Abstract
Cytonuclear signaling is essential for long-term alterations of cellular properties. Several pathways involving regulated nuclear accumulation of Ser/Thr kinases have been described but little is known about cytonuclear trafficking of tyrosine kinases. Proline-rich tyrosine kinase 2 (Pyk2) is a cytoplasmic non-receptor tyrosine kinase enriched in neurons and involved in functions ranging from synaptic plasticity to bone resorption, as well as in cancer. We previously showed the Ca2+-induced, calcineurin-dependent, nuclear localization of Pyk2. Here, we characterize the molecular mechanisms of Pyk2 cytonuclear localization in transfected PC12 cells. The 700–841 linker region of Pyk2 recapitulates its depolarization-induced nuclear accumulation. This region includes a nuclear export motif regulated by phosphorylation at residue S778, a substrate of cAMP-dependent protein kinase and calcineurin. Nuclear import is controlled by a previously identified sequence in the N-terminal domain and by a novel nuclear targeting signal in the linker region. Regulation of cytonuclear trafficking is independent of Pyk2 activity. The region regulating nuclear localization is absent from the non-neuronal shorter splice isoform of Pyk2. Our results elucidate the mechanisms of Ca2+-induced nuclear accumulation of Pyk2. They also suggest that Pyk2 nuclear accumulation is a novel type of signaling response that may contribute to specific long-term adaptations in neurons.
Keywords: Non-receptor tyrosine kinase, Cytonuclear localization, Nucleus, Phosphorylation, Protein phosphatase, Calcineurin
Introduction
Proline-rich tyrosine kinase 2 (Pyk2) is a Ca2+-activated non-receptor tyrosine kinase closely related to focal adhesion kinase (FAK) [1]. Pyk2 is enriched in adult neurons and plays an important role in neuronal plasticity [2–6]. In non-neuronal cells, Pyk2 is involved in osteoclast function [7], macrophage migration [8], and focal adhesion disassembly [9]. Neuronal Pyk2 is 5 kDa larger than in cells from the hematopoietic lineage, due to the retention of an additional exon coding for a peptide between the kinase and focal adhesion targeting (FAT) domains [10–12]. The functional differences between these isoforms are not known. Although the precise mechanism of Pyk2 activation remains unclear, increase in intracellular free Ca2+ can directly or indirectly induce Pyk2 dimerization and trans-autophosphorylation at Y402 creating a Src-homology-2 binding site that recruits Src family kinases and activates various signaling pathways [1, 5, 13–15].
In most cells, Pyk2 is localized in the cytoplasm [16, 17] and in neurons, in perikarya and dendritic shafts [18, 19]. We have previously demonstrated that Pyk2 activation in neuronal cells is concomitant to its Ca2+-induced, calcineurin-dependent nuclear accumulation following membrane depolarization [20]. Although Pyk2 nuclear accumulation has been serendipitously observed in various cell types or following mutations [21–27], the physiological relevance and mechanisms of its cytonuclear shuttling are not known. Cytonuclear trafficking of proteins larger than 40 kDa results from an active transport mediated by karyopherins, which facilitate translocation of cargos through the nuclear pore and release them in the nucleus, in the case of importins, or in the cytoplasm in the case of exportins [28, 29]. The best characterized mechanisms for nuclear import and export are based on the association of cargos that contain nuclear localization signal (NLS) or a hydrophobic nuclear export sequence (NES) with importins and the exportin chromosome region maintenance 1 (CRM1), respectively [30–32]. Here we characterize the role of NLS, NES, and other sequences in Pyk2 cytonuclear trafficking and its control by S778 phosphorylation by PKA and dephosphorylation by calcineurin. Moreover, we show that these regulatory mechanisms are specific for Pyk2 long-splice isoform expressed in neurons.
Materials and methods
Reagents
Leptomycin B and KN93 were from Calbiochem, cyclosporin A, FK506, forskolin, IBMX, H89, and myristoylated 14–22 fragment of protein kinase A inhibitory peptide (myrPKI14–22) from Sigma. Rabbit anti-Pyk22–18 antibodies were described [33] or from Sigma, anti-pY402-Pyk2 from Invitrogen, anti-GFP from Roche and anti-DARPP-32 was a gift from P. Greengard (Rockefeller University). Anti-pS778 was raised in rabbit against a pS778-Pyk2773–784 peptide by Eurogentec with a 28-day protocol. Alexa-488- or Cy3-coupled secondary antibodies were from Molecular Probes (Sunnyvale, CA).
Cell cultures
PC12 cells were grown on type I collagen (BD Biosciences) in RPMI medium containing 10 % horse serum and 5 % fetal calf serum (v/v). COS7 cells were grown in DMEM medium containing 10 % fetal calf serum. Transfections were done with Lipofectamine 2000 (Invitrogen) in cells at about 70 % confluence. For fluorescence analyses, PC12 cells were grown in RPMI on type I collagen-coated glass coverslips after incubation with poly-l-lysine (Sigma). Depolarization was performed by isosmotic replacement of 40 mM NaCl by 40 mM KCl in the extracellular medium as described previously [20].
Immunofluorescence
Cells were fixed 15 min in a 4 % (w/v) paraformaldehyde solution, permeabilized 12 min on ice with 1/1 methanol/acetone (v/v). Cells were washed with 20 mM sodium phosphate, pH 7.5, 150 mM NaCl (PBS), blocked and incubated 2 h with primary antibodies. After washes, cells were incubated 45 min with Alexa-488- or Cy3-coupled secondary antibodies, washed and mounted in Vectashield (Vector Laboratories) with 4′,6′-diamidino-2-phenylindole (DAPI). PC12 cells transfected with GFP constructs were fixed, washed, and mounted in Vectashield with DAPI. Images were acquired with a Micromax numerical CCD camera (Roper Scientific).
Immunoblot analysis
PC12 cells were lysed in a preheated (at 100 °C) solution of 1 % (w/v) SDS, 1 mM Na3VO4, and incubated 5 min at 100 °C. Equal amounts of protein were separated by SDS-PAGE (7–10 % acrylamide, w/v) before electrophoretic transfer onto a nitrocellulose membrane (Hybond Pure; GE Healthcare). Membranes were fixed in 10 % (v/v) acetic acid, 10 % (v/v) isopropanol and washed and blocked 1 h at room temperature in Tris-buffered saline (100 mM NaCl and 10 mM Tris, pH 7.5) with 5 % (w/v) non-fat dry milk for protein detection or with 0.1 % (v/v) Tween 20, 3 % (w/v) BSA for detection of pY402 or pS778. Membranes were then incubated overnight at 4 °C with primary antibodies. Bound antibodies were detected with anti-rabbit IgG IRdye800CW-coupled and anti-mouse IgG IRdye700DX-coupled antibodies (Rockland Immunochemicals). Fluorescence was analyzed at 680 and 800 nm using the Odyssey infrared imager (Li-Cor) and quantified using Odyssey software. Data were normalized to the mean value of untreated controls in the same gels.
GST protein cloning
Pyk2 700–841 WT or S778A and DARPP-32 sequence were inserted into pGEX-6P-2 (Amersham) and expressed in BL-21-competent bacterial cells. The resultant GST-fusion proteins were affinity-purified on glutathione-Sepharose beads (Pharmacia) as described [34].
In vitro phosphorylation/dephosphorylation assays
Phosphorylation reactions were carried out 10–45 min at 30 °C in 50 mM HEPES (pH 7.4), 10 mM magnesium acetate, 1 mM EGTA, 5 μM ATP, 3 μl [γ-32P]ATP (3 Ci/mmol, 10 μCi/ml), and 10 ng of cAMP-dependent protein kinase catalytic subunit (Millipore). Reactions were stopped by the addition of 25 μl of a stop solution (150 g/l SDS, 0.3 M Tris–Cl, pH 6.8, 25 % (v/v) glycerol, traces of pyronine Y) and heated at 95 °C. For dephosphorylation GST fusion proteins bound to glutathione-Sepharose were washed in PBS containing 1 % Triton-X-100 (v/v), 0.5 mM DTT, and 0.5 mM PMSF, and incubated in a dephosphorylation buffer (Tris 20 mM, NaCl 100 mM, MgCl2 5 mM, DTT 1 mM, BSA 0.5 mg/ml, CaCl2 1 mM) with recombinant calcineurin (Calbiochem) and 1 μM calmodulin. After electrophoresis, polyacrylamide gels were dried and incorporated 32P measured with Fuji Phosphoimager, FLA7000, together with [γ-32P]ATP spotted on bench coat paper for quantification.
Cloning and directed mutagenesis
Plasmids encoding the various truncations and point mutations were prepared from full-length GFP-Pyk2 (rat sequence) [20] by site-directed mutagenesis (QuikChange, Stratagene). Briefly, the KpnI fragment of Pyk2 was subcloned into the KpnI site of pBlueScript KS (Fermentas). A BglII site was inserted upstream A421 in frame with the BglII site of pEGFP-C1 (Clontech). The BglII fragment of GFP-Pyk2 was then deleted to obtain 421–1009. The SacI fragment of Pyk2 was subcloned into the SacI site of pBlueScript KS (Fermentas). A stop codon and an EcoRI site downstream were inserted instead of L841 to delete the EcoRI fragment and obtain 1–840. A stop codon was inserted instead of R700 to obtain 1–699. Pyk2700–841 was PCR amplified and fused into the BglII–EcoRI sites of pEGFP-C1 (Clontech) to obtain GFP-(700–841). All constructs were verified by DNA sequencing. Oligonucleotides were from Sigma and MWG (Table 1).
Table 1.
Oligonucleotide sequences
| 5P-R184AR185A | 5′-GGCTGTCTGGAGCTCACGACATTCTTCAAGGACATGCCC-3′ |
| 3P-R184AR185A | 5′-GGGCATGTCCTTGAAGAATGTCGTGAGCTCCAGACAGCC-3′ |
| 5P-(1–420) | 5′-CCAGGAGGCCCACAGTAAGATCTTGCCCGTGAAGATGTA-3′ |
| 3P-(1–420) | 5′-TACATCTTCACGGGCAAGATCTTACTGTGGGCCTCCTGG-3′ |
| 5P-(1–699) | 5′-GGAACGCTCGTTACTGACCCCCCAAAATATTGG-3′ |
| 3P-(1–699) | 5′-CCAATATTTTGGGGGGTCAGTAACGAGCGTTCC-3′ |
| 5P-LFV3A | 5′-GCTGGCTCCTAAGGCGCAGGCCCAGGCCCCTGAGGGTCTGTG-3′ |
| 3P-LFV3A | 5′-CACAGACCCTCAGGGGCCTGGGCCTGCGCCTTAGGAGCCAGC-3′ |
| 5P-Δ(738–780) | 5′-GGCTCCTAAGCTGCAGTTCGAGGAGGACTTCATCCGGCCC-3′ |
| 3P-Δ(738–780) | 5′-GGGCCGGATGAAGTCCTCCTCGAACTGCAGCTTAGGAGCC-3′ |
| 5P-(700–745) | 5′-CCTGAGGGTCTGTGTGCCTGATCTCCTACACTTACG-3′ |
| 3P-(700–745) | 5′-CGTAAGTGTAGGAGATCAGGCACACAGACCCTCAGG-3′ |
| 5P-S745A/T747A | 5′-GTGTGCCAGCGCTCCTGCACTTACGAGCCC-3′ |
| 3P-S756A/T758A | 5′-GGGCTCGTAAGTGCAGGAGCGCTGGCACAC-3′ |
| 5P-(700–758) | 5′-CCTATGGAGTATCCATGACCAGTTAACTCGCTGC-3′ |
| 3P-(700–758) | 5′-GCAGCGAGTTAACTGGTCATGGATACTCCATAGG-3′ |
| 5P-S758A | 5′-CCTATGGAGTATCCAGCTCCAGTTAACTCG-3′ |
| 3P-S758A | 5′-CGAGTTAACTGGAGCTGGATACTCCATAGG-3′ |
| 5P-S762A | 5′-CCATCTCCAGTTAACGCGCTGCACACCCCG-3′ |
| 3P-S762A | 5′-CGGGGTGTGCAGCGCGTTAACTGGAGATGG-3′ |
| 5P-T765A | 5′-CGCTGCACGCCCCGCCTCTGCACC-3′ |
| 3P-T765A | 5′-GGTGCAGAGGCGGGGCGTGCAGCG-3′ |
| 5P-(700–767) | 5′-CTGCACACCCCGCCTTAGCACCGACACAATGTC-3′ |
| 3P-(700–767) | 5′-GACATTGTGTCGGTGCTAAGGCGGGGTGTGCAG-3′ |
| 5P-K775A/R776A | 5′-CGACACAATGTCTTCGCGGCCCACAGCATGCGGGAGG-3′ |
| 3P-K775A/R776A | 5′-CCTCCCGCATGCTGTGGGCCGCGAAGACATTGTGTCG-3′ |
| 5P-S778A | 5′-GCGCCACGCGATGCGGGAGGAGG-3′ |
| 3P-S778A | 5′-CCTCCTCCCGCATCGCGTGGCGC-3′ |
| 5P-S788A | 5′-GGACTTCATCCGGCCCGCTAGTCGAGAAGAGG-3′ |
| 3P-S788A | 5′-CCTCTTCTCGACTAGCGGGCCGGATGAAGTCC-3′ |
| 5P-(700–793) | 5′-CGAGAAGAGGCCTAGCAGCTCTGGGAGGC-3′ |
| 3P-(700–793) | 5′-GCCTCCCAGAGCTGCTAGGCCTCTTCTCG-3′ |
| 5P-(1–840) | 5′-GACAAGTCCCCATGAATTCAGAGAAGGAGGCCGGC-3′ |
| 3P-(1–840) | 5′-GCCGGCCTCCTTCTCAGTATTCATGGGGACTTGTC-3′ |
| 5P-GFP-(700–841) | 5′-AATTCTCGACCCCCCAAAATATTGG-3′ |
| 3P-GFP-(700–841) | 5′-ATTGAATTCTGGGGACTTGTCATTC-3′ |
| 5P-GST-D32 | 5′-AATTAAGGATCCATGGACCCCAAGGACCGCAAG-3′ |
| 3P-GST-D32 | 5′-TATTGAATTCTTTTATGTGCCGGACTCAGGGGG-3′ |
Quantifications and statistical analysis
Pyk2 is usually present in both the cytoplasm and the nucleus, with some heterogeneity in both compartments. This distribution combined with different levels of expression in various cells precluded the use of automatic detection of nuclear localization. To circumvent this difficulty, we have previously shown that it is possible to estimate the cytonuclear distribution of Pyk2 by quantifying the number of cells in which the nuclear fluorescence is superior or equal to cytoplasmic fluorescence (n ≥ c), with results similar to those obtained by confocal microscopy [20]. Thus, cells were classified by an observer blind to the treatment and/or mutation, using micrographs obtained by epifluorescence microscopy. In some experiments, the nuclear/cytoplasmic fluorescence ratio (n/c) was calculated for each cell after measuring the fluorescence intensity at several locations in the nucleus and cytoplasm. Nuclear limits were identified by DAPI staining. The percentage of cells in each category was determined in each coverslip (approximately 50–100 cells per coverslip in 10–20 fields). Data were from three or more independent experiments, each in duplicate. Statistical analysis was done using GraphPad Prism 3.02.
Results
The 700–841 region of Pyk2 recapitulates its depolarization-regulated nuclear localization
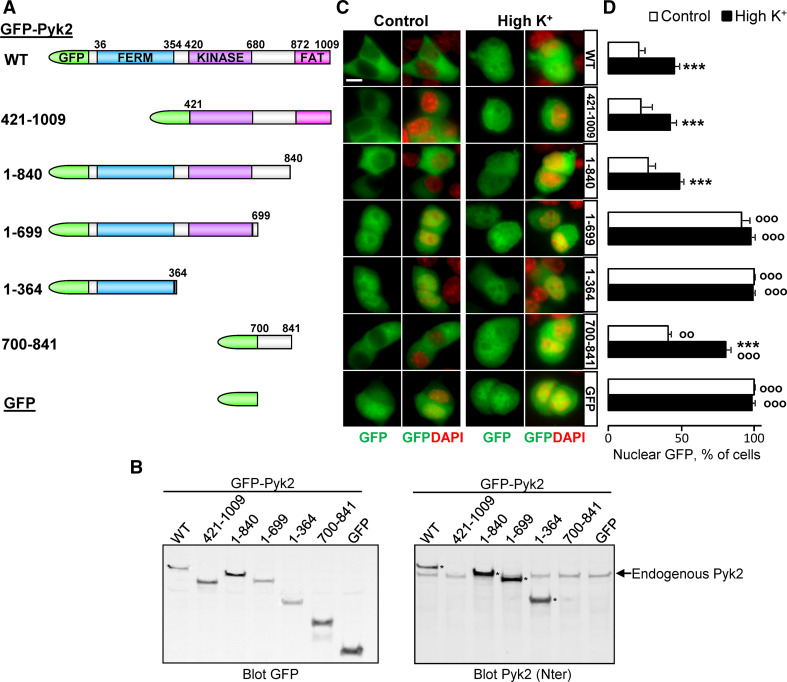
To determine which regions are important for regulated nuclear localization, we generated a set of deletions in Pyk2 fused to EGFP (Fig. 1a, b). We examined their intracellular distribution in basal conditions and following a transient depolarization induced by placing the cells in a medium in which 40 mM of extracellular NaCl were replaced by the same concentration of KCl. GFP-Pyk2 was expressed as a 135-kDa protein (Fig. 1a, b). Subcellular distribution of Pyk2 was classified into two classes (n < c and n ≥ c) depending on whether nuclear labeling intensity was lower than or equal to, or greater than cytoplasmic labeling, respectively. As previously described [20], the proportion of PC12 cells with a predominantly nuclear localization of GFP-Pyk2 was ~20 % under basal conditions (control) and ~40 % after depolarization (High K+, Fig. 1c, d). The N-terminus-deleted form, GFP-Pyk2421–1009, had a similar pattern of intracellular localization in basal conditions and after depolarization (Fig. 1c, d). This showed that the N-terminal region, including the 4.1-ezrin-radixin-moesin (FERM) domain, was not crucial for the regulation of Pyk2 intracellular localization. This result was important since it revealed that the nuclear localization sequence (NLS) located in the FERM domain of FAK and Pyk2 [23, 35] was not necessary for the regulated nuclear accumulation of Pyk2. The deletion of the C-terminal FAT domain had also no effect on the basal and depolarization-induced intracellular distribution of GFP-Pyk21–840 (Fig. 1c, d). Further truncation of GFP-Pyk2 (GFP-Pyk21–699, and GFP-Pyk21–364) resulted in a strong GFP-fluorescence in the nucleus in the absence or presence of depolarizing medium (Fig. 1c, d). We therefore focused on the 700–841 region of Pyk2 whose deletion abolished the regulated nuclear localization. GFP-Pyk2700–841 was excluded from the nucleus in ~60 % of cells in basal conditions, but was strongly relocalized in the nucleus after depolarization (Fig. 1c, d). These results demonstrated that GFP–Pyk2700–841 localization was regulated by membrane depolarization similar to the full-length protein.
Fig. 1.
Pyk2 700–841 region recapitulates cytonuclear regulation. a Schematic representation of Pyk2 constructs. b Expression levels of the various GFP-Pyk2 constructs in PC12 cells. Left panel anti-GFP immunoblot of PC12 cells transfected with GFP-Pyk2 (WT), deletion mutants (GFP-Pyk2421–1009, GFP-Pyk21–840, GFP-Pyk21–699, GFP-Pyk21–364, GFP-Pyk2700–841, as indicated) or GFP. Right panel immunoblot with an antibody reacting with the N-terminal region of the protein. GFP-Pyk2 is indicated by an asterisk when visible (i.e. N-terminus present). c Transfected PC12 cells were treated with high K+ (High K+) or control solution for 3 min. GFP fluorescence and nuclei stained with DAPI were analyzed with a fluorescence microscope. d Quantification of the number of cells in c with n ≥ c GFP fluorescence. Values are means + SEM, two-way ANOVA: depolarization effect F (6,42) = 9.93, p < 0.0001, deletion effect F (6.42) = 175.95, p < 0.0001, interaction F (1,42) = 73.71, p < 0.0001. Newman–Keuls test: ***p < 0.001 versus control. °°p < 0.01, °°°p < 0.001 versus WT. Scale bar 5 μm
Pyk2 700–841 region contains a nuclear targeting sequence (NTS) that plays an accessory role in the nuclear import of the full-length protein
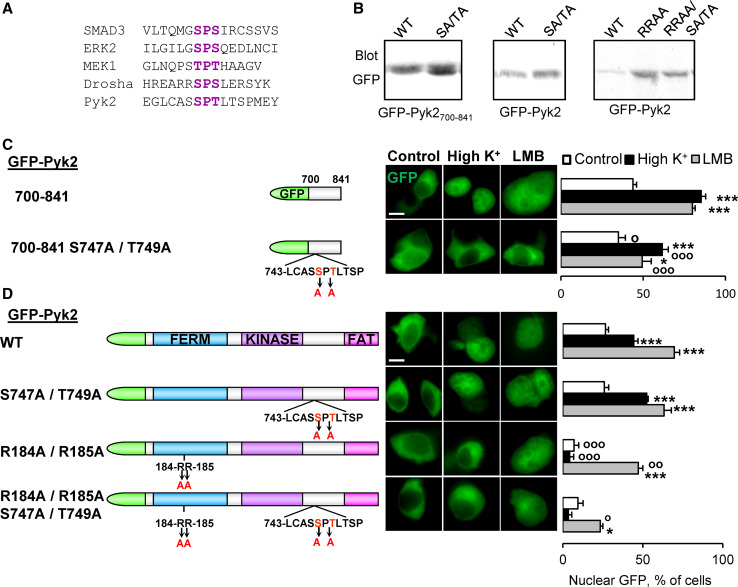
The regulated cytonuclear localization of GFP-Pyk2700–841 suggested that this protein of ~42 kDa could interact with karyopherins. We first investigated the mechanisms of import of GFP-Pyk2700–841. Importins (the karyopherins that mediate transport from the cytoplasm to the nucleus) usually interact with their cargo through basic residue-rich NLS sequences (see [29, 36] for reviews). However, GFP-Pyk2700–841 did not contain such motif. A different motif, termed nuclear translocation signal (NTS) was shown to play a role in the nuclear import of extracellular signal-regulated kinase (ERK), MAP-kinase/ERK kinase (MEK), SMAD3 [37], and RNase III Drosha [38]. The NTS consists in an S/T-P-S/T motif, which when phosphorylated binds to importin 7. The Pyk2 700–841 region contains a S747PT motif corresponding to a putative NTS (Fig. 2a). To test the role of this motif, we first analyzed the effects of mutating S747 and T749 into Ala in GFP-Pyk2700–841 (Fig. 2b, c). This mutation strongly reduced the nuclear accumulation of Pyk2 700–841 induced by depolarization (Fig. 2c). This double mutation similarly decreased the nuclear accumulation of GFP-Pyk2700–841 induced by leptomycin B (LMB) (Fig. 2c) a toxin that blocks the nuclear export by competing with the binding of NES to CRM1 [39, 40]. These findings strongly supported the idea that S747PT was a bona fide NTS involved in the control of GFP-Pyk2700–841 nuclear import.
Fig. 2.
Pyk2700–841 contains an atypical nuclear import motif. a Sequence alignment showing the NTS identified in SMAD3, ERK2, MEK1, Drosha, and the putative Pyk2 NTS. b Anti-GFP immunoblotting of PC12 cells transfected with GFP-Pyk2700–841 or GFP-Pyk2, with the indicated mutations (S747A, T749A, R184A, and R185A). PC12 cells transfected with GFP-Pyk2700–841 (c) or GFP-Pyk2 (d) with the indicated mutations (scheme on the left) were treated with high K+ (High K+, 40 mM, 3 min), LMB (LMB, 11 ng/ml, 3 h), or control solution. GFP fluorescence and nuclei stained with DAPI (middle) were analyzed and the number of cells with n ≥ c GFP quantified (right). Values are means + SEM. c Two-way ANOVA: mutation effect F (1,23) = 26.92, p < 0.0001, treatment effect F (1,23) = 118.57, p < 0.0001, interaction F (1,23) = 5.48, p < 0.05. Newman–Keuls test: *p < 0.05, **p < 0.01, ***p < 0.001 versus control. °p < 0.05, °°p < 0.01, °°°p < 0.001 versus WT. d Two-way ANOVA: p < 0.0001, mutation effect F (3,44) = 63.13, p < 0.0001, treatment effect F (1,44) = 14.55, p < 0.0005, interaction F (3,44) = 11.29. Newman–Keuls test: *p < 0.05, **p < 0.01, ***p < 0.001 versus control. °p < 0.05, °°p < 0.01, °°°p < 0.001 versus WT. Scale bar 5 μm
We then investigated the role of this putative NTS motif in the nuclear import of full-length Pyk2 by generating the same double mutation (S747A/T749A) in GFP-Pyk2 (Fig. 2b, d). This mutation did not alter the nuclear accumulation of GFP-Pyk2 induced by membrane depolarization or LMB (Fig. 2d). Since the FERM domain of Pyk2 and FAK contains a classical NLS [23, 35] we inferred that the effects of this NLS might mask those of the NTS. We tested the role of Pyk2 NLS in our experimental conditions by generating a double mutation of two of its basic residues (R184A/R185A). This mutation prevented the nuclear accumulation of GFP-Pyk2 in response to a short depolarization (Fig. 2c, d). This observation revealed that although the FERM domain was not necessary for depolarization-induced nuclear accumulation of GFP-Pyk2421–1009 (Fig. 1c, d), the NLS mutation was sufficient to block the rapid effects of depolarization in the context of the full-length protein. In contrast, the nuclear accumulation of R184A/R185A-GFP-Pyk2 was attenuated but still observed after a 3-h LMB treatment (Fig. 2d), suggesting the existence of additional weaker pronuclear sequence(s). We therefore examined the LMB-induced nuclear accumulation of GFP-Pyk2 containing mutations of both the FERM NLS and the NTS (R184A/R185A/S747A/T749A, Fig. 2b). The LMB-induced nuclear localization of this protein was dramatically reduced as compared to the simple NLS R184A/R185A mutant GFP-Pyk2 (Fig. 2d). This last experiment revealed that Pyk2 NTS in the linker region between the kinase and FAT domains plays an accessory role for the nuclear import of the full-length protein. This role appears critical only following mutation of the NLS motif or in the truncated form of the protein, GFP-Pyk2700–841.
The 700–841 region of Pyk2 contains a nuclear export sequence LL-LFV
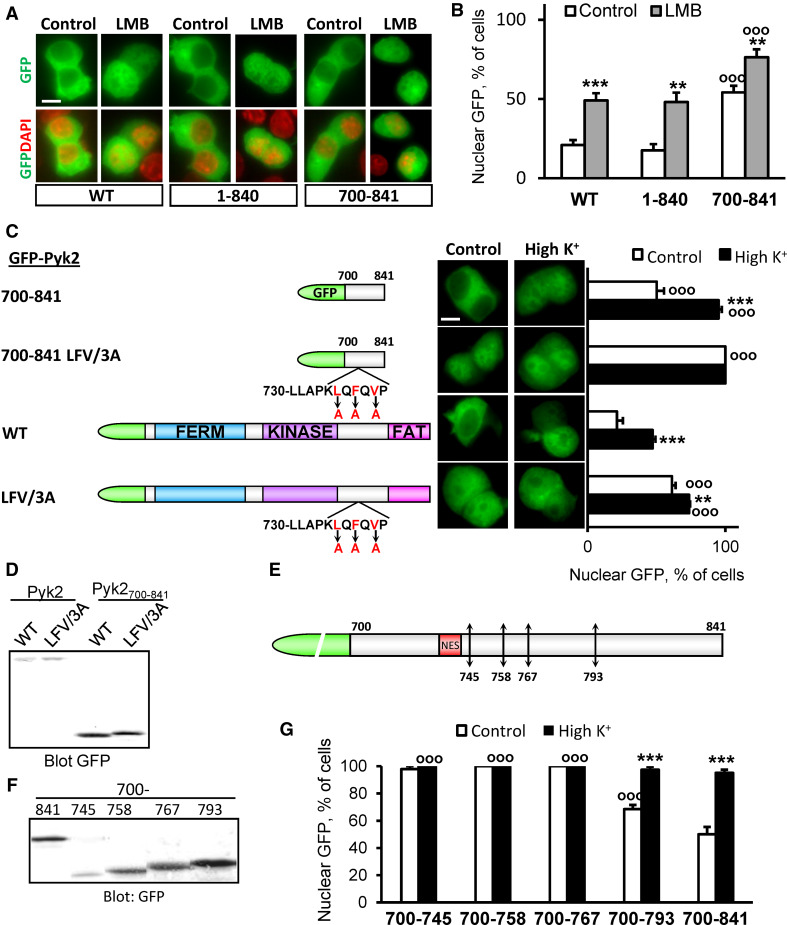
We then investigated the mechanisms of GFP-Pyk2700–841 nuclear export. We examined the localization of full-length and truncated forms of GFP-Pyk2 following LMB treatment (Fig. 3a, b). LMB increased the nuclear fluorescence of PC12 cells expressing GFP-Pyk2 or GFP-Pyk21–840 (Fig. 3a, b). It also had a significant effect on GFP-Pyk2700–841 (Fig. 3a, b). This indicated the presence of one or several LMB-sensitive NES in GFP-Pyk2700–841. The L730LAPKLQFQVP sequence of Pyk2A was identified as putative NES by the NetNES 1.1 server [41]. We mutated the 3 hydrophobic residues (L735A/F737A/V739A, i.e. LFV/3A) in GFP-Pyk2700–841 and full-length GFP-Pyk2 (Fig. 3c, d). LFV/3A-GFP-Pyk2700–841 displayed a nuclear labeling in 100 % of the cells (Fig. 3c) demonstrating the role of these residues in the regulation of Pyk2700–841 localization. This nuclear labeling was not modified by depolarization (Fig. 3c). In the context of the whole protein, GFP-Pyk2, the LFV/3A mutation also resulted in an increased number of cells displaying the fluorescence in the nucleus (Fig. 3c). However, the effect was not as pronounced as in GFP-Pyk2700–841 and a small further increase in the number of nuclear-positive cells was observed after depolarization (Fig. 3c). The incomplete effect of the mutation in full-length Pyk2 suggested the existence of additional export mechanisms and/or that the import mechanisms were not sufficiently strong to drive all the molecules into the nucleus during the LMB treatment. Altogether these data demonstrated that the LQFQV motif was necessary for the nuclear export of GFP-Pyk2700–841 and that it played an important role in the cytoplasmic localization of full-length GFP-Pyk2.
Fig. 3.
Pyk2700–841 contains a nuclear export motif. a PC12 cells transfected with GFP-Pyk2 (WT), 1–840 or 700–841 were incubated in the absence or presence of LMB (11 ng/ml, 3 h). GFP-Pyk2 fluorescence and nuclei stained with DAPI were analyzed. b Percentage of cells with nuclear GFP. Values are means + SEM, two-way ANOVA: deletion effect F (2,20) = 31.09, p < 0.0001, LMB effect F (1,20) = 48.88, p < 0.0001, interaction F (2,20) = 0.39, p > 0.05. Newman–Keuls test: **p < 0.01, ***p < 0.001 versus control. °°°p < 0.001 compared to WT. c PC12 cells transfected with WT or LFV/3A GFP-Pyk2 or GFP-Pyk2700–841 (scheme on the left) were treated with high K+ (High K+) or control solution for 3 min. GFP fluorescence and nuclei stained with DAPI were analyzed (middle) and the percentage of cells with n ≥ c GFP quantified (right). Values are means + SEM, two-way ANOVA: deletion effect F (4,30) = 158.77, p < 0.0001, depolarization effect F (1,30) = 113.22, p < 0.0001, interaction F (4,30) = 21.71, p < 0.0001. Newman–Keuls test: **p < 0.01, ***p < 0.001 versus control. °°°p < 0.001 versus WT. d Immunoblot analysis of constructs as in c with anti-GFP or anti-Pyk2 antibody. e Positions of stop codons inserted in GFP-Pyk2700–841 (745, 758, 767 and 793). f PC12 cells transfected with the constructs presented in e were analyzed by immunoblotting with anti-GFP. g Percentage of transfected cells with n ≥ c GFP. Values are means + SEM, two-way ANOVA: deletions effect F (4,25) = 47.88, p < 0.0001, depolarization effect F (1,25) = 82.58, p < 0.0001, interaction F (4,25) = 32.49, p < 0.0001. Newman–Keuls test: ***p < 0.001 versus control. °°°p < 0.001 versus 700–841. Scale bar 5 μm
We then examined whether the LQFQV motif was sufficient for the nuclear export of GFP-Pyk2700–841 by inserting stop codons in GFP-Pyk2700–841 and generating various truncated forms (GFP-Pyk2700–745, 700–758, 700–767, and 700–793, Fig. 3e, f). Only GFP700–793 was partly excluded from the nucleus but to a lesser extent than the entire GFP-Pyk2700–841 (Fig. 3g). These results suggested that (an) additional residue(s), presumably in the 767–793 sequence, was/were involved in the nuclear export activity of the 700–841 region of Pyk2 and that these residues may be involved in the Ca2+- and calcineurin-dependent regulation of GFP-Pyk2700–841.
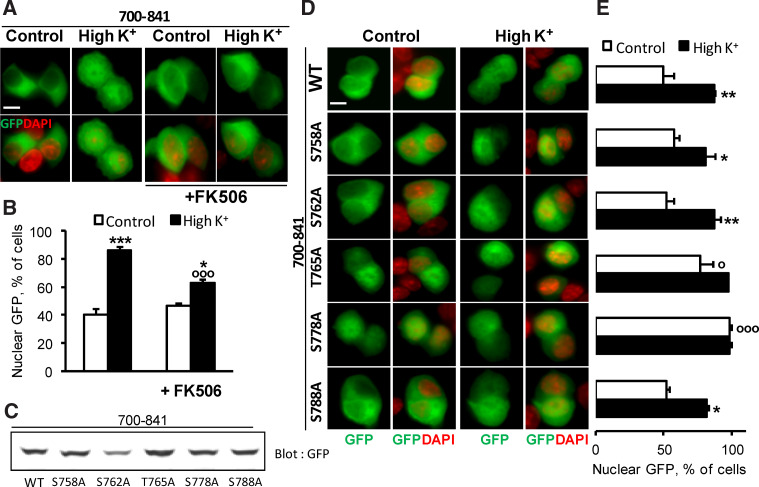
The cytonuclear localization of Pyk2-700–841 region is regulated by calcineurin
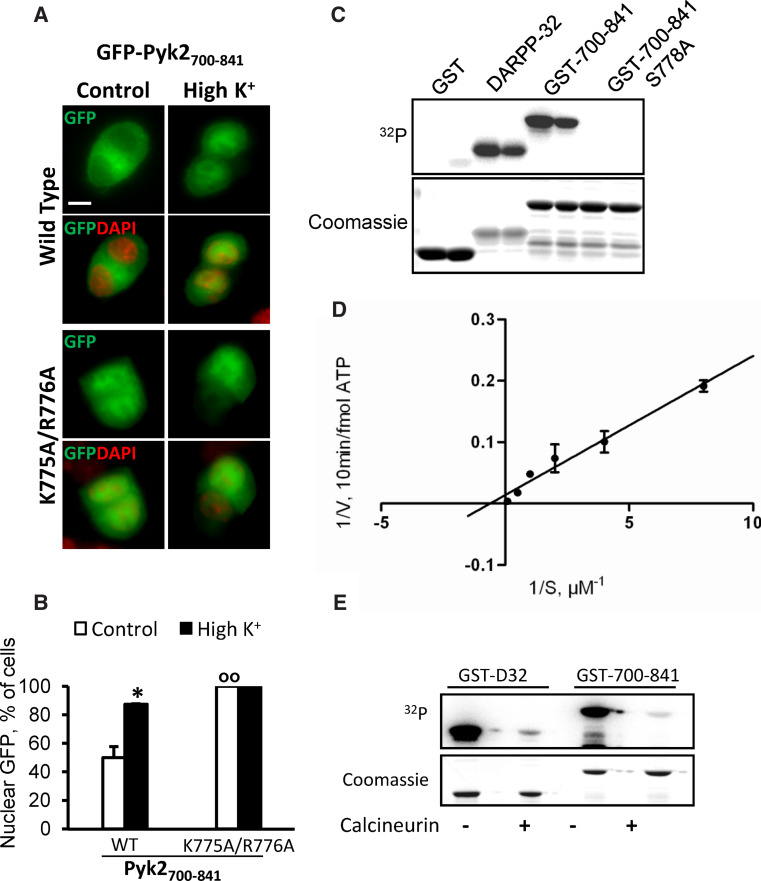
Cyto-nuclear shuttling of many proteins is regulated by S/T phosphorylation [42, 43]. Our previous results revealed that calcineurin plays a critical role in the regulation of Pyk2 nuclear redistribution [20] suggesting that calcineurin might dephosphorylate S/T residues in Pyk2 or in associated proteins. We first examined whether the depolarization-induced regulation of GFP-Pyk2700–841 cytonuclear localization was mediated by calcineurin by testing the effects of a calcineurin inhibitor, FK506. Pretreatment of PC12 cells with FK506 (2 μM, 20 min before high K+) partly prevented the depolarization-induced increase in Pyk2700–841 nuclear accumulation (Fig. 4a, b). This result indicated that calcineurin may target (a) S/T residue(s) in the 700–841 sequence. We performed site-directed mutagenesis (Ala-scan) of the S/T residues located in 758–793 region that we replaced by A (S758A, S762A, T765A, S778A, S788A) (Fig. 4c). There was no change in the localization of S758A, S762A, and S788A when compared to Pyk2700–841 (Fig. 4d, e). By contrast, in T765A-transfected cells a slight increase in the number of cells with nuclear GFP was observed in basal conditions. This effect was much more dramatic with the S778A-mutated form, which was accumulated in the nucleus in 100 % of the cells (Fig. 4d, e). We therefore focused on the role of S778.
Fig. 4.
Pyk2700–841 cytonuclear localization is regulated by calcineurin. a PC12 cells were transfected with GFP-Pyk2700–841 and treated with high K+ (High K+ 40 mM) or control solution for 3 min in the absence or presence of FK506 (2 μM, 20 min before high K+). GFP fluorescence and nuclei stained with DAPI were analyzed. b Quantification of the number of cells in a with n ≥ c GFP. Values are means + SEM, two-way ANOVA: depolarization effect F (1,14) = 80.92, p < 0.0001, FK506 effect F (1,14) = 5.88, p < 0.05, interaction F (1,14) = 17.49, p < 0.001. Newman–Keuls test: *p < 0.05, ***p < 0.001 versus control. °°°p < 0.001 versus high K+. c Anti-GFP immunoblotting of PC12 cells transfected with GFP-Pyk2700–841, S758A, S762A, T765A, S778A or S788A. d GFP fluorescence and DAPI nuclear staining of PC12 cells transfected as in c and treated with high K+ (High K+ 40 mM) or control solution for 3 min. e Percentage of cells in d with n ≥ c GFP. Values are means + SEM, two-way ANOVA: p < 0.05, mutation effect F (5,12) = 14.33, p < 0.0001, depolarization effect F (1,12) = 72.7, p < 0.0001, interaction F (5,12) = 3.82. Newman–Keuls test: *p < 0.05 versus control. °p < 0.05, °°°p < 0.001 versus 700–841 WT. Scale bar 5 μm
S778 is a substrate of PKA and is dephosphorylated by calcineurin in vitro
S778 belongs to a KRHSM sequence forming a consensus recognition motif for PKA (R/K-R/K-X-S/T-Φ) where Φ is a hydrophobic residue [44]. We tested the effects of mutating the two basic residues involved in a putative PKA recognition site (K775A/R776A) and found that K775A/R776A-GFP-Pyk2700–841 exhibited the same constitutive nuclear accumulation as the S778A-GFP-Pyk2700–841 supporting the possible role of phosphorylation by PKA (Fig. 5a, b). We then tested whether S778 is a substrate for PKA in vitro using a phosphorylation assay with [γ32P]ATP and purified PKA catalytic subunit (Fig. 5c). We produced wild type (WT) and S778A-Pyk2700–841 fused to glutathione S-transferase (GST) and used GST fused to dopamine- and cAMP-regulated phosphoprotein Mr ~32,000 (DARPP-32), a protein known to be phosphorylated on T34 by PKA [45], as a positive control. Strong 32P incorporation was observed with GST-Pyk2700–841 and DARPP-32, whereas GST-Pyk2700–841 S778A or GST alone was not radiolabeled (Fig. 5c). To further characterize this phosphorylation we studied the kinetics of phosphorylation of GST-Pyk2700–841 by PKA, using increasing amounts of substrate (0.25–10 μM) and found a K m of about 2 μM (2.32 ± 0.57, mean ± SEM, n = 3, Fig. 5d). We also determined that the stoichiometry of the reaction was close to 1. These results demonstrated that S778 was an excellent substrate for PKA. We then examined whether this residue was dephosphorylated by calcineurin. GST-Pyk2700–841 and GST-DARPP-32 were first phosphorylated by PKA and then incubated with recombinant calcineurin (Fig. 5e). The decrease in 32P labeling induced by the addition of calcineurin was similar for phospho-GST-Pyk2700–841 and phospho-GST-DARPP-32 (Fig. 5e), in which phosphoT34 is a substrate of calcineurin [46]. This demonstrated that S778 can be dephosphorylated by calcineurin in vitro.
Fig. 5.
S778 is a substrate of PKA and is dephosphorylated by calcineurin in vitro. a PC12 cells transfected with WT or K775A/R776A GFP-Pyk2700–841 were treated with high K+ (High K+ 40 mM) or control solution for 3 min. GFP fluorescence and nuclei stained with DAPI were analyzed. b Percentage of cells in a with n ≥ c GFP. Values are means + SEM. Two-way ANOVA: mutation effect F (1,4) = 63.45, p < 0.005, depolarization effect F (1,4) = 22.64, p < 0.01, interaction F (1,4) = 22.64, p < 0.01. Newman–Keuls test: **p < 0.01 versus control. °°p < 0.01 versus 700–841 WT. c Purified GST, DARPP-32, WT or S778A GST700–841 were incubated with PKA catalytic subunit and [γ-32P]ATP. 32P incorporation was analyzed with phosphorimager (upper panel) and proteins with Coomassie staining (lower panel). d Lineweaver–Burk plot of 32P incorporation with increasing concentrations (0.25–10 μM) of GST700–841. f Purified GST, alone or fused to DARPP-32 (GST-DARPP-32) or Pyk2700–841 (GST-700–841) were phosphorylated on glutathione-Sepharose beads with PKA and [γ-32P]ATP as in a, washed and incubated for 30 min in the absence (−) or presence (+) of calcineurin (PP2B). Scale bar 5 μm
S778 is phosphorylated by PKA and dephosphorylated by calcineurin in cells
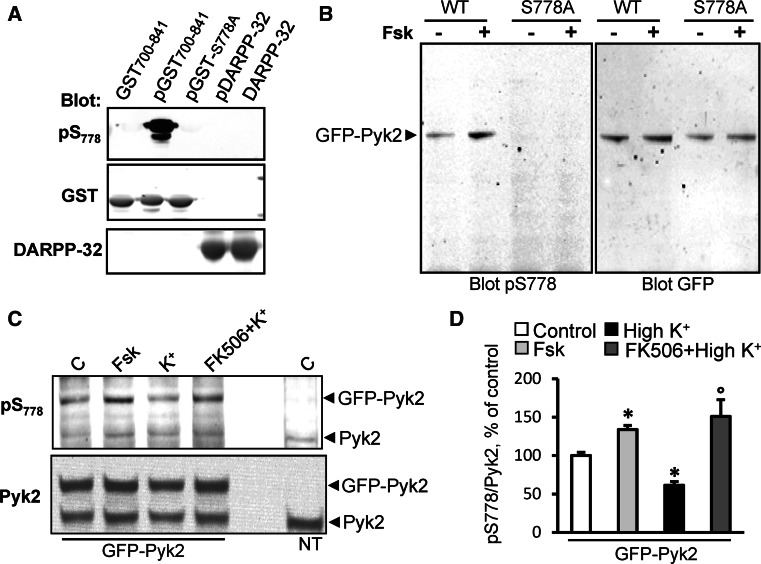
We raised an antiserum specific to the phosphorylated form of S778 (pS778). On immunoblots this antiserum only revealed the phosphorylated form of GST-Pyk2700–841 (pGST-Pyk2700–841) but neither unphosphorylated GST-Pyk2700–841 nor S778A GST-Pyk2700–841 nor pDARPP-32 (Fig. 6a). To test the specificity of the pS778 antibody we transfected WT and S778A GFP-Pyk2 in COS7 cells and incubated the cells in the absence or presence of forskolin (10 μM, 10 min) to increase cAMP production. Immunoblot with pS778 antibody showed only one major band corresponding to the molecular weight of GFP-Pyk2, whose intensity was increased by forskolin, and no label of GFP-Pyk2 S778A (Fig. 6b), showing the specificity of this antibody in COS7 cells. We then examined the regulation of S778 phosphorylation in PC12 cells transfected with GFP-Pyk2 (Fig. 6c). Forskolin treatment increased S778 phosphorylation of GFP-Pyk2, whereas depolarization decreased it (Fig. 6c, d). The effects of depolarization were blocked in the presence of FK506 (Fig. 6c, d). Although the signal observed for pS778 on endogenous Pyk2 was lower than with the transfected protein and could not be reliably quantified, a similar trend was observed (Fig. 6c). These results showed that S778 phosphorylation is regulated by both PKA and calcineurin in PC12 cells.
Fig. 6.
Regulation of S778 phosphorylation by PKA and calcineurin in cells. a The pS778 antibody reacts specifically with Pyk2 phosphorylated by PKA. Wild type GST-Pyk2700–841 incubated in the absence of PKA and ATP (lane 1), WT or S778A GST-Pyk2700–841 incubated in the presence of PKA and ATP (lane 2 pGST700–841, lane 3 pGST-S778A). DARPP-32 incubated in the presence (lane 4 pDARPP-32) or absence of PKA and ATP (lane 5 DARPP-32) was used as a specificity control. Samples were analyzed by immunoblotting with anti-pS778, (upper panel), anti-GST (middle) or anti-DARPP-32 antibody (lower). b COS7 cells transfected with WT or S778A GFP-Pyk2 and treated or not with forskolin (Fsk, 10 μM, 10 min) were analyzed by immunoblotting with anti-pS778 antibody (left panel) or anti-GFP antibody (right panel). c PC12 cells were transfected with GFP-Pyk2 and incubated in the absence (control, C) or presence of forskolin (Fsk, 10 μM, 10 min) or high K+ (40 mM, 5 min) with or without calcineurin inhibitor FK506 (2 μM, 15 min before K+) were analyzed by immunoblotting with anti-pS778 antibody (upper panel) or anti-Pyk2 antibody (bottom panel). Non-transfected cells (NT) are also shown. d Quantification of the results in c for Pyk2-GFP. Values are means + SEM. One-way ANOVA: F (3,8) = 10.41 (GFP), p < 0.05. Newman–Keuls test: *p < 0.05 versus control, °p < 0.05 versus high K+
S778 phosphorylation controls Pyk2 nuclear export but not its autophosphorylation on Y402
To investigate the role of S778 phosphorylation in full-length Pyk2, PC12 cells were transfected with WT or S778A-GFP-Pyk2, and treated or not with high K+. In the absence of depolarization, in contrast to WT, S778A-GFP-Pyk2 was predominantly nuclear (Fig. 7a, b). High K+ increased the nuclear localization of GFP-Pyk2 but had no further effect on S778A-GFP-Pyk2 (Fig. 7a, b). This showed that S778 was crucial for the regulation of Pyk2 localization. To determine whether the effects of depolarization were mediated by dephosphorylation of pS778, we immunostained transfected cells with the pS778-specific antibody Fig. 7a, middle panels). Immunoreactivity was detected in the cytoplasm of ~60 % of the cells transfected with WT GFP-Pyk2 and no nuclear immunoreactivity was detected with this antibody (Fig. 7a). pS778 labeling decreased after membrane depolarization in roughly the same proportion as the nuclear translocation of GFP-Pyk2 (Fig. 7a, b). As expected, there was virtually no pS778 immunofluorescence in S778A-GFP-Pyk2 transfected cells (Fig. 7a, b). To further characterize S778 phosphorylation upon the depolarization, cells were sorted in four subclasses depending on their pS778 labeling and GFP cyto/nuclear localization (Fig. 7c). In the absence of depolarization (control), approximately half of the cells displayed a cytoplasmic pS778 staining together with a predominantly cytoplasmic localization of GFP-Pyk2 (pS778+, n < c, Fig. 7c). Membrane depolarization strongly decreased this category of cells and increased the number of pS778-immunonegative cells with predominantly nuclear GFP-Pyk2 (pS778−, n < c, Fig. 7c). These results were consistent with the hypothesis of S778 dephosphorylation being the trigger for the nuclear accumulation of full-length Pyk2.
Fig. 7.
S778 phosphorylation controls Pyk2 nuclear export. a GFP fluorescence, pS778 immunoreactivity and DAPI staining in PC12 cells transfected with WT or S778A GFP-Pyk2 and incubated in the presence of a control or depolarizing (High K+ 40 mM) solution for 3 min. b Percentage of c > n GFP-positive cells in a. Values are means + SEM. Two-way ANOVA: mutation effect F (3,8) = 128.58, p < 0.0001, depolarization effect F (1,8) = 23.99, p < 0.005, interaction F (3,8) = 6.01, p < 0.05. Newman–Keuls test: ***p < 0.001 versus control, °°°p < 0.001 versus WT. c Class distribution of GFP-Pyk2-transfected cells in a depending on cytoplasmic pS778 immunoreactivity and GFP localization with schematic representation of each class (cytoplasm and nucleus: green, compartment with highest GFP concentration; red pS778 immunoreactivity, yellow colocalization). Values are means + SEM. One-way ANOVA: F (7,8) = 27.96, p < 0.0001. Newman–Keuls test: ***p < 0.001, **p < 0.01 high K+ versus control. d GFP fluorescence, pS778 immunoreactivity, and DAPI staining of PC12 cells transfected with GFP-Pyk2 and incubated in the presence of vehicle (control), or H89 (100 μM, 30 min), or myrPKI14–22 (10 μM, 10 min), or a depolarizing solution (High K+ 40 mM, 3 min). e Quantification of cells with GFP fluorescence stronger in the nucleus than in the cytoplasm in d. Values are means + SEM. One-way ANOVA: F (5,12) = 43.41, p < 0.0001. Newman–Keuls test: ***p < 0.001 versus control, °°p < 0.01 versus High K+. f PC12 cells transfected with WT or S778A GFP-Pyk2 were treated with LMB (LMB, 11 ng/ml, 3 h) as in Fig. 3a. Results are expressed as % increase in nuclear/cytoplasmic fluorescence ratio (n/c) in response to LMB-treatment. Values are means + SEM. Student t test: ***p < 0.01. g PC12 cells transfected with GFP-Pyk2 WT or S778A treated with high K+ (High K+ 40 mM) or control solution for 5 min were analyzed by immunoblotting with anti-pY402 antibody (upper) or anti-GFP antibody (lower). h Values are means + SEM. Two-way ANOVA: mutation effect F (1,6) = 1.94, p > 0.05, treatment effect F (2,6) = 88.37, p < 0.0001, interaction F (2,6) = 0.32, p > 0.05. Newman–Keuls test: **p < 0.01 versus control, ***p < 0.001 versus control. Scale bar 5 μm
To further investigate the contribution of PKA to the cellular localization of Pyk2, we examined the effects of two unrelated pharmacological inhibitors of PKA, H89 (100 μM, 30 min) and a cell-permeable form of the protein kinase A inhibitory peptide (myrPKI14–22, 10 μM, 30 min). These two molecules increased the number of cells with Pyk2 in the nucleus from 15 to approximately 55 % and pS778 labeling was virtually absent in these conditions (Fig. 7d, e). The effects of PKA inhibitors were almost as pronounced as those of depolarization (Fig. 7d, e). These results showed that the cyto/nuclear localization of Pyk2 is strongly dependent on the activity of PKA in these experimental conditions.
Although our results provided strong evidence that S778 dephosphorylation was necessary to allow Pyk2 nuclear accumulation, this effect could result from a regulation of Pyk2 nuclear export or import. To address this point, we reasoned that the inhibition of the nuclear export by LMB should have less effect on S778A-GFP-Pyk2 localization if S778 phosphorylation is necessary for Pyk2 nuclear export. Conversely, if S778 phosphorylation increases Pyk2 nuclear import, LMB effects and S778A mutation should be additive and lead to a further nuclear accumulation of S778A which has actually been observed for other mutants of Pyk2 [21]. PC12 cells transfected with WT or S778A-GFP-Pyk2 were treated or not with LMB. LMB induced a much stronger increase in the nuclear localization of GFP-Pyk2, than of S778A-GFP-Pyk2 localization (Fig. 7f), suggesting a regulation of Pyk2 nuclear export through S778 phosphorylation.
Since calcineurin is required for both Pyk2 nuclear translocation and autophosphorylation [20], it was important to determine whether the regulation of S778 phosphorylation was also involved in the regulation of Pyk2 Y402 autophosphorylation, the essential step in its functional activity. We measured Pyk2 autophosphorylation by pY402-specific immunoblotting in PC12 cells transfected with WT or S778A-GFP-Pyk2, treated or not with forskolin or high K+. Membrane depolarization induced phosphorylation of S778A-GFP-Pyk2 on Y402 to the same extent as WT GFP-Pyk2 (Fig. 7g, h). Forskolin did not alter this phosphorylation. These results indicated that the role of calcineurin in Pyk2 autophosphorylation is not mediated by S778 dephosphorylation.
Pyk2 short-splice isoform does not undergo Ca2+-induced cytonuclear shuttling
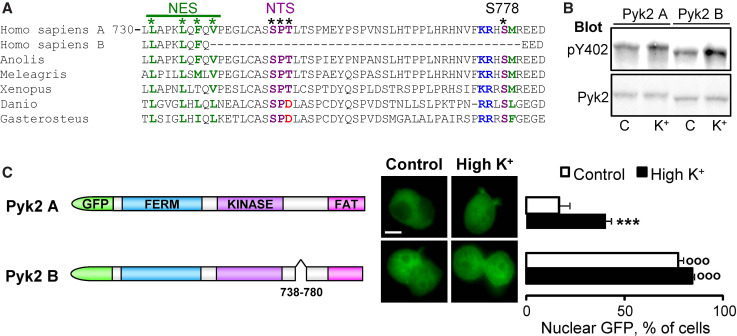
An indirect manner to evaluate the functional importance of regulatory motifs is to examine their conservation during evolution. To determine the interspecies conservation of the NES, NTS, and S778, we aligned Pyk2 sequences from Homo sapiens, Anolis carolinensis (green American lizard), Meleagris gallopavo (turkey), Xenopus silurana (Western clawed frog), Danio rerio (zebra fish), and Gasterosteus aculeatus (three-spined stickleback fish) (Fig. 8a). The NES, the NTS, and S778 were very well conserved between species. Minor differences were observed in Xenopus NES sequence which nevertheless remained hydrophobic and in fish NTS in which the T residue was replaced by a D, which has been proposed to be also efficient for nuclear import [37]. In the short isoform of Pyk2 (isoform B, aligned for Homo sapiens in Fig. 8a) 42 amino acids are deleted in the region important for regulated intracellular localization. The NTS and S778 are localized in the sequence that is spliced out in the short isoform. Moreover, in Pyk2 isoform B, the last hydrophobic residue we have shown to be important for nuclear export (V739) is missing (Fig. 8a). These motifs are not conserved in FAK sequence (not shown). Since sequence comparisons suggested that the missing exon in isoform B should alter the cytonuclear trafficking of Pyk2, we tested this hypothesis. We generated GFP-Pyk2 isoform B that migrated slightly faster than GFP-Pyk2 on SDS-PAGE (Fig. 8b). Membrane depolarization similarly increased Y402 phosphorylation of the two Pyk2 isoforms (Fig. 8b). We then examined the intracellular localization of Pyk2 isoform B. As expected, its localization was dramatically different from that of the long isoform under basal conditions, with ~80 % of cells exhibiting nuclear GFP-Pyk2 fluorescence (Fig. 8c). This localization was similar to that of GFP-Pyk2 LFV/3A (see Fig. 3c). Depolarization did not modify the localization of Pyk2 isoform B. These results show that the intracellular localization of the two isoforms of Pyk2 is different and that isoform B is constitutively nuclear. In contrast, the Ca2+-dependent regulation of nuclear localization is specific to the long isoform found in neurons.
Fig. 8.
The NES, NTS, and S778 are conserved between species and absent in Pyk2 isoform B. a Alignment of Pyk2 protein sequences from Homo sapiens, Anolis carolinensis, Meleagris gallopavo, Xenopus silurana, Danio rerio, Gasterosteus aculeatus together with the Homo sapiens isoform B lacking one exon. The NES, NTS, and S778 are shown with asterisk. The residues in the PKA consensus site are indicated in blue (−3, −2 basic) and green (+1 hydrophobic). b PC12 cells transfected with GFP-Pyk2 isoform A (long) or B (short) were treated with high K+ (K+, 40 mM) or control solution (C) for 5 min and analyzed by immunoblotting with anti-pY402 antibody (upper panel) or anti-Pyk2 antibody (lower panel). c PC12 cells transfected with GFP-Pyk2 isoform A or B (scheme on the left) were treated with high K+ (High K+) or control solution for 3 min. The percentage of cells with n ≥ c GFP fluorescence (middle) was quantified (right panel). Values are means + SEM, two-way ANOVA: isoform effect F (1,12) = 261.83, p < 0.0001, K+ effect F (1,12) = 118.57, p < 0.0005 interaction F (1,12) = 6.42, p < 0.05. Newman–Keuls test: ***p < 0.001 versus control, °°°p < 0.001 versus WT. Scale bar 5 μm
Discussion
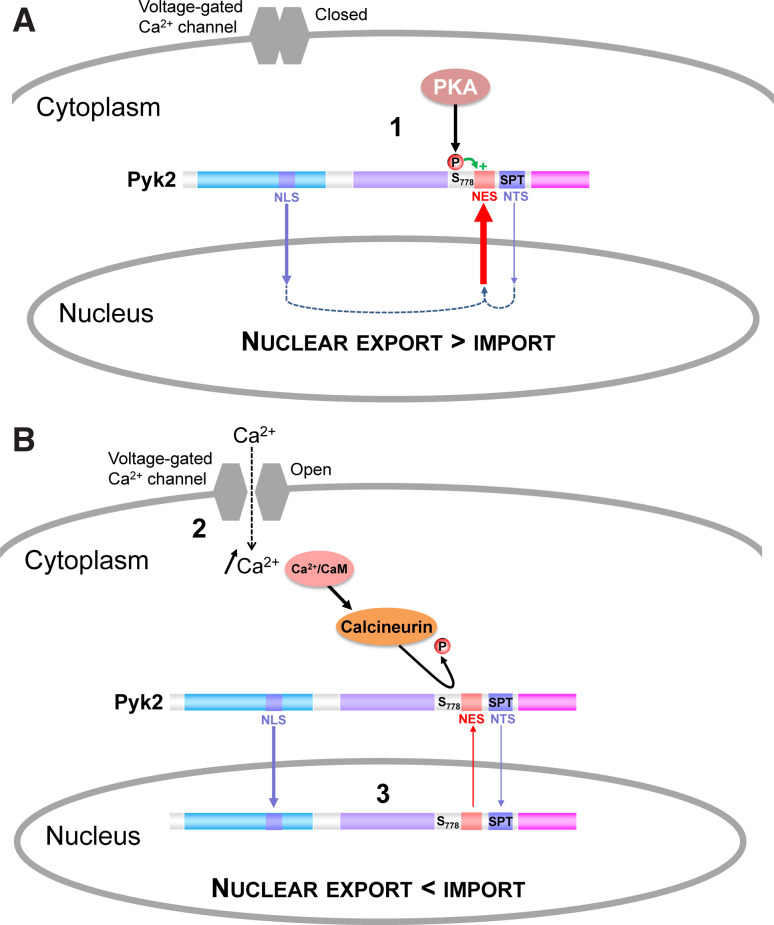
In the present study, we describe a novel mechanism by which Pyk2 intracellular localization is regulated. Using various deletions we show that depolarization-induced nuclear accumulation of Pyk2 is recapitulated by a region located in the kinase-FAT linker region, encompassing residues 700–841. Fused to GFP this polypeptide was excluded from the nucleus, whereas it was predominantly nuclear following depolarization or LMB treatment. Conversely, deletion of this polypeptide from Pyk2 resulted in a predominantly nuclear localization of the enzyme. The LMB-sensitivity suggested a CRM1-mediated nuclear export, and we identified a sequence possibly corresponding to a NES, L730LAPKLQFQVP. Mutation of L735, F737, and V739 induced a complete relocalization of GFP-Pyk2700–841 to the nucleus. This result demonstrated that L735QFQV is necessary to maintain Pyk2 in the cytoplasm in basal conditions. However, a region encompassing residues 768–793 was also necessary for the nuclear export to be active. Since we suspected that nuclear trafficking was regulated by phosphorylation/dephosphorylation, we systematically mutated Ser/Thr residues in this region and found that S778 played a critical role. S778 is located in a canonical PKA phosphorylation site and mutation of either S778 or the two basic residues involved in kinase interaction resulted in GFP-Pyk2700–841 nuclear relocalization. Large-scale analysis of protein phosphorylation using mass spectrometry has found that S778 is one of the phosphorylated residues of Pyk2 in vivo [47, 48]. However, until now, nothing was known about the specific function of S778 and this is the first time a role of Pyk2 S/T phosphorylation has been identified. We found that S778 was an excellent substrate for PKA and was dephosphorylated by calcineurin. The S778A mutation resulted in a depolarization-insensitive nuclear localization of full-length Pyk2. The “occlusion” of the LMB effects in this mutant further indicates that S778 regulates nuclear export of Pyk2. Our results suggest that S778 phosphorylation is necessary for the NES activity of the L730LAPKLQFQV motif, and that its dephosphorylation by calcineurin is a major trigger of the nuclear localization of Pyk2. A strong and delayed activation of nuclear PKA occurs following stimulation of membrane receptors in neurons [49]. Thus, PKA may phosphorylate S778 in the nucleus allowing the export of Pyk2 to the cytoplasm. Experiments with a phospho-specific antibody targeting this residue supported this hypothesis since phosphorylated S778 was found only in the cytoplasm of unstimulated cells and depolarization-induced nuclear relocalization was concomitant with the disappearance of pS778 immunocytofluorescence and with a decrease in the pS778 signal in immunoblots. Although we demonstrated that S778 is phosphorylated by PKA in vitro and in response to forskolin in intact cells, we cannot exclude that other kinase(s) is(are) also active on this residue. In fact the predominantly cytoplasmic localization of Pyk2 and the cytoplasmic pS778 immunofluorescence suggest a basal S778 phosphorylation, which may result from the activity of other kinase(s) and/or to a remarkable resistance of pS778 to dephosphorylation by phosphatases other than calcineurin. S778 also belongs to a consensus sequence for casein kinase 2 (CK2) (3 acidic residues at positions +3–5). However, CK2 inhibition did not interfere with Pyk2 localization (Faure and Girault, unpublished observation). In contrast, mutation of the basic residues defining PKA consensus recognition site (K775R776) or pharmacological inhibition of PKA activity both induced a nuclear localization of Pyk2, supporting the important role of PKA in PC12 cells. It is worth mentioning that induction of nuclear localization of other proteins such as NFAT by calcineurin involves dephosphorylation of several residues [50]. The partial increase of nuclear localization induced by the T765A mutation suggests that T765 might also contribute to nuclear exclusion of Pyk2.
Thus, our results provide strong evidence that regulation of Pyk2 nuclear export through S778 dephosphorylation is a major regulator of its intracellular localization. What then drives Pyk2 into the nucleus? A NLS, in the F2 lobe of the FERM domain is involved in its nuclear targeting [23, 35] and its mutation prevented depolarization-induced accumulation of GFP-Pyk2. Surprisingly the experiments with GFP-Pyk2421–1009 showed that the FERM domain was not necessary for depolarization- or LMB-induced nuclear accumulation. These apparently contradictory results could indicate that the FERM domain contain additional NES, as previously suggested [21], counterbalancing the effects of the NLS. However the FERM domain by itself is predominantly expressed in the nucleus and mutation of the putative FERM NES did not enhance Pyk2 nuclear localization (Faure and Girault, unpublished observation). Therefore we suggest that the apparent contradiction between the localization of the FERM-deleted and NLS-mutated Pyk2 indicate interactions between the FERM domain and the other domains of Pyk2 in the full-length enzyme which modify the relative strength of the various nuclear import and export signals. Such intra- or inter-molecular interactions provide potential additional regulatory steps yet to be explored. We also identified a NTS sequence in Pyk2, S747PT, which plays an important role to promote GFP-Pyk2700–841 nuclear accumulation. This motif has to be phosphorylated to be active [37] and additional experiments are needed to identify the regulation of the phosphorylation of this motif. In the full-length protein, however, the role of this NTS appeared minor in our experimental conditions. Mutation of the FERM NLS (R184A/R185A) dramatically decreased Pyk2 nuclear localization in basal conditions and following depolarization. It is only in cells treated with LMB that we found that the GFP-Pyk2 R184A/R185A mutant was still able to accumulate in the nucleus and that this accumulation was abrogated by the NTS mutation. We concluded from these experiments that the NTS plays an accessory role in the full-length protein. In summary, our results suggest a model for the regulation of Pyk2 subcellular localization (Fig. 9). In basal conditions, Pyk2 is phosphorylated on S778, the NES in the linker region between the kinase and the FAT domain is active, and Pyk2 is predominantly cytoplasmic (Fig. 9A). After a rise of Ca2+, calcineurin is activated and dephosphorylates S778 leading to the inactivation of the NES. Pyk2 then accumulates in the nucleus due to the predominant activity of the NLS and to a lesser extent of the NTS (Fig. 9b).
Fig. 9.
Working model of Pyk2 intracellular localization regulation. a In basal conditions, Pyk2 is phosphorylated on S778 by PKA, the NES in the linker region between the kinase and the FAT domain, is active, and Pyk2 is predominantly cytoplasmic. b After a rise of Ca2+, for example through opening of voltage-sensitive Ca2+ channels, calcineurin is activated and dephosphorylates S778 leading to the inactivation of the NES. Pyk2 then accumulates in the nucleus due to the predominant activity of the NLS and possibly the NTS
It is interesting to note that the NLS is conserved between FAK and Pyk2, suggesting that the ability to be enriched in the nucleus is common to these kinases. In contrast, the region containing the NES, NTS, and S778 is specific to Pyk2 and these motifs are conserved in Pyk2 from various species from fish to mammals, suggesting that the Ca2+/calcineurin regulated nuclear localization is an important and specific feature of Pyk2. It is remarkable that these sequences are absent from the short isoform of Pyk2, which is predominantly found in hematopoietic cells, whereas the long isoform is highly expressed in the brain [10, 12]. In PC12 cells the short isoform was found in the nucleus in basal conditions. Thus the regulated nuclear export of Pyk2 appears to be specific to the neuronal isoform.
What is the function of Pyk2 in the nucleus? Interesting clues are available from various models. Nuclear Pyk2 activates keratinocytes differentiation by increasing the expression of Fra-1 and JunD transcription factors [24]. The autophosphorylated, active form of Pyk2 accumulates in the nucleus of depolarized neurons [20], although Pyk2 nuclear accumulation is independent of its autophosphorylation and kinase activity ([20] and present study). Pyk2 could directly regulate tyrosine phosphorylated resident proteins of the nucleus such as histone variant H2AX [51] or H3/H4 [52]. Pyk2 associates with Src-family kinases, which localize to the nucleus in specific conditions, regulating euchromatin decondensation [53] and protection against oxidative stress through phosphorylation of Nrf2 by Fyn [54, 55]. Nuclear Pyk2 may also play a role of a scaffolding protein for p53 [35]. Our findings will allow designing the tools necessary for addressing these possibilities.
In conclusion, the present study shows that the Pyk2 long isoform expressed in neurons can undergo a Ca2+- and calcineurin-regulated nuclear accumulation. Evidence from other cell types indicates possible functional targets of nuclear Pyk2 and suggests that its regulated cyto-nuclear traffic may be involved in various brain functions or dysfunctions.
Acknowledgments
This work was supported by Inserm, UPMC, and grants from Agence nationale de la recherche (ANR-08-BLAN-0287-02), Fondation pour la recherche médicale (FRM), Association pour la recherche contre le cancer (ARC), Fondation pour la recherche sur le cerveau (FRC), Framework Program 7 (SynSys), and European research council (ERC). The group of JAG is affiliated with the Paris School of Neuroscience (ENP).
Conflict of interest
None.
References
- 1.Lev S, et al. Protein tyrosine kinase Pyk2 involved in Ca(2+)-induced regulation of ion channel and MAP kinase functions. Nature. 1995;376(6543):737–745. doi: 10.1038/376737a0. [DOI] [PubMed] [Google Scholar]
- 2.Salter MW, Kalia LV. Src kinases: a hub for NMDA receptor regulation. Nat Rev Neurosci. 2004;5(4):317–328. doi: 10.1038/nrn1368. [DOI] [PubMed] [Google Scholar]
- 3.Girault JA, et al. FAK and PYK2/CAK in the nervous system, a link between neuronal activity, plasticity and survival? Trends Neurosci. 1999;22:257–263. doi: 10.1016/S0166-2236(98)01358-7. [DOI] [PubMed] [Google Scholar]
- 4.Hsin H, et al. Proline-rich tyrosine kinase 2 regulates hippocampal long-term depression. J Neurosci. 2010;30(36):11983–11993. doi: 10.1523/JNEUROSCI.1029-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartos JA, et al. Postsynaptic clustering and activation of Pyk2 by PSD-95. J Neurosci. 2010;30(2):449–463. doi: 10.1523/JNEUROSCI.4992-08.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Y, et al. CAKbeta/Pyk2 kinase is a signaling link for induction of long-term potentiation in CA1 hippocampus. Neuron. 2001;29(2):485–496. doi: 10.1016/S0896-6273(01)00220-3. [DOI] [PubMed] [Google Scholar]
- 7.Gil-Henn H, et al. Defective microtubule-dependent podosome organization in osteoclasts leads to increased bone density in Pyk2(−/−) mice. J Cell Biol. 2007;178(6):1053–1064. doi: 10.1083/jcb.200701148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okigaki M, et al. Pyk2 regulates multiple signaling events crucial for macrophage morphology and migration. Proc Natl Acad Sci USA. 2003;100(19):10740–10745. doi: 10.1073/pnas.1834348100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashido M, et al. Ca2+ lightning conveys cell–cell contact information inside the cells. EMBO Rep. 2006;7(11):1117–1123. doi: 10.1038/sj.embor.7400821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dikic I, Schlessinger J. Identification of a new Pyk2 isoform implicated in chemokine and antigen receptor signaling. J Biol Chem. 1998;273(23):14301–14308. doi: 10.1074/jbc.273.23.14301. [DOI] [PubMed] [Google Scholar]
- 11.Li X, et al. A calcium-dependent tyrosine kinase splice variant in human monocytes. Activation by a two-stage process involving adherence and a subsequent intracellular signal. J Biol Chem. 1998;273:9361–9364. doi: 10.1074/jbc.273.16.9361. [DOI] [PubMed] [Google Scholar]
- 12.Xiong WC, Macklem M, Parsons JT. Expression and characterization of splice variants of PYK2, a focal adhesion kinase-related protein. J Cell Sci. 1998;111(Pt 14):1981–1991. doi: 10.1242/jcs.111.14.1981. [DOI] [PubMed] [Google Scholar]
- 13.Kohno T, et al. Protein-tyrosine kinase CAKbeta/PYK2 is activated by binding Ca2+/calmodulin to FERM F2 alpha2 helix and thus forming its dimer. Biochem J. 2008;410(3):513–523. doi: 10.1042/BJ20070665. [DOI] [PubMed] [Google Scholar]
- 14.Park SY, Avraham HK, Avraham S. RAFTK/Pyk2 activation is mediated by trans-acting autophosphorylation in a Src-independent manner. J Biol Chem. 2004;279(32):33315–33322. doi: 10.1074/jbc.M313527200. [DOI] [PubMed] [Google Scholar]
- 15.Avraham H, et al. RAFTK/Pyk2-mediated cellular signalling. Cell Signal. 2000;12(3):123–133. doi: 10.1016/S0898-6568(99)00076-5. [DOI] [PubMed] [Google Scholar]
- 16.Schaller MD, Sasaki T. Differential signaling by the focal adhesion kinase and cell adhesion kinase beta. J Biol Chem. 1997;272(40):25319–25325. doi: 10.1074/jbc.272.40.25319. [DOI] [PubMed] [Google Scholar]
- 17.Zheng C, et al. Differential regulation of Pyk2 and focal adhesion kinase (FAK). The C-terminal domain of FAK confers response to cell adhesion. J Biol Chem. 1998;273(4):2384–2389. doi: 10.1074/jbc.273.4.2384. [DOI] [PubMed] [Google Scholar]
- 18.Corvol JC, et al. Depolarization activates ERK and proline-rich tyrosine kinase 2 (PYK2) independently in different cellular compartments in hippocampal slices. J Biol Chem. 2005;280(1):660–668. doi: 10.1074/jbc.M411312200. [DOI] [PubMed] [Google Scholar]
- 19.Menegon A, et al. FAK+ and PYK2/CAKbeta, two related tyrosine kinases highly expressed in the central nervous system: similarities and differences in the expression pattern. Eur J Neurosci. 1999;11(11):3777–3788. doi: 10.1046/j.1460-9568.1999.00798.x. [DOI] [PubMed] [Google Scholar]
- 20.Faure C, et al. Calcineurin is essential for depolarization-induced nuclear translocation and tyrosine phosphorylation of PYK2 in neurons. J Cell Sci. 2007;120(Pt 17):3034–3044. doi: 10.1242/jcs.009613. [DOI] [PubMed] [Google Scholar]
- 21.Aoto H, et al. Nuclear translocation of cell adhesion kinase beta/proline-rich tyrosine kinase 2. Cell Struct Funct. 2002;27(1):47–61. doi: 10.1247/csf.27.47. [DOI] [PubMed] [Google Scholar]
- 22.Arcucci A, Montagnani S, Gionti E. Expression and intracellular localization of Pyk2 in normal and v-src transformed chicken epiphyseal chondrocytes. Biochimie. 2006;88(1):77–84. doi: 10.1016/j.biochi.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 23.Lim ST, et al. Nuclear FAK promotes cell proliferation and survival through FERM-enhanced p53 degradation. Mol Cell. 2008;29(1):9–22. doi: 10.1016/j.molcel.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schindler EM, et al. The role of proline-rich protein tyrosine kinase 2 in differentiation-dependent signaling in human epidermal keratinocytes. J Invest Dermatol. 2007;127(5):1094–1106. doi: 10.1038/sj.jid.5700662. [DOI] [PubMed] [Google Scholar]
- 25.Sun L, et al. Role of the Pyk2-MAP kinase-cPLA2 signaling pathway in shear-dependent platelet aggregation. Ann Biomed Eng. 2004;32(9):1193–1201. doi: 10.1114/B:ABME.0000039353.97347.28. [DOI] [PubMed] [Google Scholar]
- 26.Ding L, Guo D, Homandberg GA. Fibronectin fragments mediate matrix metalloproteinase upregulation and cartilage damage through proline rich tyrosine kinase 2, c-src, NF-kappaB and protein kinase Cdelta. Osteoarthr Cartil. 2009;17(10):1385–1392. doi: 10.1016/j.joca.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 27.Farshori PQ, et al. Activation and nuclear translocation of PKCdelta, Pyk2 and ERK1/2 by gonadotropin releasing hormone in HEK293 cells. J Steroid Biochem Mol Biol. 2003;85(2–5):337–347. doi: 10.1016/S0960-0760(03)00226-7. [DOI] [PubMed] [Google Scholar]
- 28.Gorlich D, Mattaj IW. Nucleocytoplasmic transport. Science. 1996;271(5255):1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 29.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8(3):195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 30.Yoneda Y. Nucleocytoplasmic protein traffic and its significance to cell function. Genes Cells. 2000;5(10):777–787. doi: 10.1046/j.1365-2443.2000.00366.x. [DOI] [PubMed] [Google Scholar]
- 31.Kutay U, Guttinger S. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 2005;15(3):121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 32.Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6(3):187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 33.Siciliano JC, et al. Differential regulation of proline-rich tyrosine kinase 2/cell adhesion kinase beta (PYK2/CAKbeta) and pp 125(FAK) by glutamate and depolarization in rat hippocampus. J Biol Chem. 1996;271(46):28942–28946. doi: 10.1074/jbc.271.46.28942. [DOI] [PubMed] [Google Scholar]
- 34.Smith AG, et al. Enhancement by iron of hepatic neoplasia in rats caused by hexachlorobenzene. Carcinogenesis. 1993;14(7):1381–1387. doi: 10.1093/carcin/14.7.1381. [DOI] [PubMed] [Google Scholar]
- 35.Lim ST, et al. Pyk2 inhibition of p53 as an adaptive and intrinsic mechanism facilitating cell proliferation and survival. J Biol Chem. 2010;285(3):1743–1753. doi: 10.1074/jbc.M109.064212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 37.Chuderland D, Konson A, Seger R. Identification and characterization of a general nuclear translocation signal in signaling proteins. Mol Cell. 2008;31(6):850–861. doi: 10.1016/j.molcel.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Tang X, et al. Phosphorylation of the RNase III enzyme Drosha at Serine300 or Serine302 is required for its nuclear localization. Nucleic Acids Res. 2010;38(19):6610–6619. doi: 10.1093/nar/gkq547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fornerod M, et al. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90(6):1051–1060. doi: 10.1016/S0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 40.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278(5335):141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 41.la Cour T, et al. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel. 2004;17(6):527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 42.Poon IK, Jans DA. Regulation of nuclear transport: central role in development and transformation? Traffic. 2005;6(3):173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- 43.Nardozzi JD, Lott K, Cingolani G. Phosphorylation meets nuclear import: a review. Cell Commun Signal. 2010;8:32. doi: 10.1186/1478-811X-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kennelly PJ, Krebs EG. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J Biol Chem. 1991;266(24):15555–15558. [PubMed] [Google Scholar]
- 45.Hemmings HC, Jr, et al. DARPP-32, a dopamine- and adenosine 3′:5′-monophosphate- regulated neuronal phosphoprotein. I. Amino acid sequence around the phosphorylated threonine. J Biol Chem. 1984;259:14486–14490. [PubMed] [Google Scholar]
- 46.King MM, et al. Mammalian brain phosphoproteins as substrates for calcineurin. J Biol Chem. 1984;259:8080–8083. [PubMed] [Google Scholar]
- 47.Wissing J, et al. Proteomics analysis of protein kinases by target class-selective prefractionation and tandem mass spectrometry. Mol Cell Proteomics. 2007;6(3):537–547. doi: 10.1074/mcp.T600062-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Oppermann FS, et al. Large-scale proteomics analysis of the human kinome. Mol Cell Proteomics. 2009;8(7):1751–1764. doi: 10.1074/mcp.M800588-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gervasi N, et al. Dynamics of protein kinase A signaling at the membrane, in the cytosol, and in the nucleus of neurons in mouse brain slices. J Neurosci. 2007;27(11):2744–2750. doi: 10.1523/JNEUROSCI.5352-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crabtree GR. Generic signals and specific outcomes: signaling through Ca2+, calcineurin, and NF-AT. Cell. 1999;96(5):611–614. doi: 10.1016/S0092-8674(00)80571-1. [DOI] [PubMed] [Google Scholar]
- 51.Cook PJ, et al. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458(7238):591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh RK, Gunjan A. Histone tyrosine phosphorylation comes of age. Epigenetics. 2011;6(2):153–160. doi: 10.4161/epi.6.2.13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi A, et al. Nuclear localization of Src-family tyrosine kinases is required for growth factor-induced euchromatinization. Exp Cell Res. 2009;315(7):1117–1141. doi: 10.1016/j.yexcr.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 54.Jain AK, Jaiswal AK. Phosphorylation of tyrosine 568 controls nuclear export of Nrf2. J Biol Chem. 2006;281(17):12132–12142. doi: 10.1074/jbc.M511198200. [DOI] [PubMed] [Google Scholar]
- 55.Jain AK, Jaiswal AK. GSK-3beta acts upstream of Fyn kinase in regulation of nuclear export and degradation of NF-E2 related factor 2. J Biol Chem. 2007;282(22):16502–16510. doi: 10.1074/jbc.M611336200. [DOI] [PubMed] [Google Scholar]