Abstract
Rme1p, a repressor of meiosis in the yeast Saccharomyces cerevisiae, acts as both a transcriptional repressor and activator. Rme1p is a zinc-finger protein with no other homology to any protein of known function. The C-terminal DNA binding domain of Rme1p is essential for function. We find that mutations and progressive deletions in all three zinc fingers can be rescued by fusion of RME1 to the DNA binding domain of another protein. Thus, structural integrity of the zinc fingers is not required for the Rme1p-mediated effects on transcription. Using a series of mutant Rme1 proteins, we have characterized domains responsible for repression and activation. We find that the minimal transcriptional repression and activation domains completely overlap and lie in an 88-amino-acid N-terminal segment (aa 61–148). An additional transcriptional effector determinant lies in the first 31 amino acids of the protein. Notwithstanding the complete overlap between repression and activation domains of Rme1p, we demonstrated a functional difference between repression and activation: Rgr1p and Sin4p are absolutely required for repression but dispensable for activation.
INTRODUCTION
Control of transcription is central to the regulation of cell growth and differentiation. Transcriptional control is achieved through the activities of two types of site-specific DNA binding proteins: activators and repressors, and through the action of the Mediator complex of RNA polymerase II, which is implicated in positive as well as negative regulation of transcription (Myers et al., 1999). Some proteins act as activators in one context and as repressors in another. The precise mechanism of action of these proteins is unknown.
Rme1p can exert either a positive or negative effect on gene expression. Rme1p blocks meiosis in haploid yeast cells in response to starvation by preventing transcription of IME1, which encodes a positive regulator of several early meiotic genes (Kassir and Simchen, 1976; Mitchell and Herskowitz, 1986; Kassir et al., 1988; Kupiec et al., 1997). Rme1p is a zinc-finger protein with no other similarity to known repressor proteins. Rme1p binds to two sites that lie at −2030 and −1950 bp upstream of the IME1 gene (Covitz and Mitchell, 1993; Shimizu et al., 1998). The binding sites are contained within a 404-bp DNA segment called the repression cassette (RC). The RC confers Rme1p-dependent repression to the heterologous CYC1 promoter when inserted adjacent to the CYC1 upstream activating sequence (UAS) (Covitz and Mitchell, 1993; Shimizu et al., 1997b). It has been proposed that Rme1p represses transcription through an activator exclusion mechanism. Transcriptional activators Hap1p and Hap2p are unable to bind to their DNA recognition sites at a Rme1p-repressed hybrid promoter (Shimizu et al., 1997b).
Repression by RME1 depends on Rgr1p and Sin4p (Covitz et al., 1994). These proteins are best known as subunits of the Mediator complex of RNA polymerase II (Li et al., 1995; Carlson, 1997). These subunits of the Mediator were identified in several additional genetic screens as negative effectors of transcription (Sakai et al., 1990; Stillman et al., 1994; Jiang et al., 1995). However, they are also required for maximal induction of particular sets of genes. In addition, mutations in RGR1 and SIN4 confer phenotypes common to histone and spt mutations, namely, decreased plasmid superhelicity and activation of UAS-less promoters (Jiang and Stillman, 1992; Jiang et al., 1995), suggesting that the genes are involved in determining chromatin structure.
When an Rme1p binding site is situated 5′ of CLN2 or other reporter genes, it can also activate transcription (Toone et al., 1995). Activation or repression depends on the context and flanking regions of the binding site: the presence of RC causes repression, and the absence of RC causes activation (Covitz and Mitchell, 1993).
All rme1 mutations obtained so far affect zinc fingers and confer deficiencies in both repression and activation (Covitz, 1993). Thus, it is not clear whether zinc-finger function is restricted to DNA binding or whether zinc fingers participate in repression/activation as well. Mammalian YY1 protein is a precedent for a later model: it is capable of either activating or repressing transcription. The repression domain of YY1 is embedded within the zinc-finger regions, although the normal structure of zinc fingers is not required for repression (Bushmeyer et al., 1995).
The following structure-functional analysis of Rme1p was performed to delineate the repression and activation domains of Rme1 to see whether or not they overlap and to test whether the role of the zinc fingers is restricted to DNA binding. We show that Rme1p can be dissected into two domains: a minimal transcriptional effector domain, which resides in an 88-amino-acid segment (aa 61–148) at the N-terminus of the protein; and the C-terminal DNA binding domain, which can be replaced by other DNA binding domains. Thus, zinc-finger integrity is not required for either activation or repression by the effector domain. Additional effector determinants exist within the first 31 amino acids of Rme1p. Thus, the Rme1p effector domain is composed of multiple subdomains that contribute synergistically to efficient repression/activation. Although the repression and activation domains of Rme1p overlap, only repression, not activation, depends on Rgr1p (Covitz et al., 1994) and Sin4p.
MATERIALS AND METHODS
Growth Media, Strains, and RME1 Alleles
Yeast cells were grown and media were prepared according to standard techniques (Rose et al., 1990). Yeast strains were isogenic to SK-1 (Kane and Roth, 1974); genotypes are listed in Table 1.
Table 1.
Yeast strains used in this study
| Strains | Genotype |
|---|---|
| AMP108 | α GAL80 RME1 |
| AMP714 | a his3 met4 |
| AMP764 | α his4-G IME2-lacZ-URA3 |
| AMP1122 | α rme1::PGAL1-S53-RME1::TRP1 |
| AMP1396 | α rme1::PGAL1-S53-RME1::TRP1 sin4Δ::TRP1 arg6 |
| AMP1420 | α rme1::PGAL1-S53-RME1::TRP1 his3ΔSK rgr1-100 |
| AMP1615 | a his3 |
| WL447 | a trp1::PGAL1-lexA-RME1::TRP1 |
| WL448 | a trp1::PGAL1-lexA::TRP1 |
| WL450 | a trp1::PGAL1-lexA-rme1-213::TRP1 |
| WL457 | a trp1::PGAL1-lexA-rme1-Cla179::TRP1 |
| WL458 | a trp1::PGAL1-lexA-rme1-Cla210::TRP1 |
| WL459 | a trp1::PGAL1-lexA-rme1-Cla239::TRP1 |
| WL460 | a trp1::PGAL1-lexA-rme1-Cla239,269::TRP1 |
| WL471 | a trp1::PGAL1-lexA-rme1Δ4-179::TRP1 |
| WL472 | a trp1::PGAL1-lexA-rme1Δ148-300::TRP1 |
| WL473 | a trp1::PGAL1-lexA-rme1Δ179-300::TRP1 |
| WL474 | a trp1::PGAL1-lexA-rme1Δ120-179::TRP1 |
| WL475 | a trp1::PGAL1-lexA-rme1Δ148-179::TRP1 |
| WL489 | a trp1::PGAL1-lexA-rme1Δ4-31::TRP1 |
| WL491 | a trp1::PGAL1-lexA-rme1Δ31-90::TRP1 |
| WL492 | a trp1::PGAL1-lexA-rme1Δ31-120::TRP1 |
| WL494 | a trp1::PGAL1-lexA-rme1Δ61-148::TRP1 |
| WL495 | a trp1::PGAL1-lexA-rme1Δ210-300::TRP1 |
| BPA71 | a/α trp1::PGAL1-lexA-rme1-213::TRP1/trp1 lexO-IME1/ime1Δ12-TRP1 IME2-lacZ-URA3/IME2 his3/HIS3 met4/MET4 |
| BPA80 | a/α trp1::PGAL1-lexA-RME1::TRP1/trp1 lexO-IME1/ime1Δ12-TRP1 IME2-lacZ-URA3/IME2 his3/HIS3 met4/MET4 |
| BPA86 | a/α trp1::PGAL1-lexA::TRP1/trp1 lexO-IME1/ime1Δ12-TRP1 IME2-lacZ-URA3/IME2 his3/HIS3 met4/MET4 |
| BPA101 | a/α trp1::PGAL1-lexA-rme1-Δ179-300::TRP1/trp1 met4/MET4 lexO-IME1/ime1Δ12-TRP1 IME2-lacZ-URA3/IME2 his3/HIS3 |
| BPA103 | a/α trp1::PGAL1-lexA-rme1-Δ148-179::TRP1/trp1 met4/MET4 lexO-IME1/ime1Δ12-TRP1 IME2-lacZ-URA3/IME2 his3/HIS3 |
| BPA105 | a/α trp1::PGAL1-lexA-rme1-Δ120-179::TRP1/trp1 met4/MET4 lexO-IME1/ime1Δ12-TRP1 IME2-lacZ-URA3/IME2 his3/HIS3 |
| BPA107 | a/α trp1::PGAL1-lexA-rme1-Δ148-300::TRP1/trp1 met4/MET4 lexO-IME1/ime1Δ12-TRP1 IME2-lacZ-URA3/IME2 his3/HIS3 |
| BPA110 | a/α trp1::PGAL1-lexA-rme1-Δ31-90::TRP1/trp1 met4/MET4 lexO-IME1/ime1Δ12-TRP1 IME2-lacZ-URA3/IME2 his3/HIS3 |
| BPA112 | a/α trp1::PGAL1-lexA-rme1-Δ31-120::TRP1/trp1 met4/MET4 lexO-IME1/ime1Δ12-TRP1 IME2-lacZ-URA3/IME2 his3/HIS3 |
| BPA113 | a/α trp1::PGAL1-lexA-rme1-Δ210-300::TRP1/trp1 met4/MET4 lexO-IME1/ime1Δ12-TRP1 IME2-lacZ-URA3/IME2 his3/HIS3 |
| BPA131 | a his3 met4 trp1::PGAL1-rme1Δ1-31::TRP1 |
| BPA132 | a/α trp1::PGAL1-rme1Δ1-31::TRP1/trp1 IME2-lacZ-URA3/IME2 his3/HIS3 his4-G/HIS4 met4/MET4 |
| BPA134 | a his3 met4 trp1::PGAL1-rme1Δ1-61::TRP1 |
| BPA135 | a/α trp1::PGAL1-rme1Δ1-61::TRP1/trp1 IME2-lacZ-URA3/IME2 his3/HIS3 his4-G/HIS4 met4/MET4 |
| BPA137 | a his3 met4 trp1::PGAL1-rme1Δ1-90::TRP1 |
| BPA138 | a/α trp1::PGAL1-rme1Δ1-90::TRP1/trp1 IME2-lacZ-URA3/IME2 his3/HIS3 his4-G/HIS4 met4/MET4 |
| BPA140 | a his3 met4 trp1::PGAL1-rme1Δ1-120::TRP1 |
| BPA141 | a/α trp1::PGAL1-rme1Δ1-120::TRP1/trp1 IME2-lacZ-URA3/IME2 his3/HIS3 his4-G/HIS4 met4/MET4 |
| BPA143 | a his3 met4 trp1::PGAL1-rme1Δ1-148::TRP1 |
| BPA144 | a/α trp1::PGAL1-rme1Δ1-148::TRP1/trp1 IME2-lacZ-URA3/IME2 his3/HIS3 his4-G/HIS4 met4/MET4 |
| BPA238 | a/α trp1::PGAL1-lexA-rme1-Cla179::TRP1/trp1 met4/MET4 lexO-IME1/ime1Δ12-TRP1 IME2-lacZ-URA3/IME2 his3/HIS3 |
| BPA240 | a/α trp1::PGAL1-lexA-rme1-Cla239,269::TRP1/trp1 met4/MET4 lexO-IME1/ime1Δ12-TRP1 IME2-lacZ-URA3/IME2 his3/HIS3 |
| PJ69-4A | a trp1-901 leu2-3,112 ura3-52 his3-2000 gal4Δ gal80Δ LYS::GAL1-HIS3 GAL2-ADE2 GAL7-lacZ RME1 (James et al., 1996) |
All the strains are SK-1 derivatives and carry the mutations leu2::hisG, trp1::hisG, lys2 ura3, ho::LYS2, gal80::LEU2, and rme1Δ5::LEU2, except where indicated otherwise.
The mutations gal80::LEU2, rme1Δ5::LEU2, IME2-lacZ-URA3, and rme1::PGAL1-S53-RME1::TRP1 have been described previously (Neigeborn and Mitchell, 1991; Covitz and Mitchell, 1993; Su and Mitchell, 1993).
All lexA-RME1 and truncated RME1 alleles were generated by integrating the corresponding plasmid at the TRP1 locus of strain AMP1615 or AMP714. The integration plasmid was digested with Bsu36I to target the integration at the chromosomal TRP1 locus. The resulting transformants were purified as single colonies. All integrations were confirmed by Southern analysis.
Expression of the lexA-Rme1p derivatives was scored by Western analysis with anti-lexA antibodies. By this criterion, all lexA-Rme1p derivatives were expressed at similar levels.
Expression of N-terminal deletions of Rme1p was confirmed by comparing the phenotypes of GAL80 strains, in which there is no expression of rme1 alleles, and gal80::LEU2 strains, in which the rme1 alleles are expressed.
RME1-Derived Plasmids
The plasmid carrying PGAL1-lexA-RME1 (pWL126) was constructed as follows. The RME1 coding sequence flanked by BamHI sites was amplified by PCR using oligos RME1–5′-Bam and RME1–3′-Bam (see below). The PCR product was ligated into the EcoRV site in pBS-SK to generate pWL105. A BamHI-BamHI fragment containing RME1 ORF was excised from pWL105 and inserted into the BamHI site in pBTM116 (Ruden et al., 1991) to generate pWL107. This resulted in a fusion of lexA (1–200) to the Rme1p start codon. A HindIII-SalI fragment containing lexA-RME1 was inserted between the HindIII-SalI site in pSV150 (Vidan and Mitchell, 1997) to generate pWL123 with lexA-RME1 driven by the Gal1 promoter. A PstI-SalI fragment containing PGAL1-lexA-RME1 from pWL123 was inserted into PstI-SalI site in pRS304 (Sikorski and Hieter, 1989).
The PGAL1-lexA plasmid (pWL137) was constructed as follows. A HindIII-SalI fragment containing lexA (1–200) from pBTM116 (Ruden et al., 1991) was cloned between the HindIII-SalI sites in pSV150 to produce pWL133. Then, PGAL1-lexA from pWL133 was excised and moved into the TRP1 integration vector pWL131 (pRS304 with the BamHI site removed by filling in).
The PGAL1-lexA-rme1–213 plasmid (pWL139) carries the previously characterized RME1 zinc-finger mutation (Covitz et al., 1991). The plasmid was derived from pWL126 by site-directed mutagenesis using oligos RMESER213 (Covitz et al., 1991).
ClaX plasmids were generated by site-directed mutagenesis using pWL126 as a template. Clones were screened by ClaI digest, and the candidates were sequenced to confirm the existence of the mutation. Deletions between ClaI sites were used to create deletion derivatives such as PGAL1-LexA-rme1Δ14148–300.
The plasmids carrying N-terminal deletions of rme1 were constructed as follows. To obtain the truncated version of Rme1p under control of the PGAl1 promoter, pWL126 derivatives with the appropriate ClaI site were digested with the restriction enzymes PstI and ClaI. This removed PGAL1-lexA and unwanted parts of RME1. Next, the remaining plasmids were ligated to the PstI-HindIII fragment of plasmid pSV150 (Vidan and Mitchell, 1997), carrying PGAL1. The ligation was performed using a HindIII/ClaI, which introduced Ser-Pro-His6 at the beginning of each protein. The amino acids serine and proline were chosen because they are the first amino acids of wild-type Rme1p. Adaptor oligos were HIII-ClaI sense: 5′-AGCTATGTCACCGCACCACCACCATCATCATAT-3′; HIII-ClaI antisense: 5′-CGATATGATGATGATGGTGGTGGTGCGGTGACAT-3′.
Reporter Plasmids
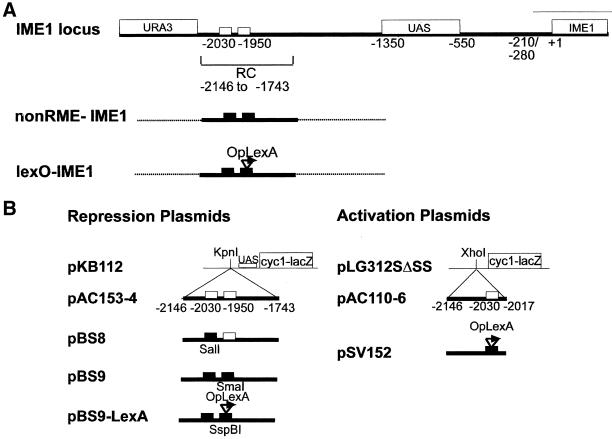
Plasmid pLS312SΔSS carries the ΔUAS-CYC1-lacZ reporter gene (Guarente and Mason, 1983). Plasmids PAC153–4 and PAC110–6 carry RC-CYC1-lacZ and RRE-CYC1-lacZ reporter genes, respectively (Covitz and Mitchell, 1993), and plasmid pSV152 carries lexA sites inserted upstream of the ΔUAS-CYC1-lacZ (Vidan and Mitchell, 1997) (Figure 1B).
Figure 1.
Schematic of structural modifications of the IME1 upstream region (A) and plasmids used in this study (B). The diagram shows Rme1p binding sites, the UAS, IME1, and URA3 coding regions (labeled rectangles), and RNA start sites (−280 to −210). Small white rectangles indicate the Rme1 binding sites. Small black rectangles indicate the mutations in the Rme1 binding sites. Arrow indicates the lexA operator. End points are numbered with respect to the IME1 translation start.
The plasmids to test repression were pBS8, pBS9, and pBS9-LexA (Figure 1B). They were derived from PAC153–4 by site-directed mutagenesis. The plasmid pBS8 was created using the bottom-strand oligonucleotide IME1-R23 (Covitz and Mitchell, 1993). This resulted in a deletion of sequences from −2044 to −2025 bp and inserted a C to generate a SalI restriction site instead of the Rme1 binding site at −2030 bp. The plasmid pBS9, which carries mutations of both Rme1 sites, was created using oligonucleotides IME1-R23 and IME1-BS1 5′-ATTTTATGCTCCCGGGGTACAGC-3′. The IME1-BS1 replaces the Rme1 binding site at −1956 to −1951 bp upstream of IME1 with 5′-CCGGGA-3′ and generates a SmaI restriction site instead of an Rme1 site.
The pBS9-LexA reporter plasmids were created by site-directed mutagenesis of pBS9 using an oligonucleotide containing the lexA binding site: 5′-TCGAGTACTGTATGTACATACAGTAC-3′ (Brent and Ptashne, 1984). The resulting plasmids pBS9-LexA1 and pBS9-LexA3 contain one or two lexA binding sites in place of the Rme1 binding site at −1950.
Modifications in the IME1 Upstream Region
Three types of modification were done. In all cases, the URA3 gene was placed upstream of −2146 bp (Figure 1A). In the first case, the RC was left intact. In the second case, both Rme1 binding sites were destroyed within the RC, generating an IME1 allele that was not repressible by Rmep1 (nonRme1-IME1 allele). In the third, both Rme1 binding sites were destroyed, and the −1950 binding site was replaced by the lexA site, generating the lexO-IME1 allele (Figure 1A).
The modifications were introduced by transforming yeast with PCR products. The PCR product contained the URA3 gene, followed by the desired type of RC. These were flanked by sequences homologous to the native sequences flanking the RC in the IME1-upstream region (60 bp on each side of the PCR product). The plasmids PAC153–4 (Covitz and Mitchell, 1993), pBS9, and pBS9-LexA were templates for PCR, using primers ime1–2207-yep2 5′-AAAAATCAATTCATATCATATATTATCTATATCATGCTGTTCTTTCCGCCACGGCCCGTAAAGCTTTTCAATTCAATTCAT-3′ and ime1–1747 5′-GGCCAAAAAATAGTTCAAATT-3′. Correct integration was confirmed by Southern analysis and by PCR using URA3 and IME1-R6 (Covitz, 1993) as primers. PCR products were digested with the restriction enzymes SmaI and SalI to verify the presence of the RC with both Rme1 sites destroyed and with SspBI to verify the presence of the lexA site at position −1950 bp (Figure 1B).
To test different PGAL1-lexA-rme1 derivatives for their ability to repress through the lexA binding site, strains marked by the URA3 gene and bearing modified RC were crossed to strain AMP108 GAL80+ ura3. Diploids were sporulated, spores were dissected, and GAL80+ URA3 segregants with the modified RC were obtained. These were used to transfer RC modification to any other desired strain. The presence of the modified RC was verified by PCR after each cross.
Two-Hybrid–Based Plasmids
The plasmid pGBDU-C1 (James et al., 1996), which contains the GAL4 DNA binding domain, was used to check the activation ability of different domains of Rme1p. The plasmids pGBDU-C1-rme1–90-210, pGBDU-C1-rme1–120-210, pGBDU-C1-rme1–148-210 were constructed by the following steps.
The desired part of the RME1 coding sequence, flanked by a BamHI site at its 5′ end and by SalI at its 3′ end, was amplified by PCR. Oligonucleotides for the upper strand were BamHI-rme1–90: 5′-GCAGGATCCGGTACAGCACCTCAATTACGG-3′, for pGBDU-C1-rme1–90-210; BamHI-rme1–120: 5′-GCAGGATCCAATTATGGACGTCAAAAAGGA-3′, for pGBDU-C1-rme1–120-210; and BamHI-rme1–148: 5′-GCAGGATCCTATCCCCAAAAATCGCACGTG-3′, for pGBDU-C1-rme1–148-210.
The rme1–210-SalI 5′-GTCGTCGACTTGCTCTATGGGACACTTACA-3′ primer was used as a bottom-strand oligonucleotide. Next, the PCR product was ligated into the EcoRV site in pBS-SK. Third, a BamHI-SalI fragment containing part of the RME1 ORF was excised from pBS-SK-rme1 and inserted between the BamHI-SalI sites in pGBDU-C1 to generate an in-frame fusion of the GAL4 binding domain and part of RME1.
To construct plasmid pGBDU-C1-rme1–61-148, the rme1–61-148 was removed from the appropriate pWL126 derivative by ClaI digestion and then ligated into the ClaI site of plasmid pGBDU-C1.
To create in-frame fusions of Rme1-Δ31–90p and Rme1Δ31–120p to the GAL4 DNA binding domain, the RME1-Δ31–90 and RME1-Δ31–120 alleles were amplified from plasmid templates by PCR using the following primers, which introduced BamHI sites on both sides of RME1: RME-5′-Bam primer: 5′-GCAGGATCCTTATGTCACCGTGTTATGG-3′; RME-3′-Bam primer: 5′-ACAGGATCCACAAGAGTTTCATGGGGTAC-3′.
The PCR products were cloned into vector pGEM-T (Promega). BamHI fragments containing rme1-Δ31–90 and rme1-Δ31–120 were excised from pGEM-T-rme1 and were then ligated into pGBU-C2 at the BamHI site, resulting in in-frame fusion of rme1-Δ31–90 and rme1-Δ31–120 to the GAL4-DNA binding domain.
β-Galactosidase and Sporulation Assays
The liquid β-galactosidase assays were conducted as described elsewhere (Covitz and Mitchell, 1993). For liquid sporulation assays, cells were grown for 24–29 h at 30°C in synthetic medium containing 0.5% glucose and lacking uracil, filtered, washed once in water, and transferred at the same cell density to 2% potassium acetate supplemented with lysine. After 24 h at 30°C, half of each culture was taken for IME2-lacZ assays to monitor IME1 regulation (Smith et al., 1990). The level of sporulation was scored by counting the number of asci per 200 cells after 2 d in 2% potassium acetate liquid medium or on standard Spo plates (Rose et al., 1990). The reported values are the average of at least three determinations from three independent transformants. β-Galactosidase was measured in permeabilized cells as previously described (Smith et al., 1990).
RESULTS
Rme1p Effector Domain Acts Independently from the DNA-Binding Domain
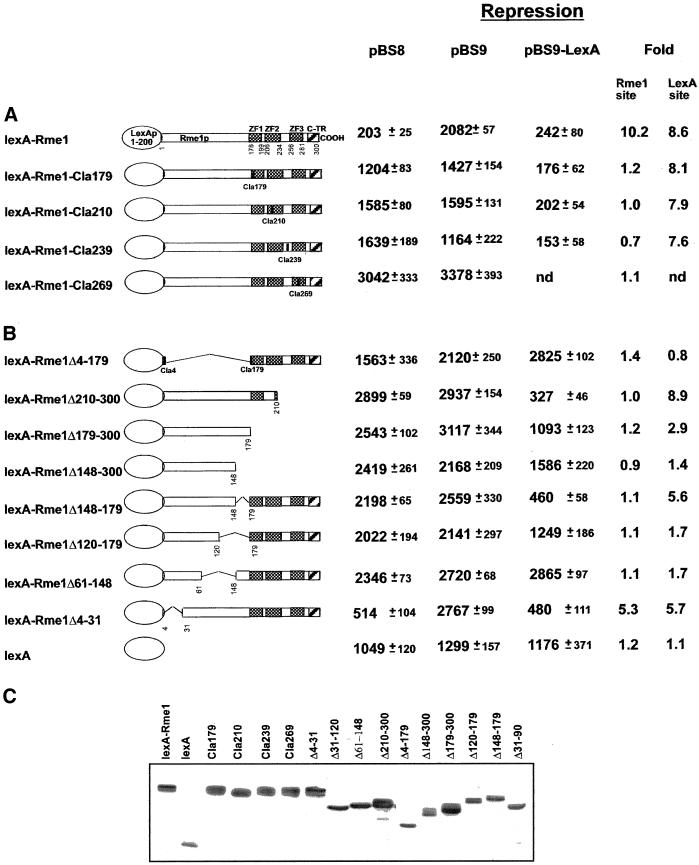
The Rme1p zinc fingers are located in the C-terminal part of the protein. To study the repression and activation functions of Rme1p independently of its DNA binding activity, RME1 was fused to the lexA DNA binding domain (1–200 bp). The ability of fusion protein lexA-Rme1p to repress and to activate transcription was tested via both the Rme1 binding site and the lexA binding site using different reporter genes that carried wild-type or altered RC from the 5′ region of the IME1 gene (Figure 1). To repress via the Rme1 binding site, lexA-Rme1p needs both the repression and DNA-binding domain from Rme1p. To test repression via the Rme1 site, we compared β-galactosidase activity expressed from plasmid pBS9, which lacks Rme1p sites, and plasmid pBS8, which has an intact RC. To repress via the lexA site, lexA-Rme1p needs only the repression domain of Rme1p. To test repression via the lexA site, we compared β-galactosidase activity from a plasmid that lacks the lexA sites, pBS9, and pBS9-lexA, which contains the lexA site within the RC (Figure 1B). Figure 2A shows that lexA-Rme1p can repress via both the Rme1 and lexA sites. Next, ClaI restriction site insertion mutations, which disrupt the integrity of the zinc fingers, were created at four places in the C terminus of the RME1 coding region by site-directed mutagenesis of lexA-RME1. We will refer to these alleles as lexA-RME1-ClaX, where X indicates the number of the amino acid after which the insertion occurred. lexA-RME1-Cla179, lexA-RME1-Cla210, and lexA-RME1-Cla269 carry insertions that disrupt zinc-finger structures, lexA-RME1-Cla239 carries the insertion between the second and the third zinc fingers. All these mutant derivatives (Figure 2A) failed to repress via the Rme1p sites located in the promoter of reporter plasmid pBS8. Nevertheless, all four Cla mutants retained the wild-type ability to repress via the lexA site located in the promoter of reporter plasmid pBS9-lexA. In addition, Rme1–213p, the zinc-finger mutant that has been shown to be incapable of binding to the Rme1 sites (Covitz and Mitchell, 1993; Shimizu et al., 1997b), and Rme1Δ210–300p, with the last two zinc fingers deleted, display wild-type levels of repression through the lexA site.
Figure 2.
Properties of lexA-Rme1p derivatives carrying different alterations in the zinc fingers (A) and in regions proximal to zinc fingers (B). A schematic of lexA-Rme1p is shown at the top. The shaded regions represent zinc-finger like domains. Numbers below the Rme1p derivatives indicate positions (in amino acids) of insertion mutation or site of other alteration. Rme1 site repression fold was calculated as β-galactosidase units produced by cells with pBS9 (carrying RC that lacks Rme1 sites) divided by β-galactosidase units of cells with pBS8 (carrying RC with Rme1 site). lexA site repression was calculated as β-galactosidase units of pBS9 divided by β-galactosidase units of pBS9-lexA (carrying RC with lexA site in place of Rme1 site). (C) Western blot showing the expression of lexA-Rme1p derivatives in transformed yeast cells.
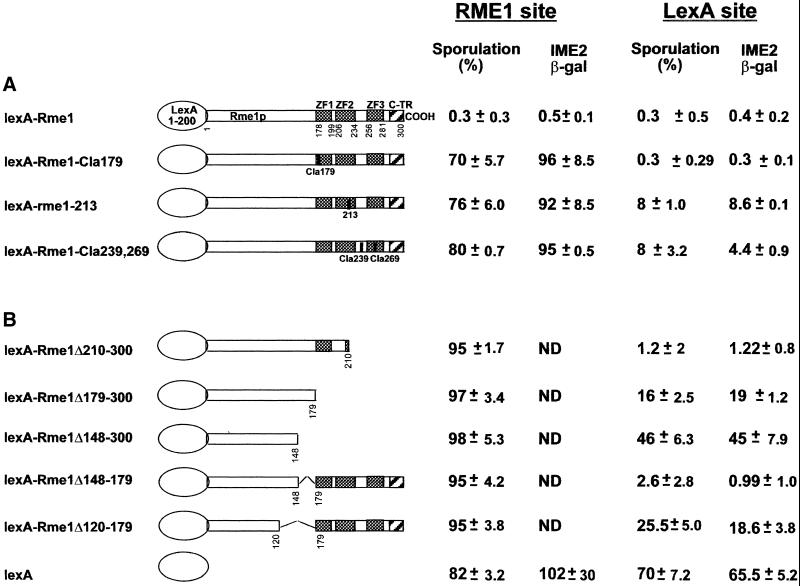
To test whether repression of reporter plasmids by lexA-Rme1p reflects repression by Rme1p in the natural context of its DNA binding site, we examined repression of both the wild-type and a modified IME1 chromosomal region. The wild-type IME1 has the natural Rme1p binding sites, so lexA-Rme1p derivatives must have both functional DNA binding and repression domains from Rme1p to exert repression of IME1. The modified IME1 has a deletion of the two Rme1p sites and an insertion of a single lexA site at position −1950 bp. We call this altered allele lexO-IME1 (Figure 1A). Thus, lexA-Rme1p derivatives need to have only a functional repression domain from Rme1p to repress lexO-IME1. We assayed expression of IME1 and lexO-IME1 by sporulation ability and by expression of an ime2-lacZ meiotic reporter gene (Smith et al., 1990). To permit expression of lexA-Rme1p hybrid proteins in sporulating cells, the proteins were expressed from the GAL1 promoter in diploids homozygous for a gal80 mutation. This genotype causes high-level GAL1 promoter activity in sporulation medium even without the addition of galactose. Our assays of sporulation in strains expressing lexA-Rme1p derivatives are shown in Figure 3. None of the mutant lexA-Rme1p derivatives repressed the natural IME1 locus. However, all of the insertion and deletion derivatives with perturbations of the zinc-finger region repressed lexO-IME1 and thus blocked sporulation and ime2-lacZ expression (Figure 3A). Although lexA-Rme1–213p and lexA-Rme1-Cla239-Cla269p repress less efficiently than wild-type, the Rme1p derivative carrying the deletion of the two last zinc fingers represses very efficiently. Therefore, these mutations might interfere with the protein secondary structure and not with repression itself. In conclusion, this analysis confirms that the structural integrity of Rme1p zinc fingers and the last two zinc fingers are needed only for DNA binding, not for repression itself.
Figure 3.
The effect on repression of the IME1 chromosomal region by lexA-Rme1p derivatives carrying different alterations in the zinc fingers (A) and alterations in the regions of RME1 that are proximal to zinc-finger regions (B). The Rme1 site repression was tested by the ability of different rme1 derivatives to repress sporulation in gal80Δ diploid strains carrying the wild-type RC. The lexA site repression was tested by the ability of different rme1 derivatives to repress sporulation via the lexA site placed at position −1950 bp in RC. Sporulation was scored after 48 h in liquid sporulation medium. IME2-LacZ expression was calculated after 24 h in sporulation medium.
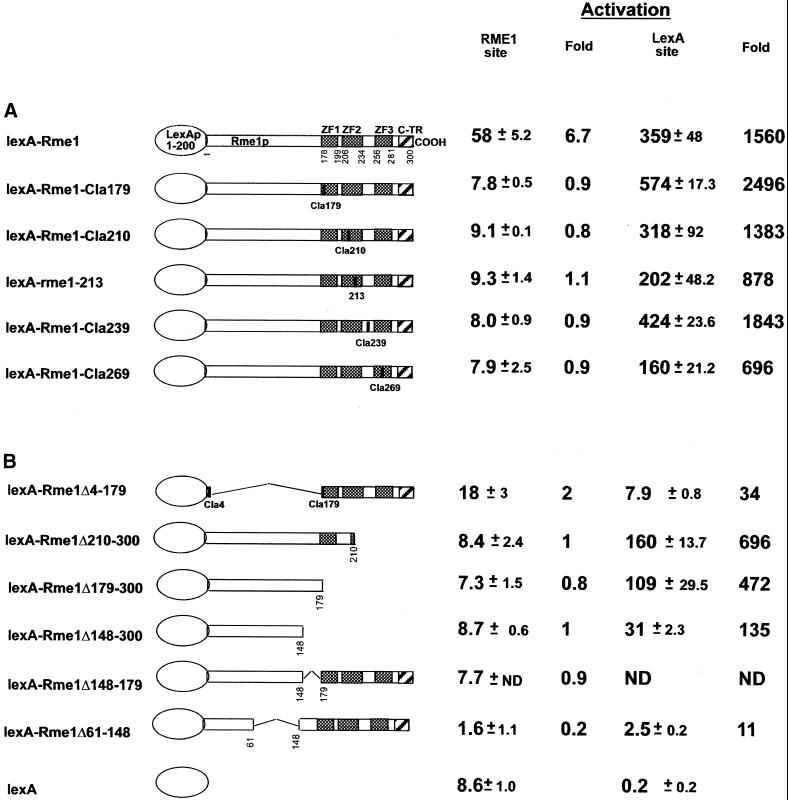
Activation ability of the same lexA-Rme1p derivatives was assayed by the ability to activate transcription of an UAS-less reporter gene with the Rme1 site (pAC110–6) or a reporter with the lexA site replacing the UAS (pSV152) (Figure 1B). All the mutants were able to activate only via the lexA site (Figure 4). We conclude that structural integrity of the Rme1p zinc-finger region is not required for either repression or activation.
Figure 4.
Activation by lexA-Rme1p derivatives carrying different alterations in the zinc fingers (A) and in regions proximal to zinc fingers (B). Activation ability was determined by liquid β-galactosidase assays using pAC110–6 as a reporter for the Rme1 site and pSV152 as a reporter for the lexA site activation.
Rme1p Effector Domain Lies in the N-Terminus of the Protein
C-Terminal Extent of the Effector Domain
To elucidate the role of the N-terminal region of Rme1p in repression and/or activation, a lexA-Rme1p derivative lacking amino acids 4–179 was constructed. This mutant protein was defective in both activation and repression in assays of either the Rme1 or lexA binding sites (Figures 2B and 4). Therefore, the N-terminal region of Rme1p is required for both activation and repression.
To determine the C-terminal boundary of the repression domain, we created increasingly large deletions of the C-terminal part of the protein and various deletions internal to the zinc fingers. These deletions were tested for their ability to repress expression from reporter plasmid and to repress sporulation (Figures 2B and 3B). All these alleles failed to repress expression from the reporter plasmid and sporulation via the Rme1 site. Via the lexA site, lexA-Rme1Δ210–300p repressed efficiently (it showed 8.9-fold repression and allowed only 1.2% sporulation). However, lexA-Rme1Δ179–300p was partially defective in repression of lexA-IME1 (permitting 16% sporulation, with 2.9-fold repression), whereas lexA-Rme1Δ148–300p showed a significant reduction in repression ability (46% sporulation, with 1.4-fold repression). Therefore, residues 179–210, which lie within the first zinc-finger domain, may contribute to repression. This region is composed of charged and hydrophobic amino acids. It contains a long stretch of hydrophobic residues at 187–197 (FATLVEFAAHL). Moreover, this region is predicted to form an α-helix with very prominent clusters of hydrophobic amino acids at both sides of the helix.
To elucidate the role of amino acids 120–179 in repression, two additional deletion derivatives were tested for their ability to repress via the lexA site. Figures 2B and 3B show that lexA-Rme1-Δ148–179p repressed very efficiently (5.6-fold repression, 2.6% sporulation), whereas a protein with a deletion of 28 more amino acids (lexA-Rme1-Δ120–179p) repressed only weakly (1.7-fold repression, 23% sporulation). These results indicate that the amino acid residues 120–148 are required for repression activity, whereas amino acids 148–179 are not.
To determine the C-terminal boundary of the activation domain, the same lexA-Rme1p derivatives were tested for their ability to activate transcription of the reporter plasmids. Figure 4 demonstrates that constructs lexA-Rme1Δ210–300, lexA-Rme1Δ179–300, and lexA-Rme1Δ148–300 were unable to activate transcription of the reporter gene via the Rme1 site, whereas they activated via the lexA site. This again demonstrates that amino acids 148–300 are required for DNA binding and not for activation. Activation of transcription was more efficient by lexA-Rme1Δ210–300 and lexA-Rme1Δ179–300 than by lexA-Rme1Δ148–300. This can mean that amino acids 148–179 contribute to but are not necessary for activation. Thus, the C-terminal boundary of the minimal activation domain lies proximal to amino acid 148.
N-Terminal Extent of the Effector Domain
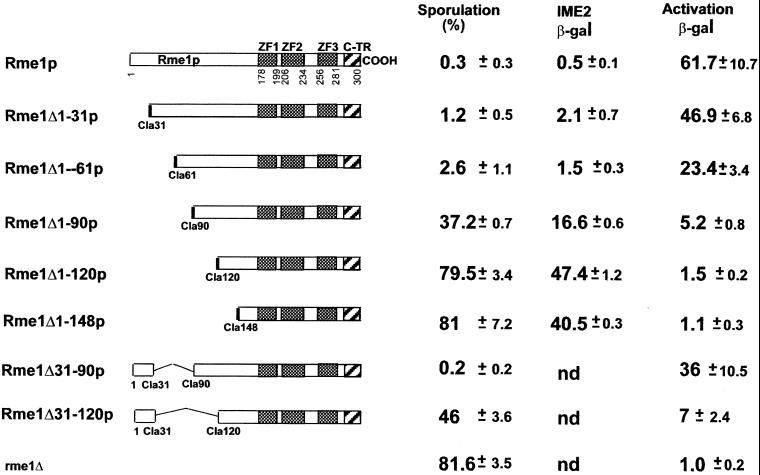
To map the N-terminal boundary of the region required for repression, we constructed six ClaI restriction site insertion mutations at 30 codon intervals in the N-terminus. Deletions between these insertions were then constructed and assayed for repression of IME1 by sporulation and by ime2-lacZ expression (Figure 5). Rme1Δ1–31p and Rme1Δ1–61p repressed almost as efficiently as wild-type Rme1p. However, Rme1Δ1–90p, Rme1Δ1–120p, and Rme1Δ1–148p had little or no ability to repress. Therefore, amino acids 1–60 of Rme1p are dispensable for repression, and the region between amino acids 61 and 89 contains the N-terminal boundary of the repression domain. Together with data from the previous section, our results indicate that amino acids 61–148 of Rme1p are required for full repression. In agreement, the protein deleted for 61–148 amino acids is completely incapable of repression (Figure 2B).
Figure 5.
Effect of a series of N-terminal deletions on the ability to repress the endogenous IME1 gene and to activate a reporter plasmid. The Rme1 site repression was tested by the ability of gal80Δ diploids carrying different Rme1p derivatives to repress sporulation in strains with wild-type RC. Sporulation was scored after 48 h in liquid sporulation medium. IME2-LacZ expression was determined after 24 h in sporulation medium. Activation ability was determined by liquid β-galactosidase assays using pAC110–6 as a reporter.
Repression and activation may be entirely independent functions, involving different domains of Rme1p. Alternatively, they may be interdependent and reside in the same region of the protein. To distinguish between these alternatives, we tested the series of N-terminal Rme1p deletion derivatives for activation ability (Figure 5). We saw a gradual reduction in activation ability of different N-terminal deletions of Rme1p. Proteins deleted for 30 or 60 residues retained a strong ability to activate transcription; proteins deleted for more than 90 residues did not possess activation ability. Therefore, the minimal activation and repression functions of Rme1p are independent of amino acids 1–60 and depend on residues between 61 and 89. Together with the results presented above, our data show that both activation and repression by Rme1p depend on amino acids 61–148.
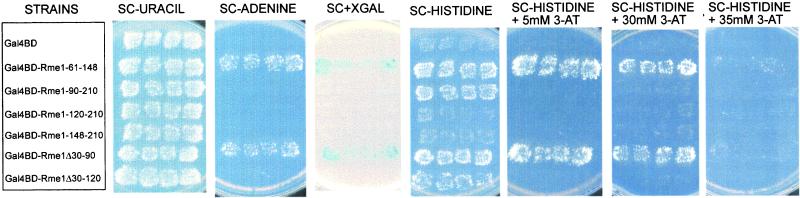
To confirm the mapping of the Rme1p minimal activation domain, we assayed activation by fusions of RME1 N-terminal segments to the GAL4 DNA binding domain (GBD). Activation ability was determined by expression of GAL1-HIS3 and GAL1-ADE2 reporter genes (James et al., 1996), which allowed the cells to grow on media lacking histidine or adenine, respectively. Figure 6 shows that Rme1p amino acids 61–148 were sufficient to activate both reporter genes, whereas residues 90–210 activated transcription of only one reporter gene (HIS3), and did so weakly. The addition of 5 mM 3-aminotriazole (3-AT) completely inhibited the ability of cells carrying amino acids 90–210 of Rme1p to grow on synthetic medium lacking histidine. The shorter segments, with amino acids 120–210 and 148–210, did not activate either reporter. These results indicate that Rme1p residues 61–148 comprise a transcriptional activation domain, which can be transferred to a heterologous DNA-binding domain. Thus, we have demonstrated that the 88 amino acids 61–148 of Rme1p contain a potent effector of transcription function that confers activation and, possibly in cooperation with amino acids 179–210, repression of high-level transcription.
Figure 6.
Transcriptional activation by Gal4-Rme1p fusion derivatives. The GAL4 DNA-binding domain was expressed as a fusion protein to the indicated Rme1p derivatives. The ability to activate transcription of ADE2 and HIS3 reporter genes in strain PJ69–4A was tested on SC medium (Rose et al., 1990) lacking adenine and on SC medium lacking histidine with or without addition of 3-AT.
For certain activators, the minimal activation domain is not always the only region of the protein that possesses the ability to affect transcription (Hope et al., 1988; Drysdale et al., 1995; Jackson et al., 1996). In such cases, high-level activation can occur with only a portion of minimal activation domain if other parts of the protein contain functionally redundant activation subdomains (Hardwick et al., 1992; Regier et al., 1993; Walker et al., 1993; Blair et al., 1994; Jackson et al., 1996). The progressive N-terminal deletions of such proteins show gradual reduction in activation ability (Hope et al., 1988). Moreover, the effect of mutation in residues, which are important to activation function, often can be seen only when several regions important for activation are mutated (Jackson et al., 1996). Progressive N-terminal deletions of Rme1p display a gradual reduction in repression/activation ability rather than a sudden complete loss of activity (Figure 5). Moreover, we were unable to obtain a single mutation that affects Rme1p function as an effector of transcription, except mutations that interfere with binding to DNA (Covitz, 1993). These results suggest that additional activation/repression determinants could exist in the N-terminal region of Rme1p. To test this possibility, we constructed two additional fusion proteins on the basis of Rme1Δ1–90p and Rme1Δ1–120p alleles, adding back the first 31 amino-acid residues to each of these proteins. These two proteins were tested for their ability to repress sporulation and to activate transcription of the reporter gene CYC1-lacZ via the Rme1 site (Figure 5). In addition, these two proteins were tested for their ability to activate transcription of ADE2 and HIS3 reporter genes via the GAL4-DNA binding site (Figure 6). The addition of the first 31 amino acids to Rme1Δ1–90p renders it a potent effector of transcription. Cells carrying Rme1Δ31–90p do not sporulate, and Rme1Δ31–90p efficiently activates transcription of all reporter genes. The addition of the first 31 residues to the effector-deficient Rme1Δ1–120p converts it to a weak transcriptional effector. This lowers sporulation of strains carrying this allele from 79.5% to 46% and is associated with a very weak activation of CYC1-lacZ gene (Figure 5). Rme1Δ31–120 fused to the GAL4 DNA binding domain activated transcription of the HIS3 reporter gene, but not the ADE2 reporter gene. The activation of the HIS3 transcription is known to require weaker interactions between the activation domain and basic transcription machinery compared with other reporter genes (James et al., 1996). This weak ability to activate transcription of the HIS3 was inhibited by the addition of 5 mM of 3-AT (Figure 6). Thus, the first 31 amino acids of Rme1p represent an additional effector module, which can restore activation/repression when the minimal effector domain of Rme1p is altered. Accordingly, lexA-Rme1Δ4–31 repression fold is lower then that of the wild-type lexA-Rme1p (Figure 2B). Moreover, it is possible that amino acids 31–60 of Rme1p also contribute redundantly to repression and activation, because the progressive deletion of these amino acids results in an additional reduction in activation ability. The significance of this region is also suggested by the comparison of the activation and repression abilities of proteins Rme1Δ1–31 and Rme1Δ1–61 in Figure 5. The progressive C-terminal deletions of Rme1p also display a gradual reduction in repression/activation ability. Indeed, compare activation and repression abilities of proteins lexA-Rme1Δ210–300, lexA-Rme1Δ179–300, and lexA-Rme1Δ148–300 via the lexA sites in Figures 2B, 3B, and 4B. Moreover, the amino acids 148–179 are dispensable for repression in the otherwise wild-type protein but contribute to the repression in lexA-Rme1Δ179–300 (Figure 2B and 3B). These data suggest that Rme1p may include multiple redundant determinants that can contribute synergistically to repression.
In summary, the data presented above demonstrated that structural integrity of zinc fingers of Rme1p is not required for Rme1p-mediated effects on transcription. Amino acids 61–148 of the N-terminus comprise a minimal activation domain, which efficiently activates transcription when transferred to a heterologous DNA binding domain. Moreover, these amino acids are absolutely required for repression as well. However, it seems that the efficient repression depends on additional amino acids of the C terminus (179–210). Additional effector determinants exist within the first 31 amino acids of the N-terminal part of the Rme1 protein. These additional effector determinants contribute both to repression and to activation.
Sin4p and Rgr1p Are Not Required for Rme1p-Mediated Activation
The finding that Rme1p repression and activation domains overlap suggests that Rme1p may have a single biochemical activity or interaction that influences transcription. If so, other gene products must determine whether that activity results in repression or activation. This model predicts that such gene products will be required only for activation or repression, but not for both activities. Covitz et al. (1994) showed previously that the rgr1–100 mutation causes a defect in repression of the IME1 and reporter genes, but not in activation of reporter genes. However, rgr1–100 is not a simple loss-of-function mutation: whereas the rgr1Δ mutation is lethal (Stillman et al., 1994), the rgr1–100 is not, and is partially dominant (Covitz et al., 1994). This could mean that rgr1–100 mutation causes a defect only in repression, whereas rgr1Δ may be defective in both repression and activation. It was also found previously that a sin4Δ mutation causes a defect in repression (Covitz et al., 1994). Thus, using reporter genes, we quantified activation and repression activities of Rme1p in sin4Δ, rgr1–100, and control strains (Figure 1B). It has been shown that rgr1 and sin4 mutations permit some expression of genes lacking UAS regions (Jiang and Stillman, 1992; Stillman et al., 1994). Indeed, the expression of all reporter plasmids was higher in the rgr1–100 and the sin4Δ strains (Table 2). The rgr1–100 and the sin4Δ strains were both defective in repression of reporter plasmids, as expected. On the other hand, the activation fold was 53 for the wild-type strain and 89 and 47 for sin4Δ and rgr1–100 strains carrying the wild-type RME1 gene, respectively. In conclusion, Sin4p is not required for Rme1p-dependent activation, just as Rgr1p is not required for this mode of transcriptional activation.
Table 2.
Effects of rgr1-100 and sin4Δ on Rme1p-mediated repression and activation
| Host genotype | Repression
|
Activation
|
||||
|---|---|---|---|---|---|---|
| pBS9 | pBS8 | Fold | pLG312SΔSS | pAC110-6 | Fold | |
| Wild type | 854 | 108 | 8 | 1 | 74 | 53 |
| sin4Δ | 1601 | 1375 | 1 | 4 | 356 | 89 |
| rgr1-100 | 2196 | 1622 | 1 | 7 | 317 | 47 |
DISCUSSION
Rme1p functions both as a repressor and an activator of transcription. The three zinc fingers in the Rme1p C-terminal region are required for DNA binding (Figure 7) (Covitz and Mitchell, 1993; Shimizu et al., 1997a; Shimizu et al., 2001). We have shown here that the Rme1p N-terminal region is necessary and sufficient for both repression and activation (Figures 2 and 4). Thus, the role of Rme1p as an effector of transcription is not simply to displace proteins that bind to overlapping DNA sites.
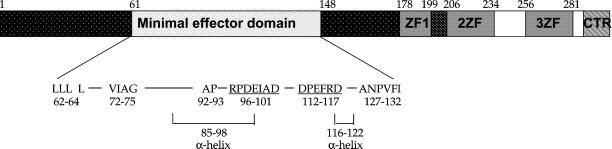
Figure 7.
Structure-functional summary of Rme1p. The C-terminal part of Rme1p contains three zinc fingers (ZF boxes) and C-terminal segment (CTR) with properties of α-helix. This part of the protein is important for DNA- binding (Shimizu et al., 2001). The N-terminal part of Rme1p contains minimal effector domain (hatched region), which spans between amino acids 61 and 148, and additional effector subdomains (dotted regions), which augment repression and activation. The minimal effector domain contains stretches of bulky hydrophobic amino acids at positions 62–64,66; 72–77; 92–93; and 127–132 and stretches of charged residues at positions 96–101 and 112–117 (underline). The regions 85–98 and 116–122 are predicted to form α-helixes. Amino acids are presented by their one-letter codes.
The minimal regions required for repression and activation overlap completely and lie between amino acids 61 and 148 (Figure 7). It is unusual for a protein to have a single effector region that directs both repression and activation. For example, Ume6p has a central repression domain that interacts with Sin3p and Rdp3p (Kadosh and Struhl, 1997) and an N-terminal activation domain that interacts with Rim11p and Ime1p (Bowdish et al., 1995; Rubin-Bejerano et al., 1996; Malathi et al., 1997). Rap1p has neighboring but separable repression and activation domains (Sussel and Shore, 1991). It is possible that the single Rme1p transcriptional effector domain interacts with a single target protein whose activity—repression or activation—is dictated by neighboring proteins or the chromatin environment. A second possibility is that the N-terminal region of Rme1p can interact with two different proteins or complexes that individually yield exclusively repression or activation.
The Rme1p N-terminal effector domain shows no extensive homology to known activation or repression domains, and has no significant primary sequence identity to other proteins in current databases. However, the effector domains, does contain residues and possible secondary structures that are implicated in activation or repression by other transcription factors. For example, it contains stretches of bulky hydrophobic amino acids and charged residues (Figure 7). Two regions (85–98 and 116–122) are predicted to form α-helices, which may facilitate protein–protein interactions. These patches of similarity to other repressors and activators are consistent with the possibility that Rme1p has interdigitated residues that contribute only to repression or activation. This model predicts that mutational alteration of specific Rme1p N-terminal residues may impair only repression or activation, in contrast to the broad effects of the deletions studied here. However, we note that random mutagenesis of RME1 has yielded numerous mutations that impair DNA binding, but none that specifically impair repression. It is possible that the individual Rme1p effector segments function redundantly, so that multiple point mutations would be necessary to inactivate a specific effector function. Our deletion analysis here is consistent with such a model, in that regions flanking the effector domain can augment repression and activation (Figure 7).
The observation that Rme1p activation and repression domains overlap brings to the foreground the question of whether identical protein complexes form at Rme1p-repressed and Rme1p-activated promoters. One simple possibility is that the Mediator or a smaller Rgr1p-Sin4p complex is the Rme1p-interacting target, because these proteins act as both positive and negative regulators of transcription (Stillman et al., 1994). However, we have shown clearly that Sin4p and Rgr1p are not required for Rme1p-mediated activation. Thus, if RNA polymerase II holoenzyme subcomplexes are direct Rme1p targets, there must be distinct subcomplexes that are brought to the Rme1p-repressed and Rme1p-activated promoters (Myers et al., 1999). Perhaps recruitment of a Mediator subcomplex lacking Sin4p and Rgr1p prompts RNA polymerase II to activate transcription, as occurs when the Rme1p-binding site is situated in place of a UAS.
We have favored the model that Rgr1p-Sin4p is recruited by Rme1p at repressed promoters because it explains genetic relationships simply. However, we have recently observed that lexA-Rgr1p does not repress the lexO-IME1 test gene (Blumental-Perry, 2001), whereas lexA-Rme1p derivatives are effective repressors. In addition, the fact that Rme1p repression excludes nearby transcriptional activators from DNA (Shimizu et al., 1997b) is not an expected consequence of direct interaction between Rme1p and the Mediator. Thus, more complex biochemical relationships must be considered. One possibility, discussed previously (Shimizu et al., 1997b), is that Rme1p repression depends on a nucleosome structure or density that is unachievable in rgr1 or sin4 mutants. This model predicts that other mutations with similar effects on nucleosome structure will also impair Rme1p repression specifically. A second possibility is that a gene specifying the hypothetical Rme1p corepressor is not expressed in rgr1 or sin4 mutants. Candidate corepressor genes may then be identified through genome-wide expression surveys. A direct approach to this question is to identify proteins that interact with the Rme1p effector domain. Overlap between Rme1p repression and activation regions precludes the use of conventional two-hybrid cloning, but we expect that biochemical identification of Rme1p effector region–interacting proteins, combined with chromatin immunoprecipitation of the RC, will provide a direct route to address these mechanistic questions.
ACKNOWLEDGMENTS
We thank Dr. D. Zenvirth and Dr. S. Klein for comments on the manuscript and Dr. R. Kornberg for fruitful discussion. This work was supported by Human Frontier Science Program grant RG0379/1997-M and National Institutes of Health grant GM-39531 to A.P.M. and by a research grant from the Israel Science Foundation to G.S.
Abbreviations used:
- RC
repression cassette
- UAS
upstream activating sequence
- GBD
GAL4 DNA binding domain
- 3-AT
3-aminotriazole
Footnotes
DOI: 10.1091/mbc.01–09–0468.
REFERENCES
- Blair WS, Bogerd HP, Madore SJ, Cullen BR. Mutational analysis of the transcription activation domain of RelA: identification of a highly synergistic minimal acidic activation module. Mol Cell Biol. 1994;14:7226–7234. doi: 10.1128/mcb.14.11.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumental-Perry A. Negative Regulation of Meiosis by RME1, and CDC25 Gene Products. Ph.D. Thesis. Jerusalem, Israel: The Hebrew University of Jerusalem; 2001. [Google Scholar]
- Bowdish KS, Yuan HE, Mitchell AP. Positive control of yeast meiotic genes by the negative regulator UME6. Mol Cell Biol. 1995;15:2955–2961. doi: 10.1128/mcb.15.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent R, Ptashne M. A bacterial repressor protein or a yeast transcriptional terminator can block upstream activation of a yeast gene. Nature. 1984;312:612–615. doi: 10.1038/312612a0. [DOI] [PubMed] [Google Scholar]
- Bushmeyer S, Park K, Atchison ML. Characterization of functional domains within the multifunctional transcription factor, YY1. J Biol Chem. 1995;270:30213–30220. doi: 10.1074/jbc.270.50.30213. [DOI] [PubMed] [Google Scholar]
- Carlson M. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu Rev Cell Dev Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- Covitz P. Limitation of the Repression Cassette and Structural Features of RME1. Ph.D. Thesis. New York, NY: Columbia University; 1993. [Google Scholar]
- Covitz PA, Herskowitz I, Mitchell AP. The yeast RME1 gene encodes a putative zinc finger protein that is directly repressed by a1–a2. Genes Dev. 1991;5:1982–1989. doi: 10.1101/gad.5.11.1982. [DOI] [PubMed] [Google Scholar]
- Covitz PA, Mitchell AP. Repression by the yeast meiotic inhibitor RME1. Genes Dev. 1993;7:1598–1608. doi: 10.1101/gad.7.8.1598. [DOI] [PubMed] [Google Scholar]
- Covitz PA, Song W, Mitchell AP. Requirement for RGR1 and SIN4 in RME1-dependent repression in Saccharomyces cerevisiae. Genetics. 1994;138:577–586. doi: 10.1093/genetics/138.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale C, Duenas E, Jackson BM, Reusser U, Braus GH, Hinnebusch AG. The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol Cell Biol. 1995;15:1220–1233. doi: 10.1128/mcb.15.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L, Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983;32:1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Hardwick JM, Tse L, Applegren N, Nicholas J, Veliuona MA. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J Virol. 1992;66:5500–5508. doi: 10.1128/jvi.66.9.5500-5508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope IA, Mahadevan S, Struhl K. Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature. 1988;333:635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- Jackson BM, Drysdale CM, Natarajan K, Hinnebusch AG. Identification of seven hydrophobic clusters in GCN4 making redundant contributions to transcriptional activation. Mol Cell Biol. 1996;16:5557–5571. doi: 10.1128/mcb.16.10.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YW, Dohrmann PR, Stillman DJ. Genetic and physical interactions between yeast RGR1 and SIN4 in chromatin organization and transcriptional regulation. Genetics. 1995;140:47–54. doi: 10.1093/genetics/140.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YW, Stillman DJ. Involvement of the SIN4 global transcriptional regulator in the chromatin structure of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4503–4514. doi: 10.1128/mcb.12.10.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- Kane SM, Roth R. Carbohydrate metabolism during ascospore development in yeast. J Bacteriol. 1974;118:8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassir Y, Granot D, Simchen G. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell. 1988;52:853–862. doi: 10.1016/0092-8674(88)90427-8. [DOI] [PubMed] [Google Scholar]
- Kassir Y, Simchen G. Regulation of mating and meiosis in yeast by the mating-type region. Genetics. 1976;82:187–206. doi: 10.1093/genetics/82.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec M, Byers B, Esposito R, Mitchell AP. Meiosis and sporulation in Saccharomyces cerevisiae. In: Pringle JR, Broach JR, Jones EW, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 889–1036. [Google Scholar]
- Li Y, Bjorklund S, Jiang YW, Kim Y, Lane WS, Stillman DJ, Kornberg RD. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K, Xiao Y, Mitchell AP. Interaction of yeast repressor-activator protein Ume6p with glycogen synthase kinase 3 homolog Rim11p. Mol Cell Biol. 1997;17:7230–7236. doi: 10.1128/mcb.17.12.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AP, Herskowitz I. Activation of meiosis and sporulation by repression of the RME1 product in yeast. Nature. 1986;319:738–742. doi: 10.1038/319738a0. [DOI] [PubMed] [Google Scholar]
- Myers LC, Gustafsson CM, Hayashibara KC, Brown PO, Kornberg RD. Mediator protein mutations that selectively abolish activated transcription. Proc Natl Acad Sci USA. 1999;96:67–72. doi: 10.1073/pnas.96.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn L, Mitchell AP. The yeast MCK1 gene encodes a protein kinase homolog that activates early meiotic gene expression. Genes Dev. 1991;5:533–548. doi: 10.1101/gad.5.4.533. [DOI] [PubMed] [Google Scholar]
- Regier JL, Shen F, Triezenberg SJ. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc Natl Acad Sci USA. 1993;90:883–887. doi: 10.1073/pnas.90.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winstone F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Rubin-Bejerano I, Mandel S, Robzyk K, Kassir Y. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional repressor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol Cell Biol. 1996;16:2518–2526. doi: 10.1128/mcb.16.5.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruden DM, Ma J, Li Y, Wood K, Ptashne M. Generating yeast transcriptional activators containing no yeast protein sequences. Nature. 1991;350:250–252. doi: 10.1038/350250a0. [DOI] [PubMed] [Google Scholar]
- Sakai A, Shimizu Y, Kondou S, Hibazakura T, Hishinuma F. Structure and molecular analysis of RGR1, a gene required for glucose repression of Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:4130–4138. doi: 10.1128/mcb.10.8.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M, Hara M, Murase A, Shindo H, Mitchell AP. Dissection of the DNA binding domain of yeast Zn-finger protein Rme1p, a repressor of meiotic activator IME1. Nucleic Acids Symp Ser. 1997a;37:175–176. [PubMed] [Google Scholar]
- Shimizu M, Li W, Covitz PA, Hara M, Shindo H, Mitchell AP. Genomic footprinting of the yeast zinc finger protein Rme1p and its roles in repression of the meiotic activator IME1. Nucleic Acids Res. 1998;26:2329–2336. doi: 10.1093/nar/26.10.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M, Li W, Shindo H, Mitchell AP. Transcriptional repression at a distance through exclusion of activator binding in vivo. Proc Natl Acad Sci USA. 1997b;94:790–795. doi: 10.1073/pnas.94.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M, Murase A, Hara M, Shindo H, Mitchell AP. A C-terminal segment with properties of α-helix is essential for DNA binding, and in vivo function of zinc finger protein Rme1p. J Biol Chem. 2001;276:37680–37685. doi: 10.1074/jbc.M105342200. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HE, Su SS, Neigeborn L, Driscoll SE, Mitchell AP. Role of IME1 expression in regulation of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6103–6113. doi: 10.1128/mcb.10.12.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman DJ, Dorland S, Yu Y. Epistasis analysis of suppressor mutations that allow HO expression in the absence of the yeast SW15 transcriptional activator. Genetics. 1994;136:781–788. doi: 10.1093/genetics/136.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su SS, Mitchell AP. Identification of functionally related genes that stimulate early meiotic gene expression in yeast. Genetics. 1993;133:67–77. doi: 10.1093/genetics/133.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Shore D. Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc Natl Acad Sci USA. 1991;88:7749–7753. doi: 10.1073/pnas.88.17.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toone WM, Johnson AL, Banks GR, Toyn JH, Stuart D, Wittenberg C, Johnston LH. Rme1, a negative regulator of meiosis, is also a positive activator of G1 cyclin gene expression. EMBO J. 1995;14:5824–5832. doi: 10.1002/j.1460-2075.1995.tb00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidan S, Mitchell AP. Stimulation of yeast meiotic gene expression by the glucose-repressible protein kinase Rim15p. Mol Cell Biol. 1997;17:2688–2697. doi: 10.1128/mcb.17.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S, Greaves R, O'Hare P. Transcriptional activation by the acidic domain of Vmw65 requires the integrity of the domain and involves additional determinants distinct from those necessary for TFIIB binding. Mol Cell Biol. 1993;13:5233–5244. doi: 10.1128/mcb.13.9.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]