Abstract
The target of rapamycin (TOR) is a central regulator controlling cell growth. TOR is highly conserved from yeast to mammals, and is deregulated in human cancers and diabetes. TOR complex 1 (TORC1) integrates signals from growth factors, cellular energy status, stress, and amino acids to control cell growth, mitochondrial metabolism, and lipid biosynthesis. The mechanisms of growth factors and cellular energy status in regulating TORC1 have been well established, whereas the mechanism by which amino acid induces TORC1 remains largely unknown. Recent studies revealed that Rag GTPases play a central role in the regulation of TORC1 activation in response to amino acids. In this review, we will discuss the recent progress in our understanding of Rag GTPase-regulated TORC1 activation in response to amino acids. Particular focus will be given to the function of Rag GTPases in TORC1 activation and how Rag GTPases are regulated by amino acids.
Keywords: Target of rapamycin, TORC1, Amino acid, Rag GTPase, Cell growth
Introduction
The target of rapamycin (TOR) is a central regulator controlling cell growth and is highly conserved from yeast to human. TOR is a Ser/Thr protein kinase and exists in two structurally and functionally distinct protein complexes: TOR complex 1 (TORC1) and TOR complex 2 (TORC2). TORC1 integrates extracellular and intracellular signals and functions as a central regulator of cell growth (mass and size) and proliferation through regulation of a variety of cellular processes including translation, lipid synthesis, autophagy, and metabolism [1–9]. Compared to TORC1, our understanding of TORC2 is very limited. Deregulation of mammalian TORC1 (mTORC1) has been found in many human diseases, such as cancer and type 2 diabetes [10–14]. Thus, mTORC1 is an attractive drug target and its inhibitors (rapamycin, rapalogues, and ATP-competitive inhibitors) have been clinically used for the treatment of organ transplantation and solid tumors [15].
The protein kinase activity of mTORC1 is regulated by growth factors, cellular energy levels, stress, and amino acids. The mechanisms of mTORC1 regulation by growth factors and energy have been studied extensively. However, although amino acids serve as an essential signaling input for mTORC1 activation [16], how amino acids regulate mTORC1 is largely unknown. In 2008, two independent studies revealed that Ras-associated GTPases (Rag GTPases) play a central role in amino acid-induced mTORC1 activation [17, 18]. Recently, Ragulator (a trimeric protein complex p18/p14/MP1) and v-ATPase were described to work together with Rag GTPases, recruiting mTORC1 to the surface of lysosome for activation by Ras-homolog enriched in brain (Rheb) [19, 20]. Structural studies of Rag GTPases and Ragulator orthologs in yeast further deepen the understanding of Rag GTPase function in mTORC1 regulation [21, 22]. Vam6 was identified to interact with and function as a guanine nucleotide-exchange factor (GEF) for Gtr1p, homologue of RagA GTPase in yeast [23]. Leucyl-tRNA synthetase directly binds to Rag GTPases in response to amino acids and functions as a GTPase-activating protein (GAP) for RagD GTPases [24, 25]. Most recently, SH3BP4 was shown to directly associate with Rag GTPases and inhibit the formation of active form Rag GTPases complex, thereby negatively regulating mTORC1 activity [26]. These studies advanced our understanding of Rag GTPase function in the activation of TORC1. This review will highlight the recent research that delineates Rag GTPase-regulated TORC1 activation in response to amino acids.
Components of TOR complexes
Target of rapamycin was initially identified from genetic screening in yeast in which mutations of two genes TOR1 and TOR2 conferred resistance to rapamycin [27]. FKBP12 (FK506-binding protein, 12 kDa), an ubiquitous protein, is the intracellular rapamycin receptor in all Eukaryotes [28]. RAFT1, now termed mTOR (mammalian TOR), was found to be a direct TOR-FKBP12 complex and a mammalian homolog of yeast TOR protein [29]. mTOR is a 289-kDa Ser/Thr kinase that belongs to the family of phosphoinositide-3-kinase-related kinases (PIKK) and is functionally conserved from yeast to mammals [30].
mTOR exists and functions as the catalytic subunit in two distinct multiprotein complexes: rapamycin-sensitive mTOR complex 1 (mTORC1) and rapamycin-insensitive mTOR complex 2 (mTORC2) [2–4, 6, 31]. Besides mTOR, mTORC1 and mTORC2 share two other core components: mammalian lethal with SEC13 protein 8 (mLST8, also known as GβL) and DEP-domain-containing mTOR-interacting protein (Deptor). mTORC1 contains two unique subunits: regulatory-associated protein of mTOR (Raptor) and proline-rich AKT substrate 40 kDa (PRAS40). mTORC2 has three unique subunits: rapamycin insensitive companion of mTOR (Rictor), protein observed with Rictor (Protor) and mammalian stress-activated protein kinase-interacting protein (mSin1). The function of, and relationship between, each component are summarized in Table 1. For simplicity, TORC1 is referred to as TOR complex 1 in yeast and mTORC1 as TOR complex 1 in mammals hereafter.
Table 1.
TORC components and proteins involved in Rag GTPase regulation
| Mammals | Yeast | Function |
|---|---|---|
| mTOR | TOR1, TOR2 | TOR is the catalytic subunit of TOR complexes and belongs to the phosphoinositide-3-kinase-related kinases (PIKK) family of kinases |
| mLST8 | LST8 | LST8 is a shared subunit of mTORC1 and mTORC2. mLST8 deletion has no effect on mTORC1 activity in vivo or in vitro [103], while may be involved in modulating both the integrity and the kinase activity of mTORC2 [104] |
| Deptor | Deptor contains one PDZ and two DEP domains and interacts with the mTOR FAT domain through its PDZ domain. Deptor depletion in cells activates mTORC1 and mTORC2 signaling and in vitro kinase activity, indicating Deptor is a negative regulator of mTOR [105, 106] | |
| Raptor | KOG1 | Raptor functions as scaffold for complex assembly and substrate recruitment in mTORC1. Raptor is also involved in mTORC1 localization regulated by amino acid [18, 107, 108] |
| Rictor | AVO3 | Rictor functions as a scaffold for complex assembly and substrate recruitment in mTORC2 [104, 109] |
| PRAS40 | PRAS40 binds to mTORC1 via Raptor and kinase domain of mTORC1, and is the substrate of mTORC1. Phosphorylated PRAS40 inhibits mTORC1 autophosphorylation and mTORC1 kinase activity toward 4E-BP and PRAS40 itself [55, 110, 111] | |
| Protor | Protor (also known as PRR5) binds specifically to mTORC2 via Rictor and/or mSin1. Protor is not required for mTORC2 integrity or kinase activity, but Protor silencing inhibits Akt and S6K1 phosphorylation and reduces cell proliferation rates [55, 110, 112, 113] | |
| mSin1 | AVO1 | mSin1 disrupts the interaction between Rictor and mTOR, which is necessary for the assembly of mTORC2. mSin1 is also required for the phosphorylation of substrate Akt/PKB by mTORC2. Knockdown of Sin decreases Akt phosphorylation in both Drosophila and mammalian cells and diminishes Akt function in vivo [114, 115] |
| Rheb | Ras-homolog enriched in brain (Rheb) is a small GTPase, which is essential for mTORC1 activation. GTP bound Rheb stimulates mTOCR1 kinase activity [53, 54] | |
| TSC1/2 | Tuberous Sclerosis Complex 1/2 (TSC1/2) functions as a GTPase-activating protein to inactivate Rheb. TSC1/2 is a tumor suppressor and negative regulator of mTORC1 [48–52] | |
| RagA/B | Gtr1p | Rag GTPases form heterodimers and function as the central regulator of amino acid-induced mTORC1 activation through recruitment of mTORC1 to lysosomal surface. RagA/B plays a major role in TORC1 association, with RagA/BGTP and RagC/DGDP heterodimer as the fully active form [17, 18, 20, 21] |
| RagC/D | Gtr2p | |
| p18 | EGO1 | The trimeric protein complex, named Ragulator, localizes on the surface of the lysosome with p18 directly associated with membrane. Ragulator interacts with Rag GTPases and is essential for mTORC1 localization and activation by amino acids [20, 22, 87, 88] |
| p14,MP1 | EGO3 | |
| v-ATPase | ATP hydrolysis and associated rotation of v-ATPase are required to regulate the interaction between v-ATPase and Ragulator, and Rag GTPases regulated lysosomal localization for mTORC1 activation [19] | |
| hVam6 | Vam6 | Vam6 is a subunit of Class C Vps complex (HOPS) and a known GEF for Ypt7. Vam6 functions as a GEF for Gtr1p and leads to TORC1 activation in response to amino acids [23]. Mammalian Vam6 (hVPS39) has not been verified to be GEF for Rag GTPases so far |
| LRS | LRS | Leucyl-tRNA synthetase serves as a leucine sensor that couples amino acids stimulation to TORC1 activation by modulating Rag GTPases. In mammals, LRS functions as a GAP for RagD GTPase, while no evidence shows that cdc6 in yeast is GAP for Gtr1p or Gtr2p [24, 25] |
| p62 | p62 interacts with, and favors formation of, the Rag GTPase heterodimers and is also required for the interaction of mTORC1 with Rag GTPases for lysosomal localization in response to amino acids [74] | |
| SH3BP4 | SH3BP4 directly interacts with the inactive Rag GTPases complex. SH3BP4 negatively regulates amino acids-mTORC1 pathway by inhibiting Rag GTPases and Raptor binding, thereby preventing the translocation of mTORC1 to the lysosome [26] |
Downstream targets of mTORC1
mTORC1 regulates cell growth by controlling protein synthesis primarily through phosphorylation of two well-characterized targets: p70-S6 Kinase 1 (S6K1) and eukaryotic initiation factor 4E (eIF4E) binding protein 1 (4E-BP1) [32–34] (Fig. 1). Unphosphorylated 4E-BP1 binds to the translation initiation factor eIF4E and prevents its association with cap-binding complex, thus blocking mRNA translation [35]. mTORC1 phosphorylates 4E-BP1, releases eIF4E, and allows translational initiation [36, 37]. Phosphorylation of S6K1 by mTORC1 stimulates the subsequent phosphorylation and activation of S6K1 by phosphoinositide-dependent kinase 1 (PDK1) [38]. Active S6K1 can in turn promote mRNA translation by phosphorylation of several translational machinery proteins, such as ribosomal protein S6 (PRS6), S6K1 aly/REF-like target (SKAR), programmed cell death 4 (PDCD4), eukaryotic elongation factor 2 kinase (eEF-2K), eukaryotic initiation factor 4B (eIF4B), and cap-binding protein 80 (CBP80) [39]. These proteins collectively enhance translation efficiency and cell growth upon S6K1 phosphorylation. On the other hand, activated S6K1 can also phosphorylate insulin receptor substrate 1 (IRS1) and reduce mTORC1 activity, forming a negative feedback loop to attenuate the magnitude of growth factor signaling [40].
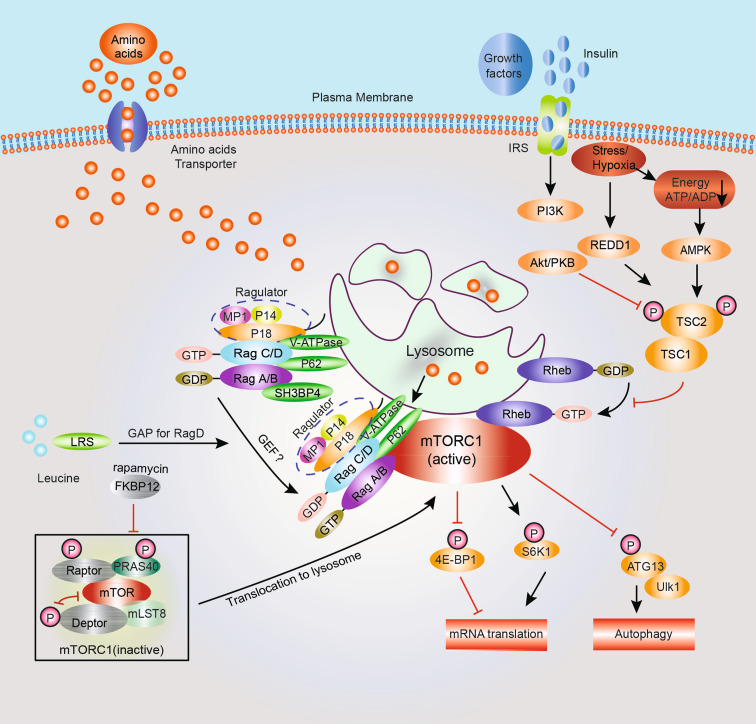
Fig. 1.
Rag GTPases in activation of mTORC1 signaling pathway in mammals. The mammalian target of rapamycin complex 1 (mTORC1) is a central regulator controlling cell growth through integrating signals from growth factors, cellular energy levels, stress, to amino acids. mTORC1 promotes mRNA translation by phosphorylation of S6K1 and 4E-BP1 and inhibits autophagy by phosphorylation of ATG13 and Ulk1. Growth factors stimulate mTORC1 through phosphorylation and inactivation of TSC1/2 complex. Akt phosphorylates TSC2 and inactivates the GAP activity of TSC1/2, leading to the activation of Rheb, which is essential for mTORC1 activation. AMPK is activated when cells are exposed to low energy (low ATP:ADP ratio). Stresses inhibit mTORC1 in part by reducing cellular ATP levels and leading to AMPK activation. Hypoxia also induces the expression of DNA damage response 1(REDD1), which activates TSC1/2 and inhibits mTORC1. Activated AMPK phosphorylates TSC2 and leads to activation of TSC1/2 GAP activity, Rheb inhibition, and mTORC1 inactivation. Rag GTPases play a central role in amino acid-induced mTORC1 activation. Rag GTPases form heterodimers and are localized on the late endosomal or lysosomal surface through the interaction with Ragulator complex (p18/MP1/p14). The heterodimerization of Rag GTPases does not depend on amino acids, whereas the nucleotide loading status of Rag GTPases is regulated by amino acids through proteins as indicated. LRS is a leucine sensor and functions as a GAP for RagD GTPase to stimulate mTORC1 activity. Moreover, amino acid accumulation in lysosomal lumen promotes v-ATPase-mediated regulation of nucleotide loading on Rag GTPases. Active Rag GTPase heterodimers (RagA/BGTP-RagC/DGDP) work together with Ragulator and v-ATPases to recruit mTORC1 to the lysosomal surface where Rheb is localized for mTORC1 activation. Adaptor protein p62 interacts with Rag GTPases to form a complex distinct from Ragulator-Rag, and may recruit mTORC1 to lysosomal surface for activation. SH3BP4 is a negative regulator of Rag GTPase, which prevents active Rag GTPases formation through interaction with RagAGDP, and thereby inhibits the interaction between Rag GTPases and Raptor for mTORC1 activation
TORC1 is one of the most important regulators of autophagy, a degradative process to maintain fundamental cellular activities upon stress and nutrient starvation. Activation of mTORC1 suppresses autophagy, and inhibition of mTORC1 significantly activates autophagy [41]. In Saccharomyces cerevisiae, TORC1 phosphorylates Atg13 and disrupts the Atg13-Atg1 complex formation, thus inhibiting Atg1 and suppressing autophagy [42] (Fig. 1). Nutrient starvation relieves this inhibition and allows autophagy initiation. In mammals, mTORC1 phosphorylates ATG13 and Ulk1 (mammalian homologues of Atg13 and Atg1) and inhibits autophagosome initiation through a mechanism that does not involve Ulk1 complex disruption, distinguishing it from yeast TORC1 inhibition [43–45]. Besides the prominent role in protein synthesis and autophagy, mTORC1 also regulates lipid synthesis, mitochondria metabolism, and lysosome biogenesis, which will not be discussed in this review [8, 46, 47].
Upstream signaling of mTORC1
TORC1 integrates signals from four major inputs: growth factors, energy status, stress, and amino acids, which collectively regulate cell growth (Fig. 1). In this review, we will describe studies of growth factors, stress, and energy status very briefly, and focus on amino acid-induced Rag GTPase regulation of TORC1/mTORC1 activity in yeast and mammals. Several reviews are referred to for detailed descriptions of previous studies on upstream signaling [1–5].
Growth factors stimulate mTORC1 through phosphorylation and inactivation of Tuberous Sclerosis Complex 1/2 (TSC1/2) complex by several signaling pathways, including PI3K-Akt, extracellular-signal-regulated kinase 1/2 (ERK1/2), and p90 ribosomal S6 kinase 1 (RSK1). As a negative regulator of mTORC1, the TSC1/2 complex functions as a GAP to inactivate the small GTPase Ras homolog Rheb [48–50]. Akt phosphorylates TSC2 and inactivates the GAP activity of TSC1/2, leading to the activation of Rheb [48, 51, 52]. Rheb is essential for mTORC1 activity; when loaded with GTP, it promotes mTORC1 activation by a yet-to-be identified mechanism [53, 54]. Alternatively, Akt activation by growth factors can activate mTORC1 through phosphorylation and disruption of the inhibition of PRAS40 on mTORC1 in a TSC1/2-independent manner [55].
Energy status is sensed by AMP-activated protein kinase (AMPK) [56]. When cells are exposed to low energy (low ATP:ADP ratio), AMPK is activated and phosphorylates TSC2, which leads to activation of TSC1/2 GAP activity, Rheb inhibition, and mTORC1 inactivation [57–59]. Moreover, in response to energy depletion, AMPK can directly phosphorylates Raptor, a mTORC1 component, and inactivate mTORC1 [60].
Stresses, like hypoxia and DNA damage, negatively regulate mTORC1 activity through multiple mechanisms. Hypoxia inhibits mTORC1 in part by reducing cellular ATP levels, thereby leading to AMPK activation [57]. Hypoxia also induces the expression of DNA damage response 1(REDD1), which activates TSC1/2 and inhibits mTORC1 via a poorly characterized mechanism [61, 62]. DNA damage communicates with mTORC1 in a p53-dependent manner. In response to DNA damage, p53 up-regulates the transcription levels of TSC2, REDD1, and PTEN to suppress mTORC1 activity [63–65].
Amino acids are required for mTORC1 activation and its depletion can completely block mTORC1 activity, which cannot be compensated by stimulation from growth factors or energy [16–18, 66]. Although amino acids play an essential role in mTORC1 activation, how amino acids stimulate mTORC1 remains largely unknown. Several studies reported that two proteins, hVps34, a class III PI3K kinase [67–70] and Ste20 family member, MAP4K3 [71, 72] are involved in amino acid-induced mTORC1 activation. However, the function of these proteins in the mTORC1 signaling pathway remains to be further elucidated [70, 73]. In 2008, two independent studies identified that Rag GTPases, a family of small GTPases, are essential for amino acid-induced mTORC1 activation [17, 18]. Rag GTPases are regulated by amino acid stimulation, and recruit mTORC1 to the lysosomal surface for activation [20]. Recently, several important advances have been made in the study of amino acid-stimulated Rag GTPases [19, 21–25, 74]. The mechanism of amino acid-regulated Rag GTPases and mTORC1 activation will be discussed in detail below.
Amino acid sensing
Amino acids are basic building blocks of protein and exist in l- or d-optical isomers [75]. Biological sensing of l-amino acids plays a key role linking amino acid metabolism to appropriate physiological responses such as exocrine, endocrine, and nutrient utilization. For example, the release of amino acids, especially aromatic amino acids in the gut, supports the integrity and defense of the gastrointestinal mucosa through coupling protein ingestion to responses such as gastric acid and pancreatic enzyme release [76–78]. When amino acid supply is limited, mammalian cells employ homeostatic mechanisms to rapidly inhibit protein synthesis and other metabolic processes.
The first study that linked amino acids and mTOR signal was reported in 1998. Withdrawal of amino acids from the nutrient medium of CHO-IR cells results in a rapid deactivation of p70 S6 kinase and dephosphorylation of eIF-4E BP1 indicating that mTOR is required for the response to amino acids [16]. Later, glutamine, leucine, and arginine were reported to be important upstream regulators of the TORC1 signaling pathway [66, 79, 80]. However, how intracellular amino acids regulated mTORC1 activation was a mystery for a long time until the discovery of the function of Rag GTPases in amino acid-induced mTORC1 activation in 2008 [17, 18].
Discovery of Rag GTPases
The first Rag GTPase gene, GTR1, was identified in S. cerevisiae in an effort to determine the sequence of PHO84, which encodes an inorganic phosphate transporter. Gtr1p null cells grow slower than WT cells, but is not lethal, indicating that Gtr1p is important but not essential for cell growth in yeast [81]. The first functional study of Gtr1p came from a yeast genetic screen for proteins associated with RCC1, a GEF for nuclear GTPase Ran/Gsp1p. Gtr1p was identified to interact with the RCC1 and function as a negative regulator for Ran GTPase cycle [82]. Gtr2p was later identified as a Gtr1p-associating protein and the two proteins form a heterodimer and negatively regulate the Ran cycle [83]. However, elucidation of the precise role that the Gtr1p/Gtr2p complex plays in the regulation of the Ran/Gsp1p cycle in yeast requires further investigation.
RagA and RagB were identified as the mammalian homologues of Gtr1p by a PCR-based cloning in 1995 [84]. Both RagA and RagB can rescue the growth defect caused by Gtr1p mutation in S. cerevisiae. Dominant negative RagA (RagAGDP) was distributed in discrete speckles in the nucleus while the WT and GTP bound RagA localized to the cytoplasm [85]. RagC and RagD were identified in a yeast two-hybrid screen using RagA as bait. RagC/D interacts with RagA/B and forms a heterodimer through their C terminus. The subcellular localization of RagC/D changes depending on the nucleotide-bound form of RagA, suggesting that RagA may shuttle between the nucleus and cytoplasm [86].
Rag GTPases are small guanosine triphosphatases (GTPases) and highly conserved from yeast to human. There are four Rag GTPases in human: RagA, RagB, RagC, and RagD. RagA and RagB are closely related to each other with 98 % sequence identity with the exception of 33 additional residues at the N terminus of Rag B. RagC and RagD are closely related to each other with 81 % sequence identity. The homology between RagA/B and RagC/D is limited. Yeast has only two Rag GTPases, Gtr1p, the ortholog of RagA/B, and Gtr2p, the ortholog of RagC/D. Rag GTPases form a heterodimer consisting of one Gtr1p-like GTPase (RagA or RagB) and one Gtr2p-like GTPase (RagC or RagD) [86]. The heterodimerization is characteristic of Rag GTPases and unique from other Ras family small GTPases, which usually function in monomeric form. Sequence analyses indicate that Rag GTPases contain a canonical N-terminal Ras-like GTPase domain and a unique C-terminal region with potential leucine zipper motif and coiled coil [86].
In an attempt to identify proteins involved in the exit from rapamycin-induced growth arrest, Dubouloz et al. [87] found that Gtr1p/Gtr2p interact with the fungal-specific proteins Ego1, Ego3, and form a vacuolar membrane-associated EGO (exit from growth arrest) complex. EGO complex localizes to vacuole through N-terminal myristoylation of Ego1 (Fig. 2). In conjunction with TOR, the EGO complex positively regulates rapamycin-induced macroautophagy [87]. The EGO complex (named GSE complex with an additional protein Ltv1) is required for proper sorting of Gap1p from the late endosome to the plasma membrane in response to amino acids. Gap1p, a general amino-acid permease in S. cerevisiae, is regulated by amino acid abundance and is thus often used as a model to study the intracellular sorting of membrane proteins. Gtr2p interacts with the C-terminal cytosolic domain of Gap1p, within which a tyrosine-containing motif is necessary to bind Gtr2p and sort Gap1p to the plasma membrane [88]. These results provide evidence that the Gtr1p/Gtr2p-containing complex functions upstream of TORC1 and is involved in amino acid signaling. However, the exact role of Rag GTPases in TORC1 regulation is still largely unknown.
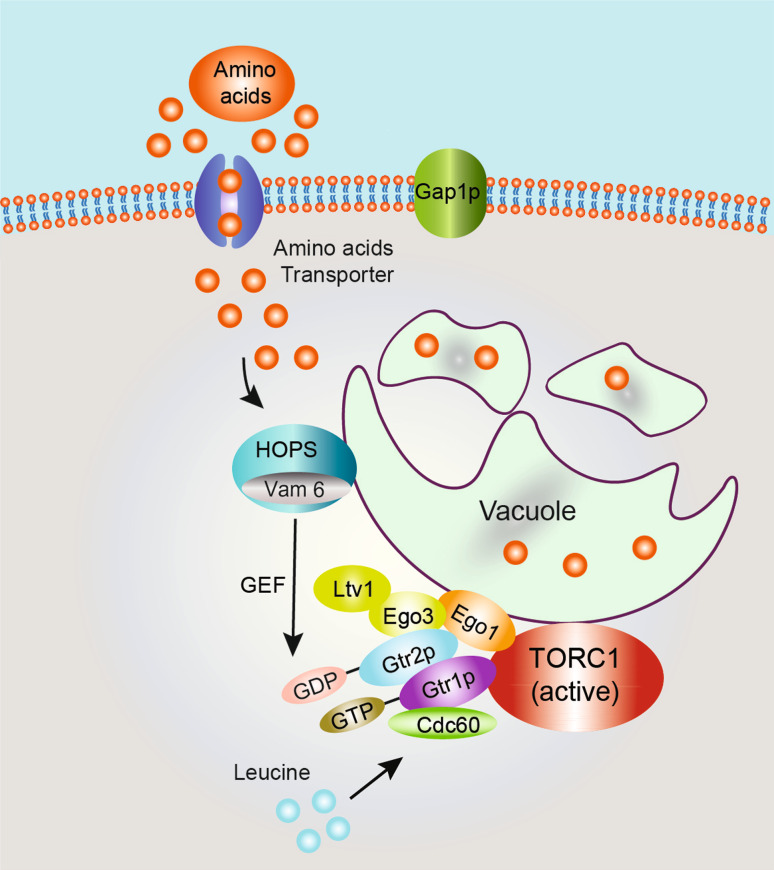
Fig. 2.
Amino acid-induced TORC1 activation in yeast. Gtr1p and Gtr2p in yeast are orthologs of mammalian Rag A/B and Rag C/D GTPases, respectively. Gtr1pGTP-Gtr2pGDP forms an active heterodimer to mediate amino acid-induced TORC1 activation. Gtr1pGTP-Gtr2pGDP heterodimer interacts with Ltv1, Ego1, and Ego3, and forms an EGO/GSE complex, which is localized to the vacuole surface via N-terminal myristoylation of Ego1. Vam6 interacts with Gtr1p and functions as a GEF for Gtr1p. Cdc60, a leucine sensor, binds to Gtr1pGDP and prevents GTP from hydrolysis, and therefore activates TORC1
Rag GTPases as a TORC1 regulator
The first direct link between Rag GTPases and the TORC1 signaling pathway was from two studies in 2008 [17, 18]. Using genetic and biochemical approaches, two groups independently demonstrated that Rag GTPases mediate mTORC1 activation upon amino acid stimulation in both Drosophila and mammalian cells (Fig. 1). Kim et al. used an RNA interference (RNAi) approach to screen 132 annotated Drosophila GTPases in Drosophila S2 cells and identified that dRag GTPases activate TORC1 in response to amino acids. Further studies in mammalian cells indicated a conserved mechanism whereby Rag GTPases promote cell growth and inhibit autophagy by activating mTORC1 [17]. Sancak et al. identified that RagC GTPase is an mTORC1-associating protein using a protein complex purification strategy. Rag GTPases interact with mTORC1 via Raptor in an amino acid-sensitive manner and are necessary for the activation of the mTORC1 by amino acids [18].
Further, both groups found that a RagA/B mutant that is constitutively bound to guanosine triphosphate (RagA/BGTP) activates mTORC1 in amino acid-free conditions. Conversely, expression of a guanosine diphosphate-bound RagA/B mutant (RagA/BGDP) prevents mTORC1 activation by amino acid stimulation. The heterodimer formation is required for mTORC1 activation and only when RagA/B is in GTP form (RagA/BGTP) and RagC/D in GDP form (RagC/DGDP), the heterodimer, such as RagAGTP/RagCGDP, is fully active [17, 18]. Sancak et al. [18] also found that the active Rag heterodimer directly interacts with Raptor, a key component of mTORC1 and proposed a model that the Rag GTPases promote the intracellular localization of mTORC1 from discrete locations in the cytoplasm to the late endosomal and/or lysosomal compartments that contain Rheb for mTORC1 activation.
Ragulator
The identification of Rag GTPase provides important clues for the mechanism of amino acid-induced mTORC1 activation. However, there are still some questions to be addressed. For example, sequence analyses indicate that Rag GTPases, unlike other small GTPases, do not have membrane-anchoring motifs. How do Rag GTPases recruit mTORC1 to the endosomal or lysosomal membrane? In 2010, Sancak et al. identified a trimeric protein complex named “Ragulator”, which directly interacts with and tethers the Rag GTPases heterodimer to the lysosomal surface. The interaction between Ragulator and Rag GTPases is not affected by amino acids but is absolutely required for proper intracellular localization and activation of mTORC1 [20]. The Ragulator complex contains three proteins: p18, p14, and MP1 (mitogen-activated protein kinase scaffold protein 1). In the trimeric protein complex, p18 localizes to the lysosomal membrane through N-terminal myristoylation and palmitoylation sites and serves as a platform for associating MP1 and p14 to the lysosome [89]. Although MP1 and p14 do not directly anchor to the lysosomal membrane, both proteins are required for the recruitment and activation of mTORC1 [20] (Fig. 1). Previous studies have demonstrated that p18, p14, and MP1 interact with each other and function as a positive regulator in the MEK-ERK signaling pathway [89–92]. Thus, the Ragulator serves as scaffolds in both mTORC1 and MEK-ERK pathways. Collectively, these studies demonstrate that Ragulator is required for amino acid-induced mTORC1 activation. In response to amino acids, Rag GTPases recruit mTORC1 to lysosomal membranes for activation through interacting with trimeric protein complex Ragulator (p18/p14/MP1) in mammalian cells. Rag GTPases interact with mTORC1 depending on amino acid availability, while the interaction between Rag GTPases and Ragulator is not regulated by amino acids.
The Ragulator proteins, p18, p14, and MP1, are conserved from fly to human, but could not be found in yeast based on sequence homology. EGO complex proteins Ego1 and Ego3 have been reported to interact with and localize Gtr1p and Gtr2p to vacuolar membrane in budding yeast, and are functionally important for the exit from growth arrest induced by rapamycin [87]. The crystal structure of Ego3 revealed a similar fold to that of MP1/p14, suggesting that Ego1/Ego3 complex is the structural and functional homologue of the Ragulator in yeast [22]. The observation that EGO mutants are involved in glutamate-induced cell growth, which is also dependent on TORC1 function, strongly indicates that the EGO complex may function upstream of TORC1 to mediate amino acid signaling. The GSE complex (GTPase-containing complex for sorting in the endosomes and is comprised of the EGO complex plus Ltv1) is also reported to be involved in proper sorting of Gap1p from the late endosome to the plasma membrane in response to amino acids availability. It is tempting to hypothesize that amino acids act upstream of the EGO complex to control Gap1p trafficking [88]. In support of the above speculation, Binda et al. [23] provided compelling evidence describing the EGO complex as a central factor in the amino acid responsiveness of TORC1 in yeast. Deletion of any component of the EGO complex suppresses TORC1 activation induced by intracellular amino acid increase.
As a component of the EGO complex, Gtr1p directly interacts with TORC1 and regulates its activity in an amino acid-sensitive manner. Interestingly, GTP-bound Gtr1p combined with wild-type Gtr2p or GDP-bound Gtr2p increases TORC1 activity. The Gtr1p/Gtr2p heterodimer is fully active only if Gtr1p is GTP loaded and Gtr2p is GDP loaded, which is consistent with previous observations [88]. Moreover, amino acid availability influences the nucleotide-loading state of Gtr1p. Leucine deprivation results in GTP hydrolysis of Gtr1p, causing decreased interaction between Gtr1p and TORC1, leading to TORC1 inactivation [23]. It would be interesting to know how amino acids are sensed by the EGO complex and whether other signaling events integrate amino acids signal to TORC1.
Notably, although the functions of Rag GTPases and the Ragulator in TORC1 regulation seem to be conserved in yeast and human, differences do exist. For example, unlike RagA/BGTP, which constitutively activates mTORC1 even in the absence of amino acids, Gtr1pGTP can only lead to partial activation of yeast TORC1, suggesting other signaling pathways may exist to transmit amino acids signal to TORC1 in yeast. Moreover, unlike in mammalian cells, the subcellular localization of TORC1 is not affected by amino acids in yeast [23].
v-ATPase
mTORC1 is recruited by Rag GTPases to the lysosomal surface in response to amino acids, suggesting a critical role for this organelle in mTORC1 activation. To identify whether lysosome-associated proteins are involved in this process, Zoncu et al. used RNAi to knockdown lysosomal proteins in Drosophila S2 cells, using phosphorylation of dS6K (T398) as a readout for dTORC1 activation. Surprisingly, the authors found that loss of vacuolar H+-adenosine triphosphatase ATPase (v-ATPase) components significantly suppressed amino acid-induced mTORC1 activation. v-ATPases are multisubunit complexes formed by a membrane-embedded V0 domain (a, d, e, c, c’, and c” subunits) responsible for proton translocation, and a cytosolic V1 domain (A–H subunits) providing energy through ATP hydrolysis [93, 94]. Knockdown of v-ATPase component genes leads to a decreased S6K1 phosphorylation and cell size, similar to dsRNA targeting dRagC. Two v-ATPase inhibitors also inhibit amino acid-induced mTORC1 activation in a concentration-dependent manner. v-ATPase is required for mTORC1 recruitment to the lysosomal membrane and functions downstream of amino acids but upstream of the regulation of nucleotide loading on Rag GTPases. v-ATPase physically interacts with Ragulator, and indirectly associates with Rag GTPases. The interaction between the V1 domain of v-ATPase, Ragulator and Rag GTPases is strengthened by amino acid deprivation, and weakened by amino acid stimulation, suggesting that amino acids induce conformational changes of the v-ATPase-Ragulator-Rag GTPase complexes by an unknown mechanism. An in vitro cell-free system was developed to recapitulate the recruitment of mTORC1 to Rag GTPases induced by amino acids on the lysosomal surface. Taking advantage of this in vitro system, the authors demonstrated that mTORC1 activation by amino acids initiates within the lysosomal lumen, where accumulated amino acids are sensed. Amino acid stimulation enhanced the binding of Raptor and purified lysosomes containing Rag GTPases. This suggests all the components responsible for signaling from amino acids sensing to Rag GTPase activation reside in lysosomes. ATP hydrolysis and the resulting rotation of v-ATPase, but not the lysosomal pH gradient set by v-ATPase, are required for amino acid-stimulated interaction between Raptor and RagB. As indicated in Fig. 1, the authors proposed an inside-out model in which amino acid accumulation in lysosomal lumen initiates mTORC1 activation through v-ATPase-mediated regulation of nucleotide loading on Rag GTPases [19].
Interestingly, mTORC1 was also found to regulate v-ATPase expression both in cells and in mice. The transcription factor TFEB is a target of mTORC1 and TFEB phosphorylation promotes expression of v-ATPase lysosomal genes [95]. It is tempting to speculate that mTORC1 and v-ATPases form a positive feedback loop, in which mTORC1 promotes expression of lysosomal v-ATPases and v-ATPases further activate mTORC1 in response to amino acids.
p62
p62, an adaptor protein, was previously found to interact with different signaling proteins and regulate multiple cellular functions, including cell survival, inflammation, apoptosis, and autophagy [96]. In an attempt to identify p62-interacting proteins, Duran et al. [74] found that p62 specifically interacts with Raptor and is essential for amino acid-induced mTORC1 activity. In p62-deficient cells, decreased levels of S6K1 and 4EBP1 phosphorylation were both observed in response to amino acid stimulation. Like Rag GTPases, p62 localizes to the lysosomal surface independent of amino acid availability and is required for proper translocation of mTORC1 to the lysosome in response to amino acids. p62 interacts with and favors formation of the active Rag GTPase heterodimer and is required for the interaction of mTORC1 with Rag GTPases. However, the immune-staining results indicate that only a small portion of cellular p62 interacts with Raptor and is involved in mTORC1 regulation, likely reflecting a complicated crosstalk between mTORC1 and other signaling pathways that p62 is involved in, such as autophagy. It is also surprising that p62 only associates with a small portion of RagC/D. Interestingly, p62 does not interact with Ragulator and loss of p62 has no effect on the integrity of Rag-Ragulator complex, suggesting that p62 may serve as an alternative platform with Rag GTPases on the lysosome for the recruitment of mTORC1 [74]. Several key issues remain to be addressed [97]. What are the functional differences between the two distinct complexes, p62/Rag and Ragulator/Rag? How does p62 regulate Rag GTPases and mTORC1 activation in response to amino acids?
Structure of Rag GTPases
To reveal how Rag GTPases interact with TORC1/mTORC1 in response to amino acid stimulation, our group determined the crystal structure of the yeast Rag GTPases complex: Gtr1p-Gtr2p in GTP-bound form [21]. The crystal structure revealed that heterodimeric GTPases adopt a pseudo-twofold symmetric organization. Functional analyses of RagA-RagC showed that both the GTPase domain and C-terminal dimerization are important for mTORC1 association upon amino acid stimulation. The switch regions of the GTPase domain in RagA are indispensable for interaction with Raptor, and hence mTORC1 activation, whereas the nucleotide-loading status of GTPase domain of RagC/D plays a minor role in mTORC1 interaction. One possible model is that mTORC1 mainly interacts with RagA GTPase domain, and GDP-bound RagC GTPase domain may fine-tune the interaction and thus provide specific recognition and regulation [21]. The dimerized C-terminal domains interact with p18 and mediate the localization of Rag GTPases. These results are consistent with the notion that the nucleotide-loading status of Rag GTPases affects mTORC1 interaction, but not cellular localization [20]. Interestingly, the dimerized C-terminal domains of Gtr1p-Gtr2p display a remarkable structural similarity to MP1/p14 and Ego3, which may facilitate the interaction between p18 and Rag GTPases in a yet-to-be-identified mechanism [21, 22, 91, 92].
Functional and structural studies of Rag GTPases shed light on understanding the Rag GTPases function in mTORC1 regulation. However, there are still important questions to be addressed. For example, how do RagA/B and RagC/D coordinately interact with Raptor and why is RagAGTP/RagCGDP required for full activation of mTORC1? The structure of Rag GTPases in complex with Raptor or mTORC1 may reveal the underlying mechanism. More importantly, how the Rag GTPases are regulated by amino acids is a key question to be addressed in studies of mTORC1. The very recent identification of potential GEF, GAP, and regulator proteins advanced our current understanding of the Rag GTPases function in mTORC1 activation [23–25].
Vam6: GEF for Gtr1p
In a yeast genetic screening, Binda et al. [23] found that Vam6 (also known as Vps39) functions as a GEF for Gtr1p. Vam6 is a subunit of homotypic fusion and vacuole protein sorting complex (HOPS, also referred to as Class C Vps complex), which regulates tethering, SNARE pair formation and fusion of vacuolar compartment in yeast, and delivers vesicle contents to the lysosome in metazoans [98]. HOPS complex has been shown to be required to provide intracellular amino acid homeostasis for proper Tor1 signaling in S. cerevisiae [99]. Vam6 is a known GEF for Ypt7, a homolog of mammalian Rab7 GTPase, which is involved in intracellular trafficking. Binda et al. found that overexpression of Vam6 suppresses the defect resulting from the overexpression of Gtr1p-GDP mutant. The recombinant Vam6 stimulates GDP release from both Gtr1p and Ypt7, but not a control Ras GTPase using an in vitro nucleotide exchange experiment. Like the loss of Gtr1p, loss of Vam6 resulted in a failure to exit from growth arrest induced by rapamycin and constitutively decreased TORC1 activity. Gtr1p interacts with Ego1 in a manner dependent on the Gtr1p GTP loading. The authors found that the interaction between Gtr1p and Ego1 is significantly decreased upon loss of Vam6 in yeast, suggesting an essential role for Vam6 in Gtr1p function in vivo. The authors concluded that Vam6 functions as a GEF for Gtr1p, activating Gtr1p in response to amino acids, which in turn results in TORC1 activation [23]. A recent study in Schizosaccharomyces pombe also indicated that Gtr1p/Gtr2p functions downstream of Vam6 and upstream of TORC1 in response to amino acid signals, which in turn stimulates cell growth and inhibits sexual differentiation [100]. However, several key questions remain to be addressed. For example, in contrast to the observation in mammalian cells, amino acids do not affect TORC1 localization in yeast [23]. How does Gtr1p/Gtr2p activate TORC1 in response to amino acid and how is Vam6 regulated by amino acid in yeast? In addition, comparing to Ego1 or Ego3 knockout strains, vacuolar morphology is greatly altered in Vam6 knockout strains [23, 87]. Since cellular localization of mTORC1 is essential for amino acid-induced mTORC1 activation in mammals, deletion of Vam6 in yeast may affect the TORC1 signaling through altering intracellular vacuolar physiology.
The function of human hVPS39 in the mTORC1 pathway was reported recently [101, 102]. As a GEF for Rab7 GTPase, hVPS39 regulates maturation of the late endosome. Flinn et al. showed that knockdown of hVPS39 blocked early/late endosomal conversion and inhibited amino acid-stimulated mTORC1 activation. The authors concluded that the compartmental integrity of the late endosome is critical for the amino acid-induced mTORC1 activation. Thus, in mammals, VPS39 regulation of the mTORC1 pathway may be non-specific, as the disruption of a key cellular process such as endosomal maturation is very likely to alter mTORC1 activation. Interestingly, mTORC1 is still localized to hybrid early/late endosome in the VPS39 knockdown cells, suggesting that VPS39 may not be a GEF for RagA/B since the late endosomal or lysosomal localization of mTORC1 is dependent on nucleotide loading of RagA/B GTPases [18, 20].
LRS: GAP for RagD GTPase
While the discovery of Rag GTPases, the Ragulator and Vam6 in mediating amino acid-induced TORC1/mTORC1 activation is exciting, the issue of how amino acids are sensed and how the loading state of Rag GTPases are regulated by amino acids remains elusive. Very recently, two groups independently reported that leucyl-tRNA synthetase (LRS) serves as a leucine sensor and mediates leucine-stimulated mTORC1 or TORC1 activation in both mammalian cells and yeast [24, 25]. LRS is primarily responsible for the synthesis of leucyl-tRNA, which is utilized for protein synthesis.
Han et al. [24] have recently found that LRS specifically interacts with mTORC1 in a leucine-dependent manner in mammalian cells. LRS is also required for amino acid-induced mTORC1 translocation to the lysosomal surface for activation. LRS mutants that disrupt leucine binding abolish leucine-induced S6K phosphorylation, suggesting LRS is a direct leucine sensor for cell growth. Specifically, LRS directly interacts with the very C terminus of RagD, preferentially in GTP bound form, and this interaction is leucine sensitive. Knockdown of RagD significantly suppressed leucine-induced S6K phosphorylation, suggesting that LRS functions upstream of Rag GTPases and downstream of amino acids. LRS enhances RagD GTPase activity and activates mTORC1 upon leucine stimulation, consistent with the fact that the GDP-bound RagD is the active form [17, 18]. Surprisingly, the authors found that LRS functions as a GAP for RagD, but not RagC, although both proteins share high sequence identity (Fig. 1).
Independently, Bonfils et al. [25] found that cdc60, the yeast LRS, is essential for leucine-stimulated TORC1 activation via the EGO complex, which is a structurally related mammalian Ragulator-Rag GTPase complex. However, strikingly different from mammalian LRS interacting with RagD, cdc60 physically interacts with Gtr1p in a leucine-dependent manner. Cdc60 can decrease GTP loading upon leucine starvation, in accordance with the previous data that amino acid stimulation increases the GTP loading of RagB [18]. Given the fact that leucine is one of the most potent single-amino acid stimuli for mTOR activation [16], LRS may serve as the major amino acid sensor for TORC1/mTORC1 activation. Interestingly, in yeast, cdc60 binds to Gtr1p but not Gtr2p and protects it from GTP hydrolysis, whereas mammalian LRS binds to RagD and functions as a GAP for RagD. Although amino acids are the most evolutionarily conserved signal input for TORC1, it is likely that the mechanism by which LRS conveys amino acid signaling to TORC1 might have diverged throughout evolution. The discovery that LRS functions as both a leucine sensor and a GAP for RagD opens a door for the understanding of amino acid-induced Rag GTPases activation. However, many questions remain to be addressed. For example, are there any other amino acids sensors that exist besides LRS? Is there any GAP for other Rag GTPases? Since RagA/BGTP/RagC/DGDP is the fully active form of the Rag GTPases, the regulation of Rag GTPases is likely rather complex and some other regulators may also be involved in Rag-dependent TORC1/mTORC1 activation in response to amino acids.
SH3BP4: a negative regulator of active Rag GTPase complex
Most recently, SH3BP4 was demonstrated to be a new member of the Rag GTPases binding proteins that are involved in amino acid-mTORC1 signaling [26]. SH3BP4 directly associates with Rag GTPases in a manner dependent on the nucleotide loading status. SH3BP4 preferentially interacts with the GDP-bound RagB in complex with RagC, and the binding affinity is further increased when RagC is charged with GDP rather than GTP. Leucine starvation enhanced the interaction of SH3BP4 with GDP-bound RagB, but not with GTP bound RagB. Knockdown of SH3BP4 results in a substantial increase in leucine-induced S6K phosphorylation, while SH3BP4 overexpression robustly inhibits S6K phosphorylation in TSC2 null cells, demonstrating that SH3BP4 is a specific negative regulator of amino acid-mTORC1 signaling. The effect of both knockdown and overexpression of SH3BP4 on mTORC1 signaling are compromised by dominant negative RagBGDP-RagCGTP complex or constitutively active RagBGTP-RagCGDP complex, respectively, indicating that SH3BP4 is a regulator of mTORC1 upstream of Rag GTPases. Overexpression of SH3BP4 blocks mTORC1 translocation to the lysosome by inhibiting Rag GTPases binding to Raptor. Through suppression of GDP dissociation, SH3BP4 prevents the amino acid-induced GTP loading of RagB GTPase. Consistent with this, SH3BP4 also inhibits amino acid-induced GTP changing of RagB. These data establish that SH3BP4 is a negative regulator for amino acid-mTORC1 signaling by directly associating with the inactive Rag GTPases heterodimer and preventing it from activation.
Nevertheless, many aspects with regard to how SH3BP4 regulates Rag GTPases and how SH3BP4 is regulated by amino acids remain unresolved. First, does SH3BP4 inhibit both RagB and RagC’s GDP release? Moreover, how is SH3BP4’s inhibition of Rag GTPases released under nutrient-rich conditions? It is possible that SH3BP4 might prevent the accessibility of Rag GTPases to their potentials GEFs. Amino acid stimulation triggers the dissociation of SH3BP4 from Rag GTPases and then allows GEFs to function on Rag GTPases. In addition, what are the relationships between SH3BP4 and other components, including Ragulator, v-ATPases, P62, and LRS that connects amino acids sensing to Rag GTPases activation? Is there a SH3BP4 homolog in yeast that plays a similar role in inhibiting Gtr1p/Gtr2p complex activation? Further investigation on these issues will shed light on the detailed mechanisms of how SH3BP4 regulates amino acid-mTORC1 signaling.
Conclusions
The role of Rag GTPases in TORC1 activation has just been established. Rag GTPases form heterodimers and are localized on the lysosomal surface through the interaction with Ragulator (p18/MP1/p14). The heterodimerization of Rag GTPases and the Ragulator-Rag GTPases interaction do not depend on amino acids, whereas the nucleotide-loading status of Rag GTPases is regulated by amino acids. Active Rag GTPase heterodimers (RagA/BGTP/RagC/DGDP) recruit TORC1 to the lysosomal surface, where Rheb is localized for TORC1 activation. In this process, ATP hydrolysis by the v-ATPase is necessary for amino acids to regulate the v-ATPase-Ragulator interaction and promote mTORC1 translocation. Vam6 functions as a GEF for Gtr1p in yeast and leucyl-tRNA synthetase interacts with RagD GTPase and may function as a GAP to regulate nucleotide-loading status of Rag GTPases in response to amino acids. Adaptor protein p62 interacts with Rag GTPases to form a complex distinct from Ragulator-Rag, and may recruit TORC1 to lysosomal surface for activation. SH3BP4 negatively regulates the amino acid-mTORC1 pathway by inhibiting Rag GTPases and Raptor binding, thereby preventing the translocation of mTORC1 to lysosome (Fig. 1).
Despite this exciting progress, our understanding of Rag GTPases in TORC1 activation is far from complete and there are still many important questions that remain to be addressed. For example, although leucyl-tRNA synthetase was just found to function as a GAP for RagD GTPase, the mechanism of amino acid sensing by Rag GTPases still remains unknown. Another important question is why active Rag GTPase heterodimers contain RagA/B in GTP form and RagC/D in GDP form. What is the functional difference between RagA/B and RagC/D? How does the nucleotide-loading status of Rag GTPases change in response to amino acids? Is this process regulated by the same or different GEF and GAP? Addressing these questions will advance our understanding of Rag GTPase-regulated mTORC1 activation and cell growth stimulated by amino acids. Since mTORC1 plays an important role in many human diseases and Rag GTPases are essential for mTORC1 activation, these findings may also open the door to new therapeutic strategies.
Acknowledgments
We apologize to those authors whose excellent work we did not reference directly in this review due to the limit of text space. We thank Professor Kun-liang Guan and Dr. Ryan C. Russell at UCSD for critical reading of the manuscript. This work was supported by grants from National Basic Research Program of China (2011CB918600, 2009CB918600) and the National Natural Science Foundation of China (31030019, 11079016, 30870493).
Footnotes
H. Yang and R. Gong contributed equally to this work.
References
- 1.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 2.Yang Q, Guan KL. Expanding mTOR signaling. Cell Res. 2007;17(8):666–681. doi: 10.1038/cr.2007.64. [DOI] [PubMed] [Google Scholar]
- 3.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J, Guan KL. Amino acid signaling in TOR activation. Annu Rev Biochem. 2011;80:1001–1032. doi: 10.1146/annurev-biochem-062209-094414. [DOI] [PubMed] [Google Scholar]
- 6.Russell RC, Fang C, Guan KL. An emerging role for TOR signaling in mammalian tissue and stem cell physiology. Development. 2011;138(16):3343–3356. doi: 10.1242/dev.058230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tormos KV, Anso E, Hamanaka RB, et al. Mitochondrial complex III ROS regulate adipocyte differentiation. Cell Metab. 2011;14(4):537–544. doi: 10.1016/j.cmet.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laplante M, Sabatini DM. An emerging role of mTOR in lipid biosynthesis. Curr Biol. 2009;19(22):R1046–R1052. doi: 10.1016/j.cub.2009.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duran RV, Hall MN. Regulation of TOR by small GTPases. EMBO Rep. 2012;13(2):121–128. doi: 10.1038/embor.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tee AR, Blenis J. mTOR, translational control and human disease. Semin Cell Dev Biol. 2005;16(1):29–37. doi: 10.1016/j.semcdb.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Inoki K, Corradetti MN, Guan KL. Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet. 2005;37(1):19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- 12.Harrington LS, Findlay GM, Lamb RF. Restraining PI3K: mTOR signalling goes back to the membrane. Trends Biochem Sci. 2005;30(1):35–42. doi: 10.1016/j.tibs.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Polak P, Hall MN. mTOR and the control of whole body metabolism. Curr Opin Cell Biol. 2009;21(2):209–218. doi: 10.1016/j.ceb.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 14.Dazert E, Hall MN. mTOR signaling in disease. Curr Opin Cell Biol. 2011;23(6):744–755. doi: 10.1016/j.ceb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Benjamin D, Colombi M, Moroni C, et al. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov. 2011;10(11):868–880. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- 16.Hara K, Yonezawa K, Weng QP, et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273(23):14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 17.Kim E, Goraksha-Hicks P, Li L, et al. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10(8):935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sancak Y, Peterson TR, Shaul YD, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320(5882):1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zoncu R, Bar-Peled L, Efeyan A, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334(6056):678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sancak Y, Bar-Peled L, Zoncu R, et al. Ragulator–Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141(2):290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gong R, Li L, Liu Y, et al. Crystal structure of the Gtr1p–Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation. Genes Dev. 2011;25(16):1668–1673. doi: 10.1101/gad.16968011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kogan K, Spear ED, Kaiser CA, et al. Structural conservation of components in the amino acid sensing branch of the TOR pathway in yeast and mammals. J Mol Biol. 2010;402(2):388–398. doi: 10.1016/j.jmb.2010.07.034. [DOI] [PubMed] [Google Scholar]
- 23.Binda M, Peli-Gulli MP, Bonfils G, et al. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell. 2009;35(5):563–573. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 24.Han JM, Jeong SJ, Park MC, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149(2):410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 25.Bonfils G, Jaquenoud M, Bontron S, et al. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol Cell. 2012;46(1):105–110. doi: 10.1016/j.molcel.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Kim YM, Stone M, Hwang TH, et al. SH3BP4 is a negative regulator of amino acid-rag GTPase-mTORC1 signaling. Mol Cell. 2012;46(6):833–846. doi: 10.1016/j.molcel.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253(5022):905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 28.Siekierka JJ, Wiederrecht G, Greulich H, et al. The cytosolic-binding protein for the immunosuppressant FK-506 is both a ubiquitous and highly conserved peptidyl-prolyl cis-trans isomerase. J Biol Chem. 1990;265(34):21011–21015. [PubMed] [Google Scholar]
- 29.Sabatini DM, Erdjument-Bromage H, Lui M, et al. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78(1):35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 30.Peng T, Golub TR, Sabatini DM. The immunosuppressant rapamycin mimics a starvation-like signal distinct from amino acid and glucose deprivation. Mol Cell Biol. 2002;22(15):5575–5584. doi: 10.1128/MCB.22.15.5575-5584.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40(2):310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holz MK, Ballif BA, Gygi SP, et al. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123(4):569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 33.Holz MK, Blenis J. Identification of S6 kinase 1 as a novel mammalian target of rapamycin (mTOR)-phosphorylating kinase. J Biol Chem. 2005;280(28):26089–26093. doi: 10.1074/jbc.M504045200. [DOI] [PubMed] [Google Scholar]
- 34.Gingras AC, Gygi SP, Raught B, et al. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13(11):1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pause A, Belsham GJ, Gingras AC, et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371(6500):762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 36.Hara K, Yonezawa K, Kozlowski MT, et al. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem. 1997;272(42):26457–26463. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- 37.Beamish H, Williams R, Chen P, et al. Rapamycin resistance in ataxia-telangiectasia. Oncogene. 1996;13(5):963–970. [PubMed] [Google Scholar]
- 38.Pullen N, Thomas G. The modular phosphorylation and activation of p70s6k. FEBS Lett. 1997;410(1):78–82. doi: 10.1016/S0014-5793(97)00323-2. [DOI] [PubMed] [Google Scholar]
- 39.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10(5):307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 40.Harrington LS, Findlay GM, Gray A, et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol. 2004;166(2):213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12(Suppl 2):1509–1518. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- 42.Kamada Y, Funakoshi T, Shintani T, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150(6):1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jung CH, Jun CB, Ro SH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20(7):1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ganley IG, du Lam H, Wang J, et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284(18):12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schieke SM, Finkel T. Mitochondrial signaling, TOR, and life span. Biol Chem. 2006;387(10–11):1357–1361. doi: 10.1515/BC.2006.170. [DOI] [PubMed] [Google Scholar]
- 47.Schieke SM, Phillips D, McCoy JP, Jr, et al. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem. 2006;281(37):27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- 48.Inoki K, Li Y, Xu T, et al. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17(15):1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garami A, Zwartkruis FJ, Nobukuni T, et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11(6):1457–1466. doi: 10.1016/S1097-2765(03)00220-X. [DOI] [PubMed] [Google Scholar]
- 50.Tee AR, Manning BD, Roux PP, et al. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13(15):1259–1268. doi: 10.1016/S0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 51.Manning BD, Tee AR, Logsdon MN, et al. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10(1):151–162. doi: 10.1016/S1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 52.Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol. 2002;4(9):658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 53.Stocker H, Radimerski T, Schindelholz B, et al. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila . Nat Cell Biol. 2003;5(6):559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- 54.Saucedo LJ, Gao X, Chiarelli DA, et al. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5(6):566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- 55.Sancak Y, Thoreen CC, Peterson TR, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25(6):903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 56.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8(10):774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 57.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115(5):577–590. doi: 10.1016/S0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 58.Inoki K, Ouyang H, Zhu T, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126(5):955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 59.Corradetti MN, Inoki K, Bardeesy N, et al. Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 2004;18(13):1533–1538. doi: 10.1101/gad.1199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gwinn DM, Shackelford DB, Egan DF, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30(2):214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brugarolas J, Lei K, Hurley RL, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18(23):2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reiling JH, Hafen E. The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes Dev. 2004;18(23):2879–2892. doi: 10.1101/gad.322704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ellisen LW, Ramsayer KD, Johannessen CM, et al. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol Cell. 2002;10(5):995–1005. doi: 10.1016/S1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- 64.Feng Z, Zhang H, Levine AJ, et al. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 2005;102(23):8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stambolic V, MacPherson D, Sas D, et al. Regulation of PTEN transcription by p53. Mol Cell. 2001;8(2):317–325. doi: 10.1016/S1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 66.Wang X, Campbell LE, Miller CM, et al. Amino acid availability regulates p70 S6 kinase and multiple translation factors. Biochem J. 1998;334(Pt 1):261–267. doi: 10.1042/bj3340261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280(38):33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 68.Gulati P, Gaspers LD, Dann SG, et al. Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metab. 2008;7(5):456–465. doi: 10.1016/j.cmet.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nobukuni T, Kozma SC, Thomas G. hvps34, an ancient player, enters a growing game: mTOR Complex1/S6K1 signaling. Curr Opin Cell Biol. 2007;19(2):135–141. doi: 10.1016/j.ceb.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 70.Juhasz G, Hill JH, Yan Y, et al. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila . J Cell Biol. 2008;181(4):655–666. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Findlay GM, Yan L, Procter J, et al. A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. Biochem J. 2007;403(1):13–20. doi: 10.1042/BJ20061881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yan L, Mieulet V, Burgess D, et al. PP2A T61 epsilon is an inhibitor of MAP4K3 in nutrient signaling to mTOR. Mol Cell. 2010;37(5):633–642. doi: 10.1016/j.molcel.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 73.Yan Y, Flinn RJ, Wu H, et al. hVps15, but not Ca2+/CaM, is required for the activity and regulation of hVps34 in mammalian cells. Biochem J. 2009;417(3):747–755. doi: 10.1042/BJ20081865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duran A, Amanchy R, Linares JF, et al. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44(1):134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pisarewicz K, Mora D, Pflueger FC, et al. Polypeptide chains containing d-gamma-hydroxyvaline. J Am Chem Soc. 2005;127(17):6207–6215. doi: 10.1021/ja050088m. [DOI] [PubMed] [Google Scholar]
- 76.McArthur KE, Isenberg JI, Hogan DL, et al. Intravenous infusion of l-isomers of phenylalanine and tryptophan stimulate gastric acid secretion at physiologic plasma concentrations in normal subjects and after parietal cell vagotomy. J Clin Invest. 1983;71(5):1254–1262. doi: 10.1172/JCI110875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Conigrave AD, Quinn SJ, Brown EM. L-amino acid sensing by the extracellular Ca2+-sensing receptor. Proc Natl Acad Sci USA. 2000;97(9):4814–4819. doi: 10.1073/pnas.97.9.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.San Gabriel A, Uneyama H (2012) Amino acid sensing in the gastrointestinal tract. Amino Acids [DOI] [PubMed]
- 79.Crespo JL, Powers T, Fowler B, et al. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc Natl Acad Sci USA. 2002;99(10):6784–6789. doi: 10.1073/pnas.102687599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Iiboshi Y, Papst PJ, Kawasome H, et al. Amino acid-dependent control of p70(s6k). Involvement of tRNA aminoacylation in the regulation. J Biol Chem. 1999;274(2):1092–1099. doi: 10.1074/jbc.274.2.1092. [DOI] [PubMed] [Google Scholar]
- 81.Bun-Ya M, Harashima S, Oshima Y. Putative GTP-binding protein, Gtr1, associated with the function of the Pho84 inorganic phosphate transporter in Saccharomyces cerevisiae . Mol Cell Biol. 1992;12(7):2958–2966. doi: 10.1128/mcb.12.7.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakashima N, Hayashi N, Noguchi E, et al. Putative GTPase Gtr1p genetically interacts with the RanGTPase cycle in Saccharomyces cerevisiae . J Cell Sci. 1996;109(Pt 9):2311–2318. doi: 10.1242/jcs.109.9.2311. [DOI] [PubMed] [Google Scholar]
- 83.Nakashima N, Noguchi E, Nishimoto T. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics. 1999;152(3):853–867. doi: 10.1093/genetics/152.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schurmann A, Brauers A, Massmann S, et al. Cloning of a novel family of mammalian GTP-binding proteins (RagA, RagBs, RagB1) with remote similarity to the Ras-related GTPases. J Biol Chem. 1995;270(48):28982–28988. doi: 10.1074/jbc.270.48.28982. [DOI] [PubMed] [Google Scholar]
- 85.Hirose E, Nakashima N, Sekiguchi T, et al. RagA is a functional homologue of S. cerevisiae Gtr1p involved in the Ran/Gsp1-GTPase pathway. J Cell Sci. 1998;111(Pt 1):11–21. doi: 10.1242/jcs.111.1.11. [DOI] [PubMed] [Google Scholar]
- 86.Sekiguchi T, Hirose E, Nakashima N, et al. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem. 2001;276(10):7246–7257. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- 87.Dubouloz F, Deloche O, Wanke V, et al. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell. 2005;19(1):15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 88.Gao M, Kaiser CA. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat Cell Biol. 2006;8(7):657–667. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]
- 89.Nada S, Hondo A, Kasai A, et al. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. EMBO J. 2009;28(5):477–489. doi: 10.1038/emboj.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wunderlich W, Fialka I, Teis D, et al. A novel 14-kilodalton protein interacts with the mitogen-activated protein kinase scaffold mp1 on a late endosomal/lysosomal compartment. J Cell Biol. 2001;152(4):765–776. doi: 10.1083/jcb.152.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kurzbauer R, Teis D, de Araujo ME, et al. Crystal structure of the p14/MP1 scaffolding complex: how a twin couple attaches mitogen-activated protein kinase signaling to late endosomes. Proc Natl Acad Sci USA. 2004;101(30):10984–10989. doi: 10.1073/pnas.0403435101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lunin VV, Munger C, Wagner J, et al. The structure of the MAPK scaffold, MP1, bound to its partner, p14. A complex with a critical role in endosomal map kinase signaling. J Biol Chem. 2004;279(22):23422–23430. doi: 10.1074/jbc.M401648200. [DOI] [PubMed] [Google Scholar]
- 93.Qi J, Wang Y, Forgac M. The vacuolar (H+)-ATPase: subunit arrangement and in vivo regulation. J Bioenerg Biomembr. 2007;39(5–6):423–426. doi: 10.1007/s10863-007-9116-8. [DOI] [PubMed] [Google Scholar]
- 94.Jefferies KC, Forgac M. Subunit H of the vacuolar (H+) ATPase inhibits ATP hydrolysis by the free V1 domain by interaction with the rotary subunit F. J Biol Chem. 2008;283(8):4512–4519. doi: 10.1074/jbc.M707144200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pena-Llopis S, Vega-Rubin-de-Celis S, Schwartz JC, et al. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 2011;30(16):3242–3258. doi: 10.1038/emboj.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137(6):1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Proud CG. A new link in the chain from amino acids to mTORC1 activation. Mol Cell. 2011;44(1):7–8. doi: 10.1016/j.molcel.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 98.Zlatic SA, Tornieri K, L’Hernault SW, et al. Metazoan cell biology of the HOPS tethering complex. Cell Logist. 2011;1(3):111–117. doi: 10.4161/cl.1.3.17279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zurita-Martinez SA, Puria R, Pan X, et al. Efficient Tor signaling requires a functional class C Vps protein complex in Saccharomyces cerevisiae . Genetics. 2007;176(4):2139–2150. doi: 10.1534/genetics.107.072835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Valbuena N, Guan KL, Moreno S (2012) The Vam6-Gtr1/Gtr2 pathway activates TORC1 in response to amino acids in fission yeast. J Cell Sci [DOI] [PMC free article] [PubMed]
- 101.Flinn RJ, Yan Y, Goswami S, et al. The late endosome is essential for mTORC1 signaling. Mol Biol Cell. 2010;21(5):833–841. doi: 10.1091/mbc.E09-09-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Flinn RJ, Backer JM. mTORC1 signals from late endosomes: taking a TOR of the endocytic system. Cell Cycle. 2010;9(10):1869–1870. doi: 10.4161/cc.9.10.11679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim DH, Sarbassov DD, Ali SM, et al. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11(4):895–904. doi: 10.1016/S1097-2765(03)00114-X. [DOI] [PubMed] [Google Scholar]
- 104.Wullschleger S, Loewith R, Oppliger W, et al. Molecular organization of target of rapamycin complex 2. J Biol Chem. 2005;280(35):30697–30704. doi: 10.1074/jbc.M505553200. [DOI] [PubMed] [Google Scholar]
- 105.Peterson TR, Laplante M, Thoreen CC, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137(5):873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao Y, Xiong X, Sun Y. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF(betaTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Mol Cell. 2011;44(2):304–316. doi: 10.1016/j.molcel.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim DH, Sarbassov DD, Ali SM, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110(2):163–175. doi: 10.1016/S0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 108.Hara K, Maruki Y, Long X, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110(2):177–189. doi: 10.1016/S0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 109.Sarbassov DD, Ali SM, Kim DH, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14(14):1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 110.Thedieck K, Polak P, Kim ML, et al. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS ONE. 2007;2(11):e1217. doi: 10.1371/journal.pone.0001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vander Haar E, Lee SI, Bandhakavi S, et al. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9(3):316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 112.Woo SY, Kim DH, Jun CB, et al. PRR5, a novel component of mTOR complex 2, regulates platelet-derived growth factor receptor beta expression and signaling. J Biol Chem. 2007;282(35):25604–25612. doi: 10.1074/jbc.M704343200. [DOI] [PubMed] [Google Scholar]
- 113.Pearce LR, Huang X, Boudeau J, et al. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J. 2007;405(3):513–522. doi: 10.1042/BJ20070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Frias MA, Thoreen CC, Jaffe JD, et al. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2 s. Curr Biol. 2006;16(18):1865–1870. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 115.Yang Q, Inoki K, Ikenoue T, et al. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006;20(20):2820–2832. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]