Abstract
Eukaryotic cells store excess fatty acids as neutral lipids, predominantly triacylglycerols and sterol esters, in organelles termed lipid droplets (LDs) that bulge out from the endoplasmic reticulum. LDs are highly dynamic and contribute to diverse cellular functions. The catabolism of the storage lipids within LDs is channeled to multiple metabolic pathways, providing molecules for energy production, membrane building blocks, and lipid signaling. LDs have been implicated in a number of protein degradation and pathogen infection processes. LDs may be linked to prevalent human metabolic diseases and have marked potential for biofuel production. The knowledge accumulated on LDs in recent years provides a foundation for diverse, and even unexpected, future research. This review focuses on recent advances in LD research, emphasizing the diverse physiological roles of LDs in the model system of budding yeast.
Keywords: Endoplasmic reticulum, Triacylglycerol, Sterol ester, Phospholipid, Membrane, Metabolism
Introduction
Lipid droplets (LDs), also termed as lipid particles, lipid bodies, fat bodies, oil bodies, or adiposomes, are ubiquitously found in virtually all eukaryotes and even some bacteria. Although they were long thought to be inert fat storage depots, recent discoveries of LD-specific molecules have attracted much of the scientific interests. LDs are now perceived as metabolically active and highly dynamic organelles with a specialized function in regulating cellular lipid homeostasis (reviewed in [1–3]). The contents of LDs, predominantly triacylglycerols (TAGs) and sterol esters (SEs), provide cells with membrane and energy sources, and also protect cells against lipotoxicity. The accumulating evidence suggests that the surface of LDs may create an environment of temporal protein inactivation. Several pathogens hijack this energy-rich organelle for use in their life cycles. In addition, LDs are involved in prevalent lipid metabolic disorders and biofuel production, making LD-related research an attractive field of modern cell biology.
Unraveling the biogenesis, maintenance, and physiological roles of LDs among all systems are important for understanding the nature of this organelle. The budding yeast Saccharomyces cerevisiae is instrumental for biomedical research. This amenable model system offers powerful genetic, cell biology, and biochemical manipulation tools necessary for dissecting scientific questions at the cellular and molecular levels. This organism accumulates LDs, most prominently when cells enter the stationary phase or encounter environmental stresses. Consistent with observations in other eukaryotes, the yeast LDs appear to be well controlled for their size and number. The detailed knowledge of biosynthetic pathways and signaling networks provide the yeast system with essential information for current and future LD studies. Here, I review recent advances and highlight the dynamic roles of LDs in yeast.
LD morphology and dynamics
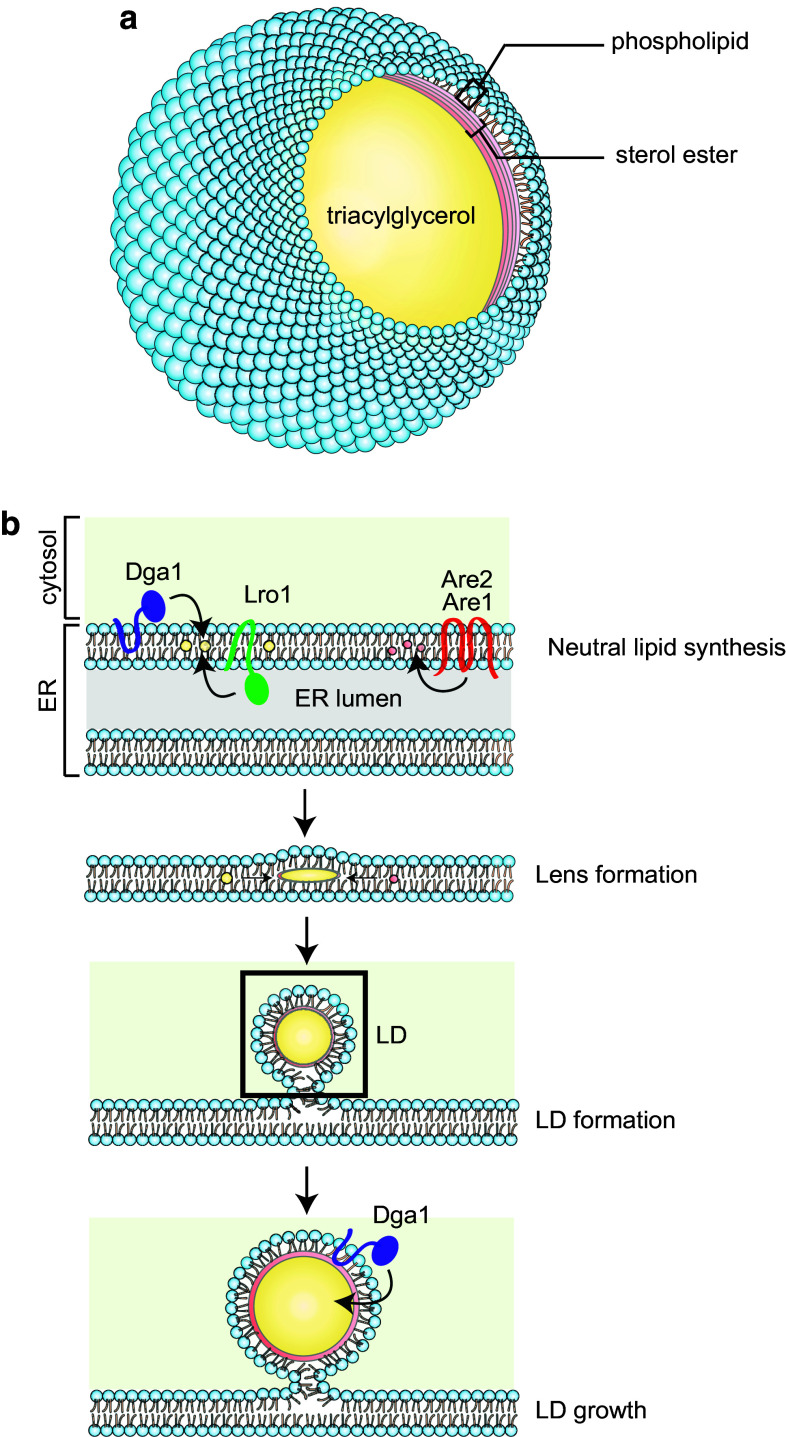
The basic structure of cellular LDs in eukaryotes is composed of a hydrophobic core and a phospholipid monolayer (Fig. 1a). The most prominent neutral lipids in the core are TAGs and SEs, but other hydrophobic lipids such as retinyl esters [4], wax, ether lipids [5], and squalene [6] are also possible constituents, depending on the cell types and growth conditions. For example, LDs in white adipocytes contain mostly TAGs, while those in macrophages contain mostly SEs. In wild-type yeast cells grown in normal laboratory conditions with glucose as the carbon source, TAGs and SEs accumulate equally and are typically stored within the same LDs. Differential scanning calorimetry and X-ray studies suggest that randomly packed fluid-state TAGs in the center of LDs are surrounded by several rigidly packed SE shells beneath the phospholipid monolayer (Fig. 1a) [7]. The composition of the LD phospholipid monolayer is distinct from its origin, the endoplasmic reticulum (ER), indicative of a selective partitioning event coordinated with LD biogenesis. In general, the LD monolayer is enriched in anionic phospholipids. Mammalian LDs contain mostly phosphatidylcholine (PC) and phosphatidylethanolamine (PE), but little phosphatidylserine (PS) and phosphatidic acid (PA) [5]. In yeast, LDs comprise mostly of PC, phosphatidylinositol (PI), and PE and are enriched in double-unsaturated fatty acid species [8–10]. Lyso-PE, lyso-PC, and PC, but not sphingomyelin, PS, and PA, are enriched in the yeast LD monolayers relative to the total membranes [5]. The biophysical properties and roles of these phospholipids during droplet emulsion were reviewed by Thiam et al. in 2013 [11].
Fig. 1.
The formation of LD. a LD composition. b A simple model of LD formation and growth. Yeast contains four neutral lipid synthesis enzymes. Are1 and Are2 catalyze the synthesis of sterol ester (SE), while Dga1 and Lro1 catalyze the synthesis of triacylglycerol (TAG). Neutral lipids accumulated within the ER bilayer on reaching a critical threshold form lens, and subsequently a nascent LD buds into the cytoplasm. LDs can grow by recruiting Dga1 to the surface of LDs for TAG synthesis
LDs in many organisms, including yeast, are visible structures by light, particularly with differential interference contrast, microscopy. The size, number, and distribution of LDs vary under different physiological states, implying that they are dynamically regulated in response to physiological cues. Several cells, such as adipocytes, accumulate LDs up to 100 μm in diameter, while in most cells LDs are small and often less than 1 μm wide. Yeast cells normally form LDs of less than 250 nm, but supersized LDs of more than 1 μm are formed in some mutants [12–14]. LDs in yeast are highly dynamic. In the exponential growth phase, small and highly mobile LDs disperse and are connected to the ER, including the perinulcear ER and cortical ER [15]. When cells reach the diauxic shift, LDs typically surround the perinuclear ER and expand. LDs continue to grow and become even more prominent during the stationary phase. The size and contents of LDs can change rapidly. Yeast cells treated with oleic acids had larger LDs, with only TAGs, not SEs, being synthesized and loaded into the LD core [16]. It is important to note that the quality and quantity of the varying LD components may influence the LDs’ stability and behavior. Thus, experimental parameters, such as growth media and conditions, might readily impact the LD analyses.
Neutral lipid synthesis and the underlying lipid metabolic pathways
LD formation is solely dependent on the synthesis of neutral lipids, TAGs, and SEs [17] (Fig. 2). In mammalian cells, the acyl-coenzyme A (acyl-CoA):diacylglycerol acyltransferase (DGAT) enzymes DGAT1 and DGAT2 catalyze TAG synthesis by transferring a fatty acyl chain to diacylglycerol (DAG). The major DGAT enzyme in yeast is Dga1 (the mammalian DGAT2 ortholog) [18]. In addition to Dga1, yeast contains Lro1, a mammalian lecithin-cholesterol acyltransferase (LCAT) homolog, responsible for the synthesis of the other TAG pool [19, 20]. The preferred acyl donors for Lro1 are phospholipids, particularly PC and PE, suggesting that the major role of Lro1 is in remodeling membrane phospholipids [21, 22]. The two TAG synthesis enzymes vary in their functions under different growth conditions. Lro1 activity is more prominent in the exponential growth phase, whereas Dga1 significantly contributes to TAG synthesis during the stationary phase [23]. The mammalian SE is synthesized by two major acyl-CoA:cholesterol acyltransferase (ACAT) enzymes, ACAT1 and ACAT2. In yeast, Are1 and Are2 are the ACAT homologs that catalyze the esterification of sterol intermediates [21, 24]. The preferred substrate for Are2 is ergosterol, while Are1 largely synthesizes the lanosterol esters [25]. Are2 catalyzes most of the SE synthesis in yeast grown under normal laboratory conditions, whereas Are1 appears to be activated under anaerobic conditions. These two enzymes may also contribute to the residual TAG synthesis in cells lacking Dga1 and Lro1, as Are2 is able to perform DGAT activity in vitro [23]. Importantly, the synthesis of neutral lipids in cells is tightly connected to, and thus is affected by, the metabolism of fatty acids (FAs), glycerolipids, and sterols (Fig. 2). These lipid metabolic pathways are discussed below.
Fig. 2.
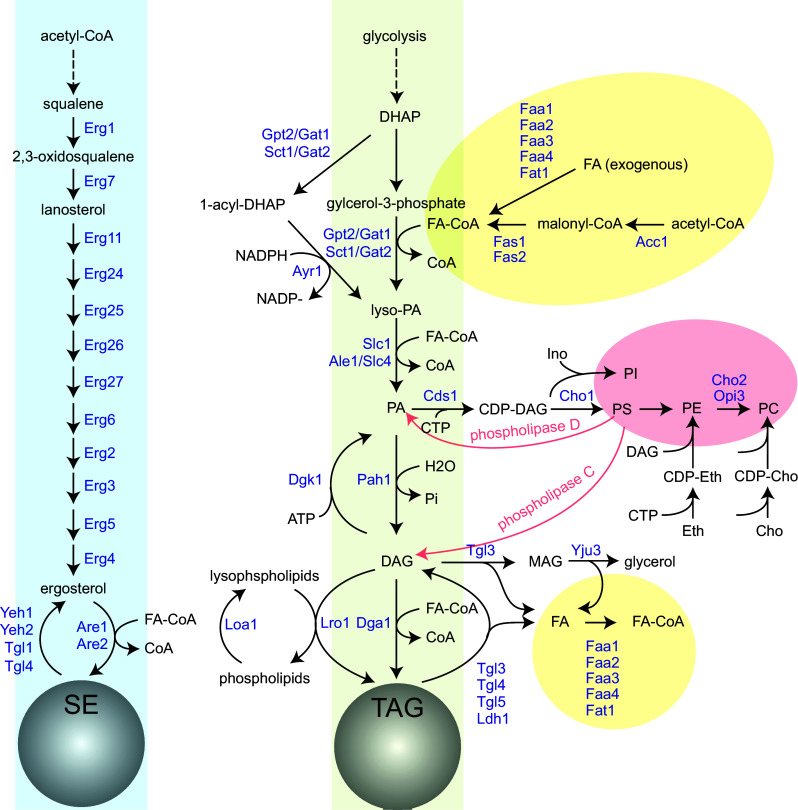
The metabolic pathways of SE and TAG in S. cerevisiae (adopted from Rajakumari et al. [26] and Currie et al. [105]). Enzymes are indicated in blue. Cyan zone formation of SE. Green zone formation of TAG. Yellow zone formation of FA-CoA. Red zone formation of phospholipids. FA fatty acid, CoA coenzyme A, FA-CoA fatty acyl-CoA, DHAP dihydroxyacetone phosphate, lyso-PA 1-acylglycerol-3-phosphate, PA phosphatidic acid, DAG diacylglycerol, CDP-DAG cytidine diphosphate-diacylglycerol, Ino inositol, PI phosphatidylinositol, PS phosphatidylserine, PE phosphatidylethanolamine, PC phosphatidylcholine, CDP-Eth cytidine diphosphate-ethanolamine, CDP-Cho cytidine diphosphate-choline, Eth ethanolamine, Cho choline, MAG monoacylglycerol, CTP cytidine triphosphate, Pi inorganic phosphate
Cells can synthesize FAs de novo. This pathway is important because no exogenous FAs are added in typical yeast growth media. The first step of FA synthesis requires the activity of acetyl-CoA carboxylase Acc1 to convert acetyl-CoA to malonyl-CoA (Fig. 2). The fatty acid synthases (Fas1 and Fas2) subsequently elongate the acyl chain step by step to form medium-chain fatty acids, mostly C16 and C18 in yeast. Acetyl-CoA carboxylase is the rate-limiting enzyme for FA synthesis, and it is regulated by complex transcriptional controls in conjunction with phospholipid synthesis [27] and post-translational controls via Snf1/mammalian AMP-activated protein kinase [28]. In addition to the de novo pathway, free FAs may be derived from an exogenous supply or from lipolysis. The activation of FAs in yeast is mediated through five acyl-CoA synthetases that differ in their localization and substrate specificities. Three of these enzymes, Faa1, Faa4, and Fat1, localize to the LDs and likely function there to activate the FAs derived from TAG and SE breakdown. In the exponential phase, the catabolism of TAG in yeast cells allows for the rapid synthesis of membrane lipids for use in cell growth and division [29, 30].
Excess FAs are toxic to cells and, thus, the metabolic flux of FAs needs to be incorporated either into phospholipids or the biologically inert TAGs. TAG synthesis depends on two major precursors, PA and DAG, and both are central intermediates for phospholipid synthesis [22, 31] (Fig. 2). The PA precursor 1-acylglycerol-3-phosphate (lyso-PA) is synthesized either from the acylation of glycerol-3-phosphate by the glycerol-3-phosphate acyltransferases (GPATs), Gpt2/Gat1 and Sct1/Gat2, or from the acylation of dihydroxyacetone phosphate by a two-step reaction involving Gpt2/Gat1 and Sct1/Gat2 followed by Ayr1 [32]. The two GPAT enzymes exhibit substrate specificity, thus contributing differentially to the synthesis of TAG species [33, 34]. Lyso-PA is then acylated at the sn-2 position by the two lyso-PA acyltransferases, Slc1 and Ale1/Slc4, to yield PA. The two enzymes display different substrate specificities and may also be involved in the FA exchange at the sn-2-position of mature glycerophospholipids [35]. In addition to Slc1 and Ale1/Slc4, the LD-localized protein Loa1/Vps66 also functions as a lyso-PA acyltransferase with a specificity for oleoyl-CoA [36].The dephosphorylation of PA in yeast is performed by the phosphatidate phosphatase (PAP), Pah1, an ortholog of mammalian lipin, to give rise to DAG production [37–39]. Finally, the esterification of DAG by Dga1 and Lro1 generates TAGs that are stored within LDs.
In addition to synthesizing TAG, PA also supplies cells with phospholipids via the activation of CTP by Cds1 to produce CDP-DAG, a precursor for PI, PS, PE, phosphatidylglycerol, cardiolipin, and PC (reviewed in [40, 41]) (Fig. 2). The PA level in the ER is monitored by Scs2 and Opi1. When the PA level in the membrane elevates, the INO1 repressor Opi1 is trapped by Scs2 in the ER and the synthesis of INO1 activates PI synthesis by reducing PA and CDP-DAG levels [42, 43]. PA can also be derived from phospholipids by the action of phospholipase D or by the phosphorylation of DAG. The DAG kinase Dgk1, which counteracts Pah1 activity, is thought to be particularly important for consuming storage lipids during growth resumption [44, 45]. In addition to the CDP-DAG pathway, in the Kennedy pathway choline or ethanolamine is first activated with CTP and ultimately attached to DAG to form PC or PE, providing an alternative route for membrane lipid biosynthesis derived from DAG [46–48]. In general, the core precursors of phospholipid and TAG synthesis are PA and DAG.
Yeast cells, like mammalian cells, utilize most of the sterols (ergosterols in yeast and cholesterols in mammals) in the plasma membrane [49]. Sterol biosynthesis is strictly aerobic and requires heme in yeast. The major precursor for sterol synthesis is squalene, which is modified by a sequential reaction from acetyl-CoA (Fig. 2). Squalene is converted to 2,3-oxidosqualene by the squalene epoxidase Erg1 [50, 51]. The next step in sterol biosynthesis involves the oxidosqualene cyclase Erg7 to synthesize lanosterols [52, 53], followed by several steps to generate ergosterols. Eukaryotic cells have evolved sophisticated mechanisms at the transcriptional, translational, and post-translational levels to fine-tune the cellular sterol levels. One such example is that the enzymatic activity of Erg1 is regulated by its dual localization between the ER and LDs [51] and that the sterol abundance is sensed and subjects Erg1 to degradation by a branch of the ER-associated protein degradation (ERAD) pathway [54]. Similar to TAGs that play an important role in buffering excess FAs, the conversion of sterols to biologically inert SEs potentially prevents the accumulation of excess sterols, which is harmful to the cells.
Biogenesis of LDs
In eukaryotic cells, LDs form de novo from the outer leaflet of the ER (Fig. 1b). Inducible LD biogenesis has been studied using yeast cells genetically modified to express a sole neutral lipid synthesis enzyme under an inducible promoter [55]. In this system, LDs arise from the perinuclear ER upon induction of neutral lipid synthesis and once formed associate with the ER for most of the time. Many LD proteins partition freely between ER and LDs and re-localize from the ER to nascent LDs during LD biogenesis. Several models have been proposed to explain LD biogenesis from the ER (reviewed in [2, 56]). The most popular model hypothesizes that the neutral lipids between two leaflets of the ER membrane are organized into a lens that bulges out toward the cytosolic face of the ER to form LDs (Fig. 1b). However, a pure physical model proposes that TAGs and the phospholipid surfactants accumulate until they reach levels sufficient for spontaneous LD emulsification and budding [57]. It remains unclear how the orientation of LD budding is determined, as chylomicrons, TAG-containing lipoprotein particles in the absorptive cells of small intestines, are packaged and bud toward the luminal site of the ER [58]. Although the exact mechanisms of LD biogenesis remain largely unknown, the process is likely driven by the availability of lipids, enzymes, and structural proteins.
In yeast, neutral lipids are synthesized by the four acyltransferases Dga1, Lro1, Are1, and Are2 (Fig. 2). Mutants lacking all four of these enzymes have a complete loss of LD formation, indicating that the synthesis of neutral lipids is a prerequisite for LD formation [59]. Wild-type yeast cells store equal amount of TAGs and SEs in their LDs. In dga1Δ lro1Δ cells, LDs contain mostly SEs but their numbers are drastically reduced [18].In contrast, the are1Δ are2Δ cells produce LDs mostly containing TAGs, but the number of LDs was not significantly affected [17, 23]. Thus, LD formation is correlated with the activity of these acyltransferases, particularly Dga1 and Lro1, which synthesize TAGs. Many neutral lipid synthesis enzymes in eukaryotic cells are found in the ER. For example, Lro1, Are1, and Are2 are exclusively localized to the ER, and not to the LDs, in yeast. Fluorescence microscopy studies indicate that the phospholipid-remodeling acyltransferase Lro1 in yeast is associated with a highly dynamic subdomain that moves along the ER and may transiently interact with the LDs [60]. Intriguingly, topological studies on Lro1 suggest that its active site is localized within the ER lumen [61]. Similarly, the conserved active sites of Are1 and Are2 are exposed to the luminal side of the ER [62]. How exactly the neutral lipids are organized into nascent LDs is currently unknown. The mammalian DGAT2 and its ortholog yeast Dga1 can be recruited to, and function on, the LD surface, indicating that TAG synthesis can also occur directly on the LD surface [55, 63] (Fig. 1b). It has been proposed that the LDs’ recruitment of Dga1 may use a stretch of its hydrophobic residues. Moreover, LD formation by Dga1 in yeast is independent of temperature and energy, thus ruling out vesicle formation [55].
In addition to TAGs, DAGs and PAP, an enzyme required for DAG synthesis, are critical factors for LD biogenesis. The disruption of the yeast PAP, Pah1, largely impairs LD formation, concomitant with the accumulation of neutral lipids in the ER [64]. Pah1 is crucial for LD formation even in cells synthesizing only SEs. Interestingly, the LD biogenesis defect caused by pah1Δ can be bypassed by the absence of the Dgk1 activity that converts DAGs to PA. Thus, the DAGs generated by Pah1 is important for LD biogenesis. DAGs displaying a negative curvature in the membrane may either offer a structural advantage or serve as a protein-binding platform during LD formation. The binding of DAGs to LD proteins, such as perilipin 3 [65] and CTP:phosphocholine cytidylyltransferase (CCT) [66], has been reported in other eukaryotes.
A critical issue for LD biogenesis is how cells couple the expansion of the LD phospholipid monolayer to the growth of the neutral lipid core. In mammalian cells, the major phospholipid in the LD monolayer is PC, which serves as a surfactant and prevents LDs from coalescing [11]. Additional PC loading onto the LD surface is mediated through the Kennedy pathway [66]. In mice and Drosophila, the rate-limiting enzyme CCT in the Kennedy pathway is targeted to the growing LDs for LD expansion during the FA-induced LD formation. CCT can be activated by DAG and PA, and the protein prefers binding with membranes deficient in PC. Thus, de novo PC synthesis might also modulate the PC level on the LD surface. An alternative pathway that contributes to PC synthesis on the LD surface is the Lands cycle that forms PC from lyso-PC and fatty acyl-CoA. Lyso-PC transferase 1 and 2 are recruited to, and function on, LDs [67]. In Drosophila cells, the enzyme glycerol-3-phosphate acyltransferase 4 in the TAG synthesis pathway is relocalized from the ER to the population of large, but not small, LDs [63], supporting the hypothesis that PC and TAG synthesis are coordinated with the LD growth. Although a similar mechanism has not been reported in yeast, many enzymes involved in TAG synthesis localize at least partially to LDs [34, 68].
In addition to the requirements of specific lipids and enzymes, structural proteins might also be involved in LD biogenesis. Although yeast genome-wide genetic screens did not identify a single gene essential for LD formation [12, 69, 70], studies did uncover Sei1/Fld1 and Ldb16, which are molecules targeted specifically to the ER/LD contact sites [14, 69]. Sei1/Fld1 is an ortholog of the human lipodystrophy protein seipin, also known as Berardinelli–Seip congenital lipodystrophy type 2 [71]. Seipin homologs have been identified widely in eukaryotes and their deficiencies result in an irregular LD morphology. In yeast cells lacking Sei1/Fld1, LDs become either small clusters tangled with the ER or supersized [12, 69], suggesting an assembly defect. The mutant cells accumulate supersized LDs under inositol starvation, a condition that suppresses phospholipid synthesis [48], and accumulate small clustered LDs when exogenous inositol is supplied [13], indicating that the phospholipid metabolism is altered in the absence of seipin. Whether Sei1/Fld1 provides a structural or lipid regulatory role for LD assembly is currently under debate; however, the protein is assembled into a higher-ordered complex with Ldb16 at the ER/LD contact sites [14, 72]. Additionally, an Ldb16 deficiency also results in aberrant LDs in yeast. Topological studies have indicated that the large domain of seipin, which houses most lipodystrophy mutations, is exposed to the luminal side of the ER, but its function is still unclear [71]. In an inducible LD formation system, an Sei1/Fld1 deficiency and an Sei1/Fld1 mutant lacking an N-terminal peptide delay LD formation with a concomitant accumulation of neutral lipids in the ER [73], suggesting a role for Sei1/Fld1 in LD biogenesis. Whether Sei1/Fld1 controls the partitioning of enzymes and/or lipids during LD biogenesis requires further investigation. However, the localization of acyltransferase Dga1 to the LD surface is not affected by the lack of Sei1/Fld1 [55]. Sei1/Fld1 and human seipin may also control enzyme activity levels. Recent findings in mammalian cells indicate that seipin physically interacts with 1-acylglycerol-3-phosphate O-acyltransferase 2 and lipin [74]. Thus, seipin may act as a docking site for multiple enzymes or even regulate enzyme activity during LD biogenesis. In another line of evidence, seipin has been shown to interact with sarcoplasmic/endoplasmic reticulum calcium ATPase, an ER-localized calcium pump, and thus is proposed to function in moving calcium from the cytosol to the ER [75]. It is currently unclear whether Sei1/Fld1 acts similarly in yeast.
The fat-inducing transcript (FIT) proteins, Fit1 and Fit2, may have roles in LD biogenesis [76, 77]. The overexpression of these proteins in mouse liver results in LD accumulation, whereas fewer LDs and TAGs are seen in Fit2-deficient cells. FIT proteins are transmembrane proteins residing in the ER and are thought to mediate the filling of LDs with TAGs, rather than regulating TAG synthesis. Yeast cells contain two FIT protein homologs, Scs3 and Yft2. Cells lacking Scs3 exhibit inositol auxotrophy, indicative of an altered phospholipid metabolism, a phenotype that can be complemented by expressing human Fit2. Scs3 and Yft2 have both shared and unique functions in lipid metabolism, vesicular trafficking, and membrane biogenesis [78]. However, it remains unclear whether the two molecules affect TAG metabolism or LD biogenesis in yeast.
Most cells, including yeast, maintain a discrete LD size for yet unknown reasons. Fusion is a common phenomenon for most organelles, but not for LDs. However, LD fusion can occur under specific conditions, such as the supersized LDs formed in cells lacking Sei1/Fld1, which may be a result of LD fusion [12]. The level of PC on the LD surface is important for LD maintenance, as LDs are prone to fuse in cells containing a knockdown of the PC biosynthesis enzyme CCT [79]. Consistently, compromised PC synthesis in yeast also resulted in supersized LDs [13]. The most well-known protein participating in LD growth is the fat-specific protein Fsp27 (also known as CIDEC), which is involved in adipocyte differentiation. The protein forms a stable pore between the LD/LD contact sites to facilitate the transfer of TAGs from the small to the large LDs [80], a process known as LD ripening. The LD protein perilipin 1 interacts with Fsp27 to promote large LD formation [81]. However, there is no perilipins or Fsp27 homologsin yeast.
Regression of LDs
TAGs and SEs are degraded by TAG lipases and SE hydrolases, respectively. Lipolysis, the process that mobilizes TAGs to generate energy and membrane lipids, is best studied in 3T3-L1 adipocytes. During the first step of adipocyte lipolysis, activated adipose triacylglyceride lipase (ATGL) on LDs hydrolyzes TAGs to generate DAGs and FAs, and several LD-localized proteins, including perilipins and CGI-58, modulate the ATGL activity [82–85]. Hormone-sensitive lipase, regulated by hormones and PKA phosphorylation, catalyzes the second step of lipolysis on LDs. Its products, monoacylglycerols (MAGs), undergo subsequent breakdown by the cytosolic MAG lipase to produce glycerol and FAs [86, 87]. Lipolysis occurs at the expense of LD volume, but the fates of the LD membrane monolayer and proteins have not been well studied.
In yeast, Yju3 is the MAG lipase ortholog, and its deletion results in marked MAGs accumulation [88]. Unlike the cytosolic MAG lipase in humans, Yju3 predominately associates with membranes and LDs. The LD-localized enzymes Tgl3, Tgl4, and Tgl5, containing a GXSXG motif at their active sites, are characterized as members of the conserved patatin domain-containing hydrolases [89–91]. Tgl3 and Tgl4, but not Tgl5, deletions lead to increases in LD size, thereby serving as major TAG lipases in yeast [89, 90]. Tgl3 activity is relatively unspecific and it hydrolyzes not only TAGs, but also DAGs. The carboxyl terminus of Tgl3 that faces the inside of the LDs is important for its stability [92]. Tgl4 has been characterized as the yeast ATGL [93]. It shows a preference for TAG species with C14 and C16 acyl chains and may also function as an SE hydrolase or phospholipase A2 [90]. In addition, the protein may have acyltransferase activity that catalyzes the acyl-CoA-dependent acylation of lyso-PA to PA [94]. Similarly, lyso-PA and lyso-PE acyltransferase activities have also been reported for Tgl3 and Tgl5. Cells deficient in Tgl5 accumulate TAGs with C26 acyl-chain, indicating that the function of this lipase is specific to the breakdown of very long-chain FAs [90]. A residual TAG lipase activity exists in cells lacking all three of these lipases when grown under oleic acid conditions, which have led to the discovery of Ayr1 as another TAG lipase in yeast [95]. Moreover, Ldh1 localizes to yeast LDs and also functions as a TAG lipase. In the absence of Ldh1, oleate-treated cells form giant LDs and accumulate neutral lipids and phospholipids [96, 97]. Overall, yeast cells have multiple TAG lipases that exhibit substrate specificity for different TAG species. These properties may lead to the production of various types of phospholipids with specific lengths or types of acyl chains and, thus, may be used by cells for different purposes. Deciphering the timing and the signaling pathways that activate these lipases should advance our knowledge of LD physiology.
SE hydrolysis in yeast involves three enzymes, Tgl1, Yeh1, and Yeh2, which are homologs of mammalian acid lipases with the conserved GXSXG motif at the active sites [98]. Yeh2 is a plasma membrane-localized enzyme, whereas Tgl1 and Yeh1 are localized to the LDs. The highest SE hydrolase activities were found in the plasma membrane and secretory vesicle fractions [49]. However, the bulk SE mobilization rate in the yeh2Δ strain is similar to that of the wild type; thus other enzymes likely have overlapping functions [99]. Mutant analysis results indicate that tgl1Δ causes a marked accumulation of SEs, while yeh1Δ and yeh2Δ cells show no effects. However, Yeh1 appears to be the most important SE hydrolase in hem1Δ cells that mimic anaerobic growth condition [100]. The three SE hydrolases exhibit preferences for certain SE substrates. Yeh1, similar to Yeh2, has a preference for zymosterol, lanosterol, and ergosterol esters [99, 101], while Tgl1 seems to prefer ergosterol and zymosterol esters [101]. Defects in the SE hydrolases, however, do not appear to affect the morphology or the number of cellular LDs.
How LD regression is regulated at the cellular level has just begun to be understood. Lipolysis can be activated upon growth resumption, as the LD size shrinks when the stationary phase yeast cells are switched back to fresh medium [29, 30]. The Dga1 targeted to LD surface synthesizes most of the TAGs during the stationary phase [23, 55]. Therefore, substrate DAGs for Dga1 must be supplied from the ER to the LDs during that stage. Conversely, DAGs derived from TAG lipolysis by lipases on LDs must be returned to the ER for membrane synthesis during growth resumption. A multi-spanning ER membrane protein, Ice2, is important for the efficient utilization of LDs during growth resumption [102]. Ice2 dispersed in the ER during the exponential phase accumulates into punctate structures adjacent to LDs during the stationary phase. During growth resumption, Ice2 quickly leaves the punctate structures and returns to the ER, preceding Dga1 relocalization. By binding to LDs through a cytosolic loop at the carboxyl terminus, Ice2 facilitates efficient DAG shuttling between the ER and LDs, thus promoting the phospholipid synthesis necessary for growth resumption. Additionally, the protein suppresses a futile cycle of TAG synthesis and degradation on the LDs by promoting Dga1 movement from the LDs to the ER [102].
Targeting of proteins to LDs
LDs can be purified by simple flotation methods, and the LD proteomes have been studied in various species. In higher eukaryotic cells, the perilipins belong to a family of conserved LD proteins [103]. However, no perilipin-like proteins exist in yeast. The LD proteins found in yeast proteomes are mostly enzymes involved in lipid metabolism [9, 46, 104–106] (Table 1). LDs contain the enzymes Ayr1, Slc1, Gpt2/Gat1, and Dga1, which are involved in the stepwise synthesis of glycerolipids. The SE hydrolases Yeh1 and Tgl1, the TAG lipases Tgl3, Tgl4, and Tgl5, and the MAG lipase Yju3, used for the breakdown of SE and TAG, are found in LDs. The FA-activation enzymes Faa1, Faa4, and Fat1, and several sterol biosynthesis enzymes, namely Erg1, Erg6, Erg7, and Erg27, are abundant in LD proteins. The presence of these enzymes in LDs suggests that LDs are the sites of action. However, LD might also serve as places for protein inactivation. For example, Erg1 displays dual localization pattern, while it is active only in the ER and not in the LDs [51].
Table 1.
LD-associated proteins in S. cerevisiae
| Gene | Systemic name | A | B | G | C | N | Localization (SGD) | Function (SGD) |
|---|---|---|---|---|---|---|---|---|
| ACH1 | YBL015W | V | C/M | Protein with CoA transferase activity | ||||
| ACS1 | YAL054C | V | C/M | Acetyl-coA synthetase | ||||
| ACT1 | YFL039C | V | Actin | Actin | ||||
| ADH1 | YOL086C | V | C/PM | Alcohol dehydrogenase | ||||
| ADH2 | YMR303C | V | C | Glucose-repressible alcohol dehydrogenase II | ||||
| ADY2 | YCR010C | V | M/PM | Acetate transporter | ||||
| ALD4 | YOR374W | V | M | Mitochondrial aldehyde dehydrogenase | ||||
| ALG9 | YNL219C | V | ER | Mannosyltransferase | ||||
| ANR2 | YKL047W | V | C/LP | Unknown | ||||
| ARF1 | YDL192W | V | G | ADP-ribosylation factor | ||||
| ATF1 | YOR377W | V | LP | Alcohol acetyltransferase | ||||
| ATP1 | YBL099W | V | M | Alpha subunit of the F1 sector of mitochondrial F1F0 ATP synthase | ||||
| ATP2 | YJR121W | V | V | M | Beta subunit of the F1 sector of mitochondrial F1F0 ATP synthase | |||
| ATP4 | YPL078C | V | M | Subunit b of the stator stalk of mitochondrial F1F0 ATP synthase | ||||
| ATP6 | Q0085 | V | M | Subunit a of the F0 sector of mitochondrial F1F0 ATP synthase | ||||
| ATP7 | YKL016C | V | M | Subunit d of the stator stalk of mitochondrial F1F0 ATP synthase | ||||
| AYR1 | YIL124W | V | V | V | V | V | ER/C/LP/M | Bifunctional triacylglycerol lipase and 1-acyl DHAP reductase |
| BAT1 | YHR208W | V | M | Mitochondrial branched-chain amino acid (BCAA) aminotransferase | ||||
| BMH2 | YDR099W | V | C/N/PM | 14-3-3 protein | ||||
| BSC2 | YDR275W | V | LP | Unknown | ||||
| CAB5 | YDR196C | V | ER/M/LP | Subunit of the CoA-synthesizing protein complex (CoA-SPC) | ||||
| CAT2 | YML042W | V | M/P | Carnitine acetyl-CoA transferase | ||||
| CIT1 | YNR001C | V | M | Citrate synthase | ||||
| COR1 | YBL045C | V | M | Core subunit of the ubiquinol cytochrome-c reductase complex | ||||
| COY1 | YKL179C | V | G | Golgi membrane protein with similarity to mammalian CASP | ||||
| CPR5/CYP5 | YDR304C | V | ER/C | Peptidyl-prolylcis-trans isomerase (cyclophilin) of the ER | ||||
| CRM1 | YGR218W | V | N | Major karyopherin | ||||
| CSR1/SFH2 | YLR380W | V | C/E/M/LP | Phosphatidylinositol transfer protein | ||||
| CST26/PSI1 | YBR042C | V | V | LP | Putative transferase involved in phospholipid biosynthesis | |||
| CTA1 | YDR256C | V | M/P | Catalase A | ||||
| CWH43 | YCR017C | V | PM | Putative sensor/transporter protein involved in cell wall biogenesis | ||||
| DFM1 | YDR411C | V | ER | ERAD | ||||
| DGA1 | YOR245C | V | V | V | ER/LP | Diacylglycerol acyltransferase | ||
| DLD1 | YDL174C | V | M | D-lactate dehydrogenase | ||||
| DPL1 | YDR294C | V | ER | Dihydrosphingosine phosphate lyase | ||||
| DPM1 | YPR183W | V | V | ER/M | Dolichol phosphate mannose (Dol-P-Man) synthase | |||
| ECM29 | YHL030W | V | C/N | Scaffold protein | ||||
| EFT1/2 | YOR133W/YDR385W | V | ribosome | Elongation factor 1/2 | ||||
| EHT1 | YBR177C | V | V | V | V | V | LP/M | Acyl-coenzymeA:ethanol O-acyltransferase |
| ENO2 | YHR174W | V | PM/M/V | Enolase II | ||||
| ENV9 | YOR246C | V | M/LP | Protein proposed to be involved in vacuolar functions | ||||
| ERG1 | YGR175C | V | V | V | V | V | ER/LP | Squalene epoxidase |
| ERG2 | YMR202W | V | ER | C-8 sterol isomerase | ||||
| ERG27 | YLR100W | V | V | V | V | ER/LP/M | 3-keto sterol reductase | |
| ERG6 | YML008C | V | V | V | V | V | ER/LP/M | Delta(24)-sterol C-methyltransferase |
| ERG7 | YHR072W | V | V | V | V | V | LP | Lanosterol synthase |
| ERV14 | YGL054C | V | ER/vesicles | COPII-coated vesicle protein | ||||
| FAA1 | YOR317W | V | V | V | V | ER/LP/M/PM | Long-chain fatty acyl-CoA synthetase | |
| FAA2 | YER015W | V | P/M | Medium-chain fatty acyl-CoA synthetase | ||||
| FAA3 | YIL009W | V | Unknown | Long-chain fatty acyl-CoA synthetase | ||||
| FAA4 | YMR246W | V | V | V | V | V | LP/C | Long-chain fatty acyl-CoA synthetase |
| FAS1 | YKL182W | V | C/LP/M | Beta subunit of fatty acid synthetase | ||||
| FAT1 | YBR041W | V | V | V | V | V | ER/LP/P | Very long-chain fatty acyl-CoA synthetase and fatty acid transporter |
| FBA1 | YKL060C | V | C/M | Fructose 1,6-bisphosphate aldolase | ||||
| FMP52 | YER004W | V | V | ER/M | Unknown | |||
| FOX2 | YKR009C | V | P | 3-hydroxyacyl-CoA dehydrogenase and enoyl-CoA hydratase | ||||
| GPI8 | YDR331W | V | ER | ER membrane glycoprotein subunit of the GPI transamidase complex | ||||
| GPT2/GAT1 | YKR067W | V | ER/LP/C | Glycerol-3-phosphate/dihydroxyacetone phosphate sn-1 acyltransferase | ||||
| GTT1 | YIR038C | V | V | V | ER/M/PM | ER-associated glutathione S-transferase | ||
| GUT2 | YIL155C | V | M | Mitochondrial glycerol-3-phosphate dehydrogenase | ||||
| GVP36 | YIL041W | V | V | G/C | BAR domain protein that localizes to early and late Golgi vesicles | |||
| HFD1 | YMR110C | V | V | V | LP/E/M | Hexadecenal dehydrogenase | ||
| HHF1/2 | YBR009C/YNL030W | V | N | Histone H4 | ||||
| HOM3 | YER052C | V | C | Aspartate kinase | ||||
| HSC82 | YMR186W | V | C/M/PM | Cytoplasmic chaperone of the Hsp90 family | ||||
| HSP104 | YLL026W | V | C/N | Disaggregase | ||||
| HSP12 | YFL014W | V | C/E/PM/N | Plasma membrane protein involved in maintaining membrane organization | ||||
| HSP60 | YLR259C | V | M | Tetradecameric mitochondrial chaperonin | ||||
| HTB1/2 | YDR224C/YBL002 W | V | N | Histone H2B | ||||
| HXT6/7 | YDR343C/YDR342C | V | PM/M | High-affinity glucose transporter | ||||
| IDP3 | YNL009W | V | C/P/M | Peroxisomal NADP-dependent isocitrate dehydrogenase | ||||
| ILV5 | YLR355C | V | M | Acetohydroxyacid reductoisomerase and mtDNA binding protein | ||||
| KAR2 | YJL034W | V | V | ER | ATPase involved in protein import into the ER | |||
| KES1/OSH4 | YPL145C | V | C/G | oxysterol binding protein | ||||
| KGD1 | YIL125W | V | M | Subunit of the mitochondrial alpha-ketoglutarate dehydrogenase complex | ||||
| LAP4 | YKL103C | V | C/V | Vacuolar aminopeptidase | ||||
| LDB16 | YCL005W | V | LP/M | Unknown | ||||
| LDH1 | YBR204C | V | V | V | LP | Serine hydrolase | ||
| LDS1* | YAL018C | LP | Protein involved in spore wall assembly | |||||
| LDS2* | YOL047C | C/LP | Protein involved in spore wall assembly | |||||
| LOA1/VPS66 | YPR139C | V | ER/LP/C | Lysophosphatidic acid acyltransferase | ||||
| LPL1 | YOR059C | V | V | V | V | LP | Phospholipase | |
| MIR1 | YJR077C | V | M | Mitochondrial phosphate carrier | ||||
| MSC1 | YML128C | V | V | M/ER/PM | Unknown | |||
| NCE2 | YPR149W | V | C/ER/M/PM | Unknown | ||||
| NDE1 | YMR145C | V | M | Mitochondrial external NADH dehydrogenase | ||||
| NDE2 | YDL085W | V | M | Mitochondrial external NADH dehydrogenase | ||||
| NDI1 | YML120C | V | M | NADH:ubiquinone oxidoreductase | ||||
| NTE1 | YML059C | V | ER | Serine esterase | ||||
| NUS1 | YDL193W | V | V | V | V | ER/LP | Putative prenyltransferase | |
| OM45 | YIL136W | V | M | Mitochondrial outer membrane protein | ||||
| OSW5 | YMR148W | V | Unknown | Unknown | ||||
| PCS60 | YBR222C | V | P/C | Oxalyl-CoA synthetase | ||||
| PDC1 | YLR044C | V | C/N | Major of three pyruvate decarboxylase isozymes | ||||
| PDI1 | YCL043C | V | V | ER | Protein disulfide isomerase | |||
| PDR16/SFH3 | YNL231C | V | V | V | LP/C/PM | Phosphatidylinositol transfer protein (PITP) | ||
| PET10 | YKR046C | V | V | V | V | V | LP | Unknown |
| PET9 | YBL030C | V | M | Major ADP/ATP carrier of the mitochondrial inner membrane | ||||
| PGC1 | YPL206C | V | V | LP/M | Phosphatidyl glycerol phospholipase C | |||
| PGK1 | YCR012W | V | M/C/PM | 3-phosphoglycerate kinase | ||||
| PHB1 | YGR132C | V | M/P | Subunit of the prohibitin complex | ||||
| PHO81 | YGR233C | V | N/C | Cyclin-dependent kinase (CDK) inhibitor | ||||
| PIL1 | YGR086C | V | C/M/PM | Eisosome core component | ||||
| PMA1 | YGL008C | V | PM/M | Plasma membrane P2-type H+-ATPase | ||||
| PMA2 | YPL036W | V | PM/M | Plasma membrane H+-ATPase | ||||
| PMT1 | YDL095W | V | V | ER | Protein O-mannosyltransferase | |||
| PMT2 | YAL023C | V | ER | Protein O-mannosyltransferase | ||||
| POR1 | YNL055C | V | V | M/C | Mitochondrial porin | |||
| POX1 | YGL205W | V | V | P | Fatty-acyl-coenzyme A oxidase | |||
| QCR2 | YPR191W | V | M | Subunit 7 of ubiquinol cytochrome-c reductase (Complex III) | ||||
| QCR7 | YDR529C | V | M | Subunit 7 of ubiquinol cytochrome-c reductase (complex III) | ||||
| RER2 | YBR002C | V | ER/LP | Cis-prenyltransferase involved in dolichol synthesis | ||||
| RHO1 | YPR165W | V | V | M/P/G/PM | GTP-binding protein of the rho subfamily of Ras-like proteins | |||
| RIB1 | YBL033C | V | C/N | GTP cyclohydrolase II | ||||
| RPL10 | YLR075W | V | Ribosome | Ribosomal 60S subunit protein L10 | ||||
| RPL4A | YBR031W | V | Ribosome | Ribosomal 60S subunit protein L4A | ||||
| RPL5 | YPL131W | V | V | Ribosome | Ribosomal 60S subunit protein L5 | |||
| RPL7A | YGL076C | V | Ribosome | Ribosomal 60S subunit protein L7A | ||||
| RPS19B | YNL302C | V | Ribosome | Protein component of the small (40S) ribosomal subunit | ||||
| RPS1B | YML063W | V | Ribosome | Ribosomal protein 10 (rp10) of the small (40S) subunit | ||||
| RPS3 | YNL178W | V | Ribosome | Protein component of the small (40S) ribosomal subunit | ||||
| RPS31 | YLR167W | V | Ribosome | Fusion protein cleaved to yield ribosomal protein S31 and ubiquitin | ||||
| RPS5 | YJR123W | V | Ribosome | Protein component of the small (40S) ribosomal subunit | ||||
| RPS7A | YOR096W | V | Ribosome | Protein component of the small (40S) ribosomal subunit | ||||
| RRT8 | YOL048C | V | PM/LP | Protein involved in spore wall assembly | ||||
| RTN1 | YDR233C | V | ER/G/M | Reticulon | ||||
| RTN2 | YDL204W | V | ER/C/N | Reticulon | ||||
| SAR1 | YPL218W | V | ER/vesicles | GTP-binding protein of the ARF family | ||||
| SAY1 | YGR263C | V | ER/LP | Sterol deacetylase | ||||
| SDH1 | YKL148C | V | M | Flavoprotein subunit of succinate dehydrogenase | ||||
| SEC21 | YNL287W | V | COPI vesicles | Gamma subunit of coatomer | ||||
| SEC61 | YLR378C | V | ER | Conserved ER protein translocation channel | ||||
| SEC63 | YOR254C | V | V | ER/M | Essential subunit of Sec63 complex | |||
| SHE10 | YGL228W | V | Unknown | Protein involved in outer spore wall assembly | ||||
| SLC1 | YDL052C | V | V | V | V | V | LP | 1-acyl-sn-glycerol-3-phosphate acyltransferase |
| SNA2 | YDR525W-A | V | C/V | Unknown | ||||
| SNX41 | YDR425W | V | E | Sorting nexin | ||||
| SPS19 | YNL202W | V | P | Peroxisomal 2,4-dienoyl-CoA reductase | ||||
| SRT1 | YMR101C | V | LP | Cis-prenyltransferase | ||||
| SSA1 | YAL005C | V | N/C/PM/V | ATPase involved in protein folding and NLS-directed nuclear transport | ||||
| SSA3 | YBL075C | V | C | ATPase involved in protein folding and the response to stress | ||||
| SSB1 | YDL229W | V | C/PM | Cytoplasmic ATPase that is a ribosome-associated molecular chaperone | ||||
| SSO1 | YPL232W | V | V/PM | Plasma membrane t-SNARE | ||||
| TAZ1 | YPR140W | V | M | Lyso-phosphatidylcholine acyltransferase | ||||
| TDH1 | YJL052W | V | V | C/LP/M/PM | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | |||
| TDH2 | YJR009C | V | V | C/LP/M/PM | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | |||
| TDH3 | YGR192C | V | V | C/LP/M/PM | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | |||
| TEF1/2 | YPR080W/YBR118W | V | V | C/M/ribosome | Translational elongation factor EF-1 alpha | |||
| TES1 | YJR019C | V | P/M | Peroxisomal acyl-CoA thioesterase | ||||
| TGL1 | YKL140W | V | V | V | V | V | LP | Steryl ester hydrolase |
| TGL3 | YMR313C | V | V | V | V | V | LP | Bifunctional triacylglycerol lipase and LPE acyltransferase |
| TGL4 | YKR089C | V | V | V | LP | Multifunctional lipase/hydrolase/phospholipase | ||
| TGL5 | YOR081C | V | V | V | LP | Bifunctional triacylglycerol lipase and LPA acyltransferase | ||
| TIF1/2 | YKR059W/YJL138C | V | C/ribosome | Translation initiation factor eIF4A | ||||
| TOM40 | YMR203W | V | M | Component of the TOM complex | ||||
| TSC10 | YBR265W | V | V | LP/C/ER/M | 3-ketosphinganine reductase | |||
| TUB2 | YFL037W | V | microtubules | Beta-tubulin | ||||
| UBX2 | YML013W | V | V | ER/M/LP | Bridging factor involved in ER-associated protein degradation | |||
| USE1 | YGL098W | V | ER | Essential SNARE protein localized to the ER | ||||
| WBP1 | YEL002C | V | ER | Beta subunit of the oligosaccharyltransferase glycoprotein complex | ||||
| YDR018C | YDR018C | V | N | Unknown | ||||
| YEH1 | YLL012W | V | LP | Steryl ester hydrolase | ||||
| YGR038C-B | YGR038C-B | V | N | Retrotransposon TYA Gag and TYB Pol genes | ||||
| YHB1 | YGR234W | V | C/M/N | Nitric oxide oxidoreductase | ||||
| YIM1 | YMR152W | V | V | V | V | LP/C/M | Unknown | |
| YJU3 | YKL094W | V | V | V | V | LP/M/C/PM | Monoglyceride lipase | |
| YNL134C | YNL134C | V | C/N | Unknown | ||||
| YNL208 W | YNL208W | V | M/ribosome | Unknown | ||||
| YPR127 W | YPR127W | V | C/N | Putative pyridoxine 4-dehydrogenase | ||||
| YPR147C | YPR147C | V | LP/C | Unknown | ||||
| YPT7 | YML001W | V | V/M | Rab family GTPase | ||||
| ZEO1 | YOL109W | V | PM/M | Peripheral membrane protein of the plasma membrane |
LD proteomes also revealed proteins without known enzymatic functions. Ubx2 sorts into LDs upon LD formation [60]. It is needed for LD maintenance, as removing or forcing an Ubx2 association with LDs results in aberrant LDs. Ubx2 has been postulated to modulate TAG synthesis by regulating Lro1. However, the protein has other functions, acting as the Cdc48 adaptor for ERAD and for the processing of the transcription factors Spt23 and Mga2 to control the expression of the Δ9-desaturase Ole1 [108–110]. Whether these pathways involve LDs awaits further investigation, but LD formation is dispensable for ERAD [111]. Ldb16, the molecule assembled together with Sei1/Fld1 at the ER/LD contact site, also appears in the LD proteome. LDs lacking Ldb16, like Sei1/Fld1, become irregular [14]. In addition to supersized LDs, both mutants form small LDs entangled with the ER, probably resulting from inefficient segregation during cytokinesis [112]. The LD morphologies in the mutants are hypersensitive to the phospholipid precursor supplements, such as inositol, suggesting that the ratio of phospholipids to neutral lipids is important for LD maintenance [13]. Ldb16 requires Sei1/Fld1 for stability; otherwise it is targeted for degradation by a branch of ERAD, termed ERAD-C [14]. Pdr16/Sft3 is a PI-binding protein that localizes to LDs. Cells lacking Pdr16/Sft3 are sensitive to azole-based antifungals, suggesting a role for the protein in sterol synthesis and/or mobilization [113]. Pdr16/Sfh3 associates with and modulates the mobilization of LDs during yeast sporulation [114]. Pet10 is found in all yeast LD proteomes. The protein interacts with many proteins in LDs, mitochondria, and peroxisomes [115], but no functional insight is currently available. Many other enzymes and proteins involved in diverse cellular activities in the cytosol, ER, Golgi, peroxisomes, endosomes, vacuoles, and plasma membrane have been reported in the yeast LD proteome (Table 1). Their association with and importance for LDs await future investigations. It should be noted that LDs often interact with other organelles, such that contamination from other organelles during LD purification needs to be carefully ruled out. In addition, protein expression and stability might be regulated differentially during various physiological stages. For example, Lds1 and Lds2 are induced and localized to LDs during yeast sporulation [116]. A list of LD proteins identified in yeast is summarized in Table 1.
LDs are distinct from other organelles in that their surface consists of a phospholipid monolayer. Therefore, it is conceivable that proteins stabilized on the LD surface require special determinants. This concept has attracted many studies focusing on protein targeting to LDs in various eukaryotic systems. These mechanisms may include (a) binding to intrinsic amphipathic helices within the protein, such as CCT [66] and the hepatitis C virus core protein [117]; (b) binding to internal protein domain containing a hairpin, such as the oleosins [118] and caveolins [119–121]; (c) binding to short hydrophobic stretches of the protein, such as in AAM-B [122] and perilipin 1 [123]; (d) lipid conjugation of the protein, such as for Rab18 [124]; and (e) binding through interactions with other LD proteins, such as hormone-sensitive lipase binding to LDs through perilipin 1 [125]. Given the expanding knowledge on LDs, other mechanisms are likely to emerge in the future.
Physiological roles of LDs
LDs protect cells against lipotoxicity
The major function of LDs is to prevent lipotoxicity by converting excess FAs into neutral lipids (TAGs and SEs). Cells lacking SEs increase the sensitivity to sterol synthesis inhibitors, such as terbinafine, and grow poorly after several generations [25]. TAG formation is particularly important in buffering the excess FAs in cells. Cells lacking TAGs have reduced LD numbers and show a prolonged lag phase [18, 23]. In addition, genes involved in phospholipid metabolism are repressed in the mutants, which indicates that FA homeostasis is crucial for the control of membrane lipid synthesis and proliferation [126]. When the PA phosphatase PAH1 is deleted, cells accumulate elevated levels of PA and FAs and reduced levels of DAG and TAG [127]. The mutants are sensitive to exogenous supplies of unsaturated FAs [45]. Similarly, are1Δ are2Δ dga1Δ lro1Δ LD-deficient cells accumulate a 2.5-fold higher amount of FAs than normal [17]. The quadruple mutant is sick, especially under nutrient-limited or stress conditions, while the cell loses viability under nitrogen starvation conditions [17]. An exogenous supply of oleic acids to the quadruple mutant causes a rapid block of the secretory pathway, an up-regulated unfolded protein response, and ultimately necrotic cell death with elevated levels of reactive oxygen species [128, 129]. The oleate-induced cell death of the quadruple mutant can be rescued by adaptation after the lag phase [16], and the suppression appears to involve mitochondrial DNA mutations [129]. As in yeast, TAG synthesis also plays a protective role against lipotoxicity in mammalian cells [128].
Regulations of LDs during the cell cycle
Both TAG synthesis and lipolysis are subjected to cell cycle control. Pah1 and Tgl4 are lipid enzymes known to be substrates for Cdc28, the main cyclin-dependent kinase governing the cell cycle in yeast. Pah1 is the key enzyme in TAG and phospholipid synthesis. The phosphorylation of Pah1 by Cdc28 occurs at the G2/M phase to inactivate Pah1 activity [130]. The phosphorylated form of Pah1 in the cytosol can be recruited to the ER membrane upon dephosphorylation by the ER-localized phosphatase complex Spo7–Nem1 [131, 132]. In addition to Cdc28, the cyclin-dependent kinase Pho85 also phosphorylates Pah1 to inhibit Pah1 phosphatase activity [133]. On the other hand, Cdc28 phosphorylates the major TAG lipase Tgl4 at the G1/S phase [93]. Cells lacking TAG lipolysis delay cell cycle progression at the stage of bud emergence. It remains unclear how the temporal control of lipolysis is monitored and how the lipolysis products actually contribute to the cell cycle. Overall, the catabolism of the LDs is likely coupled to the FAs and/or phospholipids supply for cell growth and division.
Delivery of LDs to the vacuoles by autophagy
Autophagy is a recycling pathway known to target damaged or excess organelles for breakdown in the lytic compartments, yeast vacuoles/mammalian lysosomes. Lysosomal targeting of LDs by autophagy was first discovered in hepatocytes [134], providing an alternative route for the mobilization of the LDs’ contents. In yeast, two microautophagy pathways that involve the direct vacuolar engulfment of LDs have been reported [15, 135]. Yeast cells incubated with oleate [135], a condition that increases TAG synthesis, triggers LD uptake by the vacuole [16]. This LD autophagy pathway uses a unique subset of the autophagy machinery, including Atg15, the vacuolar lipase needed for TAG breakdown inside the vacuole [135]. A distinct type of LD autophagy pathway, termed stationary phase lipophagy, exists in the quiescent yeast cells [15]. When glucose becomes limited in the culture medium, yeast LDs gradually move from the ER toward the vacuole. LDs in the quiescent cells are engulfed by the vacuole and accumulated inside the vacuole lumen, a process that requires core autophagy components and a subset of selective autophagy proteins. This pathway is connected to a lipid phase partitioning event that segregates vacuolar proteins to either one of the forming vacuolar micro domains during the stationary phase [136]. LDs are specifically taken up by the sterol-enriched, termed liquid-ordered, vacuolar microdomain, which in turn provides the vacuolar membranes with sterols to sustain the lipid phase partitioning [15]. Thus, yeast cells may use the feedforward loop to recycle sterols and LDs by autophagy during quiescence.
Dynamics of LDs during yeast sporulation
During sporulation, de novo biogenesis of the prospore membranes defines a robust program in which a flux of FAs and membrane lipids are remodeled toward the ordered sequestration of the haploid genome produced by meiosis [137]. However, little is known about the molecular mechanisms that govern the prospore membrane nucleation, elongation, and completion. Several lines of evidence have implied that LDs may be crucial for sporulation. The deletion of Are2 alone, or in combination with Are1, reduces the sporulation efficiency [21]. The diploid yeast strains lacking the TAG lipase Tgl3 or Tgl4 and Tgl5 also bear sporulation defects [90]. However, the enzymatic function of Tgl3 needed for efficient sporulation is its acyltransferase, rather than its lipase activity [138]. It is not clear which steps in sporulation require these enzymes. Intriguingly, at the cellular level, a discrete population of LDs is found to associate with the ascal side (outer, in contrast to the inner, which engulfs the nuclei) of the prospore membrane, and these LDs recruit a unique set of protein components [139, 140]. The deletion of these LD proteins results in spore wall assembly defect by an unknown mechanism [140]. Tgl3 and the PI-binding protein Pdr16/Sfh3 selectively localize to the pool of LDs associated with the prospore membrane [114]. Pdr16/Sfh3 is thought to reduce the rate of LD lipid mobilization and thus tune the formation of the prospore membrane. However, the functional connection between the prospore membrane and the pool of prospore membrane-associated LDs remains to be determined. It is important to decipher whether LDs provide fuel, are directly involved in prospore membrane morphogenesis, or even play an active role during spore wall assembly.
Interplay between LDs and other organelles
LDs are often found in close proximity with other organelles. In differentiating 3T3-L1 cells, ER, LDs, mitochondria, and peroxisomes form constellations, suggesting a coupling of these organelles for efficient lipid metabolism [141]. In yeast, physical associations between LDs and peroxisomes have been observed [104]. Peroxisomes can form stable structures termed “pexopodia” that penetrate LDs in oleate-cultured cells, a phenomenon that likely also occurs transiently under normal yeast growth conditions. The mechanisms and importance of these interactions are currently unknown, but probably involve lipid exchange. Presumably, LDs can transfer lipids into peroxisomes and mitochondria for oxidation, and the establishment of organelle contact sites may be dependent on specific protein–protein and/or protein–lipid interactions to mediate the binding. These notions await further supporting molecular evidence.
Potential contribution of LD studies in yeast to human metabolic diseases
LDs store fats. Excesses or deficiencies in fat storage lead to many different types of human diseases (reviewed in [142]). Pathological evidence has shown that patients suffering from obesity and other metabolic syndromes, such as hepatic steatosis and cardiovascular diseases, accumulate excess TAGs and LDs. A rare autosomal disorder, neutral lipid storage disease, is characterized by the accumulation of excess neutral lipids in multiple tissues and is associated with mutations in ATGL and CGI-58 genes that are involved in lipolysis [143]. In addition, several forms of human lipodystrophy, characterized by a loss of body fat storage, have been connected to mutations in the genes of 1-acylglycerol-3-phosphate O-acyltransferase 2 for TAG synthesis and Berardinelli–Seip congenital lipodystrophy type 2 for LD maintenance (reviewed in [144]). While it remains unclear how mutations in these genes with roles in LD formation and regulation cause lipodystrophy, it is possible that lipid perturbation inhibits adipocyte differentiation by affecting peroxisome proliferator-activated receptor γ signaling (reviewed in [145]).
Lipid-related metabolic disorders in humans involve various types of differentiated cells and organs. Nevertheless, the fundamental processes of TAG metabolism and LD formation and maintenance are evolutionarily conserved, and studies in yeast can further improve our knowledge, which can be applied to human diseases. Moreover, most metabolic syndromes in humans are caused by complex genetic and environmental factors that are difficult to resolve. Parallel studies of LD biology in yeast will contribute to the better understanding of the major regulatory pathways that govern feedback controls and signaling in TAG metabolism and lipid homeostasis, thus advancing our knowledge of the disease etiology in humans.
Conclusion
Tremendous progress has been made in the past several years in establishing the roles of LDs as versatile organelles rather than inert fat storage depots. However, many fundamental questions regarding the nature of LDs remain unanswered. In fact, we have learned that the biogenesis and regression of LDs are driven by complex mechanisms that are coupled to lipid homeostasis. Meanwhile, more knowledge is required to understand the mechanism behind lipid packaging into nascent LDs, the organization and the gating mechanism at the ER/LD contact sites, the actual parameters of LD size and number necessary for maintenance, and even the spatial and temporal cues that execute LD dynamics. From the physiological perspective, we have learned that LDs are metabolically active and can exchange lipids with other organelles to facilitate timely and efficient lipid metabolism. However, most of these interactions at the molecular levels and their significance have not yet been determined. FAs, phospholipids, and many other lipid metabolites are found to play diverse roles in various cellular activities, and it is conceivable that some might be coupled to the catabolism of storage lipids within LDs. Overall, it is evident that the field of LD biology will be driven by insights derived from fundamental LD research to increase our knowledge of disease pathology and to benefit growing bioindustries.
Acknowledgments
I thank Dr. Rey-Huei Chen at the Institute of Molecular Biology, Academia Sinica, for reading the manuscript and her helpful commentary and Miss Yu-Chun Weng at the Institute of Plant and Microbial Biology, Academia Sinica, for help with art and design work. This work was supported by an intramural fund from Academia Sinica and the grants NSC 101-2311-B-001-028-MY3 and 103-2633-B-001-003 from the Ministry of Science and Technology, Taiwan.
Abbreviations
- ACAT
Acyl-CoA:cholesterol acyltransferase
- ATGL
Adipose triacylglyceride lipase
- CCT
CTP:phosphocholine cytidylyltransferase
- CoA
Coenzyme A
- CDP-DAG
Cytidine diphosphate-diacylglycerol
- CTP
Cytidine triphosphate
- DAG
Diacylglycerol
- DGAT
Acyl-CoA:diacylglycerol acyltransferase
- ER
Endoplasmic reticulum
- ERAD
ER-associated protein degradation
- FA
Fatty acid
- FIT
Fat-inducing transcript
- GPAT
Glycerol-3-phosphate acyltransferase
- LD
Lipid droplet
- lyso-PA
1-acylglycerol-3-phosphate
- MAG
Monoacylglycerol
- PA
Phosphatidic acid
- PAP
Phosphatidate phosphatase
- PC
Phosphatidylcholine
- PE
Phosphatidylethanolamine
- PI
Phosphatidylinositol
- PS
Phosphatidylserine
- SE
Sterol ester
- TAG
Triacylglycerol
References
- 1.Pol A, Gross SP, Parton RG. Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J Cell Biol. 2014;204(5):635–646. doi: 10.1083/jcb.201311051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walther TC, Farese RV., Jr Lipid droplets and cellular lipid metabolism. Annu Rev Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohlwein SD, Veenhuis M, van der Klei IJ. Lipid droplets and peroxisomes: key players in cellular lipid homeostasis or a matter of fat–store ‘em up or burn ‘em down. Genetics. 2013;193(1):1–50. doi: 10.1534/genetics.112.143362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaner WS, O’Byrne SM, Wongsiriroj N, Kluwe J, D’Ambrosio DM, Jiang H, Schwabe RF, Hillman EM, Piantedosi R, Libien J. Hepatic stellate cell lipid droplets: a specialized lipid droplet for retinoid storage. Biochim Biophys Acta. 2009;1791(6):467–473. doi: 10.1016/j.bbalip.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartz R, Li WH, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RG, Liu P, Chapman KD. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res. 2007;48(4):837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- 6.Spanova M, Zweytick D, Lohner K, Klug L, Leitner E, Hermetter A. Daum G (2012) Influence of squalene on lipid particle/droplet and membrane organization in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 1821;4:647–653. doi: 10.1016/j.bbalip.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czabany T, Wagner A, Zweytick D, Lohner K, Leitner E, Ingolic E, Daum G. Structural and biochemical properties of lipid particles from the yeast Saccharomyces cerevisiae. J Biol Chem. 2008;283(25):17065–17074. doi: 10.1074/jbc.M800401200. [DOI] [PubMed] [Google Scholar]
- 8.Schneiter R, Brugger B, Sandhoff R, Zellnig G, Leber A, Lampl M, Athenstaedt K, Hrastnik C, Eder S, Daum G, Paltauf F, Wieland FT, Kohlwein SD. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol. 1999;146(4):741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grillitsch K, Connerth M, Kofeler H, Arrey TN, Rietschel B, Wagner B, Karas M, Daum G. Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: lipidome meets proteome. Biochim Biophys Acta. 2011;1811(12):1165–1176. doi: 10.1016/j.bbalip.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radulovic M, Knittelfelder O, Cristobal-Sarramian A, Kolb D, Wolinski H, Kohlwein SD. The emergence of lipid droplets in yeast: current status and experimental approaches. Curr Genet. 2013;59(4):231–242. doi: 10.1007/s00294-013-0407-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thiam AR, Farese RV, Jr, Walther TC. The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol. 2013;14(12):775–786. doi: 10.1038/nrm3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fei W, Shui G, Gaeta B, Du X, Kuerschner L, Li P, Brown AJ, Wenk MR, Parton RG, Yang H. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J Cell Biol. 2008;180(3):473–482. doi: 10.1083/jcb.200711136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fei W, Shui G, Zhang Y, Krahmer N, Ferguson C, Kapterian TS, Lin RC, Dawes IW, Brown AJ, Li P, Huang X, Parton RG, Wenk MR, Walther TC, Yang H. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 2011;7(7):e1002201. doi: 10.1371/journal.pgen.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang CW, Miao YH, Chang YS. Control of lipid droplet size in budding yeast requires the collaboration between Fld1 and Ldb16. J Cell Sci. 2014;127(Pt 6):1214–1228. doi: 10.1242/jcs.137737. [DOI] [PubMed] [Google Scholar]
- 15.Wang CW, Miao YH, Chang YS. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J Cell Biol. 2014;206(3):357–366. doi: 10.1083/jcb.201404115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connerth M, Czabany T, Wagner A, Zellnig G, Leitner E, Steyrer E, Daum G. Oleate inhibits steryl ester synthesis and causes liposensitivity in yeast. J Biol Chem. 2010;285(35):26832–26841. doi: 10.1074/jbc.M110.122085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandager L, Gustavsson MH, Stahl U, Dahlqvist A, Wiberg E, Banas A, Lenman M, Ronne H, Stymne S. Storage lipid synthesis is non-essential in yeast. J Biol Chem. 2002;277(8):6478–6482. doi: 10.1074/jbc.M109109200. [DOI] [PubMed] [Google Scholar]
- 18.Sorger D, Daum G. Synthesis of triacylglycerols by the acyl-coenzyme A:diacyl-glycerol acyltransferase Dga1p in lipid particles of the yeast Saccharomyces cerevisiae. J Bacteriol. 2002;184(2):519–524. doi: 10.1128/JB.184.2.519-524.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S. Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci USA. 2000;97(12):6487–6492. doi: 10.1073/pnas.120067297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oelkers P, Tinkelenberg A, Erdeniz N, Cromley D, Billheimer JT, Sturley SL. A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J Biol Chem. 2000;275(21):15609–15612. doi: 10.1074/jbc.C000144200. [DOI] [PubMed] [Google Scholar]
- 21.Yu C, Kennedy NJ, Chang CC, Rothblatt JA. Molecular cloning and characterization of two isoforms of Saccharomyces cerevisiae acyl-CoA:sterol acyltransferase. J Biol Chem. 1996;271(39):24157–24163. doi: 10.1074/jbc.271.39.24157. [DOI] [PubMed] [Google Scholar]
- 22.Kohlwein SD. Triacylglycerol homeostasis: insights from yeast. J Biol Chem. 2010;285(21):15663–15667. doi: 10.1074/jbc.R110.118356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oelkers P, Cromley D, Padamsee M, Billheimer JT, Sturley SL. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J Biol Chem. 2002;277(11):8877–8881. doi: 10.1074/jbc.M111646200. [DOI] [PubMed] [Google Scholar]
- 24.Yang H, Bard M, Bruner DA, Gleeson A, Deckelbaum RJ, Aljinovic G, Pohl TM, Rothstein R, Sturley SL. Sterol esterification in yeast: a two-gene process. Science. 1996;272(5266):1353–1356. doi: 10.1126/science.272.5266.1353. [DOI] [PubMed] [Google Scholar]
- 25.Zweytick D, Leitner E, Kohlwein SD, Yu C, Rothblatt J, Daum G. Contribution of Are1p and Are2p to steryl ester synthesis in the yeast Saccharomyces cerevisiae. Eur J Biochem FEBS. 2000;267(4):1075–1082. doi: 10.1046/j.1432-1327.2000.01103.x. [DOI] [PubMed] [Google Scholar]
- 26.Rajakumari S, Grillitsch K, Daum G. Synthesis and turnover of non-polar lipids in yeast. Prog Lipid Res. 2008;47(3):157–171. doi: 10.1016/j.plipres.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Hasslacher M, Ivessa AS, Paltauf F, Kohlwein SD. Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J Biol Chem. 1993;268(15):10946–10952. [PubMed] [Google Scholar]
- 28.Shirra MK, Patton-Vogt J, Ulrich A, Liuta-Tehlivets O, Kohlwein SD, Henry SA, Arndt KM. Inhibition of acetyl coenzyme A carboxylase activity restores expression of the INO1 gene in a snf1 mutant strain of Saccharomyces cerevisiae. Mol Cell Biol. 2001;21(17):5710–5722. doi: 10.1128/MCB.21.17.5710-5722.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurat CF, Natter K, Petschnigg J, Wolinski H, Scheuringer K, Scholz H, Zimmermann R, Leber R, Zechner R, Kohlwein SD. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J Biol Chem. 2006;281(1):491–500. doi: 10.1074/jbc.M508414200. [DOI] [PubMed] [Google Scholar]
- 30.Zanghellini J, Natter K, Jungreuthmayer C, Thalhammer A, Kurat CF, Gogg-Fassolter G, Kohlwein SD, von Grunberg HH. Quantitative modeling of triacylglycerol homeostasis in yeast–metabolic requirement for lipolysis to promote membrane lipid synthesis and cellular growth. The FEBS journal. 2008;275(22):5552–5563. doi: 10.1111/j.1742-4658.2008.06681.x. [DOI] [PubMed] [Google Scholar]
- 31.Athenstaedt K, Daum G. Phosphatidic acid, a key intermediate in lipid metabolism. Eur J Biochem FEBS. 1999;266(1):1–16. doi: 10.1046/j.1432-1327.1999.00822.x. [DOI] [PubMed] [Google Scholar]
- 32.Zheng Z, Zou J. The initial step of the glycerolipid pathway: identification of glycerol 3-phosphate/dihydroxyacetone phosphate dual substrate acyltransferases in Saccharomyces cerevisiae. J Biol Chem. 2001;276(45):41710–41716. doi: 10.1074/jbc.M104749200. [DOI] [PubMed] [Google Scholar]
- 33.Zaremberg V, McMaster CR. Differential partitioning of lipids metabolized by separate yeast glycerol-3-phosphate acyltransferases reveals that phospholipase D generation of phosphatidic acid mediates sensitivity to choline-containing lysolipids and drugs. J Biol Chem. 2002;277(41):39035–39044. doi: 10.1074/jbc.M207753200. [DOI] [PubMed] [Google Scholar]
- 34.Marr N, Foglia J, Terebiznik M, Athenstaedt K, Zaremberg V. Controlling lipid fluxes at glycerol-3-phosphate acyltransferase step in yeast: unique contribution of Gat1p to oleic acid-induced lipid particle formation. J Biol Chem. 2012;287(13):10251–10264. doi: 10.1074/jbc.M111.314112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benghezal M, Roubaty C, Veepuri V, Knudsen J, Conzelmann A. SLC1 and SLC4 encode partially redundant acyl-coenzyme A 1-acylglycerol-3-phosphate O-acyltransferases of budding yeast. J Biol Chem. 2007;282(42):30845–30855. doi: 10.1074/jbc.M702719200. [DOI] [PubMed] [Google Scholar]
- 36.Ayciriex S, Le Guedard M, Camougrand N, Velours G, Schoene M, Leone S, Wattelet-Boyer V, Dupuy JW, Shevchenko A, Schmitter JM, Lessire R, Bessoule JJ, Testet E. YPR139c/LOA1 encodes a novel lysophosphatidic acid acyltransferase associated with lipid droplets and involved in TAG homeostasis. Mol Biol Cell. 2012;23(2):233–246. doi: 10.1091/mbc.E11-07-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han GS, Wu WI, Carman GM. The Saccharomyces cerevisiae Lipin homolog is a Mg2+ -dependent phosphatidate phosphatase enzyme. J Biol Chem. 2006;281(14):9210–9218. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Hara L, Han GS, Peak-Chew S, Grimsey N, Carman GM, Siniossoglou S. Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+ -dependent phosphatidate phosphatase. J Biol Chem. 2006;281(45):34537–34548. doi: 10.1074/jbc.M606654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carman GM, Han GS. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem Sci. 2006;31(12):694–699. doi: 10.1016/j.tibs.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carman GM, Zeimetz GM. Regulation of phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. J Biol Chem. 1996;271(23):13293–13296. doi: 10.1074/jbc.271.23.13293. [DOI] [PubMed] [Google Scholar]
- 41.Carman GM, Henry SA. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog Lipid Res. 1999;38(5–6):361–399. doi: 10.1016/s0163-7827(99)00010-7. [DOI] [PubMed] [Google Scholar]
- 42.Hirsch JP, Henry SA. Expression of the Saccharomyces cerevisiae inositol-1-phosphate synthase (INO1) gene is regulated by factors that affect phospholipid synthesis. Mol Cell Biol. 1986;6(10):3320–3328. doi: 10.1128/mcb.6.10.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loewen CJ, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 2004;304(5677):1644–1647. doi: 10.1126/science.1096083. [DOI] [PubMed] [Google Scholar]
- 44.Han GS, O’Hara L, Siniossoglou S, Carman GM. Characterization of the yeast DGK1-encoded CTP-dependent diacylglycerol kinase. J Biol Chem. 2008;283(29):20443–20453. doi: 10.1074/jbc.M802866200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fakas S, Qiu Y, Dixon JL, Han GS, Ruggles KV, Garbarino J, Sturley SL, Carman GM. Phosphatidate phosphatase activity plays key role in protection against fatty acid-induced toxicity in yeast. J Biol Chem. 2011;286(33):29074–29085. doi: 10.1074/jbc.M111.258798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Natter K, Leitner P, Faschinger A, Wolinski H, McCraith S, Fields S, Kohlwein SD. The spatial organization of lipid synthesis in the yeast Saccharomyces cerevisiae derived from large scale green fluorescent protein tagging and high resolution microscopy. Mol Cell Proteomics MCP. 2005;4(5):662–672. doi: 10.1074/mcp.M400123-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Gibellini F, Smith TK. The Kennedy pathway–De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life. 2010;62(6):414–428. doi: 10.1002/iub.337. [DOI] [PubMed] [Google Scholar]
- 48.Henry SA, Kohlwein SD, Carman GM. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012;190(2):317–349. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zinser E, Paltauf F, Daum G. Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J Bacteriol. 1993;175(10):2853–2858. doi: 10.1128/jb.175.10.2853-2858.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jandrositz A, Turnowsky F, Hogenauer G. The gene encoding squalene epoxidase from Saccharomyces cerevisiae: cloning and characterization. Gene. 1991;107(1):155–160. doi: 10.1016/0378-1119(91)90310-8. [DOI] [PubMed] [Google Scholar]
- 51.Leber R, Landl K, Zinser E, Ahorn H, Spok A, Kohlwein SD, Turnowsky F, Daum G. Dual localization of squalene epoxidase, Erg1p, in yeast reflects a relationship between the endoplasmic reticulum and lipid particles. Mol Biol Cell. 1998;9(2):375–386. doi: 10.1091/mbc.9.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi Z, Buntel CJ, Griffin JH. Isolation and characterization of the gene encoding 2,3-oxidosqualene-lanosterol cyclase from Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1994;91(15):7370–7374. doi: 10.1073/pnas.91.15.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Milla P, Athenstaedt K, Viola F, Oliaro-Bosso S, Kohlwein SD, Daum G, Balliano G. Yeast oxidosqualene cyclase (Erg7p) is a major component of lipid particles. J Biol Chem. 2002;277(4):2406–2412. doi: 10.1074/jbc.M104195200. [DOI] [PubMed] [Google Scholar]
- 54.Foresti O, Ruggiano A, Hannibal-Bach HK, Ejsing CS, Carvalho P. Sterol homeostasis requires regulated degradation of squalene monooxygenase by the ubiquitin ligase Doa10/Teb4. eLife. 2013;2:e00953. doi: 10.7554/eLife.00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Sci. 2011;124(Pt 14):2424–2437. doi: 10.1242/jcs.076836. [DOI] [PubMed] [Google Scholar]
- 56.Guo Y, Cordes KR, Farese RV, Jr, Walther TC. Lipid droplets at a glance. J Cell Sci. 2009;122(Pt 6):749–752. doi: 10.1242/jcs.037630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zanghellini J, Wodlei F, von Grunberg HH. Phospholipid demixing and the birth of a lipid droplet. J Theor Biol. 2010;264(3):952–961. doi: 10.1016/j.jtbi.2010.02.025. [DOI] [PubMed] [Google Scholar]
- 58.Hussain MM. A proposed model for the assembly of chylomicrons. Atherosclerosis. 2000;148(1):1–15. doi: 10.1016/s0021-9150(99)00397-4. [DOI] [PubMed] [Google Scholar]
- 59.Sorger D, Athenstaedt K, Hrastnik C, Daum G. A yeast strain lacking lipid particles bears a defect in ergosterol formation. J Biol Chem. 2004;279(30):31190–31196. doi: 10.1074/jbc.M403251200. [DOI] [PubMed] [Google Scholar]
- 60.Wang CW, Lee SC. The ubiquitin-like (UBX)-domain-containing protein Ubx2/Ubxd8 regulates lipid droplet homeostasis. J Cell Sci. 2012;125(Pt 12):2930–2939. doi: 10.1242/jcs.100230. [DOI] [PubMed] [Google Scholar]
- 61.Choudhary V, Jacquier N, Schneiter R. The topology of the triacylglycerol synthesizing enzyme Lro1 indicates that neutral lipids can be produced within the luminal compartment of the endoplasmatic reticulum: implications for the biogenesis of lipid droplets. Commun Integr Biol. 2011;4(6):781–784. doi: 10.4161/cib.17830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pagac M, de la Mora HV, Duperrex C, Roubaty C, Vionnet C, Conzelmann A. Topology of 1-acyl-sn-glycerol-3-phosphate acyltransferases SLC1 and ALE1 and related membrane-bound O-acyltransferases (MBOATs) of Saccharomyces cerevisiae. J Biol Chem. 2011;286(42):36438–36447. doi: 10.1074/jbc.M111.256511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Frohlich F, Liu X, Buhman KK, Coleman RA, Bewersdorf J, Farese RV, Jr, Walther TC. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell. 2013;24(4):384–399. doi: 10.1016/j.devcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adeyo O, Horn PJ, Lee S, Binns DD, Chandrahas A, Chapman KD, Goodman JM. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J Cell Biol. 2011;192(6):1043–1055. doi: 10.1083/jcb.201010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skinner JR, Shew TM, Schwartz DM, Tzekov A, Lepus CM, Abumrad NA, Wolins NE. Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J Biol Chem. 2009;284(45):30941–30948. doi: 10.1074/jbc.M109.013995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krahmer N, Guo Y, Wilfling F, Hilger M, Lingrell S, Heger K, Newman HW, Schmidt-Supprian M, Vance DE, Mann M, Farese RV, Jr, Walther TC. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 2011;14(4):504–515. doi: 10.1016/j.cmet.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moessinger C, Kuerschner L, Spandl J, Shevchenko A, Thiele C. Human lysophosphatidylcholine acyltransferases 1 and 2 are located in lipid droplets where they catalyze the formation of phosphatidylcholine. J Biol Chem. 2011;286(24):21330–21339. doi: 10.1074/jbc.M110.202424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Athenstaedt K, Daum G. 1-Acyldihydroxyacetone-phosphate reductase (Ayr1p) of the yeast Saccharomyces cerevisiae encoded by the open reading frame YIL124w is a major component of lipid particles. J Biol Chem. 2000;275(1):235–240. doi: 10.1074/jbc.275.1.235. [DOI] [PubMed] [Google Scholar]
- 69.Szymanski KM, Binns D, Bartz R, Grishin NV, Li WP, Agarwal AK, Garg A, Anderson RG, Goodman JM. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci USA. 2007;104(52):20890–20895. doi: 10.1073/pnas.0704154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fei W, Alfaro G, Muthusamy BP, Klaassen Z, Graham TR, Yang H, Beh CT. Genome-wide analysis of sterol-lipid storage and trafficking in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7(2):401–414. doi: 10.1128/EC.00386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cartwright BR, Goodman JM. Seipin: from human disease to molecular mechanism. J Lipid Res. 2012;53(6):1042–1055. doi: 10.1194/jlr.R023754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Binns D, Lee S, Hilton CL, Jiang QX, Goodman JM. Seipin is a discrete homooligomer. Biochemistry. 2010;49(50):10747–10755. doi: 10.1021/bi1013003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cartwright BR, Binns DD, Hilton CL, Han S, Gao Q, Goodman JM. Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Mol Biol Cell. 2014 doi: 10.1091/mbc.E14-08-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Talukder MM, Sim MF, O’Rahilly S, Edwardson JM, Rochford JJ. Seipin oligomers can interact directly with AGPAT2 and lipin 1, physically scaffolding critical regulators of adipogenesis. Mol Metab. 2015;4(3):199–209. doi: 10.1016/j.molmet.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bi J, Wang W, Liu Z, Huang X, Jiang Q, Liu G, Wang Y, Huang X. Seipin promotes adipose tissue fat storage through the ER Ca(2)(+)-ATPase SERCA. Cell Metab. 2014;19(5):861–871. doi: 10.1016/j.cmet.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 76.Kadereit B, Kumar P, Wang WJ, Miranda D, Snapp EL, Severina N, Torregroza I, Evans T, Silver DL. Evolutionarily conserved gene family important for fat storage. Proc Natl Acad Sci USA. 2008;105(1):94–99. doi: 10.1073/pnas.0708579105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gross DA, Snapp EL, Silver DL. Structural insights into triglyceride storage mediated by fat storage-inducing transmembrane (FIT) protein 2. PLoS One. 2010;5(5):e10796. doi: 10.1371/journal.pone.0010796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moir RD, Gross DA, Silver DL, Willis IM. SCS3 and YFT2 link transcription of phospholipid biosynthetic genes to ER stress and the UPR. PLoS Genet. 2012;8(8):e1002890. doi: 10.1371/journal.pgen.1002890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453(7195):657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gong J, Sun Z, Wu L, Xu W, Schieber N, Xu D, Shui G, Yang H, Parton RG, Li P. Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites. J Cell Biol. 2011;195(6):953–963. doi: 10.1083/jcb.201104142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun Z, Gong J, Wu H, Xu W, Wu L, Xu D, Gao J, Wu JW, Yang H, Yang M, Li P. Perilipin1 promotes unilocular lipid droplet formation through the activation of Fsp27 in adipocytes. Nat Commun. 2013;4:1594. doi: 10.1038/ncomms2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, Lass A, Neuberger G, Eisenhaber F, Hermetter A, Zechner R. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306(5700):1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 83.Subramanian V, Rothenberg A, Gomez C, Cohen AW, Garcia A, Bhattacharyya S, Shapiro L, Dolios G, Wang R, Lisanti MP, Brasaemle DL. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J Biol Chem. 2004;279(40):42062–42071. doi: 10.1074/jbc.M407462200. [DOI] [PubMed] [Google Scholar]
- 84.Gruber A, Cornaciu I, Lass A, Schweiger M, Poeschl M, Eder C, Kumari M, Schoiswohl G, Wolinski H, Kohlwein SD, Zechner R, Zimmermann R, Oberer M. The N-terminal region of comparative gene identification-58 (CGI-58) is important for lipid droplet binding and activation of adipose triglyceride lipase. J Biol Chem. 2010;285(16):12289–12298. doi: 10.1074/jbc.M109.064469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang H, Bell M, Sreenivasan U, Hu H, Liu J, Dalen K, Londos C, Yamaguchi T, Rizzo MA, Coleman R, Gong D, Brasaemle D, Sztalryd C. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J Biol Chem. 2011;286(18):15707–15715. doi: 10.1074/jbc.M110.207779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schweiger M, Schreiber R, Haemmerle G, Lass A, Fledelius C, Jacobsen P, Tornqvist H, Zechner R, Zimmermann R. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J Biol Chem. 2006;281(52):40236–40241. doi: 10.1074/jbc.M608048200. [DOI] [PubMed] [Google Scholar]
- 87.Taschler U, Radner FP, Heier C, Schreiber R, Schweiger M, Schoiswohl G, Preiss-Landl K, Jaeger D, Reiter B, Koefeler HC, Wojciechowski J, Theussl C, Penninger JM, Lass A, Haemmerle G, Zechner R, Zimmermann R. Monoglyceride lipase deficiency in mice impairs lipolysis and attenuates diet-induced insulin resistance. J Biol Chem. 2011;286(20):17467–17477. doi: 10.1074/jbc.M110.215434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heier C, Taschler U, Rengachari S, Oberer M, Wolinski H, Natter K, Kohlwein SD, Leber R, Zimmermann R. Identification of Yju3p as functional orthologue of mammalian monoglyceride lipase in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2010;1801(9):1063–1071. doi: 10.1016/j.bbalip.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Athenstaedt K, Daum G. YMR313c/TGL3 encodes a novel triacylglycerol lipase located in lipid particles of Saccharomyces cerevisiae. J Biol Chem. 2003;278(26):23317–23323. doi: 10.1074/jbc.M302577200. [DOI] [PubMed] [Google Scholar]
- 90.Athenstaedt K, Daum G. Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles. J Biol Chem. 2005;280(45):37301–37309. doi: 10.1074/jbc.M507261200. [DOI] [PubMed] [Google Scholar]
- 91.Daum G, Wagner A, Czabany T, Athenstaedt K. Dynamics of neutral lipid storage and mobilization in yeast. Biochimie. 2007;89(2):243–248. doi: 10.1016/j.biochi.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 92.Koch B, Schmidt C, Ploier B, Daum G. Modifications of the C terminus affect functionality and stability of yeast triacylglycerol lipase Tgl3p. J Biol Chem. 2014;289(28):19306–19316. doi: 10.1074/jbc.M114.556944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kurat CF, Wolinski H, Petschnigg J, Kaluarachchi S, Andrews B, Natter K, Kohlwein SD. Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol Cell. 2009;33(1):53–63. doi: 10.1016/j.molcel.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 94.Rajakumari S, Rajasekharan R, Daum G. Triacylglycerol lipolysis is linked to sphingolipid and phospholipid metabolism of the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2010;1801(12):1314–1322. doi: 10.1016/j.bbalip.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 95.Ploier B, Scharwey M, Koch B, Schmidt C, Schatte J, Rechberger G, Kollroser M, Hermetter A, Daum G. Screening for hydrolytic enzymes reveals Ayr1p as a novel triacylglycerol lipase in Saccharomyces cerevisiae. J Biol Chem. 2013;288(50):36061–36072. doi: 10.1074/jbc.M113.509927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thoms S, Debelyy MO, Connerth M, Daum G, Erdmann R. The putative Saccharomyces cerevisiae hydrolase Ldh1p is localized to lipid droplets. Eukaryot Cell. 2011;10(6):770–775. doi: 10.1128/EC.05038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Debelyy MO, Thoms S, Connerth M, Daum G, Erdmann R. Involvement of the Saccharomyces cerevisiae hydrolase Ldh1p in lipid homeostasis. Eukaryot Cell. 2011;10(6):776–781. doi: 10.1128/EC.05040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koffel R, Tiwari R, Falquet L, Schneiter R. The Saccharomyces cerevisiae YLL012/YEH1, YLR020/YEH2, and TGL1 genes encode a novel family of membrane-anchored lipases that are required for steryl ester hydrolysis. Mol Cell Biol. 2005;25(5):1655–1668. doi: 10.1128/MCB.25.5.1655-1668.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mullner H, Deutsch G, Leitner E, Ingolic E, Daum G. YEH2/YLR020c encodes a novel steryl ester hydrolase of the yeast Saccharomyces cerevisiae. J Biol Chem. 2005;280(14):13321–13328. doi: 10.1074/jbc.M409914200. [DOI] [PubMed] [Google Scholar]
- 100.Koffel R, Schneiter R. Yeh1 constitutes the major steryl ester hydrolase under heme-deficient conditions in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5(7):1018–1025. doi: 10.1128/EC.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wagner A, Grillitsch K, Leitner E, Daum G. Mobilization of steryl esters from lipid particles of the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2009;1791(2):118–124. doi: 10.1016/j.bbalip.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 102.Markgraf DF, Klemm RW, Junker M, Hannibal-Bach HK, Ejsing CS, Rapoport TA. An ER protein functionally couples neutral lipid metabolism on lipid droplets to membrane lipid synthesis in the ER. Cell Reports. 2014;6(1):44–55. doi: 10.1016/j.celrep.2013.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J Lipid Res. 2010;51(3):468–471. doi: 10.1194/jlr.R000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Binns D, Januszewski T, Chen Y, Hill J, Markin VS, Zhao Y, Gilpin C, Chapman KD, Anderson RG, Goodman JM. An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol. 2006;173(5):719–731. doi: 10.1083/jcb.200511125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Currie E, Guo X, Christiano R, Chitraju C, Kory N, Harrison K, Haas J, Walther TC, Farese RV., Jr High confidence proteomic analysis of yeast LDs identifies additional droplet proteins and reveals connections to dolichol synthesis and sterol acetylation. J Lipid Res. 2014;55(7):1465–1477. doi: 10.1194/jlr.M050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Athenstaedt K, Zweytick D, Jandrositz A, Kohlwein SD, Daum G. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J Bacteriol. 1999;181(20):6441–6448. doi: 10.1128/jb.181.20.6441-6448.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Athenstaedt K, Zweytick D, Jandrositz A, Kohlwein SD, Daum G. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J Bacteriol. 1999;181(20):6441–6448. doi: 10.1128/jb.181.20.6441-6448.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat Cell Biol. 2005;7(10):993–998. doi: 10.1038/ncb1298. [DOI] [PubMed] [Google Scholar]
- 109.Surma MA, Klose C, Peng D, Shales M, Mrejen C, Stefanko A, Braberg H, Gordon DE, Vorkel D, Ejsing CS, Farese R, Jr, Simons K, Krogan NJ, Ernst R. A lipid E-MAP identifies Ubx2 as a critical regulator of lipid saturation and lipid bilayer stress. Mol Cell. 2013;51(4):519–530. doi: 10.1016/j.molcel.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kolawa N, Sweredoski MJ, Graham RL, Oania R, Hess S, Deshaies RJ. Perturbations to the ubiquitin conjugate proteome in yeast deltaubx mutants identify Ubx2 as a regulator of membrane lipid composition. Mol Cell Proteomics MCP. 2013;12(10):2791–2803. doi: 10.1074/mcp.M113.030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Olzmann JA, Kopito RR. Lipid droplet formation is dispensable for endoplasmic reticulum-associated degradation. J Biol Chem. 2011;286(32):27872–27874. doi: 10.1074/jbc.C111.266452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wolinski H, Kolb D, Hermann S, Koning RI, Kohlwein SD. A role for seipin in lipid droplet dynamics and inheritance in yeast. J Cell Sci. 2011;124(Pt 22):3894–3904. doi: 10.1242/jcs.091454. [DOI] [PubMed] [Google Scholar]
- 113.van den Hazel HB, Pichler H, do Valle Matta MA, Leitner E, Goffeau A, Daum G. PDR16 and PDR17, two homologous genes of Saccharomyces cerevisiae, affect lipid biosynthesis and resistance to multiple drugs. J Biol Chem. 1999;274(4):1934–1941. doi: 10.1074/jbc.274.4.1934. [DOI] [PubMed] [Google Scholar]
- 114.Ren J, Pei-Chen Lin C, Pathak MC, Temple BR, Nile AH, Mousley CJ, Duncan MC, Eckert DM, Leiker TJ, Ivanova PT, Myers DS, Murphy RC, Brown HA, Verdaasdonk J, Bloom KS, Ortlund EA, Neiman AM, Bankaitis VA. A phosphatidylinositol transfer protein integrates phosphoinositide signaling with lipid droplet metabolism to regulate a developmental program of nutrient stress-induced membrane biogenesis. Mol Biol Cell. 2014;25(5):712–727. doi: 10.1091/mbc.E13-11-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pu J, Ha CW, Zhang S, Jung JP, Huh WK, Liu P. Interactomic study on interaction between lipid droplets and mitochondria. Protein Cell. 2011;2(6):487–496. doi: 10.1007/s13238-011-1061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lafer G, Szolderits G, Paltauf F, Daum G. Isolation of a phosphatidylserine transfer protein from yeast cytosol. Biochim Biophys Acta. 1991;1069(2):139–144. doi: 10.1016/0005-2736(91)90115-o. [DOI] [PubMed] [Google Scholar]
- 117.Barba G, Harper F, Harada T, Kohara M, Goulinet S, Matsuura Y, Eder G, Schaff Z, Chapman MJ, Miyamura T, Brechot C. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc Natl Acad Sci USA. 1997;94(4):1200–1205. doi: 10.1073/pnas.94.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Abell BM, Holbrook LA, Abenes M, Murphy DJ, Hills MJ, Moloney MM. Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell. 1997;9(8):1481–1493. doi: 10.1105/tpc.9.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Monier S, Parton RG, Vogel F, Behlke J, Henske A, Kurzchalia TV. VIP21-caveolin, a membrane protein constituent of the caveolar coat, oligomerizes in vivo and in vitro. Mol Biol Cell. 1995;6(7):911–927. doi: 10.1091/mbc.6.7.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fujimoto T, Kogo H, Ishiguro K, Tauchi K, Nomura R. Caveolin-2 is targeted to lipid droplets, a new “membrane domain” in the cell. J Cell Biol. 2001;152(5):1079–1085. doi: 10.1083/jcb.152.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ostermeyer AG, Paci JM, Zeng Y, Lublin DM, Munro S, Brown DA. Accumulation of caveolin in the endoplasmic reticulum redirects the protein to lipid storage droplets. J Cell Biol. 2001;152(5):1071–1078. doi: 10.1083/jcb.152.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zehmer JK, Bartz R, Bisel B, Liu P, Seemann J, Anderson RG. Targeting sequences of UBXD8 and AAM-B reveal that the ER has a direct role in the emergence and regression of lipid droplets. J Cell Sci. 2009;122(pt 20):3694–3702. doi: 10.1242/jcs.054700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Turro S, Ingelmo-Torres M, Estanyol JM, Tebar F, Fernandez MA, Albor CV, Gaus K, Grewal T, Enrich C, Pol A. Identification and characterization of associated with lipid droplet protein 1: a novel membrane-associated protein that resides on hepatic lipid droplets. Traffic. 2006;7(9):1254–1269. doi: 10.1111/j.1600-0854.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 124.Ozeki S, Cheng J, Tauchi-Sato K, Hatano N, Taniguchi H, Fujimoto T. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J Cell Sci. 2005;118(Pt 12):2601–2611. doi: 10.1242/jcs.02401. [DOI] [PubMed] [Google Scholar]
- 125.Sztalryd C, Xu G, Dorward H, Tansey JT, Contreras JA, Kimmel AR, Londos C. Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J Cell Biol. 2003;161(6):1093–1103. doi: 10.1083/jcb.200210169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Garbarino J, Sturley SL. Homoeostatic systems for sterols and other lipids. Biochem Soc Trans. 2005;33(Pt 5):1182–1185. doi: 10.1042/BST20051182. [DOI] [PubMed] [Google Scholar]
- 127.Han GS, Siniossoglou S, Carman GM. The cellular functions of the yeast lipin homolog PAH1p are dependent on its phosphatidate phosphatase activity. J Biol Chem. 2007;282(51):37026–37035. doi: 10.1074/jbc.M705777200. [DOI] [PubMed] [Google Scholar]
- 128.Petschnigg J, Wolinski H, Kolb D, Zellnig G, Kurat CF, Natter K, Kohlwein SD. Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J Biol Chem. 2009;284(45):30981–30993. doi: 10.1074/jbc.M109.024752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rockenfeller P, Ring J, Muschett V, Beranek A, Buettner S, Carmona-Gutierrez D, Eisenberg T, Khoury C, Rechberger G, Kohlwein SD, Kroemer G, Madeo F. Fatty acids trigger mitochondrion-dependent necrosis. Cell Cycle. 2010;9(14):2836–2842. doi: 10.4161/cc.9.14.12267. [DOI] [PubMed] [Google Scholar]
- 130.Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 2005;24(11):1931–1941. doi: 10.1038/sj.emboj.7600672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Carman GM, Han GS. Phosphatidic acid phosphatase, a key enzyme in the regulation of lipid synthesis. J Biol Chem. 2009;284(5):2593–2597. doi: 10.1074/jbc.R800059200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Karanasios E, Han GS, Xu Z, Carman GM, Siniossoglou S. A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc Natl Acad Sci USA. 2010;107(41):17539–17544. doi: 10.1073/pnas.1007974107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Choi HS, Su WM, Morgan JM, Han GS, Xu Z, Karanasios E, Siniossoglou S, Carman GM. Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of SER(602), THR(723), AND SER(744) as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J Biol Chem. 2011;286(2):1486–1498. doi: 10.1074/jbc.M110.155598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.van Zutphen T, Todde V, de Boer R, Kreim M, Hofbauer HF, Wolinski H, Veenhuis M, van der Klei IJ, Kohlwein SD. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2014;25(2):290–301. doi: 10.1091/mbc.E13-08-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Toulmay A, Prinz WA. Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J Cell Biol. 2013;202(1):35–44. doi: 10.1083/jcb.201301039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Neiman AM. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics. 2011;189(3):737–765. doi: 10.1534/genetics.111.127126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rajakumari S, Daum G. Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions. Mol Biol Cell. 2010;21(4):501–510. doi: 10.1091/mbc.E09-09-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lam C, Santore E, Lavoie E, Needleman L, Fiacco N, Kim C, Neiman AM. A visual screen of protein localization during sporulation identifies new components of prospore membrane-associated complexes in budding yeast. Eukaryot Cell. 2014;13(3):383–391. doi: 10.1128/EC.00333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lin CP, Kim C, Smith SO, Neiman AM. A highly redundant gene network controls assembly of the outer spore wall in S. cerevisiae. PLoS Genet. 2013;9(8):e1003700. doi: 10.1371/journal.pgen.1003700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Novikoff AB, Novikoff PM, Rosen OM, Rubin CS. Organelle relationships in cultured 3T3-L1 preadipocytes. J Cell Biol. 1980;87(1):180–196. doi: 10.1083/jcb.87.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Krahmer N, Farese RV, Jr, Walther TC. Balancing the fat: lipid droplets and human disease. EMBO Mol Med. 2013;5(7):905–915. doi: 10.1002/emmm.201100671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schweiger M, Lass A, Zimmermann R, Eichmann TO, Zechner R. Neutral lipid storage disease: genetic disorders caused by mutations in adipose triglyceride lipase/PNPLA2 or CGI-58/ABHD5. Am J Physiol Endocrinol Metab. 2009;297(2):E289–E296. doi: 10.1152/ajpendo.00099.2009. [DOI] [PubMed] [Google Scholar]
- 144.Vigouroux C, Caron-Debarle M, Le Dour C, Magre J, Capeau J. Molecular mechanisms of human lipodystrophies: from adipocyte lipid droplet to oxidative stress and lipotoxicity. Int J Biochem Cell Biol. 2011;43(6):862–876. doi: 10.1016/j.biocel.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 145.Fei W, Du X, Yang H. Seipin, adipogenesis and lipid droplets. Trend Endocrinol Metab TEM. 2011;22(6):204–210. doi: 10.1016/j.tem.2011.02.004. [DOI] [PubMed] [Google Scholar]