Abstract
Mechanotransduction encompasses the role of mechanical forces in controlling cell behavior by activating signal transduction pathways. Most forces at a cellular level are caused by myosin II, which contracts and cross-links actin. Myosin II-dependent forces are transmitted through the actin cytoskeleton to molecular endpoints that promote specific cellular outcomes, e.g., cell proliferation, adhesion, or migration. For example, most adhesive and migratory phenomena are mechanically linked by a molecular clutch comprised of mechanosensitive scaffolds. Myosin II activation and mechanosensitive molecular mechanisms are finely tuned and spatiotemporally integrated to coordinate morphogenetic events during development. Mechanical events dependent on myosin II also participate in tumor cell proliferation, invasion, and metastatic dissemination. Specifically, tumor cells alter the mechanical properties of the microenvironment to create favorable conditions for proliferation and/or dissemination. These observations position myosin II-dependent force generation and mechanotransduction at the crossroads between normal development and cancer.
Keywords: Mechanotransduction, Force, Myosin, Actin, Tumor, Migration, Adhesion
Introduction
The building blocks of development, organogenesis, and homeostasis are cell proliferation, migration, differentiation, and death. For a long time, most biologists have tried to explain these phenomena solely in terms of biochemical reactions, e.g., binding of a hormone to its membrane-bound receptor and the signal transduction cascades that follow (biochemical paradigm). These cascades involve modifications of a variable number of proteins, complexes, and other macromolecules, such as conformational alterations, changes of complex composition, and/or post-translational modifications, e.g., phosphorylation, acetylation, etc. [1]. Signal transduction cascades converge into the activation of endpoint molecules. Endpoints function as effectors that elicit cellular responses to the initial signal, e.g., cell division (proliferation), migration, differentiation, or apoptosis.
The biochemical paradigm has been very successful in explaining the responses of different cell types to stimulation. It has also provided fundamental knowledge for the development of drugs against many diseases, e.g., cancer, viral and bacterial infections, etc. However, this approach overlooks the mechanics of the molecules and cells during these processes. Increasing evidence, particularly from the development and cell biology fields, indicates that mechanical forces also govern the fate of cells and tissues. They modulate how cells respond to environmental cues, both chemical and mechanical. Furthermore, alterations in the mechanical properties of a tissue may facilitate or interfere with the development of infection, degenerative syndromes, e.g., dystrophies, or proliferative diseases, e.g., cancer.
Here, we offer a brief summary of experiments and data leading to a mechano-biochemical paradigm that accommodates the mechanical properties of cells and their microenvironment. We review evidence of mechanical modulation of the cellular behavior at a molecular level, including those events in which biochemical processes and mechanical forces converge. This is illustrated by discussion of several mechanosensitive molecules, i.e. those that “sense” the forces and activate, or inhibit, cellular effectors in response to mechanical inputs. We also describe the crucial role of myosin II in force generation and cellular plasticity in response to chemical or mechanical inputs. Based on recent evidence, we postulate that cancer growth and metastasis involve the subversion of mechanically controlled signaling pathways similar to those that control certain stages of normal development and organogenesis. Alterations of these relays participate in the abnormal responses of tumor cells and promote the modification of the physical properties of the microenvironment to perpetuate and amplify malignancy and promote cancer cell dissemination.
From action to reaction: mechanical forces control different signal transduction pathways through myosin II
Multiple studies have shown that mechanical inputs modulate signaling pathways that control the behavior of the cell under different conditions. For example, the mechanical properties of the substrate, i.e. its compliance or stiffness, determine force generation and distribution along the adhesive surface of the cell. These mechanical properties are sufficient to direct mesenchymal stem cell differentiation to a certain lineage [2]. Commitment is related to the rigidity of the target tissue: stiff substrates drive stem cells to lineages from stiff tissues, e.g., bone. Conversely, soft substrates steer them to become cells from soft tissues, e.g., brain. Substrate rigidity also controls cell proliferation and migration; stiff substrates increase proliferation [3] and promote migration [4]. Furthermore, in areas of variable rigidity, cells migrate towards the stiffer regions, a property called durotaxis [4, 5].
The regulatory mechanisms that control these responses are poorly characterized, but myosin II emerges as a major regulator and integrator. Myosin II is a motor protein that binds actin. Structurally, myosin II is a hexamer made of two long heavy chains (MHCII) that contain a head domain and a long coiled-coil rod domain, two regulatory light chains (RLC) and two essential light chains (ELC) that bind to the neck area that separates the head and rod domains (reviewed in [6]). MHCII molecules dimerize through the rod domain, forming the backbone of the hexamer. Myosin II hexamers assemble into multimers through interaction of the end portion of the rod domain, forming myosin mini-filaments. The head domain of MHCII is a globular domain that binds actin filaments and has ATPase activity. ATP hydrolysis induces a conformational movement that slides the actin filament, generating contractile force [7]. Major insights on the function of the ATPase domain in actin motility have resulted from use of synthetic drugs, most notably blebbistatin. Blebbistatin was identified in a small molecule screening and first reported to impair cytokinesis and cell migration [8]. Detailed study of its molecular mechanism revealed that blebbistatin does not block the ATPase activity of myosin II, but it slows down phosphate release from the active pocket, lowering its affinity for actin [9].
There are three major variants of myosin II: muscle, smooth muscle (reviewed elsewhere [10, 11]), and non-muscle (NMII). In mammals, there are three main isoforms: NMII-A, II-B, and II-C. Whenever isoform-specific functions have been identified, it is indicated in the text. NMII without reference to the isoform is used throughout the text for organisms that do not express NMII isoforms (e.g., D. discoideum or D. melanogaster), or when specificity has not been determined experimentally. Specificity is based on the isoform of the MHCII, and heterodimerization has not been observed in vivo. Three different isoforms of the MHCII are encoded by three separate genes: Myh9 (MHCII-A), Myh10 (MHCII-B), and Myh14 (MHCII-C). In addition to the isoform of the heavy chain, another determinant of the molecular properties of each hexamer is the phosphorylation state of the RLC. RLC binds to the three isoforms; thus, it seems unlikely that this constitutes a general mechanism of differential regulation of the isoforms, although Rho-kinase has been suggested to show a modest preference for RLC bound to NMII-A [12]. A major mechanism of NMII isoform regulation is the differential phosphorylation of specific sequences residing in the coiled-coil domain and the non-helical tail of the heavy chains. Several kinases have been reported to phosphorylate the tail domains of MHCII, most notably PKCζ, and TRPM6 and TRPM7, which are members of the TRP calcium channels bearing kinase activity. PKCζ phosphorylates the non-helical tail domain of MHCII-B in a PAK-1-dependent manner [13], whereas TRPM6 and TRPM7 phosphorylate MHCII-A and MHCII-B in the coiled-coil and non-helical tail domain, potentially regulating their localization [14, 15].
NMII is central in the cellular response to mechanical stimulation due to its ability to generate mechanical force. When attached to a fixed point, e.g., a cell–cell or cell–matrix adhesion, NMII-induced sliding of the actin filaments generates contractile force that is transmitted to the plasma membrane, the extracellular matrix and/or other cells. Interestingly, NMII also reacts to mechanical forces, e.g., by changing its localization. Application of mechanical force to the plasma membrane recruits NMII, which assembles thick, stable actomyosin filaments, likely to counter the tension generated in the membrane [16]. Other actin-binding proteins, e.g., cortexillin-I, form a complex with actomyosin that constitutes a “mechanosensory system” that “feels” the mechanical forces applied on the plasma membrane and mediates the cellular response to such stimuli. One example is the change in cellular shape during cytokinesis, which is driven by NMII [17–19].
In addition to its role in the generation of contractile force, NMII is an actin cross-linker [20, 21]. This function is, in principle, force-independent. The mechanism of action of myosin II and the regulation of its different tissue-specific isoforms and splicing variants have been reviewed elsewhere [22, 23]. Here, we focus on two major questions: (1) the activation of myosin II, particularly NMII, by extracellular signals, mechanical and chemical, and (2) whether myosin II mechanically regulates different endpoints to control cellular behavior, focusing on cell migration.
NMII activation by biochemical and mechanical signals
NMII activity is controlled by phosphorylation of the regulatory light chain (RLC). RLC contains several regulatory sites. The most important is Ser19, which enhances the ATPase activity of the heavy chain head domain, and also promotes a conformational change of NMII from folded into extended, assembly-competent [24]. MLCK was the first kinase described to phosphorylate Ser19 [25], but several others, including ROCK-I, ROCK-II, MRCK, PAK kinases, and citron kinase, also phosphorylate it. Some of these kinases also phosphorylate Thr18, which is a synergy site [26]. Thr18 and Ser19 are dephosphorylated by the phosphatase MYPT1. Also, PKC phosphorylates RLC in Ser1 and Ser2 [27], but these phosphorylations inhibit NMII function, likely by preventing its normal assembly [24]. The expression and function of the group of kinases that activate NMII in a given cell in response to specific stimulation is, in general, not well defined.
Different extracellular signals activate NMII through phosphorylation of the RLC, including integrin activation by extracellular matrix and cellular ligands, growth factors acting through kinase-type receptors, e.g., EGF [28], and cytokines and chemokines [29]. The kinases that phosphorylate NMII in response to an extracellular signal may be cell type-dependent or exhibit preference for one isoform; also, different extracellular signals may activate NMII through different kinases, or preferentially activate one isoform. It is important to explore and define these redundancies if these pathways are to be exploited therapeutically.
NMII assembly is also regulated by phosphorylation of the heavy chain of NMII by proteins of the PKC family as well as stretch-activated calcium channels (TRPMs) (reviewed in [22]). This mechanism negatively regulates the nucleation process that forms NMII mini-filaments. Different NMII isoforms have different nucleation kinetics that, together with the ATPase activity of the head domain, determines their force bearing and actin cross-linking capabilities [30].
In addition to chemical signaling, NMII may also be activated directly by mechanical forces. Pipette microaspiration experiments revealed that NMII locates to the point of mechanical perturbation [19]. This is critically dependent on actin cross-linking by additional non-contractile proteins, but also on the ATPase activity of myosin II as well as the lever arm, which controls the threshold of the external force that triggers NMII accumulation at the perturbation site [17]. Externally applied forces lead to mechanical strain on the NMII lever arm when bound to actin, preventing the motor from undergoing full cycle and extending its interaction period with actin. Importantly, these experiments showed that the external force required to stall the motor cycle of the NMII depends on the length of the lever arm, providing a geometric explanation for the mechanical activation of NMII [17]. This may underlie the fact that NMII is required for certain cellular responses to mechanical stimulation, e.g., stiffness-induced stem cell differentiation [2].
Interestingly, external forces can replace NMII in certain scenarios. For example, zipper-deficient embryos (zipper encodes Drosophila non-muscle myosin II heavy chain, or MHCII) have impaired dorsal closure, head involution, and axon patterning [31]. Mutants of zipper exhibit cellular and organization defects during dorsal closure and decreased rate of closure at end stages, and disorganization of the actin at the leading edge cells as well as the amnioserosa and the boundary between them [32]. In zipper mutants, expression of GFP-zip in the amnioserosa alone rescues the organization of zipper-null leading edge cells during this process [32]. From the point of view of the leading edge cells, NMII-dependent contraction generated in the amnioserosa acts as an external force. This suggests that NMII-generated force is transmitted to adjoining cells (presumably through cell–cell adhesions) to reorganize the actin and distribute morphogenetic forces at a distance. Another example of external force replacement of NMII occurs in Drosophila mutants of the snail gene, in which apical NMII accumulation is impaired; in these cells, external force application is sufficient to restore apical NMII clustering [33]. Likewise, in isolated cells, application of mechanical force using a micropipette tip is sufficient to induce focal adhesion elongation [34], suggesting that external forces can substitute actomyosin contractility to mediate adhesion maturation.
NMII controls the mechanics of the molecular clutch that integrates cell adhesion and migration
NMII is emerging as a master integrator of the cellular response to mechanical forces due to its ability to generate forces that in turn induce activation of other downstream effectors. An example of this is integrin-based adhesion. Integrin localize to adhesive structures where the extracellular matrix (ECM) and actin connect. Adhesions are also crossroads of biochemical and mechanical signals that contain multiple mechanorreacting elements.
Fibroblast adhesion to the extracellular matrix protein fibronectin is mainly mediated by the integrin α5β1 [35, 36]. Fibronectin is an ECM protein that interacts with the α5β1 receptor via its RGD motif (reviewed in [37]). However, other motifs (synergy sites) in fibronectin cooperate to α5β1-RGD binding [38]. Importantly, some of these sites are conformationally concealed (cryptic). An elegant study from the Burridge group in the late 1990s showed that fibronectin has a mechanorreactive cryptic site that is exposed upon mechanical stretching of the fibronectin module [39]. Importantly, the cells themselves expose the fibronectin cryptic site by applying mechanical force on the fibronectin fibers in an NMII-dependent manner. NMII creates large adhesions that serve as traction points [40] and generate the contractile force that is transmitted to fibronectin and opens its conformation [41].
Forces are directly implicated in integrin activation. Actomyosin-generated forces expose the domain of α5β1 that interacts with the synergy site on fibronectin [42]. A similar force-dependent mechanism has been proposed for activation of the leukocyte integrin αLβ2 (LFA-1) [43]. Force applied to the tail of the integrin β chain through its tethering to the actin cytoskeleton contributes to the separation of the tail domains of the α and β domains (Fig. 1), causing complete conformational extension of LFA-1 (Fig. 1b, c). This is the high-affinity conformation that supports full leukocyte arrest during extravasation [44].
Fig. 1.
Mechanical regulation of the molecular clutch that controls cell adhesion and migration. Diagrams show the mechanical regulation of talin that controls its interaction with vinculin and force-dependent integrin activation. a α and β integrin chains are represented in an extended, intermediate affinity conformation that binds to RGD motifs in ECM proteins. Integrin is bound to non-stretched talin (represented as a coiled spring), through the tail of the β chain. Talin is bound to actin. Also shown is non-muscle myosin II (NMII). Arrows point to the prospective sliding direction of the actin filaments upon force generation induced by NMII. Numbers represent the critical points of the mechanotransduction point prior to force application (b). b Conformation change of the NMII head (1′) slides actin filaments in the indicated directions. Actin bound to the talin tail stretches the molecule (represented as an extended spring; a two-headed arrow signals the extension) and exposes a binding site (2′) for binding to vinculin. Vinculin binds to actin and strengthens the integrin-actin linkage. Also, mechanical force separates the cytoplasmic domains of the α and β integrin chain (3′) and evokes a conformational movement that extends the head domain of the integrin (4′), increasing its affinity. Force is transmitted to the extracellular matrix and stretches fibronectin, exposing a cryptic site (5′, shown in green) that cooperates to binding. c Same as (b), except vinculin is shown recruiting an extra actin bundle, increasing actin cross-linking at adhesions. d The integrin-actin linkage. Some of the mechanosensitive scaffolds are represented. Black dotted lines represent the talin-integrin force-dependent binding and activation pathway outlined in (a). Red dotted lines indicate the possible routes of mechanical activation of p130CAS. CAS has a cryptic, stretch-dependent Src-phosphorylable Tyr residue (Tyr165. NMII-dependent sliding of the actin filaments generates mechanical force that is transmitted to CAS through zyxin, exposing Tyr165 in CAS. Also depicted, focal adhesion kinase (FAK), which interacts with talin and activates Src, Crk-II, and paxillin, which link FAK-CAS to vinculin and could constitute an alternative pathway of force transmission from actomyosin to CAS
Integrin activation triggers the recruitment of a complex array of adaptor proteins that form the adhesome [45]. Among these, talin promotes integrin conformational activation through its interaction with the tail of the β chain through its N-terminus [46]. The C-terminus of talin interacts with actin and vinculin, which is another adhesion adaptor that also binds actin (reviewed in [47]). Thus, talin functions as a “bridge” between integrins and actin. This function of talin is force-dependent. Sheetz et al. [48] showed that the linkage between fibronectin and actin through integrins could be disassembled by application of a 2-pN mechanical force per binding unit, and that this was dependent on talin. In an in vitro setting, it has been shown that mechanical stretch “opens-up” talin, exposing its site of interaction with vinculin, thus reinforcing its linkage with the actin [49]. The actin/vinculin-binding domain is required for full integrin activation [50]. A model emerges in which force-activated talin exposes its vinculin binding domain, which reinforces the interaction of the integrin complex with actin (Fig. 1b). This model requires talin to incorporate to adhesions in a force-independent manner; talin will bind to integrin and actin in the absence of vinculin, promoting a partial activation of the integrin (Fig. 1a). NMII generates a pulling force that is transmitted to talin through the actin and stretches it. In this model, NMII-generated force is not, by itself, enough to displace the integrin domain to complete activation. Conformational opening of talin recruits vinculin, which reinforces the bonds of the whole complex to actin via formation of additional bonds to the same actin filament (Fig. 1b) or through association to an additional actin filament (Fig. 1c). Increased tethering to actin likely increases the efficiency (or the module) of the force transmitted to the integrin, promoting full activation. A FRET-based force sensor has recently been used to show that vinculin transmits force when incorporated to adhesions, particularly in adhesions within protrusive regions, supporting this model [51].
Mechanical forces also activate other adhesion proteins, although the mechanisms are not always well characterized. For example, mechanical forces stretch p130CAS [52], exposing a phosphorylation site for Src [53]. The physiological mechanism by which CAS is stretched is unclear, since it does not bind actin directly (Fig. 1b). However, it interacts with several adaptor and signaling proteins that bind actin, e.g., zyxin-α-actinin (Fig. 1d); therefore, these second-order interactions may be sufficient to support actomyosin-dependent stretching. Mechanical forces regulate the preferential association of zyxin to adhesions or actin filaments [54]. Likewise, it has been proposed that Src is activated by mechanical stretch [55]; however, it remains undetermined whether the enzymatic activity of the kinase is directly activated by force, or mechanical forces increase Src substrate availability or accessibility.
Mechanosensitive molecules are crucial regulators of cell migration. Integrin ligation promotes the recruitment of talin and vinculin to form stable structures called adhesions. In adhesions, Src phosphorylates CAS and several other substrates, and the clustering and activation of different effectors, amplify the recruitment to include >150 proteins that form an interactive lattice including >600 protein–protein interactions [45].
The molecular makeup of the adhesions and the number and strength of the integrin–ECM interactions regulate cell adhesion and migration [56]. This means that more integrin–ECM bonds result in stronger adhesion. Conversely, migration displays an optimal threshold of integrin–ECM bonds. Below this threshold, cells do not attach properly. Above the threshold, they attach too strongly and do not move [56].
Adhesive strength also depends on a “molecular clutch” that connects the ECM with the actin cytoskeleton [57–59]. The clutch components interact strongly (grip) or weakly (slip). These interactions determine the strength of the adhesive bonds and their ability to transmit traction to support cell migration [60]. Migration is also enabled within an optimal threshold of traction force. When forces are within the optimal transmission threshold, cells migrate. Above the traction threshold, cells adhere too strongly, contract the matrix and do not move; below the threshold, the cells slip, and the traction force generated is often insufficient to move the cell body [61].
A simplified view of this molecular clutch includes the interaction of the ECM ligand with the integrin, the integrin with the adhesion complex (talin, vinculin, etc.) and the adhesion complex with actin. The efficiency of the clutch depends of the integrin ligand, the integrin type, and the recruitment of the components of the adhesive complex. It also depends on the type and number of molecules within adhesive complexes and their interacting affinities [59]. Ultimately, transmission efficiency depends on how adhesion complexes bind actin. In general, strong interactions will transmit forces better than weak bonds that may be destroyed upon force application.
NMII controls the efficiency of the adhesion clutch by applying force to the adhesions. Of note, NMII is not physically present at adhesions, but generates contractile forces that pass through the actin cables and are applied on the extracellular matrix through adhesive contacts (“action at a distance”), which thus act as traction points. Therefore, NMII-generated mechanical force relies on good transmission, i.e., a clutch in which every piece is bound to the next with enough affinity to support mechanical force passing through the linkages without breaking the interactions. At the same time, the clutch must allow some slippage to enable disassembly, which is required to restart the assembly/turnover cycle that occurs in migrating cells [62]. NMII also regulates adhesion growth and dynamics through actin cross-linking, which brings together actin-associated adhesion proteins [20]. This is force-independent, although tension enhances cross-linking.
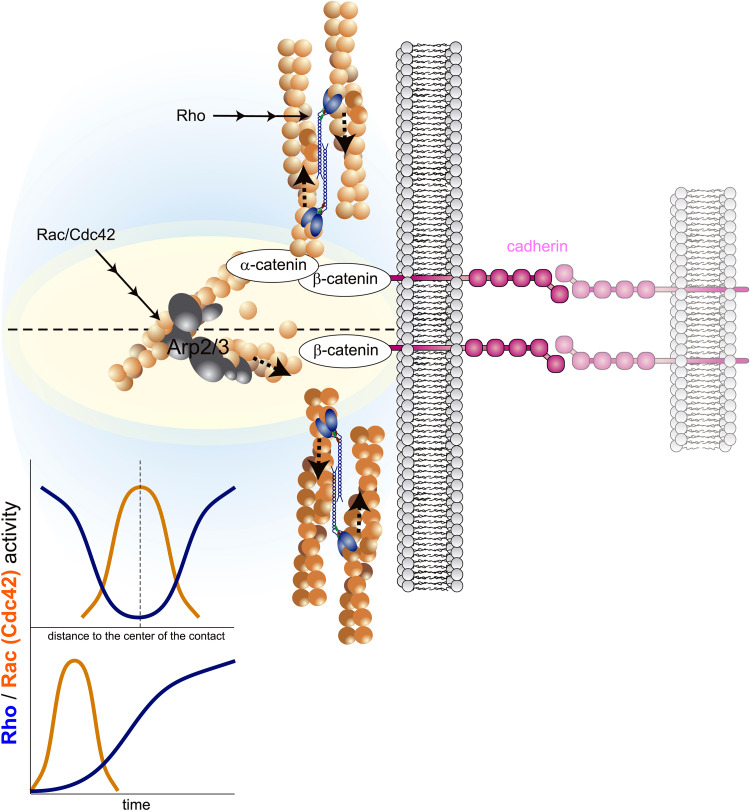
NMII not only regulates cell–matrix adhesions, but also cadherin-mediated cell–cell contacts. NMII is recruited to cell–cell contacts upon cadherin engagement and seems to stabilize the complex from the sides (Fig. 2) [63]. Epithelial cells in which NMII-A is downregulated using interfering RNA do not form stable contacts [64], and mice depleted of the heavy chain of isoform II-A (MHCII-A−/−) die very early (E6.5) due to massive defects in cell–cell contacts and epithelial layering [65]. It is unclear whether cadherin engagement triggers a biochemical signal that recruits NMII, or mechanical tension on the plasma membrane at the junction promotes NMII nucleation [63], similar to what has been observed in Drosophila [66]. The function of NMII in cell–cell interactions is quite different from cell–matrix adhesions during migration. In migration, adhesions are dynamic and may assemble, disassemble, and reform elsewhere to enable traction at new gripping points [62]. Cell–cell adhesions are more stable, and disassemble only in response to massive reorganizations of the cell layers, e.g., during morphogenesis. In the next section, we discuss these differences in light of our emerging understanding of the molecular regulation of the role of mechanical forces and the function of NMII.
Fig. 2.
NMII participates in cell–cell contact formation and stabilization. Diagram shows actin polymerization and NMII-mediated contraction and bundling in forming cell–cell contacts. Cadherins recruit different proteins, most notably β and α-catenin, which links to actin and may act as a force transducer [115]. Upon cadherin engagement (right), an early, transient wave of Rac/Cdc42-mediated actin polymerization pushes the membrane to form the cell–cell contact. Later, Rho-dependent NMII activation in the periphery of the initial contact extends the contact area and solidifies the cell–cell contact. Bottom left, graphs are non-scale, schematic representations of the Rac/Cdc42 and Rho localization (top) and kinetics (bottom)
NMII shapes development and homeostasis
Mechanical forces contribute to shaping cells and tissues during development. Other excellent reviews have dissected the role of mechanical forces in development in anatomical detail [67–69]. The powerful genetics of development systems rapidly identified NMII as a central integrator of morphogenesis [31]. In most morphogenetic movements, NMII localizes to areas that undergo constriction [70], e.g., during spiracle invagination in Drosophila, or during gastrulation. These areas often coincide with the apical pole of an epithelial sheet. Interestingly, NMII clustering in the apical pole cannot be described using a cumulative distribution function, but it displays a dynamic oscillatory behavior that enables tissue elongation by causing periodic contractions of the underlying actomyosin cytoskeleton [71]. These systems have revealed a pattern: deactivators of Rho function (GTPase Activating Proteins) localize to the basolateral face of epithelial cells, whereas Rho activators (GTP Exchange Factors) translocate to the apical pole and activate Rho1 (homologue of RhoA) [72], which activates zipper (homologue of NMII) through the kinase DRok (Drosophila Rho-Associated Kinase, homologue of ROCK) (Fig. 3a) [73]. NMII-based actin cross-linking and contraction constrict the apical pole, whereas deactivation of Rho and accumulation of Rac/Cdc42 activators (GEFs) at the basolateral pole enable Rac/Cdc42 activation and induce actin polymerization-based extension (Fig. 3b) [72]. The segregation of Rho and Rac/Cdc42 activities to different poles is a spatial mechanism that supports the observation that Rac/Cdc42 and Rho activities antagonize each other [74]. Several recent studies revealed that the Rho/Rac antagonism is more complicated than originally thought. At the leading edge of migrating cells, the activation pattern of small Rho GTPases is tightly regulated spatially and temporally as follows: an initial spike of RhoA is closely followed by Rac/Cdc42, and when Rac starts to decline inside the leading edge, RhoA promotes actin reorganization into thicker bundles [75]. Mechanistically, Rac activates p190RhoGAP, blocking RhoA [76]. Conversely, RhoA promotes Rac inactivation via ARHGAP22 [77], providing a molecular switch that underlies reciprocal inhibition. However, which signals induce predominance of one or other pathway remains unclear. In principle, areas where NMII accumulate will show predominance of RhoA signaling, whereas protrusive areas where NMII is not found would be dominated by Rac signaling.
Fig. 3.
Role of NMII and actin polymerization in morphogenetic epithelial movements. a Apical constriction mediated by NMII. The initial signal is represented by a morphogenetic hormone or peptide, but other signals may accomplish the same effect. The hormone binds a G protein-coupled receptor that, acting through G proteins, activates RhoGEFs, which in turn activate Rho, which induces NMII phosphorylation and activation via ROCK [116]. NMII-mediated actin contraction mediates constriction of the apical pole. b Basolateral extension in the direction of migration is induced by actin polymerization. Constriction at this area is prevented by accumulation of RhoGAPs, which inactivate Rho and locally deactivate NMII. This is simultaneous to the clustering of Rac/Cdc42 activators (GEFs), which depends of the presence of the polarity proteins Par3/Par6 and may require atypical PKC activation [72]. Rac/Cdc42 promote actin polymerization through the Arp2/3 complex via WASP/WAVE proteins
In intercalation, NMII mainly localizes to dorso-ventral junctions and away from lateral junctions [78], where bazooka (Drosophila homologue of mammalian Par3) localize [79]. This decreases tension in the lateral junctions and allows lateral displacements, such as those required for cell intercalation [80]. In general, NMII localizes to areas that undergo constriction and subcellular structures resistant to deformation. NMII may also recruit additional signaling elements through cross-linking, or by inducing force-dependent conformational changes that create binding sites for other signaling components. A recent study has identified the GIT–PIX–PAK complex as a key component of hemidesmosomes that reacts to mechanical forces [81]. NMII may control the recruitment of this complex to hemidesmosomes by activating mechanosensitive kinases (e.g., Src) and exposing phosphorylable sites in upstream regulators to create new docking sites for the complex [82].
The signal that initially recruits the NMII activating machinery remains elusive. Early signaling can be tracked to polarity determinants, e.g., Par proteins, which may recruit and activate NMII-activating or -inhibiting signals; but what triggers their initial polarization is unclear. A hypothesis is that gradients of secreted factors, hormones or morphogens, induce the localized activation of polarity determinants. This would create an asymmetric signal that would initiate cellular polarization. One such molecule is Fog (folded gastrulation), which is secreted apically by the mesoderm precursor cells in a polarized fashion in response to tissue identity genes, e.g., Twist. Fog acts in an autocrine manner to recruit and activate NMII, producing apical constriction [70]. Wnt signaling is also involved in activating NMII, but the connections are not clear [83].
NMII also mediates more sophisticated morphogenetic movements, e.g., cell rotation during ommatidia formation in eye development. In this type of movement, groups of preformed cells rotate inside the layer to form polarized photorreceptors [84]. NMII localizes to the interface between ommatidial and undifferentiated cells and causes local constriction and retraction, weakening the boundary between the ommatidial cells and surrounding undifferentiated cells, and allowing the cells to rotate. The direction of rotation depends on the localization of the cell clusters within the eye, but ultimately depends on unknown reasons. Several possibilities include: polarized morphogen gradients, asymmetric forces exerted by the surrounding, undifferentiated cells, or the intrinsic chirality of the cells [85], which is under the control of the isoform B of NMII in mammalian cells [86].
Another role of NMII in morphogenesis resides in its ability to control retrograde flow within the cell [87]. An asymmetric distribution of NMII generates tension gradients that create contractile flows of actomyosin [88, 89]. These flow movements exert friction as the actin moves. Friction has been shown to direct some morphogenetic movements such as epiboly [90]. This is similar to the retrograde flow of actin observed in migrating cells, which causes friction on substrate-bound adhesions and may underlie force-dependent adhesion growth [91].
NMII participates in mechanotransduction during malignant transformation and metastasis
Growing evidence indicates that cancer cells modify themselves and their microenvironment to promote their own survival and proliferation [92]. They also undergo different transitions that enhance cell invasion and migration [93]. Together, these changes facilitate their adaptation against external aggressions, e.g., chemotherapy. A major difference with normal cells is that, whereas normal cells die as part of differentiation and renewal programs, tumors develop mechanisms to avoid programmed death and grow uncontrollably [94]. As part of this “super-adaptation” program, cancer cells can modify the mechanical properties of their microenvironment as well as their own to enhance proliferation and/or to migrate away from the primary tumor and generate distal metastases.
Physiological stiffness acts a proliferation inhibitor: in vitro increased stiffness relieves growth inhibition, and even normal cells increase their proliferation [3]. Tumor cells increase tissue stiffness, likely to enhance proliferation and tumor growth [92]. Stiffness enhances proliferation through several molecular mechanisms. These may be dependent on the cell type and its context. For example, increased stiffness promotes integrin clustering, which leads to an increase in FAK/Src kinase activity and promotes recruitment of the Ras/MAPK pathway scaffolds SHC and Grb2 [95]. As outlined in previous sections, p130CAS phosphorylation by Src depends on its mechanical extension [53]. Src-dependent phosphorylation of p130CAS facilitates its interaction with the adaptor Nck, which also serves as a MAPK/ERK scaffold [96]. Therefore, it seems that the mechanical properties of the microenvironment modulate cell proliferation through different pathways that converge on the activation of the MAPK/ERK pathway.
How do tumor cells increase the stiffness of their microenvironment? Enhanced proliferation increases the cell density per volume unit, packing the cells more tightly, which increases the overall resistance of the tumor mass. This phenomenon also enhances resistance against some forms of chemotherapy [97]. Another mechanism is extracellular matrix deposition and reorganization. Most carcinomas of epithelial origin display increased matrix deposition and also reorganize collagen fibers by several mechanisms, including secretion of matrix metalloproteinases (reviewed in [98]), post-translational modifications, e.g., oxidation by lysyl oxidases [99], and NMII-mediated cell contraction [39, 100]. Tumor cells reorganize the matrix using focal adhesions and stress fibers as traction devices that pull the matrix in a RhoA- and NMII-dependent manner [40]. In fact, some solid tumors display elevated rigidity that depends on the activity of RhoA and ERK; countering RhoA decreases cellular stiffness and reverts some of the cellular traits associated with malignant transformation [101]. The in vivo role of Rho-mediated contractility in promoting tumorigenesis was recently illustrated in a rodent model in which conditional induction of ROCK, a downstream effector of RhoA that activates NMII, promotes tumorigenesis, whereas its inhibition blocked tumor formation and progression [102].
Importantly, recent progress using atomic force microscopy (AFM) has revealed that tissue becomes heterogeneously rigid during breast malignant transformation. Whereas the overall stiffness of breast tumors increases likely due to ECM stiffening, the actual tumor cells become “softer” or less rigid. Also, tumors display discrete areas of increased stiffness towards the outer edge of the tumors [103]. Interestingly, similar increases in peripheral stiffness have been recorded using AFM in transgenic mice in which the ROCK–NMII axis was activated [102], suggesting that NMII is at least partially responsible for the local changes in tissue rigidity during tumor formation.
The mechanical properties of the microenvironment also play a central role in how cells break away from the primary tumor and invade the surrounding healthy tissue. The initial process can be envisioned as a “perversion” of the apical constriction/basolateral extension observed in morphogenesis, which is likely enhanced by the increased rigidity of the tumor microenvironment and the reorganization of the collagen fibers. In this case, the “apical” side of the tumor cells would be facing the core of the tumor, whereas the basolateral area would be oriented outwards, towards the host tissue. How these cells acquire a polarized, migratory morphology is unclear. One possibility is that chemoattractant signals from outside the tumor induce polarized actin polymerization at the front and switch signaling from cadherin-based to integrin-based, promoting motility [104]. Another possibility is based on an initial constriction of the rear. This would result from mechanical and cell–cell contact cues, similar to what is observed in gastrulation and morphogenesis (reviewed in [105]).
The change of cellular behavior from epithelial-like to migratory and invasive can also be explained in terms of a possible differentiation of migratory cancer cells from a hypothetical “cancer stem cell”. This has yet to be fully demonstrated in vivo, and implies that the migratory cells would not need to be located to the periphery of the tumor; but they would still require to break free from their microenvironment using mechanisms similar to those described.
Dissociation from the primary tumor may occur in two different ways: as single cells that become migratory, or as collective “chains” of cells that migrate together. Both situations have been observed in vivo [106]. Important outstanding questions are whether different types of tumor cells display preference for one mechanism of egress or the other, and whether NMII regulates their “choice”.
There is emerging evidence that increased stiffness promotes the acquisition of migratory traits by tumor cells [92, 107]. Although the picture is not complete, the current data suggest that rigidity promotes an epithelial–mesenchymal transition (EMT) [101], which means that the cells undergo morphological changes from epithelial-like to mesenchymal, becoming more migratory [108]. Interestingly, some genes that control matrix rigidity and metastasis through post-translational modification of the ECM components, e.g., lysyl oxidase (LOX)-2 [109], also control EMT from inside cancer cells [110]. Tumor cells follow reorganized collagen bundles emanating from the tumor in a radial manner and move towards the basement membrane [111].
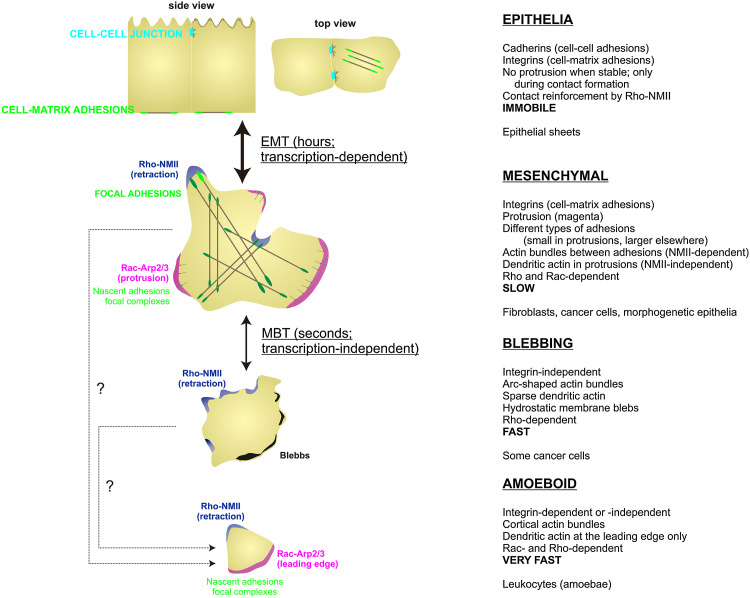
Cancer cells adopt varied morphologies during migration. EMT postulates that most invasive cells display mesenchymal morphology and behavior, but different morphologies have been observed both in vitro and in vivo (Fig. 4). These morphologies are part of a continuum that includes cell shapes from non-migratory epithelial, to mesenchymal, to amoeboid-like, or blebbing [106]. Unlike mesenchymal cells, blebbing cells do not generally degrade the matrix as they go and their migration depends on oscillatory, NMII-dependent contractions of the cellular cortex [112], similar to leukocytes [113]. Importantly, this process depends on the microenvironment of the migrating cell and occurs in a matter of seconds, suggesting that, unlike EMT, this is a transcription-independent process [114].
Fig. 4.
Morphology relates to the migratory properties of the cell. The four most typical morphologies related to cell migration are depicted: non-migratory epithelial, mesenchymal, bleb-based, and amoeboid. Magenta represents Rac/Cdc42-dependent protrusive areas, blue RhoA-NMII-dependent retraction areas, black RhoA-NMII-dependent membrane blebs, green cell–matrix adhesions, and brown lines actin bundles. Text next to each of the morphologies describes receptors implicated in adhesion, protrusiveness, and predominant geometry of the actin, small Rho GTPase usage, and migration speed (in bold) and cell types that most commonly display the indicated morphology. Known transitions between the different migratory modes are indicated between morphologies; dashed lines with question marks indicate transitions that have been postulated but lack definitive experimental proof
Another type of migratory morphology is that of very fast, amoeboid cells, e.g., leukocytes. This modality shares traits of the mesenchymal mode (front–back polarization, small adhesive contacts, Rho/Rac dependence) and blebbing mode (high speed, possible integrin independence), but whether mesenchymal or blebbing cells can adopt amoeboid morphologies is currently undetermined.
Future perspectives
Despite the abundance of data, current research is only scratching the surface of the regulatory role of mechanical forces and NMII on cellular behavior. Progress has been more evident in development. This is due to the size of the experimental systems, which enables observing large movements caused by forces of a relatively high magnitude. But advances in physical detection and measurement methods, microscopy, and cell biology, have allowed a rapid development of the field of mechanobiology at a molecular level. In this review, we have illustrated relatively well-characterized molecular phenomena, but it is unlikely these are unique, and the list of mechanorreactive molecules will likely extend over subsequent years.
Over 90 % of cancer-related deaths are due to metastatic dissemination of cells of the original tumor. Although the connection of these molecular events with normal development and malignancy is still tenuous, it opens up novel avenues of possible therapeutic intervention to arrest malignant cells. Careful understanding of how the forces implicated in the conversion of normal into tumor cells, and the acquisition of motile, invasive features, may prompt the discovery of novel therapeutic targets to control metastasis.
Acknowledgments
We apologize for those studies not cited due to space constraints. Also, we are grateful to Professors Michel Labouesse and Francisco Sánchez-Madrid for critical reading of the manuscript. M.V.-M. acknowledges funding from the Ramón y Cajal program (RyC-2010-06094, MINECO, Spain), Grant SAF2011-24953 from the MINECO, Spain, a Marie Curie Career Integration Grant (CIG-293719) and a grant from the Fundación Ramón Areces (Spain).
References
- 1.Scott JD, Pawson T. Cell signaling in space and time: where proteins come together and when they’re apart. Science. 2009;326(5957):1220–1224. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 3.Klein EA, Yin L, Kothapalli D, Castagnino P, Byfield FJ, Xu T, Levental I, Hawthorne E, Janmey PA, Assoian RK. Cell-cycle control by physiological matrix elasticity and in vivo tissue stiffening. Curr Biol. 2009;19(18):1511–1518. doi: 10.1016/j.cub.2009.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79(1):144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raab M, Swift J, PC PD, Shah P, Shin JW, Discher DE (2012) Crawling from soft to stiff matrix polarizes the cytoskeleton and phosphoregulates myosin-II heavy chain. J Cell Biol. doi:10.1083/jcb.201205056 [DOI] [PMC free article] [PubMed]
- 6.Mooseker MS, Foth BJ (2007) The structural and functional diversity of the myosin family of actin-based molecular motors. In: Coluccio LM (ed) Myosins: a superfamily of molecular motors. Springer, Watertown, pp 1–34
- 7.Spudich JA. The myosin swinging cross-bridge model. Nat Rev Mol Cell Biol. 2001;2(5):387–392. doi: 10.1038/35073086. [DOI] [PubMed] [Google Scholar]
- 8.Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299(5613):1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 9.Kovacs M, Toth J, Hetenyi C, Malnasi-Csizmadia A, Sellers JR. Mechanism of blebbistatin inhibition of myosin II. J Biol Chem. 2004;279(34):35557–35563. doi: 10.1074/jbc.M405319200. [DOI] [PubMed] [Google Scholar]
- 10.Cremo CR, Hartshorne DJ (2007) Smooth-muscle myosin II. In: Coluccio LM (ed) Myosins: a superfamily of molecular motors. Springer, Watertown, pp 171–222
- 11.El-Mezgueldi M, Bagshaw CR (2007) The myosin family: biochemical and kinetic properties. In: Coluccio LM (ed) Myosins: a superfamily of molecular motors. Springer, Watertown, pp 55–93
- 12.Sandquist JC, Swenson KI, Demali KA, Burridge K, Means AR. Rho kinase differentially regulates phosphorylation of nonmuscle myosin II isoforms A and B during cell rounding and migration. J Biol Chem. 2006;281(47):35873–35883. doi: 10.1074/jbc.M605343200. [DOI] [PubMed] [Google Scholar]
- 13.Even-Faitelson L, Ravid S. PAK1 and aPKCzeta regulate myosin II-B phosphorylation: a novel signaling pathway regulating filament assembly. Mol Biol Cell. 2006;17(7):2869–2881. doi: 10.1091/mbc.E05-11-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark K, Middelbeek J, Dorovkov MV, Figdor CG, Ryazanov AG, Lasonder E, van Leeuwen FN. The alpha-kinases TRPM6 and TRPM7, but not eEF-2 kinase, phosphorylate the assembly domain of myosin IIA, IIB and IIC. FEBS Lett. 2008;582(20):2993–2997. doi: 10.1016/j.febslet.2008.07.043. [DOI] [PubMed] [Google Scholar]
- 15.Clark K, Middelbeek J, Lasonder E, Dulyaninova NG, Morrice NA, Ryazanov AG, Bresnick AR, Figdor CG, van Leeuwen FN. TRPM7 regulates myosin IIA filament stability and protein localization by heavy chain phosphorylation. J Mol Biol. 2008;378(4):790–803. doi: 10.1016/j.jmb.2008.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo T, Mohan K, Srivastava V, Ren Y, Iglesias PA, Robinson DN. Understanding the cooperative interaction between myosin II and actin cross-linkers mediated by actin filaments during mechanosensation. Biophys J. 2012;102(2):238–247. doi: 10.1016/j.bpj.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren Y, Effler JC, Norstrom M, Luo T, Firtel RA, Iglesias PA, Rock RS, Robinson DN. Mechanosensing through cooperative interactions between myosin II and the actin crosslinker cortexillin I. Curr Biol. 2009;19(17):1421–1428. doi: 10.1016/j.cub.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reichl EM, Ren Y, Morphew MK, Delannoy M, Effler JC, Girard KD, Divi S, Iglesias PA, Kuo SC, Robinson DN. Interactions between myosin and actin crosslinkers control cytokinesis contractility dynamics and mechanics. Curr Biol. 2008;18(7):471–480. doi: 10.1016/j.cub.2008.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Effler JC, Kee YS, Berk JM, Tran MN, Iglesias PA, Robinson DN. Mitosis-specific mechanosensing and contractile-protein redistribution control cell shape. Curr Biol. 2006;16(19):1962–1967. doi: 10.1016/j.cub.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell Biol. 2008;10(9):1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu XS, Lee E, Chen T, Kuczmarski E, Chisholm RL, Knecht DA. During multicellular migration, myosin ii serves a structural role independent of its motor function. Dev Biol. 2001;232(1):255–264. doi: 10.1006/dbio.2000.0132. [DOI] [PubMed] [Google Scholar]
- 22.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10(11):778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conti MA, Adelstein RS. Nonmuscle myosin II moves in new directions. J Cell Sci. 2008;121(Pt 1):11–18. doi: 10.1242/jcs.007112. [DOI] [PubMed] [Google Scholar]
- 24.Ikebe M. Regulation of the function of mammalian myosin and its conformational change. Biochem Biophys Res Commun. 2008;369(1):157–164. doi: 10.1016/j.bbrc.2008.01.057. [DOI] [PubMed] [Google Scholar]
- 25.Adelstein RS, Conti MA. Phosphorylation of platelet myosin increases actin-activated myosin ATPase activity. Nature. 1975;256(5518):597–598. doi: 10.1038/256597a0. [DOI] [PubMed] [Google Scholar]
- 26.Ikebe M, Hartshorne DJ, Elzinga M. Identification, phosphorylation, and dephosphorylation of a second site for myosin light chain kinase on the 20,000-dalton light chain of smooth muscle myosin. J Biol Chem. 1986;261(1):36–39. [PubMed] [Google Scholar]
- 27.Komatsu S, Ikebe M. The phosphorylation of myosin II at the Ser1 and Ser2 Is critical for normal platelet-derived growth factor induced reorganization of myosin filaments. Mol Biol Cell. 2007;18(12):5081–5090. doi: 10.1091/mbc.E06-12-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwabu A, Smith K, Allen FD, Lauffenburger DA, Wells A. Epidermal growth factor induces fibroblast contractility and motility via a protein kinase C delta-dependent pathway. J Biol Chem. 2004;279(15):14551–14560. doi: 10.1074/jbc.M311981200. [DOI] [PubMed] [Google Scholar]
- 29.Vicente-Manzanares M, Cabrero JR, Rey M, Perez-Martinez M, Ursa A, Itoh K, Sanchez-Madrid F. A role for the Rho-p160 Rho coiled-coil kinase axis in the chemokine stromal cell-derived factor-1alpha-induced lymphocyte actomyosin and microtubular organization and chemotaxis. J Immunol. 2002;168(1):400–410. doi: 10.4049/jimmunol.168.1.400. [DOI] [PubMed] [Google Scholar]
- 30.Kim KY, Kovacs M, Kawamoto S, Sellers JR, Adelstein RS. Disease-associated mutations and alternative splicing alter the enzymatic and motile activity of nonmuscle myosins II-B and II-C. J Biol Chem. 2005;280(24):22769–22775. doi: 10.1074/jbc.M503488200. [DOI] [PubMed] [Google Scholar]
- 31.Young PE, Richman AM, Ketchum AS, Kiehart DP. Morphogenesis in Drosophila requires nonmuscle myosin heavy chain function. Genes Dev. 1993;7(1):29–41. doi: 10.1101/gad.7.1.29. [DOI] [PubMed] [Google Scholar]
- 32.Franke JD, Montague RA, Kiehart DP. Nonmuscle myosin II generates forces that transmit tension and drive contraction in multiple tissues during dorsal closure. Curr Biol. 2005;15(24):2208–2221. doi: 10.1016/j.cub.2005.11.064. [DOI] [PubMed] [Google Scholar]
- 33.Pouille PA, Ahmadi P, Brunet AC, Farge E (2009) Mechanical signals trigger myosin II redistribution and mesoderm invagination in Drosophila embryos. Sci Signal 2 (66):ra16. doi:10.1126/scisignal.2000098 [DOI] [PubMed]
- 34.Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, Kam Z, Geiger B, Bershadsky AD. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J Cell Biol. 2001;153(6):1175–1186. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamkun JW, DeSimone DW, Fonda D, Patel RS, Buck C, Horwitz AF, Hynes RO. Structure of integrin, a glycoprotein involved in the transmembrane linkage between fibronectin and actin. Cell. 1986;46(2):271–282. doi: 10.1016/0092-8674(86)90744-0. [DOI] [PubMed] [Google Scholar]
- 36.Horwitz AR. The origins of the molecular era of adhesion research. Nat Rev Mol Cell Biol. 2012;13(12):805–811. doi: 10.1038/nrm3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110(6):673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 38.Dufour S, Duband JL, Humphries MJ, Obara M, Yamada KM, Thiery JP. Attachment, spreading and locomotion of avian neural crest cells are mediated by multiple adhesion sites on fibronectin molecules. EMBO J. 1988;7(9):2661–2671. doi: 10.1002/j.1460-2075.1988.tb03119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong C, Chrzanowska-Wodnicka M, Brown J, Shaub A, Belkin AM, Burridge K. Rho-mediated contractility exposes a cryptic site in fibronectin and induces fibronectin matrix assembly. J Cell Biol. 1998;141(2):539–551. doi: 10.1083/jcb.141.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chrzanowska-Wodnicka M, Burridge K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J Cell Biol. 1996;133(6):1403–1415. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beningo KA, Dembo M, Kaverina I, Small JV, Wang YL. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J Cell Biol. 2001;153(4):881–888. doi: 10.1083/jcb.153.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedland JC, Lee MH, Boettiger D. Mechanically activated integrin switch controls alpha5beta1 function. Science. 2009;323(5914):642–644. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- 43.Schurpf T, Springer TA. Regulation of integrin affinity on cell surfaces. EMBO J. 2011;30(23):4712–4727. doi: 10.1038/emboj.2011.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carman CV, Springer TA. Integrin avidity regulation: are changes in affinity and conformation underemphasized? Curr Opin Cell Biol. 2003;15(5):547–556. doi: 10.1016/j.ceb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat Cell Biol. 2007;9(8):858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horwitz A, Duggan K, Buck C, Beckerle MC, Burridge K. Interaction of plasma membrane fibronectin receptor with talin–a transmembrane linkage. Nature. 1986;320(6062):531–533. doi: 10.1038/320531a0. [DOI] [PubMed] [Google Scholar]
- 47.Critchley DR. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu Rev Biophys. 2009;38:235–254. doi: 10.1146/annurev.biophys.050708.133744. [DOI] [PubMed] [Google Scholar]
- 48.Jiang G, Giannone G, Critchley DR, Fukumoto E, Sheetz MP. Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature. 2003;424(6946):334–337. doi: 10.1038/nature01805. [DOI] [PubMed] [Google Scholar]
- 49.del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323(5914):638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Calderwood DA, Yan B, de Pereda JM, Alvarez BG, Fujioka Y, Liddington RC, Ginsberg MH. The phosphotyrosine binding-like domain of talin activates integrins. J Biol Chem. 2002;277(24):21749–21758. doi: 10.1074/jbc.M111996200. [DOI] [PubMed] [Google Scholar]
- 51.Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, Schwartz MA. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466(7303):263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sawada Y, Sheetz MP. Force transduction by Triton cytoskeletons. J Cell Biol. 2002;156(4):609–615. doi: 10.1083/jcb.200110068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127(5):1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshigi M, Hoffman LM, Jensen CC, Yost HJ, Beckerle MC. Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J Cell Biol. 2005;171(2):209–215. doi: 10.1083/jcb.200505018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, Chien S. Visualizing the mechanical activation of Src. Nature. 2005;434(7036):1040–1045. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 56.Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature. 1997;385(6616):537–540. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 57.Mitchison T, Kirschner M. Cytoskeletal dynamics and nerve growth. Neuron. 1988;1(9):761–772. doi: 10.1016/0896-6273(88)90124-9. [DOI] [PubMed] [Google Scholar]
- 58.Brown CM, Hebert B, Kolin DL, Zareno J, Whitmore L, Horwitz AR, Wiseman PW. Probing the integrin-actin linkage using high-resolution protein velocity mapping. J Cell Sci. 2006;119(Pt 24):5204–5214. doi: 10.1242/jcs.03321. [DOI] [PubMed] [Google Scholar]
- 59.Hu K, Ji L, Applegate KT, Danuser G, Waterman-Storer CM. Differential transmission of actin motion within focal adhesions. Science. 2007;315(5808):111–115. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- 60.Jurado C, Haserick JR, Lee J. Slipping or gripping? Fluorescent speckle microscopy in fish keratocytes reveals two different mechanisms for generating a retrograde flow of actin. Mol Biol Cell. 2005;16(2):507–518. doi: 10.1091/mbc.E04-10-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gardel ML, Sabass B, Ji L, Danuser G, Schwarz US, Waterman CM. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J Cell Biol. 2008;183(6):999–1005. doi: 10.1083/jcb.200810060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, Horwitz AF. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol. 2004;6(2):154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 63.Yamada S, Nelson WJ. Localized zones of Rho and Rac activities drive initiation and expansion of epithelial cell–cell adhesion. J Cell Biol. 2007;178(3):517–527. doi: 10.1083/jcb.200701058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shewan AM, Maddugoda M, Kraemer A, Stehbens SJ, Verma S, Kovacs EM, Yap AS. Myosin 2 is a key Rho kinase target necessary for the local concentration of E-cadherin at cell–cell contacts. Mol Biol Cell. 2005;16(10):4531–4542. doi: 10.1091/mbc.E05-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Conti MA, Even-Ram S, Liu C, Yamada KM, Adelstein RS. Defects in cell adhesion and the visceral endoderm following ablation of nonmuscle myosin heavy chain II-A in mice. J Biol Chem. 2004;279(40):41263–41266. doi: 10.1074/jbc.C400352200. [DOI] [PubMed] [Google Scholar]
- 66.Fernandez-Gonzalez R, Simoes Sde M, Roper JC, Eaton S, Zallen JA. Myosin II dynamics are regulated by tension in intercalating cells. Dev Cell. 2009;17(5):736–743. doi: 10.1016/j.devcel.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Keller R, Davidson LA, Shook DR. How we are shaped: the biomechanics of gastrulation. Differentiation. 2003;71(3):171–205. doi: 10.1046/j.1432-0436.2003.710301.x. [DOI] [PubMed] [Google Scholar]
- 68.Heisenberg CP, Tada M. Zebrafish gastrulation movements: bridging cell and developmental biology. Semin Cell Dev Biol. 2002;13(6):471–479. doi: 10.1016/s1084952102001003. [DOI] [PubMed] [Google Scholar]
- 69.Lecuit T, Lenne PF, Munro E. Force generation, transmission, and integration during cell and tissue morphogenesis. Annu Rev Cell Dev Biol. 2011;27:157–184. doi: 10.1146/annurev-cellbio-100109-104027. [DOI] [PubMed] [Google Scholar]
- 70.Dawes-Hoang RE, Parmar KM, Christiansen AE, Phelps CB, Brand AH, Wieschaus EF. Folded gastrulation, cell shape change and the control of myosin localization. Development. 2005;132(18):4165–4178. doi: 10.1242/dev.01938. [DOI] [PubMed] [Google Scholar]
- 71.He L, Wang X, Tang HL, Montell DJ. Tissue elongation requires oscillating contractions of a basal actomyosin network. Nat Cell Biol. 2010;12(12):1133–1142. doi: 10.1038/ncb2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simoes S, Denholm B, Azevedo D, Sotillos S, Martin P, Skaer H, Hombria JC, Jacinto A. Compartmentalisation of Rho regulators directs cell invagination during tissue morphogenesis. Development. 2006;133(21):4257–4267. doi: 10.1242/dev.02588. [DOI] [PubMed] [Google Scholar]
- 73.Winter CG, Wang B, Ballew A, Royou A, Karess R, Axelrod JD, Luo L. Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell. 2001;105(1):81–91. doi: 10.1016/s0092-8674(01)00298-7. [DOI] [PubMed] [Google Scholar]
- 74.Sander EE, ten Klooster JP, van Delft S, van der Kammen RA, Collard JG. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J Cell Biol. 1999;147(5):1009–1022. doi: 10.1083/jcb.147.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461(7260):99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nimnual AS, Taylor LJ, Bar-Sagi D. Redox-dependent downregulation of Rho by Rac. Nat Cell Biol. 2003;5(3):236–241. doi: 10.1038/ncb938. [DOI] [PubMed] [Google Scholar]
- 77.Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135(3):510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 78.Bertet C, Sulak L, Lecuit T. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature. 2004;429(6992):667–671. doi: 10.1038/nature02590. [DOI] [PubMed] [Google Scholar]
- 79.Zallen JA, Wieschaus E. Patterned gene expression directs bipolar planar polarity in Drosophila. Dev Cell. 2004;6(3):343–355. doi: 10.1016/s1534-5807(04)00060-7. [DOI] [PubMed] [Google Scholar]
- 80.Rauzi M, Verant P, Lecuit T, Lenne PF. Nature and anisotropy of cortical forces orienting Drosophila tissue morphogenesis. Nat Cell Biol. 2008;10(12):1401–1410. doi: 10.1038/ncb1798. [DOI] [PubMed] [Google Scholar]
- 81.Zhang H, Landmann F, Zahreddine H, Rodriguez D, Koch M, Labouesse M. A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature. 2011;471(7336):99–103. doi: 10.1038/nature09765. [DOI] [PubMed] [Google Scholar]
- 82.Nayal A, Webb DJ, Brown CM, Schaefer EM, Vicente-Manzanares M, Horwitz AR. Paxillin phosphorylation at Ser273 localizes a GIT1-PIX-PAK complex and regulates adhesion and protrusion dynamics. J Cell Biol. 2006;173(4):587–589. doi: 10.1083/jcb.200509075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zimmerman SG, Thorpe LM, Medrano VR, Mallozzi CA, McCartney BM. Apical constriction and invagination downstream of the canonical Wnt signaling pathway require Rho1 and myosin II. Dev Biol. 2010;340(1):54–66. doi: 10.1016/j.ydbio.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fiehler RW, Wolff T. Drosophila Myosin II, Zipper, is essential for ommatidial rotation. Dev Biol. 2007;310(2):348–362. doi: 10.1016/j.ydbio.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu J, Van Keymeulen A, Wakida NM, Carlton P, Berns MW, Bourne HR. Polarity reveals intrinsic cell chirality. Proc Natl Acad Sci USA. 2007;104(22):9296–9300. doi: 10.1073/pnas.0703153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vicente-Manzanares M, Zareno J, Whitmore L, Choi CK, Horwitz AF. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J Cell Biol. 2007;176(5):573–580. doi: 10.1083/jcb.200612043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rauzi M, Lenne PF. Cortical forces in cell shape changes and tissue morphogenesis. Curr Top Dev Biol. 2011;95:93–144. doi: 10.1016/B978-0-12-385065-2.00004-9. [DOI] [PubMed] [Google Scholar]
- 88.Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell. 2004;7(3):413–424. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 89.Mayer M, Depken M, Bois JS, Julicher F, Grill SW. Anisotropies in cortical tension reveal the physical basis of polarizing cortical flows. Nature. 2010;467(7315):617–621. doi: 10.1038/nature09376. [DOI] [PubMed] [Google Scholar]
- 90.Behrndt M, Salbreux G, Campinho P, Hauschild R, Oswald F, Roensch J, Grill SW, Heisenberg CP. Forces driving epithelial spreading in zebrafish gastrulation. Science. 2012;338(6104):257–260. doi: 10.1126/science.1224143. [DOI] [PubMed] [Google Scholar]
- 91.Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM. Mechanical integration of actin and adhesion dynamics in cell migration. Annu Rev Cell Dev Biol. 2010;26:315–333. doi: 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat Rev Cancer. 2009;9(2):108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3(5):362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 94.Weinberg R (2006) The biology of cancer. Garland, New York
- 95.Schlaepfer DD, Hanks SK, Hunter T, van der Geer P. Integrin-mediated signal transduction linked to Ras pathway by GRB2 binding to focal adhesion kinase. Nature. 1994;372(6508):786–791. doi: 10.1038/372786a0. [DOI] [PubMed] [Google Scholar]
- 96.Rivera GM, Antoku S, Gelkop S, Shin NY, Hanks SK, Pawson T, Mayer BJ. Requirement of Nck adaptors for actin dynamics and cell migration stimulated by platelet-derived growth factor B. Proc Natl Acad Sci USA. 2006;103(25):9536–9541. doi: 10.1073/pnas.0603786103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tredan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst. 2007;99(19):1441–1454. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- 98.Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25(1):9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 99.Lucero HA, Kagan HM. Lysyl oxidase: an oxidative enzyme and effector of cell function. Cell Mol Life Sci. 2006;63(19–20):2304–2316. doi: 10.1007/s00018-006-6149-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Meshel AS, Wei Q, Adelstein RS, Sheetz MP. Basic mechanism of three-dimensional collagen fibre transport by fibroblasts. Nat Cell Biol. 2005;7(2):157–164. doi: 10.1038/ncb1216. [DOI] [PubMed] [Google Scholar]
- 101.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8(3):241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 102.Samuel MS, Lopez JI, McGhee EJ, Croft DR, Strachan D, Timpson P, Munro J, Schroder E, Zhou J, Brunton VG, Barker N, Clevers H, Sansom OJ, Anderson KI, Weaver VM, Olson MF. Actomyosin-mediated cellular tension drives increased tissue stiffness and beta-catenin activation to induce epidermal hyperplasia and tumor growth. Cancer Cell. 2011;19(6):776–791. doi: 10.1016/j.ccr.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Plodinec M, Loparic M, Monnier CA, Obermann EC, Zanetti-Dallenbach R, Oertle P, Hyotyla JT, Aebi U, Bentires-Alj M, Lim RY, Schoenenberger CA. The nanomechanical signature of breast cancer. Nat Nanotechnol. 2012;7(11):757–765. doi: 10.1038/nnano.2012.167. [DOI] [PubMed] [Google Scholar]
- 104.de Rooij J, Kerstens A, Danuser G, Schwartz MA, Waterman-Storer CM. Integrin-dependent actomyosin contraction regulates epithelial cell scattering. J Cell Biol. 2005;171(1):153–164. doi: 10.1083/jcb.200506152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Quintin S, Gally C, Labouesse M. Epithelial morphogenesis in embryos: asymmetries, motors and brakes. Trends Genet. 2008;24(5):221–230. doi: 10.1016/j.tig.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 106.Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2010;188(1):11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Egeblad M, Rasch MG, Weaver VM. Dynamic interplay between the collagen scaffold and tumor evolution. Curr Opin Cell Biol. 2010;22(5):697–706. doi: 10.1016/j.ceb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nieto MA, Cano A. The epithelial-mesenchymal transition under control: global programs to regulate epithelial plasticity. Semin Cancer Biol. 2012;22(5–6):361–368. doi: 10.1016/j.semcancer.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 109.Erler JT, Bennewith KL, Nicolau M, Dornhofer N, Kong C, Le QT, Chi JT, Jeffrey SS, Giaccia AJ. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440(7088):1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 110.Moreno-Bueno G, Salvador F, Martin A, Floristan A, Cuevas EP, Santos V, Montes A, Morales S, Castilla MA, Rojo-Sebastian A, Martinez A, Hardisson D, Csiszar K, Portillo F, Peinado H, Palacios J, Cano A. Lysyl oxidase-like 2 (LOXL2), a new regulator of cell polarity required for metastatic dissemination of basal-like breast carcinomas. EMBO Mol Med. 2011;3(9):528–544. doi: 10.1002/emmm.201100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sahai E, Wyckoff J, Philippar U, Segall JE, Gertler F, Condeelis J. Simultaneous imaging of GFP, CFP and collagen in tumors in vivo using multiphoton microscopy. BMC Biotechnol. 2005;5:14. doi: 10.1186/1472-6750-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Charras GT, Yarrow JC, Horton MA, Mahadevan L, Mitchison TJ. Non-equilibration of hydrostatic pressure in blebbing cells. Nature. 2005;435(7040):365–369. doi: 10.1038/nature03550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lammermann T, Bader BL, Monkley SJ, Worbs T, Wedlich-Soldner R, Hirsch K, Keller M, Forster R, Critchley DR, Fassler R, Sixt M. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453(7191):51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 114.Bergert M, Chandradoss SD, Desai RA, Paluch E. Cell mechanics control rapid transitions between blebs and lamellipodia during migration. Proc Natl Acad Sci USA. 2012;109(36):14434–14439. doi: 10.1073/pnas.1207968109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Maiden SL, Hardin J. The secret life of alpha-catenin: moonlighting in morphogenesis. J Cell Biol. 2011;195(4):543–552. doi: 10.1083/jcb.201103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nikolaidou KK, Barrett K. A Rho GTPase signaling pathway is used reiteratively in epithelial folding and potentially selects the outcome of Rho activation. Curr Biol. 2004;14(20):1822–1826. doi: 10.1016/j.cub.2004.09.080. [DOI] [PubMed] [Google Scholar]