Abstract
The Hippo pathway is emerging as a critical nexus that balances self-renewal of progenitors against differentiation; however, upstream elements in vertebrate Hippo signalling are poorly understood. High expression of Fat1 cadherin within the developing neuroepithelium and the manifestation of severe neurological phenotypes in Fat1-knockout mice suggest roles in neurogenesis. Using the SH-SY5Y model of neuronal differentiation and employing gene silencing techniques, we show that FAT1 acts to control neurite outgrowth, also driving cells towards terminal differentiation via inhibitory effects on proliferation. FAT1 actions were shown to be mediated through Hippo signalling where it activated core Hippo kinase components and antagonised functions of the Hippo effector TAZ. Suppression of FAT1 promoted the nucleocytoplasmic shuttling of TAZ leading to enhanced transcription of the Hippo target gene CTGF together with accompanying increases in nuclear levels of Smad3. Silencing of TAZ reversed the effects of FAT1 depletion thus connecting inactivation of TAZ-TGFbeta signalling with Hippo signalling mediated through FAT1. These findings establish FAT1 as a new upstream Hippo element regulating early stages of differentiation in neuronal cells.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-015-1955-6) contains supplementary material, which is available to authorized users.
Keywords: FAT1 cadherin, Cadherin, Differentiation, Hippo pathway, Neurite outgrowth, Neuronal differentiation, SMAD transcription factor, TAZ, TGFβ signalling
Introduction
The vertebrate Fat cadherins [1] comprise a family of four structurally similar genes homologous to the Drosophila tumour suppressor Fat cadherin [2] and the related Fat2 (Ft2) gene [3]. Knockout of the Fat1 gene in mice caused lethal defects in kidney development with a proportion of Fat1−/− newborns displaying deformed eyes and craniofacial malformations, while others were cannibalised at birth due to appearance of exencephaly [4]. The latter phenotype was independently confirmed by Saburi and colleagues who reported that the manifestation of exencephaly in Fat1 knockout mice appeared strain dependent [5]. In mouse developing brain, Fat1 expression was prominent in the proliferating ventricular zones and down-regulated as cells ceased dividing [6]. In the rat no expression of Fat1 was found in adult tissues except in the CNS, where the remnant of the germinal zones, as well as the dentate gyrus, continued to express Fat1 [7]. Collectively these findings promote a role for Fat1 during brain formation and moreover, its continued expression at sites of neurogenesis in the adult brain also suggests that it may be required for tissue homeostasis.
Neurogenesis collectively describes the complex events required for the development and maintenance of the brain. This involves a multiplicity of processes where cells undergo proliferation, migration, fate determination, differentiation, maturation, and functional integration into existing networks [8]. At the apex of these processes lie neural stem cells (NSCs), pluripotent cells that can be triggered to differentiate into neurons, astrocytes, and oligodendrocytes to maintain brain function. A pool of these cells exists in the brain throughout life of the organism and is maintained through a programme of self-renewal. The balance between self-renewal and differentiation is considered a critical factor controlling brain homeostasis [9]. It is widely thought that harnessing the potential of NSCs can be utilised to address neural damage and degeneration [10, 11]. Towards this, some progress has been made to understand the signalling processes that regulate NSC homeostasis, but much remains to be determined [12].
A number of classical signalling pathways have been linked to neuronal differentiation including Wnt/β-catenin pathway [13], Notch-Hes1 pathway [14], and Sonic hedgehog [15]. More recently the Salvador–Warts–Hippo (SWH) or Hippo pathway (a fundamental pathway conserved from Drosophila to mammals) has also been associated with the control of neurogenesis [16, 17]. The pathway is involved in the cell density-dependent control of cell growth and is conceptually related to the control of organ size [18–20]. The pathway comprises three main components; upstream regulators, a core kinase cassette and downstream transcriptional activators. Under conditions of low cell density, YAP (Yes-associated protein) and TAZ (transcriptional coactivator with PDZ-binding motif) control a transcriptional programme of genes primarily involving targets of TEAD and SMAD transcriptional factors [21–23]. Reaching high density triggers cells to activate the core kinase cassette which in mammals involves Salvador, LATS1/2 and MST1/2, homologous to Drosophila kinases Salvador, Warts and Hippo, respectively. Such activation prevents the nucleocytoplasmic shuttling of YAP and TAZ and inhibits their transcriptional activities [18, 19]. In Drosophila, upstream regulation of the Hippo pathway involves several components, for example, Merlin and Expanded [24], but the only event occurring at the cell surface presently known to activate Hippo signalling involves the binding between Fat cadherin and its ligand Dachsous (Ds) (reviewed in [18]). This interaction has been shown to be important in the developing optic lobe where Fat promotes neural differentiation, acting through Hippo signalling [25]. However, potential links between the Fat cadherins and Hippo signalling in mammals are presently limited [26, 27].
Emerging evidence from a variety of sources indicates that the Hippo pathway acts to control the balance between self-renewal of progenitors and cell differentiation [28, 29]. For example, YAP expression decreases during differentiation in the mouse retina and acts to inhibit progenitor proliferation [17]. In the chick model of neural tube development, ectopic expression of YAP and TEAD or inhibition of upstream Hippo kinases all increased the numbers of neural progenitors [16]. Forced over-expression of TAZ in mouse neurosphere cultures also caused reprogramming of NSCs, by interrupting differentiation into glia and neurons to promote mesenchymal differentiation [30]. Most of these studies have focused on the roles of the core kinases and downstream transcriptional programme but less attention has been paid to the upstream elements. Based on the proposed role of Fat1 during brain development and homeostasis, we hypothesised that the human FAT1 may play a role in differentiation of neurons, possibly via the Hippo signalling pathway. Towards this notion, we used in vitro differentiation of human SH-SY5Y neuroblastoma cells, a widely accepted model of neuronal differentiation [31]. Our study found that induction of FAT1 expression was required for the induction of both morphological and biochemical changes occurring during neuronal differentiation. Moreover, we provide evidence that FAT1 engages the Hippo signalling pathway and regulates the differentiation process.
Materials and methods
Cell culture
Human SH-SY5Y neuroblastoma cells (ATCC) were routinely cultured in DMEM containing 4.5 g/L glucose and l-glutamine (Lonza) supplemented with 10 % foetal bovine serum (v/v) (Sigma-Aldrich) and 25 mM HEPES buffer (Lonza). Cells were maintained at 37 °C in a humidified atmosphere of 95 % air/5 % CO2. Cultures were subcultivated by trypsinization (0.25 % trypsin/EDTA) (Lonza). Based on the protocols developed by Ota et al. [32], cells were subjected to differentiation using 80 nM 12-O-tetradecanoyl-phorbol-13-acetate (0.1 mM stock in DMSO; Sigma-Aldrich). TPA-containing media was added on day 0 and then replaced with fresh TPA-containing media after 3 days thereby exposing the cells to the differentiation agent for a total of 6 days. Control (undifferentiated) cells omitting TPA were prepared under identical conditions. Alternatively TPA was substituted with 10 µM retinoic acid (Sigma-Aldrich) using the same treatment protocol where indicated. Cell number and viability were determined using an automatic cell counter (Digital Bio, ADAM-MC).
Microarray analysis
The GDS2125 dataset deposited by Peddada and colleagues was sourced from NCBI’s gene expression omnibus (GEO) database (http://www.ncbi.nlm.nlm.nih.gov/geo/). The dataset contains expression microarray experiments of untreated SH-SY5Y cells and cells treated for 48 h with 16 nM TPA [33]. For each gene of interest, the RMA algorithm was used to transform logarithmic signal intensities (SI) to linear values as previously described [34]. The SI values of three biological replicates were averaged to compare relative mRNA expression. Similar analyses were conducted on gene expression datasets derived from sequential analyses of murine embryonic brain development (GSE8091; Hartl et al. [35]), neuronal differentiation of human induced pluripotent stem cells (iPSCs) (GSE25542; Pasca et al. [36]) and neuronal differentiation of human embryonic stem cells (hESCs) (GSE40593; Lafaille et al. [37]).
Quantitative real time PCR
Total RNA was extracted (Illustra RNAspin Mini RNA Isolation Kit; GE Healthcare) and concentrations determined using a NanoPhotometer™ Pearl spectrophotometer (Implen). Total RNA (500 ng) was used to prepare cDNA (Transcriptor First Strand cDNA Synthesis Kit; Roche) and reactions performed using SYBR chemistry (iQ™ SYBR® Green Supermix; BioRad) with data collected using an Applied Biosystems 7500 system. Each reaction was performed in triplicate and data analysis performed using the ΔΔCt method with the SDS system software (7500 system v 1.4.0). Target primer sequences are listed in Table S1 with all results normalised against the average of three housekeeping genes (RPS18, GusB and HMBS).
Western blotting
Lysates were prepared from cell monolayers using NDE lysis buffer supplemented with protease and phosphatase inhibitors (Roche Applied Science) as previously described [38]. Protein concentrations were estimated (BCA protein; Thermo Scientific) and equal protein amounts (25–35 µg) resolved by electrophoresis using 3–8 % Tris acetate gels for FAT1 or 4–12 % Bis–Tris gels for other proteins (Novex®, Invitrogen). Transfer to nitrocellulose membranes (iBlot®, Invitrogen) and Western blotting was conducted against FAT1 as previously described [38]. Antibodies against GAPDH, cell cycle components and elements of the Hippo pathway were purchased from Cell Signalling Technologies (GAPDH (2118), Cell Cycle Regulation Kit (9932), Hippo Signalling Antibody Kit (8579), respectively). Where indicated, bands were quantitated using the MultiGuage v3.0 software package (Fujifilm Life Science Systems) and band densities normalised against appropriate loading controls.
Knockdown of gene expression using shRNA-miR
MicroRNA-based short hairpin (shRNA-miR) constructs directed against FAT1 together with an empty control vector were purchased from Thermo Scientific (Open Biosystems GIPZ Lentiviral shRNA library; Table S2). Constructs were used to prepare lentiviral particles in 293FT according to the suppliers’ protocol before ‘spinfection’ [39] of SH-SY5Y cells in the presence of 8 µg/ml hexadimethrine bromide (Sigma). After transduction, cells were selected with 4 µg/ml puromycin (Astral Scientific) before sorting against co-expressed turboGFP (tGFP) using a FACSAria™ II (BD Biosciences). Populations of cells expressing >90 % tGFP+ were derived through repeated sorting (Fig. S1).
Analysis of neurite outgrowth
Phase contrast digital micrographs of cell monolayers were collected (AxioCamHR camera system; Zeiss) and analysed using Axiovision v4.7 software. The number of primary neurites per cell was determined according to standard criteria [40, 41] where a neurite is defined as a cellular projection as long or wide as the cell soma. Five fields per experiment were used from three different experiments. Differences in the frequency of neurites between cell populations was analysed using Fisher’s exact test. Data for the length of neurites was not normally distributed and the Mann–Whitney U non-parametric test was employed.
Cell proliferation assay
Proliferation over time was determined using a fluorescence-based assay involving measurements of tGFP in virally transduced SH-SY5Y cells. Cells were seeded into 96 well plates (3595, Corning Incorporated) on day 0 using six wells for each cell density. Fluorescent images of the entire plate were captured on sequential days using a Typhoon Trio multimode fluorescence scanner (GE Healthcare) using constant settings for all readings (emission filter 520BP 40CY2, ECL+, blue FAM laser, high sensitivity, PMT 300, 50 micron pixel size and focal plane +3 mm). The fluorescence output of each well was quantified using MultiGuage v3.0 software package (Fujifilm Life Science Systems), range scope at 0–2500. Raw data were exported to Microsoft Excel and the data normalised to the levels of fluorescence determined on day 1.
Sub-cellular fractionation
Nuclear and cytoplasmic cell fractions were prepared using the detergent lysis method as previously described [42]. Western blotting was then performed on each amounts of protein against TAZ, Smad2/3, Smad4 and p-Smad1/5/8 [YAP/TAZ Antibody (8418), Smad2/3 Antibody (5678), Smad4 antibody (9515) and Smad 1/5/8 Antibody Sampler Kit (12656), respectively; Cell Signalling Technologies]. Antibodies against Lamin A/C [a constituent of the nuclear lamina (sc-6215); Santa Cruz] and GAPDH [cytoplasmic marker protein (sc-25778); Santa Cruz] were used to validate the efficiency of each fractionation.
Small interfering RNA (siRNA) knockdown
Cells were seeded in 6-well plates at 400,000 cells/well. On the day of transfection, media were replaced by Opti-MEM® reduced serum media before the addition of 50 nM siRNA complexes prepared using Lipofectamine RNAiMAX (#13778; Invitrogen) according to the manufacturer’s instructions. Duplexes used are listed in Table S3. After 18 h the cells were trypsinized and re-cultured at 150,000 cells/well prior to further experiments.
Results
FAT1 expression is increased during neuronal differentiation
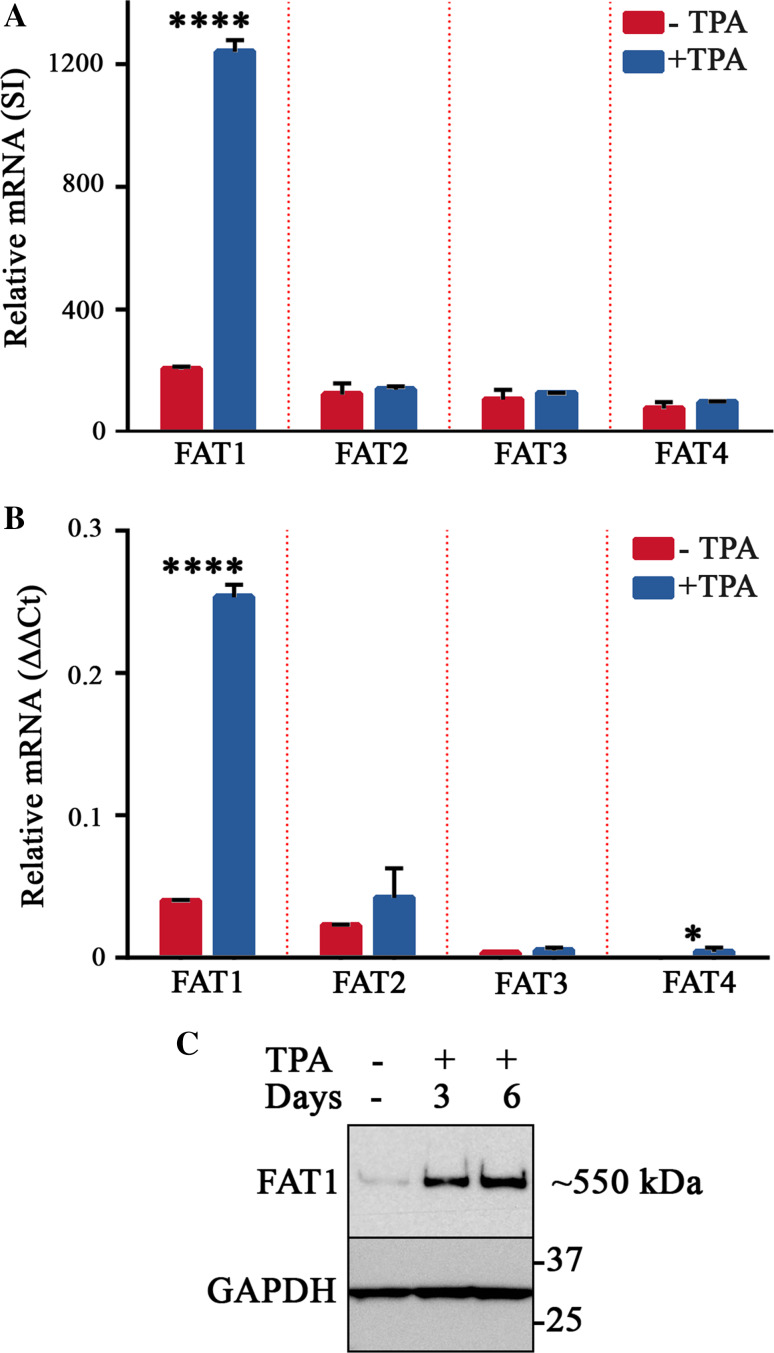
We first assessed the expression of all four members of the FAT cadherin subfamily in SH-SY5Y cells prior to and after differentiation induced with 12-O-tetradecanoyl-phorbol-13-acetate (TPA). Analysis of publically available expression microarrays (GDS2125) showed that the relative levels of FAT1, FAT2, FAT3 and FAT4 mRNA appeared low in undifferentiated SH-SY5Y cells. However, after 2 days of treatment with TPA, FAT1 mRNA was observed to be markedly increased whereas the expression of the other FAT cadherins remained unchanged (Fig. 1a). The mRNA expression profiles of FAT cadherins were confirmed using qPCR in cells treated with TPA over 6 days (Fig. 1b), a protocol resulting in phenotypic differentiation (see “Materials and methods”). These results suggested that amongst all FAT cadherins, FAT1 was selectively induced during the differentiation of SH-SY5Y cells.
Fig. 1.
FAT1 expression is strongly induced by differentiation of SH-SY5Y cells. a Expression microarray analysis of the relative mRNA levels of FAT family cadherins comparing untreated cells with those treated with TPA for 2 days. The mRNA levels of each FAT cadherin are expressed as the mean signal intensity (SI) ± SD of three biological replicates (****p ≤ 0.0001, t test). b Analysis of the relative mRNA levels of FAT family cadherins in SH-SY5Y cells left untreated for 6 days or treated with 80 nM TPA for 6 days using qPCR. The mRNA levels of each FAT cadherin are expressed as the mean ΔΔCt value ± SD from 3 replicates (see “Materials and methods”). Similar results were obtained in two experiments (*p ≤ 0.05; ****p ≤ 0.0001, t test). c Western blot analysis of FAT1 protein levels in cell lysates of SH-SY5Y cells untreated for 6 days or treated with 80 nM TPA for 3 and 6 days, respectively. Blots were probed with using affinity purified polyclonal antibodies raised against the cytoplasmic tail of FAT1 with similar results obtained in five experiments
To determine if the levels of FAT1 mRNA corresponded to similar levels of protein, we undertook Western blotting of cell lysates using antibodies directed against the cytoplasmic tail of FAT1. Examination of untreated SH-SY5Y cells revealed a weak band ~550 kDa consistent with the expected size of FAT1 [38]. Relative to the levels of GAPDH used as a loading control protein, FAT1 expression increased after 3 and 6 days of TPA-treatment (Fig. 1c) mirroring the changes observed in FAT1 mRNA levels. Employing retinoic acid (RA) to differentiate SH-SY5Y cells also induced marked expression of FAT1 (Fig. S2). Additionally, the selective induction of FAT1 was also noted in RA-treated NTera2-clone D2 cells, an independent model of neuronal differentiation (data not shown). Collectively, this suggested that upregulation of FAT1 was not associated with specific agonists but rather the neuronal differentiation process itself.
FAT1 is required for neurite outgrowth
The selective induction of FAT1 cadherin during TPA treatment of SH-SY5Y cells suggested that it might drive aspects of differentiation. To investigate the possible roles of FAT1, we transduced SH-SY5Y cells with lentiviral particles containing microRNA-adapted short hairpin RNAs (shRNA-miRs) as described in the “Materials and methods”. Two populations containing independent sequences targeting FAT1 were prepared along with comparable control cells containing a non-targeting shRNA sequence [43]. After sequential selection with puromycin and multiple rounds of cell sorting against co-transduced tGFP, all populations used contained ~95 % tGFP positive cells (Fig. S1A). Evaluation of FAT1 expression by Western blotting in these cells demonstrated that induction of FAT1 during differentiation with TPA was effectively suppressed by FAT1-targeting shRNA-miRs (Fig. 2a and see Fig. 3).
Fig. 2.
Suppression of FAT1 induction during differentiation of SH-SY5Y cells decreases neurite outgrowth. a Western blotting analyses against cell lysates prepared from untreated cells or those treated with 80 nM TPA for 3 and 6 days, respectively. Comparison of cells transduced with control shRNA-miR (non-targeting sequence; NTS) or FAT1-targeting shRNA-miR demonstrates effective silencing of FAT1 protein expression. GAPDH was used as a loading control. b Representative phase contrast photomicrographs demonstrating phenotypic differences between neuritogenesis in control and FAT1-shRNA knockdown cells observed after 6 days of treatment with TPA. Bar represents 50 μm. c Quantitative comparison of the frequency and length of neurites in 6 day differentiated SH-SY5Y cell populations bearing NTS and FAT1-shRNA (n = 562 and 683 cells analysed, respectively). Neurites were defined as cellular projections as long or wide as the soma with absolute neurite lengths normalised against cell body measurements for each cell. Neurites were divided into six length categories ranging from 1 to 1.25 through to >5 cell body lengths as shown. d The percentage of cells from C recorded as having one or more neurites in either NTS or FAT1-shRNA populations (****p ≤ 0.0001, 2-sided Fisher’s exact test). e Box-Whisker plot showing neurite lengths for NTS and FAT1-shRNA populations. The box limits indicate the 25th and 75th percentiles with the internal line showing median values. Whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles (**p ≤ 0.01, Mann–Whitney U test). Plots were prepared using the BoxPlotR software package
Fig. 3.

Suppression of FAT1 expression following TPA-treatment affects cell density-dependent proliferation and survival. Cell growth rates were compared for NTS or FAT1-shRNA SH-SY5Y cells treated with 80 nM TPA. Cells were seeded at (a) low and (b) high densities (3750 and 15,000 cells/well, respectively) and total cell number estimated as fluorescence units determined through detection of co-transduced tGFP. Measurements conducted over consecutive days were normalised to day 1 with values representing relative fluorescence units (RFU) ± SD of six biological replicates. Similar results were obtained in three experiments (*p ≤ 0.05; ***p ≤ 0.001; ****p ≤ 0.0001, t test). c Comparative Western blotting analyses of FAT1 and cell cycle regulatory proteins (cyclin D1 and CDK2). Blots were performed on SH-SY5Y cell lysates prepared from NTS and two independent populations of FAT1-shRNA cells (refer Figure S2 and Table S2). GAPDH was used as a loading control
SH-SY5Y cell populations were then evaluated for the production of neurites that represents the key feature of neuronal differentiation [44]. The cell phenotype examined after 6 days of TPA treatment showed that a large proportion of the NTS-control cells displayed multiple neurites whereas these processes appeared less abundant in the FAT1 shRNA cell populations (Fig. 2b). To provide an objective measurement of neurite outgrowth, over 500 cells were measured in each cell population to determine both the length and number of cell projections. Referencing the length of the neurites against the cell body, both the number and length of neurites appeared to be substantially decreased in FAT1 shRNA cells (Fig. 2c). Statistical analyses confirmed the number of cells with neurites has decreased from 45 % in NTS shRNA controls compared to 8.7 % in FAT1 shRNA cells (p ≤ 0.0001, Fig. 2d). Similarly neurite lengths in FAT1 knockdown cultures were confirmed to be significantly shorter than controls (p ≤ 0.01, Fig. 2e). These data, therefore, indicate that expression of FAT1 is required for efficient neuritogenesis during differentiation of SH-SY5Y cells.
Suppression of FAT1 during differentiation affects SH-SY5Y cell density-dependent proliferation
In parallel with the decreased neurite outgrowth that accompanied suppression of FAT1 expression, it was also observed that there were relatively increased cell numbers in FAT1-shRNA cells compared to NTS-control SH-SY5Y cells after 6 days (Fig. 2b). To evaluate this further, proliferation rates and the levels of cell cycle proteins were compared throughout differentiation of the cells.
NTS-control and FAT1-shRNA cells were initially seeded at both low and high densities (3750 and 15,000 cells/well, respectively) and cell number changes measured using the relative levels of co-transduced tGFP. Changes were in cell growth were apparent comparing NTS- and FAT1-shRNA after treatment with TPA to induce differentiation. In cells seeded at low density, a small but significant difference occurred after 4 days of culture where the relative number of FAT1-shRNA knockdown cells was increased compared to NTS controls (Fig. 3a). The differences were more profound in cells initially seeded at high density where relatively increased numbers of FAT1-shRNA knockdown cells were apparent after 3 days of culture with NTS control cell number reaching a plateau (Fig. 3b). Thus, these assays confirm the increased numbers of FAT1-shRNA knockdown cells visualised in culture (Fig. 2b).
Western blotting analyses were then undertaken to measure the levels of FAT1 along with cell cycle regulatory proteins. Cell lysates were prepared from NTS-control and FAT1-shRNA cells after 6 days of TPA treatment. Western blotting confirmed the induction of FAT1 protein in control cells and that it was effectively suppressed in the two independent populations of FAT1-shRNA cells (Fig. 3c). Blotting the same samples for cyclin D1 and CDK2 showed that both markers were relatively increased in FAT1-shRNA cells compared to NTS-control cultures (Fig. 3c). Given these proteins are considered to be markers of cell cycle activation, these data suggest suppressing the induction of FAT1 in cells triggered to differentiate results in cells displaying a higher proliferative state. Moreover, these effects are imparted through cell density where in the absence of FAT1, cells overcome contact inhibition and reach higher densities without succumbing to death signals. Together with the preceding results showing the requirement for efficient neuritogenesis, the results collectively point to a role for FAT1 in regulating neuronal differentiation.
FAT1 acts upstream of Hippo signalling to regulate nucleocytoplasmic shuttling of the transcriptional cofactor TAZ
The major question arising from the data concerns how the expression of FAT1 imparts effects on neuronal differentiation. FAT1 is one of the four genes homologous to Drosophila Fat cadherin, a receptor known to influence Hippo signalling and the density-dependent control of cell proliferation. To examine the possibility that Hippo signalling is involved in differentiation, and that FAT1 is engaged in this process, we first examined the expression of the two major vertebrate effectors of Hippo signalling, the transcriptional cofactors YAP and TAZ. Western blotting of SH-SY5Y cell lysates showed that TAZ was the predominant Hippo effector expressed by undifferentiated cells (Fig. 4a). After subjecting the cells to TPA treatment, TAZ expression levels were notably increased after 3 days of differentiation protocol with high levels sustained at 6 days. Differentiation also resulted in increased in the levels of YAP albeit at much lower levels than TAZ. To determine if FAT1 expression was required for the induction of YAP and/or TAZ, their expression was compared in NTS-control and FAT1-shRNA cells. As shown in Fig. 4b the levels of YAP and TAZ appeared unchanged after 6 days of TPA treatment, suggesting that the expression levels of the Hippo effectors were not affected by FAT1.
Fig. 4.
Expression of the Hippo pathway effectors YAP and TAZ during SH-SY5Y differentiation. a SH-SY5Y cells were treated with 80 nM TPA as described for Fig. 1c and cell lysate samples analysed for YAP and TAZ expression by Western blotting using a dual specificity polyclonal antibody recognising both YAP and TAZ. Anti- GAPDH was used as a loading control. b Transduced SH-SY5Y cells (NTS-control and FAT1 shRNA) populations were subjected to 80 nM TPA treatment or cultured without TPA as indicated. Cell lysate samples were subjected to Western blotting against YAP/TAZ. Thereafter, the membrane was reprobed with anti-GAPDH as a loading control
Although the cellular levels of YAP/TAZ were not affected by FAT1 expression, the cell density effects of FAT1 on SH-SY5Y differentiation did not altogether rule out regulatory effects through the Hippo pathway. Indeed, the major transcriptional activities of YAP and TAZ are enacted through their localisation to the cell nucleus (e.g., see [18]). To test this sub-cellular fractionation studies were performed to isolate fractions enriched for the nuclear and cytoplasmic compartments in NTS-control versus FAT1-shRNA cells. On this basis of the strong induction of TAZ relative to YAP during differentiation, our investigations focused on the expression and activity of TAZ. In cells subjected to TPA for 3 days, there was a marked increase in the amount of TAZ protein localising to the nucleus in FAT1-shRNA but not control shRNA cells (Fig. 5a). Blotting against the nuclear matrix protein Lamin-A/C and the cytosolic marker protein GAPDH was used to verify the isolation procedure.
Fig. 5.
FAT1 acts through the core Hippo kinase cassette to inhibit nucleocytoplasmic shuttling of TAZ with regulatory effects on the levels and localisation of Smad3. a Subcellular fractionations enriched for nuclear or cytoplasmic proteins were prepared from NTS or FAT1-shRNA SH-SY5Y cells after 3 days of TPA treatment. Western blotting analyses were then performed using antibodies directed against TAZ, Smad2/3, p-Smad1/5/8 and Smad4. Lamin A/C and GAPDH were used as nuclear and cytoplasmic markers, respectively. b qPCR analysis of the Hippo pathway target gene, CTGF, was conducted on NTS or FAT1-shRNA SH-SY5Y cells after 3 days of TPA treatment. Prior to the assay, cells were pre-treated with either negative control (NC) or TAZ-targeted siRNA duplexes. c Western blotting analyses were performed against SH-SY5Y cell lysates using antibodies directed against the Hippo kinase cassette components (Mst1, Sav1 and p-MOB1) together with GAPDH as a loading control. d Western blotting analyses were performed against cell lysates prepared from NTS or FAT1-shRNA SH-SY5Y cells after 3 days of TPA treatment. Samples were blotted against Smad2/3 together with GAPDH as a loading control
Towards validating the comparative distribution of TAZ in differentiating NTS-control versus FAT1 shRNA cells, we also performed immunofluorescence (IF) staining combined with confocal microscopy. Here SH-SY5Y cells adhered to glass coverslips were stained with antibodies against TAZ using methodology previously described [38]. In untreated controls, we observed strong nuclear staining in ~5 % of cells but such staining was not diminished after TAZ siRNA (data not shown). This suggested that another protein was responsible for the strong reactivity. As anticipated, overall staining for TAZ increased following differentiation in ~50 % of cells (data not shown). However, in the context of comparing shRNA controls with FAT1 shRNA cells, it was not possible to distinguish between cells with genuine TAZ staining and those with high background nuclear reactivity. Notwithstanding issues with IF staining, other biochemical findings provided additional evidence for the FAT1-dependent changes in the subcellular localization of TAZ during neuronal differentiation.
The translocation of TAZ to the nucleus is known to lead to the activation of TEAD transcription factors with a well-known transcriptional target being the CTGF gene, also considered to be a representative target gene of Hippo signalling [45, 46]. Corresponding analyses of CTGF gene expression using qPCR after 3 days of TPA treatment showed that mRNA levels were significantly higher in FAT1-shRNA cells compared to NTS control shRNA cells (Fig. 5b). In concert with TPA to initiate differentiation, treatment of cells with small interfering RNA (siRNA) to deplete the cellular levels of TAZ markedly decreased CTGF levels in FAT1-shRNA cells. Together these findings suggest that FAT1 expression can control the cellular distribution of TAZ, preventing its translocation to the nucleus and transcription of TAZ-dependent target genes.
One key function of the core kinase cassette of the Hippo signalling pathway involves control of the cellular levels and localisation of YAP and TAZ. To determine if Hippo pathway activation occurs during neuronal differentiation and whether FAT1 expression influences this process, Western blotting was performed against key kinase components and associated adapter proteins (Fig. 5c). In control cells, the levels of Mst1 kinase and the adaptor molecules Salvador and phosphorylated MOB-1 (p-MOB1) were all relatively increased after 6 days of differentiation compared with untreated cells. Comparative analyses showed that these increases were attenuated in FAT1 shRNA knockdown cells, especially the increased expression of p-MOB1 (Fig. 5c). Salvador levels were relatively higher in untreated FAT1 shRNA knockdown cells compared to controls but its level did not further increase following TPA treatment. Taken together, these results indicate that FAT1 expression serves to activate the core kinase cassette of the Hippo pathway that further influences the nucleocytoplasmic shuttling of TAZ and activation of downstream target genes.
FAT1 acts as a driver of differentiation largely through antagonising TAZ function
We next sought to better understand how FAT1 acts as a driver of the differentiation, particularly how antagonising the nuclear activities of TAZ may be important to control differentiation. Previously, a role has also been established for TAZ in the regulation of TGFβ signalling through its direct binding to Smad transcriptional cofactors [23]. TAZ was shown to stabilise heteromeric transcriptional complexes of the receptor-regulated (R)-Smads, Smad2 and Smad3, which in turn facilitated their nuclear translocation and activity [23]. In the context of human embryonic stem cells (hESCs), it was demonstrated that TAZ expression was required for maintain self-renewal and depletion of TAZ inhibited TGFβ signalling resulting in differentiation towards the neuroectoderm lineage [23]. Indeed, as described in the Introduction, the investigation of YAP and TAZ in a variety of systems have shown these act to inhibit stem cell differentiation and to guide distinct fate decisions (reviewed in [28, 29]). Based on this knowledge, the possible connection between FAT1, TAZ and Smads were investigated in SH-SY5Y cells.
To examine if the increased levels of nuclear TAZ in SH-SY5Y FAT-shRNA cells were associated with altered levels of Smads, we returned to analyse the nuclear and cytoplasmic fractions isolated from differentiated SH-SY5Y cells. From this analysis, it was evident that the amount of Smad3 was increased in FAT1-depleted cells compared to levels detected in control cells (Fig. 5a). There was a relatively small increase in cytoplasmic Smad3 but a more prominent increase occurred in the nuclear fraction from the FAT1 knockdown cells. In contrast, the levels of Smad2 appeared relatively unchanged in both nuclear and cytoplasmic fractions. Examination of total cellular lysates revealed increased levels of Smad3 in FAT1-shRNA cells after TPA-treatment (Fig. 5d) consistent with the results obtained in fractions. The nuclear/cytoplasmic samples were also blotted with antibodies against p-Smad1/5/8 (associated with BMP signalling) and the cooperating (co)-Smad, Smad4, common to both TGFβ and BMP branches [47] (Fig. 5a). Blotting signals for p-Smad1/5/8 were unchanged in FAT1-shRNA cells with reactivity largely restricted to nuclear fractions. Similarly, the bulk of reactivity for Smad4 was found in the cell nucleus with a modest increase in FAT1-shRNA cells. Since Smad3 along with Smad2 are known to form transcriptionally active complexes with Smad4 [48], the increased nuclear levels of these proteins likely indicates higher transcriptional activity of TGFβ pathway targets.
Given that TAZ along with FAT1 is induced during differentiation, the question then arose as to whether there was any interdependency in their regulation. To gain further insights, we employed siRNA to deplete TAZ levels alone and in combination with FAT1 suppression using shRNA. Control NTS and FAT1 shRNA-transduced cells were pre-treated with control (NC) or TAZ siRNA and thereafter subjected to TPA-induced differentiation for 3 days. Western blotting of cellular lysates revealed that suppressing FAT1 alone had no effect on the levels of TAZ induced in TPA-treated cells (Fig. 6a) consistent with the results of Fig. 4b. We then turned to examine the relative effects of FAT and TAZ on cellular levels of Smad2/3. Consistent with prior experiments (Fig. 5d), FAT1-shRNA cells displayed greater expression of Smad3 than NTS-controls (Fig. 6a). When TAZ protein levels were reduced by 60–70 % using siRNA, there was profound reduction in the total levels of Smad3 and to a lesser extent Smad2 in both the shRNA-control and FAT1-shRNA cells (Fig. 6a). The relative reduction in Smad3 after TAZ siRNA in either the shRNA-control or FAT1-shRNA cells appeared proportional to the levels observed in corresponding siRNA controls. In comparison, Smad2 levels were less dependent on the presence of TAZ. Thus, there is a strong dependency between the expression of TAZ and Smad3 levels during the differentiation of SH-SY5Y cells. Conversely FAT1 negatively regulates the levels of Smad3.
Fig. 6.
The growth promoting effect and inhibition of neuritogenesis observed after FAT1 depletion are regulated through TAZ. a Control or FAT1-shRNA SH-SY5Y cells pre-treated with negative control (NC) or TAZ-targeted siRNA duplexes were treated with TPA for a period of 3 days. Western blotting analyses of cell lysates were performed against TAZ, Smad2/3 and the cell cycle regulatory proteins cyclin D1, CDK2 and CDK4. GAPDH was used as a loading control. b SH-SY5Y cell populations bearing NTS and FAT1-shRNA were pre-treated with control (NC) or TAZ-targeting siRNA duplexes prior to 6d of treatment with TPA. Thereafter neurite length was determined from >600 cells. Neurite lengths were normalised to cell body measurements for each cell and sub-divided into six length categories (1.000–1.25, 1.26–1.5, 1.51–2.00, 2.01–3.00, 3.01–5.00, >5.00) and plotted as histograms. c The percentage of cells recorded as having one or more neurites in indicated populations from B (a p ≤ 0.0001, b p ≤ 0.0001, c p ≤ 0.0001, Chi squared test). d Box-Whisker plot depicting neurite lengths from populations analysed in b. The box limits indicate the 25th and 75th percentiles with the internal line showing median values. Whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles [a p ≤ 0.0001, b p ≤ 0.01, c p ≤ 0.0001, d p ≤ 0.0001, e p ≤ 0.01, Kruskal–Wallis ANOVA by ranks test followed by post hoc analysis corrected for multiple comparisons (two-tailed)]. Plots were prepared using the BoxPlotR software package
Analyses were also undertaken to determine whether the alterations occurring in cell cycle regulator expression after FAT1-suppression were also dependent on TAZ. Consistent with the prior experiments (Fig. 3c and data not shown), there were increased levels of cyclin D1, CDK2 and CDK4 apparent in FAT1 shRNA knockdown cells after TPA treatment (Fig. 6c). Depletion of TAZ in control shRNA cells resulted in some inhibition of the levels of all three cell cycle regulatory proteins. In contrast silencing of TAZ in FAT1 shRNA cells entirely obviated the increased levels of these markers. Therefore, high levels of TAZ appear necessary for the increased levels of cyclins and CDKs observed after suppression of FAT1. These findings again support the notion that the effects of FAT1 are manifested through TAZ, and notably as shown through the preceding experiments, the likely importance of FAT1 inhibiting the nuclear translocation of TAZ.
Finally, we returned to explore the effects of TAZ depletion on neuritogenesis. As shown in Fig. 6b, the frequency and length of neurites was determined in cells after 6 days of TPA treatment. As expected, in cultures treated with control siRNA, cell populations carrying FAT1 shRNA displayed markedly diminished numbers of cells with neurites (Fig. 6b, c; 42 versus 9.5 %, respectively, p ≤ 0.0001) with also significant reductions in neurite length (Fig. 6d, p ≤ 0.0001). Transfection of FAT1 knockdown cells with TAZ siRNA was able to rescue the production of neurites, albeit conditionally. TAZ depletion was able to restore the density of neurites to levels observed in control cells (Fig. 6b, c, 43 versus 42 %, respectively, p = 0.7661) but the length of neurites remained significantly shorter compared to controls (Fig. 6d, p = 0.012). Additionally, TAZ depletion in control cells increased the numbers of cell with neurites (Fig. 6b, c, 42 versus 64 %, respectively, p ≤ 0.0001) although neurite length was not significantly affected (Fig. 6d, p = 1.000).
Together these results suggest that TAZ, in the context of neuronal differentiation, is involved in positively promoting cell proliferation and in suppressing the production of neurites. On the other hand, the preceding mechanistic data also shows that FAT1 controls the cellular distribution of TAZ. The further functional correlates provided by these experiments suggest that the control FAT1 exerts on the differentiation process fundamentally occurs though antagonising these actions of TAZ. Based on this and the preceding results, a model depicting how FAT1 engages the Hippo pathway to effect control of neuronal differentiation is illustrated in Fig. 7.
Fig. 7.
Model depicting how FAT1 engages with the Hippo pathway to effect control of neuronal differentiation through antagonising TGFβ signalling. FAT1 is induced in cells triggered to differentiate where it promotes dual effects. First, FAT1 activates the core Hippo kinase cassette that serves to suppress the nuclear shuttling of TAZ. Second, FAT1 serves to both initiate and extend neurites (left panel). Conversely in the absence of FAT1 (or inhibiting its induction during differentiation using shRNA), Hippo kinases are not activated, thereby releasing constraints on TAZ. This serves to promote higher levels of Smad2/3 and both TAZ and Smad2/3 translocate to nucleus where they form transcriptionally active complexes involving TEADs and Smads. Activating gene targets of TEADs and Smad/TGFβ signalling invokes a transcriptional programme known to promote proliferation and self-renewal (right panel)
Fat1 and TAZ are induced in physiological models of neuronal differentiation
The preceding results have established that FAT1 was selectively induced during SH-SY5Y neuronal differentiation along with the increased expression of TAZ. To determine if these phenomena are consistent with more physiological examples of gene regulation, expression changes in Fat1 and TAZ were analysed in microarray datasets obtained from a variety of neuronal differentiation systems.
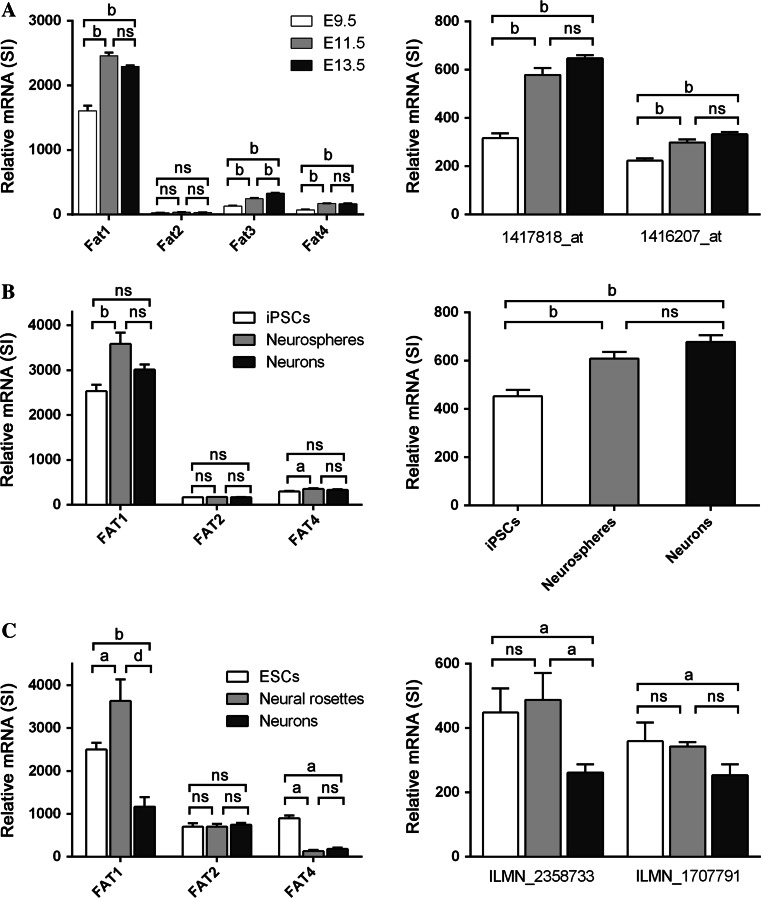
The sequential expression of the Fat family of cadherins was first examined during the mouse brain development from embryos at E9.5, E11.5 and E13.5, respectively (GSE8091; [6]). Each stage corresponds to a key phase of neuronal development: E9.5 represents the phase prior to neurogenesis where the brain tissue is largely comprised of NSCs, neurogenesis begins around E11–E12 with neural progenitor cells (NPCs) and from E13 to E17 further progresses where different types of mature neurons are formed [6, 7]. Thus, the three time points analysed (E9.5, E11.5 and E13.5) can be considered to represent the pre-neural, intermediate and later stages of differentiation. Analysis of the relative expression of each of the four Fat cadherin members in brain tissue showed that the expression of Fat1 at E9.5 was more than tenfold higher than that measured for Fat2, Fat3 and Fat4 (Fig. 8a, left panel). As neuronal differentiation progressed, the levels of Fat1 were further increased at E11.5 with levels remaining high at E13.5. Some significant increases also occurred in Fat3 and Fat4 but their relative expression remained low compared to Fat1. TAZ mRNA was also highly expressed with two independent probes ranking in the top ~85–90 % of genes analysed. TAZ levels were observed to increase as differentiation progressed (Fig. 8a, right panel).
Fig. 8.
Expression analyses of Fat cadherins and TAZ in alternate models of neuronal differentiation. a Relative mRNA expression of Fat family cadherins (left) and TAZ (right) in murine embryonic brain tissue collected at the E9.5, E11.5 and E13.5 stages of development (GSE8091 dataset; [35]). The data represent the mean ± SD of 4-6 biological replicates per developmental stage. Each Fat cadherin was represented by one probe on the array while the results of two independent probes are shown for TAZ. b Gene expression analyses of FAT family cadherins (left) and TAZ (right) conducted on human iPSCs, neurospheres and neurons (GSE25542; [36]). The data represent the mean ± SD of 12 biological replicates per group. Probes for FAT3 were absent on the array. c Gene expression analyses of FAT family cadherins (left) and TAZ (right) conducted on human ESCs, neural rosettes and mature neurons (GSE40593; [37]). The data represent the mean ± SD of 3 biological replicates per differentiation stage. Probes for FAT3 were absent on the array while results for two independent probes are shown for TAZ. Statistical analyses were conducted by ANOVA followed by Tukeys Post HOC Test (ns not significant, a p < 0.05, b p < 0.01, c p < 0. 001, d p < 0. 0001)
We next turned to examine neuronal differentiation in human iPSCs cells where neurospheres are used as intermediates prior to derivation of neurons [9]. Using global gene expression profiling data generated by Pasca and colleagues [10], gene expression analyses were conducted in data from iPSCs, neurospheres and neurons. It was observed that the levels of FAT1 were high in iPSCs and were further increased in neurospheres. In contrast, the levels of FAT2 and FAT4 were comparably low with no appreciable changes occurring throughout differentiation (Fig. 8b, left panel). As indicated by the signal intensity values, the relative expression levels of TAZ were also was moderately high in these samples with a significant increase from iPSCs towards neurons (Fig. 8b, right panel).
A third dataset produced by Lafaille and colleagues examined neuronal differentiation of human ESCs using neural rosettes as intermediates to neurons [11]. As above, analysis of the relative expression of the FAT cadherins showed that FAT1 was the most abundant in hESCs with three–four fold higher expression than either FAT2 or FAT4 (Fig. 8c, left panel). FAT1 expression increased between the progression of hESCs to neural rosettes but interestingly decreased in mature neurons (refer “Discussion”). In comparison FAT2 levels remained unchanged, while FAT4 expression was reduced in both neural rosettes and neurons. The results from two independent probes for TAZ indicated its relative expression was ranked in top 85 % of highly expressed genes (Fig. 8c, right panel). During differentiation towards neurons, it was observed that the TAZ level in neural rosettes remained high before reduction at the neuron stage. Taken together the results derived from the three different models show good concordance with the selective induction of Fat1 and the increased expression of TAZ during neuronal differentiation.
Discussion
Conservation of Hippo signalling from Drosophila to vertebrates?
While many of the Hippo pathway elements discovered in Drosophila have clear homologues in vertebrates, some uncertainty exists in the conservation of Fat signalling functions between arthropods and vertebrates [1, 18, 20]. Fat4 is considered to be the orthologue of Ft and binds the Ds orthologue, Dchs1 [49]. However, considered from the Drosophila perspective, it was recently proposed that the Ft-Hippo signalling axis represents an evolutionary gain in Drosophila which is absent in Fat4 [50]. On the other hand, Fat1 is most closely related to Drosophila Ft2 [1, 51] and currently available studies suggest that Ft2 does not engage in Hippo signalling [1, 51]. However, despite there being no readily transposable mechanism to explain the involvement of vertebrate Fat cadherins in Hippo signalling, there are precedents to support this linkage, particularly the function of Fat4 in the context of neuronal differentiation.
Mutations in FAT4 or its ligand DCHS1 were causally linked to Van Maldergem syndrome (VMS), a recessive condition characterised by distinctive craniofacial features, intellectual disability together with auditory, renal, skeletal and limb malformations [52]. Manipulating mouse embryos in utero recapitulated aspects of VMS where depletion of Fat4 and/or Dchs1 resulted in accumulation of neuronal progenitors [52]. Knockdown of the Hippo effector Yap in the VMS model rescued the Fat4 or Dchs1 phenotypes, thus providing evidence for engagement with Hippo signalling. Similarly the ablation of Fat4 in the developing chicken spinal cord increased the numbers of neural progenitors, an effect also functionally dependent upon Yap expression [26]. Like FAT4, FAT1 has also been linked to neurological conditions in humans where single nucleotide polymorphisms (SNPs) were identified across multiple families affected by bipolar disorder [53] and autism [54]. However, it is less clear whether Fat1 fulfils any role in Hippo signalling, the main evidence provided by studies in zebrafish where developmental kidney defects occurring after Fat1 knockdown were mediated through YAP [27]. More work is, therefore, required to address how Fat1 functions in different tissue-specific contexts particularly in the brain.
New evidence linking FAT1 to neuronal differentiation, acting through the Hippo signalling pathway
Fat1 is expressed in the developing and adult rat brain with strongest expression in areas engaged in neurogenesis [7], implying that Fat1 plays some role in neuronal differentiation. Towards gauging the role of FAT1 in the neuronal compartment, we turned to human SH-SY5Y cells, a tractable widely used in vitro model of neuronal differentiation. Here, we observed both FAT1 mRNA and protein were massively increased during differentiation into neuronal cells. Moreover, as depicted in Fig. 7, limiting the induction of FAT1 expression counteracted the differentiation programme resulting in markedly decreased neuritogenesis and accompanying increases in cell proliferation. This suggests that expression of FAT1 is a requirement for differentiation, at least in this model system. We, therefore, also considered whether Fat1 was induced during neuronal differentiation in alternative, more physiological models of neurogenesis. Indeed our analysis of different models (mouse NSCs, human iPSCs and ESCs; Fig. 8) provided clear evidence that Fat1 was selectively induced during the process of neural differentiation, thereby providing strong support for the concept that Fat1 induction reflects a normal physiological event. Nevertheless, despite the general similarity between these data, one apparent difference between the physiological systems versus the SH-SY5Y model was the time point at which Fat1 expression peaked.
In SH-SY5Y cells, the expression of FAT1 increased as differentiation progressed; while in the physiological models, Fat1 expression peaked at the ‘intermediate’ stages with reduction of levels in the differentiated neuron samples. Moreover, while the same pattern of expression changes was reproduced in all three models, the reduction of FAT1 expression in the hESC-derived neurons was clearly more evident. One distinguishing feature of the hESC data was the implementation of cell sorting to isolate neurons [37]. As NSCs and NPCs are multi-potent, sorting for CD44/EGFR served to remove cells with radial glial/astrocytic markers resulting in a highly purified population of mature neurons. The reduction in FAT1 expression in mature neurons has notable implications. Firstly, this implies that other brain cell lineages likely express significant levels of Fat1 and, therefore, its expression may have broader importance in neural development. Indeed it has been found that FAT1 is expressed by immortalised human astrocytes and acts to inhibit growth [55] but whether FAT1 influences differentiation of these cells is not known. Secondly, if peak expression occurs during the intermediate stages of neuronal differentiation, this suggests that Fat1 functions during early stages of differentiation. Since SH-SY5Y cells do not develop into fully mature neurons, the strong induction of FAT1 can be viewed to correlate with earlier stages of neuronal differentiation. Following on from this, our findings now provides new evidence of how FAT1 is involved in regulating neuronal differentiation, namely through effects on neuritogenesis and cell proliferation.
One known function of Fat1 that may explain its effects on neuritogenesis involves its role in the regulation of actin dynamics. It has been shown that the cytoplasmic tail of Fat1 contains several EVH motifs that are functional in binding to Ena/VASP proteins [56, 57]. These proteins in turn directly regulate the actin cytoskeleton [1] and in the context of neurons, initiate filopodial formation from which neurites are subsequently formed [58]. This proposes that FAT1 may be directly involved in the initiation and extension of neurites, with both processes being controlled through the regulation of actin dynamics. Although we did not test this, it, therefore, seems likely that FAT1 may act through Ena/VASP proteins to directly initiate neurite outgrowth. However, it is intriguing to consider how the process of neurite formation may be linked to the control of proliferation. As explained further below, our results favour the notion that FAT1 acts beyond the establishment of neurites to initiate differentiation signals, acting through the Hippo pathway.
FAT1-knockdown cells were also able to reach higher densities implying that FAT1 may attenuate proliferation through contact-dependency. Recently it was reported that FAT1 acts to inhibit apoptosis by directly interacting with elements of the apoptotic machinery [59], although how this relates to the process of neuronal differentiation remains to be established. Suppressing the induction of FAT1 during differentiation also resulted in concordant increases in the expression of cyclin D1, CDK2 and CDK4, effectors that generally promote cell cycle progression through G1/S. It has been previously shown that lengthening of the G1 phase in neural progenitors acts as a switch to promote neurogenesis and such G1 lengthening can be perturbed by overexpression of cyclinD1/CDK4 [60]. The increased levels of cyclinD1/CDK4 in turn serve to increase the expansion of neural progenitors. This regulatory concept is supported through studies of vertebrate development where forced over-expression of cyclin D in the chick spinal cord promoted cycling of neural progenitor cells and delayed differentiation [61, 62] and similarly ectopic expression of cyclin D1/CDK4 in the developing mouse cerebral cortex was found to inhibit differentiation and to stimulate proliferation [60]. Our results appear consistent with these reports where FAT1-mediated effects on attenuate high levels of cell cycle regulators that in turn influences cell proliferation. Relationships between Hippo signalling and the control of G1-cyclin expression are well established with Cyclin E a critical downstream target in Drosophila Hippo signalling [63, 64]. Similarly in vertebrates, the increased proliferation of neural progenitors observed after manipulating YAP and TEAD expression in the chick neural tube was correlated with high expression of Cyclin D [16]. Finally, links between Fat1 and cyclin D1 expression are established, at least in primary smooth muscle cells where knockdown of endogenous Fat1 increased the expression of cyclin D1 [65]. As the effects of FAT1 on SH-SY5Y differentiation bore many of the established hallmarks of Hippo pathway regulation, including dependency on cell density [18–20], we sought to confirm the link implied between FAT1 and Hippo signalling.
Our analysis of the Hippo effectors YAP and TAZ during SH-SY5Y differentiation indicated that TAZ in particular was strongly induced. We also recorded alterations in some of the Hippo core kinase components during the differentiation response, namely Salvador [20] and its regulatory component MOB1 [66]. In the scheme of canonical Hippo pathway [18] activation of these components would predict that YAP and TAZ levels would be reduced in differentiated cells. Nevertheless, TAZ levels accumulate during differentiation and this is also reflected in the early stages of neuronal differentiation in different model systems (Fig. 8). While this suggests induction of TAZ expression is a bona fide physiological feature during early neuronal differentiation, it also suggests that this does not reflect canonical Hippo signalling. Indeed, recent studies have demonstrated that the regulation of YAP/TAZ can be independent of Hippo kinases, e.g., in the context of mechanotransduction [67, 68], leading to the identification of ‘non-canonical’ Hippo signalling (reviewed in [69, 70]). The function of FAT1 within the SH-SY5Y system is particularly notable because preventing its expression during differentiation was necessary for the changes in Salvador and pMOB1. Moreover while FAT1 did not affect the cellular levels of TAZ, suppressing FAT1 expression promoted TAZ nuclear localisation and measurable changes in the Hippo target gene, CTGF. We then investigated if these effects were mediated through TAZ by its depletion using siRNA. Here it was found that the FAT-dependent induction of CTGF was ameliorated when TAZ was reduced. Moreover, this also acted to reverse the changes in cell cycle markers, together establishing a novel link between FAT1 and Hippo signalling in the control of TAZ function. The findings of this study have important implications for both the control of neurogenesis together with providing a general paradigm for more comprehensive investigations of the vertebrate Hippo signalling pathway.
FAT1-mediated inhibition of crosstalk between Hippo and TGFβ signalling
Prior studies have built up a picture of the function of TAZ in the cell nucleus where it not only interacts with TEADs to regulate classical Hippo targets [30, 71] but it also engages in cross-talk with elements of TGFβ signalling [22, 23]. Here TAZ binds to Smad2/3 [72] and this interaction was subsequently linked to the nuclear retention and activation of Smad-mediated transcription [23]. Our Western blotting analyses showed that following depletion of FAT1 there was a striking spatial and temporal re-distribution of TAZ to the nucleus in cells triggered to differentiate. We endeavoured to confirm the re-distribution of TAZ to the nucleus using confocal microscopy but because of vagaries in TAZ staining we could not utilise this technique. Nevertheless, other data involving Smad2/3 were consistent with the notion that FAT1 influences the cellular localisation and function of TAZ. Preventing FAT1 expression during differentiation caused up-regulation of the total levels of Smad3 and notably the increased Smad levels were dependent on TAZ. This finding together with the known regulatory association between TAZ and Smads provides clear support for the relationship between FAT1 expression and TAZ regulation.
If FAT1 serves to disable the Hippo-TGFβ signalling axis during neuronal differentiation what are the likely physiological implications of these findings? Based on the existing knowledge that Hippo [16, 17] and TGFβ pathways [73] act to promote the renewal of progenitors, our study suggests that FAT1 signalling acts to inhibit this process and drive aspects of neuronal differentiation. Here the known ability of FAT1 to regulate actin dynamics through binding to Ena/Vasp provides a ready explanation for its effects on neuritogenesis, but how FAT1 communicates with the Hippo core kinases is less clear. New reports indicating the importance of actin in Hippo signalling may provide the answer. Contractile forces generated through the actomyosin cytoskeleton can inhibit the translocation of both YAP and TAZ from the cytoplasm to the nucleus, thereby inactivating their downstream transcriptional programme [68, 74, 75]. Thus, through the same physical process, FAT1 enacts both inhibition of Hippo signalling and promotes neuritogenesis. Our data also provided a further intriguing link between FAT1 and TAZ.
During cell differentiation, FAT1 was required for efficient neuritogenesis since reduced levels of FAT1 decreased both neurite initiation (number) and extension (length). Depletion of TAZ increased neurite initiation but in the context of FAT1 knockdown cells, TAZ depletion was able to rescue the reduction in total neurites but did not restore neurite length. The implication from these findings is that TAZ does not affect the process of neurite extension per se but does counteract neurite initiation in addition to its role in proliferation and self-renewal. Most studies have presently focused on the transcriptional activities of Hippo effectors although significant amounts of YAP [76] and TAZ can occur in the cell cytoplasm [77]. These data now raise the possibility that the interplay between FAT1 and TAZ represents a control mechanism for the initiation of neurites and presumably a new cytoplasmic role for TAZ. One possible intermediate linking FAT1 to Hippo in this schema is the polarity regulator Scribble. Cordenonsi and colleagues demonstrated that Scribble acts as a membrane scaffold to recruit the Hippo kinases and TAZ at the cell membrane [78]. It has also been shown in zebrafish that Scribble binds to the cytoplasmic tail of Fat1 [27]. It is not known if FAT1 also binds Scribble but the conservation of the same binding motif in the human protein suggest that it is likely [1]. Nevertheless, more work needs to be done to identify the critical binding partners of FAT1 and to establish the mechanisms involved. On the other hand, our study provides mechanistic insights into the downstream consequences of FAT1 signalling and the overall process of neurogenesis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- CTGF
Connective tissue growth factor
- NC
Negative control
- NSC
Neuronal stem cell
- NTS
Non-targeting sequence
- qPCR
Quantitative real time PCR
- RA
Retinoic acid
- TEAD
Transcriptional enhancer activator (TEA) domain
- TPA
12-O-tetradecanoyl-phorbol-13-acetate
References
- 1.Sadeqzadeh E, de Bock CE, Thorne RF. Sleeping giants: emerging roles for the fat cadherins in health and disease. Med Res Rev. 2014;34(1):190–221. doi: 10.1002/med.21286. [DOI] [PubMed] [Google Scholar]
- 2.Dunne J, Hanby AM, Poulsom R, Jones TA, Sheer D, Chin WG, Da SM, Zhao Q, Beverley PC, Owen MJ. Molecular cloning and tissue expression of FAT, the human homologue of the Drosophila fat gene that is located on chromosome 4q34-q35 and encodes a putative adhesion molecule. Genomics. 1995;30(2):207–223. doi: 10.1006/geno.1995.9884. [DOI] [PubMed] [Google Scholar]
- 3.Matsui S, Utani A, Takahashi K, Mukoyama Y, Miyachi Y, Matsuyoshi N. Human Fat2 is localized at immature adherens junctions in epidermal keratinocytes. J Dermatol Sci. 2007;48(3):233–236. doi: 10.1016/j.jdermsci.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Ciani L, Patel A, Allen ND, Ffrench-Constant C. Mice lacking the giant protocadherin mFAT1 exhibit renal slit junction abnormalities and a partially penetrant cyclopia and anophthalmia phenotype. Mol Cell Biol. 2003;23(10):3575–3582. doi: 10.1128/MCB.23.10.3575-3582.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saburi S, Hester I, Goodrich L, McNeill H. Functional interactions between Fat family cadherins in tissue morphogenesis and planar polarity. Development. 2012;139(10):1806–1820. doi: 10.1242/dev.077461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox B, Hadjantonakis AK, Collins JE, Magee AI. Cloning and expression throughout mouse development of mfat1, a homologue of the Drosophila tumour suppressor gene fat. Dev Dyn. 2000;217(3):233–240. doi: 10.1002/(SICI)1097-0177(200003)217:3<233::AID-DVDY1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 7.Ponassi M, Jacques TS, Ciani L, Ffrench Constant C. Expression of the rat homologue of the Drosophila fat tumour suppressor gene. Mech Dev. 1999;80(2):207–212. doi: 10.1016/S0925-4773(98)00217-2. [DOI] [PubMed] [Google Scholar]
- 8.Gage FH. Mammalian neural stem cells. Science. 2000;287(5457):1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- 9.Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6(10):777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 10.Gincberg G, Arien-Zakay H, Lazarovici P, Lelkes PI. Neural stem cells: therapeutic potential for neurodegenerative diseases. Br Med Bull. 2012;104:7–19. doi: 10.1093/bmb/lds024. [DOI] [PubMed] [Google Scholar]
- 11.van Velthoven CT, Kavelaars A, van Bel F, Heijnen CJ. Nasal administration of stem cells: a promising novel route to treat neonatal ischemic brain damage. Pediatr Res. 2010;68(5):419–422. doi: 10.1203/PDR.0b013e3181f1c289. [DOI] [PubMed] [Google Scholar]
- 12.Shoae-Hassani A, Mortazavi-Tabatabaei SA, Sharif S, Rezaei-Khaligh H, Verdi J. DHEA provides a microenvironment for endometrial stem cells neurogenesis. Med Hypotheses. 2011;76(6):843–846. doi: 10.1016/j.mehy.2011.02.033. [DOI] [PubMed] [Google Scholar]
- 13.Hirabayashi Y, Itoh Y, Tabata H, Nakajima K, Akiyama T, Masuyama N, Gotoh Y. The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development. 2004;131(12):2791–2801. doi: 10.1242/dev.01165. [DOI] [PubMed] [Google Scholar]
- 14.Guo Y, Wang P, Sun H, Cai R, Xia W, Wang S. Advanced glycation end product-induced astrocytic differentiation of cultured neurospheres through inhibition of Notch-Hes1 pathway-mediated neurogenesis. Int J Mol Sci. 2013;15(1):159–170. doi: 10.3390/ijms15010159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin YT, Ding JY, Li MY, Yeh TS, Wang TW, Yu JY. YAP regulates neuronal differentiation through Sonic hedgehog signaling pathway. Exp Cell Res. 2012;318(15):1877–1888. doi: 10.1016/j.yexcr.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Cao X, Pfaff SL, Gage FH. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 2008;22(23):3320–3334. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Deo M, Thompson RC, Uhler MD, Turner DL. Negative regulation of Yap during neuronal differentiation. Dev Biol. 2012;361(1):103–115. doi: 10.1016/j.ydbio.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24(9):862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, Makita R, Kurihara H, Morin-Kensicki EM, Nojima H, Rossant J, Nakao K, Niwa H, Sasaki H. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16(3):398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Zhao B, Li L, Guan KL. Hippo signaling at a glance. J Cell Sci. 2010;123(Pt 23):4001–4006. doi: 10.1242/jcs.069070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beyer TA, Weiss A, Khomchuk Y, Huang K, Ogunjimi AA, Varelas X, Wrana JL. Switch enhancers interpret TGF-beta and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep. 2013;5(6):1611–1624. doi: 10.1016/j.celrep.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 22.Wrighton KH, Dai F, Feng XH. A new kid on the TGFbeta block: tAZ controls Smad nucleocytoplasmic shuttling. Dev Cell. 2008;15(1):8–10. doi: 10.1016/j.devcel.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol. 2008;10(7):837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- 24.Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8(1):27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 25.Kawamori H, Tai M, Sato M, Yasugi T, Tabata T. Fat/Hippo pathway regulates the progress of neural differentiation signaling in the Drosophila optic lobe. Dev Growth Differ. 2011;53(5):653–667. doi: 10.1111/j.1440-169X.2011.01279.x. [DOI] [PubMed] [Google Scholar]
- 26.Van Hateren NJ, Das RM, Hautbergue GM, Borycki AG, Placzek M, Wilson SA. FatJ acts via the Hippo mediator Yap1 to restrict the size of neural progenitor cell pools. Development. 2011;138(10):1893–1902. doi: 10.1242/dev.064204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skouloudaki K, Puetz M, Simons M, Courbard JR, Boehlke C, Hartleben B, Engel C, Moeller MJ, Englert C, Bollig F, Schafer T, Ramachandran H, Mlodzik M, Huber TB, Kuehn EW, Kim E, Kramer-Zucker A, Walz G. Scribble participates in Hippo signaling and is required for normal zebrafish pronephros development. Proc Natl Acad Sci USA. 2009;106(21):8579–8584. doi: 10.1073/pnas.0811691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Jiang D, Chi F, Zhao B. The Hippo pathway regulates stem cell proliferation, self-renewal, and differentiation. Protein Cell. 2012;3(4):291–304. doi: 10.1007/s13238-012-2919-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiemer SE, Varelas X. Stem cell regulation by the Hippo pathway. Biochim Biophys Acta 1830. 2013;2:2323–2334. doi: 10.1016/j.bbagen.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 30.Bhat KP, Salazar KL, Balasubramaniyan V, Wani K, Heathcock L, Hollingsworth F, James JD, Gumin J, Diefes KL, Kim SH, Turski A, Azodi Y, Yang Y, Doucette T, Colman H, Sulman EP, Lang FF, Rao G, Copray S, Vaillant BD, Aldape KD. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25(24):2594–2609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leli U, Cataldo A, Shea TB, Nixon RA, Hauser G. Distinct mechanisms of differentiation of SH-SY5Y neuroblastoma cells by protein kinase C activators and inhibitors. J Neurochem. 1992;58(4):1191–1198. doi: 10.1111/j.1471-4159.1992.tb11328.x. [DOI] [PubMed] [Google Scholar]
- 32.Ota A, Shen-Orr Z, Roberts CT, Jr, LeRoith D. TPA-induced neurite formation in a neuroblastoma cell line (SH-SY5Y) is associated with increased IGF-I receptor mRNA and binding. Brain Res Mol Brain Res. 1989;6(1):69–76. doi: 10.1016/0169-328X(89)90030-2. [DOI] [PubMed] [Google Scholar]
- 33.Peddada S, Yasui DH, LaSalle JM. Inhibitors of differentiation (ID1, ID2, ID3 and ID4) genes are neuronal targets of MeCP2 that are elevated in Rett syndrome. Hum Mol Genet. 2006;15(12):2003–2014. doi: 10.1093/hmg/ddl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Bock CE, Ardjmand A, Molloy TJ, Bone SM, Johnstone D, Campbell DM, Shipman KL, Yeadon TM, Holst J, Spanevello MD, Nelmes G, Catchpoole DR, Lincz LF, Boyd AW, Burns GF, Thorne RF. The Fat1 cadherin is overexpressed and an independent prognostic factor for survival in paired diagnosis-relapse samples of precursor B-cell acute lymphoblastic leukemia. Leukemia. 2012;26(5):918–926. doi: 10.1038/leu.2011.319. [DOI] [PubMed] [Google Scholar]
- 35.Hartl D, Irmler M, Romer I, Mader MT, Mao L, Zabel C, de Angelis MH, Beckers J, Klose J. Transcriptome and proteome analysis of early embryonic mouse brain development. Proteomics. 2008;8(6):1257–1265. doi: 10.1002/pmic.200700724. [DOI] [PubMed] [Google Scholar]
- 36.Pasca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Pasca AM, Cord B, Palmer TD, Chikahisa S, Nishino S, Bernstein JA, Hallmayer J, Geschwind DH, Dolmetsch RE. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. 2011;17(12):1657–1662. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lafaille FG, Pessach IM, Zhang SY, Ciancanelli MJ, Herman M, Abhyankar A, Ying SW, Keros S, Goldstein PA, Mostoslavsky G, Ordovas-Montanes J, Jouanguy E, Plancoulaine S, Tu E, Elkabetz Y, Al-Muhsen S, Tardieu M, Schlaeger TM, Daley GQ, Abel L, Casanova JL, Studer L, Notarangelo LD. Impaired intrinsic immunity to HSV-1 in human iPSC-derived TLR3-deficient CNS cells. Nature. 2012;491(7426):769–773. doi: 10.1038/nature11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sadeqzadeh E, de Bock CE, Zhang XD, Shipman KL, Scott NM, Song C, Yeadon T, Oliveira CS, Jin B, Hersey P, Boyd AW, Burns GF, Thorne RF. Dual processing of FAT1 cadherin protein by human melanoma cells generates distinct protein products. J Biol Chem. 2011;286(32):28181–28191. doi: 10.1074/jbc.M111.234419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berggren WT, Lutz M, Modesto V (2008) General spinfection protocol. In: Berggren WT, Lutz M, Modesto V (eds) StemBook: 2012. Cambridge [PubMed]
- 40.Sontag JM, Nunbhakdi-Craig V, Mitterhuber M, Ogris E, Sontag E. Regulation of protein phosphatase 2A methylation by LCMT1 and PME-1 plays a critical role in differentiation of neuroblastoma cells. J Neurochem. 2010;115(6):1455–1465. doi: 10.1111/j.1471-4159.2010.07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dehmelt L, Smart FM, Ozer RS, Halpain S. The role of microtubule-associated protein 2c in the reorganization of microtubules and lamellipodia during neurite initiation. J Neurosci. 2003;23(29):9479–9490. doi: 10.1523/JNEUROSCI.23-29-09479.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michaelson D, Abidi W, Guardavaccaro D, Zhou M, Ahearn I, Pagano M, Philips MR. Rac1 accumulates in the nucleus during the G2 phase of the cell cycle and promotes cell division. J Cell Biol. 2008;181(3):485–496. doi: 10.1083/jcb.200801047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2(6):525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 44.Perez-Polo JR, Werbach-Perez K, Tiffany-Castiglioni E. A human clonal cell line model of differentiating neurons. Dev Biol. 1979;71(2):341–355. doi: 10.1016/0012-1606(79)90174-X. [DOI] [PubMed] [Google Scholar]
- 45.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, Guan KL. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22(14):1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai D, Ho KC, Hao Y, Yang X. Taxol resistance in breast cancer cells is mediated by the hippo pathway component TAZ and its downstream transcriptional targets Cyr61 and CTGF. Cancer Res. 2011;71(7):2728–2738. doi: 10.1158/0008-5472.CAN-10-2711. [DOI] [PubMed] [Google Scholar]
- 47.Massague J, Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 2000;19(8):1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Derynck R. Regulation of Smad signalling by protein associations and signalling crosstalk. Trends Cell Biol. 1999;9(7):274–279. doi: 10.1016/S0962-8924(99)01579-2. [DOI] [PubMed] [Google Scholar]
- 49.Mao Y, Mulvaney J, Zakaria S, Yu T, Morgan KM, Allen S, Basson MA, Francis-West P, Irvine KD. Characterization of a Dchs1 mutant mouse reveals requirements for Dchs1-Fat4 signaling during mammalian development. Development. 2011;138(5):947–957. doi: 10.1242/dev.057166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bossuyt W, Chen CL, Chen Q, Sudol M, McNeill H, Pan D, Kopp A, Halder G. An evolutionary shift in the regulation of the Hippo pathway between mice and flies. Oncogene. 2013 doi: 10.1038/onc.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Castillejo-Lopez C, Arias WM, Baumgartner S. The fat-like gene of Drosophila is the true orthologue of vertebrate fat cadherins and is involved in the formation of tubular organs. J Biol Chem. 2004;279(23):24034–24043. doi: 10.1074/jbc.M313878200. [DOI] [PubMed] [Google Scholar]
- 52.Cappello S, Gray MJ, Badouel C, Lange S, Einsiedler M, Srour M, Chitayat D, Hamdan FF, Jenkins ZA, Morgan T, Preitner N, Uster T, Thomas J, Shannon P, Morrison V, Di Donato N, Van Maldergem L, Neuhann T, Newbury-Ecob R, Swinkells M, Terhal P, Wilson LC, Zwijnenburg PJ, Sutherland-Smith AJ, Black MA, Markie D, Michaud JL, Simpson MA, Mansour S, McNeill H, Gotz M, Robertson SP. Mutations in genes encoding the cadherin receptor-ligand pair DCHS1 and FAT4 disrupt cerebral cortical development. Nat Genet. 2013;45(11):1300–1308. doi: 10.1038/ng.2765. [DOI] [PubMed] [Google Scholar]
- 53.Blair IP, Chetcuti AF, Badenhop RF, Scimone A, Moses MJ, Adams LJ, Craddock N, Green E, Kirov G, Owen MJ, Kwok JB, Donald JA, Mitchell PB, Schofield PR. Positional cloning, association analysis and expression studies provide convergent evidence that the cadherin gene FAT contains a bipolar disorder susceptibility allele. Mol Psychiatry. 2006;11(4):372–383. doi: 10.1038/sj.mp.4001784. [DOI] [PubMed] [Google Scholar]
- 54.Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, Polak P, Yoon S, Maguire J, Crawford EL, Campbell NG, Geller ET, Valladares O, Schafer C, Liu H, Zhao T, Cai G, Lihm J, Dannenfelser R, Jabado O, Peralta Z, Nagaswamy U, Muzny D, Reid JG, Newsham I, Wu Y, Lewis L, Han Y, Voight BF, Lim E, Rossin E, Kirby A, Flannick J, Fromer M, Shakir K, Fennell T, Garimella K, Banks E, Poplin R, Gabriel S, DePristo M, Wimbish JR, Boone BE, Levy SE, Betancur C, Sunyaev S, Boerwinkle E, Buxbaum JD, Cook EH, Jr, Devlin B, Gibbs RA, Roeder K, Schellenberg GD, Sutcliffe JS, Daly MJ. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485(7397):242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morris LG, Kaufman AM, Gong Y, Ramaswami D, Walsh LA, Turcan S, Eng S, Kannan K, Zou Y, Peng L, Banuchi VE, Paty P, Zeng Z, Vakiani E, Solit D, Singh B, Ganly I, Liau L, Cloughesy TC, Mischel PS, Mellinghoff IK, Chan TA. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat Genet. 2013;45(3):253–261. doi: 10.1038/ng.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moeller MJ, Soofi A, Braun GS, Li X, Watzl C, Kriz W, Holzman LB. Protocadherin FAT1 binds Ena/VASP proteins and is necessary for actin dynamics and cell polarization. EMBO J. 2004;23(19):3769–3779. doi: 10.1038/sj.emboj.7600380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanoue T, Takeichi M. New insights into Fat cadherins. J Cell Sci. 2005;118(Pt 11):2347–2353. doi: 10.1242/jcs.02398. [DOI] [PubMed] [Google Scholar]
- 58.Dent EW, Kwiatkowski AV, Mebane LM, Philippar U, Barzik M, Rubinson DA, Gupton S, Van Veen JE, Furman C, Zhang J, Alberts AS, Mori S, Gertler FB. Filopodia are required for cortical neurite initiation. Nat Cell Biol. 2007;9(12):1347–1359. doi: 10.1038/ncb1654. [DOI] [PubMed] [Google Scholar]
- 59.Kranz D, Boutros M. A synthetic lethal screen identifies FAT1 as an antagonist of caspase-8 in extrinsic apoptosis. EMBO J. 2014;33(3):181–197. doi: 10.1002/embj.201385686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5(3):320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 61.Lobjois V, Bel-Vialar S, Trousse F, Pituello F. Forcing neural progenitor cells to cycle is insufficient to alter cell-fate decision and timing of neuronal differentiation in the spinal cord. Neural Dev. 2008;3:4. doi: 10.1186/1749-8104-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lobjois V, Benazeraf B, Bertrand N, Medevielle F, Pituello F. Specific regulation of cyclins D1 and D2 by FGF and Shh signaling coordinates cell cycle progression, patterning, and differentiation during early steps of spinal cord development. Dev Biol. 2004;273(2):195–209. doi: 10.1016/j.ydbio.2004.05.031. [DOI] [PubMed] [Google Scholar]
- 63.Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16(21):2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 64.Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol. 2006;16(21):2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 65.Hou R, Liu L, Anees S, Hiroyasu S, Sibinga NE. The Fat1 cadherin integrates vascular smooth muscle cell growth and migration signals. J Cell Biol. 2006;173(3):417–429. doi: 10.1083/jcb.200508121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hergovich A. MOB control: reviewing a conserved family of kinase regulators. Cell Signal. 2011;23(9):1433–1440. doi: 10.1016/j.cellsig.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154(5):1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 68.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474(7350):179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 69.Hergovich A. Regulation and functions of mammalian LATS/NDR kinases: looking beyond canonical Hippo signalling. Cell Biosci. 2013;3(1):32. doi: 10.1186/2045-3701-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Varelas X (2013) Non-canonical roles for the Hippo pathway. In: Oren M, Aylon Y (eds) The Hippo signaling pathway and cancer. Springer Science & Business Media, pp 327
- 71.Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, Xiong Y, Lei QY, Guan KL. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284(20):13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, Luga V, Przulj N, Robinson M, Suzuki H, Hayashizaki Y, Jurisica I, Wrana JL. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307(5715):1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 73.Singh AM, Reynolds D, Cliff T, Ohtsuka S, Mattheyses AL, Sun Y, Menendez L, Kulik M, Dalton S. Signaling network crosstalk in human pluripotent cells: a Smad2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell. 2012;10(3):312–326. doi: 10.1016/j.stem.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26(1):54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wada K, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138(18):3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- 76.Mohler PJ, Kreda SM, Boucher RC, Sudol M, Stutts MJ, Milgram SL. Yes-associated protein 65 localizes p62(c-Yes) to the apical compartment of airway epithelia by association with EBP50. J Cell Biol. 1999;147(4):879–890. doi: 10.1083/jcb.147.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC, Yaffe MB. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19(24):6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, Daidone MG, Dupont S, Basso G, Bicciato S, Piccolo S. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147(4):759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.