Abstract
Members of the tristetraprolin (TTP/TIS11) family are important RNA-binding proteins initially characterized as mediators of mRNA degradation. They act via their interaction with AU-rich elements present in the 3′UTR of regulated transcripts. However, it is progressively appearing that the different steps of mRNA processing and fate including transcription, splicing, polyadenylation, translation, and degradation are coordinately regulated by multifunctional integrator proteins that possess a larger panel of functions than originally anticipated. Tristetraprolin and related proteins are very good examples of such integrators. This review gathers the present knowledge on the functions of this family of RNA-binding proteins, including their role in AU-rich element-mediated mRNA decay and focuses on recent advances that support the concept of their broader involvement in distinct steps of mRNA biogenesis and degradation.
Keywords: Tristetraprolin, AU-rich element, mRNA stability, mRNA processing
Overview of the tristetraprolin (TTP/TIS11) family
Description of tristetraprolin (TTP) and related proteins started in the early 1990s with the cloning of an immediate–early response gene induced by the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA) in murine fibroblasts, which was thereafter named TPA-induced sequence 11 (TIS11) [1]. At that time, several other groups that were deciphering the complex transcriptional response of various cell types to stimulation by mitogens reported the induction of mRNAs sharing structural similarities with TIS11 and contributed to the definition of a new family of early response genes, the TTP/TIS11 family [2–6].
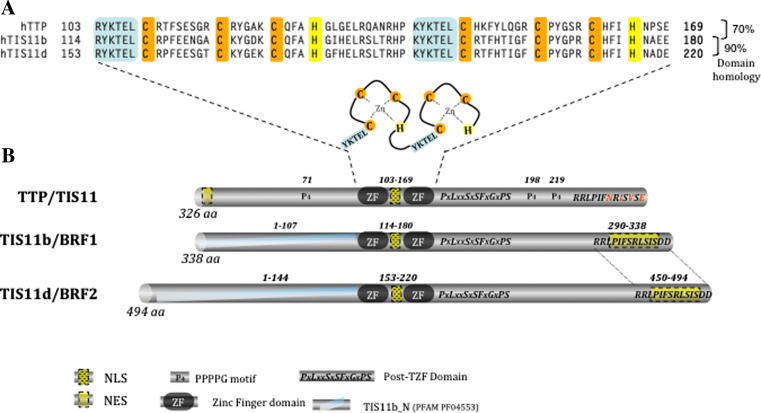
In mammals, beside TTP/TIS11 (HGNC alias: ZFP36), which is the most thoroughly studied member, the family contains two other members called TIS11b/BRF1 (HGNC alias: ZFP36L1) and TIS11d/BRF2 (HGNC alias: ZFP36L2). In addition to these well-described members, rodents possess an additional member (HGNC alias: ZFP36L3) specifically expressed in the placenta [7, 8]. Like TTP, all members of the family are characterized by the presence in their coding sequence of a very particular tandem zinc-finger domain (TZF), which can be found in almost all branches of eukaryotic evolution from lower unicellular eukaryotes to invertebrates and mammals [9–12]. This structural domain is composed of a double zinc-finger motif of the CCCH type, each one preceded by a leader sequence, and bears the RNA-binding property (see Fig. 1a).
Fig. 1.
Similarities in the protein domains of the TTP family members. a Sequence of the tandem zinc-finger domain (TZF), which is characteristic of the TTP family. TIS11b amino-acid residues inside the TZF share 70 and 90 % of sequence identity with TIS11d and TTP/TIS11, respectively. b Remarkable features in TTP/TIS11, TIS11b/BRF1, and TIS11d/BRF2 protein. Tristetraprolin has been named after the particular repeated motif (PPPPG) of the protein. Each protein of the family contains a nuclear localization signal (NLS) inside the much-conserved TZF domain and a conserved post-TZF domain of unknown function. Nuclear export signals (NES) are located in the C-termini of TIS11b and TIS11d and in the N-terminus of TTP. The N-terminal domains of TIS11b and TIS11d are closely related, but differ from that of TTP
It is worth mentioning that the TZF domain is of very high sequence homology between the TTP family members, whereas the N-terminal and C-terminal regions differ much more. Figure 1b, which depicts the similarities and differences in the sequences of the human members of the TTP family illustrates the remarkable features of these proteins. Indeed, in addition to the TZF motif, only two other domains of relative high homology are present in the three proteins. One is a stretch of 14 amino acids that contains a functional nuclear export sequence (NES). This sequence is C-terminal in the TIS11b and TIS11d sequences, whereas for TTP/TIS11, the NES is located instead in the N-terminus of the protein. The second similarity motif is located immediately downstream of the TZF domain and is composed of the PxLxxSxSFxGxPS sequence, where x represents one member of a closely related family of amino acids. The exact function of this domain is unknown, but contains (at least for TTP and TIS11b) a binding site for 14-3-3 proteins [13, 14]. Moreover, the presence of three serine and one proline residues at both ends suggests a loop conformation for this motif. In fact, sequence alignments suggest that TIS11b and TIS11d are more closely related to each other than to TTP. This is particularly true at the N-terminal part of these proteins, where domain similarities between TIS11b and TIS11d define a specific submotif in the protein family (PFAM PF04553). Besides these similarities, the weaker protein homology observed outside the TZF motif suggests that the specific protein–protein interactions mediating the distinct functions and regulations of the family members are borne by these divergent domains.
The notion that these proteins are not physiologically redundant is clearly illustrated by the differences in the phenotypes of the knock-out mice generated by ubiquitous genetic deletion of the distinct family members (presented in Table 1). TTP/TIS11 KO mice appear normal at birth, but rapidly develop a complex syndrome of cachexia, arthritis, and general inflammation, which can be almost completely prevented by injection of antibodies against TNF-α [15]. In contrast, disruption of the TIS11b/BRF1 gene induces embryonic lethality around E10.5 with neural tube abnormalities, failure of chorioallantoic fusion, and angiogenesis defects [16, 17]. Concerning TIS11d, deletion of the 29 N-terminal amino acids of the protein results in female infertility, whereas the complete inactivation of the gene induces lethality within 2 weeks after birth due to diverse hemorrhages, probably caused by defective definitive hematopoiesis [18–20]. These studies showed that (at least for some of their physiological functions) members of the TIS11 family cannot compensate for each other. However, compensation between TIS11b and TIS11d proteins has been particularly well described in a recent study, where inducible and concomitant deletions of both TIS11b and TIS11d genes was realized in the thymus during thymopoiesis. This was reported to trigger lymphoblastic leukemia, whereas disruption of either one of these genes was not sufficient to induce this phenotype [21]. Altogether, these studies suggest that possible compensation might exist when two or three members are expressed in the same organ during the same developmental window. The activities of the TTP family proteins must therefore be analyzed in a cell- and context-dependent manner.
Table 1.
Phenotypes of the knock-out mice for the three members of the TTP family
| 8Name (alias) | Reference | Lethality | Main characteristics | Comments |
|---|---|---|---|---|
| ZFP36 (TTP,TIS11,G0S24, NUP475, RNF162A) | [15] | Post-natal (depending on the severity of the syndrome) | Inflammatory syndrome (cachexia, arthritis) | Rescued by anti-TNF-α antibodies injection first identification of TTP implication in AU-rich mediated decay |
| ZFP36L1 (BRF1, TIS11b, Berg36, ERF-1, ERF1, cMG1) | [17] | E11 | Neural tube and chorioallantoic fusion defects | |
| [16] | E10.5 | Neural tube and vascular angiogenesis defects | Identification of potential role of Tis11b/BRF1 in the control of AU-rich-mediated translation | |
| ZFP36L2 (TIS11d, BRF2, ERF-2, ERF2, RNF162C) | [18, 19] | None | Female infertility | Knock-out is incomplete resulting in an amino-terminal truncation of the protein |
| [20] | 2 weeks after birth | Definitive hematopoiesis deficiency | Identification of 293 up-regulated transcripts in KO fetal livers, but no apparent effect on mRNA stability in KO fibroblasts | |
| ZFP36L1-ZFP36L2-inducible double KO in the thymus | [21] | 90 % of lethality by 6 months of age | Lymphoblastic, leukemia | First direct identification of malignant transformation due to deficiency in Tis11 proteins expression |
Localization, expression, and regulation of TTP and related proteins
TTP and related proteins are nucleocytoplasmic shuttling proteins. They all contain a specific nuclear localization sequence (NLS) located between the two zinc fingers [22, 23]. In addition to these import sequences, nuclear export signals are present in the N-terminus of TTP or in the C-termini of both TIS11b and TIS11d. Nucleocytoplasmic shuttling of TTP family members is dependent on these signals and on the activity of the nuclear export receptor CRM1 [23]. Moreover, TTP directly associates with the nucleoporin NUP214 [24]. Interestingly, a modification of the C-terminal part of ZFP36L3, the murine-specific member of the family specifically expressed in the placenta, appears to result in the loss of its shuttling capacity and in an enforced cytoplasmic localization [8]. Subcellular localization studies indicated that the nuclear or cytoplasmic localizations of TTP-related proteins differ from one cell type to another and are regulated by extracellular signals, which thereby modulate their respective functions [25–29].
Post-translational modifications strongly contribute to the regulation of TTP family protein activities. TTP family members are phosphorylated on several distinct sites in response to multiple signaling pathways [28, 30–34]. Initially, it has been speculated that phosphorylation of TTP-related proteins would modulate their RNA-binding properties and therefore their mRNA-destabilizing properties. Nevertheless, experimental results have been contradictory, with some studies showing an increase in TTP RNA-binding capacities in phosphatase-treated cellular extracts, whereas in vitro phosphorylation of either TTP or TIS11b had no effect on RNA binding [35–37]. To date, phosphorylations of either TTP or TIS11b have been reported to impair their mRNA decay-promoting activities (reviewed in [38]). The most extensively studied kinases are the p38-MAPK (p38-mitogen-activated protein kinase) and its downstream target MAPK-activated protein kinase 2 (MK2), both of which appear to play a pivotal role in the regulation of ARE-mediated mRNA decay (AMD). Inhibition of p38 MAPK signaling pathway has been reported to promote AMD [25], whereas, on the contrary, expression of constitutively active upstream activators of p38 MAPK increases stability of ARE-containing transcripts [39]. p38 and MK2 phosphorylate TTP protein at two critical sites, Ser52 and Ser178, leading to an increased TTP protein stability and TTP cytoplasmic localization [13, 25, 32]. Similarly, phosphorylations of TIS11b at Ser92 and Ser203 by protein kinase B (PKB) and MK2 stabilize TIS11b protein [14, 37, 40]. For both TTP and TIS11b, the above-mentioned phosphorylated serines are docking sites for 14-3-3 protein isoforms [14, 41]. 14-3-3 protein binding can indeed modulate the cytoplasmic localization and protein stability of both TTP and TIS11b [25, 40]. Moreover, interaction with 14-3-3 inhibits TTP protein dephosphorylation by protein phosphatase 2A (PP2A) and therefore modulates TTP function [42]. It has also been shown that phosphorylation of TTP favors its ubiquitinylation and that TTP and TIS11b proteins are regulated by proteasome activity depending on their phosphorylation status [25, 40, 43, 44]. Phosphorylations of TTP protein also have a direct impact on its mRNA-destabilizing activity by preventing the recruitment of mRNA deadenylases onto the target mRNA. This inhibitory effect appears to be, at least partially, independent of 14-3-3 protein sequestration and unrelated to TTP RNA-binding [45, 46]. Overall, the observation that TTP or TIS11b protein stability was regulated by the same phosphosites which were originally identified as regulatory sites for their mRNA decay-promoting activity revealed an unexpected link between TTP or TIS11b protein turnover and their function in AMD.
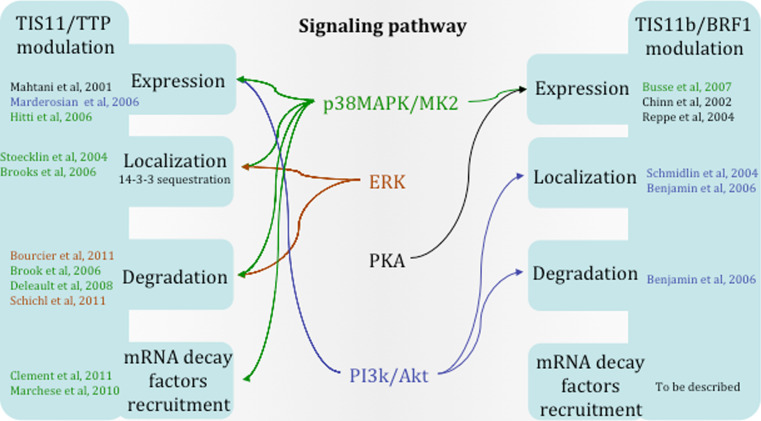
The signaling pathways that regulate expression, localization, degradation, or function of TTP family members are well described in recent reviews [38, 47–49] and are illustrated in Fig. 2. It has been speculated that, by regulating the stability, localization and function of TTP family proteins, distinct signaling pathways can coordinate the decay rate of specific subsets of target mRNAs and therefore the expression of the corresponding proteins [32, 40].
Fig. 2.
Signaling pathways acting on TTP and TIS11b/BRF1 proteins. Schematic representation of the diverse signaling pathways modulating either TTP/TIS11 or TIS11b/BRF1 protein expression, localization, degradation, or capacity to recruit mRNA decay factors
Not only AU-rich RNA binding proteins
AU-rich elements (AREs) are consensus mRNA cis-acting sequences containing a high proportion of adenylate or uridylate bases, which have been previously described as RNA-destabilizing motifs [50]. These regulatory motifs have been characterized in early response genes or short-lived mRNAs and classified depending on the absence (class III) or the presence of isolated (Class I) or clustered (Class II) pentameric “AUUUA” motifs [51]. Since then, computational analyses have been used to cluster human and mouse ARE-containing transcripts. A comprehensive database (ARED database, [52, 53]) has been built based on the presence in the transcript 3′-UTR of the minimal motif WWWT[ATTTA]TWWW (with one allowed mismatch outside of the bracketed region and where W is either an adenylate or an uridylate). Indeed, these analyses showed that almost 8 % of the human transcriptome contain AREs in their mRNA 3′-UTRs and that conservation between species is high for a large portion of them (ARED database, [52, 53]). This suggests that the role of AMD in gene regulation is widespread among the living species. Recently, an algorithm (termed AREScore) has been described, which uses the only core “AUUUA” motif and its surrounding sequence characteristics to identify and score the chance of AMD regulation for a given mRNA [54]. Interestingly, this approach, which uses a less-stringent criterion, can identify potential ARE-controlled mRNAs that are not integrated in the ARED database.
As mentioned above, TTP and its family members have been shown to specifically bind AREs via their highly conserved double zinc-finger domain [55]. Identification and analysis of RNA sequences selectively bound by TTP in RNA SELEX experiments confirmed this preferential recognition of the core AUUUA element and showed a strong specificity for the extended motif UAUUUAU [56]. Further studies confirmed that this extended motif is sufficient to allow formation of an RNA–protein complex [57]. Indeed, NMR structures of the TIS11d double zinc finger in complex with RNA showed that each zinc finger binds symmetrically to adjacent 5′-UAUU-3′ sub-sites on the single-stranded RNA [58]. Of note, a previous NMR structure analysis of the single first zinc finger of TTP showed slight differences in folding, probably due to the absence of the RNA ligand [59, 60].
Despite their high specificity for AREs, it has been suggested that TTP family members can bind non-ARE-like motifs [61]. When identifying TTP mRNA targets in human dendritic cells, Emmons et al. [61] characterized a subset of mRNA bound and regulated by TTP which contained a CTTGTG motif. They further demonstrated that deletion of a 36-nt portion of the 3′-UTR containing this motif was sufficient to abrogate the TTP response in reporter gene experiments. Moreover, within the same year, a genome-wide screening identified 137 potential TTP-bound mRNAs, with only some of them containing well-defined AREs, suggesting that TTP binding might not be restricted to AREs or might associate with other mRNA-binding proteins that target distinct sequences [62].
In addition to these experiments showing binding to non-ARE sequences, some reports also suggested that TTP can modulate mRNA expression independently of its mRNA-binding activity. Indeed, regulation of iNOS mRNA stability seems to involve a complex interplay between several ARE-binding proteins including HuR and KH-type splicing regulatory protein (KSRP) [63]. A direct interaction between TTP and KSRP inhibits the recruitment of the latter onto ARE and therefore prevents mRNA degradation via the exosome [64]. Moreover, it has recently been shown that binding of different isoforms of AUF1 to TTP via its zinc-finger domain can modulate TTP-binding affinity to its target mRNA sequence, thereby promoting TTP-dependent mRNA degradation [65]. These studies suggest that TTP may also act indirectly on non-ARE sequences by participating in a network of interactions with other RNA-binding proteins.
Functions of the TTP/TIS11 proteins in mRNA destabilization
mRNA destabilization historically remains the best-characterized function of the TTP family members. The first description of TTP involvement in transcript degradation came from the description of the mouse phenotype resulting from its genetic deletion [66]. In this study, Blackshear and collaborators showed that the induction of TNF-α production observed in the TTP-KO mice was due to increased TNF-α mRNA stability in macrophages. Further studies from the same group indicated that TTP and related proteins are able to bind to TNF-α AREs via their double zinc-finger motif, to promote deadenylation of the TNF-α transcript and thereby to induce mRNA destabilization [55, 67–69]. In vitro experiments showed that TTP and its family members can promote deadenylation of ARE-containing mRNAs by the deadenylase PARN, while having no impact on ARE-devoid transcripts [70]. Besides this deadenylation-promoting effect, no direct interaction could be identified between TTP and PARN, suggesting that modification of the RNA conformation induced upon TTP binding to AREs might favor the recruitment of this deadenylase [45, 70, 71].
On the other hand, more recently, TTP and TIS11b/BRF1 have been found to be involved in the recruitment of several mRNA decay enzymes (including deadenylases, decapping enzyme, and exosome complex components) onto ARE-containing transcripts [71]. Coimmunoprecipitation assays using TTP or TIS11b as bait revealed an interaction of these proteins with the CCR4 deadenylase. CCR4 is a component of a large cytoplasmic deadenylation complex containing another deadenylase, CAF1, and the scaffolding protein NOT1. Later on, the involvement of the CCR4–CAF1 complex in TTP-dependent deadenylation was further confirmed by the identification of an interaction between TTP and CAF1 by GST pull-down assays and by the observation that depletion of either the CCR4 or CAF1 protein abrogates in vitro TTP-directed deadenylation [46]. It appears that TTP is associated with multiple deadenylases as reported by Clement and collaborators [45], even though they confirmed that TTP-directed deadenylation principally relies on the CCR4–CAF1 complex. Indeed, recruitment of CAF1 seems to depend on the scaffolding protein NOT1, which interacts with the C-terminal domain of TTP, the presence of which is necessary for TTP-induced mRNA decay in HeLa cells [72]. Whether TTP directly interacts with these distinct proteins or is indirectly associated with them through a multi-protein complex remains to be determined. However, these three aforementioned studies all showed that recruitment of the CCR4–CAF1–NOT1 complex by TTP or its related proteins is mandatory for AMD and thereby highly contributes to this process [45, 46, 72]. Moreover, regulation of deadenylase recruitment has been shown to be dependent on the TTP phosphorylation status, reinforcing the notion that post-translational modifications of TTP family members are key events in the regulation of their activities [45, 46].
Deadenylation is a key step in AMD and it has been shown that poly(A) binding protein (PABP), which protects the mRNA poly(A) tail from degradation and is involved in mRNA circularization upon binding to eukaryotic translation initiation factor 4G (eIF4G), could indeed inhibit TTP-directed deadenylation [45, 73]. This TTP–PABP interaction has been further confirmed and characterized by yeast two-hybrid analyses and shown to be RNA-independent [74]. Therefore, it has been speculated that TTP binding to PABP can induce its displacement from the poly(A) tail of ARE-containing mRNAs and thereby promote 3′ to 5′degradation.
Once the poly(A) tail has been removed, further mRNA degradation can occur either in a 3′ to 5′ or in a 5′ to 3′ way. The exosome, a conserved protein complex with a 3′ to 5′ exonuclease activity is responsible for the 3′-end trimming of RNA and has been shown to be involved in AMD, among other functions [75, 76]. Exosome integrity is required for AMD and it has been suggested that components of the exosome can bind ARE-containing transcripts [76, 77]. Interestingly, association between the purified exosome complex and TTP has been detected and, in the same study, TTP appears to promote exosome-directed degradation of ARE-containing mRNAs [75]. Since then, interactions with several components of the exosome have been shown by co-immunoprecipitation. TTP has been reported to specifically recruit the functional exosome onto ARE-containing mRNAs and thereby to promote mRNA decay [78]. In addition to this 3′–5′ mRNA decay, ARE-containing transcripts are subjected to 5′–3′ degradation as well. TTP and TIS11b/BRF1 interact with decapping enzymes and the cytoplasmic 5′-3′exoribonuclease Xrn1, thereby promoting their recruitment onto the mRNA, mRNA decapping as well as 5′ mRNA degradation [71, 79]. It is difficult to determine the relative importance of 5′- versus 3′-AMD. Specific depletion of either the decapping enzyme Xrn1 or a component of the exosome complex inhibits AMD, suggesting the participation of both mechanisms in living cells [80].
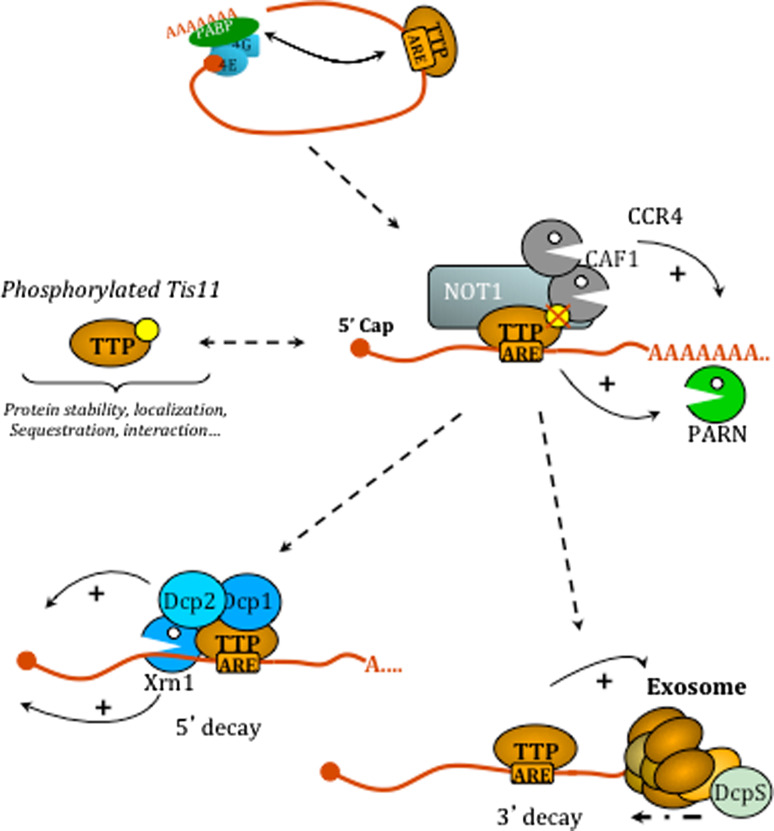
Together, these studies support a model of mRNA stability regulation by TTP or its related proteins, where these zinc-finger proteins act as docking proteins for components of the mRNA decay machinery, as depicted in Fig. 3.
Fig. 3.
Involvement of TTP family proteins in the mRNA degradation pathway. Upon binding to ARE-containing transcripts, TTP proteins may interfere with mRNA circularization via its interaction with PABP. Subsequently, deadenylases are recruited onto the transcript either by direct interaction with TTP proteins (Crr4–Caf–Not complex) or indirectly (PARN). This step is modulated via TTP phosphorylation. Interactions of TTP and related proteins with decapping enzymes (Dcp1/2) may then promote decapping of the transcript, which can thereafter be trimmed by the 5′-exoribonuclease Xrn1. On the 3′-end, TTP proteins can also interact with the exosome complex which associates with the scavenger decapping enzyme (DcpS) to achieve 3′-decay of the transcript
Alternative functions of the TTP/TIS11 proteins throughout the life of an mRNA
In addition to their well-known effects on mRNA stability, emerging experimental evidence indicates that the TTP family of RNA binding proteins might participate in the regulation of almost all the key steps controlling mRNA from biogenesis to decay.
Functions of the TTP/TIS11 family members in mRNA transcription and 3′-end processing
At the onset of their discovery, because of the presence of a TZF domain in their sequence, it had been speculated that TTP and related proteins might be transcription factors. Two recent studies seem to eventually sustain this hypothesis. TIS11 possesses the potential to activate transcription when fused to the GAL4 DNA binding domain [81]. TIS11b/BRF1 was also shown to directly interact with the transcription factor HNF1α (hepatocyte nuclear factor 1α) as shown by pull-down assays, and this interaction seems to attenuate the HNF1α transcriptional activity [82].
The regulation of 3′-end mRNA processing and consecutive alternative polyadenylation is becoming an important challenge in RNA research as it appears that more than half of all human transcripts contain more than one poly(A) signal [83]. In yeast cells, Cth2, one of the homologues of human TTP and TIS11b, can interfere, beyond its activity in ARE-dependent degradation, with poly(A) site selection when the AREs are located in close proximity with the poly(A) signal [84]. Deletion of a specific domain of Cth2 allows the synthesis of a 3′-extended transcript, due to selection of a distal poly(A) signal in the target transcript.
Recent evidence has identified a similar mechanism in mammalian cells. In endothelial cells, TIS11b has been shown to modulate 3′-end processing of an ARE-containing mRNA (encoding Dll4) that contained TIS11b binding sites embedded in the actual poly(A) signal [27]. In these cells, specific inhibition of TIS11b, which appears to be localized in the nucleus, had no effect at all on mRNA stability, suggesting that the functions of TIS11b might highly depend on its nucleocytoplasmic localization. Since then, a high-throughput analysis has identified and characterized a direct interaction between TTP and poly(A)-binding protein nuclear 1 (PABPN1). This interaction induces the inhibition of the polyadenylation of an ARE-containing reporter transcript. Interestingly, this interaction seems to be dependent on the phosphorylation status of TTP with better PABPN1 binding for hypophosphorylated TTP [85].
Interference of TTP family members with mRNA cleavage and polyadenylation is a very new finding that deserves further characterization. It may be an important nuclear function for these nucleocytoplasmic shuttling proteins as it has been observed that regulation of 3′-UTR length (one of the main consequences of alternative polyadenylation) highly contributes to the control of gene expression [86].
Functions of the TTP/TIS11 family members in mRNA transport and/or localization
The subcellular compartment in which TTP or its related proteins are actually recruited onto ARE-containing mRNAs is not exactly known. At least in some circumstances, it has been suggested that this recruitment occurs in the nucleus. Indeed, the yeast TTP homolog Cth2 seems to be co-transcriptionally recruited onto nascent ARE-containing transcripts [27, 84, 87]. It has been shown that disruption of yeast Cth2 shuttling impaired AMD, suggesting a model whereby TTP and related proteins would be able to recruit mRNAs in the nucleus and deliver them to the cytoplasm for degradation [87]. These data suggested that co-transcriptional recruitment of these RBPs can occur and that yeast Cth2 might contribute to the nuclear export of mRNAs. Such a mechanism, however, remains to be validated in mammalian cells.
Following transcription, whether bound or unbound by TTP family members, mRNAs translocate to the cytoplasm where they are directed towards translation, storage, or decay. Processing bodies (PB) are RNA granules located in discrete cytoplasmic foci that contain decapping and 5′ to 3′ decay enzymes (Xrn1) and deadenylases among other proteins. These small structures constitute a specialized compartment for mRNA degradation given the promiscuity of multiple components of the decay machinery even though mRNA decay is not restricted to PBs [88]. As mentioned above, TTP can interact with decapping enzymes and exoribonucleases promoting transcript degradation and is found to be part of PBs [79, 89]. Actually, it has been shown that TTP and its related proteins can direct the localization of ARE-containing transcripts to PBs for decay and thereby can induce PB nucleation by aggregation of ribonucleoproteins [90].
The composition of PBs is highly dynamic, with transcripts and proteins able to move in and out of these structures. It has been shown that TTP and its related proteins can shuttle between PBs and stress granules (SG). SGs are aggregates of mRNAs stalled in the preinitation complex of ribosome assembly induced under stress conditions [89]. The presence of TTP and related proteins in SGs therefore suggests that they are involved in translation repression. Binding of TTP and its family members onto ARE-containing transcripts may allow escorting them to SGs or PBs for translation suppression or degradation, respectively, and may thereby contribute to mRNA sorting. TTP translocation between these RNA granules seems to rely on TTP interaction with transportin (TRN), a member of the importin-β family involved in nucleocytoplasmic transport of macromolecules [91]. TTP and its associated mRNAs interact with transportin, as it was shown that silencing of transportin sequesters TTP in PBs and therefore reduces their translocation to SGs. Moreover, the phosphorylation status of TTP modulates its binding to 14-3-3 protein and it was shown that formation of this molecular complex is able to exclude TTP and associated mRNAs from SGs [13]. These last two studies also showed that regulated trafficking of TTP between PBs and SGs induces an alteration of AMD, suggesting that TTP can select ARE-containing mRNAs stalled in translation in SGs to direct them to PBs for decay [13, 91]. These data argue in favor of a model suggesting that TTP and related proteins play a critical role in determining the fate of ARE transcripts depending on the environmental and stress conditions.
Functions of the TTP/TIS11 family members in mRNA translation
In addition to their role in mRNA decay, AREs have been known for a long time to be involved in the regulation of translation via the binding of specific AU-binding proteins such as TIA-1, TIAR-1, and HuR [92]. Furthermore, translational regulation of ARE-containing transcripts might be a means to control gene expression, in particular during serum starvation [93].
Indeed, an increasing number of studies suggest that TTP and its related proteins are involved in ARE-mediated regulation of translation under certain circumstances. First, following LPS stimulation, TTP can be found associated with polysomes in macrophages, suggesting a possible involvement in translation regulation [94, 95]. Second, a recent analysis showed that TTP specifically inhibits translation of an ARE-containing reporter transcript by excluding the targeted transcript from heavy polyribosomal fractions [96]. The same study further showed that TTP can associate directly with the helicase RCK/P54, which plays a major role in translation repression. Both this interaction and the helicase activity are necessary for ARE-mediated repression of translation. Interestingly, a previous study has shown a similar interaction in yeast between TTP and RCK/P54 homologues, supporting the idea that in yeast and mammals TTP-related proteins may have convergent functions even though translational inhibition has not been addressed in the latter report [97].
However, the involvement of TTP in ARE-dependent repression of translation is not clearly understood. A recent screening for TTP protein partners identified a specific interaction with Cullin 4B (Cul4B), which is a scaffolding component of a ubiquitin E3 ligase complex [98]. Depletion of Cul4B in macrophages decreased TNF-α mRNA stability and, in the meantime, inhibited its loading to polysomes upon LPS induction. The authors then suggested that recruitment of Cul4B to TTP promotes translation of the transcript, indicating that TTP function in translational regulation is more complex than anticipated and needs further characterization. Moreover, regulation of RNA translation is not a specific feature of TTP. Indeed, genetic disruption of TIS11b/BRF1 in mice revealed vascular defects and associated increased expression of VEGF, a potent angiogenic factor [16]. VEGF mRNA had been previously shown to be a target of both TIS11b and TTP, with a marked effect on mRNA stability observed in in vitro experiments [99–101]. However, very convincing data from the analysis of TIS11b−/− fibroblasts showed that VEGF up-regulation in this in vivo situation was due to modification of mRNA translation efficiency and not related to changes in transcript stability [16]. Altogether, these data suggested that the physiological functions of TTP and related proteins might highly depend on the cell type and on the extracellular environment (hormones, metabolic nutrients, oxygenation, stress).
Functions of the TTP/TIS11 family members in microRNA-regulated pathways
mRNA 3′-UTRs are docking sites for both RNA-binding proteins and microRNAs (miRNAs), which both regulate stability and/or translation of the transcripts. Recent experimental data suggest numerous similarities and connections between the miRNA-regulated pathway and the ARE-mediated control of gene expression [102]. Concerning TTP and its related proteins, cooperation between TTP, miR-16, and AGO2, which are part of the RNA-induced silencing complex (RISC), has been described in ARE-mediated control of the TNF-α transcript [103]. This work identified a requirement for the miRNA pathway in TTP-mediated ARE-dependent decay. TTP cooperation with AGO2, without direct binding to miR-16, may assist the miRNA to specifically target the AU-rich element. This indicated that ARE-dependent decay may rely, at least partially, on the miRNA pathway. However, this interaction with miRNAs seems dispensable as depletion of components of the miRNA pathway does not prevent AMD in Drosophila and mouse cells [104]. Interestingly, the best evidence for a role played by TTP or related proteins in miRNA- or siRNA-regulated pathways came from an analysis in Drosophila cells, where only one TTP homologue is present [105]. A screening set up to identify factors involved in the RNAi pathway indicated that Drosophila TTP is required to achieve complete siRNA silencing, but the mechanism of this action remains to be characterized. Recently, a report showed that TTP could induce ARE-mediated decay of the Lin28 transcript, which is a negative modulator of the let7 microRNA, and thereby stimulate the biogenesis of the let7 microRNA in cancer cells [106].
Taken together, these studies suggest that the connection between ARE-dependent degradation and the miRNA pathway may be quite complex and needs further investigation.
RNA-binding independent functions of the TTP/TIS11 family members
In addition to the aforementioned functions of the TTP family members, recent data showed that, under some circumstances, their RNA-binding properties are not absolutely required for their activity. Two independent reports published back to back in 2009 identified TTP as a modulator of NF-κB signaling [107, 108]. Following NF-κB activation, TTP binding to the p65 subunit of this transcription factor seems able to impair either its nucleocytoplasmic shuttling or its acetylation and thereby to contribute to attenuation of NF-κB signaling.
Functions of TTP and related proteins in biological processes and diseases
Regulation of inflammation is one example of a biological process for which involvement of TTP and its related proteins is well described. As mentioned above, genetic inactivation of TTP in mice induces a complex inflammatory syndrome that is mainly due to TNF-α overproduction [66]. Following this study, granulocyte–macrophage colony-stimulating factor (GM-CSF) is also overexpressed in TTP−/− macrophages, reinforcing the key player role of TTP in mediating the inflammatory response [109]. Moreover, a high proportion of ARE-containing RNAs are transcripts encoding proteins involved in immune functions and the list of mediators of inflammation experimentally validated as targets for ARE-mediated decay keeps growing (for review, see [48]). As a matter of fact, TTP regulates mRNA stability of both inflammatory and anti-inflammatory cytokines, attenuates NF-κB signaling, and thereby coordinately modulates inflammation.
Because of the vast diversity of their potential targets, it is not surprising to find alterations of TTP family member expression in cancer [110]. For instance, it has been shown that the level of TIS11b/BRF1 expression is associated with cisplatin sensitivity in head and neck squamous cell carcinoma and that specific silencing of TIS11b contributes to the development of cisplatin resistance [111]. In fact, converging data argue in favor of a tumor suppressor role for TTP and its related proteins. Indeed, overexpression of TTP in a v-H-ras-dependent mast cell tumor mouse model notably delayed tumor progression [112]. Recently, it was reported that TTP expression levels are inversely correlated with aggressiveness, metastatic potential, and resistance to antitumorigenic treatment in breast cancer [113]. Moreover, a direct role of ARE-mediated regulation in cancer progression has been firmly established by a study showing that inducible deletion of both TIS11b and TIS11d in thymus leads to almost 100 % of mice developing lymphoblastic leukemia [21]. This study is the first to show that malignant development could be a direct consequence of dysregulated AMD. Since then, loss of TTP and TIS11b expression has been shown to be a hallmark of Myc-induced malignancy in mice and it was shown that TTP overexpression could override this malignant state, clearly confirming its tumor suppressor function [114]. Very recent reviews analyze in depth the molecular mechanisms that link TTP and related proteins with cancer [48, 49, 115].
TIS11b/BRF1 genetic knock-out in mice induced numerous vascular defects, suggesting its direct involvement in angiogenesis [16]. The control of potent angiogenic factor expression by TTP proteins had been previously documented and the list of ARE-regulated transcripts related to angiogenesis regulation is constantly growing [99, 100, 116]. Beside its role in neovascularization, TIS11b is also involved in the maintenance of stem cell pluripotency. Indeed, in embryonic stem (ES) cells, TIS11b gene expression is under the control of the key transcriptions factors that are also known to control pluripotency and silencing of TIS11b expression, promoting cell differentiation into cardiomyocytes [117, 118].
Therapeutic potentials of the TTP/TIS11 proteins
As mentioned above, TTP and related proteins are involved in numerous biological processes during which their dysregulation might contribute to established pathologies. Due to the numerous ARE-containing mRNA targets involved in carcinogenesis, tumoral angiogenesis, and inflammation, enforced overexpression of TTP proteins in tumor cells has been tested as a novel anti-tumoral therapeutic approach and shown to efficiently slow down tumor growth in mouse tumorigenesis models [100, 101]. Similarly, in an experimental periodontitis mouse model, adenovirus-delivered TTP was reported to inhibit bone loss through simultaneous down-regulation of the expression of several inflammatory cytokines [119]. Altogether, these data provide a proof of concept that the mRNA-destabilizing properties of TTP family members can be used to design multi-target therapies of inflammatory diseases and cancer.
Concluding remarks: TTP proteins and AU-rich elements are working partners from the synthesis to the decay of an mRNA
Since the first description of the function of TTP in AMD, accumulating experimental data have sustained that this RNA-binding protein and its homologues are actually involved in many different biological processes. It has been suggested that TTP proteins belong to a family of negative feedback regulators that contribute to the attenuation of growth factor signaling and therefore contribute to the coordination of gene expression upon changes of the cellular environment [120]. Expression and regulation of TTP and related proteins are indeed highly dependent on the cell type considered and this might, at least in part, explain the major differences in the phenotypes of the different knock-out mice and the reason why, under some circumstances, the TTP family members can compensate for one another. As an example, some of the ARE-regulated transcripts which have been described as up-regulated in TTP−/− fibroblasts are not modified in TIS11b/BRF1−/− fibroblasts [121]. It is therefore essential to distinguish physiological targets for each TTP protein and to be aware that target specificity is dependent on the physiological context.
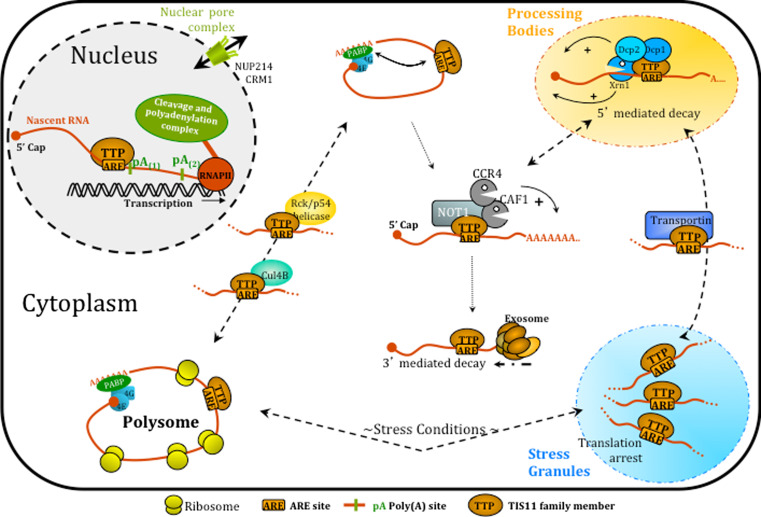
Adding to this complexity, it is now increasingly clear that the molecular function of this family of RNA-binding proteins is not restricted to AMD. Poly(A) site selection, translation regulation or mRNA transport are key steps in mRNA life, which are all regulated by TTP family members and will ultimately determine the fate of the transcript, as depicted in Fig. 4. AU-rich elements (or other yet-to-be-identified binding sites for the TTP proteins) will therefore mark transcripts for specific regulation. Depending on the local environment of this cis-regulating element, TTP proteins might trigger different mechanisms of regulation. Recruitment of TTP and related proteins within the poly(A) signal during transcription in the nucleus or in close proximity to an miRNA-binding site in the cytoplasm have different consequences in terms of transcript regulation.
Fig. 4.
Multiple functions of TTP proteins in mRNA life. TTP and TIS11b have been shown to modulate 3′-end mRNA maturation and poly(A) site selection in the cell nucleus. Recruitment of TTP proteins onto ARE-containing transcripts may occur under some circumstances in the nucleus and interactions with the nuclear pore complex may modulate mRNA delivery to the cytoplasm. After destabilization of the circularized mRNA, recruitment of deadenylases may direct AU-rich mRNAs to either 5′- or 3′-decay. TIS11 proteins are component of Processing Bodies (PB, in yellow) as well as Stress Granules (SG, in blue) induced in stress conditions. They are involved in PBs nucleation and promote 5′-mediated decay of target transcripts as well as their sorting and escorting between PBs and SG (interaction with transportin). TTP also regulates AU-rich mediated translation as it has been shown to exclude ARE-containing RNAs from heavy polysomes. Specific protein interactions with TTP seem to increase (Cul4B) or decrease (Rck/p54) polysome loading of ARE-containing transcripts. See text for detailed description of these and other functions of TTP/TIS11 proteins
The existence of such interconnected functions for a specific RNA-binding protein is now emerging as a general concept. The list of RNA-binding proteins with multiple functions keeps growing and recent high-throughput sequencing of binding sites for these proteins allowed identification and characterization of new functions. For example HuR, which was previously known as an mRNA stabilizing factor, has been reported to modulate mRNA splicing. Similarly, NOVA, previously described as a splicing factor, can modulate poly(A) site selection and alternative polyadenylation [122–124]. In light of these recent observations, one could predict that further analyses will define a broader range of regulatory functions for TTP proteins in the life of an mRNA than originally anticipated.
Acknowledgments
This work was supported by Institut National de la Santé et de la Recherche Médicale (U1036), Université Joseph Fourier, Commissariat à l’Energie Atomique et aux Energies Alternatives, Association pour la Recherche sur le Cancer (ARC) and Groupement des Entreprises Françaises pour la Lutte contre le Cancer (GEFLUC-Comité Dauphiné-Savoie). DC was supported by post-doctoral grants from ARC and Fondation Lefoulon Delalande. We are indebted to Dr. Sabine Bailly for helpful discussions and friendly support.
References
- 1.Varnum BC, Lim RW, Sukhatme VP, Herschman HR. Nucleotide sequence of a cDNA encoding TIS11, a message induced in Swiss 3T3 cells by the tumor promoter tetradecanoyl phorbol acetate. Oncogene. 1989;4:119–120. [PubMed] [Google Scholar]
- 2.DuBois RN, McLane MW, Ryder K, Lau LF, Nathans D. A growth factor-inducible nuclear protein with a novel cysteine/histidine repetitive sequence. J Biol Chem. 1990;265:19185–19191. [PubMed] [Google Scholar]
- 3.Gomperts M, Pascall JC, Brown KD. The nucleotide sequence of a cDNA encoding an EGF-inducible gene indicates the existence of a new family of mitogen-induced genes. Oncogene. 1990;5:1081–1083. [PubMed] [Google Scholar]
- 4.Lai WS, Stumpo DJ, Blackshear PJ. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J Biol Chem. 1990;265:16556–16563. [PubMed] [Google Scholar]
- 5.Nie XF, Maclean KN, Kumar V, McKay IA, Bustin SA. ERF-2, the human homologue of the murine Tis11d early response gene. Gene. 1995;152:285–286. doi: 10.1016/0378-1119(94)00696-P. [DOI] [PubMed] [Google Scholar]
- 6.Varnum BC, Ma QF, Chi TH, Fletcher B, Herschman HR. The TIS11 primary response gene is a member of a gene family that encodes proteins with a highly conserved sequence containing an unusual Cys-His repeat. Mol Cell Biol. 1991;11:1754–1758. doi: 10.1128/mcb.11.3.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackshear PJ, Phillips RS, Ghosh S, Ramos SB, Richfield EK, Lai WS. Zfp36l3, a rodent X chromosome gene encoding a placenta-specific member of the tristetraprolin family of CCCH tandem zinc finger proteins. Biol Reprod. 2005;73:297–307. doi: 10.1095/biolreprod.105.040527. [DOI] [PubMed] [Google Scholar]
- 8.Frederick ED, Ramos SB, Blackshear PJ. A unique C-terminal repeat domain maintains the cytosolic localization of the placenta-specific tristetraprolin family member ZFP36L3. J Biol Chem. 2008;283:14792–14800. doi: 10.1074/jbc.M801234200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30:945–952. doi: 10.1042/BST0300945. [DOI] [PubMed] [Google Scholar]
- 10.De J, Lai WS, Thorn JM, Goldsworthy SM, Liu X, Blackwell TK, Blackshear PJ. Identification of four CCCH zinc finger proteins in Xenopus, including a novel vertebrate protein with four zinc fingers and severely restricted expression. Gene. 1999;228:133–145. doi: 10.1016/S0378-1119(98)00617-9. [DOI] [PubMed] [Google Scholar]
- 11.Ma Q, Wadleigh D, Chi T, Herschman H. The Drosophila TIS11 homologue encodes a developmentally controlled gene. Oncogene. 1994;9:3329–3334. [PubMed] [Google Scholar]
- 12.Thompson MJ, Lai WS, Taylor GA, Blackshear PJ. Cloning and characterization of two yeast genes encoding members of the CCCH class of zinc finger proteins: zinc finger-mediated impairment of cell growth. Gene. 1996;174:225–233. doi: 10.1016/0378-1119(96)00084-4. [DOI] [PubMed] [Google Scholar]
- 13.Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P. MK2-induced tristetraprolin:14–3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidlin M, Lu M, Leuenberger SA, Stoecklin G, Mallaun M, Gross B, Gherzi R, Hess D, Hemmings BA, Moroni C. The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J. 2004;23:4760–4769. doi: 10.1038/sj.emboj.7600477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, Blackshear PJ. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/S1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 16.Bell SE, Sanchez MJ, Spasic-Boskovic O, Santalucia T, Gambardella L, Burton GJ, Murphy JJ, Norton JD, Clark AR, Turner M. The RNA binding protein Zfp36l1 is required for normal vascularisation and post-transcriptionally regulates VEGF expression. Dev Dyn. 2006;235:3144–3155. doi: 10.1002/dvdy.20949. [DOI] [PubMed] [Google Scholar]
- 17.Stumpo DJ, Byrd NA, Phillips RS, Ghosh S, Maronpot RR, Castranio T, Meyers EN, Mishina Y, Blackshear PJ. Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the tristetraprolin family. Mol Cell Biol. 2004;24:6445–6455. doi: 10.1128/MCB.24.14.6445-6455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramos SB. Characterization of DeltaN-Zfp36l2 mutant associated with arrest of early embryonic development and female infertility. J Biol Chem. 2012;287:13116–13127. doi: 10.1074/jbc.M111.330837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramos SB, Stumpo DJ, Kennington EA, Phillips RS, Bock CB, Ribeiro-Neto F, Blackshear PJ. The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development. 2004;131:4883–4893. doi: 10.1242/dev.01336. [DOI] [PubMed] [Google Scholar]
- 20.Stumpo DJ, Broxmeyer HE, Ward T, Cooper S, Hangoc G, Chung YJ, Shelley WC, Richfield EK, Ray MK, Yoder MC, Aplan PD, Blackshear PJ. Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood. 2009;114:2401–2410. doi: 10.1182/blood-2009-04-214619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodson DJ, Janas ML, Galloway A, Bell SE, Andrews S, Li CM, Pannell R, Siebel CW, MacDonald HR, De Keersmaecker K, Ferrando AA, Grutz G, Turner M. Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat Immunol. 2010;11:717–724. doi: 10.1038/ni.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murata T, Yoshino Y, Morita N, Kaneda N. Identification of nuclear import and export signals within the structure of the zinc finger protein TIS11. Biochem Biophys Res Commun. 2002;293:1242–1247. doi: 10.1016/S0006-291X(02)00363-7. [DOI] [PubMed] [Google Scholar]
- 23.Phillips RS, Ramos SB, Blackshear PJ. Members of the tristetraprolin family of tandem CCCH zinc finger proteins exhibit CRM1-dependent nucleocytoplasmic shuttling. J Biol Chem. 2002;277:11606–11613. doi: 10.1074/jbc.M111457200. [DOI] [PubMed] [Google Scholar]
- 24.Carman JA, Nadler SG. Direct association of tristetraprolin with the nucleoporin CAN/Nup214. Biochem Biophys Res Commun. 2004;315:445–449. doi: 10.1016/j.bbrc.2004.01.080. [DOI] [PubMed] [Google Scholar]
- 25.Brook M, Tchen CR, Santalucia T, McIlrath J, Arthur JS, Saklatvala J, Clark AR. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Mol Cell Biol. 2006;26:2408–2418. doi: 10.1128/MCB.26.6.2408-2418.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cherradi N, Lejczak C, Desroches-Castan A, Feige JJ. Antagonistic functions of tetradecanoyl phorbol acetate-inducible-sequence 11b and HuR in the hormonal regulation of vascular endothelial growth factor messenger ribonucleic acid stability by adrenocorticotropin. Mol Endocrinol. 2006;20:916–930. doi: 10.1210/me.2005-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desroches-Castan A, Cherradi N, Feige JJ, Ciais D. A novel function of Tis11b/BRF1 as a regulator of Dll4 mRNA 3′-end processing. Mol Biol Cell. 2011;22:3625–3633. doi: 10.1091/mbc.E11-02-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gringhuis SI, Garcia-Vallejo JJ, van Het Hof B, van Dijk W. Convergent actions of I kappa B kinase beta and protein kinase C delta modulate mRNA stability through phosphorylation of 14–3-3 beta complexed with tristetraprolin. Mol Cell Biol. 2005;25:6454–6463. doi: 10.1128/MCB.25.15.6454-6463.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor GA, Thompson MJ, Lai WS, Blackshear PJ. Mitogens stimulate the rapid nuclear to cytosolic translocation of tristetraprolin, a potential zinc-finger transcription factor. Mol Endocrinol. 1996;10:140–146. doi: 10.1210/me.10.2.140. [DOI] [PubMed] [Google Scholar]
- 30.Bourcier C, Griseri P, Grepin R, Bertolotto C, Mazure N, Pages G. Constitutive ERK activity induces downregulation of tristetraprolin, a major protein controlling interleukin8/CXCL8 mRNA stability in melanoma cells. Am J Physiol Cell Physiol. 2011;301:C609–C618. doi: 10.1152/ajpcell.00506.2010. [DOI] [PubMed] [Google Scholar]
- 31.Graham JR, Hendershott MC, Terragni J, Cooper GM. mRNA degradation plays a significant role in the program of gene expression regulated by phosphatidylinositol 3-kinase signaling. Mol Cell Biol. 2010;30:5295–5305. doi: 10.1128/MCB.00303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hitti E, Iakovleva T, Brook M, Deppenmeier S, Gruber AD, Radzioch D, Clark AR, Blackshear PJ, Kotlyarov A, Gaestel M. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol Cell Biol. 2006;26:2399–2407. doi: 10.1128/MCB.26.6.2399-2407.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahtani KR, Brook M, Dean JL, Sully G, Saklatvala J, Clark AR. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol Cell Biol. 2001;21:6461–6469. doi: 10.1128/MCB.21.9.6461-6469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marderosian M, Sharma A, Funk AP, Vartanian R, Masri J, Jo OD, Gera JF. Tristetraprolin regulates cyclin D1 and c-Myc mRNA stability in response to rapamycin in an Akt-dependent manner via p38 MAPK signaling. Oncogene. 2006;25:6277–6290. doi: 10.1038/sj.onc.1209645. [DOI] [PubMed] [Google Scholar]
- 35.Cao H, Dzineku F, Blackshear PJ. Expression and purification of recombinant tristetraprolin that can bind to tumor necrosis factor-alpha mRNA and serve as a substrate for mitogen-activated protein kinases. Arch Biochem Biophys. 2003;412:106–120. doi: 10.1016/S0003-9861(03)00012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carballo E, Cao H, Lai WS, Kennington EA, Campbell D, Blackshear PJ. Decreased sensitivity of tristetraprolin-deficient cells to p38 inhibitors suggests the involvement of tristetraprolin in the p38 signaling pathway. J Biol Chem. 2001;276:42580–42587. doi: 10.1074/jbc.M104953200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maitra S, Chou CF, Luber CA, Lee KY, Mann M, Chen CY. The AU-rich element mRNA decay-promoting activity of BRF1 is regulated by mitogen-activated protein kinase-activated protein kinase 2. RNA. 2008;14:950–959. doi: 10.1261/rna.983708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baou M, Jewell A, Murphy JJ. TIS11 family proteins and their roles in posttranscriptional gene regulation. J Biomed Biotechnol. 2009;2009:634520. doi: 10.1155/2009/634520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winzen R, Kracht M, Ritter B, Wilhelm A, Chen CY, Shyu AB, Muller M, Gaestel M, Resch K, Holtmann H. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 1999;18:4969–4980. doi: 10.1093/emboj/18.18.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benjamin D, Schmidlin M, Min L, Gross B, Moroni C. BRF1 protein turnover and mRNA decay activity are regulated by protein kinase B at the same phosphorylation sites. Mol Cell Biol. 2006;26:9497–9507. doi: 10.1128/MCB.01099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson BA, Stehn JR, Yaffe MB, Blackwell TK. Cytoplasmic localization of tristetraprolin involves 14–3-3-dependent and -independent mechanisms. J Biol Chem. 2002;277:18029–18036. doi: 10.1074/jbc.M110465200. [DOI] [PubMed] [Google Scholar]
- 42.Sun L, Stoecklin G, Van Way S, Hinkovska-Galcheva V, Guo RF, Anderson P, Shanley TP. Tristetraprolin (TTP)-14-3-3 complex formation protects TTP from dephosphorylation by protein phosphatase 2a and stabilizes tumor necrosis factor-alpha mRNA. J Biol Chem. 2007;282:3766–3777. doi: 10.1074/jbc.M607347200. [DOI] [PubMed] [Google Scholar]
- 43.Deleault KM, Skinner SJ, Brooks SA. Tristetraprolin regulates TNF TNF-alpha mRNA stability via a proteasome dependent mechanism involving the combined action of the ERK and p38 pathways. Mol Immunol. 2008;45:13–24. doi: 10.1016/j.molimm.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 44.Schichl YM, Resch U, Lemberger CE, Stichlberger D, de Martin R. Novel phosphorylation-dependent ubiquitination of tristetraprolin by mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase 1 (MEKK1) and tumor necrosis factor receptor-associated factor 2 (TRAF2) J Biol Chem. 2011;286:38466–38477. doi: 10.1074/jbc.M111.254888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clement SL, Scheckel C, Stoecklin G, Lykke-Andersen J. Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol Cell Biol. 2011;31:256–266. doi: 10.1128/MCB.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marchese FP, Aubareda A, Tudor C, Saklatvala J, Clark AR, Dean JL. MAPKAP kinase 2 blocks tristetraprolin-directed mRNA decay by inhibiting CAF1 deadenylase recruitment. J Biol Chem. 2010;285:27590–27600. doi: 10.1074/jbc.M110.136473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandler H, Stoecklin G. Control of mRNA decay by phosphorylation of tristetraprolin. Biochem Soc Trans. 2008;36:491–496. doi: 10.1042/BST0360491. [DOI] [PubMed] [Google Scholar]
- 48.Sanduja S, Blanco FF, Young LE, Kaza V, Dixon DA. The role of tristetraprolin in cancer and inflammation. Front Biosci. 2012;17:174–188. doi: 10.2741/3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baou M, Norton JD, Murphy JJ. AU-rich RNA binding proteins in hematopoiesis and leukemogenesis. Blood. 2011;118:5732–5740. doi: 10.1182/blood-2011-07-347237. [DOI] [PubMed] [Google Scholar]
- 50.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen CY, Shyu AB. Selective degradation of early-response-gene mRNAs: functional analyses of sequence features of the AU-rich elements. Mol Cell Biol. 1994;14:8471–8482. doi: 10.1128/mcb.14.12.8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bakheet T, Williams BR, Khabar KS. ARED 2.0: an update of AU-rich element mRNA database. Nucleic Acids Res. 2003;31:421–423. doi: 10.1093/nar/gkg023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spasic M, Friedel CC, Schott J, Kreth J, Leppek K, Hofmann S, Ozgur S, Stoecklin G. Genome-wide assessment of AU-rich elements by the ARE score algorithm. PLoS Genet. 2012;8:e1002433. doi: 10.1371/journal.pgen.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J Biol Chem. 2000;275:17827–17837. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 56.Worthington MT, Pelo JW, Sachedina MA, Applegate JL, Arseneau KO, Pizarro TT. RNA binding properties of the AU-rich element-binding recombinant Nup475/TIS11/tristetraprolin protein. J Biol Chem. 2002;277:48558–48564. doi: 10.1074/jbc.M206505200. [DOI] [PubMed] [Google Scholar]
- 57.Blackshear PJ, Lai WS, Kennington EA, Brewer G, Wilson GM, Guan X, Zhou P. Characteristics of the interaction of a synthetic human tristetraprolin tandem zinc finger peptide with AU-rich element-containing RNA substrates. J Biol Chem. 2003;278:19947–19955. doi: 10.1074/jbc.M301290200. [DOI] [PubMed] [Google Scholar]
- 58.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 59.Amann BT, Worthington MT, Berg JM. A Cys3His zinc-binding domain from Nup475/tristetraprolin: a novel fold with a disklike structure. Biochemistry. 2003;42:217–221. doi: 10.1021/bi026988m. [DOI] [PubMed] [Google Scholar]
- 60.Brown RS. Zinc finger proteins: getting a grip on RNA. Curr Opin Struct Biol. 2005;15:94–98. doi: 10.1016/j.sbi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 61.Emmons J, Townley-Tilson WH, Deleault KM, Skinner SJ, Gross RH, Whitfield ML, Brooks SA. Identification of TTP mRNA targets in human dendritic cells reveals TTP as a critical regulator of dendritic cell maturation. RNA. 2008;14:888–902. doi: 10.1261/rna.748408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, Blackshear PJ, Anderson P. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J Biol Chem. 2008;283:11689–11699. doi: 10.1074/jbc.M709657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Linker K, Pautz A, Fechir M, Hubrich T, Greeve J, Kleinert H. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 2005;33:4813–4827. doi: 10.1093/nar/gki797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fechir M, Linker K, Pautz A, Hubrich T, Forstermann U, Rodriguez-Pascual F, Kleinert H. Tristetraprolin regulates the expression of the human inducible nitric-oxide synthase gene. Mol Pharmacol. 2005;67:2148–2161. doi: 10.1124/mol.104.008763. [DOI] [PubMed] [Google Scholar]
- 65.Kedar VP, Zucconi BE, Wilson GM, Blackshear PJ. Direct binding of specific AUF1 isoforms to tandem zinc finger domains of tristetraprolin (TTP) family proteins. J Biol Chem. 2012;287:5459–5471. doi: 10.1074/jbc.M111.312652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 67.Lai WS, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA: tristetraprolin-mediated AU-rich element-dependent mRNA degradation can occur in the absence of a poly (A) tail. J Biol Chem. 2001;276:23144–23154. doi: 10.1074/jbc.M100680200. [DOI] [PubMed] [Google Scholar]
- 68.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lai WS, Carrick DM, Blackshear PJ. Influence of nonameric AU-rich tristetraprolin-binding sites on mRNA deadenylation and turnover. J Biol Chem. 2005;280:34365–34377. doi: 10.1074/jbc.M506757200. [DOI] [PubMed] [Google Scholar]
- 70.Lai WS, Kennington EA, Blackshear PJ. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly (A) ribonuclease. Mol Cell Biol. 2003;23:3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sandler H, Kreth J, Timmers HT, Stoecklin G. Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 2011;39:4373–4386. doi: 10.1093/nar/gkr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rowlett RM, Chrestensen CA, Schroeder MJ, Harp MG, Pelo JW, Shabanowitz J, DeRose R, Hunt DF, Sturgill TW, Worthington MT. Inhibition of tristetraprolin deadenylation by poly (A) binding protein. Am J Physiol Gastrointest Liver Physiol. 2008;295:G421–G430. doi: 10.1152/ajpgi.00508.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kedar VP, Darby MK, Williams JG, Blackshear PJ. Phosphorylation of human tristetraprolin in response to its interaction with the Cbl interacting protein CIN85. PLoS ONE. 2010;5:e9588. doi: 10.1371/journal.pone.0009588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/S0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 76.Mukherjee D, Gao M, O’Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Dijk EL, Schilders G, Pruijn GJ. Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways. RNA. 2007;13:1027–1035. doi: 10.1261/rna.575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hau HH, Walsh RJ, Ogilvie RL, Williams DA, Reilly CS, Bohjanen PR. Tristetraprolin recruits functional mRNA decay complexes to ARE sequences. J Cell Biochem. 2007;100:1477–1492. doi: 10.1002/jcb.21130. [DOI] [PubMed] [Google Scholar]
- 79.Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE-binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 80.Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murata T, Hikita K, Kaneda N. Transcriptional activation function of zinc finger protein TIS11 and its negative regulation by phorbol ester. Biochem Biophys Res Commun. 2000;274:526–532. doi: 10.1006/bbrc.2000.3182. [DOI] [PubMed] [Google Scholar]
- 82.Dudziak K, Mottalebi N, Senkel S, Edghill EL, Rosengarten S, Roose M, Bingham C, Ellard S, Ryffel GU. Transcription factor HNF1beta and novel partners affect nephrogenesis. Kidney Int. 2008;74:210–217. doi: 10.1038/ki.2008.149. [DOI] [PubMed] [Google Scholar]
- 83.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Prouteau M, Daugeron MC, Seraphin B. Regulation of ARE transcript 3′ end processing by the yeast Cth2 mRNA decay factor. EMBO J. 2008;27:2966–2976. doi: 10.1038/emboj.2008.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Su YL, Wang SC, Chiang PY, Lin NY, Shen YF, Chang GD, Chang CJ. Tristetraprolin inhibits poly (A)-tail synthesis in nuclear mRNA that contains AU-rich elements by interacting with poly (A)-binding protein nuclear 1. PLoS ONE. 2012;7:e41313. doi: 10.1371/journal.pone.0041313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vergara SV, Puig S, Thiele DJ. Early recruitment of AU-rich element-containing mRNAs determines their cytosolic fate during iron deficiency. Mol Cell Biol. 2011;31:417–429. doi: 10.1128/MCB.00754-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 89.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007;21:719–735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chang WL, Tarn WY. A role for transportin in deposition of TTP to cytoplasmic RNA granules and mRNA decay. Nucleic Acids Res. 2009;37:6600–6612. doi: 10.1093/nar/gkp717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Espel E. The role of the AU-rich elements of mRNAs in controlling translation. Semin Cell Dev Biol. 2005;16:59–67. doi: 10.1016/j.semcdb.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 93.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brooks SA, Connolly JE, Diegel RJ, Fava RA, Rigby WF. Analysis of the function, expression, and subcellular distribution of human tristetraprolin. Arthr Rheum. 2002;46:1362–1370. doi: 10.1002/art.10235. [DOI] [PubMed] [Google Scholar]
- 95.Rigby WF, Roy K, Collins J, Rigby S, Connolly JE, Bloch DB, Brooks SA. Structure/function analysis of tristetraprolin (TTP): p38 stress-activated protein kinase and lipopolysaccharide stimulation do not alter TTP function. J Immunol. 2005;174:7883–7893. doi: 10.4049/jimmunol.174.12.7883. [DOI] [PubMed] [Google Scholar]
- 96.Qi MY, Wang ZZ, Zhang Z, Shao Q, Zeng A, Li XQ, Li WQ, Wang C, Tian FJ, Li Q, Zou J, Qin YW, Brewer G, Huang S, Jing Q. AU-rich-element-dependent translation repression requires the cooperation of tristetraprolin and RCK/P54. Mol Cell Biol. 2011;32:913–928. doi: 10.1128/MCB.05340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pedro-Segura E, Vergara SV, Rodriguez-Navarro S, Parker R, Thiele DJ, Puig S. The Cth2 ARE-binding protein recruits the Dhh1 helicase to promote the decay of succinate dehydrogenase SDH4 mRNA in response to iron deficiency. J Biol Chem. 2008;283:28527–28535. doi: 10.1074/jbc.M804910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pfeiffer JR, Brooks SA. Cullin 4B is recruited to tristetraprolin-containing messenger ribonucleoproteins and regulates TNF-alpha mRNA polysome loading. J Immunol. 2012;188:1828–1839. doi: 10.4049/jimmunol.1102837. [DOI] [PubMed] [Google Scholar]
- 99.Ciais D, Cherradi N, Bailly S, Grenier E, Berra E, Pouyssegur J, Lamarre J, Feige JJ. Destabilization of vascular endothelial growth factor mRNA by the zinc-finger protein TIS11b. Oncogene. 2004;23:8673–8680. doi: 10.1038/sj.onc.1207939. [DOI] [PubMed] [Google Scholar]
- 100.Essafi-Benkhadir K, Onesto C, Stebe E, Moroni C, Pages G. Tristetraprolin inhibits Ras-dependent tumor vascularization by inducing vascular endothelial growth factor mRNA degradation. Mol Biol Cell. 2007;18:4648–4658. doi: 10.1091/mbc.E07-06-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Planel S, Salomon A, Jalinot P, Feige JJ, Cherradi N. A novel concept in antiangiogenic and antitumoral therapy: multitarget destabilization of short-lived mRNAs by the zinc finger protein ZFP36L1. Oncogene. 2010;29:5989–6003. doi: 10.1038/onc.2010.341. [DOI] [PubMed] [Google Scholar]
- 102.von Roretz C, Gallouzi IE. Decoding ARE-mediated decay: is microRNA part of the equation? J Cell Biol. 2008;181:189–194. doi: 10.1083/jcb.200712054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 104.Helfer S, Schott J, Stoecklin G, Forstemann K. AU-rich element-mediated mRNA decay can occur independently of the miRNA machinery in mouse embryonic fibroblasts and Drosophila S2-cells. PLoS ONE. 2012;7:e28907. doi: 10.1371/journal.pone.0028907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dorner S, Lum L, Kim M, Paro R, Beachy PA, Green R. A genomewide screen for components of the RNAi pathway in Drosophila cultured cells. Proc Natl Acad Sci USA. 2006;103:11880–11885. doi: 10.1073/pnas.0605210103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim CW, Vo MT, Kim HK, Lee HH, Yoon NA, Lee BJ, Min YJ, Joo WD, Cha HJ, Park JW, Cho WJ. Ectopic over-expression of tristetraprolin in human cancer cells promotes biogenesis of let-7 by down-regulation of Lin28. Nucleic Acids Res. 2012;40:3856–3869. doi: 10.1093/nar/gkr1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liang J, Lei T, Song Y, Yanes N, Qi Y, Fu M. RNA-destabilizing factor tristetraprolin negatively regulates NF-kappaB signaling. J Biol Chem. 2009;284:29383–29390. doi: 10.1074/jbc.M109.024745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schichl YM, Resch U, Hofer-Warbinek R, de Martin R. Tristetraprolin impairs NF-kappaB/p65 nuclear translocation. J Biol Chem. 2009;284:29571–29581. doi: 10.1074/jbc.M109.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- 110.Brennan SE, Kuwano Y, Alkharouf N, Blackshear PJ, Gorospe M, Wilson GM. The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res. 2009;69:5168–5176. doi: 10.1158/0008-5472.CAN-08-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee SK, Kim SB, Kim JS, Moon CH, Han MS, Lee BJ, Chung DK, Min YJ, Park JH, Choi DH, Cho HR, Park SK, Park JW. Butyrate response factor 1 enhances cisplatin sensitivity in human head and neck squamous cell carcinoma cell lines. Int J Cancer. 2005;117:32–40. doi: 10.1002/ijc.21133. [DOI] [PubMed] [Google Scholar]
- 112.Stoecklin G, Gross B, Ming XF, Moroni C. A novel mechanism of tumor suppression by destabilizing AU-rich growth factor mRNA. Oncogene. 2003;22:3554–3561. doi: 10.1038/sj.onc.1206418. [DOI] [PubMed] [Google Scholar]
- 113.Griseri P, Bourcier C, Hieblot C, Essafi-Benkhadir K, Chamorey E, Touriol C, Pages G. A synonymous polymorphism of the Tristetraprolin (TTP) gene, an AU-rich mRNA-binding protein, affects translation efficiency and response to Herceptin treatment in breast cancer patients. Hum Mol Genet. 2012;20:4556–4568. doi: 10.1093/hmg/ddr390. [DOI] [PubMed] [Google Scholar]
- 114.Rounbehler RJ, Fallahi M, Yang C, Steeves MA, Li W, Doherty JR, Schaub FX, Sanduja S, Dixon DA, Blackshear PJ, Cleveland JL. Tristetraprolin impairs myc-induced lymphoma and abolishes the malignant state. Cell. 2012;150:563–574. doi: 10.1016/j.cell.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ross CR, Brennan-Laun SE, Wilson GM (2012) Tristetraprolin: roles in cancer and senescence. Ageing Res Rev [DOI] [PMC free article] [PubMed]
- 116.Chang SH, Hla T. Gene regulation by RNA binding proteins and microRNAs in angiogenesis. Trends Mol Med. 2011;17:650–658. doi: 10.1016/j.molmed.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wegmuller D, Raineri I, Gross B, Oakeley EJ, Moroni C. A cassette system to study embryonic stem cell differentiation by inducible RNA interference. Stem Cells. 2007;25:1178–1185. doi: 10.1634/stemcells.2006-0106. [DOI] [PubMed] [Google Scholar]
- 119.Patil CS, Liu M, Zhao W, Coatney DD, Li F, VanTubergen EA, D’Silva NJ, Kirkwood KL. Targeting mRNA stability arrests inflammatory bone loss. Mol Ther. 2008;16:1657–1664. doi: 10.1038/mt.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, Amariglio N, Vaisman N, Segal E, Rechavi G, Alon U, Mills GB, Domany E, Yarden Y. A module of negative feedback regulators defines growth factor signaling. Nat Genet. 2007;39:503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- 121.Lai WS, Parker JS, Grissom SF, Stumpo DJ, Blackshear PJ. Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol Cell Biol. 2006;26:9196–9208. doi: 10.1128/MCB.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 123.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]