Abstract
The Streptococcus thermophilus CRISPR3-Cas (StCas9) system has been shown to mediate DNA cleavage in its original host and in E. coli as well as in vitro. Here, we have reconstituted the StCas9 system in yeast and conducted a systematic optimization of the sgRNA structure, including the minimal length of tracrRNA, loop structure, Match II region, Bulge motif, the minimal length of guide sequence within the crRNA, tolerance of mismatches and target sequence preference. The optimal sgRNA design for the StCas9 system achieved up to 12 and 40 % targeting efficiencies in yeast and human cells, respectively. This study provides important insight into the sequence and structural requirements necessary to develop a targeted and highly efficient eukaryotic gene editing platform using CRISPR-Cas systems.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-014-1679-z) contains supplementary material, which is available to authorized users.
Keywords: S. thermophilus Cas9, Single guide RNA, Surrogate reporter, DNA repair, Genome targeting
Introduction
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated (Cas) genes provide bacteria and archaea with an adaptive immune system that defends against invading DNA [1–3]. The system has evolved with the ability to incorporate the “protospacer” sequences from invading bacteriophages or plasmids as “spacers” within CRISPR loci. Subsequent transcription of the “spacers” as specific small-interfering CRISPR RNA (crRNA) directs the degradation of homologous sequences within invading foreign DNA [2, 3], conferring adaptive immunity. Three types (types I, II and III) of CRISPR-Cas systems have been classified based on the structural organization and functions of nucleoprotein complexes involved in crRNA-mediated silencing of foreign nucleic acids [4]. All types of CRISPR-Cas systems share a common ability to destroy invading nucleic acids that is guided by short CRISPR RNAs (crRNAs). Engineering the guiding RNAs instead of proteins for targeted gene editing would confer many advantages over existing gene editing tools, including zinc finger nucleases (ZFNs) and TAL Effector Nucleases (TALENs).
In the type II CRISPR-Cas system [5–7], three minimal components, the CRISPR-associated nuclease Cas9, a specificity-determining crRNA, and an auxiliary trans-activating crRNA (tracrRNA), have been used to successfully reconstitute the system from Streptococcus pyogenes (S. pyogenes) both in vitro [8] and in mammalian cells [9, 10]. Previous studies demonstrated that the tracrRNA and crRNA duplexes can be fused with a 5′-GAAA-3′ loop to generate a single chimeric guiding RNA (sgRNA or gRNA). Importantly, both crRNA:tracrRNA and sgRNA have been demonstrated to direct SpCas9 target cleavage activity in eukaryotic cells [10]. As a result, the sgRNA in CRISPR-Cas system has subsequently become the focus of intense development for eukaryotic genome editing application.
Almost all of the work reported thus far has focused on the CRISPR-Cas system derived from a single S. pyogenes species. Exploiting different CRISPR-Cas systems may help with overcoming the restrictions including off-target frequency [11–13], PAM-required limitation [8–10] and low activity of single sgRNA guided Cas9 transcriptional activator [14–17]. Recently, a new Neisseria meningitides (N. meningitides) CRISPR-Cas system with a different requirement for the PAM sequence (5′-NNNNGATT-3′) has been reported to function in hPSC lines [18, 19]. In vitro investigation demonstrated that Cas9 orthologs from Streptococcus thermophilus (S. thermophilus) LMD-9 and Listeria innocua (L. innocua) Clip11262 could support S. pyogenes crRNA:tracrRNA to direct DNA cleavage [8]. Although previous studies have shown that S. thermophilus CRISPR3-Cas system [20] enables to mediate DNA cleavage in its original host [21, 22], E. coli [23] as well as in vitro [24], systematic engineering of S. thermophilus CRISPR3-Cas system for eukaryotic usage has not been reported.
Here, we reported the development of a sensitive yeast assay for reconstitution of the S. thermophilus CRISPR3-Cas (StCas9) system. We characterized the sgRNA structure, including the minimal length of tracrRNA, Loop, Match II region, Bulge motif, the minimal length of guide sequence, tolerance of mismatches and the target sequence preference. An optimal design of the sgRNA structure was finally drawn and tested in both yeast and human cells. Using this optimal sgRNA design for the StCas9 system, we achieved efficient genome targeting in human cells. Importantly, this work improves our ability to engineer the CRISPR-Cas system by designing efficient sgRNA for targeted eukaryotic gene editing.
Results
Engineering the StCas9 system and functional assay in yeast
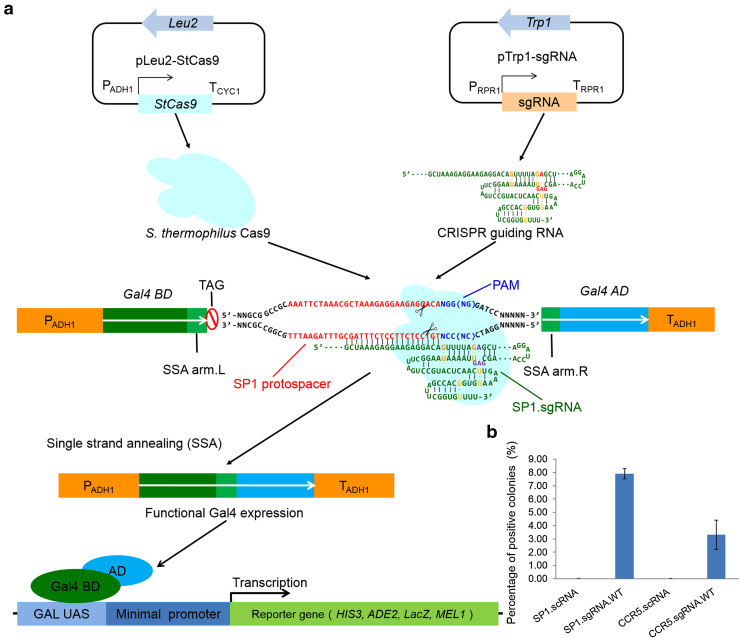
In order to reconstitute the StCas9 system in eukaryotic cells, we designed a yeast assay system. The system consists of a yeast strain AH109 and three expression vectors; the StCas9, the sgRNA and Gal4 reporter. The Gal4 reporter was designed by inserting the target sequence flanked by direct repeats as SSA arms into the middle of Gal4 gene, resulting in a disrupted open reading frame (Sequence S3). The yeast strain AH109 contains four chromosome integrated reporter genes (HIS3, ADE2, LacZ and MEL1), whose expression is induced by the Gal4 protein. Conceptually, if the StCas9 system functions, it will cleave the target sequence. The double strand break (DSB) will trigger the cellular DNA repair system leading to restoration of Gal4 function via single strand annealing (SSA) repair pathway. Functional Gal4 will activate the expression of the four chromosomal integrated reporter genes (Fig. 1a).
Fig. 1.
Schematic of yeast assay for the engineered StCas9 system. a Illustration of the yeast assay system. b Comparison of the scRNA and sgRNA.WT designs of SP1 protospacer and human CCR5 gene for guiding the StCas9 activity in yeast. Yeast transformation assay was performed with expression vectors for bStCas9, scRNA/sgRNA.WT bearing SP1 or CCR5 guiding sequence and corresponding surrogate reporter plasmids
After failed with a short chimeric RNA (scRNA; Figure S1) design for CCR5 or SP1 based on Jinek’s result [8], we then modified the chimeric RNA structure. A long chimeric guiding RNA (wild type design, sgRNA.WT; Fig. 2a) bearing a full length of crRNA and tracrRNA was designed. The new sgRNAs generated significant numbers of colonies in histidine and adenine dropout plate, and the activity was calculated as 7.90 % (Fig. 1b; Figure S2C).
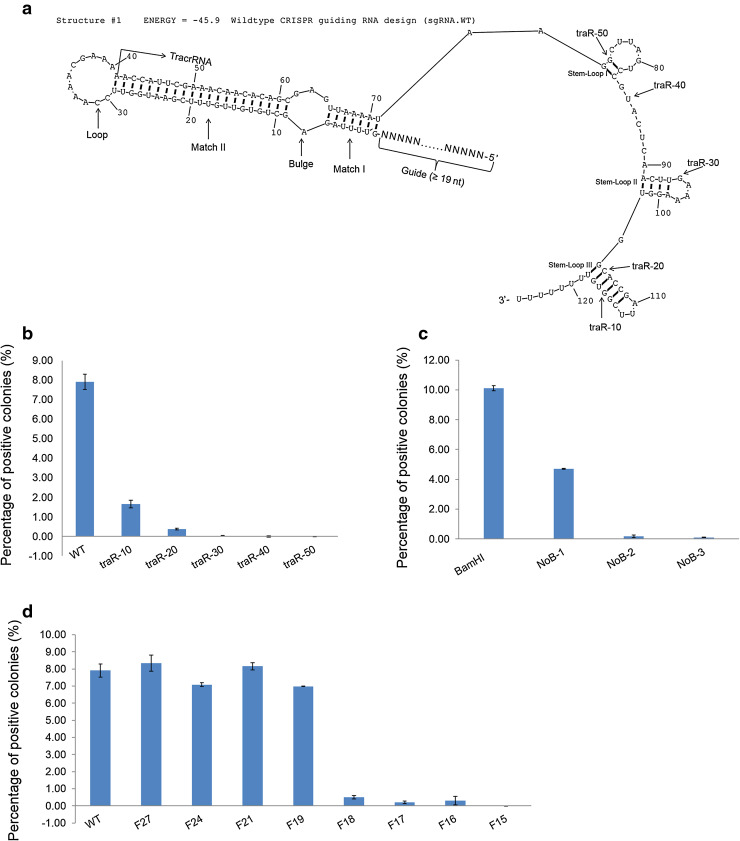
Fig. 2.
Optimization of the StCas9 system in yeast. a Secondary structure of the wild type guiding RNA (sgRNA.WT) design. The structure was drawn by RNAstructure Version 4.6 not accounting for the influence of the guide sequence. Different motifs and truncations of tracrRNA 3′ ends are indicated by arrows. b Examination of different tracrRNA length for guiding the StCas9 activity. c Effect of the Bulge mutants on the StCas9 activity. d Minimum length requirement of the guide sequence for the StCas9 activity. Transformation assays were conducted with bStCas9 expression vector, the surrogate reporter pRep.20.SP1 and corresponding sgRNA mutant vectors
To confirm the results obtained above, we conducted further β-galactosidase assay indicating the expression of the LacZ reporter gene (Figure S3C). PCR assay was conducted to examine the repair of the Gal4 gene. The results clearly demonstrated that the Gal4 gene was repaired via the SSA mechanism (Figure S3D). All of these results indicated that the engineered StCas9 system functions in yeast.
Optimization of the StCas9 system in yeast
Encouraged by the results obtained above, we leveraged our simple and sensitive Gal4 reporter assay system to systematically characterize the engineered StCas9 system. RNAstructure analysis identifies several motifs on sgRNA.WT (Fig. 2a), including TracrRNA (Stem-Loop I/II/III), Loop, Match I, Bulge, Match II and Guide sequence. We carried out a series of experiments to characterize the contributions of these motifs to the StCas9 system.
Full-length tracrRNA is required for maximum StCas9 activity
To determine the functions of the TracrRNA motif, we generated five mutants; each mutant was progressively shorter by 10 nt from its 3′ end (Fig. 2a, Figure S4A). The functional assay demonstrated that successive deletion greatly reduced the StCas9 activity (Fig. 2b). Activity of the mutant lacking 30 nt was virtually zero, suggesting that the 3′ end sequence of tracrRNA is important to maintain the maximum StCas9 activity.
Loop sequence is changeable
For the purpose of easy manipulation, we generated several Loop mutants based on the Pst I or BamH I sites (Figure S4B). Transformation assay showed that these Loop mutants did not significantly affect the StCas9 activity (Figure S5A), suggesting that the loop sequence can be altered without loss of function.
Minimum length of Match II sequence
The Match II region contains a stem of 21 bp in the wild type sgRNA design (Fig. 2a). We generated a series of Match II mutants; each mutant was progressively shorter by 3 nt from the direction of the Loop motif (Figure S4C). All mutants showed similar activities (Figure S5B), suggesting that the Match II motif does not play an important role in maintaining the StCas9 activity. Interestingly, RNA structure prediction suggests that the mutants Mat-18 and Mat-21 likely affected the Bulge structure (Figure S6), yet still showed comparable activity with the full length of Match II.
Bulge is essential for the StCas9 activity
The Bulge motif is essential in the StCas9 system. To determine its function, we generated three mutants that destroy the Bulge structure (Figure S4D). Transformation assays demonstrated that the mutants NoB-2 and NoB-3 almost abolished the StCas9 activity (Fig. 2c). However, the mutant NoB-1 still showed some activity, although it was reduced by more than 50 %. These data suggested that the wild type Bulge structure seems essential for the maximum activity of the StCas9 system.
Minimum length requirement of the guide sequence
To investigate the minimum length of guide sequence required for sgRNAs to direct the StCas9 activity, we first generated five mutants, each mutant progressively shorter by 3 nt from the 5′ end (Figure S4E). Functional assays indicated that mutants with <18 bp almost completely lost activity (Fig. 2d). We further generated three additional mutants, SP1.sgRNA.F19/F17/F16. The results demonstrated that the 19 nt was enough for the StCas9 system (Fig. 2d).
Tolerance of mismatches
To study the tolerance of mismatches, we designed a set of 18 sgRNA mutants with mutations in the SP1 guide sequence (Figure S4F). Results from the functional assay revealed that the StCas9 system showed some degree of tolerance for the mismatches in a position-dependent manner (Fig. 3a). The mismatches within the first five positions adjacent to the PAM region abolished the StCas9 activity, suggesting that these positions are crucial for determining the StCas9 system specificity. Interestingly, the mutation A → G in sgRNA at position 8 and 15 did not significantly affect the StCas9 activity.
Fig. 3.
Investigation of target mismatch tolerance and sequence preference. a Effect of mismatches on StCas9 activity. b The StCas9 system functions on different targets with distinct activities. c Functional analysis of target sequence preference for the StCas9 system. The significant up (G04T) and down (G10T) regulations are indicated by arrows
The StCas9 system functions on different targets with distinct activities
We initially tested the target sequences derived from one human gene CCR5 and three yeast genes ERG6, TPD1 and YAP1 (Figure S7A). The data indicated that, although the StCas9 functions on these target sequences, the activities showed significant difference among these four targets (Fig. 3b). This result suggested that the StCas9 system apparently has target preferences.
Target sequence preference
By aligning the 12 spacer sequences (SP1-12) within the S. thermophilus DGCC7710 CRISPR3-Cas locus, we tried to find out any sequence preference for targeting (Table S1). The nucleotides that occurred with a frequency of at least 40 % are as follow: 5′-NATNNN(A/T)NANNA(A/T)NN(A/T)AAAANAA(T/G)T(G/A)NAGA-3′. Based on this alignment, we constructed a series of sgRNA (Figure S4F) and target (Figure S7B) mutants. These mutants were designed either the same as the consensus or employing the bases with the lowest frequency in alignment. Although the results were not entirely as anticipated (Fig. 3c), the StCas9 activity was significantly increased for target SP1.Mu-04 (G04T) and decreased for SP1.Mu-10 (G10T). However, further investigation is required to reveal the target preference for the StCas9 system.
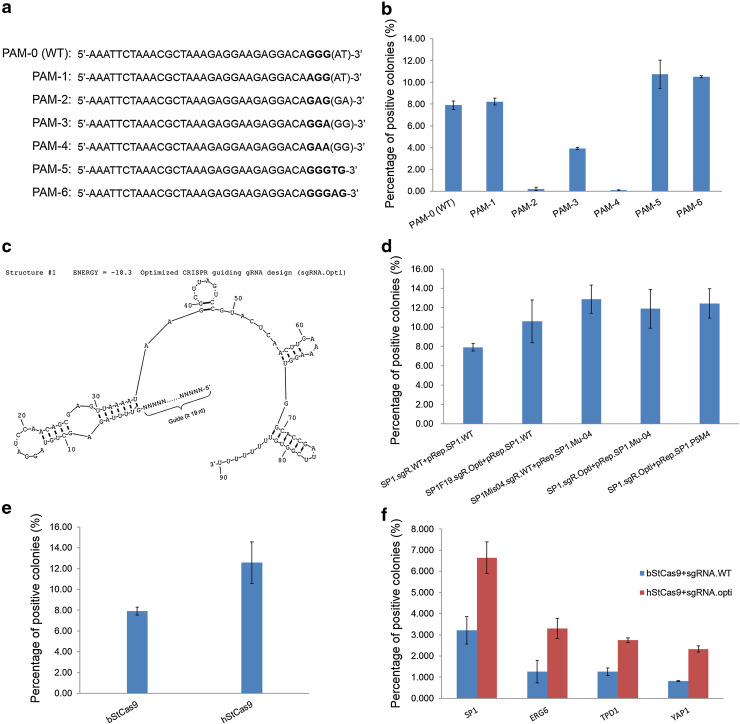
The G on the second position within PAM is essential
We initially used the 5′-GGG-3′ PAM according the SpCas9 system [8]. We generated a set of mutants (PAM-1/2/3/4) to define the PAM pattern (Fig. 4a). Functional assays indicated that the G on the second position is essential for the StCas9 activity (Fig. 4b). As previous studies suggested that the StCas9 system recognizes a 5′-NGGNG-3′ PAM [21, 23, 24], additional two PAM patterns (PAM-5/6) were investigated. The mutant (PAM-3) for the G on third position reduced colony numbers by half, and the G on the fifth position (PAM-5/6) did increase the positive colonies slightly, implying that the G on the third or fifth position is not essential, but does help with the StCas9 activity.
Fig. 4.
The optimized StCas9 system functions in yeast. a Different PAM patterns designed in reporter vectors for SP1 protospacer targeting. b Comparison of different PAM patterns for the StCas9 activity. c Secondary structure of the optimized CRISPR guiding RNA (sgRNA.Opti) design. The structure was drawn by RNAstructure Version 4.6 not accounting for the influence of the guide sequence. d Comparison of wild type and optimized sgRNA designs for SP1 protospacer targeting. e Comparison of the bacterial origin bStCas9 and codon-optimized hStCas9 in yeast assay. f Functional analysis of the StCas9 system on different chromosome integrated targets
Optimized sgRNA and humanized Cas9 function with better performances
After the series of experiments conducted for characterizing the sgRNA as shown above, we proposed an optimized sgRNA structure (sgRNA.Opti; Fig. 4c). For functional assay, the sgRNA.Opti design showed a better performance for the wild type SP1 protospacer targeting. While for the protospacer mutants with G04T, the optimized and wild type sgRNA designs exhibited similar activities (Fig. 4d). In addition, we later humanized the Cas9 gene codons (hStCas9; Sequence S4) and compared it with the bacterial origin Cas9 sequence (bStCas9; Sequence S1). Transformations with hStCas9 showed significantly higher activity (Fig. 4e).
Furthermore, we employed this optimized structure for designing three other sgRNAs for the mouse ROSA26 locus targeting. Functional assay in yeast revealed that the optimized sgRNAs could function well on different targets (Figure S10A and B).
The StCas9 system functions on targets in yeast genome
To test if this optimized StCas9 system cleaves targets within an actual chromosome, we integrated the Gal4 reporter cassettes at the yeast HO gene locus (Figure S8A-C). Two SSA arms with length of 20 and 150 bp both produced a similar number of positive colonies (Figure S8D and E), indicating that the SSA arms of 20 bp were sufficient for maximal effect in the context of integration.
For the three chromosome integrated reporter strains containing the yeast endogenous ERG6, TPD1 and YAP1 protospacers, the optimized StCas9 system showed activities ranging from 2.3 to 3.3 % (Fig. 4f), suggesting that the StCas9 system can be used to efficiently target yeast chromosome.
The StCas9 system functions in human 293T cells
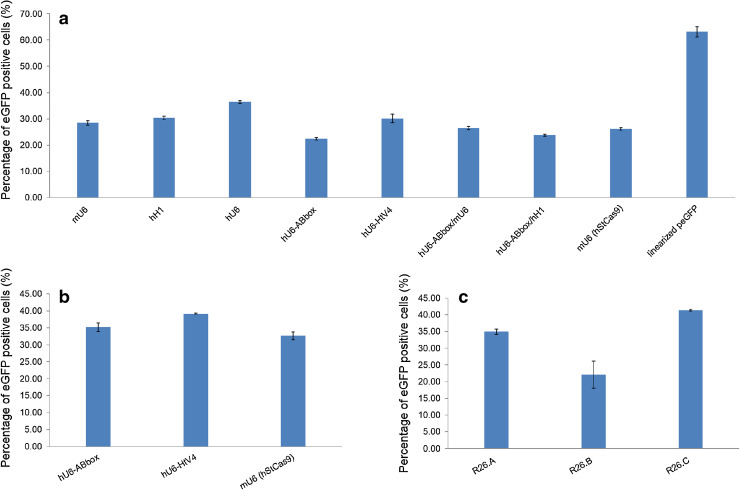
To quickly test the StCas9 system in mammalian cell, we constructed eGFP reporter vectors designed similarly as the yeast Gal4 reporter (Figure S9A). In addition, several strategies were employed to express the wild type sgRNA design (Figure S9B). Transient transfection assays also indicated that the highest activity (36.46 %) was generated by the human U6 promoter (Fig. 5a). We initially fused the human tRNA variant (HtV4) sequence [25] and the leader sequence (ABbox) from yeast RPR1 gene [26] with sgRNA driven by the human U6 promoter, respectively, expecting to enhance the sgRNA expression and stability. However, the hU6-HtV4-sgRNA did not show a significant difference, and activity of the hU6-ABbox-sgRNA was dramatically decreased.
Fig. 5.
Examination of the StCas9 system in human 293T cells. a Comparison of different sgRNA expressing strategies in 293T cells. Transfection assay was conducted with the hStCas9 expression vector and the surrogate reporter pRep.eGFP.SP1.PAM5 combining with different vectors for the wild type sgRNA design. For the linearized pRep.eGFP control, the reporter plasmid was linearized with BamH I before transfection. b Functional analysis of the SP1.sgRNA.Opti design for guiding StCas9 activity in 293T cells. Transfection assay was conducted with corresponding SP1.sgRNA.Opti and hStCas9 expression vectors and the surrogate reporter pRep.eGFP.SP1.P5M4. c Validation of the StCas9 activity on targets from mouse ROSA26 locus in 293T cells. Transfection assay was carried out with the sgRNA-StCas9 expression vectors (mU6.sgRNA.Opti-CMV.hStCas9) and corresponding mammalian cell surrogate reporters, pRep.eGFP.R26A/B/C
We further tested the optimized sgRNA design with three expressing strategies. Transfection assay with SP1.sgRNA.Opti driven by hU6-HtV4 generated 39.19 % of eGFP positive cells, the highest for the tested SP1 protospacer targeting in 293T cells (Fig. 5b). On the other hand, as expected, the hStCas9 showed significant higher activities than bacterial origin Cas9 sequence for both the wild type and optimized sgRNA designs (data shown partially, Figure S10C).
To further verify the results, we examined the three sgRNAs designed for the mouse ROSA26 locus targeting with our optimized StCas9 system in 293T cells. The results revealed even higher activity (41.3 % for R26.C; Fig. 5c).
The StCas9 system functions on targets in 293T genome
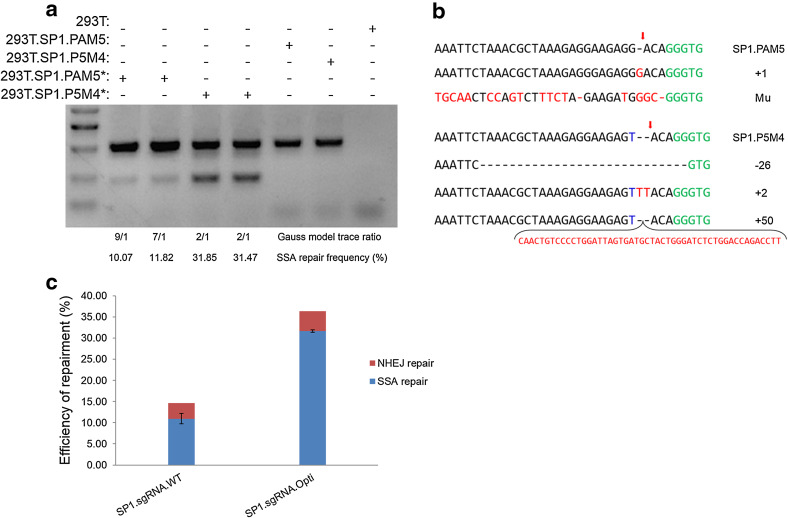
We next examined the optimized StCas9 system on the chromosomal locus by generating reporter-integrated stable cell lines. PCR assay generated two products, 494 and 246 bp bands. The 246 bp PCR product was derived from the SSA-mediated repair, while the 494 bp PCR product contains both NHEJ-mediated repair and the wild type target sequence. To determine the frequency of NHEJ-mediated repair, we recovered the 494 bp band from gel for sequencing. The results revealed that SSA-mediated DNA repair frequency was 10.95 % for SP1.sgRNA.WT and 31.66 % for SP1.sgRNA.Opti (Fig. 6a, c) and NHEJ-mediated repair frequency was 3.71 and 4.66 %, respectively (Fig. 6b, c. These data suggested that the activity of the optimized StCas9 system could reach up to more than 36 %, and if considering the precise-repair product, the cleavage activity may be even higher. We further tested three other target sequences from human AAVS1 [9] and CCR5 [27] loci and demonstrated our optimized StCas9 system activities in both surrogate reporter assay and endogenous gene targeting with T7EI assay (Figure S11, Table S2).
Fig. 6.
The StCas9 system functions on targets integrated in 293T genome. a Detection of SSA-mediated repair frequencies for integrated reporter constructs. 293T.SP1.PAM5* and 293T.SP1.P5M4* represent cells transfected with corresponding sgRNA and hStCas9 expression vectors. b Mutations detected in the integrated reporter constructs caused by NHEJ repair pathway. 2 of 48 and 3 of 44 for the targets SP1.PAM5 and SP1.P5M4, respectively. c Comparison of wild type (SP1.gRNA.WT) and optimized (SP1.gRNA.Opti) designs for guiding the StCas9 activity on integrated reporter targets in 293T cells
Discussion
Differently with the smaller Cas9 from the S. thermophilus CRISPR1 exploited for genome editing by Esvelt et al. [28], we cloned the larger Cas9 gene (bStCas9) from the S. thermophilus CRISPR3 locus and successfully reconstituted the StCas9 system in yeast with a wild type sgRNA design (sgRNA.WT). We initially engineered the SP1-crRNA and the tracrRNA from S. thermophilus CRISPR3 as a single guiding RNA (sgRNA) referring to previous in vitro study of the SpCas9 system [8], and later demonstrated that the two stem-loop structures (Fig. 2a) at the 3′ end of tracrRNA were essential for a maximum activity of the programmed StCas9 system in yeast (Fig. 2b). However, in vitro study with the pre-crRNA:tracrRNA:StCas9 complex [22] suggested that only the Stem-Loop II is responsible for the cleavage activity. And studies with the SpCas9 system used a truncated tracrRNA still showed some activity, too [8, 27, 29]. Nevertheless, further crystal structure analysis of SpCas9 in complex revealed that all the stem-loops function to reinforce the interaction between Cas9 and sgRNA [30, 31].
Up to now, all of the sgRNAs reported with the SpCas9 system were engineered with a 5′-GAAA-3′ Loop. We found that a 5′-AGGAUCCA-3′ Loop with a BamH I site exhibited the best performance in our study (Figure S5A). This Loop design makes it easier for constructing sgRNA expression vectors. It appears that the Match II motif does not affect the system activity (Figure S5B). This observation is also consistent with previous report for the SpCas9 nuclease [11]. However, the Bulge mutation data (Fig. 2c) indicated that the wild type Bulge motif is essential for StCas9 maximum activity, but the sgRNA structure maybe flexible (Figure S6). The significance of a Bulge motif and the flexibility of the sgRNA structure were also observed for the SpCas9 system [31].
We proposed our own optimized sgRNA design, which is almost the same with the sgRNA used for SpCas9 system [9, 10] except some nucleotides. It seems that a Bulge motif and the two stem-loops at the 3′ end of tracrRNA are necessary for a maximum activity to both systems. Indeed, in vitro investigation demonstrated that Cas9 orthologs from S. thermophilus LMD-9 and L. innocua Clip11262 could support S. pyogenes crRNA:tracrRNA to direct DNA cleavage partially [8], and the two components are interchangeable only between closely related type II systems [6].
The specificity of the CRISPR-Cas system is determined by the complementarity between the guide sequence of sgRNA and target. The StCas9 system requires 19 nt of guide sequence for its maximum activity in yeast (Fig. 2d). This finding was later also confirmed in human 293T cells (data not shown). Surprisingly, we found that the guide sequence with 15 nt almost abolished the StCas9 nuclease activity in yeast (Fig. 2d), while the guide sequence with 14 nt was demonstrated to be able to direct the SpCas9 activity in vitro [8]. Another interesting discovery from our data is that the extra-sequence (ABbox and restriction enzyme site) added to the 5′ end of the guide sequence does not affect the StCas9 activity. This finding was further confirmed by the subsequent mammalian cell assays with 5′ end extra-sequence (Htv4, ABbox and restriction enzyme sites) introduced (Fig. 5a, b; Figure S9B). A recent report for the SpCas9 system demonstrated that sgRNAs with extended guide sequence is processed to be short [32]. Similar processing maybe employed for the StCas9 system. Perhaps, the minimal length of the guide sequence could be explained to be determined by the precrRNA maturation process as occurs within the nature form bacterial CRISPR-Cas systems [5].
Consistent with previous findings in the SpCas9 system [8, 10–14, 33], we observed that the StCas9 system tolerates single mismatches in the PAM-proximal region less than in the PAM-distal region (Fig. 3a). However, investigation of the StCas9 system in E. coli indicated that the PAM-distal mismatches were much more tolerated than our yeast assay (Sapranauskas et al. [23]). Nevertheless, a seed region of 5 bp proximal to the PAM is essential to determine the target specificity for the StCa9 system. Interestingly, two single mismatches with rG:dT RNA:DNA interaction caused by the A → G mutation at the position 8 and 15 in sgRNA did not affect the StCas9 activity. Actually, many serious off-target events for genome targeting mainly caused by the rG:dT/rU:dG interaction have been observed for the SpCas9 system [11–13], and similar cases may happen to the StCas9 system.
Previous studies suggested that CRISPR-Cas system has target preference with various Cas9 activity [9, 10]. Our results also support this proposal (Fig. 3b, c). However, the number of protospacer sequences used is limited. We cannot make any strong conclusion about the StCas9 target sequence preference. From our series of mutation experiments, we only confirmed the hypothesis that StCas9 system has protospacer sequence preference as some mutations significantly affect its activity. Rules for target sequence preference remain unclear, and factors including melting temperature, GC proportion, nucleotide distribution, and the influence of target sequence on sgRNA secondary structure remain to be determined.
The PAM is used for CRISPR-Cas system to distinguish self versus non-self target. We initially employed the 5′-NGG-3′ PAM pattern, and determined that the G on the second position was essential for the StCas9 system in yeast, arguing with the possible 5′-NAG-3′ PAM mutation for the SpCas9 system [11, 33]. Previous studies of the S. thermophilus CRISPR3-Cas system in prokaryotic cells and in vitro suggested a PAM pattern of 5′-NGGNG-3′ [21–24]. Investigation in E. coli demonstrated that all three conserved G residues are equally important [23]. Unfortunately, our yeast assay indicated that the G on the third or fifth position was not essential to maintain the StCas9 activity, but they were important for the maximum activity (Fig. 4a, b).
For yeast genome DNA targeting, we initially tried both T7EI nuclease assay and Sequencing method (100 clones for each gene), but unfortunately we failed to detect any indels. This result was later confirmed by the report that the induced SpCas9 mutagenesis experiment only produced knockout rates of at most 0.07 % [34], which may be caused by the low NHEJ-mediated indel frequency in yeast of the blunt break caused by type II CRISPR-Cas systems [8, 24]. Hence, we integrated the Gal4 reporter construct containing endogenous gene target sequence into yeast chromosome HO locus and confirmed that the StCas9 system functions on yeast chromosome.
We compared 8 different methods to express the sgRNA (Figure S9B) and found that both human U6 alone and its fusion with HtV4 exhibited a similarly high targeting activity of around 40 % in 293T (Fig. 5a). Interestingly, fusion of the HtV4 to the 5′ end of the sgRNA did not affect the activity, suggesting that extra sequence at the 5′ end does not contribute its specificity and efficiency, which could be explained by the post-transcriptional processing [32]. Similar with the finding that the crRNA:tracrRNA:SpCas9 complex functions without the host factor RNaseIII introduced in 293FT [10], our crRNA:tracrRNA:StCas9 design (sgRNA.Bi, Figure S9B) generated comparable activity with the StCas9:sgRNA designs (Fig. 5a).
The SP1.sgRNA.Opti design showed twofold higher than SP1.sgRNA.WT for directing the StCas9 activity (Fig. 6c) in our stable cell assays. Unexpectedly, SSA-mediated double strand DNA repair is dominant, in contrary to the previous report in Chinese hamster ovary cells [35]. However, our finding is supported by a recent report with fluorescent genome editing reporters [36].
One of the most important concerns about the newly emerged CRISPR-Cas9 nuclease technology is its off-target effects. To improve the specificity, several strategies have been suggested (1) avoiding potential off-target sequences by considering the tolerance of mismatches and PAM mutations [11–14]; (2) titrating the dosage of Cas9 and sgRNA to minimize the off-target activity [11–13]; (3) utilizing sgRNA with shorter guide sequence [37] or truncated tracrRNA [13] that is less active but more specific; (4) delivering CRISPR-Cas9 components as in vitro transcribed and short-lived RNAs instead of DNA plasmids [38]; (5) nicking target DNA through Cas9 nickases to mediate the homology directed repair (HDR) with appropriate repair donors [9, 10]; (6) generating targeted DSBs by introducing offset nicking via Cas9 nickases and paired sgRNAs [14, 32]; (7) exploiting the family of CRISPR-Cas systems for more-specific Cas9 orthologs [19, 28, 39] or new enzymes, such as dCas9-FokI nucleases [40]. Although most of the literatures cited above were focused on the SpCas9 system, our StCas9 system would benefit from the very strategies. Actually, we have generated our hStCas9 nickases and dead Cas9 (dCas9) by introducing the mutations with D10A in RuvC or/and H847A in HNH [23, 24], all of the mutants abolished the DNA cleavage activity for hStCas9 in both yeast AH109 and human 293T cells (data not shown), suggesting additional potential applications of our hStCas9 for dCas9-FokI nuclease [40], DNA nickases [14, 32], CRISPRi [41, 42] and crisprTFs [14–17, 42–44].
In conclusion, we reconstituted and optimized the StCas9 system in yeast. This system has allowed us to systematically investigate how each part of the CRISPR3-Cas system contributes to optimal activity. Our works provide important insight into the sequence and structural requirements necessary to develop a targeted and highly efficient eukaryotic gene editing platform by programming different type II CRISPR-Cas systems [7].
Materials and methods
Yeast assay for the programmed StCas9 system
Yeast strain and media
The Saccharomyces cerevisiae strain used in this study was AH109 (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2 : : GAL1 UAS-GAL1 TATA-HIS3, GAL2 UAS- GAL2 TATA-ADE2, URA3 : : MEL1 UAS-MEL1 TATA-LacZ MEL1) (Clontech), in which different Gal4-responsive upstream activating sequences (UASs) and TATA boxes control four integrated reporter genes, HIS3, ADE2, LacZ and MEL1. The AH109 strain was cultured with YPDA (Clontech) medium before transformation. Plasmids transformed AH109 or reporter construct integrated AH109 strains were selected or cultured with SD media supplemented with G418 antibiotic, leucine, tryptophan, adenine or/and histidine (Sigma) if necessary.
Yeast transformation and manipulation
Yeast transformation (1–10 μg for different experiment series per transformation) was carried out using the LiAc method as Gietz et al. [45, 46] described. Yeast protein extracts were prepared with the TCA method referring to the Yeast Protocols Handbook (Clontech), and western blotting for StCas9 expression was conducted with Monoclonal ANTI-FLAG (Sigma) as primary antibody (Figure S2A). Yeast plasmids and DNA were extracted via Yeast Plasmid Kit and Yeast DNA Kit (OMEGA Bio-Tek) by standard procedure, respectively.
Engineering the StCas9 system and functional assay in yeast
Cloning the S. thermophilus Cas9 gene
The bacterial S. thermophilus Cas9 gene (bStCas9) was obtained by PCR from S. thermophilus stains isolated from yogurt. Primers (Y.bStCas9.F/R) were designed according to the S. thermophilus strain DGCC7710 CRISPR3-Cas gene locus (GenBank: HQ712120.1). The PCR product was cloned into a yeast expression vector (with a LEU2 marker gene, pLeu2) driven by ADH1 promoter and the bStCas9 gene sequence was tagged with SV40 NLS and FLAG nucleotide sequence (pLeu2-bStCas9; Sequence S1).
Yeast RNA expression vector construction
The backbone vector for RNA expression in yeast was constructed based on the yeast RNase P RNA (RPR1) gene (GenBank: M27035.1). The RPR1 gene promoter is one of RNA polymerase III promoters [26]. The RPR1 promoter including an 84 bp leader sequence after the +1 position and its terminator were cloned by PCR with two pairs of primers (P.RPR1.F/R and T.RPR1.F/R) using AH109 genomic DNA as template and inserted into a yeast cloning vector with the TRP1 marker gene to generate the pTrp1 plasmid (Sequence S2).
CRISPR guiding RNA design
Initially, we designed a short chimeric RNA (scRNA) based on Jinek et al.’s [8] result. The scRNA containing a 32 nt crRNA (20 nt SP1 spacer or CCR5 guide sequence and 12 nt repeat sequence from direct repeat region) and a 26 nt tracrRNA sequence of 5′ end of tracrRNA linked by the 5′-GAAA-3′ loop. The mimicked structure was drawn by RNAstructure Version 4.6 (Figure S1). A long chimeric sgRNA was designed to include a 66 nt crRNA (30 nt SP1 spacer sequence and 36 nt direct repeat sequence) and a 85 nt tracrRNA (for DNA sequences of crRNA and tracrRNA, refer to GenBank: HQ712120.1) linked by the 5′-GAAA-3′ loop sequence. The modified long chimeric RNA was named as SP1.sgRNA.WT. DNA fragments for expressing the scRNAs and SP1.sgRNA.WT were synthesized directly (Genscript) and inserted into pTrp1 with Xho I/EcoR I sites to generate corresponding vectors.
Yeast surrogate reporter vector construction
To test targeting efficiency, we constructed surrogate reporters based on the Gal4 transcription factor. A Gal4-based reporter gene driven by the ADH1 promoter was designed such that GAL4 gene was interrupted by multiple cloning site (Not I/BamH I) flanked with 20 or 30 bp direct repeats of Gal4 sequence as SSA arms (Fig. 1a; Sequence S3). Sequences of Gal4 BD and AD were amplified by PCR (for primer information, refer to Table S3) from two Y2H plasmids, pGBKT7 and pGADT7, respectively (Clontech) and cloned into a yeast expression vector with the KanMX4 marker gene to generate PADH1-Gal4BD-AD-TADH1 reporter vector (pRep.20/pRep.30). The SP1 protospacer and CCR5 target fragments with a 5′-GGG-3′ PAM pattern prepared via directly annealing oligonucleotides [47] were subsequently inserted into the Not I/BamH I sites to generate corresponding reporter vectors (pRep.20.SP1/pRep.30.SP1/pRep.20.CCR5).
Detection for the spontaneous repair frequency of surrogate reporter
The AH109 strain was transformed with pRep.20.SP1 and pRep.30.SP1 (2 μg per transformation), respectively. 100 μL of transformation mixtures were spread on SD/−Ade/−His/+Leu/+Trp/+G418 agar plates for detecting recombination while 100 μL of 1/10,000 dilutions on SD/+Ade/+His/+Leu/+Trp/+G418 agar plates for examining transformation efficiency. The plates were maintained at 30 °C for 4 days. For each transformation, three parallel experiments were conducted. Plates (10 mm) with colonies were photographed by ChemiDoc™ MP System (BIO-RAD), and colonies were counted by Quantity One 4.6.2 (BIO-RAD) with Sensitivity set at 5.0 or directly counted when colonies were <500. Colonies on SD/-Ade/-His/+Leu/+Trp/+G418 plates were considered as Gal4 positive clones with spontaneous repair of the reporter vectors, and the numbers of colonies on SD/+Ade/+His/+Leu/+Trp/+G418 plates were used to calculate the total numbers of transformed clones. The spontaneous repair frequencies of the reporter vectors were estimated by the percentages of positive colonies compared with transformed colonies (Figure S2B).
Yeast assay of the StCas9 system with surrogate reporters
AH109 strain was transformed with different experiment or control plasmid groups (molar ratio = pLeu2-bStCas9/pLeu2:pTrp1-SP1.sgRNA.WT/pTrp1 : pRep.20.SP1/pRep.30.SP1 = 1:1:1;8–9.4 μg total plasmid DNA per transformation. Because the bStCas9 was almost 4.3 kb in length, the total DNA used for transformations with pLeu2-bStCas9 was increased). 100 μL of transformation mixtures were spread on SD/-Ade/-His/-Leu/−Trp/+G418 agar plates, while 100 μL of 1/10 dilutions on SD/+Ade/+His/−Leu/−Trp/+G418 agar plates used for examination of the transformation efficiency. The plates were maintained at 30 °C for 4 days. For each transformation, three independent repeats were performed. Gal4 positive clones and transformed colonies were counted as described above. The percentage of positive colonies compared with transformed colonies was calculated to estimate the activity of the programmed StCas9 system (Figure S2C). Different AH109 transformants were further streaked on adenine and histidine auxotrophic plates to confirm the Ade+His+ phenotype (Figure S3A, S3B).
Confirmation of LacZ+ phenotype
To conduct the β-galactosidase assay, 20 μL cultures of different colonies were transferred into a 96-well plate containing a mixture of 35 μL of Y-PER (Thermo Fisher) and 85 μL of 1.1 mg/mL ONPG (Sigma). Samples were incubated at RT for 30–90 (t) min until the positive control turned to be yellow, then 60 μL of 1 mol/L Na2CO3 was added to stop the reaction. The absorbance of each well at 405 nm (A 405) and 590 nm (A 590) was determined using the microtiter plate reader. For each colony, three repeats were conducted. 1 unit of β-galactosidase is defined as the amount which hydrolyses 1 µmol of ONPG to o-nitrophenol and d-galactose min−1 cell−1. The β-gal activity was calculated as follows: β-gal activity = 200 × (A 405 − 1.18 × A 590)/(t × 0.02 × OD600). This method was modified as described by Kohei Ichikawa [48]. Data from the positive control (AH109.pGal4), experimental groups and the other controls were adjusted by subtracting the result of the negative control (AH109).
Identification of the restoration of Gal4 sequence within the reporter plasmids
Half of the cultures of positive control AH109.pGal4 and StCas9 positive yeast transformants were prepared for β-galactosidase assay, and the other half of the cultures was used to extract yeast plasmids. These yeast plasmids were used as templates to amplify the PADH1-Gal4BD-AD-TADH1 fragments by PCR with primers ADH1P.F and ADH1T.R. Plasmids pRep.SP1.20 and pRep.SP1.30 were used as negative controls and pGal4 as positive control. All of the PCR products were digested subsequently with Not I/BamH I (NEB). For the negative controls, the fragments should be cut into three fragments, two of which should be visible in agarose gel electrophoresis assay, while for positive control and the StCas9 positive transformants only one fragment of 1980 bp should be detectable in the gel, because the Not I/BamH I sites (Fig. 1a, Sequence S3) should be removed if repaired.
Optimization of the StCas9 system in yeast
CRISPR guiding RNA optimization
For SP1.sgRNA.WT design, besides the 30 nt SP1 guide sequence (Guide), the locations of the various motifs recognized, including Match I, Bulge, Match II, Loop and TracrRNA are indicated (Fig. 2a). To investigate the sgRNA structure, including the minimal length of tracrRNA, loop structure, Match II region, Bulge motif, the minimal length of guide sequence, we designed series of SP1.sgRNA mutants (Figure S4) and constructed corresponding pTrp1 expression vectors through PCR method (for primer information, refer to Table S3). Yeast transformation assays for these SP1.sgRNA mutants coupled with pLeu2-bStCas9 and pRep.20.SP1 were conducted as described above. To prove that the programmed StCas9 system functions on different targets, the target sequences derived from one human gene CCR5 and three yeast genes ERG6, TPD1 and YAP1 (Figure S7A) were chosen. Corresponding sgRNA expression vectors and reporter vectors (pRep.20s) were constructed (for primer information, refer to Table S3), and yeast transformation assays coupled with pLeu2-bStCas9 were conducted as described above. Data (percentages of positive colonies) for each experiment group was adjusted relative to those of the controls when preparing histograms.
Target mismatch tolerance and sequence preference
To investigate the target mismatch tolerance and sequence preference for the StCas9 system, a series of SP1.sgRNA (Figure S4F) and target (Figure S7B) mutants were designed and corresponding vectors were constructed as above (for primer information, refer to Table S3). Yeast transformation assay with these SP1.sgRNA mutants for target mismatch tolerance was conducted with pLeu2-bStCas9 and pRep.20.SP1, while for target sequence preference was performed with pLeu2-bStCas9 and corresponding reporter vector mutants.
Investigation of the PAM pattern with the target
We initially used the 5′-GGG-3′ PAM according the SpCas9 system [8]. To define the PAM pattern, four mutants were firstly designed. As previous studies suggested that StCas9 system recognizes a 5′-NGGNG-3′ PAM [21–24], another two PAM patterns were later introduced. The reporter vectors for SP1 protospacer with different PAM patterns (Fig. 4a) were constructed as above (for primer information, refer to Table S3), and yeast transformation assay was performed with pLeu2-bStCas9 and SP1.sgRNA.WT expression vector.
Optimized CRISPR guiding RNA design and function assay
Based on the results of the experiments as shown above, we proposed an optimized sgRNA structure (sgRNA.Opti; Fig. 4c). The optimized sgRNA includes the full-length tracrRNA, a Match II motif of 5 bp, the Loop sequence containing a BamH I site, and if available, the guide sequence with the consensus N9VN5UACA (V: A/C/G, at least 19 nt). For functional assay, we generated the SP1.sgRNA.Opti design with 19 nt SP1 protospacer (specially, the G04U mutation was introduced, refer to Figure S4F) immediately adjacent to the PAM as guide sequence. Corresponding DNA fragment was synthesized directly (Genscript) and inserted into pTrp1 with Xho I/EcoR I sites to construct the expression vector. For SP1.sgRNA.Opti targeting, a G04T mutation was also introduced into two target sequences (SP1.Mu-04/SP1.P5M4) with different PAM patterns (Figure S7A), and corresponding reporter vectors were constructed via directly annealing oligonucleotides as described above. For further studies, the sgRNA.Opti design were employed for targeting the SP1.WT (nature SP1 protospacer without G04T mutation; corresponding sgRNA named as SP1F19.sgRNA.Opti), the target sequences from three yeast genes ERG6, TPD1 and YAP1 and another three from mouse ROSA26 locus (Figure S7A). Corresponding sgRNA expression vectors and reporter vectors were both constructed through the method of directly annealing oligonucleotides (for primer information, refer to Table S3). Yeast transformation assay was performed with pLeu2-bStCas9, sgRNA.Opti expression vectors and corresponding reporter vectors.
Codon humanized optimization for S. thermophilus Cas9 gene
We later codon optimized the S. thermophilus Cas9 gene (hStCas9). As studies for SpCas9 system suggested that two nuclear localization signals (NLS) are more efficient at targeting the nucleus [10], the same NLSs were introduced when synthetizing the hStCas9 DNA (Sequence S4, Genscript). The hStCas9 DNA fragment was cloned into pLeu2 to generate the yeast expression vector pLeu2-hStCas9. Yeast transformation assay for comparing bStCas9 and hStCas9 was performed with SP1.sgRNA.WT expression vector and the reporter vector pRep.20.SP1.
Yeast assay of the StCas9 system with target-integrated AH109 reporter strains
Yeast integrating reporter vector construction
Two yeast integrating reporter vectors pInt.20.SP1 and pInt.150.SP1 for integration of the PADH1-Gal4BD-AD-TADH1 reporter cassette was targeted at the HO gene locus (Figure S8A) via homologous recombination. These two plasmids were generated by several steps. The KanMX4 marker gene expression cassette, the homologous recombination (HR) arms (HO.L 906 bp and HO.R 500 bp) [49] and the reporter constructs with SP1 protospacer flanked by 20 or 150 bp SSA arms, were cloned by PCR (for primer information, refer to Table S3) and inserted into a cloning vector pBlue successively. For three other targets from yeast endogenous genes, ERG6, TPD1 and YAP1, target sequence fragments with PAM and sticky ends (Not I/BamH I) were generated by directly annealing oligonucleotides and inserted between the Not I/BamH I sites of pInt.20.SP1 replacing the SP1 protospacer to construct the integrating reporter vectors.
Selection and identification of the target-integrated AH109 reporter strains
The DNA fragments (about 5.2 kb) containing the KanMX4 gene expression cassette, the HR arms and the PADH1-Gal4BD-AD-TADH1 reporter constructs were cut from the integrating reporter vectors prepared as above by double digestion with Nhe I/Sac I (NEB), and were used to transform AH109 yeast strain (1 μg DNA per transformation). Positive colonies were selected on SD/+Ade/+His/+Leu/+Trp/+G418 agar plates, picked and incubated in YPDA/+G418 liquid medium. Genome DNAs were extracted from the cultures and were used as PCR templates with primers (Figure S8A) 5′ AD (designed according to the Gal4 AD sequence) and 3′ HO (designed to the downstream of HO.R arm according to HO locus sequence) for a fragment of 914 bp to confirm the integration. Positive integrated AH109 strains were stored in YPDA with 20 % glycerol at −70 °C.
Transformation of integrated AH109 reporter strains
Target-integrated AH109 reporter strains were streaked from the glycerol stocks on SD/+Ade/+His/+Leu/+Trp/+G418 agar plates and colonies were picked and incubated in YPDA/+G418 liquid medium before transformation. Transformations were conducted with the StCas9 and sgRNA expression vectors or different control plasmid groups (molar ratio = StCas9/pLeu2:sgRNA/pTrp1 = 1:1; 5.1–6.5 μg total plasmid DNA per transformation). As described above, the percentage of Gal4 positive colonies compared with transformed colonies was calculated to estimate the activity of the StCas9 system.
Identification for the restoration of Gal4 sequence within the yeast genome
Colonies of the StCas9 positive reporter-integrated strains were picked from the SD/−Ade/−His/−Leu/−Trp/+G418 selection plates and cultured in YPDA/+G418 liquid medium for genome DNA extraction. The genomic DNA was used as PCR templates and amplified with primers ADH1P.F/ADH1T.R for PADH1-Gal4BD-AD-TADH1 fragments. The PCR products were digested with Not I/BamH I to confirm the SSA-mediated DSB repair within the integrated reporter constructs.
Mammalian cell assay for the programmed StCas9 system
Cell line and medium
293T cells derived from human embryonic kidney 293 cells were purchased from the Cell bank of Chinese Academy of Sciences, and were routinely maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10 % fatal bovine serum (FBS, Gibco), 100 μg/mL penicillin and streptomycin at 37 °C with 5 % CO2.
Cell transfection and DNA extraction
293T cells were transfected with the StCas9 system related plasmids (total of 0.9–2.05 μg plasmid DNA per transfection) within 24-well plates by Sofast transfection reagent (Xiamen Sunma Biotechnology Co., Ltd. China). Genomic DNA was prepared with E.Z.N.A. DNA Kit (OMEGA Bio-Tek) by standard procedure.
Mammalian plasmid construction
Strategies for Cas9 and sgRNA expression
The bStCas9 and hStCas9 sequences were, respectively cloned and inserted into pLL3.7 by replacing the eGFP gene driven by the CMV promoter. Different sgRNA expression vectors (Figure S9B) were constructed by replacing mU6 (Hpa I/Xho I)-CMV.eGFP construct of the vector pLL3.7 with sgRNA fragments driven by different RNA polymerase III promoters (human U6, human H1 and mouse U6) or their modified versions. Among these sgRNA expression designs, a 82 bp processing-defective human tRNA variant (HtV4) sequence [25] and the 84 bp leader sequence (ABbox) from yeast RPR1 promoter [26] were introduced after the human U6 promoter, with the expectation that this would help with the stabilization of transcribed sgRNAs. Two additional designed sgRNA (sgRNA.Bi, crRNA:tracrRNA) vectors were constructed with the 66 nt crRNA and 85 nt tracrRNA independently expressed from different promoters. As well, another sgRNA vector (sgRNA-StCas9) was generated with the expression of sgRNA and Cas9 from the same plasmid.
Mammalian surrogate reporter vector construction
Similar with the yeast assay system, an eGFP reporter cassette was designed such that the multiple clone site (Not I-EcoR I-BamH I) flanked by 215 bp of direct repeats was inserted into the middle of eGFP gene, leading to the disruption of the open reading frame (Sequence S5; Figure S9A). For the integrating reporter vector (pInt.eGFP), the eGFP expression cassette was cloned between the Hind III/Xho I sites into a PiggyBac based vector with a puromycin resistance maker. For the surrogate reporter vector (pRep.eGFP), the puromycin resistance gene cassette was cut out by the restriction enzymes Spe I/Bgl II and replaced by a DsRed gene expression cassette (Xba I/BamH I). The proto-spacer sequences were cloned into Not I/BamH I sites by directly annealing oligonucleotides to generate different reporter vectors. The wild type or optimized SP1 (G04T) proto-spacer sequences were designed with a 5′-GGGTG-3′ (PAM5) PAM pattern (Figure S9A) to generate the reporter vectors, pRep.eGFP.SP1.PAM5/P5M4 and pInt.eGFP.SP1.PAM5/P5M4.
Mammalian cell assay of the StCas9 system with surrogate reporters in 293T
293T cells were transfected with different experiment (Cas9 and sgRNA expression and surrogate reporter vectors) or control plasmid groups within 24-well plates. Three different control transfections were performed, the StCas9 control, the sgRNA control and the linearized reporter vector control. For each transfection, three independent replicates were performed. Percentages of eGFP positive cells were calculated to estimate the activity of the programmed StCas9 system in 293T cells. Data were firstly corrected by the transfection efficiencies (DsRed positive cells) and then adjusted by subtracting the spontaneous repair frequencies generated by the controls.
Mammalian cell assay of the StCas9 system in target-integrated 293T reporter cell lines
Construction of the target-integrated 293T stable cell lines
293T cells were co-transfected with pInt.eGFP.SP1.PAM5/P5M4 and a transposase plasmid pBSII-hs-orf. Transfected cells were selected in medium containing 1 μg/mL puromycin (Sigma), and the medium was changed every 3 days. After selecting for 2 weeks, positive clones were picked and expanded for experiments. Genomic DNA of the stable cell clones were extracted and integration of the reporter constructs was confirmed by genome walking.
Mammalian cell assay with target-integrated 293T reporter cell lines
293T.SP1.PAM5 and 293T.SP1.P5M4, two target-integrated stable cell lines, were, respectively co-transfected with hStCas9 expression vector, an mRFP expression vector and the corresponding sgRNA expression vectors. The mRFP positive cells were collected by flow cytometry and used for genomic DNA preparation. A PCR assay was conducted with primers (eGFP.F and eGFP.R; Figure S9A; Sequence S5). The PCR products should generate two fragments of about 494 bp for the unrepaired or NHEJ repaired eGFP construct and 246 bp for the SSA repaired eGFP gene. Agarose gel electrophoresis analysis of the PCR products was carried out subsequently with the gel photographed. The bands were analyzed using Quantity One 4.6.2 software (BIO-RAD) with Gauss Model. The repair frequencies induced by SSA were calculated as follows: (R: Gauss model trace ratio; Fig. 6a). Both the fragments were recovered from the gel, inserted into the pGEM-T vector by T-A cloning and sequenced.
Endogenous gene targeting with the StCas9 system in 293T
For endogenous gene targeting, we selected three targets from human AASV1 and CCR5 loci (Table S2). We employed the strategy for expressing sgRNA.Opti and hStCas9 with one plasmid (Figure S9B). Besides, another SSA-RPG surrogate reporter vector was developed, containing a mRFP marker gene, a target/SSA arms-interrupted puromycin resistant gene (Puro r) and a downstream T2A-eGFP reporter gene, which is similar as Suresh Ramakrishna et al. [50] have recently reported. 293T cells were transfected with the sgRNA.Opti-hStCas9 vectors and corresponding SSA-RPG surrogate reporter vectors, selected with puromycin for 3 days for enrichment, collected and subjected to T7EI nuclease assay.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors would like to thank the colleagues in Professor Zhang’s lab for their excellent technical assistance and helpful discussions. This work was supported by National Science and Technology Major Project of China [2014ZX0801009B].
Footnotes
K. Xu and C. Ren contributed equally to this work.
References
- 1.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 2.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 3.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 4.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9(6):467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fonfara I, Le Rhun A, Chylinski K, Makarova KS, Lecrivain AL, Bzdrenga J, Koonin EV, Charpentier E. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42(4):2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013;10(5):726–737. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31(9):839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31(9):833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farzadfard F, Perli SD, Lu TK. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth Biol. 2013 doi: 10.1021/sb400081r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10(10):977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, Gersbach CA. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10(10):973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Heidrich N, Ampattu BJ, Gunderson CW, Seifert HS, Schoen C, Vogel J, Sontheimer EJ. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis . Mol Cell. 2013;50(4):488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis . Proc Natl Acad Sci USA. 2013;110(39):15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus . J Bacteriol. 2008;190(4):1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magadan AH, Dupuis ME, Villion M, Moineau S. Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3-Cas system. PLoS One. 2012;7(7):e40913. doi: 10.1371/journal.pone.0040913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karvelis T, Gasiunas G, Miksys A, Barrangou R, Horvath P, Siksnys V. crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus . RNA Biol. 2013;10(5):841–851. doi: 10.4161/rna.24203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli . Nucleic Acids Res. 2011;39(21):9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109(39):E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato Y, Sano M, Taira K. Analysis of processing-defective variants of human tRNA(Val) Nucleic Acids Res Suppl. 2003;3:283–284. doi: 10.1093/nass/3.1.283. [DOI] [PubMed] [Google Scholar]
- 26.Good PD, Engelke DR. Yeast expression vectors using rna-polymerase-Iii promoters. Gene. 1994;151(1–2):209–214. doi: 10.1016/0378-1119(94)90658-0. [DOI] [PubMed] [Google Scholar]
- 27.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 28.Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10(11):1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. ELife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, Iavarone AT, Charpentier E, Nogales E, Doudna JA. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343(6176):1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31(3):233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41(7):4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mansour WY, Schumacher S, Rosskopf R, Rhein T, Schmidt-Petersen F, Gatzemeier F, Haag F, Borgmann K, Willers H, Dahm-Daphi J. Hierarchy of nonhomologous end-joining, single-strand annealing and gene conversion at site-directed DNA double-strand breaks. Nucleic Acids Res. 2008;36(12):4088–4098. doi: 10.1093/nar/gkn347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuhar R, Gwiazda KS, Humbert O, Mandt T, Pangallo J, Brault M, Khan I, Maizels N, Rawlings DJ, Scharenberg AM, Certo MT. Novel fluorescent genome editing reporters for monitoring DNA repair pathway utilization at endonuclease-induced breaks. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiro R, Shitrit D, Qimron U. Efficient engineering of a bacteriophage genome using the type I-E CRISPR-Cas system. RNA Biol. 2014;11(1):42–44. doi: 10.4161/rna.27766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32(6):569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41(15):7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500(7463):472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gietz RD, Schiestl RH. Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2(1):38–41. doi: 10.1038/nprot.2007.15. [DOI] [PubMed] [Google Scholar]
- 46.Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2(1):31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 47.Wang L, Lin J, Zhang T, Xu K, Ren C, et al. (2013) Simultaneous screening and validation of effective zinc finger nucleases in yeast. PLoS One 8:e64687 [DOI] [PMC free article] [PubMed]
- 48.Ichikawa K, Eki T. A novel yeast-based reporter assay system for the sensitive detection of genotoxic agents mediated by a DNA damage-inducible LexA-GAL4 protein. J Biochem. 2006;139(1):105–112. doi: 10.1093/jb/mvj011. [DOI] [PubMed] [Google Scholar]
- 49.Voth WP, Richards JD, Shaw JM, Stillman DJ. Yeast vectors for integration at the HO locus. Nucleic Acids Res. 2001;29(12):E59. doi: 10.1093/nar/29.12.e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramakrishna S, Cho SW, Kim S, Song M, Gopalappa R, Kim JS, Kim H. Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nuclease-induced mutations. Nat Commun. 2014;5:3378. doi: 10.1038/ncomms4378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.