Abstract
The establishment of neuronal polarity and axonal outgrowth are key processes affecting neuronal migration and synapse formation, their impairment likely leading to cognitive deficits. Here we have found that the apoptotic protease activating factor 1 (Apaf1), apart from its canonical role in apoptosis, plays an additional function in cortical neurons, where its deficiency specifically impairs axonal growth. Given the central role played by centrosomes and microtubules in the polarized extension of the axon, our data suggest that Apaf1-deletion affects axonal outgrowth through an impairment of centrosome organization. In line with this, centrosomal protein expression, as well as their centrosomal localization proved to be altered upon Apaf1-deletion. Strikingly, we also found that Apaf1-loss affects trans-Golgi components and leads to a robust activation of AMP-dependent protein kinase (AMPK), this confirming the stressful conditions induced by Apaf1-deficiency. Since AMPK hyper-phosphorylation is known to impair a proper axon elongation, our finding contributes to explain the effect of Apaf1-deficiency on axogenesis. We also discovered that the signaling pathways mediating axonal growth and involving glycogen synthase kinase-3β, liver kinase B1, and collapsing-response mediator protein-2 are altered in Apaf1-KO neurons. Overall, our results reveal a novel non-apoptotic role for Apaf1 in axonal outgrowth, suggesting that the neuronal phenotype due to Apaf1-deletion could not only be fully ascribed to apoptosis inhibition, but might also be the result of defects in axogenesis. The discovery of new molecules involved in axonal elongation has a clinical relevance since it might help to explain neurological abnormalities occurring during early brain development.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-015-1927-x) contains supplementary material, which is available to authorized users.
Keywords: Centrosome, Golgi, Rab GTPases, NF1, Mitochondria, Neuro-rehabilitation
Introduction
The apoptotic protease activating factor 1 (Apaf1) is a key player in mitochondria-mediated apoptosis since it allows apoptosome formation upon the release of the cytochrome c from mitochondria. The assembling of the apoptosome, in turn, elicits caspase activation and the final stages of the apoptotic process [1]. It has recently been shown that Apaf1 is necessary not only for the death of the cells, but also for their survival [2, 3]. In particular, we have found that Apaf1 contributes to the correct functioning of the centrosome and, as a consequence, influences all the processes depending on centrosomes and microtubules, such as cell division and migration. Its deletion causes a higher responsiveness of cells to stressful conditions [3].
It is widely accepted that both cytoskeleton structuring and remodeling play a critical role in neuronal differentiation and, in particular, in axon specification and outgrowth [4, 5]. In most cells, the centrosomes are the main microtubule nucleation center [6]. Therefore, despite some controversy as to whether they might directly control neuronal polarity [7], centrosomes are considered crucial for neuronal differentiation and migration since they are a primary source of polarized microtubule nucleation which could specifically support axon outgrowth. In fact, in differentiating neurons, the disruption of centrosomal activity and the downregulation of centrosomal proteins impair microtubule organization, trafficking, and axon extension [8]. Moreover, mutations in genes coding for centrosomal proteins cause severe neurodevelopmental disorders [5]. In addition, centrosomes are closely connected to the Golgi apparatus [9], thus supporting an intense membrane traffic directed to the growing axon. The fast elongating axon requires the addition of a large amount of plasma membrane at the growth cone. Although the molecular events involved in the vesicle trafficking underlying axonal development need to be clarified, it has been found that many members of the Rab (Ras-related in brain) family of small GTPases play a key role in regulating transport vesicle formation, translocation, docking, and fusion needed for axonal growth [10, 11].
Although the mechanisms underlying cytoskeleton-driven axonal elongation are poorly understood, several molecules have been found to participate to the signaling orchestrating neuronal polarization and neurite outgrowth, including the glycogen synthase kinase-3β (GSK3β) [12, 13]. GSK3β is a serine-threonine kinase, which can be phosphorylated in Ser9 and inactivated by several kinases. During axonal growth, the phosphorylated-inactive form of GSK3β becomes enriched at the tip of axons with the subsequent dephosphorylation of its substrates, such as the collapsing-response mediator protein-2 (CRMP2) and the structural microtubule-associated proteins (MAPs), including Tau [4]. These proteins, in their dephosphorylated form, stabilize microtubules. Axon growth depends on the localized inactivation of GSK-3β and the accumulation of active dephosphorylated CRMP2 in the nascent axon tip. Dephosphorylated CRMP2 transports tubulin heterodimers via kinesin-1 into the axon and plays a critical role in axonal outgrowth by promoting microtubule assembly [14, 15]. Another key molecule involved in neuronal polarization is the kinase liver kinase B1 (LKB1). LKB1 downregulation inhibits axonal differentiation and impairs neuronal migration [16, 17], while LKB1 overexpression leads to the formation of multiple axons. LKB1 deletion in mammalian cerebral cortex causes the loss of axonal specification [18]. LKB1 acts on kinases essential for maintaining the dynamic state of microtubules necessary for axon outgrowth [19]. In addition, it has recently been reported that the over-activation of the regulator of cell energy homeostasis AMP-activated protein kinase (AMPK) also impairs axogenesis [20, 21].
Here we provide evidence that Apaf1 plays a key non-apoptotic role in cortical axon outgrowth in mammals, possibly by acting on centrosome, microtubule assembly, and trans-Golgi trafficking. We also demonstrate that Apaf1-deficiency causes a robust activation of AMPK, a reduction in LKB1 activity, and also CRMP2 hyper-phosphorylation, all contributing to the impairment of axonal growth.
Materials and methods
Cell culture and treatments
ETNA+/+ (wild-type, WT) and ETNA−/− (knockout, KO) cells were obtained as described elsewhere [22, 23]. They were routinely grown in DMEM (Life Technologies Ltd, Paisley, UK) + 10 % FBS (Life Technologies), at 33 °C in an atmosphere of 5 % CO2 in air. To induce differentiation, ETNA cells were treated with 50 μM Forskolin (Sigma-Aldrich Inc., St. Louis, MO, USA) 250 μM IBMX (Sigma-Aldrich), 200 nM TPA (Sigma-Aldrich), 10μM dopamine (Sigma-Aldrich), 10 ng/ml aFGF (Sigma-Aldrich) for 24 and 48 h. The differentiation medium was replaced every 24 h.
Primary mouse cortical neurons
Mouse primary cortical neurons (PCN) were obtained from cerebral cortices of Apaf1 +/+ (WT) and Apaf1 −/− (KO) E14.5 embryos [24]. All the experiments were performed according to the Animal Research Guidelines of the European Communities Council Directive (86/609/EEC). Dissected cortices were digested with trypsin 0.25 %/EDTA (Sigma-Aldrich) at 37 °C for 7 min, washed, and triturated to give a single cell suspension. Viable cells were counted by means of trypan blue staining and 1 × 105 cells were plated on poly-d-lysine (Sigma-Aldrich) coated coverslips (18 mm Ø) placed into 12 well plates in 25 mM glucose-containing MEM (Sigma-Aldrich) medium supplemented with 10 % fetal bovine serum (Life Technologies), 2 mM glutamine (Life Technologies), and 0.1 mg/ml gentamicin (Life Technologies). After 1 h, the medium was completely replaced with Neurobasal medium (Life Technologies) containing B 27 supplement (Life Technologies), 2 mM glutamine, and 0.1 mg/ml gentamicin. Cell cultures were kept at 37 °C in a humidified atmosphere containing 5 % CO2 and were analyzed after different days in vitro (DIV). Cell death was evaluated by direct cell count using optic microscopy following Trypan blue (Life Technologies) staining and by evaluating size and granulosity features by cytofluorimetric analysis on a FACScalibur flow cytometer (Becton-Dickinson).
Real-time PCR
RNA was isolated by using RNeasy Micro Kit (Qiagen Inc., Valencia, CA, USA), according to the manufacturer’s instructions. For reverse transcriptase reaction, first strand cDNA was synthesized with esaprimers by adding 1 μg of RNA with M-MLV Reverse Transcriptase (Life Technologies). Real-time PCR was performed by using SYBR Green PCR master mix (Applied Biosystems, Life Technologies). Real-time quantitation was performed by using the StepOne real-time PCR System (Applied Biosystems, Life Technologies). Data were normalized to L34. Resulting data were analyzed by the StepOne™ Software (v2.3) and fold-change was determined by using the method. All reactions were performed in triplicate.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Apaf1 Ct | 5′-GCTTGTCTCTGCTGGAGGATA-3′ | 5′-GTCTGTGAGGAGTCCCCAGT-3′ |
| Apaf1 Nt | 5′-CTTCCAGTGGCAAGGACAC-3′ | 5′-TCTGGGGTACTCCACCTTCA-3′ |
| GAP43 | 5′-CCAACGGAGACTGCAGAAA-3′ | 5′-GGTTTGGCTTCGTCTACAGC-3′ |
| L34 | 5′-GGTGCTCAGAGGCACTCAGGATG-3′ | 5′-GTGCTTTCCCAACCTTCTTGGTGT-3′ |
| MAP2 | 5′-GCTCCAAGTTTCACAGAAGGAG-3′ | 5′-AGGTTGGTTCAGATCAATATAAATAGG-3′ |
| PSD95 | 5′-CGCTACCAAGATGAAGACACG-3′ | 5′-CAATCACAGGGGGAGAATTG-3′ |
| Synaptophysin | 5′-AACAACAAAGGGCCAATGAT-3′ | 5′-TAGCCACATGAAAGCGAACA-3′ |
| Tau | 5′-GGCTCTACTGAGAACCTGAAGC-3′ | 5′-TCCAGCTTCTTATTAATTATCTGCAC-3′ |
| Tubb3 | 5′-CGGCAACTATGTAGGGGACT-3′ | 5′-CCTGGGCACATACTTGTGAG-3′ |
Immunocytochemistry
PCN were cultured as above described. Cells cultured on 12-well plates at 1 × 105 per well in 1 ml. After 48 h from seeding, cells were washed with phosphate-buffered saline (PBS) and fixed with methanol/acetone at −20 °C for 20 min or with 4 % paraformaldehyde (PFA) in PBS for 15 min. After permeabilization with 0.5 % Triton X-100 in PBS for 5 min (performed only after PFA fixation), cells were blocked in 2 % horse serum in PBS and incubated for 1 h at 37 °C with primary antibodies. We used anti-SMI312 R-100 (Covance Assay Designs), anti-γ-tubulin ab27074 (Abcam), anti-pericentrin ab4448 (Abcam), anti-NEDD1 sc-100961 (Santa Cruz), anti-GM130 610823 (BD Biosciences), anti-Diva sc-8739 (Santa Cruz), anti-Rab10 ab104859 (Abcam), and anti-HCA66 (kind gift form A. Merdes, CNRS-Pierre-Fabre, Toulouse, France). Cells were then washed in blocking buffer and incubated for 1 h at room temperature (RT) with labeled anti-mouse (Alexa Fluor 488; Molecular Probes, Life Technologies) or anti-rabbit 711-035-152 (FITC; Jackson ImmunoResearch) secondary antibodies. Nuclei were stained with Hoechst 33342 (Sigma-Aldrich). The samples were mounted in SlowFade Gold-mounting media. The images were acquired with a Leica TCS SP5 confocal microscope.
Time-lapse video microscopy
Cells cultured in 35-mm Petri dishes were washed in PBS. A Zeiss Axiovert-35 microscope equipped with a JVC digital CCD camera and the IAS2000 software (Deltasistemi, Rome, Italy) were used to take images every 5 min for an observation period of 20 h. Applying the “visualize” mode, these series of photographs were displayed as continuous time-lapse movies. Cells were seeded the day before recording into 35-mm dishes. Temperature was adjusted to 33 or 37 °C with a Peltier apparatus. A 50× magnification was applied.
Western blotting
WT and KO PCN were seeded on 12-well plates at 1 × 105 per well and were washed twice in ice-cold PBS and lysed at 4 °C in lysis buffer in RIPA buffer (50 mM Tris–HCl pH 7.4, 1 % NP40, 150 mM NaCl, 0.1 % SDS, 0.5 % deoxycholic acid) containing Protease Inhibitor Cocktail (Roche) and Phosphatase Inhibitor Cocktail (Sigma-Aldrich). A clear supernatant was obtained by centrifugation of lysates at 13,000g for 20 min at 4 °C. Protein concentration in the supernatant was determined by BCA Bradford protein assay (Bio-Rad, Hercules, CA, USA). Aliquots of total cell lysate were then separated on SDS-PAGE. Proteins were transferred to nitrocellulose membranes (Hybond, C Extra Amersham Biosciences). and were blocked overnight at 4 °C with 5 % non-fat milk in TTBS (TBS with 0.05 % Tween 20). Incubation with primary specific antibodies and horseradish peroxidase-conjugated secondary antibodies was performed in blocking solution for 1 h at room temperature. Equal loading of samples was confirmed by Tubulin or Gapdh normalization. We used anti-Actin A2066 (Sigma-Aldrich), Apaf1 AB16941 (Millipore), SMI312 R-100 (Covance), PSD95 MAB1598 (Millipore), Tau-1 MAB3420 (Millipore), Tau-5 AHB0042 (Life Technologies), p-Tau (ser231) 55313 (Anaspec), p-Tau (ser422) ab9664 (Chemicon), Diva sc-8739 (Santa Cruz), Rab8 D22D8 (Cell Signaling), Rab10 ab104859 (Abcam), Pan-cadherin C3678 (Sigma), Gapdh MAB374 (Millipore), Tubulin T5168 (Sigma-Aldrich), γ-tubulin T3559 (Sigma-Aldrich), p-AMPK 2535 (Cell Signaling), AMPK 2603P (Cell Signaling), p-GSK3β (ser9) ab75814 (Abcam), p-GSK3β (tyr216) GWB-516365, GSK3β ab93926 (Abcam), HCA66 (kind gift form A. Merdes, CNRS-Pierre-Fabre, Toulouse, France), Histone H1 05-457 (Millipore), pericentrin ab4448 (Abcam), NEDD1 sc-100961 (Santa Cruz), p-LKB1 sc-28465 (Santa Cruz), LKB1 ab58786 (Abcam), p-CRMP2 9397S (Cell Signaling), CRMP2 9393 (Cell Signaling), p-ACC 3661P (Cell Signaling), ACC 3676P (Cell Signaling), GM130 610823 (BD Biosciences), cleaved Caspase-3 9661 (Cell Signaling), and Tom20 sc-11415 (Santa Cruz), p-MARK 4836S (Cell Signaling) antibodies. Immunoreactive bands were visualized by SuperSignal West Pico Chemioluminescent substrate kit (Pierce). For loading control, anti-Tubulin or anti-Gapdh antibodies were used.
Nuclear–cytosolic fractionation
Cells were detached from the dish with trypsin and washed with PBS. Pellets were incubated in 5 volumes ice-cold hypotonic buffer (10 mM Hepes pH 7.4, 5 mM MgCl2, 10 mM NaCl, 1 mM DTT) containing a protease inhibitor cocktail (Sigma-Aldrich) and the phosphatase inhibitors Na3VO4 (1 mM) and NaF (1 mM), and homogenized in a glass–glass homogenizer (Dounce homogeniser) using 100 strokes of a tight-fitting pestle. The soluble cytoplasmic and the insoluble nuclear fractions were separated by centrifugation at 2000g for 10 min at 4 °C. The pellets containing the nuclei were extracted in the same buffer containing either 1 % Triton X-100 or 1 % Triton X-100 plus 250 mM NaCl. The extracts were centrifuged at 15,000g for 10 min at 4 °C and supernatants and pellets were analyzed by Western blotting.
Membrane isolation
PCN were incubated in 50 mM Tris–HCl pH 7.4, 10 mM MgCl2, 100 mM NaCl, 1 mM EDTA, 20 mM EGTA, and centrifugated at 700g for 10 min at 4 °C. The supernatant containing the membrane and cytosolic fractions was fractionated by ultracentrifugation at 100,000g for 10 min at 4 °C and the pellet containing the membranes was analyzed by Western blotting.
Assessment of ΔΨm
ΔΨm was measured using tetramethylrhodamine ethyl ester (TMRE, Molecular Probes). Cells were incubated at 37 °C for 15 min in media containing TMRE (50 nM). As a control for ΔΨm dissipation, cells were treated with 10 μM carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone (FCCP). Cells were then rinsed in fresh medium and detached from the dish. TMRE fluorescence was detected by flow cytometry on a FACScalibur flow cytometer (Becton-Dickinson).
Measurement of reactive oxygen species production
The reactive oxygen species (ROS)-sensitive probe H2DCF-DA (2′,7′-dichlorodihydrofluorescein-diacetate; 30 μM) was added directly to the culture medium for 1 h. Cells were washed with PBS and DCF fluorescence analyzed immediately upon reaction with ROS by flow cytometry using the FITC channel on a FACScalibur flow cytometer (Becton-Dickinson).
Histology
E12.5 and E14.5 embryos were fixed in 4 % paraformaldehyde (PFA) followed by cryoprotection in 30 % sucrose in PBS. The embryo heads were sectioned coronally (20-µm thick) with a cryostat (Leica; CM 3050S). Cryosections were permeabilized and blocked with PBS containing 0.1 % Triton X-100 and 5 % NGS. Primary antibodies were incubated in PBS containing 0.1 % Triton X-100 and 5 % NGS (mouse anti-Monoclonal Anti-Neurofilament 160/200 (1:500; Sigma-Aldrich, N2912)). Immunostaining was detected using Alexa fluorescent secondary antibody (1:600; Invitrogen) in PBS containing 5 % NGS. Slices were counterstained with Neurotrace Nissl 640/660 (1:100; Invitrogen Corporation, Carlsbad, CA, USA). Samples were mounted in Vectashield (Vector Laboratories, H 1000). For neurofilament analysis, the images from sections counterstained with Neurotrace Nissl were acquired on a confocal laser-scanning microscope (TCS SP5; Leica Microsystems, Milan, Italy) equipped with a 10× immersion objective (numerical aperture (NA) 0.3) for the low magnification and 63× immersion objective (NA 1.4) for high magnification. Confocal images (20-µm-thick z-stacks) were acquired, and Z-series were projected to two-dimensional representations. The contrast of the images was adjusted to enhance the fluorescence of neurofilaments.
Statistical analysis
All experiments were performed at least three times, unless otherwise indicated. Data are expressed as mean ± SEM. Data were compared by using Student’s t tests. Differences were considered as significant for p < 0.05.
Results
Apaf1 is highly expressed during the early phases of neuronal differentiation
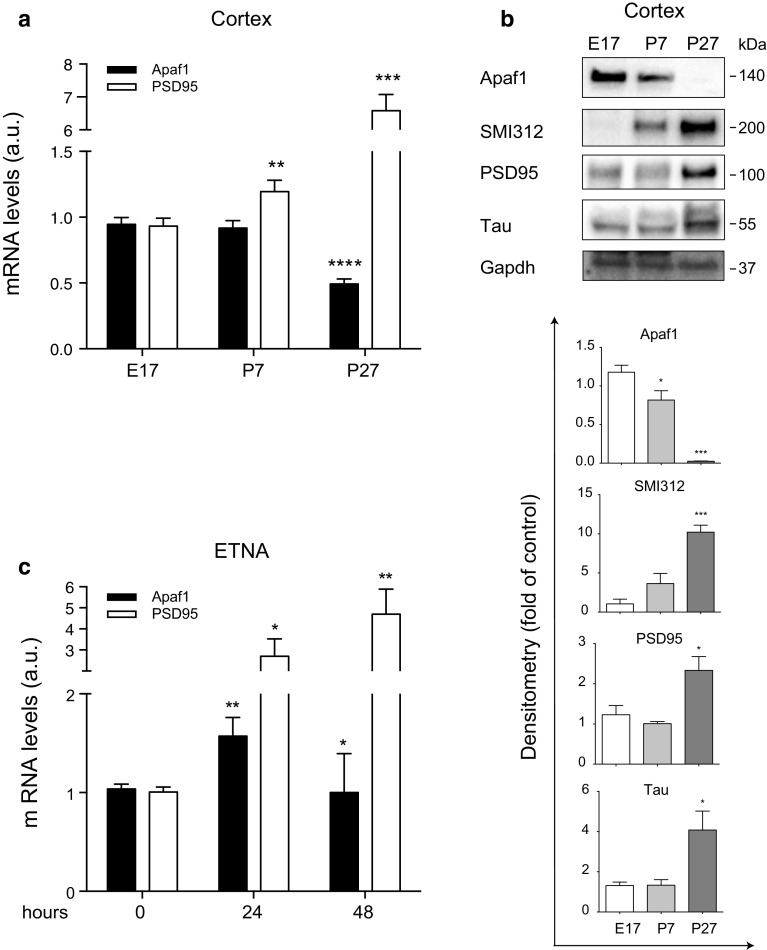
As previously reported, Apaf1 expression in mammalian adult brain decreases progressively during development from embryonic to early postnatal stages, with different timings depending on the neuron populations [25–27]. Here we have confirmed that both Apaf1 mRNA and protein levels are more abundant in embryonic brains (embryonic day 17; E17) compared to postnatal and adult brains (postnatal days 7 and 27; P7 and P27) (Fig. 1a, b). As control of proper brain development, we evaluated the expression of the synaptic marker PSD95, and of the axonal markers SMI312 and Tau (Fig. 1a, b), which increase in the adult versus the embryo along with the enhanced complexity of the inter-neuronal connections occurring during development [28]. Consistently, we observed that the Apaf1 mRNA levels also increase in neuronal precursor embryonic telencephalic naïve Apaf1 (ETNA) cells during the first 24 h of differentiation, while they tend to decrease later on (48 h) (Fig. 1c), when PSD95 keeps increasing, so confirming that ETNA cell differentiation proceeds regularly. Both these findings suggest that Apaf1 might play a role during early neuronal development, while it might be less crucial at later stages.
Fig. 1.
Apaf1 is highly expressed during early neuronal differentiation. a Mouse cortex extracts of embryonic stage 17 (E17), postnatal day 7 and 27 (P7 and P27) were assayed for Apaf1 and PSD95 mRNAs by quantitative real-time PCR. mRNA levels were normalized to L34 mRNA used as internal control. Data display the fold-changes of Apaf1 or PSD95 mRNA relative to E17 brains and are shown as the mean ± SEM; n = 3, **P ≤ 0.01, ***P ≤ 0.005, ****P ≤ 0.001 with respect to E17. b Mouse cortex extracts of E17, P7, and P27 were assayed for Apaf1, SMI312, PSD95, and Tau protein levels, and Gapdh was used as loading control. Density of immunoreactive bands was calculated using the software Image Lab (Bio-Rad), normalized for Gapdh, and reported as arbitrary units (shown as the mean ± SEM); n = 3, *P ≤ 0.05, ***P ≤ 0.005 with respect to E17. c ETNA+/+ cells differentiated in vitro for 24 and 48 h were assayed for Apaf1 and PSD95 mRNAs by quantitative real-time PCR. mRNA levels were normalized to L34 mRNA used as internal control. Data display the fold-changes of Apaf1 or PSD95 mRNA relative to undifferentiated cells (0 h) and are shown as the mean ± SEM; n = 3, *P ≤ 0.05, **P ≤ 0.01 with respect to 0 h
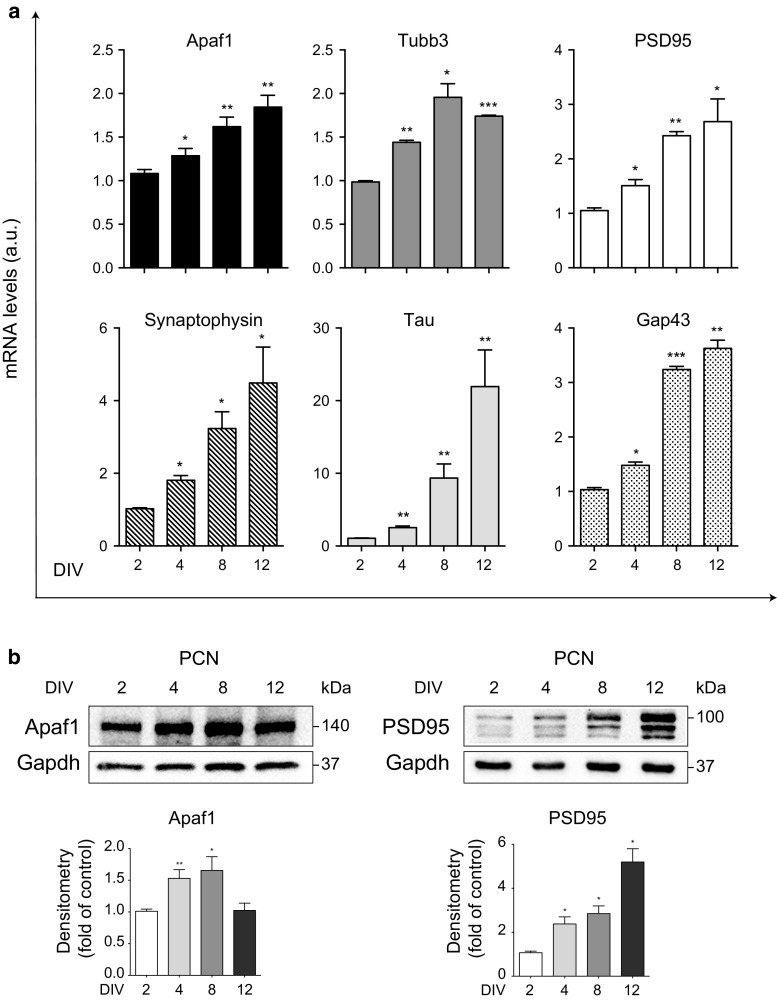
Since immortalized cell lines have several limitations as neuronal models, we adopted primary cortical neurons (PCN) to confirm and to circumstantiate our observations. We evaluated the expression of Apaf1 during PCN maturation in vitro, and we confirmed that Apaf1 mRNA increases during the first stages of differentiation (Fig. 2a; from 2 to 12 day in vitro, DIV) in parallel with the enhanced expression of the neuronal markers Tubb3, PSD95, Synaptophysin, Tau, and Gap43 (Fig. 2a). In addition, Apaf1 protein levels also increase during early PCN maturation (Fig. 2b; 2, 4, 8 DIV), although they start decreasing later on (Fig. 2b, 12 DIV) when, as expected, PSD95 keeps increasing (Fig. 2b). This evidence further corroborates our hypothesis that Apaf1 is necessary mostly for the early phases of neuronal differentiation.
Fig. 2.
Apaf1 increases during early differentiation of primary cortical neurons (PCN) in vitro. a Primary cortical neurons (PCN) from WT mouse embryos at E13.5 were cultured for 2, 4, 8, and 12 day in vitro (DIV) and assayed for Apaf1, Tubb3, PSD95, Synaptophysin, Tau, and Gap43 mRNAs by quantitative real-time PCR. mRNA levels were normalized to L34 mRNA used as internal control. Data display the fold-changes of Apaf1, Tubb3, PSD95, Synaptophysin, Tau, and Gap43 mRNAs relative to 2 DIV PCN and are shown as the mean ± SEM.; n = 3, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005 with respect to 2 DIV. b PCN from WT mouse embryos at E13.5 were cultured for 2, 4, 8, and 12 DIV and assayed for Apaf1 and PSD95 protein levels, using Gapdh as loading control. Density of immunoreactive bands was calculated using the software Image Lab (Bio-Rad), normalized for Gapdh, and reported as arbitrary units (shown as the mean ± SEM). *P ≤ 0.05, **P ≤ 0.01 with respect to 2 DIV
Apaf1-deletion impairs neuronal differentiation
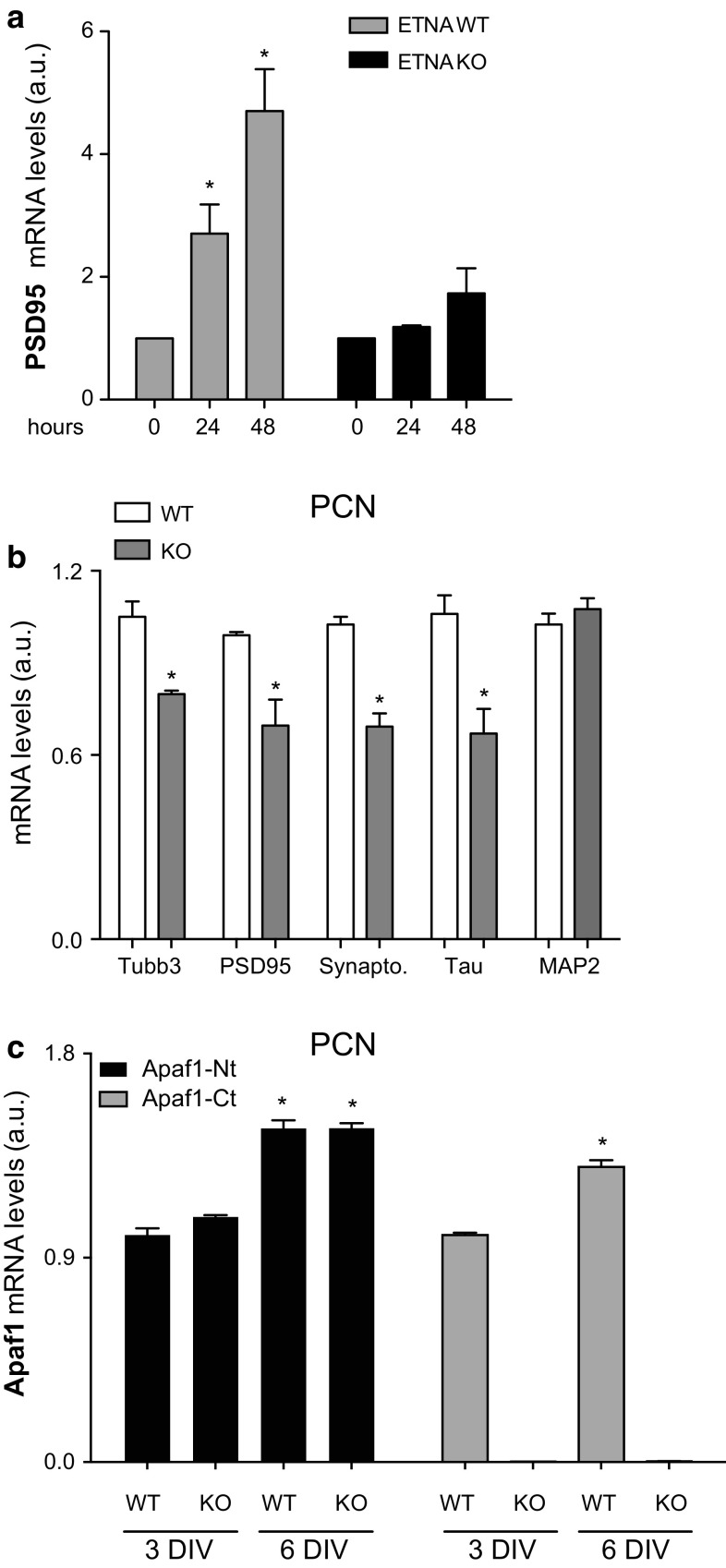
To decipher if the high level of Apaf1 expression detected during PCN maturation might indicate a critical role for Apaf1 in the first stages of neuronal differentiation, we analyzed ETNA cells devoid of Apaf1, i.e., ETNA-knockout (ETNA-KO). Results shown in Fig. 3 indicate a lower expression of the neuronal marker PSD95 (Fig. 3a), suggesting that ETNA-KO differentiation is reduced compared to wild-type (WT) ETNA. To substantiate our hypothesis, we performed a deep analysis of PCN devoid of Apaf1 by analyzing a number of markers expressed during neuronal differentiation. In particular, we observed that the neuronal markers Tubb3, PSD95, Synaptophysin, and Tau were significantly less expressed at mRNA levels in in vitro differentiating Apaf1-KO PCN compared with their WT counterpart (Fig. 3b, 3 DIV). These findings clearly demonstrate that Apaf1 is necessary to guarantee a proper neuronal differentiation.
Fig. 3.
Apaf1-deficiency leads to decreased expression of neuronal markers in ETNA cells and PCN. a ETNA+/+ and ETNA−/− cells in vitro differentiated for 24 and 48 h were assayed for PSD95 mRNAs by quantitative real-time PCR. mRNA levels were normalized to L34 mRNA used as internal control. Data display the fold-changes of PSD95 mRNA relative to undifferentiated cells (0 h) and are shown as the mean ± SEM; n = 3, *P ≤ 0.05 with respect to 0 h. b, c PCN from Apaf1 +/+ (WT) and Apaf1 −/− (KO) mouse embryos at E13.5 were cultured for 3 DIV and 6 DIV and assayed for Tubb3, PSD95, Synaptophysin, Tau, and Map2 (b) or Apaf1 (c) mRNAs by quantitative real-time PCR. mRNA levels were normalized to L34 mRNA used as internal control. b Data display the fold-changes of Tubb3, PSD95, Synaptophysin, Tau, and Map2 mRNAs relative to WT PCN at 3 DIVand are shown as the mean ± SEM; n = 3, *P ≤ 0.05 with respect to WT. c Data display the fold-changes of Apaf1 N-terminal (Nt) or C-terminal (Ct) mRNAs relative to WT PCN at 3 and 6 DIV and are shown as the mean ± SEM; n = 3, *P ≤ 0.05 with respect to 3 DIV. Apaf1-KO embryos produce a fusion transcript (Apaf1 trapped gene-LacZ) in which Apaf1 N-terminus is still present, while the Apaf1 C-terminus (and, consequently, the complete mRNA) is not [24]
Interestingly, we also found that, although the levels of neuronal markers decrease in Apaf1-null PCN, the transcription rate of the Apaf1 mRNA (Apaf1-N-terminus; Apaf1-Nt) in PCN genetically ablated of Apaf1 is comparable to that of WT PCN during neuronal differentiation (3 DIV versus 6 DIV) (Fig. 3c). This further reinforces our finding since it shows that the decreased expression of neuronal markers has no impact on Apaf1 transcription, which continued to be required and induced (Fig. 3c), while, vice versa, Apaf1-absence impairs neuronal transcription (Fig. 3b).
Axonal outgrowth is affected in Apaf1-deficient cortical neurons
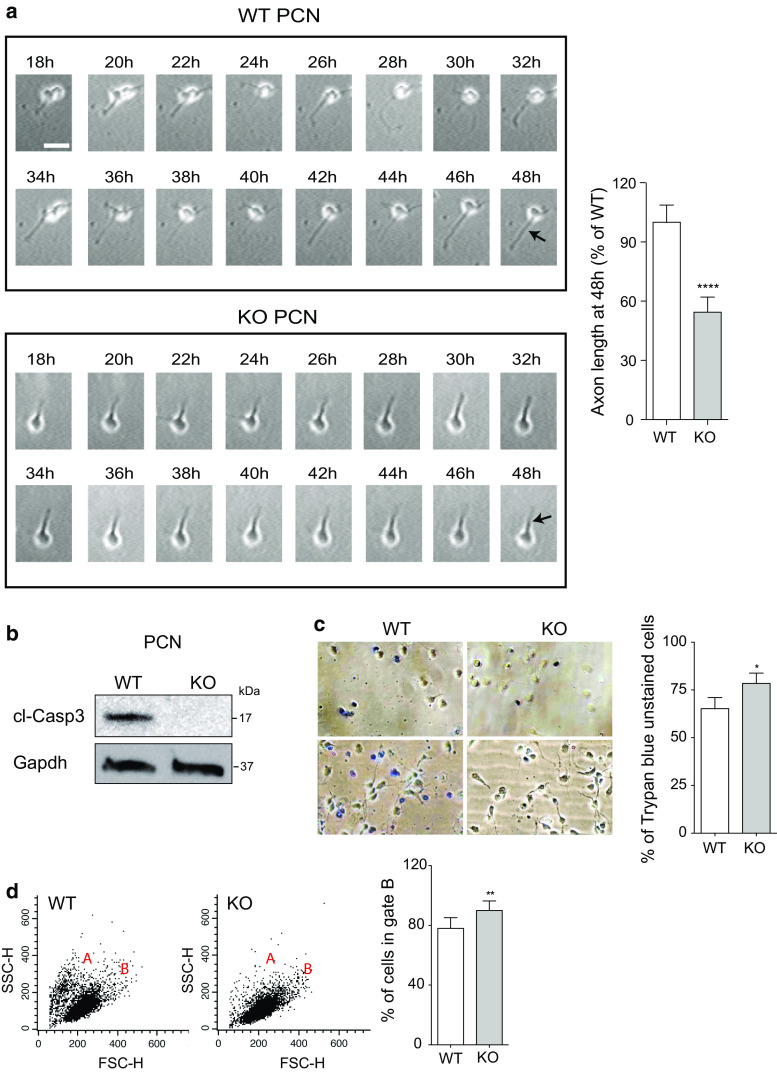
A careful evaluation of the markers expressed during neuronal differentiation revealed that, in contrast to Tau, Synaptophysin, and PDS95, MAP2 is not affected by Apaf1 deficiency (Fig. 3b). It is worthwhile noting that MAP2 is mostly expressed in dendrites, whereas Tau, Synaptophysin, and PSD95 are axonal and synaptic markers [29–31]. We thus hypothesized that Apaf1 was necessary specifically for a proper axonal differentiation. To investigate Apaf1’s potential role in axon specification and elongation, we evaluated the length of axons in PCN seeded in an appropriate number so as to enable us to analyze single neurons. Apaf1-WT and Apaf1-KO PCN were fixed at 2 DIV, and immunofluorescence of the axonal marker SMI312 was performed. Results obtained argued for axon length of WT PCN being significantly higher compared to that of Apaf1-deficient neurons (Fig. 4a). To investigate a possible impairment of axonal specification dependent on Apaf1-deficiency, we also counted the number of neurons with multiple axon-like neurites; we found out that their percentage is very small in our PCN cultures and tends to increase in Apaf1-null neurons, although not significantly so (Fig. 4a). We then quantified by Western blot (WB) the amount of specific axonal markers. In line with our previous observations, we found that the protein levels of SMI-213 and Tau were reduced in Apaf1-KO PCN (Fig. 4b). In order to rescue the axon phenotype, we transfected the Apaf1 transgene in Apaf1-KO PCN. However, as previously reported by others [32, 33], we obtained a high level of cell death which prevented an efficient re-expression of the protein in neurons (data not shown). This was likely due to the enormous amount of the pro-apoptotic protein Apaf1 accumulating in each neuron upon transfection. Finally, we performed a time-lapse analysis of cortical neurons during differentiation: We compared axons elongation in Apaf1-WT and Apaf1-KO PCN between 2 DIV and 3 DIV (namely, from 18 to 48 h after plating). Results shown in Fig. 5a clearly demonstrate that the axons become longer and that their elongation is more dynamic in WT PCN than in KO neurons.
Fig. 4.
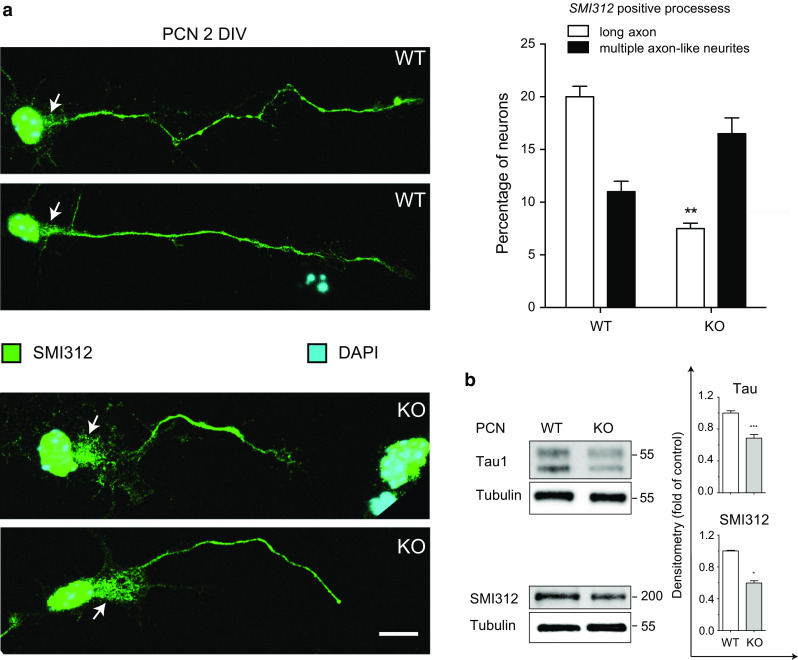
Axon formation is impaired in Apaf1-deficient PCN in vitro. a Representative fluorescence microscopy images of PCN isolated from E13.5 WT and Apaf1-KO mice embryos, cultured for 2 DIV, and stained with anti-SMI312 and DAPI to highlight neuron axons and nuclei, respectively. Determination of the percentage of single neurons whose axons are more than five times longer than cell body (long axon) and of the percentage of single neurons with more than two SMI312 positive neurites (multiple axon-like neurites) was performed in WT and KO PCN at 2 DIV and were shown as the mean ± SEM; the axonal length of, at least, 40 neurons was measured in several images collected in 3 independent experiments, **P ≤ 0.01 with respect to WT. Arrows point to the cell body area from where the axon originates. Scale bar represents 10 μm. b WT and KO PCN at 3 DIV were assayed for Tau and SMI312 protein levels, and Tubulin was used as loading control. Density of immunoreactive bands was calculated using the software Image Lab (Bio-Rad), normalized for Tubulin and reported as arbitrary units (shown as the mean ± SEM; *P ≤ 0.05, ***P ≤ 0.005 with respect to WT
Fig. 5.
Slow axonal growth and reduced cell death in Apaf1-KO PCN. a Time-lapse analysis of PCN isolated from E13.5 WT and Apaf1-KO mice embryos and seeded at the same density was performed. 30 single neurons for each genotype were recorded in three independent experiments, one representative example for each genotype being reported. The axon elongation was recorded starting from 18 h after plating (18 h) and followed for 30 h (48 h), as indicated by the arrows. Scale bar represents 50 μm. Determination of the axon length of isolated KO neurons with respect to the WT ones performed at 48 h after plating is shown as the mean ± SEM; n = 30, ****P ≤ 0.001 with respect to WT. b WT and KO PCN at 2 DIV were assayed for cleaved Caspase3 (cl-Casp3) protein levels, and Gapdh was used as loading control. c WT and KO PCN at 2 DIV were stained with Trypan blue and unstained cells were counted and shown as percentage of total cells. At least 2000 cells, in three independent experiments, were counted for each genotype. d Flow cytometric analysis of WT and KO PCN at 2 DIV. Dot plots show cell size (x axis) and granulosity (y axis) which allow identification of living cells (population B). The percentage of cells in population B with respect to the total number of cells analyzed is shown. *P ≤ 0.05, **P ≤ 0.01 with respect to WT
Next, we excluded that the impairment of axonal growth occurring in KO PCN was due to a higher level of cell death and a consequent lower density of these cells. Figure 5b shows that KO PCN do not undergo apoptosis (as indicated by the absence of caspase-3 cleavage) and experience non-apoptotic cell death to a lesser extent than WT PCN, as indicated by Trypan blue staining (Fig. 5c) and by flow cytometric analysis (Fig. 5d).
Taken together, these experiments show that Apaf1-deficient PCN are still able to emit and elongate axons, but with a lower efficiency compared with WT PCN.
Impairment of axonal elongation dependent on Apaf1-deficiency correlates with centrosomal alterations
We previously found that Apaf1 is crucial for a correct centrosome assembly and for centrosome-dependent activities in immortalized mouse embryonic fibroblasts (I-MEFs) and ETNA cells [3]. Centrosomes and microtubules play a key role in neuronal process formation and axonal differentiation by determining the site of axogenesis and by controlling axonal elongation [5, 6, 8]. Therefore, we hypothesized that the consequences of Apaf1-deficiency in axonal outgrowth might be correlated with its detrimental effect on centrosomes.
To investigate this issue, we analyzed centrosome composition in Apaf1-deficient PCN. First, we performed immunofluorescence of 2 DIV PCN stained with anti-SMI312 antibody and with an antibody recognizing the centrosomal marker γ-tubulin; as expected, we found that the centrosomes of Apaf1-deficient PCN were smaller when compared with those of WT PCN (Fig. S1 and Fig. 6a). We also found that the assembly of other centrosomal proteins (namely pericentrin and NEDD1) is impaired in Apaf1-deficient PCN, where centrosome staining appears fainter than in WT cells (Fig. 6a). Moreover, in line with our previous observations [3], we confirmed that the absence of Apaf1 impairs the localization of its co-activator hepatocellular carcinoma-associated antigen 66 (HCA66) [34, 35] to the centrosomes also in PCN (Fig. 6a).
Fig. 6.
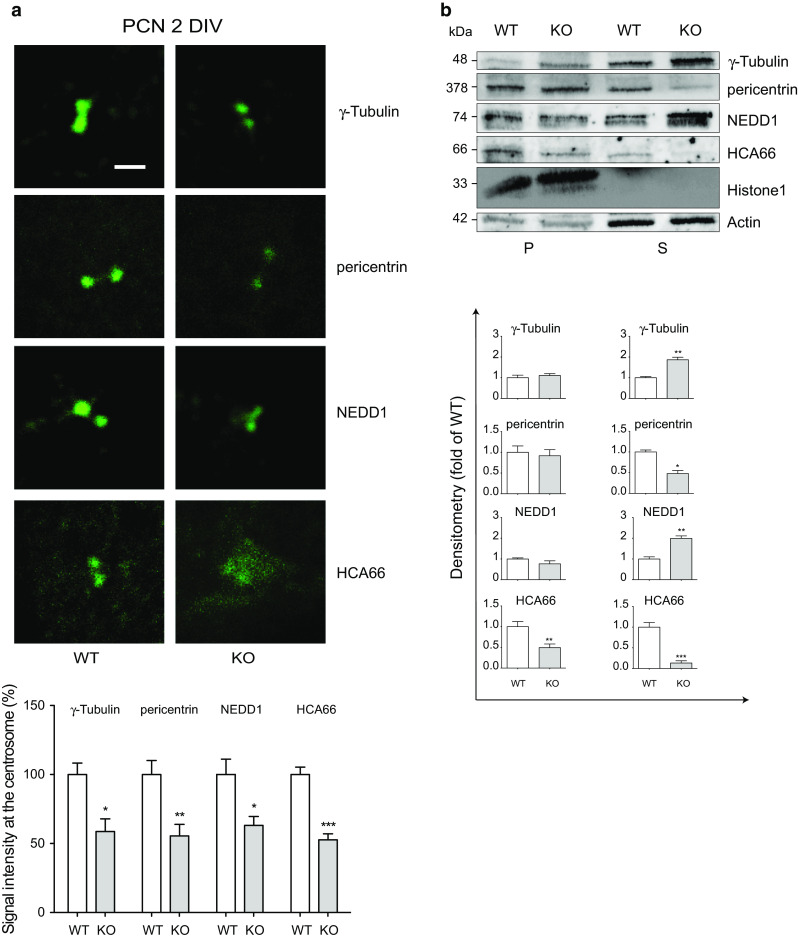
Centrosomes are impaired in Apaf1-deficient PCN. a Representative fluorescence microscopy images of WT and KO PCN at 2 DIV upon staining with anti-γ-tubulin, pericentrin, NEDD1, and HCA66 (green). Scale bar represents 1 μm. Quantitative analysis of the intensity of immunofluorescence at the centrosomes was performed using ImageJ software and reported as percentage of WT. At least 30 centrosomes were analyzed for each marker in several collected images. Values are mean ± SEM. b Solubilization properties of centrosome proteins in WT and KO PCN at 3 DIV. Purified nuclei were isolated by nuclear-cytosolic fractionation and were then extracted in the same buffer containing 1 % Triton X-100. The extracts were centrifuged and supernatants (S) and pellets (P) were analyzed by WB to determine the location of γ-tubulin, pericentrin, NEDD1, and HCA66. Histone H1 and Actin were used as loading control for pellets and supernatants, respectively. Density of immunoreactive bands was calculated using the software Image Lab (Bio-Rad), normalized for Histone H1 (pellets) and Actin (supernatants) and reported as arbitrary units (shown as the mean ± SEM). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005 with respect to WT
Since centrosomes are intimately connected with the nuclear envelope, we analyzed by WB analysis the association of HCA66 and other centrosomal proteins with the nuclear envelope. To this end, we isolated nuclei of 3 DIV WT and Apaf1-deficient PCN by nuclear–cytosolic fractionation. Nuclei were extracted with detergents at low-salt concentration before centrifugation, and supernatants (S) and pellets (P) were analyzed by WB to determine the distribution of proteins of interest [3, 36] (Fig. 6b). Pellet localization indicates that a protein is resistant to solubilization and is an integral membrane protein. Vice versa, centrosomal proteins are bound to the outer nuclear envelope membrane but are easily solubilized under the mild extraction conditions used; in fact, we found γ-tubulin, pericentrin, NEDD1, and HCA66 both in the pellet and in the soluble fractions. Interestingly, we observed, in Apaf1-deficient PCN, both a reduction of HCA66 expression and of its association to the nuclear envelope. By contrast, pericentrin and NEDD1 association to nuclear membranes is comparable in the two genotypes. However, their expression is severely altered in KO compared to WT PCN: Indeed, soluble pericentrin is less abundant in KO PCN, whereas soluble NEDD1 and γ-tubulin are far more abundant in KO compared to WT PCN (Fig. 6b). These experiments show that Apaf1-deletion alters centrosome organization.
Apaf1-deletion leads to Golgi morphology and trans-Golgi network alterations which correlate with defects in axogenesis
Altered centrosomal composition affects cell fate by impairing mechanisms regulated by these organelles; e.g., the assembly and morphology of the Golgi apparatus depend on centrosomes and microtubules [37]. Moreover, the morphology of the Golgi strongly correlates with axon elongation. Compacted Golgi is, indeed, associated to long axons, whereas fragmented Golgi is associated to shorter axons [37–39]. The immunofluorescence analysis, previously performed, (Fig. 4a, arrows) indicates that the cell body area where Golgi is localized and from where the axon originates, is larger and less compact in KO than in WT neurons (see also Fig. 7a). We therefore assessed the Golgi morphology by performing immunofluorescence for the cis-Golgi marker GM130 and we found that the Golgi complex is less condensed in KO neurons (Fig. 7b). The central role played by centrosomes and microtubules in the polarized extension of the axon [5, 6, 8] strongly suggests a correlation between the alteration in centrosome composition and in Golgi organization found in Apaf1-deleted neurons and the defects in axonal elongation caused by Apaf1-deficiency.
Fig. 7.
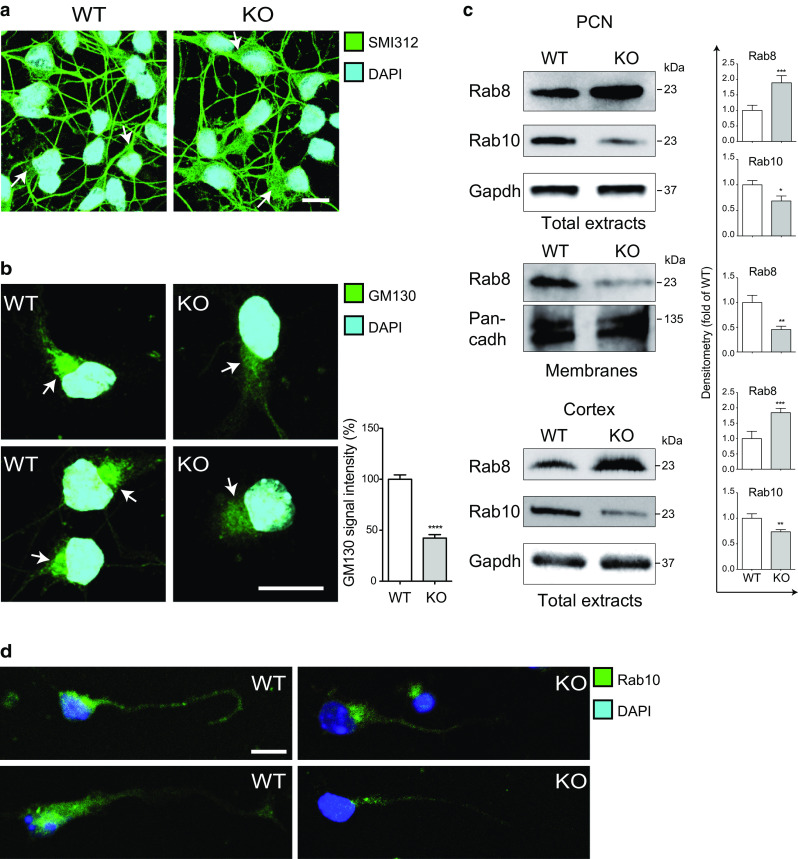
Apaf1 depletion affects Golgi morphology and trans-Golgi Rab GTPases in PCN. Representative fluorescence microscopy images of WT and KO PCN at 2 DIV upon staining with anti-SMI312 (a) and GM130 (b) antibodies to highlight axons and Golgi (arrows) morphology, respectively. DAPI was used to show nuclei. Scale bar represents 10 μm. Quantitative analysis of the intensity of immunofluorescence for GM130 was performed using ImageJ software and reported as percentage of WT. At least 30 cells were analyzed for each genotype in two independent experiments. Values are mean ± SEM. c WT and KO from PCN at 2 DIV or their membrane fractions or E14.5 brain cortices were assayed for Rab8 and Rab10 protein levels. Gapdh (PCN or cortices) or Pan-cadherin (membranes) were used as loading control. Density of immunoreactive bands was calculated using the software Image Lab (Bio-Rad), normalized for Gapdh or Pan-cadherin, and reported as arbitrary units (shown as the mean ± SEM). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005 with respect to WT. d Representative fluorescence microscopy images of WT and KO PCN at 2 DIV upon staining with anti-Rab10. DAPI was used to show nuclei. Scale bar represents 10 µm
To further investigate this issue, we analyzed the function of selected RabGTPases belonging to the trans-Golgi network, which were shown to regulate the secretory pathway and found to be crucial for axogenesis [40–42]. Indeed, the exocytic membrane trafficking from the trans-Golgi to the plasma membrane is essential for the transport of transmembrane elements and is a source of membrane for the elongating axons. Interestingly, we found evident alterations in the expression level of the GTPases Rab8 and Rab10 in KO PCN and cortices (Fig. 7c). The protein levels of Rab10 were markedly reduced in KO specimens and Rab10 was not enriched in the distal part or in the growth cone of axons as it normally occurs in WT neurons (Fig. 7c, d). Since it has been shown that Rab10 downregulation by specific siRNA impairs the axonal elongation in cultured neurons [40, 41], its low expression in Apaf1–deficient neurons might contribute to the impaired axonal phenotype. By contrast, Rab8, also known to be necessary for axonal elongation [40–42], was overexpressed in KO neurons as well as cortices (Fig. 7c). Since this finding contradicts the data obtained for Rab10, we decided to evaluate the extent of Rab8 activation by assessing its membrane attachment [40, 43]. The activity of Rab proteins is regulated both by positive and negative modulators of the GDP/GTP exchange, and by membrane attachment and GDP dissociation inhibitor (GDI) dissociation which promote Rab activation [11]. In accordance with our results on Rab10, we found that, although Rab8 is overexpressed in KO neurons, the level of membrane-associated Rab8 is greatly reduced with respect to WT PCN (Fig. 7c). These experiments strongly suggest that Apaf1-deficiency impacts on Rab8 and Rab10 activity and on trans-Golgi trafficking which is critical for axonal elongation. This correlates well with the reduced elongation of axons in KO cells.
Apaf1-deficiency leads to mitochondrial impairment and AMPK hyper-activation
The alteration of centrosome-associated activities is highly stressful for the cell, this matching evidence that ETNA and I-MEFs deprived of Apaf1 are more fragile and display a higher responsiveness to stressful conditions as well as an altered cell metabolism compared to WT cells [3, 23, 44]. To assess this issue at a molecular level, we decided to analyze the AMP-activated protein kinase (AMPK), which becomes phosphorylated at Thr172 upon metabolic stress [45, 46]. We discovered that the level of AMPK activation is strongly increased in Apaf1-deficient specimens compared to WT, which is accompanied by a higher phosphorylation level of Acetyl-CoA Carboxylase (ACC), one of its substrates (Fig. 8a, b and Fig. S2).
Fig. 8.
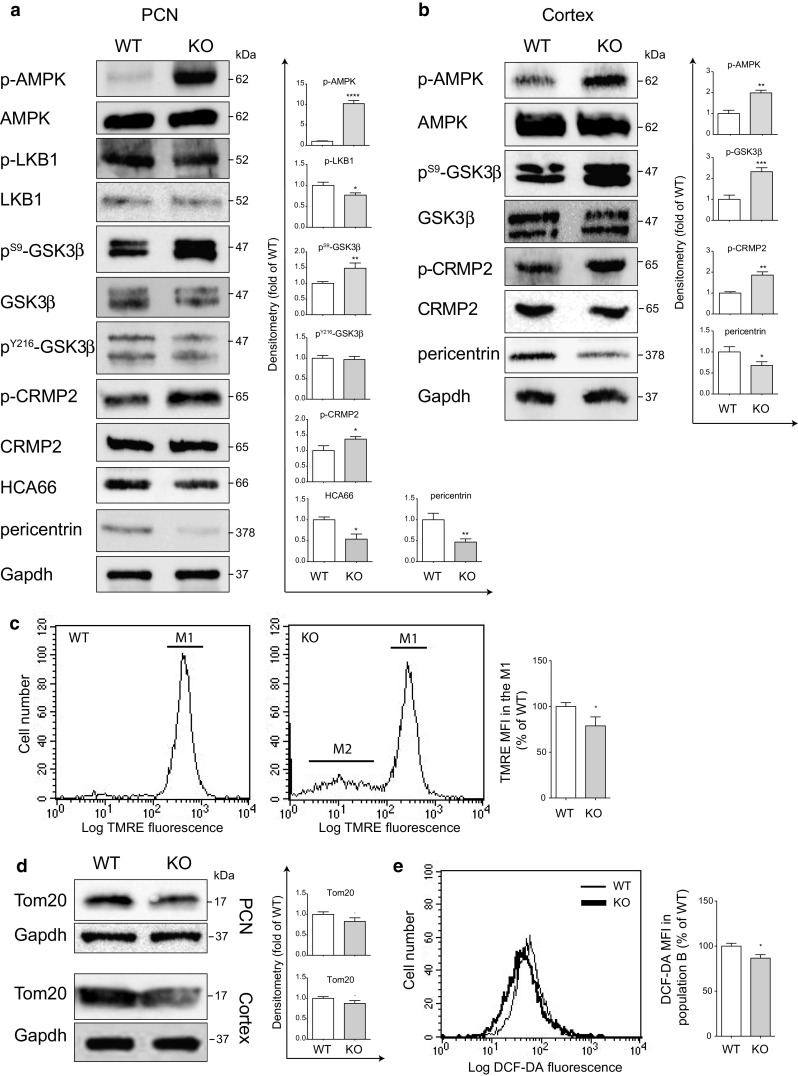
Molecular pathways involved in axonal growth and mitochondrial membrane potential are altered in conditions of Apaf1 deficiency both in vitro and in vivo. a WT and KO PCN at 3 DIV were assayed for p-AMPK, AMPK, p-LKB1, LKB1, pS9-GSK3β, pY216-GSK3β, GSK3β, p-CRMP2, HCA66, and pericentrin protein levels. Gapdh was used as loading control. Density of immunoreactive bands was calculated using the software Image Lab (Bio-Rad), normalized for Gapdh and reported as arbitrary units (shown as the mean ± SEM). b WT and KO mouse cortex extracts (E14.5) were assayed for p-AMPK, AMPK, pS9-GSK3β, GSK3β, p-CRMP2, and pericentrin protein levels. Gapdh was used as loading control. Density of immunoreactive bands was calculated using the software Image Lab (Bio-Rad), normalized for Gapdh, and reported as arbitrary units (shown as the mean ± SEM). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005, ****P ≤ 0.001 with respect to WT. c Analysis of mitochondrial transmembrane potential (ΔΨm) of WT and KO PCN at 2 DIV was performed by incubation with TMRE and FACS analysis of the population of living cells (gating on population B previously shown in Fig. 5d). M1 indicates a single population of cells with normal ΔΨm, M2 indicates a population with depolarized mitochondria (representing the 15–20 % of the gated living cells previously indicated as B). M2 is presumably formed by living PCN that do not use mitochondria but glycolysis for energy production, as found for other cell lines by Ferraro and coworkers [23]. The graph shows the TMRE mean fluorescence intensity (MFI) reported as percentage of TMRE MFI in WT cells. *P ≤ 0.05 with respect to WT. d WT and KO PCN at 2 DIV or mouse cortex extracts (E14.5) were assayed for Tom20 protein levels. Gapdh was used as loading control. Density of immunoreactive bands was calculated using the software Image Lab (Bio-Rad), normalized for Gapdh, and reported as arbitrary units (shown as the mean ± SEM). *P ≤ 0.05, with respect to WT. e Cytofluorimetric analysis of ROS in WT and KO PCN at 2 DIV upon 2′,7′-dihydrodichlorofluorescein-diacetate (H2DCF-DA) staining. The graph shows the DCF-DA MFI reported as percentage of DCF-DA MFI in WT cells. *P ≤ 0.05 with respect to WT
Since AMPK is over-activated in KO PCN, we partly evaluated the mitochondrial homeostasis of Apaf1-deficient PCN by dosing ROS production and by measuring the mitochondrial membrane potential (ΔΨm) by TMRE staining. Cytofluorometric analyses showed that ROS levels are not increased in KO PCN, in fact, they are slightly lower (Fig. 8e). However, both ΔΨm and the mitochondrial mass (as measured by WB analyses of the mitochondrial protein Tom 20) (Fig. 8c, d) are significantly reduced in Apaf1-KO neurons; indeed, while WT PCN are formed by a single population of cells with normal ΔΨm (M1), in KO cells is also present a population of neurons with depolarized mitochondria (M2) (Fig. 8c). Notably, KO PCN with normal ΔΨm (M1) have a lower mean of fluorescence compared to that of WT PCN (Fig. 8c, graph), which suggests that mitochondrial respiration is reduced in KO also in M1. This indicates a partially impaired mitochondrial metabolism in KO PCN. An affected mitochondrial metabolism might generate an energetic unbalance in conditions of high energy requirement, leading to AMPK over-activation.
The signaling pathways mediating axonal growth are altered in Apaf1-KO neurons
Results hitherto obtained show that the absence of Apaf1 affects axonal growth; this prompted us to explore the activation state of canonical pathways underlying axonal differentiation. First, we analyzed the kinase LKB1, a molecule involved in axonal elongation and neuronal migration and which has also been found to act through centrosomal regulation [16–18]. Once phosphorylated, LKB1 mediates neuronal polarization and axonal outgrowth. We therefore analyzed LKB1 phosphorylation at Ser431. In line with the defects in axonal elongation found in Apaf1-KO cortical neurons, we detected lower levels of phosphorylated LKB1 and of its substrate MARK (microtubule affinity-regulating kinase) in conditions of Apaf1-deficiency (Fig. S2). Notably, despite AMPK being a target for LKB1 in several tissues, LKB1 is not the major regulator of AMPK phosphorylation in neurons [19, 21].
Another key regulator of the axonal growth is GSK3β, a protein kinase whose activity is inversely correlated to the phosphorylation state of Ser9. WB analyses showed that the levels of p-GSK3β increase in Apaf1-KO PCN (Fig. 8a), so suggesting a possible compensative effect aimed at counterbalancing the observed inhibition of axonal growth. To confirm reduced activity of GSK3β, we also analyzed the phosphorylation level of Tau at Thr231 and Ser422, specifically targeted by GSK3β. As shown in Fig. S2, a decrease in pT231- and pS422-Tau was observed in KO versus WT PCN and brain cortices. Since phosphorylation of Tyr216 might increase the activity of GSK3β, we evaluated the extent of GSK3β phosphorylation at this residue. Tyr216 phosphorylation was, however, unchanged in KO PCN compared to WT neurons (Fig. 8a). Data shown in Fig. 8a further confirm the reduction of HCA66 and pericentrin protein levels in PCN total extracts. These results were also confirmed by WB analysis performed in lysates of brain cortices dissected at E14.5, in which we observed that the levels of p-AMPK and p-GSK3β increase in Apaf1-KO conditions along with a decrease of the expression level of pericentrin (Fig. 8b).
Finally, we analyzed a key molecule specifically involved in the regulation of axonal elongation, i.e., CRMP2. CRMP2 dephosphorylation is required for axonal growth. Interestingly, we found that levels of phosphorylated CRMP2 are higher in KO specimens compared with their WT counterpart (Fig. 8b); this strongly correlates with the impairment of axonal elongation caused by Apaf1-deficiency.
Discussion
In this paper, we show that the apoptotic protein Apaf1 is required for proper cortical neuron differentiation since its deletion specifically impairs axonal outgrowth. This is in accordance with accumulating evidence suggesting that apoptotic molecules also control processes other than apoptotic cell death. Previous studies reported that, in C. elegans, CED-4/Apaf1 and also CED-3 caspase activity are needed for efficient regeneration of severed axons [33] and that, in mice, Apaf1 and caspase-9 mediate a non-apoptotic caspase signaling required for the proper axonal projection of olfactory sensory neurons [47].
Strikingly, our study also reveals an interesting link between cellular bioenergetic homeostasis and neuronal maturation. Indeed, we have found that Apaf1 deficiency leads to over-activation of AMPK. AMPK is a major regulator of cell energy homeostasis, so acting as a metabolic sensor for energy deprivation [45, 46]. It has previously been shown that stroke and hypoxic-ischemic encephalopathy promote AMPK activation in the brain [48, 49]. Analogously, AMPK phosphorylation in Apaf1-KO PCN confirms that Apaf1 deficiency is highly stressful for the neurons, this highlighting its relevant non-apoptotic role: We have previously shown that Apaf1-depletion impairs cell performance and causes a higher responsiveness to stressful conditions [3, 23, 44]. Accordingly, here we have found that Apaf1 KO cells display a lower ΔΨm and lower mitochondrial mass indicating an altered mitochondrial homeostasis that, under the high energy demanding process of axonal growth, might lead to AMPK over-activation. Notably, it has also been reported that AMPK over-activation impairs axonal growth [20, 21, 49]. This provides a mechanistic connection between Apaf1 deficiency-induced stress and axonal differentiation, and could explain the axonal phenotype we observed. Our data suggest an intriguing model in which Apaf1 is necessary for the maintenance of cellular homeostasis; its absence might perturb centrosome, trans-Golgi, and microtubule-dependent mechanisms affecting axonal elongation and also mitochondria. Mitochondria impairment causes an energetic unbalance that is sensed by AMPK, which becomes phosphorylated. p-AMPK, in turn, would likely trigger compensation mechanisms against unfavorable conditions, i.e., p-AMPK slows down high energy demanding mechanisms, including axonal growth, in order to allow cell survival. The absence of high ROS production in KO neurons (possibly due to lower respiratory chain activity indicated by low ΔΨm) excludes this toxic factor as a cause of perturbed axogenesis.
Along with AMPK over-activation, we detected reduced levels of p-LKB1, which correlates very well with the impairment of axonal outgrowth occurring in the absence of Apaf1. Although AMPK is a canonical LKB1-target, LKB1 cannot be the AMPK-activating kinase in our system due to its being down-phosphorylated. This is coherent with the knowledge that LKB1 is not the major regulator of AMPK phosphorylation in neurons [19] where alternative upstream kinases mediate AMPK phosphorylation at Thr-172 (e.g., calmodulin-dependent protein kinase kinases; CaMKKs or TAK-1) [49]. LKB1 is critical for the control of centrosomal positioning and dynamics, which allows proper axon specification and neuronal migration [50]. AMPK has also been localized into centrosomes [45]. This raises the possibility that the impairment of centrosome organization detected in Apaf1-depleted cells might be linked to the alteration of LKB1 and AMPK modulation found in Apaf1-deficient cells, even though the mechanisms underlying this modulation need further elucidation.
Surprisingly, we found that the levels of Ser9-phosphorylated and inactive GSK3β in Apaf1 KO neurons are high, a condition normally associated with axon elongation stimulation. Our hypothesis is that GSK3β hyper-phosphorylation might be part of a compensation mechanism aimed at overcoming LKB1 activity reduction, AMPK and CRMP2 hyper-phosphorylation, and the centrosome assembly and Golgi alterations caused by Apaf1-deficiency, all of which impair axonal growth. The scenario we propose is that, differently from WT PCN, Apaf1-deficient PCN need to overcome a big hurdle in order to allow axonal polarization and that this would increase the levels of pS9GSK3β, so allowing the formation of axons, albeit of shorter length. The balance between these two opposite triggers, one inhibiting and the other one activating axonal elongation, would determine the phenotype of Apaf1-depleted neurons. This balance might vary depending on the environmental conditions, such as nutrient availability and cell–cell interactions; this would help explain the incomplete penetrance of the Apaf1-KO phenotype in terms of macroscopic brain malformations (unpublished observations). Consistent with our results, a simultaneous increase of both pS9GSK3β and p-AMPK in cerebral cortex under stress conditions has already been reported, although it is not clear if they are related or independent events [51–54].
While we largely documented some downstream events triggered by Apaf1 deficiency and impacting on cytoskeleton-associated processes, we did not unravel the upstream molecular mechanisms underlying Apaf1 function in axogenesis. In our attempts, we evaluated the possible involvement of the Apaf-1 interactor Diva/BclB which was reported to negatively modulate neurite extension in PC12 cells [55] and to be down-regulated during neuronal differentiation. However, we found that the expression of Diva does not vary in KO PCN (Fig. S3), indicating that impairment of axon elongation caused by Apaf1-deficiency does not depend on Diva levels, however, not excluding that DIVA could act upstream of Apaf1.
It is noteworthy that the Apaf1 interactor HCA66, whose expression is altered in Apaf1-KO PCN (Figs. 6, 8), is one of the genes heterozygously deleted in neurofibromatosis type I (NF1) microdeletion syndrome [56, 57]. NF1 is characterized by nervous system tumors and impaired CNS functions caused by mutations of the Nf1 gene encoding the protein neurofibromin [58–60]. An intriguing aspect of this work is the fact that neurofibromin regulates neurite length [61, 62], interacts with CRMP2, and is required for CRMP2 dephosphorylation and for axonal growth [63]. Moreover, most NF1 microdeleted patients, where HCA66 is less expressed, show a higher incidence of learning disabilities than do NF1 non-microdeleted patients [58, 61] while the low expression of HCA66 might be associated with the worsening of the phenotype characterizing NF1 microdeletion syndrome. Therefore, given that HAC66 expression and centrosomal localization are particularly reduced in Apaf1-KO neurons, and since CRMP2 has been localized at the centrosomes [64], it is tempting to speculate that the relevance of Apaf1 in axonal growth might be associated with the abnormal regulation of HCA66; this might interfere with NF1 activity, CRMP2 activation and, consequently, with axon elongation. Future studies on the mechanism of action of Apaf1 and HCA66 will likely reveal new insights into the regulation of axonal growth.
The role of Apaf1 in regulating axonal elongation is also supported by the high similarity between the brain phenotype of Apaf1-KO (Fig. S4a) [24] and JNK1/2 double mutant embryos (JNK−/−/JNK2+/−) [65], both displaying exencephaly, abnormal folding of the neuroepithelium, obliteration of the cerebral ventricles and protrusion of brain tissues. Indeed, JNK1 is required for neurite outgrowth and for cytoskeletal regulation and maintenance of neuronal microtubule homeostasis, and its deficiency leads to progressive degeneration of long nerve fibers [66]. Moreover, notably, apoptosis does indeed occur in the brain of the JNK1/2 double mutant embryos [65]. The marked disorganization of the brain of Apaf1-KO embryos and their embryonic lethality (E16.5) [24] made the analysis of axonal growth and migration impairment in vivo technically challenging. Moreover, assessing the specific role of Apaf1 on axonal growth in Apaf1-KO embryos was prevented by the fact that defects in axon patterning are also associated to apoptosis; in fact, impaired neurons unable to die because of the absence of Apaf1 do not innervate proper targets, this making it impossible to discriminate between the two potential roles of this protein. We have found a low intensity of Neurofilament 160/200 staining in KO versus WT cortices (Fig. S4c, d), indicating less/shorter neuronal projections and being in line with our discovery of axogenesis impairment in Apaf1-KO PCN. However, as above assessed, since axonal outgrowth and neuronal migration in vivo depend not only on the intrinsic genetic characteristics of the neuron, but also on environmental molecules and interactions with other cells (both factors altered by the absence of apoptosis), this staining does not definitively clarify the role of Apaf1 in axogenesis in vivo.
Our finding that Apaf1 exerts a role in cortical axon outgrowth suggests that the severe brain malformations displayed by Apaf1-deficient mice due to apoptosis deficiency [24, 67] might also be partially ascribed to the alteration of centrosome, Golgi and axonal elongation caused by Apaf1-deletion. Interestingly, after many years, the mechanism that explains how apoptosis inhibition results in brain malformations in Apaf1-KO mice has been partly revised, thus confirming the possibility of alternative or additional explanations [68]. Indeed, Nonomura and colleagues have shown that the absence of Apaf1 or Caspase-9-mediated apoptosis in the brain causes the persistence of Fgf8-expressing non-proliferative cells in a specific area of the developing brain’s anterior neural ridge. Permanent Fgf8 expression induces a profound alteration of the signaling pathways triggered in ventral forebrain cells which leads to a dysregulation of brain development at early stages and, finally, to failure in brain ventricle expansion and incomplete closure of the cranial neural tube [68]. Therefore, the derangement of brain morphology, typical of Apaf1-KO embryos, is not only due to the over-proliferation of cells not undergoing to apoptosis, but it is also mainly caused by alterations in the signaling pathways triggered by cells unable to die, through secreted factors and cell–cell interactions [68]. Moreover, the role of Apaf1 in allowing a correct axogenesis might not necessarily be related to its canonical function in apoptosome-dependent caspase-9 activation, and caspases might not be involved in this pro-survival role of Apaf1. Interestingly, functions not involving all the components of the apoptosome have been demonstrated in the axons; in particular, it has been shown that the selective degeneration of the axon (pruning) occurring during neural plasticity and not leading to neuronal death is mediated by caspases, whereas Apaf1 is not required [69]. This indicates that Apaf1 might have distinct functions in neurons depending on the specific phase of neuron-life, and further supports our discovery of Apaf1’s non-apoptotic role in axonal outgrowth.
Although further investigations are needed to fully elucidate the sequence of events triggered by Apaf1-deficiency and leading to axonal elongation defects, the discovery of additional molecular players involved in axonal growth has a clinical relevance in that it might help to explain neurological abnormalities caused by stressful conditions during early brain development.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by the Italian Ministry of Health (RF-2010-2318508 to E Ferraro, Institutional research–Ricerca corrente and GR-2008-1138121 to G Filomeni). We wish to thank MW Bennett for the valuable editorial work, V Frezza for technical support, and M Canossa, L. Cancedda, E. Santonico, N. Canu, L.Vitiello and M Racaniello for helpful discussions. We are also grateful to A Merdes (CNRS-Pierrre-Fabre, Toulouse, France) for kindly providing the HCA66 antibody.
Conflict of interest
The authors declare no conflict of interest.
Abbreviations
- ACC
Acetyl-CoA carboxylase
- AMPK
AMP-dependent protein kinase
- Apaf1
Apoptotic protease activating factor 1
- CRMP2
Collapsing-response mediator protein-2
- DIV
Day in vitro
- Diva
Death inducer binding to vBcl2 and Apaf1
- ETNA
Embryonic telencephalic naïve Apaf1
- Gap43
Growth associated protein 43
- GDI
GDP dissociation inhibitor
- GM130
cis-Golgi marker
- GSK3β
Glycogen synthase kinase-3β
- HCA66
Hepatocellular carcinoma-associated antigen 66
- I-MEFs
Immortalized mouse embryonic fibroblasts
- LKB1
Liver kinase B1
- MAP2
Microtubule-associated protein 2
- MAPs
Microtubule-associated proteins
- MARK
Microtubule affinity-regulating kinase
- NEDD1
Neural precursor cell expressed developmentally down-regulated protein 1
- NF1
Neurofibromatosis type I
- PCN
Primary cortical neurons
- PSD95
Postsynaptic density protein 95
- Rab8
Ras-related in brain 8
- Rab10
Ras-related in brain 10
- SMI312
Pan-axonal neurofilament marker
- Tau
Tau protein
- Tom20
Translocase of outer membrane 20
- Tubb3
Tubulin, beta 3 class III
References
- 1.Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91(4):479–489. doi: 10.1016/S0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 2.Mouhamad S, Galluzzi L, Zermati Y, Castedo M, Kroemer G. Apaf-1 deficiency causes chromosomal instability. Cell Cycle. 2007;6(24):3103–3107. doi: 10.4161/cc.6.24.5046. [DOI] [PubMed] [Google Scholar]
- 3.Ferraro E, Pesaresi MG, De Zio D, Cencioni MT, Gortat A, Cozzolino M, Berghella L, Salvatore AM, Oettinghaus B, Scorrano L, Perez-Paya E, Cecconi F. Apaf1 plays a pro-survival role by regulating centrosome morphology and function. J Cell Sci. 2011;124(Pt 20):3450–3463. doi: 10.1242/jcs.086298. [DOI] [PubMed] [Google Scholar]
- 4.Stiess M, Bradke F. Neuronal polarization: the cytoskeleton leads the way. Dev Neurobiol. 2011;71(6):430–444. doi: 10.1002/dneu.20849. [DOI] [PubMed] [Google Scholar]
- 5.Kuijpers M, Hoogenraad CC. Centrosomes, microtubules and neuronal development. Mol Cell Neurosci. 2011;48(4):349–358. doi: 10.1016/j.mcn.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Higginbotham HR, Gleeson JG. The centrosome in neuronal development. Trends Neurosci. 2007;30(6):276–283. doi: 10.1016/j.tins.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Distel M, Hocking JC, Volkmann K, Koster RW. The centrosome neither persistently leads migration nor determines the site of axonogenesis in migrating neurons in vivo. J Cell Biol. 2010;191(4):875–890. doi: 10.1083/jcb.201004154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Anda FC, Meletis K, Ge X, Rei D, Tsai LH. Centrosome motility is essential for initial axon formation in the neocortex. J Neurosci. 2010;30(31):10391–10406. doi: 10.1523/JNEUROSCI.0381-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutterlin C, Colanzi A. The Golgi and the centrosome: building a functional partnership. J Cell Biol. 2010;188(5):621–628. doi: 10.1083/jcb.200910001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz SL, Cao C, Pylypenko O, Rak A, Wandinger-Ness A. Rab GTPases at a glance. J Cell Sci. 2007;120(Pt 22):3905–3910. doi: 10.1242/jcs.015909. [DOI] [PubMed] [Google Scholar]
- 11.Villarroel-Campos D, Gastaldi L, Conde C, Caceres A, Gonzalez-Billault C. Rab-mediated trafficking role in neurite formation. J Neurochem. 2014;129(2):240–248. doi: 10.1111/jnc.12676. [DOI] [PubMed] [Google Scholar]
- 12.Trivedi N, Marsh P, Goold RG, Wood-Kaczmar A, Gordon-Weeks PR. Glycogen synthase kinase-3beta phosphorylation of MAP1B at Ser1260 and Thr1265 is spatially restricted to growing axons. J Cell Sci. 2005;118(Pt 5):993–1005. doi: 10.1242/jcs.01697. [DOI] [PubMed] [Google Scholar]
- 13.Hur EM, Zhou FQ. GSK3 signalling in neural development. Nat Rev Neurosci. 2010;11(8):539–551. doi: 10.1038/nrn2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukata Y, Itoh TJ, Kimura T, Menager C, Nishimura T, Shiromizu T, Watanabe H, Inagaki N, Iwamatsu A, Hotani H, Kaibuchi K. CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat Cell Biol. 2002;4(8):583–591. doi: 10.1038/ncb825. [DOI] [PubMed] [Google Scholar]
- 15.Kimura T, Watanabe H, Iwamatsu A, Kaibuchi K. Tubulin and CRMP-2 complex is transported via Kinesin-1. J Neurochem. 2005;93(6):1371–1382. doi: 10.1111/j.1471-4159.2005.03063.x. [DOI] [PubMed] [Google Scholar]
- 16.Shelly M, Cancedda L, Heilshorn S, Sumbre G, Poo MM. LKB1/STRAD promotes axon initiation during neuronal polarization. Cell. 2007;129(3):565–577. doi: 10.1016/j.cell.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Bony G, Szczurkowska J, Tamagno I, Shelly M, Contestabile A, Cancedda L. Non-hyperpolarizing GABAB receptor activation regulates neuronal migration and neurite growth and specification by cAMP/LKB1. Nat Commun. 2013;4:1800. doi: 10.1038/ncomms2820. [DOI] [PubMed] [Google Scholar]
- 18.Barnes AP, Lilley BN, Pan YA, Plummer LJ, Powell AW, Raines AN, Sanes JR, Polleux F. LKB1 and SAD kinases define a pathway required for the polarization of cortical neurons. Cell. 2007;129(3):549–563. doi: 10.1016/j.cell.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 19.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9(8):563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amato S, Liu X, Zheng B, Cantley L, Rakic P, Man HY. AMP-activated protein kinase regulates neuronal polarization by interfering with PI 3-kinase localization. Science. 2011;332(6026):247–251. doi: 10.1126/science.1201678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams T, Courchet J, Viollet B, Brenman JE, Polleux F. AMP-activated protein kinase (AMPK) activity is not required for neuronal development but regulates axogenesis during metabolic stress. Proc Natl Acad Sci USA. 2011;108(14):5849–5854. doi: 10.1073/pnas.1013660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cozzolino M, Ferraro E, Ferri A, Rigamonti D, Quondamatteo F, Ding H, Xu ZS, Ferrari F, Angelini DF, Rotilio G, Cattaneo E, Carri MT, Cecconi F. Apoptosome inactivation rescues proneural and neural cells from neurodegeneration. Cell Death Differ. 2004;11(11):1179–1191. doi: 10.1038/sj.cdd.4401476. [DOI] [PubMed] [Google Scholar]
- 23.Ferraro E, Pulicati A, Cencioni MT, Cozzolino M, Navoni F, di Martino S, Nardacci R, Carri MT, Cecconi F. Apoptosome-deficient cells lose cytochrome c through proteasomal degradation but survive by autophagy-dependent glycolysis. Mol Biol Cell. 2008;19(8):3576–3588. doi: 10.1091/mbc.E07-09-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cecconi F, Alvarez-Bolado G, Meyer BI, Roth KA, Gruss P. Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell. 1998;94(6):727–737. doi: 10.1016/S0092-8674(00)81732-8. [DOI] [PubMed] [Google Scholar]
- 25.Johnson CE, Huang YY, Parrish AB, Smith MI, Vaughn AE, Zhang Q, Wright KM, Van Dyke T, Wechsler-Reya RJ, Kornbluth S, Deshmukh M. Differential Apaf-1 levels allow cytochrome c to induce apoptosis in brain tumors but not in normal neural tissues. Proc Natl Acad Sci USA. 2007;104(52):20820–20825. doi: 10.1073/pnas.0709101105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright KM, Smith MI, Farrag L, Deshmukh M. Chromatin modification of Apaf-1 restricts the apoptotic pathway in mature neurons. J Cell Biol. 2007;179(5):825–832. doi: 10.1083/jcb.200708086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leveille F, Papadia S, Fricker M, Bell KF, Soriano FX, Martel MA, Puddifoot C, Habel M, Wyllie DJ, Ikonomidou C, Tolkovsky AM, Hardingham GE. Suppression of the intrinsic apoptosis pathway by synaptic activity. J Neurosci. 2010;30(7):2623–2635. doi: 10.1523/JNEUROSCI.5115-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng S, Gray EE, Chawla G, Porse BT, O’Dell TJ, Black DL. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat Neurosci. 2012;15(3):381–388, S381. doi: 10.1038/nn.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandell JW, Banker GA. A spatial gradient of tau protein phosphorylation in nascent axons. J Neurosci. 1996;16(18):5727–5740. doi: 10.1523/JNEUROSCI.16-18-05727.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caceres A, Banker GA, Binder L. Immunocytochemical localization of tubulin and microtubule-associated protein 2 during the development of hippocampal neurons in culture. J Neurosci. 1986;6(3):714–722. doi: 10.1523/JNEUROSCI.06-03-00714.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradke F, Dotti CG. Differentiated neurons retain the capacity to generate axons from dendrites. Curr Biol. 2000;10(22):1467–1470. doi: 10.1016/S0960-9822(00)00807-1. [DOI] [PubMed] [Google Scholar]
- 32.Shaham S, Horvitz HR. Developing Caenorhabditis elegans neurons may contain both cell-death protective and killer activities. Genes Dev. 1996;10(5):578–591. doi: 10.1101/gad.10.5.578. [DOI] [PubMed] [Google Scholar]
- 33.Pinan-Lucarre B, Gabel CV, Reina CP, Hulme SE, Shevkoplyas SS, Slone RD, Xue J, Qiao Y, Weisberg S, Roodhouse K, Sun L, Whitesides GM, Samuel A, Driscoll M. The core apoptotic executioner proteins CED-3 and CED-4 promote initiation of neuronal regeneration in Caenorhabditis elegans . PLoS Biol. 2012;10(5):e1001331. doi: 10.1371/journal.pbio.1001331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piddubnyak V, Rigou P, Michel L, Rain JC, Geneste O, Wolkenstein P, Vidaud D, Hickman JA, Mauviel A, Poyet JL. Positive regulation of apoptosis by HCA66, a new Apaf-1 interacting protein, and its putative role in the physiopathology of NF1 microdeletion syndrome patients. Cell Death Differ. 2007;14(6):1222–1233. doi: 10.1038/sj.cdd.4402122. [DOI] [PubMed] [Google Scholar]
- 35.Fant X, Gnadt N, Haren L, Merdes A. Stability of the small gamma-tubulin complex requires HCA66, a protein of the centrosome and the nucleolus. J Cell Sci. 2009;122(Pt 8):1134–1144. doi: 10.1242/jcs.035238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, Gotzmann J, Foisner R, Karakesisoglou I. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci. 2005;118(Pt 15):3419–3430. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- 37.Hanus C, Ehlers MD. Secretory outposts for the local processing of membrane cargo in neuronal dendrites. Traffic. 2008;9(9):1437–1445. doi: 10.1111/j.1600-0854.2008.00775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beffert U, Dillon GM, Sullivan JM, Stuart CE, Gilbert JP, Kambouris JA, Ho A. Microtubule plus-end tracking protein CLASP2 regulates neuronal polarity and synaptic function. J Neurosci. 2012;32(40):13906–13916. doi: 10.1523/JNEUROSCI.2108-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller PM, Folkmann AW, Maia AR, Efimova N, Efimov A, Kaverina I. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat Cell Biol. 2009;11(9):1069–1080. doi: 10.1038/ncb1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang T, Liu Y, Xu XH, Deng CY, Wu KY, Zhu J, Fu XQ, He M, Luo ZG. Lgl1 activation of rab10 promotes axonal membrane trafficking underlying neuronal polarization. Dev Cell. 2011;21(3):431–444. doi: 10.1016/j.devcel.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 41.Sann S, Wang Z, Brown H, Jin Y. Roles of endosomal trafficking in neurite outgrowth and guidance. Trends Cell Biol. 2009;19(7):317–324. doi: 10.1016/j.tcb.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 42.Huber LA, Dupree P, Dotti CG. A deficiency of the small GTPase rab8 inhibits membrane traffic in developing neurons. Mol Cell Biol. 1995;15(2):918–924. doi: 10.1128/MCB.15.2.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfeffer S. A model for Rab GTPase localization. Biochem Soc Trans. 2005;33(Pt 4):627–630. doi: 10.1042/BST0330627. [DOI] [PubMed] [Google Scholar]
- 44.Sancho M, Gortat A, Herrera AE, Andreu-Fernandez V, Ferraro E, Cecconi F, Orzaez M, Perez-Paya E. Altered mitochondria morphology and cell metabolism in Apaf1-deficient cells. PLoS ONE. 2014;9(1):e84666. doi: 10.1371/journal.pone.0084666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vazquez-Martin A, Oliveras-Ferraros C, Menendez JA. The active form of the metabolic sensor: AMP-activated protein kinase (AMPK) directly binds the mitotic apparatus and travels from centrosomes to the spindle midzone during mitosis and cytokinesis. Cell Cycle. 2009;8(15):2385–2398. doi: 10.4161/cc.8.15.9082. [DOI] [PubMed] [Google Scholar]
- 46.Fogarty S. Hardie DG (2010) Development of protein kinase activators: AMPK as a target in metabolic disorders and cancer. Biochim Biophys Acta. 1804;3:581–591. doi: 10.1016/j.bbapap.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 47.Ohsawa S, Hamada S, Kuida K, Yoshida H, Igaki T, Miura M. Maturation of the olfactory sensory neurons by Apaf-1/caspase-9-mediated caspase activity. Proc Natl Acad Sci USA. 2010;107(30):13366–13371. doi: 10.1073/pnas.0910488107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Coven DL, Miller EJ, Hu X, Young ME, Carling D, Sinusas AJ, Young LH. Activation of AMPK alpha- and gamma-isoform complexes in the intact ischemic rat heart. Am J Physiol Heart Circ Physiol. 2006;291(4):H1927–1934. doi: 10.1152/ajpheart.00251.2006. [DOI] [PubMed] [Google Scholar]
- 49.Amato S, Man HY. AMPK signaling in neuronal polarization: putting the brakes on axonal traffic of PI3-kinase. Commun Integr Biol. 2012;5(2):152–155. doi: 10.4161/cib.18968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asada N, Sanada K, Fukada Y. LKB1 regulates neuronal migration and neuronal differentiation in the developing neocortex through centrosomal positioning. J Neurosci. 2007;27(43):11769–11775. doi: 10.1523/JNEUROSCI.1938-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Son HS, Kwon HY, Sohn EJ, Lee JH, Woo HJ, Yun M, Kim SH, Kim YC. Activation of AMP-activated protein kinase and phosphorylation of glycogen synthase kinase3 beta mediate ursolic acid induced apoptosis in HepG2 liver cancer cells. Phytother Res. 2013;27(11):1714–1722. doi: 10.1002/ptr.4925. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L, Jouret F, Rinehart J, Sfakianos J, Mellman I, Lifton RP, Young LH, Caplan MJ. AMP-activated protein kinase (AMPK) activation and glycogen synthase kinase-3beta (GSK-3beta) inhibition induce Ca2+-independent deposition of tight junction components at the plasma membrane. J Biol Chem. 2011;286(19):16879–16890. doi: 10.1074/jbc.M110.186932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki T, Bridges D, Nakada D, Skiniotis G, Morrison SJ, Lin JD, Saltiel AR, Inoki K. Inhibition of AMPK catabolic action by GSK3. Mol Cell. 2013;50(3):407–419. doi: 10.1016/j.molcel.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi SH, Kim YW, Kim SG. AMPK-mediated GSK3beta inhibition by isoliquiritigenin contributes to protecting mitochondria against iron-catalyzed oxidative stress. Biochem Pharmacol. 2010;79(9):1352–1362. doi: 10.1016/j.bcp.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 55.Lim JQ, Lu J, He BP. Diva/BclB regulates differentiation by inhibiting NDPKB/Nm23H2-mediated neuronal differentiation in PC-12 cells. BMC Neurosci. 2012;13:123. doi: 10.1186/1471-2202-13-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jenne DE, Tinschert S, Dorschner MO, Hameister H, Stephens K, Kehrer-Sawatzki H. Complete physical map and gene content of the human NF1 tumor suppressor region in human and mouse. Genes Chromosom Cancer. 2003;37(2):111–120. doi: 10.1002/gcc.10206. [DOI] [PubMed] [Google Scholar]
- 57.Gutmann DH, Parada LF, Silva AJ, Ratner N. Neurofibromatosis type 1: modeling CNS dysfunction. J Neurosci. 2012;32(41):14087–14093. doi: 10.1523/JNEUROSCI.3242-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pasmant E, Sabbagh A, Spurlock G, Laurendeau I, Grillo E, Hamel MJ, Martin L, Barbarot S, Leheup B, Rodriguez D, Lacombe D, Dollfus H, Pasquier L, Isidor B, Ferkal S, Soulier J, Sanson M, Dieux-Coeslier A, Bieche I, Parfait B, Vidaud M, Wolkenstein P, Upadhyaya M, Vidaud D. NF1 microdeletions in neurofibromatosis type 1: from genotype to phenotype. Hum Mutat. 2010;31(6):E1506–1518. doi: 10.1002/humu.21271. [DOI] [PubMed] [Google Scholar]
- 59.Listernick R, Louis DN, Packer RJ, Gutmann DH. Optic pathway gliomas in children with neurofibromatosis 1: consensus statement from the NF1 Optic Pathway Glioma Task Force. Ann Neurol. 1997;41(2):143–149. doi: 10.1002/ana.410410204. [DOI] [PubMed] [Google Scholar]
- 60.Rosenfeld A, Listernick R, Charrow J, Goldman S. Neurofibromatosis type 1 and high-grade tumors of the central nervous system. Child’s Nerv Syst. 2010;26(5):663–667. doi: 10.1007/s00381-009-1024-2. [DOI] [PubMed] [Google Scholar]
- 61.Brown JA, Diggs-Andrews KA, Gianino SM, Gutmann DH. Neurofibromatosis-1 heterozygosity impairs CNS neuronal morphology in a cAMP/PKA/ROCK-dependent manner. Mol Cell Neurosci. 2012;49(1):13–22. doi: 10.1016/j.mcn.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown JA, Gianino SM, Gutmann DH. Defective cAMP generation underlies the sensitivity of CNS neurons to neurofibromatosis-1 heterozygosity. J Neurosci. 2010;30(16):5579–5589. doi: 10.1523/JNEUROSCI.3994-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patrakitkomjorn S, Kobayashi D, Morikawa T, Wilson MM, Tsubota N, Irie A, Ozawa T, Aoki M, Arimura N, Kaibuchi K, Saya H, Araki N. Neurofibromatosis type 1 (NF1) tumor suppressor, neurofibromin, regulates the neuronal differentiation of PC12 cells via its associating protein, CRMP-2. J Biol Chem. 2008;283(14):9399–9413. doi: 10.1074/jbc.M708206200. [DOI] [PubMed] [Google Scholar]
- 64.Jauffred B, Llense F, Sommer B, Wang Z, Martin C, Bellaiche Y. Regulation of centrosome movements by numb and the collapsin response mediator protein during Drosophila sensory progenitor asymmetric division. Development. 2013;140(13):2657–2668. doi: 10.1242/dev.087338. [DOI] [PubMed] [Google Scholar]
- 65.Sabapathy K, Jochum W, Hochedlinger K, Chang L, Karin M, Wagner EF. Defective neural tube morphogenesis and altered apoptosis in the absence of both JNK1 and JNK2. Mech Dev. 1999;89(1–2):115–124. doi: 10.1016/S0925-4773(99)00213-0. [DOI] [PubMed] [Google Scholar]
- 66.Chang L, Jones Y, Ellisman MH, Goldstein LS, Karin M. JNK1 is required for maintenance of neuronal microtubules and controls phosphorylation of microtubule-associated proteins. Dev Cell. 2003;4(4):521–533. doi: 10.1016/S1534-5807(03)00094-7. [DOI] [PubMed] [Google Scholar]
- 67.Yoshida H, Kong YY, Yoshida R, Elia AJ, Hakem A, Hakem R, Penninger JM, Mak TW. Apaf1 is required for mitochondrial pathways of apoptosis and brain development. Cell. 1998;94(6):739–750. doi: 10.1016/S0092-8674(00)81733-X. [DOI] [PubMed] [Google Scholar]
- 68.Nonomura K, Yamaguchi Y, Hamachi M, Koike M, Uchiyama Y, Nakazato K, Mochizuki A, Sakaue-Sawano A, Miyawaki A, Yoshida H, Kuida K, Miura M. Local apoptosis modulates early mammalian brain development through the elimination of morphogen-producing cells. Dev Cell. 2013;27(6):621–634. doi: 10.1016/j.devcel.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 69.Cusack CL, Swahari V, Hampton Henley W, Michael Ramsey J, Deshmukh M. Distinct pathways mediate axon degeneration during apoptosis and axon-specific pruning. Nat Commun. 2013;4:1876. doi: 10.1038/ncomms2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.