Abstract
Zinc-finger nucleases (ZFNs) are engineered site-specific DNA cleavage enzymes that may be designed to recognize long target sites and thus cut DNA with high specificity. ZFNs mediate permanent and targeted genetic alteration via induction of a double-strand break at a specific genomic site. Compared to conventional homology-based gene targeting, ZFNs can increase the targeting rate by up to 100,000-fold; gene disruption via mutagenic DNA repair is similarly efficient. The utility of ZFNs has been shown in many organisms, including insects, amphibians, plants, nematodes, and several mammals, including humans. This broad range of tractable species renders ZFNs a useful tool for improving the understanding of complex physiological systems, to produce transgenic animals, cell lines, and plants, and to treat human disease.
Keywords: Zinc-finger nucleases, Homology-directed repair, Transgenic animals, Gene knockout, Targeting efficiency
Introduction
Genetic modification often starts with the creation of a double-strand break (DSB) in DNA. The efficiency of targeted genetic modification can be significantly enhanced by creation of a site-specific DSB [1]. An effective tool for the induction of specific DNA cleavage is the ZFN, as the variable DNA binding domain of a ZFN can be designed to bind to an investigator-specified DNA sequence. By selecting for different outcomes of DNA repair, either gene knockout (KO) or targeted transgene insertion (KI) can be obtained. In this review, we describe zinc-finger nucleases and their application to genetic modification in a variety of organisms and cell types. We begin with the discovery of the zinc-finger domain and the invention of the ZFN, and then proceed to describe how to modify a genome with ZFNs. Further, we discuss different methods to generate ZFNs. Finally, we survey the growing literature describing the utility of ZFNs for targeted genome modification.
Structural aspects of zinc-fingers (ZFs)
The first zinc-finger (ZF) protein with a specific binding affinity to DNA was discovered as part of transcription factor IIIa in Xenopus oocytes [2]. A typical zinc-finger (Cys2 His2) consists of ~30 amino acids that form two anti-parallel β sheets opposing an α-helix (Fig. 1) [3]. The domain is stabilized by two cysteine and two histidine residues binding a zinc ion, thus forming a compact globular domain. The zinc-finger motif uses residues in the α-helix to bind to approximately 3 specific bp in the major groove of the DNA [4]. ZFs can be designed for binding to almost any triplet [3]. Several ZFs can be combined to form a larger DNA-recognition domain.
Fig. 1.
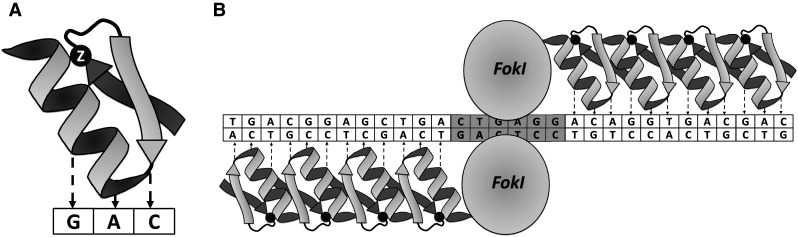
a The zinc-finger molecule consists of one α-helix and two β sheets. The zinc ion (black ball) is bound by two cysteines of the β sheets and two histidines of the α-helix leading to a stabilization of the fold. Residues of the α-helix contact 3 bp of the DNA. b Four ZF molecules fused to a nuclease form a ZFN. Two ZFNs have to bind the targeted region in tail to tail direction to allow dimerization and cleavage of the FokI nuclease domain
The invention of zinc-finger nucleases (ZFNs)
While the zinc-finger motif was discovered in the 1980s [2], ZFNs have a shorter history. Prior to the invention of ZFNs, naturally occurring restriction enzymes were the main class of site-specific nucleases. These endonucleases cut many times in even the smallest genome due to their short recognition sites (8 bp or less). Meganucleases such as the naturally occurring I-Sce-I recognize very long target sequences (as much as 18 bp), but have proven difficult to engineer to recognize new DNA targets. A ZFN consists of a site-specific zinc-finger DNA binding domain fused to the nonspecific cleavage domain of the FokI endonuclease first described in 1996 by the group of Dr. Chandrasegaran [5]. Since the FokI nuclease must dimerize to cut the DNA, two ZFN molecules are usually required, doubling the number of specifically recognized bp [6]. Moreover, for productive dimerization and cleavage, the two ZFN molecules bind to the targeted DNA on opposite strands in a tail-to-tail orientation separated by 5–7 bp, with double-stranded DNA cleavage occurring in this spacer region (Fig. 2). As described below, ZFNs are regularly used as tools to introduce double-strand breaks (DSBs) at specific sites of the genome to induce mutations via non homologous end joining (NHEJ) or to insert exogenous DNA via homology-directed repair (HDR). Initial studies on the ZFN function in multicellular organisms were performed in Xenopus oocytes in 2001 [7]. A plasmid containing the ZFN target sequence was co-injected with ZFN mRNA resulting in an extraordinary high homologous recombination (HR) efficiency of 46 %. These first preliminary experiments showed that ZFNs could work in a eukaryotic cell when the target sequence was supplied on a plasmid. Successful endogenous gene targeting mediated by ZFNs was accomplished in Drosophila melanogaster (the drosophilas yellow (y) gene) and was reported in 2002 and led to a functional gene knockout by small deletions and/or insertions at the targeted site [8]. The following years saw a burst of research activity in this field.
Fig. 2.
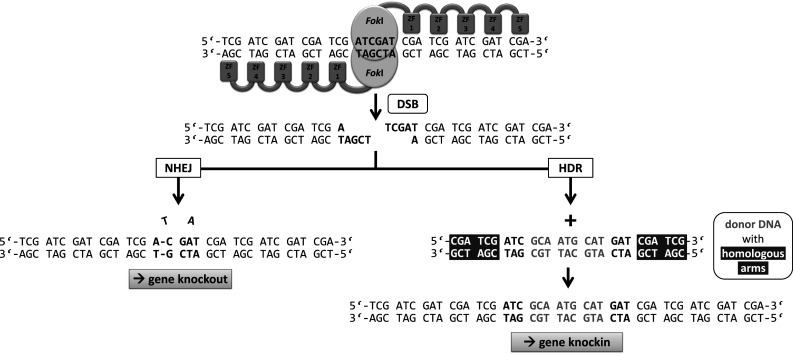
After two ZFN molecules, each consisting of five zinc fingers, bind specifically to their target sequences, the nuclease domains dimerize and cut the DNA. Double-strand break repair by non homologous end joining (NHEJ) can induce mutations leading to a gene knockout by frame shift. If a donor DNA (grey) is added with homologous arms (white letters on grey background) to the targeted region, the sequence information between the homology arms of the donor DNA is copied into the genome and a gene knockin occurs
Modification of a genome using ZFNs
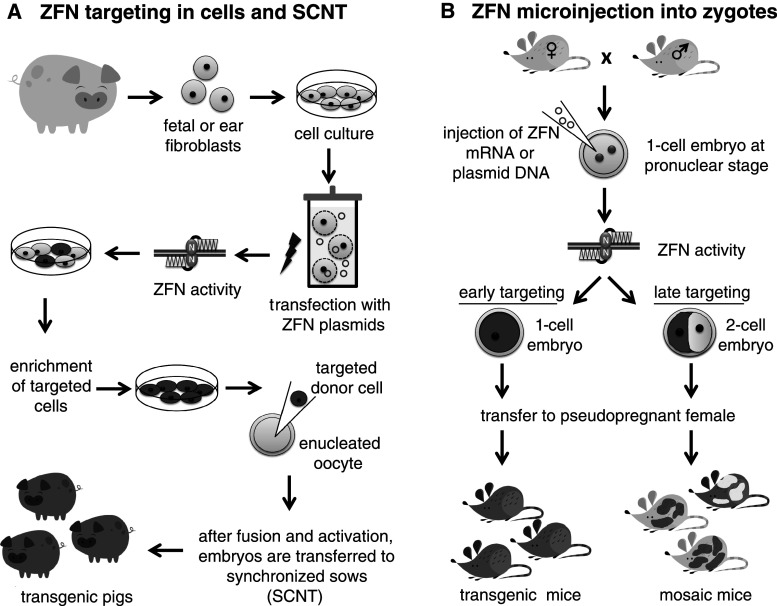
To employ a ZFN for genetic engineering, the plasmid DNA or mRNA encoding a specific ZFN is introduced into cells or embryos via microinjection or transfection (Fig. 3). After translation, the ZFN pair binds to its specific target facilitating the DNA-binding dependent dimerization of the FokI catalytic domains, and resulting in DNA cleavage. ZFN activity can be enhanced by incubating transfected cells at 30 °C for a few days as proven in human cells for several ZFN pairs [9]. Possible explanations for this observation could be a decreased mRNA and protein degradation after hypothermia, and induction of a cellular stress response, which might protect modified cells [10]. Thus, a ZFN pair induces a site-specific DSB at the unique site for which the molecule was designed.
Fig. 3.
a ZFN targeting in pig cells: fetal or ear fibroblasts are obtained for cell culture. After expansion, cells are transfected (electroporation) with ZFN plasmids or mRNA. ZFN activity leads to gene targeting in cell culture and targeted cells are enriched (FACS, bead selection etc.) and injected into enucleated oocytes (SCNT). After fusion and activation, the embryos are transferred to synchronized sows and transgenic animals are born. b Zygotes at the pronuclear stage are obtained and injected with ZFN mRNA or ZFN plasmid DNA. If the ZFN cleaves both alleles in one-cell embryos, the targeted gene will be uniformly mutated throughout the embryo, leading to transgenic animals after transfer to a pseudopregnant female. If the ZFN does not cleave both alleles in the one-cell embryo, mosaic animals will result
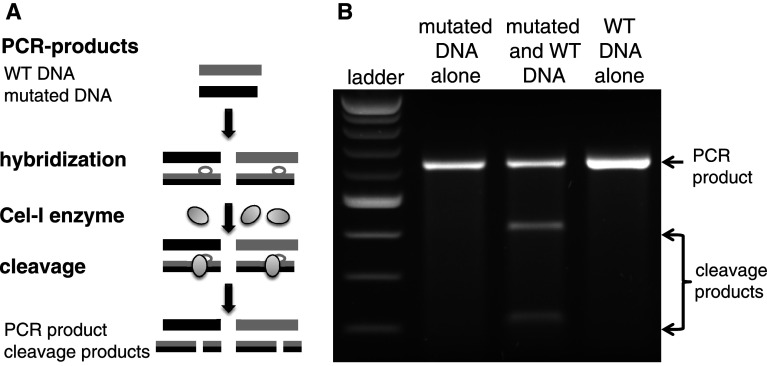
After ZFN-mediated DNA cleavage in eukaryotic cells, double-strand break repair is initiated (Fig. 2). There are two main DNA repair pathways, NHEJ and HDR. NHEJ is inherently error-prone and often creates insertions or deletions of a few base pairs (10–20 bp) in the repaired chromosome [8]. Such mutations can cause a frame-shift resulting in the disruption of a gene, leading to a knockout of the targeted gene. The Cel-I assay (Fig. 4, Surveyor nuclease assay) is a suitable in vitro system to evaluate ZFN-mediated DNA modification by NHEJ [11]. The enzyme used in this assay recognizes and cuts base pair mismatches (loops) formed when wild-type DNA and mutated DNA are hybridized. From the extent of digestion (ratio of cleaved to uncleaved product) one can calculate the NHEJ frequency in a given cell population. Since the frequency of ZFN-mediated gene modification is generally >1 % even in the absence of selection for the desired events, isolation of knockout cells is readily achieved by interrogation of cell clones generated by limiting dilution [12–15]. For non-immortalized and other poorly clonable cell lines, fluorescence activated cell sorting (FACS) or magnetic bead selection have been successfully employed to enrich the targeted cells [16–18]. Such custom schemes are target-specific and require the ability to select or enrich cells based on a phenotype of the knockout cell.
Fig. 4.
a Cel-I assay (Surveyor nuclease assay) scheme: PCR products of wild-type DNA and mutated DNA of the ZFN-targeted locus are hybridized, allowing individual DNA strands to reassort. Incubation with the Surveyor nuclease cleaves the resulting base mismatches or loops. b The digestion products are electrophoresed; cleavage products are only seen when wild-type and mutant DNA are mixed (50 % PCR product, 25 % cleavage product each). When performed on ZFN-modified samples, the diversity of alleles generated by NHEJ allows the Cel-I assay to report the degree of gene modification
The alternate DNA repair pathway, HDR, occurs routinely when the cell uses the sister chromosome as a template to repair the DSB [19]. When a donor DNA molecule containing regions homologous to both sides of the DSB is co-transfected with the ZFNs, this molecule can be used as a template instead of the sister chromosome. Critically, exogenous DNA sequence placed between the two regions of homology will be copied into the chromosome during DNA repair [20]. In the absence of a site-specific break, the donor DNA must contain a large region (6–7 kb) homologous to the targeted region so as to maximize the chance of capturing the donor via a very infrequent spontaneous break [21]. In contrast, the ZFN-based targeting strategy is compatible with a significantly shorter stretch of homologous DNA. Typically, 500–1,500 bp are used and even 50 bp on each side can be sufficient for site-specific integration [22].
Pristine genome editing results when only the donor sequence information between the homologous regions is inserted into the genome. Transient transfection of plasmid DNA is ideal for this purpose since both the ZFN and donor DNA plasmids will be rapidly diluted and lost from the treated cells. Delivery of the ZFNs as an mRNA completely eliminates the possibility of ZFN plasmid DNA integration. The ability to exploit transient delivery approaches and yet generate a permanent genetic modification is a major advantage of ZFN targeting. As in cells transfected with a donor, but not ZFNs, random integration of the donor DNA still likely occurs at some level, but targeted integration of the donor DNA is typically much more frequent in the presence of ZFNs.
ZFN design
In its simplest form new zinc-finger proteins (ZFP) DNA binding domains can be generated via the “mix and match” combination of several individual pre-characterized ZFs (each finger recognizing 3 bp) to design a specific and efficient ZFN pair. However, the design of a specific ZFN by such modular assembly, in which individual fingers with known recognition sites are joined together, fails to take into account potential interactions between the different ZFs [23, 24]. The success of modular assembly is therefore largely a function of the quality and interoperability of the modules used. A large-scale test of three zinc-finger proteins (recognizing 9 bp) assembled from publicly available modules succeeded in creating a single site-specific DNA binding protein only 24 % (25/104) of the time [25]. To overcome this limitation, a selection-free context-dependent assembly system has recently been demonstrated in the facile generation of three-finger ZFNs targeting genes in organisms as diverse as zebrafish and Arabidopsis [26].
An alternate public platform for constructing zinc-finger proteins is OPEN (oligomerized pool engineering) provided by the ZF consortium. This method has been shown to produce more active ZFN than by assembly of publicly available modules, potentially because context-dependent effects on DNA-binding among adjacent ZFs are considered during selection [25, 27]. The new tool ZFN Genome identifies potential targeting sites for ZFN generation by OPEN [28]. Engineered ZFN pairs are also commercially available from the Sigma-Aldrich Corporation’s CompoZr ZFN platform. There are two main advantages of CompoZr ZFNs; (i) these are routinely longer proteins (4-, 5-, and 6-finger proteins), which recognize longer and thus rarer target sites in the genome - thus improving the specificity of these reagents; and (ii) the investigator receives ZFNs that have been confirmed to be active in cells, which saves investigator time and eliminates ZFN quality as a potential limitation in achieving the desired goal of gene knockout, target transgene integration, and/or other downstream experiments.
Off-target site (OTS) mutations
ZFNs bind their target sites and induce a DSB at the desired genomic position. With 4–6 fingers per ZFN molecule a total of 24–36 bases are recognized, making it very unlikely that the identical sequence exists elsewhere in the genome. The specificity of a given ZFN pair can be assessed using a combined biochemical and bioinformatic approach - identifying the most highly related sequences genome-wide (using either the intended or experimentally derived consensus binding site motif for each ZFP) and then interrogating the integrity of these specific locations via direct sequencing or by using the Cel-I assay. This approach has proven very useful in confirming the specificity of ZFN action in a range of settings and delivers fast and clear results [18, 29, 30].
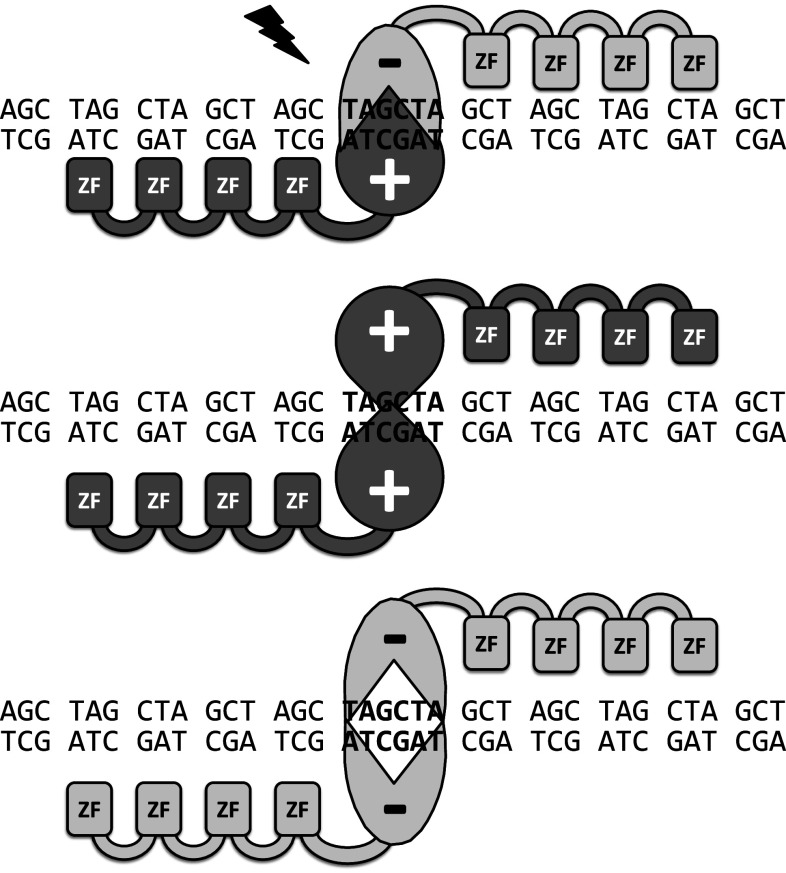
The studies reviewed here (Table 1) used either self-assembled ZFNs or ZFNs provided by Sigma-Aldrich or Sangamo BioSciences. Approximately half of all studies tested mutations at OTS, which represent the highest similarity to the targeting sequence. Half of the studies that included OTS analysis found unwanted mutations at some OTSs (varying from 0.5 to 20 % of analyzed OTSs), with cleavage occurring from 10 [31] to 770-times less frequently [32] than at the intended target site. In addition to identifying ZFs with high target specificity, significant effort has been devoted to maximizing the specificity of ZFN cleavage via engineering of the FokI portion of the molecules. While intended to bind to their target site as heterodimers, in the FokI domain’s “wild-type” form, off-target homodimer formation by individual ZFNs can occur, potentially resulting in off-target cleavage. Modified FokI nuclease variants cut the DNA only if two different ZFNs bind to the target (Fig. 5). This “obligate heterodimer” strategy has been demonstrated to markedly improve the specificity of ZFN action [33–35]. The latest generation of FokI modifications retains the specificity of the obligate heterodimers and the full activity of the wild-type domain [36]. Creation of a second, independent and non-cross-reactive set of FokI domains has allowed the simultaneous use of two independent ZFN pairs without off-target cleavage [35, 36]. Thus, several ZFN pairs can be used in parallel, if care is taken to ensure that only the intended pairs bind and cleaves their targets.
Table 1.
Listing of organisms treated with ZFNs against specific genes, leading either to a targeted integration (by HDR) or a gene knockout (by NHEJ), with efficiencies of ZFN targeting and OTS (off-target site) analysis
| Organism, cell type | Method/ZFN source | Targeted gene | Targeted integration (HR) | Knockout (KO) by NHEJ | OTS analysis | Aim | Reference |
|---|---|---|---|---|---|---|---|
| (a) Model organisms | |||||||
| Frog, oocytes | Microinjection of DNA/using an established ZF combination | Injected plasmid targetable by established ZFNs | Up to 46 % of recombination | – | – | ZFNs find recognition site in plasmids in oocytes and cleave target | [7] |
| Frog, oocytes | Microinjection of mRNA/Sangamo modular assembly | Noggin and GFP | – | 10–47 % | OTS mutation tested and found negligible | Generally applicable protocol for gene disruption in Xenopus | [47] |
| Fruit fly, embryos | Microinjection of DNA/established ZFs were modified | Yellow gene (y) | – | 46 % (mosaicism), Germline transmission in 0.44 % | – | Monitoring targeted mutation by ZFNs | [8] |
| Zebrafish, one-cell embryos (heterozygous gol-KO) | Microinjection of mRNA/Sangamo modular assembly | Golden/slc24a5 (gol) and no tail/Brachyury (ntl) | – | On avg. 12 %, up to 32 % (dose dependent) | 5 OTS analyzed, no mutation | Investigate feasibility of applying designed ZFN in zebrafish | [29] |
| Zebrafish, one-cell embryos | Microinjection of mRNA/modular assembly plus selection | Kdra exon 2 (vascular endothelial growth factor-2 receptor) | – | 10 % | 20 % of 41 OTS were mutated; 770-fold less likely than targeted mutation | Gene KO →display specific embryonic defects | [32] |
| Nematode | mRNA injection/Sangamo modular assembly | ben-1, rex-1, sdc-2 | – | Up to 5 % | 39 OTS analyzed, no mutation | First KO in nematodes | [49] |
| (b) Laboratory animals and human cells | |||||||
| Hamster, CHO cells | Transfection (lipofection and nucleofection)/Sangamo modular assembly | Dihydrofolate reductase (DHFR) | – | On average 5, 2 % biallelic | – | Proof of principle; DHFR is a selectable marker gene | [13] |
| Rat, embryos | Microinjection of mRNA and plasmid/Sangamo modular assembly | eGFP (hemizygous), IgM and Rab38 | – | On avg. 12 %, mosaicism | 20 OTS analyzed, no mutation | Investigate feasibility of applying designed ZFN in rat | [30] |
| Rat, embryos | Microinjection of mRNA/Sigma-Aldrich modular assembly | Il2Rγ locus (X chromosome) | – | 24 %, mosaicism | X-SCID rats showed immunodeficiency | [57] | |
| Rat, embryos | Microinjection of mRNA | Mdr1a P-glycoprotein (multidrug-resistant protein) | – | 33 %, mosaicism | No OTS mutations occurred (at least 4 bp different to targeting site) | Studying the role of Mdr1a P in brain penetration of drugs | [67] |
| Mouse, fertilized oocytes | Microinjection of mRNA/Sigma-Aldrich modular assembly | Mdr1a, Jag1, and Notch3 | – | 20–75 % | No OTS mutation in analyzed regions | [50] | |
| Mouse, one-cell embryos | Microinjection/Sigma-Aldrich modular assembly | Rosa26 | 1.7–4.5 % (integration of venus reporter gene (1.1 kb) and beta-galactosidase (4.2 kb)) | 22 %, mosaicism | – | Using Rosa26 locus as integration site for steady expression | [51] |
| Mouse, in vivo (expressed in liver) | Intravenous/Sangamo modular assembly | AAV8 site | 1–3 % (hFIX (human blood coagulation factor IX)) | 34–47 % | – | Hemophilia B model treatment | [55] |
| Rabbit, fertilized oocytes | Microinjection of mRNA/Sangamo modular assembly | Immunoglobulin IgM | On avg. 1.2 % targeted replacement (PGK neo cassette (1.9 kb)) | 31, 6 % biallelic, mosaicism | – | KO of IgM is essential step for production of therapeutic human polyclonal antibodies in rabbit | [58] |
| Human, K562 | Transfection (nucleofection)/Sangamo modular assembly | Il2Rγ (X chromosome) | 20 % (donor DNA: same gene with point mutation), 6, 6 % biallelic | – | – | Therapeutic usage (severe combined immunodeficiency (SCID)) | [39] |
| Human, primary CD4+ cells | Transfection (electroporation)/Sangamo modular assembly | CCR5 | – | 23, 8 % biallelic | 1 of 15 OTS analyzed showed mutation (ten times less efficient) | HIV-1-resistant cells | [31] |
| Human, hESCs and hiPSCs | Transfection (electroporation)/Sangamo modular assembly | OCT4, AAVS1 and PITX3 | Up to 94 % of selected cells | 7 % of clones with HR show NHEJ on second allele | 46 OTS analyzed, 0.5 % mutated | Monitoring pluripotency | [43] |
| (c) Large-animal models | |||||||
| Pig, fetal fibroblasts | Electroporation, ZFN-encoding mRNA/Sigma-Aldrich modular assembly | eGFP (ten copies) | – | 15 % (complete KO of all ~ 10 copies) | – | Show principle of ZFN in porcine system | [64] |
| Pig, fetal fibroblasts | Cotransfection (ZFN pair and red fluorescence plasmid)/Sigma-Aldrich modular assembly | Hemizygous eGFP | – | 5 % of selected (2 %) cells; 6 of 7 pigs mutated | – | Show principle of ZFN in porcine system | [17] |
| Pig, (a) fibroblasts, (b) embryos | (a) Cotransfection (with plasmid for selection), (b) Microinjection of mRNA/Sigma-Aldrich modular assembly | Peroxisome proliferator-activated receptor-γ(Ppar-γ) | – | (a) 4 % of selected cells carried mutation, (b) 0 % | 37 OTS analyzed, 2 mutated | Model for role of PPAR-γ in cardio vascular diseases | [37] |
| Pig, fetal fibroblasts | Electroporation with ZFN plasmids/Sangamo modular assembly | α-1,3-galactosyltransferase (GGTA1) | – | About 1 % biallelic KO | 10 OTS analyzed, no mutation | Xenotransplantation | [18] |
| Pig, liver-derived cells | Transfection of ZFN plasmids/Sigma-Aldrich modular assembly | α-1,3-galactosyltransferase (GGTA1) | – | 6.48 % | – | Xenotransplantation | [66] |
| Cattle, fetal fibroblasts | Transfection, nucleofection, ZFN-encoding mRNA/no information | beta-lactoglobulin (BLG) | – | On avg. 15 % of the cells showed mutations, only 3 % biallelic | Targeting of cells with SNP showed dramatic decrease in efficiency | Transgenic animal for production of milk with less allergens | [16] |
Fig. 5.
Mutated FokI nucleases only dimerize and cut (flash) the DNA if matching ZFNs (ZFN±) bind to its target DNA (obligate heterodimer). If the same ZFNs bind to an off-target site (homodimer formation: ZFN+/+or ZFN−/−) no cleavage occurs (modified from [33])
If OTS mutations are induced in transgenic animals, it is possible to get rid of these unwanted mutations by backcrossing ZFN-mutated individuals to wild-type animals [30, 37]. Overall, while the possibility of off-target cleavage may cause concerns, one has to take into account that off-target cleavage is very rarely found to interfere with the biology of engineered cells or organisms. Indeed, many more mutations may be introduced by the growth of the cells during their maintenance ex vivo than by nuclease action itself [38].
ZFN-mediated targeting
The following paragraphs summarize the current state of the art using ZFNs in cell cultures, various model organisms and mammalian species. We provide the characteristic features of ZFN-mediated targeting in the various organisms.
Mammalian and pluripotent cells
Human cells
ZFN-mediated targeting has been successfully employed to modify many genes in human cells. The first to be modified was the interleukin-2 receptor common γ-chain (IL2Rγ) gene, a mutation of which causes x-linked severe combined immune deficiency (SCID) [39]. The introduction of donor DNA consisting of the IL2Rγ gene with a point mutation resulted in correction of a mutant allele. Targeting efficiency was 20 % and 7 % (1/3 of mutated cells) carried a biallelic genetic modification [39]. Similar or slightly lower efficiencies were reported when the IL2Rγ locus plus donor DNA consisting of either a 12 bp tag, a 900 bp green fluorescence protein (GFP) open reading frame (ORF) or a 1.5 kb promoter-transcription unit, each flanked by locus-specific homologous arms, was used (15, 6 and 5 % efficiencies, respectively). Gene targeting was substantially more efficient when stimulated by ZFN cleavage (1.4 vs. <0.06 %) [20].
ZFN-mediated targeting was successfully used for the production of HIV-1-resistant T cells. A well-known homozygous mutation, the delta32 mutation in the CCR5 (human chemokine receptors 5) gene, confers resistance to HIV-1 infection. ZFN targeting of the CCR5 gene upstream of the natural delta32 mutation led to a specific disruption in about 50 % of the alleles in primary human CD4+ T lymphocytes, which conferred robust protection against HIV-1 infection, both in an in vitro and in an in vivo mouse model. A total of 12 out of 52 (23 %) alleles carried a mutation, of which four (33 %) showed biallelic mutations [31]. ZFN-mediated disruption of the CCR5 gene in hematopoietic stem progenitor cells (HSPCs) is desirable for clinical use. Modified HSPCs could provide a long-term antiviral effect and give rise to CCR5-KO cells in the lymphoid and myeloid compartments that HIV-1 infects. The mean disruption frequency of the CCR5 gene in HSPCs cell population was ~17 % (n = 21; ranging from 0.69 to 44 %). ZFN-mediated mutations mainly consisted of deletions ranging from 1 to 45 bp and less frequent insertions of 2 and 5 bp [40]. Transplantation of ZFN-treated and untreated (control) human CD34+ HSPCs into one day old mice resulted into 40 % human CD45+ leukocytes in the peripheral blood 8 weeks after transplantation. Infection of those animals with CCR5-tropic virus HIV-1BAL led to normal CD4/CD8 ratio in case of ZFN-treated donor cells, while untreated donor cell mice showed a loss of CD4+ cells, which is typical for an HIV infection. HIV-1 RNA was undetectable in intestinal samples from ZFN-treated donor cell mice, showing that modification of only a minority of human CD34+ HSPCs may provide a strong antiviral benefit [40].
ES and iPS cells
Embryonic stem (ES) cells and induced pluripotent stem cells (iPS) are a promising source of patient-specific autologous therapeutic cells in regenerative medicine and gene therapy, as they preclude the risk of transplant rejection. Because iPS cells are derived from somatic cells and no embryos are destroyed, their use in human medicine is favored for ethical reasons over ES cells [41]. To treat a disease, the patients’ own cells (host-derived cells), could be genetically modified by ZFN-mediated gene targeting ex vivo and then transplanted back into the recipient [42, 43]. Successful gene targeting in human iPS cells is extremely difficult, because they do not proliferate well compared with mouse ES cells. An increase in targeting efficiency may improve both the therapeutic and experimental potential of these cells [44].
The first ZFN application in pluripotent stem cells entailed a gene-delivery approach based on IDLV (integrase-defective lentivirus) to express ZFNs and to provide the template DNA for gene correction in different cell types. IDLV-mediated delivery was compatible with high rates (13–39 %) of editing of the IL2Rγ gene in different cell types. Targeting of the CCR5 gene in two human ES cell lines (HUES-3 and HUES-1) led to ~3.5 % GFP-positive cells versus 0.3 % GFP-positive cells without ZFN [45]. Using nucleic acid delivery, a ZFN specific for the OCT4 locus was used to produce OCT4-eGFP reporter cells to monitor the pluripotent status of hESCs. The OCT4-eGFP-targeted cells maintained a pluripotent status as indicated by the expression of the endogenous pluripotency markers OCT4, NANOG, SOX2, Tra-1-60, and SSEA4. The same was achieved in hESCs with a drug-inducible system when the transgene was integrated into the AAVS1 locus. About 50 % of the cells were puromycin-resistant and carried the targeted insertion in one or both alleles. Targeting of the PITX3 gene (nonexpressed gene) was also achieved in hESCs and iPSCs [43]. ZFN-mediated gene targeting was successful in two human iPS cell for the PIG-A locus which is required for retention of glycosyl-phosphatidyl-inositol anchored proteins (GPI-AP). Targeting efficiency was enhanced >2,400-fold over conventional HR and adverse effects on the pluripotent status of human ES cells were not observed [44]. Together these results demonstrate that ZFNs can be used successfully for the induction of precise genetic modifications in human ES and iPS cells.
More recently, ZFNs have been used to correct a mutant allele of the α1-antitrypsin (A1AT) gene in human iPS cells [38]. In this work the authors used ZFNs that cleave at the site of a common mutation in A1AT coupled with a donor DNA that contains a selectable marker within a Piggybac transposon. Following ZFN-mediated gene correction, modified cells were selected and the transposon excised, leaving behind only the corrected A1AT gene. As described earlier, exome sequencing of these modified cells revealed no ZFN induced changes beyond the intended correction of the “Z” mutation (Glu342Lys).
Classical gene targeting has been most successful in mouse ES cells, where both positive and negative selection approaches and a presumed higher capacity for HR combine to enhance the recovery of desired modifications. Even in this setting, however, ZFN-mediated correction of a mutated eGFP gene located in Rosa26 locus was achieved with an efficiency of >1,000-fold higher when using the ZFN with the absolute targeting efficiency differing between cell lines: ROSA-3T3 cells (1.8–6.7 %), primary adult (2 %) and embryonic (1.8 %) fibroblasts, primary astrocytes (up to 0.17 %). The donor DNA containing the correct GFP sequence led to fluorescence in correctly targeted cells. Gene corrected cells were transplanted back into a recipient mouse and transplanted cells retained their gene-corrected phenotype. Injection of cells into nude mice led to teratoma formation showing that these cells were indeed pluripotent [42]. These results indicate ZFN-mediated gene targeting is possible in a variety of different cell types with efficiencies significantly higher than conventional HR.
CHO cells
Gene modification in Chinese hamster ovary (CHO) cells has been done for several genes. Disruption of the dihydrofolate reductase (DHFR) which is essential for purine and thymidylate synthesis and therefore cell growth, by ZFN-mediated targeting led to 7 % of the cell clones showing a mutated DHFR allele; 2 % carried a biallelic mutation. Most mutations consisted of small deletions of less than 20 bp. The resulting DHFR −/− cells only grew in specific medium supplemented with hypoxanthine and thymidine, indicating a functional gene knockout [13].
A double gene knockout was produced in CHO cells by sequential deletion of the Cricetelus griseus BAX1 and BAK1 orthologues, creating a cell line resistant to apoptosis [46]. A triple gene knockout was induced in CHO cells with deletion of the glutamine synthetase (GS), DHFR and the α-1,6-fucosyltransferase (FUT8) genes by targeting with the aid of ZFNs [14]. Disruption of the GS gene was successful in 19 % of the cells, with many cells carrying biallelic mutations and lacking the GS protein. These cells were then targeted for DHFR knockout, with 2 % of all cells harboring biallelic mutations. The double knockout cell line was then used for the third gene targeting. CHO GS −/−/DHFR −/− were treated with a ZFN pair targeting the FUT8 gene leading to 2 % of the cells totally lacking FUT8 enzymatic activity [15]. This study shows that it is possible to generate multiple-KO cell lines by consecutive ZFN-mediated targeting and demonstrate the suitability of ZFNs for CHO cell engineering [14].
Model organisms
Frog
ZFN-mediated gene knockout in Xenopus tropicalis carrying one copy of the eGFP transgene was obtained by mRNA injection into early embryos at the two-cell stage. Injection of 20 pg ZFN-DNA led to mosaic loss of fluorescence in healthy tadpoles. Higher doses increased the amount of targeted cells in tadpoles. ZFN-mediated gene knockout has also been used to reveal the essential role of the Noggin locus which contributes to dorsal/ventral patterning during gastrulation in Xenopus in amphibian development. The ZFN-mediated mutations in tadpoles covered a broad range of insertions and deletions ranging from 5 to 195 bp in size, with the frequency of mutant amplicons between 10 and 47 % depending on the specific ZFN pair. After reaching sexual maturity and mating to wild-type (WT) animals, 18 % of tadpoles carried a mutated Noggin locus, showing that ZFN-induced knockout of an endogenous gene is transmitted to the next generation without deleterious side effects [47].
Fruit fly
ZFNs were also used to achieve mediated gene knockout of an endogenous gene in Drosophila targeted the yellow (y) gene located on the X chromosome. The ZFN open reading frame was combined with the Hsp70 heat-shock promoter and integrated into the genome via transposases. The DNA encoding the ZFN was injected into early embryos and the resulting adults were mated so that offspring had both ZFN genes in their genome. Incubation of the flies at 35 °C for 1 h induced expression of ZFNs and cleavage of the DNA at the expected chromosomal site. NHEJ led to deletions of a few base pairs (~10 bp) and some large deletions (~800 bp). Adult male flies carried 46 % y patches indicating a mutation of the y + gene, clearly illustrating that ZFNs are capable of inducing somatic mutations at their designated target [8].
Zebrafish
The golden/slc24a5 (gol) pigment locus encodes a transmembrane protein which results in coloration of the zebrafish embryo. Conversion to homozygosity for a gol mutation facilitates detection of ZFN activity by loss of function due to the loss of embryonic pigmentation. One-cell zebrafish embryos heterozygous for the gol b1 mutation were injected with ZFN mRNA targeting the gol locus. After 2 days, up to 32 % had nonpigmented eye cells, with the intensity of pigmentation being dose dependent. Deletions induced by NHEJ after ZFN cleavage ranged from 7 to 65 bp in size and the insertions were 4–12 bp in size. A significant proportion of mosaicism was observed, likely due to the embryonic injection approach [29].
Another gene that was successfully knocked out in zebrafish is the ntl gene (T-box transcription factor) which is crucial for mesoderm formation. The ntl b195 heterozygous fish embryos were injected and 16–27 % of injected embryos displayed an ntl-like phenotype with 5–32 % having germline mutations [29]. Further zebrafish genes successfully targeted by ZFNs include kdra exon 2 coding for vascular endothelial growth factor-2 receptor with 10 % NHEJ frequency [32]. Finally, ZFN-mediated gene knockout of the following genes: dopamine transporter (dat), hypoxia-inducible factor 1α (hif1a), telomerase, transferrin receptor 2 (tfr2) and gridlock led to mutagenesis rates of 3–20 % [48]. Collectively, these data show that ZFN-mediated gene targeting has emerged as useful tool for studying basic biology in this important model organism.
Nematodes
Despite their extensive use in the laboratory, gene disruption was not possible in Caenorhabditis elegans until 2011 when nuclease-mediated gene knockout was adapted for this organism [49]. Using 5′ and 3′ untranslated regions known to enhance translation in the germline, ZFN mRNA was injected into the worm gonad. Up to 5 % of the progeny of injected worms contained mutations at the ZFN target site. To monitor off-target cleavage, the authors sequenced the entire genome of the mutant worm; no off-target ZFN activity was detected.
Laboratory animals
Mouse
ZFN driven genetic targeting in the laboratory mouse was predominantly achieved by microinjection of DNA or mRNA directly into early fertilized oocytes. The first genome-engineered mice using ZFN technology targeted three genes, incl. the multidrug-resistant 1a (Mdr1a), jagged 1 (Jag1), and notch homolog 3 (Notch3) and yielded mutation rates in offspring ranging from 20 to 75 %. The founder animals transmitted the genetic modifications through the germline. Mating of a targeted animal with a wild-type mouse revealed that the wild-type allele was present in the germline, but was not represented in the toe or tail samples (Mdr1a) [50]. A disadvantage of microinjection into one-cell embryos is the potential production of mosaicism. ZFNs however, can remain active and cleave the DNA several times leading to different mutations in different cell types [50, 51]. Targeted integration using HDR can also be performed directly in the mouse embryo [52].
Transgenes can be introduced into a so called safe harbor site, (e.g., AAVS (human) or Rosa26 (mouse)), which is known to be able to both effectively express the transgene and to avoid problems arising from the positional effects associated with the random integration of a transgene. Targeting the Rosa26 locus was compatible with ubiquitous transgene expression and had no adverse consequences on mouse viability or phenotype [51]. In contrast, random integration of the transgene can have confounding effects on gene expression by activation or disruption of endogenous genes at or near the site of integration, and by unstable expression of the transgene due to epigenetic regulation [53]. A comparative study between random integration and targeted integration of eGFP, mediated by ZFN co-transfection, demonstrated the positive effects of targeted integration. The mean fluorescence intensity of each clone was stable when the transgene was integrated by ZFNs compared to a conventional, random targeting protocol [54].
ZFNs also can be employed to model gene therapy of human genetic diseases, by introducing the healthy variant of a mutated gene into the designated organ. Using a mouse model for hemophilia B, which is caused by congenital deficiency of blood coagulation factor IX, encoded by the FIX gene, Li et al. [55] employed in vivo gene targeting to correct the gene defect and expression of active human factor IX (hFIX) gene in situ. In the first step a transgenic mouse was produced carrying an hFIX mini-gene to mimic the congenital mutation that resulted in the absence of factor IX protein in blood. Then, neonatal hFIX−/− mice were injected with a cDNA containing exons 2–8 of the wild-type hFIX gene and the AAV8-ZFN DNA. The hFIX-specific ZFN pair was exclusively expressed in liver cells of neonatal mice with a targeting efficiency of 1–3 % (homology-directed repair) leading to a production of 2–3 % of normal level of circulating hFIX. There was no obvious toxicity in mice derived from ZFN-mediated gene mutation. The animals had a normal growth rate and weight gain [55].
Rat
The production of knockout rats using ZFNs has been demonstrated for an exogenous gene (GFP) and two endogenous genes, the Immunoglobulin M (IgM) and Rab38. Embryos were injected either with the mRNA or a plasmid coding for ZFNs. On average, 12 % of the founder animals carried the targeted mutations. ZFN activity was dose-dependent. The deletions induced by ZFN-mediated NHEJ ranged from 3 to 187 bp, and 1 of 32 IgM mutants carried a biallelic gene disruption. Breeding experiments with ZFN-mutated animals and wild-type animals revealed that mutations were transmitted through the germline [30]. The Ig-deficient rats are a useful model for analyzing the role of specific antibodies in a variety of pathophysiological situations. By producing several KOs (Ig-J, kappa and lamda chain loci) and introducing an exogenous human Ig heavy or light chain, rats with a fully humanized humoral immune response can be produced [56]. ZFN knockout rats could be created within a 4–6 months period with high efficiency of 20 %. This is faster than ES-cell-based methods for mice that usually take 12–18 months [57].
Rabbit
The rabbit is an important research model due to its short generation time, the large number of offspring produced, and its ability to be raised under special pathogen-free conditions. Further, inactivation of the endogenous IgM locus is an essential step for the production of therapeutic human polyclonal antibodies in rabbit [58]. Two ZFN pairs were employed for targeting the rabbit IgM locus. The ZFN mRNA was coinjected with donor DNA (~1.9 kb) into rabbit fertilized oocytes. Targeted replacement was achieved with an efficiency of 1.2 %. With the optimal ZFN mRNA and donor DNA concentration, up to 17 % showed a targeted gene replacement. About 30 % of the rabbits showed mutations by NHEJ and 6 % had a biallelic gene disruption. A functional knockout of the immunoglobulin heavy chain was confirmed by the absence of IgM and IgG in serum and lack of IgM+ and IgG+ B lymphocytes in compound heterozygous (biallelic KO) rabbits.
Large-animal models
The domestic pig
Transgenic farm animals, specifically the domestic pig, increasingly serve as model for human diseases. Pigs are an important complement to the laboratory mouse where it has been shown that the murine disease models often do not fully mimic the human disease. Pigs share many anatomical and physiological features with humans, and have recently emerged as a suitable model for specific diseases, including cystic fibrosis, diabetes, and several neurological disorders [59–61]. Pigs are also considered as suitable organ donor for xenotransplantation to reduce or even eliminate the shortage of suitable human organs [62]. This requires genetic modification of the donor pigs to overcome the severe immunological rejection responses after pig-to-primate xenotransplantation.
Conventional HR targeting is extremely inefficient and normally does not lead to a biallelic KO. Moreover, true pluripotent cells are not yet available from pigs and the other domestic animals, preventing selection for rare HR events as is done with mice. The production of transgenic farm animals is significantly facilitated and improved by effective somatic cell nuclear transfer (SCNT) protocols [63]. This cell-mediated transgenesis is compatible with screening for the genetic modification and analysis of the transgenic genotype in the laboratory rather than in animals ‘on the farm’. These cells are then used to produce the modified genotype in whole animals. While cell-mediated transgenesis is more labour intensive than direct transgenesis, in vitro genetic manipulation of cells followed by detailed genome analysis offers significant advantages. First, it reduces the total number of animals required to generate a useful transgenic offspring. Second, it dramatically increases the number of independent transgene integration events that can be screened and investigated. Third, it facilitates the engineering of precisely controlled genetic alterations (gene targeting) by allowing selection and isolation of rare integration events resulting from HDR.
ZFN targeting of the transgenic eGFP (pCX-eGFP) locus in pig, with ~10 genomic integration sites, decreased fluorescence intensity due to mutation of some of the multiple ZFN targets. After targeting, the rate of nonfluorescent cells increased from 6 % (control) to 21 % (ZFN-targeted cells), showing that in ~15 % of the cells nearly all copies of the eGFP gene were disrupted [64].
To achieve ZFN-mediated knockout of a hemizygous transgenic eGFP, porcine fibroblasts were co-transfected with a pair of ZFN plasmids and a red-fluorescent CAG-tomato plasmid (transient selectable fluorophore). Two percent of the cells showed red fluorescence and could be sorted by FACS. A second round of selection for green cells by FACS led to 5 % eGFP-negative cells. Selected cells used in SCNT led to the delivery of six out of seven piglets without the specific eGFP fluorescence. Sequencing revealed several deletions and insertions [17]. A third litter with six piglets was completely eGFP-negative. One piglet had an unusual large deletion of 700 bp that had removed nearly the entire eGFP coding sequence [65].
Peroxisome proliferator-activated receptor-γ (Ppar-γ) knockout animals could be useful for studies on cardiovascular diseases. To generate Ppar-γ knockout animals using ZFNs, male fibroblasts were co-transfected with a Ppar-γ-specific ZFN pair and a neomycin-resistance gene. After selection with G418, 4 % of screened cell clones carried one mutated Ppar-γ gene and served as donor cells in SCNT. Two live piglets carried a mutation in one of the Ppar-γ alleles. Western blotting analysis confirmed the successful production of heterozygous Ppar-γ KO animals [37].
The first live pigs carrying a biallelic knockout via single-step ZFN-mediated targeting of an endogenous gene were generated by our laboratory [18]. Transfection of fetal fibroblasts with a pair of ZFN plasmids directed against exon 9 of the α1,3-galactosyltransferase (GGTA1, Gal) gene, induced biallelic mutations in about 1 % of the cells. With the aid of magnetic beads, Gal-negative cells could be successfully enriched in a much more gentle manner than by sorting via FACS; >99 % of the cells were Gal-negative. After use of the selected cells in SCNT, Gal-negative fetuses were obtained 25 days after transfer of the reconstructed embryos to recipients. In total, 9 live GGTA1-KO piglets were produced from fetal fibroblasts. Sequencing of eight female pigs revealed five different haplotypes with two homozygous and three heterozygous (individual mutations on each allele) mutations. The GGTA1 gene showed deletions from 1 to 7 bp in size and one large deletion of 96 bp. The GGTA1-KO fibroblasts derived from ZFN treatment were protected in an in vitro complement-mediated lysis assay to a similar extent as existing HR-Gal-KO cells. The results show that the production of homozygous GGTA1-KO pigs can be significantly accelerated compared to conventional gene targeting, giving knockout pigs in as few as 6 months.
Li et al. [66] reported the production of GGTA1 biallelic KO pigs obtained via ZFN targeting. The main difference to the study from our laboratory [18] was the usage of porcine liver-derived cells for transfection, which proliferate faster than the fetal fibroblasts used in our study [18]. These results emphasize the significance of the generation of ZFN-mediated biallelic GGTA1-KO pigs and demonstrate that such a gene knockout can easily be reproduced, even with a different ZFN pair.
Cattle
In cattle, ZFN-mediated gene targeting was conducted for the production of beta-lactoglobulin (BLG) knockout animals. BLG is the major whey protein in bovine milk and is one of the major milk allergens. Bovine fetal fibroblasts were transfected with mRNA coding for ZFNs designed against the BLG gene. Sequencing revealed that ~15 % of the cells carried a mutated variant of the BLG gene and 3 % of the single-cell colonies showed a biallelic BLG gene knockout. The biallelic-KO cells were used in SCNT and 8 cloned animals were born, but only one survived the postnatal period. The mutated BLG gene was shorter (9 and 15 bp deletion, no frame shift) than the wild-type version. However, the milk could not be analyzed for the absence of BLG. Potential off-target mutations were also analyzed in this study. While a one base pair mismatch with the targeting sequence leads to 7 % gene targeting (single-nucleotide polymorphism in cattle), 3 and 7 base pair mismatches did not result in ZFN-mediated mutations in sheep and pigs. Results suggest that ZFN-mediated targeting is promising for specific gene editing in large domestic animals [16].
Conclusions and perspectives
ZFNs have emerged as useful tool for precise genetic modifications in a variety of organisms. Biallelic targeting can be achieved in one step, which is generally not possible with conventional homology-based knockout techniques. Methods which avoid exogenous DNA being integrated into the genome can be exploited and therefore negative side effects such as gene silencing (due to backbone integration) are negligible. Further, due to the high frequency of ZFN-mediated cleavage, antibiotic selection is not necessary, a step which is required for selection of targeting events by conventional means. ZFNs are particularly valuable for primary somatic cells which are usually not compatible with the extended selection process needed to isolate rare spontaneous targeting events. ZFNs thus have made feasible gene targeting in species in which pluripotent cells are not yet available, enabling the production of transgenic domestic animals via SCNT. ZFN-mediated targeting has shown to be successful using self-made and commercially obtained ZFNs; off-target mutations rarely occur and do not seem to be a major hurdle for a broader application of ZFNs.
Abbreviations
- BKG
Beta-lactoglobulin
- CCR5
Human chemokine receptors 5 gene
- CHO
Chinese hamster ovary
- Dat
Dopamine transporter
- DHFR
Dihydrofolate reductase
- DSB
Double-strand break
- ES cells
Embryonic stem cells
- FACS
Fluorescence-activated cell sorting
- FUT8
α-1,6-fucosyltransferase
- GFP
Green fluorescence protein
- GGTA1
α1,3-galactosyltransferase (Gal)
- GPI-AP
Glycosyl-phosphatidyl-inositol anchored proteins
- GS
Glutamine synthetase
- HDR
Homology-direct repair
- hFIX
Human factor IX
- hif1 α
Hypoxia-inducible factor 1α
- HR
Homologous recombination
- HSPCs
Hematopoietic stem progenitor cells
- IDLV
Integrase-defective lentivirus
- IgM
Immunoglobulin M
- IL2Rγ
Interleukin-2 receptor common γ-chain
- iPS cells
Induced pluripotent stem cells
- Jag1
Jagged 1
- KO
Knockout
- KI
Knockin
- Mdr1a
Multidrug-resistant 1a
- NHEJ
Non-homologous end joining
- Notch3
Notch homolog 3
- OPEN
Oligomerized pool engineering
- ORF
Open reading frame
- OTS
Off-target site
- Ppar-γ
Peroxisome proliferator-activated receptor-γ
- SCID
Severe combined immune deficiency
- SCNT
Somatic cell nuclear transfer
- tfr2
Telomerase, transferrin receptor 2
- WT
Wild type
- ZF
Zinc-finger
- ZFN
Zinc-finger nucleases
- ZFP
Zinc-finger proteins
References
- 1.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci USA. 1994;91(13):6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 4.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 5.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93(3):1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S, Carroll D. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28(17):3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21(1):289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161(3):1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doyon Y, Choi VM, Xia DF, Vo TD, Gregory PD, Holmes MC. Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat Methods. 2010;7(6):459–460. doi: 10.1038/nmeth.1456. [DOI] [PubMed] [Google Scholar]
- 10.Roobol A, Carden MJ, Newsam RJ, Smales CM. Biochemical insights into the mechanisms central to the response of mammalian cells to cold stress and subsequent rewarming. FEBS J. 2009;276(1):286–302. doi: 10.1111/j.1742-4658.2008.06781.x. [DOI] [PubMed] [Google Scholar]
- 11.Qiu P, Shandilya H, D’Alessio JM, O’Connor K, Durocher J, Gerard GF. Mutation detection using surveyor nuclease. Biotechniques. 2004;36(4):702–707. doi: 10.2144/04364PF01. [DOI] [PubMed] [Google Scholar]
- 12.Kim S, Lee HJ, Kim E, Kim JS (2010) Analysis of targeted chromosomal deletions induced by zinc finger nucleases. Cold Spring Harb Protoc 2010 (8):pdb prot5477. doi:10.1101/pdb.prot5477 [DOI] [PubMed]
- 13.Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, Holmes MC, Guschin D, Waite A, Miller JC, Rebar EJ, Gregory PD, Klug A, Collingwood TN. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105(15):5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu PQ, Chan EM, Cost GJ, Zhang L, Wang JB, Miller JC, Guschin DY, Reik A, Holmes MC, Mott JE, Collingwood TN, Gregory PD. Generation of a triple-gene knockout mammalian cell line using engineered zinc-finger nucleases. Biotechnol Bioeng. 2010;106(1):97–105. doi: 10.1002/bit.22654. [DOI] [PubMed] [Google Scholar]
- 15.Malphettes L, Freyvert Y, Chang J, Liu PQ, Chan E, Miller JC, Zhou Z, Nguyen T, Tsai C, Snowden AW, Collingwood TN, Gregory PD, Cost GJ. Highly efficient deletion of FUT8 in CHO cell lines using zinc-finger nucleases yields cells that produce completely nonfucosylated antibodies. Biotechnol Bioeng. 2010;106(5):774–783. doi: 10.1002/bit.22751. [DOI] [PubMed] [Google Scholar]
- 16.Yu S, Luo J, Song Z, Ding F, Dai Y, Li N. Highly efficient modification of beta-lactoglobulin (BLG) gene via zinc-finger nucleases in cattle. Cell Res. 2011;21(11):1638–1640. doi: 10.1038/cr.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whyte JJ, Zhao J, Wells KD, Samuel MS, Whitworth KM, Walters EM, Laughlin MH, Prather RS. Gene targeting with zinc finger nucleases to produce cloned eGFP knockout pigs. Mol Reprod Dev. 2011;78(1):2. doi: 10.1002/mrd.21271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauschild J, Petersen B, Santiago Y, Queisser AL, Carnwath JW, Lucas-Hahn A, Zhang L, Meng X, Gregory PD, Schwinzer R, Cost GJ, Niemann H. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc Natl Acad Sci USA. 2011;108(29):12013–12017. doi: 10.1073/pnas.1106422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gandhi M, Evdokimova VN, TC K, Nikiforova MN, Kelly LM, Stringer JR, Bakkenist CJ, Nikiforov YE. Homologous chromosomes make contact at the sites of double-strand breaks in genes in somatic G0/G1-phase human cells. Proc Natl Acad Sci USA. 2012;109(24):9454–9459. doi: 10.1073/pnas.1205759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, Urnov FD, Holmes MC. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci USA. 2007;104(9):3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng C, Capecchi MR. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol Cell Biol. 1992;12(8):3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orlando SJ, Santiago Y, DeKelver RC, Freyvert Y, Boydston EA, Moehle EA, Choi VM, Gopalan SM, Lou JF, Li J, Miller JC, Holmes MC, Gregory PD, Urnov FD, Cost GJ. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Res. 2010;38(15):e152. doi: 10.1093/nar/gkq512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pruett-Miller SM, Connelly JP, Maeder ML, Joung JK, Porteus MH. Comparison of zinc finger nucleases for use in gene targeting in mammalian cells. Mol Ther. 2008;16(4):707–717. doi: 10.1038/mt.2008.20. [DOI] [PubMed] [Google Scholar]
- 24.Isalan M, Choo Y, Klug A. Synergy between adjacent zinc fingers in sequence-specific DNA recognition. Proc Natl Acad Sci USA. 1997;94(11):5617–5621. doi: 10.1073/pnas.94.11.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, Cathomen T, Voytas DF, Joung JK. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008;5(5):374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, Pierick CJ, Hoffman E, Maeder ML, Khayter C, Reyon D, Dobbs D, Langenau DM, Stupar RM, Giraldez AJ, Voytas DF, Peterson RT, Yeh JR, Joung JK. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat Methods. 2011;8(1):67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, Unger-Wallace E, Sander JD, Muller-Lerch F, Fu F, Pearlberg J, Gobel C, Dassie JP, Pruett-Miller SM, Porteus MH, Sgroi DC, Iafrate AJ, Dobbs D, McCray PB, Jr, Cathomen T, Voytas DF, Joung JK. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31(2):294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reyon D, Kirkpatrick JR, Sander JD, Zhang F, Voytas DF, Joung JK, Dobbs D, Coffman CR. ZFNGenome: a comprehensive resource for locating zinc finger nuclease target sites in model organisms. BMC Genomics. 2011;12:83–91. doi: 10.1186/1471-2164-12-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26(6):702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, Vincent A, Lam S, Michalkiewicz M, Schilling R, Foeckler J, Kalloway S, Weiler H, Menoret S, Anegon I, Davis GD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jacob HJ, Buelow R. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325(5939):433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, Guschin DY, Rupniewski I, Waite AJ, Carpenito C, Carroll RG, Orange JS, Urnov FD, Rebar EJ, Ando D, Gregory PD, Riley JL, Holmes MC, June CH. Establishment of HIV-1 resistance in CD4+T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26(7):808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26(6):695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, Gregory PD, Pabo CO, Rebar EJ. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25(7):778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 34.Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25(7):786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 35.Sollu C, Pars K, Cornu TI, Thibodeau-Beganny S, Maeder ML, Joung JK, Heilbronn R, Cathomen T. Autonomous zinc-finger nuclease pairs for targeted chromosomal deletion. Nucleic Acids Res. 2010;38(22):8269–8276. doi: 10.1093/nar/gkq720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011;8(1):74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 37.Yang D, Yang H, Li W, Zhao B, Ouyang Z, Liu Z, Zhao Y, Fan N, Song J, Tian J, Li F, Zhang J, Chang L, Pei D, Chen YE, Lai L. Generation of PPARgamma mono-allelic knockout pigs via zinc-finger nucleases and nuclear transfer cloning. Cell Res. 2011;21(6):979–982. doi: 10.1038/cr.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yusa K, Rashid ST, Strick-Marchand H, Varela I, Liu PQ, Paschon DE, Miranda E, Ordonez A, Hannan NR, Rouhani FJ, Darche S, Alexander G, Marciniak SJ, Fusaki N, Hasegawa M, Holmes MC, Di Santo JP, Lomas DA, Bradley A, Vallier L. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478(7369):391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435(7042):646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 40.Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, Cannon PM. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28(8):839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bobis-Wozowicz S, Osiak A, Rahman SH, Cathomen T. Targeted genome editing in pluripotent stem cells using zinc-finger nucleases. Methods. 2011;53(4):339–346. doi: 10.1016/j.ymeth.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 42.Connelly JP, Barker JC, Pruett-Miller S, Porteus MH. Gene correction by homologous recombination with zinc finger nucleases in primary cells from a mouse model of a generic recessive genetic disease. Mol Ther. 2010;18(6):1103–1110. doi: 10.1038/mt.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, Meng X, Miller JC, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27(9):851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, Porteus MH, Joung JK, Cheng L. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5(1):97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, Holmes MC, Naldini L. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25(11):1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 46.Cost GJ, Freyvert Y, Vafiadis A, Santiago Y, Miller JC, Rebar E, Collingwood TN, Snowden A, Gregory PD. BAK and BAX deletion using zinc-finger nucleases yields apoptosis-resistant CHO cells. Biotechnol Bioeng. 2010;105(2):330–340. doi: 10.1002/bit.22541. [DOI] [PubMed] [Google Scholar]
- 47.Young JJ, Cherone JM, Doyon Y, Ankoudinova I, Faraji FM, Lee AH, Ngo C, Guschin DY, Paschon DE, Miller JC, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Harland RM, Zeitler B. Efficient targeted gene disruption in the soma and germ line of the frog Xenopus tropicalis using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2011;108(17):7052–7057. doi: 10.1073/pnas.1102030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foley JE, Yeh JR, Maeder ML, Reyon D, Sander JD, Peterson RT, Joung JK. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by oligomerized pool engineering (OPEN) PLoS One. 2009;4(2):e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Meyer BJ. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333(6040):307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carbery ID, Ji D, Harrington A, Brown V, Weinstein EJ, Liaw L, Cui X. Targeted genome modification in mice using zinc-finger nucleases. Genetics. 2010;186(2):451–459. doi: 10.1534/genetics.110.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyer M, de Angelis MH, Wurst W, Kuhn R. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc Natl Acad Sci USA. 2010;107(34):15022–15026. doi: 10.1073/pnas.1009424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui X, Ji D, Fisher DA, Wu Y, Briner DM, Weinstein EJ. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol. 2011;29(1):64–67. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- 53.Uren AG, Kool J, Berns A, van Lohuizen M. Retroviral insertional mutagenesis: past, present and future. Oncogene. 2005;24(52):7656–7672. doi: 10.1038/sj.onc.1209043. [DOI] [PubMed] [Google Scholar]
- 54.Perez-Pinera P, Ousterout DG, Brown MT, Gersbach CA. Gene targeting to the ROSA26 locus directed by engineered zinc finger nucleases. Nucleic Acids Res. 2011;40(8):3741–3752. doi: 10.1093/nar/gkr1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS, Malani N, Anguela XM, Sharma R, Ivanciu L, Murphy SL, Finn JD, Khazi FR, Zhou S, Paschon DE, Rebar EJ, Bushman FD, Gregory PD, Holmes MC, High KA. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475(7355):217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Remy S, Tesson L, Menoret S, Usal C, Scharenberg AM, Anegon I. Zinc-finger nucleases: a powerful tool for genetic engineering of animals. Transgenic Res. 2010;19(3):363–371. doi: 10.1007/s11248-009-9323-7. [DOI] [PubMed] [Google Scholar]
- 57.Mashimo T, Takizawa A, Voigt B, Yoshimi K, Hiai H, Kuramoto T, Serikawa T. Generation of knockout rats with X-linked severe combined immunodeficiency (X-SCID) using zinc-finger nucleases. PLoS One. 2010;5(1):e8870. doi: 10.1371/journal.pone.0008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flisikowska T, Thorey IS, Offner S, Ros F, Lifke V, Zeitler B, Rottmann O, Vincent A, Zhang L, Jenkins S, Niersbach H, Kind AJ, Gregory PD, Schnieke AE, Platzer J. Efficient immunoglobulin gene disruption and targeted replacement in rabbit using zinc finger nucleases. PLoS One. 2011;6(6):e21045. doi: 10.1371/journal.pone.0021045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rogers CS, Stoltz DA, Meyerholz DK, Ostedgaard LS, Rokhlina T, Taft PJ, Rogan MP, Pezzulo AA, Karp PH, Itani OA, Kabel AC, Wohlford-Lenane CL, Davis GJ, Hanfland RA, Smith TL, Samuel M, Wax D, Murphy CN, Rieke A, Whitworth K, Uc A, Starner TD, Brogden KA, Shilyansky J, McCray PB, Jr, Zabner J, Prather RS, Welsh MJ. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science. 2008;321(5897):1837–1841. doi: 10.1126/science.1163600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van den Heuvel M, Sorop O, Koopmans SJ, Dekker R, de Vries R, van Beusekom HM, Eringa EC, Duncker DJ, Danser AH, van der Giessen WJ. Coronary microvascular dysfunction in a porcine model of early atherosclerosis and diabetes. Am J Physiol Heart Circ Physiol. 2012;302(1):H85–H94. doi: 10.1152/ajpheart.00311.2011. [DOI] [PubMed] [Google Scholar]
- 61.Mikkelsen M, Moller A, Jensen LH, Pedersen A, Harajehi JB, Pakkenberg H. MPTP-induced Parkinsonism in minipigs: a behavioral, biochemical, and histological study. Neurotoxicol Teratol. 1999;21(2):169–175. doi: 10.1016/S0892-0362(98)00037-3. [DOI] [PubMed] [Google Scholar]
- 62.Cooper DK, Ayares D. The immense potential of xenotransplantation in surgery. Int J Surg. 2011;9(2):122–129. doi: 10.1016/j.ijsu.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 63.Petersen B, Lucas-Hahn A, Oropeza M, Hornen N, Lemme E, Hassel P, Queisser AL, Niemann H. Development and validation of a highly efficient protocol of porcine somatic cloning using preovulatory embryo transfer in peripubertal gilts. Cloning Stem Cells. 2008;10(3):355–362. doi: 10.1089/clo.2008.0026. [DOI] [PubMed] [Google Scholar]
- 64.Watanabe M, Umeyama K, Matsunari H, Takayanagi S, Haruyama E, Nakano K, Fujiwara T, Ikezawa Y, Nakauchi H, Nagashima H. Knockout of exogenous EGFP gene in porcine somatic cells using zinc-finger nucleases. Biochem Biophys Res Commun. 2010;402(1):14–18. doi: 10.1016/j.bbrc.2010.09.092. [DOI] [PubMed] [Google Scholar]
- 65.Whyte JJ, Prather RS. Zinc finger nucleases to create custom-designed modifications in the swine (Sus scrofa) genome. J Anim Sci. 2011;90(4):1111–1117. doi: 10.2527/jas.2011-4546. [DOI] [PubMed] [Google Scholar]
- 66.Li P, Estrada JL, Burlak C, Tector AJ. Biallelic knockout of the alpha-1,3 galactosyltransferase gene in porcine liver-derived cells using zinc finger nucleases. J Surg Res. 2012 doi: 10.1016/j.jss.2012.06.035. [DOI] [PubMed] [Google Scholar]
- 67.Chu X, Zhang Z, Yabut J, Horwitz S, Levorse J, Li XQ, Zhu L, Lederman H, Ortiga R, Strauss J, Li X, Owens KA, Dragovic J, Vogt T, Evers R, Shin MK. Characterization of multidrug resistance 1a/P-glycoprotein knockout rats generated by zinc finger nucleases. Mol Pharmacol. 2012;81(2):220–227. doi: 10.1124/mol.111.074179. [DOI] [PubMed] [Google Scholar]