4, Q8WZ75), dog hemicentin (XP_548414), and human NCAM1 (P13591) and NCAM2 (O15394). Residues identical to human MMP23 are colored red, conserved residues are blue, and cysteine residues involved in disulphide bonds are green. Secondary structural elements based on the X-ray crystal structure of human ROBO1 (PDB ID: 2V9R) are shown above the sequence alignment where arrows represent β-strands and cylinders are α-helices. Adapted from Rangaraju et al. [39]
Toxin-like domain (MMP23-TxD)
The MMP23 toxin-like domain (MMP23-TxD), which lies immediately after the catalytic domain and is joined to the C-terminal IgCAM domain by a short Pro-rich linker (Fig. 7), is highly conserved from humans to hydra (Fig. 9a) [39]. The MMP23b isoform of MMP23 contains a short 25-residue Gly/Pro-rich linker joining the catalytic and MMP23-TxD domains that is not found in the other two isoforms (Fig. 7). The MMP23-TxD domain shares homology with the ShKT family of proteins from worms, cnidarians, and plants as well as the ion channel regulatory (ICR) domains of CRISPs (Fig. 9b). Phylogenetic analysis places the MMP23 TxDs, the sea anemone toxins, and the ICR-CRISP domains in distinct but related clades (Fig. 9c). MMP23 TxD is also closely related to ShKT domains in MFAP2 [101], Caenorhabditis elegans proteins (astacin metalloprotease NAS14, tyrosinase Try3, ligand-gated channel Igc22 and Mab7) [102], hydra and jellyfish astacin metalloproteases (HMP2 and PMP1) [103, 104], and plant oxidoreductases [2OG-Fe(II)] and propyl-4-hydroxylases (Fig. 9c). The TxDs in MMP23 and the ICR domains in CRISPs are encoded by a single exon, suggesting that these domains originate from an ancient exon. MMP23-TxD is most similar to the sea anemone toxin BgK, with identical or equivalent residues at 14 of 36 positions [39, 105–107]. The solution structure of a synthetic 37-residue peptide corresponding to rat MMP23-TxD also shows close similarity to the sea anemone toxins BgK [105] and ShK [107] (Fig. 9d) [39]. Rat MMP23-TxD possesses three short α-helices encompassing residues 10–14, 23–29, and 31–34, and a salt-bridge between Asp5, which is conserved in the ShKT domain family, and Arg32 (Ser in human MMP23 and Lys in ShK and BgK) that has been shown to be important for the correct folding of several sea anemone toxins [107–109]. Other MMP-TxDs contain conserved Ser or Thr residues at positions 32 and 33 (Fig. 9b) and may form a hydrogen bond to the side chain hydroxyl or the peptide backbone of either of these residues. MMP23-TxD shares greater structural similarity with BgK than ShK (r.m.s. deviation of backbone heavy atoms of 2.28 and 2.77 Å, respectively) (Fig. 9d). MMP23-TxD, BgK, and ShK have a turn involving the fifth Cys residue that is followed by a short α-helix (residues 31–34) in MMP23-TxD and BgK but not ShK [39]. The main differences between MMP23-TxD and BgK are in the length of the first two helices, with the first helix of MMP23-TxD being shorter and the second longer than in BgK.
Fig. 9.
The toxin-like domain (TxD) of MMP23 is conserved across species and is similar to the cysteine-rich ShKT family of proteins and the ion channel regulatory (ICR) domains of CRISPS. a Multiple sequence alignment of MMP23 TxDs from diverse species. Human (UniProt O75900), monkey (I0FJ41), pig (B0FXJ8), cow (Q2TBM7), dog (E2R4V4), rat (O88272), mouse (O88676), ferret (M3YSR1), opossum (F7CDE4), chicken (E1BX58), zebra finch (H0YXW0), xenopus (F6RIT9), turtle (K7FBH1), stickleback fish (G3NNG1), puffer fish (H3CZY7), zebra fish (F1R0R0), lancelet (C3Y5G9), C. elegans (Q9XTD6), sea urchin (H3IL27), Nematostella vectensis (starlet sea anemone, A7RW59), and hydra (Q9XZG0). Arrows point to Asp5, Ser32 and Ser33. b Multiple sequence alignment of representative ShKT domains with the ICR domains of CRISPs. Residues identical to human MMP23-TXD are colored red, conserved residues are blue and cysteine residues involved in disulphide bonds are yellow. Sea anemone toxins include BgK (UniProt P29186), ShK (P29187), AeTX-K (Q0EAE5), AsKS (Q9TWG1), AeK (P81897), HmKT (O16846). Nas14-C. elegans, nematode astacin metalloprotease NAS14 (Q19269). Tyr3-C. elegans, tyrosinase 3 (Q19673). LGC-22-C. elegans, ligand-gated channel 22 (NP_500538). HMP2-Hydra, Hydra metalloprotease 2 (AAD33860). MFAP2-human, microfibril associated protein 2 (P55001). MAB-7, male abnormal protein 7 (NP_508174). PMP1-Jellyfish, Podocoryne metalloprotease 1 [103]. Arrows point to the conserved Lys/aromatic residue dyad found in many Kv channel-blocking toxins and Lys residue that is important for correct folding of several sea anemone toxins. c Evolutionary relationship of MMP23 TxD with sea anemone toxins, ShKT domains and ion channel regulatory (ICR) domains of cysteine-rich secretory proteins (CRISPs). The phylogenetic tree (PHYLIP) [175] was generated using the alignment in (c) and sequences for oxidoreductase, 2OG-Fe(II) oxygenase family protein from A. thaliana (NP_189490) and propyl-4-hydroxylase α-subunit, O. sativa japonica group (AAT77286). d Comparison of the solution structure of the MMP23 TxD (PDB id: 2K72) with those for sea anemone Kv channel blockers BgK (1BGK) and ShK (1ROO) where α-helices are colored red and loops are green. Disulfide bonds are colored yellow and the pore-occluding residue is blue and labeled. e Structure of ShK docked to a model of the pore vestibule region of Kv1.3 [108]. Kv1.3 is represented as a light brown ribbon, while ShK is shown as a molecular surface with all residues colored red except Lys22 (blue) which occludes the channel pore. Adapted from Rangaraju et al. [39]
BgK and ShK are potent voltage-gated potassium (Kv) channel blockers, which contain a positively-charged Lys residue (Lys25 in BgK, Lys22 in ShK) that occludes the channel pore (Fig. 9d, e) [110]. MMP23-TxD contains an evolutionarily conserved, similarly charged residue at this position (Arg276) and also blocks several Kv channels (Kv 1.1, 1.3, 1.4, 1.6, and 3.2) with IC50 values in the nanomolar to low micromolar range while having no effect on others (Kv1.2, 1.5, 1.7, and KCa3.1) [39]. MMP23-TxD blocked human Kv1.6 channels expressed in COS7 cells with an IC50 of 370 nM and Kv1.3 expressed in L929 fibroblasts and T cells with an IC50 of 2.7 μM, compared to 10–20 pM [108, 111] for ShK [39, 108, 109, 112, 113]. The critical Lys/aromatic residue dyad found in many potassium channel-blocking toxins [114] is missing in MMP23-TxD (Fig. 9b), which may help explain its lower potency, although mutation of the corresponding residues in MMP23-TxD (Arg24/Leu25) to the conserved dyad motif (Lys/Tyr) did not improve potency against Kv1.3 (IC50 of 2.7 μM) [39]. Previous studies have demonstrated that replacement of Lys22 in ShK with Ala abolishes channel blocking activity while replacement with shorter or longer positively charged residues retains activity, albeit with a lower potency [108, 109, 112, 113].
Potassium channel regulation of Kv1.3 channel trafficking
TxD-containing proteins may play a role in regulating the intracellular trafficking of Kv1 channels [115–117]. It has been suggested that the preponderance of intracellular Kv1 channels in many tissue types may be due to the interaction of a protein containing a TxD with Kv1 channels, retaining them within the ER [115–117]. This was supported by evidence showing that heterologous over-expression of ER-luminal dendrotoxin (DTX), which presumably competes with this TxD-containing protein, allows these channels to escape the ER and migrate to the cell surface [116]. This resulted in increased levels of cell-surface Kv1 channel, and patch-clamp analyses showed increased Kv1 current amplitudes, suggesting that the DTX does not remain bound during passage to the cell surface [116]. Recent evidence suggests that the TxD-containing protein may be MMP23 [39].
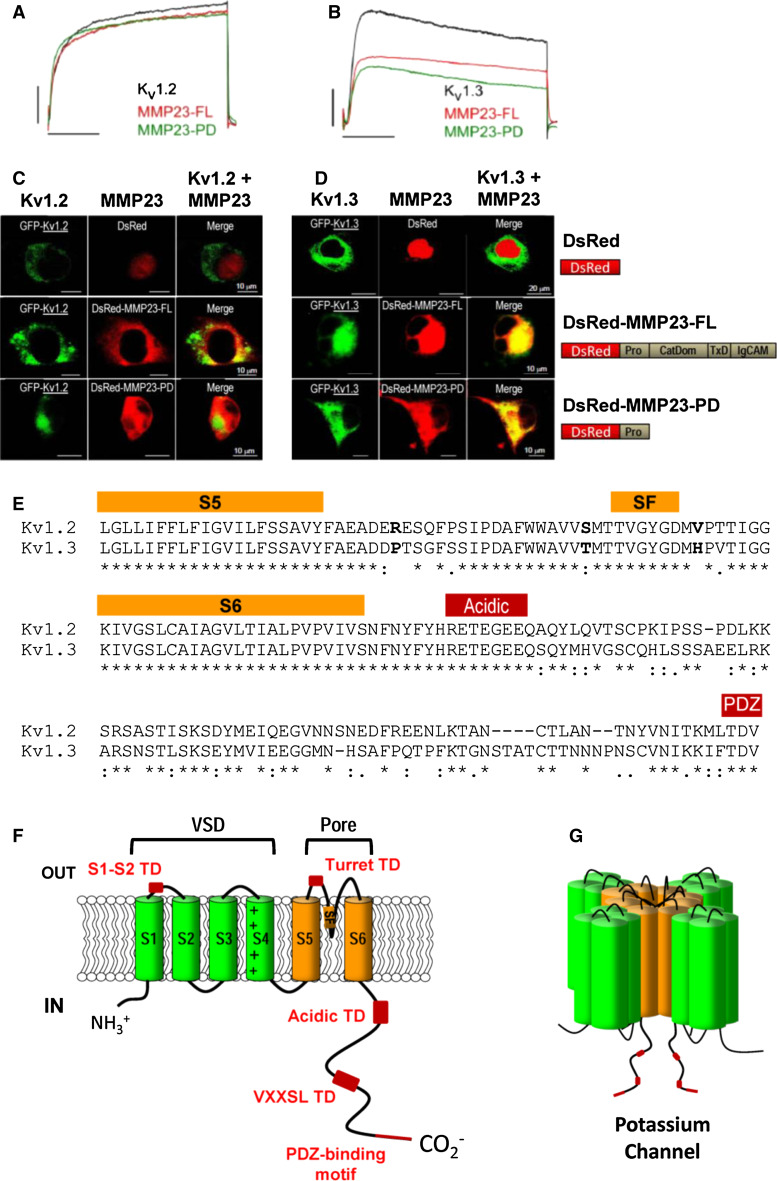
Full-length MMP23, which is found predominantly in perinuclear and ER membranes in mammalian cells [51, 53], has been shown to co-localize with Kv1.3, but not the closely-related Kv1.2 channels, in these organelles (Fig. 10c, d) [38, 39]. This specificity suggests that the observed attenuation in potassium channel activity may result from intracellular retention of the channels upon interaction with MMP23 (Fig. 10a, b) [38, 39].
Fig. 10.
The pro-domain of MMP23 specifically interacts with Kv1.3 and not the closely-related Kv1.2 channel. a, b Whole cell patch-clamp currents showing dose-dependent block of Kv1.2 (a) and Kv1.3 (b) by full-length MMP23 (MMP23-FL) and the pro-domain of MMP23 (MMP23-PD). c, d DsRed-MMP23-FL (full-length) and DsRed-MMP23-PD (pro-domain of MMP23) co-localise with Kv1.3-GFP (d) but not Kv1.2-GFP (c). Adapted from Nguyen et al. [38]. e, f Structural elements involved in Kv channel trafficking. e Sequence alignment of the S5 to C-terminal regions of Kv1.2 (UniProt P16389) and Kv1.3 (P22001) required for the intracellular trapping of Kv channels. Residues corresponding to the S5 and S6 transmembrane helices and specificity filter (SF) are highlighted by an orange rectangle above the sequences while the acidic and PDZ trafficking determinant (TD) motifs are denoted by red rectangles. Sequence alignments were performed using the program ClustalW [174]. f Cartoon representation of a Kv channel where the voltage-sensor domain (VSD) and pore domain TM helices are colored green and orange, respectively. Various TDs are denoted by red boxes. TDs include acidic and VXXSL (where X is any residue) motifs in the unstructured C-terminal tail of the Kv channel [118, 119], an ER retention TD in the turret domain external to the channel pore [115] and a motif in the extracellular loop between the TM helices S1 and S2 [123]. g Cartoon model of the assembled Kv channel where TDs of the C-terminal tail are denoted by red boxes
It was assumed initially that the intracellular retention of Kv1.3 channels was mediated by the MMP23-TxD [39]. However, truncation analyses showed that the pro-domain of MMP23 (MMP23-PD) alone could suppress Kv1.3, although not Kv1.2, currents to the same extent as the full-length protein (Fig. 10a, b) [38]. MMP23-PD and Kv1.3 were shown to be co-localized predominantly in the ER of COS-7 cells, similar to the full-length protein, indicating that the pro-domain of MMP23 mediates the intracellular trapping of Kv1.3 channels (Fig. 10c, d) [38].
Examination of the interaction of MMP23-PD with several Kv1.2/Kv1.3 chimeras showed that the Kv1.3 region from the S5 transmembrane helix to the C-terminal end of the channel is required for MMP23-PD-dependent channel modulation [38]. However, it remains to be determined whether this is mediated by a direct interaction of MMP23-PD with the channel. The sequences of the S5 and S6 transmembrane helices and selectivity filter for Kv1.2 and Kv1.3 are highly homologous, while the C-terminal tail differs between these proteins (Fig. 10e). The unstructured C-terminal tail contains several trafficking determinant (TD) motifs that have been shown to regulate the localization of Kv channels (Fig. 10f, g). These TD motifs include acidic and VXXSL motifs [118, 119] and a PDZ-binding motif at the end of the C-terminal tail that binds to the postsynaptic density 95 protein (PSD95) [120–122]. Other TD motifs include an ER retention TD in the turret domain external to the channel pore [115] and a motif in the extracellular loop between S1 and S2 [123].
Possible mechanisms for the modulation of ion channel activity
Intra- or extracellular helical segments adjacent to TMDs, similar to the juxta-membrane helix α2 (Ala47–Leu58) in the MMP23 pro-domain, are not uncommon [124, 125] and often play a functional role [126]. KCNE1, another potassium channel-modulating protein, contains an α-helical trans-membrane domain linked to two membrane-interacting juxta-membrane helical domains [125]. KCNE1 and the related KCNE2 protein both suppress currents generated by homomeric Kv1.4, Kv3.3, and Kv3.4 channels by trapping them early in the secretory pathway [127, 128]. Furthermore, the closely-related protein KCNE4 suppresses Kv1.3 currents by inducing partial ER retention of the channel [129]. KCNE1 has additional channel-modulating effects on Kv7.1 channels mediated by the C-terminal juxta-membrane helical domain, which is not present in MMP23-PD [130]. These effects are mediated by a direct interaction of the TMD and C-terminal juxta-membrane helical domains of KCNE1 with the Kv channel or other accessory proteins [131]. It is possible that the TMD within MMP23-PD mediates a similar interaction with Kv1.3 channels but this remains to be confirmed.
Proteolytic cleavage by MMP23 may also regulate Kv channel activity. Protease-mediated regulation of ion channel activity can occur either directly by proteolytic cleavage and activation (or inactivation) of the channel [e.g., serine protease cleavage of epithelial sodium channels (ENaC)] [132, 133] or indirectly via cleavage of an upstream regulator of channel activity [e.g., serine protease cleavage of protease-activated receptor 2 (PAR2) leads to the phosphorylation and activation of transient receptor potential (TRP) ion channels] [134, 135].
MMP23 shares several features with voltage-gated sodium channel β1 subunits (NaVβ1). Navβ1 subunits contain an N-terminal extracellular Ig-like domain, a single helical type-I transmembrane domain that anchors the protein to the plasma membrane and a C-terminal flexible intracellular domain [136]. NaVβ1 subunits normally form an integral component of NaV channels and can act as cell adhesion molecules and modulate the cell surface expression of NaV channels, enhancing sodium channel density and cell excitability [137]. However, NaVβ1 subunits can also co-assemble with and modulate the biophysical properties of Kv1 and Kv7 channels, although not Kv3 channels [138]. The extracellular Ig domain of NaVβ1 was required for the acceleration of Kv1.3 channel activation while the full-length protein was required for acceleration of Kv1.2 activity and slowing of Kv1.1 deactivation [138]. These interactions were mediated by an association of the NaVβ1 TM domain with the Kv1 channel pore and voltage-sensor domains, while the Ig domain forms extensive interactions with the S1–S2, S5–P and P–S6 extracellular loops of the channel [138].
Biological function
MMP23 and Kv1.3 exhibit overlapping tissue expression in many tissues where they may interact: lung, heart, uterus, placenta, ovary, testis seminiferous tubules, prostate, intestine, pancreatic islets, cingulated cortex, adrenal cortex, osteoblasts, chondroblasts, cartilage, synovium, natural killer cells, dendritic cells, and tendons [39, 51, 54, 139–146] [see BioGPS Web site (http://biogps.org)].
The physiological function of MMP23 is unknown, although there is evidence it may play a role in various physiological processes. Human MMP23 is expressed predominantly in ovarian, testis and prostate tissues [54]. MMP23 mRNA is also expressed in chrondrocytes and osteoblasts, implying involvement in cartilage and bone formation [139]. ADAM12 and MMP23 have also been linked to inflammatory disorders as they are co-expressed in painful tendinopathy [147]. Human MMP23 is also expressed in the synovium and cartilage [140], and in cranial sutures [148] and human aminochorion [141]. Expression of rat MMP23 occurs mainly in heart, spleen, and lung [52], while expression is spatially and temporally regulated in a cell-type-specific manner during follicular development [51]. In addition, porcine MMP23B was strongly expressed in ovaries and heart [149]. Differential expression of human MMP23 has been reported in breast [142] and prostate [150] cancers, as well as in multiple myelomas [151] and synovial sarcomas [152].
Microarray studies using a zebrafish liver cancer model have shown that mmp23b is significantly down-regulated in liver cancer, whereas other MMPs are up-regulated [153]. More recently it was reported that mmp23b is expressed in hepatocytes during normal development in zebrafish and that mmp23b deficiency leads to a reduction in liver size due to defective hepatocyte proliferation [154]. There is evidence that MMP23B functions through the tumor necrosis factor (TNF) signaling pathway [154]. TNF is a 26-kDa membrane-anchored pro-inflammatory cytokine that is a key regulator of liver homeostasis in mammals [155]. Hepatic TNF is derived mainly from Kupffer cells and regulates liver homeostasis by controlling hepatocyte proliferation and cell death [156, 157]. MMP23B directly interacts with TNF and mediates its release (shedding) from the cell membrane [154]. In vitro immunoprecipitation experiments have demonstrated that human MMP23B binds to human TNF, although it is not known if MMP23B cleaves TNF or whether this is mediated by another protein such as TACE/ADAM17 [TNF-converting enzyme transarterial chemoembolization, a member of the disintegrin family of metalloproteases (ADAMs)] [158].
We have recently shown expression of MMP23 and Kv1.3 in normal colonic epithelium with overlapping staining of both proteins in human colorectal cancers [38]. MMP23-PD may regulate the surface expression of Kv1.3 channels in primary and metastatic colorectal cancer cells and thereby regulate cellular function. Colon cancer cells secreting processed active MMP23 enzyme containing the Kv1.3 channel-blocking toxin domain (TxD) may block Kv1.3 channels on infiltrating anti-tumor T cells [38], and suppress them as a means of immune evasion. Tumor cells would be protected from the MMP23 TxD because their Kv1.3 channels are trapped intracellularly upon interaction with the MMP23 pro-domain. In support of this hypothesis, human malignant melanoma tumors with higher MMP23 expression contain significantly fewer tumor-infiltrating lymphocytes, and are associated with a greater risk of recurrence among patients treated with immune biologics [159]. However, we cannot exclude a role for Kv1.3 in the endoplasmic reticulum of tumors.
Overview and future directions
MMP23 is a unique member of the matrix metalloprotease family of proteins with a distinctive domain architecture and function. Moreover, MMP23 appears to be involved in the regulation of Kv1 channel trafficking.
Kv1 channels are abnormally expressed in cancer cells and have been identified as potential targets for the treatment of various cancers [160–164] and autoimmune diseases [165]. Inhibition of Kv1 channels has been shown to induce Bax/Bak-independent death of cancer cells [166] and down-regulate the activity of disease-associated T cells in various autoimmune diseases [167]. Kv channel activity is regulated by mechanisms that control the flow of ions through the pore as well as the trafficking of channels to the cell surface. Dysfunction of Kv channel trafficking can have detrimental consequences. For example, Kv channels play a fundamental role in determining intrinsic neuronal excitability, and channel trafficking defects can lead to aberrant intrinsic excitability and seizures (such as in epilepsy) [168].
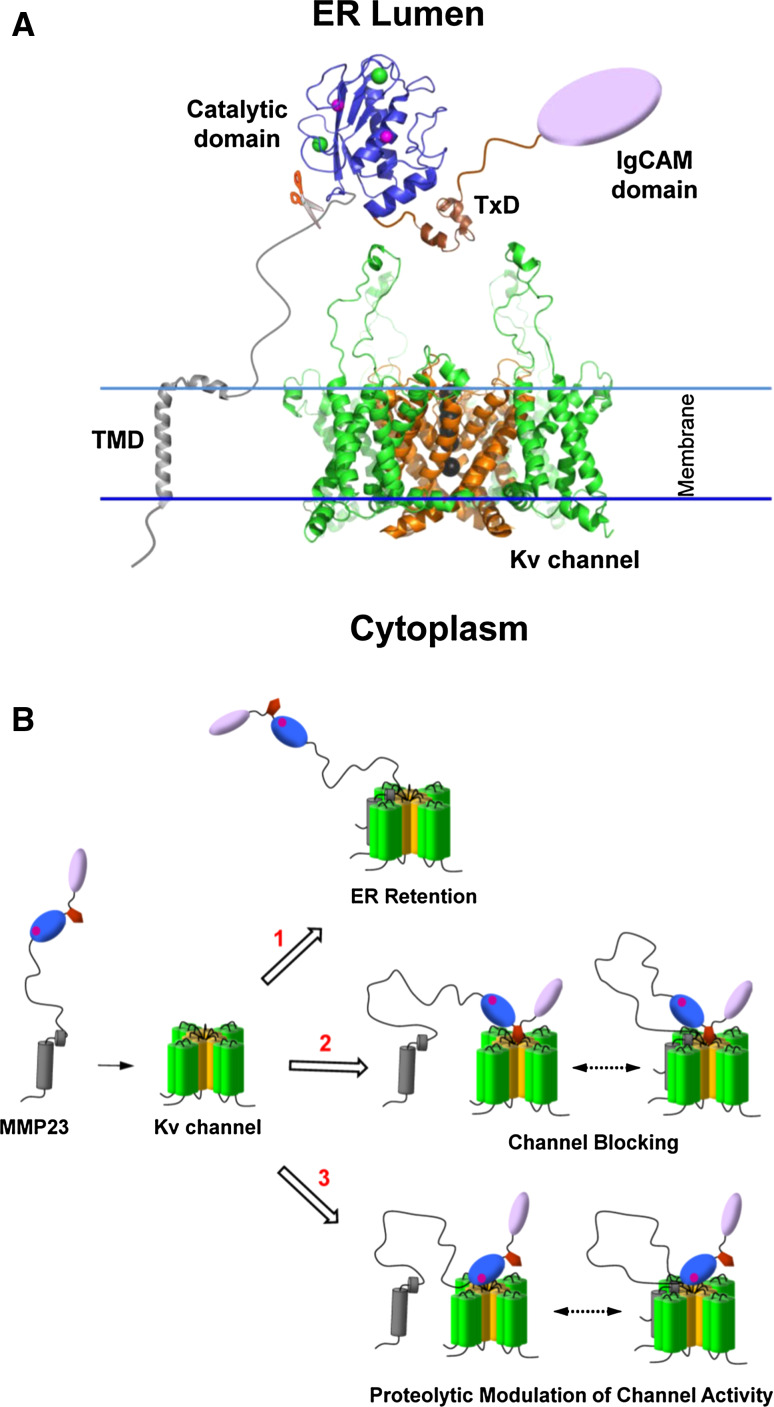
MMP23 has been shown to play a role in regulating Kv1 channel activity (Fig. 11) [38, 39]. The MMP23 pro-domain can regulate Kv1 channel trafficking by trapping them in the ER, although it is not known whether this requires a direct interaction or occurs through other unknown mediators. Moreover, the toxin-like domain of MMP23 can block the Kv1.3 pore, obstructing the passage of potassium ions through the channel, although it remains to be determined whether this occurs intracellularly or following cleavage and secretion of the active enzyme.
Fig. 11.
Possible role of various MMP23 domains in modulating Kv channel activity. a Proposed model of MMP23 and Kv1 channels in the ER. The X-ray crystal structure for Kv1.2 (PDB ID: 3LUT) was used. The Kv1 VSD is colored green and the pore domain is orange. The large flexible S1-S2 loops are shown extending out from the Kv1.2 channel. Importantly, the topology of Kv1.2, particularly in the external vestibule region where toxins bind, is similar to that of other Kv1 channels. Black spheres depict potassium ions within the conduction pore. The TMD of the MMP23 pro-domain (gray) transverses the ER membrane while the juxtamembrane helix (α2) is located in the membrane facing the ER lumen. The metalloprotease domain (blue) is represented by the MMP8 catalytic domain structure. Zinc and calcium ions are depicted by magenta and green spheres, respectively. The MMP23 TxD (brown) is situated directly above the outer vestibule of the Kv1 channel. The IgCAM domain is represented by a purple oval. The position of the furin cleavage motif is indicated by a pair of scissors. Adapted from Rangaraju et al. [39]. b Possible modes of MMP23-mediated regulation of Kv1 channel activity. Modulation of Kv1 channel activity may occur by one or more of following mechanisms either individually or in combination. Domains are colored according to (a) while the catalytic zinc is represented by a magenta sphere. (1) The MMP23 pro-domain regulates intracellular trafficking of Kv1 channels by trapping them in the ER. (2) The MMP23 TxD blocks the Kv1 channel pore, attenuating channel activity. (3) Cleavage of loops on the exposed surface of Kv1 channels may lead to modulation of channel activity. It is possible that channel modulation may also occur indirectly by MMP23-mediated proteolytic activation of another protein which then indirectly modulates Kv1 channel activity (not shown). It is possible that interaction of the MMP23 pro-domain with the Kv1 channel may also be required for (2) and (3)
MMPs degrade a wide range of substrates, including proteins of the ECM, signalling molecules and cell-surface receptors [2, 3, 169, 170]. Although Kv channels have not been identified previously as a substrate for MMPs, it is possible that MMP23 may regulate Kv1 activity either indirectly by cleavage of an upstream regulator of channel activity, e.g., serine protease cleavage of protease-activated receptor 2 (PAR2) leads to the phosphorylation and activation of transient receptor potential (TRP) ion channels [98, 124], while MMP1 and MMP13 have been shown to activate the closely-related protein PAR1 [171]) or directly by cleavage of the channel itself (e.g., serine protease cleavage of epithelial sodium channels) (Fig. 11b) [132, 133, 172].
The IgCAM domain of MMP23 shares homology with Ig-like C2-type domains of the immunoglobulin superfamily, which mediate protein–protein or protein–lipid interactions [88, 89, 91]. The MMP23 IgCAM domain may act in a manner similar to the Ig domain of NaVβ1 subunits by mediating interactions with other proteins or interacting with and modulating the activity of the Kv1.3 channel [137, 138]. Further work is warranted to determine whether the MMP23 IgCAM domain targets substrate proteins analogous to the hemopexin domain of other MMPs or plays a role in modulating intracellular Kv1 trafficking or activity.
MMP23 is widely expressed and is involved in a diverse range of physiological processes [139, 147]. Aberrant MMP23 expression has been found in a number of human cancers [142, 150–152], and we have recently demonstrated the co-localization of MMP23 and Kv1.3 in colon cancer cells [38]. Importantly, Kv1.3 channels have been identified recently as a therapeutic target for the treatment of human cancers [163, 164, 166].
Further studies are now required to (1) identify in vivo binding partners and proteolytic substrates of MMP23, (2) clarify whether the MMP23 pro-domain and MMP23-TxD act independently or cooperatively in binding to Kv1 channels, (3) determine the possible involvement of the IgCAM domain in substrate processing or Kv1 channel modulation, (4) identify MMP23 binding partners that regulate enzyme activity or Kv1 channel trafficking, and (5) define the role (if any) of the N-terminal cytoplasmic tail. Kv1 channels play an essential cellular role and their dysregulation has been implicated in a number of devastating diseases. Characterization of the function and mechanism of MMP23, a unique member of the MMP family, should yield significant insights into physiology and disease.
Acknowledgments
The authors’ work described herein was supported in part by grants from the National Institutes of Health (NIH) NS48252 (K.G.C.), and the Australian Research Council (DP1093909 to R.S.N., B.J.S., and K.G.C.). R.S.N. acknowledges fellowship support from the Australian National Health and Medical Research Council.
References
- 1.Butler GS, Overall CM. Updated biological roles for matrix metalloproteinases and new “intracellular” substrates revealed by degradomics. Biochemistry. 2009;48:10830–10845. doi: 10.1021/bi901656f. [DOI] [PubMed] [Google Scholar]
- 2.Morrison CJ, Butler GS, Rodriguez D, Overall CM. Matrix metalloproteinase proteomics: substrates, targets, and therapy. Curr Opin Cell Biol. 2009;21:645–653. doi: 10.1016/j.ceb.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez D, Morrison CJ, Overall CM. Matrix metalloproteinases: what do they not do? New substrates and biological roles identified by murine models and proteomics. Biochim Biophys Acta. 2010;1803:39–54. doi: 10.1016/j.bbamcr.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 4.Gururajan R, Grenet J, Lahti JM, Kidd VJ. Isolation and characterization of two novel metalloproteinase genes linked to the Cdc2L locus on human chromosome 1p36.3. Genomics. 1998;52:101–106. doi: 10.1006/geno.1998.5401. [DOI] [PubMed] [Google Scholar]
- 5.Gill SE, Parks WC. Metalloproteinases and their inhibitors: regulators of wound healing. Int J Biochem Cell Biol. 2008;40:1334–1347. doi: 10.1016/j.biocel.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mott JD, Werb Z. Regulation of matrix biology by matrix metalloproteinases. Curr Opin Cell Biol. 2004;16:558–564. doi: 10.1016/j.ceb.2004.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagase H, Visse R, Murphy G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc Res. 2006;69:562–573. doi: 10.1016/j.cardiores.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodeling. Nat Rev Mol Cell Biol. 2007;8:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vu TH, Werb Z. Matrix metalloproteinases: effectors of development and normal physiology. Genes Dev. 2000;14:2123–2133. doi: 10.1101/gad.815400. [DOI] [PubMed] [Google Scholar]
- 11.Hadler-Olsen E, Fadnes B, Sylte I, Uhlin-Hansen L, Winberg JO. Regulation of matrix metalloproteinase activity in health and disease. FEBS J. 2011;278:28–45. doi: 10.1111/j.1742-4658.2010.07920.x. [DOI] [PubMed] [Google Scholar]
- 12.Sbardella D, Fasciglione GF, Gioia M, Ciaccio C, Tundo GR, Marini S, Coletta M. Human matrix metalloproteinases: an ubiquitarian class of enzymes involved in several pathological processes. Mol Aspects Med. 2012;33:119–208. doi: 10.1016/j.mam.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 13.Piperi C, Papavassiliou AG. Molecular mechanisms regulating matrix metalloproteinases. Curr Top Med Chem. 2012;12:1095–1112. doi: 10.2174/1568026611208011095. [DOI] [PubMed] [Google Scholar]
- 14.Mannello F, Medda V. Nuclear localization of matrix metalloproteinases. Prog Histochem Cytochem. 2012;47:27–58. doi: 10.1016/j.proghi.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Murphy G, Nagase H. Localizing matrix metalloproteinase activities in the pericellular environment. FEBS J. 2011;278:2–15. doi: 10.1111/j.1742-4658.2010.07918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bourboulia D, Stetler-Stevenson WG. Matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs): positive and negative regulators in tumor cell adhesion. Semin Cancer Biol. 2010;20:161–168. doi: 10.1016/j.semcancer.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brew K, Nagase H. The tissue inhibitors of metalloproteinases (TIMPs): an ancient family with structural and functional diversity. Biochim Biophys Acta. 2010;1803:55–71. doi: 10.1016/j.bbamcr.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu X, Parks WC, Heinecke JW. Activation and silencing of matrix metalloproteinases. Semin Cell Dev Biol. 2008;19:2–13. doi: 10.1016/j.semcdb.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Murphy G. Tissue inhibitors of metalloproteinases. Genome Biol. 2011;12:233. doi: 10.1186/gb-2011-12-11-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gustafsson T. Vascular remodeling in human skeletal muscle. Biochem Soc Trans. 2011;39:1628–1632. doi: 10.1042/BST20110720. [DOI] [PubMed] [Google Scholar]
- 21.Kraiem Z, Korem S. Matrix metalloproteinases and the thyroid. Thyroid. 2000;10:1061–1069. doi: 10.1089/thy.2000.10.1061. [DOI] [PubMed] [Google Scholar]
- 22.Ortega N, Behonick D, Stickens D, Werb Z. How proteases regulate bone morphogenesis. Ann NY Acad Sci. 2003;995:109–116. doi: 10.1111/j.1749-6632.2003.tb03214.x. [DOI] [PubMed] [Google Scholar]
- 23.Parks WC, Shapiro SD. Matrix metalloproteinases in lung biology. Respir Res. 2001;2:10–19. doi: 10.1186/rr33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clutterbuck AL, Asplin KE, Harris P, Allaway D, Mobasheri A. Targeting matrix metalloproteinases in inflammatory conditions. Curr Drug Targets. 2009;10:1245–1254. doi: 10.2174/138945009789753264. [DOI] [PubMed] [Google Scholar]
- 25.Decock J, Thirkettle S, Wagstaff L, Edwards DR. Matrix metalloproteinases: protective roles in cancer. J Cell Mol Med. 2011;15:1254–1265. doi: 10.1111/j.1582-4934.2011.01302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gialeli C, Theocharis AD, Karamanos NK. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011;278:16–27. doi: 10.1111/j.1742-4658.2010.07919.x. [DOI] [PubMed] [Google Scholar]
- 27.Hua H, Li M, Luo T, Yin Y, Jiang Y. Matrix metalloproteinases in tumorigenesis: an evolving paradigm. Cell Mol Life Sci. 2011;68:3853–3868. doi: 10.1007/s00018-011-0763-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korpos E, Wu C, Sorokin L. Multiple roles of the extracellular matrix in inflammation. Curr Pharm Des. 2009;15:1349–1357. doi: 10.2174/138161209787846685. [DOI] [PubMed] [Google Scholar]
- 29.Rucci N, Sanita P, Angelucci A. Roles of metalloproteases in metastatic niche. Curr Mol Med. 2011;11:609–622. doi: 10.2174/156652411797536705. [DOI] [PubMed] [Google Scholar]
- 30.Siasos G, Tousoulis D, Kioufis S, Oikonomou E, Siasou Z, Limperi M, Papavassiliou AG, Stefanadis C. Inflammatory mechanisms in atherosclerosis: the impact of matrix metalloproteinases. Curr Top Med Chem. 2012;12:1132–1148. doi: 10.2174/1568026611208011132. [DOI] [PubMed] [Google Scholar]
- 31.Stellas D, Patsavoudi E. Inhibiting matrix metalloproteinases, an old story with new potentials for cancer treatment. Anticancer Agents Med Chem. 2012;12:707–717. doi: 10.2174/187152012802650246. [DOI] [PubMed] [Google Scholar]
- 32.Aldonyte R, Brantly M, Block E, Patel J, Zhang J. Nuclear localization of active matrix metalloproteinase-2 in cigarette smoke-exposed apoptotic endothelial cells. Exp Lung Res. 2009;35:59–75. doi: 10.1080/01902140802406059. [DOI] [PubMed] [Google Scholar]
- 33.Eguchi T, Kubota S, Kawata K, Mukudai Y, Uehara J, Ohgawara T, Ibaragi S, Sasaki A, Kuboki T, Takigawa M. Novel transcription-factor-like function of human matrix metalloproteinase 3 regulating the CTGF/CCN2 gene. Mol Cell Biol. 2008;28:2391–2413. doi: 10.1128/MCB.01288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ip YC, Cheung ST, Fan ST. Atypical localization of membrane type 1-matrix metalloproteinase in the nucleus is associated with aggressive features of hepatocellular carcinoma. Mol Carcinog. 2007;46:225–230. doi: 10.1002/mc.20270. [DOI] [PubMed] [Google Scholar]
- 35.Kwan JA, Schulze CJ, Wang W, Leon H, Sariahmetoglu M, Sung M, Sawicka J, Sims DE, Sawicki G, Schulz R. Matrix metalloproteinase-2 (MMP-2) is present in the nucleus of cardiac myocytes and is capable of cleaving poly (ADP-ribose) polymerase (PARP) in vitro. FASEB J. 2004;18:690–692. doi: 10.1096/fj.02-1202fje. [DOI] [PubMed] [Google Scholar]
- 36.Limb GA, Matter K, Murphy G, Cambrey AD, Bishop PN, Morris GE, Khaw PT. Matrix metalloproteinase-1 associates with intracellular organelles and confers resistance to lamin A/C degradation during apoptosis. Am J Pathol. 2005;166:1555–1563. doi: 10.1016/S0002-9440(10)62371-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchenko ND, Marchenko GN, Weinreb RN, Lindsey JD, Kyshtoobayeva A, Crawford HC, Strongin AY. Beta-catenin regulates the gene of MMP-26, a novel metalloproteinase expressed both in carcinomas and normal epithelial cells. Int J Biochem Cell Biol. 2004;36:942–956. doi: 10.1016/j.biocel.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen HM, Galea CA, Schmunk G, Smith BJ, Edwards RA, Norton RS, Chandy KG. Intracellular trafficking of the Kv1.3 potassium channel is regulated by the prodomain of a matrix metalloprotease. J Biol Chem. 2013;288:6451–6464. doi: 10.1074/jbc.M112.421495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rangaraju S, Khoo KK, Feng ZP, Crossley G, Nugent D, Khaytin I, Chi V, Pham C, Calabresi P, Pennington MW, Norton RS, Chandy KG. Potassium channel modulation by a toxin domain in matrix metalloprotease 23. J Biol Chem. 2010;285:9124–9136. doi: 10.1074/jbc.M109.071266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Si-Tayeb K, Monvoisin A, Mazzocco C, Lepreux S, Decossas M, Cubel G, Taras D, Blanc JF, Robinson DR, Rosenbaum J. Matrix metalloproteinase 3 is present in the cell nucleus and is involved in apoptosis. Am J Pathol. 2006;169:1390–1401. doi: 10.2353/ajpath.2006.060005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W, Schulze CJ, Suarez-Pinzon WL, Dyck JR, Sawicki G, Schulz R. Intracellular action of matrix metalloproteinase-2 accounts for acute myocardial ischemia and reperfusion injury. Circulation. 2002;106:1543–1549. doi: 10.1161/01.cir.0000028818.33488.7b. [DOI] [PubMed] [Google Scholar]
- 42.Yang Y, Candelario-Jalil E, Thompson JF, Cuadrado E, Estrada EY, Rosell A, Montaner J, Rosenberg GA. Increased intranuclear matrix metalloproteinase activity in neurons interferes with oxidative DNA repair in focal cerebral ischemia. J Neurochem. 2010;112:134–149. doi: 10.1111/j.1471-4159.2009.06433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao YG, Xiao AZ, Newcomer RG, Park HI, Kang T, Chung LW, Swanson MG, Zhau HE, Kurhanewicz J, Sang QX. Activation of pro-gelatinase B by endometase/matrilysin-2 promotes invasion of human prostate cancer cells. J Biol Chem. 2003;278:15056–15064. doi: 10.1074/jbc.M210975200. [DOI] [PubMed] [Google Scholar]
- 44.Bode W, Maskos K (2011) Matrix metalloproteinases. In: Encyclopedia of inorganic and bioinorganic chemistry. Wiley, New York. doi:10.1002/9781119951438.eibc0495
- 45.Klein T, Bischoff R. Physiology and pathophysiology of matrix metalloproteases. Amino Acids. 2011;41:271–290. doi: 10.1007/s00726-010-0689-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zitka O, Kukacka J, Krizkova S, Huska D, Adam V, Masarik M, Prusa R, Kizek R. Matrix metalloproteinases. Curr Med Chem. 2010;17:3751–3768. doi: 10.2174/092986710793213724. [DOI] [PubMed] [Google Scholar]
- 47.Huxley-Jones J, Clarke TK, Beck C, Toubaris G, Robertson DL, Boot-Handford RP. The evolution of the vertebrate metzincins; insights from Ciona intestinalis and Danio rerio . BMC Evol Biol. 2007;7:63. doi: 10.1186/1471-2148-7-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fanjul-Fernandez M, Folgueras AR, Cabrera S, Lopez-Otin C. Matrix metalloproteinases: evolution, gene regulation and functional analysis in mouse models. Biochim Biophys Acta. 2010;1803:3–19. doi: 10.1016/j.bbamcr.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 49.Van Wart HE, Birkedal-Hansen H. The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc Natl Acad Sci USA. 1990;87:5578–5582. doi: 10.1073/pnas.87.14.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ra HJ, Parks WC. Control of matrix metalloproteinase catalytic activity. Matrix Biol. 2007;26:587–596. doi: 10.1016/j.matbio.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohnishi J, Ohnishi E, Jin M, Hirano W, Nakane D, Matsui H, Kimura A, Sawa H, Nakayama K, Shibuya H, Nagashima K, Takahashi T. Cloning and characterization of a rat ortholog of MMP-23 (matrix metalloproteinase-23), a unique type of membrane-anchored matrix metalloproteinase and conditioned switching of its expression during the ovarian follicular development. Mol Endocrinol. 2001;15:747–764. doi: 10.1210/mend.15.5.0638. [DOI] [PubMed] [Google Scholar]
- 52.Pei D. CA-MMP: a matrix metalloproteinase with a novel cysteine array, but without the classic cysteine switch. FEBS Lett. 1999;457:262–270. doi: 10.1016/s0014-5793(99)01046-7. [DOI] [PubMed] [Google Scholar]
- 53.Pei D, Kang T, Qi H. Cysteine array matrix metalloproteinase (CA-MMP)/MMP-23 is a type II transmembrane matrix metalloproteinase regulated by a single cleavage for both secretion and activation. J Biol Chem. 2000;275:33988–33997. doi: 10.1074/jbc.M006493200. [DOI] [PubMed] [Google Scholar]
- 54.Velasco G, Pendas AM, Fueyo A, Knauper V, Murphy G, Lopez-Otin C. Cloning and characterization of human MMP-23, a new matrix metalloproteinase predominantly expressed in reproductive tissues and lacking conserved domains in other family members. J Biol Chem. 1999;274:4570–4576. doi: 10.1074/jbc.274.8.4570. [DOI] [PubMed] [Google Scholar]
- 55.Sounni NE, Noel A. Membrane type-matrix metalloproteinases and tumor progression. Biochimie. 2005;87:329–342. doi: 10.1016/j.biochi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 56.Zucker S, Pei D, Cao J, Lopez-Otin C. Membrane type-matrix metalloproteinases (MT-MMP) Curr Top Dev Biol. 2003;54:1–74. doi: 10.1016/s0070-2153(03)54004-2. [DOI] [PubMed] [Google Scholar]
- 57.Koziol A, Martin-Alonso M, Clemente C, Gonzalo P, Arroyo AG. Site-specific cellular functions of MT1-MMP. Eur J Cell Biol. 2012;91:889–895. doi: 10.1016/j.ejcb.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 58.Labrecque L, Nyalendo C, Langlois S, Durocher Y, Roghi C, Murphy G, Gingras D, Beliveau R. Src-mediated tyrosine phosphorylation of caveolin-1 induces its association with membrane type 1 matrix metalloproteinase. J Biol Chem. 2004;279:52132–52140. doi: 10.1074/jbc.M409617200. [DOI] [PubMed] [Google Scholar]
- 59.Artym VV, Zhang Y, Seillier-Moiseiwitsch F, Yamada KM, Mueller SC. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 2006;66:3034–3043. doi: 10.1158/0008-5472.CAN-05-2177. [DOI] [PubMed] [Google Scholar]
- 60.Cauwe B, Van den Steen PE, Opdenakker G. The biochemical, biological, and pathological kaleidoscope of cell surface substrates processed by matrix metalloproteinases. Crit Rev Biochem Mol Biol. 2007;42:113–185. doi: 10.1080/10409230701340019. [DOI] [PubMed] [Google Scholar]
- 61.Gingras D, Michaud M, Di Tomasso G, Beliveau E, Nyalendo C, Beliveau R. Sphingosine-1-phosphate induces the association of membrane-type 1 matrix metalloproteinase with p130Cas in endothelial cells. FEBS Lett. 2008;582:399–404. doi: 10.1016/j.febslet.2007.12.029. [DOI] [PubMed] [Google Scholar]
- 62.Gonzalo P, Guadamillas MC, Hernandez-Riquer MV, Pollan A, Grande-Garcia A, Bartolome RA, Vasanji A, Ambrogio C, Chiarle R, Teixido J, Risteli J, Apte SS, del Pozo MA, Arroyo AG. MT1-MMP is required for myeloid cell fusion via regulation of Rac1 signaling. Dev Cell. 2010;18:77–89. doi: 10.1016/j.devcel.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roghi C, Jones L, Gratian M, English WR, Murphy G. Golgi reassembly stacking protein 55 interacts with membrane-type (MT) 1-matrix metalloprotease (MMP) and furin and plays a role in the activation of the MT1-MMP zymogen. FEBS J. 2010;277:3158–3175. doi: 10.1111/j.1742-4658.2010.07723.x. [DOI] [PubMed] [Google Scholar]
- 64.Sakamoto T, Seiki M. A membrane protease regulates energy production in macrophages by activating hypoxia-inducible factor-1 via a non-proteolytic mechanism. J Biol Chem. 2010;285:29951–29964. doi: 10.1074/jbc.M110.132704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith-Pearson PS, Greuber EK, Yogalingam G, Pendergast AM. Abl kinases are required for invadopodia formation and chemokine-induced invasion. J Biol Chem. 2010;285:40201–40211. doi: 10.1074/jbc.M110.147330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tapia T, Ottman R, Chakrabarti R. LIM kinase1 modulates function of membrane type matrix metalloproteinase 1: implication in invasion of prostate cancer cells. Mol Cancer. 2011;10:6. doi: 10.1186/1476-4598-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Knauper V, Kramer S, Reinke H, Tschesche H. Characterization and activation of procollagenase from human polymorphonuclear leucocytes. N-terminal sequence determination of the proenzyme and various proteolytically activated forms. Eur J Biochem. 1990;189:295–300. doi: 10.1111/j.1432-1033.1990.tb15489.x. [DOI] [PubMed] [Google Scholar]
- 68.Nagase H, Suzuki K, Enghild JJ, Salvesen G. Stepwise activation mechanisms of the precursors of matrix metalloproteinases 1 (tissue collagenase) and 3 (stromelysin) Biomed Biochim Acta. 1991;50:749–754. [PubMed] [Google Scholar]
- 69.Nagase H, Suzuki K, Morodomi T, Enghild JJ, Salvesen G. Activation mechanisms of the precursors of matrix metalloproteinases 1, 2 and 3. Matrix Suppl. 1992;1:237–244. [PubMed] [Google Scholar]
- 70.Springman EB, Angleton EL, Birkedal-Hansen H, Van Wart HE. Multiple modes of activation of latent human fibroblast collagenase: evidence for the role of a Cys73 active-site zinc complex in latency and a “cysteine switch” mechanism for activation. Proc Natl Acad Sci USA. 1990;87:364–368. doi: 10.1073/pnas.87.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li SS. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J. 2005;390:641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saksela K, Permi P. SH3 domain ligand binding: what’s the consensus and where’s the specificity? FEBS Lett. 2012;586:2609–2614. doi: 10.1016/j.febslet.2012.04.042. [DOI] [PubMed] [Google Scholar]
- 73.Gomis-Ruth FX, Trillo-Muyo S, Stocker W. Functional and structural insights into astacin metallopeptidases. Biol Chem. 2012;393:1027–1041. doi: 10.1515/hsz-2012-0149. [DOI] [PubMed] [Google Scholar]
- 74.Mohrlen F, Hutter H, Zwilling R. The astacin protein family in Caenorhabditis elegans . Eur J Biochem. 2003;270:4909–4920. doi: 10.1046/j.1432-1033.2003.03891.x. [DOI] [PubMed] [Google Scholar]
- 75.Rachamim T, Sher D. What Hydra can teach us about chemical ecology—how a simple, soft organism survives in a hostile aqueous environment. Int J Dev Biol. 2012;56:605–611. doi: 10.1387/ijdb.113474tr. [DOI] [PubMed] [Google Scholar]
- 76.Guevara T, Yiallouros I, Kappelhoff R, Bissdorf S, Stocker W, Gomis-Ruth FX. Proenzyme structure and activation of astacin metallopeptidase. J Biol Chem. 2010;285:13958–13965. doi: 10.1074/jbc.M109.097436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maskos K. Crystal structures of MMPs in complex with physiological and pharmacological inhibitors. Biochimie. 2005;87:249–263. doi: 10.1016/j.biochi.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 78.Tallant C, Marrero A, Gomis-Ruth FX. Matrix metalloproteinases: fold and function of their catalytic domains. Biochim Biophys Acta. 2010;1803:20–28. doi: 10.1016/j.bbamcr.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 79.Andreini C, Banci L, Bertini I, Luchinat C, Rosato A. Bioinformatic comparison of structures and homology-models of matrix metalloproteinases. J Proteome Res. 2004;3:21–31. doi: 10.1021/pr0340476. [DOI] [PubMed] [Google Scholar]
- 80.Bode W, Gomis-Rüth F-X, Stöckler W. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the ‘metzincins’. FEBS Lett. 1993;331:134–140. doi: 10.1016/0014-5793(93)80312-i. [DOI] [PubMed] [Google Scholar]
- 81.Stocker W, Grams F, Baumann U, Reinemer P, Gomis-Ruth FX, McKay DB, Bode W. The metzincins–topological and sequential relations between the astacins, adamalysins, serralysins, and matrixins (collagenases) define a superfamily of zinc-peptidases. Protein Sci. 1995;4:823–840. doi: 10.1002/pro.5560040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Steffensen B, Wallon UM, Overall CM. Extracellular matrix binding properties of recombinant fibronectin type II-like modules of human 72-kDa gelatinase/type IV collagenase. High affinity binding to native type I collagen but not native type IV collagen. J Biol Chem. 1995;270:11555–11566. doi: 10.1074/jbc.270.19.11555. [DOI] [PubMed] [Google Scholar]
- 83.Lambert E, Dasse E, Haye B, Petitfrere E. TIMPs as multifacial proteins. Crit Rev Oncol Hematol. 2004;49:187–198. doi: 10.1016/j.critrevonc.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 84.Marrero A, Duquerroy S, Trapani S, Goulas T, Guevara T, Andersen GR, Navaza J, Sottrup-Jensen L, Gomis-Ruth FX. The crystal structure of human α2-macroglobulin reveals a unique molecular cage. Angew Chem Int Ed Engl. 2012;51:3340–3344. doi: 10.1002/anie.201108015. [DOI] [PubMed] [Google Scholar]
- 85.Sottrup-Jensen L. α-macroglobulins: structure, shape, and mechanism of proteinase complex formation. J Biol Chem. 1989;264:11539–11542. [PubMed] [Google Scholar]
- 86.Strickland DK, Ashcom JD, Williams S, Burgess WH, Migliorini M, Argraves WS. Sequence identity between the alpha 2-macroglobulin receptor and low density lipoprotein receptor-related protein suggests that this molecule is a multifunctional receptor. J Biol Chem. 1990;265:17401–17404. [PubMed] [Google Scholar]
- 87.Willenbrock F, Thomas DA, Amour A. Kinetic analysis of the inhibition of matrix metalloproteinases: lessons from the study of tissue inhibitors of metalloproteinases. Methods Mol Biol. 2010;622:435–450. doi: 10.1007/978-1-60327-299-5_25. [DOI] [PubMed] [Google Scholar]
- 88.Murphy G, Knauper V. Relating matrix metalloproteinase structure to function: why the “hemopexin” domain? Matrix Biol. 1997;15:511–518. doi: 10.1016/s0945-053x(97)90025-1. [DOI] [PubMed] [Google Scholar]
- 89.Bode W. A helping hand for collagenases: the haemopexin-like domain. Structure. 1995;3:527–530. doi: 10.1016/s0969-2126(01)00185-x. [DOI] [PubMed] [Google Scholar]
- 90.Clark IM, Cawston TE. Fragments of human fibroblast collagenase. Purification and characterization. Biochem J. 1989;263:201–206. doi: 10.1042/bj2630201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Overall CM. Molecular determinants of metalloproteinase substrate specificity: matrix metalloproteinase substrate binding domains, modules, and exosites. Mol Biotechnol. 2002;22:51–86. doi: 10.1385/MB:22:1:051. [DOI] [PubMed] [Google Scholar]
- 92.Chung L, Dinakarpandian D, Yoshida N, Lauer-Fields JL, Fields GB, Visse R, Nagase H. Collagenase unwinds triple-helical collagen prior to peptide bond hydrolysis. EMBO J. 2004;23:3020–3030. doi: 10.1038/sj.emboj.7600318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer. 2004;4:118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- 94.Hohenester E. Structural insight into Slit-Robo signalling. Biochem Soc Trans. 2008;36:251–256. doi: 10.1042/BST0360251. [DOI] [PubMed] [Google Scholar]
- 95.Morlot C, Thielens NM, Ravelli RB, Hemrika W, Romijn RA, Gros P, Cusack S, McCarthy AA. Structural insights into the Slit-Robo complex. Proc Natl Acad Sci USA. 2007;104:14923–14928. doi: 10.1073/pnas.0705310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rasmussen KK, Kulahin N, Kristensen O, Poulsen JC, Sigurskjold BW, Kastrup JS, Berezin V, Bock E, Walmod PS, Gajhede M. Crystal structure of the Ig1 domain of the neural cell adhesion molecule NCAM2 displays domain swapping. J Mol Biol. 2008;382:1113–1120. doi: 10.1016/j.jmb.2008.07.084. [DOI] [PubMed] [Google Scholar]
- 97.Sanchez-Arrones L, Cardozo M, Nieto-Lopez F, Bovolenta P. Cdon and Boc: two transmembrane proteins implicated in cell-cell communication. Int J Biochem Cell Biol. 2012;44:698–702. doi: 10.1016/j.biocel.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 98.Law RH, Lukoyanova N, Voskoboinik I, Caradoc-Davies TT, Baran K, Dunstone MA, D’Angelo ME, Orlova EV, Coulibaly F, Verschoor S, Browne KA, Ciccone A, Kuiper MJ, Bird PI, Trapani JA, Saibil HR, Whisstock JC. The structural basis for membrane binding and pore formation by lymphocyte perforin. Nature. 2010;468:447–451. doi: 10.1038/nature09518. [DOI] [PubMed] [Google Scholar]
- 99.Lopez JA, Brennan AJ, Whisstock JC, Voskoboinik I, Trapani JA. Protecting a serial killer: pathways for perforin trafficking and self-defence ensure sequential target cell death. Trends Immunol. 2012;33:406–412. doi: 10.1016/j.it.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 100.Voskoboinik I, Dunstone MA, Baran K, Whisstock JC, Trapani JA. Perforin: structure, function, and role in human immunopathology. Immunol Rev. 2010;235:35–54. doi: 10.1111/j.0105-2896.2010.00896.x. [DOI] [PubMed] [Google Scholar]
- 101.Faraco J, Bashir M, Rosenbloom J, Francke U. Characterization of the human gene for microfibril-associated glycoprotein (MFAP2), assignment to chromosome 1p36.1-p35, and linkage to D1S170. Genomics. 1995;25:630–637. doi: 10.1016/0888-7543(95)80004-6. [DOI] [PubMed] [Google Scholar]
- 102.Tsang SW, Nguyen CQ, Hall DH, Chow KL. mab-7 encodes a novel transmembrane protein that orchestrates sensory ray morphogenesis in C. elegans . Dev Biol. 2007;312:353–366. doi: 10.1016/j.ydbio.2007.09.037. [DOI] [PubMed] [Google Scholar]
- 103.Pan T, Groger H, Schmid V, Spring J. A toxin homology domain in an astacin-like metalloproteinase of the jellyfish Podocoryne carnea with a dual role in digestion and development. Dev Genes Evol. 1998;208:259–266. doi: 10.1007/s004270050180. [DOI] [PubMed] [Google Scholar]
- 104.Yan L, Fei K, Zhang J, Dexter S, Sarras MP., Jr Identification and characterization of Hydra metalloproteinase 2 (HMP2): a meprin-like astacin metalloproteinase that functions in foot morphogenesis. Development. 2000;127:129–141. doi: 10.1242/dev.127.1.129. [DOI] [PubMed] [Google Scholar]
- 105.Cotton J, Crest M, Bouet F, Alessandri N, Gola M, Forest E, Karlsson E, Castaneda O, Harvey AL, Vita C, Menez A. A potassium-channel toxin from the sea anemone Bunodosoma granulifera, an inhibitor for Kv1 channels. Revision of the amino acid sequence, disulfide-bridge assignment, chemical synthesis, and biological activity. Eur J Biochem. 1997;244:192–202. doi: 10.1111/j.1432-1033.1997.00192.x. [DOI] [PubMed] [Google Scholar]
- 106.Norton RS. Structures of sea anemone toxins. Toxicon. 2009;54:1075–1088. doi: 10.1016/j.toxicon.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 107.Tudor JE, Pallaghy PK, Pennington MW, Norton RS. Solution structure of ShK toxin, a novel potassium channel inhibitor from a sea anemone. Nat Struct Biol. 1996;3:317–320. doi: 10.1038/nsb0496-317. [DOI] [PubMed] [Google Scholar]
- 108.Kalman K, Pennington MW, Lanigan MD, Nguyen A, Rauer H, Mahnir V, Paschetto K, Kem WR, Grissmer S, Gutman GA, Christian EP, Cahalan MD, Norton RS, Chandy KG. ShK-Dap22, a potent Kv1.3-specific immunosuppressive polypeptide. J Biol Chem. 1998;273:32697–32707. doi: 10.1074/jbc.273.49.32697. [DOI] [PubMed] [Google Scholar]
- 109.Tudor JE, Pennington MW, Norton RS. Ionisation behaviour and solution properties of the potassium-channel blocker ShK toxin. Eur J Biochem. 1998;251:133–141. doi: 10.1046/j.1432-1327.1998.2510133.x. [DOI] [PubMed] [Google Scholar]
- 110.Lanigan MD, Kalman K, Lefievre Y, Pennington MW, Chandy KG, Norton RS. Mutating a critical lysine in ShK toxin alters its binding configuration in the pore-vestibule region of the voltage-gated potassium channel, Kv1.3. Biochemistry. 2002;41:11963–11971. doi: 10.1021/bi026400b. [DOI] [PubMed] [Google Scholar]
- 111.Pennington MW, Harunur Rashid M, Tajhya RB, Beeton C, Kuyucak S, Norton RS. A C-terminally amidated analogue of ShK is a potent and selective blocker of the voltage-gated potassium channel Kv1.3. FEBS Lett. 2012;586:3996–4001. doi: 10.1016/j.febslet.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Alessandri-Haber N, Lecoq A, Gasparini S, Grangier-Macmath G, Jacquet G, Harvey AL, de Medeiros C, Rowan EG, Gola M, Menez A, Crest M. Mapping the functional anatomy of BgK on Kv1.1, Kv1.2, and Kv1.3. Clues to design analogs with enhanced selectivity. J Biol Chem. 1999;274:35653–35661. doi: 10.1074/jbc.274.50.35653. [DOI] [PubMed] [Google Scholar]
- 113.Rauer H, Pennington M, Cahalan M, Chandy KG. Structural conservation of the pores of calcium-activated and voltage-gated potassium channels determined by a sea anemone toxin. J Biol Chem. 1999;274:21885–21892. doi: 10.1074/jbc.274.31.21885. [DOI] [PubMed] [Google Scholar]
- 114.Dauplais M, Lecoq A, Song J, Cotton J, Jamin N, Gilquin B, Roumestand C, Vita C, de Medeiros CL, Rowan EG, Harvey AL, Menez A. On the convergent evolution of animal toxins. Conservation of a diad of functional residues in potassium channel-blocking toxins with unrelated structures. J Biol Chem. 1997;272:4302–4309. doi: 10.1074/jbc.272.7.4302. [DOI] [PubMed] [Google Scholar]
- 115.Manganas LN, Wang Q, Scannevin RH, Antonucci DE, Rhodes KJ, Trimmer JS. Identification of a trafficking determinant localized to the Kv1 potassium channel pore. Proc Natl Acad Sci USA. 2001;98:14055–14059. doi: 10.1073/pnas.241403898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vacher H, Mohapatra DP, Misonou H, Trimmer JS. Regulation of Kv1 channel trafficking by the mamba snake neurotoxin dendrotoxin K. FASEB J. 2007;21:906–914. doi: 10.1096/fj.06-7229com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhu J, Watanabe I, Gomez B, Thornhill WB. Determinants involved in Kv1 potassium channel folding in the endoplasmic reticulum, glycosylation in the Golgi, and cell surface expression. J Biol Chem. 2001;276:39419–39427. doi: 10.1074/jbc.M107399200. [DOI] [PubMed] [Google Scholar]
- 118.Li D, Takimoto K, Levitan ES. Surface expression of Kv1 channels is governed by a C-terminal motif. J Biol Chem. 2000;275:11597–11602. doi: 10.1074/jbc.275.16.11597. [DOI] [PubMed] [Google Scholar]
- 119.Manganas LN, Akhtar S, Antonucci DE, Campomanes CR, Dolly JO, Trimmer JS. Episodic ataxia type-1 mutations in the Kv1.1 potassium channel display distinct folding and intracellular trafficking properties. J Biol Chem. 2001;276:49427–49434. doi: 10.1074/jbc.M109325200. [DOI] [PubMed] [Google Scholar]
- 120.Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 121.Magidovich E, Orr I, Fass D, Abdu U, Yifrach O. Intrinsic disorder in the C-terminal domain of the Shaker voltage-activated K+ channel modulates its interaction with scaffold proteins. Proc Natl Acad Sci USA. 2007;104:13022–13027. doi: 10.1073/pnas.0704059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ogawa Y, Horresh I, Trimmer JS, Bredt DS, Peles E, Rasband MN. Postsynaptic density-93 clusters Kv1 channels at axon initial segments independently of Caspr2. J Neurosci. 2008;28:5731–5739. doi: 10.1523/JNEUROSCI.4431-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.McKeown L, Burnham MP, Hodson C, Jones OT. Identification of an evolutionarily conserved extracellular threonine residue critical for surface expression and its potential coupling of adjacent voltage-sensing and gating domains in voltage-gated potassium channels. J Biol Chem. 2008;283:30421–30432. doi: 10.1074/jbc.M708921200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Klammt C, Maslennikov I, Bayrhuber M, Eichmann C, Vajpai N, Chiu EJ, Blain KY, Esquivies L, Kwon JH, Balana B, Pieper U, Sali A, Slesinger PA, Kwiatkowski W, Riek R, Choe S. Facile backbone structure determination of human membrane proteins by NMR spectroscopy. Nat Methods. 2012;9:834–839. doi: 10.1038/nmeth.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Van Horn WD, Vanoye CG, Sanders CR. Working model for the structural basis for KCNE1 modulation of the KCNQ1 potassium channel. Curr Opin Struct Biol. 2011;21:283–291. doi: 10.1016/j.sbi.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Barrett PJ, Song Y, Van Horn WD, Hustedt EJ, Schafer JM, Hadziselimovic A, Beel AJ, Sanders CR. The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science. 2012;336:1168–1171. doi: 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kanda VA, Abbott GW. KCNE regulation of K+ channel trafficking—a sisyphean task? Front Physiol. 2012;3:231. doi: 10.3389/fphys.2012.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kanda VA, Lewis A, Xu X, Abbott GW. KCNE1 and KCNE2 provide a checkpoint governing voltage-gated potassium channel alpha-subunit composition. Biophys J. 2011;101:1364–1375. doi: 10.1016/j.bpj.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sole L, Roura-Ferrer M, Perez-Verdaguer M, Oliveras A, Calvo M, Fernandez-Fernandez JM, Felipe A. KCNE4 suppresses Kv1.3 currents by modulating trafficking, surface expression and channel gating. J Cell Sci. 2009;122:3738–3748. doi: 10.1242/jcs.056689. [DOI] [PubMed] [Google Scholar]
- 130.McCrossan ZA, Abbott GW. The MinK-related peptides. Neuropharmacology. 2004;47:787–821. doi: 10.1016/j.neuropharm.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 131.Tapper AR, George AL., Jr MinK subdomains that mediate modulation of and association with KvLQT1. J Gen Physiol. 2000;116:379–390. doi: 10.1085/jgp.116.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Haerteis S, Krappitz M, Bertog M, Krappitz A, Baraznenok V, Henderson I, Lindstrom E, Murphy JE, Bunnett NW, Korbmacher C. Proteolytic activation of the epithelial sodium channel (ENaC) by the cysteine protease cathepsin-S. Pflugers Arch. 2012;464:353–365. doi: 10.1007/s00424-012-1138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kitamura K, Tomita K. Proteolytic activation of the epithelial sodium channel and therapeutic application of a serine protease inhibitor for the treatment of salt-sensitive hypertension. Clin Exp Nephrol. 2012;16:44–48. doi: 10.1007/s10157-011-0506-1. [DOI] [PubMed] [Google Scholar]
- 134.Liedtke W. Molecular mechanisms of TRPV4-mediated neural signaling. Ann NY Acad Sci. 2008;1144:42–52. doi: 10.1196/annals.1418.012. [DOI] [PubMed] [Google Scholar]
- 135.Poole DP, Amadesi S, Veldhuis NA, Abogadie FC, Lieu T, Darby W, Liedtke W, Lew MJ, McIntyre P, Bunnett NW. Protease-activated receptor 2 (PAR2) protein and transient receptor potential vanilloid 4 (TRPV4) protein coupling is required for sustained inflammatory signaling. J Biol Chem. 2013;288:5790–5802. doi: 10.1074/jbc.M112.438184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hanlon MR, Wallace BA. Structure and function of voltage-dependent ion channel regulatory beta subunits. Biochemistry. 2002;41:2886–2894. doi: 10.1021/bi0119565. [DOI] [PubMed] [Google Scholar]
- 137.Patino GA, Isom LL. Electrophysiology and beyond: multiple roles of Na+ channel beta subunits in development and disease. Neurosci Lett. 2010;486:53–59. doi: 10.1016/j.neulet.2010.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nguyen HM, Miyazaki H, Hoshi N, Smith BJ, Nukina N, Goldin AL, Chandy KG. Modulation of voltage-gated K+ channels by the sodium channel beta1 subunit. Proc Natl Acad Sci USA. 2012;109:18577–18582. doi: 10.1073/pnas.1209142109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Clancy BM, Johnson JD, Lambert AJ, Rezvankhah S, Wong A, Resmini C, Feldman JL, Leppanen S, Pittman DD. A gene expression profile for endochondral bone formation: oligonucleotide microarrays establish novel connections between known genes and BMP-2-induced bone formation in mouse quadriceps. Bone. 2003;33:46–63. doi: 10.1016/s8756-3282(03)00116-9. [DOI] [PubMed] [Google Scholar]
- 140.Davidson RK, Waters JG, Kevorkian L, Darrah C, Cooper A, Donell ST, Clark IM. Expression profiling of metalloproteinases and their inhibitors in synovium and cartilage. Arthr Res Ther. 2006;8:R124. doi: 10.1186/ar2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fortunato SJ, Menon R. Screening of novel matrix metalloproteinases (MMPs) in human fetal membranes. J Assist Reprod Genet. 2002;19:483–486. doi: 10.1023/A:1020362519981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hegedus L, Cho H, Xie X, Eliceiri GL. Additional MDA-MB-231 breast cancer cell matrix metalloproteinases promote invasiveness. J Cell Physiol. 2008;216:480–485. doi: 10.1002/jcp.21417. [DOI] [PubMed] [Google Scholar]
- 143.Jones GC, Corps AN, Pennington CJ, Clark IM, Edwards DR, Bradley MM, Hazleman BL, Riley GP. Expression profiling of metalloproteinases and tissue inhibitors of metalloproteinases in normal and degenerate human achilles tendon. Arthr Rheum. 2006;54:832–842. doi: 10.1002/art.21672. [DOI] [PubMed] [Google Scholar]
- 144.Okada A, Okada Y. Progress of research in osteoarthritis. Metalloproteinases in osteoarthritis. Clin Calcium. 2009;19:1593–1601. [PubMed] [Google Scholar]
- 145.Riddick AC, Shukla CJ, Pennington CJ, Bass R, Nuttall RK, Hogan A, Sethia KK, Ellis V, Collins AT, Maitland NJ, Ball RY, Edwards DR. Identification of degradome components associated with prostate cancer progression by expression analysis of human prostatic tissues. Br J Cancer. 2005;92:2171–2180. doi: 10.1038/sj.bjc.6602630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Scrideli CA, Carlotti CG, Jr, Okamoto OK, Andrade VS, Cortez MA, Motta FJ, Lucio-Eterovic AK, Neder L, Rosemberg S, Oba-Shinjo SM, Marie SK, Tone LG. Gene expression profile analysis of primary glioblastomas and non-neoplastic brain tissue: identification of potential target genes by oligonucleotide microarray and real-time quantitative PCR. J Neurooncol. 2008;88:281–291. doi: 10.1007/s11060-008-9579-4. [DOI] [PubMed] [Google Scholar]
- 147.Riley G. Tendinopathy—from basic science to treatment. Nat Clin Pract Rheumatol. 2008;4:82–89. doi: 10.1038/ncprheum0700. [DOI] [PubMed] [Google Scholar]
- 148.Gajecka M, Yu W, Ballif BC, Glotzbach CD, Bailey KA, Shaw CA, Kashork CD, Heilstedt HA, Ansel DA, Theisen A, Rice R, Rice DP, Shaffer LG. Delineation of mechanisms and regions of dosage imbalance in complex rearrangements of 1p36 leads to a putative gene for regulation of cranial suture closure. Eur J Hum Genet. 2005;13:139–149. doi: 10.1038/sj.ejhg.5201302. [DOI] [PubMed] [Google Scholar]
- 149.Zhao S, Zhao Y, Niu P, Wang N, Tang Z, Zan L, Li K. Molecular characterization of porcine MMP19 and MMP23B genes and its association with immune traits. Int J Biol Sci. 2011;7:1101–1113. doi: 10.7150/ijbs.7.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Acevedo VD, Gangula RD, Freeman KW, Li R, Zhang Y, Wang F, Ayala GE, Peterson LE, Ittmann M, Spencer DM. Inducible FGFR-1 activation leads to irreversible prostate adenocarcinoma and an epithelial-to-mesenchymal transition. Cancer Cell. 2007;12:559–571. doi: 10.1016/j.ccr.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 151.Carrasco DR, Sukhdeo K, Protopopova M, Sinha R, Enos M, Carrasco DE, Zheng M, Mani M, Henderson J, Pinkus GS, Munshi N, Horner J, Ivanova EV, Protopopov A, Anderson KC, Tonon G, DePinho RA. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11:349–360. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell. 2007;11:375–388. doi: 10.1016/j.ccr.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 153.Lam SH, Gong Z. Modeling liver cancer using zebrafish: a comparative oncogenomics approach. Cell Cycle. 2006;5:573–577. doi: 10.4161/cc.5.6.2550. [DOI] [PubMed] [Google Scholar]
- 154.Qi F, Song J, Yang H, Gao W, Liu NA, Zhang B, Lin S. MMP23b promotes liver development and hepatocyte proliferation through the tumor necrosis factor pathway in zebrafish. Hepatology. 2010;52:2158–2166. doi: 10.1002/hep.23945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wajant H, Pfizenmaier K, Scheurich P. Tumor necrosis factor signaling. Cell Death Differ. 2003;10:45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- 156.Schwabe RF, Brenner DA. Mechanisms of Liver Injury. I. TNF-alpha-induced liver injury: role of IKK, JNK, and ROS pathways. Am J Physiol Gastrointest Liver Physiol. 2006;290:G583–G589. doi: 10.1152/ajpgi.00422.2005. [DOI] [PubMed] [Google Scholar]
- 157.Wullaert A, van Loo G, Heyninck K, Beyaert R. Hepatic tumor necrosis factor signaling and nuclear factor-kappaB: effects on liver homeostasis and beyond. Endocr Rev. 2007;28:365–386. doi: 10.1210/er.2006-0031. [DOI] [PubMed] [Google Scholar]
- 158.Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, Nelson N, Boiani N, Schooley KA, Gerhart M, Davis R, Fitzner JN, Johnson RS, Paxton RJ, March CJ, Cerretti DP. A metalloproteinase disintegrin that releases tumor-necrosis factor-alpha from cells. Nature. 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 159.Krogsgaard M, Ma W, Friedman EB, Vega-Saenz de Miera E, Darvishian F, Perez-Garcia RS, Berman RS, Sharpio RL, Christos PJ, Osman I, Pavlick AC. An analysis of altered melanoma matrix metalloprotease-23 (MMP-23) expression and response to immune biologic therapy. J Clin Oncol. 2011;29(Suppl):8541. [Google Scholar]
- 160.Bielanska J, Hernandez-Losa J, Moline T, Somoza R, Cajal SR, Condom E, Ferreres JC, Felipe A. Increased voltage-dependent K(+) channel Kv1.3 and Kv1.5 expression correlates with leiomyosarcoma aggressiveness. Oncol Lett. 2012;4:227–230. doi: 10.3892/ol.2012.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bielanska J, Hernandez-Losa J, Moline T, Somoza R, Ramon Y, Cajal S, Condom E, Ferreres JC, Felipe A. Differential expression of Kv1.3 and Kv1.5 voltage-dependent K+ channels in human skeletal muscle sarcomas. Cancer Invest. 2012;30:203–208. doi: 10.3109/07357907.2012.654872. [DOI] [PubMed] [Google Scholar]
- 162.Bielanska J, Hernandez-Losa J, Perez-Verdaguer M, Moline T, Somoza R, Ramon YCS, Condom E, Ferreres JC, Felipe A. Voltage-dependent potassium channels Kv1.3 and Kv1.5 in human cancer. Curr Cancer Drug Targets. 2009;9:904–914. doi: 10.2174/156800909790192400. [DOI] [PubMed] [Google Scholar]
- 163.Felipe A, Bielanska J, Comes N, Vallejo A, Roig S, Ramon YCS, Condom E, Hernandez-Losa J, Ferreres JC. Targeting the voltage-dependent K+ channels Kv1.3 and Kv1.5 as tumor biomarkers for cancer detection and prevention. Curr Med Chem. 2012;19:661–674. doi: 10.2174/092986712798992048. [DOI] [PubMed] [Google Scholar]
- 164.Stuhmer W, Pardo LA. K+ channels as therapeutic targets in oncology. Future Med Chem. 2010;2:745–755. doi: 10.4155/fmc.10.24. [DOI] [PubMed] [Google Scholar]
- 165.Beeton C, Wulff H, Standifer NE, Azam P, Mullen KM, Pennington MW, Kolski-Andreaco A, Wei E, Grino A, Counts DR, Wang PH, LeeHealey CJ, Andrews BS, Sankaranarayanan A, Homerick D, Roeck WW, Tehranzadeh J, Stanhope KL, Zimin P, Havel PJ, Griffey S, Knaus HG, Nepom GT, Gutman GA, Calabresi PA, Chandy KG. Kv1.3 channels are a therapeutic target for T cell-mediated autoimmune diseases. Proc Natl Acad Sci USA. 2006;103:17414–17419. doi: 10.1073/pnas.0605136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Leanza L, Henry B, Sassi N, Zoratti M, Chandy KG, Gulbins E, Szabo I. Inhibitors of mitochondrial Kv1.3 channels induce Bax/Bak-independent death of cancer cells. EMBO Mol Med. 2012;4:577–593. doi: 10.1002/emmm.201200235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chi V, Pennington MW, Norton RS, Tarcha EJ, Londono LM, Sims-Fahey B, Upadhyay SK, Lakey JT, Iadonato S, Wulff H, Beeton C, Chandy KG. Development of a sea anemone toxin as an immunomodulator for therapy of autoimmune diseases. Toxicon. 2012;59:529–546. doi: 10.1016/j.toxicon.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Vacher H, Trimmer JS. Trafficking mechanisms underlying neuronal voltage-gated ion channel localization at the axon initial segment. Epilepsia. 2012;53(Suppl 9):21–31. doi: 10.1111/epi.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Patterson NL, Iyer RP, Bras LD, Li Y, Andrews TG, Aune GJ, Lange RA, Lindsey ML. Using proteomics to uncover extracellular matrix interactions during cardiac remodeling. Proteomics Clin Appl. 2013 doi: 10.1002/prca.201200100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Stegemann C, Didangelos A, Barallobre-Barreiro J, Langley SR, Mandal K, Jahangiri M, Mayr M. Proteomic identification of matrix metalloproteinase substrates in the human vasculature. Circ Cardiovasc Genet. 2013;6:106–117. doi: 10.1161/CIRCGENETICS.112.964452. [DOI] [PubMed] [Google Scholar]
- 171.Austin KM, Covic L, Kuliopulos A. Matrix metalloproteases and PAR1 activation. Blood. 2013;121:431–439. doi: 10.1182/blood-2012-09-355958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Haerteis S, Krappitz M, Diakov A, Krappitz A, Rauh R, Korbmacher C. Plasmin and chymotrypsin have distinct preferences for channel activating cleavage sites in the gamma subunit of the human epithelial sodium channel. J Gen Physiol. 2012;140:375–389. doi: 10.1085/jgp.201110763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Schrodinger LLC (2010) The PyMOL Molecular Graphics System, Version 1.3r1
- 174.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Brodskii LI, Ivanov VV, Ia Kalaidzidis Ia L, Leontovich AM, Nikolaev VK, Feranchuk SI, Drachev VA. GeneBee-NET: an Internet based server for biopolymer structure analysis. Biokhimiia. 1995;60:1221–1230. [PubMed] [Google Scholar]