Abstract
Cell density is one of the extrinsic factors to which cells adapt their physiology when grown in culture. However, little is known about the molecular changes which occur during cell growth and how cellular responses are then modulated. In many cases, inhibitors, drugs or growth factors used for in vitro studies change the rate of cell proliferation, resulting in different cell densities in control and treated samples. Therefore, for a comprehensive data analysis, it is essential to understand the implications of cell density on the molecular level. In this study, we have investigated how lipid composition changes during cell growth, and the consequences it has for transport of Shiga toxin. By quantifying 308 individual lipid species from 17 different lipid classes, we have found that the levels and species distribution of several lipids change during cell growth, with the major changes observed for diacylglycerols, phosphatidic acids, cholesterol esters, and lysophosphatidylethanolamines. In addition, there is a reduced binding and retrograde transport of Shiga toxin in high density cells which lead to reduced intoxication by the toxin. In conclusion, our data provide novel information on how lipid composition changes during cell growth in culture, and how these changes can modulate intracellular trafficking.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-013-1441-y) contains supplementary material, which is available to authorized users.
Keywords: Shiga toxin, Gb3, Lipidomics, Endocytosis, Retrograde transport
Introduction
Cell lines are widely used in molecular biology and genetic studies. However, various factors, such as growth conditions, passage number, cell cycle and cell-to-cell interactions, may lead to significant changes in cells, thus affecting the data obtained. For instance, cell density in the culture modulates cell sensitivity to drugs [1], cytokines [2], and viruses [3], as well as bacterial and plant toxins [4, 5] in various cell types, both normal and cancer-originated. Nevertheless, cell density is often underestimated during evaluation of results, although cell growth is affected by various widely used inhibitors and drugs which lead to different cell densities in the control and treated samples, and thus may affect results and their interpretation. Therefore, a better knowledge of cell density induced molecular changes is required for proper experimental planning and data interpretation in most cell culture-based studies.
Shiga toxins (Stx), bacterial protein toxins produced by the bacteria Shigella dysenteriae and certain strains of Escherichia coli, have been proven to be useful tools for studying mechanisms of intracellular transport [6, 7]. Interestingly, cell sensitivity to Stx has been shown to be dependent on cell density in the culture. Confluent HUVEC cells were found to be up to 50-fold less sensitive to Stx than cells in non-confluent culture [4]. Stx binds the carbohydrate moiety of the glycosphingolipid globotriaosylceramide (Gb3; abbreviations of all lipid classes are given under the section “Annotations of lipid species” in “Materials and methods ”) in the extracellular leaflet of the plasma membrane in target-cells [8–10]. Interestingly, both the biosynthesis [11–13] and the content of Gb3 [11, 14] have been shown to increase with increasing cell density, which would suggest increasing cell susceptibility to the toxin in high density cultures. However, the binding of Stx is not only influenced by the number of receptors but also depends on Gb3 species [15–17] and/or the membrane environment of the receptor [17]. While the sphingosine chain in the ceramide part of Gb3 is relatively invariable, the fatty acyl chain varies both in length and saturation [17, 18]. Since Gb3 species vary in their ability to bind Stx [15], the distribution of the different Gb3 species in the membranes may affect Stx-binding to target-cells. Moreover, other membrane lipids, such as cholesterol [19–22], affect Stx binding to Gb3, probably by modulating the presentation of the carbohydrate head of glycosphingolipids [22, 23]. Although surface expression of Gb3 is a prerequisite for Stx toxicity, it is in itself not sufficient for cytotoxic action of the toxin [24–26]. Gb3-bound Stx needs to be endocytosed and transported retrogradely to the endoplasmic reticulum (ER) where its enzymatically active A1-moiety (StxA1) is released into the cytosol and inhibits protein synthesis [7]. Therefore, not only binding but also changes in multiple transport steps may modulate cell sensitivity to Stx. It has been shown that Gb3 association with detergent-resistant membranes (DRMs) is necessary for efficient Stx intracellular transport and toxicity [19, 27], suggesting that Stx transport is affected by membrane composition in the cells.
Increasing amounts of data now support the idea that not only proteins but also lipids are involved in controlling vesicular transport. The localization of different phosphoinositides is strictly regulated within certain vesicular membranes, and therefore function as specific markers and docking sites recognized by dedicated transport proteins [28]. Furthermore, DAG has been shown to be required for retrograde Golgi-to-ER transport by facilitating COPI-coated vesicle formation [29], and for protein transport from the trans-Golgi network to the plasma membrane [30], presumably by participating in membrane bending and neck formation [31]. Importantly, lipid phosphate phosphatase 3 (LPP3, also known as PAP2b) was recently found to be essential for both COPI-dependent and COPI-independent retrograde transport in HeLa cells [32]. MS analysis is now providing increasing knowledge about eukaryotic lipidomes and their flexibility [33], but additional studies are needed to understand the biological functions of different lipids in the cells, and how the lipid composition is regulated in response to intra- and inter-cellular signals.
In the present study, we have employed two different approaches to study cell density-induced changes: (1) we performed quantitative MS analyzes to study the effects on lipid composition, and (2) we analyzed intracellular trafficking by using Stx as a tool. Firstly, we found that increasing cell density induced multiple changes in the lipidome of HEp-2 cells, with most profound effects on PA, DAG, LPE, neutral glycosphingolipids, and CE. Secondly, we tested Stx binding and transport in HeLa and HEp-2 cells, and found that high cell density significantly reduced Stx binding to the cells, without affecting either efficiency of Stx endocytosis or the rate of Stx transport to the Golgi. However, the release of StxA1 from the ER into the cytosol was reduced, suggesting that either Stx transport to the ER and/or translocation of StxA1 across the ER membrane was impaired in high density cells. Interestingly, increased cell density had opposite effects on HeLa and HEp-2 cells with regards to Gb3 content, since there was a significant reduction in total Gb3 in high density HeLa cells, but rather a small increase in HEp-2 cells. However, both these cell types showed identical cell density-dependent responses to Stx treatment, supporting the hypothesis that it is not the number of the receptors but the distribution of different species and/or membrane microenvironment that regulates Stx binding and intracellular transport. Although additional studies are required to fully understand cell density-induced changes in the cultured cells, our data provide the most detailed analysis so far on how lipids and certain steps of cellular trafficking are affected by cell density.
Materials and methods
If not specified otherwise, the reagents were purchased from Sigma Aldrich, MO, USA. Purified Stx was a gift from J.E. Brown (USAMRIID, Fort Detrick, MD, USA). Glycosphingolipids extracted from human erythrocytes was a kind gift from Prof. Dr. Johannes Müthing (Institute for Medical Physics and Biophysics, University of Münster, Germany). HPTLC (high-performance thin-layer chromatography) silica gel 60 glass plates were purchased from Merck Millipore (Germany). For activation, the plates were heated for 30 min at 110 °C, and cooled down before use.
Cells
HeLa and HEp-2 cells were grown at 37 °C under 5 % CO2 in DMEM + GlutaMAX™ medium supplemented with 10 % (v/v) fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin (all Gibco®, Life Technologies, UK). If not specified otherwise, cells were seeded 1 day prior to the experiment. At the day of the experiment, cells were examined by phase contrast microscopy to roughly evaluate cell density: cell confluency was in the range of 80–90 % in high density samples, and in the range of 20–30 % in low density samples. This difference was achieved either by seeding similar number of cells to different culture dishes (approximately five times difference in growth area), or by seeding different numbers of cells into similar dishes. Using the second approach, the cell number in low and high density cultures is different, and it might interfere with some of the treatments. Therefore, all the experiments where reproduced at least once by using the first seeding approach where equal starting number of the cells was seeded into different size culture dishes.
Shiga toxin toxicity
Cells were seeded in 24-well culture plates with different starting number of the cells, which led to high- and low density cultures at the day of the experiment. Cells were washed with leucine-free HEPES-buffered medium and incubated with tenfold serial dilutions of Stx in HEPES-buffered medium for 2 h at 37 °C. Then, the cells were washed and incubated with leucine-free HEPES-buffered medium containing 1 μCi/ml [3H]leucine (PerkinElmer, MA, USA) for 20 min at 37 °C. The proteins were precipitated with 5 % (w/v) trichloroacetic acid (TCA), washed once with 5 % (w/v) TCA, and dissolved in 0.1 M KOH. The amount of incorporated [3H]leucine was measured by β-counting on a Tri-Carb 2100TR Liquid Scintillation Analyzer (Packard Bioscience, CT, USA). The cytotoxicity was determined as the concentration of the toxin which reduced protein synthesis by 50 % (IC50).
Analysis of Gb3 content in cells by HPTLC
Cells were seeded in 225-ml culture flasks with different starting numbers, and were of low and high density when they were harvested by trypsinization on the third day after seeding. Cells were counted using Coulter Z1 Particle Counter (Coulter International, FL, USA), and 1–2 × 107 cells/sample were transferred to methanol pre-washed centrifuge glass tubes. The cells were centrifuged at 400g for 10 min at 10 °C, and the pellet was resuspended in 10 ml methanol. The suspension was then sonicated for 15 min and centrifuged at 5,500g for 10 min at 10 °C. The supernantant was transferred to a 60-ml SampleGenie Genevac glass bottle by means of filtration. The pellet was resuspended in 10 ml 1:2 (v/v) chloroform:methanol following the sonication and centrifugation steps as described above. The procedure was repeated two more times resuspending the pellet in 1:1(v/v) and 2:1 (v/v) chloroform:methanol, and the supernatants were collected in Genevac glass bottle. The samples were dried using Genevac EZ-2 automated evaporation system (Genevac, NY, USA).
For hydrolysis of esterified lipids, dried lipid extracts were incubated with 4.5 ml 1 M NaOH for 1 h at 37 °C. Then, 450 μl 10 M HCl was added, and the samples were transferred to dialysis tubes (Spectra/Pro No4, MWCO 12-14,000) following the dialysis against water for 2 days. The extracts were then dried using Genevac EZ-2 automated evaporation system (Genevac) and resuspended in 2:1 (v/v) chloroform:methanol with a volume of 25 μl per 1 million cells.
The extracts were applied to HPTLC Silica gel 60 plate using Linomat 5 (CAMAG, Switzerland) and chromatographed with a mixture of chloroform:methanol:water (70:30:4) as the mobile phase, following the staining by 0.3 % (w/v) orcinol solution in 3 M H2SO. Developed plates were scanned using GS-800 Calibrated Densitometer (Bio-Rad Laboratories, CA, USA) and the bands were quantified by Quantity One 1-D Analysis Software (Bio-Rad Laboratories).
Quantification of protein and cholesterol content in cell lysates
Protein content in cell lysates was measured using BCA protein Assay (Thermo Scientific, MA, USA) as described by the manufacturer, and using bovine serum albumin as standard protein.
For cholesterol measurements, low and high density cells were washed with 37 °C PBS and lysed in lysis buffer (0.1 % (w/v) SDS, 0.1 M Tris, 1 mM EDTA, pH 7.4) for 5 min at 37 °C. The lysates were homogenized by passing ten times through a 19G needle, and the content of free cholesterol was measured using Amplex® Red Cholesterol Assay Kit (Invitrogen, UK) according to the manufacturer’s protocol.
Harvesting of cells for lipidomic analyzes
Cells were washed in 37 °C HEPES-buffered medium; trypsin/EDTA was added and the incubation continued at 37 °C with 5 % CO2 until the cells detached. Cells were then resuspended in HEPES-buffered medium and transferred to microfuge tubes, centrifuged for 10 min at 2,500g, washed with PBS, and centrifuged again before freezing at −80 °C.
For protein quantification, cells were lysed in cell lysis buffer [0.1 M NaCl, 10 mM Na2HPO4, 1 mM EDTA, 1 % (w/v) Triton X-100, cOmplete EDTA-free Protease Inhibitor Cocktail (1 tablet/50 ml; Roche Diagnostics, Switzerland), 60 mM octyl β-d-glucopyranoside, pH 7.4].
Annotations of lipid species
The different lipid species of phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), phosphatidylinositol (PI), phosphatidic acid (PA), phosphatidylglycerol (PG) and diacylglycerol (DAG) are listed with the two fatty acyl groups separated with a slash, e.g., PC 16:0/18:1 (sn1/sn2). LysoPC and lysoPE are abbreviated as LPC and LPE, respectively, and cholesteryl esters are abbreviated CE. The ether-linked phospholipids are shown as PC O (alkyl), PC P (alkenyl), PE O (alkyl), and PE P (alkenyl). The fatty acyl groups of ether-linked lipids and the N-amidated fatty acyl groups for SM, ceramide (Cer), and glycosphingolipids are shown after the slash. Abbreviations for glycosphingolipids are: glucosylceramide (GlcCer), lactosylceramide (LacCer), and globotriaosylceramide (Gb3).
Lipid extraction for MS analyzes
Lipids were extracted from 0.5 to 1.5 million cells containing 150–230 μg of protein using a modified Folch lipid extraction procedure [34]. Known amounts of deuterium-labeled or heptadecanoyl-based synthetic internal standards of LPC, PC, PE, PS, PG, PA, DAG, CE, Cer, HexCer, LacCer, and Gb3 were added and used for quantification of the endogenous lipid species as described [35, 36]. Following lipid extraction, samples were reconstituted in 1:2 (v/v) chloroform:methanol and stored at −20 °C prior to MS analysis of two replicates.
MS analyzes
The species of all phospholipids, SM, DAG, and CE were analyzed by shotgun analysis on a hybrid triple quadrupole/linear ion trap mass spectrometer (QTRAP 5500; AB SCIEX, MA, USA) equipped with a robotic nanoflow ion source (NanoMate HD; Advion Biosciences, NY, USA) [37]. These analyzes were performed using both positive and negative ion modes using multiple precursor ion scanning (MPIS) and neutral loss (NL) based methods [34, 38], whereas CEs were analyzed in positive ion mode [39]. Sphingolipids were analyzed by reverse phase ultra-high pressure liquid chromatography (UHPLC) as previously described [40] using an Acquity BEH C18, 2.1 × 50 mm column with a particle size of 1.7 μm (Waters, Milford, MA, USA) coupled to a hybrid triple quadrupole/linear ion trap mass spectrometer (QTRAP 5500; AB SCIEX). A 25-min gradient using 10 mM ammonium acetate in water with 0.1 % (v/v) formic acid (mobile phase A) and 10 mM ammonium acetate in 4:3 (v/v) acetonitrile:2-propanol containing 0.1 % (v/v) formic acid (mobile phase B) was used. Quantification of sphingolipids was performed using multiple reaction monitoring. Data from one such experiment are shown; similar results were obtained in another independent experiment.
The MS lipidomic analyzes were performed in a laboratory which works according to GLP (Good Laboratory Practice), and the published validation data show less than 15 % variation for most lipid species even when analyzed on three different days [36].
Data processing
The MS data files were processed as previously described [35] using Lipid Profiler™ and MultiQuant™ software for producing a list of lipid names and peak areas. A stringent cutoff was applied for separating background noise from actual lipid peaks. Masses and counts of detected peaks were converted into a list of corresponding lipid names. Lipids were normalized to their respective internal standard [35] and the concentrations of molecular lipids are presented as nmol/mg protein. Quality control samples were utilized to monitor the overall quality of the lipid extraction and mass spectrometry analyzes [36] mainly to remove technical outliers and lipid species that were detected below the limit of quantification.
Quantitative real-time reverse transcription (qRT)-PCR
Cells were grown in 6-well culture plates, washed with sterile ice-cold PBS twice, and then stored in a −80 °C freezer. Total RNA was isolated using RNeasy Plus Mini Kit and QIAcube (QIAGEN, Germany) for automated sample handling. For RNA purification, the protocol “Purification of total RNA from easy-to-lyse animal tissues and cells using gDNA Eliminator columns” (v.2, October 2007; QIAGEN) was used. If not used immediately, samples with isolated RNA were stored in a −80 °C freezer. The RNA content in the samples was measured with NanoDrop 2000 spectrophotometer (Thermo Scientific). Complementary DNA was synthesized from 0.5 μg/sample of the isolated RNA using iScript cDNA synthesis kit (Bio-Rad Laboratories). For negative control, one additional tube was prepared containing all components except for the reverse transcriptase. qRT-PCR was performed using LightCycler 480 SYBR Green I Master kit and LightCycler 480 (Roche Diagnostics, Switzerland). All samples were run in duplicate, and serial dilutions of one cDNA sample were used for standard curve plotting. Primers used: QuantiTect Primer Assay for Gb3 synthase (QIAGEN) and custom made primers for TATA-binding protein gene (TBP) (forward: 5′-GCCCGAAACGCCGAATAT-3′, reverse: 5′-CGTGGCTCTCTTATCCTCATGA-3′; MWG-Biotech, Germany). Relative quantification of the ratio of Gb3syn/TBP was performed using LightCycler 480 Analysis software.
Expression and purification of the non-toxic Shiga like toxin 1 mutant (Stx1-mut)
The Shiga like toxin 1 mutant (Stx1-mut) used in this study carries two point mutations in the active site which render it non-toxic. The tyrosine at position 77 had been changed to serine (T77S) and the glutamate at position 167 had been changed to glutamine (E167Q) [41]. For expression of Stx1-mut, E. coli DH10 (pSW09) were first grown in LB medium supplemented with ampicillin [100 μg/ml; LB (amp)] at 37 °C, 220 rpm to stationary phase. This culture was diluted 1:100 in fresh LB (amp) and incubated overnight under the same conditions. Bacteria were harvested by centrifugation at 5,000g for 10 min. The bacterial pellet was resuspended in 20 % (w/v) sucrose (in 1 mM EDTA, 20 mM Tris buffer, pH 8.0), incubated for 20 min at room temperature and centrifuged at 5,000g for 10 min. The pellet was resuspended in ice-cold de-ionized water, incubated for 10 min on ice and spun down at 13,000g for 10 min. Proteins in the supernatant were precipitated by incubation in 60 % (w/v) ammonium sulfate for 1 h at room temperature, followed by centrifugation at 13,000g for 10 min. The pellet was dissolved in a buffer containing 1 M ammonium sulfate and 50 mM sodium phosphate (pH 8.0). The sample was centrifuged at 13,000 g for 10 min. The proteins were separated on a HiTrap Butyl HP column (GE Healthcare, UK) using a linear gradient from 1 to 0 M ammonium sulfate in 50 mM sodium phosphate (pH 8.0). Fractions containing Stx1-mut were pooled, the buffer was changed by ultrafiltration using Amicon Ultra filters with molecular weight cut-off of 10 kDa (Millipore, MA, USA) and the retained sample was loaded onto a MonoQ 5/50 GL column (GE Healthcare). The proteins were separated by a linear gradient starting with 20 mM Tris (pH 8.0) and ending with 20 mM Tris, 1 M NaCl (pH 8.0). The fractions in which Stx1-mut eluted were pooled, concentrated, and loaded onto a Superdex 200 5/150 GL column (GE Healthcare). An isocratic mobile phase (20 mM Tris–Cl, pH 8.0) was used to elute the proteins. The fractions containing Stx1-mut were pooled. The purity of the Stx1-mut preparation was assessed by Coomassie staining following SDS-PAGE on a 4–20 % polyacrylamide gel. The result showed only the two bands corresponding to the A- and B-subunits of the Stx1-mut (data not shown).
Analysis of Stx1-mut transport to Golgi by immunofluorescence microscopy
HeLa cells were grown on glass coverslips in common cell culture dishes. To have a synchronous internalization of the toxin and its transport to the Golgi, cells were pre-cooled on ice for 15 min, and then incubated with 100 ng/ml Stx1-mut for 20 min on ice. Cells were washed twice with cold and once with 37 °C HEPES-buffered medium, and then fresh 37 °C HEPES-buffered medium was added. At this moment, the time counting for the toxin transport was started. Samples for the 0 min time point were fixed straight after the washing. Cells were fixed by 10 % (v/v) formalin solution (Sigma Aldrich), permeabilized by 0.1 % (w/v) Triton-X/PBS for 5 min and blocked by 10 % (v/v) FBS/PBS for 30 min at room temperature. Samples were labeled by monoclonal mouse anti-Stx antibody (STX1-3C10; Toxin Technology, FL, USA) and polyclonal rabbit anti-giantin antibody (Covance, NJ, USA). Finally, samples were incubated with fluorophore-labeled secondary antibodies (Jackson ImmunoResearch Laboratories, PA, USA) and mounted in ProLong® Gold Antifade Reagent (Molecular Probes®, Life Technologies, UK). Images were taken by Olympus Scan^R fluorescence microscope and analyzed using Olympus Scan^R Analysis software. Three individual slides with 20–50 images/slide were analyzed for each time point and condition, which resulted in more than 250 cells analyzed for each sample. Samples were also examined with a confocal fluorescence microscope LSM 710 (Carl Zeiss, Germany).
125I-labeling of Stx1-mut
Stx1-mut was 125I-labeled by the iodogen method with the IODO-GEN Iodination Reagent (Pierce Biotechnology, MA, USA) according to the manufacturer’s protocol. The yield of iodine incorporation varied from batch to batch with the lowest of 33 cpm/pg protein and the highest of 44 cpm/pg protein. The amount of free iodine was less than 1 % in all batches. To determine the amount of pre-nicked Stx A-subunit (StxA) or free StxA1 moiety in our labeled toxin preparations, 1 μl of each batch was separated by non-reducing (without dithiothreitol (DTT); −DTT) or reducing (with 0.2 M DTT; +DTT) SDS-PAGE and visualized by autoradiography. There were no detectable amounts of pre-nicked Stx1-mut or free StxA1 detected in prepared 125I-Stx1-mut fractions.
Shiga toxin nicking and StxA1 release
To obtain different cell densities but similar sample size, equal numbers of HeLa cells were seeded into 12-well culture plate and 60-mm Petri dishes 1 day prior to experiment. In 12-well plate, cells were of high density (80–90 % confluence), and in 60-mm Petri dishes, they were of low density (20–30 % confluence). In wells/dishes parallel to the ones used for sampling, cells were counted to ensure equal size of low and high density samples. Cells were treated with 10 ng/ml 125I-Stx1-mut in HEPES-buffered medium for 5 h at 37 °C. After incubation with the toxin, cells were washed once with HEPES-buffered medium and incubated with 1 mM N-ethylmaleimide (NEM) in HEPES-buffered medium for 5 min at 37 °C. Finally, cells were lysed in sample buffer, and proteins were separated by non-reducing (−DTT) or reducing (+DTT) SDS-PAGE to determine the amount of released StxA1 or the amount of nicked StxA, respectively. Proteins were transferred to polyvinylide difluoride (PVDF) membranes, visualized by autoradiography and quantified using the Quantity One 1-D Analysis Software (Bio-Rad Laboratories). The values were normalized to total StxA signal for each sample.
StxA1 release to cytosol
HeLa cells were seeded and treated with the 125I-Stx1-mut as described in the previous section. Then, the cells were incubated with fresh HEPES-buffered medium for 30 min at 37 °C to reduce the amount of surface exposed 125I-Stx1-mut. Following this, cells were treated with 1 mM NEM for 5 min at 37 °C. Cells were placed on ice and washed twice with ice-cold KOAc buffer (115 mM CH3COOK, 25 mM HEPES, 2.5 mM MgCl2, pH 8.6) supplemented with 0.9 mM CaCl2 and 0.5 mM MgCl2. At this stage, activated ice-cold Streptolysin O (SLO; Sigma Aldrich) was added at a final concentration of 5 μg/ml. The activation of SLO was performed by incubating SLO with 20 mM sodium 2-mercaptoethanesulfonate (MESNa) in KOAc buffer (pH 8.6) supplemented with 0.1 % (w/v) BSA for 30 min at 37 °C. After 10 min incubation on ice with SLO, cells were washed twice with ice-cold KOAc buffer (pH 7.4) supplemented with 0.9 mM CaCl2 and 0.5 mM MgCl2 to remove the unbound SLO. To induce SLO pore formation, warm (37 °C) transport medium [KOAc buffer supplemented with 5 mM EGTA, 2.5 mM CaCl2, 0.1 % (w/v) BSA, pH 7.4] was added, and the cells were incubated for 10 min at 37 °C. Finally, cells were placed on ice and incubated with gentle rotation for 30 min to allow cytosolic proteins to leak out into the medium. The medium (cytosolic fraction) was collected and stored on ice. The cells were lysed in cell lysis buffer (0.1 M NaCl, 10 mM Na2HPO4, 1 mM EDTA, 1 % (w/v) Triton X-100, cOmplete EDTA-free Protease Inhibitor Cocktail (1 tablet/50 ml; Roche Diagnostics), 60 mM octyl β-d-glucopyranoside, pH 7.4). The cytosolic fraction as well as the cell lysates were centrifuged for 10 min at 4 °C at 8,000 rpm in an Eppendorf centrifuge 5417R (Eppendorf, Germany) to remove cell debris. One-tenth of the samples was used to determine the efficiency and selectivity of the cell permeabilization (see below). From the remaining samples, 125I-Stx1-mut was immunoprecipitated with monoclonal mouse anti-Stx antibodies (STX1-3C10; Toxin Technology) immobilized on protein A-Sepharose CL-4B (GE Healthcare). The immunoprecipitate was washed twice with 0.35 % (w/v) Triton X-100 in PBS, separated by non-reducing SDS-PAGE (10 %) and transferred to a PVDF membrane using Trans-Blot Turbo Transfer System (Bio-Rad Laboratories). Dried membranes were exposed to Kodak BioMax MR film (Eastman Kodak, NY, USA) at room temperature. Band intensities were quantified using QUANTITY ONE 1-D Analysis Software (Bio-Rad Laboratories).
The permeabilization efficiency was estimated based on the release of a cytosolic enzyme lactate dehydrogenase (LDH) into the medium. The LDH activity in the cytosolic fractions and cell lysates (collected as described above) was analyzed using LDH Activity Assay Kit (Sigma-Aldrich) according to the manufacturer’s instructions. The fluorescence was measured by a microplate reader Synergy 2 (Biotek Instruments, VT, USA). Permeabilization efficiency was calculated as follows: [LDH Activity cytosolic fraction/(LDH Activity cytosolic fraction + LDH Activity cell lysates)] × 100 %.
Selective permeabilization of plasma membrane, but not the ER membrane, was confirmed by the absence of the ER protein calreticulin in the cytosolic fraction using the monoclonal mouse anti-calreticulin antibody (SPA-601; Enzo Life Sciences, NY, USA). LDH was detected by polyclonal goat anti-LDH-A antibody (sc-27230; Santa Cruz Biotechnology, CA, USA) and was used as loading control. Cells were counted in parallel wells/dishes and all measurements were normalized to cell number.
Results
High density cells are less sensitive to Shiga toxin
Confluent HUVEC [42] and Vero [43] cells are shown to be more resistant to Shiga-like toxin 1 (Stx1) than non-confluent cells, but little is known about the factors causing this cell density-dependent effect on Stx toxicity. Stx binding and toxicity may depend on the cell cycle [43, 44]. In order to eliminate the cell cycle as a possible variable in this study, we grew high density cells only up to 90 % confluency, and did not observe any significant difference in cell cycle phase distribution between low and high density cells (Online supplementary material 1, Fig. S1). In addition, we measured the incorporation of [3H]leucine per cell into newly synthesized proteins in low and high density HeLa cells, and found only a slight reduction (by approx. 17 %) in high compared to low density cells (Online supplementary material 1, Fig. S2), indicating that cell density had almost no effect on the rate of protein synthesis.
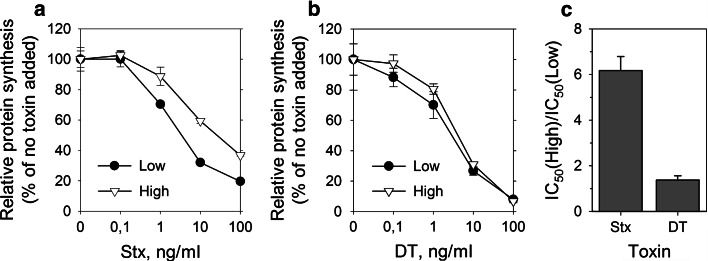
We tested toxicity of Stx, and also, for comparison, toxicity of diphtheria toxin (DT), in low and high density HeLa cells by measuring incorporation of [3H]leucine into newly synthesized proteins (Fig. 1). The concentrations of each toxin required to give 50 % inhibition of protein synthesis (IC50) was determined. Based on the IC50 values for high and low density cells, high density HeLa cells were sixfold less sensitive to Stx, while there was no difference in sensitivity to DT (Fig. 1c). Similar reduction in cell sensitivity to Stx at high cell density was also observed for HEp-2 cells (Online supplementary material 1, Fig. S3).
Fig. 1.
High density HeLa cells are less sensitive to Shiga toxin but not to diphtheria toxin. Low and high density HeLa cells were treated with tenfold serial dilutions of Shiga toxin (Stx) or diphtheria toxin (DT) for 3 h in leucine-free medium. Cells were then incubated in the presence of [3H]leucine for 20 min, and protein synthesis was measured. IC50 was determined, and the ratio between IC50 of low and high density samples was calculated. One representative experiment is shown for a Stx and b DT. Error bars represent difference between the mean value and two parallels. c The quantification of the ratio between IC50 of high and low density cells + SEM, n = 8 for Stx and n = 4 for DT
Shiga toxin binding is decreased in high density cells
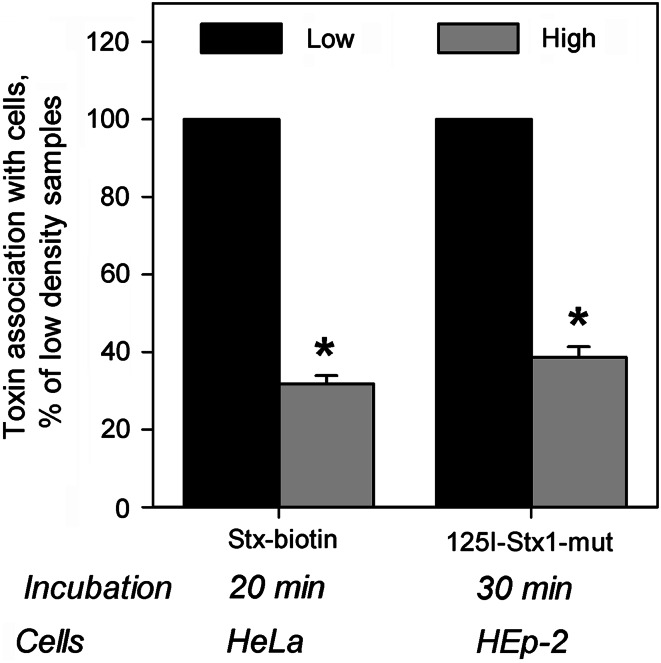
We tested binding of Stx and Stx1-mut to HeLa and HEp-2 cells grown at low and high densities in the culture. Essentially identical reduction by 60–70 % in the toxin-association per cell to high density cells compared to low density cells was observed for both toxins and cell lines tested (Fig. 2). To exclude the possibility that reduced binding in high density cells is due to toxin depletion from the medium, we quantified how much of the toxin was left in the medium after cell treatment, and saw no significant reduction in the concentration of the toxin in the medium, indicating that only a small fraction of the added toxin was associating with the cells during the incubation time used (data not shown).
Fig. 2.
Stx binding is decreased in high density HeLa and HEp-2 cells. Low and high density cells were incubated with biotinylated Stx (Stx-biotin) or iodinated Stx1-mut (125I-Stx1-mut) at 37 °C for 20 and 30 min, respectively. Non-bound toxin was washed away and cell-associated toxin was quantified in cell lysates. For Stx-biotin detection, the toxin was precipitated by streptavidin-coated beads and measured by electrochemiluminescence via a Ru(II)-tag-labeled Stx antibody, while 125I-Stx1-mut was measured directly in cell lysates by a gamma counter. Results were normalized to low density samples and plotted as mean values + SEM, n = 3, *p < 0.01, paired Student’s t test
HeLa but not HEp2 cells have a lower amount of Gb3 in high density culture
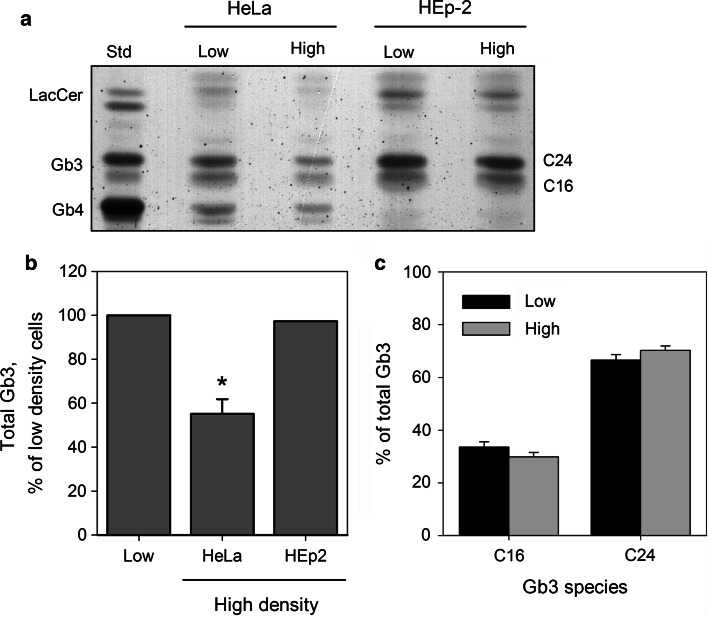
Since Gb3 is the receptor for Stx, the reduction in binding might be caused by reduced Gb3 levels in cells growing at high density. To analyze cellular levels of Gb3, we extracted glycosphingolipids from the cells grown at low and high density cultures as described in the “Materials and methods ”, and analyzed the extracts by HPTLC. Compared to low density cells, cellular Gb3 content was reduced by 45 % in high density HeLa cells (Fig. 3b), while there was no change observed in Gb3 for HEp-2 cells (Fig. 3b).
Fig. 3.
The Gb3 content is reduced in high density HeLa but not in HEp-2 cells. Glycosphingolipids were extracted from low and high density HeLa and Hep-2 cells with chloroform/methanol as described in “Materials and methods ”. Samples were then dried, resuspended in 1 M NaOH, dialyzed against water and dried again. Finally, samples were resuspended in 2:1 (v/v) chloroform:methanol, applied on a silica plate and chromatographed. The bands were visualized by orcinol staining and scanned for intensity quantification. a A representative chromatogram is shown. Gb3 separated into two bands, C16 and C24, which were named according to the number of carbon atoms in the fatty acyl chains of major Gb3 species present in each band. b Quantification of Gb3 content in HeLa and HEp-2 cells. For plotting, band intensity values were normalized to low density samples. For HeLa cells, the experiment was performed four times, and the graph shows mean value + SEM; *p < 0.01, paired Student’s t test. The lipid extraction from HEp-2 cells was performed once, and the bar shows the mean value of two separate chromatographic analyzes performed on the extract. c Quantification of major Gb3 species in low and high density HeLa cells, n = 4
During chromatography, Gb3 separates into two bands, C16 and C24 (Fig. 3a), consisting of different Gb3 species. The C16 band consists mainly of Gb3 species with 16 and 18 carbon atoms in the fatty acyl chain, while C24 mainly consists of Gb3 molecules with 22 and 24 carbon atoms in the fatty acyl chain [16]. We observed no significant changes in the ratio between C16 and C24 band intensity in Gb3 extracted from high density HeLa cells compared to low density cells, suggesting that it is rather a reduction in total cellular Gb3 than change in its species distribution that causes reduced Stx binding and toxicity to high density HeLa cells. Other factors than Gb3 itself are also likely to be involved, since high density HEp-2 cells bound less Stx even without detectable reduction in their Gb3 content.
Since we could not detect reduction in Gb3 in HEp-2 cells by HPTLC which would explain the reduced Stx binding to these cells in high density culture, we performed MS/MS-based quantitative lipidomic analysis of HEp-2 cells to investigate potential differences in the Gb3 species distribution which could lead to reduced Stx binding. HEp-2 cells were grown for 1, 2, or 3 days in culture before lipid extraction. Cells were of 30 % confluence at day 1, and these samples represent a low density culture similar to what was used for other experiments. After 2 days in the culture, cells reached 50–60 % confluence, and after 3 days, they were close to forming a confluent layer (density of approx. 90 %).
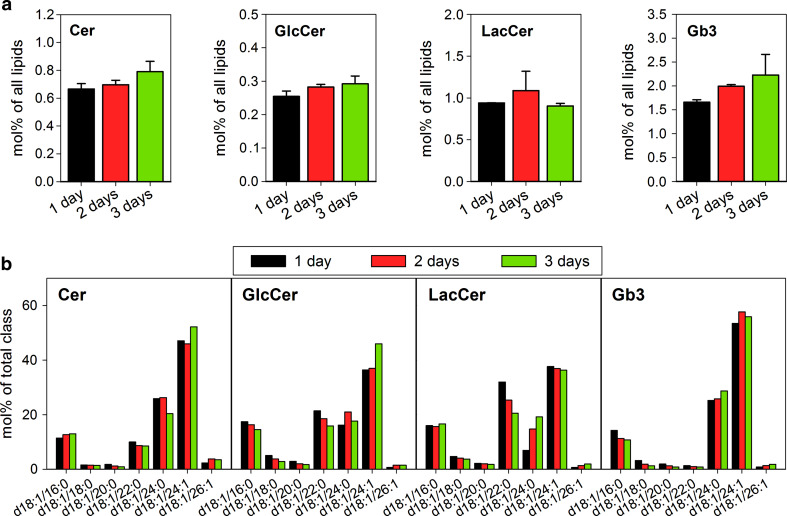
We observed a tendency to increased Gb3 and its precursors Cer and GlcCer during HEp-2 cell growth, but no effect on the total amount of LacCer (Fig. 4a). When the species composition of these lipid classes was compared between cells grown for 1, 2, or 3 days in the culture, only small changes were observed except for LacCer d18:1/22:0 and d18:1/24:0 (Fig. 4b). As there is very little Gb3 d18:1/22:0 in these cells, it is most likely that LacCer d18:1/22:0 is used for synthesis of other glycosphingolipids, e.g., gangliosides [45]. We have not analyzed gangliosides in this study, but it has previously been shown that ganglioside synthesis is also dependent on cell growth and density [13]. In agreement with our data from HPTLC-based analysis of Gb3 in low and high density HEp-2 cell cultures, MS/MS analysis did not show any significant changes in Gb3 species during cell growth from low to high density in HEp-2 cells (Fig. 4b). Thus, neither a change in the amount of cellular Gb3 nor a shift in its species content seems to explain the reduced Stx binding and toxicity to high density HEp-2 cells.
Fig. 4.
The content and species composition of Cer, GlcCer, LacCer and Gb3 in HEp-2 cells. a The content of Cer, GlcCer, LacCer and Gb3 in the HEp-2 cells after 1, 2, and 3 days growth in the culture. Data are normalized to the total lipid content detected for each time point. The error bars show the difference between the mean value and two independent biological replicas. b Species distribution of Cer, GlcCer, LacCer, and Gb3 in the HEp-2 cells after 1, 2, or 3 days growth in the culture. The content of each species is represented as a percentage of the total amount of the class
Lipid profile of HEp-2 cells is dependent on cell density
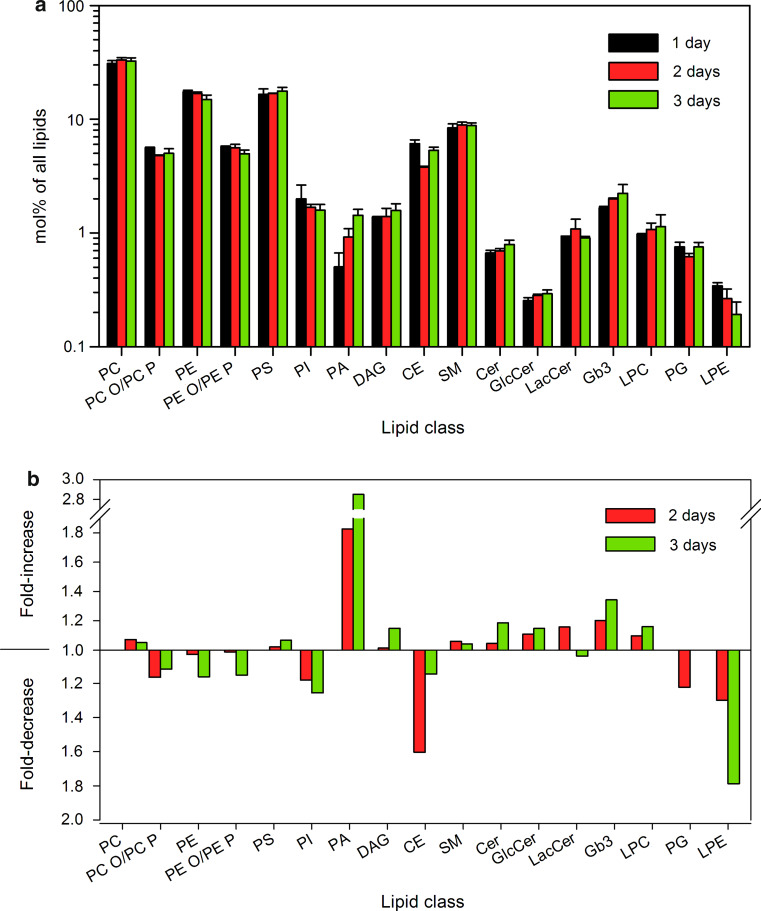
Binding and intracellular transport of Stx is dependent not only on Gb3 itself but also on other membrane lipids [19–22]. We therefore quantified 308 lipid species from 17 different lipid classes in lipid extracts from HEp-2 cells grown in the culture for 1, 2, or 3 days (Online supplementary material 1, Table S1; Online supplementary material 2). For the quantitative data shown in the figures, the minor lipid species of each class (see Online supplementary material 2) were excluded to make it easier to get the overview, and the final calculations were performed with 129 lipid species for 17 lipid classes. The lipid content was quantified as pmol lipid per μg of protein in the sample. The total amount of quantified lipids increased by 14 and 23 % in samples from cells grown for 2 and 3 days, respectively, compared to the samples obtained after 1 day (Online supplementary material 1, Table S1). The data are also presented as mol% of lipids.
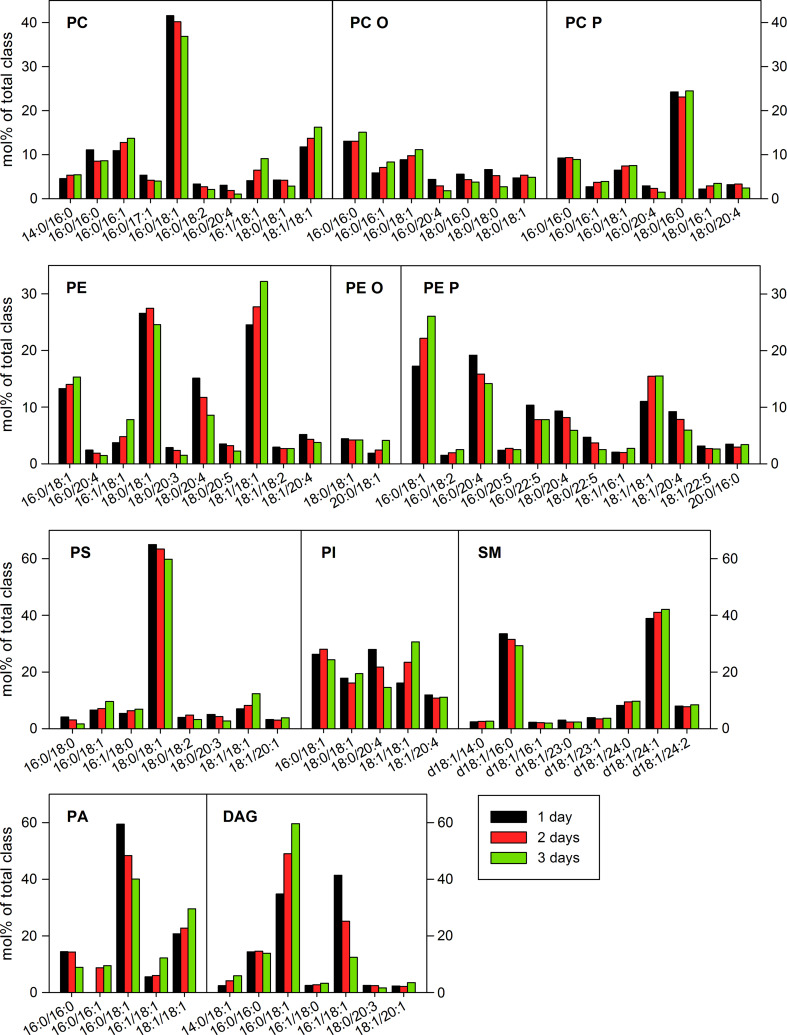
The mol% of several lipid classes changed during cell growth (Fig. 5). The highest increase, 1.8-fold and 2.8-fold after 2 and 3 days, respectively, was observed for PA (Fig. 5b). The levels of PA can be modulated by PA–DAG interconversion, where DAG is formed by phosphatidic acid phosphohydrolases (PAPs), and PA results from the activity of the DAG-consuming kinases (DAGKs) [29]. During cell growth, the levels of DAG increased only slightly by 1.15-fold (Fig. 5b), but there was a significant change in DAG species distribution with an increase in DAG 16:0/18:1 from 35 to 60 mol% of DAG and a decrease in DAG 16:1/18:1 from 41 to 12 mol% of DAG (Fig. 6). It should be noted, that the changes in DAG species were opposite to the changes observed for PA, where relative amount of PA 16:0/18:1 decreased from 59 to 40 mol% during cell growth, whereas the PA 16:1/18:1 increased from 5 to 12 mol% of PA (Fig. 6). Thus, our data indicate that the total levels and species distribution of PA and DAG change during cell growth, and that these changes seem to be interdependent.
Fig. 5.
Lipid composition is changing during HEp-2 cell growth in the culture. Samples for lipid analysis by MS/MS were collected 1, 2, or 3 days after HEp-2 cell seeding to culture plate dishes. a The content of different lipid classes in the samples. Data are normalized to total lipid content detected for each time point. The error bars show the difference between the mean value and two independent biological replicas. b Relative change in lipid classes after 2 and 3 days in the culture compared to cells grown for 1 day. Abbreviations of all lipid classes are given under the section “Annotations of lipid species” in “Materials and methods”
Fig. 6.
Species composition of major lipid classes in HEp-2 cells. Samples were collected 1, 2, or 3 days after HEp-2 cell seeding to culture plate dishes, and analyzed by MS/MS. The amount of individual lipid species was quantified as percentage of the total content of the lipid class. For clarity, minor species are not presented in the graph (for full species list and quantitative data, see Online supplementary material 2). A contamination of an isobaric alkyl species can exist for the PC P and PE P species
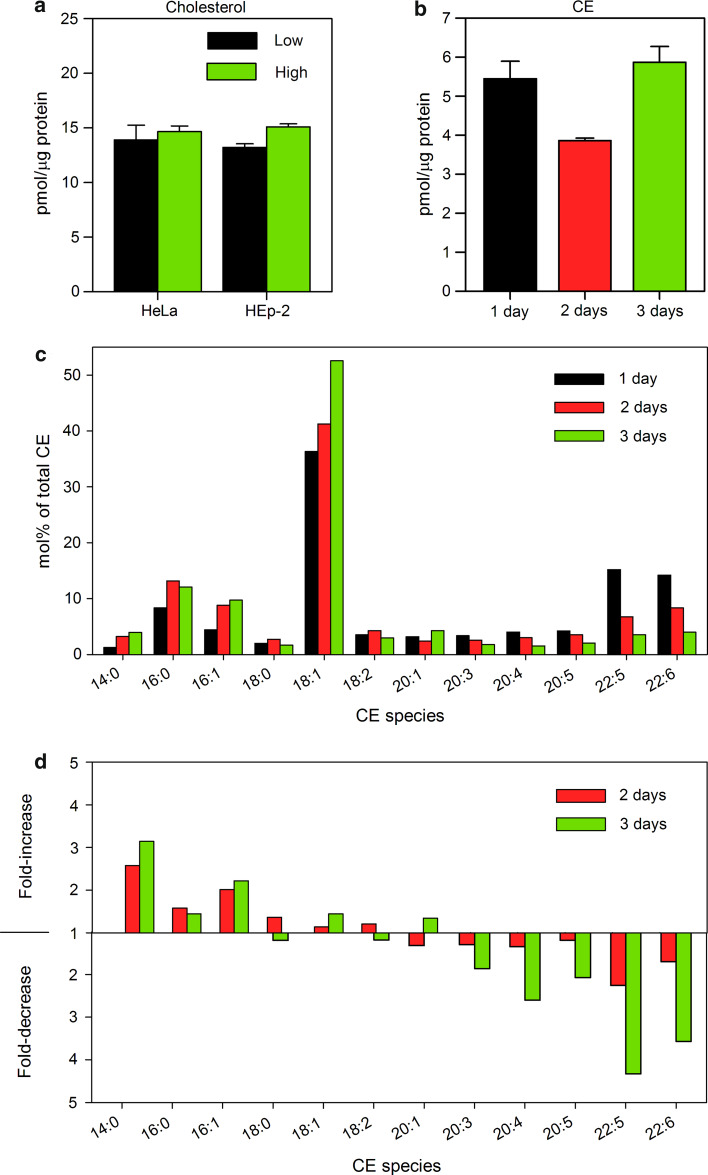
The largest reduction was observed for LPE with a 1.8-fold decrease after 3 days (Fig. 5b), which was mainly due to reduction in LPE 18:0. The level of CE showed a 1.6-fold reduction after 2 days, but it was recovered after 3 days (Fig. 5b). Interestingly, there was a dramatic change in the composition of CE species with a shift towards short fatty acids containing species during cell growth (Fig. 7d), which suggests a significant impact on cholesterol metabolism by cell density. However, we could not detect significant changes in free cholesterol levels either in HeLa or in HEp-2 cells (Fig. 7a). Since we analyzed whole cell extracts, this does not exclude the possibility that there are local changes in cholesterol levels in plasma membrane and/or internal cellular membranes.
Fig. 7.
Free cholesterol and cholesterol ester (CE) content in low and high density cells. a The content of free cholesterol was measured by Amplex® Red Cholesterol Assay Kit in cell lysates from low and high density HeLa and HEp-2 cell cultures. The graph shows mean value of two independent experiments; the error bars represent the difference between the mean value and values of the two experiments. b The content of CE quantified by MS/MS in HEp-2 cells grown for 1, 2, or 3 days in the culture. The error bars show the difference between the mean value and two independent biological replicas. c Species distribution of CE in HEp-2 cells after 1, 2, or 3 days in the culture. d Relative change in CE species after 2 and 3 days in the culture compared to cells grown for 1 day
The overall saturation and chain length of fatty acyl chains (FA) in the lipids were analyzed. The chain length of FA did not change during cell growth except for a slight reduction in the FAs with 20 carbon atoms (Online supplementary material 1, Fig. S4a). As for overall saturation, there was an increase in mono-unsaturated FAs (MUFA) by approximately 15 % in the cells grown for 3 days compared to 1 day, and an almost twofold reduction in poly-unsaturated FAs (PUFA) (Online supplementary material 1, Fig. S4b). These changes were mainly due to an increase in glycerolipid species with two MUFAs and a decrease in glycerolipid species with one PUFA (Online supplementary material 1, Fig. S4c). Interestingly, DAG was the only glycerolipid showing a decrease in the species with two MUFAs (in particular, DAG 16:1/18:1; Fig. 6), while, for all other glycerolipids, the relative amount of species with two MUFAs increased or did not change during cell growth (Online supplementary material 1, Fig. S4d).
The expression of Gb3 synthase is modulated by cell density
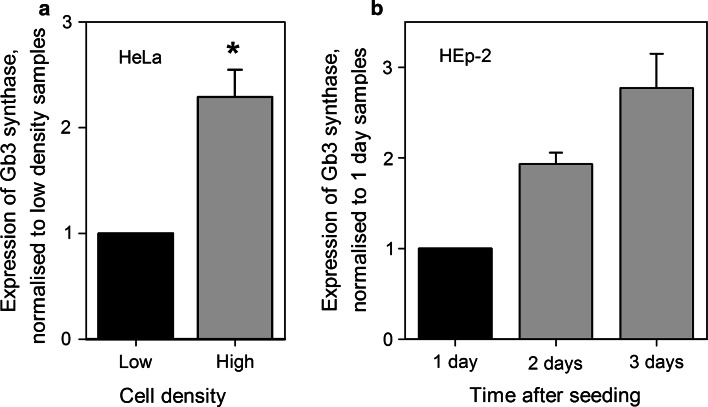
We have previously shown that newly synthesized Gb3 seems to be important for cell sensitivity to Stx [18], and the possibility existed that there was a decrease in Gb3 synthesis at increased cell density. The last step of Gb3 biosynthesis from LacCer is mediated by the enzyme α-1,4-galactosyltransferase (later referred to as Gb3 synthase) [46–48]. We therefore analyzed the expression of the Gb3 synthase gene in HeLa and HEp-2 cells at different cell densities. Total mRNA extracts were analyzed by qRT-PCR, and the house-keeping gene, TBP, was used for relative quantification since the expression of TBP was steady and did not depend on cell density (data not shown). We found that, based on mRNA levels, Gb3 synthase expression was 2.2-fold higher in high density HeLa cells compared to low density culture (Fig. 8a). For HEp-2 cells, mRNA levels for the Gb3 synthase gene were analyzed 1, 2, and 3 days after cell seeding, and there were 1.9-fold and 2.8-fold increases in Gb3 synthase expression after 2 and 3 days, respectively (Fig. 8b). Although our data only show the increase in mRNA levels for Gb3 synthase and not the activity of the enzyme itself, the results are in agreement with published data showing that both Gb3 synthase activity [12] and the biosynthesis of Gb3 [13] were increased in high density BHK cells and cultured human skin fibroblasts, respectively. Therefore, a reduced synthesis of Gb3 in high density HEp-2 cells does not seem to explain the lower sensitivity to Stx in these cells. As for HeLa cells, although the expression of Gb3 synthase gene was increased in response to high cell density, our data from HPTLC analysis suggest that there is a reduction in LacCer, a precursor for Gb3 biosynthesis (Fig. 3a), which would impair Gb3 biosynthesis and lead to the reduced Gb3 levels we have observed in these cells.
Fig. 8.
Gb3 synthase gene expression is up-regulated in high density cells. Total mRNA extracts from the cells were analyzed by qRT-PCR, and relative expression of the Gb3 synthase gene was quantified in the samples. TBP encoding gene was used as a reference for the quantification. a Expression of the Gb3 synthase gene in low and high density HeLa cells. For plotting, data were normalized to low density samples, and the graph represents mean values + SEM, n = 3, *p < 0.01, paired Student’s t test. b Expression of the Gb3 synthase gene in HEp-2 cells 1, 2, and 3 days after cell seeding. For plotting, data were normlized to 1-day samples, and the graph represents mean values of two independent experiments. The error bars show the difference between the mean value and values of the two experiments
Stx endocytosis and transport to the Golgi is not dependent on cell density
To investigate whether reduced binding of Stx to high density cells is the only determinant for the reduced sensitivity of these cells to the toxin, we analyzed Stx endocytosis and transport to the Golgi in low and high density HeLa cells. Low and high density HeLa cells were treated with biotin-labeled Stx and the ratio between endocytosed and total cell-associated toxin was quantified. As shown previously, binding of the biotin-labeled Stx to high density cells was reduced to 32 % of the binding to low density cells (Fig. 2). However, there was no effect on the efficiency of Stx endocytosis, since in both low and high density HeLa cells an equal portion of Stx (approx. 35 % of total cell-associated toxin) was internalized during 20 min incubation at 37 °C (Online supplementary material 1, Fig. S5).
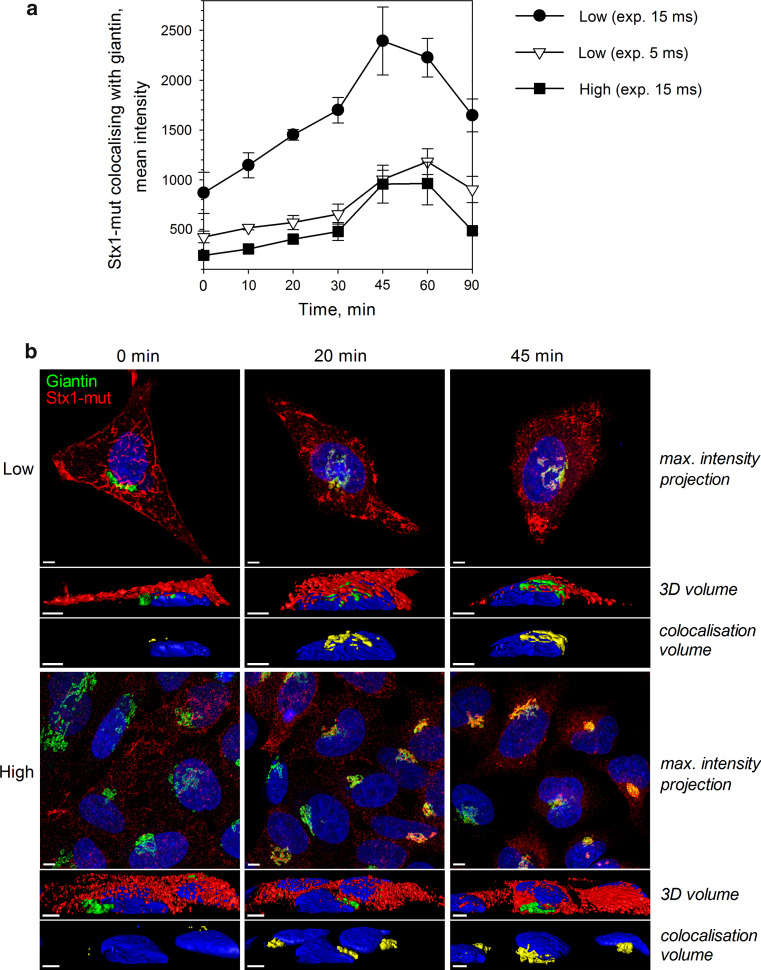
To test whether Stx retrograde transport to the Golgi is affected by cell density, we pre-incubated low and high density HeLa cells with Stx1-mut for 20 min on ice, removed the unbound toxin and started cell incubation at 37 °C (time point 0 min). Cells were washed and fixed at multiple time periods from 10 to 90 min. Cells were then immunostained by antibodies against Stx and the Golgi marker giantin, and analyzed by fluorescence microscopy. To quantify the colocalization between Stx and giantin, multiple images for each time point were taken by Olympus Scan^R microcope, and at least 250 cells were analyzed for each data point. All samples were also observed by confocal microscope to confirm the quantitative data obtained. We observed approx. three-times lower signal for Stx in high density samples at all time points (Fig. 9), which is in agreement with previous data showing the reduction in Stx binding to high density cells (Fig. 2). As for Stx1-mut transport to the Golgi, we could not observe any significant difference between low and high density cells, when not considering the difference in signal intensities. In both conditions, the toxin was transported to the Golgi at a similar rate, and the maximum colocalization between Stx1-mut and giantin was reached after 45–60 min (Fig. 9a). We also tested cellular levels of two retromer components, sorting nexins SNX1 and SNX2, because retromer complex has been previously shown to be involved in Stx sorting from endosomes to the Golgi [49–51]. We did not find any difference in protein levels of SNX1 or SNX2 between low and high density HeLa cells (data not shown), which is in agreement with the finding that Stx transport to the Golgi is not dependent on cell density.
Fig. 9.
Stx transport to the Golgi is not affected by cell density. Low and high density HeLa cells were pre-cooled on ice for 15 min, and then incubated with Stx1-mut for 20 min on ice. Non-bound toxin was washed away and 37 °C HEPES-buffered medium was added (time point 0 min). Cells were fixed, permeabilized, and imunolabeled for Stx (green) and giantin (red). a Multiple images for each time point were taken by Scan^R fluorescence microscope. For low density samples, two different camera exposure times, of 5 and 15 ms, were used. To quantify the colocalization between Stx1-mut and giantin, at least 250 cells were analyzed for each data point using Scan^R Analysis software, and the representative data from one of two independent experiments are shown here. Error bars represent standard deviation between values obtained from individual microscopy slides. b Representative images of 0, 20, and 45 min time points obtained by confocal fluorescence microscope. For visualization, the exposure time for Stx1-mut signal was increased by 3 times in high density samples. The top rows of the images show maximum intensity projection of multiple z-planes; the middle rows show 3D volume reconstruction with the cut along the xz-plane; the bottom rows show the colocalization volume (yellow) between giantin and Stx1-mut. Scale bars 5 μm. Image reconstruction was performed using Imaris x64 7.5.2 software (Bilplane)
Release and translocation of StxA1 into the cytosol is reduced in high density cells
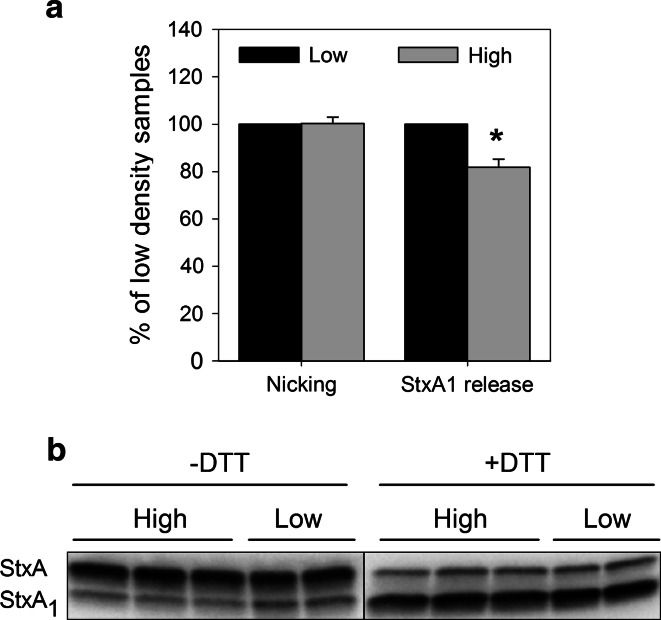
For the cytotoxic action, Stx needs to be transported from Golgi to the ER, where the enzymatically active moiety of the toxin, StxA1, is released and translocated into the cytosol [6]. Stx also requires nicking by the protease furin, which occurs in the endosomes or the Golgi apparatus [52], followed by the reduction of the disulfide bond connecting A1- and A2-moieties in the ER. To test whether these steps are affected, we incubated low and high density HeLa cells with 125I-Stx1-mut for 6 h followed by SDS-PAGE under reducing (+DTT) and non-reducing (−DTT) conditions to quantify release of the StxA1 from the holotoxin. In the autoradiogram, the bottom band represents StxA1 (27 kDa) and the top band StxA (32 kDa) (Fig. 10b). When samples were run under reducing conditions (+DTT), the disulfide bond connecting A1- and A2-moieties was reduced, and all furin-precleaved StxA1 was separated from non-cleaved StxA. In contrast, StxA1 present at non-reducing (−DTT) SDS-PAGE represents the fraction of the A1-moiety released by reducing conditions in the ER. We observed no change in Stx nicking by furin between low and high density cells (Fig. 10), but there was a significant reduction by 20 % in release of StxA1 from the holotoxin in high density HeLa cells (Fig. 10a), suggesting that less toxin has reached the ER.
Fig. 10.
Release of StxA1 is reduced in high density HeLa cells. Low and high density HeLa cells were treated with 10 ng/ml 125I-Stx1-mut for 5 h at 37 °C. Then, cells were washed and incubated with 1 mM NEM for 5 min at 37 °C and lysed. The lysates were separated by non-reducing (−DTT) or reducing (+DTT) SDS-PAGE to determine StxA1 release or nicking of StxA, respectively. A representative autoradiogram is shown in (b) (sample loading was equalized according to cpm values). The amount of released or nicked StxA1 was calculated as a percentage of total Stx (the sum of both StxA and StxA1 bands) in low and high density cells. Finally, results were normalized to low density samples and plotted as mean values + SEM, n = 4, *p < 0.01, paired Student’s t test; shown in (a)
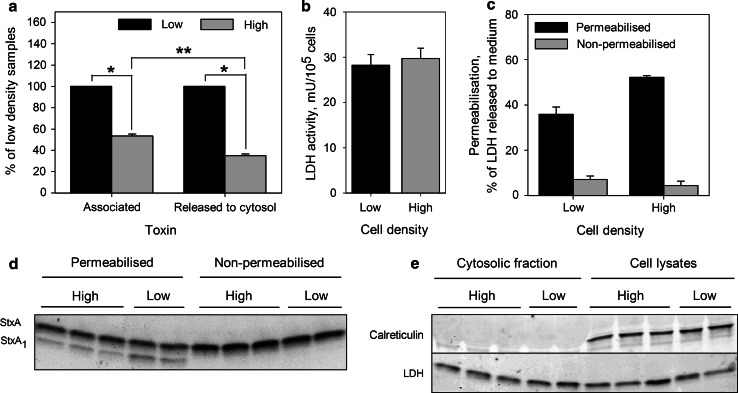
To investigate the translocation of StxA1 into the cytosol, we used Streptolysin O to permeabilize the plasma membrane in 125I-Stx1-mut treated HeLa cells, and then analyzed released cytosolic fractions from low and high density cells. The efficiency of the permeabilization was controlled for each experiment by measuring LDH activity in the cell lysates and cytosolic fractions. The released fraction of LDH varied between 20 and 50 % of total cellular LDH (Fig. 11c). The cells were regularly monitored by light microscopy, but the permeabilization did not induce cell detachment during the experiment. Furthermore, in all experiments, the specific permeabilization of plasma membrane, but not the ER membrane, was confirmed, since the latter would interfere with the results. We did not detect any traces of the ER protein calreticulin in cytosolic fractions in any of the experiments (Fig. 11e), confirming that the ER membrane was intact during plasma membrane permeabilization. Finally, to analyze the content of StxA1 in the collected cytosolic fractions, the toxin was up-concentrated by immunoprecipitation, and then separated by non-reducing (−DTT) SDS-PAGE. During quantification, the content of StxA1 per cell was also adjusted to permeabilization efficacy, since it was always 20–30 % higher in high density cells (Fig. 11c). Hence, we observed a significant reduction by 65 % in StxA1 release into the cytosol in high density HeLa cells compared to low density cells (Fig. 11a). Importantly, this reduction was significantly larger than the difference in total cell associated toxin between low and high density cells (Fig. 11a), which confirms that the release of StxA1 in the ER is impaired in high density cells.
Fig. 11.
Release of StxA1 into the cytosol is reduced in high density HeLa cells. Low and high density HeLa cells were treated with 10 ng/ml 125I-Stx1-mut for 5 h at 37 °C. Then, cells were washed and incubated with fresh HEPES-buffered medium for 30 min at 37 °C to reduce the amount of surface exposed Stx. Finally, 1 mM NEM was added to cells for 5 min at 37 °C. Cells were permeabilized with activated Streptolysin O, and then incubated with transport medium for 30 min on ice to allow the cytosol to leak out (cytosolic fraction). Transport medium was then collected, and the cells lysed. The toxin was immunoprecipitated from cytosolic fraction and then separated by non-reducing SDS-PAGE. a The amount of StxA1 released to the cytosol was quantified from the autoradiogram, and adjusted according to permeabilization efficiency and cell number of each sample. To determine total cell-associated toxin, radioactivity was measured in cell lysates from non-permeabilized samples, and normalized to cell number. Finally, results were normalized to low density samples and plotted as mean value + SEM, n = 3, *p < 0.001, **p < 0.05 Student’s t test. b LDH activity per cell in low and high density samples; representative graph from one of three experiments. Error bars show the difference between the mean value and the two parallels. c The efficiency of cell permeabilization calculated as percentage of total LDH which was released to the medium; representative graph from one of three experiments. Error bars show the difference between the mean value and the two parallels. d A representative autoradiogram of immunoprecipitated toxin from the cytosolic fractions. e One-tenth of cell lysates and cytosolic fractions were precipitated by TCA, separated by reducing SDS-PAGE and blotted to PDF membrane. A representative western blot of ER protein calreticulin and LDH levels in the cytosolic fractions and cell lysates is shown
Discussion
In this study, we have shown that the lipid composition strongly depends on cell density in the culture. Moreover, binding, intracellular transport, and the cytotoxic action of Stx are reduced at high cell density, supporting the findings that multiple changes appear in the cells during growth in culture. At least part of these changes may be related to modification in the cellular lipidome.
To study the effect of cell density on the lipidome, we performed quantitative lipidomic analysis of HEp-2 cells grown for 1, 2, or 3 days in culture and compared the total levels and species distribution of 17 lipid classes. The largest increase in total content was observed for PA with significant change in its species distribution as well. Importantly, there was an opposite tendency on changes in DAG species, suggesting that the changes in PA and DAG species was at least in part caused by interconversion between these two lipids. Both PA and DAG have been associated with COPI-dependent and COPI-independent retrograde protein transport from Golgi to the ER [29, 32]. DAG is suggested to induce membrane bending and formation of highly curved membranes due to its small and electrically neutral polar head, and it has been reported to enhance membrane fusion [53]. Decreased levels of Golgi-associated DAG have been demonstrated to inhibit retrograde (Golgi-to-ER) but not anterograde (ER-to-Golgi) transport [29, 32]. It should be noted that the effect on the retrograde transport depended on which pathway of the DAG production that was blocked. The most profound effect was observed after propanolol (inhibits DAG generation form PA) treatment, and a smaller effect by inhibitor U73122 (blocks DAG generation from phosphoinositides PI4P/PI4,5P2), but no effect by FB1 (inhibits ceramide synthase leading to lowered SM levels and reduced DAG generation from SM) were observed [29], suggesting that only certain DAG pools are involved in the retrograde transport. DAG has been proposed to facilitate the recruitment of ADP-ribosylating factor GTPase-activating protein (ARF1GAP1) at early Golgi compartments [29]. ARF1GAP1 constitutes a structural component of the COPI coat and is required for COPI-coated vesicle formation on Golgi membranes [54]. ARF proteins are shown to activate PC-specific phospholipase D (PLD), which generates PA from PC [55]. Therefore, the action of PLDs and PAPs are proposed to regulate PA and DAG levels in the membranes (including Golgi membranes). As a result, PA has also been implicated in Golgi vesicle formation and budding [56]. Interestingly, the knockdown of either COPI or ARF1 protects cells against Stx [57]. However, the protection could be due to secondary changes occurring during knockdown, as retrograde transport of Stx has been demonstrated to be COPI-independent [58–60]. Nevertheless, the balance between the lipids might be important for retrograde Stx transport. For instance, DAG has also been found to be important for COPI-independent transport [29]. Also, it is possible that not only the level of DAG but also the species play a role. As observed in this study, DAG 16:0/18:1 is increased whereas DAG 16:1/18:1 is decreased in high density cells, suggesting a role for the latter species in the retrograde transport. This is in agreement with the idea that disordering of the membrane has a functional role in the ER and Golgi [61]. Thus, high cell density-induced changes in DAG and PA could impair Golgi-to-ER transport, and therefore lead to reduced StxA1 release in the ER, as we have observed in high density cells.
Golgi transport and vesicle formation have been shown to depend on the action of phospholipase A2 [62], which cleaves membrane phospholipids into lysophospholipids and free fatty acids. We observed a reduction in LPE levels during cell growth, suggesting that phospholipase activity or localization to membranes was reduced in high density cells, and therefore could additionally impair Stx transport, since reduction in LPE levels might affect membrane curvature.
In this study, we compared how cell density modulates Gb3 levels in two well-known cancer cell lines, HeLa and HEp-2, and found striking differences between these two cells with respect to Gb3. Although both cells express Gb3 and are sensitive to Shiga toxin, at high cell density, HeLa cells showed almost a twofold reduction in Gb3 content, while there was a slight increase in Gb3 in HEp-2 cells. Since both of these cell lines up-regulate Gb3 synthase gene expression at high cell density, we speculate that the reduction in Gb3 in HeLa cells is caused by depletion of LacCer or earlier precursors for Gb3 synthesis, since we could detect a reduction in LacCer in high density HeLa cells. Although the total content of Gb3 was changing during cell growth, there was no significant change in the ratios of different Gb3 species in either HeLa (analyzed by HPTLC) or HEp-2 (analyzed both by HPTLC and MS) cells. Interestingly, the binding of Stx was reduced to the same extent in both of these cells at high density, supporting the hypothesis that other factors, rather than the number of receptors, caused reduced binding in dense cells.
One of the explanations for reduced Stx binding in HEp-2 cells, which do not have reduced Gb3 levels, could be different Gb3 localization in the plasma membrane (to DRMs or not) or within the cell. It has been demonstrated that Stx binding depends on Gb3 localization to DRMs [19, 27, 63], as well as lipid composition of DRMs [63], and the majority of the data suggest that Stx preferably binds to DRM-associated Gb3. As for localization within the cell, Gb3 could be internalized from the plasma membrane and up-concentrated in the internal vesicles/membranes, where it would not be accessible for Stx. Additionally, since we analyzed the whole cell extracts, we cannot exclude the possibility that there are local changes in Gb3 species, which would affect its function.
It has been shown that cholesterol can modulate the orientation of the carbohydrate group of several glycosphingolipids, including Gb3, and thus affect Gb3 availability for Stx binding [19–22]. We did not find any significant change in total levels of free cholesterol between low and high density cells when the whole cell lysates were analyzed. However, we observed significantly higher permeabilization efficiency by Streptolysin O, a cholesterol binding toxin, in high density cells compared to low density cells, suggesting that cholesterol levels in the plasma membrane might be increased in high density cells, and thus lead to reduced Stx binding. Furthermore, there was a surprisingly large shift of CE species towards isoforms with shorter fatty acyl chains, suggesting that cholesterol metabolism was highly affected by cell density. CE is synthesized in the ER, and one could therefore speculate that changes in CE composition might affect ER functions. It has been suggested that CE and lipid droplets might facilitate transport from the ER and into the cytosol [64].
As indicated by the change in mRNA levels for the Gb3 synthase, cellular expression of specific proteins might also be affected by cell density and thus modulate Stx toxicity. Since the translocation of the StxA1 across the ER membrane is suggested to occur via the cellular ER-associated protein degradation (ERAD) pathway and to be mediated by Sec61 protein [65], we tested the levels of Sec61β, which is a component of the translocon complex, and has been shown to be involved in Stx intoxication [66], but we found no effect on Sec61β levels by cell density (data not shown). Protein synthesis inhibitors have been shown to increase cell sensitivity to Stx [67], as hypothesized, by reducing the competition between newly synthesized proteins and the StxA1 for the components of ERAD. However, we did not observe an increase in protein synthesis in high density cells, which eliminates the possibility that the release of StxA1 in the ER was impaired due to higher competition with newly synthesized proteins in high density cells. We have no reason to believe that the small reduction actually observed in the rate of overall protein synthesis in high density cells is related to the large protection against Stx toxicity. However, this does not rule out that there are changes in the expression of specific proteins which contribute to decreased Stx toxicity. Finally, it must be noted that the reduction in StxA1 release in the ER and translocation into the cytosol that we observed in high density cells could be at least in part caused by impaired Stx transport from Golgi to the ER, and not by changes in the ER itself, but at this moment, we do not have an efficient method to evaluate the rate of Stx transport from Golgi into the ER, and to test this possibility.
In summary, we have demonstrated that both lipid composition and transport of Stx to the cytosol is modulated by cell density in the culture. The data demonstrate that important changes in lipid composition occur, changes known to affect several cellular transport steps. The results shown here may throw light on differences previously reported, and indicate that close control of culture conditions is essential for reproducibility and understanding of molecular mechanisms studied. In conclusion, our results provide important information on how lipids and changes in lipids can affect intracellular trafficking.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The work performed by the Oslo group has been supported by The Norwegian Council for Science and Humanities, The Norwegian Cancer Society, and Southern and Eastern Norway Regional Authority. The authors thank Anne Grethe Myrann for technical assistance with cell studies, Ellen Skarpen for assistance with microcopy studies, and Sirpa Sutela-Tuominen for assistance with the lipidomic analyzes.
References
- 1.Meli L, Jordan ET, Clark DS, Linhardt RJ, Dordick JS. Influence of a three-dimensional, microarray environment on human cell culture in drug screening systems. Biomaterials. 2012;33:9087–9096. doi: 10.1016/j.biomaterials.2012.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaye SA, Louise CB, Boyd B, Lingwood CA, Obrig TG. Shiga toxin-associated hemolytic uremic syndrome: interleukin-1 beta enhancement of Shiga toxin cytotoxicity toward human vascular endothelial cells in vitro. Infect Immun. 1993;61:3886–3891. doi: 10.1128/iai.61.9.3886-3891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snijder B, Sacher R, Ramo P, Damm EM, Liberali P, Pelkmans L. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature. 2009;461:520–523. doi: 10.1038/nature08282. [DOI] [PubMed] [Google Scholar]
- 4.Obrig TG, Del Vecchio PJ, Brown JE, Moran TP, Rowland BM, Judge TK, Rothman SW. Direct cytotoxic action of Shiga toxin on human vascular endothelial cells. Infect Immun. 1988;56:2373–2378. doi: 10.1128/iai.56.9.2373-2378.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandvig K. Cell density affects the binding of the toxic lectin abrin to HeLa cells in monolayer cultures. FEBS Lett. 1978;89:233–236. doi: 10.1016/0014-5793(78)80225-7. [DOI] [PubMed] [Google Scholar]
- 6.Sandvig K, Torgersen ML, Engedal N, Skotland T, Iversen TG. Protein toxins from plants and bacteria: probes for intracellular transport and tools in medicine. FEBS Lett. 2010;584:2626–2634. doi: 10.1016/j.febslet.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 7.Bergan J, Dyve Lingelem AB, Simm R, Skotland T, Sandvig K. Shiga toxins. Toxicon. 2012;60:1085–1107. doi: 10.1016/j.toxicon.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 8.Jacewicz M, Clausen H, Nudelman E, Donohue-Rolfe A, Keusch GT. Pathogenesis of shigella diarrhea. XI. Isolation of a shigella toxin-binding glycolipid from rabbit jejunum and HeLa cells and its identification as globotriaosylceramide. J Exp Med. 1986;163:1391–1404. doi: 10.1084/jem.163.6.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindberg AA, Brown JE, Stromberg N, Westling-Ryd M, Schultz JE, Karlsson KA. Identification of the carbohydrate receptor for Shiga toxin produced by Shigella dysenteriae type 1. J Biol Chem. 1987;262:1779–1785. [PubMed] [Google Scholar]
- 10.Lingwood CA, Law H, Richardson S, Petric M, Brunton JL, De GS, Karmali M. Glycolipid binding of purified and recombinant Escherichia coli produced verotoxin in vitro. J Biol Chem. 1987;262:8834–8839. [PubMed] [Google Scholar]
- 11.Desselle A, Chaumette T, Gaugler MH, Cochonneau D, Fleurence J, Dubois N, Hulin P, Aubry J, Birkle S, Paris F. Anti-gb3 monoclonal antibody inhibits angiogenesis and tumor development. PLoS One. 2012;7:e45423. doi: 10.1371/journal.pone.0045423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kijimoto S, Hakomori S. Enhanced glycolipid: á-galactosyltransferase activity in contact-inhibited hamster cells, and loss of this response in polyoma transformants. Biochem Biophys Res Commun. 1971;44:557–563. doi: 10.1016/S0006-291X(71)80119-5. [DOI] [PubMed] [Google Scholar]
- 13.Vukelic Z, Kalanj-Bognar S. Cell density-dependent changes of glycosphingolipid biosynthesis in cultured human skin fibroblasts. Glycoconj J. 2001;18:429–437. doi: 10.1023/A:1016066816457. [DOI] [PubMed] [Google Scholar]
- 14.Hakomori S. Cell density-dependent changes of glycolipid concentrations in fibroblasts, and loss of this response in virus-transformed cells. Proc Natl Acad Sci USA. 1970;67:1741–1747. doi: 10.1073/pnas.67.4.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiarash A, Boyd B, Lingwood CA. Glycosphingolipid receptor function is modified by fatty acid content. Verotoxin 1 and verotoxin 2c preferentially recognize different globotriaosyl ceramide fatty acid homologues. J Biol Chem. 1994;269:11138–11146. [PubMed] [Google Scholar]
- 16.Pellizzari A, Pang H, Lingwood CA. Binding of verocytotoxin 1 to its receptor is influenced by differences in receptor fatty acid content. Biochemistry. 1992;31:1363–1370. doi: 10.1021/bi00120a011. [DOI] [PubMed] [Google Scholar]
- 17.Lingwood CA, Binnington B, Manis A, Branch DR. Globotriaosyl ceramide receptor function—where membrane structure and pathology intersect. FEBS Lett. 2010;584:1879–1886. doi: 10.1016/j.febslet.2009.11.089. [DOI] [PubMed] [Google Scholar]
- 18.Raa H, Grimmer S, Schwudke D, Bergan J, Walchli S, Skotland T, Shevchenko A, Sandvig K. Glycosphingolipid requirements for endosome-to-Golgi transport of Shiga toxin. Traffic. 2009;10:868–882. doi: 10.1111/j.1600-0854.2009.00919.x. [DOI] [PubMed] [Google Scholar]
- 19.Tam P, Mahfoud R, Nutikka A, Khine AA, Binnington B, Paroutis P, Lingwood C. Differential intracellular transport and binding of verotoxin 1 and verotoxin 2 to globotriaosylceramide-containing lipid assemblies. J Cell Physiol. 2008;216:750–763. doi: 10.1002/jcp.21456. [DOI] [PubMed] [Google Scholar]
- 20.Khan F, Proulx F, Lingwood CA. Detergent-resistant globotriaosyl ceramide may define verotoxin/glomeruli-restricted hemolytic uremic syndrome pathology. Kidney Int. 2009;75:1209–1216. doi: 10.1038/ki.2009.7. [DOI] [PubMed] [Google Scholar]
- 21.Mahfoud R, Manis A, Binnington B, Ackerley C, Lingwood CA. A major fraction of glycosphingolipids in model and cellular cholesterol-containing membranes is undetectable by their binding proteins. J Biol Chem. 2010;285:36049–36059. doi: 10.1074/jbc.M110.110189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lingwood D, Binnington B, Rog T, Vattulainen I, Grzybek M, Coskun U, Lingwood CA, Simons K. Cholesterol modulates glycolipid conformation and receptor activity. Nat Chem Biol. 2011;7:260–262. doi: 10.1038/nchembio.551. [DOI] [PubMed] [Google Scholar]
- 23.Yahi N, Aulas A, Fantini J. How cholesterol constrains glycolipid conformation for optimal recognition of Alzheimer’s beta amyloid peptide (Abeta1-40) PLoS One. 2010;5:e9079. doi: 10.1371/journal.pone.0009079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandvig K, Garred O, Prydz K, Kozlov JV, Hansen SH, van Deurs B. Retrograde transport of endocytosed Shiga toxin to the endoplasmic reticulum. Nature. 1992;358:510–512. doi: 10.1038/358510a0. [DOI] [PubMed] [Google Scholar]
- 25.Sandvig K, Ryd M, Garred O, Schweda E, Holm PK, van Deurs B. Retrograde transport from the Golgi complex to the ER of both Shiga toxin and the nontoxic Shiga B-fragment is regulated by butyric acid and cAMP. J Cell Biol. 1994;126:53–64. doi: 10.1083/jcb.126.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacewicz MS, Mobassaleh M, Gross SK, Balasubramanian KA, Daniel PF, Raghavan S, McCluer RH, Keusch GT. Pathogenesis of Shigella diarrhea: XVII. A mammalian cell membrane glycolipid, Gb3, is required but not sufficient to confer sensitivity to Shiga toxin. J Infect Dis. 1994;169:538–546. doi: 10.1093/infdis/169.3.538. [DOI] [PubMed] [Google Scholar]
- 27.Falguieres T, Mallard F, Baron C, Hanau D, Lingwood C, Goud B, Salamero J, Johannes L. Targeting of Shiga toxin B-subunit to retrograde transport route in association with detergent-resistant membranes. Mol Biol Cell. 2001;12:2453–2468. doi: 10.1091/mbc.12.8.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayinger P. Phosphoinositides and vesicular membrane traffic. Biochim Biophys Acta. 2012;1821:1104–1113. doi: 10.1016/j.bbalip.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandez-Ulibarri I, Vilella M, Lazaro-Dieguez F, Sarri E, Martinez SE, Jimenez N, Claro E, Merida I, Burger KN, Egea G. Diacylglycerol is required for the formation of COPI vesicles in the Golgi-to-ER transport pathway. Mol Biol Cell. 2007;18:3250–3263. doi: 10.1091/mbc.E07-04-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baron CL, Malhotra V. Role of diacylglycerol in PKD recruitment to the TGN and protein transport to the plasma membrane. Science. 2002;295:325–328. doi: 10.1126/science.1066759. [DOI] [PubMed] [Google Scholar]
- 31.Shemesh T, Luini A, Malhotra V, Burger KN, Kozlov MM. Prefission constriction of Golgi tubular carriers driven by local lipid metabolism: a theoretical model. Biophys J. 2003;85:3813–3827. doi: 10.1016/S0006-3495(03)74796-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutierrez-Martinez E, Fernandez-Ulibarri I, Lazaro-Dieguez F, Johannes L, Pyne S, Sarri E, Egea G. Lipid phosphate phosphatase 3 participates in transport carrier formation and protein trafficking in the early secretory pathway. J Cell Sci. 2013 doi: 10.1242/jcs.117705. [DOI] [PubMed] [Google Scholar]
- 33.Klose C, Surma MA, Gerl MJ, Meyenhofer F, Shevchenko A, Simons K. Flexibility of a eukaryotic lipidome—insights from yeast lipidomics. PLoS ONE. 2012;7:e35063. doi: 10.1371/journal.pone.0035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ekroos K, Chernushevich IV, Simons K, Shevchenko A. Quantitative profiling of phospholipids by multiple precursor ion scanning on a hybrid quadrupole time-of-flight mass spectrometer. Anal Chem. 2002;74:941–949. doi: 10.1021/ac015655c. [DOI] [PubMed] [Google Scholar]
- 35.Ejsing CS, Duchoslav E, Sampaio J, Simons K, Bonner R, Thiele C, Ekroos K, Shevchenko A. Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal Chem. 2006;78:6202–6214. doi: 10.1021/ac060545x. [DOI] [PubMed] [Google Scholar]
- 36.Jung HR, Sylvanne T, Koistinen KM, Tarasov K, Kauhanen D, Ekroos K. High throughput quantitative molecular lipidomics. Biochim Biophys Acta. 2011;1811:925–934. doi: 10.1016/j.bbalip.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 37.Stahlman M, Ejsing CS, Tarasov K, Perman J, Boren J, Ekroos K. High-throughput shotgun lipidomics by quadrupole time-of-flight mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:2664–2672. doi: 10.1016/j.jchromb.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 38.Ekroos K, Ejsing CS, Bahr U, Karas M, Simons K, Shevchenko A. Charting molecular composition of phosphatidylcholines by fatty acid scanning and ion trap MS3 fragmentation. J Lipid Res. 2003;44:2181–2192. doi: 10.1194/jlr.D300020-JLR200. [DOI] [PubMed] [Google Scholar]
- 39.Liebisch G, Binder M, Schifferer R, Langmann T, Schulz B, Schmitz G. High throughput quantification of cholesterol and cholesteryl ester by electrospray ionization tandem mass spectrometry (ESI-MS/MS) Biochim Biophys Acta. 2006;1761:121–128. doi: 10.1016/j.bbalip.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 40.Merrill AH, Jr, Sullards MC, Allegood JC, Kelly S, Wang E. Sphingolipidomics: high-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods. 2005;36:207–224. doi: 10.1016/j.ymeth.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 41.Wen SX, Teel LD, Judge NA, O’Brien AD. Genetic toxoids of Shiga toxin types 1 and 2 protect mice against homologous but not heterologous toxin challenge. Vaccine. 2006;24:1142–1148. doi: 10.1016/j.vaccine.2005.08.094. [DOI] [PubMed] [Google Scholar]
- 42.Obrig TG, Del Vecchio PJ, Karmali MA, Petric M, Moran TP, Judge TK. Pathogenesis of haemolytic uraemic syndrome. Lancet. 1987;2:687. doi: 10.1016/S0140-6736(87)92473-1. [DOI] [PubMed] [Google Scholar]
- 43.Pudymaitis A, Lingwood CA. Susceptibility to verotoxin as a function of the cell cycle. J Cell Physiol. 1992;150:632–639. doi: 10.1002/jcp.1041500324. [DOI] [PubMed] [Google Scholar]
- 44.Majoul I, Schmidt T, Pomasanova M, Boutkevich E, Kozlov Y, Soling HD. Differential expression of receptors for Shiga and Cholera toxin is regulated by the cell cycle. J Cell Sci. 2002;115:817–826. doi: 10.1242/jcs.115.4.817. [DOI] [PubMed] [Google Scholar]
- 45.Merrill AH., Jr Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem Rev. 2011;111:6387–6422. doi: 10.1021/cr2002917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keusch JJ, Manzella SM, Nyame KA, Cummings RD, Baenziger JU. Cloning of Gb3 synthase, the key enzyme in globo-series glycosphingolipid synthesis, predicts a family of alpha 1, 4-glycosyltransferases conserved in plants, insects, and mammals. J Biol Chem. 2000;275:25315–25321. doi: 10.1074/jbc.M002630200. [DOI] [PubMed] [Google Scholar]
- 47.Kojima Y, Fukumoto S, Furukawa K, Okajima T, Wiels J, Yokoyama K, Suzuki Y, Urano T, Ohta M, Furukawa K. Molecular cloning of globotriaosylceramide/CD77 synthase, a glycosyltransferase that initiates the synthesis of globo series glycosphingolipids. J Biol Chem. 2000;275:15152–15156. doi: 10.1074/jbc.M909620199. [DOI] [PubMed] [Google Scholar]
- 48.Steffensen R, Carlier K, Wiels J, Levery SB, Stroud M, Cedergren B, Nilsson SB, Bennett EP, Jersild C, Clausen H. Cloning and expression of the histo-blood group Pk UDP-galactose: Ga1beta-4G1cbeta1-cer alpha1, 4-galactosyltransferase. Molecular genetic basis of the p phenotype. J Biol Chem. 2000;275:16723–16729. doi: 10.1074/jbc.M000728200. [DOI] [PubMed] [Google Scholar]
- 49.Bujny MV, Popoff V, Johannes L, Cullen PJ. The retromer component sorting nexin-1 is required for efficient retrograde transport of Shiga toxin from early endosome to the trans Golgi network. J Cell Sci. 2007;120:2010–2021. doi: 10.1242/jcs.003111. [DOI] [PubMed] [Google Scholar]
- 50.Popoff V, Mardones GA, Tenza D, Rojas R, Lamaze C, Bonifacino JS, Raposo G, Johannes L. The retromer complex and clathrin define an early endosomal retrograde exit site. J Cell Sci. 2007;120:2022–2031. doi: 10.1242/jcs.003020. [DOI] [PubMed] [Google Scholar]
- 51.Utskarpen A, Slagsvold HH, Dyve AB, Skanland SS, Sandvig K. SNX1 and SNX2 mediate retrograde transport of Shiga toxin. Biochem Biophys Res Commun. 2007;358:566–570. doi: 10.1016/j.bbrc.2007.04.159. [DOI] [PubMed] [Google Scholar]
- 52.Garred O, van Deurs B, Sandvig K. Furin-induced cleavage and activation of Shiga toxin. J Biol Chem. 1995;270:10817–10821. doi: 10.1074/jbc.270.18.10817. [DOI] [PubMed] [Google Scholar]
- 53.Chernomordik L, Kozlov MM, Zimmerberg J. Lipids in biological membrane fusion. J Membr Biol. 1995;146:1–14. doi: 10.1007/BF00232676. [DOI] [PubMed] [Google Scholar]
- 54.Lee SY, Yang JS, Hong W, Premont RT, Hsu VW. ARFGAP1 plays a central role in coupling COPI cargo sorting with vesicle formation. J Cell Biol. 2005;168:281–290. doi: 10.1083/jcb.200404008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong JX, Lee FJ, Patton WA, Lin CY, Moss J, Vaughan M. Phospholipid- and GTP-dependent activation of cholera toxin and phospholipase D by human ADP-ribosylation factor-like protein 1 (HARL1) J Biol Chem. 1998;273:15872–15876. doi: 10.1074/jbc.273.25.15872. [DOI] [PubMed] [Google Scholar]
- 56.Siddhanta A, Shields D. Secretory vesicle budding from the trans-Golgi network is mediated by phosphatidic acid levels. J Biol Chem. 1998;273:17995–17998. doi: 10.1074/jbc.273.29.17995. [DOI] [PubMed] [Google Scholar]
- 57.Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M, Hyman AA, Leproust EM, McManus MT, Weissman JS. A systematic Mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Girod A, Storrie B, Simpson JC, Johannes L, Goud B, Roberts LM, Lord JM, Nilsson T, Pepperkok R. Evidence for a COP-I-independent transport route from the Golgi complex to the endoplasmic reticulum. Nat Cell Biol. 1999;1:423–430. doi: 10.1038/15658. [DOI] [PubMed] [Google Scholar]
- 59.Jackson ME, Simpson JC, Girod A, Pepperkok R, Roberts LM, Lord JM. The KDEL retrieval system is exploited by Pseudomonas exotoxin A, but not by Shiga-like toxin-1, during retrograde transport from the Golgi complex to the endoplasmic reticulum. J Cell Sci. 1999;112(Pt 4):467–475. doi: 10.1242/jcs.112.4.467. [DOI] [PubMed] [Google Scholar]
- 60.White J, Johannes L, Mallard F, Girod A, Grill S, Reinsch S, Keller P, Tzschaschel B, Echard A, Goud B, Stelzer EH. Rab6 coordinates a novel Golgi to ER retrograde transport pathway in live cells. J Cell Biol. 1999;147:743–760. doi: 10.1083/jcb.147.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bigay J, Antonny B. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev Cell. 2012;23:886–895. doi: 10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 62.Cancino J, Luini A. Signaling circuits on the Golgi complex. Traffic. 2013;14:121–134. doi: 10.1111/tra.12022. [DOI] [PubMed] [Google Scholar]
- 63.Hanashima T, Miyake M, Yahiro K, Iwamaru Y, Ando A, Morinaga N, Noda M. Effect of Gb3 in lipid rafts in resistance to Shiga-like toxin of mutant Vero cells. Microb Pathog. 2008;45:124–133. doi: 10.1016/j.micpath.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 64.Ploegh HL. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature. 2007;448:435–438. doi: 10.1038/nature06004. [DOI] [PubMed] [Google Scholar]
- 65.LaPointe P, Wei X, Gariepy J. A role for the protease-sensitive loop region of Shiga-like toxin 1 in the retrotranslocation of its A1 domain from the endoplasmic reticulum lumen. J Biol Chem. 2005;280:23310–23318. doi: 10.1074/jbc.M414193200. [DOI] [PubMed] [Google Scholar]
- 66.Moreau D, Kumar P, Wang SC, Chaumet A, Chew SY, Chevalley H, Bard F. Genome-wide RNAi screens identify genes required for Ricin and PE intoxications. Dev Cell. 2011;21:231–244. doi: 10.1016/j.devcel.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 67.Sandvig K, Tonnessen TI, Olsnes S. Ability of inhibitors of glycosylation and protein synthesis to sensitize cells to abrin, ricin, Shigella toxin, and Pseudomonas toxin. Cancer Res. 1986;46:6418–6422. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.