Abstract
Nuclear localization of classical growth factors is a well-known phenomenon but still remains a molecular and cellular conundrum. Fibroblast growth factor-2 (FGF-2) is an excellent example of a protein which functions as an extracellular molecule involved in canonical receptor tyrosine kinase signaling as well as displaying intracellular functions. Paracrine and nuclear functions are two important sides of the same protein. FGF-2 is expressed in isoforms with different molecular weights from one mRNA species. In rodents, all of these isoforms become imported to the nucleus. In this review, we discuss structural and functional aspects of FGF-2 isoforms in the nervous system. The nuclear odyssey of FGF-2 is reflected by nuclear dynamics, localization to nuclear bodies such as nucleoli, binding to chromatin and engagement in various protein interactions. Recently discovered molecular partnerships of the isoforms shed light on their nuclear functions, thereby greatly extending our knowledge of the multifaceted functions of FGF-2.
Keywords: FGF signaling, Isoforms, Neurotrophic factor, Nuclear protein, SMN, Chromatin, Splicing, Spinal muscular atrophy (SMA), Parkinson´s disease
The dawn of nuclear FGF-2
The fibroblast growth factor (FGF) family represents a large group of signaling molecules and comprises 22 members [1] encoded by the same number of genes [2]. One group of FGFs was primarily identified and purified from bovine pituitary glands [3, 4] and later subdivided into acidic FGF (FGF-1) and basic FGF (FGF-2) using isoelectric focusing [5]. Based on the tissue of origin or target cell specificity, FGF-2 was given different names like basic FGF (bFGF), pituitary cationic FGF, pituitary brain FGF, cartilage-derived growth factor (CDGF), heparin-binding growth factor β (HGFβ), eye-derived growth factor 1 (EDGF-1), or astroglial growth factor 2 (AGF-2) and classified as class 2 heparin-binding growth factors (class 2 HBGFs) [6]. Later, basic FGF was renamed to fibroblast growth factor-2 [7].
FGF-2 is expressed in different isoforms from one mRNA species with distinct molecular weights. In addition to 18 kDa FGF-2 [FGF-218, denoted as low-molecular weight (LMW-FGF-2)] another two high molecular weight isoforms have been described as 21 and 23 kDa FGF-2 (FGF-221 and 23, together denoted as HMW-FGF-2) in rodents [8–10]. In humans, besides 18 kDa FGF-2, even four high molecular weight isoforms are expressed. Translation either starts at a conventional AUG start codon for 18 kDa FGF-2 or alternative upstream CUG start codons for HMW-FGF-2 isoforms from a single mRNA species [11–15]. The HMW-FGF-2 isoforms show N-terminal sequence extensions compared with the 18 kDa isoform. However, all isoforms comprise the 18 kDa core sequence (Fig. 1). The FGF-2 mRNA has interesting features permitting highly regulated translation: although this mRNA exhibits a classical cap for translational initiation, an internal ribosomal entry site (IRES) in the 5′ untranslated region allows additional means of translational control [16]. The IRES is used preferentially for regulation of protein expression, especially in brain tissue [17] and at least partly determines expression of specific isoforms. However, also the 3′ untranslated region takes part in alternative initiation of translation as well as overall destabilization of the mRNA [18, 19].
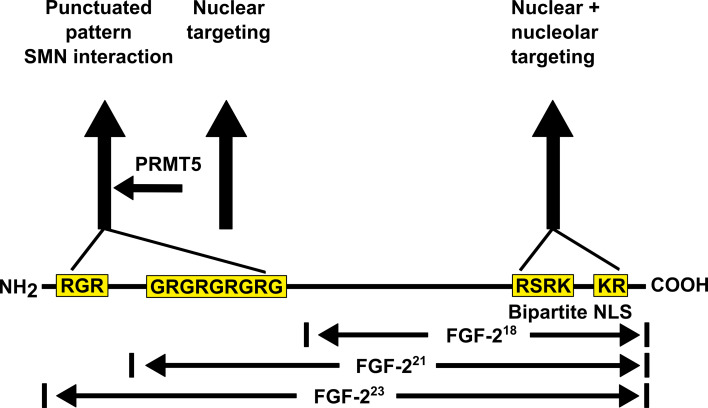
Fig. 1.
Primary structures of FGF-2 isoforms and distribution of RG motifs in rodent FGF-2 isoforms. A bipartite nuclear NLS, comprised by splitted arginine–lysine (RK) motifs, is important for nuclear targeting of all isoforms and nucleolar targeting of FGF-218 [81, 83]. Nuclear localization of HMW-FGF-2 isoforms is mediated by a large conserved arginine–glycine (RG) sequence [84, 85]. In this model, a critical threshold of positive charges in the N-terminal FGF-2 extension is required for nuclear localization and chromatin association [89] which could be eventually responsible for transcriptional modification. Furthermore, the N-terminal extension of HMW-FGF-2 promotes SMN binding [81, 118]. The FGF-223-SMN interaction is severely affected by symmetrical dimethylation by PRMT5 [89]
Release of FGF-2 from the cell into the extracellular space has been an enigma for a long time. Although lacking a conventional signal peptide sequence, 18 kDa FGF-2 is released in an energy-dependent exocytotic mechanism bypassing the Golgi/ER pathway [20, 21]. This process is induced by the binding of cytoplasmic FGF-2 to phosphoinositides at the plasma membrane [22] followed by the oligomerization of FGF-2 and formation of a lipidic membrane pore [23] and finally the release on the cell surface [24]. It should be clearly noted that export of 18 kDa FGF-2 may not be an exclusive characteristic of this low molecular weight isoform. Release of HMW-FGF-2 isoforms from the cell has not been widely considered as a mechanism providing additional functional complexity of the isoforms as paracrine factors. However, HMW-FGF-2 becomes released in a highly regulated angiotensin and caspase-1 dependent manner from cardiac non-myocytes [25, 26]. This paracrine action induces hypertrophy of cardiac myocytes and other deleterious, e.g., pro-inflammatory downstream events [27].
Depending on the isoform or more precisely on the length of the N-terminal extension, FGF-2 plays diverse roles in different tissues and cell types. In the 1970s, FGF-2 was initially purified from pituitary glands [3, 4] due to high concentrations of the growth factor in the brain indicating that FGF-2 has a specific role in the nervous system [3, 4]. FGF-2 has been then considered to be a neurotrophic factor due to in vitro and in vivo promoted survival, proliferation, and regulation of differentiation of central or peripheral neurons [28, 29]. For example, FGF-2 is retrogradely transported in the hypoglossal nerve [30]. Retrograde axonal transport was before shown for nerve growth factor (NGF) as the prototype of a neurotrophic factor [31–33]. In this review, we focus on the roles of FGF-2 isoforms in the nervous system and provide an overview about recently described molecular mechanisms of FGF-2 isoforms in the nucleus. These nuclear FGFs display a remarkable dynamics in the interchromosomal space—compared to a nuclear space odyssey—where they interact with a number of proteins which were recently identified. Identification of molecular partner proteins defines new functional roles of FGF-2 isoforms in signaling.
FGF-2 isoforms in the nervous system
The FGF-2 isoforms are not equally distributed in different brain regions. Diverse structures of the central (CNS) and peripheral nervous system (PNS) display distinct FGF-2 profiles with specific expression of isoforms in development and after tissue damage. In the adult rat cerebellum, cortex and spinal cord, specific expression patterns of FGF-2 isoforms have been found and these profiles become particularly regulated during development. This indicates diverse functions of the corresponding protein isoforms in formation, maturation and organization of the central nervous system [34].
In the CNS, FGF-2 is widely expressed in different structures [35, 36]. However, most studies address expression on mRNA level and not the expression of FGF-2 protein isoforms. Therefore, our knowledge of isoform distribution and expression is still limited in the nervous system. All three FGF-2 isoforms are expressed in the striatum and substantia nigra of adult rats [37]. This could indicate a role of FGF-2 in supporting differentiation or survival of neurons in these structures. Since degeneration of substantia nigra neurons occurs in Parkinson’s disease (PD), expression of FGF-2 or its specific isoforms could be a means of attenuating degeneration. In a model of PD, chemically lesioned mice display increased recovery of striatal dopaminergic (DA) neurons after intrastriatal application of 18 kDa FGF-2 [38, 39]. Accordingly, mice deficient of all FGF-2 isoforms show reduced numbers of DA neurons in contrast to FGF-2 transgenic mice which display an increase [40]. Moreover, HMW-FGF-2 expressing Schwann cells increase the survival of rat dopaminergic mesencephalic neurons in co-culture experiments in vitro as well as exogenously applied FGF-221 and FGF-223 [41]. Moreover, co-transplantation of HMW-FGF-2 expressing Schwann cells improve in vivo the restoration, reinnervation and survival of dopaminergic micrografts in a rat model of Parkinson’s disease, compared with FGF-218 expressing Schwann cells [42]. In the rat cortex, FGF-221 has been mainly found at embryonic day 18 (E18) and day of birth (postnatal day 0, P0). In contrast, no FGF expression is detected in the adult cortex. However, in the rat cerebellum, FGF-218 and FGF-221 have been found highly expressed between E18 and P7. After this point of time, only FGF-218 remains strongly expressed and no high molecular weight isoforms have been found in the adult cerebellum. In the spinal cord, FGF-218 and FGF-221 are expressed at E18 and P0. This pattern changes afterwards—also in contrast to the cerebellum: here, in the adult spinal cord exclusively both HMW-FGFs are highly expressed and LMW-FGF-2 displays only moderate expression levels [34]. Rat oligodendrocytes are affected by the application of FGF-2, terminal differentiation in vitro [43] as well as myelin production in vivo [44] is severely reduced. Consistently, FGF-2 deletion in mice promotes oligodendrocyte regeneration in vivo [45]. Expression of FGF-223 in pyramidal neurons of the rat hippocampus results in increased detection of two nuclei in these cells. The same has been found for postganglionic neurons and non-neuronal cells, followed by degeneration [46]. Interestingly, transfection of this isoform into postmitotic sympathetic neurons resulted in bi- or multinucleated phenotypes supporting a function of FGF-223 as a positive regulator of karyokinesis [46]. Interestingly, this phenotype has also been observed in a non-neuronal system: cardiac myocytes also exhibit binuclear morphology upon transfection with HMW-FGF-2 [47]. Furthermore, the expression profiles of FGF-2 isoforms can be pharmacologically modulated. The antidepressants desipramine and fluoxetine increase FGF-2 protein levels in vivo in rat frontal cortical neurons and shift particularly the intracellular HMW-FGF-2 localization from the nucleus into the cytoplasm [48]. The antidepressant amitriptyline increases FGF-2 mRNA transcription and equally the expression of low and high molecular weight isoforms in vitro only in rat cortical astrocytes but not in cortical neuron-enriched cultures [49].
In the PNS, FGF-2 mRNA has been found upregulated after peripheral nerve lesion in rat sensory ganglia [50, 51]. After hypoglossal nerve transection, all FGF isoforms are downregulated 3 days after lesion and then upregulated 14 days after lesion above control levels [52]. Axotomy of the rat superior cervical ganglion (SCG) results in a fivefold increased FGF-218 and FGF-223 and a threefold increased FGF-221 protein level [53]. In the lesioned rat sciatic or saphenous nerves, FGF-2 displays neurite-promoting functions [54–56]. The same improvement can be observed in the regenerating rat sciatic nerve by transplantation of HMW-FGF-2 over-expressing Schwann cells [57]. Indeed, after lesion of spinal ganglia or sciatic nerve FGF-2 protein isoforms are differentially upregulated, indicating isoform-specific physiological functions in the regeneration of injured peripheral nerves [58–60]. FGF-2 knockout in mice leads to increased regeneration and myelination close to the injury site after sciatic nerve lesion with indications of augmented Wallerian degeneration, suggesting crucial functions of FGF-2 in early peripheral nerve regeneration [61]. After sciatic nerve transection leaving a certain gap length, HMW-FGF-2 over-expressing Schwann cells promote sensory nerve recovery and long-distance myelination in vivo in contrast to inhibitory effects of FGF-218 over-expressing Schwann cells on myelination [62]. In contrast, neither LMW- nor HMW-FGF-2 overexpressing Schwann cells affect the recovery after facial nerve transection in adult rats [63]. Taken together, the results demonstrate specific roles of FGF-2 isoforms in regeneration both in the CNS and the PNS.
Pheochromocytoma cells (PC12) are an excellent model of neuronal differentiation and neurotransmitter vesicle release due to their origin from modified sympathetic ganglia cells in the adrenal gland. During postnatal development of rat adrenal medulla, a peak expression of FGF-221 at postnatal day 28 has been observed [64]. Glucocorticoids like dexamethasone are suggested to trigger the differentiation of adrenergic chromaffin cells out of noradrenergic sympathoadrenal precursor cells [65]. In the rat adrenal medulla similar to PC12 cells, but not in non-neuronal tissues, dexamethasone enhances in vivo specifically the expression of the FGF-221 isoform indicating a neuron specific physiological function [66, 67]. Overexpression of FGF-218 alone in PC12 cells is sufficient to start differentiation and outgrowth of long processes in contrast to cells expressing HMW-FGF-2 which develop a round-shaped morphology [68]. Moreover, in immortalized Schwann cells, morphology and growth are altered to shorter processes after overexpression of HMW-FGF-2, compared with Schwann cells expressing FGF-218 or control cells [68]. However, stimulation with 18 kDa FGF-2 enhances proliferation of cultured mouse and rat Schwann cells [69–72].
Besides the neurotrophic functions of FGF-2 and its isoforms, diverse functions in other organs have been observed. For example, after myocardial infarction in a rat model, FGF-218 promotes angiogenesis and cardioprotection in contrast to HMW-FGF-2 which promoted deleterious effects [27, 73]. Furthermore, FGF-218 enhances bone formation, whereas HMW-FGF-2 has an inhibitory effect [74–76]. Interestingly, HMW-FGF-2 expression in fibroblasts leads to growth reduction and lower levels of proliferating cell nuclear antigen (PCNA) [77, 78] indicating widespread isoform-specific FGF-2 functions in different tissues.
The mission of FGF-2: nuclear import and engagement
FGF-218 has been found in the nucleus as well as in the cytoplasm, whereas HMW-FGF-2 isoforms localize predominantly in the nucleus of different cell types in isoform-specific nuclear localization patterns [79, 80]. However, the distribution of nuclear FGF-218 is clearly different from the distribution of HMW-FGF-2 indicating different structural elements of the isoforms responsible for nuclear localization [79, 81]. HMW-FGF-2 isoforms show a punctuated pattern in the nucleus and a chromatin association, whereas 18 kDa FGF-2 localizes to nucleoli [79, 82].
All isoforms share a nuclear localization signal (NLS) in the C-terminal core sequence (Fig. 1). For the purpose of comparison between the isoforms, we define here the sequence of 18 kDa FGF-2 as a “core” since this primary structure is common to all isoforms. The first part of the bipartite NLS comprises Arg116, Arg118 [81] and Lys119 [83]. Together with the second part (Lys128 and Arg129), these amino acid clusters are designated as a non-classical bipartite NLS, not only mediating nuclear import but also nucleolar accumulation of FGF-218 [81, 83]. HMW-FGF-2 isoforms comprise a second nuclear localization signal in their N-terminal extensions. Fusion of the HMW extension to chloramphenicol acetyltransferase (CAT), green fluorescent protein (GFP) or β-galactosidase (β-gal) results in nuclear accumulation of the fusion proteins in contrast to control constructs [81, 84, 85]. Conserved glycine-arginine-glycine (RGR) sequence motifs in HMW-FGF-2 cause nuclear localization [81, 84, 85]. Basic peptide sequences, containing mainly arginine and lysine residues serve as nuclear localization sequences due to the potential to build an import complex with importin-α and importin-β, followed by a transfer through a nuclear core complex into the nucleus [86, 87]. Insertion of the RGR motif into FGF-218 leads to increased nuclear localization of the protein, supporting the supposed role of the amino acid sequence [88]. FGF-221 and FGF-223 differ in only one RGR motif. However, this single element is not sufficient for nuclear localization of the protein. It has been suggested that the positive charge of this sequence regulates the punctuated patterns of the HMW-FGF-2 isoform. Deletion of a hydrophobic stretch between RGR clusters and mutation of two arginine to histidine residues, retaining a positive charge, does not change the nuclear distribution of FGF-223 [89]. It has been suggested that a threshold of positive charge regulates chromatin association primarily as a result of certain number of arginine residues in the N-terminal FGF-2 extension [89] (Fig. 1). Interestingly, the chromatin-binding high-mobility group proteins A1/A2 (HMG-A1/A2) also comprise RGR motifs as extended motifs with a proline residue forming RGRP motifs. These motifs are reminiscent of the RGR-rich primary structure of HMW-FGF-2 [90]. It has been shown that these structures designated as AT-hooks in HMGA proteins bind to AT-rich DNA [91].
Arginine residues in the N-terminus of HMW-FGF-2 can be methylated, affecting the intracellular distribution of the protein [89, 92, 93]. FGF-223 becomes methylated by protein arginine methyltransferase 5 (PRMT5) [89] (Fig. 2). PRMT5 protein complexes in the cytoplasm are designated as methylosomes [94] and responsible for symmetrical methylation of Sm proteins resulting in the assembly of small nuclear ribonucleoproteins (snRNPs) by the survival of motoneuron (SMN) protein [95–97]. SnRNPs are composed of Sm proteins in a ring-like structure as well as a catalytic small uridine-rich RNA molecule (U snRNAs) [98]. Symmetrically dimethylated RG motifs are preferred binding sites for the SMN protein [97] due to an aromatic cage in the SMN Tudor domain mediating dimethyl-arginine recognition [99]. Therefore, it was supposed that methylated FGF-223 could also be a target and interaction partner of SMN like the Sm proteins. This is indeed the case: symmetrically dimethylated FGF-223 does not only show an interaction with SMN but also is additionally targeted to the nucleus. As expected, FGF-218 does not interact with SMN. This clearly confirms that the N-terminal extension of FGF-223 is the molecular interface responsible for direct molecular partnering with SMN. Inhibition of methyltransferases by 5′-deoxy-5′-methylthioadenosine (MTA) or adenosine dialdehyde (AdOx) causes a dysregulation of the nucleus/cytoplasmic distribution of FGF-2 towards a cytoplasmic localization [89, 93] showing a clear effect of FGF-2 methylation on intracellular localization [59, 62]. Due to the strong influence of FGF-2 methylation on SNM binding, it has been suggested that the FGF-223-SMN interaction influences FGF-2 nuclear import. Since FGF-218 does not interact with SMN, the nuclear transport of this isoform may be based on a different mechanism. How could the nuclear transport of FGF-218 be mediated? The protein Translokin (Tlk) has been shown to interact with FGF-218 but not with the high molecular weight isoforms. Reduced Tlk expression leads to reduced nuclear FGF-2 accumulation emphasizing the existence of FGF-2 isoform-specific nuclear import mechanisms [100]. However, treatment of Swiss 3T3 cells with FGF-218 results in importin-β-dependent internalization of the FGF receptor 1 (FGFR1) [101, 102]. Internalization and nuclear translocation of ligands with the respective growth factor receptors are not an exclusive mechanism of the FGF-2/FGFR1 interaction but can be shown for different ligand/receptor interactions like epidermal growth factor (EGF)/EGF receptor (EGFR) or interferon γ (IFNγ)/INFNγ receptor (INFGR) [103]. FGFR1 interacts directly with importin-β, but uses an adapter protein that could be HMW-FGF-2 [104]. Immortalized pancreatic stellate cell line 1 (PS1) cells in a model for pancreatic cancer display a correlation of FGF-2/FGFR1 nuclear accumulation and the ability to invade into the extracellular matrix. Inhibition or RNAi knock-down of FGFR1 leads to cytoplasmic FGF-2/FGFR1 localization and a reduction in proliferation and cell invasion, suggesting a potential novel therapeutic usability of FGFR1 inhibition in pancreatic cancer [105]. Prostaglandin F2α incubated immortalized rat osteoblastic Py1a cells display an increased binding of FGF-2 and FGFR2, a co-localization with importin-β and a nuclear translocation suggesting a role of FGF-2/FGFR2 nuclear trafficking in bone metabolism [106]. Following the translocation, different nuclear localization sequences and isoform-specific nuclear localization patterns of FGF-2 point to diverse functions of this nuclear growth factor.
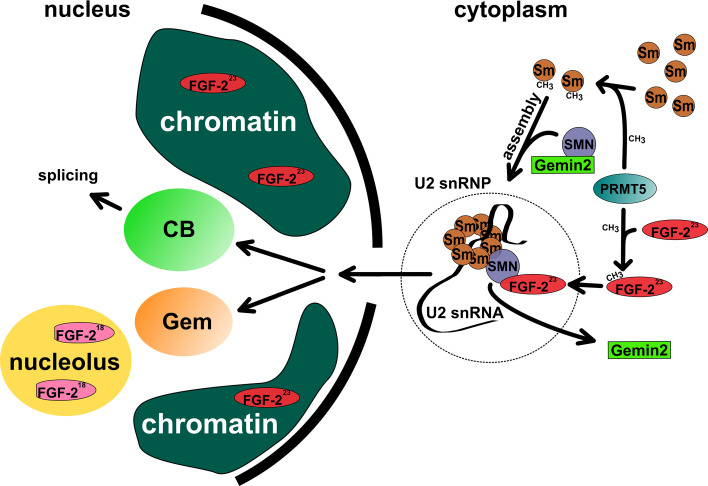
Fig. 2.
FGF-223 interaction with the Survival of Motoneuron (SMN) protein. SMN is an assembly factor for small nuclear ribonucleoprotein particles (snRNPs) composes of catalytic RNA molecules (U RNAs, shown here for the U2 snRNA) and Smith antigen (Sm) proteins. FGF-223 becomes arginine-methylated at its N-terminus by Protein Arginine Methyltransferase 5 (PRMT5) [89] as well as the Sm proteins. FGF-223 binds to SMN thereby competing with Gemin2 for binding to the N-terminus of SMN [81, 118]. The snRNP translocates to Cajal bodies (CBs) in the nucleus where different snRNPs undergo a subsequent master assembly into tri-snRNP complexes which later engage in pre-mRNA splicing. SMN dissociates from the complex forming small nuclear bodies called nuclear gems [110]. CBs and gems are highly mobile structures moving in the nuclear space between the chromatin domains [127, 136–141]. The low molecular weight FGF-218 isoform does not bind to SMN but translocates to the nucleolus, whereas FGF-223 also localizes to chromatin [81]
Another protein that is symmetrical methylated by PRMT5 and thereby affected in its affinity to SMN in a similar manner as FGF-223 is the Cajal body protein coilin [107]. Cajal bodies (CBs) are nuclear structures involved in the maturation of snRNPs, a relevant step in spliceosome assembly [98, 108, 109]. Furthermore, SMN is concentrated in a second nuclear compartment called Gemini of CBs (gem) that could have a role in RNA metabolism [110]. Both of these SMN containing nuclear bodies are sensitive for expression of FGF-223 [111, 112]. However, there are even more binding partners and proposed specific roles of different FGF-2 isoforms, in particular inside the cell nucleus.
Nuclear FGF-2 and beyond—the interaction partners
Besides the canonical signaling via cell surface receptors and signaling cascades, there is an increasing number of intracellular and intranuclear FGF-2 functions that have been recently discovered. In the nucleus, FGF-218 localizes to nucleoli of NIH 3T3 cells [79] and additionally to Cajal bodies in rat immortalized Schwann cells [81]. In contrast, HMW-FGF-2 shows a chromatin-associated localization pattern accompanied by chromatin condensation in cell lines as well as in primary cells indicating different nuclear functions of individual FGF-2 isoforms [79, 81, 82, 113].
Intracellular FGF-2 specifically interacts with different nuclear proteins providing potential involvements in multifaceted nuclear regulations. Depending on the respective FGF-2 binding domain involved, the interactions can be isoform-specific or include all isoforms. The interaction of FGF-2 with the splicing factor SF3a66 depends on the 18 kDa core sequence comprised by all FGF-2 isoforms, since a direct interaction can be shown for both FGF-223 and FGF-218 [114]. Isoform-specific binding was found for the binding to the ribosomal protein L6/TAXREB107, bearing at least one binding site for all FGF-2 isoforms but an additional binding site for HMW-FGF-2 only, strongly increasing the affinity of the larger isoforms to the ribosomal protein [115]. This is similar to high affinity interaction of HMW-FGF-2 with fibroblast growth factor-2 (FGF-2)-interacting factor (FIF). For FIF interactions, FGF-218 was not detectable after co-precipitation with FIF antibody [116] indicating a dependence of the N-terminal extension of HMW-FGF-2 for binding to FIF. FIF is also called apoptosis inhibitor-5 or antiapoptotic protein 5 (Api5) and is found to be upregulated together with HMW-FGF-2 in peripheral blood mononuclear cells (PBMCs) of patients with B cell chronic lymphoid leukemia (B-CLL) supposing a possible role of the binding partners in the pathology of the disease [117].
FGF-223 but not FGF-218 binds to the survival of motoneuron (SMN) protein in the nucleus with the arginine-rich N-terminal extension of this HMW isoform [81, 118]. Interestingly, the cellular SMN protein amount is linked to the pathogenesis of the monogenic, neurodegenerative disease spinal muscular atrophy (SMA) [119, 120]. A hallmark of SMA is the progressive degeneration of spinal cord motoneurons caused by disruptions of the Survival of Motor Neuron 1 (Smn1) gene. The selective death of motoneurons is currently still unexplained. The ubiquitously expressed SMN protein functions as an assembly factor for small nuclear ribonuclear protein particles (snRNPs) or small nucleolar RNPs (snoRNPs) involved in splicing [121]. In the nucleus, SMN is found in nuclear foci denoted as gemini of Cajal bodies (gems) which partly colocalize with Cajal bodies (CBs). Some SMN mutations identified in SMA patients disrupt in vitro the binding of SMN to Sm proteins, which are components of snRNPs. However, no splicing defects have been found in presymptomatic stages which are linked to motoneuron death [122].
FGF-223 associates with spliceosomal subunits called small nuclear ribonucleoproteins (snRNPs) without interfering with the interaction of SMN and small nuclear RNAs (snRNAs) within the snRNPs [118] (Fig. 2). The binding of FGF-223 to SMN has been mapped to the first 90 amino acid residues of N-terminus of SMN not competing with Sm protein binding to the Tudor domain [118]. However, SMN binding to nuclear proteins like gemin2 and coilin is severely affected by FGF-223 causing a nuclear phenotype including alterations in nuclear body formation. Expression of FGF-223 leads to a disruption of nuclear gems due to a competition of the growth factor with Gemin2 for binding to SMN [111]. Down-regulation of Gemin2 causes a similar phenotype showing that Gemin2 and the interaction with SMN are necessary for nuclear gem stabilization. FGF-223 can compete with this interaction as demonstrated in vitro using a cell culture model and in vivo using FGF-2 transgenic mice [89] (Fig. 2). The loss of nuclear gems has also been observed in fibroblasts derived from patients suffering from spinal muscular atrophy [123]. Interestingly, the total number of coilin positive Cajal bodies (CBs) is not affected by FGF-223 [111], but the function of CBs is as well affected by the growth factor. Coilin is a marker protein for Cajal bodies and a direct interaction partner of SMN [124–126]. The interaction of SMN and coilin is diminished by FGF-223 leading to a mobilization of the SMN protein from an immobile into a fast mobile fraction and a depletion of SMN from Cajal bodies [112]. SMN-coilin binding is an important requirement for the formation of tri-snRNPs in CBs as a crucial step of the spliceosome cycle resulting in spliceosome formation [127]. FGF-223 expression leads to inhibited tri-snRNP assembly at CBs indicated by a U4 snRNP accumulation at the nuclear complex [112] (Fig. 3). These results indicate a putative role of FGF-2 as a disease modifier of SMA. In a Drosophila model of SMA, activation of muscle-specific FGF signaling is sufficient to rescue neuromuscular junction (NMJ) defects [128], indicating a function link between FGF-signaling and SMA pathology. Furthermore, the expressional level of FGF-2 mRNA is increased in a mouse model of SMA at disease onset [36]. In a cellular model of SMA in PC12 cells, coexpression of FGF-223 antagonizes the SMN promoting effect on neurite outgrowth [129], indicating a negative regulatory effect of HMW-FGF-2 on SMA pathology. FGF-2 loss could, therefore, possibly attenuate the SMA phenotype. Similarly, FGF-2 deficiency in a mouse model for the neurodegenerative disease amyotrophic lateral sclerosis (ALS) resulted in a milder phenotype [130].
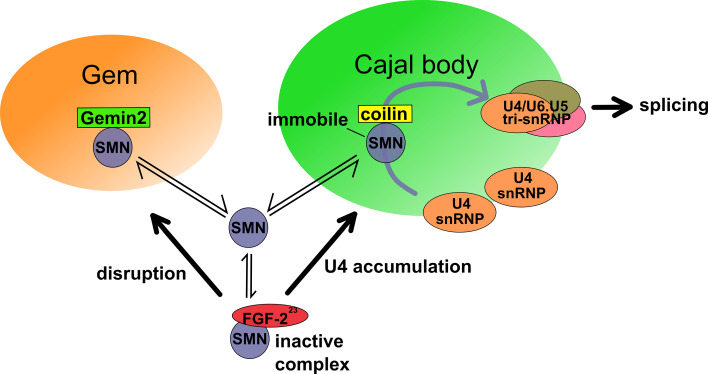
Fig. 3.
FGF-223 expression causes nuclear gem disruption and U4 snRNP accumulation at Cajal Bodies. Binding of FGF-223 to SMN [81, 118] interferes with the binding of SMN to Gemin2 resulting in a disruption of nuclear gems [111]. Moreover, FGF-223 competes with coilin for binding to SMN causing U4 snRNP accumulation in Cajal bodies [112]. The SMN/FGF-223 interaction results in an inactivating protein complex disrupting the normal functions of Coilin and Gemin2, thereby putatively modifying nuclear splicing patterns [129]
Nuclear HMW-FGF-2 induces chromatin compaction and cell death with participation of the extracellular signal-regulated kinase 1 and 2 (ERK1/2) and a mitochondrial cell death pathway [82]. Due to the high overlap of HMW-FGF-2 with DAPI stainings [81, 82], the high molecular FGF-2 isoforms can be suggested to be involved in the regulation of gene expression. Interestingly, also FGFR1 localizes to the nucleus and is part of a chromatin bound complex with CREB-binding protein (CBP) [131, 132]. Nuclear FGF-223 co-localizes with FGFR1 and is suggested to be part of the chromatin bound complex with CBP and, therefore, to be directly involved in the regulation of gene transcription, in contrast to FGF-218 that seems not to affect this mechanism [133]. Accordingly, FGF-2 is assumed to participate in a pathway denoted as Integrative Nuclear FGFR1 Signaling (INFS) controlling growth, proliferation and differentiation particularly in the nervous system by regulating gene transcription [104, 134].
Indeed, many genes are regulated in isoform-specific processes as revealed by comparing transcriptomes of HMW- with LMW-FGF-2 expressing cells, albeit in non-neuronal NIH 3T3 fibroblasts. These mechanisms include up-regulation of genes involved in growth inhibition, tumor suppression, development as well as differentiation. In addition, down-regulation of genes involved in cell proliferation, mitogenic signaling and ribosome biogenesis has been observed [78]. HMW-FGF-2 in a complex with FGFR1 is suggested to directly up-regulate FGF23 expression in bone marrow stromal cells (BMSCs) [75]. However, isoform-dependent regulation of mRNAs in neuronal systems has also been described. Gene transcription dependent on the neuronal transcription factor Nurr-1 is amplified by FGF-223 in human neuroblastoma cells [129, 135].
Outlook
The analysis of the functions of nuclear FGF-2 still remains a challenge. On the one hand, there is an increasing number of different and partly antagonistic cellular functions regulated by high and low molecular FGF-2 isoforms. The distribution of the growth factor isoforms inside the cell, the binding to proteins and the involvement in cellular processes vary depending on the length of the N-terminal extensions. On the other hand, there are specific FGF-2 functions in neuronal systems like differentiation and outgrowth of long processes that are enhanced by FGF-218 but inhibited by FGF-223. By now, it is poorly understood how molecular complexes regulated by FGF-2 isoforms are directly engaged in these processes and—also beyond the nervous system—are involved in tissue-specific functions. A model to combine both could be that differential binding affinities and specificities of low and high molecular weight FGF-2 to other nuclear proteins involved in splicing and regulation of transcription like SMN and FGFR1 could result in altered splicing and transcription of FGF-2 dependent genes. As a consequence, differential regulation of proteins could result in tissue-specific effects. Furthermore, the binding of FGF-223 to the methyl transferase PRMT5 could affect the methylation of other PRMT5 targets and, therefore, directly influence protein functions with tissue dependent effects. To further analyze nuclear FGF-2 functions, it could be helpful to screen different tissues for alterations in splicing, transcription and methylation after expression of FGF-2 isoforms to better understand the connection between cellular differences and tissue-specific functions. Another interesting aspect is the positive effect of HMW-FGF-2 expressing transplanted cells on the survival, restoration and reinnervation of dopaminergic micrografts or, in the PNS, the support of nerve recovery by HMW-FGF-2 glial cells. Taken together, it is an ambitious and promising challenge to bring together isoform-specific FGF-2 functions inside the nucleus and their impact on cellular and organ function.
Acknowledgments
This work was supported by the Niedersachsen‐Research Network on Neuroinfectiology (N‐RENNT) of the Ministry of Science and Culture of Lower Saxony.
Abbreviations
- CB
Cajal body
- SMN
Survival of motoneuron protein
- SMA
Spinal muscular atrophy
- FGF
Fibroblast growth factor
- snRNP
Small nuclear ribonucleoprotein particle
References
- 1.Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2011;2:reviews3005–reviews3005.12. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004;20:563–569. doi: 10.1016/j.tig.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Gospodarowicz D, Jones KL, Sato G. Purification of a growth factor for ovarian cells from bovine pituitary glands. Proc Natl Acad Sci USA. 1974;71:2295–2299. doi: 10.1073/pnas.71.6.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armelin HA. Pituitary extracts and steroid hormones in the control of 3T3 cell growth. Proc Natl Acad Sci USA. 1973;70:2702–2706. doi: 10.1073/pnas.70.9.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gambarini AG, Armelin HA. Purification and partial characterization of an acidic fibroblast growth factor from bovine pituitary. J Biol Chem. 1982;257:9692–9697. [PubMed] [Google Scholar]
- 6.Lobb RR, Harper JW, Fett JW. Purification of heparin-binding growth factors. Anal Biochem. 1986;154:1–14. doi: 10.1016/0003-2697(86)90487-2. [DOI] [PubMed] [Google Scholar]
- 7.Baird A, Klagsbrun M. The fibroblast growth factor family. Cancer Cells. 1991;3:239–243. [PubMed] [Google Scholar]
- 8.Moscatelli D, Joseph-Silverstein J, Manejias R, Rifkin DB. Mr 25,000 heparin-binding protein from guinea pig brain is a high molecular weight form of basic fibroblast growth factor. Proc Natl Acad Sci USA. 1987;84:5778–5782. doi: 10.1073/pnas.84.16.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Presta M, Rusnati M, Maier JA, Ragnotti G. Purification of basic fibroblast growth factor from rat brain: identification of a Mr 22,000 immunoreactive form. Biochem Biophys Res Commun. 1988;155:1161–1172. doi: 10.1016/s0006-291x(88)81262-2. [DOI] [PubMed] [Google Scholar]
- 10.Shing Y, Folkman J, Sullivan R, Butterfield C, Murray J, Klagsbrun M. Heparin affinity: purification of a tumor-derived capillary endothelial cell growth factor. Science. 1984;223:1296–1299. doi: 10.1126/science.6199844. [DOI] [PubMed] [Google Scholar]
- 11.Arnaud E, Touriol C, Boutonnet C, Gensac MC, Vagner S, Prats H, Prats AC. A new 34-kilodalton isoform of human fibroblast growth factor 2 is cap dependently synthesized by using a non-AUG start codon and behaves as a survival factor. Mol Cell Biol. 1999;19:505–514. doi: 10.1128/mcb.19.1.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Florkiewicz RZ, Sommer A. Human basic fibroblast growth factor gene encodes four polypeptides: three initiate translation from non-AUG codons. Proc Natl Acad Sci USA. 1989;86:3978–3981. doi: 10.1073/pnas.86.11.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prats H, Kaghad M, Prats AC, Klagsbrun M, Lelias JM, Liauzun P, Chalon P, Tauber JP, Amalric F, Smith JA, et al. High molecular mass forms of basic fibroblast growth factor are initiated by alternative CUG codons. Proc Natl Acad Sci USA. 1989;86:1836–1840. doi: 10.1073/pnas.86.6.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorensen V, Nilsen T, Wiedlocha A. Functional diversity of FGF-2 isoforms by intracellular sorting. BioEssays. 2006;28:504–514. doi: 10.1002/bies.20405. [DOI] [PubMed] [Google Scholar]
- 15.Chlebova K, Bryja V, Dvorak P, Kozubik A, Wilcox WR, Krejci P. High molecular weight FGF2: the biology of a nuclear growth factor. Cell Mol Life Sci. 2009;66:225–235. doi: 10.1007/s00018-008-8440-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vagner S, Gensac MC, Maret A, Bayard F, Amalric F, Prats H, Prats AC. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Audigier S, Guiramand J, Prado-Lourenco L, Conte C, Gonzalez-Herrera IG, Cohen-Solal C, Recasens M, Prats AC. Potent activation of FGF-2 IRES-dependent mechanism of translation during brain development. RNA. 2008;14:1852–1864. doi: 10.1261/rna.790608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Touriol C, Morillon A, Gensac MC, Prats H, Prats AC. Expression of human fibroblast growth factor 2 mRNA is post-transcriptionally controlled by a unique destabilizing element present in the 3′-untranslated region between alternative polyadenylation sites. J Biol Chem. 1999;274:21402–21408. doi: 10.1074/jbc.274.30.21402. [DOI] [PubMed] [Google Scholar]
- 19.Touriol C, Roussigne M, Gensac MC, Prats H, Prats AC. Alternative translation initiation of human fibroblast growth factor 2 mRNA controlled by its 3′-untranslated region involves a Poly(A) switch and a translational enhancer. J Biol Chem. 2000;275:19361–19367. doi: 10.1074/jbc.M908431199. [DOI] [PubMed] [Google Scholar]
- 20.Florkiewicz RZ, Majack RA, Buechler RD, Florkiewicz E. Quantitative export of FGF-2 occurs through an alternative, energy-dependent, non-ER/Golgi pathway. J Cell Physiol. 1995;162:388–399. doi: 10.1002/jcp.1041620311. [DOI] [PubMed] [Google Scholar]
- 21.Mignatti P, Morimoto T, Rifkin DB. Basic fibroblast growth factor, a protein devoid of secretory signal sequence, is released by cells via a pathway independent of the endoplasmic reticulum–Golgi complex. J Cell Physiol. 1992;151:81–93. doi: 10.1002/jcp.1041510113. [DOI] [PubMed] [Google Scholar]
- 22.Temmerman K, Ebert AD, Muller HM, Sinning I, Tews I, Nickel W. A direct role for phosphatidylinositol-4,5-bisphosphate in unconventional secretion of fibroblast growth factor 2. Traffic. 2008;9:1204–1217. doi: 10.1111/j.1600-0854.2008.00749.x. [DOI] [PubMed] [Google Scholar]
- 23.Steringer JP, Bleicken S, Andreas H, Zacherl S, Laussmann M, Temmerman K, Contreras FX, Bharat TA, Lechner J, Muller HM, Briggs JA, Garcia-Saez AJ, Nickel W. Phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2)-dependent oligomerization of fibroblast growth factor 2 (FGF2) triggers the formation of a lipidic membrane pore implicated in unconventional secretion. J Biol Chem. 2012;287:27659–27669. doi: 10.1074/jbc.M112.381939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steringer JP, Muller HM, Nickel W. Unconventional secretion of fibroblast growth factor 2—a novel type of protein translocation across membranes? J Mol Biol. 2014 doi: 10.1016/j.jmb.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 25.Santiago JJ, Dangerfield AL, Rattan SG, Bathe KL, Cunnington RH, Raizman JE, Bedosky KM, Freed DH, Kardami E, Dixon IM. Cardiac fibroblast to myofibroblast differentiation in vivo and in vitro: expression of focal adhesion components in neonatal and adult rat ventricular myofibroblasts. Dev Dyn. 2010;239:1573–1584. doi: 10.1002/dvdy.22280. [DOI] [PubMed] [Google Scholar]
- 26.Jiang ZS, Jeyaraman M, Wen GB, Fandrich RR, Dixon IM, Cattini PA, Kardami E. High- but not low-molecular weight FGF-2 causes cardiac hypertrophy in vivo; possible involvement of cardiotrophin-1. J Mol Cell Cardiol. 2007;42:222–233. doi: 10.1016/j.yjmcc.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Santiago JJ, McNaughton LJ, Koleini N, Ma X, Bestvater B, Nickel BE, Fandrich RR, Wigle JT, Freed DH, Arora RC, Kardami E. High molecular weight fibroblast growth factor-2 in the human heart is a potential target for prevention of cardiac remodeling. PLoS One. 2014;9:e97281. doi: 10.1371/journal.pone.0097281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sensenbrenner M. The neurotrophic activity of fibroblast growth factors. Prog Neurobiol. 1993;41:683–704. doi: 10.1016/0301-0082(93)90031-m. [DOI] [PubMed] [Google Scholar]
- 29.Barde YA. Trophic factors and neuronal survival. Neuron. 1989;2:1525–1534. doi: 10.1016/0896-6273(89)90040-8. [DOI] [PubMed] [Google Scholar]
- 30.Grothe C, Unsicker K. Basic fibroblast growth factor in the hypoglossal system: specific retrograde transport, trophic, and lesion-related responses. J Neurosci Res. 1992;32:317–328. doi: 10.1002/jnr.490320304. [DOI] [PubMed] [Google Scholar]
- 31.Stoeckel K, Schwab M, Thoenen H. Specificity of retrograde transport of nerve growth factor (NGF) in sensory neurons: a biochemical and morphological study. Brain Res. 1975;89:1–14. doi: 10.1016/0006-8993(75)90129-8. [DOI] [PubMed] [Google Scholar]
- 32.Johnson EM, Jr, Taniuchi M, Clark HB, Springer JE, Koh S, Tayrien MW, Loy R. Demonstration of the retrograde transport of nerve growth factor receptor in the peripheral and central nervous system. J Neurosci. 1987;7:923–929. doi: 10.1523/JNEUROSCI.07-03-00923.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barde YA. What, if anything, is a neurotrophic factor? Trends Neurosci. 1988;11:343–346. doi: 10.1016/0166-2236(88)90055-0. [DOI] [PubMed] [Google Scholar]
- 34.Giordano S, Sherman L, Lyman W, Morrison R. Multiple molecular weight forms of basic fibroblast growth factor are developmentally regulated in the central nervous system. Dev Biol. 1992;152:293–303. doi: 10.1016/0012-1606(92)90136-5. [DOI] [PubMed] [Google Scholar]
- 35.Ratzka A, Baron O, Grothe C. FGF-2 deficiency does not influence FGF ligand and receptor expression during development of the nigrostriatal system. PLoS One. 2011;6:e23564. doi: 10.1371/journal.pone.0023564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hensel N, Ratzka A, Brinkmann H, Klimaschewski L, Grothe C, Claus P. Analysis of the fibroblast growth factor system reveals alterations in a mouse model of spinal muscular atrophy. PLoS One. 2012;7:e31202. doi: 10.1371/journal.pone.0031202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Claus P, Werner S, Timmer M, Grothe C. Expression of the fibroblast growth factor-2 isoforms and the FGF receptor 1–4 transcripts in the rat model system of Parkinson’s disease. Neurosci Lett. 2004;360:117–120. doi: 10.1016/j.neulet.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 38.Date I, Yoshimoto Y, Imaoka T, Miyoshi Y, Gohda Y, Furuta T, Asari S, Ohmoto T. Enhanced recovery of the nigrostriatal dopaminergic system in MPTP-treated mice following intrastriatal injection of basic fibroblast growth factor in relation to aging. Brain Res. 1993;621:150–154. doi: 10.1016/0006-8993(93)90312-b. [DOI] [PubMed] [Google Scholar]
- 39.Otto D, Unsicker K. Basic FGF reverses chemical and morphological deficits in the nigrostriatal system of MPTP-treated mice. J Neurosci. 1990;10:1912–1921. doi: 10.1523/JNEUROSCI.10-06-01912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Timmer M, Cesnulevicius K, Winkler C, Kolb J, Lipokatic-Takacs E, Jungnickel J, Grothe C. Fibroblast growth factor (FGF)-2 and FGF receptor 3 are required for the development of the substantia nigra, and FGF-2 plays a crucial role for the rescue of dopaminergic neurons after 6-hydroxydopamine lesion. J Neurosci. 2007;27:459–471. doi: 10.1523/JNEUROSCI.4493-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grothe C, Schulze A, Semkova I, Muller-Ostermeyer F, Rege A, Wewetzer K. The high molecular weight fibroblast growth factor-2 isoforms (21,000 mol. wt and 23,000 mol. wt) mediate neurotrophic activity on rat embryonic mesencephalic dopaminergic neurons in vitro. Neuroscience. 2000;100:73–86. doi: 10.1016/s0306-4522(00)00247-5. [DOI] [PubMed] [Google Scholar]
- 42.Timmer M, Muller-Ostermeyer F, Kloth V, Winkler C, Grothe C, Nikkhah G. Enhanced survival, reinnervation, and functional recovery of intrastriatal dopamine grafts co-transplanted with Schwann cells overexpressing high molecular weight FGF-2 isoforms. Exp Neurol. 2004;187:118–136. doi: 10.1016/j.expneurol.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 43.Bansal R, Pfeiffer SE. FGF-2 converts mature oligodendrocytes to a novel phenotype. J Neurosci Res. 1997;50:215–228. doi: 10.1002/(SICI)1097-4547(19971015)50:2<215::AID-JNR10>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 44.Goddard DR, Berry M, Kirvell SL, Butt AM. Fibroblast growth factor-2 inhibits myelin production by oligodendrocytes in vivo. Mol Cell Neurosci. 2001;18:557–569. doi: 10.1006/mcne.2001.1025. [DOI] [PubMed] [Google Scholar]
- 45.Armstrong RC, Le TQ, Frost EE, Borke RC, Vana AC. Absence of fibroblast growth factor 2 promotes oligodendroglial repopulation of demyelinated white matter. J Neurosci. 2002;22:8574–8585. doi: 10.1523/JNEUROSCI.22-19-08574.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nindl W, Kavakebi P, Claus P, Grothe C, Pfaller K, Klimaschewski L. Expression of basic fibroblast growth factor isoforms in postmitotic sympathetic neurons: synthesis, intracellular localization and involvement in karyokinesis. Neuroscience. 2004;124:561–572. doi: 10.1016/j.neuroscience.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 47.Pasumarthi KB, Kardami E, Cattini PA. High and low molecular weight fibroblast growth factor-2 increase proliferation of neonatal rat cardiac myocytes but have differential effects on binucleation and nuclear morphology. Evidence for both paracrine and intracrine actions of fibroblast growth factor-2. Circ Res. 1996;78:126–136. doi: 10.1161/01.res.78.1.126. [DOI] [PubMed] [Google Scholar]
- 48.Bachis A, Mallei A, Cruz MI, Wellstein A, Mocchetti I. Chronic antidepressant treatments increase basic fibroblast growth factor and fibroblast growth factor-binding protein in neurons. Neuropharmacology. 2008;55:1114–1120. doi: 10.1016/j.neuropharm.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kajitani N, Hisaoka-Nakashima K, Morioka N, Okada-Tsuchioka M, Kaneko M, Kasai M, Shibasaki C, Nakata Y, Takebayashi M. Antidepressant acts on astrocytes leading to an increase in the expression of neurotrophic/growth factors: differential regulation of FGF-2 by noradrenaline. PLoS One. 2012;7:e51197. doi: 10.1371/journal.pone.0051197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ji RR, Zhang Q, Zhang X, Piehl F, Reilly T, Pettersson RF, Hokfelt T. Prominent expression of bFGF in dorsal root ganglia after axotomy. Eur J Neurosci. 1995;7:2458–2468. doi: 10.1111/j.1460-9568.1995.tb01044.x. [DOI] [PubMed] [Google Scholar]
- 51.Li GD, Wo Y, Zhong MF, Zhang FX, Bao L, Lu YJ, Huang YD, Xiao HS, Zhang X. Expression of fibroblast growth factors in rat dorsal root ganglion neurons and regulation after peripheral nerve injury. NeuroReport. 2002;13:1903–1907. doi: 10.1097/00001756-200210280-00014. [DOI] [PubMed] [Google Scholar]
- 52.Huber K, Meisinger C, Grothe C. Expression of fibroblast growth factor-2 in hypoglossal motoneurons is stimulated by peripheral nerve injury. J Comp Neurol. 1997;382:189–198. [PubMed] [Google Scholar]
- 53.Klimaschewski L, Meisinger C, Grothe C. Localization and regulation of basic fibroblast growth factor (FGF-2) and FGF receptor-1 in rat superior cervical ganglion after axotomy. J Neurobiol. 1999;38:499–506. doi: 10.1002/(sici)1097-4695(199903)38:4<499::aid-neu6>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 54.Danielsen N, Pettmann B, Vahlsing HL, Manthorpe M, Varon S. Fibroblast growth factor effects on peripheral nerve regeneration in a silicone chamber model. J Neurosci Res. 1988;20:320–330. doi: 10.1002/jnr.490200306. [DOI] [PubMed] [Google Scholar]
- 55.Aebischer P, Salessiotis AN, Winn SR. Basic fibroblast growth factor released from synthetic guidance channels facilitates peripheral nerve regeneration across long nerve gaps. J Neurosci Res. 1989;23:282–289. doi: 10.1002/jnr.490230306. [DOI] [PubMed] [Google Scholar]
- 56.Fujimoto E, Mizoguchi A, Hanada K, Yajima M, Ide C. Basic fibroblast growth factor promotes extension of regenerating axons of peripheral nerve. In vivo experiments using a Schwann cell basal lamina tube model. J Neurocytol. 1997;26:511–528. doi: 10.1023/a:1015410023132. [DOI] [PubMed] [Google Scholar]
- 57.Timmer M, Robben S, Muller-Ostermeyer F, Nikkhah G, Grothe C. Axonal regeneration across long gaps in silicone chambers filled with Schwann cells overexpressing high molecular weight FGF-2. Cell Transpl. 2003;12:265–277. doi: 10.3727/000000003108746821. [DOI] [PubMed] [Google Scholar]
- 58.Grothe C, Meisinger C, Hertenstein A, Kurz H, Wewetzer K. Expression of fibroblast growth factor-2 and fibroblast growth factor receptor 1 messenger RNAs in spinal ganglia and sciatic nerve: regulation after peripheral nerve lesion. Neuroscience. 1997;76:123–135. doi: 10.1016/s0306-4522(96)00355-7. [DOI] [PubMed] [Google Scholar]
- 59.Meisinger C, Grothe C. Differential regulation of fibroblast growth factor (FGF)-2 and FGF receptor 1 mRNAs and FGF-2 isoforms in spinal ganglia and sciatic nerve after peripheral nerve lesion. J Neurochem. 1997;68:1150–1158. doi: 10.1046/j.1471-4159.1997.68031150.x. [DOI] [PubMed] [Google Scholar]
- 60.Grothe C, Nikkhah G. The role of basic fibroblast growth factor in peripheral nerve regeneration. Anat Embryol (Berl) 2001;204:171–177. doi: 10.1007/s004290100205. [DOI] [PubMed] [Google Scholar]
- 61.Jungnickel J, Claus P, Gransalke K, Timmer M, Grothe C. Targeted disruption of the FGF-2 gene affects the response to peripheral nerve injury. Mol Cell Neurosci. 2004;25:444–452. doi: 10.1016/j.mcn.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 62.Haastert K, Lipokatic E, Fischer M, Timmer M, Grothe C. Differentially promoted peripheral nerve regeneration by grafted Schwann cells over-expressing different FGF-2 isoforms. Neurobiol Dis. 2006;21:138–153. doi: 10.1016/j.nbd.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 63.Haastert K, Grosheva M, Angelova SK, Guntinas-Lichius O, Skouras E, Michael J, Grothe C, Dunlop SA, Angelov DN. Schwann cells overexpressing FGF-2 alone or combined with manual stimulation do not promote functional recovery after facial nerve injury. J Biomed Biotechnol. 2009;2009:408794. doi: 10.1155/2009/408794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meisinger C, Grothe C. Differential expression of FGF-2 isoforms in the rat adrenal medulla during postnatal development in vivo. Brain Res. 1997;757:291–294. doi: 10.1016/s0006-8993(97)00341-7. [DOI] [PubMed] [Google Scholar]
- 65.Seidl K, Unsicker K. Survival and neuritic growth of sympathoadrenal (chromaffin) precursor cells in vitro. Int J Dev Neurosci. 1989;7:465–473. doi: 10.1016/0736-5748(89)90006-3. [DOI] [PubMed] [Google Scholar]
- 66.Meisinger C, Zeschnigk C, Grothe C. In vivo and in vitro effect of glucocorticoids on fibroblast growth factor (FGF)-2 and FGF receptor 1 expression. J Biol Chem. 1996;271:16520–16525. doi: 10.1074/jbc.271.28.16520. [DOI] [PubMed] [Google Scholar]
- 67.Grothe C, Meisinger C. The multifunctionality of FGF-2 in the adrenal medulla. Anat Embryol (Berl) 1997;195:103–111. doi: 10.1007/s004290050029. [DOI] [PubMed] [Google Scholar]
- 68.Grothe C, Meisinger C, Holzschuh J, Wewetzer K, Cattini P. Over-expression of the 18 kD and 21/23 kD fibroblast growth factor-2 isoforms in PC12 cells and Schwann cells results in altered cell morphology and growth. Brain Res Mol Brain Res. 1998;57:97–105. doi: 10.1016/s0169-328x(98)00076-x. [DOI] [PubMed] [Google Scholar]
- 69.Davis JB, Stroobant P. Platelet-derived growth factors and fibroblast growth factors are mitogens for rat Schwann cells. J Cell Biol. 1990;110:1353–1360. doi: 10.1083/jcb.110.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Watabe K, Fukuda T, Tanaka J, Toyohara K, Sakai O. Mitogenic effects of platelet-derived growth factor, fibroblast growth factor, transforming growth factor-beta, and heparin-binding serum factor for adult mouse Schwann cells. J Neurosci Res. 1994;39:525–534. doi: 10.1002/jnr.490390504. [DOI] [PubMed] [Google Scholar]
- 71.Dong Z, Sinanan A, Parkinson D, Parmantier E, Mirsky R, Jessen KR. Schwann cell development in embryonic mouse nerves. J Neurosci Res. 1999;56:334–348. doi: 10.1002/(SICI)1097-4547(19990515)56:4<334::AID-JNR2>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 72.Hansen MR, Vijapurkar U, Koland JG, Green SH. Reciprocal signaling between spiral ganglion neurons and Schwann cells involves neuregulin and neurotrophins. Hear Res. 2001;161:87–98. doi: 10.1016/s0378-5955(01)00360-4. [DOI] [PubMed] [Google Scholar]
- 73.Kardami E, Detillieux K, Ma X, Jiang Z, Santiago JJ, Jimenez SK, Cattini PA. Fibroblast growth factor-2 and cardioprotection. Heart Fail Rev. 2007;12:267–277. doi: 10.1007/s10741-007-9027-0. [DOI] [PubMed] [Google Scholar]
- 74.Sabbieti MG, Agas D, Marchetti L, Coffin JD, Xiao L, Hurley MM. BMP-2 differentially modulates FGF-2 isoform effects in osteoblasts from newborn transgenic mice. Endocrinology. 2013;154:2723–2733. doi: 10.1210/en.2013-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xiao L, Esliger A, Hurley MM. Nuclear fibroblast growth factor 2 (FGF2) isoforms inhibit bone marrow stromal cell mineralization through FGF23/FGFR/MAPK in vitro. J Bone Miner Res. 2013;28:35–45. doi: 10.1002/jbmr.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiao L, Ueno D, Catros S, Homer-Bouthiette C, Charles L, Kuhn L, Hurley MM. Fibroblast growth factor-2 isoform (low molecular weight/18 kDa) overexpression in preosteoblast cells promotes bone regeneration in critical size calvarial defects in male mice. Endocrinology. 2014;155:965–974. doi: 10.1210/en.2013-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quarto N, Talarico D, Florkiewicz R, Rifkin DB. Selective expression of high molecular weight basic fibroblast growth factor confers a unique phenotype to NIH 3T3 cells. Cell Regul. 1991;2:699–708. doi: 10.1091/mbc.2.9.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quarto N, Fong KD, Longaker MT. Gene profiling of cells expressing different FGF-2 forms. Gene. 2005;356:49–68. doi: 10.1016/j.gene.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 79.Arese M, Chen Y, Florkiewicz RZ, Gualandris A, Shen B, Rifkin DB. Nuclear activities of basic fibroblast growth factor: potentiation of low-serum growth mediated by natural or chimeric nuclear localization signals. Mol Biol Cell. 1999;10:1429–1444. doi: 10.1091/mbc.10.5.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pasumarthi KB, Doble BW, Kardami E, Cattini PA. Over-expression of CUG- or AUG-initiated forms of basic fibroblast growth factor in cardiac myocytes results in similar effects on mitosis and protein synthesis but distinct nuclear morphologies. J Mol Cell Cardiol. 1994;26:1045–1060. doi: 10.1006/jmcc.1994.1125. [DOI] [PubMed] [Google Scholar]
- 81.Claus P, Doring F, Gringel S, Muller-Ostermeyer F, Fuhlrott J, Kraft T, Grothe C. Differential intranuclear localization of fibroblast growth factor-2 isoforms and specific interaction with the survival of motoneuron protein. J Biol Chem. 2003;278:479–485. doi: 10.1074/jbc.M206056200. [DOI] [PubMed] [Google Scholar]
- 82.Ma X, Dang X, Claus P, Hirst C, Fandrich RR, Jin Y, Grothe C, Kirshenbaum LA, Cattini PA, Kardami E. Chromatin compaction and cell death by high molecular weight FGF-2 depend on its nuclear localization, intracrine ERK activation, and engagement of mitochondria. J Cell Physiol. 2007;213:690–698. doi: 10.1002/jcp.21139. [DOI] [PubMed] [Google Scholar]
- 83.Sheng Z, Lewis JA, Chirico WJ. Nuclear and nucleolar localization of 18-kDa fibroblast growth factor-2 is controlled by C-terminal signals. J Biol Chem. 2004;279:40153–40160. doi: 10.1074/jbc.M400123200. [DOI] [PubMed] [Google Scholar]
- 84.Bugler B, Amalric F, Prats H. Alternative initiation of translation determines cytoplasmic or nuclear localization of basic fibroblast growth factor. Mol Cell Biol. 1991;11:573–577. doi: 10.1128/mcb.11.1.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Quarto N, Finger FP, Rifkin DB. The NH2-terminal extension of high molecular weight bFGF is a nuclear targeting signal. J Cell Physiol. 1991;147:311–318. doi: 10.1002/jcp.1041470217. [DOI] [PubMed] [Google Scholar]
- 86.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 87.Boulikas T. Nuclear localization signals (NLS) Crit Rev Eukaryot Gene Expr. 1993;3:193–227. [PubMed] [Google Scholar]
- 88.Dono R, James D, Zeller R. A GR-motif functions in nuclear accumulation of the large FGF-2 isoforms and interferes with mitogenic signalling. Oncogene. 1998;16:2151–2158. doi: 10.1038/sj.onc.1201746. [DOI] [PubMed] [Google Scholar]
- 89.Bruns AF, Grothe C, Claus P. Fibroblast growth factor 2 (FGF-2) is a novel substrate for arginine methylation by PRMT5. Biol Chem. 2009;390:59–65. doi: 10.1515/BC.2009.001. [DOI] [PubMed] [Google Scholar]
- 90.Claus P, Schulze E, Wisniewski JR. Insect proteins homologous to mammalian high mobility group proteins I/Y (HMG I/Y). Characterization and binding to linear and four-way junction DNA. J Biol Chem. 1994;269:33042–33048. [PubMed] [Google Scholar]
- 91.Huth JR, Bewley CA, Nissen MS, Evans JN, Reeves R, Gronenborn AM, Clore GM. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat Struct Biol. 1997;4:657–665. doi: 10.1038/nsb0897-657. [DOI] [PubMed] [Google Scholar]
- 92.Burgess WH, Bizik J, Mehlman T, Quarto N, Rifkin DB. Direct evidence for methylation of arginine residues in high molecular weight forms of basic fibroblast growth factor. Cell Regul. 1991;2:87–93. doi: 10.1091/mbc.2.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pintucci G, Quarto N, Rifkin DB. Methylation of high molecular weight fibroblast growth factor-2 determines post-translational increases in molecular weight and affects its intracellular distribution. Mol Biol Cell. 1996;7:1249–1258. doi: 10.1091/mbc.7.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M, Dreyfuss G. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol. 2001;21:8289–8300. doi: 10.1128/MCB.21.24.8289-8300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell. 2001;7:1111–1117. doi: 10.1016/s1097-2765(01)00244-1. [DOI] [PubMed] [Google Scholar]
- 96.Brahms H, Meheus L, de Brabandere V, Fischer U, Luhrmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B’ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA. 2001;7:1531–1542. doi: 10.1017/s135583820101442x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Meister G, Eggert C, Buhler D, Brahms H, Kambach C, Fischer U. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr Biol. 2001;11:1990–1994. doi: 10.1016/s0960-9822(01)00592-9. [DOI] [PubMed] [Google Scholar]
- 98.Will CL, Luhrmann R. Protein functions in pre-mRNA splicing. Curr Opin Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- 99.Tripsianes K, Madl T, Machyna M, Fessas D, Englbrecht C, Fischer U, Neugebauer KM, Sattler M. Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat Struct Mol Biol. 2011;18:1414–1420. doi: 10.1038/nsmb.2185. [DOI] [PubMed] [Google Scholar]
- 100.Bossard C, Laurell H, Van den Berghe L, Meunier S, Zanibellato C, Prats H. Translokin is an intracellular mediator of FGF-2 trafficking. Nat Cell Biol. 2003;5:433–439. doi: 10.1038/ncb979. [DOI] [PubMed] [Google Scholar]
- 101.Maher PA. Nuclear translocation of fibroblast growth factor (FGF) receptors in response to FGF-2. J Cell Biol. 1996;134:529–536. doi: 10.1083/jcb.134.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Reilly JF, Maher PA. Importin beta-mediated nuclear import of fibroblast growth factor receptor: role in cell proliferation. J Cell Biol. 2001;152:1307–1312. doi: 10.1083/jcb.152.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Johnson HM, Subramaniam PS, Olsnes S, Jans DA. Trafficking and signaling pathways of nuclear localizing protein ligands and their receptors. BioEssays. 2004;26:993–1004. doi: 10.1002/bies.20086. [DOI] [PubMed] [Google Scholar]
- 104.Stachowiak MK, Fang X, Myers JM, Dunham SM, Berezney R, Maher PA, Stachowiak EK. Integrative nuclear FGFR1 signaling (INFS) as a part of a universal “feed-forward-and-gate” signaling module that controls cell growth and differentiation. J Cell Biochem. 2003;90:662–691. doi: 10.1002/jcb.10606. [DOI] [PubMed] [Google Scholar]
- 105.Coleman SJ, Chioni AM, Ghallab M, Anderson RK, Lemoine NR, Kocher HM, Grose RP. Nuclear translocation of FGFR1 and FGF2 in pancreatic stellate cells facilitates pancreatic cancer cell invasion. EMBO Mol Med. 2014;6:467–481. doi: 10.1002/emmm.201302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Marchetti L, Sabbieti MG, Agas D, Menghi M, Materazzi G, Menghi G, Hurley MM. PGF2alpha increases FGF-2 and FGFR2 trafficking in Py1a rat osteoblasts via clathrin independent and importin beta dependent pathway. J Cell Biochem. 2006;97:1379–1392. doi: 10.1002/jcb.20746. [DOI] [PubMed] [Google Scholar]
- 107.Hebert MD, Shpargel KB, Ospina JK, Tucker KE, Matera AG. Coilin methylation regulates nuclear body formation. Dev Cell. 2002;3:329–337. doi: 10.1016/s1534-5807(02)00222-8. [DOI] [PubMed] [Google Scholar]
- 108.Dundr M, Misteli T. Functional architecture in the cell nucleus. Biochem J. 2001;356:297–310. doi: 10.1042/0264-6021:3560297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sleeman JE, Lamond AI. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr Biol. 1999;9:1065–1074. doi: 10.1016/s0960-9822(99)80475-8. [DOI] [PubMed] [Google Scholar]
- 110.Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 111.Bruns AF, van Bergeijk J, Lorbeer C, Nolle A, Jungnickel J, Grothe C, Claus P. Fibroblast growth factor-2 regulates the stability of nuclear bodies. Proc Natl Acad Sci USA. 2009;106:12747–12752. doi: 10.1073/pnas.0900122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Förthmann B, Brinkmann H, Ratzka A, Stachowiak MK, Grothe C, Claus P. Immobile survival of motoneuron (SMN) protein stored in Cajal bodies can be mobilized by protein interactions. Cell Mol Life Sci. 2013;70:2555–2568. doi: 10.1007/s00018-012-1242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sun G, Doble BW, Sun JM, Fandrich RR, Florkiewicz R, Kirshenbaum L, Davie JR, Cattini PA, Kardami E. CUG-initiated FGF-2 induces chromatin compaction in cultured cardiac myocytes and in vitro. J Cell Physiol. 2001;186:457–467. doi: 10.1002/1097-4652(2000)9999:999<000::AID-JCP1044>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 114.Gringel S, van Bergeijk J, Haastert K, Grothe C, Claus P. Nuclear fibroblast growth factor-2 interacts specifically with splicing factor SF3a66. Biol Chem. 2004;385:1203–1208. doi: 10.1515/BC.2004.156. [DOI] [PubMed] [Google Scholar]
- 115.Shen B, Arese M, Gualandris A, Rifkin DB. Intracellular association of FGF-2 with the ribosomal protein L6/TAXREB107. Biochem Biophys Res Commun. 1998;252:524–528. doi: 10.1006/bbrc.1998.9677. [DOI] [PubMed] [Google Scholar]
- 116.Van den Berghe L, Laurell H, Huez I, Zanibellato C, Prats H, Bugler B. FIF [fibroblast growth factor-2 (FGF-2)-interacting-factor], a nuclear putatively antiapoptotic factor, interacts specifically with FGF-2. Mol Endocrinol. 2000;14:1709–1724. doi: 10.1210/mend.14.11.0556. [DOI] [PubMed] [Google Scholar]
- 117.Krejci P, Pejchalova K, Rosenbloom BE, Rosenfelt FP, Tran EL, Laurell H, Wilcox WR. The antiapoptotic protein Api5 and its partner, high molecular weight FGF2, are up-regulated in B cell chronic lymphoid leukemia. J Leukoc Biol. 2007;82:1363–1364. doi: 10.1189/jlb.0607425. [DOI] [PubMed] [Google Scholar]
- 118.Claus P, Bruns AF, Grothe C. Fibroblast growth factor-2(23) binds directly to the survival of motoneuron protein and is associated with small nuclear RNAs. Biochem J. 2004;384:559–565. doi: 10.1042/BJ20040801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McAndrew PE, Parsons DW, Simard LR, Rochette C, Ray PN, Mendell JR, Prior TW, Burghes AH. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Feldkotter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Terns MP, Terns RM. Macromolecular complexes: SMN—the master assembler. Curr Biol. 2001;11:R862–R864. doi: 10.1016/s0960-9822(01)00517-6. [DOI] [PubMed] [Google Scholar]
- 122.Baumer D, Lee S, Nicholson G, Davies JL, Parkinson NJ, Murray LM, Gillingwater TH, Ansorge O, Davies KE, Talbot K. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet. 2009;5:e1000773. doi: 10.1371/journal.pgen.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, Coulson SE, Androphy EJ, Prior TW, Burghes AH. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 124.Andrade LE, Chan EK, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hebert MD, Szymczyk PW, Shpargel KB, Matera AG. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev. 2001;15:2720–2729. doi: 10.1101/gad.908401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, Chan EK, Lafarga M, Conlon RA, Matera AG. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol. 2001;154:293–307. doi: 10.1083/jcb.200104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stanek D, Neugebauer KM. The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma. 2006;115:343–354. doi: 10.1007/s00412-006-0056-6. [DOI] [PubMed] [Google Scholar]
- 128.Sen A, Yokokura T, Kankel MW, Dimlich DN, Manent J, Sanyal S, Artavanis-Tsakonas S. Modeling spinal muscular atrophy in Drosophila links Smn to FGF signaling. J Cell Biol. 2011;192:481–495. doi: 10.1083/jcb.201004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Förthmann B, van Bergeijk J, Lee YW, Lubben V, Schill Y, Brinkmann H, Ratzka A, Stachowiak MK, Hebert M, Grothe C, Claus P. Regulation of neuronal differentiation by proteins associated with nuclear bodies. PLoS One. 2013;8:e82871. doi: 10.1371/journal.pone.0082871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Thau N, Jungnickel J, Knippenberg S, Ratzka A, Dengler R, Petri S, Grothe C. Prolonged survival and milder impairment of motor function in the SOD1 ALS mouse model devoid of fibroblast growth factor 2. Neurobiol Dis. 2012;47:248–257. doi: 10.1016/j.nbd.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 131.Fang X, Stachowiak EK, Dunham-Ems SM, Klejbor I, Stachowiak MK. Control of CREB-binding protein signaling by nuclear fibroblast growth factor receptor-1: a novel mechanism of gene regulation. J Biol Chem. 2005;280:28451–28462. doi: 10.1074/jbc.M504400200. [DOI] [PubMed] [Google Scholar]
- 132.Coleman SJ, Bruce C, Chioni AM, Kocher HM, Grose RP. The ins and outs of fibroblast growth factor receptor signalling. Clin Sci (Lond) 2014;127:217–231. doi: 10.1042/CS20140100. [DOI] [PubMed] [Google Scholar]
- 133.Dunham-Ems SM, Lee YW, Stachowiak EK, Pudavar H, Claus P, Prasad PN, Stachowiak MK. Fibroblast growth factor receptor-1 (FGFR1) nuclear dynamics reveal a novel mechanism in transcription control. Mol Biol Cell. 2009;20:2401–2412. doi: 10.1091/mbc.E08-06-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stachowiak MK, Maher PA, Stachowiak EK. Integrative nuclear signaling in cell development—a role for FGF receptor-1. DNA Cell Biol. 2007;26:811–826. doi: 10.1089/dna.2007.0664. [DOI] [PubMed] [Google Scholar]
- 135.Baron O, Forthmann B, Lee YW, Terranova C, Ratzka A, Stachowiak EK, Grothe C, Claus P, Stachowiak MK. Cooperation of nuclear fibroblast growth factor receptor 1 and Nurr1 offers new interactive mechanism in postmitotic development of mesencephalic dopaminergic neurons. J Biol Chem. 2012;287:19827–19840. doi: 10.1074/jbc.M112.347831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cioce M, Lamond AI. Cajal bodies: a long history of discovery. Annu Rev Cell Dev Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- 137.Sleeman JE, Trinkle-Mulcahy L, Prescott AR, Ogg SC, Lamond AI. Cajal body proteins SMN and Coilin show differential dynamic behaviour in vivo. J Cell Sci. 2003;116:2039–2050. doi: 10.1242/jcs.00400. [DOI] [PubMed] [Google Scholar]
- 138.Sleeman JE, Ajuh P, Lamond AI. snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J Cell Sci. 2001;114:4407–4419. doi: 10.1242/jcs.114.24.4407. [DOI] [PubMed] [Google Scholar]
- 139.Morris GE. The Cajal body. Biochim Biophys Acta. 2008;1783:2108–2115. doi: 10.1016/j.bbamcr.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 140.Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell. 2009;17:639–647. doi: 10.1016/j.devcel.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hebert MD. Phosphorylation and the Cajal body: modification in search of function. Arch Biochem Biophys. 2010;496:69–76. doi: 10.1016/j.abb.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]